- 1Center of Experimental & Molecular Medicine, Amsterdam University Medical Centers, Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands
- 2Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Amsterdam University Medical Centers, Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands
- 3Division of Infectious Diseases, Amsterdam University Medical Centers, Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands
Host cells undergo complex transcriptional reprogramming upon infection. Epigenetic changes play a key role in the immune response to bacteria, among which DNA modifications that include methylation have received much attention in recent years. The extent of DNA methylation is well known to regulate gene expression. Whilst historically DNA methylation was considered to be a stable epigenetic modification, accumulating evidence indicates that DNA methylation patterns can be altered rapidly upon exposure of cells to changing environments and pathogens. Furthermore, the action of proteins regulating DNA methylation, particularly DNA methyltransferases and ten-eleven translocation methylcytosine dioxygenases, may be modulated, at least in part, by bacteria. This review discusses the principles of DNA methylation, and recent insights about the regulation of host DNA methylation during bacterial infection.
Introduction
DNA methylation refers to the addition of a methyl group to the DNA cytosine residues at the fifth carbon position (5mC), which is a common epigenetic mark in many eukaryotes and often found in the sequence context CpG (i.e., regions in the DNA where a cytosine nucleotide is followed by a guanine nucleotide along the 5’ to 3’ direction) (1). The methylation process is promoted by the DNA methyltransferases (DNMTs), of which DNMT3A and DNMT3B mediate de novo DNA methylation, establishing a pattern of methylation that is then sustained by the maintenance methyltransferase, DNMT1 (2). DNMT2 is not involved in DNA methylation, but rather mediates methylation of RNA (3), and therefore is further not discussed in this review. The process of DNA methylation can be reversed passively through cell division or actively catalyzed by ten-eleven translocation (TET) methylcytosine dioxygenases family proteins, and a subsequent nucleotide excision and repair process, called DNA demethylation (4). There are three members in the TET family, namely TET1, TET2 and TET3, all sharing a conserved catalytic domain in their C terminus (5). DNA methylation is generally associated with transcriptional silencing, although this paradigm has been challenged by recent studies showing that DNA methylation can both positively and negatively regulate gene expression depending on the position where it occurred (6).
Both innate and adaptive immune responses contribute to protection of the host against bacterial pathogens (7). The innate immune system functions as the first line of defense against invading pathogens and is composed of innate immune cells (including basophils, dendritic cells, eosinophils, Langerhans cells, mast cells, monocytes, macrophages, neutrophils and natural killer cells) and some stromal cells, such as epithelial cells that sense bacteria by their surface or endosomal pathogen recognition receptors (PRRs). Toll-like receptors (TLRs), RIG-I-like receptors, NOD-like receptors and C-type lectin receptors are among the large array of PPRs that are able to detect pathogens by recognizing microbial components known as pathogen-associated molecular patterns, among which lipopolysaccharide (LPS), flagellin and lipoteichoic acid (8, 9). Upon recognition of bacteria or bacterial components, innate immune cells initiate intracellular signaling cascades to induce functional changes and to elicit the production of immune effectors, such as cytokines, chemokines and antimicrobial peptides, that directly or indirectly contribute to host antibacterial defense and inflammatory responses. When bacterial pathogens evade host innate immunity, adaptive immune responses can contribute to defense mechanisms. T and B cells are dominant players in adaptive immunity, activated through presentation of bacterial antigens by antigen-presenting cells. Innate and adaptive immune responses do not act independently, but coordinated actions of these two systems are required for efficient elimination of bacterial invaders. Furthermore, in order to prevent collateral damage both innate and adaptive immune responses need to be tightly regulated at different levels (10). Modification of DNA methylation in host cells, induced by infectious agents, has been implicated in the induction and regulation of the immune response to bacteria.
DNA methylation has been considered to be relatively stable when compared with other epigenetic modifications, such as those involving histones, but recent findings have documented that DNA methylation can occur faster than previously thought, particularly when cells are exposed to changing environments, including contact with pathogens during infection (11). Importantly, accumulating evidence indicates that pathogens can alter DNA methylation and/or regulate the expression and function of DNA methylation modifiers such as TETs and DNMTs, resulting in altered expression of important host genes involved in immune responses (11). These alterations in DNA methylation or its related factors can either contribute to protective host immunity to eliminate pathogens or benefit pathogens to evade immune responses for persistence within the host. This review summarizes current understanding of the effects of DNA methylation on host immune responses and pathogen elimination during infection.
DNA Methylation
Two families of proteins directly contribute to the DNA methylation pathway: the DNMTs promote and maintain DNA methylation, while the TETs catalyze demethylation via multiple steps (Figure 1). DNA methylation is established by the de novo methyltransferases DNMT3A and DNMT3A with the help of catalytically inactive DNMT3L in mammals, whilst the maintenance of DNA methylation is mediated by DNMT1 and its obligate partner ubiquitin-like plant homeodomain and RING finger domain 1 (UHRF1), which preferentially recognizes hemimethylated CpGs during cell division (12).
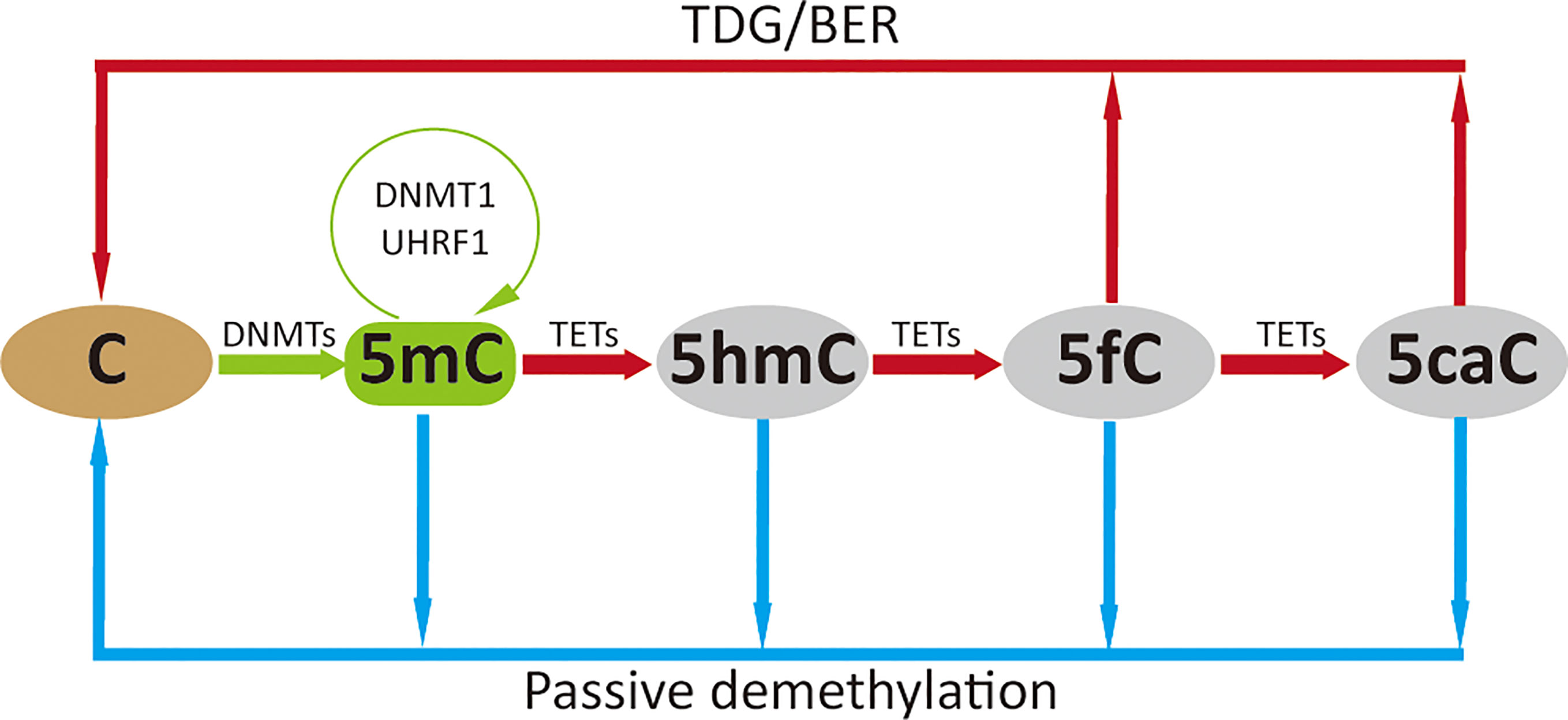
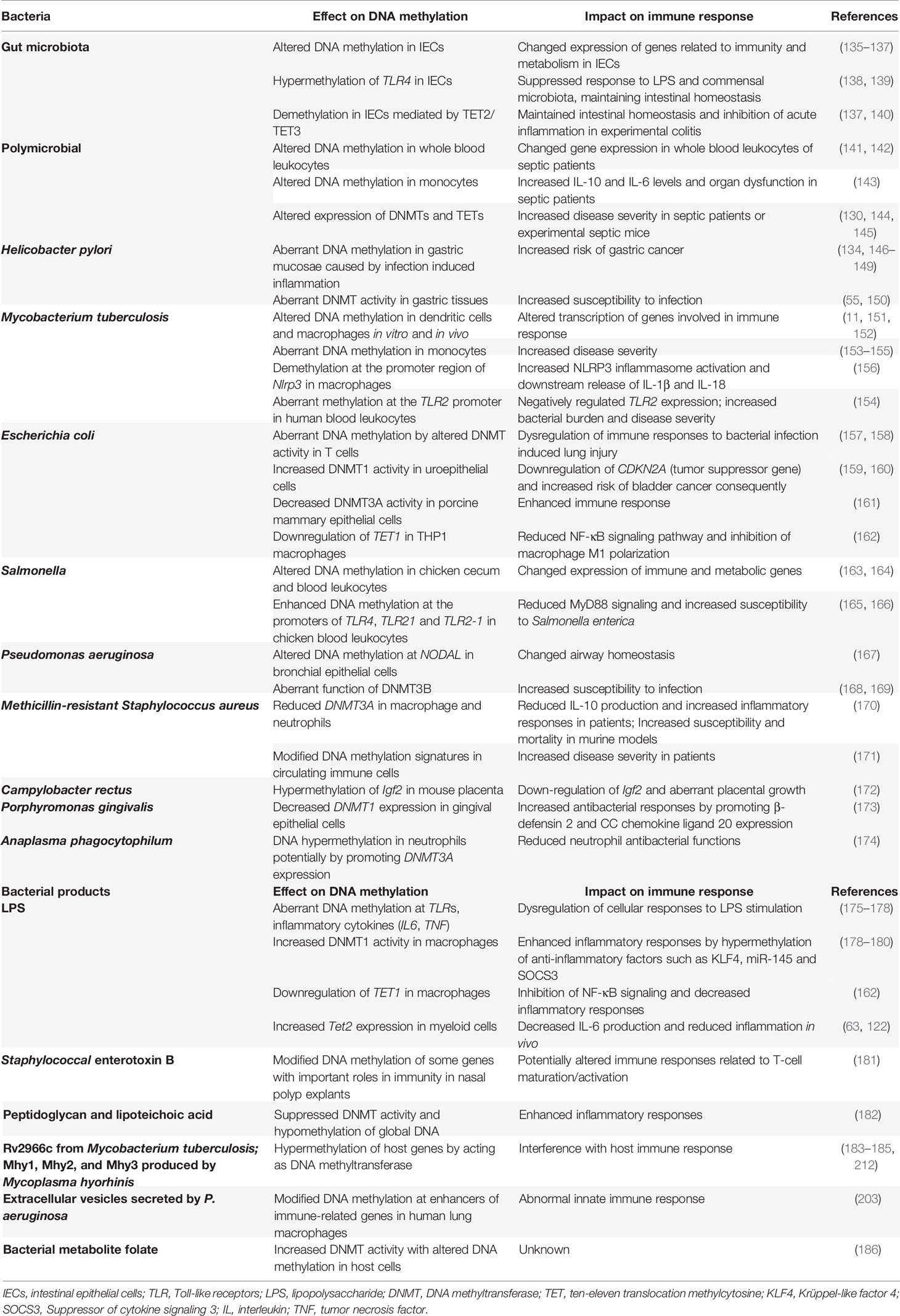
Figure 1 DNA methylation cycle. DNMTs catalyze the addition of a methyl group to the fifth carbon position of cytosine to generate methylated cytosine (5mC), which is maintained by DNMT1 (green arrow); 5mC is oxidized to 5-hydroxymethylcytosine (5hmC), which can be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by TETs. The higher oxidized cytosine bases 5fC and 5caC can then be converted back to their unmodified state directly by thymine DNA glycosylase (TDG) and subsequently base excision repair (BER) processing; these oxidative steps contribute to active demethylation (red arrow). Passive demethylation removes 5mC from all forms of methylcytosine due to absence or reduction in DNMT levels and function (blue arrow).
Although DNA methylation is reported to be stable, DNA demethylation has been widely observed during development and activation of mammalian cells. Possible mechanisms underlying DNA demethylation have been reviewed by other researchers (13–16); we here only briefly introduce the broadly recognized passive and active routes. Passive demethylation occurs in the absence of the DNA methylation maintenance machinery (DNMT1/UHRF1) during DNA replication, which leads to dilution of 5mC, or removal of 5mC due to absence or reduction in DNMT levels and function (17). Active demethylation is mostly dependent on the oxidation of 5mC by TETs, that oxidize 5mC to 5-hydroxymethylcytosine (5hmC), which can be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). These oxidized cytosine bases (5hmC/5fC/5caC) may facilitate DNA demethylation by impairing the binding and/or activity of enzymes regulating the maintenance methylation machinery (DNMT1/UHRF1) which impairs remethylation during DNA replication (13). The higher oxidized cytosine bases (5fC/5caC) can be efficiently excised by thymine DNA glycosylase (TDG), followed by the base-excision-repair (BER) pathway, which accounts for the major DNA demethylation mechanism. Interestingly, TETs might not decrease methylation levels, but specifically prevent aberrant methylation spreading into CpG islands (CGIs) (18), and DNMTs might also contribute to active DNA demethylation in conditions of low methyl group sources (19).
