- Pengiran Anak Puteri Rashidah Sa'adatul Bolkiah (PAPRSB) Institute of Health Sciences, Universiti Brunei Darussalam, Bandar Seri Begawan, Brunei
COVID-19 pandemic remains an on-going global health and economic threat that has amassed millions of deaths. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of this disease and is constantly under evolutionary pressures that drive the modification of its genome which may represent a threat to the efficacy of current COVID-19 vaccines available. This article highlights the pressures that facilitate the rise of new SARS-CoV-2 variants and the key mutations of the viral spike protein – L452R, E484K, N501Y and D614G– that promote immune escape mechanism and warrant a cautionary point for clinical and public health responses in terms of re-infection, vaccine breakthrough infection and therapeutic values.
Introduction
Since the outbreak of the COVID-19 pandemic in late 2019, SARS-CoV-2 virus has caused significant mortality and morbidity worldwide. While some regions of the world are seeing a dwindling of infected cases, others, especially densely populated regions are still recording high levels of infection. Efficient control of viral spread and decrease in mortality has been attributed to rigorous public health measures and global mass immunisation campaigns that have begun in December 2019. Manifestations of COVID-19 may range from mild to severe and life-threatening. While the majority of cases are subclinical, a significant portion of COVID-19 patients develop severe or fatal illness, which is often characterised by acute respiratory distress syndrome and hyper-inflammatory immune responses (1, 2). Epidemiological and public health surveillance efforts utilise viral genome information to track and monitor the growth of the pandemic. There is a growing concern for emerging viral strains that can potentially hamper public health strategies, post-vaccine or natural immunity and antiviral treatments. The World Health Organisation (WHO) has classified emerging variants with reference to the original Wuhan strain into different categories according to its hazard grade, with Variants of Concern (VOCs) as the ones needing urgent monitoring of. Currently, there are four VOCs which include Alpha (B.1.1.7), Beta (B.1351), Gamma (P.1) and Delta (B.1.617.2) variants that are at the front of the pandemic wave, with the latter seemingly outperforming the others. In response to this, unprecedented scale of sequencing and tracking of SARS-CoV-2 genome evolution has been initiated to accommodate the rapid accumulation of viral mutations and to understand the evolutionary adaptation of the virus in humans in hope to better design effective COVID-19 vaccines and treatment options.
Evolutionary Pressure Driving the Emergence of SARS-CoV-2 Variants
SARS-CoV-2 virus is a large RNA virus with a 30-kb genome that encodes for four structural proteins: spike glycoprotein, nucleocapsid, membrane and envelope proteins (3). The genome of SARS-CoV-2 has been reported to accumulate two nucleotide substitutions per month, which is relatively slow for an RNA virus owing to its proofreading 3’–5’exoribonuclease (4). While most chance mutations are often silent that lead to no changes at biological level or deleterious that compromise viral fitness, some may confer selective advantage for its fitness. This leads to their propagation in subsequent viral populations, which carry advantageous phenotypes and are often subjected to a purifying selection. This is evident by a genetic drift reported in SARS-CoV-2 variants particularly in their spike and nucleocapsid gene sequences which are most variable (5). Additionally, evidence of independently co-occurring or converging mutations in SARS-CoV-2 genome also suggests that there exists a persistent and increasing selection pressure on the virus, which can occur both at population and individual patient levels.
In an infected individual, variants can independently inhabit different tissues and up to four variants have reportedly been identified within a patient (6, 7). However, it is unlikely that minor intra-host mutations will propagate and become fixed due to their low frequencies and natural bottleneck effects (8, 9). While minor viral genetic mutations within infected individuals are expected, a genetic drift may raise concern at the population level. Currently, the pandemic has already infected over 200 million cases globally, leaving behind convalescent individuals with natural immunity against the virus. Additionally, as immunisation has begun globally, a growing portion of the population is now carrying vaccine-induced immunity against the circulating virus. The increasing degree of immunity in the human population is inevitably conferring a great deal of selective pressure on the virus that promotes the rise of antibody escape mutants. The emergence of immune escape mutants is perhaps most apparent in chronic COVID-19 patients as documented in multiple reports. Persistent infections with SARS-CoV-2 seen in immunocompromised individuals who cannot effectively fight infection accelerates viral evolution which gives rise to large genomic diversity and mutations in the viral spike, ORF1ab, ORF8 and nsp1 proteins (10, 11). These mutations were observed to recur and independently emerge in these patients (10, 11).
In addition to vaccination, immunotherapies have also been used as COVID-19 disease interventions. These include repurposed drugs such as antiviral remdesivir and corticosteroids, convalescent plasma therapy that carry neutralising antibodies, and antiviral monoclonal antibodies (12, 13). Unfortunately, these may also pose as drivers for advantageous mutations. Reports on the use of convalescent plasma has shown to contribute to the generation of antibody escape variants (14–16). The use of sub-optimal antibodies in convalescent therapy and re-infection at the face of a decaying or a partial primary immunity may provide a selective pressure for immune escape mutations.
Spike Variants Pose a Threat to Current Vaccines and Therapeutics
Spike protein of SARS-CoV-2 defines the tropism of the virus, facilitates its spread, and modulates host immune function. It is the viral entry point of the host cell by which it binds to host angiotensin-converting enzyme 2 (ACE2) receptor. It is composed of 1273 amino acids and harbours two subunits that are bridged by a furin cleavage site. Subunit 1 contains two important domains: N terminal domain (NTD) and receptor binding domain (RBD) (Figure 1). It is also the main target of vaccination efforts as the majority of serum neutralising activity is directed on its RBD (17, 18).
