
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 09 December 2021
Sec. Viral Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.740223
This article is part of the Research Topic Ubiquitin and Ubiquitin-like Modifications in Viral Infection and Innate Immunity View all 12 articles
 Haoran Cui1,2,3†
Haoran Cui1,2,3† Yaxian Zhang 2,3†and
Yaxian Zhang 2,3†and  Leiliang Zhang1,2,3*
Leiliang Zhang1,2,3*Poxviruses have evolved a variety of innate immunity evasion mechanisms, some of which involve poxvirus-encoded E3 ubiquitin ligases and adaptor proteins. Based on their functional domains and ubiquitin transfer mechanisms, these poxvirus-encoded E3 ubiquitin ligases and adaptor proteins can be divided into five categories: PRANC, ANK/BC, BBK, P28/RING, and MARCH proteins. Although the substrates of many poxvirus E3 ubiquitin ligases remain to be discovered, most of the identified substrates are components of the innate immune system. In this review, we discuss the current research progress on poxvirus-encoded E3 ubiquitin ligases and adaptor proteins to provide mechanistic insights into the interplay between these viruses and their hosts.
Members of the family Poxviridae are double-stranded DNA viruses that replicate in the cytoplasm of the host cell. After infecting the host, poxviruses usually cause local or systemic purulent skin damage. Poxviridae is divided into two subfamilies: Chordopoxvirinae and Entomopoxvirinae. The typical members of the Chordopoxvirinae subfamily are variola virus (VARV), cowpox virus (CPXV), monkeypox virus (MPXV), vaccinia virus (VACV), orf virus (ORFV), myxoma virus (MYXV), and ectromelia virus (ECTV) (1).
The host inhibits the replication and spread of the virus through both the innate and adaptive immune systems. Therefore, immune escape mechanisms are particularly important for the survival of the virus (2). Poxviruses have gradually developed a variety of immune escape strategies during their evolution. For instance, poxviruses encode ubiquitination pathway components that modify the host proteins, directly affecting viral recognition, the generation of antiviral signals and inflammation, and the elimination of the virus.
Ubiquitin consists of 76 amino acids and can be attached to target proteins. The ubiquitination process occurs through an enzymatic cascade. First, ubiquitin needs to be activated by an E1 ubiquitin-activating enzyme; then, the activated ubiquitin is transferred to an E2 ubiquitin-conjugating enzyme; and finally, an E3 ubiquitin ligase transfers ubiquitin to a lysine residue on the target protein (Figure 1A) (3). Deubiquitylating enzymes (DUBs) maintain the dynamic state of the cellular ubiquitome by releasing conjugated ubiquitin from proteins and recycling it to maintain the cellular level of free ubiquitin (4). In the above series of enzymatic cascade reactions, the E3 ubiquitin ligase enzyme plays a vital role in the specific recognition of target substrates. At present, 8 types of polyubiquitination modification linkages have been reported. Seven of them involve connections of the glycine at the C-terminus of the ubiquitin molecule to a lysine in the ubiquitin chain, specifically, K6, K11, K27, K29, K33, K48 or K63 (Figure 1B). Among those seven polyubiquitination modifications, those at K48 and K63 are the best studied. At least four ubiquitins linked together via their Lys48 residues form the ubiquitin chain that triggers degradation by the 26S proteasome (Figure 1C). In addition, monoubiquitination or Lys63-linked polyubiquitination functions as a nonproteolytic signal in intracellular trafficking, DNA repair, and signal transduction pathways (Figure 1C). The eighth type of ubiquitin linkage is linear ubiquitination, in which the amino group of the methionine residue on ubiquitin is connected to the carboxy group of the glycine residue of another ubiquitin. Ubiquitin-like proteins (UbLs), including NEDD8, SUMO, and ISG15, are biochemically similar to Ub and are also covalently attached to the lysines of their substrates. The conjugation of ubiquitin to UbL, and vice versa, can also occur, forming hybrid chains (5).
