- 1Department of Hepatobiliary and Pancreatic Surgery, The Center for Integrated Oncology and Precision Medicine, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Cell Biology and Department of Cardiology of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Key Laboratory for Biomedical Engineering of Ministry of Education, State Key Laboratory for Modern Optical Instrumentation, College of Biomedical Engineering and Instrument Science, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang University, Hangzhou, China
Plasma membrane provides a biophysical and biochemical platform for immune cells to trigger signaling cascades and immune responses against attacks from foreign pathogens or tumor cells. Mounting evidence suggests that the biophysical-chemical properties of this platform, including complex compositions of lipids and cholesterols, membrane tension, and electrical potential, could cooperatively regulate the immune receptor functions. However, the molecular mechanism is still unclear because of the tremendous compositional complexity and spatio-temporal dynamics of the plasma membrane. Here, we review the recent significant progress of dynamical regulation of plasma membrane on immune receptors, including T cell receptor, B cell receptor, Fc receptor, and other important immune receptors, to proceed mechano-chemical sensing and transmembrane signal transduction. We also discuss how biophysical-chemical cues couple together to dynamically tune the receptor’s structural conformation or orientation, distribution, and organization, thereby possibly impacting their in-situ ligand binding and related signal transduction. Moreover, we propose that electrical potential could potentially induce the biophysical-chemical coupling change, such as lipid distribution and membrane tension, to inevitably regulate immune receptor activation.
Introduction
The plasma membrane (PM) of cells, mainly consisting of lipid, cholesterol, and protein, is a lipid bilayer structure. Its outer leaflet enriches phosphatidylcholine, sphingolipid, and cholesterol, and the inner leaflet mainly contains cholesterol and acidic phospholipids (e.g. phosphatidylserine, phosphatidylinositol, and phosphatidic acid) (1–3). The asymmetry mobility and dynamic organization of lipid and membrane proteins have been proposed in the Fluid-Mosaic model (4–7). In this model, the PM is a very dynamic structure, where lipid-protein, lipid-lipid, and protein-protein interactions occur at all times, and all of these interactions regulate membrane receptor’s ligand recognition and triggering (6, 8). Moreover, sphingolipid and cholesterol contribute to the formation of nanodomains or lipid rafts, which are highly dynamic in many receptor-activated cellular processes (9–12). It has been reported that the biophysical-chemical properties of the PM, including the asymmetry of lipid and protein distribution, the membrane curvature and mechanical tension, and the membrane electrical potential, could dynamically regulate diverse cellular processes (13–16).
For immune cells, the PM tunes their essential physiological processes. For example, the cholesterol accumulation could increase T cell differentiation and proliferation, whereas it also induces T cell exhaustion through T-cell receptor (TCR) signaling (17–20). Phosphatidylserine directly tunes T cell migration, adhesion, tissue infiltration, and rapid inflammatory response (21, 22). Moreover, PM’s mechanical tension, driven by the cytoskeleton and its associated molecular motors, dynamically shapes PM morphology and regulates T cell adhesion, migration, and activation cooperatively via many immune receptors (e.g. TCR and integrin) (23–25). Also, PM morphology (e.g. microvilli) facilitates the discrimination of peptide major histocompatibility complex (pMHC) for TCR (26–28). Membrane potential, another PM biophysical property, might also regulate T cell proliferation and cytotoxicity through TCR activation (29, 30).
Here, we review how PM couples with biophysical and biochemical factors to regulate the functions of immune cells (e.g. T cell, B cell, and natural killer cell) through the respective immune receptor activation, such as TCR, B-cell receptor (BCR) or Fc receptor (FcR), and further discuss and propose the potential molecular mechanism.
Double-Edged Regulation of Cholesterol
It has been reported that many receptors (e.g. acetylcholine receptor and G protein-coupled receptor) contain the cholesterol recognition/interaction amino acid consensus (CARC, mainly containing Valine, Isoleucine, Alanine, Methionine, and Serine amino acids) motif in the transmembrane domain (TMD) which directly interacts with cholesterol (31, 32). This CARC motif might be conserved for many immune receptors. Based on previous findings, we propose a double-edged model of cholesterol regulation on receptor activation: 1) cholesterol directly binds TMD to keep immune receptors in an inactive (close conformation) state (33); 2) once the immune cell is stimulated, cholesterol indirectly mediates the clustering of immune receptors (34, 35), which might be in an activation (open conformation) state. Cholesterol might finely tune the activation threshold to avoid perturbations from non-specific noise signals. Once strong stimulation activates the conformational change of receptor TMD, cholesterol could facilitate immune receptors clustering to launch and amplify downstream signaling cascades. Therefore, whether and which residues of receptor TMD mediate direct interaction with cholesterol, and if so, how cholesterol keeps immune receptors in the close conformation or resting state, and how strong stimulation (e.g. ligand binding) could trigger the conformational change of immune receptors to form the cholesterol-mediated nano or micro clusters, need to be further investigated with atomic resolutions.
