- 1Department of Pharmacology of Traditional Chinese Medicine, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, China
- 2Department of Pathogen Biology, The Chinses Center for Disease Control and Prevention, Beijing, China
- 3Department of Pharmacy, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, China
- 4School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
- 5Section of Immunology, Guangdong Provincial Academy of Chinese Medical Sciences, Guangzhou, China
- 6Department of Dermatology, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
- 7Department of Pathology, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, China
N6-methyladenosine (m6A) modification, the addition of a methylation decoration at the position of N6 of adenosine, is one of the most prevalent modifications among the over 100 known chemical modifications of RNA. Numerous studies have recently characterized that RNA m6A modification functions as a critical post-transcriptional regulator of gene expression through modulating various aspects of RNA metabolism. In this review, we will illustrate the current perspectives on the biological process of m6A methylation. Then we will further summarize the vital modulatory effects of m6A modification on immunity, viral infection, and autoinflammatory disorders. Recent studies suggest that m6A decoration plays an important role in immunity, viral infection, and autoimmune diseases, thereby providing promising biomarkers and therapeutic targets for viral infection and autoimmune disorders.
Introduction
N6-methyladenosine (m6A) modification, one of the most prevalent RNA chemical modifications, refers to the methylation of adenosine A at the position of N6. Despite the fact that the m6A modification of RNA was identified in the 1970s (1), the activation mechanism and biological function of m6A modification remained unclear until recently. Thanks to the advances in high-throughput sequencing technology, two research groups independently and simultaneously used methyl-RNA-immunoprecipitation-sequencing (MeRIP-seq) or m6A-seq to exploit the distribution and function of m6A modification in human genome during 2012, leading to better global methods for studying the RNA m6A methylation (2, 3). From then on, the biological and pathological functions of m6A have been extensively studied. Here we will highlight recent advances in understanding the biological process of m6A modification. Then we will discuss the critical functions of m6A modification in innate/adaptive immune responses, immune system development, and viral infection. Finally, we will address the contributory effects of m6A methylation on various autoinflammation or autoimmune diseases.
The Dynamic and Reversible Process of m6A Modification
The modification of m6A is dynamic and reversible, because it can be catalyzed by methyltransferase (also known as the m6A “writers”) or removed by demethylase (also known as the m6A “erasers”). The recognition of m6A modification is mediated by several nuclear and cytoplasmic RNA binding proteins (RBPs), including YTH-domain containing family proteins, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) and heterogeneous nuclear ribonucleoproteins (HNRNPs) (Figure 1).

Figure 1. The dynamic methylation process of RNA m6A modification. The m6A decoration can be dynamically regulated by several m6A methyltransferases and demethylase.
m6A Writers
The decoration of m6A is mediated by a big methyltransferase complex consisting of Methyltransferase-like 3 (METTL3), METTL14, RNA binding motif protein 15 (RBM15), Wilms tumor 1 associated protein (WTAP), HAKAI (also known as Casitas B-lineage lymphoma-transforming sequence-like protein 1, CBLL1), zinc finger CCCH domain-containing protein 13 (ZC3H13), and Vir-like M6A Methyltransferase Associated (VIRMA, also known as KIAA1429) subunits. METTL3 is a predominant catalytic subunit of the writer complex, while METTL14 functions as an allosteric activator to enhance the catalytic activity of METTL3 (4). WTAP, a regulatory component of this complex, can bind to the target RNA and then recruit the METTL3 and METTL14 to form the catalytic core components (METTL3/METTL14/WTAP) that install the m6A methylation onto the target RNA (5). RBM15, which is an interacting partner of WTAP, serves as an adapter protein recruiting the m6A methyltransferase complex to U-rich regions (6). VIRMA, another important subunit of the writer complex, also plays an important role in guiding m6A modification in 3′ untranslated regions (3′ UTRs) and near stop codon (7). HAKAI acts synergistically with the other core components of the methyltransferase complex to form the m6A methylation (8). ZC3H13 exerts vital effects on anchoring and mediating nuclear m6A modification (9). This methylation writer complex usually installs the m6A modification on a specific and consensus RNA sequence, RRACH (R = G or A; H = U, A or C). Additionally, the m6A methylation is found to be enriched in 3′ UTRs, long exons, and near stop codons.
m6A Erasers
The m6A decoration can be removed by demethylase, such as fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5). FTO, which belongs to the non-heme Fe (II)- and α-KG-dependent dioxygenase AlkB family of proteins, has been identified as the first demethylase of m6A modification by Chuan He and his colleagues in 2011 (10). This study provided the first evidence that RNA m6A modification is reversible and dynamic, which is similar to the processes of methylation of DNA and histones. ALKBH5, which was identified to be another m6A demethylase in 2013, binds to specific m6A-modified single-stranded RNA, thereby catalyzing the demethylation of m6A (11).
m6A Readers
RNA-binding proteins, which can bind to m6A decoration and execute m6A-mediated biological functions, are referred to as m6A readers. According to their location, the m6A-binding proteins can be further divided into nuclear and cytoplasmic m6A readers: ① The nuclear m6A readers, including YTH domain containing 1-2 (YTHDC1-2), HNRNPA2B1 HNRNPC, HNRNPG, and Fragile X mental retardation protein (FMRP), play multiple roles in modulating mRNA splicing (12), epigenetic silencing (6), nuclear export of mRNA (13), regulation of non-coding RNA (14), and RNA structure switching (15); ② The cytoplasmic m6A readers, such as YTHDC2, YTH domain family 1-3 (YTHDF1-3), IGF2BPs, FMRP, and Proline rich coiled-coil 2 A (Prrc2a), exert regulatory effects on mRNA stability (16), mRNA translation (17), mRNA degradation (18–20), and phase separation (21, 22) (Figure 2).

Figure 2. The diverse biological functions of m6A methylation exerted by different m6A-binding proteins. The nuclear m6A modification regulates mRNA splicing, RNA structure switching, epigenetic silencing, non-coding RNA modulation and nuclear export of RNA, while the cytoplasmic m6A methylation modulates RNA stability, RNA translation, and phase separation.
The m6A-Mediated Biological Functions
Emerging evidence has shown that the decoration of m6A and m6A-binding proteins regulates various post-transcriptional aspects of the mRNA lifecycle (Figure 2).
The nuclear m6A readers can affect several nuclear processes of RNA (mRNA and non-coding RNA) metabolism. For example, Xu et al. found that METTL3-mediated m6A decoration contributes to spermatogenesis through regulating the alternative splicing of spermatogenesis-associated genes (23). In addition, Pan et al. and Chang et al. found that m6A modification alters the secondary structure of mRNA and long non-coding RNA, ultimately increasing the binding of RNA and RBPs (15, 23, 24). They named this m6A-mediated RNA structural remodeling that enhanced RNA–RBPs interactions as “m6A-switch” (15). During 2016, P. Patil et al. further demonstrated that m6A methylation promotes long non-coding RNA X-inactive specific transcript (XIST)-mediated epigenetic gene silencing by stabilizing the silencing proteins on XIST (6). Additionally, growing evidence indicates that m6A readers promote nuclear export of mRNA. Roundtree et al. found that m6A reader YTHDC1 facilitates the export of m6A-modified mRNAs through interaction with serine and arginine rich splicing factor 3 (SRSF3)—nuclear RNA export factor 1 (NXF1) protein complexes (13). FMRP, another m6A reader, can modulate nuclear export of m6A-decorated mRNAs via interacting with nuclear RNA export factors (NXF2) or chromosomal maintenance 1 (CRM1) (25, 26). Moreover, Sohail F. Tavazoie's group demonstrated that m6A decoration acts as an important post-transcriptional regulator for miRNA biogenesis. They found that DGCR8 can recognize and efficiently process METTL3-methylated pri-miRNA (27). Further evidence indicated that the reorganization of m6A mark by DGCR8 is mediated by HNRNPA2B1 (14). Collectively, these studies indicate that m6A modification is a critical regulatory factor for RNA splicing, structure remodeling, epigenetic silencing, nuclear export, and non-coding RNAs regulation in the nucleus.
