- 1Aquaculture and Fisheries Group, Department of Animal Sciences, Wageningen University & Research, Wageningen, Netherlands
- 2Department of Microbial Ecology, Netherlands Institute of Ecology-The Royal Netherlands Academy of Arts and Sciences, (NIOO-KNAW), Wageningen, Netherlands
- 3Animal Nutrition Group, Department of Animal Sciences, Wageningen University & Research, Wageningen, Netherlands
Dietary supplementation of fish with β-glucans has been commonly associated with immunomodulation and generally accepted as beneficial for fish health. However, to date the exact mechanisms of immunomodulation by β-glucan supplementation in fish have remained elusive. In mammals, a clear relation between high-fibre diets, such as those including β-glucans, and diet-induced immunomodulation via intestinal microbiota and associated metabolites has been observed. In this study, first we describe by 16S rRNA sequencing the active naive microbiota of common carp intestine. Based on the abundance of the genus Bacteroides, well known for their capacity to degrade and ferment carbohydrates, we hypothesize that common carp intestinal microbiota could ferment dietary β-glucans. Indeed, two different β-glucan preparations (curdlan and MacroGard®) were both fermented in vitro, albeit with distinct fermentation dynamics and distinct production of short-chain fatty acids (SCFA). Second, we describe the potential immunomodulatory effects of the three dominant SCFAs (acetate, butyrate, and propionate) on head kidney leukocytes, showing effects on both nitric oxide production and expression of several cytokines (il-1b, il-6, tnfα, and il-10) in vitro. Interestingly, we also observed a regulation of expression of several gpr40L genes, which were recently described as putative SCFA receptors. Third, we describe how a single in vivo oral gavage of carp with MacroGard® modulated simultaneously, the expression of several pro-inflammatory genes (il-1b, il-6, tnfα), type I IFN-associated genes (tlr3.1, mx3), and three specific gpr40L genes. The in vivo observations provide indirect support to our in vitro data and the possible role of SCFAs in β-glucan-induced immunomodulation. We discuss how β-glucan-induced immunomodulatory effects can be explained, at least in part, by fermentation of MacroGard® by specific bacteria, part of the naive microbiota of common carp intestine, and how a subsequent production of SFCAs could possibly explain immunomodulation by β-glucan via SCFA receptors present on leukocytes.
Introduction
Effects of immunomodulation by β-glucans have been widely studied in teleost fish. Regardless of administration route or fish species, β-glucans are often associated with immunomodulatory effects and increased resistance to both viral and bacterial infections [as reviewed by (1–3)]. Although supplementation of β-glucans may have become common practise in aquaculture, modulation routes and mechanisms explaining the observed immunomodulatory effects of β-glucans in the fish intestine have remained largely obscure.
In mammalian vertebrates, direct recognition of β-glucans is mediated by at least the C-type lectin receptor (CLR) Dectin-1 (4–6), whereas in invertebrates β-glucans can be recognised by β-glucan-binding proteins (7). For both mammals (8, 9) and invertebrates (7, 10), several non-exclusive pathways play a downstream role in the response to β-glucans, often mediating activation of the transcription factor NF-κB (11). Although in vertebrate fish genomes no clear homologue of mammalian Dectin-1 could be identified so far (6, 12), we could show activation of carp macrophages with curdlan (13, 14), considered a Dectin-1-specific (1,3)-β-glucan stimulus. We therefore proposed that the immuno-modulatory effects of β-glucan in carp macrophages could include regulation of a downstream signalling pathway typical of CLR activation and confirmed our hypothesis by pathway analysis of differentially expressed genes (DEGs) (14). Among the regulated genes were card9 and bcl10, both members of the signalling complex downstream of mammalian Dectin-1. Not only did we observe a regulation of the CLR signalling pathway, several candidate receptors could be identified based on regulation of expression upon stimulation with β-glucans, or based on the conservation of β-glucan-binding motifs in their predicted translations to protein (14). As there is clear evidence for presence of macrophages and other antigen-sampling cells in the fish intestine [as reviewed by (15)], it is possible that recognition of β-glucans in the intestine occurs via a direct recognition mechanism of β-glucan receptors.
Another mechanism of immune modulation by β-glucans could be indirect, relying on prebiotic effects inducing a shift in intestinal microbiota which would influence the microenvironment of the intestine. The two mechanisms explaining immunomodulation by β-glucans (direct recognition and indirect prebiotic effects) should not necessarily be exclusive. The intestinal microbiota is of well-known importance to the host, for example via degradation and fermentation of non-starch polysaccharides and/or resultant production of metabolites, which in turn affect the host immune system [as reviewed by (16–20)]. Also for fish, the importance of microbial communities for their health and the mitigation of diseases has been acknowledged [as reviewed by (19–21)]. Several studies have shown the ability of certain fish microbiota to degrade carbohydrates, showing growth of specific intestinal bacterial monocultures on different carbohydrates, among which is β-glucan (22), and observing the in vitro fermentation of several carbohydrates and production of short-chain fatty acids (SCFAs) by intestinal microbes of common carp (Cyprinus carpio) and Nile tilapia (Oreochromis niloticus) (23, 24). In a comparison of the fermentability of different carbohydrates and produced SCFAs between Nile tilapia and European sea bass, both their gut microbiota were found capable of fermenting carbohydrates but with differing end-product profiles (25). Recently, a systematic review extensively described the capacity of tilapia to digest carbohydrates (26). Still, documentation on the effects of dietary β-glucan supplementation on the composition of the intestinal microbiota is relatively scarce. In common carp, a 2-week feeding with β-glucan-enriched diets induced shifts in the intestinal microbiota, manifested either as a reduction in species richness and absolute number of operational taxonomic units (OTUs) (27) or as an increase in microbial diversity (28). In seabass, a 4-week feeding with β-glucan-enriched diets induced a shift toward more Methylobacterium (29), whereas in rainbow trout and feral common carp, an 8-week feeding with yeast-derived probiotics rich in β-glucans induced a shift toward more lactic acid bacteria (30, 31). Recently, a study investigating the effect of feeding β-glucans for 7 weeks to common carp showed, through DGGE analysis, a possible shift in bacterial species richness alongside a downregulation of il-1b expression (32). Also, addition of β-glucans to the water could affect the composition of intestinal microbiota, at least of Nile tilapia (33). Overall, possibly owing to differences in diet formulation, sampling, microbial analysis, and fish species, no consistent and distinct shift in intestinal microbial composition induced by β-glucans has been discerned. This leaves undecided if immunomodulation by β-glucans is based on a) receptor-mediated recognition or b) fermentation of β-glucans facilitating a subsequent shift in composition of intestinal microbiota with an associated difference in SCFA metabolic profile, or both.
In the present study, we investigated the role of β-glucan as prebiotic first based on the hypothesis that common carp intestinal microbiota would be able to ferment β-glucans. We further hypothesised that, as a result of this fermentation process, in vivo treatment of carp with β-glucans would induce a shift in intestinal microbial composition and subsequently might result in a shift in production of short-chain fatty acid (SCFA) metabolites. Finally, we hypothesised that this series of processes would contribute to the commonly observed immunomodulation by β-glucans of the host. To test our hypotheses, we first characterised the naive, active intestinal microbiota of common carp by 16S rRNA transcript sequencing. We also analysed the SCFA profile generated during in vitro fermentation and noticed β-glucan-dependent shifts in SCFA production. To this purpose, we used an in vitro batch culture method to assess the potential fermentability of feed ingredients (25, 34, 35). With this method, an inoculum is prepared under strictly anaerobic conditions using freshly collected intestinal contents. The inoculum is incubated with a fermentable substrate of choice and with a medium to support bacterial growth. Subsequently, gas accumulating during fermentation indicates the kinetics of fermentation. At the end of the fermentation period, samples are taken from the fermentation fluid in order to analyse fermentation end-products and substrate utilization (36). Subsequently, we performed further in vitro studies into the modulation by SCFAs of leukocyte function and possible involvement of G-protein-coupled receptor (GPR) 40 family members. Our recent study of phylogeny and synteny of several cDNA sequences for carp gpr40L genes (37) showed a division into three subclasses (a, b, and c), all structurally conserved. Here, we present a first investigation of the importance of these putative SCFA receptors in the route to immunomodulation by β-glucans after microbial fermentation by studying the regulation of gpr40L gene expression. Last but not least, common carp were treated with a single oral gavage with β-glucans to analyse the in vivo effects of β-glucans on intestinal microbiota and effects on regulation of local gene expression. Overall, our data suggest that dietary β-glucan administration may have indirect immunomodulatory effects, associated with a shift in SCFA profile and detection of these SCFAs by gpr40L SCFA receptors. The interconnection between SCFA profile shifts and detection of such by the appropriate receptors warrants further investigation.
Material and Methods
Animals
European common carp (Cyprinus carpio carpio L.) of the R3 × R8 strain were used, which originated from a cross between the Hungarian R8 strain and the Polish R3 strain (38). Carp were bred and raised in the aquatic research facility of Wageningen University at 24°C in recirculating UV-treated water and fed pelleted dry food (Skretting, Nutreco) twice daily. Fish were housed in rectangular 60-l tanks connected to the same recirculation system, resulting in common water supply. All experiments were performed with the approval of the animal experiment committee of Wageningen University (DEC number 2015098).
Collection of Intestinal Contents for 16s rRNA Profiling
Nine-month-old fish (20–40 g, n = 5) were starved for 24 h and subsequently euthanised in 0.3 g/l tricaine methane sulfonate (TMS) (Crescent Research Chemicals, Phoenix, USA) in aquarium water buffered with 0.6 g/l sodium bicarbonate and bled via the caudal vein. The third segment of the intestine was isolated, as this segment shows a stable microbiota composition (39), and intestinal content of these samples was collected by scraping. Samples were snap-frozen in liquid nitrogen and stored at -80°C.
Total Bacterial RNA Isolation
Prior to isolation of RNA from intestinal content, all samples were weighed. Total RNA from intestinal content was isolated using the RNeasy PowerMicrobiome Kit (Qiagen, 26000-50), following the manufacturer’s instructions. Agarose gel electrophoresis and a ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) were used to control RNA yield and quality (260/280: >2.0, 260/230: >2.0). RNA samples were stored at -80°C until further use.
16S rRNA Profiling
Total community analyses were performed on initially five replicates per treatment similar as described for Perez-Jaramillo et al. (40). Briefly, total community RNA was used for amplification and sequencing of the 16S rRNA, targeting the variable V3–V4 regions resulting in amplicons of approximately ~460 bp. Illumina 16S rRNA gene amplicon libraries were generated and sequenced at BaseClear (Leiden, the Netherlands). In short, barcoded amplicons from the V3–V4 region of 16S rRNA genes were generated using a 2-step approach. Ten ng RNA was used as template for the One-Step RT-PCR Kit (Qiagen®) employed according to the manufacturer’s instructions with a total volume of 50 μl using the 341F (5′-CCTACGGGNGGCWGCAG-3′) and the 785R (5′-GACTACHVGGGTATCTAATCC-3′) primers appended with Illumina adaptor sequences. Control PCR reactions were performed alongside each separate amplification without addition of template. PCR products were purified using AMPure XP beads according to the manufacturer’s instructions, and the sizes of the PCR products were checked on a Fragment Analyzer (Advanced Analytical) and quantified by fluorometric analysis. Purified PCR products were used for the 2nd PCR in combination with sample-specific barcoded primers (Nextera XT Index Kit, Illumina). Subsequently, PCR products were purified, checked on a Fragment Analyzer (Advanced Analytical), and quantified, followed by multiplexing, clustering, and sequencing on an Illumina MiSeq with the paired-end (2×) 300-bp protocol and indexing. For each independent sample, at least 25,000 raw reads were sequenced. The sequencing run was analysed with the Illumina CASAVA pipeline (v1.8.3) with demultiplexing based on sample-specific barcodes. The raw sequencing data produced were processed removing the sequence reads of too low quality (based on “failed-chastity<=1”, using a chastity threshold of 0.6, on the first 25 cycles) and discarding reads containing adaptor sequences or PhiX control with an in-house filtering protocol. A quality assessment on the remaining reads was performed using the FASTQC quality control tool version 0.10.0.
Bacterial Community Analyses
The RDP extension to PANDASeq (41) named Assembler (42) was used to merge paired-end reads with a minimum overlap of 10 bp and at least a PHRED score of 25. Primer sequences were removed from the paired sample FASTQ files using FLEXBAR version 2.5 (43). Sequences were converted to FASTA format and concatenated into a single file. All reads were clustered at 97% similarity into OTUs using the UPARSE strategy by dereplication, sorting by abundance with at least two sequences and clustering using the UCLUST smallmem algorithm (44). These steps were performed with VSEARCH version 1.0.10 (45), which is an open-source and 64-bit multithreaded compatible alternative to USEARCH. Next, chimeric sequences were detected using the UCHIME algorithm (46) implemented in VSEARCH. All reads before the dereplication step were mapped to OTUs using the usearch_global method implemented in VSEARCH to create an OTU table and converted to BIOM-Format 1.3.1 (47). Finally, taxonomic information for each OTU was added to the BIOM file by using the RDP Classifier version 2.10 (48). Representative OTU sequences were assigned to a taxonomic classification via BLAST against the Silva database (version 128). All steps were implemented in a Snakemake workflow (49), which is available on GitHub1. For downstream analysis, we took the obtained OTU table and prepared a “filtered table” using QIIME (1.9.1) custom scripts (50). First, we extracted from the OTU table the Bacteria domain using the command split_otu_table_by_taxonomy.py. Next, we discarded singletons, doubletons, Chloroplast, and Mitochondria sequences using the command filter_otus_from_otu_table.py.