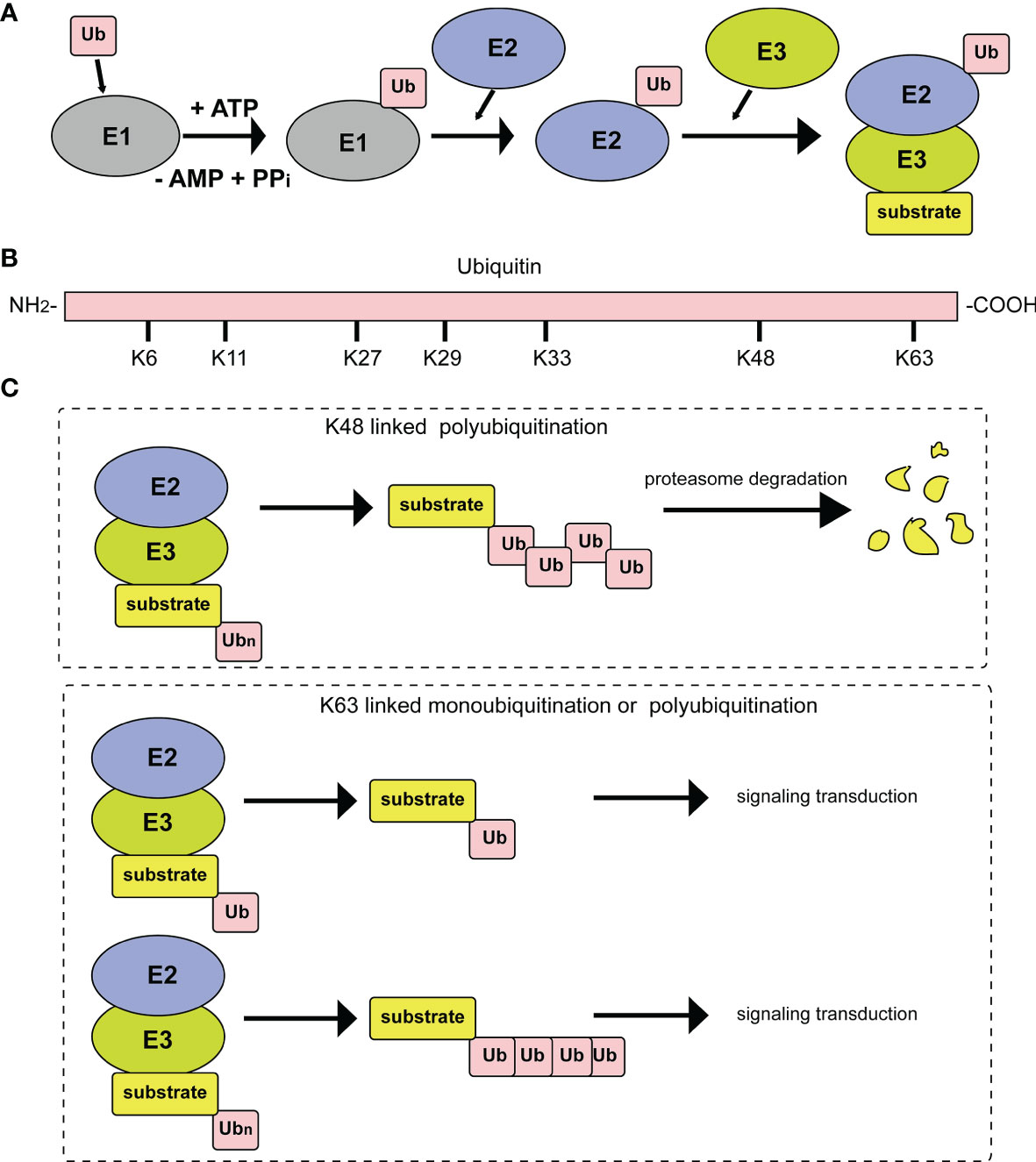
Figure 1 Diagrammatic sketch of the ubiquitin cascade and E3 classification. (A) Illustration of the ubiquitin/26S proteome pathway. E1 is activated through ATP hydrolysis and adenosylates the ubiquitin molecule. Then, ubiquitin is successfully transferred to E3 from E1 by the catalysis of E2. With the participation of E3, substrates are modified by the ubiquitin molecule in different linkage modes. (B) The seven lysine residues on the ubiquitin molecule that can be ubiquitinated. (C) Different ubiquitin molecule linkage types. Upper panel, ubiquitin molecules connect with each other through K48 to form a polyubiquitin chain. Substrates modified by the polyubiquitin chain are degraded by the proteasome pathway. Lower panel, substrate modification by ubiquitin molecules through K63 type monoubiquitylation or polyubiquitylation, which plays important roles in signal transduction.
The E3 ubiquitin ligases in eukaryotes are mainly classified into three types based on their functional domains and ubiquitin transfer mechanisms (6). The most abundant type of ubiquitin ligase is Really Interesting New Gene (RING) E3s. They are characterized by a zinc-binding domain, called a RING domain, or a U-box domain (7). Some RING E3s, such as cullin-RING ligases (CRLs), are composed of multiple subunits. CRLs are composed of a cullin scaffold with a RING-box domain at its N-terminus, an adaptor protein and a substrate receptor. There are several subtypes of CRL ligases. One subtype contains Casitas B-Lineage Lymphoma Proto-Oncogene C (c-Cbl), Mouse Double Minute 2 (Mdm2), and Inhibitor of Apoptosis (IAP) and RING finger proteins (8). Another CRL ligase subtype comprises large protein complexes that include a minimal core element composed of cullin and RING-H2. Another subtype of CRL ligase is the Membrane-Associated RING-CH (MARCH) type. The E3 ligases of the Homologous to the E6AP Carboxyl Terminus (HECT) domain family are another E3 ligase type and are characterized by a conserved HECT domain located at the C-terminus of the protein (9). The other E3 ligase type is RING-between RING-RING (RBR) E3s, named after their two predicted RING domains (RING1 and RING2) and an in-between-RING domain (IBR). In the RBR catalytic process, the RING1 domain recruits ubiquitin-charged E2, and the RING2 domain possesses a catalytic cysteine. In this review, we focus on poxvirus-encoded E3 ubiquitin ligases and adaptor proteins, which are CRL ligases.
Poxviruses encode many proteins with E3 ubiquitin ligase functions, including pox protein repeats of ankyrin-C-terminal domain (PRANC), ankyrin repeat (ANK)/Elongin B/Elongin C (BC), BTB/Kelch (BBK), P28/RING, and MARCH. Diagrams of the E3 ubiquitin ligases and adaptor proteins encoded by poxvirus are illustrated in Figure 2 and summarized in Tables 1, 2.
