
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 19 October 2021
Sec. Comparative Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.716289
The genes of the leukocyte immunoglobulin-like receptor (LILR) family map to the leukocyte receptor complex (LRC) on chromosome 19, and consist of both activating and inhibiting entities. These receptors are often involved in regulating immune responses, and are considered to play a role in health and disease. The human LILR region and evolutionary equivalents in some rodent and bird species have been thoroughly characterized. In non-human primates, the LILR region is annotated, but a thorough comparison between humans and non-human primates has not yet been documented. Therefore, it was decided to undertake a comprehensive comparison of the human and non-human primate LILR region at the genomic level. During primate evolution the organization of the LILR region remained largely conserved. One major exception, however, is provided by the common marmoset, a New World monkey species, which seems to feature a substantial contraction of the number of LILR genes in both the centromeric and the telomeric region. Furthermore, genomic analysis revealed that the killer-cell immunoglobulin-like receptor gene KIR3DX1, which maps in the LILR region, features one copy in humans and great ape species. A second copy, which might have been introduced by a duplication event, was observed in the lesser apes, and in Old and New World monkey species. The highly conserved gene organization allowed us to standardize the LILR gene nomenclature for non-human primate species, and implies that most of the receptors encoded by these genes likely fulfill highly preserved functions.
The human immunoglobulin superfamily (IgSF) represents more than 700 cell-surface and secreted receptors, which are characterized by the presence of one or more immunoglobulin-like (Ig) domains (1). Several IgSF subfamilies are encoded within the leukocyte receptor complex (LRC), which spans approximately 900 kb on chromosome 19 (Figure 1A) (2). This complex encodes the leukocyte immunoglobulin-like receptors (LILR), the killer immunoglobulin-like receptors (KIR) and the leukocyte-associated immunoglobulin-like receptors (LAIR) (Figure 1B) (3–5). Other immune-related genes embedded in the LRC are those encoding for the natural cytotoxicity receptor 1 (NCR1) and the Fc-alpha receptor (FcAR) (2, 6). The extended LRC region is located centromeric of the LRC, and was formed by multiple duplication events, eventually resulting in the formation of additional gene families, including sialic acid-binding immunoglobulin-type lectins (SIGLEC), neonatal Fc receptor (FcGRT), the carcinoembryonic antigen-related cell adhesion molecule (CEACAM/CD66), and pregnancy-specific glycoprotein (PSG) (6). Although the extended LRC region encompasses multiple IgSF subfamilies, only the genomic organization of the LRC region, encoding the LILR, KIR, and LAIR gene families, has been reviewed on a regular basis (2, 6, 7).
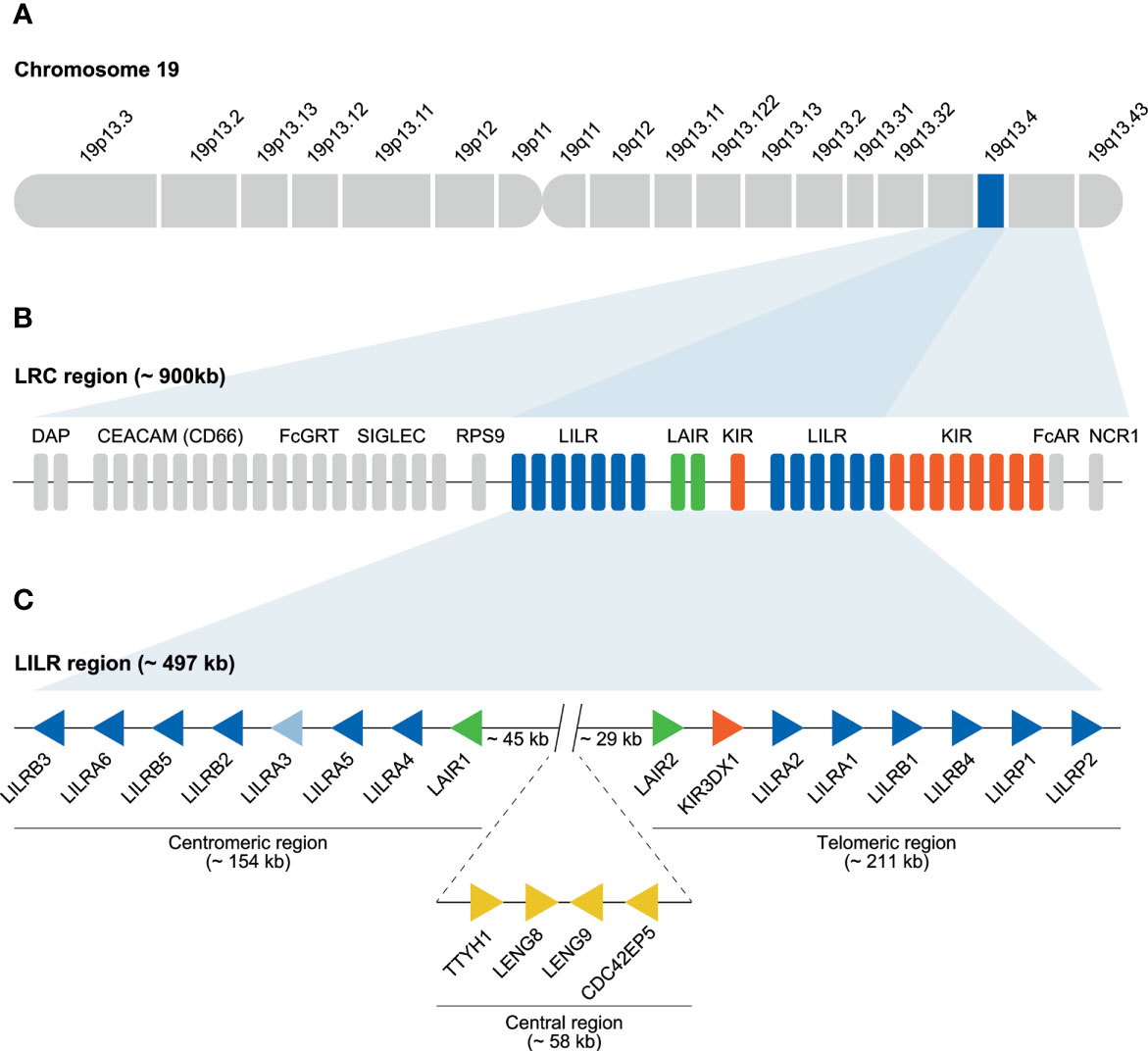
Figure 1 Human LRC region and genomic organization of the LILR gene family. (A) The LRC region located on chromosome 19q13.4. (B) LILR (blue bars), KIR (red bars), and LAIR (green bars) gene families located within the LRC. The remaining genes located in the LRC are indicated by grey bars. (C) The LILR region (~ 497 kb), schematic illustration of the division of the 11 functional LILR genes on the centromeric (~154 kb) and telomeric side of the region (~211 kb) as well as the pseudogenes LILRP1 and LILRP2 located on the telomeric side. LILRA3 is light blue to indicate the null haplotype observed in humans. The regions are separated by a central region of approximately 58 kb that includes the genes TTYH1, LENG8, LENG9, and CDC42EP5. Between the centromeric and central region, and the central and telomeric region, a stretch of 45 and 29 kb, respectively, is observed. Representing different genes, the arrows are aligned in such a way that they point in the direction of transcription.
The KIR region, located on the telomeric side of the LILR gene cluster, is known to be highly dynamic, and, at population level, haplotypes may show considerable diversity in gene architecture and allelic content (5, 8, 9). The diversity of the KIR region is a result of substantial homologous recombination and unequal crossing-over events (10–13). The KIR receptors are expressed by NK cells and subsets of T lymphocytes, and play a key role in immune regulation by interacting with polymorphic epitopes on major histocompatibility complex (MHC) class I molecules, designated in humans as human leukocyte antigen (HLA) (14–17). Furthermore, KIR receptors play a pivotal role in the recognition and elimination of cells lacking the expression of MHC class I molecules (18, 19).
The LAIR gene family consists of two genes that encode a cell-surface (LAIR1) and a soluble (LAIR2) receptor, and are located in the center of the LILR region (6, 20, 21). The expression of LAIR is broadly confined to peripheral blood lymphocytes, including NK cells, T and B lymphocytes, neutrophils, monocytes, and macrophages (21–25). LAIR1 and LAIR2 gene products are both collagen-binding receptors, and play a key role in controlling tissue inflammation (24, 26–29).
In contrary to the KIR gene family, the organization of the LILR gene content is conserved in humans (9, 30). A conventional LILR haplotype contains 13 genes, 11 of which encode a functional protein, and two are classified as pseudogenes (Figure 1C) (31). The human LILRP1 gene has an apparent 5’ acceptor splice site in front of the exon encoding the third Ig-like domain, resulting in a pseudo-exon, while LILRP2 became inactivated due to a 7 bp insert as evidenced of a tandem repeat in the exon encoding the second Ig-like domain (32).
LILR gene products are widely expressed by immune cell populations of both myeloid and lymphoid lineages, and several members interact with HLA class I molecules (33–35). In contrast to the KIR receptors, LILR receptors do not interact with polymorphic epitopes on the alpha 1 and 2 domains of HLA molecules, but engage with conserved epitopes on the alpha 3 domain and/or with the highly conserved β2-microglobulin structure (a component of the MHC class I dimer) (33). The KIR region has been extensively studied in different primate species, including humans. The LILR region, on the other hand, has only been thoroughly studied in humans, and its equivalents in mice, chicken, and other vertebrates such as cattle and pigs (36–43). In this communication, we aim to provide a comprehensive overview of the LILR region in humans and of the available genomic data in different non-human primate species.
Information regarding the ancestry of any genetic system can be recovered by comparing its presence or absence in different indicator species that once shared common ancestors. Approximately 65 million years ago (mya), the first ancestral mammalian species started to roam the earth. As mentioned previously, the presence of a LILR system has been documented in humans and several other species, including mouse and cattle. These data suggest that the emergence of the LILR cluster predates radiation of the mammalian lineage (Figure 2) (44).
Paired Ig-like receptors (PIR) are encountered in mice, and the genomic architecture of the PIR region is comparable to the LILR region in humans (39, 45). The order Artiodactyla, which includes cattle, goat, and pig, emerged approximately 65 mya, with cattle and goat diversifying approximately 20 mya. The LILR region in pigs turned out to be similar to the human equivalent, but the region itself shows an expansion of the LILR genes in the centromeric region (41). In goats, the LILR region exhibits a contraction in the number of genes as compared to its human counterpart (42–44). These observations highlight not only that the LILR system is subject to purifying selection but also that specialization may have emerged during vertebrate evolution.
The marsupials radiated approximately 100 mya and is represented here by the opossum (Figure 2). In opossums, 124 Ig-like domains with similarity to KIR and LILR Ig-like domains were identified (46). The avian lineage, which emerged approximately 300 mya, provided the next major informative event (Figure 2) (44). The chicken immunoglobulin-like receptor (CHIR) gene system is characterized by massive expansion and diversification in comparison to the human LILR region. Nonetheless, the highly similar structures found in both humans and chicken suggest that the emergence of the LILR cluster might date from before the avian lineage (40, 47, 48). The genes encoding for CHIR and marsupial Ig-like domains have, however, a different transcriptional direction as compared to their evolutionary equivalents in mammalian species.
Data obtained from the class of ray-finned fish (Actinopterygii) evidenced that the origin of the LILR system might even date back to approximately 450 mya (Figure 2). Ray-finned fish comprise the largest class of vertebrates (~25000 marine and fresh-water species) including, for instance, channel catfish (Ictalurus punctatus) (44). In this species, leukocyte immune-type receptors (LITR) have been identified, and have an evident orthologous relationship to human LILR receptors (49).
In summary, there is compelling evidence that a LILR-like system, in a way similar to that of the MHC complex, emerged before the major expansion of the vertebrate lineage, approximately 450 mya. Some of the LILR receptors have MHC class I molecules as their ligands (Table S1), and it is tempting to speculate that both systems co-evolved and had an impact on each other. During vertebrate evolution, the LILR complex was subjected to a modest number of expansions and contractions. Some of the receptors may have experienced purifying selection, and therefore still interact with their original ligands. Alternatively, certain LILR receptors of different species may have diverged and specialized, and thereby acquired novel functions. Aside from that, pathogens may have evolved strategies to misuse these types of receptors: for instance, to invade the cell or escape the immune system. An ancestral KIR gene existed approximately 50-100 mya, but the primate KIR gene cluster arose approximately 30-45 mya (3, 5). The most parsimonious explanation is that initially the presence/absence of MHC class I was scanned by LILR receptors that can diagnose the presence of conserved epitopes. Later, when the MHC complex expanded and staged extensive allelic polymorphism, more sophisticated systems – like the KIR gene system – arose, which are able to scan for the presence of polymorphic epitopes on MHC class I molecules.
Several receptors encoded within the LRC region, including FcAR, NCR1, and various LILR and KIR receptors, function as activating receptors (34, 50–52). The complexity of the immune response and the need to control dangerous immune reactions probably drove the onset of a large arsenal of inhibiting receptors.
It has been proposed that an ancestral gene encoding Ig domains acted as progenitor for LILR, KIR, and other immune-related genes such as FcAR and NCR1 (2). This progenitor gene presumably encoded an activating receptor, as it likely featured a positively charged arginine residue in the transmembrane region, because the evolutionarily old FCAR, NCR1, and LILR genes share this feature. The progenitor of LILRA1 may have acted as the founder gene of the LILR cluster (2, 53, 54). Several LILRA1 duplication events shaped the first primitive LILR organization, which contained LAIR2, LILRA1, LILRB1, and LILRP1 (Figure 3A). This event was followed by a relatively stable period, after which time the region duplicated entirely, and was reversely incorporated into the genome, forming the centromeric LILR region. The genes in this region are LILRB3, LILRB2, LILRA3, and LAIR1 (Figure 3B). Several autonomous tandem duplication events occurred within the centromeric and telomeric region, and gave rise to the contemporary organization of the LILR region in humans, as illustrated in Figure 3C (2, 9, 31). The human LILR region (~ 497 kb) is divided into the centromeric region (~ 154 kb) and the telomeric region (~ 211 kb). Notably, KIR3DX1, previously known as KIR3DL0 or LENG12, is embedded between LAIR2 and LILRA2 in the telomeric region (Figure 3C). It represents an independent ancestral and highly conserved KIR lineage with orthologs in human, chimpanzee, gorilla, rhesus monkey, and common marmoset (55). As the KIR gene family arose later in evolution than the LILR gene family, it is likely that the KIR3DX1 gene became inserted into the telomeric section after duplication of the entire LILR region.
