
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 13 May 2021
Sec. Mucosal Immunity
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.683022
This article is part of the Research TopicImmunoregulation at Mucosal SurfacesView all 36 articles
The innate immune system is the oldest protection strategy that is conserved across all organisms. Although having an unspecific action, it is the first and fastest defense mechanism against pathogens. Development of predominantly the adaptive immune system takes place after birth. However, some key components of the innate immune system evolve during the prenatal period of life, which endows the newborn with the ability to mount an immune response against pathogenic invaders directly after birth. Undoubtedly, the crosstalk between maternal immune cells, antibodies, dietary antigens, and microbial metabolites originating from the maternal microbiota are the key players in preparing the neonate’s immunity to the outer world. Birth represents the biggest substantial environmental change in life, where the newborn leaves the protective amniotic sac and is exposed for the first time to a countless variety of microbes. Colonization of all body surfaces commences, including skin, lung, and gastrointestinal tract, leading to the establishment of the commensal microbiota and the maturation of the newborn immune system, and hence lifelong health. Pregnancy, birth, and the consumption of breast milk shape the immune development in coordination with maternal and newborn microbiota. Discrepancies in these fine-tuned microbiota interactions during each developmental stage can have long-term effects on disease susceptibility, such as metabolic syndrome, childhood asthma, or autoimmune type 1 diabetes. In this review, we will give an overview of the recent studies by discussing the multifaceted emergence of the newborn innate immune development in line with the importance of maternal and early life microbiota exposure and breast milk intake.
Our understanding of how microbial communities can influence health and disease of their host has significantly improved in the last decades. This boost of scientific discoveries in the microbiome field has been facilitated mainly by the availability and affordability of different techniques, such as gnotobiology, next-generation sequencing (NGS), and metatranscriptomics. Evidence linking the microbiome with the pathophysiology of diseases, for instance, such as inflammatory bowel diseases (IBD) (1–3), cancer (4, 5), obesity (6, 7), type 2 diabetes (8), and neurological disorders (9, 10) is increasing and future research in the field will in-depth dissect how the microbiota and changes in its composition lead to multiple effects in the host.
Birth marks the start of colonization with microbial communities. Many factors are known to influence the microbiota composition in these early days, e.g. birth mode (11), antibiotic treatment during pregnancy (12, 13), or infancy (14, 15), maternal diet (16), breast- or formula feeding (17, 18), and the introduction to solid food (19), while the host genetic background is estimated to only shape about 9% of the intestinal microbiota (20, 21). The microbiome of an infant in its first three years of life is clearly distinguishable from an adult microbiome by a lower diversity index reflected in half as many operational taxonomic units (OTUs) compared to adults and higher interindividual variability (22, 23). Dominant bacterial taxa in the first weeks of life of a newborn include Enterococcacae, Clostridiaceae, Lactobacillaceae, Bifidobacteriaceae, and Streptococcaceae. In the first months of life, Bifidobacteriaceae thrive since they feed on oligosaccharides, which are highly abundant in maternal milk, the main energy source of newborn babies. During weaning, when solid food is introduced, the abundance of Bifidobacteriaceae decreases, while Bacteroides, Ruminococcus, and Clostridium become more prevalent (Figure 1) (24).
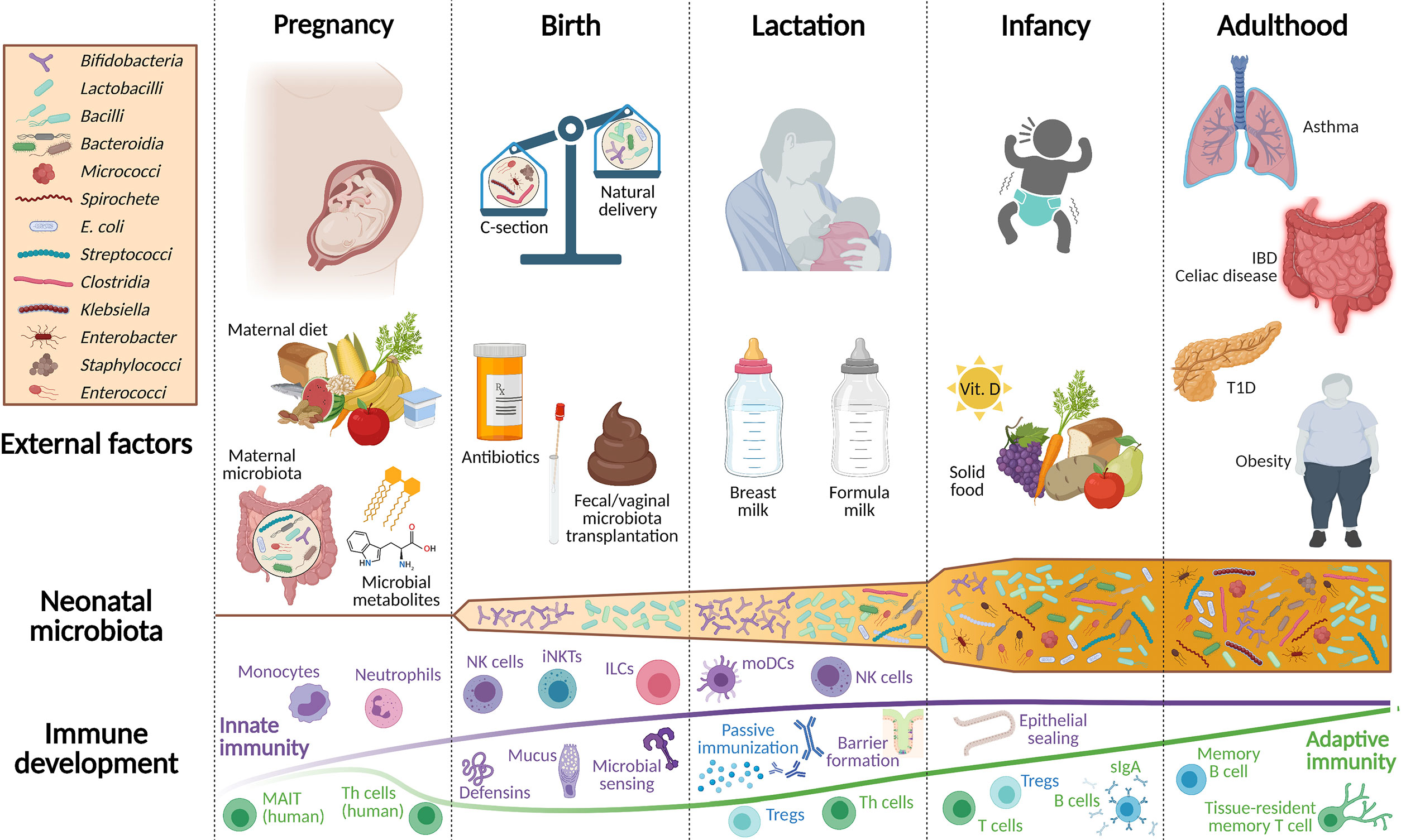
Figure 1 Overview of environmental factors shaping the development of the newborn microbiota and mucosal immune system. Throughout pregnancy, fetal immune development is supported by microbial metabolites originating from the maternal microbiota and by dietary compounds. Innate immune cell populations, such as monocytes, ILCs, and neutrophils belong to the most affected immune cells at this stage. Only at birth, the emerging immune system of the newborn is confronted with live bacteria and thus, is still dependent on maternal protection, which is ensured through breastfeeding. Apart from passive immunization via breast milk, neonatal iNKTs, NK cells, ILCs, and the gastrointestinal epithelial barrier protect against invading pathogens and promote the beneficial interplay with the neonatal microbiota. The adaptive immune system in mice develops primarily postnatally, while a component of adaptive immunity is already present in the human fetus. Birth mode, feeding, and the intake of antibiotics are additional factors that shape the early life microbiota and the neonatal immune system. During the subsequent weaning reaction, when solid food is introduced to the infant’s diet, a tremendous shift occurs in its intestinal bacterial community composition. Consequently, the microbiota is no longer represented by Bifidobacteria and Lactobacilli, but increases in metabolomic diversity evolving to a more adult-like microbiota that is established during this early period of life. This period is often also called the window of opportunity since particularly during this time, external cues have a profound impact on life-long health, the evolving microbiota, and the mucosal immune system. IBD, inflammatory bowel disease; sIgA, secretory immunoglobulin A; ILCs, innate lymphoid cells; iNKTs, invariant natural killer T cells; MAIT, mucosal-associated invariant T cells; moDCs, monocyte-derived dendritic cells; NK, natural killer cells; Th cells, T helper cells; Treg, regulatory T cells; T1D, type 1 diabetes.
The enormous impact of the microbiota on the development of the immune system was in the spotlight early on. Pioneers like René Dubos, Russell Schaedler, and Dwayne Savage have revealed the importance of the gastrointestinal microbiota and its interaction with the host immune system (25–28). Here, we want to guide through the manifold changes that occur in the microbiota during early life and how this leads to a temporally layered postnatal establishment of the intestinal immune system, starting from gestation, via birth, followed by lactation, and the first years of life (Figure 1). A strong focus has been put on reviewing literature on innate immune development.
The first line of defense against invaders in the gastrointestinal tract includes the mucus layer, the intestinal epithelial cell layer, and hematopoietic immune cells, either scattered throughout the lamina propria or settled as intraepithelial lymphocytes, all of which extensively interact with the microbiota as well as with microbial and diet-derived metabolites.
Mucins build a physical wall that separates the host tissue from the microbial community in the lumen. The large, highly O-glycosylated mucin proteins are either covering the apical surface of enterocytes or secreted into the lumen by goblet cells as gel-forming mucus (Figure 2). The small intestine has a single unattached mucus layer, whereas in the colon and the stomach, two layers exist. There, the inner layer is attached to the epithelium, while the outer layer is unattached and less dense. The mucus fulfills different important functions: (1) It protects against the invasion of pathogens, (2) builds a physical barrier between microbial consortia and the host tissue, (3) protects against self-digestion (for example in the highly acidic environment of the stomach), and (4) directly modulates the expansion of different bacterial strains and the composition of the whole community (Figure 2). Mucins do not only protect from too close contact to the commensal consortia but also serve as a nutrient source for some bacteria that possess glycosidases; These glycosidases enable bacteria to cleave mucin 2 (MUC2) proteins and use them as an energy source. In turn, bacteria release short-chain fatty acids (SCFAs) that are beneficial for the host (29–31). The mucus is bathed with defensins, different antimicrobial peptides (AMPs) originating from Paneth cells, and secretory immunoglobulin type A (sIgA) deriving from plasma cells in the lamina propria. On the other hand, the microbiota affects mucus property and function (32, 33).
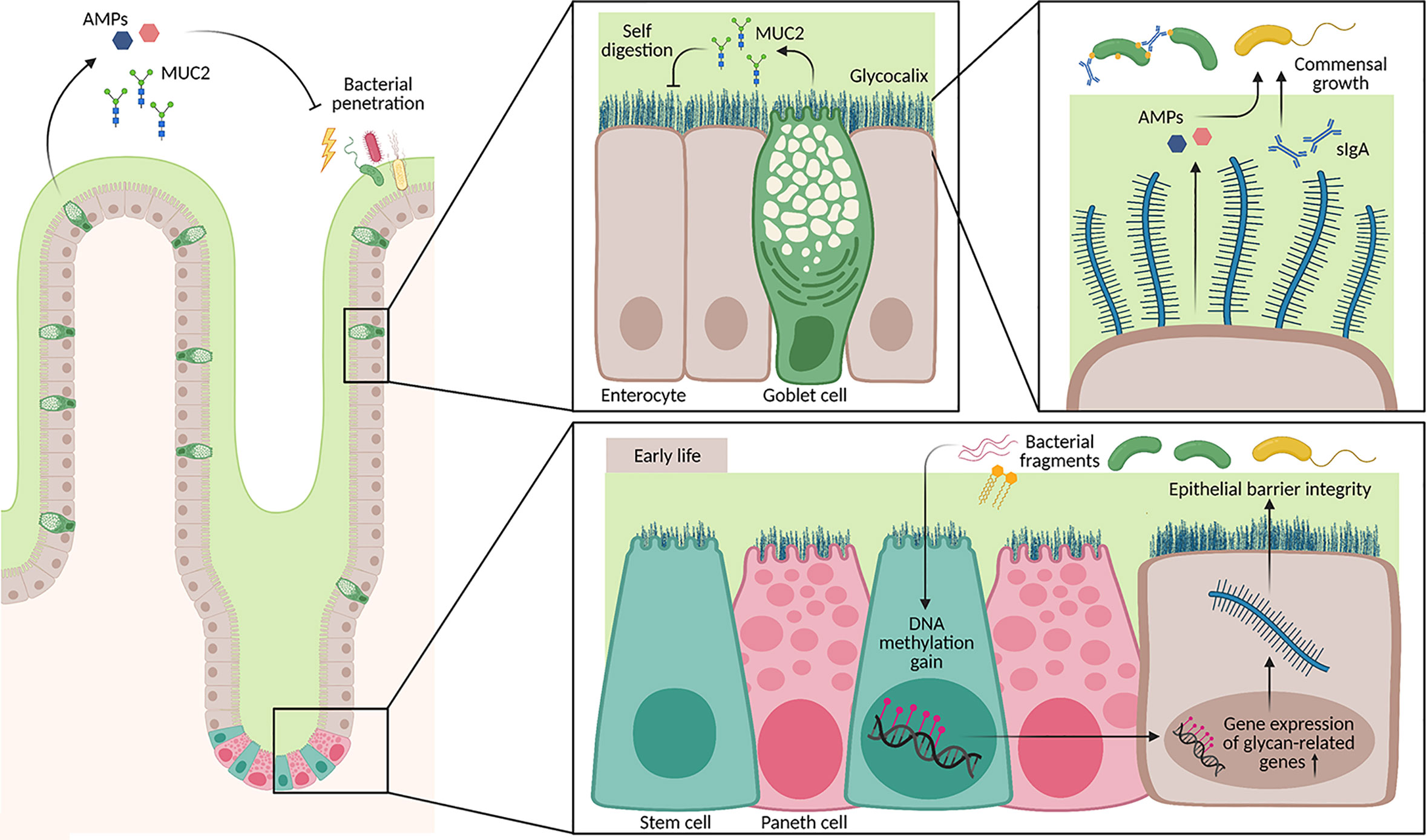
Figure 2 Architecture of the mucus layer in the small intestine. The small intestine has a loose mucus layer (illustrated in green), which keeps commensals at distance. In addition, the epithelium is covered by a glycocalyx, a dense layer composed of secreted mucus proteins (MUC2) that is attached to epithelial cells via the transmembrane part. These layers do not only protect from bacterial penetration, but also from self-digestion of host intestinal tissue. Secretion of AMPs by Paneth cells and other epithelial cells as well as sIgA by plasma cells regulate the growth of different commensal bacterial strains. Furthermore, signals from the neonatal microbiota shape the DNA methylome in intestinal stem cells from birth until weaning. Genes associated with cell glycosylation are particularly affected by a DNA methylation gain, which also correlates with an increase in gene expression. Hence, barrier integrity during early life is additionally ensured through epigenetic remodeling triggered by the microbiota.
The mucosal immune system further comprises the single epithelial cell layer, the underlying lamina propria, and the organized lymphoid structures, such as Peyer’s Patches, isolated lymphoid follicles, and mesenteric lymph nodes (MLNs). Different hematopoietic cells of the innate immune system can be found in the mucosa including mononuclear phagocytes, intestinal macrophages, intestinal dendritic cells (DCs), eosinophils, mast cells, and innate lymphoid cells (ILCs) (Figure 1). They sense the presence of microbes through pattern recognition receptors (PRRs) both in the context of an infection with a pathogen and under homeostatic conditions. Many such PRRs and their signaling pathways have been identified in the last decades, including Toll-like receptors (TLRs), C-type lectins (CTLs), nucleotide-binding oligomerization (NOD)-like receptors (NLRs), RIG-I-like receptors (RLRs), and others. These PRRs bind to pathogen-associated molecular patterns (PAMPs) for example lipopolysaccharides (LPS), an outer cell wall component of gram-negative bacteria (Figure 3).