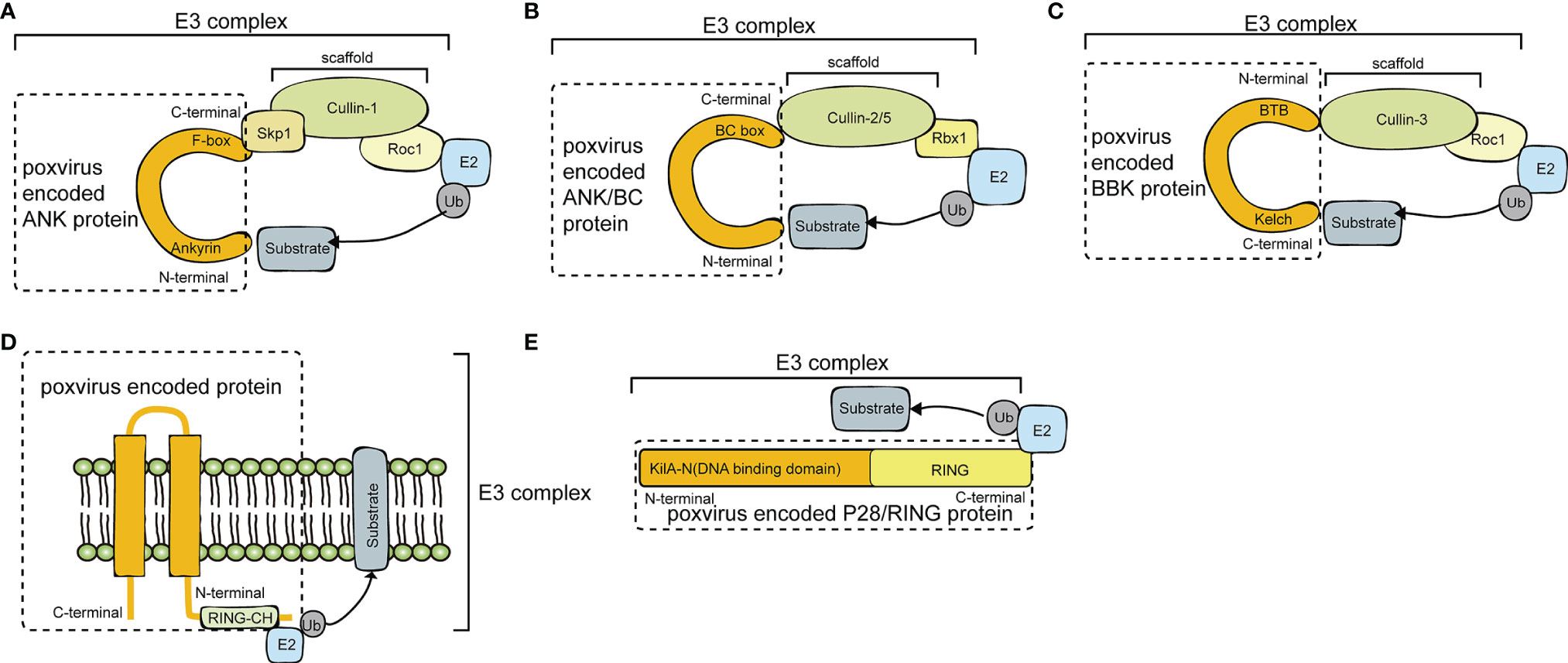
Figure 2 A diagram of poxvirus E3 ubiquitin ligases and adaptor proteins. (A) Schematic diagram of Cullin-1 E3 Ub ligases and poxvirus-encoded pox protein repeats of ankyrin-C-terminal domain (PRANC) protein. (B) Schematic diagram of Cullin-2/Cullin-5 E3 Ub ligases and poxvirus-encoded ankyrin repeat (ANK)/Elongin B/Elongin C (BC) protein. (C) Schematic diagram of Cullin-3 E3 Ub ligases and poxvirus-encoded BTB/Kelch (BBK) protein. (D) Schematic diagram of P28/RING protein. (E) Schematic diagram of membrane-associated RING-CH (MARCH) protein.
Poxviruses usually encode 4 to 5 ANK proteins. The ANK repeat domain, composed of 33 amino acid residues, mainly exists in Chordopoxvirinae, often functioning as a protein-protein interaction motif. Approximately 80% of ANK proteins have an F-box domain at the C-terminus, and this F-box domain is shorter than the typical F-box domain in the host cells (17). Specifically, the F-box domain encoded by poxviruses is similar in length to one or two of the three alpha helices of the cellular F-box protein. The M-T5 protein of MYXV; the VACV protein of B18R; the ORFV008, ORFV123, ORFV126, ORFV128, and ORFV129 proteins of ORFV; and EVM002, EVM005, EVM154, and EVM165 of ECTV all contain F-boxes. The Skp, Cullin, F-box (SCF) motif is the recognition motif of E3 ubiquitin ligases that mediate the ubiquitination and degradation of substrates via the 26S proteasome (8). In the viral ANK/F-box protein, the F-box motif binds to the N-terminus of the scaffold protein cullin-1 (Cul-1) via the adaptor protein S-Phase Kinase Associated Protein 1 (SKP1) (Figure 2A). The substrate is then ubiquitinated and degraded by binding to the substrate receptor region of the F-box protein through the degradation sequence. The SCF complex can selectively degrade regulatory proteins, thus modulating a variety of cell activities, such as signal transduction and cell cycle regulation (26).