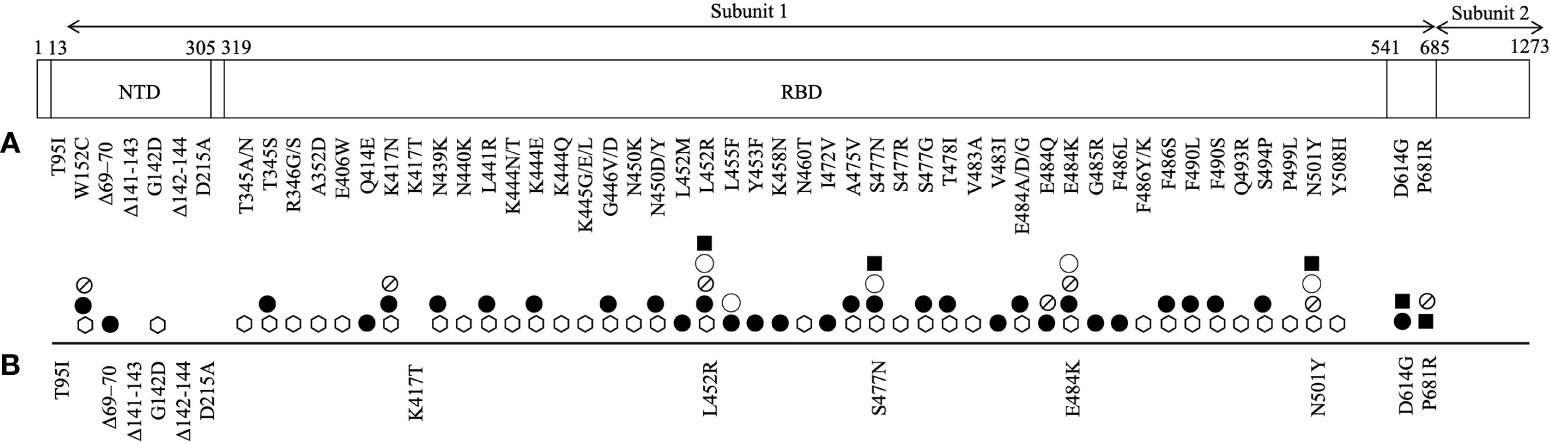
Figure 1 An illustration of SARS-CoV-2 spike protein mutations identified in its subunit 1 that have been (A) shown to promote resistance to neutralizing monoclonal antibodies (⎔) and serum antibodies (from convalescent (●) and/or vaccinated (⦰) individuals), and increase the protein affinity to ACE2 receptor (◯) and viral infectivity (■) and (B) reported in breakthrough and/or secondary infections.
A primary infection with SARS-CoV-2 provokes immune responses that generate potent anti-SARS-CoV-2 neutralising antibodies and CD4+ and CD8+ T cell responses which clear the infection and culminate with an immune response memory to fight future infection. As the pandemic continues to sweep across the globe, it has gradually become apparent that a primary infection with SARS-CoV-2 does not provide a full protection. Although rare, cases of re-infection were documented as early as August 2020 and many are expected to be underreported as it can be difficult to distinguish between re-infection and prolonged viral shedding (19–22). Additionally, a comprehensive whole genome sequencing is necessary to identify two separate episodes of infection. A natural exposure to SARS-CoV-2 generate anti-SARS-CoV-2 IgG antibodies that are largely protective for at least 6 months post-infection (23, 24). Repeat infection can be caused by a low potency and quality of immune memory, or a decline in anti-SARS-CoV-2 antibody titre over time. However, a report of a re-infection case indicates that a secondary infection with SARS-CoV-2 occurred in the presence of intact anti-SARS-CoV-2 antibody titre, suggesting that a seropositivity does not necessarily guarantee immunity and raising concern about immune escape variants that can overcome previous immunity (25).
Reports on vaccine breakthrough infections in vaccinated individuals are also accumulating, raising concern of developing escape mutants in the face of immune selection (26–30). A breakthrough infection is defined as a detection of SARS-CoV-2 antigen or RNA more than 14 days after receiving a COVID-19 vaccine. To date, over 10, 000 breakthrough infections have been reported (31). The durability of vaccine-induced antibodies is yet to be determined, but vaccines against SARS-CoV-2 elicit more robust antibody responses than a natural infection which may suggest a more stable protection (32). Nevertheless, both natural- and vaccine-induced anti-SARS-CoV-2 antibodies demonstrate comparable neutralising power (33).
As of June 14th, a total of 6,238 nucleotide substitutions, 56 insertions and 278 deletions in the spike protein have been recorded in clinical isolates (34, 35). The most profound mark of SARS-CoV-2 evolution is perhaps the D614G mutation that emerged as early as 6 months into the outbreak. D614G mutation is located outside of the RBD (Figure 1). D614G variant bypassed natural bottlenecks due to its selective advantages that lead to improved viral infectivity and transmissibility and reduced viral sensitivity to neutralising convalescent sera (36–38). It immediately outcompeted other sequence groups and became ubiquitous worldwide. All VOCs that are driving the current pandemic waves harbour this dominant mutation.
The main target of serum neutralising antibodies is the RBD of the virus spike protein, which upon recognition prevents the virus from gaining entry to the host cell. A growing list of mutations in the RBD and NTD of the spike protein has been reported to resist neutralisation by therapeutic monoclonal antibodies, convalescent sera and vaccinee sera (Figure 1). Among them, a few merit concern as they have also recently been identified in re-infection and breakthrough infections with VOCs, likely for their location in the immune epitope region of the RBD that may impact antigenicity, hinder immune neutralising antibody binding and promote pathogenesis (Table 1) (17, 18).