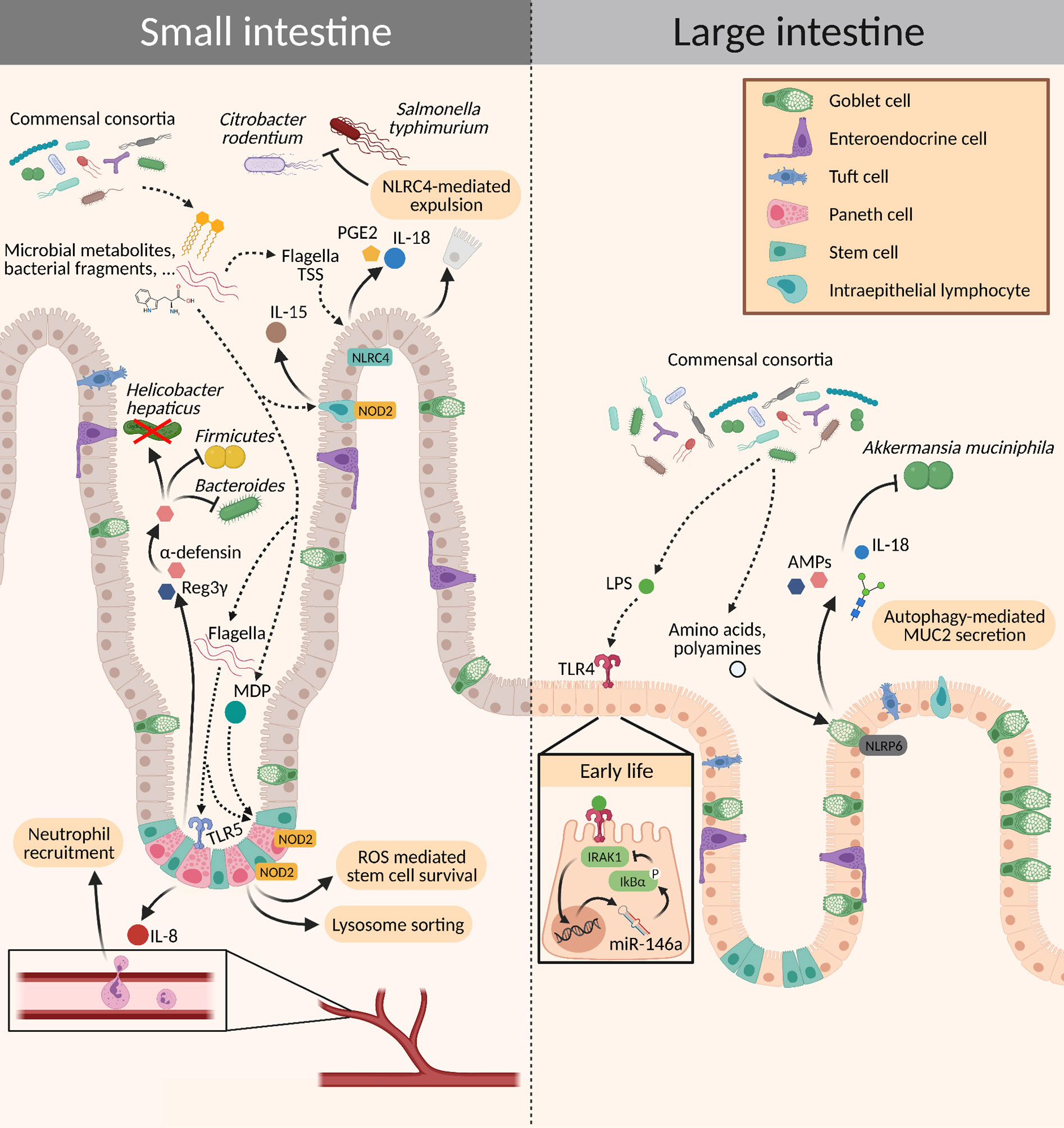
Figure 3 Crosstalk between the microbiota and intestinal epithelial cells. Different signaling molecules from the microbiota bind to PRRs expressed on the epithelium, which subsequently activate innate immune mechanisms. This activation can elicit a wide range of effects: It may trigger a pro-inflammatory state for the elimination of pathogens or induce tolerance to commensals by increasing the production of mucins and AMPs, promoting epithelial cell turnover, and mediating stem cell survival. AMPs, antimicrobial peptides; MDP, muramyl dipeptide; PGE2, prostaglandin E2; ROS, reactive oxygen species; TSS, type secretion system.
First hints of the possible existence of a “window of opportunity” in early life during which environmental influences can have long-lasting effects on microbiota composition, immune regulation, and disease susceptibility of the host, came from epidemiological studies. Scientists observed a positive correlation between higher hygiene standards in industrialized countries and a rising incidence of autoimmune and allergic diseases (34, 35), which was followed by studies that suggested a reduced exposure to a microbial-rich environment as a possible cause. For example, growing up on a farm, being representative for a microbial-rich environment, prevents the development of allergic asthma (36–39) and this effect has been shown to be long-lasting (40). Furthermore, optimal nutrition is pivotal for both the pregnant mother and her unborn child, as perturbations in this critical time pose the fetus at risk of later developing a plethora of chronic disorders, such as metabolic syndrome, type 2 diabetes, coronary heart disease, adiposity, and osteoporosis (41–44). Until now, therapeutic interventions – for example via fecal transplantation – were of limited success, one reason why much effort is taken to further dissect the causal mechanisms of how such early events can lead to later disease onset. The microbiota certainly is a key player during this window of opportunity. Early colonization during this critical time window by a microbial consortium is crucial for the proper development of the immune system and has been demonstrated in various mouse models (14, 45–48). It seems that the window of opportunity in mice closes around the time of weaning. In humans, we know that a stable microbiota composition is established at the age of 2-3 years. However, no clear data are yet available to assess the exact time when the window of opportunity closes. While the immune system of the child predominantly matures in the first few years after birth, it is still strongly shaped later during childhood. Due to the distinct needs at different stages in life, the structural organization of various immune layers at the intestine has been identified. In neonatal mice, there is no established crypt-villous axis, and their epithelium is characterized by a lower turnover compared to adult mice (48–50). Additionally, there are almost no mature Paneth cells present in neonatal mice and the mucus layer is much thinner – it is only around the time of weaning when a reliable mucus shield against invaders is established (48). Further, PRRs are expressed in an age-dependent manner. For example, TLR4 expression is increased in the prenatal period and decreases at term, whereas TLR9 expression is reduced during gestation and increases after birth. This pattern of expression for TLR4 and TLR9 is inversed in tissue from infants suffering from necrotizing enterocolitis (NEC) and might be of pathophysiological relevance (51). TLR3 expression is low at birth and increases during the postnatal period (52). Sensing of LPS in the neonatal intestine via TLR4 leads to expression of the microRNA miR-146a which maintains phosphorylation of IκBα, subsequently inhibiting IRAK expression and resulting in LPS tolerance (53, 54) (Figure 3). One could hypothesize that age-dependent expression of PRRs may represent a strategy of the host to push colonization of the mucosal sites towards beneficial commensals and it is possible that altering expression levels of different PRRs in a timely manner helps to fine-tune age-dependent requirements of the host, e.g., in terms of nutrients provided by certain bacteria.
What has started as epidemiological correlation-based observations has developed into in-depth experimental research. These scientific investigations have revealed mechanistic insights into how events during this critical window affect long-term health of the host. The window of opportunity not only opens after birth but already during pregnancy, having manifold effects on the developing fetus, in ways that we will discuss next.
Adaptations of the female body during pregnancy are remarkable, affecting all organ systems and including the development of the placenta, a highly specialized organ that provides an anatomical separation of the fetus and the mother for preventing immunogenicity of the mother against the fetus and vice versa. At the same time, this complex organ ensures the maternofetal exchange of molecules, including those originating from the maternal microbiota (55). In recent years, much attention has been drawn to the possible existence of a placental microbiome itself.
Whether the placenta harbors a microbial community is a matter of debate and has started when a study from the Versalovic group in 2014 has challenged the paradigm of a sterile womb. They performed 16S sequencing on human placental samples and detected a microbial community (56). Before that, others have found bacteria in the human placenta during term (57, 58) and preterm deliveries (58). Bacteria have also been identified from human umbilical cord blood (59), meconium (60), and amniotic fluid (57). Additionally, genetically labeled E. faecium was administered orally to pregnant mice and subsequently isolated from the culture of amniotic fluid (59) and meconium (60), many studies followed claiming the existence of a placental microbiome (61–64).
Immediately after the Versalovic group published their paper in 2014, Harvey Kliman pointed out that the sole detection of DNA does not provide evidence for the existence of living microbes (65). Over time, it became more and more obvious that contamination issues (66, 67) and the Test-Kit’s own “microbiome”, the so-called “kitome” (68), represent big challenges in the search for a microbiota inhabiting the placenta. Subsequently, several groups had a closer look and performed even more careful evaluations (1) by adding controls at every step of the process, (2) only including samples of cesarian (C)- section-derived tissue to reduce the risk for contamination during the birth process, (3) combining high-throughput sequencing with qPCR and bacterial culture, (4) comparing the bacterial taxa from those found in the close environment (e.g. the processing room), and (5) subtracting the taxa that overlapped with the kitome. Scientists were not able to detect a placental microbiome by including the above-mentioned precautions (68–73). However, the issue does not seem settled, as a recently published paper claims to have detected bacterial DNA and viable bacteria in the fetal intestine by using 16S rRNA gene sequencing, qPCR, electron microscopy, and bacterial culture (74), which re-sparked the controversy whether a placental microbiome exists (74–76).
From a biological perspective, we doubt the presence of an established microbial community in the placenta (55, 77). As elegantly reasoned by Walter & Hornef, «multi-layered contextual evidence» has not been taken into account by proponents of the in utero colonization hypothesis, but most studies have focused on sequencing techniques only (78). They further emphasized that there is no overlap between the bacterial taxa detected in utero in the different sequencing studies but almost a congruence between the bacterial taxa identified in utero and the controls (78).
Even though the existence of a placental microbiota is questionable, the transport of commensal bacteria-derived metabolites via the placental barrier is well established. The gut maternal microbiota plays an important role in this maternofetal molecular transfer and is thereby able to modulate fetal development (79). Our group showed that reversible colonization of germ-free females with a genetically modified E. coli strain (80) during pregnancy induced distinct changes in the intestinal innate immune system of the offspring (77). This was dependent on the transfer of maternal microbiota-derived aryl hydrocarbon receptor (AhR) ligands (81), which stimulated the proliferation of innate lymphoid cells type 3 (ILC3s) (77). ILC3s are crucial for maintaining the gut epithelial barrier and host defense by the production of IL-22 and IL-17 and subsequently inducing the secretion of AMPs (82, 83). Moreover, the expression of genes involved in epithelial cell differentiation, integrity, and homeostasis was altered in small intestinal epithelial cells of the offspring born to mothers who had experienced reversible colonization during pregnancy (77). Our group recently reviewed how manifold metabolites processed by the maternal microbiota can reach the fetus and affect its development and physiology (55).
Antibiotics and other drugs can indirectly influence the maternal microbiota-fetus crosstalk, by altering the microbiota composition and subsequently the metabolite-pool derived from the microbiota. Perinatal antibiotic exposure reduced Nrp-1-RORγt-Foxp3+ regulatory T cells (Tregs) in the offspring and was irreversible after weaning (84). Another study found lower levels of IL-17 and granulocyte colony-stimulating factor (G-CSF) in the intestine of antibiotic-treated dams. Their neonates exhibited decreased numbers of circulating and bone marrow neutrophils as well as granulocyte/macrophage-restricted progenitor cells in the bone marrow (82). However, the offspring of antibiotic-treated non-obese diabetic dams were protected against the development of type 1 diabetes through mechanisms of alteration in the microbiota composition and induction of tolerogenic antigen-presenting cells (APCs) (85, 86). In humans, a population-based Danish cohort study found a positive correlation between antibiotic exposure during pregnancy and a risk for severe infections in children (87). Antibiotic treatment during pregnancy was further associated with an increased risk for very early onset of IBD in the offspring (88).
An important side note is the observation that a healthy pregnancy leads to changes in the microbiota composition that resembles a dysbiotic composition – however, in the context of pregnancy with its unique needs and requirements, these adaptations are physiological (89, 90). For example, Faecalibacterium, which is a SCFA producer, decreases in abundance in the last trimester of pregnancy. This decline of Faecalibacterium has also been observed in populations with metabolic syndrome (91). Overall, pregnancy is associated with a decrease in microbial diversity and richness, and an increase in bacterial load with an expansion in Proteobacteria and Actinobacteria (92, 93). This shift in the microbiota of pregnant women is subjected to adjustments in dietary habits, which are accompanied by changes in the pool of bacterial metabolites to fully support the development of the fetal immune system. A fiber-rich diet during pregnancy protected the offspring against the onset of asthma, probably via inhibition of histone deacetylase 9 (HDAC9) mediated by acetate resulting in higher gene transcription of Foxp3 in Tregs. They further lowered frequencies of eosinophils and macrophages in the blood and bronchoalveolar lavage fluid as well as serum IgE levels of the offspring (94).
1,25-dihydroxyvitamin D3 plays an important role in epithelial barrier integrity. Mice with vitamin D deficiency and C. rodentium challenge demonstrated increased colonic hyperplasia and epithelial barrier dysfunction (95). Malnourished pregnant mothers, specifically in 1,25-dihydroxyvitamin D3, might be at higher risk for developing intestinal infections, which poses a substantial risk to the unborn child. Another study found that lymphocytes isolated from the cord blood downregulated TLR1, -2, -4, -6, and -9 upon supplementation with high doses of 1,25-dihydroxyvitamin D3 during pregnancy (96). In mice, maternal dietary-derived retinoic acid, the active form of vitamin A, influences secondary lymphoid development in the offspring as lymphoid tissue inducer (LTi) cells, a subset of ILC3s (97). Mechanistic insights on a molecular level for the relation between prenatal nutrition and intrauterine immune development come from a study investigating metastable alleles in a Gambian human population with seasonal variations in food supply. A metastable allele, VTRNA2-1, was differentially methylated between offspring from mothers, which were either at conception when food was available in adequate or insufficient amounts. Strikingly, VTRNA2-1 has been identified to play a role in viral immunity (98), which reflects common observations linking undernutrition with higher infection rates.
While the mentioned examples illustrate that the maternal microbiota can affect the development of the offspring’s immune system already in utero, additional studies are needed in the future to better understand this crosstalk.
When it comes to the development and maturation of the newborn’s immune system to guarantee lifelong health, the immediate period after birth is as important as the gestational stage. Although several key steps regarding innate immune development take place already in utero, many others require postnatal antigen exposure to evolve. NK cells and ILCs are present already at birth and subsequently expand and even reach higher frequencies than in adulthood. This ensures that the newborn is prepared against immediate threads and protected against infections early in life (99–103). Even though the neonate’s innate immune system is capable to mount an immediate response against potential pathogens right after birth, it still has to mature in coordination with the microbiota and many other environmental factors (48, 104–106). In contrast, the murine adaptive immune system develops predominately postnatally. This is not completely transferrable to the human organism. Data are available that demonstrate the presence of adaptive immune cells in the developing fetus. Teichmann and colleagues detected thymic T cells in the fetus as early as 7 weeks post conception (107), while effector CD4+ T cells were described in the intestine of the developing fetus during the second trimester of pregnancy (108). In addition, mucosal-associated invariant T (MAIT) cells are already present during in utero development of humans and can protect the newborn baby from infections (109).
The event of birth represents the change from the sterile environment in utero to the rapid colonization of all body surfaces. For a vaginally born baby, this is initiated by vertical transmission of microbes when passing the birth canal and primarily includes microbes inhabiting the maternal gut lumen (110). A thorough metagenomic shotgun sequencing analysis of fecal samples collected at different timepoints from full-term infants during their first year of life points out the dynamics and importance of the microbiota early in life. As previously mentioned, the complexity of the gut microbiota increases during the first year after birth (Figure 1). Simultaneously, the composition of the gut microbiota progressively resembles the maternal gut microbiota and ultimately develops into the adult gut microbiota (13, 23, 111).
This process of early life colonization can be perturbed by external factors, for example, when a baby is delivered by C-section. Babies born via surgical delivery share around 30% less bacterial species with their mother than naturally born babies, indicating different sources of gut colonizers. Indeed, infants born via C-section harbor an increased number of species usually colonizing the skin (Staph. saprophyticus) or circulating in the hospital (Enterococcus faecalis, Enterobacter cloacae, Klebsiella pneumoniae, and Clostridium perfringens) (13, 112). Most often, acquired from the hospital environment, these strains are opportunistic pathogens, also relevant in nosocomial infections and harboring antimicrobial resistance genes (112). At the phylum level, the microbiota of babies born via C-section is dominated by Firmicutes and Proteobacteria, with a shift to fewer Bacteriodetes and Actinobacteria (113). Additionally to the missing passage through the birth canal, also the use of antibiotics, which is the first-line pharmacological therapy intrapartum, disturbs the microbial colonization of the neonate at birth. Within the first year of life, the microbiota of C-section babies is able to recover but may also persist for longer (13, 111, 113–116). Therefore, it is still highly debated whether surgical delivery has life-long consequences. A comprehensive study performed in Denmark associated a multitude of inflammatory diseases to C-section delivery. Over 2.5 million candidates were followed from birth up to 40 years of age. Indeed, participants born via C-section were at a higher risk to develop diabetes, arthritis, celiac disease, or IBD. Nonetheless, a correlation between a distinct microbiota early in life as a consequence of cesarean section and the onset of immune-mediated etiologies later in life could not be disentangled (117). In a recently published study, Stockholm and colleagues addressed this relationship (110). They could demonstrate that infants with a long-term C-section-associated microbiota composition suffer from a higher susceptibility to childhood asthma or an increased risk of allergic sensitization marked by high IgE. Children who retained a C-section microbiota profile at the age of one year also mounted a different immune response during episodes of acute airway symptoms, determined by lower levels of immune mediators, such as TNF-α, IL-4, IL-13, or IL-1β. It is not yet clear, which changes specifically in the gut microbiota are the key factors driving these allergic phenotypes (113). In contrast, children born via C-section, who could normalize their microbial colonization pattern during their first year of life, were not affected by either a higher risk for childhood asthma or allergic sensitization early in life. Hence, other impacts on gut microbial composition, such as the contact with the maternal microbiota from body sites other than the gut or having older siblings in the family might play a pivotal role in maturing the neonatal microbiota towards a less sensitizing composition (110, 113, 118, 119).
Since the immune system is largely evolving during this window of opportunity, a C-section-associated composition could interfere with the healthy development of the immune system and conclusively explain the threefold risk to develop childhood asthma or other immune-related disorders after cesarean delivery (113, 117). Specifically, the development of the regulatory immune system is affected by the birth mode, as shown in several murine studies. Mice born via C-section had stunted levels of Tregs in the spleen and MLNs and reduced systemic IL-10 levels until adulthood (120, 121). On the other hand, the proportion of invariant natural killer T cells (iNKT) was increased in the colon, as well as their expression of Il4 and Il15. A prebiotic diet was able to reestablish reduced numbers of iNKTs similarly to vaginally delivered mice. However, the levels of Tregs were unaffected and remained low (121). The causative effect of high iNKTs and the early life microbial colonization pattern after C-section in humans has yet to be elucidated. Notably, normal iNKT cell levels in the colonic lamina propria could only be restored when germ-free mice were colonized in the first few weeks of life but not at later time points (46). Additionally, germ-free mice and mice delivered via C-section were more susceptible to iNKT cell-mediated oxazolone-induced colitis, a murine model for IBD (46, 122).
