- 1Department of Gastroenterology, Khoo Teck Puat Hospital, Singapore, Singapore
- 2Department of Hematopathology and Microbiology, Aga Khan University Hospital, Karachi, Pakistan
- 3Division of Infectious Diseases, Medical College of Wisconsin, Milwaukee, WI, United States
- 4Division of Hematology/Oncology, Medical College of Wisconsin, Milwaukee, WI, United States
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), resulted in an unprecedented global crisis. Although primarily a respiratory illness, dysregulated immune responses may lead to multi-organ dysfunction. Prior data showed that the resident microbial communities of gastrointestinal and respiratory tracts act as modulators of local and systemic inflammatory activity (the gut–lung axis). Evolving evidence now signals an alteration in the gut microbiome, brought upon either by cytokines from the infected respiratory tract or from direct infection of the gut, or both. Dysbiosis leads to a “leaky gut”. The intestinal permeability then allows access to bacterial products and toxins into the circulatory system and further exacerbates the systemic inflammatory response. In this review, we discuss the available data related to the role of the gut microbiome in the development and progression of COVID-19. We provide mechanistic insights into early data with a focus on immunological crosstalk and the microbiome’s potential as a biomarker and therapeutic target.
Introduction
Since it was first recognized, Coronavirus Disease 2019 (COVID-19), caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), remains a global affliction (1). Although vaccines offer hope in curbing the pandemic (2), an improved understanding of its pathogenesis and concurrent efforts to explore preventive and therapeutic strategies remain a priority to consolidate the success of mass vaccination and herd immunity.
The clinical spectrum of COVID-19 ranges from asymptomatic to severe, life-threatening disease (3). Current understanding of pathogenesis postulates a rapid and intense hyperactivation of the immune system, resulting in critical illness and mortality (4). Older age, burden of comorbidities, obesity, immunocompromised states, malignancy or ongoing cancer treatment, and being a transplant recipient, have been strongly linked with severe, and sometimes fatal, outcomes (5–9). Evolving data suggest that a state of chronic inflammation or baseline activation of the immune system might influence the course of COVID-19 more than direct cytopathic effects of the SARS-CoV-2. Furthermore, a subgroup of patients have been noted to develop auto-inflammatory symptoms (such as Kawasaki-like disease in children and multi-system inflammatory syndrome) long after clearance of the SARS-CoV-2 virus from body, suggesting an immune dysregulation (8, 10).
In the human body, the gastrointestinal tract (GIT) is the largest immune organ (11). The pool of resident microorganisms (bacteria, viruses, and fungi) in the GIT, collectively known as the gut microbiota, not only supports mucosal immunity but also modulates the systemic immune response of the host (12). Current evidence from other respiratory illnesses indicates that the gut microbiota affects the immunity and inflammation in the lungs (13, 14). Lately, some studies have examined the association between gut microbiota and SARS-CoV-2. In this review, we present the existing data related to the intersection of gut microbiome and the host’s immune response to SARS-CoV-2. We further explore the role of gut microbiome diversity and its compositional differences as diagnostic biomarkers, and the potential of the gut microbiome as an interventional target in modifying COVID-19 outcomes.
Significance of Gut Microbiota
The human GIT is home to about 104–105 bacteria per millimeter of content in the small intestine, and 1011 bacteria per gram of colonic content (15). In a healthy person, the gut microbiota comprises more than 100 bacterial phyla and the majority of bacteria belong to Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria, with Firmicutes and Bacteroidetes phyla constituting over 90% of the entire gut microbiota (16). The microorganisms and their combined genetic material make up the gut microbiome, which outnumber the human genome by about 150 times (17). The proportion of the various phyla remains quasi-stable and unique for an individual, although a shift can be observed during a change in health status. For example, the gut microbiome in the elderly has been observed to drift away from Firmicutes and towards Proteobacteria and Alistipes (18).
The gut microbiota exists in a symbiotic relationship with its host. It facilitates the synthesis of vitamins and fermentation of carbohydrates and other undigested nutrients and aids in the delivery of essential nutrients like short-chain fatty acids (SCFAs) to colonic epithelial cells. In addition, it also regulates mucosal permeability and provides deterrence against pathogenic microbes. More importantly, the microbiota plays an indispensable role in the preservation of intestinal homeostasis by modulating local and systemic immune responses of the host (19). The microbiota protects the GIT by (a) acting as a competitor against binding of pathogenic microbes, (b) neutralizing pathogens with their anti-microbial metabolites, (c) keeping the local immune system in a perpetual vigilant state, and (d) regulating the innate and adaptive immunity.
In a healthy person, the proportion of the various phyla mostly remains quasi-stable and unique for an individual (18). An imbalanced state is described as “gut dysbiosis”, a condition characterized by an alteration in the abundance or composition of the microbiota. Gut dysbiosis may occur with aging, dietary effects, drugs, gastrointestinal infections, and anatomical alterations of the GIT. A significantly dysbiotic state may predispose to the diseases of GIT, such as Clostridioides difficile enterocolitis, which is associated with prolonged and recurrent broad-spectrum antibiotic usage (20). Since gut microbiota modulates the fine balance between pro- and anti-inflammatory systemic responses, a dysbiotic state has also been associated with non-gastrointestinal systemic illnesses such as malignancy (12), type 2 diabetes mellitus (21), non-alcoholic steatohepatitis (22), coronary artery disease (23), neurodegenerative diseases (24), and depression (25).
The Gut–Lung Axis
The GI and respiratory tracts share a common mucosal immune system, known as the gut–lung axis (26, 27). From birth, both tracts receive their quota of microbiota via the oral route (28), and subsequently establish a differing but internally quasi-stable genre of microorganisms or microbiota (29). Although the microbiota of both tracts consists of similar phyla, they differ at the level of species in composition and density. Understandably, studies on respiratory microbiota have been complicated by tedious and invasive methods for collection of uncontaminated lower respiratory samples, and most data have been derived from mice models where lung tissue can be aseptically obtained (30). Consequently, there is growing excitement in understanding this complex immunological intersection.
Throughout the lifespan of an individual, established microbiota of both tracts contribute to the gut–lung axis, modulating both local and systemic immune responses when faced with a pathogenic threat. The axis is believed to be bidirectional, affecting the immune response of either tract when one site is activated (31). Using a germ-free murine model, Ichinohe et al. demonstrated potentially deleterious effects on respiratory immune responses after alteration of the gut microbiota with antibiotics (32). Other studies have also found that gut microbiota alterations result in abnormal activation of the immune system, predisposing to respiratory illnesses such as asthma, lung allergic responses, and chronic respiratory diseases (27, 33). Conversely, animal studies have also revealed an alteration in the gut microbiota after respiratory viral and bacterial infections (34–36). This distant effect is believed to be communicated by activation of the systemic immune system, with dysbiosis of either tract feeding into the other.
Role of Gut Microbiota in Respiratory Virus Infection
While the immune-related interactions between resident gut and respiratory tract microbiota are yet to be explored, a conceptual understanding of the impact of the gut microbiota in patients with COVID-19 may be extrapolated by examining the existing evidence of its role in non-SARS-CoV-2 respiratory virus infections.
