
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 19 October 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.703936
This article is part of the Research Topic The Role of Epigenetics in Infectious Diseases View all 14 articles
 Ousman Tamgue*
Ousman Tamgue* Cybelle Fodieu Mezajou
Cybelle Fodieu Mezajou Natacha Njike Ngongang
Natacha Njike Ngongang Charleine Kameni
Charleine Kameni Jubilate Afuoti Ngum
Jubilate Afuoti Ngum Ulrich Stephane Fotso Simo
Ulrich Stephane Fotso Simo Fabrice Junior Tatang
Fabrice Junior Tatang Mazarin Akami
Mazarin Akami Annie Ngane Ngono
Annie Ngane NgonoNon-coding RNAs (ncRNAs) including microRNAs (miRs) and long non-coding RNAs (lncRNAs) have emerged as key regulators of gene expression in immune cells development and function. Their expression is altered in different physiological and disease conditions, hence making them attractive targets for the understanding of disease etiology and the development of adjunctive control strategies, especially within the current context of mitigated success of control measures deployed to eradicate these diseases. In this review, we summarize our current understanding of the role of ncRNAs in the etiology and control of major human tropical diseases including tuberculosis, HIV/AIDS and malaria, as well as neglected tropical diseases including leishmaniasis, African trypanosomiasis and leprosy. We highlight that several ncRNAs are involved at different stages of development of these diseases, for example miR-26-5p, miR-132-3p, miR-155-5p, miR-29-3p, miR-21-5p, miR-27b-3p, miR-99b-5p, miR-125-5p, miR-146a-5p, miR-223-3p, miR-20b-5p, miR-142-3p, miR-27a-5p, miR-144-5p, miR-889-5p and miR-582-5p in tuberculosis; miR-873, MALAT1, HEAL, LINC01426, LINC00173, NEAT1, NRON, GAS5 and lincRNA-p21 in HIV/AIDS; miR-451a, miR-let-7b and miR-106b in malaria; miR-210, miR-30A-5P, miR-294, miR-721 and lncRNA 7SL RNA in leishmaniasis; and miR-21, miR-181a, miR-146a in leprosy. We further report that several ncRNAs were investigated as diseases biomarkers and a number of them showed good potential for disease diagnosis, including miR-769-5p, miR-320a, miR-22-3p, miR-423-5p, miR-17-5p, miR-20b-5p and lncRNA LOC152742 in tuberculosis; miR-146b-5p, miR-223, miR-150, miR-16, miR-191 and lncRNA NEAT1 in HIV/AIDS; miR-451 and miR-16 in malaria; miR-361-3p, miR-193b, miR-671, lncRNA 7SL in leishmaniasis; miR-101, miR-196b, miR-27b and miR-29c in leprosy. Furthermore, some ncRNAs have emerged as potential therapeutic targets, some of which include lncRNAs NEAT1, NEAT2 and lnr6RNA, 152742 in tuberculosis; MALAT1, HEAL, SAF, lincRNA-p21, NEAT1, GAS5, NRON, LINC00173 in HIV/AIDS; miRNA-146a in malaria. Finally, miR-135 and miR-126 were proposed as potential targets for the development of therapeutic vaccine against leishmaniasis. We also identify and discuss knowledge gaps that warrant for increased research work. These include investigation of the role of ncRNAs in the etiology of African trypanosomiasis and the assessment of the diagnostic potential of ncRNAs for malaria, and African trypanosomiasis. The potential targeting of ncRNAs for adjunctive therapy against tuberculosis, leishmaniasis, African trypanosomiasis and leprosy, as well as their targeting in vaccine development against tuberculosis, HIV/AIDS, malaria, African trypanosomiasis and leprosy are also new avenues to explore.
Non-translated or non-coding RNAs (ncRNAs) are the transcripts of the genome that are not meant to be translated into proteins (1). They represent about 98% of total RNAs content within the human cells (2, 3) and were initially thought to be byproducts of transcription, therefore referred to as “Junk RNAs”. However, a growing body of evidence have unveiled the role of certain ncRNAs including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as key regulators of gene expression, that is they can alter genes expression in a reversible, transmissible, and adaptative way, without modifying the DNA sequence (4). MicroRNAs are an abundant class of highly conserved small (18-25 nucleotides long) RNA species that generally downregulate the expression of their target genes at the post-transcriptional level. Mechanically, miRNAs bind in a sequence-specific manner to complementary regions in the 3’ untranslated region (3’UTR) of their target mRNAs, thereby triggering mRNA degradation or translation inhibition. In this way, a single miRNA can control the expression of several genes and a single gene expression can be controlled by several different miRNAs (5). Contrary to miRNAs, lncRNAs (at least 200 nucleotides long) are less studied, display poor sequence conservation and regulate the expression of their nearby proximal genes (Cis regulation) as well as distant genes (Trans regulation) at the chromatin, transcription and translation levels (6–8).
Several miRNAs and lncRNAs are emerging as key regulators of immune cells differentiation, activation, and function, including macrophages, dendritic cells and T lymphocytes (6). Some have been associated with specific disease conditions such as Cancer, cardiovascular, developmental (1, 9–12),neurodegenerative (13) and major infectious diseases such as tuberculosis and HIV/AIDS (14–16). There are however few or no studies addressing the role of ncRNAs in the etiology, diagnosis, treatment, or vaccine development for neglected human tropical diseases (NTDs) which are a group of less investigated infectious diseases especially common in tropical areas such as Africa and Southeast Asia where people do not have proper access to clean water and adequate means to discard their waste. In this review, we summarize most recent findings on the role of miRNAs and lncRNAs on major human tropical diseases including tuberculosis, HIV/AIDS and malaria. We also provide a first-time summary of our current understanding of the role of these ncRNAs in the etiology and control of neglected tropical diseases including leishmaniasis, African trypanosomiasis and leprosy. We also identify and discuss knowledge gaps that warrant for increased research effort.
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (Mtb) which has topped HIV as the deadliest infectious agent worldwide since 2017. Developing countries are highly burdened by this disease and further threatened by the emergence of multi-drug resistant and extensively drug resistant Mtb strains. Accurate diagnosis and effective treatment are the key elements to interrupt TB transmission (17). Several studies have linked miRNAs and lncRNAs to the onset and progression of TB, and some of those ncRNAs were identified as biomarkers for TB diagnosis or treatment (Table 1).
To fight a bacterial infection, host innate and/or adaptive immunity has to be activated. Mycobacterium tuberculosis like many other successful pathogens has evolved mechanisms to avoid the host immune system and ensure its intracellular survival and persistence. This is possible through the subversion of key ncRNAs that control the cellular and humoral processes enacted in host innate and adaptive immune response against Mtb (Figure 1).
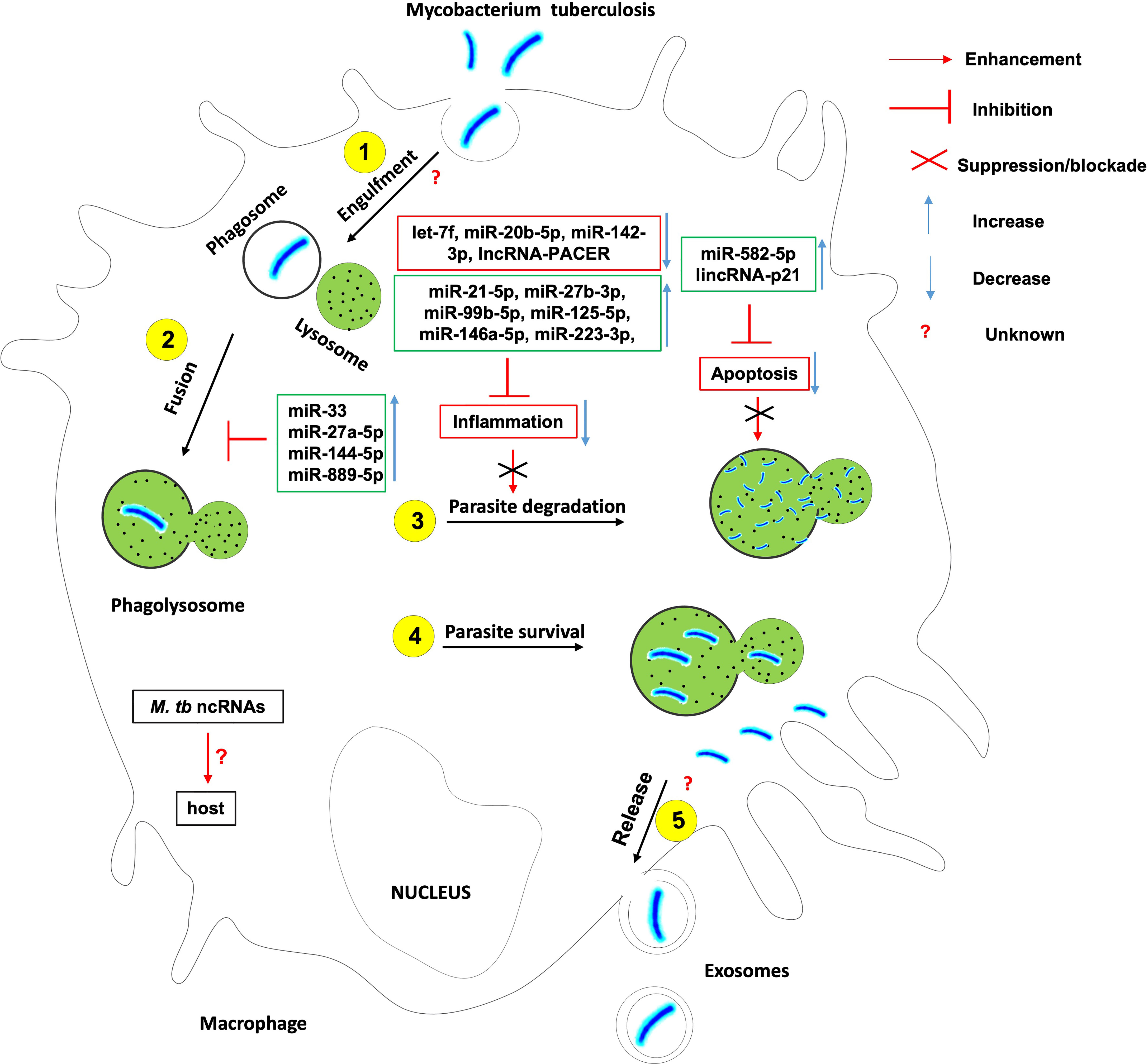
Figure 1 The role of non-coding RNAs in the etiology of tuberculosis. Several ncRNAs enhance host effector killing functions against mycobacterium tuberculosis and favor the bacterium survival and persistence within the infected host. There is knowledge gap about 1- host ncRNAs that regulate the bacterium engulfment within phagosomes and release in extracellular vesicles such as exosomes; 2- the role of Mtb-derived ncRNAs on the outcome of Mtb-macrophage interaction.
