
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 26 November 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.766401
Probiotic administration is a potential strategy against enteric pathogen infection in either clinical treatment or animal nutrition industry, but the administration duration of probiotics varied and the underlying mechanisms remain unclear. A strain (YC) affiliated to Pediococcus pentosaceus, a commonly used probiotic, was isolated from fish gut and the potential role of YC against Aeromonas hydrophila was detected in zebrafish. We found that 3- or 4-week YC administration (YC3W or YC4W) increased the resistance against A. hydrophila while 1- or 2-week treatment (YC1W or YC2W) did not. To determine the possible reason, intestinal microbiota analysis and RNAseq were conducted. The results showed that compared with CON and YC1W, YC4W significantly increased the abundance of short-chain fatty acids (SCFAs) producing bacteria and elevated the gene expression of nlrp3. Higher butyrate content and enhanced expression of IL1β were subsequently found in YC4W. To identify the causal relationship between butyrate and the higher pathogen resistance, different concentrations of sodium butyrate (SB) were supplemented. The results suggested that 10 mmol/kg SB addition mirrored the protective effect of YC4W by increasing the production of IL-1β. Furthermore, the increased IL-1β raised the percentage of intestinal neutrophils, which endued the zebrafish with A. hydrophila resistance. In vivo knockdown of intestinal il1b eliminated the anti-infection effect. Collectively, our data suggested that the molecular mechanism of probiotics determined the administration duration, which is vital for the efficiency of probiotics. Promoting host inflammation by probiotic pretreatment is one potential way for probiotics to provide their protective effects against pathogens.
Probiotic supplementation is commonly used to protect the host against enteropathogenic bacteria in human or animals and the mechanisms of different probiotics varied (1). Probiotics increased the host resistance against pathogen infection and could be attributed to host immunomodulation-independent and immunomodulation-dependent types (2). In the immunomodulation-independent type, probiotics mainly inhibit pathogens by nutrient competition, niche occupancy, or bacteriocin production (3–5). In the immunomodulation-dependent type, probiotics could modulate the immune responses of the host in direct (bacterial compounds or metabolites) or indirect (gut microbiota modulation) ways (6). It has been reported that some probiotic strains including Lactobacillus and Bacillus directly inhibit pathogen adhesion by producing bacteriocin or interfering quorum sensing (7, 8), while others could modulate the gut microbiota shift to a healthier status to protect the host. For example, Lactobacilli has been reported to protect the pig from pathogen invasion by improving the abundance and number of Lactobacilli and other indigenous probiotic bacteria (9). Bacillus clausii is reported to control the development of Clostridium difficile infection in clinical trials and the anti-infection activity of B. clausii is gut microbiota dependent (10). Faecalibacterium prausnitzii ameliorated gut dysbiosis by increasing the abundance of short-chain fatty acids (SCFA)-producing bacteria (11). All these researches suggested that regulating the intestinal microbiota is an effective way of probiotics to protect the host.
Pediococcus pentosaceus is one common probiotic and multiple strains have been reported to play antibacterial roles in hosts (12, 13). P. pentosaceus has been applied in food industry (14) and used as a growth promoter in animal nutrition (15). P. pentosaceus can produce bacteriocins to directly inhibit pathogen proliferation, and it can also regulate intestinal bacteria to protect the host against pathogens (16, 17). For example, oral administration of P. pentosaceus LI05 for 14 days enhanced the survival rate of C. difficile infected mice by increasing the abundance of beneficial microbial taxa and restraining the opportunistic pathogens in mice gut (18). In a recent study, 30-day pretreatment of P. pentosaceus SL001 is reported to alter the intestinal microbiota composition and upregulate the expression level of immune-related genes to protect the carp against A. hydrophila infection (19). P. pentosaceus was also reported to elevate the intestinal propionate and butyrate by increasing the relative abundance of SCFA-producing bacteria (20). However, the exact mechanism by which P. pentosaceus play its protective effect against pathogens remains unclear.
Pattern recognition receptors (PRR) are important in maintaining the intestinal homeostasis (21). NOD-like receptors (NLRs) are known for pathogen-associated molecular pattern (PAMP) recognition and play essential roles in the innate immune response against pathogen infection. When sensing PAMPs, NLRs were activated to increase the expression of pro-inflammatory cytokines. Among all NLRs, the activation of the pyrin domain-containing NLRP3 recruits adaptor protein ASC and caspase-1 for NLRP3 inflammasome formation, which results in IL-1β maturation and secretion (22). Higher expression of IL-1β could enhance the migration and recruitment of neutrophils in the inflammation or infected sites (23). Previous studies suggested that probiotics suppressed the activation of the inflammasome and reduced the inflammation caused by pathogen infection (24), while probiotic is also reported to trigger the activation of the inflammasome; for example, incubation of Caco-2 cells with probiotic Escherichia coli Nissle 1917 resulted in lower inflammasome activation and subsequent secretion of IL-18 (25). An in vivo study showed that Lactobacillus reuteri B1/1-treated chicken exhibited higher gene expression of NLRP3 and higher Campylobacter jejuni resistance (26), but how the activated inflammasome increased pathogen resistance still needs more evidence.
The duration of treatment is important for probiotics to exert their beneficial effects. It was reported that 6–8 h Enterococcus faecium NCIMB 10415 pretreatment significantly increased the transepithelial electrical resistance of Caco-2 cells in vitro, while a shorter (<6 h) or longer (>8 h) treatment did not (27). It was found that compared with the control group and 1-week treatment group, 2-week Kocuria SM1 addition significantly reduced the mortality of rainbow trout challenged with Vibrio anguillarum (28). Different administration duration influences the probiotics efficiency and the underlying mechanism should be addressed.
Zebrafish is an excellent model for studying host–gut microbe–pathogen interactions because it is optical transparency during early development status and it shares the most orthologous genes with mice (29, 30). Furthermore, it can be used for generating a series of mutant lines in a short time because its rapid reproduction and it has a conserved innate immune system with mammals (31). Nowadays, functions of many probotics, including Lactobacillus sp., Bacillus sp., and Bifidobacterium sp., have been investivated in zebrafish (32).
In the present study, a bacterial strain (YC) affiliated to P. pentosaceus was used to administrate zebrafish for up to 4 weeks. Aeromonas hydrophila, a commonly used pathogen for fish, was used to infect zebrafish. The pathogen-resistant effect of YC was detected and the possible mechanism was identified. We found that 4-week but not 1-week YC administration increased the level of intestinal butyrate and subsequently increased the production of IL-1β via NLRP3 inflammasome activation, which contributed to the higher resistance against A. hydrophila of zebrafish.
All experiments were carried out according to the guidelines for the care and use of laboratory animals in China. This work was approved by the Committee on the Ethics of Animal Experiments of East China Normal University (assurance no. F20201002).
A strain (designated as YC) was isolated from the intestine of Nile tilapia by using MRS broth (De Man, Rogosa, Sharpe) at 28°C. Briefly, the gut contents of Nile tilapia were collected and diluted with PBS. After 700×g centrifugation, the supernatant was inoculated on the MRS agar plate. According to the 16S rRNA sequencing, the most enriched bacteria were affiliated to P. pentosaceus. One clone was randomly picked and designated as YC. The phylogenetic analysis was performed with MEGA 7.0. The nearest neighbors of YC were identified by 16S rRNA gene sequencing (BioProject PRJNA732367) and RDP SEQMATCH. A. hydrophila ATCC 7966 was purchased from China General Microbiological Culture Collection Center (resource no. 1511C0002100007469), and cultured in LB broth at 37°C, 180 rpm. Bacteria were centrifuged at 3,000 rpm for 10 min at 4°C and resuspended in PBS to a final concentration of 109 CFU/ml before the administration.
Adult zebrafish (3 months old) line AB (male:female = 1:1) were obtained from the Chinese National Zebrafish Resource Center (Wuhan, China). Zebrafish maintenance and breeding were conducted according to the standard methods (33). The water pH was maintained at 7.0 to 8.0 and the temperature was maintained at 28°C. The dissolved oxygen in the water was 5–8 mg/L. In the YC administration trial, zebrafish were randomly divided into five experimental groups and each group has three replicate tanks in a partial-reuse system. YC was administrated in water at a concentration of 106 CFU/ml for 0 week (CON), 1 week (YC1W), 2 weeks (YC2W), 3 weeks (YC3W), and 4 weeks (YC4W). The water was renewed (50%) every day. YC was re-provided after water renewal. The fish were maintained under a 14-h day and 10-h dark photoperiod.
For the sodium butyrate supplementation trial, zebrafish were fed with a commercial diet containing 50% protein and 8% lipid (Shengsuo, China) supplemented with 10 (SB10), 20 (SB20), and 40 mmol/kg (SB40) sodium butyrate (Sigma, USA) for 7 days using a spraying method. Zebrafish fed with a commercial diet were designed as the control group. Zebrafish were fed with the diets at a ratio of 4% of their average body weight twice daily at 10:00 a.m. and 18:00 p.m. during the feeding period.
After each feeding trail, five adult zebrafish (3 months old) from each treatment were randomly sampled and dissected under MS222 (325 mg/L) anesthesia. The whole intestinal contents were collected for short-chain fatty acids analysis and 16S rRNA gene sequencing. The whole intestine was collected for the gene expression level detection, Western blot, RNAseq, and flow cytometry analysis. All samples were kept at −80°C until use.