MYXV encodes M-T5, M148R, M149R and M150, all of which are considered PRANC proteins, as they include N-terminal ANK repeats and a C-terminal F-box structure. The association of M-T5 and Cul-1 reduces P27 expression levels through 26S proteasomal degradation mediated by M-T5/SCF1. The association of M-T5 and Cul-1 maintains the protein level of Akt instead of reducing it through M-T5/SCF1-mediated 26S proteasome degradation. M-T5 can mimic the cellular protein PIKE-A, forming a complex with Akt to induce its activity. M-T5 interacts with Akt to promote the phosphorylation of P27 to regulate apoptosis and cell growth. P27/kip1 belongs to the cell cycle control protein Cip/Kip family and is a negative cell cycle regulator. The continuous phosphorylation and ubiquitination-associated degradation of P27 promote the cell cycle to cross the G0/G1 checkpoint, thereby avoiding programmed cell death due to virus infection (13, 17). The M148 protein encoded by MYXV contains 10 ankyrin repeats and is located in the cytoplasm and nucleus. M149 contains 9 of these ankyrin repeats and is unevenly distributed in the cytoplasm in the form of dots. Neither M148 nor M149 is required for virus replication in tissue culture. However, when MYXV infects rabbits, it acts as a virulence factor (14). The identification of the proteins and target substrates that interact with M148, M149, and M150 requires further research. M150 is another viral protein with an ankyrin repeat sequence, containing nine ankyrin repeats in its N-terminus and an F-box in its C-terminus. This structure is necessary for the pathogenic mechanism of MYXV. The M150 protein localizes to dot-like structures in the nucleus. However, deletion of the eighth ankyrin repeat disrupts its nuclear localization. Therefore, the nuclear localization of M150 depends on the eighth ankyrin repeat. M150 colocalized with p50 of NF-κB in the nucleus in cells stimulated by tumor necrosis factor (TNF). This indicates that M150 may interfere with the NF-κB signaling pathway, but the details of this mechanism remain unclear (14).
The CPXV protein CP77 is a PRANC protein that contains nine ankyrin repeats and a 13 amino acid F-box motif at its C-terminus (15). CP77 blocks TNF-mediated nuclear translocation and activation of the NF-κB subunit p65, while it does not block IκBα phosphorylation. CP77 binds to the NF-κB subunit p65 through six ankyrin repeats in the N-terminus and binds to Cul-1 and SKP1 of the SCF complex through the C-terminal 13 amino acid F-box-like sequence. Through these two pathways, poxvirus CP77 inhibits NF-κB activation and weakens the signal transduction of the natural immune response in cells (15).
In CPXV and other orthopoxviruses, viral inducer of RIPK3 degradation (vIRD) was identified to trigger the ubiquitination and proteasome-mediated degradation of receptor interacting protein kinase 3 (RIPK3) and inhibit necroptosis (21).
ORFV belongs to the genus Parapoxvirus and causes local skin infections in goats, sheep and humans (29). The five proteins ORFV008, ORFV123, ORFV126, ORFV128 and ORFV129 encoded by ORFV all contain F-box domains (18). ORFV008, ORFV123, ORFV126, ORFV128 and ORFV129 interact with the SKP1, Cul-1 and Roc1 proteins of host cells in an F-box-dependent manner, and the interaction of ORFV008 with SCF does not inhibit the E3 ubiquitin ligase function of cellular SCF (17). In summary, poxviruses are likely to use ANK/F-box proteins to recruit target proteins to SCF1 ubiquitin ligase and degrade specific cellular proteins through the ubiquitin protease system of the host to support virus replication.
ECTV causes lethal mousepox in infected mice. The four proteins EVM002, EVM005, EVM154 and EVM165 encoded by ECTV all contain an F-box domain in the C-terminus that interacts with the host SCF complex (30). EVM005 colocalizes with Cul-1 and interacts with Cul-1, SKP1 and Roc1. Deletion of the F-box domain eliminates the interactions among EVM005, Cul-1 and SKP1 (30). One of the key steps in NF-κB activation is the ubiquitination and degradation of IκBα by the cellular SCF β-TrCP-ubiquitin ligase complex. The overexpression of EVM002, EVM005, EVM154 and EVM165 inhibited TNF- or IL-1β-stimulated IκBα degradation and NF-κB subunit p65 nuclear translocation. The inhibition of the NF-κB pathway by EVM005 depends on its F-box domain and the interaction between EVM005 and the SCF complex (19, 20). In A/NCR and C57BL/6 mouse models, virus lacking EVM005 exhibited significantly weakened virulence, indicating that EVM005 is necessary for the toxicity and immune regulation of ECTV (19).
VACV B18R is highly conserved in orthopoxviruses, containing ANK repeats and an F-box domain (16). B18R, together with two other VACV proteins, M2 and C5, has functions in uncoating and in viral DNA replication (31). Yeast two-hybrid screening experiments showed that SKP1A binds to B18R, and this interaction was confirmed by immunoprecipitation experiments, which further showed that the binding of SKP1A to B18R was dependent on the F-box (16).
VACV C9 contains 6 ANK repeats and an F-box domain near the C-terminus, which antagonizes the antiviral state induced by type I interferons (IFNs) in the early stage of VACV replication (32). When VACV infects host cells, pattern recognition receptors (PRRs) bind to pathogen-associated molecular patterns (PAMPs), activating a series of signal cascades to induce the transcription of IFN-encoding genes. Secreted IFNs act on the originating cell or on neighboring cells to activate the JAK-STAT pathway, inducing the transcription of more than 300 interferon-stimulated genes (ISGs). These genes induced by IFNs can protect the host against infection by viruses and other pathogens. Studies have found that C9 interacts with SCF and SCN (COP9 signal body/deubiquitination) complexes. C9 may interact with related proteins through both the F-box and SKP1 to mediate the proteasomal degradation of specific host proteins, such as ISGs. The degradation of ISGs antagonizes the host antiviral status induced by IFNs (32). In addition, the N- and C-terminal portions of C9 bind interferon-induced proteins with tetratricopeptide repeats (IFITs) and ubiquitin regulatory complexes, respectively. Ectopic expression of C9 rescues IFN-induced inhibition of viral DNA replication in IFIT KO cell lines (33).
