- 1Department of Rheumatology and Immunology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Department of Epidemiology and Biostatistics, School of Public Health, Anhui Medical University, Anhui Provincial Laboratory of Inflammatory and Immune Diseases, Hefei, China
Abnormal vitamin D metabolism is involved in the pathogenesis of rheumatoid arthritis (RA). In this study, we evaluated the association of single nucleotide polymorphisms (SNPs) and methylation levels in vitamin D metabolic pathway genes with RA susceptibility. Ten SNPs in vitamin D metabolic pathway genes (CYP2R1, CYP24A1, VDR, CYP27B1) were genotyped in 477 RA patients and 496 controls by improved multiple ligase detection reaction (iMLDR). The methylation levels of the promoter regions of these genes were detected in 122 RA patients and 123 controls using Illumina Hiseq platform. We found that the CYP2R1 rs1993116 GA genotype, CYP27B1 rs4646536 GA genotype, rs4646536 A allele frequencies were significantly increased in RA patients when compared to controls. The decreased risk of rs1993116, rs4646536 was found under the dominant mode in RA patients. However, no significant association was found between CYP2R1 rs7936142, rs12794714, CYP24A1 rs2762934, rs6068816, rs2296239, rs2296241, VDR rs11574129, rs3847987 polymorphism, and RA susceptibility. The VDR, CYP27B1 methylation levels in RA patients were significantly lower than those in controls, while CYP2R1, CYP24A1 methylation levels were not associated with RA. There were no statistical associations between CYP2R1, CYP24A1, VDR, CYP27B1 methylation levels and their respective genotype in RA patients. In addition, plasma 25OHD level in RA patients was significantly lower than that in healthy controls. In summary, our results showed that CYP2R1, CYP27B1 genetic variations were associated with the genetic background of RA, while altered VDR, CYP27B1 methylation levels were related to the risk of RA.
Introduction
Rheumatoid arthritis (RA) is a common autoimmune, chronic inflammatory disease, with a prevalence of 1% around the world (1). The disease is characterized primarily by affecting peripheral joints of the hands, wrists, and feet and can eventually lead to the accumulation of joint damage and irreversible disability in patients (2). At present, the pathogenesis of RA has not been fully elucidated, and individual progress is highly variable during the development of RA; thence, early diagnosis and treatment to maximize the chance of inducing remission is important to prevent permanent disabling joint damage. Many studies have explored the influence of genetic susceptibilities, epigenomic features, abnormal immune response, and environmental factors on the onset, disease activity, and prognosis of RA (3–5). A large number of genes/loci and even some specific variants underlying RA in different races have been identified, with different study protocols like genome-wide association study (GWAS), and candidate gene approaches, while these genes/loci only account for a fraction of phenotypic variance in RA (6, 7). Therefore, it is necessary to study whether immune-modulating gene variation is related to RA susceptibility.
With the development of molecular biology, the regulation of epigenetic changes in gene expression and disease progression has received more and more attention (8). DNA methylation, which is an important key epigenetic trait, involves the addition of a methyl group to the cytosine of CpG dinucleotides and is associated with many biological processes, including X-chromosome inactivation, genomic imprinting, aging, and canceration (9). According to recent studies, epigenetic changes by DNA methylation were dynamic, individual, and highly important in inflammatory processes, and influencing mechanisms of DNA methylation such as DNA methyltransferases activity could directly affect the RA development and might be a very promising therapeutic target for RA (10, 11).
In another context, the association between DNA methylation and inflammation-regulating immune pathways might play a significant role in the pathogenesis of RA (11). Recent studies had suggested that the vitamin D metabolic pathway might be a potential contributor to RA (12). In vitro, vitamin D metabolites modulate inflammation by altering the function of T helper and regulatory T cell (12, 13). In vivo, vitamin D metabolites including the 25-hydroxyvitamin D (25OHD, the main circulating metabolite) and 1,25-dihydroxyvitamin D (1,25(OH)2D, the active form of vitamin D) had been reported to be associated with RA disease progression, and vitamin D supplementation might have beneficial effects for RA patients (14–16). Moreover, some studies also analyzed the correlation between vitamin D metabolism gene polymorphisms and RA, although the results were inconsistent (17, 18).
The methylation levels of several key genes in the vitamin D metabolic pathway were found to associate with the risk and prognosis of tuberculosis (19). However, the role of vitamin D metabolic pathway gene methylation levels in RA had not been systematically studied. Thus, we performed this molecular epidemiological study to explore the effect of genetic variation, aberrant DNA methylation in four key vitamin D metabolic pathway genes (CYP2R1, CYP24A1, VDR, CYP27B1) on the risk of RA in a Chinese population.
Materials and Methods
RA Patients and Normal Controls
In this case-control study, a total of 973 subjects including 477 RA patients and 496 normal controls were consecutively included to explore the relationship between CYP2R1, CYP24A1, VDR, CYP27B1 gene polymorphisms and RA susceptibility. Then, 122 RA patients and 123 normal controls were enrolled to detect their promoter methylation levels. The RA patients’ diagnosis was based on the 1987 American College of Rheumatology revised criteria (20), and peripheral blood samples and clinical information of all subjects were obtained from Anhui Provincial Laboratory of Inflammatory and Immune Diseases. The clinical data [including anticyclic citrullinate peptide (anti-CCP), rheumatoid factor (RF), etc.] and drug treatment (including glucocorticoid, methotrexate) of RA patients were collected. The normal controls did not have a history of inflammatory/autoimmune diseases or cancer.
SNP Selection
We first sought the genotype data of Han Chinese people in Beijing from CHBS_1000g and Ensembl Genome Browser 85. Then, we selected the tag SNPs, capturing all the common SNPs located in the chromosome locus transcribed into vitamin D metabolic pathway genes (CYP2R1, CYP24A1, VDR, CYP27B1) and their flanking 2000 bp region. The selection was performed with Haploview 4.0 software (Cambridge, MA, USA). Moreover, the existing studies on the association between CYP2R1, CYP24A1, VDR, CYP27B1 gene polymorphism and RA susceptibility were also reviewed to search for significant SNP. Finally, we selected one tagSNP (rs4646536) in CYP27B1, three tagSNPs (rs12794714, rs7936142, rs1993116) in CYP2R1, four tagSNPs (rs2762934, rs6068816, rs2296239, rs2296241) in CYP24A1, and two tagSNPs (rs11574129, rs3847987) in VDR for genotyping in the present study. Above SNPs accorded with MAF ≥ 0.05 in CHB and r2 threshold > 0.8.
Genotyping and Methylation Analysis
The genomic DNA was extracted from the peripheral blood leukocytes by the Flexi Gene-DNA Kit (Qiagen, Valencia, CA, USA). Improved multiple ligase detection reaction (iMLDR) genotyping assay was used for genotyping with technical support from the Center for Genetic & Genomic Analysis, Genesky Biotechnologies (Inc., Shanghai). Those individuals with 100% genotyping success rate for the above SNPs were included for final analysis.