On the other hand, the cytosolic m6A readers also modulate many cytoplasmic processes of RNA metabolism. One of the most established functions of m6A is degradation of RNA. Compelling evidence demonstrated that m6A-methylated mRNAs can be degraded through YTHDF2/YTHDF3-mediated degradative pathway (18–20). Mechanistically, YTHDF2 directly recruits the CCR4-NOT deadenylase complex to its target transcripts, thereby resulting in degradation of m6A-containing mRNA through CCR4/NOT-induced deadenylation. Additionally, m6A-methylated mRNAs can be also decayed by YTHDF2-Heat-responsive protein 12 (HRSP12)-RNase P/MRP-mediated endoribonucleolytic cleavage (20). Moreover, YTHDF3 can induce m6A-decorated mRNAs deadenylation and sequent decay by recruiting PAN2-PAN3 deadenylase complex (28). Nonetheless, accumulating evidence also indicate that several m6A-readers, including IGF2BPs, FMRP and Prrc2a, can conversely enhance stability of m6A-containg mRNA (29–31). These studies provide valuable insights into the complex role of m6A modification in modulating mRNA stability and degradation. Interestingly, mounting evidence also demonstrates that m6A modification plays a contributory role in translation through distinct cellular mechanisms. One regulatory mechanism is that YTHDF1 interacts with eIF3 and consequently recruits the ribosomes to m6A-modified mRNAs to promote their translation (32). Another modulatory mechanism is that m6A modification within the 5′ UTR of mRNAs enhances translation via directly interacting with eIF3 but not cap-binding factor eIF4E, especially upon heat shock stress (33, 34). In addition, cytoplasmic METTL3 can directly associate with eIF3 subunit h (eIF3h) independent of its catalytic activity and ultimately induce translation of various oncogenic mRNAs (35, 36). Furthermore, m6A modification contributes to circRNAs-regulated extensive translation through recruiting initiation factor eIF4G2 and YTHDF3 (37). Remarkably, three independent research group simultaneously revealed that m6A RNA methylation enhances phase separation. They showed that poly-methylated mRNA can function as a scaffold that recruits and juxtaposes several m6A-binding proteins, such as YTHDF1, YTHDF2, and YTHDF3, finally forming liquid droplets composed of m6A-modified mRNA–YTHDF complexes and resulting in phase separation (21, 22, 38). Together, these studies provide the important experimental evidence that m6A modification plays a key role in regulating multiple cytoplasmic biological processes, such mRNA stability or destabilization, mRNA translation, and phase separation.
The Key Role of m6A Modification in the Immune System
The Role of m6A Modification in Innate Immune System
The innate immunity is an arm of the immune system that comprises the cells and mechanisms that provide the first line of defense against infections in a non-specific manner. Innate immune cells, such as monocytes, macrophages, neutrophils, natural killer cells, can sense invading pathogens (such as virus and bacteria) and exogenous RNAs and subsequently respond to pathogens infections. Additionally, dendritic cells (DCs), which function as messengers between the innate and the adaptive immune systems, can process antigen materials from pathogens and present them on the cell surface to activate T cells. Accumulative evidence indicates that m6A modification and m6A-related proteins participate in innate immunity via regulating the recognitions of and responses to unmodified transcribed RNA (tRNA), invading pathogens, exogenous RNAs and aberrant endogenous double-stranded RNA (dsRNA). Additionally, m6A-methylation also plays a complex role in the DCs activation and functions.
Recognition of invading pathogens is the initial step of innate immunity. The detection of invading pathogens is dependent on a number of pattern-recognition receptors, including plasma membrane receptors (Toll-like receptors, TLRs) and cytosolic sensor (RIG-I-like receptors, RIG-I and NLR proteins). During 2005, Karikó et al. firstly demonstrated that nucleoside modifications, such as m5C, m5U, Ψ, s2U, and m6A modification, reduce or eliminate the TLR3, TLR7, or TLR8 activation in monocyte-derived DCs (MDDCs) (39). This finding represents the first indication of an interferential effect of m6A modification on the process of RNA recognition. As m6A modification is also widely distributed in viral mRNA, it's possible that viruses use m6A-modification to evade host immune recognition. In line with this speculation, Jianrong Li and Lee Gehrke demonstrated that m6A-methylated virion RNA, such as HMPV (human metapneumovirus) and HCV (hepatitis C virus), bound RIG-I with a very low affinity, finally disturbing the conformational change of RIG-I and reducing the sequent RIG-I-mediated innate immune responses in A549 cells and Huh7 cells (40, 41). These discoveries further proved that viruses can take advantage of m6A-modification as camouflage to escape from host immune system. Similar to linear RNAs, circRNAs, a class of loop non-coding RNAs, can also utilize m6A-modification to avoid immune detection. Endogenous circRNAs, in which introns transfer and deposit the m6A modifications onto the exons before their final back-splicing, can bypass host immune recognition. However, synthetic circRNAs, which lack m6A modifications, can be discriminated by RIG-I in Hela cells (42). Moreover, endogenous ssRNA (single-stranded RNA) with m6A modifications can prevent abnormal immune recognition and keep the innate immune response at steady state. Gao et al. demonstrated that endogenous ssRNA without m6A modifications in hematopoietic stem cells can be recognized by RIG-I as foreign dsRNA and subsequently trigger a deleterious innate immune response and hematopoietic failure (43). Taken together, these studies demonstrate that the m6A modification in tRNA, viral RNA, host circRNAs, and normal endogenous ssRNA can serve as a critical factor to allow innate immune system to avoid abnormal immune recognition (Figures 3A–D).
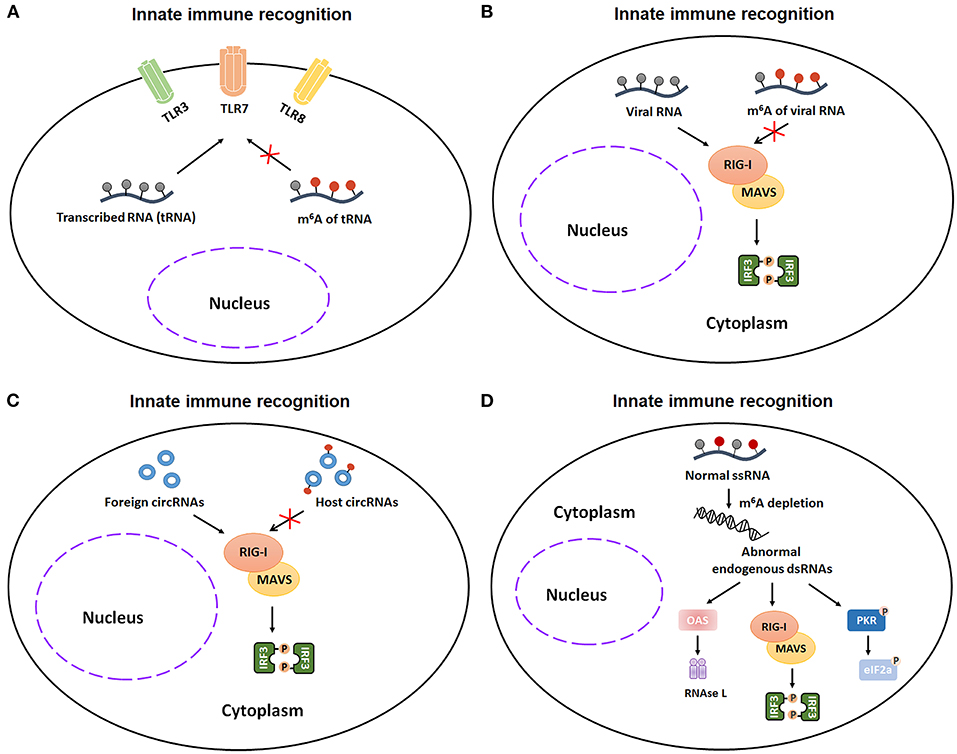
Figure 3. m6A modification participates in innate immunity via regulating the recognitions of unmodified transcribed RNA (tRNA), invading pathogens, exogenous RNAs, and aberrant endogenous double-stranded RNA (dsRNA). (A) m6A-modified tRNA reduces or eliminates the TLR3, TLR7, or TLR8 activation in monocyte-derived DCs (MDDCs). (B) m6A-methylation favors virion RNA escaping from the innate immunity recognitions in Huh7 cells and A549 cells. (C) Exogenous circRNAs, which lack m6A modifications, can be recognized by RIG-I signaling pathway in Hela cells. (D) Aberrant endogenous dsRNA in hematopoietic stem cells, which is formed by loss of m6A addition in ssRNA, can be recognized by RIG-I as foreign dsRNA and subsequently trigger a deleterious innate immune response. TLR3, Toll-like receptor 3; TLR7, Toll-like receptor 7; TLR8, Toll-like receptor 8; tRNA, transcribed RNA; RIG-I, retinoic acid-inducible gene I; MAVS, mitochondrial antiviral signaling protein; IRF3, interferon regulatory transcription factor 3; ssRNA, single-stranded RNA; dsRNA, double-stranded RNA; OAS, 2',5'-oligoadenylate synthetase gene; PKR, protein kinase R.
Once RNA recognition happens, innate immunity is immediately activated and releases multiple cytokines, such as type I interferons (IFNs) (44, 45). The role of m6A modifications in innate immune response is much more complex and controversial. Several researches illustrated that m6A modifications contributes to innate defense against viral infection (Figures 4A–C). The host innate responses will be suppressed by erasing the m6A modification from several antiviral transcripts. Mechanistically, DDX46, which forms a complex with ALKBH5, inhibits antiviral innate responses against VSV infection by reducing m6A modification-mediated nucleus export and translation of MAVS/TRAF3/TRAF6 (46). Consistently, impeding demethylation can amplify the innate immune responses. Wang et al. reported that hnRNPA2B1 enhances m6A-dependent nucleocytoplasmic trafficking and translation of STING, CGAS, and IFI16 mRNAs by preventing FTO-mediated demethylation, ultimately leading to amplifying the host innate immune response to HSV-1 infection (47). Moreover, m6A modifications-mediated cellular metabolic reprogram facilitates host immunity against viral infection. Mechanistically, the enzymatic activity of ALKBH5 is decreased upon viral infection, which consequently increases m6A modifications on the mRNA of α-ketoglutarate dehydrogenase (OGDH). The m6A modifications then downregulate the OGDH and its downstream metabolite itaconate, finally reprograming host cell metabolism and suppressing viral replication. Thus, in these studies, m6A modifications exerts contributory effects on antiviral responses through increasing export and translation of some key antiviral components and rewiring cellular metabolism. However, Noam Stern-Ginossar's and Ian Mohr's researches illustrated that m6A modifications plays an obstructive role in innate immunity response (44, 45) (Figure 4D). METTL3/METTL14-mediated m6A modification decreases the production and stability of IFNB mRNA, eventually promoting HCMV replication in vitro and in vivo. In contrast, demethylation by ALKBH5 promotes the production of IFNB mRNA, ultimately elevating type I interferon level. These results indicate that m6A modification acts as a negative modulator of type I interferon-mediated antiviral responses. Future studies are warranted to further exploit the precise role of m6A modification in innate immune responses.
