- 1Women’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Cancer Hospital of University of Chinese Academy of Sciences, Zhejiang Cancer Hospital, Hangzhou, China
- 3Key Laboratory of Women’s Reproductive Health of Zhejiang Province, Hangzhou, China
- 4Key Laboratory of Reproductive Genetics, Ministry of Education, Zhejiang University, Hangzhou, China
Recurrent pregnancy loss (RPL), especially the unexplained RPL, is associated with the disruption of maternal immune tolerance. However, little is known about the immune status at the decidua of RPL with embryonic chromosomal aberrations. Herein, mass cytometry (CyTOF) was used to interrogate the immune atlas at the decidua which was obtained from 15 RPL women—six with normal chromosome and nine with chromosomal aberrations—and five controls. The total frequency of CCR2−CD11chigh macrophages increased, while CD39high NK cells and CCR2−CD11clow macrophages decrease significantly in RPL when RPLs were stratified, compared with controls. Pro-inflammatory subsets of CD11chigh macrophages increased, while less pro-inflammatory or suppressive subsets decreased statistically in RPL decidua whenever RPLs were stratified or not. However, CD11chigh NK and CD161highCD8+ T cells increased only in RPL with normal chromosome, while the inactivated and naive CD8+/CD4+ T cells were enriched only in RPL with chromosomal aberrations. A pro-inflammatory signature is observed in RPL decidua; however, differences exist between RPL with and without chromosomal abnormalities.
1 Introduction
Recurrent pregnancy loss (RPL) is defined as two or more consecutive pregnancy losses before the 20th week of gestation and occurs in about 1%–3% of reproductive women (1). With the number of miscarriages increased, the recurrent risk of pregnancy loss raises quickly from about 11% at one loss to 40% after three or more losses (2). Though possible causes including uterine malformations, maternal infection, endocrine disorders, and genetic abnormalities have been identified, only maternal age and the number of prior pregnancy losses are considered as prognostic factors for majority of RPL women (1). No obvious cause can be found in more than 50% of RPL even after comprehensive investigation, and they are called as unexplained RPL (3).
It has been proposed that RPL, especially the unexplained RPL, arises from the disruption of maternal immune homeostasis (4). Innate lymphoid cells (ILCs), myeloid cells, T cells and B cells contribute to the maintenance of tolerance in pregnancy (5–7). ILCs, including NK cells and non-NK ILCs, are the most abundant immune cells in a pregnant uterus. Decidual NK cells (dNKs) possess a CD56brightCD16− phenotype and account for 50%–70% of leukocytes in the first trimester. They not only modulate trophoblast invasion by producing IL-8 and IL-10 but also initiate artery remodeling through secreting vascular endothelial growth factor (VEGF) and arginase-1 (5, 6). dNK plays an important role in extravillous trophoblast (EVT) cells acquiring invasive phenotype and invasive EVT gaining endothelial-like properties (8). Decidual non-NK ILCs are mainly ILC3 and participate in tissue remodeling by expressing IL-22 and IL-8 or inducing decidual stromal cell (DSC) to express adhesion molecules (9).
Myeloid cells, mainly including macrophages and dendritic cells (DCs), remain relatively high and constant during gestation. Decidual macrophages play an essential role in inducing maternal immune tolerance, modulating vascular modeling and trophoblast invasion (10). DCs are the most efficient type of antigen-presenting cells and can drive naive CD4+ T-cell polarization to a Th2 phenotype (11).
Decidual CD4+/CD8+ T cells are associated with recognizing fetal antigens and regulating decidual immune microenvironment, and activated CD4+/CD8+ T cells and Foxp3+ Tregs are essential in mediating immunological tolerance to the fetus (12). B cells, particularly Breg cells, can induce maternal tolerance by producing IL-10 or the CD39/CD73 pathway (13). Immune dysregulations from each type of cells are associated with RPL.
In unexplained RPL women aged less than 35 years of age, the proportion of blastocyst aneuploidy increased, but the clinical miscarriage rate is higher when they have euploid embryos transferred by preimplantation genetic testing (PGT) than that in women with PGT just for monogenic defects (14). In women over 35 years old aneuploidy rates neither in embryo nor in blastocysts are not significantly different between sporadic abortion and RPL or idiopathic RPL (14, 15). These findings indicate that embryonic chromosomal abnormalities are not the only cause for RPL and may occur no matter how old women are.
Trisomy 21 (T21) is one of the most common chromosomal abnormalities in RPL and is also the most widespread survival autosomal aneuploidy. The incidence of leukemia is approximately 50-fold higher in children with T21 compared with normal children in the first 5 years of life (16), while adults with T21 are predisposed to autoimmune diseases and global immune dysregulations (17). These imply that the extra copy of chromosome 21 in embryo may induce immune variations. Thus, we hypothesized that immune abnormalities might exist in RPL with embryonic chromosomal aberrations.
Single-cell technologies such as CyTOF and single-cell RNA sequencing (scRNA-seq) have been used to explore the immune-related physiological mechanism in early or peripartum decidual tissue and endometrium across the menstrual cycle (5, 18, 19). Though a few studies using scRNA-seq have demonstrated a systematic pro-inflammatory feature in RPL with normal embryonic chromosome (20–23), the global immune landscape of RPL, especially those with embryonic chromosomal aberrations, is still unknown.
Herein, we harnessed the power of CyTOF and tried to reveal the immune phenotypic characteristics of RPL with or without embryonic chromosomal aberrations. We observed a pro-inflammatory signature in RPL whether the embryonic chromosome is normal or not, and we also noticed differences between RPL with and without chromosomal abnormalities.
2 Materials and Methods
2.1 Subjects
Fifteen RPL women, six with normal chromosome and nine with chromosome aberrations, and five controls were enrolled from May 2020 to October 2020 at Women’s Hospital, Zhejiang University School of Medicine. The baseline characteristics were summarized in Table 1. RPLs were diagnosed according to the criteria of ESHRE 2017 (1). RPL women with autoimmune disorders such as systemic lupus erythematosus, antiphospholipid antibody syndrome, and those prescribed with immunosuppressor, aspirin, or low molecular heparin were excluded from this study. Five women undergoing artificial abortions with a similar gestation served as controls. They should not have the above autoimmune disorders and prior miscarriages and stillbirth. The protocol was approved by the ethics committee of the hospital (IRB-20200126-R) and informed consents were obtained from all participants.
2.2 Villus Separation and Embryonic Chromosome Analyses
Villus was obtained and separated as previously described (24) and was stored at −80°C before use. Chromosomal microarray analysis (CMA) or copy number variation sequencing (CNV-seq) was used to analyze the karyotypes of villus.
2.3 Mass Cytometry
2.3.1 Decidual Tissue Single-Cell Dissociation and Sample Preparation
Decidual tissues were washed with saline, collected in ice-cold MACS Tissue storage solution (catalog no. 130-100-008, Miltenyi, Germany) and dissociated into single cells. CyTOF analyses were performed by PLT Tech Inc. (Hangzhou, China). In brief, cells were stained 5 min with 5 µM 194Pt cisplatin (Fluidigm) for viability in phosphate buffered saline (PBS). After being blocked with PBS containing 5% goat serum and 30% bovine serum albumin (BSA) for 30 min at 4°C, cells were stained with cell-surface antibodies for 30 min at 4°C and washed twice with PBS containing 2.5% BSA. The Foxp3 Fixation and Permeabilization kit (eBioscience) was used according to the instructions of the manufacturer, and cells were incubated overnight in 1.6% paraformaldehyde (PFA) PBS with 100 nM iridium nucleic acid intercalator (Fluidigm) and lastly incubated with intracellular antibodies in permeabilization buffer for 30 min at 4 C after being washed twice with Foxp3 permeabilization buffer and twice with FACS buffer. Cells were washed, resuspended at a concentration of one million cells/ml with water containing EQ Four Element Calibration Beads (Fluidigm), and analyzed within 12 h on a Helios CyTOF System (Fluidigm) at an event rate of <500 events/s.
2.3.2 Panel Design, CyTOF Data Acquisition, and Visualization
A list of 42 metal-tagged monoclonal antibodies including their label, clone, source, cat. number, dilution, and staining information was provided in Supplementary Table S1. All data were produced on a Helio3 CyTOF Mass Cytometer (Fluidigm) and normalized by the bead-based normalization software, which used the intensity values of a sliding window of these bead standards to correct for instrument fluctuations over time and among samples. The normalized FCS files were loaded into Cytobank (www.cytobank.org) to define CD45+ viable single living cells and other subpopulations. The gating strategies were shown in Supplementary Figure S1. Then FCS files were loaded into R and analyzed with an in-house written script. Cluster-colored tSNE maps and phenotype heatmaps were generated by the expression levels of markers with arcsinh transformation from 5,000 cells of each sample except Tregs and B cells (all cells). Hyperparameters were set as perplexity of 30, theta of 0.5, and iterations of 1,000 per 100,000 analyzed cells.
2.4 Statistical Analyses
RPLs were firstly stratified by embryonic chromosome: RPL-NK was composed of six RPLs with normal chromosome and RPL-AK was composed of nine RPLs with chromosome aberrations, while NC-NK was composed of five controls. One-way ANOVA analysis was applied followed by the Benjamini, Krieger, and Yekutieli test, and the adjusted false discovery rate (FDR) <0.05 was considered significant. Then, RPLs were compared with controls without stratification and Student’s t-test was used with P-value <0.05 considered as statistical significance. GraphPad Prism version 9 software was used for the statistical analyses.
3 Results
3.1 The General Immune Landscapes of RPL
CD45+ leukocytes were clustered by phenograph (Figures 1A–C and Supplementary Figure S1). The percentages of M2 macrophages statistically decreased in both RPL subgroups when they were stratified, compared with controls (Figure 1D), while variation existed among the individuals (Figures 1E, F). The M1 macrophage LC14 (individually labeled) increased more than two-fold and the M2 cluster LC2 decreased statistically but less than half in each RPL subgroup. NK cluster LC33 decreased more than half in both RPL subgroups, while clusters LC5 and LC12 increased (no more than two-fold and more than two-fold, respectively) only in RPL with normal chromosome (Figure 1G and Supplementary Table S2). When RPLs were not stratified, NK cluster LC33 and M1 cluster LC14 remained statistically different (Figure 1H).