Regulation of DNMTs
DNMT proteins are recruited to certain locations in the genome where they catalyze the transfer of methyl groups from S-adenosyl-L-methionine (SAM) to the C5 of cytosine to establish 5mC. During this process, the activity of DNMTs can be regulated at the following levels (Figure 2).
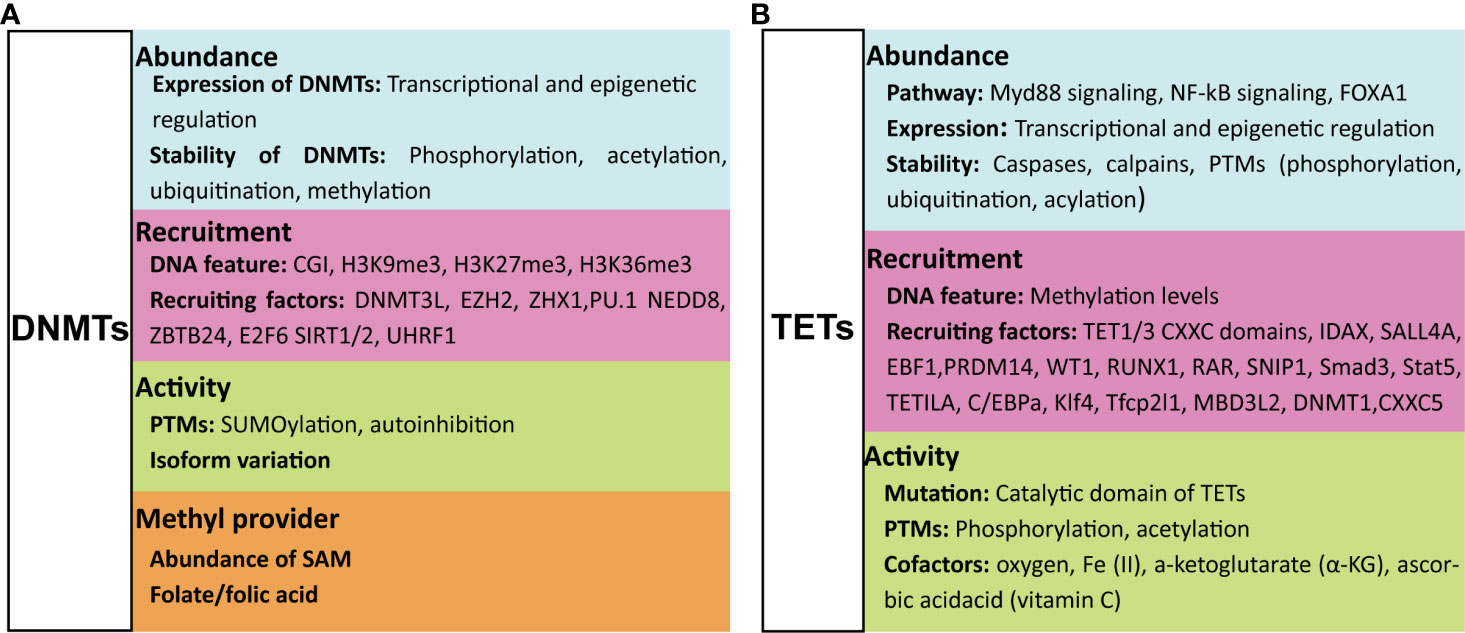
Figure 2 Factors that regulate the function of DNMTs and TETs. The function of DNMTs can be influenced at four levels: their abundance, their recruitment to DNA, their catalytic activity, and the methyl group source (A). The function of TETs is regulated at three levels: their abundance, their recruitment to DNA and their catalytic activity (B). For details see text. DNMTs, DNA methyltransferases; TETs, ten-eleven translocation methylcytosine dioxygenases; PTMs, post-translational modifications; CGI, CpG islands; SAM, S-adenosyl-L-methionine.
First, by the Abundance of DNMTs
The expression and stability of DNMTs can be regulated by transcriptional regulation and post-translational modifications (PTMs), respectively. Numerous pathways have been shown to induce or inhibit expression of DNMTs, and the extent of their expression can be further regulated by multiple epigenetic regulatory mechanisms (20). Proteolytic degradation of DNMT proteins can be promoted or inhibited by PTMs. Acetylation and ubiquitination of DNMT1 either protect from or promote proteolytic degradation (21, 22). Phosphorylation of Ser143 stabilizes DNMT1 (23), whilst methylation of Lys142 and Lys1096 promotes its proteolytic degradation (24, 25).
Second, Through the Function/Activity of DNMTs
DNA methylation by DNMTs is dependent on their catalytic activity, which is largely regulated by PTMs or isoform variation of DNMTs. SUMOylation of DNMT1 increases the catalytic activity of this enzyme on genomic DNA (26); SUMOylation of DNMT3A, however, abolishes its capacity to interact with histone deacetylases (HDACs) (27). DNMT1 is an auto-inhibitory protein that is activated upon binding to unmethylated cytosines (28, 29). The same auto-inhibitory characteristic was also found for DNMT3A, the activation of which is induced by histone H3 (30); this is might be the reason why the histone H3 N-terminal tail with an unmethylated Lys4 (H3K4) is required for de novo DNA methylation (31). In addition, the activity of DNMTs can be affected by isoform variation (32, 33), and other regulatory proteins, such as the microprocessor component DROSHA that interacts with DNMT1 to ensure its full methyltransferase activity (34).
Third, Through Recruitment of DNMTs to the Genome
To successfully perform DNA methylation, DNMTs are first recruited to the targeted DNA motif, and this recruitment is affected by both the features of the target DNA motif and factors that influence DNMT recruitment to the genome. DNMTs can be specifically recruited to DNA marked with unmethylated H3K4 via interacting with the ADD domain of DNMTs (35), while methylated H3K4 repulses the binding of de novo methyltransferases resulting in maintaining the hypomethylated state of CGIs (36). CGIs marked by H3K27me3 are more susceptible to de novo DNA methylation during differentiation and in disease states such as cancer (37, 38). Gene body enriched with H3K9me3 or H3K36 tri-methylation (H3K36me3) is also reported to be favorable for DNMT3B recruitment, leading to hypermethylation at these regions that functionally relate to gene transcription initiation, proper splicing and compact chromatin at active genes (37, 39, 40). The affinity of DNMT3A and DNMT3B for DNA can be further enhanced by DNMT3L through the formation of heterotetrametric complexes with either DNMT3A or DNMT3B, resulting in more efficient DNA methylation (41, 42). A large class of proteins, including polycomb group protein enhancer of zeste homolog 2 (EZH2) (43), Zinc-fingers and homeoboxes 1 (ZHX1) (44), ubiquitin-like protein modifier NEDD8 (45), zinc-finger protein ZBTB24, transcription factor E2F6 and PU.1, and Sirtuins 1 and 2 (SIRT1/2), were reported to recruit DNMTs to genes targeted for DNA methylation mediated gene silencing (46–49). The binding of DNMT1 to hemimethylated cytosines is selectively promoted by UHRF1 (50), but this binding is prevented by a DNA aptamer named Apt. #9 that competes with the hemiDNA for binding to DNMT1 (51). Besides protein molecules discussed above, some RNAs were also reported to affect the recruitment of DNMTs (52–54).
Fourth, the Methyl Group Donors Determine the Direction of the DNA Methylation Pathway
SAM is the major source of methyl groups for DNA methylation. The addition of folate/folic acid to provide methyl groups was reported to maintain DNA methylation and/or prevent the loss of global DNA methylation in health and disease (55, 56). However, factors that lead to less SAM decreases the transfer of methyl groups to DNA and RNA (57). In the absence of SAM, DNMT3a and DNMT3b can exhibit DNA dehydryoxymethylase activity, by directly converting 5hmC and 5caC, but not 5fC, to unmodified cytosines (58, 59). In some cases, DNMT1 is able to mediate oxidation of cytosine with formaldehyde, forming 5hmC (60), which further can participate in the DNA methylation cycle.
Regulation of TETs
The presence and catalytic activity of TETs are necessary for DNA demethylation, but their function is affected by multiple regulatory mechanisms that (amongst others) modulate substrate accessibility, enzymatic activity, expression levels and genomic targeting of TETs. Factors that are of importance for the regulation of activity of TETs are the following.
First, the Abundance of TETs Can Be Regulated at Transcriptional and Post-Transcriptional Levels
The expression of TETs can be induced by multiple signaling pathways, such as hydrogen sulfide (61), Myd88 signaling (62), NF-kB signaling (63) and Forkhead box A1 (FOXA1) (64), and frequently regulated at transcriptional level. IDAX (also known as CXXC4) and lysine demethylase KDM2A (65) negatively regulate whilst transcription factors Oct4 and CEBPα positively regulate TET2 protein expression (66–68). TET3 can be negatively regulated by nuclear receptor TLX (69). More recently, TETs were shown to be regulated by epigenetic modifications involving long non-coding RNA’s or microRNA’s (70–73). The abundance of TETs can also be regulated at protein level. TETs can be directly cleaved by caspases (68) and calpains (74) or degraded through PTMs. For instance, all three TET proteins can be monoubiquitinated by the VprBP-DDB1-CUL4-ROC1 E3 ubiquitin ligase (CRL4VprBP) (75), whilst MAPK-mediated phosphorylation at Serine-99 of TET2 stabilizes this enzyme (76, 77). Moreover, the 14-3-3 proteins bind phosphorylated TET2 and protect Serine-99 phosphorylation (78). Other modifications like (de)acetylation of TETs have also been reported; for example, acetylation of TET2 by p300 stabilizes this enzyme by inhibiting ubiquitination (79), whilst deacetylation of TET2 by the deacetylase SIRT1 promotes its ubiquitination degradation as well as enhances its catalytic activity (80, 81).
Second, the Binding of TETs to Genomic DNA Sequences Can Be Modulated
Similar to DNMTs, TET proteins also need to be recruited to the genome for implementing their functions. TET1 and TET3 can be recruited to genomic target sites through direct binding of their respective CXXC domains to DNA (82). This binding process can be influenced by several proteins. For instance, Lin28A recruits TET1 to common genomic loci to regulate DNA methylation and gene expression (83), thyroid hormone receptors stabilize the association of TET3 to chromatin depending on the catalytic activity of TET3 (84). In contrast to TET1 and TET3, TET2 is recruited to genomic DNA by a distinct CXXC domain-independent mechanism since TET2 does not have any discernable domains that bind directly to DNA. Indeed, numerous proteins have been discovered that promote or inhibit binding of TET2 to DNA. IDAX/CXXC4, originally encoded within an ancestral TET2 gene but separated from TET2 during evolution, recruits TET2 to DNA sequences containing unmethylated CpG dinucleotides located at promoters and CGIs in genomic DNA (68, 85). Other molecules such as Wilms tumor protein 1 (WT1) (86), early B-cell factor 1 (EBF1) (87), PRDM14 (88), RUNX1 (89), retinoic acid receptor (RAR) (90), SNIP1 (91), Smad3 and Stat5 (61), TET2 interacting long noncoding RNA (TETILA) (92) and transcription factors C/EBPa, Klf4, and Tfcp2l1 (93) can interact with TETs and enhance the recruitment of TETs to target loci. In addition, some proteins like Methyl-CpG binding domain protein 3-like 2 (MBD3L2) (94), DNMT1 (79), CXXC5 (95) and SALL4A (96) can further strengthen or stabilize the binding between TETs and methylated DNA targets. Besides factors modifying the recruitment of TETs, the character of target DNA sequences can also affect the binding of TETs. For example, low-methylated regions (LMRs) of CpG-poor distal regulatory regions that are occupied with DNA-binding factors are favorable for TET binding, thereby maintaining low methylation levels in these regions (97).
Third, Dioxygenase Activity of TETs Is Tightly Regulated
The dioxygenase activity of TETs is largely dependent on their catalytic domain and any mutation or modification within this region is likely to lead to a change in their function. Enzymatic reactions mediated by TETs highly rely on the cofactors oxygen, Fe (II), and a-ketoglutarate (α-KG) (98). Therefore, any modification in the production or activity of these cofactors is expected to lead to a functional change of TETs. Mutations in the genes encoding the metabolic enzymes isocitrate dehydrogenases 1 and 2 (IDH1/2), succinate dehydrogenase, and fumarate hydratase, result in aberrant accumulation of metabolites such as 2-hydroxyglutarate (2-HG), succinate and fumarate, respectively, which act as competitors of α-KG to broadly inhibit the α-KG-dependent enzymatic activity of TETs (99–101). Hypoxia, such as frequently occurs in tumor tissues, leads to loss of TET activity (102). On the other hand, addition of ascorbic acid (vitamin C), which is needed to reduce the oxidized iron species, enhances the catalytic activity of TETs (103–105). Additionally, TETs activity has also suggested to be affected by PTMs. Acetylation enhances TET2 function (79) and phosphorylation of TET3 at the highly conserved Serine-1310 and -1379 residues within its catalytic domain by cyclin-dependent kinase 5 (cdk5) is required for its dioxygenase activity (106). Moreover, the phosphorylation of TETs can be suppressed via O-GlcNAcylation by the glycosyltransferase OGT (107).