As a major biochemical component of the PM, cholesterol can bifunctionally regulate TCR dynamics and functions (Figure 1A). On one hand, it associates with the TMD of the TCR β chain and keeps TCR in a resting and inactive conformation, preventing CD3 phosphorylation and recruitment of downstream signaling components, such as ZAP70 and ERK (33). On the other hand, it can also enhance TCR nanoclustering to promote T cell activation (36–38). Moreover, cholesterol can regulate TCR clustering and signaling through dynamic lipid rafts (35, 39, 40). For example, the increased cholesterol level in the PM by inhibiting cholesterol esterification of CD8+ T cell in vivo can promote TCR clustering, enhance immune synapse formation, and amplify the phosphorylation of CD3, ZAP70, and ERK to produce more cytokine, leading to T cell proliferation (41). Consistently, cholesterol sulfate can inhibit CD3 immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation by replacing cholesterol to disrupt the formation of TCR nano-clustering (42). The depletion of cholesterol in T cells also drastically reduces in-situ TCR/pMHC binding affinities and association rates, potentially through regulating the conformation or orientation of TCR’s TMD and ectodomains to impair TCR antigen recognition (43). In brief, cholesterol possibly tunes TCR initial allosteric switch and subsequent clustering, respectively. The detailed regulation mechanism remains ambiguous, which requires further investigation with atomic resolution to reveal how exactly cholesterol dynamically associates with the TCR/CD3 complex. The cryo-EM structure of TCR/CD3 complex with membrane lipid and cholesterol will provide us more meaningful insights.
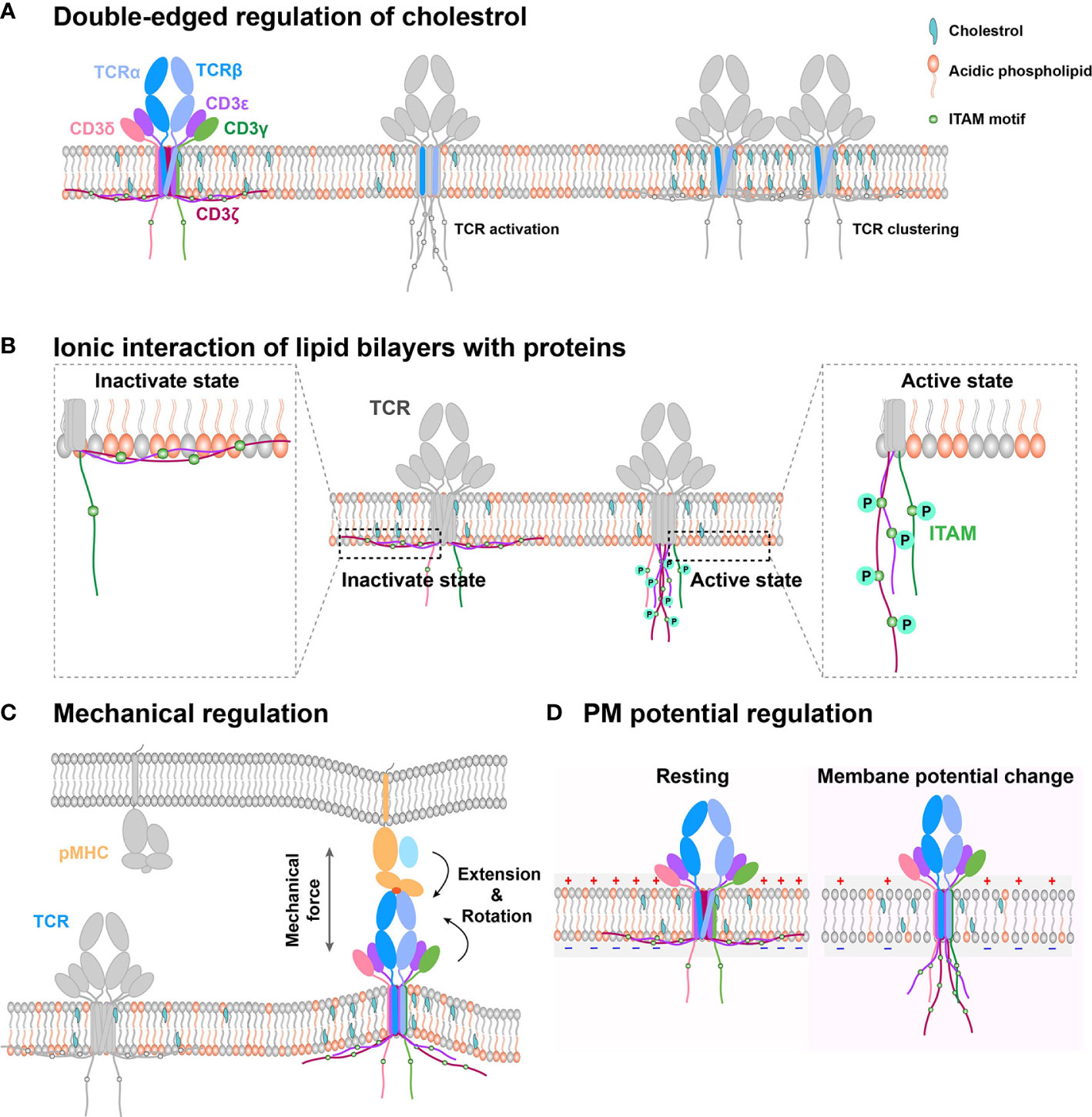
Figure 1 Schematic models of PM regulation on TCR complex signaling. (A) Double-edged regulation of cholesterol on TCR activation. Cholesterol could directly bind with the TMD of the TCR β chain to keep TCR in an inactive state in the resting T cell. Cholesterol disassociation from the TCR β chain can switch the TCR complex to the activation state. Meanwhile, cholesterol also indirectly mediates TCR clustering, following TCR initial activation. (B) The interaction between negatively charged lipid and basic motif regulates CD3 ITAM motif exposure. TCR cytoplasmic domains contain polybasic regions, which directly interact with the negatively charged lipid in the membrane inner leaflet to embed the ITAM motif in hydrophobic core of the PM in resting cell. The disruption of this interaction can expose the signaling motif to amplify downstream signaling. (C) PM provides a platform to sense outside cues for immune receptors. On this platform, mechanical force regulates TCR/pMHC recognition through conformation change. (D) Electrical potential might directly trigger TCR signaling. Since TCR TMD contains several charged residues, PM potential depolarization might induce TMD titling conformation to further allosterically regulate dissociation of CD3 tails from inner leaflet and activate intracellular downstream signaling.