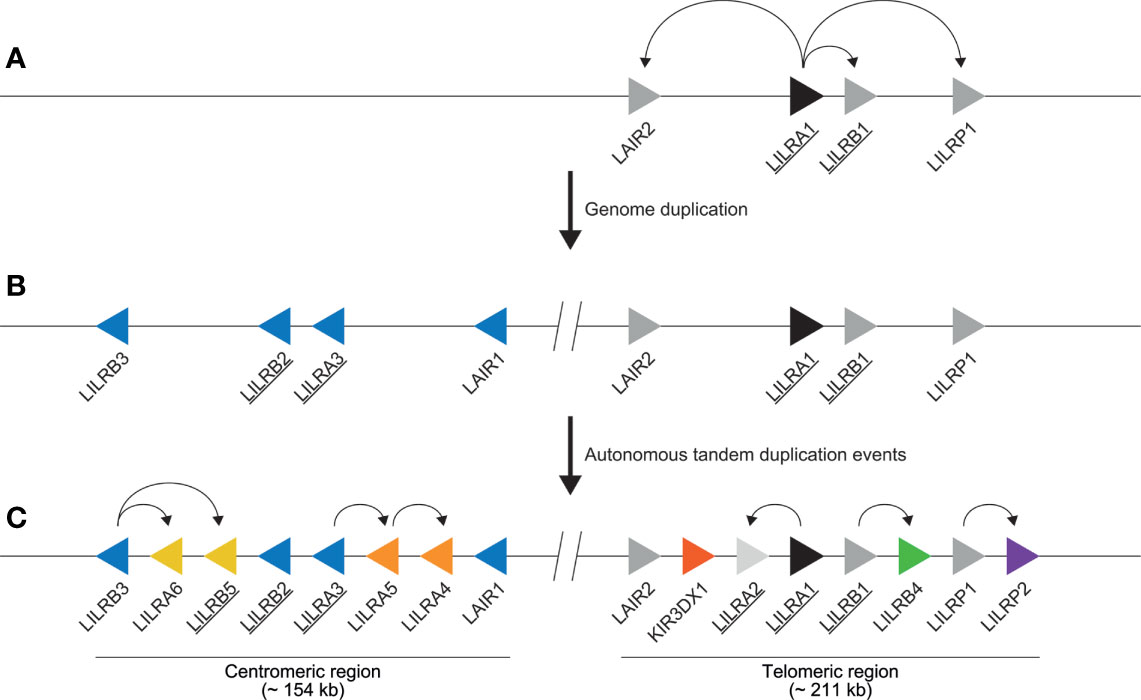
Figure 3 Schematic illustration of the evolution of the human LILR region. (A) As indicated by the curved arrows, the first primitive LILR region arose due to independent LILRA1 duplication events, resulting in the emergence of LAIR2, LILRB1, and LILRP1 (depicted by grey arrows). LILRA1 is denoted as the ancestral LILR gene and is therefore depicted by a black arrow. Genes encoding for receptors interacting with HLA epitopes are underlined. (B) The primitive LILR region was subsequently duplicated and reversely incorporated into the genome, shaping the centromeric region containing LILRB3, LILRB2, LILRA3, and LAIR1, depicted by blue arrows. (C) Next, several independent duplication events occurred in both regions, as indicated by the curved arrows, modeling both a centromeric and a telomeric make-up. Representing the different genes, the arrows are aligned in such a way that they point in the direction of transcription. The colored arrows highlight the different autonomous tandem duplication events. The location of KIR3DX1 gene is indicated (red arrow). The interception with two vertical lines indicates the central region.
In humans, most gene products of the telomeric LILR region interact with HLA class I ligands, except for receptor LILRB4. Gene products of the centromeric region, however, may dock on HLA and non-HLA molecules (Figure 3C) (35). This suggests that the LILR receptors encoded in the telomeric region maintained more or less the original functions, whereas receptors encoded by genes in the centromeric region further specialized and acquired novel functions. Receptor LILRB2, for example, mapping in the centromeric region, mainly docks on HLA molecules but may also interact with angiopoietin-like (ANGPTL) molecules, β-amyloid, and Nogo-66, suggesting that this receptor gained new functions as well (35, 56–59). The genomic architecture of the LILR region in humans appears to be relatively stable, and the most common haplotype organization found world-wide is as presented in Figure 3C. However, a haplotype with a relative high frequency lacking LILRA3 as result of a 6.7 kb deletion is also known (referred to as a null haplotype) (60–63). The centromeric LILR region has an average gene distance of 18 kb (8 kb – 27 kb) between the stop codon and the start codon of adjacent genes, while the telomeric region has an average gene distance of 23 kb (9 kb – 30 kb) (Figure S1A). When the UTR regions are included, the centromeric region has an average gene distance of 16 kb (7 kb – 26 kb), while the telomeric region has an average gene distance of 18 kb (9 kb – 27 kb) (Figure S1B). Some LILR genes have long intergenic regions: for example, the stretch between LILRB1 and LILRB4. Despite these relatively long intergenic regions, no recombination events in the LILR region have been recorded (3, 7). This is in sharp contrast to the situation for the neighboring KIR region, which is characterized as being highly dynamic and with variable gene numbers and gene combinations, and consists of highly polymorphic genes (5, 12, 64).
In the past, the terms Ig-like transcripts (ILT), leukocyte Ig-like receptors (LIR), and CD85 were different names used to denote LILR genes in humans (65, 66). In 2004, however, the LILR gene nomenclature was officially standardized and accepted by the HUGO Gene Nomenclature Committee (HGNC) (33, 67). LILR family members are categorized as activating (termed LILRA1 to LILRA6) or inhibitory receptors (termed LILRB1 to LILRB5). For LILR in non-human primates (NHP), the human LILR gene nomenclature is loosely followed by annotation algorithms such as those from NCBI and ENSEMBL. Due to the high levels of similarity between the different LILR genes, difficulties may arise in phasing the genomic regions, and therefore it may accidently happen that orthologous genes are not given the corresponding name. To root out such errors, we have selected the latest and freely accessible genomes, which were sequenced by the latest next generation platforms (Table S2). The LILR region including RPS9 and KIR was extracted from NCBI genome data viewer, and, if available, ENSEMBL database. In Table S3, the genes located within the LILR region are listed, and the coordinates of these genes on the reference genome are indicated with specific LOC numbers. Available genomic DNA and mRNA sequences were downloaded from the NCBI database and compared to the human LILR sequences using Geneious Prime 2020.2.4. Non-human primate genomic DNA sequences were aligned using the MUSCLE method (standard settings, eight iterations) against human genomic sequences. However, different intron length resulted in alignment issues. Therefore, all available primate mRNA sequences were aligned, and a phylogenetic tree analysis was performed using the Geneious tree builder (Jukes-Cantor, Neigbor-Joining, no outgroup, resample tree, bootstrap, random seed = 724,574, number of replicates = 100, create consensus tree, support threshold % = 50). Each cluster with mRNA sequences was aligned and compared to identify transcripts with a potentially incorrect gene name. These sequences were then aligned against the relevant human reference transcripts to sort out whether the genes were annotated correctly or incorrectly in the public database. Next the genomic gene location was defined in relation to the human LILR region, before adjusting the gene nomenclature in the relevant non-human primate species. Using this approach, we have explored all available LILR data on different nonhuman primate species: namely, chimpanzee, bonobo, gorilla, orangutan, gibbon, rhesus and cynomolgus macaque, and common marmoset, and have introduced a naming of LILR genes based on the orthologous positions in the human genome, which was taken as a reference (Tables S2, S3). Moreover, during this study we have reannotated several of the LOC ID’s that originally comprised two gene entities (Table S3). For the chimpanzee and bonobo, we have renamed an additional LILRA2-like gene probably encoding for a bona fide gene product (LOC 100612450 and LOC117974252, respectively). This gene locates in the centromeric region between LILRB2 and LILRA3, and shares 99.3% similarity between the two species. Comparative sequence analysis revealed that the domain and intermediate intron sequences of this additional LILRA2-like gene are approximately 94% similar to the adjacent LILRB2 gene. The sequence encoding the domains is comparable with LILRB2, while the stem and transmembrane region are more similar to activating LILR receptors; therefore, we have designated this gene as LILRAB2. Although, the stem and transmembrane region is comparable to activating LILR receptors, we were not able to pinpoint which recombination event(s) generated the LILRAB2 gene. There is no evidence for the presence of an additional, maybe disrupted, human LILRA2 gene in the centromeric region. The reannotation of LOC109024105 for gorilla, LOC100432416 for orangutan, and LOC718403 for rhesus macaque revealed evidence for the presence of a LILRAB2 gene in these species (Table S3). For gorilla, however, the transcription status for this gene is questionable because the current genomic sequence shows a mutation in the transcription initiation site. At this stage we are not certain about the presence or absence of the LILRB2 and LILRAB2 genes in the cynomolgus macaque genome. Additionally, in the non-human primates that we have analyzed LILR genes that might encode for activating receptors are located at the same position as human pseudogenes LILRP1 and LILRP2. These activating genes belong to a divergent lineage of the ancestral LILRA1 gene (Figure 3C), and we have denoted them as LILRA7 and LILRA8 in non-human primates (2).
The KIR nomenclature has already been standardized in humans and NHP (68–70). In humans and great and lesser apes, the first KIR gene located at the boundary of the KIR and LILR region is KIR3DL3. In rhesus and cynomolgus macaques, KIR3DL20, which was previously designated as KIR3DL2, is located at the corresponding position and depicted in relevant schemes (12, 71). Furthermore, the LAIR gene family consists of two genes that are easily differentiated, and therefore no nomenclature conflicts are reported.
The chimpanzee and bonobo, gorilla, and orangutan are all great ape species, and they shared a common ancestor with the human lineage approximately 4.5-6, 6-8, and 12-16 mya, respectively (Figure 4) (72–75). For the purpose of this communication, the reference genomes of the chimpanzee, bonobo, western lowland gorilla, and Sumatran orangutan were extracted from the NCBI database and their LILR gene organization was thoroughly compared and analyzed using both genomic and mRNA sequences with the human LILR region as reference (Tables S2, S3) (76, 77).
In great apes, the LILR gene region is fixed on chromosome 19, and includes the flanking genes RPS9 and KIR3DL3. In general, the human (~ 600 kb) and chimpanzee LILR region (~ 605 kb) are highly similar (Figure 5), and orthologs have been reported; they include LILRA2, LILRA4, LILRB4, and LILRB5 (78, 79). In contrast to humans, however, the chimpanzee LILR region contains a LILRAB2 gene in the centromeric region. At the same position as the human pseudogenes LILRP1 and -P2, two genes are identified in chimpanzees, named LILRA7 and LILRA8 (Figure 5). These sequences are, however, defined as low quality and modeled from the genome sequence (Table S3), and thus might feature inactive genes, comparable to the human pseudogene tandem. In the bonobo, the LILR organization (~ 546 kb) is similar to that of the chimpanzee, but the transcription status of several genes, including LILRA6, LILRB5 and LILRA7, could not be confirmed. The LILRA8 gene appears to be non-functional, as sequence analysis revealed two frameshift mutations that result in the introduction of a premature stop codon, and, therefore, this gene is considered to be a pseudogene (Figure 5 and Table S3). The centromeric LILR region in gorillas seems to lack LILRB5, while the transcription status for four other LILR genes is uncertain. The start codon of LILRA1, for instance, seems to be interrupted by an insertion. Inactive orthologs of the LILRA7 and LILRA8 genes are identified in the telomeric region. The inactivation of LILRA7 is a result of a mutation in the exon encoding the first Ig-like domain, resulting in a premature stop codon, while LILRA8 became non-functional due to a similar inactivation event as described in human LILRP2. The orangutan centromeric region lacks a LILRA6 gene, while LILRB5, LILRA3, LILRA7 and LILRA8 are defined as low quality sequences (Figure 5 and Table S3). In the telomeric region near the haplotype center, three additional KIR genes are annotated in the orangutan reference genome, and a FCAR gene could be identified adjacent to them. This region (~ 240 kb) most likely represents an assembly error introduced by the shotgun approach that is used to sequence the reference genome and is therefore not illustrated in Figure 5.