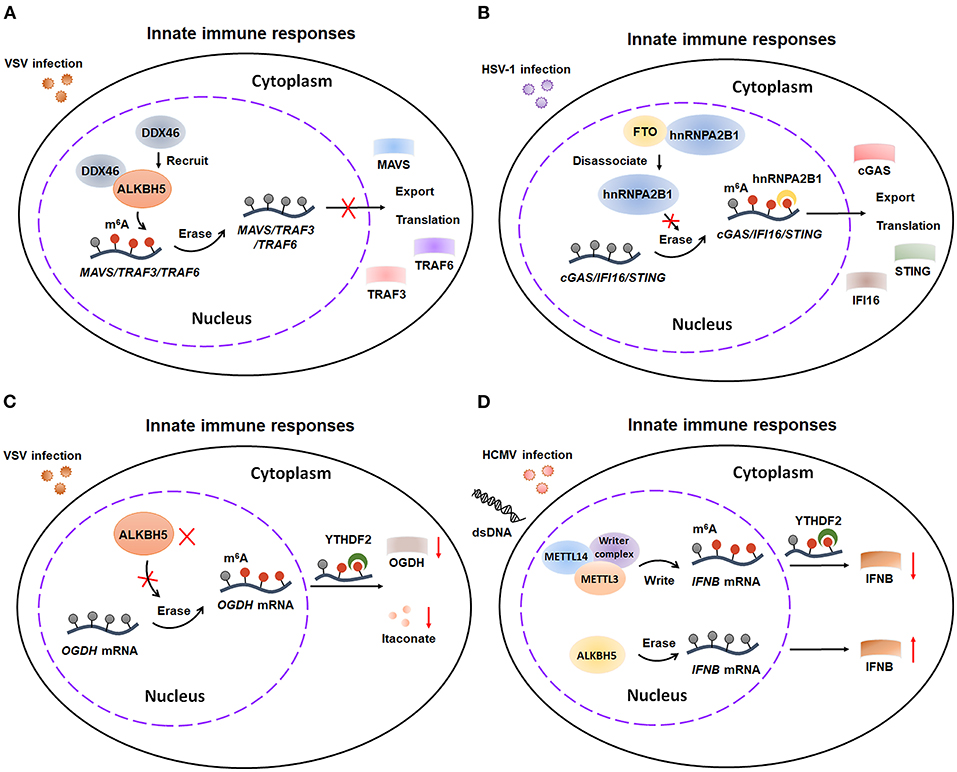
Figure 4. m6A modification is involved in antiviral responses of innate immunity. (A) DDX46, which forms a complex with ALKBH5, inhibits antiviral responses by reducing m6A modification-mediated nucleus export and translation of MAVS, TRAF3, and TRAF6 transcripts. (B) hnRNPA2B1 enhances antiviral responses by preventing FTO-mediated demethylation on STING, CGAS, and IFI16 transcripts. (C) During VSV infection, the enzymatic activity of ALKBH5 is decreased, which results in increasing m6A level. The upregulation of m6A modification leads to decreasing the expression of OGDH transcripts and reprograming host cell metabolism, which consequently enhancing antiviral responses. (D) m6A modification suppresses antiviral responses by repressing the production and stability of IFNB mRNA stability. VSV, vesicular stomatitis virus; TRAF3, TNF receptor-associated factor 3; TRAF6, TNF receptor-associated factor 6; HSV-1, herpes simplex virus-1; STING, stimulator of interferon response CGAMP interactor 1; CGAS, cyclic GMP-AMP synthase; IFI16, interferon gamma inducible protein 16; OGDH, α-ketoglutarate dehydrogenase; HCMV, Human cytomegalovirus; dsDNA, double stranded DNA; IFNB, IFN-beta.
Mounting evidence has shown that m6A methylation is critical for DCs activation and functions (Figure 5). On one hand, m6A methylation promotes DCs maturation. Huamin et al. showed that METTL3-mediated mRNA m6A methylation is essential for dendritic cell (DC) maturation and function (48). Mechanistically, METTL3 enhances the translation of CD40, CD80, and TLR4 signaling adaptor Tirap in DCs through an m6A catalytic activity-dependent manner, ultimately promoting DC maturation and strengthening TLR4/NF-κB signaling pathway signaling during DC activation and DC-mediated T cells–priming (Figure 5). On the other hand, m6A methylation may decrease the antigen-presentation capacity of DCs. Dali and colleagues demonstrated that the cross-presentation of tumor antigens and the cross-priming capacity of tumor-associated DCs are enhanced in Ythdf1−/− mice. Moreover, the CD8+ T cell-mediated tumor antigens-specific immune response is elevated in Ythdf1−/− mice owing to the enhancement of cross-presentation of DCs (49). Mechanistically, m6A-binding protein YTHDF1 can elevate the translation of lysosomal cathepsins in DCs, which consequently degrade the tumor antigens and suppress their antigen-presentation as well as CD8+ T cells-priming ability (Figure 5). Taken together, these studies indicated that m6A modification may play dual roles in enhancing or suppressing DCs activation and function in a context dependent way. Further studies are needed to expand our understanding and uncover the potential role of m6A methylation in DCs maturation, activation and function in future.
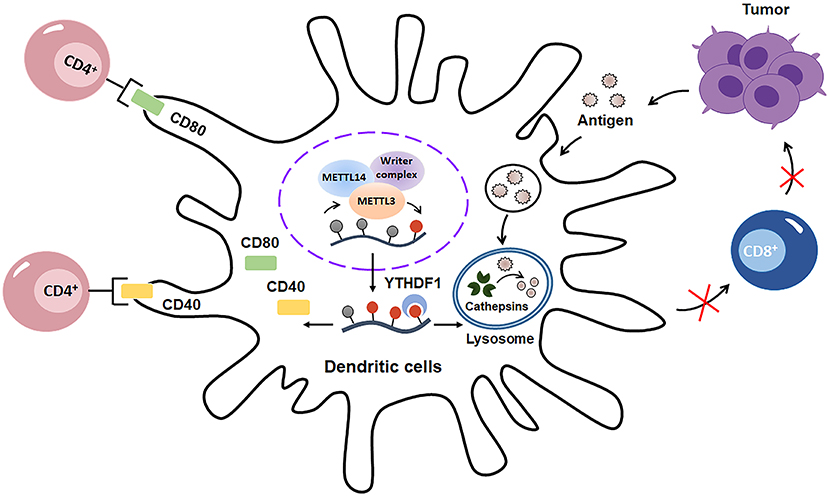
Figure 5. m6A modification has double-sided regulatory effects on dendritic cells (DCs). On one hand, m6A methylation promotes DCs maturation and activation through enhancing the translation of CD40, CD80. On the other hand, m6A modification decreases the antigen-presentation capacity of DCs by increasing the translation of cathepsins in lysosomes, which consequently decays the tumor antigens and suppress sequent CD8+-associated anti-tumor immunity.
The Role of m6A Methylation in Adaptive Immune System
The adaptive immunity is another arm of the immune system that specializes in the clearance of specific pathogens. It is mediated by the activation of antigen-specific T/B lymphocytes, ultimately establishing long-lasting immunological memory against the given antigen. Emerging evidence indicates that m6A exerts a vital effect on adaptive immunity (Figure 6).
