
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol., 06 December 2021
Sec. Alloimmunity and Transplantation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.788769
This article is part of the Research TopicThe Biological and Clinical Aspects of HLA-G, Volume IIView all 16 articles
 Aifen Lin1,2*
Aifen Lin1,2* Wei-Hua Yan2,3*
Wei-Hua Yan2,3*COVID-19, the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has threatened public health worldwide. Host antiviral immune responses are essential for viral clearance and disease control, however, remarkably decreased immune cell numbers and exhaustion of host cellular immune responses are commonly observed in patients with COVID-19. This is of concern as it is closely associated with disease severity and poor outcomes. Human leukocyte antigen-G (HLA-G) is a ligand for multiple immune inhibitory receptors, whose expression can be upregulated by viral infections. HLA-G/receptor signalling, such as engagement with immunoglobulin-like transcript 2 (ILT-2) or ILT-4, not only inhibit T and natural killer (NK) cell immune responses, dendritic cell (DC) maturation, and B cell antibody production. It also induces regulatory cells such as myeloid-derived suppressive cells (MDSCs), or M2 type macrophages. Moreover, HLA-G interaction with CD8 and killer inhibitory receptor (KIR) 2DL4 can provoke T cell apoptosis and NK cell senescence. In this context, HLA-G can induce profound immune suppression, which favours the escape of SARS-CoV-2 from immune attack. Although detailed knowledge on the clinical relevance of HLA-G in SARS-CoV-2 infection is limited, we herein review the immunopathological aspects of HLA-G/receptor signalling in SARS-CoV-2 infection, which could provide a better understanding of COVID-19 disease progression and identify potential immunointerventions to counteract SARS-CoV-2 infection.
COVID-19, the disease caused by the highly contagious virus “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2), has become a serious global public health concern (1). Although unprecedented comprehensive virus transmission prevention measures have been strictly implemented, such as travel restrictions, public social distancing, personal hygiene, and patient quarantine requirements, the virus has spread widely and caused more than 5,054,267 deaths worldwide since its outbreak in December 2019 (2–4).
The clinical manifestation of COVID-19 can be asymptomatic, mild to moderate, or severe, or critical pneumonia with symptoms of acute respiratory distress syndrome, multi-organ failure, and/or shock (5). Among patients with COVID-19, risk factors such as advanced age and pre-existing conditions are associated with increased disease severity (6). From an immunological perspective, a host’s innate and adaptive immune responses are indispensable in controlling viral infection and disease progression. However, abnormal host immune responses are common in patients with severe COVID-19, including cytokine storm resulting from hyper-inflammatory immunological humoral reactions, as well as impairment of cellular antiviral immune responses (7, 8). Our previous studies revealed that total lymphocytes, CD3+, CD4+, and CD8+ T cells were dramatically lower among patients with severe COVID-19 than among non-severe patients at admission. These cells returned to normal levels by the second week after discharge, however, lower CD8+ T cell count is an independent risk factor for longer viral positivity duration and is related to an increased risk for discharged patients with SARS-CoV-2 re-positivity (9–11). Pro-inflammatory cytokines and chemokines, such as interleukin (IL)-1β, IL-6, IL-8, tumour necrosis factor (TNF)-α, macrophage inflammatory protein (MIP) 1α/CCL3, interferon (IFN)-γ-induced protein 10 (IP10), monocyte chemoattractant protein 1 (MCP1), granulocyte colony-stimulating factor (G-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF), were substantially increased (12, 13). Moreover, antiviral cellular immune responses were compromised because of the following: (a) the dramatically decreased absolute number of CD3+ lymphocytes, subpopulations of CD4+ and CD8+ T cells, CD3+CD56+ NKT cells, B cells, and natural killer (NK) cells (14, 15); (b) various functions were impaired and/or exhausted, such as the cytotoxicity of these immune effectors (16–18); and (c) immune regulatory cells, including myeloid-derived suppressor cells, were notably expanded in severe cases (19).
Although an increasing number of clinical and immunological findings on the immunopathological features of COVID-19 are being reported, the molecular mechanisms involved in the dysregulation of cellular immune responses against SARS-CoV-2 infection are yet to be discovered (20). During viral infection, various strategies to escape the host antiviral immune attack and to favour replication and disease progression have been developed by the virus particles (21, 22). Alteration and intervention of human leukocyte antigen (HLA) and/or its receptor expression is one of the strategies applied by viruses (23). In patients with COVID-19, HLA-E receptor CD94/NK group 2 member A (NKG2A), a member of the immune inhibitory receptors, is remarkably increased in CD8+ T and NKT cells, resulting in their functional exhaustion. Notably, high levels of NKG2A expression are significantly reduced when patients recover from the disease (18).
HLA-G is a non-classical HLA class I antigen, which is a strong immune inhibitory mediator via receptor signalling. Because of the alternative splicing its primary transcript, at least seven HLA-G isoforms have been identified, which include four membrane-bound (HLA-G1, HLA-G2, HLA-G3, and HLA-G4) and three soluble (HLA-G5, HLA-G6, and HLA-G7) isoforms (24). HLA-G can be upregulated by various viral infections, including SARS-CoV-2, which can render comprehensive immunosuppressive roles in favouring virus immune evasion and subsequent disease progression (25, 26). Several immune cell surface-expressed receptors have been identified that bind to HLA-G, including immunoglobulin-like transcripts-2 (ILT-2)/CD85j/LIR1, ILT-4/CD85d/LIR2, killer inhibitory receptor (KIR) 2DL4/CD158d, CD8, and CD160 (24). The glycosylphosphatidylinositol-anchored transmembrane glycoprotein receptor CD160 is closely related to the KIR2DL4, although their homology is rather weak only with 29% identity and 44% similarity (27). In this scenario, HLA-G/receptor signalling among various immune cells is important in COVID-19 pathogenesis and progression (Figure 1).
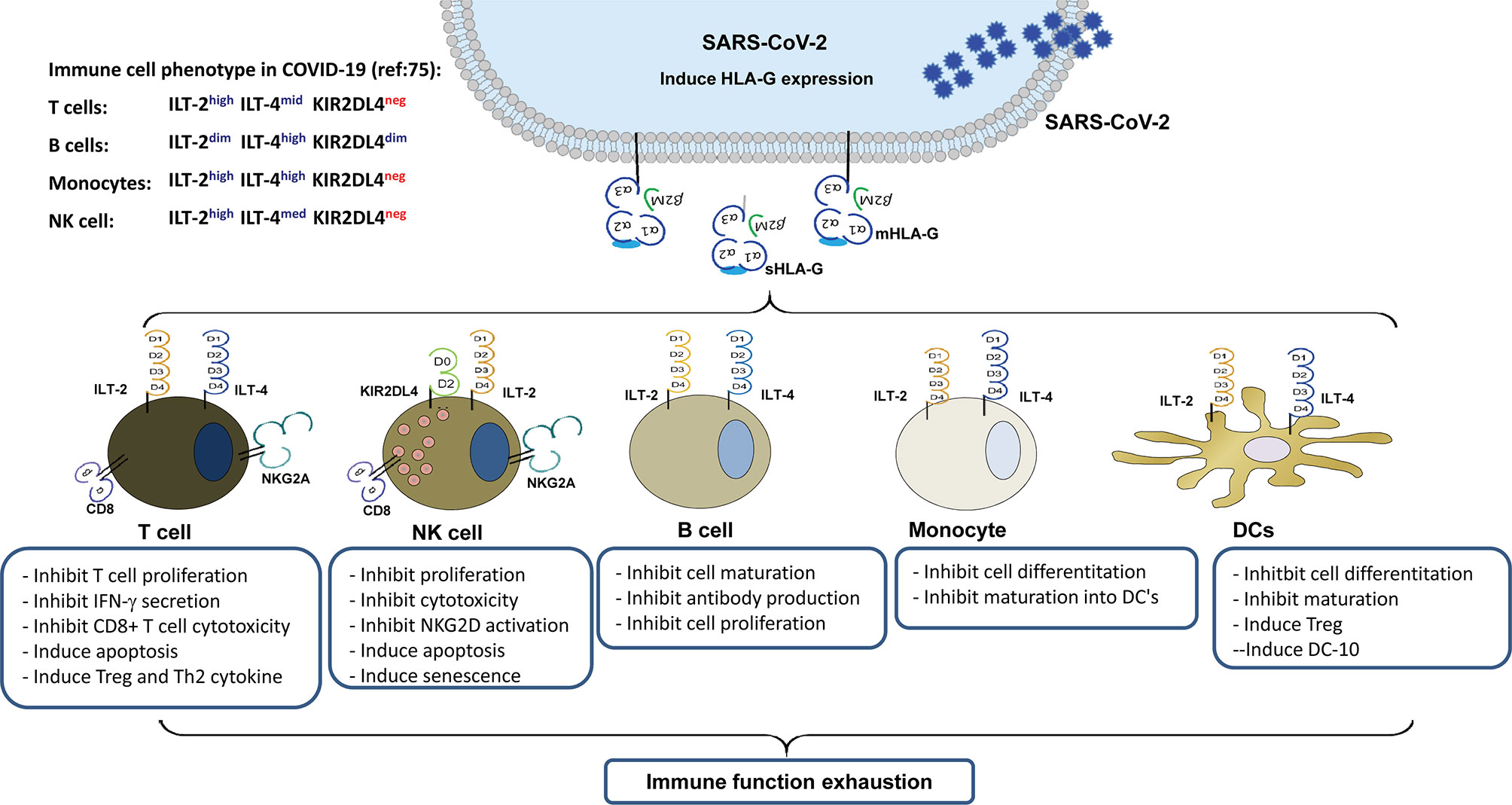
Figure 1 Immune suppression mediated by up-regulated HLA-G expression in virus infected cells and its interaction with immune receptors expressed on different types of immune cells during SARS-CoV-2 infection.
