- Department of Biology, Bryn Mawr College, Bryn Mawr, PA, United States
Transmembrane protein engulfment receptors expressed on the surface of phagocytes engage ligands on apoptotic cells and debris to initiate a sequence of events culminating in material internalization and immunologically beneficial outcomes. Engulfment receptors are modular, comprised of functionally independent extracellular ligation domains and cytosolic signaling motifs. Cognate kinases, adaptors, and phosphatases regulate engulfment by controlling the degree of receptor activation in phagocyte plasma membranes, thus acting as receptor-proximal signaling modules. Here, we review recent efforts to reprogram phagocytes using modular synthetic receptors composed of antibody-based extracellular domains fused to engulfment receptor signaling domains. To aid the development of new phagocyte reprogramming methods, we then define the kinases, adaptors, and phosphatases that regulate a conserved family of engulfment receptors. Finally, we discuss current challenges and opportunities for the field.
Introduction
The Megf10/Draper/CED-1 receptors, an ancient family of single-pass transmembrane proteins, enable phagocytes across phyla to initiate the internalization of apoptotic cells, synapses, and cell debris (1–3). Ligation of engulfment receptor extracellular domains to “eat me” signals on the target, including the complement protein C1q and the lipid phosphatidylserine (PS), induces receptor phosphorylation by “writer” kinases. Phosphorylation recruits “reader” proteins that initiate a series of cytoskeletal and membrane-remodeling events that enable the phagocytes to ingest large targets and digest internalized material in intracellular compartments. “Eraser” phosphatases then dephosphorylate engulfment receptors to return the receptor to its unphosphorylated resting state (4). Writer-reader-eraser modules that regulate receptor activation are common components of vertebrate immune cell signaling pathways (Figure 1A). Notable examples include the Src-Syk-CD45 writer-reader-eraser network that controls Fc-receptor signaling and the Lck-ZAP70-CD45 module that tunes activation the T cell receptor (TCR) (5).
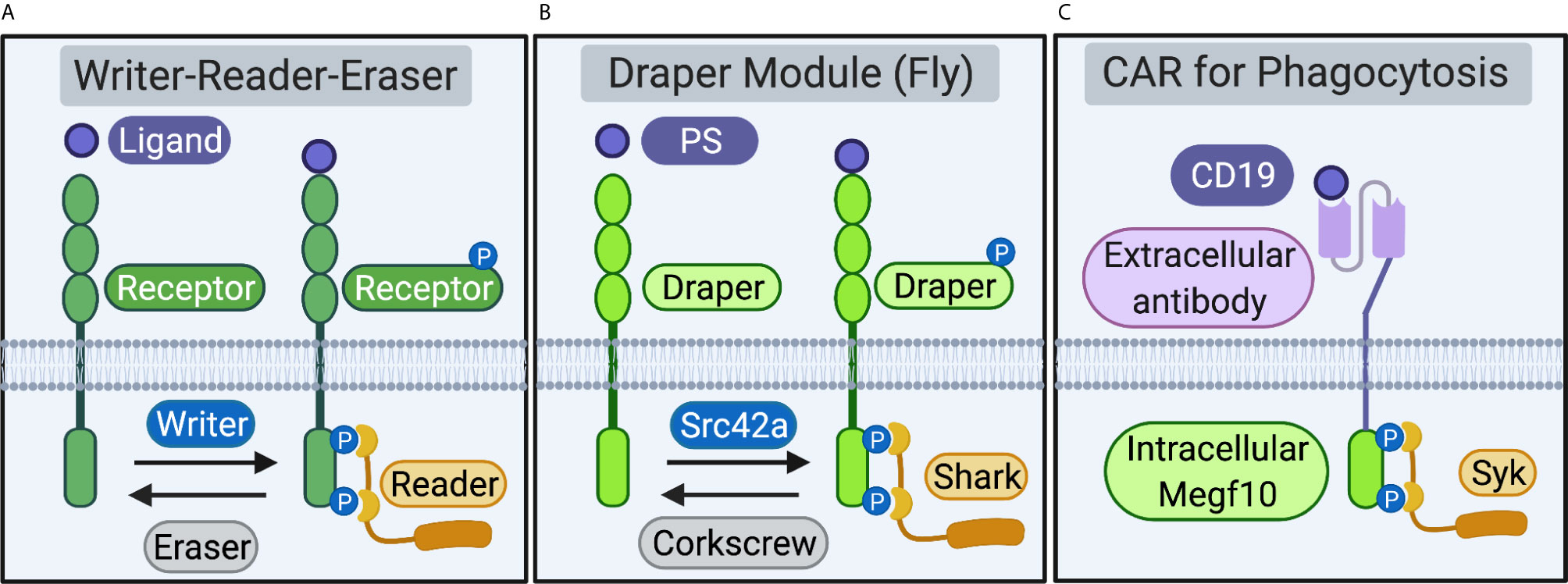
Figure 1 Receptors and proximal writer-reader-eraser modules. (A) Schematic of a generalized receptor and writer-reader-eraser module. Receptor ligation induces phosphorylation of a receptor intracellular signaling domain by a writer kinase. The phosphorylated receptor recruits a reader adaptor protein. An eraser phosphatase removes phosphorylation marks on the receptor intracellular domain, returning the system to its resting state. (B) A writer-reader-eraser module regulates signal transduction through the Draper receptor. Draper, expressed on fruit fly phagocytes, recognizes phosphatidylserine (PS) or other ligands. Phosphorylation of the Draper intracellular domain by the kinase Src42a induces recruitment of the adaptor kinase Shark. The phosphatase Corkscrew erases phosphorylation marks on the intracellular domain of Draper. (C) Chimeric Antigen Receptor for Phagocytosis (CAR-P). Synthetic CAR-Ps bind cancer antigens (e.g. the B cell antigen CD19) via an extracellular single-chain antibody fragment (scFv). Extracellular antibody ligation induces phosphorylation of engulfment receptor signaling domains (e.g. the intracellular domain of mouse Megf10). Phosphorylation of the Megf10 receptor signaling domain on CAR-P recruits the Syk adaptor, a cognate reader for murine Megf10. Schematic created in BioRender (biorender.com).