In Vitro Gas Production Technique
Collection of Intestinal Contents for In Vitro Fermentation
Naive carp were euthanised with 0.3 g/l TMS in aquarium water buffered with 0.6 g/l sodium bicarbonate and bled via the caudal vein. Subsequently, fish were put on ice and the intestine, without bulbus, was removed. The content of the intestine was collected in pre-weighed plastic tubes that were filled with CO2 to ensure anaerobic conditions. The intestinal content of ten fish was pooled, and five independent pools were used for subsequent in vitro fermentation.
Substrates for In Vitro Fermentation
Four different substrates were used for in vitro gas production analyses: glucose, (D-glucose monohydrate; Merck, Darmstadt, Germany), PBS (cell culture grade, Lonza), curdlan (β-(1,3)-glucan extracted from Alcaligenes faecalis, Sigma-Aldrich), and MacroGard® [a cell wall preparation of S. cerevisiae comprising at least 60% β-(1,3/1,6)-glucan (Zilor, São Paulo, Brazil)]. Glucose was included as a readily fermentable substrate for comparative purposes (25).
Inoculum Preparation and Measurement of Cumulative Gas Production
Pooled intestinal content was immediately transported to the laboratory after collection where it was weighed and transferred to a beaker. The contents of the beaker were stirred and flushed with a constant gentle stream of CO2. Pre-warmed (25°C) anaerobic, sterile saline (9 g/l NaCl) was added in a ratio of 1:5 (W/V) to ensure sufficient amount of inoculate. The diluted material was homogenised using a vortex mixer and strained through a double layer of cheese cloth with 16 threads per cm in both directions. From the resulting inoculate, 5 ml was then dispensed into a pre-warmed 300-ml fermentation bottle, containing 0.5 g substrate and 82 ml of medium. Three replicate bottles for each substrate per inoculate were used. The medium consisted of a basal solution containing micronutrients required by the microbial population for growth, a bicarbonate solution, a vitamin/phosphate solution, and a reducing agent. The composition of the medium is described in detail by Williams et al. (51). Subsequently, bottles were immediately attached to an automated gas production system (52). Within this system, pressure sensors detect a fixed pressure, after which a computer software program allows opening of a valve to release gas, the time at which this occurred was recorded. Bottles were incubated for 168 h at 25°C, equal to the body temperature of carp.
Curve Fitting and Statistics of Cumulative Gas Production
A monophasic model as described by Groot et al. (53), was fitted to the profile of the cumulative gas production of each fermentation bottle according to the equation G = A/(1 + (C/t)B), where G is the total millilitre gas produced per gram organic matter (OM) at time t; A is the asymptotic gas production (mL/g OM); B is the switching characteristic of the curve; C is the half time (time at which half of the asymptote is reached); and t is the time (h).
The maximum rate of gas production (Rmax) and the time at which it occurs (Tmax) were calculated according to (54) as
For each bottle, curve fitting was done using the non-linear least-square regression procedure NLIN (SAS Inst. Inc., Cary, NC, USA). One-way analysis of variance using the GLM procedure of SAS (SAS Inst. Inc.) was used to test the effect of substrate on gas production parameters and fermentation end products. The average of replicate bottles per substrate per inoculum was considered as the experimental unit. The effect of replicate bottles was tested separately and was not significant for any of the parameters. It was therefore excluded from the model and thus became part of the error term.
Sampling and Analyses of Fermentation Liquid
At the end of the incubation period, pH of the fermentation fluids was recorded and samples were collected for analysis of ammonia (NH3), SCFAs, and lactate. Analyses of NH3 and SCFA were determined as described previously (55), using gas chromatography for SCFA analysis (GC; Fisons HRGC Mega 2, CE Instruments, Milan, Italy). Lactate was measured using the Lactate Colorimetric Assay Kit II (K627, BioVision) according to the manufacturer’s instructions, including the optional filtration step with Amicon 10K spin columns (Z677108-96EA, Sigma-Aldrich, centrifugation for 20 min at 21.100× g), as described previously (56).
Head Kidney Leukocyte Isolation
Carp were euthanised with 0.3 g/l TMS (Crescent Research Chemicals) in aquarium water buffered with 0.6 g/l sodium bicarbonate and bled using vacuettes (BD Vacutainer SST with 21-G needles, Becton Dickinson) via the caudal vein. The head kidney was isolated, and total head kidney leukocytes (HKLs) were separated on a 1.02–1.08-g/ml Percoll (GE Healthcare, Thermo Fisher Scientific) density gradient, as previously described (13).
Nitrogen Radical Production
Production of NO was determined as nitrite accumulation using the Griess reaction, as previously described (57). HKLs were seeded at a density of 1 × 106 per well in 96-well culture plates (CORN3596; Corning) and stimulated with one of the following: RPMI (control) or LPS (L2880, 30 µg/ml, Sigma-Aldrich) in the presence or absence of different concentrations of sodium acetate (0.5–25 mM, S2889-250G, Sigma-Aldrich), sodium butyrate (0.05–25 mM, 303410-100G, Sigma-Aldrich), or sodium propionate (0.05–25 mM, P1880-100G, Sigma-Aldrich) or in the presence or absence of a mix of acetate (62.5%), butyrate (10%), or propionate (27.5%) in the proportion of produced SCFAs after in vitro fermentation of MacroGard® at 0.1–25 mM. After 96 h at 27°C in the presence of 5% CO2, nitrite production was measured at OD540, using a FilterMax F5 Multi-Mode Microplate Reader and quantified using a sodium nitrite (NaNO2) standard curve.
Reactive Oxygen Species (ROS) Production
Production of reactive oxygen species (ROS) was determined by a real-time luminol-based ECL assay, as previously described (58). HKLs were seeded at a density of 1 × 106 per well in white 96-well plates (CLS3912; Corning) and stimulated with one of the following: RPMI (control), zymosan (tlrl-zyd, 50 µg/ml, InvivoGen) in the presence or absence of different concentrations of sodium acetate (0.5 and 2.5 mM, S2889-250G, Sigma-Aldrich), sodium butyrate (0.1–0.5 mM, 303410-100G, Sigma-Aldrich), or sodium-propionate (0.1–0.5 mM, P1880-100G, Sigma-Aldrich) or in the presence or absence of a mix of acetate (62.5%), butyrate (10%), or propionate (27.5%) in the proportion of produced SCFAs after in vitro fermentation of MacroGard® at 0.5–5 mM. Stimulation with PMA (P8139, 1 mg/ml, stimulated control; Sigma-Aldrich) was used as a technical control for the assay (data not shown). SCFA concentrations were based on the outcomes of the nitrogen radical production analysis, and thus fewer SCFA concentrations were included the ROS analysis. Chemiluminescence emission was measured in real time (every 2 min for 120 min) with a FilterMax F5 Multi-Mode Microplate Reader at 27°C and expressed as area under the curve, as previously described (56). Fold changes were calculated as the area under the curve of stimulated HKLs relative to unstimulated HKLs (RPMI without zymosan).
Head Kidney Leukocyte Stimulation for Gene Expression Analysis
For gene expression analysis, HKLs were seeded at a density of 4.5 × 106 per well in a 24-well plates (CORN3524; Corning) and stimulated with RPMI and LPS (L2880, 30 µg/ml, Sigma-Aldrich) in the presence or absence of a mix of acetate (62.5%), butyrate (10%), or propionate (27.5%) in the proportion of produced SCFAs after in vitro fermentation of MacroGard® at 2.5 mM or of sodium acetate (1.55 mM, S2889-250G, Sigma-Aldrich), sodium butyrate (0.25 mM, 303410-100G, Sigma-Aldrich), or sodium propionate (0.765 mM, P1880-100G, Sigma-Aldrich); at the relative concentration, each was included in the 2.5-mM SCFA mix. Each stimulation was performed in technical triplicate and in n = 5 independent experiments. At 3 and 6 h post stimulation, 13.5 × 106 cells were lysed in the lysis buffer from the RNeasy Kit (RLT buffer) and stored at -80°C until RNA isolation.
Oral Gavage and Tissue Sampling
Carp of 9 months (20–40 g, n = 20 per group) were starved overnight and anaesthetised with one-third the killing dose of TMS (i.e., 0.10 g/l TMS in aquarium water buffered with 0.2 g/l sodium bicarbonate) to allow manipulation without killing. Anaesthetised fish received an oral gavage with 100 µl of PBS (n = 20, Lonza, LO BE17-516F) or MacroGard® dissolved in PBS (n = 20, 10 mg/ml, 1 mg per fish) using a 200-μl pipette. Fish feeding (twice-daily) was resumed at 12 h post gavage. At 3, 7, 11, and 14 days post gavage, before morning feeding, n = 5 fish were euthanised in 0.3 g/l TMS in aquarium water buffered with 0.6 g/l sodium bicarbonate and bled via the caudal vein (data for 3, 11, and 14 days are not shown). The third segment of the intestine was isolated, and intestinal content of the third segment was collected by scraping, snap-frozen in liquid nitrogen, and stored at -80°C for 16S rRNA sequencing. A tissue sample of the third segment of the intestine was snap-frozen in liquid nitrogen and stored at -80°C for gene expression analysis.
Total RNA Isolation and cDNA Synthesis
Total RNA from isolated tissue samples (25–50 mg per sample) and cells (13.5 × 106 cells per sample) was isolated using the RNeasy Mini Kit (Qiagen, 74106), including on-column DNase treatment, according to the manufacturer’s instructions, and stored at −80°C. Prior to cDNA synthesis, total RNA was treated with DNase I, Amplification Grade (Invitrogen), and cDNA was synthesised using random primers (300 ng) and Superscript III First-Strand Synthesis for RT-PCR (Invitrogen). cDNA samples were diluted 25× in nuclease-free water prior to real-time quantitative PCR (RT-qPCR) analysis.
Gene Expression Analysis
Gene expression was measured with RT-qPCR using ABsolute qPCR SYBR Green Mix (Thermo Scientific) in a Rotor-Gene Q (Qiagen) as previously described (59). Fluorescence data were analysed using Rotor-Gene Analysis software version 1.7. The relative expression ratio (R) of each sample was calculated according to the Pfaffl method (60) based on the take-off deviation of sample versus each of the PBS controls and normalised relative to the s11 protein of the 40-s subunit as reference gene (see Table 1 for primer information).
Statistical Analyses
Statistical analyses were performed using SPSS (v23.0), and differences were considered significant if p ≤ 0.05. Data presented as fold change (gene expression analyses) were first transformed using natural log transformation [Y = LN(Y)] before analysis. All data were tested for Gaussian distribution using the Shapiro–Wilk test.
Differences in cumulative gas production were analysed with two-way ANOVA followed by the Bonferroni post hoc test. Differences in pH of the fermentation liquid after in vitro fermentation were assessed by Welch’s ANOVA followed by the Games–Howell test, as the data did not have equal variances. Differences in NH3 and short-chain fatty acid profiles in the fermentation liquid after in vitro fermentation were assessed by one-way ANOVA followed by Tukey post hoc test. Data for comparisons of NO production, ROS production, and gene expression in vitro were analysed using repeated measures and sometimes had some data points missing; therefore, analyses were performed with a linear mixed model, followed by a Bonferroni post hoc test. Comparison of gene expression in vivo was assessed by one-way ANOVA.
Results
Characterization of Intestinal Microbial Composition Implies β-Glucan Fermentation Capacity
Analysis of 16S ribosomal RNA rather than DNA provided insight into the active intestinal microbial communities. Identification of bacterial microbiota up to family level allowed us to investigate the presence of bacteria with theoretical fermenting capacities. The intestinal content of the third segment of the intestine from n = 5 individual fish was analysed by 16S rRNA sequencing, revealing a total of n = 55 active operational taxonomic units (OTUs) in the intestinal content of unhandled carp.
By far, the most represented phyla in the carp intestinal microbiota were Fusobacteria (71% ± 5.8% (mean ± SD, n = 5) of total reads), whereas Bacteroidetes (21% ± 5.3%), Proteobacteria (5% ± 0.5%), and Firmicutes (2% ± 0.5%) were considerably less frequent (Figure 1A). The majority of Proteobacteria belonged to the order of Gammaproteobacteria (95%), while the rest belonged to Betaproteobacteria (4%). Looking at the distribution per family in the active microbiota, the most abundant family were Fusobacteriaceae (71% ± 5.8% [mean ± SD, n = 5)], followed by Bacteroidaceae (16% ± 4%), Porphyromonadaceae (5% ± 2%), Vibrionaceae (3% ± 0.75%), Aeromonadaceae (1.5% ± 0.4%), and Erysipelotrichaceae (1.4% ± 0.1%) (Figure 1B). Finally, looking at the putative genus level, it becomes clear that all Fusobacteriaceae are member of the Cetobacterium genus, as 71% ± 5.8% (1 OTU [mean ± SD, n = 5)] of the total reads belong to this genus. Following Cetobacterium genus, the most abundant genera based on 16S RNA sequencing are Bacteroides (16% ± 4.1%, 2 OTUs), Vibrio (3% ± 0.75%, 1 OTU), Aeromonas (1.5% ± 0.4%, 1 OTU), and two different uncultured genera of the families Porphyromonadaceae (4.5% ± 4%, 3 OTUs) and Erysipelotrichaceae (1.1% ± 0.1%, 1 OTU). Although the sequencing effort could not identify bacteria at the species level, the abundant presence of the Bacteroides genus well known to express β-glucan-degrading genes (61) suggested the ability of carp microbiota to ferment β-glucans.