The induction of a robust yet controlled inflammatory response plays a key role in the containment and eradication of the infection at an early stage. It was found that Mtb suppresses inflammation via the upregulation of miR-21-5p, miR-27b-3p, miR-99b-5p, miR-125-5p, miR-146a-5p, miR-223-3p, and the downregulation of let-7f, miR-20b-5p and miR-142-3p (18). Mtb HN878 infection of monocyte-derived macrophages (MDMs) induces the expression of the pro-inflammatory lncRNA-PACER (also known as lncRNA-Cox-2) which is a positive regulator of its proximal pro-inflammatory gene Ptgs-2 (Also known as Cox-2) (25).
Macrophage and other phagocytes are the first immune cells to encounter Mtb in the lungs and make use of their phagocytic activity to engulf and destroy the invading Mtb using different cell death mechanisms. Mtb has developed several strategies to avoid killing within phagocytes. Mtb inhibits the phagosome maturation and autophagy via upregulation of miR-33 locus (18, 26), miR-27a-5p (18, 20), miR-144-5p and miR-889-5p. Mtb also evades the host defense by Inhibiting macrophage apoptosis (27) via the upregulation of miR-582-5p (28). It was also observed that Mtb HN878 infection of MDMs induces the expression of lincRNA-p21, a positive regulator of p53-dependent cell cycle arrest and apoptosis in numerous cell types (25, 29–32).
Much research is warranted to understand the contribution of lncRNA-PACER and lincRNA-p21 to the onset and progression of TB. Also there are growing body of evidence suggesting that Mtb-derived ncRNAs may be delivered to the host immune cells and affect their function. The contribution of such mechanisms to host immune evasion needs in-depth investigation.
Several miRNAs are decreased in the plasma of Mycobacterium tuberculosis infected patients compared to healthy individuals and are described as biomarkers for the diagnosis of tuberculosis (33). Amongst those miRNAs, miR-769-5p, miR-320a and miR-22-3p subsequently showed higher specificity (4, 33) at 90% sensitivity (92%), AUC (95%) and lower heterogeneity (I= 0%) in ethological-confirmation validation sets (17). Also, miR-423-5p, miR-17-5p, and miR-20b-5p were reported to be significantly increased in the serum of patients with tuberculosis and had the potential to be used to diagnose TB with an accuracy of 78.18% (21). The level of long noncoding RNA LOC152742 was found to be high in sputum and plasma of infected patients, hence could serve as novel biomarker for the diagnosis of active tuberculosis (22).
Sputum-negative pulmonary tuberculosis cases showing no clinical or microbial evidence contribute to the development and spread of active tuberculosis (34). Accurate diagnosis of sputum smear-negative cases of pulmonary TB remains very challenging. It was found that lncRNAs ENST00000429730.1 and MSTRG.93125.4 were upregulated in lung tissue samples collected from patients with sputum-negative pulmonary TB with high metabolic activity as compared to low metabolic activity according to FDG-PET/CT(Positron emission tomography with computed tomography (PET/CT) using fluorine-18-fluoro-deoxyglucose (FDG)) classification. Hence these lncRNAs might be potential biological indicators of metabolic activity in tuberculosis lesions for sputum-negative tuberculosis (24).
The emergence of multidrug-resistant strains of Mtb has further complicated the control and eradication of this disease. It was found that the plasma levels of miR-320a were decreased in drug-resistant TB patients as compared to pan-susceptible TB patients (33). Therefore, this miR-320a may serve as a biomarker for drug-resistant TB. Also, lncRNAs CTD-2331D11.3 and AC079779.5 were found to be increased in the Peripheral Blood Monocytic Cells (PBMCs) from patients infected with Multi-drug resistant Mtb strains (MDR-TB) when compared to patients infected with drug-sensitive strains, indicating this lncRNAs may be potential biomarkers for multi-drug resistant TB (35).
The role of ncRNAs as potential host-directed therapeutical targets has been reviewed before (36). As a complement to that review article, recent studies have reported an increase in the expression of lncRNAs NEAT1 and NEAT2 in macrophages during Mtb infection. Their expression level was decreased during drug treatment, which was associated with improvement of the disease (23). The same observation was made with lnr6RNA 152742 which was upregulated in the plasma and sputum of patients and gradually downregulated in the course of the treatment (22).
Successful treatment of pulmonary tuberculosis is generally declared after absence of Mtb in sputum smear under microscopy and under culture. However, pulmonary TB lesions may still be harboring persisting slow growing, metabolically active but non-culturable bacilli that are less sensitive to chemotherapy agent and may cause TB relapse (37, 38) lncRNAs ENST00000429730.1 and MSTRG.93125.4 described as indicators of metabolic activity in tuberculosis lesions for sputum-negative tuberculosis (24), hence are potential biomarkers of complete inactivation of tuberculosis lesions, thus of complete cure of tuberculosis (24).
Malaria is a mosquito-borne disease caused by parasites of the genus Plasmodium. It is transmitted through the bites of infected female Anopheles mosquitoes. Five parasite species cause malaria in humans: P. knowlesi, P.malariae, P.ovale, P. vivax and P. falciparum1. The two last pose the greatest threat2. In 2019, around 229 million cases of malaria were recorded in the world with approximately 409 000 deaths3. Symptoms of malaria comprise fever, shaking chills, headache, muscle aches, and tiredness. Nausea, vomiting, and diarrhea may also be involved4. Due to the non-specificity of its symptoms, it is difficult to distinguish malaria from other acute febrile illnesses. Non-coding RNAs, which are specific, can be of great help in resolving this problem.
Despite the growing recognition of the contribution of ncRNAs in the etiology of infectious diseases, only a handful studies have specifically associated ncRNAs with the onset or progression of any clinical form of malaria, be it uncomplicated or cerebral (Figure 2).
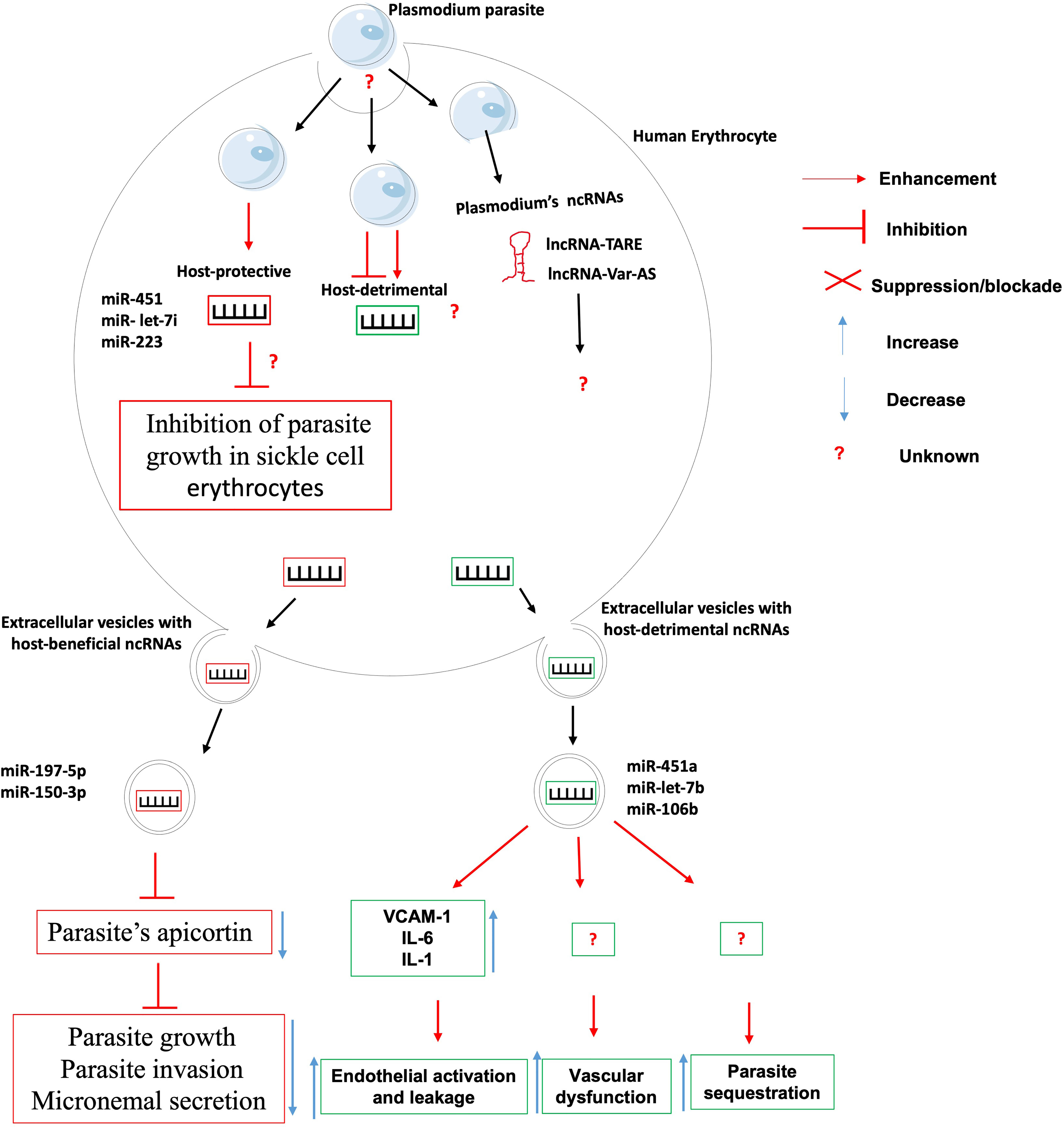
Figure 2 The role of non-coding RNAs in the etiology of malaria. Very few ncRNAs known to involve in the etiology of Malaria. There is knowledge gap about 1- host ncRNAs that regulate the uptake/entry of plasmodium within erythrocytes; 2- host-detrimental ncRNAs and their mechanisms of action; and 3- the role of plasmodium-derived ncRNAs on the outcome of plasmodium-erythrocyte interaction.
The clinical outcome of persons infected with Plasmodium falciparum parasites depends on many factors including parasite sequestration in tissues, host systemic inflammatory responses, and vascular dysfunction. It was found that Plasmodium falciparum-infected red blood cells release extracellular vesicles (EV) loaded with functional host miR-451a, miR-let-7b and miR-106b. These miRNAs-loaded EVs are internalized by endothelial cells within which they induce the production of surface receptor vascular cell adhesion protein 1 (VCAM-1) and proinflammatory cytokines such as interleukin-6 (IL-6) and interleukin-1 (IL-1). This will lead to the promotion of endothelial activation, leakage and parasite sequestration as well as vascular dysfunction and pathology during malaria infection (39). Contrary to the abovementioned microRNAs that promote malaria pathology, it was found that erythrocytes-derived miR-197-5p and miR-150-3p reduced the parasite growth, invasion and micronemal secretion via a mechanism involving the inhibition of the expression of apicortin, a Plasmodium falciparum’s protein with putative microtubule-stabilizing properties (40). In addition, the resistance of sickle cell erythrocytes (carrying the HbS hemoglobin allele variant in either the heterozygous or the homozygous form) to infection by Plasmodium falciparum was associated to the expression of miR-451, miR- let-7i and miR-223 which were translocated into the parasite during its intraerythrocytic life cycle and negatively regulated the parasite growth (41). The mRNA targets of those microRNAs were however not investigated, neither was their contribution to the onset and maintenance of epigenetic marks such as DNA methylation and histone post-translational modifications on the promoter of possible target genes.
