
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 14 July 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.720393
This article is part of the Research TopicThe Role of Dietary Interventions in The Regulation of Host-Microbe InteractionsView all 24 articles
Atopic dermatitis (AD) is a public health concern and is increasing in prevalence in urban areas. Recent advances in sequencing technology have demonstrated that the development of AD not only associate with the skin microbiome but gut microbiota. Gut microbiota plays an important role in allergic diseases including AD. The hypothesis of the “gut-skin” axis has been proposed and the cross-talk mechanism between them has been gradually demonstrated in the research. Probiotics contribute to the improvement of the intestinal environment, the balance of immune responses, regulation of metabolic activity. Most studies suggest that probiotic supplements may be an alternative for the prevention and treatment of AD. This study aimed to discuss the effects of probiotics on the clinical manifestation of AD based on gut microbial alterations. Here we reviewed the gut microbial alteration in patients with AD, the association between gut microbiota, epidermal barrier, and toll-like receptors, and the interaction of probiotics and gut microbiota. The potential mechanisms of probiotics on alleviating AD via upregulation of epidermal barrier and regulation of immune signaling had been discussed, and their possible effective substances on AD had been explored. This provides the supports for targeting gut microbiota to attenuate AD.
Atopic dermatitis (AD) is an inflammatory skin disease characterized by recurrence, dry skin, erythema and itchiness. The incidence of AD gradually increases with the development of industrialization and urbanization, affecting 15-30% of children and 10% of adults all over the world (1, 2). The clinical symptoms will disappear with growing up in some children with AD, but about 1/2 of children may develop into allergic asthma and 2/3 of children have a risk of allergic rhinitis in the future. The process is called the “atopic march” (3). The scratching induced by intense itchiness leads to skin barrier destruction, which disturbs the immune responses and microbial ecology in the local area and makes patients fall into the circle of “itch-scratch-severe itch”. The itch and recurrence of AD result in a poor quality of sleep, which may be closely associated with the inferiority complex, anxiety, depression, and other psychological diseases in patients with AD based on the brain-skin connection (4). Additionally, it brings a great economic burden to patients due to the long-term treatment for AD and significantly decreases the life quality of their families (5).
The causes of AD are complex and include genetic and environmental factors. Genetic linkage analysis has been identified AD locus on chromosomes 1q21, 17q25 and 20p (6). Furthermore, evidence has been shown that loss and mutations in the gene encoding filaggrin are closely related to AD onset and development (2). Although genetics play an important role in the onset of AD, changes in environmental factors are significantly associated with the increase in prevalence in recent years. Individuals with AD are commonly stimulated by allergens including pollen, dust mite and animal dander around them (7, 8). Skin flora, especially, Staphylococcus aureus and Malassezia blooms in the lesions results in more severe AD clinical symptoms (9, 10). The pathogenesis mechanism of atopic dermatitis has not been fully demonstrated but T helper type 2 (Th2) - and Th17-skewed immune dysregulation is predominant in the acute phase and chronic phase, respectively (11). Air pollutions, such as polycyclic aromatic hydrocarbons, have been reported to cause Th2 cell-related skin disorders including atopic dermatitis (1). Interleukin (IL) 4 and IL-13 are excessively released after Th2 cell activation and increase immunoglobulin (Ig) E class switching and specific IgE production in B cells (12). The specific IgE binds with the high-affinity receptor FcϵRI expressed by mast cells and basophils, resulting in degranulation of these cells and release of inflammatory mediators and inducing clinical symptoms of AD (13). IL-31, as the product of Th2 cells and immature dendritic cells, activates IL-31 receptor A/oncostatin M receptor to stimulate itch and neuronal outgrowth (14). It has been identified as a critical cytokine involving neuroimmune communication and thus affects the development of pruritus closely associated with nerves in AD patients. The humanized monoclonal antibody against IL-31 receptor A, Nemolizumab, has been reported to result in a significant decrease in pruritus in patients after a 16-week intervention (15). These studies suggest that immune response balance is a critical factor to protect the host from suffering AD. Therefore, regulation for immune response is an effective approach to alleviate atopic dermatitis in patients.
Intestinal microecology is a dynamic and unique ecosystem and affected by diet, living habits and mental stress. The imbalance of gut microbial diversity and composition causes intestinal microecological disorder and results in the shifts of gut microbial metabolism and immune responses. These alterations are closely associated with physiological and pathological activities and important for human health. The maintenance for structural diversity of gut microbiota resists the invasion of pathogen bacteria and reduces the nutritional competition between the potentially harmful bacteria and commensal bacteria. Gut microbiota involved in short-chain fatty acid (SCFA), amino acid, vitamin and bile acid metabolism induces the mature of the innate and adaptive immune system (16). Therefore, the gut microbiota is a potential target for regulating immune responses in the host. With the development of sequencing technology, the correlation between gut microbiota and human diseases including allergic asthma, atopic dermatitis, is revealed in many studies (17–19). The “gut-skin” axis has been proposed and is recognized as a new target to prevent and treat AD. Gut and skin have several similar characteristics and are parts of the overall immune and endocrine systems (20). They are the major compositions of mucosal immunity and directly contact the environmental antigens including foods, commensal bacteria and pathogens. The development of gut diseases is commonly accompanied by cutaneous lesional manifestations and this implies the association between them may affect each other’s states (21). Therefore, targeting gut microbial alterations may be an alternative to regulate immune responses and ameliorate cutaneous health in AD patients. Probiotics and/or prebiotics, as the common regulator for gut microbiota, have been used to alleviate AD clinical symptoms, but with controversial outcomes (positive or ineffective). This is related to the complex interaction between immune response, gut microbiota, and metabolic activity in the host. With a focus on gut microbial alteration, this review discusses the beneficial role of probiotics in the prevention and treatment of AD and the possible underlying mechanisms.
Numerous studies have shown that the development of allergic diseases such as asthma and AD is closely associated with changes in gut microbial diversity and composition (22–25). Over 1000 different species reside in the gastrointestinal tract (26), and the number of bacterial cells is about 10 times larger than that of eukaryotic cells in the human body (27). The bacteria have evolved with the human, and it becomes a mutualistic relationship. Therefore, gut microbiota may involve in the development of certain diseases, and the role of gut microbiota is worth exploring in the development of AD. Before birth, microbial compositions have been found in the placenta and meconium, suggesting microbial colonization in early life (28). Following birth, gut microbiota and mucous membranes begin to establish and have been affected by delivery modes such as natural birth or caesarean section (29). Lactobacillus, Prevotella and Sneathia spp. are dominant bacterial communities in the gut of naturally delivered infants, resembling their own mother’s vaginal microbiota, but the microbial communities in caesarean delivery infants are similar to skin microbiota, dominated by Staphylococcus, Corynebacterium and Propionibacterium spp (30), implying the shaping role of delivery mode in the diversity and structure of the initial microbiota. After that, the gut microbial diversity increases rapidly and dietary factors including breast- and formula-fed become the important perturbations for shaping gut microbial diversity and composition. Lactobacillus and Bifidobacterium are dominant gut microbiota in breast-fed infants at 12 months of age, but Roseburia, Clostrium and Anaerostipes, that belonging to Clostirdia, are enriched in the gut microbiota of no longer breast-fed children (31). Lactobacillus, Bifidobacterium and Bacteroides can degrade oligosaccharides from breast milk into small sugars and utilize them to obtain an advantage for growth (32). Therefore, they are the most abundant bacterial communities in the gut of breast-fed infants. Enterococci and Clostridia are dominant bacteria in formula-fed infants (33), and the intestinal tract contains fewer bacterial cells and more species than in breast-fed infants (34). At 3 years of age, gut microbial composition toward a more stable shift and resemble that of the adult (31).
Bacterial diversity and composition are closely associated with the onset and development of various diseases such as acute infective diarrhea, constipation, obesity, and depression (35–38), underlining the importance of bacterial diversity and colonization in early life for future health. Table 1 shows gut microbial alteration in patients with AD. Compared to healthy individuals, gut microbial diversity decreased, and the relative abundances of the beneficial microbes such as Lactobacillus, Bifidobacterium significantly reduced but the proportions of Escherichia coli, Clostridium difficile and Staphylococcus aureus increased in patients. Especially, gut microbial colonization and alteration were demonstrated prior to any clinical manifestations in early life, indicating gut microbial dysbiosis as one of the causes of AD (54). Infants with less gut microbial diversity seem to be susceptible to atopic dermatitis. A cross-sectional study among 1440 children showed that the α diversity of gut microbiota was closely associated with a decreased risk of eczema (55). The α diversity was not different in adult patients suffering from allergic asthma comparing the healthy controls (56). Although the relative abundance of bifidobacteria was reduced, Bifidobacterium adolescentis species prevailed within the bifidobacterial population (56). The results showed bifidobacterial composition, especially the proportion of B. adolescentis, had special effects on the development of allergic disease. A clinical trial showed that the diversity of Bifidobacterium species in allergic infants was similar to non-allergic infants (57). However, in a study from rural Japan infants, allergic infants had a higher abundance of B. catenulatum and B. bifidum than healthy controls in different stages of age (58). These controversial outcomes further show that the association between allergic disease and gut microbiota is complex and maybe not be restricted to Bifidobacterium species. In early life, regulation of gut microbial diversity and composition may reduce the onset and development of allergic symptoms including AD. Therefore, this is an alternative to reduce the adverse reactions of drugs for AD.
The “hygiene hypothesis” suggesting that decreased early life microbial exposure and diversity result in loss of immunological tolerance and this is being linked to an increased prevalence of allergic diseases in the urban environment (59, 60). Gut microbiota is the most important component of microbial exposures. Therefore, gut microbial communities affect shaping the host immune development, and dysbiosis of gut microbiota is closely associated with immune disorders (61–63). Throughout the lifespan, the host’s immune system is constantly regulated by gut microbiota. The maternal gut microbial alterations in pregnancy affect the early postnatal immunity of offsprings. Comparing to the offsprings of germ-free mice, gestation colonization with Escherichia coli HA107 significantly altered the numbers of postnatal intestinal leukocytes in offsprings and regulated the development of the innate immune system in early life (64). Furthermore, the interactions of gut microbiota with T cells and B cells can lead to systemic outcomes that are distal to the site of the gut. For example, the strains belonging to Clostridia induce expansion and differentiation of regulatory T cells (Treg) and alleviate the clinical symptoms of colitis and allergic diarrhea in mice (65). Segmented filamentous bacteria induce the T helper 17 cells (Th17) in the small intestinal lamina propria to drive autoimmune arthritis (66).