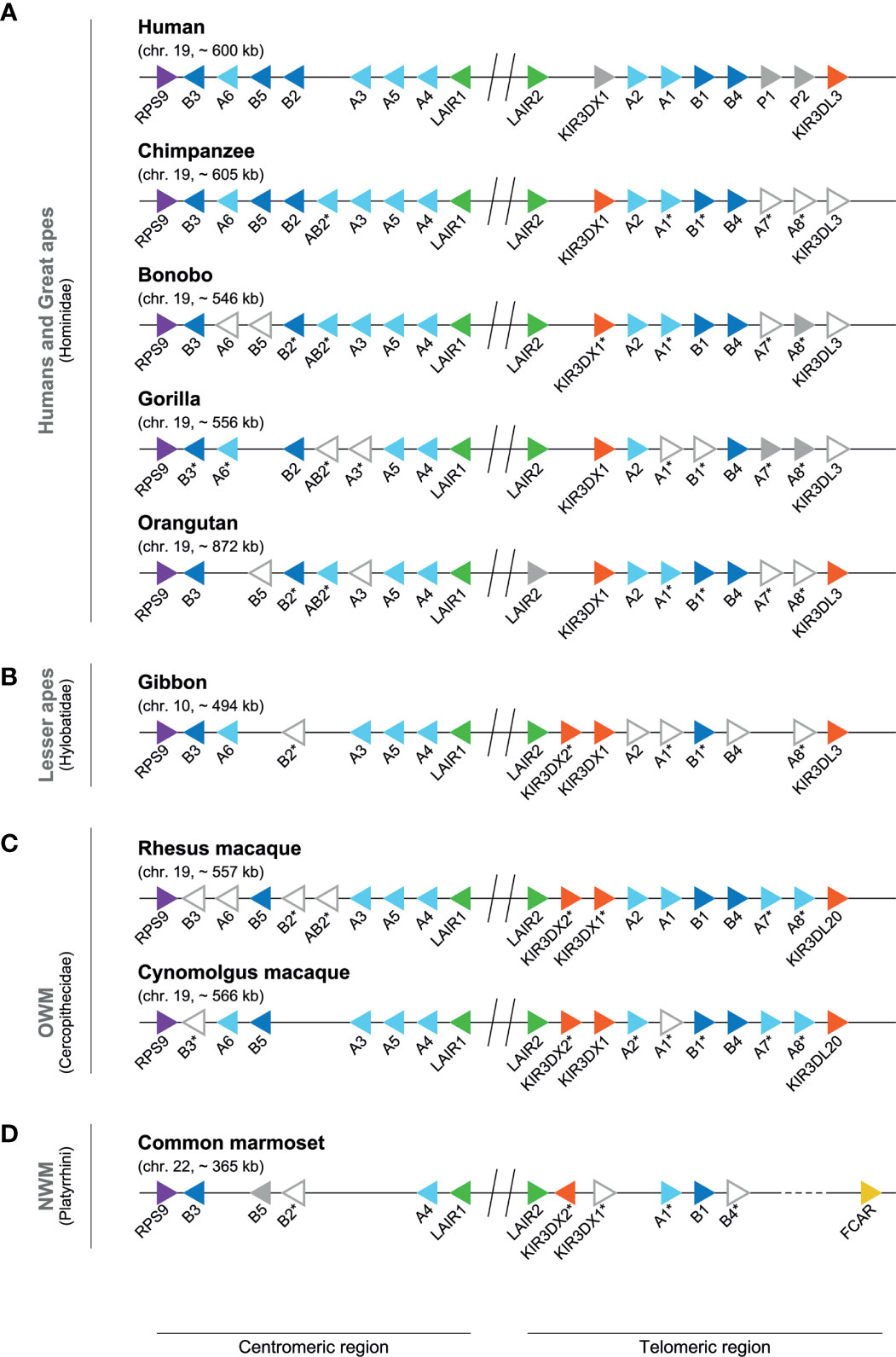
Figure 5 Genomic organization of the LILR region in different primate species. Genomic organization of the LILR region in human and great apes (A), lesser ape (B), and representatives of the OWM (C) and NWM (D) species (Table S2). Each different LILR gene present on a genome is indicated by a colored arrow aligned in such a way that it points in the direction of transcription: RPS9 (purple), LILRA (light blue), LILRB (dark blue), LAIR (green), KIR (red), FCAR (yellow), and pseudogenes or genes indicated by NCBI as non-functional, such as bonobo LILRA8 (grey). The arrows with only a gray outside line represent genes that are indicated by NCBI with low quality protein (Table S3), and the expression status of these genes is currently uncertain. The sequence of cynomolgus macaque LILRA1, found after reannotation of LILRA2 (Table S3), shows only a correct alignment up to the exon encoding for the third domain structure, and most likely represents a pseudogene. The absence of LILRB2 and LILRAB2 on the reference genome of cynomolgus macaques might be an assembly error, as a stretch of approximately 35 kb is identified in this region, which contains an unrelated gene (Table S3). The central region is indicated by an interception with two vertical lines. The dotted line (common marmoset) illustrates a large genomic region between LILRB4 and the FCAR lacking annotation. The genes indicated with an asterisk have been renamed using standardized LILR gene nomenclature (previously used names are listed in Table S3), and particular of these genes were found by re-annotation (Table S3).
The hylobatidae – also known as the lesser apes, and to which the Northern white-cheeked gibbon belongs – shared a common ancestor with humans approximately 18-20 mya (Figure 4) (75). The gibbon LILR region is located on chromosome 10, and comprises approximately 494 kb. As compared to other primate species, gibbons frequently feature chromosome re-arrangements (80, 81). In the gibbon, part of an ancestral chromosome 19 was re-arranged and incorporated in a reversed form into chromosome 10 (81), which probably resulted in shifting the LILR region toward chromosome 10. In comparison to humans, the centromeric region in the gibbon lacks LILRB5, while LILRA7 is absent in the telomeric region (Figure 5). For five LILR genes, the transcription status is uncertain. Moreover, a second KIR3DX gene is observed within the telomeric region, and has been denoted as KIR3DX2.
The cercopithecidae, also known as Old World monkeys (OWM), shared a common ancestor with humans approximately 25-33 mya (Figure 4) (75, 82). The majority of the OWM species can be found in Africa. Most macaque species, however, inhabit various parts of Asia, and one species, the Barbary macaque, lives on the island of Gibraltar in Europe as well as in sections of Northern Africa. Genomes of the Indian-origin rhesus macaque and cynomolgus macaque originating from Tinjil island were available at the NCBI Genome data viewer and Ensembl release 103 and 102, respectively, and were manually explored and annotated for the LILR region make-up (Tables S2, S3) (76, 77). Both the NCBI and Ensembl developed their own unique automatic annotation pipeline, which may result in minor differences between the assemblies. The LILR region of rhesus and cynomolgus macaques is located on chromosome 19, and spans approximately 557 and 566 kb, respectively. In rhesus macaques and humans, the make-up of the centromeric LILR region appears to be highly similar. The rhesus macaque genome seems to contain, however, a LILRAB2 gene that might be an orthologue of the chimpanzee and bonobo LILRAB2, while it is not certain whether this gene, as well as LILRB2, is present or absent in cynomolgus macaques (Figure 5). The LILRA7 and LILRA8 genes, located at the same position as the equivalent pseudogenes in humans, seem to encode bona-fide activating receptors in macaques, as in-frame transcripts are expected to be transcribed from these genes. In rhesus macaques the transcription status of LILRB3, LILRA6, and LILRB2 is uncertain. In cynomolgus macaques, the LILRA1 gene may be disrupted by an insertion subsequent to exon 5, which indicates a secreted gene product or an inactive copy, and the transcription status of LILRB3 is uncertain. In addition, like found in the gibbon, also the telomeric region of macaques contains an additional gene that belongs to the KIRDX lineage, termed KIR3DX2.
Platyrrhini, or New World monkeys (NWM), shared with humans a common ancestor that lived approximately 27-52 mya (Figure 4) (75, 82). The common marmoset is likely the most prominently studied NWM organism; its genome is available at the NCBI Genome data viewer, and it was manually explored and annotated for the LILR region (Table S2, S3) (76, 77). The common marmoset LILR region is located on chromosome 22, and spans approximately 365 kb. Comparative karyotyping of the chromosomes of common marmosets and humans showed the homology of marmoset chromosome 22 with human chromosome 19 (83). In marmosets, both LILR regions, centromere and telomere, show a substantial contraction with regard to the number of genes as compared to the human LILR region (Figure 5). The centromeric region contains LILRB3, LILRB5 (which became a pseudogene), LILRB2, and LILRA4. In the telomeric region LILRA1, LILRB1, and LILRB4 were encountered. LILRB2 and LILRB4 are, however, defined as low quality sequence. Furthermore, a tandem of KIR3DX1 and KIR3DX2 genes was observed. Adjacent to LILRB4, an unannotated stretch of ~ 280 kb is identified, which does not contain a LILR or KIR equivalent. This might reflect an error in the assembly of the reference genome.
To the best of our knowledge, we present here the first comprehensive comparison of the LILR region in different primate species. We should note, however, that for most non-human primate species only one or a few complete genomes have been sequenced. The LILR organization in this study is based on a single reference genome per species. Although the LILR cluster seems to be an ancient and conserved set of genes, subtle variation in the gene organization might exist per individual or per population, as is demonstrated for the null haplotype in humans that lacks the presence of LILRA3. Moreover, the transcription status of several genes was also not confirmed on the current reference genomes and requires additional characterization studies before definitive conclusions can be drawn.
In primates and cattle, two distinct KIR gene clades, KIR3DL and KIR3DX, have been defined (55, 84). The KIR3DL lineage is duplicated, and generated a KIR gene family in simian primates, while the KIR3DX lineage in cattle was subjected to expansion, resulting in a functional KIR gene family (84, 85). It is hypothesized that an ancestral KIR gene emerged approximately 135 mya before radiation of the placental mammal resulting in KIR3DL and KIR3DX daughter genes. In primates, the KIR3DX gene is embedded within the LILR region, while the KIR gene family is located telomeric of the LILR gene family (Figure 1). In humans, KIR3DX1 is regarded as a pseudogene due to a 7 bp deletion at the end of exon 5, resulting in the introduction of a premature stop codon in exon 7, and the frameshift was confirmed in 86 healthy individuals (55). Although human KIR3DX1 is classified as a pseudogene by HUGO, KIR3DX1 cDNA could be cloned from a human NK cell line (NK-92), suggesting that transcription of the gene may occur (55).
In the genomes of several NHP species analyzed, we observed a second KIR3DX gene that most likely arose by an ancient duplication event of KIR3DX1, and the sister gene has been termed KIR3DX2 (Figures 5, 6). The NCBI database classifies the KIR3DX2 as a protein coding gene, but it lacks the exons encoding the transmembrane and signaling regions, and therefore may encode a secreted gene product (Figure 6B). In marmoset, the two gene copies are arranged head-to-head (Figure 5), and it is postulated that this orientation arose due to a species-specific duplication event (55). However, we observed two KIR3DX copies arranged head-to-tail in the lesser apes and OWMs, suggesting that both gene entities might already be present in a common ancestor with NWM and remained conserved, while one of the KIR3DX genes was lost during evolution of the human and great ape lineage. From this perspective, the head-to-head arrangement in common marmoset might be a genome assembly failure, and the orientation should be head-to-tail as well. Additional genome analysis must be performed to sort out if this arrangement is a species-specific duplication event or an assembly failure. The presence of two related KIR3DX genes is, however, not specific for macaques residing in Asia, but can be found in the NCBI database available genomes of olive baboon and green monkey as well (data not shown), both of which inhabit Africa.
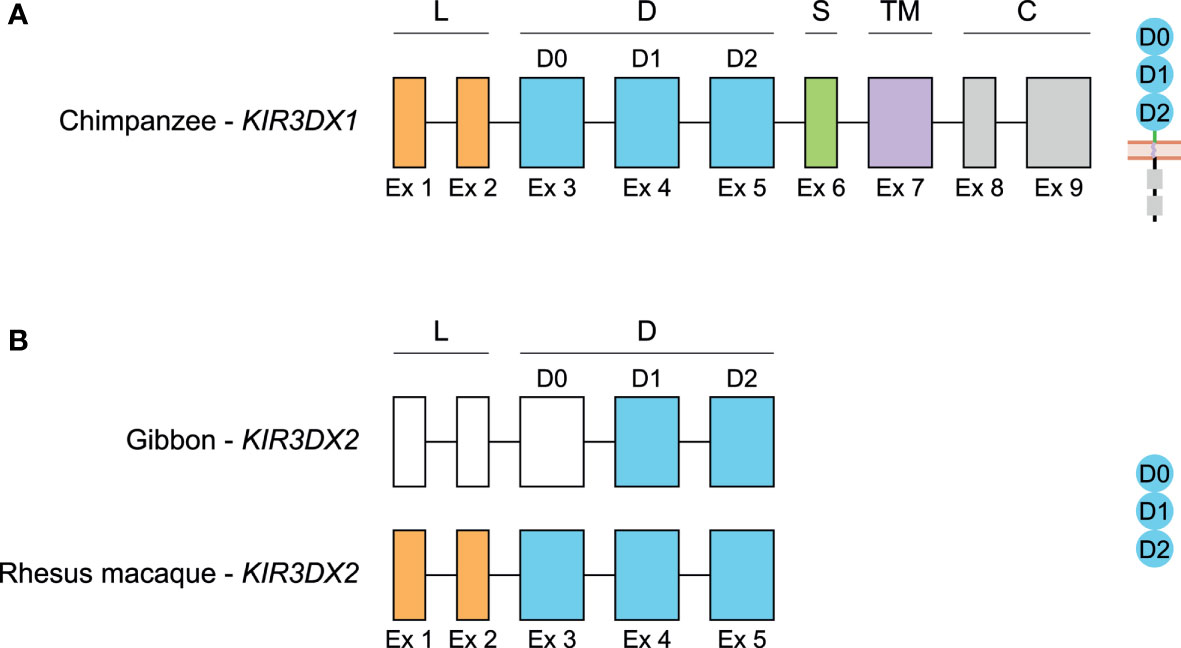
Figure 6 Genomic organization of KIR3DX1 and KIR3DX2 genes in primates. Schematic illustration of the genomic organization of the chimpanzee KIR3DX1 gene that is representative for human, bonobo, gorilla, orangutan, gibbon, rhesus and cynomolgus macaque, and common marmoset (A), and of the KIR3DX2 gene in gibbon and rhesus macaque, of which the latter is representative for cynomolgus macaque and common marmoset (B). Exons 1 and 2, encoding the leader peptide (L), are depicted in orange; exons 3-5, encoding Ig-like domains (D), are in blue; exon 6, encoding the stem (S), is in green; exon 7, encoding the transmembrane (TM) region, is in purple; and exons 8 and 9, encoding the cytoplasmic tail (C), are in grey. Even though the start codon of gibbon KIR3DX2 is identified, the first three exons could not be defined, and are therefore represented by white boxes. The putative corresponding protein structures are schematically depicted adjacent to the genomic organization.
