- 1Division of Pharmacogenomics and Personalized Medicine, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
- 2Laboratory for Pharmacogenomics, Somdech Phra Debaratana Medical Center (SDMC), Ramathibodi Hospital, Bangkok, Thailand
- 3Division of General Pharmacy Practice, Department of Pharmaceutical Care, College of Pharmacy, Rangsit University, Pathum Thani, Thailand
- 4Department of Clinical Chemistry, Faculty of Medical Technology, Huachiew Chalermprakiet University, Samut Prakan, Thailand
- 5The Skin and Allergy Research Unit, Chulalongkorn University, Bangkok, Thailand
- 6Division of Dermatology, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 7Division of Allergy and Clinical Immunology, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 8King Chulalongkorn Memorial Hospital, Thai Red Cross Society, Bangkok, Thailand
- 9Department of Pharmacology, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand
- 10Biocatalyst and Environmental Biotechnology Research Unit, Department of Biochemistry, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 11Program in Bioinformatics and Computational Biology, Graduated School, Chulalongkorn University, Bangkok, Thailand
- 12Pharmacy Unit, Udon Thani Hospital, Udon Thani, Thailand
- 13Unit of Excellence on Pharmacogenomic Pharmacokinetic and Pharmacotherapeutic Researches (UPPER), School of Pharmaceutical Sciences, University of Phayao, Phayao, Thailand
- 14Department of Medicine, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand
- 15Division of Dermatology, Department of Internal Medicine, Khon Kaen Hospital, Khon Kaen, Thailand
- 16Division of Dermatology, Department of Pediatrics, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 17Division of Allergy Immunology and Rheumatology, Department of Medicine, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
- 18Department of Dermatology, Drug Hypersensitivity Clinical and Research Center, Chang Gung Memorial Hospital (CGMH), Taipei, Taiwan
- 19Cancer Vaccine and Immune Cell Therapy Core Laboratory, Chang Gung Memorial Hospital, Linkou, Taiwan
- 20Department of Dermatology, Xiamen Chang Gung Hospital, Xiamen, China
- 21Department of Molecular and Clinical Pharmacology, MRC Centre for Drug Safety Science, University of Liverpool, Liverpool, United Kingdom
- 22Skin Center, Ruampaet Dr.ANAN Hospital, Surin, Thailand
- 23Department of Pediatrics, Prapokklao Hospital, Chantaburi, Thailand
- 24Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan
- 25Genomic Medicine Core Laboratory, Chang Gung Memorial Hospital, Linkou, Taiwan
- 26The Thai Severe Cutaneous Adverse Drug Reaction (THAI-SCAR) Research Group, Bangkok, Thailand
HLA-B*13:01 allele has been identified as the genetic determinant of dapsone hypersensitivity syndrome (DHS) among leprosy and non-leprosy patients in several studies. Dapsone hydroxylamine (DDS-NHOH), an active metabolite of dapsone, has been believed to be responsible for DHS. However, studies have not highlighted the importance of other genetic polymorphisms in dapsone-induced severe cutaneous adverse reactions (SCAR). We investigated the association of HLA alleles and cytochrome P450 (CYP) alleles with dapsone-induced SCAR in Thai non-leprosy patients. A prospective cohort study, 16 Thai patients of dapsone-induced SCARs (5 SJS-TEN and 11 DRESS) and 9 Taiwanese patients of dapsone-induced SCARs (2 SJS-TEN and 7 DRESS), 40 dapsone-tolerant controls, and 470 general Thai population were enrolled. HLA class I and II alleles were genotyped using polymerase chain reaction-sequence specific oligonucleotides (PCR-SSOs). CYP2C9, CYP2C19, and CYP3A4 genotypes were determined by the TaqMan real-time PCR assay. We performed computational analyses of dapsone and DDS-NHOH interacting with HLA-B*13:01 and HLA-B*13:02 alleles by the molecular docking approach. Among all the HLA alleles, only HLA-B*13:01 allele was found to be significantly associated with dapsone-induced SCARs (OR = 39.00, 95% CI = 7.67–198.21, p = 5.3447 × 10−7), SJS-TEN (OR = 36.00, 95% CI = 3.19–405.89, p = 2.1657 × 10−3), and DRESS (OR = 40.50, 95% CI = 6.38–257.03, p = 1.0784 × 10−5) as compared to dapsone-tolerant controls. Also, HLA-B*13:01 allele was strongly associated with dapsone-induced SCARs in Asians (OR = 36.00, 95% CI = 8.67–149.52, p = 2.8068 × 10−7) and Taiwanese (OR = 31.50, 95% CI = 4.80–206.56, p = 2.5519 × 10−3). Furthermore, dapsone and DDS-NHOH fit within the extra-deep sub pocket of the antigen-binding site of the HLA-B*13:01 allele and change the antigen-recognition site. However, there was no significant association between genetic polymorphism of cytochrome P450 (CYP2C9, CYP2C19, and CYP3A4) and dapsone-induced SCARs (SJS-TEN and DRESS). The results of this study support the specific genotyping of the HLA-B*13:01 allele to avoid dapsone-induced SCARs including SJS-TEN and DRESS before initiating dapsone therapy in the Asian population.
Introduction
Dapsone (4, 4’-diaminodiphenylsulfone, DDS) is wildly used for treatment of infection and inflammation including of leprosy, Pneumocystis jiroveci pneumonia (PJP), or Toxoplasma gondii encephalitis in human immunodeficiency virus (HIV) prophylaxis, neutrophilic dermatoses, dermatitis herpetiformis, and autoimmune bullous disease (1). However, the most frequent adverse drug reactions of dapsone are dose-dependent adverse effects (hemolytic anemia and methemoglobinemia) and rarely dose-independent adverse effects (dapsone hypersensitivity syndrome) (2). Dapsone hypersensitivity syndrome (DHS) or dapsone-induced hypersensitivity reactions (DIHRs) is a life-threatening drug reaction and usually manifested between the 4 and 6 weeks after initiation of treatment. The clinically characterized through fever, rash, hepatitis or systemic involvement, lymphadenopathy, and abnormal hematologic system (eosinophilia or atypical lymphocytosis) (3). This entity is also termed DHS and DIHRs has been considered a manifestation of drug reaction with eosinophilia and systemic symptoms (DRESS). There was found approximately 0.5–3.6% of patients treated with dapsone have been reported to develop DHS and the mortality rate of 9.9% (4). Especially, about 2% of leprosy patients treated with dapsone have a DHS and 12.5% of mortality (5, 6). According to data from the King Chulalongkorn Memorial Hospital, Thailand reported during 2004–2014, dapsone is the 5th ranked common culprit drug causing DRESS in Thai patients (7).
Severe cutaneous adverse drug reactions (SCARs) is a type of adverse drug reactions (ADRs) that remains a rare but potentially severe life-threatening adverse effect and major problems for both clinical treatment and pharmaceutical industry (8). SCARs comprise a heterogeneous groups of distinct clinical manifestation, including of Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug reaction with eosinophilia and systemic symptoms (DRESS), and acute generalized exanthematous pustulosis (AGEP) (9). Clinical characteristic of SJS, SJS/TEN overlap, and TEN are acute and rapid progression of mucous detachment and systemic symptoms. They are differentiated by the severe of skin detachment, involving <10% of body surface area (BSA) in SJS, 10–30% of BSA in SJS/TEN overlap, and >30% of BSA in TEN (10). According to the RegiSCARs study, SJS has a mortality rates in the range from about 10% and more than 40% for TEN (11). The main causes of SJS-TEN are medicines and risk factors such as HIV infection, renal disease, liver disease, and active systemic autoimmune disease (12). Drug reaction with eosinophilia and systemic symptoms (DRESS) are characterized by a skin rash usually occurring more than 2 weeks after drug initiation with fever, hepatitis or internal organ involvement, lymphadenopathy, and hematological abnormalities (eosinophilia or atypical lymphocytosis) (13). The mortality rate of DRESS is approximately 10% (14).
Although the exact mechanism of SCARs remains unclear, numerous studies have described the associations between human leukocyte antigen (HLA) and cytochrome P450 genes with the specific drug hypersensitivity reaction (15, 16). For example, HLA-B*15:02 with carbamazepine-induced SJS/TEN is recommended for Han Chinese, Malaysia, India, and Thailand (17–20). On the contrary, HLA-A*31:01 is the main genetic determinant for carbamazepine-induced SJS, TEN, and DRESS in Japanese and Europeans (21, 22). Thus, HLA-B*15:02 is phenotype-specific with carbamazepine-induced SJS/TEN in each population. Additionally, there were important discovered the drug metabolism enzymes of phenytoin-induced SJS-TEN. The metabolize processes of phenytoin to p-HPPH (inactive form), arene oxides were cause of phenytoin hypersensitivity reactions by poor metabolizer (PM) alleles of mutation CYP2C9 gene consist of CYP2C9*2 and CYP2C9*3 in Asian (23, 24).
In previous studies, only HLA-B*13:01 was strongly associated with DHS in leprosy Han Chinese (odds ratio 122.1, p-value = 6.038 × 10−12 and odds ratio 20.53, p-value = 6.84 × 10−25) and dapsone-induced DRESS in non-leprosy Thais (odds ratio = 60.75, p-value = 0.0001) (25–27). Furthermore, Dapsone is metabolized through acetylation and N-hydroxylation. In human study, they found a relation between the rate of N-hydroxylation and clearance of dapsone by cytochrome P450 (28). Genetic polymorphisms of CYP2C9, CYP2C19, and CYP3A4 influenced the dapsone metabolism and cause of DHS through by DDS-NHOH (dapsone hydroxylamine) (29). Nevertheless, there are no data describing whether HLA class I, II alleles and cytochrome P450 is a valid marker for prediction of dapsone-induced SCARs in non-leprosy patients in addition to HLA-B*13:01. Consequently, the aim of this study was to investigate the contributing pharmacogenetics markers association between HLA class I, II, cytochrome P450, and dapsone-induced SCARs in Thai non-leprosy patients.