A human intervention trial of manipulating urban environmental biodiversity showed that skin and gut microbial diversity of children in nature-oriented daycare centers were increased and this was closely associated with an overall more healthy immune system (67). The ratio of IL-10:IL-17A was increased in plasma samples of these children. The decrease in IL-17A expression was related to and decreased Romboutsia and Dorea, increased Anaerostipes, and higher Faecalibacterium Otu00007 in the gut. Gut microbial diversity contributes to the education of the immune system and decreases the prevalence of immune-mediated diseases such as allergies. Additionally, gut microbial colonization promoted the development of microbiota-T cells in the thymus via migration of microbial antigens from the gut to the thymus by intestinal dendritic cells (DCs) (68). This not only expanded T cells but increased the capability of thymic T cells to identify gut microbiota and pathogens. It further indicates that gut microbial colonization affects the development of adaptive immune responses and educates the immune system. Therefore, gut microbial alterations are closely associated with the immune responses and play a crucial role in the development of diseases involved in aberrant immune functions.
As a systemic disease, AD may have the aberrant barrier function across multiple organ sites including skin, lung and gut. In an epithelial barrier dysfunctions study in AD, epidermal barrier disruption led to allergen sensitization and pathogens colonization. This induced inflammatory response and increased barrier breakdown at distant sites such as the gut and respiratory tract (69). It is suggested that there is a crosstalk mechanism between skin and gut, and thus gut microbial alteration may be associated with epidermal barrier function in AD. In AD, the epidermal barrier may be an important component of the innate immune system because it protects from the invasion of pathogens and allergens and prevents water loss in the skin. Filaggrin (FLG) is an important epidermal protein, and FLG deficiency results in the epidermal barrier defect and increases the risk of the microbiome and virus invasions. It has been demonstrated that FLG deficiency is closely associated with AD and plays a crucial role in the pathogenesis of AD (70). In FLG-deficient (flg-/-) mice model, AD symptoms were induced using calcipotriol. Comparing to wild-type mice, the flg-/- mice exerted severer clinical symptoms characterized by increased ear thickness, mast cells and CD3+ T cells infiltration, and the level of thymic stromal lymphopoietin, interleukin (IL)-4, IL-6 and IL-13 (71). This is implied that FLG is an important predisposing factor in the pathogenesis of AD. In a genetic correlation study consisting of 386 whole-genome sequencing samples, there was a significant association between FLG function mutation and age of onset of AD (72). L. plantarum LM1004 significantly improved the AD-like symptoms, decreased Th2 and Th17 cell transcription factor levels, and increased the transcription factors of Treg and Th1 cells, galactin-9 and FLG (73). This is implied that there is an interaction between probiotics, gut microbiota, and the epidermal barrier. Furthermore, the important feature of AD is an itch, which contributes to damage of the epidermal barrier. Surfaces of skin, lung and gut can act as a shared immunological interface, and environmental stimulation such as gut microbial alteration affects interactions of immune responses among them. This immunological interface as part of the mucosal membrane is the first line to combat infection. Innate lymphoid cells (ILCs) play a key role in the homeostasis and pathology of mucosal membranes and affect the interactions of the “gut-lung” axis. It has been identified that inflammatory type 2 ILCs from the intestine are recruited to the lung by IL-25 and mediate type 2 immune responses (74). Gut inflammation and gut barrier leakage increased the activation of skin epithelial cells and recruitment of T cells to the skin in patients with Omenn syndrome, and this exacerbated the skin inflammation (75). This had been verified in Rag2R229Q mice which simulated the clinical symptoms of Omenn syndrome. These results further identify skin, lung and gut share the immunological interface and it mainly consists of the mucosal membrane from these sites. Therefore, the integrity of the epidermal barrier is essential for maintaining the immune responses in the skin.
Toll-like receptors (TLRs), a superfamily of pattern-recognition receptors bridge innate and adaptive immunity (Figure 1). TLRs are a class of transmembrane non-catalytic proteins and can recognize molecules with conserved structures from microorganisms. These molecules are known as the pathogen-associated molecular pattern (PAMP) such as lipopolysaccharide, peptidoglycan and zymosan. When TLRs bind PAPM, they initiate a signal transduction cascade to activate innate immune responses to eliminate pathogens that break through the skin or mucosa barrier (76). Most TLRs (TLR1, TLR2, TLR4, TLR5, TLR6, TLR10, TLR11 and TLR12) are expressed on the cell surface to recognize PAMPs, but TLR3, TLR7, TLR8 and TLR9 are intracellular to detect nucleic acids (77). As yet, 13 and 10 members of TLRs have been identified in mice and humans, respectively, and their respective ligands also have been revealed (78). The skin harbors various cells expressing TLRs and directly expose to microbes and pathogens in the environment. Therefore, invading pathogens-inducing aberrant TLRs responses may result in skin diseases including AD (79). S. aureus colonizes skin lesions of AD patients and can be recognized by TLR2 due to its cell wall components. Compared to healthy volunteers, the expression of TLR2 was decreased on Langerhans cells (LC) in AD patients with high colonization by in situ analysis. TLR2 ligand induced maturation and migratory activity of LC and decreased IL-6 and IL-10 production of skin samples from AD patients (80). This suggested that TLR2-mediated immunoregulation signal pathways had been impaired in AD patients. Additionally, macrophages are known to express TLR2 and accumulate in acute and chronic stages of AD in skin lesions. Compared to healthy controls, macrophages from peripheral blood monocytes of AD patients expressed decreased TLR2 and pro-inflammatory cytokines including IL-6, IL-8 and IL-1β after TLR2 ligands intervention (81). In primary human keratinocytes, TLR2 agonists such as S. aureus-derived peptidoglycan and Pam3CSK4 significantly improved the tight junction barrier and increased the expression of tight junction proteins. Therefore, the epidermal barrier in AD patients was restored after a TLR2 agonist intervention. TLR2-/- mice also exerted a delayed barrier recovery indicating that TLR2 signaling played a critical role in epidermal barrier integrity (82).
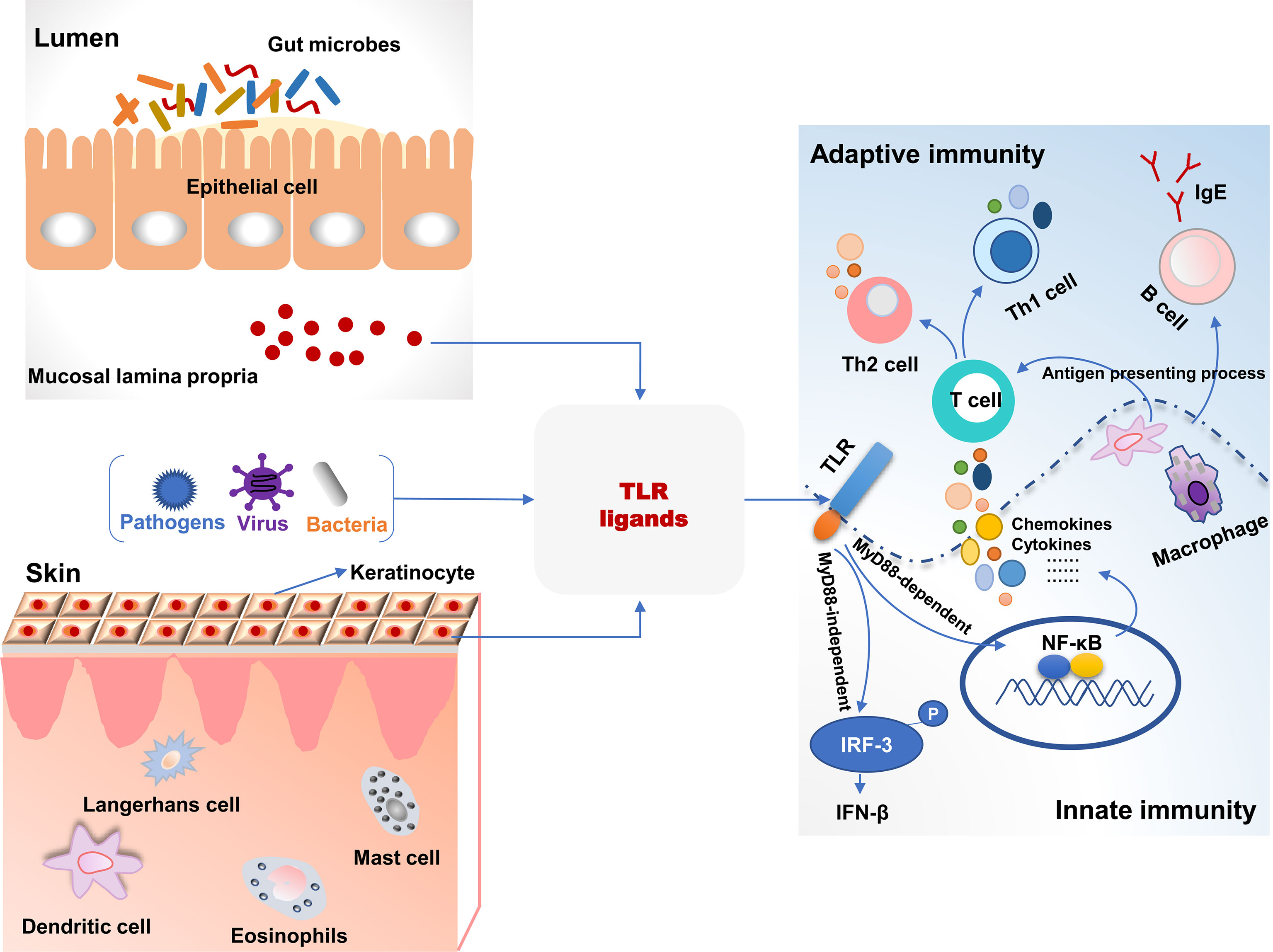
Figure 1 The association of toll-like receptor signaling and immune responses in the intestine and skin. TLR ligands from bacteria, viruses, and pathogens were recognized and activated TLR signaling pathways, which bridged the innate and adaptive immunity in the intestine and skin. TLR, toll-like receptors; MyD88, myeloid differentiation factor 88; P, phosphorylation.
