
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 26 August 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.695373
This article is part of the Research TopicThe Skin Immune Response to Infectious AgentsView all 18 articles
 Aislyn Oulee1
Aislyn Oulee1 Feiyang Ma1,2,3
Feiyang Ma1,2,3 Rosane M. B. Teles1
Rosane M. B. Teles1 Bruno J. de Andrade Silva1
Bruno J. de Andrade Silva1 Matteo Pellegrini3
Matteo Pellegrini3 Eynav Klechevsky4
Eynav Klechevsky4 Andrew N. Harman5,6
Andrew N. Harman5,6 Jake W. Rhodes5,6
Jake W. Rhodes5,6 Robert L. Modlin1,2*
Robert L. Modlin1,2*Langerhans cells (LCs) reside in the epidermis where they are poised to mount an antimicrobial response against microbial pathogens invading from the outside environment. To elucidate potential pathways by which LCs contribute to host defense, we mined published LC transcriptomes deposited in GEO and the scientific literature for genes that participate in antimicrobial responses. Overall, we identified 31 genes in LCs that encode proteins that contribute to antimicrobial activity, ten of which were cross-validated in at least two separate experiments. Seven of these ten antimicrobial genes encode chemokines, CCL1, CCL17, CCL19, CCL2, CCL22, CXCL14 and CXCL2, which mediate both antimicrobial and inflammatory responses. Of these, CCL22 was detected in seven of nine transcriptomes and by PCR in cultured LCs. Overall, the antimicrobial genes identified in LCs encode proteins with broad antibacterial activity, including against Staphylococcus aureus, which is the leading cause of skin infections. Thus, this study illustrates that LCs, consistent with their anatomical location, are programmed to mount an antimicrobial response against invading pathogens in skin.
Dendritic cells (DCs) are the key antigen presenting cells (APCs) that control both immunity and tolerance (1). DCs are localized in most tissues and surface barriers, where they function as sentinels for pathogen recognition. Stimulation of innate signaling receptors induce DCs to migrate from the periphery to secondary lymphoid organs, where they present antigens that drive adaptive immunity. DCs are divided into distinct subsets characterized by their unique expression of surface receptors and transcription factors, pathogen sensors and cytokines secretion profiles that contribute to their specialized capacities in activating different modules of immunity (2–6).
Human skin harbors multiple types of dendritic-appearing cells including Langerhans cells (LCs) that exclusively reside in the epidermis as well as conventional DCs (cDCs) in the underlying dermis. In addition to their localization to the epidermis, LCs are distinguished by their high expression of CD1a, the C-type lectin langerin (CD207) which induces the formation of a LC-specific organelle, the Birbeck granule, and lower expression of CD11c than dermal DCs (7). Through their dendrites, they form an extensive cellular network patrol the interface between the skin and the outside environment for pathogens (8–10), bind microbial ligands via toll-like receptors (TLRs) and CD207 and taking up pathogens via endocytosis (11–13).
LCs, although derived in mice from similar precursors as macrophages, have antigen presentation capacities similar to DCs (14, 15). Upon antigen capture, LCs undergo phenotypic changes during maturation and migrate to regional lymph nodes where they activate adaptive responses (12, 16–18). During infection, LC emigration from the epidermis is significantly enhanced by inflammatory cytokines such as IL-1 and TNF (19, 20). Migrating LCs express the DC-specific transcription factor ZBTB46 (21, 22), IL-15 (23) and IRF4, which is important for their ability to prime and cross-present antigens to CD8+ T cells at that site (24, 25). Additionally, LCs enhances cellular immunity by inducing Th1 and Th2 differentiation of CD4+ T cells (3, 4), they are the main skin DC subset responsible for directing IL-17 and IL-22-mediated responses (12, 26, 27), indicative of skin inflammatory and antimicrobial diseases. A subset of migratory LCs express CD5 with an even greater capacity to amplify these T cell responses (6). Moreover, a unique aspect of human LC is their ability to present antigen via CD1a both autoreactive (28, 29) and Mycobacterium leprae- and M. tuberculosis-reactive CD1a-restricted T cell responses have been reported (30).
Although it has been previously shown that LCs contribute to cutaneous host defense against pathogens including viruses (13, 31, 32), bacteria and fungi (33), only a few genes have been identified that directly mediate the antimicrobial response. In order to more broadly define the mechanisms by which LCs potentially contribute to an antimicrobial response, we mined public LC transcriptomes and surveyed the literature to identify “antimicrobial genes”, defined as genes encoding proteins with direct antimicrobial activity.
We surveyed Gene Expression Omnibus (GEO) (34) for transcriptomes of human skin-derived LCs and Langerhans-like dendritic cells (LCDCs), which are derived from CD34+ stem cells, using the key terms “(Langerhans AND skin) AND Homo sapiens[Organism]”. Our search was for the period before August 2020 and include those studies in which the LCs were activated with pro-inflammatory stimuli and/or as compared to other myeloid populations. This search yielded 24 series, nine of which met the criteria that n ≥ 3 samples for the LC and comparison group and did not contain only Langerhans cell histiocytosis samples. We then used GEO2R, an R-based web application, to obtain a list of genes that were differentially expressed in LCs. After obtaining the list of genes, we then filtered the comparisons by logFC>1 and adj. p-value<0.05. Of the nine series which the described criteria, one (GSE32648) did not yield any recognizable gene names on GEO2R and therefore we contacted the authors who provided us with their new RNA-seq data instead of the microarray data currently deposited in GEO2R and for a second dataset (GSE120386), GEO2R was not available. We used DESeq2 to run differential expression analysis of both of the bulk RNA-seq data with the default parameters. Genes with an adj. p value <0.05 were considered significantly differentially expressed.
We curated our direct antimicrobial list based on the 105 antimicrobial peptides listed in the Antimicrobial Peptide Database (APD) (35). The criteria for data registration into APD are the following: the peptides must be from natural sources, their antimicrobial activities must have been demonstrated (MIC <100 ug/ml), and their amino acid sequences elucidated. We also supplemented this list with literature findings of eight genes encoding peptides with direct antimicrobial activity not yet registered into the database including CCL2 (36), CCL14, CCL15 (37), CXCL7 (38), CXCL17 (39), MPEG1 (40), S1008A (41), and S1009A (42) yielding a total of 113 genes. To identify which genes encoded peptides with direct antimicrobial activity, we overlapped the results with our curated direct antimicrobial list using Venny 2.1 (43).
We also reviewed the literature for direct antimicrobial genes using the key terms “(Langerhans [Title/Abstract]) AND (antimicrobial [Title/Abstract])” which yielded 40 results of which five studies contained evidence for eight genes encoding peptides with direct antimicrobial activity in LCs. Our search “(Langerhans [Title/Abstract]) AND (antibacterial [Title/Abstract])” yielded 21 results, none of which included genes encoding peptides with direct antimicrobial in LCs.
IPA Upstream Regulator Analysis was used to identify upstream regulators and predict whether they are activated or inhibited, given the observed gene expression changes in our experimental dataset. The analysis examines the known targets of each upstream regulator in a dataset, compares the targets’ actual direction of change to expectations derived from the literature, then generates a prediction for each upstream regulator. Briefly, IPA uses an ‘enrichment’ score [Fisher’s exact test (FET) P-value] that measures the overlap of observed and predicted regulated gene sets.
To identify potential mechanisms by which LCs mount an antimicrobial response, we queried GEO and identified seven microarray series that permitted the mining of the LC transcriptome data using GEO2R. In addition, there was one bulk RNA-seq series for which GEO2R was not available (GSE120386) and another bulk-RNA seq data series not yet deposited in GEO2R and therefore we used DESeq2 on RStudio to compute the differential gene expression for both data series (Supplementary Table S1).
In three of nine series, LCs were directly isolated from skin specimens by enzymatic digestion, and the transcriptomes measured immediately. In one study, CD11c+ DDCs were directly isolated from skin and monocyte derived DCs and CD1c+ DCs from blood (44). In another study, plasmacytoid DCs (pDCs) and myeloid DCs (mDCs) were isolated from peripheral blood (45), and in the third study, pDCs were isolated from spleen and dermal macrophages from skin (46). Five of the nine transcriptomes were derived from LCs isolated by migration, in order to represent those LCs that are in the process of migration to lymph nodes, albeit this leads to an altered phenotype. In one study each, CD14+ DCs and CD14+ macrophages (47), or CD141+ dermal DCs, CD14+ dermal DCs, and CD141-CD14- dermal DCs (21), or dermal langerin- type 2 conventional dendritic cell (cDC2), dermal langerin+ cDC2, dermal CD14+CD1c- monocyte-derived macrophages, and dermal CD14+CD1c+ monocyte-derived dendritic cells (48) were isolated from skin by enzymatic digestion. In two of the migratory LC studies, the LC transcriptomes were measured at time zero and various timepoints following stimulation by TNF at (24, 49). In the same transcriptome, CD11c+ dermal DCs were also isolated by migration (49). In the last series, Langerhans-like dendritic cells (LCDCs) were generated in vitro and infected with the live mosquito-derived third-stage larvae (L3) of the parasitic nematode Brugia malayi (50).
