- Department of Pharmacology and Regenerative Medicine, University of Illinois at Chicago College of Medicine, Chicago, IL, United States
Lifelong mammalian hematopoiesis requires continuous generation of mature blood cells that originate from Hematopoietic Stem and Progenitor Cells (HSPCs) situated in the post-natal Bone Marrow (BM). The BM microenvironment is inherently complex and extensive studies have been devoted to identifying the niche that maintains HSPC homeostasis and supports hematopoietic potential. The Notch signaling pathway is required for the emergence of the definitive Hematopoietic Stem Cell (HSC) during embryonic development, but its role in BM HSC homeostasis is convoluted. Recent work has begun to explore novel roles for the Notch signaling pathway in downstream progenitor populations. In this review, we will focus an important role for Notch signaling in the establishment of a T cell primed sub-population of Common Lymphoid Progenitors (CLPs). Given that its activation mechanism relies primarily on cell-to-cell contact, Notch signaling is an ideal means to investigate and define a novel BM lymphopoietic niche. We will discuss how new genetic model systems indicate a pre-thymic, BM-specific role for Notch activation in early T cell development and what this means to the paradigm of lymphoid lineage commitment. Lastly, we will examine how leukemic T-cell acute lymphoblastic leukemia (T-ALL) blasts take advantage of Notch and downstream lymphoid signals in the pathological BM niche.
Introduction
Notch signaling is a highly conserved pathway activated through cell-to-cell, ligand-receptor interactions. There are five Notch ligands in mammals: Delta like (Dll) 1, 3 and 4 and Jagged (Jag) 1 and 2 which are presented on the surface of multiple cells and tissues (1). When the ligand interacts with one of the 4 mammalian Notch receptors, (Notch1-4) a series of proteolytic cleavages releases the Notch receptor from the plasma membrane (2). Subsequently, the intracellular Notch (ICN) domain translocates to the nucleus, where it binds to Recombining Binding Protein Suppressor of Hairless (RBP-J) and co-activator Mastermind-like (MAML) (2, 3). Ultimately, it is this tri-molecular complex that binds to enhancer and promoter elements to initiate transcriptional activation of target genes. Along with Wtn, Hedgehog, and Bone Morphogenic Peptide/TGF- β, Notch signaling is one of the fundamental pathways essential for mammalian embryogenesis (4). Notch signaling plays a multitude of roles in the differentiation, proliferation, self-renewal, and survival in diverse cell types across many tissues (5). Particularly well studied are the roles of Notch1 in somite segmentation (6, 7), in angiogenesis and vascular development (8, 9), and the emergence of the definitive hematopoietic stem cell (HSC) in the aorta-gonad-mesonephros (10, 11). In the post-natal murine BM, HSC cell-autonomous and non-cell-autonomous Notch signaling has been implicated in several contexts including aging, regeneration, and mobilization, reviewed in these studies (12–15). Though loss-of-function studies in adult mice do not support a requirement for HSC cell-autonomous Notch activation during homeostasis (16, 17) and in one more recent study in regenerative hematopoiesis (18), Notch signaling has been implicated in the development of several different blood lineages, including megakaryocytes (19), NK cells (20), and erythrocytes (21). Yet, it is the role of Notch signaling in the cell fate determination of the T cell lineage that remains as the archetypic function of the pathway in adult hematopoiesis (22, 23).
The developmental progression from the BM HSC to the production of functional peripheral T cells is physiologically continuous but can be delineated using surface markers and expression of key transcriptional regulators. In both mouse and human, BM lymphoid progenitors give rise to thymic precursors, which progress through well-defined developmental stages in the thymus to become naïve T cells (24). Progression through distinct stages of thymic T cell development requires the careful coordination of several lineage regulatory transcription factors, including: Ikaros, Gfi1, Myb, Runx family proteins, E2A, HEB, TCF1, GATA3, Bcl11b, LEF1, and of course Notch1 (25). In the thymus, the roles of Notch signaling have been well studied. After homing to the thymus, early progenitors activate Notch signaling, which is required for thymocyte development (26–28). Notch is implicated in a variety of functions such as inhibition of progenitor apoptosis, induction of T cell lineage master regulators Gata3, Tcf7, and Bcl11b, as well as activation of genes involved in functional T cell receptor (TCR) production such as Ptcrα (29–31). Notch signaling becomes dispensable for T cell differentiation after β-selection occurs, at which point subsequent development is dependent on signals from the pre-TCR complex (29, 32). The main receptor expressed by thymocytes is Notch1, while the major Notch ligand expressed by cortical Thymic Epithelial Cells (TECs) is Delta-like 4 (Dll4) (30, 33). While the role of Notch in T cell development in undeniable, the temporal and spatial aspects of the first requirement for Notch in driving T cell fate have not been fully established. Recently, several findings have begun to address this issue by suggesting that pre-thymic Notch signals influence the ability of primitive BM lymphoid progenitors to produce thymus-seeding cells (18, 34, 35). Here, we will review the work which encompasses our current understanding of the BM populations that give rise to thymic progenitors, and the role of Notch signaling as a niche component in driving this process. Under this new paradigm of pre-thymic Notch activation, we will then examine the pathological Notch-dependent mechanisms of the lymphoid niche in the leukemic BM environment.
BM Thymocyte Progenitors
In adult mammals, the hematopoietic system is maintained via the production of functional blood cells and hematopoietic progenitors by self-renewing HSCs in the BM (36). The BM microenvironment is composed of osteoprogenitors, stromal cells, endothelial cells, and multiple hematopoietic cell types (Figure 1) (37, 38). At the apex of BM hematopoiesis is the HSC, which is defined best by its self-renewal and functional capacity to produce all the lineages of blood rather than by a specific set of markers. Even so, for isolation purposes the HSC has been classified by surface markers as Lin-cKit+Sca1+CD150+CD48- (39), by the presence of efflux pumps (40) and by the expression of intracellular proteins including Hoxb5 and α-catulin (41, 42). Next in the hematopoietic hierarchy are the HSPCs, which in murine hematopoiesis are generally classified by the combination of Lin-cKit+Sca1+ and become increasingly lineage committed. This differentiation potential arises at the expense of the capacity to self-renew (43, 44). As the HSPCs gain lineage specific potential, they begins to express surface proteins which have been used to define specific progenitor populations in the BM, termed Multipotent Progenitor Populations (MPP) (45–47).
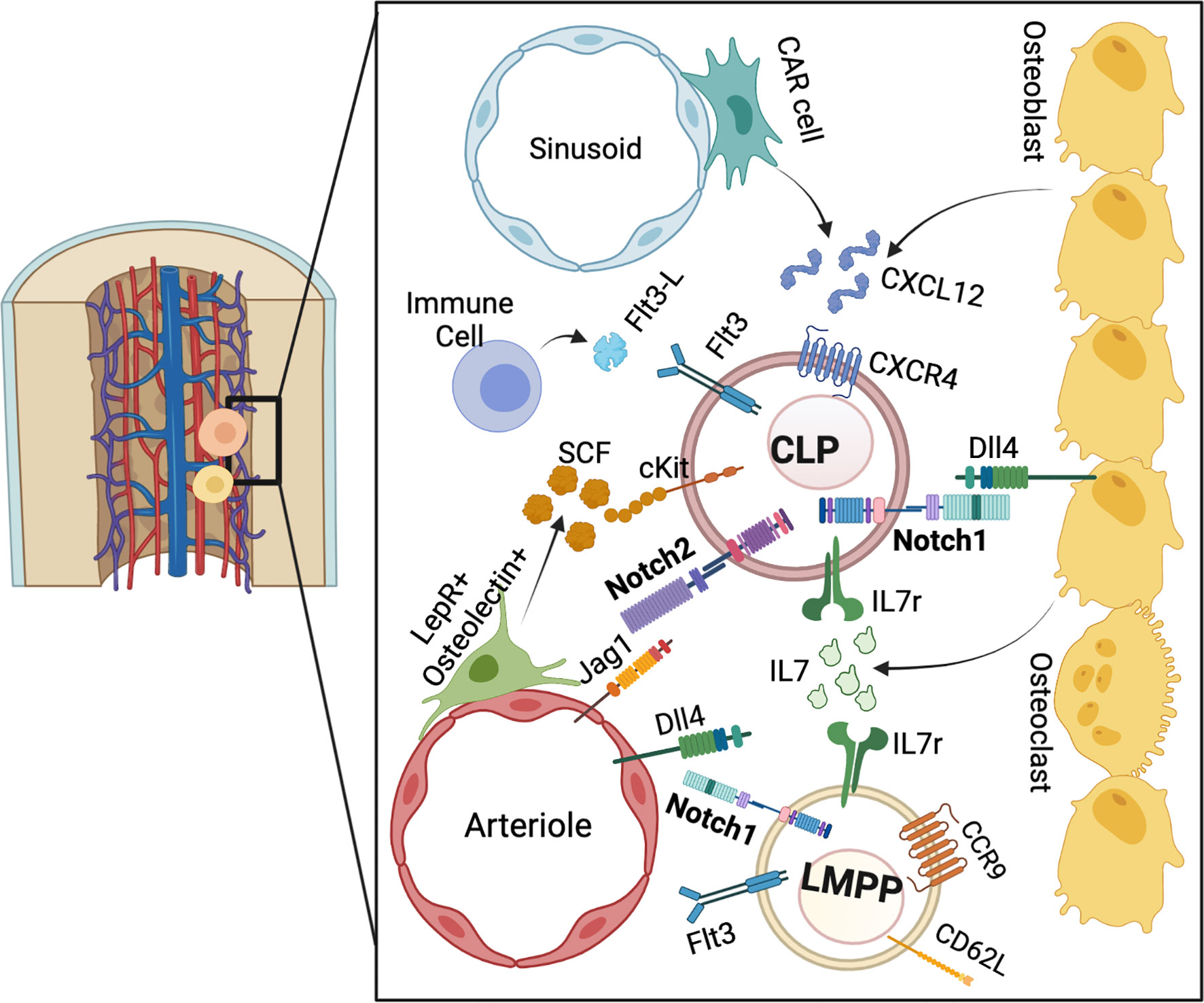
Figure 1 The BM niche for pre-thymic T cell progenitor development. LMPPs and CLPs reside in the endosteal niche. Notch, IL7r, and CXCR4 ligands are derived from the osteoblastic and stromal niche, while SCF is provided from peri-arteriolar cells. Flt3L is provided by mature immune cells. Overall, these signaling pathways converge to stimulate lymphoid progenitors to the T cell lineage.
In the case of early BM lymphopoiesis, several progenitor populations have been described. Cells within the HSPC pool which express the tyrosine kinase receptor Fms-like tyrosine kinase 3 (Flt3) have been labeled as lymphoid-primed MPPs (LMPP), also termed MPP4 (45, 48–50). LMPP lineage output is functionally distinct from myeloid biased MPP2 (Flt3-CD48+CD150+) and MPP3 (Flt3-CD48+CD150-) populations as determined via murine transplantation experiments (50). LMPPs were shown to have equivalent B and T cell potential, retain some granulocyte and monocyte potential, but lack the ability to produce erythroid and megakaryocyte lineages (49, 51). LMPPs can be further segregated into lymphoid biased cells through expression of a selection of surface proteins. The Interleukin 7 receptor (IL7r), which is required for lymphoid development, is expressed on a subset of LMPPs which efficiently generate T cells and innate lymphoid cells in a murine transplantation setting (52, 53). L-selectin (CD62L) is involved in the trafficking of naïve lymphocytes to peripheral lymphoid organs by binding to a selection of different glycan residues and can be used to specify T lineage progenitors in the BM (54–57). Expression of CD62L separates LMPPs which have transient B cell potential and can yield rapid thymocyte production, but lack the ability to produce cells of myeloid lineages (58). Furthermore, CD62L upregulation has been shown to be an early event in the lymphoid priming of human BM progenitors (59).
Additionally, Vascular Cell Adhesion Molecule 1 (VCAM1) and Flt3 expression can be used to segregate MPPs with combined lymphoid/myeloid (Flt3hiVCAM1+), erythroid (Flt3loVCAM1+), or B and T cell potential (Flt3hiVCAM1-) (60). VCAM1 is a cell adhesion molecule with roles in vascular adhesion and transendothelial migration of leukocytes (61, 62). Originally identified on the surface of endothelial cells (63, 64), VCAM1 has since been found to be expressed on the surface of multiple cell types including hematopoietic progenitors, macrophages, and BM fibroblasts (65). It is through ligand binding, specifically the α4β1 integrin (CD49d/CD29) and α4β7 integrins expressed on the surface of leukocytes, that VCAM1 mediates adhesion and transmigration of T cells and macrophages (61, 66). The VCAM1- LMPP population in the BM homogeneously expresses Flt3, and expression of C-C chemokine Receptor type 9 (CCR9), which also is suggested to play a major role in the recruitment of BM derived cells to the thymus, further delineates a subset of T cell progenitors (67–69). Taken together, MPPs expressing Flt3, IL7r, CD62L, CCR9 and lacking VCAM1 appear to be the main HSPC component contributing to B and T cell development.