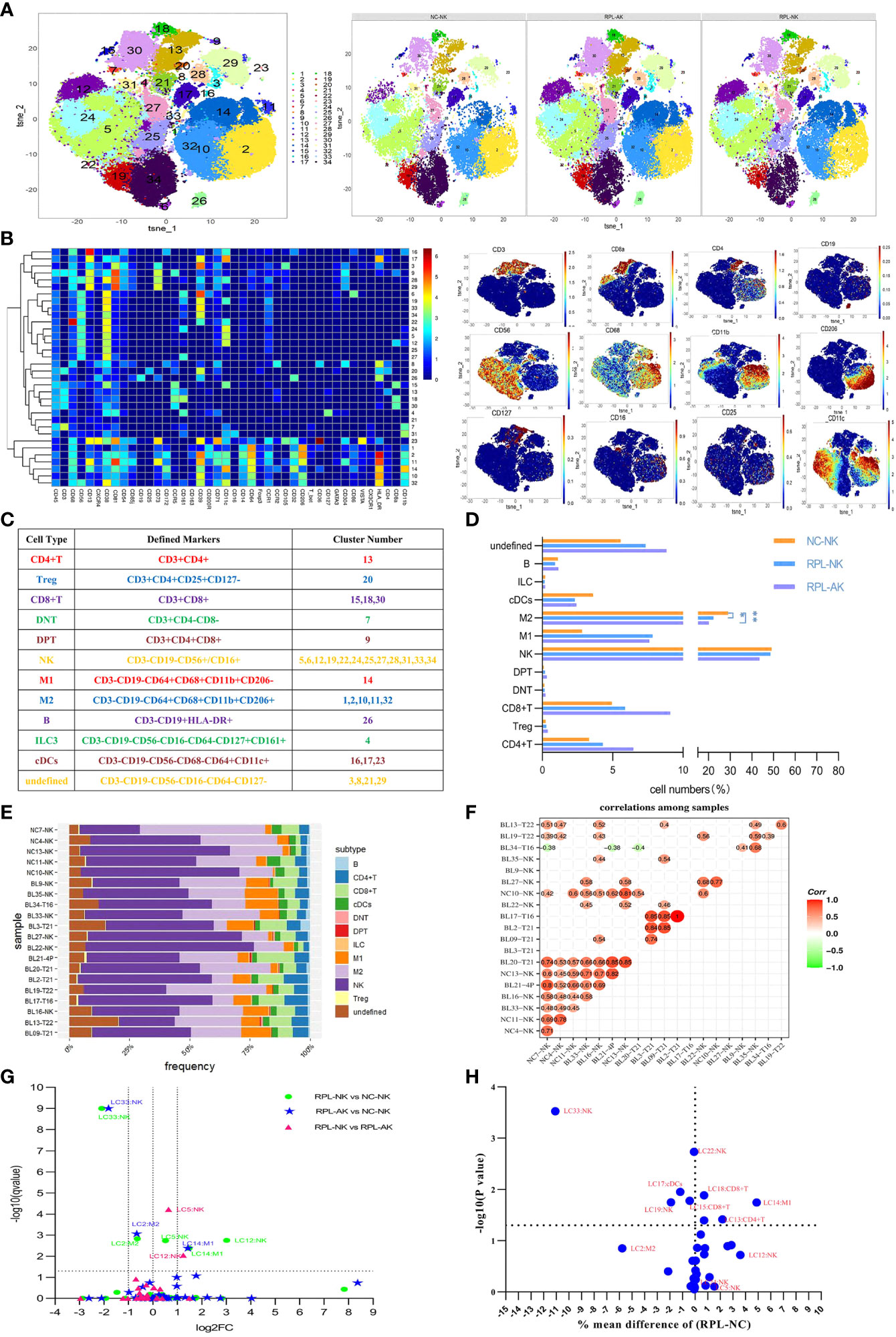
Figure 1 The general immune landscape of RPL. (A) CD45+ t-distributed stochastic neighbor embedding (tSNE) with all samples combined (left) and composite tSNE for groups (right). (B) Heatmap with clusters and the expression of 42 markers (left) and expression plots of the key markers (right). (C) Sum of cell subtypes, clusters, and their defined markers. (D) Graph showing cell numbers (%) in CD45+ cell population, * adjusted P < 0.05, ** adjusted P < 0.001. (E) Stack graph showing the frequency of subtypes of each sample. Samples were named by their code and embryonic chromosome result. (F) Pearson’s correlation coefficient used to evaluate the correlations among samples. (G) Volcano plot showing differential clusters when RPL was stratified. Fold change (FC) = the mean percent ratio of RPL-NK over NC-NK in RPL-NK vs. NC-NK, and so on. A dotted grid line Y = −log10 (0.05) = 1.30103 showing no statistical significance. Two dotted grid lines at X = 1, −1 showing increased two-fold or decreased half. (H) Volcano plot showing the differential clusters between RPL and controls when compared without stratification.
3.2 CD39high NK Cells Decreased in RPL
ILCs were clustered from manually gated CD45+CD3−CD19−HLA-DR− subpopulation (Figures 2A–C and Supplementary Figure S1). The overall frequency of CD39highNK cells and two CD39high NK cell subsets (NKC15, NKC26) decreased significantly in each RPL subgroup when RPLs were stratified, compared with controls (Figures 2D–F and Supplementary Table S2). If not stratified by embryonic chromosome, differences of NKC26, NKC13, and NKC25 between RPL and controls were obvious (Figure 2G). Only one NK ILC cluster (cluster 24) was identified and no difference was detected (data not shown). The phenotype signature of NKC26 was mirrored to LC33 (Figures 8A, B).
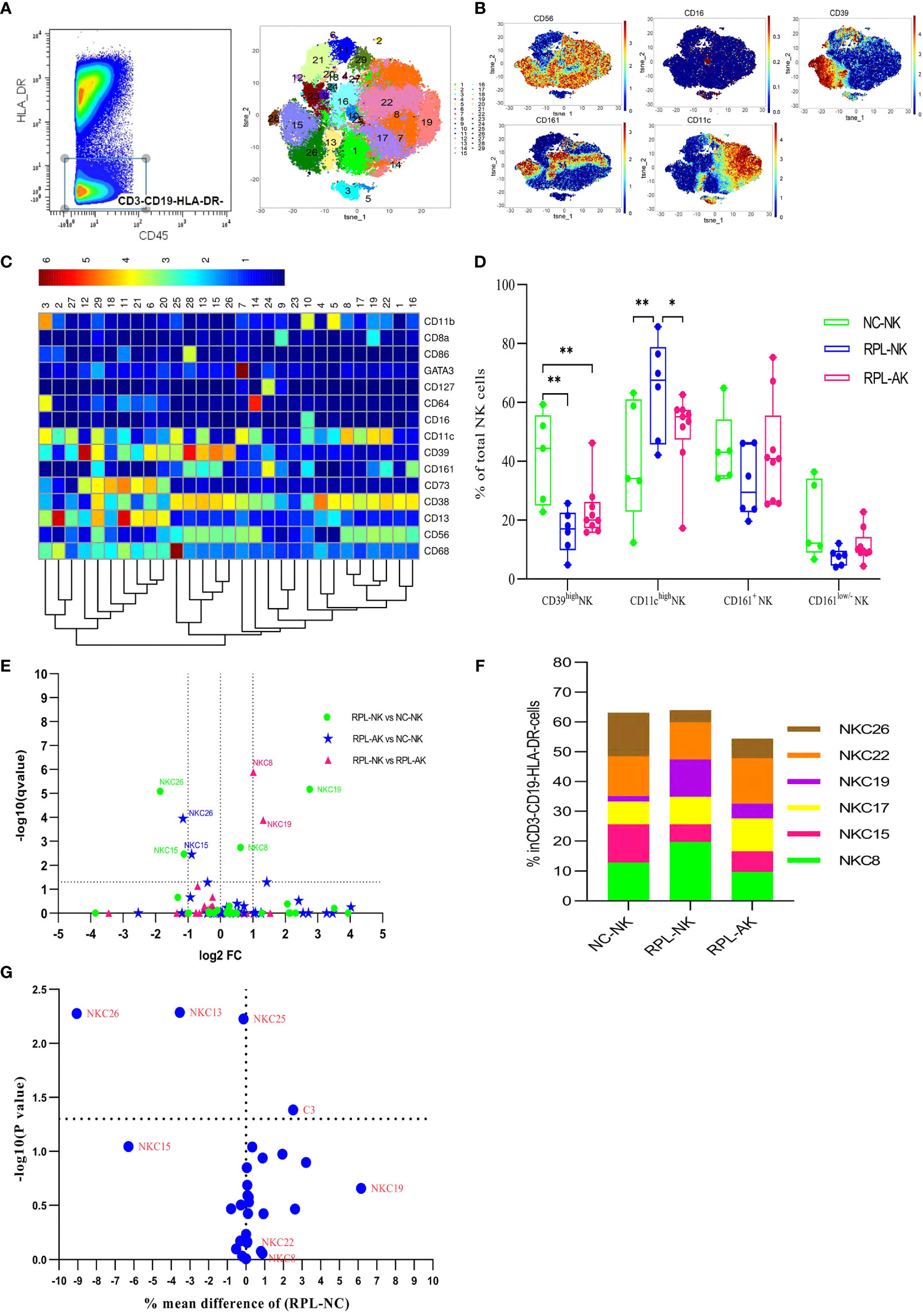
Figure 2 Cluster analysis of ILCs in the CD3−CD19−HLA-DR− population. (A) CD3−CD19−HLA-DR− population was manually gated (left) and tSNE in all samples combined (right). (B) Heatmap showing the expression profile of key markers. (C) Heatmap used for the cluster classification with the expression of markers in the CD45+CD3−CD19−HLA-DR− population. (D) Box plot showing the comparison of CD39high NK cells and CD11chigh NK cells. (E) Volcano plot showing the differential clusters when RPLs were stratified by embryonic chromosome. (F) Map with cumulative mean values for differential clusters and/or the most abundant three clusters. (G) Volcano plot showing the differential clusters when RPLs were not stratified. * adjusted P < 0.05, ** adjusted P < 0.001.