DNA Methylation and Gene Expression
DNA Methylation, DNA Demethylation and Gene Expression
DNA methylation plays a critical role in the regulation of many cellular processes, including X chromosome inactivation, genomic imprinting, stem cell differentiation, chromosomal conformation, chromatin structure, developmental stages and transcriptional activation/repression of genes (108). DNA methylation in the genome is not uniformly distributed: both promoter and CGIs typically are hypomethylated, whereas the extent of methylation in gene bodies is higher than that in intergenic regions (2). While early studies suggested that DNA methylation represses gene expression, a growing body of evidence has indicated that DNA methylation has a dual role, both inhibitory and permissive, depending on the genomic region at which DNA methylation occurs (2). DNA methylation of CpGs at promoters and enhancers that usually remain unmethylated is mainly coupled with transcriptional silencing (108, 109), but DNA methylation at the gene body has been associated with enhanced gene transcription or elongation (39, 110). DNA methylation can also indirectly regulate gene expression by altering the chromatin accessibility for transcription factors or by recruiting repressive proteins with methyl-binding domains (111). For instance, DNA methylation changes the accessibility of B cell enhancers for transcription factors E2A and PU.1 and blocks the binding of transcription factor erythroblastosis 1 (ETS1) at Ets binding site during B cells development (112, 113). In addition, DNA methylation closely cooperates with other regulatory machineries to modify gene expression, especially with histone modifications, which can partially be mediated through methylcytosine-binding proteins, such as MECP2 or MBD2, that are capable of recruiting histone deacetylases or transcriptional repressors to methylated regions (111, 114). DNA demethylation, on the other hand, is normally positively correlated with gene transcription (13). However, the precise relationship between DNA (de)methylation and gene expression is complex and requires further investigation. For instance, it is reported that microbe-induced changes in the expression of some genes can occur prior to modification of DNA methylation at their sites (11, 115) and that elevated DNA methylation outside of gene promoters has been shown to facilitate gene transcription to a larger extent than promoter DNA methylation (116, 117).
DNMT Related Gene Expression
DNMTs can repress gene expression by increasing DNA methylation at promoters and enhancers, resulting in reduced binding of transcriptional factors to these positions or inducing changes in the chromatin structure to make it less accessible for transcription (2, 111). For instance, DNMT3B mediated DNA methylation at the promoter regions of NF-κB responsive genes decreases NF-κB recruitment to the promoters, suppressing the expression of downstream genes (33). H3k6me3 selectively recruits DNMT3B to gene bodies of actively transcribed genes, thereby promoting DNA methylation and gene expression (37, 39, 110, 118). DNMTs can regulate gene expression not only via directly modifying DNA methylation, but also through mechanisms that are unrelated to DNA methylation but achieved by cooperating with other regulatory machineries. All three DNMTs (DNMT1, 3A and 3B) have been reported to repress gene transcription through interacting with HDACs independent of their catalytic activity (27, 119). DNMT3A-mediated DNA methylation increases HDAC9 transcription by repressing the inhibitory histone mark H3K27me3 at its distal promoter (116). DNMTs work together with polycomb group proteins for repression of their common target loci (43). The tricarboxylic acid cycle metabolites succinate and fumarate determine the catalytic activity of DNMTs; in turn, DNMT3B has been reported to modulate mitochondrial metabolism for maintaining articular cartilage homeostasis (120).
TET Related Gene Expression
TETs regulate gene expression directly by demethylation, dependent on their catalytic activity, or indirectly through interaction with other regulatory mechanisms, mostly independent of their catalytic activity. All three TETs contribute to dynamic demethylation during development, activation and oncologic transformation, linked with wide transcription reprogramming in cells during these processes (5, 121). In recent years, more and more DNA methylation independent functions of TETs have been discovered, indicating that TETs closely work together with other epigenetic regulatory mechanisms in the setting of infection. TET2 and TET3 have been shown to inhibit proinflammatory cytokine expression by recruiting HDAC1/2 to the promoters of cytokine encoding genes during bacterial and viral infection, respectively (122–124). TET2 also mediated transcriptional repression by facilitating the recruitment of the polycomb Repressive Complex 2 to CpG dinucleotide-rich gene promoters (125). TET1 can be incorporated in the SIN3A co-repressor complex, resulting in transcriptional effects independent of 5hmC (126), and this might be the underlying mechanisms of TET1 mediated inhibition of IL1B transcription (127). The same mechanism applies to TET3 regulated inhibition of type I interferon production during viral infection or poly(I:C) stimulation (124). TET2 and TET3 facilitate OGT-dependent histone O-GlcNAcylation by interacting with the enzyme O-linked b-N-acetylglucosamine (O-GlcNAc) transferase (OGT) (128, 129). Beyond oxidation of methylated cytosine in DNA, TET2 has also been reported to promote mRNA oxidation during infection derived sepsis, thereby destabilizing target mRNA (130); TET2 can suppress expression of endogenous retroviruses through a similar mechanism (131).
Modification of DNA Methylation Associated With Infection
The host response to an infection involves transcriptional changes in different types of immune cells, which can affect their function to either promote host defense against invading pathogens or benefit pathogen persistence. The transcriptional reprogramming during infection is highly regulated and epigenetic regulatory mechanisms are involved herein (132, 133) (Figure 3). Until recently, the extent of DNA methylation was thought to be stable and resistant to environmental stimulation. However, it is now well recognized that DNA methylation can be altered in a brief time frame in response to inflammation or infection and that these modifications in DNA methylation can influence immune cell responsiveness (11). Two possible mechanisms underlie infection induced alterations in DNA methylation: infection can directly alter DNA methylation by inducing or repressing DNA methylation enzymes (DNMTs and TETs), and/or indirectly through inflammatory mediators induced by the infection (134). Modification of host DNA methylation associated with bacterial infection and the consequent effects on immune responses were summarized in Table 1 and detailed below.
Figure 3 Regulation of host DNA methylation of immune responses during infection. Figure representing a general overview of how infection can affect DNA methylation. Note: not all infection modify DNA methylation; an overview of changes induced by specific pathogens is provided in the table. ① Infection induces DNA (de)methylation at target genes; ② Infection alters the transcription of DNA methylation modifiers TETs and DNMTs; ③ Loss of DNMTs promotes infection induced DNA demethylation at target genes; ④ TET proteins promote infection induced DNA demethylation at target genes; ⑤ TET proteins recruit HDACs for histone modification at IL1B and IL6 promoters; ⑥ TET proteins oxidize 5-methylcytosine (5-mC) on SOCS3 messenger RNA (mRNA); ⑦ Infection alter metabolic products that regulate the activity of TET proteins. “arrow” symbol represents promotion, “bar-headed arrow” symbol represents inhibition. DNMTs, DNA methyltransferases; TETs, ten-eleven translocation methylcytosine dioxygenases; HDACs, Histone deacetylases; TFs, transcription factors; IL, interleukin; SOCS3, Suppressor of cytokine signaling 3; ATP, Adenosine triphosphate; α-KG, a-ketoglutarate.
Gut Microbiota and Intestinal Pathogens
Commensal bacteria contribute to the maintenance of intestinal symbiosis by shaping host gene expression via epigenetic modification (187). Gut microbiota-dependent and -independent processes act together to form the postnatal development of the transcriptome and DNA methylation signatures of intestinal epithelial cells (IECs) early after birth. The formation of microbiota related “functional” methylation sites might impact long-term gene expression signatures in IECs (135, 136). Furthermore, some intestinal genes, related to innate immunity, phagocytosis, endothelial homeostasis and tissue metabolism are influenced by microbiota through DNA methylation (136). For instance, exposure of colonic epithelial cells to commensal bacteria results in Toll-like receptor (TLR)4 gene hypermethylation and transcriptional downregulation, thereby suppressing responsiveness to LPS (138, 139). More importantly, TET2/3 in IECs contribute to enhanced demethylation induced by microbiota under homeostasis and during acute inflammation (137). Besides IECs, the development and function of immune cells at nonmucosal sites, such as the bone marrow, peripheral lymph nodes and spleen, are also suggested to be regulated by microbiota via DNA methylation (188). On the other hand, TET2 deficiency in hematopoietic cells can lead to a microbiota-dependent impairment of gut barrier (140).
Many intestinal pathogenic bacteria have been suggested to cause aberrant DNA methylation in host cells. In this context. Helicobacter (H.) pylori is one of the most investigated enteric pathogens. H. pylori is able to change DNA methylation directly. High levels of aberrant DNA methylation in H. pylori–infected gastric mucosae have been associated with gastric cancer risk (146). Indeed, several tumor suppressing genes were found downregulated in gastric mucosae through H. pylori–infection induced hypermethylation. DNA methylation at the promoter region of trefoil factors, which regulate mucosal repair and suppress tumor formation in the stomach, was found increased early after H. pylori infection and throughout gastric tumor progression (189). Similarly, hypermethylation of DNA repair protein O6-methylguanine DNA methyltransferase (MGMT) and reduced levels of MGMT were common in the gastric epithelium of H. pylori infected patients, increasing mutagenesis in H. pylori-infected gastric mucosa (190). Other important genes like CX32 and CX43 were also repressed by H. pylori induced hypermethylation (191). DNA hypermethylation in the context of H. pylori infection was partially reversible after eradication of this bacterium or administration of a DNA demethylating agent, 5-aza-2-deoxycytidine, resulting in decreased the incidence of gastric cancers induced by H. pylori infection (190, 192). Single nucleotide polymorphisms in DNMT1 were reported to be genotypic markers for predicting genetic susceptibility to H. pylori infection (150), whilst the addition of folic acid to promote the activity of DNMTs was able to counteract H. pylori induced DNA demethylation (55), suggesting a direct role for methylation related factors herein. More recent evidence suggests that H. pylori induced inflammatory responses rather than the bacteria itself cause aberrant DNA methylation in the gastric mucosa (147). DNA hypermethylation induced by H. pylori infection was associated with down-regulation of genes involved in cell cycle progression control and DNA repair, thereby increasing the risk for gastric cancer (148). Mechanisms implicated in DNA hypermethylation during H. pylori infection include inflammation associated with the infection (134, 149) and altered expression or activity of DNA methylation related enzymes (62); as an example, IL-1β is able to induce TET2 expression in macrophages via IL-1R-Myd88 signaling (62).
Polymicrobial Infection and Sepsis
Sepsis is defined as life-threatening organ dysfunction resulting from a dysregulated host response to infection (193) and one of the leading causes of death globally (194). Sepsis is associated with changes in DNA methylation patterns in blood leukocytes of critically ill patients, and the majority of the differentially methylated region-associated genes were differentially expressed (141). Functional analysis showed that these sepsis related alterations in DNA methylation involved inflammatory pathways participating in both the innate and adaptive immune response, as well as in cell adhesion and cell junctions (141, 195). Likewise, the altered DNA methylation profiles in monocytes of septic patients correlated with increased IL-10 and IL-6 levels, as well as with organ dysfunction (143). Analysis of the CpG methylation status in blood cells of neonates with sepsis showed differential methylation of several CpGs located in functionally important genes including a group of PCDHB genes that play vital roles in leukocyte cell adhesion and the Wnt signaling pathway when compared to health (142). Another investigation indicated that the DNA methylation pattern of CpG sites in the promoter region of the calcitonin-related polypeptide α (CALCA) gene might be used as an epigenetic biomarker for bacterial sepsis in preterm newborns (196). Sepsis associated DNA methylation signatures in either specific genes or at genome-wide level have potential as diagnostic tools for predicting sepsis outcome or distinguishing sepsis subtypes. For instance, methylation of the NF-κB binding site in the Aquaporin5 (AQP5) promoter diminishes the binding of NF-κB and increased the expression of AQP5 in blood cells of septic patients is associated with substantially greater 30-day mortality (197). Similarly, DNA methylation signatures in critically ill adults can distinguish septic and nonseptic patients, and can associate with clinical traits including severity of illness, need for vasopressors, and length of stay (141). These changes in DNA methylation likely at least in part are caused by sepsis-induced changes in the levels of enzymes mediating DNA methylation, as indicated by decreased DNMT1 and increased TET2 mRNA levels in blood leukocytes of sepsis patients (144). However, de novo DNMT mRNAs (DNMT3A and DNMT3B) in extracellular vesicles in blood were much higher than in healthy controls and strongly correlated with disease severity; DNMT mRNA levels were higher in septic shock patients than in sepsis patients without shock (145). In sepsis models, the inhibition of DNA methyltransferases by Decitabine attenuated NF-κB activation, downregulated inflammatory cytokine levels, inhibited the progression of sepsis and improved survival in mice with severe sepsis induced by cecal ligation and puncture (198). The presence of TET2 impaired survival in mice with sepsis by promoting emergency myelopoiesis and a cytokine storm through oxidation of 5-mC in Socs3 mRNA resulting in destabilization of this mRNA (130). Collectively, DNA methylation could be a potential diagnostic tool or biomarker for sepsis, and manipulation of DNA methylation enzymes might be a novel strategy in the treatment of sepsis.
Specific Pathogens
Mycobacterium tuberculosis
Mycobacterium tuberculosis (MTB) infection has been reported to change DNA methylation at global level and at specific target CpGs both in vivo and in vitro. An in vitro study showed that MTB infection can lead to rapid changes in DNA methylation in non-proliferating cells, in parallel with the transcriptional response (11). Altered DNA methylation in macrophages was predominantly found at non-CpG dinucleotide sites during MTB infection (151), and the mycobacterial protein Rv2966c might be responsible for this type of DNA methylation change (183). Macrophages isolated from MTB infected patients also showed altered DNA methylation profiles of the promoter sequences of many cytokines and their receptors (152). For instance, demethylation at the promoter region of NLRP3 by MTB infection activates the NLRP3 inflammasome and increases IL-1β and IL-18 release (156). Peripheral blood mononuclear cells from TB patients are characterized by DNA hyper-methylation of genes critical to mycobacterial immunity resulting in decreased mycobacteria-specific and non-specific immune responsiveness (153). Aberrant methylation of certain CpG sites over the TLR2 promoter negatively regulated TLR2 expression in NK cells/monocytes of patients with active pulmonary TB and correlated with the bacterial burden and disease severity (154); likewise, increased DNA methylation in monocytes from tuberculosis patients was suggested to reflect disease severity (155). Collectively, these results suggest that DNA methylation profiles of leukocyte subsets might be used as clinically prognostic tools for TB.