P. pentosaceus YC was cultured in MRS medium at 28°C till the mid-exponential phase before labeling. Then, TAMRA (tetramethylrhodamine)-bearing FDAA (TADA) was added to the medium to a final concentration of 300 µM (34). The bacteria were labeled for 6 h and added to the water at a final concentration of 106 CFU/ml. Five zebrafish larvae at 4 days post-fertilization (dpf) were then immersed in the water for 24 h. Zebrafish larvae were anesthetized using MS-222 (Sigma, USA) at a concentration of 325 mg/L and washed three times before the fluorescence microscopy analysis. Confocal images were obtained using a Multiphoton Microscope Leica TCS SP8 DIVE (Leica, Germany) with a 555-nm laser. Images were processed by Imaris (Bitplane, UK) and ImageJ (NIH, USA) software.
At the end of the experiment, adult zebrafish (3 months old, 30 fish/group, three replicates per tank) were bathed in the water (28°C, pH 7.0–8.0) containing 108 CFU/ml A. hydrophila according to the method previously described (35), and the mortality was recorded for 7 days. For the in vivo il1b siRNA trial, zebrafish were challenged with A. hydrophila at a concentration of 109 CFU/ml, and the mortality was recorded for 48 h. Each curve represents the sum of three independent tanks.
At the end of the YC administration experiments, 30 fish from each group underwent anesthetization using MS-222 (Sigma, USA) at a concentration of 325 mg/L. The intestinal contents of five zebrafish were pooled as one sample and total bacterial genomic DNA was isolated from the mixture of intestinal contents of zebrafish using the OMEGA DNA Kit (Omega, USA). PCR amplification of the bacterial 16S rRNA genes of the V3–V4 region was carried out using the forward primer 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and the reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’). The amplifications were pooled in equal amounts and subsequently submitted for sequencing using an Illumina Miseq platform. Sequencing reads were analyzed with QIIME2 (36). In brief, raw sequence data were demultiplexed using the demux, then the sequences were quality filtered, denoised, merged, and chimera-removed using the DADA2 plugin. Non-singleton amplicon sequence variants (ASVs) were aligned with MAFFT software. Alpha diversity metrics (Chao 1, Shannon, and Simpson) and beta diversity metrics (weighted UniFrac distances) were calculated using the diversity plugin after sequencing depth was downsized to 2,996 sequences per sample. The distance matrices were demonstrated by two-dimensional principal coordinates analysis (PCoA) plots. The raw data were available in GenBank as Sequence Read Archive (SRA) (BioProject PRJNA674514).
For SCFA analysis, adult zebrafish intestinal content was collected. The intestine samples from five fish were pooled and resuspended with 200 µl of distilled water. Samples were then acidified with 50 µl of 50% sulfuric acid and well mixed using a vortex mixer (DLAB Scientific, China). SCFAs were extracted with 250 µl of diethyl ether and the concentration was measured by a Gas chromatography Nexis GC-2030 (SHIMADZU, Japan). The oven temperature was programmed from 60°C to 100°C at a rate of 5°C/min, with a 2-min hold; to 180°C at 5°C/min, with a 2-min hold. The temperature of the injection port and fame-ionization detector was 200°C. The external standard method was used for SCFA concentration calculation.
Total RNA was isolated from the whole intestine of zebrafish using the RNAiso Plus (TaKaRa, Japan). Complementary DNA (cDNA) was synthesized using PrimerScript TM RT reagent Kit (TaKaRa, Japan). Quantitative PCR (qPCR) was performed on a QuantStudio® 5 Real-Time PCR Instrument (Applied Biosystems, USA) thermocycler with the following cycle parameters: 94°C for 3 min followed by 40 cycles of 94°C for 10 s and 60°C for 30 s. Gene expression levels were normalized to the expression of elongation factor 1 alpha gene Ef1a and β-actin. The primer sequences used in the present study are shown in Table S3. The relative mRNA expression levels of genes were estimated by using the method of 2−ΔΔCt thereof, ΔCt = Cttarget − (CtEF1α + Ctβ-actin)/2.
Total RNA was isolated from the whole intestine of zebrafish and mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and divalent cations under elevated temperature for fragmentation. Then, the first- and the second-strand cDNA was synthesized. The library fragments were purified using the AMPure XP system (Beckman, USA) and amplified using Illumina PCR Primer Cocktail in a 15-cycle PCR reaction. Products were quantified by the Agilent High Sensitivity DNA Kit (Agilent, USA) on a Bioanalyzer 2100 system (Agilent, USA). Paired-end sequencing was subsequently performed on a Hiseq platform (Illumina, USA). The raw data were available under the BioProject accession number PRJNA674905.
For RNAseq analysis, the filtered data were obtained by removing adaptor-containing or low-quality reads and then mapped to the Danio rerio reference genome (Zv9, Ensembl) using HISAT2 software. HTSeq was used to generate the read count number of each mapped gene. Gene expression level normalization was compiled using Fragments Per Kilobases per Million fragments (FPKM). Differential expression analysis between CON and YC4W, or YC1W and YC4W was conducted using DEGSeq R package (37). p-value < 0.05 and |log2 Fold Change| > 1 gene was considered differentially expressed. Differentially expressed genes (DEGs) were functionally assorted and characterized using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (38).
The intestinal tissue without intestinal contents of zebrafish from each group was collected and homogenized. The level of IL-1β (JianglaiBio, JL48855-96T) was determined using Zebrafish Interleukin 1β (IL-1 β) Kit according to the manufacturer’s instructions.
For zebrafish intestinal immunohistochemistry, OCT (Sakura Finetek, Japan)-embedded frozen sections were fixed using anhydrous methanol (Servicebio, China) for 20 min. Antigen retrieval was carried out using 10 mM sodium citrate (pH 6) in a microwave for 15 min. Endogenous peroxidases were quenched with 3% H2O2 for 25 min. All slides were blocked with 10% nonspecific antigen with normal rabbit serum (BosterBio, AR1010, USA) for 30 min at room temperature and then incubated with 1:50 diluted cleaved-IL-1β (Affinity Biosciences, AF4006, USA) antibody overnight at 4°C. After that, slides were incubated with 1:200 diluted horseradish peroxidase-conjugated secondary antibodies (Serivcebio, GB23303, China) at room temperature for 30 min and developed with 3,3’-diaminobenzidine (DAB) solution (DAKO, K5007, Denmark). Slides were counterstained with hematoxylin. Images of stained slides were processed by Nikon Eclipse TS100 (Nikon, Japan).
The intestine of zebrafish was lysed using RIPA buffer (Beyotime, China) supplemented with protease and phosphatase inhibitor cocktail (Thermo Scientific, USA) for total protein extraction. The concentration was measured using Bicinchoninic acid (BCA) assay Protein Assay Kit (Beyotime, China). Equivalent amounts of total protein were run on an SDS-PAGE gel and then transferred onto a nitrocellulose membrane (Millipore, USA). After 1 h incubation in 5% BSA blocking solution at room temperature, the membranes were incubated with 1:600 diluted anti-GAPDH antibody (Affinity Biosciences, AF0911, USA) or anti-cleaved IL-1 beta antibody (Affinity Biosciences, AF4006, USA) overnight at 4°C. Subsequently, the membranes were washed in TBST and incubated for 1 h at room temperature with 1:10,000 diluted IRDye® 800CW secondary antibody (LI-COR, 925-32211, USA). Visualization was carried out using Odyssey Clx (LI-COR, USA) and the densitometric quantification was performed using Image Studio (LI-COR, USA) software.
Chemically modified siRNA (Table S4) were delivered into the zebrafish intestine using an oral intubation method reported before (39). In brief, zebrafish were anesthetized and placed on a wet sponge. Ten microliters of PBS, non-targeting siRNA, 5 µM, 25 µM, and 50 µM il1b siRNA1 (Target site1), and 5 µM, 25 µM, and 50 µM il1b siRNA2 (Target site2) were delivered into the zebrafish intestine using a microinjector via oral gavage. After 12 and 24 h, the gut was collected for il1b quantification.
For flow cytometry analysis, adult zebrafish (3 months old) intestine was collected for the generation of single-cell suspension according to the protocols reported previously (40, 41). Intestinal single-cell suspension was double-stained with 1:400 diluted FITC anti-CD11b (Biolegend, 101206, USA) and APC anti-Ly6G (Biolegend, 127613, USA) antibodies on ice for 30 min. After washing for three times, cells were resuspended in 2% FBS-containing PBS and analyzed using a CytoFLEX flow cytometer (Beckman coulter, USA).
All experimental data are presented as the mean ± standard error of the mean (SEM). Data were analyzed for normal distribution using D’Agostino and Pearson normality test and F test was used to compare variances. One-way analysis of variance followed by Dunnett’s multiple comparisons test was used to compare the differences among groups and unpaired Student’s t-test was used for analysis of differences between groups. Mantel–Cox test was used for analysis of survival curve differences. GraphPad Prism 8 software (GraphPad, USA) was used to perform all statistical analysis.
A bacterial strain (designated as YC) was isolated from the fish intestine and the 16S rRNA gene sequencing showed that the nearest neighbor of the isolated bacterium is P. pentosaceus (Figure 1A). YC was directly added to the water where the zebrafish were cultured. To determine whether the water-administrated YC could successfully reach the zebrafish gut, YC was labeled with fluorescent D-amino acids (FDAA). The result revealed that YC was successfully detected in the intestine of zebrafish larvae (Figure 1B), suggesting that the water-administrated YC could reach the zebrafish intestine.
