
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 12 July 2021
Sec. Multiple Sclerosis and Neuroimmunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.679055
This article is part of the Research TopicGut Microbiota-Brain Axis in Enteric and Central Neuronal Functions in Health and Neuropsychiatric DisordersView all 10 articles
The peripheral nervous system consists of sensory circuits that respond to external and internal stimuli and effector circuits that adapt physiologic functions to environmental challenges. Identifying neurotransmitters and neuropeptides and the corresponding receptors on immune cells implies an essential role for the nervous system in regulating immune reactions. Vice versa, neurons express functional cytokine receptors to respond to inflammatory signals directly. Recent advances in single-cell and single-nuclei sequencing have provided an unprecedented depth in neuronal analysis and allowed to refine the classification of distinct neuronal subsets of the peripheral nervous system. Delineating the sensory and immunoregulatory capacity of different neuronal subsets could inform a better understanding of the response happening in tissues that coordinate physiologic functions, tissue homeostasis and immunity. Here, we summarize current subsets of peripheral neurons and discuss neuronal regulation of immune responses, focusing on neuro-immune interactions in the gastrointestinal tract. The nervous system as a central coordinator of immune reactions and tissue homeostasis may predispose for novel promising therapeutic approaches for a large variety of diseases including but not limited to chronic inflammation.
The nervous system in multi-cellular organisms consists of a complex network of neurons, which can rapidly and precisely transmit signals. Signal transmission within the nervous system is mediated by the release of neurotransmitters or neuropeptides from the presynaptic neuron into the synaptic cleft, the engagement of the cognate receptor on the postsynaptic neuron, and the elicitation of a signaling cascade within the postsynaptic neuron. The nervous system is organized in circuits linking afferent sensory input with a broad array of reactions at effector sites. In this way, the nervous system coordinates physiological functions, such as behavior, motor functions, blood pressure and hormone release (1–4). The nervous system is classified into the central nervous system (CNS) and the peripheral nervous system (PNS). The CNS, which includes the brain and the spinal cord, is enclosed by the dura mater. The PNS, placed outside of the dura mater, is sub-classified into the somatic and autonomic nervous systems. The somatic nervous system consists of peripheral somatosensory nerves, which convey afferent signals and efferent nerves controlling motor functions, e.g. regulating movements of extremities. The autonomic nervous system can be functionally distinguished into the sympathetic, the parasympathetic, and the enteric nervous system (5). The parasympathetic and sympathetic nervous system are typical functional counter players for opposing physiological functions: the rest-and-digest (parasympathetic nervous system) and the fight-or-flight reaction (sympathetic nervous system). These functions are highly conserved across species and build the basis for survival during external threats (6). The third component of the autonomic nervous system is the gut intrinsic enteric nervous system (ENS), which controls intestinal movement, mixing of ingested food and the secretion of fluids. In the gastrointestinal tract, intrinsic neurons are those whose cell bodies lie within the organ, whereas extrinsic nerves (e.g. sensory nerve fibers) have their cell bodies outside the innervated organ. Typically, the soma of extrinsic sensory afferents is located within dorsal root ganglia, celiac ganglia, superior or inferior mesenteric ganglia or the nodose/jugular ganglia. The intrinsic and independent coordinator ability of the ENS has been underlined in experiments following extrinsic denervation, where a lack of extrinsic signals only impaired physiologic functions of the intestine to a minor degree. Conversely, loss of the intrinsic ENS can be disastrous, as shown in Hirschsprung or Chagas disease, where intestinal motor functions are significantly reduced or absent (7, 8).
The ENS is composed of different neuronal populations, which fulfil specific physiologic functions. The traditional classification of neurons solely according to their chemical signature is not entirely sufficient to define a functional type of neuron because similar neurotransmitters seem to exert different physiologic functions. In the same line, anatomical/morphological classifications do not adhere to specific neuronal functions because of similar shapes of enteric neurons in distinct functional classes. Thus, current neuronal classifications need to be extended by a broader array of markers to better understand the peripheral nervous system in detail. Single-cell RNA-sequencing is a powerful tool, which provides several vital transcripts per cell and can zoom in at the potential correct resolution.
Here we review the current understanding of the function of different neuronal subsets that relay the signals to subsets of immune cells in the peripheral nervous system, which regulate intestinal physiology as a response to environmental challenges and physiologic perceptions.
The intestine has its own nervous system, the ENS, which operates to a large degree autonomously and outside of voluntary control, despite being innervated by extrinsic nerve fibers. The ENS is the largest collection of neurons outside the CNS comprising hundreds of millions of neurons. It is the major coordinator of physiologic bowel functions, including but not limited to peristaltic movement. The importance of the autonomous function of the ENS has been demonstrated by experiments using extrinsic denervation of the intestine, in which the bowel function was only mildly affected by the cessation of extrinsic signals (9).
The ENS has a characteristic spider web structure, is embedded in the intestinal wall and closely associated with the muscle layers. The somas of enteric neurons mainly cluster in two anatomically distinct but strongly interconnected ganglionated plexuses (10). The outer, myenteric plexus (Auerbach Plexus) lies within the longitudinal and the circular muscle layer, and the inner, submucosal plexus (Meissner Plexus) is located below the muscle layers (11).
While a classification based on morphological aspects has been proposed, this classification does not take into account that functionally similar neurons can vary in their morphology. Using open-end single-cell or single-nuclei sequencing approaches, several studies have revealed the transcriptomic landscape of the ENS and thus provide a classification, which is more functionally substantiated. A classification based on a transcriptional code, as discussed in the following paragraph, includes marker genes encoding for receptors, ion channels and neuropeptides and can thus broaden our understanding of physiologic and immunologic functions.
The intrinsic micro-circuitry regulation of intestinal function consists of five neuronal classes with distinct functional specializations. Sensory neurons, also referred to as intrinsic primary afferent neurons (IPANs), detect chemical and physical alterations in the intestine and transmit the signal via interneurons to excitatory motor neurons, inhibitory motor neurons or secretomotor/vasodilator neurons (Figure 1). Enteric neurons are replenished from neuronal stem cells of the intestine. While the traditional sub-classification was based on the neuronal type, the anatomic location and axonal projection as well as neurochemical signature, recent advances in single-cell sequencing has enabled a comprehensive clustering based on gene expression.
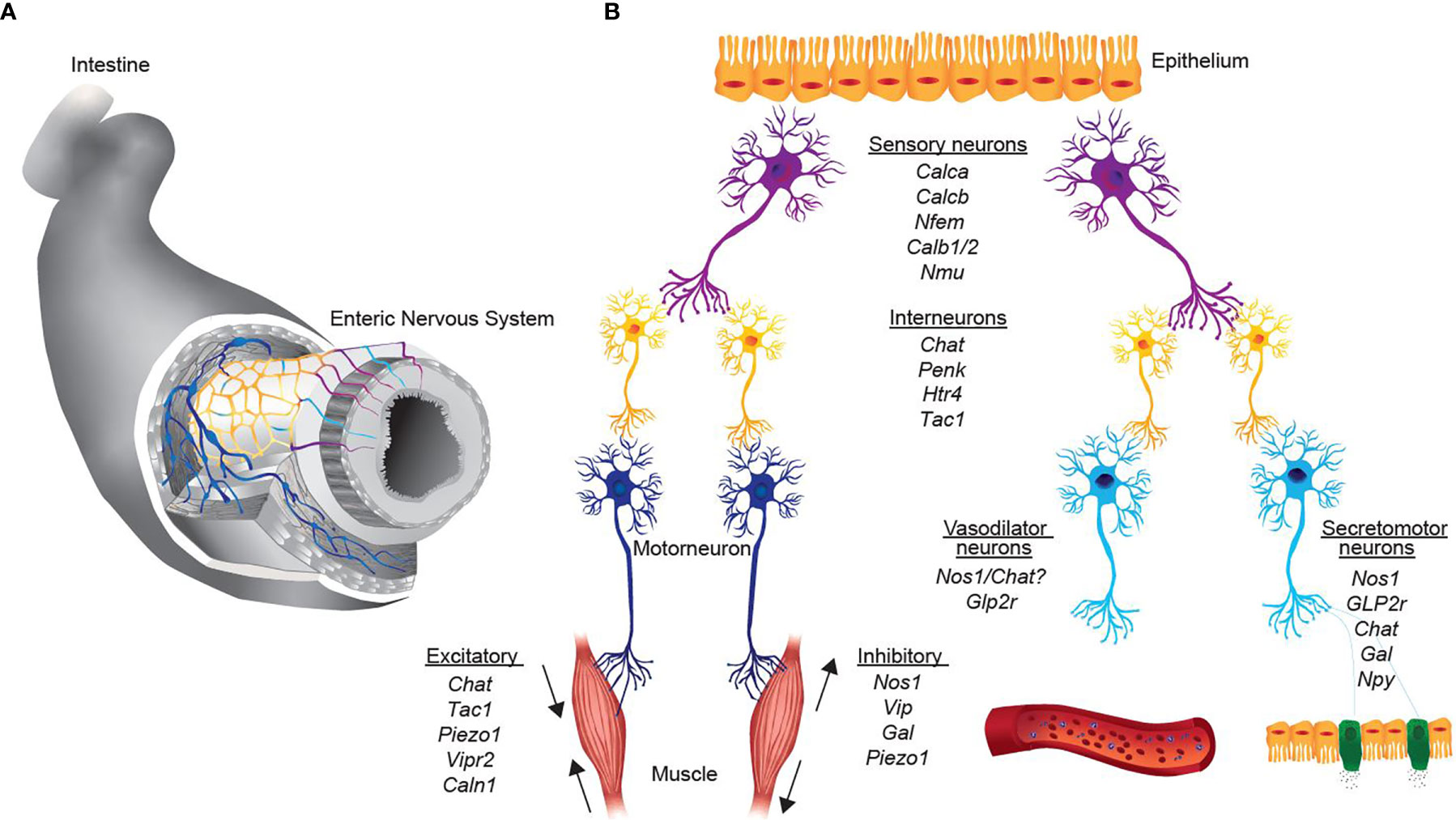
Figure 1 Representation of neurons, key transcripts and functional assignments in the intrinsic enteric nervous system. (A) 2-Dimensional scheme of the enteric nervous system. Colors represent the schematic representation of distinct functional neuronal groups. (B) Simplified representation from sensing of the environment of different stimuli (sensory neurons, purple), signaling transmission to interneurons (yellow) and excitation of effector neurons (motor neurons, dark blue; vasodilator and secretomotor, light blue). Italics are the key transcripts of the respective neuronal group.
Single-cell sequencing of the ENS using fluorescence-activated sorting of Wnt1-Cre; R26Tomato mice revealed 1105 enteric neurons (with ~ 3066 genes detected on average) and eventually found nine clusters of neurons in the muscular sheet of the intestine (12). Based on neuro-transmitters nitric oxide synthase 1 (NOS1) and choline acetyltransferase (ChAT), two main groups can be classified, NOS1+ neurons and ChAT+ neurons. The authors assigned three distinct neuronal clusters of nitrergic neurons, which comprise inhibitory motor neurons and secretomotor/vasodilator neurons (Table 1). Further, 6 clusters of cholinergic neurons, which express ChAT and the solute carrier family 5 member 7 (Slc5a7, gene encodes for an ion transporter) could be distinguished (12). ChAT+ neurons are further subdivided into excitatory motor neurons, sensory neurons and interneurons according to their gene expression (Table 1).
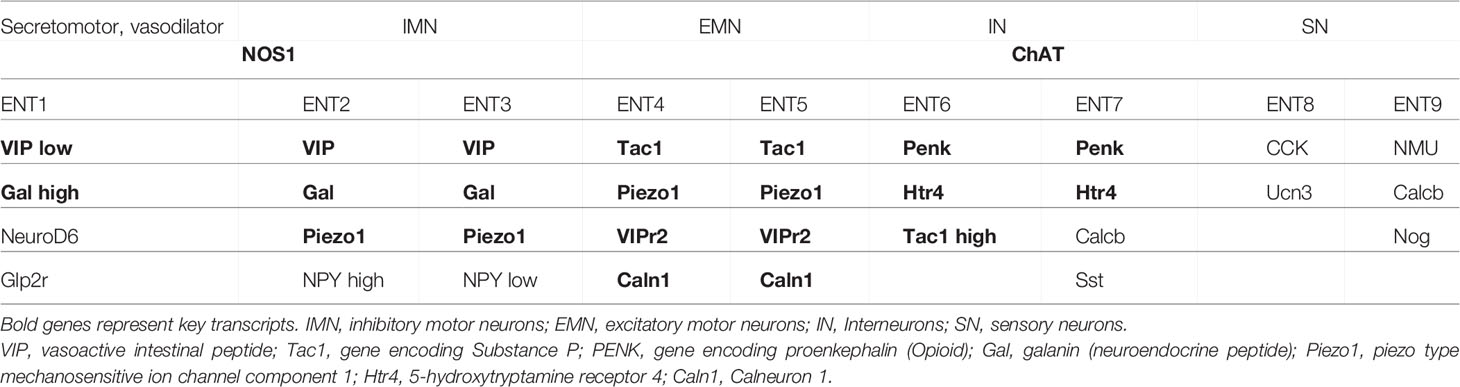
Table 1 Reference genes for the identification of neuronal subsets based on unbiased single-cell RNA-sequencing (12–14).
Using the pan-neuronal Baf53b-cre deleter mice crossed to R26R-tomato to enable sort-purification of enteric neurons from the myenteric plexus of the small intestine combined with single-cell RNA-sequencing, 4,892 high-quality enteric neurons have been analyzed and 12 classes of neurons could be identified based on their gene expression (13). Matching these results to known entities of enteric neurons based on functional properties, the authors could propose the following classification: Populations 1-4 can be classified as excitatory motor neurons predominantly defined via the expression of Tac1 and Calb2. Class 8 and 9 have been assigned to inhibitory motor neurons and are characterized by the expression of Nos1/Gal/VIP/Npy. Different classes (Populations 6, 7, 11) of sensory intrinsic primary afferent neurons have been identified based on the known markers Calca/Calcb/Nfem/Calb1/Calb2 and the selective expression for NMU, Ucn-3/Cck or Nxph2. Interneurons are represented with a mixed neurochemical signature, such as Nos1/ChAT for Interneurons group 1, Sst/Calcb/Calb2 for interneurons group 2. The co-expression of Nos1 and ChAT was also found in the dataset of Zeisel and, thus, may reflect the profile of interneurons, which connect different neuronal groups (12). While all the single-cell RNA-sequencing studies nicely delineate distinct populations of enteric neurons based on specific transcripts, dissociation and sort-purification of entire neurons from the muscular sheet of the intestine might not equally represent all neurons in the ENS, some of which might be sensitive to the isolation or sorting procedure. It should be noted that even for tissue-resident immune cells, which are presumably easier to release from the fabric of the tissue after digestion, a discrepancy between cells recovered after digestion and those detected in situ was reported (15).