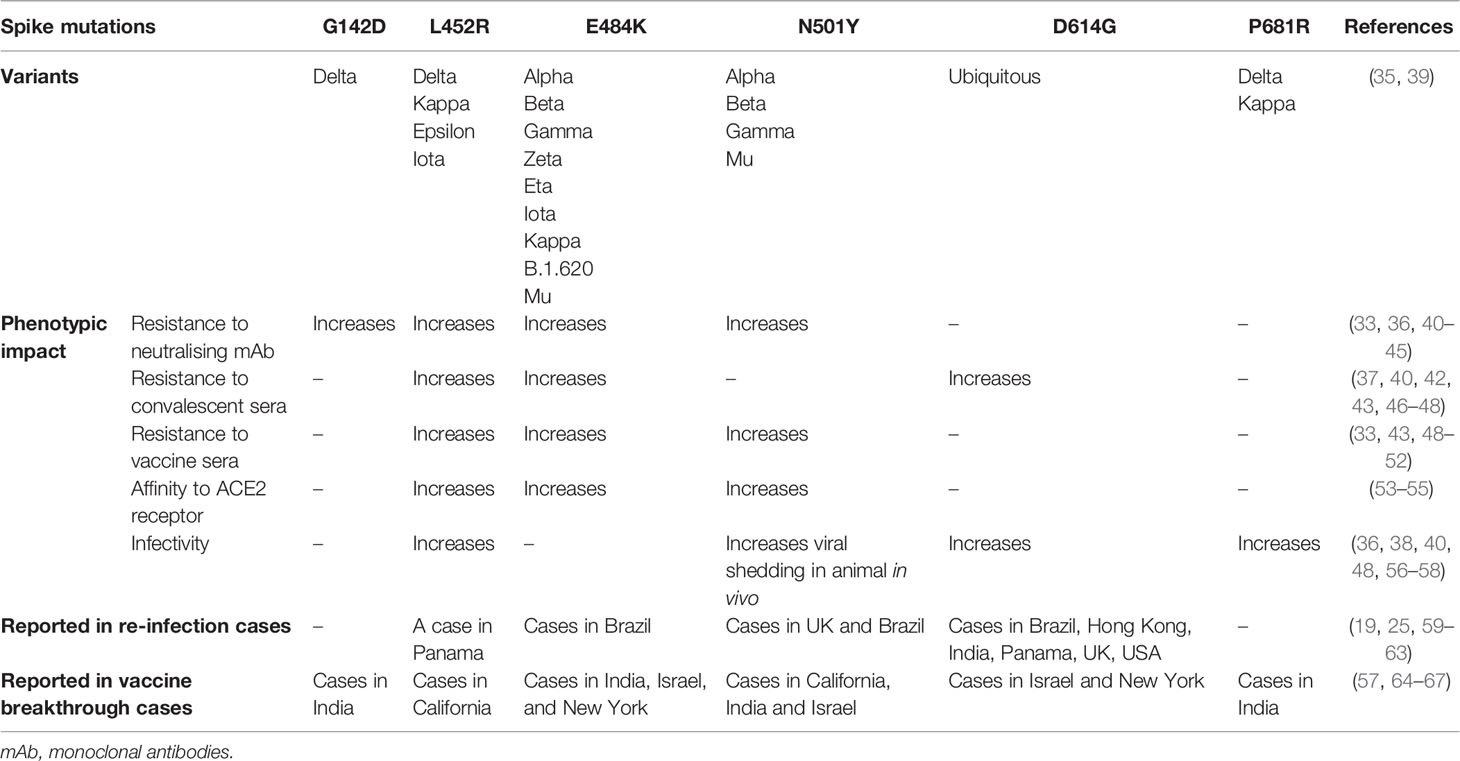
Table 1 Mutations of SARS-CoV-2 spike protein, their phenotypic impact in vitro, in vivo or in silico, and reports in re-infection and vaccine breakthrough infection cases.
E484K mutation first emerged in late 2020 and has now been gaining prevalence among the circulating strains. It has been acquired by at least 9 strains including VOCs Alpha, Beta and Gamma, and other variants under monitoring Zeta (P.2), Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1), B.1.620 and Mu (B.1.621) (39). It is noteworthy to mention that five reinfection cases of E484K-carrying variants (Gamma and Zeta), which primary infections were of non-E484K variants, have been reported in Brazil as early as December 2020. Two of the cases reported the presence of anti-SARS-CoV-2 IgG antibodies during re-infection suggesting the possibility of E484K mutation in aiding antibody evasion (59). Furthermore, breakthrough infections in vaccinated individuals are also growing in number. A cohort study identified a slight reduction of protection by vaccines against subsequent infection with E484K-carrying Alpha and Beta variants of concern (64). Another report also documented an infection of a vaccinated individual with E484K-carrying variant (65). In another study, a full vaccination elicited a high level of neutralising antibodies that were capable of inhibiting an E484K variant in vitro but failed to pre-empt a high viral replication (66).