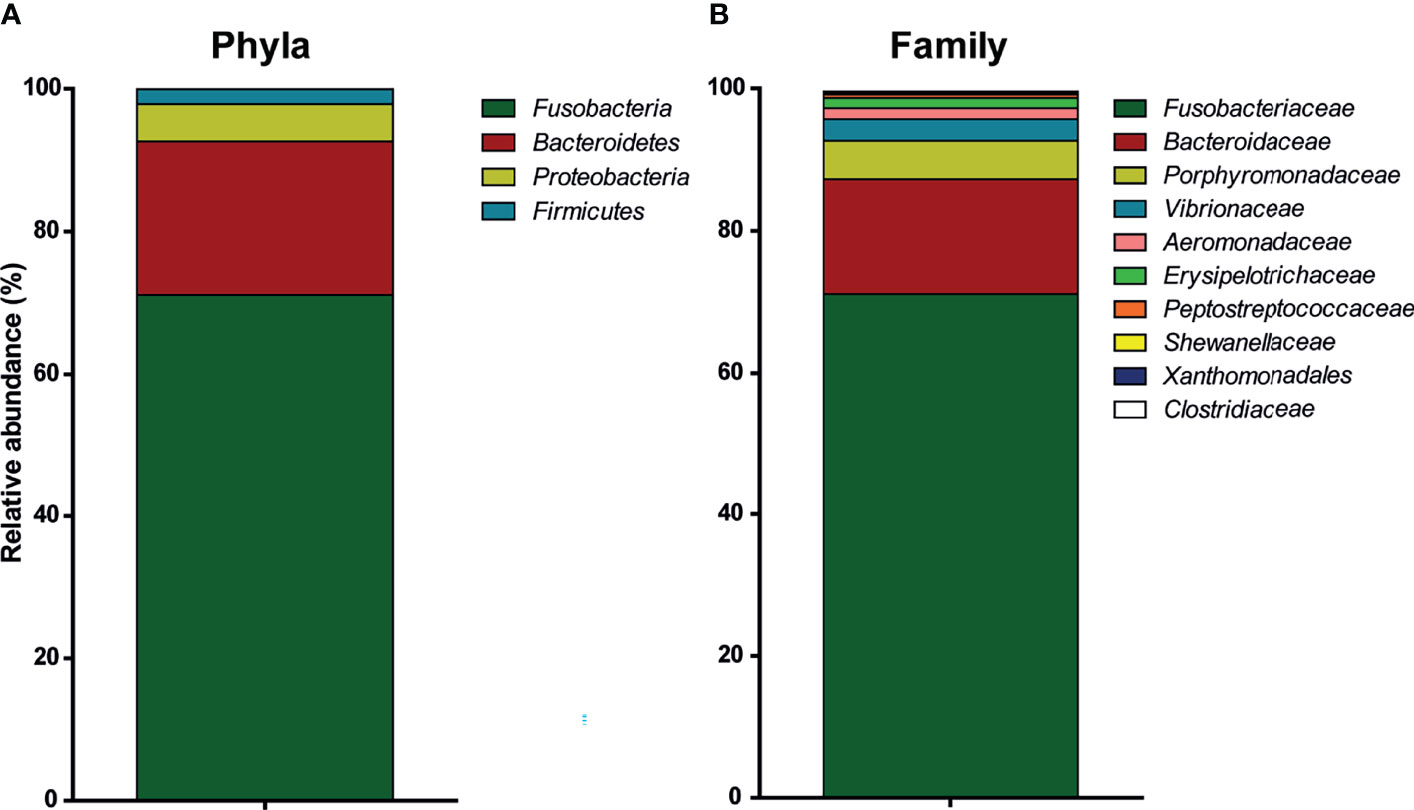
Figure 1 Relative abundance of active bacteria in the intestinal microbiota of common carp. Characterization with 16S rRNA sequencing of the intestinal microbiota of naive, unhandled common carp [(bars indicate relative abundance (mean, n = 5)]. (A) Relative abundance of phyla. (B) Relative abundance of family. Xanthomonadales refers to family Xanthomonadales Incertae sedis.
In Vitro Batch Analysis Confirms Fermentation of β-Glucans
In order to further investigate the ability of the naive intestinal microbiota of carp to ferment β-glucans, in vitro fermentation was performed (Figure 2A). Through an in vitro batch culture system, fermentation of different substrates could be analysed. As a measure for fermentation kinetics, cumulative gas production was measured over time. As a negative control, minor cumulative gas production was measured in the PBS group, similar to gas production in the negative control without substrate (data not shown). As a positive control, fermentation of the readily digestible monosaccharide glucose was measured, resulting in the highest cumulative gas production. Two different β-glucan preparations were studied: curdlan, a high molecular weight linear polymer consisting of β-1-3-linked glucose residues from Alcaligenes faecalis, and MacroGard®, a branched polymer β-(1,3/1,6)-glucose feed additive. In comparison, fermentation of curdlan resulted in the most continuous and highest cumulative gas production, while fermentation of MacroGard® showed an intermediate plateau around 24–36 h. Gas production differed significantly between MacroGard® and curdlan from 44 to 86 h after start of the in vitro fermentation (as assessed with a two-way ANOVA followed by Bonferroni post hoc, p ≤ 0.05). No matter the difference in kinetics of gas production between curdlan and MacroGard®, naive carp intestinal microbes were able to ferment β-glucans.
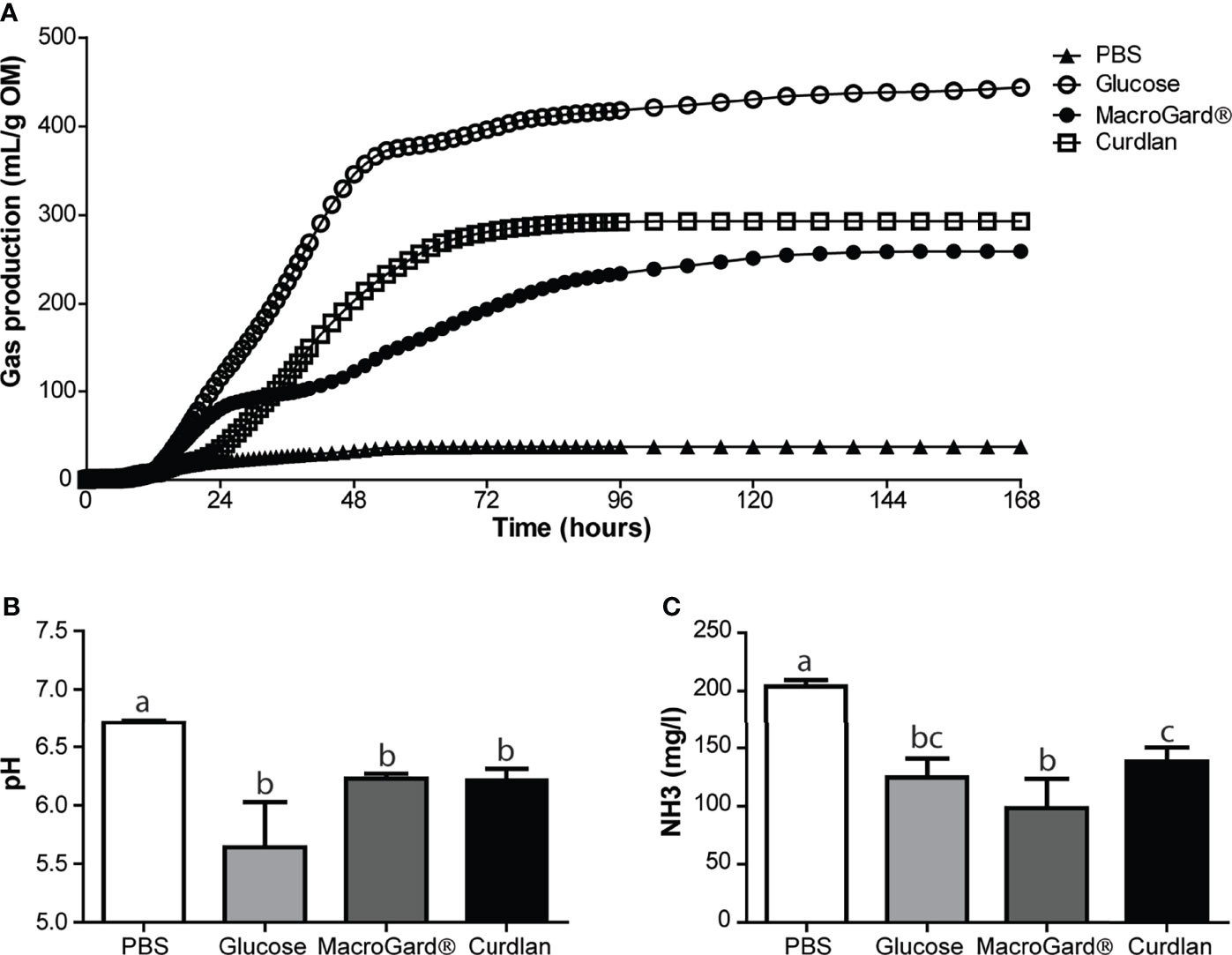
Figure 2 Carp naive microbiota is capable of in vitro fermentation of different β-glucans. (A) Cumulative gas production over 168 h as a result of in vitro fermentation of PBS, glucose, MacroGard® and curdlan by carp intestinal microbiota. Data shown as mean of n = 5 independent intestinal pools. Significant difference between cumulative gas production between MacroGard® and curdlan from 44–86 h [two-way ANOVA followed by Bonferroni post hoc (not shown)]. (B) pH of fermentation fluids after 168 h of in vitro fermentation (mean ± SD, n = 5). Significant differences between groups were assessed by Welch’s ANOVA followed by Games–Howell test. (C) Ammonia (NH3) accumulation in fermentation fluids after 168 h of in vitro fermentation (mean ± SD, n = 5). Significant differences between groups were assessed by one-way ANOVA followed by Tukey test. Groups with different letters are statistically different from one another.
Finally, analysis of the fermentation liquid after in vitro fermentation revealed differences between substrates in pH and NH3, whereas these parameters did not differ at start (data not shown). In comparison, a (non-significant) trend toward lower pH was observed in the glucose group, compared to the two β-glucan-treated groups. No difference in pH was observed between MacroGard® and curdlan treatment (Figure 2B). Ammonia production was significantly higher in the PBS group compared to all other groups (Figure 2C). Interestingly, NH3 production was different between curdlan and MacroGard®, providing further evidence for differences in fermentation between the two β-glucans.
In Vitro Fermentation of β-Glucans Results in Significantly Increased SCFA Levels
Results from the in vitro batch culture analysis provided evidence for the ability of intestinal microbiota of carp to ferment β-glucans, at least to a certain degree. In general, metabolites produced by microbiota during fermentation such as SCFA are often considered necessary components for immune homeostasis (62). We therefore measured the accumulation of SCFA metabolites in the in vitro fermentation liquid, produced during β-glucan fermentation. Significant differences in SCFA profiles between the readily fermentable substrate glucose and the two different β-glucan preparations could be observed (Figures 3A–E). Overall, a clear production of acetate, butyrate, and propionate was observed after fermenting either curdlan or MacroGard® in vitro. Comparing the two different β-glucan preparations showed no differences in acetate production but revealed an interesting inverted trend for butyrate and propionate levels. After fermentation of curdlan, higher butyrate concentrations were measured, while after fermentation of MacroGard® higher propionate concentrations were measured (Figures 3B, C). Profiles of other SCFA; formic, isobutyric, valeric, and iso-valeric acid were also measured, but concentrations were considered negligible (<0.1 mM, data not shown) and therefore excluded during the follow-up experiments. In both β-glucan groups, total SCFA levels were higher than in the glucose group; however, no differences were observed between both β-glucan groups (Figure 3E). Overall, differential production of SCFAs could be observed in both β-glucan groups, with fermentation of MacroGard® specifically resulting in an increase in propionate. With the practical and commercial applicability of MacroGard® being higher than of curdlan, our follow-up studies focused on the effects of fermentation of MacroGard®.
