- 1Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College, Kunming, China
- 2Yunnan Key Laboratory of Vaccine Research & Development on Severe Infectious Diseases, Kunming, China
- 3State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, Ningbo, China
Dengue virus is a significant public health threat worldwide; however, the pathogenesis of dengue disease remains poorly understood due to lack of appropriate small animal models. Tree shrews are an emerging experimental animal model for the study of human diseases due to their resemblance of genetic characteristics to primate animals. Herein we report that dengue infection in tree shrews elicits resemble clinical symptoms as in humans. Dengue fever (△2°C> normal body temperature) developed in ~22% healthy Chinese tree shrews from 2 through 33 days after infection with a low dose (1 ∗ 104 PFU/animal) of dengue virus serotype 2 or 3 intravenously or subcutaneously. The dengue genomic RNA and neutralizing antibodies were detected in ~78% of animals at days 7 and 15 post infection respectively. The serum levels of liver enzymes including aspartate transaminase, alanine aminotransferase and alkaline phosphatase were elevated with peaks at day 7 after infection. Modest thrombocytopenia and a slight decrease in the white blood cell count were observed. Intriguingly, although viral RNA was barely detectable in the liver by 48 days after infection, it was still evident in the brain. The intra-brain bleeding lesions in the intravenous infection group were more severe than those in the subcutaneous infection group. Our data demonstrate that primary dengue virus infection in tree shrews causes resemble clinical disease as in humans and thus tree shrews may be a suitable model for the study of dengue disease pathogenesis.
Introduction
Dengue infection is one of the major public burdens in the tropical and subtropical areas worldwide, particularly in south Asia, Africa, central and south America. Approximately 3.6 billion people in these areas are at risk for epidemic transmission. Approximately 400 million people in more than 100 countries are infected with dengue each year (1). This significant public health threat is no longer confined to the traditional districts, and autochthonous dengue transmission has now been recorded in several European countries (2). In 2014, Japan reported its first outbreak of dengue in 70 years (3).
Despite the public health significance of dengue disease, there are still no highly effective vaccines and antiviral drugs. A major obstacle to the development of vaccines and therapeutics is the lack of validated animal models that recapitulate human infection. Primates are the only natural vertebrate hosts for DENVs. DENV infection in non-human primates (NHPs) results in acute viremia and a strong neutralizing antibody response; however, no overt clinical symptoms develop, such as hemorrhage, fever and shock (4, 5). Even many recent efforts have been made to improve the NHP models (6), the majority of the studies have not shown promising clinical disease (7). Moreover, the ethical concerns and high cost significantly make it unfeasible to apply NHPs to widespread DENV research.
Chinese tree shrews are mainly distributed across southwestern China (including the Yunnan, Sichuan, Hainan, Guizhou, and Guangxi provinces) and have been classified as Tupaia belangeri. Phylogenetic analysis of the Chinese tree shrew supports its close relationship to primates, and this animal is more resemble to humans than murine species, such as mice, marmots, and rats, in terms of its genomics (8). Moreover, Chinese tree shrews have other advantages, such as low cost, availability and easy maintenance. Thus, the Chinese tree shrew may be a promising animal model for human diseases. The tree shrew model is more frequently used in biomedical research as a replacement for primates in many human disease research, such as HCV, HBV, HEV, HSV, depressive disorder, thrombus and glioblastoma (9–11). The tree shrew T cells are genetically closely related to those of humans. A member of the flaviviruses, Zika virus, has been recently shown to induce clinical symptoms in tree shrews (12, 13), suggesting that tree shrews may be a good model for dengue virus research.
In this study, we explore the feasibility of applying tree shrews to dengue research. We observe clinical symptoms including fever, viral infection, high antibody titers and pathology in the central nervous system after primary infection. These symptoms largely recapitulate dengue infection in humans.
Materials and Methods
Phylogenetic and Identity Analysis of STING Gene Among Tree Shrew, Homo and Rattus Species
The reference STING (stimulator of interferon genes), TLR3 (Toll-like receptors) and MDA5 (melanoma differentiation-associated protein) genes sequences that were used to construct the distinct phylogenetic branches and identity analysis were collected from the GenBank sequence database of National Center for Biotechnology Information under the following accession numbers: Homo sapiens (MF622062.1, DQ360815.1 and AF095844.1), Rattus norvegicus (NM_001109122.1, NM_001109199.1 and XM_008771267.2), T. belangeri (KU998263.1, XM_006160265.3 and XM_006160465.2), Macaca mulatta (MF622060.1, NM_001036685.1 and DQ875603.1), Sus scrofa (MF358967.1, KT735340.1 and FJ455509.1) and Mus musculus (KR154221.1, NM_001357317.1 and NM_001164477.1). STING gene of tree shrew in this study was amplified from spleen tissue, followed by sequencing.
The phylogenetic analyses of STING and MDA5 gene were performed using the MEGA version 7 software and the phylogenetic trees were assembled using the Neighbor-Joining method of the Maximum Composite Likelihood model (14). The branching pattern was statistically evaluated by bootstrap analysis of 1,000 replicates. The identity analyses among tree shrew, Homo and Rattus species were performed using the Bioedit software.
Culture of Primary Tupaia Bone Marrow Mesenchymal Stem Cells and DENV Infection
DENV-2 (KR870423.1) and DENV-3 (KY911981.1) were isolated from Guangzhou City, Guangdong Province, China and propagated in Aedes albopictus mosquito (C6/36, at 16th generations). The cell line was maintained in RPMI-1640 medium (1640, BI) supplemented with 10% fetal bovine serum (FBS, SJQ), 100 μg/ml streptomycin and 100 U/ml penicillin at 28°C in 5% CO2. The bone marrow mesenchymal stem cells (BM-MSCs) isolated and cultured from femur and tibia of tree shrew were as previously described (15). MSC plated in 24-well plates at 5 × 105 cells per ml were cultivated in DMEM medium and incubated 24 h at 37°C in 5% CO2 in a humidified incubator. Twenty-four hours later, the DENV-3 was added in BM-MSCs at a multiplicity of infection (MOI) of 0.1. Cells were infected in triplicate and collected at 0, 12, 24, 36, 48, 60, 72, 84 and 96 h post infection. Vero cells were used as positive controls. The culture supernatants were collected for DENV RNA load determination using qRT-PCR. qRT-PCR was performed using SYBR Premix Ex Taq II (TaKaRa Bio, China). The standard curve of qRT-PCR was y = −0.3062 x + 12.054 (y = log10, x = Ct).
Tree Shrews
Adult wild male (N = 21) and female (N = 24) Chinese tree shrews (N = 45, weighing 110−200 g and normal body temperature 36–37.5°C were obtained from Lufeng, Yunnan, China at the primate of medicine research center, Institute of Medical Biology, Chinese Academy of Medical Sciences. All animals were provided free access to food and water. All animal care and experimental protocols were approved by the Animal Care and Use Committee of Institute of Medical Biology, Chinese Academy of Medical Sciences, China.