3.3 CCR2−CD11chigh Macrophages Increased, While Less Inflammatory CCR2−CD11clow Decreased in RPL
The CD45+CD3−CD19−CD56− subpopulation was gated manually to analyze myeloid macrophages and DCs (Supplementary Figure S1). Macrophages were phenotyped based on the expression of CD64, CD68, and CD11b, while DCs were characterized by CD64−CD11c+ (Figures 3A–C). Compared with controls, total CD11chigh macrophages increased, while CD11clow or CCR2−CD11clow macrophages decreased with MC12 increased more than two-fold, but MC3 and MC19 decreased less than half in RPL subgroups (Figures 3D–F and Supplementary Table S2). MC19 and MC12 were still obviously different when RPLs were not stratified (Figure 3G). Six DC clusters were identified with no significant difference noticed in RPL (data not shown). The phenotype signature of MC3 was mirrored to LC2 (Figures 8A, C).
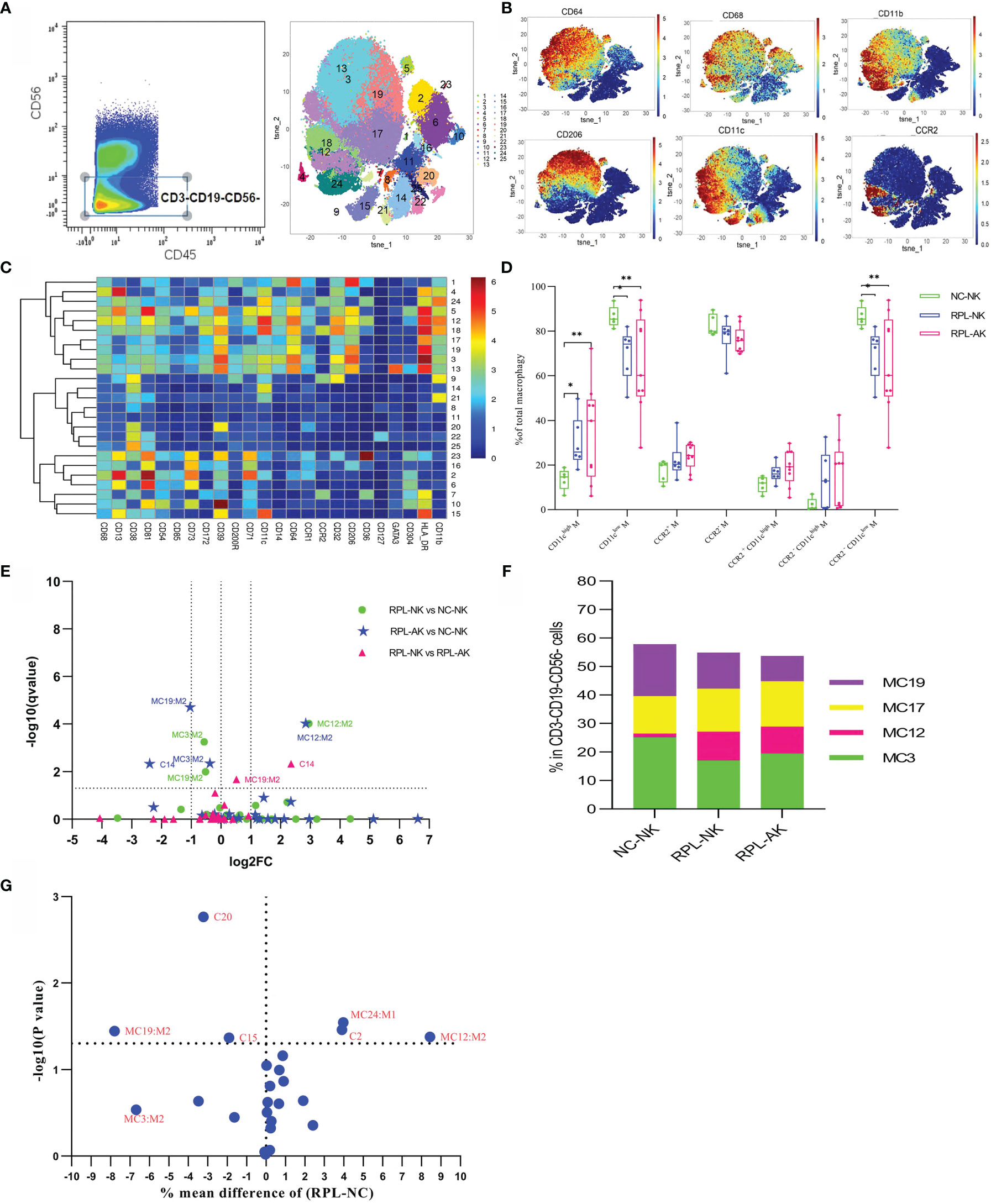
Figure 3 Cluster analysis of myeloid macrophages and DC in the CD3−CD19−CD56− population. (A) Cells manually gated from the CD45+CD3−CD19−CD56− population (left) and tSNE in all samples combined (right). (B) Marker expression profile of the key markers. (C) Heatmap with the expression of markers in the CD45+CD3−CD19−CD56− population. (D) Box plot showing the comparison of total CD11chigh, CD11clow, and/or CCR2−CD11clow when RPLs were stratified. (E) Volcano plot showing the differential clusters when RPLs were stratified by embryonic chromosome. (F) Map with cumulative mean values for differential clusters and/or the most abundant three clusters. (G) Volcano plot showing the differential clusters when RPLs were not stratified. * adjusted P < 0.05, ** adjusted P < 0.001.
3.4 CD38highHLA-DRhighCCR5highCD8+ T and CD8+CD56+ NKT Cells Decreased in RPL
CD3+ T cells were firstly clustered (Figures 4A–D). The CD8+ T-cell cluster TC3, characterized by CD38highHLA-DRhighCCR5high, decreased nearly by half, while CD8+CD56+ NKT cells (TC20) reduced more than half in each RPL subgroup, compared with controls (Figures 4E, F). Except TC3 and TC20, three CD4−CD8− T (DNT) subsets were significantly different between RPL and controls if RPLs were compared without stratification (Figure 4G).
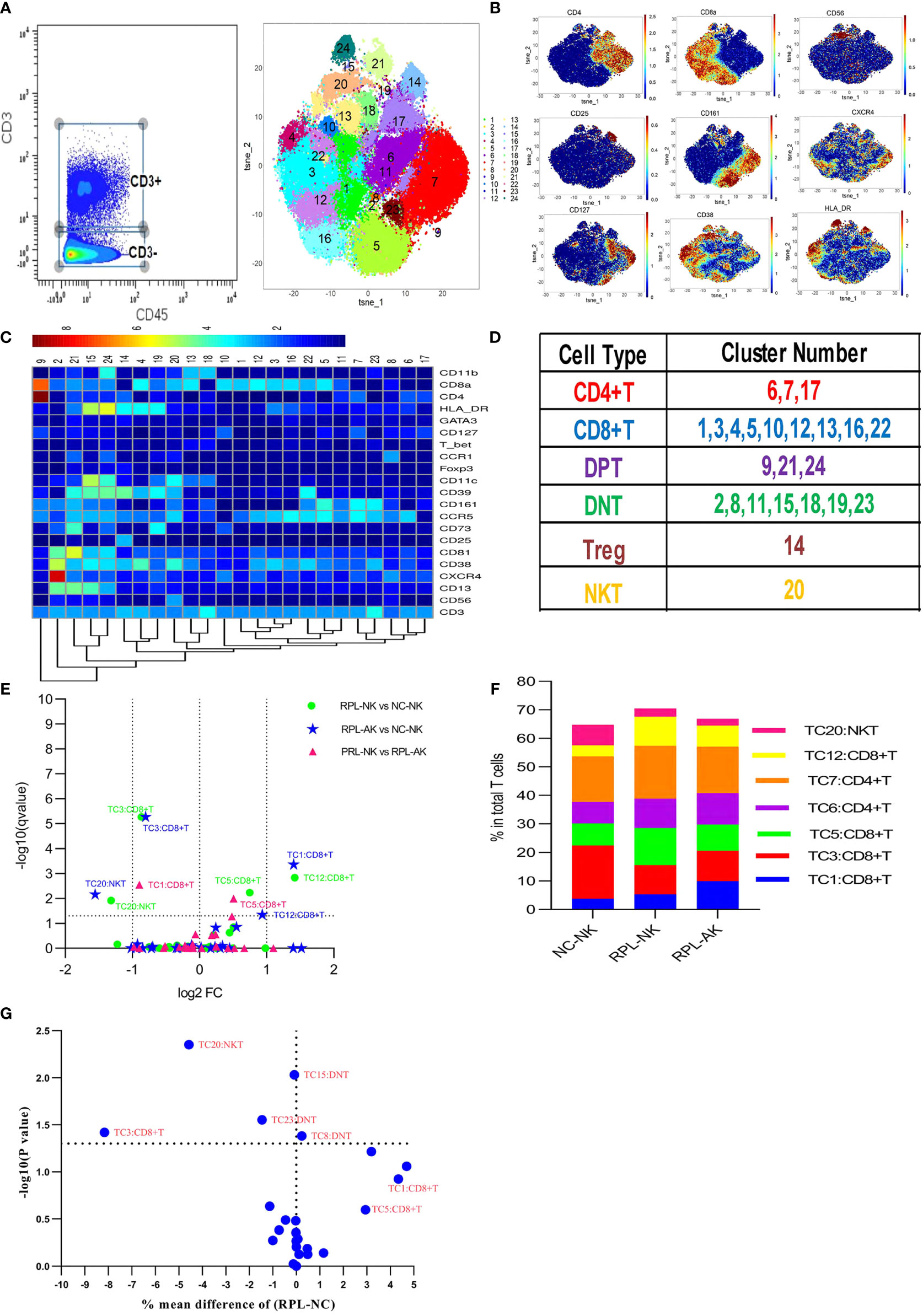
Figure 4 CD3+ T-cell cluster analysis. (A) CD3+ T cells manually gated on CD45+ leukocytes by CD3 expression (left) and CD3+ T-cell tSNE in all samples combined (right). (B) Expression profile of the key markers. (C) Heatmap with clusters and the expression of markers in CD3+ T cell. (D) Sum of the clusters for each subset. (E) Volcano plot showing the differential clusters when RPLs were stratified by embryonic chromosome. (F) Map with cumulative mean values for differential clusters and/or the most abundant three clusters. (G) Volcano plot showing the differential clusters when not stratified.