Materials and Methods
Subjects
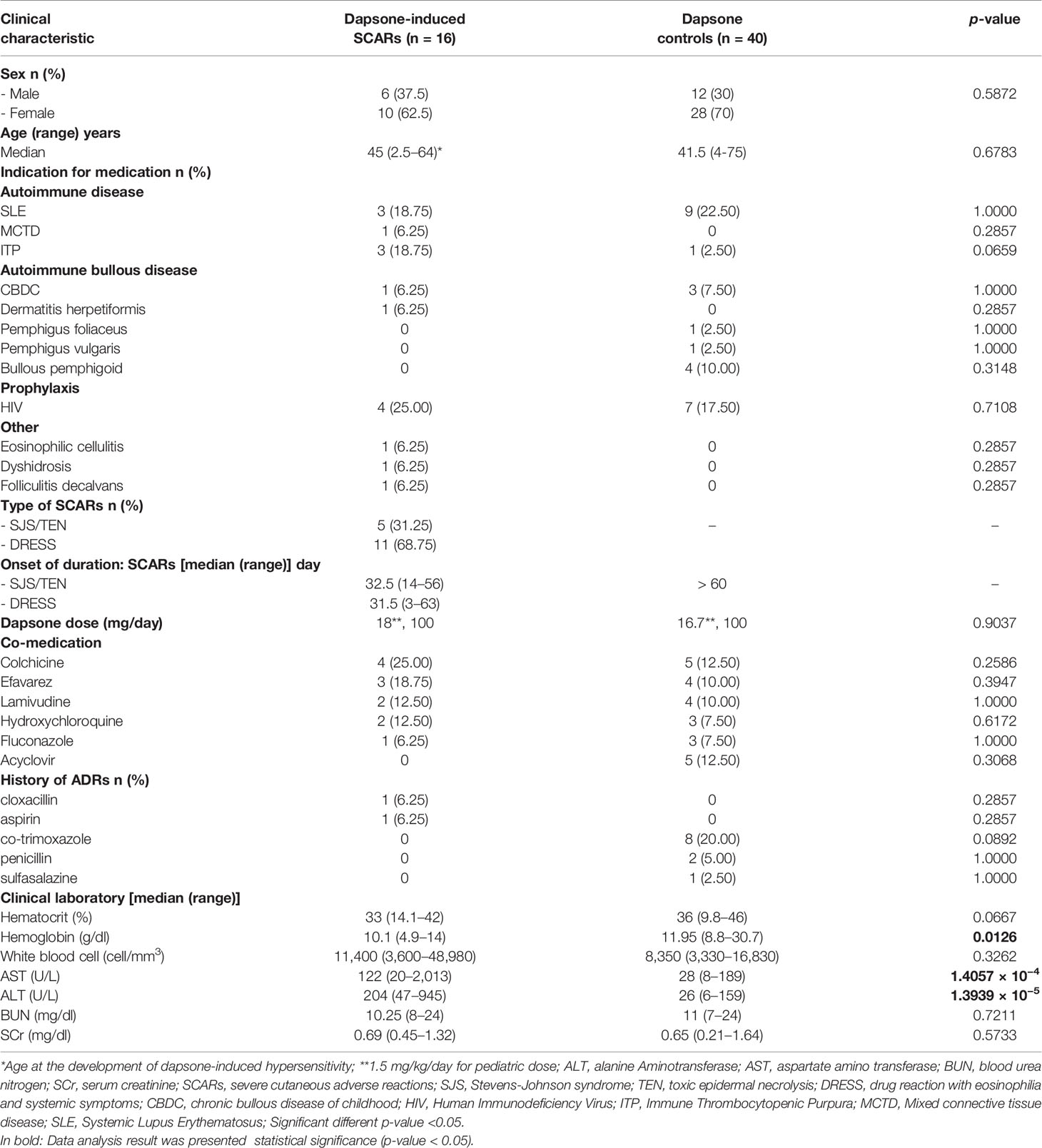
We enrolled 16 non-leprosy Thai patients with dapsone-induced SCARs consist of 5 SJS-TEN patients and 11 DRESS patients were classified by RegiSCAR criteria. SJS is defined as skin detachment less than 10% of BSA, SJS/TEN overlap has 10–30% of BSA involved, and TEN as skin detachment more than 30% of BSA (30). Moreover, SJS-TEN with severe ocular surface complications (SOC) was diagnosis with history of acute-onset high fever, serious mucocutaneous illness with skin eruption, and the involvement of at least two mucosal sites (oral cavity and ocular surface) (31). DRESS was defined by the triad of skin eruption, hematological involvement, and internal organ involvements according to the RegiSCAR Group Diagnosis Score (13). All patients with dapsone-induced SCARs were accessed through review of photographs, pathologic slides, and medical records by two dermatologists. Furthermore, there were two cases with SJS-TEN and seven cases with DRESS in the Taiwan population. Forty dapsone-tolerant controls who had been non-leprosy Thai patients and received dapsone more than 6 months without any cutaneous adverse reaction.
All of participants in this study from the Faculty of Medicine Ramathibodi Hospital, Mahidol University, Faculty of Medicine, Chulalongkorn University; Faculty of Medicine, Khon Kaen University; Udon Thani Hospital and the Thai Severe Cutaneous Adverse Drug Reaction (THAI-SCAR) research group. In addition, 470 unrelated healthy Thai population were recruited for this study. The study was approved by the ethics committee of Ramathibodi Hospital (MURA2016/105), Khon Kaen University (HE510837) and Udon Thani Hospital (22/2563). Written informed consent was obtained from each patients before enrollment.
There were collected the clinical data of dapsone-induced SCARs and controls consist of age, gender, indication for dapsone treatment, dapsone dose (mg/day), co-medication, complete blood cell count (CBC), blood urea nitrogen (BUN), serum creatinine (SCr), aspartate aminotransferase (AST) or serum glutamic oxaloacetic transaminase (SGOT), and alanine aminotransferase (ALT) or serum glutamic pyruvic transaminase (SGPT).
HLA Class I and II Genotyping
HLA class I and II alleles were genotyped using sequence-specific oligonucleotides (PCR-SSOs). Diluted DNA sample was amplified polymerase chain reaction (PCR) by GeneAmp®PCR System 9700 (Applied Biosystems, Waltham, USA). The PCR product was then hybridized against a panel of oligonucleotide probes on coated polystyrene microspheres that had sequences complementary to stretches of polymorphism within the target HLA class I and II alleles using the Lifecodes HLA SSO typing kits (Immucor, West Avenue, Stamford, USA) and detection by the Luminex®IS 100 system (Luminex Corporation, Austin, TX, USA). HLA class I and II alleles were performed using MATCH IT DNA software version 3.2.1 (One Lambda, Canoga Park, CA, USA).
CYP2C9, 2C19, and 3A4 Genotyping
The genotyping of candidate genes [CYP2C9*2 (430C > T, rs1799853), CYP2C9*3 (1075A > C, rs1057910), CYP2C19*2 (681G > A, rs4244285), CYP2C19*3 (636G > A, rs4986893), CYP2C19*17 (-806C > T, rs12248560), CYP3A4*1B (c.-392A > G, rs2740574), and CYP3A4*18 (c.878T > C, rs28371759)] were genotyped by the TaqMan real time PCR assay (ABI, Foster City, CA, USA). The SNPs genotyping will be conducted using the real-time PCR ViiA7 (ABI, Foster City, CA, USA).
In Silico Model of Dapsone, DDS-NHOH, and HLA-B*13:01 Complex
The 3D structures of HLA-B*13:01 and HLA-B*13:02 were modeled by using HLA-B*5201 from Protein Data Bank (3W39.PDB) as the template structure. The protonation states of all ionizable amino acids were assigned at pH 7.0 using PROPKA 3.0 (32). The structural geometries of Dapsone and DDS-NHOH were generated and fully optimized by the HF/6-31 G(d) level of theory using Gaussian09 program (33). Then, each drug was docked into the binding pocket of specific HLA with 100 independent docking runs using the CDOCKER module implemented in Discovery Studio 2.5 (Accelrys, Inc.).
Statistical Analysis
Chi-square test and Fisher’s exact test were used to analyze the association between dapsone-induced SCARs, dapsone controls, and healthy Thai population. Statistical analysis was performed using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA). The association was estimated by calculating the odds ratio (OR) with a 95% confidence interval (CI). Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated. The corrected P-values (Pc) for the multiple comparison of HLA alleles (16 for HLA-A, 22 for HLA-B, 20 for HLA-C, 18 for HLA-DRB1, 9 for HLA-DQA1, and 11 for HLA-DQB1) were calculated using Bonferroni’s correction. P-values were less than 0.05 (two-tailed) was considered to indicate statistically significant.
Results
Clinical Characteristic
The demographic and clinical data of patients with dapsone-induced SCARs and controls are listed in Table 1. Patients who were diagnosed with SJS, TEN, and DRESS were validated as “probable” and “definite” case by dermatologists using RegiSCAR criteria and all of dapsone-induced SJS-TEN patients without severe ocular complications (SOC). The 16 patients with dapsone-induced SCARs consisted of 10 females (62.5%) and 6 males (37.5%), with a median age of 45 (range 2.5–64) years. Meanwhile, 28 (70%) dapsone controls were females with a median age of 41.5 (range 4–75) years. The median onset time of SJS-TEN and DRESS was 32.5 (14–56) and 31.5 (3–63) days, respectively, after exposure to dapsone. The median onset time of SJS-TEN and DRESS were not significantly different. Dapsone was used among the cases and controls for the HIV prophylaxis (25.00% of cases, 17.50% of controls), systemic lupus erythematosus (SLE) (18.75% of cases, 22.50% of controls), chronic bullous disease of childhood (CBDC) (6.25% of cases, 7.50% of controls), and immune thrombocytopenic purpura (ITP) (18.75% of cases, 2.50% of controls). Eight patients (20.00%) had a previous history of cotrimoxazole-induced hypersensitivity reaction in the dapsone-tolerant group. Dapsone dosages used were 100 mg/day, while two patients (2.5 and 4 years old) received 18 and 16.7 mg/day, respectively. The hematological abnormalities and hepatitis were more prominent among the dapsone cases, as shown in Table 1. Furthermore, the most common of co-medication used among the dapsone cases and controls were colchicine, efavirenz, lamivudine, and acyclovir.