KIR3DX1 exon sequences are compared with chimpanzee KIR3DX1 using Geneious Prime 2020.2.4, since the full-length human KIR3DX1 transcripts would contain the 7 bp deletion, and the chimpanzee is most closely related to humans (Table S4) (86). Comparative analysis of the exons revealed a high level of interspecies sequence similarity to chimpanzee KIR3DX1 for gorilla (97.9%), orangutan (95.1%), gibbon (94.3%), rhesus macaque (90.8%), and cynomolgus macaque (91.0%). In contrast, KIR3DX1 in common marmoset is 68.4% similar to chimpanzee KIR3DX1. Furthermore, the coding region of KIR3DX1 in rhesus and cynomolgus macaque is 99.4% similar, indicating that this gene is highly conservated in macaque species. When introns are included in the sequence similarity comparison, higher levels of diversity are determined. In orangutans, for instance, the genomic KIR3DX1 sequence displays 83.5% similarity to chimpanzees, and the difference are mainly explained by insertions and deletions in the introns. The genomic sequence of chimpanzee KIR3DX1 is 76.6% and 78.7% similar to the sequences of rhesus and cynomolgus macaques, respectively. These observations suggest a selective pressure to largely conserve the KIR3DX1 exons, whereas the introns might be more prone to diversification, which may impact the regulation of expression levels and transcript splicing. Overall, the KIR3DX1 coding sequence is highly conserved between human, great apes, lesser apes, and OWM, whereas the KIR3DX1 gene in NWM diverged during evolution. The function of KIR3DX1 is still unknown. However, it has an inhibitory potential due to the presence of two ITIM motifs in the cytoplasmic tail (55). Full-length KIR3DX1 transcripts were identified in peripheral blood mononuclear cells of rhesus macaques, in addition to an isoform that lacked exon 5, but quantification indicated low expression levels (55). To date, no ligand for KIR3DX1 has been reported.
Comparing the sequences of KIR3DX2, up to exon 5 that encodes the third extracellular Ig-like domain, more species-specific variation is observed. Taking the rhesus macaque sequence as most well-defined reference, the gibbon KIR3DX2 only shows 73.1% and 35.9% similarity at the exons and genomic level, respectively. Cynomolgus macaques, which shares a relatively close common ancestor with the rhesus macaques approximately 1-3 mya, have almost an identical KIR3DX2 gene in the coding (99.9%) and genomic (99.5%) sequence regions. In marmosets, the KIR3DX2 gene largely deviated at the genomic DNA level, with 57.5% similarity to rhesus macaques, whereas the exon sequences were more conserved (80.4%).
The most likely explanation for an additional KIR3DX gene in lesser apes, OWM and NWM is a duplication of KIR3DX1 in a common ancestor that is lost during radiation towards human and great apes. The KIR3DX2 sequence in gibbons is, however, far from similar to KIR3DX1 in chimpanzees (59.4% on exons and 40.6% on gDNA). In contrast, the coding regions of KIR3DX2 in both macaque species and in the common marmoset seem to be more conserved, with 83.7/83.8% and 85.7% sequence similarity to the coding sequence of KIR3DX1 in chimpanzees, respectively. This substantiates an ancient KIR3DX1 duplication event, which remained more conserved in OWM, while the duplicated gene diverged during evolution in gibbons.
In humans, LILR receptors are expressed both on myeloid and lymphoid immune cells, including monocytes, B and T lymphocytes, natural killer (NK) cells, neutrophils, and dendritic cells (DC), and it is generally accepted that they control a variety of immune responses and maintain homeostasis (54, 59, 87, 88). Each LILR receptor is expressed on a unique repertoire of cell populations (Table 1). Inhibitory LILR receptors probably function as immune checkpoints by screening and eliminating manipulated immune cells, which lack HLA class I expression, as depicted in the “missing self” hypothesis (19, 59, 107). For example, LILRB1 interaction with MHC class I molecules may regulate cell phenotype and function (108). Several immune checkpoint receptors are present in the human immune system, including programmed cell death protein-1 (PD-1). PD-1 is not a member of the IgSF superfamily, but the intracellular signal transduction is equal to inhibitory LILR receptors, resulting in negative regulation of the immune system. Furthermore, the inhibitory receptor LILRB2 regulates neuronal functions such as axonal regeneration and synaptic plasticity (57–59). Interaction between LILRB2 and ANGPTL molecules might play a role in angiogenesis, although its precise role is at present poorly understood (56). Both LILRB1 and LILRB2 interact with HLA-G, a non-classical MHC class I molecule, resulting in a dominant immunosuppressive effect that plays a role in pregnancy as well as in transplant tolerance, infection, and cancer (6, 109–111).
Activating and inhibitory LILR receptors consist of two or four extracellular Ig-like domains (Figure 7A) (33). In addition, LILR receptors are classified based on amino acid sequence similarity in the ligand binding sites, distinguishing class I and II type receptors (Table 1). Class I includes LILRA1, LILRA2, LILRA3, LILRB1, and LILRB2, and interacts with classical and non-classical HLA class I molecules (88). Class II includes LILRA4, LILRA5, LILRA6, LILRB3, LILRB4, and LILRB5, and seems to have ligands other than HLA molecules. An exception is formed by LILRB5, which interacts with ANGPTL, but binding to HLA class I heavy chains was recently reported as well (88, 113). As is known so far, class II receptors appear to interact with one or two specific ligands, like bone marrow stromal antigen 2 (BST2/CD317) and an activated leukocyte cell adhesion molecule (ALCAM/CD166), while class I receptors can interact with a broader repertoire of HLA-ligands (35, 88, 114–116). Caution should be exercised in this regard, however, because the ligands of LILRA5, LILRA6, and LILRB3 are unknown at present.
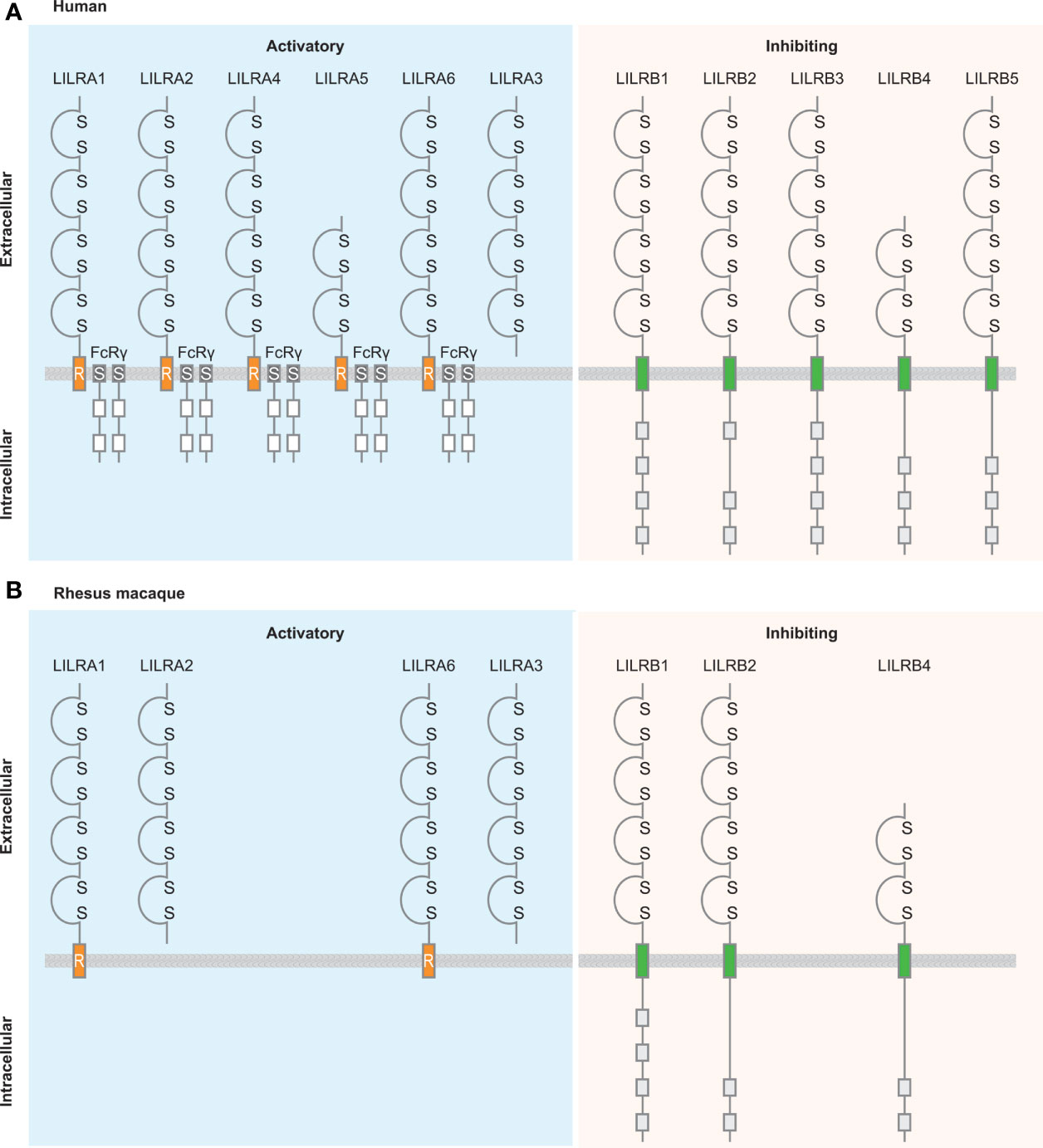
Figure 7 Schematic illustration of the structure of LILR family members in human and rhesus macaque. A structural comparison between LILR family members in humans (A) and rhesus macaques (B) (112). LILR receptors are classified based on the presence or absence of the transmembrane region and the cytoplasmic tail. Activating receptors associate with the γ-chain of Fc receptors, while the inhibitory receptors contain cytoplasmic immunoreceptor tyrosine-based inhibition motifs. Based on the conserved nature of the LILRA receptors it is plausible that also in rhesus macaque these types of receptors associate with FcRγ, however, because this is not officially documented it is not illustrated in the figure. LILRA3 is a soluble receptor lacking the transmembrane region and cytoplasmic tail. Ig-like domains are indicated by a domain structure containing disulfide bridges; transmembrane region is indicated by orange boxes including an R, which refers to the presence of the positively charged arginine residue, for the activating LILR receptors and by green boxes for the inhibitory LILR receptors; Fc receptor transmembrane by dark grey boxes containing an S; immunoreceptor tyrosine-based activation motifs by white boxes; and immunoreceptor tyrosine-based inhibition motifs by light-grey boxes.
Activating LILR receptors do not possess a cytoplasmic tail that has the capacity to transduce a signal and therefore associate with the γ-chain of Fc receptors via a positively charged arginine residue in the transmembrane region of the LILRA receptor (114, 117). Immunoreceptor tyrosine-based activation motifs (ITAM) located in the γ-chain of Fc receptors are phosphorylated, and activate downstream activation pathways (118). Inhibitory LILR receptors have a cytoplasmic tail containing three or four immunoreceptor tyrosine-based inhibitory motifs (ITIM), which downregulate cell activation by recruiting phosphatases of the Src kinase family (119–121). Phosphorylated ITIM motifs serve as docking sites for other enzymes such as Src homology region 2 domain-containing phosphatase-1 (SHP-1), Src homology region 2 domain-containing phosphatase-2 (SHP-2), and Src homology 2 domain containing inositol polyphosphate 5-phosphatase 1 (SHIP), resulting in the negative regulation of immune cell activation. The fine balance between activating and inhibitory LILR receptors is necessary for the modulation of immune responses and the maintenance of immune cell homeostasis.
To date, nine chimpanzee LILR genes have been thoroughly characterized, of which eight seem to encode for a functional protein, seven with four Ig-like domains and one with two Ig-like domains (78). As expected, the position of the cysteine residue essential for disulfide bridge formation in the domains is conserved between humans and chimpanzees. Comparable with humans, the activating LILR receptors in chimpanzees contain an arginine residue in the transmembrane region, while the inhibitory LILR receptors in chimpanzees have a cytoplasmic tail containing three or four ITIM motifs.
In rhesus macaques, LILR receptors comprise the same characteristics as human LILR receptors, including two or four Ig-like domains, the presence of a positively charged arginine residue in the transmembrane region of activating LILR, and the intracellular region of inhibitory LILR containing two or four ITIM motifs (Figure 7B) (112).
As far as we know, ligand-binding studies have not been conducted for chimpanzee and rhesus macaques. LILR/MHC dynamics was, however, studied recently in HIV infection in humans and compared to an early SIV-infection model in cynomolgus macaques (122). A monoclonal antibody specific for the extracellular part of human LILRB2 showed cross-reactivity with cynomolgus macaque cells. In humans and cynomolgus macaques, the expression of LILRB2 and a LILRB2-like protein, respectively, was shown on a similar immune cell subset, which included monocytes, classical DC, plasmacytoid DC, and polymorphonuclear leucocytes. Overall, the data illustrated that during a SIV/HIV-infection the cynomolgus macaques LILRB2-like protein seems to negatively regulate the same immune cell population as human LILRB2. So far, MHC class I is the only known ligand engaging with cynomolgus macaque LILRB2.
Considering the conserved LILR function in humans, and the structural similarities reported for chimpanzees and rhesus macaques, it is tempting to speculate that NHP LILR receptors engage with MHC equivalents of HLA ligands (45, 78, 112, 122).
In healthy individuals and during pregnancy, the modulation of immune activity is tightly regulated by a complex mechanism that involves different members of the HLA family and multiple regulatory gene systems, including LILR, KIR, and NKG2. Pregnancy is an exquisite example of a balanced immune regulation and adaption to protect the embryo against an unintended maternal immune response. Expression of HLA-A and -B is limited during pregnancy to avoid alloreactivity by B and T cells, whereas classical HLA-C and non-classical HLA-E and -G are expressed on fetal trophoblasts (123, 124). During the early stages of pregnancy, a distinctive subset of uterine NK (uNK) cells is involved in placental formation. Different combinations of KIR and HLA-C allotypes regulate these uNK cells, in which the extensive genetic variation of both gene systems associates with successful pregnancy or with complications, such as recurrent miscarriage (125). The role of the KIR gene family is reflected by interactions of KIR2DL4 with soluble HLA-G, which has been described (126, 127). In short, these interactions probably modulate the production of cytokines and chemokines to promote vascular remodeling in early pregnancy (128). Activating receptors NKG2D, as well as DNAM-1- and NKp44 mediate the regulation of NK cell and modulate NK cell activity (126, 129). HLA-G expression is restricted to trophoblast cells and might also be recognized by LILRB1, which is highly expressed on NK cells found in the maternal decidua, and by LILRB2, which is expressed on maternal decidual macrophages (124, 130, 131). These mechanisms protect the embryo against NK- directed cell lysis (131). Furthermore, multiple LILR receptors are described as beneficial, resulting in individuals with a protective phenotype against multiple sclerosis, or in individuals who can control virus infections such as HIV-1 (132, 133). However, LILR receptors might play a negative role as well, and are associated with the outcome of several diseases. In some diseases, LILR receptors can be regarded as a genetic risk factor (Table 2). Therefore, LILR receptors might be useful as diagnostic markers and a target for immunotherapies (58, 59). The role of LILRs in different kinds of diseases is briefly discussed in the following paragraphs. We would like to emphasize, however, that these mainly concern diseases with an immunological component that we have highlighted here exclusively in the context of LILR.
