- 1State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3Hubei Hongshan Laboratory, Chinese Academy of Sciences, Wuhan, China
Polyploidy and subsequent diploidization provide genomic opportunities for evolutionary innovations and adaptation. The researches on duplicated gene evolutionary fates in recurrent polyploids have seriously lagged behind that in paleopolyploids with diploidized genomes. Moreover, the antiviral mechanisms of Viperin remain largely unclear in fish. Here, we elaborate the distinct antiviral mechanisms of two viperin homeologs (Cgviperin-A and Cgviperin-B) in auto-allo-hexaploid gibel carp (Carassius gibelio). First, Cgviperin-A and Cgviperin-B showed differential and biased expression patterns in gibel carp adult tissues. Subsequently, using co-immunoprecipitation (Co-IP) screening analysis, both CgViperin-A and CgViperin-B were found to interact with crucian carp (C. auratus) herpesvirus (CaHV) open reading frame 46 right (ORF46R) protein, a negative herpesvirus regulator of host interferon (IFN) production, and to promote the proteasomal degradation of ORF46R via decreasing K63-linked ubiquitination. Additionally, CgViperin-B also mediated ORF46R degradation through autophagosome pathway, which was absent in CgViperin-A. Moreover, we found that the N-terminal α-helix domain was necessary for the localization of CgViperin-A and CgViperin-B at the endoplasmic reticulum (ER), and the C-terminal domain of CgViperin-A and CgViperin-B was indispensable for the interaction with degradation of ORF46R. Therefore, the current findings clarify the divergent antiviral mechanisms of the duplicated viperin homeologs in a recurrent polyploid fish, which will shed light on the evolution of teleost duplicated genes.
Introduction
Genome sequencing explosion has highlighted a profound impact of polyploidy and subsequent post-polyploid diploidization (PPD) on evolutionary innovations and adaptation. As the impact might lead to genomic diversity, variability, and complexity, polyploidy is generally considered to have far-reaching consequences in shaping species speciation, diversification, and ecological adaption (1–10). The evolutionary fates of duplicated genes are dynamic and complex and include pseudogenization or gene deletion, subfunctionalization, or neofunctionalization under relaxed purifying selection (3). In animals, the evolution of duplicated genes has been documented in more easily discernible paleopolyploids, which are characterized by highly diploidized genomes after several rounds of whole-genome duplications (WGD) at the root of the vertebrate (11, 12). However, research on recurrent polyploids, which are much more difficult to disentangle bioinformatically and experimentally, has seriously lagged behind that on animal paleopolyploids (13).
Viperin (also known as Vig1 in rainbow trout) was first identified as an antiviral protein induced by human cytomegalovirus in 2001 (14, 15). It belongs to the radical S-adenosylmethionine (SAM) enzyme family (14, 16, 17) and is composed of three distinct domains (18). Its N-terminal domain contains an amphipathic α-helix, which localizes Viperin to the ER, lipid droplets, or mitochondria (19–21). The central SAM domain has a characteristic CxxxCxxC motif responsible for binding proteins containing an iron-sulfur cluster (22, 23). The C-terminal domain is highly conserved, but its role remains poorly defined (24). This domain has been proposed to be involved in protein–protein interactions (19).
Viperin is known as an interferon (IFN)-stimulated gene (ISG) with broad-spectrum antiviral activities. Its expression can be induced by lipopolysaccharide (LPS), polyinosinic:polycytidylic acid [poly (I:C)], or various viruses (19, 25). Studies on mammalian Viperin have shown that its antiviral mechanisms appear to be virus-specific, owing to the distinct infection routes and replication strategies of different viruses (26). Recently, Ghosh and Marsh (2020) listed the different viruses reported to be restricted by viperin and the known mechanisms of restriction, and summarized two major ways of the antiviral activities of viperin (27). Viperin has been found to interact with a wide variety of host and viral proteins, and the complex network of interactions inhibits viral RNA transcription and replication (28–30), interrupts viral particle assembly and maturation (14), or impairs viral budding and release (20, 31, 32). Another way relies on the catalytic activity of Viperin to generate 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP), which can directly interfere with RNA synthesis and the replication of flaviviruses (33). Similar to mammal, fish viperin is considered an important antiviral gene because its expression is also induced by LPS, poly(I:C), or viruses (15, 34–40). However, little is known about the molecular mechanism underlying the antiviral effect of fish Viperin.
Gibel carp (Carassius gibelio), widely distributed across the Eurasian continent, can reproduce by unisexual gynogenesis or sexual reproduction (41–43). Along with a large-scale culture of several improved varieties, such as allogynogenetic gibel carp “CAS III” (clone A+) and “CAS III” (clone F) (44, 45), gibel carp has become one of the most important aquaculture species in China with about 3 million tons of annual production capacity (46). However, a serious outbreak of an epizootic disease caused by CaHV has resulted in enormous economic losses since 2012 (47). In previous studies, we identified distinct immune responses and the differential expression of innate and adaptive immune genes among three clones (A+, F, and H) after CaHV challenge (48–51). Eight IFN system genes, including viperin, were identified as candidate resistant-related genes for disease-resistance breeding of gibel carp. Viperin was sharply upregulated among three clones at post infection (52). Additionally, gibel carp has over 150 chromosomes and is considered an auto-allo-hexaploid in comparison with goldfish (Carassius auratus) with 100 chromosomes (53, 54). It had been speculated that two rounds (2R) of polyploidy events, an allotetraploidy and an autotriploidy, occurred during its evolution (55). Viperin unigenes in transcriptomic data have been classified into two divergent homeologs with biased expression after CaHV infection (52). It is necessary to further analyze expression and function divergence of the two viperin homeologs. Therefore, in this study, we first characterized two gibel carp viperin homeologs (Cgviperin-A and Cgviperin-B), and then investigated their divergent antiviral mechanisms through subcellular localization, overexpression, Co-IP, and in vitro ubiquitination assay.
Materials and Methods
Cells, Gibel Carp, and Virus
Fish cell line Carassius auratus L. blastulae embryonic (CAB) cell was cultured at 28°C in 5% CO2 in medium 199 (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) as described previously (56–58). Female gibel carp clone F (about 67.68 ± 2.16 g) were obtained from the National Aquatic Biological Resource Center (NABRC), Institute of Hydrobiology (IHB), Chinese Academy of Sciences (CAS). CaHV was provided by Prof. Q. Y. Zhang (Institute of Hydrobiology, Chinese Academy of Sciences) and propagated by intraperitoneal injection into healthy gibel carp. The isolation method of CaHV was used as previously described (48, 59). Before sampling, fish were deeply euthanized by overdosed anesthesia styrylpyridine (30–50 mg/L; Aladdin, Shanghai, China) and immediately cut off spinal cord adjacent to the head. All procedures were approved by the Institutional Animal Care and Use Committee of IHB, CAS (protocol number 2016-018).
Gene Identification and Sequence Analysis
According to the genome sequences of gibel carp clone F (CI01000363_00413629_00414546 and ENSONIP00000026093-D2), specific primers (Supplementary Table S1) were designed to amplify viperin from gibel carp clone F head kidney cDNA library by 3’ and 5’ rapid amplification of cDNA ends (RACE) (SMARTer® RACE 5’/3’ Kit, Clontech).
The cDNA sequences of six viperin transcripts were deposited in GenBank (accession numbers from MZ055409 to MZ055414). The specificity of RACE primer was confirmed by sequence analysis. The sequence assembly was performed by DNAstar software, and the assembly validation was confirmed by full-length sequence amplification and sequence analysis. The validation of open reading frame (ORF), multiple sequence alignments, and neighbor-joining (NJ) phylogeny construction were analyzed by using ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/), ClustalW program, and MEGA 7.0 software, respectively.
Chromosome Preparation and Fluorescence In Situ Hybridization
The bacterial artificial chromosome (BAC) clones containing Cgviperin-A and Cgviperin-B were screened by Polymerase Chain Reaction (PCR). Then, Cgviperin-A-BAC-DNA and Cgviperin-B-BAC-DNA were labeled by Biotin-Nick Translation Mix (Roche) and DIG-Nick Translation Mix, respectively. Chromosome preparation and FISH analyses were performed as described previously (55, 60). All metaphase chromosomes were counterstained with 4’, 6-diamidino-2-phenylindole (DAPI).
RNA Extraction and Quantitative Real-Time PCR
RNA extraction and qPCR were performed as described previously (51, 52). Total RNAs from 10 adult tissues (including intestine, gill, muscle, head-kidney, trunk kidney, liver, spleen, thymus, gonad, and brain) were isolated using SV Total RNA isolation System (Promega) according to the manufacturer’s protocols. Subsequently, the primeScript™ RT reagent Kit with gDNA Eraser (TaKaRa) was used to solve the DNA contamination of total RNA. The quantity of total RNA was detected by Nanodrop 2000 (Thermo Scientific), and the quality was assessed by agarose gel electrophoresis. One µg of total RNAs was used to synthesize first-strand cDNAs in a 20 µl reaction volume following the protocol of GoldScript cDNA synthesis Kit (Invitrogen).
qPCR was performed by using iTaq™ Universal SYBR® Green Supermix (Bio-Rad) according to the manufacturer’s protocol. The reference gene, eukaryotic translation elongation factor 1 alpha 1, like 1 (eef1a1l1) (M value=0.74 < 1.5) was selected as the normalizer for qPCR as described previously (52). The primers (Supplementary Table S1) used for qPCR analysis were designed with http://biotools.nubic.northwestern.edu/OligoCalc.html. The specificity of each pair of primers was analyzed by sequencing, and the primer efficiency and R2 were assessed by constructing standard curve using the dilutions of cDNA as the template. All samples (n = 3) were analyzed in triplicate. The relative gene expression levels were calculated using the 2−ΔΔCT method. IBM® SPSS® statistics 20 software was used for statistical analysis. A probability (p) of <0.05 was considered statistically significant.
Plasmid Constructs
According to the genome sequences of gibel carp clone F (Wang et al., unpublished data), 5’ flanking sequences and partial sequences of exon 1 of Cgviperin-A and Cgviperin-B (accession numbers from MZ055415-MZ055420) were also amplified from gibel carp clone F. For luciferase activity assays, these sequences were cloned into Kpn I/Xho I sites of pGL3-Basic luciferase reporter vector (Promega) to construct promoter-driven luciferase vector, respectively. For overexpression, the wild type (WT) Cgviperin-A ORF, Cgviperin-B ORF, and their corresponding sequences of mutants with a C-terminal hemagglutinin (HA)-tag or a C-terminal Flag-tag were cloned into pcDNA3.1(+) vector, respectively. The Cgviperin-A ORF and Cgviperin-B ORF were also inserted into pEGFP (enhanced green fluorescent protein)-N3 and pDsRed-N1 vector, respectively. The ORF of CaHV gene ORF46R with a C-terminal HA-tag, Flag-tag, or Myc-tag were cloned into pcDNA3.1(+) vector, respectively. CaIFN-Luc, IFN-stimulated response elements (ISRE)-Luc, and ER-DsRed were constructed previously (61). All constructs were confirmed by sequence analysis.
Transfection and Luciferase Activity Assays
Luciferase activity assays were performed as described previously (49, 50). CAB cells were seeded in 24-well plates overnight and co-transfected with various plasmid constructs at a ratio of 10:10:1 (promoter-driven luciferase plasmid/expression plasmid/Renilla luciferase plasmid pRL-TK) using FuGENE HD Transfection Reagent (Promega). Empty vector pGL3-Basic luciferase reporter vector (Promega) was used as control. If necessary, the cells were transfected again with poly (deoxyadenylic-deoxythymidylic) acid sodium salt [poly (dA:dT)] (Invivogen) or poly (I:C) (Sigma-Aldrich) (1μg/ml) at 24 h post-transfection. Then, the cells were harvested at 24 h post-transfection and lysed according to the Dual-Luciferase Reporter Assay System (Promega). Luciferase activities were measured by a Junior LB9509 luminometer (Berthold, Pforzheim, Germany) and normalized to the amounts of Renilla luciferase activities. The results were representative of more than three independent experiments, and each was performed in triplicate. The significant differences were calculated by IBM® SPSS® statistics 20 software.