To overcome this potential limitation, a study by the Regev lab performed single-nuclei sequencing by using sort-purification of labelled nuclei with a nuclear-tagged fluorescent protein (14). This approach is expected to result in an equal representation of neuronal nuclei present in the tissue. The authors sequenced 1´187´535 colonic and ileal nuclei and eventually profiled 2´657 neuronal nuclei (with 7´369 genes detected per nucleus). By using nuclear isolation and sequencing, Drokhlyansky and colleagues identified 21 neuronal populations based on known marker genes. These 21 identified neuronal classes could be further broadly sub-classified into 5 populations of ChAT+Nos1+ double-expressing putative excitatory motor neurons, 7 populations of Nos1+ inhibitory motor neurons (4 subsets are Nos1+Vip+), 2 populations of Glp2r+ secretomotor and vasodilator neurons, 4 populations of CGRP+ sensory neurons, 3 populations of Penk+ Interneurons (Table 1). Apart from the transcripts at the single-cell level, there is limited data available that reports a deeper characterization of neuronal subsets after sort purification. Thus, the detection limit of single-cell RNA-sequencing can miss important transcripts in certain subpopulations of cells. Purification of neuronal subpopulations may allow to assign defined functions and to delineate the neuronal subclasses in more detail in the near future. In-between the above-mentioned studies, minor discrepancies in the representation of neuronal subclasses were reported, which may be due to different reporter mouse strains used for sort purification, different isolation/sequencing techniques or differences in bioinformatic analyses. However, even though transcripts may differ in between different single-cell RNA-sequencing datasets, the known functional subsets of enteric neurons are uniformly present in all studies. Consequently, each study concluded putative functional roles of identified neuronal subclasses by associating gene expression to a certain known function, even though without experimental proof. Thus, future studies need to experimentally confirm functional coherence of the identified transcripts.
Within the enteric nervous system, only one study performed sequencing of human neurons (14). By using MIRACL-sequencing, a total of 436'202 human nuclei were profiled and 1'445 neurons clustered into 14 neuronal subsets (with ~ 4302 genes detected on average). By comparing human and mouse colon neuronal nuclei, the authors found strong congruence between species but also differences in ENS composition, such as a higher abundance of motor neurons and a lower diversity of sensory neurons, interneurons and secretomotor/vasodilator neurons in humans. The fact that the abundance of neurons is relatively low, future studies should aim to address to enrich neurons in specimens to gain deeper insights in regulated neuronal gene expression.
Understanding which genes enteric neurons have adopted for sensing the immune system is one of the burning questions in the field today. We aimed to provide an overview by re-examining published single-cell and single-nuclei datasets deposited in publicly available databases in silico for expression of cytokine receptors, chemokine receptors, NOD-like receptors (NLRs) and toll-like receptors (TLRs) (Figures 2–4) (12, 14). With regard to cytokine receptors Il11ra1, Il4Ra, Il13ra1, and Il6st (Figure 2) were detectable in enteric neurons.
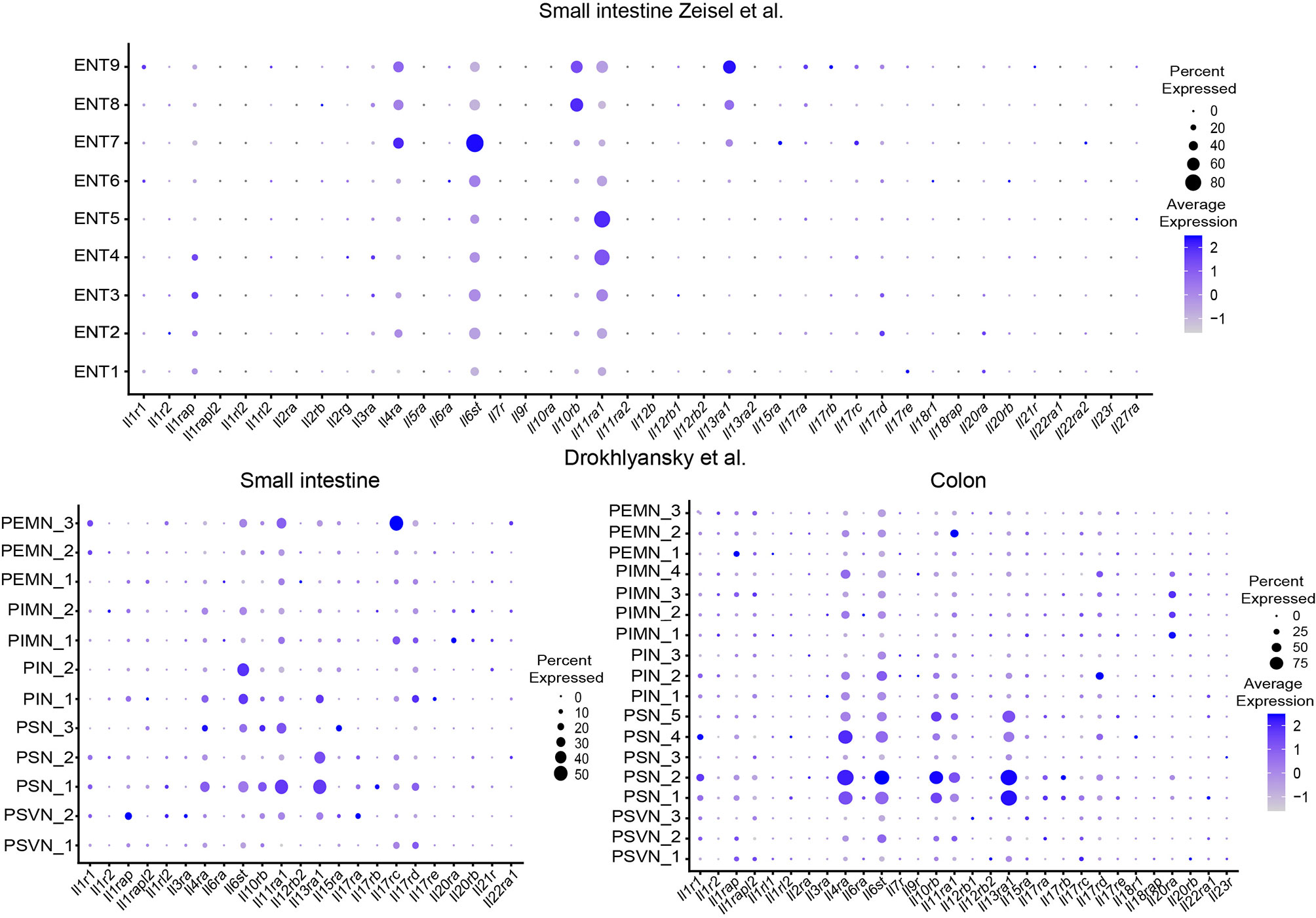
Figure 2 Expression of interleukin receptors in different ENS subsets. Dotplots showing the percentage of expressing cells as well as average expression within the indicated identified neuronal clusters for selected murine interleukin receptor genes. Data was downloaded from http://mousebrain.org/ (12) or from https://singlecell.broadinstitute.org/ (14). Genes not present in the plot showing the Drokhlyansky data have been filtered. All dataset were normalized and transformed before the plots were created using the Seurat package (16).
The expression of the type 2 cytokine receptors Il4Ra and IL13ra1 in sensory ganglia and their functional role has been studied in the context of chronic itch in mice and humans (17). Functionally, type 2 cytokines can directly activate sensory neurons, activate itch-sensory pathways and are critical players in the development of chronic itch sensations. Furthermore, type 2 cytokines induce the JAK signaling pathway in neurons and JAK inhibitors have been successfully tested in chronic itch diseases in humans (17). In the same line, the important initiator of type 2 responses thymic stromal lymphopetin (TSLP) activates TRPA1+ sensory neurons and promotes itch responses in mice (18). However, their role in the ENS needs to be delineated in future studies.
Recent studies suggest a role for pattern recognition receptors (PRR) in DRG neurons and pain sensation (19). Given its location, the ENS is constantly exposed to microbial factors. A role for the ENS in sensing microbial metabolites was found but the importance of PRR on enteric neurons is not well defined. Reanalysis of the datasets shows expression of Nlrp6 and TLR3 and is consistently found in enteric neurons among several studies (Figures 3 and 4).
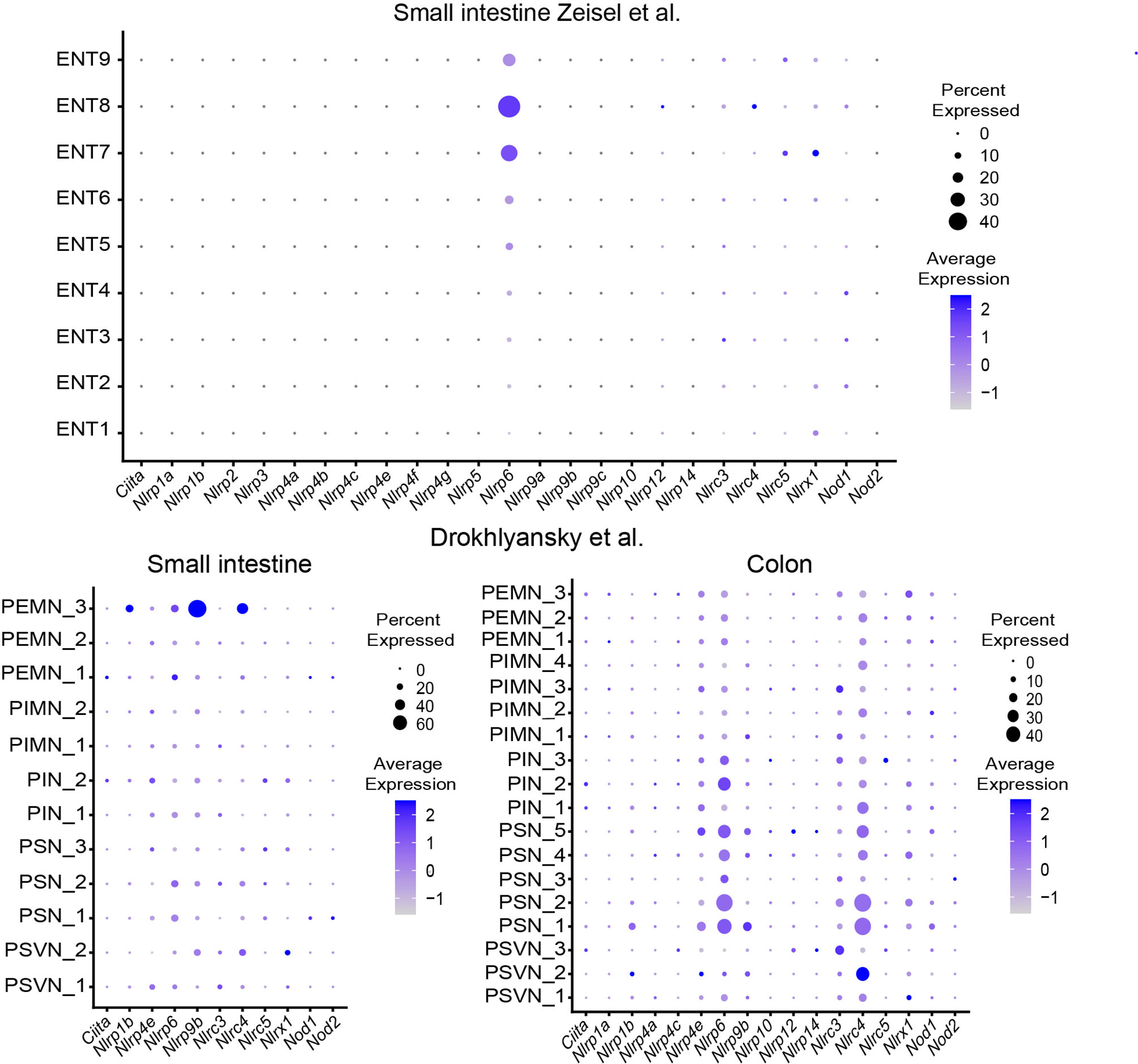
Figure 3 Expression of NOD-like receptors in different ENS subsets. Dotplots showing the percentage of expressing cells as well as average expression within the indicated identified neuronal clusters for selected murine NOD-like receptor genes. Data was downloaded from http://mousebrain.org/ (12) or from https://singlecell.broadinstitute.org/ (14). Genes not present in the plot showing the Drokhlyansky data have been filtered. All dataset were normalized and transformed before the plots were created using the Seurat package (16).
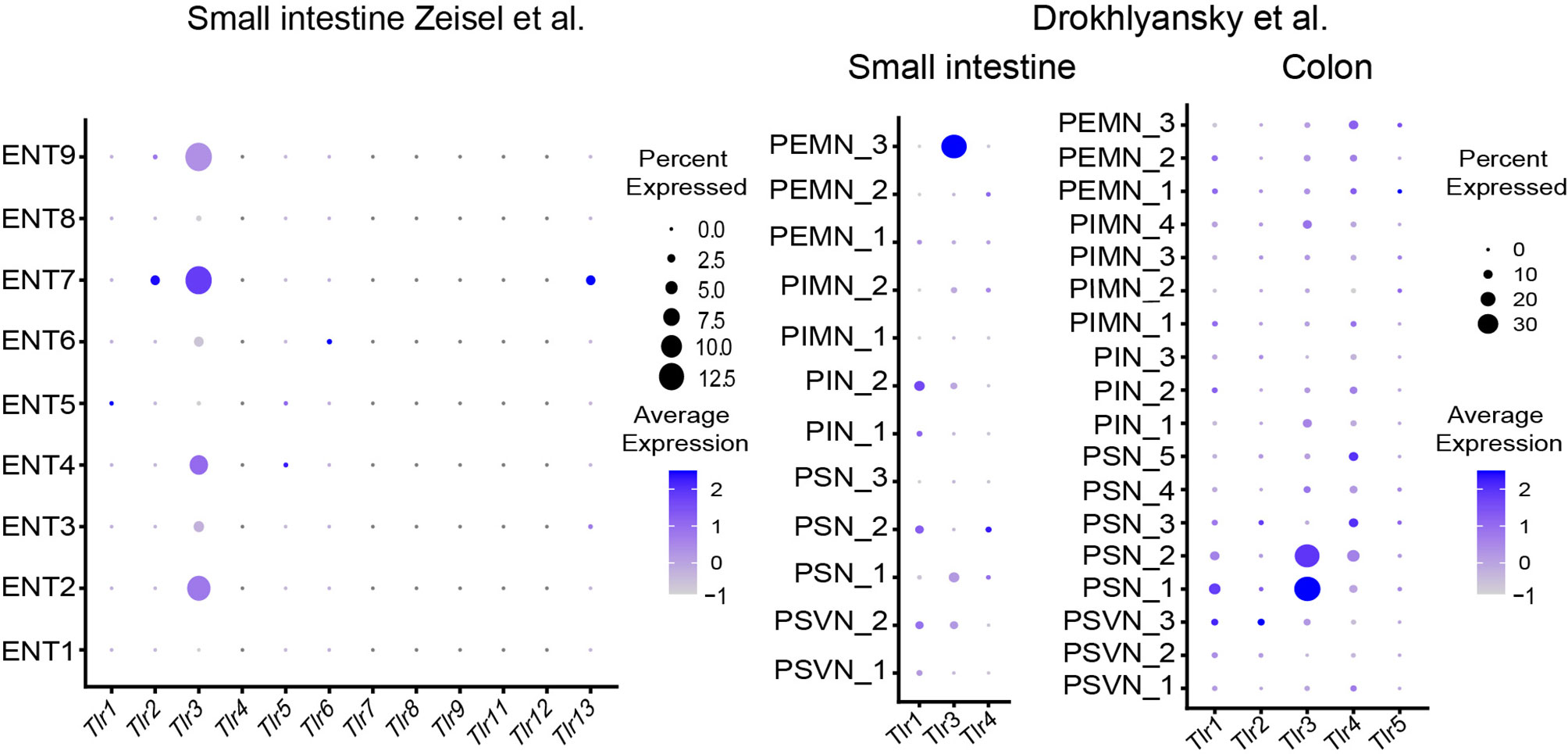
Figure 4 Expression of Toll-like receptors in different ENS subsets. Dotplots showing the percentage of expressing cells as well as average expression within the indicated identified neuronal clusters for selected murine Toll-like receptor genes. Data was downloaded from http://mousebrain.org/ (12) or from https://singlecell.broadinstitute.org/ (14). Genes not present in the plot showing the Drokhlyansky data have been filtered. All dataset were normalized and transformed before the plots were created using the Seurat package (16).