Herein, we focus on the implication of HLA-G/receptor signalling on immune response impairment during SARS-CoV-2 infection.
The HLA-G gene was identified by Geraghty et al. in 1987, and HLA-G protein expression was first found in extravillous cytotrophoblasts in 1990 (28, 29). A number of studies have focused on the basic and clinical significance of HLA-G in foetal-maternal immune tolerance. Since then, immune receptors, including KIR2DL4, ILT-2, ILT-4, CD8, and CD160, have been discovered (30–34). HLA-G induced immune suppression has been extensively documented (including direct and/or indirect as well as long- and short-term), and the immunosuppressive functions of HLA-G have been well established (29, 35). The signalling between HLA-G and receptors KIR2DL4, ILT-2, ILT-4, CD8, and CD160 expressed on different types of immune cells is a fundamental prerequisite for the aforementioned immune suppressive functions of HLA-G (36). Recently, the immune inhibitory NKG2A/CD94 receptor, a well-known receptor for HLA-E, has been reported to be a new HLA-G allele-dependent receptor (37).
By directly binding to immune cell surface receptors ILT-2 or/and ILT-4, HLA-G can inhibit T cells, NK cells, and B cell proliferation, cytotoxicity, anti-inflammatory cytokines such as interferon-chemotaxis, immunoglobulin production, or MICA/NKG2D activation or cell senescence (22). HLA-G can also induce the generation of anergic and regulatory T cells (Treg), tolerogenic DCs, polarisation of M1 to M2, and Th2-type cytokine secretion (24). Moreover, HLA-G can suppress neutrophil reactive oxygen species production and the capacity for phagocytosis (38). Indirect immune suppression induced by HLA-G could be caused by multiple intracellular transfers of HLA-G from HLA-G-bearing cells to HLA-G-negative neighbouring cells or distant cells. Both allogeneic and autologous cell membranes containing HLA-G between/among cells through the process of trogocytosis have been observed, resulting in the activation of immune effector cells (T cells, NK cells, monocytes) into suppressor cells. In addition, HLA-G can be transferred by exosomes to long-distance cells, impairing the functions of immune-competent cells (39). Furthermore, indirect immune modulation induced by HLA-G could increase HLA-E expression, thus influencing the HLA-E/CD94/NKG2 receptor signalling pathway. Cell surface HLA-E expression is dependent on the binding of TAP-associated peptides derived from other HLA class I signal sequences (40). Among the different HLA class I signal peptides, the HLA-E/G-nonameric complex has a significantly higher affinity (41). However, different HLA-G isoforms may have different effects on the enhancement of HLA-E cell surface localization (42).
HLA antigens are critical for both the innate and adaptive immune systems as they bind to T cell receptors to present HLA-restricted peptides to T cells and interact with NK cell receptors to modulate the functions of innate immune components, such as NK cells (43). Downregulation of the expression of the classical HLA class I antigens (HLA-A, -B, and -C) and HLA class II antigens (DP, DQ, and DR) is one of the most effective strategies for perpetuating viral infections, as it allows virus-infected cells to escape from the host immune attack led by virus-specific CD8+ T cells and blunt CD4+ T cells, which help B cells to produce virus-specific antibodies (44). The aberrant upregulation of the immune tolerant HLA-G expression is more common than the downregulation of the HLA-I and -II antigens during many infections, providing virus-infected cells a strategy to protect themselves from NK cell cytolysis (45, 46).
Mechanisms underlying the regulation of HLA-G protein expression is quite complex. HLA-G protein expression can be driven by specific HLA-G genetic polymorphisms, and extracellular and intracellular signals. HLA-G polymorphisms such as a rs66554220, a 14-bp insertion/deletion in the HLA-G exon 8 involved in the stability of HLA-G mRNA, where HLA-G mRNA is more stable with the 14-bp deleted allele and related to HLA-G protein expression (47–49). MicroRNAs miR-148a,miR-148b, miR-152, miR-133a, miR-628-5p, and miR-548q have been reported to regulate HLA-G expression (50). Among them, position +3142 (C>G, rs1063320) has high binding affinity to microRNAs miR-148a,miR-148b, andmiR-152, which is related to the suppression of HLA-G production (51). In addition to the microRNAs, specific RNA binding proteins, such as the heterogeneous nuclear ribonucleoprotein R (HNRNPR), which can bind HLA-G in 3’ untranslated regions (UTR) and stablize the HLA-G transcripts, and increase HLA-G expression (52). HLA-G promoter region includes several specific regulatory elements. HLA-G promoter heat shock element can response to heat shock proteins, and dexamethasone and progesterone can interact with a unique progesterone response element to regulate HLA-G protein expression (53, 54). Also, extracellular environmental factors such as cytokine IL-10, IFNs, indolamine 2, 3-dioxygenase (IDO), granulocyte-macrophage colony-stimulating factor (GM-CSF), hypoxia and demethylation condition affects HLA-G protein expression (48, 55–58). However, peripheral sHLA-G levels are not significantly different between male and female individuals (49, 59). Virus itself or viral gene products such as U94 viral gene of human herpesvirus 6 (HHV-6) can induce HLA-G activation by recognizing an HLA-G promoter consensus sequence (60). Through no specific mechanism for HLA-G up-regulation has been outlined during the SARS-CoV-2 infection, COVID-19 patients peripheral circulation highly increased cytokines such as IL-10, GM-CSF and IDO could be factors involved in the HLA-G expression modification (61, 62).
Upregulation of both virus-infected cell surface membrane-bound HLA-G and peripheral soluble HLA-G expression has been observed in various viral infectious diseases, such as human immunodeficiency virus type 1 (HIV-1), herpes simplex virus-1, rhabdovirus, human cytomegalovirus, hepatitis B and C virus, and influenza A virus (63). HLA-G expression in monocytes and T cells of HIV-1 infected individuals was much higher than that in healthy controls (64). Recent studies have shown that increased HLA-G expression on monocytes could be induced by highly active antiretroviral therapy (HAART), and that cell surface HLA-G expression is more stable than that of the other HLA molecules owing to its resistance to HIV-1 derived protein Nef degradation (65, 66). Moreover, soluble HLA-G could inhibit myeloid dendritic cell function, and higher peripheral circulating sHLA-G levels were shown to be linked to the rapid progression of HIV-1 infection (67, 68).
