- 1Department of Pediatrics, Division of Hematology, Oncology, Stem Cell Transplantation and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA, United States
- 2Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA, United States
- 3Center for Definitive and Curative Medicine (CDCM), Stanford University School of Medicine, Stanford, CA, United States
FOXP3 is the master transcription factor in both murine and human FOXP3+ regulatory T cells (Tregs), a T-cell subset with a central role in controlling immune responses. Loss of the functional Foxp3 protein in scurfy mice leads to acute early-onset lethal lymphoproliferation. Similarly, pathogenic FOXP3 mutations in humans lead to immunodysregulation, polyendocrinopathy, enteropathy, and X-linked (IPEX) syndrome, which are characterized by systemic autoimmunity that typically begins in the first year of life. However, although pathogenic FOXP3 mutations lead to overlapping phenotypic consequences in both systems, FOXP3 in human Tregs, but not mouse, is expressed as two predominant isoforms, the full length (FOXP3FL) and the alternatively spliced isoform, delta 2 (FOXP3Δ2). Here, using CRISPR/Cas9 to generate FOXP3 knockout CD4+ T cells (FOXP3KOGFP CD4+ T cells), we restore the expression of each isoform by lentiviral gene transfer to delineate their functional roles in human Tregs. When compared to FOXP3FL or FOXP3Δ2 alone, or double transduction of the same isoform, co-expression of FOXP3FL and FOXP3Δ2 induced the highest overall FOXP3 protein expression in FOXP3KOGFP CD4+ T cells. This condition, in turn, led to optimal acquisition of Treg-like cell phenotypes including downregulation of cytokines, such as IL-17, and increased suppressive function. Our data confirm that co-expression of FOXP3FL and FOXP3Δ2 leads to optimal Treg-like cell function and supports the need to maintain the expression of both when engineering therapeutics designed to restore FOXP3 function in otherwise deficient cells.
Introduction
Regulatory T cells (Tregs) are defined as a CD4+ CD25+ CD127- population and characterized by their suppressive function in both human and mice (1, 2). Forkhead Box P3 (FOXP3) is an essential transcription factor for the development of functional Tregs in both systems (3, 4). The role of FOXP3 in regulating the suppressive function of human Tregs is exemplified by loss-of-function FOXP3 mutations, which result in primary Treg dysfunction and effector T cell (Teff) abnormalities, leading to the monogenic autoimmune disease, IPEX syndrome (5–10). Similarly, mutation of Foxp3 (Foxp3SF) in mice results in fatal systemic lymphoproliferation and rapid wasting disease. The “scurfy” mice, which has many similarities to IPEX syndrome, can be rescued by wild-type (WT) Treg transfer (11).
In human T cells, FOXP3 is expressed as two predominant isoforms, the full-length isoform, FOXP3FL, and an alternatively spliced isoform, FOXP3Δ2 that lacks exon 2. Two other isoforms, FOXP3Δ2Δ7 and FOXP3Δ7, have also been described in human lymphocytes and epithelial cells, but unlike FOXP3FL and FOXP3Δ2, neither appears to play a role in Treg suppressive activity (12). Therefore, the need to maintain their expression when restoring a dysregulated immune system, such as that in IPEX patients, is not clear (13, 14). In contrast to human T cells, mice express only FOXP3FL (15). The fact that FOXP3Δ2 has not been detected in mouse hints at key evolutionary differences in T cell lineage development between the two species (16). Importantly, clearly defining the differential or complementary roles of FOXP3FL and FOXP3Δ2 in human health and disease has been challenging, due to limitations in experimental tools, including humanized mouse models, to cleanly characterize the function of each isoform, a lack of antibodies that can specifically detect the FOXP3Δ2 isoform (anti-FOXP3 antibody clone 150D/E4 binds to exon 2 and can detect its absence), and no currently identified strategies that can accurately replicate the endogenous alternative splicing of FOXP3 exon 2 (17). Notably, a recent paper reported a family with a FOXP3 mutation (c.305delT: p.Phe102fs) that led to loss of FOXP3FL but maintained the expression of FOXP3Δ2. Interestingly, affected males reached adulthood suggesting a level of immune functionality, but they did develop autoimmune manifestations including type 1 diabetes, inflammatory bowel disease, and psoriatic arthritis (17). These findings indicate that Tregs may be able to differentiate with FOXP3Δ2 in the absence of FOXP3FL, but that expression of only FOXP3Δ2 may lead to impaired suppressive function which cannot be rescued from the absence of FOXP3FL. Collectively, the data hint that both FOXP3FL and FOXP3Δ2 isoforms may be necessary for Tregs to fully acquire suppressive functionality.
In recent years, we have established several FOXP3 gene transfer approaches mainly for clinical translational purposes but which have also provided fundamental findings, clarifying the role of FOXP3 in human T cells (18). We have demonstrated that lentiviral-mediated gene transfer of FOXP3FL under a constitutive promoter in Teff cells is sufficient to convey suppressive function, upregulate several key Treg molecules, and suppress others so that the transduced Teff cells become very similar to Tregs (“CD4LVFOXP3 Treg-like cells”). Additionally, CRISPR/Cas9 FOXP3 gene editing in which we inserted either FOXP3FL or FOXP3Δ2 at the FOXP3 locus in Teff or Tregs revealed comparable activities for the two isoforms, with each inducing WT restoration of function in Teff cells, but only suboptimal repair of function in Tregs, suggesting that the simultaneous expression of both isoforms is a requirement for optimal Treg development and function (19). Further, although other published gene transfer studies have explored the roles of FOXP3FL and FOXP3Δ2, these experiments were performed in T cells expressing endogenous FOXP3, therefore never clearly dissecting the impact of each isoform, or both (15).
Here, using our established Teff to Treg “conversion system,” we elucidate the roles for each isoform in conveying Treg-cell features and functions. Using CRISPR/Cas9, we have first made CD4+ T cells knocked out (KO) for endogenous FOXP3 (FOXP3KOGFP CD4+ T cells). The FOXP3KOGFP CD4+ T cells were then transduced with lentiviral vectors expressing FOXP3FL and/or FOXP3Δ2. Each isoform was cloned into lentiviral vectors (LV-) co-expressing the truncated Epithelial Growth Factor Receptor (EGFR) or Nerve Growth Factor Receptor (NGFR) surface marker, thus enabling both simultaneous transduction and tracking of the expression of each isoform, as well as delineation of their functional roles. Our data demonstrates that co-transduction of both FOXP3FL and FOXP3Δ2 isoforms, in comparison to each alone, is optimal for the conversion of FOXP3KOGFP CD4+ T cells into functional Treg-like cells in vitro. These findings suggest that both isoforms play key roles in Treg-cell function and provide important considerations for the design of gene engineering strategies in stem cell progenitors aimed at restoring FOXP3 expression and function across different T cell lineages.
Materials and Methods
Cell Isolation and In Vitro Culture
HEK293T/17 cells were purchased from ATCC. HEK293T/17 cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10% fetal bovine serum (FBS, Gibco). MT-2 cells are human adult Treg-cell lines immortalized with HTLV-1 and were kindly provided by Dr. Donald B. Kohn (UCLA) (20, 21). MT-2 cells maintain a very high expression of FOXP3 but have the ability to proliferate without TCR-mediated restimulation, and have been elsewhere characterized (22). MT-2 cells were cultured with Roswell Park Memorial Institute medium (RPMI, Life Technologies) supplemented with 10% FBS. Mycoplasma testing (MycoAlert Quick Mycoplasma Detection Kit, Lonza) was done routinely with mycoplasma contamination not detected during the study period.
Buffy coats from healthy male donors were obtained from the Stanford Blood Center. Peripheral blood mononuclear cells (PBMCs) were separated from the Buffy coat by density gradient centrifugation using Ficoll Paque (GE Healthcare Science). CD4+ T cells were enriched from PBMCs by negative selection (Stem Cell Technologies). CD4+ T cells were cultured by X-VIVO 15 media supplemented with gentamicin (Lonza) and 5% human serum from pooled male AB plasma (Sigma-Aldrich) with the presence of 100 U/ml of recombinant human (rh) IL-2 (PeproTech). Human expanded Tregs were generated according to previous publications (23, 24). Briefly, CD4+ T cells were activated by CD3/CD28 polyclonal stimulation and cultured in the presence of rapamycin (100 nM). Expanded Tregs were harvested and analyzed on day 21 (suppression assay) and day 28 (phenotype).
FOXP3 Gene Disruption by CRISPR/Cas9-Mediated Homologous Recombination
The FOXP3 gene was disrupted in CD4+ T cells by CRISPR/Cas9-mediated homologous recombination. Freshly isolated CD4+ T cells were activated overnight by plate-bound 10 μg/ml of anti-CD3 antibody (eBioscience) and 1 μg/ml of anti-CD28 antibody (eBioscience). A ribonucleoprotein (RNP) complex was made by incubating 15 μg of Cas9 protein (Alt-R® S.p. Cas9 Nuclease V3, Integrated DNA Technologies) with 8 μg of a small guide RNA (sgRNA; TriLink BioTechnologies) targeting FOXP3 with the following sequence: 5′-2′OMe[A(ps)G(ps)G(ps)]ACCCGAUGCCCAACCCCGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUG
AAAAAGUGGCACCGAGUCGGUGCUUU 2′OMe (U(ps) U(ps)U)-3′ where ps = phosphorothioate, 2′OMe = 2′-O-methyl, at room temperature for 15 min. The RNP complex, together with an AAV6 vector (multiplicity of infection (MOI) of 100,000) containing FOXP3 homologous arms and GFP expression cassette under the PGK promoter (Vigene), was transduced into CD4+ T cells using the Amaxa 4D-Nucleofector (Lonza). The GFP-positive population was sorted by FACS 4–5 days after transduction.
LV Vector Construct and Transduction
Vectors LV-FOXP3FL (FOXP3FL-NGFR) and LV-FOXP3Δ2 (FOXP3Δ2-NGFR) were kindly provided by Dr. Megan K Levings (University of British Columbia). Vectors FOXP3FL-EGFR and FOXP3Δ2-EGFR were generated from FOXP3FL-NGFR and FOXP3Δ2-NGFR by restriction enzyme digestion (AgeI and BlpI, New England Biolabs Inc.) to remove NGFR, then EGFR was inserted using a DNA ligase (Mighty Mix DNA Ligation Kit, ClonTech; Figure S1A). Control vectors expressing only NGFR or EGFR were also constructed.
Lentivirus was produced as previously described (18, 25, 26). Briefly, each vector together with helper plasmids (gag/pol, REV, VSVG, and pAdVAntage) were transiently co-transfected into HEK293T/17 cells by Mirus-LT1 reagent (Mirus Bio LLC, Madison, WI). The supernatant was collected 30 h after transfection and concentrated by ultracentrifugation. The titer of lentivirus was determined by serial dilution of 293T cells.
