- Key Laboratory for Animal Disease-Resistance Nutrition, Animal Nutrition Institute, Sichuan Agricultural University, Chengdu, China
L-leucine (Leu), as one of the effective amino acids to activate the mTOR signaling pathway, can alleviate transmissible gastroenteritis virus (TGEV) infection. However, the underlying mechanism by which Leu alleviates the virus infection has not been fully characterized. In particular, how Leu impacts TGEV replication through mTOR signaling has yet to be elucidated. In the present study, we found that TGEV proliferated efficiently in intestinal porcine epithelial cells (IPEC-J2 cells) as evidenced by the increase in viral contents by flow cytometry, the inhibition of cell proliferation by CCK-8 assay as well as the reduction of PCNA level by western blot. Besides, western blot analysis showed that STAT1 expression was markedly reduced in TGEV-infected cells. The results of ELISA revealed the inhibition of ISGs (ISG56, MxA, and PKR) expressions by TGEV infection. TGEV-induced mTOR and its downstream p70 S6K and 4E-BP1, STAT1 and ISGs downregulation were blocked by an mTOR activator-MHY1485 but not by an mTOR inhibitor-RAPA. Concurrently, mTOR activation by MHY1485 reduced the contents of TGEV and vice versa. Furthermore, Leu reversed the inhibition of STAT1 and ISGs by activating mTOR and its downstream p70 S6K and 4E-BP1 in TEGV-infected cells. Our findings demonstrated that Leu promoted the expressions of STAT1 and ISGs via activating mTOR signaling in IPEC-J2 cells, aiming to prevent TGEV infection.
Introduction
Transmissible gastroenteritis virus (TGEV), which belongs to the genus Alphacoronavirus, is an enveloped, single-stranded, positive-sense RNA virus (1, 2). TGEV replicates in the cytoplasm of differentiated enterocytes covering the small intestinal villi and causes acute enteritis in swine of all ages (3, 4). The common clinical manifestations are anorexia, vomiting, watery diarrhea, dehydration, and weight loss in piglets. Specifically, the mortality rate of seropositive suckling piglets may reach up to 100% during epidemics (5). Despite the availability of vaccines, outbreaks can be encountered globally and cause great economic losses in the swine industry (6).
In the process of viral infection and replication, innate immunity, as the first line of defense, detects pathogen-associated molecular patterns (PAMPs) of invading viruses by different pattern recognition receptors (PRRs) and responds accordingly by producing a series of effector molecules or inflammatory elements against viral invasion (7, 8). Among most innate immune responses, IFN-I signaling is one of the most important antiviral innate immunity to antagonize virus infection. Phosphorylation of STAT1 on tyrosine 701 plays a pivotal role in activating the IFN-I signaling pathway (9–11). Even though IFNs exert antiviral activity during viral infection, the ability to suppress innate immune responses provides invading viruses with the opportunity to replicate and establish a productive infection (12–14). A previous study revealed that porcine epidemic diarrhea virus (PEDV), which is genetically related to TGEV, can inhibit STAT1 expression through the ubiquitin-proteasome targeting degradation system (15).
A great deal of evidence supports the notion of mTOR function in maintaining the structure of the intestinal mucosa and in the self-renewal of stem cells. Emerging studies have begun to shed light on the interplay between mTOR signaling and innate immune signaling transductions, arguing for their possible roles in the regulation of host antiviral responses (16, 17). Several studies have indicated that mTOR could regulate STAT1 expression (18–20). However, how mTOR regulation influences STAT1 expression in TGEV-infected cells remains unknown.
In the meantime, identifying an effective treatment for TGEV pathogens is of utmost importance in terms of the antibiotic prohibition. Because the possibility of viral gene mutation, general therapeutic drawbacks of vaccines, and effects of prescription drugs only on clinical symptoms still put the host at risk, the development of effective nutritional regimens targeting mTOR/STAT1 activation has become one of the integral means to antagonize TGEV infection (21). Recent studies on pigs have shown that branched-chain amino acids (BCAAs) improve intestinal integrity and function and modify the production of immunoregulatory cytokines to protect the host from different diseases (22, 23). Among the three BCAAs, L-leucine (Leu) earns the greatest reputation for its unique function of activating the mTOR signaling pathway (24, 25). In the present study, we investigated the role of mTOR in regulating the IFN-I response. Our results clearly demonstrated that TGEV inhibited the expressions of mTOR, p70 S6K, 4E-BP1, STAT1 and ISGs. Mechanistically, mTOR activity regulation both by its own activator and Leu could alleviate TGEV infection via increasing in STAT1 and ISGs expression. On the contrary, the specific inhibitor of mTOR promoted TGEV replication by reducing STAT1 and ISGs expressions.
Materials and Methods
Cells and Virus
TGEV strain WH-1 (GenBank accession no. HQ462571.1) was kindly offered from the College of Veterinary Medicine, Sichuan Agricultural University (26). Virus titers were determined by 50% tissue culture infective doses (TCID50) assay. Intestinal porcine epithelial cells (IPEC-J2 cells) were cultured and maintained in Dulbecco’s modified Eagle’s F12 Ham medium (DMEM-F12; Gibco, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, USA) and 1% penicillin–streptomycin (Gibco, USA) at 37°C in a humidified 5% CO2 incubator. The swine testis cells (ST-0746) were cultured and maintained in DMEM (Gibco, USA) and used to amplify TGEV.
Reagents and Antibodies
Polyclonal antibodies against STAT1, phospho-STAT1 (Tyr701) Rabbit mAb, PCNA antibody Mouse mAb, and the β-actin antibody were purchased from Cell Signaling Technology, USA. Recombinant human IFN-β was purchased from Peprotech, USA. Rapamycin (RAPA) and MHY1485 were purchased from Selleck Chemicals, USA. Leu was purchased from Sigma-Aldrich, USA. Before TGEV infection, IPEC-J2 cells were pretreated with 10 nM RAPA or 10 μM MHY1485 for 30 min. Differently, the cells were starved for 3 h in the Earle’s Balanced Salt Solution (EBSS; Gibco, USA) and then pretreated with 10 mM Leu before TGEV infection. TGE Virus Antibody (1-Q-17) was purchased from Santa Cruz Biotechnology, USA. Goat Anti-Mouse IgG H&L (Alexa Fluor® 488) was purchased from Abcam, USA.
CCK-8 Assay
IPEC-J2 cells were seeded on glass coverslips (Corning, USA) in 96-well plates (1 × 104 cells per well). After reaching 80% confluence, the medium was replaced with IPEC-J2 cell starvation medium, followed by infection with TGEV (MOI = 5) for 1 h. Then, the virus culture medium was replaced by IPEC-J2 cell starvation medium. The cells were cultured for 0, 6, 12, 24, 36, and 48 h, respectively. After that, 10 μl of CCK-8 solution was added into each well and then cultured in the cell incubator for 2 h. The absorbance at 450 nm was determined by enzyme-linked immunosorbent assay.
The Kinetic Curve of TGEV Replication in IPEC-J2 Cells
ST cells were infected with TGEV (MOI = 5) at different time points (6, 12, 24, 36, 48 h). The virus was harvested at different time points by repeated freezing and thawing for 3 times, followed by centrifugation at 3,000 r/min for 10 min. Then, the supernatant was filtered with a 0.22 μm filter to preserve at −80°C. The virus titers were measured in IPEC-J2 cells. TCID50 was calculated by the Reed–Muench method to draw the kinetic curve of TGEV replication.
Flow Cytometry Analysis
IPEC-J2 cells were collected in the EP tubes with 1 ml PBS and centrifuged for 5 min at 4°C in 350 relative centrifugal force (rcf). Then we removed the supernatant and resuspended and fixed cells with 4% paraformaldehyde for 15 min. After 500 rcf centrifugation at 4°C for 5 min, 100 μl TGEV primary antibody (0.05% Titonx-100: primary antibody = 100: 1 v/v) was added. Two hours later at room temperature, cells were centrifuged for 5 min at 4°C in 500 rcf. Next, the supernatant was removed and we added 100 μl second antibody (PBS: second antibody = 1,000:1) in the dark condition. After storage at room temperature for 1 h, we added 900 μl PBS for centrifugation. Then the supernatant was removed and 500 μl PBS was added to resuspend cells for detection.
RT-qPCR
Total cellular RNA was extracted with TRIzol Reagent (Invitrogen, USA) from TGEV-infected IPEC-J2 cells and an aliquot (1 μg) was reverse-transcribed into cDNA using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Japan). The obtained cDNA was then used as the template in the SYBR Green I PCR assay (Applied Biosystems, CA). Real-time qPCR was performed for STAT1 and a house-keeping gene (GAPDH) according to standard protocols with the primers indicated in Table S1.
