
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 15 July 2021
Sec. Molecular Innate Immunity
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.684240
This article is part of the Research TopicNanoparticle-Mediated Signaling Rewiring and Reprogramming of Immune ResponsesView all 12 articles
Systemic delivery of peptide-major histocompatibility complex (pMHC) class II-based nanomedicines can re-program cognate autoantigen-experienced CD4+ T cells into disease-suppressing T-regulatory type 1 (TR1)-like cells. In turn, these TR1-like cells trigger the formation of complex regulatory cell networks that can effectively suppress organ-specific autoimmunity without impairing normal immunity. In this review, we summarize our current understanding of the transcriptional, phenotypic and functional make up of TR1-like cells as described in the literature. The true identity and direct precursors of these cells remain unclear, in particular whether TR1-like cells comprise a single terminally-differentiated lymphocyte population with distinct transcriptional and epigenetic features, or a collection of phenotypically different subsets sharing key regulatory properties. We propose that detailed transcriptional and epigenetic characterization of homogeneous pools of TR1-like cells will unravel this conundrum.
Interleukin 10 (IL-10)-producing regulatory T cells (Tregs) are key to immune homeostasis and play opposing roles in autoimmunity versus cancer. While the FoxP3+ Treg cell subset has been thoroughly described, FoxP3 and CD25 double-negative T cells producing IL-10 in the context of low IL-4 secretion are generally known as T-regulatory type 1 (TR1) cells (1). Given the lack of specificity of these phenotypic descriptors, the literature has considered as TR1-like cells what appears to be a rather heterogeneous collection of cell types (1), thus clouding our understanding of the true lineage identity of this regulatory T-cell subset. Production of IL-10, coupled to the expression of Latency-Associated Peptide (LAP), Lymphocyte Activation Gene 3 (LAG-3) or CCR5 and Programmed cell death protein 1 (PD-1) in the absence of CD25, or CD4+ cells lacking IL-7R expression, as well as cells induced by vitamin D3 or CD46-stimulation are some of the examples of cell types identified as TR1 (1, 2). Recently, co-expression of CD49b and LAG-3, accompanied by the expression of ICOS and PD-1, has been associated, in both humans and mice, with TR1-ness (3, 4), but these markers are not sufficiently specific or sensitive. Other surface markers have been found to be variably upregulated by IL-10-producing T-cell subsets (5–8), including Cytotoxic T-Lymphocyte antigen 4 (CTLA-4), T-cell immunoglobulin and mucin-domain containing-3 (TIM-3) or TIGIT, as well as transcription factors (TFs) like T-bet, Aryl hydrocarbon receptor (AhR) or Nuclear Factor Interleukin 3-regulated (Nfil3).
Because of the lack of specific markers, it remains unclear whether the various IL-10 producing ‘TR1-like’ subsets correspond to multiple different cell types, or to cells at different stages of differentiation. Many studies implicating a role for Treg/TR1 cells in the therapeutic activity of various immunotherapies have often done so solely based on an increase in IL-10 expression by splenic CD4+ T cells. It is entirely possible that the various phenotypes associated to IL-10-producing FoxP3-negative CD4+ T-cell subsets correspond to cells at different stages of TR1 cell differentiation, or to distinct subsets of terminally differentiated cells with distinct phenotypic and/or functional properties. Unfortunately, the transcriptional and epigenetic profiles associated with true TR1-ness remain incompletely defined, a fact compounded by our incomplete knowledge on the developmental biology of the TR1 subset(s).
We have shown that treatment of various mouse models of autoimmune disease with nanoparticles (NPs) coated with disease-relevant peptide-major histocompatibility complex class II (pMHCII) molecules (9) suppresses organ inflammation and disease progression without impairing systemic immunity (10–12). This approach has shown clear therapeutic efficacy in animal models of type 1 diabetes (T1D), experimental autoimmune encephalomyelitis (EAE), collagen-induced arthritis (11), as well as primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC) and autoimmune hepatitis (AIH) (12, 13). pMHCII-NP therapy triggers the formation and expansion of TR1-like CD4+ T cells from autoantigen-experienced CD4+ T-cell precursors of as yet undefined identity. pMHCII-NPs bind directly to TCRs on cognate T cells, resulting in prolonged pMHCII-TCR interactions, the assembly of large TCR microclusters on such T cells, and rapid, robust and prolonged TCR signaling. In turn, this results in the acquisition of immunoregulatory properties, including the upregulation of the cytokines IL-10, IL-21 and Transforming Growth Factor β (TGF-β) (but neither IL-2 nor IL-4), the co-inhibitory receptors LAG-3, CTLA-4 and PD-1, the Inducible T-cell Costimulator (ICOS) and the transcription factors T-bet and c-Maf, among others, in the absence of FoxP3 expression (Figure 1).
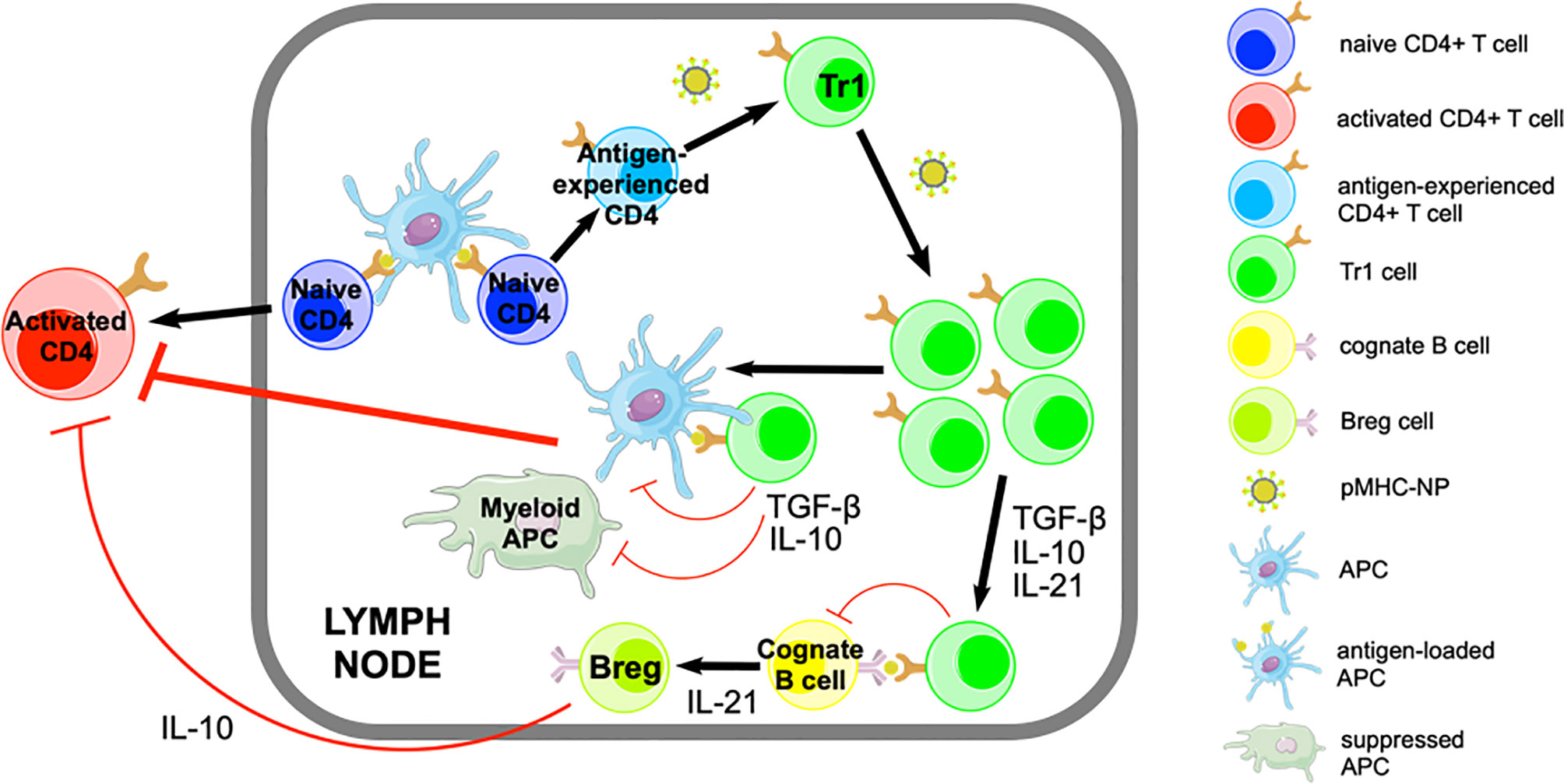
Figure 1 Pharmacodynamic activity of pMHCII-NPs. pMHCII-NPs target autoantigen-experienced CD4+ T cells and induce their differentiation into memory TR1-like cells followed by their systemic expansion. This process involves IFN-γ and IL-10 signaling, but does not require IL-27. pMHCII-NP-induced TR1-like cells carry out their regulatory function by suppressing other autoreactive T-cell specificities via IL-10, IL-21 and TGF-β. IL-10 and TGF-β have immunosuppressive effects on autoantigen-loaded APCs, inhibiting their proinflammatory function and thus avoiding the activation of other non-cognate autoreactive T cells. pMHCII-NP-induced TR1-like cells can also interact with cognate B-cells, promoting their differentiation into Bregs in part via IL-21. Figure adapted from Clemente-Casares et al. (11).
Here, we review our current understanding of the phenotype, function and development of TR1-like cells in different experimental settings, including pMHCII-NP-treated mice. We identify knowledge gaps and propose that detailed transcriptional and epigenetic characterization of homogeneous pools of TR1-like cells will help define both, a true state of TR1-ness as well as the identity of the TR1-poised cell precursors that give rise to TR1-like cells.
The TR1 cell subset was first described in 1997 by Groux et al. (14). Previously, others (15, 16) had described a suppressor T-cell population that secreted IL-10 and protected patients against graft-versus-host disease (GvHD). This population displayed a cytokine profile that was distinct from those of common T-helper cell subsets, and involved the expression of IL-10, IL-5, TGF-β, and IFN-γ in the absence of IL-4 or IL-2 secretion (17). In 1997, TR1 cells were generated in vitro and their suppressive activity was documented both in vitro and in vivo, in a model of colitis.
Currently, all the regulatory CD4+ T cells that are FoxP3-negative and secrete IL-10 and low levels or no IL-4 are considered to be ‘TR1’. Unfortunately, this characterization lacks specificity and likely includes phenotypically, functionally and developmentally heterogeneous T cells. This is compounded by the variety of protocols that can trigger the formation of IL-10-producing cells with regulatory properties. In some cases, TR1-like cells were generated from naive CD4+ T cells. For example, in vitro TCR stimulation of human naive CD4+/CD45RA+ T cells in the context of IL-10 secreted by dendritic cells (DCs) triggered their conversion into anergic, IL-10- and TGF-β-expressing T cells capable of suppressing effector T cells (14, 18). Likewise, in vitro culture of murine CD4+/CD44–/CD62L+ T cells with IL-10 or IL-27 can induce their differentiation into IL-10 producing TR1-like cells [reviewed in (19)]. Other lines of experimentation have suggested that IL-10-producing TR1-like cells can also be generated from memory CD4+ T cells, in the absence of polarizing cytokines in the culture (20, 21). In mice, induction of transplantation tolerance via anti-CD45RB mAb therapy is associated with the presence of antigen-specific IL-10-producing CD4+ T cells in the memory T-cell compartment (22, 23). There are also data supporting the view that TR1-like cells can develop from differentiated T-helper cell subsets. For example, Gagliani et al. provided evidence suggesting that a fraction of the regulatory T cells that are found in the gut arise from Th17 cells and display a TR1-like phenotype, including the production of IL-10 and some IFN-γ, and the expression of CD49b and LAG-3, while lacking expression of IL-4 and CCR6 (24). Moreover, there is also evidence that culture of Th17 cells in the presence of IL-27 and TGF-β can trigger the formation of IL-10-producing TR1-like cells in vitro (24, 25). Likewise, stimulation of Th1 cells with CXCL12 in vitro (26), or in the context of malaria infection (27), can promote their differentiation into CD4+/CD25–/FoxP3–/IL-10+ T cells. Human allergen-specific Th2 cells can also differentiate into IL-10-producing CD49b+/LAG3+ cells with regulatory properties (28, 29).
Unfortunately, these various ‘TR1-like’ cell types of different developmental origin were not thoroughly characterized at the phenotypic, transcriptional or functional levels. Accordingly, whether the various TR1-like cells that were generated in these studies correspond to one or several different cell types, or to cells at different stages of differentiation, remains unclear.
Several surface phenotypes have been attributed to TR1-like cells (Table 1). Whether all these subsets correspond to one single, incompletely characterized population, or comprise a collection of phenotypically and/or functionally distinct subsets remains to be determined.
CD4+ CD25+ Treg cells express TGF-β on their surface and one of their mechanisms of suppression involves TGF-β recognition by target cells upon cell-to-cell contact (47). Weiner et al. reported a population of regulatory T cells that suppressed murine colitis in a TGF-β-dependent manner, but where CD25-negative and LAP-positive (30). LAP is the amino-terminal domain of the TGF-β precursor peptide that contains the TGF-β peptide within its latent complex (48). CD4+/CD25–/LAP+ cells are positive for thrombospondin, which can convert latent TGF-β to its active form. CD4+/CD25–/LAP+ cells represent ~3-5% of murine splenocytes and express high levels of TGF-β and IL-10, as well as IL-2, IL-4 and IFN-γ. A similar population was generated after oral anti-CD3 treatment and had a suppressive effect against autoimmune encephalomyelitis (31).
A small population of human CD4+ T cells that produce IL-10 and TGF-β express the natural killer receptor NKG2D. These cells are FoxP3-, CD103- and LAG-3-negative. They also express Fas ligand (FasL), which appears to be a main contributor of suppression by inhibiting the growth of bystander T cells (32). Although these T cells can be found in the peripheral blood of healthy individuals (~1-3%), they appear to increase substantially in cancer patients (to ~6-70%). They have also been described in patients with rheumatoid arthritis (33). One ligand of the NKG2D receptor is the MHC class I-related chain A (MICA), which is upregulated in tissues undergoing inflammation or in epithelial tumors. The role of NKG2D with regards to the immunoregulatory properties of these cells remains unclear.
The IL-7 receptor (IL-7R) α-chain (CD127) is important for the survival of conventional CD4+ T cells (49) but is expressed at low levels in CD4+CD25+ T cells (50). Häringer et al. found a population of adaptive Treg cells that were CD25-, FoxP3- and IL-7R-negative. These cells comprised ~1% of the total CD4+ population from human peripheral blood. They expressed low levels of Bcl-2 and high levels of Ki-67 and ICOS, suggesting that they had been recently activated, and had a suppressive function mediated primarily by the secretion of IL-10 in response to potent T-cell receptor stimuli (34). However, only 10% of this T-cell pool produced IL-10 upon stimulation, compatible with the presence of a small subset of TR1-like cells within the CD25–/FoxP3–/IL-7R– pool.
Several studies have identified a population of CD4+ T cells with regulatory activity that express CD49b. These cells had anti-diabetogenic (35) and anti-arthritogenic properties in mice (36), were both FoxP3– and CD25– and secreted the regulatory cytokines IL-10 and TGF-β, as well as IFN-γ. In later studies, it was shown that these T cells suppressed CD8+ T-cell responses and IFN-γ production by CD4+ T cells, presumably via IL-10 (37, 38).
LAG-3 is known to suppress T-cell proliferation (51). Despite being required for the maximal regulatory activity of conventional CD4+CD25+ Treg cells, LAG-3 protein can hardly be detected on the surface of CD4+CD25+ T cells. In contrast, LAG-3 was found to be expressed by a subset of CD4+CD25– T cells (39) found at low frequencies in the spleen (2%) and lymph nodes (1%) but at higher frequencies in Peyer’s patches (PP) (8%). These T cells are anergic upon TCR ligation, but they secrete high quantities of IL-10, moderate amounts of IFN-γ and low amounts of IL-2 and IL-4. These cells do not express FoxP3 and, unlike CD4+/CD25–/LAP+ cells, express low levels of CD103 and LAP. They are further characterized by expression of the Early response gene 2 (Egr-2), a transcription factor that is a negative regulator of T-cell activation, inducing an anergic state (52). These CD4+/CD25–/LAG-3+ cells were also found to express the Prdm1 gene, encoding the B lymphocyte-induced maturation protein (Blimp)-1.
In 2013, Gagliani et al. provided evidence indicating that co-expression of LAG-3 and CD49b can be used to enumerate human and mouse TR1-like cells (3). CD49b had been previously described as a marker for regulatory CD25– T cells, but cannot be used in isolation to identify this T-cell subset, because it can also be expressed by Th17 cells and certain memory CD4+ T-cell subsets (53). Likewise, LAG-3 is associated with T-cell activation and IL-10 production, but its expression is not unique to any particular T-cell subset; it can be upregulated by conventional T cells upon activation and is also expressed by FoxP3+ Tregs (51).