Rheumatoid arthritis patients abundantly express LILRA2, LILRA3, LILRA5, and LILRB2, with the presence of LILRA2, LILRA5, and LILRB2 significantly correlating with disease activity (95, 134, 145). The underlying mechanisms are not yet fully understood, but it is postulated that disrupted gene expression may contribute to an excessive inflammatory immune response. In addition, insufficient inhibitory signaling as a result of single nucleotide polymorphisms (SNP) in the promotor region or of post-transcriptional regulation might contribute to rheumatoid arthritis susceptibility (147). LILRA3 is identified as a genetic risk factor for rheumatoid arthritis, systemic lupus erythematosus, and Sjogren’s syndrome (137–139). In systemic lupus erythematosus patients, disrupted LILRB1 expression and/or deficient inhibitory signaling is observed (148). This observation is comparable with the postulated cause of the excessive inflammatory immune response in rheumatoid arthritis patients. In addition, polymorphisms may impact disease association. Polymorphisms found in LILRB4 resulted in a loss of function, and, as a consequence, increased inflammatory cytokine levels in systemic lupus erythematosus were observed (162). LILRA2 splice site polymorphism affects alternative splicing, resulting in a different isoform, which is associated with systemic lupus erythematosus and microscopic polyangiitis (135).
Likewise, several LILR disease associations are reported in chronic disorders, and the majority of these associations are genetic. In a cohort of family-related atopic disease patients, a single copy of LILRA6 might be related to the development of the disease (146). LILRA3 and LILRB3 are both identified as susceptibility genes in Takayasu’s arteritis (161). Furthermore, LILRA3 is associated with ankylosing spondylitis susceptibility in different cohorts, including Han Chinese subpopulations and a Polish population, underlying the genetic differences between different ethnicities (140, 149). Associations involving disruptive gene expression are seldom reported. LILRA3 expression is increased in intestinal bowel disease patients, probably resulting in suppression of the anti-inflammatory immune response (141).
Human LILRB2 and its murine ortholog PirB interact with soluble β-amyloid, leading to enhanced cofilin signaling, which is observed in the brains of humans with Alzheimer’s disease (58, 165). In a transgenic mice Alzheimer’s disease model, memory deficits in adult mice are caused by PirB deficiency, which results in the loss of synaptic plasticity in the juvenile visual cortex. It is postulated that due to the orthologous relationship between LILRB2 and PirB, LILRB2 may contribute to Alzheimer’s disease neuropathology, and might be a suitable therapeutic target. The LILRA3 null haplotype might increase the risk of relapsing multiple sclerosis in Spanish patients (133). These findings were confirmed in German and French multiple sclerosis patients of Caucasian descent (142). In contrast, in a Polish cohort, no association for disease susceptibility was found, but it was shown instead that a LILRA3 deletion is associated with the later onset of multiple sclerosis (143). In multiple sclerosis patients, LILRB2 and HLA-G are co-expressed on central nervous system cells and in areas with microglia activation, while HLA-G expression is barely detectable in healthy controls (166). LILRB2 and HLA-G play a role in immune reactivity in the central nervous system, which might act as an inhibitory feedback mechanism to downregulate the damaging effect of T-cell infiltration in neuroinflammation.
The immune response to bacteria often results in increased expression of LILR receptors on the cell surface. For example, in lepromatous patients, LILRA2 is upregulated in the lesions, and suppresses the innate host immune response by shifting cytokine production from interleukin-12 (IL-12) toward IL-10 (136). LILRB1 expression is elevated on CD56dimCD16+ NK cells during active pulmonary tuberculosis (150). It is postulated that CD56dimCD16+ NK cells correlate with the disease severity of pulmonary tuberculosis, because CD56dimLILRB1+ NK cells are not capable of eliminating infected cells. Also, during Salmonella infection, LILRB2 and LILRB4 are upregulated, which results in the expansion of tolerogenic antigen-presenting cells owing to an insufficient response to toll-like receptor signaling (158).
Viruses developed other strategies to dysregulate the host immune response by abusing immune receptors. The most studied viral infection with regard to LILR receptors is cytomegalovirus (CMV) infection, which expresses UL18, an MHC class I homolog, on infected cells. The engagement of UL18 and LILRB1 may inhibit the clearance of CMV-infected cells, and, therefore, CMV might escape the innate immune response (99). During the adaptive immune response, CMV-infected cells expressing UL18 are lysed by CD8+ T cells, while CMV-infected cells lacking UL18 are not eliminated, and therefore CMV might escape this host immune response as well (151). Recurrent CMV infection or deficient immune response is frequently observed in transplant patients. Another example was observed during a dengue infection, where LILRB1 was shown to engage with an unknown dengue virus-related ligand, resulting in the obstruction of FcγR activation and allowing host cell entrance and viral replication (152, 153). Recently, an association with LILRB1 and malaria was observed. Plasmodium falciparum, the causative agent of malaria, produces repetitive interspersed family (RIFIN) proteins, which are displayed on infected erythrocytes (167). Some RIFINs interact with LILRB1, which could potentially result in tempering the host immune response by suppressing NK cell function response (154).
In chronic lymphocytic leukemia, a significant increase of LILRB1 expression is detected on NK cells, resulting in a lack of elimination of leukemic cells (156). In acute myeloid leukemia, LILRB4 is expressed on monocytic leukemia cells, generating an immunosuppressive microenvironment contributing to the infiltration of other tissues, including the central nervous system (163, 164). Co-expression of LILRB2 and HLA-G is observed in tissues of human primary colorectal cancer, while different expression patterns of LILRB1 and LILRB4 are observed in gastric cancer patients (157, 160). Differential expression may contribute to the proliferation, migration, and invasion of tumor cells. In addition, genetic risk factors have been reported in different types of cancer. A genome-wide association study in Chinese men revealed that LILRA3 SNP rs103294 and LILRB1 SNP rs16985478 may be a risk factor for prostate cancer and non-small cell lung cancer, respectively (144, 155).
In this communication, we provided an overview on the genomic organization of the LILR region in primates with which we illustrate that the LILR region remained largely conserved throughout primate evolution (Figure 5). Minor differences in gene content were observed, but at this stage it is not clear whether allelic variation influences the complexity of the system. Further research is necessary to arrive at a solid conclusion. By comparing channel catfish, chicken, opossum, primates, mice, cattle, goat, and pig, we estimated that the LILR gene family likely emerged more than 450 mya, probably in the same time frame as the MHC system. It is thought that the evolution of the MHC system influenced KIR gene evolution (and vice versa), but it is not evident whether it influenced LILR gene evolution as well (36, 49, 55, 168). Some LILR receptors engage with the highly conserved α3-domain of MHC class I molecules and the β2m subunit, suggesting that the main function of LILR receptors is immune surveillance by scanning immune cells for the presence or absence of MHC class I. More sophisticated systems, like that of the KIR genes, appeared later in evolution, and are able to scan for the presence of polymorphic epitopes on MHC class I molecules. In humans, KIR3DX1 is classified as a pseudogene, while KIR3DX1 in the non-human primates is seen to code for a functional gene product. A duplication of KIR3DX1 was observed in the lesser apes, OWMs, and NWMs, suggesting that this duplication might be present in the ancestor and was lost in great apes and humans. As far as we know, the function and ligand of KIR3DX1 is not yet resolved. At last, LILR receptors are placed in context for the role they may play in health and disease. It is tempting to speculate that old genes are frequently associated with diseases. However, one would expect that serious disease associations linked to old genes have been weeded out during evolution. The other issue is that disease association in a highly conserved region with limited levels are hard to pick up due to linkage phenomena. Since the LILR region in primates is remarkably conserved, non-human primates are an excellent tool to thoroughly study the functional aspects of LILR genes. This type of undertaking might enhance the available non-human primate disease models in order to improve the health both of humans and animals.
LS drafted the manuscript. JB, NG, and RB edited the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank D. Devine for editing the manuscript and F. van Hassel for preparing the figures.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.716289/full#supplementary-material
1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial Sequencing and Analysis of the Human Genome. Nature (2001) 409(6822):860–921. doi: 10.1038/35057062
2. Volz A, Wende H, Laun K, Ziegler A. Genesis of the ILT/LIR/MIR Clusters Within the Human Leukocyte Receptor Complex. Immunol Rev (2001) 181(1):39–51. doi: 10.1034/j.1600-065x.2001.1810103.x
3. Martin AM, Freitas EM, Witt CS, Christiansen FT. The Genomic Organization and Evolution of the Natural Killer Immunoglobulin-Like Receptor (KIR) Gene Cluster. Immunogenetics (2000) 51(4-5):268–80. doi: 10.1007/s002510050620
4. Robert Liu W, Kim J, Nwankwo C, Ashwort L, Arm J. Genomic Organization of the Human Leukocyte Immunoglobulin-Like Receptors Within the Leukocyte Receptor Complex on Chromosome 19q13.4. Immunogenetics (2000) 51(8-9):659–69. doi: 10.1007/s002510000183
5. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the Organization and Sequences of Human KIR/ILT Gene Families. Proc Natl Acad Sci USA (2000) 97(9):4778–83. doi: 10.1073/pnas.080588597
6. Barrow AD, Trowsdale J. The Extended Human Leukocyte Receptor Complex: Diverse Ways of Modulating Immune Responses. Immunol Rev (2008) 224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x
7. Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The Genomic Context of Natural Killer Receptor Extended Gene Families. Immunol Rev (2001) 181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x
8. Vojvodic S, Ademovic-Sazdanic D. KIR and HLA Haplotype Analysis in a Family Lacking the KIR 2DL1-2DP1 Genes. Balkan J Med Genet (2015) 18(1):55–64. doi: 10.1515/bjmg-2015-0006
9. Young NT, Canavez F, Uhrberg M, Shum BP, Parham P. Conserved Organization of the ILT/LIR Gene Family Within the Polymorphic Human Leukocyte Receptor Complex. Immunogenetics (2001) 53(4):270–8. doi: 10.1007/s002510100332
10. Vendelbosch S, de Boer M, van Leeuwen K, Pourfarzad F, Geissler J, van den Berg TK, et al. Novel Insights in the Genomic Organization and Hotspots of Recombination in the Human KIR Locus Through Analysis of Intergenic Regions. Genes Immun (2015) 16(2):103–11. doi: 10.1038/gene.2014.68
11. Roe D, Vierra-Green C, Pyo CW, Eng K, Hall R, Kuang R, et al. Revealing Complete Complex KIR Haplotypes Phased by Long-Read Sequencing Technology. Genes Immun (2017) 18(3):127–34. doi: 10.1038/gene.2017.10
12. Bruijnesteijn J, van der Wiel MKH, Swelsen WTN, Otting N, de Vos-Rouweler AJM, Elferink D, et al. Human and Rhesus Macaque KIR Haplotypes Defined by Their Transcriptomes. J Immunol (2018) 200(5):1692–701. doi: 10.4049/jimmunol.1701480
13. Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting Edge: Expansion of the KIR Locus by Unequal Crossing Over. J Immunol (2003) 171(5):2192–5. doi: 10.4049/jimmunol.171.5.2192
14. Valiante NM, Lienert K, Shilling HG, Smits BJ, Parham P. Killer Cell Receptors: Keeping Pace With MHC Class I Evolution. Immunol Rev (1997) 155:155–64. doi: 10.1111/j.1600-065x.1997.tb00948.x
15. Huard B, Karlsson L. KIR Expression on Self-Reactive CD8+T Cells is Controlledby T-Cell Receptor Engagement. Nature (2000) 403(6767):325–8. doi: 10.1038/35002105
16. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-Specific Evolution of Killer Cell Immunoglobulin-Like Receptor Recognition of Major Histocompatibility Complex Class I Molecules. Philos Trans R Soc Lond B Biol Sci (2012) 367(1590):800–11. doi: 10.1098/rstb.2011.0266
17. Parham P, Ohta T. Population Biology of Antigen Presentation by MHC Class I Molecules. Science (1996) 272(5258):67–74. doi: 10.1126/science.272.5258.67
18. Hilton HG, Parham P. Missing or Altered Self: Human NK Cell Receptors That Recognize HLA-C. Immunogenetics (2017) 69(8-9):567–79. doi: 10.1007/s00251-017-1001-y
19. Ljunggren H, Karre K. In Search of the ‘Missing Self’ MHC Molecules and NK Cell Recognition. Immunol Today (1990) 11(7):237–44. doi: 10.1016/0167-5699(90)90097-s
20. Verbrugge A, Ruiter Td T, Clevers H, Meyaard L. Differential Contribution of the Immunoreceptor Tyrosine-Based Inhibitory Motifs of Human Leukocyte-Associated Ig-Like Receptor-1 to Inhibitory Function and Phosphatase Recruitment. Int Immunol (2003) 15(11):1349–58. doi: 10.1093/intimm/dxg134
21. Lebbink RJ, van den Berg MC, de Ruiter T, Raynal N, van Roon JA, Lenting PJ, et al. The Soluble Leukocyte-Associated Ig-Like Receptor (LAIR)-2 Antagonizes the Collagen/LAIR-1 Inhibitory Immune Interaction. J Immunol (2008) 180(3):1662–9. doi: 10.4049/jimmunol.180.3.1662