We mined the LC transcriptomes by comparing either LCs to another myeloid cell type or a specific time point following stimulation. We filtered the comparisons by logFC>1 and adjusted p-value <0.05, then overlapped the results with the direct antimicrobial gene list consisting of 113 genes using Venny 2.1 (43). Using this approach, we identified 23 genes encoding proteins with direct antimicrobial activity in the LC transcriptomes (Table 1). Of these 23 genes, 11 were uniquely identified in LCs isolated by migration (then either unstimulated or cytokine activated), nine were uniquely identified in LCs derived from digested skin samples and three were presented in LCs isolated by migration as well as from digested skin samples. Although there were more genes identified in LCs obtained by migration from skin samples as compared to digested skin samples, as LCs isolated by enzymatic digestion are immature compared to those isolated by migration which are in a mature state, and that the migrated LCs were sometimes activated with cytokines whereas the digested LCs were not (55).
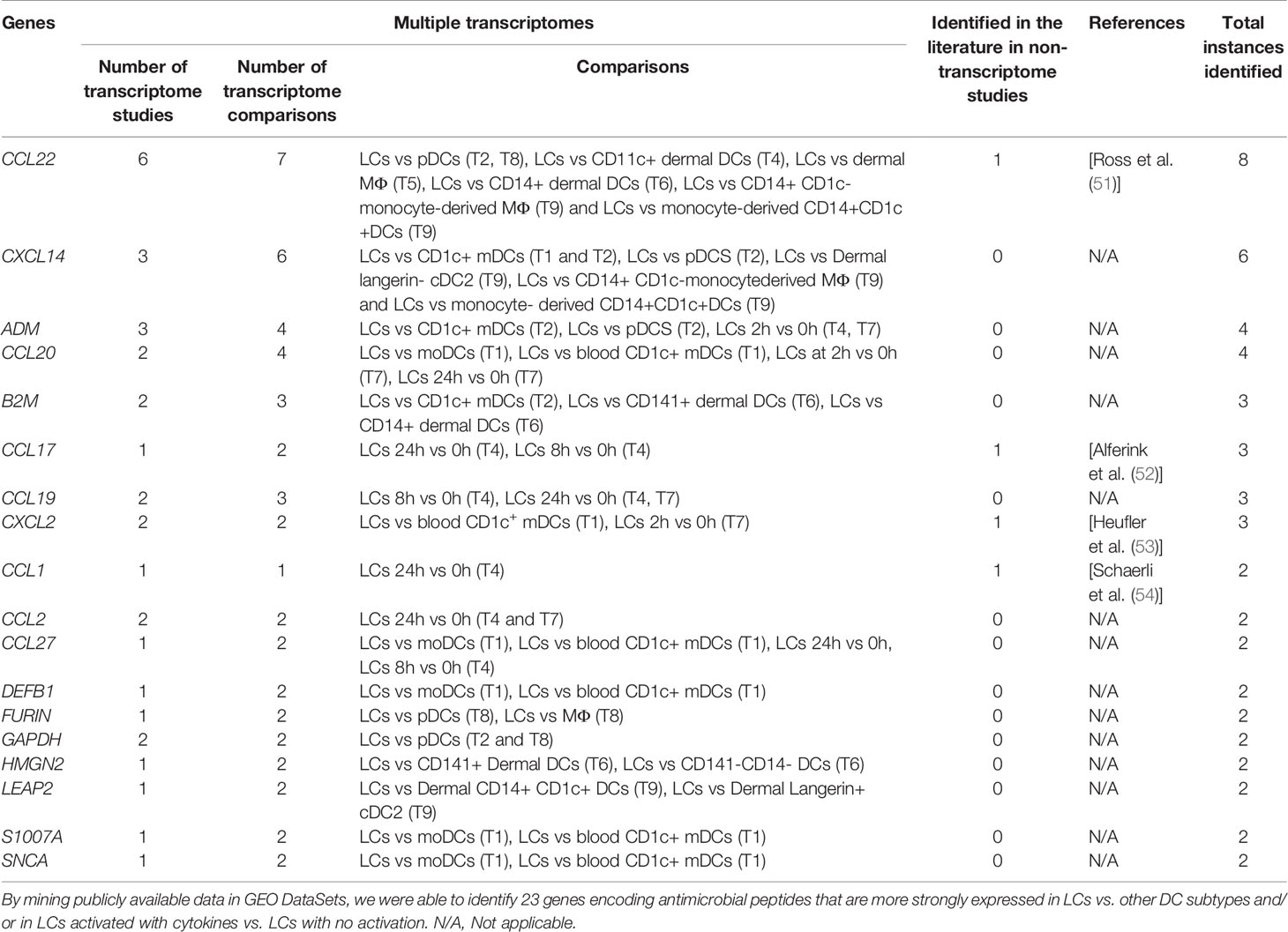
Table 1 Identification of genes encoding peptides with antimicrobial activity in LCs in transcriptome studies.
We examined the transcriptomes of LCs activated in vitro by cytokines or microbes. We identified eight genes that encode proteins with direct antimicrobial activity by mining the two transcriptomes of TNF-activated migratory LCs (Transcriptomes 4 and 7), this was the greatest number in any of the comparisons performed (Supplementary Table S2). There were six genes encoding chemokines that were upregulated in migratory LCs after stimulation with TNF: CCL1, CCL2, CCL17, CCL19, CCL20, and CXCL2. In addition, we detected two other genes, ADM and IL26 in LCs stimulated with TNF (Figure 1). Of the eight total genes, CCL2, CCL19 and ADM were detected in both transcriptomes of TNF treated LCs. We did not identify any genes encoding peptides with direct antimicrobial genes upregulated in LCs stimulated with live mosquito-derived third-stage larvae (L3) of B. malayi, which is consistent with the previous finding that the live mosquito-derived third-stage larvae (L3) fails to activate LCs compared to known activators (50). By analyzing the comparisons of LCs to other cell types, we identified 16 antimicrobial genes, of which only ADM was identified in the transcriptomes of TNF treated LCs.
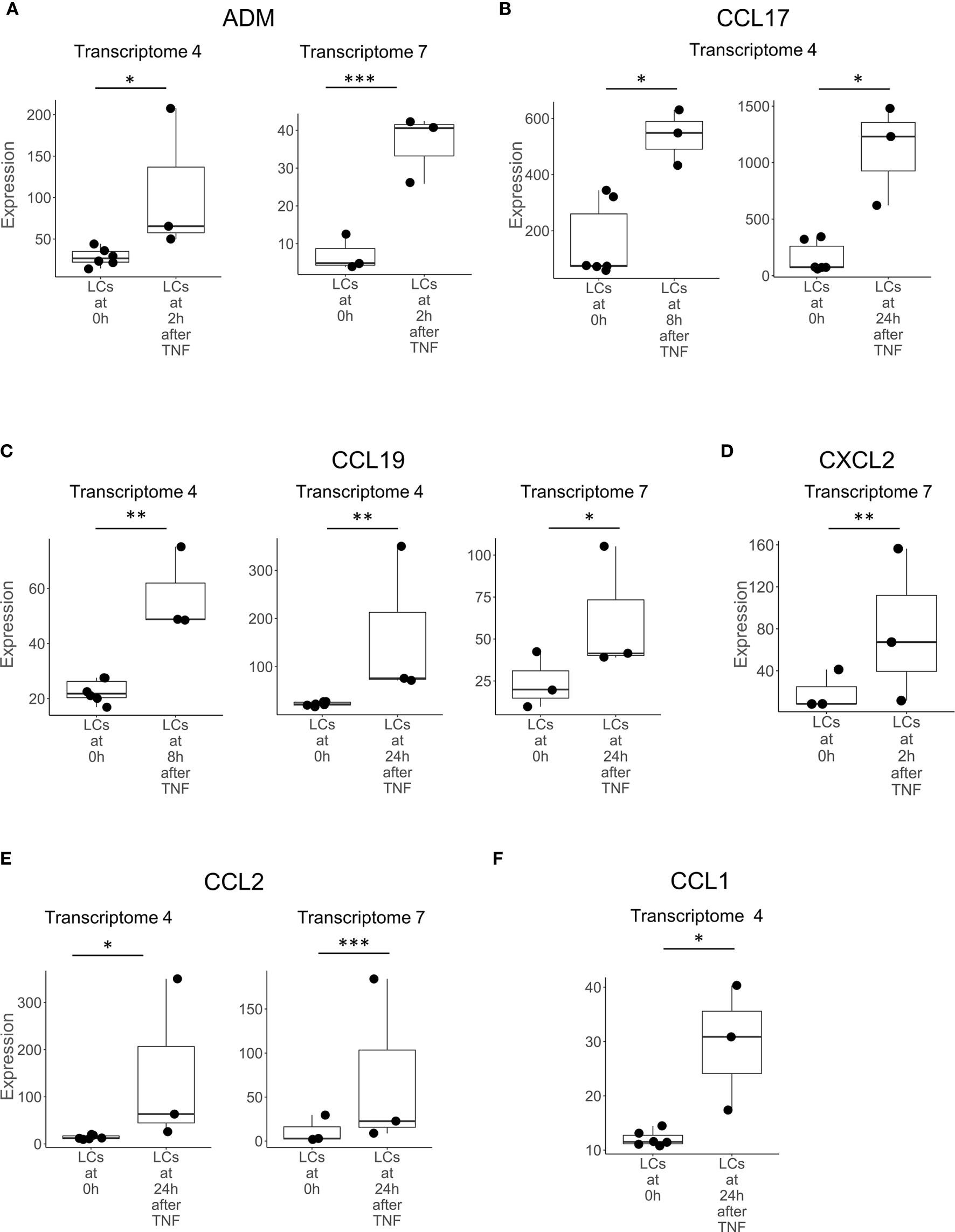
Figure 1 Genes upregulated in LCs after activation with TNF. Boxplots showing the expression of (A) ADM, (B) CCL17, (C) CCL19, (D) CXCL2, (E) CCL2, and (F) CCL1, encoding antimicrobial peptides in LCs prior to (0h) and at various time points following activation with TNF in transcriptomes 4 and 7. Genes shown above were identified in at least two instances, either in multiple transcriptomes experiments or in one transcriptome experiment but also identified in LCs in non-transcriptome experiments in the literature. *p < 0.05, **p < 0.01, ***p < 0.001.
