
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 15 February 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.619362
This article is part of the Research Topic Strategies Played by Immune Cells and Mycobacteria in the Battle between Antimicrobial Activity and Bacterial Survival View all 13 articles
 Gang Zhao1,2
Gang Zhao1,2 Xifang Zhu1,2
Xifang Zhu1,2 Hui Zhang1,2
Hui Zhang1,2 Yingyu Chen1,2,3,4,5,6
Yingyu Chen1,2,3,4,5,6 Elise Schieck7
Elise Schieck7 Changmin Hu1
Changmin Hu1 Huanchun Chen1,2,3,4,5,6
Huanchun Chen1,2,3,4,5,6 Aizhen Guo1,2,3,4,5,6*
Aizhen Guo1,2,3,4,5,6*Mycoplasma bovis causes important diseases and great losses on feedlots and dairy farms. However, there are only a few measures to control M. bovis-related diseases. As in other mycoplasma species, this is predominantly because the virulence related factors of this pathogen are largely unknown. Therefore, in this study, we aimed to identify novel virulence-related factors among the secreted proteins of M. bovis. Using bioinformatic tools to analyze its secreted proteins, we preliminarily predicted 39 secreted lipoproteins, and then selected 11 of them for confirmation based on SignalP scores >0.6 or SceP scores >0.8 and conserved domains. These 11 genes were cloned after gene modification based on the codon bias of Escherichia coli and expressed. Mouse antiserum to each recombinant protein was developed. A western blotting assay with these antisera confirmed that MbovP280 and MbovP475 are strongly expressed and secreted proteins, but only MbovP280 significantly reduced the viability of bovine macrophages (BoMac). In further experiments, MbovP280 induced the apoptosis of BoMac treated with both live M. bovis and MbovP280 protein. The conserved coiled-coil domain of MbovP280 at amino acids 210–269 is essential for its induction of apoptosis. Further, immunoprecipitation, mass spectrometry, and coimmunoprecipitation assays identified the anti-apoptosis regulator αB-crystallin (CRYAB) as an MbovP280-binding ligand. An αβ-crystallin knockout cell line BoMac-cryab−, Mbov0280-knockout M. bovis strain T9.297, and its complemented M. bovis strain CT9.297 were constructed and the apoptosis of BoMac-cryab− induced by these strains was compared. The results confirmed that CRYAB is critical for MbovP280 function as an apoptosis inducer in BoMac. In conclusion, in this study, we identified MbovP280 as a novel secreted protein of M. bovis that induces the apoptosis of BoMac via its coiled-coil domain and cellular ligand CRYAB. These findings extend our understanding of the virulence mechanism of mycoplasmal species.
Mycoplasma bovis is a member of the class Mollicutes, a group of the smallest self-replicating wall-less prokaryotes. It causes several important diseases, including pneumonia, mastitis, and arthritis, in cattle throughout the world (1–3). In addition, it usually co-infects cattle together with other pathogens, such as Pasteurella multocida, Mannheimia haemolytica, bovine viral diarrhea virus (BVDV), bovine respiratory syncytial virus (BRSV), Bovine herpes virus 1 (BHV-1), etc to cause bovine respiratory disease complex (BRD) (4). Despite its minimal genome, M. bovis is a successful pathogen capable of developing both persistent infections and clinical diseases in cattle. As is well-known, M. bovis, like other mycoplasma species, lacks conventional toxins and its virulence mechanism is still poorly understood.
Although many previous studies have focused on the membrane and membrane-associated proteins of mycoplasmal species that are involved in virulence-related processes, such as adhesion (5), invasion (6, 7), and the inflammatory response (8), secreted proteins haven't yet attracted considerable attention until only recent years. However, secreted proteins often function as virulence-related factors or important antigens in pathogenic bacteria. Several studies have shown that the culture supernatant of M. bovis (9) induces the expression of several cytokines in different types of host cells and that live M. bovis behaves differently from the killed bacterium in inducing cytokine expression (10). Furthermore, several proteins of Mycoplasma species, including P80 of M. hominis (11), P102 of M. hyopneumoniae (12), Mpn491 of M. pneumoniae (13), CARDS toxin of M. pneumoniae (14), and a nuclease encoded by MBOV_RS02825 of M. bovis (15), have been shown to be secreted proteins. More recently, the secretomes and extracellular vesicles of several mycoplasmas species have been investigated with proteomic approaches, such as two-dimensional electrophoresis and liquid chromatography–tandem mass spectrometry (LC–MS/MS), isobaric tags for relative and absolute quantitation (iTRACK), and label-free proteomic analyses (16–18). However, the progress is relatively slow because it is usually difficult to confirm the secretomes and secreted proteins of mycoplasma species based on following reasons: (1) Mycoplasma species grow slowly and it is difficult to get sufficient proteins in short time; (2) Mycoplasma species growth requires rich medium supplemented with high concentrations of serum and yeast extract and it is difficult to exclude the contamination of abundant foreign proteins from the secretome in the culture supernatant; (3) There is no efficient genetic tools to manipulate gene expression of mycoplasma species by knock-out and knock-in to verify the predicted secreted proteins; (4) Most of the genes in mycoplasmal genomes are functionally unknown. One way dealing with this awkward situation tactfully is to combine the prediction of secreted proteins with bioinformatic tools, such as SignalP, SecretomeP, PSORT-B, and PrediSi (17–19) and identification of the secreted proteins with proteomic methods.
Therefore, in this study, we aimed to determine the novel secreted proteins of M. bovis and examine their association with M. bovis virulence. The secreted lipoproteins were first predicted with online softwares, and then the predicted secreted proteins were expressed and verified. Among the 11 predicted proteins, MbovP280 was identified as a secreted protein that induces apoptosis in a bovine macrophage cell line (BoMac) via the anti-apoptosis regulator αB-crystallin (CRYAB).
The protocols for the mouse experiments in this study were approved by the Experimental Animal Ethics Committee of Huazhong Agricultural University (Wuhan, China) (Permit number: HZAUMO-2018-027) and were performed in accordance with the Hubei Regulations for the Administration of Affairs Concerning Experimental Animals.
Mycoplasma bovis strain HB0801 (GenBank accession no. NC_018077.1) was isolated from the lesioned lung of an infected beef cattle from Yingcheng city in Hubei province, China and characterized by this laboratory (20). The strain was grown in pleuropneumonia-like organism (PPLO) medium (BD Company, Sparks, MD, USA), as previously described (20). Mycoplasma bovis mutants were grown in the same PPLO medium but supplemented with 100 μg/mL gentamycin or 10 μg/mL puromycin, depending upon the resistance genes the mutants carried. Recombinant Escherichia coli strains DH5α and BL21 (TransGen, Beijing, China) were grown in Luria–Bertani (LB) medium with antibiotics as necessary.
The BoMac cell line was kindly provided by Prof. Judith R. Stabel from the Johne's Disease Research Project at the United States Department of Agriculture in Ames, Iowa, and grown in RPIM 1640 medium (HyClone, UT, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Sydney, Australia) as described previously (21). The HEK293T cell line was purchased from the China Center for Type Culture Collection and cultured in high-glucose Dulbecco's modified Eagle's medium (HyClone) supplemented with 10% heat-inactivated FBS (Gibco).
Classical secreted proteins were predicted with SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), while non-classical secreted proteins with SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/) as described previously (19). The conserved domains in the predicted proteins were analyzed online with the National Center for Biotechnology Information (NCBI) Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Proteins homologous to MbovP280 were identified with MolliGen 3.0 (http://services.cbib.u-bordeaux.fr/molligen/). The coiled-coil domain of MbovP280 was predicted with COILS (https://embnet.vital-it.ch/software/COILS_form.html). The homology models of CRYAB, caspase 3, and MbovP280 were generated with SWISS-MODEL (https://swissmodel.expasy.org/), and the interactions between CRYAB, caspase 3, and MbovP280 were analyzed with ClusPro 2.0 (https://cluspro.bu.edu/login.php).
The putative secreted lipoproteins with the highest prediction values and carrying conserved domains were selected for further analysis. First, the sequences of the selected genes were site-directedly edited by replacing the TGA codon with TGG to ensure that M. bovis tryptophan was correctly translated in E. coli. The sequences were synthesized by Beijing Tianyi Huiyuan Bioscience & Technology Inc. (Wuhan, China) and ligated into the pET-30a vector (Novagen, Darmstadt, Germany) (Supplementary Table 1). The modified genes were individually cloned into the pET-30a vector after digestion with restriction endonucleases BamHI and XhoI. The MbovP280 mutant in which amino acids 210–269 (predicted to form a coiled-coil domain by the COILS software) were deleted was similarly cloned into pET-30a to generate pET-30a-Mbov_0280Δ210−269. Escherichia coli strain BL21 (TransGen, Beijing, China) was then transformed with each of the constructed plasmids individually, and the recombinant proteins were expressed after the cells were treated with isopropyl β-D-1-thiogalactopyranoside (IPTG) (0.8 mM). The proteins were purified with nickel affinity chromatography (GE Healthcare, NJ, USA), as described previously (7).
Mouse antisera against eight recombinant secreted proteins (rMbovP280, rMbovP290, rMbovP475, rMbovP458, rMbovP468, rMbovP537, rMbovP682, and rMbovP838) were produced in this study with a previously described method (7). Briefly, 4-week-old female BALB/c mice were purchased from China Hubei Provincial Center for Disease Control and Prevention (Wuhan, China) and raised in the Animal Facility of Huazhong Agriculture University. For each protein, five mice were immunized with 100 μg of each purified protein emulsified in an equal volume of Freund's complete adjuvant (Sigma, USA) for the priming immunization or with Freund's incomplete adjuvant for the subsequent immunization. Immunizations were performed by subcutaneous injection at an interval of 2 weeks. When the antiserum titers peaked, the mice were euthanized and bled. The antisera were collected and stored at −20°C for further use, and the preimmunization sera were stored for use as the negative controls.