The Megf10/Draper/CED-1 receptors are composed of functionally separable extracellular ligation and intracellular signaling modules. The modularity of engulfment receptors enables phagocyte reprogramming, as replacement of the extracellular domain with a new recognition element directs phagocytes to identify and ingest new targets (6–8). The organization of engulfment signaling is also modular at the level of the receptor proximal signaling networks. Engulfment receptors function in concert with effector modules comprised of kinases, adaptors, and phosphatases that initiate the engulfment signaling program. Here, we review recent work that highlights the promise of reprogramming phagocytosis to understand basic immunology in pre-clinical settings. We then use the modular structure of synthetic engulfment receptors as an inlet to define and compare the writers, readers, and erasers that regulate initiation of the engulfment signaling program through the Megf10/Draper/CED-1 receptors (Figure 1B).
Reprogramming Phagocytosis Using Chimeric Antigen Receptors
In multiple immune cell types, including T cells, phagocytes, and natural killer cells, replacement of an extracellular domain with a single-chain antibody fragment (scFv) can reprogram an immune cell towards the cognate antigen of a specific scFv (7, 9, 10). Reprogramming immune cell signaling has ushered in a new suite of methods to engineer the immune response to target cancer (11). Analogous to Chimeric Antigen Receptors in T cells (CAR-T), the modular organization of engulfment receptors enables investigators to reprogram phagocytes to identify and eliminate targets of therapeutic interest.
The modular organization of the T cell, Fc-, and phagocyte receptors enables the reprogramming of immune cells to recognize and respond to non-native targets of therapeutic interest, including cancer antigens (6, 7, 9). Multiple groups have successfully reprogrammed macrophages to recognize cancer antigens by introducing synthetic Chimeric Antigen Receptors for Phagocytosis (CAR-Ps) (6, 7). Fusing the Megf10 or Fc-receptor signaling domains to an scFv recognizing the B-lymphocyte antigen CD19 programmed mouse macrophages to ingest antigen-coated beads and cancer cells (Figure 1C) (7). Surprisingly, chimeric receptors expressed in macrophages composed of extracellular scFvs fused to the T cell receptor signaling domain CD3ζ also drive engulfment (6, 7). Importantly, Klichinsky et al. also demonstrated the following: anti-cancer activity in humanized mouse models; induction of a pro-inflammatory tumor microenvironment; and antigen cross presentation to boost anti-tumor T cell responses (6). Collectively, coupling scFv-based extracellular modules to ITAM-based intracellular signaling domains from Megf10, Fc-receptor, and the TCR, presents an attractive strategy to reprogram macrophages towards therapeutically relevant targets linked to both hematopoietic malignancies and solid tumors (6, 7).