Western Blot Analysis
Briefly, IPEC-J2 cells were collected in the 1.5 ml EP tube for centrifugation at 500 rcf for 5 min at 4°C. After removing the supernatant, cells were mixed with RIPA lysis buffer (Beyotime, China) containing PMSF (Sigma-Aldrich, US) and kept on the ice for 30 min. Ultrasonication was then performed to break cells, followed by centrifugation at 10,000 rcf for 15 min at 4°C. The proteins in the supernatant containing with 4× Laemmli Sample Buffer (Bio-Rad, USA) were denatured in the 98°C-metal bath for 10 min. Equal amounts of samples were then subjected to SDS-PAGE, and the expressions of STAT1 and p-STAT1 protein were examined by western blot analysis using the indicated antibodies. The expression of β-actin was detected to verify equal protein sample loading.
Enzyme-Linked Immunosorbent Assay (ELISA)
The supernatants of different treatments were collected to determine the concentrations of ISGs (ISG15, ISG56, MxA and PKR) using spectrophotometric kits in line with the manufacturer’s instructions (MEIMIAN, Jiangsu, China). The protein concentrations were expressed pg/ml or ng/ml.
Statistical Analysis
All data were expressed as means ± standard error of means (SEMs). The statistical significance was tested by unpaired two-tailed Student’s T test and/or one-way analysis of variance (ANOVA) using IBM SPSS Statistics version 20.0 (IBM, USA). When there was a significant interaction, post hoc testing was conducted using Tukey’s multiple comparison test. A p-value of <0.05 was considered statistically significant.
Results
IPEC-J2 Cells Are Susceptible to TGEV
Firstly, to determine whether TGEV could proliferate in IPEC-J2 cells, cells were incubated with TGEV (MOI = 5) for 0, 6, 12, 24, 36, and 48 h. We observed a massive viral replication at 24 h post-infection (hpi). Moreover, the contents of TGEV increased in a time-dependent manner (Figure 1A). Then, the percentage of TGEV infected IPEC-J2 cells was examined. At first, virus particles were assembled and released into the extracellular matrix; however, the titer of TGEV was very low. During 24 to 48 h, the virus replicated rapidly. In particular, at 48 hpi, the TCID50 of virus reached the highest level (10−6.57/100 μl), which is close to the TCID50 (10−6.8/100 μl) measured by TGEV in ST cells (Figure 1B).
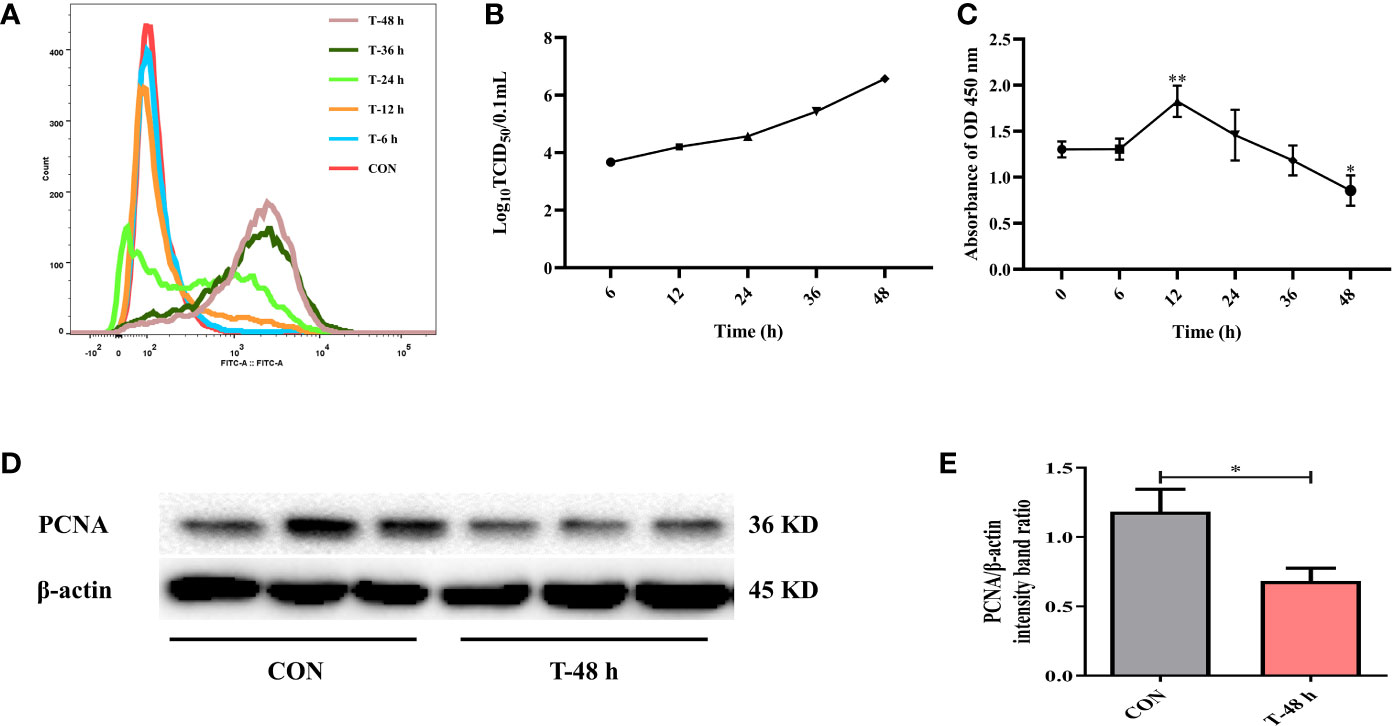
Figure 1 TGEV infection in IPEC-J2 cells. (A) The contents of TGEV at 0, 6, 12, 24, 36, and 48 hpi were analyzed via flow cytometry. (B) The viral titers of TGEV infected IPEC-J2 cells at 6, 12, 24, 36, and 48 hpi. (C) CCK-8 assay was used to detect the cell proliferation after TGEV infection at 6, 12, 24, 36, and 48 hpi. Data express the mean ± SEM (n = 3). (D) The PCNA level was detected by western blot. (E) Quantitation of bands to demonstrate the protein level of PCNA. Data express the mean ± SEM (n = 3). The symbol * indicates statistically significant differences (P < 0.05), and the symbol ** indicates statistically very significant differences (P < 0.01).
Next, CCK-8 assays were performed to assess cell proliferation at different time points after TGEV infection (MOI = 5). The results revealed that cell proliferation was boosted at 12 hpi, but was remarkably suppressed afterward (Figure 1C). Besides, the protein expression of PCNA was dramatically inhibited by TGEV infection at 48 hpi (Figures 1D, E).
TGEV Replication Counteracts IFN-I Signaling Pathway
To assess the effect of TGEV infection on STAT1 activation, the levels of phosphorylated STAT1 (p-STAT1) and STAT1 were examined in TGEV-infected cells. Since the ability of IFN-β to induce STAT1 phosphorylation has been well documented previously, recombinant IFN-β was exogenously administered in the positive control (27). Western blot analysis revealed that compared with the positive control, TGEV significantly decreased the expressions of p-STAT1 and STAT1 (Figures 2A, B). Similarly, the concentrations of ISGs (ISG56, MxA, and PKR) related to STAT1 activation were also decreased by TGEV (Figure 2C). Taken together, TGEV could inhibit the activation of STAT1 regulated by IFN-β and its downstream ISGs to block the IFN-I signaling pathway.