Geginat and coworkers used the C-C chemokine receptor type 5 (CCR5) and PD-1 as markers to purify TR1-like IL-10- and IFN-γ-producing cells from the human intestine (2, 40). They demonstrated that the majority of IL-10+/CD4+/CD25–/IL-7R– T cells found in the lamina propria co-expressed CCR5+ and PD-1+ (2). Despite expressing Lag3 mRNA, only a small percentage of cells displayed LAG-3 protein in the steady state. In vitro stimulation triggered the upregulation of surface LAG-3 protein expression (2).
TR1-like cells express several other surface molecules and transcription factors, albeit none of them specifically. For example, both murine and human IL-10 producing TR1-like cells can express the immune checkpoint molecules TIGIT (5) and TIM-3 (8), but conventional FoxP3+ Treg cells and T-follicular regulatory (Tfr) cells (54, 55) can also express these markers. CD226, presumably involved in the cytotoxic activity of at least some TR1-like cells, is another example of such lack of specificity (3, 41).
With regards to transcription factors, ROG (the repressor of GATA-3), a regulator of Th differentiation and cytokine production upon activation (56), has also been described in TR1-like cells (42). Since expression of Egr-2 in CD4+ T cells induces IL-10 production by binding to the Blimp-1 promoter (57), Okamura et al. proposed that this transcription factor might be involved in the acquisition of a suppressor phenotype by CD4+/CD25–/LAG-3+ T cells (43). However, purified TR1-like cells from the gut of anti-CD3 mAb-treated mice, as well as those induced in vitro, express levels of Egr-2 that are no different than those seen in effector T cells (3). Likewise, the transcription factors c-Maf and AhR, which are expressed by IL-27-induced TR1 cells and bind to the Il10 promoter in TR1 cells (44, 45), are also expressed by non-TR1 cell types, including human and murine Th17 subsets (58, 59). The interferon regulatory factor 4 (IRF4) is yet another non-TR1 cell-specific transcription factor that presumably plays a role in the developmental biology of TR1-like cells, as a downstream effector of the inducible tyrosine kinase (ITK) (60). Since IRF4 regulates Blimp-1, it is probably involved in the regulation of IL-10 expression in these cells, along with other transcription factors. Activin A-induced IRF4 activation has been suggested to promote human TR1-like cell formation in vitro (46). The liver X receptor (LXR) and Bhlhe40 are other transcription factors found to be expressed in at least some TR1-like cells (7).
When compared to other TR1-like subsets, the IL-10-producing CD44hi/CD62Llo/IL-7R–/CD25–/FoxP3– TR1-like cells that arise in vivo in response to pMHCII-NP therapy co-express several of the markers previously identified in different TR1-like cell subsets, including LAG-3, CD49b, ICOS, LAP, c-Maf, T-bet, and Blimp-1. These cells produce the cytokines IL-10, IL-21 and, to a lesser extent, IFN-γ, but no or very low levels of IL-2, IL-4 or IL-17 (11). Thus, these cells appear to embody the phenotypic properties of most other TR1-like cells, begging the question of whether different IL-10-expressing CD4+CD25– TR1-like cell subsets, as described in the literature, correspond to one single cell subset rather than to a phenotypically heterogenous collection of distinct cell types.
In order to affect regulatory activity, TR1 cells need to be activated by antigen recognition. Upon activation, they target effector T cells and/or professional APCs via cytokines, direct cell contact, metabolic disruption and/or cytolysis (Figure 2). Although TCR activation is antigen-specific, TR1-mediated suppression of APCs or neighboring T cells is antigen-agnostic (bystander immunoregulation).
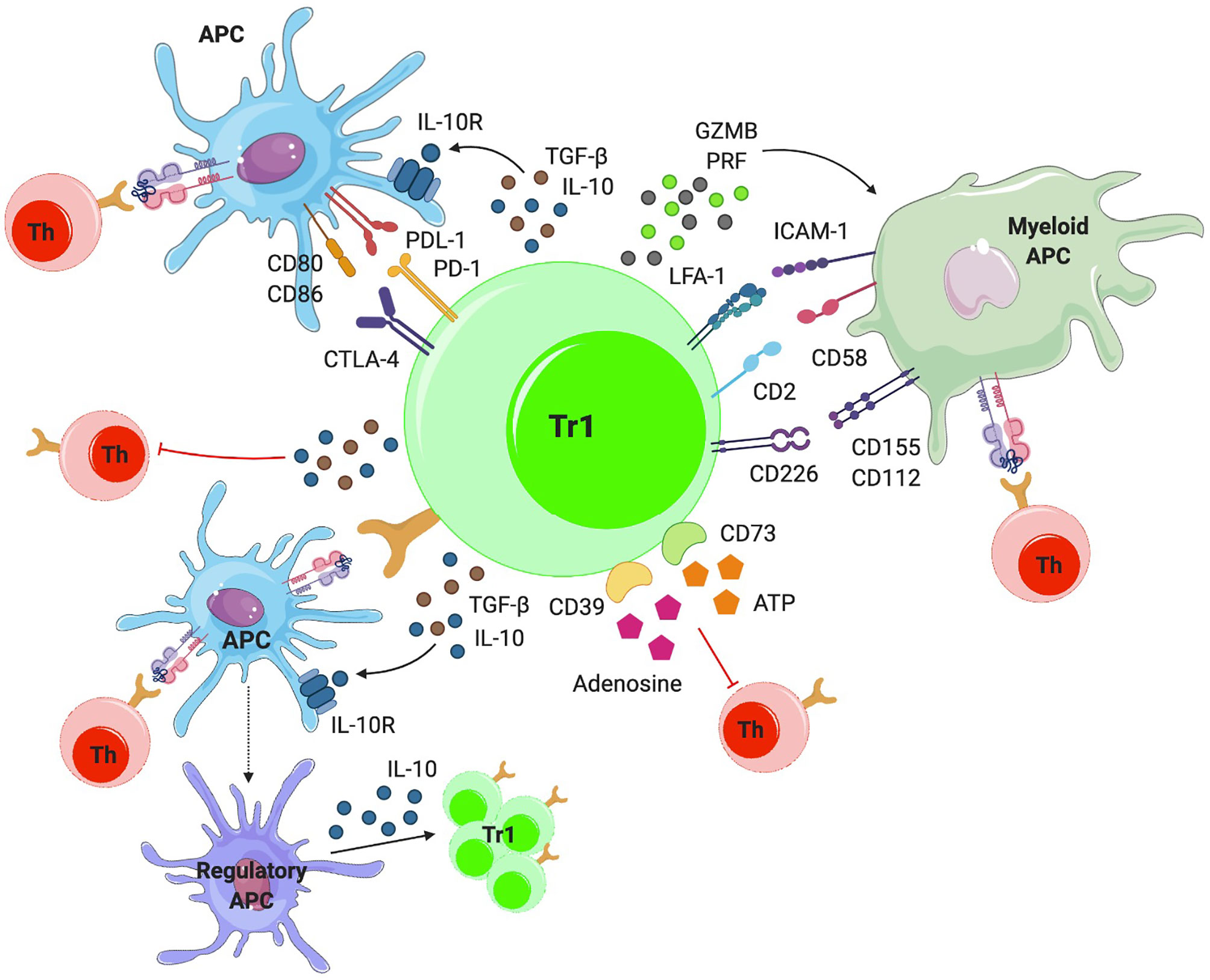
Figure 2 Mechanisms of action. TR1-like cells can suppress effector T cells directly or indirectly, by modulating the phenotype and function of APCs via IL-10 and TGF-β. IL-10 also induces a regulatory phenotype in APCs, by triggering the upregulation of tolerogenic molecules (i.e. ILT3, ILT4 and HLA-G) and the production of IL-10, further amplifying TR1-like cell formation. TR1-like cells can also make direct cell-to-cell contacts with APCs via cell surface CTLA-4 and PD-1, inhibiting APC-induced effector T-cell activation. TR1-like cells can kill myeloid APCs via granzyme-B (GZMB) and perforin (PRF). In addition, they can suppress T-cell activation via metabolic disruption.
Upon activation, TR1 cells secrete the immunoregulatory cytokines IL-10 and TGF-β (Figure 2). IL-10 has effects on different cell populations. Although IL-10 expression is a hallmark of TR1-like cells, this cytokine can also be produced by other CD4+ T-cell subsets, as well as CD8+ T cells, macrophages, DCs and B cells (61). IL-10 suppresses T-cell responses by inhibiting T-cell proliferation (62) and cytokine production by effector T cells, including IL-2, IFN-γ, IL-4, IL-5 and TNF-α. Moreover, IL-10 can downregulate MHC class II and costimulatory molecule expression in APCs, and reduce the production of pro-inflammatory cytokines (IL-1α and -β, IL-6, IL-12, IL-18, and TNF-α) and chemokines (CCL2, CCL5, CCL12, CXCL2, CXCL10, and IL-8) by these APCs (61). In humans, IL-10 can elicit the generation of tolerogenic DCs by upregulating immunoglobulin-like transcripts 3 and 4 (ILT3, ILT4) and the non-classical HLA-G molecule (63). On B-cells, IL-10 promotes proliferation, MHC II expression and isotype switching to IgG4 (64). IL-10 also amplifies regulatory T-cell formation. IL-10 stimulation of CD4+ T cells can induce the expression of IL-10, T-cell anergy or TR1-like cell differentiation in a STAT3-dependent manner. STAT3 promotes IL-10 expression and represses pro-inflammatory cytokine expression (65). It is unclear whether the phenotype of full-fledged (i.e. fully differentiated) TR1-like cells is stable. However, pMHCII-NP-induced, antigen-specific TR1-like cells can persist for several months post-treatment withdrawal without any obvious loss of key phenotypic properties or acquisition of pathogenic activity (11).
Like IL-10, TGF-β inhibits APC function and T-cell proliferation, differentiation and cytokine production (Figure 2). TGF-β suppresses T-cell proliferation by inhibiting IL-2 production and downregulating cyclins while upregulating cyclin-dependent kinase (CDK) inhibitors. It also inhibits the differentiation of both CD4+ and CD8+ T cells into effectors, by inhibiting master transcriptional regulators of each phenotype (GATA-3, T-bet, IL-12Rβ2). The main effect of TGF-β on APCs involves inhibition of their maturation, in part by upregulating indoleamine 2,3-dioxygenase (IDO) expression and by inhibiting MyD88-mediated TLR signaling (66). As shown in (11, 12), the therapeutic effects of pMHCII-NP-induced TR1-like cells are dependent on IL-10 and TGF-β. The blockade of these cytokines with monoclonal antibodies abrogates the suppression of autoantigen crosspresentation by pMHCII-NP-expanded TR1-like cells and thus the therapeutic properties of pMHCII-NP treatment in several models, including T1D, EAE and liver autoimmunity.
TR1-like cells can also inhibit APCs in a cell contact-dependent manner upon engagement of co-inhibitory receptors such as CTLA-4, PD-1, LAG-3 or TIGIT and the costimulatory molecule ICOS (Figure 2).
Like other members of the CD28 family, CTLA-4 can bind CD80/86, but it does so with higher affinity than the co-stimulatory molecule CD28. In the presence of CTLA-4, CD80/86 engagement by CD28 on T cells is inhibited. In addition, CTLA-4 can signal into T cells through Src homology region 2-contatining protein tyrosine phosphatase 2 (SHP-2), dephosphorylating TCR and CD28 signaling intermediates and promoting T-cell inactivation (67). However, engagement of CTLA-4 on T cells by its ligands on APCs can also have inhibitory effects on the latter, such as by triggering the downregulation of CD80 and CD86 (68–70), or by upregulating IDO expression by APCs (71).
LAG-3 is another negative regulator of T-cell activation. This molecule is structurally similar to CD4 and binds MHC class II molecules with higher affinity than CD4 (39). Okazaki’s work has recently shown that LAG-3 does not universally bind to all MHC class II molecules, but rather recognizes stable pMHC class II complexes (72). LAG-3 signals intracellularly, transducing inhibitory signals that hinder T-cell activation (72). Inhibitory signals through the LAG-3 intracytoplasmic region are mediated by a FXXL motif in the membrane-proximal region and the EX repeat in the C-terminal region (73). In addition, the LAG-3-pMHCII interaction inhibits DC activation (74).
PD-1 is a co-inhibitory receptor that belongs to the Ig superfamily containing ITIM and ITSM motifs and signals after interacting with its ligands PD-L1 or PD-L2. PD-L1 is expressed on leukocytes, non-hematopoietic cells and non-lymphoid tissues, and can be induced in parenchymal cells by inflammatory cytokines (e.g. IFN-γ) or tumorigenic signaling pathways. PD-L1 expression is also found on different tumor types and is associated with an increased number of tumor-infiltrating lymphocytes (TILs) and poor prognosis. PD-L2 is primarily expressed on professional APCs (DCs and monocytes) but can be induced in other immune and non-immune cell types. PD-1 has a higher binding affinity for PD-L2 than for PD-L1, a difference that might be responsible for the differential contributions of these two ligands to immune responses. It has an inhibitory function similar to that of CTLA-4, by recruiting SHP-1 and SHP-2 phosphatases, reducing T-cell activation and inducing Treg differentiation (75). There is also emerging evidence for ‘reverse signaling’ through PD-L into DCs. PD-1 binding to PD-L2 decreases the expression of DC maturation markers, such as CD40, CD80 and CD86, and increases IL-10 production by DCs, resulting in a suppressive DC phenotype (76).
TIGIT is another immune checkpoint inhibitor that interferes with the activation of T and NK cells. It has an extracellular IgV domain and an intracellular ITT domain that recruits SHIP-1 to mediate T-cell inactivation (77). TIGIT competes with the immunoactivator receptor CD226 (DNAM-1) for the same ligands: CD155 (poliovirus receptor, PVR) and CD112 (Nectin-2 or PVRL2), expressed on APCs, T cells and some non-hematopoietic cell types like tumor cells (78). TIGIT binding to its ligands on APCs has an effect on DC cytokine production, inducing IL-10 expression and inhibiting the expression of IL-12, reducing T-cell activation (79).
ICOS is a costimulatory molecule with structural homology to CD28 and CTLA-4 that binds to ICOS-L on DCs, B cells, and macrophages. ICOS-ICOS-L engagement regulates antigen presentation and secretion of regulatory cytokines such as IL-10 by APCs (80–82).
TR1-like cells can also inhibit effector T cells via metabolic disruption mechanisms, similar to those used by FoxP3+ Tregs. In TR1-like cells, the main proteins involved in this process are the ectoenzymes ectonucleoside triphosphate diphosphohydrolase 1 (CD39) and ecto-5’-nucleotidase (CD73). These enzymes hydrolyze extracellular 5’-adenosine triphosphate (ATP) to adenosine, disrupting the metabolic state of T cells. ATP released during T-cell activation (83) has an effect on T-cell and APC activation (Figure 2). First, CD39 degrades ATP and ADP into AMP (84), which is then further degraded to adenosine by CD73 (85). Adenosine can bind to A2A receptors, inhibiting T-cell proliferation and cytokine production of effector T cells (86). Binding of adenosine to these receptors on APCs inhibits their maturation and the secretion of pro-inflammatory cytokines, while inducing the secretion of IL-10 (87).
Another mechanism via which TR1-like cells can inhibit T-cell responses is by killing APCs, particularly APCs of myeloid origin. TR1-like cells can express both granzyme A and B proteins, which, together with perforin, mediate cell-mediated cytotoxicity (88) (Figure 2). In humans, granzyme expression has been shown to be induced by IL-10 signaling (89). Unlike NK-mediated killing, which takes place when target cells lack or downregulate MHC class I, TR1-mediated cytolysis is antigen-dependent and only takes place when there is TCR engagement with cognate pMHC on the APC (it also requires recognition of other surface molecules expressed by the APC, including CD54 (ICAM-1), CD58, CD155 and CD112) (41). In addition to direct effects on the activation of antigen-specific CD4+ T-cell responses, APC killing indirectly impairs the activation of bystander T cells. Although pMHCII-NP-induced TR1-like cells can express granzymes, they lack cytolytic activity against peptide-pulsed B-cells or DC cells (11).
TCR stimulation is essential, but not sufficient for the generation of TR1-like cells. pMHC multimers (90–92) or superantigens (93, 94) have been found to induce IL-10-production in some T-cell populations, although it is not clear whether the resulting cells were bona fide TR1-like cells. Several studies have suggested that the strength of the TCR interaction plays an important role; high avidity interactions favor IL-10 production (95), in particular the number of IL-10-producing cells and the cells’ suppressive properties (96). The dose of antigen appears to play a lesser role, as high doses of ligands were not enough to induce IL-10 unless they were administered simultaneously with IL-12 (97, 98). Nevertheless, repeated high-dose stimulation was indeed sufficient to induce IL-10. One study pointed to Nfil3 as a transcription factor involved in the upregulation of IL-10 production in response to repeated antigenic stimulation (99). However, as noted above, it is unclear whether these cells were true TR1-like cells or simply Th1 cells that have acquired the ability to produce IL-10. Singha et al. have shown that the ability of pMHCII-NP to elicit TR1 cell formation is dependent on high pMHCII densities onto the NPs. High densities promote sustained pMHC-NP-TCR interactions and formation of TCR microclusters, amplifying the duration and magnitude of TCR signaling, which is associated with their pro-TR1-like cell-differentiation properties (9).