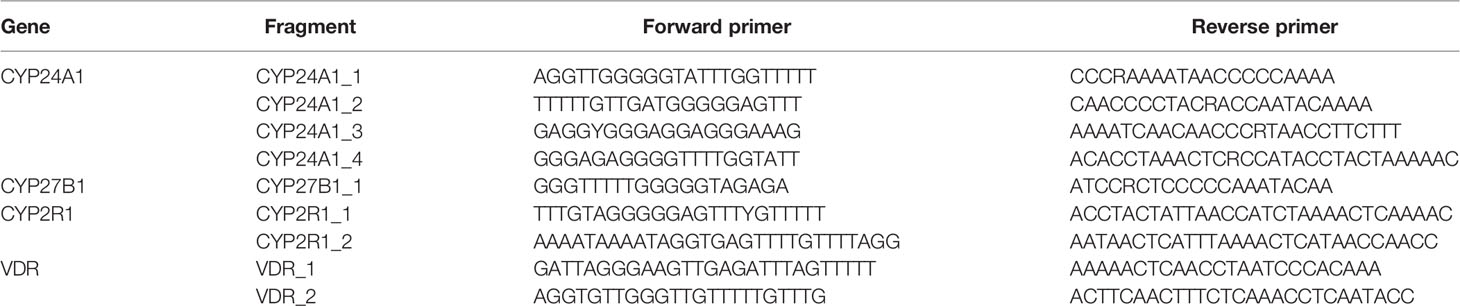
The methylation level of vitamin D metabolic pathway genes was detected using MethylTarget® with technical support from the Center for Genetic & Genomic Analysis, Genesky Biotechnologies (Inc., Shanghai). We sequenced the CpG islands in the promoter region of CYP2R1, CYP24A1, VDR, CYP27B1 by the Illumina Hiseq platform. Primers were designed to amplify the specific sites of interest from the bisulfite-converted DNA (Table 1), and the mean methylation level of all CpG sites on the fragment was calculated as the methylation level of the specific sites of each gene.
Enzyme-Linked Immunosorbent Assay
In this study, an additional 2 ml peripheral blood was collected from 84 RA patients and 84 normal patients by EDTA anticoagulant tube, and then plasma was obtained by Ficoll-Hypaque density gradient centrifugation. Plasma 25OHD level was determined by ELISA kits (MyBioSource Inc., USA), and the result was expressed as nanograms per milliliter.
Statistical Analysis
Statistical analysis was conducted with the SPSS 23.0 (Armonk, NY: IBM Corp, USA). Hardy-Weinberg equilibrium test was performed in normal controls with Chi-square (χ2). Logistic regression analysis was used to calculate the association between genotype, allele distribution frequencies of each SNP and RA risk. Two genetic models (dominant model, recessive model) were also analyzed, and SHEsis software was used to calculate haplotype analysis. The methylation levels of candidate genes were shown as median value and interquartile range, and the differences in methylation levels between two groups and three groups were analyzed by Mann-Whitney U test and Kruskal-Wallis H test, respectively. The diagnostic value of candidate gene methylation levels in RA patients was calculated by receiver operating characteristic (ROC) analysis. P value < 0.05 was considered as statistically significant.
Results
Association of Vitamin D Metabolic Pathway Gene Polymorphisms With RA Susceptibility
In this step, we included 477 RA patients and 496 controls. The RA patients consisted of 389 females and 88 males, with an average age of 52.70 ± 12.25 years, and 384 females and 112 males were enrolled in controls with a mean age of 50.61 ± 14.76 years. Table 2 shows the allele and genotype frequency distributions of all SNPs in RA patients and normal controls, and all SNPs were conformed to Hardy Weinberg equilibrium in controls.
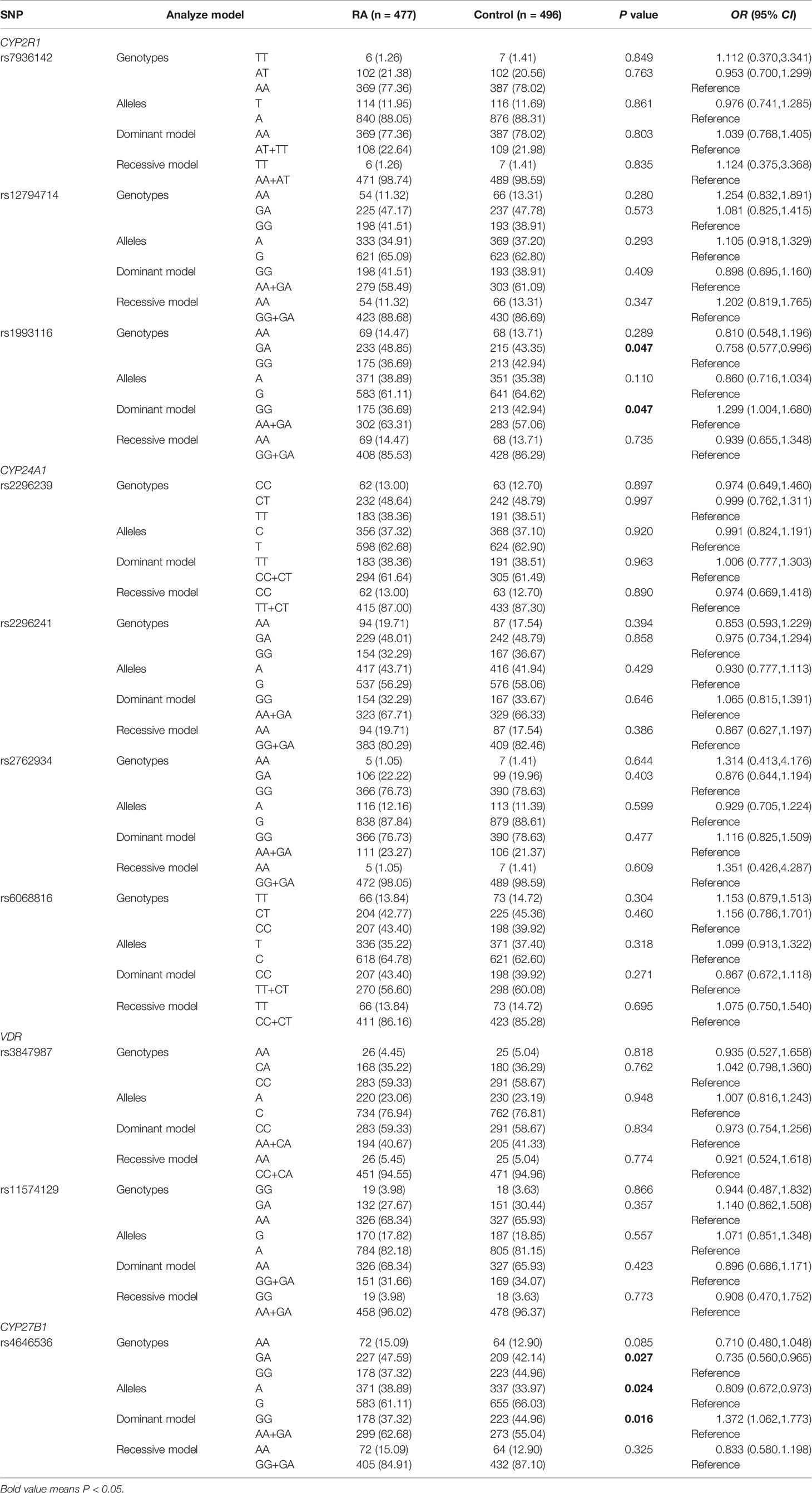
Table 2 Genotypes and allele frequencies of vitamin D metabolic pathway genes in RA patients and controls.
We found that CYP2R1 rs1993116 GA genotype frequency was significantly higher in RA patients than that in normal controls (GA versus GG: P = 0.047), while a decreased risk of rs1993116 was observed under the dominant mode (GG versus AA+GA: P = 0.047). In the CYP27B1 gene, the rs4646536 GA genotype and A allele frequencies were significantly increased in RA patients in comparison to normal controls (GA versus GG: P = 0.027; A versus G: P = 0.024, respectively). Moreover, a decreased risk of rs4646536 variant was found under dominant mode (GG versus AA+GA: P = 0.016). However, there was no significant difference in allele and genotype distribution of the CYP2R1 rs7936142 and rs12794714 between RA patients and normal controls (all P>0.05). Similarly, CYP24A1 gene rs2762934, rs6068816, rs2296239, rs2296241, and VDR gene rs11574129, rs3847987 polymorphisms were not significantly associated with RA.