At present, methods primarily rely on extracellular antibody-based modules that recognize antigens with high affinity. However, high-affinity CAR-Ps may signal in a manner that does not reflect endogenous receptor mechanisms. Recent work demonstrates that T cells transduce signals through CAR-T independently of a key T cell signaling protein required by physiological T cells called Linker of Activated T cells (LAT) (12). Future work for the field will require engineering molecules that more closely reflect the biology of native engulfment receptors, perhaps by developing new CARs that combine multiple signaling modules to orient cellular responses towards desired immunological outcomes. To learn how engulfment receptors interface with proximal signaling modules, we systematically defined the writer, reader, and erasers that regulate the Megf10/Draper/CED-1 family.
Modular Organization of Engulfment Signaling Networks Across Phyla
Application of the writer-reader-eraser framework enables conserved rules and regulatory principles underlying engulfment to emerge from this complex collection of receptors and their proximal signaling networks. Here, writers refer to protein tyrosine kinases that create reversible binding sites for readers. The readers, SH2- and PTB-domain-containing proteins, initiate the engulfment program by activating “downstream” signaling modules that remodel the actin cytoskeleton and cell membranes (13). The cycle is completed by erasers, protein tyrosine phosphatases that return the system to its deactivated state.
Megf10: Clearing Corpses and Shaping Synapses in the Mouse Brain
Megf10, a receptor expressed on phagocytic astrocytes in the mouse brain, promotes synapse elimination (14) and apoptotic cell clearance (1). Megf10 also plays roles in retinal patterning (15) and elimination of amyloid-β (16). After recognition of its ligand, the complement protein C1q (1), Src-family kinase writers phosphorylate Tyrosine (Tyr) residues within the Megf10 Immunoreceptor Tyrosine-based Activation Motif (ITAM) (17). In addition to its ITAM, the cytosolic tail of Megf10 encodes an NPxY motif (18), a domain capable of recruiting cytosolic signaling proteins via direct interaction with phosphotyrosine binding (PTB) domains. Consistent with a writer function for Src-family kinases, treating HeLa cells that overexpress Megf10 with the Src-family kinase inhibitor PP2 decreases engulfment of microspheres by HeLa cells transfected with Megf10 (17). Thus, multiple Src-family kinases can create binding sites for readers on the cytosolic tail of Megf10.
The tandem SH2-domain-containing kinase Syk is a Megf10 reader that interacts with phosphorylated ITAM residues on the cytosolic domain of Megf10 (17). Though the specific actions of Syk that enable phagocytosis in glia are unclear, Syk is an important engulfment effector in macrophages and plays a role in remodeling the actin cytoskeleton when bound to the Fc-receptor (19). The PTB-containing protein GULP is upregulated in reactive astrocytes after transient ischemic injury, an expression pattern similar to Megf10 (20). Thus, in addition to Syk, GULP is a candidate to interact with the intracellular domain of Megf10 via an NPxY motif.
A specific eraser for Megf10 is not reported, though mouse astrocytes express SHP-2 (PTPN11) mRNA (21). SHP-2 is a cytosolic protein tyrosine phosphatase that dephosphorylates the ITAM-bearing C-type lectin receptor that governs anti-fungal immunity (22). In sum, Megf10 is an invariant single-pass transmembrane engulfment receptor. A module comprised of multiple Src-family kinases and Syk enables Megf10 to initiate the clearance of apoptotic cells and synapses.
Draper and a Proximal Writer-Reader-Eraser Module Initiate Engulfment by Fly Phagocytes
Phagocytes in the fruit fly Drosophila melanogaster clear apoptotic corpses and axonal debris to maintain tissue homeostasis and repair the nervous system after injury. Engulfment in flies is carried out by an array of cell types including hemocytes (23), epithelial cells (24), and glia (2, 25). The Draper receptor, expressed in each of these cell types, is comprised of an extracellular emlin (EMI) domain that facilitates protein-protein interactions (26) and nimrod (NIM) repeats, EGF-like domains found on multiple fruit fly innate immune receptors (27). The intracellular signaling domain of Draper contains two phosphotyrosine-based signaling sequences that serve as candidate sites for phosphorylation by Src-family kinase writers: an NPxY and an ITAM.
