
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 30 August 2021
Sec. Cancer Immunity and Immunotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.697026
 Chao Quan1†
Chao Quan1† Othmane Belaydi1†
Othmane Belaydi1† Jiao Hu1
Jiao Hu1 Huihuang Li1
Huihuang Li1 Anze Yu1,2
Anze Yu1,2 Peihua Liu1
Peihua Liu1 Zhenglin Yi1
Zhenglin Yi1 Dongxu Qiu1
Dongxu Qiu1 Wenbiao Ren3
Wenbiao Ren3 Hongzhi Ma4
Hongzhi Ma4 Guanghui Gong5
Guanghui Gong5 Zhenyu Ou1
Zhenyu Ou1 Minfeng Chen1
Minfeng Chen1 Yin Sun3
Yin Sun3 Jinbo Chen1*
Jinbo Chen1* Xiongbing Zu1*
Xiongbing Zu1*N6-methylation of adenosine (m6A), a post-transcriptional regulatory mechanism, is the most abundant nucleotide modification in almost all types of RNAs. The biological function of m6A in regulating the expression of oncogenes or tumor suppressor genes has been widely investigated in various cancers. However, recent studies have addressed a new role of m6A modification in the anti-tumor immune response. By modulating the fate of targeted RNA, m6A affects tumor-associated immune cell activation and infiltration in the tumor microenvironment (TME). In addition, m6A-targeting is found to affect the efficacy of classical immunotherapy, which makes m6A a potential target for immunotherapy. Although m6A modification together with its regulators may play the exact opposite role in different tumor types, targeting m6A regulators has been shown to have wide implications in several cancers. In this review, we discussed the link between m6A modification and tumor with an emphasis on the importance of m6A in anti-tumor immune response and immunotherapy.
The expression of genetic information is regulated at multiple levels. RNA modification, a post-transcriptional regulatory mechanism, is thought to alter the RNA function through changing the chemical-structural features of RNA (1). One hundred and seventy-two kinds of RNA modifications have been reported, most of which are restricted to non-coding RNA (ncRNA) instead of messenger RNA (mRNA) (2, 3). Unlike most other RNA modifications, m6A, which is first identified in the 1970s (4, 5), is an abundant nucleotide modification in almost all types of RNAs, including mRNA and ncRNA, and is involved in various physiological and pathological processes (6). Generally, the rise in m6A methylation in cancer cells plays a tumor-suppressor role with only a couple of studies showing the opposite (7–9). By targeting specific RNA, m6A triggers certain alterations to tumor-specific mRNA/ncRNA behavior and biologic activity, thereby influencing tumor progression.
Compared with the conventional therapies (radiation and chemotherapy) for cancers that directly target tumor cells, anti-tumor immunotherapy, which promotes a systemic immune response against tumors, exhibits great potential in cancer treatment (10). m6A, whose intrinsic function in tumor cells has been widely studied and discussed, is proved to influence the anti-tumor immune response and can be a novel target in anti-tumor immunotherapy (11–14). In this review, we briefly introduce the regulation and function of m6A, with an emphasis on the role of m6A in anti-tumor immune response and a summary of prominent m6A-related functions and strategies in immunotherapy.
The m6A modification, a dynamic and reversible methyl-modification occurring at the N6 position of adenine (A) bases in RNA, is enriched in the internal exons, 3’ untranslated region (3’UTR) and the stop codon and restricted to a consensus sequence DRACH (D=G, A, or U; R=G or A; H=A, C, or U) (15–17). Generally, the regulation and function of m6A modification involve three kinds of protein factors: methyltransferases (writers), binding proteins (readers), and demethylases (erasers).
‘Writers’ mediate m6A modifications by adding methylation to RNA. The classic ‘writer complex’ includes a core heterodimer consisted of methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14), together with several accessory proteins (Wilms tumor 1-associated protein (WTAP), zinc finger CCCH domain-containing protein 13 (ZC3H13), RNA binding motif protein 15 (RBM15) or RBM15B and Vir like m6A methyltransferase associated (VIRMA) (11). METTL3 is responsible for catalyzing methyl-translocation from methyl donor [S-adenosylmethionine (SAM)] to RNA, while METTL14 mediates m6A deposition through recognizing and binding to the target RNA (18, 19). As a regulatory subunit, WTAP interacts with the METTL3/14 heterodimer and mediates the localization of the heterodimer in nuclear speckles (20). ZC3H13, RBM15 or RBM15B, and VIRMA are all WTAP-related and facilitate the nuclear localization and specificity of the ‘writer complex’ (21–23). In addition to the classic ‘writer complex’, several other enzymes independently catalyze the methyl transfer to target RNA, including METTL16 targeting U6 small nuclear RNAs, zinc finger CCHC-type containing 4 (ZCCHC4) targeting ribosomal RNAs, METTL5 targeting 18S ribosomal RNAs and PDX1 C-terminal inhibiting factor 1 (PCIF1) targeting 7-methyl guanosine (m7G)-capped mRNAs (11).
‘Readers’ bind to m6A on modified RNAs and induce various biological effects (24). YT521-B homology (YTH) domain-containing ‘readers’, which bind RNA in an m6A-dependent manner, are categorized into three types: YTHDC1, YTHDC2 and YTHDF family proteins (YTHDFs) (11). YTHDC1 mainly affects mRNA splicing within the nucleus and participates in the nuclear export of mRNA (25, 26), whereas YTHDC2 promotes the translation as well as degradation of the targeted mRNA in/out the nucleus (27–29). There are three members in the YTHDF family: YTHDF 1, 2 and 3. However, their functions remain unclear, and further studies are needed (30). Some other RNA-binding proteins which preferentially bind to m6A-containing RNA are thought to be ‘readers’ too (11). However, more studies are also needed to decipher their regulatory functions and determine whether their preference for m6A-containing RNA is due to specific recognition for m6A (31).
‘Erasers’ are responsible for removing m6A from mRNA. The fat-mass and obesity-associated protein (FTO) and Alpha-ketoglutarate-dependent dioxygenase ALK B homologue 5 (ALKBH5) are two demethylases that proved to remove methylation on adenosine from RNA (17, 32, 33). FTO removes both m6A and N6,2′-O-dimethyladenosine (m6Am), whereas ALKBH5 specifically demethylates m6A (17, 32). Though FTO and ALKBH5 both are ‘erasers’, their biological function can be quite different. They act as either an oncogene or a tumor suppressor depending on tumor types (6, 34). It is clear that the different biological effect of ‘erasers’ is the result of demethylation of specific target mRNAs, but what leads to the mRNA selectivity still need to be addressed.
The regulatory function of m6A on mRNA and ncRNA is very critical in a lot of key biological processes (Figure 1).
m6A is deposited on mRNA under the mediation of writers and then enrolls in the whole lifecycle of mRNA. Mediated by the readers, m6A affects mRNA splicing and export within the nucleus and influences the stability, translation and localization of mRNAs in the cytoplasm. It has been proved that m6A on precursor mRNAs (pre-mRNAs) promotes the mRNA splicing by directly recruiting splicing factor or indirectly altering mRNA structure and subsequently increasing the accessibility of flanking RNA sequence to splicing factors (35–38). Moreover, the m6A writers, readers, and erasers are all associated with mRNA splicing (17, 20, 25, 33, 39). The m6A modification also promotes mRNA export from the nucleus to the cytoplasm, which is mediated by writers or readers (40, 41). Although m6A modification is suggested to accelerate and promote the mRNA translation through several mechanisms, it still shows a negative correlation with mRNA half-life (42–44). This indicates that m6A may enhance mRNA translation by depending more on its readers rather than lengthening mRNA lifetime. A closed-loop model, in which eukaryotic translation initiation factor 3 (eIFs) subunit at the 5’ end of the mRNA interacts with METTL3 or YTHDF1 that is bound to m6A sites near the stop codon, is proposed to explain how m6A regulates mRNA translation (45, 46). YTHDFs, which specifically recognize m6A, are shown to regulate mRNA translation and degradation. YTHDF1 recruits eukaryotic translation initiation factor 3 (eIF-3) complex, which leads to enhanced mRNA translation (45), while YTHDF2 mediates the degradation of mRNA through recruiting Ccr4-Not deadenylase complex (42, 47). YTHDF3’s function is variable, which is linked with either mRNA translation promotion or degradation acceleration (48, 49). However, the insulin-like growth factor-2 mRNA-binding protein (IGF2BP) enhances the mRNA stability by recognizing m6A and facilitate mRNA translation (50–52). METTL3 promotes mRNA translation through directly recruiting eIF3 in the cytoplasm (46, 53). In addition, translational enhancement of m6A is partially mediated by the direct binding of eIF3 to m6A in the 5’ UTR (54).