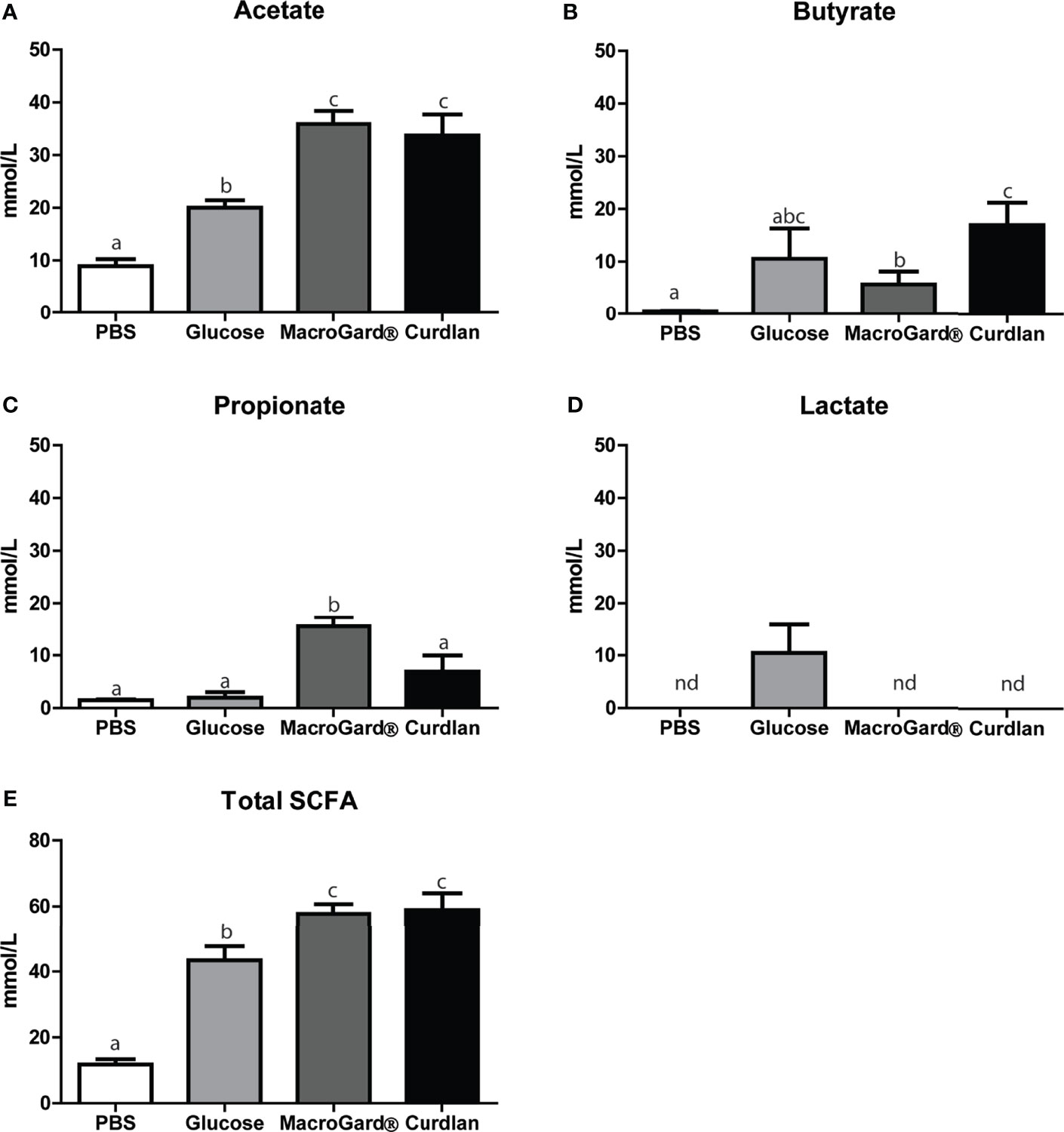
Figure 3 Differential SCFA profiles after in vitro fermentation of β-glucan. SCFA levels in the fermentation liquid following 168 h in vitro fermentation of PBS, glucose, MacroGard®, or curdlan. (A–E) Acetate, butyrate, propionate, and lactate, or total sum. Total SCFA levels were calculated by adding up all analysed SCFA levels including branched chain short-chain fatty acids. Bars indicate mean ± SD of n = 5 independent intestinal pools. Significant differences between groups were assessed by one-way ANOVA followed by Tukey test (A, D, E) or by Welch’s ANOVA followed by the Games–Howell test (B, C). Groups with different letters are statistically different from one another. (D) The annotation “nd” indicates that lactate was not detected in these samples.
Presence of SCFAs Modulates Radical Production by Head Kidney Leukocytes In Vitro
In order to investigate the potential immunoregulatory effects of SCFAs, head kidney leukocytes (HKLs) were stimulated with LPS as a well-known stimulator of pro-inflammatory responses and stimulator of nitric oxide (NO) production or with zymosan as a well-known stimulator of pro-inflammatory responses and stimulator of ROS production in common carp, while different concentrations of SCFAs were added. Analysis of NO production at 96 h post stimulation revealed differential modulating potencies of the various SCFAs (Figure 4). Clear dose-dependent modulating effects could be observed for the three SCFAs investigated, but only in the presence of LPS. Butyrate was the most potent SCFA, followed by propionate and lastly acetate.
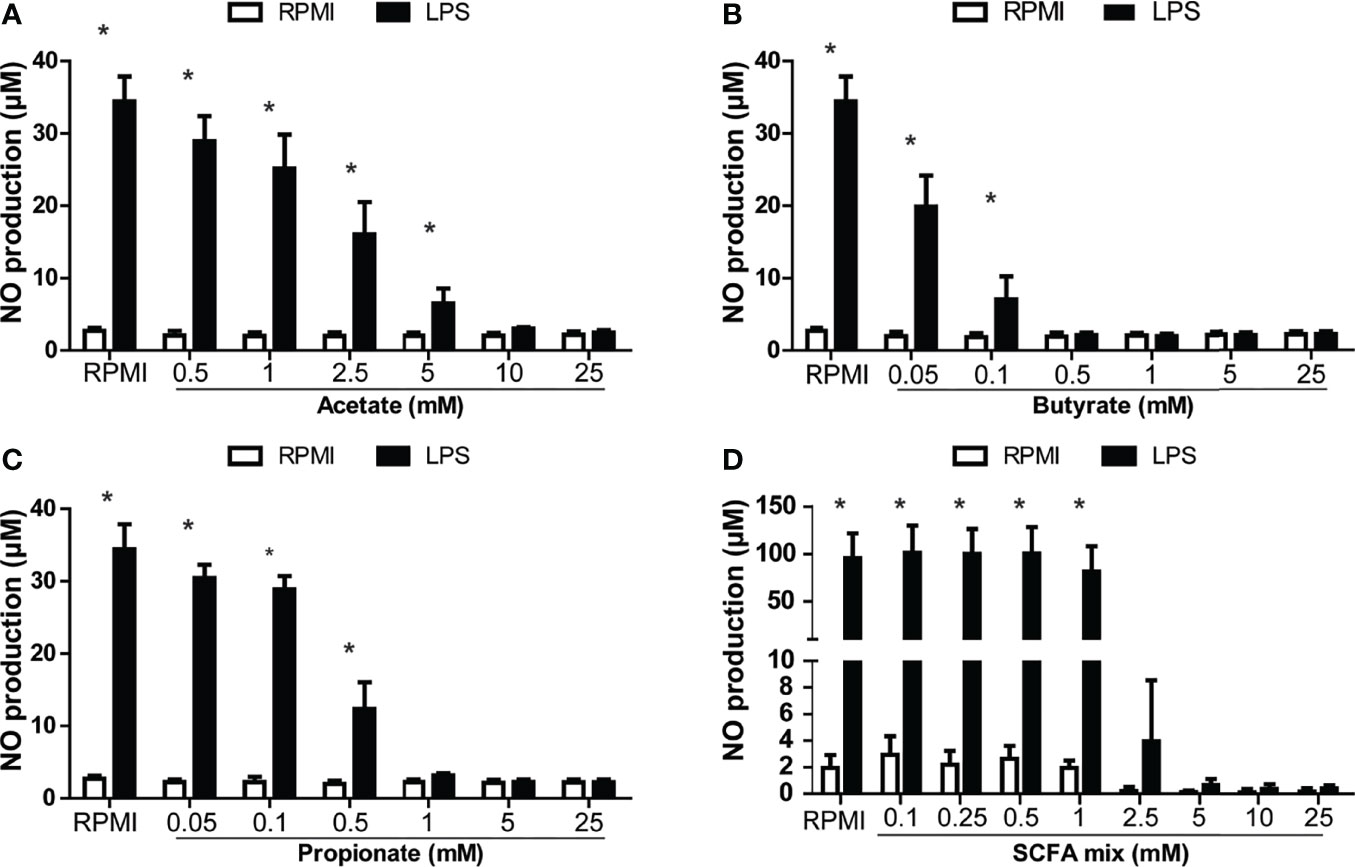
Figure 4 SCFA reduced LPS-induced production of nitric oxide (NO) in head kidney leukocytes (HKL). Cells were stimulated with medium control (RPMI) or LPS (30 µg/ml) for 96 h in the presence or absence of the SCFAs, acetate, butyrate, or propionate or a mix in the relative proportion produced by in vitro fermenting MacroGard® (62.5%/10%/27.5%, acetate/butyrate/propionate). (A) NO production of LPS-stimulated HKL in the presence of acetate. (B) NO production of LPS-stimulated HKL in the presence of butyrate. (C) NO production of LPS-stimulated HKL in the presence of propionate. (A–C) Bars indicate mean ± SD of n = 3 independent experiments. (D) NO production of LPS-stimulated HKL in the presence of SCFA mix, indicating that the concentration is determined by the total amount of acetate, butyrate, and propionate (25 mM (15.625 mM acetate/2.5 mM butyrate/6.875 mM propionate). Bars indicate mean ± SD of n = 5 independent experiments. Note the difference in scale for the Y-axis in (D). Asterisk (*) indicates significant difference between NO production induced in the presence (solid black bars) or absence of LPS (open white bars) assessed by the linear mixed model followed by the Bonferroni post hoc test.
Furthermore, adding the SCFAs mixed in the same ratio as measured after in vitro fermentation of MacroGard® (see Figure 3) showed similar reductions in in vitro induced NO production in HKLs (Figure 4D). Finally, titrating down this SCFA mixture revealed a significant reduction of LPS-induced NO production even at a concentration as low as 2.5 mM.
Interestingly, when analysing the production of ROS, SCFAs appeared to increase the ROS production in a dose-dependent manner, although this trend was only significant for acetate and propionate (Figure 5). Furthermore, adding the SCFAs mixed in the same ratio as measured after in vitro fermentation of MacroGard® (see Figure 3) showed similar modulations of in vitro production of ROS by HKLs. Both with and without the presence of a strong ROS inducer (zymosan), a clear increase could be observed in the ROS production of HKLs.
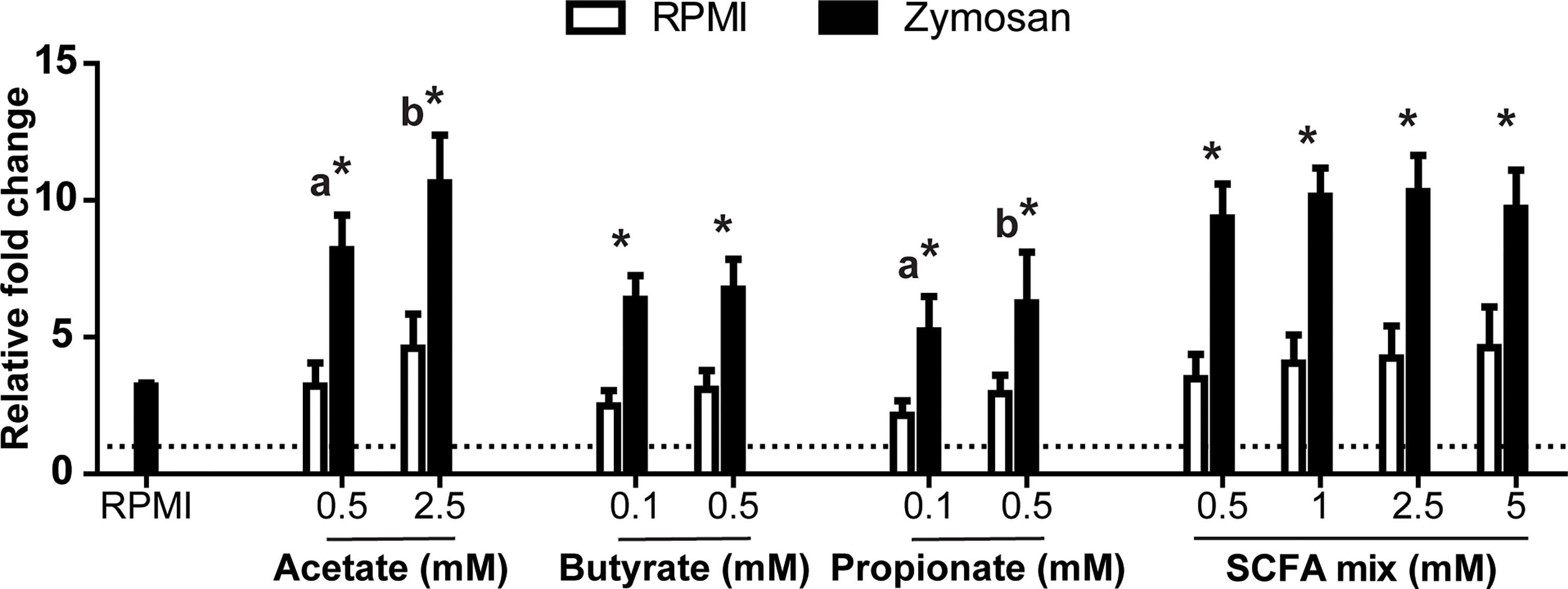
Figure 5 SCFA increased oxidative burst in head kidney leukocytes (HKL). Total production of reactive oxygen species (ROS) relative to unstimulated HKL (dotted line). Cells were exposed to different concentrations of SCFAs in the absence (medium control (RPMI), open bars) or presence (black bars) of zymosan (50 µg/ml). Immediately following exposure, ROS production was measured real-time over a period of 2 h. Bars indicate mean ± SD of n = 5 experiments performed independently. Asterisk (*) indicates significant difference between ROS production induced in the presence (solid black bars) or absence of Zymosan (open white bars) assessed by the linear mixed model followed by the Bonferroni post hoc test. Groups with different letters are statistically different from one another, within one treatment (acetate, butyrate, propionate or SCFA mix) where “a” indicates the lowest value.
Presence of SCFAs Modulates Cytokine and SCFA Receptor Gene Expression In Vitro
Studying the gene expression can provide preliminary insights in the effects of SCFAs on inflammation, via gpr40L SCFA receptors. To this end, HKLs were stimulated in vitro with LPS, a mix of SCFAs in the same ratio as measured after in vitro fermentation of MacroGard®, both LPS and the mix, or with different SCFAs (acetate, butyrate, or propionate) separately. Subsequently, at 3 and 6 h post stimulation, cytokine gene expression (Figure 6) and gpr40L (Figure 7) gene expression were studied.