Injection
The tree shrews were anesthetized with ketamine. Then, 150 µl of DENV (DENV-3: 1 × 104 PFU/ml, DENV-2: 1 × 104 PFU/ml, diluent with RPMI-1640 medium (1640, BI)) was injected through intravenous injection or multisite subcutaneous injection (Abdomen, random of five sites, 30 µl/site). Complete the operation, the tree shrew was put into the ventilation area to restore, and all tree shrews were provided good care and recovered well (Table 1).
Weight and Temperature
The weight and temperature of tree shrews were measured on the 2nd, 7th, 15th, 21st, 28th, 35th, 41st and 48th days through electronic scales in a muffler and infrared thermometer on anus with three repetitions.
Blood and Tissue Sample Collection
Blood samples were collected on the 2nd, 7th, 15th, 21st, 28th, 35th, 41st and 48th days after DENV-2/-3 infection (tail vein blood, 0.4 ml each time). The DENV detection in liver and brain biopsies were performed on the 48th day under anesthesia with ketamine hydrochloride and 1% pentobarbital. Serum samples were separated from the collected blood through centrifugation at 3,500 rpm at 4°C. Viral RNA was extracted from 150 μl of tree shrew serum or liver and brain tissue extracts using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) and eluted in 50 μl of nuclease-free water. The extracted RNA was used for RT-PCR amplification to calculate viral RNA copy number titration (16). The residue of each liver and brain biopsy sample was frozen immediately by immersion in liquid nitrogen and then stored at −80°C before use.
Neutralizing Titer Examination of Tree Shrew Serum
The standard method of antibody titer determination was used. Briefly, twofold serial dilutions of DENV2/3 immune serum (collected on the 2nd, 7th and 15th days) were prepared with RPMI-1640 medium (1640, BI). A mixture of 0.05 ml of each dilution degree and DENV-3/2 (200 TCID50) was maintained at 37°C for 2 h and then added to every well of a 96-well plate, with saturability for 95% of the C6/36 cell line. The cells were cultured for 7 days at 28°C and 5% CO2, and examinations were conducted at 5 and 7 days with a microscope. The endpoint is to quantify viral titer by determining the concentration at which 50% of the infected cells display cytopathic effect.
Biochemical Markers and Routine Blood Examination
The serum of tree shrews was collected on the 2nd, 7th, 15th, 21st, 28th, 35th, 41st, and 48th days after intravenous and multisite subcutaneous injection. The biochemical markers and routine blood examination were conducted by the Ministry of Experimental Animals, Institute of Medical Biology, Chinese Academy of Medical Sciences (Automatic biochemical analyzer (BS-200) and Automatic blood analyzer (XT-2000i)).
Histological Analysis in the Brain and Liver of Tree Shrews After Infection With DENV-3
The tree shrews brain tissues were collected from anesthetized animals at 48th day after intravenous and multisite subcutaneous injection of DENV virus, and then fixed with 10% neutral buffered formalin, embedded in paraffin, sectioned at 4 μm and stained with hematoxylin and eosin (H&E) after dehydration (17). The pathological sections were examined and photographed under a microscope (Nikon, ECLIPSE Ni).
Ethics Statement
All animal care and experimental protocols were approved by the Animal Care and Use Committee of Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College, China (NO. ky201110).
Statistical Analysis
The statistical data was analyzed using software version 17.0 (ANOVA) (IBM, Chicago, USA). Animal groups were compared by the nonparametric Mann–Whitney Test. The relationship between histological scores for the duration of DENV infection and the titer of DENV RNA in liver and brain tissue was analyzed by Spearman’s rank. The bilateral significance level was α = 0.05.
Results
Phylogenetic Analysis of Tree Shrew STING, MDA5 and TLR3 Gene
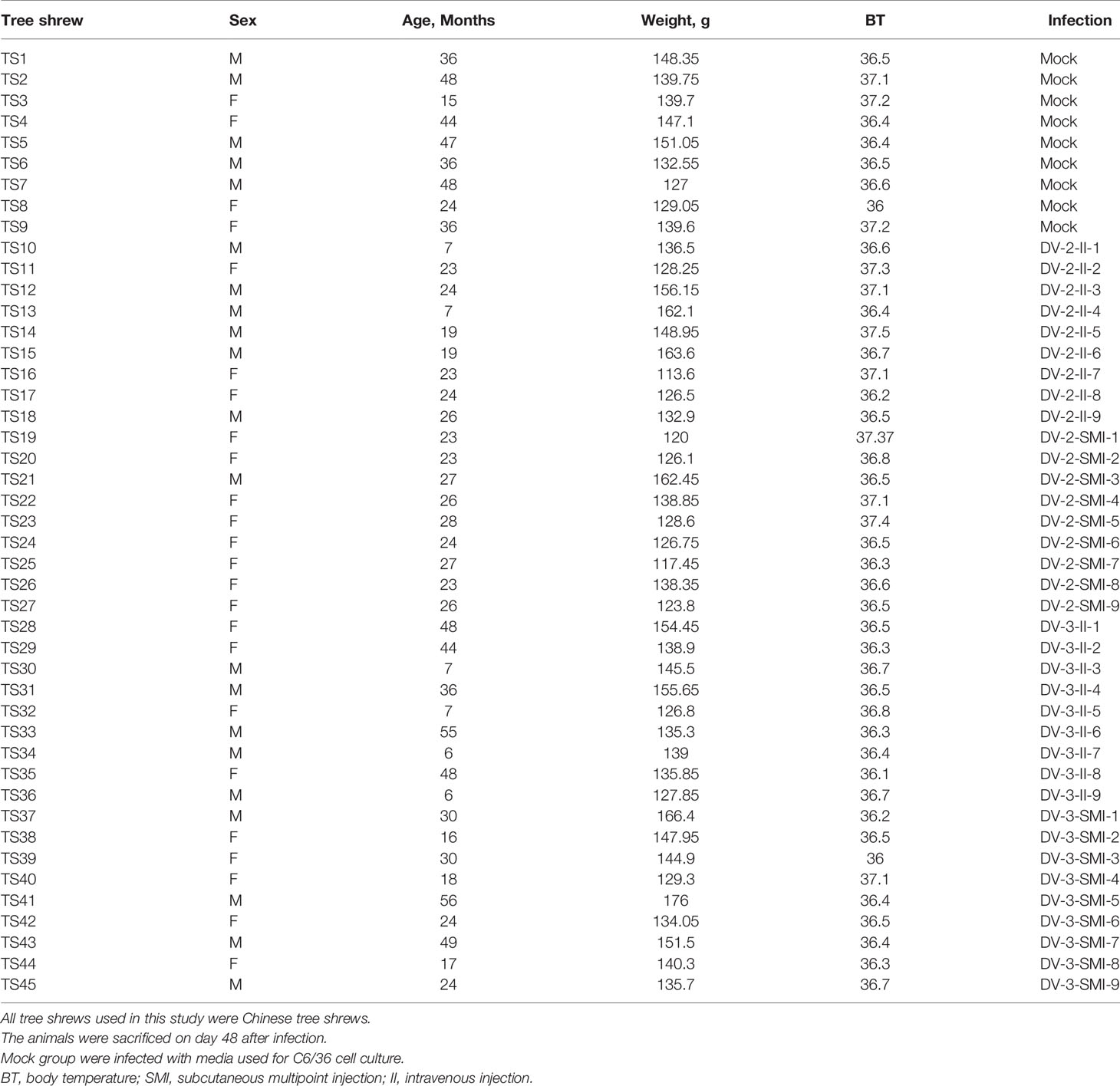