In the context of SARS-CoV-2 infection, there is currently limited information on the biological and clinical significance of HLA-G. In a previous study using global transcriptomic analysis, Josset et al. (26) found that SARS-CoV-2 and Middle East respiratory syndrome coronavirus (MERS-CoV) differentially activated HLA-G transcription in the human lung epithelial cell line Calu-3 in vitro. This study revealed that HLA-G transcripts could be specifically downregulated by MERS-CoV, whereas HLA-G was upregulated by SARS-CoV-2 infection. With a larger case/control cohort (2244/10220), a recent genome-wide association study (GWAS) by Pairo-Castineira et al. (69) showed that HLA-G (rs9380142) is a novel genetic locus, which is strongly associated with the severity of COVID-19. Association between HLA-G polymorphism and viral infection has been documented in a number of studies. HLA-G*01:01:08 was reported to be a risk factor for HIV-1 infection in Zimbabwean while 3’UTR 14-bp In/14-bp In was a risk factor for HIV infection in South Africans of African ancestry women (70, 71). 14-bp del/14-bp del and HLA-G*01:04:01/14-bp del genotypes, and UTR-2 and UTR-3 haplotypes were found to be the risk factors for hepatitis C virus (72, 73).
In a patient recovered from COVID-19 for four weeks, induced HLA-G expression was observed in the intestinal mucosa epithelial cells and lymphocytes at the sites corresponding to SARS-CoV-2 positivity (74). In our case report, the dynamics of HLA-G and expression of its receptors ILT-2, ILT4, and KIR2DL4 on peripheral immune cell subpopulations in a critical COVID-19 case to convalescence were analysed. Our data showed that HLA-G expression in peripheral immune cells fluctuates along with the status of the disease that the percentage of HLA-G-positive T cells, B cells and monocytes presented a high-low-high pattern, while the percentage of receptors ILT2-, ILT4- and KIR2DL4-expressing immune cells remained relatively stable (75). Moreover, sHLA-G has been found to be significantly elevated in patients with COVID-19 and is related to disease severity (76). Notably, a very recent study by Bortolotti et al. (77) reported that an increased peripheral blood sHLA-G level was associated with an improved outcome in patients with COVID-19, which might be a result of reduced neutrophil adhesion to activated endothelia by sHLA-G as well as interaction with the receptor CD160 (Table 1).
However, the clinical significance of HLA-G and its receptor expression on immune cells among patients with COVID-19 remains largely unknown.
The marked manifestations of immunopathology during SARS-CoV-2 infection, particularly in patients with severe COVID-19, is a salient reduction in immune-competent cells and an overregulated production of pro-inflammatory cytokines and chemokines (78). Dramatically impaired antiviral cellular immune responses and uncontrolled pro-inflammatory humoral immunity lead to SARS-CoV-2 immune evasion and collateral local or systemic tissue damage (79). In severe COVID-19 cases, cellular immune functions are not compromised by dramatically decreased CD3+ lymphocytes, CD4+, CD8+ T, CD3+CD56+ NK T, B, and NK cells, but by the impaired and/or exhausted functions of these immune cells, which is accompanied by the expansion of myeloid-derived suppressor cells (80–82).
Cytotoxic lymphocytes, such as NK cells and CD8+ T lymphocytes, can directly target virus-infected cells, and virus-specific antibody-producing B cells are essential for viral clearance and infectious disease control. In severe COVID-19 cases, not only the absolute number but also the cytotoxicity of both NK cells and CD8+ T lymphocytes are remarkably reduced. Mazzoni et al. (16) found that peripheral circulating NK cell intracellular granzyme and perforin levels were dramatically lower in patients with severe COVID-19 than in healthy controls. At the same time, there are much higher frequencies of senescent phenotype TEMRA+ CD57+ CD8+ T cells but reduced antiviral cytokine production and cytotoxicity of CD4+, CD8+ T, and NK cells in patients with COVID-19. NK cell immune function impairment could be due to the significantly increased expression of the inhibitory receptor NKG2A, as reported by Zheng et al. (18). In that study, the functions of NK and CD8+ T cells were exhausted and exhibited lower CD107a, IFN-γ, IL-2, granzyme B, and TNF-α production, which were accompanied by highly increased expression of the inhibitory receptor NKG2A on both cells during SARS-CoV-2 infection. Importantly, in most patients with COVID-19, the reduced number of NK and CD8+ T cells was restored and the initially high level of NKG2A expression was reduced during the convalescent period after antiviral therapy.
Although detailed information on the significance of HLA-G/receptor signalling in SARS-CoV-2 infection is lacking, the multifaceted immune suppression induced by HLA-G engagement with the aforementioned receptors has provided accumulating evidence that HLA-G/receptor signalling induces immune impairment and exhaustion, and cytokine release could be of critical importance in COVID-19. Previous studies have described the profound immune suppression mediated by HLA-G interaction with ILT-2/4 in a wide range of contexts (83). The interaction and signalling could inhibit the cytotoxicity of NK and CD8+ T cells, allo-proliferation of CD4+ T cells, maturation of DCs, differentiation, proliferation, and immunoglobulin (IgA, IgG, and IgM) production by B cells (84–88). In line with this, the activation of DCs and B cells was hampered in patients with severe COVID-19, as indicated by Wang et al. (89). Moreover, HLA-G/CD8 interaction could induce the apoptosis of CD8+ T cells through the Fas/FasL pathway, which may also occur in subsets of CD8+ NK T cells (90, 91). In contrast, HLA-G/ILT-2/4 engagement could also induce the generation of CD8+CD28+ or CD4+CD25+CTLA-4+ regulatory T cells (Tregs), expansion of MDSCs, tolerogenic DC-10 induced adaptive type 1 regulatory T cells, and M2 type macrophages (92–95). A study by Tomić et al. (96) revealed that the expansion of PD-L1, ILT-3, and IDO-1-expressing monocytic MDSCs was related to the accumulation of regulatory B and T cells and poor T cell immune responses in patients with severe COVID-19. Other studies have shown that CD4+CD25+CD127low Treg cells were significantly increased in patients with both mild or severe COVID-19, regardless of recovery, and that the proportion of IL-10 producing Treg was significantly increased in patients with severe COVID-19 (97, 98). However, the expression status of HLA-G and its receptors on these immune cells remains to be investigated.
Moreover, HLA-G expression in swine endothelial cells can protect them from human macrophage-mediated cytotoxicity (99, 100). Based on our preliminary study on HLA-G receptor expression in circulating immune subpopulations in a critical patient with COVID-19, the data showed that T cells can be phenotyped as ILT-2highILT-4 midKIR2DL4dim, B cells as ILT-2 midILT-4highKIR2DL4dim, and monocytes as ILT-2highILT-4highKIR2DL4dim. However, the cell surface expression levels of these receptors remained relatively stable from the critical stage to convalescent stage and irrespective of the viral load in SARS-COV-2 infection (75). The marginal KIR2DL4 expression observed in T cells, B cells, and monocytes in our study is in agreement with previous reports and is mainly located intracellularly but detectable upon IL-2 activation of NK cells (101). The activation signal resulting from HLA-G/KIR2DL4 interaction not only initiates the production of robust pro-inflammatory cytokines and chemokines, such as IFN-γ, TNF-α, IL-1β, IL-6, IL-8, MIP-3α, MIP-1δ, MIP-1α, and MIP-2β, but also leads to cell senescence and cell cycle arrest in NK cells (102, 103).
HLA-G allelic products also affect the interaction with its receptors, caused by different amino acid residues, and consequently alter its biological functions. Celik et al. (104) indicated that a single amino acid difference in the α two domains of HLA-G could affect the lysis of target cells by NK cells. The data showed that a much stronger immune suppressive function was observed for the HLA-G*01:04 allele than for the HLA-G*01:01 and HLA-G*01:03 alleles. It seems reasonable with a recent finding that the binding of the HLA-G*01:04 product to NKG2A/CD94 has a higher affinity than that of HLA-G*01:01 and HLA-G*01:03 products (37). As NKG2A expression is highly associated with the severity of COVID-19, these findings indicate that the genetic variation of HLA-G could be linked to susceptibility to disease and host immune response regulation during SARS-CoV-2 infection (105).
Since the outbreak of the worldwide COVID-19 pandemic in December 2019, it has claimed more than 5,054,267 lives (4). More insights into the ever-increasing clinical characteristics and laboratory findings on COVID-19 are being reported, which show that immune-competent cell function impairment and/or exhaustion is one of the major features of COVID-19 pathogenesis (7). However, the mechanisms underlying immunological abnormalities remain largely unknown. As reported in previous studies, viruses have developed effective strategies to hide from host antiviral immune responses and survive during infection (106). One strategy successfully deployed by viruses for immune evasion is the impairment of the classical HLA class I and II antigens to hide infected cells from T cell recognition, and the induction of non-classical HLA class I antigen HLA-G, a ligand for immune inhibitory receptors differentially expressed on almost all subsets of immune cells. Consequently, differential alteration in HLA antigen expression by viral infection makes the host antiviral immune system vulnerable (22, 107).