Ligation of Draper to phosphatidylserine induces phosphorylation of the cytosolic signaling domain of Draper (8, 28). Intriguingly, Draper recognizes a diverse array of additional ligands in other contexts including the proteins Pretaporter (29) and DmCaBP1 (30) and the bacterial surface molecule Lipoteichoic acid (31). Once ligand-receptor binding occurs, clustered Draper is rapidly phosphorylated in phagocyte plasma membranes (8). At least 5 residues on the cytoplasmic signaling domain of Draper, including at least 4 outside the ITAM, are phosphorylated by the kinase Src42a (8, 32). The kinase Src42a is the writer in this system, creating a binding site for the tandem SH2-domain kinase Shark (32) (Figure 1B).
The cytosolic protein tyrosine phosphatase Corkscrew, which binds an alternatively spliced isoform of Draper called Draper-II via an Immunoreceptor Tyrosine-based Inhibitory Motif (ITIM), is the eraser for Draper (33). Corkscrew is capable of dephosphorylating Draper and, intriguingly, loss of Corkscrew in vivo inhibits the ability of glia to respond to secondary injury (33). The interdependence of the writer, reader, and eraser in the fly system underscores the importance of returning this immune cell signaling system to its ground state to respond to future insults.
Collectively, a wealth of genetic, biochemical, and cell biological evidence supports the model that engulfment through Draper is carried out by a writer-reader-eraser module comprised of a writer kinase Src42a, an SH2-domain-containing reader Shark, and an eraser Corkscrew. As writers, readers, and erasers remain to be determined in other organisms, the fruit fly represents an exceptional model system for defining the mechanisms underlying engulfment through the Megf10/Draper/CED-1 receptors.
CED-1 in the Worm: A Divergent Engulfment Receptor
Genetic work in the nematode Caenorhabditis elegans, including the discovery of the CED-1 receptor (3), founded the engulfment field. C. elegans phagocytes clear dead and dying cells using two pathways governed by engulfment factors expressed by ced genes. One pathway includes ced-2, ced-5, and ced-12 while the other pathway includes ced-1, ced-6, and ced-7 (34). These pathways converge on actin to facilitate rearrangements that enable engulfment (35).
Corpse clearance by phagocytic cells in the worm is initiated when cells neighboring the apoptotic cell release the secretory protein Transthyretin-related 52 (TTR-52) (36). TTR-52 binds PS on the corpse and serves as a bridging molecule to the receptor CED-1. CED-1 ligation to TTR-52 induces activation of the CED-6 and CED-7 signaling pathway. Like Draper and Megf10, the extracellular domain of CED-1 contains EMI and EGF-like domains (3, 26). The cytosolic signaling tail on CED-1 contains a possible Tyrosine phosphorylation site on a single YxxL motif (3). The tail also contains an NPxY motif with a Tyr residue. However, in vitro evidence indicates that NPxY motifs may bind PTB-domain-containing proteins such as CED-6 in the absence of Tyr phosphorylation (37). Thus, our application of the writer-reader-eraser framework to the C. elegans CED-1/CED-6/CED-7 engulfment module reveals that while worms do encode candidate writer kinases and eraser phosphatases, the CED-1 receptor may perform engulfment in the absence of Tyr phosphorylation.
Conclusions, Current Challenges, and New Opportunities
Comparison of Writer-Reader-Eraser Modules Used by Phagocytes Across Phyla
Our application of the writer-reader-eraser framework to define the proximal signaling networks that regulate the Megf10/Draper/CED-1 receptor family revealed broad mechanistic similarities and intriguing differences (Table 1). All three receptors contain at least one cytoplasmic Tyr residue in an ITAM or NPxY motif, potential phosphorylation sites for kinase writers. At least one reader protein interacts with Megf10, Draper, or CED-1 to initiate the complex series of cytoskeletal and membrane-remodeling events that power material internalization. Megf10 and Draper recruit a Syk-family kinase reader and CED-6/GULP adaptor proteins. CED-1 binds the reader CED-6. Collectively, the proximal writers and readers governing Megf10 and Draper function are highly similar to one another. Because the CED-6 adaptor protein may bind CED-1 independently of Tyr phosphorylation, CED-1 may function via a distinct activation mechanism that does not require a writer kinase. Less is known about the eraser phosphatases that negatively regulate the Megf10/Draper/CED-1 receptors. The eraser for Draper, a SHP-2 tyrosine phosphatase ortholog called Corkscrew, works by binding an inhibitory splice isoform of Draper (33). The cognate phosphatases for Megf10 and CED-1, though presumably expressed by the relevant phagocytes, remain unreported.