To explore the potential relationship between CYP2R1, CYP24A1, VDR, and CYP27B1 genetic variation and anti-CCP, RF status in RA patients, we also conducted a case-only analysis (Table 3). We found that allele and genotype frequencies of all SNPs did not have statistically significant differences between anti-CCP-positive RA patients and anti-CCP-negative RA patients, as well as RA patients with RF-positive and with RF-negative.
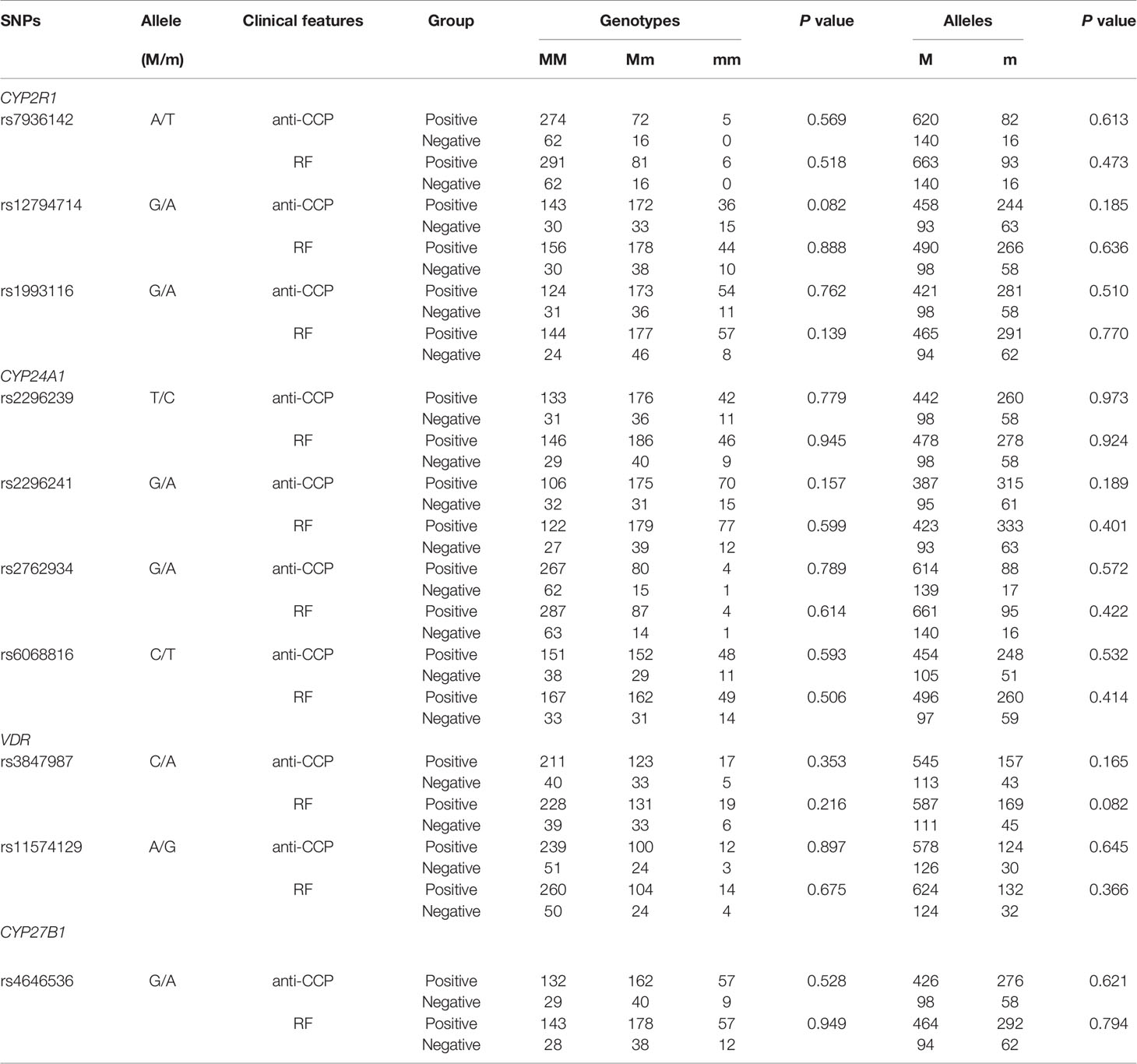
Table 3 Association between vitamin D metabolic pathway gene polymorphisms and anti-CCP, RF in RA patients.
Haplotype Analysis
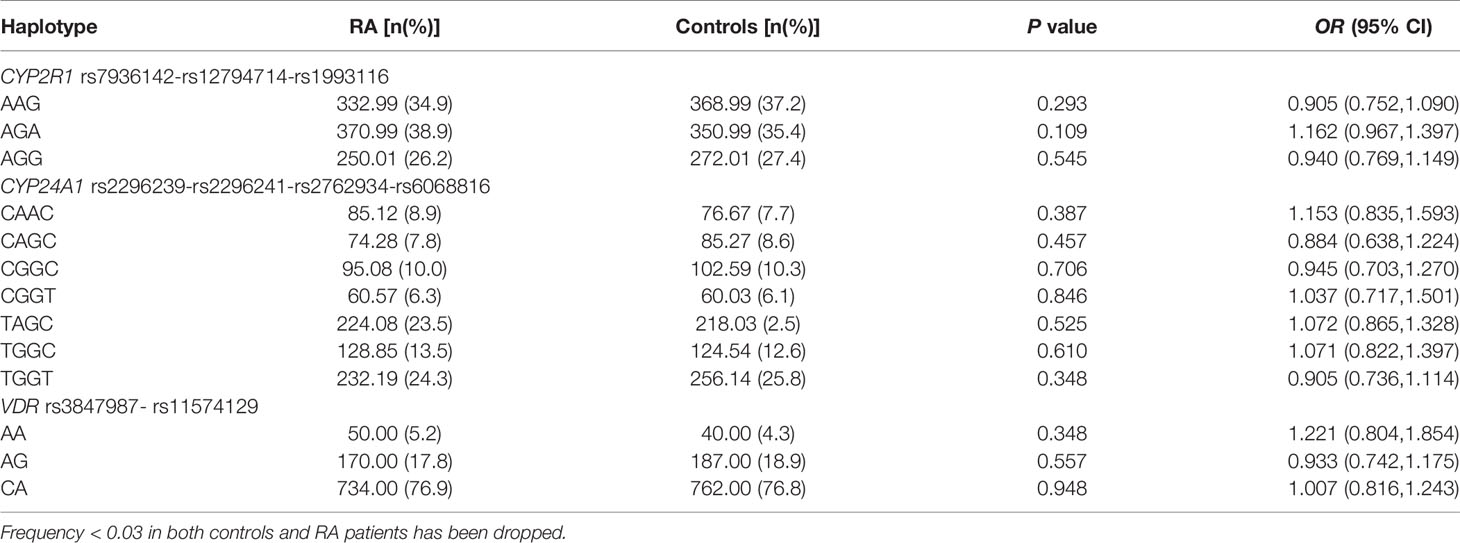
We constructed the haplotypes of CYP2R1, CYP24A1, VDR through SHEsis software and analyzed the relationship between these haplotypes and RA susceptibility. Seven main haplotypes (CAAC, CAGC, CGGC, CGGT, TAGC, TGGC, TGGT) for CYP24A1, three main haplotypes (AAG, AGA, AGG) for CYP2R1, and three main haplotypes (AA, AG, CA) for VDR were detected by SHEsis in this study (Table 4). The results demonstrated that there was no statistically significant difference in the frequency distribution of the above haplotypes between RA patients and normal controls (all P>0.05).
The Methylation Levels of Vitamin D Metabolic Pathway Genes in RA Patients and Normal Controls
In this step, the RA group included 100 females and 22 males, with a mean age of 52.61±13.05 years, and the control group included 82 females and 41 males, with an average age of 46.93±14.29 years. The methylation levels of specific sites between RA patients and controls are shown in Table 5. The results demonstrated that CYP24A1_1, CYP27B1_1, and VDR_1 methylation levels were significantly lower in RA patients than that in normal controls (P=0.032, P<0.001, P<0.001, respectively).
