
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol., 10 March 2021
Sec. T Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.613591
This article is part of the Research TopicThe Role of Biomembranes and Biophysics in Immune Cell SignalingView all 11 articles
T cells communicate with the environment via surface receptors. Cooperation of surface receptors regulates T-cell responses to diverse stimuli. Recently, finger-like membrane protrusions, microvilli, have been demonstrated to play a role in the organization of receptors and, hence, T-cell activation. However, little is known about the morphogenesis of dynamic microvilli, especially in the cells of immune system. In this review, I focus on the potential role of lipids and lipid domains in morphogenesis of microvilli. Discussed is the option that clustering of sphingolipids with phosphoinositides at the plasma membrane results in dimpling (curved) domains. Such domains can attract phosphoinositide-binding proteins and stimulate actin cytoskeleton reorganization. This process triggers cortical actin opening and bundling of actin fibres to support the growing of microvilli. Critical regulators of microvilli morphogenesis in T cells are unknown. At the end, I suggest several candidates with a potential to organize proteins and lipids in these structures.
T lymphocytes, important supervisors of the immune system, are activated and regulated through receptors expressed on their surface. Surface of lymphocytes is densely covered by membrane protrusions, mainly microvilli (1, 2), which allow for a more complex three-dimensional (3D) organization of receptors compared to a flat membrane. Indeed, critical receptors of T-cell activation, T cell receptor (TCR), CD2, CD4 and CD28 were shown to accumulate at the tips of microvilli in recent studies benefitting from 3D imaging at high resolution (1, 3–7). On the contrary, CD45 is excluded from these areas (6, 7). It was suggested that non-random 3D distribution of receptors is important for optimisation of signalling and cellular responses (8–10). However, little is known about the origin of microvilli and molecules involved in their formation and homeostasis in T lymphocytes. Insight into molecular biophysics and structural details of these membrane protrusions can help to better understand T-cell function in health and disease.
In this work, I suggest the role of lipids and lipid domains in deformation of membranes and their potential role in the formation and organization of microvilli. I start with a brief introduction to microvilli structure and function. These data almost exclusively originate from studies of microvilli in epithelial cells. It is thus important to note here that microvilli of epithelial cells are more stable and may differ in structural details when compared to microvilli on leukocytes. In the central sections, I hypothezise a role of curved lipid domains in microvilli formation and describe regulatory role of lipids for the function of proteins localized prevalently to these structures. I finish by discussing a handful of molecules with a potential role in morphogenesis of T-cell microvilli. Like lipid domains, function of these proteins in T cells needs to be determined.
Microvilli are finger-like membrane protrusions at the surface of metazoan cells (11). Microvilli consist of the tip, shaft, and base, which connects these structures to the plasma membrane and cortical actin (Figure 1). Actin bundles determine a shape of microvilli and are responsible for their stability, but also a dynamic character. In the shaft, the membrane is tightly linked to actin bundles via actin- and membrane-binding proteins [e.g., myosins and ERM proteins; see Figure 1 (12)]. At the base, at least in epithelial cells, actin bundle terminates in the network of intermediate filaments known as ‘terminal web’ (13, 14). The size of microvilli is regulated by the growth of actin fibres at the tip (15). Microvilli on the surface of polarized epithelial cells covering organs in direct contact with the exterior are rather stable and long (11). Microvilli on T cells are smaller and highly dynamic (1, 2, 16, 17). With ~100 nm in a diameter and a length of 0.5–5 µm, they represent rather small surface structures [Figure 1 in (17) and Figure 1 in (1)]. It is their abundance and flexibility, which makes these structures important for T-cell function. For example, vesicles with receptors and other effector molecules can be shed off the microvilli tips. This phenomenon was observed in epithelia, as well as in T cells, and can be part of complex regulatory mechanisms in multicellular organisms (1, 18).
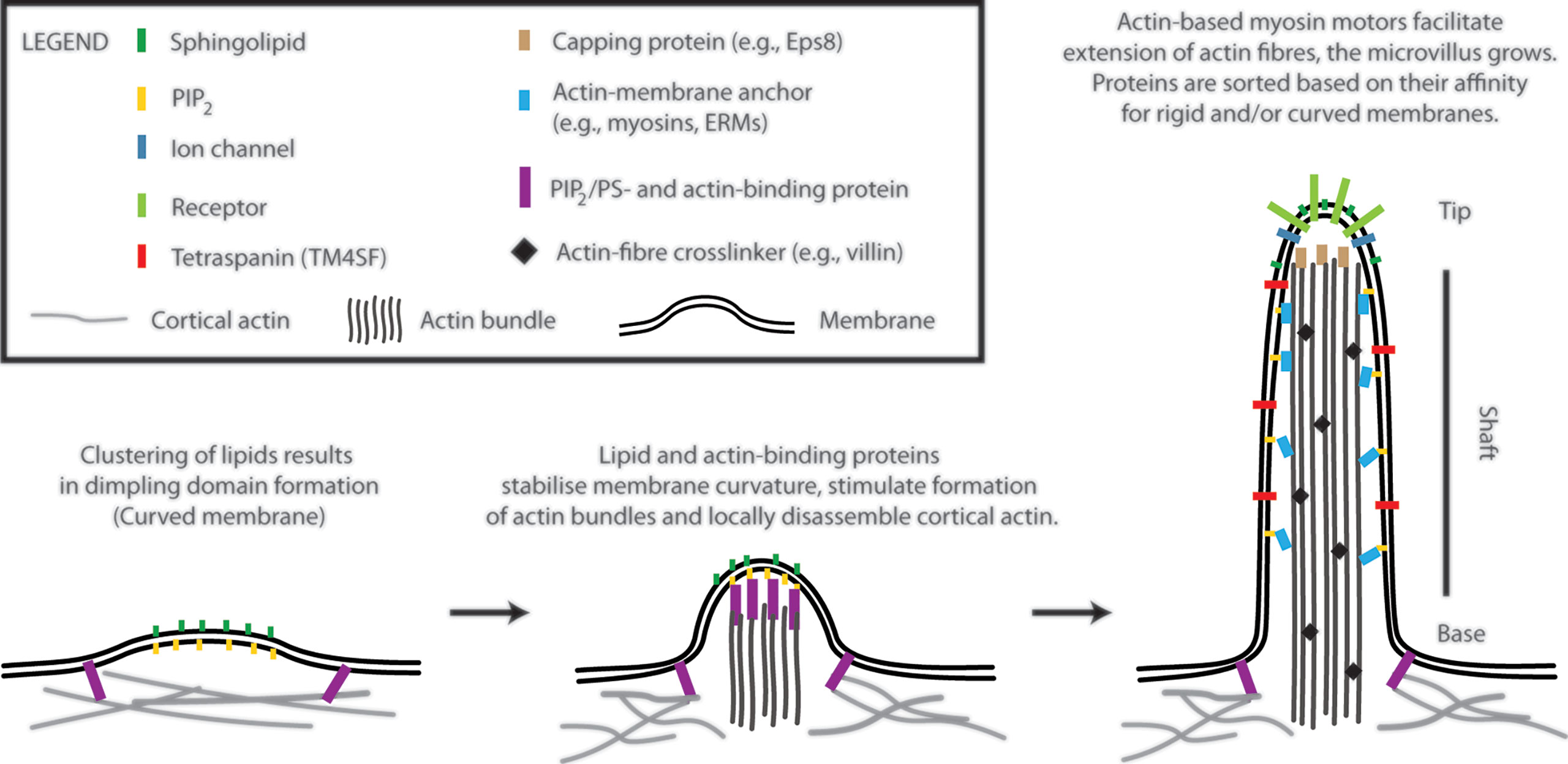
Figure 1 A model of T-cell microvilli morphogenesis. The onset of microvilli can be triggered by transient formation of dimpling lipid domains (see also Figure 2). Sphingolipids together with phosphoinositides (e.g., phosphatidylinositol 4,5-bisphosphate; PIP2) own a high potential to form dimpling domains in asymmetric membranes. Later, actin-binding proteins, which associate with plasma membrane via PIP2 (or phosphatidylserine), induce cortical actin opening and stabilize dimpling domains. Similar proteins can stimulate bundling of actin fibres. The growth of microvilli is driven by polymerization of actin at the plus end of the fibres (distal end of microvilli). Myosins dynamically anchor actin bundles to the membrane at the shaft. ERM (ezrin, radixin, and moesin) proteins function in a similar fashion (membrane anchor) and regulate stability of microvilli. Proteins with affinity for rigid (sphingolipid-enriched) and/or for curved membranes accumulate at the tip or shaft of microvilli. Little is known about T-cell signalling molecules in the lumen of microvilli. The existence of terminal web in microvilli of leukocytes remains unknown. Components of microvilli are not drawn in scale.
The accumulation of receptors at the tip of microvilli is beneficiary for an easy access to ligands, substrates or mechanical forces and can determine cellular responses to such stimuli (8, 9, 11). The shaft and the base potentially function as a selecting region, segregating molecules to different membrane environments. However, physico-chemical basis of such selection remains unknown. Importantly, it is still unclear what defines a local onset and chemical composition of microvilli. In the following sections, I suggest a model (Figure 1), in which membrane lipids and their physico-chemical properties trigger the onset of microvilli formation.
In our review on membrane lipid nanodomains [(19), Section 8.4], we discussed a role of curvature in stabilization of domains and prevention of their fusion. In general, formation of a lipid domain with different properties (e.g., rigidity and thickness) compared to the adjacent membrane results in line tension at the boundary (borderline) between the two ‘phases’ (Figure 2A). In a growing domain, the length of boundary increases, and line tension rises. However, lipid membranes prefer to minimize tensions associated with their organization (20, 21). Since elastic properties (bending modulus) of membranes are not changing significantly, the size of a domain can reach the point, at which membrane starts to bend and form dimpling domains [Figure 2B (22, 23)]. This is caused by the fact that line tension at the boundary exceeds the bending energy (resistance) of a membrane required for its deformation. Membrane bending reduces the boundary length and, thus, line tension. Further growth of a domain is enabled by enhanced curvature, which can result in membrane tubulation. The length of boundary and line tension remain constant for such growing domain/protrusion (21).