It was shown that knocking-out/down miR-155 led to reduced endothelial activation, reduced microvascular leak and preservation of blood-brain-barrier, reduced disease severity and improved survival in an experimental mouse model of cerebral malaria and an engineered human endothelialized microvessel system (42). Similarly, It was found that miR-19a-3p, miR-19b-3p, miR-540-5p, miR-142-3p and miR-223-3p were significantly upregulated in the brain of mice displaying cerebral malaria (infected with Plasmodium berghei ANKA) as compared to those with severe but non-cerebral malaria (infected with Plasmodium yoelii). These miRNA are involved in the control of TGF-β and endocytosis pathways which are known to be relevant to cerebral malaria (43). These works on gene-deficient mice models need to be repeated using humanized mice models and in-vitro human infection models for those data to have any translational value. Lastly, a mutation in the miRNA−146a was reported to be linked with increased odds for P. falciparum infection in first-time pregnant women, thus providing an indirect evidence of miRNA-146a protective role against P. falciparum infection (44).
Although no host lncRNA has been associated with malaria etiology to date, however, high throughput analysis of Plasmodium falciparum transcriptome have uncovered several lncRNAs including lncRNA-TARE (45, 46) and lncRNA var-AS (47, 48) that play important role in the development and virulence of the parasite. Research is warranted to investigate the role of these lncRNAs in the parasite immune evasion, host cell invasion and development within the human host.
The patients infected with P. falciparum manifest malaria of differing severities and clinical outcomes, such as uncomplicated malaria (UM), severe malaria, and cerebral malaria (CM). To date, few research have focused on investigating ncRNAs as biomarkers for the diagnosis of malaria. It was found that the plasma level of miR-451 and miR-16 were significantly lower in malaria patients compared to uninfected individuals, thus suggesting that plasma miR-451 and miR-16 are potentially relevant biomarkers for malaria infection (49).
Many miRNAs including miR-3135b, miR-6780b-5p, miR-1246, miR-6126, miR-3613-5p, miR-4532 and miR-6068 are upregulated in humans during the blood phase of P. falciparum infection as compared to negative controls. This upregulation was as the result of activation of host innate immunity (50). These miRNAs could be further investigated as potential blood biomarkers of the immunopathological state, thus helping in the early diagnosis of the disease. To date no lncRNA has been investigated as potential biomarker for the diagnosis of malaria.
MiRNA-146a is involved in innate immune response through a negative feedback loop comprising two key molecules downstream of the TLR machinery: the kinase associated with the interleukin -1 receptor (IRAK) -1 and the factor associated with the receptor of TNF (TRAF) -6. Recent studies have shown the potential of using miRNA-146a as a biopharmaceutical agent; The results of a current study suggest that miRNA-146a is involved in innate immunity against malaria, demonstrating its potency as a biopharmaceutical target (44).
HIV remains a major global public health issue despite the increasing access to effective HIV prevention, diagnosis, treatment, and care, including for opportunistic infections. Approximately 38.0 million people were living with HIV at the end of 20195. The Human Immunodeficiency Virus (HIV) targets the immune system and weakens people’s defense against many infections and some types of cancer. As the virus destroys and impairs the function of immune cells, immunodeficiency gradually sets in the infected individual. The most advanced stage of HIV infection is Acquired Immunodeficiency Syndrome (AIDS), which can take many years to develop if not treated, depending on the individual. With the introduction of Highly Active Antiretroviral Therapy (HAART), HIV infection has become a manageable chronic health condition. There is still no cure or vaccine against HIV infection, which has been known for about forty years to date and for which research is restless (51). Many studies reported ncRNAs as novel targets for new drugs (Table 2). These ncRNAs influence the replication cycle of the virus.
It is known that HIV interacts with the host in order to complete its replication cycle, escape immune response and persist within infected hosts. Such interactions involve both host ncRNAs and HIV-produced ncRNAs amongst other factors (Figure 3).
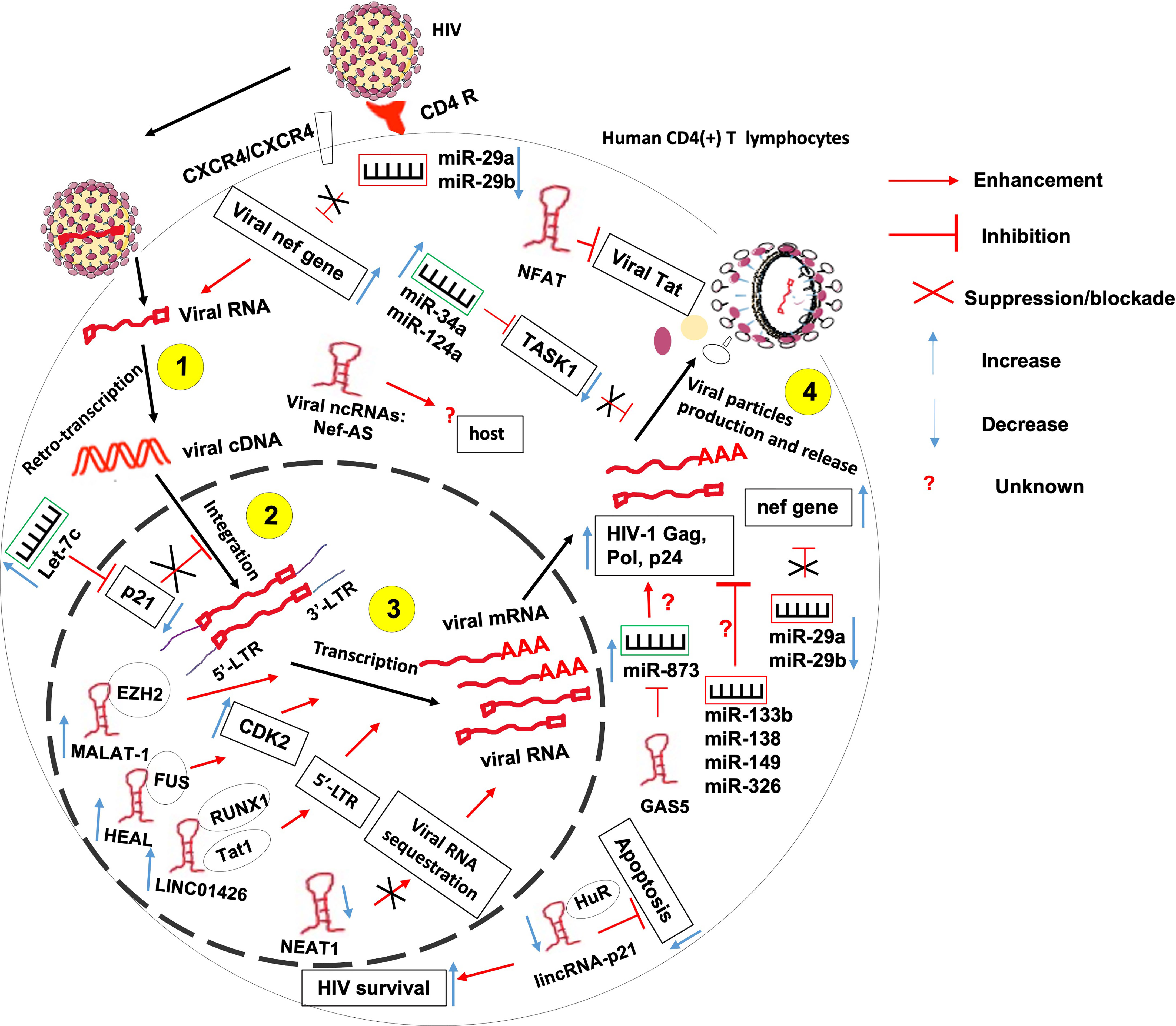
Figure 3 The role of non-coding RNAs in the etiology of HIV/AIDS. ncRNAs control several steps of HIV life cycle including viral RNA retro-transcription, cDNA integration, transcription and viral particles production. More research needed to identify 1- host ncRNAs that regulate viral particles release from the host and 2- the role of HIV-derived ncRNAs on the outcome of HIV-T lymphocyte interaction.
HIV hijacks host ncRNAs to promote its replication within the host. Indeed, it was observed that at the peak of HIV-1 replication, the virus downregulates the expression of miR-29a and miR-29b in CD4(+) CD8 (–) PBMCs (67). These two miRNAs were previously reported to inhibit viral replication through direct targeting of a conserved site within the viral nef gene (68, 69). Also, miR-873 was shown to promote HIV-1 replication in an in-vitro infection model of Jurkat and 293T cells. miR-873 promotes the production of HIV-1 gag, pol and p24 proteins through yet undefined mechanisms (52). HIV also upregulates the expression of host miRNAs let-7c, miR-34a, and miR-124a to promote its propagation. Let-7c post-transcriptionally inhibits the expression of p21 which is a known negative regulator of viral integration and RNA expression within the infected host cell (70). miR-34a and miR-124a decrease the mRNA level of TASK1 (70), which is a mammalian potassium channel known to counteract the viral Vpu -induced release of HIV virions (71).
Besides miRNAs, HIV also hijacks numbers of lncRNAs to its advantage, including Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1), HIV-1 enhanced lncRNA (HEAL), LINC01426, lincRNA-p21 and Nuclear-Enriched Abundant Target 1 (NEAT1).
Indeed, induction of MALAT1 expression in T lymphocytes contributes to initial viral replication and to disease progression. Mechanistically, MALAT1 sequesters EZH2, the Histone H3K27 trimethylase of the polycomb repressive complex 2, hence releasing the epigenetic inhibition of the HIV-1 LTR promoter responsible for the latency (54).
HIV-1 infection-induced upregulation of lncRNA HEAL promotes the transcription of HIV-1 in both Monocytes-derived macrophages and in primary CD4+ T cells. Indeed, acting as a scaffold, HEAL recruits fused in sarcoma (FUS) RNA-binding protein to the promoter of CDK2 and HIV-1 LTR Which are known as activators of multiple proteins essential for HIV-1 transcription (55).
HIV1 infection-induced LINC01426 enhances HIV-1 replication thanks to its interaction with both the host RUNX1a transcription factor and viral Tat1 factors that mediate the lncRNA binding to the 5′ LTR of HIV-1 (56).
HIV-1 induces the complexation of the apoptosis-promoting lincRNA-p21 with the host protein human antigen R (HuR) and its subsequent degradation. This will lead to apoptosis inhibition and enhanced HIV survival within infected macrophages but not lymphocytes (62). It was observed that HIV-1 infection downregulates the expression of NEAT1 lncRNA leading to the reduction of the number of host-protective paraspeckle bodies, hence increased HIV-1 expression within CD4+ T lymphocytes (58). NEAT1 is also downregulated during viral reactivation from a resting state in CD4(+) T cells through an unknown mechanism leading to the promotion of HIV-1 transcription, and potentially HIV-1 dissemination (58, 59).
Contrary to the above-mentioned ncRNAs, some host ncRNAs were reported to repress the replication of HIV. These include miR-29a, miR-133b, miR-138, miR-149 and miR-326, NEAT1, noncoding repressor of Nuclear Factor of Activated T cells (NFAT or NRON), growth arrest-specific transcript 5 (GAS5), lincRNA-p21, 7SK and NEAT1.