In addition to mRNA, m6A and its regulators has also been found to affect the biological activity of other ncRNAs (55). METTL3-mediated m6A methylation on primary microRNA (pri-miRNA) are proved to promote the microRNA (miRNA) maturation in various pathophysiological processes (35, 56, 57). Mechanically, METTL3 methylates pri-miRNAs marking them for recognition and processing by DGCR8, which is the first step in the biogenesis of miRNAs (58). METTL3 may also promote the synthesis of mature miRNAs by increasing the splicing of pre-miRNAs by Dicer (59). As for long ncRNA (lncRNA), METTL3-mediated m6A modification on lung cancer associated transcript 3 (LCAT3), a long ncRNA (lncRNA), leads to the lncRNA stabilization (60), while METTL14-mediated m6A modification on lncRNA XIST facilitates the degradation of XIST which is attributable to YTHDF2 (61). In addition, m6A modification on circRNA has been identified and characterized (62). Furthermore, m6A modifications are believed to be involved in the biogenesis, translation, export and endoribonucleolytic cleavage of circRNAs, which has been discussed (63).
The role of intrinsic m6A modifications in modulating tumor fate through regulating specific target genes in different cancers has been thoroughly discussed (6, 62, 64). However, the link between aberrant m6A modifications and the anti-tumor immune response has received less attention, which has been indicated to be a potential therapeutic target in anti-tumor immunotherapy (Figure 2) (13, 65, 66).
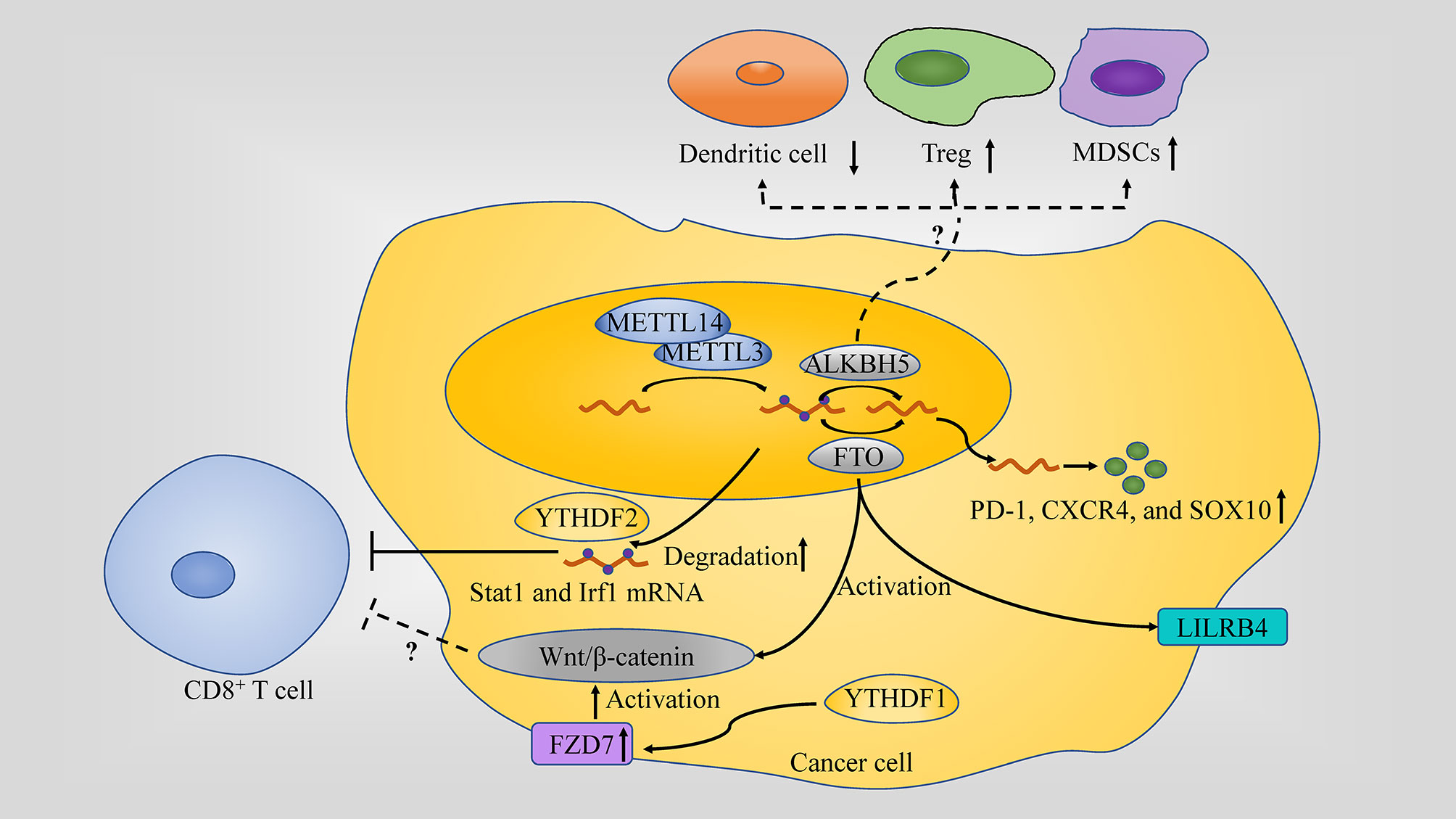
Figure 2 Role of m6A in the anti-tumor immune response. m6A regulators play different role in anti-tumor immune response: (1) METTL3 or METTL14 inhibit IFN-γ-Stat1-Irf1 signaling through decaying the Stat1 and Irf1 mRNA via Ythdf2. (2) FTO directly upregulate leukocyte Ig-like receptor B4 (LILRB4) which is an immune checkpoint gene expression via an m6A-dependent mechanism in AML; Through targeting PD-1, CXCR4, and SOX10FTO in cancer cell, FTO causes the resistance to the killing activity. (3) ALKBH5 affects the tumor infiltration of Tregs, MDSCs and DCs in TME. (4) Wnt/β-catenin hyperactivation through FZD7 overexpression which is mediated by YTHDF1 or by FTO is associated with non-T cell inflamed TME (a lack of CD8+ T cells along with DCs).
The TME plays an important role in cancer progression and is also largely responsible for immunotherapeutic responsiveness. This is highlighted in tumor with T-cell-inflamed TME, containing abundant infiltration of CD8+ T cells (67), and thus predicting a better clinical outcome due to priming for the efficacy of immunotherapy (68), unlike the non-T-cell-inflamed TME. Interestingly, bioinformatical analysis shows that m6A modifications and its regulators are associated with the TME response and immune checkpoint blockade (ICB) efficacy in several tumor types. Moreover, it has been found that m6A modification influence the infiltration, activation and function of immune cell in TME, which makes m6A-targeting a promising therapy (65, 69).
Many studies have confirmed the association between m6A modifications and TME condition in cancers with different histogenesis and organ location.
In renal clear-cell carcinoma (RCC), low m6A score group was characterized by a hot/inflammatory TME, which may be more sensitive to anti-tumor immunotherapy (70). Similar result was found in a RCC cohort that m6A score was negatively correlated with antigen processing machinery (APM) (71). Moreover, low m6A score, characterized by increased mutation burden and activation of immunity, indicates an inflamed TME phenotype and enhanced response to anti-PD-1/L1 immunotherapy in gastric cancer (GC) (65). In addition, m6A-related signature has been identified as biomarker for tumor immune phenotypes and anti-PD-1 immunotherapy treatment response in lung adenocarcinoma (LADC) (72, 73), stomach adenocarcinomas (STADs) (74), Esophageal squamous cell carcinoma (ESCC) (75, 76), Renal Papillary Cell Carcinoma (RPCC), Hepatocellular Carcinoma (HCC) (77, 78). These statistic results together indicate that m6A modification may reflect TME status and predict immunotherapy efficacy in pan-cancers instead of limited to specific cancer types. However, heterogeneity between these analysis limits the application in clinical practice, since each study focused on single cancer type and included different m6A indicator for analysis.