The practice of C-section, specifically without any urgent medical indication, is increasing worldwide, and in wealthy countries even with a skyrocketing prevalence (123). Hence, it is also of interest to overcome the imbalanced mother-to-neonate transmission after surgical delivery. In a proof-of-concept safety study, Korpela and colleagues orally transplanted fecal microbiota, collected from a total of 7 mothers, to their C-section-born neonates. Fecal microbiota transplantation (FMT) could shift the gut microbiota towards a vaginally born colonization pattern, including the restoration of a healthy Bacteroides level (124). Additionally, FMT alleviated the abundance of opportunistic pathogens characteristic of a C-section microbiota (112, 124). Even though the proof-of-concept safety study displayed benefits for FMT after cesarean delivery, such practices should be regarded with caution. The mother could harbor potential pathogens or viruses, with which her immune system is able to cope, but not the immature immune system of the newborn. Therefore, thorough microbiota profiling beforehand is crucial. In the mentioned study, for example, only 7 out of 17 mothers were selected after careful examination (124). A different approach for microbiota restoration in newborns is vaginal microbiota transfer at birth. Nevertheless, the major source for neonatal gut colonizers is the maternal intestinal microbiota, hence, not surprisingly vaginal swabbing at birth was unable to durably establish a microbiota similar to vaginally born babies (110, 124). These findings stand in contrast with the conclusions drawn by Dominguez and colleagues. Exposure of newborns with vaginal fluids at birth, exhibited a vaginal microbiome-like signature during the first week of life, which was similar to vaginally born babies and to the vaginal microbiota of the mother (125). Hence, even though the effects of C-section on the development of the newborn microbiota and its immune maturation are well established, the options to adjust the imbalance during the window of opportunity and particularly its consequences on long-term microbiota composition and human health are still a matter of debate and extensive research.
The postnatal maturation of the immune system is highly sensitive to environmental factors. Preterm babies are particularly weak and complications at this stage are still a major cause of neonatal death (126). Preterm babies often need to undergo empirical antibiotic treatment in the neonatal intensive care unit (NICU), leading to nosocomial late-onset sepsis (LOS), which can appear 3 days after birth or later (127, 128). Empirical antibiotic treatments include generally broad-spectrum antibiotics, including Vancomycin and thus, the increasing prevalence of antimicrobial-resistant pathogens, such as the multidrug-resistant Staphylococcus capitis clone (NRCS-A) is alarming. Not only because NRCS-A is exceptionally disseminating in NICUs, but also due to its probable relevance in LOS pathogenesis (129, 130). Approaches, such as the MinION seq platform, aim to rapidly (in less than 5 h) identify the pathogenic species and their antimicrobial-resistant genes to reduce broad-spectrum antibiotic treatment early in life and specifically target LOS in the newborn patient (131). Neonatal antibiotic treatment impedes the building microbiota and may set the basis for pathological colonization and finally LOS. Likewise, the maturation of the immune system is impaired due to its indispensable relationship with the microbiota. In a murine model, Niu et al. could unravel the effects of antibiotic use and LOS on the innate immune system (83). Specific pathogen-free (SPF) dams were treated with antibiotics either shortly (3 d) or for a prolonged period (7 d) while they were nursing their neonates, resulting in antibiotic exposure of the neonates through their mother. This experimental design mimics the situation of preterm babies receiving empirical antibiotic treatments resulting in a stunted microbiota. Both, in short, and in prolonged antibiotic exposure and similar to human babies born via C-section, Proteobacteria expanded persistently, whereas Enterobacter and Enterococcus species could only translocate to the spleen and the liver after long-term antibiotic exposure. Due to this shift in neonatal microbiota, K. pneumoniae was able to colonize the neonatal gut and additionally translocate systemically, resulting in sepsis. Reduced bacterial signals diminished epithelial TLR2 and TLR4 gene expression, which commonly drive ILC3 expansion. Ultimately, long-term antibiotics also diminished the ILC3 population in the lamina propria. Since microbiota restoration reestablished ILC3s and rescued the antibiotic-treated pups from sepsis, this innate immune population contributes to the important protection of the neonate after birth. However, changes in the abundance of ILC3s through neonatal antibiotic exposure by treating the dams might also be explained by changes in the maternal microbiota or breast milk itself (83). The importance of maternal microbiota-induced intestinal ILC3s in regulating early life colonization was also demonstrated by our group as discussed earlier (77).
Vitamin D deficiency during the first years after birth was previously associated with an increased risk to develop asthma, eczema, or atopic sensitization during childhood (132, 133). One of the determinants of risk for childhood asthma development might be early nasopharyngeal colonization with pathogenic Streptococcus species (132). The immunological mechanism behind, most probably involves the development of tolerogenic DCs and Tregs after repeated exposure to aeroallergens, which affects normal maturation of pulmonary function, followed by cumulative airway tissue damage (134, 135). Under healthy conditions, CD11b+ migratory DCs are sensitized after house dust mite and microbial LPS exposure, which prompts them to dampen type 2 T helper (Th2) cell differentiation through TNF-α signaling. This process is well-developed in adult mice, in contrast to younger mice, where the threshold for sensitization is decreased. Thus, young mice are more prone to develop asthmatic inflammation due to the weakened response of DCs towards microbial low-dose LPS stimuli, which then insufficiently suppress Th2 cell development (136). Latest studies report altered differentiation and activation of several T cell subsets, including Th1 and Th2 cells after oral vitamin D supplementation or UV exposure in neonates. Especially the differentiation of naïve CD4+ T cells into Th2 cells was reduced by lower IL-2 production (137, 138).
Apart from vitamin D deficiency, childhood asthma has also been associated with respiratory viral infections early in life. Therefore, vitamin D supplementation could alleviate asthma risk by reducing viral infections, explained through distinct neonatal IFN-γ production and abnormal neutrophil responses (139, 140). A study performed in Vietnam confirmed that an 8-month supplementation of vitamin D during infancy diminished the frequency of respiratory infections caused by non-influenza viruses, such as rhinoviruses. However, the influenza infection rate remained unaffected by vitamin D administration (141). Collectively, neonatal vitamin D is crucial for the healthy development of the respiratory immune system and notably to improve the communication between respiratory microbial signals, the innate and the adaptive immune system with life-long consequences on asthma susceptibility (133, 136, 137).
Gut commensals play a key role in processing indigestible food components to provide essential vitamins and SCFAs to the host. The period when solid food is gradually introduced coincides with a burst in microbial changes and immune regulatory processes, also known as the weaning reaction. In case of an absent weaning reaction, pathological imprinting occurs, having life-long consequences on host allergy and cancer susceptibility (19). The weaning reaction is dependent on the microbiota and additionally on a specific time window since the reconstitution of germ-free mice at later stages could not rescue from pathological imprinting in the small intestine (19). During a preweaning interval, goblet cell-associated antigen passages (GAPs) are formed and specifically deliver antigens from epithelium-adhering bacteria to the colonic lamina propria. These encounters prime Foxp3+ Tregs to protect the neonate from dextran sulfate sodium (DSS)-induced colitis later in life. Essentially, blocking GAPs before weaning inhibited Treg development in the colonic lamina propria and resulted in an impaired establishment of early life bacterial-induced tolerance (142). The microbiota produces SCFAs from dietary fibers, which promote the expansion of RORγt+ Tregs in the small intestine and thereby prevent pathological imprinting. Nevertheless, SCFAs alone were not able to induce Tregs in germ-free mice, nor was exclusive bacterial colonization. Hence, additional signals are required and we will display later in this review that the weaning reaction is not solely happening because of the digestion of dietary fibers by gut microbes, but that it depends further on components in the breast milk, which delay the weaning reaction (19).
Dietary fibers are found in plant-derived food. Apart from vegetables and fruits, also barley, rye, and wheat are vital fiber sources. These different grain varieties contain gluten, an important food antigen, which can cause celiac disease in genetically predisposed individuals. Since not all individuals with the celiac-relevant HLA antigen genotype develop celiac disease, environmental factors seem to play an essential role. One currently discussed environmental aspect is the effect of early life gluten intake because the disease often manifests during infancy (143–145). Several studies highlight the association of extensive gluten intake during childhood with an increased risk of developing celiac disease if genetically predisposed (146–148). It is hypothesized that shifts in the microbiota composition during the first year of life precede the disease pathology. These include high abundance of Bifidobacterium breve and Enterococcus spp., whereas increases in Firmicutes and Bifidobacterium longum are correlated with a reduced risk. Even though the overall changes in the commensal community were minute, children who later developed celiac disease exhibited a premature microbial diversity and complexity, followed by increased IL-6, reduced sIgA, and TNF-α levels in the feces (149). However, the cause for the microbial shift and its consequences on immune regulation and response to the gluten antigen could again be attributed to different cues in utero or after birth, such as the feeding practice (150).
Celiac disease onset as a result of early life development summarizes the importance of a balanced interplay between food intake, gut microbial metabolism, and mucosal immune function, which altogether maintain gastrointestinal epithelial barrier integrity, an innate immune mechanism we will highlight in the next section (149–152).
The gastrointestinal epithelium acts as a physical barrier to prevent translocation of pathogens, but it also senses microbial antigens to maintain host-microbial mutualism (153). The oral epithelium is a multilayered, stratified epithelium similar to the cutaneous epithelium, whereas the lower parts of the intestine are aligned with a single layer of cells. Therefore, the epithelial establishment and its crosstalk with immune cells and the microbiota evolve to some extent differently in the oral cavity, the small intestine, and the colon (49, 50, 154, 155).
The oral cavity belongs to the earliest microbially exposed body surfaces. Nevertheless, little is known about the development of its mucosal immune system. Only recently, light was shed on the mutual interplay between microbiota development and local immunity. In contrast to the intestine, neutrophils are highly abundant in the neonatal oral mucosa and due to the increased permeability of the oral epithelium before weaning, they play an important role in the first line of defense against the high microbial load acquired at birth. Upon microbial stimulation, γδ T cells produce IL-17 and induce the recruitment of prenatally established neutrophils. At weaning, these γδ T cells diminish and simultaneously the oral epithelial barrier strengthens by reducing its permeability and upregulating saliva production. With the higher levels of salivary AMPs, the microbial load decreases and the oral epithelium finally matures, having a greater turnover and a reduced expression of microbial recognition receptors, including TLR2 and TLR4, and lower expression of antimicrobial defensins. Conclusively, the early defense mechanisms of the innate immune system against the oral microbiota enable the development of a tolerogenic oral epithelium, which during the neonatal phase is most vulnerable (49).
In the lower intestine, where microbial density increases, luminal metabolites change dramatically during weaning. This newly introduced metabolome not only affects the development of the immune system but also its regulation through epithelial cells by strengthening the physical barrier and supporting epithelial maturation to an absorptive phenotype (19, 156). Identical to the oral cavity, the small intestinal and cecal epithelia reduce the transcription of TLRs and AMPs at the suckling-to-weaning transition (156–158). A recent study by Hornef and colleagues found an age-dependent expression of TLR5, where relative expression was 200-fold higher in newborns compared to adult mice. Importantly, this difference in TLR5 expression was specific to the intestinal epithelium and had a lifelong impact on microbial composition in the gut (158). Specifically, stimulation of Reg3γ production mediated by TLR5 controlled counter-selection of flagellated bacteria and thereby modulated the intestinal microbiota until adulthood (Figure 3).
Apart from microbial sensing, the microbiota shapes the functionality and performance of the intestinal epithelial stem cell niche during development. Bacterial exposure at weaning affected gene expression of the erythroid differentiation regulator-1 (Erdr1), which is important for stem cell proliferation and regeneration after epithelial damage in the neonatal colon (159). However, already before weaning, microbial metabolites influence intestinal epithelial renewal. Small intestinal organoids stimulated with sterile filtered stool supernatant obtained from term-babies exhibited higher proliferation and accelerated maturation than untreated organoids. Stool supernatants from pre-term babies, having a stunted microbiota composition, were unable to stimulate organoid development (160). From this mixture of microbial metabolites produced by a healthy microbiota, it is the ambitious goal of mucosal immunologists to identify substances to use as possible therapeutic agents to modulate the imbalanced mucosal homeostasis in susceptible newborns. To narrow down the myriad of possible metabolites, analyses on less diverse systems are performed. Monocolonization of murine pups with a particular Bifidobacterium breve strain (UCC2003) was able to boost epithelial regeneration by supporting the stem cell niche and increasing epithelial differentiation in the small intestine. Additionally, upregulation of integrins, tight-junction molecules, and increased mucus production in pups after Bifidobacterium breve treatment supported its role in strengthening the physical intestinal barrier (161).
The turnover rate of the small intestinal epithelium is slower in neonates than in adult mice (49, 50). This might explain the protection of neonatal mice against inflammation-induced cell shedding after LPS administration. Since apoptotic signaling cascades, including TNF-α production, were intact in neonates, it appears that innate immune mechanisms guided through IFN-γ provide the necessary protection against pathological cell shedding. Again, gut commensals were crucial to orchestrate the immune system towards a tolerogenic phenotype with elevated IL-10 levels to protect from increased LPS-induced epithelial cell death (162).
Long-lasting epithelial homeostasis is partially driven by epigenetic remodeling as a response to microbial stimuli early in life. The most intensively studied metabolite with epigenetic relevance is butyrate, a product from bacterial fiber metabolism. Its supportive role on epithelial sealing, which coincides with the implementation of dietary fibers in combination with its inhibitory effect on histone deacetylases, suggests an important role in regulating epithelial maturation to ensure lifelong mucosal homeostasis (156). Nonetheless, epigenetic remodeling on the level of DNA methylation appears to be more central early in life. Already in 2015, Yu et al. demonstrated that a DNA methylation gain in intestinal stem cells from birth until weaning had lifelong consequences on barrier integrity (163). Genes associated with a methylation gain were involved in intestinal maturation and glycosphingolipid biosynthetic processes. Also, the expression of these genes affecting glycosylation was increased, indicating that the DNA methylation gain is affecting regulatory regions and further supporting the role of epigenetic remodeling in early life barrier formation (163). Glycosphingolipids contribute to the epithelial cell barrier integrity by shaping the glycocalyx of intestinal epithelial cells (Figure 2). Furthermore, the glycosylation pattern of intestinal epithelial cells has been directly associated with beneficial effects on age susceptibility to pathogenic bacterial infections, IBD, and cancer metastasis (164, 165). The DNA methylation gain is microbiota-dependent since mice raised germ-free showed a dysregulated DNA methylation development in the colon from birth until adulthood when compared to conventionally raised mice (CNV). Conventionalizing germ-free pups with FMT at postnatal day 25 partially restored the gain in DNA methylation in adulthood (163). The impact of the microbiota on the DNA methylome in colonic or small intestinal epithelial cells is most probably restricted to specific gene sets and does not affect the DNA methylation pattern on a global scale (157, 163). Enhancer elements known to be lowly methylated, so-called low-methylated regions (LMRs) are strongly affected by the microbiota. This DNA methylation analysis stays in agreement with the transcriptomic analysis and indicates that gut colonization integrates DNA demethylation in LMRs early in life with consequences on gene transcription, which finally drives intestinal development and homeostasis. The epigenomic differences between germ-free and CNV mice had functional consequences on gastrointestinal health, confirmed by increased chromatin accessibility after induction of DSS colitis in genes playing a role in the inflammatory response of the colonic epithelium in CNV mice (166). These findings indicate a profound effect of the microbiota on intestinal epithelial cell homeostasis through epigenetic regulation early in life, certainly, an area that requires further analytic investigations in the future (157, 163, 166).
Breastfeeding is one of the most meaningful exposures during early life. It is considered the best nutrition source for the infant as it contains the perfect balance of lipids, proteins, and carbohydrates, as well as high amounts of micronutrients that are crucial for neonatal growth (167–169). Many of the breast milk components are not only a nutrient source but are additionally biologically active, protect the neonate against pathogen- and immune-mediated diseases (Figure 4) (168, 170–174), and drive the maturation of its immune system (175, 176). Recently, a human study showed that breast milk promotes neonatal immune tolerance in response to antigenic stimulation by increasing the proportion of Tregs while reducing the proliferation of T helper cells and cytokine production (177). This may be one mechanism of considerable importance to ensure lifelong health.
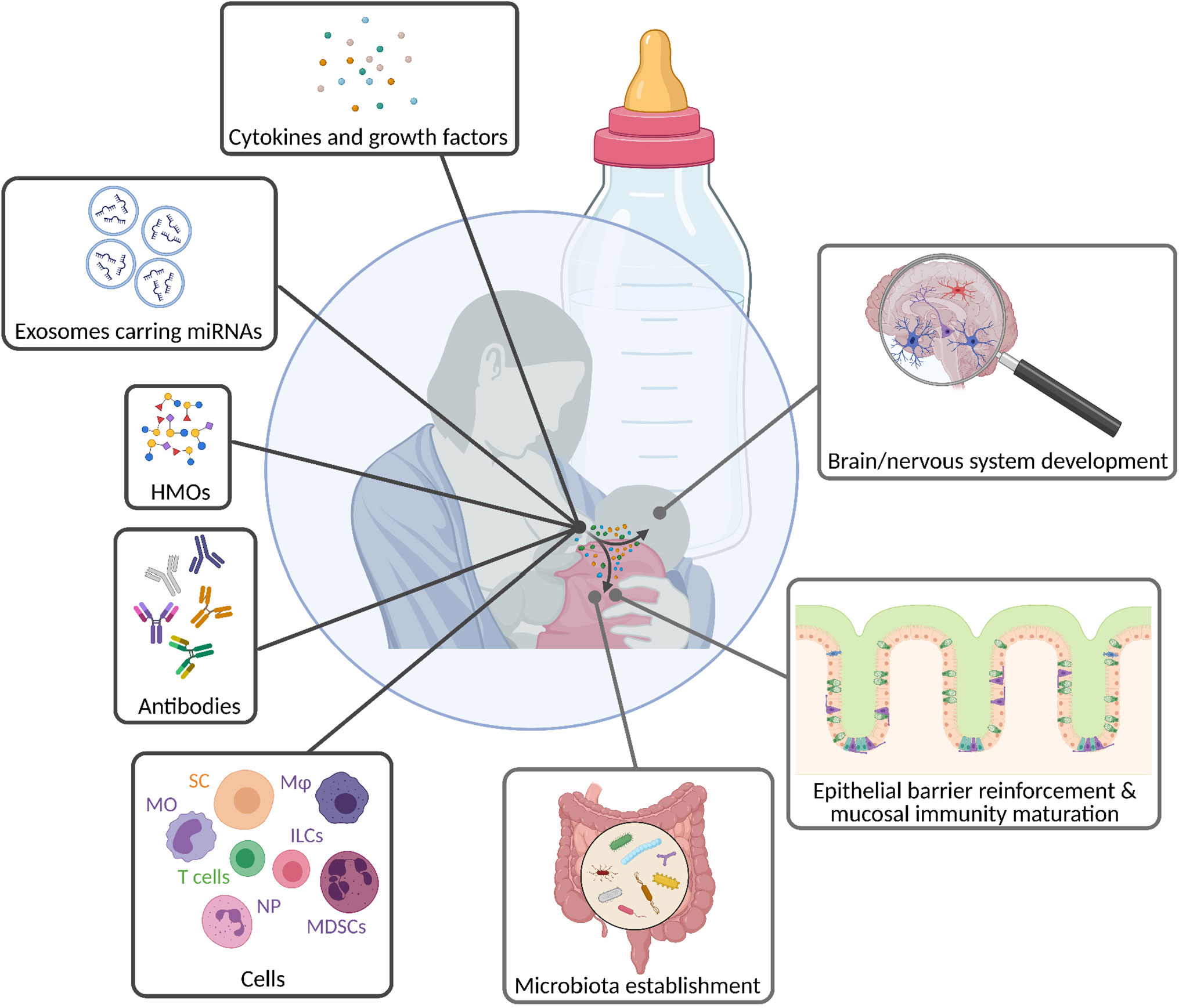
Figure 4 Breastfeeding mediates the transfer of biologically active molecules from the mother to the infant. Breast milk contains a huge diversity of components, ranging from simple sugars, antibodies, and cells to molecules that directly trigger reactions in target cells and/or tissues, such as cytokines, growth factors, and exosomes. Thereby, milk molecules ensure the infant’s well-being by driving innate and adaptive immune maturation, and further contributing to the development of its mucosal and nervous system. HMOs, human milk oligosaccharides; ILCs, innate lymphoid cells; Mφ, macrophages; MDSCs, mononuclear-derived suppressor cells; MO, monocytes; NP, neutrophils; SC, stem cells.