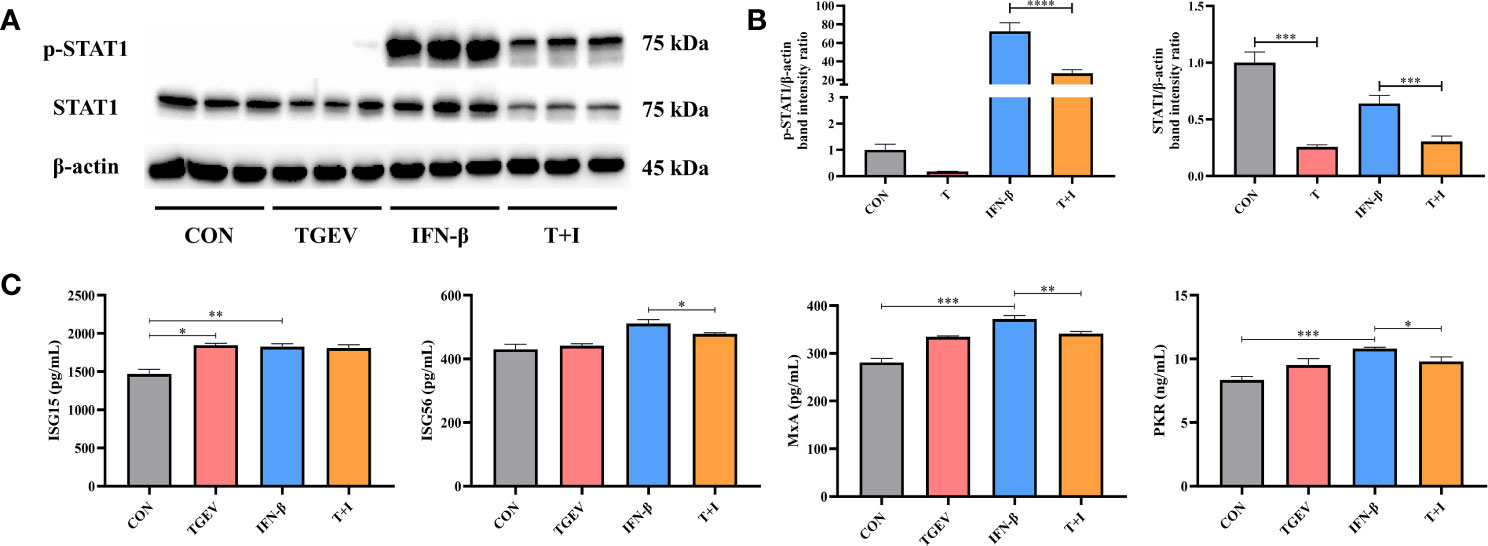
Figure 2 IFN-I signaling pathway was blocked by TGEV infection. (A) Both p-STAT1 and T-STAT1 levels were detected by western blot. (B) Quantitation of bands to demonstrate the protein level of p-STAT1 and T-STAT1. Data express the means ± SEMs (n = 3). (C) The expressions of ISGs (ISG15, ISG56, MxA and PKR) were analyzed via ELISA. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01, ***P < 0.001 and ****P < 0.0001.
mTOR Regulation Is Involved in Control of TGEV Replication
Our research demonstrated that the level of phosphorylation of mTOR and its downstream p70 S6K and 4E-BP1 was generally turned down by TGEV infection (Figure 3A). Given that mTOR acts as the regulatory center of various innate immune responses, we wonder whether mTOR could modulate TGEV replication. IPEC-J2 cells were treated with mTOR specific activator-MHY1485 and inhibitor-RAPA, followed by infection with TGEV. We observed that the inhibition of cell proliferation induced by TGEV was alleviated by MHY1485 pretreatment (Figure 3A), but not RAPA pretreatment (Figure 4A). At the same time, MHY1485 pretreatment markedly curbed the replication of TGEV (Figure 3B). By contrast, RAPA pretreatment promoted the viral replication at 48 hpi in IPEC-J2 cells (Figure 4B).
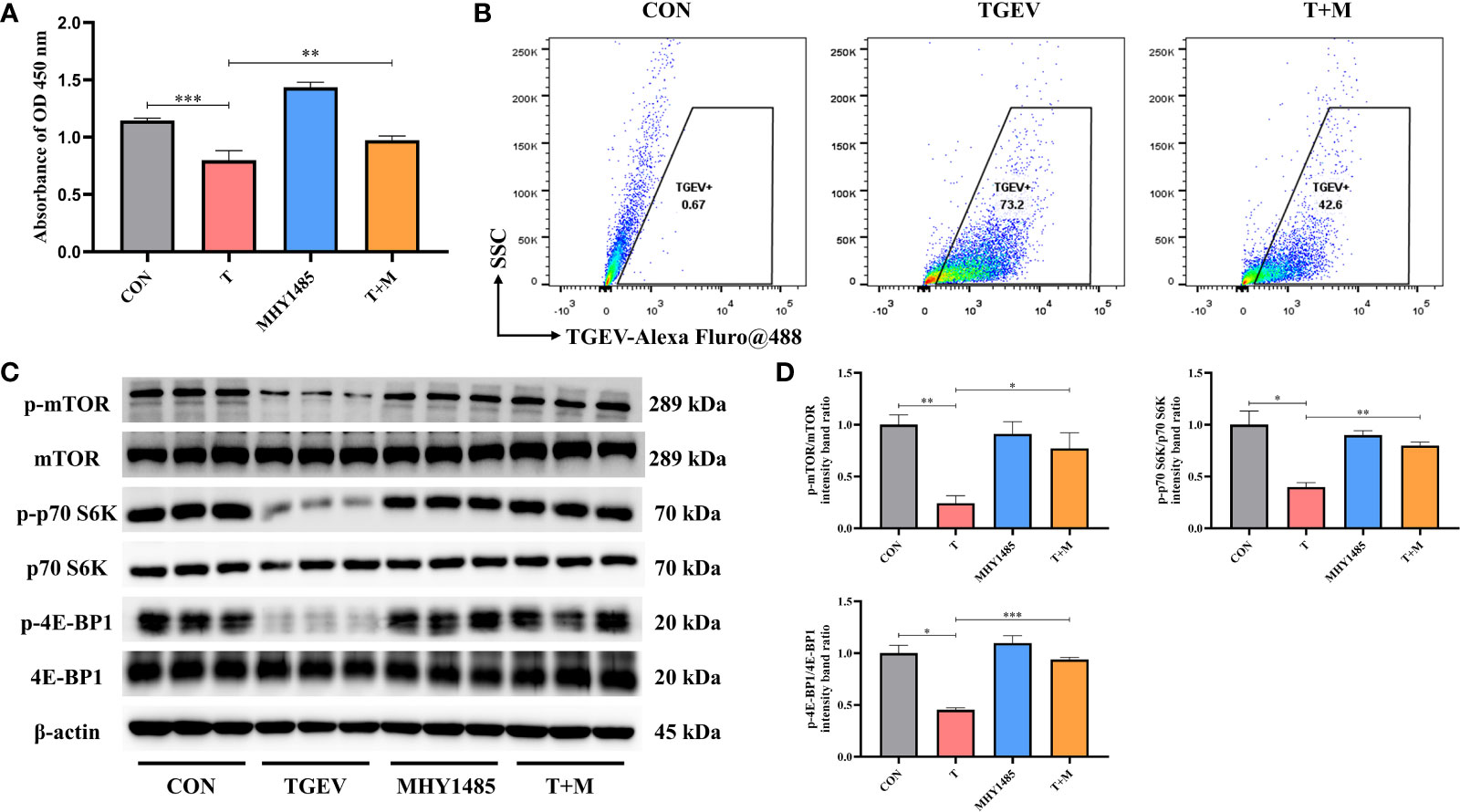
Figure 3 MHY1485 attenuated the TGEV infection by activating mTOR signaling. (A) CCK-8 assay was used to detect the proliferation of IPEC-J2 cells pretreated with 10 μM MHY1485, followed by TGEV infection. Data express the mean ± SEM (n = 3). (B) The contents of TGEV were analyzed via flow cytometry in the IPEC-J2 cells pretreated with 10 μM MHY1485, followed by TGEV infection. (C) The protein levels of p-mTOR, mTOR, p-p70 S6K, p70 S6K, p-4E-BP1 and 4E-BP-1 were detected by western blot. (D) Quantitation of bands to demonstrate the protein level of p-mTOR/mTOR, p-p70 S6K/p70 S6K and p-4E-BP1. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01 and ***P < 0.001.