3.5 CD38highHLA-DRhighCD161high Teff Decreased in RPL
To better phenotype CD4+ T cells, they were clustered from gated CD8− T cells (Figures 5A–C and Supplementary Figure S1). CD4+CCR2+CCR5+ defined an effector T-cell (Teff) phenotype. CD38highHLA-DRhighCD161high Teff subset (CD4+TC15) decreased more than half, while CXCR4+CD161high Teffs (CD4+TC12) increased more than two-fold in each RPL subgroup, compared with controls (Figures 5D, E and Supplementary Table S2). However, the difference of CXCR4+CD161high Teffs was obvious only when RPLs were stratified (Figure 5F).
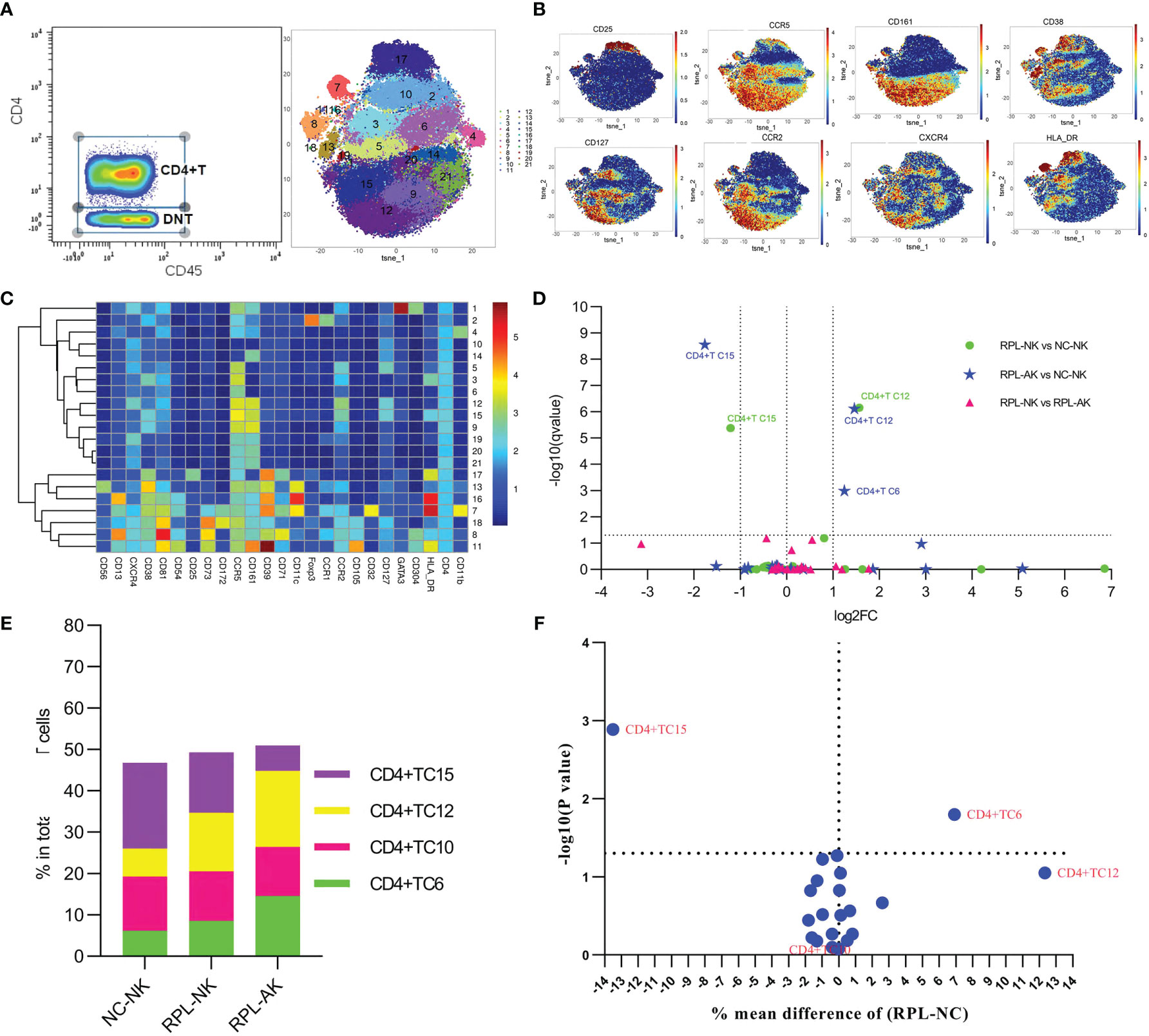
Figure 5 In-depth analysis of CD4+ T cells. (A) CD4+ T cells manually gated on concatenated CD45+CD3+CD8− population (left) and CD4+ T-cell tSNE in all samples combined (right). (B) Expression profile of the key markers. (C) Heatmap with clusters and the expression of markers in CD4+ T-cell clusters. (D) Volcano plot showing the differential clusters when RPLs were stratified by embryonic chromosome. (E) Map with cumulative mean values for differential clusters and/or the most abundant three clusters. (F) Volcano plot showing the differential clusters when RPLs were not stratified.
3.6 CD39−CCR5high Tregs Might Be Enhanced in RPL
Tregs were gated as CD4+CD25+CD127− and clustered automatically (Figures 6A–C and Supplementary Figure S1). The most abundant cluster TregC3, characterized by CD39−CCR5high, increased more than two-fold in each RPL subgroup (Figures 6D, E and Supplementary Table S2). While this difference was not significant between RPL and controls if they were not stratified (Figure 6F).
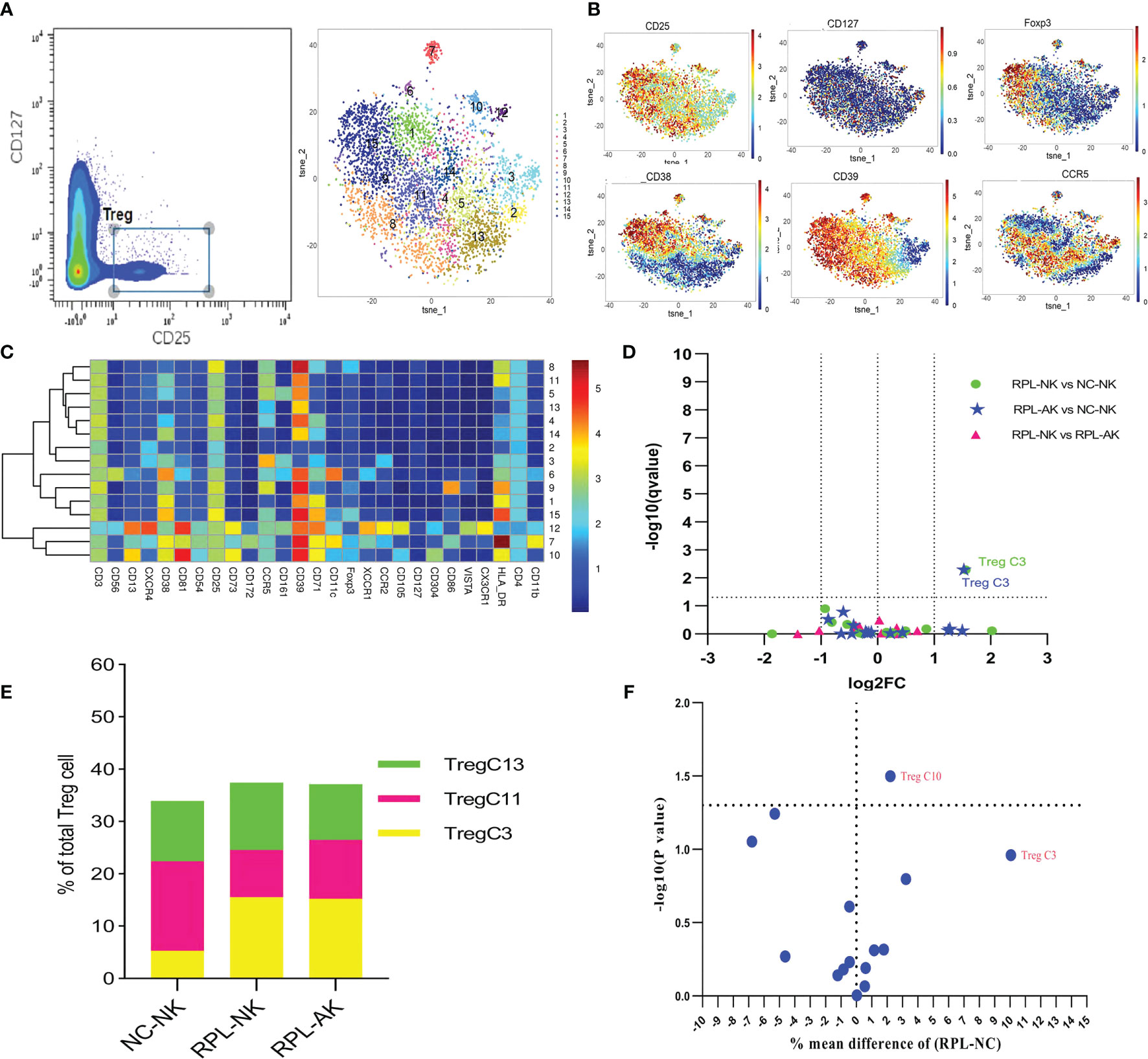
Figure 6 In-depth analysis of Tregs. (A) Tregs manually gated on concatenated CD45+CD25+CD127− population (left) and Tregs tSNE in all samples combined (right). (B) Expression profile of the key markers. (C) Heatmap with clusters and the expression of markers in Tregs. (D) Volcano plot showing the differential clusters when RPLs were stratified by embryonic chromosome. (E) Map with cumulative mean values for differential clusters and/or the most abundant three clusters. (F) Volcano plot showing differential clusters when RPLs were compared without stratification.