Human milk contains substantial amounts of HMOs, which are fucosylated or sialylated. These complex carbohydrates are one of the most important biologically active compounds that are present in human milk playing essential roles in the development of the newborn (178, 179). They act as prebiotics in the establishment of the infant gut microbiota and inhibit for example the expansion of B. Streptococci, which can cause invasive infections in neonates (180, 181). HMOs also protect against NEC, a widespread and serious gastrointestinal condition affecting predominantly premature infants, leading to the destruction of the intestinal barrier (182, 183). Interestingly, the level of HMOs in breast milk was recently shown to negatively correlate with the incidence of this disease (184, 185). HMOs increase the intestinal mucin level thereby reducing bacterial attachment to the gut epithelium and the risk to develop NEC (186). Remarkably, not only the incidence of pathogen-mediated illnesses is influenced by HMOs, but also the onset of immune-mediated etiologies, such as autoimmune type 1 diabetes (187). Overall, the protective capacity of HMOs raises questions such as: Could additional HMO supplementation during early life be used as a therapeutic strategy for the treatment or prevention of diseases?
HMOs are known to support the development of neonatal immunity. They positively affect the expansion of intestinal commensals, which is part of our first line defense strategy by providing colonization resistance and contributing to mucosal homeostasis (179, 188). Furthermore, HMOs directly support the intestinal barrier function by affecting the maturation of epithelial cells (179). In vitro studies revealed that they protect the gut barrier in a dose-dependent manner by conferring resistance against inflammation-induced epithelial cell dysfunction (189).
The contribution of HMOs to immune cell development and function is also well established (179). For instance, it was recently published that they drive the maturation of monocyte-derived dendritic cells (moDCs), which in turn stimulate the generation of Tregs. Hence, HMOs enhance immune tolerance, which may be one of the central mechanisms by which they contribute to the prevention of immune-mediated diseases in the newborn (187, 190).
Apart from the immune-related and prebiotic functions of HMOs, which have long been recognized and accepted, they are also crucial for the infant’s nutrition. Recent publications display associations between the breast milk HMO composition and infant growth (191). Moreover, its supplementation during childhood has been proposed to be a promising tool to support the development and even improve growth in undernourished infants (192, 193). Additionally, HMOs were suggested to have beneficial effects on brain development by altering the expression of several genes relevant to improve recognition memory (194).
To sum up, HMOs are fundamental breast milk components for the newborn when it comes to the development of its first line defense mechanism by promoting intestinal barrier function and contributing to immune cell maturation, both key factors for guaranteeing lifelong health.
A broad range of peptides and proteins is found in human milk, which are involved in nutrient absorption (amylase, α1-antitrypsin), have immune and antimicrobial properties (immunoglobulins, lactoferrin, cytokines), and possess the ability to promote growth (e.g. epidermal growth factor (EGF)). Among all, α-lactalbumin, casein, lysozyme, and sIgA are the most abundant ones (167, 195, 196). As previously discussed, the neonatal immune system is inexperienced and immature. Thus, neonates rely on passive immunization through breast milk by maternally derived antibodies that offer effective and specific protection against pathogens (171). Lately, it has been demonstrated that maternal IgA prevents NEC in preterm infants by binding to intestinal bacteria (172). Hence, breast milk additionally sustains host-microbiota homeostasis, which is key for the establishment and maintenance of an equilibrated immune system. However, the binding of maternal antibodies to bacteria in the neonatal intestine not only prevents infections but also diminishes immune responses towards commensals by limiting T cell-mediated reactions early in life. An in vivo study found that commensal specific antibodies are transferred from the mother to the offspring via breast milk and persist in the offspring until weaning. In addition, mice born to antibody-deficient mothers had higher numbers of activated follicular T helper cells, which was accompanied by an increase in germinal center B cells in MLNs and Peyer’s Patches. This demonstrated that maternal commensal-specific antibodies delivered to the newborn via breast milk dampen host-mediated commensal specific T cell responses in the offspring, thereby contributing to mucosal homeostasis (197).
Generally, milk-derived proteins are considered essential contributors to the first line defense strategy. For instance, lactoferrin, which is part of the innate immune system, has important antimicrobial and immunomodulatory properties that support health and prevent disorders in the neonate (198). It elicits beneficial effects in a disease state on the intestinal barrier by stimulating the proliferation of epithelial cells and reducing the expression of pro-inflammatory cytokines in innate immune cells (199, 200). β-defensin 1, another protein in human milk, has antimicrobial functions and further promotes the differentiation of moDCs from neonatal cord-blood-derived monocytic precursors. This further drives the maturation of DCs, which thereby obtain their characteristic antigen-presenting capacity (201, 202). Moreover, a study revealed that milk fat globule epidermal growth factor VIII (MFGE8), also known as lactadherin, prevents NEC by limiting intestinal permeability and thus reinforces the barrier function (203). Accordingly, the amount of MFGE8 in breast milk correlated with the infant’s inflammatory state, with higher levels being associated with an anti-inflammatory gut environment (204).
Cytokines are essential contributors to the immune response, which stimulate the differentiation and maturation of various immune cells. The cytokine TGF-β is found in breast milk and is ingested by the neonate, where it stimulates mucosal IgA production and inhibits the synthesis of pro-inflammatory cytokines (205). TGF-β supplementation by oral gavage during the suckling period promoted immune maturation. It lowered NK cell frequency in the MLNs and altered the cytokine profile in the neonate (206). The same group showed that oral administration of TGF-β during this period of life further modified the splenic lymphocyte composition, suggesting effects on systemic immunity (207). Furthermore, breast milk levels of TGF-β negatively correlated with the occurrence of eczema in neonates (208). This supports the assumed potential of maternally derived cytokines to drive the maturation of the neonatal systemic and mucosal immune system. IL-7 in breast milk correlated with thymic development in the offspring (209).
Last, milk contains EGF, which was shown to prevent the weaning reaction (19). This finding suggests that breast milk may be involved in determining the duration of the previously discussed window of opportunity. Additionally, milk-derived EGF has been attributed protective features by inhibiting the formation of GAPs during the early postnatal phase, which in turn prevents the translocation of gut bacteria and thus, systemic pathogen dissemination (210). Nevertheless, and as previously mentioned, the formation of GAPs must also happen during a precise time window before weaning to develop lifelong tolerance to the gut bacteria (142). This switch to inhibit GAP formation around weaning may be fine-tuned by breast milk since the levels of milk EGF decrease throughout lactation. Overall, breast milk may not only protect the neonate but also timely regulate the different immune developmental steps (142, 210).
Exosomes are endosome-derived extracellular vesicles that are 30 - 100 nm in size and circulate in body fluids, including blood, saliva, and breast milk (211–213). They are involved in physiological and pathophysiological immune-related processes, such as antigen-presentation, immune activation, and suppression, as well as intercellular communication. Overall, they are carriers that mediate communication between different parts of the body by transferring proteins, lipids, miRNAs, and other substances (213, 214). miRNAs are small non-coding single-stranded RNA molecules, about 22 nucleotides long, which regulate gene expression and protein translation (215, 216). It is well established that they have immunoregulatory functions by interfering with inflammatory responses thereby playing a role in health and disease. Aberrant expression of miRNAs is associated with severe consequences, ranging from cell death to autoimmunity and cancer (217–220). About a decade ago, several immune-related miRNAs were discovered in breast milk and found to be highly enriched in milk-derived exosomes, suggesting that they may influence the development of the offspring (221, 222). At this time, in vitro studies revealed the uptake of milk-derived exosomes by human macrophages. Nevertheless, their role upon cellular absorption remains to be elucidated (223, 224). Although the capacity of cells to ingest those vesicles was demonstrated, it remained unknown whether they survive the digestive processes and whether they are eventually absorbed by intestinal cells. The latter would mean that breast milk-derived exosomes may transfer cellular components from the mother to the offspring, implying a role in the infant’s development. In more recent studies, scientists investigated whether those vesicles survive digestion by mimicking the infant’s gastric and pancreatic digestion by adjusting the pH and the addition of digestive enzymes to in vitro cultures. Here, milk-derived vesicles were resistant to proteolysis and survived digestion in vitro. They further showed that exosomal miRNAs content remained stable and were absorbed by human intestinal cells in vitro, suggesting the maternal-neonatal transfer of nucleic acids via breast milk-derived exosomes (225, 226).
The transfer of miRNA from the mother to the offspring was further investigated in vivo. A study conducted with pigs showed that colostrum contains higher levels of miRNA compared to mature milk and the serum of pigs, which were only fed with colostrum, had higher levels of colostrum-derived miRNAs, indicating absorption of maternally derived miRNAs (227). Another study further discovered a dose-depended increase in miRNAs in human serum post-cow milk consumption (228). In contrast, a third study did not show any evidence of miRNA absorption after milk consumption (229). These studies did not differentiate between endogenous and exogenous miRNAs. Therefore, Title et al. generated KO mice for specific miRNAs, which were then fostered by wild-type (miRNA sufficient) mothers and their results revealed no evidence for mother-to-offspring transfer of these particular miRNAs (230). In the meanwhile, a publication showed that bovine miRNAs were found in human plasma after bovine milk consumption, insinuating uptake of milk-derived miRNAs (231, 232).
A recent study using newborn calves examined the postprandial ingestion of colostrum-derived miRNAs. Colostrum, as well as maternal and calf blood, were sampled and bioavailability of colostrum-derived vesicles in calf blood and miRNA expression profiles in the different samples were assessed by small RNA-Seq. Although colostrum-derived vesicles were detected in the blood of calves, the miRNA expression profiles of the neonatal blood did not match that of colostrum. The authors consequently proposed two possible mechanisms: First, a disassembly of extracellular vesicles and a release of miRNAs, which may take place during their uptake into epithelial cells, leading to an unequal availability of vesicles and miRNAs in the circulation. Second, an imbalanced absorption of vesicle subpopulations within the colostrum. Other tissues were not analyzed for the presence of milk-derived miRNAs (233). Hence, the transfer of maternal miRNAs into the neonatal systemic circulation remains elusive and a highly discussed topic, which still needs further investigation.
The role of milk-derived exosomes and miRNAs may be of particular interest during neonatal development. The nutritional hypothesis was rapidly completed by a functional hypothesis, suggesting that they may regulate gene expression and immune processes in the newborn (234). Recently, the focus was set on investigating the ability of milk-derived exosomes to protect the mucosal epithelium during infection. Intestinal epithelial cells were incubated with H2O2, which increases oxidative stress-mediated cell death and mimics an inflammatory environment, and simultaneously treated with human breast milk-derived exosomes. The treatment had a protective effect on intestinal epithelial cells by attenuating cell death (235). Another study investigated the effect of breast milk-derived exosomes on goblet cell activity by incubating human colonic epithelial cells with bovine milk-derived exosomes. Interestingly, those vesicles promoted the expression of mucin-related genes. Moreover, they assessed whether this effect was of clinical relevance. Therefore, they induced experimental NEC in murine pups by exposing them to hypoxia and LPS and supplemented them with breast milk-derived exosomes. The treatment with milk-derived exosomes prevented intestinal injury and the reduction of goblet cells, which is a hallmark of NEC (236). Lately, human milk-derived exosomes attenuated intestinal damage and protected intestinal stem cells from undergoing apoptosis due to oxidative stress in vitro (237, 238). Altogether, breast milk-derived exosomes seem to protect the mucosal environment against injury and inflammation-mediated cell death by positively affecting different cell types of the intestinal epithelium.
As previously reviewed, the intestinal epithelium is protected against injury through breast milk-derived extracellular vesicles also in the context of NEC (236). An additional study could show that the administration of human breast milk-derived exosomes decreased the incidence of NEC, confirming their ability to prevent this disease (239). Furthermore, mice that were administered with milk-derived exosomes showed reduced signs of inflammation induced by DSS, which was accompanied by a lower expression of IL-6 and TNF-α, suggesting a protective effect in the context of colitis (240). Overall, existing in vivo studies indicate the possibility of using breast milk-derived exosomes as a potential treatment for infants with intestinal injury and NEC, or for patients with IBD.
Finally, a study investigated the direct effect on the immune system by analyzing the influence of breast milk-derived exosomes on immune cells. Human-derived peripheral blood mononuclear cells (PBMCs) were treated with milk-derived exosomes and stimulated in vitro. This resulted in stronger activation of NK cells as well as γδ T cells, but only in the presence of IL-2 and IL-12. This indicates that while milk exosomes alone may not activate immune cells, they may do so under inflammatory conditions (241). This could be an additional mechanism by which maternal exosomes in breast milk contribute to the prevention of immune-mediated diseases in the offspring.
The presence of immune cells in breast milk, including neutrophils, macrophages, and lymphocytes, is well established. Many animal studies demonstrated the transfer of those maternal cells to the neonate, and it is assumed that they contribute to the maturation of the offspring innate immune system (175, 242–244). Furthermore, flow cytometric analysis revealed the presence of ILCs in human milk, with ILC1s being the most abundant subset followed by ILC3s and ILC2s (245). While ILC1s and ILC3s are crucial to protect against bacteria and to maintain epithelial homeostasis, ILC2s play a crucial role in the defense against parasitic infections at mucosal surfaces (82, 83, 246). The role of ILCs in breast milk remains unexplored and needs further research. Another study confirmed the presence of MAIT cells and γδ T cells in human milk (247). Both cell types display features of innate immunity and are predominantly found in the gut (248), suggesting that they may influence the development of the infant’s microbiota, and thus also its first line defense strategy at mucosal surfaces.
Myeloid-derived suppressor cells (MDSCs) have repressing effects on other immune cells of the innate and adaptive immune system, such as monocytes and T cells. There are two main subsets of MDSCs, monocytic MDSCs (MO-MDSCs) and granulocytic MDSCs (GR-MDSCs) (249). Remarkably, GR-MDSCs accumulate in breast milk and suppress neonatal T cell proliferation, suggesting that milk-derived GR-MDSCs may be able to promote immune tolerance in the offspring. The same group found that GR-MDSCs reduced TLR expression on monocytes, indicating that they regulate innate immune responses in young infants (250). They also showed that the level of GR-MDSCs correlated with gestational and postnatal age, while the levels of those cells in breast milk of mothers who had delivered preterm infants were lowest (251). The authors suggested that these low levels of GR-MDSCs may contribute to reduced immune tolerance and consequently to increased susceptibility to infections, which is in line with preterm infants being at highest risk to develop NEC. Another recent study revealed that the macrophage profile in breast milk changes in response to ongoing respiratory infections in the nursing infant. The researchers observed increased frequencies of anti-inflammatory macrophages and higher IL-6 and IL-8 concentrations in the milk of mothers whose infants had an ongoing respiratory infection, indicating that the composition of breast milk changes according to the infant’s needs to ensure neonatal protection (252).
A reduction in the frequency of IL-13 producing cells in human milk has been associated with an increased incidence of atopic dermatitis in newborns (253). This cytokine is an important mediator of atopic diseases (254), implying that maternal-derived IL-13 producing cells may be involved in protecting the infant from allergy. For instance, maternal-derived IL-13 may prevent its synthesis in the offspring, thereby avoiding the activation of eosinophils and secretion of IgE, which are both central in the pathophysiological mechanism of atopy.
Apart from immune cells, breast milk contains stem cells which are more abundant in colostrum compared to mature milk (255, 256). An in vivo study using wild-type pups that were fostered by GFP+ expressing transgenic mice detected breast milk-derived stem cells in the blood as well as in the brain of the suckling pups, confirming the transfer of milk-derived cells to the offspring. Those cells differentiated into neuronal and glial cells in the pup’s brain, indicating that they play a role in the development of the offspring’s nervous system (257). Maternal immune cells found in breast milk may therefore even be implicated in the development of the infant’s immune system. However, further research is required to prove this hypothesis. A recent study using breast milk stem cells to treat spinal cord injury showed that the administration of these cells reduced apoptosis and inflammation at the site of injury, indicating their influence on immune responses and therapeutic potential (258).