Downstream of the LMPP population, the Common Lymphoid Progenitor (CLP), which in murine hematopoiesis is isolated by surface markers Lin-cKitLoSca1LoFlt3+IL7r+, represents a canonical branching point between myeloid and lymphoid development and is restricted for lineage production of Natural Killer (NK), B cell and T cell development (70). IL7 signaling is critically involved in BM B cell and thymic T cell lymphopoiesis, and the CLP population is defined by IL7r expression (70–72). Surface expression of Lymphocyte Antigen 6 Family Member D (Ly6D) can be used to divide the CLP population into those with T cell biased potential (Ly6D-) and B cell biased potential (Ly6D+) (73–75). Downstream of the CLP in bone marrow NK development is the Pre-Natural Killer Progenitor (Pre-NKP) and Refined Natural Killer Progenitor (rNKP) (76). In B cell development, the CLP differentiates into a series of BM sub-populations traditionally referred to as the Hardy Fractions (43), of which Fraction A (B220+CD43+CD24−BP-1-) is immediately downstream of the CLP (77–79). The next well-defined downstream T cell lineage progenitor of the CLP is the early T lineage progenitor (ETP), which is the earliest T cell progenitor in the thymus (80, 81).
While both LMPPs and CLPs possess T cell lineage potential, efforts to determine the exact BM progenitor population which is the Thymic Seeding Progenitor (TSP) have yielded conflicting reports. Although transplantation of CLPs yields thymus engraftment and thymopoiesis, Ikaros-deficient mice have been shown to have thymic ETPs without a detectable CLP population in the BM (82, 83). Additionally, IL7r+ LMPPs can generate thymocytes in a CLP-independent manner post-transplant (53). A potential resolution for this issue has been proposed during pre-natal thymopoiesis, where TSPs are produced in two separate waves, the first of which resembles CLPs and a second resembling LMPPs (84). However, a caveat to these findings is the use of transplantation to determine functional kinetics of TSP generation, which requires removal of these BM resident populations and injection into a recipient’s bloodstream. Thus, while TSP generation is determined in vivo, the path of the populations in question from the BM to the bloodstream and finally the thymus is in the form of a transplant and does not necessarily mimic natural BM egress and thymic homing. This a principle commonly seen in HSC studies, where it has been recognized that transplantation leads to oligoclonal dominance that does not reflect unperturbed hematopoiesis (85, 86). In order to more accurately determine the BM source for TSP generation, additional methods such as in vivo lineage tracing and single cell RNA sequencing should be applied, such as in recent work which unveiled direct production of megakaryocyte progenitors (MkP) from long term HCSs (LT-HSCs) (87). Thus, while the exact BM population for TSP generation has yet to be specifically determined, both the LMPP and CLP populations remain viable sources.
Pre-Thymic Notch Signaling in Lymphopoiesis
Notch signaling is essential for T cell development as shown by the seminal loss-of-function studies by Radtke F et al. (28). However, similar loss of function experiments that deleted the DNA binding member of the trimolecular complex RBPJ (88–90) as well as pan-Notch inhibition with a Mastermind truncation named dominant-negative Mastermind (dnMAML) (17) all indicated that loss of Notch signaling in the BM HSC population had no effect on HSC homeostasis in adult mice. However, as we have described above, several stages of progenitor differentiation occur between the HSC in the BM and the emigrating TPS. The critical temporal question is whether Notch signaling is activated and required for development of the LMPP, CLP or the ETP. Early insight into the role of Notch in BM T lineage lymphopoiesis can be found in studies which showed that the CLP population expresses Notch1 at the mRNA level, and that Notch1 deficient CLP cells erroneously differentiate into B cells in the thymus (91, 92). BM cells transduced with dnMAML failed to produce ETP cells post-transplant, once again hinting at a pre-thymic role for Notch in ETP generation (26). It was further observed that CCR9+ T cell biased MPPs have the potential to activate Notch signaling (68). To determine the expression of individual Notch receptors in HSPC populations, an in vivo lineage tracing system has been developed. Cre-recombinase was “knocked into” the individual loci for Notch1-4, which allowed for determination of receptor expression using a fluorescent Cre-reporter mouse strain. This system revealed Notch1 expression in LMPPs, Notch1 and Notch2 expression in CLPs, and an absence of Notch3 or Notch4 in either population (21).
Abrogation of Notch signaling in the BM through inhibition or genetic deletion of Notch receptors or ligands has indicated a role for Notch-dependent T cell progenitor development in the BM. Injection of Notch ligand Dll4 neutralizing antibodies, which have been shown to block ligand specific signaling, leads to a decrease in the BM CLP population (93, 94). Consistently, deletion of either Notch ligand Dll4 or Mindbomb (Mib), which is involved in Notch ligand endocytosis, in Osteocalcin (Ocn) expressing bone cells led to a significant decrease in the CLP population (34, 95). Similar results were obtained when either RBP-J or GDP-fucose protein O-fucosyltransferase 1 (POFUT1) were deleted from BM hematopoietic progenitors (34, 96). The CLP defect observed after conditional deletion of osteoblastic Dll4 underscores the potential existence of an osteoblastic niche for Notch-dependent priming of BM lymphoid progenitors (Figure 1). Indeed, it has been shown that C-X-C Motif Chemokine Ligand 12 (Cxcl12) derived from osteoblasts, and not endothelial or hematopoietic cells, is required for CLP and LMPP maintenance in the BM niche (97). The osteoblastic niche has also been implicated in BM B cell progenitor development, through stimulation of HSCs towards the lymphoid lineage via Gsα dependent osteoblast IL7 production (98–100). Peri-arteriolar LEPR+Osteolectin+ cells have also been shown to stimulate CLP development through secretion of SCF (Figure 1) (101). This osteoblast-derived SCF secretion decreases in aged mice which have an imbalance in blood lineage output with a propensity toward myeloid populations (101, 102).
While there is mounting evidence supporting the osteoblastic microenvironment as a lymphoid sub-niche in the BM, there are also reports that implicate different niche cells in the priming of lymphoid progenitors. For example, endothelial cells which express high levels of Notch ligands Dll4 and Jag1 (103) have been suggested as an alternative niche for lymphoid progenitor development. Deletion of endothelial expression of Dll4, but not Dll1, leads to a decrease in the frequency of CLP cells, with no effect on the LMPP in the BM (104). However, a direct contribution of endothelial Notch ligand to CLP Notch receptor activation was not shown, and the potential mechanism of CLP depletion was myeloid skewing of upstream HSCs. Additionally, conditional deletion of Cxcl12 in endothelial cells lead to specific depletion of HSCs in the BM, not lymphoid progenitors (97). Furthermore, deletion of SCF derived from peri-arteriolar LEPR+ cells, and not arteriolar or sinusoidal endothelial cells, depleted CLP cells in the BM (101, 105, 106). Taken together these experiments do not support a direct contribution of endothelial derived factors in lymphoid progenitor maintenance during steady-state hematopoiesis in the adult mouse bone marrow.
In most circumstances, the proposed endothelial niche, be it sinusoidal or peri-arterial (101), has been shown to sustain stemness and support self-renewal of HSCs or HSPCs, which by virtue of their hierarchical position in BM hematopoiesis yield more downstream progenitors including CLPs (107, 108). This is evident in several experiments where regenerating or expanding endothelial compartments produce more HSCs and by connection more lineage specific progenitors (109–112). Because Notch signaling is essential for endothelial growth and regeneration, and because the endothelium is a primary niche for HSCs, the effects observed in the CLP compartment could be attributed to an increase in the general abundance in HSC numbers. Disassociation of the intrinsic role of Notch signaling in arterial cell fate and endothelial function must first be shown to determine if endothelial cells represent the key components of the BM lymphopoietic niche.
Hematopoietic derived signals have also been shown to play a role in CLP homeostasis. Flt3 and Flt3-Ligand (Flt3-L) regulate both myeloid and lymphoid hematopoiesis, and Flt3-L knock-out mice have a severe defect in CLP generation (113, 114). Interestingly, Flt3-L has been shown to be produced in the BM by immune cell populations, including CD4+ memory T cells, rather than stromal, endothelial, or osteoblastic niche cells (115, 116). Although a role for Notch in regulation of Flt3 in homeostatic lymphopoiesis has yet to be established, canonical Notch target Hes1 transcriptionally represses Flt3 expression in Acute Myeloid Leukemia (AML) (117). Additionally, lymphoma/leukemia-Related Factor (LRF), which plays a role in erythroid and late lymphoid lineage decisions, downregulates Dll4 in BM erythroblasts, thus preventing a Notch1 dependent increase in CLP generation at the expense of the HSC pool (118–120).
Confirmation of the existence of the lymphopoietic sub-niche in the BM has been supported by other studies that have shown a cell-intrinsic role for Notch receptor activation in BM lymphoid development. Hematopoietic LRF expression promotes proper B cell development through suppression of Notch signals in CLPs, and hematopoietic deletion of LRF leads to enhanced Notch activity and extra-thymic CD4+CD8+ T cell generation in the BM (121). Hypomorphic Notch signaling achieved by deletion of the Notch1 Transcriptional Activation Domain (TAD) showed a significant decrease in CLP abundance in the BM (18, 122). Furthermore, an inducible RBPj on/off genetic mouse model has confirmed a role of pre-thymic Notch signaling. Specifically, Notch signaling through RBPj is involved in CD62L+ LMPP generation, with no effect in the T cell primed Ly6D- CLP (35). Collectively, these findings confirm a cell-intrinsic role for Notch signaling in pre-thymic T cell progenitor development in the BM microenvironment.
Mechanisms of Notch in CLP Development
The activation of the Notch receptor is only the first step in the signaling pathway that eventually leads to transcriptional activation of target genes. While a direct role for Notch signaling in pre-thymic progenitor development is evident, the cell intrinsic mechanisms that prime T cell development downstream of Notch in the BM are unknown. A possible mechanism involves the regulation of receptors that are important for cellular migration and tissue retention. Recently, a genetic mouse model for inducible deletion and subsequent inducible expression of RBPj in vivo has been developed (35). In this model, floxed Rbpj can be conditionally deleted through an inducible Vav-Cre transgene, while a tetracycline responsive element-controlled hemagglutinin (HA)-tagged RBPJ transgene can be induced via doxycycline (Dox) injection. RNA sequencing of LMPP cells isolated from Rbpjf/+ control, RBPj knock-out, and Dox induced RBPj-HA expressing mice suggested that PSGL1, CCR7, and CCR9 are regulated by Notch signaling in the CD62L+ LMPP population (35). Additionally, deletion of Dll4 in the osteoblastic niche lead to a decrease in CLP cells expressing CCR7 and P-selectin glycoprotein ligand-1 (PSGL1) (34). PSGL1, CCR7 and CCR9 are all involved in the recruitment and migration of BM derived progenitor cells to the thymus (69, 123, 124). However, Notch signaling has also been shown to directly repress CCR9 expression in fetal liver derived T cell progenitors produced via co-culture with stromal cells expressing Dll1 (125). The caveat of fetal progenitor acclimation to ex vivo co-culture conditions could account for these contrary results. Overall, these observations highlight the possibility that Notch activation in BM lymphoid progenitors prepares cells for thymic migration through induction of genes involved in thymic homing.