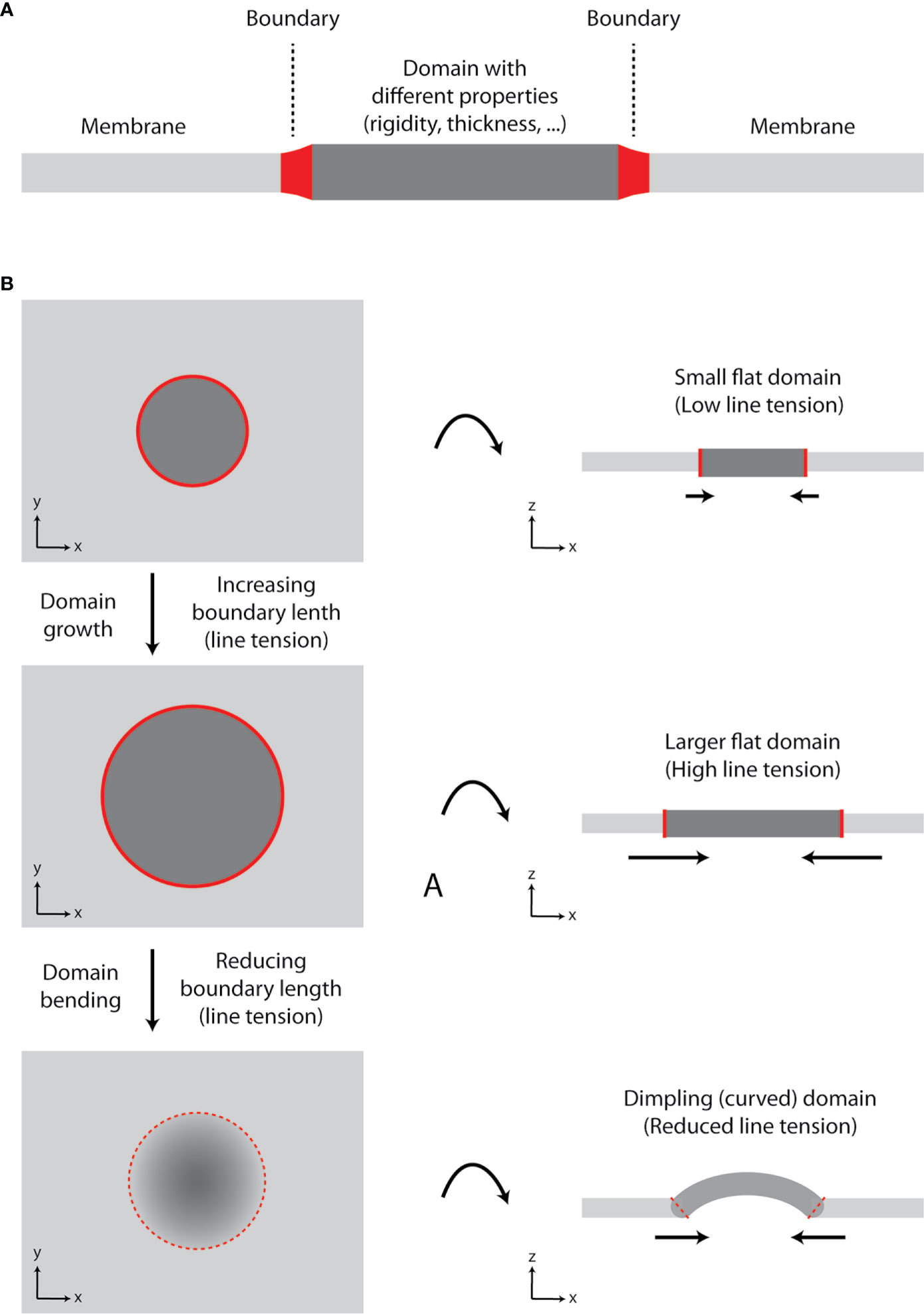
Figure 2 Growth of lipid nanodomains and dimpling (curved) domain formation. (A) Schematic illustration of lipid membrane with a domain. The domain has different physico-chemical properties (e.g., rigidity-conformational order, thickness) compared to the surrounding lipid bilayer. The two environments are separated by the boundary. (B) Schematic illustration of a domain growth and formation of a dimpling domain. Certain lipids (e.g., sphingolipids and cholesterol) tend to segregate into circular domains in synthetic membranes containing unsaturated glycerophospholipids due to their immiscibility at lower temperatures or in the presence of other clustering factors (e.g., proteins). As the domain grows, line tension at the boundary increases, until it reaches the point, at which it exceeds bending energy required for membrane deformation and dimpling domain is formed. The length of the boundary is reduced, and further growth of the membrane is accompanied by membrane tubulation, but not increase in line tension. Hence, domain formation can lead to induction of membrane curvature and its tubulation.
In flat membranes, small domains diminish due to their fusion into larger entities, as observed in model, phase-separated giant unilamellar vesicles (24). Fusion of small domains reduces the length of boundary and line tension (23). However, large lipid domains are not frequent in cells. It is currently agreed that the plasma membrane is highly heterogenous due to the presence of small (nanometric) domains (25, 26). One can thus speculate that plasma membrane is prone to form dimpling domains, which cannot fuse due to repulsive forces at their boundaries (19, 22).
Cellular membranes are composed of a large variety of lipid species. Among those, sphingolipids, with their long and saturated acyl chains and affinity to cholesterol, are prone to segregate from unsaturated glycerophospholipids and form nanodomains (19, 27–29). Ikenouchi and colleagues suggested that sphingolipids are required for the existence of microvilli and, potentially, also initiation of their formation in epithelial cells (30). Conversion of sphingomyelin to ceramides by acidic sphingomyelinase in these cells led to impaired microvilli. In untreated cells, sphingolipids accumulated on microvilli (30). Accumulation of sphingomyelin (and cholesterol) in microvilli was confirmed in another study, which employed lysenin labeling of sphingomyelin (perfolysin O for cholesterol) and sensitive nanoSIMS imaging in CHO-K1 epithelial-like cells (31). Of note, only freely accessible lipids could be detected using this method. In another study, interference with sphingolipid or cholesterol synthesis lead to reduced presence of microvilli on epithelial cells (32). All these studies indicate that sphingolipids are essential for the morphogenesis of microvilli.