Breast milk is categorized into colostrum and mature milk, although the general components of the milk remain stable throughout lactation. As previously mentioned, the composition of milk is believed to alter, depending on the neonate’s needs. An example is that newborns are at the greatest risk to develop diseases just after birth and thus rely on maternal-derived protection. Accordingly, colostrum contains significantly higher amounts of maternally derived antibodies than mature milk. Therefore, it is widely accepted that colostrum has a principal immunologic function, while mature milk plays a more nutritional role for the neonate. Correspondingly, in humans and mice, the fat content increases throughout lactation. Hence, the milk composition adapts not only to the protective need but also to the nutritional requirements of the growing infant (195, 259).
Studies further revealed that the disease state of the mother also influences the composition of breast milk. Recently, aberrant levels of miRNAs in breast milk exosomes of diabetic mothers were found (260). Furthermore, the milk of mothers with IBD contained significantly lower levels of IgA and higher concentrations of pro-inflammatory cytokines (261). Therefore, breast milk may lead to differential immune priming of the neonate in a disease context. However, more studies are needed to investigate how breast milk affects the development of the infant’s immune system in such situations. Moreover, we have previously shown that bacteria-derived metabolites, such as AhR ligands from the maternal microbiota translocate into breast milk and contribute to the development of innate immune cells in the offspring, namely ILC3s and F4/80+CD11b+ mononuclear cells (77). Hence, the maternal intestinal microbiota can also alter breast milk composition, an area still largely elusive.
In this review, we have covered the period from the very beginning of the developing embryo and fetus in utero up to the weaning of the young offspring and have given an overview on recent publications that show an impact of the commensal microbiota on the emerging immune system. Although we have focused on findings regarding the innate immune system, we also highlighted few key studies that demonstrated an impact on the adaptive immune system.
We hope we have convinced the reader that already during gestation the maternal microbiota can efficiently influence innate immune maturation in the developing fetus, despite the inborn character of the innate immune system. We have spread light on early life environmental factors, including birth mode and the intake of antibiotics in regard to microbiota and immune development in the neonate. We have further emphasized that the maturation of the intestinal microbiota and the evolving immune system go hand in hand and influence each other during the first few weeks (mice) or years (human) after birth. Last, we have tried to give an extensive overview on classical and non-classical immunological compounds present in breast milk and how those affect innate immune maturation in the offspring.
While evidence for an influence of the maternal microbiota, the neonate’s own microbiota or the breast milk during this critical time window is undoubtedly increasing, many aspects remain to be understood: What are the molecular mechanisms underlying the described phenotypes? How is long-term persistence into adulthood achieved? Why does the window of opportunity close around weaning, could it be reopened later in life, and how? To which extent does the maternal microbiota influence the composition of breast milk and thus immune development in the offspring? Finally, how do the maternal diet and later the offspring’s diet, and additional factors (e.g. exposure to environmental toxins or drugs) interact with the microbiota and mutually or independently influence immune maturation in the growing organism?
With our current and future research projects, we aim to complement the understanding of the window of opportunity and to use this knowledge in a preventive or therapeutic setting to improve human health from the neonatal age on.
CK, NF, SC, and SG-V wrote the manuscript and generated figures. All authors contributed to the article and approved the submitted version.
This work was funded through a Peter Hans Hofschneider Professorship provided by the Stiftung Molekulare Biomedizin to SG-V. CK was funded through a European Research Council Starting Grant (H2020 ERC-2016-ADG HHMM_Neonates, Grant Agreement: 742195) provided to Andrew Macpherson. SC received an MD-PhD scholarship of the Swiss National Science Foundation (323530_199385).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Andrew Macpherson for helpful scientific discussions and support. All figures were created with Biorender.com.
1. Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc Natl Acad Sci (2007) 104(34):13780–5. doi: 10.1073/pnas.0706625104
2. Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe (2014) 15(3):382–92. doi: 10.1016/j.chom.2014.02.005
3. Ni J, Shen T-CD, Chen EZ, Bittinger K, Bailey A, Roggiani M, et al. A Role for Bacterial Urease in Gut Dysbiosis and Crohn’s Disease. Sci Trans Med (2017) 9(416):eaah6888. doi: 10.1126/scitranslmed.aah6888
4. Kostic Aleksandar D, Chun E, Robertson L, Glickman Jonathan N, Gallini Carey A, Michaud M, et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
5. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The Human Tumor Microbiome is Composed of Tumor Type–Specific Intracellular Bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
6. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An Obesity-Associated Gut Microbiome With Increased Capacity for Energy Harvest. Nature (2006) 444(7122):1027–31. doi: 10.1038/nature05414
7. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A Core Gut Microbiome in Obese and Lean Twins. Nature (2009) 457(7228):480–4. doi: 10.1038/nature07540
8. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature (2012) 490(7418):55–60. doi: 10.1038/nature11450
9. Cryan JF, O’Riordan KJ, Sandhu K, Peterson V, Dinan TG. The Gut Microbiome in Neurological Disorders. Lancet Neurol (2020) 19(2):179–94. doi: 10.1016/S1474-4422(19)30356-4
10. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated With Neurodevelopmental Disorders. Cell (2013) 155(7):1451–63. doi: 10.1016/j.cell.2013.11.024
11. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota Across Multiple Body Habitats in Newborns. Proc Natl Acad Sci (2010) 107(26):11971–5. doi: 10.1073/pnas.1002601107
12. Dierikx TH, Visser DH, Benninga MA, van Kaam AHLC, de Boer NKH, de Vries R, et al. The Influence of Prenatal and Intrapartum Antibiotics on Intestinal Microbiota Colonisation in Infants: A Systematic Review. J Infect (2020) 81(2):190–204. doi: 10.1016/j.jinf.2020.05.002
13. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome During the First Year of Life. Cell Host Microbe (2015) 17(6):852. doi: 10.1016/j.chom.2015.05.012
14. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early Life Antibiotic-Driven Changes in Microbiota Enhance Susceptibility to Allergic Asthma. EMBO Rep (2012) 13(5):440–7. doi: 10.1038/embor.2012.32
15. Cox Laura M, Yamanishi S, Sohn J, Alekseyenko Alexander V, Leung Jacqueline M, Cho I, et al. Altering the Intestinal Microbiota During a Critical Developmental Window has Lasting Metabolic Consequences. Cell (2014) 158(4):705–21. doi: 10.1016/j.cell.2014.05.052
16. García-Mantrana I, Selma-Royo M, González S, Parra-Llorca A, Martínez-Costa C, Collado MC. Distinct Maternal Microbiota Clusters are Associated With Diet During Pregnancy: Impact on Neonatal Microbiota and Infant Growth During the First 18 Months of Life. Gut Microbes (2020) 11(4):962–78. doi: 10.1080/19490976.2020.1730294
17. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, et al. Comparison of Gut Microbiota in Exclusively Breast-Fed and Formula-Fed Babies: A Study of 91 Term Infants. Sci Rep (2020) 10(1):15792. doi: 10.1038/s41598-020-72635-x
18. Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-Analysis of Effects of Exclusive Breastfeeding on Infant Gut Microbiota Across Populations. Nat Commun (2018) 9(1):4169–. doi: 10.1038/s41467-018-06473-x
19. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, et al. A Weaning Reaction to Microbiota is Required for Resistance to Immunopathologies in the Adult. Immunity (2019) 50(5):1276–88.e5. doi: 10.1016/j.immuni.2019.02.014
20. Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in Gut Microbiota Composition is a Complex Polygenic Trait Shaped by Multiple Environmental and Host Genetic Factors. Proc Natl Acad Sci (2010) 107(44):18933–8. doi: 10.1073/pnas.1007028107
21. Goodrich Julia K, Davenport Emily R, Beaumont M, Jackson Matthew A, Knight R, Ober C, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe (2016) 19(5):731–43. doi: 10.1016/j.chom.2016.04.017
22. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of Microbial Consortia in the Developing Infant Gut Microbiome. Proc Natl Acad Sci (2011) 108(Supplement 1):4578–85. doi: 10.1073/pnas.1000081107
23. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human Gut Microbiome Viewed Across Age and Geography. Nature (2012) 486(7402):222–7. doi: 10.1038/nature11053
24. Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The Intestinal Microbiome in Early Life: Health and Disease. Front Immunol (2014) 5:427(427):1–18. doi: 10.3389/fimmu.2014.00427
25. Dubos R, Schaedler RW, Costello R, Hoet P. Indigenous, Normal, and Autochthonous Flora of The Gastrointestinal Tract. J Exp Med (1965) 122(1):67–76. doi: 10.1084/jem.122.1.67
26. Savage DC, Dubos R, Schaedler RW. The Gastrointestinal Epithelium and its Autochthonous Bacterial Flora. J Exp Med (1968) 127(1):67–76. doi: 10.1084/jem.127.1.67
27. Schaedler RW, Dubos R, Costello R. The Development of the Bacterial Flora in the Gastrointestinal Tract of Mice. J Exp Med (1965) 122(1):59–66. doi: 10.1084/jem.122.1.59
28. Schaedler RW, Dubos R, Costello R. Association of Germfree Mice With Bacteria Isolated From Normal Mice. J Exp Med (1965) 122(1):77–82. doi: 10.1084/jem.122.1.77
29. Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe (2008) 4(5):447–57. doi: 10.1016/j.chom.2008.09.007
30. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell (2016) 167(5):1339–53.e21. doi: 10.1016/j.cell.2016.10.043
31. Martens EC, Neumann M, Desai MS. Interactions of Commensal and Pathogenic Microorganisms With the Intestinal Mucosal Barrier. Nat Rev Microbiol (2018) 16(8):457–70. doi: 10.1038/s41579-018-0036-x
32. Johansson Malin EV, Jakobsson Hedvig E, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro Ana M, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe (2015) 18(5):582–92. doi: 10.1016/j.chom.2015.10.007
33. Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, et al. The Composition of the Gut Microbiota Shapes the Colon Mucus Barrier. EMBO Rep (2015) 16(2):164–77. doi: 10.15252/embr.201439263
34. Bach J-F. The Effect of Infections on Susceptibility to Autoimmune and Allergic Diseases. New Engl J Med (2002) 347(12):911–20. doi: 10.1056/NEJMra020100
35. Strachan DP. Hay Fever, Hygiene, and Household Size. BMJ Br Med J (1989) 299(6710):1259–60. doi: 10.1136/bmj.299.6710.1259
36. Ege MJ, Mayer M, Normand A-C, Genuneit J, Cookson WOCM, Braun-Fahrländer C, et al. Exposure to Environmental Microorganisms and Childhood Asthma. New Engl J Med (2011) 364(8):701–9. doi: 10.1056/NEJMoa1007302
37. Braun-Fahrlander C, Gassner M, Grize L, Neu U, Sennhauser FH, Varonier HS, et al. Prevalence of Hay Fever and Allergic Sensitization in Farmer’s Children and Their Peers Living in the Same Rural Community. SCARPOL Team. Swiss Study on Childhood Allergy and Respiratory Symptoms With Respect to Air Pollution. Clin Exp Allergy J Br Soc Allergy Clin Immunol (1999) 29(1):28–34. doi: 10.1046/j.1365-2222.1999.00479.x
38. Braun-Fahrländer C, Riedler J, Herz U, Eder W, Waser M, Grize L, et al. Environmental Exposure to Endotoxin and Its Relation to Asthma in School-Age Children. New Engl J Med (2002) 347(12):869–77. doi: 10.1056/NEJMoa020057
39. Riedler J, Braun-Fahrländer C, Eder W, Schreuer M, Waser M, Maisch S, et al. Exposure to Farming in Early Life and Development of Asthma and Allergy: A Cross-Sectional Survey. Lancet (2001) 358(9288):1129–33. doi: 10.1016/S0140-6736(01)06252-3
40. Eriksson J, Ekerljung L, Lötvall J, Pullerits T, Wennergren G, Rönmark E, et al. Growing Up on a Farm Leads to Lifelong Protection Against Allergic Rhinitis. Allergy (2010) 65(11):1397–403. doi: 10.1111/j.1398-9995.2010.02397.x
41. Barker DJ, Thornburg KL. The Obstetric Origins of Health for a Lifetime. Clin Obstet Gynecol (2013) 56(3):511–9. doi: 10.1097/GRF.0b013e31829cb9ca
42. Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, et al. Developmental Plasticity and Human Health. Nature (2004) 430(6998):419–21. doi: 10.1038/nature02725
43. Fleming TP, Watkins AJ, Velazquez MA, Mathers JC, Prentice AM, Stephenson J, et al. Origins of Lifetime Health Around the Time of Conception: Causes and Consequences. Lancet (2018) 391(10132):1842–52. doi: 10.1016/S0140-6736(18)30312-X
44. Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of In Utero and Early-Life Conditions on Adult Health and Disease. New Engl J Med (2008) 359(1):61–73. doi: 10.1056/NEJMra0708473
45. Cahenzli J, Köller Y, Wyss M, Geuking Markus B, McCoy Kathy D. Intestinal Microbial Diversity During Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe (2013) 14(5):559–70. doi: 10.1016/j.chom.2013.10.004
46. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science (2012) 336(6080):489. doi: 10.1126/science.1219328
47. Al Nabhani Z, Eberl G. Imprinting of the Immune System by the Microbiota Early in Life. Mucosal Immunol (2020) 13(2):183–9. doi: 10.1038/s41385-020-0257-y
48. Hornef MW, Torow N. ‘Layered Immunity’ and the ‘Neonatal Window of Opportunity’ - Timed Succession of non-Redundant Phases to Establish Mucosal Host-Microbial Homeostasis After Birth. Immunology (2020) 159(1):15–25. doi: 10.1111/imm.13149
49. Koren N, Zubeidat K, Saba Y, Horev Y, Barel O, Wilharm A, et al. Maturation of the Neonatal Oral Mucosa Involves Unique Epithelium-Microbiota Interactions. Cell Host Microbe (2021) 29(2):197–209.e5. doi: 10.1016/j.chom.2020.12.006
50. Muncan V, Heijmans J, Krasinski SD, Büller NV, Wildenberg ME, Meisner S, et al. Blimp1 Regulates the Transition of Neonatal to Adult Intestinal Epithelium. Nat Commun (2011) 2(1):452. doi: 10.1038/ncomms1463
51. Gribar SC, Sodhi CP, Richardson WM, Anand RJ, Gittes GK, Branca MF, et al. Reciprocal Expression and Signaling of TLR4 and TLR9 in the Pathogenesis and Treatment of Necrotizing Enterocolitis. J Immunol (2009) 182(1):636–46. doi: 10.4049/jimmunol.182.1.636
52. Pott J, Stockinger S, Torow N, Smoczek A, Lindner C, McInerney G, et al. Age-Dependent TLR3 Expression of the Intestinal Epithelium Contributes to Rotavirus Susceptibility. PloS Pathog (2012) 8(5):e1002670. doi: 10.1371/journal.ppat.1002670
53. Chassin C, Kocur M, Pott J, Duerr CU, Gütle D, Lotz M, et al. Mir-146a Mediates Protective Innate Immune Tolerance in the Neonate Intestine. Cell Host Microbe (2010) 8(4):358–68. doi: 10.1016/j.chom.2010.09.005
54. Lotz M, Gütle D, Walther S, Ménard S, Bogdan C, Hornef MW. Postnatal Acquisition of Endotoxin Tolerance in Intestinal Epithelial Cells. J Exp Med (2006) 203(4):973–84. doi: 10.1084/jem.20050625
55. Ganal-Vonarburg SC, Hornef MW, Macpherson AJ. Microbial–Host Molecular Exchange and its Functional Consequences in Early Mammalian Life. Science (2020) 368(6491):604–7. doi: 10.1126/science.aba0478
56. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Trans Med (2014) 6(237):237ra65–ra65. doi: 10.1126/scitranslmed.3008599
57. Rautava S, Collado MC, Salminen S, Isolauri E. Probiotics Modulate Host-Microbe Interaction in the Placenta and Fetal Gut: A Randomized, Double-Blind, Placebo-Controlled Trial. Neonatology (2012) 102(3):178–84. doi: 10.1159/000339182
58. Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, et al. Identification of Intracellular Bacteria in the Basal Plate of the Human Placenta in Term and Preterm Gestations. Am J Obstet Gynecol (2013) 208(3):226.e1–.e7. doi: 10.1016/j.ajog.2013.01.018
59. Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, et al. Isolation of Commensal Bacteria From Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section. Curr Microbiol (2005) 51(4):270–4. doi: 10.1007/s00284-005-0020-3
60. Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, et al. Is Meconium From Healthy Newborns Actually Sterile? Res Microbiol (2008) 159(3):187–93. doi: 10.1016/j.resmic.2007.12.007
61. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human Gut Colonisation may be Initiated In Utero by Distinct Microbial Communities in the Placenta and Amniotic Fluid. Sci Rep (2016) 6(1):23129. doi: 10.1038/srep23129
62. Bassols J, Serino M, Carreras-Badosa G, Burcelin R, Blasco-Baque V, Lopez-Bermejo A, et al. Gestational Diabetes is Associated With Changes in Placental Microbiota and Microbiome. Pediatr Res (2016) 80(6):777–84. doi: 10.1038/pr.2016.155
63. Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The Preterm Placental Microbiome Varies in Association With Excess Maternal Gestational Weight Gain. Am J Obstet Gynecol (2015) 212(5):653.e1–.e16. doi: 10.1016/j.ajog.2014.12.041
64. Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J. The Placental Microbiome Varies in Association With Low Birth Weight in Full-Term Neonates. Nutrients (2015) 7(8):6924–37. doi: 10.3390/nu7085315
65. Kliman HJ. Comment on “The Placenta Harbors a Unique Microbiome”. Sci Trans Med (2014) 6(254):254le4–le4. doi: 10.1126/scitranslmed.3009864
66. de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, et al. Recognizing the Reagent Microbiome. Nat Microbiol (2018) 3(8):851–3. doi: 10.1038/s41564-018-0202-y
67. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and Laboratory Contamination can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol (2014) 12(1):87. doi: 10.1186/s12915-014-0087-z
68. Olomu IN, Pena-Cortes LC, Long RA, Vyas A, Krichevskiy O, Luellwitz R, et al. Elimination of “Kitome” and “Splashome” Contamination Results in Lack of Detection of a Unique Placental Microbiome. BMC Microbiol (2020) 20(1):157. doi: 10.1186/s12866-020-01839-y
69. de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human Placenta has No Microbiome But can Contain Potential Pathogens. Nature (2019) 572(7769):329–34. doi: 10.1038/s41586-019-1451-5
70. Kuperman A, Zimmerman A, Hamadia S, Ziv O, Gurevich V, Fichtman B, et al. Deep Microbial Analysis of Multiple Placentas Shows No Evidence for a Placental Microbiome. BJOG: Int J Obstet Gynaecol (2020) 127(2):159–69. doi: 10.1111/1471-0528.15896
71. Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of Placenta Samples With Contamination Controls Does Not Provide Evidence for a Distinct Placenta Microbiota. Microbiome (2016) 4(1):29. doi: 10.1186/s40168-016-0172-3
72. Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, et al. Does the Human Placenta Delivered At Term Have a Microbiota? Results of Cultivation, Quantitative Real-Time PCR, 16s rRNA Gene Sequencing, and Metagenomics. Am J Obstet Gynecol (2019) 220(3):267.e1–.e39. doi: 10.1016/j.ajog.2018.10.018
73. Li Y, Toothaker JM, Ben-Simon S, Ozeri L, Schweitzer R, McCourt BT, et al. In Utero Human Intestine Harbors Unique Metabolome, Including Bacterial Metabolites. JCI Insight (2020) 5(21):e138751. doi: 10.1172/jci.insight.138751
74. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Corroborating Evidence Refutes Batch Effect as Explanation for Fetal Bacteria. Microbiome (2021) 9(1):10. doi: 10.1186/s40168-020-00948-0
75. de Goffau MC, Charnock-Jones DS, Smith GCS, Parkhill J. Batch Effects Account for the Main Findings of an In Utero Human Intestinal Bacterial Colonization Study. Microbiome (2021) 9(1):6. doi: 10.1186/s40168-020-00949-z
76. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable Bacterial Colonization is Highly Limited in the Human Intestine In Utero. Nat Med (2020) 26(4):599–607. doi: 10.1038/s41591-020-0761-3
77. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The Maternal Microbiota Drives Early Postnatal Innate Immune Development. Science (2016) 351(6279):1296–302. doi: 10.1126/science.aad2571
78. Walter J, Hornef MW. A Philosophical Perspective on the Prenatal In Utero Microbiome Debate. Microbiome (2021) 9(1):5. doi: 10.1186/s40168-020-00979-7
79. Macpherson AJ, de Aguero MG, Ganal-Vonarburg SC. How Nutrition and the Maternal Microbiota Shape the Neonatal Immune System. Nat Rev Immunol (2017) 17(8):508–17. doi: 10.1038/nri.2017.58
80. Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible Microbial Colonization of Germ-Free Mice Reveals the Dynamics of IgA Immune Responses. Science (2010) 328(5986):1705–9. doi: 10.1126/science.1188454
81. Stockinger B, Meglio PD, Gialitakis M, Duarte JH. The Aryl Hydrocarbon Receptor: Multitasking in the Immune System. Annu Rev Immunol (2014) 32(1):403–32. doi: 10.1146/annurev-immunol-032713-120245
82. Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, et al. The Microbiota Regulates Neutrophil Homeostasis and Host Resistance to Escherichia Coli K1 Sepsis in Neonatal Mice. Nat Med (2014) 20(5):524–30. doi: 10.1038/nm.3542
83. Niu X, Daniel S, Kumar D, Ding EY, Savani RC, Koh AY, et al. Transient Neonatal Antibiotic Exposure Increases Susceptibility to Late-Onset Sepsis Driven by Microbiota-Dependent Suppression of Type 3 Innate Lymphoid Cells. Sci Rep (2020) 10(1):12974. doi: 10.1038/s41598-020-69797-z
84. Zhang X, Borbet TC, Fallegger A, Wipperman MF, Blaser MJ, Müller A. An Antibiotic-Impacted Microbiota Compromises the Development of Colonic Regulatory T Cells and Predisposes to Dysregulated Immune Responses. mBio (2021) 12(1):e03335–20. doi: 10.1128/mBio.03335-20
85. Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong FS, et al. Maternal Antibiotic Treatment Protects Offspring From Diabetes Development in Nonobese Diabetic Mice by Generation of Tolerogenic Apcs. J Immunol (2015) 195(9):4176–84. doi: 10.4049/jimmunol.1500884
86. Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different Immunological Responses to Early-Life Antibiotic Exposure Affecting Autoimmune Diabetes Development in NOD Mice. J Autoimmun (2016) 72:47–56. doi: 10.1016/j.jaut.2016.05.001
87. Miller JE, Wu C, Pedersen LH, de Klerk N, Olsen J, Burgner DP. Maternal Antibiotic Exposure During Pregnancy and Hospitalization With Infection in Offspring: A Population-Based Cohort Study. Int J Epidemiol (2018) 47(2):561–71. doi: 10.1093/ije/dyx272
88. Örtqvist AK, Lundholm C, Halfvarson J, Ludvigsson JF, Almqvist C. Fetal and Early Life Antibiotics Exposure and Very Early Onset Inflammatory Bowel Disease: A Population-Based Study. Gut (2019) 68(2):218–25. doi: 10.1136/gutjnl-2017-314352
89. Neuman H, Koren O. Chapter 1 - The Microbiome in a Healthy Pregnancy. In: Koren O, Rautava S, editors. The Human Microbiome in Early Life. Cambridge, Massachusettes, USA: Academic Press (2021). p. 3–20.