3.7 CD85highCD39highCD73− B Cells Might Decrease in RPL
As subsets of B cells could mediate immune tolerance, CD19+HLA-DR+ B cells were clustered (Figures 7A–C). The overall frequency of CD39high B cells decreased in RPL subgroups, compared with controls (Figure 7D). Cluster BC11, characterized by CD85highCD39highCD73−, decreased statistically though less than half, while cluster BC13, characterized by CD39lowCD73high, increased much more in RPL with embryonic chromosomal aberrations (Figures 7E, F and Supplementary Table S2). When RPLs were compared as an entirety, only the CD85highCD39highCD73− B subset was statistically significant (Figure 7G).
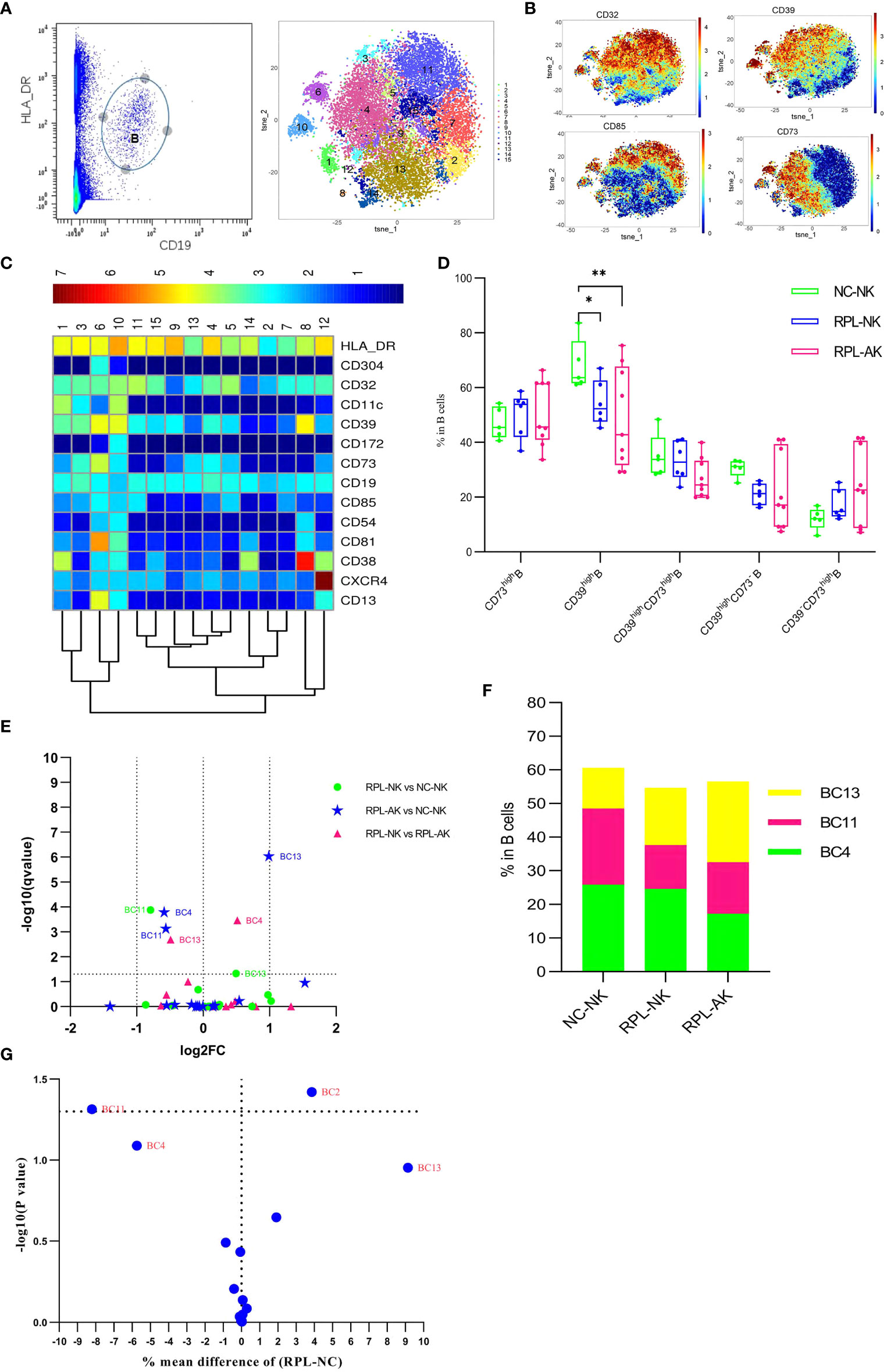
Figure 7 B-cell cluster analysis. (A) B cell manually gated on concatenated CD45+CD3− population (left) and B-cell tSNE in all samples combined (right). (B) Expression profile of key markers. (C) Heatmap with clusters and the expression of markers in CD45+CD3−CD19+HLA-DR+ population. (D) Box plot showing the comparison of total CD39high Breg cell when RPLs were stratified by embryonic chromosome. (E) Volcano plot showing the differential clusters when RPLs were stratified. (F) Map with cumulative mean values for differential clusters and/or the most abundant three clusters. (G) Volcano plot showing differential clusters when RPLs were compared as an entirety. * adjusted P < 0.05, ** adjusted P < 0.001.
3.8 CD11chigh NK and CD161highCD8+ T Cells Were Enriched Only in RPL With Normal Embryonic Chromosome
The overall frequency of CD11chigh NK cells (Figure 2D) and two CD11chigh NK subsets enriched significantly with NKC8 increased less than and NKC19 more than two-fold enriched in RPL with normal embryonic chromosome (Figures 2C, D and Supplementary Table S2).
CD161highCD8+ T cells (TC5) also increased statistically but less than two-fold, which might indicate local inflammatory activation (Figures 4A–E and Supplementary Table S2). The phenotype signature of NKC19 and NKC22 was mirrored to LC12 and LC24, respectively (Figures 8A, C).
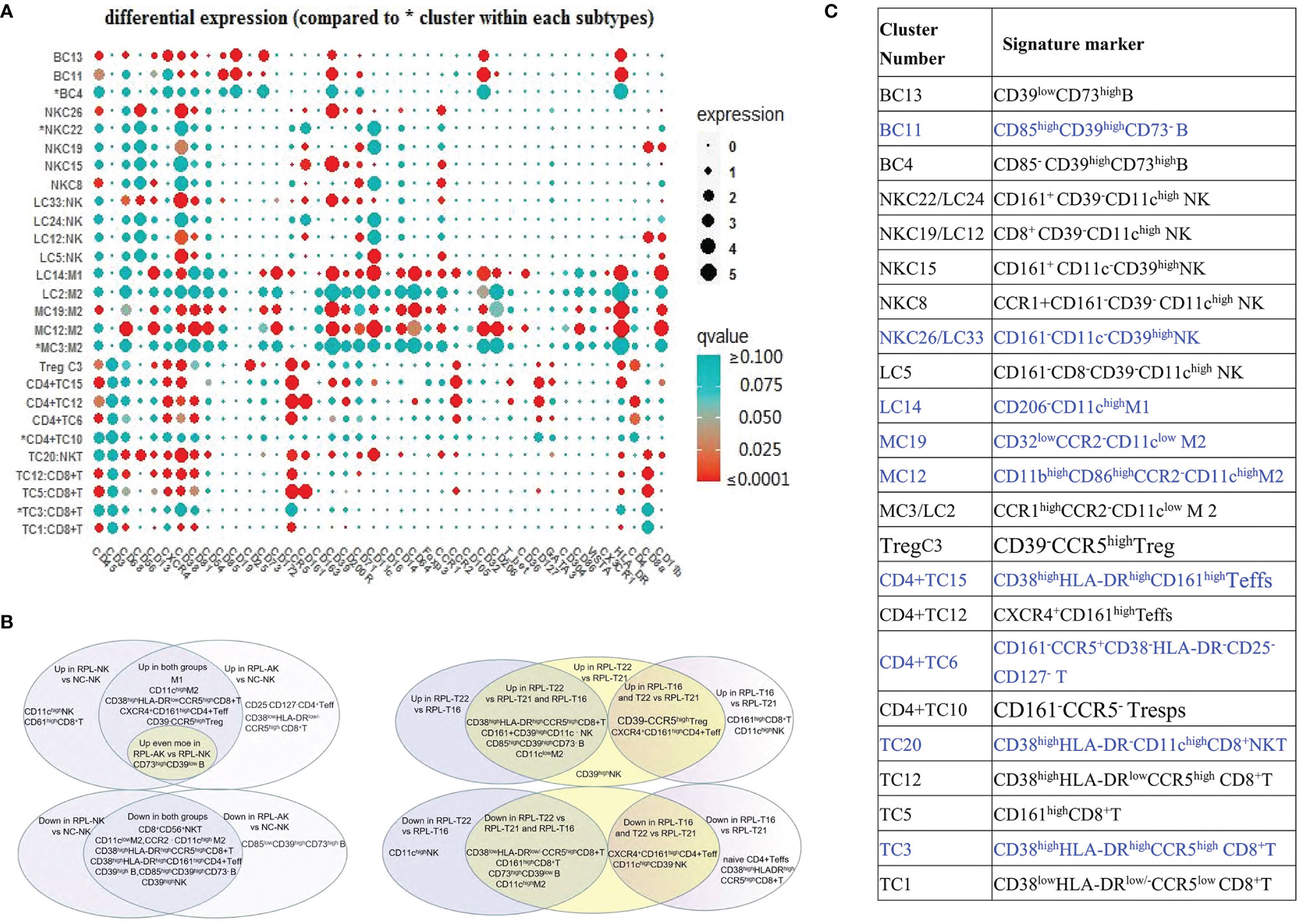
Figure 8 Sum of differential clusters. (A) Bubble map of all markers, compared with the most abundant cluster indicated by *. Circle size showing the expression level (arcsinh) and its colors showing P-value [range from <0.0001 (red) to >0.100 (green)]. (B) Sum of the characteristics of differential clusters and clusters with blue font showing they were differential whenever RPLs were stratified or not. (C) Venn diagram summarizing the differential clusters when RPLs were stratified by embryonic chromosome (left) or the differences among three types of fetal aneuploidy (right).
