- 1Institute of Marine Sciences and Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou, China
- 2STU-UMT Joint Shellfish Research Laboratory, Shantou University, Shantou, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
Shrimp aquaculture is an essential economic venture globally, but the industry faces numerous challenges, especially pathogenic infections. As invertebrates, shrimp rely mainly on their innate immune system for protection. An increasing number of studies have shown that ubiquitination plays a vital role in the innate immune response to microbial pathogens. As an important form of posttranslational modification (PTM), both hosts and pathogens have exploited ubiquitination and the ubiquitin system as an immune response strategy to outwit the other. This short review brings together recent findings on ubiquitination and how this PTM plays a critical role in immune modulation in penaeid shrimps. Key findings inferred from other species would help guide further studies on ubiquitination as an immune response strategy in shrimp-pathogen interactions.
Introduction
Shrimp aquaculture has become one of the world’s most extensive and most valuable farming practices due to increased shrimp consumption. However, the rapid development of shrimp aquaculture comes with several challenges, especially pathogenic infections, which therefore hamper the development of the shrimp aquaculture industry. Penaeid shrimps are particularly vulnerable to various microorganisms, including viruses, bacteria, fungi, and other parasites (1, 2) due to the nature of their environment. Moreover, as invertebrates, penaeid shrimps mainly rely on their innate immune system for protection against pathogens (3).
The innate immune system of shrimp comprises cellular and humoral immune responses, which work in concert to offer protection against microbial infections (4, 5). The cellular arm of the innate immune response is involved several immune functions such as phagocytosis and apoptosis through immune cells such as hemocytes (6–8). On the other hand, humoral immune responses involve non-specific enzymes or factors in hemolymph, such as phenoloxidase, lectins, antimicrobial peptides (AMPs), etc., which eliminate pathogens by direct killing or inhibit their growth and spread (2, 3, 9–11). In addition to the direct elimination of pathogens through immune factors in shrimp body fluids, shrimp can also identify pathogens through pattern recognition receptors (PRRs) to activate a series of downstream immune signaling pathways that mediate the activation of cellular and humoral immune responses (12, 13). For example, the carbohydrate recognition domain (CTLD) of C-type lectins (CTLs) in Marsupenaeus japonicus recognizes bacterial glycans, while the coiled-coil domain (CCD) interacts with the surface receptor, Domeless, to activate JAK/STAT signaling pathway, which then regulates AMPs production to clear bacterial pathogens (14). Similarly, the Toll and immune deficiency (IMD) pathways are essential in shrimp antimicrobial response due to their major role in regulating AMPs expression via the NF-κB/Relish signaling pathways (15, 16).
A growing number of studies have shown that PTMs play an essential role in regulating immune responses (17–20). For instance, phosphorylation of Ser349 on tumor necrosis factor receptor-related protein 3 (TRAF3) by the serine-threonine kinase CK1ϵ facilitates the production of antiviral cytokines. Thus, deficiency of CK1ϵ in mice attenuates its ability to effectively produce antiviral factors during viral infection (21). In M. japonicus, the phosphorylation of focal adhesion kinase (FAK) promotes the activation of multiple immune signaling pathways, including antiviral response during white spot syndrome virus (WSSV) infection (22). The envelope proteins, gp116 and gp64, of yellow head virus (YHV), possess N-linked glycosylation (23), which accelerates the formation and release of YHV virions into the hemolymph of infected shrimp (Penaeus monodon), hence increasing the severity of the disease (24). Similarly, we previously revealed that the glycosylation of Penaeus vannamei hemocyanin increases its antibacterial and agglutination activity against Vibrio alginolyticus and Vibrio fluvialis (25). In mammals, infection of mouse embryo fibroblasts (MEFs) with herpes simplex virus type 1 (HSV-1) and vesicular stomatitis virus (VSV) induces acetylation of the tumor suppressor p53 at Lys379, which is indispensable for the transcriptional activation of p53-dependent genes in response to viral infection and replication (26). Acetylation of interferon regulatory factor 3 (IRF3) at Lys359 by lysine acetyltransferase 8 (KAT8) results in the inhibition of the transcriptional activity of IRF3 in mouse macrophages, hence, decreasing type I interferons (IFN-I) production and antiviral responses (27).
Recent studies have implicated other PTMs such as small ubiquitin-related modifier (SUMO) modification (SUMOylation) and ubiquitination in immune response in both vertebrates and invertebrates (28–31). For instance, in the retinoic acid–inducible gene I (RIG-I)-like receptors (RLRs) signaling pathway in mice, ubiquitination of RIG-I and mitochondrial antiviral signaling protein (MAVS) affects downstream TNF receptor-associated factors (TRAFs) and IFN regulatory factors, thereby stimulating IFN-I production for antiviral immunity (32–34). In zebrafish, tripartite motif proteins (TRIMs) when induced during viral hemorrhagic septicemia virus (VHSV) infection, display E3 ubiquitin ligase activity, which suggest that TRIMs could regulate antiviral response via ubiquitination (35). The viral immediate early (IE) proteins of WSSV can be modified by SUMOylation, thus promoting viral gene transcription and replication in Procambarus clarkii (36). During Staphyloccocus aureus and Vibrio parahaemolyticus infection in Chinese mitten crab Eriocheir sinensis, the ubiquitin-like protein, neural precursor cells expresses developmental downregulation 8 (NEDD8) is inhibited to prevent its conjugation with Cullin4 (37), which attenuates the E3 ubiquitin ligase activity of Cullin4 (38). For penaeid shrimps, there is limited information on the role of ubiquitination in immune response. This paper reviews recent findings on the significance of ubiquitination during immune response to microbial pathogens and identify areas that could be explored to understand the role played by ubiquitination in penaeid shrimp immune defense.
Ubiquitination in Host-Pathogen Interactions
Ubiquitination is an essential type of PTM, whereby ubiquitin (76 amino acid polypeptides) units are attached to a target protein through a series of three steps catalyzed by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligase (E3) (39). In this cascade reaction, E1 activates and transfers ubiquitin to E2 using ATP, followed by the interaction between E2 and E3, allowing the transfer of ubiquitin to the target protein. The seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) on ubiquitin or its N-terminal methionine are the residues through which other ubiquitin units bind to increase the number of ubiquitin molecules on the target protein (40). Ubiquitination of target proteins allows these proteins to be recognized by other enzyme complexes or organelles in the cell (40), thereby executing their functions (Figure 1), such as cell cycle, proliferation, differentiation, DNA repair, energy metabolism, etc. (41–44).
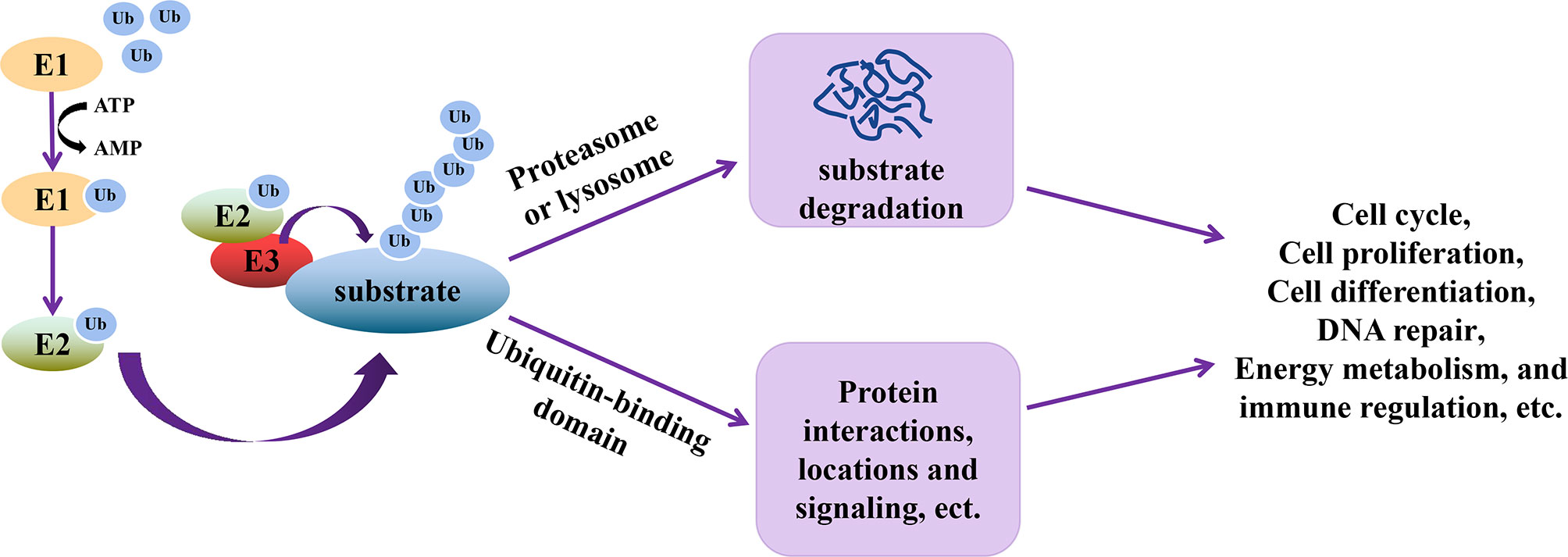
Figure 1 Schematic representation of the functional relevance of ubiquitination. During ubiquitination, E1 is first activated to transfer ubiquitin (Ub) to E2 using ATP. Through the interactions of E2 and E3, E3 and ubiquitinated substrates, Ub is transferred to the substrates, which allows the substrates to be recognized by enzyme complex or organelles. The ubiquitinated substrate can then perform multiple functions, including cell cycle, proliferation, differentiation, DNA repair, energy metabolism, signal transduction and immune regulation, etc.