Association Between Dapsone-Induced SCARs and HLA Class I, II Alleles
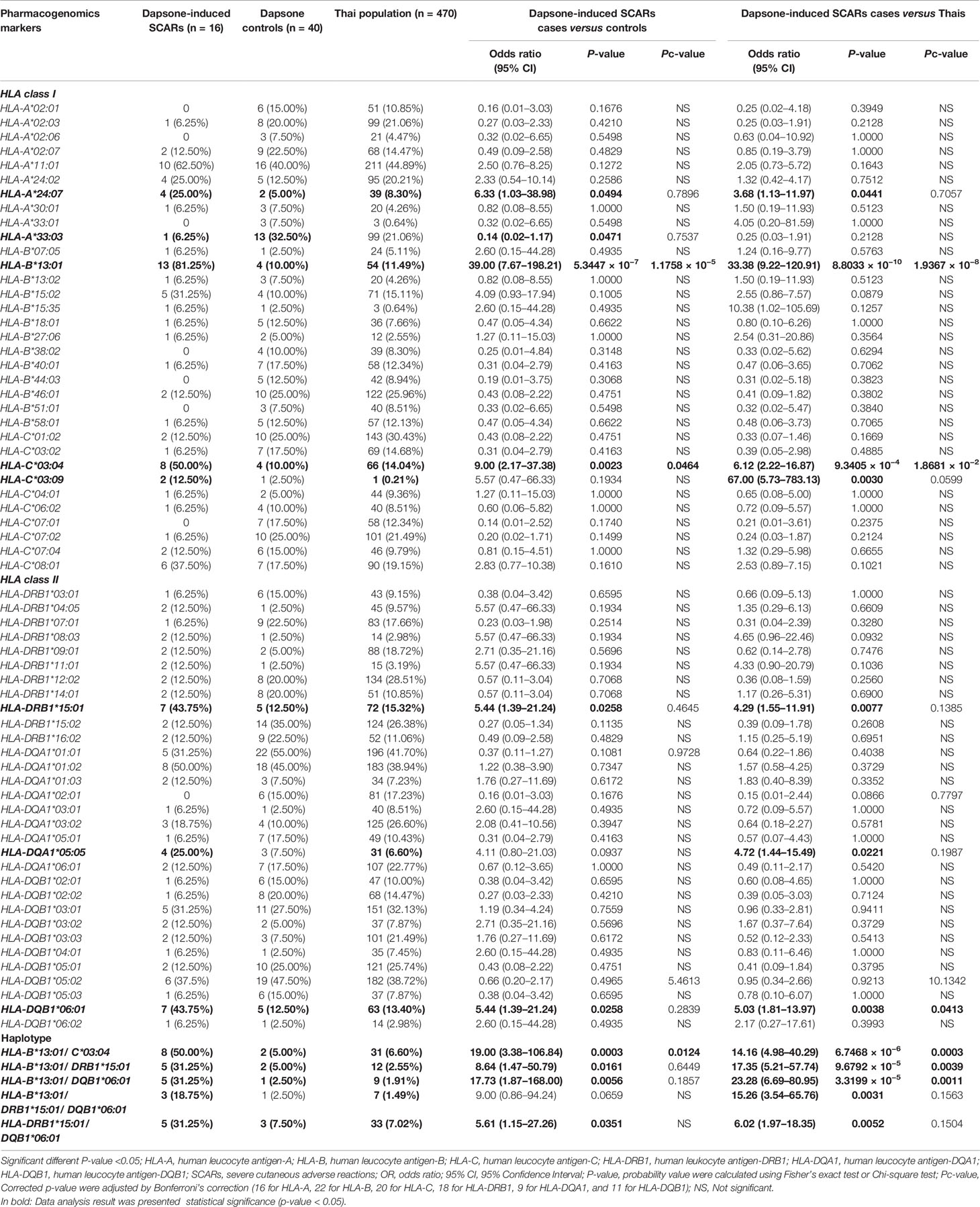
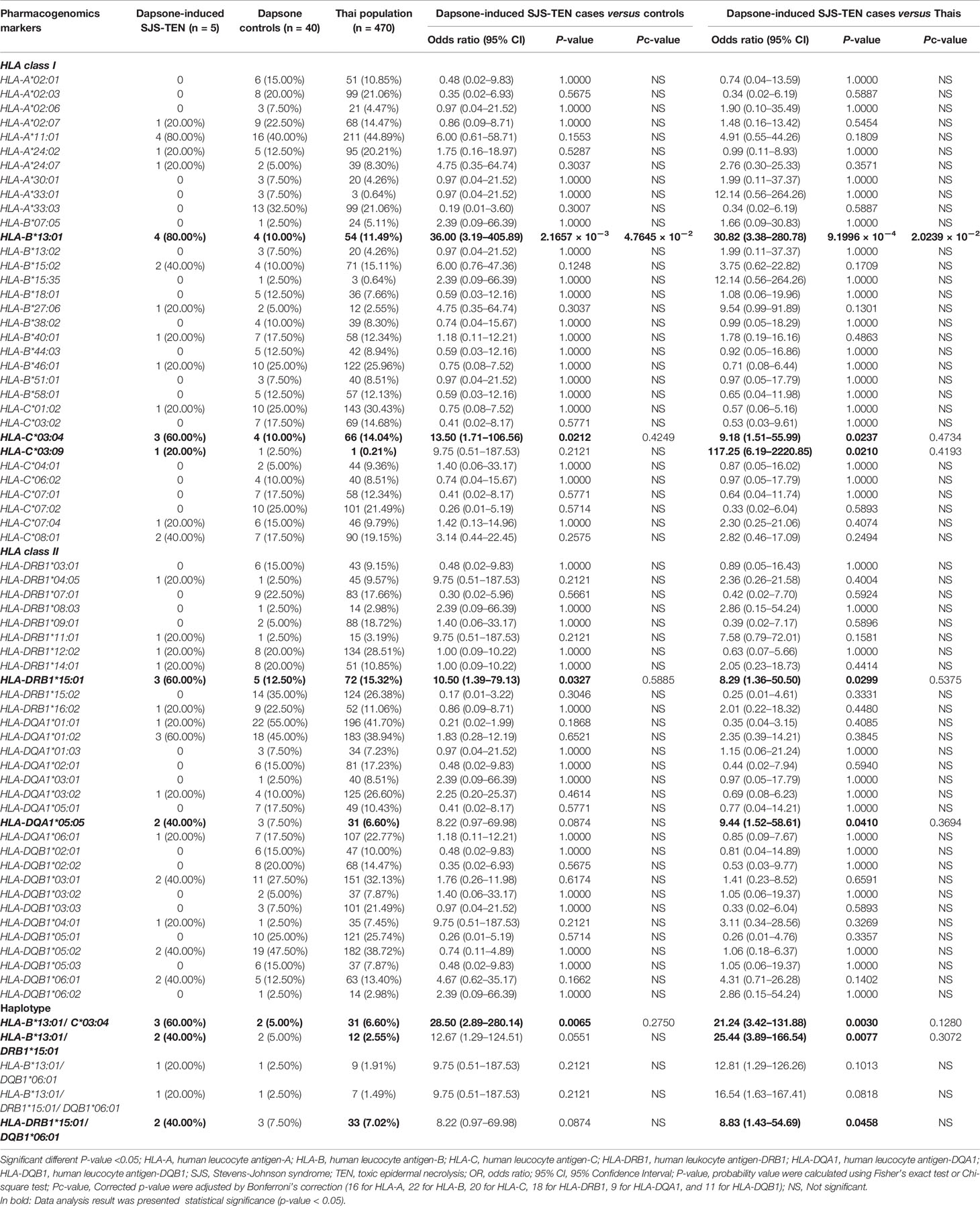
The association between HLA class I and II alleles and dapsone-induced SCARs were evaluated by comparing the SCARs group with the dapsone-tolerant controls group and the Thai general population. The number of HLA-B*13:01 carriers were 13 of 16 (81.25%) in dapsone-induced SCARs, 4 of 40 (10.00%) in dapsone-tolerant controls, and 54 of 470 (11.49%) in Thai population. The frequency of HLA-B*13:01 was significantly associated with dapsone-induced SCARs when compared with dapsone controls (OR: 39.00; 95% CI: 7.67–198.21 and p-value = 5.3447 × 10−7) and general Thai population (OR: 33.38; 95% CI: 9.22–120.91 and p-value = 8.8033 × 10−10) as shown in Table 2. Also, other HLA alleles were significant association with dapsone-induced SCARs including of HLA-A*24:07, HLA-C*03:04, HLA-DRB1*15:01, and HLA-DQB1*06:01 by p-value = 0.0494, 0.0023, 0.0258, and 0.0258, respectively (Table 2). In this study, HLA-B*15:02 was not significantly associated with dapsone-induced SCARs (p-value = 0.1005). The HLA-B*13:01-C*03:04, HLA-B*13:01-DRB1*15:01, HLA-B*13:01-DQB1*06:01, and HLA-DRB1*15:01-DQB1*06:01 haplotypes showed significant association when compared between dapsone-induced SCARs and tolerant controls (Table 2).
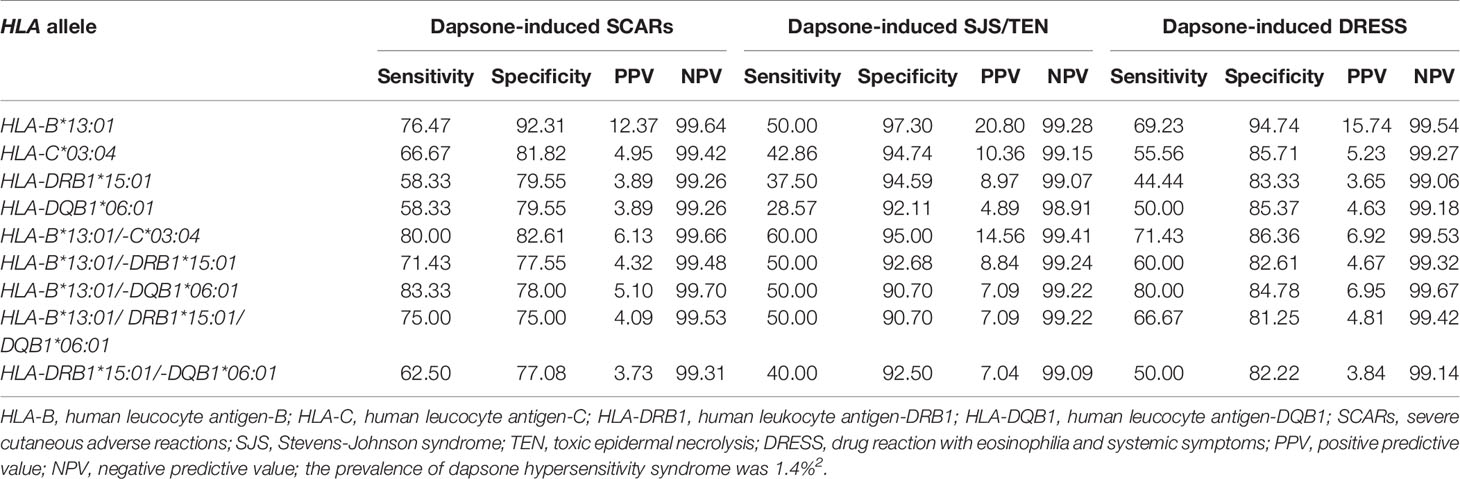
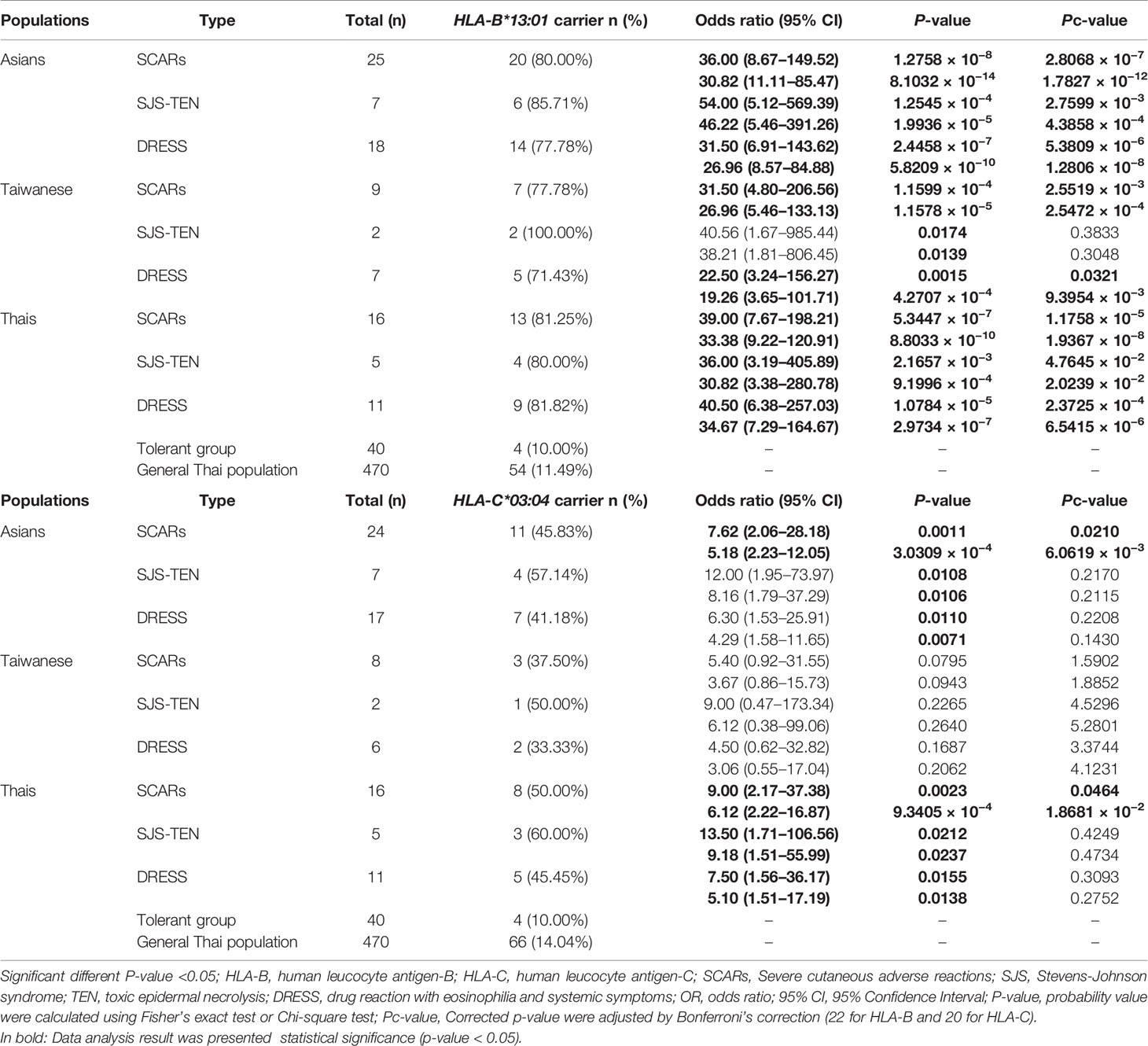
When p-values were adjusted by Bonferroni’s correction (16 for HLA-A, 22 for HLA-B, 20 for HLA-C, 18 for HLA-DRB1, 9 for HLA-DQA1, and 11 for HLA-DQB1), only HLA-B*13:01 allele was strongly associated in dapsone-induced SCARs when compared with tolerant controls and general Thai population. Also, HLA-B*13:01–C*03:04 haplotype was significantly associated with dapsone-induced SCARs with corrected p-value = 0.0124. The sensitivity, specificity, PPV, and NPV of HLA–B*13:01 allele for prediction of dapsone-induced SCARs were 76.47, 92.31, 12.37, and 99.64%, respectively (Table 3). We then examined the carrier rate of HLA-B*13:01 and HLA-C*03:04 alleles among the study population (Thais and Taiwanese) with dapsone-induced SCARs. We found HLA-B*13:01 was significantly associated with dapsone-induced SCARs when compared with dapsone controls (OR: 36.00; 95% CI: 8.67–149.52 and Pc-value = 2.8068 × 10−7) and with general Thai population (OR: 30.82; 95% CI:11.11–85.47 and Pc-value = 1.7827 × 10−12) (Table 4). Furthermore, there was a statistical significance between HLA-C*03:04 and dapsone-induced SCARs in Asian patients.