3.9 Naive CD8+ T/CD4+ T Increased Only in RPL With Embryonic Chromosomal Aberrations
The inactivated or naive CD38lowHLA-DRlow/−CCR5lowCD8+ T cells (TC1) enriched more than two-fold in RPL with embryonic chromosomal aberrations, compared with controls or RPL without chromosomal aberrations (Figure 4E and Supplementary Table S2). A similar tendency was observed in the CD38−HLA-DR−CD25−CD127−CD4+ T subset (CD4+ TC6) and its differential remained obviously when RPLs were not stratified (Figures 5D–F and Supplementary Table S2). B-cell cluster BC4, characterized by CD85−CD39highCD73high, decreased significantly RPL with chromosomalaberrations, but the difference was not obvious if RPLs were compared without stratification (Figures 7C–E and Supplementary Table S2).
3.10 The Immune Atlas Among RPLs With Embryonic Aneuploid Is Heterogeneous
Compared with RPL with embryo T21, pro-inflammatory subsets including CD8+CD39−CD11chigh NK, CXCR4+CD161high Teffs, and CD39−CCR5high Tregs increased more than two-fold, while suppressive or less pro-inflammatory subsets decreased nearly half in RPL with T16 (Figures 8B, 9A, D and Supplementary Table S3). Some suppressive or pro-inflammatory subsets increased more than two-fold, while part of the pro-inflammatory subsets decreased more than half in RPL with T22 (Figures 9B, D and Supplementary Table S3). Suppressive subsets increased, while pro-inflammatory decreased in RPL with T22, when compared with RPL with T16 (Figures 8B, 9C, D and Supplementary Table S3). The total CD11chigh macrophages decreased, while CD11clow or CCR2-CD11clow macrophages increased in RPL with T16 and T22 compared with RPL-T21 (Figure 9E). These results should be assessed with more participants.
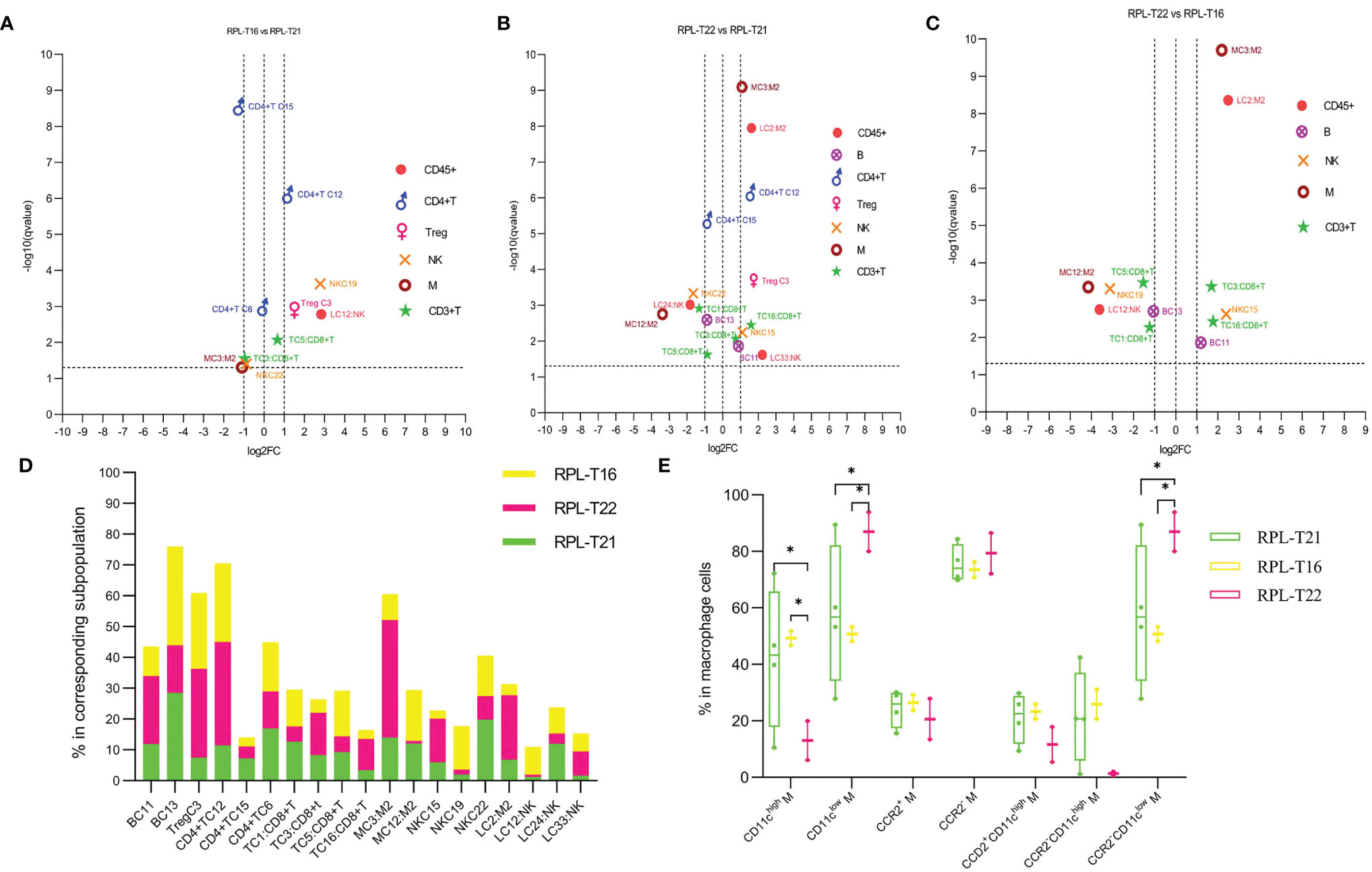
Figure 9 Immune atlas of RPL with embryonic aneuploidy. (A) Volcano plot showing differential clusters in RPL with embryo T16 (RPL-T16), compared with RPL with T21 (RPL-T21). log2FC >1 indicating this cluster increased more than two-fold in RPL-T16 compared with RPL-T21, and so on. (B) Volcano plot showing differential clusters in RPL with T22 (RPL-T22), compared with RPL-T21. (C) Volcano plot showing differential clusters in RPL-T16, compared with RPL-T22. (D) Map with cumulative mean values for differential clusters of RPL with embryos T21, T22, and T16. (E) Frequencies of macrophage subsets in each aneuploidy. * adjusted P < 0.05.
4 Discussion
Previous attention has been mainly focused on the immune mechanism of RPL with normal embryonic chromosome. In the present study, stratification analysis showed that the immune status of RPL with chromosomal aberrations was pro-inflammatory, as suppressive CD39highdNK1 and CCR2−CD11clow macrophages decreased, while pro-inflammatory CCR2−CD11chigh and CD206+M2 subset increased, which were found in RPL without chromosomal aberrations in previous studies and in our study. Secondly, we observed that the fully activated CD8+ T/CD161highCD4+ Teff cells and CD8+CD56+ NKT cells decreased in RPL whether stratified or not. Moreover, the inactivated and naive CD8+/CD4+ T cells were enriched statistically only in RPL with chromosomal aberrations, while CD11chigh NK and CD161highCD8+ T cells increased only in RPL without aberrations.
By far, five decidual NK-cell subsets are identified and only dNK1 cells express CD39 (20, 25). CD39highdNK1 cells, confined to late luteal phase and pregnant uterus, mediate immune tolerance and vascularization by producing galectin-1 and galectin-9 (25). Their frequency decreases significantly in RPL with normal embryonic chromosome (20), and we detect similar results both in RPL with and without chromosomal aberrations The above research also indicates RPL women have obviously increased frequency of dNK3 (20). dNK3 may be mirrored to CD11chigh NK cells because they are both pro-inflammatory and negative for CD39 expression (25). However, CD11chigh NK cells increase only in RPL without chromosomal aberrations by stratification analysis. These data support the fact that decreased CD39highdNK1 cells are involved in RPL occurrence, while whether NK cells from RPL with normal chromosome have a defective differentiation process deserve further discussion because dNK cells differentiate with sequential expression of killer cell immunoglobulin-like receptors (KIRs) and CD39 but lose CD11c expression eventually (25).