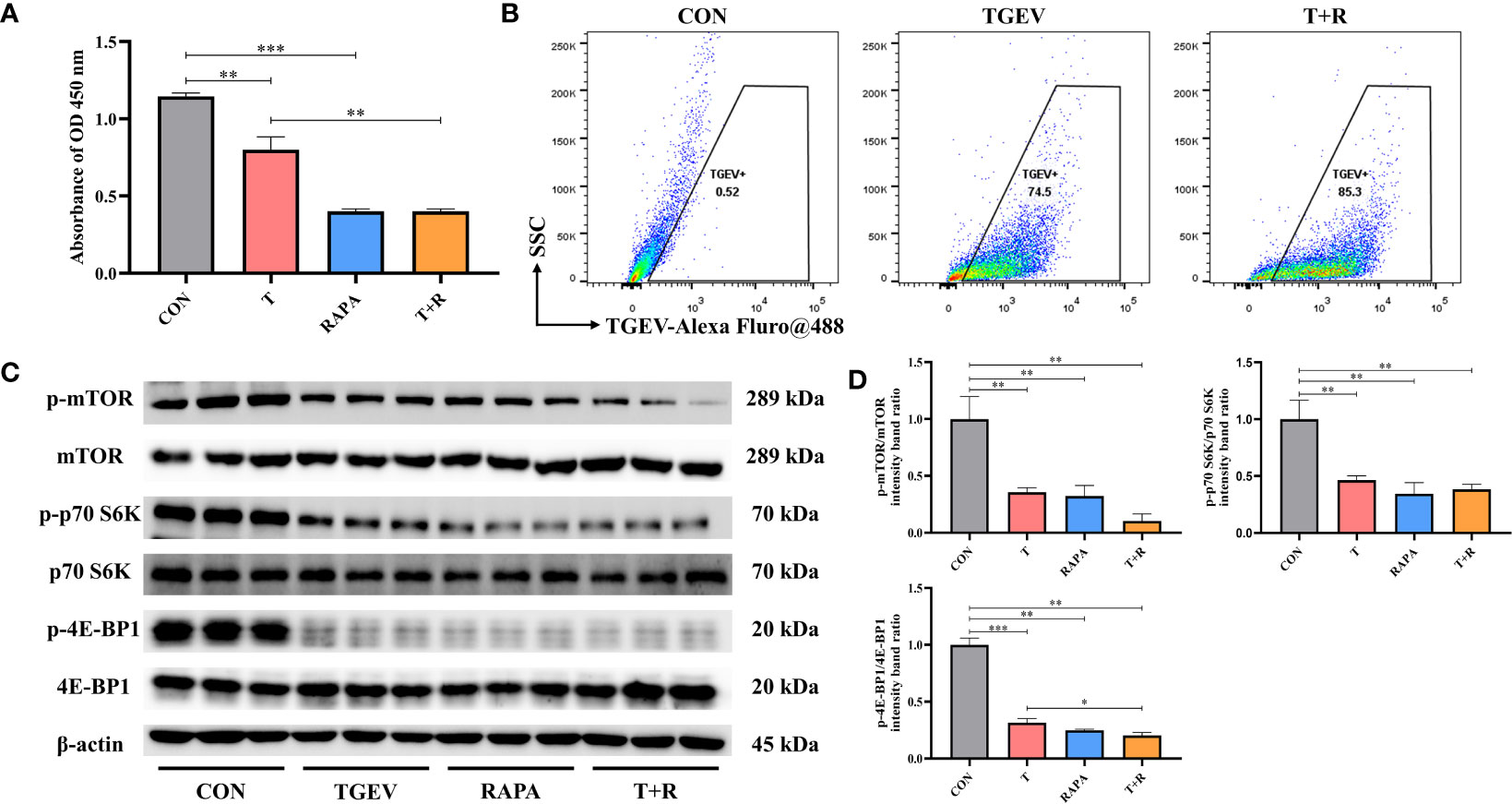
Figure 4 RAPA promoted the TGEV replication by inhibiting mTOR signaling. (A) CCK-8 assay was used to detect the proliferation of IPEC-J2 cells pretreated with 10 nM RAPA, followed by TGEV infection. Data express the mean ± SEM (n = 3). (B) The contents of TGEV were analyzed via flow cytometry in the IPEC-J2 cells pretreated with 10 nM RAPA, followed by TGEV infection. (C) The protein levels of p-mTOR, mTOR, p-p70 S6K, p70 S6K, p-4E-BP1 and 4E-BP-1 were detected by western blot. (D) Quantitation of bands to demonstrate the protein level of p-mTOR/mTOR, p-p70 S6K/p70 S6K and p-4E-BP1. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01 and ***P < 0.001.
With further research, we found that the protein expression of p-mTOR and its downstream p-p70 S6K and p-4E-BP1 significantly increased with MHY1485 pretreatment (Figures 3C, D), while RAPA inhibited the p-mTOR, p-p70 S6K and p-4E-BP1 protein expressions regardless of the presence or absence of viral infection (Figures 4C, D). These data indicated that pharmacological manipulation of mTOR could control the yield of TGEV.
mTOR Activator Augments STAT1 and ISGs Expressions in TGEV-Infected IPEC-J2 Cells
To investigate how mTOR modulates STAT1 activity, IPEC-J2 cells were pretreated with mTOR activator-MHY1485, followed by TGEV infection. Western blot analysis revealed that 10 μM MHY1485 significantly elevated the protein expression of p-STAT1 (Figures 5A, B). Similarly, the concentrations of ISGs, including ISG15, ISG56, MxA, and PKR, increased under MHY1485 pretreatment (Figure 5C). The evidence showed that mTOR activation could upregulate the expressions of p-STAT1 and downstream ISGs.
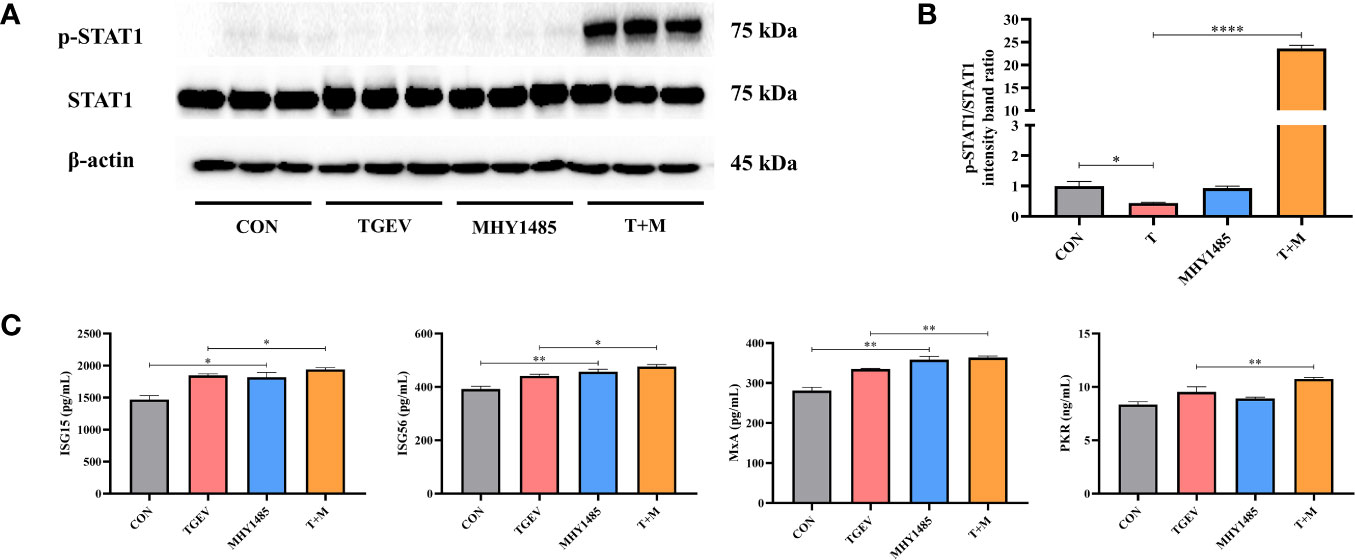
Figure 5 The inhibition of STAT1 and ISGs induced by TGEV infection was alleviated by MHY1485 pretreatment. (A) Both p-STAT1 and STAT1 levels were detected by western blot. (B) Quantitation of bands to demonstrate the protein level of p-STAT1/STAT1. Data express the means ± SEMs (n = 3). (C) The expressions of ISGs (ISG15, ISG56, MxA and PKR) were detected by ELISA. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01 and ****P < 0.0001.
mTOR Inhibitor Reduces STAT1 and ISGs Expressions in TGEV-Infected IPEC-J2 Cells
Considering the promotion of TGEV replication triggered by mTOR inhibitor-RAPA, we clarified whether RAPA could suppress STAT1 and ISGs expressions to interrupt innate immune response against TGEV infection. It is shown that p-STAT1 level was not decreased by 10 nM RAPA pretreatment in TGEV-infected cells (Figures 6A, B). However, compared with the control group, 10 nM RAPA significantly inhibited p-STAT1 expression with non-infection (Figures 6A, B). To further confirm the effect of RAPA on STAT1 expression, 1,000 U/ml IFN-β was treated in the IPEC-J2 cells. The result showed that 10 nM RAPA markedly blocked the STAT1 activation by IFN-β (Figure 6C). Similarly, the concentrations of ISG56, PKR, and MxA were significantly decreased in RAPA-pretreated cells in response to IFN-β (Figure 6D). These observations suggested that mTOR inhibition could downregulate the expressions of p-STAT1 and downstream ISGs.