Tree shrews are much genetically closer to primates than rodents. We validated this with several important immune genes including STING, MAD5 and TLR3 that play important role in the innate immunity against DENV. STING has dual functions in host defense, it can regulate protein synthesis to prevent RNA virus infection; on the other hand, it can regulate IFN expression to restrict DNA viruses (18). The nonstructural protein of DENV, NS2B-NS3 protease complex cleaves STING in permissive human primary cells; while, murine STING is resistant to the NS2B3 cleavage (19, 20). The length of STING gene is approximately 1,050 bp in tree shrew, primate and rodent species. Melanoma differentiation-associated gene 5 (MDA5) is a viral RNA receptor and triggers robust innate immune responses to DENV. Phylogenetically, STING, TLR3 and MDA5 genes of tree shrew is more closely related to H. sapien and M. mulatta than S. scrofa in the ML tree (maximum likelihood tree). Indeed, STING and MDA5 genes of tree shrew had a much high similarity to humans than rat. The homology of STING and MDA5 gene between human and tree shrew was as high as 85%, while that is only 75–76% between human and rat (Figures 1A–D, Tables S1–3).
Figure 1 The key innate immune genes of tree shrew are genetically close to Homo sapiens. (A), The variation of coding amino acid of STING gene in tree shrew (Tupaia belangeri chinensis), and Homo sapiens. (B–D), Phylogenetic tree of STING (B), TLR3 (C) and MDA5 (D) gene of T. belangeri, H. sapiens, Macaca mulatta, Sus scrofa and Mus. The phylogenetic tree was constructed using the Maximum Likelyhood phylogeny test. Bootstrap values were set for 1,000 repetitions. (E, F) Viral replication of DENV-3 in Vero (E) and BM-MSCs (F) cells. DENV-3 genomic RNA was tested from 0 to 96 h post-inoculation in culture supernatants of Vero cells and tupaia BM-MSCs.
DENV-3 Infection of Primary Tupaia Bone Marrow Mesenchymal Stem Cells
The proliferative of DENV-3 viruses in tupaia BM-MSCs (Bone marrow mesenchyml stem cell) was investigated every 12 h. As shown in Figures 1E, F, DENV virus began to productively proliferate in tupaia BM-MSCs at 4 days after infection. Vero cells were used as positive controls (Figures 1E, F). Using the reverse-transcription quantitative (RT-qPCR) methods, the presence of DENV-3 genomic RNA were tested from 0 to 96 h post-inoculation in the culture supernatants of tupaia BM-MSCs and Vero cells. As shown in Figures 1E, F, the DENV-3 RNA was positive in all the culture supernatants of tupaia BM-MSCs and Vero cells beginning from 60 h post-infection and steadily increase thereafter (Figures 1E, F).
Fever and Death Induced by DENV
In this study, thirty-six tree shrews were injected with 150 µl (1 × 104 PFU/ml) of DENV through intravenous injection (II) and subcutaneous multipoint injection (SMI) (performed on the abdomen of the tree shrew that three points close to each other) (Figure 2A). The normal temperature of the tree shrew is 36.5 ± 0.5°C. The temperature was measured among different time periods early (2nd d), middle (7th d) and later (15th d) after injection, and it was generated through infrared thermometer for each animal. The results indicate that there were no significant differences among the DV-2-II, DV-2-SMI, DV-3-II, and DV-3-SMI groups. The DV-2-II group had a maximum mean temperature; DV-2-SMI was the minimum temperature group. The body temperature of four tree shrews was over 39 °C (DV2-II, DV2-SMI, DV3-II, DV3-SMI) from 2nd to 8th days and that of eight tree shrews was over 38 °C (DV2-II (3), DV2-SMI (1), DV3-II (2), DV3-SMI (2)) after the virus injection from 3rd to 10th days. One tree shrew presented with dengue fever on the next day after DENV-2 injection, while up to six tree shrews presented with dengue fever on the 28th day. Statistically significant differences in the body temperature between the infected and non-infected animals were observed at different time points (**P <0.01) (Figure 2B). A total of five deaths occurred within 48 days after the injection (DV2-II (1), DV3-II (2), DV3-SMI (2)), three males and two females died on days 3, 7, 21, 34 and 37 (Table 2).