Co-IP Assay and Western Blotting
Co-IP and western blotting were performed as described previously (62). Briefly, the CAB or 293T cells were seeded into 10 cm2 dishes overnight, and transfected with different plasmids (a total of 10 μg) indicated on the Figure using FuGENE HD Transfection Reagent (Promega). At 24 h post-transfection, the cells were lysed by radioimmunoprecipitation (RIPA) lysis buffer with protease inhibitor cocktail (Sigma-Aldrich). After removing cellular debris, the supernatants were incubated with 15 μl anti-Flag or anti-Myc affinity gel (Sigma-Aldrich) overnight at 4°C. Immunoprecipitated proteins were resuspended in 100 μl SDS-PAGE protein loading buffer (Beyotime) after collecting by centrifugation and washing three times with lysis buffer. The whole cell lysates (WCL) or immunoprecipitated proteins were separated by 10–15% SDS-PAGE and then transferred to polyvinylidene fluoride membranes (Bio-Rad). The membranes were incubated with anti-β-actin (Cell Signaling Technology) at 1:3,000, anti-Flag/HA (Sigma-Aldrich) at 1:3,000, or anti-Myc (Santa Cruz Biotechnology) at 1:3,000, and then hybridized with the secondary HRP-conjugated anti-mouse IgG or anti-rabbit IgG (Thermo Scientific) at 1:5,000. Results are captured by using an ImageQuant LAS 4000 system (GE Healthcare) and are representative of three independent experiments.
In Vitro Ubiquitination Assay
Ubiquitination assay was performed as described previously (62, 63). Briefly, CAB cells were transiently co-transfected with different overexpression plasmids (a total of 10 μg) indicated on the Figure and 1 μg HA-Ub or HA-Ub-K63O expression plasmids. At 18 h post-transfection, the cells were treated with 20 mM MG132 (Sigma-Aldrich) for 6 h and then were lysed using a RIPA lysis buffer containing 1% SDS and denatured by heating for 10 min. After denaturing and diluting, the supernatants were immunoprecipitated overnight at 4°C with 30 μl anti-Flag or anti-Myc agarose conjugate (Sigma-Aldrich). The whole cell lysates or immunoprecipitated proteins were analyzed by western blot as above described.
Results
Molecular Characterization and Biased Expression Patterns of Cgviperin Homeologs
Six Cgviperin transcripts were cloned from the head-kidney of gibel carp clone F using RACE-PCR (Supplementary Figure S1). Analysis of multiple nucleotide alignments and their locations in the assembled genome (Wang et al., unpublished data) showed that they were clustered into two homeologs, each possessing three alleles (Supplementary Figure S1). The average identities among the three Cgviperin-A and Cgviperin-B alleles are 99.5 ± 0.3 and 99.4 ± 0.1%, respectively, while the average identity between the Cgviperin-A and Cgviperin-B homeologs is 91.3 ± 0.1%. The cDNAs of Cgviperin-A and Cgviperin-B were predicted to encode one CgViperin-A protein (359aa, CgViperin-A1/A2/A3) and three CgViperin-B proteins (371aa, CgViperin-B1, CgViperin-B1 and CgViperin-B3), respectively. Only two amino acid differences were observed among the three CgViperin-B proteins. CgViperin-A and CgViperin-B have about 90.1 ± 0.3% amino acid identity (Supplementary Figure S2). Phylogenetic analysis (Figure 1B) confirmed that Cgviperin-A and Cgviperin-B might be a pair of homeologs derived from early allotetraploidy (4R), and they both have three alleles because of late autotriploidy (5R).
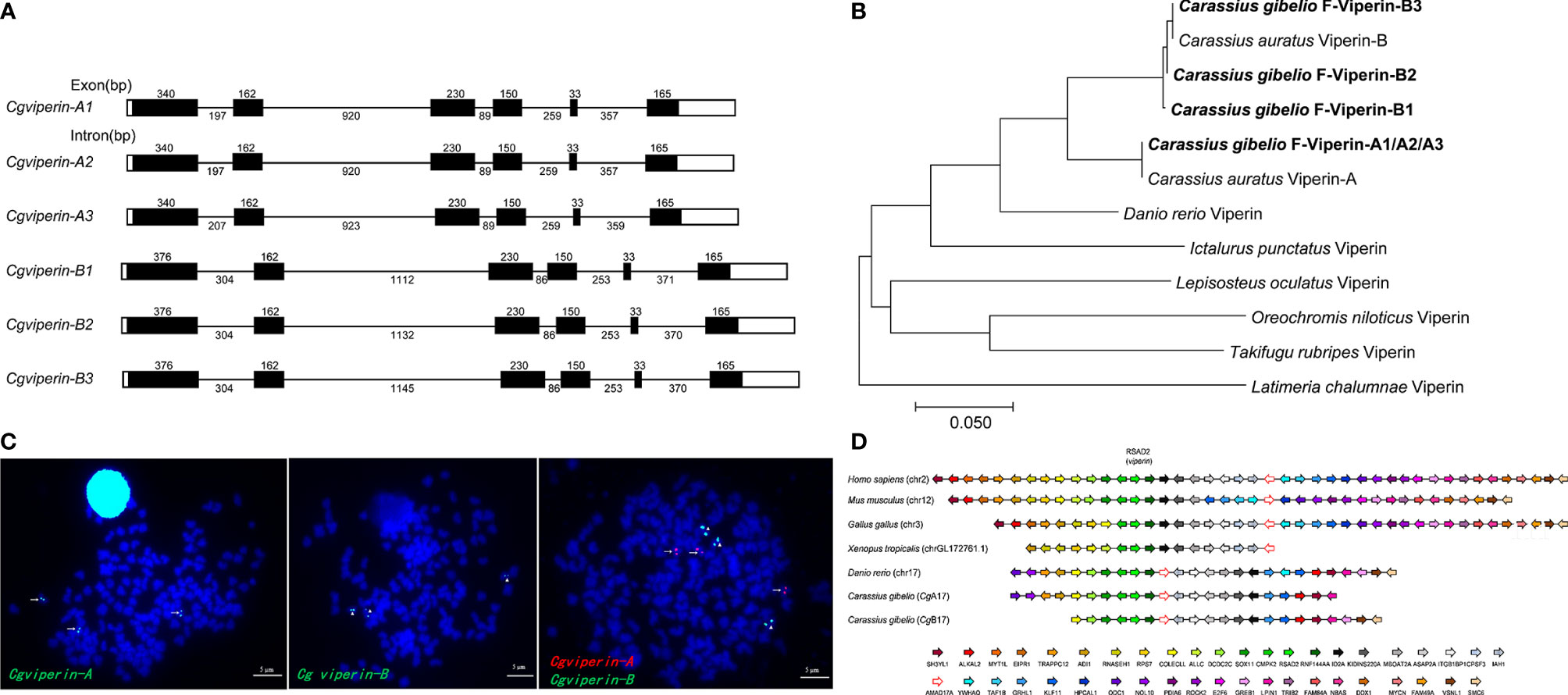
Figure 1 Molecular characterization of Cgviperin-A and Cgviperin-B in gibel carp. (A) Genomic structure of six Cgviperin genes. Exons and introns are depicted by rectangle boxes and thick lines, respectively, and ORFs are highlighted by black boxes. Sizes (bp) are indicated upon or below themselves. (B) Phylogenetic tree of vertebrate Viperin. (C) Localization of Cgviperin-A and Cgviperin-B on gibel carp metaphase chromosomes. Cgviperin-A-BAC-DNA probe was labeled with DIG (green signals) or Biotin (red signals) (indicated by arrows), and Cgviperin-B-BAC-DNA probe was labeled with DIG (indicated by arrowheads). All metaphase chromosomes (blue) were counterstained with DAPI. (D) Gene synteny of vertebrate viperin. Conserved gene blocks are represented in matching colors. Transcription orientations are indicated by arrows.
Subsequently, the genomic structure and syntenic alignment were analyzed. Cgviperin-As and Cgviperin-Bs all contain six exons and five introns, and major differences between Cgviperin-A and Cgviperin-B homeologs exist in the introns (Figure 1A). The lengths of five introns vary between the two homeologs, especially the length of first and second introns. The average identity of the corresponding introns among three alleles is 99.3 ± 0.3%, but the average identity between two homeologs is 75.1 ± 0.1%, which is remarkably lower than that of the cDNAs (91.3 ± 0.1%). Cgviperin-As and Cgviperin-Bs are located on chromosome CgA17 and CgB17 of the gibel carp assembled genome, respectively, and the three alleles of each homeolog in three homologous chromosomes were confirmed by FISH. Three red Cgviperin-As signals and three green Cgviperin-Bs signals were observed on three different homologous chromosomes when Cgviperin-A-BAC-DNA and Cgviperin-B-BAC-DNA were simultaneously used as probes (Figure 1C). The Cgviperin homologous genes were found in zebrafish chromosome (Danio rerio) 17 (chr17), human (Homo sapiens) chr2, mouse (Mus musculus) chr12, chicken (Gallus gallus) chr3, and tropical clawed frog (Xenopus tropicalis) chrLG172761.1 with a conserved gene cluster (CMPK2-RSAD2/viperin-RNF144AA) (Figure 1D). Compared to zebrafish chr17, gibel carp CgA17 and CgB17 both retain most of the zebrafish homologous genes except ywhaq.
To confirm the potential antivirus activities of Cgviperin-A and Cgviperin-B, we compared their expression differences in the adult tissues of healthy individuals (Figure 2). Cgviperin-A and Cgviperin-B were constitutively expressed in the 10 tissues examined. However, obvious expression differences between Cgviperin-A and Cgviperin-B were observed. Both Cgviperin-A and Cgviperin-B showed high expression in immune tissue spleen. Additionally, Cgviperin-A and Cgviperin-B had differentially biased expression among different tissues. Cgviperin-A was abundantly expressed and showed higher expression than Cgviperin-B in the spleen and liver, while Cgviperin-B was highly expressed in the gill.
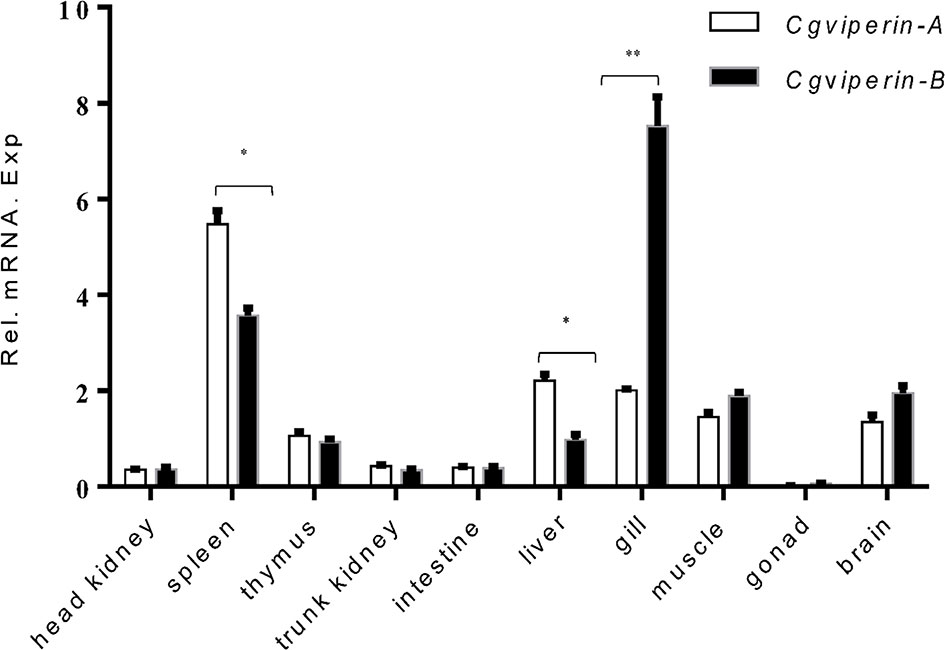
Figure 2 qPCR analysis of Cgviperin-A and Cgviperin-B expression in adult tissue from healthy gibel carp clone F. Eukaryotic translation elongation factor 1 alpha 1, like 1(eef1a1l1) was used as control. Each bar represents mean ± standard deviation (SD) (n = 3). *P < 0.05, **P < 0.01.