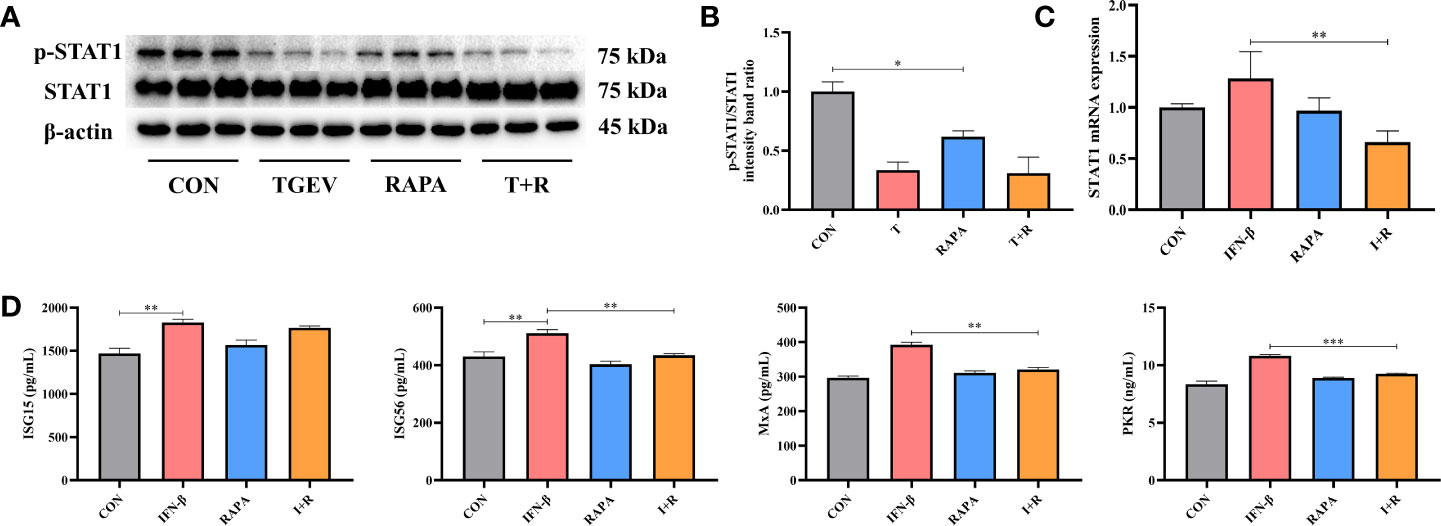
Figure 6 The expressions of STAT1 and ISGs was lessened by RAPA pretreatment. (A) Both p-STAT1 and T-STAT1 levels were detected by western blot. (B) Quantitation of bands to demonstrate the protein level of p-STAT1/STAT1. Data express the means ± SEMs (n = 3). (C) The mRNA expression of STAT1 were analyzed via RT-qPCR. Data express the means ± SEMs (n = 4). (D) The expressions of ISGs (ISG15, ISG56, MxA and PKR) were detected by ELISA. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01 and ***P < 0.001.
Leu Activated mTOR Signaling and Prevents TGEV Replication
As shown in Figure 7A, Leu pretreatment mitigated the reduction of cell proliferation by TGEV infection. At the same time, we observed that Leu significantly reduced the contents of TGEV (Figure 7B) and alleviated the inhibition of p-mTOR, p-p70 S6K and p-4E-BP1 with TGEV infection (Figures 7C, D). Therefore, Leu may protect IPEC-J2 cells from TGEV replication by activating mTOR signaling.
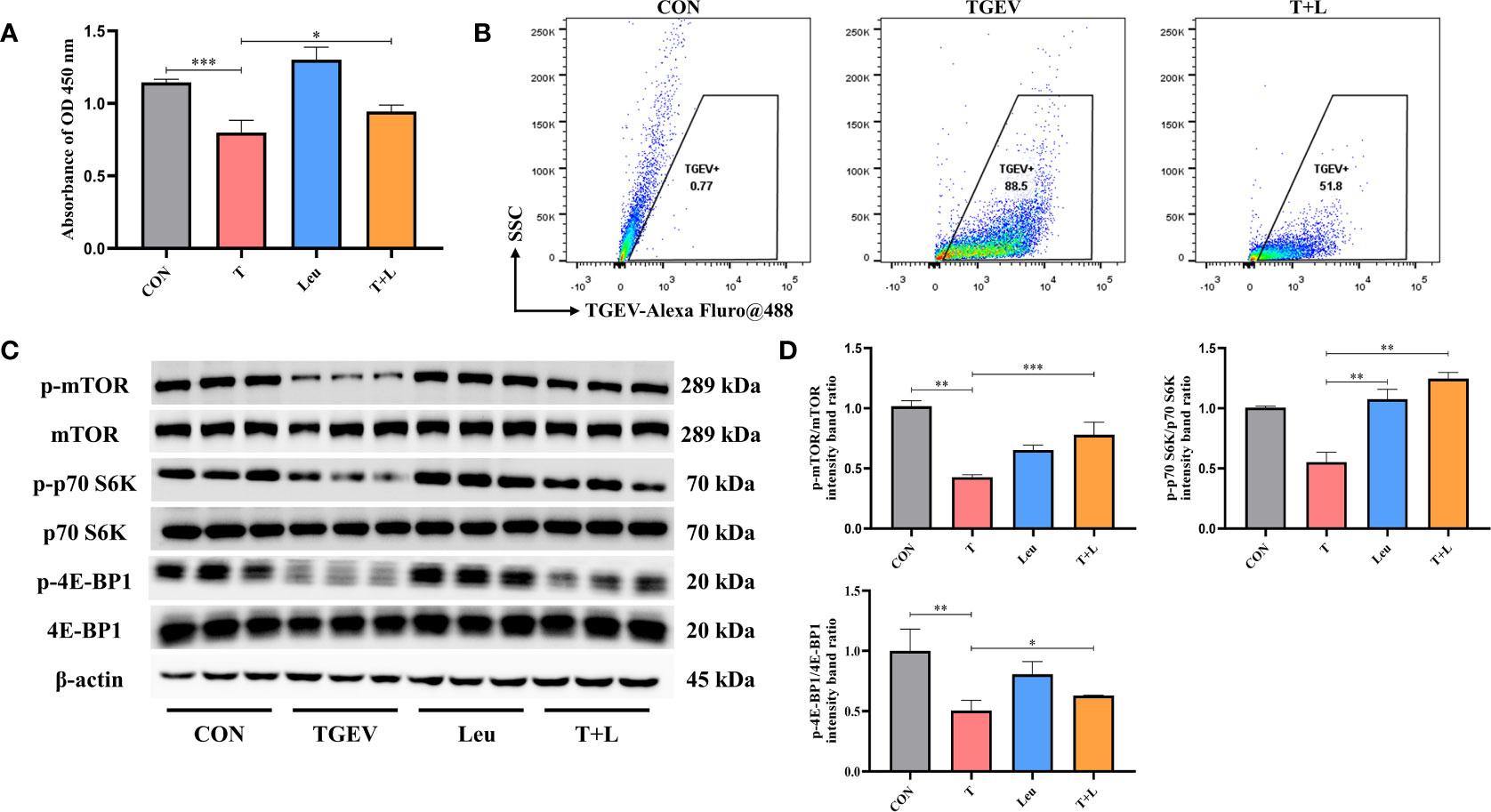
Figure 7 TGEV replication was regulated by Leu treatment. (A) CCK-8 assay was used to detect the proliferation of IPEC-J2 cells pretreated with 10 mM Leu, followed by TGEV infection. Data express the mean ± SEM (n = 3). (B) The contents of TGEV were analyzed via flow cytometry in the IPEC-J2 cells pretreated with 10 mM Leu, followed by TGEV infection. (C) The protein levels of p-mTOR, mTOR, p-p70 S6K, p70 S6K, p-4E-BP1 and 4E-BP-1 were detected by western blot. (D) Quantitation of bands to demonstrate the protein level of p-mTOR/mTOR, p-p70 S6K/p70 S6K and p-4E-BP1. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01 and ***P < 0.001.
Leu Boosts STAT1 and ISGs Expressions in TGEV-Infected IPEC-J2 Cells
In this study, we investigated whether Leu could increase the STAT1 and ISGs expressions to counteract virus infection. As shown in Figures 8A, B, the reduction of p-STAT1 was significantly alleviated by Leu in TGEV-infected cells under the RAPA pretreatment. As expected, Leu remarkably enhanced the concentrations of ISG56, MxA, and PKR in infected IPEC-J2 cells (Figure 8C).