The role of the inflammasome components Nlrp6 and caspase 11 has recently been highlighted to control enteric neuronal cell death and glucose metabolism in a microbiota-dependent manner (20). In fact, microbiota-depletion with antibiotics leads to loss of enteric associated CART+ neurons in a Nlrp6 and Caspase 11 dependent manner. Analysis of SPF-colonized and GF mice revealed reduction in blood glucose levels, which was linked to CART+ neurons suggesting the regulation of blood glucose level independent from CNS control (20). The role of TLRs in neurons is intriguing because it may suggest direct microbial sensing of enteric neurons (11). However, the role of TLR3 in the ENS remains elusive.
The expression pattern of cytokine receptors, NLRs and TLRs, and chemokine receptors can guide the readers to design their research projects accordingly (Figures 2–4, Supplementary Figure 1).
Within the group of motor neurons, a sub-classification proposed by Furness and colleagues (21) distinguishes five main classes of motor neurons present in the intestine including excitatory motor neurons, inhibitory motor neurons, secretomotor/vasodilator neurons, secretomotor neurons that are not vasodilator and neurons to enteroendocrine cells (Figure 1).
Excitatory motor neurons use the main neurotransmitter acetylcholine (Ach) for signal transmission and are thus distinguishable from inhibitory motor neurons, which use nitric oxide (NO) as the main neurotransmitter (5). This difference in signal transmission of excitatory versus inhibitory motor neurons becomes evident in anticholinergic medications, such as atropine or antidepressants, that typically lead to constipation due to the lack of signals from excitatory motor neurons (22). Furthermore, conditional deletion of ChAT in neural-crest derived neurons by using the Wnt1-cre driver in mice resulted in gastrointestinal dysmotility, dysbiosis and eventually death of the mice at post-natal day 30. This phenotype highlights the extraordinary role of ChAT for host physiology and its role in the coordination of motor functions (23).
Even though Ach is the dominant neurotransmitter used by excitatory motor neurons, these neurons show a residual excitation after muscarinic block suggesting that other neurotransmitters are involved (24, 25). The residual excitation is assumed to be mediated by tachykinins, in particular Substance P (gene Tac1), which engage on NK1 and NK2 receptors on muscle cells (26). Altogether, these data indicate that excitation of motor neurons is mediated by cholinergic and non-cholinergic neurotransmitters and each excitation may be fine-tuned dependent on the transmitter involved.
On the contrary, experiments in mice with the deletion of NOS1 revealed that neurons normally expressing NOS remain intact (apart from grossly enlarged stomach due to pyloric stenosis) and respective mice do not show evident histopathological abnormalities (27). These results suggest that inhibitory neurons are also co-regulated by other neurotransmitters to terminate the excitatory signal. Several transmitters/neuropeptides have been described to regulate inhibitory motor neurons including adenosine triphosphate (ATP) (28), vasoactive intestinal peptide (VIP) (29), pituitary adenylyl cyclase activating peptide (PACAP) and carbon monoxide (30) in addition to NO as neuromuscular transmitters (31).
In summary, excitatory and inhibitory motor neurons in the ENS control muscle contraction and relaxation in an autonomous manner (Figure 1). In this way, ingested food is squirted, mixed with digestive enzymes and eventually transported aborally. Based on gene profiles detected in single-cell RNA-sequencing studies, neurons can be associated to either excitatory or inhibitory motor neurons. However, the exact functional role of the subpopulations within excitatory or inhibitory motor neurons remains elusive.
In this context, putative intrinsic but also extrinsic neurons are equipped with mechanosensory ion channels. Different cell types have been described to use mechanosensitive ion channels to detect alterations in mechanical forces. In this way, neurons can adapt their functions dependent on mechanical perturbations. Piezo ion channels are important signaling cationic ion channels for mechanosensation and their role in the extrinsic nervous system and the enteric nervous system are emerging (32). Functionally in the peripheral nervous system, PIEZO ion channels have been linked to the sensation of nociceptive signals, proprioception and touch. An interesting finding from an immunologic perspective is that mechanical processes can also regulate immune cell activity (33). The authors found that the mechanosensory ion channel PIEZO1 is able to mount a proinflammatory reprogramming in macrophages. Upon conditional deletion of PIEZO1 in myeloid cells in the context of P. aeruginosa infection in the lung, the PIEZO1-mediated mechanosensation protected against bacterial infection (33). This data argues for similar mechanisms used by the nervous and the immune system to adapt physiological functions to mechanical forces. Thus, mechanosensory ion channels may be important for neuro-immune disturbances in ileus and other gastrointestinal diseases. However, their exact role, in particular the role of PIEZO1 in the gastrointestinal tract, needs further investigation.
The intestine is constantly exposed to potential external threats, such as the commensal microbiota, in addition to the needs to control bowel absorption and to avoid electrolyte disturbances. Intrinsic secretomotor neurons have their cell bodies predominantly in the submucosa and play an important role in the regulation of these functions (Figure 1). In the guinea-pig small intestine, three different ChAT expressing secretomotor/vasodilator neurons have been described. Co-expression of ChAT and Calretinin, both of which innervate glands and arterioles, has been linked to vasomotor function. Functionally, excitation of vasomotor neurons lead to vasodilation and, thus, increase local blood flow (34). This is interesting because the topical application of Ach on vascular smooth muscle normally causes vasoconstriction and thus Ach seems to exert context-dependent effects. The two other neuronal classes described in the guinea-pig can be functionally assigned to different secretomotor neuronal populations. These neurons and their associated regulatory mechanisms are of great importance in host physiology because interference with secretory mechanisms may lead to constipation or diarrhea. Secretomotor neurons are activated via intrinsic primary afferent neurons following chemical or physical interactions with luminal contents (34). Such stimuli can activate cAMP or Ca2+ activated chloride ion channels, which regulate the movements of chloride towards the intestinal lumen or through epithelial cells into the lamina propria and cause water to diffuse along (35, 36). Based on expressed neuro-transmitters, two types of secretomotor neurons can be distinguished; the first group co-expresses ChAT and neuropeptide Y, whereas the second group is characterized by the expression of ChAT only (37). However, future studies need to delineate the functional role of each neuro-transmitter in detail, which will allow to correctly assign the transcriptional profiles to neuronal classes.
The ENS needs to monitor mechanical and chemical perturbations in order to react to incoming physiologic or pathologic signals and to tune local cellular components (Figure 1). Extrinsic sensory innervation includes spinal and vagal afferent neurons, whose cell bodies lie outside the intestine, e.g. in the Dorsal Root Ganglia or in the nodose/jugular ganglia. While anatomically defined, distinct afferent neurons originating from the intestine project into dorsal root ganglia, the vagus nerve innervates the entire intestine with a denser innervation in the small intestine as compared to the large intestine. Both types of extrinsic axons, DRG-derived or vagal, project into the inner and outer layers of the intestinal wall and perceive signals that are important to guide homeostatic functions. The respective neuronal population and its functional role are discussed in the next paragraph (Dorsal Root Ganglia section of the review).
Intrinsic innervation is characterized by cell bodies lying inside the gut wall. Neurons that perceive and integrate sensory information in the ENS are called intrinsic primary afferent neurons (IPANs). IPANs are integrated in the neuronal architecture to act in concert with motor neurons, interneurons and secretomotor neurons to direct homeostatic functions depending on external stimuli and the needs of digestive functions (38). Similar to the ENS, sensory neurons can be classified according to their neurochemical signature. Sensory IPANs have been identified based on the known markers Calca/Calcb/Nfem/Calb1/Calb2 and the selective expression for NMU, Ucn-3/Cck or Nxph2.
Depending on the function of the neurons, three types of IPANs are identified using small intestine of the guinea pig as a model organism. First, chemosensitive IPANs respond to chemicals present on the surface of the small intestine (39). Because nerve fibers do not reach the surface of the intestine, and thus, do not come into direct contact with luminal contents, chemical changes in the intestinal lumen have to be sensed indirectly via signals from epithelial cells (40). Enteroendocrine cells are specialized epithelial cells, which for instance release 5-Hydroxytryptamine (5-HT) upon mucosal chemical or mechanical stimulation, which is a potent stimulator of IPANs and act as a signal transducer (41). However, knowledge concerning how endothelial cells communicate with the ENS remains a black box. Analysis of single-cell RNA-sequencing data from epithelial cells suggest the presence of potential stimulating peptides, such as Substance P, cholecystokinin, ghrelin, and synthesizing enzymes of signaling amines (5-HT) in these cells (42). In line with these observations, receptors for the respective stimulators, such as the 5-HT3 receptor and Substance P receptor 1, is expressed in IPANs suggesting an enteroendocrine to ENS signaling hub. Thus, sensory neurons functionally assigned to chemosensitive IPANs could be identified by a distinct gene profile for receptors of stimulating peptides and amines. However, the stimulation of other classes of IPANs, the mucosal mechanoreceptors, is also mostly indirect via 5-HT released from enterochromaffine cells (43). The second functional class described are stretch-sensitive IPANs that react to mechanical tension/distortion (38). Interestingly, these neurons seem to be not only mechanosensitive, but can also directly act as inhibitory motor neurons (44). Thus, their gene profile may be mixed and complicated for a functional assignment. The third group of IPANs are mucosal mechanoreceptors, which may be identified by putative mechanosensitive ion channels (e.g. Piezo2). However, as stated above, the stimulation of mucosal mechanoreceptors is mostly indirect and further studies need to unravel their profiles in more detail.
The interneurons, as the name suggests, are neurons, which connect functionally diverse neuronal populations in order to complete a neuronal circuit of varying complexities. For instance, interneurons receive signal from sensory neurons (which sense environmental perturbations) and relay the signal to either inhibitory or excitatory motor neurons to trigger an effector function to the sensed stimulus (Figure 1). The interneurons, as a type of 'bridging neurons', are multipolar and can be excitatory or inhibitory. They are primarily located within the myenteric plexus forming uniaxonal chains along the length of the gut with the ascending interneurons projecting orally and the descending interneurons projecting anally. There are different interneurons within the myenteric plexus with distinct neurochemical signatures, which can differ between gut regions. Using the guinea pig as a model organism, one class of excitatory ascending interneurons and three classes of descending interneurons have been described in the small intestine (45). In the colon on the other hand, three neurochemical classes of ascending interneurons and four classes of descending interneurons have been identified (46). These anatomic differences may come from the distinct local environment (e.g. microbiota) and the functional role it has to execute in different anatomic regions. The method applied by the authors was to morphologically distinguish interneuronal subpopulations. Whereas, upon distinguishing the subpopulations of interneurons based on their transcriptional signature, Morarach et al. was able to identify two subpopulations: one expressing motor-neuron-like inhibitory and excitatory neuropeptides Nos1/ChAT and the other expressing sensory-neuron-like neuropeptides Sst/Calcb/Calb2 (13). The discrepancy in the findings/characterization of these studies might be a result of difference in either their method of evaluation (morphological vs transcriptomic) or the anatomic locations within the gut studied. However, several studies demonstrate that neurochemical markers of the interneurons overlap with other neuronal populations and, as such, do not have their unique neurochemical signature.
Since nerve fibers do not reach the lumen of the intestine under homeostatic conditions, microbiota and metabolites could directly stimulate neurons by penetrating the epithelial barrier or indirectly through epithelial cells or other cell types. Epithelial enteroendocrine cells were shown to be directly innervated by neurons and transduce signals to the CNS within milliseconds after being exposed to sugar (47). Although the finding that epithelial cells are innervated is still controversial (48), sensing of secondary signals released from epithelial cells by enteric neurons is likely to contribute to the regulation of intestinal homeostasis.
Using AAV particles for neuronal-specific deletion of aryl hydrocarbon receptor (Ahr) in enteric neurons, Obata et al. could demonstrate a pivotal functional for enteric neurons in metabolite sensing (49). Genetic ablation of Ahr resulted in reduced peristalsis and increased intestinal transit times. Further, Ahr expression was altered in germ-free mice suggesting a role for commensal microbiota in regulating Ahr expression in neurons. Several studies have reported that the absence of an intact microbiota resulted in activation of neurons and alterations of neuronal composition in ENS (48–50) as well as effects on CNS function were reported (51, 52). This leads to the question if and how neurons sense microorganisms. In vitro data show that neurons are able to respond to PAMPs, for example LPS via an TLR4-independent, or excretory secretory product of helminth in an Myd88-dependent manner (53, 54). Analysis of enteric neuronal populations in germ-free mice has revealed alterations in the enteric nervous system, in particular of NOS1+ neurons, although there is some discrepancy between the studies whether NOS1+ neurons are over- or underrepresented (50, 55, 56). Similar findings were reported in Myd88-deficient mice arguing for a role of TLRs in microbial sensing and development of the ENS. However, it remains unclear if the phenotype is explained by direct sensing of neurons via TLRs or indirect mechanisms via secondary messengers. Conditional gene targeting of TLR4 using WNT1-Cre resulted in altered ENS development. However, it should be mentioned that this targeting strategy does not exclude a major role for glial cells in TLR4 microbial sensing instead of or in addition to enteric neurons (13, 50).
Investigating neuronal-specific gene expression in germ-free animals by using the Ribotag system activated by SNP25Cre, Muller et al. could detect cell death of CART+ neurons, which are involved in regulation of blood glucose levels, mediated by NLRP6 and Casape-11-dependent pathway (20). Further, the absence of an intact commensal microbiota is detected in parts via SCFA and GPR41 resulted in an increased sympathetic neuronal activity, as measured by c-FOS expression in the coeliac-superior mesenteric ganglia, activation of glutamatergic sensory neurons in the brainstem and decreased intestinal transit time (48). Similarly, cell death controlled by a NLRP6 and Casape-11-dependent pathway as well as sympathetic activation have been reported following Salmonella enterica infection resulting in decreased intestinal motility. In this context, activation of the sympathetic nervous system and release of norepinephrine can instruct a tissue-protective program in muscularis macrophages characterized by Arginase 1 and BMP-2 expression and responsiveness to enteric neuron-derived CSF-1 (48, 57).
Altogether, the signaling circuits sensing mucosal homeostasis are emerging and the integration of these signals in the CNS can provide on more holistic view on how intestinal homeostasis is regulated.