Besides the primary functions of ubiquitination, recent studies have implicated ubiquitination as an essential regulatory mechanism in immune response (45, 46). For instance, in the orange-spotted grouper Epinephelus coioides, the E3 ubiquitin ligase TRIM8 can inhibit the replication of Singapore grouper iridovirus (SGIV) and red-spotted grouper nervous necrosis virus (RGNNV) by enhancing the expression of IR3, IR7 and interferon-related factors (47). During bacterial infection, Drosophila E3 ubiquitin ligase LUBEL catalyzes the conjugation of IκB kinase γ (IKKγ) Kenny to form M1-linked linear ubiquitin (M1-Ub) chains, which activates Relish-mediated AMPs gene expression to clear the bacteria (48). In Scylla paramamosain, an E3 ubiquitin ligase casitas B-lineage lymphoma (CBL) protein has been found to decrease WSSV proliferation through hemocytes apoptosis (49).
Ubiquitination and Shrimp Antiviral Response
The key to successful virus infection is the entry of the virus into cells, hence, most viruses use host receptor proteins to enter cells by endocytosis followed by exploiting host nutrients for its replication, proliferation, etc. For example, the WSSV envelope protein, VP24, interacts with polymeric immunoglobulin receptor (pIgR) to enable endocytosis of WSSV into cells, through a pIgR-Calmodulin-Clathrin mediated mechanism in M. japonicus (50). Similarly, the WSSV protein kinase 1 interacts with ferritin to block the binding of ferritin to free iron, which facilitates the use of iron by the virus for its proliferation in M. japonicus (51). Some viruses also exploit or hijack the immune signaling pathways in shrimp, such as Toll/IMD-nuclear factor-κB (NF-κB), JAK-STAT, and Wnt/β-catenin signaling pathways, for their proliferation (7, 52). For instance, upon WSSV stimulation, NF-κB factor Relish binds to the promoter of the immediate early gene of WSSV, ie1, to induce its expression and WSSV replication (7). Toll4 is reported to mediate the production of AMPs through the Toll4-dorsal pathway as an antiviral immune response in P. vannamei (16). After JAK/STAT silencing in P. vannamei, WSSV copies decreased significantly, indicating that the JAK/STAT signal pathway plays an important role in the antiviral activity of shrimp (53, 54).
In mammals and Drosophila, ubiquitination plays a role in mediating innate immune response regulation (46, 55). The E3 ubiquitin ligase RNF125 in mammals, promote K48-linked polyubiquitination and degradation of the mitochondrial adaptor TRIM14 to suppress IFN-I production upon Sendai virus infection (56). Similarly, polyubiquitination of IMD at Lys137 and Lys153 by the E2 ubiquitin-conjugating enzymes (i.e., Effete (Ubc5) and Bendless (Ubc13)-Uev1a complex) and E3 ubiquitin ligase DIAP2 in Drosophila, triggers the removal of K63-linked polyubiquitin chains, thereby increasing K48 polyubiquitination to degrade IMD via proteasome degradation as part of immune response homeostasis (55). Some emerging evidence suggests the involvement of ubiquitination in mediating antiviral immune response in shrimp. For instance, transcriptome analysis revealed dysregulation in ubiquitin-proteasome pathway genes in shrimp hemocytes (57), while ubiquitination in gills (58) was found to alter WSSV infection of P. vannamei, which suggest the modulation involvement of the ubiquitin system in shrimp during WSSV infection. In F. chinensis, the ubiquitin-conjugating enzyme FcUbc, was shown to ubiquitinate the really interesting new gene (RING) domains (WRDs) of WSSV viral proteins WSSV277 and WSSV304, to inhibit viral replication and reduces shrimp mortality upon WSSV infection (59). Thus, ubiquitination can play a direct antiviral role in shrimp.
Analysis of the hepatopancreas transcriptome of WSSV infected shrimp (P. vannamei) revealed decreased expression of Wnt signaling pathway genes (60). Indeed, ubiquitination is reported to regulate the Wnt/β-catenin pathway in shrimp, as β-catenin undergoes ubiquitination after treatment with the proteasome inhibitor MG132 followed by WSSV infection (61). Given that Pvβ-catenin positively regulates AMPs production (52), interacts with WSSV069, and attenuates the expression of viral genes (61), it indicates that ubiquitination plays a positive antiviral role in shrimp through β-catenin in the Wnt/β-catenin pathway, especially during early viral replication (Figure 2). Besides, ubiquitination is involved in antiviral immune response through other proteins by indirectly affecting immune signaling pathway. For instance, the E3 ubiquitin ligase tripartite motif 9 protein (PvTRIM9) interacts with the NF-κB pathway inhibitor beta-transducin repeat-containing protein (Pvβ-TrCP) in P. vannamei, hence knockdown of PvTRIM9 followed by WSSV infection increases PvRelish expression and AMPs production, coupled with a decrease in viral copy number (64). This indicates that WSSV inhibits the NF-κB pathway and AMPs production by annexing the E3 ubiquitin ligase PvTRIM9 in shrimp (64).
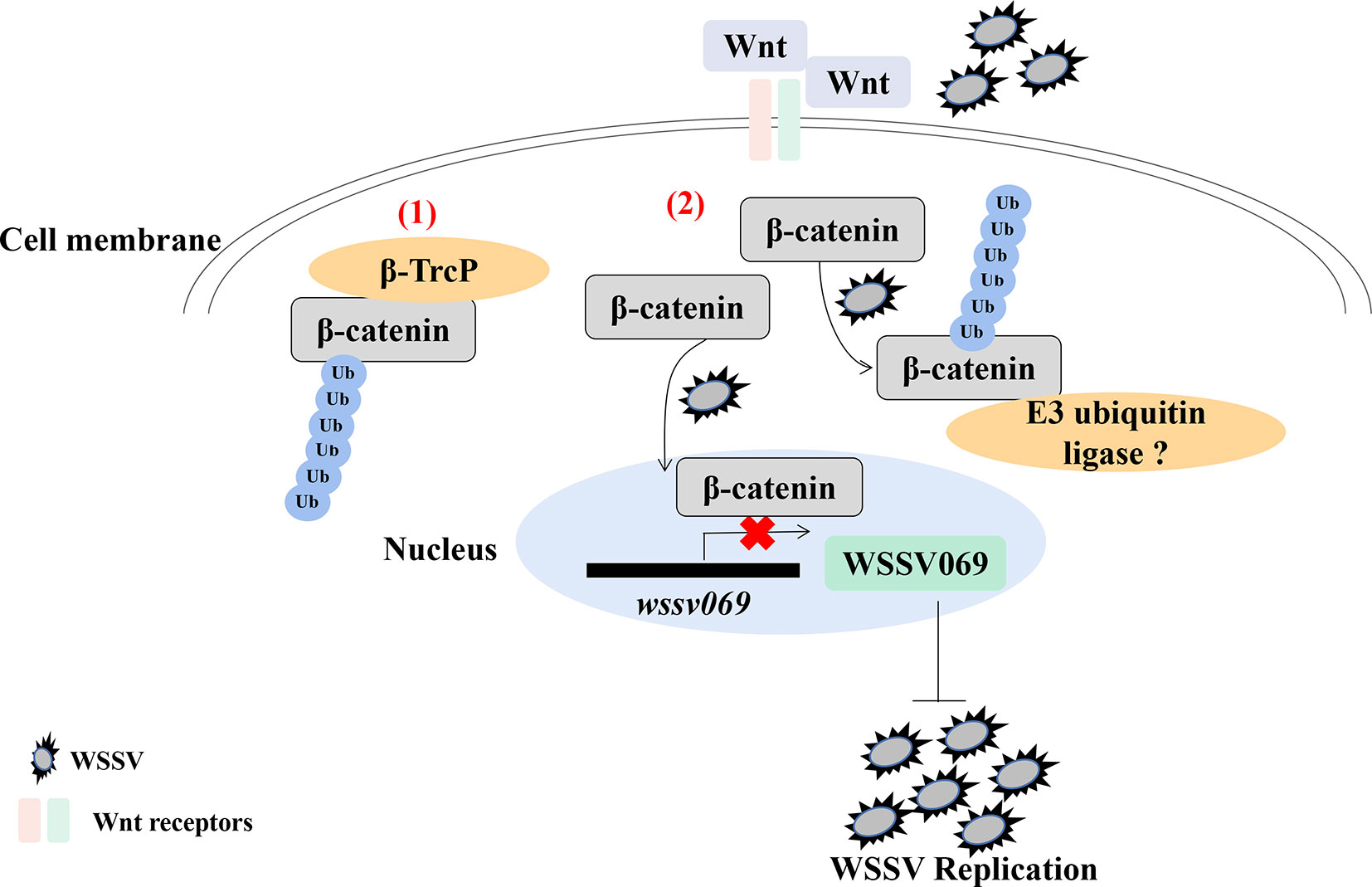
Figure 2 Ubiquitination of β-catenin is important for penaeid shrimp antiviral immune response. β-catenin is a key regulator in the Wnt/β-catenin signaling pathway. (1) In mammals, β-catenin in cytoplasm could be ubiquitinated by an E3 ubiquitin ligase β-TrcP for degradation (62, 63). (2) In the cytoplasm of shrimp, β-catenin is ubiquitinated by WSSV infection, which also promotes the translocation of β-catenin into nucleus to inhibit the expression of virus immediate early gene wssv069. β-catenin can also interact with wssv069 (61). Thus, β-catenin plays a positive role through ubiquitination to inhibit WSSV replication during infection. However, which protein could ubiquitinate β-catenin in penaeid shrimp has not been found.