For the single transduction of each LV construct, FOXP3KOGFP CD4+ T cells were activated overnight by TransAct (Miltenyi Biotec) with 100 U/ml rhIL-2 (PeproTech) and 10 ng/ml rhIL-7 (PeproTech). On day 1, lentivirus was added to the activated T cells at the MOI of 50. Cells were split every 2–3 days. On days 9–10, the transduced FOXP3KOGFP CD4+ T cells were enriched using the MACSelect LNGFR System (NGFR; for vector constructs co-expressing NGFR). For the sequential transduction, FOXP3KOGFP CD4+ were sequentially transduced with a second LV construct after the activation and enriched by the MACS PE beads with an anti-human EGFR antibody conjugated with PE (EGFR; for vector constructs expressing EGFR) (Miltenyi Biotec).
We have confirmed isoform expression in HEK293T/17 cells by transducing lentiviral vectors expressing 1) FOXP3FL (NGFR); 2) FOXP3Δ2 (EGFR), or 3) FOXP3FL (NGFR)+FOXP3Δ2 (EGFR) (Figures S1B, D). FACS analysis confirmed that all three conditions gave a robust FOXP3 expression. Notably, the HEK293T/17 cells double transduced with both FOXP3 isoforms gave the highest FOXP3 expression (Figures S1C, E).
Vector Copy Number
Genomic DNA was extracted from 1 × 106 transduced FOXP3KOGFP CD4+ T cells by DNeasy Blood and Tissue Kit (Qiagen). Lentiviral copy numbers were measured by Woodchuck Hepatitis Virus Posttranscriptional Responsive Element (WPRE) qPCR, as previously described (27). Primer sequences for copy number qPCR were WPRE (forward primer, 5′-CCGTTGTCAGGCAACGTG-3′; reverse primer, 5′-AGCTGACAGGTGGTGGCAAT-3′; probe, 5′-FAM TGCTGACGCAACCCCCACTGGT-TAMRA-3) and ALB (forward primer, 5′-TGAAACATACGTTCCCAAAGAGTTT-3′; reverse primer, 5′-CTCTCCTTCTCAGAAAGTGTGCATAT-3′; probe, -FAM- TGCTGAAACATTCACCTTCCATGCAGATAMRA-3′). The copy number using WPRE/ALB primers was determined using 10 ng of genomic DNA. VCN was calculated by WPRE/ALB copy number and shown as copy number per genome.
RT-PCR
RNA was extracted from 1 × 106 FOXP3KOGFP MT-2 or CD4+ T cells by RNeasy Mini Kit (Qiagen). One hundred nanograms of total RNA was used to synthesize cDNA using SuperScript IV VILO (Thermo Fisher Scientific). RT-PCR was done using 1 μl of synthesized cDNA amplified by PrimeSTAR GXL DNA Polymerase (Takara Bio) as described in (15). Primer sequences were FOXP3 (forward primer, 5′-TTCCTTGGCCCTTGGCCCATC-3′; reverse primer, 5′-CATTTGCCAGCAG TGGGTAGGA-3′) and ACTB (forward primer, 5′-CCTCGCCTTTGCCGATCC-3′; reverse primer, 5′-GGATCTTCATGAGGTAGTCAGTC-3′). PCR products were analyzed by 2% agarose gel electrophoresis.
qPCR
Total RNA was extracted from 1 million CD4+ T cells by RNeasy Plus Mini Kit (Qiagen). One hundred nanograms of RNA was used to synthesize cDNA by SuperScript IV VILO (Thermo Fisher Scientific, Waltham, MA). Q-PCR was done by TaqMan Gene Expression Assay and TaqMan Universal Master Mix II (Thermo Fisher). Gene expression was calculated by ΔCT where ΔCT = CTtarget - CThousekeeping. The internal control housekeeping gene is HPRT (Hs99999909_m1). The expressions of the target gene were measured by q-PCR primers (TaqMan Gene Expression Assay) targeting FOXP3 (Hs01085834_m1), NGFR (Hs00609976_m1), EGFR (Hs01076088_m1), GARP (Hs00194136_m1), and LAP (Hs00998133_m1).
Flow Cytometry
CD4+ T cells were resuspended in flow cytometry (FACS) buffer (PBS supplemented with 0.5% BSA and 2 mM EDTA) and stained with antibody cocktails (CD4, CD25, CD127, NGFR, and EGFR) for 30 min. After surface staining, intracellular staining for FOXP3 and GFP was done using the Foxp3/Transcription Factor Staining buffer set (eBioscience). Antibodies used are listed in Table S1. Data were acquired by (BD Biosciences) and analyzed by FlowJo 10.4 software (FlowJo LLC).
Immunoblotting
Immunoblotting was done to analyze FOXP3 isoform expression. Briefly, total protein was extracted from transduced FOXP3KOGFP MT-2 or CD4+ T cells by RIPA buffer (Life Technologies) and protein concentration was measured by Pierce BC Protein Assay Kit (Thermo Fisher Scientific). Ten micrograms of protein per condition was loaded on the 4%–15% gradient SDS-PAGE gel (Bio-Rad) and transferred to a PVDF membrane (Bio-Rad). The primary antibody (Anti-FOXP3 antibody Rabbit, Cell Signaling Technologies) was added overnight at the concentration of 1:1,000. The secondary antibody (Anti-Rabbit IgG antibody-HRP conjugated, Cell Signaling Technologies) was added for 1 h at the concentration of 1:10,000. Antibodies used are listed in Table S2. Chemiluminescent substrate (Clarity Western ECL Substrate, Bio-Rad) was added to the membrane, and the signal was captured using the ChemiDoc XRS+ system.
ELISA
Transduced FOXP3KOGFP CD4+ T cells were cultured at the density of 1 × 106 cells/ml in a round-bottom 96-well plate and activated by Dynabeads Human T-cell Activator CD3/CD28 (Gibco) at a 1:25 bead: cell ratio. Culture supernatant was collected at 24 h (IL-2) and 72 h (IL-4, IFN-γ, IL-17A, IL-22) after stimulation. Cytokine concentrations, including IL-2, IL-4, and IFN-γ; were measured by Human OptEIA ELISA Kit (BD Biosciences). Cytokine concentrations, including IL-17A and IL-22 concentrations, were measured by Human DuoSet ELISA Kit (R&D).
Suppression Assay
Suppression assay for transduced FOXP3KOGFP CD4+ T cells was done according to the previous publication (18). Briefly, responder CD4+ T cells (50,000 cells) labeled by CellTrace CFSE (Life Technologies) and transduced FOXP3KOGFP suppressor cells (12,500–50,000 cells) labeled by CellTrace Violet (Life Technologies) were cocultured at different concentrations (1:0.25–1:1) after activation by Dynabeads Human T-cell Activator CD3/CD28 (Gibco) at the 1:25 bead: cell ratio. Ninety-six hours after stimulation, proliferations of responder CD4+ T cells were counted by FACS.
Statistical Analysis
GraphPad Prism Software ver8.0 (GraphPad software) was used for statistical analyses. All statistical analyses were performed with the two-tailed Student t-test or Mann–Whitney U-test. One-way ANOVA followed by Tukey’s multiple-comparison tests was performed when comparing multiple conditions. p values <0.05 were considered as significant. Data are shown as mean ± SEM.
Results
FOXP3 Expression Can Be Restored in FOXP3KOGFP MT-2 Cells by LV-Mediated FOXP3 Isoform Gene Transfer
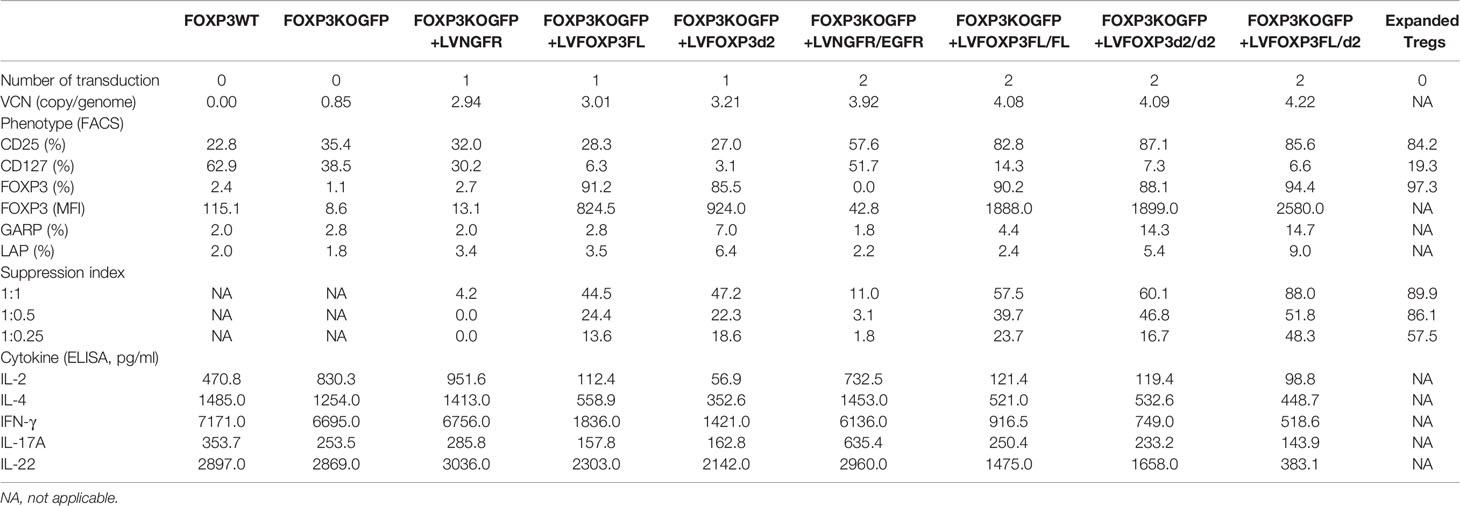
To first test and optimize the FOXP3KO and ectopic FOXP3 gene re-expression strategy, we used the MT-2 cell-line, which grows easily in culture and aberrantly expresses FOXP3, in an activation-independent manner. We disrupted the FOXP3 open reading frame in MT2 cells using CRISPR/Cas9 and an sgRNA targeting the FOXP3 locus. A GFP open reading frame (expressed via the constitutive PGK promoter and stabilized by the WPRE element which follows) was inserted at the cut site, after delivery to cells using an AAV6 vector; GFP expression was used to purify the edited cells (Figures 1A, B). Using this strategy, we generated GFP-positive FOXP3KO MT-2 cells (FOXP3KOGFP MT-2: targeting efficiency 20.6 ± 1.04%, MEAN ± SEM, n = 4) which were enriched by FACS sorting (Figures 1C, D) to produce a pure population of the edited MT-2 cells. RT-PCR and immunoblotting (Figure 1E) confirmed that the FOXP3WT MT-2 cells highly expressed both isoforms of FOXP3, prior to gene editing, but that neither were detectable in the sorted GFP+ MT-2 cells, following the CRISPR/Cas9-mediated disruption of the endogenous FOXP3 gene.