Antibodies directed against the predicted secreted proteins rMbovP116, rMbovP275, and rMbovP739, which were previously developed by this laboratory (22), were also used in this study.
The secretomes and whole-cell proteins of M. bovis were extracted to confirm the secreted nature of the predicted proteins with western blotting assays. The secretome of M. bovis was extracted with a previously described method (18). Briefly, M. bovis was cultured for 36 h to late log phase and harvested by centrifugation at 140,000 × g for 20 min. The bacterial debris in the supernatant was removed with a 0.22 μm filter. The filtered solution was then precipitated with 10% trichloroacetic acid (TCA), stored at 4°C overnight, and pelleted by centrifugation at 16,000 × g for 20 min at 4°C. The pellet was washed three times with cold acetone (−20°C) and resuspended in lysis buffer [8 M urea, 4% CHAPS, 2 M thiourea, 60 mM dithiothreitol, 2% amidosulfobetaine-14, 40 mM Tris-HCl (pH 8.8)]. The whole-cell proteins of M. bovis were prepared by sonicating the cells (200 W) on ice for 5 min. The protein concentrations of the secretome and whole-cell proteins were determined with the 2D Quant Kit (GE Healthcare, Sweden) and the BCA Protein Assay Kit (Cellchip Biotechnology Company, Beijing, China), respectively.
The western blotting assay was performed as follows. The secretome and whole-cell proteins were resolved with SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Darmstadt, Germany). The mouse antisera directed against the 11 proteins and negative control sera were prediluted to 1:500 and separately overlaid onto the blotted PVDF strips. The proteins that specifically interacted with the antisera were visualized with WesternBright™ ECL (Advansta, CA, USA).
To detect the secretion process of MbovP280 and MbovP475 in vitro, the cultural supernatant of M. bovis HB0801 at 6, 12, 24, and 36 h was collected by centrifugation (15,400 g, 20 min, 4°C). Then, the supernatant (10 mL) was concentrated to 1 mL in an Amicon Ultra-4 Centrifugal Filter Unit (15 mL, 10 kDa) (Millipore). The equal volume of PPLO medium was concentrated as the negative control. The equal volume of supernatant (10 uL) from culture at each time point was resolved with SDS-PAGE and then transferred onto PVDF membranes (Millipore). The whole-cell proteins served as positive control and the membrane-associated protein NOX (7) of M. bovis was used as negative control. The bands were visualized by western blotting assay using the method described above.
BoMac cells (1 ×105) were propagated overnight on a microscope coverslip in each well of a 12-well-plate. To observe the binding of MbovP280 to BoMac, 0.5 μM MbovP280 was added to each well and incubated for 1 h at 37 °C. Phosphate-buffered saline (PBS) was used as the negative control. After the medium was removed, all the cells on the coverslips were washed three times with PBS, fixed with 4% paraformaldehyde in PBS for 10 min, and permeabilized with 0.5% Triton X-100 for 5 min at room temperature. All the cells were then blocked with 1% (w/v) bovine serum albumin (BSA) in PBS for 2 h. The cells were immunolabeled with mouse antiserum directed against rMbovP280 (1:500), and an Alexa-Fluor-488-conjugated goat anti-mouse IgG (H+L) secondary antibody (1:1,000) (Southern Biotech, MI, USA). Finally, the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) 5 mg/ml (Beyotime, Shanghai, China), and the polymerized form of actin was labeled with rhodamine phalloidin (100 nM) (Cytoskeleton, CO, USA). Finally, the slides were cover-slipped and observed with a confocal laser fluorescence microscope (Olympus FV1000 and IX81, Tokyo, Japan).
The relative viability of BoMac and RAW264.7 cells after treatment with either recombinant MbovP280 (rMbovP280) or rMbovP475 was determined with a Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan). The cells were seeded at a density of 5,000 cells/well in 96-well-plates and incubated overnight at 37°C. They were then treated in triplicate with either rMbovP280 or rMboP475 at a concentration of 1 μM for 24 h. Cells treated with PBS were used as the negative control. CCK-8 (10 μl) was then added to each well, and the samples incubated for 2 h. The optical density at a wavelength of 450 nm (OD450) was measured and the relative cell viability was calculated as:
The cells were then treated with either rMbovP280 or its mutant rMbovP280Δ210−269 at a concentration of 0.25, 0.5, or 1 μM for 24 h and the relative cell viability of BoMac was determined with the CCK-8 assay, as described above. Each treatment was carried out in five repeat and all experiments were performed independently three times.
An IP–MS method was used to screen for MbovP280-binding proteins. Briefly, BoMac cells were cultured, harvested, and lysed in RIPA buffer supplemented with cOmplete™ Protease Inhibitor Cocktail (Roche, Mannheim, Germany). The whole-cell lysates (400 μg) were incubated with 10 μg of either rMbovP280 or rMbovP280Δ210−269 for 1 h at 4°C. Mouse antiserum (5 μg) directed against MbovP280 was added to the lysates for 12 h at 4°C, and then 50 μl of Protein A/G Agarose Beads (Beyotime) were added and incubated for an additional 1 h. The immunoprecipitates were extensively washed with NP-40 buffer and eluted with SDS loading buffer by boiling them for 5 min. The cellular proteins co-precipitated with rMbovP280 or rMbovP280Δ210−269 were resolved with SDS-PAGE and stained with silver staining. The specific bands that bound only rMbovP280 were commercially sequenced and analyzed with MS by Applied Protein Technology (Shanghai, China).
To confirm the interaction between MbovP280 and α-crystallin (CRYAB) in BoMac cells, the CRYAB gene (GenBank accession no. AF029793.2) was commercially synthesized at Beijing Tianyi Huiyuan Bioscience & Technology Inc. and cloned into pCAGGS-Flag, kindly provided by Prof. Xiao Shaobo (23). The Mbov_0280 gene (GenBank accession no. AFM51648.1) of M. bovis was synthesized and cloned into pCAGGS-HA, kindly provided by Prof. Xiao Shaobo at Huazhong Agricultural University, Wuhan, China (23). Escherichia coli strain DH5α (TransGen Biotech, Beijing, China) was transformed with the individual constructed plasmids, pCAGGS-Flag–CRYAB or pCAGGS-HA–Mbov_0280. The endotoxin-free plasmids were prepared with the E.Z.N.A.® Endo-Free Plasmid Mini Kit II (Omega Bio-tek, GA, USA) and then stored at −20°C until use. For the Co-IP assay, HEK-293T cells were cotransfected with both pCAGGS-Flag–CRYAB (8 μg) and pCAGGS-HA–Mbov_0280 (8 μg) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in a 10 cm dish. After 32 h, the cells were harvested and lysed in 1.5 ml of NP-40 buffer supplemented with cOmplete™ Protease Inhibitor Cocktail (Roche). The cell lysates (600 μl) were immunoprecipitated with 2 μg of commercial antibody directed against one of the recombinant tags [either Flag or hemagglutinin (HA)] for 12 h at 4°C. After the addition of Protein A/G Agarose Beads for 1 h, the immunoprecipitates were washed extensively with NP-40 buffer and eluted with SDS loading buffer by boiling for 5 min. A western blotting assay was then performed. The samples were resolved with SDS-PAGE and transferred to PVDF membranes (Millipore). The proteins were immunodetected with antibodies directed against either Flag (MBL, Nagoya, Japan) or HA (MBL, Nagoya, Japan). MbovP280 and CRYAB reacted with the antibodies directed against the corresponding tags on the PVDF membrane and were visualized with WesternBright™ ECL (Advansta).
The Mbov_0280-knockout mutant T9.297 was identified from a transposon-mediated M. bovis mutant library previously constructed in this laboratory (24). The mutated site was at nucleotide (nt) 418 of the Mbov_0280 coding sequence (CDS) or nt 323, 346 of the M. bovis HB0801 genome.
To construct a strain to complement the mutant T9.297, the sequence of the M. agalactiae P40 promoter followed by the intact Mbov_0280 CDS was synthesized at Beijing Tianyi Huiyuan Bioscience & Technology Inc. and ligated into the pOH/P plasmid after digestion with restriction enzyme NotI, generating the recombinant plasmid pOH/P–Mbov_0280. T9.297 cells were transfected with pOH/P–Mbov_0280 to generate the complementing strain CT9.297, with a previously described method (25). Single colonies were selected with puromycin (10 μg/ml) in the medium and then confirmed with DNA sequencing. The T9.297 and CT9.297 strains were cultured in s medium containing 100 μg/ml gentamycin and 10 μg/ml puromycin, respectively, and their growth curves were determined with a colony counting method.
MbovP280 expression in mutant strain T9.297 and complementary strain CT9.297 was tested with western blotting assay. First, both strains were cultured in 20 ml of PPLO medium with the necessary antibiotics for 36 h and precipitated by centrifugation. The pellet of each strain was then suspended in 1 ml of PBS and lysed by sonication (200 W) on ice for 5 min. The proteins in the lysate were then separated with SDS-PAGE (10%) and transferred onto PVDF membrane (Millipore). The membrane was incubated with mouse antiserum (1:500) directed against rMbovP280 or rMbovP579 at room temperature for 1 h. After the membrane was washed, it was reacted with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody (1:5,000; Southern Biotech) for 1 h at room temperature. The bands on the membrane were visualized with WesternBright™ ECL (Advansta).