We found nine studies in which the LC transcriptome was compared to other DC subtypes, including dermal DCs, peripheral blood DCs and cytokine-derived DCs, as well as to macrophage subpopulations. The nomenclature used to define DC subpopulations has evolved with changing technologies, such that different studies use different markers to define subpopulations. Dermal DCs have been identified based on the expression of various cell surface markers including XCR1+, CD141+, CD1c+, CD1a+ and CD14+ (3, 21, 47, 56–58), which may vary according to the method of isolation, digestion vs. migration (55). The analysis of DC subpopulations in human blood by single cell RNA sequencing has led to a revised gene-based classification (59). In reporting the comparison of transcriptomes in LCs to other cell types, we have maintained the nomenclature in the original citation.
In comparing LCs to other DC and myeloid cell types, CCL22 was the most frequently detected gene, expressed in seven of the nine studies and in eight separate comparisons (Figure 2). CXCL14 was detected as upregulated in six instances in three LCs transcriptomes (Figure 3). B2M was identified in the transcriptomes of LCs compared to other cell types in three different instances (Supplementary Figure S1). GAPDH was more highly expressed in LCs in two different transcriptomes (Supplementary Figure S2). CCL27, DEFB1, FURIN, LEAP2, SNCA, and S100A7 were each identified as preferentially expressed in LCs in two instances but always in a single LC transcriptome as compared to other cell types (Supplementary Table S3). HMGN2 was preferentially expressed in LCs compared to CD141+ and CD141-CD14- dermal DCs in one transcriptome (Supplementary Figure S3). SAA2, FAM3A, and RARRES2 were identified as upregulated in LCs in one instance each. Heat maps showing the expression of each gene in the different transcriptome comparisons are shown in Supplementary Figures 4–6.
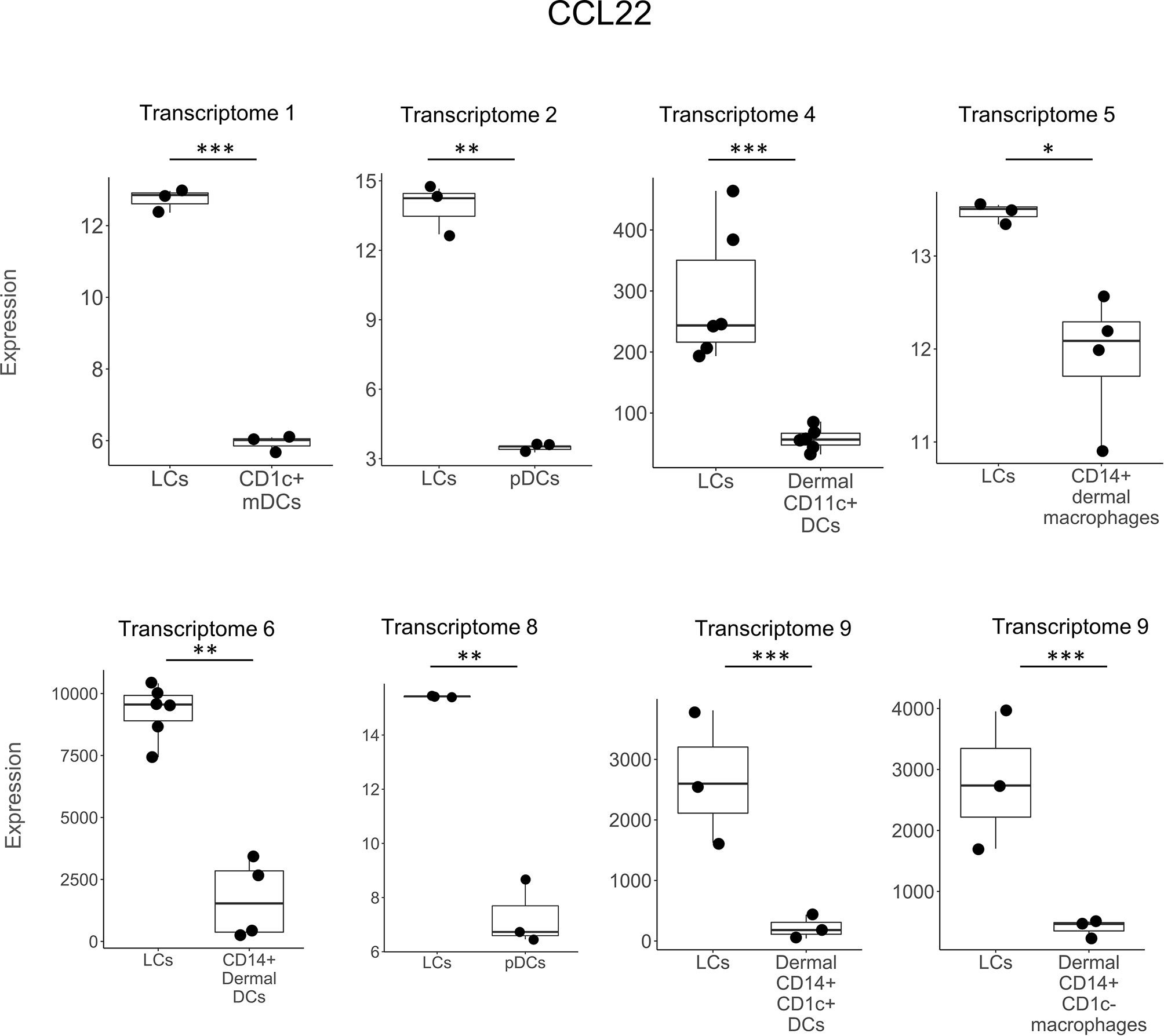
Figure 2 CCL22 expression in LCs vs other DC subtypes. CCL22 was preferentially expressed in LCs vs other DC types in 7 out of the 9 transcriptomes in a total of 8 instances. CCL22 was the most frequently detected gene in transcriptomes and was also previously reported in LCs in non-transcriptome experiments in the literature. *p < 0.05, **p < 0.01, ***p < 0.001.