22. Meyaard L, Adema GJ, Chang C, Woollatt E, Sutherland GR, Lanier LL, et al. LAIR-1, a Novel Inhibitory Receptor Expressed on Human Mononuclear Leukocytes. Immunity (1997) 7(2):283–90. doi: 10.1016/s1074-7613(00)80530-0
23. Jin J, Wang Y, Ma Q, Wang N, Guo W, Jin B, et al. LAIR-1 Activation Inhibits Inflammatory Macrophage Phenotype In Vitro. Cell Immunol (2018) 331:78–84. doi: 10.1016/j.cellimm.2018.05.011
24. Meyaard L. LAIR And Collagens in Immune Regulation. Immunol Lett (2010) 128(1):26–8. doi: 10.1016/j.imlet.2009.09.014
25. Verbrugge A, de Ruiter T, Geest C, Coffer PJ, Meyaard L. Differential Expression of Leukocyte-Associated Ig-Like Receptor-1 During Neutrophil Differentiation and Activation. J Leukoc Biol (2006) 79(4):828–36. doi: 10.1189/jlb.0705370
26. Lebbink RJ, de Ruiter T, Adelmeijer J, Brenkman AB, van Helvoort JM, Koch M, et al. Collagens are Functional, High Affinity Ligands for the Inhibitory Immune Receptor LAIR-1. J Exp Med (2006) 203(6):1419–25. doi: 10.1084/jem.20052554
27. Lebbink RJ, Raynal N, de Ruiter T, Bihan DG, Farndale RW, Meyaard L. Identification of Multiple Potent Binding Sites for Human Leukocyte Associated Ig-Like Receptor LAIR on Collagens II and III. Matrix Biol (2009) 28(4):202–10. doi: 10.1016/j.matbio.2009.03.005
28. Carvalheiro T, Garcia S, Pascoal Ramos MI, Giovannone B, Radstake T, Marut W, et al. Leukocyte Associated Immunoglobulin Like Receptor 1 Regulation and Function on Monocytes and Dendritic Cells During Inflammation. Front Immunol (2020) 11:1793. doi: 10.3389/fimmu.2020.01793
29. Martinez-Esparza M, Ruiz-Alcaraz AJ, Carmona-Martinez V, Fernandez-Fernandez MD, Anton G, Munoz-Tornero M, et al. Expression of LAIR-1 (CD305) on Human Blood Monocytes as a Marker of Hepatic Cirrhosis Progression. J Immunol Res (2019) 2019:2974753. doi: 10.1155/2019/2974753
30. Middleton D, Gonzelez F. The Extensive Polymorphism of KIR Genes. Immunology (2010) 129(1):8–19. doi: 10.1111/j.1365-2567.2009.03208.x
31. Wende H, Volz A, Ziegler A. Extensive Gene Duplications and a Large Inversion Characterize the Human Leukocyte Receptor Cluster. Immunogenetics (2000) 51(8-9):703–13. doi: 10.1007/s002510000187
32. Torkar M, Norgate Z, Colonna M, Trowsdale J, Wilson MJ. Isotypic Variation of Novel Immunoglobulin-Like Transcript:Killer Cel Inhibitory Receptor Loci in the Leukocyte Receptor Complex. Eur J Immunol (1998) 28:3959–67. doi: 10.1002/(SICI)1521-4141(199812)28:12<3959::AID-IMMU3959>3.0.CO;2-2
33. Brown D, Trowsdale J, Allen R. The LILR Family: Modulators of Innate and Adaptive Immune Pathways in Health and Disease. Tissue Antigens (2004) 64(3):215–25. doi: 10.1111/j.0001-2815.2004.00290.x
34. Hirayasu K, Arase H. Functional and Genetic Diversity of Leukocyte Immunoglobulin-Like Receptor and Implication for Disease Associations. J Hum Genet (2015) 60(11):703–8. doi: 10.1038/jhg.2015.64
35. Burshtyn DN, Morcos C. The Expanding Spectrum of Ligands for Leukocyte Ig-Like Receptors. J Immunol (2016) 196(3):947–55. doi: 10.4049/jimmunol.1501937
36. Wroblewski EE, Parham P, Guethlein LA. Two to Tango: Co-Evolution of Hominid Natural Killer Cell Receptors and MHC. Front Immunol (2019) 10:177. doi: 10.3389/fimmu.2019.00177
37. Guethlein LA, Norman PJ, Hilton HG, Parham P. Co-Evolution of MHC Class I and Variable NK Cell Receptors in Placental Mammals. Immunol Rev (2015) 267(1):259–82. doi: 10.1111/imr.12326
38. Augusto DG, Norman PJ, Dandekar R, Hollenbach JA. Fluctuating and Geographically Specific Selection Characterize Rapid Evolution of the Human KIR Region. Front Immunol (2019) 10:989. doi: 10.3389/fimmu.2019.00989
39. Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-Like Receptor Complex (LRC) in Mice and Men. Trends Immunol (2002) 23(2):81–8. doi: 10.1016/s1471-4906(01)02155-x
40. Nikolaidis N, Makalowska I, Chalkia D, Makalowski W, Klein J, Nei M. Origin and Evolution of the Chicken Leukocyte Receptor Complex. Proc Natl Acad Sci USA (2005) 102(11):4057–62. doi: 10.1073/pnas.0501040102
41. Schwartz JC, Hammond JA. The Unique Evolution of the Pig LRC, a Single KIR But Expansion of LILR and a Novel Ig Receptor Family. Immunogenetics (2018) 70(10):661–9. doi: 10.1007/s00251-018-1067-1
42. Schwartz JC, Sanderson ND, Bickhart DM, Smith TPL, Hammond JA. The Structure, Evolution, and Gene Expression Within the Caprine Leukocyte Receptor Complex. Front Immunol (2019) 10:2302. doi: 10.3389/fimmu.2019.02302
43. Hogan L, Bhuju S, Jones DC, Laing K, Trowsdale J, Butcher P, et al. Characterisation of Bovine Leukocyte Ig-Like Receptors. PLoS One (2012) 7(4):e34291. doi: 10.1371/journal.pone.0034291
44. Kumar S, Hedges SB. A Molecular Timescale for Vertebrate Evolution. Nature (1998) 392(6679):917–20. doi: 10.1038/31927
45. Takai T, Ono M. Activating and Inhibitory Nature of the Murine Paired Immunoglobulin-Like Receptor Family. Immunol Rev (2001) 181:215–22. doi: 10.1034/j.1600-065x.2001.1810118.x
46. Belov K, Sanderson C, Deakin J, Wong E, Assange D, McColl K, et al. Characterization of the Opossum Immune Genome Provides Insights Into the Evolution of the Mammalian Immune System. Genome Res (2007) 17(7):982–91. doi: 10.1101/gr.6121807
47. Laun K, Coggill P, Palmer S, Sims S, Ning Z, Ragoussis J, et al. The Leukocyte Receptor Complex in Chicken is Characterized by Massive Expansion and Diversification of Immunoglobulin-Like Loci. PLoS Genet (2006) 2(5):e73. doi: 10.1371/journal.pgen.0020073
48. Magor KE, Miranzo Navarro D, Barber MR, Petkau K, Fleming-Canepa X, Blyth GA, et al. Defense Genes Missing From the Flight Division. Dev Comp Immunol (2013) 41(3):377–88. doi: 10.1016/j.dci.2013.04.010
49. Stafford JL, Bengten E, Du Pasquier L, Miller NW, Wilson M. Channel Catfish Leukocyte Immune-Type Receptors Contain a Putative MHC Class I Binding Site. Immunogenetics (2007) 59(1):77–91. doi: 10.1007/s00251-006-0169-3
50. Launay P, Lehuen A, Kawakami T, Blank U, Monteiro R. IgA Fc Receptor (CD89) Activation Enables Coupling to Syk and Btk Tyrosine Kinase Pathways Differential Signaling After IFN-G or Phorbol Ester Stimulation. J Leuk Biol (1998) 63(5):636–42. doi: 10.1002/jlb.63.5.636
51. Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, et al. Molecular Cloning of NKp46 A Novel Memberof the Immunoglobulin Superfamily Involved in Triggering of Natural Cytotoxicity. J Exp Med (1998) 188(5):953–60. doi: 10.1084/jem.188.5.953
52. Middleton D, Curran M, Maxwell L. Natural Killer Cells and Their Receptors. Transplant Immunol (2002) 10(2–3):147–64. doi: 10.1016/s0966-3274(02)00062-x
53. Jones DC, Kosmoliaptsis V, Apps R, Lapaque N, Smith I, Kono A, et al. HLA Class I Allelic Sequence and Conformation Regulate Leukocyte Ig-Like Receptor Binding. J Immunol (2011) 186(5):2990–7. doi: 10.4049/jimmunol.1003078
54. Allen RL, Raine T, Haude A, Trowsdale J, Wilson MJ. Leukocyte Receptor Complex-Encoded Immunomodulatory Receptors Show Differing Specificity for Alternative HLA-B27 Structures. J Immunol (2001) 167(10):5543–7. doi: 10.4049/jimmunol.167.10.5543
55. Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, et al. Identification of the Ancestral Killer Immunoglobulin-Like Receptor Gene in Primates. BMC Genomics (2006) 7:209. doi: 10.1186/1471-2164-7-209
56. Zheng J, Umikawa M, Cui C, Li J, Chen X, Zhang C, et al. Inhibitory Receptors Bind ANGPTLs and Support Blood Stem Cells and Leukaemia Development. Nature (2012) 485(7400):656–60. doi: 10.1038/nature11095
57. Cao Q, Shin WS, Chan H, Vuong CK, Dubois B, Li B, et al. Inhibiting Amyloid-Beta Cytotoxicity Through its Interaction With the Cell Surface Receptor LilrB2 by Structure-Based Design. Nat Chem (2018) 10(12):1213–21. doi: 10.1038/s41557-018-0147-z
58. Kim T, Vidal GS, Djurisic M, William CM, Birnbaum ME, Garcia KC, et al. Human LilrB2 is a Beta-Amyloid Receptor and its Murine Homolog PirB Regulates Synaptic Plasticity in an Alzheimer’s Model. Science (2013) 341(6152):1399–404. doi: 10.1126/science.1242077
59. Takeda K, Nakamura A. Regulation of Immune and Neural Function via Leukocyte Ig-Like Receptors. J Biochem (2017) 162(2):73–80. doi: 10.1093/jb/mvx036
60. Torkar M, Haude A, Milne S, Beck S, Trowsdale J, Wilson MJ. Arrangement of the ILT Gene Cluster: A Common Null Allele of the ILT6 Gene Results From a 6.7-Kbp Deletion. Eur J Immunol (2000) 30(12):3655–62. doi: 10.1002/1521-4141(200012)30:12<3655::AID-IMMU3655>3.0.CO;2-Y
61. Hirayasu K, Ohashi J, Tanaka H, Kashiwase K, Ogawa A, Takanashi M, et al. Evidence for Natural Selection on Leukocyte Immunoglobulin-Like Receptors for HLA Class I in Northeast Asians. Am J Hum Genet (2008) 82(5):1075–83. doi: 10.1016/j.ajhg.2008.03.012
62. Norman PJ, Carey BS, Stephens HAF, Vaughan RW. DNA Sequence Variation and Molecular Genotyping of Natural Killer Leukocyte Immunoglobulin-Like Receptor, LILRA3. Immunogenetics (2003) 55(3):165–71. doi: 10.1007/s00251-003-0561-1
63. Norman PJ, Cook MA, Carey BS, Carrington CV, Verity DH, Hameed K, et al. SNP Haplotypes and Allele Frequencies Show Evidence for Disruptive and Balancing Selection in the Human Leukocyte Receptor Complex. Immunogenetics (2004) 56(4):225–37. doi: 10.1007/s00251-004-0674-1
64. Hsu KC, Chida S, Geraghty DE, Dupont B. The Killer Cell Immunoglobulin-Like Receptor (KIR) Genomic Region: Gene-Order, Haplotypes and Allelic Polymorphism. Immunol Rev (2002) 190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x
65. Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, et al. A Novel Immunoglobulin Superfamily Receptor for Cellular and Viral MHC Class I Molecules. Immunity (1997) 7(2):273–82. doi: 10.1016/s1074-7613(00)80529-4
66. Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, et al. A Common Inhibitory Receptor for Major Histocompatibility Complex Class I Molecules on Human Lymphoid and Myelomonocytic Cells. J Exp Med (1997) 186(11):1809–18. doi: 10.1084/jem.186.11.1809
67. Wain HM, Lush MJ, Ducluzeau F, Khodiyar VK, Povey S. Genew: The Human Gene Nomenclature Database, 2004 Updates. Nucleic Acids Res (2004) 32(Database issue):D255–7. doi: 10.1093/nar/gkh072
68. Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-Cell Immunoglobulin-Like Receptor (KIR) Nomenclature Report, 2002. Immunogenetics (2003) 55(4):220–6. doi: 10.1007/s00251-003-0571-z
69. Robinson J, Guethlein LA, Maccari G, Blokhuis J, Bimber BN, de Groot NG, et al. Nomenclature for the KIR of non-Human Species. Immunogenetics (2018) 70(9):571–83. doi: 10.1007/s00251-018-1064-4
70. Bruijnesteijn J, de Groot NG, Otting N, Maccari G, Guethlein LA, Robinson J, et al. Nomenclature Report for Killer-Cell Immunoglobulin-Like Receptors (KIR) in Macaque Species: New Genes/Alleles, Renaming Recombinant Entities and IPD-NHKIR Updates. Immunogenetics (2020) 72(1-2):37–47. doi: 10.1007/s00251-019-01135-8
71. de Groot N, Blokhuis J, Otting N, Doxiadis G, Bontrop R. Co-Evolution of the MHC Class I and KIR Gene Families in Rhesusmacaques Ancestry and Plasticity. Immunol Rev (2015) 267:228–45. doi: 10.1111/imr.12313
72. Fujiyama A, Watanabe H, Toyoda A, Taylor TD, Itoh T, Tsai SF, et al. Construction and Analysis of a Human-Chimpanzee Comparative Clone Map. Science (2002) 295(5552):131–4. doi: 10.1126/science.1065199
73. Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, et al. Comparative and Demographic Analysis of Orang-Utan Genomes. Nature (2011) 469(7331):529–33. doi: 10.1038/nature09687
74. Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, et al. Insights Into Hominid Evolution From the Gorilla Genome Sequence. Nature (2012) 483(7388):169–75. doi: 10.1038/nature10842
75. Glazko GV, Nei M. Estimation of Divergence Times for Major Lineages of Primate Species. Mol Biol Evol (2003) 20(3):424–34. doi: 10.1093/molbev/msg050
76. Coordinators NR. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res (2018) 46(D1):D8–D13. doi: 10.1093/nar/gkx1095
77. Yates A, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res (2020) 48(D1):D682–D8. doi: 10.1093/nar/gkz966
78. Canavez F, Young NT, Guethlein LA, Rajalingam R, Khakoo SI, Shum BP, et al. Comparison of Chimpanzee and Human Leukocyte Ig-Like Receptor Genes Reveals Framework and Rapidly Evolving Genes. J Immunol (2001) 167(10):5786–94. doi: 10.4049/jimmunol.167.10.5786
79. Dennis G Jr., Kubagawa H, Cooper MD. Paired Ig-Like Receptor Homologs in Birds and Mammals Share a Common Ancestor With Mammalian Fc Receptors. Proc Natl Acad Sci USA (2000) 97(24):13245–50. doi: 10.1073/pnas.230442897
80. Muller S, Hollatz M, Wienberg J. Chromosomal Phylogeny and Evolution of Gibbons (Hylobatidae). Hum Genet (2003) 113(6):493–501. doi: 10.1007/s00439-003-0997-2
81. Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, et al. Reconstruction of Genomic Rearrangements in Great Apes and Gibbons by Chromosome Painting. Proc Natl Acad Sci USA (1992) 89(18):8611–5. doi: 10.1073/pnas.89.18.8611
82. Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, et al. A Molecular Phylogeny of Living Primates. PLoS Genet (2011) 7(3):e1001342. doi: 10.1371/journal.pgen.1001342
83. Sherlock J, Griffin D, Delhanty J, Parringtong J. Homologies Between Human and Marmoset (Callithrix Jacchus) Chromosomes Revealed by Comparative Chromosome Painting. Genomics (1996) 33:214–9. doi: 10.1006/geno.1996.0186
84. Sanderson ND, Norman PJ, Guethlein LA, Ellis SA, Williams C, Breen M, et al. Definition of the Cattle Killer Cell Ig-Like Receptor Gene Family: Comparison With Aurochs and Human Counterparts. J Immunol (2014) 193(12):6016–30. doi: 10.4049/jimmunol.1401980
85. Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The Expanded Cattle KIR Genes are Orthologous to the Conserved Single-Copy KIR3DX1 Gene of Primates. Immunogenetics (2007) 59(6):517–22. doi: 10.1007/s00251-007-0214-x