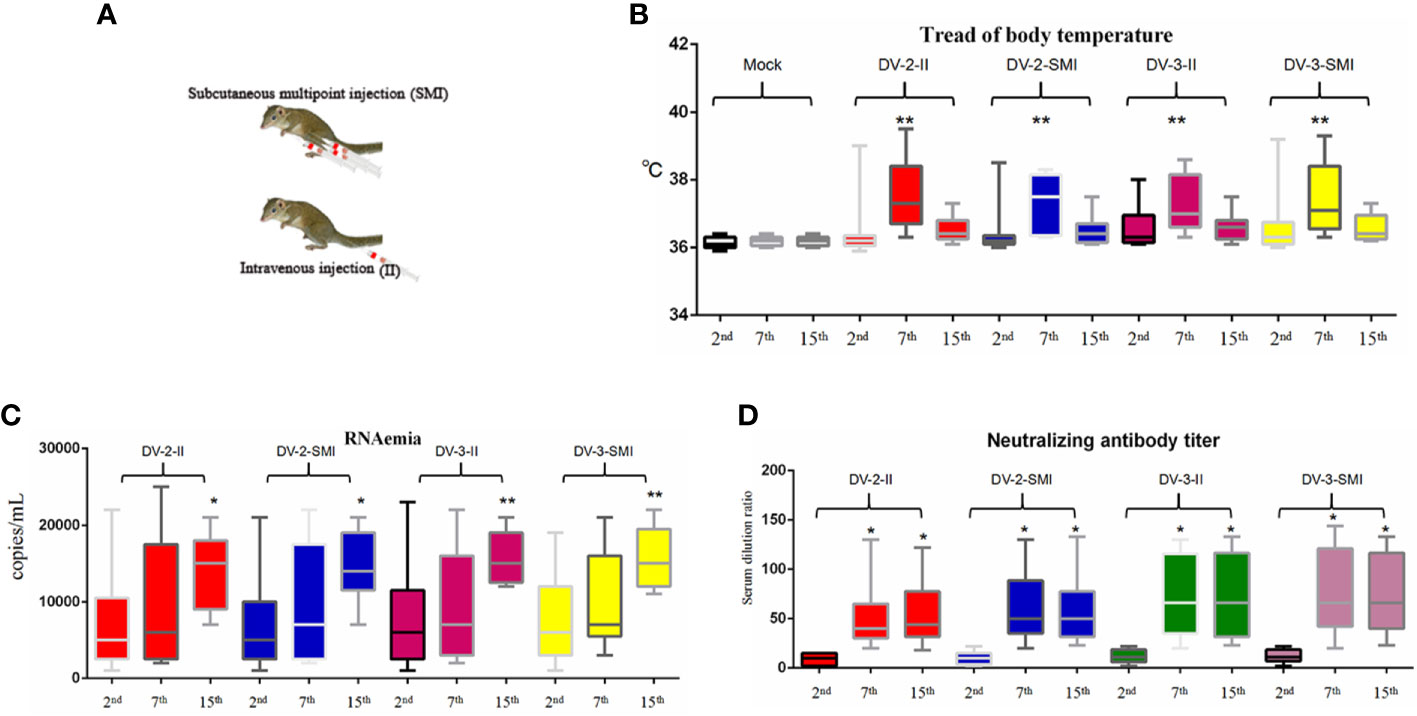
Figure 2 Fever, viremia and neutralizing antibody level of DENV-2 and DENV-3-inoculated tree shrew (n = 9 per group). (A) Schematic diagram of subcutaneous multipoint injection (SMI) and intravenous injection (II) in tree shrew. (B) The fever situations of the entire experimental process. The results from each group were compared using Student’s t test. **P <0.01. (C) The viral load in the serum. The viral RNA copies wERE tested at the day after blood collection on the 2nd, 7th and 15th days post injection. (D) The neutralizing titer in the serum after injection was observed on the 2nd, 7th and 15th days. The results from each group were compared using Student’s t test. *P <0.05, **P <0.01, ***P <0.001.
DENV Viremia
The dengue viral RNA in the plasma of infected tree shrews was quantified using real-time RT-PCR. Half of the tree shrews had shown viremia at the 7th and 15th days after injection (Figure 2C). The dengue viral RNA was not detected in the control group. The viremia of DV3-II and DV3-SMI groups were slightly higher than DV2-II and DV2-SMI groups. Using statistical analysis software, significant differences in viremia of tree shrew serum were observed at different time points among DV2-II and DV2-SMI groups (**P <0.01) (Figure 2C).
Neutralizing Antibodies
Production of neutralization antibodies is a major indicator of host resistance to pathogens. Most tree shrews presented high neutralizing antibody titers at 7 and 15 days after injection. The neutralizing titers in the DV-3-II and DV-3-SMI groups were slightly higher than those in the DV-2-II and DV-2-SMI groups. There was no difference between the DV-3-II and DV-3-SMI groups, and the lowest neutralizing titer was observed in the DV-2-II group. A much higher neutralizing antibody titer was observed on the 7th and 15th days than that on the 2nd day. Using statistical analysis software, significant differences in the neutralizing titer of tree shrews were observed at different times among DV2-II and DV2-SMI groups (*P <0.05) (Figure 2D). The neutralizing titer was not detected in the control group and the groups of DV3-II and DV3-SMI were slightly higher than DV2-II and DV2-SMI.
Blood Chemistry
The WBC (white blood cell), RBC (red blood cell), HGB (hemoglobin), HCT (hematocrit), PLT (Platelets), MPV (mean platelet volume), PCT (thrombocytocrit), MCV (mean corpuscular volume), MCH (mean corpuscular hemoglobin), MCHC (mean corpuscular hemoglobin concentration), RDW-SD (red blood cell distribution width), RDW-CV (red blood cell-CV), PDW (platelet distribution width) and P-LCR (platelet large cell ratio) were measured in fourteen tree shrews. No significant changes were observed in RBC, HGB, HCT, MPV, MCV, MCH, MCHC, RDW-SD, RDW-CV and PDW, but changes were observed in WBC, PLT, PCT and P-LCR among different time periods early (2nd d), middle (7th d) and later (15th d). The WBC and P-LCR increased during the middle periods of infection and fell thereafter when compared to uninfected controls; however, the PCT and PLT showed opposite trends (Figures 3B–E). Clinically, approximately one-third of dengue fever patients show mildly or moderately elevated alanine aminotransferase (ALT) levels. At three blood collection times including early (2nd d), middle (7th d) and later (15th d) phases after DENV injection, the biochemical indicators AST, ALT and ALP were detected in the sera of tree shrews. AST and ALT levels were elevated in the second and third weeks of infection when compared to uninfected controls and wend down to basal levels thereafter (Figure 3A).