VACV A49 is phosphorylated at serine 7 but not serine 12, and this phosphorylation is necessary and sufficient for its binding to β-TrCP to antagonize NF-κB (34). A49 inhibits NF-κB activation by molecular mimicry and has a motif near the N-terminus that is conserved in IκBα, β-catenin, HIV Vpu, and some other proteins (34). Phosphorylation of A49 S7 occurs when NF-κB signaling is activated by the addition of IL-1β or overexpression of TRAF6 or IKKβ, the kinase needed for IκBα phosphorylation (34). Thus, A49 is an elegant biological regulation because it becomes an NF-κB antagonist upon activation of NF-κB signaling (34). Interestingly, A49 encodes a second, smaller polypeptide that is expressed via leaky scanning translation from methionine 20 and is unable to block NF-κB activation (35). Viruses engineered to express either only the large protein or only the small A49 protein both have lower virulence than wild-type virus and greater virulence than an A49L deletion mutant (35).
In summary, poxviruses encode PRANC proteins that can bind SKP1. The cullin-based E3 ubiquitin ligase consists of three functional components: a catalytic component consisting of a small RING domain protein to recruit the ubiquitin-conjugating enzyme, a cullin scaffold component, and a substrate recognition component that can bind the substrate and recruit it to the vicinity of the catalytic component.
In contrast to ANK repeat/F-box proteins that associate with Cul-1, noncanonical ANK protein 010 encoded by ECTV EVM010 interacts with host cullin-2 (Cul-2) via a C-terminal BC box (22). Bioinformatics and mass spectrometry approaches revealed that poxviral ANK ortholog groups IV and VI represent a novel class of viral ANK proteins targeting host Cul-2. In ECTV, ANK/BC proteins suppress the production of CXCL10, CCL5, and IFN, thus inhibiting innate immune signaling (22). Thus, it is speculated that the potential substrate of EVM010 is an innate immune signaling component.
MCTV MC132 was identified to associate with NF-κB subunit p65 by unbiased affinity purification. MC132 targeted p65 for ubiquitin-dependent proteasomal degradation by recruiting p65 to a host Cullin-5 (Cul-5)/BC complex, leading to effective suppression of NF-κB activity and thus benefiting virus replication (23). Poxvirus ANK/BC E3 ligase is illustrated in Figure 2B.
The molecular composition and function of the Cul-3-dependent E3 ligase complex has been revealed. In this complex, the binding of BTB domain-containing proteins to their substrates is mediated by Cul-3. Poxvirus is the only virus family known to encode BBK proteins, indicating that poxvirus may interact with Cul-3 to regulate the host ubiquitination pathway.
The BTB domains of the ECTV virus BBK proteins EVM150 and EVM167 bind to the N-terminal region of Cul-3 (Figure 2C). Moreover, EVM150 and EVM167 bind to E2-bound ubiquitin and Rocl, and the RING finger protein has the effect of an E3 ubiquitin ligase. In summary, EVM150 and EVM167 rely on Cul-3 to activate their ubiquitin ligase functions, recruiting as-yet unknown substrates for ubiquitination (24). In addition, the BBK proteins EVM18 and EVM27 also interact with Cul-3. It is not clear whether all BBK proteins in poxviruses are related to the ubiquitination pathway.
Studies have found that knocking out the BBK gene in VACV leads to a decrease in the number of mature progeny viruses (36). VACV bearing a BBK gene knockout mutation showed reduced toxicity in the nasal cavities of infected mice. VACV with BBK knockout also exhibited reduced cytopathic changes and formed fewer cell protuberances in vitro. The sheeppox virus BBK protein (SPPV)-019 is an important virulence factor. Calcium-independent cell adhesion is reduced in sheep infected with SPPV-019 knockout virus, indicating that SPPV-019 may regulate cell adhesion (25). When injected into sheep through the nasal cavity or an intradermal route, the SPPV-019-knockout virus was very weak, indicating that the SPPV-019 protein plays an important role in the life cycle of the poxvirus (25).
VACV encodes three BBK family proteins: C2L, F3L and A55R (26). The VACV production in mice subcutaneously injected with BBK-knockout VACV was very weak. However, the role of BBK in the ubiquitination pathway of host cells is still unclear. Previous studies showed that the N-terminal BTB-BACK (BB) domain of A55 binds directly to the Cul3 N-terminal domain (Cul3-NTD) (37). According to sequence analysis, D11L, C18L, G3L, and A57R encoded by VACV and M006, M008, M009, M014, and M140 encoded by MYXV also belong to the BBK family of proteins (27, 28).