In vitro studies showed that E484K mutation can significantly reduce binding and resist neutralisation by convalescent and vaccine-induced sera and monoclonal antibodies (33, 40–42, 46, 47, 49–51). Additionally, in silico and cryo-EM studies have found that E484K mutation enhances the virus affinity for host ACE2 receptor binding which may account for the increased infectivity of these mutant variants (53, 54). Its impact on infectivity in vitro has been revealed in an infection assay of murine ACE2-expressing HEK29T cells with E484K-carrying pseudotyped viruses whereby 3-fold more infection was observed (68). The prevalence of E484K-carrying variants has been increasingly reported in viral isolates, presenting at low frequency in the circulating strain populations (42), likely due to the positive selection that provides for an immune escape and a greater transmissibility. This is consistent with in vitro evolutionary studies that showed that E484K mutation is readily introduced in the viral genome when cultured in the presence of anti-SARS-CoV-2 neutralising antibodies or ACE2 receptor (42, 47, 54). Furthermore, four E484 mutational changes at this residue position demonstrated an immune escape phenotype in presence of vaccinee sera, highlighting the importance of this residue as part of the dominant neutralising epitope (40).
Also gaining in frequency among circulating variant sequences is N501Y substitution that has been acquired by three VOCs Alpha, Beta and Gamma. A report has demonstrated that N501Y mutation reduces the virus sensitivity to neutralising monoclonal antibodies and vaccine-induced polyclonal antibodies (33, 51). In vitro infection assay of murine ACE2-expressing HEK293T cells with N501Y-carrying pseudotyped virus also showed that this mutation caused a 5-fold increase in viral infectivity (68). Additionally, it was shown that it is positively selected in vitro when the virus is grown in the presence of ACE2 to which it developed a higher binding affinity, and in vivo in a chronically infected immunocompromised patient (54, 69, 70). The increase in ACE2 tropism consequently promotes viral replication, transmission and shedding (69). N501Y-carrying variants (VOCs Alpha and Gamma) have been reported in four re-infection cases in which primary episode of infection agents were of non-N501Y-carrying variant (25, 59). Three of these cases described the presence of anti-SARS-CoV-2 IgG antibodies at the timepoint of secondary infection, raising speculation of antibody escape that may be mediated by this mutation. Furthermore, several independent reports of breakthrough infections with N501Y-caryying variant had been reported in vaccinated individuals suggesting an evasion of humoral immunity (65, 67).
While E484K only mildly increases the affinity of the RBD for ACE2 receptor, N501Y appears to substantially enhance it by allowing it to engage the receptor for longer (71–73). This is in agreement with an observation of pseudotyped viruses carrying N501Y mutation demonstrating a higher infectivity in vitro than those carrying E484K mutation (68). In contrast, the E484 residue in RBD engages more with antibodies than with ACE2 receptor, which may explain the relevance of E484K mutation in mediating antibody escape (72). The co-existence of these two mutations can produce a synergistic effect whereby they dramatically improve spike binding affinity to ACE2 receptor and reinforce the virus ability to evade immunity (68, 72, 73). The importance of these mutations for viral fitness is highlighted by their convergence in certain variants, including Beta, Gamma, and Mu (Table 1).
Another mutation that has gained attention is L452R substitution which can resist neutralisation by monoclonal antibodies and vaccinee and convalescent sera (36, 40, 43, 46, 48, 52). It provides a greater affinity of binding to ACE2 receptor and thus promotes viral replication and infectivity (48, 55, 56). A report of breakthrough infections in fully or partially vaccinated healthcare workers and a secondary infection with L452R-carrying variants have been described, highlighting the relevance of this mutation in mediating viral immune escape (60, 67). L425R mutation has been identified in the circulating VOC Delta and other variants including Epsilon (B.1.427/B.1.429), Iota and Kappa. Of important note, Delta variant is currently posing the most concern and threat to public health, as it has displaced other variants owing to its pronounced ability to transmit and escape antibodies (monoclonal antibodies and convalescent and vaccinee sera) (74–76). Increasing numbers of breakthrough infection with Delta variant have been reported (57, 77). Other mutations, in addition to L452R of the RBD, have been attributed to its explosive spread. Upstream NTD mutation G142D has been shown to resist binding by monoclonal antibodies which may be mediated by the change of the conformation of the spike protein that deters antibody binding (44, 45). Downstream P681R mutation in furin cleavage site promotes cleavage and processing of the spike protein and hence the entry efficiency and infectivity of the virus in vitro (57, 58).
Although current available vaccines are engineered based on the spike protein of the Wuhan reference strain, vaccinated individuals are expected to carry potent neutralising antibodies that are cross-reactive which can circumvent the issue of variants (78). Nevertheless, the growing evidence for immune evasion by emerging spike variants prompts a heavier consideration for a more adequate and effective vaccine design guided by genomic surveillance that can stimulate a more durable protection from SARS-CoV-2 infection.
Research Gaps and Prospects
SARS-CoV-2 spike and nucleocapsid proteins are hotspots of genetic modification owing to the host immune selection pressure. Genomic surveillance is increasingly becoming a powerful tool in guiding public health responses and health interventions and therefore in controlling the trajectory of this pandemic. This will provide a wealth of databases that can be used to infer decisions on evaluating health interventions, vaccine efficacy, inform therapeutic development and design, assess the risk of re-infection and breakthrough infections, and tackle the issue of immune and diagnostic escape variants.
The growing relevance of a select spike mutations urges immediate investigation, particularly E484K, to discern whether it will become the next consensus sequence (as did D614G). Not explored in this article is the impact of spike or nucleocapsid mutations on primary diagnosis which have been documented elsewhere (79–81). Other clinical indicators should therefore accompany molecular-based COVID-19 diagnostic tests – more so in the wake of a growing trend of re-infection and breakthrough infection – to guide not only clinical evaluation, but also isolation and discharge decisions.