It has been studied that TLR2 rs5743708 and TLR4 rs4986790 polymorphisms are associated with susceptibility to AD (83). In neonates, the incidence of AD was significantly associated with twofold lower TLR4-mediated IL-10 production and resulted in an impaired Th1 type polarizing immune response (84). Furthermore, the single nucleotide polymorphisms (SNPs) related to oxidative stress and inflammation indicating that there was a close association between TLR2, TLR4, and TNF and traffic-related air pollution, and this revealed the gene-environment interactions in the development of AD (85). In TLR4-/- mice, hapten (2,4-dinitrochlorobenzene)-induced AD symptoms and Th2-type inflammatory responses were more severe than wild-type mice and increased the migration of DCs into draining lymph nodes (86). This indicated that TLR4 mediated immune responses associated with AD development. TLR4 ligands increased IL-23 production in skin lesions and resulted in the migration of skin DCs that induced IL-22 expression by naïve CD4+ T cells. IL-22 increased keratinocyte proliferation and inflammatory infiltration from Th22 cells in AD mice (87). These studies suggest TLR4 activation contributes to the balance between Th1- and Th2-type immune responses and is one of the targets to treat AD symptoms.
Itch is an important symptom of AD and is associated with TLRs signaling pathways. TLR3 is expressed by small-sized primary sensory neurons and plays a role in modulating sensory neuronal excitability and central sensitization. TLR3 knockdown alleviated pruritus in wild-type mice. In TLR3-/- mice, excitatory synaptic transmission was impaired, and scratching behaviors were significantly decreased after histamine and pruritogens challenges (88). This demonstrated the potential anti-itch role of TLR3 in AD. House dust mite (HDM) is a common allergin and is related to exacerbation of AD. HDM-induced Th2-type immune responses were closely associated with the expressions of IL-25 and IL-33 via activation of TLR1 and TLR6 signaling (89). Additionally, in a total of 1063 children cohort study, prenatal contact with farm animals and cats significantly decreased the risk of incidence of AD. This was closely associated with increased expression of TLR5 and TLR9 in cord blood (90). Collectively, TLRs signaling pathways play a critical role in innate immune responses in the development of AD, and impaired TLRs signaling leads to an aberrant balance between Th1- and Th2-type immune responses.
Gut microbiota and their metabolites can be recognized by TLRs, and the interactions between bacteria and TLRs contribute to systemic immune homeostasis (Figure 1) (91). Perturbations in gut microbiota lead to the invasion of microbes and their metabolites into circulation and affect pathological symptoms of the distant site organs such as the brain, liver, kidney, lung, and skin via TLRs signaling pathways. In a cohort study with 957 children, there was a significant multiplicative interaction between TLR4 SNP rs10759932 and E. coli regarding allergic sensitization in the first 2 years of life (92). It demonstrated the effect of TLR4 genetic variations on allergy development in early life and the modulating role of gut microbe in immune responses in relation to TLRs signaling. An evaluation of gut microbiota and innate immune responses in IgE-associated eczema showed that Ruminococcaceae in fecal samples was lower in atopic eczema infants than that in healthy controls and was negatively related to TLR2-induced IL-6 and TNF-α. Enterobacteriaceae (a genus of Proteobacteria phylum) was negatively related to TLR4-induced TNF-α, and α-diversity of Bacteroidetes and Actinobacteria were lower in atopic eczema infants versus the controls (93). The administration of a food allergen increased specific IgE and histamine levels and induced allergic symptoms in TLR4-mutant or -deficient mice. However, after antibiotic treatment, gut microbial composition and structure were disturbed in TLR4 wild-type mice, and they were susceptible to the induction of food allergy like the TLR4-mutant mice (94). It indicated that microbes and TLRs signaling were necessary for the development of the immune system. Dysbiosis of gut microbiota results in immune disorder and increases the risk of allergy. TLRs are not only able to recognize invading pathogens and provoke immune responses but play a critical role in the cross-talk between commensal bacteria that inducing immune tolerance and host. Therefore, these studies provide a potential approach to improve immune-mediated diseases such as allergy, based on gut microbial alteration.
As mentioned earlier, the onset and development of AD are closely associated with gut microbial alterations, and beneficial bacteria such as Bifidobacterium and Lactobacillus are in shortage in patients. Probiotics consumption may be an effective alternative to supply beneficial bacteria and restore intestinal dysfunction. The gut microbial environment can be reshaped with long-term consumption of probiotics and contributes to the balance of gut microbiota and systemic immune responses. Probiotics promote the synthesis of nutrients such as amino acids and vitamins in the host and increase the content of SCFA in the intestinal lumen. Especially, SCFA including acetate, propionate, and butyrate leads to an intestinal environment with a low pH value to inhibit the growth of pathogens. Additionally, probiotics compete against pathogens including competition for the nutrient substrates and ecological niches, and these interactions contribute to suppression for the excess proliferation of pathogens in the intestine. Therefore, probiotics may alleviate AD clinical manifestation via affecting the gut microbial composition, metabolic functions, and immune responses. Table 2 shows the effects of probiotics on clinical manifestations of patients from pregnant, infant, children to adult and the potential to alleviate AD, although there are some controversial outcomes. Most probiotics reduced the SCORAD (scoring atopic dermatitis index) scores and even decreased the risk of developing AD. The controversial conclusions are associated with many factors such as environment and diet, and in the future, larger samples and more precise experimental design are necessary for clinical trials to verify the effectiveness of probiotics on AD.
Based on the “hygiene hypothesis”, bacterial stimulation is required for the maturation of the gut immune system in early life. Most probiotics, derived from the commensal bacteria in the intestine, have been demonstrated to contribute to education for immune tolerance and maintenance of the intestinal immune responses. Immunoglobulin (Ig) A is a crucial antibacterial protein in the intestinal mucosal defense. It blocks pathogens to adhere the intestinal epithelium and increases bacterial entrapment in mucus (110). Bifidobacterium is known to stimulate Peyer’s patches to induce IgA production and maintain the integrity of the gut barrier. Administration of Lactobacillus GG and Saccharomyces boulardii affects the cytokine release and mucosal milieu, and this increases IgA production in the intestine (111). Regulation of the balance between Th1- and Th2-type immune responses is one way to improve clinical symptoms in allergic diseases. B. animalis subspecies lactis Bb12 increased IgA response in serum and IgG1 and IgG2 response in the ileal fluid in Ascaris suum infected pigs (112). B. animalis subspecies lactis Bb12 treatment improved expression of genes related to Th1/Th2 cells, inflammatory cells, Treg, and physiological function in the gut and reduced Th2 type immune responses. In β-lactoglobulin-induced allergic mice, L. plantarum ZDY2013, L. plantarum WLPL04 and L. rhamnosus GG increased Th1 cells differentiation and inhibited the Th2-biased immune response (113). Furthermore, Treg differentiation not only regulates Th1/Th2 immune balance but suppresses Th17-biased response. L. paracasei KBL382 significantly improved the pathological features and altered the gut microbial composition in AD mice (114). It regulated immune balance via increasing the expression of IL-10 and transforming growth factor-β and enhancing the differentiation of CD4+ CD25+ Foxp3+ Treg in mesenteric lymph nodes. L. sakei WIKIM30 enhanced Treg differentiation in mesenteric lymph nodes via inducing DCs tolerance and ameliorated AD-like skin lesions (11). It increased the proportion of Ruminococcus, which was positively associated with Treg-related immune responses and might contribute to the alleviation of AD.
Probiotics also contribute to the reduction in the expression of pro-inflammatory cytokines such as IL-13, thymic stromal lymphopoietin (TSLP), and IL-5. The differentiation of eosinophils is closely associated with allergic diseases such as AD, but IL-5 is the critical cytokine to increase the development and survival of eosinophils (115). L. chungangensis CAU 28(T) significantly reduced the expression of IL-5, TNF-α, and thymus- and activation-regulated chemokine and alleviated the inflammatory infiltration in AD mice (116). IL-13, like the IL-4, is the key driver to activate Th2-type immune response and shares a common receptor subunit with IL-4. IL-13 and IL-4 bind receptors to activate JAK -STAT6 (Janas kinase-signal transducers and activators of transcription 6) pathways and lead to the decrease in the expression of structural protein such as FLG, involucrin, and lipid composition in the skin (117). Tralokinumab, a monoclonal antibody to neutralize IL-13, had been reported to improve the clinical in adults with AD in randomized, double-blind, multicenter, and placebo-controlled phase III trials (118). Pediococcus acidilactici intake reduced the mRNA expression of IL-4, TNF-α, and IL-13 in dorsal skin and improved the clinical severity of AD (119). Levels of TSLP are high in the lesions of AD patients and TSLP is a key protein in the development of AD (120). TSLP is expressed by epithelial cells of the gut, lung, and skin and increases Th2 cell differentiation and Th2-type inflammation through interacting with immune cells such as DCs, natural killer T cells, and CD4+ T cells (121). In a murine model with AD, skin-specific overexpression of TSLP led to the increases in Th2 CD4+ T cells and serum IgE levels (122). Tezepelumab is a monoclonal antibody targeting TSLP and has been reported to treat AD. In a phase 2a study, tezepelumab plus topical corticosteroids (TCS) treatment resulted in a 64.7% reduction in the eczema area and severity index versus 48.2% of that in the placebo plus TCS treatment (123). L. rhamnosus Lcr35 significantly reduced the expression of IL-4 and TSLP and prevented the development of AD (124). Collectively, probiotics have the great potential to modulate the immune function in AD and may be a microbial alternative strategy to improve AD.