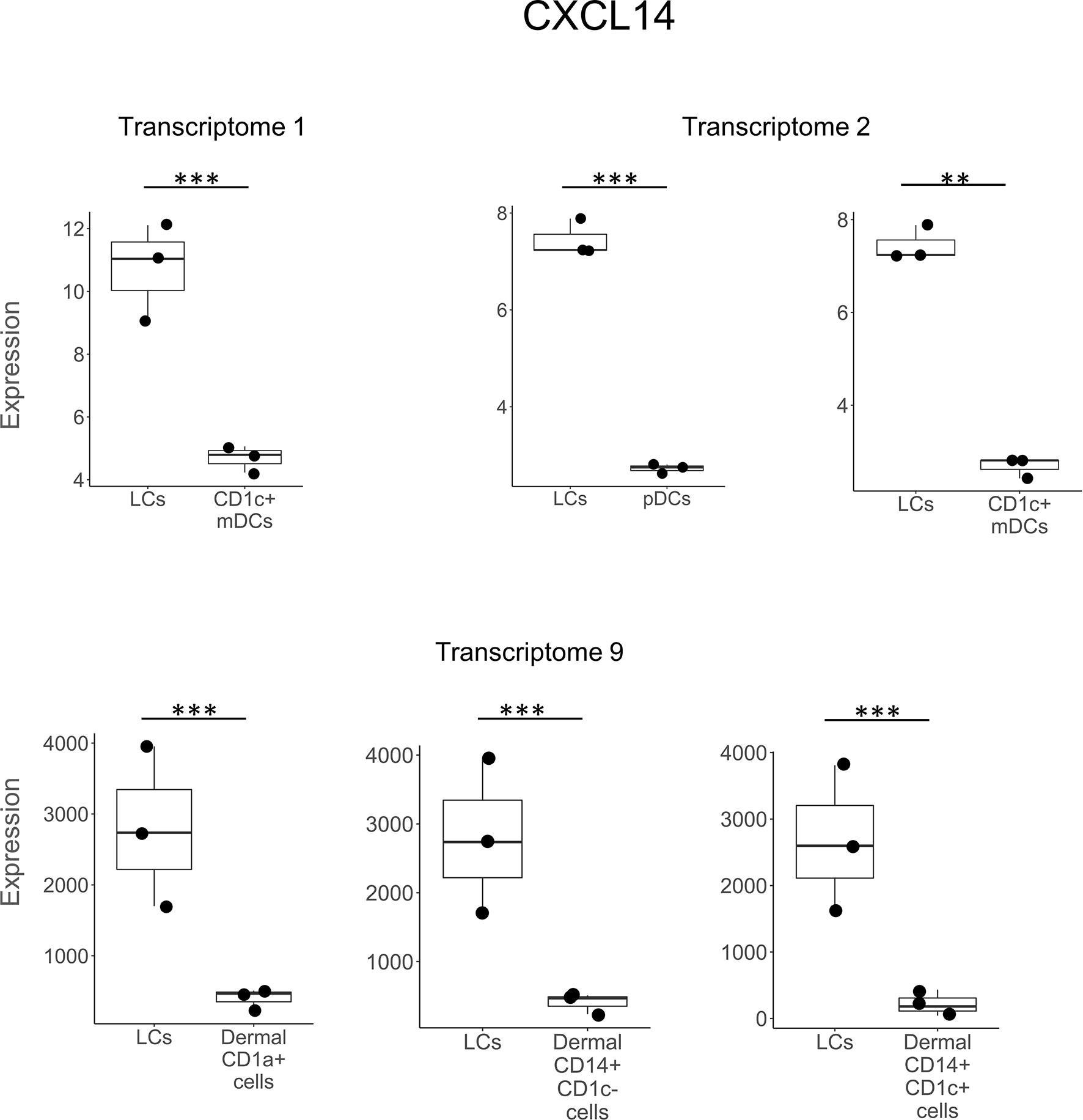
Figure 3 CXCL14 expression in LCs vs other DC subtypes. CXCL14 was preferentially expressed in LCs vs other DC types in 3 out of the 9 transcriptomes in a total of 6 different instances. **p < 0.01, ***p < 0.001.
We found corroborating evidence in the literature that four of the 23 direct antimicrobial genes were expressed in LCs. These included the CCL17-encoded peptide in cytokine activated LCs (52), CXCL2 mRNA in freshly isolated LCs (53), CCL22 mRNA during maturation of LCs (51), and the CCL1-encoded peptide in epidermal LCs in situ (54). We found reports indicating expression of eight genes encoding directly antimicrobial peptides and/or the antimicrobial proteins themselves in activated LCs that were not detected in any of transcriptomes. These include CXCL9, CXCL10, CXCL11 (60), POMC (61) and NPY (62) mRNAs, as well as CAMP, DEFB4 (33) and DEFB103 (63, 64) encoded antimicrobial peptides (Supplementary Table S4). Thus, a total of 31 antimicrobial genes/proteins were identified in LCs from analysis of LCs transcriptomes and published studies.
Overall, we found that ten of the 23 antimicrobial genes identified in the LC transcriptomes were cross-validated in at least two separate studies in the nine LC transcriptomes and/or four additional published studies. Six of the ten antimicrobial genes were cross validated by detection in two separate LC transcriptomes each, in each instance comparing LCs to the same other DC or myeloid cell type. CXCL14 was upregulated in LCs vs. blood CD1c+ DCs (Transcriptomes 1 and 2), CCL22 and GAPDH in LCs vs pDCs (Transcriptomes 2 and T8), and B2M in LCs compared to different DC populations in Transcriptomes 2 and 6. CCL2 and CCL19 were each upregulated in LCs treated with TNF for 24 hours vs 0 hours (Transcriptomes 4 and 7). ADM was upregulated in LCs treated with TNF for 2 hours vs 0 hours (Transcriptomes 4 and 7) and was also more strongly expressed in LCs vs blood CD1c+ DCs. CCL22 and CXCL2 expression was greater in LCs compared to other cell types in seven and two different transcriptomes, respectively, and validated by reverse transcriptase-polymerase chain reaction in additional studies (51, 53).
We also examined which antimicrobial genes were differentially expressed in LCs vs keratinocytes (KCs). We surveyed GEO DataSets for datasets containing both LCs and KCs using the key terms “Langerhans AND keratinocytes” and found two datasets (GSE168167 and GSE72104), both data sets containing LCs (n=3) and KCs (n=2) although our original criteria required n≥3 for each cell type. We found the expression of CCL22 was greater in LCs than KCs for both datasets, showing a 6.4- and 3.9-fold change. In one dataset (GSE72104), CCL17 expression was 4.3-fold greater in LCs than KCs and was identified in transcriptome 4 as being upregulated in LCs by TNF at 8 and 24 hours and validated at the protein level in a reporter mouse (CCL17) (52). CCL1 was identified in a single LC transcriptome upregulated by TNF after 24 hours and the protein validated by immunohistochemistry (CCL1) (54).
Using Ingenuity Pathways Analysis, we investigated the canonical pathways in LCs compared to other cell types, focusing on the three “Noah’s ark like” instances in which LCs were compared to the identical cell type in two transcriptomic studies. Thus, there were two studies each comparing LCs to pDCs, blood CD1c+ DCs and CD14+ dermal DCs. From the top 100 canonical pathways in each comparison, we identified one pathway present in all six comparisons and 23 in 5/6 comparisons (Supplementary Table S7), noting that there were fewer genes and hence pathways identified in LCs vs. CD14+ dermal DCs from Transcriptome 5. Overall, 21/23 pathways were identified as “signaling” pathways, including RANK, CD40, CXCR4, IL6 and IL8 signaling, consistent with the known functional properties of LCs.
We used Ingenuity Pathways Analysis and its knowledge database to identify the predicted upstream regulators of the 31 antimicrobial genes identified in LCs. Of the genes that encode cytokines, the top upstream regulator was IL1B, (p= 7.07x10-18) (Figure 4). The top 5 upstream regulator genes encode IL-1β, IFN-γ and TNF, all have been reported to induce one or more of the 30 LC antimicrobial genes in vitro (52). TNF was identified as the upstream regulator of 20 antimicrobial genes, followed by IFNG as the upstream regulator of 19 antimicrobial genes and IL1B as the upstream regulator of 18 antimicrobial genes. Together, the three cytokine genes were identified as upstream regulators for 25 of the 31 antimicrobial genes (Figure 5). In addition, we examined the target genes for other top upstream regulators that are known to contribute to the pathogenesis of skin disease: IL-10 (n=14 downstream genes), IL-22 (n=10), IL-13 (n=9), IL-17A (n=9). Thus, the antimicrobial gene response would likely be influenced by the local cytokine environment.
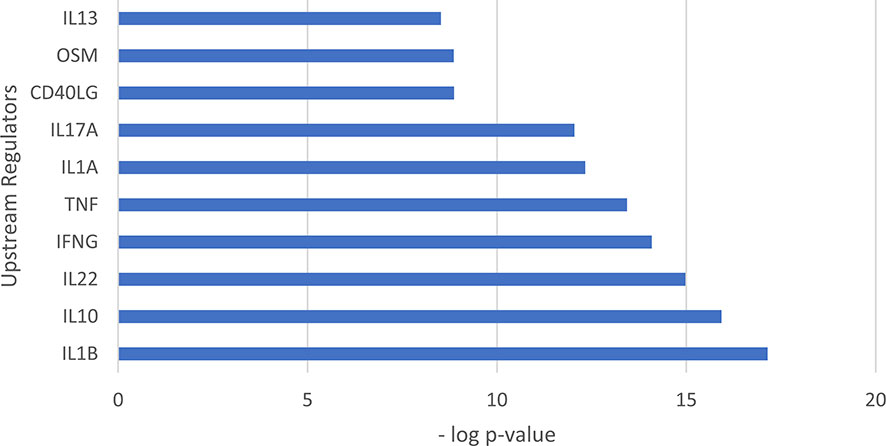
Figure 4 Upstream regulation of genes encoding antimicrobial peptides identified in LCs. Bar graph showing the top 10 upstream regulators ranked by p value. The top upstream regulator was IL1B (p= 7.07x10-18).