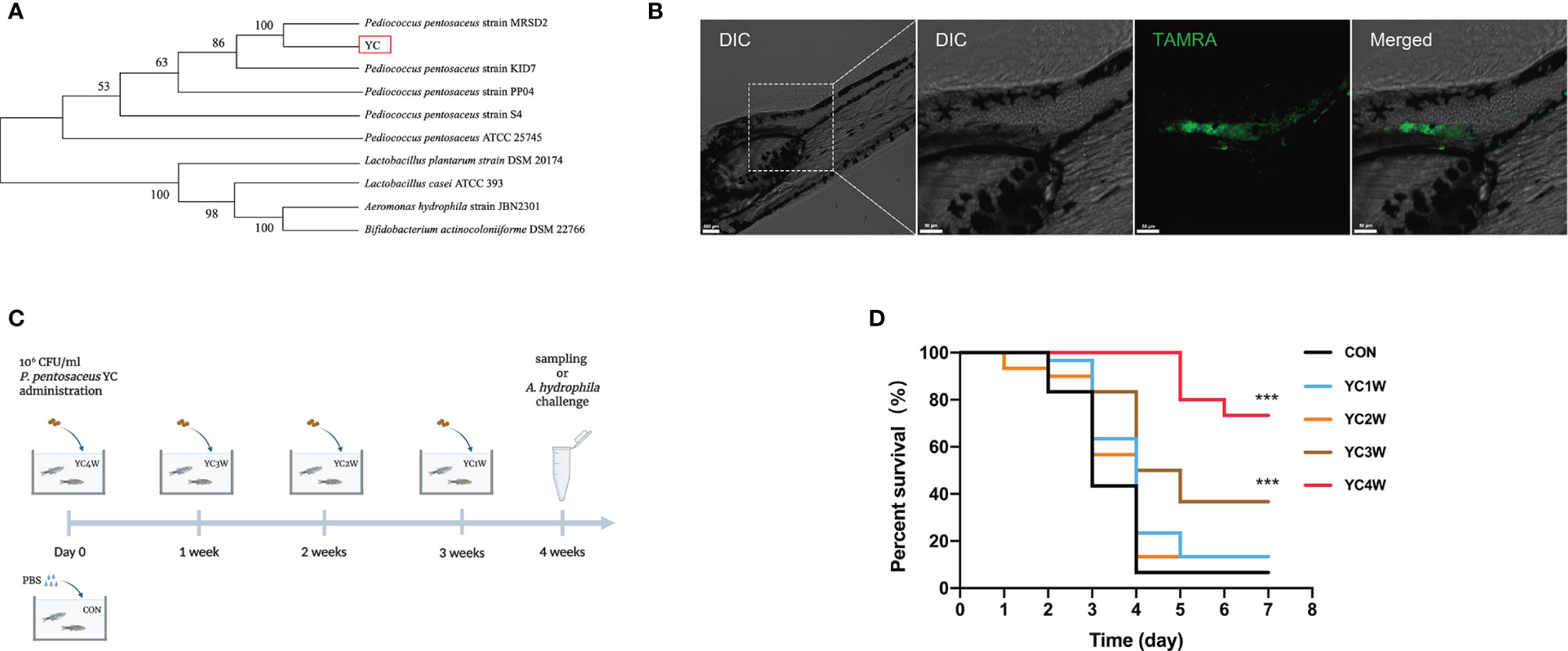
Figure 1 Administration of the isolated bacteria YC and its protective effect on the resistance against A. hydrophila of zebrafish. (A) Phylogenetic tree of the isolated bacteria bacterium. Distance was calculated by using 16S rRNA gene based on the neighbor-joining criterion and the bootstrap confidence values were 100 replicates. (B) Confocal fluorescence imaging of zebrafish larvae (4 dpf) immersed in water containing 106 CFU/ml P. pentosaceus YC (pre-treated with 300 µM TAMRA) for 24 h. (C) Experimental design for accessing the protective effect of P. pentosaceus YC administration. YC was administrated into water at a concentration of 106 CFU/ml for 1 week (YC1W), 2 weeks (YC2W), 3 weeks (YC3W), and 4 weeks (YC4W). PBS treatment as control group (CON). YC or PBS were provided every 24 h for different durations. (D) Survival curve of zebrafish from different groups following 108 CFU/ml A. hydrophila challenge. Each curve represents the sum of three independent tanks. ***p < 0.001 by Mantel–Cox test.
To determine the protective effect of YC against A. hydrophila, zebrafish were treated with YC for different durations from 1 week to 4 weeks (Figure 1C). At the end of the trial, the A. hydrophila challenge test was conducted. The results indicated that zebrafish of YC4W and YC3W had a significantly higher survival rate compared with CON on the seventh day post A. hydrophila challenge while zebrafish from YC2W and YC1W showed no significant difference with CON (Figure 1D).
Several studies reported that probiotics exert anti-pathogen benefits by modulating the host–gut microbiota composition (42). Considering the different protective effects between 1-week and 4-week YC administration, samples from CON; YC1W, which has no significant difference with CON in the challenge test; and YC4W, which showed the significant difference with CON in the challenge test, were collected for intestinal bacterial composition analysis. The results showed that 4-week YC administration significantly increased the community richness and diversity of gut microbiota, while YC1W showed no difference compared with CON (Figure 2A). At the phylum level, Fusobacteria and Proteobacteria, two dominant phyla in the zebrafish gut, showed no obvious difference between CON and YC1W, while Fusobacteria significantly decreased and Proteobacteria significantly increased in YC4W compared to CON (Figure 2B). The ASV-based principal coordinate analysis plot also indicated that samples in YC4W differed from those in CON and YC1W (Figure 2C). Compared to CON and YC1W groups, 4-week YC administration significantly increased the proportions of Hydrogenophaga, Rhizobiales, Alphaproteobacteria, Rhodobacter, Rhodobacteraceae, Devosia, Agrobacterium, and Shinella (Figure 2D). In summary, 4-week YC administration alters the zebrafish intestinal microbial community structure.
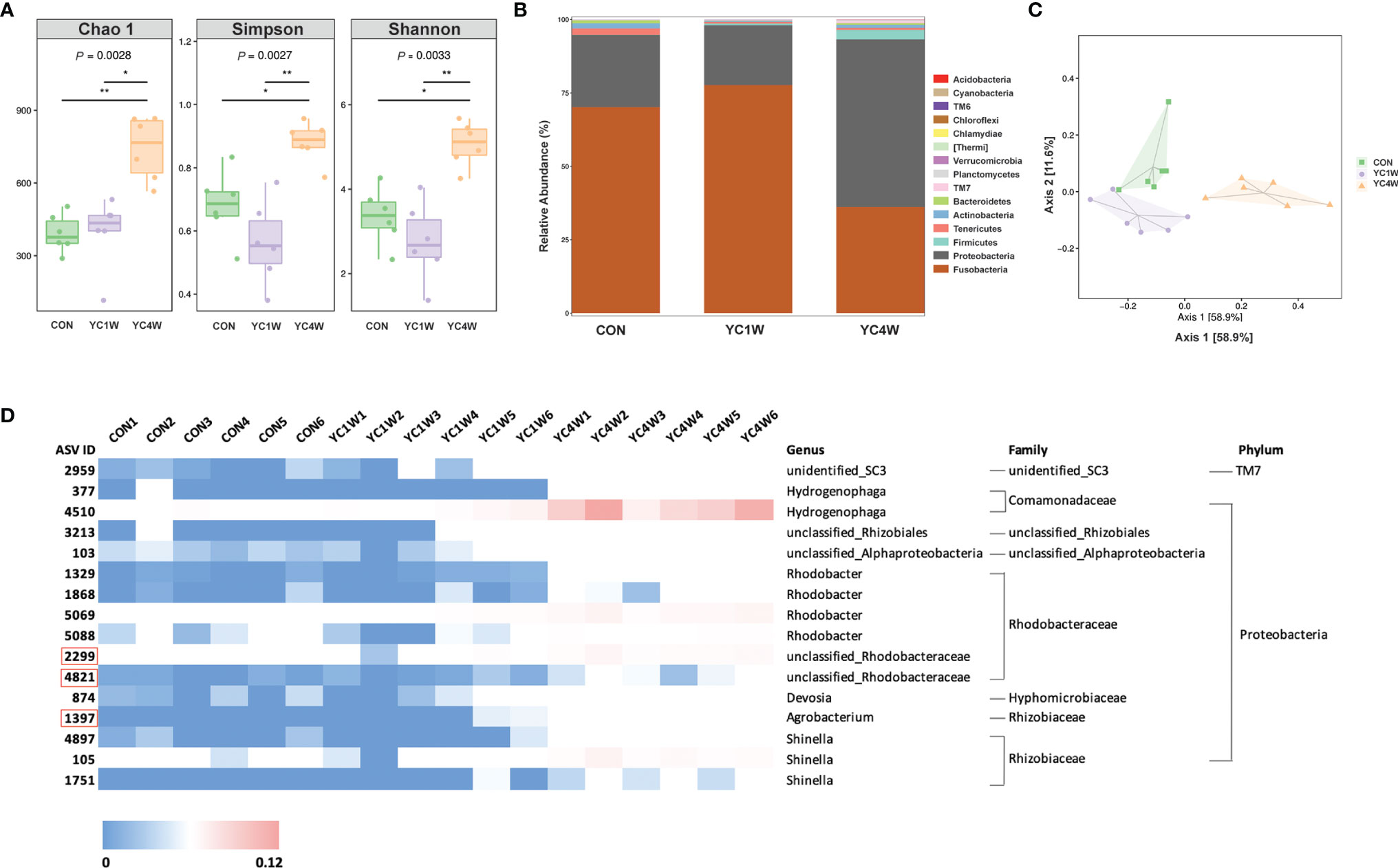
Figure 2 Administration of P. pentosaceus YC for 4 weeks alerts gut microbiota composition. 16S rRNA gene sequencing of zebrafish intestinal contents from each group (n = 6). (A) Alpha diversity metrics, p-values were identified with Kruskal–Wallis rank sum test. (B) Gut microbiota community of each group at the phylum level. (C) Principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarity matrices to show beta diversity among groups at ASV level. (D) Heatmap analysis of 16 significantly changed ASVs in the YC4W group compared with the CON and YC1W group. The color bar of each ASV in each treatment is shown. The taxonomy of the ASVs (genus, family, and phylum) is described on the right.