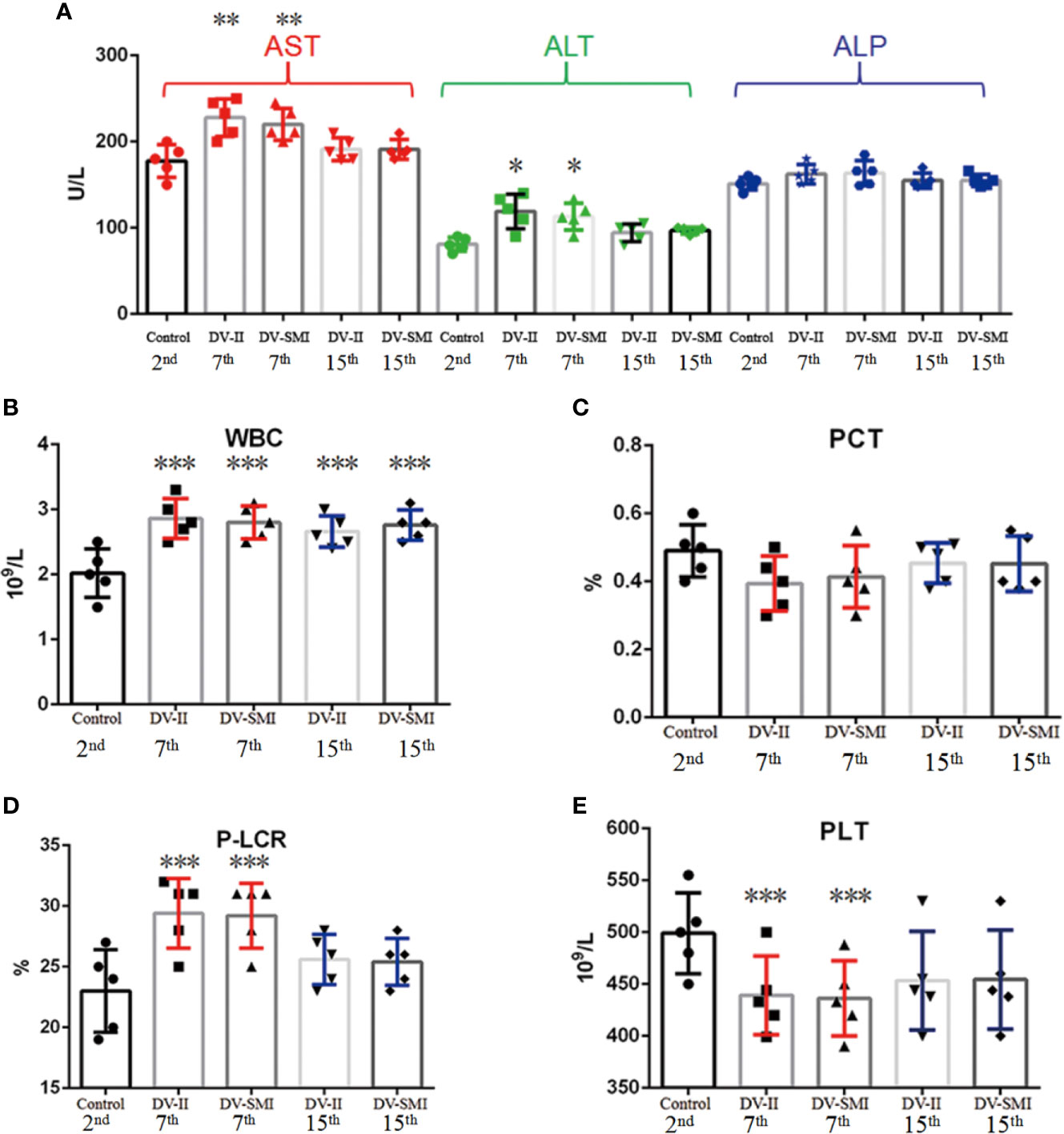
Figure 3 Hematological analysis and concentration of liver enzymes in the blood of DENV-2 and DENV-3-inoculated tree shrew (n = 9 per group). (A) The biochemical indicators AST, ALT and ALP on the different time periods early (2nd d), middle (7th d) and later (15th d) after injection. (B–E) The blood tests for WBC, PCT, P-LCR and PLT on the different time periods early (2nd d), middle (7th d) and later (15th d) after injection. *P <0.05, **P <0.01.
DENV Dissemination to Liver and Brain
Routine examination of the cerebrospinal fluid is usually normal, and intracranial hemorrhage or brain edema in DENV patients is rare. From cerebrospinal fluid, the dengue virus can be isolated, and the viral nucleic acid or specific virus antigen can be detected. The liver histopathology of dengue fever cases shows nonspecific lesions. The cytoplasm of most of the liver cells shows a balloon-like and slight eosinophilic change, accompanied by an individual eosinophilic body.
The DENV RNA of tree shrew liver and brain were examined at the 48th day after injection. Viral RNA was detected in the brain of tree shrews infected with either DENV-2 or DENV-3. However, DENV-2 and DENV-3 RNA were barely detected in the liver. Using statistical analysis software, significant differences in the viral load were observed in the liver and brain of tree shrews among DV2-II and DV2-SMI groups (*P <0.05) (Figure 4A).
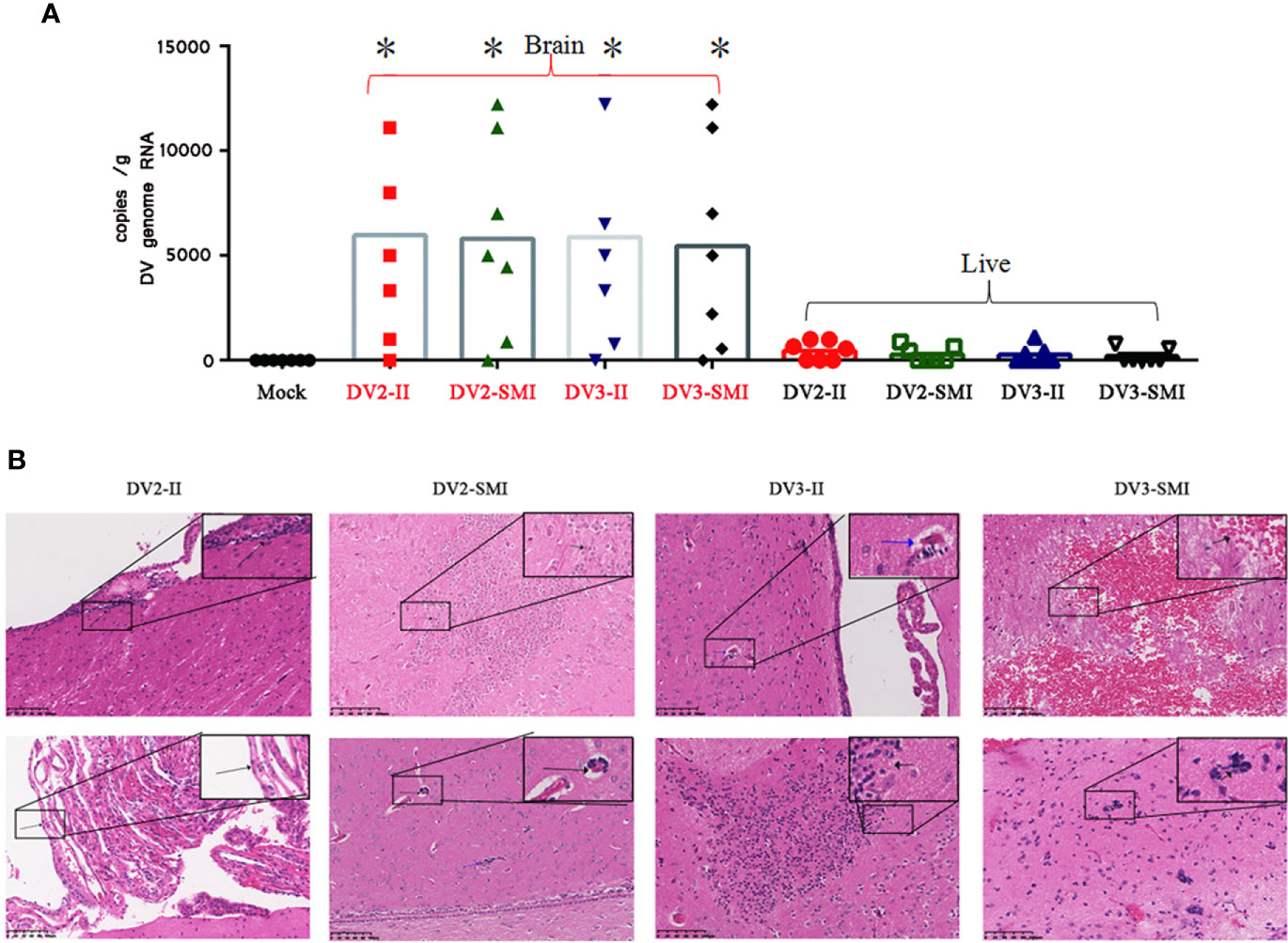
Figure 4 Viral RNA copies and H&E analysis in tissues of DENV-2 and DENV-3 infected tree shrew. (A) Genome RNA of DENV in the liver and brain. Circle dots represent the control group. 48 days post infection. High viral RNA copies were detected in the brain. The results from each group were compared using Student’s t test. *P <0.05. (B) Histopathology aspects of Tupaia chinensis brain tissue 48 days post infection of DENV-2 (DV2) and DENV-3 (DV3). DV2-II, a small number of inflammatory cells were seen locally near the ventricle in brain tissue (Black arrow) and local proliferation of arachnoid endothelial cells in brain tissue (Black arrow) were observed. The Blood vessel sleeve (Black arrow) and glial nodule (Blue arrow) were observed in brain tissue of DV2-SMI. DV3-II, glial nodules (Black arrow), individual vascular sleeves (Blue arrow) and proliferation of keratinocytes (Black arrow) in brain tissue were observed. DV3-SMI, hemorrhagic foci of brain tissue (Black arrow) and multiple glial nodules in brain tissue (Black arrow) were observed. *P <0.05.