90. Sato Y, Sakurai K, Tanabe H, Kato T, Nakanishi Y, Ohno H, et al. Maternal Gut Microbiota is Associated With Newborn Anthropometrics in a Sex-Specific Manner. J Dev Origins Health Dis (2019) 10(6):659–66. doi: 10.1017/S2040174419000138
91. Haro C, Garcia-Carpintero S, Alcala-Diaz JF, Gomez-Delgado F, Delgado-Lista J, Perez-Martinez P, et al. The Gut Microbial Community in Metabolic Syndrome Patients is Modified by Diet. J Nutr Biochem (2016) 27:27–31. doi: 10.1016/j.jnutbio.2015.08.011
92. Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct Composition of Gut Microbiota During Pregnancy in Overweight and Normal-Weight Women. Am J Clin Nutr (2008) 88(4):894–9. doi: 10.1093/ajcn/88.4.894
93. Koren O, Goodrich Julia K, Cullender Tyler C, Spor A, Laitinen K, Kling Bäckhed H, et al. Host Remodeling of the Gut Microbiome and Metabolic Changes During Pregnancy. Cell (2012) 150(3):470–80. doi: 10.1016/j.cell.2012.07.008
94. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence That Asthma is a Developmental Origin Disease Influenced by Maternal Diet and Bacterial Metabolites. Nat Commun (2015) 6(1):7320. doi: 10.1038/ncomms8320
95. Assa A, Vong L, Pinnell LJ, Avitzur N, Johnson-Henry KC, Sherman PM. Vitamin D Deficiency Promotes Epithelial Barrier Dysfunction and Intestinal Inflammation. J Infect Dis (2014) 210(8):1296–305. doi: 10.1093/infdis/jiu235
96. Akhtar E, Mily A, Haq A, Al-Mahmud A, El-Arifeen S, Hel Baqui A, et al. Prenatal High-Dose Vitamin D3 Supplementation has Balanced Effects on Cord Blood Th1 and Th2 Responses. Nutr J (2016) 15(1):75. doi: 10.1186/s12937-016-0194-5
97. van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal Retinoids Control Type 3 Innate Lymphoid Cells and Set the Offspring Immunity. Nature (2014) 508(7494):123–7. doi: 10.1038/nature13158
98. Silver MJ, Kessler NJ, Hennig BJ, Dominguez-Salas P, Laritsky E, Baker MS, et al. Independent Genomewide Screens Identify the Tumor Suppressor VTRNA2-1 as a Human Epiallele Responsive to Periconceptional Environment. Genome Biol (2015) 16(1):118. doi: 10.1186/s13059-015-0660-y
99. Forkel M, Berglin L, Kekäläinen E, Carlsson A, Svedin E, Michaëlsson J, et al. Composition and Functionality of the Intrahepatic Innate Lymphoid Cell-Compartment in Human Nonfibrotic and Fibrotic Livers. Eur J Immunol (2017) 47(8):1280–94. doi: 10.1002/eji.201646890
100. Phillips JH, Hori T, Nagler A, Bhat N, Spits H, Lanier LL. Ontogeny of Human Natural Killer (NK) Cells: Fetal NK Cells Mediate Cytolytic Function and Express Cytoplasmic CD3 Epsilon,Delta Proteins. J Exp Med (1992) 175(4):1055–66. doi: 10.1084/jem.175.4.1055
101. Popescu D-M, Botting RA, Stephenson E, Green K, Webb S, Jardine L, et al. Decoding Human Fetal Liver Haematopoiesis. Nature (2019) 574(7778):365–71. doi: 10.1038/s41586-019-1652-y
102. Sagebiel AF, Steinert F, Lunemann S, Körner C, Schreurs RRCE, Altfeld M, et al. Tissue-Resident Eomes+ NK Cells are the Major Innate Lymphoid Cell Population in Human Infant Intestine. Nat Commun (2019) 10(1):975. doi: 10.1038/s41467-018-08267-7
103. Stras SF, Werner L, Toothaker JM, Olaloye OO, Oldham AL, McCourt CC, et al. Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev Cell (2019) 51(3):357–73.e5. doi: 10.1016/j.devcel.2019.09.008
104. Renz H, Adkins BD, Bartfeld S, Blumberg RS, Farber DL, Garssen J, et al. The Neonatal Window of Opportunity - Early Priming for Life. J Allergy Clin Immunol (2017) 141(4):1212–4. doi: 10.1016/j.jaci.2017.11.019
105. Torow N, Hornef MW. The Neonatal Window of Opportunity: Setting the Stage for Life-Long Host-Microbial Interaction and Immune Homeostasis. J Immunol (2017) 198(2):557–63. doi: 10.4049/jimmunol.1601253
106. Park JE, Jardine L, Gottgens B, Teichmann SA, Haniffa M. Prenatal Development of Human Immunity. Science (2020) 368(6491):600–3. doi: 10.1126/science.aaz9330
107. Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, et al. A Cell Atlas of Human Thymic Development Defines T Cell Repertoire Formation. Science (2020) 367(6480):eaay3224. doi: 10.1126/science.aay3224
108. Schreurs R, Baumdick ME, Sagebiel AF, Kaufmann M, Mokry M, Klarenbeek PL, et al. Human Fetal TNF-Alpha-Cytokine-Producing CD4(+) Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity (2019) 50(2):462–76 e8. doi: 10.1016/j.immuni.2018.12.010
109. Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of Innate-Like Microbial Reactivity in Mucosal Tissues During Human Fetal MAIT-cell Development. Nat Commun (2014) 5:3143. doi: 10.1038/ncomms4143
110. Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-Infant Microbial Transmission From Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe (2018) 24(1):133–45.e5. doi: 10.1016/j.chom.2018.06.005
111. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, Birth Mode, and Diet Shape Microbiome Maturation During Early Life. Sci Trans Med (2016) 8(343):343ra82. doi: 10.1126/scitranslmed.aad7121
112. Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted Microbiota and Opportunistic Pathogen Colonization in Caesarean-Section Birth. Nature (2019) 574(7776):117–21. doi: 10.1038/s41586-019-1560-1
113. Stokholm J, Thorsen J, Blaser MJ, Rasmussen MA, Hjelmsø M, Shah S, et al. Delivery Mode and Gut Microbial Changes Correlate With an Increased Risk of Childhood Asthma. Sci Trans Med (2020) 12(569):eaax9929. doi: 10.1126/scitranslmed.aax9929
114. Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, et al. Impact of Maternal Intrapartum Antibiotics, Method of Birth and Breastfeeding on Gut Microbiota During the First Year of Life: A Prospective Cohort Study. BJOG: Int J Obstet Gynaecol (2016) 123(6):983–93. doi: 10.1111/1471-0528.13601
115. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased Gut Microbiota Diversity, Delayed Bacteroidetes Colonisation and Reduced Th1 Responses in Infants Delivered by Caesarean Section. Gut (2014) 63(4):559. doi: 10.1136/gutjnl-2012-303249
116. Stokholm J, Thorsen J, Chawes BL, Schjørring S, Krogfelt KA, Bønnelykke K, et al. Cesarean Section Changes Neonatal Gut Colonization. J Allergy Clin Immunol (2016) 138(3):881–9.e2. doi: 10.1016/j.jaci.2016.01.028
117. Andersen V, Möller S, Jensen PB, Møller FT, Green A. Caesarean Delivery and Risk of Chronic Inflammatory Diseases (Inflammatory Bowel Disease, Rheumatoid Arthritis, Coeliac Disease, and Diabetes Mellitus): A Population Based Registry Study of 2,699,479 Births in Denmark During 1973-2016. Clin Epidemiol (2020) 12:287–93. doi: 10.2147/CLEP.S229056
118. Laursen MF, Zachariassen G, Bahl MI, Bergström A, Høst A, Michaelsen KF, et al. Having Older Siblings is Associated With Gut Microbiota Development During Early Childhood. BMC Microbiol (2015) 15(1):154. doi: 10.1186/s12866-015-0477-6
119. Penders J, Gerhold K, Thijs C, Zimmermann K, Wahn U, Lau S, et al. New Insights Into the Hygiene Hypothesis in Allergic Diseases. Gut Microbes (2014) 5(2):239–44. doi: 10.4161/gmic.27905
120. Hansen CHF, Andersen LSF, Krych Ł, Metzdorff SB, Hasselby JP, Skov S, et al. Mode of Delivery Shapes Gut Colonization Pattern and Modulates Regulatory Immunity in Mice. J Immunol (2014) 193(3):1213. doi: 10.4049/jimmunol.1400085
121. Zachariassen LF, Krych L, Rasmussen SH, Nielsen DS, Kot W, Holm TL, et al. Cesarean Section Induces Microbiota-Regulated Immune Disturbances in C57BL/6 Mice. J Immunol (2019) 202(1):142. doi: 10.4049/jimmunol.1800666
122. Zachariassen LF, Hansen AK, Krych L, Nielsen DS, Holm TL, Tougaard P, et al. Cesarean Section Increases Sensitivity to Oxazolone-Induced Colitis in C57BL/6 Mice. Mucosal Immunol (2019) 12(6):1348–57. doi: 10.1038/s41385-019-0207-8
123. Boatin AA, Schlotheuber A, Betran AP, Moller A-B, Barros AJD, Boerma T, et al. Within Country Inequalities in Caesarean Section Rates: Observational Study of 72 Low and Middle Income Countries. BMJ (2018) 360:k55. doi: 10.1136/bmj.k55
124. Korpela K, Helve O, Kolho K-L, Saisto T, Skogberg K, Dikareva E, et al. Maternal Fecal Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell (2020) 183(2):324–34.e5. doi: 10.1016/j.cell.2020.08.047
125. Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial Restoration of the Microbiota of Cesarean-Born Infants Via Vaginal Microbial Transfer. Nat Med (2016) 22(3):250–3. doi: 10.1038/nm.4039
126. Clements E, Schlichting LE, Clyne A, Vivier PM. Underlying Causes and Distribution of Infant Mortality in a Statewide Assessment From 2005 to 2016 by Infant, Maternal, and Neighborhood Characteristics. R I Med J (2013) (2019) 102(9):15–22.