Exploitation of Ubiquitination by Viruses to Outwit Penaeid Shrimp Immune Response
Viruses invade their host through interactions between host and viral proteins, especially via the use of viral envelope proteins for recognition and entry (7, 65), recruiting cellular proteins in order to invade the cytoplasm (7, 66), etc. Furthermore, viruses can manipulate and exploit the host ubiquitin system to elude host immune response (67). For instance, the immediate-early 1 protein (IE1) of congenital human cytomegalovirus (HCMV) promotes the ubiquitination and degradation of Hairy and Enhancer of Split 1 (Hes1), an essential downstream effector of Notch signaling (68). By this, IE1 acts as an E3 ubiquitin ligase to dysregulate Notch signaling, thereby resulting in aberrant differentiation of in neural progenitor cells (NPCs) (68, 69). Similarly, proteins that contain the RING domain are reported to act as an E3 ubiquitin ligase, such as TRIM proteins (70, 71). In E. coioides, the TRIM32 protein, which has a deletion of the RING domain, cannot positively regulate the expression of IFN-stimulated genes, hence attenuating antiviral response to SGIV or RGNNV infection, an indication of the importance of the RING domain in host antiviral response (72). Recent studies show that some WSSV proteins, such as WSSV199, WSSV222, WSSV249, and WSSV403, contain the RING domain; hence, they modulate the ubiquitin system of shrimp to benefit (73). For instance, in the presence of shrimp ubiquitin-conjugating enzyme UbcH6, WSSV222 acts as E3 ubiquitin ligase to mediate ubiquitination and degradation of shrimp tumor suppressor-like protein (TSL), which aids viral replication (74). Furthermore, knockdown of WSSV222 by small interfering RNA (siRNA) reduces the severity of WSSV infection by delaying WSSV replication in shrimp (75). The WSSV222 viral protein, therefore, promotes WSSV replication by annexing the ubiquitin system in shrimp during WSSV infection. Similarly, the viral protein, WSSV403, is reported to exert its E3 ubiquitin ligase activity with shrimp ubiquitin-conjugating enzyme E2 (76). WSSV403 can interact with protein phosphatase (PPs) in shrimp, which suggests that WSSV403 uses its E3 ubiquitin ligase activity to regulate WSSV latent infection (76). However, whether PPs are the ubiquitinated substrate of WSSV403 under WSSV infection has not been reported. Details on this would identify new strategies or potential targets for preventing and treating WSSV infection in shrimp.
Ubiquitination as an Immune Response to Other Microorganisms or Environmental Cues in Penaeid Shrimp
During host-pathogen interactions, the host can regulate its ubiquitin system to eliminate the pathogens (30). For example, a 74 amino acid residue containing ubiquitin obtained from gills of Pacific oyster Crassostrea giga shows strong antimicrobial activity against Gram-positive (e.g., Streptococcus iniae) and Gram-negative bacteria (e.g., Vibrio parahemolyticus) (77). Host cells can eliminate pathogens through ubiquitination and degradation mediated by proteasome, phagosome, and autophagosome (30, 78). In MEFs, an E3 ubiquitin ligase, neuronal precursor cell expressed, developmentally downregulated 4 (NEDD4), promotes autophagy to clear bacteria via ubiquitination of BECN1 (79). During infection with fungi or Gram-positive bacteria, Drosophila Spätzle activates the Toll receptor, resulting in the ubiquitination and proteasome degradation of the Toll receptor adaptor protein Cactus, thereby enhancing AMPs production to clear the pathogens (31). Apart from the modulation of the ubiquitin system by the host during an immune response, some pathogens also exploit or interfere with the host ubiquitin system to replicate and escape immune surveillance (80). For example, the NleL ligase (Non-Lee encoded effector ligase), a virulent protein of enterohemorrhagic Escherichia coli (EHEC), acts as an E3 ubiquitin ligase that ubiquitinates the Lys68 of human c-Jun NH2-terminal kinases (JNK), which results in the dephosphorylation and deactivation of JNK to enhance EHEC infection (81).
In shrimp, few studies have explored ubiquitination as an immune response to invasion by pathogens other than viruses. Nonetheless, transcriptomic analysis of hemocytes from lipopolysaccharide (LPS) treated P. vannamei revealed the upregulation of some ubiquitin-proteasome pathway genes including ubiquitin, ubiquitin-conjugating enzyme E2C, ubiquitin-conjugating enzyme H1 and ubiquitin-conjugating enzyme H5b (82). Similarly, ubiquitin-related genes, such as ubiquitin, ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, various E3 ubiquitin ligases and deubiquitinating enzymes, were significantly changed in the transcriptome of hemocytes from P. vannamei infected with acute hepatopancreas necrosis disease (AHPND) V. parahemolyticus (Figure 3) (83). Significant changes in the expression of ubiquitin-mediated pathway genes have also been found in the transcriptomic study of hemocytes from V. parahaemolyticus infected mud crab S. paramamosain (84). All these studies indicate a strong association between the ubiquitin system and bacterial infection in crustaceans. Although there is currently no sufficient evidence in penaeid shrimps relating the use of the ubiquitin system by host to clear invading bacteria or it being hijacked by pathogens to invade immune clearance, a number of studies including in vivo knockdown of the E3 ubiquitin ligase Pellino in P. vannamei, have shown decreased AMPs production and increase shrimp mortality upon V. parahaemolyticus challenge (85). This suggests that PvPellino plays a positive role in the antibacterial response in shrimp. However, the specific role of Pellino in shrimp antibacterial response and its ubiquitinated substrate has not been reported. In any case, Pellino proteins are involved in the TLR signaling pathway, acting as conserved scaffold proteins and also function as an E3 ubiquitin ligase (86, 87). In mouse macrophages, Pellino is induced by LPS stimulation, thus promoting the ubiquitination of TANK-binding kinase 1 (TBK1) and TRAF6 to regulate the TLR signaling pathway (88). Similarly, Pellino can also interact with and ubiquitinate MyD88 to maintain innate immune homeostasis (89). All these pieces of evidence can be used as references to explore the ubiquitinated substrates of Pellino and its response to microorganisms in shrimp (Figure 4).