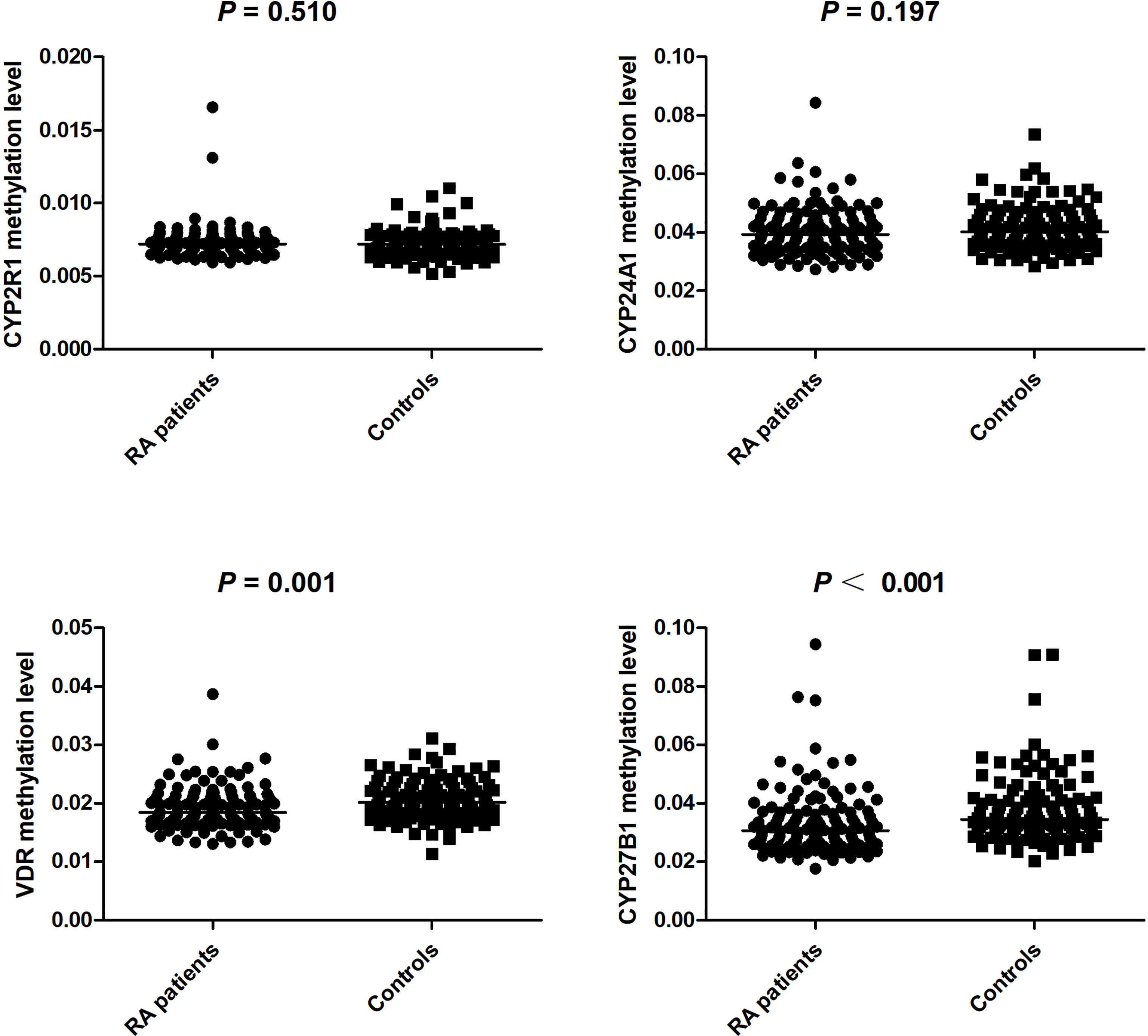
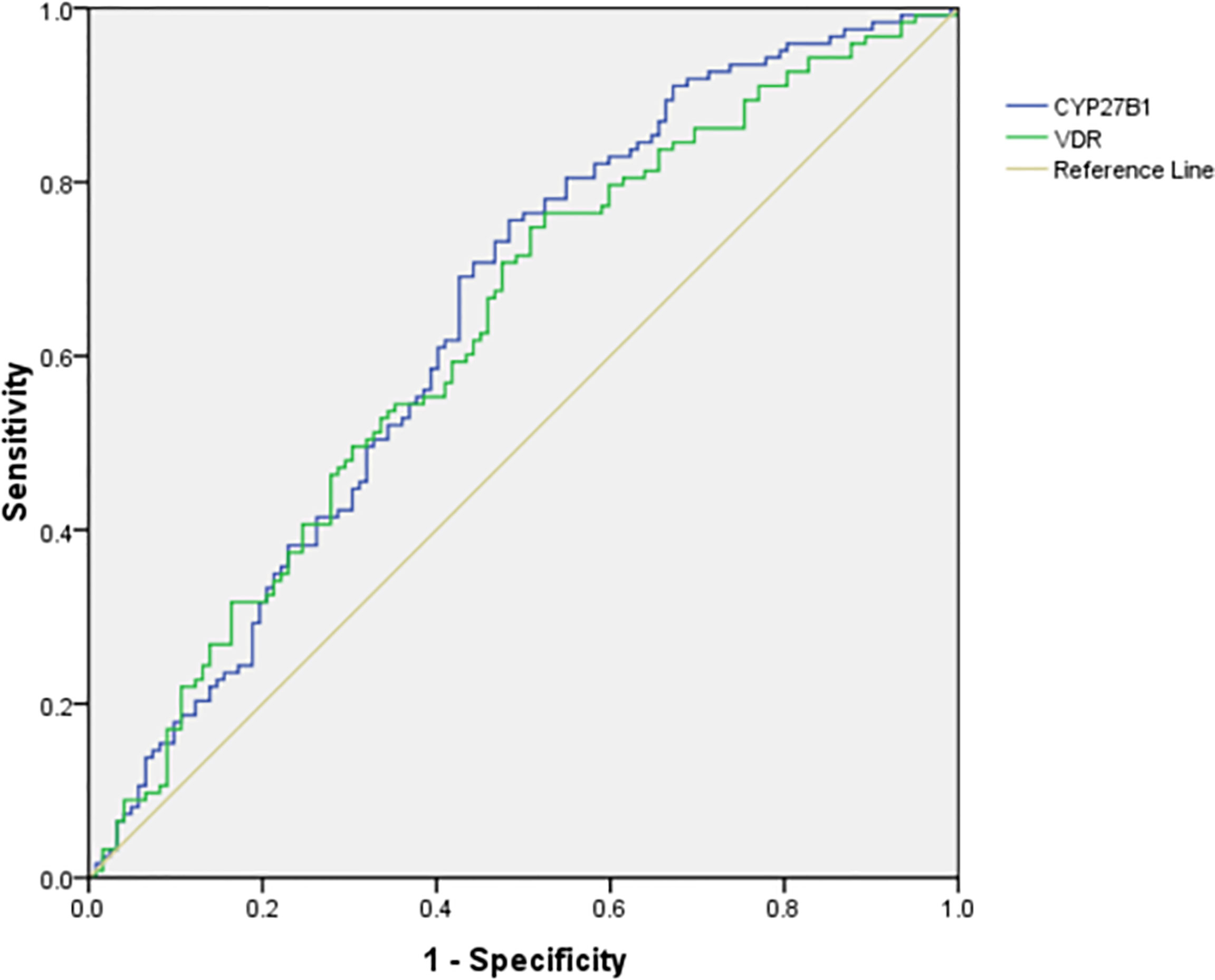
We further calculated the cumulative methylation levels of each gene by calculating the mean methylation levels of all CpG sites on the included fragments. The methylation levels of VDR and CYP27B1 were significantly reduced when compared to normal controls, while the differences in CYP2R1 and CYP24A1 methylation levels between RA patients and controls were not statistically significant (Figure 1). The diagnostic value of VDR and CYP27B1 methylation levels for RA diagnosis was also assessed, and the AUCs of VDR and CYP27B1 were 0.628 (0.559–0.698) and 0.645 (0.575–0.714), respectively (Figure 2). Moreover, the optimal cutoff value of VDR for RA diagnosis was 0.018, and the corresponding sensitivity and specificity were 74.8 and 49.2%, respectively. The optimal cutoff value of CYP27B1 for RA diagnosis was 0.031, and the corresponding sensitivity and specificity were 75.6 and 51.2%, respectively.