CgViperin-A and CgViperin-B Both Interact and Colocalize With ORF46R at ER
To further uncover their antiviral mechanisms, we analyzed the roles of different CgViperin domains and tried to determine their viral interactome. Firstly, we analyzed subcellular the localization of CgViperin-A and CgViperin-B in CAB cells (Figure 3). After co-transfection of CgViperin-A- EGFP or CgViperin-B-EGFP with ER-DsRed, confocal microscopy revealed that both CgViperin-A and CgViperin-B (green) mainly overlapped the red ER signals (Figure 3A). Then, we performed domain mapping to determine which domain was necessary for CgViperin-A and CgViperin-B ER localization. Three mutants of CgViperin-A and CgViperin-B were constructed, i.e., Vip-A-ΔN (lacking aa 1-74 in the N-terminal α-helix domain), Vip-A-ΔM (lacking CxxxCxxC motif in the middle SAM domain), and Vip-A-ΔC (339–359) (lacking aa 339-359 in the C terminus) (Figure 3B). Similar to WT CgViperin-A, Vip-A-ΔM and Vip-A-ΔC could localize at the ER, but Vip-A-ΔN was diffusely distributed in the cytoplasm and nucleus (Figure 3C). Similar subcellular localization was observed in the mutants of CgViperin-B (Figure 3D). The results indicate that the N-terminal α-helix domain is necessary for the localization of CgViperin-A and CgViperin-B at the ER.
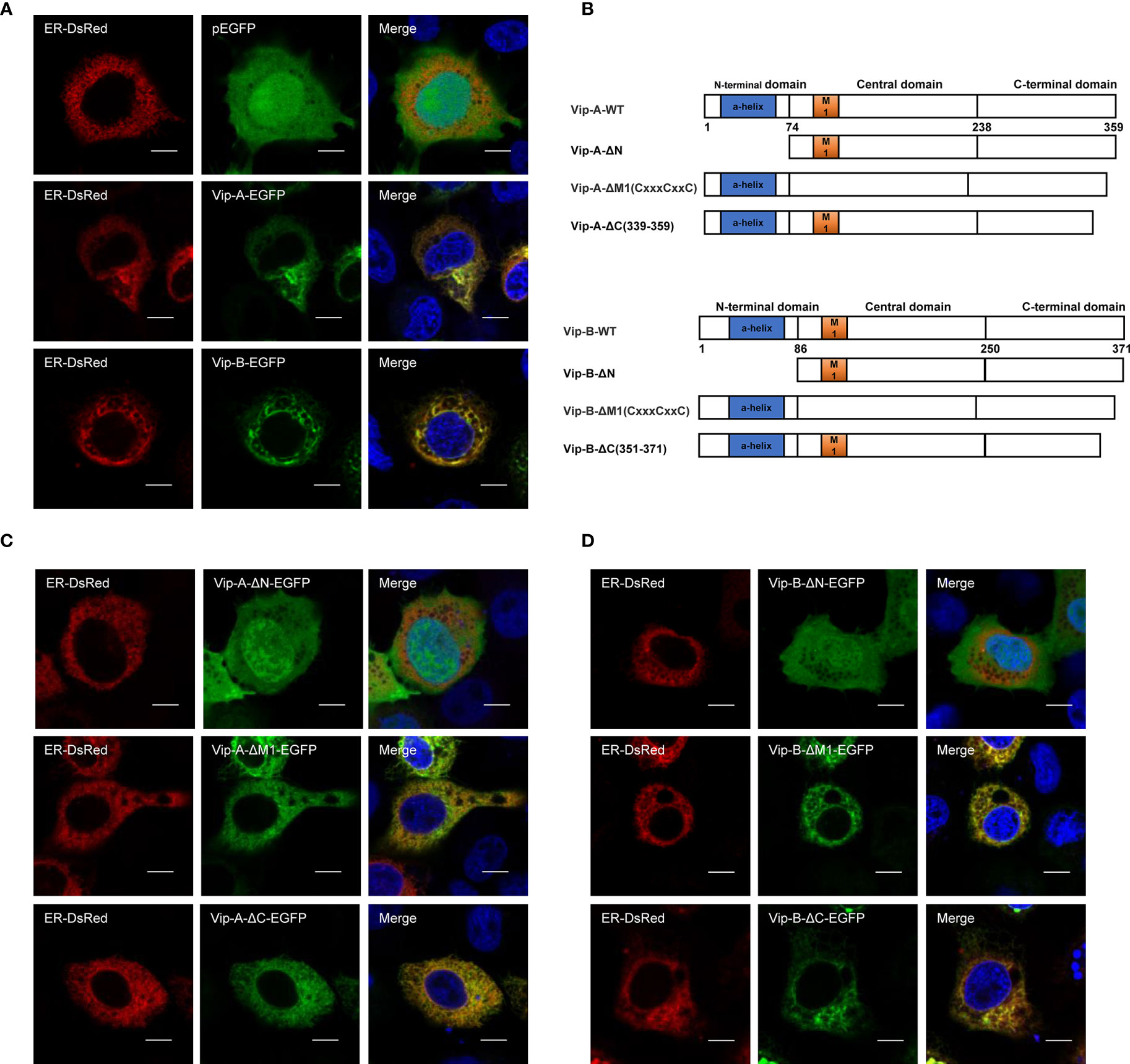
Figure 3 CgViperin-A and CgViperin-B colocalize at ER through N-terminus domain. (A) Subcellular localization of WT CgViperin-A, CgViperin-B, and ER-DsRed. CAB cells were plated onto coverslips in six-well plates and co-transfected CgViperin-A-EGFP or CgViperin-B-EGFP with ER-DsRed. After 24 h, the cells were fixed and observed under confocal microscopy. Nuclear was stained with DAPI. Scale bars = 5 μm. (B) Diagrammatic representation of WT and three mutants of CgViperin-A or CgViperin-B. (C, D) Subcellular localization of three mutants of CgViperin-A (C) or CgViperin-B (D) with ER-DsRed.
Subsequently, to clarify the relevance of CgViperins and CaHV, the Co-IP experiments were performed, which demonstrated that both CgViperin-A and CgViperin-B were efficiently associated with CaHV ORF46R (Figure 4A). Next, the subcellular locations of CgViperins and ORF46R were monitored in CAB cells. Confocal microscopy revealed that the signals of ORF46R were mainly overlapped with those of CgViperin-A and CgViperin-B (Figure 4B), indicating that both CgViperin-A and CgViperin-B partly colocalize with ORF46R at the ER. Finally, we analyzed which domain was essential for the colocalization of CgViperins with ORF46R. Interestingly, the signals of ORF46R almost overlapped with those of Vip-A-ΔN and partly colocalized with Vip-A-ΔM, but was not with Vip-A-ΔC (Figure 4C), and similar subcellular localization was observed among three mutants of CgViperin-B (Figure 4D). These data imply that the C-terminal domain of CgViperin-A and CgViperin-B is indispensable for the colocalization with ORF46R.
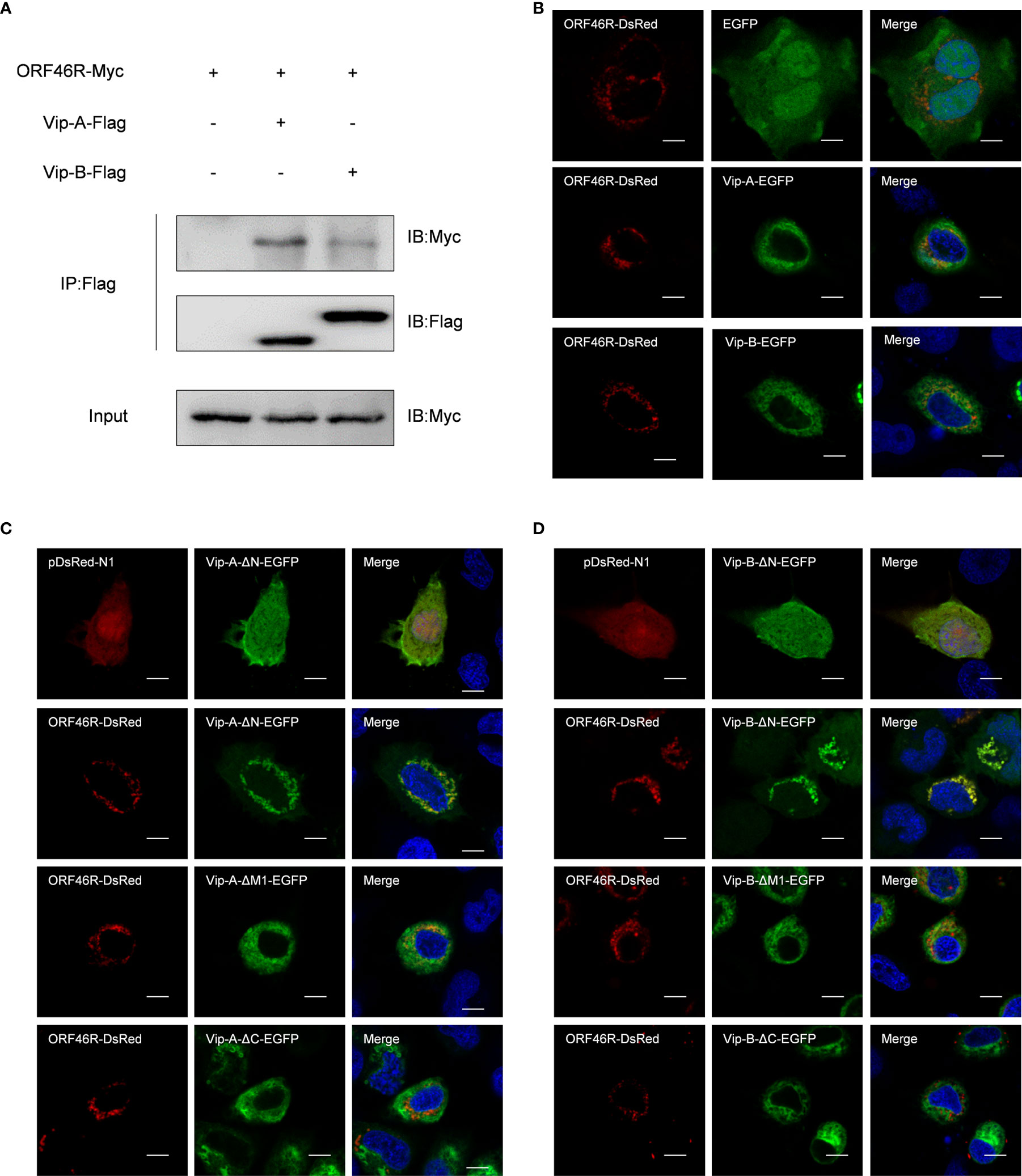
Figure 4 Both CgViperin-A and CgViperin-B interact with ORF46R of CaHV. (A) Co-IP of Vip-A-Flag or Vip-B-Flag with Myc-ORF46R in CAB cells transfected with the indicated plasmids. Anti-Flag Ab was used for Co-IP. (B) Subcellular localization of WT CgViperin-A, CgViperin-B, and ORF46R. CAB cells were plated onto coverslips in six-well plates and co-transfected CgViperin-A-EGFP or CgViperin-B-EGFP with ORF46R-DsRed. After 24 h, the cells were fixed and observed under confocal microscopy. Nuclear was stained with DAPI. Scale bars = 5 μm. (C, D) Subcellular localization of three mutants of CgViperin-A or CgViperin-B with ORF46R-DsRed. Scale bars = 5 μm.
CaHV ORF46R Blocks IFN Induction
To obtain a clear picture of the functions of CgViperin-A and CgViperin-B in the cellular antiviral immunity, the role of ORF46R for immune response was investigated. As shown in Figure 5A, poly(dA:dT) and poly(I:C) could induce the activation of the crucian carp CaIFN promoter (64). However, such induction was significantly impeded by ORF46R in a dose-dependent manner. ISRE, a transcription factor binding motif in the IFN promoter region, facilitates gene transcription (64). Consistently, the same results were also observed in ISRE (Figure 5B). For viperin, which was focused in this study, had been identified as an antiviral factor, their promoter activities were checked under ORF46R overexpression as well. The six Cgviperin promoters stimulated by poly(dA:dT) or poly(I:C) were blunted ORF46R as expected (Figures 5C, D). These results suggest that CaHV ORF46R serves as a negative regulator to block host IFN production or this suppression happened following an induction.