Association Between Dapsone-Induced SJS-TEN and HLA Class I, II Alleles
The association between HLA class I and II alleles and dapsone-induced SJS-TEN is shown in Table 5. HLA-B*13:01 showed a significant association with dapsone-induced SJS-TEN in Thais. HLA-B*13:01 was observed in 80.00% (4/5) of patients with dapsone-induced SJS-TEN, but only in 10.00% (4/40) of tolerant controls (OR: 36.00; 95% CI: 3.19–405.89 and p-value = 2.1657 × 10−3) and 11.49% (54/470) of general Thai population (OR: 30.82; 95% CI: 3.38–280.78 and p-value = 9.199 × 10−4). HLA-B*15:02 allele was found in 40.00% (2/5) of patients with dapsone-induced SJS-TEN, 10% (4/40) of tolerant controls, and 15.11% (71/470) of the general Thai population. There was no significant association between the HLA-B*15:02 allele and the dapsone-induced SJS-TEN (Table 5). We also observed a significant association of HLA-DRB1*15:01, HLA-B*13:01–C*03:04, and HLA-B*13:01–DRB1*15:01 with dapsone-induced SJS-TEN when compared with tolerant controls and general Thai population (p < 0.05). After taking corrected p-values into account, only HLA-B*13:01 was significantly associated with dapsone-induced SJS-TEN in Thais. HLA-B*13:01 had a sensitivity of 50.00% and specificity of 97.30% as a predictor for dapsone-induced SJS-TEN in Thais. Also, the PPV and NPV of the HLA-B*13:01 were 20.80 and 99.28%, respectively (Table 3).
When we compared the frequency of HLA-B*13:01 allele of seven Asian patients with dapsone-induced SJS-TEN and dapsone-tolerant control Thais and the general Thai population, HLA-B*13:01 allele was strongly associated with dapsone-induced SJS-TEN among Asians compared to the dapsone-tolerant control Thais (OR: 54.00; 95% CI: 5.12–569.39 and Pc-value = 2.7599 × 10−3) and general Thai population (OR: 46.22; 95% CI: 5.46–391.26 and Pc-value = 4.3858 × 10−4) respectively (Table 4). For the Taiwanese study, the results showed a significant association between HLA-B*13:01 allele and dapsone-induced SJS/TEN when compared with Thai tolerant control groups with an OR of 40.56 (95% CI = 1.67–985.44; p = 0.0174).
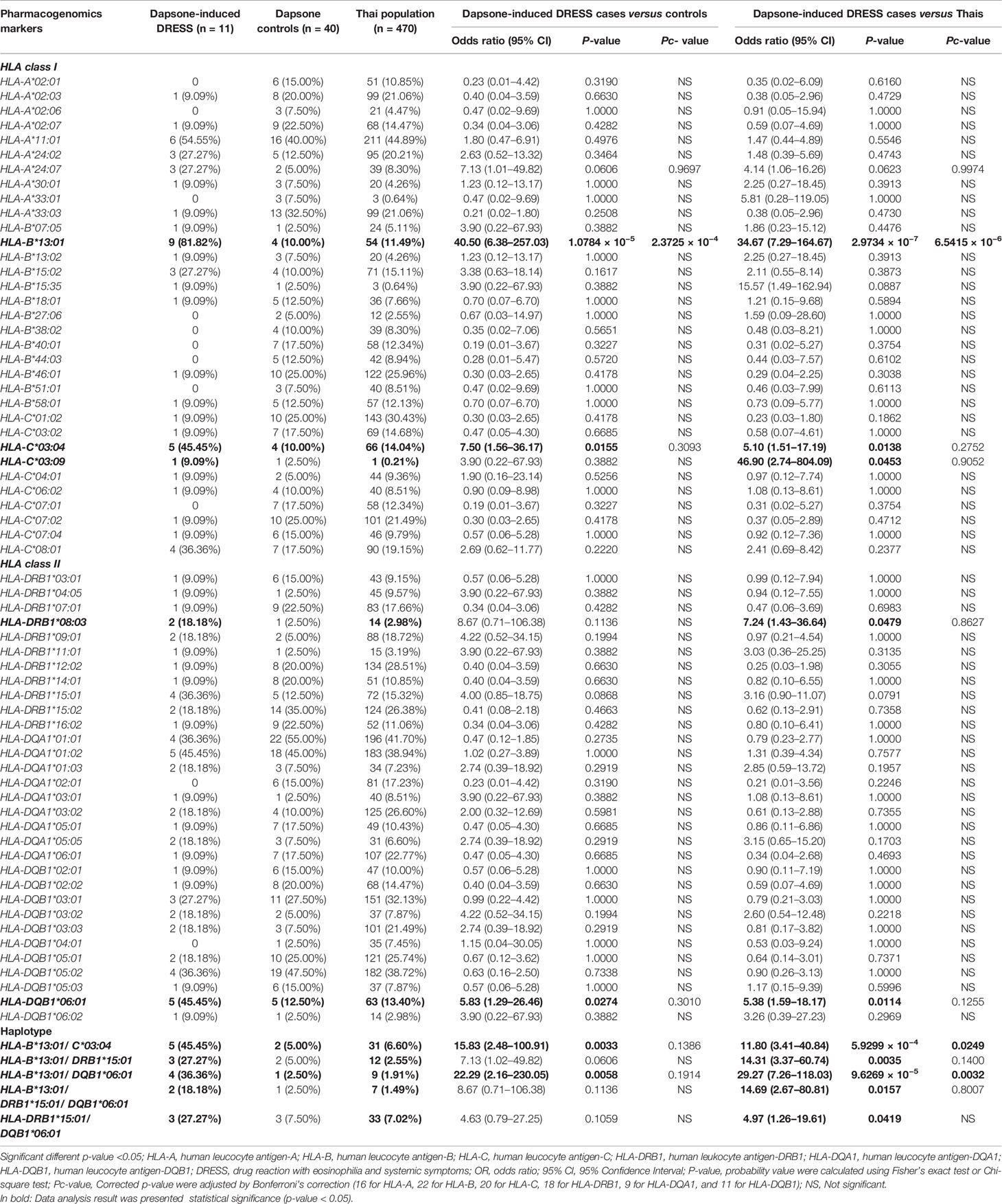
Association Between Dapsone-Induced DRESS and HLA Class I, II Alleles
The association of HLA class I and II alleles with dapsone-induced DRESS were shown in Table 6. We found that 81.82% (9/11) of dapsone-induced DRESS cases carried HLA-B*13:01, while 10.00% (4/40) of dapsone-tolerant controls and 11.49% (54/470) of the general Thai population carried HLA-B*13:01 allele. The HLA-B*13:01 allele was significantly associated with dapsone-induced DRESS when compared with dapsone-tolerant controls (OR: 40.50; 95% CI: 6.38–257.03 and p-value = 1.0784 × 10−5) and general Thai population (OR: 34.67; 95% CI: 7.29–164.67 and p-value = 2.9734 × 10−7). These results were confirmed by corrected p-value of HLA-B alleles (2.3725 × 10−4 and 6.5415 × 10−6, respectively) as presented in the Table 6. The sensitivity, specificity, PPV, and NPV of HLA-B*13:01 allele and dapsone-induced DRESS patients was 69.23, 94.74, 15.74, and 99.54%, respectively (Table 3).
On comparing, 11 dapsone-induced DRESS cases with 40 tolerant controls and 470 general Thai population, the frequencies of HLA-C*03:04, HLA-DQB1*06:01, HLA-B*13:01–C*03:04, and HLA-B*13:01–DQB1*06:01 were significantly associated with dapsone-induced DRESS (p-value < 0.05). However, HLA-B*15:02 allele was not statistically significant association with dapsone-induced DRESS when compared with tolerant controls and Thai population by p-value of 0.1617 and 0.3873, respectively. When the frequencies of HLA alleles in Asian and Taiwanese group were compared with those in Thai dapsone-tolerant controls and the general Thai population, only the HLA-B*13:01 allele was associated with dapsone-induced DRESS (Table 4). Whereas the HLA-C*03:04 allele was not statistically significant in this subgroup.