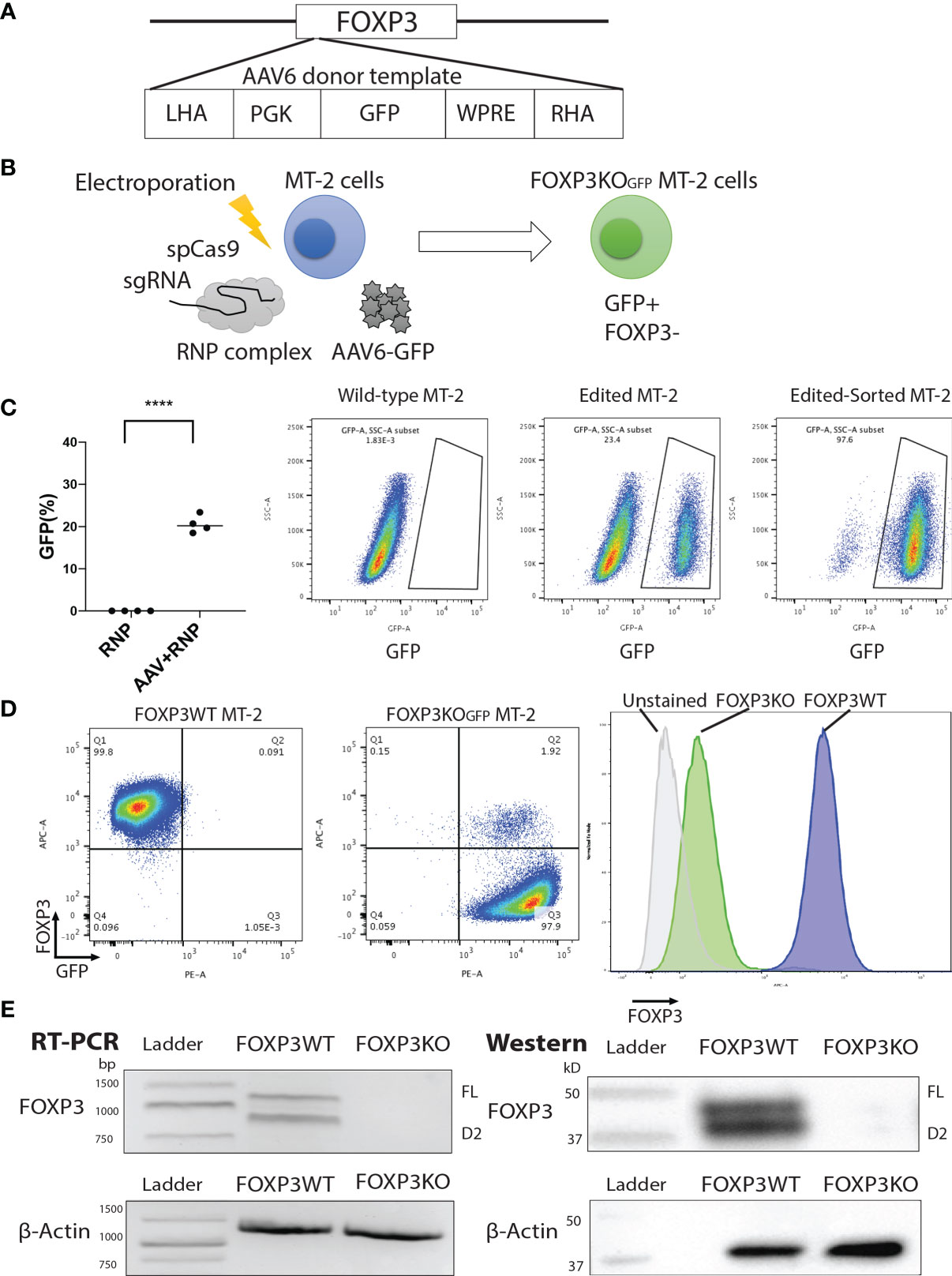
Figure 1 FOXP3KOGFP MT-2 cells can be generated by CRISPR/Cas9-mediated homologous recombination. The FOXP3 locus of MT-2 cells was knocked out by CRISPR/Cas9-mediated homologous recombination. (A) Design of the PGK-GFP AAV6 donor template, left homology arm (LHA), Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element (WPRE), and right homology arm (RHA). (B) FOXP3 Editing strategy of MT-2 cells. (C) FOXP3 editing efficiency of MT-2 cells (n = 4). (D) Representative FACS staining of FOXP3KOGFP MT-2 cells. (E) RT-PCR and immunoblotting of FOXP3KOGFP MT-2 cells. ****P < 0.0001.
FOXP3KOGFP MT-2 cells were then transduced with lentiviral vectors expressing FOXP3FL-NGFR or FOXP3Δ2-EGFR, or both, and protein expression examined by FACS analysis (Figure 2A and Figures S2A, B). Single transduction of either FOXP3FL or FOXP3Δ2 restored FOXP3 expression in FOXP3KOGFP MT-2 cells. Notably, FOXP3 expression from cells expressing FOXP3FL or FOXP3Δ2 was not as high as that of FOXP3WT MT-2 cells. In contrast, we obtained an almost full restoration of FOXP3 expression, when both isoforms (FOXP3FL+ FOXP3Δ2) were transduced (Figures 2B, C). We also transduced the two isoforms into FOXP3WT MT-2 cells (i.e., expressing endogenous FOXP3) but saw no increase in FOXP3 expression, suggesting that expression levels of this gene are tightly regulated or that FOXP3 expression cannot be increased beyond the endogenous level (Figures S3A–C). Taken together, these data show that LV-mediated FOXP3 gene transfer can restore FOXP3 expression in FOXP3KOGFP MT-2 cells and that dual transduction of both isoforms is feasible and traceable with the different marker genes. Those results support our strategy for further testing in CD4+ T cells.
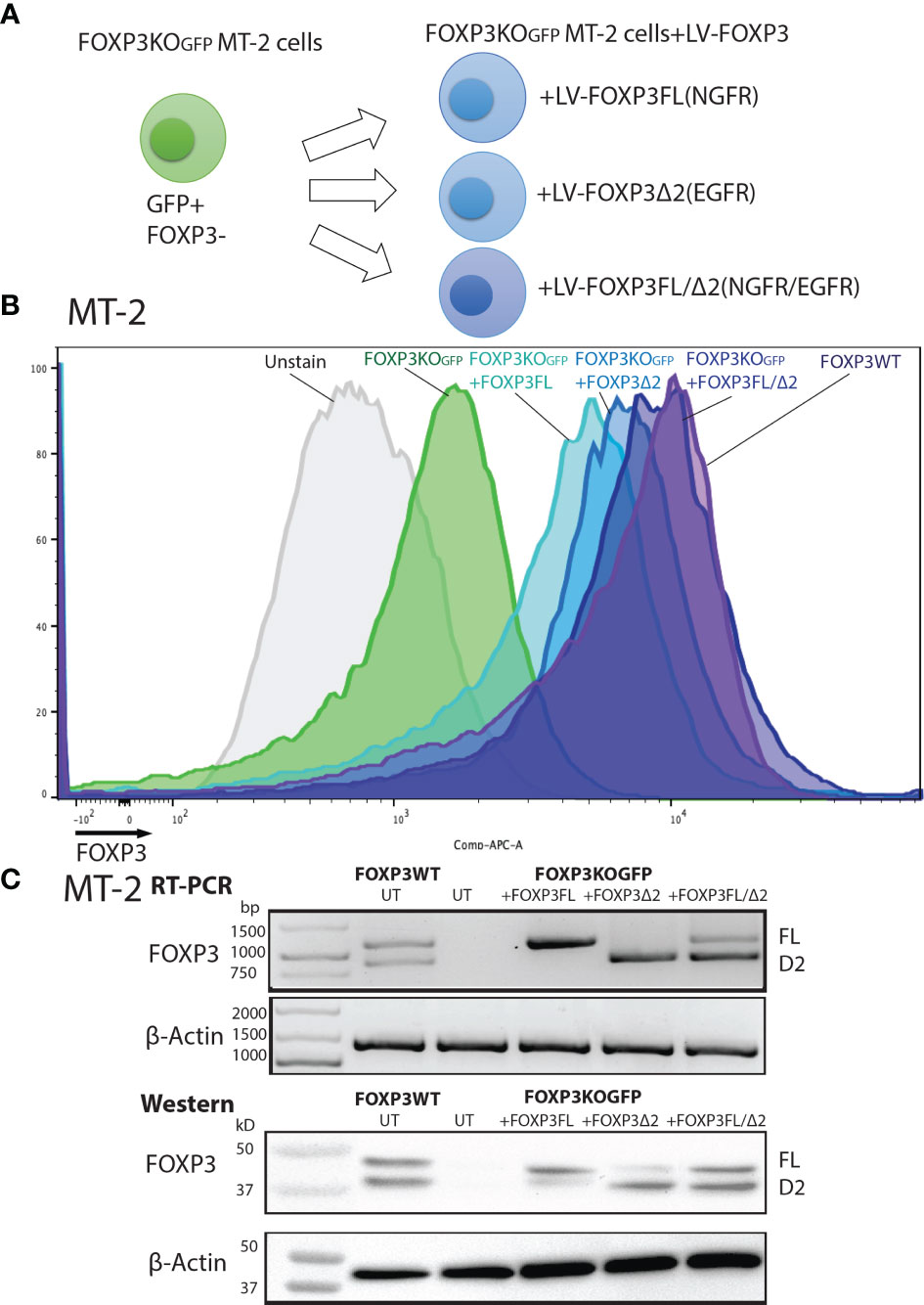
Figure 2 FOXP3KOGFP MT-2 cells transduced with FOXP3 isoforms. FOXP3KOGFP. MT-2 cells were transduced with FOXP3FL and/or FOXP3Δ2. (A) LV transduction strategy of FOXP3KOGFP MT-2 cells. (B) FACS staining of FOXP3 knockout MT-2 cells. (C) RT-PCR and immunoblotting of FOXP3KOGFP MT-2 cells.
Single FOXP3 Isoform Ectopic Expression in FOXP3KOGFP CD4+ T Cells
To assess the roles of each FOXP3 isoform alone, or together, in the conversion of Teff into Tregs, we used our previously established robust experimental system, in which transduction of LV-FOXP3FL converts CD4+ T cells into Treg-like cells (25, 26). To use this experimental system, we first disrupted the endogenous FOXP3 gene and inserted GFP at the cut site of CD4+ T cells (FOXP3KOGFP CD4+ T cells), as was done in the MT-2 cells. We generated GFP-positive FOXP3 KO CD4+ T cells (targeting efficiency 14.4 ± 1.93%, MEAN ± SEM, n = 4) and enriched the GFP+ cells by FACS sorting (Figures 3A, B). The FOXP3KOGFP CD4+ T cells were then used to test the functional impact of LV-FOXP3FL or LV-FOXP3Δ2 gene transfer. The efficiency of LV transduction into the FOXP3KOGFP CD4+ T cells was considerably lower than our previous experience using similar vectors in primary CD4+ T cells that have not undergone CRISPR/Cas9 editing (18). However, selection of the transduced cells by detection of the co-expressed NGFR or EGFR surface markers allowed us to purify the transduced cells and evaluate the expression and impact of each isoform on cell functions (Figure 3C). Notably, there was no significant difference in the LV vector copy number (VCN) between the FOXP3FL- and FOXP3Δ2-transduced cells (FOXP3FL 3.01 ± 0.59 copy/cell, FOXP3Δ2 3.21 ± 0.42 copy/cell, NGFR 2.93 ± 0.37 copy/cell, MEAN ± SEM, n = 4) (Figure 3D). At the same time, it was shown that FOXP3KOGFP CD4+ T cells have a VCN of 0.85 ± 0.03 copy/cell, which corresponds to the WPRE element inserted in the FOXP3 locus. Further, RT-PCR and immunoblotting demonstrated equivalent FOXP3 mRNA and protein in cells transduced with each isoform (Figure 3E). Moreover, as expected, activation-induced FOXP3 expression observed in the FOXP3WT CD4+ T cells was not detected in the FOXP3KOGFP CD4+ T cells. These data do confirm that LV-mediated gene transfer of either FOXP3FL or FOXP3Δ2 into FOXP3KOGFP CD4+ T cells restores the expression of each FOXP3 isoform.