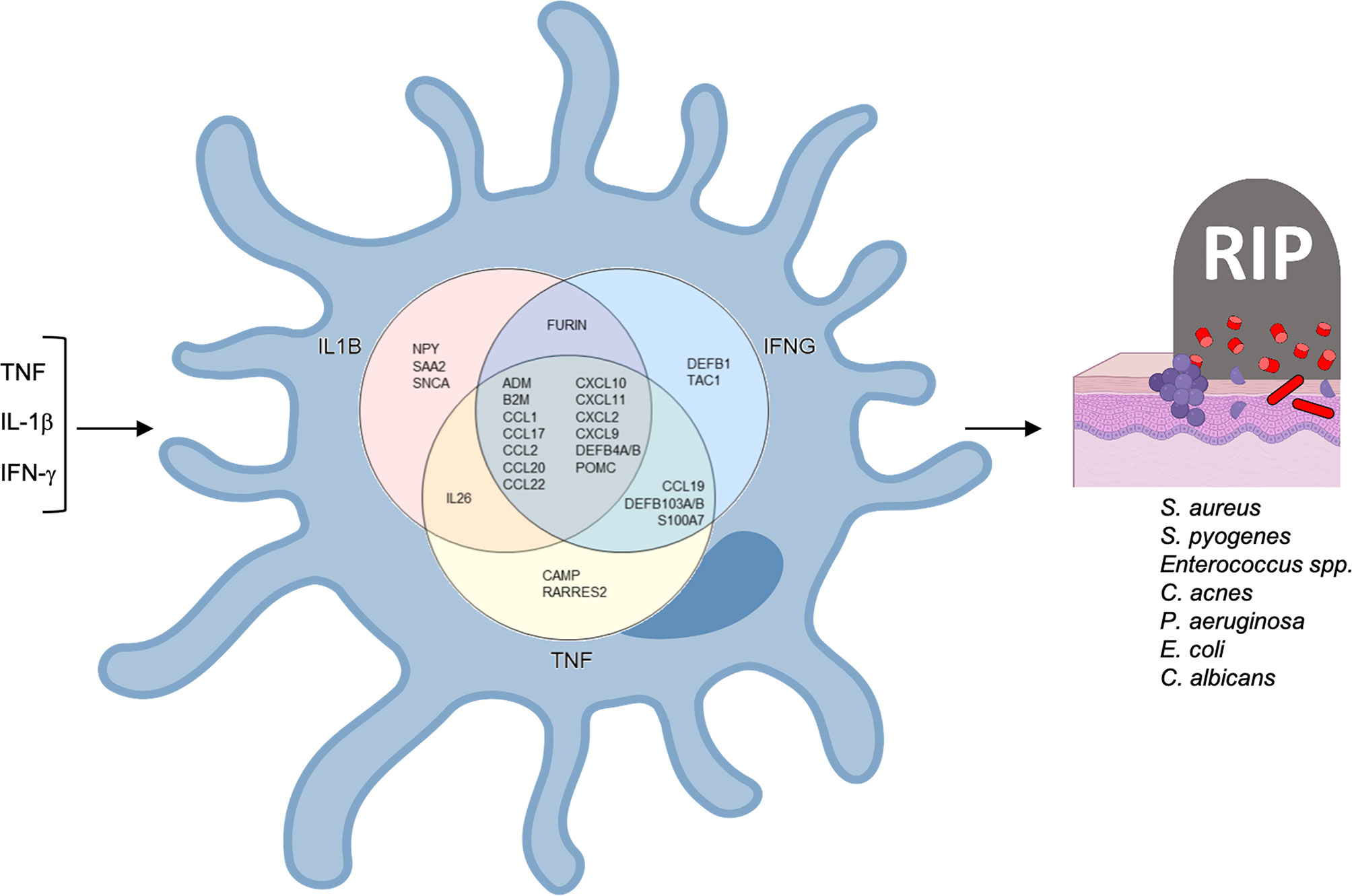
Figure 5 Antimicrobial network induced by TNF, IFNG and IL1B in LCs. TNF and IFNG were each identified as upstream regulators of 19 antimicrobial genes and IL1B as the upstream regulator of 18 antimicrobial genes.
Of the 20 genes predicted to be induced by TNF, we detected nine genes, ADM, CXCL2, CCL17, CCL27, IL26, CCL19, CCL2, CCL20 and CCL1, that were also upregulated in the transcriptomes of TNF-treated LCs. Of these, CCL17 protein has been validated to be induced by TNF in vitro (52). Although the Ingenuity pathways analysis did not predict TNF as an upstream regulator of CXCL10, TNF induced LCs to secrete CXCL10 in vitro (65).
Ingenuity pathways analysis identified IL1B as the upstream regulator for 18 of the 31 antimicrobial genes we identified in LC transcriptomes and/or the literature. For one these genes, the IL-1 family member, IL-1α, induces CCL17 encoded peptide in LCs (52). was validated to induce CCL17 peptide (Alferink et al., 2003). The addition of IFN-γ to LCs leads to the induction of CAMP and DEFB4 encoded peptides (33), as well as CXCL9, CXCL10, and CXCL11 mRNAs (60).
Overall, we identified 31 antimicrobial genes in LCs, of which eight genes were induced by activation with TNF in transcriptomes, 16 additional genes by comparison of LCs to other cell types, of which all but one gene were unique, and eight additional genes were identified in LCs in publications. Of the 31 genes, 12 genes belonged to the chemokine superfamily and making it the largest family of antimicrobial genes identified in LCs. Additionally, according to the Antimicrobial Peptide Database (APD) (35), of the 31 antimicrobial genes identified in LCs, 29 encode proteins that are antibacterial. Of the 29 genes, 23 encode peptides with activity against gram-positive bacillus Staphylococcus aureus, which is the leading cause of skin and soft tissue infections (66–68) (Supplementary Table S6). A total of 18 of the 31 genes encode proteins that are antifungal, six are antiviral, and five are antiparasitic (Supplementary Table S5).
The localization of LCs to the epidermis provides a first line of defense for the innate immune system to defend the host against microbial pathogens invading the skin. Surprisingly, few pathways have been identified by which LCs mediate antimicrobial responses against viruses (31, 32), bacteria, and fungi (33). Here, in order to gain insight into the breadth of mechanisms by which LCs are equipped to mount an antimicrobial response, we searched publicly available databases for LC transcriptomes and also reviewed the literature to identify genes which encode proteins with direct antimicrobial activity against cutaneous pathogens. Overall, we identified 31 genes encoding proteins with direct antimicrobial activity, ten of which were identified in at least two different experiments, thus representing a core set of genes that comprise the LC antimicrobial gene program. Seven of these ten antimicrobial genes encode chemokines, CCL1, CCL17, CCL19, CCL2, CCL22, CXCL14 and CXCL2, which mediate both antimicrobial and inflammatory responses. CCL22 was identified in seven of nine transcriptomes in eight total comparisons, as well as validated in cultured LCs by PCR (51). As such, LCs are armed with an antimicrobial gene program to combat microbial pathogens.
Chemokines were the largest family of antimicrobial genes identified in LCs, accounting for 12 of the 31 genes, including seven of the ten genes that were cross-validated in at least two studies. Of the 12 genes, seven belonged to the chemokine family with a “CC” structure and five to the family with the “CXC” structure. Chemokines are pro-inflammatory, such that as part of host defense against microbial pathogens their trigger the migration of immune cells to the site of infection (69). However, many chemokines have a dual function, as they possess direct microbicidal activity (36, 37, 39, 70). Of the chemokines, CCL22 was the most frequently detected antimicrobial gene, expressed in six different LC transcriptomes when compared to other cell types. CCL22 was also previously identified in mature LCs cocultured with keratinocytes (51). CCL22 is one of the natural ligands for CCR4, along with CCL17 and CCL2. Both CCL17 and CCL22 were also upregulated in TNF treated LCs, with CCL17 protein induced in LCs by TNF in vitro (52). CCR4 is highly expressed by skin-infiltrating lymphocytes (71) and is involved in skin homing (72–74) of Th2 T cells, Th17 cells, Th22 cells and Tregs (75–79). LCs, by expression of CCL22, CCL17, and CCL2 have the potential to recruit a range of functional CCR4+ T cell subpopulations to the site of disease.
Three of the top five upstream regulators of the 31 antimicrobial genes detected in LCs, TNF, IL1B and IFNG, have been corroborated by in vitro studies in which the cytokine was directly added to LCs. In the two data series in which TNF was added to activate migratory LCs in vitro, eight antimicrobial genes were identified (24, 49), all consistent with the TNF-downstream genes in the Ingenuity knowledge database. TNF is known to induce the maturation and migration of LCs (19, 80), increasing the number of LCs (65), and induce the expression of inflammatory genes in LCs (49, 65, 81).
Of the eight TNF inducible genes in migrating LCs, six encode chemokines, CCL1, CCL2, CCL17, CCL19 and CCL20, which along with IL26 were only detected in the transcriptomes of TNF activated LCs but not in LCs compared to transcriptomes of other myeloid cell types. Three of these antimicrobial genes have been corroborated in published papers; CCL1 protein has been identified in epidermal LCs in situ (54), CCL17 protein in IL-1α or TNF-activated LCs in vivo in mice (52) and CXCL2 mRNA in freshly isolated murine LC cells (53). In addition to TNF, other inflammatory stimuli have been reported to induce the expression of genes in LCs encoding directly antimicrobial peptides. CAMP and DEFB4 encoded peptides are induced in LCs by IFN-γ (33). CXCL9, CXCL10, and CXCL11 mRNAs are induced in LCs by stimuli including IFN-γ, LPS, and poly I:C (32, 60). NPY mRNA expression in LCs is enhanced by GM-CSF and LPS (62). In addition, LCs have been shown to express POMC mRNA upon activation (61). Therefore, the activation and/or maturation of LCs triggers expression of multiple antimicrobial genes.