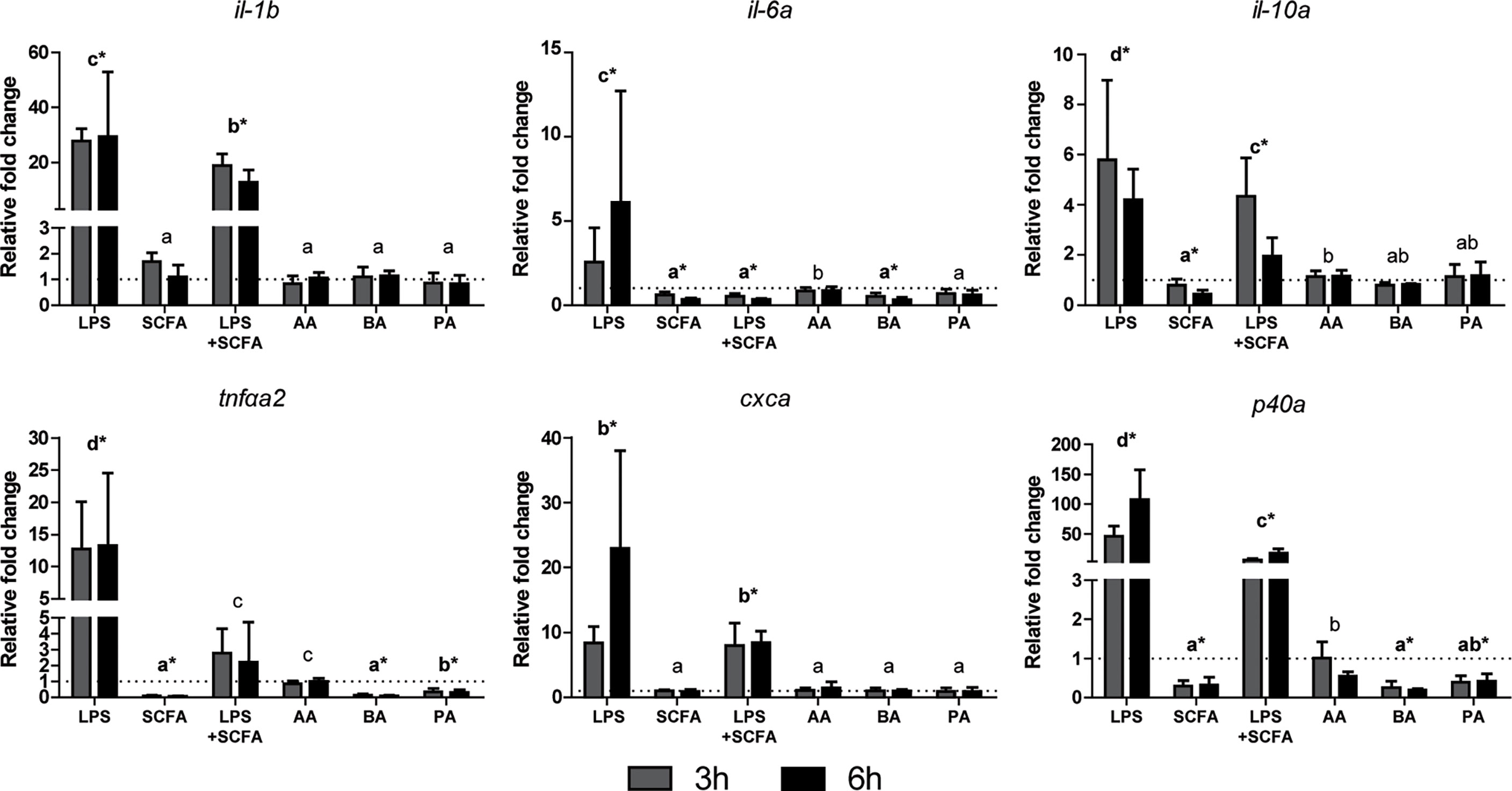
Figure 6 SCFAs reduce the expression of several cytokines ex vivo stimulated head kidney leukocytes. Gene expression after 3 (grey bars) and 6 (closed bars) h of stimulation with LPS (30 µg/ml), SCFA mix (2.5 mM), LPS, and SCFA mix or the three SCFAs at the concentration they were included in the mix; acetate 1.55 mM (AA), butyrate 0.25 mM (BA), and propionate 0.765 mM (PA), compared to unstimulated HKLs (dotted line). Bars indicate mean ± SD of n = 5 experiments performed independently. Significant differences between groups were assessed by the linear mixed model followed by the Bonferroni post hoc test. Groups with different letters are statistically different from one another where “a” indicates the lowest value, groups indicated with asterisk (*) are significantly different from unstimulated HKLs.
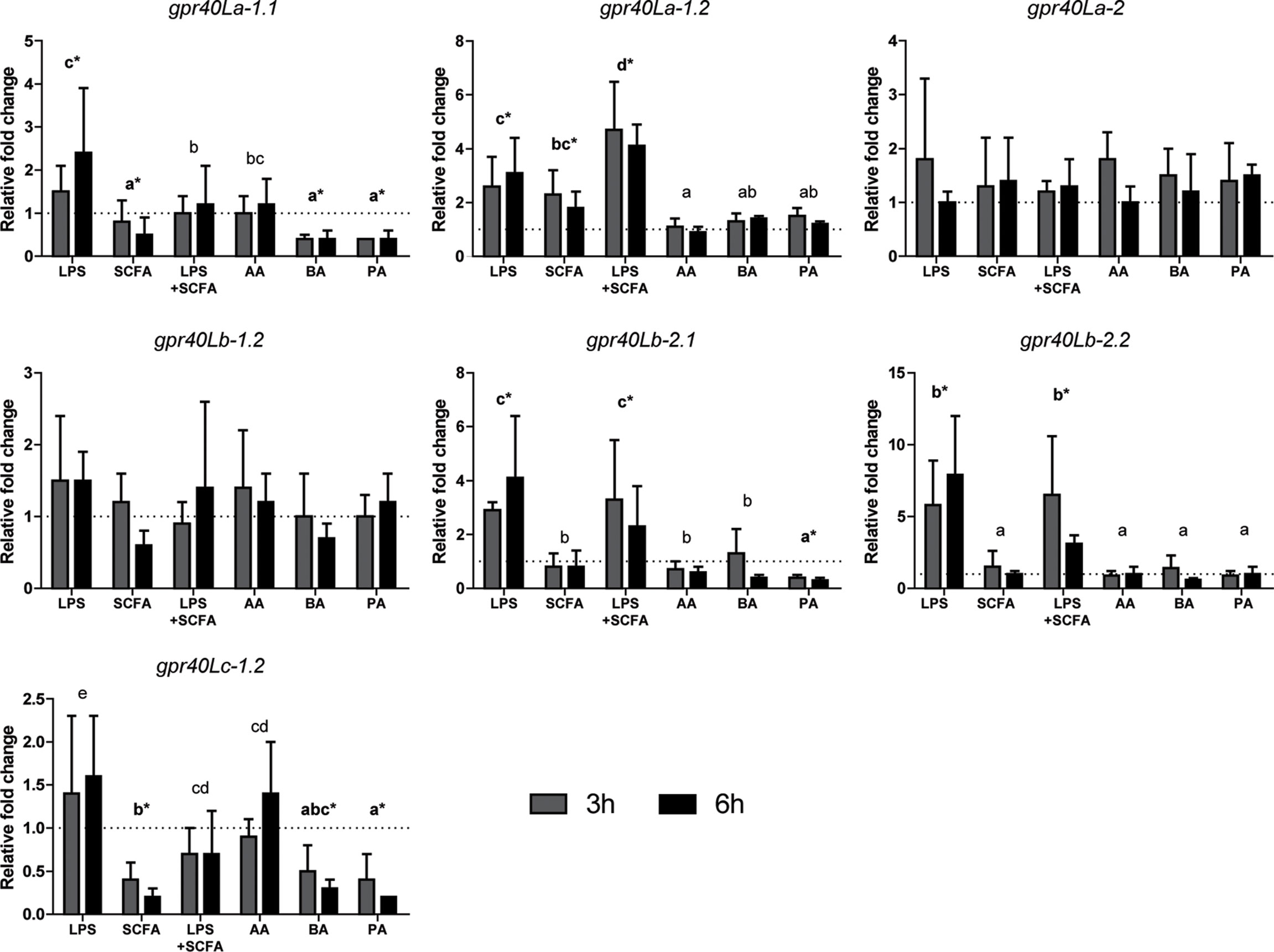
Figure 7 SCFAs regulate the expression of several gpr40L receptors ex vivo stimulated head kidney leukocytes. Gene expression after 3 (grey bars) and 6 (closed bars) h of stimulation with LPS (30 µg/ml), SCFA mix (2.5 mM), LPS, and SCFA mix or the three SCFAs at the concentration they were included in the mix; acetate 1.55 mM (AA), butyrate 0.25 mM (BA), and propionate 0.765 mM (PA), compared to unstimulated HKLs (dotted line). Bars indicate mean ± SD of n = 5 experiments performed independently. Significant differences between groups were assessed by the linear mixed model followed by the Bonferroni post hoc test. Groups with different letters are statistically different from one another, where “a” indicates the lowest value; groups indicated with asterisk (*) are significantly different from unstimulated HKLs.
Addition of SCFAs generally resulted in downregulation of cytokine gene expression (Figure 6), either pro-inflammatory (il-1b, il-6, tnfα, cxca, p40) or anti-inflammatory (il-10), especially when first induced by presence of LPS. In the absence of LPS, addition of SCFAs resulted in a downregulation of cytokine gene expression of il-6, tnfa, and p40. Separately, SCFAs acetate, butyrate, and propionate showed slightly different levels of regulation, with acetate barely regulating gene expression. In general, addition of SCFAs as a mix showed comparable regulation as addition of SCFAs separately. Gene duplicates were also studied (il-6b, il-10b, tnfαa1, tnfαb1, tnfαb2, cxcb, p40b, and p40c, data not shown) and were regulated similarly to the genes presented in Figure 6).
Addition of SCFAs resulted in a differential regulation of gene expression of different grp40L receptors (Figure 7). Here, we studied gene expression of ten gpr40L genes and found three (gpr40Lb-1.1, gpr40Lc-1.1, and gpr40Lc-2) not expressed in HKLs. For the seven expressed gpr40L receptors, four were upregulated by stimulation with LPS (grp40La-1.1, gpr40La-1.2, gpr40Lb-2.1, gpr40Lb-2.2).
Regulation of the gpr40L expression in HKL by SCFAs was only stimulatory for gpr40La-1.2 while inhibitory for gpr40La-1.1, gpr40Lb-2.1, and gpr40Lc-1.2. Again, of the different SCFAs analysed in this study, addition of acetate barely regulated gene expression.
Oral Gavage With β-Glucans Modulates SCFA Receptor and Cytokine Gene Expression In Vivo
To follow up on the above-described in vitro modulating effects of SCFAs, we subsequently analysed the in vivo gene expression locally in the posterior intestine in response to a single oral gavage with β-glucans (MacroGard®). Based on the in vitro fermentation results (see Figures 2, 3) and the assumption that also in vivo the active naive microbiota of common carp can ferment β-glucans and lead to the production of specific SCFAs, we measured whether the effects of an oral gavage with β-glucans could be measured by a change in gene expression of gpr40L and other genes. Analysis of microbiota composition on day 7 and day 14 showed no significant differences between the control (PBS) and the β-glucan-treated group (data not shown). Gene expression regulation was most clear at 7 days post gavage (Figure 8), while gene expression profiles on 3, 11, and 14 days post gavage did not show distinct trends (data not shown). Similar to the in vitro observations on HKL, we found a significant downregulation in the posterior intestine of cxca, il-1b, il-6, and tnfa expression at 7 days post gavage (Figure 8A). Analysis of the expression response of type I IFN-related genes (tlr3.1, tlr3.2, and mx1-5) revealed a clear downregulation of tlr3.1 and a marginal but significant downregulation of mx3 expression (Figure 8B). Further, both in vitro and in vivo gpr40La-1.1 gene expression was downregulated (Figure 8C), but differently, gpr40La-2 expression was significantly upregulated only in vivo. Of interest, all ten gpr40L receptors were expressed in the posterior intestine.