Another chemokine pathway involved in BM hematopoiesis is CXCR4/CXCL12, which regulates migration, survival, and quiescence of various progenitor populations (126–130). CXCL12 is expressed by several cell types in the BM, including endothelial cells, osteoblasts, stromal cells, and hematopoietic cells (131). Interestingly, stromal CXCL12 production and HSC release from the BM have been shown to be influenced by circadian neural release of noradrenaline, which activates AdrB3 receptor on Nestin+ osteoprogenitors (132, 133). Although migration of mature leukocyte populations in the BM is regulated in part by circadian rhythms, a direct role for circadian influences on BM lymphoid progenitor biology has yet to be established (134). In humans, there is evidence that MCAM+CD146+ subendothelial stromal cells express CXCL12 (135). Hematopoietic deletion of CXCR4 results in a reduction of the BM stem cell pool. Specifically affected are HSCs in close contact with CXCL12-abundant reticular cells which surround sinusoidal endothelial cells in the BM (136). CXCR4 has also been shown to regulate the integrity of the vascular barrier in the BM, which further modulates hematopoietic trafficking (137). Work in multiple cell types has revealed dynamic regulation of CXCR4 by the Notch pathway in both mouse and human mesenchymal and endothelial cells (138–142). In the BM, Notch2 has been shown to directly activate CXCR4 expression in HSPCs, while stromal production of CXCL12 has been shown to play a role in CLP maintenance (143, 144). It should be noted, however, that blockade with a Notch2 specific antibody yields only a modest reduction of the CLP population compared to a 30% decrease with antibody blockade of Notch1 (144). Furthermore, mice expressing CXCR4 mutations derived from patients with Warts, Hypogammaglobulinemia, Infections, and Myelokathexis (WHIM) syndrome, which prevent receptor internalization and desensitization, have decreased LMPP and CLP populations (145). These findings suggest a potential role for the CXCR4/CXCL12 axis in the CLP population by placing the CLP near CXCL12-abundant reticular cells, which have further been shown to provide CLPs with the pro-lymphoid cytokine IL7 (72, 126, 146). These observations highlight the potential for the Notch-CXCR4 pathway in lymphoid progenitor development and trafficking within the BM niche.
Lineage commitment of BM hematopoietic progenitors is a complex process involving coordination of cell fate determining transcriptional networks, which contribute to the heterogeneity of the progenitor pool (44, 86, 147). Given that Notch signaling is essential in the differentiation and maturation of thymic T cells, a direct functional role for Notch signaling in the early BM hematopoietic lineage decisions is a strong possibility (5, 25). Indeed, Dll4 expressed by vascular cells has been shown to suppress the myeloid transcriptional program in HSCs (104). Abrogation of Notch within hematopoietic progenitors leads to an altered myeloid differentiation program, with an increase in GMP production at the expense of MEPs and CMPs (19, 148). In the context of lymphoid progenitors, RNA sequencing has shown that Notch inhibits the myeloid program in both the thymic DN1a/b population and the CD62L+PSGL1+CCR9+ subset of LMPPs, which constitutes a putative TSP population (35, 58). Additionally, deletion of Dll4 in Ocn+ BM osteoblasts yields specific depletion of the T lineage primed Ly6D- CLP population, hinting at a role for endosteal Notch signaling in influencing B cell vs T cell fates in BM CLPs (34, 73). Such a role has also been established in the thymus, as thymocyte Notch signaling inhibits expression of B-lineage specific factors EBF1 and Pax5 (80). Taken together, there are likely multiple distinct roles for the Notch pathway in the priming and generation of lymphoid progenitors in the BM, including activation of receptors involved in niche trafficking and repression of alternative lineage potential.
Notch in BM Leukemic Niche
Notch signaling has been implicated in progression of various types of cancer, including Acute Lymphoblastic Leukemia (ALL) (149). Notch signaling plays a well-established role in T-cell acute lymphoblastic leukemia (T-ALL), which is a neoplasm of T-cell blasts accounting for 25% of adult ALL (150). Greater than 60% of patient samples contain mutations in the Notch pathway, with several gain of function mutations in the Notch1 gene, and inactivating mutations in negative regulators of Notch signaling, including FBW7 (151–158). Notch3, which is a Notch1 target gene, has also been shown to play a role in T-ALL (159–161). Mechanisms of Notch signaling in T-ALL oncogenesis include promotion of anabolic cell growth and chemoresistance, activation of the PI3K-AKT-mTOR pathway, and induction of genes involved in G1/S cell cycle progression (158, 162, 163). Notch has also been implicated in B cell leukemias. Hyperactive Notch1 and Notch2 have been shown to sustain B cell Chronic lymphocytic leukemia (B-CLL) (164–167). Conversely, all Notch receptors and the Notch target Hes5 have been shown to act as tumor suppressors in B cell ALL (B-ALL). Even so, Notch3 and Notch4 can prevent apoptosis of human B-ALL cells cultured on human stromal cells ex vivo (168–170).
In the context of the BM microenvironment, leukemic cells have been shown to modulate the hematopoietic niche to form a pro-leukemic microenvironment at the expense of homeostatic hematopoiesis (Figure 2) (171). Interestingly, and unlike homeostatic lymphoid progenitors, Notch driven T-ALL cells are not maintained by a specific BM niche, but lead to remodeling of the endosteal niche and loss of osteoprogenitors (172). Such remodeling leads to perturbations in BM hematopoiesis, including reduced quiescence of HSCs and more severe leukemia progression (173). Additionally, a multitude of cell extrinsic signaling molecules have been implicated in the pathogenesis of T-ALL (174). The CXCL12/CXCR4 pathway is involved in homing of T-ALL cells to the bone marrow and in Leukemia Initiating Cell (LIC) activity (175). LICs propagate leukemia progression via their ability to both self-renew and produce clonal daughter blasts (176). Similarly to homeostatic HSPCs, T-ALL cells are subject to increased CXCR4 expression and activity downstream of Notch activation (144, 175, 177, 178). In human Chronic Lymphoid Leukemia (CLL) and Multiple Myeloma (MM), CXCR4 is also a direct transcriptional target of Notch1 (179, 180). Furthermore, CXCL12 receptor CXCR7 has been shown to be transcriptionally activated by Notch signaling in T-ALL and potentiates CXCR4 signaling and migration (181, 182). This pathway has become clinically relevant since CXCR4 inhibition has shown therapeutic potential in T-ALL (183). Specifically, direct CXCR4 antagonism prevents migration of CD4+/CD8+ leukemic cells from the thymus to the bone marrow in hypermorphic Notch3 transgenic mice (178). Furthermore, CXCR4 deletion in Notch1-induced T-ALL cells or CXCL12 deletion in endothelial, but not perivascular cells, limits T-ALL progression in mice through induction of cell death (177, 183). Thus, the CXCR4/CXCL12 axis, regulated in part by a hyperactive Notch pathway, is involved in the homing and progression of several leukemia subtypes in the BM niche.
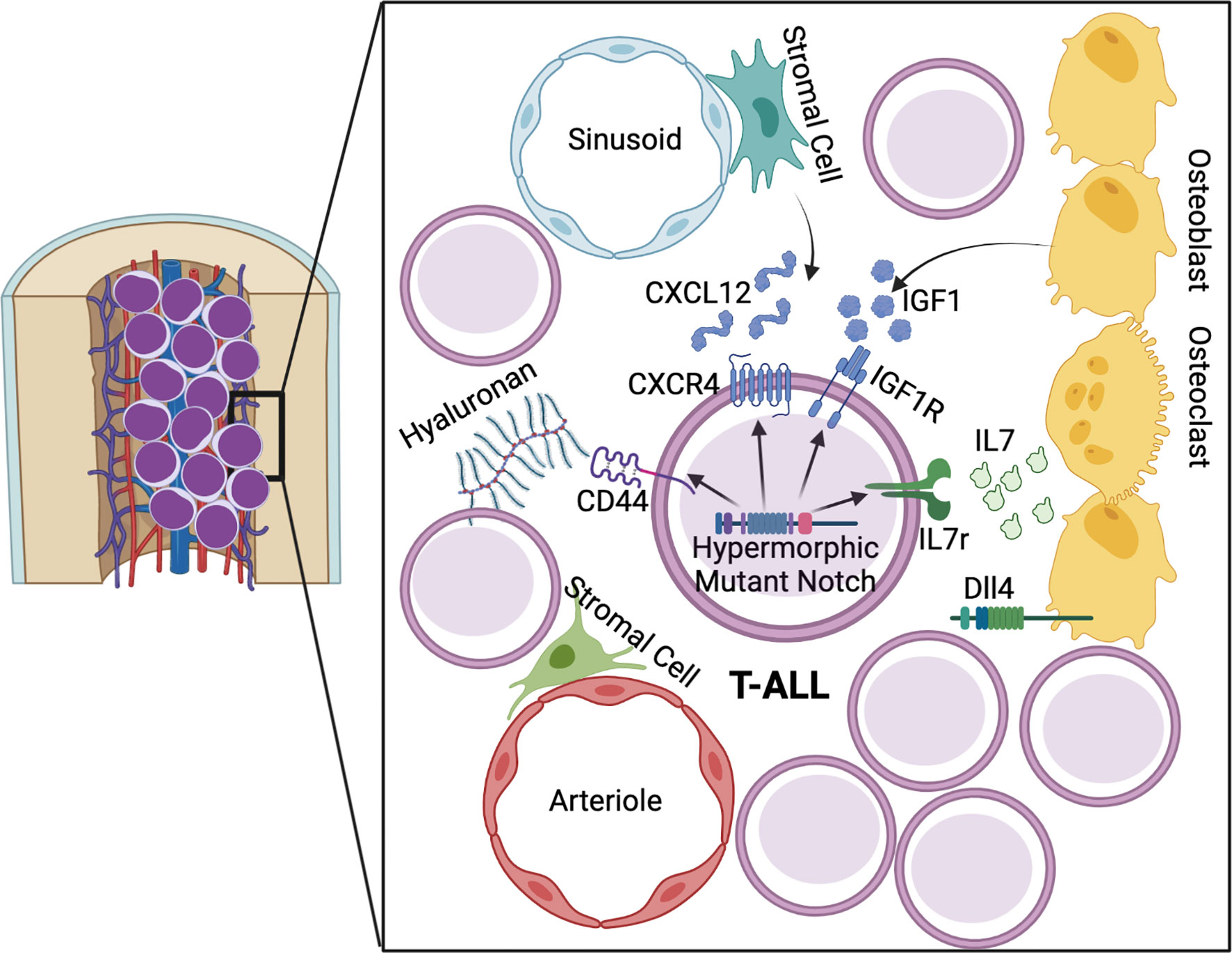
Figure 2 Notch driven mechanisms of T-ALL in the lymphoid niche. Hypermorphic Notch signaling promotes T-ALL progression and amplification of pathways involved in early BM lymphopoiesis. Growth factor signaling from IL7 and IGF1 are augmented via Notch driven expression of IL7r and IGF1R. CXCR4 and CD44 promote maintenance of LIC blasts in the BM microenvironment.
IL7 signaling is critical for lymphopoiesis and, in the context of T-ALL, it plays a role in activation of the JAK/STAT5 and PI3K/Akt/mTOR pathways, with 10% of patient leukemia samples containing activating IL7r mutations (184–186). Stromal derived IL7 has been shown to activate the PI3K/Akt pathway, which is the dominant pathway mediating the proliferative and pro-survival signals downstream of IL7 in T-ALL cells (187–189). Notch1 activates IL7r transcription in human hematopoietic progenitors, as well as in murine leukemia cells (190–192). Additionally, in a human T cell leukemia cell line, IL7r has been shown to be directly co-regulated by Notch1 and RUNX1 (182). Thus, hyperactive Notch signaling contributes to the IL7 dependent proliferation of BM T-ALL cells. Another growth factor involved in BM lymphopoiesis is Insulin-like growth factor 1 (IGF1), which is released from osteoblasts, osteoclasts, and stromal cells in BM and is critical for bone growth (193, 194). Bone marrow levels of IGF1 decrease with age, resulting in increased myeloid bias of HSCs, while temporary IGF1 stimulation of murine hematopoietic progenitors ex vivo promotes lymphoid differentiation post-transplant in recipient mice (195). Notch1 has been shown to directly activate expression of Insulin-like growth factor 1 receptor (IGF1R) in T-ALL, which contributes to leukemia survival through the PI3K/Akt pathway (196, 197). IGF1R inhibition yields therapeutic benefits in several solid tumor types and leukemias (198). However, not all T-ALL cell lines are sensitive to IGF1R inhibition, with co-expression of surface IGF1R and tumor-suppressor PTEN indicating IGF1 dependence (196, 199). Interestingly, miR-233 has also been shown to be a Notch target which separately regulates IGF1R expression via targeting of the 3′ UTR and reduction of IGF1R protein levels in T-ALL (200). Taken together, Notch signaling in T-ALL allows for optimal signaling of BM derived growth factors, through regulation of their receptors.