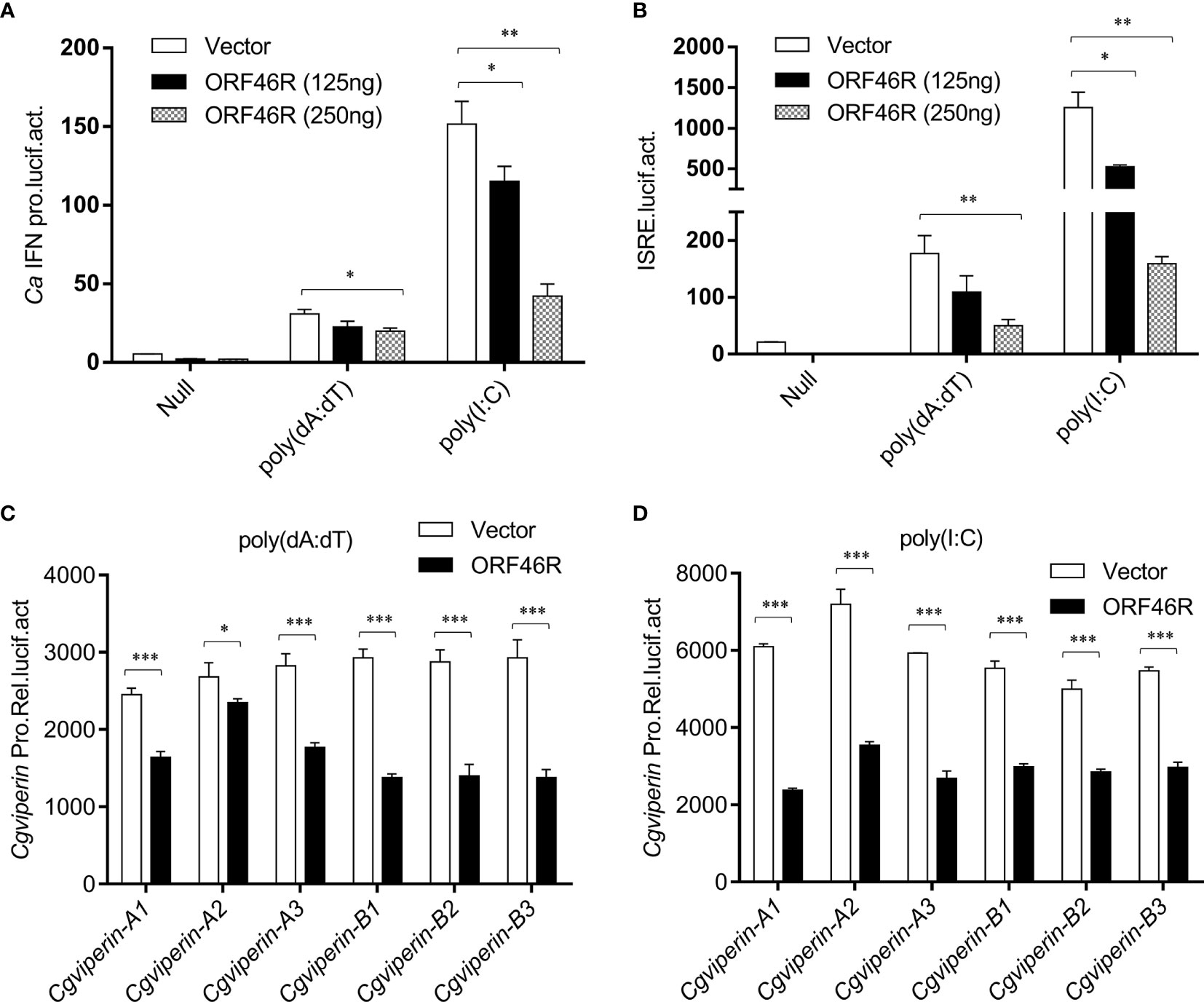
Figure 5 CaHV ORF46R blocks poly (dA:dT) or poly (I:C)-triggered activation of CaIFN promoter (A), ISRE luciferase reporter (B), and six Cgviperin promoters (C, D). CAB cells seeded overnight were co-transfected the empty vector or ORF46R (250 ng) and 25 ng pRL-TK plus 250 ng CaIFNpro-luc or ISRE-luc. At 24 h post-transfection, cells were untreated (null) or transfected with 1 μg poly (dA:dT) or poly (I:C). The luciferase activities were monitored 24 h after stimulation. Each bar represents mean ± standard deviation (SD) (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
CgViperin-A and CgViperin-B Promote CaHV ORF46R Proteasomal Degradation via Decreasing Lysine 63 (K63)-Linked Ubiquitination
To determine the effect of CgViperin-A and CgViperin-B on the ORF46R of CaHV, Vip-A-Flag or Vip-B-Flag was co-transfected with ORF46R-HA. The overexpression of CgViperin-A and CgViperin-B caused a significant reduction of ORF46 protein in a dose-dependent manner, respectively (Figures 6A, B). We also used the mutants to determine which domain was indispensable for the capacity of CgViperin-A or CgViperin-B to influence the stability of ORF46R. Consistent with WT CgViperin-A, Vip-A-ΔN and Vip-A-ΔM still could reduce the expression of ORF46R, whereas such an effect was abrogated in the Vip-A-ΔC group (Figure 6C). Similar results were obtained in the three mutants of CgViperin-B (Figure 6D), implying that the C-terminus of CgViperin is necessary for the regulation on ORF46R stability.
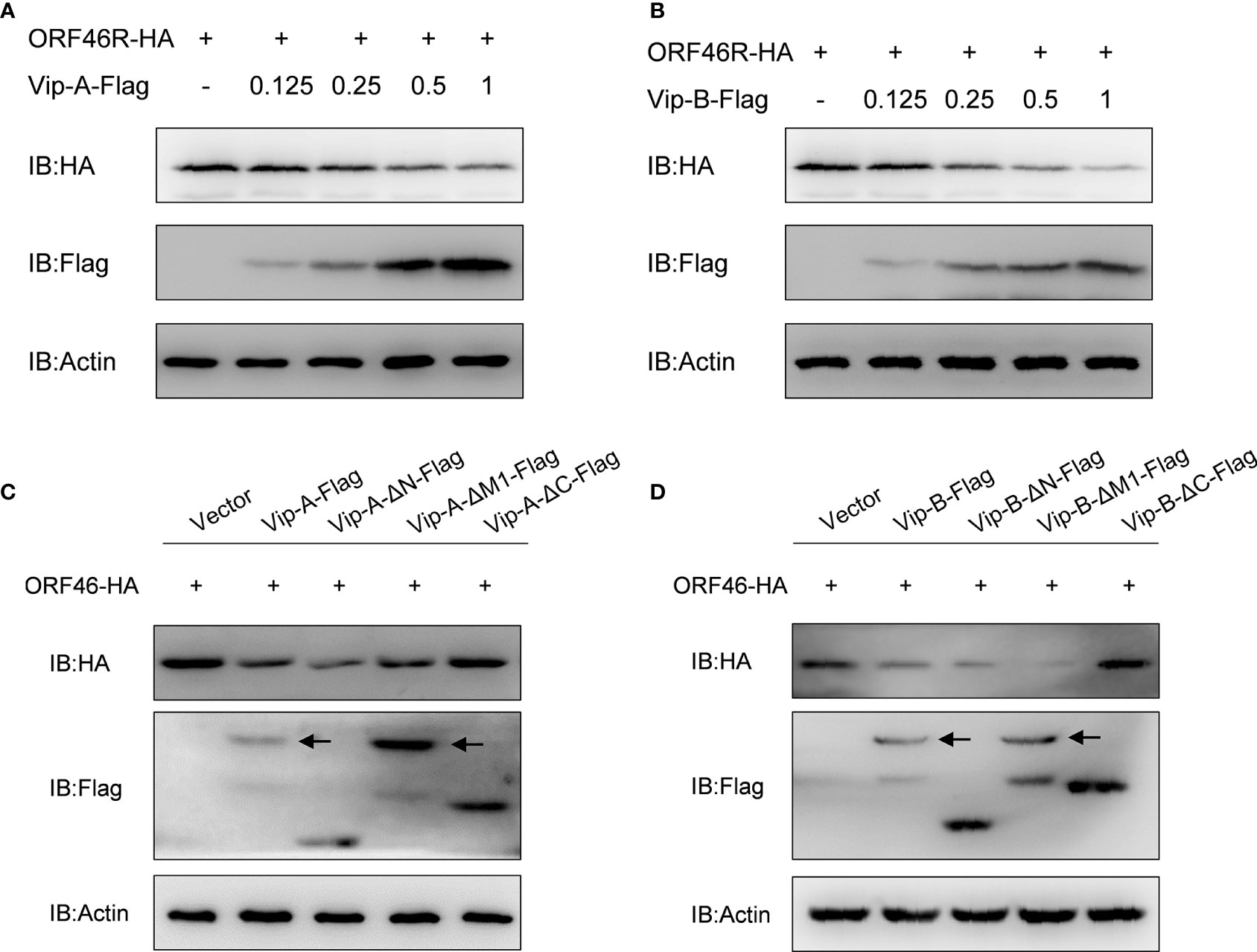
Figure 6 Overexpression of CgViperin-A and CgViperin-B induce the reduction of CaHV ORF46R protein via its C-terminus. (A, B) Overexpression of CgViperin-A (A) or CgViperin-B (B) induced the reduction of ORF46 protein in a dose-dependent manner in CAB cells. CAB cells were transfected with the indicated expression vectors and harvested for western blot analysis after 18–24 h (C, D) Domain mapping for CgViperin-A (C) or CgViperin-B (D) domains acting on ORF46R. CAB cells were transfected with the indicated expression vectors (2 μg/well) and harvested for western blot analysis after 18–24 h Diagrammatic representation of three mutants of CgViperin-A or CgViperin-B is shown in Figure 3B. Data are presented based on three independent experiments.
To determine how CgViperin-A and CgViperin-B regulate ORF46R at the protein level, a proteasome inhibitor (MG132), two lysosomal inhibitors [NH4Cl and CQ (Chloroquine)], and an autophagosome inhibitor 3-methyladenine (3-MA) were used to examine the manner of CgViperin-A and CgViperin-B-mediated ORF46R degradation. In comparison with the control group (DMSO treatment), MG132 could effectively inhibit the degradation of ORF46R induced by CgViperin-A (Figure 7A). Significantly, the reduction of ORF46R was rescued by MG132 dose-dependently (Figure 7B). Similar results were also observed in CgViperin-B group (Figures 7C, D). These observations suggest that both CgViperin-A and CgViperin-B could promote the proteasomal degradation of ORF46R. Protein ubiquitination is critical for the proteasomal degradation pathway (65). Whether CgViperin-A and CgViperin-B induce the polyubiquitination of ORF46R was further investigated. As shown in Figure 7E, both CgViperin-A and CgViperin-B suppressed the polyubiquitination of ORF46R. Previous studies suggested that target proteins were stabilized by K63-linked polyubiquitin chains (66, 67). Therefore, we tested a ubiquitin mutant, K63-only (K63O) (in which lysine at position 63 was ubiquitinated and linked with polyubiquitin chains, while all other lysine residues were mutated to arginine residues), to determine whether the attenuation of ORF46R was mediated by the decrease of K63-ubiquitination. Consistent with prediction, the K63-linked ubiquitination of ORF46R was significantly impaired under CgViperin-A regulation (Figure 7F). Similar results were also obtained in CgViperin-B group (Figure 7G). The above results suggest that both CgViperin-A and CgViperin-B promote ORF46R proteasomal degradation via decreasing K63-linked ubiquitination.