Synergistic suppression effects induced by HLA-G/receptor signalling are well recognised. These effects include the inhibition of cell proliferation and differentiation and the induction of cell apoptosis and senescence, which could be involved in significant decrease or even exhaustion of immune-competent cells such as T cells, NK cells, B cells, and macrophages in patients with COVID-19. Other effects include the inhibition of T and NK cell cytotoxicity, antibody production by B cells, and induction of regulatory cells and expansion of MDSCs, which might be linked to the functional impairment of effector cells, such as T, NK, and B cells, during SARS-CoV-2 infection. However, more information on HLA-G and its receptor status is necessary for future clinical investigations and basic science studies.
More evidence can be accumulated to solidify the basic and clinical aspects of HLA-G in COVID-19 progression and outcome. Aspects expected to be explored include (a) HLA-G expression is reported to be correlated with the progression of various infectious diseases (108, 109). We hypothesise that cell surface HLA-G and circulating soluble HLA-G levels are related to the severity, outcome, or viral load in patients with COVID-19. (b) The upregulation of HLA-G expression by cytokines, such as IFN-γ, IL-6, and IL-10, is dramatically increased in patients with severe COVID-19 (110, 111). We hypothesise that HLA-G expression is related to cytokines in patients with COVID-19. (c) Recently identified HLA-G allelic product-dependent receptor NKG2A has been observed to be dramatically increased in patients with COVID-19 (18). What is the status of other HLA-G receptors, such as ILT-2, ILT-4, and KIR2DL4, and their relationship with disease progression? and (d) Given HLA-E-CD94/NKG2A axis plays critical roles in COVID-19 and HLA-E cell surface expression depends other leader sequence peptides, particularly derived from HLA-G (112), what is the relationship between HLA-G and HLA-E expression? (Figure 2).
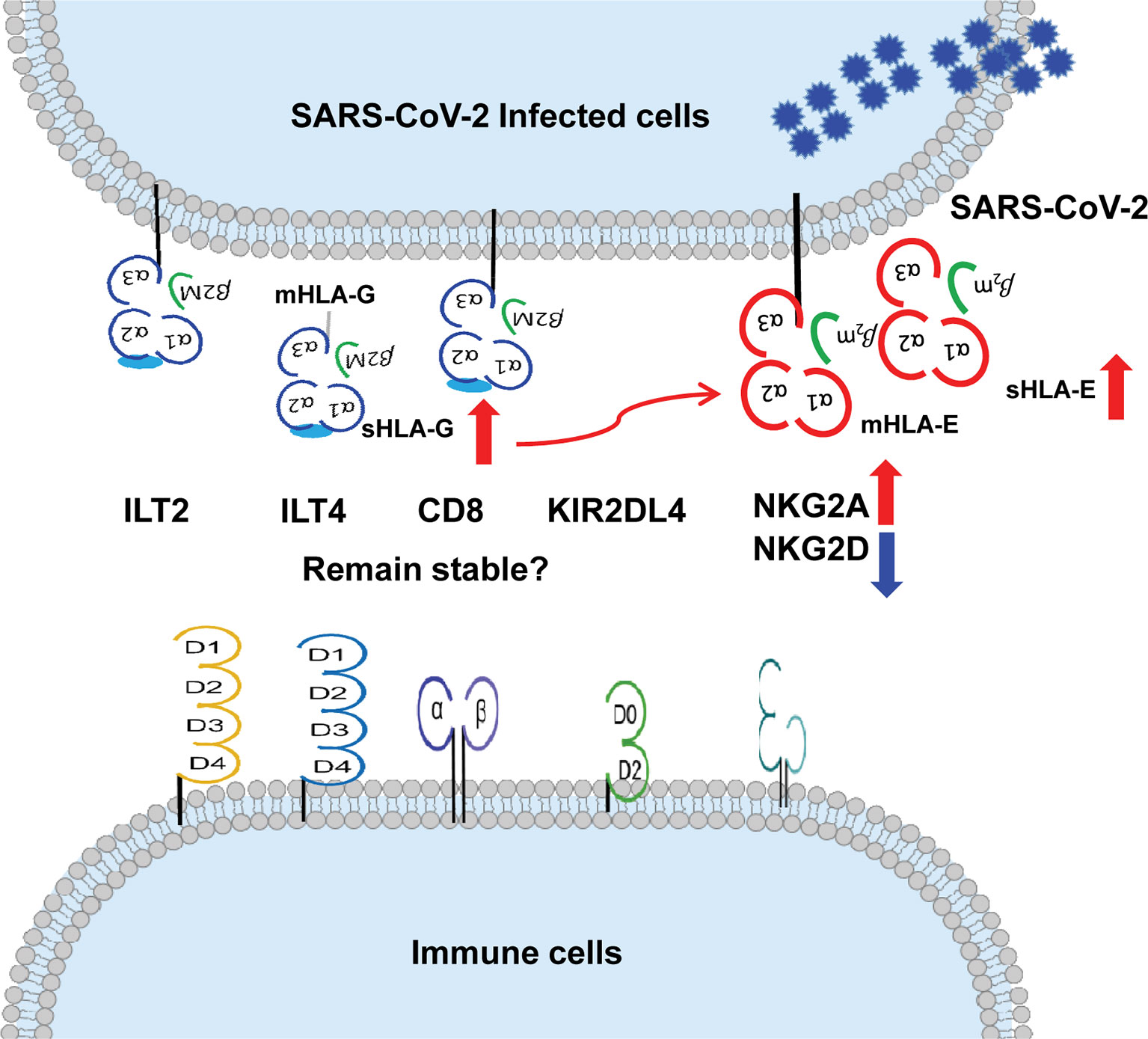
Figure 2 More evidence can be expected on the relationship between up-regulation of HLA-G and HLA-E expression and their immune receptors on immune cells during SARS-CoV-2 infection. ↑up-regulation. ↓down-regulation.
In this context, the clinical trial “HLA-G Immuno-Inhibitor Checkpoint Study in Patients With COVID-19 Infection: Molecular and Cellular Assessment (HLA-G-COVID) (NCT04613297)” has been started to evaluate the clinical significance of HLA-G and receptors ILT-2 expression on CD4+ and CD8+ lymphocytes, and the levels of peripheral sHLA-G and plasma HLA-G-bearing microvesicles among COVID-19 uninfected patients, non-hospitalized COVID-19 infected patients and hospitalized COVID-19 infected patients. With this clinical trial, further understanding of the significance of HLA-G and its receptors in COVID-19 patients can be expected.
The impaired immune functions of NK and T cells resulting from ILTs and NKG2A expression have been reported to be involved in virus immune evasion and related infectious disease progression (113–116). Fortunately, ILTs, NKG2A, and HLA-G targeted immunotherapy and signalling pathway blockades are already in development in clinical trials for cancer immunotherapy. In previous preclinical investigations, blocking tumour cell-expressed HLA-G or immune cell surface ILT2/4 with specific antibodies could restore the functions of NK cells or T cells against target cells (36). Furthermore, blocking NKG2A with monalizumab, an anti-NKG2A monoclonal blocking antibody, can significantly restore the cytotoxic function of NKG2A+ NK and T cells (117–119). Along with these findings, the application of ILTs and NKG2A targeted blocking antibodies could be an additional intervention to mitigate the severity of COVID-19. Finally, a clinical phase I trial with an HLA-G blockade antibody, TTX-80, was launched in July 2020 for patients with advanced solid cancer (120), which shed new light on the restoration of exhausted immune responses induced by HLA-G in diseases such as cancers or viral infections.
We hope that our review will provide a much better understanding of the immune pathogenesis of COVID-19, and thereby help in the development of immunointerventions to counteract SARS-CoV-2 infection.