Considering the potential effect of m6A on TME, it is promising to remolding TME through enhancing or reducing m6A modification by targeting m6A regulator.
METTL14, along with METTL3, affects the number of CD8+ T cells infiltrating into tumors since its depletion led to an increase of CD8+ T cells, in addition to as well as IFN‐γ, CXCL9 and CXCL10 in colorectal cancer (CRC) (13). Moreover, depletion of Mettl3 and Mettl14 enhanced the response to anti-PD-1 treatment in pMMR-MSI-L CRC and melanoma (13).
FTO, prompted by metabolic stress and starvation through the NF-κB signaling pathway and autophagy, increases melanoma growth and decreases response to anti-PD-1 blockade immunotherapy by protecting tumor cell-intrinsic genes (PD-1 (PDCD1), CXCR4 and SOX10) mRNA from decaying. In addition, in the case of endometrial cancer, FTO is also found to be changing the TME through regulating matrix metalloproteinase (MMPs), which are key player enzymes in promoting cancer growth (79, 80). Moreover, FTO can also directly upregulate leukocyte Ig-like receptor B4 (LILRB4) which is an immune checkpoint gene expression via an m6A-dependent mechanism in AML. CS1/CS2 (FTO inhibitor) treatment decreased the expression of LILRB4 in AML cells and substantially increased the sensitivity of AML cells to the cytotoxicity of activated T cells (81).
ALKBH5 alteration in tumor cell modulating its lactate content affects the tumor infiltration of Tregs and myeloid-derived suppressor cells (MDSCs) in TME, which are notorious immunosuppressants in anti-tumor immunity. ALKBH5 also regulates Mct4/Slc16a3 expression and DCs abundance. These regulations were highlighted when knocking down ALKBH5 along with administering GVAX/anti–PD-1 treatment resulted in Tregs and MDSCs infiltration of tumors to go down, DCs to increase and Mct4/Slc16a3 expression and lactate content in TME to decrease. ALKBH5, unlike FTO, is not changing the TME by affecting the IFN-γ pathway since the population of CD8+ T lymphocytes remains unchanged when the demethylase is depleted (82). In addition, single-cell mass cytometry analysis unveiled a role of ALKBH5 in TME by promoting the expression of PD-L1 on monocytes/macrophages and decreasing the infiltration of MDSCs (83).
In addition, Wnt/β-catenin pathway has been linked to m6A modifications and cancer in many studies. For example, Wnt/β-catenin signaling, which is, at least partially, overexpressed due to frizzled class receptor (FZD) 10’s m6A modification, contributes to epithelial ovarian cancers (EOC) resistance to poly ADP-ribose polymerase inhibitor (PARPi) (84). Wnt/β-catenin hyperactivation through FZD7 overexpression, a Wnt receptor, which has its elevation mediated by YTHDF1, leads to GC carcinogenesis (85) and Wnt pathway activation by FTO promotes endometrial cancer (86). Activation of the Wnt/β-catenin pathway is considered as a potential cause in developing the non-T cell inflamed TME since it is usually associated with poor T cell infiltration (a lack of CD8+ T cells along with DCs) among most human cancers. This activation also renders the tumors resistant to immune checkpoint therapy, in addition to resistance to vaccination and adoptive T cell transfer in the case of β-catenin-expressing melanomas. Therefore, targeting the Wnt/β-catenin pathway is a high priority for reversing non-T cell inflamed TME and sensitizing patients to different types of therapy (68).
As an important part of anti-tumor response, tumor infiltrating lymphocytes (TILs) and other immune cells is vital for shaping the TME and anti-tumor immunotherapy. While few published research on the effect of m6A on immune cell function is available (Figure 3).
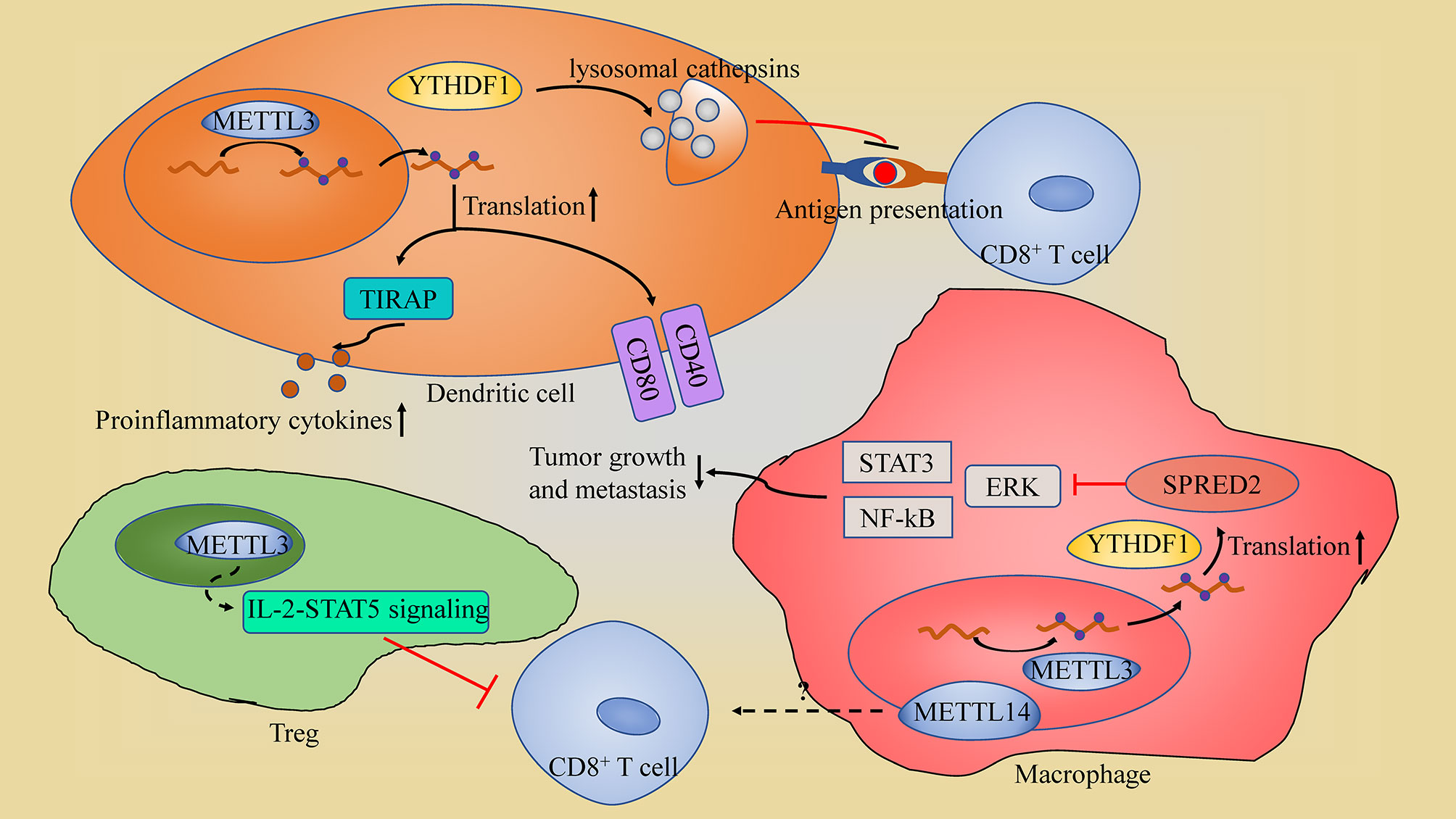
Figure 3 The schematic of m6A in immune cell. (1) In dendritic cells (DCs), YTHDF1 suppresses the antigen-presentation by DCs through maintain the expression of lysosomal cathepsins; METTL3-mediated m6A modification promotes DCs activation through enhancing the translation efficiency of CD40, CD80 and TIRAP. (2) METTL3 in Tregs sustains the suppression roles of regulatory T cells through IL-2-STAT5 signaling pathway impairing CD8+ T cells tumor killing ability. (3) In macrophages, METTL3 sustains the YTHDF1-mediated translation of SPRED2, which inhibits NF-kB and STAT3 through the ERK pathway, restricting tumor growth and metastasis; METTL14-deficiency in macrophage impairs CD8+ T cells to eliminate tumors.