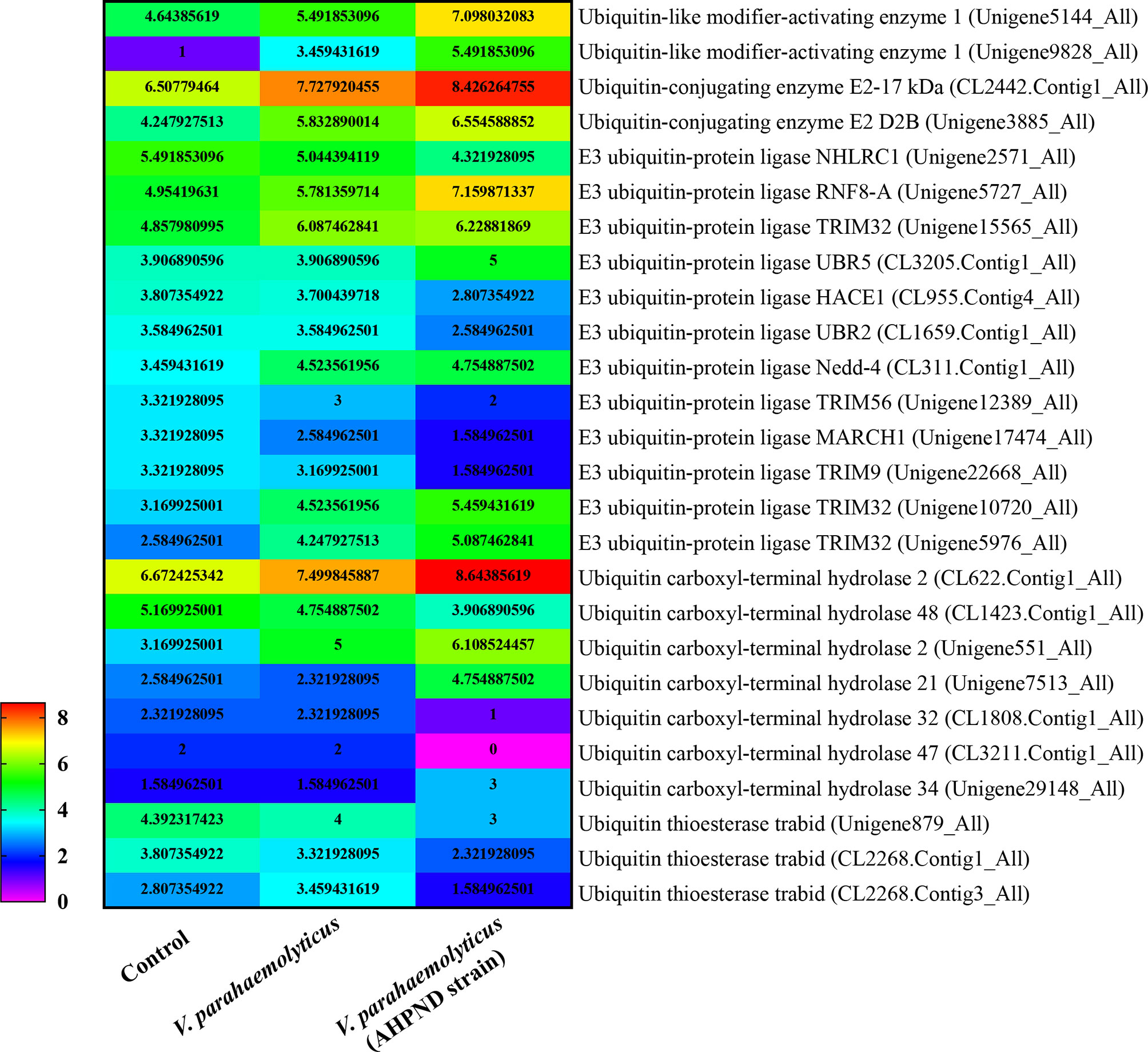
Figure 3 |eat map showing changes in the expression pattern of ubiquitin-related genes in penaeid shrimp (Penaeus vannamei) hemocytes infected with V. parahaemolyticus and V. parahemolyticus (AHPND strain). The numbers represent Log2 fold change. Hemocytes samples were pulled from 30 individual shrimps (n=30). Data used for the figure was culled from the transcriptome data of (83).
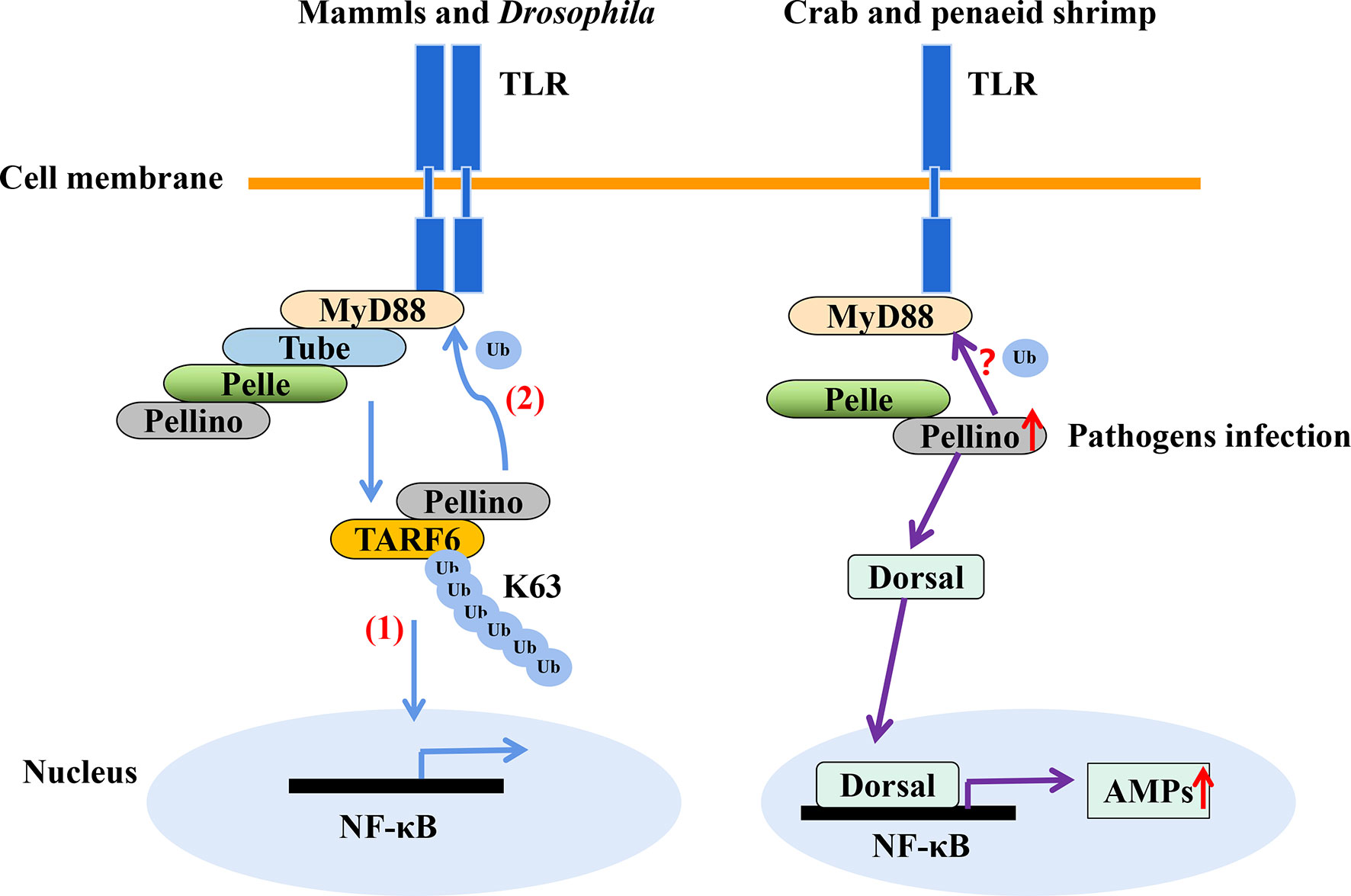
Figure 4 The E3 ubiquitin ligase Pellino plays a positive role in shrimp antibacteria response. Pellino acts as a conserved scaffold protein and E3 ubiquitin ligase in the TLR signaling pathway. (1) Pellino promotes the K63-linked ubiquitination of TARF6 (88). (2) In addition, Pellino can ubiquitinate MyD88 for degradation to negatively regulate TLR signaling and maintain innate immune homeostasis (89). Pellino can also interact with Pelle in shrimp. The expression of Pellino is up-regulated upon pathogens stimulation, and to increase the activity of Dorsal, thus enhancing AMPs expression with V. parahaemolyticus challenge (85), in which Dorsal is also involved in defense against gram-positive bacterial in S. paramamosain (90). However, the ubiquitinated substrates of Pellino in shrimp (such as whether MyD88 is ubiquitinated) is currently unknown.
Prokaryotes have acquired a complex system for hijacking host cells in evolution. As much as most bacterial genome components are used for host infections (91), some bacterial proteins can modulate the host’s immune response as E3 ubiquitin ligases. For instance, in mice infected with Shigella flexneri, bacterial E3 ubiquitin ligase invasion plasmid Ag H (IpaH) protein IpaH4.5 promotes K48-linked polyubiquitination of TBK1 for degradation, which inhibits IFN production and contributes to bacteria colonization (92). The ubiquitin-like protein NEDD88 in Drosophila modifies Cullin family. The effector protein Cif, derived from EHEC, deaminates NEDD8, thus disrupting Cullins’ modification, which leads to accumulation of cell cycle regulatory factor and cell cycle retardation of the host (93, 94). In addition, 5’ expressed sequence tags (ESTs) analysis of Perkinsus marinus, a protozoan parasite of the eastern oyster Crassostrea virginica, revealed ubiquitin-specific proteases, which are thought to be involved in the degradation of host protein substrates to obtain normal cell function and proliferation necessary for nutrition (95). Ubiquitin components, proteasome, and autophagy pathway are involved in the defense of C. elegans against microsporidian Nematocida parisii infection (96). Thus far, no bacterial proteins or proteins from other pathogens have been identified as E3 ubiquitin ligase or found to interfere with the ubiquitin system in shrimps to mediate immune response. It, therefore, indicates that few studies have explored ubiquitination as an immune response feature of shrimp to bacteria, fungi, or other parasites infections.