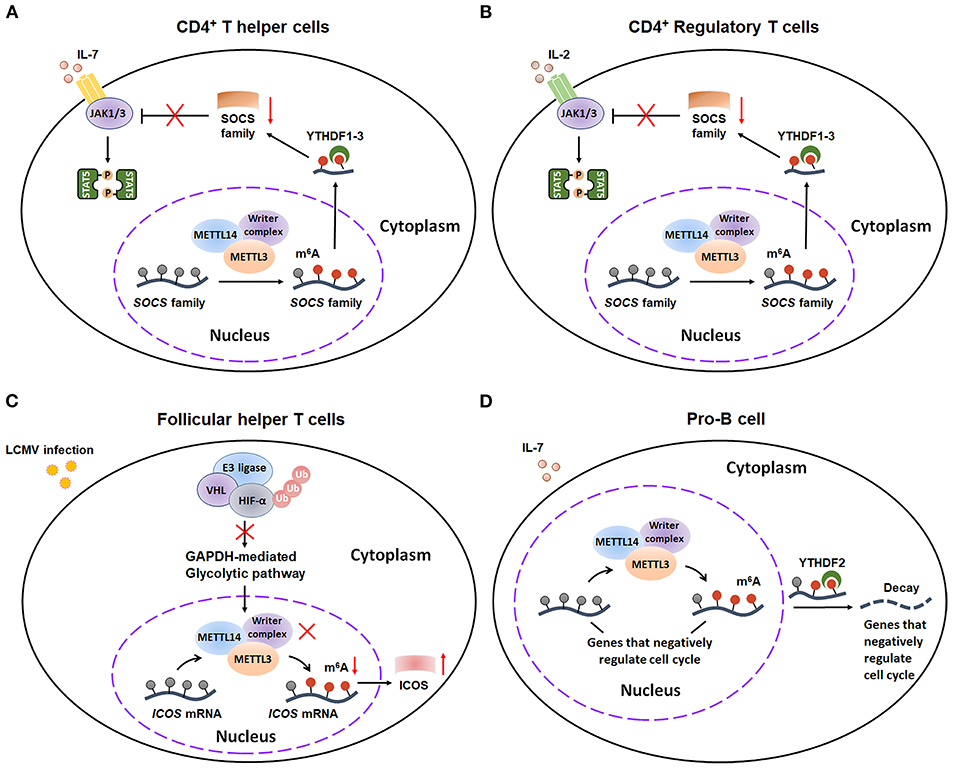
Figure 6. m6A methylation plays a critical role in adaptive immunity. (A) m6A modification can promote the CD4+ T helper cells proliferation and differentiation by decaying SOCS mRNAs and consequently activating IL-7-STAT5 pathway. (B) m6A methylation plays an essential role in sustaining the suppressive functions of CD4+ T Regulator cells by decaying SOCS mRNAs and consequently activating IL-2-STAT5 pathway. (C) E3 ligase VHL promotes the early initiation of Tfh cells development via suppressing HIF-1α-GAPDH–mediated glycolysis, ultimately enhancing ICOS expression by reducing m6A-labed ICOS mRNA. (D) METTL14-mediated m6A modification facilitates IL-7-induced pro-B cell proliferation by decreasing a group of transcripts that negatively regulate cell cycle. SOCS, suppressor of cytokine signaling; JAK, Janus Kinase; STAT5, Signal transducer and activator of transcription 5; VHL, Von Hippel–Lindau; HIF-1α, hypoxia-inducible factor 1α; ICOS, inducible T cell costimulatory; Ub, Ubiquitination.
m6A modification serves as a critical modulator for the differentiation and function of different subsets of T cells, including CD4+ T helper cells (Th1 and Th17 cells), CD4+ Regulatory T cells (Treg cells) and Follicular helper T cells (Tfh cells). By using CD4-Cre conditional Mettl3lox/lox mice, Richard Flavell's group found that m6A modification controls T cell homeostatic expansion, especially affecting the proliferation and differentiation of CD4+ Th1 and Th17 cells (50). Mechanistically, m6A modification has an inducible degradative effect on SOCS mRNAs, consequently promoting T cell homeostatic proliferation and differentiation via increasing IL-7-mediated STAT5 activation (Figure 6A). Additionally, Richard A Flavell, Hua-Bing Li, and coworkers gave another evidence that m6A mRNA methylation is also essential for sustaining the suppressive functions of Treg (51). Similarly, m6A decoration also plays an important role in degradation of SOCS mRNAs, thus activating IL-2-STAT5 signaling pathway to maintain the suppressive functions and cell stability of Treg cells (Figure 6B). On contrary, m6A mRNA methylation might exert an inhibitory effect on the Tfh cells differentiation (52). Recently, Yun-Cai Liu et al. illustrated that E3 ligase Von Hippel–Lindau (VHL) promotes the early initiation of Tfh cells development via suppressing hypoxia-inducible factor 1α (HIF-1α)—mediated glycolysis. Mechanistically, VHL deficiency results in activation of HIF-1α-GAPDH glycolytic pathway and consequently reduction in ICOS (inducible costimulator) expression by enhancing m6A modification on ICOS mRNA, ultimately leading to attenuated Tfh cell differentiation (Figure 6C). Taken together, these studies extend our understanding of crucial regulatory effects of m6A decoration on the differentiation of T cells.
Additionally, m6A also has essential effects on early B cell development. By using Mb1cre/+Mettl14fl/fl mice, Haochu Huang et al. found that METTL14 deficiency dramatically reduced the B cell numbers (53). Then the researchers further divided the CD19+B220midIgk/l population into pro-B cells, early/late large pre-B cells, and small pre-B cells. Further study indicated that loss of METTL14 led to inhibition of the pro-B cell proliferation and large-to-small-pre-B transition. Mechanistical study demonstrated that METTL14 promotes B cell development through two distinct ways: ① METTL14-mediated m6A modification that facilitates IL-7-triggered pro-B cell proliferation by decreasing a series of YTHDF2-bound m6A-labeled mRNA (Figure 6D); ② The large- to small-pre-B transition, which is independent of m6A-regconized proteins YTHDF1/2. Actually, this transition is largely dependent on the METTL14-mediated proper transcriptional activation of several critical transcription factors (TFs). Collectively, this finding represents the first indication that m6A modification participates in the development of B cells. However, future studies are needed to further explore the mechanism by which METTL14 regulates the transcriptional activation of these key TFs.
m6A Modification in Viral Infection
Despite the critical roles of m6A decoration in host anti-viral responses, emerging evidence also indicate that m6A modification might play a vital role in viral lifecycle and infection. In fact, the presence of m6A addition in several viruses, such as Influenza A virus (IAV) (54), B77 avian sarcoma virus (55), and Rous Sarcoma Virus (56), has been reported several decades ago. However, the potential regulatory effects of m6A modification on viral life cycle remains unclear until recently.
Human Immunodeficiency Virus 1 (HIV-1)
Human immunodeficiency virus 1 (HIV-1), a lentivirus belonging to the Retroviridae family, can cause acquired immunodeficiency syndrome (AIDS). Several studies tried to identify the potential role of m6A modification in HIV infection. Firstly, Lichinchi et al. found that the m6A levels of host and viral mRNAs are dramatically increased in MT4 T-cells during HIV-1 LAI strain infection. In addition, this m6A installation is critical for replication and nuclear export of HIV-1 (57). Depletion of METTL3/METTL14 inhibits the HIV replication by inducing a decrease in gp120 mRNA and CAp24 protein levels, whereas depletion of ALKBH5 results in enhanced HIV replication with a remarkable increase of the HIV-1 env gene. Moreover, researchers found that two m6A-labeled adenosines (A7883 and A7877) within the stem loop II region of HIV-1 Rev response element (RRE) RNA can promote the interaction between RRE and HIV-1 Rev protein, finally enhancing the nuclear export of HIV RNA. Later Kennedy et al. identified four to six m6A clusters in the 3′ UTR region of HIV-1 NL4.3 Genome in human CD4+ CEMSS T-cells by using photo-crosslinking-assisted m6A sequencing (PA-m6A-seq) techniques. Additionally, they demonstrated that the YTHDF1-3 proteins, especially YTHDF2, can bind to these m6A decoration and consequently increase the expression of HIV-1 genomic RNA (gRNA) and p24 capsid protein, ultimately enhancing virus proteins translation at 48 h post-infection (hpi) (58). However, there is contradictory evidence regarding whether viral m6A modification positively regulates HIV replication or not. In the contrast with Kennedy's results, Chuan He and Li Wu's group indicated that YTHDF1–3 proteins, which can bind to m6A-modified HIV-1 NL4.3 genome in HeLa/CD4+ T cells, suppress HIV-1 post-entry infection via degrading HIV-1 gRNA and inhibiting both early and late viral reverse transcription at 24 h post-infection (hpi) (59). Moreover, Chuan He and Li Wu's groups recently confirmed that YTHDF1-3 can bind to two m6A-modified GGACU motifs (positions 198 and positions 242) within 5′ UTR of HIV-1 NL4.3 gRNA and then form a complex with HIV-1 Gag and viral RNAs, resulting in an enhanced release of HIV-1 particles. However, these progeny HIV-1 viruses have lower infectivity (60). Collectively, these studies suggest that m6A modification may have a complex function on HIV life cycle in the host cells: ① Upon early entry, YTHDF1–3 proteins bind to m6A-modified HIV-1 NL4.3 genome and induce degradation of the HIV gRNA, finally impeding reverse transcription of HIV-1; ②Once integrated, m6A modification favors the Rev-mediated nuclear export of HIV RNA and YTHDF1–3 regulated translation of HIV-related protein; ③ Moreover, YTHDF1–3 can form a complex with HIV-1 Gag and viral RNAs, culminating in an enhancement of progeny HIV-1 particles release and an inhibition of progeny HIV-1 viruses infectivity (Figure 7A).