We also analyzed the influences of the main drug treatment on the methylation levels of these genes, and there were no significant associations regarding CYP24A1, CYP27B1, CYP2R1, and VDR methylation levels between RA patients being treated with glucocorticoid and without, as well as the patients being treated with methotrexate and without (Table 6). In addition, the results revealed that CYP24A1, CYP27B1, CYP2R1, and VDR methylation levels were not associated with anti-CCP, RF in RA patients (Table 6). Nevertheless, the CYP2R1 methylation level was positively associated with erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) in RA patients (P = 0.003, P = 0.018, respectively) (Table 7). No significant correlations were observed about CYP27B1, CYP2R1, and VDR methylation levels and ESR and CRP in RA patients.
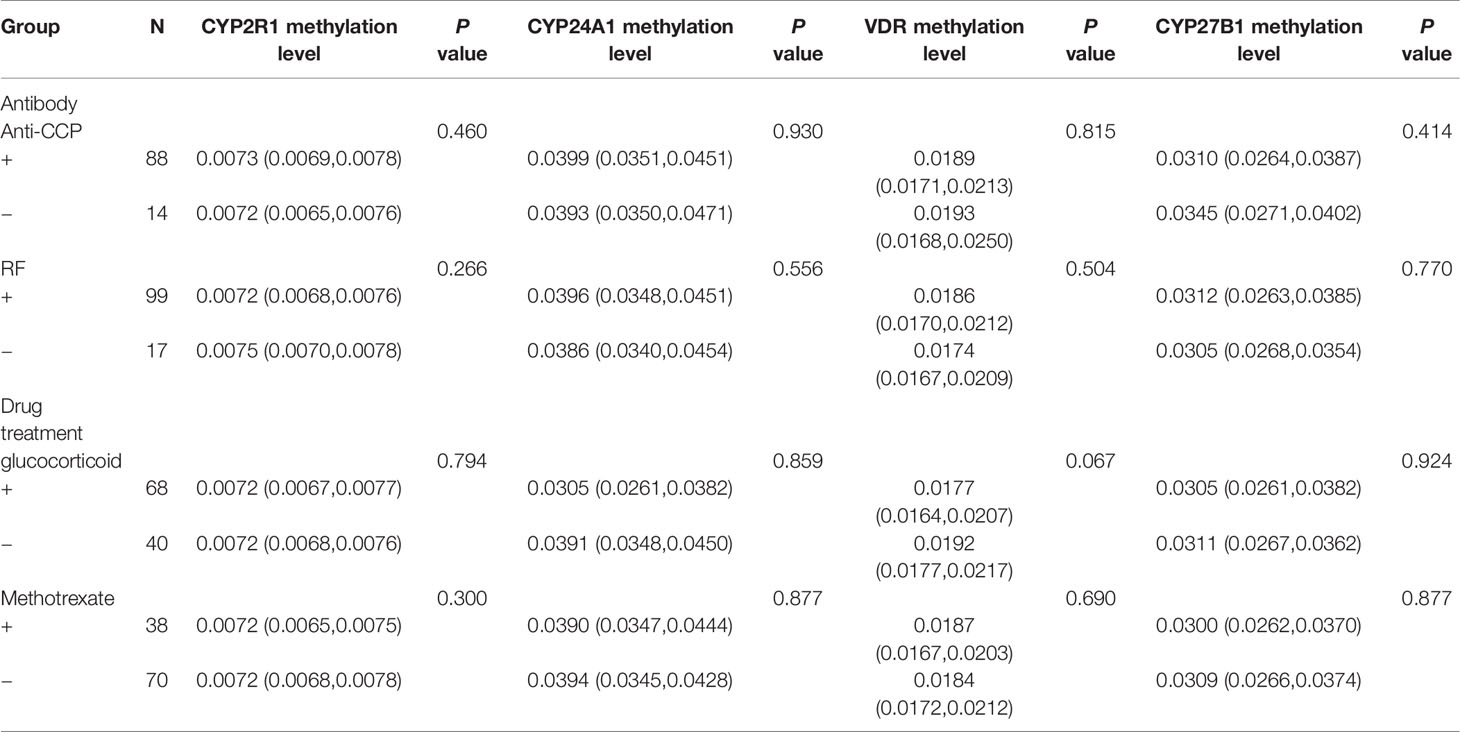
Table 6 The association between CYP24A1, CYP27B1, CYP2R1, VDR methylation levels and antibody, drug treatment in RA patients.
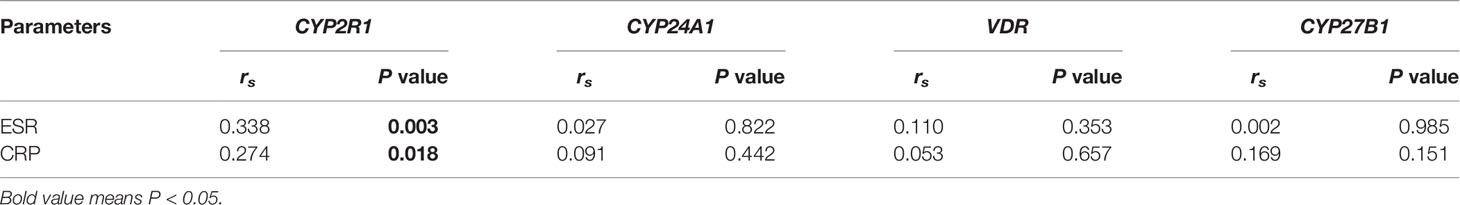
Table 7 The relationship between CYP24A1, CYP27B1, CYP2R1, VDR methylation levels and ESR and CRP in RA patients.
Associations Between Vitamin D Metabolic Pathway Gene Polymorphisms With Their Methylation Levels in RA Patients
To explore the associations between the genotype frequencies of CYP2R1, CYP24A1, VDR, and CYP27B1 genes and their methylation levels among RA patients, we included 122 RA patients for analysis. The results showed no statistical associations between CYP2R1, CYP24A1, VDR, CYP27B1 methylation levels and their respective genotype in RA patients (Table 8).
