- Department of Molecular Microbiology and Immunology, School of Medicine, University of Missouri, Columbia, MO, United States
CD8 positive, tissue resident memory T cells (TRM) are a specialized subset of CD8 memory T cells that surveil tissues and provide critical first-line protection against tumors and pathogen re-infection. Recently, much effort has been dedicated to understanding the function, phenotype and development of TRM. A myriad of signals is involved in the development and maintenance of resident memory T cells in tissue. Much of the initial research focused on the roles tissue-derived signals play in the development of TRM, including TGFß and IL-33 which are critical for the upregulation of CD69 and CD103. However, more recent data suggest further roles for antigenic and pro-inflammatory cytokines. This review will focus on the interplay of pro-inflammatory, tissue and antigenic signals in the establishment of resident memory T cells.
Introduction
Over the course of an infection, naïve CD8 T cells become activated in the lymphoid tissues and differentiate into CD8 effector T cells. As effector T cells abandon the secondary lymphoid organs and migrate to tissue, they need to integrate a multitude of signals coming from cytokines, chemokines and antigen in order to gain access to infected cells, clear the pathogen and differentiate into memory T cells. Among the T cell responders with effector function, the vast majority die and only a few persist as memory T cells. We do not yet fully understand what endows T cells with the potential to become memory T cells, although we do know that the level of exposure to antigenic and pro-inflammatory signals play an important role (1–6). We also know that a balance in the level of a set of transcription factors is crucial (i.e. Eomes/T-bet, Bcl-6/Blimp-1, Id-2/Id-3, ZEB1/ZEB2, BACH/AP-1, NR4A1/IRF4) (7, 8); that specific costimulatory and homeostatic cytokines signals impart maturing memory cells with longevity properties (9, 10); and that dramatic metabolic and epigenetic changes are essential (11, 12). Precursors of memory T cells (or MPECs) have been well defined as KLRG1lo and IL-7Rhi (2) and are readily present early in the immune response albeit at small frequencies. Yet, as most of antigen specific-T cell responders progress through the immune response and die off (Short lived effectors/SLECS KLRG1hi IL-7Rlo expressors), MPECs continue their process of maturation towards memory. Consequently, T cell memory is the result of a combination of early signals which configure the transcriptional/epigenetic memory program, and late signals that during the same immune response help to fully execute this program (13, 14). T cell memory differentiation becomes even more complex when considering that memory T cells come in different “flavors” (T cell memory subsets) and with different benefits (T cell memory functions and locations). Thus, a T cell transitioning to memory, may become a central memory (TCM), an effector memory, (TEM), a stem-cell memory (TSCM), or a resident memory (TRM). Each population has evolved to fill a specific niche required to protect the host. TCM (CCR7+ CD62L+ expressors) circulate between the blood and secondary lymphoid tissues and retain an extraordinary proliferative potential. TEM (CCR7-CD62L-), in turn, circulate between the blood and peripheral tissues and are very efficient at exerting immediate effector functions upon antigen restimulation [reviewed recently in (15)]. TSCM have been described in humans (CD122+, CD95+, CCR7+, CD62L+, CD45RA+, CXCR3+) and share the proliferative, self-renewal and pluripotency potential of TCM cells (16).
Tissue resident memory T cells persist in the peripheral tissues following infection and act as front-line sentries against pathogen re-infection. The response of CD8 TRM triggers fast innate (17–19) and adaptive immune responses in the site of re-infection (20). Furthermore, CD8 TRM have also been linked to defense against tumors, with its presence correlating with good prognosis (19, 21, 22). CD8 TRM are present in almost every tissue, including secondary lymphoid organs (23). However, there is also phenotypic diversity of the TRM subset depending on the tissue. This suggests that local tissue signals may play a critical role in positioning TRM in specific locations to perform specialized functions (24). In spite of how much we have learned in recent years about TRM, there is still little known about how cytokines, antigens and other tissue signals “crosstalk” intracellularly to program the generation and maintenance of CD8 TRM (Figure 1). In this review article we will discuss how much the field has advanced in this aspect and point out to the gaps that still remain uncovered.
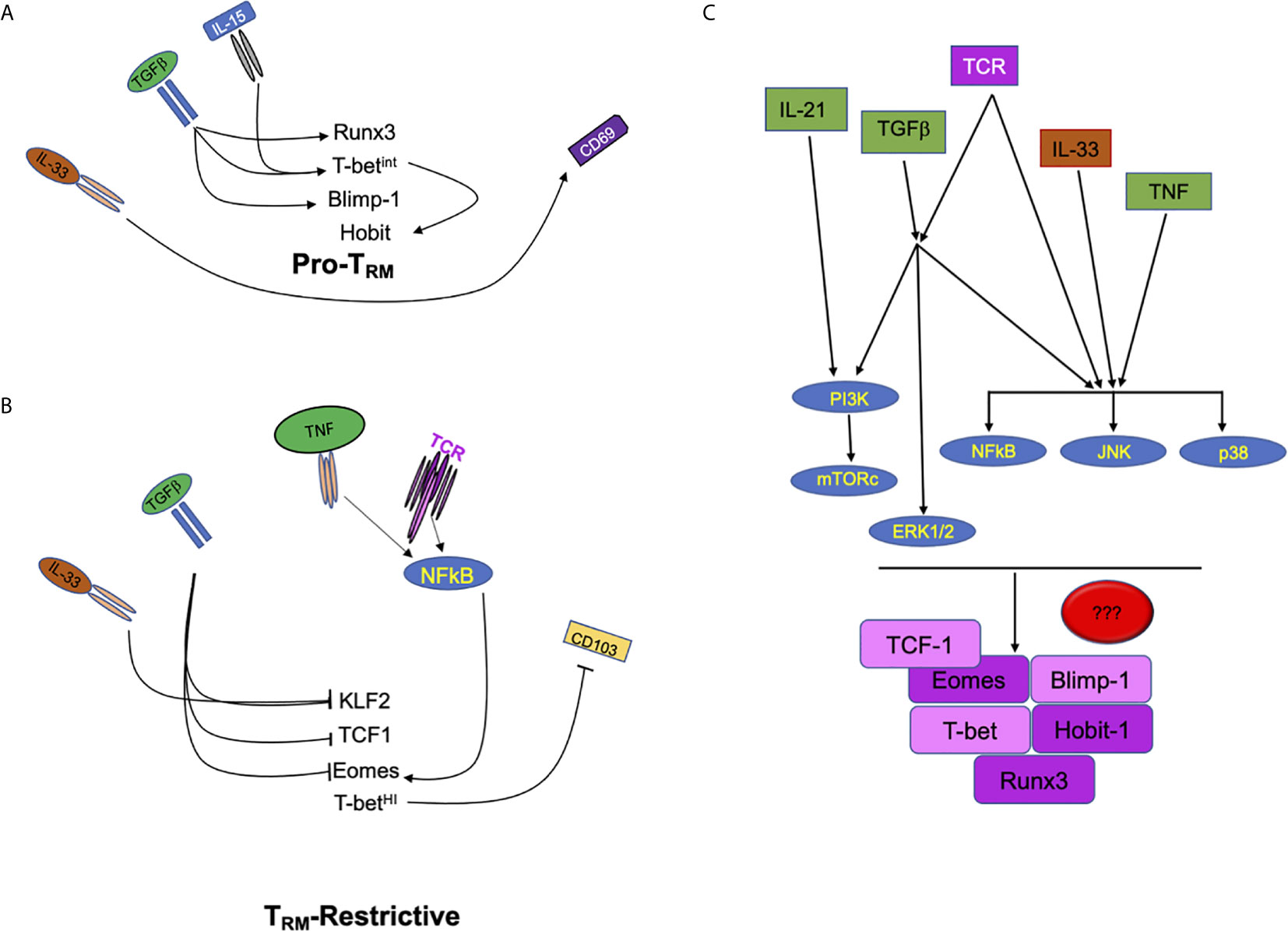
Figure 1 Extracellular factors regulate multiple signals in CD8 T cells to drive or repress TRM development. (A) Schematic of signals including IL-33, TGFβ, and IL-15 which promote the development of tissue resident T cell memory through the increase of transcription factors Runx3, Hobit, Blimp1, and the tuning of T-bet expression. (B) Tissue cytokines such as IL-33 and TGFβ also inhibit transcription factors (KLF2, TCF1, and Eomes) that can restrict the development of CD8 TRM. In contrast, pro-inflammatory cytokine and antigenic/T cell receptor signals can modulate the expression of Eomes which can, then, interfere with CD8 TRM development. (C) Signaling crosstalk between pro-inflammatory, tissue and antigenic signals. PI3K, MAPKs (ERK, JNK and p38 MAPK) and NFκB are potential nodes where extracellular cues converge to tune CD8 TRM programming, differentiation and maintenance.
Tissue Resident Memory CD8 T Cells
As mentioned before, tissue resident memory CD8 T cells have been found in peripheral healthy tissues such as lung, brain, gut, liver, skin, oral, nasal and female reproductive tract mucosal tissue, and also in tumors, transplants and organs subjected to autoimmune reactions (23). Most interestingly, tissue resident memory T cells also re-populate tissue draining lymph nodes upon antigen recall. Even at the memory stage, tissue TRM can occupy local draining lymph nodes, most likely, to warrant extended protection (25, 26). All together this puts TRM as the most abundant memory T cell in our bodies and especially so as we age. In mice, it is difficult to evaluate the lifespan of TRM beyond one year. However, in humans, it has been shown that TRM are stably maintained from childhood well into old age, at levels that are tissue specific (27, 28). Surprisingly and in contrast to mice (where naïve T cells largely reside in lymphoid organs), in humans naïve T cells are also long-term resident of tissues, although they are quickly outnumbered by memory T cells in mucosal sites (29). Resident memory T cells are extremely efficient at mounting protective innate and adaptive secondary responses upon re-infection (17, 30) and can control pathogen spread without the need of other T cell memory subsets (31). Yet whether this helps to spare the naïve and central memory population in lymph nodes from activation, and further maintain diversity in the T cell repertoire remains to be shown.