CRISPR/cas9 gene editing was used to mutate the CRYAB gene in the BoMac cell line as described below. Single guide RNA (sgRNA) oligonucleotides to the CRYAB gene were designed with CCTop (https://cctop.cos.uni-heidelberg.de/) (Table 1). Ten microliter mixture of 100 μM CRYAB sgRNA oligonucleotides 1 (CRYABsgRNAoligo1) and 2 (CRYABsgRNAoligo2) (each 1 μl) (Table 1), NEB buffer 2 (1 μl), and ddH2O (7 μl) were prepared, and then annealed at 37°C for 30 min, heated at 95°C for 5 min, and cooled to 25°C. The lentiCRISPRv2 plasmid (Addgene plasmid #52961; http://n2t.net/addgene:52961; RRID: Addgene_52961) (26) was cut with BsmBI and purified with the EasyPure® PCR Purification Kit (TransGen Biotech). Then 1 μl of the diluted (1:50) and annealed oligonucleotides was cloned into lentiCRISPRv2 with T4 ligase (New England Biolabs, Beijing, China) at 16°C for 12 h. Competent E. coli DH5α cells were transformed with the constructed plasmid plentiCRISPRv2-CRYABsgRNA and the construct was verified by sequencing with the primer U6-F (Table 1).
The constructed plasmid plentiCRISPRv2-CRYABsgRNA and packaging plasmids pMD2.G (Addgene plasmids #12259; http://n2t.net/addgene:12259; RRID: Addgene_12259) and psPAX2 (Addgene plasmids #12260; http://n2t.net/addgene:12260; RRID: Addgene_12260) were amplified and purified with an Endo-free Plasmid Mini Kit II (Omega Bio-tek). The lentivirus was rescued as described previously (26) with some modification as follows. HEK293T cells were transfected with 12 μg of plentiCRISPRv2-CRYABsgRNA, 8 μg of psPAX2, and 4 μg of pMD2.G in 500 μl of jetPRIME® buffer (PolyPlus, Ilkirch, France) containing 48 μl of jetPRIME® reagent (PolyPlus). After 6 h, the medium was changed to DMEM containing 10% FBS and the cells were further cultured for 48 h. The lentivirus was harvested by centrifugation at 3,000 × g at 4°C for 10 min and then filtered through a 0.45 μm membrane (Millipore), concentrated 10-folds with Lenti-Pac™ Lentivirus Concentration Solution (GeneCopoeia, MD, USA), purified as described in the manual, and resuspended in 1 ml of PBS.
The BoMac cells with 50% confluency in each well of a 24-well-plate were infected with 100 μl of the purified lentivirus for 12 h. The medium was then changed to RPMI 1640 containing 10% FBS and 1% penicillin–streptomycin, and the cells were incubated for 36 h more. The media in the infected and uninfected wells were changed to fresh RPMI 1640, as described above, but with the addition of 2 μg/ml puromycin. After 3 days, the adherent cells were propagated in one well of a six-well-plate and cultured in RPMI 1640 (10% FBS, 1% penicillin–streptomycin) with 2 μg/ml puromycin for 3 days. The cells in the wells of the six-well-plate were then counted and 10-fold serially diluted in RPMI 1640 to a final concentration of 1 cell per 100 μl. The diluted cells (100 μl) were seeded to the wells of two 96-well-plates. The clonal cell lines were isolated from the 96-well-plates after 7 days, and expanded for 14 days.
The genome of the selected mutated cell line in each well was isolated with the TIANamp Genomic DNA Kit (Tiangen, Beijing, China). Q5 High-Fidelity 2 × Master Mix (NEB, Beijing, China) and the primers CRYAB-C-F and CRYAB-C-R (Table 1) were then used to amplify the sequences edited by CRISPR/cas9. The whole-cell proteins of each mutated cell line and those of the wild type (WT) were prepared in RIPA lysis buffer (Beyotime) and then resolved with SDS-PAGE, transferred onto PVDF membrane (Millipore), and incubated with a mouse monoclonal antibody directed against CRYAB (1:1,000) (Abcam, ON, Canada) at room temperature for 1 h. The membranes were then incubated with a HRP-conjugated goat anti-mouse IgG antibody (1:5,000) (Southern Biotech) for 1 h at room temperature. The bands on the membrane were visualized with Western Bright™ ECL (Advansta).
To examine the specific effect of rMbovP280 on the induction of BoMac apoptosis, the WT BoMac and mutant BoMac cryab− cells were seeded at a density of 5 ×105 cells per well in six-well-plates and incubated overnight at 37°C. They were then treated with rMbovP280 or its mutant rMbovP280Δ210−269 at three concentrations (0.25, 0.5, or 1 μM) for 24 h.
To observe the effect of MbovP280 expression in M. bovis strains on cell apoptosis, 5 ×105 BoMac cells were seeded into each well of a six-well-plate and incubated overnight. The cells were then infected with WT M. bovis HB0801, the mutant T9.297, or complementary strain CT9.297, respectively, at an MOI of 1,000 for 24 h. The cells were harvested and stained with the Annexin V-FITC Apoptosis Detection Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions (15). An Apoptosis Inducer Kit (Beyotime) was used as the positive control. Flow cytometry (BD Company, NJ, USA) was used to detect the apoptotic cells, and the data were analyzed with the FlowJo VX software. The experiments were performed independently three times.
To confirm the apoptosis determined by Annexin V-FITC Apoptosis Detection Kit, the levels of cleaved caspase-3 in BoMac cells infected with M. bovis or treated with rMbvoP280 and rMbovP280Δ210−269 as described above were detected by western blotting assay. The cells were washed with cold PBS, resuspended with RIPA buffer containing proteinase and phosphatase inhibitors, and lysed at 4°C for 1 h. The proteins in the lysates were then separated with SDS-PAGE (10%) and transferred onto PVDF membrane (Millipore). The membrane was incubated with cleaved caspase-3 (Asp175) antibody (Cell Signaling Technology, DANVERS, MA, USA) (1:1,000) at 4°C overnight after blocking with 5% skimmed milk. After the membrane was washed, it reacted with HRP-conjugated goat anti-mouse IgG antibody (1:5,000) (Southern Biotech) for 1 h at room temperature. The bands on the membrane were visualized with WesternBright™ ECL (Advansta).
Data were expressed as means ± standard error of mean (SEM). Student's t-test was used for single comparison and one-way ANOVA for multiple comparisons with the GraphPad Prism version 5 software (GraphPad Software, La Jolla, CA, USA).
A total of 39 secreted lipoproteins were predicted with a genome-wide analysis of M. bovis based on the SignalP-TM score and the SecP score (Supplementary Table 1). Among these lipoproteins, 19 were non-classical secreted proteins, four were classical secreted proteins, and 16 were non-classical/classical secreted proteins. Using the criteria of a SignalP score >0.6 or SceP score >0.8 and the prediction of conserved domains, 11 of the predicted secreted lipoproteins were selected for further investigation, including two classical, four non-classical, and five non-classical/classical secreted proteins, and seven of which contained conserved domains. For example, MbovP537 contains the aromatic cluster surface protein domain, which suggests its surface expression; MbovP458 contains the PAP1 superfamily domain, which may regulate the transcription of antioxidant genes in response to H2O2; and MbovP682 contains the PRK08581 domain, which may function as an N-acetylmuramoyl-L-alanine amidase (Table 2).
The 11 selected genes of M. bovis were modified appropriately and cloned into E. coli. The recombinant proteins were shown to be correctly expressed, with the expected molecular masses, with SDS-PAGE (Figure 1A). The antiserum against each recombinant protein was used to detect the presence of each protein in the secretome and whole-cell proteins of M. bovis HB0801 with western blotting assays. The results indicated that all the proteins were present in large amounts in the whole-cell proteins, but only MbovP280 and MbovP475 were abundant in the secretome, whereas the other proteins were represented by only variously weak bands in the secretome (Figure 1B, Supplementary Figure 1). In order to detect the secretion process of MbovP280 and MbovP475 in vitro, the presence of MbovP280 and MbovP475 in the culture supernatant of M. bovis was kinetically examined, while the membrane-associated protein NOX served as the negative control. The results indicated the MbovP475 was detected in the supernatant after M. bovis was cultured for 24 h. But MbovP280 wasn't detected in the supernatant until M. bovis was cultured for 36 h. The membrane-associated protein NOX of M. bovis wasn't detected in the culture supernatant as expected (Figure 1C, Supplementary Figure 2).