IL-10 has been associated with the induction and maintenance of TR1-like cells (14, 100), although some studies have suggested that it is dispensable (101). In the absence of IL-10 (in Il10 gene knockout mice) pMHCII-based nanomedicines could readily trigger the expansion of cognate T cells, but these cells upregulated IL-4, suggesting a role for IL-10 in the acquisition of the full-fledged TR1-like cell phenotype (11). Tolerogenic DCs are the main source of IL-10 in vivo and they may play a role in the induction of TR1-like cells under physiological conditions (102, 103). Indeed, human IL-10-producing DCs have the ability to induce TR1-like cell differentiation in vitro in an IL-10-dependent manner (63, 104).
IL-27, largely produced by activated APCs (105), can support the generation of IL-10-producing TR1-like cells and CD8+ T cells (106, 107). It is a member of the IL-12 family and is a heterodimer composed by the Epstein-Barr virus-induced gene 3 (Ebi3)-encoded IL-12-related p40 and the IL-27 p28 (or IL-27α) chains. IL-27 binds to the IL-27 receptor (IL-27R) on DCs, monocytes, macrophages, T and B lymphocytes, NK cells, mast cells, and endothelial cells. This receptor is a heterodimer composed by the orphan cytokine receptor WSX-1 (also known as T-cell cytokine receptor (TCCR)) and a signal-transducing chain, the glycoprotein 130 (gp130).
IL-27 has inhibitory effects on Th1, Th2 and Th17 subsets as well as on APCs (108–110). Several studies have shown that it is capable of inducing both murine (111, 112) and human (106, 113) IL-10-producing T cells. Signaling through the IL-27R primarily induces STAT1 and STAT3 activation, promoting the expression of AhR and c-Maf transcription factors, which in turn control IL-10 and IL-21 production, hallmarks of the TR1-like cell phenotype (44). STAT3 further upregulates Egr-2, which as noted above contributes to IL-10 production by promoting Blimp-1 expression (57) (Figure 3).
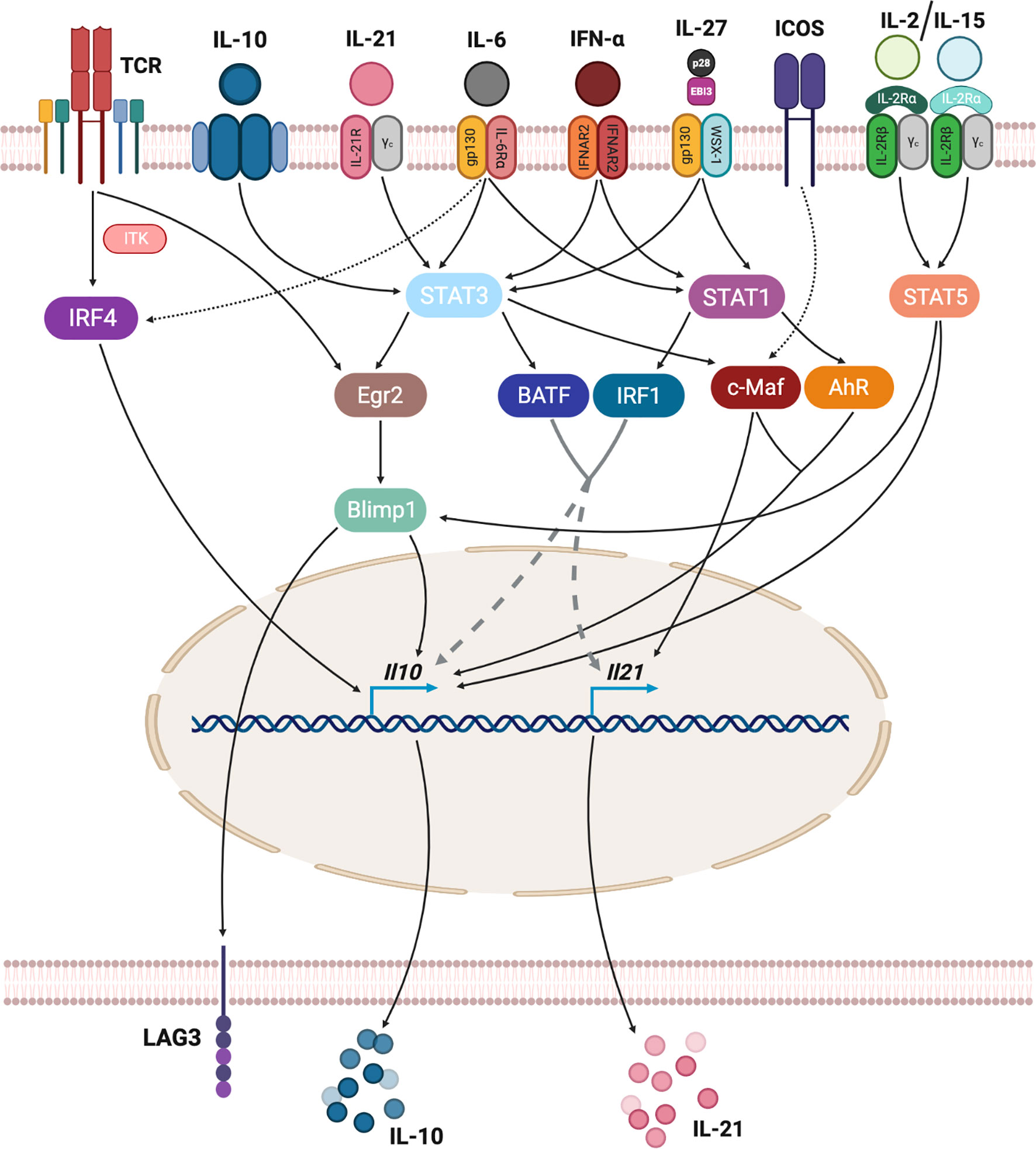
Figure 3 Transcriptional regulation of TR1-like cell formation. TR1-like cell differentiation requires the integration of different stimuli. TCR signaling, through IRF4, can activate IL-10 expression. Many cytokines, including IL-10, IL-21, IL-6, type-I interferons and IL-27 signal via STAT1 and/or STAT3 proteins, activating several transcription factors that regulate IL-10 and IL-21 expression. ICOS signaling is also a direct regulator of IL-21 expression, while IL-2 or IL-15 cytokines can induce IL-10 directly via STAT5 binding to Il10 or via STAT5-mediated activation of Blimp-1.
Notwithstanding the positive role of IL-27-IL-27R signaling in TR1-like cell differentiation in vitro, pMHCII-NP-induced TR1-like cell formation in vivo is IL-27R-independent (11).
IL-21 is a type I cytokine that is produced by antigen-stimulated CD4+ T cells as well as NKT cells, and it has pleiotropic effects targeting T, B, NK, and myeloid cells (114). IL-21 binds to a heterodimeric receptor that is composed by the IL-21Rα chain (115) and the common cytokine receptor γc chain and signals through STAT3 and, to a lesser extent, STAT1 and STAT5. IL-21 plays a critical role in the regulation of Ig production and in the differentiation of B-cells into antibody-producing plasma cells (116, 117), in part by inducing T-follicular helper (TFH) cells (118), and has been implicated in the promotion of CD8+ T-cell and NK cell responses (119). IL-21 can also have negative effects on immune responses, such as by inducing B-cell apoptosis (120) and inhibiting DC maturation and function (121). c-Maf, expressed by TR1-like cells, contributes to IL-21 expression (59), and IL-27 promotes IL-21 expression in TR1-like cells by upregulating c-Maf (44). Furthermore, IL-21 functions as an autocrine growth factor that facilitates the expansion and homeostasis of IL-27-derived TR1-like cells (44), in part by promoting the upregulation of IL-10 (122) and, in turn, c-Maf expression. Like their IL-27-induced counterparts, pMHCII-NP-induced TR1-like cells express and secrete high levels of IL-21 upon recognition of cognate pMHCII on professional APCs (11, 123), which then plays a critical role in the TR1-like cell-induced differentiation of conventional B-cells into IL-10/IL-35-producing Breg cells and in the recruitment/re-programming of neutrophils into myeloid-derived suppressor-like cells, as downstream effectors of pMHCII-NP-induced immunoregulation (11, 123).
The ICOS molecule, a member of the B7 superfamily, is a glycosylated disulfide-linked homodimer that is expressed by certain T-cell subsets, including TFH- and TR1-like cells, upon productive TCR ligation. The ICOS-L is expressed on a wide range of lymphoid and non-lymphoid cells types, including APCs (124). ICOS signaling has been implicated in IL-10 production (80), as well as in IL-6-induced TFH cell specification (125, 126), although it can also stimulate the production of Th1 and Th2 cytokines in vivo. There is also evidence that c-Maf is a downstream target of ICOS engagement (59, 127), suggesting that ICOS engagement on TR1-like cells plays a role in the stabilization of the TR1-like cell phenotype, in part by sustaining IL-21 and IL-10 expression.
IL-6 is a pleotropic cytokine with both pro- and anti-inflammatory effects. It has been associated with the development/progression of certain autoimmune diseases, such as EAE, rheumatoid arthritis and psoriasis (128–130), in part by promoting Th17, TFH and B-cell responses (131). However, it has suppressive effects on the development of T1D, dextran sodium sulfate (DSS)-induced colitis and inflammatory bone destruction (132–134). IL-6, together with TGF-β, was found to induce expression of IL-10 in Th17 cells without suppressing IL-17 production (112, 135). It has also been shown that IL-6 can upregulate IL-21 production and, together with IL-2, can induce IL-10 expression and thus promote TR1-like cell generation (136), even in the absence of IL-27 or TGF-β. It is worth noting that IL-6 shares certain structural homology with IL-27 and that, like IL-27, binds to the gp130 receptor. Both cytokines signal through STAT1 and STAT3. IL-6 can upregulate the TR1-like transcription factors c-Maf, AhR and IRF4 which, as noted above, play a role in IL-10 and IL-21 production (136).
The type I interferons IFN-α and -β, constitute the first barrier against viral infections by inducing an ‘antiviral state’ in target cells which seeks to blunt protein synthesis, degrade mRNA and promote cell death in order to prevent viral replication. Type I interferons also induce upregulation of MHC I and adhesion molecules to enhance cytotoxic T lymphocyte (CTL)-mediated killing of virus-infected cells. However, IFN-α also has anti-inflammatory properties, such as the suppression of IL-8 and IL-1 production or the upregulation of the IL-1R antagonist (IL-1RA). By signaling via STAT1, STAT2 and STAT3 (137, 138), type I IFNs can promote the expression of IL-10 by CD4+ T cells (139–142), including memory T cells. When administered with anti-CD3 and IL-10, IFN-α promoted the development of TR1-like cells (100).
IL-2 and IL-15 function as T-cell growth factors (143, 144). IL-15 was initially shown to play a critical role in the preservation of the memory repertoire, by preventing T-cell apoptosis (145) and promoting the survival of resting memory T cells (146, 147). Some years later, Bacchetta et al. described IL-15 as a growth factor capable of inducing and supporting TR1-like cell proliferation in the absence of TCR ligation (148). Culture of TR1-like cell clones with IL-15 supported their in vitro proliferation. A recent report has suggested that IL-15 may inhibit the production of IL-10 by DCs, thus preventing the generation of IL-10-producing CD4+ T cells (149).
The IL-15R shares its β and γ chains with the IL-2R (150, 151). Although similar, IL-2 and IL-15 have non-overlapping functions. While IL-2 is mainly produced by T cells and plays a major role in the homeostasis of IL-2Rα (CD25)-expressing T cells, like activated T cells or nTregs (143, 152), IL-15 is produced during the innate immune response by cell types other than T cells (151). Stimulation with IL-2 can reverse clonal anergy (153). IL-2 and other γ-chain cytokines, such as IL-15 or IL-21, signal through STAT5. The presence of a STAT5-responsive intronic enhancer in the Il10 locus suggests that these cytokines might also contribute to IL-10 expression by CD4+ T cells (154, 155).
The pMHCII-NP-induced TR1 population specifically develops from autoantigen-experienced precursors (11). For example, whereas diabetic NOD.G6pc2−/− mice (which lack IGRP) responded to BDC2.5mi/IAg7-NPs like wild-type NOD mice, they did not respond to IGRP4–22/IAg7-NPs. In vitro, BDC2.5 TCR-transgenic anti-CD3/anti-CD28 mAb-activated but not naïve T cells upregulate both CD49b, LAG-3 and IL-10 in response to BDC2.5mi/IAg7-NPs, indicating that ligation of cognate TCRs by NP-bound pMHCII complexes can trigger these events only in antigen-experienced cells.
Studies using diabetic NOD.Ifng−/− and NOD.Il10−/− mice revealed that development of the TR1 precursors and/or TR1-like cells that expand in response to this therapy requires IFN-γ in addition to IL-10 (11). The memory-like phenotype and the upregulation of T-bet mRNA in the expanded TR1-like cells, coupled with the inability of pMHC-NPs to trigger expansion of cognate TR1-like cells in non-diseased mice or NOD.Ifng−/− mice suggested that the TR1 precursors might be autoantigen-experienced effector/memory T cells of an as yet unknown identity.
As noted above, although IL-27 plays a role in the induction of TR1-like cells from naive T-cell precursors, where it triggers expression of the transcription factor c-Maf, IL-21 and ICOS (44), IL-27 is dispensable for pMHCII-NP-induction of TR1-like cells (11). Since, unlike IL-27, pMHC class II-NPs can only trigger TR1-like cell formation from antigen-experienced but not naive T cells (11), these observations are compatible with the possibility that pMHCII-NPs operate downstream of IL-27.
The specific roles of ICOS, IL-2, IL-6, IL-15 and type I IFNs in the development of pMHCII-NP-induced TR1-like cells remains to be determined.
Transcription factors translate different TR1-like cell-promoting stimuli into transcriptional regulation of key TR1-like cell genes, thus playing a critical role in TR1-like cell specification (Figure 3).
The IL-2 inducible T-cell kinase (ITK) plays an essential role in T-cell activation, differentiation and function in response to TCR ligation (156). Although the lack of ITK impairs the development of IL-27-induced TR1-like cells, constitutive expression of the IRF4 (a downstream target of ITK signaling) overcomes this effect (60). Of note, IRF4 expression has been linked to the expression of IL-4 and IL-10 in Th2 cells (157), IL-21, Blimp-1 and Bcl-6 in TFH cells (158) and IL-10 expression in Treg cells (159) or Th1 cells (157).
c-Maf has context-dependent effects on IL-4, IL-10 and IL-21 expression. c-Maf positively regulates IL-4 production in both TFH and Th2 cells (160, 161), induces IL-21 expression in both TFH and Th17 cells (59) and contributes to the expression of CXCR5 (162). c-Maf is expressed early on during IL-27-induced TR1-like cell induction, and its expression progressively increases with time (44). IL-27 stimulation also upregulates the expression of AhR, implicated in FoxP3+ Treg and Th17 differentiation (44). c-Maf and AhR have been shown to transactivate both Il10 and Il21 gene expression in TR1-like cells (45).
Egr-2 is a transcription factor that plays a role in T-cell anergy (163) and has been associated with the acquisition of regulatory activity by CD4+ T cells (52). Egr-2 expression can be induced by TCR ligation in the absence of costimulation, as well as by IL-27 stimulation (via STAT3). In turn, Egr-2 promotes IL-10 and LAG-3 expression via Blimp-1 (57).
The Blimp-1 protein, encoded by the Prdm1 gene, is a zinc finger-containing transcriptional regulator of plasma cell differentiation (164), but has also been implicated in IL-10 production by CD4+ T cells (165), including both TR1-like (25, 57, 166) and FoxP3+ Treg cells (159).
Whereas IL-27R signaling promotes TR1-like cell formation, in part via the transcription factors c-Maf, AhR, Egr-2 and Blimp-1, access of these transcription factors to their binding sites on target genes, such as Il10 or Il21, is enabled by pioneering transcription factors, such as BATF and IRF1 (167). BATF had been previously defined as a pioneer factor for Th2, Th17 and effector CD8+ T-cell differentiation, by modifying the chromatin landscape of precursor cells (168–172). BATF also plays a role in TFH differentiation, by regulating Bcl-6 and c-Maf expression (173).
Other transcription factors, such as Eomes (174, 175) and Rora (176), have also been proposed to transactivate the Il10 gene in CD4+ T cells in a context-dependent manner. For example, Eomes requires co-expression of T-bet, the key Th1 transcription factor.
Figure 3 summarizes the main stimuli leading to TR1-like cell induction, integrating transcriptional regulation of the key TR1-associated genes, Il10 and Il21.
Although pMHCII-NP-induced, antigen-specific TR1-like cells can persist for several months post-treatment withdrawal without any obvious loss of key phenotypic properties or acquisition of pathogenic activity, the cues responsible for their homeostatic survival remain unclear. Cytokines produced by the TR1-like cells themselves or by downstream regulatory cell types (e.g. Breg cells), including IL-10, IL-21 and IL-35, may play a role. Studies employing cell-specific cytokine receptor knock-out mice should help address this knowledge gap.