Probiotics alter the gut microbial composition and simultaneously affect their metabolic activities that may lead to a decreased risk for allergy (Figure 2). The metabolites of B. breve C50 and Streptococcus thermophilus 065 increased the proportion of CD4+ and CD8+ T cells secreting Th1-type cytokine IFN-γ and restored Th1/Th2 immune balance in IL-10-deficient mice (125). Bacteriocins of B. animalis subspecies lactis Bb12 and B. longum Bb46 significantly inhibited the growth of S. aureus and E. coli in the intestine (126), and these harmful bacteria were associated with the development of AD and the proportion of them was increased in patients. SCFA is produced by the gut microbial fermentation of indigestible carbohydrates and is closely associated with the alleviation of AD clinical manifestations. Furthermore, SCFA has been demonstrated to regulate the size and function of the Treg pool in the intestine (127). In a cohort study, the severity of AD was negative with the proportion of butyrate-producing bacteria in infants and suggesting that butyrate had a potential role in improving AD symptoms (128). High levels of propionate and butyrate in feces reduced atopic sensitization in early life and administration of butyrate decreased the severity of allergic inflammation in mice (129). Antibiotics-induced gut microbial dysbiosis resulted in a decrease in SCFA production and an increase in the levels of inflammatory cells, and these alterations were highly associated with aggravated AD-like skin lesions (130). However, fecal microbial transplantation significantly reduced the clinical score of AD-like lesions via increasing SCFA levels and regulating the numbers of immune cells. SCFA contributes to the balance of gut microbiota and is closely associated with levels of immune cells. Therefore, increasing SCFA production in the intestine via probiotics consumption may be an effective way to alleviate AD-like symptoms.
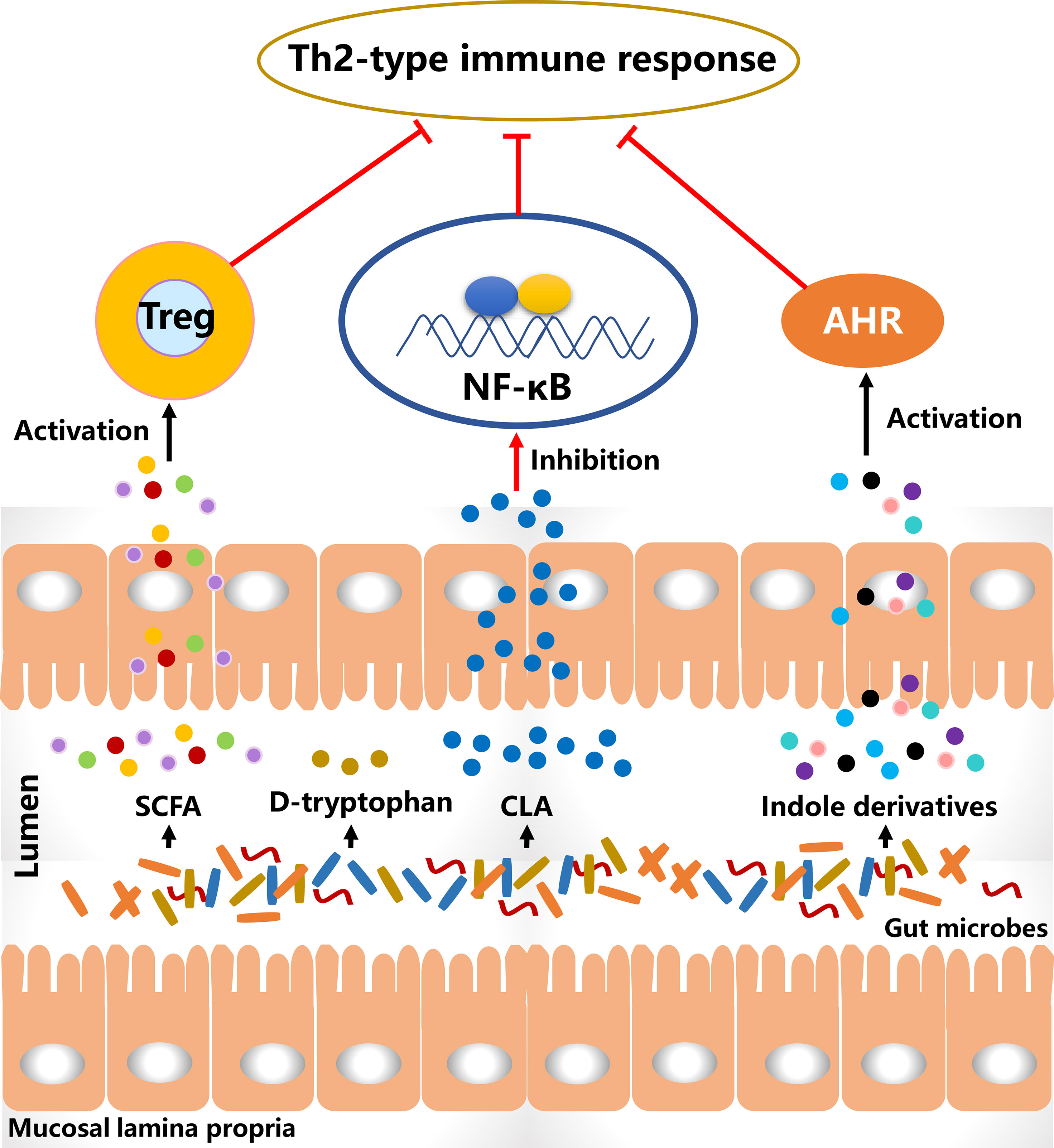
Figure 2 The diagram of the potential effective substances for suppressing Th2-type immune responses. CLA, conjugated linoleic acid; SCFA, short-chain fatty acid; Treg, regulatory T cells.
D-tryptophan, as a metabolite of Bifidobacterium, Lactobacillus and Lactococcus, suppressed the expression of Th2-associated CCL17 in KM-H2 cells (131). It significantly increased IL-10 production and decreased IL-12, IL-5 and IFN-g in human DCs. After supplementation with D-tryptophan in mice with allergic airway inflammation, the clinical manifestations were alleviated and Th2-biased immune responses were significantly reversed. Conjugated linoleic acid (CLA), as a natural unsaturated fatty acid, can inhibit the release of histamine, which induces an increase in vascular permeability and is associated with the development of AD. Bifidobacterium, Lactobacillus and Roseburia spp. metabolize polyunsaturated fatty acids including omega-3 and omega-6 fatty acids to CLA (132). B. breve and B. pseudocatenulatum are CLA-producing bacteria and have been reported to alleviate colitis via modulating gut microbiota and TLR4/NF-κB signaling (133, 134). It is suggested that probiotics consumption increases CLA production in the intestine and affects the systemic immune responses. L. plantarum JBCC105645 and JBCC105683, isolated from the salted fermented seafood according to CLA-producing activity, significantly alleviated the pathological symptoms of AD via reducing IL-4 levels and increasing IFN-γ levels (135). This suggested that CLA might be the material basis of probiotics to alleviate AD. Oral administration of CLA significantly attenuated AD-like skin lesions via inhibition of COX-2/5-LOX and TLR4/NF-κB signaling pathways (136). The results showed with the anti-inflammatory effect of CLA had a strong potential to alleviate AD.
Aryl hydrocarbon receptor (AHR) has been reported to be closely associated with the development of AD (137). Indole-3-aldehyde (IAld), a metabolite of tryptophan, was lower in AD lesional skin than that of healthy controls and significantly alleviated skin inflammation via activating AHR (138). Coal tar is usually used to improve the clinical symptoms of AD. In the skin models with primary keratinocytes, it activated AHR to induce epidermal differentiation and interfered with Th2 cytokine signaling (139). These results suggest the activation of AHR plays an important role in the treatment of AD. Indole derivatives including IAld, tryptamine, indole acetic acid, indole-3-acetaldehyde, indole acrylic acid, and indole-3-propionic acid, are metabolites from tryptophan metabolism of gut microbiota and have been demonstrated as the ligands to activate AHR (140). In a study involving the gut-brain axis, gut microbial metabolites of tryptophan affected the activation of microglia and modulated the central nervous system inflammation via a mechanism mediated by AHR (141). Additionally, the metabolites of tryptophan such as indoxyl sulfate and indole-3-propionic acid have been found in blood circulation (142). This suggests that gut microbiota-produced ligands of AHR have the potential to regulate systemic inflammation, including skin inflammation. In a meta-analysis, probiotics consumption significantly regulated the ratio of kynurenine: tryptophan and mediated the tryptophan metabolism (143). Therefore, these studies imply that probiotics regulate tryptophan metabolism in the intestine, and the metabolites as AHR ligands may mediate skin inflammation via AHR signaling.
There are some limitations about the effects of probiotics on the alleviation of AD in this review. The effectiveness of probiotics on the improvement of clinical symptoms of AD needs the larger scale and more rigorous clinical trials to demonstrate in different groups of patients stratified by age, sex, and concurrent diseases. The interactions between probiotics and gut microbiota are complex and lead to difficulties in revealing the precise alleviating mechanisms on AD. Furthermore, the immunomodulation of probiotics is strain-specific and they may activate different signaling pathways to improve the clinical manifestations of AD. The substance basis of probiotics to alleviate AD is still to be elucidated whether it is from the component of probiotic itself and the metabolites from probiotic or gut microbiota.
In summary, although the cross-talk mechanism between gut microbiota and skin needs to be explored, the gut microbiota is closely associated with dermatology and may serve as a target for the prevention and treatment of AD. Probiotics supplementation alters the intestinal environment, including modulating gut microbial composition, preventing pathogens colonization, affecting bacterial metabolism, and restoring immune balance. These alterations may contribute to the decrease in inflammation and improvement of clinical manifestation in AD. Although the effects of probiotics on AD have been investigated in numerous clinical trials, the effective substance basis of probiotics to alleviate AD remains unclear. To precisely manipulate gut microbiota to attenuate clinical manifestation of AD, the mechanism of interactions between probiotics, gut microbiota, and skin needs to be elucidated. Combination with metatranscriptomics, metagenomics and metabolomics, effects of probiotics on functional gene alteration, specific gut microbe, metabolic pathway, and specific metabolite could be revealed in the future and overall analyze the alleviating mechanism of probiotics targeting the gut microbiota. Future results of probiotic clinical trials on AD may support a microbiome replacement strategy.