Because a waning and heterogeneous protective immunity has been suggested as risk factors of secondary and breakthrough infections, the identification of correlate of protection with a consideration of immune escape mutations, is crucial in identifying the most optimal vaccine design that promises a more preserved antibody titre and protection. One can expect the need for periodic reformulation of the vaccines in the future as ‘boosters’ to adapt to the most common variant at the time, recover the declining immunity and mitigate the risk of future emerging variants. Future interventions could consider harnessing other facet of protective immune responses, in addition to humoral immunity, to relieve selective pressure for immune escape variants. Notably, T cell-mediated immune responses are reported to be relatively more stable following SARS-CoV-2 infections and are potent at targeting the spike protein (82). Additionally, the heterogeneity of resistance profile of spike variants to monoclonal antibodies suggests that administering a cocktail of antibodies would be a more beneficial approach at targeting these variants (42). Other mutations outside of the spike gene should also be warranted surveillance in studying the evolution of SARS-CoV-2 and its interaction with human host to pre-empt potential novel variants that may affect current efforts of controlling the pandemic.
Author Contributions
LA conducted the study and agrees to be accountable for the content of the work.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med (2020) 382:1708–20. doi: 10.1101/2020.02.06.20020974
2. Wang Y, Liu Y, Liu L, Wang X, Luo N, Li L. Clinical Outcomes in 55 Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Who Were Asymptomatic at Hospital Admission in Shenzhen, China. J Infect Dis (2020) 221:1170–4. doi: 10.1093/infdis/jiaa119
3. Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell (2020) 183:730–8. doi: 10.1101/2020.07.08.192104
4. Worobey M, Pekar J, Larsen BB, Nelson MI, Hill V, Joy JB, et al. The Emergence of SARS-CoV-2 in Europe and North America. Science (2020) 370(6516):564–70. doi: 10.1101/2020.05.21.109322
5. Vilar S, Isom DG. One Year of SARS-CoV-2: How Much has the Virus Changed? Biol (Basel) (2021) 10:91. doi: 10.1101/2020.12.16.423071
6. Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis (2020) 71:713–20. doi: 10.1093/cid/ciaa203
7. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological Assessment of Hospitalized Patients With COVID-2019. Nature (2020) 581:465–9. doi: 10.1038/s41586-020-2196-x.
8. Popa A, Genger JW, Nicholson MD, Penz T, Schmid D, Aberle SW, et al. Genomic Epidemiology of Superspreading Events in Austria Reveals Mutational Dynamics and Transmission Properties of SARS-CoV-2. Sci Transl Med (2020) 12. doi: 10.1101/2020.07.15.204339
9. Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, et al. SARS-CoV-2 Within-Host Diversity and Transmission. Science (2021) 372. doi: 10.1126/science.abg0821.
10. Hensley MK, Bain WG, Jacobs J, Nambulli S, Parikh U, Cillo A, et al. Intractable Coronavirus Disease 2019 (COVID-19) and Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Replication in a Chimeric Antigen Receptor-Modified T-Cell Therapy Recipient: A Case Study. Clin Infect Dis (2021) 73:e815–21. doi: 10.1093/cid/ciab072.
11. Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding From an Asymptomatic Immunocompromised Individual With Cancer. Cell (2020) 183:1901–12.e9. doi: 10.2139/ssrn.3699798
12. Stasi C, Fallani S, Voller F, Silvestri C. Treatment for COVID-19: An Overview. Eur J Pharmacol (2020) 889:173644. doi: 10.1016/j.ejphar.2020.173644
13. Masoomikarimi M, Garmabi B, Alizadeh J, Kazemi E, Azari Jafari A, Mirmoeeni S, et al. Advances in Immunotherapy for COVID-19: A Comprehensive Review. Int Immunopharmacol (2021) 93:107409. doi: 10.1016/j.intimp.2021.107409
14. Chen L, Zody MC, Mediavilla JR, Cunningham MH, Composto K, Fai Chow K, et al. Emergence of Multiple SARS-CoV-2 Antibody Escape Variants in an Immunocompromised Host Undergoing Convalescent Plasma Treatment 2 3. medRxiv (2021) 6:e00480–21. doi: 10.1101/2021.04.08.21254791. 2021.04.08.21254791.
15. Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, et al. SARS-CoV-2 Evolution During Treatment of Chronic Infection. Nature (2021) 592:277–82. doi: 10.1038/s41586-021-03291-y.
16. Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, et al. Deployment of Convalescent Plasma for the Prevention and Treatment of COVID-19. J Clin Invest (2020) 130:2757–65. doi: 10.1172/JCI138745
17. Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell (2020) 183:1024–42.e21. doi: 10.1016/j.cell.2020.09.037.