TRM ontogeny is also still poorly understood as well as the relationship of the TRM subset with the other T memory subsets. Initially MacKay, Carbone and Gebhardt described KLRG1lo epithelium expressors that encounter IL-15 and TGFβ signals as precursors of skin TRM. This led to the idea that TRM cells deviate from the T effector differentiation path once in tissue (32, 33). More recently, other studies have confirmed that even before tissue entrance circulating T cells can commit to the TRM fate. This is readily concluded when considered that: (1) TCM and TRM share a common clonal origin (34); (2) even at the naïve stage, T cells can be pre-condition to “walk” the TRM differentiation journey (35) and (3) that circulating effectors with a skewed TRM transcriptional profile that preferably become TRM exist (36). Whether this also applies to the ontogeny of TRM in other tissues is still uncertain. Indeed, in contrast to the skin TRM studies, scRNA sequencing studies in the gut have identified TRM precursors in tissue very early upon infection (37). From all these data, one thing is still clear, regardless of the potential for becoming TRM, circulating effectors will not be able to fulfill this potential unless exposed to tissue signals.
At the point T cells commit to the TRM fate, are they deadlock in this identity? or on the contrary, do they retain pluripotency to generate other T cell memory subsets upon recall? Fonseca et al. answered this question recently and provided evidence supporting the idea that TRM cells are not completely locked into the resident lineage. Upon rechallenge, ex-TRM cells epigenetically retained the potential to become TCM and TEM (38), however, they preferentially re-differentiate into TEM and TRM that homed back to their original tissue (38, 39).
Another important issue in the field is TRM diversity of heterogeneity. TRM diversity is defined by changes in transcription profile, phenotype, location and function (37). However, despite the heterogeneity within the TRM compartment, all TRM share a specific transcriptional profile characterized by expression of Runx3, Blimp-1, and Hobit and reduction of Eomes, T-bet, and KLF-2 levels (40–43) (Figure 1). This transcriptional profile enables the expression of molecules that permit recruitment and lodging to tissue in addition to special adaptation to unique tissue signals for TRM survival. What is less known is how the different signals a T cell encounters in its journey to TRM regulate this transcriptional program.
A more precise view of TRM development is arising. Cumulative evidence supports a multistep differentiation process where T cells have the potential to enter in the TRM path at different stages (naïve, in circulation, in tissue). Yet how much the quality or amount of signals a TRM precursor receives conditions its resident potential is unclear. Additionally, it is still ill-defined whether the same signals regulate TRM development, maintenance, function, retrograde migration to draining lymph nodes and/or pluripotency upon recall. Initial findings pointed to various cytokine signals and antigen within local tissues as main triggers to support CD8 effectors to CD8 TRM differentiation. TGFβ has been shown to be a major contributor to this pathway along with IL-33 and IL-15. Roles for both antigenic stimulations along with inflammatory signals such as IL-12, IL-21, and TNF have been linked to the regulation of CD8 TRM development as well (Figure 1).
Tissue Signals Involved in Cd8 Trm Development
Tissue cytokines have been shown to act synergistically in establishing the resident memory phenotype in tissues such as the gut, skin, brain, and the lungs (40, 44–49). Hereafter, we will discuss what it is known of how each one of these signals contribute to TRM development and maintenance and discuss the synergism of the signaling pathways they trigger.
TGFβ Signaling
TGFβ is a crucial cytokine for T cell development and differentiation. TGFβ is involved in thymic development, in the maintenance of naïve T cells, and also in CD8 T cell effector activation (50, 51). Seemingly, TGFβ has also been linked to the formation of CD8 TRM in different organs such as skin, the gut and lung (32, 44, 45, 52, 53).
Although TGFβ and its receptor are ubiquitous in many cells, TGFβ activity is tightly controlled at multiple levels. At the extracellular level, TGFβ activity depends on induced cleavage of latent TGFβ that is associated to the extracellular matrix or presentation by cells (such as T regs, epithelial cells, fibroblasts, keratinocytes or DCs). Large latent TGFβ can be cleaved by ECM proteases. Alternatively, it can bind to integrin receptors in the membrane of cells, which via the actin cytoskeleton promote a conformational change in TGFβ that enables the mature TGFβ release process (54). TGFβ modulates TRM in a manner that is contingent on the presence of immune cells expressing a specific set of integrin receptors. Thus, in the draining lymph nodes of the skin, specialized migratory DCs that express αv integrins present active TGFβ to naïve T cells and pre-condition them to become epithelial CD8 TRM (35). More recently, Hirai et al. provided data showing that keratinocytes activation and presentation of TGFβ to fully matured skin CD8 TRM is crucial for their maintenance. Especially, if these TRM had been generated in a bystander manner. Even more striking is that skin CD8 TRM produce their own TGFβ, thereby, contributing to their own maintenance (55). These new compelling roles of TGFβ in skin CD8 TRM add to the already known role of TGFβ in CD8 TRM differentiation (32, 40). However, they also open up new exciting questions. For instance, do these new roles of TGFβ apply to TRM in other tissues? Or what is the relative contribution of autocrine CD8 TRM TGFβ to TRM lineage identity versus TRM survival?
CD103 is one of the most thoroughly described targets of TGFβ in TRM cells (32, 44, 45, 52, 56, 57). CD103 is an integrin (alpha E) that associates with integrin beta 7. The αEβ7 integrin complex binds to E cadherin and facilitates migration and retention of CD8 T cells (32, 58, 59). While not exclusively required for development of all TRM cells, CD103 has an important role in the establishment of tissue residency within certain tissues, such as gut and skin. Sheridan et al., showed that upon oral Listeria monocytogenes infection, the majority of the intestinal effector cells rapidly upregulated CD103, but this population was lost when TGFβ signals were blocked (52). In the lung, it has been reported that CD1c+DCs control CD103 expression on CD8 T cells, enabling their accumulation in lung epithelia through a membrane-bound TGFβ dependent process (60). Lack of access to active TGFβ from fully matured skin CD8 TRM also lead to a loss of CD103 expression, although this loss appears to correlate better with the amount of active TGFβ than with a defect in CD8 TRM differentiation (55). This raises the question as to whether CD103 only provides signals for localization or whether it also activates signal transduction pathways that promote TRM lineage stability. The former is supported by the fact that in several tissues (female reproductive tract, liver, lung, and lamina propria) CD103 is not expressed by all resident memory cells (23, 61). It is also important to mention that CD103 is an integrin able to trigger bidirectional signaling and that it can cooperate with TCR signals to enable T cell migration and effector function (62). This suggests that synergism between antigenic and integrin signaling at the epithelium may be relevant for TRM maturation.
Despite the important role of CD103 in CD8 TRM adhesion, migration and retention in TGFβ rich environments, TGFβ receptor deficient cells are more compromised than CD103 deficient T cells for tissue long-term retention (44). Thus, the TGFβ role in CD8 TRM development must be broader than CD103 regulation. Indeed, several studies have pointed to other roles. TGFβ has been found to induce apoptosis of short-lived effector cells (SLECs) by antagonizing the survival effects of IL-15 (63). Since CD8 TRM maintenance in some tissues depends on both cytokines, it is possible that TGFβ contributes to the removal of SLECS, thereby favoring MPEC survival and retention in tissue (Figure 1). Comparative in vitro analysis also demonstrates a great overlapping between TRM and TGFβ transcriptional signatures (64). More precisely, TGFβ signaling regulates the expression of transcription factors involved in TRM development, such as Runx3 (65) and Blimp1 (66) and repress transcription factors (Eomes, TCF1, and T-bet) (40, 46), which are classically associated with CD8 terminal effector and central memory differentiation (5, 67–70). Achieving the right balance in the levels of all of these transcription factors appears to be crucial for the development of CD8 TRM. Thus, while some T-bet expression is necessary for the expression of IL-15Rβ to receive sufficient IL-15 signals to lodge and survive in tissue (40, 47), over activation of T-bet can also result in the loss of CD103 expression (40, 71). Similarly, high levels of Eomes have been shown to repress TRM development (40). It is still unclear how these transcriptions factors cooperate to establish the TRM program. Yet, they seem to operate under different transcriptional rules than those regulating effector CTL differentiation (where all transcription factors work together in a synergistic way) (68).
Another role of TGFβ is to control tissue lodging by suppressing the expression of Krupple-Like Factor 2 (KLF2), which in turn regulates the expression of S1PR1 (42). Skon et al. reported that TGFβ can control the lodging of CD8 TRM by downregulating KLF2 in a PI3K/Akt dependent manner (42). Curiously, canonical TGFβ signaling classically occurs through the induction of the SMAD pathway and involves formation of activated Smad2/3/4 complexes (54). However, Smad4 appears to be dispensable for CD8 TRM development (72, 73). This implies that non canonical TGFβ signaling may be more important than anticipated for CD8 TRM. TGFβR engagement can activate MAPKs p38, JNK, and ERK, NFκB, PI3K, and mTOR signaling pathways independently of Smad proteins (72–74), although the role of these pathways in CD8 TRM remains elusive. MAPKs (Figure 1), in particular, might be especially relevant as recent transcriptional studies have found an association between JunB and FosL and TRM differentiation (37).
Lastly, it is important not to underestimate the crosstalk of TGFβ with other tissue signals which may further tune TGFβ signaling and pay attention of how these signals interaction may account for further diversity or differences in CD8 TRM longevity and/or function (54, 74).
IL-33 Signaling
Along with TGFβ, IL-33 has also been involved in the establishment of CD8 resident memory. IL-33 is a part of the IL-1 family of cytokines. It is expressed by non-hematopoietic cells, constitutively in epithelial cells and inducible in activated DCs, necrotic cells, and tumor cells. It works as an alarmin in response to infection or injury [reviewed in (75, 76)]. CD8 T cells express low levels of the IL-13R or ST2 but IL-33 signaling is still important for effector function (77) and antiviral protective responses (78). Following the initial characterization of CD8 TRM, Casey et al. showed in in vitro experiments, that IL-33 could act synergistically with TGFβ to induce CD69 among CD8 T cells in the gut (45). The role of IL-33 was further defined to include the down regulation of KLF2, again in synergism with TGFβ (42). More recently, Harty’s group explored the role of IL-33 in the formation and maintenance of lung CD8 TRM in vivo. They found that when ST2 was blocked with a neutralizing antibody, the accumulation of influenza specific CD8 TRM was significantly reduced. Yet no effect on conversion to a TRM phenotype was observed (79). In another study, McLaren et al. also showed a loss of CD8 and CD4 TRM (CD69+CD103- or CD69+CD103+) in the lungs and salivary glands of IL-33-deficient mice upon MCMV infection (49). Collectively, these data strongly support a critical role of IL-33 in the establishment of the TRM pool in the lung, although whether this role impinges on CD8 TRM differentiation, maintenance and/or recruitment is unclear. Similarly, it is still unknown whether IL-33 impacts CD8 TRM in a CD8 T cell intrinsic manner or through an indirect mechanism. The in vitro experiments mentioned above (45), however, point out to a direct role in synergism with TGFβ.