86. Geneious. Geneious Prime 2020.2.4 . Available at: https://www.geneious.com.
87. Young NT, Waller EC, Patel R, Roghanian A, Austyn JM, Trowsdale J. The Inhibitory Receptor LILRB1 Modulates the Differentiation and Regulatory Potential of Human Dendritic Cells. Blood (2008) 111(6):3090–6. doi: 10.1182/blood-2007-05-089771
88. Willcox BE, Thomas LM, Bjorkman PJ. Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral Major Histocompatibility Complex Receptor. Nat Immunol (2003) 4(9):913–9. doi: 10.1038/ni961
89. Tedla N, Lee CW, Borges L, Geczy CL, Arm JP. Differential Expression of Leukocyte Immunoglobulin-Like Receptors on Cord-Blood-Derived Human Mast Cell Progenitors and Mature Mast Cells. J Leukoc Biol (2008) 83(2):334–43. doi: 10.1189/jlb.0507314
90. Borges L, Hsu M, Fanger N, Kubin M, Cosman D. A Family of Human Lymphoid and Myeloid Ig-Like Receptors, Some of Which Bind to MHC Class I Molecules. J Immunol (1997) 159:5192–6.
91. Tedla N, Bandeira-Melo C, Tassinari P, Sloane DE, Samplaski M, Cosman D, et al. Activation of Human Eosinophils Through Leukocyte Immunoglobulin-Like Receptor 7. Proc Natl Acad Sci USA (2003) 100(3):1174–9. doi: 10.1073/pnas.0337567100
92. Lu HK, Mitchell A, Endoh Y, Hampartzoumian T, Huynh O, Borges L, et al. LILRA2 Selectively Modulates LPS-Mediated Cytokine Production and Inhibits Phagocytosis by Monocytes. PLoS One (2012) 7(3):e33478. doi: 10.1371/journal.pone.0033478
93. Sloane DE, Tedla N, Awoniyi M, Macglashan DW Jr., Borges L, Austen KF, et al. Leukocyte Immunoglobulin-Like Receptors: Novel Innate Receptors for Human Basophil Activation and Inhibition. Blood (2004) 104(9):2832–9. doi: 10.1182/blood-2004-01-0268
94. Lewis Marffy AL, McCarthy AJ. Leukocyte Immunoglobulin-Like Receptors (LILRs) on Human Neutrophils: Modulators of Infection and Immunity. Front Immunol (2020) 11:857. doi: 10.3389/fimmu.2020.00857
95. An H, Chandra V, Piraino B, Borges L, Geczy C, McNeil HP, et al. Soluble LILRA3, a Potential Natural Antiinflammatory Protein, is Increased in Patients With Rheumatoid Arthritis and is Tightly Regulated by Interleukin 10, Tumor Necrosis Factor-Alpha, and Interferon-Gamma. J Rheumatol (2010) 37(8):1596–606. doi: 10.3899/jrheum.091119
96. Ju XS, Hacker C, Scherer B, Redecke V, Berger T, Schuler G, et al. Immunoglobulin-Like Transcripts ILT2, ILT3 and ILT7 are Expressed by Human Dendritic Cells and Down-Regulated Following Activation. Gene (2004) 331:159–64. doi: 10.1016/j.gene.2004.02.018
97. Borges L, Kubin M, Kuhlman T. LIR9, an Immunoglobulin-Superfamily-Activating Receptor, is Expressed as a Transmambrane and as a Secreted Molecule. Blood (2003) 101(4):1484–6. doi: 10.1182/blood-2002-
98. Bashirova AA, Apps R, Vince N, Mochalova Y, Yu XG, Carrington M. Diversity of the Human LILRB3/A6 Locus Encoding a Myeloid Inhibitory and Activating Receptor Pair. Immunogenetics (2014) 66(1):1–8. doi: 10.1007/s00251-013-0730-9
99. Berg L, Riise GC, Cosman D, Bergstrom T, Olofsson S, Karre K, et al. LIR-1 Expression on Lymphocytes, and Cytomegalovirus Disease in Lung-Transplant Recipients. Lancet (2003) 361(9363):1099–101. doi: 10.1016/S0140-6736(03)12855-3
100. Young NT, Uhrberg M, Phillips JH, Lanier LL, Parham P. Differential Expression of Leukocyte Receptor Complex-Encoded Ig-Like Receptors Correlates With the Transition From Effector to Memory CTL. J Immunol (2001) 166(6):3933–41. doi: 10.4049/jimmunol.166.6.3933
101. Inui M, Hirota S, Hirano K, Fujii H, Sugahara-Tobinai A, Ishii T, et al. Human CD43+ B Cells are Closely Related Not Only to Memory B Cells Phenotypically But Also to Plasmablasts Developmentally in Healthy Individuals. Int Immunol (2015) 27(7):345–55. doi: 10.1093/intimm/dxv009
102. Lichterfeld M, Kavanagh DG, Williams KL, Moza B, Mui SK, Miura T, et al. A Viral CTL Escape Mutation Leading to Immunoglobulin-Like Transcript 4-Mediated Functional Inhibition of Myelomonocytic Cells. J Exp Med (2007) 204(12):2813–24. doi: 10.1084/jem.20061865
103. Manavalan JS, Rossi PC, Vlad G, Piazza F, Yarilina A, Cortesini R, et al. High Expression of ILT3 and ILT4 is a General Feature of Tolerogenic Dendritic Cells. Transplant Immunol (2003) 11(3-4):245–58. doi: 10.1016/s0966-3274(03)00058-3
104. Park M, Liu RW, An H, Geczy CL, Thomas PS, Tedla N. A Dual Positive and Negative Regulation of Monocyte Activation by Leukocyte Ig-Like Receptor B4 Depends on the Position of the Tyrosine Residues in its ITIMs. Innate Immun (2017) 23(4):381–91. doi: 10.1177/1753425917699465
105. Cella M, Dohring C, Samaridis J, Dessing M, Brockhaus M, Lanzavecchia A, et al. A Novel Inhibitory Receptor (ILT3) Expressed on Monocytes, Macrophages, and Dendritic Cells Involved in Antigen Processing. J Exp Med (1997) 185(10):1743–51. doi: 10.1084/jem.185.10.1743
106. Hogan LE, Jones DC, Allen RL. Expression of the Innate Immune Receptor LILRB5 on Monocytes is Associated With Mycobacteria Exposure. Sci Rep (2016) 6:21780. doi: 10.1038/srep21780
107. Freeman G, Long A, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 ImmunoinhibitoryReceptor by a Novel B7 Family Member Leadsto Negative Regulation of Lymphocyte Activation. J Exp Med (2000) 192(7):1027–34. doi: 10.1084/jem.192.7.1027
108. Hudson LE, Allen RL. Leukocyte Ig-Like Receptors - A Model for MHC Class I Disease Associations. Front Immunol (2016) 7:281. doi: 10.3389/fimmu.2016.00281
109. Anderson KJ, Allen RL. Regulation of T-Cell Immunity by Leucocyte Immunoglobulin-Like Receptors: Innate Immune Receptors for Self on Antigen-Presenting Cells. Immunology (2009) 127(1):8–17. doi: 10.1111/j.1365-2567.2009.03097.x
110. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A Class I Antigen, HLA-G, Expressed in Human Trophoblasts. Science (1990) 248(4952):220–3. doi: 10.1126/science.2326636
111. Cross JC, Werb Z, Fisher SJ. Implantation and the Placenta: Key Pieces of the Development Puzzle. Science (1994) 266(5190):1508–18. doi: 10.1126/science.7985020
112. Slukvin II, Grendell RL, Rao DS, Hughes AL, Golos TG. Cloning of Rhesus Monkey LILRs. Tissue Antigens (2006) 67(4):331–7. doi: 10.1111/j.1399-0039.2006.00579.x
113. Zhang Z, Hatano H, Shaw J, Olde Nordkamp M, Jiang G, Li D, et al. The Leukocyte Immunoglobulin-Like Receptor Family Member LILRB5 Binds to HLA-Class I Heavy Chains. PLoS One (2015) 10(6):e0129063. doi: 10.1371/journal.pone.0129063
114. Cao W, Bover L. Signaling and Ligand Interaction of ILT7: Receptor-Mediated Regulatory Mechanisms for Plasmacytoid Dendritic Cells. Immunol Rev (2010) 234(1):163–76. doi: 10.1111/j.0105-2896.2009.00867.x
115. Bego MG, Miguet N, Laliberte A, Aschman N, Gerard F, Merakos AA, et al. Activation of the ILT7 Receptor and Plasmacytoid Dendritic Cell Responses are Governed by Structurally-Distinct BST2 Determinants. J Biol Chem (2019) 294(27):10503–18. doi: 10.1074/jbc.RA119.008481
116. Xu Z, Chang CC, Li M, Zhang QY, Vasilescu EM, D’Agati V, et al. ILT3.Fc-CD166 Interaction Induces Inactivation of P70 S6 Kinase and Inhibits Tumor Cell Growth. J Immunol (2018) 200(3):1207–19. doi: 10.4049/jimmunol.1700553
117. Nakajima H, Samaridis J, Angman L, Colonna M. Human Myeloid Cells Epxress an Activating ILT Receptor (ILT1) That Associates With Fc Receptor Y-Chain. J Immunol (1999) 162):5–8.