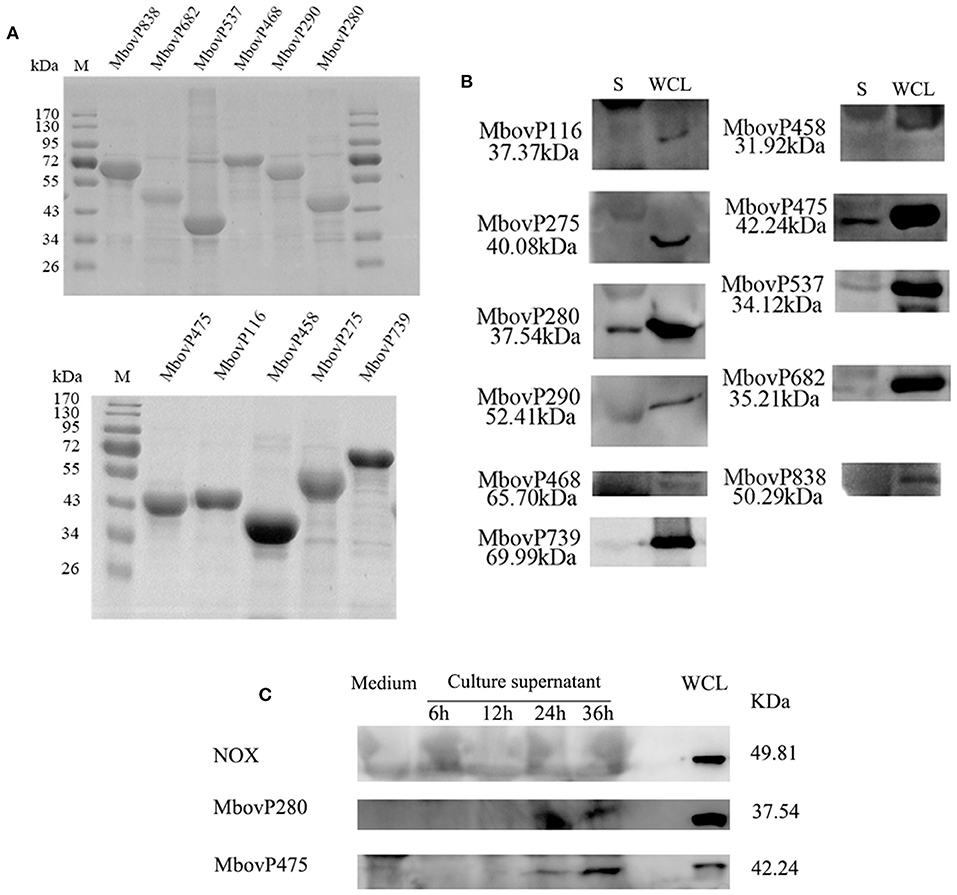
Figure 1. Secretion of MbovP280. (A) Purification of 11 predicted secreted proteins. The proteins were purified with nickel affinity chromatography and resolved with SDS-PAGE. (B) Confirmation of the predicted secreted proteins with western blotting assays. Secretome (S) and whole-cell lysate (WCL) of M. bovis were resolved with SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Polyclonal antibodies directed against rMbovP280, rMbovP290, rMbovP475, rMbovP468, rMbovP838, rMbovP537, rMbovP458, rMbovP682, rMbovP116, rMbovP275, and rMbovP739 were used to detect the proteins in the secretome. (C) Visualization of the secreted MbvoP280 in culture supernatant. M. bovis HB0801 was cultured in PPLO medium. The culture supernatant was collected and concentrated at 6, 12, 24, and 36 h after incubation. The antiserum against rMbovP280 and rMbovP475 were used to detect the proteins in the supernatant, while M. bovis NOX known as the membrane-associated protein served as the negative control (NC).
The effects of rMbovP280 and rMboP475, at concentrations of 1 μM, on the viability of BoMac and RAW264.7 cells were determined with a CCK-8 kit after the cells were treated for 24 h. The viability of mock-treated cells as the negative control was taken as 100%. As a result, the rMbovP280 significantly reduced cell viability in a cell-type-dependent way. The rMbovP280 caused the viability a reduction of ~77.3% in BoMac cells (p <0.001), but a reduction of only about 12.9% in RAW264.7 cells (p <0.05). This is in agreement with our expectation that the macrophages of bovine origin is more sensitive to MbovP280 of M. bovis, the bovine pathogen, than the cells of mouse origin. However, rMbovP475 had no significant effect on the viability of either BoMac or RAW cells (Figures 2A,B).
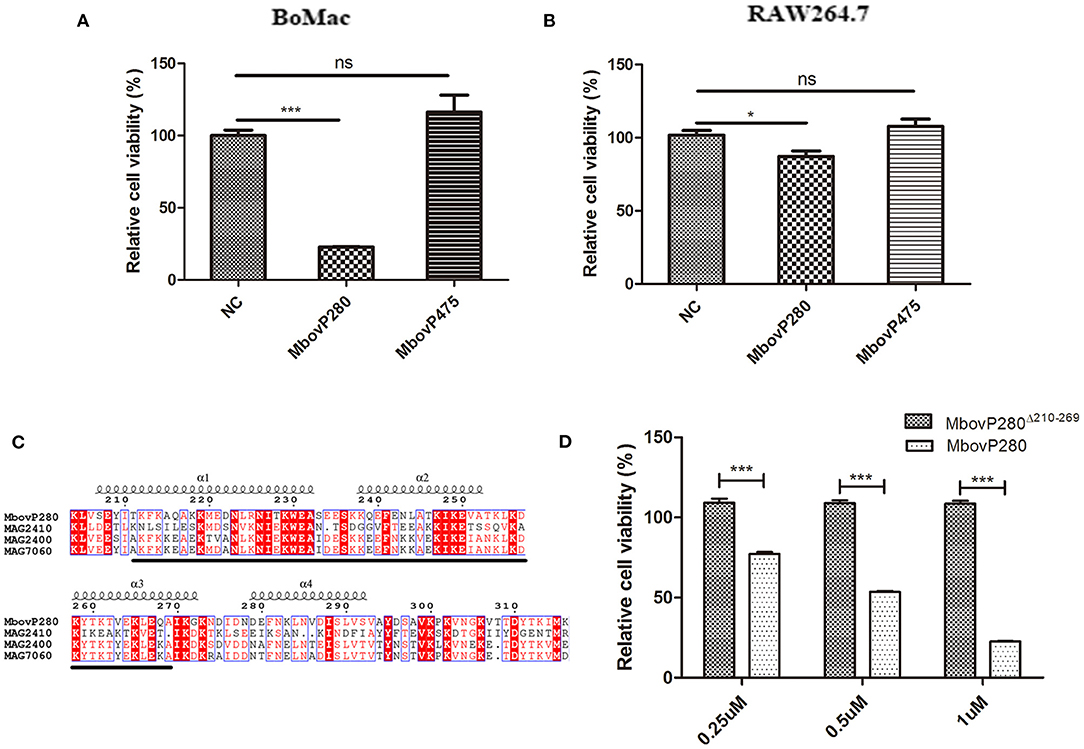
Figure 2. Viability change of macrophages stimulated with the recombinant proteins of M.bovis. (A) BoMac cells (5 ×103 per well) or (B) RAW264.7 cells (1 ×104) were treated with either 1 μM MbovP280 or 1 μM MbovP475 for 24 h, and then CCK-8 was added to each well to determine cell viability. Wells treated with PBS were used as the negative control (NC). (C) Proteins homologous to MbovP280 in Mycoplasma agalactiae were identified with MolliGen 3.0 (http://services.cbib.u-bordeaux.fr/molligen/). Amino acids 210–269 (black underline) were predicted to form a coiled-coil domain. A protein alignment was constructed with the ClustalW multiple sequence alignment programs (https://www.ebi.ac.uk/Tools/msa/clustalw2/) combined with ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/). (D) Relative cell viability of BoMac (5 ×103 cells per well) treated with MbovP280 or MbovP280Δ210−269 at concentrations of 0.25, 0.5, or 1 μM for 24 h was tested with a CCK-8 assay. *p <0.05, ***p <0.001 indicate statistically significant differences; while ns indicates no difference.
A BLAST analysis with MolliGen 3.0 revealed that MbovP280 only shares some homology with genes in M. agalactiae strain PG2, including MAG2400 (GenBank accession no. CAL58938.1, similarity = 62%), MAG2410 (GenBank accession no. CAL58939.1, similarity = 36%), and MAG7060 (GenBank accession no. CAL59406.1, similarity = 31%). MbovP280 was also predicted to contain a conserved coiled-coil domain in the fragment defined by amino acids 210–269, which was predicted with COILS. This domain is potentially involved in protein–protein interactions (Figure 2C). To verify the role of the coiled-coil domain in reducing cell viability, the Mbov_0280 gene was mutated by deleting the coiled-coil domain, and the resultant recombinant protein MbovP280Δ210−269 was expressed. The relative viability of BoMac treated with different concentrations of rMbovP280 and rMbovP280Δ210−269 was determined in parallel, with a CCK-8 kit, and decreased significantly as the rMbovP280 concentration increased from 0.25 μM to 1 μM (p <0.001). However, treatment with rMbovP280Δ210−269 at any concentration (0.25, 0.50, or 1 μM) had no effect on BoMac viability (Figure 2D).
To investigate the mechanism underlying the reduction in cell viability induced by MbovP280, the ability of rMbovP280 to reduce cell viability of BoMac was assayed with CCK-8. This assay demonstrated that rMbovP280 significantly reduced the cell viability of BoMac in a dose-dependent way (between 0.25 and 1.00 μM) (Figure 3A). In contrast, the mutant MbovP280Δ210−269 at 0.25 and 0.50 μM did not apparently reduce the cell viability of BoMac presenting the levels of cell viability similar to those in the negative control. However, 1.00 μM MbovP280Δ210−269 reduced cell viability, but the level was much lower than that in the rMbovP280-treated group (Figure 3B).