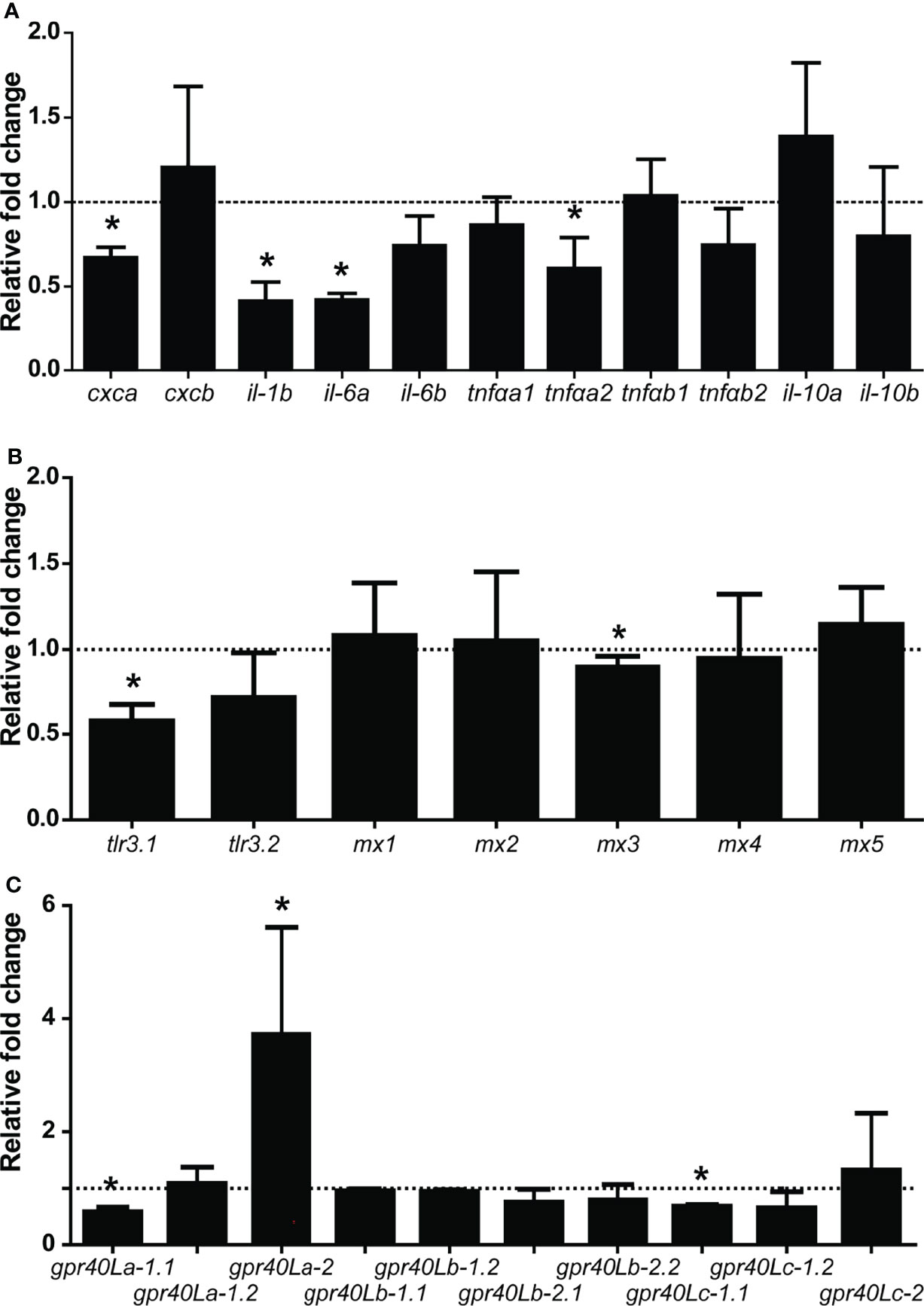
Figure 8 Oral gavage with β-glucans alters cytokine and gpr40L gene expression in the posterior intestine of carp. Gene expression of selected cytokines (A), Type I IFN-related genes (B), and gpr40L genes (C) in the posterior intestine of carp treated with the β-glucan group compared to the control group (dotted line), as measured by RT-qPCR 7 days post-gavage. (A) Significantly lower expressions of cxca, il-1b, il-6a, and tnfαa2. A trend toward a lower expression of il-6b (p = 0.062) and tnfαb2 (p = 0.090) could be observed in the treated group. (B) Significantly lower expressions of tlr3.1 and mx3 (C) Significantly lower expressions of gpr40La-1.1 and gpr40Lc-1.1, but significantly higher expression of gpr40La-2. Bars indicate mean ± SD of n = 5 animals. Asterisk (*) indicates significant difference in expression between the group orally gavaged with PBS and the group orally gavaged with 1 mg MacroGard® as assessed by one-way ANOVA.
Discussion
To investigate whether dietary β-glucans can alter the composition or activity of the intestinal microbiota, we first characterised the active intestinal microbiota of unhandled carp or naive intestinal microbiota. We observed, among others, a prominent presence of the genus Bacteroides, of interest because some species of this genus have been shown able to degrade β-glucans (61). Subsequently, we confirmed that the carp naive intestinal microbiota has β-glucan fermenting ability by using an in vitro batch culture system to test fermentation of two different β-glucan preparations (curdlan and MacroGard®). The in vitro fermentation of these two β-glucan preparations led to the production of several and different short-chain fatty acids (SCFAs), of which an increase in propionate could be specifically linked to the fermentation of MacroGard®. We first confirmed the hypothesis that SCFAs might have immunomodulatory activity by titrating different SCFAs on head kidney leukocytes in vitro, measuring the modulation of induction of NO production and oxidative burst capacity. We also measured the immunomodulatory activity of SCFAs by determining gene expression and could show a clear and mostly downregulation of cytokine expression and putative SCFA receptor gpr40L genes. In vivo, administration of β-glucans by oral gavage with MacroGard® also modulated the gene expression of several cytokines and gpr40L genes in the posterior intestine. Of particular interest for follow-up studies could be the upregulation of one particular gpr40L receptor sub-variant (gpr40La-2). Collectively, our data provide support for the hypotheses that β-glucans can act as substrate for fermentation by intestinal microbiota of common carp, thereby possibly inducing an alteration of local intestinal microbial composition and activity with associated changes in production of SCFA. Altered ratios of SCFA could then be sensed by gpr40L receptor(s) and following downstream signalling, modulating immune responses. Such a line of events could provide a logical explanation for the modulation of immune responses by β-glucans, at least in common carp.
We identified four abundant phyla in the naive intestinal microbiota of common carp (Fusobacteria, Bacteroidetes, Gammaproteobacteria, Firmicutes) using 16S rRNA analysis. Of the phylum Fusobacteria, the genus Cetobacterium clearly dominated in abundance and appears to do so in omnivorous and carnivorous species more so than in herbivorous fishes (63). The genus Bacteroides appeared as the second most abundant and is particularly interesting for its linkage to fermentation of carbohydrates and capability of degrading even highly complex carbohydrates (64–66). Degradation of carbohydrates is achieved with so-called polysaccharide utilization loci (PUL) of which there can be many different ones, each related to the degradation of a specific glycan or group of related glycans (67). At least in humans, members of the Bacteroides found in the gastrointestinal tract express such PUL and have been shown able to degrade several different β-glucans, among which barley-derived β-1,4-glucans (65) and fungus-derived β-1,6-glucans (61). Interestingly, analysis of microbiota composition following oral gavage with β-glucans in vivo revealed no alteration of the composition, arguing that a single oral gavage with β-glucans will not alter the composition, but can alter the metabolism. In contrast to other studies investigating the effects of β-glucans on fish microbiota composition, those studies investigated the effect of constant exposure during dietary supplementation (27–33). Our microbiota sequencing data confirm the presence of bacterial genera commonly associated with β-glucan-fermenting capacity. Future metagenomic approaches are needed to confirm and study in more detail the presence of bacterial species with β-glucan-fermenting ability in the naive microbiota of carp.
Ours is not the first study suggesting that fish microbiota could have the ability to degrade carbohydrates; others already observed fermentation of several carbohydrates and production of SCFAs by intestinal microbes of tilapia, European sea bass, and common carp (23, 25), or showed growth of fish intestinal bacteria on different carbohydrate sources, including β-glucan (22). To verify the β-glucan fermenting capacity of naive carp intestinal microbiota, we performed an in vitro batch culture experiment using two different β-glucans as substrates: MacroGard®, a branched β-1,3/1,6-glucan isolated from the yeast Saccharomyces cerevisiae and commonly used in aquaculture practices, and curdlan, a linear β-1,3-glucan isolated from the bacteria Alcaligenes faecalis. Analysis of gas production after fermentation of both β-glucans revealed distinct fermentation curves. Gas production differed significantly between the two β-glucan substrates in the period of 44–86 h after the start of fermentation. Fermentation also led to specific SCFA profiles, with higher butyrate production associated with curdlan and higher propionate production associated with MacroGard®. It cannot be fully excluded that some of the differences in gas production and SCFA profiles between the two β-glucan substrates might be caused by “contamination” of MacroGard® with products such as chitin and mannose (10). Mannose can inhibit carbohydrate uptake of bacteria under specific conditions (68), and chitin can also be fermented, which could muddle SCFA profiles, since, at least in tilapia, chitin is fermented in acetate and propionate but not in butyrate (23). Regardless of these considerations, in vitro fermentation of β-glucans by the intestinal microbiota of carp resulted in increased gas and in the production of SCFA, especially acetate, butyrate, and propionate.
Also in mammals, the main SCFAs produced as metabolites of fermentation of resistant starches, fibre, and polysaccharides are acetate, butyrate, and propionate. Butyrate and propionate have been associated with clear immunomodulatory effects such as cancer treatment in humans (69). Further, butyrate and propionate are known to regulate inflammatory responses in epithelial cells and leukocytes (70–74). These encompass, but are not restricted to, effects on leukocyte recruitment, cytokine production, lymphocyte activation, or even phagocytosis or oxygen radical production [as reviewed by (75)]. Interestingly, there are also studies which show promotion of inflammatory responses via SCFAs (76, 77). Within an immunological context, butyrate and propionate are currently considered the two most potent SCFAs [as reviewed by (77)]. For example, both have been shown in humans to inhibit LPS-induced TNF-α production via inhibition of NF-κB and increased IL-10 production (78, 79) or to inhibit antigen-specific cytotoxic T-lymphocyte activation via the regulation of IL-12 production in dendritic cells (DCs) (80). Several studies in mice have connected oral intake of fibres to increased SCFA levels, both local and systemic, and linked these increased SCFA levels with subsequent reductions in pathogenesis. For example, mice fed with oat bran-derived β-glucan-enriched diets had higher SCFA levels in the cecum, especially after having been fed diets with practically insoluble β-glucan of heavy molecular weight (81). Further, mice fed with high-fibre diets showed systemic increases in propionate levels, while oral supplementation with propionate reduced allergy-induced inflammation in the lungs (82). Often, such studies highlight important signalling roles for SCFAs receptors such as the G-protein-coupled receptors GPR41 and GPR43 (also known as free fatty acid receptor 2 or FFAR2) (73), and to a lesser extent for GPR109A and OLFR78, or mention SCFA mediation via their effect on histone deacetylases (HDACs) [as reviewed by (77)]. Up to date, for fish a direct link between fermentation of immunomodulating products such as β-glucans, and production of SCFAs with subsequent reduction of pathogenesis has not been clearly shown. Nevertheless, over the past years, incorporation of SCFA-related products in fish feed has received increasing interest (83). Several studies have investigated the effects of SCFA salts on several immune parameters in fish. For example, incorporation of sodium propionate into the diets of zebrafish for 8 weeks induced expression of il-1b and tnfα (84). Further, supplementation of the diet of common carp with encapsulated sodium butyrate affected il-1b, tnfα, and tgf-β expression after feeding for 8 weeks (85). Also, incorporation of calcium propionate or calcium lactate in the diet of Nile tilapia increased fish performance and affected haematological parameters (86). Last but not least, a recent study in zebrafish showed that immersion of zebrafish larvae in butyrate reduced migration to a wound area of both neutrophils and pro-inflammatory macrophages, while specific knockdown of hydroxycarboxylic acid receptor 1 (hcar1) abolished the reduced migration of the neutrophils (87). The study in zebrafish will support further research into the immunomodulating role of SCFAs, not only in this model animal but also in fish species of commercial interest.
Recently, we used genomic resources and cDNA cloning to identify and validate ten coding sequences for gpr40L genes in common carp (37). Phylogenetic analysis showed a division into three subclasses and a closer relationship to mammalian GPR43 than to mammalian GPR41. Synteny analysis revealed a clear conservation of syntenic relationships between carp gpr40L and the relevant region in the human genome. Here, we provide a first but important step toward a further understanding of the role and function of these gpr40L receptors and link with immunomodulation in fish. In our study, leukocytes isolated from a primary lymphoid organ (head kidney derived) expressed seven out of ten gpr40L receptors; the expression of some but not all could be stimulated by LPS, or inhibited by SCFA. Expression of gpr40L receptors may differ between leukocytes from different organs and may change with maturation stage; therefore, in our analysis leukocytes from a primary lymphoid organ were used. The posterior intestine expressed all ten gpr40L receptors, but expression of only one particular SCFA receptor (gpr40La-2) was upregulated after oral gavage with β-glucans. It remains to be proven whether modulation of the gene expression of specific gpr40L receptors in the fish intestine could be a reliable read-out for immunomodulating effects of products such as β-glucans.
Although we hypothesised that the change in SCFA profile associated with fermentation of β-glucans and subsequent regulation of gpr40L as SCFA receptors might explain the downregulation of cytokine gene expression observed in the carp intestine, there might be alternative explanations. For example, in mice, commensal lactic acid bacteria can induce an antiviral IFN response induced by recognition of shedded, double-stranded RNA by intestinal DCs. Sensing via a TLR3-mediated mechanism then leads to the production of IFNβ, which in turn dampens the expression of inflammatory cytokines such as Il-6 and Tnfα (88). Of interest, in a previous study we described a concurrent increase in mx and tlr3 expression in the intestine of carp fed with β-glucans for 25 days, and clear downregulation of il-1b, il-10, and tnfα (89). In our present study, we observed a similar inhibition of il-1b, il-6, and tnfα in carp intestine but could not repeat all the observations by Falco and colleagues; i.e., a downregulation of tlr3.1 and mx3 was observed in the current study in contrast to the significant upregulation of tlr3 and mx by Falco and colleagues. This leaves the presence of dsRNA as an alternative explanation for the downregulation of cytokine gene expression in the intestine to be confirmed. No matter the presence of dsRNA and/or altered ratios of SCFA detected by their receptors, regulation of pro-inflammatory gene expression in the fish intestine by supplementation with β-glucans was evident.