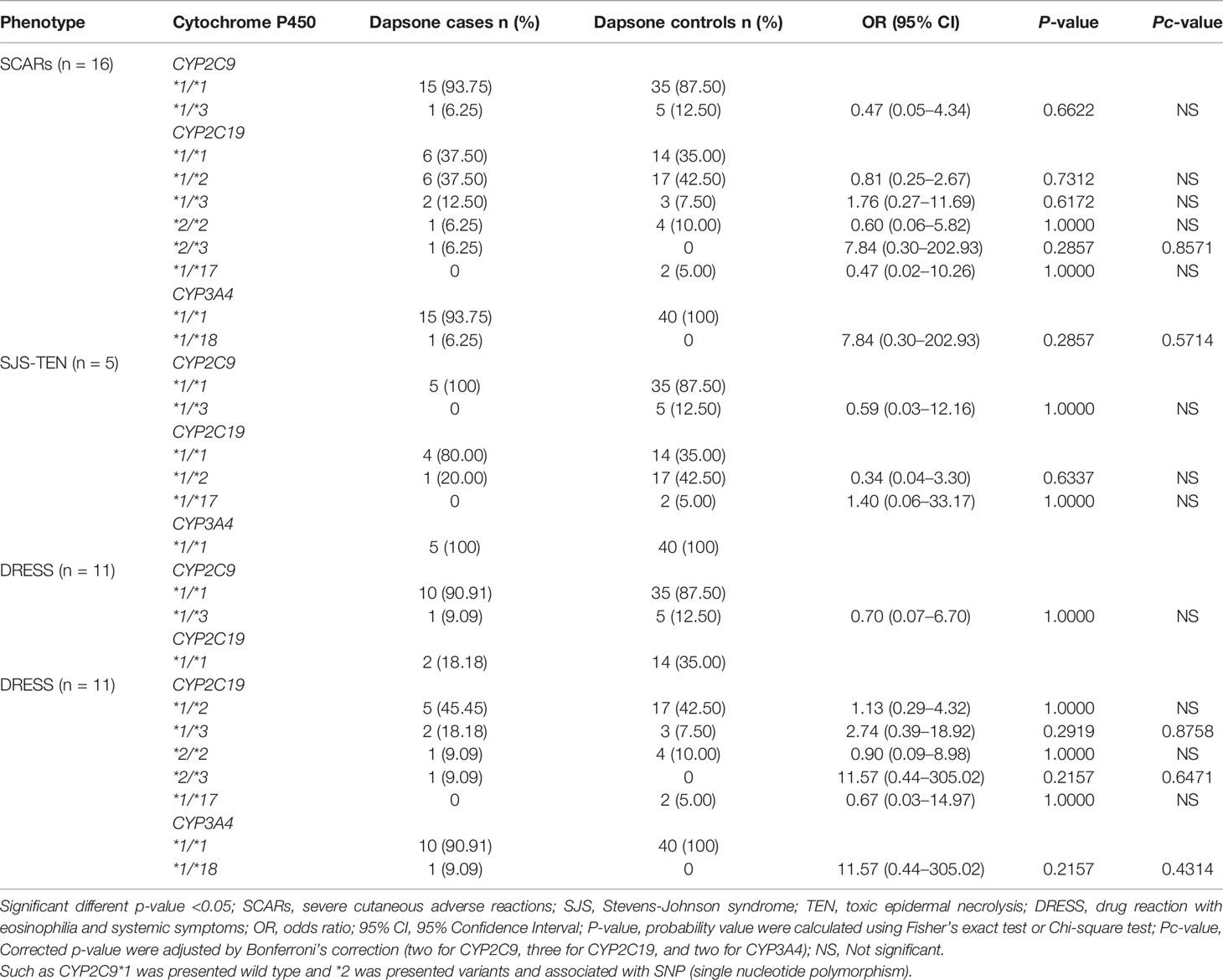
Association Between Dapsone-Induced SCARs and Cytochrome P450 (CYP2C9, CYP2C19, and CYP3A4 Variants)
In this study, none of the dapsone-induced SCARs and subgroups carried CYP2C9*2 variant along with tolerant controls. CYP2C9*3 variant (intermediate metabolizer, IM) was found in 6.25% (1/16) of the patients with dapsone-induced SCARs and 12.50% (5/40) of the dapsone-tolerant controls. Dapsone-induced SCARs and subgroups were not significantly associated with CYP2C9*3 variant (p-value = 0.6622 and 1.0000) as shown in the Table 7. There were no significant association between CYP2C19 variant and dapsone-induced SCARs and subgroup. CYP3A4*1B variant was absent in this study population. We found one individual of dapsone-induced SCARs carrying CYP3A4*1/*18. There were not significantly associated between CY3A4 variant and dapsone-induced SCARs and subgroups in Thais.
Structure Activity Relationship of Dapsone and DDS-NHOH With HLA-B*13:01 by In Silico Model
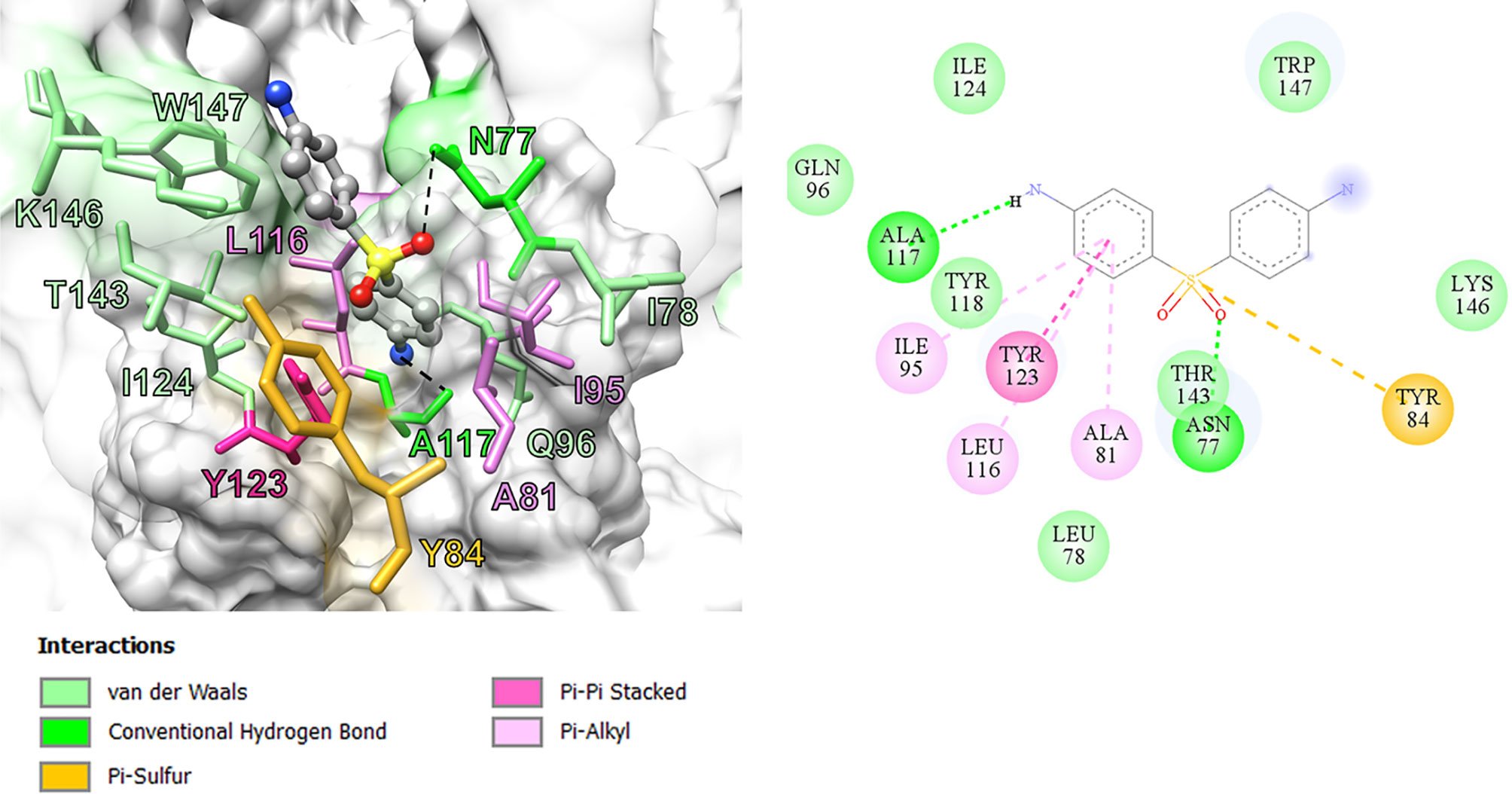
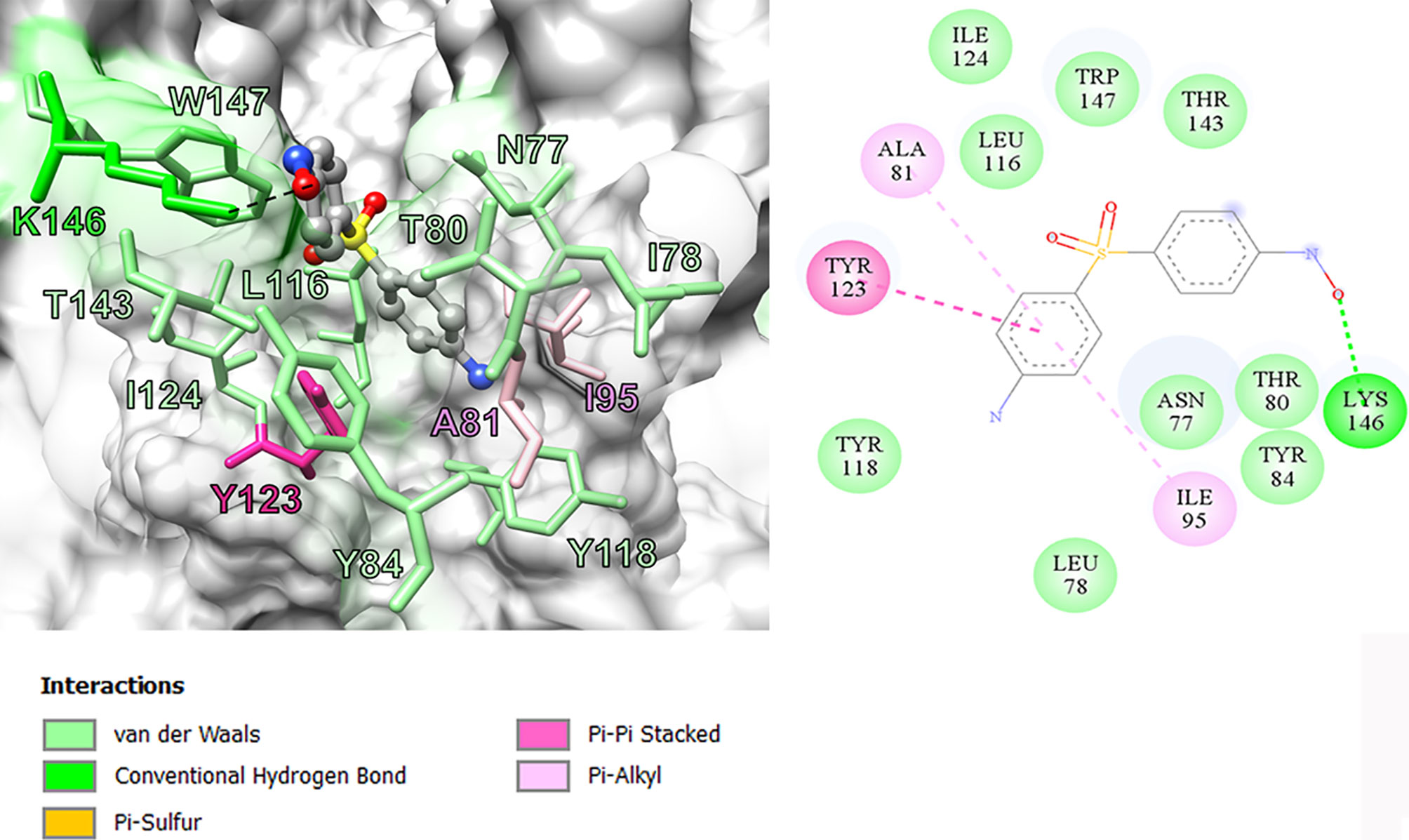
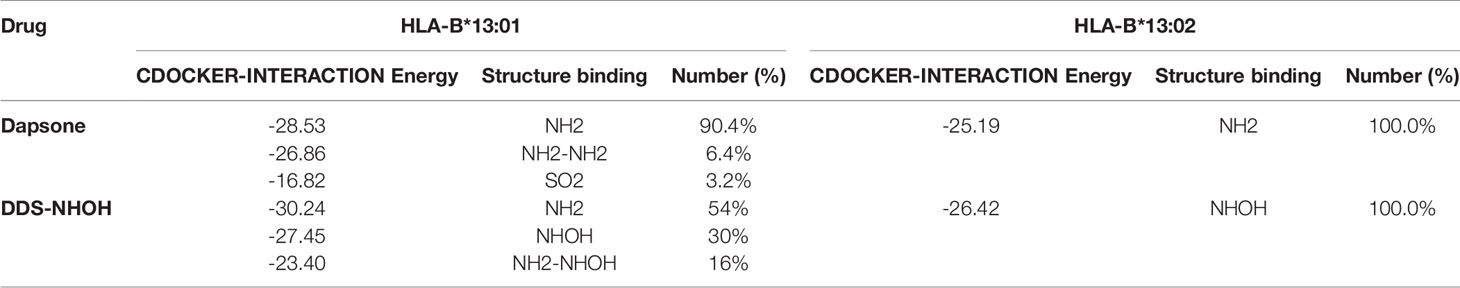
In this study, we performed computational analyses of dapsone and DDS-NHOH interacting with HLA-B*13:01 and -13:02 allele using the molecular docking approach by CDOCKER in Discovery Studio 2.5 program package. The homology models of HLA-B*13:01 and -13:02 were constructed by using HLA-B*5201 (PDB ID: 3W39) as the template structure. The 3D structure of either HLA-B*13:01 or -13:02 was deposited as a heterodimer containing α-domain and β-domain. In comparison between these two proteins, there are three different amino acids in the antigen-binding site of α-domain (I94, I95, and R97 in HLA-B*13:01, and T94, W95, and T97 in HLA-B*13:02). As a result, HLA-B*13:01 had an extra deep sub-pocket around the F-pocket at antigen-binding site, in which both drugs favorably occupied (Figures 1 and 2). The docking results in Table 8 showed that although dapsone likely interacted with both proteins via an insertion of its –NH2 group into the F-pocket (90.4 and 100.0% for HLA-B*13:01 and -13:02), it preferred to bind with HLA-B*13:01 (−28.53 kcal/mol) more than HLA-B*13:02 (−25.19 kcal/mol). The functional substitution on one of –NH2 groups to the –NHOH group in DDS-NHOH could lead to a more stable complex with HLA-B*13:01 (−30.24 kcal/mol for the conformation with –NH2 insertion, 54.0%), however the complex with the –NHOH insertion was also possible (30.0%) but it was less stable (−27.45 kcal/mol). This is in contrast for DDS-NHOH/HLA-B*13:02 in which only the conformation with –NHOH insertion was detected (−26.42 kcal/mol) in the F-pocket at antigen-binding site.