To date, the TR1-ness of specific T-cell types has generally been ascribed to IL-10 expression. However, IL-10 can be expressed by differentiated Th subsets without the need to invoke a true TR1/regulatory phenotype.
For instance, whereas the Il10 locus lies in a closed conformation in naive CD4+ T cells (177), all differentiated T-helper subsets expose accessible regions along the locus (178), together with deposition of H3K4me3 in the absence of H3K27me3 marks (171, 179), promoting a transcriptionally-competent state. The chromatin remodeling processes that lead to a poised or active Il10 transcription state in Th subsets is mediated by pioneering transcription factors.
In TR1-like cells, BATF and IRF1 are thought to function as pioneering factors responsible for eliciting some of the chromatin accessibility changes that are required for TR1-like cell differentiation. Only after certain loci, such as Il10, become accessible, other TR1-like cell-associated transcription factors, such as AhR and c-Maf, can then bind the Il10 promoter (167). In Th17 cells, BATF, in association with IRF4, induces Il10 transcription (180). IRF4 is also involved in eliciting Il10 expression in Th2 (157, 181) and Treg cells (159). STAT proteins also contribute to Il10 expression in various Th cell subsets, such as by priming the locus with H3K4me1. STAT4, and STAT6 and GATA-3, induce IL-10 production in Th1 and Th2 cells, respectively (98, 182). GATA-3 induces H3 and H4 acetylation and an increase in chromatin accessibility in the Il10 locus (183). In Tregs, FoxP3 regulates IL-10 expression, but this process is independent of DNA binding (184). Rather, FoxP3 recruits HAT1 complexes to the locus where they induce the acetylation of H4K5 and H5K12 at the Il10 promoter, making it more permissive for STAT3 binding (185). Nfil3 is another transcription factor linked to IL-10 production in Th1, Th2, Treg and NK cells, by promoting acetylation of H3 in the Il10 locus (99). In contrast, in both Th1 and Th2 cells, Ets1 suppresses IL-10 production, by recruiting the de-acetylase HDAC1 to the Il10 promoter and enhancer regions (186, 187). Importantly, transcription factors involved in T-helper subset specification, such as T-bet, GATA-3 or RORγt can enhance IL-10 expression.
Thus, IL-10 expression per se is not a cell subset- but rather a functional state-defining property and IL-10 expression can co-exist with an effector cell program within a given T-cell subset.
Transcriptional features alone do not invariably define a final differentiated cell state. Like in other T-cell developmental or differentiation steps, cell fate decisions require both transcriptional changes and epigenetic remodeling of the chromatin.
For example, during development, the epigenome of the parental gametes progressively evolves to acquire the specific epigenome of the zygote (188). The chromatin of the zygote further undergoes additional waves of epigenetic changes, including DNA demethylation and methylation, modifications in histones, and changes in chromatin accessibility (189, 190). Epigenetic reprogramming is also essential for differentiation of embryonic stem cells (ESCs) into distinct cell populations (190–192). Pluripotent cells display an open chromatin configuration that is progressively restricted during development (193), accompanied by an increase in DNA methylation and the redistribution of histone marks. The gene expression changes that are associated with such chromatin remodeling processes are not unique to the germline and also take place in somatic cells in response to stimuli. For example, cytokine stimulation induces chromatin changes in APCs, such as DCs or macrophages (194). This phenomenon has also been reported for cytokine-challenged pancreatic β-cells (195), where cytokine stimulation triggers the appearance of new regulatory elements (neo-IREs).
It is becoming increasingly clear that susceptibility of the chromatin to undergo certain epigenetic modifications is affected by the underlying nucleotide sequence. A significant number of disease-associated single nucleotide polymorphisms (SNPs) lie in fact in non-coding, regulatory regions (196). For example, T1D-associated variants appear to be enriched in T- and B-cell enhancers (196, 197), in some cases promoting a three dimensional chromatin architecture that facilitates changes in gene expression in immune cells that might be able to promote the autoimmune pathology (198). Type 2 diabetes (T2D) is another example of a disease whose genetic susceptibility is commonly associated with non-coding variants (199, 200). In this case, many risk variants locate in enhancers or super-enhancers of genes involved in islet cell function and differentiation (201–204).
T cells are known to undergo extensive epigenome remodeling in response to activation/differentiation cues, enabling the acquisition of phenotypic and functional stability (Figure 4). The first epigenetic decision takes place when the T-cell fate is defined in developing thymocytes (205). T-cell activation (206) and T-helper cell polarization also involve epigenetic modifications along with changes in transcription factor expression. For example, Th1 development involves the upregulation of STAT1 in response to IFN-γ and IL-27, leading to the expression of T-bet, which upregulates the expression of IFN-γ, H2.0-like homeobox (HLX) transcription factors and Runt-related transcription factor 3 (Runx3), and suppresses the expression of GATA-3 (207–209). In turn, T-bet and Runx3 repress the Il4 gene to prevent Th2 differentiation. The Ifng gene harbors multiple regulatory elements around the locus, including enhancers at conserved non-coding sequences and an insulator. This locus is found in a poised, de-methylated state marked by bivalent histone modifications (poised for either expression or silencing) in naive CD4+ T cells, which produce low levels of this cytokine. Th1 differentiation involves H3K4me2, H3 and H4 acetylation and the creation of accessible chromatin at regulatory elements within the Ifng locus, together with loss of H3K27me3 throughout the locus, followed by DNA demethylation (210–213). T-bet transactivates expression of Ifng by binding to its promoter as well as several enhancers and by recruiting histone acetyltransferases (HATs) (214) and histone demethylases (HDMs) (215).
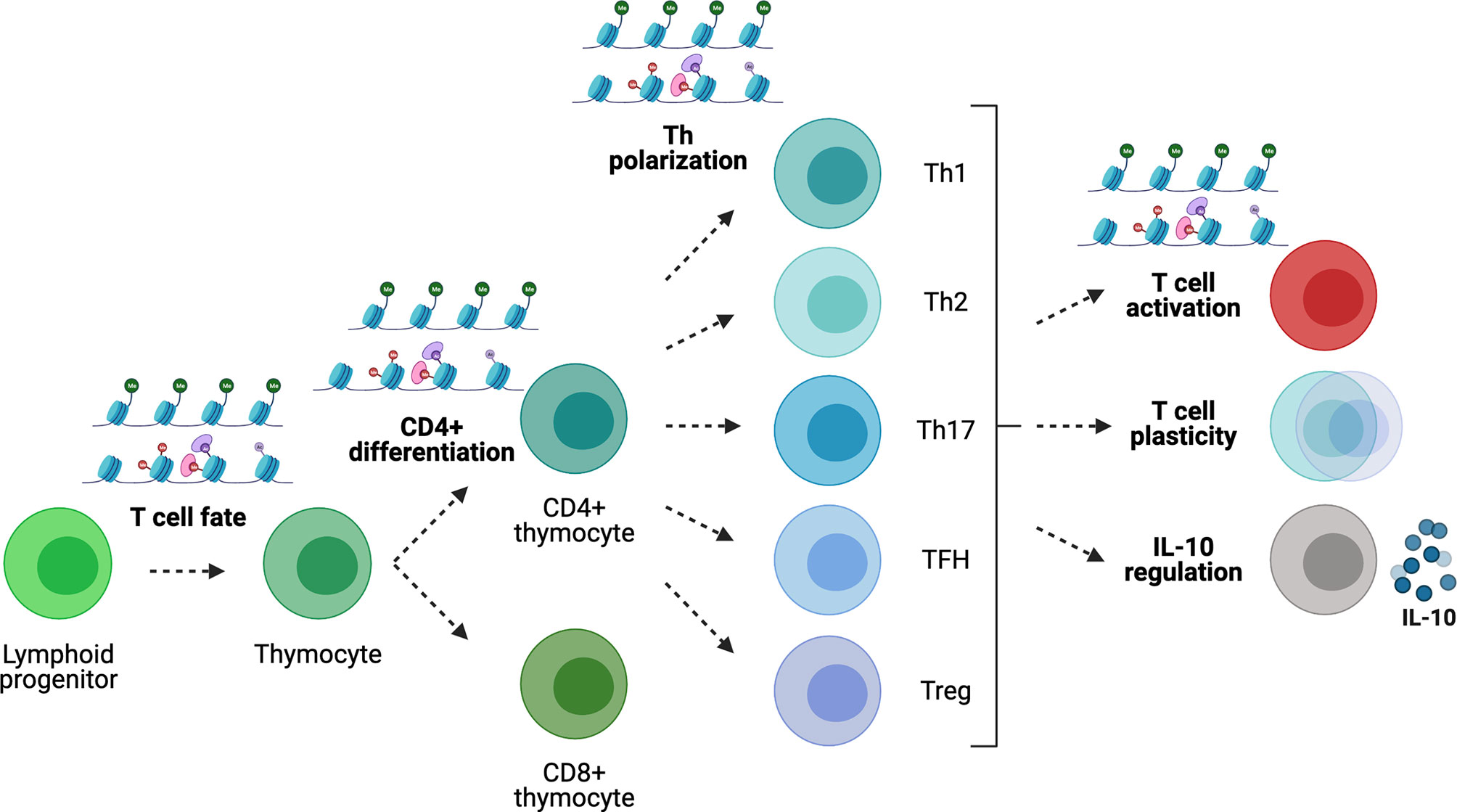
Figure 4 Gene regulation in T cells. Epigenetic modifications play key roles during T-cell development, differentiation and Th polarization. In the periphery, the epigenome regulates Th cell lineage stability/plasticity as well as IL-10 expression competency.
In contrast, activation of the Th2 program results in the loss of permissive histone modifications and H3K27 trimethylation along the Ifng locus, coupled to DNA methylation (210, 216, 217). The Th2 program is induced by IL-4-mediated activation of STAT6, which in turn activates GATA-3 (218). GATA-3 induces the expression of c-Maf, regulating IL-4 expression, and together with STAT6 enhances the transcription of Il4, Il5 and Il13 (218). In mice, Il4, Il5 and Il13 (encoding Th2 cytokines), together with the Rad50 gene, co-localize near a Locus Control Region (LCR). Expression of the Il4 gene is regulated by enhancers (overlapping with DNAse I hypersensitive sites) that bind NFAT and Th2-promoting transcription factors. In naive T cells, there are few accessibility and histone modifications at these DNAse I hypersensitive sites, and the cytokine gene promoters and enhancers are hyper-methylated (219). Upon Th2-polarizing stimulation, the loci acquire permissive histone modifications and lose H3K27me3 (220, 221). In Th1 cells, the Th2-cytokine locus is all covered with H3K27me3 (222). GATA-3 induces most of these epigenetic modifications, as it can recruit HATs and histone H3K4 methyltransferases (218, 223), inhibit histone deacetylases (HDACs) (224) and DNA (cytosine-5)-methyltransferase 1 (DNMT1) (219, 225), and recruit chromatin-remodeling factors (226). In addition, the Ifng locus in Th2 cells is silenced by H3K27me3 deposition (217).
TGF-β induction of Th17 and Treg lineage formation represents another example. This cytokine induces both the expression of FoxP3 and retinoic-acid-receptor-related orphan receptor-γt (RORγt) (227–230). The context determines if the Treg or the Th17 program is induced: in the absence of IL-6, FoxP3 inhibits RORγt and leads to Treg formation. If IL-6 is present, STAT3 is activated, inhibiting the expression of FoxP3 and enhancing Th17 formation. IL-17A and IL-17F are both co-expressed by Th17 cells and the genes encoding these cytokines co-localize and may be regulated by shared regulatory elements. The Il17 locus contains eight different gene regulatory elements (231). When naïve CD4+ T cells are cultured under Th17-polarizing conditions, STAT3 (227) and RORγt (232) induce the appearance of permissive H3 acetylation changes in the Il17a and Il17f gene regulatory elements (231), enabling their expression.
The fate of TFH and non-TFH (Th1, Th2, Th17) effector cells is regulated by Bcl-6 and Blimp-1, which are reciprocal regulators of each other (233). Bcl-6 binds promoters and enhancers regulating genes involved in T-cell migration (Ebi2, CCR6, CCR7, S1pr1, Klf-2, PSGL-1, CXCR5, CXCR4, PD-1 and SAP) (126, 234). Ascl-2 also controls TFH differentiation by upregulating CXCR5 and CXCR4, while downregulating CCR7 and PSGL-1 expression. The Bcl6 locus in TFH cells displays positive histone modifications, but it also contains permissive marks in Th1, Th2 and Th17 cells (235). Other TFH-related genes, such as c-Maf, BATF and IRF4, are also associated with H3K4me3 in all subsets. In contrast, the Ascl2 locus is uniquely marked with the active chromatin mark H3K4me3 in TFH cells. Prdm1 (encoding BLIMP-1), which is downregulated in TFH cells, displays bivalent modifications, allowing re-programming between TFH and other Th effector subsets (236).
Acquisition of Treg-cell-specific epigenetic marks during thymocyte development (237, 238), along with FoxP3 expression, determines the regulatory phenotype of nTregs (239). Once in the periphery, Treg cells can be divided into subpopulations that locate in different tissues, and each acquires an additional level of epigenetic modification that defines a tissue-specific epigenetic footprint (240). The expression of some Treg-function associated molecules, such as CTLA-4 or CD25, is clearly associated with DNA de-methylation and can occur in the absence of FoxP3. In contrast, expression of Il2, Ifng or Zap70 is lost if FoxP3 is not present. Several regulatory elements control Foxp3 gene expression (237). The Foxp3 promoter is de-methylated upon TCR signaling, facilitating the binding of FoxP3-inducing transcription factors (241).
Moreover, there is increasing evidence that Th subsets are plastic. To name a few examples, T-cell populations have been described that stably express both T-bet and GATA-3 and produce both IFN-γ and IL-4 (242), produce both Th1 and Th17 cytokines (243, 244), or have a Th1/Th17 phenotype but can switch to Th2 during helminth infections (245). This observed plasticity can be regulated by different mechanisms. First, certain environmental stimuli may be able to modify epigenetic marks responsible for maintaining lineage stability; for example, prolonged activation of Treg cells in vitro can lead to demethylation of the Rorc locus in FoxP3+ Tregs and allow IL-17 production (246). Second, Th subsets display an intrinsic plasticity potential. Although the various lineage-specific cytokines present active histone marks in the corresponding cell lineages and repressive marks in the others, some transcription factors are not so strictly marked. For example, in Th1 cells, Tbx21, encoding T-bet, bears activating H3K4me3 marks in the promoter. In other Th cell subsets, on the other hand, the Tbx21 promoter bears bivalent modifications. Likewise, the Gata3 promoter carries H3K4me3 marks in Th2 cells, but bivalent marks in other Th cell subsets. The same is true for Rorc or Bcl6 genes in non-Th17 or non-TFH cells, respectively (235, 247). Third, polarized T-cell types might represent stable lineages yet comprise a continuum of different epigenotypes with differential susceptibility for lineage conversion in response to external signals.
To date, the study of TR1-like cell specification is largely based on phenotypic and transcriptional studies. Based on the data summarized above, it is reasonable to suspect that the cues responsible for TR1-like cell formation operate on a precursor cell type that either has a TR1-poised epigenome or responds to TR1-inducing signals by undergoing further epigenetic modifications enabling the acquisition of a stable TR1-like cell phenotype, including DNA hypomethylation (248–250). Thus, detailed characterization of TR1-like cells at the transcriptional and epigenetic levels, including analysis of their chromatin status, three-dimensional structure and interactions, as well as DNA methylation status (Table 2), should provide unique clues about the true identity of this cell lineage and the identity of their cellular precursors.
Currently, TR1-like cells are defined as a regulatory CD4+ T-cell subset that lacks FoxP3 expression (unlike conventional FoxP3+ Treg cells) and secretes IL-10 and low levels or no IL-4. However, this Treg cell subset lacks cell-specific markers and their developmental origin remains a mystery. Moreover, the signaling, genetic and epigenetic mechanisms that are responsible for the acquisition of the TR1-like cell phenotype in vivo remain unclear.
We posit that detailed transcriptional and epigenetic studies will enable a better understanding of the role of this T-cell subset in immunity, autoimmunity and cancer, the identification of biomarkers capable of accurately track its development in vivo, as well as a detailed understanding of gene regulatory mechanisms responsible for TR1-like cell specification. In turn, this will help pinpoint specific areas of the genome that might be impacted by genetic polymorphisms associated with susceptibility and/or resistance to specific immune-mediated diseases, and the development and testing of compounds capable of triggering the formation of antigen-specific TR1-like cells in vivo, for therapeutic purposes.