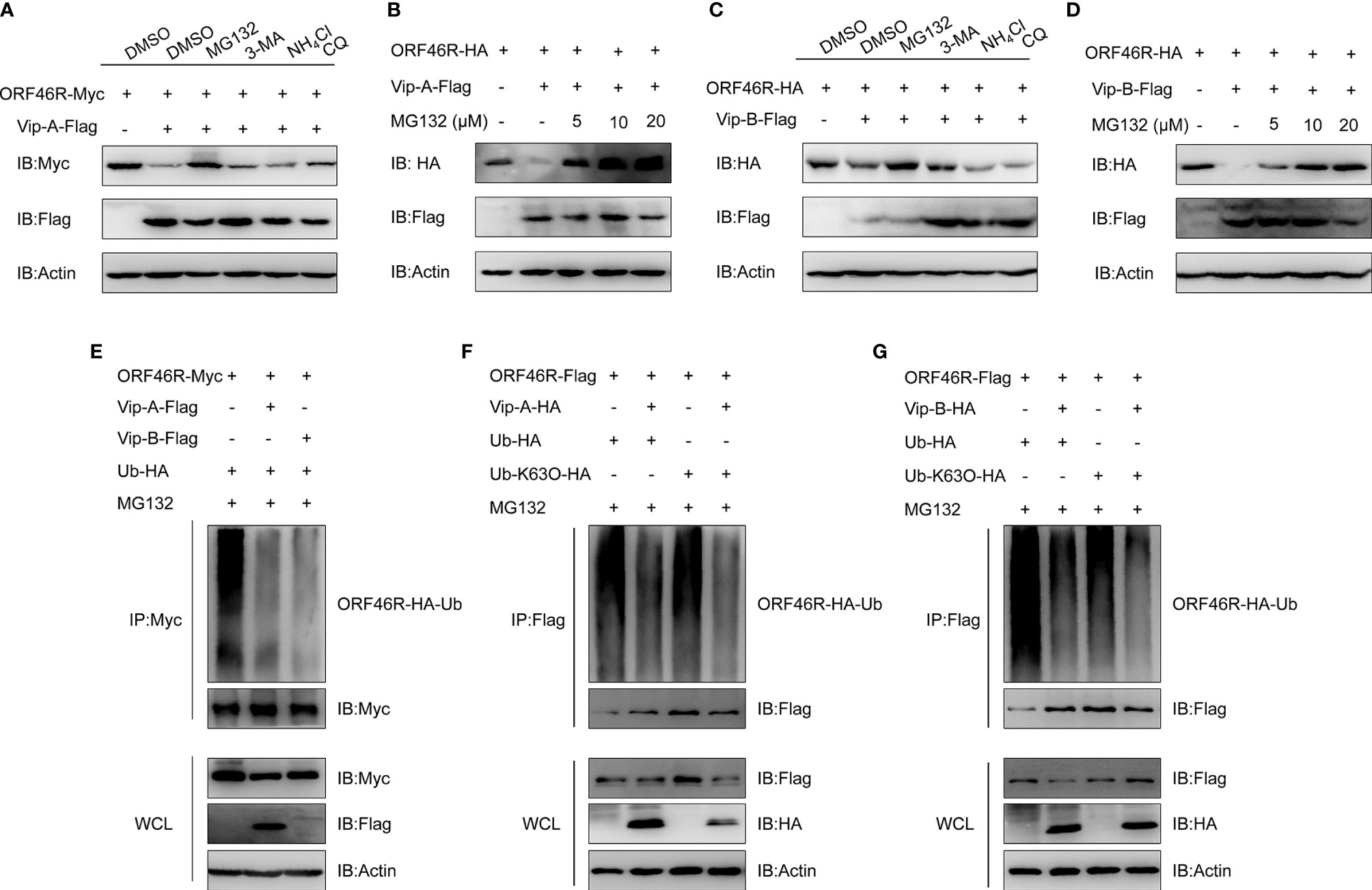
Figure 7 CgViperin-A and CgViperin-B promote CaHV ORF46R proteasomal degradation via decreasing K63-linked ubiquitination. (A, C) Effects of inhibitors on CgViperin-A (A) or CgViperin-B-mediated (C) destabilization of ORF46R. CAB cells were seeded in six-well overnight and transfected with the indicated expression vectors (2 μg/well). The cells were treated with the indicated inhibitors at 18 h post-transfection and harvested for western blot analysis with the anti-Flag and anti-β-actin Abs. (B, D) ORF46R degradation induced by CgViperin-A (B) or CgViperin-B (D) was rescued by MG132 in a dose-dependent manner. CAB cells were transfected with the indicated expression vectors (2 μg/well) and treated with DMSO or MG132 (5, 10, or 20 μM) for 6 h at 18 h post-transfection. Then, the WCL were subjected to western blot analysis with the indicated Abs. (E) CgViperin-A and CgViperin-B decreased the ubiquitination of ORF46R. CAB cells were seeded in 10 cm2 dishes and transfected with 5 μg ORF46R-Myc, 4 μg CgViperin-A-Flag or CgViperin-B-Flag, and 1 μg HA-Ub and were treated with DMSO or MG132 for 6 h at 18 h post-transfection. After IP with anti-Myc-affinity gels, the immunoprecipitates and WCL were analyzed by western blot analysis with the indicated Abs. (F, G) CgViperin-A (F) and CgViperin-B (G) decreased the K63-linked ubiquitination of ORF46R. CAB cells were seeded in 10 cm2 dishes were co-transfected with the indicated expression vectors (5 μg ORF46R-Flag, 4 μg CgViperin-A-HA, or CgViperin-B-HA) and 1 μg HA-Ub or Ub-K63O-HA, and were treated with MG132 for 6 h at 18 h post-transfection. After IP with anti-Flag-affinity gels, the immunoprecipitates and WCL were analyzed by western blot analysis with the indicated Abs. Data are presented based on three independent experiments.
CgViperin-B Mediates Autophagic Degradation of CaHV ORF46R
In Figure 7C, we noticed that 3-MA could efficiently block ORF46R degradation induced by CgViperin-B, implying that CgViperin-B might promote ORF46R degradation through autophagosome pathway, which was distinguished with CgViperin-A, meaning that the CgViperin-A and CgViperin-B functional divergence occurs during the progression of ORF46R degradation. To identify our hypothesis, 3-MA treatment was employed for ORF46R degradation experiments subsequently. Consistent with the results above, increasing doses of 3-MA could not block the ORF46R degradation induced by CgViperin-A (Figure 8A), but CgViperin-B-mediated ORF46R degradation was obviously rescued by 3-MA in a dose-dependent manner (Figure 8B). These results suggest that CgViperin-B degrade ORF46R via an autophagic pathway that is absent in CgViperin-A. To further illustrate the exact mechanism in this process, we identified that the conversion of Microtubule-associated Protein 1A/1B-Light Chain 3 (LC3)-I to LC3-II was promoted by CgViperin-B, which means that autophagy is enhanced by the overexpression of CgViperin-B (Figure 8C). Co-IP assays showed that CgViperin-B interacted with many autophagic components, such as LC3, Beclin1, and autophagy-related gene 5 (ATG5) (Figure 8D). Meanwhile, interactions between ORF46R and these autophagic components were also confirmed (Figure 8E). Confocal immunofluorescence analysis showed that CgViperin-B overexpression caused the aggregation of LC3-GFP, demonstrating that cellular autophagy is enhanced (Figure 8F). These results suggest that CgViperin-B mediates the autophagic degradation of ORF46R, which is not possessed by CgViperin-A.
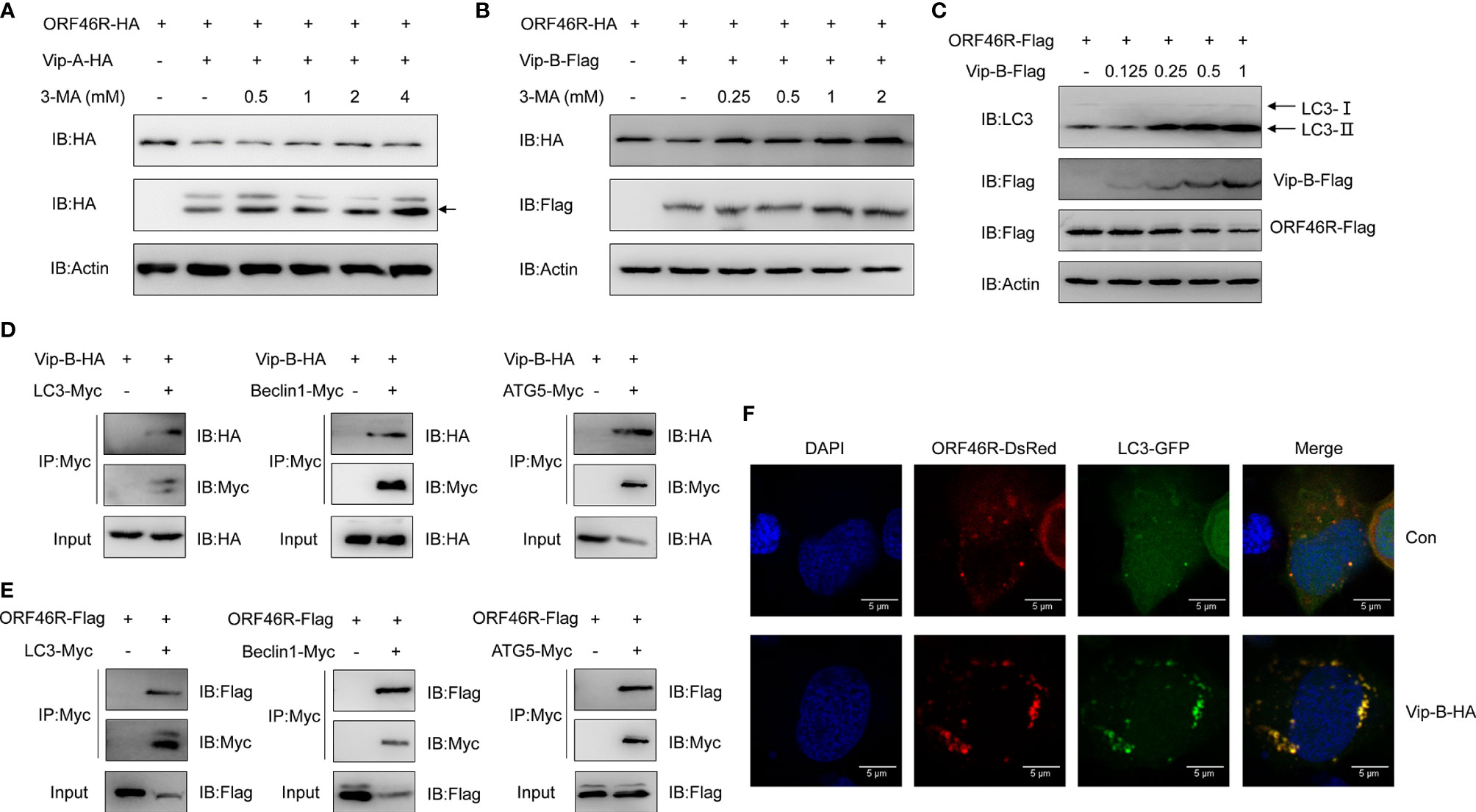
Figure 8 CgViperin-B mediates degradation of CaHV ORF46R via an autophagic pathway. (A, B) Effects of 3-MA on CgViperin-A- (A) or CgViperin-B-mediated (B) destabilization of ORF46R. CAB cells were transfected with the indicated expression vectors (2 μg/well) and treated with DMSO or 3-MA (0.5, 1, 2, or 4 mM in A and 0.25, 0.5, 1, or 2 mM in B) for 6 h at 18 h post-transfection. Then, the WCL were subjected to western blot analysis with the indicated Abs. (C) Effects of CgViperin-B on the conversion of LC3B-I to LC3B-II. CAB cells were transfected with increasing amounts of CgViperin-B-Flag (0.125, 0.25, 0.5, and 1 μg), analyzed by western blot analysis with the indicated Abs. (D, E) Co-IP of Vip-B-HA (D) or ORF46R-Flag (E) with LC3-Myc, Beclin1-Myc, and ATG5-Myc in HEK 293T cells transfected with the indicated plasmids. Anti-Myc Ab was used for Co-IP. (F) Subcellular colocalization of ORF46R and CgViperin-B with LC3-DsRed. CAB cells were plated onto coverslips in six-well plates and co-transfected with the indicated plasmids. After 24 h, the cells were fixed and observed under confocal microscopy. Nuclear was stained with DAPI. Scale bars = 5 μm.