18. Steffen TL, Stone ET, Hassert M, Geerling E, Grimberg BT, Espino AM, et al. The Receptor Binding Domain of SARS-CoV-2 Spike Is the Key Target of Neutralizing Antibody in Human Polyclonal Sera. bioRxiv (2020). doi: 10.1101/2020.08.21.261727
19. To KK-W, Hung IF-N, Ip JD, Chu AW-H, Chan W-M, Tam AR, et al. Coronavirus Disease 2019 (COVID-19) Re-Infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin Infect Dis (2020). doi: 10.1093/cid/ciaa1275
20. Dao TL, Hoang VT, Gautret P. Recurrence of SARS-CoV-2 Viral RNA in Recovered COVID-19 Patients: A Narrative Review. Eur J Clin Microbiol Infect Dis (2021) 40:13–25. doi: 10.1007/s10096-020-04088-z
21. Letizia AG, Ge Y, Vangeti S, Goforth C, Weir DL, Kuzmina NA, et al. SARS-CoV-2 Seropositivity and Subsequent Infection Risk in Healthy Young Adults: A Prospective Cohort Study. Lancet Respir Med (2021) 9:712–20. doi: 10.1101/2021.01.26.21250535
22. Pilz S, Chakeri A, Ioannidis JPA, Richter L, Theiler-Schwetz V, Trummer C, et al. SARS-CoV-2 Re-Infection Risk in Austria. Eur J Clin Invest (2021) 51:e13520. doi: 10.1101/2021.02.08.21251362
23. Lumley SF, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N Engl J Med (2021) 384(6):533–40. doi: 10.1056/NEJMoa2034545
24. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological Memory to SARS-CoV-2 Assessed for Up to 8 Months After Infection. Science (2021) 371. doi: 10.1126/science.abf4063
25. Harrington D, Kele B, Pereira S, Couto-Parada X, Riddell A, Forbes S, et al. Confirmed Reinfection With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variant VOC-202012/01. Clin Infect Dis (2021). doi: 10.1093/cid/ciab014
26. Butt AA, Khan T, Yan P, Shaikh OS, Omer SB, Mayr F. Rate and Risk Factors for Breakthrough SARS-CoV-2 Infection After Vaccination. J Infect (2021) 83:237–79. doi: 10.1016/j.jinf.2021.05.021
27. Teran RA, Walblay KA, Shane EL, Xydis S, Gretsch S, Gagner A, et al. Postvaccination SARS-CoV-2 Infections Among Skilled Nursing Facility Residents and Staff Members — Chicago, Illinois, December 2020–March 2021. Am J Transplant (2021) 21(6):2290–7. doi: 10.1111/ajt.16634
28. Ulhaq ZS, Soraya GV, Indriana K. Breakthrough COVID-19 Case After Full-Dose Administration of CoronaVac Vaccine. Indian J Med Microbiol (2021). doi: 10.1016/j.ijmmb.2021.05.017
29. Niyas VKM, Arjun R. Correspondence: Breakthrough COVID-19 Infections Among Health Care Workers After Two Doses of ChAdOx1 Ncov-19 Vaccine. QJM Int J Med (2021). doi: 10.1093/qjmed/hcab167/6297399
30. Tyagi K, Ghosh A, Nair D, Dutta K, Singh Bhandari P, Ahmed Ansari I, et al. Breakthrough COVID19 Infections After Vaccinations in Healthcare and Other Workers in a Chronic Care Medical Facility in New Delhi, India. Diabetes Metab Syndr Clin Res Rev (2021) 15(3):1007–8. doi: 10.1016/j.dsx.2021.05.001
31. CDC COVID-19 Vaccine Breakthrough Case Investigations Team. COVID-19 Vaccine Breakthrough Infections Reported to CDC - United States, January 1-April 30, 2021. MMWR Morb Mortal Wkly Rep (2021) 70(21):792–3. doi: 10.15585/mmwr.mm7021e3
32. Assis R, Jain A, Nakajima R, Jasinskas A, Kahn S, Palma A, et al. Substantial Differences in SARS-CoV-2 Antibody Responses Elicited by Natural Infection and mRNA Vaccination. bioRxiv (2021). doi: 10.1101/2021.04.15.440089
33. Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA Vaccine-Elicited Antibodies to SARS-CoV-2 and Circulating Variants. Nature (2021) 592:616–22. doi: 10.3410/f.739524179.793585051
34. CoV-GLUE. CoV-GLUE (2021). Available at: http://cov-glue.cvr.gla.ac.uk/.
35. GISAID. GISAID (2021). Available at: https://www.gisaid.org.
36. Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell (2020) 182:1284–94.e9. doi: 10.1016/j.cell.2020.07.012
37. Hu J, He CL, Gao Q, Zhang GJ, Cao XX, Long QX, et al. The D614G Mutation of SARS-CoV-2 Spike Protein Enhances Viral Infectivity and Decreases Neutralization Sensitivity to Individual Convalescent Sera. bioRxiv (2020). doi: 10.1101/2020.06.20.161323
38. Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, et al. SARS-CoV-2 Spike D614G Change Enhances Replication and Transmission. Nature (2021) 592:122–7. doi: 10.1038/s41586-021-03361-1
39. CoVariants. CoVariants - Shared Mutations (2021). Available at: https://covariants.org.
40. Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 Spike Mutations That Attenuate Monoclonal and Serum Antibody Neutralization. Cell Host Microbe (2021) 29:477–88.e4. doi: 10.2139/ssrn.3725763
41. Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain That Escape Antibody Recognition. Cell Host Microbe (2021) 29:44–57.e9. doi: 10.1101/2020.09.10.292078
42. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JCC, et al. Escape From Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants. Elife (2020) 9:e61312. doi: 10.7554/eLife.61312.
43. McCallum M, Bassi J, Marco A, Chen A, Walls AC, Iulio J, et al. SARS-CoV-2 Immune Evasion by Variant B.1.427/B.1.429. BioRxiv Prepr Serv Biol (2021) 373:648–54. doi: 10.1126/science.abi7994
44. Shen L, Triche TJ, Bard JD, Biegel JA, Judkins AR, Gai X. Spike Protein NTD Mutation G142D in SARS-CoV-2 Delta VOC Lineages Is Associated With Frequent Back Mutations, Increased Viral Loads, and Immune Evasion. medRxiv (2021). doi: 10.1101/2021.09.12.21263475v1. 2021.09.12.21263475.