127. Boghossian NS, Page GP, Bell EF, Stoll BJ, Murray JC, Cotten CM, et al. Late-Onset Sepsis in Very Low Birth Weight Infants From Singleton and Multiple-Gestation Births. J Pediatr (2013) 162(6):1120–4.e1. doi: 10.1016/j.jpeds.2012.11.089
128. Clark RH, Bloom BT, Spitzer AR, Gerstmann DR. Empiric Use of Ampicillin and Cefotaxime, Compared With Ampicillin and Gentamicin, for Neonates At Risk for Sepsis is Associated With an Increased Risk of Neonatal Death. Pediatrics (2006) 117(1):67. doi: 10.1542/peds.2005-0179
129. Carter GP, Ussher JE, Da Silva AG, Baines SL, Heffernan H, Riley TV, et al. Genomic Analysis of Multiresistant ≪Span Class=&Quot;Named-Content Genus-Species&Quot; Id=&Quot;named-content-1"<Staphylococcus Capitis≪/Span< Associated With Neonatal Sepsis. Antimicrob Agents Chemother (2018) 62(11):e00898–18. doi: 10.1128/AAC.00898-18
130. Wirth T, Bergot M, Rasigade J-P, Pichon B, Barbier M, Martins-Simoes P, et al. Niche Specialization and Spread of Staphylococcus Capitis Involved in Neonatal Sepsis. Nat Microbiol (2020) 5(5):735–45. doi: 10.1038/s41564-020-0676-2
131. Leggett RM, Alcon-Giner C, Heavens D, Caim S, Brook TC, Kujawska M, et al. Rapid MinION Profiling of Preterm Microbiota and Antimicrobial-Resistant Pathogens. Nat Microbiol (2020) 5(3):430–42. doi: 10.1038/s41564-019-0626-z
132. Hollams EM, Teo SM, Kusel M, Holt BJ, Holt KE, Inouye M, et al. Vitamin D Over the First Decade and Susceptibility to Childhood Allergy and Asthma. J Allergy Clin Immunol (2017) 139(2):472–81.e9. doi: 10.1016/j.jaci.2016.07.032
133. Thorsteinsdottir F, Cardoso I, Keller A, Stougaard M, Frederiksen P, Cohen AS, et al. Neonatal Vitamin D Status and Risk of Asthma in Childhood: Results From the D-Tect Study. Nutrients (2020) 12(3):842–55. doi: 10.3390/nu12030842
134. Chambers ES, Suwannasaen D, Mann EH, Urry Z, Richards DF, Lertmemongkolchai G, et al. 1α,25-Dihydroxyvitamin D3 in Combination With Transforming Growth Factor-β Increases the Frequency of Foxp3+ Regulatory T Cells Through Preferential Expansion and Usage of Interleukin-2. Immunology (2014) 143(1):52–60. doi: 10.1111/imm.12289
135. Penna G, Amuchastegui S, Giarratana N, Daniel KC, Vulcano M, Sozzani S, et al. 1,25-Dihydroxyvitamin D3 Selectively Modulates Tolerogenic Properties in Myeloid But Not Plasmacytoid Dendritic Cells. J Immunol (2007) 178(1):145. doi: 10.4049/jimmunol.178.1.145
136. Bachus H, Kaur K, Papillion AM, Marquez-Lago TT, Yu Z, Ballesteros-Tato A, et al. Impaired Tumor-Necrosis-Factor-α-Driven Dendritic Cell Activation Limits Lipopolysaccharide-Induced Protection From Allergic Inflammation in Infants. Immunity (2019) 50(1):225–40.e4. doi: 10.1016/j.immuni.2018.11.012
137. Rueter K, Jones AP, Siafarikas A, Lim E-M, Bear N, Noakes PS, et al. Direct Infant UV Light Exposure is Associated With Eczema and Immune Development. J Allergy Clin Immunol (2019) 143(3):1012–20.e2. doi: 10.1016/j.jaci.2018.08.037
138. Youssef MAM, Zahran AM, Hussien AM, Elsayh KI, Askar EA, Farghaly HS. In Neonates With Vitamin D Deficiency, Low Lymphocyte Activation Markers are Risk Factors for Infection. Paediatr Int Child Health (2019) 39(2):111–8. doi: 10.1080/20469047.2018.1528755
139. Hornsby E, Pfeffer PE, Laranjo N, Cruikshank W, Tuzova M, Litonjua AA, et al. Vitamin D Supplementation During Pregnancy: Effect on the Neonatal Immune System in a Randomized Controlled Trial. J Allergy Clin Immunol (2018) 141(1):269–78.e1. doi: 10.1016/j.jaci.2017.02.039
140. Thysen AH, Waage J, Larsen JM, Rasmussen MA, Stokholm J, Chawes B, et al. Distinct Immune Phenotypes in Infants Developing Asthma During Childhood. Sci Trans Med (2020) 12(529):eaaw0258. doi: 10.1126/scitranslmed.aaw0258
141. Loeb M, Dang AD, Thiem VD, Thanabalan V, Wang B, Nguyen NB, et al. Effect of Vitamin D Supplementation to Reduce Respiratory Infections in Children and Adolescents in Vietnam: A Randomized Controlled Trial. Influenza Other Respir Viruses (2019) 13(2):176–83. doi: 10.1111/irv.12615
142. Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, et al. Microbial Antigen Encounter During a Preweaning Interval is Critical for Tolerance to Gut Bacteria. Sci Immunol (2017) 2(18):eaao1314. doi: 10.1126/sciimmunol.aao1314
143. Husby S, Koletzko S, Korponay-Szabó IR, Mearin ML, Phillips A, Shamir R, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition Guidelines for the Diagnosis of Coeliac Disease. J Pediatr Gastroenterol Nutr (2012) 54(1):136–60. doi: 10.1097/MPG.0b013e31821a23d0
144. Kemppainen KM, Lynch KF, Liu E, Lönnrot M, Simell V, Briese T, et al. Factors That Increase Risk of Celiac Disease Autoimmunity After a Gastrointestinal Infection in Early Life. Clin Gastroenterol Hepatol (2017) 15(5):694–702.e5. doi: 10.1016/j.cgh.2016.10.033
145. Romanos J, Rosén A, Kumar V, Trynka G, Franke L, Szperl A, et al. Improving Coeliac Disease Risk Prediction by Testing non-HLA Variants Additional to HLA Variants. Gut (2014) 63(3):415. doi: 10.1136/gutjnl-2012-304110
146. Andrén Aronsson C, Lee H-S, Hård af Segerstad EM, Uusitalo U, Yang J, Koletzko S, et al. Association of Gluten Intake During the First 5 Years of Life With Incidence of Celiac Disease Autoimmunity and Celiac Disease Among Children At Increased Risk. JAMA (2019) 322(6):514–23. doi: 10.1001/jama.2019.10329
147. Andrén Aronsson C, Lee H-S, Koletzko S, Uusitalo U, Yang J, Virtanen SM, et al. Effects of Gluten Intake on Risk of Celiac Disease: A Case-Control Study on a Swedish Birth Cohort. Clin Gastroenterol Hepatol (2016) 14(3):403–9.e3. doi: 10.1016/j.cgh.2015.09.030
148. Crespo-Escobar P, Mearin ML, Hervás D, Auricchio R, Castillejo G, Gyimesi J, et al. The Role of Gluten Consumption At an Early Age in Celiac Disease Development: A Further Analysis of the Prospective PreventCD Cohort Study. Am J Clin Nutr (2017) 105(4):890–6. doi: 10.3945/ajcn.116.144352
149. Olivares M, Walker AW, Capilla A, Benítez-Páez A, Palau F, Parkhill J, et al. Gut Microbiota Trajectory in Early Life may Predict Development of Celiac Disease. Microbiome (2018) 6(1):36. doi: 10.1186/s40168-018-0415-6
150. Olivares M, Benítez-Páez A, de Palma G, Capilla A, Nova E, Castillejo G, et al. Increased Prevalence of Pathogenic Bacteria in the Gut Microbiota of Infants At Risk of Developing Celiac Disease: The PROFICEL Study. Gut Microbes (2018) 9(6):551–8. doi: 10.1080/19490976.2018.1451276
151. Håkansson Å, Andrén Aronsson C, Brundin C, Oscarsson E, Molin G, Agardh D. Effects of Lactobacillus Plantarum and Lactobacillus Paracasei on the Peripheral Immune Response in Children With Celiac Disease Autoimmunity: A Randomized, Double-Blind, Placebo-Controlled Clinical Trial. Nutrients (2019) 11(8):1925–37. doi: 10.3390/nu11081925
152. Primec M, Klemenak M, Di Gioia D, Aloisio I, Bozzi Cionci N, Quagliariello A, et al. Clinical Intervention Using Bifidobacterium Strains in Celiac Disease Children Reveals Novel Microbial Modulators of TNF-α and Short-Chain Fatty Acids. Clin Nutr (2019) 38(3):1373–81. doi: 10.1016/j.clnu.2018.06.931
153. Allaire JM, Crowley SM, Law HT, Chang S-Y, Ko H-J, Vallance BA. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol (2018) 39(9):677–96. doi: 10.1016/j.it.2018.04.002
154. Renz H, Brandtzaeg P, Hornef M. The Impact of Perinatal Immune Development on Mucosal Homeostasis and Chronic Inflammation. Nat Rev Immunol (2012) 12(1):9–23. doi: 10.1038/nri3112
155. Arnaud AP, Rome V, Richard M, Formal M, David-Le Gall S, Boudry G. Post-Natal Co-Development of the Microbiota and Gut Barrier Function Follows Different Paths in the Small and Large Intestine in Piglets. FASEB J (2020) 34(1):1430–46. doi: 10.1096/fj.201902514R
156. Beaumont M, Paës C, Mussard E, Knudsen C, Cauquil L, Aymard P, et al. Gut Microbiota Derived Metabolites Contribute to Intestinal Barrier Maturation At the Suckling-to-Weaning Transition. Gut Microbes (2020) 11(5):1268–86. doi: 10.1080/19490976.2020.1747335
157. Pan W-H, Sommer F, Falk-Paulsen M, Ulas T, Best P, Fazio A, et al. Exposure to the Gut Microbiota Drives Distinct Methylome and Transcriptome Changes in Intestinal Epithelial Cells During Postnatal Development. Genome Med (2018) 10(1):27. doi: 10.1186/s13073-018-0534-5
158. Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, et al. Neonatal Selection by Toll-like Receptor 5 Influences Long-Term Gut Microbiota Composition. Nature (2018) 560(7719):489–93. doi: 10.1038/s41586-018-0395-5
159. Abo H, Chassaing B, Harusato A, Quiros M, Brazil JC, Ngo VL, et al. Erythroid Differentiation Regulator-1 Induced by Microbiota in Early Life Drives Intestinal Stem Cell Proliferation and Regeneration. Nat Commun (2020) 11(1):513. doi: 10.1038/s41467-019-14258-z
160. Dougherty MW, Kudin O, Mühlbauer M, Neu J, Gharaibeh RZ, Jobin C. Gut Microbiota Maturation During Early Human Life Induces Enterocyte Proliferation Via Microbial Metabolites. BMC Microbiol (2020) 20(1):205. doi: 10.1186/s12866-020-01892-7
161. Kiu R, Treveil A, Harnisch LC, Caim S, Leclaire C, van Sinderen D, et al. Bifidobacterium Breve UCC2003 Induces a Distinct Global Transcriptomic Program in Neonatal Murine Intestinal Epithelial Cells. iScience (2020) 23(7):101336. doi: 10.1016/j.isci.2020.101336
162. Hughes KR, Schofield Z, Dalby MJ, Caim S, Chalklen L, Bernuzzi F, et al. The Early Life Microbiota Protects Neonatal Mice From Pathological Small Intestinal Epithelial Cell Shedding. FASEB J (2020) 34(5):7075–88. doi: 10.1096/fj.202000042R
163. Yu D-H, Gadkari M, Zhou Q, Yu S, Gao N, Guan Y, et al. Postnatal Epigenetic Regulation of Intestinal Stem Cells Requires DNA Methylation and is Guided by the Microbiome. Genome Biol (2015) 16(1):211. doi: 10.1186/s13059-015-0763-5
164. Dai D, Nanthkumar NN, Newburg DS, Walker WA. Role of Oligosaccharides and Glycoconjugates in Intestinal Host Defense. J Pediatr Gastroenterol Nutr (2000) 30:S23–33. doi: 10.1097/00005176-200000002-00005
165. Campbell BJ, Yu L-G, Rhodes JM. Altered Glycosylation in Inflammatory Bowel Disease: A Possible Role in Cancer Development. Glycoconjugate J (2001) 18(11):851–8. doi: 10.1023/A:1022240107040
166. Ansari I, Raddatz G, Gutekunst J, Ridnik M, Cohen D, Abu-Remaileh M, et al. The Microbiota Programs DNA Methylation to Control Intestinal Homeostasis and Inflammation. Nat Microbiol (2020) 5(4):610–9. doi: 10.1038/s41564-019-0659-3
167. Ballard O, Morrow AL. Human Milk Composition: Nutrients and Bioactive Factors. Pediatr Clin North Am (2013) 60(1):49–74. doi: 10.1016/j.pcl.2012.10.002
168. Walker A. Breast Milk as the Gold Standard for Protective Nutrients. J Pediatr (2010) 156(2 Suppl):S3–7. doi: 10.1016/j.jpeds.2009.11.021
169. Dror DK, Allen LH. Overview of Nutrients in Human Milk. Adv Nutr (2018) 9(suppl_1):278s–94s. doi: 10.1093/advances/nmy022
170. M’Rabet L, Vos AP, Boehm G, Garssen J. Breast-Feeding and its Role in Early Development of the Immune System in Infants: Consequences for Health Later in Life. J Nutr (2008) 138(9):1782s–90s. doi: 10.1093/jn/138.9.1782S
171. Caballero-Flores G, Sakamoto K, Zeng MY, Wang Y, Hakim J, Matus-Acuña V, et al. Maternal Immunization Confers Protection to the Offspring Against an Attaching and Effacing Pathogen Through Delivery of IgG in Breast Milk. Cell Host Microbe (2019) 25(2):313–23.e4. doi: 10.1016/j.chom.2018.12.015
172. Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R, et al. Maternal IgA Protects Against the Development of Necrotizing Enterocolitis in Preterm Infants. Nat Med (2019) 25(7):1110–5. doi: 10.1038/s41591-019-0480-9
173. El-Khuffash A, Jain A, Lewandowski AJ, Levy PT. Preventing Disease in the 21st Century: Early Breast Milk Exposure and Later Cardiovascular Health in Premature Infants. Pediatr Res (2020) 87(2):385–90. doi: 10.1038/s41390-019-0648-5
174. Poudel RR, Shrestha D. Breastfeeding for Diabetes Prevention. J Pak Med Assoc (2016) 66(9 Suppl 1):S88–90.
175. Laouar A. Maternal Leukocytes and Infant Immune Programming During Breastfeeding. Trends Immunol (2020) 41(3):225–39. doi: 10.1016/j.it.2020.01.005
176. Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front Immunol (2018) 9:361. doi: 10.3389/fimmu.2018.00361
177. Wood H, Acharjee A, Pearce H, Quraishi MN, Powell R, Rossiter A, et al. Breastfeeding Promotes Early Neonatal Regulatory T-cell Expansion and Immune Tolerance of non-Inherited Maternal Antigens. Allergy (2021). doi: 10.1111/all.14736
178. Bode L. Human Milk Oligosaccharides: Every Baby Needs a Sugar Mama. Glycobiology (2012) 22(9):1147–62. doi: 10.1093/glycob/cws074
179. Donovan SM, Comstock SS. Human Milk Oligosaccharides Influence Neonatal Mucosal and Systemic Immunity. Ann Nutr Metab (2016) 69 Suppl 2(Suppl 2):42–51. doi: 10.1159/000452818
180. Ackerman DL, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. Human Milk Oligosaccharides Exhibit Antimicrobial and Antibiofilm Properties Against Group B Streptococcus. ACS Infect Dis (2017) 3(8):595–605. doi: 10.1021/acsinfecdis.7b00064
181. Lin AE, Autran CA, Szyszka A, Escajadillo T, Huang M, Godula K, et al. Human Milk Oligosaccharides Inhibit Growth of Group B Streptococcus. J Biol Chem (2017) 292(27):11243–9. doi: 10.1074/jbc.M117.789974
182. Rich BS, Dolgin SE. Necrotizing Enterocolitis. Pediatr Rev (2017) 38(12):552–9. doi: 10.1542/pir.2017-0002
183. Wang C, Zhang M, Guo H, Yan J, Liu F, Chen J, et al. Human Milk Oligosaccharides Protect Against Necrotizing Enterocolitis by Inhibiting Intestinal Damage Via Increasing the Proliferation of Crypt Cells. Mol Nutr Food Res (2019) 63(18):e1900262. doi: 10.1002/mnfr.201900262
184. Autran CA, Kellman BP, Kim JH, Asztalos E, Blood AB, Spence ECH, et al. Human Milk Oligosaccharide Composition Predicts Risk of Necrotising Enterocolitis in Preterm Infants. Gut (2018) 67(6):1064–70. doi: 10.1136/gutjnl-2016-312819
185. Sodhi CP, Wipf P, Yamaguchi Y, Fulton WB, Kovler M, Niño DF, et al. The Human Milk Oligosaccharides 2’-Fucosyllactose and 6’-Sialyllactose Protect Against the Development of Necrotizing Enterocolitis by Inhibiting Toll-Like Receptor 4 Signaling. Pediatr Res (2021) 89(1):91–101. doi: 10.1038/s41390-020-0852-3
186. Wu RY, Li B, Koike Y, Määttänen P, Miyake H, Cadete M, et al. Human Milk Oligosaccharides Increase Mucin Expression in Experimental Necrotizing Enterocolitis. Mol Nutr Food Res (2019) 63(3):e1800658. doi: 10.1002/mnfr.201800658
187. Xiao L, Van’t Land B, Engen PA, Naqib A, Green SJ, Nato A, et al. Human Milk Oligosaccharides Protect Against the Development of Autoimmune Diabetes in NOD-Mice. Sci Rep (2018) 8(1):3829. doi: 10.1038/s41598-018-22052-y
188. Lawley TD, Walker AW. Intestinal Colonization Resistance. Immunology (2013) 138(1):1–11. doi: 10.1111/j.1365-2567.2012.03616.x
189. Natividad JM, Rytz A, Keddani S, Bergonzelli G, Garcia-Rodenas CL. Blends of Human Milk Oligosaccharides Confer Intestinal Epithelial Barrier Protection In Vitro. Nutrients (2020) 12(10):3047–60. doi: 10.3390/nu12103047
190. Xiao L, van De Worp WR, Stassen R, van Maastrigt C, Kettelarij N, Stahl B, et al. Human Milk Oligosaccharides Promote Immune Tolerance Via Direct Interactions With Human Dendritic Cells. Eur J Immunol (2019) 49(7):1001–14. doi: 10.1002/eji.201847971
191. Lagström H, Rautava S, Ollila H, Kaljonen A, Turta O, Mäkelä J, et al. Associations Between Human Milk Oligosaccharides and Growth in Infancy and Early Childhood. Am J Clin Nutr (2020) 111(4):769–78. doi: 10.1093/ajcn/nqaa010
192. Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JCC, Barratt MJ, et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell (2016) 164(5):859–71. doi: 10.1016/j.cell.2016.01.024