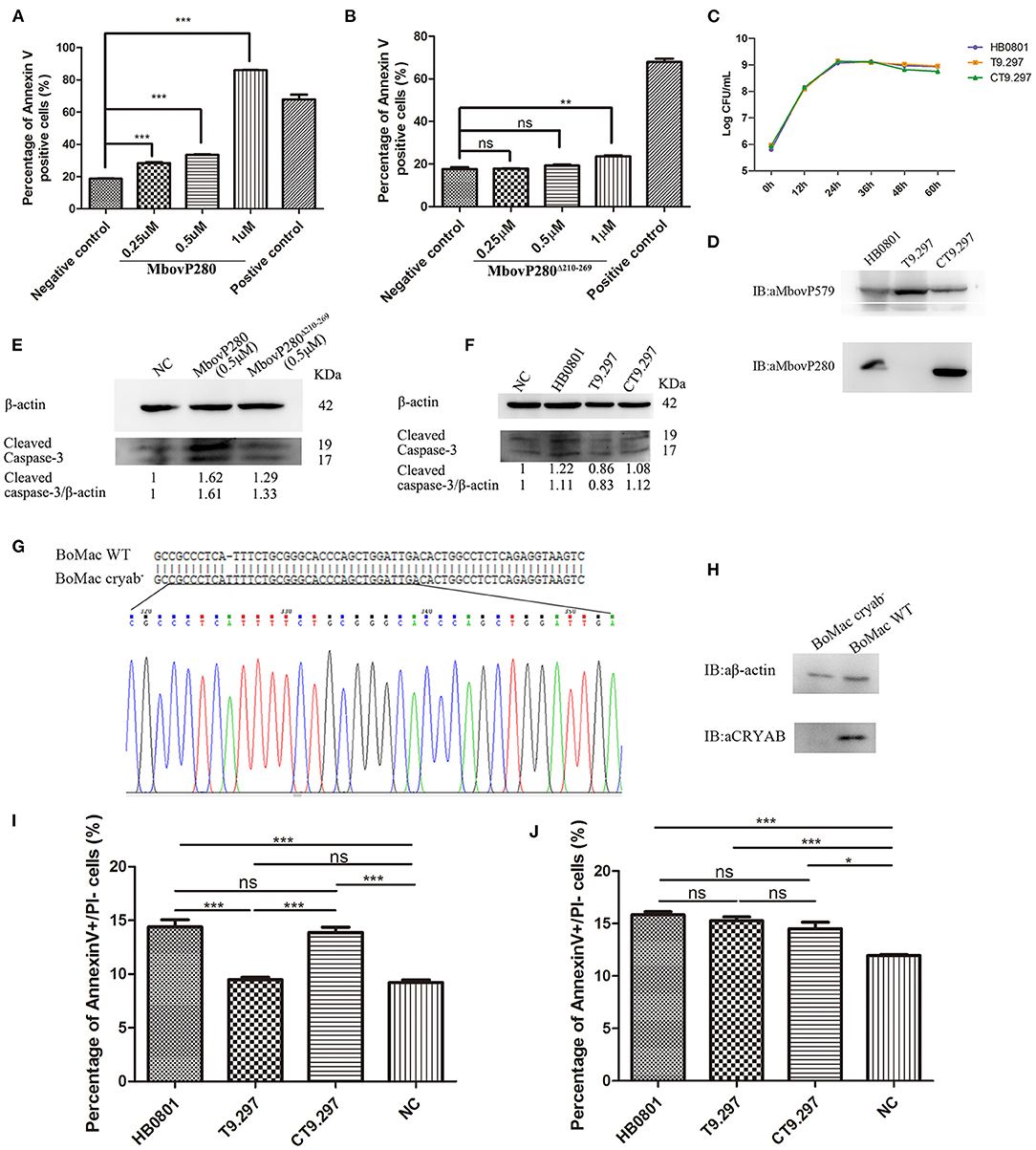
Figure 3. Apoptosis of BoMac cells induced by MbovP280 in mycoplasmas cells or as the recombinant protein. Cells (5 ×105 per well) were treated with MbovP280 (A) or MbovP280Δ210−269 (B) at concentrations of 0.25, 0.5, or 1 μM for 24 h. The cells were then stained with annexin V and propidium iodide (PI) and detected with flow cytometry. (C) Growth curves of strains HB0801, the Mbov_0280 mutant T9.297, and its complement CT9.297. Growth of M. bovis at each time point was determined with a plating assay. (D) Visualization of MbovP280 expression in T9.297 and CT9.297 strains with a western blotting assay. Wild-type strain HB0801 was used as the positive control. (E) The cleaved caspase-3 of BoMac cells treated with rMbovP280. The cell lysates of BoMac treated with 0.5 μM rMbovP280 or rMbovP280Δ210−269 were resolved with SDS-PAGE, transferred onto membrane, and then immunodetected with an antibody directed against cleaved caspase-3. β-actin was used as the internal control. The ratio of the amount of cleaved caspase-3 to the amount of β-actin was calculated and normalized to the NC. (F) The cleaved caspase-3 of BoMac cells infected with M. bovis. The cell lysates of BoMac infected with HB0801, T9.297, or CT9.297 (MOI = 1,000) were resolved with SDS-PAGE, transferred onto PVDF membrane, and then immunodetected with the antibody directed against cleaved caspase-3. β-actin was used as the internal control. The ratio of the amount of cleaved caspase-3 to the amount of β-actin was calculated and normalized to the NC. (G) Single peak around the protospacer adjacent motif (PAM) in the sequence of BoMac cryab− clonal cell line. BLAST analysis of sequences around the PAM sequence of WT BoMac (BoMac WT) and BoMac cryab− cell lines. (H) Expression of CRYAB in BoMac WT and BoMac cryab− cells. Whole-cell lysates of BoMac WT and BoMac cryab− cells were resolved with SDS-PAGE, transferred onto membrane, and then immunodetected with the antibody directed against CRYAB. β-actin was used as the internal control. (I) Early apoptosis of BoMac induced by HB0801, T9.297, or CT9.297 was compared. BoMac (5 ×105 cells per well) were treated with 5 ×108 CFU of HB0801, T9.297, or CT9.297, or with PBS for 24 h. The cells only stained with annexin V are defined as undergoing early apoptosis. (J) Early apoptosis of BoMac-cryab− cell line induced by M. bovis. BoMac (5 ×105 cells per well) were infected with HB0801 (5 ×108 CFU), T9.297 (5 ×108 CFU), or CT9.297 (5 ×108 CFU), or treated with PBS (NC) for 24 h. *p <0.05, **p <0.01, ***p <0.001 indicate statistically significant differences; ns indicates no difference.
Further, the Mbov_0280 knockout mutant T9.297 and its complementary strain CT9.297 in which the fragment of the whole gene Mbov_0280 was inserted into the mutant T9.297, were constructed and confirmed with DNA sequencing. Although the growth curves of the three strains (HB0801, CT9.297, and T9.297) did not differ significantly (Figure 3C), the western blotting assay demonstrated that MbovP280 was expressed in both M.bovis HB0801 and CT9.297 strains, but not in the mutant T9.297 strain (Figure 3D).
In order to further verify the apoptosis of BoMac specifically induced by MbovP280, the levels of cleaved caspase-3 in BoMac cells either infected with M. bovis strains or treated with rMbovP280 or its mutant were detected by western blot assays. As a result, the rMbovP280 increased the amount of cleaved caspase-3, while the rMbovP280Δ210−269 did not (Figure 3E, Supplementary Figure 3). For the infection with M. bovis strains, the wild type strain HB0801 and complementary strain CT9.297 increased the levels of cleaved caspase-3 in BoMac, however the Mbov_0280 knock-out mutant T9.297 didn't (Figure 3F, Supplementary Figure 3).
The CRYAB gene of BoMac was knocked out with the CRISPR-Cas9 lentiviral system. Then the genome of the CRYAB-knockout cell line (BoMac-cryab−) was isolated and used as the template to amplify the edited sequence. The sequencing results indicated that the CRYAB gene was correctly mutated by the insertion of an additional T at nt 159 in the CDS of the CRYAB gene (Figure 3G). The deficiency of CRYAB expression in the clonal cell line (BoMac-cryab−) was confirmed with a western blotting assay (Figure 3H).
Further, the flow cytometry was used to test the apoptosis of BoMac-cryab− cell line and its WT BoMac induced by the three strains of M. bovis: HB0801, the Mbov_0280 knock-out mutant T9.297, and its complementary strain CT9.297 after stained with annexin V and PI. As a result, the proportions of the cells at the early stage of apoptosis stained only by annexin V were apparently associated with MbovP280 stimulation. In WT BoMac, the apoptosis levels induced by HB0801, T9.297, CT9.297, and PBS were 14.40, 9.48, 13.87, and 9.21%, respectively. There is no difference in the apoptosis levels between HB0801/CT9.297; and T9.297/NC (PBS) groups (p > 0.05); However, there is a significant difference between HB0801/T9.297, CT9.297/T9.297, HB0801/NC, and CT9.297/NC (p <0.001) (Figure 3I). In contrast, in BoMac-cryab− cells, the levels of apoptosis induced by HB0801 (15.83%), T9.297 (15.27%), and CT9.297 (14.5%) did not change significantly (p > 0.05), although all three strains indeed generated some degree of cellular apoptosis compared to NC (p<0.05) suggesting there is some other mechanism in M. bovis to induce BoMac apoptosis independent of MbovP280/CRYAB (Figure 3J). Taken altogether, these results revealed that the induction of apoptosis by MbovP280 depends on the expression of CRYAB and the amino acids 210–269 in MbovP280 constitute an essential domain for this function.
Confocal laser microscopy was used to observe the binding of MbovP280 to BoMac. As shown in Figure 4A, rMbovP280 bound strongly to BoMac cells after it was incubated with the cells for 1 h at 37°C, demonstrated by the co-localization (merged yellow signal) of rMbovP280 (green) and F-actin (red).