Histopathology in the Brain of Tree Shrews After Infection With DENV-3 and DENV-2
The bleeding lesions in the intravenous infection group were more severe than those in the subcutaneous infection group in brain tissues. Local subarachnoid hemorrhage, local proliferation of glial cells in the cortex and parenchymal hemorrhage of brain tissue were found in the brain tissues of tree shrews infected by DENV-3 (Figure 4B). Individual glial nodules and individual vascular sleeves were observed in local ventricle. Severe bleeding was observed in three tree shrews of intravenous infection group, and only one showed severe bleeding in the subcutaneous infection group. Moreover, one tree shrew showed edema of choroid plexus and ependymal epithelial cells in the intravenous infection group.
Discussion
There have been some studies on the DENV infection model in NHPs. Development of a suitable animal model for DENV disease is important for understanding pathogenesis (7). These studies included those related to the effect of experimental and natural infections, studies of the immune response of animals infected with DENV, and the evaluation of several candidate dengue viral vaccine formulations (21–25). The kinetics of dengue viremia and the generation of dengue virus specific antibody in these monkeys have been shown to generally resemble to that observed in human dengue virus infection (22). However, even many recent efforts have been made to improve the NHP models, no overt clinical symptoms develop, could sustain viral replication but at a low level, and do not develop vascular leak or syndromes resembling DHF or DSS (6, 26, 27).
As for mouse models of DENV infection, neurological disease and antibody dependent enhancement (ADE) could be developed in AG129 mice, which lack IFN-α/β and -γ receptors (28, 29). The AG129 mice infected with mouse-adapted DENV strains could also lead to a lethal vascular leak syndrome (30–32). SCID mice have also been used to assess vaccine effectiveness after DENV challenge (33–35). Symptoms of human related clinical diseases have not been observed in immunocompetent mouse models, but neurotropic disease could be developed (36, 37). A research group also tried to develop new mouse models by knocking in human STING, which is susceptible to cleavage by the DENV NS2B-NS3 protease, and more susceptible to DENV infection (19, 20, 38, 39).
Several virus infection models have been established in tree shrews, such as HEV, HAV, HBV, HSV, HCV and HDV (40). It is also reported that infection model of Zika virus had been established in tree shrew (12, 13). Here, we reported an infection tree shrew animal model of DENV. In this study, tree shrews experienced fever, death, viremia and abnormal aminotransferase levels after infection with DENV, and showed resemble clinical manifestations as in humans.
Notably, viremia was detected in tree shrews at a lower level, which may account for the relatively benign overall disease course in these animals, whereas the lower viral load in tree shrews is sufficient to induce mild hemorrhage. Lower viremia could lead to their prolonging the infectious reservoir (41). The presence of antibodies was almost non-detectable on the second day, and the purpose of antibody detection on the second day was to conduct a control experiment. The antibody titers range from 160 to 320 in mice animal model, contrastive, the antibody titers range from 20 to 150 in tree shrew animal model (42). Neutralizing antibodies and viremia were detected in the same time frame for the reason of that haven’t produced enough high levels of neutralizing antibodies, in addition, there was a good multiplication in tree shrews of DENV infection at early stage (Figures 2C, D). A similar phenomenon happens to human, DENV viremia can be seen in both asymptomatic and symptomatic patients and the viral decay rate was slower in asymptomatic patients (41). Thus, this tree shrew model of DENV infection may not only provide a valuable tool for further studies on the various pathophysiologic effects of dengue virus infection, but also provide a comprehensive analysis of host–virus interactions. In addition, this model is also an important tool for the testing and evaluation of potential dengue virus vaccines and therapeutic drugs. Since antibody dependent enhancement (ADE) of second infection of DENV could be develop in AG129 mice, so it is also a promising work to develop ADE model in tree shrew in the future.
Vascular leakage is an important aspect of dengue model system (43). Severe vascular pathology of extensive microvascular permeability and plasma leakage into tissues and organs were observed in DENV infection of patients (44). It is very important and necessary to examine blood vascular leakage in tree shrews. The vascular leakage of tree shrew in DENV infection was pending experimental validation.
Central nervous system (CNS) impairment was reported in DENV infection of mice and human (36). The bleeding lesions, local subarachnoid hemorrhage, local proliferation of glial cells in the cortex and parenchymal hemorrhage of brain tissue were found in the brain tissues of tree shrews infected by DENV-3 (Figure 4B). In addition, viral RNA were detected in the brain, but not detected in the liver. The breaching mechanism and hyperpermeability of blood–brain barrier by DENV were primarily due to direct consequences of viral infection of endothelial cells (45).
The tree shrews used in this study were collected from the wild which possess complex genetic background. The complexity and diversity of results may be caused by the diversity of tree shrews. Gratifying, there were fever, viremia, DV proliferation and pathological changes in brain and other clinical symptoms among many tree shrews. The technology of artificial propagation of tree shrews has been realized, so in the future, when the genetic background is simpler and clear, the disease symptoms of tree shrews DENV infection model will be more uniform.