Dorsal root ganglia (DRGs) comprise of neuronal cell bodies of sensory neurons, whose nerve fibers innervate different anatomic regions (e.g. thorax, small intestine, colon and extremities). The cell bodies represent the somas of the first-order sensory neurons and constitute an integral part of the somatosensory system. DRGs process sensory information transduced from afferent nerves, which may include nociceptive information, in addition to signals generated in steady-state conditions such as the feeling of discomfort and satiety (Figure 5). The afferent neurons of DRGs integrate sensory information from distinct body regions and transmit the signal to the CNS (58). Sensory afferent signals from the thoracolumbar region predominantly originate from the small intestine and lumbosacral DRGs mainly project the lower extremities, but also to the large intestine (59–61). Different types of sensory neurons within DRGs allow sensing of distinct stimuli. Initially, sensory neurons were categorized based on their degree of myelination and the associated conduction velocity. This classification led to four main classes of neurons, namely thinly myelinated Aδ fibers, unmyelinated C fibers, heavily and moderately A myelinated fibers. Because of a significant heterogeneity in the degree of myelination and conduction velocity in functionally similar classes, this classification insufficiently reflects function. To gain further insight into different populations and to functionally discern the neurons within DRGs, there are now many studies available, which have performed single-cell RNA-sequencing and associated gene expression with a function in mice, primates and humans (12, 62–68). The respective datasets can be accessed via online tools and screened for genes of interest (as outlined in Table 2).
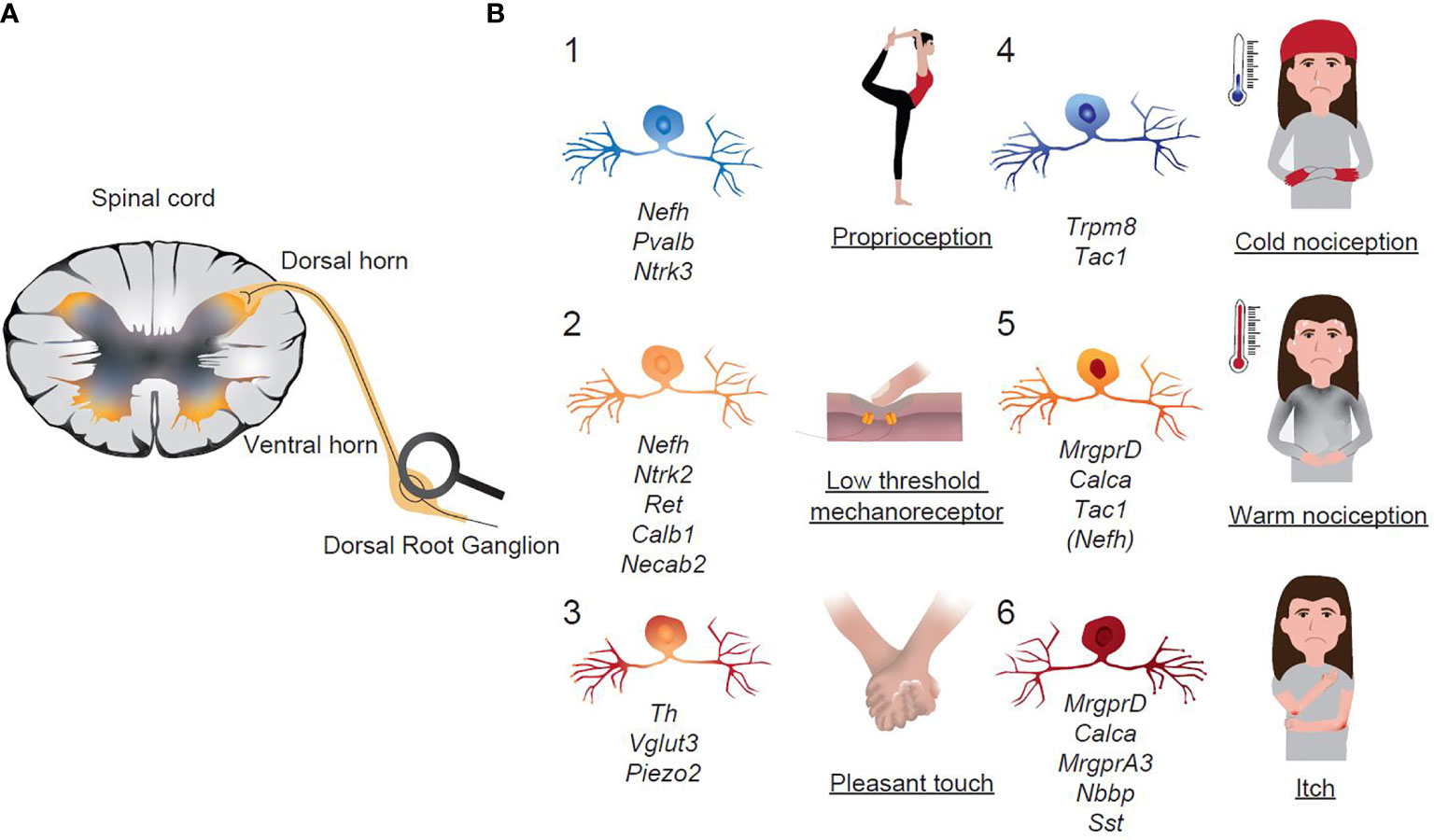
Figure 5 Functional classification of sensory neurons in Dorsal Root Ganglia. (A) Schematic representation of extrinsic sensory afferent neurons that transmit signals from the periphery to the CNS. (B) Schematic representation of sensory neuronal groups in DRGs with assigned functions. Italics are the key transcripts of the respective neuronal group.
Single-cell RNA-sequencing, if not used with any selection, is an untargeted and unbiased approach that allows identification of neuronal cell types based on transcripts expressed by the cells. However, there is a certain noise in between studies that complicates the interpretation of the results. Furthermore, a clear limitation of current techniques is the rather superficial sequencing depth at a single-cell level and the required dissociation of cells, which may omit large and long axonal neurons during sort-purification. In future studies, such limitation may be overcome by the single-nuclei sequencing approach as described above (14).
However, with the available data, neurons can be classified into neurofilamentous, peptidergic and non-peptidergic neurons, which are uniformly detected in all the above-mentioned studies. A recent study used a machine-learning approach for three available nomenclatures/classification of DRG neurons (12, 63, 68) in finding the corresponding cell types in different datasets (Table 3) (64). The Usoskin classification showed the least 'noise' and highest prediction score when comparing different datasets and may therefore be the classification of choice (64). However, it has to be mentioned that the Usoskin study detected fewer neurons (622 neurons) compared to the work of Zeisel (1'580 neurons) or Sharma (10´922 neurons). The Usoskin-classification proposed a classification of DRG neurons into a total of 11 groups: 5 subgroups within the neurofilamentous group (NF1-5), 4 non-peptidergic neurons including TH+ neurons (NP1-3, TH), 2 peptidergic neurons (PEP) including Nav1.8+ neurons (Table 3) (63). Another study by Zeisel et al. was consistent with the results obtained by Usoskin, but due to more sequenced neurons, sub-clusters within peptidergic neurons (PEP1, 2, Trpm8), non-peptidergic neurons (NP 1, 2) were identified (12). One has to consider that available single-cell RNA-sequencing studies may compare DRGs that receive afferents from different anatomic locations and the subtype composition may vary across axial levels, which therefore could explain minor discrepancies when comparing neuronal population datasets (68, 70). Thus, sequencing of neurons isolated from either thoracic, lumbar or sacral regions may unravel anatomic fingerprints of gene expression in future studies.
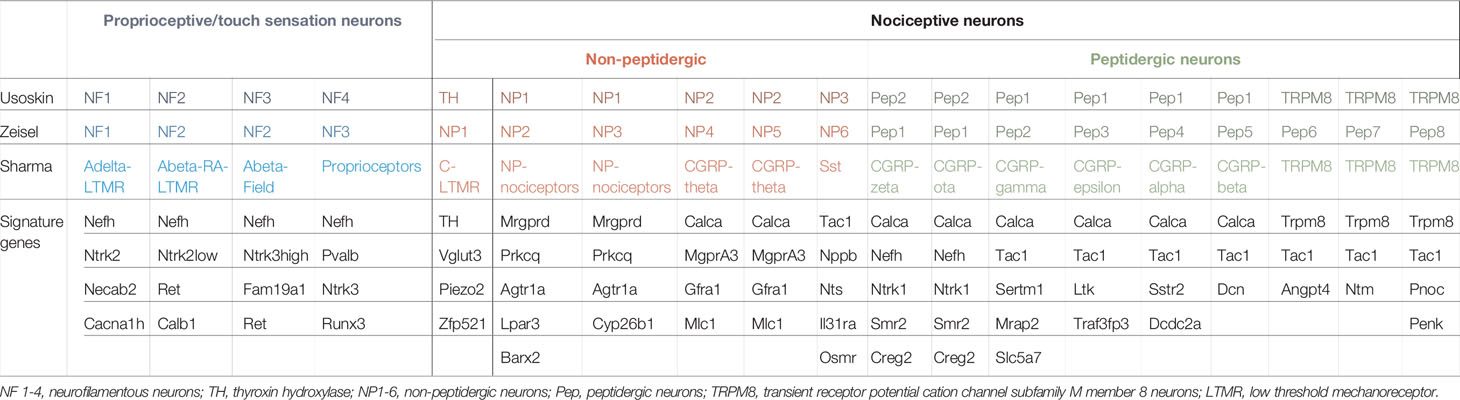
Table 3 Reference genes for the identification of neuronal subsets in DRGs based on unbiased single-cell RNA sequencing (adapted from (12, 58, 63, 64, 68).
Another recent study identified a total of 12 classes of DRG neurons, which consists of Aβ-rapidly adapting (RA) low threshold mechanoreceptors (LTMR), Aδ-LTMR, C-LTMR, 6 groups of Calcitonin Gene-related Peptide (CGRP) neurons, Mrgrd+ polymodal nociceptors, Somatostatin+ and cold thermosensors (68). Interestingly, a direct comparison of the three main classifications (Usoskin et al. vs Zeisel et al. vs Sharma et al.) by the before mentioned machine-learning approach found a high probability of finding a distinct cell across different datasets (64). Thus, the same cell can be found in different datasets and the difference between studies relies on the annotation and is not based on biologic differences. All the above-mentioned DRG-related studies have been performed in naïve animals and no specific disease model was profiled in detail. Therefore, it is of great interest to decipher transcriptional regulators in disease states such as nerve injury and concomitant neuronal regeneration at a single-cell resolution. A recent single-cell sequencing paper describes interesting features associated with different nerve injury models (69). First, a reduced expression of neuronal-subtype marker genes, such as Tac1, Mrgprd, and Nefh is described. Second, but not unexpected, genes involved in axon guidance, axogenesis and cell migration have overall increased. Of particular interest are specific transcripts found in nociceptive neurons because of their role in the development of neuropathic pain (69). Genes of interest can be browsed in the available online tool (see Table 2).
A large set of publications has studied different neuronal molecules, which help to propose a relation of the molecular profile of the respective neuron with modality-specific function within DRG neurons. These studies allow to functionally predict the different neuronal populations found in single-cell RNA-sequencing studies (Figure 5). Generally, neurofilamentous and TH populations are proprioceptors (control of body positioning and balance) and LTMRs (touch sensations), whereas non-peptidergic and peptidergic neurons are nociceptors (damage/potential damage signals), which represent the majority of all DRG neurons. Within the group of nociceptors and based on the molecular signature, a broad functional sub-classification is made: TRP cation channel subfamily member 8 (TRPM8) neurons are cold-sensory neurons, peptidergic neurons detect heat and pain, isolectin4-binding non-pepdidergic neurons can sense noxious touch, itch and chemical signals (Figure 5).
Because of available top gene transcripts, the populations can be identified and targeted, either via immunohistochemistry, potentially flow cytometry or used for a conditional deletion by using specific Cre-drivers in mice that allow deleting genes within a neuronal subpopulation.
A recent single-cell RNA-sequencing study on retrogradely traced colonic sensory neurons in the mouse identified seven neuronal subtypes (70). The authors identified 5 specific subtypes in the thoracolumbar region and seven subtypes in the lumbosacral region, two of them being exclusively found in the lumbosacral region. The populations identified in both anatomic regions included Neurofilament-a and Neurofilament-b, which express genes typically associated with myelinated DRG neurons, such as neurofilament heavy chain (Nefh) and lactate dehydrogenase B (Ldhb). The third subtype was classified as non-peptidergic neurons and showed an expression pattern of non-peptidergic nociceptors, such as the purinergic receptors P2X3 or glial-cell line derived neurotrophic factor family receptor alpha2 (Gfrα2). The last two subtypes express Calca, Tac1 and TrkA and have been termed as peptidergic nociceptors. The subtypes of neurons exclusively found in the lumbosacral region included a neurofilament and a peptidergic subtype, both of which show a relatively similar basic expression pattern (neurofilament subgroup: Piezo2, Nefh, Ldhb; peptidergic subgroup: Calca, Tac1) but are furthermore characterized by a specific gene set suggesting that neurons have an anatomic fingerprint.
Apart from sensory neurons directly originating from organs such as the intestine, lymph nodes show a spatial distribution of sensory and sympathetic neurons (72). A more comprehensive view by using tracing experiments via the injection of Cre-expressing viruses into iliac lymph nodes and single-cell RNA-sequencing of labelled DRG neurons reveals 4 transcriptionally distinct neuronal subtypes (72). The vast majority of detected neurons express Nav1.8 and only few co-expressed TH. By comparing the detected neurons to publicly available datasets (63, 68), the study revealed that lymph nodes are equipped with transcriptionally heterogenous, predominant peptidergic nociceptors. Of great interest in inguinal-lymph nodes are particularly enriched genes involved in inflammation and neuro-immune-communication, such as Il33, TLRs or Ptgir. The same authors also observed sensory neuronal remodeling after LPS application suggesting modular sensing of danger signals by the nervous system (72). Future studies should focus on the sensory DRGs of mesenteric lymph nodes to delineate transcriptional profiles associated with the primary hubs exposed to constant PRR signals.
Apart from DRGs located in close proximity to the spinal cord, vagal ganglia receive afferent signal input and mediate host protection against a broad variety of sensations. The upper airways are densely innervated by the vagus nerve with the fused primary order neurons located in the nodose/jugular/petrosal superganglia clustering near the jugular foramen. These sensory neurons play a pivotal role in swallowing reflex pathways and coordinate the upper airways to prevent aspiration pneumonia or dysphagia (73). Single-cell RNA-sequencing of nodose/jugular/petrosal ganglia with a coverage of 25'117 sensory neurons (spanning 2293 genes) revealed a total of 37 classes of neurons. P2RY1+ neurons seem to be particularly relevant with respect to swallow reflexes because conditional ablation of P2RY1 neurons led to impaired swallowing responses (73). Given the wide variety of neurons located in vagal ganglia, the role of other subclasses remains elusive and needs to be studied in future projects.
Neurofilaments (NF, gene Nefh) are 10nm bundles of fibrils within neurons (74). In contrast to the perikarya, NF can be highly abundant in axons (75). In concert with microtubule and microfilaments, NF form the neuronal cytoskeleton and are aligned in parallel along the axonal axis. The filaments primarily support neuronal structure and regulate the axonal diameter, which is a critical determinant of neuronal conduction velocity (76, 77). Based on the marker NF200, two distinct neuronal subtypes have been identified that allow the discrimination between large myelinated or thinly myelinated neurons with fast conduction velocities (NF200 positive) and small unmyelinated neurons with low conduction velocities (NF200 negative) (78). Because of their high conduction velocities, NF neurons are involved in signals that require fast signal transmission, such as touch sensations and proprioception. Apart from NF, spinal proprioceptive neurons, which innervate muscle spindles and Golgi tendon organs, are further characterized by the expression of neurotrophin-3 (Ntf3) and parvalbumin (Pvalb) (79). The expression of other marker genes, such as Tropomyosin receptor kinase A and B (TrkA, TrkB), Ret, calbindin (Calb), functional predictions can be made of NF neurons and allow for a certain functional classification (58).