Membrane lipid composition considerably differs between various cell types. Though sphingolipids consistently constitute 20-40% of plasma membrane lipids (33, 34). Local concentration of sphingolipids is even higher due to chemical asymmetry of the plasma membrane (lipid bilayer). Such high content of sphingolipids in the outer leaflet can lead to their transient clustering and, occasionally, formation of dimpling domains. Indeed, bilayer asymmetry reduces bending modulus of a membrane and, thus, facilitates its deformation (22, 35). In cells, phosphatidylinositol 4,5-bisphosphate (PIP2) molecules were found to cluster underneath sphingolipid domains during membrane deformation induced by viral proteins [virion budding (36, 37)]. PIP2 was also found to accumulate in microvilli (30). But comprehensive analysis of lipids in microvilli has not been performed to date (38). Therefore, it is unclear what is the content of PIP2, sphingolipids and other lipids (e.g., cholesterol) in these structures.
The presence of PIP2 in the apical membrane of epithelial cells, but of phosphatidylinositol 3,4,5-trisphosphate (PIP3) in basolateral membrane, further supports the involvement of this lipid in microvilli formation (39). Microvilli can be found only on the apical surface of epithelial cells. Apical membrane of polarized cells is also enriched in sphingomyelin and cholesterol (40). Moreover, PIP2 accumulates in the uropod of motile cells, whereas PIP3 can be found in the leading edge. Microvilli are often observed at the back of motile cells, including T cells (17, 41, 42). In analogy to sphingomyelin domains, cholesterol facilitates clustering of PIP2 (43). Due to its high lateral and transbilayer mobility (44, 45), cholesterol is expected to freely access dimpling domains. Interestingly, cholesterol does not influence bending modulus of synthetic membranes with diverse lipid composition (46, 47). Thus, cholesterol does not directly raise the energy required for membrane deformation and establishment of dimpling membranes, but the effect can depend on its intramembrane orientation and distribution between the outer and inner leaflet (45).
To further highlight the importance of lipids in microvilli morphogenesis, I will describe three examples where lipid metabolism determines the function of critical proteins in microvilli. The examples were selected based on the depth of our understanding of these regulatory processes. As in the case of microvilli structure, this knowledge comes from microvilli of epithelial cells, but similar regulatory mechanisms can be expected in T-cells.
ERM family proteins (ezrin, radixin, and moesin) tightly anchor actin-bundles to the membrane of microvilli. This is facilitated by binding of their FERM domain to PIP2 (48). The process is regulated by a local lipid environment. Conversion of sphingomyelin to ceramide and of sphingosine to sphingosine-1-phosphate negatively and positively, respectively, regulate membrane-association of ERM proteins and, thus, stability of microvilli (49, 50). The role of ERM proteins for microvilli is evidently critical, since their knock-down leads to their reduced size and number (51, 52).
Podocalyxin-1 accumulates in microvilli of epithelial cells. Podocalyxin-1 interacts with ERM proteins via EBP50 (53). It further interacts with phosphoinositide-4-phosphate 5-kinase (PI5K) β and delivers this critical enzyme to microvilli. The formation of podocalyxin-1 multiprotein complex with PI5K leads to a local increase in PIP2 synthesis and stability of microvilli (30). Interestingly, podocalyxin-1 associates with sphingolipid domains, probably upon its palmitoylation (54). The crosstalk of diverse lipids in the regulation of this protein remains unknown.
Another protein associating with sphingolipid domains on microvilli is prominin-1 [also called CD133 (55)]. Overexpression of prominin-1 increases a number of microvilli (56). This protein directly binds cholesterol and GM1 ganglioside (57). These lipid-protein interactions were found essential for fine tuning of microvillar structure. The protein is further regulated by phosphorylation of its regulatory tyrosines [Y817/Y828 (56)]. Phosphorylation of these tyrosines regulates interaction of prominin-1 with phosphoinositide 3-kinase (PI3K). In contrast to PI5K, PI3K locally reduces available PIP2 by its conversion to PIP3 and destabilizes the anchorage of actin bundles to the membrane (56).
I have argued above that lipid domains induce curvature in flat regions of the plasma membrane. Such domains would be transient in the absence of supporting proteins (Figure 1). The process is well described for the endocytosis or viral budding (36, 37, 58–61). For example, matrix proteins (e.g., Gag of HIV-1) form a dome-like structure under the curved membrane of nascent viral particles.
Proteins stabilizing dimpling domains at the sites of newly assembling microvilli have not been described yet. A few proteins (e.g., prominin-1/CD133, podocalyxin-1) reported to regulate microvilli morphogenesis in epithelial cells (30, 55), are not expressed in T cells or at highly variable levels in diverse T-cell subsets. Their role in microvilli morphogenesis in T cells is thus questionable. Here, I will focus on four proteins (protein families), which exhibit great potential to induce or stabilize microvilli in T cells.