Escherichia coli
Escherichia (E.) coli is a Gram-negative and common causative pathogen in gastroenteritis, urinary tract infection, neonatal meningitis, hemorrhagic colitis, peritonitis and pneumonia. Several studies have documented modifications of DNA methylation in host cells during E.coli infection. DNA methylation within the promoters of a core set of CD4+ T-cell pathway genes attenuated neonatal immune responses to pneumonia-induced injury (157). Yet, DNMT inhibition by 5-aza-2-deoxycytidine (DAC) augmented the number and function of regulatory T cells thereby accelerating the repair of experimental lung injury (158), suggesting that the altered DNA methylation might be caused by the changes in the abundance or activity of regulatory enzymes during E.coli infection. Moreover, E. coli induced alterations in DNA methylation are frequently accompanied by changes in the expression of genes encoding proteins that are required for controlling bacterial infection. Uropathogenic E. coli infection induces de novo methyltransferase activity and DNMT1 expression causing increased methylation of CDKN2A exon 1 and downregulation of this tumor suppressor gene in uroepithelial cells, which may increase the risk of bladder cancer (159, 160). However, downregulation of de novo methyltransferase DNMT3A by E. coli was accompanied by hypomethylation of some immune response genes in porcine mammary epithelial cells (161). Additionally, knockdown of TET1 in THP1 macrophages downregulated the activity of the NF-κB signaling pathway activated by E. coli, thus inhibiting macrophage M1 polarization (162). Avian pathogenic E. coli infection led to changes of DNA methylation at gene body regions in the spleen, which negatively correlated with the expression of genes involved in the host inflammatory response and other networks and pathways related to injury/survival (199).
Salmonella
Salmonella is the most frequently detected causative agent in foodborne outbreaks worldwide. Salmonella (S.) typhimurium and S. enteritidis are the most common serotypes associated with foodborne diseases (200). The domestic chicken is an important host of S. enterica, and some studies showed that S. enterica infection alters DNA methylation in immune and metabolism related genes in chicken cecum and blood leukocytes (163, 164). Furthermore, enhanced DNA methylation levels at the promoters of Tlr4, Tlr21 and Tlr2-1 of blood leukocytes is related to reduced expression of genes in the MyD88 signaling pathway and increased susceptibility to S. enterica infection (165, 166). Notably, although Salmonella is an important pathogen in humans, knowledge of its capacity to modify DNA methylation in human cells is lacking.
Pseudomonas aeruginosa
P. aeruginosa is one of the main causative pathogens in hospital-acquired pneumonia and chronic airway infection associated with cystic fibrosis (201). Bronchial epithelial cells (BECs) are activated by and required for host defense against P. aeruginosa infection (202). Recently P. aeruginosa was shown to inhibit NODAL expression in BECs through methylation modification of its promoter. Nodal is vital for regulating proliferation of BECs and BEC-induced differentiation of T helper (Th) cells from Th1 to Th2 and Th17, thus regulating the immunological balance of the airway microenvironment (167). DNA methylation in human lung macrophages can be modified by P. aeruginosa secreted extracellular vesicles; DNA methylation modifications particularly occurred at distal DNA regulatory elements, including enhancer regions and DNase hypersensitive sites, and some CpGs associated with cytokines such as CSF3 displayed strong negative correlations between DNA methylation and gene expression (203). DNA methylation enzymes are important for regulating host immune responses against this bacterium infection, as indicated by the association between genetic variants of DNMT3B and P. aeruginosa infection in children (168). We recently identified a role for DNMT3B in bronchial epithelial cells during P. aeruginosa pneumonia (169). DNMT3B deficient human bronchial epithelial cells produced more CXCL1 and related chemokines than control cells when stimulated with P. aeruginosa. Mechanistically, DNMT3B deficiency reduced DNA methylation at exon 1 of CXCL1 and increased NF-ĸB p65 binding to the CXCL1 promoter. These in vitro findings were corroborated by studies in mice with bronchial epithelial Dntm3b deficiency infected with viable P. aeruginosa via the airways, which showed increased Cxcl1 expression in bronchial epithelium and CXCL1 protein release together with enhanced neutrophil recruitment and accelerated bacterial clearance. Additional studies using purified flagellin (an important virulent factor expressed by Pseudomonas) and a flagellin-deficient P. aeruginosa strain demonstrated that bronchial epithelial DNMT3b impaired host defense during Pseudomonas induced pneumonia at least in part by diminishing mucosal responses to flagellin (169). In separate investigations we showed that the DNA methylation eraser TET2 maintains epithelium barrier function during acute P. aeruginosa infection in mice (204).
Burkholderia pseudomallei
B. pseudomallei is an intracellular Gram-negative pathogen causing melioidosis, a common cause of sepsis in Southeast Asia and Australia. B. pseudomallei induced changes in DNA methylation of human macrophage-like U937 cells in vitro, particularly in the vicinity of genes involved in inflammatory responses, intracellular signaling and apoptosis (205).
Methicillin-Resistant Staphylococcus aureus (MRSA)
MRSA infection significantly decreased DNMT3A in blood leukocytes in vivo and in macrophage and neutrophils in vitro. DNMT3A knockdown increased S. aureus induced IL-10 production by macrophages in vitro and pretreatment with DAC increased mortality in a S. aureus murine sepsis model. However, a DNMT3A polymorphism increased the capacity to resolve MRSA bacteremia, potentially by reducing IL-10 production though a DNA methylation dependent mechanism (170). Indeed, persistent and resolving MRSA bacteremia were associated with different DNA methylation signatures in circulating immune cells of patients, particularly in neutrophils, and this distinct DNA methylation patterns were able to predict persistent MRSA bacteremia (171).
Campylobacter rectus
Placental and fetal infection with C. rectus in mice caused hypermethylation in the promoter region of Igf2 in the placenta, resulting in down-regulation of Igf2, which affects the growth of the fetus by controlling both the placental supply of, and the genetic demand for, maternal nutrients to the fetus (172).
Porphyromonas gingivalis
P. gingivalis, the major pathogen in chronic periodontits, modifies DNMT1 expression and changes methylation at the promoter region of several genes implicated in the innate immune response against bacteria and during tissue remodeling, whilst the DNMTs inhibitor DAC restores the expression of these genes in infected gingival epithelial cells (173).
Anaplasma phagocytophilum
A. phagocytophilum is a Gram-negative bacterium with a strong tropism for neutrophils that causes human granulocytic anaplasmosis, a zoonosis transmitted by ticks. A. phagocytophilum infection induces genome-wide hypermethylation in neutrophils potentially by promoting DNMT3A expression (174). Furthermore, inhibition of DNMTs by 5-azacytidine resulted in a partially recovery of neutrophil antibacterial functions and decreased bacterial growth (174).
Bacterial Products
DNA methylation of immune cells can affect their responsiveness to microbial products, as illustrated by strong correlations between DNA methylation in human peripheral blood mononuclear cells and IL-6 production elicited by various TLR agonists (206). LPS is one of the major virulence factors of Gram-negative bacteria and the most used molecule for studying mechanisms underlying cellular immune responses. Recent evidence has indicated that changes in DNA methylation regulate LPS-induced immune responses and that modifying DNMT activity influences cellular responses to LPS (175). One way by which DNA methylation might influence LPS responsiveness is by affecting the expression of TLR4, the LPS receptor, as has been documented in intestinal epithelial cells (207). However, the most frequently reported mechanisms by which DNA methylation regulates LPS induced responses are associated with the function of DNA methylation modifiers. Increasing the methyl donor for DNA methylation by adding the S-adenosylmethionine (SAM) precursor methionine attenuated LPS-induced inflammatory responses in macrophages, whilst the DNMTs inhibitor DAC partially suppressed inflammatory responses induced by LPS in macrophages and other cell types (208, 209). Furthermore, DAC reduced lung inflammation and injury by inhibiting M1 macrophage activation in vivo (210). DNMTs were altered in bovine endometrial cells and microglia upon LPS stimulation and the expression of some inflammatory cytokines such as IL-1β, IL-6 and IL-8 were negatively regulated by methylation at their promoters (176, 177). Similarly, DNMT3B was reported to inhibit pro-inflammatory cytokine production by hypermethylation at their promoters or by downregulation of PPARγ expression (33, 211). Conversely, DNMTs mediated hypermethylation at promoters of anti-inflammatory factors, such as SOCS1, KLF4 and miR-145 – and as a consequence thereof – their downregulation, exacerbates inflammatory responses either in vivo or in vitro (178–180). The role of TET proteins in LPS induced activation of immune cells was intensively studied, revealing both inhibitory and stimulatory functions. TET1 is able to interfere with the NF-κB signaling pathway and knockdown of TET1 resulted in decreased production of proinflammatory markers by LPS/IFN-γ-induced M1 macrophages (162). TET2 functions downstream of the NF-κB signaling pathway by recruiting HDACs to the IL6 promoter resulting in reduced IL6 expression in macrophages and attenuation of inflammatory responses in murine endotoxemia model (63, 122). Besides LPS, there are few other bacterial compounds reported to affect DNA methylation in host cells. Staphylococcus aureus enterotoxin B altered the DNA methylation pattern in nasal polyp explants, most notably in IKBKB and STAT5B, genes encoding proteins with important roles in immunity (181). Likewise, peptidoglycan and lipoteichoic acid from this bacterium are able to suppress DNMT activity, resulting in enhanced inflammatory responses in bovine mammary epithelial cells (182). While the majority of bacterial compounds alter host DNA methylation by modifying the expression and activity of DNA methylation enzymes, mycobacterial protein Rv2966c by itself acts as a DNA methyltransferase that binds to host specific DNA sequences and methylates cytosines predominantly in a non-CpG context (183). Likewise, the swine pneumonia pathogen Mycoplasma hyorhinis produces Mhy1, Mhy2 and Mhy3, which can serve as mammalian DNMTs able to modify host DNA methylation (184, 185, 212). Besides bacterial components, bacterial metabolites might also affect host cell DNA methylation after uptake by these cells. For instance, folate produced by the commensal bacteria Bifidobacterium and Lactobacillus contributes to the generation of SAM resulting in increased DNMT activity and altered DNA methylation in host cells (186).
Conclusion and Perspectives
Bacterial infection can alter the DNA methylation pattern of host cells, which may represent a strategy of pathogens to modify host gene expression to avoid clearance and facilitate colonization (213, 214). Changes in DNA methylation may also contribute to short-term memory in innate immune cells (215). Most of our current understanding of DNA methylation is derived from research fields outside infection immunity, in particular cancer and developmental immunology. Whilst awareness of the crucial role of DNA methylation and the proteins involved herein in regulating host immune defense against bacterial infection has increased, much remains to be learned about the mechanisms by which bacterial infection alters host DNA methylation and how this interferes with immune responses. Additionally, compared to a broad spectrum of bacteria that can modify host DNA methylation, thus far only few bacterial components or products have been reported to alter host DNA methylation, through mechanisms that are incompletely understood. Therefore, further research is warranted to reveal which bacterial effectors and mechanisms are involved in modification of host DNA methylation in bacterial infection. Expanding our knowledge of the role of variations in the methylation of DNA in host immune cells may not only enhance our understanding of host defense and the pathogenesis of bacterial infection, but also may provide clues for the development of novel therapeutics.
Author Contributions
WQ and TP wrote the first draft of the article, with subsequent input from BC. All authors contributed to the article and approved the submitted version.