According to the Usoskin-classification, gene expression profiles of NF group 1-3 is characterized by a pattern of Nefh, Ntrk2, Ret, Calb1 and Ntrk3 (63). These groups can be functionally assigned to low threshold mechanoreceptors (LTMRs). The expression of Ntrk3 and Pvalb in NF groups 4/5 suggests that these neurons have a proprioceptive role (Table 3) (63). In general, proprioceptors are located within the musculoskeletal system and relay the information to the CNS concerning body position and movements (Figure 5).
A more comprehensive view into the subgroups and the associated gene expression suggests that NF group 1 are lightly myelinated Aδ LTMRs and very sensitive velocities detectors tuned to the deflection of body hairs (80). NF group 2 are rapid adapting LTMRs that end in Meissner corpuscle and longitudinal lanceolate endings and are critical for the perception of skin movement and vibration (81). Because of the high expression of Ntrk3, NF group 3 can be assigned to slowly adapting LTMRs, which control the perception of stretch and indentation (81).
TH-expressing DRG neurons have been shown to be C-LTMRs (82). One of the top genes expressed in this population, Vglut3, has been identified to be uniquely found in C-LTMRs. Functionally, absence of Vglut3 in Vglut3-/- mice has been linked to defects in acute mechanical pain sensations upon intense noxious stimuli (83). An additional feature of these neurons is the high expression of the mechanosensitive ion channel, Piezo2. Apart of mechanical pain, C-LTMRs perceive low mechanical forces such as pleasant touch of the skin.
Slow conducting DRG neurons are classified into peptidergic and non-peptidergic neurons. Peptidergic neurons are defined based on the expression of the neuropeptides substance P, calcitonin gene-related peptide (CGRP), and somatostatin, whereas non-peptidergic neurons bind the plant lectin IB4, express the Mrg family of G-protein coupled receptors and P2X3 (84, 85). However, one must take into account that all of these markers are only partially selective and some overlap can exist (86). Basically, peptidergic and non-peptidergic neurons are unmyelinated, primary afferent neurons assigned to nociceptors and thus respond to damage and potential damage stimuli (Figure 5).
From an immunologic perspective, peptidergic neurons and their secreted neuropeptides, including Substance P, CGRP and NMU, have a marked immunoregulatory potential, which has been shown by many studies (54, 87–91) and will be discussed in detail in a later paragraph.
The expression of the Mas-related G-protein coupled receptor member D (MrgprD) in non-peptidergic neurons group 1 (NP1) suggests its role in the perception of noxious mechanical and thermal stimuli as well as in the perception of itch (92, 93). NP2, expressing MrgprA3 and Calca, are polymodal nociceptors and evoke itch responses (94). Both groups, NP1 and NP2, have been linked to the development of neuropathic pain (63, 95, 96). Even though characterized by a different set of key genes, NP3 have similar functions than NP2 and are also itch-sensing neurons. Relevant genes in this neuronal subset include the natriuretic polypeptide B (Nbbp) and Somatostatin (Sst). Injection of Nbbp intrathecal triggered itch responses in mice whereas itch-responses were blocked upon ablation of Nbbp-receptor-expressing cells (97). In the same line, Sst co-expressed with Nbbp neurons and Sst+ neurons triggered itch behavior (98).
Pep1 neurons can be largely grouped by the expression of Tac1, which represents the gene for substance P. In terms of neuronal function, this neuronal population is involved in thermosensation and thus reacts to noxious heat and cold stimuli (63, 99, 100). However, substance P has also a major role in neuro-immune interactions, which will be discussed in a later paragraph. Similarly, these neurons express CGRP (Calca), which has also been implicated in regulating immune functions. Pep2 neurons express neurofilament heavy chain (Nefh) as well as Ntrk1, suggesting that these neurons are lightly myelinated Aδ nociceptors, which have a higher conduction velocity than unmyelinated neurons (63). Thus, these neurons respond to dangerously intense mechanical or mechanothermal stimuli. TRPM8 neurons are involved in thermal perception of temperatures <20°C and cold-triggered nociception (101).
Somatosensory sensations, as described above, can be activated by a large variety of stimuli. Excitatory and inhibitory afferent neurons transmit the signal to DRGs, where a first processing of the signal input occurs. Afferents from DRGs relay the information to the CNS via interneurons located in the dorsal horn of the spinal cord. The incoming information is further processed within the dorsal spinal cord and ultimately relayed to different brain areas (102). A recent study using single-cell RNA-sequencing of the dorsal spinal cord identified that all neurons either express the vesicular glutamate receptor 2 (Vglut2) or the vesicular GABA transporter (Vgat), thus, representing glutamatergic excitatory or GABAergic inhibitory neurons. To enrich neurons, the authors used the reporter mouse lines VgattdTom and Vglut2tdTom and FACS-enriched the samples for the respective neuronal subsets (71). The final dataset of 1'545 neurons revealed 15 glutamatergic and 15 GABAergic neuronal subsets and based on literature, the expressed transcripts have been linked to certain physiologic functions (71). For detailed information, we kindly refer to the original articles since the complexity of spinal cord neuronal population is beyond the scope of this review (71, 103). A very interesting novel feature that allows to monitor activation of cell types involved in sensory signal transmission is the sensory-transcription coupling. By using triple in-situ hybridization and the combination of the immediate-early gene expression of Arc with markers for distinct neuronal cell types revealed that noxious heat and cold activate different sets of excitatory and inhibitory neurons (71). This finding underlines the complexity of neuronal networks and argues for a broad activation of neurons in response to one distinct modality.
Enteric and DRG neurons express the neuro-peptides VIP, CGRP, Substance P, and NMU, and the neuro-transmitters norepinephrine and acetylcholine. Many studies identified the respective receptor expression on a large set of immune cells, which implies the regulation of immune responses by the nervous system. Furthermore, the close co-localization of hematopoietic cells with neuronal fibers suggests a bidirectional signaling exchange. These neuro-immune modules constantly interact in order to adapt physiologic processes. The following paragraph reviews neuro-immune interactions based on neuro-peptides or transmitters.
VIP engages on two subsets of receptors, Vipr1 and Vipr2. Thus, solely studying the experimental effects of VIP may be difficult to interpret because of receptor-dependent responses. Furthermore, mechanisms behind such studies do not allow to conclude specific effects on a cellular level. This fact has been observed in the following experimental designs: On one hand and in the context of gastrointestinal inflammation, VIP knock-out mice showed a more severe phenotype in DNBS- and DSS-induced colitis in mice and VIP has been linked to the maintenance of intestinal integrity (104). On the other hand, models of DSS-induced colitis in Vipr1-/- and Vipr2-/- mice displayed opposite results. In fact, Vipr1-/- mice showed a milder disease score compared to wild type mice, whereas Vipr2-/- developed a more severe colitis (105, 106). Thus, studies addressing the role of VIP need to take into account the distinct affinity of VIP onto its receptors and the distinct receptor expression on a broad array of immune cells. Vipr2 is expressed by a large set of immune cells including innate lymphoid cells type 3 (ILC3), which play a crucial role in host defense mechanism against bacteria. Recent studies have now described Vipr2 expression by ILC3s and binding of the ligand VIP regulates the release of IL-22 (106, 107). Because IL-22 controls the antimicrobial peptide production and regulates lipid absorption, the VIP-IL-22 axis has been proposed to shift the balance in host defense mechanisms and lipid uptake (107). Furthermore, VIP and IL-22 is released in a feeding-dependent manner and, thus, underlines the role of neuro-immune signaling for adaptations of barrier mechanisms dependent on food-uptake. However, the role played by VIP on ILC3s seems to be context dependent acknowledged by controversial results published by different authors (106, 107). VIP has also been shown to regulate type 2 immune reactions in the lung (108). By using ovalbumin-induced lung inflammation, the authors showed an increase of VIP transcripts in nodose ganglia of inflamed lungs, which was dependent on nociceptive neuronal firing. Upon release, VIP engages to Vipr2 and activated the release of type 2 cytokines from ILC2 and CD4+ T cells (108). In the same context, nociceptors express FcϵR1 and directly sense IgE-OVA complexes to initiate type 2 immune reactions (109). In summary, VIP appears as an important regulator of type 2 and type 3 immune responses.
By using Capsaicin-induced denervation in neonatal mice and immunoassays, early studies already suggested that the vast majority of CGRP neurons have an extrinsic source in the intestine (110). According to current knowledge, the main source of CGRP are the unmyelinated sensory C fibers in DRGs (111). As reviewed above, CGRP-expressing neurons are assigned to nociceptive neurons and, thus, sense damage and potential damage stimuli and are involved in the perception of pain. Because CGRP expressing nerve fibers have been found in many immunologic organs including bone marrow, spleen, lymph nodes, skin and the intestine, and the receptor is expressed on many different immune cells, the immunoregulatory potential of CGRP has been repeatedly shown (54, 88, 89, 111–113). Following the release, CGRP engages the calcitonin receptor-like receptor (CALCRL) and the receptor-modifying protein 1 (RAMP1). It is important to note that immune cells have also been shown to express CGRP, which suggests a bi-directional crosstalk between immune cells and the nervous system (114, 115). In general, CGRP seems to have anti-inflammatory effects in the GI-tract. This has been highlighted in gain-of function experiments by modelling inflammatory bowel disease and the systemic administration of CGRP in rats, which resulted in an amelioration of TNBS-induced colitis (116, 117). In line with these results, loss-of-function experiments by using CGRP antagonists in rats or CGRP knock-out mice revealed increased susceptibility to colitis (118, 119). These results suggest that CGRP agonists may be potential therapeutic targets for the treatment of inflammatory diseases. The anti-inflammatory role of CGRP has also been highlighted in the context of Streptococcus pyogenes infection in the skin, which is the leading cause of the life-threatening necrotizing fasciitis. The authors found that S. pyogenes directly secretes streptolysin S and promotes the release of CGRP from nociceptors, which inhibits neutrophil recruitment and phagocytic killing (120). In the same line and in the context of Salmonella enterica infection, TRPV1+ nociceptors release CGRP and modulate M cells to eventually control host defense against Salmonella (121). In terms of type 2 immune reactions, innate lymphoid cells type 2 (ILC2) express the receptors CALCRL/Ramp1 and binding of CGRP modulate ILC2 activation, whereas deletion of this signaling cascade elevated ILC2 responsiveness and type 2 immunity (88, 89, 113). Thus, the prevailing view regarding the role of the neuropeptide CGRP is its overall anti-inflammatory properties. With regard to many overwhelming immune reactions, topic or systemic application of CGRP may be a valuable treatment strategy. However, further studies need to investigate the related mechanisms in more detail.
Substance P (SP) is a member of the tachykinin family of neuropeptides and is encoded by the Tachykinin 1 (Tac1) gene. SP exerts its function via the engagement on G-protein coupled neurokinin receptors but predominantly binds on the neurokinin 1 receptor (NK1R) (122). SP is expressed in the central and peripheral nervous system and intrinsic neurons seem to be the major source in the intestine (123, 124). Once synthesized, SP is transported in large dense-core vesicle and released via exocytosis where it exerts its function on the same cell or the adjacent cell (125). Apart from neurons, immune cells have been implicated to express both, SP and its receptor NK1R (126). The relative broad expression of the receptor on immune cells in the lamina propria, such as mast cells, eosinophils, neutrophils, macrophages, dendritic cells and natural killer cells implies its tight regulatory immune function. In general, SP has a pro-inflammatory role via the induction of pro-inflammatory cytokines in immune cells (127). In particular, it has been highlighted that SP and its receptor NK1R increase the susceptibility to DSS- and TNBS-induced colitis (128, 129). A newly developed intestinal organ culture system observed an anti-correlation of Tac1 and its receptor TacR1 after the application of different microbiota strains into the in vitro system. This negative correlation has then been linked to alterations of Rorgt+Tregs. Such observations suggest the potential of microbiota-sensing by Tac1+ neurons and the consecutive modulation of immunologic reactions (130). In the allergic setting, nociceptors release SP after allergen exposure and promote migration and activation of adaptive type 2 immune responses (131). However, because of its broad expression in several tissues, the exact cellular role of SP/NK1R has to be further studied in conditional deletion models in order to decipher the mechanism behind the observed phenotypes.
NMU is a short neuropeptide with highly conserved amidated C-terminus required for receptor binding. Neurons of the CNS, the pituitary gland and the ENS express NMU (132). Within the ENS, NMU labels together with CGRP a subset of ChAT+ sensory neurons (12, 53, 132). Further, NMU expression is regulated by the commensal microbiota and modulated by secretory excretory products of helminths (48, 53, 133). To mediate its biological function, NMU binds to two large G-protein coupled receptors Nmur1 and Nmur2. Nmur2 is expressed in neurons in particular in the CNS and regulates feeding-behavior, circadian rhythm as well as pain perception and bone formation (132). The immunomodulatory functions of NMU were recognized years ago (134–136), however, before ILCs were emerging. Thus, the mechanism remained elusive until several publications demonstrated that NMU acts specifically via ILC2s in different organs (53, 133, 137). By binding to Nmur1, NMU triggers a signal cascade via Gαq - PLC, activation of NFAT, resulting in activation of ILC2, proliferation and cytokine production. Upon NMU stimulation, ILC2 promote the type 2 immune response characterized by eosinophil recruitment, goblet cell hyperplasia and mucus production, resulting in increased worm expulsion during N. brasiliensis infection or enhanced airway inflammation following papain challenge. Altogether, these data indicate that sensory neurons regulate ILC2 activation via NMU and CGRP with downstream effects on various immune cell types participating in type 2 inflammation at mucosal barriers.
Norepinephrine has a dual function in mammalian hosts. On one hand, it is a stress or danger hormone, which is released during a fight-or-flight reaction and results in a concomitant increase in blood pressure, heart rate, glucose mobilization and other stress reactions. On the other hand, norepinephrine acts as a neurotransmitter released from sympathetic nerves located in sympathetic ganglia. Once released, norepinephrine binds and activates α- and β-adrenergic receptors, which are G protein-coupled receptors and thus exert their effect via a second messenger system. In general, β-adrenergic receptors seem to be immunologically more important given their anti-inflammatory effects in neutrophils, macrophages and ILCs (138–140). More specifically, β-adrenergic stimulation mediates the polarization of intestinal macrophages, which reside in close proximity to sympathetic neurons (138). After the activation of the β2-receptor, macrophages upregulate tissue protective-programs (138). Reciprocally, macrophages upregulate neuro-protective programs through an arginase1-polyamine axis, and, thus limit neuronal damage (57).
β2-adrenergic receptors are expressed in ILC2s and argue for a regulatory role of the sympathetic nervous system in controlling type 2 immune reactions. Indeed, norepinephrine has been shown to inhibit ILC2s, whereas ILC2 specific ablation of the β2 receptor magnified type 2 immune reactions and improved worm clearance in the context of N. brasiliensis infection (139). In summary, sympathetic neurons have an anti-inflammatory role in a broad range of immune cells.