Dengue fever and neutralizing antibodies are consistently observed in the tree shrew model of dengue virus infection. These findings provide a scientific basis to establish a future mature dengue animal model as a scientific platform for the in-depth understanding of the disease pathogenesis and the development of effective therapeutic drugs and vaccines against dengue fever.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Institute of Medical Biology, Chinese Academy of Medical Sciences.
Author Contributions
LJ: Performed the experiments, acquisition and analysis of data, drafting of the manuscript, critical revision of the manuscript. CL and QS: Technical or material support, study concept and design, critical revision of the manuscript, obtained funding, study supervision. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Foundation of the CAMS Initiative for Innovative Medicine (CAMS-I2M) (grant no. 2017-I2M-2-006), the National Natural Science Foundation of China (31970868), Major Projects and Key Research and Development Plans of Yunnan Province (2019ZF004), the Natural Science Foundation of Yunnan Province (2016FA029), Yunnan Health Training Project of High Level Talents (D-201605).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Jiejie Dai of Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College, for providing tree shrew and technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.621164/full#supplementary-material
Abbreviations
DENV, dengue virus; NHP: nonhuman primates; DF, dengue fever.
References
1. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature (2013) 496:504–7. doi: 10.1038/nature12060
2. Schaffner F, Mathis A. Dengue and dengue vectors in the WHO European region: past, present, and scenarios for the future. Lancet Infect Dis (2014) 14:1271–80. doi: 10.1016/S1473-3099(14)70834-5
3. Kutsuna S, Kato Y, Moi ML, Kotaki A, Ota M, Shinohara K, et al. Autochthonous dengue fever, Tokyo, Japan 2014. Emerg Infect Dis (2015) 21:517–20. doi: 10.3201/eid2103/141662
4. Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol (2009) 9:523–40. doi: 10.1016/j.meegid.2009.02.003
5. Lin YP, Luo YS, Chen Y, Lamers MM, Qiang Z, Han YX, et al. Clinical and epidemiological features of the 2014 large-scale dengue outbreak in Guangzhou city, China. BMC Infect Dis (2016) 16:1–8. doi: 10.1186/s12879-016-1379-4
6. Clark KB, Onlamoon N, Hsiao HM, Perng GC, Villinger F. Can non-human primates serve as models for investigating dengue disease pathogenesis. Front Microbiol (2013) 4:1–17. doi: 10.3389/fmicb.2013.00305
7. Chan KWK, Watanabe S, Kavishna R, Alonso S, Vasudevan SG. Animal models for studying dengue pathogenesis and therapy. Antivir Res (2015) 123:5–14. doi: 10.1016/j.antiviral.2015.08.013
8. Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, et al. Genome of the Chinese tree shrew. Nat Commun (2013) 4:1–9. doi: 10.1038/ncomms2416
9. Wang J, Zhou QX, Lü LB, Xu L, Yang YX. A depression model of social defeat etiology using tree shrews. Zool Res (2012) 33:92–8. doi: 10.3724/SP.J.1141.2012.0109
10. Yaohui T, Junjun H, Qiu T, Hualin Y, Lanzhen Y, Yuan L. A tree shrew glioblastoma model recapitulates features of human glioblastoma. Oncotarget (2017) 8:17897–907. doi: 10.18632/oncotarget.15225
11. Li L, Li Z, Li X, Wang E, Lang F, Xia Y, et al. Reactivation of HSV-1 following explant of tree shrew brain. J Neurovirol (2016) 22:293–306. doi: 10.1007/s13365-015-0393-4
12. Zhang L, Shen ZL, Feng Y, Li DQ, Zhang NN, Deng YQ, et al. Infectivity of Zika virus on primary cells support tree shrew as animal model. Emerg Microbes Infect (2019) 8:232–41. doi: 10.1080/22221751.2018.1559707
13. Zhang NN, Zhang L, Deng YQ, Feng Y, Ma F, Wang Q, et al. Zika Virus Infection in Tupaia belangeri Causes Dermatological Manifestations and Confers Protection against Secondary Infection. J Virol (2019) 29:1–14. doi: 10.1128/JVI.01982-18
14. Felsenstein J. Confidence-limits on phylogenies: an approachusing the bootstrap. Evolution (1985) 39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x
15. Lu CX, Li XF, Wang WG, Sun XM, Tong PF. Isolation, culture, adipocgenic and osteogenic induction of Tupaia bone marrow mesenchymal stem cells. Chin J Comparartive Med (2014) 24:10–3. doi: 10. 3969.j.issn.1671.7856
16. Drosten C, Göttig S, Schilling S, Asper M, Panning M, Schmitz H, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol (2002) 40:2323–30. doi: 10.1128/JCM.40.7.2323-2330.2002
17. van den Brand JM, Stittelaar KJ, van Amerongen G, Reperant L, de Waal L, Osterhaus AD, et al. Comparison of temporal and spatial dynamics of seasonal H3N2, pandemic H1N1 and highly pathogenic avian influenza H5N1 virus infections in ferrets. PLoS One (2012) 7:1–21. doi: 10.1371/journal.pone.0042343
18. Franz KM, Neidermyer WJ, Tan YJ, Whelan SPJ, Kagan JC. STING-dependent translation inhibition restricts RNA virus replication. PNAS (2018) 115:2058–67. doi: 10.1073/pnas.1716937115
19. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, et al. DENV Inhibits Type I IFN Production in Infected Cells by Cleaving Human STING. PLoS Pathog (2012) 8:1–14. doi: 10.1371/journal.ppat.1002934
20. Kevin M, Ana FS. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev (2014) 25:669–79. doi: 10.1016/j.cytogfr.2014.08.004
21. Habib M, Yaqub T, Nazir J, Shehzad W, Aziz-Ul-Rahman, Sohail T, et al. Genomic and biological characterization of Newcastle disease viruses isolated from migratory mallards (Anas platyrhynchos). Arch Virol (2018) 163(8):2179–88. doi: 10.1007/s00705-018-3840-8
22. Charnay C, Burdin N, Dumas R, Lang J. Evaluation of interferences between dengue vaccine serotypes in a monkey model. Am J Trop Med Hyg (2009) 80:302–11. doi: 10.4269/ajtmh.2009.80.302
23. Whitehead RH, Chaicumpa V, Olson LC, Russell PK. Sequential dengue virus infections in the white-handed gibbon (Hylobates lar). Am J Trop Med Hyg (1970) 19:94–102. doi: 10.4269/ajtmh.1970.19.94
24. Goncalvez AP, Engle RE, St Claire M, Purcell RH, Lai CJ. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc Natl Acad Sci U S A (2007) 104:9422–7. doi: 10.1073/pnas.0703498104
25. McArthur JH, Durbin AP, Marron JA, Wanionek KA, Thumar B, Pierro DJ, et al. Phase I clinical evaluation of rDEN4Delta30-200,201: a live attenuated dengue 4 vaccine candidate designed for decreased hepatotoxicity. Am J Trop Med Hyg (2008) 79:678–84. doi: 10.4269/ajtmh.2008.79.678
26. Wastika CE, Sasaki M, Yoshii K, Anindita PD, Hang'ombe BM, Mweene AS, et al. Serological evidence of Zika virus infection in non−human primates in Zambia. Arch Virol (2019) 164:2165–70. doi: 10.1007/s00705-019-04302-0
27. Althouse BM, Durbin AP, Hanley KA, Halstead SB, Weaver SC, Cummings DAT. Viral kinetics of primary dengue virus infection in non-human primates: A systematic review and individual pooled analysis. Virology (2014) 452-453:237–46.