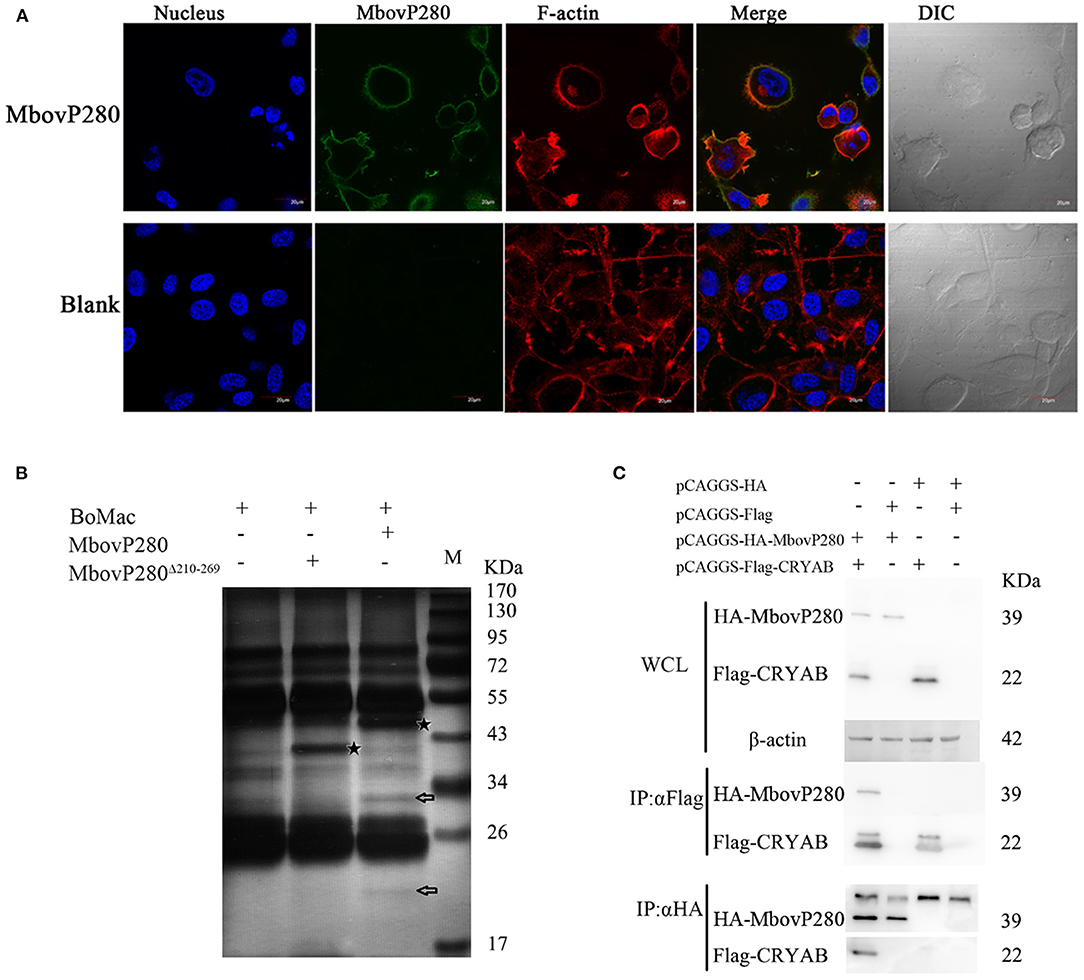
Figure 4. Interaction between MbovP280 and BoMac. (A) Binding assay of MbovP280 to BoMac cells. BoMac cells (1 ×105) were incubated with 0.5 μM MbovP280 for 1 h at 37°C. MbovP280 was probed with polyclonal antibody directed against MbovP280 (1:500) and Alexa-Fluor-488-conjugated anti-IgG antibody (1:1,000) (green). Nuclei were stained with DAPI (5 μg/ml) and actin filaments with rhodamine phalloidin (100 nM). (B) Identification of MbovP280-binding ligands with mass spectrometry. BoMac cell lysates were incubated with MbovP280 (10 μg), MbovP280Δ210−269 (10 μg), or PBS, and immunoprecipitated (IP) with a polyclonal antibody directed against MbovP280 (5 μg) and agarose bead (50 μl) labeled with Protein A/G. Arrows indicate specific MbovP280-binding ligands resolved with SDS-PAGE. And the asterisks represent either MbovP280 or MbovP280Δ210−269. (C) Interaction between MbovP280 and CRYAB. At 32 h post transfection with plasmids encoding HA, HA–MbovP280, Flag, or Flag–CRYAB, HEK293T cells were harvested and the whole-cell lysates were immunoprecipitated with commercial antibodies directed against either the HA or Flag tag, and Protein A/G agarose beads. MbovP280 and CRYAB were then detected with western blotting assays using the antibodies against either the HA or Flag tags.
Further the IP–MS method was to screen for MbovP280-binding proteins by using the antisera against rMbovP280 and rMbovP280Δ210−269 to catch the interactive components from lysates of the BoMac cells, separated by Protein A/G Agarose Beads and resolved by SDS-PAGE and specific bands were analyzed with mass spectrometry (MS). As a result, two specific bands (arrows) from BoMac were precipitated by rMbovP280, but not by rMbovP280Δ210−269. And the asterisks represent MbovP280 or MbovP280Δ210−269 (Figure 4B). Then the bands were subject to be assayed with MS and the resultant proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022080. From the MS data, CRYAB, endothelin 2, lipocalin 2, THO complex subunit 4, and DCN1-like protein were preliminarily identified. Among these, CRYAB had the highest percentage coverage (12.57%) (Table 3). CRYAB is a known anti-apoptosis protein, which is consistent with our previous finding that MbovP280 induced the apoptosis of its host cells. Therefore, further experiments were performed to verify the interaction between CRYAB and MbovP280.
HEK293T cells were cotransfected with plasmids expressing Flag–CRYAB and HA–MbovP280, and a Co-IP assay was performed. The total cellular proteins were prepared and the components immunoprecipitated with either the anti-Flag or anti-HA antibodies. The precipitates were resolved with SDS-PAGE and analyzed with either the anti-HA or anti-Flag antibody by western blotting assays. As shown in Figure 4C, Supplementary Figure 4, the coprecipitated MbovP280 was detected in the compounds precipitated with anti-Flag antibody, and the coprecipitated CRYAB was detected in the compounds precipitated with anti-HA antibody. These results confirmed that MbovP280 specifically interacted with CRYAB.
A number of software are usually used to predict the secreted proteins of Mycoplasma species in silico. The SignalP4.1 server is used to identify the classically secreted proteins, such as P80 of M. hominis (11), which are secreted after the cleavage of the signal peptide, and the SecretomeP 2.0 server is used to predict the non-classical secreted proteins, which may be secreted independently of any signal peptide (27), for example via the extracellular vesicles recently identified in M. mycoides subsp. mycoides (16) and Acholeplasma laidlawii PG8 (28). However, these predictions should be confirmed with other methods, such as western blotting assays. During this study, only two of 11 predicted secreted proteins (MbovP280 and MbovP475) were confirmed as both highly expressed and immunogenic (Figure 1B) suggesting the difficulties in identifying the secreted proteins of mycoplasmal species. Several factors might affect this process, including the reliability of prediction, the relative times and levels of expression and the immunogenicity of the secreted proteins, the complex background of proteins in the media (derived from serum and yeast extracts), and the strong contamination of the secretomes with cytoplasmic proteins during the extraction process (29). Because the lipoproteins of mycoplasmas usually function as virulence factors, such as OppA, a lipoprotein of both M. agalactiae and M. fermentans that induces apoptosis (16, 30), the secreted lipoproteins MbovP280 and MbovP475 could play critical roles in the pathogenesis of M. bovis.
It has previously been reported that M. bovis functions as an inducer or inhibitor of the apoptosis of infected cells, probably dependent on the strain or cell type infected (31, 32). For example, M. bovis induced the apoptosis of bovine peripheral blood mononuclear cells (PBMCs) (31) and macrophages (33), but is also reported to inhibit the apoptosis of PBMCs, monocytes, primary bovine macrophages, and bovine macrophages (32, 34–36). However, few studies have determined the specific mechanism responsible for this effect.
In this study, we first demonstrated that MbovP280 significantly reduced cell viability and then confirmed it induced the apoptosis of BoMac when applied as either mycoplasmal cells or the purified recombinant protein. However, there is a large discrepancy in proportions of apoptotic cells between induction of purified rMbovP280 (25–80%) and M. bovis infection (about 15%). We think this discrepancy might be caused by following reasons. Firstly, the amount of purified rMbovP280 used to treat BoMac is larger than the amount of MbovP280 naturally expressed by the HB0801 strains. Secondly, in M. bovis strains, there may be other proteins except MbovP280 function as either inducers or inhibitors of the apoptosis. Therefore, the overall effect of MbovP280 in the strain on apoptosis might be partially contradicted. Since the secreted proteins in the cultural supernatant is very low, the further study should verify whether the concentration of secreted MbovP280 of M. bovis increases or not under natural infection and the increase concentration of MbovP280 is necessary to induce apoptosis.
CRYAB is a molecular chaperone that is induced by stress and suppresses the aggregation of denatured proteins (37). It belongs to the conserved small heat shock protein family and is highly constitutively expressed in human cancers. Recent studies have shown that CRYAB inhibits apoptosis during myogenic differentiation (38) or differentiation induced by a wide range of stimuli, such as staurosporine, tumor necrosis factor α (TNF-α), UVA irradiation, okadaic acid, hydrogen peroxide, and TNF-related apoptosis-inducing ligand (TRAIL) (39–44). Several studies have demonstrated that CRYAB inhibits apoptosis by suppressing the activation of caspase 3 by interacting with its precursor (38, 45), and by interacting with Bcl-XS and Bax to suppress their translocation (41, 46). In the present study, IP-MS, and Co-IP methods were used to identify the interaction between MbovP280 and CRYAB in BoMac, and confirmed that CRYAB is a specific ligand of MbovP280. A homology model of CRYAB, MbovP280, and caspase 3 was established with SWISS-MODEL, and protein–protein docking was examined with ClusPro 2.0. The results suggested that MbovP280 and the caspase 3 precursor competitively bind to the surface pocket of the CRYAB protein (Supplementary Figure 5) to suppress the anti-apoptosis effect of CRYAB (Figure 5).