ZF: writing-original draft. LL: editing. HZ, JZ, and WC: writing-review and funding acquisition. WL: wring-review and editing, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (No. 31820103010), the national first-class discipline program of Food Science and Technology (No. JUFSTR20180102), the Fundamental Research Funds for the Central Universities (No. JUSRP51903B), the Postdoctoral Research Funding Scheme of Jiangsu Province (No. 2021K018A).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Thanks for the help of the Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
1. Hidaka T, Ogawa E, Kobayashi EH, Suzuki T, Funayama R, Nagashima T, et al. The Aryl Hydrocarbon Receptor Ahr Links Atopic Dermatitis and Air Pollution Via Induction of the Neurotrophic Factor Artemin. Nat Immunol (2017) 18(1):64–73. doi: 10.1038/ni.3614
2. Langan SM, Irvine AD, Weidinger S. Atopic Dermatitis. Lancet (2020) 396(10247):345–60. doi: 10.1016/s0140-6736(20)31286-1
3. Ricci G, Patrizi A, Baldi E, Menna G, Tabanelli M, Masi M. Long-Term Follow-Up of Atopic Dermatitis: Retrospective Analysis of Related Risk Factors and Association With Concomitant Allergic Diseases. J Am Acad Dermatol (2006) 55(5):765–71. doi: 10.1016/j.jaad.2006.04.064
4. Paus R, Theoharides TC, Arck PC. Neuroimmunoendocrine Circuitry of the ‘Brain-Skin Connection’. Trends Immunol (2006) 27(1):32–9. doi: 10.1016/j.it.2005.10.002
5. Drucker AM, Wang AR, Li WQ, Sevetson E, Block JK, Qureshi AA. The Burden of Atopic Dermatitis: Summary of A Report for the National Eczema Association. J Invest Dermatol (2017) 137(1):26–30. doi: 10.1016/j.jid.2016.07.012
6. Cookson WO, Ubhi B, Lawrence R, Abecasis GR, Walley AJ, Cox HE, et al. Genetic Linkage of Childhood Atopic Dermatitis to Psoriasis Susceptibility Loci. Nat Genet (2001) 27(4):372–3. doi: 10.1038/86867
7. Jedrychowski W, Perera F, Maugeri U, Mrozek-Budzyn D, Miller RL, Flak E, et al. Effects of Prenatal and Perinatal Exposure to Fine Air Pollutants and Maternal Fish Consumption on the Occurrence of Infantile Eczema. Int Arch Allergy Immunol (2011) 155(3):275–81. doi: 10.1159/000320376
8. Goon A, Leow YH, Chan YH, Ng SK, Goh CL. Atopy Patch Testing With Aeroallergens in Patients With Atopic Dermatitis and Controls in Singapore. Clin Exp Dermatol (2005) 30(6):627–31. doi: 10.1111/j.1365-2230.2005.01916.x
9. Reginald K, Westritschnig K, Linhart B, Focke-Tejkl M, Jahn-Schmid B, Eckl-Dorna J, et al. Staphylococcus Aureus Fibronectin-Binding Protein Specifically Binds Ige From Patients With Atopic Dermatitis and Requires Antigen Presentation for Cellular Immune Responses. J Allergy Clin Immunol (2011) 128(1):82–91.e8. doi: 10.1016/j.jaci.2011.02.034
10. Reginald K, Westritschnig K, Werfel T, Heratizadeh A, Novak N, Focke-Tejkl M, et al. Immunoglobulin E Antibody Reactivity to Bacterial Antigens in Atopic Dermatitis Patients. Clin Exp Allergy (2011) 41(3):357–69. doi: 10.1111/j.1365-2222.2010.03655.x
11. Kwon MS, Lim SK, Jang JY, Lee J, Park HK, Kim N, et al. Lactobacillus Sakei WIKIM30 Ameliorates Atopic Dermatitis-Like Skin Lesions by Inducing Regulatory T Cells and Altering Gut Microbiota Structure in Mice. Front Immunol (2018) 9:1905. doi: 10.3389/fimmu.2018.01905
12. Gandhi NA, Bennett BL, Graham NM, Pirozzi G, Stahl N, Yancopoulos GD. Targeting Key Proximal Drivers of Type 2 Inflammation in Disease. Nat Rev Drug Discovery (2016) 15(1):35–50. doi: 10.1038/nrd4624
13. Werfel T, Allam JP, Biedermann T, Eyerich K, Gilles S, Guttman-Yassky E, et al. Cellular and Molecular Immunologic Mechanisms in Patients With Atopic Dermatitis. J Allergy Clin Immunol (2016) 138(2):336–49. doi: 10.1016/j.jaci.2016.06.010
14. Datsi A, Steinhoff M, Ahmad F, Alam M, Buddenkotte J. Interleukin-31: The “Itchy” Cytokine in Inflammation and Therapy. Allergy (2021) 00:1–16. doi: 10.1111/all.14791
15. Kabashima K, Matsumura T, Komazaki H, Kawashima M. Trial of Nemolizumab and Topical Agents for Atopic Dermatitis With Pruritus. N Engl J Med (2020) 383(2):141–50. doi: 10.1056/NEJMoa1917006
16. O’Hara AM, Shanahan F. The Gut Flora as a Forgotten Organ. EMBO Rep (2006) 7(7):688–93. doi: 10.1038/sj.embor.7400731
17. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging Pathogenic Links Between Microbiota and the Gut-Lung Axis. Nat Rev Microbiol (2017) 15(1):55–63. doi: 10.1038/nrmicro.2016.142
18. Salem I, Ramser A, Isham N, Ghannoum MA. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front Microbiol (2018) 9:1459. doi: 10.3389/fmicb.2018.01459
19. Stefanovic N, Flohr C, Irvine AD. The Exposome in Atopic Dermatitis. Allergy (2020) 75(1):63–74. doi: 10.1111/all.13946
20. O’Neill CA, Monteleone G, McLaughlin JT, Paus R. The Gut-Skin Axis in Health and Disease: A Paradigm With Therapeutic Implications. Bioessays (2016) 38(11):1167–76. doi: 10.1002/bies.201600008
21. Saarialho-Kere U. The Gut-Skin Axis. J Pediatr Gastroenterol Nutr (2004) 39(Suppl 3):S734–5. doi: 10.1097/00005176-200406003-00009
22. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the Gut Microbiome and Risk of Asthma in Childhood. Nat Commun (2018) 9(1):141. doi: 10.1038/s41467-017-02573-2
23. Fujimura KE, Lynch SV. Microbiota in Allergy and Asthma and the Emerging Relationship With the Gut Microbiome. Cell Host Microbe (2015) 17(5):592–602. doi: 10.1016/j.chom.2015.04.007
24. Kim JE, Kim HS. Microbiome of the Skin and Gut in Atopic Dermatitis (AD): Understanding the Pathophysiology and Finding Novel Management Strategies. J Clin Med (2019) 8(4):444. doi: 10.3390/jcm8040444
25. Lopez-Santamarina A, Gonzalez EG, Lamas A, Mondragon ADC, Regal P, Miranda JM. Probiotics as a Possible Strategy for the Prevention and Treatment of Allergies. A Narrative Review. Foods (2021) 10(4):701. doi: 10.3390/foods10040701
26. Albenberg L, Kelsen J. Advances in Gut Microbiome Research and Relevance to Pediatric Diseases. J Pediatr (2016) 178:16–23. doi: 10.1016/j.jpeds.2016.08.044
27. Guarner F, Malagelada JR. Gut Flora in Health and Disease. Lancet (2003) 361(9356):512–9. doi: 10.1016/s0140-6736(03)12489-0
28. Nuriel-Ohayon M, Neuman H, Koren O. Microbial Changes During Pregnancy, Birth, and Infancy. Front Microbiol (2016) 7:1031. doi: 10.3389/fmicb.2016.01031
29. Lindberg M, Söderquist B. Atopic Dermatitis and Gut Microbiota. Br J Dermatol (2017) 176(2):297–98. doi: 10.1111/bjd.15276
30. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota Across Multiple Body Habitats in Newborns. Proc Natl Acad Sci USA (2010) 107(26):11971–5. doi: 10.1073/pnas.1002601107
31. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome During the First Year of Life. Cell Host Microbe (2015) 17(6):690–703. doi: 10.1016/j.chom.2015.04.004
32. Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, et al. An Infant-Associated Bacterial Commensal Utilizes Breast Milk Sialyloligosaccharides. J Biol Chem (2011) 286(14):11909–18. doi: 10.1074/jbc.M110.193359
33. Balmer SE, Wharton BA. Diet and Faecal Flora in the Newborn: Breast Milk and Infant Formula. Arch Dis Child (1989) 64(12):1672–7. doi: 10.1136/adc.64.12.1672
34. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota Profile in Feces of Breast- and Formula-Fed Newborns by Using Fluorescence in Situ Hybridization (Fish). Anaerobe (2011) 17(6):478–82. doi: 10.1016/j.anaerobe.2011.03.009
35. Kedia S, Rampal R, Paul J, Ahuja V. Gut Microbiome Diversity in Acute Infective and Chronic Inflammatory Gastrointestinal Diseases in North India. J Gastroenterol (2016) 51(7):660–71. doi: 10.1007/s00535-016-1193-1
36. Parthasarathy G, Chen J, Chen X, Chia N, O’Connor HM, Wolf PG, et al. Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation. Gastroenterology (2016) 150(2):367–79.e1. doi: 10.1053/j.gastro.2015.10.005
37. Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut Microbiome and Serum Metabolome Alterations in Obesity and After Weight-Loss Intervention. Nat Med (2017) 23(7):859–68. doi: 10.1038/nm.4358
38. Stower H. Depression Linked to the Microbiome. Nat Med (2019) 25(3):358. doi: 10.1038/s41591-019-0396-4
39. Björkstén B, Naaber P, Sepp E, Mikelsaar M. The Intestinal Microflora in Allergic Estonian and Swedish 2-Year-Old Children. Clin Exp Allergy (1999) 29(3):342–6. doi: 10.1046/j.1365-2222.1999.00560.x
40. Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct Patterns of Neonatal Gut Microflora in Infants in Whom Atopy Was and Was Not Developing. J Allergy Clin Immunol (2001) 107(1):129–34. doi: 10.1067/mai.2001.111237
41. Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy Development and the Intestinal Microflora During the First Year of Life. J Allergy Clin Immunol (2001) 108(4):516–20. doi: 10.1067/mai.2001.118130
42. Watanabe S, Narisawa Y, Arase S, Okamatsu H, Ikenaga T, Tajiri Y, et al. Differences in Fecal Microflora Between Patients With Atopic Dermatitis and Healthy Control Subjects. J Allergy Clin Immunol (2003) 111(3):587–91. doi: 10.1067/mai.2003.105
43. Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut Microbiota Composition and Development of Atopic Manifestations in Infancy: The KOALA Birth Cohort Study. Gut (2007) 56(5):661–7. doi: 10.1136/gut.2006.100164
44. Gore C, Munro K, Lay C, Bibiloni R, Morris J, Woodcock A, et al. Bifidobacterium Pseudocatenulatum Is Associated With Atopic Eczema: A Nested Case-Control Study Investigating the Fecal Microbiota of Infants. J Allergy Clin Immunol (2008) 121(1):135–40. doi: 10.1016/j.jaci.2007.07.061
45. Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced Diversity in the Early Fecal Microbiota of Infants With Atopic Eczema. J Allergy Clin Immunol (2008) 121(1):129–34. doi: 10.1016/j.jaci.2007.09.011
46. Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered Early Infant Gut Microbiota in Children Developing Allergy Up to 5 Years of Age. Clin Exp Allergy (2009) 39(4):518–26. doi: 10.1111/j.1365-2222.2008.03156.x
47. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low Diversity of the Gut Microbiota in Infants With Atopic Eczema. J Allergy Clin Immunol (2012) 129(2):434–40, 40.e1-2. doi: 10.1016/j.jaci.2011.10.025
48. Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Licciardi PV, Robins-Browne RM, et al. Reduced Gut Microbial Diversity in Early Life Is Associated With Later Development of Eczema But Not Atopy in High-Risk Infants. Pediatr Allergy Immunol (2012) 23(7):674–81. doi: 10.1111/j.1399-3038.2012.01328.x
49. Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium Prausnitzii Subspecies-Level Dysbiosis in the Human Gut Microbiome Underlying Atopic Dermatitis. J Allergy Clin Immunol (2016) 137(3):852–60. doi: 10.1016/j.jaci.2015.08.021
50. Lee MJ, Kang MJ, Lee SY, Lee E, Kim K, Won S, et al. Perturbations of Gut Microbiome Genes in Infants With Atopic Dermatitis According to Feeding Type. J Allergy Clin Immunol (2018) 141(4):1310–19. doi: 10.1016/j.jaci.2017.11.045
51. Melli L, Carmo-Rodrigues MSD, Araújo-Filho HB, Mello CS, Tahan S, Pignatari ACC, et al. Gut Microbiota of Children With Atopic Dermatitis: Controlled Study in the Metropolitan Region of São Paulo, Brazil. Allergol Immunopathol (Madr) (2020) 48(2):107–15. doi: 10.1016/j.aller.2019.08.004
52. Ye S, Yan F, Wang H, Mo X, Liu J, Zhang Y, et al. Diversity Analysis of Gut Microbiota Between Healthy Controls and Those With Atopic Dermatitis in A Chinese Population. J Dermatol (2021) 48(2):158–67. doi: 10.1111/1346-8138.15530
53. Su YJ, Luo SD, Hsu CY, Kuo HC. Differences in Gut Microbiota Between Allergic Rhinitis, Atopic Dermatitis, and Skin Urticaria: A Pilot Study. Med (Baltimore) (2021) 100(9):e25091. doi: 10.1097/md.0000000000025091
54. Zachariassen LF, Krych L, Engkilde K, Nielsen DS, Kot W, Hansen CH, et al. Sensitivity to Oxazolone Induced Dermatitis is Transferable With Gut Microbiota in Mice. Sci Rep (2017) 7:44385. doi: 10.1038/srep44385
55. Hu C, van Meel ER, Medina-Gomez C, Kraaij R, Barroso M, Kiefte-de Jong J, et al. A Population-Based Study on Associations of Stool Microbiota With Atopic Diseases in School-Age Children. J Allergy Clin Immunol (2021) S0091-6749(21):00563–7. doi: 10.1016/j.jaci.2021.04.001
56. Hevia A, Milani C, López P, Donado CD, Cuervo A, González S, et al. Allergic Patients With Long-Term Asthma Display Low Levels of Bifidobacterium Adolescentis. PloS One (2016) 11(2):e0147809. doi: 10.1371/journal.pone.0147809
57. Waligora-Dupriet AJ, Campeotto F, Romero K, Mangin I, Rouzaud G, Ménard O, et al. Diversity of Gut Bifidobacterium Species is Not Altered Between Allergic and Non-Allergic French Infants. Anaerobe (2011) 17(3):91–6. doi: 10.1016/j.anaerobe.2011.04.003
58. Suzuki S, Shimojo N, Tajiri Y, Kumemura M, Kohno Y. Differences in the Composition of Intestinal Bifidobacterium Species and the Development of Allergic Diseases in Infants in Rural Japan. Clin Exp Allergy (2007) 37(4):506–11. doi: 10.1111/j.1365-2222.2007.02676.x
59. Sbihi H, Boutin RC, Cutler C, Suen M, Finlay BB, Turvey SE. Thinking Bigger: How Early-Life Environmental Exposures Shape the Gut Microbiome and Influence the Development of Asthma and Allergic Disease. Allergy (2019) 74(11):2103–15. doi: 10.1111/all.13812
60. Noverr MC, Huffnagle GB. The ‘Microflora Hypothesis’ of Allergic Diseases. Clin Exp Allergy (2005) 35(12):1511–20. doi: 10.1111/j.1365-2222.2005.02379.x
61. Johnson CC, Ownby DR. The Infant Gut Bacterial Microbiota and Risk of Pediatric Asthma and Allergic Diseases. Transl Res (2017) 179:60–70. doi: 10.1016/j.trsl.2016.06.010
62. Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity (2020) 52(2):241–55. doi: 10.1016/j.immuni.2020.01.007
63. Lee SY, Lee E, Park YM, Hong SJ. Microbiome in the Gut-Skin Axis in Atopic Dermatitis. Allergy Asthma Immunol Res (2018) 10(4):354–62. doi: 10.4168/aair.2018.10.4.354
64. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The Maternal Microbiota Drives Early Postnatal Innate Immune Development. Science (2016) 351(6279):1296–302. doi: 10.1126/science.aad2571
65. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg Induction by A Rationally Selected Mixture of Clostridia Strains From the Human Microbiota. Nature (2013) 500(7461):232–6. doi: 10.1038/nature12331
66. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis Via T Helper 17 Cells. Immunity (2010) 32(6):815–27. doi: 10.1016/j.immuni.2010.06.001
67. Roslund MI, Puhakka R, Grönroos M, Nurminen N, Oikarinen S, Gazali AM, et al. Biodiversity Intervention Enhances Immune Regulation and Health-Associated Commensal Microbiota Among Daycare Children. Sci Adv (2020) 6(42):eaba2578. doi: 10.1126/sciadv.aba2578
68. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WH, Saldana-Morales FB, et al. Thymic Development of Gut-Microbiota-Specific T Cells. Nature (2021) 594(863):413–17. doi: 10.1038/s41586-021-03531-1
69. Zhu TH, Zhu TR, Tran KA, Sivamani RK, Shi VY. Epithelial Barrier Dysfunctions In Atopic Dermatitis: A Skin-Gut-Lung Model Linking Microbiome Alteration and Immune Dysregulation. Br J Dermatol (2018) 179(3):570–81. doi: 10.1111/bjd.16734
70. Cabanillas B, Novak N. Atopic Dermatitis and Filaggrin. Curr Opin Immunol (2016) 42:1–8. doi: 10.1016/j.coi.2016.05.002
71. Xiao C, Sun Z, Gao J, Bai Y, Zhang C, Pang B, et al. Enhanced Phenotype of Calcipotriol-Induced Atopic Dermatitis in Filaggrin-Deficient Mice. FASEB J (2021) 35(5):e21574. doi: 10.1096/fj.202002709R
72. Smieszek SP, Welsh S, Xiao C, Wang J, Polymeropoulos C, Birznieks G, et al. Correlation of Age-Of-Onset of Atopic Dermatitis With Filaggrin Loss-Of-Function Variant Status. Sci Rep (2020) 10(1):2721. doi: 10.1038/s41598-020-59627-7
73. Kim IS, Lee SH, Kwon YM, Adhikari B, Kim JA, Yu DY, et al. Oral Administration of β-Glucan and Lactobacillus Plantarum Alleviates Atopic Dermatitis-Like Symptoms. J Microbiol Biotechnol (2019) 29(11):1693–706. doi: 10.4014/jmb.1907.07011
74. Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, et al. S1P-Dependent Interorgan Trafficking of Group 2 Innate Lymphoid Cells Supports Host Defense. Science (2018) 359(6371):114–19. doi: 10.1126/science.aam5809
75. Rigoni R, Fontana E, Dobbs K, Marrella V, Taverniti V, Maina V, et al. Cutaneous Barrier Leakage and Gut Inflammation Drive Skin Disease in Omenn Syndrome. J Allergy Clin Immunol (2020) 146(5):1165–79.e11. doi: 10.1016/j.jaci.2020.04.005
76. McGirt LY, Beck LA. Innate Immune Defects in Atopic Dermatitis. J Allergy Clin Immunol (2006) 118(1):202–8. doi: 10.1016/j.jaci.2006.04.033