Considering Rhodobacteraceae and Agrobacterium, which were significantly increased in the YC4W group, are closely related to the production of SCFAs (43, 44), SCFA content of zebrafish intestine in CON, YC1W, and YC4W was detected. The results indicated that there was no significant difference in acetate, propionate, and butyrate level between CON and YC1W, while the concentration of butyrate significantly increased in YC4W compared with CON (Figure 3A). Consistent with the butyrate content, the expression level of butyrate-sensing receptor gpr109a and SCFAs transporter mct1 was significantly enhanced in YC4W compared with CON but the expression level of these two genes showed no significant difference in YC1W compared with CON (Figures 3B, C). We tested the butyrate-producing ability of YC; however, we did not detect an increase in butyrate in YC culture medium (Figure S1). Together, our data suggested that 4-week YC administration induced the accumulation of butyrate in the intestine of zebrafish while YC did not produce butyrate in vitro.
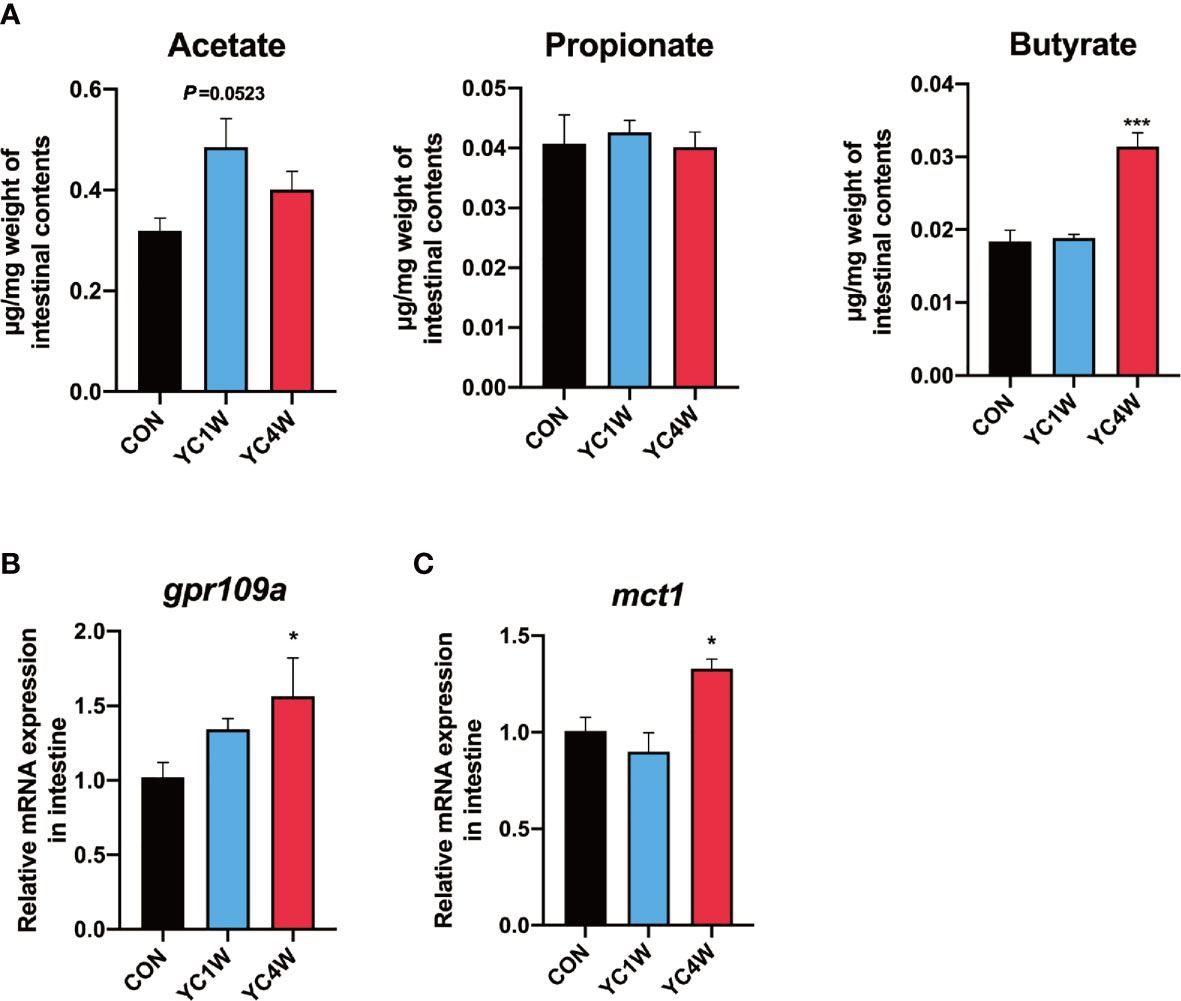
Figure 3 Gut microbiota alteration induces the accumulation of gut butyrate. (A) Intestinal short-chain fatty acid levels in CON, YC1W, and YC4W groups (n = 3). (B) Relative mRNA expression of butyrate-sensing receptor gene gpr109a and (C) butyrate transporter gene mct1 in zebrafish intestine (n = 6). Data are presented as mean ± SEM. *p < 0.05; ***p < 0.001 by one-way analysis of variance followed by Dunnett’s multiple comparisons test.
To investigate the possible mechanism by which YC regulated zebrafish resistance to the pathogen, RNAseq was conducted in CON, YC1W, and YC4W. DEGs in the comparison of CON vs. YC4W and YC1W vs. YC4W were selected based on KEGG pathway enrichment analysis (Tables S1, S2). We screened all immune system-related pathways and found three pathways significantly changed in both comparisons CON vs. YC4W and YC1W vs. YC4W, including NOD-like receptor signaling pathway, intestinal immune network for IgA production, and C-type lectin receptor signaling pathway (Figure 4A). Because the equivalents of IgA gene segments have not been found in zebrafish and IgM is the best-characterized teleost immunoglobulin isotype that represents the prevalent immunoglobulin in systemic immune responses (45), the concentrations of intestinal and serum IgM were measured. However, no significant difference was found among treatments (Figure S2). In the other two pathways, we found that the significantly changed genes were involved in NLRP3 inflammasome activation (Figure 4B). The expression level of nlrp3 was measured to validate RNAseq results, and consistent with the transcriptomic data, the expression level of nlrp3 in YC4W was significantly increased compared with CON (Figure 4C). IL-1β was cleaved and secreted after NLRP3 inflammasome activation (46). Thus, the intestinal IL-1β was assessed by ELISA, Western blot, and immunohistochemical analysis. The results suggested that IL-1β significantly increased in the intestine of zebrafish administrated YC for 4 weeks, while YC1W showed no significant difference compared with CON (Figures 4D–F). Taken together, 4-week YC administration promoted the activation of NLRP3 inflammasome and increased the secretion of pro-inflammatory cytokine IL-1β.