TR1 cell formation in response to pMHCII-NPs afford a unique opportunity to address the above knowledge gaps. These compounds trigger the formation of relatively large numbers of mono-specific TR1-like cells, thus enabling this type of studies with unprecedented resolution. Transcriptional studies at the bulk and single cell levels should help determine the homogeneity or heterogeneity of the resulting cognate T-cell pools, and pinpoint developmentally-related cell subsets with poised TR1-like cell transcriptional programs. Epigenetic studies shall include screening for histone modifications, chromatin accessibility and 3D chromatin maps, to enumerate the genome-wide distribution of active promoters and enhancers, define the epigenomic architecture underpinning the TR1-like cell state, describe the various steps underlying TR1-like cell re-programming, identify key TR1-like cell epigenetic signatures, and potentially expose new targets for therapeutic intervention.
Collectively, these studies should provide a comprehensive set of functional, phenotypic, transcriptional and epigenetic markers capable of specifically identifying TR1 cells. These markers would likely play a pivotal role in guiding the clinical translation of compounds capable of promoting TR1 cell formation in vivo for the treatment of autoimmunity, including pMHCII-based nanomedicines. They should also prove useful to enumerate the contribution of this cell type to tumor progression in the context of cancer.
PSo and PSa co-wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors’ work is funded by the Canadian Institutes of Health Research (CIHR), Diabetes Canada, REEM (Red Española de Esclerosis Múltiple), the Praespero Foundation, the Ministerio de Economia y Competitividad of Spain (MINECO; RD16/0015/0020), and Generalitat de Catalunya (SGR and CERCA Programmes). The JMDRC is supported by Diabetes Canada.
PSa is scientific founder of Parvus Therapeutics Inc. and has a financial interest in the company.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the members of the PSa’s laboratories for their contributions.
1. Fujio K, Okamura T, Yamamoto K. The Family of IL-10-Secreting CD4+T Cells. In: Advances in Immunology. Elsevier Inc. (2010). p. 99–130. doi: 10.1016/S0065-2776(10)05004-2
2. Alfen JS, Larghi P, Facciotti F, Gagliani N, Bosotti R, Paroni M, et al. Intestinal IFN-γ–Producing Type 1 Regulatory T Cells Coexpress CCR5 and Programmed Cell Death Protein 1 and Downregulate IL-10 in the Inflamed Guts of Patients With Inflammatory Bowel Disease. J Allergy Clin Immunol (2018) 142:1537–47.e8. doi: 10.1016/j.jaci.2017.12.984
3. Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limon P, et al. Coexpression of CD49b and LAG-3 Identifies Human and Mouse T Regulatory Type 1 Cells. Nat Med (2013) 19:739–46. doi: 10.1038/nm.3179
4. Roncarolo MG, Gregori S, Bacchetta R, Battaglia M, Gagliani N. The Biology of T Regulatory Type 1 Cells and Their Therapeutic Application in Immune-Mediated Disease. Immunity (2018) 49:1004–19. doi: 10.1016/j.immuni.2018.12.001
5. Burton BR, Britton GJ, Fang H, Verhagen J, Smithers B, Sabatos-Peyton CA, et al. Sequential Transcriptional Changes Dictate Safe and Effective Antigen-Specific Immunotherapy. Nat Commun (2014) 5:4741. doi: 10.1038/ncomms5741
6. White AM, Wraith DC. Tr1-Like T Cells - An Enigmatic Regulatory T Cell Lineage. Front Immunol (2016) 7:355. doi: 10.3389/fimmu.2016.00355
7. Brockmann L, Soukou S, Steglich B, Czarnewski P, Zhao L, Wende S, et al. Molecular and Functional Heterogeneity of IL-10-Producing CD4+ T Cells. Nat Commun (2018) 9:5457. doi: 10.1038/s41467-018-07581-4
8. Zhu C, Sakuishi K, Xiao S, Sun Z, Zaghouani S, Gu G, et al. An IL-27/NFIL3 Signaling Axis Drives Tim-3 and IL-10 Expression and T Cell Dysfunction. Nat Commun (2015) 6:1–14. doi: 10.1038/ncomms7072
9. Singha S, Shao K, Yang Y, Clemente-Casares X, Solé P, Clemente A, et al. Peptide–MHC-Based Nanomedicines for Autoimmunity Function as T-Cell Receptor Microclustering Devices. Nat Nanotechnol (2017) 12:701–10. doi: 10.1038/nnano.2017.56
10. Tsai S, Shameli A, Yamanouchi J, Clemente-Casares X, Wang J, Serra P, et al. Reversal of Autoimmunity by Boosting Memory-Like Autoregulatory T Cell. Immunity (2010) 32:568–80. doi: 10.1016/j.immuni.2010.03.015
11. Clemente-Casares X, Blanco J, Ambalavanan P, Yamanouchi J, Singha S, Fandos C, et al. Expanding Antigen-Specific Regulatory Networks to Treat Autoimmunity. Nature (2016) 530:434–40. doi: 10.1038/nature16962
12. Umeshappa CS, Singha S, Blanco J, Shao K, Nanjundappa RH, Yamanouchi J, et al. Suppression of a Broad Spectrum of Liver Autoimmune Pathologies by Single Peptide-MHC-Based Nanomedicines. Nat Commun (2019) 10:1–17. doi: 10.1038/s41467-019-09893-5
13. Umeshappa CS, Mbongue J, Singha S, Mohapatra S, Yamanouchi J, Lee JA, et al. Ubiquitous Antigen-Specific T Regulatory Type 1 Cells Variably Suppress Hepatic and Extrahepatic Autoimmunity. J Clin Invest (2020) 130:1823–9. doi: 10.1172/JCI130670
14. Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE. Roncarolo MG. A CD4+ T-Cell Subset Inhibits Antigen-Specific T-Cell Responses and Prevents Colitis. Nature (1997) 389:737–42. doi: 10.1038/39614
15. Roncarolo MG, Yssel H, Touraine JL, Betuel H, De Vries JE, Spits H. Autoreactive T Cell Clones Specific for Class I and Class II HLA Antigens Isolated From a Human Chimera. J Exp Med (1988) 167:1523–34. doi: 10.1084/jem.167.5.1523
16. Vieira P, De Waal-Malefyt R, Dang MN, Johnson KE, Kastelein R, Fiorentino DF, et al. Isolation and Expression of Human Cytokine Synthesis Inhibitory Factor cDNA Clones: Homology to Epstein-Barr Virus Open Reading Frame BCRF. Proc Natl Acad Sci USA (1991) 88:1172–6. doi: 10.1073/pnas.88.4.1172
17. Bacchetta R, Bigler M, Touraine JL, Parkman R, Tovo PA, Abrams J, et al. High Levels of Interleukin 10 Production In Vivo Are Associated With Tolerance in SCID Patients Transplanted With HLA Mismatched Hematopoietic Stem Cells. J Exp Med (1994) 179:493–502. doi: 10.1084/jem.179.2.493
18. Bacchetta R, Gregori S, Serafini G, Sartirana C, Schulz U, Zino E, et al. Molecular and Functional Characterization of Allogantigen Specific Anergic T Cells Suitable for Cell Therapy. Haematologica (2010) 95:2134–43. doi: 10.3324/haematol.2010.025825
19. Roncarolo MG, Gregori S, Bacchetta R, Battaglia M. Tr1 Cells and the Counter-Regulation of Immunity: Natural Mechanisms and Therapeutic Applications. Curr Top Microbiol Immunol (2014) 380:39–68. doi: 10.1007/978-3-662-43492-5_3
20. Bollyky PL, Wu RP, Falk BA, Lord JD, Long SA, Preisinger A, et al. ECM Components Guide IL-10 Producing Regulatory T-Cell (TR1) Induction From Effector Memory T-Cell Precursors. Proc Natl Acad Sci USA (2011) 108:7938–43. doi: 10.1073/pnas.1017360108
21. Yao Y, Vent-Schmidt J, McGeough MD, Wong M, Hoffman HM, Steiner TS, et al. Tr1 Cells, But Not Foxp3 + Regulatory T Cells, Suppress NLRP3 Inflammasome Activation via an IL-10–Dependent Mechanis. J Immunol (2015) 195:488–97. doi: 10.4049/jimmunol.1403225
22. Gagliani N, Gregori S, Jofra T, Valle A, Stabilini A, Rothstein DM, et al. Rapamycin Combined With Anti-CD45RB mAB and IL-10 or With G-CSF Induces Tolerance in a Stringent Mouse Model of Islet Transplantation. PloS One (2011) 6:1–12. doi: 10.1371/journal.pone.0028434
23. Gagliani N, Jofra T, Valle A, Stabilini A, Morsiani C, Gregori S, et al. Transplant Tolerance to Pancreatic Islets Is Initiated in the Graft and Sustained in the Spleen. Am J Transplant (2013) 13:1963–75. doi: 10.1111/ajt.12333
24. Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, et al. TH17 Cells Transdifferentiate Into Regulatory T Cells During Resolution of Inflammation. Nature (2015) 523:221–5. doi: 10.1038/nature14452
25. Heinemann C, Heink S, Petermann F, Vasanthakumar A, Rothhammer V, Doorduijn E, et al. IL-27 and IL-12 Oppose Pro-Inflammatory IL-23 in CD4+ T Cells by Inducing Blimp1. Nat Commun (2014) 5:1–13. doi: 10.1038/ncomms4770
26. Meiron M, Zohar Y, Anunu R, Wildbaum G, Karin N. CXCL12 (SDF-1α) Suppresses Ongoing Experimental Autoimmune Encephalomyelitis by Selecting Antigen-Specific Regulatory T Cells. J Exp Med (2008) 205:2643–55. doi: 10.1084/jem.20080730
27. Lönnberg T, Svensson V, James KR, Fernandez-Ruiz D, Sebina I, Montandon R, et al. Single-Cell RNA-Seq and Computational Analysis Using Temporal Mixture Modeling Resolves TH1/TFH Fate Bifurcation in Malaria. Sci Immunol (2017) 2(9):eaal2192. doi: 10.1126/sciimmunol.aal2192
28. Pacciani V, Gregori S, Chini L, Corrente S, Chianca M, Moschese V, et al. Induction of Anergic Allergen-Specific Suppressor T Cells Using Tolerogenic Dendritic Cells Derived From Children With Allergies to House Dust Mites. J Allergy Clin Immunol (2010) 125:727–36. doi: 10.1016/j.jaci.2009.12.004
29. Pellerin L, Jenks JA, Chinthrajah S, Dominguez T, Block W, Zhou X, et al. Peanut-Specific Type 1 Regulatory T Cells Induced In Vitro From Allergic Subjects Are Functionally Impaired. J Allergy Clin Immunol (2018) 141:202–213.e8. doi: 10.1016/j.jaci.2017.05.045
30. Oida T, Zhang X, Goto M, Hachimura S, Totsuka M, Kaminogawa S, et al. CD4+ CD25– T Cells That Express Latency-Associated Peptide on the Surface Suppress CD4+ CD45RB High -Induced Colitis by a TGF-β-Dependent Mechanis. J Immunol (2003) 170:2516–22. doi: 10.4049/jimmunol.170.5.2516
31. Ochi H, Abraham M, Ishikawa H, Frenkel D, Yang K, Basso AS, et al. Oral CD3-Specific Antibody Suppresses Autoimmune Encephalomyelitis by Inducing CD4+CD25–LAP+ T Cells. Nat Med (2006) 12:627–35. doi: 10.1038/nm1408
32. Groh V, Smythe K, Dai Z, Spies T. Fas Ligand-Mediated Paracrine T Cell Regulation by the Receptor NKG2D in Tumor Immunity. Nat Immunol (2006) 7:755–62. doi: 10.1038/ni1350
33. Groh V, Brühl A, El-Gabalawy H, Nelson JL, Spies T. Stimulation of T Cell Autoreactivity by Anomalous Expression of NKG2D and Its MIC Ligands in Rheumatoid Arthritis. Proc Natl Acad Sci USA (2003) 100:9452–7. doi: 10.1073/pnas.1632807100
34. Häringer B, Lozza L, Steckel B, Geginat J. Identification and Characterization of IL-10/IFN-γ-Producing Effector-Like T Cells With Regulatory Function in Human Blood. J Exp Med (2009) 206:1009–17. doi: 10.1084/jem.20082238
35. Gonzalez A, Andre-Schmutz I, Carnaud C, Mathis D, Benoist C. Damage Control, Rather Than Unresponsiveness, Effected by Protective DX5+T Cells in Autoimmune Diabetes. Nat Immunol (2001) 2:1117–25. doi: 10.1038/ni738
36. Charbonnier L-M, van Duivenvoorde LM, Apparailly F, Cantos C, Han WGH, Noël D, et al. Immature Dendritic Cells Suppress Collagen-Induced Arthritis by In Vivo Expansion of CD49b + Regulatory T Cell. J Immunol (2006) 177:3806–13. doi: 10.4049/jimmunol.177.6.3806
37. Han WGH, Schuurhuis DH, Fu N, Camps M, van Duivenvoorde LM, Louis-Plence P, et al. DC-Induced CD8+ T-Cell Response Is Inhibited by MHC Class II-Dependent DX5+CD4+ Treg. Eur J Immunol (2009) 39:1765–73. doi: 10.1002/eji.200838842
38. El Bannoudi H, Han WGH, Stoop JN, Louis-Plence P, Huizinga TWJ, Toes REM. DX5+CD4+ T Cells Modulate CD4+ T-Cell Response via Inhibition of IL-12 Production by DC. Eur J Immunol (2013) 43:439–46. doi: 10.1002/eji.201242796
39. Workman CJ, Rice DS, Dugger KJ, Kurschner C, Vignali DAA. Phenotypic Analysis of the Murine CD4-Related Glycoprotein, CD223 (LAG-3). Eur J Immunol (2002) 32:2255–63. doi: 10.1002/1521-4141(200208)32:8<2255::AID-IMMU2255>3.0.CO;2-A
40. Facciotti F, Gagliani N, Häringer B, Alfen JS, Penatti A, Maglie S, et al. IL-10-Producing Forkhead Box Protein 3-Negative Regulatory T Cells Inhibit B-Cell Responses and Are Involved in Systemic Lupus Erythematosus. J Allergy Clin Immunol (2016) 137:318–21. doi: 10.1016/j.jaci.2015.06.044
41. Magnani CF, Alberigo G, Bacchetta R, Serafini G, Andreani M, Roncarolo MG, et al. Killing of Myeloid APCs via HLA Class I, CD2 and CD226 Defines a Novel Mechanism of Suppression by Human Tr1 Cells. Eur J Immunol (2011) 41:1652–62. doi: 10.1002/eji.201041120
42. Cobbold SP, Nolan KF, Graca L, Castejon R, Le Moine A, Frewin M, et al. Regulatory T Cells and Dendritic Cells in Transplantation Tolerance: Molecular Markers and Mechanisms. Immunol Rev (2003) 196:109–24. doi: 10.1046/j.1600-065X.2003.00078.x
43. Okamura T, Fujio K, Shibuya M, Sumitomo S, Shoda H, Sakaguchi S, et al. CD4+CD25-LAG3+ Regulatory T Cells Controlled by the Transcription Factor Egr-2. Proc Natl Acad Sci (2009) 106:13974–9. doi: 10.1073/pnas.0906872106
44. Pot C, Jin H, Awasthi A, Liu SM, Lai C-Y, Madan R, et al. Cutting Edge: IL-27 Induces the Transcription Factor C-Maf, Cytokine IL-21, and the Costimulatory Receptor ICOS That Coordinately Act Together to Promote Differentiation of IL-10-Producing Tr1 Cell. J Immunol (2009) 183:797–801. doi: 10.4049/jimmunol.0901233
45. Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, et al. The Aryl Hydrocarbon Receptor Interacts With C-Maf to Promote the Differentiation of Type 1 Regulatory T Cells Induced by IL-27. Nat Immunol (2010) 11:854–61. doi: 10.1038/ni.1912
46. Tousa S, Semitekolou M, Morianos I, Banos A, Trochoutsou AI, Brodie TM, et al. Activin-A Co-Opts IRF4 and AhR Signaling to Induce Human Regulatory T Cells That Restrain Asthmatic Responses. Proc Natl Acad Sci USA (2017) 114:2891–900. doi: 10.1073/pnas.1616942114
47. Nakamura K, Kitani A, Strober W. Cell Contact-Dependent Immunosuppression by CD4+CD25+ Regulatory T Cells Is Mediated by Cell Surface-Bound Transforming Growth Factor β. J Exp Med (2001) 194:629–44. doi: 10.1084/jem.194.5.629
48. Nakajima U, Miyazono K, Kato M, Takase M, Yamagishi T, Nakamura H. Extracellular Fibrillar Structure of Latent Tgfβ Binding Protein-1: Role in Tgfβ-Dependent Endothelial-Mesenchymal Transformation During Endocardial Cushion Tissue Formation in Mouse Embryonic Heart. J Cell Biol (1997) 136:193–204. doi: 10.1083/jcb.136.1.193
49. Bradley LM, Haynes L, Swain SL. IL-7: Maintaining T-Cell Memory and Achieving Homeostasis. Trends Immunol (2005) 26:172–6. doi: 10.1016/j.it.2005.01.004
50. Liu W, Putnam AL, Xu-yu Z, Szot GL, Lee MR, Zhu S, et al. CD127 Expression Inversely Correlates With FoxP3 and Suppressive Function of Human CD4+ T Reg Cells. J Exp Med (2006) 203:1701–11. doi: 10.1084/jem.20060772
51. Huang C-TT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, et al. Role of LAG-3 in Regulatory T Cells. Immunity (2004) 21:503–13. doi: 10.1016/j.immuni.2004.08.010
52. Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, et al. Egr-2 and Egr-3 Are Negative Regulators of T Cell Activation. Nat Immunol (2005) 6:472–80. doi: 10.1038/ni1193
53. Kassiotis G, Gray D, Kiafard Z, Zwirner J, Stockinger B. Functional Specialization of Memory Th Cells Revealed by Expression of Integrin CD49. J Immunol (2006) 177:968–75. doi: 10.4049/jimmunol.177.2.968
54. Gupta S, Thornley TB, Gao W, Larocca R, Turka LA, Kuchroo VK, et al. Allograft Rejection Is Restrained by Short-Lived TIM 3+ PD1+ Foxp3+ Treg. J Clin Invest (2012) 122:2395–404. doi: 10.1172/JCI45138
55. Wu H, Chen Y, Liu H, Xu LL, Teuscher P, Wang S, et al. Follicular Regulatory T Cells Repress Cytokine Production by Follicular Helper T Cells and Optimize IgG Responses in Mice. Eur J Immunol (2016) 46:1152–61. doi: 10.1002/eji.201546094
56. Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, Repressor of GATA, Regulates the Expression of Cytokine Genes. Immunity (2000) 12:323–33. doi: 10.1016/s1074-7613(00)80185-5
57. Iwasaki Y, Fujio K, Okamura T, Yanai A, Sumitomo S, Shoda H, et al. Egr-2 Transcription Factor Is Required for Blimp-1-Mediated IL-10 Production in IL-27-Stimulated CD4+T Cells. Eur J Immunol (2013) 43:1063–73. doi: 10.1002/eji.201242942
58. Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld J-C, et al. The Aryl Hydrocarbon Receptor Links TH17-Cell-Mediated Autoimmunity to Environmental Toxins. Nature (2008) 453:106–9. doi: 10.1038/nature06881
59. Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho I-CC, Sharpe AH, et al. The Costimulatory Molecule ICOS Regulates the Expression of C-Maf and IL-21 in the Development of Follicular T Helper Cells and TH -17 Cells. Nat Immunol (2009) 10:167–75. doi: 10.1038/ni.1690
60. Huang W, Solouki S, Koylass N, Zheng S-GG, August A. ITK Signalling via the Ras/IRF4 Pathway Regulates the Development and Function of Tr1 Cells. Nat Commun (2017) 8:15871. doi: 10.1038/ncomms15871
61. Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the Interleukin-10 Receptor. Annu Rev Immunol (2001) 19:683–765. doi: 10.1146/annurev.immunol.19.1.683