W-HY and AL conceived and designed the review. W-HY made the figures. AL and W-HY drafted and revised the manuscript and approved it for publication. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Science and Technology Bureau of Taizhou (1901ky01; 1901ky04, 1901ky05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wilder-Smith A, Chiew CJ, Lee VJ. Can We Contain the COVID-19 Outbreak With the Same Measures as for SARS? Lancet Infect Dis (2020) 20:e102–7. doi: 10.1016/S1473-3099(20)30129-8
2. Redelmeier DA, Shafir E. Pitfalls of Judgment During the COVID-19 Pandemic. Lancet Public Health (2020) 5:e306–8. doi: 10.1016/S2468-2667(20)30096-7
3. Hong LX, Lin A, He ZB, Zhao HH, Zhang JG, Zhang C, et al. Mask Wearing in Pre-Symptomatic Patients Prevents SARS-CoV-2 Transmission: An Epidemiological Analysis. Travel Med Infect Dis (2020) 36:101803. doi: 10.1016/j.tmaid.2020.101803
4. Available at: https://covid19.who.int/ (Accessed on 9th Nov, 2021).
5. Machhi J, Herskovitz J, Senan AM, Dutta D, Nath B, Oleynikov MD, et al. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J Neuroimmune Pharmacol (2020) 15:359–86. doi: 10.1007/s11481-020-09944-5
6. Tian W, Jiang W, Yao J, Nicholson CJ, Li RH, Sigurslid HH, et al. Predictors of Mortality in Hospitalized COVID-19 Patients: A Systematic Review and Meta-Analysis. J Med Virol (2020) 92:1875–83. doi: 10.1002/jmv.26050
7. García LF. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front Immunol (2020) 11:1441. doi: 10.3389/fimmu.2020.01441
8. Jesenak M, Brndiarova M, Urbancikova I, Rennerova Z, Vojtkova J, Bobcakova A, et al. Immune Parameters and COVID-19 Infection - Associations With Clinical Severity and Disease Prognosis. Front Cell Infect Microbiol (2020) 10:364. doi: 10.3389/fcimb.2020.00364
9. Jin XH, Zhou HL, Chen LL, Wang GF, Han QY, Zhang JG, et al. Peripheral Immunological Features of COVID-19 Patients in Taizhou, China: A Retrospective Study. Clin Immunol (2021) 222:108642. doi: 10.1016/j.clim.2020.108642
10. Lin A, He ZB, Zhang S, Zhang JG, Zhang X, Yan WH. Early Risk Factors for the Duration of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Positivity in Patients With Coronavirus Disease 2019. Clin Infect Dis (2020) 71:2061–5. doi: 10.1093/cid/ciaa490
11. Hong LX, Liu L, Lin A, Yan WH. Risk Factors for SARS-CoV-2 Re-Positivity in COVID-19 Patients After Discharge. Int Immunopharmacol (2021) 95:107579. doi: 10.1016/j.intimp.2021.107579
12. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
13. Jia R, Wang X, Liu P, Liang X, Ge Y, Tian H, et al. Mild Cytokine Elevation, Moderate Cd4+ T Cell Response and Abundant Antibody Production in Children With COVID-19. Virol Sin (2020) 35:734–43. doi: 10.1007/s12250-020-00265-8
14. Huang W, Berube J, McNamara M, Saksena S, Hartman M, Arshad T, et al. Lymphocyte Subset Counts in COVID-19 Patients: A Meta-Analysis. Cytometry A (2020) 97:772–6. doi: 10.1002/cyto.a.24172
15. Shi H, Wang W, Yin J, Ouyang Y, Pang L, Feng Y, et al. The Inhibition of IL-2/IL-2R Gives Rise to CD8+ T Cell and Lymphocyte Decrease Through JAK1-STAT5 in Critical Patients With COVID-19 Pneumonia. Cell Death Dis (2020) 11:429. doi: 10.1038/s41419-020-2636-4
16. Mazzoni A, Salvati L, Maggi L, Capone M, Vanni A, Spinicci M, et al. Impaired Immune Cell Cytotoxicity in Severe COVID-19 Is IL-6 Dependent. J Clin Invest (2020) 130:4694–703. doi: 10.1172/JCI138554
17. Bordoni V, Sacchi A, Cimini E, Notari S, Grassi G, Tartaglia E, et al. An Inflammatory Profile Correlates With Decreased Frequency of Cytotoxic Cells in Coronavirus Disease 2019. Clin Infect Dis (2020) 71:2272–5. doi: 10.1093/cid/ciaa577
18. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional Exhaustion of Antiviral Lymphocytes in COVID-19 Patients. Cell Mol Immunol (2020) 17:533–5. doi: 10.1038/s41423-020-0402-2
19. Agrati C, Sacchi A, Bordoni V, Cimini E, Notari S, Grassi G, et al. Expansion of Myeloid-Derived Suppressor Cells in Patients With Severe Coronavirus Disease (COVID-19). Cell Death Differ (2020) 27:3196–207. doi: 10.1038/s41418-020-0572-6
20. Catanzaro M, Fagiani F, Racchi M, Corsini E, Govoni S, Lanni C. Immune Response in COVID-19: Addressing a Pharmacological Challenge by Targeting Pathways Triggered by SARS-CoV-2. Signal Transduct Target Ther (2020) 5:84. doi: 10.1038/s41392-020-0191-1
21. Forte E, Zhang Z, Thorp EB, Hummel M. Cytomegalovirus Latency and Reactivation: An Intricate Interplay With the Host Immune Response. Front Cell Infect Microbiol (2020) 10:130. doi: 10.3389/fcimb.2020.00130
22. Lin A, Xu H, Yan W. Modulation of HLA Expression in Human Cytomegalovirus Immune Evasion. Cell Mol Immunol (2007) 4:91–8.
23. Halenius A, Gerke C, Hengel H. Classical and Non-Classical MHC I Molecule Manipulation by Human Cytomegalovirus: So Many Targets—But How Many Arrows in the Quiver? Cell Mol Immunol (2015) 12:139–53. doi: 10.1038/cmi.2014.105
24. Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
25. Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V. Recent Advances in Our Understanding of HLA-G Biology: Lessons From a Wide Spectrum of Human Diseases. J Immunol Res (2016) 2016:4326495. doi: 10.1155/2016/4326495
26. Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, et al. Cell Host Response to Infection With Novel Human Coronavirus EMC Predicts Potential Antivirals and Important Differences With SARS Coronavirus. mBio (2013) 4:e00165–13. doi: 10.1128/mBio.00165-13
27. Anumanthan A, Bensussan A, Boumsell L, Christ AD, Blumberg RS, Voss SD, et al. Cloning of BY55, a Novel Ig Superfamily Member Expressed on NK Cells, CTL, and Intestinal Intraepithelial Lymphocytes. J Immunol (1998) 161:2780–90.
28. Geraghty DE, Koller BH, Orr HT. A Human Major Histocompatibility Complex Class I Gene That Encodes a Protein With a Shortened Cytoplasmic Segment. Proc. Natl Acad Sci USA (1987) 84:9145–9. doi: 10.1073/pnas.84.24.9145
29. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A Class I Antigen, HLA-G, Expressed in Human Trophoblasts. Science (1990) 248:220–3. doi: 10.1126/science.2326636
30. Rajagopalan S, Long EO. A Human Histocompatibility Leukocyte Antigen (HLA)-G-Specific Receptor Expressed on All Natural Killer Cells. J Exp Med (1999) 189:1093–100. doi: 10.1084/jem.189.7.1093
31. Navarro F, Llano M, Bellón T, Colonna M, Geraghty DE, López-Botet M. The ILT2(LIR1) and CD94/NKG2A NK Cell Receptors Respectively Recognize HLA-G1 and HLA-E Molecules Co-Expressed on Target Cells. Eur J Immunol (1999) 29:277–83. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4
32. Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O’Callaghan CA, et al. Human Myelomonocytic Cells Express an Inhibitory Receptor for Classical and Nonclassical MHC Class I Molecules. J Immunol (1998) 160:3096–100.