Indeed, miR-29a is highly induced in HIV-1-infected Jurkat cells where it represses HIV replication through direct targeting of the HIV-1 nef 3’ UTR region. However, the expression of this miR-29a is significantly downregulated at the peak of HIV-1 replication as already mentioned, thus highlighting its host-protective effect against HIV-1 infection (68, 69). In addition to miR-29a, it was found that overexpression of in-silico predicted miR-133b, miR-138, miR-149 and miR-326 decrease HIV replication in various primary T cells and T cell lines. It was further shown that miR-326 acts by direct targeting of a sequence within HIV-1 (72).
As mentioned above, HIV-1 infection downregulates the expression of NEAT1 which would otherwise restrict HIV-1 replication through sequestration of unspliced viral RNA into paraspeckles (58).
The Noncoding Repressor of Nuclear Factor of Activated T cells (NFAT, or NRON) was shown to inhibit HIV-1 transcription and induce HIV-1 latency via induction of a proteasome-mediated HIV-1 Tat degradation, and in an NFAT-independent manner (60). This findings corroborate the previous observation that NRON is highly expressed in resting CD4(+) T lymphocytes and HIV-proteins Nef and Vpu downregulate NRON expression, hence increase HIV-1 transcription via mechanisms that involved NFAT transcription factor but are still not fully understood (61).
The lncRNA Growth Arrest-Specific Transcript 5 (GAS5) inhibits HIV-1 replication by acting as a competing endogenous RNA (ceRNA), suppressing the effects of the host-detrimental miR-873 (52).
It is now recognized that there are ncRNAs originating from virus genomes. For instance, the antisense transcript originating from the Nef region in the HIV-1 genome (73, 74) which plays important role in the transcriptional control of HIV-1, notably via epigenetics mechanisms (75). There is currently no studies investigating the role of HIV-1-originating ncRNAs in the regulation of the outcome of host-virus interaction.
miR-146b-5p, miR-223, miR-150, miR-16, and miR-191 were found to be down regulated during HIV infection and plentifully expressed in B and T-lymphocytes, confirming a positive disease status (64, 65). Furthermore, some authors suggested that the presence of NEAT1 in plasma is a potential biomarker of HIV-1 infection (76).
Some ncRNAs have been associated to the treatment of HIV infection. miR-29a can be used as an indicator for the on-treatment disease evolution CD4 count or zenith HIV viral load (77). Its expression is associated to markers of HIV infection in long-term survivors, treatment-experienced patients (77). 7SK, by its pseudouridylation, can inhibit HIV transcription and escape from latency, suggesting it may be a new target for eliminating latent viral reservoirs (63, 78).
Many lncRNAs can be related to therapeutic research of HIV infection. MALAT1, as a promoter of HIV transcription, is a potential therapeutic target (54, 63). uc002yug.2, by activating latent HIV and HIV replication, can be a potential therapeutic target (56, 63). HEAL may be an attractive therapeutic target to inhibit HIV-1 latency, particularly considering that it is only upregulated in infected CD4+ and macrophages (53, 55, 63). SAF, involved in the resistance of HIV-1–infected macrophages to activation of apoptotic caspases, is a potential therapeutic target specifically intended for HIV cellular reservoirs (63, 66). lincRNA-p21 can constitute a novel therapeutic intervention strategy for HIV infection in macrophages (62, 63). NEAT1 is a feasible target for HIV treatment that involves the reactivation of latent HIV (58, 59, 63). GAS5, by suppressing miR-873, may be a novel biomarker for antiviral drugs and potential target for HIV treatment (52, 63). NRON, as an actor of HIV-1 latency, may be a novel target for reversing viral latency (60, 61, 63). TAR-gag, as an “RNA machine” of HIV genetic regulation, is a novel therapeutic target to reverse viral latency (63, 79). HIV-encoded lncRNA, as an epigenetic brake to regulate viral transcription, is a novel therapeutic target to inhibit the emergence of viral latency (51, 63, 75). LINC00173, which regulates cytokine levels in T cells, is a new therapeutic target for immunotherapy (57, 63).
Leishmaniasis is a neglected tropical disease caused by infection with Leishmania parasites, which are spread by the bite of phlebotomine sand flies. There are different forms of leishmaniasis in people and the most common are Cutaneous Leishmaniasis (CL), which causes skin sores, and Visceral Leishmaniasis (VL), which affects several internal organs (generally spleen, liver, and bone marrow)6.
Leishmaniasis is prevalent on every continent except Australia and Antarctica. It is difficult to estimate the number of new cases that may vary over time. For CL, estimates of the number of new cases per year have ranged from approximately 700,000 to 1.2 million or more. For VL, the estimated number of new cases per year may have decreased to <100,000, but previous estimates ranged up to 400,000 or more cases7. If not treated, severe cases of visceral leishmaniasis typically are fatal.
Before considering treatment of leishmaniasis, it is essential to make sure the diagnosis is correct. It can be done by detecting Leishmania parasites (or DNA) in tissue specimens from skin lesions (CL) or bone marrow (VL). This tissue sampling is an invasive method. Conversely, the diagnosis of disease with the help of a biomarker is a non-invasive tool that has shown to have an important function in early diagnosis of infection. There is no cure for leishmaniasis and chemotherapy is threatened by limited efficacy coupled with the development of resistance and other side effects (80). Leishmania parasites elude the host defensive. Some ncRNAs have been reported as biomarkers for the diagnosis and the treatment of leishmaniasis (Table 3).
Like many sophisticated intracellular pathogens, Leishmania has evolved mechanisms to modify the host responses to ensure their intracellular differentiation and multiplication. The parasite does so through the manipulation of the host factors including several miRNAs (Figure 4).
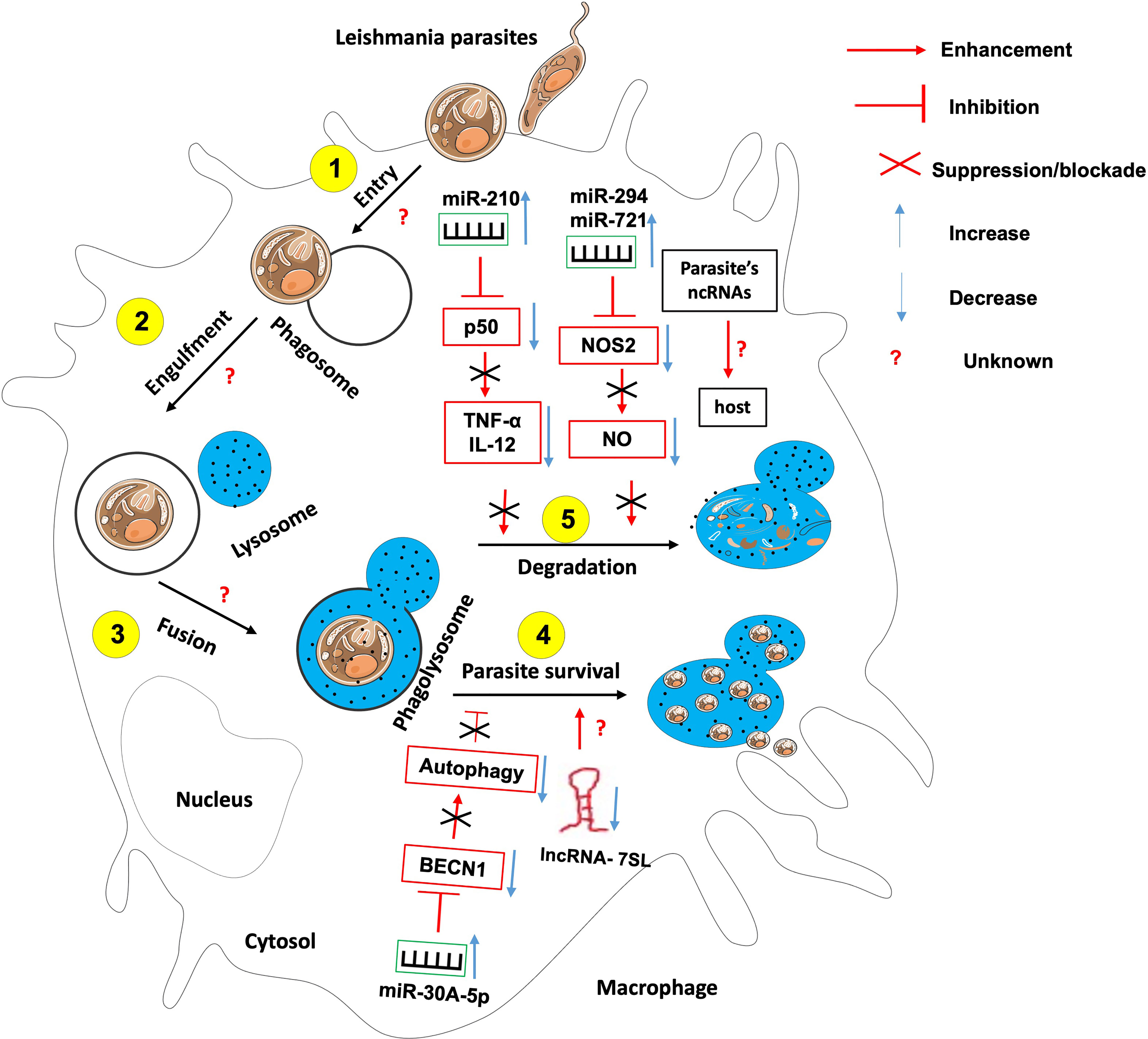
Figure 4 The role of non-coding RNAs in the etiology of leishmaniasis. Most studies have identified ncRNAs that enhance host effector killing functions against leishmania parasites as well as those that favor parasite survival and persistence within the infected host. There is knowledge gap about 1- host ncRNAs that regulate the parasite entry within the host, its engulfment within phagosome and the fusion between phagosome and lysosomes; 2- the role of Leishmania-derived ncRNAs on the outcome of leishmania-macrophage interaction.
For instance, Leishmania donovani infection creates hypoxic conditions leading to HIF-1a-mediated induction of miR-210 in infected macrophages (81, 86). This miR-210 was shown to promote the survival of the parasite within the host by targeting the NFkb subunit p50 and subsequently downregulating the expression of pro-inflammatory cytokines TNF-α and IL-12 while upregulating the anti-inflammatory cytokine IL-10 within the infected macrophage (81). Also, L. donovani interferes with host autophagy by inducing the expression of host miR-30A-5P which in turn will downregulate the expression of the pro-autophagic BECN1 protein, hence the increased survival of the parasite within the infected macrophage (87). Leishmania amazonensis promotes its survival within the infected host by upregulating miR-294 and miR-721. These miRNAs target the nitric oxide synthase 2 (NOS2) mRNA at the post-transcriptional level, then leading to decreased nitric oxide production and increased arginase activity within the infected macrophage (88). miR-21 and miR-146b-5p are significantly downregulated in monocytes-derived dendritic cells following L. donovani and L. major infection. In-silico prediction have identified SMAD7 and TRAF6, two members of the TGF-β signaling pathway as targets for these two miRNAs (89). It was found that L. major infection led to the down-regulation of miR-10a in Foxp3+ Treg cells. This miR-10a decreased IFNγ and enhanced the suppressive function of Treg cells (90).