Besides microbes, environmental factors, such as temperature and ammonia nitrogen stress, can directly affect protein modification. For instance, phosphorylation of p38MAPK in gills of P. vannamei increased rapidly under acute cold stress (97). Under chronic low-salinity stress, the acetylation of histone H4 changed significantly in the hepatopancreas of P. vannamei (98). In P. monodon, the expression of ubiquitin-conjugating enzyme E2 increased significantly in muscle but decreased in gut tissue in response to low salinity stress (99). Moreover, ubiquitin-related proteins, oxygen-free radicals, and oxidative stress-related proteins all changed significantly under long-term low salinity stress in the hepatopancreas of P. vannamei (100), which suggest that changes in the ubiquitination pathway in shrimp may be related to detoxification and immunity (100). It has also been reported that environmental factors affect the survival and proliferation of microorganisms. For instance, water temperature changes could promote Vibrio abundance in aquatic environments (101, 102). Similarly, low salinity levels could decrease the immune response of P. vannamei and attenuate their resistance to pathogens (103). As discussed in this short synthesis, microbial invasion could elicit host immune response via the ubiquitin system in penaeid shrimp. It is therefore conceivable that aquatic environmental factors such as temperature and salinity could also impact on ubiquitination thereby directly or indirectly affecting microorganisms and their interaction with hosts.
Summary and Future Perspective
Aquatic pathogens are still a threat to shrimp aquaculture. Ubiquitination regulates many biological processes in cells, including host immune response to pathogens. The synthesis presented in this paper provides a snapshot of ubiquitination as a host-pathogen immune response strategy in shrimp. Most current studies have shown that microbial infection of shrimp could induce ubiquitination through ubiquitin-conjugating enzymes, ubiquitin ligases, or ubiquitinated substrates (64, 85, 104). On the one hand, the ubiquitin system in shrimp could be exploited as a mechanism to resist pathogen invasion or annexed by microbial pathogens for replication and survival. For instance, WSSV viral proteins WSSV277 and WSSV304 are ubiquitinated by FcUbc, thereby inhibiting WSSV replication in Chinese white shrimp F. chinensis (59), while ubiquitination of β-catenin in shrimp promotes viral replication (61). Pathogens can also exploit the ubiquitin system for their benefit. Some viral proteins act as E3 ubiquitin ligases to promote viral infection and survival using the host ubiquitin system. The RING proteins of WSSV, including WSSV199, WSSV222, WSSV249, and WSSV403, could serve as E3 ligases to coopt the ubiquitin-proteasome pathway of shrimp for viral replication (74, 76, 104). Furthermore, WSSV249 acts as an E3 ligase through interaction of ubiquitin-conjugating enzyme E2 for viral pathogenesis in P. vannamei (105). Besides, the ubiquitination pathway might also be related to detoxification and shrimp immunity with salinity stress (100).
Although an increasing number of studies show that ubiquitination plays a vital role in immune response to pathogens, the detailed mechanisms and key players have not been elucidated in penaeid shrimps. For instance, the E3 ubiquitin ligases of some ubiquitinated proteins have not been identified. Although ubiquitination of Pvβ-catenin is reported to inhibit WSSV replication, the corresponding E3 ubiquitin ligase of Pvβ-catenin in shrimp remains unknown. Furthermore, it is unclear whether penaeid shrimp β-catenin also requires phosphorylation to initiate the ubiquitination process as in mammals. Besides, if phosphorylation of β-catenin does occur in shrimp, whether the regulation of phosphorylation and ubiquitination modification affects the antimicrobial immune function of β-catenin remains to be delineated. Currently, the specific functions of some identified E3 ubiquitin ligases in shrimps remains unknown. For instance, while the E3 ubiquitin ligase activity of PvTRIM9 in P. vannamei has been demonstrated, PvTRIM9 involvement in shrimp antiviral and antibacterial response is still unknown. Most reports on the relationship between ubiquitination and immune response in penaeid shrimp have mainly focused on WSSV infection. However, numerous studies have established links between ubiquitination and immune response to several pathogens in other species. For example, depletion of the E3 ubiquitin ligase TRIM62 in mice increases their susceptibility to fungal infection (106). Similarly, TRIM8 (47) and TRIM32 (72) are found to inhibit the replication of DNA (e.g., SGIV) and RNA (e.g., RGNNV) viruses in E. coioidest. Treating human peripheral blood mononuclear cells (PBMCs) with the proteasome inhibitor MG132 inhibits the ubiquitin-proteasome system and attenuates the replication of Plasmodium falciparum (107).
Given that the ubiquitin-proteasome system is conserved, it is conceivable that ubiquitination also serves as an immune response strategy to fungal and parasite infections in shrimp. Studies could therefore explore the significance and relevance of ubiquitination in penaeid shrimp immune response. A better understanding of the specific role of ubiquitination in host-pathogen interaction as an immune response strategy in penaeid shrimps would provide new avenues for improving shrimp immunity and, therefore, disease prevention. Some key components of the ubiquitin system, such as E1, E2, E3, and the ubiquitinated substrates, could be explored as potential targets to enhance shrimp immunity or treatment (108). Focused studies on the interplay between the ubiquitin system of penaeid shrimp and microbial pathogens would help unravel the molecular mechanism of shrimp immune response and how resistance to pathogens can be improved, which is vital for the development of penaeid shrimp aquaculture industry.
Author Contributions
JA and YZ conceived the idea. ZXZ and JA performed the literature search, wrote and revised the paper. YZ and JA obtained funding and provided supervision. DY, ZHZ, NT, and SL provided literature input and suggestions. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by the 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (No. 2020LKSFG01E), National Natural Science Foundation of China (Nos. 31872596 & 32073008), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. GML2019ZD0606) and Shantou University Scientific Research Foundation for Talents (No. NTF19005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Shrimp Disease and Immunology Laboratory for the exchange of ideas and for providing constructive critique of the manuscript.
References
1. Lightner DV. Virus Diseases of Farmed Shrimp in the Western Hemisphere (the Americas): A Review. J Invertebr Pathol (2011) 106:110–30. doi: 10.1016/j.jip.2010.09.012
2. Tassanakajon A, Somboonwiwat K, Supungul P, Tang S. Discovery of Immune Molecules and Their Crucial Functions in Shrimp Immunity. Fish Shellfish Immunol (2013) 34:954–67. doi: 10.1016/j.fsi.2012.09.021
3. Li F, Xiang J. Recent Advances in Researches on the Innate Immunity of Shrimp in China. Dev Comp Immunol (2013) 39:11–26. doi: 10.1016/j.dci.2012.03.016
4. Iwanaga S, Lee BL. Recent Advances in the Innate Immunity of Invertebrate Animals. J Biochem Mol Biol (2005) 38:128–50. doi: 10.5483/bmbrep.2005.38.2.128