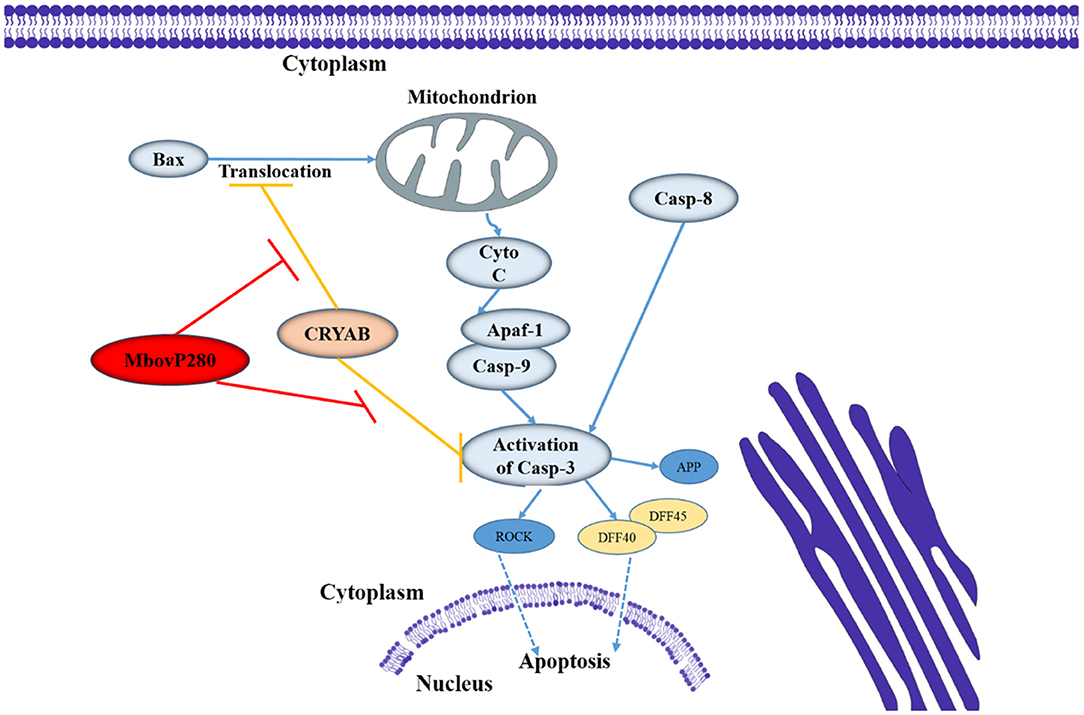
Figure 5. Diagram of the possible signaling pathway of MbovP280-induced apoptosis in BoMac. MbovP280 binds to CRYAB and induces apoptosis of BoMac cells through CRYAB.
In conclusion, in this study, we have demonstrated that MbovP280 is a highly expressed and immunogenic secreted protein that induces the apoptosis of BoMac through its ligand CRYAB and the functional domain is located at amino acids 210–269 which form a coiled-coil domain.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Experimental Animal Ethics Committee of Huazhong Agricultural University.
GZ, HZ, and AG: study design. GZ and XZ: study conduct. YC, CH, and HC: data analysis and interpretation. GZ, AG, and ES: wrote the manuscript with all authors providing feedback.
This work was supported by National Natural Science Foundation of International (Regional) Cooperation Projects (#31661143015), Special Fund for Chinese Agricultural Research System (Beef/yaks) (#CARS-37), the Special Fund for National Distinguished Scholars in Agricultural Research and Technical Innovative Team and the Fundamental Research Funds for the Central Universities (2013QC001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.619362/full#supplementary-material
Supplementary Figure 1. Secretion of predicted secreted proteins was detected with western blotting assays. Secretome (S) and whole-cell lysate (WCL) of M. bovis were resolved with SDS-PAGE, transferred onto the polyvinylidene difluoride membranes, and immunodetected with polyclonal antibodies in the antisera directed against each predicted protein.
Supplementary Figure 2. Verification of secreted MbovP280 and MbovP475 in culture supernatant with western blotting assays. M. bovis HB0801 was cultured in PPLO medium and culture supernatant was collected and concentrated at 6, 12, 24, and 36 h. Polyclonal antibodies in the antisera against rMbovP280 and rMbovP475 were used to detect the proteins in the supernatant, while the known M. bovis membrane-associated protein NOX served as negative control.
Supplementary Figure 3. MbovP280 increased the levels of cleaved caspase-3. The cell lysates of BoMac cells treated with 0.5 μM rMbovP280 or rMbovP280Δ210−269 or infected with M. bovis strains (MOI = 1,000) were resolved with SDS-PAGE, transferred onto the polyvinylidene difluoride membrane, and then immunodetected with the antibody directed against cleaved caspase-3. β-actin was used as the internal control.
Supplementary Figure 4. Interaction between MbovP280 and CRYAB. (A) Expression of MbovP280 and CRYAB in HEK293T cells at 32 h after transfection with the plasmids encoding HA or HA–MbovP280 together with the plasmids encoding Flag or Flag–CRYAB. (B) Interaction between MbovP280 and CRYAB was detected with a western blotting assay.The cell lysates were immunoprecipitated with the antibody against the Flag tag and immunoblotted with the antibody against the HA tag. (C) Interaction between MbovP280 and CRYAB was detected with a western blotting assay. The cell lysates were immunoprecipitated with the antibody against the HA tag and immunoblotted with the antibody against the Flag tag.
Supplementary Figure 5. Images of protein–protein docking. (A) Homology model of CRYAB and the MbovP280 functional domain (amino acids 206–294) was generated with SWISS-MODEL. Protein–protein docking between CRYAB and MbovP280 (amino acids 206–294) was established with ClusPro 2.0. (B) Homology model of CRYAB and caspase 3 was generated with SWISS-MODEL. Protein–protein docking between CRYAB and caspase 3 was established with ClusPro 2.0.
Supplementary Table 1. The predicted secreted lipoproteins of Mycoplasma bovis.
1. Caswell JL, Bateman KG, Cai HY, Castillo-Alcala F. Mycoplasma bovis in respiratory disease of feedlot cattle. Vet Clin North Am Food Anim Pract. (2010) 26:365–79. doi: 10.1016/j.cvfa.2010.03.003
2. Nicholas RA. Bovine mycoplasmosis: silent and deadly. Vet Rec. (2011) 168:459–62. doi: 10.1136/vr.d2468
3. Zhang R, Han X, Chen Y, Mustafa R, Qi J, Chen X, et al. Attenuated Mycoplasma bovis strains provide protection against virulent infection in calves. Vaccine. (2014) 32:3107–14. doi: 10.1016/j.vaccine.2013.12.004
4. Arcangioli MA, Duet A, Meyer G, Dernburg A, Bezille P, Poumarat F, et al. The role of Mycoplasma bovis in bovine respiratory disease outbreaks in veal calf feedlots. Vet J. (2008) 177:89–93. doi: 10.1016/j.tvjl.2007.03.008
5. Sachse K, Helbig JH, Lysnyansky I, Grajetzki C, Muller W, Jacobs E, et al. Epitope mapping of immunogenic and adhesive structures in repetitive domains of Mycoplasma bovis variable surface lipoproteins. Infect Immun. (2000) 68:680–7. doi: 10.1128/IAI.68.2.680-687.2000
6. Guo Y, Zhu H, Wang J, Huang J, Khan FA, Zhang J, et al. TrmFO, a fibronectin-binding adhesin of Mycoplasma bovis. Int J Mol Sci. (2017) 18:1732. doi: 10.3390/ijms18081732
7. Zhao G, Zhang H, Chen X, Zhu X, Guo Y, He C, et al. Mycoplasma bovis NADH oxidase functions as both a NADH oxidizing and O2 reducing enzyme and an adhesin. Sci Rep. (2017) 7:44. doi: 10.1038/s41598-017-00121-y
8. Wang Y, Liu S, Li Y, Wang Q, Shao J, Chen Y, et al. Mycoplasma bovis-derived lipid-associated membrane proteins activate IL-1beta production through the NF-kappaB pathway via toll-like receptor 2 and MyD88. Dev Comp Immunol. (2016) 55:111–8. doi: 10.1016/j.dci.2015.10.017
9. Gondaira S, Higuchi H, Iwano H, Nakajima K, Kawai K, Hashiguchi S, et al. Cytokine mRNA profiling and the proliferative response of bovine peripheral blood mononuclear cells to Mycoplasma bovis. Vet Immunol Immunopathol. (2015) 165:45–53. doi: 10.1016/j.vetimm.2015.03.002
10. Zbinden C, Pilo P, Frey J, Bruckmaier RM, Wellnitz O. The immune response of bovine mammary epithelial cells to live or heat-inactivated Mycoplasma bovis. Vet Microbiol. (2015) 179:336–40. doi: 10.1016/j.vetmic.2015.07.007
11. Hopfe M, Hoffmann R, Henrich B. P80, the HinT interacting membrane protein, is a secreted antigen of Mycoplasma hominis. BMC Microbiol. (2004) 4:46. doi: 10.1186/1471-2180-4-46
12. Djordjevic SP, Cordwell SJ, Djordjevic MA, Wilton J, Minion FC. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect Immun. (2004) 72:2791–802. doi: 10.1128/IAI.72.5.2791-2802.2004
13. Yamamoto T, Kida Y, Sakamoto Y, Kuwano K. Mpn491, a secreted nuclease of Mycoplasma pneumoniae, plays a critical role in evading killing by neutrophil extracellular traps. Cell Microbiol. (2017) 19:e12666. doi: 10.1111/cmi.12666
14. Kannan TR, Krishnan M, Ramasamy K, Becker A, Pakhomova ON, Hart PJ, et al. Functional mapping of community-acquired respiratory distress syndrome (CARDS) toxin of Mycoplasma pneumoniae defines regions with ADP-ribosyltransferase, vacuolating and receptor-binding activities. Mol Microbiol. (2014) 93:568–81. doi: 10.1111/mmi.12680
15. Zhang H, Zhao G, Guo Y, Menghwar H, Chen Y, Chen H, et al. Mycoplasma bovis MBOV_RS02825 encodes a secretory nuclease associated with cytotoxicity. Int J Mol Sci. (2016) 17:628. doi: 10.3390/ijms17050628
16. Gaurivaud P, Ganter S, Villard A, Manso-Silvan L, Chevret D, Boule C, et al. Mycoplasmas are no exception to extracellular vesicles release: revisiting old concepts. PLoS ONE. (2018) 13:e0208160. doi: 10.1371/journal.pone.0208160