In the future, it can be of interest to analyse immunomodulatory effects of β-glucans on sterile intestinal samples, for example using ex vivo stimulation of gut tissue, to further investigate the contribution of the active naive microbiota to β-glucan-induced regulation of immune gene expression. Such an experimental setup could also allow for selectively testing immunomodulatory effects of SCFAs and other microbial metabolites. The present study provides support for a line of reasoning that suggests β-glucans can be fermented by intestinal microbiota and lead to changes in production of SCFA. Certainly, investigating the role of gpr40L receptor(s) in such processes will be of great interest.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found as follows: https://www.ncbi.nlm.nih.gov/, PRJNA766179.
Ethics Statement
All animals were handled in accordance with good animal practice as defined by the European Union guidelines for the handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm), local experimental animal committee (DEC number 2015098).
Author Contributions
JP, IdB, WP, EvdB, MF, and GW contributed to the design of the experiments. JP, MG, and EvdB contributed to the acquisition of samples and analysis of data. GW acquired funding. WP contributed to the in vitro digestion analysis. IdB contributed to the analysis of microbiota. JP, IdB, MG, WP, MF, and GW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Research leading to this review was funded by the Netherlands Organisation for Scientific Research and São Paulo Research Foundation, Brazil (FAPESP), as part of the Joint Research Projects BioBased Economy NWO-FAPESP Programme (project number 729.004.002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Adrià López Nadal from the Cell Biology and Immunology group and Saskia van Laar from the Animal Nutrition group are gratefully acknowledged for their help during the in vitro batch culture experiment. Johan Schrama from the Aquaculture and Fisheries group is gratefully acknowledged for his suggestions and helpful advice on measuring SCFAs.
Footnotes
References
1. Dalmo RA, Bøgwald J. ß-Glucans as Conductors of Immune Symphonies. Fish Shellfish Immunol (2008) 25:384–96. doi: 10.1016/j.fsi.2008.04.008
2. Meena DK, Das P, Kumar S, Mandal SC, Prusty AK, Singh SK, et al. Beta-Glucan: An Ideal Immunostimulant in Aquaculture (a Review). Fish Physiol Biochem (2013) 39:431–57. doi: 10.1007/s10695-012-9710-5
3. Petit J, Wiegertjes GF. Long-Lived Effects of Administering β-Glucans: Indications for Trained Immunity in Fish. Dev Comp Immunol (2016) 64:93–102. doi: 10.1016/j.dci.2016.03.003
4. Ariizumi K, Shen GL, Shikano S, Xu S, Ritter R 3rd, Kumamoto T, et al. Identification of a Novel, Dendritic Cell-Associated Molecule, Dectin-1, by Subtractive cDNA Cloning. J Biol Chem (2000) 275:20157–67. doi: 10.1074/jbc.M909512199
5. Brown GD, Gordon S. Immune Recognition. A New Receptor for Beta-Glucans. Nature (2001) 413:36–7. doi: 10.1038/35092620
6. Zelensky AN, Gready JE. The C-Type Lectin-Like Domain Superfamily. FEBS J (2005) 272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x
7. Cerenius L, Kawabata S-I, Söderhäll K. Biological and Immunological Aspects of Innate Defence Mechanisms Activated by (1, 3)-β-Glucans and Related Polysaccharides in Invertebrates. In: Bacic A, Fincher GB, Stone BA eds. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides. Academic Press (2009). p. 563–77. doi: 10.1016/B978-0-12-373971-1.00017-0
8. Brown GD, Williams DL. (1, 3)-β-Glucans in Innate Immunity: Mammalian Systems. In: Bacic A, Fincher GB, Stone BA eds. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides. Academic Press (2009). p. 579–619. doi: 10.1016/B978-0-12-373971-1.00018-2
9. Legentil L, Paris F, Ballet C, Trouvelot S, Daire X, Vetvicka V, et al. Molecular Interactions of β-(1→ 3)-Glucans With Their Receptors. Molecules (2015) 20:9745–66. doi: 10.3390/molecules20069745
10. Soltanian S, Francois JM, Dhont J, Arnouts S, Sorgeloos P, Bossier P. Enhanced Disease Resistance in Artemia by Application of Commercial Beta-Glucans Sources and Chitin in a Gnotobiotic Artemia Challenge Test. Fish Shellfish Immunol (2007) 23:1304–14. doi: 10.1016/j.fsi.2007.07.004
11. Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, et al. Card9 Controls a Non-TLR Signalling Pathway for Innate Anti-Fungal Immunity. Nature (2006) 442:651–6. doi: 10.1038/nature04926
12. Zelensky AN, Gready JE. C-Type Lectin-Like Domains in Fugu Rubripes. BMC Genomics (2004) 5:51. doi: 10.1186/1471-2164-5-51
13. Pietretti D, Vera-Jimenez N, Hoole D, Wiegertjes G. Oxidative Burst and Nitric Oxide Responses in Carp Macrophages Induced by Zymosan, MacroGard® and Selective Dectin-1 Agonists Suggest Recognition by Multiple Pattern Recognition Receptors. Fish shellfish Immunol (2013) 35:847–57. doi: 10.1016/j.fsi.2013.06.022
14. Petit J, Bailey EC, Wheeler RT, De Oliveira CAF, Forlenza M, Wiegertjes GF. Studies Into β-Glucan Recognition in Fish Suggests a Key Role for the C-Type Lectin Pathway. Front Immunol (2019) 10. doi: 10.3389/fimmu.2019.00280
15. Lokka G, Koppang EO. Antigen Sampling in the Fish Intestine. Dev Comp Immunol (2016) 64:138–49. doi: 10.1016/j.dci.2016.02.014
16. Round JL, Mazmanian SK. The Gut Microbiota Shapes Intestinal Immune Responses During Health and Disease. Nat Rev Immunol (2009) 9:313–23. doi: 10.1038/nri2515
17. Purchiaroni F, Tortora A, Gabrielli M, Bertucci F, Gigante G, Ianiro G, et al. The Role of Intestinal Microbiota and the Immune System. Eur Rev Med Pharmacol Sci (2013) 17:323–33.
18. Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell (2014) 157:121–41. doi: 10.1016/j.cell.2014.03.011
19. Montalban-Arques A, De Schryver P, Bossier P, Gorkiewicz G, Mulero V, Gatlin DM 3rd, et al. Selective Manipulation of the Gut Microbiota Improves Immune Status in Vertebrates. Front Immunol (2015) 6:512. doi: 10.3389/fimmu.2015.00512
20. López Nadal A, Ikeda-Ohtsubo W, Sipkema D, Peggs D, Mcgurk C, Forlenza M, et al. Feed, Microbiota, and Gut Immunity: Using the Zebrafish Model to Understand Fish Health. Front Immunol (2020) 11. doi: 10.3389/fimmu.2020.00114
21. De Bruijn I, Liu Y, Wiegertjes GF, Raaijmakers JM. Exploring Fish Microbial Communities to Mitigate Emerging Diseases in Aquaculture. FEMS Microbiol Ecol (2018) 94:1–12. doi: 10.1093/femsec/fix161
22. Rurangwa E, Laranja JL, Van Houdt R, Delaedt Y, Geraylou Z, Van De Wiele T, et al. Selected Nondigestible Carbohydrates and Prebiotics Support the Growth of Probiotic Fish Bacteria Mono-Cultures. vitro. J Appl Microbiol (2009) 106:932–40. doi: 10.1111/j.1365-2672.2008.04034.x
23. Kihara M, Sakata T. Fermentation of Dietary Carbohydrates to Short-Chain Fatty Acids by Gut Microbes and its Influence on Intestinal Morphology of a Detritivorous Teleost Tilapia (Oreochromis Niloticus). Comp Biochem Physiol Part A: Physiol (1997) 118:1201–7. doi: 10.1016/S0300-9629(97)00052-2
24. Kihara M, Sakata T. Production of Short-Chain Fatty Acids and Gas From Various Oligosaccharides by Gut Microbes of Carp (Cyprinus Carpio L.) in Micro-Scale Batch Culture. Comp Biochem Physiol A Mol Integr Physiol (2002) 132:333–40. doi: 10.1016/S1095-6433(02)00029-6
25. Leenhouwers JI, Pellikaan WF, Huizing HFA, Coolen ROM, Verreth JAJ, Schrama JW. Fermentability of Carbohydrates in an In Vitro Batch Culture Method Using Inocula From Nile Tilapia (Oreochromis Niloticus) and European Sea Bass (Dicentrarchus Labrax). Aquaculture Nutr (2008) 14:523–32. doi: 10.1111/j.1365-2095.2007.00558.x
26. Maas RM, Verdegem MCJ, Wiegertjes GF, Schrama JW. Carbohydrate Utilisation by Tilapia: A Meta-Analytical Approach. Rev Aquaculture (2020) 12:1851–66. doi: 10.1111/raq.12413
27. Kuhlwein H, Emery MJ, Rawling MD, Harper GM, Merrifield DL, Davies SJ. Effects of a Dietary Beta-(1,3)(1,6)-D-Glucan Supplementation on Intestinal Microbial Communities and Intestinal Ultrastructure of Mirror Carp (Cyprinus Carpio L.). J Appl Microbiol (2013) 115:1091–106. doi: 10.1111/jam.12313
28. Jung-Schroers V, Adamek M, Jung A, Harris S, Dóza Ö.-S., Baumer A, et al. Feeding of β-1,3/1,6-Glucan Increases the Diversity of the Intestinal Microflora of Carp (Cyprinus Carpio). Aquaculture Nutr (2016) 22:1026–39. doi: 10.1111/anu.12320
29. Carda-Dieguez M, Mira A, Fouz B. Pyrosequencing Survey of Intestinal Microbiota Diversity in Cultured Sea Bass (Dicentrarchus Labrax) Fed Functional Diets. FEMS Microbiol Ecol (2014) 87:451–9. doi: 10.1111/1574-6941.12236
30. Nikkhoo M, Yousefian M, Safari R, Nikkhoo M. The Influence Probiotic of Aqualase on the Survival, Growth, Intestinal Microflora and Challenge Infection in Wild Carp (Cyprinius Carpio L.). Res J Fisheries Hydrobiology (2010) 5:168–72.