MYXV causes widespread lethal multiple myxomatosis in European rabbits (38). The M153R gene of MYXV encodes the E3 ubiquitin ligase M153, which contains a RING-CH domain and two transmembrane domains in its N-terminus (Figure 2D). Proteins with such features are referred to as MARCH proteins. In the early stage of MYXV infection, M153 is translocated from the cytoplasm to the endoplasmic reticulum, which in turn upregulates the plasma membrane levels of MHC-I, CD4, activated leukocyte cell adhesion molecule (ALCAM) and the proapoptotic factor Fas/CD95. Those ubiquitinated substrates are then degraded by the lysosomal pathway. M153 inhibits the recognition of MHC-I and death signals by CD8 T lymphocytes and inhibits the recognition of MHC-II molecules by CD4 T lymphocytes (10). During MYXV infection, the RING-CH domain of M153 acts as a ubiquitin ligase to recognize and ubiquitinate the lysine residue in the cytoplasmic tail of CD4, which is responsible for the recognition of MHC-II molecules. Therefore, CD4 ubiquitination inhibits the recognition of MHC-II molecules. The ubiquitination and degradation of immune molecules on the cell surface induced by M153 is an important mechanism by which MYXV suppresses the immune response (10).
ECTV causes fatal skin damage in mice, and the P28 protein plays a key role in this process (39). P28 is a virulence factor with E3 ubiquitin ligase activity. P28 contains two functional domains, the DNA binding domain in the N-terminus and the RING domain in the C-terminus. The DNA binding domain is also called the KilA-N domain. The KilA-N domain plays an important role in the cytoplasmic localization of P28 (Figure 2E). Fowlpox virus (FWPV) encodes 2 functional P28 ubiquitin ligases, FWPV150 and FWPV157 (40). P28 completely loses its E3 ubiquitin ligase function when the RING domain of P28 is mutated. Therefore, the RING domain plays a role in maintaining its E3 ligase activity. Both the KilA-N domain and RING domain are crucial for the function of P28 ubiquitin ligases. ECTV and VARV P28 interact with the E2 ubiquitin-conjugating enzymes Ubc4 and UbcH5c to degrade substrates. When Ubc13/Uev1A is present, P28 can catalyze the Lys63 ubiquitination of multiple protein substrates (41).
Homologs of P28-containing proteins have also been found in other orthopoxviruses, such as VARV, CPXV, MPXV, VACV, fibroma virus (SFV) and MYXV. P28 in ECTV, M143R in MYXV (11), P28 in VACV (11) and other P28 E3 ubiquitin ligases all contain a RING finger structure. The structure of the P28/RING protein is relatively conserved among different poxviruses. Although the P28/RING protein is not essential for virus replication in cell culture, it is important for the pathogenic mechanism in virus-infected mice (12).
During coevolution with their hosts, poxviruses have incorporated host cell genes into their genomes and adapted them to promote the viral life cycle. In recent years, with in-depth research on the mechanisms of host–virus interactions, research on the function of the E3 ubiquitin ligases encoded by poxviruses has made significant progress. Recently, many new proteins and protein substrates have been discovered using different approaches. For example, bioinformatics analysis and quantitative proteomics were applied to discover that ECTV protein 010 is a noncanonical ANK protein that binds to Cul-2 (22). In contrast, a small interfering RNA (siRNA) approach was used in the discovery of RIPK3 (21). In an extensive crystal structure study, the Cul2-Rbx1-EloBC-VHL complex was revealed, extending the classification of viruses encoding RING E3s (42). Many poxvirus ubiquitin E3 ligases and adapter proteins play important roles in the viral life cycle, including in replication and productive infection. The main functions of poxvirus-encoded E3 ligases are related to host immune evasion (Tables 1, 2).
However, the current understanding of poxvirus E3 ubiquitin ligases is limited. More E3 ligases encoded by poxviruses will be discovered, and many poxvirus E3 ubiquitin ligase substrates remain to be explored. The homologies between genes from different poxviruses and between poxvirus genes and human genes have aided the rational discovery of new poxvirus-encoded E3 ligases, which will facilitate future studies in this field.
The known poxvirus E3 ubiquitin ligases involved in host immune invasion could be developed as potential inhibitors of the host immune system, providing new antiviral strategies. In view of the complex interactions between poxviruses and their hosts, the interplay between poxviruses and the ubiquitin system needs further exploration in the future. The identification of natural substrates of poxvirus E3 ubiquitin ligases will facilitate our understanding of host–virus interactions, and particularly the role of poxvirus E3 ubiquitin ligases in virus infection.