Discussion
The immunopathogenesis of SCARs are associated with expression of specific HLA allele, T-lymphocyte, structure of drug and peptide molecules (34, 35). In this study, we presented the highly specific association of HLA-B*13:01 allele and dapsone-induced SCARs (OR = 39.00, p-value = 5.3447 × 10−7), dapsone-induced SJS-TEN (OR = 36.00, p-value = 2.1657 × 10−3), and dapsone-induced DRESS (OR = 40.50, p-value = 1.0784 × 10−5) in Thai population. The frequency of HLA-B*13:01 was found in 81.25% of dapsone-induced SCARs, 10.0% of tolerant controls and 11.49% of general Thai population. The HLA-B*13:01 has a sensitivity of 76.47% and a specificity of 92.31% for predicted dapsone-induced SCARs with the prevalence of dapsone hypersensitivity syndrome was 1.4% (2). Previous study, we found the incidence of DHS among non-leprosy patients (1.66%) was compatible to that observed among leprosy patients (1.0%) (2). Meanwhile, HLA-B*13:01 allele sensitively and specifically predicted DHS in Han Chinese leprosy patients (85.5 and 85.7%, respectively). Furthermore, DHS in Han Chinese leprosy patients were found to carry HLA-B*13:01 (OR 122.1, p-value = 6.038 × 10−12 and OR 20.53, p-value = 6.84 × 10−25), Indonesian leprosy patients (OR 233.46, p-value = 7.11 × 10−9), and Korean patients (OR 73.67) (25, 26, 36, 37). When we used corrected p-values for multiple comparison, the only HLA-B*13:01 has a statistically significant association when compared between dapsone-induced SCARs and tolerant controls and general Thai population and significantly reduce the incidence of DHS in the Chinese population (38). Moreover, the risk of dapsone-induced SCARs was significantly associated with Asian patients (Thais and Taiwanese) with the HLA-B*13:01 allele, with an OR of 36.00, 95% CI = 8.67–149.52, and Pc-value = 2.8068 × 10−7. Thus, HLA-B*13:01 is strongly associated with dapsone-induced SCARs including of SJS-TEN and DRESS in leprosy and non-leprosy Asian patients. The allele frequency of HLA-B*13:01 distribution was 2–20% of Chinese, 28% of Papuans and Australian aborigines, 1–12% of Indians, 18.2% of Turkey, 8.72% of Korean, 2–4% of Southeast Asians, 1.5% of Japanese, 5.60% in Taiwanese, 5.96% of Thais, and 0% of Europeans and Africans (2, 39–41) (http://www.allelefrequencies.net/hla6006a.asp?hla_population=2842). Certainly, HLA-B*13:01 with dapsone-induced SCARs (SJS-TEN and DRESS) was strongly associated of ethnic-specific genetic in different populations. Correspondingly, HLA-B*15:02 and HLA-A*31:01 have been identified as predictive genetic markers for carbamazepine hypersensitivity in Asian and European patients (42). The biogeographical ancestry has important role in express a range of pharmacogenetics alleles and several type of SCARs. Further studies should investigate the association of pharmacogenetics marker and dapsone-induced SCARs in other population, especially Europeans and Africans.
We observed a significant association between HLA alleles such as HLA-A*24:07, HLA-C*03:04, HLA-DRB1*15:01, and HLA-DQB1*06:01 and dapsone-induced SCARs. The HLA-DRB1*15:01 allele was significantly associated with dapsone-induced SJS-TEN, whereas HLA-C*03:04 and HLA-DQB1*06:01 were significantly associated with dapsone-induced DRESS (p-value <0.05). Previous genome-wide association study had reported the association between HLA-C*03:04 and DHS in Han Chinese leprosy patients with OR = 9.00 and p-value = 2.23 × 10−19 (26). In the present study, we also found association between HLA-C*03:04 and dapsone-induced SCARs (OR = 9.00, p-value = 0.0023), SJS-TEN (OR = 13.50, p-value = 0.0212), and DRESS (OR= 7.50, p-value = 0.0155). The distribution of HLA-C*03:04 allele has been reported in different populations such as 4.37% in African Americans, 7.27% in Hispanics, 8.11% in Caucasians, 11.23% in North Americans, 10.03% in Asians, 13.70% in Japanese, 12.20% in Taiwanese, 8.09% in Thais, and 9.90% in Han Chinese (41, 43) (http://www.allelefrequencies.net/hla6006a.asp?hla_population=2842). This possibly suggests that HLA-C*03:04 allele might be a pharmacogenetics marker for dapsone-induced SCARs in many populations. Frequencies of several HLA haplotypes such as HLA-B*13:01–C*03:04, HLA-B*13:01–DRB1*15:01, and HLA-B*13:01–DQB1*06:01 were higher in dapsone-induced SCARs group compared to dapsone-tolerant controls and general Thai population. When the p-values were adjusted for multiple comparisons, associations were lost except HLA-B*13:01–C*03:04 haplotype in dapsone-induced SCARs. Nonetheless, individual HLA-B*13:01 genotypes had a high risk for dapsone-induced SCARs (SJS-TEN and DRESS) when compared with haplotypes. Although, in this study was found all dapsone-induced SJS-TEN patients without severe ocular complications (SOC), HLA-A*02:06 and HLA-B*44:03 were strong risk factor of cold medicine-induced SJS-TEN with SOC in Japanese population (31). With the presence of these alleles, further study should be conducted on these HLA alleles and culprit drugs-induced SJS-TEN with SOC in Thai population.
The sulfonamide structure is the basis of many drugs. Base on the sulfonamides structure can be divided into three types, consisting of sulfonylarylamines, non-sulfonylarylamines, and sulfonamide moiety-containing drugs (44). Consequently, the cross-reactivity of sulfonamide hypersensitivity reactions have been reported among sulfonylarylamines (antimicrobial sulfonamides) (45). Co-trimoxazole (sulfamethoxazole, SMX: trimethoprim, TMP) is commonly used for antibiotic, Pneumocystis jiroveci pneumonia (PJP) for HIV prophylaxis, organ transplantation, and cancer chemotherapy. Nevertheless, co-trimoxazole has been reported as the most common culprit drug for SJS/TEN in several countries and Thailand (46, 47). According to the data from the spontaneous reports during 1984 to 2014 by the Health Product and Vigilance Center of Thailand, co-trimoxazole is the most common culprit drug causing SJS and TEN, whereas dapsone is the 20th ranked culprit drug who suffered from SJS and TEN in Thailand (http://thaihpvc.moph.go.th/thaihvc/Public/News/uploads/hpvc_5_13_0_100526.pdf). Particularly, structure of dapsone is comprised of the simplest of the sulfones, there is considerable cross-reactivity among various sulfonamide structure. The previous study showed a significant association of HLA-B*15:02, HLA-C*06:02, and HLA-C*08:01 alleles with co-trimoxazole-induced SJS-TEN in Thai patients (48). The HLA-B*15:02 allele was strongly associated with co-trimoxazole-induced SJS-TEN in Thai patients with (OR = 3.91, p-value = 0.0037). However, our results from this study were not consistent with the results of co-trimoxazole-induced SJS-TEN regarding the HLA-B*15:02 allele, although the frequency of HLA-B*15:02 allele in Thai population and Han Chinese is approximately 10–20% (49). Recent studies from meta-analysis and molecular dynamic simulation between HLA-B*13:01 and dapsone structure proposed that dapsone would fit within the structure of the antigen-recognition site and may change the self-peptides that bind to HLA-B*13:01 causing dapsone hypersensitivity syndrome (50, 51). The association of HLA-B*15:02 or HLA-B*13:01 alleles with cross-reactivity between sulfonamide structure and different types of SCARs needs further exploration.
In addition to the HLA alleles, drug-metabolizing enzymes have been found to play a role in the pathogenesis of SCARs. The genetic variants of cytochrome P450 (CYP2C9), encoding an enzyme responsible for metabolic clearance of phenytoin are strongly associated with phenytoin-induced SCARs in Taiwanese, Japanese, and Malaysians (23). CYP2C9*3 was significantly associated with phenytoin-induced SJS/TEN (OR: 4.30; 95% CI: 1.41–13.09 and p-value = 0.0133) in Thais (24). Dapsone is metabolized in the liver by nitrogen (N)-acetylation and N- hydroxylation. The N-hydroxylation is mediated by cytochrome P450 (CYP2E1, CYP2C9, CYP2C19, and CYP3A4) (29). N-hydroxylated metabolites consist of DDS-NHOH and monoacetyl dapsone hydroxylamine (MADDS-NHOH). DDS-NHOH is responsible for fever, rash, and internal organ involvement in dapsone hypersensitivity reactions (52). CYP2C9 extensively metabolizes co-trimoxazole and influences reactive metabolites induced cytotoxicity (53, 54). In this study, we did not find the significant association between genotypes and phenotypes of CYP2C9, CYP2C19, and CYP3A4 variants and dapsone-induced SCARs (SJS-TEN and DRESS). There is an association of mucosal involvement, hepatitis, higher age, and disease occurrence with a higher risk of fatal outcome of dapsone hypersensitivity syndrome (55). Our results suggest that the severity of internal organ involvement (hepatitis) and hematological abnormalities may correlate with dapsone-induced SCARs, but dapsone dosage does not seem to affect the incidence of dapsone-induced SCARs (SJS-TEN and DRESS) in the Thai population. Nevertheless, the number of subjects in this study may not be sufficient enough to confirm all the assumptions. Further studies using a large number of samples are required for better comprehension.