Funding
WQ is supported by a Scholarship from the Chinese Scholarship Council (CSC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Laird PW. Principles and Challenges of Genomewide DNA Methylation Analysis. Nat Rev Genet (2010) 11(3):191–203. doi: 10.1038/nrg2732
2. Jones PA. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat Rev Genet (2012) 13(7):484–92. doi: 10.1038/nrg3230
3. Jeltsch A, Ehrenhofer-Murray A, Jurkowski TP, Lyko F, Reuter G, Ankri S, et al. Mechanism and Biological Role of Dnmt2 in Nucleic Acid Methylation. RNA Biol (2017) 14(9):1108–23. doi: 10.1080/15476286.2016.1191737
4. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science (2009) 324(5929):930–5. doi: 10.1126/science.1170116
5. Rasmussen KD, Helin K. Role of TET Enzymes in DNA Methylation, Development, and Cancer. Genes Dev (2016) 30(7):733–50. doi: 10.1101/gad.276568.115
6. Lyko F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat Rev Genet (2018) 19(2):81–92. doi: 10.1038/nrg.2017.80
7. Medzhitov R. Recognition of Microorganisms and Activation of the Immune Response. Nature (2007) 449(7164):819–26. doi: 10.1038/nature06246
8. Kumar H, Kawai T, Akira S. Pathogen Recognition by the Innate Immune System. Int Rev Immunol (2011) 30(1):16–34. doi: 10.3109/08830185.2010.529976
9. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu Rev Immunol (2015) 33:257–90. doi: 10.1146/annurev-immunol-032414-112240
10. Zhang Q, Cao X. Epigenetic Regulation of the Innate Immune Response to Infection. Nat Rev Immunol (2019) 19(7):417–32. doi: 10.1038/s41577-019-0151-6
11. Pacis A, Tailleux L, Morin AM, Lambourne J, MacIsaac JL, Yotova V, et al. Bacterial Infection Remodels the DNA Methylation Landscape of Human Dendritic Cells. Genome Res (2015) 25(12):1801–11. doi: 10.1101/gr.192005.115
12. Kilin V, Gavvala K, Barthes NP, Michel BY, Shin D, Boudier C, et al. Dynamics of Methylated Cytosine Flipping by UHRF1. J Am Chem Soc (2017) 139(6):2520–8. doi: 10.1021/jacs.7b00154
13. Wu H, Zhang Y. Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions. Cell (2014) 156(1-2):45–68. doi: 10.1016/j.cell.2013.12.019
14. Bhutani N, Burns DM, Blau HM. DNA Demethylation Dynamics. Cell (2011) 146(6):866–72. doi: 10.1016/j.cell.2011.08.042
15. Bochtler M, Kolano A, Xu GL. DNA Demethylation Pathways: Additional Players and Regulators. Bioessays (2016) 39(1):1–13. doi: 10.1002/bies.201600178
16. Wu X, Zhang Y. TET-Mediated Active DNA Demethylation: Mechanism, Function and Beyond. Nat Rev Genet (2017) 18(9):517–34. doi: 10.1038/nrg.2017.33
17. Kohli RM, Zhang Y. TET Enzymes, TDG and the Dynamics of DNA Demethylation. Nature (2013) 502(7472):472–9. doi: 10.1038/nature12750
18. Jin C, Lu Y, Jelinek J, Liang S, Estecio MR, Barton MC, et al. TET1 is a Maintenance DNA Demethylase That Prevents Methylation Spreading in Differentiated Cells. Nucleic Acids Res (2014) 42(11):6956–71. doi: 10.1093/nar/gku372
19. van der Wijst MG, Venkiteswaran M, Chen H, Xu GL, Plösch T, Rots MG. Local Chromatin Microenvironment Determines DNMT Activity: From DNA Methyltransferase to DNA Demethylase or DNA Dehydroxymethylase. Epigenetics (2015) 10(8):671–6. doi: 10.1080/15592294.2015.1062204
20. Lin RK, Wang YC. Dysregulated Transcriptional and Post-Translational Control of DNA Methyltransferases in Cancer. Cell Biosci (2014) 4:46. doi: 10.1186/2045-3701-4-46
21. Bronner C. Control of DNMT1 Abundance in Epigenetic Inheritance by Acetylation, Ubiquitylation, and the Histone Code. Sci Signal (2011) 4(157):pe3. doi: 10.1126/scisignal.2001764
22. Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. DNMT1 Stability is Regulated By Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination. Sci Signal (2010) 3(146):ra80. doi: 10.1126/scisignal.2001462
23. Estève PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, et al. A Methylation and Phosphorylation Switch Between an Adjacent Lysine and Serine Determines Human DNMT1 Stability. Nat Struct Mol Biol (2011) 18(1):42–8. doi: 10.1038/nsmb.1939
24. Estève PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, et al. Regulation of DNMT1 Stability Through SET7-mediated Lysine Methylation in Mammalian Cells. Proc Natl Acad Sci USA (2009) 106(13):5076–81. doi: 10.1073/pnas.0810362106
25. Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The Lysine Demethylase LSD1 (KDM1) is Required for Maintenance of Global DNA Methylation. Nat Genet (2009) 41(1):125–9. doi: 10.1038/ng.268
26. Lee B, Muller MT. Sumoylation Enhances DNA Methyltransferase 1 Activity. Biochem J (2009) 421(3):449–61. doi: 10.1042/BJ20090142
27. Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Modification of De Novo DNA Methyltransferase 3a (Dnmt3a) by SUMO-1 Modulates its Interaction With Histone Deacetylases (Hdacs) and its Capacity to Repress Transcription. Nucleic Acids Res (2004) 32(2):598–610. doi: 10.1093/nar/gkh195
28. Fatemi M, Hermann A, Pradhan S, Jeltsch A. The Activity of the Murine DNA Methyltransferase Dnmt1 is Controlled by Interaction of the Catalytic Domain With the N-terminal Part of the Enzyme Leading to an Allosteric Activation of the Enzyme After Binding to Methylated DNA. J Mol Biol (2001) 309(5):1189–99. doi: 10.1006/jmbi.2001.4709
29. Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA Complex Reveals a Role for Autoinhibition in Maintenance DNA Methylation. Science (2011) 331(6020):1036–40. doi: 10.1126/science.1195380
30. Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, et al. Structural Insight Into Autoinhibition and Histone H3-induced Activation of DNMT3A. Nature (2015) 517(7536):640–4. doi: 10.1038/nature13899
31. Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL. The N-terminus of Histone H3 is Required for De Novo DNA Methylation in Chromatin. Proc Natl Acad Sci USA (2009) 106(52):22187–92. doi: 10.1073/pnas.0905767106
32. Manzo M, Wirz J, Ambrosi C, Villaseñor R, Roschitzki B, Baubec T. Isoform-Specific Localization of DNMT3A Regulates DNA Methylation Fidelity at Bivalent CpG Islands. EMBO J (2017) 36(23):3421–34. doi: 10.15252/embj.201797038
33. Atsumi T, Suzuki H, Jiang JJ, Okuyama Y, Nakagawa I, Ota M, et al. Rbm10 Regulates Inflammation Development Via Alternative Splicing of Dnmt3b. Int Immunol (2017) 29(12):581–91. doi: 10.1093/intimm/dxx067
34. Stathopoulou A, Chhetri JB, Ambrose JC, Estève PO, Ji L, Erdjument-Bromage H, et al. A Novel Requirement for DROSHA in Maintenance of Mammalian CG Methylation. Nucleic Acids Res (2017) 45(16):9398–412. doi: 10.1093/nar/gkx695
35. Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA Methylation Pathways and Their Crosstalk With Histone Methylation. Nat Rev Mol Cell Biol (2015) 16(9):519–32. doi: 10.1038/nrm4043
36. Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. DNMT3L Connects Unmethylated Lysine 4 of Histone H3 to De Novo Methylation of DNA. Nature (2007) 448(7154):714–7. doi: 10.1038/nature05987
37. Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, et al. The Histone Mark H3K36me2 Recruits DNMT3A and Shapes the Intergenic DNA Methylation Landscape. Nature (2019) 573(7773):281–6. doi: 10.1038/s41586-019-1534-3
38. Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, et al. Lineage-Specific Polycomb Targets and De Novo DNA Methylation Define Restriction and Potential of Neuronal Progenitors. Mol Cell (2008) 30(6):755–66. doi: 10.1016/j.molcel.2008.05.007
39. Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, et al. Intragenic DNA Methylation Prevents Spurious Transcription Initiation. Nature (2017) 543(7643):72–7. doi: 10.1038/nature21373
40. Jeziorska DM, Murray R, De Gobbi M, Gaentzsch R, Garrick D, Ayyub H, et al. DNA Methylation of Intragenic CpG Islands Depends on Their Transcriptional Activity During Differentiation and Disease. Proc Natl Acad Sci USA (2017) 114(36):E7526–7526E7535. doi: 10.1073/pnas.1703087114
41. Veland N, Lu Y, Hardikar S, Gaddis S, Zeng Y, Liu B, et al. DNMT3L Facilitates DNA Methylation Partly by Maintaining DNMT3A Stability in Mouse Embryonic Stem Cells. Nucleic Acids Res (2019) 47(1):152–67. doi: 10.1093/nar/gky947
42. Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L Stimulates the DNA Methylation Activity of Dnmt3a and Dnmt3b Through a Direct Interaction. J Biol Chem (2004) 279(26):27816–23. doi: 10.1074/jbc.M400181200
43. Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb Group Protein EZH2 Directly Controls DNA Methylation. Nature (2006) 439(7078):871–4. doi: 10.1038/nature04431
44. Kim SH, Park J, Choi MC, Kim HP, Park JH, Jung Y, et al. Zinc-Fingers and Homeoboxes 1 (ZHX1) Binds DNA Methyltransferase (DNMT) 3B to Enhance DNMT3B-mediated Transcriptional Repression. Biochem Biophys Res Commun (2007) 355(2):318–23. doi: 10.1016/j.bbrc.2007.01.187
45. Shamay M, Greenway M, Liao G, Ambinder RF, Hayward SD. De Novo DNA Methyltransferase DNMT3b Interacts With NEDD8-modified Proteins. J Biol Chem (2010) 285(47):36377–86. doi: 10.1074/jbc.M110.155721
46. Velasco G, Hubé F, Rollin J, Neuillet D, Philippe C, Bouzinba-Segard H, et al. Dnmt3b Recruitment Through E2F6 Transcriptional Repressor Mediates Germ-Line Gene Silencing in Murine Somatic Tissues. Proc Natl Acad Sci USA (2010) 107(20):9281–6. doi: 10.1073/pnas.1000473107
47. Thompson JJ, Kaur R, Sosa CP, Lee JH, Kashiwagi K, Zhou D, et al. ZBTB24 is a Transcriptional Regulator That Coordinates With DNMT3B to Control DNA Methylation. Nucleic Acids Res (2018) 46(19):10034–51. doi: 10.1093/nar/gky682
48. Li T, Garcia-Gomez A, Morante-Palacios O, Ciudad L, Özkaramehmet S, Van Dijck E, et al. SIRT1/2 Orchestrate Acquisition of DNA Methylation and Loss of Histone H3 Activating Marks to Prevent Premature Activation of Inflammatory Genes in Macrophages. Nucleic Acids Res (2020) 48(2):665–81. doi: 10.1093/nar/gkz1127
49. de la Rica L, Rodríguez-Ubreva J, García M, Islam AB, Urquiza JM, Hernando H, et al. PU.1 Target Genes Undergo Tet2-coupled Demethylation and DNMT3b-mediated Methylation in Monocyte-to-Osteoclast Differentiation. Genome Biol (2013) 14(9):R99. doi: 10.1186/gb-2013-14-9-r99
50. Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, et al. UHRF1 Targets DNMT1 for DNA Methylation Through Cooperative Binding of Hemi-Methylated DNA and Methylated H3K9. Nat Commun (2013) 4:1563. doi: 10.1038/ncomms2562
51. Wang L, Lee JY, Gao L, Yin J, Duan Y, Jimenez LA, et al. A DNA Aptamer for Binding and Inhibition of DNA Methyltransferase 1. Nucleic Acids Res (2019) 47(22):11527–37. doi: 10.1093/nar/gkz1083
52. Zhang G, Estève PO, Chin HG, Terragni J, Dai N, Corrêa IR Jr, et al. Small RNA-mediated Dna (cytosine-5) Methyltransferase 1 Inhibition Leads to Aberrant DNA Methylation. Nucleic Acids Res (2015) 43(12):6112–24. doi: 10.1093/nar/gkv518
53. Holz-Schietinger C, Reich NO. RNA Modulation of the Human DNA Methyltransferase 3A. Nucleic Acids Res (2012) 40(17):8550–7. doi: 10.1093/nar/gks537
54. Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-Interacting RNAs Block Gene-Specific DNA Methylation. Nature (2013) 503(7476):371–6. doi: 10.1038/nature12598
55. Gonda TA, Kim YI, Salas MC, Gamble MV, Shibata W, Muthupalani S, et al. Folic Acid Increases Global DNA Methylation and Reduces Inflammation to Prevent Helicobacter-associated Gastric Cancer in Mice. Gastroenterology (2012) 142(4):824–33.e7. doi: 10.1053/j.gastro.2011.12.058
56. Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate’s Role. Adv Nutr (2012) 3(1):21–38. doi: 10.3945/an.111.000992
57. Maddocks OD, Labuschagne CF, Adams PD, Vousden KH. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation Through De Novo ATP Synthesis in Cancer Cells. Mol Cell (2016) 61(2):210–21. doi: 10.1016/j.molcel.2015.12.014
58. Chen CC, Wang KY, Shen CK. The Mammalian De Novo DNA Methyltransferases DNMT3A and DNMT3B are Also DNA 5-Hydroxymethylcytosine Dehydroxymethylases. J Biol Chem (2012) 287(40):33116–21. doi: 10.1074/jbc.C112.406975
59. Liutkevičiūtė Z, Kriukienė E, Ličytė J, Rudytė M, Urbanavičiūtė G, Klimašauskas S. Direct Decarboxylation of 5-Carboxylcytosine by DNA C5-Methyltransferases. J Am Chem Soc (2014) 136(16):5884–7. doi: 10.1021/ja5019223
60. Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases Add Aldehydes to DNA. Nat Chem Biol (2009) 5(6):400–2. doi: 10.1038/nchembio.172
61. Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y, et al. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity (2015) 43(2):251–63. doi: 10.1016/j.immuni.2015.07.017
62. Pan W, Zhu S, Qu K, Meeth K, Cheng J, He K, et al. The DNA Methylcytosine Dioxygenase Tet2 Sustains Immunosuppressive Function of Tumor-Infiltrating Myeloid Cells to Promote Melanoma Progression. Immunity (2017) 47(2):284–97.e5. doi: 10.1016/j.immuni.2017.07.020
63. Cull AH, Snetsinger B, Buckstein R, Wells RA, Rauh MJ. Tet2 Restrains Inflammatory Gene Expression in Macrophages. Exp Hematol (2017) 55:56–70.e13. doi: 10.1016/j.exphem.2017.08.001
64. Yang YA, Zhao JC, Fong KW, Kim J, Li S, Song C, et al. FOXA1 Potentiates Lineage-Specific Enhancer Activation Through Modulating TET1 Expression and Function. Nucleic Acids Res (2016) 44(17):8153–64. doi: 10.1093/nar/gkw498
65. Chen JY, Luo CW, Lai YS, Wu CC, Hung WC. Lysine Demethylase KDM2A Inhibits TET2 to Promote DNA Methylation and Silencing of Tumor Suppressor Genes in Breast Cancer. Oncogenesis (2017) 6(8):e369. doi: 10.1038/oncsis.2017.71
66. Wu Y, Guo Z, Liu Y, Tang B, Wang Y, Yang L, et al. Oct4 and the Small Molecule Inhibitor, SC1, Regulates Tet2 Expression in Mouse Embryonic Stem Cells. Mol Biol Rep (2013) 40(4):2897–906. doi: 10.1007/s11033-012-2305-5
67. Kallin EM, Rodríguez-Ubreva J, Christensen J, Cimmino L, Aifantis I, Helin K, et al. Tet2 Facilitates the Derepression of Myeloid Target Genes During Cebpα-Induced Transdifferentiation of Pre-B Cells. Mol Cell (2012) 48(2):266–76. doi: 10.1016/j.molcel.2012.08.007
68. Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, et al. Modulation of TET2 Expression and 5-Methylcytosine Oxidation by the CXXC Domain Protein IDAX. Nature (2013) 497(7447):122–6. doi: 10.1038/nature12052
69. Cui Q, Yang S, Ye P, Tian E, Sun G, Zhou J, et al. Downregulation of TLX Induces TET3 Expression and Inhibits Glioblastoma Stem Cell Self-Renewal and Tumorigenesis. Nat Commun (2016) 7:10637. doi: 10.1038/ncomms10637
70. Ren S, Xu Y. Ac016405.3, a Novel Long Noncoding RNA, Acts as a Tumor Suppressor Through Modulation of TET2 by microRNA-19a-5p Sponging in Glioblastoma. Cancer Sci (2019) 110(5):1621–32. doi: 10.1111/cas.14002
71. Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J, Shi BJ, et al. MicroRNA-191 Acts as a Tumor Promoter by Modulating the TET1-p53 Pathway in Intrahepatic Cholangiocarcinoma. Hepatology (2017) 66(1):136–51. doi: 10.1002/hep.29116
72. Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, et al. The Oncogenic microRNA miR-22 Targets the TET2 Tumor Suppressor to Promote Hematopoietic Stem Cell Self-Renewal and Transformation. Cell Stem Cell (2013) 13(1):87–101. doi: 10.1016/j.stem.2013.06.003
73. Zhaolin Z, Jiaojiao C, Peng W, Yami L, Tingting Z, Jun T, et al. OxLDL Induces Vascular Endothelial Cell Pyroptosis Through miR-125a-5p/TET2 Pathway. J Cell Physiol (2019) 234(5):7475–91. doi: 10.1002/jcp.27509
74. Wang Y, Zhang Y. Regulation of TET Protein Stability by Calpains. Cell Rep (2014) 6(2):278–84. doi: 10.1016/j.celrep.2013.12.031
75. Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D’Alessio AC, et al. CRL4(Vprbp) E3 Ligase Promotes Monoubiquitylation and Chromatin Binding of TET Dioxygenases. Mol Cell (2015) 57(2):247–60. doi: 10.1016/j.molcel.2014.12.002
76. Wu D, Hu D, Chen H, Shi G, Fetahu IS, Wu F, et al. Glucose-Regulated Phosphorylation of TET2 by AMPK Reveals a Pathway Linking Diabetes to Cancer. Nature (2018) 559(7715):637–41. doi: 10.1038/s41586-018-0350-5
77. Lv L, Wang Q, Xu Y, Tsao LC, Nakagawa T, Guo H, et al. Vpr Targets TET2 for Degradation by CRL4VprBP E3 Ligase to Sustain Il-6 Expression and Enhance Hiv-1 Replication. Mol Cell (2018) 70(5):961–70.e5. doi: 10.1016/j.molcel.2018.05.007
78. Kundu A, Shelar S, Ghosh AP, Ballestas M, Kirkman R, Nam H, et al. 14-3-3 Proteins Protect AMPK-phosphorylated Ten-Eleven Translocation-2 (TET2) From PP2A-mediated Dephosphorylation. J Biol Chem (2020) 295(6):1754–66. doi: 10.1074/jbc.RA119.011089
79. Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, et al. Acetylation Enhances Tet2 Function in Protecting Against Abnormal Dna Methylation During Oxidative Stress. Mol Cell (2017) 65(2):323–35. doi: 10.1016/j.molcel.2016.12.013
80. Li X, Liu T, Wu TT, Feng Y, Peng SJ, Yin H, et al. Sirt1 Deacetylates TET2 and Promotes its Ubiquitination Degradation to Achieve Neuroprotection Against Parkinson’s Disease. Front Neurol (2021) 12:652882. doi: 10.3389/fneur.2021.652882
81. Sun J, He X, Zhu Y, Ding Z, Dong H, Feng Y, et al. Sirt1 Activation Disrupts Maintenance of Myelodysplastic Syndrome Stem and Progenitor Cells by Restoring Tet2 Function. Cell Stem Cell (2018) 23(3):355–69.e9. doi: 10.1016/j.stem.2018.07.018
82. Pastor WA, Aravind L, Rao A. Tetonic Shift: Biological Roles of TET Proteins in DNA Demethylation and Transcription. Nat Rev Mol Cell Biol (2013) 14(6):341–56. doi: 10.1038/nrm3589
83. Zeng Y, Yao B, Shin J, Lin L, Kim N, Song Q, et al. Lin28A Binds Active Promoters and Recruits Tet1 to Regulate Gene Expression. Mol Cell (2016) 61(1):153–60. doi: 10.1016/j.molcel.2015.11.020
84. Guan W, Guyot R, Samarut J, Flamant F, Wong J, Gauthier KC. Methylcytosine Dioxygenase TET3 Interacts With Thyroid Hormone Nuclear Receptors and Stabilizes Their Association to Chromatin. Proc Natl Acad Sci USA (2017) 114(31):8229–34. doi: 10.1073/pnas.1702192114
85. Aravind L, Abhiman S, Iyer LM. Natural History of the Eukaryotic Chromatin Protein Methylation System. Prog Mol Biol Transl Sci (2011) 101:105–76. doi: 10.1016/B978-0-12-387685-0.00004-4
86. Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, et al. WT1 Recruits TET2 to Regulate its Target Gene Expression and Suppress Leukemia Cell Proliferation. Mol Cell (2015) 57(4):662–73. doi: 10.1016/j.molcel.2014.12.023
87. Guilhamon P, Eskandarpour M, Halai D, Wilson GA, Feber A, Teschendorff AE, et al. Meta-Analysis of IDH-mutant Cancers Identifies EBF1 as an Interaction Partner for TET2. Nat Commun (2013) 4:2166. doi: 10.1038/ncomms3166
88. Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, et al. PRDM14 Promotes Active DNA Demethylation Through the Ten-Eleven Translocation (TET)-Mediated Base Excision Repair Pathway in Embryonic Stem Cells. Development (2014) 141(2):269–80. doi: 10.1242/dev.099622
89. Suzuki T, Shimizu Y, Furuhata E, Maeda S, Kishima M, Nishimura H, et al. RUNX1 Regulates Site Specificity of DNA Demethylation by Recruitment of DNA Demethylation Machineries in Hematopoietic Cells. Blood Adv (2017) 1(20):1699–711. doi: 10.1182/bloodadvances.2017005710
90. Hassan HM, Kolendowski B, Isovic M, Bose K, Dranse HJ, Sampaio AV, et al. Regulation of Active Dna Demethylation Through RAR-Mediated Recruitment of a TET/TDG Complex. Cell Rep (2017) 19(8):1685–97. doi: 10.1016/j.celrep.2017.05.007
91. Chen LL, Lin HP, Zhou WJ, He CX, Zhang ZY, Cheng ZL, et al. Snip1 Recruits TET2 to Regulate C-MYC Target Genes and Cellular Dna Damage Response. Cell Rep (2018) 25(6):1485–500.e4. doi: 10.1016/j.celrep.2018.10.028
92. Zhou L, Ren M, Zeng T, Wang W, Wang X, Hu M, et al. TET2-Interacting Long Noncoding RNA Promotes Active DNA Demethylation of the MMP-9 Promoter in Diabetic Wound Healing. Cell Death Dis (2019) 10(11):813. doi: 10.1038/s41419-019-2047-6
93. Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell (2018) 23(5):727–41.e9. doi: 10.1016/j.stem.2018.08.016
94. Peng L, Li Y, Xi Y, Li W, Li J, Lv R, et al. MBD3L2 Promotes Tet2 Enzymatic Activity for Mediating 5-Methylcytosine Oxidation. J Cell Sci (2016) 129(5):1059–71. doi: 10.1242/jcs.179044
95. Ma S, Wan X, Deng Z, Shi L, Hao C, Zhou Z, et al. Epigenetic Regulator CXXC5 Recruits DNA Demethylase Tet2 to Regulate TLR7/9-elicited IFN Response in Pdcs. J Exp Med (2017) 214(5):1471–91. doi: 10.1084/jem.20161149
96. Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, et al. Cooperative Action Between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Mol Cell (2016) 64(5):913–25. doi: 10.1016/j.molcel.2016.10.013
97. Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-Binding Factors Shape the Mouse Methylome at Distal Regulatory Regions. Nature (2011) 480(7378):490–5. doi: 10.1038/nature10716
98. Moen EL, Mariani CJ, Zullow H, Jeff-Eke M, Litwin E, Nikitas JN, et al. New Themes in the Biological Functions of 5-Methylcytosine and 5-Hydroxymethylcytosine. Immunol Rev (2015) 263(1):36–49. doi: 10.1111/imr.12242
99. Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of α-KG-dependent Histone and DNA Demethylases by Fumarate and Succinate That are Accumulated in Mutations of FH and SDH Tumor Suppressors. Genes Dev (2012) 26(12):1326–38. doi: 10.1101/gad.191056.112
100. Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell (2010) 18(6):553–67. doi: 10.1016/j.ccr.2010.11.015
101. Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations is a Neomorphic Enzyme Activity Converting Alpha-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell (2010) 17(3):225–34. doi: 10.1016/j.ccr.2010.01.020
102. Thienpont B, Steinbacher J, Zhao H, D’Anna F, Kuchnio A, Ploumakis A, et al. Tumour Hypoxia Causes DNA Hypermethylation by Reducing TET Activity. Nature (2016) 537(7618):63–8. doi: 10.1038/nature19081
103. Minor EA, Court BL, Young JI, Wang G. Ascorbate Induces Ten-Eleven Translocation (Tet) Methylcytosine Dioxygenase-Mediated Generation of 5-Hydroxymethylcytosine. J Biol Chem (2013) 288(19):13669–74. doi: 10.1074/jbc.C113.464800
104. Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, et al. Vitamin C Induces Tet-dependent DNA Demethylation and a Blastocyst-Like State in ES Cells. Nature (2013) 500(7461):222–6. doi: 10.1038/nature12362
105. Shenoy N, Bhagat TD, Cheville J, Lohse C, Bhattacharyya S, Tischer A, et al. Ascorbic Acid-Induced TET Activation Mitigates Adverse Hydroxymethylcytosine Loss in Renal Cell Carcinoma. J Clin Invest (2019) 130:1612–25. doi: 10.1172/JCI98747
106. Rao VK, Swarnaseetha A, Tham GH, Lin WQ, Han BB, Benoukraf T, et al. Phosphorylation of Tet3 by Cdk5 is Critical for Robust Activation of BRN2 During Neuronal Differentiation. Nucleic Acids Res (2020) 48(3):1225–38. doi: 10.1093/nar/gkz1144
107. Bauer C, Göbel K, Nagaraj N, Colantuoni C, Wang M, Müller U, et al. Phosphorylation of TET Proteins is Regulated Via O-GlcNAcylation by the O-linked N-Acetylglucosamine Transferase (OGT). J Biol Chem (2015) 290(8):4801–12. doi: 10.1074/jbc.M114.605881
108. Schübeler D. Function and Information Content of DNA Methylation. Nature (2015) 517(7534):321–6. doi: 10.1038/nature14192
109. Anastasiadi D, Esteve-Codina A, Piferrer F. Consistent Inverse Correlation Between DNA Methylation of the First Intron and Gene Expression Across Tissues and Species. Epigenet Chromatin (2018) 11(1):37. doi: 10.1186/s13072-018-0205-1
110. Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene Body Methylation can Alter Gene Expression and is a Therapeutic Target in Cancer. Cancer Cell (2014) 26(4):577–90. doi: 10.1016/j.ccr.2014.07.028
111. Cedar H, Bergman Y. Linking DNA Methylation and Histone Modification: Patterns and Paradigms. Nat Rev Genet (2009) 10(5):295–304. doi: 10.1038/nrg2540
112. Lio CW, Zhang J, González-Avalos E, Hogan PG, Chang X, Rao A. Tet2 and Tet3 Cooperate With B-lineage Transcription Factors to Regulate DNA Modification and Chromatin Accessibility. Elife (2016) 5:e18290. doi: 10.7554/eLife.18290
113. Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the Early B-cell-specific Mb-1 (Ig-Alpha) Gene by Pax-5 is Dependent on an Unmethylated Ets Binding Site. Mol Cell Biol (2003) 23(6):1946–60. doi: 10.1128/MCB.23.6.1946-1960.2003
114. Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional Repression by the methyl-CpG-binding Protein MeCP2 Involves a Histone Deacetylase Complex. Nature (1998) 393(6683):386–9. doi: 10.1038/30764