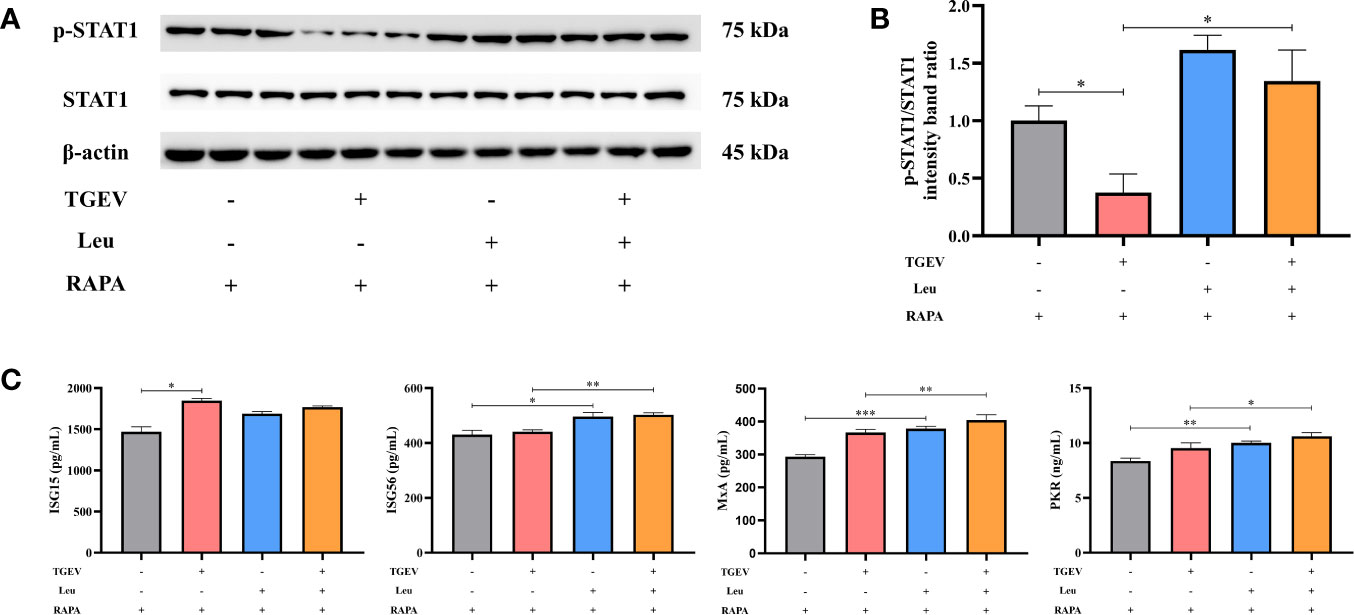
Figure 8 The expressions of STAT1 and ISGs increased by Leu and RAPA pretreatment in TGEV-infected cells. (A) Both p-STAT1 and T-STAT1 levels were detected by western blot. (B) Quantitation of bands to demonstrate the protein level of p-STAT1/STAT1. Data express the means ± SEMs (n = 3). (C) The expressions of ISGs (ISG15, ISG56, MxA and PKR) were detected by ELISA. Data express the means ± SEMs (n = 3). The symbol * indicates statistically significant differences (P < 0.05), **P < 0.01 and ***P < 0.001.
Discussion
TGEV is known to the main etiological agent to cause diarrhea, dehydration, and high mortality of piglets, and leads to devastating economic losses in the swine industry. Once piglets are infected, the virus enters the digestive tract and destroys the small intestinal epithelial cells of piglets, affecting the absorption of nutrients (28). The construction of TGEV-infected IPEC-J2 cells, which emulates the in vivo intestinal environment of piglets infected by TGEV, is an ideal model for studying the mechanism of viral infection (29). In the present study, a large number of necrotic cells were observed in the culture plate, and virulence reached the highest level at 48 hpi. Moreover, the results of flow cytometry showed that the contents of TGEV increased in a time-dependent manner. Consequently, it provides a reference for the selection of appropriate time points for our subsequent experiments.
Virus infection activates the host innate and adaptive immunity to resist virus invasion. Innate immunity is the first line of defense in host protection (30). The host utilizes PRRs to detect the PAMPs of the invading virus and induces the expressions of some effector molecules through a series of signal transduction (14). ISGs play an important role in the process of direct resistance to viral infection, and JAK/STAT signaling pathway mediates the expressions of ISGs (31). STAT1, a member of the STAT family, is an important effector molecule that mediates type I IFN response (32). However, IFN-β has multi-function of antiviral infection and immunosuppression but shows less cytotoxicity. Thus, IFN-β is inclined to clinical application (33). Considering its superiority, IFN-β was selected as a positive control to activate STAT1. In order to circumvent the host innate immunity, the virus has developed a variety of strategies to inhibit the activation of antiviral effector molecules, especially to reduce the expressions of IFNs and inhibit the IFN signaling pathway (34, 35). In the present study, TGEV infection significantly inhibited the expression of STAT1 under normal conditions and the expressions of p-STAT1 and STAT1 induced by IFN-β at 48 hpi. At the same time, the expressions of ISGs, including ISG56, MxA, PKR, and OASL were also inhibited. Therefore, these results indicate that STAT1 is a vital signal transduction target against TGEV infection. TGEV invasion can block IFN signal transduction by suppressing STAT1 activity.
The mTOR signaling pathway, as the center of various important physiological processes, has been a research hotspot (36). A great deal of evidence supports the notion that the mTOR signaling pathway plays an important role in viral infection, replication, particle assembly and release (37, 38). Our study showed that the mTOR activator (MHY1485) markedly inhibited TGEV contents and mitigated the suppression of p-mTOR, p-p70 S6K and p-4E-BP1 induced by TGEV infection. Conversely, mTOR inhibitor (RAPA) promoted the TGEV infection and has an adverse impact on the expressions of p-mTOR, p-p70 S6K and p-4E-BP1 induced by TGEV infection. In addition, recent reports have revealed the points that mTOR is able to regulate STAT1 activity, suggesting the possibility of a specific cell type cascade between mTOR and STAT1 pathways to synergistically regulate the host immune response (39, 40). A recent study demonstrated that RAPA pretreatment could inhibit STAT1 nuclear translocation in primary human fetal astrocytes infected with neurotropic polyomavirus JC (JCV) (41). Therefore, we hypothesize that the resistance of TGEV infection by mTOR upregulation is mediated by STAT1 regulation. In this study, IPEC-J2 cells were pretreated with mTOR specific activator and inhibitor, respectively, to detect the STAT1 activity and the concentrations of downstream ISGs in TGEV-infected cells. It is reported that mTOR activator-MHY1485 significantly alleviated the inhibitory effect of TGEV on p-STAT1 expression, and increased the expressions of ISGs to achieve the purpose of inhibiting TGEV proliferation. On the contrary, compared with the control group, the mTOR inhibitor-RAPA inhibited the expression of p-STAT1. However, p-STAT1 level was not decreased by RAPA pretreatment in TGEV-infected cells. It is speculated that TGEV and RAPA both act as antagonists of STAT1. Under the circumstances of STAT1 suppression by TGEV infection to evade innate immunity, the negative effect of RAPA on STAT1 was not obvious in TGEV-infected cells. Moreover, RAPA significantly inhibited the expression of STAT1 in IFN-β-driven cells, consistent with the trend of expressions of ISGs determined by ELISA. This is a unique example of the positive regulation of STAT1 activity by mTOR, mechanistically highlighting the multifaceted control of TGEV replication.
BCAAs enhance the intestinal immune defense system by maintaining mucosal immune homeostasis and increasing the level of immunoregulatory cytokines (42). Leu participates in various cellular signal transduction mechanisms to regulate intestinal growth, integrity and immune function against virus invasion. Rag guanosine triphosphatases (GTPase) located on lysosomes can sense the changes in amino acids and transfer them to the mTORC1 pathway. With sufficient amino acids, mTORC1 is recruited to play a role in lysosomes (43). Our research showed that Leu could alleviate the inhibition of p-mTOR, p-p70 S6K and p-4E-BP1 and reduce the content of TGEV. The results above indicated that mTOR activation significantly increased the expression of p-STAT1 and attenuated the TGEV infection. Furthermore, we confirmed that Leu could regulate the expressions of STAT1 and ISGs by enhancing the activity of mTOR, which is coherent with the results caused by MHY1485. Overall, our findings clearly demonstrate that Leu may promote STAT1 and ISGs expressions through activating mTOR signaling, with the consequence of alleviation of TGEV infection in enterocytes.
In conclusion, mTOR regulation is involved in the process of innate immunity against TGEV invasion. The ability of IPEC-J2 cells to prevent TGEV infection can be altered by regulating mTOR signaling. The mechanism by which Leu alleviates TGEV infection is related to its activation of mTOR signaling and promotion of STAT1 and ISGs expressions (Figure 9). Harnessing an effective nutrient strategy provides a novel theoretical basis for targeting mTOR/STAT1 activation in the prevention of transmissible gastroenteritis.