5. Kurata S, Ariki S, Kawabata S-I. Recognition of Pathogens and Activation of Immune Responses in Drosophila and Horseshoe Crab Innate Immunity. Immunobiology (2006) 211:237–49. doi: 10.1016/j.imbio.2005.10.016
6. Stuart LM, Ezekowitz RA. Phagocytosis and Comparative Innate Immunity: Learning on the Fly. Nat Rev Immunol (2008) 8:131–41. doi: 10.1038/nri2240
7. Wang P, Huang T, Zhang X, He J. Antiviral Defense in Shrimp: From Innate Immunity to Viral Infection. Antiviral Res (2014) 108:129–41. doi: 10.1016/j.antiviral.2014.05.013
8. Wang W, Zhang X. Comparison of Antiviral Efficiency of Immune Responses in Shrimp. Fish Shellfish Immunol (2008) 25:522–7. doi: 10.1016/j.fsi.2008.07.016
9. Marques MRF, Barracco MA. Lectins, as Non-Self-Recognition Factors, in Crustaceans. Aquaculture (2000) 191:23–44. doi: 10.1016/S0044-8486(00)00417-8
10. Wang X, Xu W, Zhang X, Zhao X, Yu X, Wang J. A C-Type Lectin is Involved in the Innate Immune Response of Chinese White Shrimp. Fish Shellfish Immunol (2009) 27:556–62. doi: 10.1016/j.fsi.2009.07.011
11. Tassanakajon A, Amparyup P, Somboonwiwat K, Supungul P. Cationic Antimicrobial Peptides in Penaeid Shrimp. Mar Biotechnol (2011) 13:639–57. doi: 10.1007/s10126-011-9381-8
12. Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz R. Phylogenetic Perspectives in Innate Immunity. Science (1999) 284:1313–7. doi: 10.1126/science.284.5418.1313
13. Li C, Wang S, He J. The Two Nf-κb Pathways Regulating Bacterial and WSSV Infection of Shrimp. Front Immunol (2019) 10:1785. doi: 10.3389/fimmu.2019.01785
14. Sun JJ, Lan JF, Zhao XF, Vasta GR, Wang JX, Schneider DS. Binding of a C-Type Lectin’s Coiled-Coil Domain to the Domeless Receptor Directly Activates the JAK/STAT Pathway in the Shrimp Immune Response to Bacterial Infection. PloS Pathog (2017) 13:e1006626. doi: 10.1371/journal.ppat.1006626
15. Feng N, Wang D, Wen R, Li F. Functional Analysis on Immune Deficiency (Imd) Homolog Gene in Chinese Shrimp Fenneropenaeus Chinensis. Mol Biol Rep (2014) 41:1437–44. doi: 10.1007/s11033-013-2988-2
16. Li H, Yin B, Wang S, Fu Q, Xiao B, Lǚ K, et al. Rnai Screening Identifies a New Toll From Shrimp Litopenaeus Vannamei That Restricts WSSV Infection Through Activating Dorsal to Induce Antimicrobial Peptides. PloS Pathog (2018) 14:e1007109. doi: 10.1371/journal.ppat.1007109
17. Liu J, Qian C, Cao X. Post-Translational Modification Control of Innate Immunity. Immunity (2016) 45:15–30. doi: 10.1016/j.immuni.2016.06.020
18. Cameron AM, Lawless SJ, Pearce EJ. Metabolism and Acetylation in Innate Immune Cell Function and Fate. Semin Immunol (2016) 28:408–16. doi: 10.1016/j.smim.2016.10.003
19. Gong T, Jiang W, Zhou R. Control of Inflammasome Activation by Phosphorylation. Trends Biochem Sci (2018) 43:685–99. doi: 10.1016/j.tibs.2018.06.008
20. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in Health and Disease. Nat Rev Nephrol (2019) 15:346–66. doi: 10.1038/s41581-019-0129-4
21. Zhou Y, He C, Yan D, Liu F, Liu H, Chen J, et al. The Kinase Ck1ε Controls the Antiviral Immune Response by Phosphorylating the Signaling Adaptor TRAF3. Nat Immunol (2016) 17:397–405. doi: 10.1038/ni.3395
22. Lu H, Ruan L, Xu X. An Immediate-Early Protein of White Spot Syndrome Virus Modulates the Phosphorylation of Focal Adhesion Kinase of Shrimp. Virology (2011) 419:84–9. doi: 10.1016/j.virol.2011.07.021
23. Soowannayan C, Cowley JA, Pearson RD, Wallis TP, Gorman JJ, Michalski WP, et al. Glycosylation of gp116 and Gp64 Envelope Proteins of Yellow Head Virus of Penaeus Monodon Shrimp. J Gen Virol (2010) 91:2463–73. doi: 10.1099/vir.0.022111-0
24. Soowannayan C, Chanarpakorn N, Phanthura M, Deekhlai N, Kunasol C, Sriurairatana S. N-Linked Glycosylation Is Essential for the Yellow Head Virus Replication Cycle. J Gen Virol (2013) 94:2458–68. doi: 10.1099/vir.0.054379-0
25. Zhang Z, Wang F, Chen C, Zheng Z, Aweya JJ, Zhang Y. Glycosylation of Hemocyanin in Litopenaeus Vannamei Is an Antibacterial Response Feature. Immunol Lett (2017) 192:42–7. doi: 10.1016/j.imlet.2017.10.008
26. Muñoz-Fontela C, González D, Marcos-Villar L, Campagna M, Gallego P, González-Santamaría J, et al. Acetylation Is Indispensable for P53 Antiviral Activity. Cell Cycle (2011) 10:3701–5. doi: 10.4161/cc.10.21.17899
27. Huai W, Liu X, Wang C, Zhang Y, Chen X, Chen X, et al. KAT8 Selectively Inhibits Antiviral Immunity by Acetylating IRF3. J Exp Med (2019) 216:772–85. doi: 10.1084/jem.20181773
28. Adorisio S, Fierabracci A, Muscari I, Liberati AM, Ayroldi E, Migliorati G, et al. Sumo Proteins: Guardians of Immune System. J Autoimmun (2017) 84:21–8. doi: 10.1016/j.jaut.2017.09.001
29. Koltun B, Shackelford E, Bonnay F, Matt N, Reichhart JM, Orian A. The SUMO-targeted Ubiquitin Ligase, Dgrn, Is Essential for Drosophila Innate Immunity. Int J Dev Biol (2017) 61:319–27. doi: 10.1387/ijdb.160250ao
30. Jiang X, Chen ZJ. The Role of Ubiquitylation in Immune Defence and Pathogen Evasion. Nat Rev Immunol (2012) 12:35–48. doi: 10.1038/nri3111
31. Zhou R, Silverman N, Hong M, Liao DS, Chung Y, Chen ZJ, et al. The Role of Ubiquitination in Drosophila Innate Immunity. J Biol Chem (2005) 280:34048–55. doi: 10.1074/jbc.M506655200
32. Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, et al. Trim25 RING-finger E3 Ubiquitin Ligase Is Essential for RIG-I-Mediated Antiviral Activity. Nature (2007) 446:916–20. doi: 10.1038/nature05732
33. Wang Y, Tong X, Ye X. Ndfip1 Negatively Regulates RIG-I–dependent Immune Signaling by Enhancing E3 Ligase Smurf1-Mediated MAVS Degradation. J Immunol (2012) 189:5304–13. doi: 10.4049/jimmunol.1201445
34. Kuniyoshi K, Takeuchi O, Pandey S, Satoh T, Iwasaki H, Akira S, et al. Pivotal Role of RNA-binding E3 Ubiquitin Ligase MEX3C in RIG-I–mediated Antiviral Innate Immunity. Proc Natl Acad Sci USA (2014) 111:5646–51. doi: 10.1073/pnas.1401674111
35. van der Aa LM, Jouneau L, Laplantine E, Bouchez O, Van Kemenade L, Boudinot P. Fintrims, Fish Virus-Inducible Proteins With E3 Ubiquitin Ligase Activity. Dev Comp Immunol (2012) 36:433–41. doi: 10.1016/j.dci.2011.08.010
36. Chen AJ, Gao L, Wang XW, Zhao XF, Wang JX. Sumo-Conjugating Enzyme E2 UBC9 Mediates Viral Immediate-Early Protein Sumoylation in Crayfish to Facilitate Reproduction of White Spot Syndrome Virus. J Virol (2012) 87:636–47. doi: 10.1128/jvi.01671-12
37. Yang L, Ruan Z, Li X, Li L, Wang Q, Li W. Nedd8-Conjugated Cullin4 Positive Regulates Antimicrobial Peptides Expression in Eriocheir Sinensis. Fish Shellfish Immunol (2019) 84:1041–9. doi: 10.1016/j.fsi.2018.10.077
38. Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural Insights Into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell (2008) 134:995–1006. doi: 10.1016/j.cell.2008.07.022
39. Ciechanover A. The Unravelling of the Ubiquitin System. Nat Rev Mol Cell Biol (2015) 16:322–4. doi: 10.1038/nrm3982
40. Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci (2017) 42:873–86. doi: 10.1016/j.tibs.2017.09.002
41. Nakayama KI, Nakayama K. Ubiquitin Ligases: Cell-Cycle Control and Cancer. Nat Rev Cancer (2006) 6:369–81. doi: 10.1038/nrc1881
42. Reinstein E, Ciechanover A. Narrative Review: Protein Degradation and Human Diseases: The Ubiquitin Connection. Ann Intern Med (2006) 145:676–84. doi: 10.7326/0003-4819-145-9-200611070-00010
43. Naujokat C, Šarić T. Concise Review: Role and Function of the Ubiquitin-Proteasome System in Mammalian Stem and Progenitor Cells. Stem Cells (2007) 25:2408–18. doi: 10.1634/stemcells.2007-0255
44. Lavie J, De Belvalet H, Sonon S, Ion AM, Dumon E, Melser S, et al. Ubiquitin-Dependent Degradation of Mitochondrial Proteins Regulates Energy Metabolism. Cell Rep (2018) 23:2852–63. doi: 10.1016/j.celrep.2018.05.013