115. Pacis A, Mailhot-Léonard F, Tailleux L, Randolph HE, Yotova V, Dumaine A, et al. Gene Activation Precedes DNA Demethylation in Response to Infection in Human Dendritic Cells. Proc Natl Acad Sci USA (2019) 116(14):6938–43. doi: 10.1073/pnas.1814700116
116. Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K, et al. Methyltransferase Dnmt3a Upregulates HDAC9 to Deacetylate the Kinase TBK1 for Activation of Antiviral Innate Immunity. Nat Immunol (2016) 17(7):806–15. doi: 10.1038/ni.3464
117. Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, et al. Dnmt3a-dependent Nonpromoter DNA Methylation Facilitates Transcription of Neurogenic Genes. Science (2010) 329(5990):444–8. doi: 10.1126/science.1190485
118. Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, et al. Genomic Profiling of DNA Methyltransferases Reveals a Role for DNMT3B in Genic Methylation. Nature (2015) 520(7546):243–7. doi: 10.1038/nature14176
119. Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 Forms a Complex With Rb, E2F1 and HDAC1 and Represses Transcription From E2F-responsive Promoters. Nat Genet (2000) 25:338–42. doi: 10.1038/77124
120. Shen J, Wang C, Li D, Xu T, Myers J, Ashton JM, et al. DNA Methyltransferase 3b Regulates Articular Cartilage Homeostasis by Altering Metabolism. JCI Insight (2017) 2:e93612. doi: 10.1172/jci.insight.93612
121. Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, et al. Global Loss of DNA Methylation Uncovers Intronic Enhancers in Genes Showing Expression Changes. Genome Biol (2014) 15:469. doi: 10.1186/s13059-014-0469-0
122. Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is Required to Resolve Inflammation by Recruiting Hdac2 to Specifically Repress IL-6. Nature (2015) 525:389–93. doi: 10.1038/nature15252
123. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal Hematopoiesis Associated With TET2 Deficiency Accelerates Atherosclerosis Development in Mice. Science (2017) 355:842–7. doi: 10.1126/science.aag1381
124. Xue S, Liu C, Sun X, Li W, Zhang C, Zhou X, et al. Tet3 Inhibits Type I Ifn Production Independent of DNA Demethylation. Cell Rep (2016) 16:1096–105. doi: 10.1016/j.celrep.2016.06.068
125. Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual Functions of Tet1 in Transcriptional Regulation in Mouse Embryonic Stem Cells. Nature (2011) 473:389–93. doi: 10.1038/nature09934
126. Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and Hydroxymethylcytosine in Transcription and DNA Methylation Fidelity. Nature (2011) 473:343–8. doi: 10.1038/nature10066
127. Neves-Costa A, Moita LF. TET1 is a Negative Transcriptional Regulator of IL-1β in the THP-1 Cell Line. Mol Immunol (2013) 54:264–70. doi: 10.1016/j.molimm.2012.12.014
128. Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 Promotes Histone O-GlcNAcylation During Gene Transcription. Nature (2013) 493:561–4. doi: 10.1038/nature11742
129. Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, et al. TET2 and TET3 Regulate GlcNAcylation and H3K4 Methylation Through OGT and SET1/COMPASS. EMBO J (2013) 32:645–55. doi: 10.1038/emboj.2012.357
130. Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y, et al. Tet2 Promotes Pathogen Infection-Induced Myelopoiesis Through mRNA Oxidation. Nature (2018) 554:123–7. doi: 10.1038/nature25434
131. Guallar D, Bi X, Pardavila JA, Huang X, Saenz C, Shi X, et al. RNA-Dependent Chromatin Targeting of TET2 for Endogenous Retrovirus Control in Pluripotent Stem Cells. Nat Genet (2018) 50:443–51. doi: 10.1038/s41588-018-0060-9
132. Bierne H, Hamon M, Cossart P. Epigenetics and Bacterial Infections. Cold Spring Harb Perspect Med (2012) 2:a010272. doi: 10.1101/cshperspect.a010272
133. Takahashi K. Influence of Bacteria on Epigenetic Gene Control. Cell Mol Life Sci (2014) 71:1045–54. doi: 10.1007/s00018-013-1487-x
134. Hur K, Niwa T, Toyoda T, Tsukamoto T, Tatematsu M, Yang HK, et al. Insufficient Role of Cell Proliferation in Aberrant DNA Methylation Induction and Involvement of Specific Types of Inflammation. Carcinogenesis (2011) 32:35–41. doi: 10.1093/carcin/bgq219
135. Pan WH, Sommer F, Falk-Paulsen M, Ulas T, Best P, Fazio A, et al. Exposure to the Gut Microbiota Drives Distinct Methylome and Transcriptome Changes in Intestinal Epithelial Cells During Postnatal Development. Genome Med (2018) 10:27. doi: 10.1186/s13073-018-0534-5
136. Pan X, Gong D, Nguyen DN, Zhang X, Hu Q, Lu H, et al. Early Microbial Colonization Affects DNA Methylation of Genes Related to Intestinal Immunity and Metabolism in Preterm Pigs. DNA Res (2018) 25:287–96. doi: 10.1093/dnares/dsy001
137. Ansari I, Raddatz G, Gutekunst J, Ridnik M, Cohen D, Abu-Remaileh M, et al. The Microbiota Programs DNA Methylation to Control Intestinal Homeostasis and Inflammation. Nat Microbiol (2020) 5:610–9. doi: 10.1038/s41564-019-0659-3
138. Takahashi K, Sugi Y, Nakano K, Tsuda M, Kurihara K, Hosono A, et al. Epigenetic Control of the Host Gene by Commensal Bacteria in Large Intestinal Epithelial Cells. J Biol Chem (2011) 286:35755–62. doi: 10.1074/jbc.M111.271007
139. Tolg C, Bägli DJ. Uropathogenic Escherichia Coli Infection: Potential Importance of Epigenetics. Epigenomics (2012) 4:229–35. doi: 10.2217/epi.12.5
140. Meisel M, Hinterleitner R, Pacis A, Chen L, Earley ZM, Mayassi T, et al. Microbial Signals Drive Pre-Leukaemic Myeloproliferation in a Tet2-deficient Host. Nature (2018) 557:580–4. doi: 10.1038/s41586-018-0125-z
141. Binnie A, Walsh CJ, Hu P, Dwivedi DJ, Fox-Robichaud A, Liaw PC, et al. Epigenetic Profiling in Severe Sepsis: A Pilot Study of DNA Methylation Profiles in Critical Illness. Crit Care Med (2020) 48:142–50. doi: 10.1097/CCM.0000000000004097
142. Dhas DB, Ashmi AH, Bhat BV, Kalaivani S, Parija SC. Comparison of Genomic DNA Methylation Pattern Among Septic and non-Septic Newborns - An Epigenome Wide Association Study. Genom Data (2015) 3:36–40. doi: 10.1016/j.gdata.2014.11.004
143. Lorente-Sorolla C, Garcia-Gomez A, Català-Moll F, Toledano V, Ciudad L, Avendaño-Ortiz J, et al. Inflammatory Cytokines and Organ Dysfunction Associate With the Aberrant DNA Methylome of Monocytes in Sepsis. Genome Med (2019) 11:66. doi: 10.1186/s13073-019-0674-2
144. Hopp L, Loeffler-Wirth H, Nersisyan L, Arakelyan A, Binder H. Footprints of Sepsis Framed Within Community Acquired Pneumonia in the Blood Transcriptome. Front Immunol (2018) 9:1620. doi: 10.3389/fimmu.2018.01620
145. Dakhlallah DA, Wisler J, Gencheva M, Brown CM, Leatherman ER, Singh K, et al. Circulating Extracellular Vesicle Content Reveals De Novo DNA Methyltransferase Expression as a Molecular Method to Predict Septic Shock. J Extracell Vesicles (2019) 8:1669881. doi: 10.1080/20013078.2019.1669881
146. Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, et al. High Levels of Aberrant DNA Methylation in Helicobacter Pylori-Infected Gastric Mucosae and its Possible Association With Gastric Cancer Risk. Clin Cancer Res (2006) 12:989–95. doi: 10.1158/1078-0432.CCR-05-2096
147. Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory Processes Triggered by Helicobacter Pylori Infection Cause Aberrant DNA Methylation in Gastric Epithelial Cells. Cancer Res (2010) 70:1430–40. doi: 10.1158/0008-5472.CAN-09-2755
148. Nakajima T, Enomoto S, Yamashita S, Ando T, Nakanishi Y, Nakazawa K, et al. Persistence of a Component of DNA Methylation in Gastric Mucosae After Helicobacter Pylori Eradication. J Gastroenterol (2010) 45:37–44. doi: 10.1007/s00535-009-0142-7
149. Ushijima T, Hattori N. Molecular Pathways: Involvement of Helicobacter Pylori-Triggered Inflammation in the Formation of an Epigenetic Field Defect, and its Usefulness as Cancer Risk and Exposure Markers. Clin Cancer Res (2012) 18:923–9. doi: 10.1158/1078-0432.CCR-11-2011
150. Jiang J, Jia Z, Cao D, Jin MS, Kong F, Suo J, et al. Polymorphisms of the DNA Methyltransferase 1 Associated With Reduced Risks of Helicobacter Pylori Infection and Increased Risks of Gastric Atrophy. PloS One (2012) 7:e46058. doi: 10.1371/journal.pone.0046058
151. Sharma G, Sowpati DT, Singh P, Khan MZ, Ganji R, Upadhyay S, et al. Genome-Wide non-CpG Methylation of the Host Genome During M. Tuberculosis Infect Sci Rep (2016) 6:25006. doi: 10.1038/srep25006
152. Zheng L, Leung ET, Wong HK, Lui G, Lee N, To KF, et al. Unraveling Methylation Changes of Host Macrophages in Mycobacterium Tuberculosis Infection. Tuberculosis (Edinb) (2016) 98:139–48. doi: 10.1016/j.tube.2016.03.003
153. DiNardo A, Rajapakshe K, Nishiguchi T, Mtetwa G, Grimm SL, Dlamini Q, et al. DNA Hyper-Methylation During Tuberculosis Dampens Host Immune Responsiveness. J Clin Invest (2020) 130(6):3113–23. doi: 10.1172/JCI134622
154. Chen YC, Hsiao CC, Chen CJ, Chao TY, Leung SY, Liu SF, et al. Aberrant Toll-like Receptor 2 Promoter Methylation in Blood Cells From Patients With Pulmonary Tuberculosis. J Infect (2014) 69:546–57. doi: 10.1016/j.jinf.2014.08.014
155. Frantz FG, Castro RC, Fontanari C, Bollela VR. Dna Methylation Impairs Monocyte Function in Tuberculosis Leading to Disease Progression. Am Assoc Immnol (2019) 202(1 Supplement):125.10.
156. Wei M, Wang L, Wu T, Xi J, Han Y, Yang X, et al. Nlrp3 Activation was Regulated by DNA Methylation Modification During Mycobacterium Tuberculosis Infection. BioMed Res Int (2016) 2016:4323281. doi: 10.1155/2016/4323281
157. McGrath-Morrow SA, Ndeh R, Helmin KA, Chen SY, Anekalla KR, Abdala-Valencia H, et al. DNA Methylation Regulates the Neonatal CD4+ T-Cell Response to Pneumonia in Mice. J Biol Chem (2018) 293:11772–83. doi: 10.1074/jbc.RA118.003589
158. Singer BD, Mock JR, Aggarwal NR, Garibaldi BT, Sidhaye VK, Florez MA, et al. Regulatory T Cell DNA Methyltransferase Inhibition Accelerates Resolution of Lung Inflammation. Am J Respir Cell Mol Biol (2015) 52:641–52. doi: 10.1165/rcmb.2014-0327OC
159. Tolg C, Sabha N, Cortese R, Panchal T, Ahsan A, Soliman A, et al. Uropathogenic E. Coli Infection Provokes Epigenetic Downregulation of CDKN2A (p16INK4A) in Uroepithelial Cells. Lab Invest (2011) 91:825–36. doi: 10.1038/labinvest.2010.197
160. Sabha N, Aitken K, Toelg C, Panchal T, Bagli D. Increased Dnmt1 Expression And Activity in Uroepithelial Cells Following Uropathogenig E.Coli Infection. J Pediatr Urol (2009) 5:S21–2. doi: 10.1016/j.jpurol.2009.02.011
161. Sajjanar B, Trakooljul N, Wimmers K, Ponsuksili S. DNA Methylation Analysis of Porcine Mammary Epithelial Cells Reveals Differentially Methylated Loci Associated With Immune Response Against Escherichia Coli Challenge. BMC Genomics (2019) 20:623. doi: 10.1186/s12864-019-5976-7
162. Huang Y, Tian C, Li Q, Xu Q. Tet1 Knockdown Inhibits Porphyromonas Gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization Through the NF-κb Pathway in THP-1 Cells. Int J Mol Sci (2019) 20:2023. doi: 10.3390/ijms20082023
163. Wang Y, Liu L, Li M, Lin L, Su P, Tang H, et al. Chicken Cecal DNA Methylome Alteration in the Response to Salmonella Enterica Serovar Enteritidis Inoculation. BMC Genomics (2020) 21(1):814. doi: 10.1186/s12864-020-07174-w
164. Wang F, Li J, Li Q, Liu R, Zheng M, Wang Q, et al. Changes of Host DNA Methylation in Domestic Chickens Infected With Salmonella Enterica. J Genet (2017) 96(4):545–50. doi: 10.1007/s12041-017-0818-3
165. Li X, Zhang P, Jiang X, Du H, Yang C, Zhang Z, et al. Differences in Expression of Genes in the MyD88 and TRIF Signalling Pathways and Methylation of TLR4 and TRIF in Tibetan Chickens and DaHeng S03 Chickens Infected With Salmonella Enterica Serovar Enteritidis. Vet Immunol Immunopathol (2017) 189:28–35. doi: 10.1016/j.vetimm.2017.05.003
166. Gou Z, Liu R, Zhao G, Zheng M, Li P, Wang H, et al. Epigenetic Modification of TLRs in Leukocytes is Associated With Increased Susceptibility to Salmonella Enteritidis in Chickens. PloS One (2012) 7(3):e33627. doi: 10.1371/journal.pone.0033627
167. Wang L, Wu G, Qin X, Ma Q, Zhou Y, Liu S, et al. Expression of Nodal on Bronchial Epithelial Cells Influenced by Lung Microbes Through Dna Methylation Modulates the Differentiation of T-Helper Cells. Cell Physiol Biochem (2015) 37:2012–22. doi: 10.1159/000438561