By comparing the expression of antimicrobial genes in LCs to other cell types, we identified 23 genes that arm LCs with the capacity to combat cutaneous pathogens and eight additional genes described in the literature to be expressed by LCs. Of these 31 genes, 23 genes encode peptides with activity against gram-positive bacillus Staphylococcus aureus, which is the leading cause of skin and soft tissue infections (66–68). LC expression of CAMP and DEFB4 results in an antimicrobial activity against the cutaneous pathogens including M. leprae, S. aureus, Streptococcus pyogenes and Candida albicans (33). In addition, LC have been previously shown to mediate an antiviral activity (32, 82, 83), although the mechanisms involved are not clear.
We previously found that the antimicrobial activity of LCs leads to killing and subsequent processing of microbial antigens facilitating antigen presentation to T cells (33). Some of the antimicrobial peptides expressed by migratory LCs have been shown to be pro-inflammatory, such as CCL22 and CCL17, which both act as a chemoattractant for CCR4-expressing T cells promoting LC:T cell interaction (84). Thus, the ability of LCs, in particular migratory LCs, to upregulate antimicrobial peptides links the innate and adaptive immune response, defending the host against cutaneous pathogens. There are at least two possible contributions of antimicrobial gene expression in migrating LCs.
We found that ten of the antimicrobial genes expressed in LCs were cross-validated by various methodologies, identifying a core set of genes by which LCs can contribute to host defense, that provide a basis for further functional studies. Any one antimicrobial gene may be sufficient to mediate an antimicrobial response, given our published data that IFN-γ upregulation of CAMP was required for antimicrobial activity in LCs (33). This was demonstrated by knockdown of the CAMP gene and the use of neutralizing monoclonal antibodies to IFN-γ (33). These strategies provide a strategy to determine whether the upregulation of multiple antimicrobial genes by cytokines and cell surface receptors such as Toll-like receptor ligands leads to a more potent antimicrobial response. It should be possible to identify key LC pathways that could be leveraged by immune therapy augmenting LC antimicrobial responses to combat cutaneous infection.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conceptualization: AO and RM. Data curation: AH and JR. Formal analysis: AO, RM, RT, FM, and EK. Funding acquisition: RM. Investigation: AO and RM. Methodology: AO and RM. Project administration: AO and RM. Resources: RM, FM, and MP. Software: FM and MP. Supervision: RM. Validation: EK. Visualization: AO and RM. Writing—original draft preparation: AO and RM. Writing—review and editing: AO, RM, AH, RT, FM, and BA. All authors contributed to the article and approved the submitted version.
NIH grants R01 AI022553 (RLM, MP), R01 AR040312 (RLM, MP), R01 AR073252 (RLM, MP), R01 AR074302 (RLM, MP), NHMRCIdeas Grant APP1181482 (AH), 5R21EB024767-02 (EK), 1R01AR075959-01 (EK) and 1R01CA245277-01A1 (EK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.695373/full#supplementary-material
1. Banchereau J, Steinman RM. Dendritic Cells and the Control of Immunity. Nature (1998) 392:245–52. doi: 10.1038/32588
2. Klechevsky E, Banchereau J. Human Dendritic Cells Subsets as Targets and Vectors for Therapy. Ann N Y Acad Sci (2013) 1284:24–30. doi: 10.1111/nyas.12113
3. Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, et al. Functional Specializations of Human Epidermal Langerhans Cells and CD14+ Dermal Dendritic Cells. Immunity (2008) 29(3):497–510. doi: 10.1016/j.immuni.2008.07.013
4. Banchereau J, Zurawski S, Thompson-Snipes L, Blanck JP, Clayton S, Munk A, et al. Immunoglobulin-Like Transcript Receptors on Human Dermal CD14+ Dendritic Cells Act as a CD8-Antagonist to Control Cytotoxic T Cell Priming. Proc Natl Acad Sci USA (2012) 109(46):18885–90. doi: 10.1073/pnas.1205785109
5. Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, et al. Dendritic Cell Subsets in Health and Disease. Immunol Rev (2007) 219:118–42. doi: 10.1111/j.1600-065X.2007.00551.x
6. Korenfeld D, Gorvel L, Munk A, Man J, Schaffer A, Tung T, et al. A Type of Human Skin Dendritic Cell Marked by CD5 Is Associated With the Development of Inflammatory Skin Disease. JCI Insight (2017) 2(18). doi: 10.1172/jci.insight.96101
7. Bertram KM, Botting RA, Baharlou H, Rhodes JW, Rana H, Graham JD, et al. Identification of HIV Transmitting CD11c(+) Human Epidermal Dendritic Cells. Nat Commun (2019) 10(1):2759. doi: 10.1038/s41467-019-10697-w
8. Valladeau J, Ravel O, Dezutter-Dambuyant C, Moore K, Kleijmeer M, Liu Y, et al. Langerin, a Novel C-Type Lectin Specific to Langerhans Cells, is an Endocytic Receptor That Induces the Formation of Birbeck Granules. Immunity (2000) 12(1):71–81. doi: 10.1016/s1074-7613(00)80160-0
9. Fithian E, Kung P, Goldstein G, Rubenfeld M, Fenoglio C, Edelson R. Reactivity of Langerhans Cells With Hybridoma Antibody. Proc Natl Acad Sci USA (1981) 78(4):2541–4. doi: 10.1073/pnas.78.4.2541
10. Kubo A, Nagao K, Yokouchi M, Sasaki H, Amagai M. External Antigen Uptake by Langerhans Cells With Reorganization of Epidermal Tight Junction Barriers. J Exp Med (2009) 206(13):2937–46. doi: 10.1084/jem.20091527
11. Osorio F, Reis e Sousa C. Myeloid C-Type Lectin Receptors in Pathogen Recognition and Host Defense. Immunity (2011) 34(5):651–64. doi: 10.1016/j.immuni.2011.05.001
12. de Jong MA, Geijtenbeek TB. Langerhans Cells in Innate Defense Against Pathogens. Trends Immunol (2010) 31(12):452–9. doi: 10.1016/j.it.2010.08.002
13. Nasr N, Lai J, Botting RA, Mercier SK, Harman AN, Kim M, et al. Inhibition of Two Temporal Phases of HIV-1 Transfer From Primary Langerhans Cells to T Cells: The Role of Langerin. J Immunol (2014) 193(5):2554–64. doi: 10.4049/jimmunol.1400630
14. Kashem SW, Haniffa M, Kaplan DH. Antigen-Presenting Cells in the Skin. Annu Rev Immunol (2017) 35(1):469–99. doi: 10.1146/annurev-immunol-051116-052215
15. Klechevsky E. Functional Diversity of Human Dendritic Cells. Adv Exp Med Biol (2015) 850:43–54. doi: 10.1007/978-3-319-15774-0_4
16. Merad M, Ginhoux F, Collin M. Origin, Homeostasis and Function of Langerhans Cells and Other Langerin-Expressing Dendritic Cells. Nat Rev Immunol (2008) 8(12):935–47. doi: 10.1038/nri2455
17. Larsen CP, Steinman RM, Witmer-Pack M, Hankins DF, Morris PJ, Austyn JM. Migration and Maturation of Langerhans Cells in Skin Transplants and Explants. J Exp Med (1990) 172(5):1483–93. doi: 10.1084/jem.172.5.1483
18. Harman AN, Wilkinson J, Bye CR, Bosnjak L, Stern JL, Nicholle M, et al. HIV Induces Maturation of Monocyte-Derived Dendritic Cells and Langerhans Cells. J Immunol (2006) 177(10):7103–13. doi: 10.4049/jimmunol.177.10.7103
19. Kimber I, Cumberbatch M. Stimulation of Langerhans Cell Migration by Tumor Necrosis Factor Alpha (TNF-Alpha). J Invest Dermatol (1992) 99(5):48s–50s. doi: 10.1111/1523-1747.ep12668986
20. Wang B, Amerio P, Sauder DN. Role of Cytokines in Epidermal Langerhans Cell Migration. J Leukoc Biol (1999) 66(1):33–9. doi: 10.1002/jlb.66.1.33
21. Artyomov MN, Munk A, Gorvel L, Korenfeld D, Cella M, Tung T, et al. Modular Expression Analysis Reveals Functional Conservation Between Human Langerhans Cells and Mouse Cross-Priming Dendritic Cells. J Exp Med (2015) 212(5):743–57. doi: 10.1084/jem.20131675
22. Wu X, Briseño CG, Durai V, Albring JC, Haldar M, Bagadia P, et al. Mafb Lineage Tracing to Distinguish Macrophages From Other Immune Lineages Reveals Dual Identity of Langerhans Cells. J Exp Med (2016) 213(12):2553–65. doi: 10.1084/jem.20160600