Figure 3 FOXP3KOGFP CD4+ T cells can be transduced with FOXP3 isoforms. FOXP3KOGFP CD4+ T cells were transduced with FOXP3FL (NGFR) or FOXP3δ2 (NGFR). (A) LV transduction strategy of FOXP3 knock-out CD4+ T cells. (B) Editing efficiency of CD4+ T cells (n = 4). (C) Transduction efficiency of FOXP3KOGFP CD4+ T cells (n = 4). (D) VCN analysis of FOXP3KOGFP CD4+ T cells transduced with FOXP3 isoforms (n = 4). (E) RT-PCR and immunoblotting of FOXP3KOGFP CD4+ T cells transduced with FOXP3 isoforms. ***P < 0.001, NS, not significant.
In addition to restored FOXP3 expression, FOXP3KOGFP CD4+ T cells transduced with LV-FOXP3FL or LV-FOXP3Δ2 displayed a Treg-like phenotype (CD25high CD127low FOXP3positive), which was comparable to that of expanded Tregs, evaluated as a control (Figures 4A and S4A), and data are summarized in Table 1. Further and consistent with the suggested role for FOXP3Δ2 in inducing the expression of LAP (latency-associated peptide) and GARP (glycoprotein A repetitions predominant) (28–31), we found that these Treg-related proteins were significantly higher in activated FOXP3δ2-transduced FOXP3KOGFP CD4+ T cells than in FOXP3FL-transduced FOXP3KOGFP CD4+ T cells (Figures 4B and S5A, B). LAP and GARP mRNA expressions were significantly higher in the FOXP3Δ2-transduced FOXP3KOGFP CD4+ T cells than in FOXP3FL-transduced FOXP3KOGFP CD4+ T cells (Figure 4C). FOXP3KOGFP CD4+ T cells transduced with either FOXP3 isoform had a comparable suppressive activity in inhibitions of primary CD4+ T cell proliferation upon activation (suppression index FOXP3FL 44.5 ± 4.0 (R:S = 1:1), FOXP3Δ2 47.2 ± 5.3 (R:S = 1:1), MEAN ± SEM, n = 4) (Figures 5A, B). The suppression level of the FOXP3KOGFP CD4+ T cells transduced with either FOXP3 isoform was lower than that observed with expanded WT Tregs (suppression index expanded WT Tregs 89.9 ± 1.0 (R:S = 1:1), MEAN ± SEM, n = 4) (Figure S4B), expanded WT Tregs in the literature (32), and also lower than the previously reported WT CD4+ T cells converted into Treg with FOXP3 FL only (25, 26). Finally, and consistent with the known FOXP3-mediated suppression of specific cytokines, the activated FOXP3KOGFP CD4+ T cells transduced with each isoform had a significantly reduced production of IL-2, IL-4, and IFN-γ when compared to untransduced or LV-NGFR-transduced cells (Figure 5C). Further, although not statistically significant, IL-17A and IL-22 expression also trended down, when compared to untransduced or LV-NGFR-transduced cells. These results indicate that the LV-mediated transfer of either FOXP3FL or FOXP3Δ2 into FOXP3KOGFP CD4+ T cells can induce a Treg-like phenotype including suppression of cytokine production but that the suppressive function induced by each isoform alone is significantly less than that observed in expanded WT Tregs.
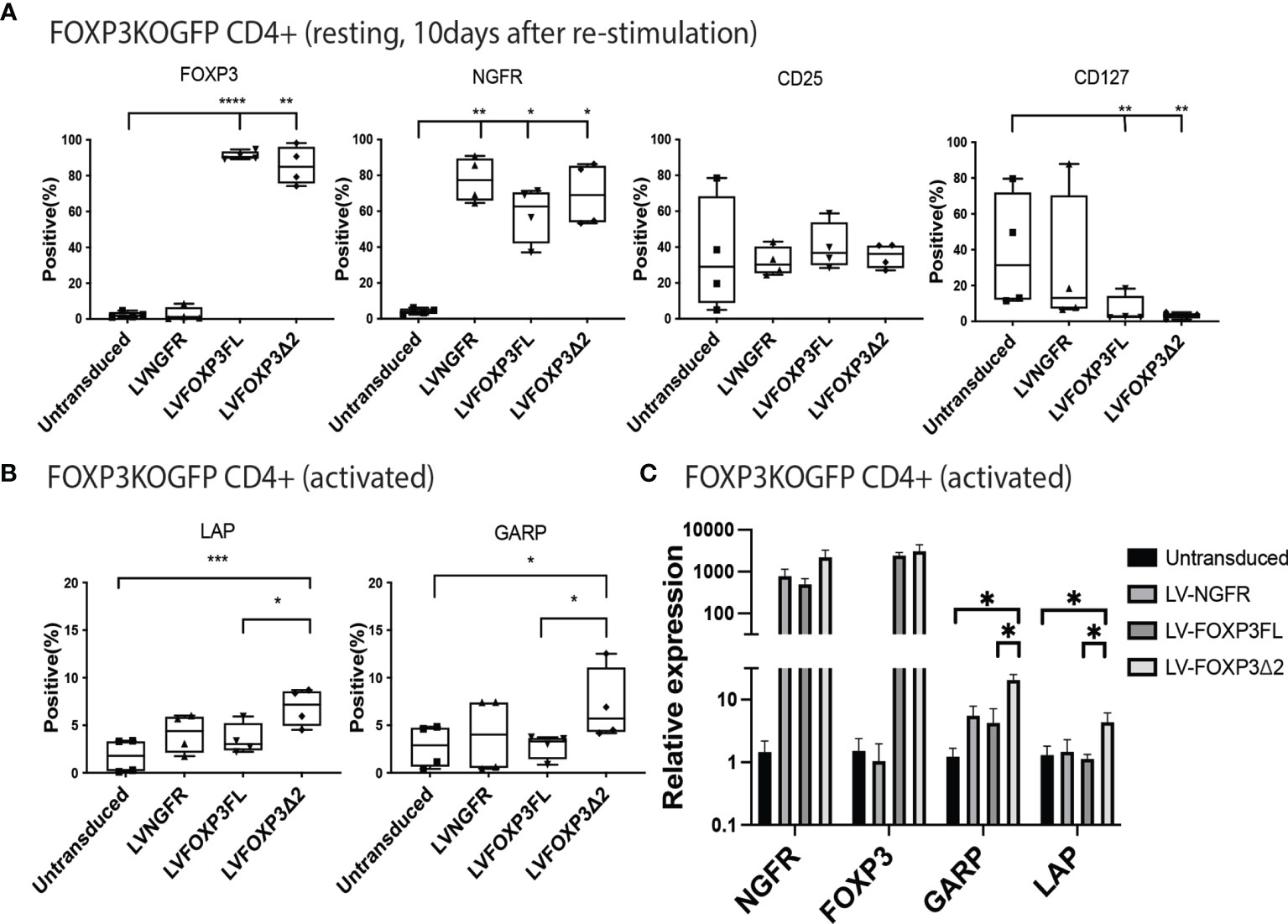
Figure 4 Single transduction of different isoforms provides a Treg-like phenotype in FOXP3KOGFP CD4+ T cells. (A, B) FACS staining of FOXP3KOGFP CD4+ T cells transduced with single FOXP3 isoforms (n = 4). (C) qPCR analysis of FOXP3KOGFP CD4+ T cells transduced single FOXP3 isoforms (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
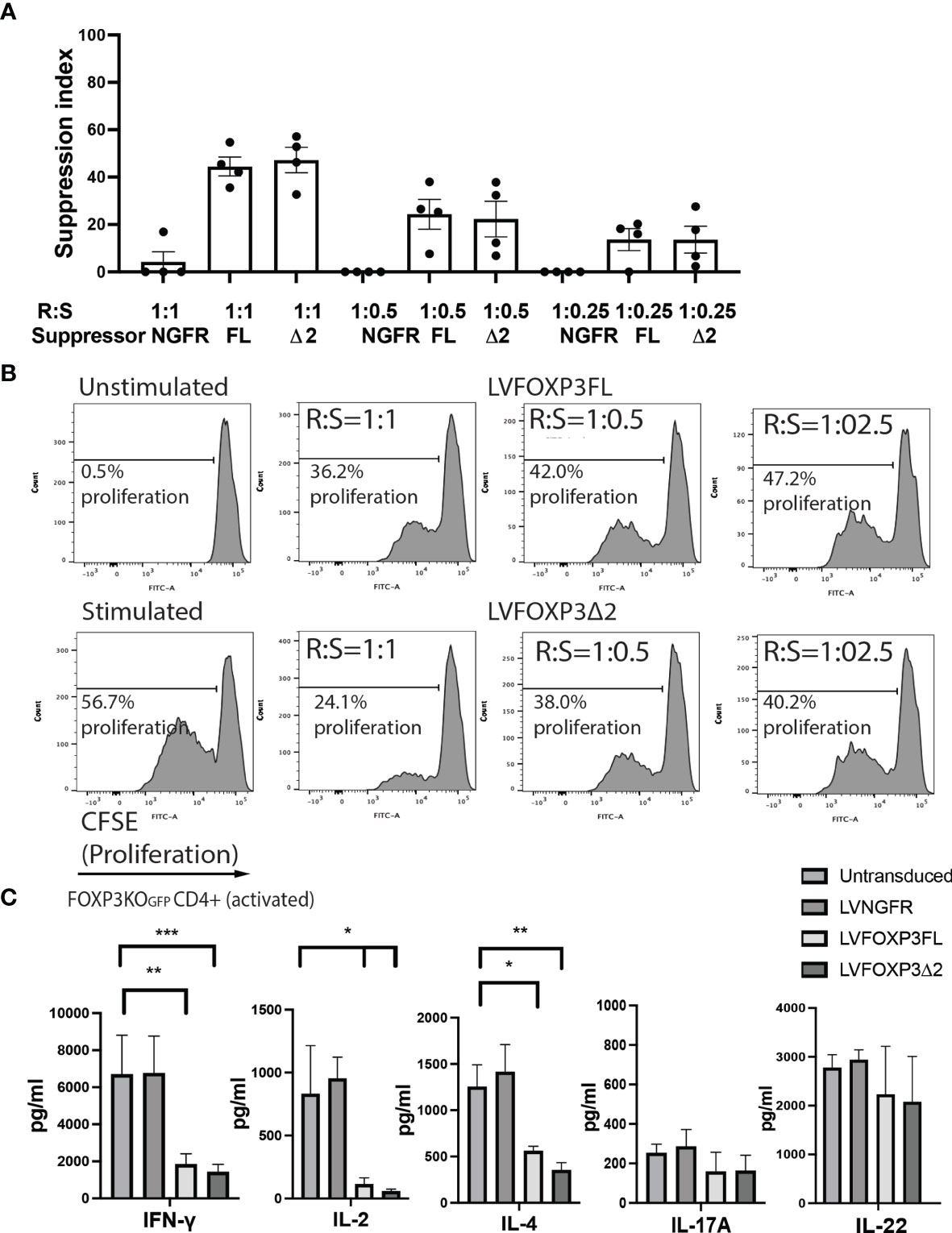
Figure 5 Single transduction of different isoforms provides Treg-like function in FOXP3KOGFP CD4+ T cells. (A, B) Suppressive function of FOXP3KOGFP CD4+ T cells transduced with single FOXP3 isoforms (n = 4). (C) Cytokine production profile of FOXP3KOGFP CD4+ T cells transduced with FOXP3 isoforms (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001.