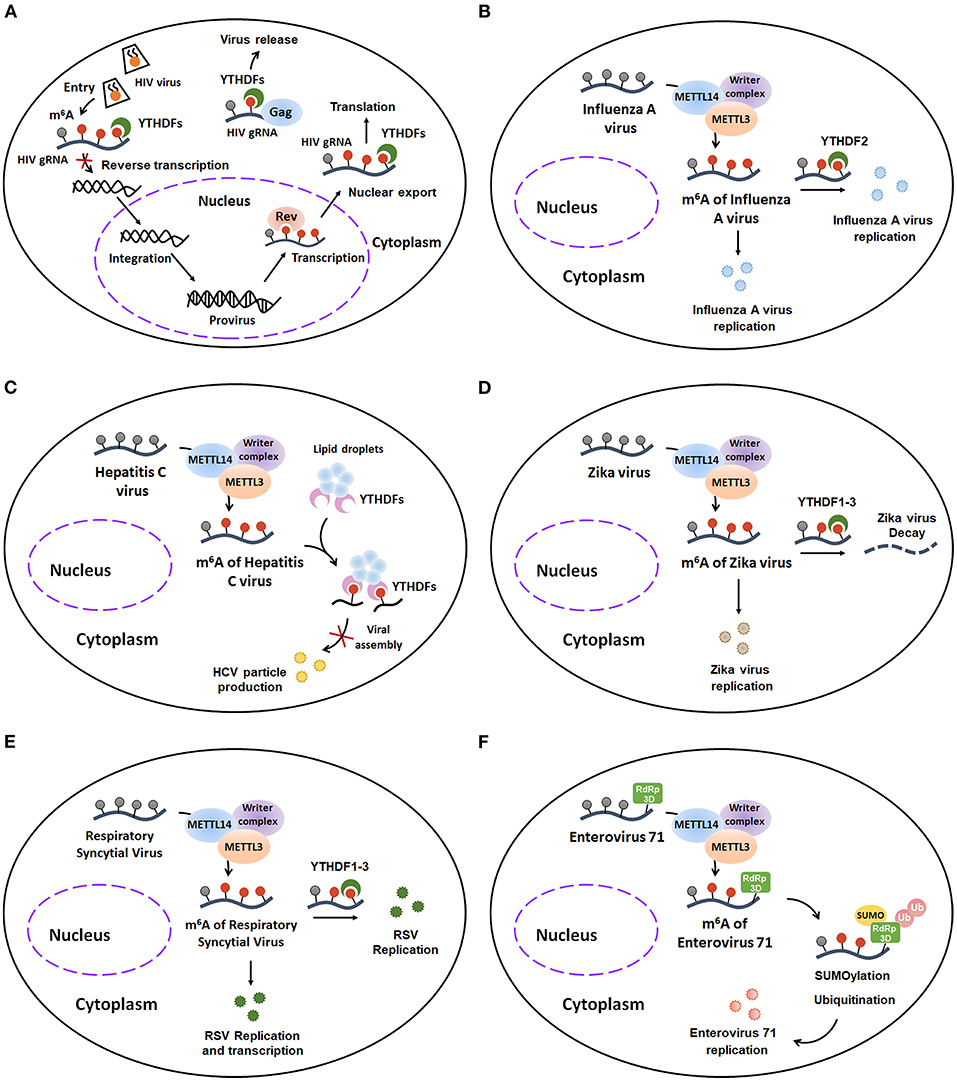
Figure 7. The host m6A machinery regulates viral RNA metabolism during viral infection. (A) m6A modification plays a complex role in HIV infection. Early upon entry, YTHDF1–3 proteins bind to m6A-modified HIV-1 NL4.3 Genome and induce degradation of the HIV gRNA, finally impeding reverse transcription of HIV-1. Once integrated, m6A modification favors the Rev-mediated nuclear export of HIV RNA and YTHDF1–3 regulated translation of HIV-related protein. Moreover, YTHDF1–3 can form a complex with HIV-1 Gag and viral RNAs, culminating in an enhancement of progeny HIV-1 particles release and an inhibition of progeny HIV-1 virus infectivity. (B) m6A addition contributes to Influenza A virus expression and replication. (C) m6A decoration negatively regulates HCV particle production by interrupting with viral assembly. (D) m6A installation promotes Zika virus replication. However, the m6A-labeled Zika virus RNA can be degraded when it binds to YTHDF1-3 proteins. (E) m6A methylation enhances the replication and transcription of Respiratory Syncytial Virus. (F) m6A modification facilitates Enterovirus type 71 replication by co-localizing with and further enhancing the SUMOylation and K63-linked ubiquitination of EV71 RNA-dependent RNA polymerase 3D (RdRp 3D) protein. HIV, Human immunodeficiency virus 1; HIV gRNA, HIV genomic RNA; HCV, Hepatitis C virus; RSV, Respiratory Syncytial Virus; RdRp 3D protein, RNA-dependent RNA polymerase 3D protein; SUMO, SUMOylation; Ub, Ubiquitination.
Influenza A Virus (IAV)
Influenza A virus (IAV), an enveloped negative-sense RNA virus belonging to the Orthomyxoviridae family, is one of the major etiologic agent of human respiratory tract infections and might result in severe infection and even death. Although previous studies have identified that IAV contains numerous m6A modification (61), the specific functions of m6A addition in IAV are poorly understood. Recently, Courtney et al. further demonstrated that m6A modifications preferentially deposit on the IAV plus (mRNA) and minus (vRNA) strands which encode viral structural proteins, including HA, M, NP and NA. Additionally, m6A modification is critical for IAV gene expression and replication (62). They found that the gene expression and viral titer of IAV are dramatically reduced in the METTL3 knockout A549 cells, whereas the expression and virion production of IAV are obviously increased in A549 cells with overexpression of YTHDF2. To further clarify the role of m6A modification in regulating IAV pathogenicity in vivo, the authors generated IAV mutants with m6A site mutations on plus strand or minus strand of the hemagglutinin (HA) segment. They found that these m6A-deficient IAV mutants attenuate their pathogenicity in mice. Taken together, these results indicate that the m6A modification is a positive regulatory factor for IAV expression and replication in vitro and in vivo (Figure 7B).
Hepatitis C Virus (HCV)
Hepatitis C virus, a small enveloped positive-sense single-stranded RNA virus of the Flaviviridae family, is the major cause of hepatitis C and hepatitis C-mediated liver cancer. Accumulating evidence indicates that the m6A modification plays a vital regulatory role in life cycle of HCV (Figure 7C). Gokhale et al. demonstrated that m6A addition, which is installed within HCV during its infection, can negatively regulate HCV particle production (63). The production of HCV RNA and viral titer were enhanced after METTL3 and METTL14 depletion. Conversely, depletion of FTO reduced the expression of HCV RNA and the viral titer of infectious HCV. Consistently, HCV m6A mutants within the viral E1 gene increased the HCV particle production. Mechanistically, the m6A-mediated decrease of HCV particle production was caused by the m6A-binding YTHDF proteins. During HCV infection, the cytosolic YTHDF proteins re-localize to lipid droplets and then directly interact with m6A-labeled HCV mRNA through DRAmCH motif. The interaction of YTHDF proteins and HCV mRNA leads to the abrogation of binding between Core protein and HCV RNA genomes, ultimately resulting in interrupting with viral assembly and suppressing HCV particle production. In summary, these studies indicate that m6A installation exerts a negative effect on HCV particle production.
Zika Virus
Zika virus (ZIKV), another enveloped positive-sense single-stranded RNA virus of the Flaviviridae family, typically causes fever, rash, headache, conjunctivitis in adults, and microcephaly in infants. Recently, the relevance of ZIKV to m6A modification is reported by Chuan He and Tariq M Rana's research groups (64). Using methylated RNA immunoprecipitation sequencing (MeRIP-seq), they discovered that there are 12 m6A peaks in the full-length of ZIKV RNA. Furthermore, they illustrated that the m6A modifications promote ZIKV replication. In addition, YTHDF family proteins, especially YTHDF2, bind directly to and further degrade ZIKV RNA. Interestingly, ZIKV infection also changes the m6A deposition and methylation motifs of host RNA. Taken together, unlike HCV, m6A decoration positively regulates Zika virus replication (Figure 7D).
Respiratory Syncytial Virus (RSV)
Respiratory Syncytial Virus (RSV), which belongs to non-segmented negative-sense (NNS) RNA viruses, is a leading cause of respiratory disease in infants, the elderly and immunocompromised individuals. To date, there is only one research article illustrating the relevance of RSV to m6A addition. Xue et al. demonstrated that m6A nucleosides are abundant in both RSV genome and antigenome. In addition, RSV can use the host m6A machinery to methylate itself. Interestingly, the m6A modification exerts promotive effects on RSV genome replication, gene expression and virus production (65). They found that the gene expression and viral titer of RSV are significantly decreased when m6A writer proteins, such as METTL3 and METTL14, are knocked down. In contrast, RSV replication is notably increased upon knockdown of m6A eraser proteins. Additionally, m6A reader proteins YTHDF1-3, especially YTHDF2, can directly bind to RSV RNAs and subsequently enhance replication and transcription of RSV. Furthermore, recombinant RSVs with abrogation of m6A sites in G gene and G mRNA are defective in replication in cultured human airway epithelial (HAE) cells and in respiratory tracts of cotton rats. Together, these findings reveal the pro-viral effects of m6A modification on RSV replication, transcription, and particle production, suggesting that targeting m6A modification may be a good way to develop antiviral drugs or vaccine for RSV (Figure 7E).