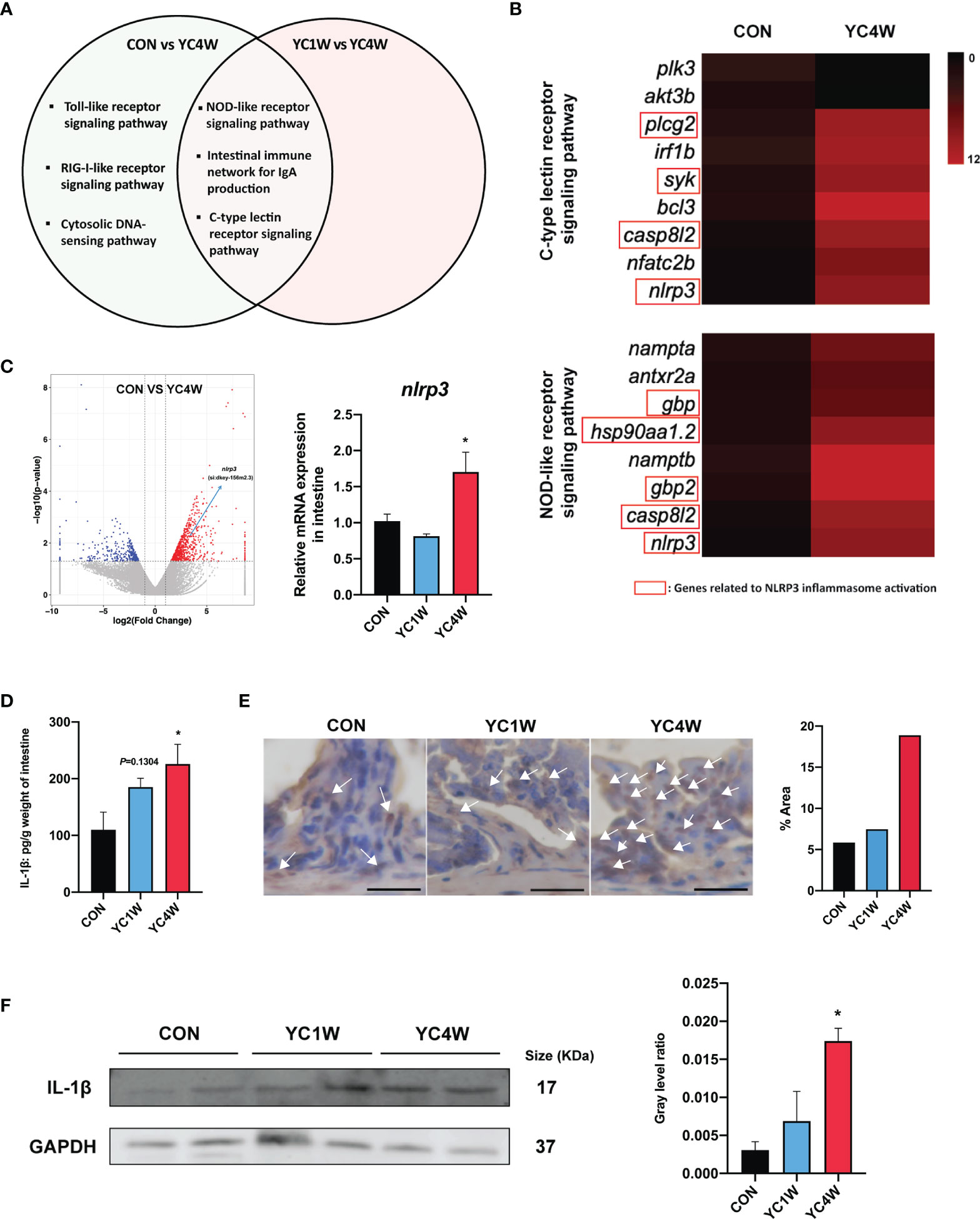
Figure 4 P. pentosaceus YC contributes to NLRP3 inflammasome activation after a 4-week administration. Differentially expressed genes (CON vs. YC4W, YC1W vs. YC4W) of zebrafish intestine were analyzed and used for pathway classification based on KEGG enrichment analysis. (A) Immune-related pathways changed in both comparison CON vs. YC4W and YC1W vs. YC4W. (B) Heatmap of differentially expressed genes of C-type lectin receptor signaling pathway and NOD-like receptor signaling pathway in comparison to CON vs. YC4W. (C) Differentially expressed intestinal genes in comparison to CON vs. YC4W and relative nlrp3 mRNA quantification of zebrafish intestine by qPCR (n = 6). (D) ELISA analysis of intestinal IL-1β level of zebrafish from CON, YC1W, and YC4W groups (n = 6). Representative (E) immunohistochemical and (F) Western blot analysis of intestinal IL-1β of zebrafish from CON, YC1W, and YC4W groups. Scale bars represents 20 µm. Data are represented as mean ± SEM. *p < 0.05 by one-way analysis of variance followed by Dunnett’s multiple comparisons test.
To determine whether the accumulation of butyrate in the intestine was the main factor of YC, zebrafish were fed with a commercial diet supplemented with sodium butyrate (SB) in three different concentrations to imitate the intestinal butyrate level induced by 4-week YC administration. The result indicated that 1 week of 10 mmol/kg SB (SB10) addition enhanced the intestinal butyrate to a comparable level of 4-week YC administration (Figure 5A and Figure S3A) and then a challenge test with A. hydrophila was conducted. Consistent with the result of 4-week YC administration, 10 mmol/kg SB addition significantly improved the survival rate of zebrafish challenged with A. hydrophila compared to CON (Figure 5B). We measured the intestinal IL-1β in SB10 to verify the activation of NLRP3 inflammasome. Similarly, ELISA and Western blot results showed that 10 mmol/kg SB significantly enhanced the intestinal IL-1β compared to CON (Figures 5C, D).
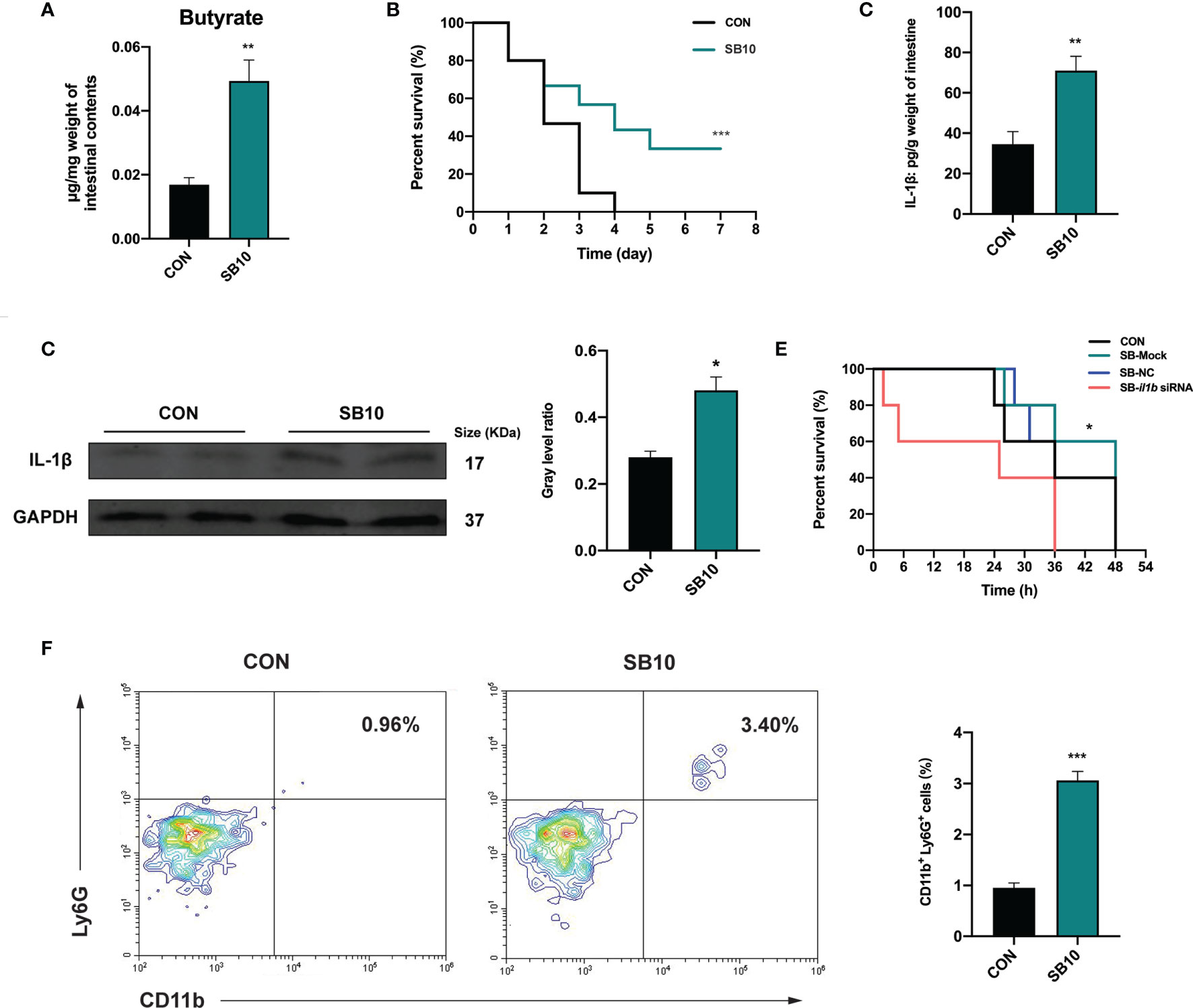
Figure 5 Sodium butyrate addition shows similar protective effect of P. pentosaceus YC in pathogen resistance on zebrafish. (A) Intestinal butyrate levels of zebrafish fed on commercial diet supplemented with 0 mmol/kg (CON) and 10 mmol/kg (SB10) sodium butyrate (n = 3). (B) Survival curve of zebrafish from CON and SB10 groups following 108 CFU/ml A. hydrophila challenge. Each curve represents the sum of three independent tanks. ***p < 0.001 by Mantel–Cox test. (C) ELISA analysis of intestinal IL-1β level of zebrafish from CON and SB10 groups (n = 6). (D) Representative Western blot of intestinal IL-1β of zebrafish from the CON and SB10 group. (E) Survival curve of zebrafish following 109 CFU/ml A. hydrophila challenge. Zebrafish were pretreated with 10 mmol/kg sodium butyrate for 1 week and then gavaged with PBS (SB-Mock), 50 µM non-targeting (SB-NC), or il1b (SB-il1b siRNA) siRNA. Asterisks indicate significant differences between SB-Mock and SB-il1b siRNA. *p < 0.05 by Mantel–Cox test. (F) Flow cytometry analysis of zebrafish intestinal neutrophils stained with anti-CD11b and anti-Ly6G antibodies (n = 3). Numbers indicate the percentage of CD11b+Ly6G+ neutrophils in intestinal cells. Data are represented as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001 by Student’s t-test.