118. Mocsai A, Ruland J, Tybulewicz VL. The SYK Tyrosine Kinase: A Crucial Player in Diverse Biological Functions. Nat Rev Immunol (2010) 10(6):387–402. doi: 10.1038/nri2765
119. Veillette A, Latour S, Davidson D. Negative Regulation of Immunoreceptor Signaling. Annu Rev Immunol (2002) 20:669–707. doi: 10.1146/annurev.immunol.20.081501.130710
120. Billadeau DD, Leibson PJ. ITAMs Versus ITIMs: Striking a Balance During Cell Regulation. J Clin Invest (2002) 109(2):161–8. doi: 10.1172/JCI14843
121. Zhang F, Zheng J, Kang X, Deng M, Lu Z, Kim J, et al. Inhibitory Leukocyte Immunoglobulin-Like Receptors in Cancer Development. Sci China Life Sci (2015) 58(12):1216–25. doi: 10.1007/s11427-015-4925-1
122. Alaoui L, Palomino G, Zurawski S, Zurawski G, Coindre S, Dereuddre-Bosquet N, et al. Early SIV and HIV Infection Promotes the LILRB2/MHC-I Inhibitory Axis in cDCs. Cell Mol Life Sci (2018) 75(10):1871–87. doi: 10.1007/s00018-017-2712-9
123. Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human Leucocyte Antigen (HLA) Expression of Primary Trophoblast Cells and Placental Cell Lines, Determined Using Single Antigen Beads to Characterize Allotype Specificities of Anti-HLA Antibodies. Immunology (2009) 127(1):26–39. doi: 10.1111/j.1365-2567.2008.03019.x
124. Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, et al. Protein Expression and Peptide Binding Suggest Unique and Interacting Functional Roles for HLA-E, F, and G in Maternal-Placental Immune Recognition. J Immunol (2003) 171(3):1376–84. doi: 10.4049/jimmunol.171.3.1376
125. Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of Maternal Killer-Cell Immunoglobulin-Like Receptors and Parental HLA-C Genotypes With Recurrent Miscarriage. Hum Reprod (2008) 23(4):972–6. doi: 10.1093/humrep/den011
126. Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and Immune Tolerance in Pregnancy. FASEB J (2005) 19(7):681–93. doi: 10.1096/fj.04-2078rev
127. Rajagopalan S, Long EO. KIR2DL4 (CD158d): An Activation Receptor for HLA-G. Front Immunol (2012) 3:258. doi: 10.3389/fimmu.2012.00258
128. Wallace AE, Whitley GS, Thilaganathan B, Cartwright JE. Decidual Natural Killer Cell Receptor Expression is Altered in Pregnancies With Impaired Vascular Remodeling and a Higher Risk of Pre-Eclampsia. J Leukoc Biol (2015) 97(1):79–86. doi: 10.1189/jlb.2A0614-282R
129. Vacca P, Cantoni C, Prato C, Fulcheri E, Moretta A, Moretta L, et al. Regulatory Role of NKp44, NKp46, DNAM-1 and NKG2D Receptors in the Interaction Between NK Cells and Trophoblast Cells. Evidence for Divergent Functional Profiles of Decidual Versus Peripheral NK Cells. Int Immunol (2008) 20(11):1395–405. doi: 10.1093/intimm/dxn105
130. Petroff MG, Sedlmayr P, Azzola D, Hunt JS. Decidual Macrophages are Potentially Susceptible to Inhibition by Class Ia and Class Ib HLA Molecules. J Reprod Immunol (2002) 56(1-2):3–17. doi: 10.1016/s0165-0378(02)00024-4
131. Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C, et al. Inhibitory Receptors Sensing HLA-G1 Molecules in Pregnancy: Decidua-Associated Natural Killer Cells Express LIR-1 and CD94/NKG2A and Acquire P49, an HLA-G1-Specific Receptor. Proc Natl Acad Sci USA (1999) 96(10):5674–9. doi: 10.1073/pnas.96.10.5674
132. Huang J, Burke PS, Cung TD, Pereyra F, Toth I, Walker BD, et al. Leukocyte Immunoglobulin-Like Receptors Maintain Unique Antigen-Presenting Properties of Circulating Myeloid Dendritic Cells in HIV-1-Infected Elite Controllers. J Virol (2010) 84(18):9463–71. doi: 10.1128/JVI.01009-10
133. Ordonez D, Sanchez AJ, Martinez-Rodriguez JE, Cisneros E, Ramil E, Romo N, et al. Multiple Sclerosis Associates With LILRA3 Deletion in Spanish Patients. Genes Immun (2009) 10(6):579–85. doi: 10.1038/gene.2009.34
134. Tedla N, An H, Borges L, Vollmer-Conna U, Bryant K, Geczy C, et al. Expression of Activating and Inhibitory Leukocyte Immunoglobulin-Like Receptors in Rheumatoid Synovium: Correlations to Disease Activity. Tissue Antigens (2011) 77(4):305–16. doi: 10.1111/j.1399-0039.2011.01633.x
135. Mamegano K, Kuroki K, Miyashita R, Kusaoi M, Kobayashi S, Matsuta K, et al. Association of LILRA2 (ILT1, LIR7) Splice Site Polymorphism With Systemic Lupus Erythematosus and Microscopic Polyangiitis. Genes Immun (2008) 9(3):214–23. doi: 10.1038/gene.2008.5
136. Bleharski J, Li H, Meinken C, Graeber T, Ochoa M-T, Yamamura M, et al. Use of Genetic Profiling in Leprosy to Discriminate Clinical Forms of the Disease. Science (2003) 301:1527–30. doi: 10.1126/science.1087785
137. Du Y, Su Y, He J, Yang Y, Shi Y, Cui Y, et al. Impact of the Leucocyte Immunoglobulin-Like Receptor A3 (LILRA3) on Susceptibility and Subphenotypes of Systemic Lupus Erythematosus and Sjogren’s Syndrome. Ann Rheum Dis (2015) 74(11):2070–5. doi: 10.1136/annrheumdis-2013-204441
138. Du Y, Cui Y, Liu X, Hu F, Yang Y, Wu X, et al. Contribution of Functional LILRA3, But Not Nonfunctional LILRA3, to Sex Bias in Susceptibility and Severity of Anti-Citrullinated Protein Antibody-Positive Rheumatoid Arthritis. Arthritis Rheumatol (2014) 66(4):822–30. doi: 10.1002/art.38308
139. Kabalak G, Dobberstein SB, Matthias T, Reuter S, The YH, Dorner T, et al. Association of Immunoglobulin-Like Transcript 6 Deficiency With Sjogren’s Syndrome. Arthritis Rheum (2009) 60(10):2923–5. doi: 10.1002/art.24804
140. Wang H, Wang Y, Tang Y, Ye H, Zhang X, Zhou G, et al. Frequencies of the LILRA3 6.7-Kb Deletion Are Highly Differentiated Among Han Chinese Subpopulations and Involved in Ankylosing Spondylitis Predisposition. Front Genet (2019) 10:869. doi: 10.3389/fgene.2019.00869
141. Lan X, Liu F, Ma J, Chang Y, Lan X, Xiang L, et al. Leukocyte Immunoglobulin-Like Receptor A3 is Increased in IBD Patients and Functions as an Anti-Inflammatory Modulator. Clin Exp Immunol (2021) 203(2):286–303. doi: 10.1111/cei.13529
142. Koch S, Goedde R, Nigmatova V, Epplen JT, Muller N, de Seze J, et al. Association of Multiple Sclerosis With ILT6 Deficiency. Genes Immun (2005) 6(5):445–7. doi: 10.1038/sj.gene.6364187
143. Wisniewski A, Wagner M, Nowak I, Bilinska M, Pokryszko-Dragan A, Jasek M, et al. 6.7-Kbp Deletion in LILRA3 (ILT6) Gene is Associated With Later Onset of the Multiple Sclerosis in a Polish Population. Hum Immunol (2013) 74(3):353–7. doi: 10.1016/j.humimm.2012.12.006
144. Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, et al. Genome-Wide Association Study in Chinese Men Identifies Two New Prostate Cancer Risk Loci at 9q31.2 and 19q13.4. Nat Genet (2012) 44(11):1231–5. doi: 10.1038/ng.2424
145. Mitchell A, Rentero C, Endoh Y, Hsu K, Gaus K, Geczy C, et al. LILRA5 Is Expressed by Synovial Tissue Macrophages in Rheumatoid Arthritis, Selectively Induces Pro-Inflammatory Cytokines and IL-10 and Is Regulated by TNF-Alpha, IL-10 and IFN-Gamma. Eur J Immunol (2008) 38(12):3459–73. doi: 10.1002/eji.200838415
146. Lopez-Alvarez MR, Jiang W, Jones DC, Jayaraman J, Johnson C, Cookson WO, et al. LILRA6 Copy Number Variation Correlates With Susceptibility to Atopic Dermatitis. Immunogenetics (2016) 68(9):743–7. doi: 10.1007/s00251-016-0924-z
147. Kuroki K, Tsuchiya N, Shiroishi M, Rasubala L, Yamashita Y, Matsuta K, et al. Extensive Polymorphisms of LILRB1 (ILT2, LIR1) and Their Association With HLA-DRB1 Shared Epitope Negative Rheumatoid Arthritis. Hum Mol Genet (2005) 14(16):2469–80. doi: 10.1093/hmg/ddi247
148. Monsivais-Urenda A, Nino-Moreno P, Abud-Mendoza C, Baranda L, Layseca-Espinosa E, Lopez-Botet M, et al. Analysis of Expression and Function of the Inhibitory Receptor ILT2 (CD85j/LILRB1/LIR-1) in Peripheral Blood Mononuclear Cells From Patients With Systemic Lupus Erythematosus (SLE). J Autoimmun (2007) 29(2-3):97–105. doi: 10.1016/j.jaut.2007.05.003
149. Majorczyk E, Wisniewski A, Zon-Giebel A, Chlebicki A, Wiland P, Kusnierczyk P. The Effect of LILRB1 But Not LILRA3 Gene Polymorphism in Immunopathology of Ankylosing Spondylitis-A Parallel to KIR Genes. Int J Immunogenet (2019) 46(3):146–51. doi: 10.1111/iji.12422
150. Wang X, Meng X, Zheng Y, Jiang J, Yang B, Liu Y, et al. Increased Frequency of ILT2-Expressing CD56(dim)CD16(+) NK Cells Correlates With Disease Severity of Pulmonary Tuberculosis. Tuberculosis (Edinb) (2014) 94(5):469–74. doi: 10.1016/j.tube.2014.03.009
151. Saverino D, Ghiotto F, Merlo A, Bruno S, Battini L, Occhino M, et al. Specific Recognition of the Viral Protein UL18 by CD85j/LIR-1/ILT2 on CD8+ T Cells Mediates the non-MHC-Restricted Lysis of Human Cytomegalovirus-Infected Cells. J Immunol (2004) 172(9):5629–37. doi: 10.4049/jimmunol.172.9.5629
152. Nimmerjahn F, Lux A. LILR-B1 Blocks Activating FcgammaR Signaling to Allow Antibody Dependent Enhancement of Dengue Virus Infection. Proc Natl Acad Sci USA (2014) 111(7):2404–5. doi: 10.1073/pnas.1324286111
153. Chan KR, Ong EZ, Tan HC, Zhang SL, Zhang Q, Tang KF, et al. Leukocyte Immunoglobulin-Like Receptor B1 is Critical for Antibody-Dependent Dengue. Proc Natl Acad Sci USA (2014) 111(7):2722–7. doi: 10.1073/pnas.1317454111
154. Harrison TE, Morch AM, Felce JH, Sakoguchi A, Reid AJ, Arase H, et al. Structural Basis for RIFIN-Mediated Activation of LILRB1 in Malaria. Nature (2020) 587(7833):309–12. doi: 10.1038/s41586-020-2530-3
155. Wisniewski A, Kowal A, Wyrodek E, Nowak I, Majorczyk E, Wagner M, et al. Genetic Polymorphisms and Expression of HLA-G and its Receptors, KIR2DL4 and LILRB1, in non-Small Cell Lung Cancer. Tissue Antigens (2015) 85(6):466–75. doi: 10.1111/tan.12561
156. Villa-Alvarez M, Sordo-Bahamonde C, Lorenzo-Herrero S, Gonzalez-Rodriguez AP, Payer AR, Gonzalez-Garcia E, et al. Ig-Like Transcript 2 (ILT2) Blockade and Lenalidomide Restore NK Cell Function in Chronic Lymphocytic Leukemia. Front Immunol (2018) 9:2917. doi: 10.3389/fimmu.2018.02917
157. Zhang Y, Lu N, Xue Y, Zhang M, Li Y, Si Y, et al. Expression of Immunoglobulin-Like Transcript (ILT)2 and ILT3 in Human Gastric Cancer and its Clinical Significance. Mol Med Rep (2012) 5(4):910–6. doi: 10.3892/mmr.2012.744
158. Brown DP, Jones DC, Anderson KJ, Lapaque N, Buerki RA, Trowsdale J, et al. The Inhibitory Receptor LILRB4 (ILT3) Modulates Antigen Presenting Cell Phenotype and, Along With LILRB2 (ILT4), Is Upregulated in Response to Salmonella Infection. BMC Immunol (2009) 10:56. doi: 10.1186/1471-2172-10-56
159. Bashirova AA, Martin-Gayo E, Jones DC, Qi Y, Apps R, Gao X, et al. LILRB2 Interaction With HLA Class I Correlates With Control of HIV-1 Infection. PLoS Genet (2014) 10(3):e1004196. doi: 10.1371/journal.pgen.1004196
160. Cai Z, Wang L, Han Y, Gao W, Wei X, Gong R, et al. Immunoglobulinlike Transcript 4 and Human Leukocyte antigenG Interaction Promotes the Progression of Human Colorectal Cancer. Int J Oncol (2019) 54(6):1943–54. doi: 10.3892/ijo.2019.4761
161. Renauer PA, Saruhan-Direskeneli G, Coit P, Adler A, Aksu K, Keser G, et al. Identification of Susceptibility Loci in IL6, RPS9/LILRB3, and an Intergenic Locus on Chromosome 21q22 in Takayasu Arteritis in a Genome-Wide Association Study. Arthritis Rheumatol (2015) 67(5):1361–8. doi: 10.1002/art.39035
162. Jensen MA, Patterson KC, Kumar AA, Kumabe M, Franek BS, Niewold TB. Functional Genetic Polymorphisms in ILT3 are Associated With Decreased Surface Expression on Dendritic Cells and Increased Serum Cytokines in Lupus Patients. Ann Rheum Dis (2013) 72(4):596–601. doi: 10.1136/annrheumdis-2012-202024
163. Deng M, Gui X, Kim J, Xie L, Chen W, Li Z, et al. LILRB4 Signalling in Leukaemia Cells Mediates T Cell Suppression and Tumour Infiltration. Nature (2018) 562(7728):605–9. doi: 10.1038/s41586-018-0615-z
164. Bergstrom CP, Dahiya S, Chen W, Zhang CC, Zhu H, Yan J, et al. The Association of Leukocyte Immunoglobulin-Like Receptor Subfamily B-4 Expression in Acute Myeloid Leukemia and Central Nervous System Involvement. Leuk Res (2021) 100:106480. doi: 10.1016/j.leukres.2020.106480
165. Atwal J, Pinkston-Gosse J, Syken J, Stawicki S, Wu Y, Shatz C, et al. PirB is a Functional Receptor for Myelin Inhibitors of Axonal Regeneration. Science (2008) 332:967–70. doi: 10.1126/science.1161151
166. Wiendl H, Feger U, Mittelbronn M, Jack C, Schreiner B, Stadelmann C, et al. Expression of the Immune-Tolerogenic Major Histocompatibility Molecule HLA-G in Multiple Sclerosis: Implications for CNS Immunity. Brain (2005) 128(Pt 11):2689–704. doi: 10.1093/brain/awh609
167. Saito F, Hirayasu K, Satoh T, Wang CW, Lusingu J, Arimori T, et al. Immune Evasion of Plasmodium Falciparum by RIFIN via Inhibitory Receptors. Nature (2017) 552(7683):101–5. doi: 10.1038/nature24994
Keywords: leukocyte receptor complex (LRC), leukocyte immunoglobulin-like receptor (LILR), killer immunoglobulin-like receptor (KIR), immunoglobulin Ig-like superfamily (IgSF), non-human primates (NHP), human
Citation: Storm L, Bruijnesteijn J, de Groot NG and Bontrop RE (2021) The Genomic Organization of the LILR Region Remained Largely Conserved Throughout Primate Evolution: Implications for Health And Disease. Front. Immunol. 12:716289. doi: 10.3389/fimmu.2021.716289
Received: 28 May 2021; Accepted: 01 October 2021;
Published: 19 October 2021.
Edited by:
Pedro Jose Esteves, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), PortugalReviewed by:
Petr Horin, University of Veterinary and Pharmaceutical Sciences Brno, CzechiaCopyright © 2021 Storm, Bruijnesteijn, de Groot and Bontrop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisanne Storm, c3Rvcm1AYnByYy5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.