Enterovirus 71 (EV71)
Enterovirus type 71 (EV71), a non-enveloped single-stranded RNA virus belonging to the Picornaviridae family, is one of the main pathogens that causes hand-foot-and-mouth disease (HFMD) in infants and young children. The relevance between EV71 and m6A modification is poorly known. Recently, using UHPLC-MS/MS, Hao et al. confirmed that EV71 RNA contains m6A modification. In addition, the host m6A machinery can install the m6A modifications into the EV71 mRNA in the cytoplasm during EV71 infection. Furthermore, these m6A decorations are critical for EV71 replication. The viral titer and the viral RNA copy numbers of EV71 are significantly decreased in the METTL3-deficient Vero cells, whereas these inhibitory effects can be reserved when the demethylases FTO is knocked down. Additionally, the replication of EV71 is suppressed when the EV71 RNA contains m6A site mutations at 3,056 ~ 4,556 NT in C residues. Mechanistically, METTL3 facilitates EV71 replication by co-localizing with and further enhancing the SUMOylation and K63-linked ubiquitination of EV71 RNA-dependent RNA polymerase 3D (RdRp 3D) protein (66). Collectively, these studies suggest that m6A decoration contributes to EV71 replication (Figure 7F). Targeting m6A addition might be a good therapeutic measure to treat the EV71 infection.
m6A Modification in Autoimmune Diseases
Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE) is a typical multi system autoinflammatory disorder characterized by the presence of pathogenic autoantibodies and immune complex deposition in skin, joints, kidneys, and serosal membranes (67). Previous studies demonstrated that epigenetic changes, such as histone modification and DNA methylation, participate in the progression of SLE (68, 69). However, the role of RNA modification in SLE remained unclear until recently. Firstly, Li et al. mentioned a potential link between m6A modification and SLE in a comprehensive review (70). Then, Luo et al. found that the mRNA expression levels of METTL14?ALKBH5 and YTHDF2 were significantly decreased in peripheral blood mononuclear cells (PBMCs) of SLE patients (71, 72). Importantly, downregulation of ALKBH5 and YTHDF2 was found to be risk factors for SLE after multivariate logistic regression analysis. However, it still lacks direct mechanistical data to confirm the precise role of m6A methylation in the pathogenesis of SLE. Future studies are needed to further explore the potential effect of m6A decoration on SLE and underlying mechanisms.
Rheumatoid Arthritis
Rheumatoid arthritis (RA) is a systemic and disabling autoimmune disorder characterized by chronic and progressive joint erosion. The main pathological features of RA are synovial inflammation, cartilage injury, and bone erosion (73). Previous studies indicated that genetic and epigenetic factors contribute to the pathogenesis of RA. However, the exact cause of RA is poorly known. Using large scale Genome-Wide Association Studies (GWAS), Mo et al. identified 37 RA-associated m6A-SNPs in Asian or European populations (74). To figure out the underlying mechanisms of these RA-associated m6A-SNPs, researchers further investigated the associations of m6A-SNPs with the expression of local genes. Finally, they formed 23 SNP-Gene-RA trios after integrating the m6A-SNPs and local gene expression data. However, more experimental studies are required to further confirm whether these RA-associated m6A-SNPs are really involved in the pathogenesis of RA. Recently, Wang et al. reported that METTL3 is significantly elevated in the PBMCs of RA patients. Interestingly, the upregulation of METTL3 is positively associated with the level of CRP (C-reactive protein) and ESR (Erythrocyte sedimentation rate) in serum, indicating that the level of METTL3 in PBMCs could be used to predict the disease activity of RA. Mechanistically, upregulated METTL3 by LPS can trigger pTHP-1 macrophages activation and subsequent inflammatory response through NF-κB signaling pathway (75). This study illustrates that upregulation of METTL3 expression in macrophage may contribute to RA. However, it's still unknown whether METTL3-mediated m6A addition in macrophage is implicated in the pathogenesis of RA or not. Further studies are warranted to fully classify the potential role of m6A modification in RA.
Multiple Sclerosis
Multiple sclerosis (MS), a chronic immune-regulated disease of the central nervous system (CNS), is characterized by various CNS lesions, including demyelination, neurodegeneration, optic neuritis, and chronic axonal damage (76). Growing evidence suggests that genetic changes may result in MS. However, the key susceptibility genes of MS are largely unknown. Using GWAS and integrating m6AVar database, Xing-Bo and coworkers found that rs2288481 (p.Glu183Lys) in DKKL1 gene and rs923829 in METTL21B might be novel susceptibility loci for MS. Furthermore, these authors validated the association of rs923829~METTL21B and rs2288481~DKKL1 in the PBMCs of 40 unrelated Chinese Han individuals. They found that rs923829 is significantly associated with METTL21B expression levels, whereas the association between rs2288481 and DKKL1 is not statistically significantly (77). This research represents the first effort to illustrate the potential causal effects of m6A-related proteins on MS. However, we still lack the clear evidence that the dysfunctional m6A modification directly participates in the pathogenesis of MS. Future studies should firstly detect the level of m6A modification in MS and further elucidate its modulatory effect and underlying mechanism on MS.
Inflammatory Bowel Disease
Inflammatory bowel disease (IBD), including Crohn's disease and ulcerative colitis, is characterized by chronic relapsing intestinal inflammation. Although the etiology of IBD is ill-defined, the dysregulation of proinflammatory Th17 cells and anti-inflammatory Treg cells contributes to the pathogenesis of IBD (78, 79). Using CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice, Thomas X et al. firstly demonstrated that METTL14 deficiency in T cells causes the development of spontaneous colitis by dramatically increasing the release of Th17 cell-related proinflammatory cytokines (80). Given the important role of m6A modification in maintaining Treg cell stability and their suppressive functions, it is reasonable to find that deletion of METTL14 impairs induction of Treg cells and consequently results in the imbalance between Th17 and Treg cells, ultimately inducing spontaneous colitis development. This study provides novel insights into recognizing the pathogenesis of IBD. Further studies are warranted to prove whether targeting m6A addition is a good therapeutic approach to the treatment of IBD.
Potential Diagnostic and Therapeutic Application of m6A Modification
Since virus utilizes m6A methylation as a strategy to mark as “self” RNA and evade the detection by innate immunity system (41), targeting viral m6A decoration might be a good measure to detect viral infection and to enhance innate immune responses to viruses.
Given the stability of circRNAs, they can be developed as an effective strategy to deliver agents. However, as mentioned above, delivery of exogenous circRNAs might induce circRNA immunogenicity in vivo. Therefore, researchers should be very careful when they use exogenous circRNA as vehicles for gene delivery. On the other side, the synthetic circRNAs-induced immunogenicity can also be exploited to induce anti-tumor immunity (42).
The m6A modification does open a new door for anti-tumor immunity. Besides the synthetic circRNAs-mediated anti-tumor Immunity, targeting FTO also contributes to anti-tumor Immunity. Su et al. found that inhibition of FTO by genetic depletion or pharmacological antagonist can sensitizes acute myeloid leukemia (AML) cells to T cell cytotoxicity by suppressing Leukocyte Immunoglobulin Like Receptor B4 (LILRB4) (81). However, there is also conflicting evidence about the role of m6A methylation in anti-tumor immunity. As described above, m6A decoration represses the antigen-presentation capacity of DCs by promoting the expression of lysosomal proteases, finally reducing the antigen-specific anti-tumor immunity (49). Taken together, these studies indicate that m6A modification may be a double-edged sword in anti-tumor immune response. More studies will be needed to further illustrate the precise role and the underlying mechanism of m6A methylation in anti-cancer immunity.
The identification of the potential contributory role of m6A modification in several autoimmune diseases is of great significance to develop effective targeted therapeutics. However, the current knowledge of the effects of m6A methylation on autoinflammatory disorders is still in its infancy. A better understanding of the promotive effects of m6A decoration on the development of autoimmune disorders is warranted.
Conclusions and Future Perspectives
Our understanding of the biological and pathological functions of m6A modification has rapidly progressed due to the advances in the m6A-related high-throughput sequencing technology. These novel findings demonstrate that RNA m6A methylation can function as an important post-transcriptional regulator that modulates innate/adaptive immunity responses. In addition, aberrant expression of m6A modification may play a contributory role in the pathogenesis of autoimmune diseases.
As we know, the RNA m6A methylation is dynamic. It will be very interesting to investigate the real expression level of m6A modification in certain physiological or pathological contexts. Taking advantage of complete loss-of-function mutants of m6A writers, erasers and readers may illustrate how the m6A machinery proteins regulate immune responses as well as immune-related diseases. We can use high-throughput sequencing technologies to forecast the potential methylation sites (82). Nevertheless, the deposition specificity of m6A methylation is still poorly understood. Further studies are needed to investigate the mechanism underlying the modification specificity of m6A methylation.
Although FTO was firstly identified as the first RNA demethylase of m6A modification, emerging controversial studies report that FTO also plays an important role in m6Am and m1A demethylation. Recently, Wei et al. demonstrated that the subcellular location of FTO, which is different between distinct cells, determines the differential modulatory pattern of FTO demethylation (83). Nuclear FTO prefers to mediate internal m6A demethylation in polyadenylated mRNA, m6A in U6 RNA, internal and cap m6Am in small nuclear RNAs (snRNAs) and m1A demethylation in tRNA, whereas cytoplasmic FTO preferentially regulates internal m6A and cap m6Am demethylation in polyadenylated mRNA as well as m1A demethylation in tRNA. Collectively, these studies indicate that FTO likely mediates m6A, m6Am, and m1A demethylation through a spatial regulatory spectrum.