45. Park Y, Jin H, Aki D, Lee J, Liu Y. The Ubiquitin System in Immune Regulation. Adv Immunol (2014) 124:17–66. doi: 10.1016/B978-0-12-800147-9.00002-9
46. Zohaib A, Duan X, Zhu B, Ye J, Wan S, Chen H, et al. The Role of Ubiquitination in Regulation of Innate Immune Signaling. Curr Issues Mol Biol (2015) 18:1–10. doi: 10.21775/cimb.018.001
47. Huang Y, Yu Y, Yang Y, Yang M, Zhou L, Huang X, et al. Fish TRIM8 Exerts Antiviral Roles Through Regulation of the Proinflammatory Factors and Interferon Signaling. Fish Shellfish Immunol (2016) 54:435–44. doi: 10.1016/j.fsi.2016.04.138
48. Aalto AL, Mohan AK, Schwintzer L, Kupka S, Kietz C, Walczak H, et al. M1-Linked Ubiquitination by LUBEL Is Required for Inflammatory Response to Oral Infection in. Drosophila Cell Death Differ (2019) 26:860–76. doi: 10.1038/s41418-018-0164-x
49. Kong T, Lin S, Gong Y, Tran NT, Zhang Y, Zheng H, et al. Sp-CBL Inhibits White Spot Syndrome Virus Replication by Enhancing Apoptosis in Mud Crab (Scylla Paramamosain). Dev Comp Immunol (2020) 105:103580. doi: 10.1016/j.dci.2019.103580
50. Niu GJ, Wang S, Xu JD, Yang MC, Sun JJ, He ZH, et al. The Polymeric Immunoglobulin Receptor-Like Protein From Marsupenaeus Japonicus Is a Receptor for White Spot Syndrome Virus Infection. PloS Pathog (2019) 15:e1007558. doi: 10.1371/journal.ppat.1007558
51. Lin SJ, Lee DY, Wang HC, Kang ST, Hwang PP, Kou GH, et al. White Spot Syndrome Virus Protein Kinase 1 Defeats the Host Cell’s Iron-Withholding Defense Mechanism by Interacting With Host Ferritin. J Virol (2015) 89:1083–93. doi: 10.1128/jvi.02318-14
52. Zhang S, Shi L, Lǚ K, Li H, Wang S, He J, et al. Cloning, Identification and Functional Analysis of a β-Catenin Homologue From Pacific White Shrimp, Litopenaeus Vannamei. Fish Shellfish Immunol (2016) 54:411–8. doi: 10.1016/j.fsi.2016.03.162
53. Wen R, Li F, Li S, Xiang J. Function of Shrimp Stat During Wssv Infection. Fish Shellfish Immunol (2014) 38:354–60. doi: 10.1016/j.fsi.2014.04.002
54. Zhu G, Li S, Wu J, Li F, Zhao XM. Identification of Functional Gene Modules Associated With STAT-Mediated Antiviral Responses to White Spot Syndrome Virus in Shrimp. Front Physiol (2019) 10:212. doi: 10.3389/fphys.2019.00212
55. Chen L, Paquette N, Mamoor S, Rus F, Nandy A, Leszyk J, et al. Innate Immune Signaling in Drosophila is Regulated by Transforming Growth Factor β (Tgfβ)-Activated Kinase (Tak1)-Triggered Ubiquitin Editing. J Biol Chem (2017) 292:8738–49. doi: 10.1074/jbc.M117.788158
56. Jia X, Zhou H, Wu C, Wu Q, Ma S, Wei C, et al. The Ubiquitin Ligase RNF125 Targets Innate Immune Adaptor Protein TRIM14 for Ubiquitination and Degradation. J Immunol (2017) 198:4652–8. doi: 10.4049/jimmunol.1601322
57. Fugmann SD, Xue S, Liu Y, Zhang Y, Sun Y, Geng X, et al. Sequencing and De Novo Analysis of the Hemocytes Transcriptome in Litopenaeus Vannamei Response to White Spot Syndrome Virus Infection. PloS One (2013) 8:e76718. doi: 10.1371/journal.pone.0076718
58. Peruzza L, Shekhar MS, Kumar KV, Swathi A, Karthic K, Hauton C, et al. Temporal Changes in Transcriptome Profile Provide Insights of White Spot Syndrome Virus Infection in Litopenaeus Vannamei. Sci Rep (2019) 9:13509. doi: 10.1038/s41598-019-49836-0
59. Chen A, Wang S, Zhao X, Yu X, Wang J. Enzyme E2 From Chinese White Shrimp Inhibits Replication of White Spot Syndrome Virus and Ubiquitinates Its RING Domain Proteins. J Virol (2011) 85:8069–79. doi: 10.1128/JVI.00487-11
60. Chen X, Zeng D, Chen X, Xie D, Zhao Y, Yang C, et al. Transcriptome Analysis of Litopenaeus Vannamei in Response to White Spot Syndrome Virus Infection. PloS One (2013) 8:e73218. doi: 10.1371/journal.pone.0073218
61. Sun JZ, Ruan LW, Zhou CZ, Shi H, Xu X. Characterization and Function of a β-Catenin Homolog From Litopenaeus Vannamei in WSSV Infection. Dev Comp Immunol (2017) 76:412–9. doi: 10.1016/j.dci.2017.07.003
62. Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin Is a Target for the Ubiquitin-Proteasome Pathway. EMBO J (1997) 16:3797–804. doi: 10.1093/emboj/16.13.3797
63. Hillesheim A, Nordhoff C, Boergeling Y, Ludwig S, Wixler V. β-Catenin Promotes the Type I IFN Synthesis and the IFN-Dependent Signaling Response But Is Suppressed by Influenza a Virus-Induced Rig-I/Nf-κb Signaling. Cell Commun Signal (2014) 12:29. doi: 10.1186/1478-811X-12-29
64. Sun M, Li S, Yu K, Xiang J, Li F. An E3 Ubiquitin Ligase TRIM9 is Involved in WSSV Infection Via Interaction With β-Trcp. Dev Comp Immunol (2019) 97:57–63. doi: 10.1016/j.dci.2019.03.014
65. Spaan W, Cavanagh D, Horzinek MC. Coronaviruses: Structure and Genome Expression. J Gen Virol (1988) 69:2939–52. doi: 10.1099/0022-1317-69-12-2939
66. Klasse PJ, Bron R, Marsh M. Mechanisms of Enveloped Virus Entry Into Animal Cells. Adv Drug Delivery Rev (1998) 34:65–91. doi: 10.1016/S0169-409X(98)00002-7
67. Shekhar MS, Ponniah AG. Recent Insights Into Host-Pathogen Interaction in White Spot Syndrome Virus Infected Penaeid Shrimp. J Fish Dis (2015) 38:599–612. doi: 10.1111/jfd.12279
68. Liu X, Yang B, Huang S, Wu C, Li X, Cheng S, et al. Human Cytomegalovirus Ie1 Downregulates Hes1 in Neural Progenitor Cells as a Potential E3 Ubiquitin Ligase. PloS Pathog (2017) 13:e1006542. doi: 10.1371/journal.ppat.1006542
69. Li X, Liu X, Yang B, Fu Y, Zhao F, Shen Z, et al. Human Cytomegalovirus Infection Dysregulates the Localization and Stability of NCD1 and Jag1 in Neural Progenitor Cells. J Virol (2015) 89:6792–804. doi: 10.1128/jvi.00351-15
70. Freemont PS. Ubiquitination: RING for Destruction? Curr Biol (2000) 10:R84–7. doi: 10.1016/S0960-9822(00)00287-6
71. Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional Interactions Between Ubiquitin E2 Enzymes and TRIM Proteins. Biochem J (2011) 434:309–19. doi: 10.1042/BJ20101487
72. Yu Y, Huang X, Liu J, Zhang J, Hu Y, Yang Y, et al. Fish TRIM32 Functions as a Critical Antiviral Molecule Against Iridovirus and Nodavirus. Fish Shellfish Immunol (2017) 60:33–43. doi: 10.1016/j.fsi.2016.11.036
73. Yang F, He J, Lin X, Li Q, Pan D, Zhang X, et al. Complete Genome Sequence of the Shrimp White Spot Bacilliform Virus. J Virol (2001) 75:11811–20. doi: 10.1128/JVI.75.23.11811-11820.2001
74. He F, Fenner BJ, Godwin AK, Kwang J. White Spot Syndrome Virus Open Reading Frame 222 Encodes a Viral E3 Ligase and Mediates Degradation of a Host Tumor Suppressor Via Ubiquitination. J Virol (2006) 80:3884–92. doi: 10.1128/JVI.80.8.3884-3892.2006
75. He F, Syed SM, Hameed ASS, Kwang J. Viral Ubiquitin Ligase WSSV222 is Required for Efficient White Spot Syndrome Virus Replication in Shrimp. J Gen Virol (2009) 90:1483–90. doi: 10.1099/vir.0.008912-0
76. He F, Kwang J. Identification and Characterization of a New E3 Ubiquitin Ligase in White Spot Syndrome Virus Involved in Virus Latency. Virol J (2008) 5:151. doi: 10.1186/1743-422X-5-151
77. Seo JK, Lee MJ, Go HJ, Kim GD, Jeong HD, Nam BH, et al. Purification and Antimicrobial Function of Ubiquitin Isolated From the Gill of Pacific Oyster, Crassostrea Gigas. Mol Immunol (2013) 53:88–98. doi: 10.1016/j.molimm.2012.07.003
78. Rytkönen A, Holden DW. Bacterial Interference of Ubiquitination and Deubiquitination. Cell Host Microbe (2007) 1:13–22. doi: 10.1016/j.chom.2007.02.003
79. Pei G, Buijze H, Liu H, Moura-Alves P, Goosmann C, Brinkmann V, et al. The E3 Ubiquitin Ligase Nedd4 Enhances Killing of Membrane-Perturbing Intracellular Bacteria by Promoting Autophagy. Autophagy (2017) 13:2041–55. doi: 10.1080/15548627.2017.1376160