62. Taga K, Tosato G. IL-10 Inhibits Human T Cell Proliferation and IL-2 Production. J Immunol (1992) 148:1143–8.
63. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, et al. Differentiation of Type 1 T Regulatory Cells (Tr1) by Tolerogenic DC-10 Requires the IL-10-Dependent ILT4/HLA-G Pathway. Blood (2010) 116:935–44. doi: 10.1182/blood-2009-07-234872
64. Satoguina JS, Weyand E, Larbi J, Hoerauf A. T Regulatory-1 Cells Induce IgG4 Production by B Cells: Role of IL-10. J Immunol (2005) 174:4718–26. doi: 10.4049/jimmunol.174.8.4718
65. Murray PJ. The Primary Mechanism of the IL-10-Regulated Antiinflammatory Response Is to Selectively Inhibit Transcription. Proc Natl Acad Sci USA (2005) 102:8686–91. doi: 10.1073/pnas.0500419102
66. Bourque J, Hawiger D. Immunomodulatory Bonds of the Partnership Between Dendritic Cells and T Cells. Crit Rev Immunol (2018) 38:379–401. doi: 10.1615/CritRevImmunol.2018026790
67. Lee KM, Chuang E, Griffin M, Khattri R, Hong DK, Zhang W, et al. Molecular Basis of T Cell Inactivation by CTLA-4. Science (1998) 282:2263–6. doi: 10.1126/science.282.5397.2263
68. Serra P, Amrani A, Yamanouchi J, Han B, Thiessen S, Utsugi T, et al. CD40 Ligation Releases Immature Dendritic Cells From the Control of Regulatory CD4+CD25+ T Cell. Immunity (2003) 19:877–89. doi: 10.1016/S1074-7613(03)00327-3
69. Oderup C, Cederbom L, Makowska A, Cilio CM, Ivars F. Cytotoxic T Lymphocyte Antigen-4-Dependent Down-Modulation of Costimulatory Molecules on Dendritic Cells in CD4+ CD25+ Regulatory T-Cell-Mediated Suppression. Immunology (2006) 118:240–9. doi: 10.1111/j.1365-2567.2006.02362.x
70. Cederbom L, Hall H, Ivars F. Stimulatory Molecules on Antigen-Presenting Cell. Cell (2000) 30:1538–43. doi: 10.1002/1521-4141(200006)30:6<1538::AID-IMMU1538>3.0.CO;2-X
71. Finger EB, Bluestone JA. When Ligand Becomes Receptor — Tolerance via B7 Signaling on DC. Nat Immunol (2002) 3:1056–7. doi: 10.1038/ni1102-1056
72. Maruhashi T, Okazaki II mi, Sugiura D, Takahashi S, Maeda TK, Shimizu K, et al. LAG-3 Inhibits the Activation of CD4 + T Cells That Recognize Stable pMHCII Through Its Conformation-Dependent Recognition of pMHCII. Nat Immunol (2018) 19:1415–26. doi: 10.1038/s41590-018-0217-9
73. Maeda TK, Sugiura D, Okazaki II mi, Maruhashi T, Okazaki T. Atypical Motifs in the Cytoplasmic Region of the Inhibitory Immune Co-Receptor LAG-3 Inhibit T Cell Activation. J Biol Chem (2019) 294:6017–26. doi: 10.1074/jbc.RA119.007455
74. Liang B, Workman C, Lee J, Chew C, Dale BM, Colonna L, et al. Regulatory T Cells Inhibit Dendritic Cells by Lymphocyte Activation Gene-3 Engagement of MHC Class II. J Immunol (2008) 180:5916–26. doi: 10.4049/jimmunol.180.9.5916
75. Sharpe AH, Pauken KE. The Diverse Functions of the PD1 Inhibitory Pathway. Nat Rev Immunol (2018) 18:153–67. doi: 10.1038/nri.2017.108
76. Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The Function of Programmed Cell Death 1 and Its Ligands in Regulating Autoimmunity and Infection. Nat Immunol (2007) 8:239–45. doi: 10.1038/ni1443
77. Liu S, Zhang H, Li M, Hu D, Li C, Ge B, et al. Recruitment of Grb2 and SHIP1 by the ITT-Like Motif of TIGIT Suppresses Granule Polarization and Cytotoxicity of NK Cells. Cell Death Differ (2013) 20:456–64. doi: 10.1038/cdd.2012.141
78. Zhang B, Zhao W, Li H, Chen Y, Tian H, Li L, et al. Immunoreceptor TIGIT Inhibits the Cytotoxicity of Human Cytokine-Induced Killer Cells by Interacting With CD155. Cancer Immunol Immunother (2016) 65:305–14. doi: 10.1007/s00262-016-1799-4
79. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The Surface Protein TIGIT Suppresses T Cell Activation by Promoting the Generation of Mature Immunoregulatory Dendritic Cells. Nat Immunol (2009) 10:48–57. doi: 10.1038/ni.1674
80. Hutloff A, Dittrich AM, Beier KC, Eljaschewitsch B, Kraft R, Anagnostopoulos I, et al. ICOS Is an Inducible T-Cell Co-Stimulator Structurally and Functionally Related to CD28. Nature (1999) 397:263–6. doi: 10.1038/16717
81. Witsch EJ, Peiser M, Hutloff A, Bchner K, Dorner BG, Jonuleit H, et al. ICOS and CD28 Reversely Regulate IL-10 on Re-Activation of Human Effector T Cells With Mature Dendritic Cells. Eur J Immunol (2002) 32:2680–6. doi: 10.1002/1521-4141(200209)32:9<2680::AID-IMMU2680>3.0.CO;2-6
82. Dong C, Juedes AE, Temann UA, Shresta S, Allison JP, Ruddle NH, et al. ICOS Co-Stimulatory Receptor Is Essential for T-Cell Activation and Function. Nature (2001) 409:97–101. doi: 10.1038/35051100
83. Junger WG. Immune Cell Regulation by Autocrine Purinergic Signalling. Nat Rev Immunol (2011) 11:201–12. doi: 10.1038/nri2938
84. Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological Roles for Ecto-5′-Nucleotidase (CD73). Purinergic Signal (2006) 2:351–60. doi: 10.1007/s11302-005-5302-5
85. Allard B, Longhi MS, Robson SC, Stagg J. The Ectonucleotidases CD39 and CD73: Novel Checkpoint Inhibitor Targets. Immunol Rev (2017) 276:121–44. doi: 10.1111/imr.12528
86. Mandapathil M, Lang S, Gorelik E, Whiteside TL. Isolation of Functional Human Regulatory T Cells (Treg) From the Peripheral Blood Based on the CD39 Expression. J Immunol Methods (2009) 346:55–63. doi: 10.1016/j.jim.2009.05.004
87. Ben Addi A, Lefort A, Hua X, Libert F, Communi D, Ledent C, et al. Modulation of Murine Dendritic Cell Function by Adenine Nucleotides and Adenosine: Involvement of the A2B Receptor. Eur J Immunol (2008) 38:1610–20. doi: 10.1002/eji.200737781
88. Grossman WJ, Verbsky JW, Tollefsen BL, Kemper C, Atkinson JP, Ley TJ. Differential Expression of Granzymes A and B in Human Cytotoxic Lymphocyte Subsets and T Regulatory Cells. Blood (2004) 104:2840–8. doi: 10.1182/blood-2004-03-0859
89. Kawamura K, Kadowaki N, Kitawaki T, Uchiyama T. Virus-Stimulated Plasmacytoid Dendritic Cells Induce CD4+ Cytotoxic Regulatory T Cells. Blood (2006) 107:1031–8. doi: 10.1182/blood-2005-04-1737
90. Casares S, Hurtado A, McEvoy RC, Sarukhan A, von Boehmer H, Brumeanu TD. Down-Regulation of Diabetogenic CD4+ T Cells by a Soluble Dimeric Peptide-MHC Class II Chimera. Nat Immunol (2002) 3:383–91. doi: 10.1038/ni770
91. Masteller EL, Warner MR, Ferlin W, Judkowski V, Wilson D, Glaichenhaus N, et al. Peptide-MHC Class II Dimers as Therapeutics to Modulate Antigen-Specific T Cell Responses in Autoimmune Diabete. J Immunol (2003) 171:5587–95. doi: 10.4049/jimmunol.171.10.5587
92. Li L, Yi Z, Wang B, Tisch R. Suppression of Ongoing T Cell-Mediated Autoimmunity by Peptide-MHC Class II Dimer Vaccinatio. J Immunol (2009) 183:4809–16. doi: 10.4049/jimmunol.0901616
93. Grundström S, Cederbom L, Sundstedt A, Scheipers P, Ivars F. Superantigen-Induced Regulatory T Cells Display Different Suppressive Functions in the Presence or Absence of Natural CD4 + CD25 + Regulatory T Cells In Vivo. J Immunol (2003) 170:5008–17. doi: 10.4049/jimmunol.170.10.5008
94. Taylor AL, Llewelyn MJ. Superantigen-Induced Proliferation of Human CD4 + CD25 – T Cells Is Followed by a Switch to a Functional Regulatory Phenotyp. J Immunol (2010) 185:6591–8. doi: 10.4049/jimmunol.1002416
95. Metzler B, Wraith DC. Inhibition of Experimental Autoimmune Encephalomyelitis by Inhalation But Not Oral Administration of the Encephalitogenic Peptide: Influence of MHC Binding Affinity. Int Immunol (1993) 5:1159–65. doi: 10.1093/intimm/5.9.1159
96. Gabryšová L, Wraith DC. Antigenic Strength Controls the Generation of Antigen-Specific IL-10-Secreting T Regulatory Cells. Eur J Immunol (2010) 40:1386–95. doi: 10.1002/eji.200940151
97. Chang HD, Helbig C, Tykocinski L, Kreher S, Koeck J, Niesner U, et al. Expression of IL-10 in Th Memory Lymphocytes Is Conditional on IL-12 or IL-4, Unless the IL-10 Gene is Imprinted by GATA-3. Eur J Immunol (2007) 37:807–17. doi: 10.1002/eji.200636385
98. Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O’Garra A. Interleukin-10 Production by Th1 Cells Requires Interleukin-12-Induced STAT4 Transcription Factor and ERK MAP Kinase Activation by High Antigen Dos. Immunity (2009) 31:209–19. doi: 10.1016/j.immuni.2009.05.012
99. Motomura Y, Kitamura H, Hijikata A, Matsunaga Y, Matsumoto K, Inoue H, et al. The Transcription Factor E4BP4 Regulates the Production of IL-10 and IL-13 in CD4+ T Cells. Nat Immunol (2011) 12:450–9. doi: 10.1038/ni.2020
100. Levings MK, Sangregorio R, Galbiati F, Squadrone S, de Waal Malefyt R, Roncarolo M-G. IFN-α and IL-10 Induce the Differentiation of Human Type 1 T Regulatory Cell. J Immunol (2001) 166:5530–9. doi: 10.4049/jimmunol.166.9.5530
101. Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, et al. Regulatory T Cells Expressing Interleukin 10 Develop From Foxp3+ and Foxp3- Precursor Cells in the Absence of Interleukin 10. Nat Immunol (2007) 8:931–41. doi: 10.1038/ni1504
102. Levings MK, Gregori S, Tresoldi E, Cazzaniga S, Bonini C, Roncarolo MG. Differentiation of Tr1 Cells by Immature Dendritic Cells Requires IL-10 But Not CD25+CD4+ Tr Cells. Blood (2005) 105:1162–9. doi: 10.1182/blood-2004-03-1211
103. Shiokawa A, Tanabe K, Tsuji NM, Sato R, Hachimura S. IL-10 and IL-27 Producing Dendritic Cells Capable of Enhancing IL-10 Production of T Cells are Induced in Oral Tolerance. Immunol Lett (2009) 125:7–14. doi: 10.1016/j.imlet.2009.05.002
104. Comi M, Amodio G, Gregori S. Interleukin-10-Producing DC-10 Is a Unique Tool to Promote Tolerance Via Antigen-Specific T Regulatory Type 1 Cell. Front Immunol (2018) 9:682. doi: 10.3389/fimmu.2018.00682
105. Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J, et al. IL-27, a Heterodimeric Cytokine Composed of EBI3 and P28 Protein, Induces Proliferation of Naive CD4+T Cells. Immunity (2002) 16:779–90. doi: 10.1016/S1074-7613(02)00324-2
106. Murugaiyan G, Mittal A, Lopez-Diego R, Maier LM, Anderson DE, Weiner HL. IL-27 Is a Key Regulator of IL-10 and IL-17 Production by Human CD4 + T Cell. J Immunol (2009) 183:2435–43. doi: 10.4049/jimmunol.0900568
107. Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. CD4+ T Cell Help and Innate-Derived IL-27 Induce Blimp-1-Dependent IL-10 Production by Antiviral CTL. Nat Immunol (2011) 12:327–35. doi: 10.1038/ni.1996
108. Wang S, Miyazaki Y, Shinozaki Y, Yoshida H. Augmentation of Antigen-Presenting and Th1-Promoting Functions of Dendritic Cells by WSX-1(IL-27r) Deficienc. J Immunol (2007) 179:6421–8. doi: 10.4049/jimmunol.179.10.6421
109. Yoshimoto T, Yoshimoto T, Yasuda K, Mizuguchi J, Nakanishi K. IL-27 Suppresses Th2 Cell Development and Th2 Cytokines Production From Polarized Th2 Cells: A Novel Therapeutic Way for Th2-Mediated Allergic Inflammatio. J Immunol (2007) 179:4415–23. doi: 10.4049/jimmunol.179.7.4415
110. Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S, et al. Interleukin 27 Limits Autoimmune Encephalomyelitis by Suppressing the Development of Interleukin 17-Producing T Cells. Nat Immunol (2006) 7:929–36. doi: 10.1038/ni1375
111. Awasthi A, Carrier Y, Peron JPS, Bettelli E, Kamanaka M, Flavell RA, et al. A Dominant Function for Interleukin 27 in Generating Interleukin 10-Producing Anti-Inflammatory T Cells. Nat Immunol (2007) 8:1380–9. doi: 10.1038/ni1541
112. Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, et al. Interleukins 27 and 6 Induce STAT3-Mediated T Cell Production of Interleukin 10. Nat Immunol (2007) 8:1363–71. doi: 10.1038/ni1537
113. Wang H, Meng R, Li Z, Yang B, Liu Y, Huang F, et al. IL-27 Induces the Differentiation of Tr1-Like Cells From Human Naive CD4+T Cells via the Phosphorylation of STAT1 and STAT3. Immunol Lett (2011) 136:21–8. doi: 10.1016/j.imlet.2010.11.007
114. Spolski R, Leonard WJ. Interleukin-21: Basic Biology and Implications for Cancer and Autoimmunit. Annu Rev Immunol (2008) 26:57–79. doi: 10.1146/annurev.immunol.26.021607.090316
115. Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, et al. Interleukin 21 and Its Receptor Are Involved in NK Cell Expansion and Regulation of Lymphocyte Function. Nature (2000) 408:57–63. doi: 10.1038/35040504
116. Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, et al. A Critical Role for IL-21 in Regulating Immunoglobulin Production. Science (2002) 298:1630–4. doi: 10.1126/science.1077002
117. Ozaki K, Spolski R, Ettinger R, Kim H-P, Wang G, Qi C-F, et al. Regulation of B Cell Differentiation and Plasma Cell Generation by IL-21, A Novel Inducer of Blimp-1 and Bcl-6. J Immunol (2004) 173:5361–71. doi: 10.4049/jimmunol.173.9.5361
118. Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A Fundamental Role for Interleukin-21 in the Generation of T Follicular Helper Cell. Immunity (2008) 29:127–37. doi: 10.1016/j.immuni.2008.06.001
119. Zeng R, Spolski R, Finkelstein SE, Oh SK, Kovanen PE, Hinrichs CS, et al. Synergy of IL-21 and IL-15 in Regulating CD8+ T Cell Expansion and Function. J Exp Med (2005) 201:139–48. doi: 10.1084/jem.20041057
120. Jin H, Carrio R, Yu A, Malek TR. Distinct Activation Signals Determine Whether IL-21 Induces B Cell Costimulation, Growth Arrest, or Bim-Dependent Apoptosi. J Immunol (2004) 173:657–65. doi: 10.4049/jimmunol.173.1.657
121. Brandt K, Bulfone-Paus S, Foster DC, Rückert R. Interleukin-21 Inhibits Dendritic Cell Activation and Maturation. Blood (2003) 102:4090–8. doi: 10.1182/blood-2003-03-0669
122. Spolski R, Kim H-P, Zhu W, Levy DE, Leonard WJ. IL-21 Mediates Suppressive Effects via Its Induction of IL-10. J Immunol (2009) 182:2859–67. doi: 10.4049/jimmunol.0802978
123. Umeshappa CS, Solé P, Surewaard BGJ, Mohopatra S, Yamanouchi J, Uddin MM, et al. Liver-Specific T-Regulatory Type 1 Cells Program Local Neutrophils to Suppress Hepatic Autoimmunity via Cram. Cell Rep (2021) 34(13):108919. doi: 10.1016/j.celrep.2021.108919