METTL3-mediated m6A modification is found to be promoting DCs activation and maturation, leading to the induction of T cell activation. This promotion is through METTL3 enhancement of the translation efficiency of CD40, CD80 and TIR domain containing adaptor protein (TIRAP: a crucial TLR4/NF-κB signaling pathway adaptor, which induces proinflammatory cytokines secretion) (87). METTL3 depletion in macrophages contributed to the formation of an immunosuppressive microenvironment with increased Treg infiltration and reduced Th1 cells and IFN-γ+CD8+ cells, and subsequently facilitated tumor growth and metastasis (88). Moreover, METTL3-deficiency in macrophages impairs the YTHDF1-mediated translation of SPRED2, which enhances the activation of NF-kB and STAT3 through the ERK pathway, leading to increased tumor growth and metastasis (88). However, METTL3 also impairs CD8+ T cells tumor killing ability through sustaining the suppression roles of regulatory T cells (Tregs) (89). Depletion of Mettl3 in Tregs suppresses the IL-2-STAT5 signaling pathway which is essential to Treg functions and stability (89). Thus, METTL3 in Tregs may suppress the anti-tumor immune response by maintaining the immunosuppressive function of Tregs. METTL14-mediated m6A modification in tumor-associated macrophages (TAMs) were found to modulate tumor-infiltrating CD8+ T cells, since macrophage-specific knockout of Mettl14 drives CD8+ T cell differentiation along a dysfunctional trajectory, impairing CD8+ T cells to eliminate tumors (90).
YTHDF1 in DCs influences the anti-tumor immune response through impairing DCs’ lysosomal cathepsins. In summary, inhibition of YTHDF1 makes DCs more efficient at antigen-presentation and, at the same time, improves CD8+ T cross-priming; thus, giving them more ability to repress tumors than the non-lacking YTHDF1 cells (12). Moveover, YTHDF1 inhibition also increases PD-L1 checkpoint blockade potency in cancer regression (12).
To date, role of m6A modification in immune cells was given little attention and most studies was conducted in tumor model, with a few exceptions focus on other pathophysiological process, including autoimmune disease (91), T cell homeostasis and differentiation (92) and macrophage polarization (93). In addition, one single regulator may play the opposite role on immune response according to cell type, like METTL3 in DCs and Treg. Due to the inadequate data and confusing result, it’s hard to develop a strategy to promote anti-tumor immunity by targeting m6A in immune cell for now.
As discussed above, m6A modifications and its regulators affect tumor-associated immune cell activation and infiltration in TME. However, only a few m6A regulators has been investigated in several tumor or cell types. Moreover, m6A and its regulators play quite different role in the anti-tumor immune response according to tumor or cell type. Thus, more researches need to be done to identify the effect of aberrant m6A modification and exact biological function of each m6A regulators in different cancers or cell type context, especially in immune cells. In addition, m6A-targeting is found to affect the efficacy of classical immunotherapy preclinically, which further indicates the potential therapeutic value of m6A-targeting in tumor treatment.
Although m6A modification together with its regulators may play the exact opposite role in different types of tumors, targeting m6A regulators has been shown to have wide implications in several cancers. Since there is no clinical research investigating the efficacy of m6A-targeting strategy, we propose several pathways we might utilize to implement m6A area of research in the clinical settings of treatment, which is usually done by trying to selectively identify and then to inhibit the enzymes responsible for tumor exacerbation in specific cancers (Table 1).
METTL3 and METTL14, as discussed earlier, play an oncogenic role and specifically targeting them can lead to better clinical outcomes. Examples of this include: (1) Depletion of the methyltransferases METLL3 in Tregs which enhances the anti-tumor ability of CD8+ T cells can be used as a strategy alongside immunotherapy for effective treatment (89). (2) Suppressing METTL3 and METTl14 increases CD8+ tumor-infiltrating lymphocytes in addition to the upregulation of IFN-γ, CXCL9, and CXCL10. Although this inhibition alone does not repress tumor growth, it is able to synergize with the immune checkpoint inhibitors such as anti-PD-1 therapy in both CRC and melanoma (13). Unfortunately, selective METTL3/METTL14 inhibitors for cancer therapies are not yet available, but bisubstrate inhibitors are promising small-molecules in that they can be developed to be effective METTL3/METTL14 targeting agents (103, 105).
YTHDF1 targeting can be a viable option for cancer therapy, as its elevation promotes cancer growth in different cancers, including GC (85), ovarian cancer (106, 107), CRC (108), and HCC (109). There are different pathways by which YTHDF1 escalates tumor aggressiveness, one of which is through influencing DCs to be less efficient in presenting tumor antigens to T cells. As a result, implementing small molecules or DC vaccines that can suppress YTHDF1 would enhance immunotherapy against cancers (12).
FTO inhibitors can be effective in sensitizing melanoma tumor tissues to anti-PD-1 blockade immunotherapy and at the same time in promoting IFN-γ induced killing, which is also essential for the ICB response (110, 111). In addition, anti-PD-1 immunotherapy effective at the beginning (112), does not have long-lasting effects (110) due to FTO mediated resistance. Therefore, the combination uses of ICB and FTO inhibitors will likely block this resistance in a host with an adaptive immunity presence. m6Am6AA recent study has also demonstrated that FTO knockdown sensitizes tumors to anti-PD-1 immunotherapy, except that this sensitization effect is not as robust when compared to ALKBH5 depletion (82). Examples of promising FTO inhibitors include the non-steroidal, anti-inflammatory drug, meclofenamic acid (MA), which is a selective FTO inhibitor (95), the ethyl ester form of meclofenamic acid (MA2) (113–115), and FB23-2 (99). In addition, R-2-hydroxyglutarate (R-2HG), an oncometabolite produced by the isocitrate dehydrogenase (IDH) mutations, inhibit FTO activity, leading to the increase of m6A abundance and decreased stability of MYC and CEBPA mRNAs, which subsequently caused growth inhibition, cell cycle arrest and apoptosis of leukemia cells (116).
ALKBH5 has been shown to be responsible for tumor proliferation in different cancers, including breast cancer (117), glioblastoma (118) and LADC (119), so its inhibition can yield good clinical outcomes if implemented. ALKBH5 knockdown has been shown to sensitize melanoma and colon cancer to overcome the resistance for immunotherapy through downregulating prominent cells (Tregs and MDSCs), which immunosuppresses the anti-tumor immunity, while at the same time upregulating DCs (82). One promising ALKBH5 targeting agent is the imidazobenzoxazin-5-thione MV1035 that shows an ALKBH5 inhibition and repression of GBM cell lines (U87-MG cells) through downregulating CD73 expression (101).
The mechanism by which Wnt/β-catenin signaling promotes tumors includes reducing T cells infiltration in the tumors and subsequently rendering immunotherapeutic approaches ineffective (e.g. anti-PD-L1/anti-CTLA-4 monoclonal antibody therapy) (120). On the other hand, blocking it would prime T cells (e.g., CD8+ T cells) against the tumor and enhance both the DCs’ antigen presentation and immunotherapy effectiveness (121–123). This means that targeting this oncogenic pathway can act as a valuable adjunct to immunotherapy. Thus, several inhibitors that either work through Wnt signals’ membrane transduction blockade as in the case of porcupine inhibitor or through β-catenin phosphorylation promotion as in the case of tankyrase inhibitors were developed and have entered preclinical trials but none of them has yet advanced to clinical use. Some of these inhibitors already show promising results, such as β-catenin inhibitor, PKF115-584, which can synergize with immunotherapies by enhancing DCs activation of T cells; therefore, overcoming the immunosuppression that may be induced by cancers (124).
Though ICB (such as anti-PD-1/PD-L1 treatment) have achieved significant clinical benefits in cancer patients receiving ICB, the immunotherapeutic outcomes exhibited individual heterogeneity. Thus, there are two problems that needs to be solved: (1) finding markers to predict the outcomes of immunotherapy; (2) figure out strategy to improve efficacy. Different m6A signatures model has been proposed to predict the response of anti-PD-1 immunotherapy (See section”m6A is associated with TME and immunotherapy efficacy”). Moreover, m6A-targeting treatment synchronizing with anti-PD-1 therapy have already shown improved outcome in preclinical studies (Figure 4).