Evidence From SARS-CoV-1 Infection
Many respiratory viral illnesses are commonly accompanied by GI symptoms. Previous studies during the severe acute respiratory syndrome (SARS) outbreak in 2002 showed that diarrhea was a common symptom and occurred in 16%–73% of patients. The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-1) was not only known to infect the lung epithelial cells but also the immune cells, triggering an intense immune response with elevation in Th2 cytokines (37). It was postulated that high levels of circulating pro-inflammatory cytokines altered the gut microbiota and compromised intestinal integrity. The resultant “leaky” gut allowed translocation of bacterial products and antigens into the circulation, further exacerbating the illness (38). Due to the bidirectional nature of the gut–lung axis, an alteration in gut microbiota further augments the respiratory immune responses, conceivably resulting in a vicious perpetuation of systemic inflammatory response (32).
Evidence From Other Community Respiratory Viruses
Using a mouse-model, Deriu et al. demonstrated that respiratory viral infection due to influenza resulted in gut dysbiosis predisposing to secondary Salmonella infection via circulatory type I interferons (34). Similarly, Wang et al. demonstrated indirect intestinal inflammation with influenza infection in a mouse-model occurring via microbiota-mediated Th17 cell-dependent inflammation (36). Several studies have reported gut dysbiosis after respiratory viral infection (39, 40). Groves et al. showed that gut dysbiosis, in the form of an increase in Bacteroidetes and a decrease in Firmicutes phyla abundance, occurred in mice models with respiratory syncytial and influenza virus infections, but not in those vaccinated with live attenuated influenza viruses (41). Furthermore, elevated levels of colonic Muc5ac and fecal lipocalin-2 in the pathogenic infection group suggest the presence of low-grade gut inflammation during respiratory virus infection.
Respiratory virus infection may also cause dysbiosis in lung microbiota, modulating both local immune responses within the lung parenchyma and systemically. Due to the difficulties in sampling of lung microbiota, only a few studies have examined the role of respiratory pathogens in altering lung microbiota. Molyneaux et al. reported an increased proportion of Proteobacteria and potentially pathogenic Haemophilus influenzae in the lower respiratory tract microbiota in rhinovirus-infected patients with chronic obstructive pulmonary disease (42). Using a mouse-model inoculated with intranasal H1N1 influenza virus, Gu et al. demonstrated a bacterial class shift in the lung microbiota, which persisted even during the recovery period (43). Notwithstanding the limitations in available studies, a common theme has emerged showing a link between respiratory virus infection and an alteration in the gut and respiratory tract microbiota, with the presence of inflammation of the GIT.
Role of Gut Microbiota in the Pathogenesis of COVID-19
Although no specific interaction between any gut microbial species and SARS-CoV-2 has been identified to date, there is indirect evidence (44, 45) that gut microbiota may have a role in the overall pathogenesis of COVID-19, as summarized in Figure 1. Taking the corollary further from non-SARS-Cov-2 virus-mediated gut dysbiosis, it is conceivable that infection with SARS-CoV-2 may also be impacted by immunological interactions with the gut microbiota.
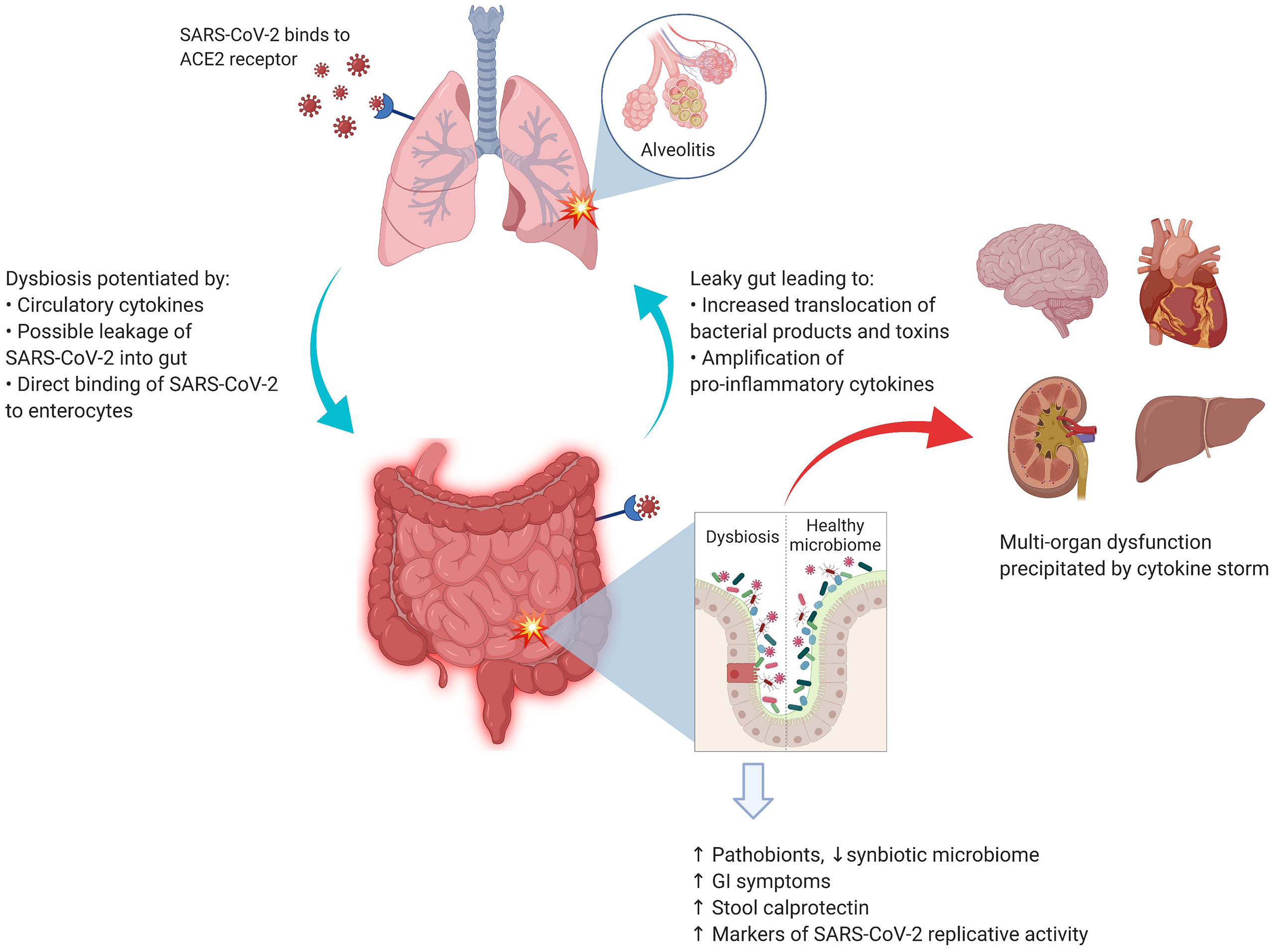
Figure 1 SARS-CoV-2 and the lung–gut axis: SARS-CoV-2 virus enters the alveolar cells by binding with ACE2 receptors, which are also abundant on the surface of enterocytes. The implication of direct infection of enterocytes by SARS-CoV-2 is still being explored. The circulatory cytokines from alveolitis (and/or direct viral infection of the enterocytes) cause the GI dysbiosis with resultant alterations in GI mucosal barrier. The entry of bacterial products and toxins from the GIT floods the circulatory system with more pro-inflammatory cytokines. Image was created with Biorender.com.