124. Greenwald RJ, Freeman GJ, Sharpe AH. The B7 Family Revisite. Annu Rev Immunol (2005) 23:515–48. doi: 10.1146/annurev.immunol.23.021704.115611
125. Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T Follicular Helper Cells Is Mediated by Interleukin-21 But Independent of T Helper 1, 2, or 17 Cell Lineage. Immunity (2008) 29:138–49. doi: 10.1016/j.immuni.2008.05.009.Generation
126. Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, et al. ICOS Receptor Instructs T Follicular Helper Cell Versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity (2011) 34:932–46. doi: 10.1016/j.immuni.2011.03.023
127. Nurieva RI, Duong J, Kishikawa H, Dianzani U, Rojo JM, Ho IC, et al. Transcriptional Regulation of Th2 Differentiation by Inducible Costimulator. Immunity (2003) 18:801–11. doi: 10.1016/S1074-7613(03)00144-4
128. Serada S, Fujimoto M, Mihara M, Koike N, Ohsugi Y, Nomura S, et al. IL-6 Blockade Inhibits the Induction of Myelin Antigen-Specific Th17 Cells and Th1 Cells in Experimental Autoimmune Encephalomyelitis. Proc Natl Acad Sci USA (2008) 105:9041–6. doi: 10.1073/pnas.0802218105
129. Fonseca JE, Santos MJ, Canhão H, Choy E. Interleukin-6 as a Key Player in Systemic Inflammation and Joint Destruction. Autoimmun Rev (2009) 8:538–42. doi: 10.1016/j.autrev.2009.01.012
130. Lindroos J, Svensson L, Norsgaard H, Lovato P, Moller K, Hagedorn PH, et al. IL-23-Mediated Epidermal Hyperplasia Is Dependent on IL-6. J Invest Dermatol (2011) 131:1110–8. doi: 10.1038/jid.2010.432
131. Hiramatsu Y, Suto A, Kashiwakuma D, Kanari H, Kagami S, Ikeda K, et al. C-Maf Activates the Promoter and Enhancer of the IL-21 Gene, and TGF-β Inhibits C-Maf-Induced IL-21 Production in CD4 + T Cells. J Leukoc Biol (2010) 87:703–12. doi: 10.1189/jlb.0909639
132. Dicosmo BF, Picarella D, Ravell RA. Local Production of Human IL-6 Promotes Insulitis But Retards the Onset of Insulin-Dependent Diabetes Mellitus in Non-Obese Diabetic Mice. Int Immunol (1994) 6:1829–37. doi: 10.1093/intimm/6.12.1829
133. Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL-6 and Stat3 Are Required for Survival of Intestinal Epithelial Cells And Development of Colitis-Associated Cance. Cancer Cell (2009) 15:103–13. doi: 10.1016/j.ccr.2009.01.001
134. Balto K, Sasaki H, Stashenko P. Interleukin-6 Deficiency Increases Inflammatory Bone Destruction. Infect Immun (2001) 69:744–50. doi: 10.1128/IAI.69.2.744-750.2001
135. McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, et al. TGF-β and IL-6 Drive the Production of IL-17 and IL-10 by T Cells and Restrain TH-17 Cell-Mediated Pathology. Nat Immunol (2007) 8:1390–7. doi: 10.1038/ni1539
136. Jin JO, Han X, Yu Q. Interleukin-6 Induces the Generation of IL-10-Producing Tr1 Cells and Suppresses Autoimmune Tissue Inflammation. J Autoimmun (2013) 40:28–44. doi: 10.1016/j.jaut.2012.07.009
137. Novick D, Cohen B, Rubinstein M. The Human Interferon α β Receptor: Characterization and Molecular Cloning. Cell (1994) 77:391–400. doi: 10.1016/0092-8674(94)90154-6
138. Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, et al. Type I Interferon Inhibits Interleukin-1 Production and Inflammasome Activatio. Immunity (2011) 34:213–23. doi: 10.1016/j.immuni.2011.02.006
139. Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, et al. IFN-γ Suppresses IL-10 Production and Synergizes With TLR2 by Regulating GSK3 and CREB/AP-1 Protein. Immunity (2006) 24:563–74. doi: 10.1016/j.immuni.2006.02.014
140. Garcia CA, Benakanakere MR, Alard P, Kosiewicz MM, Kinane DF, Martin M. Antigenic Experience Dictates Functional Role of Glycogen Synthase Kinase-3 in Human CD4 + T Cell Response. J Immunol (2008) 181:8363–71. doi: 10.4049/jimmunol.181.12.8363
141. Aman MJ, Tretter T, Eisenbeis I, Bug G, Decker T, Aulitzky WE, et al. Interferon-Alpha Stimulates Production of Interleukin-10 in Activated CD4+ T Cells and Monocytes. Blood (1996) 87:4731–6. doi: 10.1182/blood.V87.11.4731.bloodjournal87114731
142. McRae BL, Semnani RT, Hayes MP, van Seventer GA. Type I IFNs Inhibit Human Dendritic Cell IL-12 Production and Th1 Cell Development. J Immunol (1998) 160:4298–304.
143. Ma A, Boone DL, Lodolce JP. The Pleiotropic Functions of Interleukin 15: Not So Interleukin 2-Like After All. J Exp Med (2000) 191:753–5. doi: 10.1084/jem.191.5.753
144. Waldmann T, Tagaya Y, Bamford R. Interleukin-2, Interleukin-15, and Their Receptors. Int Rev Immunol (1998) 16:205–26. doi: 10.3109/08830189809042995
145. Bulfone-Paus S, Ungureanu D, Pohl T, Lindner G, Paus R, Rückert R, et al. Interleukin-15 Protects From Lethal Apoptosis In Vivo. Nat Med (1997) 3:1124–8. doi: 10.1038/nm1097-1124
146. Ku CC, Murakami M, Sakamoto A, Kappler J, Marrack P. Control of Homeostasis of CD8+ Memory T Cells by Opposing Cytokines. Science (2000) 288:675–8. doi: 10.1126/science.288.5466.675
147. Tough DF, Sun S, Zhang X, Sprent J. Stimulation of Memory T Cells by Cytokines. Vaccine (2000) 18:1642–8. doi: 10.1016/S0264-410X(99)00500-9
148. Bacchetta R, Sartirana C, Levings MK, Bordignon C, Narula S, Roncarolo MG. Growth and Expansion of Human T Regulatory Type 1 Cells Are Independent From TCR Activation But Require Exogenous Cytokines. Eur J Immunol (2002) 32:2237–45. doi: 10.1002/1521-4141(200208)32:8<2237::AID-IMMU2237>3.0.CO;2-2
149. Chow KPN, Lee JM, Qiu JT, Liao SK, Lin SC, Hsu SL, et al. Enhanced IL-10 Production by CD4 + T Cells Primed in IL-15rα-Deficient Mice. Eur J Immunol (2011) 41:3146–56. doi: 10.1002/eji.201141746
150. Giri JG, Kumaki S, Ahdieh M, Friend DJ, Loomis A, Shanebeck K, et al. Identification and Cloning of a Novel IL-15 Binding Protein That Is Structurally Related to the Alpha Chain of the IL-2 Receptor. EMBO J (1995) 14:3654–63. doi: 10.1002/j.1460-2075.1995.tb00035.x
151. Waldmann TA, Tagaya Y. The Multifaced Regulation of Interleukin-15 Expression an the Role of This Cytokine in NK Cell Differentiation and Host Response to Intracellular Pathogens. Annu Rev Immunol (1999) 17:19–49. doi: 10.1146/annurev.immunol.17.1.19
152. Papiernik M. Natural CD4+ CD25+ Regulatory T Cells. Their Role in the Control of Superantigen Responses. Immunol Rev (2001) 182:180–9. doi: 10.1034/j.1600-065X.2001.1820114.x
153. Anderson PO, Sundstedt A, Yazici Z, Minaee S, Woolf R, Nicolson K, et al. IL-2 Overcomes the Unresponsiveness But Fails to Reverse the Regulatory Function of Antigen-Induced T Regulatory Cell. J Immunol (2005) 174:310–9. doi: 10.4049/jimmunol.174.1.310
154. Tsuji-Takayama K, Suzuki M, Yamamoto M, Harashima A, Okochi A, Otani T, et al. The Production of IL-10 by Human Regulatory T Cells Is Enhanced by IL-2 Through a STAT5-Responsive Intronic Enhancer in the IL-10 Locu. J Immunol (2008) 181:3897–905. doi: 10.4049/jimmunol.181.6.3897
155. Kasprzycka M, Zhang Q, Witkiewicz A, Marzec M, Potoczek M, Liu X, et al. γc-Signaling Cytokines Induce a Regulatory T Cell Phenotype in Malignant CD4 + T Lymphocyte. J Immunol (2008) 181:2506–12. doi: 10.4049/jimmunol.181.4.2506
156. Andreotti AH, Schwartzberg PL, Joseph RE, Berg LJ. T-Cell Signaling Regulated by the Tec Family Kinase, It. Cold Spring Harb Perspect Biol (2010) 2:1–22. doi: 10.1101/cshperspect.a002287
157. Ahyi A-NN, Chang H-C, Dent AL, Nutt SL, Kaplan MH. IFN Regulatory Factor 4 Regulates the Expression of a Subset of Th2 Cytokine. J Immunol (2009) 183:1598–606. doi: 10.4049/jimmunol.0803302
158. Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, et al. Analysis of Interleukin-21-Induced Prdm1 Gene Regulation Reveals Functional Cooperation of STAT3 and IRF4 Transcription Factor. Immunity (2009) 31:941–52. doi: 10.1016/j.immuni.2009.10.008
159. Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, et al. The Transcription Factors Blimp-1 and IRF4 Jointly Control the Differentiation and Function of Effector Regulatory T Cells. Nat Immunol (2011) 12:304–12. doi: 10.1038/ni.2006
160. Kim JI, Ho IC, Grusby MJ, Glimcher LH. The Transcription Factor C-Maf Controls the Production of Interleukin-4 But Not Other Th2 Cytokines. Immunity (1999) 10:745–51. doi: 10.1016/S1074-7613(00)80073-4
161. Zhu J, Yamane H, Paul WE. Differentiation of Effector CD4 T Cell Population. Annu Rev Immunol (2010) 28:445–89. doi: 10.1146/annurev-immunol-030409-101212
162. Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, et al. Bcl6 and Maf Cooperate To Instruct Human Follicular Helper CD4 T Cell Differentiatio. J Immunol (2012) 188:3734–44. doi: 10.4049/jimmunol.1103246
163. Harris JE, Bishop KD, Phillips NE, Mordes JP, Greiner DL, Rossini AA, et al. Early Growth Response Gene-2, a Zinc-Finger Transcription Factor, Is Required for Full Induction of Clonal Anergy in CD4 + T Cell. J Immunol (2004) 173:7331–8. doi: 10.4049/jimmunol.173.12.7331
164. Crotty S, Johnston RJ, Schoenberger SP. Effectors and Memories: Bcl-6 and Blimp-1 in T and B Lymphocyte Differentiation. Nat Immunol (2010) 11:114–20. doi: 10.1038/ni.1837
165. Martins G, Calame K. Regulation and Functions of Blimp-1 in T and B Lymphocyte. Annu Rev Immunol (2008) 26:133–69. doi: 10.1146/annurev.immunol.26.021607.090241
166. Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi MF, Ahlers J, et al. Role of Blimp-1 in Programing Th Effector Cells Into IL-10 Producers. J Exp Med (2014) 211:1807–19. doi: 10.1084/jem.20131548
167. Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, et al. Critical Role of IRF1 and BATF in Forming Chromatin Landscape During Type 1 Regulatory Cell Differentiation. Nat Immunol (2017) 18:412–21. doi: 10.1038/ni.3683
168. Schraml BU, Hildner K, Ise W, Lee WL, Smith WAE, Solomon B, et al. The AP-1 Transcription Factor Batf Controls T H 17 Differentiation. Nature (2009) 460:405–9. doi: 10.1038/nature08114
169. Ellyard JI, Vinuesa CG. A BATF-Ling Connection Between B Cells and Follicular Helper T Cells. Nat Immunol (2011) 12:519–20. doi: 10.1038/ni.2042
170. Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, et al. Batf Coordinates Multiple Aspects of B and T Cell Function Required for Normal Antibody Responses. J Exp Med (2010) 207:933–42. doi: 10.1084/jem.20091548
171. Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A Validated Regulatory Network for Th17 Cell Specification. Cell (2012) 151:289–303. doi: 10.1016/j.cell.2012.09.016
172. Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, Diiorio MA, Lemieux ME, et al. The Transcription Factor BATF Operates as an Essential Differentiation Checkpoint in Early Effector CD8 + T Cells. Nat Immunol (2014) 15:373–83. doi: 10.1038/ni.2834
173. Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, et al. The Transcription Factor BATF Controls the Global Regulators of Class-Switch Recombination in Both B Cells and T Cells. Nat Immunol (2011) 12:536–43. doi: 10.1038/ni.2037
174. Zhang P, Lee JS, Gartlan KH, Schuster IS, Comerford I, Varelias A, et al. Eomesodermin Promotes the Development of Type 1 Regulatory T (TR1) Cells. Sci Immunol (2017) 2. doi: 10.1126/sciimmunol.aah7152
175. Dejean AS, Joulia E, Walzer T. The Role of Eomes in Human CD4 T Cell Differentiation: A Question of Context. Eur J Immunol (2019) 49:38–41. doi: 10.1002/eji.201848000
176. Farez MFF, Mascanfroni IDD, Méndez-Huergo SPP, Yeste A, Murugaiyan G, Garo LPP, et al. Melatonin Contributes to the Seasonality of Multiple Sclerosis Relapse. Cell (2015) 162:1338–52. doi: 10.1016/j.cell.2015.08.025
177. Im SH, Hueber A, Monticelli S, Kang KH, Rao A. Chromatin-Level Regulation of the IL10 Gene in T Cells. J Biol Chem (2004) 279:46818–25. doi: 10.1074/jbc.M401722200
178. Miraldi ER, Pokrovskii M, Watters A, Castro DM, De Veaux N, Hall JA, et al. Leveraging Chromatin Accessibility for Transcriptional Regulatory Network Inference in T Helper 17 Cell. Genome Res (2019) 29:449–63. doi: 10.1101/gr.238253.118
179. Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, et al. Discrete Roles of STAT4 and STAT6 Transcription Factors in Tuning Epigenetic Modifications and Transcription During T Helper Cell Differentiation. Immunity (2010) 32:840–51. doi: 10.1016/j.immuni.2010.06.003
180. Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, et al. BATF-JUN Is Critical for IRF4-Mediated Transcription in T Cells. Nature (2012) 490:543–6. doi: 10.1038/nature11530
181. Lee CG, Hwang W, Maeng KE, Kwon HK, So JS, Sahoo A, et al. IRF4 Regulates IL-10 Gene Expression in CD4+ T Cells Through Differential Nuclear Translocation. Cell Immunol (2011) 268:97–104. doi: 10.1016/j.cellimm.2011.02.008
182. Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, et al. STATs Shape the Active Enhancer Landscape of T Cell Populations. Cell (2012) 151:981–93. doi: 10.1016/j.cell.2012.09.044
183. Shoemaker J, Saraiva M, O’Garra A. GATA-3 Directly Remodels the IL-10 Locus Independently of IL-4 in CD4 + T Cell. J Immunol (2006) 176:3470–9. doi: 10.4049/jimmunol.176.6.3470
184. Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 Exploits a Pre-Existent Enhancer Landscape for Regulatory T Cell Lineage Specification. Cell (2012) 151:153–66. doi: 10.1016/j.cell.2012.06.053
185. Hossain DMS, Panda AK, Manna A, Mohanty S, Bhattacharjee P, Bhattacharyya S, et al. FoxP3 Acts as a Cotranscription Factor With STAT3 in Tumor-Induced Regulatory T Cell. Immunity (2013) 39:1057–69. doi: 10.1016/j.immuni.2013.11.005
186. Lee C-G, Kwon H-K, Sahoo A, Hwang W, So J-S, Hwang J-S, et al. Interaction of Ets-1 With HDAC1 Represses IL-10 Expression in Th1 Cell. J Immunol (2012) 188:2244–53. doi: 10.4049/jimmunol.1101614
187. Grenningloh R, Bok YK, Ho IC. Ets-1, A Functional Cofactor of T-Bet, is Essential for Th1 Inflammatory Responses. J Exp Med (2005) 201:615–26. doi: 10.1084/jem.20041330