Leishmania promastigotes and amastigotes infection represses the expression of the lncRNA 7SL RNA, an important component of the signal recognition particle in macrophages. This will convert these cells into permissible hosts favorable to the establishment and hiding of the parasite within the macrophages’ phagolysosomes (91).
It is still unknown whether and which host ncRNAs control leishmania parasites entry, engulfment within phagosomes and fusion of phagosomes with lysosomes.
An investigation on Leishmania major genome revealed that there are 1884 uniquely expressed ncRNAs in that parasite, some of which were recently shown to possess protein coding potential (92). The contribution of these parasite-derived ncRNA to the outcome of parasite-host interaction still need investigation.
The 7SL RNA gene can be used for diagnosis of human leishmaniasis (93). Diagnosis with the help of 7SL RNA is rapid, sensitive, specific, and simple (93).
Some authors suggested that miR-361-3p is a prognostic biomarker in cutaneous leishmaniasis lesions caused by Leishmania braziliensis (82). miR-193b and miR-671 were associated with a good response to treatment of Human localized cutaneous leishmaniasis caused by Leishmania braziliensis (83). Targeting of let-7a with Locked Nucleic Acid (LNA) Antisense Oligonucleotides (ASOs) was shown to increase L. major-infected MDMs apoptosis and necrosis, therefore, targeting let-7a was suggested as a potential therapeutic approach (94).
miRNA21 has been recently shown to negatively associate with IL-12 production and priming of protective Th1 response, suggesting declining levels of miRNA-21 as a potential biomarker of safety and immunogenicity in anti-leishmanial vaccines (84, 95).
Therapeutic vaccines may be developed by targeting miRNA-135 and−126 that bias the Th2 response toward protective Th1 type (84, 85).
Also known as sleeping sickness, Human African Trypanosomiasis (HAT) is a neglected tropical disease caused by microscopic parasites of the species Trypanosoma brucei whose vector is an insect of the genus Glossina: the tsetse fly8. Two subspecies of Trypanosoma brucei are responsible of human disease: T. b. gambiense in 24 countries in west and central Africa, and T. b. rhodesiense in 13 countries in eastern and southern Africa9
HAT is curable with medication but is fatal if left untreated. Diagnosis must be made as early as possible to avoid progressing to the neurological stage in order to prevent complicated and risky treatment procedures (95). Diagnosis is made clinically or by light microscopy; which are both insensitive and require certain skills10. The use of biomarkers such as ncRNAs could enhance sensitivity.
The alteration of nine miRNAs Including miR-193b, miR-338 (upregulated), miR-199a-3p, miR-27b and miR-126* (downregulated) has been identified in the peripheral blood of HAT patients (96). They were non-specific and some of them were previously reported changed during other infectious diseases or cancer. They might be a mirror lymphocyte activation or inflammation observed in HAT (96). However, the mechanisms of action of these microRNAs are still to be investigated.
The SL-RNA was described as an attractive molecular target of the sleeping sickness (97). Later, the small RNA derived from the non-coding 7SL RNA was detected at high levels in the serum of infected cattle (98). This ncRNA is highly sensitive and can be detected before the onset of parasitemia as well as during periods where there is subpatent parasitemia by microscopy (98). It can also make the difference between infections with Trypanosoma brucei, Trypanosoma congolense and Trypanosoma vivax; providing the basis for the development of a cheap, non-invasive and highly effective diagnostic test for trypanosomiasis (98).
The therapy of trypanosomiasis is currently based on anti-trypanosome drugs. No therapeutic field with ncRNAs has yet been investigated. Research should investigate this domain and see if ncRNAs might be useful for monitoring the treatment of this disease.
Also called Hansen’s disease, leprosy is a chronic infectious disease caused by Mycobacterium leprae. This slow-growing, obligate intracellular bacterium is the only known bacterium that infects Schwann cells of peripheral nerves. In addition, M. leprae infects macrophages and dendritic cells (99). M leprae mainly affects the skin, the peripheral nerves, mucosa of the upper respiratory tract, and the eyes11 12. According to official figures from 159 countries from the 6 WHO Regions, 208 619 new leprosy cases were globally registered in 201813.
Leprosy presents in many clinical forms with the two extremes being the Tuberculoid and the Lepromatous forms. In the tuberculoid forms (TT and BT), bacilli are absent or rarely found in neural branches, macrophages, or mononuclear cells of the papillary dermis. the disease is self-limited. On the other hand, in lepromatous forms (BL and LL), the bacilli are abundant and can parasitize practically all tissues, hence the disease is disseminated. These clinical forms can be recognized with the naked eye. To confirm the diagnosis, a sample of skin or nerve can be examined under the microscope and tests may also be done to differentiate it from other skin diseases14. These sampling techniques are invasive. Alternatively, to diagnose an infection early, biomarkers are useful and non-invasive. Some ncRNAs have been reported as biomarkers for the diagnosis of leprosy (Table 4).
It is now recognized that ncRNAs play important roles in the deregulation of the immune response in the varied and polymorphic cells targeted in the leprosy skin lesions onset (99, 104, 105). Specific miRNAs regulated during infection either stimulate the immune response or facilitate immune evasion by pathogens (Figure 5).
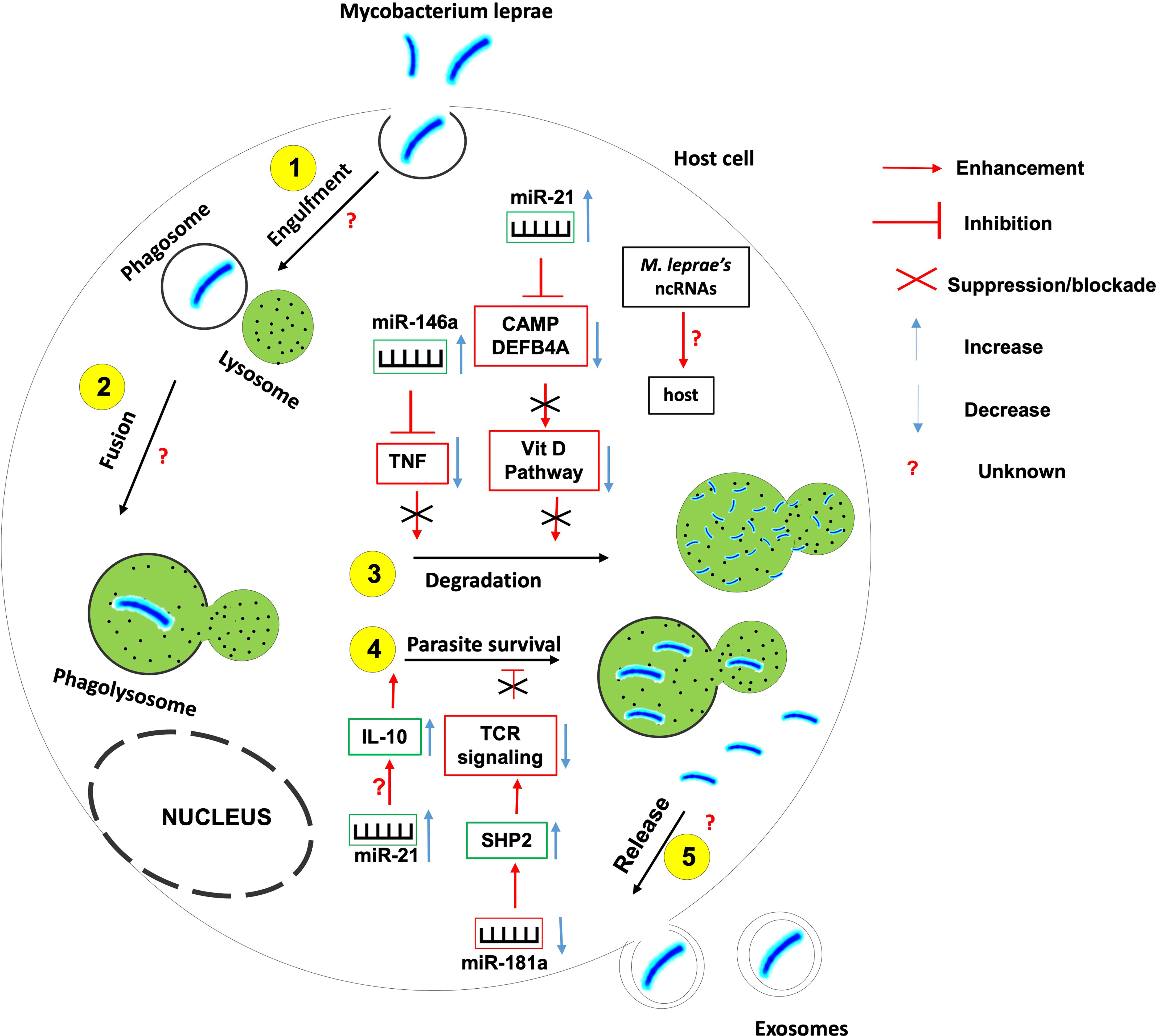
Figure 5 The role of non-coding RNAs in the etiology of leprosy. Several ncRNAs are found to be instrumental in the induction or inhibition of the host effector killing functions against Mycobacterium leprae. More research needed to identify 1- host ncRNAs that regulate the parasite entry within the host, its engulfment within phagosome and the fusion between phagosome and lysosomes; 2- the role of M. leprae-derived ncRNAs on the outcome of mycobacterium-macrophage interaction.
For instance, miR-21 is upregulated in M. leprae- infected monocytes in which it downregulates the expression of genes encoding 2 vitamin D-dependent antimicrobial peptides CAMP and DEFB4A, hence used by the mycobacterium to evade the vitamin D-antimicrobial pathway (102). Also, miR-21 involves in the indirect upregulation of IL-10 and is differentially expressed in humans with progressive lepromatosis.
miR-146a expression is upregulated in hosts infected with live M. leprae. miR-146a bears a single nucleotide polymorphism associated with the risk of developing leprosy, hence the expression of this miR is dependent on the host genotype (101). Carriers of the miR-146a C allele have been shown to express high levels of mature miR-146a coupled to a reduced expression of TNF (Tumor Necrosis Factor) with a susceptibility to leprosy; suggesting that miR-146a negatively influences the secretion of TNF by controlling its level of expression.
miR-181a expression is downregulated in M leprae-infected T lymphocytes. This downregulation correlates with the increased expression of miR-181a target SHP2, a phosphatase involved in the inhibition of T cell receptor signaling (106). Indeed, higher miR-181a expression correlates with greater T cell sensitivity in immature T cells (107) suggesting that the downregulation of miR-181a expression in M. leprae-infected T lymphocytes involves in the prevailing T cell hyporesponsiveness during leprosy progression.
Recent studies have identified differentially expressed piwiRNAs (piRNAs) in leprosy skin lesions from tuberculoid tissue, lepromatous tissue, and healthy subject tissues (108). This class of small ncRNAs is closely related to miRNAs and its study will provide additional clues on the contribution of ncRNAs to the onset, development, and progression of leprosy. The contribution of M. leprae-derived ncRNAs also need to be explored.