IL-33 signals through MyD88/NFκB can inhibit TGFβ signals through Smad6/7 (74). Furthermore, IL-33 can synergize with IL-12 to promote the expression of T-bet and Blimp-1 while repressing Eomes and TCF-1 (77) (all transcription factors linked to CD8 TRM differentiation) (Figure 1). Taking all together (Figure 1), it is tempting to speculate that CD8 TRM differentiation and maintenance will be likely dependent on the relative levels of these cytokines in tissue and how their signaling networks crosstalk.
Inflammatory Signals and Resident Memory
Tumor Necrosis Factor
TNF is a cytokine that has pro- and anti- inflammatory functions. TNF is first expressed as a biological active transmembrane homotrimer, which can either be released after cleavage and bind to TNFR1 or TNFR2 or remain bound to the membrane and signal upon binding to TNFR2. TNFR1 is expressed universally on almost all cell types, whereas TNFR2 is mainly restricted to immune cells and some tumor cells. TNF, by contrast, can be produced by T and B cells and innate immune cells (dendritic cells, monocytes, neutrophils, mast cells). TNF is an inflammatory mediator that is heavily induced upon infections such as influenza or tuberculosis but their long -term effects are frequently associated with pulmonary diseases such as asthma, COP, ALI, and ARDS (80). In T cells, TNF can promote the activation and proliferation of naïve and effector T cells, but it also promotes cell death of highly activated effector T cells, further determining the size of the memory T cell pool (81). In vitro studies have shown that TNF can synergize with TGFβ and IL-33 to regulate the expression of molecules associated with a TRM signature (CD103, CD69 and Ly6C) in the gut, as well as regulate the expression of the transcription factor KLF-2 (facilitating the retention of TRM in tissue) (42, 45, 82). Additionally, in experiments aiming to test the role for cytokines in the conversion of circulating memory T cells to lung TRM, the authors found that neutralizing TNF levels resulted in a significant reduction in the frequency of CD8 TRM in the parenchyma (79). Altogether, these studies strongly support a role for TNFα in the establishment of TRM, however, whether TNF effects act directly on CD8 TRM precursors via their TNFR1 or TNFR2 or indirectly via other cells it is still unclear. A study showed that mice lacking TNFR1 expression were inefficient at controlling vaccinia virus in the skin, rather due to defects in resident innate cells and not to the generation of skin memory T cells (82). On the other side, other studies have implicated both TNFR1 and TNFR2 in survival of airway CD8 effectors during influenza infection (83) and also in the generation of memory T cells (81, 84). Thus, when considering the multifaceted roles of TNF signals in the progressive differentiation of CD8 T cells, more studies are needed to assess when and how TNF impacts CD8 TRM and if this happens for all tissues.
Members of the TNF superfamily OX-40 (85), 4-1BB (86, 87) and LIGHT (88) have also been linked to the establishment of CD8 TRM. 4-1BB and LIGHT appear to be crucial for the survival of effector CD8 T cells as they differentiate to TRM (86–88), whereas OX40 signals rather seem to impact the generation of effector and, therefore, accumulation of memory T cells in tissue. One feature in common among all members of the TNF superfamily (TNF included) is the activation of NFκB PI3K, Akt, MAPK and JNK pathways (89), which most likely allow for enhanced survival. However, all TNF superfamily members are also notorious for their dependence on TCR (for costimulatory functions or expression) or cytokine signals (i.e. TNF synergism with TGFβ signals). This points to a more complex picture regarding how all these factors play together in tissue as T cells differentiate and are maintained as CD8 TRM (Figure 1). Given the therapeutic value of neutralizing antibodies and fusion proteins targeting TNF family members to decrease inflammation, addressing these gaps of knowledge will aid to improve current strategies directed to boost CD8 T cell immunity in organs or tumors. Similarly, and because anti-TNF treatments are often administered to diminish inflammation in diseases such as Crohn’s and rheumatoid arthritis (90–92), knowing the impact of these treatments in the generation and maintenance of the TRM pool in patients is also important.
Interleukin 12, Type I IFN, IL-18, IL-21, and IL-6
Both IL-12 and Type I IFN are the prototypic pro-inflammatory cytokines that provide signal 3, which with signal 2 (costimulation) and signal 1 (antigen/TCR) enable full effector and memory differentiation (93–96). It has also been shown that high levels of these pro-inflammatory cytokines skew effector T cells away from memory (2, 97, 98). Intestinal proinflammatory microenvironments have elevated IFN-β and IL-12 and several studies have shown that both cytokines are critical drivers of CD8 TRM in the gut. Bergsbaken et al. identified intestinal CCR2+ macrophages as the main source of both pro-inflammatory cytokines in the gut and showed that either deletion of these innate population or deletion of the receptors for IL-12 or Type I IFN on CD8 T cells could severely reduce the differentiation and persistence of gut CD103-CD69+ CD8 TRM cells. Importantly, this was not a consequence of defects in expansion or survival of effector CD8 T cells early in the infection, but rather it was connected to the integration of pro-inflammatory cytokine signals (IL-12, IFNβ, or IL-18) and TGFβ signals in tissue (99). Another report has also shown that IL-12 acting together with IL-15 and CD24 signals is essential for the development of potent CD8 resident memory responses in the skin. In this case, a migratory BATF3+ dendritic cell population was the main source of IL-12. When tissue IL-12 signaling was inhibited using antibody blockade, sub-optimal CD8 TRM generation was observed in the skin of vaccinia virus-infected mice (100).
IL-12 can also contribute to the establishment of skin CD8 TRM through the expression of the adhesion receptor CD49a, which is specifically critical for CD8 TRM persistence and IFNγ production upon recall (101). At the transcriptional level, IL-12 is a known regulator of master regulators of CD8 TRM Eomes, T-bet and Blimp-1 (102, 103). T-bet is required for the expression of CD122 and input of IL-15 signals necessary for CD8 TRM survival (40, 47), suggesting that IL-12 indirectly facilitates CD8 TRM survival. At the same time, high levels of T-bet may be detrimental for CD8 TRM (40). Since all the studies so far have evaluated the blockade of IL-12 signals to test the role of this cytokine in CD8 TRM, it would be interesting to test whether high levels of IL-12 (which can naturally occur in cytokine storms) could be detrimental, perhaps by exceeding the T-bet threshold that transcriptionally supports TRM (40, 104).
IL-21 is another pro-inflammatory cytokine that is primarily expressed by CD4 T cells, although macrophages, NKT, B, DC, and CD8 T cells can express it at low levels (105). Recently, it has been shown that IL-21R CD8 T cell intrinsic signaling is important for the development of lung and brain CD8 TRM via oxidative metabolism (106, 107). IL-21 has been shown to synergize with other cytokines (IL-2, IL-15, IL-10) and TCR signals for regulating CD8 T cell differentiation (108). IL-21R, in turn, transduces signals via STAT-1/3/5, but it also shares the activation of PI3K and MAPK with other tissue signals (antigen, TGFβ, TNF), establishing in this way a potential system of check and balances that warranties CD8 TRM [reviewed in (105)] (Figure 1).
IL-6 shares functional features with IL-21, and it is produced in certain tissues (bone, lung, liver, adipose tissue, muscle) to fulfill homeostatic functions as well as in response to infection, cancer and tissue injury (109–111). IL-6 signals through STAT3 and together with TGFβ is primordial for Th17 differentiation (112). Furthermore, IL-6 stimulates the production of IL-21 by CD4 T cells (113) and exerts a pro-survival role that can impact the effector/memory population in the context of infection (114, 115). In CD8 T cells, IL-6, together with IL-15 and IL-7, contributes to CD8 T cell proliferation and effector function (116) and to the generation of super IL-21 producer CD8 T cells that can then, help B cells in the lung (117). The connection between IL-6 and tissue resident T cell memory is still poorly understood, although a recent report has identified a distinct population of memory helper CD8 T cells in humans that singularly express IL-6R and exhibit a skin TRM transcriptional signature (118). Interestingly, these IL-6R CD8 memory T cell population is altered in psoriasis (118) and asthma (119), although a role for these type of T cells during infection is still lacking.
Experimental evidence supports that an interaction between local tissue signals and pro-inflammatory cytokines is essential for the establishment of CD8 TRM during infection. Yet, often in systemic infections, cancer therapies (CART) and autoimmunity (rheumatoid arthritis, psoriasis), levels of these pro-inflammatory cytokines or signaling can become dysregulated and cause disease. IL-6 is, indeed, together with TNF, IL-1, IL-18, IL-33, IFNγ a soluble mediator of cytokine storms (120) in mucosal tissues, although whether high levels of inflammatory cytokines are beneficial for CD8 TRM establishment or maintenance still remains to be investigated.
Homeostatic Signals IL-7, IL-15 and IL-10
Dendritic cells are key to initiating immune responses and often for directing those responses to the appropriate tissues via delivery of antigen, co-stimulation and pro-inflammatory cytokines. What is less studied is how their contribution to homeostatic signals shape the immune response. Iborra et al. recently showed that DNGR-1+ dendritic cells cross present antigen and produce IL-12, IL-15 and CD24 signals which were required for CD8 TRM formation in the skin and lungs (100). IL-15, together with IL-7, is a homeostatic cytokine whose role in TCM and TEM cell memory maintenance is well established (121–123).