Pathogenesis of GI Symptoms in COVID-19
About half of patients with COVID-19 develop GI symptoms, which often precede the respiratory symptoms (45). In the respiratory tract, the SARS-CoV-2 infects the alveolar cells by binding to angiotensin-converting enzyme 2 (ACE-2) receptors. Interestingly, these receptors are also abundantly expressed on the surface of enterocytes (46), where they play an important role in maintaining the homeostasis of microbiota and mucosal inflammation (47). In one patient with COVID-19, Xiao et al. detected SARS-CoV-2 RNA, viral nucleocapsid protein, and ACE-2 in the epithelial cells of the esophagus, stomach, duodenum, and rectum (48). Several authors have also reported detection of SARS-CoV-2 RNA fragments, but not the whole virus, in stool samples (49, 50). A study by Lamers et al. also demonstrated that SARS-CoV-2 was able to infect enterocyte lineage cells in a human intestinal organoid model (51). Unfortunately, due to the scarcity of autopsy studies and pragmatic restrictions on endoscopic examinations of the GIT, there are only limited data to support this hypothesis in vivo. It also remains unknown how SARS-CoV-2 may survive the acidic gastric environment to directly infect enterocytes. Overall, it is unclear whether the GI symptoms of COVID-19 are a result of primary infection of the GIT or derived from other indirect mechanisms mentioned below.
Fecal Shedding of SARS-CoV-2
SARS-CoV-2 is principally transmitted by respiratory droplets, but evidence is accumulating for fecal-oral transmission. The hypothesis is supported by the presence of GI symptoms, detection of SARS-CoV-2 nuclear fingerprints in the GI mucosa, and detection of viral fragments in fecal samples. Interestingly, the first COVID-19 patient in the United States tested positive for SARS-CoV-2 via stool samples (52), and subsequent studies have consistently documented shedding of viral RNA in the stool samples in COVID-19 patients (53, 54). Furthermore, viral shedding in stool samples has been observed to persist longer than that in respiratory samples (55, 56).
Hypercytokinemia and the Leaky Gut
Elevated serum levels of pro-inflammatory markers, such as interleukin-6 (IL-6) and interleukin-10 (IL-10), are hallmarks of severe COVID-19 infection (57). These cytokines predispose to dysbiosis, which consequently alters intestinal permeability, a state known as the “leaky gut”. This enables further entry of a multitude of bacterial products and toxins, activating a pro-inflammatory cascade. In a study in 204 patients with COVID-19, Pan et al. reported that GI symptoms worsened with increasing severity of COVID-19 (58). In another study, fecal calprotectin levels (a marker of GI mucosal inflammation) were elevated in patients who had diarrhea during COVID-19 illness (59). A recent study by Prasad et al. measured several markers of gut permeability in the plasma (60). Levels of FABP2, PGN, and LPS were significantly higher among COVID-19 patients compared to healthy subjects, suggesting translocation of pro-inflammatory antigens from a leaky gut.
Association Between Gut Dysbiosis and Systemic Inflammation
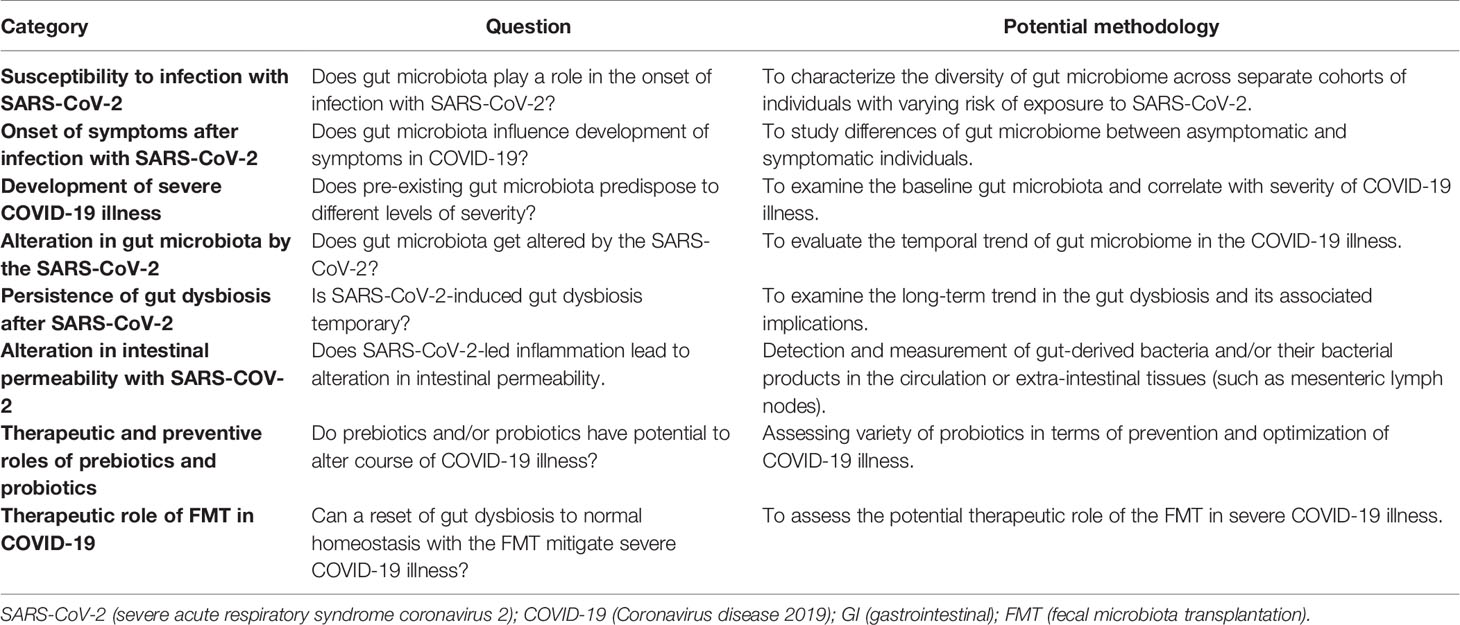
Gu et al. first presented evidence of an altered gut microbiota in COVID-19 patients by using high-throughput sequencing of 16S ribosomal RNA to compare the gut taxa of patients with COVID-19, H1N1 influenza, and healthy controls (61). Compared to healthy controls, COVID-19 patients had significantly reduced bacterial diversity, increased abundance of opportunistic pathogens (such as Streptococcus, Rothia, Veillonella, and Actinomyces), and significantly less diverse symbiotic species. Interestingly, the altered microbial signature in COVID-19 was different from patients with the H1N1 strain.