Discussion
After WGD, duplicated genes step into complex and dynamic evolutionary routes under relaxed purifying selection. One of the two duplicates may be fractionated (deleted), or both may be retained, or diverge and evolve (sub- or neofunctionalization) (3). Recently, we revealed the biased expression and sub-/neofunctionalization of multiple duplicated foxl2 homeologs and alleles in gibel carp (60). Innate immunity plays a vital role in the first barrier protecting the host from invading viruses (68). In this study, we focused on viperin, which is increasingly considered as a central player in the mammal antiviral response (27), and elucidated the divergent antiviral mechanisms of two homeologs in a polyploid fish.
In addition to the “fish-specific” (3R) WGD event about 320–350 million years ago (Mya), gibel carp was supposed to be experienced two extra rounds of polyploidy [an early allo-polyploidy (4R) and a late auto-polyploidy (5R)] at approximately 18.49 and 0.51 Mya, respectively (55, 60, 69, 70). In this study, we identified two homeologs of Cgviperins with about 91% identity, and each homeolog has three alleles with high identities (≥99.0%) and located on three homologous chromosomes separately (Supplementary Figure S1 and Figure 1C). Compared to zebrafish, our results (Figures 1B, D) indicated that the duplication of Cgviperin, same as gibel carp other genes (i.e., dmrt1, foxl2, nanos2 and bmp15) (55, 60, 69, 70), may derive from the early allotetraploidy (4R) and a late autotriploidy (5R) event. In addition, Cgviperin-A and Cgviperin-B showed a different tissue-dependent constitutive expressions, suggesting A or B homologs might occur dominant or biased expression of homeologs during gibel carp evolution. In our previous study, we also found that Cgviperin-A and Cgviperin-B exhibited a biased expression pattern after CaHV infection. Significantly, the upregulation folds of them in clone A+, F, and H were related to their resistance ability to CaHV, progressively increasing from susceptible clone to resistant clone at 1 dpi (52), which implies that Cgviperin-A and Cgviperin-B might play an important role in the battle between gibel carp and CaHV.
Consistent with other vertebrate Viperin, both CgViperin-A and CgViperin-B contain three domains. The N-terminal domain shows considerable variability between species and aims at the localization of Viperin to the ER, lipid droplets, or mitochondria (19–21). Major sequence differences between CgViperin-A and CgViperin-B also exist in their N-terminal region (Supplementary Figure S2), and the two N-terminal truncated mutants (Vip-A-ΔN and Vip-B-ΔN) both result in the delocalization of Viperin from the ER membrane (Figure 3). The other two domains, the middle SAM and C-terminal domains, are highly conserved across species. The role of the C-terminal domain remains poorly defined (24). Mutating the C-terminal tryptophan residue of human VIPERIN could prevent its binding to iron-sulfur cluster–installing protein CIAO1 (71, 72). In this study, the C-terminal truncated mutants (Vip-A-ΔC and Vip-B-ΔC) failed to colocalize with and degrade CaHV ORF46R (Figures 4C, D, 6C, D), implying that the C-terminus is necessary for CgViperin-ORF46R interactions.
Mammalian Viperin has been found to interact with three groups of proteins: host proteins involved in innate immune signaling [i.e., Interleukin 1 Receptor Associated Kinase 1 (IRAK1), TNF Receptor Associated Factor 6 (TRAF6), Stimulator of Interferon Genes (STING), and TBK1], host metabolic enzymes [i.e., Farnesyl Pyrophosphate (FPPS) and Microsomal Triglyceride Transfer Protein (MTP)], and viral structural and non-structural proteins [i.e., Non-structural Protein 3 (NS3) and NS5A of flaviviruses] (27). The interactions between mammalian Viperin and viral proteins appear idiosyncratic. Viperin can inhibit viral RNA replication by interfering with the interactions between host proteins and viral non-structural proteins (28–30), inducing the degradation of viral proteins (27, 73), or acting through Fe‐S cluster‐dependent enzymatic activity (72). In this study, we found that both CgViperin-A and CgViperin-B could interact and promote CaHV ORF46R protein degradation (Figures 4A and 6). Our preliminary study showed that CaHV ORF46R is a negative regulator of host IFN production because the overexpression of ORF46R could inhibit activation of the CaIFN promoter and ISRE luciferase reporter activity stimulated by poly (dA:dT) (Figure 5). Up to now, the domain and exact function of ORF46R are unknown. By bioinformatics analysis, we did not find any known domain in ORF46R and any viral protein similar to ORF46R that had been studied. Further research will be required to determine which region can interact with CgViperins and to reveal the exact function of ORF46R in CaHV invasion, replication, assembly, or release, especially in CaHV evasion from the host IFN response.
Among the diverse antiviral mechanisms, one emerging theme is that Viperin seems to promote the ubiquitin-dependent proteasomal degradation of some target proteins (27). For example, human VIPERIN can promote the degradation of viral NS5A through the proteasomal degradation pathway (27). It was also found to be capable of stimulating K63-linked polyubiquitination of IRAK1, which phosphorylated interferon regulatory factor (IRF) 7 and ultimately led to IFN production (74). In this study, we found that both CgViperin-A and CgViperin-B could promote CaHV ORF46R proteasomal degradation, most likely by decreasing the K63-linked ubiquitination of ORF46R (Figure 7). Our findings suggest that CgViperin-A and CgViperin-B, like their human homologous genes, may have a general role in engaging with E3 ubiquitin ligases, which are responsible for tagging proteins with polyubiquitin chains. The ubiquitin molecule contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and an amino terminus (M1), which can conjugate with other ubiquitin molecules to form polymeric chains (66, 67). Among them, the K63-based polymeric chain is one of the best described codes and mainly involves in signal transduction, protein sorting and trafficking, and enzymatic activity (67). A growing number of studies suggest that ubiquitin chains play an important role in the modulation of innate immunity (75). Altogether, our results suggest that the overexpression of CgViperin-A or CgViperin-B can decrease the K63-linked ubiquitination of endogenous ORF46R protein, which reduces the stability of ORF46R protein and ultimately results in its degradation by the proteasome. There is still much to study on this topic, including which of the ubiquitin ligases CgViperin and CgViperin-B interact with and how they cooperate to degrade viral ORF46R.
Eukaryote organisms possess two major cellular degradation machineries: the ubiquitin–proteasome system (UBS) and autophagy-lysosome pathway (ALP) (76, 77). As described above, CgViperin-A and CgViperin-B could both promote CaHV ORF46R proteasomal degradation in CAB cells (Figure 7). In this study, CgViperin-B-mediated ORF46R degradation is blocked by autophagy inhibitor 3-MA, and both of them are interacted with the same autophagic components; meanwhile the transformation of LC3 from type I to type II is increased, demonstrating autophagy is one mechanism of CgViperin-B to degrade viral protein. On the other hand, our data also suggest that ORF46R can also be eliminated by CgViperin-B during the ubiquitination. Whether there is association between these two mechanisms is still unclear. Which one exerts the major antiviral function is also unknown. Both of that need to be clarified in further study. For unknown reasons, the CAB cells and other cell lines used in our laboratory failed to proliferate CaHV. Therefore, we could not perform the antiviral studies in vitro where the proteins are overexpressed or knocked out, followed by a virus infection. In the further study, we will mutate Cgviperin-A and Cgviperin-B, respectively, in gibel carp by using CRISPR/Cas9 and compare the antiviral differences between them, which would help to complete the story and highlight their antiviral activity.
Based on the above results of functional analysis in vitro, we propose a schematic diagram for divergent antiviral mechanisms of CgViperin-A and CgViperin-B in the gibel carp resistance against herpesvirus CaHV (Figure 9). After CaHV invasion, both CgViperin-A and CgViperin-B could promote the proteasomal degradation of CaHV ORF46R via decreasing K63-linked ubiquitination. Additionally, CgViperin-B also mediated ORF46R degradation through autophagosome pathway. In conclusion, the current findings shed light on the antiviral activities of teleost Viperin as well as the divergent functions and regulative mechanisms of two duplicated viperin homeologs. Meanwhile, the above data also imply that the two viperin homeologs have subfunctionalized and cooperate to regulate antiviral response.
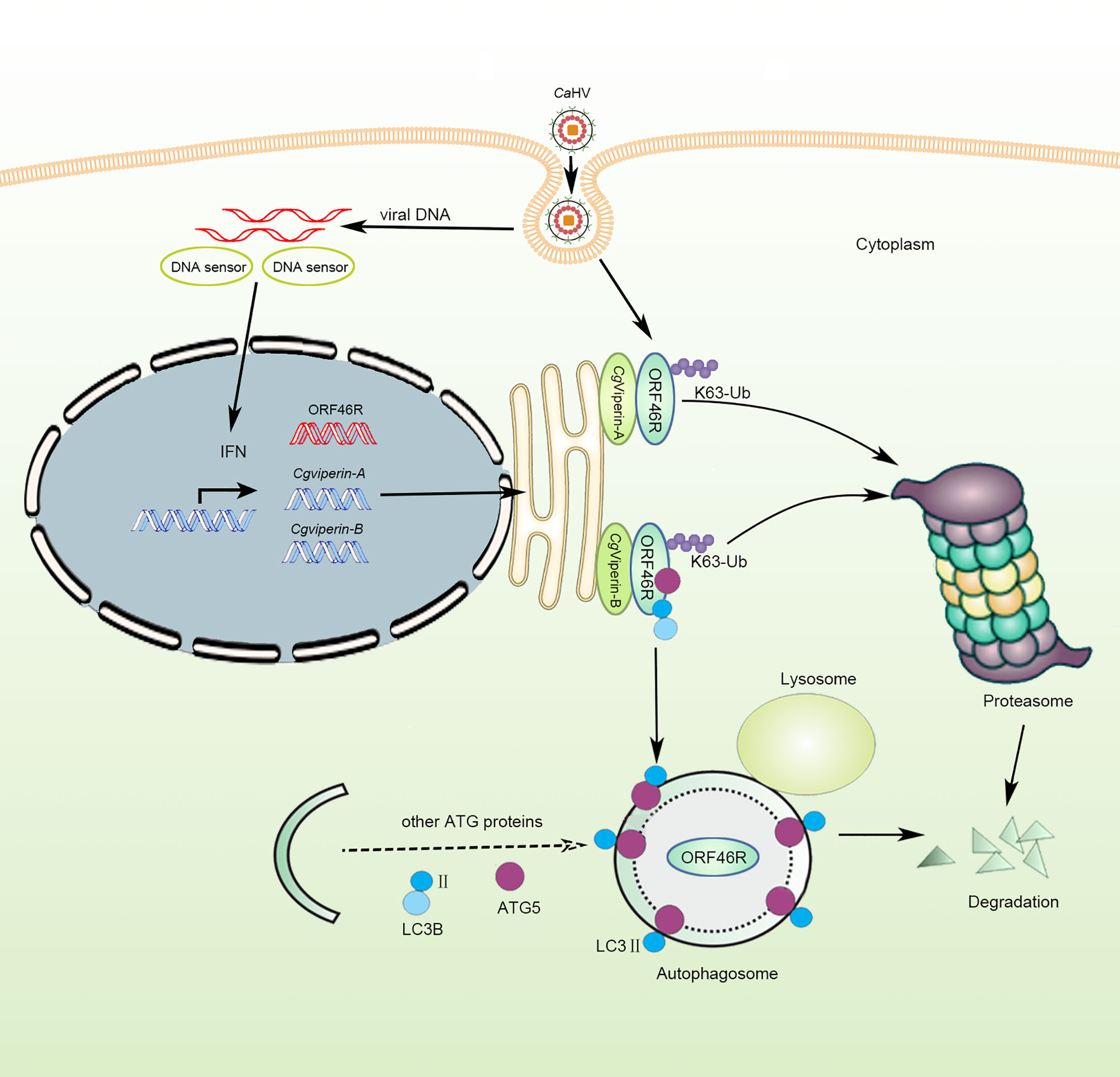
Figure 9 Schematic diagram of divergent antiviral mechanisms of CgViperin-A and CgViperin-B in gibel carp resistance against herpesvirus CaHV. Both CgViperin-A and CgViperin-B directly interact and promote proteasomal degradation of CaHV ORF46R via decreasing K63-linked ubiquitination. In addition, CgViperin-B induces ORF46R degradation through autophagosome pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: MZ055409 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055409); MZ055410 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055410); MZ055411 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055411); MZ055412 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055412); MZ055413 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055413); MZ055414 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055414); MZ055415 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055415); MZ055416 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055416); MZ055417 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055417); MZ055418 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055418); MZ055419 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055419); MZ055420 (https://www.ncbi.nlm.nih.gov/nuccore/MZ055420).