In the context of resident memory, IL-7 is almost dispensable while IL-15 has been shown fundamental for survival of CD8 TRM in some tissues (such as skin, kidney, lung and salivary glands but not in FRT, gut, pancreas) (32, 47, 124). In the skin, IL-15 contributes to lodging and maintenance of CD8 TRM by keeping balanced levels of T-bet and the transcription factor Hobit (40, 43). Hobit, in turn, is expressed exclusively in the resident memory population and has the potential to bind to regulatory regions of TCF1, KLF2 and S1PR1, all crucial for CD8 T cell tissue migration (43). In the liver, skin, and small intestine, Hobit has been shown to act in conjunction with Blimp-1 to drive TRM development as well (43). However, in the lung, Blimp-1, rather than Hobit drives TRM formation (125). This is despite the fact that persistence of a subset of lung CD8 TRM (CD103+CD69+) is completely dependent on IL-15 (40). Interestingly, the patterns of Hobit expression and function in mice and humans are different (126), but whether the results in the mouse models remain true in humans will require further investigation. Contrary to Hobit, Blimp-1 promotes CD8 TRM development in the lung while reducing the generation of CD8 TCM. This is particularly critical for CD103+ CD25+, but not CD103– CD25- lung TRM (125). While this points out to a potential role of IL-2 and IL-15 in regulating the levels of Blimp-1 the evidence remains controversial. In vitro studies have attributed a role for IL-2, but not IL-15, in the induction of Blimp-1 (127). By contrast, in vivo studies delivering IL-15 complexes have clearly shown that acute exposure (but not prolonged) to IL-15 signals can promote Blimp-1 expression (128). As IL-12 is also an inducer of Blimp-1 (103), it is possible that specialized DCs able to produce IL-15 and IL-12 (100), together with IL-2, contribute to the induction of Blimp-1 and generation of lung CD8 TRM in sites with residual inflammation.
Another cytokine that is often induced in response to infection is IL-10. CD4 regulatory T cells (Tregs) are producers of IL-10 (129). Both, Tregs and IL-10, play a critical role late in the immune response in the generation of memory CD8 T cells (130). Similarly, Type 1 Tregs (T-bet-) also promote the generation of CD8 TRM. In this case a distinct role for IL-10 was not clearly identified. Instead, the authors found that CD4 Tregs express CXCR3 and by positioning themselves close to CD8 T cells make functional TGFβ available to promote their TRM differentiation (131).These findings were consistent with previous studies indicating that TGFβ-dependent production of TGFβ resulted in increased expression of CD103 on brain CD8 T cells upon CNS infection (132).
T Cell Receptor Signals and Resident Memory CD8 T Cells
T cells recognize pathogenic or self-antigens via their T Cell Receptors (TCRs). TCR signaling is critical for memory T cells (5). Strikingly though, while T cell proliferation and some effector functions are supported by strong antigenic signals, T cell memory ensues regardless, in response to both strong and weak antigens (1, 6). These studies mainly looked at central and effector memory differentiation and found that weak TCR signals specifically favor central memory development via expression of high levels of Eomes. Moreover, TCR signal strength inversely regulated the input of inflammation by controlling the expression of inflammatory cytokine receptors and enabling a higher frequency of CD8 T cells that have been stimulated by weak antigens to become central memory T cells (1, 133). In the case of resident memory differentiation, the role of TCR signaling has been largely overlooked until recently. Fiege et al. have shown that while both high and low affinity TCR stimulation support the formation of CD8 TRM, low affinity TCR signals favored the resident memory population (134) mirroring what happens for central memory (1).
Among the signaling cascades the engaged TCR can trigger, the ones able to provide a digital type of signaling, such as Itk/Calcium and ERK (which regulate transcription factors, IRF4 and AP-1 family members) seem to be preferentially involved in promoting terminal effector differentiation (133, 135, 136). Their role in CD8 TRM remains unknown. By contrast, signaling pathways/networks leading to transcription factors that do not strictly fit the rules of TCR signal strength, appear to favor T cell memory fate (BACH2, TCF-1, Eomes) by repressing transcription factors that favor terminal effector differentiation (BACH2 represses AP-1 binding while NR4A1 represses IRF4) (1, 137–146). One of these signals is the NFκB pathway, which appears to be especially critical to the regulation of T cell memory (5, 67, 147). Both, strong and weak TCR signals use this pathway, at least to regulate central memory differentiation (147). NFκB, however, does not seem to regulate the T cell effector versus central memory decision but rather, it controls the survival of CD8 T cells during the transition to memory via maintenance of high levels of Eomes and Bcl2, which are crucial for central memory (67, 69, 70). This is possible thanks to a feedback loop where NFκB-Pim1K- Eomes drive a continuum of NFκB signals that extend beyond the peak of the immune response. These proteins also ensure memory maintenance, as memory T cells devoid of either of these failed to survive and respond (67). Whether NFκB signaling has a distinct way to regulate resident memory is unknown. NFκB signaling is also an important driver of inflammation with broad effects. From the induction of pro-inflammatory cytokines (IL-6, etc) to the signaling by inflammatory cytokines (i.e. TNF etc), NFκB holds the potential to inhibit [TGFβ (74)] or potentiate [IL-33 (148)] tissue signals that are essential for CD8 TRM [reviewed in (149)]. Although still unexplored, our previous findings and the fact that Eomes negatively modulates CD8 resident memory development (40), strongly suggest that NFκB may be an important regulator of CD8 TRM.
It is also important to mention that TCR signals are not sufficient for CD8 T cell memory and are often tuned by other environmental signals (Figure 1). This is the case of inflammatory cytokines IL-12 (102), IL-10 (150) or IL-21 (108) and metabolic signals (151). The metabolic signaling pathway, mTOR, which can also be activated by TCR and IL-12 (152), has been linked to CD8 TRM (153). Although, whether mTOR impacts on migration to tissue and/or TRM survival is still unclear.
Another important question to answer is when antigenic signals are required for establishing resident T memory. Besides the obvious need for antigenic signals to activate naïve T cells, it is widely accepted now that effector T cells that migrate from the draining lymph node to the tissue need to receive a second antigenic hit in the tissue and then, further differentiate into TRM (33, 154). Yet, depending on the tissue the continuous need to maintain antigenic signals to avoid the erosion of TRM remains contentious. Thus, several studies support that antigenic signals are required in brain, lung, female reproductive tract and skin (155–159) to accumulate TRM while in other tissues, re-exposure to antigen may be dispensable (42, 45, 157, 160). These studies only referred to cognate pathogenic antigen and did not address whether local antigenic signals were required once TRM had already been established. Moreover, while it has been shown that CD8 T cell memory does not require self-peptide-MHC signals for its maintenance or establishment (9, 161, 162), the role of self-peptide-MHC in the context of resident memory has not been sufficiently explored yet.
Conclusion
CD8 TRM are a critical first line of defense against pathogen infections and a promising tool in the fight against tumors. However, the development of CD8 resident memory requires a complex milieu of signals both from the tissues such as TGFβ, IL-33, and IL-15 and from inflammatory cytokines including IL-12 and TNF. Not only are multiple signals required, as this review discusses, specific quantities and timing of the signals are likely to be necessary. While these signals contribute to the development of CD8 resident memory, excessive amounts of some inflammatory cytokines may also limit the differentiation of CD8 TRM. Moreover, pharmaceutical treatments such as TNF blockade or other anti-inflammatory regimes may interfere with the development of the regulation of these signals and could possibly alter the development of CD8 TRM. As the transcriptional and epigenetic mechanisms that regulate CD8 TRM are becoming clearer, it is also critical that the field puts the effort to fully understand biochemically how tuning antigen, inflammatory and local tissue signals in time affect TRM. This information can be extremely valuable to the treatment of diseases where TRM are involved (infection, cancer, autoimmunity, allergies and transplantation).
Author Contributions
CJP wrote and edited the manuscript as well as organized the review. MAD edited and contributed to the discussion of the manuscript. ET wrote, edited, and contributed to the discussion of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Institutes of Health (NIH AI110420-01A1, NCI CA244314).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Michael Quaney, Dezzarae Luera and Yue Guan for critical reading of the manuscript. We apologize for the citations we did not include due to space or time limitations.