Acetylcholine (Ach) is a neurotransmitter predominantly used in the autonomous nervous system and the major neurotransmitter of the parasympathetic nervous system. In addition, excitatory motor neurons, sensory neurons and interneurons within the ENS are capable of producing Ach. Ach mediates biological effects by binding to two different families of cholinergic receptors, the nicotinic and the muscarinic Ach receptors. Among immune cells, the nicotinic α7-Ach receptor is expressed on macrophages and ILC subsets and was investigated in detail due to its anti-inflammatory effects in various disease settings including but not limited to sepsis, IBD and arthritis. This intriguing finding, termed ‘cholinergic anti-inflammatory pathway’, uncovered a vagal regulated release of Ach, which binds on the nicotinic α7-Ach receptor on macrophages to suppress release of pro-inflammatory cytokines and in particular TNF-α, which is mediating immunopathology during sepsis or intestinal inflammation (141, 142). The cholinergic anti-inflammatory pathway has profound and rapid anti-inflammatory properties. The potent effects are illustrated by the prevention of septic shock in mice mediated by this pathway (143). Furthermore, stimulation of the vagal nerves using a medical device was successfully used in patients suffering from chronic arthritis to mitigate disease symptoms, which have been proven resistant to glucocorticoid treatment (144).
α7-nicotinic Ach receptor agonists were also shown to suppress ILC2 activation during allergic asthma (145). These data suggest a broad role of α7-Ach receptor signaling in damping chronic inflammation in various settings. On the contrary, vagotomy was shown to delay the resolution of inflammation following peritoneal E. coli infection. In this disease model, vagal stimulation promoted biosynthesis of resolvins in ILC3s (146). In addition, ILC3 might also serve as a source of Ach as described during allergic airway inflammation (147). In summary, Ach emerges as an important checkpoint for immune activation but the multifaceted actions as mucosal barriers require further investigation.
The identification of different neuronal subpopulations within the intrinsic ENS, as well as the extrinsic sensory compartment, provides the basis for studying the peripheral nervous system in detail. We noticed that certain genes are more ubiquitously expressed by different neuronal subpopulations and may therefore be involved in different functions depending on the neuronal subtype. However, our review combines several datasets and allows for the identification of neuronal subtypes based on genes expressed, morphology and function. The identification of key transcripts of neuronal populations now allows to design specific Cre-driver mouse lines to study neuronal programs in health and disease. There are three technical strategies available to study gene expression in neuronal populations: The desired Cre-driver mouse line can be crossed to a mouse with conditional expression of a fluorescent protein. Fluorescent cells can then be FACS-sorted und analyzed for gene expression (12). The other option is the nuclear dissociation and FACS-enrichment of neuronal nuclei as described recently (14). And the third option is the use of Ribotag mice, which have a hemagglutinin-tagged ribosomal protein specifically active activated following Cre-recombination. Such an approach allows to study ribosomal transcripts specifically active for the desired cre-driver (148). To gain more fundamental insights into expressed genes, their regulation in health and disease as well as more definite view into neuronal subpopulations, the readers are kindly encouraged to upload their sequencing datasets into public available bioportals. This will allow the scientific community to browse for their genes of interest. Future studies should especially focus on comparing steady state gene expression to different diseases to unravel important transcripts in certain neuronal subpopulations.
If we consider the central role of the nervous system in regulating immune functions, the potential is remarkable. The work of Tracey and colleagues has underlined its potential by the discovery of the ‘cholinergic anti-inflammatory’ pathway (143, 149). Such profound anti-inflammatory properties are of need for overwhelming inflammatory diseases, such as inflammatory bowel disease, sepsis, rheumatoid arthritis, and many more. Of note, the therapeutic potential of the anti-inflammatory properties of the nervous system has been successfully tested in clinical trials in humans (144, 150, 151). Even more impressive is the notion that patients in one clinical trial did no longer respond to conventional anti-inflammatory treatments, whereas vagal nerve stimulation improved clinical symptoms of rheumatoid arthritis patients (144). However, there is an emerging need to identify neuronal mechanisms that construct anti- as well as pro-inflammatory reactions in more detail. Such insights may uncover key neuro-immune cues that can potentially be harnessed by drugs or biologics in the near future.
MJ: writing of manuscript. MK-B: bioinformatic analysis and critical revision of the manuscript. DD: writing of the manuscript. SM: design of figures and critical revision of the manuscript. CK: writing of manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Swiss National Science foundation (Grant-ID: 184425 to MJ), the German Research Foundation (DFG; Neuromac SFB TR project B17, FOR2599 project 5 - KL 2963/5-2, KL 2963/2-1 and KL 2963/3-1 to CK), (DFG; SPP1937-DI764/9 to MK-B), and the European Research Council Starting Grant (ERCEA; 803087 to CK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.679055/full#supplementary-material
Supplementary Figure 1 | Expression of chemokine receptors, C-type leptin receptors and other genes of interest in different ENS subsets. Dotplots showing the percentage of expressing cells as well as average expression within the indicated identified neuronal clusters for selected murine chemokine receptor, C-type leptin receptor and other genes of interest. Data was downloaded from http://mousebrain.org/ (12) or from https://singlecell.broadinstitute.org/ (14). Genes not present in the plot showing the Drokhlyanski data have been filtered. All dataset were normalized and transformed before the plots were created using the Seurat package (16).
1. Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central Nervous System Control of Food Intake and Body Weight. Nature (2006) 443(7109):289–95. doi: 10.1038/nature05026
2. Stifani N. Motor Neurons and the Generation of Spinal Motor Neuron Diversity. Front Cell Neurosci (2014) 8:293. doi: 10.3389/fncel.2014.00293
3. Guyenet PG. The Sympathetic Control of Blood Pressure. Nat Rev Neurosci (2006) 7(5):335–46. doi: 10.1038/nrn1902
4. Ulrich-Lai YM, Herman JP. Neural Regulation of Endocrine and Autonomic Stress Responses. Nat Rev Neurosci (2009) 10(6):397–409. doi: 10.1038/nrn2647
5. Rao M, Gershon MD. Enteric Nervous System Development: What Could Possibly Go Wrong? Nat Rev Neurosci (2018) 19(9):552–65. doi: 10.1038/s41583-018-0041-0
6. Jansen AS, Nguyen XV, Karpitskiy V, Mettenleiter TC, Loewy AD. Central Command Neurons of the Sympathetic Nervous System: Basis of the Fight-or-Flight Response. Science (1995) 270(5236):644–6. doi: 10.1126/science.270.5236.644
7. Rao M, Gershon MD. The Bowel and Beyond: The Enteric Nervous System in Neurological Disorders. Nat Rev Gastroenterol Hepatol (2016) 13(9):517–28. doi: 10.1038/nrgastro.2016.107
8. Furness JB, Callaghan BP, Rivera LR, Cho HJ. The Enteric Nervous System and Gastrointestinal Innervation: Integrated Local and Central Control. Adv Exp Med Biol (2014) 817:39–71. doi: 10.1007/978-1-4939-0897-4_3
9. Furness JB. The Enteric Nervous System: Normal Functions and Enteric Neuropathies. Neurogastroenterol Motil (2008) 20 Suppl 1:32–8. doi: 10.1111/j.1365-2982.2008.01094.x
10. Furness JB, Kunze WA, Clerc N. Nutrient Tasting and Signaling Mechanisms in the Gut. II. The Intestine as a Sensory Organ: Neural, Endocrine, and Immune Responses. Am J Physiol (1999) 277(5):G922–8. doi: 10.1152/ajpgi.1999.277.5.G922
11. Jakob MO, Murugan S, Klose CSN. Neuro-Immune Circuits Regulate Immune Responses in Tissues and Organ Homeostasis. Front Immunol (2020) 11:308. doi: 10.3389/fimmu.2020.00308
12. Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular Architecture of the Mouse Nervous System. Cell (2018) 174(4):999–1014 e22. doi: 10.1016/j.cell.2018.06.021
13. Morarach K, Mikhailova A, Knoflach V, Memic F, Kumar R, Li W, et al. Diversification of Molecularly Defined Myenteric Neuron Classes Revealed by Single-Cell RNA Sequencing. Nat Neurosci (2021) 24(1):34–46. doi: 10.1038/s41593-020-00736-x
14. Drokhlyansky E, Smillie CS, Van Wittenberghe N, Ericsson M, Griffin GK, Eraslan G, et al. The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell (2020) 182(6):1606–22.e23. doi: 10.1016/j.cell.2020.08.003
15. Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyarto BZ, et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell (2015) 161(4):737–49. doi: 10.1016/j.cell.2015.03.031
16. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated Analysis of Multimodal Single-Cell Data. Cell (2020) 184(13):3573–87.e29. doi: 10.1101/2020.10.12.335331
17. Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ, et al. Sensory Neurons Co-Opt Classical Immune Signaling Pathways to Mediate Chronic Itch. Cell (2017) 171(1):217–28 e13. doi: 10.1016/j.cell.2017.08.006
18. Wilson SR, The L, Batia LM, Beattie K, Katibah GE, McClain SP, et al. The Epithelial Cell-Derived Atopic Dermatitis Cytokine TSLP Activates Neurons to Induce Itch. Cell (2013) 155(2):285–95. doi: 10.1016/j.cell.2013.08.057
19. Kato J, Agalave NM, Svensson CI. Pattern Recognition Receptors in Chronic Pain: Mechanisms and Therapeutic Implications. Eur J Pharmacol (2016) 788:261–73. doi: 10.1016/j.ejphar.2016.06.039
20. Muller PA, Matheis F, Schneeberger M, Kerner Z, Jove V, Mucida D. Microbiota-Modulated CART(+) Enteric Neurons Autonomously Regulate Blood Glucose. Science (2020) 370(6514):314–21. doi: 10.1126/science.abd6176
21. Furness JB. Types of Neurons in the Enteric Nervous System. J Auton Nerv Syst (2000) 81(1-3):87–96. doi: 10.1016/S0165-1838(00)00127-2
22. Collamati A, Martone AM, Poscia A, Brandi V, Celi M, Marzetti E, et al. Anticholinergic Drugs and Negative Outcomes in the Older Population: From Biological Plausibility to Clinical Evidence. Aging Clin Exp Res (2016) 28(1):25–35. doi: 10.1007/s40520-015-0359-7
23. Johnson CD, Barlow-Anacker AJ, Pierre JF, Touw K, Erickson CS, Furness JB, et al. Deletion of Choline Acetyltransferase in Enteric Neurons Results in Postnatal Intestinal Dysmotility and Dysbiosis. FASEB J (2018) 32(9):4744–52. doi: 10.1096/fj.201701474RR
24. Taylor GS, Bywater RA. Antagonism of non-Cholinergic Excitatory Junction Potentials in the Guinea-Pig Ileum by a Substance P Analogue Antagonist. Neurosci Lett (1986) 63(1):23–6. doi: 10.1016/0304-3940(86)90006-6
25. Holzer P, Schluet W, Maggi CA. Ascending Enteric Reflex Contraction: Roles of Acetylcholine and Tachykinins in Relation to Distension and Propagation of Excitation. J Pharmacol Exp Ther (1993) 264(1):391–6.