45. Suryadevara N, Shrihari S, Gilchuk P, VanBlargan LA, Binshtein E, Zost SJ, et al. Neutralizing and Protective Human Monoclonal Antibodies Recognizing the N-Terminal Domain of the SARS-CoV-2 Spike Protein. Cell (2021) 184:2316–31.e15. doi: 10.1101/2021.01.19.427324
46. Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive Mapping of Mutations in the SARS-CoV-2 Receptor-Binding Domain That Affect Recognition by Polyclonal Human Plasma Antibodies. Cell Host Microbe (2021) 29:463–76.e6. doi: 10.1101/2020.12.31.425021
47. Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 Escape In Vitro From a Highly Neutralizing COVID-19 Convalescent Plasma. BioRxiv Prepr Serv Biol (2020) 118. doi: 10.1101/2020.12.28.424451
48. Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, Infectivity, and Antibody Neutralization of an Emerging SARS-CoV-2 Variant in California Carrying a L452R Spike Protein Mutation. MedRxiv Prepr Serv Heal Sci (2021) 184:3426–37.e8. doi: 10.1016/j.cell.2021.04.025. 2021.03.07.21252647.
49. Collier DA, De Marco A, Ferreira IATM, Meng B, Datir RP, Walls AC, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA Vaccine-Elicited Antibodies. Nature (2021) 593:136–41. doi: 10.1038/s41586-021-03412-7.
50. Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 Vaccine Induces Neutralizing Antibodies Against Spike Mutants From Global SARS-CoV-2 Variants. BioRxiv Prepr Serv Biol (2021). doi: 10.1101/2021.01.25.427948
51. Garcia-Beltran WF, Lam EC, St. Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell (2021) 184:2372–83.e9. doi: 10.1101/2021.02.14.21251704
52. Ferreira I, Datir R, Kemp S, Papa G, Rakshit P, Singh S, et al. SARS-CoV-2 B.1.617 Emergence and Sensitivity to Vaccine-Elicited Antibodies. bioRxiv (2021), 2021.05.08.443253. doi: 10.1101/2021.05.08.443253
53. Nelson G, Buzko O, Patricia S, Niazi K, Rabizadeh S, Soon-Shiong P. Molecular Dynamic Simulation Reveals E484K Mutation Enhances Spike RBD-ACE2 Affinity and the 1 Combination of E484K, K417N and N501Y Mutations (501Y.V2 Variant) Induces Conformational 2 Change Greater Than N501Y Mutant Alone, Potentially Resulting in an Escape Mutant 3 4. bioRxiv (2021), 2021.01.13.426558. doi: 10.1101/2021.01.13.426558
54. Zahradník J, Marciano S, Shemesh M, Zoler E, Harari D, Chiaravalli J, et al. SARS-CoV-2 Variant Prediction and Antiviral Drug Design Are Enabled by RBD In Vitro Evolution. Nat Microbiol (2021) 6:1188–98. doi: 10.1038/s41564-021-00954-4
55. Chen J, Wang R, Wang M, Wei GW. Mutations Strengthened SARS-CoV-2 Infectivity. J Mol Biol (2020) 432:5212–26. doi: 10.1016/j.jmb.2020.07.009
56. Motozono C, Toyoda M, Zahradnik J, Ikeda T, Seng Tan T, Ngare I, et al. An Emerging SARS-CoV-2 Mutant Evading Cellular Immunity and Increasing 1 Viral Infectivity 2 3. bioRxiv (2021) 29:1124. doi: 10.1101/2021.04.02.438288. 2021.04.02.438288.
57. Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Mishra S, et al. SARS-CoV-2 B.1.617.2 Delta Variant Emergence and Vaccine Breakthrough. Nature (2021). doi: 10.1038/s41586-021-03944-y
58. Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, et al. Delta Spike P681R Mutation Enhances SARS-CoV-2 Fitness Over Alpha Variant. bioRxiv (2021). doi: 10.1101/2021.08.12.456173v3. 2021.08.12.456173.
59. Naveca F, Costa C, Nascimento V, Souza V, Corado A, Nascimento F, et al. Three SARS-CoV-2 Reinfection Cases by the New Variant of Concern (VOC) P.1/501y.V3. Res Sq (2021). doi: 10.21203/rs.3.rs-318392/v1
60. Díaz Y, Ortiz A, Weeden A, Castillo D, González C, Moreno B, et al. SARS-CoV-2 Reinfection With a Virus Harboring Mutation in the Spike and the Nucleocapsid Proteins in Panama. Int J Infect Dis (2021) 108:588–91. doi: 10.1016/j.ijid.2021.06.004
61. Adrielle dos Santos L, Filho PG de G, Silva AMF, Santos JVG, Santos DS, Aquino MM, et al. Recurrent COVID-19 Including Evidence of Reinfection and Enhanced Severity in Thirty Brazilian Healthcare Workers. J Infect (2021) 82:399–406. doi: 10.1016/j.jinf.2021.01.020
62. Gupta V, Bhoyar RC, Jain A, Srivastava S, Upadhayay R, Imran M, et al. Asymptomatic Reinfection in 2 Healthcare Workers From India With Genetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2. Clin Infect Dis (2020). doi: 10.1093/cid/ciaa1451
63. Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, et al. Genomic Evidence for Reinfection With SARS-CoV-2: A Case Study. Lancet Infect Dis (2021) 21:52–8. doi: 10.2139/ssrn.3680955
64. Kustin T, Harel N, Finkel U, Perchik S, Harari S, Tahor M, et al. Evidence for Increased Breakthrough Rates of SARS-CoV-2 Variants of Concern in BNT162b2-mRNA-Vaccinated Individuals. Nat Med (2021) 27:1379–84. doi: 10.1038/s41591-021-01413-7
65. Philomina JB, Jolly B, John N, Bhoyar RC, Majeed N, Senthivel V, et al. Genomic Survey of SARS-CoV-2 Vaccine Breakthrough Infections in Healthcare Workers From Kerala, India. J Infect (2021) 83:237–79. doi: 10.1016/j.jinf.2021.05.018.
66. Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, Conlon EG, et al. Vaccine Breakthrough Infections With SARS-CoV-2 Variants. N Engl J Med (2021) 384:2212–8. doi: 10.1056/NEJMoa2105000
67. Jacobson KB, Pinsky BA, Rath MEM, Wang H, Miller JA, Skhiri M, et al. Post-Vaccination SARS-CoV-2 Infections and Incidence of the B.1.427/B.1.429 Variant Among Healthcare Personnel at a Northern California Academic Medical Center. MedRxiv Prepr Serv Heal Sci (2021). doi: 10.1101/2021.04.14.21255431
68. Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H, et al. SARS-CoV-2 501y.V2 Variants Lack Higher Infectivity But do Have Immune Escape. Cell (2021) 184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042
69. Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, et al. The N501Y Spike Substitution Enhances SARS-CoV-2 Transmission. BioRxiv (2021). doi: 10.1101/2021.03.08.434499
70. Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med (2020) 383:2291–3. doi: 10.1056/NEJMc2031364
71. Tian F, Tong B, Sun L, Shi S, Zheng B, Wang Z, et al. N501Y Mutation of Spike Protein in SARS-CoV-2 Strengthens Its Binding to Receptor ACE2. Elife (2021) 10:e69091. doi: 10.7554/eLife.69091
72. Alenquer M, Ferreira F, Lousa D, Valério M, Medina-Lopes M, Bergman ML, et al. Signatures in SARS-CoV-2 Spike Protein Conferring Escape to Neutralizing Antibodies. PloS Pathog (2021) 17:e1009772. doi: 10.1371/journal.ppat.1009772
73. Laffeber C, de Koning K, Kanaar R, Lebbink JHG. Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants. J Mol Biol (2021) 433:167058. doi: 10.1101/2021.02.22.432357
74. Li B, Deng A, Li K, Hu Y, Li Z, Xiong Q, et al. Viral Infection and Transmission in a Large Well-Traced Outbreak Caused by the Delta SARS-CoV-2 Variant. medRxiv (2021). doi: 10.1101/2021.07.07.21260122. 2021.07.07.21260122.
75. Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature (2021) 596:276–80. doi: 10.1038/s41586-021-03777-9
76. Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Mishra S, et al. SARS-CoV-2 B.1.617.2 Delta Variant Emergence, Replication and Sensitivity to Neutralising Antibodies. bioRxiv (2021). doi: 10.1101/2021.05.08.443253v4. 2021.05.08.443253.
77. Chia PY, Ong SWX, Chiew CJ, Ang LW, Chavatte J-M, Mak T-M, et al. Virological and Serological Kinetics of SARS-CoV-2 Delta Variant Vaccine-Breakthrough Infections: A Multi-Center Cohort Study. medRxiv (2021). doi: 10.1101/2021.07.28.21261295v1. 2021.07.28.21261295.
78. Moyo-Gwete T, Madzivhandila M, Makhado Z, Ayres F, Mhlanga D, Oosthuysen B, et al. Cross-Reactive Neutralizing Antibody Responses Elicited by SARS-CoV-2 501y.V2 (B.1.351). N Engl J Med (2021) 384:2161–3. doi: 10.1056/NEJMc2104192
79. Bourassa L, Perchetti GA, Phung Q, Lin MJ, Mills MG, Roychoudhury P, et al. A SARS-CoV-2 Nucleocapsid Variant That Affects Antigen Test Performance. medRxiv (2021) 141:104900. doi: 10.1101/2021.05.05.21256527
80. Kidd M, Richter A, Best A, Cumley N, Mirza J, Percival B, et al. S-Variant SARS-CoV-2 Lineage B1.1.7 Is Associated With Significantly Higher Viral Load in Samples Tested by TaqPath Polymerase Chain Reaction. J Infect Dis (2021) 223:1666–70. doi: 10.1093/infdis/jiab082
81. Vanaerschot M, Mann SA, Webber JT, Kamm J, Bell SM, Bell J, et al. Identification of a Polymorphism in the N Gene of SARS-CoV-2 That Adversely Impacts Detection by Reverse Transcription-PCR. J Clin Microbiol (2021) 59. doi: 10.1128/JCM.02369-20
Keywords: spike protein, SARS-CoV-2, secondary infection, breakthrough infection, vaccine, COVID-19, variants, immune evasion
Citation: Ahmad L (2021) Implication of SARS-CoV-2 Immune Escape Spike Variants on Secondary and Vaccine Breakthrough Infections. Front. Immunol. 12:742167. doi: 10.3389/fimmu.2021.742167
Received: 19 July 2021; Accepted: 14 October 2021;
Published: 03 November 2021.
Edited by:
Timothy Cardozo, New York University, United StatesReviewed by:
Rosemarie Mason, Vaccine Research Center (NIAID), United StatesLauren Young, University of Chicago Medicine, United States
Copyright © 2021 Ahmad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liyana Ahmad, bGl5YW5hLmFobWFkQHViZC5lZHUuYm4=
 Liyana Ahmad
Liyana Ahmad