33. Sanders SK, Giblin PA, Kavathas P. Cell-Cell Adhesion Mediated by CD8 and Human Histocompatibility Leukocyte Antigen G, a Nonclassical Major Histocompatibility Complex Class 1 Molecule on Cytotrophoblasts. J Exp Med (1991) 174:737–40. doi: 10.1084/jem.174.3.737
34. Fons P, Chabot S, Cartwright JE, Lenfant F, L’Faqihi F, Giustiniani J, et al. Soluble HLA-G1 Inhibits Angiogenesis Through an Apoptotic Pathway and by Direct Binding to CD160 Receptor Expressed by Endothelial Cells. Blood (2006) 108:2608–15. doi: 10.1182/blood-2005-12-019919
35. González A, Rebmann V, LeMaoult J, Horn PA, Carosella ED, Alegre E. The Immunosuppressive Molecule HLA-G and Its Clinical Implications. Crit Rev Clin Lab Sci (2012) 49:63–84. doi: 10.3109/10408363.2012.677947
36. Würfel FM, Winterhalter C, Trenkwalder P, Wirtz RM, Würfel W. European Patent in Immunoncology: From Immunological Principles of Implantation to Cancer Treatment. Int J Mol Sci (2019) 20:1830. doi: 10.3390/ijms20081830
37. Hò GT, Celik AA, Huyton T, Hiemisch W, Blasczyk R, Simper GS, et al. NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain. Int J Mol Sci (2020) 21:4362. doi: 10.3390/ijms21124362
38. Baudhuin J, Migraine J, Faivre V, Loumagne L, Lukaszewicz AC, Payen D, et al. Exocytosis Acts as a Modulator of the ILT4-Mediated Inhibition of Neutrophil Functions. Proc Natl Acad Sci USA (2013) 110:17957–62. doi: 10.1073/pnas.1221535110
39. Lin A, Yan WH. Intercellular Transfer of HLA-G: Its Potential in Cancer Immunology. Clin Transl Immunol (2019) 8:e1077. doi: 10.1002/cti2.1077
40. Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E Surface Expression Depends on Binding of TAP-Dependent Peptides Derived From Certain HLA Class I Signal Sequences. J Immunol (1998) 160:4951–60.
41. Llano M, Lee N, Navarro F, García P, Albar JP, Geraghty DE, et al. HLA-E-Bound Peptides Influence Recognition by Inhibitory and Triggering CD94/NKG2 Receptors: Preferential Response to an HLA-G-Derived Nonamer. Eur J Immunol (1998) 28:2854–63. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W
42. Teklemariam T, Zhao L, Hantash BM. Full-Length HLA-G1 and Truncated HLA-G3 Differentially Increase HLA-E Surface Localization. Hum Immunol (2012) 73:898–905. doi: 10.1016/j.humimm.2012.06.007
43. Jiang J, Natarajan K, Margulies DH. MHC Molecules, T Cell Receptors, Natural Killer Cell Receptors, and Viral Immunoevasins-Key Elements of Adaptive and Innate Immunity. Adv Exp Med Biol (2019) 1172:21–62. doi: 10.1007/978-981-13-9367-9_2
44. Jasinski-Bergner S, Mandelboim O, Seliger B. Molecular Mechanisms of Human Herpes Viruses Inferring With Host Immune Surveillance. J Immunother Cancer (2020) 8:e000841. doi: 10.1136/jitc-2020-000841
45. Xu HH, Yan WH, Lin A. The Role of HLA-G in Human Papillomavirus Infections and Cervical Carcinogenesis. Front Immunol (2020) 11:1349. doi: 10.3389/fimmu.2020.01349
46. Amiot L, Vu N, Samson M. Biology of the Immunomodulatory Molecule HLA-G in Human Liver Diseases. J Hepatol (2015) 62:1430–7. doi: 10.1016/j.jhep.2015.03.007
47. Svendsen SG, Hantash BM, Zhao L, Faber C, Bzorek M, Nissen MH, et al. The Expression and Functional Activity of Membrane-Bound Human Leukocyte Antigen-G1 Are Influenced by the 3’-Untranslated Region. Hum Immunol (2013) 74:818–27. doi: 10.1016/j.humimm.2013.03.003
48. Hviid TV, Rizzo R, Christiansen OB, Melchiorri L, Lindhard A, Baricordi OR. HLA-G and IL-10 in Serum in Relation to HLA-G Genotype and Polymorphisms. Immunogenetics (2004) 56:135–41. doi: 10.1007/s00251-004-0673-2
49. Chen XY, Yan WH, Lin A, Xu HH, Zhang JG, Wang XX. The 14 Bp Deletion Polymorphisms in HLA-G Gene Play an Important Role in the Expression of Soluble HLA-G in Plasma. Tissue Antigens (2008) 72:335–41. doi: 10.1111/j.1399-0039.2008.01107.x
50. Poras I, Yaghi L, Martelli-Palomino G, Mendes-Junior CT, Muniz YC, Cagnin NF, et al. Haplotypes of the HLA-G 3’ Untranslated Region Respond to Endogenous Factors of HLA-G+ and HLA-G- Cell Lines Differentially. PloS One (2017) 12:e0169032. doi: 10.1371/journal.pone.0169032
51. Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, et al. Allele-Specific Targeting of microRNAs to HLA-G and Risk of Asthma. Am J Hum Genet (2007) 81:829–34. doi: 10.1086/521200
52. Reches A, Nachmani D, Berhani O, Duev-Cohen A, Shreibman D, Ophir Y, et al. HNRNPR Regulates the Expression of Classical and Nonclassical MHC Class I Proteins. J Immunol (2016) 196:4967–76. doi: 10.4049/jimmunol.1501550
53. Yie SM, Xiao R, Librach CL. Progesterone Regulates HLA-G Gene Expression Through a Novel Progesterone Response Element. Hum Reprod (2006) 21:2538–44. doi: 10.1093/humrep/del126
54. Moreau P, Faure O, Lefebvre S, Ibrahim EC, O’Brien M, Gourand L, et al. Glucocorticoid Hormones Upregulate Levels of HLA-G Transcripts in Trophoblasts. Transplant Proc (2001) 33:2277–80. doi: 10.1016/s0041-1345(01)01990-x
55. Ugurel S, Rebmann V, Ferrone S, Tilgen W, Grosse-Wilde H, Reinhold U. Soluble Human Leukocyte Antigen–G Serum Level Is Elevated in Melanoma Patients and Is Further Increased by Interferon-Alpha Immunotherapy. Cancer (2001) 92:369–76. doi: 10.1002/1097-0142(20010715)92:2<369::aid-cncr1332>3.0.co;2-u
56. Le Rond S, Gonzalez A, Gonzalez AS, Carosella ED, Rouas-Freiss N. Indoleamine 2,3 Dioxygenase and Human Leucocyte Antigen-G Inhibit the T-Cell Alloproliferative Response Through Two Independent Pathways. Immunology (2005) 116:297–307. doi: 10.1111/j.1365-2567.2005.02224.x
57. Onno M, Le Friec G, Pangault C, Amiot L, Guilloux V, Drénou B, et al. Modulation of HLA-G Antigens Expression in Myelomonocytic Cells. Hum Immunol (2000) 61:1086–94. doi: 10.1016/s0198-8859(00)00191-9