28. Sarathy VV, Milligan GN, Bourne N, Barrett AD. Mouse models of dengue virus infection for vaccine testing. Vaccine (2015) 33:7051–60. doi: 10.1016/j.vaccine.2015.09.112
29. Martínez Gómez JM, Ong LC, Lam JH, Binte Aman SA, Libau EA, Lee PX, et al. Maternal Antibody-Mediated Disease Enhancement in Type I Interferon-Deficient Mice Leads to Lethal Disease Associated with Liver Damage. PLoS Neglected Trop Dis (2016) 10:1–22. doi: 10.1371/journal.pntd.0004536
30. Tan GK, Ng JK, Trasti SL, Schul W, Yip G, Alonso S. A Non Mouse-Adapted Dengue Virus Strain as a New Model of Severe Dengue Infection in AG129 Mice. PLoS Neglected Trop Dis (2010) 4:1–10. doi: 10.1371/annotation/03774b36-c453-404a-b295-7b91bfd9cebd
31. Sarathy VV, White M, Li L, Gorder SR, Pyles RB, Campbell GA, et al. A Lethal Murine Infection Model for Dengue Virus 3 in AG129 Mice Deficient in Type I and II Interferon Receptors Leads to Systemic Disease. J Virol (2015) 89:1254–66. doi: 10.1128/JVI.01320-14
32. Milligan GN, Sarathy VV, White MM, Greenberg MB, Campbell GA, Pyles RB, et al. A lethal model of disseminated dengue virus type 1 infection in AG129 mice. J Gen Virol (2017) 98:2507–19. doi: 10.1099/jgv.0.000923
33. Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue Fever in Humanized NOD/SCID Mice. J Virol (2005) 79:13797–9. doi: 10.1128/JVI.79.21.13797-13799.2005
34. Zompi S, Harris E. Animal Models of Dengue Virus Infection. Viruses (2012) 4:62–82. doi: 10.3390/v4010062
35. Frias-Staheli N, Dorner M, Marukian S, Billerbeck E, Labitt RN, Rice CM, et al. Utility of Humanized BLT Mice for Analysis of Dengue Virus Infection and Antiviral Drug Testing. J Virol (2014) 88:2205–18. doi: 10.1128/JVI.03085-13
36. Amorim JFS, Azevedo AS, Costa SM, Trindade GF, Basílio-de-Oliveira CA, Gonçalves AJS, et al. Dengue infection in mice inoculated by the intracerebral route: neuropathological effects and identification of target cells for virus replication. Sci Rep (2019) 9(1):17926. doi: 10.1038/s41598-019-54474-7
37. Teterina NL, Maximova OA, Kenney H, Liu G, Pletnev AG. MicroRNA-based control of tick-borne flavivirus neuropathogenesis: Challenges and perspectives. Antiviral Res (2016) 127:57–67. doi: 10.1016/j.antiviral.2016.01.003
38. Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. Inhibition of the Type I Interferon Response in Human Dendritic Cells by Dengue Virus Infection Requires a Catalytically Active NS2B3 Complex. J Virol (2010) 84:9760–74. doi: 10.1128/JVI.01051-10
39. Aguirre S, Fernandez-Sesma A. Collateral Damage during Dengue Virus Infection: Making Sense of DNA by cGAS. J Virol (2017) 91:1–5. doi: 10.1128/JVI.01081-16
40. Yu W, Yang C, Bi Y, Long F, Li Y, Wang J, et al. Characterization of hepatitis E virus infection in tree shrew (Tupaia belangeri chinensis). BMC Infect Dis (2016) 16:1–7. doi: 10.1186/s12879-016-1418-1
41. Matangkasombut P, Manopwisedjaroen K, Pitabut N, Thaloengsok S, Suraamornkul S, Yingtaweesak T, et al. Dengue viremia kinetics in asymptomatic and symptomatic infection. Int J Infect Dis (2020) 101:90–7. doi: 10.1016/j.ijid.2020.09.1446
42. Prompetchara E, Ketloy C, Keelapang P, Sittisombut N, Ruxrungtham K. Induction of neutralizing antibody response against four dengue viruses in mice by intramuscular electroporation of tetravalent DNA vaccines. PLoS One (2014) 9(6):e92643. doi: 10.1371/journal.pone.0092643. eCollection 2014.
43. Trung DT, Wills B. Systemic vascular leakage associated with dengue infections - the clinical perspective. Curr Top Microbiol Immunol (2010) 338:57–66. doi: 10.1007/978-3-642-02215-9_5
44. Rathore AP, Mantri CK, Aman SA, Syenina A, Ooi J, Jagaraj CJ, et al. Dengue virus-elicited tryptase induces endothelial permeability and shock. J Clin Invest (2019) 129(10):4180–93. doi: 10.1172/JCI128426
Keywords: dengue virus, tree shrew, animal model, dengue fever, viremia
Citation: Jiang L, Lu C and Sun Q (2021) Tree Shrew as a New Animal Model for the Study of Dengue Virus. Front. Immunol. 12:621164. doi: 10.3389/fimmu.2021.621164
Received: 25 October 2020; Accepted: 01 March 2021;
Published: 25 March 2021.
Edited by:
S. Swaminathan, Independent Researcher, Hyderabad, IndiaReviewed by:
Gerardo Guillen, Center for Genetic Engineering and Biotechnology (CIGB), CubaZifeng Yang, Guangzhou Institute of Respiratory Health, China
Copyright © 2021 Jiang, Lu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiangming Sun, cXN1bkBpbWJjYW1zLmNvbS5jbg==
†Present address: Qiangming Sun, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College (CAMS & PUMC), Kunming, China
 Liming Jiang
Liming Jiang Caixia Lu1,2
Caixia Lu1,2 Qiangming Sun
Qiangming Sun