Early diagnosis of leprosy is very important to control the disease and put in place preventive measures. Currently, the diagnosis of leprosy is based on clinical examination and skin biopsy. Techniques based on PCR and serological analysis have been developed but have not made it possible to diagnose leprosy with acceptable specificity and sensitivity given the different clinical forms and/or the bacterial load. However, the identification of biomarkers allows the diagnosis of leprosy with greater sensitivity and specificity. Due to the influence of the host’s genetic makeup on the development of leprosy and the genetic variants associated with it. The expression profile of ncRNAs and more precisely miRNAs is a key element exploited in the development of reliable diagnostic and prognostic biomarkers. miR-101, miR-196b, miR-27b and miR-29c have been differentially expressed in different cell types: macrophages, LT, epithelial cells, dendritic cells, mast cells with establishment of an immune/inflammatory microenvironment in M. leprea infection. These miRNAs are linked to immune genetic targets and could modulate the host immune response in leprosy influencing its outcome. Thus, miR-101, miR-196b, miR-27b and miR-29c were identified as good discriminators in the polar forms of leprosy (LL: lepromatous leprosy and TT: tuberculoid tuberculoids) and between physiological state and pathological state (103) with high levels of sensitivity and specificity.
Although drug treatment has been successful, leprosy still affects people all over the world. The treatment of leprosy is based mainly on polychemotherapy, which has so far remained the only strategy for the treatment and elimination of leprosy (109). hsa-miR-142-3p, hsa-miR-142-5p, hsa-miR-146b-5p, hsa-miR-342-3p, hsa-miR-361-3p, hsa-miR-3653 hsa-miR-484 and hsa-miR-1290 were reported deregulated in leprosy and could serve as therapeutic markers (7, 110).
No ncRNA-based vaccine has been developed so far against leprosy. Researchers can explore this new research avenue.
There is a growing interest in the role of host miRNAs and lncRNAs in the etiology of major human tropical diseases and the prospect of targeting these ncRNAs species as biomarkers for the early diagnosis, treatment response and vaccine development against these diseases. Although most research have shed light on the involvement of these ncRNAs in the onset and development of TB, HIV/AIDS and Malaria, but few have yet attempted to assess the potential of these ncRNAs as diagnosis biomarkers, adjunctive therapeutic targets and vaccine candidates. There is also a lack of information about the contribution of pathogen-released ncRNAs to host immune evasion and disease onset. There is a knowledge gap on the role of host miRNAs and lncRNAs in the etiology, diagnosis and vaccine development against neglected human tropical diseases. Especially, more research is warranted to understand the role of these ncRNAs in the etiology of African trypanosomiasis and the assessment of the diagnostic potential of ncRNAs for African trypanosomiasis. The potential targeting of ncRNAs for adjunctive therapy and vaccine development against leishmaniasis, African trypanosomiasis and leprosy are also new avenues to explore.
OT designed the writing plan and drew all the figures. CM and JAN did literature search on Leishmaniasis and HIV/AIDS. CK and USFS did literature search on Tuberculosis and Malaria. NN and FJT did literature search on Leprosy and Trypanosomiasis. OT, CM, CK, and NN wrote the draft. AM and AN proofread the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Figures were created using images from smart.servier.com.
1. Santosh B, Varshney A, Yadava PK. Non-Coding RNAs: Biological Functions and Applications. Cell Biochem Funct (2015) 33(1):14–22. doi: 10.1002/cbf.3079
2. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial Sequencing and Analysis of the Human Genome. Nature (2001) 409(6822):860–921. doi: 10.1038/35057062
3. Consortium EP. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature (2012) 489(7414):57–74. doi: 10.1038/nature11247
4. Chen J, Wang Y, Wang C, Hu J-F, Li W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front Genet (2020) 11(277). doi: 10.3389/fgene.2020.00277
5. Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell (2004) 116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5
6. Chen YG, Satpathy AT, Chang HY. Gene Regulation in the Immune System by Long Noncoding RNAs. Nat Immunol (2017) 18(9):962–72. doi: 10.1038/ni.3771
7. Heward JA, Lindsay MA. Long Non-Coding RNAs in the Regulation of the Immune Response. Trends Immunol (2014) 35(9):408–19. doi: 10.1016/j.it.2014.07.005
8. Flores-Concha M, Onate AA. Long Non-Coding RNAs in the Regulation of the Immune Response and Trained Immunity. Front Genet (2020) 11:718. doi: 10.3389/fgene.2020.00718
9. Esteller M. Non-Coding RNAs in Human Disease. Nat Rev Genet (2011) 12(12):861–74. doi: 10.1038/nrg3074
10. Harries LW. Long Non-Coding RNAs and Human Disease. Biochem Soc Trans (2012) 40(4):902–6. doi: 10.1042/BST20120020
11. Kopp F. Molecular Functions and Biological Roles of Long Non-Coding RNAs in Human Physiology and Disease. J Gene Med (2019) 21(8):e3104. doi: 10.1002/jgm.3104
12. Rogoyski OM, Pueyo JI, Couso JP, Newbury SF. Functions of Long Non-Coding RNAs in Human Disease and Their Conservation in Drosophila Development. Biochem Soc Trans (2017) 45(4):895–904. doi: 10.1042/BST20160428
13. Xiao Y, Xu J, Yin W. Aberrant Epigenetic Modifications of Non-Coding RNAs in Human Disease. Adv Exp Med Biol (2018) 1094:65–75. doi: 10.1007/978-981-13-0719-5_7
14. Wei L, Liu K, Jia Q, Zhang H, Bie Q, Zhang B. The Roles of Host Noncoding RNAs in Mycobacterium Tuberculosis Infection. Front Immunol (2021) 12:664787. doi: 10.3389/fimmu.2021.664787
15. Kundu M, Basu J. The Role of microRNAs and Long Non-Coding RNAs in the Regulation of the Immune Response to Mycobacterium Tuberculosis Infection. Front Immunol (2021) 12:687962. doi: 10.3389/fimmu.2021.687962
16. Lazar DC, Morris KV, Saayman SM. The Emerging Role of Long Non-Coding RNAs in HIV Infection. Virus Res (2016) 212:114–26. doi: 10.1016/j.virusres.2015.07.023
17. Lyu M, Cheng Y, Zhou J, Chong W, Wang Y, Xu W, et al. Systematic Evaluation, Verification and Comparison of Tuberculosis-Related Non-Coding RNA Diagnostic Panels. J Cell Mol Med (2021) 25(1):184–202. doi: 10.1111/jcmm.15903
18. Sinigaglia A, Peta E, Riccetti S, Venkateswaran S, Manganelli R, Barzon L. Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers. Cells (2020) 9(10):1–23. doi: 10.3390/cells9102160
19. Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, et al. Identification of a Novel Role of ESAT-6-Dependent miR-155 Induction During Infection of Macrophages With Mycobacterium Tuberculosis. Cell Microbiol (2012) 14(10):1620–31. doi: 10.1111/j.1462-5822.2012.01827.x
20. Liu F, Chen J, Wang P, Li H, Zhou Y, Liu H, et al. MicroRNA-27a Controls the Intracellular Survival of Mycobacterium Tuberculosis by Regulating Calcium-Associated Autophagy. Nat Commun (2018) 9(1):4295. doi: 10.1038/s41467-018-06836-4
21. Tu H, Yang S, Jiang T, Wei L, Shi L, Liu C, et al. Elevated Pulmonary Tuberculosis Biomarker miR-423-5p Plays Critical Role in the Occurrence of Active TB by Inhibiting Autophagosome-Lysosome Fusion. Emerg Microbes Infect (2019) 8(1):448–60. doi: 10.1080/22221751.2019.1590129
22. Wang L, Xie B, Zhang P, Ge Y, Wang Y, Zhang D. LOC152742 as a Biomarker in the Diagnosis of Pulmonary Tuberculosis Infection. J Cell Biochem (2019) 120(6):8949–55. doi: 10.1002/jcb.27452
23. Huang S, Huang Z, Luo Q, Qing C. The Expression of lncRNA NEAT1 in Human Tuberculosis and Its Antituberculosis Effect. BioMed Res Int (2018) 2018:9529072. doi: 10.1155/2018/9529072
24. Wang L, Wen Z, Ma H, Wu L, Chen H, Zhu Y, et al. Long Non-Coding RNAs ENST00000429730.1 and MSTRG.93125.4 Are Associated With Metabolic Activity in Tuberculosis Lesions of Sputum-Negative Tuberculosis Patients. Aging (Albany NY) (2021) 13(6):8228–47. doi: 10.18632/aging.202634
25. Tamgue O, Chia JE, Brombacher F. Triptolide Modulates the Expression of Inflammation-Associated lncRNA-PACER and lincRNA-P21 in Mycobacterium Tuberculosis-Infected Monocyte-Derived Macrophages. Front Pharmacol (2021) 12:618462. doi: 10.3389/fphar.2021.618462
26. Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, et al. Mycobacterium Tuberculosis Induces the miR-33 Locus to Reprogram Autophagy and Host Lipid Metabolism. Nat Immunol (2016) 17(6):677–86. doi: 10.1038/ni.3434
27. Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Pathogenic Mycobacterium Tuberculosis Evades Apoptosis of Host Macrophages by Release of TNF-R2, Resulting in Inactivation of TNF-Alpha. J Immunol (1998) 161(5):2636–41.