58. Polakova K, Bandzuchova E, Tirpakova J, Kuba D, Russ G. Modulation of HLA-G Expression. Neoplasma (2007) 54:455–62.
59. Rizzo R, Hviid TV, Stignani M, Balboni A, Grappa MT, Melchiorri L, et al. The HLA-G Genotype Is Associated With IL-10 Levels in Activated PBMCs. Immunogenetics (2005) 57:172–81. doi: 10.1007/s00251-005-0788-0
60. Rizzo R, D’Accolti M, Bortolotti D, Caccuri F, Caruso A, Di Luca D, et al. Human Herpesvirus 6A and 6B Inhibit In Vitro Angiogenesis by Induction of Human Leukocyte Antigen G. Sci Rep (2018) 8:17683. doi: 10.1038/s41598-018-36146-0
61. Kong Y, Wang Y, Wu X, Han J, Li G, Hua M, et al. Storm of Soluble Immune Checkpoints Associated With Disease Severity of COVID-19. Signal Transduct Target Ther (2020) 5:192. doi: 10.1038/s41392-020-00308-2
62. Vono M, Huttner A, Lemeille S, Martinez-Murillo P, Meyer B, Baggio S, et al. Robust Innate Responses to SARS-CoV-2 in Children Resolve Faster Than in Adults Without Compromising Adaptive Immunity. Cell Rep (2021) 37:109773. doi: 10.1016/j.celrep.2021.109773
63. Fainardi E, Castellazzi M, Stignani M, Morandi F, Sana G, Gonzalez R, et al. Emerging Topics and New Perspectives on HLA-G. Cell Mol Life Sci (2011) 68:433–51. doi: 10.1007/s00018-010-0584-3
64. Lozano JM, González R, Kindelán JM, Rouas-Freiss N, Caballos R, Dausset J, et al. Monocytes and T Lymphocytes in HIV-1-Positive Patients Express HLA-G Molecule. AIDS (2002) 16:347–51. doi: 10.1097/00002030-200202150-00005
65. Cabello A, Rivero A, Garcia MJ, Lozano JM, Torre-Cisneros J, González R, et al. HAART Induces the Expression of HLA-G on Peripheral Monocytes in HIV-1 Infected Individuals. Hum Immunol (2003) 64:1045–9. doi: 10.1016/j.humimm.2003.08.353
66. Pizzato N, Derrien M, Lenfant F. The Short Cytoplasmic Tail of HLA-G Determines Its Resistance to HIV-1 Nef-Mediated Cell Surface Downregulation. Hum Immunol (2004) 65:1389–96. doi: 10.1016/j.humimm.2004.07.239
67. Huang J, Burke P, Yang Y, Seiss K, Beamon J, Cung T, et al. Soluble HLA-G Inhibits Myeloid Dendritic Cell Function in HIV-1 Infection by Interacting With Leukocyte Immunoglobulin-Like Receptor B2. J Virol (2010) 84:10784–91. doi: 10.1128/JVI.01292-10
68. Lajoie J, Fontaine J, Tremblay C, Routy JP, Poudrier J, Roger M. Persistence of High Levels of Blood Soluble Human Leukocyte Antigen-G Is Associated With Rapid Progression of HIV Infection. AIDS (2009) 23:1437–40. doi: 10.1097/QAD.0b013e32832d0825
69. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature (2021) 591:92–8. doi: 10.1038/s41586-020-03065-y
70. Matte C, Lajoie J, Lacaille J, Zijenah LS, Ward BJ, Roger M. Functionally Active HLA-G Polymorphisms Are Associated With the Risk of Heterosexual HIV-1 Infection in African Women. AIDS (2004) 18:427–31. doi: 10.1097/00002030-200402200-00008
71. Phoswa WN, Ramsuran V, Naicker T, Singh R, Moodley J. HLA-G Polymorphisms Associated With HIV Infection and Preeclampsia in South Africans of African Ancestry. BioMed Res Int (2020) 2020:1697657. doi: 10.1155/2020/1697657
72. Martinetti M, Pacati I, Cuccia M, Badulli C, Pasi A, Salvaneschi L, et al. Hierarchy of Baby-Linked Immunogenetic Risk Factors in the Vertical Transmission of Hepatitis C Virus. Int J Immunopathol Pharmacol (2006) 19:369–78. doi: 10.1177/039463200601900213
73. Zhou S, Liu M, Xia Y, Zhang L, Shao L, Wang N, et al. Association of the 3’ Untranslated Region Polymorphisms of HLA-G With Susceptibility to Chronic Hepatitis C Virus Infection in the Chinese Population. Hum Immunol (2021) S0198-8859(21) 00213-5. doi: 10.1016/j.humimm.2021.09.001
74. Rizzo R, Neri LM, Simioni C, Bortolotti D, Occhionorelli S, Zauli G, et al. SARS-CoV-2 Nucleocapsid Protein and Ultrastructural Modifications in Small Bowel of a 4-Week-Negative COVID-19 Patient. Clin Microbiol Infect (2021) 27:936–7. doi: 10.1016/j.cmi.2021.01.012
75. Zhang S, Gan J, Chen BG, Zheng D, Zhang JG, Lin RH, et al. Dynamics of Peripheral Immune Cells and Their HLA-G and Receptor Expressions in a Patient Suffering From Critical COVID-19 Pneumonia to Convalescence. Clin Transl Immunol (2020) 9:e1128. doi: 10.1002/cti2.1128
76. Al-Bayatee NT, Ad’hiah AH. Soluble HLA-G Is Upregulated in Serum of Patients With Severe COVID-19. Hum Immunol (2021) 82:726–32. doi: 10.1016/j.humimm.2021.07.007
77. Bortolotti D, Gentili V, Rizzo S, Schiuma G, Beltrami S, Spadaro S, et al. Increased sHLA-G Is Associated With Improved COVID-19 Outcome and Reduced Neutrophil Adhesion. Viruses (2021) 13:1855. doi: 10.3390/v13091855
78. Kadkhoda K. COVID-19: An Immunopathological View. mSphere (2020) 5:e00344–20. doi: 10.1128/mSphere.00344-20
79. Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen MC, et al. Immune Response to SARS-CoV-2 and Mechanisms of Immunopathological Changes in COVID-19. Allergy (2020) 75:1564–81. doi: 10.1111/all.14364
80. Chauhan AJ, Wiffen LJ, Brown TP. COVID-19: A Collision of Complement, Coagulation and Inflammatory Pathways. J Thromb Haemost (2020) 18:2110–7. doi: 10.1111/jth.14981
81. Market M, Angka L, Martel AB, Bastin D, Olanubi O, Tennakoon G, et al. Flattening the COVID-19 Curve With Natural Killer Cell Based Immunotherapies. Front Immunol (2020) 11:1512. doi: 10.3389/fimmu.2020.01512
82. Fathi N, Rezaei N. Lymphopenia in COVID-19: Therapeutic Opportunities. Cell Biol Int (2020) 44:1792–7. doi: 10.1002/cbin.11403
83. Amodio G, Sales de Albuquerque R, Gregori S. New Insights Into HLA-G Mediated Tolerance. Tissue Antigens (2014) 84:255–63. doi: 10.1111/tan.12427
84. Yao AY, Tang HY, Wang Y, Feng MF, Zhou RL. Inhibition of the Activating Signals in NK92 Cells by Recombinant GST-sHLA-G1a Chain. Cell Res (2004) 14:155–60. doi: 10.1038/sj.cr.7290215
85. Lesport E, Baudhuin J, Sousa S, LeMaoult J, Zamborlini A, Rouas-Freiss N, et al. Inhibition of Human Gamma Delta [Corrected] T-Cell Antitumoral Activity Through HLA-G: Implications for Immunotherapy of Cancer. Cell Mol Life Sci (2011) 68:3385–99. doi: 10.1007/s00018-011-0632-7
86. Lila N, Rouas-Freiss N, Dausset J, Carpentier A, Carosella ED. Soluble HLA-G Protein Secreted by Allo-Specific CD4+ T Cells Suppresses the Allo-Proliferative Response: A CD4+ T Cell Regulatory Mechanism. Proc Natl Acad Sci USA (2001) 98:12150–5. doi: 10.1073/pnas.201407398
87. Liang S, Horuzsko A. Mobilizing Dendritic Cells for Tolerance by Engagement of Immune Inhibitory Receptors for HLA-G. Hum Immunol (2003) 64:1025–32. doi: 10.1016/j.humimm.2003.08.348
88. Naji A, Menier C, Morandi F, Agaugué S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-Bearing Ig-Like Transcript 2 Receptor Suppresses B Cell Responses. J Immunol (2014) 192:1536–46. doi: 10.4049/jimmunol.1300438
89. Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M, et al. The Laboratory Tests and Host Immunity of COVID-19 Patients With Different Severity of Illness. JCI Insight (2020) 5:e137799. doi: 10.1172/jci.insight.137799
90. Puppo F, Contini P, Ghio M, Indiveri F. Soluble HLA Class I Molecules/CD8 Ligation Trigger Apoptosis of CD8+ Cells by Fas/Fas-Ligand Interaction. ScientificWorldJournal (2002) 2:421–3. doi: 10.1100/tsw.2002.122