In another study, Zuo et al. analyzed fecal samples of 15 patients with COVID-19 using shotgun metagenomic sequencing (62). The study revealed a marked increase in opportunistic pathogens and depletion of beneficial microbes as compared to healthy controls, which persisted even after clearance of SARS-CoV-2. These findings suggest an inverse correlation between gut dysbiosis and COVID-19 severity and are congruent with the conclusions of a larger multi-center study in which Yeoh et al. examined the gut microbiota in 100 patients using shotgun sequencing. Fecal samples from 27 patients were analyzed longitudinally over 30 days, showing significant dysbiosis that persisted despite clearance of SARS-CoV-2. Correlative blood samples demonstrated an association between gut dysbiosis, elevation in the inflammatory mediators, and severity of systemic inflammation (63).
Another study by Newsome et al. compared microbiota composition from stool samples of 50 COVID-19 patients with uninfected patients (64). Significant perturbations in the microbiota composition in the COVID-19 patients were observed, independent of antibiotic exposure. The gut “metabolome”, a biochemical signature derived from bacterial metabolic activity in the gut, is another method for detecting an alteration in the gut microbiota composition. In a recent study by Lv et al., fecal samples of COVID-19 patients had altered metabolomes, suggesting malnutrition and intestinal inflammation (65). These results provide new insights into the pathogenesis of COVID-19.
Gut Dysbiosis as a Biomarker of Viral Replication
Although it was previously believed that gut dysbiosis in COVID-19 was mainly driven by inflammatory mediators from respiratory tract infection, a recent study suggests that active replication of SARS-CoV-2 in the gut may be driving the dysbiosis (66). Using in vitro transcriptional analysis in a SARS-CoV-2-infected cell model (with samples obtained from stools), the 3’ end of the SARS-CoV-2 genome was detected more than the 5’ end, suggesting active viral replication. Interestingly, majority of the patients had no GI symptoms, suggesting a quiescent GI infection despite active replication of SARS-CoV-2 in the GIT with dysbiosis. Moreover, on functional analysis of the gut microbiota, fecal samples with signatures of high SARS-CoV-2 burden demonstrated high de novo nucleotide and amino acid biosynthesis, correlating with increased bacterial proliferation. Although this was a pilot study comprising only 15 patients, further studies on alterations in the functionality of the gut microbiota may unearth the pathophysiology of COVID-19 illness.
Overall, currently expanding evidence suggests that patients with COVID-19 suffer from an alteration in the gut microbiota during and after the illness. Both systemic inflammation and replicative potential of SARS-CoV-2 in the gut may contribute towards dysbiosis.
Clinical Implication of Gut Microbiota in COVID-19
Though limited in number, studies to date have consistently demonstrated gut dysbiosis in patients with COVID-19 (Table 1). A more important clinical implication lies in understanding whether, and how, gut microbiota predisposes to varying degrees of COVID-19 severity.
Potential Role of Gut Microbiota in Asymptomatic/Subclinical and Mild COVID-19
As previously mentioned, clinical spectrum of COVID-19 ranges from asymptomatic to severe, life-threatening disease (3). A recent systematic review demonstrated that about one-third of patients remain clinically asymptomatic after infection with SARS-CoV-2 (68). However, a possibility of subclinical inflammatory process remains. In a systematic review involving 231 asymptomatic COVID-19 patients, almost two-thirds (63%) had inflammatory changes in the lungs on computed tomography (CT) scan (67). Irrespective of subtle inflammatory changes, a subset of patients may not mount the intense inflammatory response that portends severe illness. As this heterogeneity in clinical severity is less likely due to the existence of less virulent strains of SARS-CoV-2, or the protection from adaptive immunity given the novel nature of the virus, the immune response of the host remains the most probable factor in determining disease severity. It is unclear if any specific pattern of gut microbiota protects individuals from mounting a severe inflammatory state when infected with SARS-CoV-2. Kumar et al. highlighted a potential link between the environmental microbiota of a population and the burden of COVID-19. With data from 122 diverse countries, lower COVID-19-associated mortality was observed in countries with a higher percentage of rural population (alluding to higher gut microbial diversity), higher proportion of population residing in slums, and a lower water quality and sanitation score (69). While such observational data can be prone to confounders, these results offer some insight into the potential role of gut microbiota on the disease burden of COVID-19.
Gut Microbiota in Severe COVID-19
There is mounting evidence that being elderly and having a chronic inflammatory state (from chronic medical conditions) predisposes to a pro-dysbiotic state (70, 71). It is unlikely a coincidence that the highest rates of morbidity and mortality from COVID-19 have also been observed in the elderly, those with underlying chronic medical conditions, and among immunosuppressed patients with cancers (5–9). COVID-19 disease severity is likely host dependent and driven by the inflammatory response. In autopsy samples from a patient with severe COVID-19, inflammatory cells were observed in the lungs (72), suggesting an intense inflammatory response (8). Furthermore, studies have also reported elevated plasma levels of pro-inflammatory cytokines such as interleukin-1 (IL-1), IL-6, and tumor necrosis factor alpha (57, 73), in severe COVID-19. In patients prone to gut dysbiosis, further inflammatory triggers may tip the balance over to a leaky gut, resulting in a self-perpetuating inflammatory feedback circle. Notably, two small studies showed a direct correlation between severe COVID-19 and gut dysbiosis (62, 63) Another two studies have shown that patients with severe COVID-19 experienced more pronounced GI symptoms, along with higher levels of stool calprotectin (an indicator of GI inflammation and disrupted mucosal integrity) (58, 59). This supports the concept of an immunological crosstalk between the lungs and gut, presumably moderated by the gut microbiota (12). There is still a lack of data assessing the role of gut microbiota in a high-risk cohort (such as elderly or cancer patients) with COVID-19, although a study on the impact of probiotics on health and immunity in elderly and diabetic patients, and response to COVID-19 vaccination, is underway (Table 3).
Therapeutic Potential of Gut Microbiome for COVID-19
Given the association between gut dysbiosis and COVID-19 severity, the therapeutic potential for modulation of the gut microbiome to modify disease outcomes holds promise. However, there is no microbiota-directed therapy that has demonstrated efficacy in preventing the development or progression of COVID-19 currently.
Potential Role of Prebiotics
Plant-based fibers exert a prebiotic effect by promoting the growth of beneficial microorganisms in the gut microbiota (e.g., Bifidobacterium and Lactobacillus spp.) while decreasing the proportion of harmful species (e.g., Clostridia) (74). Moreover, the fermentation of soluble dietary fibers by certain bacterial species yields several beneficial metabolites, such as SCFAs, which serve to maintain colonic mucosal integrity and modulate the immune system (75). By-products of SCFAs are also absorbed into the circulatory system and have anti-inflammatory effects. In mice models, a high-fiber diet with elevated SCFA levels was protective against allergic inflammation in the lungs, while a low-fiber diet with decreased SCFA levels resulted in increased allergic airway disease (76). Interestingly, studies from other respiratory diseases have demonstrated a reduction in mortality with intake of whole grains (77, 78). Although the beneficial effects of dietary fibers are intuitive, there is currently no direct evidence that any specific amount or type of dietary fiber is beneficial in COVID-19 illness.