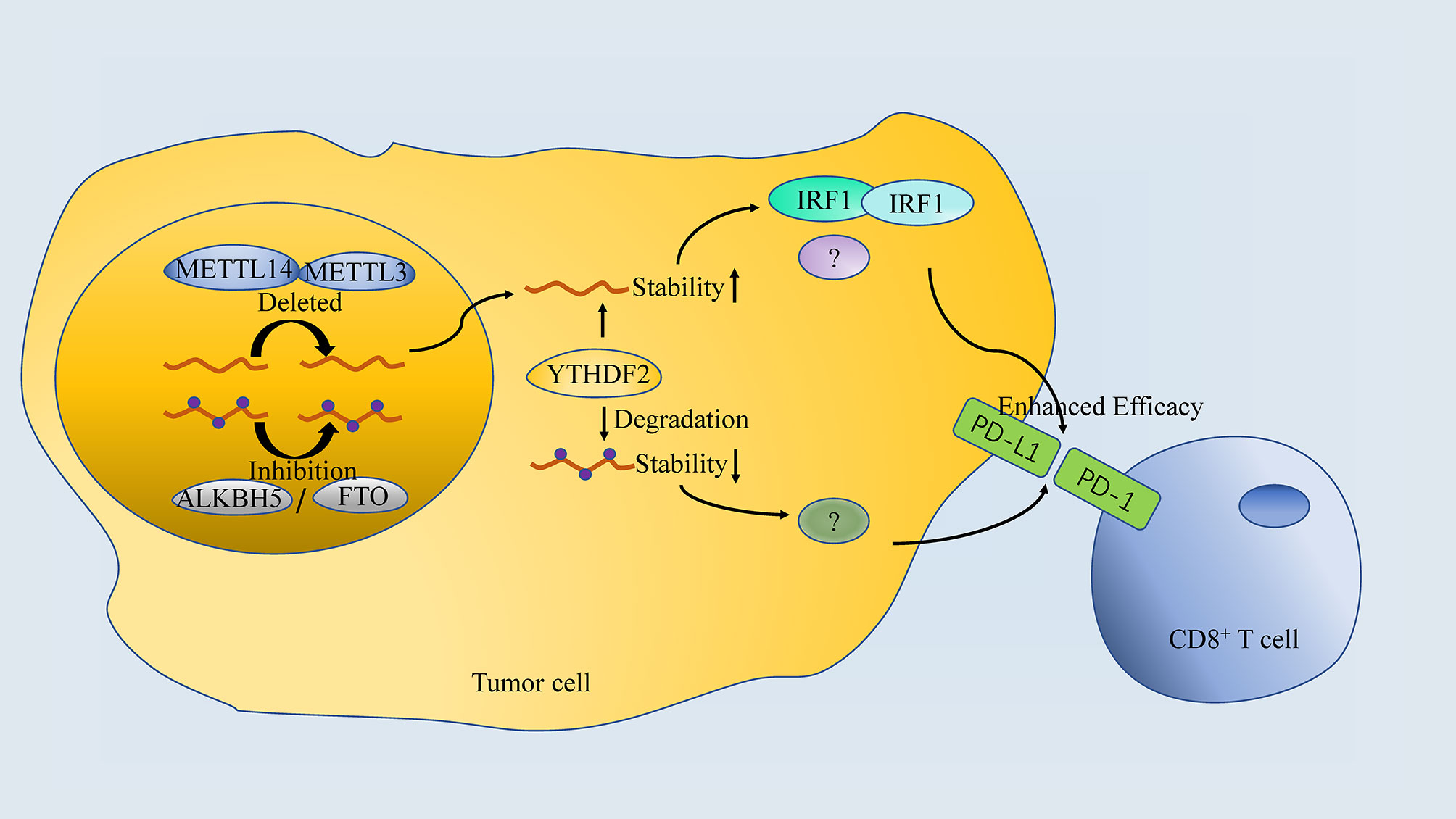
Figure 4 The association between m6A regulator and anti-PD-1 treatment. Both Inhibition or Deletion of “writers” or “erasers” enhances the efficacy of anti-PD-1 therapy, which is done by targeting specific RNA in a YTHDF2-dependant manner.
Inhibition of m6A mRNA modification by depletion of methyltransferases, Mettl3 and Mettl14, enhanced response to anti-PD-1 treatment in pMMR-MSI-L CRC and melanoma. Mechanistically, Mettl3 or Mettl14 deficiency promoted IFN-γ-Stat1-Irf1 signaling through stabilizing the Stat1 and Irf1 mRNA via YTHDF2 (13). However, both FTO and ALKBH5 in tumor cells are proved to decrease response to anti-PD-1 blockade immunotherapy. Knockdown of FTO sensitizes melanoma cells to interferon gamma (IFNγ) and sensitizes melanoma to anti-PD-1 treatment in mice (125). Tumor-intrinsic ALKBH5 inhibited the expansion and cytotoxicity of T cells by sustaining tumor cell PD-L1 expression, which was further confirmed in human intrahepatic cholangiocarcinoma (ICC) specimens (83). Importantly, a small-molecule ALKBH5 inhibitor enhanced the efficacy of cancer immunotherapy in melanoma (82), suggesting that the combination of FTO/ALKBH5 inhibition with anti-PD-1 blockade may reduce the resistance to immunotherapy in melanoma.
Intriguingly, both “writers” inhibition (METTL3 or METTL14) and “erasers” inhibition (FTO or ALKBH5) are proved to enhance the efficacy of anti-PD-1 blockade even in the same cancer (melanoma). It seem contradictory that both of the strategies work, since “writers” and “erasers” are believed to regulate m6A modification inversely. The possible explanations for this should include: (1) different targeted mRNA or ncRNA; (2) other potential pathway of “writers” and “erasers”; (3) differences in animal models (gene knockout vs. small molecular inhibitor). In addition, there is insufficient studies regarding the selectivity of target mRNA/ncRNA by “writers”, “readers” and “erasers”, which may explain the question mentioned above. As it is believed that these regulators function by affecting the translation and degradation of specific mRNA/ncRNA, it is reasonable that both “writers” and “erasers” inhibition can synchronize with anti-PD-1 immunotherapy if they act on different targets. However, future researches are needed to verify the hypothesis.
In this review, we have highlighted the roles of the different players involved in m6A modifications, their links to the TME and their potential therapeutic implementation against cancers when they are targeted, and we have particularly emphasized their actions as adjuncts to the various forms of immunotherapy. Most of these therapeutic strategies are still in the preclinical stages and haven’t entered clinical use yet. Nonetheless, these strategies show a promising outlook in impairing cancer progression, especially when bypassing the resistance to immunotherapies (e.g., primary, adaptive, or acquired resistances) through mechanisms such as reversing the immunosuppressive TME by increasing the influx of effector cells. As a consequence, these novel potential targets will be a necessary future component to combine with immunotherapy regimens, when subsequent studies further confirm their efficacy.