Another important, though understudied, component of the BM niche is the extra-cellular matrix (ECM), which has been proposed to regulate both HSPCs and leukemia cells (201–204). The ECM is a vital component of structural and signaling mechanisms in all tissues and consists of collogens, proteoglycans, and glycoproteins (205–207). One protein involved in ECM binding is CD44, which is a cell adhesion molecule that binds to hyaluronan, fibronectin, collagen, E-selectin, and is involved in migration of fetal liver HSCs to the fetal BM (208–212). CD44 is also expressed on adult HSPCs and is involved in progenitor egress from the bone marrow and entry into the thymus (213, 214). Conversely, CD44 has been proposed to play a role in HSPC retention and quiescence, and contributes to apoptosis resistance in LICs (215). A potential mechanism of CD44 mediated chemoresistance in leukemia is through induction of drug efflux (216). In human T-ALL, CD44 has been proposed as a target of Notch1 and suggested to be required for BM engraftment of early leukemic cells (217). Additionally, CD44 can be used as a marker of LICs and is positively regulated by Notch signaling (218). In many tissues, additional ECM components have been shown to influence Notch activity, and there is cross-talk between ECM mediated signaling pathways and Notch (219). Microfibril Associated Glycoprotein-2 (MAGP-2) is found in elastic fibrils and has been shown to regulate Notch activity in COS cells and endothelial cells via binding to Notch1 Epidermal Growth Factor (EGF) repeats (220–222). The Cyr61, CTGF, and NOV, (CCN) family of ECM proteins influence osteogenesis and angiogenesis by binding to and enhancing Notch1 signaling (223–226). Additionally, Epidermal Growth Factor-Like Protein 7 (EGFL7), which is secreted by endothelial cells into the vascular microenvironment, regulates angiogenesis in part through antagonization of Notch signaling (227–230). These findings support the need for further exploration into the cross-talk and direct regulation between ECM components and the Notch pathway in T-ALL.
Concluding Remarks
This review serves to highlight recent work which describes a pre-thymic niche in the BM where Notch signaling influences lymphoid, and specifically T cell, development. BM lymphoid progenitors receive Notch signals primarily in the osteoblastic niche, which also provides important signals involved in lymphoid development, including CXCL12, IL7, and SCF. Mechanisms of pre-thymic Notch signaling in BM lymphoid development include induction of molecules involved in bone egress and thymus migration. Ultimately a key outcome of this Notch signaling agenda is the early repression of the myeloid transcriptional program. However, while we know that Notch is active and functions in the BM, the downstream target genes of Notch activation in BM lymphopoiesis, particularly with respect to proliferation and survival, have yet to be fully established. We also examined the roles for aberrant Notch signaling in the BM migration, maintenance, and proliferation of T-ALL. Taken together, the works described here underscore the need for careful study of BM Notch signaling in lymphoid hematopoiesis.
Author Contributions
KS and KP wrote and edited this article. All authors contributed to the article and approved the submitted version.
Funding
NIH/NHLBI R01HL134971, NIH/NHLBI T32HL007829-27.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Steinbuck MP. Winandy s. A Review of Notch Processing With New Insights Into Ligand-Independent Notch Signaling in T-Cells. Front Immunol (2018) 9:1230–40. doi: 10.3389/fimmu.2018.01230
2. Kopan R, Ilagan MXG. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell (2009) 137:216–33. doi: 10.1016/j.cell.2009.03.045
3. Bray SJ. Notch Signalling: A Simple Pathway Becomes Complex. Nat Rev Mol Cell Biol (2006) 7:678–89. doi: 10.1038/nrm2009
4. Kelleher F, Fennelly D, Rafferty M. Common Critical Pathways in Embryogenesis and Cancer. Acta Oncol (Madr) (2006) 45:375–88. doi: 10.1080/02841860600602946
5. Andersson ER, Sandberg R, Lendahl U. Notch Signaling: Simplicity in Design, Versatility in Function. Development (2011) 138:3593–612. doi: 10.1242/dev.063610
6. Conlon RA, Reaume AG, Rossant J. Notch 1 Is Required for the Coordinate Segmentation of Somites. Development (1995) 121:1533–45. doi: 10.1242/dev.121.5.1533
7. Jiang YJ, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J. Notch Signalling and the Synchronization of the Somite Segmentation Clock. Nature (2000) 408:475–9. doi: 10.1038/35044091
8. Iso T, Hamamori Y, Kedes L. Notch Signaling in Vascular Development. Arterioscler Thromb Vasc Biol (2003) 23:543–53. doi: 10.1161/01.ATV.0000060892.81529.8F
9. Kofler NM, Shawber CJ, Kangsamaksin T, Reed HO, Galatioto J, Kitajewski J. Notch Signaling in Developmental and Tumor Angiogenesis. Genes Cancer (2011) 2:1106–16. doi: 10.1177/1947601911423030
10. Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, et al. Notch1 But Not Notch2 Is Essential for Generating Hematopoietic Stem Cells From Endothelial Cells. Immunity (2003) 18:699–711. doi: 10.1016/S1074-7613(03)00117-1
11. Gama-Norton L, Ferrando E, Ruiz-Herguido C, Liu Z, Guiu J, Islam ABMMK, et al. Notch Signal Strength Controls Cell Fate in the Haemogenic Endothelium. Nat Commun (2015) 6:1–12. doi: 10.1038/ncomms9510
12. Pajcini KV, Speck NA, Pear WS. Notch Signaling in Mammalian Hematopoietic Stem Cells. Leukemia (2011) 25:1525–32. doi: 10.1038/leu.2011.127
13. Lampreia FP, Carmelo JG, Anjos-Afonso F. Notch Signaling in the Regulation of Hematopoietic Stem Cell. Curr Stem Cell Rep (2017) 3:202–9. doi: 10.1007/s40778-017-0090-8
14. Huang C, Yang D, Ye GW, Powell CA, Guo P. Vascular Notch Signaling in Stress Hematopoiesis. Front Cell Dev Biol (2021) 8:606448. doi: 10.3389/fcell.2020.606448
15. Albakri M, Tashkandi H, Zhou L. A Review of Advances in Hematopoietic Stem Cell Mobilization and the Potential Role of Notch2 Blockade. Cell Transplant (2020) 29:1–13. doi: 10.1177/0963689720947146
16. Mancini SJC, Mantei N, Dumortier A, Suter U, MacDonald HR, Radtke F. Jagged1-Dependent Notch Signaling Is Dispensable for Hematopoietic Stem Cell Self-Renewal and Differentiation. Blood (2005) 105:2340–2. doi: 10.1182/blood-2004-08-3207
17. Maillard I, Koch U, Dumortier A, Shestova O, Xu L, Sai H, et al. Canonical Notch Signaling Is Dispensable for The Maintenance of Adult Hematopoietic Stem Cells. Cell Stem Cell (2008) 2:356–66. doi: 10.1016/J.STEM.2008.02.011
18. Shao L, Sottoriva K, Palasiewicz K, Zhang J, Hyun J, Soni SS, et al. A Tie2-Notch1 Signaling Axis Regulates Regeneration of the Endothelial Bone Marrow Niche. Haematologica (2019) 104:2164–77. doi: 10.3324/haematol.2018.208660
19. Mercher T, Cornejo MG, Sears C, Kindler T, Moore SA, Maillard I, et al. Notch Signaling Specifies Megakaryocyte Development From Hematopoietic Stem Cells. Cell Stem Cell (2008) 3:314–26. doi: 10.1016/j.stem.2008.07.010
20. Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, et al. Cutting Edge: Microrna-181 Promotes Human NK Cell Development by Regulating Notch Signaling. J Immunol (2011) 187:6171–5. doi: 10.4049/jimmunol.1100835
21. Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manent J, et al. In Vivo Mapping of Notch Pathway Activity in Normal and Stress Hematopoiesis. Cell Stem Cell (2013) 13:190–204. doi: 10.1016/j.stem.2013.05.015
22. Radtke F, Wilson A, MacDonald HR. Notch Signaling in T- and B-Cell Development. Curr Opin Immunol (2004) 16:174–9. doi: 10.1016/j.coi.2004.01.002
23. Amsen D, Helbig C, Backer RA. Notch in T Cell Differentiation: All Things Considered. Trends Immunol (2015) 36:802–14. doi: 10.1016/j.it.2015.10.007
24. Kumar BV, Connors TJ, Farber DL. Human T Cell Development, Localization, and Function Throughout Life. Immunity (2018) 48:202–13. doi: 10.1016/j.immuni.2018.01.007
25. Rothenberg EV. Programming for T-Lymphocyte Fates: Modularity and Mechanisms. Genes Dev (2019) 33:1117–35. doi: 10.1101/gad.327163.119
26. Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, et al. Notch Signaling Controls the Generation and Differentiation of Early T Lineage Progenitors. Nat Immunol (2005) 6:663–70. doi: 10.1038/ni1216
27. Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 Signals at Sequential Early Stages of Intrathymic T Cell Development. Nat Immunol (2005) 6:671–9. doi: 10.1038/ni1217
28. Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T Cell Fate Specification in Mice With an Induced Inactivation of Notch1. Immunity (1999) 10:547–58. doi: 10.1016/S1074-7613(00)80054-0
29. Hosokawa H, Rothenberg EV. Cytokines, Transcription Factors, and the Initiation of T-Cell Development. Cold Spring Harb Perspect Biol (2018) 10:a028621. doi: 10.1101/cshperspect.a028621
30. Yui MA, Feng N, Rothenberg EV. Fine-Scale Staging of T Cell Lineage Commitment in Adult Mouse Thymus. J Immunol (2010) 185:284–93. doi: 10.4049/jimmunol.1000679
31. Yui MA, Rothenberg EV. Developmental Gene Networks: A Triathlon on the Course to T Cell Identity. Nat Rev Immunol (2014) 14:529–45. doi: 10.1038/nri3702
32. Rothenberg EV. T Cell Lineage Commitment: Identity and Renunciation. J Immunol (2011) 186:6649–55. doi: 10.4049/jimmunol.1003703
33. Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, et al. Delta-Like 4 Is Indispensable in Thymic Environment Specific for T Cell Development. J Exp Med (2008) 205:2507–13. doi: 10.1084/jem.20080134
34. Yu VWC, Saez B, Cook C, Lotinun S, Pardo-Saganta A, Wang Y-HH, et al. Specific Bone Cells Produce DLL4 to Generate Thymus-Seeding Progenitors From Bone Marrow. J Exp Med (2015) 212:759–74. doi: 10.1084/jem.20141843
35. Chen ELY, Thompson PK, Zúñiga-Pflücker JC. RBPJ-Dependent Notch Signaling Initiates the T Cell Program in a Subset of Thymus-Seeding Progenitors. Nat Immunol (2019) 20:1456–68. doi: 10.1038/s41590-019-0518-7
36. Morrison SJ, Scadden DT. The Bone Marrow Niche for Haematopoietic Stem Cells. Nature (2014) 505:327–34. doi: 10.1038/nature12984
37. Morrison SJ, Spradling AC. Stem Cells and Niches: Mechanisms That Promote Stem Cell Maintenance Throughout Life. Cell (2008) 132:598–611. doi: 10.1016/j.cell.2008.01.038
38. Pinho S, Frenette PS. Haematopoietic Stem Cell Activity And Interactions With the Niche. Nat Rev Mol Cell Biol (2019) 20:303–20. doi: 10.1038/s41580-019-0103-9