In previous study, the detection of HLA‐B*13:01‐restricted dapsone and metabolite form‐responsive CD8+ clones indicates that dapsone hypersensitivity syndrome should be used as an example to discover the structural features of drug, HLA binding and interaction (56). The in silico model suggested that the 5-carboxamide group of CBZ might interact with Arg 62 of B pocket of HLA-B*15:02 (binding energy -37.104 kcal/mol) and Asn 63 contributes to the specificity in HLA recognition (57). In this study, we found three amino acid residues on an extra deep sub-pocket on F pocket within the antigen-binding site of HLA-B*13:01 and binding affinity of dapsone and DDS-NHOH for HLA-B*13:01 was much greater than HLA-B*13:02. Additionally, a docking model between dapsone and DDS-NHOH and HLA-B*13:01 allele was found to be appropriate because specific interaction triggers structural changes in the antigen-recognition site, allowing the protein to recognize peptides that are conformationally altered. Specific HLA allele plays a major immunopathogenesis role of drug hypersensitivity reactions, several hypotheses have been proposed to explain the interaction of HLA, drugs, peptides, and T cell (58). In brief, the hapten/prohapten model proposes that a chemically active drug or its metabolite forms a covalent bond with an endogenous peptide and then is intracellularly processed and presented by the particular HLA. While, the direct pharmacological interaction (p-i) model involves a non-covalent and labile interaction of the drug with HLA at the cell surface independent of antigen processing or T cell receptor. Another hypothesis, the altered peptide repertoire model, suggests the drug or its metabolites can bind non-covalent within the pocket of binding groove of certain HLA allele (34, 58). Thus, the altered peptide repertoire model involves the binding of dapsone and DDS-NHOH to HLA-B*13:01 allele and explains why the specific HLA-B*13:01 allele is a marker of dapsone-induced SCARs, despite the cytochrome P450 gene is responsible for the metabolism of dapsone to dapsone hydroxylamine.
This study confirms the specific association between HLA-B*13:01 and dapsone-induced SCARs including SJS-TEN and DRESS in the Thai and Taiwanese population. Although HLA-A*24:07, HLA-C*03:04, HLA-DRB1*15:01, and HLA-DQB1*06:01 were associated with dapsone-induced SCARs, none of these associations were considered statistically significant after Bonferroni’s correction. Furthermore, there was no association between genetic polymorphisms of CYP2C9, CYP2C19, and CYP3A4 and dapsone-induced SCARs. In addition to the specific interaction of dapsone and DDS-NHOH at the extra deep sub-pocket around the F pocket on HLA-B*13:01 allele, resulting in a change in the structure of antigen-recognition site of HLA-B*13:01 may induce altered peptides that bind to this HLA allele. Consequently, only HLA-B*13:01 might serve as a pharmacogenetics marker for screening before initiating the therapy with dapsone for the prevention of dapsone-induced SCARs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Ramathibodi Hospital (MURA2016/105), Khon Kaen University (HE510837) and Udon Thani Hospital (22/2563). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All authors helped to perform the research. PS’s contribution included sample collection, manuscript writing, drafting conception and design, performing procedures, and data analysis. JP, PR, JK, NN, TRu, PK, NS, AM, WA, UK, TT, KW, PJ, NK, TJ, TRe, C-WW, DN, WT, Ma, TRo, MP, and W-HC contributed to sample collection, data analysis and contribution to writing the manuscript. CS contributed to drafting conception, design, and contribution to writing the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer TB declared a past collaboration with the author W-HC to the handling editor.
Acknowledgments
This study was supported by grants from the (1) Faculty of Medicine, Ramathibodi Hospital, Mahidol University (2) the Health System Research Institute under Genomics Thailand Strategic Fund, (3) The Skin and Allergy Research Unit, Chulalongkorn University, and (4) The International Research Network-The Thailand Research Fund (IRN60W003). The authors thank the study participants and staffs of Pharmacogenomic and Personalized Medicine of Ramathibodi Hospital.
References
1. Zhu YI, Stiller MJ. Dapsone and Sulfones in Dermatology: Overview and Update. J Am Acad Dermatol (2001) 45:420–34. doi: 10.1067/mjd.2001.114733
2. Wang N, Parimi L, Liu H, Zhang F. A Review on Dapsone Hypersensitivity Syndrome Among Chinese Patients With an Emphasis on Preventing Adverse Drug Reactions With Genetic Testing. Am J Trop Med Hyg (2017) 96:1014–8. doi: 10.4269/ajtmh.16-0628
3. Prussick R, Shear NH. Dapsone Hypersensitivity Syndrome. J Am Acad Dermatol (1996) 35:346–9. doi: 10.1016/S0190-9622(96)90667-2
4. Rao PN, Lakshmi TS. Increase in the Incidence of Dapsone Hypersensitivity Syndrome an Appraisal. Lepr Rev (2001) 72:57–62. doi: 10.5935/0305-7518.20010009
5. Pandey B, Shrestha K, Lewis J, Hawksworth RA, Walker SL. Mortality Due to Dapsone Hypersensitivity Syndrome Complicating Multi-Drug Therapy for Leprosy in Nepal. Trop Doct (2007) 37:162–3. doi: 10.1258/004947507781524700
6. Shen J, Liu M, Zhou M, Li W. Causes of Death Among Active Leprosy Patients in China. Int J Dermatol (2011) 50:57–60. doi: 10.1111/j.1365-4632.2010.04593.x
7. Hiransuthikul A, Rattananupong T, Klaewsongkram J, Rerknimitr P, Pongprutthipan M, Ruxrungtham K. Drug-Induced Hypersensitivity Syndrome/Drug Reaction With Eosinophilia and Systemic Symptoms (Dihs/Dress): 11 Years Retrospective Study in Thailand. Allergol Int (2016) 65:432–8. doi: 10.1016/j.alit.2016.04.001
8. Harp JL, Kinnebrew MA, Shinkai K. Severe Cutaneous Adverse Reactions Impact of Immunology Genetics and Pharmacology. Semin Cutan Med Surg (2014) 33:17–27. doi: 10.12788/j.sder.0059
9. Sousa-Pinto B, Correia C, Gomes L, Gil-Mata S, Araújo L, Correia O, et al. HLA and Delayed Drug-Induced Hypersensitivity. Int Arch Allergy Immunol (2016) 170:163–79. doi: 10.1159/000448217
10. Marotti M. Severe Cutaneous Adverse Reactions (Scar) Syndromes. Rev Assoc Med Bras (2012) 58:276–8. doi: 10.1590/S0104-42302012000300003
11. Lee HY, Dunant A, Sekula P, Mockenhaupt M, Wolkenstein P, Valeyrie-Allanore L, et al. The Role of Prior Corticosteroid Use on the Clinical Course of Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis: A Case-Control Analysis of Patients Selected From the Multinational EuroSCAR and RegiSCAR Studies. Br J Dermatol (2012) 167:555–62. doi: 10.1111/j.1365-2133.2012.11074.x
12. Aihara M. Pharmacogenetics of Cutaneous Adverse Drug Reactions. J Dermatol (2011) 38:246–54. doi: 10.1111/j.1346-8138.2010.01196.x
13. Criado PR, Criado RF, Avancini JM, Santi CG. Drug Reaction With Eosinophilia and Systemic Symptoms (Dress) / Drug-induced Hypersensitivity Syndrome (Dihs): A Review of Current Concepts. Bras Dermatol (2012) 87:435–49. doi: 10.1590/S0365-05962012000300013
14. Peyriere H, Dereure O, Breton H, Demoly P, Cociglio M, Blayac JP, et al. Variability in the Clinical Pattern of Cutaneous Side-Effects of Drugs With Systemic Symptoms: Does a DRESS Syndrome Really Exist. Br J Dermatol (2006) 155:422–8. doi: 10.1111/j.1365-2133.2006.07284.x
15. Sullivan A, Watkinson J, Waddington J, Park BK, Naisbitt DJ. Implications of HLA-allele Associations for the Study of Type Iv Drug Hypersensitivity Reactions. Expert Opin Drug Metab Toxicol (2018) 14:261–74. doi: 10.1080/17425255.2018.1441285
16. Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential Role of Pharmacogenomics in Reducing Adverse Drug Reactions: A Systematic Review. JAMA (2001) 286:2270–9. doi: 10.1001/jama.286.18.2270
17. Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical Genetics: A Marker for Stevens-Johnson Syndrome. Nature (2004) 428:486. doi: 10.1038/428486a
18. Then SM, Rani ZZ, Raymond AA, Ratnaningrum S, Jamal R. Frequency of the HLA-B*1502 Allele Contributing to Carbamazepine-Induced Hypersensitivity Reactions in a Cohort of Malaysian Epilepsy Patients. Asian Pac J Allergy Immunol (2011) 29:290–3.
19. Mehta TY, Prajapati LM, Mittal B, Joshi CG, Sheth JJ, Patel DB, et al. Association of HLA-B*1502 Allele and Carbamazepine-Induced Stevens–Johnson Syndrome Among Indians. Indian J Dermatol Venereol Leprol (2009) 75:579–82. doi: 10.4103/0378-6323.57718
20. Kulkantrakorn K, Tassaneeyakul W, Tiamkao S, Jantararoungtong T, Prabmechai N, Vannaprasaht S, et al. Hla-B*1502 Strongly Predicts Carbamazepine-Induced Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in Thai Patients With Neuropathic Pain. Pain Pract (2012) 12:202–8. doi: 10.1111/j.1533-2500.2011.00479.x
21. Ozeki T, Mushiroda T, Yowang A, Takahashi A, Kubo M, Shirakata Y, et al. Genome-Wide Association Study Identifies Hla-a*3101 Allele as a Genetic Risk Factor for Carbamazepine-Induced Cutaneous Adverse Drug Reactions in Japanese Population. Hum Mol Genet (2011) 20:1034–41. doi: 10.1093/hmg/ddq537
22. McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperaviciute D, Carrington M, et al. Hla-A*3101 and Carbamazepine-Induced Hypersensitivity Reactions in Europeans. N Engl J Med (2011) 364:1134–43. doi: 10.1056/NEJMoa1013297
23. Chung WH, Chang WC, Lee YS, Wu YY, Yang CH, Ho HC, et al. Genetic Variants Associated With Phenytoin-Related Severe Cutaneous Adverse Reactions. JAMA (2014) 312:525–34. doi: 10.1001/jama.2014.14148
24. Tassaneeyakul W, Prabmeechai N, Sukasem C, Kongpan T, Konyoung P, Chumworathayi P, et al. Associations Between Hla Class I and Cytochrome P450 2c9 Genetic Polymorphisms and Phenytoin-Related Severe Cutaneous Adverse Reactions in a Thai Population. Pharmacogenet Genomics (2016) 26:225–34. doi: 10.1097/FPC.0000000000000211
25. Wang H, Yan L, Zhang G, Chen X, Yang J, Li M, et al. Association Between Hla-B*1301 and Dapsone-Induced Hypersensitivity Reactions Among Leprosy Patients in China. J Invest Dermatol (2013) 133:2642–44. doi: 10.1038/jid.2013.192
26. Zhang FR, Liu H, Irwanto A, Fu XA, Li Y, Yu GQ, et al. HLA-B*13:01 and the Dapsone Hypersensitivity Syndrome. N Engl J Med (2013) 369:1620–8. doi: 10.1056/NEJMoa1213096
27. Tempark T, Satapornpong P, Rerknimitr P, Nakkam N, Saksit N, Wattanakrai P, et al. Dapsone-Induced Severe Cutaneous Adverse Drug Reactions are Strongly Linked With Hla-B*13: 01 Allele in the Thai Population. Pharmacogenet Genomics (2017) 27:429–37. doi: 10.1097/FPC.0000000000000306
28. Tingle MD, Mahmud R, Maggs JL, Pirmohamed M, Park BK. Comparison of the Metabolism and Toxicity of Dapsone in Rat, Mouse and Man. J Pharmacol Exp Ther (1997) 283:817–23.