CQ and OB are major contributors in writing the manuscript. JC and XZ provided guidance throughout the preparation of this manuscript. PL, ZY, and DQ. WR collected and prepared the related papers. JH, HL, AZ, HM, GG, ZQ, MC, and YS reviewed and gave significant advice. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81873626, 81902592, 82070785), Hunan Natural Science Foundation (2020JJ5884), Hunan Province Key R&D Program (2019SK2202) and Xiangya Hospital Youth Fund (2018Q09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA Modification Landscape in Human Disease. RNA (2017) 23(12):1754–69. doi: 10.1261/rna.063503.117
2. Li S, Mason CE. The Pivotal Regulatory Landscape of RNA Modifications. Annu Rev Genomics Hum Genet (2014) 15:127–50. doi: 10.1146/annurev-genom-090413-025405
3. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res (2018) 46(D1):D303–D7. doi: 10.1093/nar/gkx1030
4. Desrosiers R, Friderici K, Rottman F. Identification of Methylated Nucleosides in Messenger RNA From Novikoff Hepatoma Cells. Proc Natl Acad Sci USA (1974) 71(10):3971–5. doi: 10.1073/pnas.71.10.3971
5. Perry RP, Kelley DE. Existence of Methylated Messenger RNA in Mouse L Cells. Cell (1974) 1(1):37–42. doi: 10.1016/0092-8674(74)90153-6
6. Huang H, Weng H, Chen J. M(6)A Modification in Coding and Non-Coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
7. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem (2018) 48(2):838–46. doi: 10.1159/000491915
8. Zhuang C, Zhuang C, Luo X, Huang X, Yao L, Li J, et al. N6-Methyladenosine Demethylase FTO Suppresses Clear Cell Renal Cell Carcinoma Through a Novel FTO-PGC-1α Signalling Axis. J Cell Mol Med (2019) 23(3):2163–73. doi: 10.1111/jcmm.14128
9. Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic M(6)A mRNA Methylation Reveals the Role of METTL3-M(6)A-CDCP1 Signaling Axis in Chemical Carcinogenesis. Oncogene (2019) 38(24):4755–72. doi: 10.1038/s41388-019-0755-0
10. Tang H, Qiao J, Fu YX. Immunotherapy and Tumor Microenvironment. Cancer Lett (2016) 370(1):85–90. doi: 10.1016/j.canlet.2015.10.009
11. Shulman Z, Stern-Ginossar N. The RNA Modification N(6)-Methyladenosine as a Novel Regulator of the Immune System. Nat Immunol (2020) 21(5):501–12. doi: 10.1038/s41590-020-0650-4
12. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-Tumour Immunity Controlled Through mRNA M(6)A Methylation and YTHDF1 in Dendritic Cells. Nature (2019) 566(7743):270–4. doi: 10.1038/s41586-019-0916-x
13. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. M(6) A RNA Methyltransferases METTL3/14 Regulate Immune Responses to Anti-PD-1 Therapy. EMBO J (2020) 39(20):e104514. doi: 10.15252/embj.2020104514
14. Chen YT, Shen JY, Chen DP, Wu CF, Guo R, Zhang PP, et al. Identification of Cross-Talk Between M(6)A and 5mc Regulators Associated With Onco-Immunogenic Features and Prognosis Across 33 Cancer Types. J Hematol Oncol (2020) 13(1):22. doi: 10.1186/s13045-020-00854-w
15. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
16. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and Near Stop Codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
17. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
18. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural Basis of N(6)-Adenosine Methylation by the METTL3-METTL14 Complex. Nature (2016) 534(7608):575–8. doi: 10.1038/nature18298
19. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell (2016) 63(2):306–17. doi: 10.1016/j.molcel.2016.05.041
20. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
21. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA M(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell (2018) 69(6):1028–38.e6. doi: 10.1016/j.molcel.2018.02.015
22. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the M(6)A Machinery Component Wtap/Fl(2)d. Genes Dev 32(5-6):415–29. doi: 10.1101/gad.309146.117
23. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA Mediates Preferential M(6)A mRNA Methylation in 3’UTR and Near Stop Codon and Associates With Alternative Polyadenylation. Cell Discov (2018) 4:10. doi: 10.1038/s41421-018-0019-0
24. Wang YN, Yu CY, Jin HZ. RNA N(6)-Methyladenosine Modifications and the Immune Response. J Immunol Res (2020) 2020:6327614. doi: 10.1155/2020/6327614
25. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear M(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
26. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear M6a Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing During Mouse Oocyte Development. PloS Genet (2018) 14(5):e1007412. doi: 10.1371/journal.pgen.1007412
27. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 Is an N(6)-Methyladenosine Binding Protein That Regulates Mammalian Spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
28. Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT. The M(6)A Reader Protein YTHDC2 Interacts With the Small Ribosomal Subunit and the 5’-3’ Exoribonuclease XRN1. RNA (2018) 24(10):1339–50. doi: 10.1261/rna.064238.117
29. Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of M(6)A Transcripts by the 3’→5’ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell (2017) 68(2):374–387.e12. doi: 10.1016/j.molcel.2017.09.021
30. Patil DP, Pickering BF, Jaffrey SR. Reading M(6)A in the Transcriptome: M(6)A-Binding Proteins. Trends Cell Biol (2018) 28(2):113–27. doi: 10.1016/j.tcb.2017.10.001
31. Zaccara S, Ries RJ, Jaffrey SR. Reading, Writing and Erasing mRNA Methylation. Nat Rev Mol Cell Biol (2019) 20(10):608–24. doi: 10.1038/s41580-019-0168-5
32. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible Methylation of M(6)A(m) in the 5’ Cap Controls mRNA Stability. Nature (2017) 541(7637):371–5. doi: 10.1038/nature21022
33. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 Is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
34. Liu X, Liu J, Xiao W, Zeng Q, Bo H, Zhu Y, et al. SIRT1 Regulates N(6) -Methyladenosine RNA Modification in Hepatocarcinogenesis by Inducing RANBP2-Dependent FTO SUMOylation. Hepatology (2020) 72(6):2029–50. doi: 10.1002/hep.31222
35. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of M(6)A-Dependent Nuclear RNA Processing Events. Cell (2015) 162(6):1299–308. doi: 10.1016/j.cell.2015.08.011
36. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-Dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature (2015) 518(7540):560–4. doi: 10.1038/nature14234
37. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-Methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res (2017) 45(10):6051–63. doi: 10.1093/nar/gkx141
38. Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, et al. Regulation of Co-Transcriptional Pre-mRNA Splicing by M(6)A Through the Low-Complexity Protein hnRNPG. Mol Cell (2019) 76(1):70–81.e9. doi: 10.1016/j.molcel.2019.07.005
39. Marek B, Covelo MH, Pavlina G, Dominika H, Grzegorz K, Stepanka V. N6-Methyladenosine Demethylase FTO Targets pre-mRNAs and Regulates Alternative Splicing and 3′-End Processing. Nuclc Acids Res (2017) 19:11356. doi: 10.1093/nar/gkx778
40. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 Mediates Nuclear Export of N(6)-Methyladenosine Methylated mRNAs. Elife (2017) 6:e31311. doi: 10.7554/eLife.31311
41. Lesbirel S, Viphakone N, Parker M, Parker J, Heath C, Sudbery I, et al. The M(6)A-Methylase Complex Recruits TREX and Regulates mRNA Export. Sci Rep (2018) 8(1):13827. doi: 10.1038/s41598-018-32310-8
42. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
43. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem Cells. M6a mRNA Methylation Facilitates Resolution of Naïve Pluripotency Toward Differentiation. Science (2015) 347(6225):1002–6. doi: 10.1126/science.1261417
44. Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, et al. M(6)A mRNA Modifications are Deposited in Nascent pre-mRNA and Are Not Required for Splicing But do Specify Cytoplasmic Turnover. Genes Dev (2017) 31(10):990–1006. doi: 10.1101/gad.301036.117
45. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
46. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA Circularization by METTL3-Eif3h Enhances Translation and Promotes Oncogenesis. Nature (2018) 561(7724):556–60. doi: 10.1038/s41586-018-0538-8
47. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 Destabilizes M(6)A-Containing RNA Through Direct Recruitment of the CCR4-NOT Deadenylase Complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
48. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic M(6)A Reader YTHDF3 Promotes mRNA Translation. Cell Res (2017) 27(3):444–7. doi: 10.1038/cr.2017.10
49. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 Facilitates Translation and Decay of N(6)-Methyladenosine-Modified RNA. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
50. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
51. Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, et al. Stage-Specific Requirement for Mettl3-Dependent M(6)A mRNA Methylation During Haematopoietic Stem Cell Differentiation. Nat Cell Biol (2019) 21(6):700–9. doi: 10.1038/s41556-019-0318-1
52. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA M(6)A Modification. Cell Stem Cell (2018) 22(2):191–205.e9. doi: 10.1016/j.stem.2017.11.016
53. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The M(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell (2016) 62(3):335–45. doi: 10.1016/j.molcel.2016.03.021
54. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR M(6)A Promotes Cap-Independent Translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
55. Zhao Y, Chen Y, Jin M, Wang J. The Crosstalk Between M(6)A RNA Methylation and Other Epigenetic Regulators: A Novel Perspective in Epigenetic Remodeling. Theranostics (2021) 11(9):4549–66. doi: 10.7150/thno.54967
56. Si W, Li Y, Ye S, Li Z, Liu Y, Kuang W, et al. Methyltransferase 3 Mediated miRNA M6a Methylation Promotes Stress Granule Formation in the Early Stage of Acute Ischemic Stroke. Front Mol Neurosci (2020) 13:103. doi: 10.3389/fnmol.2020.00103
57. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 Promotes Metastasis of Colorectal Cancer via miR-1246/SPRED2/MAPK Signaling Pathway. J Exp Clin Cancer Res (2019) 38(1):393. doi: 10.1186/s13046-019-1408-4
58. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine Marks Primary microRNAs for Processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
59. Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-Methyladenosine Induced miR-143-3p Promotes the Brain Metastasis of Lung Cancer via Regulation of VASH1. Mol Cancer (2019) 18(1):181. doi: 10.1186/s12943-019-1108-x
60. Qian X, Yang J, Qiu Q, Li X, Jiang C, Li J, et al. LCAT3, a Novel M6a-RegulatedPlays an Oncogenic Role in Lung Cancer via Binding With FUBP1 to Activate C-MYC. J Hematol Oncol (2021) 14(1):112. doi: 10.1186/s13045-021-01123-0
61. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-Coding RNA XIST. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
62. Yi YC, Chen XY, Zhang J, Zhu JS. Novel Insights Into the Interplay Between M(6)A Modification and Noncoding RNAs in Cancer. Mol Cancer (2020) 19(1):121. doi: 10.1186/s12943-020-01233-2
63. Tang M, Lv Y. The Role of N(6) -Methyladenosine Modified Circular RNA in Pathophysiological Processes. Int J Biol Sci (2021) 17(9):2262–77. doi: 10.7150/ijbs.60131
64. Nombela P, Miguel-Lopez B, Blanco S. The Role of M(6)A, M(5)C and Psi RNA Modifications in Cancer: Novel Therapeutic Opportunities. Mol Cancer (2021) 20(1):18. doi: 10.1186/s12943-020-01263-w
65. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. M(6)A Regulator-Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization in Gastric Cancer. Mol Cancer (2020) 19(1):53. doi: 10.1186/s12943-020-01170-0
66. Chong W, Shang L, Liu J, Fang Z, Du F, Wu H, et al. M(6)A Regulator-Based Methylation Modification Patterns Characterized by Distinct Tumor Microenvironment Immune Profiles in Colon Cancer. Theranostics (2021) 11(5):2201–17. doi: 10.7150/thno.52717
67. Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, et al. Chemokine Expression in Melanoma Metastases Associated With CD8+ T-Cell Recruitment. Cancer Res (2009) 69(7):3077–85. doi: 10.1158/0008-5472.Can-08-2281
68. Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-Catenin Pathway Activation Correlates With Immune Exclusion Across. Hum Cancers (2019) 25(10):3074–83. doi: 10.1158/1078-0432.CCR-18-1942%JClinicalCancerResearch
69. Pan Y, Xiao K, Li Y, Li Y, Liu Q. RNA N6-Methyladenosine Regulator-Mediated Methylation Modifications Pattern and Immune Infiltration Features in Glioblastoma. Front Oncol (2021) 11:632934. doi: 10.3389/fonc.2021.632934
70. Li H, Hu J, Yu A, Othmane B, Guo T, Liu J, et al. RNA Modification of N6-Methyladenosine Predicts Immune Phenotypes and Therapeutic Opportunities in Kidney Renal Clear Cell Carcinoma. Front Oncol (2021) 11:642159. doi: 10.3389/fonc.2021.642159
71. Zhong J, Liu Z, Cai C, Duan X, Deng T, Zeng G. M(6)A Modification Patterns and Tumor Immune Landscape in Clear Cell Renal Carcinoma. J Immunother Cancer (2021) 9(2):e001646. doi: 10.1136/jitc-2020-001646
72. Wu X, Sheng H, Wang L, Xia P, Wang Y, Yu L, et al. A Five-M6a Regulatory Gene Signature Is a Prognostic Biomarker in Lung Adenocarcinoma Patients. Aging (Albany NY) (2021) 13(7):10034–57. doi: 10.18632/aging.202761
73. Li Y, Gu J, Xu F, Zhu Q, Chen Y, Ge D, et al. Molecular Characterization, Biological Function, Tumor Microenvironment Association and Clinical Significance of M6a Regulators in Lung Adenocarcinoma. Brief Bioinform (2021) 22(4):bbaa225. doi: 10.1093/bib/bbaa225
74. Mo P, Xie S, Cai W, Ruan J, Du Q, Ye J, et al. N(6)-Methyladenosine (M(6)A) RNA Methylation Signature as a Predictor of Stomach Adenocarcinoma Outcomes and Its Association With Immune Checkpoint Molecules. J Int Med Res (2020) 48(9):300060520951405. doi: 10.1177/0300060520951405
75. Guo W, Tan F, Huai Q, Wang Z, Shao F, Zhang G, et al. Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and M6a RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma. Front Immunol (2021) 12:669750. doi: 10.3389/fimmu.2021.669750
76. Zhao H, Xu Y, Xie Y, Zhang L, Gao M, Li S, et al. M6a Regulators Is Differently Expressed and Correlated With Immune Response of Esophageal Cancer. Front Cell Dev Biol (2021) 9:650023. doi: 10.3389/fcell.2021.650023
77. Jiang H, Ning G, Wang Y, Lv W. Identification of an M6a-Related Signature as Biomarker for Hepatocellular Carcinoma Prognosis and Correlates With Sorafenib and Anti-PD-1 Immunotherapy Treatment Response. Dis Markers (2021) 2021:5576683. doi: 10.1155/2021/5576683
78. Li J, Wang W, Zhou Y, Liu L, Zhang G, Guan K, et al. M6a Regulator-Associated Modification Patterns and Immune Infiltration of the Tumor Microenvironment in Hepatocarcinoma. Front Cell Dev Biol (2021) 9:687756. doi: 10.3389/fcell.2021.687756
79. Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell (2010) 141(1):52–67. doi: 10.1016/j.cell.2010.03.015
80. Zhang Z, Zhou D, Lai Y, Liu Y, Tao X, Wang Q, et al. Estrogen Induces Endometrial Cancer Cell Proliferation and Invasion by Regulating the Fat Mass and Obesity-Associated Gene via PI3K/AKT and MAPK Signaling Pathways. Cancer Lett (2012) 319(1):89–97. doi: 10.1016/j.canlet.2011.12.033
81. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell (2020) 38(1):79–96.e11. doi: 10.1016/j.ccell.2020.04.017
82. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc Natl Acad Sci USA (2020) 117(33):20159–70. doi: 10.1073/pnas.1918986117
83. Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M6A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res (2021) canres.0468.2021. doi: 10.1158/0008-5472.Can-21-0468
84. Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N(6)-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer Res (2019) 79(11):2812–20. doi: 10.1158/0008-5472.Can-18-3592
85. Pi J, Wang W, Ji M, Wang X, Wei X, Jin J, et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res (2020) 81(10):2651–65. doi: 10.1158/0008-5472.Can-20-0066
86. Shang L, Wan Y, Zhang Z, Jiang Y, Lang J, Cheng W, et al. FTO Demethylates M6a Modifications in HOXB13 mRNA and Promotes Endometrial Cancer Metastasis by Activating the WNT Signaling Pathway. RNA Biol (2020) 18(9):1265–78. doi: 10.1080/15476286.2020.1841458
87. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-Mediated mRNA M6a Methylation Promotes Dendritic Cell Activation. Nat Commun (2019) 10(1):1898. doi: 10.1038/s41467-019-09903-6
88. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA M6a Methylation Orchestrates Cancer Growth and Metastasis via Macrophage Reprogramming. Nat Commun (2021) 12(1):1394. doi: 10.1038/s41467-021-21514-8
89. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. M6a mRNA Methylation Sustains Treg Suppressive Functions. Cell Res (2018) 28(2):253–6. doi: 10.1038/cr.2018.7
90. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The Loss of RNA N(6)-Adenosine Methyltransferase Mettl14 in Tumor-Associated Macrophages Promotes CD8(+) T Cell Dysfunction and Tumor Growth. Cancer Cell (2021) 39(7):945–957.e10. doi: 10.1016/j.ccell.2021.04.016
91. Zhou J, Zhang X, Hu J, Qu R, Yu Z, Xu H, et al. M(6)A Demethylase ALKBH5 Controls CD4(+) T Cell Pathogenicity and Promotes Autoimmunity. Sci Adv 7(25):eabg0470. doi: 10.1126/sciadv.abg0470
92. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)A mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
93. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N(6)-Methyladenosine (M(6)A)-Forming Enzyme METTL3 Facilitates M1 Macrophage Polarization Through the Methylation of STAT1 mRNA. Am J Physiol Cell Physiol (2019) 317(4):C762–75. doi: 10.1152/ajpcell.00212.2019
94. Aik W, Demetriades M, Hamdan MK, Bagg EA, Yeoh KK, Lejeune C, et al. Structural Basis for Inhibition of the Fat Mass and Obesity Associated Protein (FTO). J Med Chem (2013) 56(9):3680–8. doi: 10.1021/jm400193d
95. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic Acid Selectively Inhibits FTO Demethylation of M6a Over ALKBH5. Nucleic Acids Res (2015) 43(1):373–84. doi: 10.1093/nar/gku1276
96. Wang T, Hong T, Huang Y, Su H, Wu F, Chen Y, et al. Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein. J Am Chem Soc (2015) 137(43):13736–9. doi: 10.1021/jacs.5b06690
97. He W, Zhou B, Liu W, Zhang M, Shen Z, Han Z, et al. Identification of A Novel Small-Molecule Binding Site of the Fat Mass and Obesity Associated Protein (FTO). J Med Chem (2015) 58(18):7341–8. doi: 10.1021/acs.jmedchem.5b00702
98. Qiao Y, Zhou B, Zhang M, Liu W, Han Z, Song C, et al. A Novel Inhibitor of the Obesity-Related Protein FTO. Biochemistry (2016) 55(10):1516–22. doi: 10.1021/acs.biochem.6b00023
99. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell (2019) 35(4):677–91.e10. doi: 10.1016/j.ccell.2019.03.006
100. Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of Entacapone as a Chemical Inhibitor of FTO Mediating Metabolic Regulation Through FOXO1. Sci Transl Med (2019) 11(488):eaau7116. doi: 10.1126/scitranslmed.aau7116
101. Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, et al. 3D Proteome-Wide Scale Screening and Activity Evaluation of a New ALKBH5 Inhibitor in U87 Glioblastoma Cell Line. Bioorg Med Chem (2020) 28(4):115300. doi: 10.1016/j.bmc.2019.115300
102. Selberg S, Blokhina D, Aatonen M, Koivisto P, Siltanen A, Mervaala E, et al. Discovery of Small Molecules That Activate RNA Methylation Through Cooperative Binding to the METTL3-14-WTAP Complex Active Site. Cell Rep (2019) 26(13):3762–71.e5. doi: 10.1016/j.celrep.2019.02.100
103. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in Cancer: Mechanisms and Therapeutic Targeting. J Hematol Oncol (2020) 13(1):117. doi: 10.1186/s13045-020-00951-w
104. Bunse L, Pusch S, Bunse T, Sahm F, Sanghvi K, Friedrich M, et al. Suppression of Antitumor T Cell Immunity by the Oncometabolite (R)-2-Hydroxyglutarate. Nat Med (2018) 24(8):1192–203. doi: 10.1038/s41591-018-0095-6
105. Bedi RK, Huang D, Eberle SA, Wiedmer L, Śledź P, Caflisch A. Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem (2020) 15(9):744–8. doi: 10.1002/cmdc.202000011
106. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The M6a Reader YTHDF1 Promotes Ovarian Cancer Progression via Augmenting EIF3C Translation. Nucleic Acids Res (2020) 48(7):3816–31. doi: 10.1093/nar/gkaa048%JNucleicAcidsResearch
107. Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, et al. M6a-YTHDF1-Mediated TRIM29 Upregulation Facilitates the Stem Cell-Like Phenotype of Cisplatin-Resistant Ovarian Cancer Cells. Biochim Biophys Acta Mol Cell Res (2020) 118878:118878. doi: 10.1016/j.bbamcr.2020.118878
108. Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene C-Myc Promotes Epitranscriptome M(6)A Reader YTHDF1 Expression in Colorectal Cancer. Oncotarget (2018) 9(7):7476–86. doi: 10.18632/oncotarget.23554
109. Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, et al. Overexpression of YTHDF1 is Associated With Poor Prognosis in Patients With Hepatocellular Carcinoma. Cancer Biomark (2018) 21(4):859–68. doi: 10.3233/cbm-170791
110. Spranger S, Gajewski TF. Impact of Oncogenic Pathways on Evasion of Antitumour Immune Responses. Nat Rev Cancer (2018) 18(3):139–47. doi: 10.1038/nrc.2017.117
111. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell (2017) 168(4):707–23. doi: 10.1016/j.cell.2017.01.017
112. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and Activity of Anti-PD-L1 Antibody in Patients With Advanced Cancer. N Engl J Med (2012) 366(26):2455–65. doi: 10.1056/NEJMoa1200694
113. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. M(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep (2017) 18(11):2622–34. doi: 10.1016/j.celrep.2017.02.059
114. Xiao L, Li X, Mu Z, Zhou J, Zhou P, Xie C, et al. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res (2020) 80(18):3945–58. doi: 10.1158/0008-5472.Can-20-0132
115. Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, et al. FTO Regulates the Chemo-Radiotherapy Resistance of Cervical Squamous Cell Carcinoma (CSCC) by Targeting β-Catenin Through mRNA Demethylation. Mol Carcinog (2018) 57(5):590–7. doi: 10.1002/mc.22782
116. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2hg Exhibits Anti-Tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell (2018) 172(1-2):90–105.e23. doi: 10.1016/j.cell.2017.11.031
117. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia Induces the Breast Cancer Stem Cell Phenotype by HIF-Dependent and ALKBH5-Mediated M(6)A-Demethylation of NANOG mRNA. Proc Natl Acad Sci USA (2016) 113(14):E2047–E56. doi: 10.1073/pnas.1602883113
118. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. M 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31(4):591–606.e6. doi: 10.1016/j.ccell.2017.02.013
119. Chao Y, Shang J, Ji W. ALKBH5-M(6)A-FOXM1 Signaling Axis Promotes Proliferation and Invasion of Lung Adenocarcinoma Cells Under Intermittent Hypoxia. Biochem Biophys Res Commun (2020) 521(2):499–506. doi: 10.1016/j.bbrc.2019.10.145
120. Spranger S, Bao R, Gajewski TF. Melanoma-Intrinsic β-Catenin Signalling Prevents Anti-Tumour Immunity. Nature (2015) 523(7559):231–5. doi: 10.1038/nature14404
121. Ganesh S, Shui X, Craig KP, Park J, Wang W, Brown BD, et al. RNAi-Mediated β-Catenin Inhibition Promotes T Cell Infiltration and Antitumor Activity in Combination With Immune Checkpoint Blockade. Mol Ther (2018) 26(11):2567–79. doi: 10.1016/j.ymthe.2018.09.005
122. Liang X, Fu C, Cui W, Ober-Blöbaum JL, Zahner SP, Shrikant PA, et al. β-Catenin Mediates Tumor-Induced Immunosuppression by Inhibiting Cross-Priming of CD8⁺ T Cells. J Leukoc Biol (2014) 95(1):179–90. doi: 10.1189/jlb.0613330
123. Hong Y, Manoharan I, Suryawanshi A, Shanmugam A, Swafford D, Ahmad S, et al. Deletion of LRP5 and LRP6 in Dendritic Cells Enhances Antitumor Immunity. Oncoimmunology (2016) 5(4):e1115941. doi: 10.1080/2162402x.2015.1115941
124. Yaguchi T, Goto Y, Kido K, Mochimaru H, Sakurai T, Tsukamoto N, et al. Immune Suppression and Resistance Mediated by Constitutive Activation of Wnt/β-Catenin Signaling in Human Melanoma Cells. J Immunol (2012) 189(5):2110–7. doi: 10.4049/jimmunol.1102282
Keywords: N6-methyladenosine, m6A, tumor microenvironment, immune response, immunotherapy
Citation: Quan C, Belaydi O, Hu J, Li H, Yu A, Liu P, Yi Z, Qiu D, Ren W, Ma H, Gong G, Ou Z, Chen M, Sun Y, Chen J and Zu X (2021) N6-Methyladenosine in Cancer Immunotherapy: An Undervalued Therapeutic Target. Front. Immunol. 12:697026. doi: 10.3389/fimmu.2021.697026
Received: 18 April 2021; Accepted: 09 August 2021;
Published: 30 August 2021.
Edited by:
Catherine Sautes-Fridman, U1138 Centre de Recherche des Cordeliers (CRC) (INSERM), FranceReviewed by:
Gabriel Malouf, U964 Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) (INSERM), FranceCopyright © 2021 Quan, Belaydi, Hu, Li, Yu, Liu, Yi, Qiu, Ren, Ma, Gong, Ou, Chen, Sun, Chen and Zu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiongbing Zu, enV4Ynh5eXlAMTI2LmNvbQ==; Jinbo Chen, Y2hlbmppbmJvMTk4OUB5YWhvby5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.