28. Liu Y, Jiang J, Wang X, Zhai F, Cheng X. miR-582-5p Is Upregulated in Patients With Active Tuberculosis and Inhibits Apoptosis of Monocytes by Targeting FOXO1. PloS One (2013) 8(10):e78381. doi: 10.1371/journal.pone.0078381
29. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A Large Intergenic Noncoding RNA Induced by P53 Mediates Global Gene Repression in the P53 Response. Cell (2010) 142(3):409–19. doi: 10.1016/j.cell.2010.06.040
30. Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, et al. LincRNA-P21 Activates P21 in Cis to Promote Polycomb Target Gene Expression and to Enforce the G1/S Checkpoint. Mol Cell (2014) 54(5):777–90. doi: 10.1016/j.molcel.2014.04.025
31. Tu X, Zhang Y, Zheng X, Deng J, Li H, Kang Z, et al. TGF-Beta-Induced Hepatocyte lincRNA-P21 Contributes to Liver Fibrosis in Mice. Sci Rep (2017) 7(1):2957. doi: 10.1038/s41598-017-03175-0
32. Jin S, Yang X, Li J, Yang W, Ma H, Zhang Z. P53-Targeted lincRNA-P21 Acts as a Tumor Suppressor by Inhibiting JAK2/STAT3 Signaling Pathways in Head and Neck Squamous Cell Carcinoma. Mol Cancer (2019) 18(1):38. doi: 10.1186/s12943-019-0993-3
33. Cui JY, Liang HW, Pan XL, Li D, Jiao N, Liu YH, et al. Characterization of a Novel Panel of Plasma microRNAs That Discriminates Between Mycobacterium Tuberculosis Infection and Healthy Individuals. PloS One (2017) 12(9):e0184113. doi: 10.1371/journal.pone.0184113
34. Horsburgh CR Jr. Priorities for the Treatment of Latent Tuberculosis Infection in the United States. N Engl J Med (2004) 350(20):2060–7. doi: 10.1056/NEJMsa031667
35. Yan H, Xu R, Zhang X, Wang Q, Pang J, Zhang X, et al. Identifying Differentially Expressed Long Non-Coding RNAs in PBMCs in Response to the Infection of Multidrug-Resistant Tuberculosis. Infect Drug Resist (2018) 11:945–59. doi: 10.2147/IDR.S154255
36. Guler R, Brombacher F. Host-Directed Drug Therapy for Tuberculosis. Nat Chem Biol (2015) 11(10):748–51. doi: 10.1038/nchembio.1917
37. Hu Y, Mangan JA, Dhillon J, Sole KM, Mitchison DA, Butcher PD, et al. Detection of mRNA Transcripts and Active Transcription in Persistent Mycobacterium Tuberculosis Induced by Exposure to Rifampin or Pyrazinamide. J Bacteriol (2000) 182(22):6358–65. doi: 10.1128/JB.182.22.6358-6365.2000
38. Sathekge MM, Ankrah AO, Lawal I, Vorster M. Monitoring Response to Therapy. Semin Nucl Med (2018) 48(2):166–81. doi: 10.1053/j.semnuclmed.2017.10.004
39. Mantel PY, Hjelmqvist D, Walch M, Kharoubi-Hess S, Nilsson S, Ravel D, et al. Infected Erythrocyte-Derived Extracellular Vesicles Alter Vascular Function via Regulatory Ago2-miRNA Complexes in Malaria. Nat Commun (2016) 7:12727. doi: 10.1038/ncomms12727
40. Chakrabarti M, Garg S, Rajagopal A, Pati S, Singh S. Targeted Repression of Plasmodium Apicortin by Host microRNA Impairs Malaria Parasite Growth and Invasion. Dis Model Mech (2020) 13(6):1–12. doi: 10.1242/dmm.042820
41. LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, et al. Translocation of Sickle Cell Erythrocyte microRNAs Into Plasmodium Falciparum Inhibits Parasite Translation and Contributes to Malaria Resistance. Cell Host Microbe (2012) 12(2):187–99. doi: 10.1016/j.chom.2012.06.007
42. Barker KR, Lu Z, Kim H, Zheng Y, Chen J, Conroy AL, et al. miR-155 Modifies Inflammation, Endothelial Activation and Blood-Brain Barrier Dysfunction in Cerebral Malaria. Mol Med (2017) 23:24–33. doi: 10.2119/molmed.2016.00139
43. Martin-Alonso A, Cohen A, Quispe-Ricalde MA, Foronda P, Benito A, Berzosa P, et al. Differentially Expressed microRNAs in Experimental Cerebral Malaria and Their Involvement in Endocytosis, Adherens Junctions, FoxO and TGF-Beta Signalling Pathways. Sci Rep (2018) 8(1):11277. doi: 10.1038/s41598-018-29721-y
44. van Loon W, Gai PP, Hamann L, Bedu-Addo G, Mockenhaupt FP. MiRNA-146a Polymorphism Increases the Odds of Malaria in Pregnancy. Malar J (2019) 18(1):7. doi: 10.1186/s12936-019-2643-z
45. Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, et al. A Global Transcriptional Analysis of Plasmodium Falciparum Malaria Reveals a Novel Family of Telomere-Associated lncRNAs. Genome Biol (2011) 12(6):R56. doi: 10.1186/gb-2011-12-6-r56
46. Sierra-Miranda M, Delgadillo DM, Mancio-Silva L, Vargas M, Villegas-Sepulveda N, Martinez-Calvillo S, et al. Two Long Non-Coding RNAs Generated From Subtelomeric Regions Accumulate in a Novel Perinuclear Compartment in Plasmodium Falciparum. Mol Biochem Parasitol (2012) 185(1):36–47. doi: 10.1016/j.molbiopara.2012.06.005
47. Amit-Avraham I, Pozner G, Eshar S, Fastman Y, Kolevzon N, Yavin E, et al. Antisense Long Noncoding RNAs Regulate Var Gene Activation in the Malaria Parasite Plasmodium Falciparum. Proc Natl Acad Sci U S A (2015) 112(9):E982–91. doi: 10.1073/pnas.1420855112
48. Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin Associated Sense and Antisense Noncoding RNAs Are Transcribed From the Var Gene Family of Virulence Genes of the Malaria Parasite Plasmodium Falciparum. RNA (2009) 15(1):116–27. doi: 10.1261/rna.1080109
49. Chamnanchanunt S, Kuroki C, Desakorn V, Enomoto M, Thanachartwet V, Sahassananda D, et al. Downregulation of Plasma miR-451 and miR-16 in Plasmodium Vivax Infection. Exp Parasitol (2015) 155:19–25. doi: 10.1016/j.exppara.2015.04.013
50. Li JJ, Huang MJ, Li Z, Li W, Wang F, Wang L, et al. Identification of Potential Whole Blood MicroRNA Biomarkers for the Blood Stage of Adult Imported Falciparum Malaria Through Integrated mRNA and miRNA Expression Profiling. Biochem Biophys Res Commun (2018) 506(3):471–7. doi: 10.1016/j.bbrc.2018.10.072
51. Liu W, Ding C. Roles of LncRNAs in Viral Infections. Front Cell Infect Microbiol (2017) 7:205. doi: 10.3389/fcimb.2017.00205
52. Chen L, Chen L, Zuo L, Gao Z, Shi Y, Yuan P, et al. Short Communication: Long Noncoding RNA GAS5 Inhibits HIV-1 Replication Through Interaction With miR-873. AIDS Res Hum Retroviruses (2018) 34(6):544–9. doi: 10.1089/aid.2017.0177
53. Ray RM, Morris KV. Long Non-Coding RNAs Mechanisms of Action in HIV-1 Modulation and the Identification of Novel Therapeutic Targets. Noncoding RNA (2020) 6(1):1–17. doi: 10.3390/ncrna6010012
54. Qu D, Sun WW, Li L, Ma L, Sun L, Jin X, et al. Long Noncoding RNA MALAT1 Releases Epigenetic Silencing of HIV-1 Replication by Displacing the Polycomb Repressive Complex 2 From Binding to the LTR Promoter. Nucleic Acids Res (2019) 47(6):3013–27. doi: 10.1093/nar/gkz117
55. Chao TC, Zhang Q, Li Z, Tiwari SK, Qin Y, Yau E, et al. The Long Noncoding RNA HEAL Regulates HIV-1 Replication Through Epigenetic Regulation of the HIV-1 Promoter. mBio (2019) 10(5):1–18. doi: 10.1128/mBio.02016-19
56. Huan C, Li Z, Ning S, Wang H, Yu XF, Zhang W. Long Noncoding RNA Uc002yug.2 Activates HIV-1 Latency Through Regulation of mRNA Levels of Various RUNX1 Isoforms and Increased Tat Expression. J Virol (2018) 92(9):1–38. doi: 10.1128/JVI.01844-17
57. Postler TS, Pantry SN, Desrosiers RC, Ghosh S. Identification and Characterization of a Long Non-Coding RNA Up-Regulated During HIV-1 Infection. Virology (2017) 511:30–9. doi: 10.1016/j.virol.2017.08.006
58. Liu H, Hu PW, Couturier J, Lewis DE, Rice AP. HIV-1 Replication in CD4(+) T Cells Exploits the Down-Regulation of Antiviral NEAT1 Long Non-Coding RNAs Following T Cell Activation. Virology (2018) 522:193–8. doi: 10.1016/j.virol.2018.07.020
59. Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 Long Noncoding RNA and Paraspeckle Bodies Modulate HIV-1 Posttranscriptional Expression. mBio (2013) 4(1):e00596–12. doi: 10.1128/mBio.00596-12
60. Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y, et al. Long Noncoding RNA NRON Contributes to HIV-1 Latency by Specifically Inducing Tat Protein Degradation. Nat Commun (2016) 7:11730. doi: 10.1038/ncomms11730
61. Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON Modulates HIV-1 Replication in a NFAT-Dependent Manner and Is Differentially Regulated by Early and Late Viral Proteins. Sci Rep (2015) 5:8639. doi: 10.1038/srep08639
62. Barichievy S, Naidoo J, Boulle M, Scholefield J, Parihar SP, Coussens AK, et al. Viral Apoptosis Evasion via the MAPK Pathway by Use of a Host Long Noncoding RNA. Front Cell Infect Microbiol (2018) 8:263. doi: 10.3389/fcimb.2018.00263
63. Shen L, Wu C, Zhang J, Xu H, Liu X, Wu X, et al. Roles and Potential Applications of lncRNAs in HIV Infection. Int J Infect Dis (2020) 92:97–104. doi: 10.1016/j.ijid.2020.01.006
64. Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang KT. MicroRNA Profile Changes in Human Immunodeficiency Virus Type 1 (HIV-1) Seropositive Individuals. Retrovirology (2008) 5:118. doi: 10.1186/1742-4690-5-118
65. Verma P, Pandey RK, Prajapati P, Prajapati VK. Circulating MicroRNAs: Potential and Emerging Biomarkers for Diagnosis of Human Infectious Diseases. Front Microbiol (2016) 7:1274. doi: 10.3389/fmicb.2016.01274
66. Boliar S, Gludish DW, Jambo KC, Kamng’ona R, Mvaya L, Mwandumba HC, et al. Inhibition of the lncRNA SAF Drives Activation of Apoptotic Effector Caspases in HIV-1-Infected Human Macrophages. Proc Natl Acad Sci U S A (2019) 116(15):7431–8. doi: 10.1073/pnas.1818662116
67. Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K, et al. Interplay Between HIV-1 Infection and Host microRNAs. Nucleic Acids Res (2012) 40(5):2181–96. doi: 10.1093/nar/gkr961
68. Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, et al. Human Cellular microRNA hsa-miR-29a Interferes With Viral Nef Protein Expression and HIV-1 Replication. Retrovirology (2008) 5:117. doi: 10.1186/1742-4690-5-117
69. Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P Bodies Modulate Host-HIV-1 Interactions. Mol Cell (2009) 34(6):696–709. doi: 10.1016/j.molcel.2009.06.003
70. Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA-Mediated Regulation of P21 and TASK1 Cellular Restriction Factors Enhances HIV-1 Infection. J Cell Sci (2015) 128(8):1607–16. doi: 10.1242/jcs.167817