17. Paes JA, Lorenzatto KR, de Moraes SN, Moura H, Barr JR, Ferreira HB. Secretomes of Mycoplasma hyopneumoniae and Mycoplasma flocculare reveal differences associated to pathogenesis. J Proteomics. (2017) 154:69–77. doi: 10.1016/j.jprot.2016.12.002
18. Rebollo Couto MS, Klein CS, Voss-Rech D, Terenzi H. Extracellular proteins of Mycoplasma synoviae. ISRN Vet Sci. (2012) 2012:802308. doi: 10.5402/2012/802308
19. Rasheed MA, Qi J, Zhu X, Chenfei H, Menghwar H, Khan FA, et al. Comparative genomics of Mycoplasma bovis strains reveals that decreased virulence with increasing passages might correlate with potential virulence-related factors. Front Cell Infect Microbiol. (2017) 7:177. doi: 10.3389/fcimb.2017.00177
20. Qi J, Guo A, Cui P, Chen Y, Mustafa R, Ba X, et al. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese Isolate). PLoS ONE. (2012) 7:e38239. doi: 10.1371/journal.pone.0038239
21. Stabel JR, Stabel TJ. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet Immunol Immunopathol. (1995) 45:211–20. doi: 10.1016/0165-2427(94)05348-V
22. Khan F, Chao J, Liu K, Chen X, Zhao G, Menghwar H, et al. Immunoproteomic identification of MbovP579, a promising diagnostic biomarker for serological detection of Mycoplasma bovis infection. Oncotarget. (2016) 7:39376–95. doi: 10.18632/oncotarget.9799
23. Wang D, Chen J, Yu C, Zhu X, Xu S, Fang L, et al. 2019. Porcine reproductive and respiratory syndrome virus nsp11 antagonizes type I interferon signaling by targeting IRF9. J Virol. (2019) 93:e00623-19. doi: 10.1128/JVI.00623-19
24. Zhu X, Baranowski E, Dong Y, Li X, Hao Z, Zhao G, et al. An emerging role for cyclic dinucleotide phosphodiesterase and nanoRNase activities in Mycoplasma bovis: securing survival in cell culture. PLoS Pathog. (2020) 16:e1008661. doi: 10.1371/journal.ppat.1008661
25. Baranowski E, Guiral S, Sagne E, Skapski A, Citti C. Critical role of dispensable genes in Mycoplasma agalactiae interaction with mammalian cells. Infect Immun. (2010) 78:1542–51. doi: 10.1128/IAI.01195-09
26. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. (2014) 11:783–4. doi: 10.1038/nmeth.3047
27. Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. (2005) 5:58. doi: 10.1186/1471-2180-5-58
28. Chernov VM, Mouzykantov AA, Baranova NB, Medvedeva ES, Grygorieva TY, Trushin MV, et al. Extracellular membrane vesicles secreted by mycoplasma Acholeplasma laidlawii PG8 are enriched in virulence proteins. J Proteomics. (2014) 110:117–28. doi: 10.1016/j.jprot.2014.07.020
29. Weng Y, Sui Z, Shan Y, Jiang H, Zhou Y, Zhu X, et al. In-depth proteomic quantification of cell secretome in serum-containing conditioned medium. Anal Chem. (2016) 88:4971–8. doi: 10.1021/acs.analchem.6b00910
30. Hopfe M, Henrich B. OppA, the ecto-ATPase of Mycoplasma hominis induces ATP release and cell death in HeLa cells. BMC Microbiol. (2008) 8:55. doi: 10.1186/1471-2180-8-55
31. Vanden Bush TJ, Rosenbusch RF. Mycoplasma bovis induces apoptosis of bovine lymphocytes. FEMS Immunol Med Microbiol. (2002) 32:97–103. doi: 10.1111/j.1574-695X.2002.tb00540.x
32. Maina T, Prysliak T, Perez-Casal J. Mycoplasma bovis delay in apoptosis of macrophages is accompanied by increased expression of anti-apoptotic genes, reduced cytochrome C translocation and inhibition of DNA fragmentation. Vet Immunol Immunopathol. (2019) 208:16–24. doi: 10.1016/j.vetimm.2018.12.004
33. Burgi N, Josi C, Burki S, Schweizer M, Pilo P. Mycoplasma bovis co-infection with bovine viral diarrhea virus in bovine macrophages. Vet Res. (2018) 49:2. doi: 10.1186/s13567-017-0499-1
34. van der Merwe J, Prysliak T, Perez-Casal J. Invasion of bovine peripheral blood mononuclear cells and erythrocytes by Mycoplasma bovis. Infect Immun. (2010) 78:4570–8. doi: 10.1128/IAI.00707-10
35. Mulongo M, Prysliak T, Scruten E, Napper S, Perez-Casal J. In vitro infection of bovine monocytes with Mycoplasma bovis delays apoptosis and suppresses production of gamma interferon and tumor necrosis factor alpha but not interleukin-10. Infect Immun. (2014) 82:62–71. doi: 10.1128/IAI.00961-13
36. Suleman M, Prysliak T, Clarke K, Burrage P, Windeyer C, Perez-Casal J. Mycoplasma bovis isolates recovered from cattle and bison (Bison bison) show differential in vitro effects on PBMC proliferation, alveolar macrophage apoptosis and invasion of epithelial and immune cells. Vet Microbiol. (2016) 186:28–36. doi: 10.1016/j.vetmic.2016.02.016
37. Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. (1992) 89:10449–53. doi: 10.1073/pnas.89.21.10449
38. Kamradt MC, Chen F, Sam S, Cryns VL. The small heat shock protein αB-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J Biol Chem. (2002) 277:38731–36. doi: 10.1074/jbc.M201770200
39. Mao YW, Xiang H, Wang J, Korsmeyer S, Reddan J, Li DW. Human bcl-2 gene attenuates the ability of rabbit lens epithelial cells against H2O2-induced apoptosis through down-regulation of the alpha B-crystallin gene. J Biol Chem. (2001) 276:43435–45. doi: 10.1074/jbc.M102195200
40. Li DW, Xiang H, Mao YW, Wang J, Fass U, Zhang XY, et al. Caspase-3 is actively involved in okadaic acid-induced lens epithelial cell apoptosis. Exp Cell Res. (2001) 266:279–91. doi: 10.1006/excr.2001.5223
41. Mao YW, Liu JP, Xiang H, Li DW. Human alphaA- and alphaB-crystallins bind to Bax and Bcl-X(S) to sequester their translocation during staurosporine-induced apoptosis. Cell Death Differ. (2004) 11:512–26. doi: 10.1038/sj.cdd.4401384
42. Kamradt MC, Lu M, Werner ME, Kwan T, Chen F, Strohecker A, et al. The small heat shock protein alpha B-crystallin is a novel inhibitor of TRAIL-induced apoptosis that suppresses the activation of caspase-3. J Biol Chem. (2005) 280:11059–66. doi: 10.1074/jbc.M413382200
43. Adhikari AS, Singh BN, Rao KS, Rao Ch M. alphaB-crystallin, a small heat shock protein, modulates NF-kappaB activity in a phosphorylation-dependent manner and protects muscle myoblasts from TNF-alpha induced cytotoxicity. Biochim Biophys Acta. (2011) 1813:1532–42. doi: 10.1016/j.bbamcr.2011.04.009
44. Xu F, Yu H, Liu J, Cheng L. αB-crystallin regulates oxidative stress-induced apoptosis in cardiac H9c2 cells via the PI3K/AKT pathway. Mol Biol Rep. (2012) 40:2517–26. doi: 10.1007/s11033-012-2332-2
45. Kamradt MC, Chen F, Cryns VL. The small heat shock protein alpha B-crystallin negatively regulates cytochrome c- and caspase-8-dependent activation of caspase-3 by inhibiting its autoproteolytic maturation. J Biol Chem. (2001) 276:43435–45. doi: 10.1074/jbc.C100107200
Keywords: Mycoplasma bovis, MbovP280, secreted protein, apoptosis, CRYAB, macrophage
Citation: Zhao G, Zhu X, Zhang H, Chen Y, Schieck E, Hu C, Chen H and Guo A (2021) Novel Secreted Protein of Mycoplasma bovis MbovP280 Induces Macrophage Apoptosis Through CRYAB. Front. Immunol. 12:619362. doi: 10.3389/fimmu.2021.619362
Received: 20 October 2020; Accepted: 25 January 2021;
Published: 15 February 2021.
Edited by:
Maria Teresa Ochoa, University of Southern California, United StatesReviewed by:
José Ángel Gutiérrez-Pabello, National Autonomous University of Mexico, MexicoCopyright © 2021 Zhao, Zhu, Zhang, Chen, Schieck, Hu, Chen and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aizhen Guo, YWl6aGVuQG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.