31. Adel M, Lazado CC, Safari R, Yeganeh S, Zorriehzahra MJ. Aqualase®, a Yeast-Based in-Feed Probiotic, Modulates Intestinal Microbiota, Immunity and Growth of Rainbow Trout Oncorhynchus Mykiss. Aquaculture Res (2017) 48:1815–26. doi: 10.1111/are.13019
32. Harris SJ, Bray DP, Adamek M, Hulse DR, Steinhagen D, Hoole D. Effect of β-1/3,1/6-Glucan Upon Immune Responses and Bacteria in the Gut of Healthy Common Carp (Cyprinus Carpio). J Fish Biol (2020) 96:444–55. doi: 10.1111/jfb.14222
33. Souza FPD, Lima ECSD, Pandolfi VCF, Leite NG, Furlan-Murari PJ, Leal CNS, et al. Effect of β-Glucan in Water on Growth Performance, Blood Status and Intestinal Microbiota in Tilapia Under Hypoxia. Aquaculture Rep (2020) 17:100369. doi: 10.1016/j.aqrep.2020.100369
34. Davies ZS, Mason D, Brooks AE, Griffith GW, Merry RJ, Theodorou MK. An Automated System for Measuring Gas Production From Forages Inoculated With Rumen Fluid and its Use in Determining the Effect of Enzymes on Grass Silage. Anim Feed Sci Technol (2000) 83:205–21. doi: 10.1016/S0377-8401(99)00138-8
35. Coles LT, Moughan PJ, Darragh AJ. In Vitro Digestion and Fermentation Methods, Including Gas Production Techniques, as Applied to Nutritive Evaluation of Foods in the Hindgut of Humans and Other Simple-Stomached Animals. Anim Feed Sci Technol (2005) 123-124:421–44. doi: 10.1016/j.anifeedsci.2005.04.021
36. Williams BA, Verstegen MW, Tamminga S. Fermentation in the Large Intestine of Single-Stomached Animals and its Relationship to Animal Health. Nutr Res Rev (2001) 14:207–28. doi: 10.1079/NRR200127
37. Petit J, Wiegertjes GF. Conservation of Members of the Free Fatty Acid Receptor Gene Family in Common Carp. Dev Comp Immunol (2022) 126:104240. doi: 10.1016/j.dci.2021.104240
38. Irnazarow I. Genetic Variability of Polish and Hungarian Carp Lines. Aquaculture (1995) 129:215–5. doi: 10.1016/0044-8486(95)91961-T
39. Feng W, Zhang J, Jakovlic I, Xiong F, Wu S, Zou H, et al. Gut Segments Outweigh the Diet in Shaping the Intestinal Microbiota Composition in Grass Carp Ctenopharyngodon Idellus. AMB Express (2019) 9:44. doi: 10.1186/s13568-019-0770-0
40. Perez-Jaramillo JE, Carrion VJ, Bosse M, Ferrao LFV, De Hollander M, Garcia AAF, et al. Linking Rhizosphere Microbiome Composition of Wild and Domesticated Phaseolus Vulgaris to Genotypic and Root Phenotypic Traits. ISME J (2017) 11:2244–57. doi: 10.1038/ismej.2017.85
41. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: Paired-End Assembler for Illumina Sequences. BMC Bioinf (2012) 13:31. doi: 10.1186/1471-2105-13-31
42. Cole JR, Wang Q, Fish JA, Chai B, Mcgarrell DM, Sun Y, et al. Ribosomal Database Project: Data and Tools for High Throughput rRNA Analysis. Nucleic Acids Res (2014) 42:D633–642. doi: 10.1093/nar/gkt1244
43. Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biol (Basel) (2012) 1:895–905. doi: 10.3390/biology1030895
44. Edgar RC. Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
45. Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ (2016) 4:e2584. doi: 10.7717/peerj.2584
46. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
47. Mcdonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, et al. The Biological Observation Matrix (BIOM) Format or: How I Learned to Stop Worrying and Love the Ome-Ome. Gigascience (2012) 1:2047-2217X-2041-2047. doi: 10.1186/2047-217X-1-7
48. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences Into the New Bacterial Taxonomy. Appl Environ Microbiol (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
49. Koster J, Rahmann S. Snakemake–a Scalable Bioinformatics Workflow Engine. Bioinformatics (2012) 28:2520–2. doi: 10.1093/bioinformatics/bts480
50. Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to Analyze 16s rRNA Gene Sequences From Microbial Communities. Curr Protoc Microbiol (2012) 27:1E.5.1–1E.5.20. doi: 10.1002/0471250953.bi1007s36
51. Williams BA, Bosch MW, Boer H, Verstegen MWA, Tamminga S. An In Vitro Batch Culture Method to Assess Potential Fermentability of Feed Ingredients for Monogastric Diets. Anim Feed Sci Technol (2005) 123-124:445–62. doi: 10.1016/j.anifeedsci.2005.04.031
52. Cone JW, Van Gelder AH, Visscher GJW, Oudshoorn L. Influence of Rumen Fluid and Substrate Concentration on Fermentation Kinetics Measured With a Fully Automated Time Related Gas Production Apparatus. Anim Feed Sci Technol (1996) 61:113–28. doi: 10.1016/0377-8401(96)00950-9
53. Groot JCJ, Cone JW, Williams BA, Debersaques FMA, Lantinga EA. Multiphasic Analysis of Gas Production Kinetics for In Vitro Fermentation of Ruminant Feeds. Anim Feed Sci Technol (1996) 64:77–89. doi: 10.1016/S0377-8401(96)01012-7
54. Bauer E, Williams BA, Voigt C, Mosenthin R, Verstegen MWA. Microbial Activities of Faeces From Unweaned and Adult Pigs, in Relation to Selected Fermentable Carbohydrates. Anim Sci (2001) 73:313–22. doi: 10.1017/S135772980005829X
55. Vissers AM, Pellikaan WF, Bouwhuis A, Vincken JP, Gruppen H. And Hendriks, W.HLaminaria Digitata Phlorotannins Decrease Protein Degradation and Methanogenesis During In Vitro Ruminal Fermentation. J Sci Food Agric (2018) 98:3644–50. doi: 10.1002/jsfa.8842
56. Petit J, Embregts CWE, Forlenza M, Wiegertjes GF. Evidence of Trained Immunity in a Fish: Conserved Features in Carp Macrophages. J Immunol (2019) 203:216–24. doi: 10.4049/jimmunol.1900137
57. Saeij JP, Van Muiswinkel WB, Groeneveld A, Wiegertjes GF. Immune Modulation by Fish Kinetoplastid Parasites: A Role for Nitric Oxide. Parasitology (2002) 124:77–86. doi: 10.1017/S0031182001008915
58. Piazzon MC, Savelkoul HFJ, Pietretti D, Wiegertjes GF, Forlenza M. Carp Il10 Has Anti-Inflammatory Activities on Phagocytes, Promotes Proliferation of Memory T Cells, and Regulates B Cell Differentiation and Antibody Secretion. J Immunol (2015) 194:187. doi: 10.4049/jimmunol.1402093
59. Forlenza M, Walker PD, De Vries BJ, Wendelaar Bonga SE, Wiegertjes GF. Transcriptional Analysis of the Common Carp (Cyprinus Carpio L.) Immune Response to the Fish Louse Argulus Japonicus Thiele (Crustacea: Branchiura). Fish Shellfish Immunol (2008) 25:76–83. doi: 10.1016/j.fsi.2007.12.013
60. Pfaffl MW. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res (2001) 29:e45. doi: 10.1093/nar/29.9.e45
61. Temple MJ, Cuskin F, Basle A, Hickey N, Speciale G, Williams SJ, et al. A Bacteroidetes Locus Dedicated to Fungal 1,6-Beta-Glucan Degradation: Unique Substrate Conformation Drives Specificity of the Key Endo-1,6-Beta-Glucanase. J Biol Chem (2017) 292:10639–50. doi: 10.1074/jbc.M117.787606
62. Rooks MG, Garrett WS. Gut Microbiota, Metabolites and Host Immunity. Nat Rev Immunol (2016) 16:341–52. doi: 10.1038/nri.2016.42
63. Liu H, Guo X, Gooneratne R, Lai R, Zeng C, Zhan F, et al. The Gut Microbiome and Degradation Enzyme Activity of Wild Freshwater Fishes Influenced by Their Trophic Levels. Sci Rep (2016) 6:24340. doi: 10.1038/srep24340
64. Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol (2007) 5:e156. doi: 10.1371/journal.pbio.0050156
65. Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, Mcnulty NP, et al. Recognition and Degradation of Plant Cell Wall Polysaccharides by Two Human Gut Symbionts. PLoS Biol (2011) 9:e1001221. doi: 10.1371/journal.pbio.1001221
66. Ndeh D, Rogowski A, Cartmell A, Luis AS, Basle A, Gray J, et al. Complex Pectin Metabolism by Gut Bacteria Reveals Novel Catalytic Functions. Nature (2017) 544:65–70. doi: 10.1038/nature21725
67. Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe (2008) 4:447–57. doi: 10.1016/j.chom.2008.09.007
68. Degnan BA, Macfarlane GT. Carbohydrate Utilization Patterns and Substrate Preferences in Bacteroides Thetaiotaomicron. Anaerobe (1995) 1:25–33. doi: 10.1016/S1075-9964(95)80392-0
69. Fattahi Y, Heidari HR, Khosroushahi AY. Review of Short-Chain Fatty Acids Effects on the Immune System and Cancer. Food Bioscience (2020) 38:100793. doi: 10.1016/j.fbio.2020.100793
70. Voltolini C, Battersby S, Etherington SL, Petraglia F, Norman JE, Jabbour HN. A Novel Antiinflammatory Role for the Short-Chain Fatty Acids in Human Labor. Endocrinology (2012) 153:395–403. doi: 10.1210/en.2011-1457
71. Masui R, Sasaki M, Funaki Y, Ogasawara N, Mizuno M, Iida A, et al. G Protein-Coupled Receptor 43 Moderates Gut Inflammation Through Cytokine Regulation From Mononuclear Cells. Inflamm Bowel Dis (2013) 19:2848–56. doi: 10.1097/01.MIB.0000435444.14860.ea
72. Ohira H, Fujioka Y, Katagiri C, Mamoto R, Aoyama-Ishikawa M, Amako K, et al. Butyrate Attenuates Inflammation and Lipolysis Generated by the Interaction of Adipocytes and Macrophages. J Atheroscler Thromb (2013) 20:425–42. doi: 10.5551/jat.15065
73. Halnes I, Baines KJ, Berthon BS, Macdonald-Wicks LK, Gibson PG, Wood LG. Soluble Fibre Meal Challenge Reduces Airway Inflammation and Expression of GPR43 and GPR41 in Asthma. Nutrients (2017) 9:1–11. doi: 10.3390/nu9010057
74. Li M, Van Esch B, Henricks PAJ, Garssen J, Folkerts G. Time and Concentration Dependent Effects of Short Chain Fatty Acids on Lipopolysaccharide- or Tumor Necrosis Factor Alpha-Induced Endothelial Activation. Front Pharmacol (2018) 9:233. doi: 10.3389/fphar.2018.00233
75. Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of Inflammation by Short Chain Fatty Acids. Nutrients (2011) 3:858–76. doi: 10.3390/nu3100858
76. Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-Chain Fatty Acids Activate GPR41 and GPR43 on Intestinal Epithelial Cells to Promote Inflammatory Responses in Mice. Gastroenterology (2013) 145:396–406.e391-310. doi: 10.1053/j.gastro.2013.04.056
77. Li M, Van Esch B, Wagenaar GTM, Garssen J, Folkerts G, Henricks PAJ. Pro- and Anti-Inflammatory Effects of Short Chain Fatty Acids on Immune and Endothelial Cells. Eur J Pharmacol (2018) 831:52–9. doi: 10.1016/j.ejphar.2018.05.003
78. Usami M, Kishimoto K, Ohata A, Miyoshi M, Aoyama M, Fueda Y, et al. Butyrate and Trichostatin A Attenuate Nuclear Factor κb Activation and Tumor Necrosis Factor α Secretion and Increase Prostaglandin E2 Secretion in Human Peripheral Blood Mononuclear Cells. Nutr Res (2008) 28:321–8. doi: 10.1016/j.nutres.2008.02.012
79. Aoyama M, Kotani J, Usami M. Butyrate and Propionate Induced Activated or non-Activated Neutrophil Apoptosis via HDAC Inhibitor Activity But Without Activating GPR-41/GPR-43 Pathways. Nutrition (2010) 26:653–61. doi: 10.1016/j.nut.2009.07.006
80. Nastasi C, Fredholm S, Willerslev-Olsen A, Hansen M, Bonefeld CM, Geisler C, et al. Butyrate and Propionate Inhibit Antigen-Specific CD8(+) T Cell Activation by Suppressing IL-12 Production by Antigen-Presenting Cells. Sci Rep (2017) 7:14516. doi: 10.1038/s41598-017-15099-w
81. Immerstrand T, Andersson KE, Wange C, Rascon A, Hellstrand P, Nyman M, et al. Effects of Oat Bran, Processed to Different Molecular Weights of Beta-Glucan, on Plasma Lipids and Caecal Formation of SCFA in Mice. Br J Nutr (2010) 104:364–73. doi: 10.1017/S0007114510000553
82. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut Microbiota Metabolism of Dietary Fiber Influences Allergic Airway Disease and Hematopoiesis. Nat Med (2014) 20:159–66. doi: 10.1038/nm.3444
83. Tran NT, Li Z, Wang S, Zheng H, Aweya JJ, Wen X, et al. Progress and Perspectives of Short-Chain Fatty Acids in Aquaculture. Rev Aquaculture (2020) 12:283–98. doi: 10.1111/raq.12317
84. Safari R, Hoseinifar SH, Kavandi M. Modulation of Antioxidant Defense and Immune Response in Zebra Fish (Danio Rerio) Using Dietary Sodium Propionate. Fish Physiol Biochem (2016) 42:1733–9. doi: 10.1007/s10695-016-0253-z
85. Liu W, Yang Y, Zhang J, Gatlin DM, Ringø E, Zhou Z. Effects of Dietary Microencapsulated Sodium Butyrate on Growth, Intestinal Mucosal Morphology, Immune Response and Adhesive Bacteria in Juvenile Common Carp (Cyprinus Carpio) Pre-Fed With or Without Oxidised Oil. Br J Nutr (2014) 112:15–29. doi: 10.1017/S0007114514000610
86. Hassaan M, Wafa M, Soltan M, Goda A, Mogheth N. Effect of Dietary Organic Salts on Growth, Nutrient Digestibility, Mineral Absorption and Some Biochemical Indices of Nile Tilapia. Oreochromis niloticus (2014) 29:47–55. doi: 10.5829/idosi.wasj.2014.29.01.81237
87. Cholan PM, Han A, Woodie BR, Watchon M, Kurz AR, Laird AS, et al. Conserved Anti-Inflammatory Effects and Sensing of Butyrate in Zebrafish. Gut Microbes (2020) 12:1–11. doi: 10.1080/19490976.2020.1824563
88. Kawashima T, Kosaka A, Yan H, Guo Z, Uchiyama R, Fukui R, et al. Double-Stranded RNA of Intestinal Commensal But Not Pathogenic Bacteria Triggers Production of Protective Interferon-β. Immunity (2013) 38:1187–97. doi: 10.1016/j.immuni.2013.02.024
89. Falco A, Miest JJ, Pionnier N, Pietretti D, Forlenza M, Wiegertjes GF, et al. β-Glucan-Supplemented Diets Increase Poly(I:C)-Induced Gene Expression of Mx, Possibly via Tlr3-Mediated Recognition Mechanism in Common Carp (Cyprinus Carpio). Fish Shellfish Immunol (2014) 36:494–502. doi: 10.1016/j.fsi.2013.12.005
Keywords: SCFA, immunomodulation, fish, β-glucan, microbiota
Citation: Petit J, de Bruijn I, Goldman MRG, van den Brink E, Pellikaan WF, Forlenza M and Wiegertjes GF (2022) β-Glucan-Induced Immuno-Modulation: A Role for the Intestinal Microbiota and Short-Chain Fatty Acids in Common Carp. Front. Immunol. 12:761820. doi: 10.3389/fimmu.2021.761820
Received: 20 August 2021; Accepted: 08 December 2021;
Published: 06 January 2022.
Edited by:
Samuel A. M. Martin, University of Aberdeen, United KingdomReviewed by:
M. Carla Piazzon, National Research Council (CSIC), SpainJorge Galindo-Villegas, Nord University, Norway
Copyright © 2022 Petit, de Bruijn, Goldman, van den Brink, Pellikaan, Forlenza and Wiegertjes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geert F. Wiegertjes, Z2VlcnQud2llZ2VydGplc0B3dXIubmw=
 Jules Petit
Jules Petit Irene de Bruijn
Irene de Bruijn Mark R. G. Goldman
Mark R. G. Goldman Erik van den Brink
Erik van den Brink Wilbert F. Pellikaan3
Wilbert F. Pellikaan3 Maria Forlenza
Maria Forlenza Geert F. Wiegertjes
Geert F. Wiegertjes