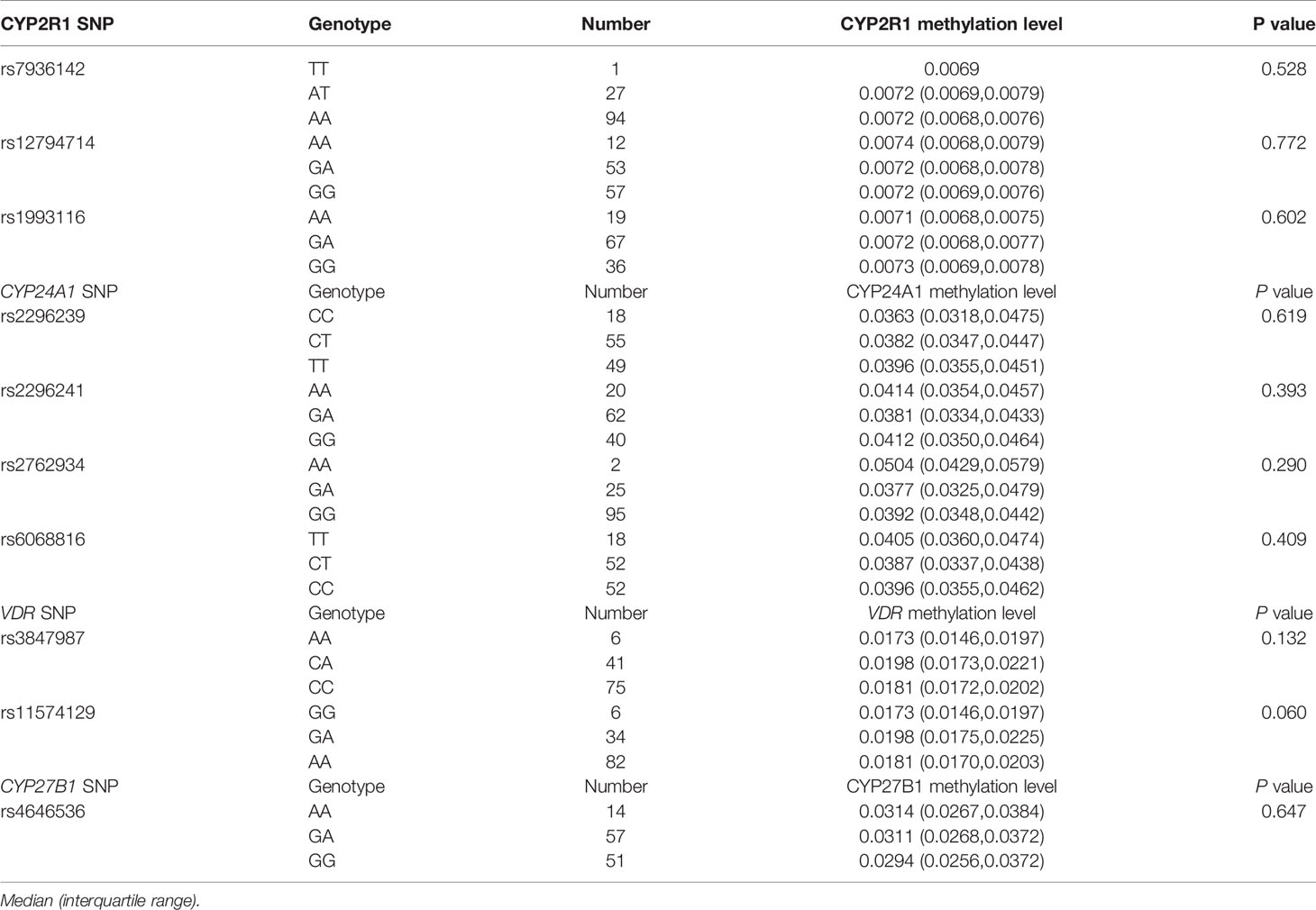
Table 8 Association between vitamin D metabolic pathway gene polymorphisms with their methylation levels in RA patients.
Plasma Level of 25OHD From RA Patients and Normal Controls
Finally, plasma 25OHD level was measured in 84 RA patients and 84 normal controls in this study. There were 76 females and 8 males in the RA group, with an average age of 53.15 ± 11.98 years. The control group included 77 females and 7 males, with a mean age of 52.57 ± 9.56 years. We found that the plasma level of 25OHD in RA patients (34.20 ± 5.15 ng/ml) was significantly lower than that in healthy controls (41.09 ± 7.52 ng/ml) (P<0.001).
Discussion
Epidemiological studies had shown a high prevalence of vitamin D deficiency in autoimmune diseases, which could lead to worse disease clinical activity and progression of RA, systemic lupus erythematosus (SLE), and multiple sclerosis (MS) (21). Vitamin D deficiency in patients with autoimmune diseases and general population might be caused by several factors, including skin pigmentation, lack of exposure to sunlight, glucocorticoids use, genetic background, and age (22). Previous studies had identified the potential role of multiple genes, which could regulate vitamin D levels, and suggested that SNPs in these genes (CYP27B1, CYP2R1, VDR, etc.) were related to decreased vitamin D level (17, 23). Therefore, in-depth exploration of the association between vitamin D metabolic pathway gene SNPs and genetic risk of autoimmune diseases was helpful to further reveal the pathogenesis of these diseases. In this study, we analyzed the relationship between 10 SNPs in vitamin D metabolic pathway genes (CYP2R1, CYP24A1, CYP27B1, VDR) and RA susceptibility in a Chinese population, and detected the methylation levels of these genes in RA patients.
Among the vitamin D metabolic pathway genes, the presence of SNPs might influence autoimmune disease genetic susceptibility through causing vitamin D deficiency, and modulate the disease activity in type 1 diabetes (TID), SLE, MS. Chen et al. investigated the association of VDR polymorphism and genetic risk of SLE in a Chinese population and found that VDR rs2228570, rs1544410 polymorphism, and their interaction were all associated with increased SLE risk (24). The role of vitamin D metabolic pathway genes in RA had also been reported in previous studies, such as CYP2R1 rs10741657 played an effect on vitamin D levels in RA patients (25). In the present study, we found that CYP2R1 rs1993116, CYP27B1 rs4646536 polymorphisms were significantly associated with RA susceptibility. In addition to this result, the decreased risk of rs1993116, rs4646536 variant was found to be related to RA risk in dominant mode. Previous studies suggested that rs1993116, rs4646536 variants were closely associated with vitamin D deficiency in human diseases (26, 27). Therefore, we assumed that rs1993116, rs4646536 might be involved in the development of RA by affecting vitamin D status, and the mechanism needed to be validated by more rigorous studies with a larger sample size and different ethnic population. CYP2R1 rs12794714, CYP24A1 rs2762934, rs6068816, and VDR rs11574129 had been reported to be involved in the genetic background of multiple diseases including diabetic ischemic stroke, gastric cancer, and other diseases (28–31). Unfortunately, this study did not demonstrate a significant association between these SNPs and RA risk. The inconsistencies might be explained by the genetic background of different diseases, sample size, different races, and experimental methods. RA patients could be divided into different genetic subsets according to the antibody status, including RF and anti-CCP (4), while we did not find any significant association with RF and anti-CCP status among RA patients.
In addition to the DNA sequence, genetic information also existed in epigenetic variation, and the role of epigenetic variation in the pathogenesis of diseases should not be overlooked (8, 32). For example, promoter methylation was involved in tumor development by silencing tumor suppressor genes (33). DNA hypomethylation was related to differentiation and proliferation of inflammatory processes and might lead to increased transcription and secretion of inflammatory proteins (34). A previous study was performed to detect the methylation status of lymphatic cells in SLE, RA patients, and found a significant hypomethylation in T cells (11, 35). The role of vitamin D metabolic pathway gene methylation in the development of human disease had been studied, and Wang et al. suggested that the methylation levels of the CYP24A1, CYP27A1, CYP27B1, CYP2R1, and VDR genes were associated with the risk and prognosis of tuberculosis (19). Another study also showed that cytochrome P450 gene silencing caused by hypermethylation in the promoter region might affect vitamin D activity (36). In the present study, we found that VDR and CYP27B1 methylation levels of RA patients were significantly decreased in comparison to normal controls, and ROC curves showed that these genes could be potential biomarkers for the diagnosis of RA. Moreover, it was necessary to further explore the combined diagnostic effect of VDR, CYP27B1, and other indicators, to improve the sensitivity and specificity of these indicators in RA diagnosis. We also found that CYP24A1_1 (one fragment) level was decreased in RA patients, and CYP2R1 methylation level was significantly associated with ESR and CRP. These suggested that CYP24A1, CYP2R1 gene might be involved in the disease process of RA, but further verification was needed. The relationship between methylation and genetic variation among individuals had also been reported in previous studies (37). We further explored the associations between the genotype frequencies of CYP2R1, CYP24A1, VDR, CYP27B1 genes and their methylation levels among RA patients; however, no statistical significance was found.