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of IHB, CAS (protocol number 2016-018).
Author Contributions
J-FG, LZ, and C-YM designed the study. C-YM, SL, L-FL, PY, ZL, J-FT, Q-YZ, Z-WW, X-JZ, and G-XW prepared the samples and carried out the experiments. C-YM, LZ, J-FG, SL, and YW analyzed and discussed the results. LZ, J-FG, SL, YW, and C-YM wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24030203 and XDA24030104), the National Natural Science Foundation (31772838), and China Agriculture Research System of MOF and MARA (CARS-45-07). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the reviewers for helpful discussion in editing this manuscript. The research was supported by the Analytical & Testing Center, IHB, CAS and Wuhan Branch, Supercomputing Centre, CAS, China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.702971/full#supplementary-material
Supplementary Figure S1 | Multiple nucleotide sequence alignment of six Cgviperin transcripts from gibel carp clone F. ORF is highlighted by red box.
Supplementary Figure S2 | Multiple amino acid sequence alignment of CgViperin proteins from gibel carp clone F.
Supplementary Table S1 | Primers used in this study.
References
1. Soltis DE, Visger CJ, Soltis PS. The Polyploidy Revolution Then … and Now: Stebbins Revisited. Am J Bot (2014) 101(7):1057–78. doi: 10.3732/ajb.1400178
2. Soltis PS, Soltis DE. Ancient WGD Events as Drivers of Key Innovations in Angiosperms. Curr Opin Plant Biol (2016) 30:159–65. doi: 10.1016/j.pbi.2016.03.015
3. Cheng F, Wu J, Cai X, Liang JL, Freeling M, Wang XW. Gene Retention, Fractionation and Subgenome Differences in Polyploid Plants. Nat Plants (2018) 4(5):258–68. doi: 10.1038/s41477-018-0136-7
4. Clark JW, Donoghue PCJ. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci (2018) 23(10):933–45. doi: 10.1016/j.tplants.2018.07.006
5. Mandakova T, Lysak MA. Post-Polyploid Diploidization and Diversification Through Dysploid Changes. Curr Opin Plant Biol (2018) 42:55–65. doi: 10.1016/j.pbi.2018.03.001
6. Van de Peer Y, Maere S, Meyer A. The Evolutionary Significance of Ancient Genome Duplications. Nat Rev Genet (2009) 10(10):725–32. doi: 10.1038/nrg2600
7. Van De Peer Y, Mizrachi E, Marchal K. The Evolutionary Significance of Polyploidy. Nat Rev Genet (2017) 18(7):411–24. doi: 10.1038/nrg.2017.26
8. Fox DT, Soltis DE, Soltis PS, Ashman TL, Van de Peer Y. Polyploidy: A Biological Force From Cells to Ecosystems. Trends Cell Biol (2020) 30(9):688–94. doi: 10.1016/j.tcb.2020.06.006
9. Carretero-Paulet L, Van de Peer Y. The Evolutionary Conundrum of Whole-Genome Duplication. Am J Bot (2020) 107(8):1101–5. doi: 10.1002/ajb2.1520
10. Zhou L, Gui JF. Natural and Artificial Polyploids in Aquaculture. Aquac Fish (2017) 3(2):103–11. doi: 10.1016/j.aaf.2017.04.003
11. Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, et al. Zebrafish Hox Clusters and Vertebrate Genome Evolution. Science. (1998) 282(5394):1711–4. doi: 10.1126/science.282.5394.1711
12. Jaillon O, Aury JM, Wincker P. “Changing by Doubling”, the Impact of Whole Genome Duplications in the Evolution of Eukaryotes. Cr Biol (2009) 332(2-3):241–53. doi: 10.1016/j.crvi.2008.07.007
13. Rodriguez F, Arkhipova IR. Transposable Elements and Polyploid Evolution in Animals. Curr Opin Genet Dev (2018) 49:115–23. doi: 10.1016/j.gde.2018.04.003
14. Chin KC, Cresswell P. Viperin (Cig5), an IFN-Inducible Antiviral Protein Directly Induced by Human Cytomegalovirus. Proc Natl Acad Sci U S A (2001) 98(26):15125–30. doi: 10.1073/pnas.011593298
15. Boudinot P, Massin P, Blanco M, Riffault S, Benmansour A. Vig-1, a New Fish Gene Induced by the Rhabdovirus Glycoprotein, has a Virus-Induced Homologue in Humans and Shares Conserved Motifs With the MoaA Family. J Virol (1999) 73(3):1846–52. doi: 10.1128/JVI.73.3.1846-1852.1999
16. Fenwick MK, Li Y, Cresswell P, Modis Y, Ealick SE. Structural Studies of Viperin, an Antiviral Radical SAM Enzyme. Proc Natl Acad Sci U S A (2017) 114(26):6806–11. doi: 10.1073/pnas.1705402114
17. Zhu H, Cong JP, Shenk T. Use of Differential Display Analysis to Assess the Effect of Human Cytomegalovirus Infection on the Accumulation of Cellular RNAs: Induction of Interferon-Responsive RNAs. Proc Natl Acad Sci U S A (1997) 94(25):13985–90. doi: 10.1073/pnas.94.25.13985
18. Mattijssen S, Pruijn GJ. Viperin, a Key Player in the Antiviral Response. Microbes Infect (2012) 14(5):419–26. doi: 10.1016/j.micinf.2011.11.015
19. Seo JY, Yaneva R, Cresswell P. Viperin: A Multifunctional, Interferon-Inducible Protein That Regulates Virus Replication. Cell Host Microbe (2011) 10(6):534–9. doi: 10.1016/j.chom.2011.11.004
20. Hinson ER, Cresswell P. The N-Terminal Amphipathic Alpha-Helix of Viperin Mediates Localization to the Cytosolic Face of the Endoplasmic Reticulum and Inhibits Protein Secretion. J Biol Chem (2009) 284(7):4705–12. doi: 10.1074/jbc.M807261200
21. Hinson ER, Cresswell P. The Antiviral Protein, Viperin, Localizes to Lipid Droplets via its N-Terminal Amphipathic Alpha-Helix. Proc Natl Acad Sci U S A (2009) 106(48):20452–7. doi: 10.1073/pnas.0911679106
22. Duschene KS, Broderick JB. The Antiviral Protein Viperin is a Radical SAM Enzyme. FEBS Lett (2010) 584(6):1263–7. doi: 10.1016/j.febslet.2010.02.041
23. Shaveta G, Shi JH, Chow VTK, Song JX. Structural Characterization Reveals That Viperin is a Radical S-Adenosyl-L-Methionine (SAM) Enzyme. Biochem Biophys Res Commun (2010) 391(3):1390–5. doi: 10.1016/j.bbrc.2009.12.070
24. Ng LFP, Hiscox JA. Viperin Poisons Viral Replication. Cell Host Microbe (2018) 24(2):181–3. doi: 10.1016/j.chom.2018.07.014
25. Helbig KJ, Beard MR. The Role of Viperin in the Innate Antiviral Response. J Mol Biol (2014) 426(6):1210–9. doi: 10.1016/j.jmb.2013.10.019
26. Lindqvist R, Overby AK. The Role of Viperin in Antiflavivirus Responses. DNA Cell Biol (2018) 37(9):725–30. doi: 10.1089/dna.2018.4328
27. Ghosh S, Marsh ENG. Viperin: An Ancient Radical SAM Enzyme Finds its Place in Modern Cellular Metabolism and Innate Immunity. J Biol Chem (2020) 295(33):11513–28. doi: 10.1074/jbc.REV120.012784
28. Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, et al. Identification of Three Interferon-Inducible Cellular Enzymes That Inhibit the Replication of Hepatitis C Virus. J Virol (2008) 82(4):1665–78. doi: 10.1128/JVI.02113-07
29. Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, Fiches G, et al. The Antiviral Protein Viperin Inhibits Hepatitis C Virus Replication via Interaction With Nonstructural Protein 5a. Hepatology (2011) 54(5):1506–17. doi: 10.1002/hep.24542
30. Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, et al. Viperin Is Induced Following Dengue Virus Type-2 (DENV-2) Infection and Has Anti-Viral Actions Requiring the C-Terminal End of Viperin. PloS Negl Trop D (2013) 7(4):e2178. doi: 10.1371/journal.pntd.0002178
31. Wang XY, Hinson ER, Cresswell P. The Interferon-Inducible Protein Viperin Inhibits Influenza Virus Release by Perturbing Lipid Rafts. Cell Host Microbe (2007) 2(2):96–105. doi: 10.1016/j.chom.2007.06.009
32. Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, et al. HIV-1 Infection of Human Macrophages Directly Induces Viperin Which Inhibits Viral Production. Blood (2012) 120(4):778–88. doi: 10.1182/blood-2012-01-407395
33. Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, et al. Publisher Correction: A Naturally Occurring Antiviral Ribonucleotide Encoded by the Human Genome. Nature (2018) 562(7725):610–4. doi: 10.1038/s41586-018-0355-0
34. Sun BJ, Nie P. Molecular Cloning of the Viperin Gene and its Promoter Region From the Mandarin Fish Siniperca Chuatsi. Vet Immunol Immunopathol (2004) 101(3-4):161–70. doi: 10.1016/j.vetimm.2004.04.013
35. Chen ZY, Lei XY, Zhang QY. The Antiviral Defense Mechanisms in Mandarin Fish Induced by DNA Vaccination Against a Rhabdovirus. Vet Microbiol (2012) 157(3-4):264–75. doi: 10.1016/j.vetmic.2011.12.025
36. Lee SH, Peng KC, Lee LH, Pan CY, Hour AL, Her GM, et al. Characterization of Tilapia (Oreochromis Niloticus) Viperin Expression, and Inhibition of Bacterial Growth and Modulation of Immune-Related Gene Expression by Electrotransfer of Viperin DNA Into Zebrafish Muscle. Vet Immunol Immunopathol (2013) 151(3-4):217–28. doi: 10.1016/j.vetimm.2012.11.010
37. Wang B, Zhang YB, Liu TK, Shi J, Sun F, Gui JF. Fish Viperin Exerts a Conserved Antiviral Function Through RLR-Triggered IFN Signaling Pathway. Dev Comp Immunol (2014) 47(1):140–9. doi: 10.1016/j.dci.2014.07.006
38. Eslamloo K, Ghorbani A, Xue X, Inkpen SM, Larijani M, Rise ML. Characterization and Transcript Expression Analyses of Atlantic Cod Viperin. Front Immunol (2019) 10:311. doi: 10.3389/fimmu.2019.00311
39. Wang F, Jiao HQ, Liu WM, Chen B, Wang YD, Chen BX, et al. The Antiviral Mechanism of Viperin and its Splice Variant in Spring Viremia of Carp Virus Infected Fathead Minnow Cells. Fish Shellfish Immunol (2019) 86:805–13. doi: 10.1016/j.fsi.2018.12.012
40. Zhang SH, Lv XY, Deng D, Zeng L, Huang W, Tang DD, et al. Gene Characterization and Expression Pattern of Mx and Viperin Genes in Dabry’s Sturgeon Acipenser Dabryanus. J Appl Ichthyol (2019) 35(2):408–19. doi: 10.1111/jai.13876
41. Gui JF, Zhou L. Genetic Basis and Breeding Application of Clonal Diversity and Dual Reproduction Modes in Polyploid Carassius Auratus Gibelio. Sci China Life Sci (2010) 53(4):409–15. doi: 10.1007/s11427-010-0092-6
42. Li XY, Gui JF. Diverse and Variable Sex Determination Mechanisms in Vertebrates. Sci China Life Sci (2018) 61(12):1503–14. doi: 10.1007/s11427-018-9415-7
43. Li XY, Liu XL, Zhu YJ, Zhang J, Ding M, Wang MT, et al. Origin and Transition of Sex Determination Mechanisms in a Gynogenetic Hexaploid Fish. Heredity (Edinb) (2018) 121(1):64–74. doi: 10.1038/s41437-017-0049-7
44. Wang ZW, Zhu HP, Wang D, Jiang FF, Guo W, Zhou L, et al. A Novel Nucleo-Cytoplasmic Hybrid Clone Formed via Androgenesis in Polyploid Gibel Carp. BMC Res Notes (2011) 4:82. doi: 10.1186/1756-0500-4-82
45. Chen F, Li XY, Zhou L, Yu P, Wang ZW, Li Z, et al. Stable Genome Incorporation of Sperm-Derived DNA Fragments in Gynogenetic Clone of Gibel Carp. Mar Biotechnol (2020) 22(1):54–66. doi: 10.1007/s10126-019-09930-w
46. Zhou L, Wang ZW, Wang Y, Gui JF. Crucian Carp and Gibel Carp Culture. In: . Aquaculture in China: Success Stories and Modern Trends. Oxford: John Wiley & Sons Ltd (2018). p. 149–57.