To further determine whether the enhanced IL-1β induced by NLRP3 inflammasome activation contributed to the pathogen resistance effect of YC, an in vivo siRNA to knock down il1b mRNA was conducted in SB10-treated zebrafish according to the previous research (47). We found that zebrafish gavaged with 50 µM il1b siRNA1 (Target site1) for 24 h exhibited lower il1b expression level compared to zebrafish gavaged with PBS (Mock) and non-targeting (NC) siRNA (Figure S4). Zebrafish gavaged with 50 µM il1b siRNA1 for 24 h were used for a 109 CFU/ml A. hydrophila challenge. The results showed that the anti-infection effect of SB10 addition was eliminated by il1b interference (SB-il1b siRNA) (Figure 5E). These data indicated that 10 mmol/kg sodium butyrate addition improved the resistance of zebrafish against A. hydrophila by enhancing the intestinal level of pro-inflammatory cytokines IL-1β.
The influence of dietary sodium butyrate supplementation on the recruitment of neutrophils in the intestine was determined. The result showed a significant increase in the percentage of intestinal CD11b+ Ly6G+ neutrophils in SB10 compared to CON (Figure 5F).
The migration of intestinal neutrophils was measured in zebrafish treated with YC, and the results indicated that the 4-week YC administration significantly enhanced the percentage of neutrophils in the intestine compared with CON, while YC1W showed no difference with CON (Figure 6A). Zebrafish treated with YC were further challenged with 108 CFU/ml A. hydrophila for 12 h and the intestinal neutrophil percentage was analyzed. Consistent with the previous finding, the intestinal neutrophil percentage of YC4W was significantly higher than CON and YC1W (Figure 6B). All these results suggested that the increased level of butyrate induced by 4-week YC administration raised the percentage of intestinal neutrophils and endued the zebrafish with A. hydrophila resistance.
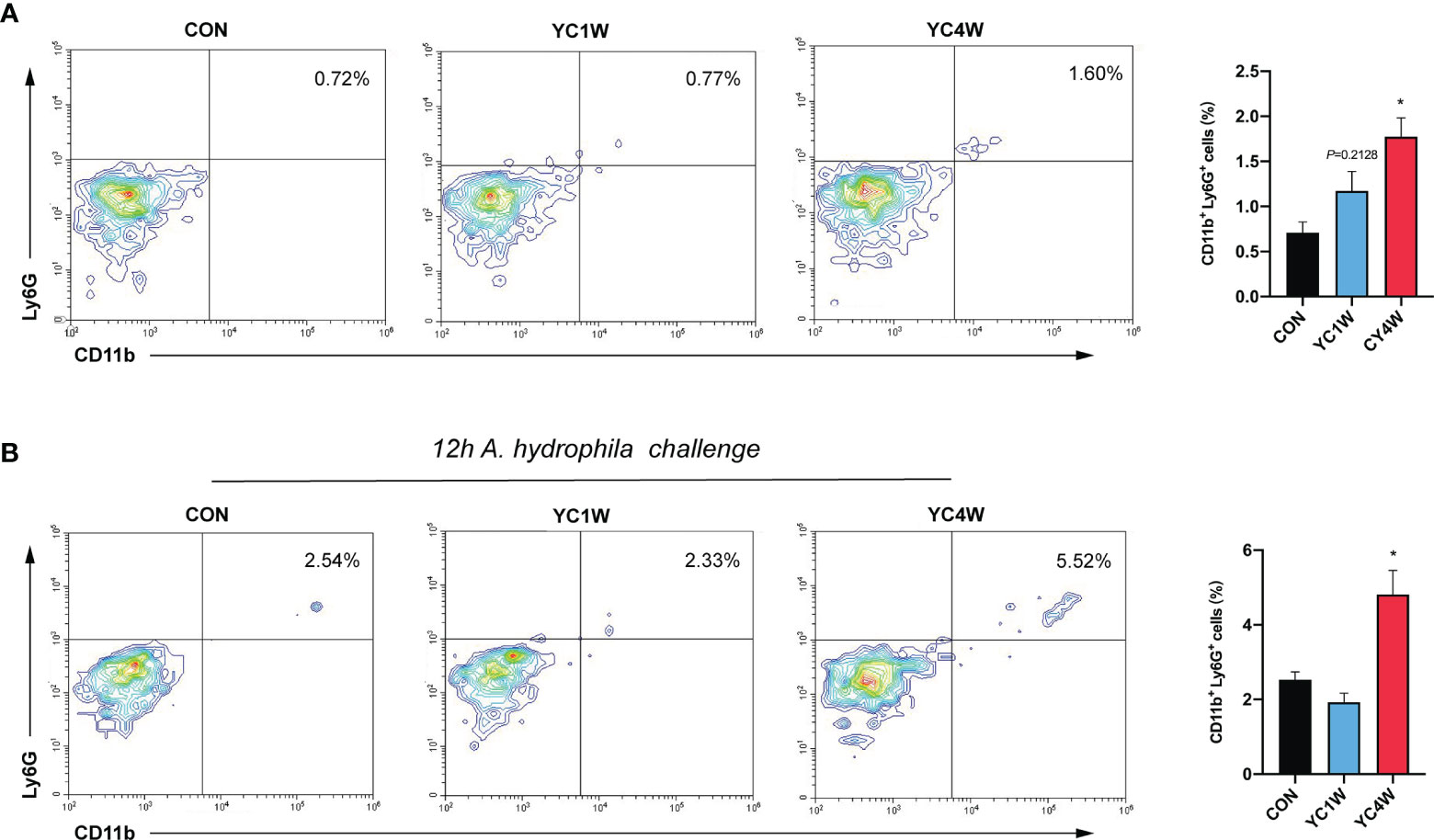
Figure 6 Four-week P. pentosaceus YC administration increases the intestinal neutrophils of zebrafish and enhances the recruitment of neutrophils under pathogen infection. (A) Flow cytometry analysis of intestinal neutrophils of zebrafish from CON, YC1W, and YC4W groups (n = 3). (B) Flow cytometry analysis of intestinal neutrophils of zebrafish from CON, YC1W, and YC4W group. All zebrafish were challenged with 108 CFU/ml A. hydrophila for 12 h before intestine collection (n = 3). Numbers indicate the percentage CD11b+Ly6G+ neutrophils in intestinal cells. Data are represented as mean ± SEM. *p < 0.05 by one-way analysis of variance followed by Dunnett’s multiple comparisons test.
Probiotic administration is a potential strategy against enteric pathogen infection. Different administration period of probiotics influenced their effectiveness, but the underlying mechanism remains unclear. In the present study, 3- or 4-week YC administration increased the pathogen resistance while 1- or 2-week YC administration did not, suggesting that the mechanism by which P. pentosaceus competes the pathogen is more complicated than secreting bacteriocin.
Considering the close relationship between the intestinal microbiota and the host immunity, intestinal bacteria in CON, YC1W, and YC4W were detected. We found that except for the bacteria related to SCFA production (ASV2299, 4821, and 1397), the abundance of ASV 4510 (Hydrogenophaga) also increased significantly in YC4W (Figure 2D). Hydrogenophaga is proved to produce poly-3-hydroxybutyrate (PHB) in vitro (48). In order to identify whether PHB was involved in the A. hydrophila resistance effect, 0.5%, 1%, or 2% of PHB were used to feed zebrafish for 7 days, but no significant difference in survival rate was detected in A. hydrophila challenge (Figure S5), suggesting that PHB may not be the key effector in the present study.
The results of 16S rRNA gene sequencing and gut SCFA determination showed that butyrate is the differential metabolite between CON and YC4W. Butyrate is an important immune regulator and is reported to suppress inflammation by inhibiting histone deacetylase, NF-κB, and Dact3 (49, 50). Butyrate has also been reported to promote inflammation by increasing the production of IL-1β (51). In the present study, 10 mmol/kg sodium butyrate (SB) raised the survival rate of A. hydrophila challenge test and increased the IL-1β production in zebrafish intestine compared with CON, while the 20 and 40 mmol/kg SB treatment group showed no difference with CON (Figures S3B, C). A previous study reported that a high concentration of butyrate decreased the pH of the culture medium and inhibited the growth of gut resident bacteria (52). In our study, higher concentrations of butyrate (20 or 40 mmol/kg SB) did not affect IL-1β production, suggesting that the immunomodulation effect of butyrate is multifaceted. Another study also found that a high dose of butyrate (8 mM) would increase the permeability of Caco-2 cell monolayer model while a low dose (2 mM) decreased the permeability (53), indicating that the effect of butyrate is determined by the concentration. IL-1β is a pro-inflammatory factor and research has proved that the secretion of IL-1β may promote the intestinal inflammasome and damage the intestine (54). However, we did not find significant difference between CON and YC4W at the histological level (Figure S6).
Furthermore, we found that the administration duration is a key factor for the beneficial effect of probiotics. A study on patients with irritable bowel syndrome (IBS) showed that treatment duration influenced the efficiency of probiotics (55), but the underlying mechanism was not demonstrated. In our study, 3- or 4-week YC treatment had a higher survival rate than 1- or 2-week treatment post A. hydrophila challenge test. The possible reason is that the shorter administration duration of YC could not efficiently increase the intestinal butyrate content, which influenced the protective effect of P. pentosaceus.