188. Lee MT, Bonneau AR, Giraldez AJ. Zygotic Genome Activation During the Maternal-To-Zygotic Transitio. Annu Rev Cell Dev Biol (2014) 30:581–613. doi: 10.1146/annurev-cellbio-100913-013027
189. Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and Epigenetic Programming of the Human Germ Line. Nat Rev Genet (2016) 17:585–600. doi: 10.1038/nrg.2016.88
190. Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic Stem Cell States: Naive to Primed Pluripotency in Rodents and Humans. Nat Rev Mol Cell Biol (2016) 17:155–69. doi: 10.1038/nrm.2015.28
191. Hackett JA, Azim Surani M. Regulatory Principles of Pluripotency: From the Ground State Up. Cell Stem Cell (2014) 15:416–30. doi: 10.1016/j.stem.2014.09.015
192. Smith ZD, Meissner A. DNA Methylation: Roles in Mammalian Development. Nat Rev Genet (2013) 14:204–20. doi: 10.1038/nrg3354
193. Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, et al. Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes. Cell (2013) 154:888–903. doi: 10.1016/j.cell.2013.07.020
194. Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, et al. Latent Enhancers Activated by Stimulation in Differentiated Cells. Cell (2013) 152:157–71. doi: 10.1016/j.cell.2012.12.018
195. Ramos-Rodríguez M, Raurell-Vila H, Colli ML, Alvelos MI, Subirana-Granés M, Juan-Mateu J, et al. The Impact of Proinflammatory Cytokines on the β-Cell Regulatory Landscape Provides Insights Into the Genetics of Type 1 Diabetes. Nat Genet (2019) 51:1588–95. doi: 10.1038/s41588-019-0524-6
196. Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, et al. Fine Mapping of Type 1 Diabetes Susceptibility Loci and Evidence for Colocalization of Causal Variants With Lymphoid Gene Enhancers. Nat Genet (2015) 47:381–6. doi: 10.1038/ng.3245
197. Farh KKH, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and Epigenetic Fine Mapping of Causal Autoimmune Disease Variants. Nature (2015) 518:337–43. doi: 10.1038/nature13835
198. Fasolino M, Goldman N, Wang W, Cattau B, Zhou Y, Petrovic J, et al. Genetic Variation in Type 1 Diabetes Reconfigures the 3D Chromatin Organization of T Cells and Alters Gene Expressio. Immunity (2020) 52:257–74.e11. doi: 10.1016/j.immuni.2020.01.003
199. Flannick J, Florez JC. Type 2 Diabetes: Genetic Data Sharing to Advance Complex Disease Research. Nat Rev Genet (2016) 17:535–49. doi: 10.1038/nrg.2016.56
200. Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, et al. The Genetic Architecture of Type 2 Diabetes. Nature (2016) 536:41–7. doi: 10.1038/nature18642
201. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell (2013) 153:307–19. doi: 10.1016/j.cell.2013.03.035
202. Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, et al. A Map of Open Chromatin in Human Pancreatic Islets. Nat Genet (2010) 42:255–9. doi: 10.1038/ng.530
203. Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman I, et al. Pancreatic Islet Enhancer Clusters Enriched in Type 2 Diabetes Risk-Associated Variants. Nat Genet (2014) 46:136–43. doi: 10.1038/ng.2870
204. Miguel-Escalada I, Bonàs-Guarch S, Cebola I, Ponsa-Cobas J, Mendieta-Esteban J, Atla G, et al. Human Pancreatic Islet Three-Dimensional Chromatin Architecture Provides Insights Into the Genetics of Type 2 Diabetes. Nat Genet (2019) 51:1137–48. doi: 10.1038/s41588-019-0457-0
205. Sellars M, Huh JR, Day K, Issuree PD, Galan C, Gobeil S, et al. Regulation of DNA Methylation Dictates Cd4 Expression During the Development of Helper and Cytotoxic T Cell Lineages. Nat Immunol (2015) 16:746–54. doi: 10.1038/ni.3198
206. Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, Jarvis RC, et al. Inducible Chromatin Priming Is Associated With the Establishment of Immunological Memory in T Cells. EMBO J (2016) 35:515–35. doi: 10.15252/embj.201592534
207. Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription Factors T-Bet and Runx3 Cooperate to Activate Ifng and Silence Il4 in T Helper Type 1 Cells. Nat Immunol (2007) 8:145–53. doi: 10.1038/ni1424
208. Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, et al. Hlx Is Induced by and Genetically Interacts With T-Bet to Promote Heritable THI Gene Induction. Nat Immunol (2002) 3:652–8. doi: 10.1038/ni807
209. Usui T, Preiss JC, Kanno Y, Zheng JY, Bream JH, O’Shea JJ, et al. T-Bet Regulates Th1 Responses Through Essential Effects on GATA-3 Function Rather Than on IFNG Gene Acetylation and Transcription. J Exp Med (2006) 203:755–66. doi: 10.1084/jem.20052165
210. Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, et al. Comprehensive Epigenetic Profiling Identifies Multiple Distal Regulatory Elements Directing Transcription of the Gene Encoding Interferon-γ. Nat Immunol (2007) 8:732–42. doi: 10.1038/ni1474
211. Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, et al. A Distal Conserved Sequence Element Controls Ifng Gene Expression by T Cells and NK Cell. Immunity (2006) 25:717–29. doi: 10.1016/j.immuni.2006.09.007
212. Lee DU, Avni O, Chen L, Rao A. A Distal Enhancer in the Interferon-γ (IFN-γ) Locus Revealed by Genome Sequence Comparison. J Biol Chem (2004) 279:4802–10. doi: 10.1074/jbc.M307904200
213. Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, et al. Evolutionarily Conserved Sequence Elements That Positively Regulate IFN-γ Expression in T Cells. Proc Natl Acad Sci USA (2004) 101:12622–7. doi: 10.1073/pnas.0400849101
214. Chen GY, Osada H, Santamaria-Babi LF, Kannagi R. Interaction of GATA-3/T-Bet Transcription Factors Regulates Expression of Sialil Lewis X Homing Receptors on Th1/Th2 Lymphocytes. Proc Natl Acad Sci USA (2006) 103:16894–9. doi: 10.1073/pnas.0607926103
215. Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated But Physically Separable Interaction With H3K27-Demethylase and H3K4-Methyltransferase Activities Are Required for T-Box Protein-Mediated Activation of Developmental Gene Expression. Genes Dev (2008) 22:2980–93. doi: 10.1101/gad.1689708
216. Jones B, Chen J. Inhibition of IFN-γ Transcription by Site-Specific Methylation During T Helper Cell Development. EMBO J (2006) 25:2443–52. doi: 10.1038/sj.emboj.7601148
217. Chang S, Aune TM. Dynamic Changes in Histone-Methylation “Marks” Across the Locus Encoding Interferon-γ During the Differentiation of T Helper Type 2 Cells. Nat Immunol (2007) 8:723–31. doi: 10.1038/ni1473
218. Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 Differentiation and Il4 Locus Accessibility. Annu Rev Immunol (2006) 24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821
219. Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sözeri O, Löhning M, et al. A Critical Control Element for Interleukin-4 Memory Expression in T Helper Lymphocytes. J Biol Chem (2005) 280:28177–85. doi: 10.1074/jbc.M502038200
220. Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. Th Cell Differentiation is Accompanied by Dynamic Changes in Histone Acetylation of Cytokine Genes. Nat Immunol (2002) 3:643–51. doi: 10.1038/ni808
221. Fields PE, Kim ST, Flavell RA. Cutting Edge: Changes in Histone Acetylation at the IL-4 and IFN-γ Loci Accompany Th1/Th2 Differentiation. J Immunol (2002) 169:647–50. doi: 10.4049/jimmunol.169.2.647
222. Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and Histone 3 Trimethyl Lysine 27 Associated With Il4 and Il13 Gene Silencing in TH1 Cells. J Biol Chem (2005) 280:31470–7. doi: 10.1074/jbc.M504766200
223. Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, et al. Crucial Role of MLL for the Maintenance of Memory T Helper Type 2 Cell Response. Immunity (2006) 24:611–22. doi: 10.1016/j.immuni.2006.03.017
224. Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, et al. Gene Silencing Quantitatively Controls the Function of a Developmental Trans-Activator. Mol Cell (2002) 10:81–91. doi: 10.1016/S1097-2765(02)00564-6
225. Makar KW, Pérez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active Recruitment of DNA Methyltransferases Regulates Interleukin 4 in Thymocytes and T Cells. Nat Immunol (2003) 4:1183–90. doi: 10.1038/ni1004
226. Wurster AL, Pazin MJ. BRG1-Mediated Chromatin Remodeling Regulates Differentiation and Gene Expression of T Helper Cell. Mol Cell Biol (2008) 28:7274–85. doi: 10.1128/mcb.00835-08
227. Ivanov II, Zhou L, Littman DR. Transcriptional Regulation of Th17 Cell Differentiation. Semin Immunol (2007) 19:409–17. doi: 10.1016/j.smim.2007.10.011
228. McGeachy MJ, Cua DJ. Th17 Cell Differentiation: The Long and Winding Roa. Immunity (2008) 28:445–53. doi: 10.1016/j.immuni.2008.03.001
229. Zhou L, Lopes JE, Chong MMW, Ivanov II, Min R, Victora GD, et al. TGF-β-Induced Foxp3 Inhibits TH17 Cell Differentiation by Antagonizing Rorγt Function. Nature (2008) 453:236–40. doi: 10.1038/nature06878
230. Dong C. TH17 Cells in Development: An Updated View of Their Molecular Identity and Genetic Programming. Nat Rev Immunol (2008) 8:337–48. doi: 10.1038/nri2295
231. Akimzhanov AM, Yang XO, Dong C. Chromatin Remodeling of Interleukin-17 (IL-17)-IL-17F Cytokine Gene Locus During Inflammatory Helper T Cell Differentiation. J Biol Chem (2007) 282:5969–72. doi: 10.1074/jbc.C600322200
232. Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T Helper 17 Lineage Differentiation Is Programmed by Orphan Nuclear Receptors Rorα and Rorγ. Immunity (2008) 28:29–39. doi: 10.1016/j.immuni.2007.11.016
233. Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science (2009) 325:1006–10. doi: 10.1126/science.1175870
234. Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, et al. In Vivo Regulation of Bcl6 and T Follicular Helper Cell Development. J Immunol (2010) 185:313–26. doi: 10.4049/jimmunol.0904023
235. Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, et al. Functional and Epigenetic Studies Reveal Multistep Differentiation and Plasticity of In Vitro-Generated and In Vivo-Derived Follicular T Helper Cell. Immunity (2011) 35:622–32. doi: 10.1016/j.immuni.2011.07.015
236. Cannons JL, Lu KT, Schwartzberg PL. T Follicular Helper Cell Diversity and Plasticity. Trends Immunol (2013) 34:200–7. doi: 10.1016/j.it.2013.01.001
237. Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, et al. Guidance of Regulatory T Cell Development by Satb1-Dependent Super-Enhancer Establishment. Nat Immunol (2017) 18:173–83. doi: 10.1038/ni.3646
238. Toker A, Engelbert D, Garg G, Polansky JK, Floess S, Miyao T, et al. Active Demethylation of the Foxp3 Locus Leads to the Generation of Stable Regulatory T Cells Within the Thymus. J Immunol (2013) 190:3180–8. doi: 10.4049/jimmunol.1203473
239. Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, et al. T Cell Receptor Stimulation-Induced Epigenetic Changes and Foxp3 Expression Are Independent and Complementary Events Required for Treg Cell Development. Immunity (2012) 37:785–99. doi: 10.1016/j.immuni.2012.09.010
240. Delacher M, Imbusch CD, Weichenhan D, Breiling A, Hotz-Wagenblatt A, Träger U, et al. Genome-Wide DNA-Methylation Landscape Defines Specialization of Regulatory T Cells in Tissues. Nat Immunol (2017) 18:1160–72. doi: 10.1038/ni.3799
241. Liu B, Tahk S, Yee KM, Fan G, Shuai K. The Ligase PIAS1 Restricts Natural Regulatory T Cell Differentiation by Epigenetic Repression. Science (2010) 330:521–5. doi: 10.1126/science.1108297
242. Hegazy AN, Peine M, Helmstetter C, Panse I, Fröhlich A, Bergthaler A, et al. Interferons Direct Th2 Cell Reprogramming to Generate a Stable GATA-3+T-Bet+ Cell Subset With Combined Th2 and Th1 Cell Function. Immunity (2010) 32:116–28. doi: 10.1016/j.immuni.2009.12.004
243. Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, et al. Fate Mapping of IL-17-Producing T Cells in Inflammatory Responses. Nat Immunol (2011) 12:255–63. doi: 10.1038/ni.1993
244. Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T Helper Type 1-Like, Foxp3+ Regulatory T Cells in Human Autoimmune Disease. Nat Med (2011) 17:673–5. doi: 10.1038/nm.2389
245. Panzer M, Sitte S, Wirth S, Drexler I, Sparwasser T, Voehringer D. Rapid In Vivo Conversion of Effector T Cells Into Th2 Cells During Helminth Infectio. J Immunol (2012) 188:615–23. doi: 10.4049/jimmunol.1101164
246. Schmidl C, Hansmann L, Andreesen R, Edinger M, Hoffmann P, Rehli M. Epigenetic Reprogramming of the RORC Locus During In Vitro Expansion Is a Distinctive Feature of Human Memory But Not Naïve Tre. Eur J Immunol (2011) 41:1491–8. doi: 10.1002/eji.201041067
247. Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global Mapping of H3K4me3 and H3K27me3 Reveals Specificity and Plasticity in Lineage Fate Determination of Differentiating CD4+ T Cell. Immunity (2009) 30:155–67. doi: 10.1016/j.immuni.2008.12.009
248. Barnett KR, Decato BE, Scott TJ, Hansen TJ, Chen B, Attalla J, et al. ATAC-Me Captures Prolonged DNA Methylation of Dynamic Chromatin Accessibility Loci During Cell Fate Transition. Mol Cell (2020) 77:1–15. doi: 10.1016/j.molcel.2020.01.004
249. Kornberg RD, Lorch Y. Chromatin Structure and Transcription. Annu Rev Cell Biol (1992) 8:563–87. doi: 10.1146/annurev.cb.08.110192.003023
Keywords: T-regulatory type 1 (TR1) cells, peptide-MHC class II-coated nanoparticles, T-cell reprogramming, interleukin 10 (IL10), autoimmune disease, therapy
Citation: Solé P and Santamaria P (2021) Re-Programming Autoreactive T Cells Into T-Regulatory Type 1 Cells for the Treatment of Autoimmunity. Front. Immunol. 12:684240. doi: 10.3389/fimmu.2021.684240
Received: 23 March 2021; Accepted: 22 June 2021;
Published: 15 July 2021.
Edited by:
Francisco Javier Quintana, Harvard Medical School, United StatesReviewed by:
Mohan Maddur, Pfizer, United StatesCopyright © 2021 Solé and Santamaria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pere Santamaria, cHNhbnRhbWFAdWNhbGdhcnkuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.