47. Zeng XT, Chen ZY, Deng YS, Gui JF, Zhang QY. Complete Genome Sequence and Architecture of Crucian Carp Carassius Auratus Herpesvirus (CaHV). Arch Virol (2016) 161(12):3577–81. doi: 10.1007/s00705-016-3037-y
48. Gao FX, Wang Y, Zhang QY, Mou CY, Li Z, Deng YS, et al. Distinct Herpesvirus Resistances and Immune Responses of Three Gynogenetic Clones of Gibel Carp Revealed by Comprehensive Transcriptomes. BMC Genomics (2017) 18(1):561. doi: 10.1186/s12864-017-3945-6
49. Gao FX, Lu WJ, Wang Y, Zhang QY, Zhang YB, Mou CY, et al. Differential Expression and Functional Diversification of Diverse Immunoglobulin Domain-Containing Protein (DICP) Family in Three Gynogenetic Clones of Gibel Carp. Dev Comp Immunol (2018) 84:396–407. doi: 10.1016/j.dci.2018.03.013
50. Lu WJ, Zhou L, Gao FX, Zhou YL, Li Z, Zhang XJ, et al. Dynamic and Differential Expression of Duplicated Cxcr4/Cxcl12 Genes Facilitates Antiviral Response in Hexaploid Gibel Carp. Front Immunol (2020) 11:2176. doi: 10.3389/fimmu.2020.02176
51. Lu WJ, Gao FX, Wang Y, Zhang QY, Li Z, Zhang XJ, et al. Differential Expression of Innate and Adaptive Immune Genes in the Survivors of Three Gibel Carp Gynogenetic Clones After Herpesvirus Challenge. BMC Genomics (2019) 20:432. doi: 10.1186/s12864-019-5777-z
52. Mou CY, Wang Y, Zhang QY, Gao FX, Li Z, Tong JF, et al. Differential Interferon System Gene Expression Profiles in Susceptible and Resistant Gynogenetic Clones of Gibel Carp Challenged With Herpesvirus CaHV. Dev Comp Immunol (2018) 86:52–64. doi: 10.1016/j.dci.2018.04.024
53. Lu M, Li XY, Li Z, Du WX, Zhou L, Wang Y, et al. Regain of Sex Determination System and Sexual Reproduction Ability in a Synthetic Octoploid Male Fish. Sci China Life Sci (2021) 64(1):77–87. doi: 10.1007/s11427-020-1694-7
54. Yu P, Wang Y, Yang WT, Li Z, Zhang XJ, Zhou L, et al. Upregulation of the PPAR Signaling Pathway and Accumulation of Lipids are Related to the Morphological and Structural Transformation of the Dragon-Eye Goldfish Eye. Sci China Life Sci (2021) 64(7):1031–49. doi: 10.1007/s11427-020-1814-1
55. Li XY, Zhang XJ, Li Z, Hong W, Liu W, Zhang J, et al. Evolutionary History of Two Divergent Dmrt1 Genes Reveals Two Rounds of Polyploidy Origins in Gibel Carp. Mol Phylogenet Evol (2014) 78:96–104. doi: 10.1016/j.ympev.2014.05.005
56. Zhang YB, Zhang QY, Xu DQ, Hu CY, Gui JF. Identification of Antiviral-Relevant Genes in the Cultured Fish Cells Induced by UV-Inactivated Virus. Chin Sci Bull (2003) 48(6):581–8. doi: 10.1360/03tb9124
57. Liu TK, Zhang YB, Liu Y, Sun F, Gui JF. Cooperative Roles of Fish Protein Kinase Containing Z-DNA Binding Domains and Double-Stranded RNA-Dependent Protein Kinase in Interferon-Mediated Antiviral Response. J Virol (2011) 85(23):12769–80. doi: 10.1128/JVI.05849-11
58. Liu Y, Zhang YB, Liu TK, Gui JF. Lineage-Specific Expansion of IFIT Gene Family: An Insight Into Coevolution With IFN Gene Family. PloS One (2013) 8(6):e66859. doi: 10.1371/journal.pone.0066859
59. Xu L, Podok P, Xie J, Lu L. Comparative Analysis of Differential Gene Expression in Kidney Tissues of Moribund and Surviving Crucian Carp (Carassius Auratus Gibelio) in Response to Cyprinid Herpesvirus 2 Infection. Arch Virol (2014) 159(8):1961–74. doi: 10.1007/s00705-014-2011-9
60. Gan RH, Wang Y, Li Z, Yu ZX, Li XY, Tong JF, et al. Functional Divergence of Multiple Duplicated Foxl2 Homeologs and Alleles in a Recurrent Polyploid Fish. Mol Biol Evol (2021) 38(5):1995–2013. doi: 10.1093/molbev/msab002
61. Lu LF, Li S, Wang ZX, Du SQ, Chen DD, Nie P, et al. Grass Carp Reovirus VP41 Targets Fish MITA To Abrogate the Interferon Response. J Virol (2017) 91(14):e00390–17. doi: 10.1128/JVI.00390-17
62. Li S, Lu LF, Liu SB, Zhang C, Li ZC, Zhou XY, et al. Spring Viraemia of Carp Virus Modulates P53 Expression Using Two Distinct Mechanisms. PloS Pathog (2019) 15(3):e1007695. doi: 10.1371/journal.ppat.1007695
63. Lu LF, Li S, Lu XB, LaPatra SE, Zhang N, Zhang XJ, et al. Spring Viremia of Carp Virus N Protein Suppresses Fish IFN Phi 1 Production by Targeting the Mitochondrial Antiviral Signaling Protein. J Immunol (2016) 196(9):3744–53. doi: 10.4049/jimmunol.1502038
64. Sun F, Zhang YB, Liu TK, Shi J, Wang B, Gui JF. Fish MITA Serves as a Mediator for Distinct Fish IFN Gene Activation Dependent on IRF3 or IRF7. J Immunol (2011) 187(5):2531–9. doi: 10.4049/jimmunol.1100642
65. Deribe YL, Pawson T, Dikic I. Post-Translational Modifications in Signal Integration. Nat Struct Mol Biol (2010) 17(6):666–72. doi: 10.1038/nsmb.1842
66. Ikeda F, Dikic I. Atypical Ubiquitin Chains: New Molecular Signals - ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep (2008) 9(6):536–42. doi: 10.1038/embor.2008.93
67. Yau R, Rape M. The Increasing Complexity of the Ubiquitin Code. Nat Cell Biol (2016) 18(6):579–86. doi: 10.1038/ncb3358
68. Feng H, Zhang YB, Gui JF, Lemon SM, Yamane D. Interferon Regulatory Factor 1 (IRF1) and Anti-Pathogen Innate Immune Responses. PloS Pathog (2021) 17(1):e1009220. doi: 10.1371/journal.ppat.1009220
69. Jiang SY, Wang Y, Zhou L, Chen F, Li Z, Gui JF. Molecular Characteristics, Genomic Structure and Expression Patterns of Diverse Bmp15 Alleles in Polyploid Gibel Carp Clone F. Acta Hydrobiol Sin (2020) 44:518–27. doi: 10.7541/2020.063
70. Zhang QQ, Zhou L, Li Z, Gan RH, Yu ZX, Gui JF, et al. Allelic Diversification, Syntenic Alignment and Expression Patterns of Nanos2 in Polyploid Gibel Carp. Acta Hydrobiol Sin (2020) 44(5):1087–96. doi: 10.7541/2020.126
71. Upadhyay AS, Stehling O, Panayiotou C, Rosser R, Lill R, Overby AK. Cellular Requirements for Iron-Sulfur Cluster Insertion Into the Antiviral Radical SAM Protein Viperin. J Biol Chem (2017) 292(33):13879–89. doi: 10.1074/jbc.M117.780122
72. Upadhyay AS, Vonderstein K, Pichlmair A, Stehling O, Bennett KL, Dobler G, et al. Viperin is an Iron-Sulfur Protein That Inhibits Genome Synthesis of Tick-Borne Encephalitis Virus via Radical SAM Domain Activity. Cell Microbiol (2014) 16(6):834–48. doi: 10.1111/cmi.12241
73. Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K, et al. Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J Virol (2018) 92(7):e02054–17. doi: 10.1128/JVI.02054-17
74. Dumbrepatil AB, Ghosh S, Zegalia KA, Malec PA, Hoff JD, Kennedy RT, et al. Viperin Interacts With the Kinase IRAK1 and the E3 Ubiquitin Ligase TRAF6, Coupling Innate Immune Signaling to Antiviral Ribonucleotide Synthesis. J Biol Chem (2019) 294(17):6888–98. doi: 10.1074/jbc.RA119.007719
75. Zheng Y, Gao CJ. E3 Ubiquitin Ligases, the Powerful Modulator of Innate Antiviral Immunity. Cell Immunol (2019) 340:103915. doi: 10.1016/j.cellimm.2019.04.003
76. Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C, Ciechanover A. The Ubiquitin-Proteasome System and Autophagy: Coordinated and Independent Activities. Int J Biochem Cell Biol (2016) 79:403–18. doi: 10.1016/j.biocel.2016.07.019
Keywords: viperin, polyploid, homeolog, herpesvirus, proteasomal degradation, autophagosome
Citation: Mou C-Y, Li S, Lu L-F, Wang Y, Yu P, Li Z, Tong J-F, Zhang Q-Y, Wang Z-W, Zhang X-J, Wang G-X, Zhou L and Gui J-F (2021) Divergent Antiviral Mechanisms of Two Viperin Homeologs in a Recurrent Polyploid Fish. Front. Immunol. 12:702971. doi: 10.3389/fimmu.2021.702971
Received: 30 April 2021; Accepted: 09 August 2021;
Published: 31 August 2021.
Edited by:
Verónica Chico Gras, Universidad Miguel Hernández de Elche, SpainReviewed by:
Sarah J. Poynter, Wilfrid Laurier University, CanadaKhalil Eslamloo, Memorial University of Newfoundland, Canada
Copyright © 2021 Mou, Li, Lu, Wang, Yu, Li, Tong, Zhang, Wang, Zhang, Wang, Zhou and Gui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhou, emhvdWxpQGloYi5hYy5jbg==; Jian-Fang Gui, amZndWlAaWhiLmFjLmNu
 Cheng-Yan Mou1,2
Cheng-Yan Mou1,2 Shun Li
Shun Li Long-Feng Lu
Long-Feng Lu Qi-Ya Zhang
Qi-Ya Zhang Zhong-Wei Wang
Zhong-Wei Wang Li Zhou
Li Zhou Jian-Fang Gui
Jian-Fang Gui