IgM exerts an important role in responding to pathogens both in systemic and mucosal compartments (45). However, there is no difference in intestinal IgM levels between CON and YC4W. PRR activation is important in recognizing and limiting microbes at the beginning of the infection. In the present study, we found that NLRP3 inflammasome is involved in PRR-related pathways and the NOD-like receptor signaling pathway. Our results revealed that 4-week YC administration promoted the production of IL-1β via NLRP3 inflammasome activation. Inflammasomes are emerging as critical regulators of the innate immune response and the activation of NLRP3 inflammasome plays an important role in pathogen clearance and gut microbiota homeostasis (56). For example, Nlrp3−/− mice were reported to exhibit decreased antimicrobial capacity (57). Inflammasomes were usually activated by pathogenic bacteria, but a recent study reported that probiotic E. faecium promoted the activation of NLRP3 inflammasome (58). E. faecium significantly evaluated pro-inflammatory cytokine IL-1β in the jejunum and ileum of 29-day-old piglets which resulted in the higher resistance against enterotoxigenic E. coli, but the causal relationship between E. coli resistance and NLRP3 activation remained unclear (58). In the present study, we found that the enhanced intestinal butyrate induced by YC administration increased IL-1β production and raised intestinal neutrophil level, which contributed to the improved resistance against A. hydrophila infection. These results suggest that promoting host inflammation is one potential way for probiotics to provide their protective effects against pathogens. Although IL-1β plays an important role in pathogen infection, overexpression of IL-1β may also cause severe gastric inflammasome to the host (59). Previous research reported that neutropenic mice that cannot recruit neutrophils efficiently were more susceptible to A. hydrophila infection (60), and consistent with their work, we found that the increased neutrophil recruitment in the intestine was related to the enhanced pathogen resistance in zebrafish.
The present study provides new insight into the protective effect of probiotic P. pentosaceus by upregulating the host inflammation level, which is a potential strategy against pathogens. The functional mechanism of probiotics determines the administration duration, and the level of microbial metabolites, butyrate, is a determining factor for P. pentosaceus exerting their beneficial effect.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA674514; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA674905.
This work was approved by the Committee on the Ethics of Animal Experiments of East China Normal University (assurance no. F20201002).
CS and M-LZ designed research. CS conducted most of the experiments. CS and M-LZ analyzed data. ML, ZL, and RX contributed to YC isolation and identification. CS and M-LZ drafted the paper. ML, FQ, Z-YD, and M-LZ revised the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (grant number 2019YFE0115000) and the National Natural Science Foundation of China (grant numbers 31972798 and 31672668).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge Dr. Wei Wang from Shanghai Jiao Tong University for providing TAMRA-bearing FDAA and Ruikun Sun from East China Normal University School of Life Sciences for technical assistance in bioinformatics analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.766401/full#supplementary-material
1. Gareau MG, Sherman PM, Walker WA. Probiotics and the Gut Microbiota in Intestinal Health and Disease. Nat Rev Gastroenterol Hepatol (2010) 7(9):503–14. doi: 10.1038/nrgastro.2010.117
2. Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. The Impact of Probiotics and Prebiotics on the Immune System. Nat Rev Immunol (2012) 12(10):728–34. doi: 10.1038/nri3312
3. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision Microbiome Reconstitution Restores Bile Acid Mediated Resistance to Clostridium Difficile. Nature (2015) 517(7533):205–8. doi: 10.1038/nature13828
4. Plichta DR, Juncker AS, Bertalan M, Rettedal E, Gautier L, Varela E, et al. Transcriptional Interactions Suggest Niche Segregation Among Microorganisms in the Human Gut. Nat Microbiol (2016) 1(11):16152. doi: 10.1038/nmicrobiol.2016.152
5. Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, et al. Bacteriocin Production Augments Niche Competition by Enterococci in the Mammalian Gastrointestinal Tract. Nature (2015) 526(7575):719–22. doi: 10.1038/nature15524
6. Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Modulation of the Gut Microbiota to Improve Innate Resistance. Curr Opin Immunol (2018) 54:137–44. doi: 10.1016/j.coi.2018.08.003
7. Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG. Bacteriocin Production as a Mechanism for the Antiinfective Activity of Lactobacillus Salivarius UCC118. Proc Natl Acad Sci USA (2007) 104(18):7617–21. doi: 10.1073/pnas.0700440104
8. Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, et al. Pathogen Elimination by Probiotic Bacillus via Signalling Interference. Nat (2018) 562(7728):532–7. doi: 10.1038/s41586-018-0616-y
9. Yang J, Qian K, Wang C, Wu Y. Roles of Probiotic Lactobacilli Inclusion in Helping Piglets Establish Healthy Intestinal Inter-Environment for Pathogen Defense. Probiotics Antimicrob Proteins (2018) 10(2):243–50. doi: 10.1007/s12602-017-9273-y
10. Mills JP, Rao K, Young VB. Probiotics for Prevention of Clostridium Difficile Infection. Curr Opin Gastroenterol (2018) 34(1):3–10. doi: 10.1097/MOG.0000000000000410
11. Zhou Y, Xu H, Xu J, Guo X, Zhao H, Chen Y, et al. F. Prausnitzii and its Supernatant Increase SCFAs-Producing Bacteria to Restore Gut Dysbiosis in TNBS-Induced Colitis. AMB Express (2021) 11(1):33–. doi: 10.1186/s13568-021-01197-6
12. Yin H, Ye P, Lei Q, Cheng Y, Yu H, Du J, et al. In Vitro Probiotic Properties of Pediococcus Pentosaceus L1 and Its Effects on Enterotoxigenic Escherichia Coli-Induced Inflammatory Responses in Porcine Intestinal Epithelial Cells. Microb Pathog (2020) 144:104163. doi: 10.1016/j.micpath.2020.104163
13. Cui S, Jiang J, Li B, Ross RP, Stanton C, Zhao J, et al. Effects of the Short-Term Administration of Pediococcus Pentosaceus on Physiological Characteristics, Inflammation, and Intestinal Microecology in Mice. Food Funct (2021) 12:1695–707. doi: 10.1039/D0FO02948C
14. Reuter G. Present and Future of Probiotics in Germany and in Central Europe. Biosci Microflora (1997) 16(2):43–51. doi: 10.12938/bifidus1996.16.43
15. Jiang S, Cai L, Lv L, Li L. Pediococcus Pentosaceus, a Future Additive or Probiotic Candidate. Microb Cell Factories (2021) 20(1):45. doi: 10.1186/s12934-021-01537-y
16. de Souza de Azevedo PO, Mendonca CMN, Moreno ACR, Bueno AVI, de Almeida SRY, Seibert L, et al. Antibacterial and Antifungal Activity of Crude and Freeze-Dried Bacteriocin-Like Inhibitory Substance Produced by Pediococcus Pentosaceus. Sci Rep (2020) 10(1):12291. doi: 10.1038/s41598-020-68922-2
17. Juturu V, Wu JC. Microbial Production of Bacteriocins: Latest Research Development and Applications. Biotechnol Adv (2018) 36(8):2187–200. doi: 10.1016/j.biotechadv.2018.10.007
18. Xu Q, Gu S, Chen Y, Quan J, Lv L, Chen D, et al. Protective Effect of Pediococcus Pentosaceus LI05 Against Clostridium Difficile Infection in a Mouse Model. Front Microbiol (2018) 9:2396. doi: 10.3389/fmicb.2018.02396
19. Gong L, He H, Li D, Cao L, Khan TA, Li Y, et al. A New Isolate of Pediococcus Pentosaceus (SL001) With Antibacterial Activity Against Fish Pathogens and Potency in Facilitating the Immunity and Growth Performance of Grass Carps. Front Microbiol (2019) 10:1384. doi: 10.3389/fmicb.2019.01384
20. Jiang X-W, Li Y-T, Ye J-Z, Lv L-X, Yang L-Y, Bian X-Y, et al. New Strain of Pediococcus Pentosaceus Alleviates Ethanol-Induced Liver Injury by Modulating the Gut Microbiota and Short-Chain Fatty Acid Metabolism. World J Gastroenterol (2020) 26(40):6224–40. doi: 10.3748/wjg.v26.i40.6224
21. Fukata M, Arditi M. The Role of Pattern Recognition Receptors in Intestinal Inflammation. Mucosal Immunol (2013) 6(3):451–63. doi: 10.1038/mi.2013.13
22. Maekawa T, Kufer TA, Schulze-Lefert P. NLR Functions in Plant and Animal Immune Systems: So Far and Yet So Close. Nat Immunol (2011) 12(9):817–26. doi: 10.1038/ni.2083
23. De Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil Migration in Infection and Wound Repair: Going Forward in Reverse. Nat Rev Immunol (2016) 16(6):378. doi: 10.1038/nri.2016.49