77. Blasius AL, Beutler B. Intracellular Toll-Like Receptors. Immunity (2010) 32(3):305–15. doi: 10.1016/j.immuni.2010.03.012
78. Kawai T, Akira S. TLR Signaling. Cell Death Differ (2006) 13(5):816–25. doi: 10.1038/sj.cdd.4401850
79. Ermertcan AT, Öztürk F, Gündüz K. Toll-Like Receptors and Skin. J Eur Acad Dermatol Venereol (2011) 25(9):997–1006. doi: 10.1111/j.1468-3083.2011.04049.x
80. Iwamoto K, Nümm TJ, Koch S, Herrmann N, Leib N, Bieber T. Langerhans and Inflammatory Dendritic Epidermal Cells in Atopic Dermatitis Are Tolerized Toward TLR2 Activation. Allergy (2018) 73(11):2205–13. doi: 10.1111/all.13460
81. Niebuhr M, Lutat C, Sigel S, Werfel T. Impaired TLR-2 Expression and TLR-2-Mediated Cytokine Secretion in Macrophages From Patients With Atopic Dermatitis. Allergy (2009) 64(11):1580–7. doi: 10.1111/j.1398-9995.2009.02050.x
82. Kuo IH, Carpenter-Mendini A, Yoshida T, McGirt LY, Ivanov AI, Barnes KC, et al. Activation of Epidermal Toll-Like Receptor 2 Enhances Tight Junction Function: Implications for Atopic Dermatitis and Skin Barrier Repair. J Invest Dermatol (2013) 133(4):988–98. doi: 10.1038/jid.2012.437
83. Zhang Y, Wang HC, Feng C, Yan M. Analysis of the Association of Polymorphisms Rs5743708 in TLR2 and Rs4986790 in TLR4 With Atopic Dermatitis Risk. Immunol Invest (2019) 48(2):169–80. doi: 10.1080/08820139.2018.1508228
84. Belderbos ME, Knol EF, Houben ML, van Bleek GM, Wilbrink B, Kimpen JL, et al. Low Neonatal Toll-Like Receptor 4-Mediated Interleukin-10 Production Is Associated With Subsequent Atopic Dermatitis. Clin Exp Allergy (2012) 42(1):66–75. doi: 10.1111/j.1365-2222.2011.03857.x
85. Hüls A, Klümper C, MacIntyre EA, Brauer M, Melén E, Bauer M, et al. Atopic Dermatitis: Interaction Between Genetic Variants of GSTP1, TNF, TLR2, and TLR4 and Air Pollution in Early Life. Pediatr Allergy Immunol (2018) 29(6):596–605. doi: 10.1111/pai.12903
86. Lin L, Xie M, Chen X, Yu Y, Liu Y, Lei K, et al. Toll-Like Receptor 4 Attenuates A Murine Model of Atopic Dermatitis Through Inhibition of Langerin-Positive Dcs Migration. Exp Dermatol (2018) 27(9):1015–22. doi: 10.1111/exd.13698
87. Yoon J, Leyva-Castillo JM, Wang G, Galand C, Oyoshi MK, Kumar L, et al. Il-23 Induced in Keratinocytes by Endogenous TLR4 Ligands Polarizes Dendritic Cells to Drive Il-22 Responses to Skin Immunization. J Exp Med (2016) 213(10):2147–66. doi: 10.1084/jem.20150376
88. Liu T, Berta T, Xu ZZ, Park CK, Zhang L, Lu N, et al. TLR3 Deficiency Impairs Spinal Cord Synaptic Transmission, Central Sensitization, and Pruritus in Mice. J Clin Invest (2012) 122(6):2195–207. doi: 10.1172/jci45414
89. Jang YH, Choi JK, Jin M, Choi YA, Ryoo ZY, Lee HS, et al. House Dust Mite Increases Pro-Th2 Cytokines Il-25 and IL-33 Via the Activation of TLR1/6 Signaling. J Invest Dermatol (2017) 137(11):2354–61. doi: 10.1016/j.jid.2017.03.042
90. Roduit C, Wohlgensinger J, Frei R, Bitter S, Bieli C, Loeliger S, et al. Prenatal Animal Contact and Gene Expression of Innate Immunity Receptors at Birth are Associated With Atopic Dermatitis. J Allergy Clin Immunol (2011) 127(1):179–85,185.e1. doi: 10.1016/j.jaci.2010.10.010
91. Yiu JH, Dorweiler B, Woo CW. Interaction Between Gut Microbiota and Toll-Like Receptor: From Immunity to Metabolism. J Mol Med (Berl) (2017) 95(1):13–20. doi: 10.1007/s00109-016-1474-4
92. Penders J, Thijs C, Mommers M, Stobberingh EE, Dompeling E, Reijmerink NE, et al. Host-Microbial Interactions in Childhood Atopy: Toll-Like Receptor 4 (TLR4), CD14, and Fecal Escherichia Coli. J Allergy Clin Immunol (2010) 125(1):231–6.e1-5. doi: 10.1016/j.jaci.2009.10.011
93. West CE, Rydén P, Lundin D, Engstrand L, Tulic MK, Prescott SL. Gut Microbiome and Innate Immune Response Patterns in IgE-Associated Eczema. Clin Exp Allergy (2015) 45(9):1419–29. doi: 10.1111/cea.12566
94. Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-Like Receptor 4 Signaling by Intestinal Microbes Influences Susceptibility to Food Allergy. J Immunol (2004) 172(11):6978–87. doi: 10.4049/jimmunol.172.11.6978
95. Enomoto T, Sowa M, Nishimori K, Shimazu S, Yoshida A, Yamada K, et al. Effects of Bifidobacterial Supplementation to Pregnant Women and Infants in the Prevention of Allergy Development in Infants and on Fecal Microbiota. Allergol Int (2014) 63(4):575–85. doi: 10.2332/allergolint.13-OA-0683
96. Rø ADB, Simpson MR, Rø TB, Storrø O, Johnsen R, Videm V, et al. Reduced Th22 Cell Proportion and Prevention of Atopic Dermatitis in Infants Following Maternal Probiotic Supplementation. Clin Exp Allergy (2017) 47(8):1014–21. doi: 10.1111/cea.12930
97. Kopp MV, Hennemuth I, Heinzmann A, Urbanek R. Randomized, Double-Blind, Placebo-Controlled Trial of Probiotics for Primary Prevention: No Clinical Effects of Lactobacillus Gg Supplementation. Pediatrics (2008) 121(4):e850–6. doi: 10.1542/peds.2007-1492
98. Dotterud CK, Storrø O, Johnsen R, Oien T. Probiotics in Pregnant Women to Prevent Allergic Disease: A Randomized, Double-Blind Trial. Br J Dermatol (2010) 163(3):616–23. doi: 10.1111/j.1365-2133.2010.09889.x
99. Plummer EL, Chebar Lozinsky A, Tobin JM, Uebergang JB, Axelrad C, Garland SM, et al. Postnatal Probiotics and Allergic Disease in Very Preterm Infants: Sub-Study to the Proprems Randomized Trial. Allergy (2020) 75(1):127–36. doi: 10.1111/all.14088
100. Wickens K, Black P, Stanley TV, Mitchell E, Barthow C, Fitzharris P, et al. A Protective Effect of Lactobacillus Rhamnosus HN001 Against Eczema in the First 2 Years of Life Persists to Age 4 Years. Clin Exp Allergy (2012) 42(7):1071–9. doi: 10.1111/j.1365-2222.2012.03975.x
101. van der Aa LB, Lutter R, Heymans HS, Smids BS, Dekker T, van Aalderen WM, et al. No Detectable Beneficial Systemic Immunomodulatory Effects of A Specific Synbiotic Mixture in Infants With Atopic Dermatitis. Clin Exp Allergy (2012) 42(4):531–9. doi: 10.1111/j.1365-2222.2011.03890.x
102. Wu YJ, Wu WF, Hung CW, Ku MS, Liao PF, Sun HL, et al. Evaluation of Efficacy and Safety of Lactobacillus Rhamnosus in Children Aged 4-48 Months With Atopic Dermatitis: An 8-Week, Double-Blind, Randomized, Placebo-Controlled Study. J Microbiol Immunol Infect (2017) 50(5):684–92. doi: 10.1016/j.jmii.2015.10.003
103. Gerasimov SV, Vasjuta VV, Myhovych OO, Bondarchuk LI. Probiotic Supplement Reduces Atopic Dermatitis in Preschool Children: A Randomized, Double-Blind, Placebo-Controlled, Clinical Trial. Am J Clin Dermatol (2010) 11(5):351–61. doi: 10.2165/11531420-000000000-00000
104. Han Y, Kim B, Ban J, Lee J, Kim BJ, Choi BS, et al. A Randomized Trial of Lactobacillus Plantarum CJLP133 for the Treatment of Atopic Dermatitis. Pediatr Allergy Immunol (2012) 23(7):667–73. doi: 10.1111/pai.12010
105. Wang IJ, Wang JY. Children With Atopic Dermatitis Show Clinical Improvement After Lactobacillus Exposure. Clin Exp Allergy (2015) 45(4):779–87. doi: 10.1111/cea.12489
106. Ahn SH, Yoon W, Lee SY, Shin HS, Lim MY, Nam YD, et al. Effects of Lactobacillus Pentosus in Children With Allergen-Sensitized Atopic Dermatitis. J Korean Med Sci (2020) 35(18):e128. doi: 10.3346/jkms.2020.35.e128
107. Navarro-López V, Ramírez-Boscá A, Ramón-Vidal D, Ruzafa-Costas B, Genovés-Martínez S, Chenoll-Cuadros E, et al. Effect of Oral Administration of a Mixture of Probiotic Strains on SCORAD Index and Use of Topical Steroids in Young Patients With Moderate Atopic Dermatitis: A Randomized Clinical Trial. JAMA Dermatol (2018) 154(1):37–43. doi: 10.1001/jamadermatol.2017.3647
108. Matsumoto M, Ebata T, Hirooka J, Hosoya R, Inoue N, Itami S, et al. Antipruritic Effects of the Probiotic Strain LKM512 in Adults With Atopic Dermatitis. Ann Allergy Asthma Immunol (2014) 113(2):209–16.e7. doi: 10.1016/j.anai.2014.05.002
109. Moroi M, Uchi S, Nakamura K, Sato S, Shimizu N, Fujii M, et al. Beneficial Effect of A Diet Containing Heat-Killed Lactobacillus Paracasei K71 on Adult Type Atopic Dermatitis. J Dermatol (2011) 38(2):131–9. doi: 10.1111/j.1346-8138.2010.00939.x
110. Chairatana P, Nolan EM. Defensins, Lectins, Mucins, and Secretory Immunoglobulin A: Microbe-Binding Biomolecules That Contribute to Mucosal Immunity in the Human Gut. Crit Rev Biochem Mol Biol (2017) 52(1):45–56. doi: 10.1080/10409238.2016.1243654
111. Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, Prebiotics and Immunomodulation of Gut Mucosal Defences: Homeostasis and Immunopathology. Nutrients (2013) 5(6):1869–912. doi: 10.3390/nu5061869
112. Solano-Aguilar G, Shea-Donohue T, Madden KB, Quinones A, Beshah E, Lakshman S, et al. Bifidobacterium Animalis Subspecies Lactis Modulates the Local Immune Response and Glucose Uptake in the Small Intestine of Juvenile Pigs Infected With the Parasitic Nematode Ascaris Suum. Gut Microbes (2018) 9(5):422–36. doi: 10.1080/19490976.2018.1460014
113. Fu G, Zhao K, Chen H, Wang Y, Nie L, Wei H, et al. Effect of 3 Lactobacilli on Immunoregulation and Intestinal Microbiota in A B;-Lactoglobulin-Induced Allergic Mouse Model. J Dairy Sci (2019) 102(3):1943–58. doi: 10.3168/jds.2018-15683
114. Kim WK, Jang YJ, Han DH, Jeon K, Lee C, Han HS, et al. Lactobacillus Paracasei KBL382 Administration Attenuates Atopic Dermatitis by Modulating Immune Response and Gut Microbiota. Gut Microbes (2020) 12(1):1–14. doi: 10.1080/19490976.2020.1819156
115. Kandikattu HK, Upparahalli Venkateshaiah S, Mishra A. Synergy of Interleukin (IL)-5 and IL-18 in Eosinophil Mediated Pathogenesis of Allergic Diseases. Cytokine Growth Factor Rev (2019) 47:83–98. doi: 10.1016/j.cytogfr.2019.05.003
116. Choi WJ, Konkit M, Kim Y, Kim MK, Kim W. Oral Administration of Lactococcus Chungangensis Inhibits 2,4-Dinitrochlorobenzene-Induced Atopic-Like Dermatitis in NC/Nga Mice. J Dairy Sci (2016) 99(9):6889–901. doi: 10.3168/jds.2016-11301
117. Bieber T. Interleukin-13: Targeting an Underestimated Cytokine in Atopic Dermatitis. Allergy (2020) 75(1):54–62. doi: 10.1111/all.13954
118. Wollenberg A, Blauvelt A, Guttman-Yassky E, Worm M, Lynde C, Lacour JP, et al. Tralokinumab for Moderate-To-Severe Atopic Dermatitis: Results From Two 52-Week, Randomized, Double-Blind, Multicentre, Placebo-Controlled Phase Iii Trials (ECZTRA 1 And ECZTRA 2). Br J Dermatol (2021) 184(3):437–49. doi: 10.1111/bjd.19574
119. Jeong DY, Ryu MS, Yang HJ, Jeong SY, Zhang T, Yang HJ, et al. Pediococcus Acidilactici Intake Decreases the Clinical Severity of Atopic Dermatitis Along With Increasing Mucin Production and Improving the Gut Microbiome in NC/Nga Mice. BioMed Pharmacother (2020) 129:110488. doi: 10.1016/j.biopha.2020.110488
120. Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, et al. Human Epithelial Cells Trigger Dendritic Cell Mediated Allergic Inflammation by Producing Tslp. Nat Immunol (2002) 3(7):673–80. doi: 10.1038/ni805
121. Indra AK. Epidermal TSLP: A Trigger Factor for Pathogenesis of Atopic Dermatitis. Expert Rev Proteomics (2013) 10(4):309–11. doi: 10.1586/14789450.2013.814881
122. Yoo J, Omori M, Gyarmati D, Zhou B, Aye T, Brewer A, et al. Spontaneous Atopic Dermatitis in Mice Expressing An Inducible Thymic Stromal Lymphopoietin Transgene Specifically in the Skin. J Exp Med (2005) 202(4):541–9. doi: 10.1084/jem.20041503
123. Simpson EL, Parnes JR, She D, Crouch S, Rees W, Mo M, et al. Tezepelumab, An Anti-Thymic Stromal Lymphopoietin Monoclonal Antibody, in the Treatment of Moderate to Severe Atopic Dermatitis: A Randomized Phase 2a Clinical Trial. J Am Acad Dermatol (2019) 80(4):1013–21. doi: 10.1016/j.jaad.2018.11.059
124. Kim HJ, Kim YJ, Kang MJ, Seo JH, Kim HY, Jeong SK, et al. A Novel Mouse Model of Atopic Dermatitis With Epicutaneous Allergen Sensitization and the Effect of. Lactobacillus Rhamnosus Exp Dermatol (2012) 21(9):672–5. doi: 10.1111/j.1600-0625.2012.01539.x
125. Ménard S, Laharie D, Asensio C, Vidal-Martinez T, Candalh C, Rullier A, et al. Bifidobacterium Breve and Streptococcus Thermophilus Secretion Products Enhance T Helper 1 Immune Response and Intestinal Barrier in Mice. Exp Biol Med (Maywood) (2005) 230(10):749–56. doi: 10.1177/153537020523001008
126. Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin Production by Bifidobacterium Spp. A Review. Biotechnol Adv (2013) 31(4):482–8. doi: 10.1016/j.biotechadv.2013.01.010
127. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
128. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of Atopic Disease Inversely Correlates With Intestinal Microbiota Diversity and Butyrate-Producing Bacteria. Allergy (2015) 70(2):241–4. doi: 10.1111/all.12549
129. Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High Levels of Butyrate and Propionate in Early Life Are Associated With Protection Against Atopy. Allergy (2019) 74(4):799–809. doi: 10.1111/all.13660
130. Kim HJ, Lee SH, Hong SJ. Antibiotics-Induced Dysbiosis of Intestinal Microbiota Aggravates Atopic Dermatitis in Mice by Altered Short-Chain Fatty Acids. Allergy Asthma Immunol Res (2020) 12(1):137–48. doi: 10.4168/aair.2020.12.1.137
131. Kepert I, Fonseca J, Muller C, Milger K, Hochwind K, Kostric M, et al. D-Tryptophan From Probiotic Bacteria Influences the Gut Microbiome and Allergic Airway Disease. J Allergy Clin Immunol (2017) 139(5):1525–35. doi: 10.1016/j.jaci.2016.09.003
132. Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J, et al. Polyunsaturated Fatty Acid Saturation by Gut Lactic Acid Bacteria Affecting Host Lipid Composition. Proc Natl Acad Sci USA (2013) 110(44):17808–13. doi: 10.1073/pnas.1312937110
133. Chen Y, Yang B, Stanton C, Ross RP, Zhao J, Zhang H, et al. Bifidobacterium Pseudocatenulatum Ameliorates Dss-Induced Colitis by Maintaining Intestinal Mechanical Barrier, Blocking Proinflammatory Cytokines, Inhibiting TLR4/NF-κb Signaling, and Altering Gut Microbiota. J Agric Food Chem (2021) 69(5):1496–512. doi: 10.1021/acs.jafc.0c06329
134. Chen Y, Jin Y, Stanton C, Paul Ross R, Zhao J, Zhang H, et al. Alleviation Effects of Bifidobacterium Breve on DSS-Induced Colitis Depends on Intestinal Tract Barrier Maintenance and Gut Microbiota Modulation. Eur J Nutr (2021) 60(1):369–87. doi: 10.1007/s00394-020-02252-x
135. Park MS, Song NE, Baik SH, Pae HO, Park SH. Oral Administration of Lactobacilli Isolated From Jeotgal, A Salted Fermented Seafood, Inhibits the Development of 2,4-Dinitrofluorobenzene-Induced Atopic Dermatitis in Mice. Exp Ther Med (2017) 14(1):635–41. doi: 10.3892/etm.2017.4536
136. Tang L, Li XL, Deng ZX, Xiao Y, Cheng YH, Li J, et al. Conjugated Linoleic Acid Attenuates 2,4-Dinitrofluorobenzene-Induced Atopic Dermatitis in Mice Through Dual Inhibition of COX-2/5-LOX and TLR4/NF-K;b Signaling. J Nutr Biochem (2020) 81:108379. doi: 10.1016/j.jnutbio.2020.108379
137. Furue M. Regulation of Filaggrin, Loricrin, and Involucrin by IL-4, Il-13, IL-17a, Il-22, AHR, and NRF2: Pathogenic Implications in Atopic Dermatitis. Int J Mol Sci (2020) 21(15):5382. doi: 10.3390/ijms21155382
138. Yu J, Luo Y, Zhu Z, Zhou Y, Sun L, Gao J, et al. A Tryptophan Metabolite of the Skin Microbiota Attenuates Inflammation in Patients With Atopic Dermatitis Through the Aryl Hydrocarbon Receptor. J Allergy Clin Immunol (2019) 143(6):2108–19.e12. doi: 10.1016/j.jaci.2018.11.036
139. van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal Tar Induces AHR-Dependent Skin Barrier Repair in Atopic Dermatitis. J Clin Invest (2013) 123(2):917–27. doi: 10.1172/jci65642
140. Roager HM, Licht TR. Microbial Tryptophan Catabolites in Health and Disease. Nat Commun (2018) 9(1):3294. doi: 10.1038/s41467-018-05470-4
141. Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao CC, Ardura-Fabregat A, et al. Microglial Control of Astrocytes in Response to Microbial Metabolites. Nature (2018) 557(7707):724–28. doi: 10.1038/s41586-018-0119-x
142. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. Proc Natl Acad Sci U S A (2009) 106(10):3698–703. doi: 10.1073/pnas.0812874106
Keywords: gut microbiota, probiotics, atopic dermatitis, immune response, effective substances
Citation: Fang Z, Li L, Zhang H, Zhao J, Lu W and Chen W (2021) Gut Microbiota, Probiotics, and Their Interactions in Prevention and Treatment of Atopic Dermatitis: A Review. Front. Immunol. 12:720393. doi: 10.3389/fimmu.2021.720393
Received: 04 June 2021; Accepted: 28 June 2021;
Published: 14 July 2021.
Edited by:
Tingtao Chen, Nanchang University, ChinaReviewed by:
Zhihong Sun, Inner Mongolia Agricultural University, ChinaCopyright © 2021 Fang, Li, Zhang, Zhao, Lu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenwei Lu, bHV3ZW53ZWlAamlhbmduYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.