23. Banchereau J, Thompson-Snipes L, Zurawski S, Blanck J-P, Cao Y, Clayton S, et al. The Differential Production of Cytokines by Human Langerhans Cells and Dermal CD14(+) DCs Controls CTL Priming. Blood (2012) 119(24):5742–9. doi: 10.1182/blood-2011-08-371245
24. Sirvent S, Vallejo AF, Davies J, Clayton K, Wu Z, Woo J, et al. Genomic Programming of IRF4-Expressing Human Langerhans Cells. Nat Commun (2020) 11(1):313. doi: 10.1038/s41467-019-14125-x
25. Balin SJ, Pellegrini M, Klechevsky E, Won ST, Weiss DI, Choi AW, et al. Human Antimicrobial Cytotoxic T Lymphocytes, Defined by NK Receptors and Antimicrobial Proteins, Kill Intracellular Bacteria. Sci Immunol (2018) 3(26). doi: 10.1126/sciimmunol.aat7668
26. Penel-Sotirakis K, Simonazzi E, Peguet-Navarro J, Rozieres A. Differential Capacity of Human Skin Dendritic Cells to Polarize CD4+ T Cells Into IL-17, IL-21 and IL-22 Producing Cells. PloS One (2012) 7(11):e45680. doi: 10.1371/journal.pone.0045680
27. Fujita H, Nograles KE, Kikuchi T, Gonzalez J, Carucci JA, Krueger JG. Human Langerhans Cells Induce Distinct IL-22-Producing CD4+ T Cells Lacking IL-17 Production. Proc Natl Acad Sci USA (2009) 106(51):21795–800. doi: 10.1073/pnas.0911472106
28. Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of Cluster of Differentiation 1 Antigens by Human CD4-CD8-Cytolytic T Lymphocytes. Nature (1989) 341:447–50. doi: 10.1038/341447a0
29. de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG, et al. CD1a-Autoreactive T Cells Recognize Natural Skin Oils That Function as Headless Antigens. Nat Immunol (2014) 15(2):177–85. doi: 10.1038/ni.2790
30. Hunger RE, Sieling PA, Ochoa MT, Sugaya M, Burdick AE, Rea TH, et al. Langerhans Cells Utilize CD1a and Langerin to Efficiently Present Nonpeptide Antigens to T Cells. J Clin Invest (2004) 113(5):701–8. doi: 10.1172/jci19655
31. van der Vlist M, Geijtenbeek TB. Langerin Functions as an Antiviral Receptor on Langerhans Cells. Immunol Cell Biol (2010) 88(4):410–5. doi: 10.1038/icb.2010.32
32. Renn CN, Sanchez DJ, Ochoa MT, Legaspi AJ, Oh CK, Liu PT, et al. TLR Activation of Langerhans Cell-Like Dendritic Cells Triggers an Antiviral Immune Response. J Immunol (2006) 177(1):298–305. doi: 10.4049/jimmunol.177.1.298
33. Dang AT, Teles RM, Liu PT, Choi A, Legaspi A, Sarno EN, et al. Autophagy Links Antimicrobial Activity With Antigen Presentation in Langerhans Cells. JCI Insight (2019) 4(8):e126955. doi: 10.1172/jci.insight.126955
34. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res (2002) 30(1):207–10. doi: 10.1093/nar/30.1.207
35. Wang G, Li X, Wang Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res (2016) 44(D1):D1087–93. doi: 10.1093/nar/gkv1278
36. Hoover DM, Boulegue C, Yang D, Oppenheim JJ, Tucker K, Lu W, et al. The Structure of Human Macrophage Inflammatory Protein-3alpha/CCL20. Linking Antimicrobial and CC Chemokine Receptor-6-Binding Activities With Human Beta-Defensins. J Biol Chem (2002) 277(40):37647–54. doi: 10.1074/jbc.M203907200
37. Kotarsky K, Sitnik KM, Stenstad H, Kotarsky H, Schmidtchen A, Koslowski M, et al. A Novel Role for Constitutively Expressed Epithelial-Derived Chemokines as Antibacterial Peptides in the Intestinal Mucosa. Mucosal Immunol (2010) 3(1):40–8. doi: 10.1038/mi.2009.115
38. Krijgsveld J, Zaat SA, Meeldijk J, van Veelen PA, Fang G, Poolman B, et al. Thrombocidins, Microbicidal Proteins From Human Blood Platelets, are C-Terminal Deletion Products of CXC Chemokines. J Biol Chem (2000) 275(27):20374–81. doi: 10.1074/jbc.275.27.20374
39. Burkhardt AM, Tai KP, Flores-Guiterrez JP, Vilches-Cisneros N, Kamdar K, Barbosa-Quintana O, et al. CXCL17 is a Mucosal Chemokine Elevated in Idiopathic Pulmonary Fibrosis That Exhibits Broad Antimicrobial Activity. J Immunol (2012) 188(12):6399–406. doi: 10.4049/jimmunol.1102903
40. Strbo N, Pastar I, Romero L, Chen V, Vujanac M, Sawaya AP, et al. Single Cell Analyses Reveal Specific Distribution of Anti-Bacterial Molecule Perforin-2 in Human Skin and its Modulation by Wounding and Staphylococcus Aureus Infection. Exp Dermatol (2019) 28(3):225–32. doi: 10.1111/exd.13870
41. Dhiman R, Venkatasubramanian S, Paidipally P, Barnes PF, Tvinnereim A, Vankayalapati R. Interleukin 22 Inhibits Intracellular Growth of Mycobacterium Tuberculosis by Enhancing Calgranulin A Expression. J Infect Dis (2014) 209(4):578–87. doi: 10.1093/infdis/jit495
42. Steinbakk M, Naess-Andresen CF, Lingaas E, Dale I, Brandtzaeg P, Fagerhol MK. Antimicrobial Actions of Calcium Binding Leucocyte L1 Protein, Calprotectin. Lancet (1990) 336(8718):763–5. doi: 10.1016/0140-6736(90)93237-j
44. Széles L, Póliska S, Nagy G, Szatmari I, Szanto A, Pap A, et al. Research Resource: Transcriptome Profiling of Genes Regulated by RXR and its Permissive and Nonpermissive Partners in Differentiating Monocyte-Derived Dendritic Cells. Mol Endocrinol (2010) 24(11):2218–31. doi: 10.1210/me.2010-0215
45. Hutter C, Kauer M, Simonitsch-Klupp I, Jug G, Schwentner R, Leitner J, et al. Notch is Active in Langerhans Cell Histiocytosis and Confers Pathognomonic Features on Dendritic Cells. Blood (2012) 120(26):5199–208. doi: 10.1182/blood-2012-02-410241
46. Lim KPH, Milne P, Poidinger M, Duan K, Lin H, McGovern N, et al. Circulating CD1c+ Myeloid Dendritic Cells Are Potential Precursors to LCH Lesion CD1a+CD207+ Cells. Blood Adv (2020) 4(1):87–99. doi: 10.1182/bloodadvances.2019000488
47. McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, et al. Human Dermal CD14+ Cells are a Transient Population of Monocyte-Derived Macrophages. Immunity (2014) 41(3):465–77. doi: 10.1016/j.immuni.2014.08.006
48. Rhodes JW, Botting RA, Bertram KM, Rana H, Baharlou H, Longmuir-Vine EE, et al. Identification of HIV-Transmitting Sub-Epithelial Mononuclear Phagocytes in Human Anogenital and Colorectal Tissues. bioRxiv (2020). doi: 10.1101/2020.05.26.117408. 2020.05.26.117408.
49. Polak ME, Thirdborough SM, Ung CY, Elliott T, Healy E, Freeman TC, et al. Distinct Molecular Signature of Human Skin Langerhans Cells Denotes Critical Differences in Cutaneous Dendritic Cell Immune Regulation. J Invest Dermatol (2014) 134(3):695–703. doi: 10.1038/jid.2013.375
50. Boyd A, Bennuru S, Wang Y, Sanprasert V, Law M, Chaussabel D, et al. Quiescent Innate Response to Infective Filariae by Human Langerhans Cells Suggests a Strategy of Immune Evasion. Infect Immun (2013) 81(5):1420–9. doi: 10.1128/iai.01301-12
51. Ross R, Ross XL, Ghadially H, Lahr T, Schwing J, Knop J, et al. Mouse Langerhans Cells Differentially Express an Activated T Cell-Attracting CC Chemokine. J Invest Dermatol (1999) 113(6):991–8. doi: 10.1046/j.1523-1747.1999.00803.x
52. Alferink J, Lieberam I, Reindl W, Behrens A, Weiss S, Hüser N, et al. Compartmentalized Production of CCL17 In Vivo: Strong Inducibility in Peripheral Dendritic Cells Contrasts Selective Absence From the Spleen. J Exp Med (2003) 197(5):585–99. doi: 10.1084/jem.20021859
53. Heufler C, Topar G, Koch F, Trockenbacher B, Kämpgen E, Romani N, et al. Cytokine Gene Expression in Murine Epidermal Cell Suspensions: Interleukin 1 Beta and Macrophage Inflammatory Protein 1 Alpha are Selectively Expressed in Langerhans Cells But Are Differentially Regulated in Culture. J Exp Med (1992) 176(4):1221–6. doi: 10.1084/jem.176.4.1221
54. Schaerli P, Ebert L, Willimann K, Blaser A, Roos RS, Loetscher P, et al. A Skin-Selective Homing Mechanism for Human Immune Surveillance T Cells. J Exp Med (2004) 199(9):1265–75. doi: 10.1084/jem.20032177
55. Botting RA, Bertram KM, Baharlou H, Sandgren KJ, Fletcher J, Rhodes JW, et al. Phenotypic and Functional Consequences of Different Isolation Protocols on Skin Mononuclear Phagocytes. J Leukoc Biol (2017) 101(6):1393–403. doi: 10.1189/jlb.4A1116-496R
56. Nestle FO, Zheng XG, Thompson CB, Turka LA, Nickoloff BJ. Characterization of Dermal Dendritic Cells Obtained From Normal Human Skin Reveals Phenotypic and Functionally Distinctive Subsets. J Immunol (1993) 151:6535–45.