Methotrexate was the first-line therapy in early RA and was often prescribed in combination with glucocorticoid, hydroxychloroquine, etc. Previous studies had suggested that the anti-inflammatory mechanism of low-dose methotrexate treatment used in RA might relate to the inhibition of key enzymes in the purine de novo synthesis pathway and release of anti-inflammatory adenosine (38). In addition, methotrexate could inhibit methionine S-adenosyltransferase (MAT), followed by the inhibition of S-adenosyl methionine (SAM) in vivo and in vitro; moreover, SAM was responsible for the donation of methyl groups required for global DNA methylation (39, 40). Therefore, methotrexate was hypothesized to affect global DNA methylation, and one study found that higher baseline global DNA methylation was associated with methotrexate non-response (40). However, we observed that the use of methotrexate, as well as glucocorticoids did not have any significant influence on the methylation levels of these genes.
In conclusion, our results demonstrated that CYP2R1 rs1993116, CYP27B1 rs4646536 polymorphisms might contribute to the genetic predisposition to RA, while CYP24A1 and VDR gene polymorphisms were not associated with RA susceptibility in a Chinese population. The methylation levels of VDR and CYP27B1 genes were significantly related to the risk of RA and might be regarded as auxiliary biomarkers for RA diagnosis. Furthermore, we also found that the plasma vitamin D level in RA patients was significantly reduced, which was consistent with previous studies. The above results implied that it was of great significance to explore the role of vitamin D metabolism abnormality in the pathogenesis of RA. However, there were some limitations in this that study should be acknowledged. First, this was a case-control study, and we were unable to evaluate the relationship between the methylation levels of these genes and disease activity, medications, clinical efficacy of RA patients over a long period. Second, this study did not analyze the potential influence of ethnic background and environmental factors, as well as the interaction between environmental factors and genetic variation, in RA patients. Finally, the genotyping and ELISA tests were not performed in the same samples, and we were unable to further analyze the relationship between gene polymorphism, methylation, and vitamin D level. The precise role of vitamin D metabolic pathway genes in RA development needed to be further explored in repetitive, functional studies in the future.
Data Availability Statement
The data presented in the study are deposited in the dbSNP repository, accession number 1063300. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of the First Affiliated Hospital of USTC. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
X-ML and T-PZ designed the study. T-PZ and H-ML conducted the experiment. H-ML performed the statistical analyses. QH and LW participated in the collection of samples. T-PZ drafted the manuscript. X-ML and LW contributed to manuscript revision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (WK9110000180, WK9110000148), National Natural Science Foundation of China (81871271), and Anhui Provincial Natural Science Foundation (2108085QH362).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rossini M, Rossi E, Bernardi D, Viapiana O, Gatti D, Idolazzi L, et al. Prevalence and Incidence of Rheumatoid Arthritis in Italy. Rheumatol Int (2014) 34:659–64. doi: 10.1007/s00296-014-2974-6
2. Imagama T, Tokushige A, Seki K, Taguchi T. Weight Bearing Joints Destruction in Rheumatoid Arthritis. Curr Rheumatol Rev (2017) 13:37–42. doi: 10.2174/1573397112666160331142548
3. Taneja V. Cytokines Pre-Determined by Genetic Factors Are Involved in Pathogenesis of Rheumatoid Arthritis. Cytokine (2015) 75:216–21. doi: 10.1016/j.cyto.2014.11.028
4. Zhang TP, Lv TT, Xu SZ, Pan HF, Ye DQ. Association of Interleukin-10 Gene Single Nucleotide Polymorphisms With Rheumatoid Arthritis in a Chinese Population. Postgrad Med J (2018) 94:284–8. doi: 10.1136/postgradmedj-2017-135441
5. Rodríguez-Ubreva J, de la Calle-Fabregat C, Li T, Ciudad L, Ballestar ML, Català-Moll F, et al. Inflammatory Cytokines Shape a Changing DNA Methylome in Monocytes Mirroring Disease Activity in Rheumatoid Arthritis. Ann Rheum Dis (2019) 78:1505–16. doi: 10.1136/annrheumdis-2019-215355
6. Kwon YC, Lim J, Bang SY, Ha E, Hwang MY, Yoon K, et al. Genome-Wide Association Study in a Korean Population Identifies Six Novel Susceptibility Loci for Rheumatoid Arthritis. Ann Rheum Dis (2020) 79:1438–45. doi: 10.1136/annrheumdis-2020-217663
7. Zhang TP, Zhu BQ, Tao SS, Fan YG, Li XM, Pan HF, et al. Long Non-Coding RNAs Genes Polymorphisms and Their Expression Levels in Patients With Rheumatoid Arthritis. Front Immunol (2019) 10:2529. doi: 10.3389/fimmu.2019.02529
8. Manjrekar J. Epigenetic Inheritance, Prions and Evolution. J Genet (2017) 96:445–56. doi: 10.1007/s12041-017-0798-3
9. Guastafierro T, Bacalini MG, Marcoccia A, Gentilini D, Pisoni S, Di Blasio AM, et al. Genome-Wide DNA Methylation Analysis in Blood Cells From Patients With Werner Syndrome. Clin Epigenet (2017) 9:92. doi: 10.1186/s13148-017-0389-4
10. Castillo-Aguilera O, Depreux P, Halby L, Arimondo PB, Goossens L. DNA Methylation Targeting: The DNMT/HMT Crosstalk Challenge. Biomolecules (2017) 7:3. doi: 10.3390/biom7010003
11. Liebold I, Grützkau A, Göckeritz A, Gerl V, Lindquist R, Feist E, et al. Peripheral Blood Mononuclear Cells Are Hypomethylated in Active Rheumatoid Arthritis and Methylation Correlates With Disease Activity. Rheumatol (Oxford) (2021) 60:1984–95. doi: 10.1093/rheumatology/keaa649
12. Jeffery LE, Raza K, Hewison M. Vitamin D in Rheumatoid Arthritis-Towards Clinical Application. Nat Rev Rheumatol (2016) 12:201–10. doi: 10.1038/nrrheum.2015.140
13. Colin EM, Asmawidjaja PS, van Hamburg JP, Mus AM, van Driel M, Hazes JM, et al. 1,25-Dihydroxyvitamin D3 Modulates Th17 Polarization and Interleukin-22 Expression by Memory T Cells From Patients With Early Rheumatoid Arthritis. Arthritis Rheum (2010) 62:132–42. doi: 10.1002/art.25043
14. Rossini M, Maddali Bongi S, La Montagna G, Minisola G, Malavolta N, Bernini L, et al. Vitamin D Deficiency in Rheumatoid Arthritis: Prevalence, Determinants and Associations With Disease Activity and Disability. Arthritis Res Ther (2010) 12:R216. doi: 10.1186/ar3195
15. Soubrier M, Lambert C, Combe B, Gaudin P, Thomas T, Sibilia J, et al. A Randomised, Double-Blind, Placebo-Controlled Study Assessing the Efficacy of High Doses of Vitamin D on Functional Disability in Patients With Rheumatoid Arthritis. Clin Exp Rheumatol (2018) 36:1056–60.