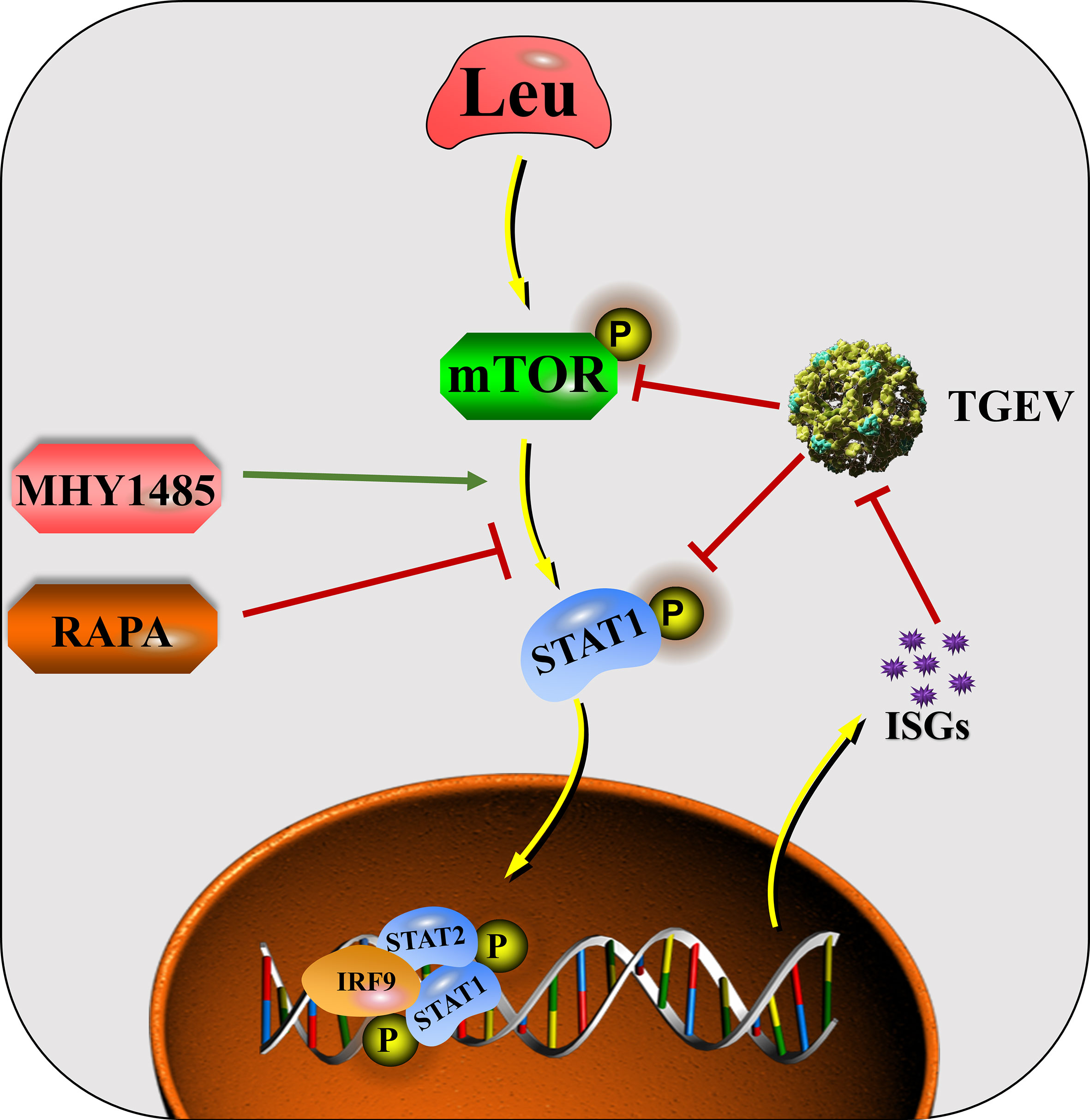
Figure 9 The mechanisms by which Leu affects TGEV infection through regulating mTOR signaling in IPEC-J2 cells.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
JD designed, performed, and analyzed the experiments and data. DC, BY, JH, JY, XM, YL, and PZ developed reagents and helped with experiments. JL acquired the funding. JD wrote the original manuscript. JL reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (31702124).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lan Wang, Kang Wang, Aimin Wu, Yao Liu, Xin Lai, Jin Wan, Linhao Zhao and Huifang He for their assistance during the experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.656573/full#supplementary-material
References
1. Cruz J, Becares M, Sola I, Oliveros JC, Enjuanes L, Zúñiga S. Alphacoronavirus Protein 7 Modulates Host Innate Immune Response. J Virol (2013) 87(17):9754–67. doi: 10.1128/JVI.01032-13
2. Sola I, Alonso S, Zúñiga S, Balasch M, Plana-Durán J, Enjuanes L. Engineering the Transmissible Gastroenteritis Virus Genome as an Expression Vector Inducing Lactogenic Immunity. J Virol (2003) 77(7):4357–69. doi: 10.1128/JVI.77.7.4357-4369.2003
3. González JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Luis E. A Comparative Sequence Analysis to Revise the Current Taxonomy of the Family Coronaviridae. Arch Virol (2003) 148(11):2207–35. doi: 10.1007/s00705-003-0162-1
4. Lia VDH, Krzysztof P, Maarten FJ, Wilma VO, Ron JMB, Katja CW, et al. Identification of a New Human Coronavirus. Nat Med (2004) 10(4):368–73. doi: 10.1038/nm1024
5. Risco C, Antón IM, Muntión M, González JM, Carrascosa JL, Luis E. Structure and Intracellular Assembly of the Transmissible Gastroenteritis Coronavirus. Adv Exp Med Biol (1998) 440:341–6. doi: 10.1007/978-1-4615-5331-1_44
6. Rossen JWA, Bekker CPJ, Voorhout WF, Strous GJAM, van der Ende A, Rottier PJM. Entry and Release of Transmissible Gastroenteritis Coronavirus Are Restricted to Apical Surfaces of Polarized Epithelial Cells. J Virol (1995) 68(12):7966–73. doi: 10.1016/0166-0934(94)90190-2
7. Zhai Y, Wang C, Jiang Z. Cross-Talk Between Bacterial Pamps and Host Prrs. Natl Sci Rev (2018) 6:791–2. doi: 10.1093/nsr/nwy103
8. Shizuo A, Satoshi U, Osamu T. Pathogen Recognition and Innate Immunity. Cell (2006) 4(124):783–801. doi: 10.1016/j.cell.2006.02.015
9. Ivashkiv LB, Donlin LT. Regulation of Type I Interferon Responses. Nat Rev Immunol (2014) 14(1):36–49. doi: 10.1038/nri3581
10. Fish EN, Platanias LC. Interferon Receptor Signaling in Malignancy: A Network of Cellular Pathways Defining Biological Outcomes. Mol Cancer Res (2014) 12(12):1691–703. doi: 10.1158/1541-7786.MCR-14-0450
11. O’Shea JJ, Holland SM, Staudt LM. Jaks and Stats in Immunity, Immunodeficiency, and Cancer. N Engl J Med (2013) 368(2):161–70. doi: 10.1056/NEJMra1202117
12. Zhang Q, Yoo D. Immune Evasion of Porcine Enteric Coronaviruses and Viral Modulation of Antiviral Innate Signaling. Virus Res (2016) 226:128–41. doi: 10.1016/j.virusres.2016.05.015
13. Du J, Luo J, Yu J, Mao X, Luo Y, Zheng P, et al. Manipulation of Intestinal Antiviral Innate Immunity and Immune Evasion Strategies of Porcine Epidemic Diarrhea Virus. BioMed Res Int (2019) 2019:1862531. doi: 10.1155/2019/1862531
14. Surapong K, Samaporn T, Phanramphoei NF, Thanathom C, Anan J. PEDV and Pdcov Pathogenesis: The Interplay Between Host Innate Immune Responses and Porcine Enteric Coronaviruses. Front Vet Sci (2019) 6:34. doi: 10.3389/fvets.2019.00034
15. Guo L, Luo X, Li R, Xu Y, Zhang J, Ge J, et al. Porcine Epidemic Diarrhea Virus Infection Inhibits Interferon Signaling by Targeted Degradation of STAT1. J Virol (2016) 90(18):8281–92. doi: 10.1128/JVI.01091-16
16. Laplante M, Sabatini DM. Mtor Signaling in Growth Control and Disease. Cell (2012) 149(2):274–93. doi: 10.1016/j.cell.2012.03.017
17. Saxton RA, Sabatini DM. Mtor Signaling in Growth, Metabolism, and Disease. Cell (2017) 168(6):960–76. doi: 10.1016/j.cell.2017.02.004
18. Azusa M, Tatsuki I, Kazuhiko N, Hisamitsu M, Kumi H, Masumi F, et al. Interferon-A-Induced Mtor Activation Is an Anti-Hepatitis C Virus Signal via the Phosphatidylinositol 3-Kinase-Akt-Independent Pathway. J Gastroenterol (2009) 44(8):856–63. doi: 10.1007/s00535-009-0075-1
19. Fielhaber JA, Han Y, Tan J, Xing S, Biggs CM, Joung K, et al. Inactivation of Mammalian Target of Rapamycin Increases STAT1 Nuclear Content and Transcriptional Activity in A4- and Protein Phosphatase 2A-Dependent Fashion. J Biol Chem (2009) 284(36):24341–53. doi: 10.1074/jbc.M109.033530
20. Kristof AS, Marks-Konczalik J, Billings E. Stimulation of Signal Transducer and Activator of Transcription-1 (STAT1)-Dependent Gene Transcription by Lipopolysaccharide and Interferon-Γ Is Regulated by Mammalian Target of Rapamycin. J Biol Chem (2003) 278(36):33637–44. doi: 10.1074/jbc.M301053200
21. Luis E, Carlos S, Fátima G, Cristian S, Ana C, Inés MA, et al. Antigen Selection and Presentation to Protect Against Transmissible Gastroenteritis Coronavirus. Vet Microbiol (1992) 33(1):249–62. doi: 10.1016/0378-1135(92)90053-v
22. Liu Y, Wang X, Hou Y, Yin Y, Qiu Y, Wu G, et al. Roles of Amino Acids in Preventing and Treating Intestinal Diseases: Recent Studies With Pig Models. Amino Acids (2017) 49(8):1277–91. doi: 10.1007/s00726-017-2450-1