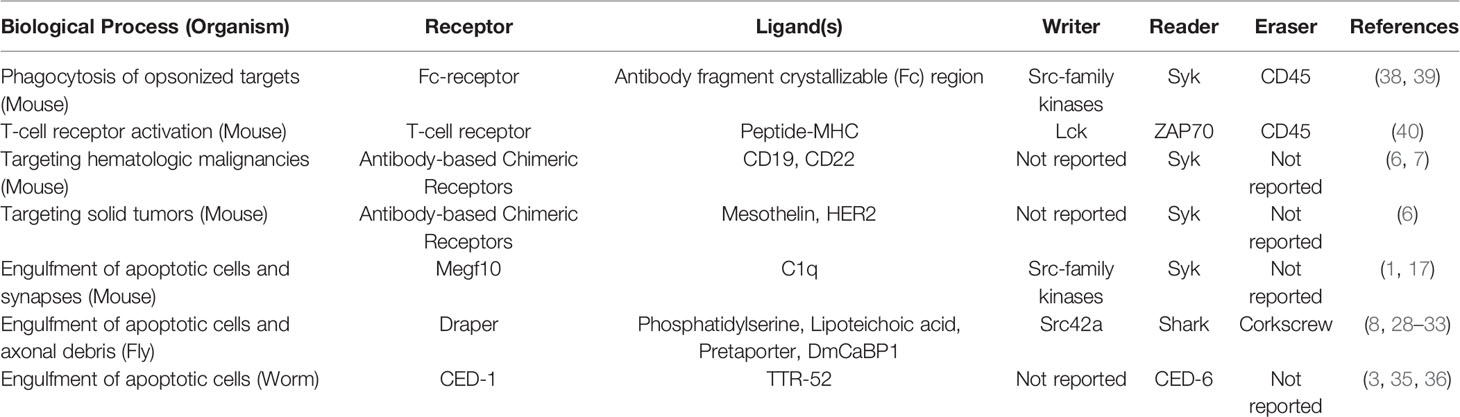
Table 1 Summary of receptors and writer-reader-eraser modules used across biological processes and organisms.
Megf10, Draper, and CED-1 differ remarkably in their ligand specificities, perhaps a result of their evolutionary divergence and the large number of different “eat me” signals presented on the surface of apoptotic cells and other targets (41). Megf10 binds the complement protein C1q to initiate the engulfment of apoptotic cells (1). Draper initiates engulfment by interacting with diverse ligands including the lipid PS and the proteins DmCaBP1 and Pretaporter (8, 28–30). Finally, CED-1 recognizes the secreted protein TTR-52 that serves as A bridge between CED-1 and PS (36). Perhaps the diversity of ligands that the Megf10/Draper/CED-1 receptors recognize provides the mechanistic basis for reprogramming the receptors using synthetic extracellular domains. We hope that further study of engulfment receptors and their proximal signaling modules will enable researchers to design a new generation of synthetic signaling systems comprised of CARs that interact with engineered intracellular signaling modules.
Reprogramming Phagocytes Toward New Targets
In theory, new CAR-Ps that enable phagocytes to bind any extracellular cell surface protein or secreted molecule should program phagocytes to ingest non-native targets. At present, however, implementing CAR-Ps in a clinical context requires laborious editing protocols to manipulate autologous cells. Thus, the use of CAR-Ps as therapies to treat human disease remains a goal for the future. For now, CAR-Ps are emerging as important tools to define basic immunology by elucidating the mechanisms used by receptors expressed across diverse phagocyte populations.
As investigators continue to design and express new receptors, it will be critical to move beyond internalization to learn how engulfed material is targeted to promote specific immunological outcomes. For example, recent work linking the receptor DNGR-1 to antigen cross-presentation suggests that expression of a DNGR-1-based CAR-P in Dendritic Cells may drive the efficient cross-presentation of cancer antigens (42). In the long term, replacing cell-based CAR-Ps with bi- or tri-specific antibodies that couple desired phagocyte populations exposing known cell surface molecules to disease-associated antigens may prove to be viable, inexpensive strategies that could democratize access to new therapies.