M1/M2 macrophage unbalance is correlated with spontaneous abortion (26, 27), and our results in RPL with and without chromosomal aberrations are consistent with previous findings. It should be noted that one of the CD206+ M2 subsets, the high expression of CD11c, increased remarkably. M2 macrophages expressing pro-inflammatory signatures have been reported from unexplained RPL decidua by scRNA-seq (22) and aorta–gonad–mesonephros (AGM) macrophages by CyTOF (28). It has been suggested that decidual macrophages can be classified into CD11chigh and CD11clow subsets or three subsets combined with CCR2 expression, of which the CCR2−CD11chigh macrophages have the highest phagocytic capacity, while CCR2−CD11clow exhibit the fewest inflammatory properties (29). Our investigations support this proposal and prove that macrophages in RPL decidua are preferentially pro-inflammatory.
Decidual CD8+CD56+ NKT cells are firstly described in a recent scRNA-seq paper (23), and they decreased significantly in our investigation whether RPLs were stratified or not. They might be immune suppressive (30), but this needs to be confirmed further (31). CD161highCD8+ T cells display a pro-inflammatory Tc1/Tc17 phenotype (32, 33), and more than 90% of them are mucosal-associated invariant T cells (MAIT) (34, 35). The roles of MAIT cells in pregnancy are evidenced in RPL peripheral blood (20, 36), while the association between decidual CD161highCD8+ T cells and RPL without chromosomal aberrations is firstly revealed. Except for the Th1/Th17 phenotype (37), CD161highCD4+ T cells can exert an inhibiting effect depending on their developed microenvironment (38). Dual expressions of CD38 and HLA-DR indicate the full activation of T cells and immune suppression (39–41); thus, CXCR4+CD161highCD4+ T cells might be pro-inflammatory, whereas fully activated CD161highCD4+ T cells are suppressive. Both the fully activated CD38highHLA-DRhighCCR5highCD8+ T and CD161highCD4+ T cells are firstly found to be decreased statistically in RPL regardless of their stratification. In a word, regulatory or suppressive CD4+ T/CD8+ T subsets play an important role in maintaining pregnancy.
The suppressive function of Tregs is governed by CD39 (42, 43), but CCR5high Tregs have pro-inflammatory potential (44); thus, CD39−CCR5high Tregs are pro-inflammatory. CD39high B cells are mainly Bregs and they possess inhibiting capacity through the adenosine pathway or secreting IL-10 (45, 46). Herein, the stratification analysis showed that CD39−CCR5high Tregs increased, while CD85highCD39highCD73− Bregs decreased both in RPL with and without aberrations. They highlight the key roles of CD39high Tregs/Bregs in successful pregnancy. However, when RPLs are not stratified, their differences are not obvious because of significant individual variations.
Stratification analysis by embryonic chromosome is very necessary. We notice that the immune atlas of RPL with chromosomal aberrations is similar in multiple cells to that of RPL without aberrations. Meanwhile, their CD32lowCCR2−CD11clow M2 and CD39lowCD73high B cells decrease or increase much more than RPL without aberrations, whereas the inactivated and naive CD8+/CD4+ T cells were enriched only in them. Whether they are associated with chromosomal alterations deserves investigating in the future. In addition, the only increased frequency of CD11chigh NK and CD161highCD8+ T cells in RPL with normal chromosome reminds us that their immune atlas might be more pro-inflammatory than RPL with aberrations.
When RPLs are not stratified, almost only differential clusters with cutoff of log2FC more than 1 both in RPL with and without chromosomal aberrations remain statistically significant. Of course, many clusters with no statistical difference during stratification analysis are of significance, whereas CXCR4+CD161high Teffs and CD39−CCR5high Tregs are no longer different though they increase more than two-fold in both. These are related with individual variations and cell counts of each cluster, and thus, it may lead to poor identification of differential clusters when RPLs were not stratified or the cutoff is more than 1. They can be proven by further immunohistochemical method or flow cytometry.
Variations among RPLs with different embryo aneuploids shall be analyzed. Decidual immune compartments of RPL with embryo T16 have been reported to be no different from those of women undergoing selective terminations (47). One limitation of this study is that only six antibodies (CD45, CD3, CD56, CD68, CD69, and CD25) have been used for immunohistological staining. Differences have been noticed in our investigation and these need further validation due to their limited participants.
In summary, we provide a comprehensive immune atlas of RPL decidual by CyTOF and discover some variations similar to the findings by scRNA-seq or flow cytometry. More importantly, we detect that immunological abnormalities of RPL with embryonic chromosomal aberrations are pro-inflammatory too, but some differences exist between RPL with and without aberrations.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Women’s Hospital, Zhejiang University School of Medicine (approval number IRB-20200126-R). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
ZW and MD conceived and designed the project. ZW, YX, MW, and XJ collected the samples and consent forms. ZW and GL analyzed the data. JQ and MD gave their constructive suggestions during data analysis. ZW wrote the draft. ZW and MD edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Natural Science Foundation of China (No. 81370726), Key Research and Development Plan of Zhejiang Province (No. 2019C03025), Natural Science Foundation of Zhejiang Province (No. LQ19H040011), Program for Education Department of Zhejiang Province (No. Y201941996), and Zhejiang Province Medical and Health Technology Project (Nos. 2019KY431 and 2020KY616).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.772729/full#supplementary-material
References
1. Bender AR, Christiansen OB, Elson J, Kolte AM, Lewis S, Middeldorp S, et al. ESHRE Guideline: Recurrent Pregnancy Loss. Hum Reprod Open (2018) 2018(2):y4. doi: 10.1093/hropen/hoy004
2. Magnus MC, Wilcox AJ, Morken NH, Weinberg CR, Haberg SE. Role of Maternal Age and Pregnancy History in Risk of Miscarriage: Prospective Register Based Study. BMJ (2019) 364:l869. doi: 10.1136/bmj.l869
3. Alijotas-Reig J, Garrido-Gimenez C. Current Concepts and New Trends in the Diagnosis and Management of Recurrent Miscarriage. Obstet Gynecol Surv (2013) 68(6):445–66. doi: 10.1097/OGX.0b013e31828aca19
4. Alecsandru D, Klimczak AM, Garcia VJ, Pirtea P, Franasiak JM. Immunologic Causes and Thrombophilia in Recurrent Pregnancy Loss. Fertil Steril (2021) 115(3):561–6. doi: 10.1016/j.fertnstert.2021.01.017
5. Huhn O, Ivarsson MA, Gardner L, Hollinshead M, Stinchcombe JC, Chen P, et al. Distinctive Phenotypes and Functions of Innate Lymphoid Cells in Human Decidua During Early Pregnancy. Nat Commun (2020) 11(1):381. doi: 10.1038/s41467-019-14123-z
6. Yang F, Zheng Q, Jin L. Dynamic Function and Composition Changes of Immune Cells During Normal and Pathological Pregnancy at the Maternal-Fetal Interface. Front Immunol (2019) 10:2317. doi: 10.3389/fimmu.2019.02317
7. Eikmans M, van der Zwan A, Claas F, van der Hoorn ML, Heidt S. Got Your Mother in a Whirl: The Role of Maternal T Cells and Myeloid Cells in Pregnancy. Hla (2020) 96(5):561–79. doi: 10.1111/tan.14055
8. Jin L, Li P, Wang YC, Feng L, Xu R, Yang DB, et al. Studies of Superb Microvascular Imaging and Contrast-Enhanced Ultrasonography in The Evaluation of Vascularization in Early Bone Regeneration. J Ultrasound Med (2019) 38(11):2963–71. doi: 10.1002/jum.15002
9. Vacca P, Vitale C, Munari E, Cassatella MA, Mingari MC, Moretta L. Human Innate Lymphoid Cells: Their Functional and Cellular Interactions in Decidua. Front Immunol (2018) 9:1897. doi: 10.3389/fimmu.2018.01897
10. Zhang Y, Liu Z, Tian M, Hu X, Wang L, Ji J, et al. The Altered PD-1/PD-L1 Pathway Delivers the 'One-Two Punch' Effects to Promote the Treg/Th17 Imbalance in Pre-Eclampsia. Cell Mol Immunol (2018) 15(7):710–23. doi: 10.1038/cmi.2017.70
11. Miyazaki S, Tsuda H, Sakai M, Hori S, Sasaki Y, Futatani T, et al. Predominance of Th2-Promoting Dendritic Cells in Early Human Pregnancy Decidua. J Leukoc Biol (2003) 74(4):514–22. doi: 10.1189/jlb.1102566
12. Huang N, Chi H, Qiao J. Role of Regulatory T Cells in Regulating Fetal-Maternal Immune Tolerance in Healthy Pregnancies and Reproductive Diseases. Front Immunol (2020) 11:1023. doi: 10.3389/fimmu.2020.01023