References
1. Knudson KM, Goplen NP, Cunningham CA, Daniels MA, Teixeiro E. Low-Affinity T Cells Are Programmed to Maintain Normal Primary Responses But Are Impaired in Their Recall to Low-Affinity Ligands. Cell Rep (2013) 4(3):554–65. doi: 10.1016/j.celrep.2013.07.008
2. Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation Directs Memory Precursor and Short-Lived Effector CD8(+) T Cell Fates Via the Graded Expression of T-Bet Transcription Factor. Immunity (2007) 27(2):281–95. doi: 10.1016/j.immuni.2007.07.010
3. Haring JS, Badovinac VP, Harty JT. Inflaming the CD8+ T Cell Response. Immunity (2006) 25(1):19–29. doi: 10.1016/j.immuni.2006.07.001
4. Badovinac VP, Messingham KAN, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-Cell Memory and Prime-Boost Response After Dendritic-Cell Vaccination. Nat Med (2005) 11(7):748–56. doi: 10.1038/nm1257
5. Teixeiro E, Daniels M, Hamilton S, Schrum A, Bragado R, Jameson S, et al. Different T Cell Receptor Signals Determine CD8+ Memory Versus Effector Development. Science (New York NY) (2009) 323(5913):502. doi: 10.1126/science.1163612
6. Zehn D, Lee SY, Bevan MJ. Complete But Curtailed T-cell Response to Very Low-Affinity Antigen. Nature (2009) 458(7235):211–4. doi: 10.1038/nature07657
7. Chen Y, Zander R, Khatun A, Schauder DM, Cui W. Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front Immunol (2018) 9:2826. doi: 10.3389/fimmu.2018.02826
8. Man K, Kallies A. Synchronizing Transcriptional Control of T Cell Metabolism and Function. Nat Rev Immunol (2015) 15(9):574–84. doi: 10.1038/nri3874
9. Surh CD, Sprent J. Homeostasis of Naive and Memory T Cells. Immunity (2008) 29(6):848–62. doi: 10.1016/j.immuni.2008.11.002
10. Hendriks J, Gravestein LA, Tesselaar K, van Lier RA, Schumacher TN, Borst J. CD27 is Required for Generation and Long-Term Maintenance of T Cell Immunity. Nat Immunol (2000) 1(5):433–40. doi: 10.1038/80877
11. van der Windt GJ, Pearce EL. Metabolic Switching and Fuel Choice During T-cell Differentiation and Memory Development. Immunol Rev (2012) 249(1):27–42. doi: 10.1111/j.1600-065X.2012.01150.x
12. Frias AB, Boi SK, Lan X, Youngblood B. Epigenetic Regulation of T Cell Adaptive Immunity. Immunol Rev (2021). doi: 10.1111/imr.12943
13. Arens R, Schoenberger SP. Plasticity in Programming of Effector and Memory CD8 T-Cell Formation. Immunol Rev (2010) 235(1):190–205. doi: 10.1111/j.0105-2896.2010.00899.x
14. Masopust D, Kaech SM, Wherry EJ, Ahmed R. The Role of Programming in Memory T-cell Development. Curr Opin Immunol (2004) 16(2):217–25. doi: 10.1016/j.coi.2004.02.005
15. Martin MD, Badovinac VP. Defining Memory CD8 T Cell. Front Immunol (2018) 9:2692. doi: 10.3389/fimmu.2018.02692
16. Gattinoni L, Speiser DE, Lichterfeld M, Bonini C. T Memory Stem Cells in Health and Disease. Nat Med (2017) 23(1):18–27. doi: 10.1038/nm.4241
17. Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, et al. T Cell Memory. Skin-resident Memory CD8(+) T Cells Trigger a State of Tissue-Wide Pathogen Alert. Science (2014) 346(6205):101–5. doi: 10.1126/science.1254803
18. Iijima N, Iwasaki A. T Cell Memory. A Local Macrophage Chemokine Network Sustains Protective Tissue-Resident Memory CD4 T Cells. Science (2014) 346(6205):93–8. doi: 10.1126/science.1257530
19. Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. T Cell Memory. Resident Memory CD8 T Cells Trigger Protective Innate and Adaptive Immune Responses. Science (2014) 346(6205):98–101. doi: 10.1126/science.1254536
20. Paik DH, Farber DL. Anti-Viral Protective Capacity of Tissue Resident Memory T Cells. Curr Opin Virol (2020) 46:20–6. doi: 10.1016/j.coviro.2020.09.006
21. Park SL, Gebhardt T, Mackay LK. Tissue-Resident Memory T Cells in Cancer Immunosurveillance. Trends Immunol (2019) 40(8):735–47. doi: 10.1016/j.it.2019.06.002
22. Rosato PC, Beura LK, Masopust D. Tissue Resident Memory T Cells and Viral Immunity. Curr Opin Virol (2017) 22:44–50. doi: 10.1016/j.coviro.2016.11.011
23. Masopust D, Soerens AG. Tissue-Resident T Cells and Other Resident Leukocytes. Annu Rev Immunol (2019) 37:521–46. doi: 10.1146/annurev-immunol-042617-053214
24. Takamura S, Kohlmeier JE. Establishment and Maintenance of Conventional and Circulation-Driven Lung-Resident Memory CD8(+) T Cells Following Respiratory Virus Infections. Front Immunol (2019) 10:733. doi: 10.3389/fimmu.2019.00733
25. Beura LK, Wijeyesinghe S, Thompson EA, Macchietto MG, Rosato PC, Pierson MJ, et al. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity (2018) 48(2):327–38.e5. doi: 10.1016/j.immuni.2018.01.015
26. Stolley JM, Johnston TS, Soerens AG, Beura LK, Rosato PC, Joag V, et al. Retrograde Migration Supplies Resident Memory T Cells to Lung-Draining LN After Influenza Infection. J Exp Med (2020) 217(8). doi: 10.1084/jem.20192197
27. Kumar BV, Kratchmarov R, Miron M, Carpenter DJ, Senda T, Lerner H, et al. Functional Heterogeneity of Human Tissue-Resident Memory T Cells Based on Dye Efflux Capacities. JCI Insight (2018) 3(22). doi: 10.1172/jci.insight.123568
28. Senda T, Dogra P, Granot T, Furuhashi K, Snyder ME, Carpenter DJ, et al. Microanatomical Dissection of Human Intestinal T-cell Immunity Reveals Site-Specific Changes in Gut-Associated Lymphoid Tissues Over Life. Mucosal Immunol (2019) 12(2):378–89. doi: 10.1038/s41385-018-0110-8
29. Thome JJ, Bickham KL, Ohmura Y, Kubota M, Matsuoka N, Gordon C, et al. Early-Life Compartmentalization of Human T Cell Differentiation and Regulatory Function in Mucosal and Lymphoid Tissues. Nat Med (2016) 22(1):72–7. doi: 10.1038/nm.4008
30. Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and Alarm Function of Resident Memory CD8(+) T Cells. Nat Immunol (2013) 14(5):509–13. doi: 10.1038/ni.2568
31. Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin Infection Generates Non-Migratory Memory CD8+ T(RM) Cells Providing Global Skin Immunity. Nature (2012) 483(7388):227–31. doi: 10.1038/nature10851
32. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The Developmental Pathway for CD103(+)CD8+ Tissue-Resident Memory T Cells of Skin. Nat Immunol (2013) 14(12):1294–301. doi: 10.1038/ni.2744
33. Muschaweckh A, Buchholz VR, Fellenzer A, Hessel C, Konig PA, Tao S, et al. Antigen-Dependent Competition Shapes the Local Repertoire of Tissue-Resident Memory CD8+ T Cells. J Exp Med (2016) 213(13):3075–86. doi: 10.1084/jem.20160888
34. Gaide O, Emerson RO, Jiang X, Gulati N, Nizza S, Desmarais C, et al. Common Clonal Origin of Central and Resident Memory T Cells Following Skin Immunization. Nat Med (2015) 21(6):647–53. doi: 10.1038/nm.3860
35. Mani V, Bromley SK, Aijo T, Mora-Buch R, Carrizosa E, Warner RD, et al. Migratory DCs Activate TGF-beta to Precondition Naive CD8(+) T Cells for Tissue-Resident Memory Fate. Science (2019) 366(6462). doi: 10.1126/science.aav5728
36. Kok L, Dijkgraaf FE, Urbanus J, Bresser K, Vredevoogd DW, Cardoso RF, et al. A Committed Tissue-Resident Memory T Cell Precursor Within the Circulating CD8+ Effector T Cell Pool. J Exp Med (2020) 217(10). doi: 10.1084/jem.20191711
37. Kurd NS, He Z, Louis TL, Milner JJ, Omilusik KD, Jin W, et al. Early Precursors and Molecular Determinants of Tissue-Resident Memory CD8(+) T Lymphocytes Revealed by Single-Cell RNA Sequencing. Sci Immunol (2020) 5(47). doi: 10.1126/sciimmunol.aaz6894
38. Fonseca R, Beura LK, Quarnstrom CF, Ghoneim HE, Fan Y, Zebley CC, et al. Developmental Plasticity Allows Outside-in Immune Responses by Resident Memory T Cells. Nat Immunol (2020) 21(4):412–21. doi: 10.1038/s41590-020-0607-7
39. Behr FM, Parga-Vidal L, Kragten NAM, van Dam TJP, Wesselink TH, Sheridan BS, et al. Tissue-Resident Memory CD8(+) T Cells Shape Local and Systemic Secondary T Cell Responses. Nat Immunol (2020) 21(9):1070–81. doi: 10.1038/s41590-020-0723-4
40. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-Box Transcription Factors Combine With the Cytokines TGF-Beta and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity (2015) 43(6):1101–11. doi: 10.1016/j.immuni.2015.11.008
41. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 Programs CD8(+) T Cell Residency in non-Lymphoid Tissues and Tumours. Nature (2017) 552(7684):253–7. doi: 10.1038/nature24993
42. Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional Downregulation of S1pr1 Is Required for the Establishment of Resident Memory CD8+ T Cells. Nat Immunol (2013) 14(12):1285–93. doi: 10.1038/ni.2745
43. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 Instruct a Universal Transcriptional Program of Tissue Residency in Lymphocytes. Science (2016) 352(6284):459–63. doi: 10.1126/science.aad2035
44. Zhang N, Bevan MJ. Transforming Growth Factor-Beta Signaling Controls the Formation and Maintenance of Gut-Resident Memory T Cells by Regulating Migration and Retention. Immunity (2013) 39(4):687–96. doi: 10.1016/j.immuni.2013.08.019
45. Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, et al. Antigen-Independent Differentiation and Maintenance of Effector-Like Resident Memory T Cells in Tissues. J Immunol (2012) 188(10):4866–75. doi: 10.4049/jimmunol.1200402
46. Wu J, Madi A, Mieg A, Hotz-Wagenblatt A, Weisshaar N, Ma S, et al. T Cell Factor 1 Suppresses CD103+ Lung Tissue-Resident Memory T Cell Development. Cell Rep (2020) 31(1):107484. doi: 10.1016/j.celrep.2020.03.048
47. Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, et al. IL-15-Independent Maintenance of Tissue-Resident and Boosted Effector Memory CD8 T Cells. J Immunol (2016) 196(9):3920–6. doi: 10.4049/jimmunol.1502337
48. Ebel ME, Kansas GS. Functions of Smad Transcription Factors in TGF-beta1-Induced Selectin Ligand Expression on Murine CD4 Th Cells. J Immunol (2016) 197(7):2627–34. doi: 10.4049/jimmunol.1600723
49. McLaren JE, Clement M, Marsden M, Miners KL, Llewellyn-Lacey S, Grant EJ, et al. IL-33 Augments Virus-Specific Memory T Cell Inflation and Potentiates the Efficacy of an Attenuated Cytomegalovirus-Based Vaccine. J Immunol (2019) 202(3):943–55. doi: 10.4049/jimmunol.1701757
50. Travis MA, Sheppard D. TGF-Beta Activation and Function in Immunity. Annu Rev Immunol (2014) 32:51–82. doi: 10.1146/annurev-immunol-032713-120257
51. Li MO, Flavell RA. TGF-Beta: A Master of All T Cell Trades. Cell (2008) 134(3):392–404. doi: 10.1016/j.cell.2008.07.025
52. Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrancois L. Oral Infection Drives a Distinct Population of Intestinal Resident Memory CD8(+) T Cells With Enhanced Protective Function. Immunity (2014) 40(5):747–57. doi: 10.1016/j.immuni.2014.03.007
53. Wakim LM, Smith J, Caminschi I, Lahoud MH, Villadangos JA. Antibody-Targeted Vaccination to Lung Dendritic Cells Generates Tissue-Resident Memory CD8 T Cells That are Highly Protective Against Influenza Virus Infection. Mucosal Immunol (2015) 8(5):1060–71. doi: 10.1038/mi.2014.133
54. Tzavlaki K, Moustakas A. TGF-Beta Signaling. Biomolecules (2020) 10(3). doi: 10.3390/biom10030487
55. Hirai T, Yang Y, Zenke Y, Li H, Chaudhri VK, De La Cruz Diaz JS, et al. Competition for Active TGFbeta Cytokine Allows for Selective Retention of Antigen-Specific Tissue- Resident Memory T Cells in the Epidermal Niche. Immunity (2021) 54(1):84–98.e5. doi: 10.1016/j.immuni.2020.10.022
56. El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, et al. TGF-{Beta}-Dependent CD103 Expression by CD8(+) T Cells Promotes Selective Destruction of the Host Intestinal Epithelium During Graft-Versus-Host Disease. J Exp Med (2005) 201(10):1647–57. doi: 10.1084/jem.20041044
57. Lee YT, Suarez-Ramirez JE, Wu T, Redman JM, Bouchard K, Hadley GA, et al. Environmental and Antigen Receptor-Derived Signals Support Sustained Surveillance of the Lungs by Pathogen-Specific Cytotoxic T Lymphocytes. J Virol (2011) 85(9):4085–94. doi: 10.1128/JVI.02493-10
58. Schon MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, et al. Mucosal T Lymphocyte Numbers are Selectively Reduced in Integrin Alpha E (CD103)-Deficient Mice. J Immunol (1999) 162(11):6641–9.