LZ conceived the work. HC and YZ draft the manuscript. LZ modified the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from National Natural Science Foundation of China [81871663 and 82072270], Academic promotion programme of Shandong First Medical University [2019LJ001], and Natural Science Foundation of Shandong Province [ZR2021QC095].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Van Vliet K, Mohamed MR, Zhang L, Villa NY, Werden SJ, Liu J, et al. Poxvirus Proteomics and Virus-Host Protein Interactions. Microbiol Mol Biol Rev (2009) 73:730–49. doi: 10.1128/MMBR.00026-09
2. Beachboard DC, Horner SM. Innate Immune Evasion Strategies of DNA and RNA Viruses. Curr Opin Microbiol (2016) 32:113–9. doi: 10.1016/j.mib.2016.05.015
3. Scheffner M, Nuber U, Huibregtse JM. Protein Ubiquitination Involving an E1-E2-E3 Enzyme Ubiquitin Thioester Cascade. Nature (1995) 373:81–3. doi: 10.1038/373081a0
4. Clague MJ, Urbé S, Komander D. Breaking the Chains: Deubiquitylating Enzyme Specificity Begets Function. Nat Rev Mol Cell Biol (2019) 20:338–52. doi: 10.1038/s41580-019-0099-1
5. Pérez Berrocal DA, Witting KF, Ovaa H, Mulder MPC. Hybrid Chains: A Collaboration of Ubiquitin and Ubiquitin-Like Modifiers Introducing Cross-Functionality to the Ubiquitin Code. Front Chem (2019) 7:931. doi: 10.3389/fchem.2019.00931
6. Morreale FE, Walden H. Types of Ubiquitin Ligases. Cell (2016) 165:248–248.e1. doi: 10.1016/j.cell.2016.03.003
7. Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem (2017) 86:129–57. doi: 10.1146/annurev-biochem-060815-014922
8. Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING Fingers Mediate Ubiquitin-Conjugating Enzyme (E2)-Dependent Ubiquitination. Proc Natl Acad Sci U States America (1999) 96:11364–9. doi: 10.1073/pnas.96.20.11364
9. Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A Family of Proteins Structurally and Functionally Related to the E6-AP Ubiquitin-Protein Ligase. Proc Natl Acad Sci USA (1995) 92:2563–7. doi: 10.1073/pnas.92.7.2563
10. Mansouri M, Bartee E, Gouveia K, Hovey Nerenberg BT, Barrett J, Thomas L, et al. The PHD/LAP-Domain Protein M153R of Myxomavirus Is a Ubiquitin Ligase That Induces the Rapid Internalization and Lysosomal Destruction of CD4. J Virol (2003) 77:1427–40. doi: 10.1128/JVI.77.2.1427-1440.2003
11. Nerenberg BT, Taylor J, Bartee E, Gouveia K, Barry M, Früh K. The Poxviral RING Protein P28 Is a Ubiquitin Ligase That Targets Ubiquitin to Viral Replication Factories. J Virol (2005) 79:597–601. doi: 10.1128/JVI.79.1.597-601.2005
12. Senkevich TG, Koonin EV, Buller RM. A Poxvirus Protein With a RING Zinc Finger Motif is of Crucial Importance for Virulence. Virology (1994) 198:118–28. doi: 10.1006/viro.1994.1014
13. Johnston JB, Wang G, Barrett JW, Nazarian SH, Colwill K, Moran M, et al. Myxoma Virus M-T5 Protects Infected Cells From the Stress of Cell Cycle Arrest Through Its Interaction With Host Cell Cullin-1. J Virol (2005) 79:10750–63. doi: 10.1128/JVI.79.16.10750-10763.2005
14. Blanie S, Mortier J, Delverdier M, Bertagnoli S, Camus-Bouclainville C. M148R and M149R Are Two Virulence Factors for Myxoma Virus Pathogenesis in the European Rabbit. Vet Res (2009) 40:11. doi: 10.1051/vetres:2008049
15. Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, et al. Poxvirus Host Range Protein CP77 Contains an F-Box-Like Domain That Is Necessary to Suppress NF-kappaB Activation by Tumor Necrosis Factor Alpha But is Independent of its Host Range Function. J Virol (2009) 83:4140–52. doi: 10.1128/JVI.01835-08
16. Sperling KM, Schwantes A, Schnierle BS, Sutter G. The Highly Conserved Orthopoxvirus 68k Ankyrin-Like Protein is Part of a Cellular SCF Ubiquitin Ligase Complex. Virology (2008) 374:234–9. doi: 10.1016/j.virol.2008.02.018
17. Herbert MH, Squire CJ, Mercer AA. Poxviral Ankyrin Proteins. Viruses (2015) 7:709–38. doi: 10.3390/v7020709
18. Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. Poxvirus Ankyrin Repeat Proteins are a Unique Class of F-Box Proteins That Associate With Cellular SCF1 Ubiquitin Ligase Complexes. Proc Natl Acad Sci USA (2008) 105:10955–60. doi: 10.1073/pnas.0802042105
19. Burles K, van Buuren N, Barry M. Ectromelia Virus Encodes a Family of Ankyrin/F-Box Proteins That Regulate Nfκb. Virology (2014) 468-470:351–62. doi: 10.1016/j.virol.2014.08.030
20. van Buuren N, Burles K, Schriewer J, Mehta N, Parker S, Buller RM, et al. EVM005: An Ectromelia-Encoded Protein With Dual Roles in NF-κb Inhibition and Virulence. PloS Pathog (2014) 10:e1004326. doi: 10.1371/journal.ppat.1004326
21. Liu Z, Nailwal H, Rector J, Rahman MM, Sam R, McFadden G, et al. A Class of Viral Inducer of Degradation of the Necroptosis Adaptor RIPK3 Regulates Virus-Induced Inflammation. Immunity (2021) 54:247–58.e7. doi: 10.1016/j.immuni.2020.11.020