The geometry and chemistry of dimpling domains delineates properties of potential supporting proteins. These must interact with negative curvature and anionic lipids. I-BAR domain proteins exhibit such properties. IRSp53 contains I-BAR domain and was shown to induce negative curvature and tubulation in synthetic vesicles (62). IRSp53 localizes to curved membranes of neuronal cells (63) and filopodia of motile fibroblasts (64). It supports membrane ruffling and protrusions in T cells (65). In epithelial cells, it is expressed at the microvilli-containing apical membrane and functionally associates with podocalyxin-1 (66). As a protein of countless functions, it will be important to characterize its specific role in microvilli of T cells. Alternatively, other I-BAR domain-containing proteins can fulfil this function in lymphoid cells.
Tetherin (also called CD317) with affinity for ordered, sphingolipid-rich membranes interacts with BAR domain-containing RICH family proteins (67). Tetherin/RICH-2 complex forms a mechanical support of epithelial microvilli (68). Its analog, RICH-1, is expressed in T-cells (Human Protein Atlas). BAR domain of RICH proteins can induce positive curvature and tubulate lipid vesicles containing PIP2 in the absence of tetherin (69). The potential of tetherin/RICH complex thus lies at the neck connecting microvilli (or dimpling domains) to membrane base via a positively curved segment (Figure 1).
Unconventional myosins (e.g., myo1a, myo7b) link actin fibres to membrane by their interaction with anionic lipids, PIP2 or phosphatidylserine (70). Myosins also contribute to the formation of a ‘hole’ in the cortical actin at the site of new microvillus formation (71). Such local depletion of cortical actin is essential for the initiation of membrane protrusions (72). This process may be also connected to the formation and stabilization of dimpling lipid domains.
Members of tetraspanin protein superfamily (TM4SF) accumulate at the microvilli of diverse cells. CD9, CD81, CD82, and TSPAN33 were shown to control the size and shape of microvilli in both, leukocytes and epithelial cells (73–75). TM4SF proteins (e.g., CD81) require highly curved membrane for their assembly into virus-like particles induced by HIV-1 Gag protein (37, 76). The main role of TM4SF is thus expected for growing or established microvilli with highly curved tubular membrane.
None of the proteins mentioned in this section was already determined as a microvilli regulator in T cells. However, I believe that intense research in this direction may soon offer interesting discoveries related not only to microvilli, but also to T-cell signaling and function.
Recent observations demonstrate that microvilli play essential role in T-cell activation. Key signalling molecules were found to accumulate in different parts of these morphological structures. Theoretical and biophysical studies indicate that sphingolipids and phosphoinositides in complex asymmetric membranes tend to generate dimpling domains. In the plasma membrane of T cells, dimpling domains can be the sites of an onset of microvilli, as indicated in the presented model. Specific lipids also fine tune behaviour of critical regulatory proteins in microvilli. These data substantiate the role of lipids in morphogenesis and function of microvilli. However, in T cells, the identity of key proteins (and lipids) in microvilli remains unknown. Future works are required to discover these important organizers of signalling receptors at the plasma membrane of T cells. Such research may open new avenues for treatment of many human diseases, which are associated with the malfunction of these critical immune cells.
The author confirms being the sole contributor of this work and has approved it for publication.
This work was supported by Czech Science Foundation (19-07043S).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I would like to thank Zuzana Kvíčalová, Harsha Mavila, Piotr Jurkiewicz and Tomáš Chum for excellent discussions, which stimulated and shaped this work.
1. Kim HR, Mun Y, Lee KS, Park YJ, Park JS, Park JH, et al. T cell microvilli constitute immunological synaptosomes that carry messages to antigen-presenting cells. Nat Commun (2018) 9(1):3630. doi: 10.1038/s41467-018-06090-8
2. Majstoravich S, Zhang J, Nicholson-Dykstra S, Linder S, Friedrich W, Siminovitch KA, et al. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require Wiskott-Aldrich syndrome protein (WASp) for their morphology. Blood (2004) 104(5):1396–403. doi: 10.1182/blood-2004-02-0437
3. Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science (2017) 356(6338):598. doi: 10.1126/science.aal3118
4. Jung Y, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci USA (2016) 113(40):E5916–24. doi: 10.1073/pnas.1605399113