59. Schlickum S, Sennefelder H, Friedrich M, Harms G, Lohse MJ, Kilshaw P, et al. Integrin Alpha E(CD103)beta 7 Influences Cellular Shape and Motility in a Ligand-Dependent Fashion. Blood (2008) 112(3):619–25. doi: 10.1182/blood-2008-01-134833
60. Yu CI, Becker C, Wang Y, Marches F, Helft J, Leboeuf M, et al. Human CD1c+ Dendritic Cells Drive the Differentiation of CD103+ CD8+ Mucosal Effector T Cells Via the Cytokine TGF-Beta. Immunity (2013) 38(4):818–30. doi: 10.1016/j.immuni.2013.03.004
61. Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, et al. Liver-Resident Memory CD8(+) T Cells Form a Front-Line Defense Against Malaria Liver-Stage Infection. Immunity (2016) 45(4):889–902. doi: 10.1016/j.immuni.2016.08.011
62. Corgnac S, Boutet M, Kfoury M, Naltet C, Mami-Chouaib F. The Emerging Role of CD8(+) Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin. Front Immunol (2018) 9:1904. doi: 10.3389/fimmu.2018.01904
63. Sanjabi S, Mosaheb MM, Flavell RA. Opposing Effects of TGF-beta and IL-15 Cytokines Control the Number of Short-Lived Effector CD8+ T Cells. Immunity (2009) 31(1):131–44. doi: 10.1016/j.immuni.2009.04.020
64. Nath AP, Braun A, Ritchie SC, Carbone FR, Mackay LK, Gebhardt T, et al. Comparative Analysis Reveals a Role for TGF-beta in Shaping the Residency-Related Transcriptional Signature in Tissue-Resident Memory CD8+ T Cells. PloS One (2019) 14(2):e0210495. doi: 10.1371/journal.pone.0210495
65. Ito Y, Miyazono K. RUNX Transcription Factors as Key Targets of TGF-Beta Superfamily Signaling. Curr Opin Genet Dev (2003) 13(1):43–7. doi: 10.1016/S0959-437X(03)00007-8
66. Salehi S, Bankoti R, Benevides L, Willen J, Couse M, Silva JS, et al. B Lymphocyte-Induced Maturation Protein-1 Contributes to Intestinal Mucosa Homeostasis by Limiting the Number of IL-17-Producing CD4+ T Cells. J Immunol (2012) 189(12):5682–93. doi: 10.4049/jimmunol.1201966
67. Knudson KM, Pritzl CJ, Saxena V, Altman A, Daniels MA, Teixeiro E. NFkappaB-Pim-1-Eomesodermin Axis is Critical for Maintaining CD8 T-Cell Memory Quality. Proc Natl Acad Sci USA (2017) 114(9):E1659–67. doi: 10.1073/pnas.1608448114
68. Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, et al. Runx3 and T-box Proteins Cooperate to Establish the Transcriptional Program of Effector CTLs. J Exp Med (2009) 206(1):51–9. doi: 10.1084/jem.20081242
69. Cho OH, Shin HM, Miele L, Golde TE, Fauq A, Minter LM, et al. Notch Regulates Cytolytic Effector Function in CD8+ T Cells. J Immunol (2009) 182(6):3380–9. doi: 10.4049/jimmunol.0802598
70. Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, et al. Cutting Edge: The Transcription Factor Eomesodermin Enables CD8+ T Cells to Compete for the Memory Cell Niche. J Immunol (2010) 185(9):4988–92. doi: 10.4049/jimmunol.1002042
71. Backer RA, Helbig C, Gentek R, Kent A, Laidlaw BJ, Dominguez CX, et al. A Central Role for Notch in Effector CD8(+) T Cell Differentiation. Nat Immunol (2014) 15(12):1143–51. doi: 10.1038/ni.3027
72. Hu Y, Lee YT, Kaech SM, Garvy B, Cauley LS. Smad4 Promotes Differentiation of Effector and Circulating Memory CD8 T Cells But is Dispensable for Tissue-Resident Memory CD8 T Cells. J Immunol (2015) 194(5):2407–14. doi: 10.4049/jimmunol.1402369
73. Derynck R, Budi EH. Specificity, Versatility, and Control of TGF-beta Family Signaling. Sci Signal (2019) 12(570). doi: 10.1126/scisignal.aav5183
74. Derynck R, Zhang YE. Smad-Dependent and Smad-independent Pathways in TGF-beta Family Signalling. Nature (2003) 425(6958):577–84. doi: 10.1038/nature02006
75. Liew FY, Girard JP, Turnquist HR. Interleukin-33 in Health and Disease. Nat Rev Immunol (2016) 16(11):676–89. doi: 10.1038/nri.2016.95
76. Griesenauer B, Paczesny S. The ST2/IL-33 Axis in Immune Cells During Inflammatory Diseases. Front Immunol (2017) 8:475. doi: 10.3389/fimmu.2017.00475
77. Yang Q, Li G, Zhu Y, Liu L, Chen E, Turnquist H, et al. IL-33 Synergizes With TCR and IL-12 Signaling to Promote the Effector Function of CD8+ T Cells. Eur J Immunol (2011) 41(11):3351–60. doi: 10.1002/eji.201141629
78. Bonilla WV, Frohlich A, Senn K, Kallert S, Fernandez M, Johnson S, et al. The Alarmin interleukin-33 Drives Protective Antiviral CD8(+) T Cell Responses. Science (2012) 335(6071):984–9. doi: 10.1126/science.1215418
79. Slutter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, Harty JT. Dynamics of Influenza-Induced Lung-Resident Memory T Cells Underlie Waning Heterosubtypic Immunity. Sci Immunol (2017) 2(7). doi: 10.1126/sciimmunol.aag2031
80. Mukhopadhyay S, Hoidal JR, Mukherjee TK. Role of TNFalpha in Pulmonary Pathophysiology. Respir Res (2006) 7:125. doi: 10.1186/1465-9921-7-125
81. Mehta AK, Gracias DT, Croft M. TNF Activity and T Cells. Cytokine (2018) 101:14–8. doi: 10.1016/j.cyto.2016.08.003
82. Tian T, Dubin K, Jin Q, Qureshi A, King SL, Liu L, et al. Disruption of TNF-alpha/TNFR1 Function in Resident Skin Cells Impairs Host Immune Response Against Cutaneous Vaccinia Virus Infection. J Invest Dermatol (2012) 132(5):1425–34. doi: 10.1038/jid.2011.489
83. Richter MV, Topham DJ. The alpha1beta1 Integrin and TNF Receptor II Protect Airway CD8+ Effector T Cells From Apoptosis During Influenza Infection. J Immunol (2007) 179(8):5054–63. doi: 10.4049/jimmunol.179.8.5054
84. Kim EY, Priatel JJ, Teh SJ, Teh HS. TNF Receptor Type 2 (p75) Functions as a Costimulator for Antigen-Driven T Cell Responses In Vivo. J Immunol (2006) 176(2):1026–35. doi: 10.4049/jimmunol.176.2.1026
85. Salek-Ardakani S, Moutaftsi M, Sette A, Croft M. Targeting OX40 Promotes Lung-Resident Memory CD8 T Cell Populations That Protect Against Respiratory Poxvirus Infection. J Virol (2011) 85(17):9051–9. doi: 10.1128/JVI.00619-11
86. Zhou AC, Batista NV, Watts TH. 4-1BB Regulates Effector CD8 T Cell Accumulation in the Lung Tissue Through a TRAF1-, mTOR-, and Antigen-Dependent Mechanism to Enhance Tissue-Resident Memory T Cell Formation During Respiratory Influenza Infection. J Immunol (2019) 202(8):2482–92. doi: 10.4049/jimmunol.1800795
87. Zhou AC, Wagar LE, Wortzman ME, Watts TH. Intrinsic 4-1BB Signals are Indispensable for the Establishment of an Influenza-Specific Tissue-Resident Memory CD8 T-Cell Population in the Lung. Mucosal Immunol (2017) 10(5):1294–309. doi: 10.1038/mi.2016.124
88. Desai P, Tahiliani V, Hutchinson TE, Dastmalchi F, Stanfield J, Abboud G, et al. The TNF Superfamily Molecule Light Promotes the Generation of Circulating and Lung-Resident Memory CD8 T Cells Following an Acute Respiratory Virus Infection. J Immunol (2018) 200(8):2894–904. doi: 10.4049/jimmunol.1701499
89. Croft M. The Role of TNF Superfamily Members in T-cell Function and Diseases. Nat Rev Immunol (2009) 9(4):271–85. doi: 10.1038/nri2526
90. Butler DM, Maini RN, Feldmann M, Brennan FM. Modulation of Proinflammatory Cytokine Release in Rheumatoid Synovial Membrane Cell Cultures. Comparison of Monoclonal Anti TNF-alpha Antibody With the Interleukin-1 Receptor Antagonist. Eur Cytokine Netw (1995) 6(4):225–30.
91. Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M. Inhibitory Effect of TNF Alpha Antibodies on Synovial Cell Interleukin-1 Production in Rheumatoid Arthritis. Lancet (1989) 2(8657):244–7. doi: 10.1016/S0140-6736(89)90430-3
92. Sandborn WJ, Feagan BG, Radford-Smith G, Kovacs A, Enns R, Innes A, et al. CDP571, a Humanised Monoclonal Antibody to Tumour Necrosis Factor Alpha, for Moderate to Severe Crohn’s Disease: A Randomised, Double Blind, Placebo Controlled Trial. Gut (2004) 53(10):1485–93. doi: 10.1136/gut.2003.035253
93. Curtsinger JM, Mescher MF. Inflammatory Cytokines as a Third Signal for T Cell Activation. Curr Opin Immunol (2010) 22(3):333–40. doi: 10.1016/j.coi.2010.02.013
94. Curtsinger JM, Valenzuela JO, Agarwal P, Lins D, Mescher MF. Type I IFNs Provide a Third Signal to CD8 T Cells to Stimulate Clonal Expansion and Differentiation. J Immunol (2005) 174(8):4465–9. doi: 10.4049/jimmunol.174.8.4465
95. Mescher MF, Curtsinger JM, Agarwal P, Casey KA, Gerner M, Hammerbeck CD, et al. Signals Required for Programming Effector and Memory Development by CD8+ T Cells. Immunol Rev (2006) 211:81–92. doi: 10.1111/j.0105-2896.2006.00382.x
96. Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. Programming for CD8 T Cell Memory Development Requires IL-12 or Type I IFN. J Immunol (2009) 182(5):2786–94. doi: 10.4049/jimmunol.0803484
97. Cui W, Joshi NS, Jiang A, Kaech SM. Effects of Signal 3 During CD8 T Cell Priming: Bystander Production of IL-12 Enhances Effector T Cell Expansion But Promotes Terminal Differentiation. Vaccine (2009) 27(15):2177–87. doi: 10.1016/j.vaccine.2009.01.088
98. Joshi NS, Kaech SM. Effector CD8 T Cell Development: A Balancing Act Between Memory Cell Potential and Terminal Differentiation. J Immunol (2008) 180(3):1309–15. doi: 10.4049/jimmunol.180.3.1309
99. Bergsbaken T, Bevan MJ, Fink PJ. Local Inflammatory Cues Regulate Differentiation and Persistence of CD8(+) Tissue-Resident Memory T Cells. Cell Rep (2017) 19(1):114–24. doi: 10.1016/j.celrep.2017.03.031
100. Iborra S, Martinez-Lopez M, Khouili SC, Enamorado M, Cueto FJ, Conde-Garrosa R, et al. Optimal Generation of Tissue-Resident But Not Circulating Memory T Cells During Viral Infection Requires Crosspriming by DNGR-1(+) Dendritic Cells. Immunity (2016) 45(4):847–60. doi: 10.1016/j.immuni.2016.08.019
101. Bromley SK, Akbaba H, Mani V, Mora-Buch R, Chasse AY, Sama A, et al. CD49a Regulates Cutaneous Resident Memory CD8(+) T Cell Persistence and Response. Cell Rep (2020) 32(9):108085. doi: 10.1016/j.celrep.2020.108085
102. Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 Inversely Regulates T-bet and Eomesodermin Expression During Pathogen-Induced CD8+ T Cell Differentiation. J Immunol (2006) 177(11):7515–9. doi: 10.4049/jimmunol.177.11.7515
103. Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, et al. A Molecular Threshold for Effector CD8(+) T Cell Differentiation Controlled by Transcription Factors Blimp-1 and T-Bet. Nat Immunol (2016) 17(4):422–32. doi: 10.1038/ni.3410
104. Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, et al. CD4+ T Cell Help Guides Formation of CD103+ Lung-Resident Memory CD8+ T Cells During Influenza Viral Infection. Immunity (2014) 41(4):633–45. doi: 10.1016/j.immuni.2014.09.007
105. Ren HM, Lukacher AE. IL-21 in Homeostasis of Resident Memory and Exhausted CD8 T Cells During Persistent Infection. Int J Mol Sci (2020) 21(18). doi: 10.3390/ijms21186966
106. Son YM, Cheon IS, Wu Y, Li C, Wang Z, Gao X, et al. Tissue-Resident CD4(+) T Helper Cells Assist the Development of Protective Respiratory B and CD8(+) T Cell Memory Responses. Sci Immunol (2021) 6(55). doi: 10.1126/sciimmunol.abb6852
107. Xin G, Schauder DM, Lainez B, Weinstein JS, Dai Z, Chen Y, et al. A Critical Role of IL-21-Induced BATF in Sustaining CD8-T-Cell-Mediated Chronic Viral Control. Cell Rep (2015) 13(6):1118–24. doi: 10.1016/j.celrep.2015.09.069
108. Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An Interleukin-21- Interleukin-10-Stat3 Pathway is Critical for Functional Maturation of Memory CD8+ T Cells. Immunity (2011) 35(5):792–805. doi: 10.1016/j.immuni.2011.09.017
109. Hirano T. Interleukin 6 in Inflammation, Autoimmunity and Cancer. Int Immunol (2020). doi: 10.1093/intimm/dxaa078
110. Garbers C, Heink S, Korn T, Rose-John S. Interleukin-6: Designing Specific Therapeutics for a Complex Cytokine. Nat Rev Drug Discov (2018) 17(6):395–412. doi: 10.1038/nrd.2018.45
111. Rincon M, Irvin CG. Role of IL-6 in Asthma and Other Inflammatory Pulmonary Diseases. Int J Biol Sci (2012) 8(9):1281–90. doi: 10.7150/ijbs.4874
112. Heink S, Yogev N, Garbers C, Herwerth M, Aly L, Gasperi C, et al. Trans-Presentation of IL-6 by Dendritic Cells is Required for the Priming of Pathogenic TH17 Cells. Nat Immunol (2017) 18(1):74–85. doi: 10.1038/ni.3632
113. Dienz O, Eaton SM, Bond JP, Neveu W, Moquin D, Noubade R, et al. The Induction of Antibody Production by IL-6 is Indirectly Mediated by IL-21 Produced by CD4+ T Cells. J Exp Med (2009) 206(1):69–78. doi: 10.1084/jem.20081571
114. Rochman I, Paul WE, Ben-Sasson SZ. IL-6 Increases Primed Cell Expansion and Survival. J Immunol (2005) 174(8):4761–7. doi: 10.4049/jimmunol.174.8.4761
115. Strutt TM, McKinstry KK, Kuang Y, Finn CM, Hwang JH, Dhume K, et al. Direct IL-6 Signals Maximize Protective Secondary CD4 T Cell Responses Against Influenza. J Immunol (2016) 197(8):3260–70. doi: 10.4049/jimmunol.1600033
116. Gagnon J, Ramanathan S, Leblanc C, Cloutier A, McDonald PP, Ilangumaran S. IL-6, in Synergy With IL-7 or IL-15, Stimulates TCR-independent Proliferation and Functional Differentiation of CD8+ T Lymphocytes. J Immunol (2008) 180(12):7958–68. doi: 10.4049/jimmunol.180.12.7958
117. Yang R, Masters AR, Fortner KA, Champagne DP, Yanguas-Casas N, Silberger DJ, et al. IL-6 Promotes the Differentiation of a Subset of Naive CD8+ T Cells Into IL-21-Producing B Helper CD8+ T Cells. J Exp Med (2016) 213(11):2281–91. doi: 10.1084/jem.20160417
118. Loyal L, Warth S, Jurchott K, Molder F, Nikolaou C, Babel N, et al. SLAMF7 and IL-6R Define Distinct Cytotoxic Versus Helper Memory CD8(+) T Cells. Nat Commun (2020) 11(1):6357. doi: 10.1038/s41467-020-19002-6
119. Lee N, You S, Shin MS, Lee WW, Kang KS, Kim SH, et al. IL-6 Receptor Alpha Defines Effector Memory CD8+ T Cells Producing Th2 Cytokines and Expanding in Asthma. Am J Respir Crit Care Med (2014) 190(12):1383–94. doi: 10.1164/rccm.201403-0601OC
120. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med (2020) 383(23):2255–73. doi: 10.1056/NEJMra2026131
121. Goldrath AW. Cytokine Requirements for Acute and Basal Homeostatic Proliferation of Naive and Memory CD8+ T Cells. J Exp Med (2002) 195(12):1515–22. doi: 10.1084/jem.20020033
122. Schluns KS, Williams K, Ma A, Zheng XX, Lefrançois L. Cutting Edge: Requirement for IL-15 in the Generation of Primary and Memory Antigen-Specific CD8 T Cells. J Immunol (2002) 168(10):4827–31. doi: 10.4049/jimmunol.168.10.4827
123. Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, et al. Interleukin 15 is Required for Proliferative Renewal of Virus-Specific Memory CD8 T Cells. J Exp Med (2002) 195(12):1541–8. doi: 10.1084/jem.20020369
124. Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, et al. Hair Follicle-Derived IL-7 and IL-15 Mediate Skin-Resident Memory T Cell Homeostasis and Lymphoma. Nat Med (2015) 21(11):1272–9. doi: 10.1038/nm.3962
125. Behr FM, Kragten NAM, Wesselink TH, Nota B, van Lier RAW, Amsen D, et al. Blimp-1 Rather Than Hobit Drives the Formation of Tissue-Resident Memory CD8(+) T Cells in the Lungs. Front Immunol (2019) 10:400. doi: 10.3389/fimmu.2019.00400
126. Vieira Braga FA, Hertoghs KM, Kragten NA, Doody GM, Barnes NA, Remmerswaal EB, et al. Blimp-1 Homolog Hobit Identifies Effector-Type Lymphocytes in Humans. Eur J Immunol (2015) 45(10):2945–58. doi: 10.1002/eji.201545650
127. Gong D, Malek TR. Cytokine-Dependent Blimp-1 Expression in Activated T Cells Inhibits IL-2 Production. J Immunol (2007) 178(1):242–52. doi: 10.4049/jimmunol.178.1.242
128. Sowell RT, Goldufsky JW, Rogozinska M, Quiles Z, Cao Y, Castillo EF, et al. IL-15 Complexes Induce Migration of Resting Memory CD8 T Cells Into Mucosal Tissues. J Immunol (2017) 199(7):2536–46. doi: 10.4049/jimmunol.1501638
129. O’Garra A, Vieira PL, Vieira P, Goldfeld AE. IL-10-producing and Naturally Occurring CD4+ Tregs: Limiting Collateral Damage. J Clin Invest (2004) 114(10):1372–8. doi: 10.1172/JCI23215