57. Zaba LC, Fuentes-Duculan J, Steinman RM, Krueger JG, Lowes MA. Normal Human Dermis Contains Distinct Populations of CD11c+BDCA-1+ Dendritic Cells and CD163+FXIIIA+ Macrophages. J Clin Invest (2007) 117(9):2517–25. doi: 10.1172/JCI32282
58. Lenz A, Heine M, Schuler G, Romani N. Human and Murine Dermis Contain Dendritic Cells. Isolation by Means of a Novel Method and Phenotypical and Functional Characterization. J Clin Invest (1993) 92:2587–96. doi: 10.1172/JCI116873
59. Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-Cell RNA-Seq Reveals New Types of Human Blood Dendritic Cells, Monocytes, and Progenitors. Science (2017) 356(6335). doi: 10.1126/science.aah4573
60. Fujita H, Asahina A, Sugaya M, Nakamura K, Gao P, Fujiwara H, et al. Differential Production of Th1- and Th2-Type Chemokines by Mouse Langerhans Cells and Splenic Dendritic Cells. J Invest Dermatol (2005) 124(2):343–50. doi: 10.1111/j.0022-202X.2004.23607.x
61. Luger TA, Scholzen T, Grabbe S. The Role of Alpha-Melanocyte-Stimulating Hormone in Cutaneous Biology. J Investig Dermatol Symp Proc (1997) 2(1):87–93. doi: 10.1038/jidsymp.1997.17
62. Lambert RW, Campton K, Ding W, Ozawa H, Granstein RD. Langerhans Cell Expression of Neuropeptide Y and Peptide YY. Neuropeptides (2002) 36(4):246–51. doi: 10.1016/s0143-4179(02)00020-3
63. Lu Q, Samaranayake LP, Darveau RP, Jin L. Expression of Human Beta-Defensin-3 in Gingival Epithelia. J Periodontal Res (2005) 40(6):474–81. doi: 10.1111/j.1600-0765.2005.00827.x
64. Pilkington SM, Dearman RJ, Kimber I, Griffiths CEM. Langerhans Cells Express Human β-Defensin 3: Relevance for Immunity During Skin Ageing. Br J Dermatol (2018) 179(5):1170–1. doi: 10.1111/bjd.16770
65. Berthier-Vergnes O, Bermond F, Flacher V, Massacrier C, Schmitt D, Péguet-Navarro J. TNF-Alpha Enhances Phenotypic and Functional Maturation of Human Epidermal Langerhans Cells and Induces IL-12 P40 and IP-10/CXCL-10 Production. FEBS Lett (2005) 579(17):3660–8. doi: 10.1016/j.febslet.2005.04.087
66. Moet GJ, Jones RN, Biedenbach DJ, Stilwell MG, Fritsche TR. Contemporary Causes of Skin and Soft Tissue Infections in North America, Latin America, and Europe: Report From the SENTRY Antimicrobial Surveillance Program (1998-2004). Diagn Microbiol Infect Dis (2007) 57(1):7–13. doi: 10.1016/j.diagmicrobio.2006.05.009
67. Ray GT, Suaya JA, Baxter R. Incidence, Microbiology, and Patient Characteristics of Skin and Soft-Tissue Infections in a U.S. Population: A Retrospective Population-Based Study. BMC Infect Dis (2013) 13:252. doi: 10.1186/1471-2334-13-252
68. Rennie RP, Jones RN, Mutnick AH. Occurrence and Antimicrobial Susceptibility Patterns of Pathogens Isolated From Skin and Soft Tissue Infections: Report From the SENTRY Antimicrobial Surveillance Program (United States and Canada, 2000). Diagn Microbiol Infect Dis (2003) 45(4):287–93. doi: 10.1016/s0732-8893(02)00543-6
69. Lukacs NW. Role of Chemokines in the Pathogenesis of Asthma. Nat Rev Immunol (2001) 1(2):108–16. doi: 10.1038/35100503
70. Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, et al. Many Chemokines Including CCL20/MIP-3alpha Display Antimicrobial Activity. J Leukoc Biol (2003) 74(3):448–55. doi: 10.1189/jlb.0103024
71. Kunkel EJ, Boisvert J, Murphy K, Vierra MA, Genovese MC, Wardlaw AJ, et al. Expression of the Chemokine Receptors CCR4, CCR5, and CXCR3 by Human Tissue-Infiltrating Lymphocytes. Am J Pathol (2002) 160(1):347–55. doi: 10.1016/s0002-9440(10)64378-7
72. Campbell JJ, Haraldsen G, Pan J, Rottman J, Qin S, Ponath P, et al. The Chemokine Receptor CCR4 in Vascular Recognition by Cutaneous But Not Intestinal Memory T Cells. Nature (1999) 400(6746):776–80. doi: 10.1038/23495
73. Reiss Y, Proudfoot AE, Power CA, Campbell JJ, Butcher EC. CC Chemokine Receptor (CCR)4 and the CCR10 Ligand Cutaneous T Cell-Attracting Chemokine (CTACK) in Lymphocyte Trafficking to Inflamed Skin. J Exp Med (2001) 194(10):1541–7. doi: 10.1084/jem.194.10.1541
74. Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, Yoshie O. The T Cell-Directed CC Chemokine TARC Is a Highly Specific Biological Ligand for CC Chemokine Receptor 4. J Biol Chem (1997) 272(23):15036–42. doi: 10.1074/jbc.272.23.15036
75. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res (2016) 44(D1):D733–45. doi: 10.1093/nar/gkv1189
76. Ghadially H, Ross XL, Kerst C, Dong J, Reske-Kunz AB, Ross R. Differential Regulation of CCL22 Gene Expression in Murine Dendritic Cells and B Cells. J Immunol (2005) 174(9):5620–9. doi: 10.4049/jimmunol.174.9.5620
77. Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface Phenotype and Antigenic Specificity of Human Interleukin 17-Producing T Helper Memory Cells. Nat Immunol (2007) 8(6):639–46. doi: 10.1038/ni1467
78. Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ Memory T Cells: Functional Modules for Tailored Immunity. Eur J Immunol (2009) 39(8):2076–82. doi: 10.1002/eji.200939722
79. Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, et al. Memory T Cells in Latent Mycobacterium Tuberculosis Infection Are Directed Against Three Antigenic Islands and Largely Contained in a CXCR3+CCR6+ Th1 Subset. PloS Pathog (2013) 9(1):e1003130. doi: 10.1371/journal.ppat.1003130
80. Ratzinger G, Baggers J, de Cos MA, Yuan J, Dao T, Reagan JL, et al. Mature Human Langerhans Cells Derived From CD34+ Hematopoietic Progenitors Stimulate Greater Cytolytic T Lymphocyte Activity in the Absence of Bioactive IL-12p70, by Either Single Peptide Presentation or Cross-Priming, Than do Dermal-Interstitial or Monocyte-Derived Dendritic Cells. J Immunol (2004) 173(4):2780–91. doi: 10.4049/jimmunol.173.4.2780
81. Polak ME, Newell L, Taraban VY, Pickard C, Healy E, Friedmann PS, et al. CD70-CD27 Interaction Augments CD8+ T-Cell Activation by Human Epidermal Langerhans Cells. J Invest Dermatol (2012) 132(6):1636–44. doi: 10.1038/jid.2012.26
82. de Witte L, Nabatov A, Pion M, Fluitsma D, de Jong MA, de Gruijl T, et al. Langerin Is a Natural Barrier to HIV-1 Transmission by Langerhans Cells. Nat Med (2007) 13(3):367–71. doi: 10.1038/nm1541
83. Ribeiro CM, Sarrami-Forooshani R, Setiawan LC, Zijlstra-Willems EM, van Hamme JL, Tigchelaar W, et al. Receptor Usage Dictates HIV-1 Restriction by Human TRIM5α in Dendritic Cell Subsets. Nature (2016) 540(7633):448–52. doi: 10.1038/nature20567
Keywords: Langerhans cells, dendritic cells, antimicrobial peptides, immunity, skin, transcriptome, bioinformatics
Citation: Oulee A, Ma F, Teles RMB, de Andrade Silva BJ, Pellegrini M, Klechevsky E, Harman AN, Rhodes JW and Modlin RL (2021) Identification of Genes Encoding Antimicrobial Proteins in Langerhans Cells. Front. Immunol. 12:695373. doi: 10.3389/fimmu.2021.695373
Received: 14 April 2021; Accepted: 06 August 2021;
Published: 26 August 2021.
Edited by:
Fatima Conceição-Silva, Oswaldo Cruz Foundation, BrazilReviewed by:
Patrick M. Brunner, Medical University of Vienna, AustriaCopyright © 2021 Oulee, Ma, Teles, de Andrade Silva, Pellegrini, Klechevsky, Harman, Rhodes and Modlin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert L. Modlin, cm1vZGxpbkBtZWRuZXQudWNsYS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.