39. Kiel MJ, Yilmaz ÖH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. Cell (2005) 121:1109–21. doi: 10.1016/j.cell.2005.05.026
40. Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and Functional Properties of Murine Hematopoietic Stem Cells That Are Replicating in Vivo. J Exp Med (1996) 183:1797–806. doi: 10.1084/jem.183.4.1797
41. Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, et al. Deep Imaging of Bone Marrow Shows non-Dividing Stem Cells Are Mainly Perisinusoidal. Nature (2015) 526:126–30. doi: 10.1038/nature15250
42. Chen JY, Miyanishi M, Wang SK, Yamazaki S, Sinha R, Kao KS, et al. Hoxb5 Marks Long-Term Haematopoietic Stem Cells and Reveals a Homogenous Perivascular Niche. Nature (2016) 530:223–7. doi: 10.1038/nature16943
43. Haas S, Trumpp A, Milsom MD. Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell (2018) 22:627–38. doi: 10.1016/j.stem.2018.04.003
44. Kato H, Igarashi K. To be Red or White: Lineage Commitment and Maintenance of the Hematopoietic System by the “Inner Myeloid”. Haematologica (2019) 104:1919–27. doi: 10.3324/haematol.2019.216861
45. Christensen JL, Weissman IL. Flk-2 Is a Marker in Hematopoietic Stem Cell Differentiation: A Simple Method to Isolate Long-Term Stem Cells. Proc Natl Acad Sci (2001) 98:14541–6. doi: 10.1073/pnas.261562798
46. Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a Lineage of Multipotent Hematopoietic Progenitors. Development (1997) 124:1929–39. doi: 10.1242/dev.124.10.1929
47. Oguro H, Ding L, Morrison SJ. SLAM Family Markers Resolve Functionally Distinct Subpopulations of Hematopoietic Stem Cells and Multipotent Progenitors. Cell Stem Cell (2013) 13:102–16. doi: 10.1016/j.stem.2013.05.014
48. Adolfsson J, Borge OJ, Bryder D, Theilgaard-Mönch K, Åstrand-Grundström I, Sitnicka E, et al. Upregulation of Flt3 Expression Within the Bone Marrow Lin-Sca1+C-Kit+ Stem Cell Compartment Is Accompanied by Loss of Self-Renewal Capacity. Immunity (2001) 15:659–69. doi: 10.1016/S1074-7613(01)00220-5
49. Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2 + Common Lymphoid Progenitors Possess Equivalent Differentiation Potential for the B and T Lineages. Blood (2008) 111:5562–70. doi: 10.1182/blood-2007-11-126219
50. Pietras EM, Reynaud D, Kang YA, Carlin D, Calero-Nieto FJ, Leavitt AD, et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell (2015) 17:35–46. doi: 10.1016/j.stem.2015.05.003
51. Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ Lympho-Myeloid Stem Cells Lacking Erythro-Megakaryocytic Potential. Cell (2005) 121:295–306. doi: 10.1016/j.cell.2005.02.013
52. Lin J, Zhu Z, Xiao H, Wakefield MR, Ding VA, Bai Q, et al. The Role of Il-7 in Immunity and Cancer. Anticancer Res (2017) 37:963–7. doi: 10.21873/anticanres.11405
53. Ghaedi M, Steer CA, Martinez-Gonzalez I, Halim TYF, Abraham N, Takei F. Common-Lymphoid-Progenitor-Independent Pathways of Innate and T Lymphocyte Development. Cell Rep (2016) 15:471–80. doi: 10.1016/j.celrep.2016.03.039
54. Rosen SD. Ligands for L-Selectin: Homing, Inflammation, and Beyond. Annu Rev Immunol (2004) 22:129–56. doi: 10.1146/annurev.immunol.21.090501.080131
55. Cho S, Spangrude GJ. Enrichment of Functionally Distinct Mouse Hematopoietic Progenitor Cell Populations Using CD62L. J Immunol (2011) 187:5203–10. doi: 10.4049/jimmunol.1102119
56. Perry SS, Welner RS, Kouro T, Kincade PW, Sun X-H. Primitive Lymphoid Progenitors in Bone Marrow With T Lineage Reconstituting Potential. J Immunol (2006) 177:2880–7. doi: 10.4049/jimmunol.177.5.2880
57. Ivetic A, Green HLH, Hart SJ. L-Selectin: A Major Regulator of Leukocyte Adhesion, Migration and Signaling. Front Immunol (2019) 10:1068. doi: 10.3389/fimmu.2019.01068
58. Perry SS, Wang H, Pierce LJ, Yang AM, Tsai S, Spangrude GJ. L-Selectin Defines a Bone Marrow Analog to the Thymic Early T-Lineage Progenitor. Blood (2004) 103:2990–6. doi: 10.1182/blood-2003-09-3030
59. Kohn LA, Hao QL, Sasidharan R, Parekh C, Ge S, Zhu Y, et al. Lymphoid Priming in Human Bone Marrow Begins Before Expression of CD10 With Upregulation of L-Selectin. Nat Immunol (2012) 13:963–71. doi: 10.1038/ni.2405
60. Lai AY, Lin SM, Kondo M. Heterogeneity of Flt3-Expressing Multipotent Progenitors in Mouse Bone Marrow. J Immunol (2005) 175:5016–23. doi: 10.4049/jimmunol.175.8.5016
61. Alon R, Kassner PD, Carr MW, Finger EB, Hemler ME, Springer TA. The Integrin VLA-4 Supports Tethering and Rolling in Flow on VCAM-1. J Cell Biol (1995) 128:1243–53. doi: 10.1083/jcb.128.6.1243
62. Muller WA. The Regulation of Transendothelial Migration: New Knowledge and New Questions. Cardiovasc Res (2015) 107:310–20. doi: 10.1093/cvr/cvv145
63. Rice GE, Bevilacqua MP. An Inducible Endothelial Cell Surface Glycoprotein Mediates Melanoma Adhesion. Science (1989) 246:1303–6. doi: 10.1126/science.2588007
64. Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, et al. Direct Expression Cloning of Vascular Cell Adhesion Molecule 1, a Cytokine-Induced Endothelial Protein That Binds to Lymphocytes. Cell (1989) 59:1203–11. doi: 10.1016/0092-8674(89)90775-7
65. van Oosten M, van de Bilt E, de Vries HE, van Berkel TJC, Kuiper J. Vascular Adhesion Molecule–1 and Intercellular Adhesion Molecule–1 Expression on Rat Liver Cells After Lipopolysaccharide Administration in Vivo. Hepatology (1995) 22:1538–46. doi: 10.1002/hep.1840220529
66. Cerutti C, Ridley AJ. Endothelial Cell-Cell Adhesion and Signaling. Exp Cell Res (2017) 358:31–8. doi: 10.1016/j.yexcr.2017.06.003
67. Lai AY, Kondo M. Asymmetrical Lymphoid and Myeloid Lineage Commitment in Multipotent Hematopoietic Progenitors. J Exp Med (2006) 203:1867–73. doi: 10.1084/jem.20060697
68. Lai AY, Kondo M. Identification of a Bone Marrow Precursor of the Earliest Thymocytes in Adult Mouse. Proc Natl Acad Sci USA (2007) 104:6311–6. doi: 10.1073/pnas.0609608104
69. Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A. CCR7 and CCR9 Together Recruit Hematopoietic Progenitors to the Adult Thymus. Blood (2010) 115:1897–905. doi: 10.1182/blood-2009-08-237784
70. Kondo M, Weissman IL, Akashi K. Identification of Clonogenic Common Lymphoid Progenitors in Mouse Bone Marrow. Cell Press (1997) 91:661–72. doi: 10.1016/S0092-8674(00)80453-5
71. Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, et al. Early Lymphocyte Expansion Is Severely Impaired in Interleukin 7 Receptor-Deficient Mice. J Exp Med (1994) 180:1955–60. doi: 10.1084/jem.180.5.1955
72. Von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SEG, Murray R. Lymphopenia in Interleukin (IL)-7 Gene-Deleted Mice Identifies IL-7 as a Nonredundant Cytokine. J Exp Med (1995) 181:1519–26. doi: 10.1084/jem.181.4.1519
73. Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, et al. Ly6d Marks the Earliest Stage of B-Cell Specification and Identifies the Branchpoint Between B-Cell and T-Cell Development. Genes Dev (2009) 23:2376–81. doi: 10.1101/gad.1836009
74. Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, et al. Single-Cell Analysis of the Common Lymphoid Progenitor Compartment Reveals Functional and Molecular Heterogeneity. Blood (2010) 115:2601–9. doi: 10.1182/blood-2009-08-236398
75. Tsapogas P, Zandi S, Åhsberg J, Zetterblad J, Welinder E, Jönsson JI, et al. IL-7 Mediates Ebf-1-Dependent Lineage Restriction in Early Lymphoid Progenitors. Blood (2011) 118:1283–90. doi: 10.1182/blood-2011-01-332189
76. Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL. Identification of the Earliest Natural Killer Cell-Committed Progenitor in Murine Bone Marrow. Blood (2011) 118:5439–47. doi: 10.1182/blood-2011-04-348912
77. Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and Characterization of Pro-B and Pre-Pro-B Cell Stages in Normal Mouse Bone Marrow. J Exp Med (1991) 173:1213–25. doi: 10.1084/jem.173.5.1213
78. Van Epps HL. Bringing Order to Early B Cell Chaos. J Exp Med (2006) 203:1389. doi: 10.1084/jem.2036fta
79. Kincade PW. Moving the Field of B Lymphopoiesis. J Immunol (2012) 189:3269–70. doi: 10.4049/jimmunol.1202245
80. Rothenberg EV. Transcriptional Control of Early T and B Cell Developmental Choices. Annu Rev Immunol (2014) 32:283–321. doi: 10.1146/annurev-immunol-032712-100024
81. Sambandam A, Bell JJ, Schwarz BA, Zediak VP, Chi AW, Zlotoff DA, et al. Progenitor Migration to the Thymus and T Cell Lineage Commitment. Immunol Res (2008) 42:65–74. doi: 10.1007/s12026-008-8035-z
82. Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, et al. Thymopoiesis Independent of Common Lymphoid Progenitors. Nat Immunol (2003) 4:168–74. doi: 10.1038/ni878
83. Serwold T, Ehrlich LIR, Weissman IL. Reductive Isolation From Bone Marrow and Blood Implicates Common Lymphoid Progenitors as the Major Source of Thymopoiesis. Blood (2009) 113:807–15. doi: 10.1182/blood-2008-08-173682
84. Ramond C, Berthault C, Burlen-Defranoux O, De Sousa AP, Guy-Grand D, Vieira P, et al. Two Waves of Distinct Hematopoietic Progenitor Cells Colonize the Fetal Thymus. Nat Immunol (2014) 15:27–35. doi: 10.1038/ni.2782
85. Busch K, Rodewald HR. Unperturbed vs. Post-Transplantation Hematopoiesis: Both In Vivo but Different. Curr Opin Hematol (2016) 23:295–303. doi: 10.1097/MOH.0000000000000250
86. Lu R, Czechowicz A, Seita J, Jiang D, Weissman IL. Clonal-Level Lineage Commitment Pathways of Hematopoietic Stem Cells in Vivo. Proc Natl Acad Sci USA (2019) 116:1447–56. doi: 10.1073/pnas.1801480116
87. Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, et al. Clonal Analysis of Lineage Fate in Native Haematopoiesis. Nature (2018) 553:212–6. doi: 10.1038/nature25168
88. Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, et al. Notch-RBP-J Signaling Is Involved in Cell Fate Determination of Marginal Zone B Cells. Nat Immunol (2002) 3:443–50. doi: 10.1038/ni793
89. Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, et al. Inducible Gene Knockout of Transcription Factor Recombination Signal Binding Protein-J Reveals its Essential Role in T Versus B Lineage Decision. Int Immunol (2002) 14:637–45. doi: 10.1093/intimm/dxf030
90. Lakhan R, Rathinam CV. Deficiency of Rbpj Leads to Defective Stress-Induced Hematopoietic Stem Cell Functions and Hif Mediated Activation of non-Canonical Notch Signaling Pathways. Front Cell Dev Biol (2021) 8:622190. doi: 10.3389/fcell.2020.622190
91. Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage Specification and Plasticity in CD19- Early B Cell Precursors. J Exp Med (2006) 203:675–87. doi: 10.1084/jem.20052444
92. Wilson A, MacDonald HR, Radtke F. Notch 1-Deficient Common Lymphoid Precursors Adopt a B Cell Fate in the Thymus. J Exp Med (2001) 194:1003–12. doi: 10.1084/jem.194.7.1003
93. Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, De Leon GP, Chen Y, et al. Therapeutic Antibody Targeting of Individual Notch Receptors. Nature (2010) 464:1052–7. doi: 10.1038/nature08878
94. Tran IT, Sandy AR, Carulli AJ, Ebens C, Chung J, Shan GT, et al. Blockade of Individual Notch Ligands and Receptors Controls Graft-Versus-Host Disease. J Clin Invest (2013) 123:1590–604. doi: 10.1172/JCI65477
95. Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, et al. Mind Bomb 1 Is Essential for Generating Functional Notch Ligands to Activate Notch. Development (2005) 132:3459–70. doi: 10.1242/dev.01922
96. Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, et al. Protein O-Fucosyltransferase 1 (Pofut1) Regulates Lymphoid and Myeloid Homeostasis Through Modulation of Notch Receptor Ligand Interactions. Blood (2011) 117:5652–62. doi: 10.1182/blood-2010-12-326074
97. Ding L, Morrison SJ. Haematopoietic Stem Cells and Early Lymphoid Progenitors Occupy Distinct Bone Marrow Niches. Nature (2013) 495:231–5. doi: 10.1038/nature11885
98. Zhu J, Garrett R, Jung Y, Zhang Y, Kim N, Wang J, et al. Osteoblasts Support B-Lymphocyte Commitment and Differentiation From Hematopoietic Stem Cells. Blood (2007) 109:3706–12. doi: 10.1182/blood-2006-08-041384
99. Wu JY, Purton LE, Rodda SJ, Chen M, Weinstein LS, McMahon AP, et al. Osteoblastic Regulation of B Lymphopoiesis Is Mediated by G Sα-Dependent Signaling Pathways. Proc Natl Acad Sci USA (2008) 105:16976–81. doi: 10.1073/pnas.0802898105
100. Visnjic D, Kalajzic Z, Rowe DW, Katavic V, Lorenzo J, Aguila HL. Hematopoiesis Is Severely Altered in Mice With an Induced Osteoblast Deficiency. Blood (2004) 103:3258–64. doi: 10.1182/blood-2003-11-4011
101. Shen B, Tasdogan A, Ubellacker JM, Zhang J, Nosyreva ED, Du L, et al. A Mechanosensitive Peri-Arteriolar Niche for Osteogenesis and Lymphopoiesis. Nature (2021) 591:438–44. doi: 10.1038/s41586-021-03298-5
102. Young K, Borikar S, Bell R, Kuffler L, Philip V, Trowbridge JJ. Progressive Alterations in Multipotent Hematopoietic Progenitors Underlie Lymphoid Cell Loss in Aging. J Exp Med (2016) 213:2259–67. doi: 10.1084/jem.20160168
103. Mack JJ, Luisa Iruela-Arispe M. NOTCH Regulation of the Endothelial Cell Phenotype. Curr Opin Hematol (2018) 25:212–8. doi: 10.1097/MOH.0000000000000425
104. Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Domínguez Á, et al. The Bone Marrow Microenvironment at Single-Cell Resolution. Nature (2019) 569:222–8. doi: 10.1038/s41586-019-1104-8
105. Comazzetto S, Murphy MM, Berto S, Jeffery E, Zhao Z, Morrison SJ. Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF From Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell (2019) 24:477–86.e6. doi: 10.1016/j.stem.2018.11.022
106. Xu C, Gao X, Wei Q, Nakahara F, Zimmerman SE, Mar J, et al. Stem Cell Factor Is Selectively Secreted by Arterial Endothelial Cells in Bone Marrow. Nat Commun (2018) 9:1–13. doi: 10.1038/s41467-018-04726-3
107. Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, et al. Tie2/Angiopoietin-1 Signaling Regulates Hematopoietic Stem Cell Quiescence in the Bone Marrow Niche. Cell (2004) 118:149–61. doi: 10.1016/j.cell.2004.07.004
108. Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and Perivascular Cells Maintain Haematopoietic Stem Cells. Nature (2012) 481:457–62. doi: 10.1038/nature10783
109. Kusumbe AP, Ramasamy SK, Itkin T, Mäe MA, Langen UH, Betsholtz C, et al. Age-Dependent Modulation of Vascular Niches for Haematopoietic Stem Cells. Nature (2016) 532:380–4. doi: 10.1038/nature17638
110. Doan PL, Russell JL, Himburg HA, Helms K, Harris JR, Lucas J, et al. Tie2 Bone Marrow Endothelial Cells Regulate Hematopoietic Stem Cell Regeneration Following Radiation Injury. Stem Cells (2013) 31:327–37. doi: 10.1002/stem.1275
111. Zhou BO, Ding L, Morrison SJ. Hematopoietic Stem and Progenitor Cells Regulate the Regeneration of Their Niche by Secreting Angiopoietin-1. Elife (2015) 2015:e05521. doi: 10.7554/eLife.05521
112. Doan PL, Himburg HA, Helms K, Russell JL, Fixsen E, Quarmyne M, et al. Epidermal Growth Factor Regulates Hematopoietic Regeneration After Radiation Injury. Nat Med (2013) 19:295–304. doi: 10.1038/nm.3070
113. Sitnicka E, Bryder D, Theilgaard-Mönch K, Buza-Vidas N, Adolfsson J, Jacobsen SEW. Key Role of Flt3 Ligand in Regulation of the Common Lymphoid Progenitor But Not in Maintenance of the Hematopoietic Stem Cell Pool. Immunity (2002) 17:463–72. doi: 10.1016/S1074-7613(02)00419-3
114. Tsapogas P, Mooney CJ, Brown G, Rolink A. The Cytokine Flt3-Ligand in Normal and Malignant Hematopoiesis. Int J Mol Sci (2017) 18:1115–37. doi: 10.3390/ijms18061115
115. Onai N, Kurosaki-Nakamura R, Ohteki T. Identification of Flt3-Ligand Producing Cells by Generating Flt3-Ligand Mcherry Reporter Mouse. Exp Hematol (2015) 43:S87. doi: 10.1016/j.exphem.2015.06.221
116. Miloud T, Fiegler N, Suffner J, Hämmerling GJ, Garbi N. Organ-Specific Cellular Requirements for In Vivo Dendritic Cell Generation. J Immunol (2012) 188:1125–35. doi: 10.4049/jimmunol.1003920
117. Kato T, Sakata-Yanagimoto M, Nishikii H, Ueno M, Miyake Y, Yokoyama Y, et al. Hes1 Suppresses Acute Myeloid Leukemia Development Through FLT3 Repression. Leukemia (2015) 29:576–85. doi: 10.1038/leu.2014.281
118. Lee SU, Maeda M, Ishikawa Y, Li SM, Wilson A, Jubb AM, et al. LRF-Mediated Dll4 Repression in Erythroblasts Is Necessary for Hematopoietic Stem Cell Maintenance. Blood (2013) 121:918–29. doi: 10.1182/blood-2012-03-418103
119. Constantinou C, Spella M, Chondrou V, Patrinos GP, Papachatzopoulou A, Sgourou A. The Multi-Faceted Functioning Portrait of LRF/ZBTB7A. Hum Genomics (2019) 13:1–14. doi: 10.1186/s40246-019-0252-0
120. Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP. Role of LRF/Pokemon in Lineage Fate Decisions. Blood (2013) 121:2845–53. doi: 10.1182/blood-2012-11-292037
121. Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B Versus T Lymphoid Lineage Fate Decision by the Proto-Oncogene LRF. Sci (80-) (2007) 316:860–6. doi: 10.1126/science.1140881
122. Gerhardt DM, Pajcini KV, D’altri T, Tu L, Jain R, Xu L, et al. The Notch1 Transcriptional Activation Domain Is Required for Development and Reveals a Novel Role for Notch1 Signaling in Fetal Hematopoietic Stem Cells. Genes Dev (2014) 28:576–93. doi: 10.1101/gad.227496.113
123. Hu Z, Lancaster JN, Ehrlich LIR. The Contribution of Chemokines and Migration to the Induction of Central Tolerance in the Thymus. Front Immunol (2015) 6:398. doi: 10.3389/fimmu.2015.00398
124. Sultana DA, Zhang SL, Todd SP, Bhandoola A. Expression of Functional P-Selectin Glycoprotein Ligand 1 on Hematopoietic Progenitors Is Developmentally Regulated. J Immunol (2012) 188:4385–93. doi: 10.4049/jimmunol.1101116
125. Krishnamoorthy V, Carr T, de Pooter RF, Akinola EO, Gounari F, Kee BL. Repression of Ccr9 Transcription in Mouse T Lymphocyte Progenitors by the Notch Signaling Pathway. J Immunol (2015) 194:3191–200. doi: 10.4049/jimmunol.1402443
126. Cordeiro Gomes A, Hara T, Lim VY, Herndler-Brandstetter D, Nevius E, Sugiyama T, et al. Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity (2016) 45:1219–31. doi: 10.1016/j.immuni.2016.11.004
127. Karpova D, Bonig H. Concise Review: CXCR4/CXCL12 Signaling in Immature Hematopoiesis - Lessons From Pharmacological and Genetic Models. Stem Cells (2015) 33:2391–9. doi: 10.1002/stem.2054
128. Nie Y, Han YC, Zou YR. CXCR4 Is Required for the Quiescence of Primitive Hematopoietic Cells. J Exp Med (2008) 205:777–83. doi: 10.1084/jem.20072513
129. Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, et al. Dependence of Human Stem Cell Engraftment and Repopulation of NOD/SCID Mice on CXCR4. Sci (80-) (1999) 283:845–8. doi: 10.1126/science.283.5403.845
130. Nagasawa T, Tachibana K, Kishimoto T. A Novel CXC Chemokine PBSF/SDF-1 and its Receptor CXCR4: Their Functions in Development, Hematopoiesis and HIV Infection. Semin Immunol (1998) 10:179–85. doi: 10.1006/smim.1998.0128
131. Chotinantakul K, Leeanansaksiri W. Hematopoietic Stem Cell Development, Niches, and Signaling Pathways. Bone Marrow Res (2012) 2012:1–16. doi: 10.1155/2012/270425
132. Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, MacArthur BD, Lira SA, et al. Mesenchymal and Haematopoietic Stem Cells Form a Unique Bone Marrow Niche. Nature (2010) 466:829–34. doi: 10.1038/nature09262
133. Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic Stem Cell Release Is Regulated by Circadian Oscillations. Nature (2008) 452:442–7. doi: 10.1038/nature06685
134. Yuan Y, Wu S, Li W, He W. A Tissue-Specific Rhythmic Recruitment Pattern of Leukocyte Subsets. Front Immunol (2020) 11:102. doi: 10.3389/fimmu.2020.00102
135. Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, et al. Self-Renewing Osteoprogenitors in Bone Marrow Sinusoids Can Organize a Hematopoietic Microenvironment. Cell (2007) 131:324–36. doi: 10.1016/j.cell.2007.08.025
136. Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the Hematopoietic Stem Cell Pool by CXCL12-CXCR4 Chemokine Signaling in Bone Marrow Stromal Cell Niches. Immunity (2006) 25:977–88. doi: 10.1016/j.immuni.2006.10.016
137. Itkin T, Gur-Cohen S, Spencer JA, Schajnovitz A, Ramasamy SK, Kusumbe AP, et al. Distinct Bone Marrow Blood Vessels Differentially Regulate Haematopoiesis. Nature (2016) 532:323–8. doi: 10.1038/nature17624
138. Wang L, Wang YC, Bin X, Zhang BF, Dou GR, He F, et al. Notch-RBP-J Signaling Regulates the Mobilization and Function of Endothelial Progenitor Cells by Dynamic Modulation of CXCR4 Expression in Mice. PloS One (2009) 4:e7572. doi: 10.1371/journal.pone.0007572
139. Williams CK, Segarra M, Sierra MDLL, Sainson RCA, Tosato G, Harris AL. Regulation of CXCR4 by the Notch Ligand Delta-Like 4 in Endothelial Cells. Cancer Res (2008) 68:1889–95. doi: 10.1158/0008-5472.CAN-07-2181
140. Yi L, Zhou X, Li T, Liu P, Hai L, Tong L, et al. Notch1 Signaling Pathway Promotes Invasion, Self-Renewal and Growth of Glioma Initiating Cells via Modulating Chemokine System CXCL12/CXCR4. J Exp Clin Cancer Res (2019) 38:1–15. doi: 10.1186/s13046-019-1319-4