24. van Zyl WF, Deane SM, Dicks LMT. Molecular Insights Into Probiotic Mechanisms of Action Employed Against Intestinal Pathogenic Bacteria. Gut Microbes (2020) 12(1):1831339. doi: 10.1080/19490976.2020.1831339
25. Becker HM, Apladas A, Scharl M, Fried M, Rogler G. Probiotic Escherichia Coli Nissle 1917 and Commensal E. Coli K12 Differentially Affect the Inflammasome in Intestinal Epithelial Cells. Digestion (2014) 89(2):110–8. doi: 10.1159/000357521
26. Karaffová V, Revajová V, Koščová J, Gancarčíková S, Nemcová R, Ševčíková Z, et al. Local Intestinal Immune Response Including NLRP3 Inflammasome in Broiler Chicken Infected With Campylobacter Jejuni After Administration of Lactobacillus Reuteri B1/1. Food Agric Immunol (2020) 31(1):954–66. doi: 10.1080/09540105.2020.1788516
27. Lodemann U, Strahlendorf J, Schierack P, Klingspor S, Aschenbach JR, Martens H. Effects of the Probiotic Enterococcus Faecium and Pathogenic Escherichia Coli Strains in a Pig and Human Epithelial Intestinal Cell Model. Scientifica (2015) 2015:235184. doi: 10.1155/2015/235184
28. Sharifuzzaman SM, Austin B. Influence of Probiotic Feeding Duration on Disease Resistance and Immune Parameters in Rainbow Trout. Fish Shellfish Immunol (2009) 27(3):440–5. doi: 10.1016/j.fsi.2009.06.010
29. Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The Use of Zebrafish to Understand Immunity. Immunity (2004) 20(4):367–79. doi: 10.1016/S1074-7613(04)00084-6
30. Kanther M, Rawls JF. Host–microbe Interactions in the Developing Zebrafish. Curr Opin Immunol (2010) 22(1):10–9. doi: 10.1016/j.coi.2010.01.006
31. Yoder JA, Nielsen ME, Amemiya CT, Litman GW. Zebrafish as an Immunological Model System. Microbes Infect (2002) 4(14):1469–78. doi: 10.1016/S1286-4579(02)00029-1
32. López Nadal A, Ikeda-Ohtsubo W, Sipkema D, Peggs D, McGurk C, Forlenza M, et al. Feed, Microbiota, and Gut Immunity: Using the Zebrafish Model to Understand Fish Health. Front Immunol (2020) 11:114. doi: 10.3389/fimmu.2020.00114
33. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Dev Dynamics (1995) 203:253–310. doi: 10.1002/aja.1002030302
34. Wang W, Lin L, Du Y, Song Y, Peng X, Chen X, et al. Assessing the Viability of Transplanted Gut Microbiota by Sequential Tagging With D-Amino Acid-Based Metabolic Probes. Nat Commun (2019) 10(1):1317. doi: 10.1038/s41467-019-09267-x
35. Qin C, Xie Y, Wang Y, Li S, Ran C, He S, et al. Impact of Lactobacillus Casei BL23 on the Host Transcriptome, Growth and Disease Resistance in Larval Zebrafish. Front Physiol (2018) 9:1245(1245). doi: 10.3389/fphys.2018.01245
36. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat Biotechnol (2019) 37(8):852–7. doi: 10.1038/s41587-019-0209-9
37. Robinson MD, McCarthy DJ, Smyth GK. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics (2010) 26(1):139–40. doi: 10.1093/bioinformatics/btp616
38. Huang da W, Sherman BT, Lempicki RA. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat Protoc (2009) 4(1):44–57. doi: 10.1038/nprot.2008.211
39. Ji J, Thwaite R, Roher N. Oral Intubation of Adult Zebrafish: A Model for Evaluating Intestinal Uptake of Bioactive Compounds. JoVE (2018) (139):e58366. doi: 10.3791/58366
40. Bresciani E, Broadbridge E, Liu PP. An Efficient Dissociation Protocol for Generation of Single Cell Suspension From Zebrafish Embryos and Larvae. MethodsX (2018) 5:1287–90. doi: 10.1016/j.mex.2018.10.009
41. Ferrer-Font L, Mehta P, Harmos P, Schmidt AJ, Chappell S, Price KM, et al. High-Dimensional Analysis of Intestinal Immune Cells During Helminth Infection. eLife (2020) 9:e51678. doi: 10.7554/eLife.51678
42. Sanchez B, Delgado S, Blanco-Miguez A, Lourenco A, Gueimonde M, Margolles A. Probiotics, Gut Microbiota, and Their Influence on Host Health and Disease. Mol Nutr Food Res (2017) 61(1):1–15. doi: 10.1002/mnfr.201600240
43. Jayachandran M, Chen J, Chung SSM, Xu B. A Critical Review on the Impacts of Beta-Glucans on Gut Microbiota and Human Health. J Nutr Biochem (2018) 61:101–10. doi: 10.1016/j.jnutbio.2018.06.010
44. Marshall CW, Ross DE, Fichot EB, Norman RS, May HD. Long-Term Operation of Microbial Electrosynthesis Systems Improves Acetate Production by Autotrophic Microbiomes. Environ Sci Technol (2013) 47(11):6023–9. doi: 10.1021/es400341b
45. Salinas I, Zhang Y-A, Sunyer JO. Mucosal Immunoglobulins and B Cells of Teleost Fish. Dev Comp Immunol (2011) 35(12):1346–65. doi: 10.1016/j.dci.2011.11.009
46. Coll RC, O’Neill L, Schroder K. Questions and Controversies in Innate Immune Research: What Is the Physiological Role of NLRP3? Cell Death Discovery (2016) 2:16019:1–5. doi: 10.1038/cddiscovery.2016.19
47. Xiao C, Wang F, Hou J, Zhu X, Luo Y, Xiong JW. Nanoparticle-Mediated siRNA Gene-Silencing in Adult Zebrafish Heart. J Vis Exp (2018) 137:1–8. doi: 10.3791/58054
48. Yoon KS, Tsukada N, Sakai Y, Ishii M, Igarashi Y, Nishihara H. Isolation and Characterization of a New Facultatively Autotrophic Hydrogen-Oxidizing Betaproteobacterium, Hydrogenophaga Sp. AH-24. FEMS Microbiol Lett (2008) 278(1):94–100. doi: 10.1111/j.1574-6968.2007.00983.x
49. Chang PV, Hao L, Offermanns S, Medzhitov R. The Microbial Metabolite Butyrate Regulates Intestinal Macrophage Function via Histone Deacetylase Inhibition. Proc Natl Acad Sci USA (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
50. Lenoir M, Martin R, Torres-Maravilla E, Chadi S, Gonzalez-Davila P, Sokol H, et al. Butyrate Mediates Anti-Inflammatory Effects of Faecalibacterium Prausnitzii in Intestinal Epithelial Cells Through Dact3. Gut Microbes (2020) 12(1):1–16. doi: 10.1080/19490976.2020.1826748
51. Ohira H, Fujioka Y, Katagiri C, Yano M, Mamoto R, Aoyama M, et al. Butyrate Enhancement of Inteleukin 1β Production via Activation of Oxidative Stress Pathways in Lipopolysaccharide -Stimulated THP 1 Cells. J Clin Biochem Nutr (2011) 50(1):59–66. doi: 10.3164/jcbn.1122
52. Ang QY, Alexander M, Newman JC, Tian Y, Cai J, Upadhyay V, et al. Ketogenic Diets Alter the Gut Microbiome Resulting in Decreased Intestinal Th17 Cells. Cell (2020) 181(6):1263–75.e16. doi: 10.1016/j.cell.2020.04.027
53. Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of Butyrate on Intestinal Barrier Function in a Caco-2 Cell Monolayer Model of Intestinal Barrier. Pediatr Res (2007) 61(1):37–41. doi: 10.1203/01.pdr.0000250014.92242.f3
54. Hasegawa M, Himpsl SD, Browne HP, Lawley TD, Mobley HL, Inohara N. Distinct Commensals Induce Interleukin-1b via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity (2015) 42:744–55. doi: 10.1016/j.immuni.2015.03.004
55. Zhang Y, Li L, Guo C, Mu D, Feng B, Zuo X, et al. Effects of Probiotic Type, Dose and Treatment Duration on Irritable Bowel Syndrome Diagnosed by Rome III Criteria: A Meta-Analysis. BMC Gastroenterol (2016) 16(1):62. doi: 10.1186/s12876-016-0470-z
56. Latz E, Xiao TS, Stutz A. Activation and Regulation of the Inflammasomes. Nat Rev Immunol (2013) 13(6):397–411. doi: 10.1038/nri3452
57. Man SM. Inflammasomes in the Gastrointestinal Tract: Infection, Cancer and Gut Microbiota Homeostasis. Nat Rev Gastroenterol Hepatol (2018) 15(12):721–37. doi: 10.1038/s41575-018-0054-1
58. Kern M, Aschenbach JR, Tedin K, Pieper R, Loss H, Lodemann U. Characterization of Inflammasome Components in Pig Intestine and Analysis of the Influence of Probiotic Enterococcus Faecium During an Escherichia Coli Challenge. Immunol Invest (2017) 46(7):742–57. doi: 10.1080/08820139.2017.1360341
59. Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B, et al. Overexpression of Interleukin-1beta Induces Gastric Inflammation and Cancer and Mobilizes Myeloid-Derived Suppressor Cells in Mice. Cancer Cell (2008) 14(5):408–19. doi: 10.1016/j.ccr.2008.10.011
Keywords: probiotic, pathogen resistance, administration duration, butyrate, NLRP3 inflammasome
Citation: Shan C, Li M, Liu Z, Xu R, Qiao F, Du Z-Y and Zhang M-L (2021) Pediococcus pentosaceus Enhances Host Resistance Against Pathogen by Increasing IL-1β Production: Understanding Probiotic Effectiveness and Administration Duration. Front. Immunol. 12:766401. doi: 10.3389/fimmu.2021.766401
Received: 29 August 2021; Accepted: 04 November 2021;
Published: 26 November 2021.
Edited by:
Zhigang Zhou, Feed Research Institute (CAAS), ChinaReviewed by:
Xin M. Luo, Virginia Tech, United StatesCopyright © 2021 Shan, Li, Liu, Xu, Qiao, Du and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei-Ling Zhang, bWx6aGFuZ0BiaW8uZWNudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.