Co-Expression of FOXP3FL and FOXP3Δ2 Provides Optimal Treg-Like Phenotype and Function in FOXP3KOGFP CD4+ T Cells
As expression of each isoform alone resulted in suboptimal suppressive function, we next evaluated whether co-expression of both FOXP3 isoforms could improve this activity. For this experiment, we generated additional LV-FOXP3 isoform constructs that replaced the NGFR sequence with the open reading frame for EGFR, thus allowing us to sequentially transduce the FOXP3KOGFP CD4+ T cells and select for cells transduced with both constructs (Figure 6A). Specifically, we generated FOXP3KOGFP CD4+ T cells expressing 1) FOXP3FL only (FOXP3FL-NGFR+ FOXP3FL-EGFR); 2) FOXP3Δ2 only (FOXP3Δ2-NGFR+ FOXP3Δ2-EGFR); 3) FOXP3FL-NGFR + FOXP3Δ2-EGFR, and control; and 4) NGFR + EGFR (Figure 6B). In all cases, the VCN was almost identical (FOXP3FL/FL 4.08 ± 0.07 copy/cell, FOXP3Δ2/Δ2 4.09 ± 0.13 copy/cell, FOXP3FL/Δ2 4.22 ± 0.10 copy/cell and NGFR/EGFR 3.92 ± 0.12 copy/cell, MEAN±SEM, n = 4) (Figure 6C). Furthermore, RT-PCR and immunoblotting showed similar expression levels of mRNA and protein for each FOXP3 isoform (Figure 6D).
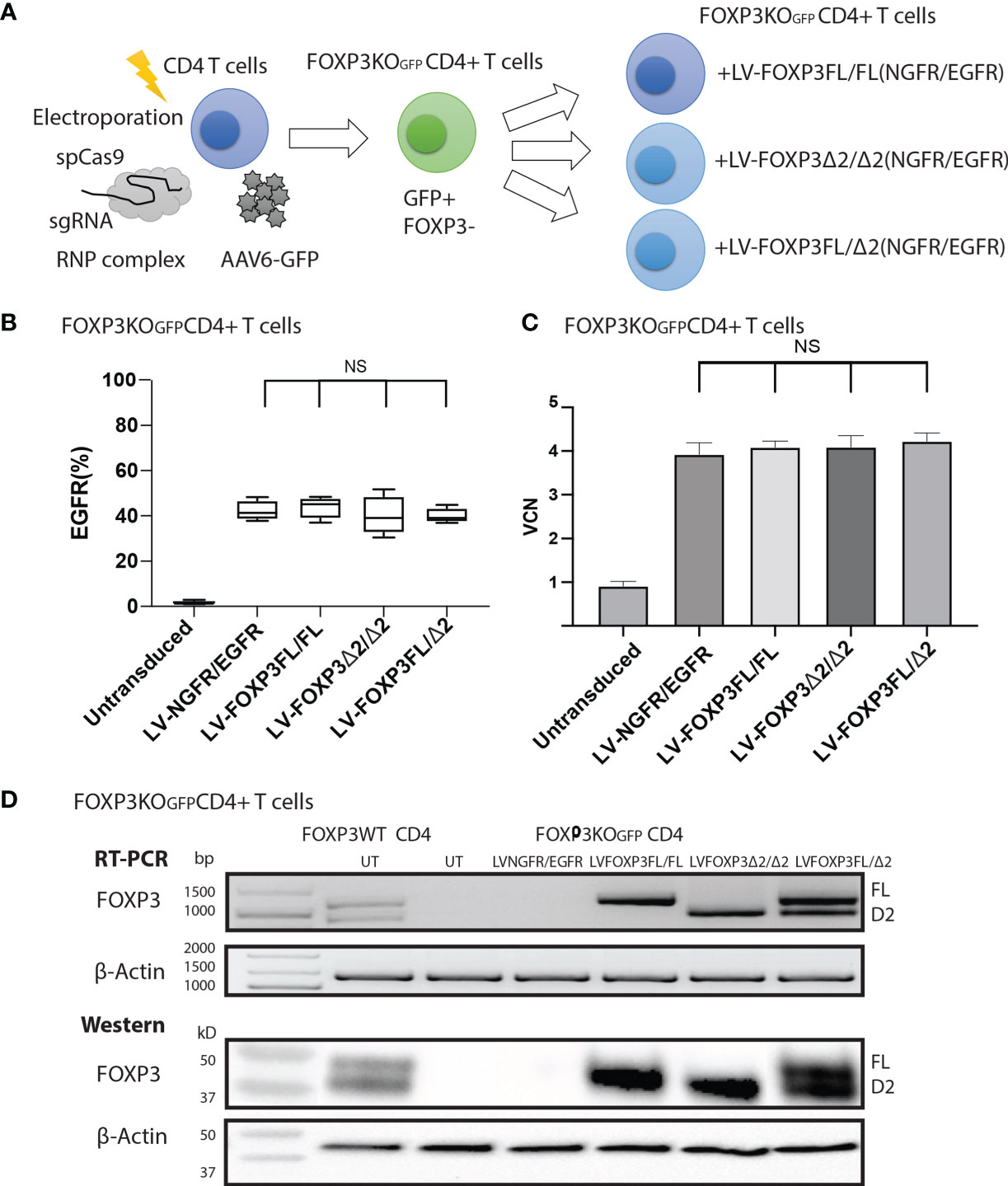
Figure 6 FOXP3KOGFP CD4+ T cells can be sequentially transduced with dual FOXP3 isoforms. FOXP3KOGFP CD4+ T cells were sequentially transduced with FOXP3FL (NGFR) and FOXP3Δ2 (EGFR). (A) Sequential LV transduction strategy of FOXP3 knockout CD4+ T cells. (B) Transduction efficiency of FOXP3KOGFP CD4+ T cells (n = 4). (C) VCN analysis of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms. (D) RT-PCR and immunoblotting of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms. NS, not significant.
We next analyzed various Treg-cell features and functions under all four transduction conditions, and the untransduced control and data are summarized in Table 1. Transduction of FOXP3FL only and FOXP3Δ2 only resulted in cells positive for FOXP3 expression. Further, and together with FOXP3FL + FOXP3Δ2, FOXP3FL only and FOXP3Δ2 only conferred Treg-like phenotypes (CD25high CD127low FOXP3 positive) to the transduced FOXP3KOGFP CD4+ T cells. Double transduction of FOXP3FL + FOXP3Δ2 led to a significantly higher overall FOXP3 protein expression, when compared with double transduction and expression of each isoform alone (Figure 7A). The control NGFR+EGFR transduced cells were indistinguishable from the untransduced cells, confirming the need for FOXP3 expression in the development of Treg features. Interestingly, LAP and GARP expression was significantly higher in the FOXP3Δ2 only and FOXP3FL + FOXP3Δ2-transduced cells (Figure 7B) when compared with FOXP3FL only or control conditions, confirming the requirement for FOXP3Δ2 for the expression of these proteins (Figures S6A, B). LAP and GARP mRNA expression, confirmed by qPCR, was significantly higher in the FOXP3Δ2 only and FOXP3FL + FOXP3Δ2-transduced cells. However, in contrast to the protein expression, FOXP3 mRNA expression was similar in all the three conditions (FOXP3FL only, FOXP3Δ2 only, and FOXP3FL + FOXP3Δ2) (Figure 7C).