Potential Role of Probiotics
Oral probiotics are live bacteria of specific species that alter the composition of gut microbiota after reaching the intestines (74). A shift to beneficial bacterial species modulates the local and systemic inflammatory balance, with several studies demonstrating a positive impact on respiratory infections and other extra-intestinal illnesses. Using a probiotic bacterium, Lactobacillus gasseri SBT2055 in mouse models, prevention of infection with respiratory syncytial virus was demonstrated (79). In another study on 30 elderly volunteers, Bifidobacterium lactis HN019 ingestion was shown to enhance the cellular immunity (80). Placebo controlled clinical trials with probiotics (using Lactobacillus rhamnosus GG, Bacillus subtilis, and Enterococcus faecalis) have also demonstrated significant improvement in patients with ventilator-associated pneumonia (81, 82).
Naturally, if dysbiosis is indeed involved in the pathogenesis of severe COVID-19, probiotics appear to be among the more convenient, efficient, and potentially safe strategies. After initial reports of gut dysbiosis in patients with severe COVID-19, the National Health Commission (of China) recommended the use of probiotics to maintain gut microbial homeostasis and prevent secondary bacterial infections (83). Given the dearth of data pertaining to SARS-CoV-2 and the relative safety of probiotics, the rapid promulgation in favor of probiotics seems reasonable while awaiting further evidence. Although there is no direct evidence yet showing the efficacy of any specific strain of probiotic against COVID-19, several registered trials are currently examining the therapeutic potential of various probiotics formulations in COVID-19 (Table 3).
Potential Role of Fecal Microbiota Transplantation
FMT is a process of actively transferring colonies of fecal bacteria from a healthy person into the GIT of another individual. The process aims to restore the composition of gut microbiota back to a healthy state. As mentioned earlier, FMT is an effective therapy for recurrent or refractory C. difficile enterocolitis (84). Given the proposed role of gut microbiota in the abnormal activation of immune responses in the COVID-19, FMT can potentially be explored as a therapeutic strategy. A recent case report (85) described two patients with rapid resolution of COVID-19 after FMT was undertaken to treat concomitant C. difficile infection. However strong may be the hypothesis and surrounding speculations, FMT should not presently be recommended as a therapy against COVID-19 due to the scarcity of large-scale studies. To explore further, a clinical trial (FeMToCOVID) is currently registered at the clinicaltrials.org (NCT04824222), though it has not yet started recruiting patients.
Unanswered Questions and Future Directions
Despite the efficient pace of clinical trials evaluating new and repurposed agents for COVID-19, success has been modest at best. Although still in nascent stages, evolving evidence signals a probable link between gut microbiota and the host’s immune response to COVID-19. However, the exact mechanism and extent of the role of gut dysbiosis in disease severity remain to be elucidated. This is further compounded by the inherent challenges associated with designing microbiome studies. Another possible angle would be to define the state of a leaky gut more clearly with biomarkers and cutoff criteria, to also enable clarification on whether patients who have developed an inflammatory cascade are still amenable to therapeutic gut microbiota modulation.
Besides exploration of any potential role in the management of COVID-19, long-term consequences of gut dysbiosis should also be explored with longitudinal follow-up (Table 2). A search of ongoing clinical trials at the US National Library of Medicine reveals 24 registered studies assessing microbiota-targeted therapeutic options (Table 3).
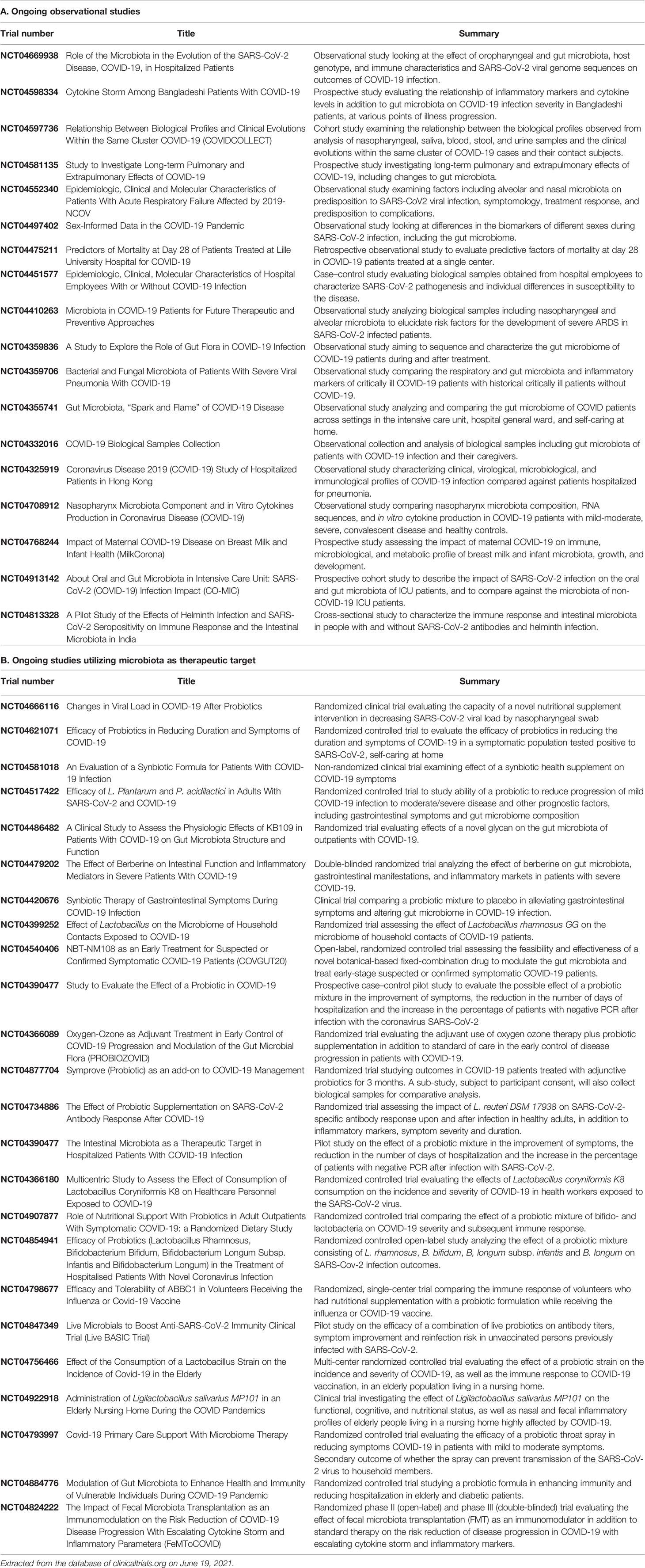
Table 3 Ongoing registered trials studying role of gut microbiota in COVID-19 (updated as of June 19, 2021).