16. Chandrashekara S, Patted A. Role of Vitamin D Supplementation in Improving Disease Activity in Rheumatoid Arthritis: An Exploratory Study. Int J Rheum Dis (2017) 20:825–31. doi: 10.1111/1756-185X.12770
17. Ruiz-Ballesteros AI, Meza-Meza MR, Vizmanos-Lamotte B, Parra-Rojas I, de la Cruz-Mosso U. Association of Vitamin D Metabolism Gene Polymorphisms With Autoimmunity: Evidence in Population Genetic Studies. Int J Mol Sci (2020) 21:9626. doi: 10.3390/ijms21249626
18. Zhang WT, Jin TF, Chen L. Associations of Four Common VDR Polymorphisms With Rheumatoid Arthritis and Systemic Lupus Erythematosus: Evidence From a Meta-Analysis. Lupus (2020) 29:364–70. doi: 10.1177/0961203320909432
19. Wang M, Kong W, He B, Li Z, Song H, Shi P, et al. Vitamin D and the Promoter Methylation of its Metabolic Pathway Genes in Association With the Risk and Prognosis of Tuberculosis. Clin Epigenet (2018) 10:118. doi: 10.1186/s13148-018-0552-6
20. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 Revised Criteria for the Classification of Rheumatoid Arthritis. Arthritis Rheum (1988) 31:315–24. doi: 10.1002/art.1780310302
21. Arnson Y, Amital H, Shoenfeld Y. Vitamin D and Autoimmunity: New Aetiological and Therapeutic Considerations. Ann Rheum Dis (2007) 66:1137–42. doi: 10.1136/ard.2007.069831
22. Brouwer-Brolsma EM, Vaes AMM, van der Zwaluw NL, van Wijngaarden JP, Swart KMA, Ham AC, et al. Relative Importance of Summer Sun Exposure, Vitamin D Intake, and Genes to Vitamin D Status in Dutch Older Adults: The B-PROOF Study. J Steroid Biochem Mol Biol (2016) 164:168–76. doi: 10.1016/j.jsbmb.2015.08.008
23. Sepulveda-Villegas M, Elizondo-Montemayor L, Trevino V. Identification and Analysis of 35 Genes Associated With Vitamin D Deficiency: A Systematic Review to Identify Genetic Variants. J Steroid Biochem Mol Biol (2020) 196:105516. doi: 10.1016/j.jsbmb.2019.105516
24. Chen XE, Chen P, Chen SS, Lu J, Ma T, Shi G, et al. A Population Association Study of Vitamin D Receptor Gene Polymorphisms and Haplotypes With the Risk of Systemic Lupus Erythematosus in a Chinese Population. Immunol Res (2017) 65:750–6. doi: 10.1007/s12026-017-8914-2
25. Rodríguez-Carrio J, Alperi-López M, Naves-Díaz M, Dusso A, López P, Ballina-García FJ, et al. Vitamin D Receptor Polymorphism and DHCR7 Contribute to the Abnormal Interplay Between Vitamin D and Lipid Profile in Rheumatoid Arthritis. Sci Rep (2019) 9:2546. doi: 10.1038/s41598-019-38756-8
26. Yu S, Li X, Wang Y, Mao Z, Xie Y, Zhang L, et al. Family-Based Association Between Allele T of Rs4646536 in CYP27B1 and Vitamin D Deficiency. J Clin Lab Anal (2019) 33:e22898. doi: 10.1002/jcla.22898
27. Arai T, Atsukawa M, Tsubota A, Koeda M, Yoshida Y, Okubo T, et al. Association of Vitamin D Levels and Vitamin D-Related Gene Polymorphisms With Liver Fibrosis in Patients With Biopsy-Proven Nonalcoholic Fatty Liver Disease. Dig Liver Dis (2019) 51:1036–42. doi: 10.1016/j.dld.2018.12.022
28. Nam HK, Rhie YJ, Lee KH. Vitamin D Level and Gene Polymorphisms in Korean Children With Type 1 Diabetes. Pediatr Diabetes (2019) 20:750–8. doi: 10.1111/pedi.12878
29. Yang W, Ma F, Wang L, He X, Zhang H, Zheng J, et al. The Association Analysis Between CYP24A1 Genetic Polymorphisms and the Risk of Ischemic Stroke in Chinese Han Population. Brain Behav (2020) 10:e01503. doi: 10.1002/brb3.1503
30. Xiong Q, Jiao Y, Yang P, Liao Y, Gu X, Hu F, et al. The Association Study Between CYP24A1 Gene Polymorphisms and Risk of Liver, Lung and Gastric Cancer in a Chinese Population. Pathol Res Pract (2020) 216:153237. doi: 10.1016/j.prp.2020.153237
31. Jia J, Ding H, Yang K, Mao L, Zhao H, Zhan Y, et al. Vitamin D Receptor Genetic Polymorphism Is Significantly Associated With Risk of Type 2 Diabetes Mellitus in Chinese Han Population. Arch Med Res (2015) 46:572–9. doi: 10.1016/j.arcmed.2015.09.006
32. Shnorhavorian M, Schwartz SM, Stansfeld B, Sadler-Riggleman I, Beck D, Skinner MK. Differential DNA Methylation Regions in Adult Human Sperm Following Adolescent Chemotherapy: Potential for Epigenetic Inheritance. PloS One (2017) 12(2):e0170085. doi: 10.1371/journal.pone.0170085
33. Gazdzicka J, Goła˛bek K, Strzelczyk JK, Ostrowska Z. Epigenetic Modifications in Head and Neck Cancer. Biochem Genet (2020) 58:213–44. doi: 10.1007/s10528-019-09941-1
34. Burmester GR, Feist E, Dorner T. Emerging Cell and Cytokine Targets in Rheumatoid Arthritis. Nat Rev Rheumatol (2014) 10:77–88. doi: 10.1038/nrrheum.2013.168
35. Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for Impaired T Cell DNA Methylation in Systemic Lupus Erythematosus and Rheumatoid Arthritis. Arthritis Rheum (1990) 33:1665–73. doi: 10.1002/art.1780331109
36. Zhu H, Wang X, Shi H, Su S, Harshfield GA, Gutin B, et al. A Genome-Wide Methylation Study of Severe Vitamin D Deficiency in African American Adolescents. J Pediatr (2013) 162:1004–9.e1. doi: 10.1016/j.jpeds.2012.10.059
37. Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, et al. Human Body Epigenome Maps Reveal Noncanonical DNA Methylation Variation. Nature (2015) 523:212–6. doi: 10.1038/nature14465
38. Brown PM, Pratt AG, Isaacs JD. Mechanism of Action of Methotrexate in Rheumatoid Arthritis, and the Search for Biomarkers. Nat Rev Rheumatol (2016) 12:731–42. doi: 10.1038/nrrheum.2016.175
39. Wang YC, E-PI C. Low-Dose Methotrexate Inhibits Methionine Sadenosyltransferase In Vitro and In Vivo. Mol Med (2012) 18:423–32. doi: 10.2119/molmed.2011.00048
Keywords: rheumatoid arthritis, autoimmune disease, vitamin D metabolic pathway, methylation, single nucleotide polymorphisms
Citation: Zhang T-P, Li H-M, Huang Q, Wang L and Li X-M (2021) Vitamin D Metabolic Pathway Genes Polymorphisms and Their Methylation Levels in Association With Rheumatoid Arthritis. Front. Immunol. 12:731565. doi: 10.3389/fimmu.2021.731565
Received: 27 June 2021; Accepted: 02 November 2021;
Published: 02 December 2021.
Edited by:
Poornima Paramasivan, Concept Life Sciences, United KingdomReviewed by:
Mikhail Kostik, Saint Petersburg State Pediatric Medical University, RussiaPrabhakar Puttachandra, Scientific Research Solution Pvt Ltd, India
Copyright © 2021 Zhang, Li, Huang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Mei Li, bGl4aWFvbWVpLmZzbXlAYWxpeXVuLmNvbQ==, bGl4aWFvbWVpQHVzdGMuZWR1LmNu; Li Wang, d2FuZ19saXlAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Tian-Ping Zhang
Tian-Ping Zhang Hong-Miao Li2†
Hong-Miao Li2† Xiao-Mei Li
Xiao-Mei Li