23. Zhang S, Zeng X, Ren M, Mao X, Qiao S. Novel Metabolic and Physiological Functions of Branched Chain Amino Acids: A Review. J Anim Sci Biotechnol (2017) 8(3):501–12. doi: 10.1186/s40104-016-0139-z
24. Hu J, Nie Y, Chen S, Xie C, Fan Q, Wang Z, et al. Leucine Reduces Reactive Oxygen Species Levels via an Energy Metabolism Switch by Activation of the Mtor-HIF-1α Pathway in Porcine Intestinal Epithelial Cells. Int J Biochem Cell B (2017) 89:42–56. doi: 10.1016/j.biocel.2017.05.026
25. Liu S, Wang L, Liu G, Tang D, Fan X, Zhao J, et al. Leucine Alters Immunoglobulin a Secretion and Inflammatory Cytokine Expression Induced by Lipopolysaccharide via the Nuclear Factor-Kb Pathway in Intestine of Chicken Embryos. Animal (2018) 12(9):1903–11. doi: 10.1017/S1751731117003342
26. Wu A, Yu B, Zhang K, Xu Z, Wu D, He J, et al. Transmissible Gastroenteritis Virus Targets Paneth Cells to Inhibit the Self-Renewal and Differentiation of Lgr5 Intestinal Stem Cells via Notch Signaling. Cell Death Dis (2020) 11(1):40. doi: 10.1038/s41419-020-2233-6
27. Seeliger C, Schyschka L, Kronbach Z, Wottge A, van Griensven M, Wildemann B, et al. Signaling Pathway STAT1 Is Strongly Activated by IFN-B in the Pathogenesis of Osteoporosis. Eur J Med Res (2015) 20(1):1. doi: 10.1186/s40001-014-0074-4
28. Pritchard GC. Transmissible Gastroenteritis in Endemically Infected Breeding Herds of Pigs in East Anglia, 1981-85. Vet Rec (1987) 120(10):226–30. doi: 10.1016/0304-4017(87)90138-5
29. Liu F, Li G, Wen K, Bui T, Cao D, Zhang Y, et al. Porcine Small Intestinal Epithelial Cell Line (IPEC-J2) of Rotavirus Infection as a New Model for the Study of Innate Immune Responses to Rotaviruses and Probiotics. Viral Immunol (2010) 23(2):135–49. doi: 10.1089/vim.2009.0088
30. Kawai T, Akira S. Innate Immune Recognition of Viral Infection. Nat Immunol (2006) 7(2):131–37. doi: 10.1038/ni1303
31. Schneider WM, Chevillotte MD, Rice CM. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu Rev Immunol (2014) 32(1):513–45. doi: 10.1146/annurev-immunol-032713-120231
32. Sandra HB, Jose LPG, Ana R, Miguel FS, Agnes LB, Ignacio M. Direct Effects of Type I Interferons on Cells of the Immune System. Clin Cancer Res (2011) 17(9):2619–27. doi: 10.1158/1078-0432.CCR-10-1114
33. Kaplan A, Michelle WL, Andrea JW, Jose JL, Courtney AB, Ding M, et al. Direct Antimicrobial Activity of IFN-B. J Immunol (2017) 198(10):4036–45. doi: 10.4049/jimmunol.1601226
34. Xue M, Fu F, Ma Y, Zhang X, Li L, Feng L, et al. The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production. J Virol (2018) 92(15):e00431. doi: 10.1128/JVI.00431-18
35. Ma Y, Wang C, Xue M, Fu F, Zhang X, Li L, et al. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response Through IRE1α-Mediated Manipulation of the Mir-30a-5p/SOCS1/3 Axis. J Virol (2018) 92(22):e00728. doi: 10.1128/JVI.00728-18
36. Saemann MD, Haidinger M, Hecking M, Hörl WH, Weichhart T. The Multifunctional Role of Mtor in Innate Immunity: Implications for Transplant Immunity. Am J Transplant (2009) 9(12):2655–61. doi: 10.1111/j.1600-6143.2009.02832.x
37. Liu Q, Miller LC, Blecha F, Sang Y. Reduction of Infection by Inhibiting Mtor Pathway Is Associated With Reversed Repression of Type I Interferon by Porcine Reproductive and Respiratory Syndrome Virus. J Gen Virol (2017) 98(6):1316–28. doi: 10.1099/jgv.0.000802
38. Kindrachuk J. Selective Inhibition of Host Cell Signaling for Rotavirus Antivirals: PI3K/Akt/Mtor-Mediated Rotavirus Pathogenesis. Virulence (2018) 9(1):5–8. doi: 10.1080/21505594.2017.1356539
39. Saleiro D, Platanias LC. Intersection of mTOR and STAT Signaling in Immunity. Trends Immunol (2015) 36(1):21–9. doi: 10.1016/j.it.2014.10.006
40. Karonitsch T, Kandasamy RK, Kartnig F, Herdy B, Dalwigk K, Niederreiter B, et al. Mtor Senses Environmental Cues to Shape the Fibroblast-Like Synoviocyte Response to Inflammation. Cell Rep (2018) 23(7):2157–67. doi: 10.1016/j.celrep.2018.04.044
41. Caocci M, Bellizzi A, Kassa WD, White MK, Wollebo HS. Interferon Alpha Mediated Suppression of JCV Is PI3K/AKT and mTOR Pathway Dependent. J Neurovirol (2018) 24:S12. doi: 10.1007/s11481-018-9786-5
42. Zhou H, Yu B, Gao J, John KH, Chen D. Regulation of Intestinal Health by Branched-Chain Amino Acids. Anim Sci J (2018) 89(1):3–11. doi: 10.1111/asj.12937
Keywords: L-leucine, transmissible gastroenteritis virus, mammalian target of rapamycin, signal transducer and activator of transcription 1, IPEC-J2 cells
Citation: Du J, Chen D, Yu B, He J, Yu J, Mao X, Luo Y, Zheng P and Luo J (2021) L-Leucine Promotes STAT1 and ISGs Expression in TGEV-Infected IPEC-J2 Cells via mTOR Activation. Front. Immunol. 12:656573. doi: 10.3389/fimmu.2021.656573
Received: 21 January 2021; Accepted: 30 June 2021;
Published: 22 July 2021.
Edited by:
R. Brad Jones, Cornell University, United StatesReviewed by:
Qinghua Yu, Nanjing Agricultural University, ChinaQiaoli Yang, Gansu Agricultural University, China
Copyright © 2021 Du, Chen, Yu, He, Yu, Mao, Luo, Zheng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junqiu Luo, NzcwMzUzMDUzQHFxLmNvbQ==
 Jian Du
Jian Du Daiwen Chen
Daiwen Chen Bing Yu
Bing Yu Jun He
Jun He Jie Yu
Jie Yu Xiangbing Mao
Xiangbing Mao Yuheng Luo
Yuheng Luo Ping Zheng
Ping Zheng Junqiu Luo*
Junqiu Luo*