29. Oliveira FR, Pessoa MC, Albuquerque RFV, Schalcher TR, Monteiro MC. Clinical Applications and Methemoglobinemia Induced by Dapsone. J Braz Chem Soc (2014) 25:1770–9. doi: 10.5935/0103-5053.20140168
30. Bastuji-Garin S, Rzany B, Stern RS, Shear NH, Naldi L, Roujeau JC. Clinical Classification of Cases of Toxic Epidermal Necrolysis, Stevens-Johnson Syndrome, and Erythema Multiforme. Arch Dermatol (1993) 129:92–6. doi: 10.1001/archderm.129.1.92
31. Ueta M, Kaniwa N, Sotozono C, Tokunaga K, Saito Y, Sawai H, et al. Independent Strong Association of HLA-A*02:06 and HLA-B*44:03 With Cold Medicine-Related Stevens-Johnson Syndrome With Severe Mucosal Involvement. Sci Rep (2014) 4:1–6. doi: 10.1038/srep04862
32. Mahalapbutr P, Darai N, Panman W, Opasmahakul A, Kungwan N, Hannongbua S, et al. Atomistic Mechanisms Underlying the Activation of the G Protein-Coupled Sweet Receptor Heterodimer by Sugar Alcohol Recognition. Sci Rep (2019) 9:1–11. doi: 10.1038/s41598-019-46668-w
33. Mahalapbutr P, Wonganan P, Chavasiri W, Rungrotmongkol T. Butoxy Mansonone G Inhibits STAT3 and Akt Signaling Pathways in Non-Small Cell Lung Cancers: Combined Experimental and Theoretical Investigations. Cancers (Basel) (2019) 11:1–20. doi: 10.3390/cancers11040437
34. Bharadwaj M, Illing P, Theodossis A, Purcell AW, Rossjohn J, McCluskey J. Drug Hypersensitivity and Human Leukocyte Antigens of the Major Histocompatibility Complex. Annu Rev Pharmacol Toxicol (2012) 52:401–31. doi: 10.1146/annurev-pharmtox-010611-134701
35. Pavlos R, Mallal S, Phillips E. HLA and Pharmacogenetics of Drug Hypersensitivity. Pharmacogenomics (2012) 13:1285–306. doi: 10.2217/pgs.12.108
36. Krismawati H, Irwanto A, Pongtiku A, Irwan ID, Maladan Y, Sitanggang YA, et al. Validation Study of HLA-B*13:01 as a Biomarker of Dapsone Hypersensitivity Syndrome in Leprosy Patients in Indonesia. PloS Negl Trop Dis (2020) 14:1–11. doi: 10.1371/journal.pntd.0008746
37. Park HJ, Park JW, Kim SH, Choi SY, Kim HK, Jung CG, et al. The HLA-B*13:01 and the Dapsone Hypersensitivity Syndrome in Korean and Asian Populations: Genotype- and Meta-Analyses. Expert Opin Drug Saf (2020) 19:1349–56. doi: 10.1080/14740338.2020.1796965
38. Liu H, Wang Z, Bao F, Wang C, Sun L, Zhang H, et al. Evaluation of Prospective Hla-B*13:01 Screening to Prevent Dapsone Hypersensitivity Syndrome in Patients With Leprosy. JAMA Dermatol (2019) 155:666–72. doi: 10.1001/jamadermatol.2019.2609
39. Schrijvers R, Gilissen L, Chiriac AM, Demoly P. Pathogenesis and Diagnosis of Delayed-Type Drug Hypersensitivity Reactions, From Bedside to Bench and Back. Clin Transl Allergy (2015) 5:5–10. doi: 10.1186/s13601-015-0073-8
40. Park HJ, Kim YJ, Kim DH, Kim J, Park KH, Park JW, et al. Hla Allele Frequencies in 5802 Koreans: Varied Allele Types Associated With SJS/TEN According to Culprit Drugs. Yonsei Med J (2016) 57:118–26. doi: 10.3349/ymj.2016.57.1.118
41. Satapornpong P, Jinda P, Jantararoungtong T, Koomdee N, Chaichan C, Pratoomwun J, et al. Genetic Diversity of HLA Class I and Class Ii Alleles in Thai Populations: Contribution to Genotype-Guided Therapeutics. Front Pharmacol (2020) 11:1–22. doi: 10.3389/fphar.2020.00078
42. Amstutz U, Ross CJ, Castro-Pastrana LI, Rieder MJ, Shear NH, Hayden MR, et al. HLA-A 31:01 and HLA-B 15:02 as Genetic Markers for Carbamazepine Hypersensitivity in Children. Clin Pharmacol Ther (2013) 94:142–9. doi: 10.1038/clpt.2013.5
43. Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández-Viña MA. Analysis of the Frequencies of HLA-A, B, and C Alleles and Haplotypes in the Five Major Ethnic Groups of the United States Reveals High Levels of Diversity in These Loci and Contrasting Distribution Patterns in These Populations. Hum Immunol (2001) 62:1009–30. doi: 10.1016/s0198-8859(01)00298-1
44. Ghimire S, Kyung E, Lee JH, Kim JW, Kang W, Kim E. An Evidence-Based Approach for Providing Cautionary Recommendations to Sulfonamide-Allergic Patients and Determining Cross-Reactivity Among Sulfonamide-Containing Medications. J Clin Pharm Ther (2013) 38:196–202. doi: 10.1111/jcpt.12048
45. Brackett CC, Singh H, Block JH. Likelihood and Mechanisms of Cross-Allergenicity Between Sulfonamide Antibiotics and Other Drugs Containing a Sulfonamide Functional Group. Pharmacotherapy (2004) 24:856–70. doi: 10.1592/phco.24.9.856.36106
46. Barvaliya M, Sanmukhani J, Patel T, Paliwal N, Shah H, Tripathi C. Drug-Induced Stevens-Johnson Syndrome (Sjs), Toxic Epidermal Necrolysis (TEN), and SJS-TEN Overlap: A Multicentric Retrospective Study. J Postgrad Med (2011) 57:115–9. doi: 10.4103/0022-3859.81865
47. Roujeau JC, Stern RS. Severe Adverse Cutaneous Reactions to Drugs. N Engl J Med (1994) 331:1272–85. doi: 10.1056/NEJM199411103311906
48. Kongpan T, Mahasirimongkol S, Konyoung P, Kanjanawart S, Chumworathayi P, Wichukchinda N, et al. Candidate HLA Genes for Prediction of Co-Trimoxazole-Induced Severe Cutaneous Reactions. Pharmacogenet Genomics (2015) 25:402–11. doi: 10.1097/FPC.0000000000000153
49. Negrini S, Becquemont L. Pharmacogenetics of Hypersensitivity Drug Reactions. Therapie (2017) 72:231–43. doi: 10.1016/j.therap.2016.12.009
50. Tangamornsuksan W, Lohitnavy M. Association Between Hla-B*1301 and Dapsone-Induced Cutaneous Adverse Drug Reactions: A Systematic Review and Meta-Analysis. JAMA Dermatol (2018) 154:441–6. doi: 10.1001/jamadermatol.2017.6484
51. Watanabe H, Watanabe Y, Tashiro Y, Mushiroda T, Ozeki T, Hashizume H, et al. A Docking Model of Dapsone Bound to HLA-B*13:01 Explains the Risk of Dapsone Hypersensitivity Syndrome. J Dermatol Sci (2017) 88:320–9. doi: 10.1016/j.jdermsci.2017.08.007
52. Wozel G, Blasum C. Dapsone in Dermatology and Beyond. Arch Dermatol Res (2014) 306:103–24. doi: 10.1007/s00403-013-1409-7
53. Ho JM, Juurlink DN. Considerations When Prescribing Trimethoprim-Sulfamethoxazole. CMAJ (2011) 183:1851–8. doi: 10.1503/cmaj.111152
54. Pirmohamed M, Alfirevic A, Vilar J, Stalford A, Wilkins EG, Sim E, et al. Association Analysis of Drug Metabolizing Enzyme Gene Polymorphisms in HIV-positive Patients With Co-Trimoxazole Hypersensitivity. Pharmacogenetics (2000) 10:705–13. doi: 10.1097/00008571-200011000-00005
55. Lorenz M, Wozel G, Schmitt J. Hypersensitivity Reactions to Dapsone: A Systematic Review. Acta Derm Venereol (2012) 92:194–9. doi: 10.2340/00015555-1268
56. Zhao Q, Alhilali K, Alzahrani A, Almutairi M, Amjad J, Liu H, et al. Dapsone- and Nitroso Dapsone-Specific Activation of T Cells From Hypersensitive Patients Expressing the Risk Allele Hla-B*13:01. Allergy (2019) 74:1533–48. doi: 10.1111/all.13769
57. Wei CY, Chung WH, Huang HW, Chen YT, Hung SI. Direct Interaction Between HLA-B and Carbamazepine Activates T Cells in Patients With Stevens-Johnson Syndrome. J Allergy Clin Immunol (2012) 129:1562–9. doi: 10.1016/j.jaci.2011.12.990
Keywords: dapsone-induced severe cutaneous adverse reactions, HLA class I and II alleles, HLA-B*13:01, cytochrome P450, Thais and Taiwaneses
Citation: Satapornpong P, Pratoomwun J, Rerknimitr P, Klaewsongkram J, Nakkam N, Rungrotmongkol T, Konyoung P, Saksit N, Mahakkanukrauh A, Amornpinyo W, Khunarkornsiri U, Tempark T, Wantavornprasert K, Jinda P, Koomdee N, Jantararoungtong T, Rerkpattanapipat T, Wang C-W, Naisbitt D, Tassaneeyakul W, Ariyachaipanich M, Roonghiranwat T, Pirmohamed M, Chung W-H and Sukasem C (2021) HLA-B*13 :01 Is a Predictive Marker of Dapsone-Induced Severe Cutaneous Adverse Reactions in Thai Patients. Front. Immunol. 12:661135. doi: 10.3389/fimmu.2021.661135
Received: 30 January 2021; Accepted: 13 April 2021;
Published: 04 May 2021.
Edited by:
Luis Eduardo Coelho Andrade, Federal University of São Paulo, BrazilReviewed by:
Mayumi Ueta, Kyoto Prefectural University of Medicine, JapanTeresa Bellon, University Hospital La Paz Research Institute (IdiPAZ), Spain
Copyright © 2021 Satapornpong, Pratoomwun, Rerknimitr, Klaewsongkram, Nakkam, Rungrotmongkol, Konyoung, Saksit, Mahakkanukrauh, Amornpinyo, Khunarkornsiri, Tempark, Wantavornprasert, Jinda, Koomdee, Jantararoungtong, Rerkpattanapipat, Wang, Naisbitt, Tassaneeyakul, Ariyachaipanich, Roonghiranwat, Pirmohamed, Chung and Sukasem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chonlaphat Sukasem, Y2hvbmxhcGhhdC5zdWtAbWFoaWRvbC5hYy50aA==
 Patompong Satapornpong
Patompong Satapornpong Jirawat Pratoomwun
Jirawat Pratoomwun Pawinee Rerknimitr5,6
Pawinee Rerknimitr5,6 Niwat Saksit
Niwat Saksit Napatrupron Koomdee
Napatrupron Koomdee Thawinee Jantararoungtong
Thawinee Jantararoungtong Ticha Rerkpattanapipat
Ticha Rerkpattanapipat Chuang-Wei Wang
Chuang-Wei Wang Munir Pirmohamed
Munir Pirmohamed Wen-Hung Chung
Wen-Hung Chung Chonlaphat Sukasem
Chonlaphat Sukasem