26. Qu ZD, Thacker M, Castelucci P, Bagyanszki M, Epstein ML, Furness JB. Immunohistochemical Analysis of Neuron Types in the Mouse Small Intestine. Cell Tissue Res (2008) 334(2):147–61. doi: 10.1007/s00441-008-0684-7
27. Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted Disruption of the Neuronal Nitric Oxide Synthase Gene. Cell (1993) 75(7):1273–86. doi: 10.1016/0092-8674(93)90615-W
28. Burnstock G. Historical Review: ATP as a Neurotransmitter. Trends Pharmacol Sci (2006) 27(3):166–76. doi: 10.1016/j.tips.2006.01.005
29. Keef KD, Shuttleworth CW, Xue C, Bayguinov O, Publicover NG, Sanders KM. Relationship Between Nitric Oxide and Vasoactive Intestinal Polypeptide in Enteric Inhibitory Neurotransmission. Neuropharmacology (1994) 33(11):1303–14. doi: 10.1016/0028-3908(94)90030-2
30. Xue L, Farrugia G, Miller SM, Ferris CD, Snyder SH, Szurszewski JH. Carbon Monoxide and Nitric Oxide as Coneurotransmitters in the Enteric Nervous System: Evidence From Genomic Deletion of Biosynthetic Enzymes. Proc Natl Acad Sci U S A (2000) 97(4):1851–5. doi: 10.1073/pnas.97.4.1851
31. Lecci A, Santicioli P, Maggi CA. Pharmacology of Transmission to Gastrointestinal Muscle. Curr Opin Pharmacol (2002) 2(6):630–41. doi: 10.1016/S1471-4892(02)00225-4
32. Alcaino C, Farrugia G, Beyder A. Mechanosensitive Piezo Channels in the Gastrointestinal Tract. Curr Top Membr (2017) 79:219–44. doi: 10.1016/bs.ctm.2016.11.003
33. Solis AG, Bielecki P, Steach HR, Sharma L, Harman CCD, Yun S, et al. Mechanosensation of Cyclical Force by PIEZO1 Is Essential for Innate Immunity. Nature (2019) 573(7772):69–74. doi: 10.1038/s41586-019-1485-8
34. Hansen MB. The Enteric Nervous System II: Gastrointestinal Functions. Pharmacol Toxicol (2003) 92(6):249–57. doi: 10.1034/j.1600-0773.2003.920601.x
35. Cooke HJ, Xue J, Yu JG, Wunderlich J, Wang YZ, Guzman J, et al. Mechanical Stimulation Releases Nucleotides That Activate P2Y1 Receptors to Trigger Neural Reflex Chloride Secretion in Guinea Pig Distal Colon. J Comp Neurol (2004) 469(1):1–15. doi: 10.1002/cne.10960
36. Bornstein JC. Physiology of the Gastrointestinal Tract, Sixth Edition. (2018). pp. 429–51, JPPF. Elsevier Inc. (2018)
37. Clerc N, Furness JB. Intrinsic Primary Afferent Neurones of the Digestive Tract. Neurogastroenterol Motil (2004) 16 Suppl 1:24–7. doi: 10.1111/j.1743-3150.2004.00470.x
38. Furness JB, Jones C, Nurgali K, Clerc N. Intrinsic Primary Afferent Neurons and Nerve Circuits Within the Intestine. Prog Neurobiol (2004) 72(2):143–64. doi: 10.1016/j.pneurobio.2003.12.004
39. Bertrand PP, Kunze WA, Bornstein JC, Furness JB, Smith ML. Analysis of the Responses of Myenteric Neurons in the Small Intestine to Chemical Stimulation of the Mucosa. Am J Physiol (1997) 273(2 Pt 1):G422–35. doi: 10.1152/ajpgi.1997.273.2.G422
40. Bellono NW, Bayrer JR, Leitch DB, Castro J, Zhang C, O’Donnell TA, et al. Enterochromaffin Cells Are Gut Chemosensors That Couple to Sensory Neural Pathways. Cell (2017) 170(1):185–98.e16. doi: 10.1016/j.cell.2017.05.034
41. Kirchgessner AL, Tamir H, Gershon MD. Identification and Stimulation by Serotonin of Intrinsic Sensory Neurons of the Submucosal Plexus of the Guinea Pig Gut: Activity-Induced Expression of Fos Immunoreactivity. J Neurosci (1992) 12(1):235–48. doi: 10.1523/JNEUROSCI.12-01-00235.1992
42. Fung C, Vanden Berghe P. Functional Circuits and Signal Processing in the Enteric Nervous System. Cell Mol Life Sci (2020) 77(22):4505–22. doi: 10.1007/s00018-020-03543-6
43. Pan H, Gershon MD. Activation of Intrinsic Afferent Pathways in Submucosal Ganglia of the Guinea Pig Small Intestine. J Neurosci (2000) 20(9):3295–309. doi: 10.1523/JNEUROSCI.20-09-03295.2000
44. Mazzuoli-Weber G, Schemann M. Mechanosensitivity in the Enteric Nervous System. Front Cell Neurosci (2015) 9:408. doi: 10.3389/fncel.2015.00408
45. Neal KB, Bornstein JC. Targets of Myenteric Interneurons in the Guinea-Pig Small Intestine. Neurogastroenterol Motil (2008) 20(5):566–75. doi: 10.1111/j.1365-2982.2007.01052.x
46. Lomax AE, Furness JB. Neurochemical Classification of Enteric Neurons in the Guinea-Pig Distal Colon. Cell Tissue Res (2000) 302(1):59–72. doi: 10.1007/s004410000260
47. Kaelberer MM, Buchanan KL, Klein ME, Barth BB, Montoya MM, Shen X, et al. A Gut-Brain Neural Circuit for Nutrient Sensory Transduction. Science (2018) 361(6408):eaat5236. doi: 10.1126/science.aat5236
48. Muller PA, Schneeberger M, Matheis F, Wang P, Kerner Z, Ilanges A, et al. Microbiota Modulate Sympathetic Neurons Via a Gut-Brain Circuit. Nature (2020) 583(7816):441–6. doi: 10.1038/s41586-020-2474-7
49. Obata Y, Castano A, Boeing S, Bon-Frauches AC, Fung C, Fallesen T, et al. Neuronal Programming by Microbiota Regulates Intestinal Physiology. Nature (2020) 578(7794):284–9. doi: 10.1038/s41586-020-1975-8
50. Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut Microbial Products Regulate Murine Gastrointestinal Motility Via Toll-Like Receptor 4 Signaling. Gastroenterology (2012) 143(4):1006–16.e4. doi: 10.1053/j.gastro.2012.06.034
51. Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell (2018) 173(7):1728–41.e13. doi: 10.1016/j.cell.2018.04.027
52. Chu C, Murdock MH, Jing D, Won TH, Chung H, Kressel AM, et al. The Microbiota Regulate Neuronal Function and Fear Extinction Learning. Nature (2019) 574(7779):543–8. doi: 10.1038/s41586-019-1644-y
53. Cardoso V, Chesne J, Ribeiro H, Garcia-Cassani B, Carvalho T, Bouchery T, et al. Neuronal Regulation of Type 2 Innate Lymphoid Cells Via Neuromedin U. Nature (2017) 549(7671):277–81. doi: 10.1038/nature23469
54. Meseguer V, Alpizar YA, Luis E, Tajada S, Denlinger B, Fajardo O, et al. TRPA1 Channels Mediate Acute Neurogenic Inflammation and Pain Produced by Bacterial Endotoxins. Nat Commun (2014) 5:3125. doi: 10.1038/ncomms4125
55. McVey Neufeld KA, Mao YK, Bienenstock J, Foster JA, Kunze WA. The Microbiome Is Essential for Normal Gut Intrinsic Primary Afferent Neuron Excitability in the Mouse. Neurogastroenterol Motil (2013) 25(2):183–e88. doi: 10.1111/nmo.12049
56. Collins J, Borojevic R, Verdu EF, Huizinga JD, Ratcliffe EM. Intestinal Microbiota Influence the Early Postnatal Development of the Enteric Nervous System. Neurogastroenterol Motil (2014) 26(1):98–107. doi: 10.1111/nmo.12236
57. Matheis F, Muller PA, Graves CL, Gabanyi I, Kerner ZJ, Costa-Borges D, et al. Adrenergic Signaling in Muscularis Macrophages Limits Infection-Induced Neuronal Loss. Cell (2020) 180(1):64–78.e16. doi: 10.1016/j.cell.2019.12.002
58. Emery EC, Ernfors P. Dorsal Root Ganglion Neuron Types and Their Functional Specialization. In: Wood JN, editor. The Oxford Handbook of the Neurobiology of Pain Oxford Handbook Online. Oxford University Press (2018).
59. Robinson DR, McNaughton PA, Evans ML, Hicks GA. Characterization of the Primary Spinal Afferent Innervation of the Mouse Colon Using Retrograde Labelling. Neurogastroenterol Motil (2004) 16(1):113–24. doi: 10.1046/j.1365-2982.2003.00456.x
60. Tan LL, Bornstein JC, Anderson CR. Distinct Chemical Classes of Medium-Sized Transient Receptor Potential Channel Vanilloid 1-Immunoreactive Dorsal Root Ganglion Neurons Innervate the Adult Mouse Jejunum and Colon. Neuroscience (2008) 156(2):334–43. doi: 10.1016/j.neuroscience.2008.06.071
61. Deshpande D, Agarwal N, Fleming T, Gaveriaux-Ruff C, Klose CSN, Tappe-Theodor A, et al. Loss of POMC-Mediated Antinociception Contributes to Painful Diabetic Neuropathy. Nat Commun (2021) 12(1):426. doi: 10.1038/s41467-020-20677-0
62. Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, et al. Somatosensory Neuron Types Identified by High-Coverage Single-Cell RNA-Sequencing and Functional Heterogeneity. Cell Res (2016) 26(8):967. doi: 10.1038/cr.2016.90
63. Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, et al. Unbiased Classification of Sensory Neuron Types by Large-Scale Single-Cell RNA Sequencing. Nat Neurosci (2015) 18(1):145–53. doi: 10.1038/nn.3881
64. Kupari J, Usoskin D, Lou D, Parisien M, Hu Y, Fatt M, et al. Single Cell Transcriptomics of Primate Sensory Neurons Identifies Cell Types Associated With Human Chronic Pain. Nat Commun (2020) 12:1510. doi: 10.1038/s41467-021-21725-z
65. Finno CJ, Peterson J, Kang M, Park S, Bordbari MH, Durbin-Johnson B, et al. Single-Cell RNA-Seq Reveals Profound Alterations in Mechanosensitive Dorsal Root Ganglion Neurons With Vitamin E Deficiency. iScience (2019) 21:720–35. doi: 10.1016/j.isci.2019.10.064
66. Faure L, Wang Y, Kastriti ME, Fontanet P, Cheung KKY, Petitpre C, et al. Single Cell RNA Sequencing Identifies Early Diversity of Sensory Neurons Forming Via Bi-Potential Intermediates. Nat Commun (2020) 11(1):4175. doi: 10.1038/s41467-020-17929-4
67. Hu G, Huang K, Hu Y, Du G, Xue Z, Zhu X, et al. Single-Cell RNA-seq Reveals Distinct Injury Responses in Different Types of DRG Sensory Neurons. Sci Rep (2016) 6:31851. doi: 10.1038/srep31851
68. Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, Ginty DD. The Emergence of Transcriptional Identity in Somatosensory Neurons. Nature (2020) 577(7790):392–8. doi: 10.1038/s41586-019-1900-1
69. Renthal W, Tochitsky I, Yang L, Cheng YC, Li E, Kawaguchi R, et al. Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes After Axonal Injury. Neuron (2020) 108(1):128–44.e9. doi: 10.1016/j.neuron.2020.07.026
70. Hockley JRF, Taylor TS, Callejo G, Wilbrey AL, Gutteridge A, Bach K, et al. Single-Cell RNAseq Reveals Seven Classes of Colonic Sensory Neuron. Gut (2019) 68(4):633–44. doi: 10.1136/gutjnl-2017-315631
71. Haring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lonnerberg P, et al. Neuronal Atlas of the Dorsal Horn Defines Its Architecture and Links Sensory Input to Transcriptional Cell Types. Nat Neurosci (2018) 21(6):869–80. doi: 10.1038/s41593-018-0141-1
72. Huang S, Ziegler CGK, Austin J, Mannoun N, Vukovic M, Ordovas-Montanes J, et al. Lymph Nodes are Innervated by a Unique Population of Sensory Neurons With Immunomodulatory Potential. Cell (2021) 184(2):441–59 e25. doi: 10.1016/j.cell.2020.11.028
73. Prescott SL, Umans BD, Williams EK, Brust RD, Liberles SD. An Airway Protection Program Revealed by Sweeping Genetic Control of Vagal Afferents. Cell (2020) 181(3):574–89 e14. doi: 10.1016/j.cell.2020.03.004
74. Yuan A, Rao MV, Veeranna, Nixon RA. Neurofilaments and Neurofilament Proteins in Health and Disease. Cold Spring Harb Perspect Biol (2017) 9(4). doi: 10.1101/cshperspect.a018309
75. Burton PR, Wentz MA. Neurofilaments Are Prominent in Bullfrog Olfactory Axons But Are Rarely Seen in Those of the Tiger Salamander, Ambystoma Tigrinum. J Comp Neurol (1992) 317(4):396–406. doi: 10.1002/cne.903170406
76. Hoffman PN, Cleveland DW, Griffin JW, Landes PW, Cowan NJ, Price DL. Neurofilament Gene Expression: A Major Determinant of Axonal Caliber. Proc Natl Acad Sci U S A (1987) 84(10):3472–6. doi: 10.1073/pnas.84.10.3472
77. Yuan A, Rao MV, Veeranna, Nixon RA. Neurofilaments at a Glance. J Cell Sci (2012) 125(Pt 14):3257–63. doi: 10.1242/jcs.104729
78. Lawson SN, Waddell PJ. Soma Neurofilament Immunoreactivity is Related to Cell Size and Fibre Conduction Velocity in Rat Primary Sensory Neurons. J Physiol (1991) 435:41–63. doi: 10.1113/jphysiol.1991.sp018497
79. Ernfors P, Lee KF, Kucera J, Jaenisch R. Lack of Neurotrophin-3 Leads to Deficiencies in the Peripheral Nervous System and Loss of Limb Proprioceptive Afferents. Cell (1994) 77(4):503–12. doi: 10.1016/0092-8674(94)90213-5
80. Rutlin M, Ho CY, Abraira VE, Cassidy C, Bai L, Woodbury CJ, et al. The Cellular and Molecular Basis of Direction Selectivity of Adelta-LTMRs. Cell (2014) 159(7):1640–51. doi: 10.1016/j.cell.2014.11.038
81. Abraira VE, Ginty DD. The Sensory Neurons of Touch. Neuron (2013) 79(4):618–39. doi: 10.1016/j.neuron.2013.07.051
82. Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, et al. The Functional Organization of Cutaneous Low-Threshold Mechanosensory Neurons. Cell (2011) 147(7):1615–27. doi: 10.1016/j.cell.2011.11.027
83. Seal RP, Wang X, Guan Y, Raja SN, Woodbury CJ, Basbaum AI, et al. Injury-Induced Mechanical Hypersensitivity Requires C-Low Threshold Mechanoreceptors. Nature (2009) 462(7273):651–5. doi: 10.1038/nature08505
84. Averill S, McMahon SB, Clary DO, Reichardt LF, Priestley JV. Immunocytochemical Localization of trkA Receptors in Chemically Identified Subgroups of Adult Rat Sensory Neurons. Eur J Neurosci (1995) 7(7):1484–94. doi: 10.1111/j.1460-9568.1995.tb01143.x
85. Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and Molecular Mechanisms of Pain. Cell (2009) 139(2):267–84. doi: 10.1016/j.cell.2009.09.028
86. Le Pichon CE, Chesler AT. The Functional and Anatomical Dissection of Somatosensory Subpopulations Using Mouse Genetics. Front Neuroanat (2014) 8:21. doi: 10.3389/fnana.2014.00021
87. Kashem SW, Riedl MS, Yao C, Honda CN, Vulchanova L, Kaplan DH. Nociceptive Sensory Fibers Drive Interleukin-23 Production From CD301b+ Dermal Dendritic Cells and Drive Protective Cutaneous Immunity. Immunity (2015) 43(3):515–26. doi: 10.1016/j.immuni.2015.08.016
88. Wallrapp A, Burkett PR, Riesenfeld SJ, Kim SJ, Christian E, Abdulnour RE, et al. Calcitonin Gene-Related Peptide Negatively Regulates Alarmin-Driven Type 2 Innate Lymphoid Cell Responses. Immunity (2019) 51(4):709–23 e6. doi: 10.1016/j.immuni.2019.09.005
89. Nagashima H, Mahlakoiv T, Shih HY, Davis FP, Meylan F, Huang Y, et al. Neuropeptide CGRP Limits Group 2 Innate Lymphoid Cell Responses and Constrains Type 2 Inflammation. Immunity (2019) 51(4):682–95.e6. doi: 10.1016/j.immuni.2019.06.009
90. Serra MC, Bazzoni F, Della Bianca V, Greskowiak M, Rossi F. Activation of Human Neutrophils by Substance PEffect on Oxidative Metabolism, Exocytosis, Cytosolic Ca2+ Concentration and Inositol Phosphate Formation. J Immunol (1988) 141(6):2118–24.