168. Asgari S, McLaren PJ, Peake J, Wong M, Wong R, Bartha I, et al. Exome Sequencing Reveals Primary Immunodeficiencies in Children With Community-Acquired Pseudomonas Aeruginosa Sepsis. Front Immunol (2016) 7:357. doi: 10.3389/fimmu.2016.00447
169. Qin W, Brands X, van’t Veer C, de Vos AF, Sirard JC, Roelofs JJTH, et al. Bronchial Epithelial DNA Methyltransferase 3b Dampens Pulmonary Immune Responses During Pseudomonas Aeruginosa Infection. PloS Pathog (2021) 17:e1009491. doi: 10.1371/journal.ppat.1009491
170. Mba Medie F, Sharma-Kuinkel BK, Ruffin F, Chan LC, Rossetti M, Chang YL, et al. Genetic Variation of DNA methyltransferase-3A Contributes to Protection Against Persistent MRSA Bacteremia in Patients. Proc Natl Acad Sci USA (2019) 116:20087–96. doi: 10.1073/pnas.1909849116
171. Chang YL, Rossetti M, Gjertson DW, Rubbi L, Thompson M, Montoya DJ, et al. Human DNA Methylation Signatures Differentiate Persistent From Resolving MRSA Bacteremia. Proc Natl Acad Sci USA (2021) 118:e2000663118. doi: 10.1073/pnas.2000663118
172. Bobetsis YA, Barros SP, Lin DM, Weidman JR, Dolinoy DC, Jirtle RL, et al. Bacterial Infection Promotes DNA Hypermethylation. J Dent Res (2007) 86:169–74. doi: 10.1177/154405910708600212
173. Yin L, Chung WO. Epigenetic Regulation of Human β-Defensin 2 and CC Chemokine Ligand 20 Expression in Gingival Epithelial Cells in Response to Oral Bacteria. Mucosal Immunol (2011) 4:409–19. doi: 10.1038/mi.2010.83
174. Sinclair SH, Yegnasubramanian S, Dumler JS. Global DNA Methylation Changes and Differential Gene Expression in Anaplasma Phagocytophilum-Infected Human Neutrophils. Clin Epigenet (2015) 7:77. doi: 10.1186/s13148-015-0105-1
175. Korkmaz FT, Kerr DE. Genome-Wide Methylation Analysis Reveals Differentially Methylated Loci That are Associated With an Age-Dependent Increase in Bovine Fibroblast Response to LPS. BMC Genomics (2017) 18:405. doi: 10.1186/s12864-017-3796-1
176. Wang J, Yan X, Nesengani LT, Ding H, Yang L, Lu W. LPS-Induces IL-6 and IL-8 Gene Expression in Bovine Endometrial Cells “Through DNA Methylation”. Gene (2018) 677:266–72. doi: 10.1016/j.gene.2018.07.074
177. Matt SM, Lawson MA, Johnson RW. Aging and Peripheral Lipopolysaccharide can Modulate Epigenetic Regulators and Decrease IL-1β Promoter DNA Methylation in Microglia. Neurobiol Aging (2016) 47:1–9. doi: 10.1016/j.neurobiolaging.2016.07.006
178. Cheng C, Huang C, Ma TT, Bian EB, He Y, Zhang L, et al. SOCS1 Hypermethylation Mediated by DNMT1 is Associated With Lipopolysaccharide-Induced Inflammatory Cytokines in Macrophages. Toxicol Lett (2014) 225:488–97. doi: 10.1016/j.toxlet.2013.12.023
179. Tang RZ, Zhu JJ, Yang FF, Zhang YP, Xie SA, Liu YF, et al. DNA Methyltransferase 1 and Krüppel-like Factor 4 Axis Regulates Macrophage Inflammation and Atherosclerosis. J Mol Cell Cardiol (2019) 128:11–24. doi: 10.1016/j.yjmcc.2019.01.009
180. Ma F, Li Z, Cao J, Kong X, Gong G. A TGFBR2/SMAD2/DNMT1/miR-145 Negative Regulatory Loop is Responsible for LPS-induced Sepsis. BioMed Pharmacother (2019) 112:108626. doi: 10.1016/j.biopha.2019.108626
181. Pérez-Novo CA, Zhang Y, Denil S, Trooskens G, De Meyer T, Van Criekinge W, et al. Staphylococcal Enterotoxin B Influences the DNA Methylation Pattern in Nasal Polyp Tissue: A Preliminary Study. Allergy Asthma Clin Immunol (2013) 9:48. doi: 10.1186/1710-1492-9-48
182. Wu Y, Chen J, Sun Y, Dong X, Wang Z, Chen J, et al. PGN and LTA From Staphylococcus Aureus Induced Inflammation and Decreased Lactation Through Regulating Dna Methylation and Histone H3 Acetylation in Bovine Mammary Epithelial Cells. Toxins (Basel) (2020) 12:238. doi: 10.3390/toxins12040238
183. Sharma G, Upadhyay S, Srilalitha M, Nandicoori VK, Khosla S. The Interaction of Mycobacterial Protein Rv2966c With Host Chromatin is Mediated Through non-CpG Methylation and Histone H3/H4 Binding. Nucleic Acids Res (2015) 43:3922–37. doi: 10.1093/nar/gkv261
184. Bierne H, Pourpre R. Bacterial Factors Targeting the Nucleus: The Growing Family of Nucleomodulins. Toxins (Basel) (2020) 12(4):220. doi: 10.3390/toxins12040220
185. Chernov AV, Reyes L, Xu Z, Gonzalez B, Golovko G, Peterson S, et al. Mycoplasma CG- and GATC-specific DNA Methyltransferases Selectively and Efficiently Methylate the Host Genome and Alter the Epigenetic Landscape in Human Cells. Epigenetics (2015) 10(4):303–18. doi: 10.1080/15592294.2015.1020000
186. Qin Y, Wade PA. Crosstalk Between the Microbiome and Epigenome: Messages From Bugs. J Biochem (2018) 163(2):105–12. doi: 10.1093/jb/mvx080
187. Ye J, Wu W, Li Y, Li L. Influences of the Gut Microbiota on DNA Methylation and Histone Modification. Dig Dis Sci (2017) 62:1155–64. doi: 10.1007/s10620-017-4538-6
188. Yu Q, Jia A, Li Y, Bi Y, Liu G. Microbiota Regulate the Development and Function of the Immune Cells. Int Rev Immunol (2018) 37:79–89. doi: 10.1080/08830185.2018.1429428
189. Peterson AJ, Menheniott TR, O’Connor L, Walduck AK, Fox JG, Kawakami K, et al. Helicobacter Pylori Infection Promotes Methylation and Silencing of Trefoil Factor 2, Leading to Gastric Tumor Development in Mice and Humans. Gastroenterology (2010) 139:2005–17. doi: 10.1053/j.gastro.2010.08.043
190. Sepulveda AR, Yao Y, Yan W, Park DI, Kim JJ, Gooding W, et al. Cpg Methylation and Reduced Expression of O6-methylguanine DNA Methyltransferase is Associated With Helicobacter Pylori Infection. Gastroenterology (2010) 138:1836–44. doi: 10.1053/j.gastro.2009.12.042
191. Wang Y, Huang LH, Xu CX, Xiao J, Zhou L, Cao D, et al. Connexin 32 and 43 Promoter Methylation in Helicobacter Pylori-Associated Gastric Tumorigenesis. World J Gastroenterol (2014) 20:11770–9. doi: 10.3748/wjg.v20.i33.11770
192. Niwa T, Toyoda T, Tsukamoto T, Mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter Pylori-Induced Gastric Cancers in Gerbils by a DNA Demethylating Agent. Cancer Prev Res (Phila) (2013) 6:263–70. doi: 10.1158/1940-6207.CAPR-12-0369
193. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA (2016) 315:801–10. doi: 10.1001/jama.2016.0287
194. Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and Septic Shock. Lancet (2018) 392:75–87. doi: 10.1016/S0140-6736(18)30696-2
195. Lorente-Pozo S, Navarrete P, Garzón MJ, Lara-Cantón I, Beltrán-García J, Osca-Verdegal R, et al. Dna Methylation Analysis to Unravel Altered Genetic Pathways Underlying Early Onset and Late Onset Neonatal Sepsis. A Pilot Study Front Immunol (2021) 12:622599. doi: 10.3389/fimmu.2021.622599
196. Tendl KA, Schulz SM, Mechtler TP, Bohn A, Metz T, Greber-Platzer S, et al. DNA Methylation Pattern of CALCA in Preterm Neonates With Bacterial Sepsis as a Putative Epigenetic Biomarker. Epigenetics (2013) 8:1261–7. doi: 10.4161/epi.26645
197. Rump K, Unterberg M, Dahlke A, Nowak H, Koos B, Bergmann L, et al. DNA Methylation of a NF-κb Binding Site in the Aquaporin 5 Promoter Impacts on Mortality in Sepsis. Sci Rep (2019) 9:18511. doi: 10.1038/s41598-019-55051-8
198. Cao L, Zhu T, Lang X, Jia S, Yang Y, Zhu C, et al. Inhibiting DNA Methylation Improves Survival in Severe Sepsis by Regulating Nf-κb Pathway. Front Immunol (2020) 11:1360. doi: 10.3389/fimmu.2020.01360
199. Xu H, Zhu X, Hu Y, Li Z, Zhang X, Nie Q, et al. Corrigendum: DNA Methylome in Spleen of Avian Pathogenic Escherichia Coli-Challenged Broilers and Integration With mRNA Expression. Sci Rep (2015) 5:8303. doi: 10.1038/srep08303
200. Shippy DC, Bearson BL, Holman DB, Brunelle BW, Allen HK, Bearson S. Porcine Response to a Multidrug-Resistant Salmonella Enterica Serovar I 4,[5],12:I:- Outbreak Isolate. Foodborne Pathog Dis (2018) 15(5):253–61. doi: 10.1089/fpd.2017.2378
201. Fujitani S, Sun HY, Yu VL, Weingarten JA. Pneumonia Due to Pseudomonas Aeruginosa: Part I: Epidemiology, Clinical Diagnosis, and Source. Chest (2011) 139:909–19. doi: 10.1378/chest.10-0166
202. Anas AA, van Lieshout MH, Claushuis TA, de Vos AF, Florquin S, de Boer OJ, et al. Lung Epithelial MyD88 Drives Early Pulmonary Clearance of Pseudomonas Aeruginosa by a Flagellin Dependent Mechanism. Am J Physiol Lung Cell Mol Physiol (2016) 311:L219–28. doi: 10.1152/ajplung.00078.2016
203. Kyung Lee M, Armstrong DA, Hazlett HF, Dessaint JA, Mellinger DL, Aridgides DS, et al. Exposure to Extracellular Vesicles From Pseudomonas Aeruginosa Result in Loss of DNA Methylation at Enhancer and DNase Hypersensitive Site Regions in Lung Macrophages. Epigenetics (2020), 1–14. doi: 10.1080/15592294.2020.1853318
204. Qin W, Brands X, van ‘t Veer C, de Vos AF, Scicluna BP, van der Poll T. Bronchial Epithelial Tet2 Maintains Epithelial Integrity During Acute Pseudomonas Aeruginosa Pneumonia. Infect Immun (2020) 89:e00603–20. doi: 10.1128/IAI.00603-20
205. Cizmeci D, Dempster EL, Champion OL, Wagley S, Akman OE, Prior JL, et al. Mapping Epigenetic Changes to the Host Cell Genome Induced by Burkholderia Pseudomallei Reveals Pathogen-Specific and Pathogen-Generic Signatures of Infection. Sci Rep (2016) 6:30861. doi: 10.1038/srep30861
206. Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, et al. Factors Underlying Variable DNA Methylation in a Human Community Cohort. Proc Natl Acad Sci USA (2012) 109 (Suppl 2):17253–60. doi: 10.1073/pnas.1121249109
207. Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic Regulation of TLR4 Gene Expression in Intestinal Epithelial Cells for the Maintenance of Intestinal Homeostasis. J Immunol (2009) 183:6522–9. doi: 10.4049/jimmunol.0901271
208. Ji J, Xu Y, Zheng M, Luo C, Lei H, Qu H, et al. Methionine Attenuates Lipopolysaccharide-Induced Inflammatory Responses Via DNA Methylation in Macrophages. ACS Omega (2019) 4:2331–6. doi: 10.1021/acsomega.8b03571
209. Green BB, Kerr DE. Epigenetic Contribution to Individual Variation in Response to Lipopolysaccharide in Bovine Dermal Fibroblasts. Vet Immunol Immunopathol (2014) 157:49–58. doi: 10.1016/j.vetimm.2013.10.015
210. Thangavel J, Malik AB, Elias HK, Rajasingh S, Simpson AD, Sundivakkam PK, et al. Combinatorial Therapy With Acetylation and Methylation Modifiers Attenuates Lung Vascular Hyperpermeability in Endotoxemia-Induced Mouse Inflammatory Lung Injury. Am J Pathol (2014) 184:2237–49. doi: 10.1016/j.ajpath.2014.05.008
211. Lei C, Jiao Y, He B, Wang G, Wang Q, Wang J. RIP140 Down-Regulation Alleviates Acute Lung Injury Via the Inhibition of LPS-induced Pparγ Promoter Methylation. Pulm Pharmacol Ther (2016) 37:57–64. doi: 10.1016/j.pupt.2016.02.001
212. Hanford HE, Von Dwingelo J, Abu Kwaik Y. Bacterial Nucleomodulins: A Coevolutionary Adaptation to the Eukaryotic Command Center. PloS Pathog (2021) 17(1):e1009184. doi: 10.1371/journal.ppat.1009184
213. Pérez-Novo CA, Bachert C. DNA Methylation, Bacteria and Airway Inflammation: Latest Insights. Curr Opin Allergy Clin Immunol (2015) 15:27–32. doi: 10.1097/ACI.0000000000000130
214. Morandini AC, Santos CF, Yilmaz Ö. Role of Epigenetics in Modulation of Immune Response at the Junction of Host-Pathogen Interaction and Danger Molecule Signaling. Pathog Dis (2016) 74:ftw082. doi: 10.1093/femspd/ftw082
Keywords: DNA methylation, immune response, bacteria, infection, mechanism, review
Citation: Qin W, Scicluna BP and van der Poll T (2021) The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection. Front. Immunol. 12:696280. doi: 10.3389/fimmu.2021.696280
Received: 16 April 2021; Accepted: 15 July 2021;
Published: 29 July 2021.
Edited by:
Smita Kulkarni, Texas Biomedical Research Institute, United StatesReviewed by:
Krishnendu Mukherjee, University Hospital Münster, GermanyXue-Ming Zhang, Jilin University, China
Zhiqin Wang, Central South University, China
Taiping Chen, University of Texas MD Anderson Cancer Center, United States
Enass Abdel-Hameed, University of Cincinnati, United States
Copyright © 2021 Qin, Scicluna and van der Poll. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanhai Qin, dy5xaW5AYW1zdGVyZGFtdW1jLm5s
 Wanhai Qin
Wanhai Qin Brendon P. Scicluna1,2
Brendon P. Scicluna1,2 Tom van der Poll
Tom van der Poll