5. Ghosh S, Di Bartolo V, Tubul L, Shimoni E, Kartvelishvily E, Dadosh T, et al. ERM-Dependent Assembly of T Cell Receptor Signaling and Co-stimulatory Molecules on Microvilli prior to Activation. Cell Rep (2020) 30(10):3434–47 e6. doi: 10.1016/j.celrep.2020.02.069
6. Franke C, Chum T, Kvíčalová Z, Glatzová D, Rodriguez A, Helmerich DA, et al. Unraveling nanotopography of cell surface receptors. Biorxiv (Preprint) (2019). doi: 10.1101/2020.08.10.244251
7. Jung Y, Wen L, Altman A, Ley K. CD45 pre-exclusion from the tips of microvilli establishes a phosphatase-free zone for early TCR triggering. Biorxiv (Preprint) (2020). doi: 10.1101/2020.05.21.109074
8. Kim HR, Jun CD. T Cell Microvilli: Sensors or Senders? Front Immunol (2019) 10:1753. doi: 10.3389/fimmu.2019.01753
9. Glatzova D, Cebecauer M. Dual Role of CD4 in Peripheral T Lymphocytes. Front Immunol (2019) 10:618. doi: 10.3389/fimmu.2019.00618
10. Garcia E, Ismail S. Spatiotemporal Regulation of Signaling: Focus on T Cell Activation and the Immunological Synapse. Int J Mol Sci (2020) 21(9):3283. doi: 10.3390/ijms21093283
11. Lange K. Fundamental role of microvilli in the main functions of differentiated cells: Outline of an universal regulating and signaling system at the cell periphery. J Cell Physiol (2011) 226(4):896–927. doi: 10.1002/jcp.22302
12. Sauvanet C, Wayt J, Pelaseyed T, Bretscher A. Structure, regulation, and functional diversity of microvilli on the apical domain of epithelial cells. Annu Rev Cell Dev Biol (2015) 31:593–621. doi: 10.1146/annurev-cellbio-100814-125234
13. Hirokawa N, Tilney LG, Fujiwara K, Heuser JE. Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J Cell Biol (1982) 94(2):425–43. doi: 10.1083/jcb.94.2.425
14. Beer AJ, Gonzalez Delgado J, Steiniger F, Qualmann B, Kessels MM. The actin nucleator Cobl organises the terminal web of enterocytes. Sci Rep (2020) 10(1):11156. doi: 10.1038/s41598-020-66111-9
15. Faust JJ, Millis BA, Tyska MJ. Profilin-Mediated Actin Allocation Regulates the Growth of Epithelial Microvilli. Curr Biol (2019) 29(20):3457–65.e3. doi: 10.1016/j.cub.2019.08.051
16. Gorelik J, Shevchuk AI, Frolenkov GI, Diakonov IA, Lab MJ, Kros CJ, et al. Dynamic assembly of surface structures in living cells. Proc Natl Acad Sci USA (2003) 100(10):5819–22. doi: 10.1073/pnas.1030502100
17. Brown MJ, Nijhara R, Hallam JA, Gignac M, Yamada KM, Erlandsen SL, et al. Chemokine stimulation of human peripheral blood T lymphocytes induces rapid dephosphorylation of ERM proteins, which facilitates loss of microvilli and polarization. Blood (2003) 102(12):3890–9. doi: 10.1182/blood-2002-12-3807
18. Shifrin DA, Jr., McConnell RE, Nambiar R, Higginbotham JN, Coffey RJ, Tyska MJ. Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr Biol (2012) 22(7):627–31. doi: 10.1016/j.cub.2012.02.022
19. Cebecauer M, Amaro M, Jurkiewicz P, Sarmento MJ, Sachl R, Cwiklik L, et al. Membrane Lipid Nanodomains. Chem Rev (2018) 118(23):11259–97. doi: 10.1021/acs.chemrev.8b00322
20. Lipowsky R. Domain-induced budding of fluid membranes. Biophys J (1993) 64(4):1133–8. doi: 10.1016/S0006-3495(93)81479-6
21. Lipowsky R. Budding of Membranes Induced by Intramembrane Domains. J Phys Ii (1992) 2(10):1825–40. doi: 10.1051/jp2:1992238
22. Ursell TS, Klug WS, Phillips R. Morphology and interaction between lipid domains. Proc Natl Acad Sci USA (2009) 106(32):13301–6. doi: 10.1073/pnas.0903825106
23. Rim JE, Ursell TS, Phillips R, Klug WS. Morphological Phase Diagram for Lipid Membrane Domains with Entropic Tension. Phys Rev Lett (2011) 106(5):057801. doi: 10.1103/PhysRevLett.106.057801
24. Veatch SL, Keller SL. Seeing spots: complex phase behavior in simple membranes. Biochim Biophys Acta (2005) 1746(3):172–85. doi: 10.1016/j.bbamcr.2005.06.010
25. Bernardino de la Serna J, Schutz GJ, Eggeling C, Cebecauer M. There Is No Simple Model of the Plasma Membrane Organization. Front Cell Dev Biol (2016) 4:106. doi: 10.3389/fcell.2016.00106
26. Levental I, Levental KR, Heberle FA. Lipid Rafts: Controversies Resolved, Mysteries Remain. Trends Cell Biol (2020) 30(5):341–53. doi: 10.1016/j.tcb.2020.01.009
27. Bjorkbom A, Rog T, Kaszuba K, Kurita M, Yamaguchi S, Lonnfors M, et al. Effect of sphingomyelin headgroup size on molecular properties and interactions with cholesterol. Biophys J (2010) 99(10):3300–8. doi: 10.1016/j.bpj.2010.09.049
28. Lonnfors M, Doux JP, Killian JA, Nyholm TK, Slotte JP. Sterols have higher affinity for sphingomyelin than for phosphatidylcholine bilayers even at equal acyl-chain order. Biophys J (2011) 100(11):2633–41. doi: 10.1016/j.bpj.2011.03.066
29. Simons K, Ikonen E. Functional rafts in cell membranes. Nature (1997) 387(6633):569–72. doi: 10.1038/42408
30. Ikenouchi J, Hirata M, Yonemura S, Umeda M. Sphingomyelin clustering is essential for the formation of microvilli. J Cell Sci (2013) 126(Pt 16):3585–92. doi: 10.1242/jcs.122325