193. Hegar B, Wibowo Y, Basrowi RW, Ranuh RG, Sudarmo SM, Munasir Z, et al. The Role of Two Human Milk Oligosaccharides, 2’-Fucosyllactose and Lacto-N-Neotetraose, in Infant Nutrition. Pediatr Gastroenterol Hepatol Nutr (2019) 22(4):330–40. doi: 10.5223/pghn.2019.22.4.330
194. Fleming SA, Mudd AT, Hauser J, Yan J, Metairon S, Steiner P, et al. Human and Bovine Milk Oligosaccharides Elicit Improved Recognition Memory Concurrent With Alterations in Regional Brain Volumes and Hippocampal mRNA Expression. Front Neurosci (2020) 14:770. doi: 10.3389/fnins.2020.00770
195. Andreas NJ, Kampmann B, Mehring Le-Doare K. Human Breast Milk: A Review on its Composition and Bioactivity. Early Hum Dev (2015) 91(11):629–35. doi: 10.1016/j.earlhumdev.2015.08.013
196. Mosca F, Giannì ML. Human Milk: Composition and Health Benefits. Pediatr Med Chir (2017) 39(2):155. doi: 10.4081/pmc.2017.155
197. Koch Meghan A, Reiner Gabrielle L, Lugo Kyler A, Kreuk Lieselotte SM, Stanbery Alison G, Ansaldo E, et al. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell (2016) 165(4):827–41. doi: 10.1016/j.cell.2016.04.055
198. Telang S. Lactoferrin: A Critical Player in Neonatal Host Defense. Nutrients (2018) 10(9):1228. doi: 10.3390/nu10091228
199. Liu J, Li B, Lee C, Zhu H, Zheng S, Pierro A. Protective Effects of Lactoferrin on Injured Intestinal Epithelial Cells. J Pediatr Surg (2019) 54(12):2509–13. doi: 10.1016/j.jpedsurg.2019.08.046
200. Wisgrill L, Wessely I, Spittler A, Förster-Waldl E, Berger A, Sadeghi K. Human Lactoferrin Attenuates the Proinflammatory Response of Neonatal Monocyte-Derived Macrophages. Clin Exp Immunol (2018) 192(3):315–24. doi: 10.1111/cei.13108
201. Wu J, Gong RL, Hu QF, Chen XT, Zhao W, Chen TX. Immunoregulatory Effect of Human β-Defensin 1 on Neonatal Cord Blood Monocyte-Derived Dendritic Cells and T Cells. Mol Immunol (2019) 109:99–107. doi: 10.1016/j.molimm.2019.03.007
202. Wang X-F, Cao R-M, Li J, Wu J, Wu S-M, Chen T-X. Identification of Sociodemographic and Clinical Factors Associated With the Levels of Human β-Defensin-1 and Human β-Defensin-2 in the Human Milk of Han Chinese. Br J Nutr (2014) 111(5):867–74. doi: 10.1017/S0007114513003292
203. Shen H, Lei Y, He X, Liu D, He Z. Role of Lactadherin in Intestinal Barrier Integrity in Experimental Neonatal Necrotizing Enterocolitis. J Cell Biochem (2019) 120(12):19509–17. doi: 10.1002/jcb.29255
204. Asaro JA, Khan Z, Brewer M, Klose K, Pesce C, Schanler RJ, et al. Relationship Between Milk Fat Globule-Epidermal Growth Factor 8 and Intestinal Cytokines in Infants Born Preterm. J Pediatr (2021) 230:71–5.e1. doi: 10.1016/j.jpeds.2020.11.014
205. Brenmoehl J, Ohde D, Wirthgen E, Hoeflich A. Cytokines in Milk and the Role of TGF-Beta. Best Pract Res Clin Endocrinol Metab (2018) 32(1):47–56. doi: 10.1016/j.beem.2018.01.006
206. Torres-Castro P, Abril-Gil M, Rodríguez-Lagunas MJ, Castell M, Pérez-Cano FJ, Franch À. Tgf-β2, EGF, and FGF21 Growth Factors Present in Breast Milk Promote Mesenteric Lymph Node Lymphocytes Maturation in Suckling Rats. Nutrients (2018) 10(9):1171. doi: 10.3390/nu10091171
207. Torres-Castro P, Grases-Pintó B, Abril-Gil M, Castell M, Rodríguez-Lagunas MJ, Pérez-Cano FJ, et al. Modulation of the Systemic Immune Response in Suckling Rats by Breast Milk Tgf-β2, EGF and FGF21 Supplementation. Nutrients (2020) 12(6):1888. doi: 10.3390/nu12061888
208. Morita Y, Campos-Alberto E, Yamaide F, Nakano T, Ohnisi H, Kawamoto M, et al. Tgf-β Concentration in Breast Milk is Associated With the Development of Eczema in Infants. Front Pediatr (2018) 6:162. doi: 10.3389/fped.2018.00162
209. Hossny EM, El-Ghoneimy DH, El-Owaidy RH, Mansour MG, Hamza MT, El-Said AF. Breast Milk Interleukin-7 and Thymic Gland Development in Infancy. Eur J Nutr (2020) 59(1):111–8. doi: 10.1007/s00394-018-01891-5
210. Knoop KA, Coughlin PE, Floyd AN, Ndao IM, Hall-Moore C, Shaikh N, et al. Maternal Activation of the EGFR Prevents Translocation of Gut-Residing Pathogenic ≪Em<Escherichia Coli≪/Em< in a Model of Late-Onset Neonatal Sepsis. Proc Natl Acad Sci (2020) 117(14):7941. doi: 10.1073/pnas.1912022117
211. Admyre C, Johansson SM, Qazi KR, Filén J-J, Lahesmaa R, Norman M, et al. Exosomes With Immune Modulatory Features are Present in Human Breast Milk. J Immunol (2007) 179(3):1969. doi: 10.4049/jimmunol.179.3.1969
212. Lönnerdal B. Human Milk Micrornas/Exosomes: Composition and Biological Effects. Nestle Nutr Inst Workshop Ser (2019) 90:83–92. doi: 10.1159/000490297
213. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Rev Proteomics (2009) 6(3):267–83. doi: 10.1586/epr.09.17
214. Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and Their Roles in Immune Regulation and Cancer. Semin Cell Dev Biol (2015) 40:72–81. doi: 10.1016/j.semcdb.2015.02.009
215. Ambros V. The Functions of Animal Micrornas. Nature (2004) 431(7006):350–5. doi: 10.1038/nature02871
216. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many Roads to Maturity: microRNA Biogenesis Pathways and Their Regulation. Nat Cell Biol (2009) 11(3):228–34. doi: 10.1038/ncb0309-228
217. O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. MicroRNA-155 Promotes Autoimmune Inflammation by Enhancing Inflammatory T Cell Development. Immunity (2010) 33(4):607–19. doi: 10.1016/j.immuni.2010.09.009
218. O’Connell RM, Rao DS, Baltimore D. microRNA Regulation of Inflammatory Responses. Annu Rev Immunol (2012) 30:295–312. doi: 10.1146/annurev-immunol-020711-075013
219. Tili E, Michaille JJ, Calin GA. Expression and Function of micro-RNAs in Immune Cells During Normal or Disease State. Int J Med Sci (2008) 5(2):73–9. doi: 10.7150/ijms.5.73
220. Xiao C, Rajewsky K. MicroRNA Control in the Immune System: Basic Principles. Cell (2009) 136(1):26–36. doi: 10.1016/j.cell.2008.12.027
221. Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a New Immune-Regulatory Agent in Breast Milk. Silence (2010) 1(1):7. doi: 10.1186/1758-907x-1-7
222. Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-Related microRNAs are Abundant in Breast Milk Exosomes. Int J Biol Sci (2012) 8(1):118–23. doi: 10.7150/ijbs.8.118
223. Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, et al. Bovine Milk Exosomes Contain microRNA and mRNA and are Taken Up by Human Macrophages. J Dairy Sci (2015) 98(5):2920–33. doi: 10.3168/jds.2014-9076
224. Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, et al. Human Saliva, Plasma and Breast Milk Exosomes Contain RNA: Uptake by Macrophages. J Transl Med (2011) 9:9. doi: 10.1186/1479-5876-9-9
225. Kahn S, Liao Y, Du X, Xu W, Li J, Lönnerdal B. Exosomal MicroRNAs in Milk From Mothers Delivering Preterm Infants Survive in Vitro Digestion and Are Taken Up by Human Intestinal Cells. Mol Nutr Food Res (2018) 62(11):1701050. doi: 10.1002/mnfr.201701050
226. Liao Y, Du X, Li J, Lonnerdal B. Human Milk Exosomes and Their microRNAs Survive Digestion In Vitro and are Taken Up by Human Intestinal Cells. Mol Nutr Food Res (2017) 61. doi: 10.1002/mnfr.201700082
227. Gu Y, Li M, Wang T, Liang Y, Zhong Z, Wang X, et al. Lactation-Related microRNA Expression Profiles of Porcine Breast Milk Exosomes. PloS One (2012) 7(8):e43691. doi: 10.1371/journal.pone.0043691
228. Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are Absorbed in Biologically Meaningful Amounts From Nutritionally Relevant Doses of Cow Milk and Affect Gene Expression in Peripheral Blood Mononuclear Cells, HEK-293 Kidney Cell Cultures, and Mouse Livers. J Nutr (2014) 144(10):1495–500. doi: 10.3945/jn.114.196436
229. Laubier J, Castille J, Le Guillou S, Le Provost F. No Effect of an Elevated miR-30b Level in Mouse Milk on its Level in Pup Tissues. RNA Biol (2015) 12(1):26–9. doi: 10.1080/15476286.2015.1017212
230. Title AC, Denzler R, Stoffel M. Uptake and Function Studies of Maternal Milk-Derived Micrornas. J Biol Chem (2015) 290(39):23680–91. doi: 10.1074/jbc.M115.676734
231. Wang L, Sadri M, Giraud D, Zempleni J. Rnase H2-Dependent Polymerase Chain Reaction and Elimination of Confounders in Sample Collection, Storage, and Analysis Strengthen Evidence That microRNAs in Bovine Milk Are Bioavailable in Humans. J Nutr (2018) 148(1):153–9. doi: 10.1093/jn/nxx024
232. Fromm B, Tosar JP, Lu Y, Halushka MK, Witwer KW. Human and Cow Have Identical miR-21-5p and Mir-30a-5p Sequences, Which Are Likely Unsuited to Study Dietary Uptake From Cow Milk. J Nutr (2018) 148(9):1506–7. doi: 10.1093/jn/nxy144
233. Kirchner B, Buschmann D, Paul V, Pfaffl MW. Postprandial Transfer of Colostral Extracellular Vesicles and Their Protein and miRNA Cargo in Neonatal Calves. PloS One (2020) 15(2):e0229606–e. doi: 10.1371/journal.pone.0229606
234. Nguyen T. Unravelling the Mysteries of microRNA in Breast Milk. Nature (2020) 582:S12–S3. doi: 10.1038/d41586-020-01768-w
235. Martin C, Patel M, Williams S, Arora H, Brawner K, Sims B. Human Breast Milk-Derived Exosomes Attenuate Cell Death in Intestinal Epithelial Cells. Innate Immun (2018) 24(5):278–84. doi: 10.1177/1753425918785715
236. Li B, Hock A, Wu RY, Minich A, Botts SR, Lee C, et al. Bovine Milk-Derived Exosomes Enhance Goblet Cell Activity and Prevent the Development of Experimental Necrotizing Enterocolitis. PloS One (2019) 14(1):e0211431. doi: 10.1371/journal.pone.0211431
237. Dong P, Zhang Y, Yan DY, Wang Y, Xu X, Zhao YC, et al. Protective Effects of Human Milk-Derived Exosomes on Intestinal Stem Cells Damaged by Oxidative Stress. Cell Transplant (2020) 29:963689720912690. doi: 10.1177/0963689720912690
238. Miyake H, Lee C, Chusilp S, Bhalla M, Li B, Pitino M, et al. Human Breast Milk Exosomes Attenuate Intestinal Damage. Pediatr Surg Int (2020) 36(2):155–63. doi: 10.1007/s00383-019-04599-7
239. Pisano C, Galley J, Elbahrawy M, Wang Y, Farrell A, Brigstock D, et al. Human Breast Milk-Derived Extracellular Vesicles in the Protection Against Experimental Necrotizing Enterocolitis. J Pediatr Surg (2020) 55(1):54–8. doi: 10.1016/j.jpedsurg.2019.09.052
240. Reif S, Elbaum-Shiff Y, Koroukhov N, Shilo I, Musseri M, Golan-Gerstl R. Cow and Human Milk-Derived Exosomes Ameliorate Colitis in DSS Murine Model. Nutrients (2020) 12(9):2589. doi: 10.3390/nu12092589
241. Komine-Aizawa S, Ito S, Aizawa S, Namiki T, Hayakawa S. Cow Milk Exosomes Activate NK Cells and γδt Cells in Human PBMCs In Vitro. Immunol Med (2020) 43(4):161–70. doi: 10.1080/25785826.2020.1791400
242. Reber A, Donovan DC, Gabbard J, Galland K, Aceves-Avila M, Holbert KA, et al. Transfer of Maternal Colostral Leukocytes Promotes Development of the Neonatal Immune System I. Effects on Monocyte Lineage Cells. Veterinary Immunol Immunopathol (2008) 123:186–96. doi: 10.1016/j.vetimm.2008.01.034
243. Reber AJ, Donovan DC, Gabbard J, Galland K, Aceves-Avila M, Holbert KA, et al. Transfer of Maternal Colostral Leukocytes Promotes Development of the Neonatal Immune System Part Ii. Effects on Neonatal Lymphocytes. Vet Immunol Immunopathol (2008) 123(3-4):305–13. doi: 10.1016/j.vetimm.2008.02.009
244. Trend S, de Jong E, Lloyd ML, Kok CH, Richmond P, Doherty DA, et al. Leukocyte Populations in Human Preterm and Term Breast Milk Identified by Multicolour Flow Cytometry. PloS One (2015) 10(8):e0135580. doi: 10.1371/journal.pone.0135580
245. Baban B, Malik A, Bhatia J, Yu JC. Presence and Profile of Innate Lymphoid Cells in Human Breast Milk. JAMA Pediatr (2018) 172(6):594–6. doi: 10.1001/jamapediatrics.2018.0148
246. Artis D, Spits H. The Biology of Innate Lymphoid Cells. Nature (2015) 517(7534):293–301. doi: 10.1038/nature14189
247. Bedin AS, Molès JP, Rutagwera D, Nagot N, Kankasa C, Tylleskär T, et al. MAIT Cells, TCR γδ+ Cells and ILCs Cells in Human Breast Milk and Blood From HIV Infected and Uninfected Women. Pediatr Allergy Immunol (2019) 30(4):479–87. doi: 10.1111/pai.13037
248. Born WK, Reardon CL, O’Brien RL. The Function of γδ T Cells in Innate Immunity. Curr Opin Immunol (2006) 18(1):31–8. doi: 10.1016/j.coi.2005.11.007
249. Gabrilovich DI, Nagaraj S. Myeloid-Derived Suppressor Cells as Regulators of the Immune System. Nat Rev Immunol (2009) 9(3):162–74. doi: 10.1038/nri2506
250. Köstlin N, Schoetensack C, Schwarz J, Spring B, Marmé A, Goelz R, et al. Granulocytic Myeloid-Derived Suppressor Cells (Gr-MDSC) in Breast Milk (Bm); GR-MDSC Accumulate in Human BM and Modulate T-Cell and Monocyte Function. Front Immunol (2018) 9:1098. doi: 10.3389/fimmu.2018.01098
251. Köstlin-Gille N, Flaig L-A, Ginzel M, Arand J, Poets CF, Gille C. Granulocytic Myeloid-Derived Suppressor Cells in Breast Milk (Bm-Mdsc) Correlate With Gestational Age and Postnatal Age and Are Influenced by Infant’s Sex. Nutrients (2020) 12(9):2571. doi: 10.3390/nu12092571
252. Zheng Y, Corrêa-Silva S, de Souza EC, Maria Rodrigues R, da Fonseca FAM, Gilio AE, et al. Macrophage Profile and Homing Into Breast Milk in Response to Ongoing Respiratory Infections in the Nursing Infant. Cytokine (2020) 129:155045. doi: 10.1016/j.cyto.2020.155045
253. Moradkhani S, Jafarzadeh A, Bazargan-Harandi N, Baneshi MR, Mohammadi MM. Association of Reduced Count of interleukin-13-producing Cells in Breast Milk With Atopic Dermatitis in Infancy. Indian J Med Res (2018) 148(3):317–22. doi: 10.4103/ijmr.IJMR_1682_16
254. Ingram JL, Kraft M. Il-13 in Asthma and Allergic Disease: Asthma Phenotypes and Targeted Therapies. J Allergy Clin Immunol (2012) 130(4):829–42. doi: 10.1016/j.jaci.2012.06.034
255. Goudarzi N, Shabani R, Ebrahimi M, Baghestani A, Dehdashtian E, Vahabzadeh G, et al. Comparative Phenotypic Characterization of Human Colostrum and Breast Milk-Derived Stem Cells. Hum Cell (2020) 33(2):308–17. doi: 10.1007/s13577-019-00320-x
256. Ninkina N, Kukharsky MS, Hewitt MV, Lysikova EA, Skuratovska LN, Deykin AV, et al. Stem Cells in Human Breast Milk. Hum Cell (2019) 32(3):223–30. doi: 10.1007/s13577-019-00251-7
257. Aydın MŞ, Yiğit EN, Vatandaşlar E, Erdoğan E, Öztürk G. Transfer and Integration of Breast Milk Stem Cells to the Brain of Suckling Pups. Sci Rep (2018) 8(1):14289–. doi: 10.1038/s41598-018-32715-5
258. Borhani-Haghighi M, Navid S, Mohamadi Y. The Therapeutic Potential of Conditioned Medium From Human Breast Milk Stem Cells in Treating Spinal Cord Injury. Asian Spine J (2020) 14(2):131–8. doi: 10.31616/asj.2019.0026
259. Görs S, Kucia M, Langhammer M, Junghans P, Metges CC. Technical Note: Milk Composition in Mice–Methodological Aspects and Effects of Mouse Strain and Lactation Day. J Dairy Sci (2009) 92(2):632–7. doi: 10.3168/jds.2008-1563
260. Mirza AH, Kaur S, Nielsen LB, Størling J, Yarani R, Roursgaard M, et al. Breast Milk-Derived Extracellular Vesicles Enriched in Exosomes From Mothers With Type 1 Diabetes Contain Aberrant Levels of Micrornas. Front Immunol (2019) 10:2543. doi: 10.3389/fimmu.2019.02543
Keywords: microbiota, innate immune system, pregnancy, gestation, early life, birth, breast milk, neonate
Citation: Kalbermatter C, Fernandez Trigo N, Christensen S and Ganal-Vonarburg SC (2021) Maternal Microbiota, Early Life Colonization and Breast Milk Drive Immune Development in the Newborn. Front. Immunol. 12:683022. doi: 10.3389/fimmu.2021.683022
Received: 19 March 2021; Accepted: 27 April 2021;
Published: 13 May 2021.
Edited by:
Christoph Siegfried Niki Klose, Charité – Universitätsmedizin Berlin, GermanyReviewed by:
Michael G. Constantinides, The Scripps Research Institute, United StatesCopyright © 2021 Kalbermatter, Fernandez Trigo, Christensen and Ganal-Vonarburg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie C. Ganal-Vonarburg, c3RlcGhhbmllLmdhbmFsQGRibXIudW5pYmUuY2g=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.