80. Angot A, Vergunst A, Genin S, Peeters N. Exploitation of Eukaryotic Ubiquitin Signaling Pathways by Effectors Translocated by Bacterial Type III and Type Iv Secretion Systems. PloS Pathog (2007) 3:e3. doi: 10.1371/journal.ppat.0030003
81. Sheng X, You Q, Zhu H, Chang Z, Li Q, Wang H, et al. Bacterial Effector Nlel Promotes Enterohemorrhagic E. Coli-Induced Attaching and Effacing Lesions by Ubiquitylating and Inactivating Jnk. PloS Pathog (2017) 13:e1006534–. doi: 10.1371/journal.ppat.1006534
82. Zhang Y, Gao G, Lin R, Aweya JJ, Tao M, Wang F. Transcriptome Analyses Reveal Litopenaeus Vannamei Hemocytes Response to Lipopolysaccharide. Fish Shellfish Immunol (2018) 76:187–95. doi: 10.1016/j.fsi.2018.03.002
83. Zheng Z, Wang F, Aweya JJ, Li R, Yao D, Zhong M, et al. Comparative Transcriptomic Analysis of Shrimp Hemocytes in Response to Acute Hepatopancreas Necrosis Disease (Ahpnd) Causing Vibrio Parahemolyticus Infection. Fish Shellfish Immunol (2018) 74:10–8. doi: 10.1016/j.fsi.2017.12.032
84. Xie C, Chen Y, Sun W, Ding J, Zhou L, Wang S, et al. Transcriptome and Expression Profiling Analysis of the Hemocytes Reveals a Large Number of Immune-Related Genes in Mud Crab Scylla Paramamosain During Vibrio Parahaemolyticus Infection. PloS One (2014) 9:e114500. doi: 10.1371/journal.pone.0114500
85. Li C, Chai J, Li H, Zuo H, Wang S, Qiu W, et al. Pellino Protein From Pacific White Shrimp Litopenaeus Vannamei Positively Regulates Nf-κb Activation. Dev Comp Immunol (2014) 44:341–50. doi: 10.1016/j.dci.2014.01.012
86. Schauvliege R, Janssens S, Beyaert R. Pellino Proteins are More Than Scaffold Proteins in TLR/IL-1R Signalling: A Role as Novel Ring E3-Ubiquitin-Ligases. FEBS Lett (2006) 580:4697–702. doi: 10.1016/j.febslet.2006.07.046
87. Schauvliege R, Janssens S, Beyaert R. Pellino Proteins: Novel Players in TLR and IL-1R Signalling. J Cell Mol Med (2007) 11:453–61. doi: 10.1111/j.1582-4934.2007.00040.x
88. Murphy M, Xiong Y, Pattabiraman G, Qiu F, Medvedev AE. Pellino-1 Positively Regulatestoll-Like Receptor (TLR) 2 and TLR4 Signaling and Is Suppressed Upon Induction of Endotoxin Tolerance. J Biol Chem (2015) 290:19218–32. doi: 10.1074/jbc.M115.640128
89. Ji S, Sun M, Zheng X, Li L, Sun L, Chen D, et al. Cell-Surface Localization of Pellino Antagonizes Toll-mediated Innate Immune Signalling by Controlling Myd88 Turnover in. Drosophila Nat Commun (2014) 5:3458. doi: 10.1038/ncomms4458
90. Deng H, Hu L, Li J, Yan W, Song E, Kuang M, et al. The NF-κb Family Member Dorsal Plays a Role in Immune Response Against Gram-Positive Bacterial Infection in Mud Crab (Scylla Paramamosain). Dev Comp Immunol (2020) 106:103581. doi: 10.1016/j.dci.2019.103581
91. Maculins T, Fiskin E, Bhogaraju S, Dikic I. Bacteria-Host Relationship: Ubiquitin Ligases as Weapons of Invasion. Cell Res (2016) 26:499–510. doi: 10.1038/cr.2016.30
92. Zheng Z, Wei C, Guan K, Yuan Y, Zhang Y, Ma S, et al. Bacterial E3 Ubiquitin Ligase IpaH4.5 of Shigella Flexneri Targets TBK1 to Dampen the Host Antibacterial Response. J Immunol (2016) 196:1199–208. doi: 10.4049/jimmunol.1501045
93. Ou CY, Lin YF, Chen YJ, Chien CT. Distinct Protein Degradation Mechanisms Mediated by Cul1 and Cul3 Controlling Ci Stability in Drosophila Eye Development. Genes Dev (2002) 16:2403–14. doi: 10.1101/gad.1011402
94. Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, et al. Glutamine Deamidation and Dysfunction of Ubiquitin/NEDD8 Induced by a Bacterial Effector Family. Science (2010) 329:1215–8. doi: 10.1126/science.1193844
95. Joseph SJ, Fernandez-Robledo JA, Gardner MJ, El-Sayed NM, Kuo CH, Schott EJ, et al. The Alveolate Perkinsus Marinus: Biological Insights From EST Gene Discovery. BMC Genomics (2010) 11:228. doi: 10.1186/1471-2164-11-228
96. Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TL, Lopez-Moyado IF, Rifkin SA, et al. Ubiquitin-Mediated Response to Microsporidia and Virus Infection in C. Elegans. PloS Pathog (2014) 10:e1004200. doi: 10.1371/journal.ppat.1004200
97. Luo Z, Huang W, Wang G, Sun H, Chen X, Luo P, et al. Identification and Characterization of p38MAPK in Response to Acute Cold Stress in the Gill of Pacific White Shrimp (Litopenaeus Vannamei). Aquacult Rep (2020) 17:100365. doi: 10.1016/j.aqrep.2020.100365
98. Chen K, Li E, Li T, Xu C, Wang X, Lin H, et al. Transcriptome and Molecular Pathway Analysis of the Hepatopancreas in the Pacific White Shrimp Litopenaeus Vannamei Under Chronic Low-Salinity Stress. PloS One (2015) 10:e0131503. doi: 10.1371/journal.pone.0131503
99. Shekhar MS, Kiruthika J, Ponniah AG. Identification and Expression Analysis of Differentially Expressed Genes From Shrimp (Penaeus Monodon) in Response to Low Salinity Stress. Fish Shellfish Immunol (2013) 35:1957–68. doi: 10.1016/j.fsi.2013.09.038
100. Xu C, Li E, Liu Y, Wang X, Qin JG, Chen L. Comparative Proteome Analysis of the Hepatopancreas From the Pacific White Shrimp Litopenaeus Vannamei Under Long-Term Low Salinity Stress. J Proteomics (2017) 162:1–10. doi: 10.1016/j.jprot.2017.04.013
101. Turner JW, Good B, Cole D, Lipp EK. Plankton Composition and Environmental Factors Contribute to Vibrio Seasonality. ISME J (2009) 3:1082–92. doi: 10.1038/ismej.2009.50
102. Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J. Environmental Occurrence and Clinical Impact of Vibrio Vulnificus and Vibrio Parahaemolyticus: A European Perspective. Env Microbiol Rep (2010) 2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x
103. Wang LU, Chen JC. The Immune Response of White Shrimp Litopenaeus Vannamei and its Susceptibility to Vibrio Alginolyticus At Different Salinity Levels. Fish Shellfish Immunol (2005) 18:269–78. doi: 10.1016/j.fsi.2004.07.008
104. Keezhedath J, Kurcheti PP, Pathan MK, Babu GP, Tripathi GT, Sudhagar A, et al. Expression Profile of Penaeus Monodon Ubiquitin Conjugating Enzyme (Pmubc) At Protein Level in White Spot Syndrome Virus Challenged Shrimp. Indian J Virol (2013) 24:48–53. doi: 10.1007/s13337-013-0131-6
105. Wang Z, Chua H, Gusti AARA, He F, Fenner B, Manopo I, et al. Ring-H2 Protein Wssv249 From White Spot Syndrome Virus Sequesters a Shrimp Ubiquitin-Conjugating Enzyme, PvUbc, for Viral Pathogenesis. J Virol (2005) 79:8764–72. doi: 10.1128/JVI.79.14.8764-8772.2005
106. Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortiz ZG, et al. Ubiquitin Ligase Trim62 Regulates CARD9-Mediated Anti-Fungal Immunity and Intestinal Inflammation. Immunity (2015) 43:715–26. doi: 10.1016/j.immuni.2015.10.005
107. Prasad R, Atul, Kolla VK, Legac J, Singhal N, Navale R, et al. Blocking Plasmodium Falciparum Development Via Dual Inhibition of Hemoglobin Degradation and the Ubiquitin Proteasome System by MG132. PloS One (2013) 8:e73530. doi: 10.1371/journal.pone.0073530
Keywords: ubiquitination, penaeid shrimp, pathogens, immune response, posttranslational modification
Citation: Zhang Z, Aweya JJ, Yao D, Zheng Z, Tran NT, Li S and Zhang Y (2021) Ubiquitination as an Important Host-Immune Response Strategy in Penaeid Shrimp: Inferences From Other Species. Front. Immunol. 12:697397. doi: 10.3389/fimmu.2021.697397
Received: 19 April 2021; Accepted: 13 May 2021;
Published: 27 May 2021.
Edited by:
Stephanie DeWitte-Orr, Wilfrid Laurier University, CanadaCopyright © 2021 Zhang, Aweya, Yao, Zheng, Tran, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueling Zhang, emhhbmd5bEBzdHUuZWR1LmNu; Jude Juventus Aweya, amphd2V5YUBzdHUuZWR1LmNu
†These authors have contributed equally to this work
 Zhaoxue Zhang1,2†
Zhaoxue Zhang1,2† Jude Juventus Aweya
Jude Juventus Aweya Defu Yao
Defu Yao Ngoc Tuan Tran
Ngoc Tuan Tran Shengkang Li
Shengkang Li Yueling Zhang
Yueling Zhang