31. He C, Hu X, Jung RS, Weston TA, Sandoval NP, Tontonoz P, et al. High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc Natl Acad Sci USA (2017) 114(8):2000–5. doi: 10.1073/pnas.1621432114
32. Poole K, Meder D, Simons K, Muller D. The effect of raft lipid depletion on microvilli formation in MDCK cells, visualized by atomic force microscopy. FEBS Lett (2004) 565(1-3):53–8. doi: 10.1016/j.febslet.2004.03.095
33. van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol (2008) 9(2):112–24. doi: 10.1038/nrm2330
34. Lorent JH, Levental KR, Ganesan L, Rivera-Longsworth G, Sezgin E, Doktorova M, et al. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat Chem Biol (2020) 16(6):644–52. doi: 10.1038/s41589-020-0529-6
35. Perlmutter JD, Sachs JN. Interleaflet interaction and asymmetry in phase separated lipid bilayers: molecular dynamics simulations. J Am Chem Soc (2011) 133(17):6563–77. doi: 10.1021/ja106626r
36. Romer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature (2007) 450(7170):670–5. doi: 10.1038/nature05996
37. Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J Virol (2011) 85(19):9749–66. doi: 10.1128/JVI.00743-11
38. Kaiser F, Huebecker M, Wachten D. Sphingolipids controlling ciliary and microvillar function. FEBS Lett (2020) 594:3652–67. doi: 10.1002/1873-3468.13816
39. Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, et al. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell (2007) 128(2):383–97. doi: 10.1016/j.cell.2006.11.051
40. Schuck S, Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J Cell Sci (2004) 117(Pt 25):5955–64. doi: 10.1242/jcs.01596
41. Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell (2002) 109(5):599–610. doi: 10.1016/s0092-8674(02)00745-6
42. Lokuta MA, Senetar MA, Bennin DA, Nuzzi PA, Chan KT, Ott VL, et al. Type Igamma PIP kinase is a novel uropod component that regulates rear retraction during neutrophil chemotaxis. Mol Biol Cell (2007) 18(12):5069–80. doi: 10.1091/mbc.e07-05-0428
43. Jiang Z, Redfern RE, Isler Y, Ross AH, Gericke A. Cholesterol stabilizes fluid phosphoinositide domains. Chem Phys Lipids (2014) 182:52–61. doi: 10.1016/j.chemphyslip.2014.02.003
44. Pinkwart K, Schneider F, Lukoseviciute M, Sauka-Spengler T, Lyman E, Eggeling C, et al. Nanoscale dynamics of cholesterol in the cell membrane. J Biol Chem (2019) 294(34):12599–609. doi: 10.1074/jbc.RA119.009683
45. Allender DW, Sodt AJ, Schick M. Cholesterol-Dependent Bending Energy Is Important in Cholesterol Distribution of the Plasma Membrane. Biophys J (2019) 116(12):2356–66. doi: 10.1016/j.bpj.2019.03.028
46. Sorre B, Callan-Jones A, Manneville JB, Nassoy P, Joanny JF, Prost J, et al. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc Natl Acad Sci USA (2009) 106(14):5622–6. doi: 10.1073/pnas.0811243106
47. Pan J, Mills TT, Tristram-Nagle S, Nagle JF. Cholesterol perturbs lipid bilayers nonuniversally. Phys Rev Lett (2008) 100(19):198103. doi: 10.1103/PhysRevLett.100.198103
48. Blin G, Margeat E, Carvalho K, Royer CA, Roy C, Picart C. Quantitative analysis of the binding of ezrin to large unilamellar vesicles containing phosphatidylinositol 4,5 bisphosphate. Biophys J (2008) 94(3):1021–33. doi: 10.1529/biophysj.107.110213
49. Adada M, Canals D, Hannun YA, Obeid LM. Sphingolipid regulation of ezrin, radixin, and moesin proteins family: implications for cell dynamics. Biochim Biophys Acta (2014) 1841(5):727–37. doi: 10.1016/j.bbalip.2013.07.002
50. Canals D, Jenkins RW, Roddy P, Hernandez-Corbacho MJ, Obeid LM, Hannun YA. Differential effects of ceramide and sphingosine 1-phosphate on ERM phosphorylation: probing sphingolipid signaling at the outer plasma membrane. J Biol Chem (2010) 285(42):32476–85. doi: 10.1074/jbc.M110.141028
51. Bonilha VL, Finnemann SC, Rodriguez-Boulan E. Ezrin promotes morphogenesis of apical microvilli and basal infoldings in retinal pigment epithelium. J Cell Biol (1999) 147(7):1533–48. doi: 10.1083/jcb.147.7.1533
52. Crepaldi T, Gautreau A, Comoglio PM, Louvard D, Arpin M. Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J Cell Biol (1997) 138(2):423–34. doi: 10.1083/jcb.138.2.423
53. Hsu YH, Lin WL, Hou YT, Pu YS, Shun CT, Chen CL, et al. Podocalyxin EBP50 ezrin molecular complex enhances the metastatic potential of renal cell carcinoma through recruiting Rac1 guanine nucleotide exchange factor ARHGEF7. Am J Pathol (2010) 176(6):3050–61. doi: 10.2353/ajpath.2010.090539
54. Marin EP, Derakhshan B, Lam TT, Davalos A, Sessa WC. Endothelial cell palmitoylproteomic identifies novel lipid-modified targets and potential substrates for protein acyl transferases. Circ Res (2012) 110(10):1336–44. doi: 10.1161/CIRCRESAHA.112.269514