141. Xiao W, Gao Z, Duan Y, Yuan W, Ke Y. Notch Signaling Plays a Crucial Role in Cancer Stem-Like Cells Maintaining Stemness and Mediating Chemotaxis in Renal Cell Carcinoma. J Exp Clin Cancer Res (2017) 36:1–13. doi: 10.1186/s13046-017-0507-3
142. Xie J, Wang W, Si JW, Miao XY, Li JC, Wang YC, et al. Notch Signaling Regulates CXCR4 Expression and the Migration of Mesenchymal Stem Cells. Cell Immunol (2013) 281:68–75. doi: 10.1016/j.cellimm.2013.02.001
143. Greenbaum A, Hsu YMS, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, et al. CXCL12 in Early Mesenchymal Progenitors Is Required for Haematopoietic Stem-Cell Maintenance. Nature (2013) 495:227–30. doi: 10.1038/nature11926
144. Wang W, Yu S, Myers J, Wang Y, Xin WW, Albakri M, et al. Notch2 Blockade Enhances Hematopoietic Stem Cell Mobilization and Homing. Haematologica (2017) 102:1785–95. doi: 10.3324/haematol.2017.168674
145. Freitas C, Wittner M, Nguyen J, Rondeau V, Biajoux V, Aknin ML, et al. Lymphoid Differentiation of Hematopoietic Stem Cells Requires Efficient Cxcr4 Desensitization. J Exp Med (2017) 214:2023–40. doi: 10.1084/jem.20160806
146. Tokoyoda K, Egawa T, Sugiyama T, Il CB, Nagasawa T. Cellular Niches Controlling B Lymphocyte Behavior Within Bone Marrow During Development. Immunity (2004) 20:707–18. doi: 10.1016/j.immuni.2004.05.001
147. Cheng H, Zheng Z, Cheng T. New Paradigms on Hematopoietic Stem Cell Differentiation. Protein Cell (2020) 11:34–44. doi: 10.1007/s13238-019-0633-0
148. Francis OL, Chaudhry KK, Lamprecht T, Klco JM. Impact of Notch Disruption on Myeloid Development. Blood Cancer J (2017) 7:e598–8. doi: 10.1038/bcj.2017.73
149. Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol Mech Dis (2017) 12:245–75. doi: 10.1146/annurev-pathol-052016-100127
150. Van Vlierberghe P, Ferrando A. The Molecular Basis of T Cell Acute Lymphoblastic Leukemia. J Clin Invest (2012) 122:3398–406. doi: 10.1172/JCI61269
151. Weng AP, Ferrando AA, Lee W, Morris JP IV, Silverman LB, Sanchez-Irizarry C, et al. Activating Mutations of NOTCH1 in Human T Cell Acute Lymphoblastic Leukemia. Sci (80-) (2004) 306:269–71. doi: 10.1126/science.1102160
152. Ferrando A. NOTCH Mutations as Prognostic Markers in T-ALL. Leukemia (2010) 24:2003–4. doi: 10.1038/leu.2010.237
153. Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 Ubiquitin Ligase Complex as a Tumor Suppressor in T Cell Leukemia. J Exp Med (2007) 204:1825–35. doi: 10.1084/jem.20070872
154. O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 Mutations in Leukemic Cells Mediate NOTCH Pathway Activation and Resistance to Γ-Secretase Inhibitors. J Exp Med (2007) 204:1813–24. doi: 10.1084/jem.20070876
155. Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, et al. NOTCH1 Extracellular Juxtamembrane Expansion Mutations in T-ALL. Blood (2008) 112:733–40. doi: 10.1182/blood-2007-12-130096
156. Eguchi-Ishimae M, Eguchi M, Kempski H, Greaves M. NOTCH1 Mutation can be an Early, Prenatal Genetic Event in T-ALL. Blood (2008) 111:376–8. doi: 10.1182/blood-2007-02-074690
157. Zheng R, Li M, Wang S, Liu Y. Advances of Target Therapy on NOTCH1 Signaling Pathway in T-Cell Acute Lymphoblastic Leukemia. Exp Hematol Oncol (2020) 9:1–9. doi: 10.1186/s40164-020-00187-x
158. Tosello V, Ferrando AA. The NOTCH Signaling Pathway: Role in the Pathogenesis of T-Cell Acute Lymphoblastic Leukemia and Implication for Therapy. Ther Adv Hematol (2013) 4:199–210. doi: 10.1177/2040620712471368
159. Bellavia D, Campese AF, Checquolo S, Balestri A, Biondi A, Cazzaniga G, et al. Combined Expression of Ptα and Notch3 in T Cell Leukemia Identifies the Requirement of Pretcr for Leukemogenesis. Proc Natl Acad Sci USA (2002) 99:3788–93. doi: 10.1073/pnas.062050599
160. Xiang J, Ouyang Y, Cui Y, Lin F, Ren J, Long M, et al. Silencing of Notch3 Using Shrna Driven by Survivin Promoter Inhibits Growth and Promotes Apoptosis of Human T-Cell Acute Lymphoblastic Leukemia Cells. Clin Lymphoma Myeloma Leuk (2012) 12:59–65. doi: 10.1016/j.clml.2011.07.005
161. Tottone L, Zhdanovskaya N, Pestaña ÁC, Zampieri M, Simeoni F, Lazzari S, et al. Histone Modifications Drive Aberrant Notch3 Expression/Activity and Growth in T-ALL. Front Oncol (2019) 9:198. doi: 10.3389/fonc.2019.00198
162. Zou J, Li P, Lu F, Liu N, Dai J, Ye J, et al. Notch1 Is Required for Hypoxia-Induced Proliferation, Invasion and Chemoresistance of T-Cell Acute Lymphoblastic Leukemia Cells. J Hematol Oncol (2013) 6:1–13. doi: 10.1186/1756-8722-6-3
163. Ferrando AA. The Role of NOTCH1 Signaling in T-ALL. Hematol Am Soc Hematol Educ Program (2009) 2009(1):353–61. doi: 10.1182/asheducation-2009.1.353
164. Duechler M, Shehata M, Schwarzmeier JD, Hoelbl A, Hilgarth M, Hubmann R. Induction of Apoptosis by Proteasome Inhibitors in B-CLL Cells Is Associated With Downregulation of CD23 and Inactivation of Notch2. Leukemia (2005) 19:260–7. doi: 10.1038/sj.leu.2403592
165. Rosati E, Sabatini R, Rampino G, Tabilio A, Di Ianni M, Fettucciari K, et al. Constitutively Activated Notch Signaling Is Involved in Survival and Apoptosis Resistance of B-CLL Cells. Blood (2009) 113:856–65. doi: 10.1182/blood-2008-02-139725
166. Hubmann R, Hilgarth M, Schnabl S, Ponath E, Reiter M, Demirtas D, et al. Gliotoxin Is a Potent NOTCH2 Transactivation Inhibitor and Efficiently Induces Apoptosis in Chronic Lymphocytic Leukaemia (CLL) Cells. Br J Haematol (2013) 160:618–29. doi: 10.1111/bjh.12183
167. Hubmann R, Düchler M, Schnabl S, Hilgarth M, Demirtas D, Mitteregger D, et al. NOTCH2 Links Protein Kinase C Delta to the Expression of CD23 in Chronic Lymphocytic Leukaemia (CLL) Cells: Research Paper. Br J Haematol (2010) 148:868–78. doi: 10.1111/j.1365-2141.2009.08024.x
168. Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, et al. Notch Signaling Is a Potent Inducer of Growth Arrest and Apoptosis in a Wide Range of B-Cell Malignancies. Blood (2005) 106:3898–906. doi: 10.1182/blood-2005-01-0355
169. Kuang SQ, Fang Z, Zweidler-McKay PA, Yang H, Wei Y, Gonzalez-Cervantes EA, et al. Epigenetic Inactivation of Notch-Hes Pathway in Human B-Cell Acute Lymphoblastic Leukemia. PloS One (2013) 8:61807. doi: 10.1371/journal.pone.0061807
170. Kamdje AHN, Mosna F, Bifari F, Lisi V, Bassi G, Malpeli G, et al. Notch-3 and Notch-4 Signaling Rescue From Apoptosis Human B-ALL Cells in Contact With Human Bone Marrow-Derived Mesenchymal Stromal Cells. Blood (2011) 118:380–9. doi: 10.1182/blood-2010-12-326694
171. Hong Z, Wei Z, Xie T, Fu L, Sun J, Zhou F, et al. Targeting Chemokines for Acute Lymphoblastic Leukemia Therapy. J Hematol Oncol (2021) 14:48. doi: 10.1186/s13045-021-01060-y
172. Hawkins ED, Duarte D, Akinduro O, Khorshed RA, Passaro D, Nowicka M, et al. T-Cell Acute Leukaemia Exhibits Dynamic Interactions With Bone Marrow Microenvironments. Nature (2016) 538:518–22. doi: 10.1038/nature19801
173. Bowers M, Zhang B, Ho Y, Agarwal P, Chen CC, Bhatia R. Osteoblast Ablation Reduces Normal Long-Term Hematopoietic Stem Cell Self-Renewal But Accelerates Leukemia Development. Blood (2015) 125:2678–88. doi: 10.1182/blood-2014-06-582924
174. Oliveira ML, Akkapeddi P, Alcobia I, Almeida AR, Cardoso BA, Fragoso R, et al. From the Outside, From Within: Biological and Therapeutic Relevance of Signal Transduction in T-Cell Acute Lymphoblastic Leukemia. Cell Signal (2017) 38:10–25. doi: 10.1016/j.cellsig.2017.06.011
175. Passaro D, Irigoyen M, Catherinet C, Gachet S, Da Costa De Jesus C, Lasgi C, et al. CXCR4 Is Required for Leukemia-Initiating Cell Activity in T Cell Acute Lymphoblastic Leukemia. Cancer Cell (2015) 27:769–79. doi: 10.1016/j.ccell.2015.05.003
176. Tan SH, Bertulfo FC, Sanda T. Leukemia-Initiating Cells in T-Cell Acute Lymphoblastic Leukemia. Front Oncol (2017) 7:218. doi: 10.3389/fonc.2017.00218
177. Pitt LA, Tikhonova AN, Hu H, Trimarchi T, King B, Gong Y, et al. CXCL12-Producing Vascular Endothelial Niches Control Acute T Cell Leukemia Maintenance. Cancer Cell (2015) 27:755–68. doi: 10.1016/j.ccell.2015.05.002
178. Ferrandino F, Bernardini G, Tsaouli G, Grazioli P, Campese AF, Noce C, et al. Intrathymic Notch3 and CXCR4 Combinatorial Interplay Facilitates T-Cell Leukemia Propagation. Oncogene (2018) 37:6285–98. doi: 10.1038/s41388-018-0401-2
179. Fabbri G, Holmes AB, Viganotti M, Scuoppo C, Belver L, Herranz D, et al. Common Nonmutational NOTCH1 Activation in Chronic Lymphocytic Leukemia. Proc Natl Acad Sci USA (2017) 114:E2911–9. doi: 10.1073/pnas.1702564114
180. Mirandola L, Apicella L, Colombo M, Yu Y, Berta DG, Platonova N, et al. Anti-Notch Treatment Prevents Multiple Myeloma Cells Localization to the Bone Marrow via the Chemokine System CXCR4/SDF-1. Leukemia (2013) 27:1558–66. doi: 10.1038/leu.2013.27
181. Melo RDCC, Longhini AL, Bigarella CL, Baratti MO, Traina F, Favaro P, et al. CXCR7 Is Highly Expressed in Acute Lymphoblastic Leukemia and Potentiates CXCR4 Response to CXCL12. PloS One (2014) 9:85926. doi: 10.1371/journal.pone.0085926
182. Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, et al. NOTCH1-RBPJ Complexes Drive Target Gene Expressionthrough Dynamic Interactions With Superenhancers. Proc Natl Acad Sci USA (2014) 111:705–10. doi: 10.1073/pnas.1315023111
183. Tsaouli G, Ferretti E, Bellavia D, Vacca A, Felli MP. Notch/CXCR4 Partnership in Acute Lymphoblastic Leukemia Progression. J Immunol Res (2019) 2019:1–11. doi: 10.1155/2019/5601396
184. Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, et al. Gain-of-Function Mutations in Interleukin-7 Receptor-α (IL7R) in Childhood Acute Lymphoblastic Leukemias. J Exp Med (2011) 208:901–8. doi: 10.1084/jem.20110580
185. Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, et al. Oncogenic IL7R Gain-of-Function Mutations in Childhood T-Cell Acute Lymphoblastic Leukemia. Nat Genet (2011) 43:932–41. doi: 10.1038/ng.924
186. Ribeiro D, Melão A, Barata JT. IL-7R-Mediated Signaling in T-Cell Acute Lymphoblastic Leukemia. Adv Biol Regul (2013) 53:211–22. doi: 10.1016/j.jbior.2012.10.005
187. Scupoli MT, Perbellini O, Krampera M, Vinante F, Cioffi F, Pizzolo G. Interleukin 7 Requirement for Survival of T-Cell Acute Lymphoblastic Leukemia and Human Thymocytes on Bone Marrow Stroma. Haematologica (2007) 92:264–6. doi: 10.3324/haematol.10356