13. Schumacher A, Ehrentraut S, Scharm M, Wang H, Hartig R, Morse HR, et al. Plasma Cell Alloantigen 1 and IL-10 Secretion Define Two Distinct Peritoneal B1a B Cell Subsets With Opposite Functions, PC1(high) Cells Being Protective and PC1(low) Cells Harmful for the Growing Fetus. Front Immunol (2018) 9:1045. doi: 10.3389/fimmu.2018.01045
14. Liu XY, Fan Q, Wang J, Li R, Xu Y, Guo J, et al. Higher Chromosomal Abnormality Rate in Blastocysts From Young Patients With Idiopathic Recurrent Pregnancy Loss. Fertil Steril (2020) 113(4):853–64. doi: 10.1016/j.fertnstert.2019.11.016
15. Rai R, Regan L. Recurrent Miscarriage. Lancet (2006) 368(9535):601–11. doi: 10.1016/S0140-6736(06)69204-0
16. Hasle H, Clemmensen IH, Mikkelsen M. Risks of Leukaemia and Solid Tumours in Individuals With Down's Syndrome. Lancet (2000) 355(9199):165–9. doi: 10.1016/S0140-6736(99)05264-2
17. Waugh KA, Araya P, Pandey A, Jordan KR, Smith KP, Granrath RE, et al. Mass Cytometry Reveals Global Immune Remodeling With Multi-Lineage Hypersensitivity to Type I Interferon in Down Syndrome. Cell Rep (2019) 29(7):1893–908. doi: 10.1016/j.celrep.2019.10.038
18. Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-Cell Reconstruction of the Early Maternal-Fetal Interface in Humans. Nature (2018) 563(7731):347–53. doi: 10.1038/s41586-018-0698-6
19. Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max K, Garzia A, et al. A Single-Cell Survey of the Human First-Trimester Placenta and Decidua. Sci Adv (2018) 4(10):u4788. doi: 10.1126/sciadv.aau4788
20. Wang F, Jia W, Fan M, Shao X, Li Z, Liu Y, et al. Single-Cell Immune Landscape of Human Recurrent Miscarriage. Genomics Proteomics Bioinf (2021) 1(19):1–35. doi: 10.1016/j.gpb.2020.11.002
21. Huang C, Zeng Y, Tu W. Single-Cell RNA Sequencing Deciphers Immune Landscape of Human Recurrent Miscarriage. Genomics Proteomics Bioinf (2021) 1(19):1–7. doi: 10.1016/j.gpb.2020.12.005
22. Chen P, Zhou L, Chen J, Lu Y, Cao C, Lv S, et al. The Immune Atlas of Human Deciduas With Unexplained Recurrent Pregnancy Loss. Front Immunol (2021) 12:689019. doi: 10.3389/fimmu.2021.689019
23. Guo C, Cai P, Jin L, Sha Q, Yu Q, Zhang W, et al. Single-Cell Profiling of the Human Decidual Immune Microenvironment in Patients With Recurrent Pregnancy Loss. Cell Discov (2021) 7(1):1. doi: 10.1038/s41421-020-00236-z
24. Lathi RB, Milki AA. Tissue Sampling Technique Affects Accuracy of Karyotype From Missed Abortions. J Assist Reprod Genet (2002) 19(11):536–8. doi: 10.1023/a:1020916003666
25. Strunz B, Bister J, Jonsson H, Filipovic I, Crona-Guterstam Y, Kvedaraite E, et al. Continuous Human Uterine NK Cell Differentiation in Response to Endometrial Regeneration and Pregnancy. Sci Immunol (2021) 6(56):1–15. doi: 10.1126/sciimmunol.abb7800
26. Tsao FY, Wu MY, Chang YL, Wu CT, Ho HN. M1 Macrophages Decrease in the Deciduae From Normal Pregnancies But Not From Spontaneous Abortions or Unexplained Recurrent Spontaneous Abortions. J Formos Med Assoc (2018) 117(3):204–11. doi: 10.1016/j.jfma.2017.03.011
27. Zhang Y, Ma L, Hu X, Ji J, Mor G, Liao A. The Role of the PD-1/PD-L1 Axis in Macrophage Differentiation and Function During Pregnancy. Hum Reprod (2019) 34(1):25–36. doi: 10.1093/humrep/dey347
28. Mariani SA, Li Z, Rice S, Krieg C, Fragkogianni S, Robinson M, et al. Pro-Inflammatory Aorta-Associated Macrophages Are Involved in Embryonic Development of Hematopoietic Stem Cells. Immunity (2019) 50(6):1439–52. doi: 10.1016/j.immuni.2019.05.003
29. Jiang X, Du MR, Li M, Wang H. Three Macrophage Subsets Are Identified in the Uterus During Early Human Pregnancy. Cell Mol Immunol (2018) 15(12):1027–37. doi: 10.1038/s41423-018-0008-0
30. Zhou L, Wang H, Zhong X, Jin Y, Mi QS, Sharma A, et al. The IL-10 and IFN-Gamma Pathways Are Essential to the Potent Immunosuppressive Activity of Cultured CD8+ NKT-Like Cells. Genome Biol (2008) 9(7):R119. doi: 10.1186/gb-2008-9-7-r119
31. Walker LJ, Kang YH, Smith MO, Tharmalingham H, Ramamurthy N, Fleming VM, et al. Human MAIT and CD8alphaalpha Cells Develop From a Pool of Type-17 Precommitted CD8+ T Cells. Blood (2012) 119(2):422–33. doi: 10.1182/blood-2011-05-353789
32. Nicol B, Salou M, Vogel I, Garcia A, Dugast E, Morille J, et al. An Intermediate Level of CD161 Expression Defines a Novel Activated, Inflammatory, and Pathogenic Subset of CD8(+) T Cells Involved in Multiple Sclerosis. J Autoimmun (2018) 88:61–74. doi: 10.1016/j.jaut.2017.10.005
33. Singh MV, Cicha MZ, Kumar S, Meyerholz DK, Irani K, Chapleau MW, et al. Abnormal CD161(+) Immune Cells and Retinoic Acid Receptor-Related Orphan Receptor Gammat-Mediate Enhanced IL-17f Expression in the Setting of Genetic Hypertension. J Allergy Clin Immunol (2017) 140(3):809–21. doi: 10.1016/j.jaci.2016.11.039
34. Mexhitaj I, Nyirenda MH, Li R, O'Mahony J, Rezk A, Rozenberg A, et al. Abnormal Effector and Regulatory T Cell Subsets in Paediatric-Onset Multiple Sclerosis. Brain (2019) 142(3):617–32. doi: 10.1093/brain/awz017
35. Kaipe H, Raffetseder J, Ernerudh J, Solders M, Tiblad E. MAIT Cells at the Fetal-Maternal Interface During Pregnancy. Front Immunol (2020) 11:1788. doi: 10.3389/fimmu.2020.01788
36. Acquaviva M, Bassani C, Sarno N, Dalla CG, Romeo M, Sangalli F, et al. Loss of Circulating CD8+ CD161(high) T Cells in Primary Progressive Multiple Sclerosis. Front Immunol (2019) 10:1922. doi: 10.3389/fimmu.2019.01922
37. Iliopoulou EG, Karamouzis MV, Missitzis I, Ardavanis A, Sotiriadou NN, Baxevanis CN, et al. Increased Frequency of CD4+ Cells Expressing CD161 in Cancer Patients. Clin Cancer Res (2006) 12(23):6901–9. doi: 10.1158/1078-0432.CCR-06-0977
38. Halkias J, Rackaityte E, Hillman SL, Aran D, Mendoza VF, Marshall LR, et al. CD161 Contributes to Prenatal Immune Suppression of IFNgamma-Producing PLZF+ T Cells. J Clin Invest (2019) 129(9):3562–77. doi: 10.1172/JCI125957
39. Chen PY, Wu CY, Fang JH, Chen HC, Feng LY, Huang CY, et al. Functional Change of Effector Tumor-Infiltrating CCR5(+)CD38(+)HLA-DR(+)CD8(+) T Cells in Glioma Microenvironment. Front Immunol (2019) 10:2395. doi: 10.3389/fimmu.2019.02395
40. Kar A, Mehrotra S, Chatterjee S. CD38: T Cell Immuno-Metabolic Modulator. Cells-Basel (2020) 9(7):1–20. doi: 10.3390/cells9071716
41. Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, et al. CD38-Mediated Immunosuppression as a Mechanism of Tumor Cell Escape From PD-1/PD-L1 Blockade. Cancer Discov (2018) 8(9):1156–75. doi: 10.1158/2159-8290.CD-17-1033
42. Zhao H, Bo C, Kang Y, Li H. What Else Can CD39 Tell Us? Front Immunol (2017) 8:727. doi: 10.3389/fimmu.2017.00727
43. Magid-Bernstein JR, Rohowsky-Kochan CM. Human CD39(+) Treg Cells Express Th17-Associated Surface Markers and Suppress IL-17 via a Stat3-Dependent Mechanism. J Interferon Cytokine Res (2017) 37(4):153–64. doi: 10.1089/jir.2016.0071
44. Butcher MJ, Filipowicz AR, Waseem TC, McGary CM, Crow KJ, Magilnick N, et al. Atherosclerosis-Driven Treg Plasticity Results in Formation of a Dysfunctional Subset of Plastic IFNgamma+ Th1/Tregs. Circ Res (2016) 119(11):1190–203. doi: 10.1161/CIRCRESAHA.116.309764
45. Kaku H, Cheng KF, Al-Abed Y, Rothstein TL. A Novel Mechanism of B Cell-Mediated Immune Suppression Through CD73 Expression and Adenosine Production. J Immunol (2014) 193(12):5904–13. doi: 10.4049/jimmunol.1400336
46. Figueiro F, Muller L, Funk S, Jackson EK, Battastini AM, Whiteside TL. Phenotypic and Functional Characteristics of CD39(high) Human Regulatory B Cells (Breg). Oncoimmunology (2016) 5(2):e1082703. doi: 10.1080/2162402X.2015.1082703
Keywords: recurrent pregnancy loss, decidua, CyTOF, embryonic chromosomal aberrations, maternal immune tolerance
Citation: Wu Z, Wang M, Liang G, Jin P, Wang P, Xu Y, Qian Y, Jiang X, Qian J and Dong M (2021) Pro-Inflammatory Signature in Decidua of Recurrent Pregnancy Loss Regardless of Embryonic Chromosomal Abnormalities. Front. Immunol. 12:772729. doi: 10.3389/fimmu.2021.772729
Received: 08 September 2021; Accepted: 17 November 2021;
Published: 08 December 2021.
Edited by:
Maria Leite-de-Moraes, Université Paris Descartes, FranceReviewed by:
Jefferson R. Victor, University of São Paulo, BrazilElba Mónica Vermeulen, Instituto de Biología y Medicina Experimental (CONICET), Argentina
Copyright © 2021 Wu, Wang, Liang, Jin, Wang, Xu, Qian, Jiang, Qian and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minyue Dong, ZG9uZ215QHpqdS5lZHUuY24=
 Zaigui Wu
Zaigui Wu Miaomiao Wang
Miaomiao Wang Guanmian Liang2
Guanmian Liang2 Yeqing Qian
Yeqing Qian Xiuxiu Jiang
Xiuxiu Jiang Minyue Dong
Minyue Dong