91. Theoharides TC, Zhang B, Kempuraj D, Tagen M, Vasiadi M, Angelidou A, et al. IL-33 Augments Substance P-Induced VEGF Secretion From Human Mast Cells and Is Increased in Psoriatic Skin. Proc Natl Acad Sci U S A (2010) 107(9):4448–53. doi: 10.1073/pnas.1000803107
92. Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, et al. Mrgprd Enhances Excitability in Specific Populations of Cutaneous Murine Polymodal Nociceptors. J Neurosci (2009) 29(26):8612–9. doi: 10.1523/JNEUROSCI.1057-09.2009
93. Liu Q, Sikand P, Ma C, Tang Z, Han L, Li Z, et al. Mechanisms of Itch Evoked by Beta-Alanine. J Neurosci (2012) 32(42):14532–7. doi: 10.1523/JNEUROSCI.3509-12.2012
94. Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, et al. A Subpopulation of Nociceptors Specifically Linked to Itch. Nat Neurosci (2013) 16(2):174–82. doi: 10.1038/nn.3289
95. Kang SA, Govindarajan R. Anti-Calcitonin Gene-Related Peptide Monoclonal Antibodies for Neuropathic Pain in Patients With Migraine Headache. Muscle Nerve (2020) 63(4):563–7. doi: 10.1002/mus.27153
96. Iyengar S, Ossipov MH, Johnson KW. The Role of Calcitonin Gene-Related Peptide in Peripheral and Central Pain Mechanisms Including Migraine. Pain (2017) 158(4):543–59. doi: 10.1097/j.pain.0000000000000831
97. Mishra SK, Hoon MA. The Cells and Circuitry for Itch Responses in Mice. Science (2013) 340(6135):968–71. doi: 10.1126/science.1233765
98. Huang J, Polgar E, Solinski HJ, Mishra SK, Tseng PY, Iwagaki N, et al. Circuit Dissection of the Role of Somatostatin in Itch and Pain. Nat Neurosci (2018) 21(5):707–16. doi: 10.1038/s41593-018-0119-z
99. McKemy DD, Neuhausser WM, Julius D. Identification of a Cold Receptor Reveals a General Role for TRP Channels in Thermosensation. Nature (2002) 416(6876):52–8. doi: 10.1038/nature719
100. Caterina MJ, Rosen TA, Tominaga M, Brake AJ, Julius D. A Capsaicin-Receptor Homologue With a High Threshold for Noxious Heat. Nature (1999) 398(6726):436–41. doi: 10.1038/18906
101. Lolignier S, Gkika D, Andersson D, Leipold E, Vetter I, Viana F, et al. New Insight in Cold Pain: Role of Ion Channels, Modulation, and Clinical Perspectives. J Neurosci (2016) 36(45):11435–9. doi: 10.1523/JNEUROSCI.2327-16.2016
102. Peirs C, Seal RP. Neural Circuits for Pain: Recent Advances and Current Views. Science (2016) 354(6312):578–84. doi: 10.1126/science.aaf8933
103. Sathyamurthy A, Johnson KR, Matson KJE, Dobrott CI, Li L, Ryba AR, et al. Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity During Behavior. Cell Rep (2018) 22(8):2216–25. doi: 10.1016/j.celrep.2018.02.003
104. Wu X, Conlin VS, Morampudi V, Ryz NR, Nasser Y, Bhinder G, et al. Vasoactive Intestinal Polypeptide Promotes Intestinal Barrier Homeostasis and Protection Against Colitis in Mice. PloS One (2015) 10(5):e0125225. doi: 10.1371/journal.pone.0125225
105. Yadav M, Huang MC, Goetzl EJ. VPAC1 (Vasoactive Intestinal Peptide (VIP) Receptor Type 1) G Protein-Coupled Receptor Mediation of VIP Enhancement of Murine Experimental Colitis. Cell Immunol (2011) 267(2):124–32. doi: 10.1016/j.cellimm.2011.01.001
106. Seillet C, Luong K, Tellier J, Jacquelot N, Shen RD, Hickey P, et al. The Neuropeptide VIP Confers Anticipatory Mucosal Immunity by Regulating ILC3 Activity. Nat Immunol (2020) 21(2):168–77. doi: 10.1038/s41590-019-0567-y
107. Talbot J, Hahn P, Kroehling L, Nguyen H, Li D, Littman DR. Feeding-Dependent VIP neuron-ILC3 Circuit Regulates the Intestinal Barrier. Nature (2020) 579(7800):575–80. doi: 10.1038/s41586-020-2039-9
108. Talbot S, Abdulnour RE, Burkett PR, Lee S, Cronin SJ, Pascal MA, et al. Silencing Nociceptor Neurons Reduces Allergic Airway Inflammation. Neuron (2015) 87(2):341–54. doi: 10.1016/j.neuron.2015.06.007
109. Crosson T, Wang JC, Doyle B, Merrison H, Balood M, Parrin A, et al. FcepsilonR1-Expressing Nociceptors Trigger Allergic Airway Inflammation. J Allergy Clin Immunol (2021) 147(4):2330–42. doi: 10.1016/j.jaci.2020.12.644
110. Sternini C, Reeve JR Jr., Brecha N. Distribution and Characterization of Calcitonin Gene-Related Peptide Immunoreactivity in the Digestive System of Normal and Capsaicin-Treated Rats. Gastroenterology (1987) 93(4):852–62. doi: 10.1016/0016-5085(87)90450-1
111. Assas BM, Pennock JI, Miyan JA. Calcitonin Gene-Related Peptide Is a Key Neurotransmitter in the Neuro-Immune Axis. Front Neurosci (2014) 8:23. doi: 10.3389/fnins.2014.00023
112. Hosoi J, Murphy GF, Egan CL, Lerner EA, Grabbe S, Asahina A, et al. Regulation of Langerhans Cell Function by Nerves Containing Calcitonin Gene-Related Peptide. Nature (1993) 363(6425):159–63. doi: 10.1038/363159a0
113. Xu H, Ding J, Porter CBM, Wallrapp A, Tabaka M, Ma S, et al. Transcriptional Atlas of Intestinal Immune Cells Reveals That Neuropeptide Alpha-CGRP Modulates Group 2 Innate Lymphoid Cell Responses. Immunity (2019) 51(4):696–708.e9. doi: 10.1016/j.immuni.2019.09.004
114. Wang X, Xing L, Xing Y, Tang Y, Han C. Identification and Characterization of Immunoreactive Calcitonin Gene-Related Peptide From Lymphocytes of the Rat. J Neuroimmunol (1999) 94(1-2):95–102. doi: 10.1016/S0165-5728(98)00230-6
115. Bracci-Laudiero L, Aloe L, Caroleo MC, Buanne P, Costa N, Starace G, et al. Endogenous NGF Regulates CGRP Expression in Human Monocytes, and Affects HLA-DR and CD86 Expression and IL-10 Production. Blood (2005) 106(10):3507–14. doi: 10.1182/blood-2004-10-4055
116. Mazelin L, Theodorou V, Fioramonti J, Bueno L. Vagally Dependent Protective Action of Calcitonin Gene-Related Peptide on Colitis. Peptides (1999) 20(11):1367–74. doi: 10.1016/S0196-9781(99)00144-8
117. Evangelista S, Tramontana M. Involvement of Calcitonin Gene-Related Peptide in Rat Experimental Colitis. J Physiol Paris (1993) 87(4):277–80. doi: 10.1016/0928-4257(93)90017-N
118. Engel MA, Khalil M, Siklosi N, Mueller-Tribbensee SM, Neuhuber WL, Neurath MF, et al. Opposite Effects of Substance P and Calcitonin Gene-Related Peptide in Oxazolone Colitis. Dig Liver Dis (2012) 44(1):24–9. doi: 10.1016/j.dld.2011.08.030
119. Reinshagen M, Flamig G, Ernst S, Geerling I, Wong H, Walsh JH, et al. Calcitonin Gene-Related Peptide Mediates the Protective Effect of Sensory Nerves in a Model of Colonic Injury. J Pharmacol Exp Ther (1998) 286(2):657–61.
120. Pinho-Ribeiro FA, Baddal B, Haarsma R, O’Seaghdha M, Yang NJ, Blake KJ, et al. Blocking Neuronal Signaling to Immune Cells Treats Streptococcal Invasive Infection. Cell (2018) 173(5):1083–97 e22. doi: 10.1016/j.cell.2018.04.006
121. Lai NY, Musser MA, Pinho-Ribeiro FA, Baral P, Jacobson A, Ma P, et al. Gut-Innervating Nociceptor Neurons Regulate Peyer’s Patch Microfold Cells and SFB Levels to Mediate Salmonella Host Defense. Cell (2020) 180(1):33–49 e22. doi: 10.1016/j.cell.2019.11.014
122. Mantyh PW. Neurobiology of Substance P and the NK1 Receptor. J Clin Psychiatry (2002) 63 Suppl 11:6–10.
123. Hokfelt T, Vincent S, Dalsgaard CJ, Skirboll L, Johansson O, Schultzberg M, et al. Distribution of Substance P in Brain and Periphery and Its Possible Role as a Co-Transmitter. Ciba Found Symp (1982) 1982(91):84–106. doi: 10.1002/9780470720738.ch6
124. Lai NY, Mills K, Chiu IM. Sensory Neuron Regulation of Gastrointestinal Inflammation and Bacterial Host Defence. J Intern Med (2017) 282(1):5–23. doi: 10.1111/joim.12591
125. De Camilli P, Jahn R. Pathways to Regulated Exocytosis in Neurons. Annu Rev Physiol (1990) 52:625–45. doi: 10.1146/annurev.ph.52.030190.003205
126. Suvas S. Role of Substance P Neuropeptide in Inflammation, Wound Healing, and Tissue Homeostasis. J Immunol (2017) 199(5):1543–52. doi: 10.4049/jimmunol.1601751
127. Shimizu Y, Matsuyama H, Shiina T, Takewaki T, Furness JB. Tachykinins and Their Functions in the Gastrointestinal Tract. Cell Mol Life Sci (2008) 65(2):295–311. doi: 10.1007/s00018-007-7148-1
128. Engel MA, Khalil M, Mueller-Tribbensee SM, Becker C, Neuhuber WL, Neurath MF, et al. The Proximodistal Aggravation of Colitis Depends on Substance P Released From TRPV1-Expressing Sensory Neurons. J Gastroenterol (2012) 47(3):256–65. doi: 10.1007/s00535-011-0495-6
129. Engel MA, Leffler A, Niedermirtl F, Babes A, Zimmermann K, Filipovic MR, et al. TRPA1 and Substance P Mediate Colitis in Mice. Gastroenterology (2011) 141(4):1346–58. doi: 10.1053/j.gastro.2011.07.002
130. Yissachar N, Zhou Y, Ung L, Lai NY, Mohan JF, Ehrlicher A, et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell (2017) 168(6):1135–48 e12. doi: 10.1016/j.cell.2017.02.009
131. Perner C, Flayer CH, Zhu X, Aderhold PA, Dewan ZNA, Voisin T, et al. Substance P Release by Sensory Neurons Triggers Dendritic Cell Migration and Initiates the Type-2 Immune Response to Allergens. Immunity (2020) 53(5):1063–77 e7. doi: 10.1016/j.immuni.2020.10.001
132. Brighton PJ, Szekeres PG, Willars GB. Neuromedin U and its Receptors: Structure, Function, and Physiological Roles. Pharmacol Rev (2004) 56(2):231–48. doi: 10.1124/pr.56.2.3
133. Klose CSN, Mahlakoiv T, Moeller JB, Rankin LC, Flamar AL, Kabata H, et al. The Neuropeptide Neuromedin U Stimulates Innate Lymphoid Cells and Type 2 Inflammation. Nature (2017) 549(7671):282–6. doi: 10.1038/nature23676
134. Moriyama M, Sato T, Inoue H, Fukuyama S, Teranishi H, Kangawa K, et al. The Neuropeptide Neuromedin U Promotes Inflammation by Direct Activation of Mast Cells. J Exp Med (2005) 202(2):217–24. doi: 10.1084/jem.20050248
135. Moriyama M, Fukuyama S, Inoue H, Matsumoto T, Sato T, Tanaka K, et al. The Neuropeptide Neuromedin U Activates Eosinophils and Is Involved in Allergen-Induced Eosinophilia. Am J Physiol Lung Cell Mol Physiol (2006) 290(5):L971–7. doi: 10.1152/ajplung.00345.2005
136. Moriyama M, Matsukawa A, Kudoh S, Takahashi T, Sato T, Kano T, et al. The Neuropeptide Neuromedin U Promotes IL-6 Production From Macrophages and Endotoxin Shock. Biochem Biophys Res Commun (2006) 341(4):1149–54. doi: 10.1016/j.bbrc.2006.01.075
137. Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour RE, Nyman J, Dionne D, et al. The Neuropeptide NMU Amplifies ILC2-Driven Allergic Lung Inflammation. Nature (2017) 549(7672):351–6. doi: 10.1038/nature24029
138. Gabanyi I, Muller PA, Feighery L, Oliveira TY, Costa-Pinto FA, Mucida D. Neuro-Immune Interactions Drive Tissue Programming in Intestinal Macrophages. Cell (2016) 164(3):378–91. doi: 10.1016/j.cell.2015.12.023
139. Moriyama S, Brestoff JR, Flamar AL, Moeller JB, Klose CSN, Rankin LC, et al. beta2-adrenergic Receptor-Mediated Negative Regulation of Group 2 Innate Lymphoid Cell Responses. Science (2018) 359(6379):1056–61. doi: 10.1126/science.aan4829
140. Nicholls AJ, Wen SW, Hall P, Hickey MJ, Wong CHY. Activation of the Sympathetic Nervous System Modulates Neutrophil Function. J Leukoc Biol (2018) 103(2):295–309. doi: 10.1002/JLB.3MA0517-194RR
141. Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susarla S, et al. Nicotinic Acetylcholine Receptor Alpha7 Subunit Is an Essential Regulator of Inflammation. Nature (2003) 421(6921):384–8. doi: 10.1038/nature01339
142. Tracey KJ. Physiology and Immunology of the Cholinergic Antiinflammatory Pathway. J Clin Invest (2007) 117(2):289–96. doi: 10.1172/JCI30555
143. Borovikova LV, Ivanova S, Zhang M, Yang H, Botchkina GI, Watkins LR, et al. Vagus Nerve Stimulation Attenuates the Systemic Inflammatory Response to Endotoxin. Nature (2000) 405(6785):458–62. doi: 10.1038/35013070
144. Koopman FA, Chavan SS, Miljko S, Grazio S, Sokolovic S, Schuurman PR, et al. Vagus Nerve Stimulation Inhibits Cytokine Production and Attenuates Disease Severity in Rheumatoid Arthritis. Proc Natl Acad Sci U S A (2016) 113(29):8284–9. doi: 10.1073/pnas.1605635113
145. Galle-Treger L, Suzuki Y, Patel N, Sankaranarayanan I, Aron JL, Maazi H, et al. Nicotinic Acetylcholine Receptor Agonist Attenuates ILC2-Dependent Airway Hyperreactivity. Nat Commun (2016) 7:13202. doi: 10.1038/ncomms13202
146. Dalli J, Colas RA, Arnardottir H, Serhan CN. Vagal Regulation of Group 3 Innate Lymphoid Cells and the Immunoresolvent PCTR1 Controls Infection Resolution. Immunity (2017) 46(1):92–105. doi: 10.1016/j.immuni.2016.12.009
147. Darby M, Roberts LB, Mackowiak C, Chetty A, Tinelli S, Schnoeller C, et al. ILC3-Derived Acetylcholine Promotes Protease-Driven Allergic Lung Pathology. J Allergy Clin Immunol (2020) 147(4):1513–6.e4. doi: 10.1016/j.jaci.2020.10.038
148. Sanz E, Bean JC, Carey DP, Quintana A, McKnight GS. Ribotag: Ribosomal Tagging Strategy to Analyze Cell-Type-Specific mRNA Expression In Vivo. Curr Protoc Neurosci (2019) 88(1):e77. doi: 10.1002/cpns.77
149. Tracey KJ. Reflex Control of Immunity. Nat Rev Immunol (2009) 9(6):418–28. doi: 10.1038/nri2566
150. Bonaz B, Sinniger V, Hoffmann D, Clarencon D, Mathieu N, Dantzer C, et al. Chronic Vagus Nerve Stimulation in Crohn’s Disease: A 6-Month Follow-Up Pilot Study. Neurogastroenterol Motil (2016) 28(6):948–53. doi: 10.1111/nmo.12792
Keywords: neuro-immune interactions, neuronal classification, peripheral nervous system, enteric nervous system, dorsal root ganglia (DRG), function of neurons
Citation: Jakob MO, Kofoed-Branzk M, Deshpande D, Murugan S and Klose CSN (2021) An Integrated View on Neuronal Subsets in the Peripheral Nervous System and Their Role in Immunoregulation. Front. Immunol. 12:679055. doi: 10.3389/fimmu.2021.679055
Received: 10 March 2021; Accepted: 15 June 2021;
Published: 12 July 2021.
Edited by:
Francesca Ronchi, University of Bern, SwitzerlandReviewed by:
Sebastien Talbot, Université de Montréal, CanadaCopyright © 2021 Jakob, Kofoed-Branzk, Deshpande, Murugan and Klose. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christoph S. N. Klose, Y2hyaXN0b3BoLmtsb3NlQGNoYXJpdGUuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.