Conclusion
The role of resident gut microbiota in other respiratory illnesses has been well recognized. Furthermore, the brunt of unfavorable COVID-19 outcomes has been on elderly patients and those with chronic medical diseases, both scenarios known to have senescence-driven gut dysbiosis. Increasingly, evidence is mounting for gut dysbiosis as a predisposing factor for severe COVID-19, through a leaky gut phenomenon and resultant spillage of bacterial products and toxins. Evidence is emerging that the degree of dysbiosis correlates with the severity of COVID-19 illness. This behooves us to explore potentially preventive and therapeutic targets, such as dietary intervention and probiotics. Several ongoing trials are evaluating various pathogenic routes and therapeutic approaches. While efforts for direct anti-viral agents and vaccines are of prime significance, the gut–lung axis could still hold therapeutic potential.
Author Contributions
IH and MBA: study concept. IH and MBA: literature search. IH, GY, and MBA: manuscript writing. MAA: critical revisions and intellectual content. MBA: supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wang C, Horby PW, Hayden FG, Gao GF. A Novel Coronavirus Outbreak of Global Health Concern. Lancet (London England) (2020) 395:470–3. doi: 10.1016/S0140-6736(20)30185-9
2. The Lancet Respiratory Medicine Realising the Potential of SARS-CoV-2 Vaccines-a Long Shot? Lancet Respir Med (2021). doi: 10.1016/S2213-2600(21)00045-X
3. Yuen K-S, Ye Z-W, Fung S-Y, Chan C-P, Jin D-Y. SARS-CoV-2 and COVID-19: The Most Important Research Questions. Cell Biosci (2020) 10:40. doi: 10.1186/s13578-020-00404-4
4. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol (2020) 11:1446. doi: 10.3389/fimmu.2020.01446
5. Abid MB, Mughal M, Abid MA. Coronavirus Disease 2019 (COVID-19) and Immune-Engaging Cancer Treatment. JAMA Oncol (2020) 6:1529–30. doi: 10.1001/jamaoncol.2020.2367
6. Sharma A, Bhatt NS, St Martin A, Abid MB, Bloomquist J, Chemaly R, et al. Clinical Characteristics and Outcomes of COVID-19 in Haematopoietic Stem-Cell Transplantation Recipients: An Observational Cohort Study. Lancet Haematol (2021) 9:117. doi: 10.1016/S2352-3026(20)30429-4
7. Jordan RE, Adab P, Cheng KK. Covid-19: Risk Factors for Severe Disease and Death. BMJ (2020) 368:m1198. doi: 10.1136/bmj.m1198
8. Abid MA, Nunley L, Abid MB. Could Coronavirus Disease 2019 (COVID-19) Render Natural Immunity to Re-Infections? A Spotlight on the Therapeutic Pipeline. Front Immunol (2020) 11:1294. doi: 10.3389/fimmu.2020.01294
9. Hoong CWS, Hussain I, Aravamudan VM, Phyu EE, Lin JHX, Koh H. Obesity Is Associated With Poor Covid-19 Outcomes: A Systematic Review and Meta-Analysis. Horm Metab Res (2021) 53:85–93. doi: 10.1055/a-1326-2125
10. Viner RM, Whittaker E. Kawasaki-Like Disease: Emerging Complication During the COVID-19 Pandemic. Lancet (London England) (2020) 395:1741–3. doi: 10.1016/S0140-6736(20)31129-6
11. Zmora N, Suez J, Elinav E. You Are What You Eat: Diet, Health and the Gut Microbiota. Nat Rev Gastroenterol Hepatol (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
12. Abid MB. Could the Menagerie of the Gut Microbiome Really Cure Cancer? Hope or Hype. J Immunother Cancer (2019) 7:92. doi: 10.1186/s40425-019-0561-9
13. Dang AT, Marsland BJ. Microbes, Metabolites, and the Gut-Lung Axis. Mucosal Immunol (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
14. McAleer JP, Kolls JK. Contributions of the Intestinal Microbiome in Lung Immunity. Eur J Immunol (2018) 48:39–49. doi: 10.1002/eji.201646721
15. Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res (2017) 77:1783–812. doi: 10.1158/0008-5472.CAN-16-2929
16. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem Across Age, Environment, Diet, and Diseases. Microorganisms (2019) 7. doi: 10.3390/microorganisms7010014
17. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
18. Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, Variability, and Temporal Stability of the Intestinal Microbiota of the Elderly. Proc Natl Acad Sci USA (2011) 108 Suppl:4586–91. doi: 10.1073/pnas.1000097107
19. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human Nutrition, the Gut Microbiome and the Immune System. Nature (2011) 474:327–36. doi: 10.1038/nature10213
20. Guh AY, Kutty PK. Clostridioides Difficile Infection. Ann Intern Med (2018) 169:ITC49–64. doi: 10.1016/j.ebiom.2019.11.051
21. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
22. Mouries J, Brescia P, Silvestri A, Spadoni I, Sorribas M, Wiest R, et al. Microbiota-Driven Gut Vascular Barrier Disruption Is a Prerequisite for non-Alcoholic Steatohepatitis Development. J Hepatol (2019) 71:1216–28. doi: 10.1016/j.jhep.2019.08.005
23. Tang WHW, Bäckhed F, Landmesser U, Hazen SL. Intestinal Microbiota in Cardiovascular Health and Disease: JACC State-of-the-Art Review. J Am Coll Cardiol (2019) 73:2089–105. doi: 10.1016/j.jacc.2019.03.024
24. Dinan TG, Cryan JF. Gut Instincts: Microbiota as a Key Regulator of Brain Development, Ageing and Neurodegeneration. J Physiol (2017) 595:489–503. doi: 10.1113/JP273106
25. Liang S, Wu X, Hu X, Wang T, Jin F. Recognizing Depression From the Microbiota–Gut–Brain Axis. Int J Mol Sci (2018) 19:1592. doi: 10.3390/ijms19061592
26. Budden KF, Gellatly SL, Wood DLA, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging Pathogenic Links Between Microbiota and the Gut-Lung Axis. Nat Rev Microbiol (2017) 15:55–63. doi: 10.1038/nrmicro.2016.142
27. Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, et al. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front Cell Infect Microbiol (2020) 10:9. doi: 10.3389/fcimb.2020.00009
28. Grier A, McDavid A, Wang B, Qiu X, Java J, Bandyopadhyay S, et al. Neonatal Gut and Respiratory Microbiota: Coordinated Development Through Time and Space. Microbiome (2018) 6:193. doi: 10.1186/s40168-018-0566-5
29. Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, et al. Serial Analysis of the Gut and Respiratory Microbiome in Cystic Fibrosis in Infancy: Interaction Between Intestinal and Respiratory Tracts and Impact of Nutritional Exposures. MBio (2012) 3. doi: 10.1128/mBio.00251-12
30. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio (2017) 8. doi: 10.1128/mBio.02287-16
31. Kraft SC, Earle RH, Roesler M, Esterly JR. Unexplained Bronchopulmonary Disease With Inflammatory Bowel Disease. Arch Intern Med (1976) 136:454–9. doi: 10.1001/archinte.1976.03630040056012
32. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota Regulates Immune Defense Against Respiratory Tract Influenza A Virus Infection. Proc Natl Acad Sci U S A (2011) 108:5354–9. doi: 10.1073/pnas.1019378108
33. Marsland BJ, Trompette A, Gollwitzer ES. The Gut-Lung Axis in Respiratory Disease. Ann Am Thorac Soc (2015) 12 Suppl 2:S150–6. doi: 10.1513/AnnalsATS.201503-133AW
34. Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, et al. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut Through Type I Interferons. PloS Pathog (2016) 12:e1005572. doi: 10.1371/journal.ppat.1005572
35. Hanada S, Pirzadeh M, Carver KY, Deng JC. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front Immunol (2018) 9:2640. doi: 10.3389/fimmu.2018.02640
36. Wang J, Li F, Wei H, Lian Z-H, Sun R, Tian Z. Respiratory Influenza Virus Infection Induces Intestinal Immune Injury via Microbiota-Mediated Th17 Cell-Dependent Inflammation. J Exp Med (2014) 211:2397–410. doi: 10.1084/jem.20140625
37. Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, et al. Multiple Organ Infection and the Pathogenesis of SARS. J Exp Med (2005) 202:415–24. doi: 10.1084/jem.20050828
38. Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the Lung Microbiome With Gut Bacteria in Sepsis and the Acute Respiratory Distress Syndrome. Nat Microbiol (2016) 1:16113. doi: 10.1038/nmicrobiol.2016.113
39. Bartley JM, Zhou X, Kuchel GA, Weinstock GM, Haynes L. Impact of Age, Caloric Restriction, and Influenza Infection on Mouse Gut Microbiome: An Exploratory Study of the Role of Age-Related Microbiome Changes on Influenza Responses. Front Immunol (2017) 8:1164. doi: 10.3389/fimmu.2017.01164
40. Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, Schmolke M. Influenza A Virus Infection Impacts Systemic Microbiota Dynamics and Causes Quantitative Enteric Dysbiosis. Microbiome (2018) 6:9. doi: 10.1186/s40168-017-0386-z
41. Groves HT, Cuthbertson L, James P, Moffatt MF, Cox MJ, Tregoning JS. Respiratory Disease Following Viral Lung Infection Alters the Murine Gut Microbiota. Front Immunol (2018) 9:182. doi: 10.3389/fimmu.2018.00182
42. Molyneaux PL, Mallia P, Cox MJ, Footitt J, Willis-Owen SAG, Homola D, et al. Outgrowth of the Bacterial Airway Microbiome After Rhinovirus Exacerbation of Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med (2013) 188:1224–31. doi: 10.1164/rccm.201302-0341OC
43. Gu L, Deng H, Ren Z, Zhao Y, Yu S, Guo Y, et al. Dynamic Changes in the Microbiome and Mucosal Immune Microenvironment of the Lower Respiratory Tract by Influenza Virus Infection. Front Microbiol (2019) 10:2491. doi: 10.3389/fmicb.2019.02491
44. Troisi J, Venutolo G, Pujolassos Tanyà M, Delli Carri M, Landolfi A, Fasano A. COVID-19 and the Gastrointestinal Tract: Source of Infection or Merely a Target of the Inflammatory Process Following SARS-CoV-2 Infection? World J Gastroenterol (2021) 27:1406–18. doi: 10.3748/wjg.v27.i14.1406
45. Han C, Duan C, Zhang S, Spiegel B, Shi H, Wang W, et al. Digestive Symptoms in COVID-19 Patients With Mild Disease Severity: Clinical Presentation, Stool Viral RNA Testing, and Outcomes. Am J Gastroenterol (2020) 115:916–23. doi: 10.14309/ajg.0000000000000664
46. Zhang H, Li H-B, Lyu J-R, Lei X-M, Li W, Wu G, et al. Specific ACE2 Expression in Small Intestinal Enterocytes may Cause Gastrointestinal Symptoms and Injury After 2019-Ncov Infection. Int J Infect Dis (2020) 96:19–24. doi: 10.1016/j.ijid.2020.04.027
47. Perlot T, Penninger JM. ACE2 - From the Renin-Angiotensin System to Gut Microbiota and Malnutrition. Microbes Infect (2013) 15:866–73. doi: 10.1016/j.micinf.2013.08.003
48. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H, et al. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology (2020) 158:1831–3.e3. doi: 10.1053/j.gastro.2020.02.055
49. Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, et al. The Presence of SARS-CoV-2 RNA in the Feces of COVID-19 Patients. J Med Virol (2020) 92:833–40. doi: 10.1002/jmv.25825
50. Park S-K, Lee C-W, Park D-I, Woo H-Y, Cheong HS, Shin H-C, et al. Detection of SARS-CoV-2 in Fecal Samples From Patients With Asymptomatic and Mild COVID-19 in Korea. Clin Gastroenterol Hepatol (2021) 19:1387–94.e2. doi: 10.1016/j.cgh.2020.06.005
51. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science (2020) 369:50–4. doi: 10.1126/science.abc1669
52. Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce C, et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med (2020) 382:929–36. doi: 10.1056/NEJMoa2001191
53. Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, et al. Prolonged Presence of SARS-CoV-2 Viral RNA in Faecal Samples. Lancet Gastroenterol Hepatol (2020) 5:434–5. doi: 10.1016/S2468-1253(20)30083-2
54. Zhang J, Wang S, Xue Y. Fecal Specimen Diagnosis 2019 Novel Coronavirus-Infected Pneumonia. J Med Virol (2020) 92:680–2. doi: 10.1002/jmv.25742
55. Xing Y-H, Ni W, Wu Q, Li W-J, Li G-J, Wang W-D, et al. Prolonged Viral Shedding in Feces of Pediatric Patients With Coronavirus Disease 2019. J Microbiol Immunol Infect (2020) 53:473–80. doi: 10.1016/j.jmii.2020.03.021
56. Xu CLH, Raval M, Schnall JA, Kwong JC, Holmes NE. Duration of Respiratory and Gastrointestinal Viral Shedding in Children With SARS-CoV-2: A Systematic Review and Synthesis of Data. Pediatr Infect Dis J (2020) 39:e249–56. doi: 10.1097/INF.0000000000002814
57. Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, et al. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 are Disease Severity Predictors. Emerg Microbes Infect (2020) 9:1123–30. doi: 10.1080/22221751.2020.1770129
58. Pan L, Mu M, Yang P, Sun Y, Wang R, Yan J, et al. Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am J Gastroenterol (2020) 115:766–73. doi: 10.14309/ajg.0000000000000620