22. Odon V, Georgana I, Holley J, Morata J, Maluquer de Motes C. Novel Class of Viral Ankyrin Proteins Targeting the Host E3 Ubiquitin Ligase Cullin-2. J Virol (2018) 92:e01374–18. doi: 10.1128/JVI.01374-18
23. Brady G, Haas DA, Farrell PJ, Pichlmair A, Bowie AG. Poxvirus Protein MC132 From Molluscum Contagiosum Virus Inhibits NF-B Activation by Targeting P65 for Degradation. J Virol (2015) 89:8406–15. doi: 10.1128/JVI.00799-15
24. Wilton BA, Campbell S, Van Buuren N, Garneau R, Furukawa M, Xiong Y, et al. Ectromelia Virus BTB/kelch Proteins, EVM150 and EVM167, Interact With Cullin-3-Based Ubiquitin Ligases. Virology (2008) 374:82–99. doi: 10.1016/j.virol.2007.11.036
25. Balinsky CA, Delhon G, Afonso CL, Risatti GR, Borca MV, French RA, et al. Sheeppox Virus Kelch-Like Gene SPPV-019 Affects Virus Virulence. J Virol (2007) 81:11392–401. doi: 10.1128/JVI.01093-07
26. Mercer AA, Fleming SB, Ueda N. F-Box-Like Domains are Present in Most Poxvirus Ankyrin Repeat Proteins. Virus Genes (2005) 31:127–33. doi: 10.1007/s11262-005-1784-z
27. Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao JX, Macaulay C, et al. The Complete DNA Sequence of Myxoma Virus. Virology (1999) 264:298–318. doi: 10.1006/viro.1999.0001
28. Kochneva G, Kolosova I, Maksyutova T, Ryabchikova E, Shchelkunov S. Effects of Deletions of Kelch-Like Genes on Cowpox Virus Biological Properties. Arch Virol (2005) 150:1857–70. doi: 10.1007/s00705-005-0530-0
29. Rohde J, Emschermann F, Knittler MR, Rziha HJ. Orf Virus Interferes With MHC Class I Surface Expression by Targeting Vesicular Transport and Golgi. BMC Vet Res (2012) 8:114. doi: 10.1186/1746-6148-8-114
30. van Buuren N, Couturier B, Xiong Y, Barry M. Ectromelia Virus Encodes a Novel Family of F-Box Proteins That Interact With the SCF Complex. J Virol (2008) 82:9917–27. doi: 10.1128/JVI.00953-08
31. Liu B, Panda D, Mendez-Rios JD, Ganesan S, Wyatt LS, Moss B. Identification of Poxvirus Genome Uncoating and DNA Replication Factors With Mutually Redundant Roles. J Virol (2018) 92:e02152–17. doi: 10.1128/JVI.02152-17
32. Liu R, Moss B. Vaccinia Virus C9 Ankyrin Repeat/F-Box Protein Is a Newly Identified Antagonist of the Type I Interferon-Induced Antiviral State. J Virol (2018) 92:e00053–18. doi: 10.1128/JVI.00053-18
33. Liu R, Olano LR, Mirzakhanyan Y, Gershon PD, Moss B. Vaccinia Virus Ankyrin-Repeat/F-Box Protein Targets Interferon-Induced IFITs for Proteasomal Degradation. Cell Rep (2019) 29:816–28.e6. doi: 10.1016/j.celrep.2019.09.039
34. Neidel S, Ren H, Torres AA, Smith GL. NF-κb Activation Is a Turn on for Vaccinia Virus Phosphoprotein A49 to Turn Off NF-κb Activation. Proc Natl Acad Sci USA (2019) 116:5699–704. doi: 10.1073/pnas.1813504116
35. Neidel S, Torres AA, Ren H, Smith GL. Leaky Scanning Translation Generates a Second A49 Protein That Contributes to Vaccinia Virus Virulence. J Gen Virol (2020) 101:533–41. doi: 10.1099/jgv.0.001386
36. Buller RM, Palumbo GJ. Poxvirus Pathogenesis. Microbiol Rev (1991) 55:80–122. doi: 10.1128/mr.55.1.80-122.1991
37. Gao C, Pallett MA, Croll TI, Smith GL, Graham SC. Molecular Basis of Cullin-3 (Cul3) Ubiquitin Ligase Subversion by Vaccinia Virus Protein A55. J Biol Chem (2019) 294:6416–29. doi: 10.1074/jbc.RA118.006561
38. Liu J, Wennier S, McFadden G. The Immunoregulatory Properties of Oncolytic Myxoma Virus and Their Implications in Therapeutics. Microbes Infect (2010) 12:1144–52. doi: 10.1016/j.micinf.2010.08.012
39. Mottet K, Bareiss B, Milne CD, Barry M. The Poxvirus Encoded Ubiquitin Ligase, P28, Is Regulated by Proteasomal Degradation and Autoubiquitination. Virology (2014) 468–470:363–78. doi: 10.1016/j.virol.2014.08.028
40. Bareiss B, Barry M. Fowlpox Virus Encodes Two P28-Like Ubiquitin Ligases That Are Expressed Early and Late During Infection. Virology (2014) 462-463:60–70. doi: 10.1016/j.virol.2014.05.018
41. Huang J, Huang Q, Zhou X, Shen MM, Yen A, Yu SX, et al. The Poxvirus P28 Virulence Factor is an E3 Ubiquitin Ligase. J Biol Chem (2004) 279:54110–6. doi: 10.1074/jbc.M410583200
Keywords: poxvirus, ubiquitin, E3 ubiquitin ligase, innate immune evasion, MARCH
Citation: Cui H, Zhang Y and Zhang L (2021) Progress on Poxvirus E3 Ubiquitin Ligases and Adaptor Proteins. Front. Immunol. 12:740223. doi: 10.3389/fimmu.2021.740223
Received: 12 July 2021; Accepted: 24 November 2021;
Published: 09 December 2021.
Edited by:
Konstantin Sparrer, Ulm University Medical Center, GermanyReviewed by:
Zheng Zhang, Second Affiliated Hospital of Southern University of Science and Technology, ChinaCopyright © 2021 Cui, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leiliang Zhang, YXJtemhhbmdAaG90bWFpbC5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.