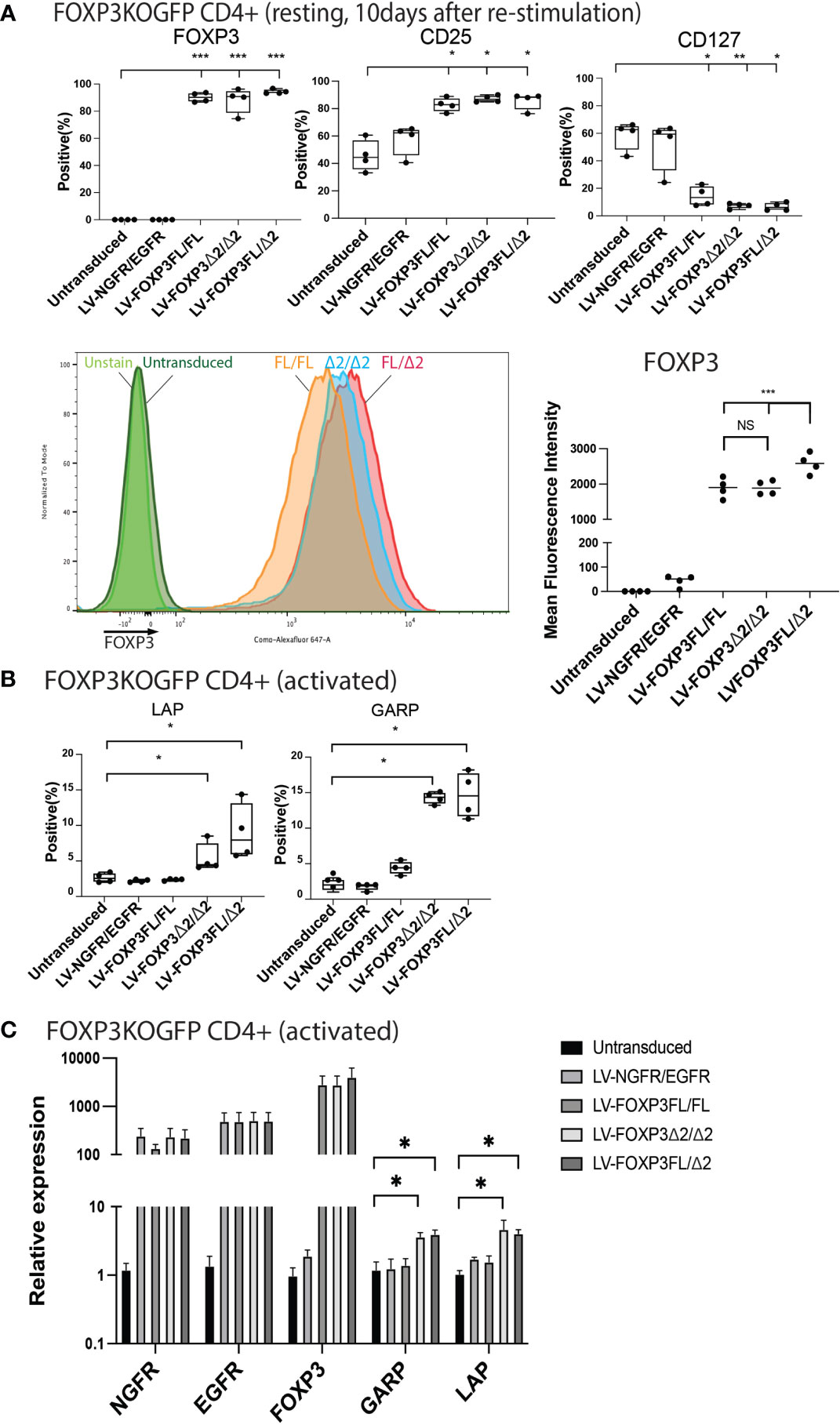
Figure 7 Dual transduction of different isoforms provides Treg-like phenotype in FOXP3KOGFP CD4+ T cells. FOXP3KOGFP CD4+ T cells were sequentially transduced with FOXP3FL(NGFR)/FOXP3FL(EGFR), FOXP3δ2(NGFR)/FOXP3δ2(EGFR), and FOXP3FL(NGFR)/FOXP3δ2(EGFR). (A, B) FACS staining of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms (n = 4). (C) qPCR analysis of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms (n = 4). NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
Finally, we analyzed the suppressive function induced by the FOXP3KOGFP CD4+ T cells sequentially transduced with either FOXP3FL, FOXP3Δ2, or FOXP3FL+FOXP3δ2. Strikingly, we found that FOXP3KOGFP CD4+ T cells transduced with FOXP3FL+FOXP3Δ2 had a significantly higher suppressive function (suppression index FOXP3FL/Δ2 88.0 ± 0.6 (R:S = 1:1), MEAN±SEM, n = 4) when compared to double transduction of FOXP3FL only or FOXP3Δ2 only (suppression index FOXP3FL/FL 57.5 ± 5.7 (R:S = 1:1), FOXP3Δ2/Δ2 60.1 ± 6.9 (R:S = 1:1), MEAN ± SEM, n = 4), or controls (Figures 8A, B). Importantly, this level of suppression was comparable to that observed with expanded WT Tregs (suppression index expanded WT Tregs 89.9 ± 1.0 (R:S = 1:1), MEAN ± SEM, n = 4) (Figure S4B), supporting our hypothesis that expression of both isoforms is required for optimal Treg-suppressive activities. All three transduction conditions, FOXP3FL only, FOXP3Δ2 only, and FOXP3FL+FOXP3Δ2, led to comparable and significant reductions in IL-2, IL-4, and IFN-γ production (Figure 8C), when compared to NGFR+EGFR-transduced or untransduced activated FOXP3KOGFP CD4+ T cell controls, suggesting that the expression of either isoform is sufficient, a view that is supported by our single transduction findings (Table 1). Moreover, when compared to transduction of FOXP3FL or FOXP3Δ2 only, FOXP3FL+FOXP3Δ2 led to a significant reduction in IL-22 production and a visible, although not statistically significant, reduction of IL-17A, suggesting that complementary activities of both isoforms are required to reduce the expression of IL-17A and IL-22. Overall, these data support the view that optimal suppressive function requires the expression of both FOXP3FL and FOXP3Δ2, suggesting that each FOXP3 isoform has distinct and key roles in conferring Treg-mediated suppression. In addition, the data suggest that while either isoform can induce a reduction in IL-2, IL-4, and IFN-γ production, both isoforms are required to reduce IL-17A and IL-22. We conclude that simultaneous expression of both FOXP3FL and FOXP3Δ2 is required for optimal overall FOXP3 protein levels, acquisition of Treg-like phenotypes, and Treg-suppressive functions.
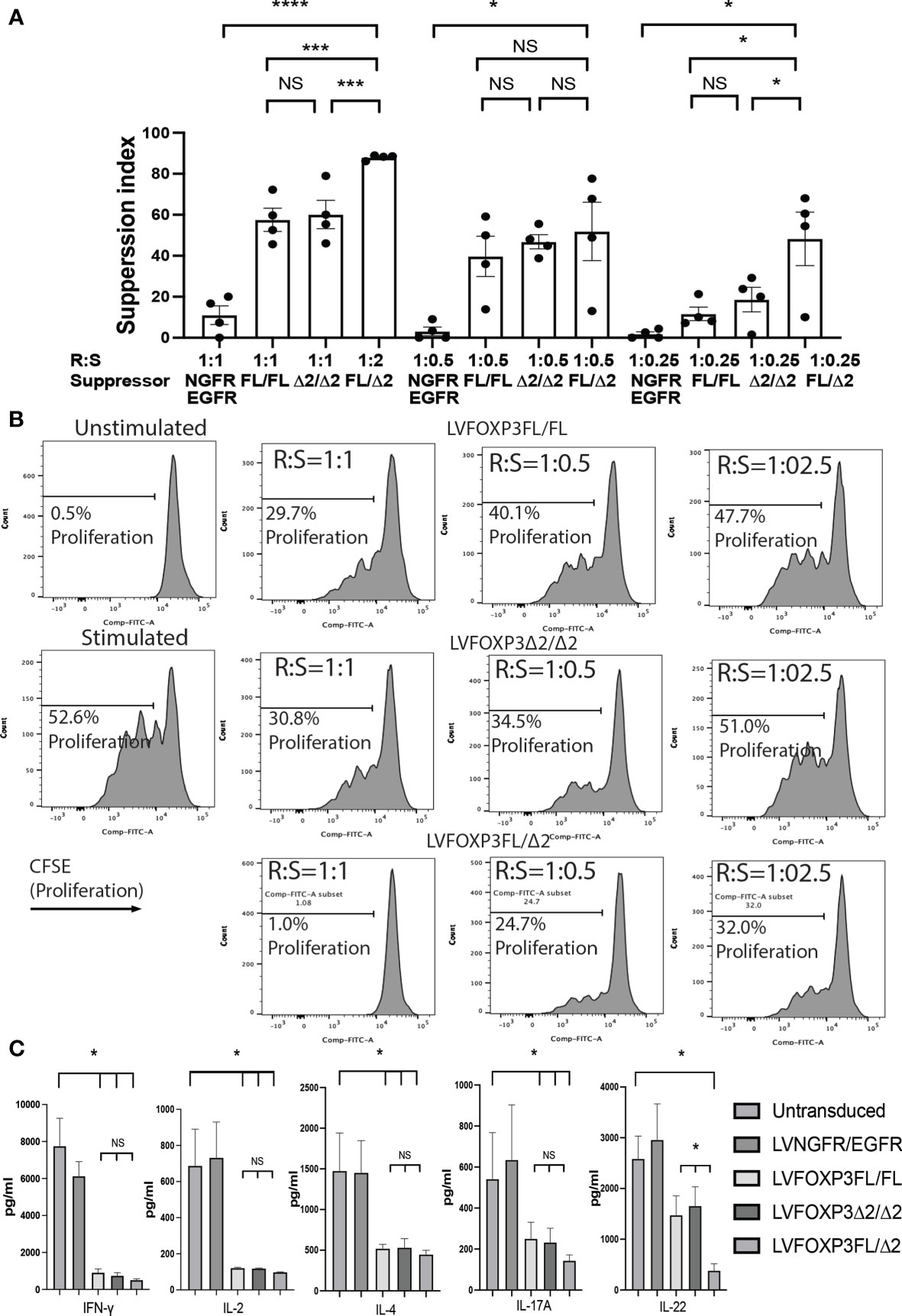
Figure 8 Dual transduction of different isoforms provides Treg-like function in FOXP3KOGFP CD4+ T cells. (A, B) Suppressive function of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms (n = 4). (C) Cytokine production profile of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms (n = 4). NS, not significant, *P < 0.05, ***P < 0.001, ****P < 0.0001.
Discussion
Human Tregs predominantly express FOXP3 as two isoforms, FOXP3FL and FOXP3Δ2 (16). While some functional similarities and differences of each FOXP3 isoform have been determined, a conclusive analysis of their respective roles in the maintenance of overall FOXP3 protein levels and the impact of each on the induction of Treg suppressive functions has not been clarified (13).
The creation of primary CD4+ T cells in which the endogenous FOXP3 gene is disrupted, followed by LV-mediated transfer of FOXPFL and/or FOXP3Δ2, has made it possible to dissect roles for each isoform, in the absence of genomic FOXP3 expression.
Using this system, we show that LV-mediated transfer of both FOXPFL and FOXP3Δ2, and not each alone, leads to the optimal acquisition of a Treg-like phenotype and suppressive functions in transduced FOXP3KOGFP CD4+ T cells in vitro. The suppressive activity achieved when both isoforms are present is comparable to that of expanded WT Tregs. Further, the expression of either isoform alone or the simultaneous expression of both leads to comparable and significant reductions in the production of hallmark cytokines IL-2, IL-4, and IFN-γ production, indicating that either FOXP3 isoform is sufficient for this reduction. In contrast, only the co-expression of FOXP3FL+ FOXP3Δ2 leads to a significant reduction in IL-22 and IL-17A production, suggesting that complementary activities of both isoforms are required to reduce the expression of these pro-inflammatory cytokines. These data support several studies that have analyzed Tregs from patients with non-monogenic autoimmune and inflammatory diseases including psoriasis (33), rheumatoid arthritis (34–37), inflammatory bowel disease (38), and coronary artery disease (39) that have demonstrated an aberrant increase in FOXP3FL relative to FOXP3Δ2, suggesting that FOXP3Δ2, or a fine balance between the two isoforms, might be important to maintain the physiological regulatory function and stability of Tregs. Previous work has also shown that FOXP3Δ2 lacks the binding sites to interact with transcriptional regulators, Signal Transducer and Activator of Transcription (STAT)-3, and retinoic acid receptor-related orphan nuclear receptor (ROR) family members, RORγT and RORα. Zhou and colleagues found that loss of RORγT binding leads to an increased expression of IL-17A, a precursor to T helper (Th) 17 lineage development, suggesting a role for regulated FOXP3Δ2 expression in determining Treg/Th17 fate lineages (40). The data we present here support these findings.