55. Roper K, Corbeil D, Huttner WB. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat Cell Biol (2000) 2(9):582–92. doi: 10.1038/35023524
56. Thamm K, Simaite D, Karbanova J, Bermudez V, Reichert D, Morgenstern A, et al. Prominin-1 (CD133) modulates the architecture and dynamics of microvilli. Traffic (2019) 20(1):39–60. doi: 10.1111/tra.12618
57. Taieb N, Maresca M, Guo XJ, Garmy N, Fantini J, Yahi N. The first extracellular domain of the tumour stem cell marker CD133 contains an antigenic ganglioside-binding motif. Cancer Lett (2009) 278(2):164–73. doi: 10.1016/j.canlet.2009.01.013
58. Romer W, Pontani LL, Sorre B, Rentero C, Berland L, Chambon V, et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell (2010) 140(4):540–53. doi: 10.1016/j.cell.2010.01.010
59. Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai W, et al. GM1 structure determines SV40-induced membrane invagination and infection. Nat Cell Biol (2010) 12(1):11–8. doi: 10.1038/ncb1999
60. Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol (2007) 8(3):185–94. doi: 10.1038/nrm2122
61. Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci USA (2009) 106(27):11090–5. doi: 10.1073/pnas.0903535106
62. Barooji YF, Rorvig-Lund A, Semsey S, Reihani SN, Bendix PM. Dynamics of membrane nanotubes coated with I-BAR. Sci Rep (2016) 6:30054. doi: 10.1038/srep30054
63. Choi J, Ko J, Racz B, Burette A, Lee JR, Kim S, et al. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J Neurosci (2005) 25(4):869–79. doi: 10.1523/JNEUROSCI.3212-04.2005
64. Nakagawa H, Miki H, Nozumi M, Takenawa T, Miyamoto S, Wehland J, et al. IRSp53 is colocalised with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J Cell Sci (2003) 116(Pt 12):2577–83. doi: 10.1242/jcs.00462
65. Rajagopal S, Ji Y, Xu K, Li Y, Wicks K, Liu J, et al. Scaffold proteins IRSp53 and spinophilin regulate localized Rac activation by T-lymphocyte invasion and metastasis protein 1 (TIAM1). J Biol Chem (2010) 285(23):18060–71. doi: 10.1074/jbc.M109.051490
66. Bisi S, Marchesi S, Rizvi A, Carra D, Beznoussenko GV, Ferrara I, et al. IRSp53 controls plasma membrane shape and polarized transport at the nascent lumen in epithelial tubules. Nat Commun (2020) 11(1):3516. doi: 10.1038/s41467-020-17091-x
67. Billcliff PG, Rollason R, Prior I, Owen DM, Gaus K, Banting G. CD317/tetherin is an organiser of membrane microdomains. J Cell Sci (2013) 126(Pt 7):1553–64. doi: 10.1242/jcs.112953
68. Rollason R, Korolchuk V, Hamilton C, Jepson M, Banting G. A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical actin cytoskeleton in polarized epithelial cells. J Cell Biol (2009) 184(5):721–36. doi: 10.1083/jcb.200804154
69. Richnau N, Fransson A, Farsad K, Aspenstrom P. RICH-1 has a BIN/Amphiphysin/Rvsp domain responsible for binding to membrane lipids and tubulation of liposomes. Biochem Biophys Res Commun (2004) 320(3):1034–42. doi: 10.1016/j.bbrc.2004.05.221
70. Weck ML, Grega-Larson NE, Tyska MJ. MyTH4-FERM myosins in the assembly and maintenance of actin-based protrusions. Curr Opin Cell Biol (2017) 44:68–78. doi: 10.1016/j.ceb.2016.10.002
71. Bisaria A, Hayer A, Garbett D, Cohen D, Meyer T. Membrane-proximal F-actin restricts local membrane protrusions and directs cell migration. Science (2020) 368(6496):1205–10. doi: 10.1126/science.aay7794
72. Welf ES, Miles CE, Huh J, Sapoznik E, Chi J, Driscoll MK, et al. Actin-Membrane Release Initiates Cell Protrusions. Dev Cell (2020) 55:P723–36. doi: 10.1016/j.devcel.2020.11.024
73. Navarro-Hernandez IC, Lopez-Ortega O, Acevedo-Ochoa E, Cervantes-Diaz R, Romero-Ramirez S, Sosa-Hernandez VA, et al. Tetraspanin 33 (TSPAN33) regulates endocytosis and migration of human B lymphocytes by affecting the tension of the plasma membrane. FEBS J (2020) 287(16):3449–71. doi: 10.1111/febs.15216
74. Runge KE, Evans JE, He ZY, Gupta S, McDonald KL, Stahlberg H, et al. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol (2007) 304(1):317–25. doi: 10.1016/j.ydbio.2006.12.041
75. Bari R, Guo Q, Xia B, Zhang YH, Giesert EE, Levy S, et al. Tetraspanins regulate the protrusive activities of cell membrane. Biochem Biophys Res Commun (2011) 415(4):619–26. doi: 10.1016/j.bbrc.2011.10.121
Keywords: T cell, microvilli, sphingolipids, phosphoinositides, lipid rafts, membrane curvature, dimpling domains, membrane-associated proteins
Citation: Cebecauer M (2021) Role of Lipids in Morphogenesis of T-Cell Microvilli. Front. Immunol. 12:613591. doi: 10.3389/fimmu.2021.613591
Received: 02 October 2020; Accepted: 13 January 2021;
Published: 10 March 2021.
Edited by:
Yan Shi, Tsinghua University, ChinaReviewed by:
Chang-Duk Jun, Gwangju Institute of Science and Technology, South KoreaCopyright © 2021 Cebecauer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marek Cebecauer, bWFyZWsuY2ViZWNhdWVyQGpoLWluc3QuY2FzLmN6
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.