71. Hsu K, Seharaseyon J, Dong P, Bour S, Marban E. Mutual Functional Destruction of HIV-1 Vpu and Host TASK-1 Channel. Mol Cell (2004) 14(2):259–67. doi: 10.1016/S1097-2765(04)00183-2
72. Houzet L, Klase Z, Yeung ML, Wu A, Le SY, Quinones M, et al. The Extent of Sequence Complementarity Correlates With the Potency of Cellular miRNA-Mediated Restriction of HIV-1. Nucleic Acids Res (2012) 40(22):11684–96. doi: 10.1093/nar/gks912
73. Ludwig LB, Ambrus JL Jr., Krawczyk KA, Sharma S, Brooks S, Hsiao CB, et al. Human Immunodeficiency Virus-Type 1 LTR DNA Contains an Intrinsic Gene Producing Antisense RNA and Protein Products. Retrovirology (2006) 3:80. doi: 10.1186/1742-4690-3-80
74. Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, et al. Detection, Characterization and Regulation of Antisense Transcripts in HIV-1. Retrovirology (2007) 4:71. doi: 10.1186/1742-4690-4-71
75. Saayman S, Ackley A, Turner AW, Famiglietti M, Bosque A, Clemson M, et al. An HIV-Encoded Antisense Long Noncoding RNA Epigenetically Regulates Viral Transcription. Mol Ther (2014) 22(6):1164–75. doi: 10.1038/mt.2014.29
76. Jin C, Peng X, Xie T, Lu X, Liu F, Wu H, et al. Detection of the Long Noncoding RNAs Nuclear-Enriched Autosomal Transcript 1 (NEAT1) and Metastasis Associated Lung Adenocarcinoma Transcript 1 in the Peripheral Blood of HIV-1-Infected Patients. HIV Med (2016) 17(1):68–72. doi: 10.1111/hiv.12276
77. Rosca A, Anton G, Botezatu A, Temereanca A, Ene L, Achim C, et al. miR-29a Associates With Viro-Immunological Markers of HIV Infection in Treatment Experienced Patients. J Med Virol (2016) 88(12):2132–7. doi: 10.1002/jmv.24586
78. Zhao Y, Karijolich J, Glaunsinger B, Zhou Q. Pseudouridylation of 7SK snRNA Promotes 7SK snRNP Formation to Suppress HIV-1 Transcription and Escape From Latency. EMBO Rep (2016) 17(10):1441–51. doi: 10.15252/embr.201642682
79. Pinto DO, Scott TA, DeMarino C, Pleet ML, Vo TT, Saifuddin M, et al. Effect of Transcription Inhibition and Generation of Suppressive Viral Non-Coding RNAs. Retrovirology (2019) 16(1):13. doi: 10.1186/s12977-019-0475-0
80. Didwania N, Shadab M, Sabur A, Ali N. Alternative to Chemotherapy-The Unmet Demand Against Leishmaniasis. Front Immunol (2017) 8:1779. doi: 10.3389/fimmu.2017.01779
81. Kumar V, Kumar A, Das S, Kumar A, Abhishek K, Verma S, et al. Leishmania Donovani Activates Hypoxia Inducible Factor-1alpha and miR-210 for Survival in Macrophages by Downregulation of NF-kappaB Mediated Pro-Inflammatory Immune Response. Front Microbiol (2018) 9:385. doi: 10.3389/fmicb.2018.00385
82. Lago TS, Silva JA, Lago EL, Carvalho EM, Zanette DL, Castellucci LC. The miRNA 361-3p, a Regulator of GZMB and TNF Is Associated With Therapeutic Failure and Longer Time Healing of Cutaneous Leishmaniasis Caused by L. (Viannia) Braziliensis. Front Immunol (2018) 9:2621. doi: 10.3389/fimmu.2018.02621
83. Nunes S, Silva IB, Ampuero MR, de Noronha ALL, de Souza LCL, Correia TC, et al. Integrated Analysis Reveals That miR-193b, miR-671, and TREM-1 Correlate With a Good Response to Treatment of Human Localized Cutaneous Leishmaniasis Caused by Leishmania Braziliensis. Front Immunol (2018) 9:640. doi: 10.3389/fimmu.2018.00640
84. Afrin F, Khan I, Hemeg HA. Leishmania-Host Interactions-An Epigenetic Paradigm. Front Immunol (2019) 10:492. doi: 10.3389/fimmu.2019.00492
85. Pandey RK, Sundar S, Prajapati VK. Differential Expression of miRNA Regulates T Cell Differentiation and Plasticity During Visceral Leishmaniasis Infection. Front Microbiol (2016) 7:206. doi: 10.3389/fmicb.2016.00206
86. Lemaire J, Mkannez G, Guerfali FZ, Gustin C, Attia H, Sghaier RM, et al. MicroRNA Expression Profile in Human Macrophages in Response to Leishmania Major Infection. PloS Negl Trop Dis (2013) 7(10):e2478. doi: 10.1371/journal.pntd.0002478
87. Singh AK, Pandey RK, Shaha C, Madhubala R. MicroRNA Expression Profiling of Leishmania Donovani-Infected Host Cells Uncovers the Regulatory Role of MIR30A-3p in Host Autophagy. Autophagy (2016) 12(10):1817–31. doi: 10.1080/15548627.2016.1203500
88. Muxel SM, Laranjeira-Silva MF, Zampieri RA, Floeter-Winter LM. Leishmania (Leishmania) Amazonensis Induces Macrophage miR-294 and miR-721 Expression and Modulates Infection by Targeting NOS2 and L-Arginine Metabolism. Sci Rep (2017) 7:44141. doi: 10.1038/srep44141
89. Geraci NS, Tan JC, McDowell MA. Characterization of microRNA Expression Profiles in Leishmania-Infected Human Phagocytes. Parasite Immunol (2015) 37(1):43–51. doi: 10.1111/pim.12156
90. Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, et al. miR-182 and miR-10a are Key Regulators of Treg Specialisation and Stability During Schistosome and Leishmania-Associated Inflammation. PloS Pathog (2013) 9(6):e1003451. doi: 10.1371/journal.ppat.1003451
91. Rana T, Misra S, Mittal MK, Farrow AL, Wilson KT, Linton MF, et al. Mechanism of Down-Regulation of RNA Polymerase III-Transcribed Non-Coding RNA Genes in Macrophages by Leishmania. J Biol Chem (2011) 286(8):6614–26. doi: 10.1074/jbc.M110.181735
92. Pawar H, Pai K, Patole MS. A Novel Protein Coding Potential of Long Intergenic Non-Coding RNAs (lincRNAs) in the Kinetoplastid Protozoan Parasite Leishmania Major. Acta Trop (2017) 167:21–5. doi: 10.1016/j.actatropica.2016.12.012
93. Nasereddin A, Jaffe CL. Rapid Diagnosis of Old World Leishmaniasis by High-Resolution Melting Analysis of the 7SL RNA Gene. J Clin Microbiol (2010) 48(6):2240–2. doi: 10.1128/JCM.00553-10
94. Hashemi N, Sharifi M, Masjedi M, Tolouei S, Hashemi M, Mortazavidehkordi N, et al. Locked Nucleic Acid -Anti- Let-7a Induces Apoptosis and Necrosis in Macrophages Infected With Leishmania Major. Microb Pathog (2018) 119:193–9. doi: 10.1016/j.micpath.2018.03.057
95. Ismail N, Kaul A, Bhattacharya P, Gannavaram S, Nakhasi HL. Immunization With Live Attenuated Leishmania Donovani Centrin(-/-) Parasites Is Efficacious in Asymptomatic Infection. Front Immunol (2017) 8:1788. doi: 10.3389/fimmu.2017.01788
96. Lueong S, Leong S, Simo G, Camara M, Jamonneau V, Kabore J, et al. The miRNA and mRNA Signatures of Peripheral Blood Cells in Humans Infected With Trypanosoma Brucei Gambiense. PloS One (2013) 8(6):e67312. doi: 10.1371/annotation/b13cc66f-93c2-4582-99c1-359276235acc
97. Gonzalez-Andrade P, Camara M, Ilboudo H, Bucheton B, Jamonneau V, Deborggraeve S. Diagnosis of Trypanosomatid Infections: Targeting the Spliced Leader RNA. J Mol Diagn (2014) 16(4):400–4. doi: 10.1016/j.jmoldx.2014.02.006
98. Chiweshe SM, Steketee PC, Jayaraman S, Paxton E, Neophytou K, Erasmus H, et al. Parasite Specific 7SL-Derived Small RNA Is an Effective Target for Diagnosis of Active Trypanosomiasis Infection. PloS Negl Trop Dis (2019) 13(2):e0007189. doi: 10.1371/journal.pntd.0007189
99. Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The Continuing Challenges of Leprosy. Clin Microbiol Rev (2006) 19(2):338–81. doi: 10.1128/CMR.19.2.338-381.2006
100. Singh PK, Singh AV, Chauhan DS. Current Understanding on Micro RNAs and Its Regulation in Response to Mycobacterial Infections. J Biomed Sci (2013) 20(1):14. doi: 10.1186/1423-0127-20-14
101. Cezar-de-Mello PF, Toledo-Pinto TG, Marques CS, Arnez LE, Cardoso CC, Guerreiro LT, et al. Pre-miR-146a (Rs2910164 G>C) Single Nucleotide Polymorphism Is Genetically and Functionally Associated With Leprosy. PloS Negl Trop Dis (2014) 8(9):e3099. doi: 10.1371/journal.pntd.0003099
102. Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, et al. MicroRNA-21 Targets the Vitamin D-Dependent Antimicrobial Pathway in Leprosy. Nat Med (2012) 18(2):267–73. doi: 10.1038/nm.2584
103. Jorge K, Souza RP, Assis MTA, Araujo MG, Locati M, Jesus AMR, et al. Characterization of MicroRNA Expression Profiles and Identification of Potential Biomarkers in Leprosy. J Clin Microbiol (2017) 55(5):1516–25. doi: 10.1128/JCM.02408-16
104. Ridley DS, Jopling WH. Classification of Leprosy According to Immunity. A Five-Group System. Int J Lepr Other Mycobact Dis (1966) 34(3):255–73.
105. Hastings RC, Gillis TP, Krahenbuhl JL, Franzblau SG. Leprosy. Clin Microbiol Rev (1988) 1(3):330–48. doi: 10.1128/CMR.1.3.330
106. Kumar S, Naqvi RA, Khanna N, Rao DN. Disruption of HLA-DR Raft, Deregulations of Lck-ZAP-70-Cbl-B Cross-Talk and miR181a Towards T Cell Hyporesponsiveness in Leprosy. Mol Immunol (2011) 48(9-10):1178–90. doi: 10.1016/j.molimm.2011.02.012
107. Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a Is an Intrinsic Modulator of T Cell Sensitivity and Selection. Cell (2007) 129(1):147–61. doi: 10.1016/j.cell.2007.03.008
108. Pinto P, da Silva MB, Moreira FC, Bouth RC, Gobbo AR, Sandoval TV, et al. Leprosy Pirnome: Exploring New Possibilities for an Old Disease. Sci Rep (2020) 10(1):12648. doi: 10.1038/s41598-020-69355-7
109. Li Z, Rana TM. Therapeutic Targeting of microRNAs: Current Status and Future Challenges. Nat Rev Drug Discov (2014) 13(8):622–38. doi: 10.1038/nrd4359
Keywords: non-coding RNAs, tuberculosis, HIV/AIDS, malaria, leishmaniasis, African trypanosomiasis, leprosy
Citation: Tamgue O, Mezajou CF, Ngongang NN, Kameni C, Ngum JA, Simo USF, Tatang FJ, Akami M and Ngono AN (2021) Non-Coding RNAs in the Etiology and Control of Major and Neglected Human Tropical Diseases. Front. Immunol. 12:703936. doi: 10.3389/fimmu.2021.703936
Received: 01 May 2021; Accepted: 09 September 2021;
Published: 19 October 2021.
Edited by:
Veron Ramsuran, University of KwaZulu-Natal, South AfricaReviewed by:
Peifeng Li, Qingdao University, ChinaCopyright © 2021 Tamgue, Mezajou, Ngongang, Kameni, Ngum, Simo, Tatang, Akami and Ngono. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ousman Tamgue, b3VzdGhlbzIwMTRAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.