Additional FOXP3 isoform-specific expression data were published by Joly et al., who showed that FOXP3Δ2, but not FOXPFL, induces transcription of GARP, a key mediator of the suppressive activities of Tregs (28–31, 41). Our data also suggest that the expression of GARP, as well as LAP, is preferentially induced by FOXP3Δ2 isoform transduction (alone or co-transduced with FOXP3FL). Wang and colleagues showed that GARP-expressing Tregs have a more potent suppressive function than those that do not express GARP (29). Furthermore, Edwards et al. found that GARP-expressing Tregs have reduced IL-17 production capacity compared to non-GARP-expressing Tregs. At the same time, they showed that expression of GARP inhibits Th17 differentiation and promotes Treg differentiation in the presence of IL-2 (30). Finally, Gandhi et al. have shown that, together with GARP, LAP is associated with a suppressive function of Tregs (42). Importantly, however, despite a role for FOXP3Δ2 in induction of GARP, and a clear function for GARP in Treg-suppressive activities, we observe the best suppressive function when the FOXP3KOGFP CD4+ T cells co-express both isoforms, supporting a role for both FOXP3FL and FOXP3Δ2 in mediating optimal Treg-suppressive function. Altogether, these previous observations and our present data imply that the presence of both FOXP3 isoforms can convert FOXP3KOGFP CD4+ T cells into more suppressive Treg-like cells with a higher expression of GARP and reduced production of IL-17A.
To date, FOXP3 engineering strategies including gene transfer and gene editing to restore protein expression have exclusively focused on FOXP3FL (43), a strategy that has been effective following different approaches (18, 25, 26, 44, 45). Our approach of autologous gene editing in hematopoietic stem cells (HSC), by which FOXP3FL cDNA is inserted at the endogenous FOXP3 initiation codon using CRISPR/Cas9, uniquely allows the expression of the inserted FOXP3FL cDNA to be expressed using the endogenous FOXP3 initiation codon and regulated by physiologic, developmental, and lineage-specific signals. Results obtained so far, in normal or IPEX patient mature Treg and Teff cells in vitro, demonstrated that this method results in the restoration of normal FOXP3 expression in Teff cells, but only 50% of normal expression in Tregs and suboptimal Treg-suppressive activity. Similarly, the T cell compartment obtained upon in vivo differentiation of FOXP3FL-edited HSCs contains a lower than expected frequency of FOXP3-expressing Tregs, when compared to unedited HSCs. These data suggest that the expression of FOXP3FL alone might not be sufficient to restore all the endogenous FOXP3 functions required for normal T cell ontogeny and a normal immune system development (18). However, we also cannot discount the possibility that, in addition to the absence of FOXP3Δ2, the endogenous FOXP3 introns that are lacking in the FOXP3 cDNA might contain key regulatory elements required for optimal FOXP3 expression (46).
As an alternative strategy, it was shown that dual transduction of FOXP3FL and Helios isoforms could convert CD4+ T cells into Treg-like cells (47), indicating the presence of additional factors that can facilitate the Treg conversion by FOXP3FL gene transfer alone.
Dual transduction of both FOXP3 isoforms could fully restore the suppressive function of FOXP3 knockout CD4+ T cells. Compared to the single transduction, FOXP3 knockout CD4+ T cells transduced with both isoforms acquired a Treg-like phenotype (CD25high CD127low FOXP3high). In vitro suppressive function was statistically higher in FOXP3 knockout CD4+ T cells transduced with dual isoforms compared to those transduced with either FOXP3FL or FOXP3Δ2 alone. These data indicate that both FOXP3FL and FOXP3Δ2 are required to convert FOXP3 knockout CD4+ T cells into functional Tregs.
Here we have successfully shown that FOXP3FL and FOXP3Δ2 have synergistic, non-overlapping effects in the conversion of FOXP3KOGFP CD4+ T cells into Treg-like cells, including the expression of molecules that have a direct role in the suppression and inhibition of inflammatory cytokines such as IL-17 and IL-22, which could not be inhibited by either isoform alone. In addition, the FOXP3FL/Δ2-transduced cells showed the highest level of suppression of proliferation of activated Teff cells. Our data also exclude the possible contribution of endogenous FOXP3 expression in the generation of the Treg function we observe and at the same time exclude that these findings are simply due to the over-transduction since the VCN in the different cell type is comparable. Our present observations highlight the synergistic effects between FOXP3FL and FOXP3Δ2 and suggest that the restoration of both FOXP3FL or FOXP3Δ2 may be required to provide fully functional Tregs. Our findings imply that restoration of both FOXP3 isoforms leads to a more physiological rescue of Treg-cell features and functions, therefore supporting further investigation as a molecular therapeutic approach for the correction of FOXP3-deficient T cells and Tregs.
In conclusion, we have shown that complementary functional regulatory activity is conferred by FOXP3FL and FOXP3Δ2 isoform gene transfer in FOXP3KOGFP CD4+ T cells. As such, maintenance of the expression of both isoforms must be further investigated in future gene editing strategies aimed at fully restoring FOXP3 functions in FOXP3-deficient cells such as those in IPEX syndrome patients.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YS conducted the experiments and wrote the manuscript. JL conducted the experiments and wrote the manuscript. EL contributed to experimental development. RP contributed to the experimental design and manuscript editing and writing. MGR and RB designed the experiments and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Stanford Maternal and Child Health Research Institute Faculty Scholar Grant to RB. YS was supported by the California Institute for Regenerative Medicine (CLIN1 11591 to RB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Megan K Levings (University of British Columbia) for providing LV-FOXP3(FOXP3Δ2). We thank Dr. Volker Wiebking and Dr. Matthew Porteus (Stanford University) for providing the EGFR construct. We thank Dr. Robertson Parkman (Stanford University) for the scientific advice and manuscript correction. Dr. Bacchetta is an endowed Anne T. and Robert M. Bass Faculty Scholar in Pediatric Cancer and Blood Diseases, Stanford University School of Medicine.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.752394/full#supplementary-material
Supplementary Figure 1 | HEK293T/17 cells transduced with FOXP3 isoforms (A) Lentiviral vector construct. Cytomegalovirus minimal promoter (mCMV), Human elongation factor-alpha promoter (EF-1α) (B) Expression of marker genes (NGFR and EGFR) (C) Expression of FOXP3. Representative FACS dot-plot of HEK293T/17 cells transduced with FOXP3 isoforms. (D) Expression of marker genes (NGFR and EGFR) (E) Expression of FOXP3.
Supplementary Figure 2 | Representative FACS dot-plot of MT-2 cells transduced with FOXP3 isoforms. (A) Expression of marker genes (NGFR and EGFR) (B) Expression of FOXP3 and GFP.
Supplementary Figure 3 | Wild-type MT-2 cells transduced with FOXP3 isoforms. (A) LV transduction strategy of WT MT-2 cells (B) FOXP3 expression of WT MT-2 cells transduced with FOXP3 isoforms (C) RT-PCR and immunoblotting of WT MT-2 cells transduced with FOXP3 isoforms.
Supplementary Figure 4 | Phenotype and suppressive function of expanded Tregs. (A) FACS staining of expanded Tregs from FOXP3WT CD4+ T cells (n=4) (B) Suppressive function of expanded Tregs from FOXP3WT CD4+ T cells (n=4).
Supplementary Figure 5 | Single transduction of different isoforms provides Treg-like phenotype in FOXP3KOGFP CD4+ T cells. FACS staining of FOXP3KOGFP CD4+ T cells transduced with single FOXP3 isoforms (n=4) shown by representative (A) dot-plot and (B) histograms.
Supplementary Figure 6 | Dual transduction of different isoforms provides Treg-like phenotype in FOXP3KOGFP CD4+ T cells. FACS staining of FOXP3KOGFP CD4+ T cells transduced with dual FOXP3 isoforms (n=4) shown by representative (A) dot-plot and (B) histograms.
References
1. Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic Self-Tolerance Maintained by Activated T Cells Expressing IL-2 Receptor Alpha-Chains (CD25). Breakdown of a Single Mechanism of Self-Tolerance Causes Various Autoimmune Diseases. J Immunol (1995) 155(3):1151–64.
2. Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, et al. CD127 Expression Inversely Correlates With FoxP3 and Suppressive Function of Human CD4+ T Reg Cells. J Exp Med (2006) 203(7):1701–11. doi: 10.1084/jem.20060772
3. Hori S, Nomura T, Sakaguchi S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science (2003) 299(5609):1057–61. doi: 10.1126/science.1079490
4. Rudensky AY. Regulatory T Cells and Foxp3. Immunol Rev (2011) 241(1):260–8. doi: 10.1111/j.1600-065X.2011.01018.x
5. Bacchetta R, Barzaghi F, Roncarolo MG. From IPEX Syndrome to FOXP3 Mutation: A Lesson on Immune Dysregulation. Ann NY Acad Sci (2018) 1417(1):5–22. doi: 10.1111/nyas.13011
6. Barzaghi F, Amaya Hernandez LC, Neven B, Ricci S, Kucuk ZY, Bleesing JJ, et al. Long-Term Follow-Up of IPEX Syndrome Patients After Different Therapeutic Strategies: An International Multicenter Retrospective Study. J Allergy Clin Immunol (2018) 141(3):1036–49.e5. doi: 10.1016/j.jaci.2017.10.041
7. Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, et al. Defective Regulatory and Effector T Cell Functions in Patients With FOXP3 Mutations. J Clin Invest (2006) 116(6):1713–22. doi: 10.1172/JCI25112
8. Levy-Lahad E, Wildin RS. Neonatal Diabetes Mellitus, Enteropathy, Thrombocytopenia, and Endocrinopathy: Further Evidence for an X-Linked Lethal Syndrome. J Pediatr (2001) 138(4):577–80. doi: 10.1067/mpd.2001.111502
9. Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, et al. X-Linked Neonatal Diabetes Mellitus, Enteropathy and Endocrinopathy Syndrome Is the Human Equivalent of Mouse Scurfy. Nat Genet (2001) 27(1):18–20. doi: 10.1038/83707
10. Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked Syndrome (IPEX) Is Caused by Mutations of FOXP3. Nat Genet (2001) 27(1):20–1. doi: 10.1038/83713
11. Ramsdell F, Ziegler SF. FOXP3 and Scurfy: How It All Began. Nat Rev Immunol (2014) 14(5):343–9. doi: 10.1038/nri3650
12. Joly AL, Liu S, Dahlberg CI, Mailer RK, Westerberg LS, Andersson J. Foxp3 Lacking Exons 2 and 7 Is Unable to Confer Suppressive Ability to Regulatory T Cells In Vivo. J Autoimmun (2015) 63:23–30. doi: 10.1016/j.jaut.2015.06.009
13. Mailer RKW. Alternative Splicing of FOXP3-Virtue and Vice. Front Immunol (2018) 9:530. doi: 10.3389/fimmu.2018.00530
14. Mailer RK. IPEX as a Consequence of Alternatively Spliced Foxp3. Front Pediatr (2020) 8:594375. doi: 10.3389/fped.2020.594375
15. Allan SE, Passerini L, Bacchetta R, Crellin N, Dai M, Orban PC, et al. The Role of 2 FOXP3 Isoforms in the Generation of Human CD4+ Tregs. J Clin Invest (2005) 115(11):3276–84. doi: 10.1172/JCI24685
16. Ziegler SF. FOXP3: Of Mice and Men. Annu Rev Immunol (2006) 24:209–26. doi: 10.1146/annurev.immunol.24.021605.090547
17. Frith K, Joly AL, Ma CS, Tangye SG, Lohse Z, Seitz C, et al. The FOXP3Delta2 Isoform Supports Treg Cell Development and Protects Against Severe IPEX Syndrome. J Allergy Clin Immunol (2019) 144(1):317–20.e8. doi: 10.1016/j.jaci.2019.03.003
18. Sato Y, Passerini L, Piening BD, Uyeda MJ, Goodwin M, Gregori S, et al. Human-Engineered Treg-Like Cells Suppress FOXP3-Deficient T Cells But Preserve Adaptive Immune Responses In Vivo. Clin Transl Immunol (2020) 9(11):e1214. doi: 10.1002/cti2.1214
19. Goodwin M, Lee E, Lakshmanan U, Shipp S, Froessl L, Barzaghi F, et al. CRISPR-Based Gene Editing Enables FOXP3 Gene Repair in IPEX Patient Cells. Sci Adv (2020) 6(19):eaaz0571. doi: 10.1126/sciadv.aaz0571
20. Miyoshi I, Kubonishi I, Yoshimoto S, Akagi T, Ohtsuki Y, Shiraishi Y, et al. Type C Virus Particles in a Cord T-Cell Line Derived by Co-Cultivating Normal Human Cord Leukocytes and Human Leukaemic T Cells. Nature (1981) 294(5843):770–1. doi: 10.1038/294770a0
21. Chen S, Ishii N, Ine S, Ikeda S, Fujimura T, Ndhlovu LC, et al. Regulatory T Cell-Like Activity of Foxp3+ Adult T Cell Leukemia Cells. Int Immunol (2006) 18(2):269–77. doi: 10.1093/intimm/dxh366
22. Hamano R, Wu X, Wang Y, Oppenheim JJ, Chen X. Characterization of MT-2 Cells as a Human Regulatory T Cell-Like Cell Line. Cell Mol Immunol (2015) 12(6):780–2. doi: 10.1038/cmi.2014.123
23. Battaglia M, Stabilini A, Migliavacca B, Horejs-Hoeck J, Kaupper T, Roncarolo MG. Rapamycin Promotes Expansion of Functional CD4+CD25+FOXP3+ Regulatory T Cells of Both Healthy Subjects and Type 1 Diabetic Patients. J Immunol (2006) 177(12):8338–47. doi: 10.4049/jimmunol.177.12.8338
24. Battaglia M, Stabilini A, Tresoldi E. Expanding Human T Regulatory Cells With the mTOR-Inhibitor Rapamycin. Methods Mol Biol (2012) 821:279–93. doi: 10.1007/978-1-61779-430-8_17
25. Allan SE, Alstad AN, Merindol N, Crellin NK, Amendola M, Bacchetta R, et al. Generation of Potent and Stable Human CD4+ T Regulatory Cells by Activation-Independent Expression of FOXP3. Mol Ther: J Am Soc Gene Ther (2008) 16(1):194–202. doi: 10.1038/sj.mt.6300341
26. Passerini L, Rossi Mel E, Sartirana C, Fousteri G, Bondanza A, Naldini L, et al. CD4(+) T Cells From IPEX Patients Convert Into Functional and Stable Regulatory T Cells by FOXP3 Gene Transfer. Sci Trans Med (2013) 5(215):215ra174. doi: 10.1126/scitranslmed.3007320
27. Charrier S, Ferrand M, Zerbato M, Precigout G, Viornery A, Bucher-Laurent S, et al. Quantification of Lentiviral Vector Copy Numbers in Individual Hematopoietic Colony-Forming Cells Shows Vector Dose-Dependent Effects on the Frequency and Level of Transduction. Gene Ther (2011) 18(5):479–87. doi: 10.1038/gt.2010.163
28. Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) Is Essential for the Surface Expression of Latent TGF-Beta on Platelets and Activated FOXP3+ Regulatory T Cells. Proc Natl Acad Sci USA (2009) 106(32):13445–50. doi: 10.1073/pnas.0901944106
29. Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP Selectively Identifies Activated Human FOXP3+ Regulatory T Cells. Proc Natl Acad Sci USA (2009) 106(32):13439–44. doi: 10.1073/pnas.0901965106
30. Edwards JP, Fujii H, Zhou AX, Creemers J, Unutmaz D, Shevach EM. Regulation of the Expression of GARP/latent TGF-Beta1 Complexes on Mouse T Cells and Their Role in Regulatory T Cell and Th17 Differentiation. J Immunol (2013) 190(11):5506–15. doi: 10.4049/jimmunol.1300199
31. Zhou AX, Kozhaya L, Fujii H, Unutmaz D. GARP-TGF-Beta Complexes Negatively Regulate Regulatory T Cell Development and Maintenance of Peripheral CD4+ T Cells In Vivo. J Immunol (2013) 190(10):5057–64. doi: 10.4049/jimmunol.1300065
32. Passerini L, Barzaghi F, Curto R, Sartirana C, Barera G, Tucci F, et al. Treatment With Rapamycin can Restore Regulatory T-Cell Function in IPEX Patients. J Allergy Clin Immunol (2020) 145(4):1262–71.e13. doi: 10.1016/j.jaci.2019.11.043
33. Chen L, Wu J, Pier E, Zhao Y, Shen Z. Mtorc2-PKBalpha/Akt1 Serine 473 Phosphorylation Axis Is Essential for Regulation of FOXP3 Stability by Chemokine CCL3 in Psoriasis. J Invest Dermatol (2013) 133(2):418–28. doi: 10.1038/jid.2012.333
34. Ryder LR, Woetmann A, Madsen HO, Odum N, Ryder LP, Bliddal H, et al. Expression of Full-Length and Splice Forms of FoxP3 in Rheumatoid Arthritis. Scand J Rheumatol (2010) 39(4):279–86. doi: 10.3109/03009740903555374
35. Ryder LR, Bartels EM, Woetmann A, Madsen HO, Odum N, Bliddal H, et al. FoxP3 mRNA Splice Forms in Synovial CD4+ T Cells in Rheumatoid Arthritis and Psoriatic Arthritis. APMIS (2012) 120(5):387–96. doi: 10.1111/j.1600-0463.2011.02848.x
36. Ryder LR, Ryder LP, Bartels EM, Woetmann A, Madsen HO, Odum N, et al. Differential Effects of Decoy Receptor- and Antibody-Mediated Tumour Necrosis Factor Blockage on FoxP3 Expression in Responsive Arthritis Patients. APMIS (2013) 121(4):337–47. doi: 10.1111/apm.12004
37. Suzuki K, Setoyama Y, Yoshimoto K, Tsuzaka K, Abe T, Takeuchi T. Decreased mRNA Expression of Two FOXP3 Isoforms in Peripheral Blood Mononuclear Cells From Patients With Rheumatoid Arthritis and Systemic Lupus Erythematosus. Int J Immunopathol Pharmacol (2011) 24(1):7–14. doi: 10.1177/039463201102400102
38. Lord JD, Valliant-Saunders K, Hahn H, Thirlby RC, Ziegler SF. Paradoxically Increased FOXP3+ T Cells in IBD Do Not Preferentially Express the Isoform of FOXP3 Lacking Exon 2. Dig Dis Sci (2012) 57(11):2846–55. doi: 10.1007/s10620-012-2292-3
39. Lundberg AK, Jonasson L, Hansson GK, Mailer RKW. Activation-Induced FOXP3 Isoform Profile in Peripheral CD4+ T Cells Is Associated With Coronary Artery Disease. Atherosclerosis (2017) 267:27–33. doi: 10.1016/j.atherosclerosis.2017.10.026
40. Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, et al. TGF-Beta-Induced Foxp3 Inhibits T(H)17 Cell Differentiation by Antagonizing RORgammat Function. Nature (2008) 453(7192):236–40. doi: 10.1038/nature06878
41. Joly AL, Seitz C, Liu S, Kuznetsov NV, Gertow K, Westerberg LS, et al. Alternative Splicing of FOXP3 Controls Regulatory T Cell Effector Functions and Is Associated With Human Atherosclerotic Plaque Stability. Circ Res (2018) 122(10):1385–94. doi: 10.1161/CIRCRESAHA.117.312340
42. Gandhi R, Farez MF, Wang Y, Kozoriz D, Quintana FJ, Weiner HL. Cutting Edge: Human Latency-Associated Peptide+ T Cells: A Novel Regulatory T Cell Subset. J Immunol (2010) 184(9):4620–4. doi: 10.4049/jimmunol.0903329
43. Ferreira LMR, Muller YD, Bluestone JA, Tang Q. Next-Generation Regulatory T Cell Therapy. Nat Rev Drug Discov (2019) 18(10):749–69. doi: 10.1038/s41573-019-0041-4
44. Masiuk KE, Laborada J, Roncarolo MG, Hollis RP, Kohn DB. Lentiviral Gene Therapy in HSCs Restores Lineage-Specific Foxp3 Expression and Suppresses Autoimmunity in a Mouse Model of IPEX Syndrome. Cell Stem Cell (2019) 24(2):309–17.e7. doi: 10.1016/j.stem.2018.12.003
45. Honaker Y, Hubbard N, Xiang Y, Fisher L, Hagin D, Sommer K, et al. Gene Editing to Induce FOXP3 Expression in Human CD4(+) T Cells Leads to a Stable Regulatory Phenotype and Function. Sci Transl Med (2020) 12(546):eaay6422. doi: 10.1126/scitranslmed.aay6422
46. Rose AB. Introns as Gene Regulators: A Brick on the Accelerator. Front Genet (2018) 9:672. doi: 10.3389/fgene.2018.00672
Keywords: Foxp3, alternative splicing, gene editing (CRISPR/Cas9), gene therapy, suppressive function, regulatory T cell (Treg)
Citation: Sato Y, Liu J, Lee E, Perriman R, Roncarolo MG and Bacchetta R (2021) Co-Expression of FOXP3FL and FOXP3Δ2 Isoforms Is Required for Optimal Treg-Like Cell Phenotypes and Suppressive Function. Front. Immunol. 12:752394. doi: 10.3389/fimmu.2021.752394
Received: 03 August 2021; Accepted: 21 September 2021;
Published: 19 October 2021.
Edited by:
Shohei Hori, The University of Tokyo, JapanReviewed by:
Dipayan Rudra, Immunobiome, South KoreaReiner Karl Walter Mailer, University Medical Center Hamburg-Eppendorf, Germany
Copyright © 2021 Sato, Liu, Lee, Perriman, Roncarolo and Bacchetta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosa Bacchetta, cm9zYWJAc3RhbmZvcmQuZWR1
 Yohei Sato
Yohei Sato Jessica Liu
Jessica Liu Esmond Lee
Esmond Lee Rhonda Perriman
Rhonda Perriman Maria Grazia Roncarolo
Maria Grazia Roncarolo Rosa Bacchetta
Rosa Bacchetta