Reprogrammed Phagocytes That Facilitate Nondestructive Immune Responses
In conclusion, further study of engineered phagocytes could spur the development of therapeutic interventions that cover a broad array of “accommodation” immune archetypes: active, nondestructive responses that, among other outcomes, promote wound healing and tissue repair (43). Phagocytes are particularly well suited to promote nondestructive responses because their endogenous functions include clearing apoptotic cells without inducing local inflammatory responses. In this review, we focused on modular receptor signaling systems that offer opportunities to reprogram phagocytes to ingest cancer cells. A key future challenge for the field is to develop phagocyte engineering strategies targeted at a broader range of diseases and cell populations. We anticipate that the development of new chimeric receptors that connect extracellular ligation to synthetic intracellular signaling modules will lead to exciting discoveries at the frontier of this nascent, expanding field.
Author Contributions
EB, VG, and AS contributed equally to this work and are listed alphabetically. EB completed an analysis of the Megf10 primary literature and drafted and edited the article. VG completed an analysis of the CED-1 primary literature and drafted and edited the article. AS completed an analysis of the Draper primary literature and drafted and edited the article. AW drafted and edited the article. All authors contributed to the article and approved the submitted version.
Funding
EB, VG, AS – Undergraduate Summer Science Research Funding, Bryn Mawr College. AW – Research Startup Funds, Bryn Mawr College.
Conflict of Interest
AW is an inventor on U.S. patent application WO2020097193 “Chimeric antigen receptors for phagocytosis” and receives licensing fees for the work described in this article.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
EB, VG, and AS thank the Bryn Mawr Undergraduate Summer Science Research program for financial support. Work in the Williamson Lab is supported by research startup funds from Bryn Mawr College. We thank Ani Dixit, Ellen Luecke, Vanessa Shehu, and Katrina Sparks for productive discussions and comments on the manuscript. We also acknowledge the efforts of two reviewers who provided helpful feedback during peer-review.
References
1. Iram T, Ramirez-Ortiz Z, Byrne MH, Coleman UA, Kingery ND, Means TK, et al. Megf10 Is a Receptor for C1Q That Mediates Clearance of Apoptotic Cells by Astrocytes. J Neurosci (2016) 36(19):5185–92. doi: 10.1523/JNEUROSCI.3850-15.2016
2. MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila Cell Corpse Engulfment Receptor Draper Mediates Glial Clearance of Severed Axons. Neuron (2006) 50(6):869–81. doi: 10.1016/j.neuron.2006.04.028
3. Zhou Z, Hartwieg E, Horvitz HR. CED-1 Is a Transmembrane Receptor that Mediates Cell Corpse Engulfment in C. elegans. Cell (2001) 104(1):43–56. doi: 10.1016/S0092-8674(01)00190-8
4. Lim WA, Pawson T. Phosphotyrosine Signaling: Evolving a New Cellular Communication System. Cell (2010) 142(5):661–7. doi: 10.1016/j.cell.2010.08.023
5. Niedergang F, Di Bartolo V, Alcover A. Comparative Anatomy of Phagocytic and Immunological Synapses. Front Immunol (2016) 7:18. doi: 10.3389/fimmu.2016.00018
6. Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol (2020) 38:947–53. doi: 10.1038/s41587-020-0462-y
7. Morrissey MA, Williamson AP, Steinbach AM, Roberts EW, Kern N, Headley MB, et al. Chimeric antigen receptors that trigger phagocytosis. eLife (2018) 7:e36688. doi: 10.7554/eLife.36688
8. Williamson AP, Vale RD. Spatial control of Draper receptor signaling initiates apoptotic cell engulfment. J Cell Biol (2018) 217(11):3977–92. doi: 10.1083/jcb.201711175
9. Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. New Engl J Med (2013) 368(16):1509–18. doi: 10.1056/NEJMoa1215134
10. Töpfer K, Cartellieri M, Michen S, Wiedemuth R, Müller N, Lindemann D, et al. DAP12-Based Activating Chimeric Antigen Receptor for NK Cell Tumor Immunotherapy. J Immunol (2015) 194(7):3201–12. doi: 10.4049/jimmunol.1400330
11. Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell (2017) 168(4):724–40. doi: 10.1016/j.cell.2017.01.016
12. Dong R, Libby KA, Blaeschke F, Fuchs W, Marson A, Vale RD, et al. Rewired signaling network in T cells expressing the chimeric antigen receptor (CAR). EMBO J (2020) 39(16):e104730. doi: 10.15252/embj.2020104730
13. Zhang Y, Hoppe AD, Swanson JA. Coordination of Fc receptor signaling regulates cellular commitment to phagocytosis. Proc Natl Acad Sci (2010) 107(45):19332–7. doi: 10.1073/pnas.1008248107
14. Chung W-S, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature (2013) 504(7480):394–400. doi: 10.1038/nature12776
15. Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature (2012) 483(7390):465–9. doi: 10.1038/nature10877
16. Singh TD, Park S-Y, Bae J, Yun Y, Bae Y-C, Park R-W, et al. MEGF10 functions as a receptor for the uptake of amyloid-β. FEBS Lett (2010) 584(18):3936–42. doi: 10.1016/j.febslet.2010.08.050
17. Scheib JL, Sullivan CS, Carter BD. Jedi-1 and MEGF10 Signal Engulfment of Apoptotic Neurons through the Tyrosine Kinase Syk. J Neurosci (2012) 32(38):13022–31. doi: 10.1523/JNEUROSCI.6350-11.2012
18. Wu H-H, Bellmunt E, Scheib JL, Venegas V, Burkert C, Reichardt LF, et al. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat Neurosci (2009) 12(12):1534–41. doi: 10.1038/nn.2446
19. Jaumouillé V, Farkash Y, Jaqaman K, Das R, Lowell CA, Grinstein S. Actin Cytoskeleton Reorganization by Syk Regulates Fcγ Receptor Responsiveness by Increasing Its Lateral Mobility and Clustering. Dev Cell (2014) 29(5):534–46. doi: 10.1016/j.devcel.2014.04.031
20. Morizawa YM, Hirayama Y, Ohno N, Shibata S, Shigetomi E, Sui Y, et al. Reactive astrocytes function as phagocytes after brain ischemia via ABCA1-mediated pathway. Nat Commun (2017) 8(28). doi: 10.1038/s41467-017-00037-1
21. Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J Neurosci (2014) 34(36):11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014
22. Deng Z, Ma S, Zhou H, Zang A, Fang Y, Li T, et al. Tyrosine phosphatase SHP-2 mediates C-type lectin receptor–induced activation of the kinase Syk and anti-fungal TH17 responses. Nat Immunol (2015) 16(6):642–52. doi: 10.1038/ni.3155
23. Wood W, Martin P. Macrophage Functions in Tissue Patterning and Disease: New Insights from the Fly. Dev Cell (2017) 40(3):221–33. doi: 10.1016/j.devcel.2017.01.001
24. Etchegaray JI, Timmons AK, Klein AP, Pritchett TL, Welch E, Meehan TL, et al. Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Development (2012) 139(21):4029–39. doi: 10.1242/dev.082776
25. Sonnenfeld MJ, Jacobs JR. Macrophages and glia participate in the removal of apoptotic neurons from theDrosophila embryonic nervous system. J Comp Neurol (1995) 359(4):644–52. doi: 10.1002/cne.903590410
26. Callebaut I, Mignotte V, Souchet M, Mornon J-P. EMI domains are widespread and reveal the probable orthologs of the Caenorhabditis elegans CED-1 protein. Biochem Biophys Res Commun (2003) 300(3):619–23. doi: 10.1016/S0006-291X(02)02904-2
27. Somogyi K, Sipos B, Penzes Z, Kurucz E, Zsamboki J, Hultmark D, et al. Evolution of Genes and Repeats in the Nimrod Superfamily. Mol Biol Evol (2008) 25(11):2337–47. doi: 10.1093/molbev/msn180
28. Tung TT, Nagaosa K, Fujita Y, Kita A, Mori H, Okada R, et al. Phosphatidylserine recognition and induction of apoptotic cell clearance by Drosophila engulfment receptor Draper. J Biochem (2013) 153(5):483–91. doi: 10.1093/jb/mvt014