188. Barata JT, Cardoso AA, Boussiotis VA. Interleukin-7 in T-Cell Acute Lymphoblastic Leukemia: An Extrinsic Factor Supporting Leukemogenesis? Leuk Lymphoma (2005) 46:483–95. doi: 10.1080/10428190400027852
189. Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K Is Indispensable for Interleukin 7-Mediated Viability, Proliferation, Glucose Use, and Growth of T Cell Acute Lymphoblastic Leukemia Cells. J Exp Med (2004) 200:659–69. doi: 10.1084/jem.20040789
190. González-García S, García-Peydró M, Martín-Gayo E, Ballestar E, Esteller M, Bornstein R, et al. CSL-MAML-Dependent Notch1 Signaling Controls T Lineage-Specifc IL-7Rα Gene Expression in Early Human Thymopoiesis and Leukemia. J Exp Med (2009) 206:779–91. doi: 10.1084/jem.20081922
191. González-García S, García-Peydró M, Alcain J, Toribio ML. Notch1 and IL-7 Receptor Signalling in Early T-Cell Development and Leukaemia. In: Current Topics in Microbiology and Immunology. Berlin, Heidelberg: Springer (2012). p. 47–73. doi: 10.1007/82_2012_231
192. Rodríguez-Caparrós A, García V, Casal Á, López-Ros J, García-Mariscal A, Tani-ichi S, et al. Notch Signaling Controls Transcription via the Recruitment of RUNX1 and MYB to Enhancers During T Cell Development. J Immunol (2019) 202:2460–72. doi: 10.4049/jimmunol.1801650
193. Govoni KE, Wergedal JE, Florin L, Angel P, Baylink DJ, Mohan S. Conditional Deletion of Insulin-Like Growth Factor-I in Collagen Type 1α2-Expressing Cells Results in Postnatal Lethality and a Dramatic Reduction in Bone Accretion. Endocrinology (2007) 148:5706–15. doi: 10.1210/en.2007-0608
194. Hayden JM, Mohan S, Baylink DJ. The Insulin-Like Growth Factor System and the Coupling of Formation to Resorption. Bone (1995) 17:S93–8. doi: 10.1016/8756-3282(95)00186-H
195. Young K, Eudy E, Bell R, Loberg MA, Stearns T, Sharma D, et al. Decline in IGF1 in the Bone Marrow Microenvironment Initiates Hematopoietic Stem Cell Aging. Cell Stem Cell (2021). doi: 10.1016/j.stem.2021.03.017
196. Gusscott S, Jenkins CE, Lam SH, Giambra V, Pollak M, Weng AP. IGF1R Derived PI3K/AKT Signaling Maintains Growth in a Subset of Human T-Cell Acute Lymphoblastic Leukemias. PloS One (2016) 11:e0161158. doi: 10.1371/journal.pone.0161158
197. Medyouf H, Gusscott S, Wang H, Tseng JC, Wai C, Nemirovsky O, et al. High-Level IGF1R Expression Is Required for Leukemia-Initiating Cell Activity in T-ALL and Is Supported by Notch Signaling. J Exp Med (2011) 208:1809–22. doi: 10.1084/jem.20110121
198. Hua H, Kong Q, Yin J, Zhang J, Jiang Y. Insulin-Like Growth Factor Receptor Signaling in Tumorigenesis and Drug Resistance: A Challenge for Cancer Therapy. J Hematol Oncol (2020) 13:1–17. doi: 10.1186/s13045-020-00904-3
199. Martelli AM, Paganelli F, Fazio A, Bazzichetto C, Conciatori F, McCubrey JA. The Key Roles of PTEN in T-Cell Acute Lymphoblastic Leukemia Development, Progression, and Therapeutic Response. Cancers (Basel) (2019) 11:629–49. doi: 10.3390/cancers11050629
200. Gusscott S, Kuchenbauer F, Humphries RK, Weng AP. Notch-Mediated Repression of Mir-223 Contributes to IGF1R Regulation in T-ALL. Leuk Res (2012) 36:905–11. doi: 10.1016/j.leukres.2012.02.013
201. Zhang P, Zhang C, Li J, Han J, Liu X, Yang H. The Physical Microenvironment of Hematopoietic Stem Cells and its Emerging Roles in Engineering Applications. Stem Cell Res Ther (2019) 10:1–13. doi: 10.1186/s13287-019-1422-7
202. Zanetti C, Krause DS. “Caught in the Net”: The Extracellular Matrix of the Bone Marrow in Normal Hematopoiesis and Leukemia. Exp Hematol (2020) 89:13–25. doi: 10.1016/j.exphem.2020.07.010
203. Sidhu I, Barwe SP, Gopalakrishnapillai A. The Extracellular Matrix: A Key Player in the Pathogenesis of Hematologic Malignancies. Blood Rev (2020) 48:100787. doi: 10.1016/j.blre.2020.100787
204. Scharff BFSS, Modvig S, Marquart HV, Christensen C. Integrin-Mediated Adhesion and Chemoresistance of Acute Lymphoblastic Leukemia Cells Residing in the Bone Marrow or the Central Nervous System. Front Oncol (2020) 10:775. doi: 10.3389/fonc.2020.00775
205. Vogel V. Unraveling the Mechanobiology of Extracellular Matrix. Annu Rev Physiol (2018) 80:353–87. doi: 10.1146/annurev-physiol-021317-121312
206. Yue B. Biology of the Extracellular Matrix: An Overview. J Glaucoma (2014) 23:S20–3. doi: 10.1097/IJG.0000000000000108
207. Bonnans C, Chou J, Werb Z. Remodelling the Extracellular Matrix in Development and Disease. Nat Rev Mol Cell Biol (2014) 15:786–801. doi: 10.1038/nrm3904
208. Cao H, Heazlewood SY, Williams B, Cardozo D, Nigro J, Oteiza A, et al. The Role of CD44 in Fetal and Adult Hematopoietic Stem Cell Regulation. Haematologica (2016) 101:26–37. doi: 10.3324/haematol.2015.135921
209. Verfaillie C, Benis A, Iida J, McGlave P, McCarthy J. Adhesion of Committed Human Hematopoietic Progenitors to Synthetic Peptides From the C-Terminal Heparin-Binding Domain of Fibronectin: Cooperation Between the Integrin Alpha 4 Beta 1 and the CD44 Adhesion Receptor. Blood (1994) 84:1802–11. doi: 10.1182/blood.v84.6.1802.1802
210. Faassen AE, Schrager JA, Klein DJ, Oegema TR, Couchman JR, McCarthy JB. A Cell Surface Chondroitin Sulfate Proteoglycan, Immunologically Related to CD44, Is Involved in Type I Collagen-Mediated Melanoma Cell Motility and Invasion. J Cell Biol (1992) 116:521–31. doi: 10.1083/jcb.116.2.521
211. Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 Is the Principal Cell Surface Receptor for Hyaluronate. Cell (1990) 61:1303–13. doi: 10.1016/0092-8674(90)90694-A
212. Dimitroff CJ, Lee JY, Rafii S, Fuhlbrigge RC, Sackstein R. CD44 Is a Major E-Selectin Ligand on Human Hematopoietic Progenitor Cells. J Cell Biol (2001) 153:1277–86. doi: 10.1083/jcb.153.6.1277
213. Protin U, Schweighoffer T, Jochum W, Hilberg F. CD44-Deficient Mice Develop Normally With Changes in Subpopulations and Recirculation of Lymphocyte Subsets. J Immunol (1999) 163:4917–23.
214. Schmits R, Filmus J, Gerwin N, Senaldi G, Kiefer F, Kundig T, et al. CD44 Regulates Hematopoietic Progenitor Distribution, Granuloma Formation, and Tumorigenicity. Blood (1997) 90:2217–33. doi: 10.1182/blood.v90.6.2217
215. Zöller M. CD44, Hyaluronan, the Hematopoietic Stem Cell, and Leukemia-Initiating Cells. Front Immunol (2015) 6:235. doi: 10.3389/fimmu.2015.00235
216. Hoofd C, Wang X, Lam S, Jenkins C, Wood B, Giambra V, et al. CD44 Promotes Chemoresistance in T-ALL by Increased Drug Efflux. Exp Hematol (2016) 44:166–71.e17. doi: 10.1016/j.exphem.2015.12.001
217. García-Peydró M, Fuentes P, Mosquera M, García-León MJ, Alcain J, Rodríguez A, et al. The NOTCH1/CD44 Axis Drives Pathogenesis in a T Cell Acute Lymphoblastic Leukemia Model. J Clin Invest (2018) 128:2802–18. doi: 10.1172/JCI92981
218. Giambra V, Jenkins CR, Wang H, Lam SH, Shevchuk OO, Nemirovsky O, et al. NOTCH1 Promotes T Cell Leukemia-Initiating Activity by RUNX-Mediated Regulation of PKC-Θ and Reactive Oxygen Species. Nat Med (2012) 18:1693–8. doi: 10.1038/nm.2960
219. LaFoya B, Munroe JA, Mia MM, Detweiler MA, Crow JJ, Wood T, et al. Notch: A Multi-Functional Integrating System of Microenvironmental Signals. Dev Biol (2016) 418:227–41. doi: 10.1016/j.ydbio.2016.08.023
220. Gibson MA, Hatzinikolas G, Kumaratilake JS, Sandberg LB, Nicholl JK, Sutherland GR, et al. Further Characterization of Proteins Associated With Elastic Fiber Microfibrils Including the Molecular Cloning of MAGP-2 (MP25). J Biol Chem (1996) 271:1096–103. doi: 10.1074/jbc.271.2.1096
221. Deford P, Brown K, Richards RL, King A, Newburn K, Westover K, et al. MAGP2 Controls Notch via Interactions With RGD Binding Integrins: Identification of a Novel ECM-Integrin-Notch Signaling Axis. Exp Cell Res (2016) 341:84–91. doi: 10.1016/j.yexcr.2016.01.011
222. Miyamoto A, Lau R, Hein PW, Shipley JM, Weinmaster G. Microfibrillar Proteins MAGP-1 and MAGP-2 Induce Notch1 Extracellular Domain Dissociation and Receptor Activation. J Biol Chem (2006) 281:10089–97. doi: 10.1074/jbc.M600298200
223. Katsube KI, Sakamoto K, Tamamura Y, Yamaguchi A. Role of CCN, a Vertebrate Specific Gene Family, in Development. Dev Growth Differ (2009) 51:55–67. doi: 10.1111/j.1440-169X.2009.01077.x
224. Katsuki Y, Sakamoto K, Minamizato T, Makino H, Umezawa A, Ikeda M, et al. Inhibitory Effect of CT Domain of CCN3/NOV on Proliferation and Differentiation of Osteogenic Mesenchymal Stem Cells, Kusa-A1. Biochem Biophys Res Commun (2008) 368:808–14. doi: 10.1016/j.bbrc.2008.02.010
225. Sakamoto K, Yamaguchi S, Ando R, Miyawaki A, Kabasawa Y, Takagi M, et al. The Nephroblastoma Overexpressed Gene (NOV/Ccn3) Protein Associates With Notch1 Extracellular Domain and Inhibits Myoblast Differentiation via Notch Signaling Pathway. J Biol Chem (2002) 277:29399–405. doi: 10.1074/jbc.M203727200
226. Chintala H, Krupska I, Yan L, Lau L, Grant M, Chaqour B, et al. The Matricellular Protein CCN1 Controls Retinal Angiogenesis bySrc Homology 2 Domain Phosphatase-1 and Notch Signaling. Dev (2015) 142:2364–74. doi: 10.1242/dev.121913
227. Lelièvre E, Hinek A, Lupu F, Buquet C, Soncin F, Mattot V. VE-Statin/Egfl7 Regulates Vascular Elastogenesis by Interacting With Lysyl Oxidases. EMBO J (2008) 27:1658–70. doi: 10.1038/emboj.2008.103
228. Fitch MJ, Campagnolo L, Kuhnert F, Stuhlmann H. Egfl7, a Novel Epidermal Growth Factor-Domain Gene Expressed in Endothelial Cells. Dev Dyn (2004) 230:316–24. doi: 10.1002/dvdy.20063
229. Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, et al. The Endothelial-Cell-Derived Secreted Factor Egfl7 Regulates Vascular Tube Formation. Nature (2004) 428:754–8. doi: 10.1038/nature02416
Keywords: lymphopoiesis, hematopoieisis, Notch signaling, T cell development, bone marrow niches
Citation: Sottoriva K and Pajcini KV (2021) Notch Signaling in the Bone Marrow Lymphopoietic Niche. Front. Immunol. 12:723055. doi: 10.3389/fimmu.2021.723055
Received: 09 June 2021; Accepted: 14 July 2021;
Published: 28 July 2021.
Edited by:
Stéphane JC Mancini, INSERM UMR1236 Microenvironnement, Différenciation cellulaire, Immunologie et Cancer, FranceReviewed by:
Fernando Afonso, Cardiff University, United KingdomLluis Espinosa, Mar Institute of Medical Research (IMIM), Spain
Edward Chen, Lunenfeld-Tanenbaum Research Institute, Canada
Copyright © 2021 Sottoriva and Pajcini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kostandin V. Pajcini, a3ZwQHVpYy5lZHU=
 Kilian Sottoriva
Kilian Sottoriva Kostandin V. Pajcini
Kostandin V. Pajcini