130. Laidlaw BJ, Cui W, Amezquita RA, Gray SM, Guan T, Lu Y, et al. Production of IL-10 by CD4(+) Regulatory T Cells During the Resolution of Infection Promotes the Maturation of Memory CD8(+) T Cells. Nat Immunol (2015) 16(8):871–9. doi: 10.1038/ni.3224
131. Ferreira C, Barros L, Baptista M, Blankenhaus B, Barros A, Figueiredo-Campos P, et al. Type 1 Treg Cells Promote the Generation of CD8(+) Tissue-Resident Memory T Cells. Nat Immunol (2020) 21(7):766–76. doi: 10.1038/s41590-020-0674-9
132. Graham JB, Da Costa A, Lund JM. Regulatory T Cells Shape the Resident Memory T Cell Response to Virus Infection in the Tissues. J Immunol (2014) 192(2):683–90. doi: 10.4049/jimmunol.1202153
133. Solouki S, Huang W, Elmore J, Limper C, Huang F, August A. TCR Signal Strength and Antigen Affinity Regulate CD8(+) Memory T Cells. J Immunol (2020) 205(5):1217–27. doi: 10.4049/jimmunol.1901167
134. Fiege JK, Stone IA, Fay EJ, Markman MW, Wijeyesinghe S, Macchietto MG, et al. The Impact of TCR Signal Strength on Resident Memory T Cell Formation During Influenza Virus Infection. J Immunol (2019) 203(4):936–45. doi: 10.4049/jimmunol.1900093
135. Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, et al. Transcription Factor Irf4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-Like T Cells During Chronic Infection. Immunity (2017) 47(6):1129–41.e5. doi: 10.1016/j.immuni.2017.11.021
136. Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, et al. Graded Levels of IRF4 Regulate CD8+ T Cell Differentiation and Expansion, But Not Attrition, in Response to Acute Virus Infection. J Immunol (2014) 192(12):5881–93. doi: 10.4049/jimmunol.1303187
137. Nowyhed HN, Huynh TR, Thomas GD, Blatchley A, Hedrick CC. Cutting Edge: The Orphan Nuclear Receptor Nr4a1 Regulates CD8+ T Cell Expansion and Effector Function Through Direct Repression of Irf4. J Immunol (2015) 195(8):3515–9. doi: 10.4049/jimmunol.1403027
138. Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, et al. BACH2 Regulates CD8(+) T Cell Differentiation by Controlling Access of AP-1 Factors to Enhancers. Nat Immunol (2016) 17(7):851–60. doi: 10.1038/ni.3441
139. Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan Q, et al. Tcf1 and Lef1 Transcription Factors Establish CD8(+) T Cell Identity Through Intrinsic HDAC Activity. Nat Immunol (2016) 17(6):695–703. doi: 10.1038/ni.3456
140. Utzschneider DT, Delpoux A, Wieland D, Huang X, Lai CY, Hofmann M, et al. Active Maintenance of T Cell Memory in Acute and Chronic Viral Infection Depends on Continuous Expression of FOXO1. Cell Rep (2018) 22(13):3454–67. doi: 10.1016/j.celrep.2018.03.020
141. Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and Persistence of Memory CD8(+) T Cells Depend on T Cell Factor 1. Immunity (2010) 33(2):229–40. doi: 10.1016/j.immuni.2010.08.002
142. Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential Role of the Wnt Pathway Effector Tcf-1 for the Establishment of Functional CD8 T Cell Memory. Proc Natl Acad Sci USA (2010) 107(21):9777–82. doi: 10.1073/pnas.0914127107
143. Kim MV, Ouyang W, Liao W, Zhang MQ, Li MO. The Transcription Factor Foxo1 Controls Central-Memory CD8+ T Cell Responses to Infection. Immunity (2013) 39(2):286–97. doi: 10.1016/j.immuni.2013.07.013
144. Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. Transcription Factor Foxo1 Represses T-bet-mediated Effector Functions and Promotes Memory CD8(+) T Cell Differentiation. Immunity (2012) 36(3):374–87. doi: 10.1016/j.immuni.2012.01.015
145. Sidwell T, Liao Y, Garnham AL, Vasanthakumar A, Gloury R, Blume J, et al. Attenuation of TCR-induced Transcription by Bach2 Controls Regulatory T Cell Differentiation and Homeostasis. Nat Commun (2020) 11(1):252. doi: 10.1038/s41467-019-14112-2
146. Jennings E, Elliot TAE, Thawait N, Kanabar S, Yam-Puc JC, Ono M, et al. Nr4a1 and Nr4a3 Reporter Mice are Differentially Sensitive to T Cell Receptor Signal Strength and Duration. Cell Rep (2020) 33(5):108328. doi: 10.1016/j.celrep.2020.108328
147. Knudson KM, Hamilton SE, Daniels MA, Jameson SC, Teixeiro E. Cutting Edge: The Signals for the Generation of T Cell Memory are Qualitatively Different Depending on TCR Ligand Strength. J Immunol (2013) 191(12):5797–801. doi: 10.4049/jimmunol.1300905
148. Dunne A, O’Neill LA. The Interleukin-1 Receptor/Toll-Like Receptor Superfamily: Signal Transduction During Inflammation and Host Defense. Sci STKE (2003) 2003(171):re3. doi: 10.1126/stke.2003.171.re3
149. Liu T, Zhang L, Joo D, Sun SC. NF-KappaB Signaling in Inflammation. Signal Transduct Target Ther (2017) 2. doi: 10.1038/sigtrans.2017.23
150. Smith LK, Boukhaled GM, Condotta SA, Mazouz S, Guthmiller JJ, Vijay R, et al. Interleukin-10 Directly Inhibits CD8(+) T Cell Function by Enhancing N-Glycan Branching to Decrease Antigen Sensitivity. Immunity (2018) 48(2):299–312 e5. doi: 10.1016/j.immuni.2018.01.006
151. Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR Kinase Determines Effector Versus Memory CD8+ T Cell Fate by Regulating the Expression of Transcription Factors T-bet and Eomesodermin. Immunity (2010). doi: 10.1016/j.immuni.2009.10.010
152. Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, et al. PDK1 Regulation of mTOR and Hypoxia-Inducible Factor 1 Integrate Metabolism and Migration of CD8+ T Cells. J Exp Med (2012) 209(13):2441–53. doi: 10.1084/jem.20112607
153. Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting Edge: Generation of Effector Cells That Localize to Mucosal Tissues and Form Resident Memory CD8 T Cells Is Controlled by mTOR. J Immunol (2014) 193(5):2067–71. doi: 10.4049/jimmunol.1400074
154. McGill J, Van Rooijen N, Legge KL. Protective Influenza-Specific CD8 T Cell Responses Require Interactions With Dendritic Cells in the Lungs. J Exp Med (2008) 205(7):1635–46. doi: 10.1084/jem.20080314
155. Takamura S, Yagi H, Hakata Y, Motozono C, McMaster SR, Masumoto T, et al. Specific Niches for Lung-Resident Memory CD8+ T Cells At the Site of Tissue Regeneration Enable CD69-independent Maintenance. J Exp Med (2016) 213(13):3057–73. doi: 10.1084/jem.20160938
156. McMaster SR, Wein AN, Dunbar PR, Hayward SL, Cartwright EK, Denning TL, et al. Pulmonary Antigen Encounter Regulates the Establishment of Tissue-Resident CD8 Memory T Cells in the Lung Airways and Parenchyma. Mucosal Immunol (2018) 11(4):1071–8. doi: 10.1038/s41385-018-0003-x
157. Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, et al. Long-Lived Epithelial Immunity by Tissue-Resident Memory T (TRM) Cells in the Absence of Persisting Local Antigen Presentation. Proc Natl Acad Sci USA (2012) 109(18):7037–42. doi: 10.1073/pnas.1202288109
158. Lee YJ, Jameson SC, Hogquist KA. Alternative Memory in the CD8 T Cell Lineage. Trends Immunol (2011) 32(2):50–6. doi: 10.1016/j.it.2010.12.004
159. Khan TN, Mooster JL, Kilgore AM, Osborn JF, Nolz JC. Local Antigen in Nonlymphoid Tissue Promotes Resident Memory CD8+ T Cell Formation During Viral Infection. J Exp Med (2016) 213(6):951–66. doi: 10.1084/jem.20151855
160. Shin YS, Takeda K, Shiraishi Y, Jia Y, Wang M, Jackson L, et al. Inhibition of Pim1 Kinase Activation Attenuates Allergen-Induced Airway Hyperresponsiveness and Inflammation. Am J Respir Cell Mol Biol (2012) 46(4):488–97. doi: 10.1165/rcmb.2011-0190OC
161. Leignadier J, Hardy M-P, Cloutier M, Rooney J, Labrecque N. Memory T-lymphocyte Survival Does Not Require T-Cell Receptor Expression. Proc Natl Acad Sci USA (2008) 105(51):20440–5. doi: 10.1073/pnas.0806289106
Keywords: resident memory, inflammation, antigenic stimulation, tissue-derived signals, memory differentiation
Citation: Pritzl CJ, Daniels MA and Teixeiro E (2021) Interplay of Inflammatory, Antigen and Tissue-Derived Signals in the Development of Resident CD8 Memory T Cells. Front. Immunol. 12:636240. doi: 10.3389/fimmu.2021.636240
Received: 01 December 2020; Accepted: 29 April 2021;
Published: 21 June 2021.
Edited by:
Nick P. Goplen, Mayo Clinic, United StatesReviewed by:
Klaas Van Gisbergen, Sanquin Diagnostic Services, NetherlandsBrian S. Sheridan, Stony Brook University, United States
Copyright © 2021 Pritzl, Daniels and Teixeiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emma Teixeiro, dGVpeGVpcm9wZXJuYXNlQG1pc3NvdXJpLmVkdQ==
 Curtis J. Pritzl
Curtis J. Pritzl Mark A. Daniels
Mark A. Daniels Emma Teixeiro
Emma Teixeiro