29. Kuraishi T, Nakagawa Y, Nagaosa K, Hashimoto Y, Ishimoto T, Moki T, et al. Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. EMBO J (2009) 28(24):3868–78. doi: 10.1038/emboj.2009.343
30. Okada R, Nagaosa K, Kuraishi T, Nakayama H, Yamamoto N, Nakagawa Y, et al. Apoptosis-dependent Externalization and Involvement in Apoptotic Cell Clearance of DmCaBP1, an Endoplasmic Reticulum Protein of Drosophila*. J Biol Chem (2012) 287(5):3138–46. doi: 10.1074/jbc.M111.277921
31. Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, et al. Identification of Lipoteichoic Acid as a Ligand for Draper in the Phagocytosis of Staphylococcus aureus by Drosophila Hemocytes. J Immunol (2009) 183(11):7451–60. doi: 10.4049/jimmunol.0901032
32. Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung Y-G, et al. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature (2008) 453(7197):935–9. doi: 10.1038/nature06901
33. Logan MA, Hackett R, Doherty J, Sheehan A, Speese SD, Freeman MR. Negative regulation of glial engulfment activity by Draper terminates glial responses to axon injury. Nat Neurosci (2012) 15(5):722–30. doi: 10.1038/nn.3066
34. Gumienny TL, Brugnera E, Tosello-Trampont A-C, Kinchen JM, Haney LB, Nishiwaki K, et al. CED-12/ELMO, a Novel Member of the CrkII/Dock180/Rac Pathway, Is Required for Phagocytosis and Cell Migration. Cell (2001) 107(1):27–41. doi: 10.1016/S0092-8674(01)00520-7
35. Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature (2005) 434(7029):93–9. doi: 10.1038/nature03263
36. Wang X, Li W, Zhao D, Liu B, Shi Y, Chen B, et al. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat Cell Biol (2010) 12(7):655–64. doi: 10.1038/ncb2068
37. Su HP, Nakada-Tsukui K, Tosello-Trampont A-C, Li Y, Bu G, Henson PM, et al. Interaction of CED-6/GULP, an Adapter Protein Involved in Engulfment of Apoptotic Cells with CED-1 and CD91/Low Density Lipoprotein Receptor-related Protein (LRP). J Biol Chem (2002) 277(14):11772–9. doi: 10.1074/jbc.M109336200
38. Freeman SA, Goyette J, Furuya W, Woods EC, Bertozzi CR, Bergmeier W, et al. Integrins Form an Expanding Diffusional Barrier that Coordinates Phagocytosis. Cell (2016) 164(1–2):128–40. doi: 10.1016/j.cell.2015.11.048
39. Greenberg S, Chang P, Wang DC, Xavier R, Seed B. Clustered syk tyrosine kinase domains trigger phagocytosis. Proc Natl Acad Sci (1996) 93(3):1103–7. doi: 10.1073/pnas.93.3.1103
40. Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol (2006) 7(8):803–9. doi: 10.1038/ni1369
41. Ravichandran KS. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J Exp Med (2010) 207(9):1807–17. doi: 10.1084/jem.20101157
42. Canton J, Blees H, Henry CM, Buck MD, Schulz O, Rogers NC, et al. The receptor DNGR-1 signals for phagosomal rupture to promote cross-presentation of dead-cell-associated antigens. Nat Immunol (2021) 22140–53. doi: 10.1038/s41590-020-00824-x
Keywords: phagocytosis, engulfment, immune receptor, signal transduction, macrophage reprogramming, chimeric antigen receptor, immunotherapy
Citation: Britt EA, Gitau V, Saha A and Williamson AP (2021) Modular Organization of Engulfment Receptors and Proximal Signaling Networks: Avenues to Reprogram Phagocytosis. Front. Immunol. 12:661974. doi: 10.3389/fimmu.2021.661974
Received: 31 January 2021; Accepted: 22 March 2021;
Published: 19 April 2021.
Edited by:
Valentin Jaumouillé, Simon Fraser University, CanadaReviewed by:
Nicolas Touret, University of Alberta, CanadaCarlos Rosales, Universidad Nacional Autónoma de México, Mexico
Copyright © 2021 Britt, Gitau, Saha and Williamson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam P. Williamson, YXdpbGxpYW1zNkBicnlubWF3ci5lZHU=
†These authors have contributed equally to this work
 Emily A. Britt
Emily A. Britt Vanessa Gitau
Vanessa Gitau Amara Saha
Amara Saha Adam P. Williamson
Adam P. Williamson