Pre-mRNA alternative splicing, a nuclear process including precise removement of introns and joining of exons, is critical for generating multiple transcript isoforms from one single gene. During alternative splicing, precise excision of introns can result in exon inclusion, whereas excision of introns and skipping of internal exons lead to exon skipping (Figure 2). Accumulative evidence has demonstrated that m6A modification acts as a key factor that regulates pre-mRNA exon inclusion or exon skipping splicing in the nucleus. For example, Xiao and coworkers illustrated that YTHDC1 selectively recruits pre-mRNA splicing factor SRSF3, which promotes exon inclusion, but blocks binding of exon-skipping SRSF10 to targeted mRNAs, ultimately resulting in alternative splicing of targeted transcripts (12). To illustrate how m6A addition regulates the pre-mRNA splicing, Yang et al. proposed a m6A-mediated alternative splicing model: (1) m6A decoration in exonic regions mainly results in exon inclusion; (2) m6A modification in intronic regions could lead to both either exon inclusion or exon skipping (84). Further studies are needed to confirm whether the different prevalence and distribution of m6A methylation in either exonic regions or intronic regions really determine the subsequent splicing mode.
Although several studies have indicated that HNRNPA2B1 can recognize the m6A modification and then execute m6A-associated biological functions, the specific binding pattern between HNRNPA2B1 and m6A decoration is poorly known. Recently, the structural and biochemical results have shown that hnRNP A2/B1 may function as an indirect m6A “reader.” Wu et al. reported that hnRNP A2/B1 does not specifically recognize m6A-labeled RNA (85). Neither full-length hnRNP A2/B1 nor tandem RRMs has a higher binding affinity to m6A-containing RNA than non-methylated RNA in vitro. Additionally, these researchers indicated that very few m6A sites exhibit proximal hnRNP A2/B1 binding in vivo. Taken together, these studies suggest that hnRNP A2/B1 may execute m6A-related biological functions indirectly.
Previously, the different m6A readers were studied separately to figure out their specific or distinct functions. A series of researches demonstrated that some m6A readers, which even belong to the same protein family, may exert opposite effects. For example, YTHDF1 is reported to enhance the translation of targeted RNA (86, 87). By contrast, YTHDF2 promotes the decay of m6A-labed RNA (88, 89). Recently, Sara Zaccara and Samie R. Jaffrey proposed a new unified model for YTHDFs mediated-m6A-depedent-functions (90). In contrast to the previous model that different DF paralogs recognize different m6A binding-sites, they demonstrated that all three DF paralogs can bind to the same m6A binding-sites. They found that all three YTHDFs proteins have the same m6A-binding structures, equivalent m6A decoration affinities, similar protein binding partners and identical intracellular localizations. Additionally, they presented conflicting evidence that all DF paralogs preferentially interact with RNA degradation machinery. The effect of m6A-mediated degradation is most pronounced when all three YTHDFs proteins are available. They also found that YTHDF1 and YTHDF3 do not regulate the m6A-mRNAs translation after reanalysis of the DF1-related ribosome profiling (32) and the DF3-associated PAR-CLIP dataset (91). Moreover, they indicated that all three DF paralogs can work together to suppress m6A-mediated differentiation. Further evidence is needed to confirm whether YTHDF proteins just lead to RNA degradation in the future.
Since YTHDC1 also contains low-complexity domains (6), it's possible that YTHDC1 also has the ability to form phase separation. Future studies are needed to explore the potential role of YTHDC1 in phase separation, which might uncover a novel biological function of YTHDC1 in promoting phase separation. Additionally, Gokhale et al. observed that the cytosolic YTHDF proteins, which re-localize to lipid droplets during HCV infection, can bind to and consequently suppress the HCV particle production (63). These studies suggest that YTHDFs-mediated phase separation may play an important role in HCV infection.
m6A modification is a double-edged sword for host immunity and virus infection. On one side, virus can take advantage of m6A methylation to escape the recognition by innate immune cells. Additionally, several viruses, such as IAV and RSV, can utilize m6A modification for further replication in the host cell. On the other side, m6A modifications contributes to host innate defense against viral infection. It's will be very interesting to figure out the precise m6A-mediated crosstalk between virus and host cells. Moreover, specifically targeting viral m6A addition and promoting host m6A-mediated antiviral type I IFN response might be promising for the treatment of viral infection.
Interestingly, Liu et al. recently identified that SARS-CoV-2 (Severe Acute Respiratory Syndrome coronavirus 2), an enveloped RNA coronavirus causing a global health emergency, is gradually m6A methylated in 3′ end of the viral genome during its infection in the host cells. However, these m6A modifications negatively modulate SARS-CoV-2 life cycle. The viral replication is dramatically increased upon knockdown of METTL3 and METTL14 (92). This study suggests that m6A decoration may be a negative regulator for SARS-CoV-2 infection, providing a potential therapeutic avenue for the development of vaccine and antiviral drugs of SARS-CoV-2.
Given that m6A methylation plays a critical role in viral infection and autoinflammatory disorders, the identification of drugs targeting m6A modification is attractive. Using 3D proteome-wide scale screening, Malacrida et al. recently illustrated that the imidazobenzoxazin-5-thione (MV1035), a newly found sodium channel blocker, is an efficient ALKBH5 inhibitor in vitro (93). In parallel, Su et al. identified two compounds (CS1 and CS2) as FTO antagonist by docking and screening 260,000 compounds. The newly identified m6A modification regulators do open novel therapeutic avenues for understanding and treating m6A methylation-associated diseases. However, further studies are needed to confirm their efficacy and specificity in vivo.
Author Contributions
LT, XW, and TL wrote the manuscript. XW, TL, and YC searched the literature. ZD, CL, and GZ edited the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Innovative and Enhancement Research Program of Guangdong Province (No. 2019KTSCX025); the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (YN2018QJ03); Medical Scientific Research Foundation of Guangdong Province (No. A2017277); and the Specific Fund of State Key Laboratory of Dampness Syndrome of Chinese Medicine (SZ2020ZZ27).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
2. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–6. doi: 10.1038/nature11112
3. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
4. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
5. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
6. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
7. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. (2018) 4:10. doi: 10.1038/s41421-018-0019-0
8. Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. (2017) 215:157–72. doi: 10.1111/nph.14586
9. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. (2018) 69:1028–38 e6. doi: 10.1016/j.molcel.2018.02.015
10. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. (2011) 7:885–7. doi: 10.1038/nchembio.687
11. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
12. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell. (2016) 61:507–19. doi: 10.1016/j.molcel.2016.01.012
13. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. (2017) 6:e31311. doi: 10.7554/eLife.31311
14. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. (2015) 162:1299–308. doi: 10.1016/j.cell.2015.08.011
15. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. (2015) 518:560–4. doi: 10.1038/nature14234
16. Zhou B, Liu C, Xu L, Yuan Y, Zhao J, Zhao W, et al. N6-methyladenosine reader protein Ythdc2 suppresses liver steatosis via regulation of mRNA Stability of lipogenic genes. Hepatology. (2020). doi: 10.1002/hep.31220. [Epub ahead of print].
17. Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B, et al. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun. (2019) 10:5332. doi: 10.1038/s41467-019-13317-9
18. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. (2016) 7:12626. doi: 10.1038/ncomms12626
19. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. (2014) 505:117–20. doi: 10.1038/nature12730
20. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell. (2019) 74:494–507 e8. doi: 10.1016/j.molcel.2019.02.034
21. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. m6A enhances the phase separation potential of mRNA. Nature. (2019) 571:424–8. doi: 10.1038/s41586-019-1374-1
22. Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, et al. Multivalent m6A motifs promote phase separation of YTHDF proteins. Cell Res. (2019) 29:767–9. doi: 10.1038/s41422-019-0210-3
23. Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, et al. N6-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol. (2016) 428:822–33. doi: 10.1016/j.jmb.2015.08.021
24. Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. (2015) 519:486–90. doi: 10.1038/nature14263
25. Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. (2019) 28:845–54 e5. doi: 10.1016/j.celrep.2019.06.072
26. Hsu PJ, Shi H, Zhu AC, Lu Z, Miller N, Edens BM, et al. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J Biol Chem. (2019) 294:19889–95. doi: 10.1074/jbc.AC119.010078
27. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. (2015) 519:482–5. doi: 10.1038/nature14281
28. Liu J, Gao M, Xu S, Chen Y, Wu K, Liu H, et al. YTHDF2/3 are required for somatic reprogramming through different RNA deadenylation pathways. Cell Rep. (2020) 32:108120. doi: 10.1016/j.celrep.2020.108120
29. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
30. Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. (2018) 27:3936–50. doi: 10.1093/hmg/ddy292
31. Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. (2019) 29:23–41. doi: 10.1038/s41422-018-0113-8
32. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
33. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. (2015) 526:591–4. doi: 10.1038/nature15377
34. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR m6A promotes cap-independent translation. Cell. (2015) 163:999–1010. doi: 10.1016/j.cell.2015.10.012
35. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. (2018) 561:556–60. doi: 10.1038/s41586-018-0538-8
36. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. (2016) 62:335–45. doi: 10.1016/j.molcel.2016.03.021
37. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
38. Fu Y, Zhuang X. m6A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol. (2020) 16:955–63. doi: 10.1038/s41589-020-0524-y
39. Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
40. Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. (2016) 7:e00833–16. doi: 10.1128/mBio.00833-16
41. Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol. (2020) 5:584–98. doi: 10.1038/s41564-019-0653-9
42. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. (2019) 76:96–109 e9. doi: 10.1016/j.molcel.2019.07.016
43. Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, et al. m6A modification prevents formation of endogenous double-stranded RNAs and deleterious innate immune responses during hematopoietic development. Immunity. (2020) 52:1007–21 e8. doi: 10.1016/j.immuni.2020.05.003
44. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol. (2019) 20:173–82. doi: 10.1038/s41590-018-0275-z
45. Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. RNA m6A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. (2018) 32:1472–84. doi: 10.1101/gad.319475.118
46. Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat Immunol. (2017) 18:1094–103. doi: 10.1038/ni.3830
47. Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. (2019) 365:eaav0758. doi: 10.1126/science.aav0758
48. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat Commun. (2019) 10:1898. doi: 10.1038/s41467-019-09903-6
49. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. (2019) 566:270–4. doi: 10.1038/s41586-019-0916-x
50. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. (2017) 548:338–42. doi: 10.1038/nature23450
51. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. (2018) 28:253–6. doi: 10.1038/cr.2018.7
52. Zhu Y, Zhao Y, Zou L, Zhang D, Aki D, Liu YC. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J Exp Med. (2019) 216:1664–81. doi: 10.1084/jem.20190337
53. Zheng Z, Zhang L, Cui XL, Yu X, Hsu PJ, Lyu R, et al. Control of early B cell development by the RNA N6-Methyladenosine methylation. Cell Rep. (2020) 31:107819. doi: 10.1016/j.celrep.2020.107819
54. Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J Virol. (1976) 20:45–53. doi: 10.1128/JVI.20.1.45-53.1976
55. Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. (1977) 16:471–8. doi: 10.1021/bi00622a021
56. Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. (1985) 5:2298–306. doi: 10.1128/MCB.5.9.2298
57. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, et al. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. (2016) 1:16011. doi: 10.1038/nmicrobiol.2016.11
58. Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, et al. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. (2016) 19:675–85. doi: 10.1016/j.chom.2016.04.002
59. Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife. (2016) 5:e15528. doi: 10.7554/eLife.15528
60. Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M, et al. N6-Methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J Biol Chem. (2018) 293:12992–3005. doi: 10.1074/jbc.RA118.004215
61. Narayan P, Ayers DF, Rottman FM, Maroney PA, Nilsen TW. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol Cell Biol. (1987) 7:1572–5. doi: 10.1128/MCB.7.4.1572
62. Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, et al. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe. (2017) 22:377–86 e5. doi: 10.1016/j.chom.2017.08.004
63. Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. (2016) 20:654–65. doi: 10.1016/j.chom.2016.09.015
64. Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. (2016) 20:666–73. doi: 10.1016/j.chom.2016.10.002
65. Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, et al. Viral N6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun. (2019) 10:4595. doi: 10.1038/s41467-019-12504-y
66. Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. (2019) 47:362–74. doi: 10.1093/nar/gky1007
67. Tsokos GC. Systemic lupus erythematosus. N Engl J Med. (2011) 365:2110–21. doi: 10.1056/NEJMra1100359
68. Karagianni P, Tzioufas AG. Epigenetic perspectives on systemic autoimmune disease. J Autoimmun. (2019) 104:102315. doi: 10.1016/j.jaut.2019.102315
69. Mazzone R, Zwergel C, Artico M, Taurone S, Ralli M, Greco A, et al. The emerging role of epigenetics in human autoimmune disorders. Clin Epigenetics. (2019) 11:34. doi: 10.1186/s13148-019-0632-2
70. Li LJ, Fan YG, Leng RX, Pan HF, Ye DQ. Potential link between m6A modification and systemic lupus erythematosus. Mol Immunol. (2018) 93:55–63. doi: 10.1016/j.molimm.2017.11.009
71. Luo Q, Fu B, Zhang L, Guo Y, Huang Z, Li J. Decreased peripheral blood ALKBH5 correlates with markers of autoimmune response in systemic lupus erythematosus. Dis Markers. (2020) 2020:8193895. doi: 10.1155/2020/8193895
72. Luo Q, Rao J, Zhang L, Fu B, Guo Y, Huang Z, et al. The study of METTL14, ALKBH5, and YTHDF2 in peripheral blood mononuclear cells from systemic lupus erythematosus. Mol Genet Genomic Med. (2020) 8:e1298. doi: 10.1002/mgg3.1298
73. Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. (2016) 388:2023–38. doi: 10.1016/S0140-6736(16)30173-8
74. Mo XB, Zhang YH, Lei SF. Genome-wide identification of N6-methyladenosine m6A SNPs associated with rheumatoid arthritis. Front Genet. (2018) 9:299. doi: 10.3389/fgene.2018.00299
75. Wang J, Yan S, Lu H, Wang S, Xu D. METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-kappaB signaling pathway. Mediators Inflamm. (2019) 2019:3120391. doi: 10.1155/2019/3120391
76. Dobson R, Giovannoni G. Multiple sclerosis - a review. Eur J Neurol. (2019) 26:27–40. doi: 10.1111/ene.13819
77. Mo XB, Lei SF, Qian QY, Guo YF, Zhang YH, Zhang H. Integrative analysis revealed potential causal genetic and epigenetic factors for multiple sclerosis. J Neurol. (2019) 266:2699–709. doi: 10.1007/s00415-019-09476-w
78. Guan Q. A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res. (2019) 2019:7247238. doi: 10.1155/2019/7247238
79. Yan JB, Luo MM, Chen ZY, He BH. The function and role of the Th17/Treg cell balance in inflammatory bowel disease. J Immunol Res. (2020) 2020:8813558. doi: 10.1155/2020/8813558
80. Lu TX, Zheng Z, Zhang L, Sun HL, Bissonnette M, Huang H, et al. A new model of spontaneous colitis in mice induced by deletion of an RNA m6A methyltransferase component METTL14 in T cells. Cell Mol Gastroenterol Hepatol. (2020) 10:747–61. doi: 10.1016/j.jcmgh.2020.07.001
81. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. (2020) 38:79–96 e11. doi: 10.1016/j.ccell.2020.04.017
82. Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, et al. Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell. (2019) 178:731–47 e16. doi: 10.1016/j.cell.2019.06.013
83. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. (2018) 71:973–85 e5. doi: 10.1016/j.molcel.2018.08.011
84. Yang Y, Hsu PJ, Chen Y-S, Yang Y-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. (2018) 28:616–24. doi: 10.1038/s41422-018-0040-8
85. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. (2018) 9:420. doi: 10.1038/s41467-017-02770-z
86. Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. (2018) 563:249–53. doi: 10.1038/s41586-018-0666-1
87. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. (2020) 48:3816–31. doi: 10.1093/nar/gkaa048
88. Huang T, Liu Z, Zheng Y, Feng T, Gao Q, Zeng W. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m6A/mRNA pathway. Cell Death Dis. (2020) 11:37. doi: 10.1038/s41419-020-2235-4
89. Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. (2017) 549:273–6. doi: 10.1038/nature23883
90. Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell. (2020) 181:1582–95 e18. doi: 10.1016/j.cell.2020.05.012
91. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
92. Liu J, Xu YP, Li K, Ye Q, Zhou HY, Sun H, et al. The m6A methylome of SARS-CoV-2 in host cells. Cell Res. (2021). doi: 10.1038/s41422-020-00465-7. [Epub ahead of print].
Keywords: N6-methyladenosine (m6A), innate immunity, adaptive immunity, autoimmune disease, viral infection
Citation: Tang L, Wei X, Li T, Chen Y, Dai Z, Lu C and Zheng G (2021) Emerging Perspectives of RNA N6-methyladenosine (m6A) Modification on Immunity and Autoimmune Diseases. Front. Immunol. 12:630358. doi: 10.3389/fimmu.2021.630358
Received: 17 November 2020; Accepted: 16 February 2021;
Published: 05 March 2021.
Edited by:
Menno de Winther, Amsterdam University Medical Center, NetherlandsReviewed by:
Eva Bartok, University Hospital Bonn, GermanyStephanie Halene, School of Medicine Yale University, United States
Copyright © 2021 Tang, Wei, Li, Chen, Dai, Lu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lipeng Tang, dGxwMDUwOUAxNjMuY29t; Zhenhua Dai, emRhaTIwMDlAb3V0bG9vay5jb20=; Chuanjian Lu, bGNqQGd6dWNtLmVkdS5jbg==; Guangjuan Zheng, emhlbmdndWFuZ2p1YW5AZ3p1Y20uZWR1LmNu
†These authors have contributed equally to this work
 Lipeng Tang
Lipeng Tang Xingyan Wei
Xingyan Wei Tong Li1,3†
Tong Li1,3† Guangjuan Zheng
Guangjuan Zheng