91. Bellesi S, Metafuni E, Hohaus S, Maiolo E, Marchionni F, D’Innocenzo S, et al. Increased CD95 (Fas) and PD-1 Expression in Peripheral Blood T Lymphocytes in COVID-19 Patients. Br J Haematol (2020) 191:207–11. doi: 10.1111/bjh.17034
92. Selmani Z, Naji A, Zidi I, Favier B, Gaiffe E, Obert L, et al. Human Leukocyte Antigen-G5 Secretion by Human Mesenchymal Stem Cells Is Required to Suppress T Lymphocyte and Natural Killer Function and to Induce CD4+CD25highFOXP3+ Regulatory T Cells. Stem Cells (2008) 26:212–22. doi: 10.1634/stemcells.2007-0554
93. Agaugué S, Carosella ED, Rouas-Freiss N. Role of HLA-G in Tumor Escape Through Expansion of Myeloid-Derived Suppressor Cells and Cytokinic Balance in Favor of Th2 Versus Th1/Th17. Blood (2011) 117:7021–31. doi: 10.1182/blood-2010-07-294389
94. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, et al. Differentiation of Type 1 T Regulatory Cells (Tr1) by Tolerogenic DC-10 Requires the IL-10-Dependent ILT4/HLA-G Pathway. Blood (2010) 116:935–44. doi: 10.1182/blood-2009-07-234872
95. Lee CL, Guo Y, So KH, Vijayan M, Guo Y, Wong VH, et al. Soluble Human Leukocyte Antigen G5 Polarizes Differentiation of Macrophages Toward a Decidual Macrophage-Like Phenotype. Hum Reprod (2015) 30:2263–74. doi: 10.1093/humrep/dev196
96. Tomić S, Đokić J, Stevanović D, Ilić N, Gruden-Movsesijan A, Dinić M, et al. Reduced Expression of Autophagy Markers and Expansion of Myeloid-Derived Suppressor Cells Correlate With Poor T Cell Response in Severe COVID-19 Patients. Front Immunol (2021) 12:614599. doi: 10.3389/fimmu.2021.614599
97. Chen X, Huang J, Huang Y, Chen J, Huang Y, Jiang X, et al. Characteristics of Immune Cells and Cytokines in Patients With Coronavirus Disease 2019 in Guangzhou, China. Hum Immunol (2020) 81:702–8. doi: 10.1016/j.humimm.2020.08.006
98. Neumann J, Prezzemolo T, Vanderbeke L, Roca CP, Gerbaux M, Janssens S, et al. Increased IL-10-Producing Regulatory T Cells Are Characteristic of Severe Cases of COVID-19. Clin Transl Immunol (2020) 9:e1204. doi: 10.1002/cti2.1204
99. Eguchi H, Maeda A, Lo PC, Matsuura R, Esquivel EL, Asada M, et al. HLA-G1, But Not HLA-G3, Suppresses Human Monocyte/Macrophage-Mediated Swine Endothelial Cell Lysis. Transplant Proc (2016) 48:1285–7. doi: 10.1016/j.transproceed.2015.10.070
100. Esquivel EL, Maeda A, Eguchi H, Asada M, Sugiyama M, Manabe C, et al. Suppression of Human Macrophage-Mediated Cytotoxicity by Transgenic Swine Endothelial Cell Expression of HLA-G. Transpl Immunol (2015) 32:109–15. doi: 10.1016/j.trim.2014.12.004
101. Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 Is an IL-2-Regulated NK Cell Receptor That Exhibits Limited Expression in Humans But Triggers Strong IFN-Gamma Production. J Immunol (2003) 171:3415–25. doi: 10.4049/jimmunol.171.7.3415
102. Rajagopalan S, Moyle MW, Joosten I, Long EO. DNA-PKcs Controls an Endosomal Signaling Pathway for a Proinflammatory Response by Natural Killer Cells. Sci Signal (2010) 3:ra14. doi: 10.1126/scisignal.2000467
103. Rajagopalan S. HLA-G-Mediated NK Cell Senescence Promotes Vascular Remodeling: Implications for Reproduction. Cell Mol Immunol (2014) 11:460–6. doi: 10.1038/cmi.2014.53
104. Celik AA, Simper GS, Huyton T, Blasczyk R, Bade-Döding C. HLA-G Mediated Immune Regulation Is Impaired by a Single Amino Acid Exchange in the Alpha 2 Domain. Hum Immunol (2018) 79:453–62. doi: 10.1016/j.humimm.2018.03.010
105. Antonioli L, Fornai M, Pellegrini C, Blandizzi C. NKG2A and COVID-19: Another Brick in the Wall. Cell Mol Immunol (2020) 17:672–4. doi: 10.1038/s41423-020-0450-7
106. Crow MS, Lum KK, Sheng X, Song B, Cristea IM. Diverse Mechanisms Evolved by DNA Viruses to Inhibit Early Host Defenses. Crit Rev Biochem Mol Biol (2016) 51:452–81. doi: 10.1080/10409238.2016.1226250
107. Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: From Biology to Clinical Benefits. Trends Immunol (2008) 29:125–32. doi: 10.1016/j.it.2007.11.005
108. Amiot L, Vu N, Rauch M, L’Helgoualc’h A, Chalmel F, Gascan H, et al. Expression of HLA-G by Mast Cells Is Associated With Hepatitis C Virus-Induced Liver Fibrosis. J Hepatol (2014) 60:245–52. doi: 10.1016/j.jhep.2013.09.006
109. Li C, Toth I, Schulze Zur Wiesch J, Pereyra F, Rychert J, Rosenberg ES, et al. Functional Characterization of HLA-G⁺ Regulatory T Cells in HIV-1 Infection. PloS Pathog (2013) 9:e1003140. doi: 10.1371/journal.ppat.1003140
110. Persson G, Bork JBS, Isgaard C, Larsen TG, Bordoy AM, Bengtsson MS, et al. Cytokine Stimulation of the Choriocarcinoma Cell Line JEG-3 Leads to Alterations in the HLA-G Expression Profile. Cell Immunol (2020) 352:104110. doi: 10.1016/j.cellimm.2020.104110
111. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol (2020) 11:1446. doi: 10.3389/fimmu.2020.01446
112. Bouayad A. Features of HLA Class I Expression and Its Clinical Relevance in SARS-CoV-2: What do We Know So Far? Rev Med Virol (2021) 31:e2236. doi: 10.1002/rmv.2236
113. Gustafson CE, Qi Q, Hutter-Saunders J, Gupta S, Jadhav R, Newell E, et al. Immune Checkpoint Function of CD85j in CD8 T Cell Differentiation and Aging. Front Immunol (2017) 8:692. doi: 10.3389/fimmu.2017.00692
114. Peña J, Frías M, Castro-Orgaz L, González R, García F, Gallart T, et al. Effects on Innate Immunity of a Therapeutic Dendritic Cell-Based Vaccine for HIV-1 Infection. Viral Immunol (2012) 25:37–44. doi: 10.1089/vim.2011.0044
115. Zhang C, Wang XM, Li SR, Twelkmeyer T, Wang WH, Zhang SY, et al. NKG2A Is a NK Cell Exhaustion Checkpoint for HCV Persistence. Nat Commun (2019) 10:1507. doi: 10.1038/s41467-019-09212-y
116. Rapaport AS, Schriewer J, Gilfillan S, Hembrador E, Crump R, Plougastel BF, et al. The Inhibitory Receptor NKG2A Sustains Virus-Specific CD8⁺ T Cells in Response to a Lethal Poxvirus Infection. Immunity (2015) 43:1112–24. doi: 10.1016/j.immuni.2015.11.005
117. André P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor That Promotes Anti-Tumor Immunity by Unleashing Both T and NK Cells. Cell (2018) 175:1731–1743.e13. doi: 10.1016/j.cell.2018.10.014
118. Kamiya T, Seow SV, Wong D, Robinson M, Campana D. Blocking Expression of Inhibitory Receptor NKG2A Overcomes Tumor Resistance to NK Cells. J Clin Invest (2019) 129:2094–106. doi: 10.1172/JCI123955
119. Li F, Wei H, Wei H, Gao Y, Xu L, Yin W, et al. Blocking the Natural Killer Cell Inhibitory Receptor NKG2A Increases Activity of Human Natural Killer Cells and Clears Hepatitis B Virus Infection in Mice. Gastroenterology (2013) 144:392–401. doi: 10.1053/j.gastro.2012.10.039
Keywords: HLA-G, immune receptor, SARS-CoV-2, COVID-19, immune suppression
Citation: Lin A and Yan W-H (2021) Perspective of HLA-G Induced Immunosuppression in SARS-CoV-2 Infection. Front. Immunol. 12:788769. doi: 10.3389/fimmu.2021.788769
Received: 03 October 2021; Accepted: 17 November 2021;
Published: 06 December 2021.
Edited by:
Raffaella Greco, San Raffaele Hospital (IRCCS), ItalyReviewed by:
Gabriele Casirati, Dana–Farber Cancer Institute, United StatesCopyright © 2021 Lin and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Hua Yan, eWFud2hjb21AeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.