59. Effenberger M, Grabherr F, Mayr L, Schwaerzler J, Nairz M, Hilbe M, et al. Faecal Calprotectin Indicates Intestinal Inflammation in COVID-19. Gut (2020) 69:1543–4. doi: 10.1136/gutjnl-2020-321388
60. Prasad R, Patton MJ, Floyd JL, Vieira CP, Fortmann S, DuPont M, et al. Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. bioRxiv Prepr Serv Biol (2021). doi: 10.1101/2021.04.06.438634
61. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin Infect Dis (2020) 71:2669–78. doi: 10.1093/cid/ciaa709
62. Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology (2020) 159:944–55.e8. doi: 10.1053/j.gastro.2020.05.048
63. Yeoh YK, Zuo T, Lui GC-Y, Zhang F, Liu Q, Li AY, et al. Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients With COVID-19. Gut (2021) 70:698–706. doi: 10.1136/gutjnl-2020-323020
64. Newsome RC, Gauthier J, Hernandez MC, Abraham GE, Robinson TO, Williams HB, et al. The Gut Microbiome of COVID-19 Recovered Patients Returns to Uninfected Status in a Minority-Dominated United States Cohort. Gut Microbes (2021) 13:1–15. doi: 10.1080/19490976.2021.1926840
65. Lv L, Jiang H, Chen Y, Gu S, Xia J, Zhang H, et al. The Faecal Metabolome in COVID-19 Patients is Altered and Associated With Clinical Features and Gut Microbes. Anal Chim Acta (2021) 1152:338267. doi: 10.1016/j.aca.2021.338267
66. Zuo T, Liu Q, Zhang F, Lui GC-Y, Tso EY, Yeoh YK, et al. Depicting SARS-CoV-2 Faecal Viral Activity in Association With Gut Microbiota Composition in Patients With COVID-19. Gut (2021) 70:276–84. doi: 10.1136/gutjnl-2020-322294
67. Tsikala Vafea M, Chander B. Chest CT Findings in Asymptomatic Cases With COVID-19: A Systematic Review and Meta-Analysis. Clin Radiol (2020) 75:876.e33–876.e39. doi: 10.1016/j.mehy.2020.110209
68. Oran DP, Topol EJ. The Proportion of SARS-CoV-2 Infections That Are Asymptomatic: A Systematic Review. Ann Intern Med (2021) 174:655–62. doi: 10.7326/M20-6976
69. Kumar P, Chander B. COVID 19 Mortality: Probable Role of Microbiome to Explain Disparity. Med Hypotheses (2020) 144:110209. doi: 10.1016/j.mehy.2020.110209
70. Bana B, Cabreiro F. The Microbiome and Aging. Annu Rev Genet (2019) 53:239–61. doi: 10.1146/annurev-genet-112618-043650
71. Wilkins LJ, Monga M, Miller AW. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci Rep (2019) 9:12918. doi: 10.1038/s41598-019-49452-y
72. Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological Findings of COVID-19 Associated With Acute Respiratory Distress Syndrome. Lancet Respir Med (2020) 8:420–2. doi: 10.1016/S2213-2600(20)30076-X
73. Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, et al. Pathogenic T-Cells and Inflammatory Monocytes Incite Inflammatory Storms in Severe COVID-19 Patients. Natl Sci Rev (2020) 7:998–1002. doi: 10.1093/nsr/nwaa041
74. Abid MB, Koh CJ. Probiotics in Health and Disease: Fooling Mother Nature? Infection (2019) 47:911–7. doi: 10.1007/s15010-019-01351-0
75. Ratajczak W, Rył A, Mizerski A, Walczakiewicz K, Sipak O, Laszczyńska M, et al. Immunomodulatory Potential of Gut Microbiome-Derived Short-Chain Fatty Acids (SCFAs). Acta Biochim Pol (2019) 66:1–12. doi: 10.18388/abp.2018_2648
76. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut Microbiota Metabolism of Dietary Fiber Influences Allergic Airway Disease and Hematopoiesis. Nat Med (2014) 20:159–66. doi: 10.1038/nm.3444
77. Tabak C, Smit HA, Heederik D, Ocké MC, Kromhout D. Diet and Chronic Obstructive Pulmonary Disease: Independent Beneficial Effects of Fruits, Whole Grains, and Alcohol (the MORGEN Study). Clin Exp Allergy (2001) 31:747–55. doi: 10.1046/j.1365-2222.2001.01064.x
78. Jacobs DR, Andersen LF, Blomhoff R. Whole-Grain Consumption is Associated With a Reduced Risk of Noncardiovascular, Noncancer Death Attributed to Inflammatory Diseases in the Iowa Women’s Health Study. Am J Clin Nutr (2007) 85:1606–14. doi: 10.1093/ajcn/85.6.1606
79. Eguchi K, Fujitani N, Nakagawa H, Miyazaki T. Prevention of Respiratory Syncytial Virus Infection With Probiotic Lactic Acid Bacterium Lactobacillus Gasseri SBT2055. Sci Rep (2019) 9:4812. doi: 10.1038/s41598-019-39602-7
80. Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. Enhancement of Immunity in the Elderly by Dietary Supplementation With the Probiotic Bifidobacterium Lactis HN019. Am J Clin Nutr (2001) 74:833–9. doi: 10.1093/ajcn/74.6.833
81. Zeng J, Wang C-T, Zhang F-S, Qi F, Wang S-F, Ma S, et al. Effect of Probiotics on the Incidence of Ventilator-Associated Pneumonia in Critically Ill Patients: A Randomized Controlled Multicenter Trial. Intensive Care Med (2016) 42:1018–28. doi: 10.1007/s00134-016-4303-x
82. Morrow LE, Kollef MH, Casale TB. Probiotic Prophylaxis of Ventilator-Associated Pneumonia: A Blinded, Randomized, Controlled Trial. Am J Respir Crit Care Med (2010) 182:1058–64. doi: 10.1164/rccm.200912-1853OC
83. Gao QY, Chen YX, Fang JY. 2019 Novel Coronavirus Infection and Gastrointestinal Tract. J Dig Dis (2020) 21:125–6. doi: 10.1111/1751-2980.12851
84. Liu X, Li Y, Wu K, Shi Y, Chen M. Fecal Microbiota Transplantation as Therapy for Treatment of Active Ulcerative Colitis: A Systematic Review and Meta-Analysis. Gastroenterol Res Pract (2021) 2021:6612970. doi: 10.1155/2021/6612970
Keywords: COVID-19, gut microbiome, cytokine release syndrome, gut–lung axis, dysbiosis, leaky gut
Citation: Hussain I, Cher GLY, Abid MA and Abid MB (2021) Role of Gut Microbiome in COVID-19: An Insight Into Pathogenesis and Therapeutic Potential. Front. Immunol. 12:765965. doi: 10.3389/fimmu.2021.765965
Received: 27 August 2021; Accepted: 20 September 2021;
Published: 14 October 2021.
Edited by:
Lucia Lopalco, San Raffaele Hospital (IRCCS), ItalyReviewed by:
Leila B. Giron, Wistar Institute, United StatesTaketoshi Mizutani, The University of Tokyo, Japan
Copyright © 2021 Hussain, Cher, Abid and Abid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Bilal Abid, YmlsYWxfYWJpZEBob3RtYWlsLmNvbQ==; Ikram Hussain, aWtyYW1faHVzc2FpbkB3aGMuc2c=
†These authors share first authorship
 Ikram Hussain
Ikram Hussain Gabriel Liu Yuan Cher
Gabriel Liu Yuan Cher Muhammad Abbas Abid2
Muhammad Abbas Abid2 Muhammad Bilal Abid
Muhammad Bilal Abid