During the antigen recognition process of B cells, the micro-cluster formation of the BCR complex is crucial to strengthen BCR activation signaling (44, 45). Cholesterol has been reported to regulate the distribution of BCRs in PM microdomains, and low cholesterol level impairs BCRs aggregation further to affect Vav and Rac1 phosphorylation (46, 47). Moreover, cholesterol may affect the formation of protein islands, nanodomains, or microvilli on the PM, which provides a platform for BCRs to form their unique signaling complex with coreceptors (28, 48–50). Meanwhile, cholesterol could also induce BCR endocytosis on anergic cells to inhibit BCR signaling (51). Therefore, cholesterol also has double-edged regulation (amplifying or attenuating) on BCR signaling. However, the detailed molecular mechanism of these two regulatory effects and their switching is still unclear.
The lipid raft, mainly consisting with cholesterol, can directly tune activating receptor FcγRIIA signaling (e.g. phosphorylation of CbI and NTAL) without ligand binding (52). The cholesterol depletion can impair FcγRIIA association with CD55, GM1, and Lyn kinase, and the related phosphorylation signaling (53). Similar to FcγRIIA, FcγRIIIA activation could also be inhibited by cholesterol depletion to reduce ERK activation and prevent IFN-γ production (54). And the intracellular tyrosine phosphorylation of inhibitory receptor, FcγRIIIB, can also be significantly attenuated when the cholesterol level reduces (55, 56). Moreover, cholesterol can directly regulate the recognition of FcγRI to IgG (57). These detailed regulation molecular mechanisms still need to be further investigated.
Signaling Motifs Protection by Negatively Charged Lipid/Basic Motif Interaction
The PM inner leaflet enriches negatively charged lipids (e.g. phosphatidylserine, phosphatidylinositol, etc.) that can interact with the polybasic regions of immune receptors to regulate their activation. Such interaction can embed the signaling motif of the immune receptors in the PM hydrophobic core. The positively charged Ca2+ ions flux, which is triggered by strong agonistic ligand stimulation, can disrupt this interaction to uncover the buried signaling site (58–61). The PM shields noise signal interference through their selective association with critical signaling motifs until strong stimulation is initiated. This mechanism of signaling shielding and amplification by regulating negatively charged lipid/basic motif interaction is potentially shared by many other immune receptors.
The inner leaflet of T cell PM mainly consists of negatively charged lipids, which associate with CD3ϵ/ζ cytoplasmic ITAM motif through electrostatic interactions (Figure 1B). These lipid and CD3 interactions protect CD3ϵ/ζ ITAMs from being recognized and phosphorylated by downstream kinase molecules, such as Lck (59, 60, 62, 63), thus keeping TCR/CD3 in the resting state and T cells in a quiescence state. As it has been extensively reviewed before (60), we here just briefly discuss it.
For the mIgG-BCR, PM’s inner side could block non-specific stimulation and keep mIgG-BCR in a resting state, providing the critical activation thresholds for mIgG-BCRs (58). As the cytoplasmic region of the mIgG (mIgG-tail) contains several basic residues, it could electrostatically bind with negatively charged acidic phospholipids of the PM’s inner leaflet to block the interference from noise signals and keep mIgG-BCR in a resting state in quiescent B cells (58). Ca2+ mobilization triggered by suitable antigenic stimulation on the mIgG-BCR complex could disrupt this protection, which further recruits pSyk, pBLNK, and pPI3K into the immunological synapse to induce a more potent Ca2+ mobilization response and B-cell hyperproliferation. Moreover, phosphatidylinositol (4,5)-bisphosphate (PIP2) triggers a signaling amplification loop to induce the initial formation of BCR micro-clusters upon B cell activation (64). PIP2 and phosphatidylinositol (3,4,5)-trisphosphate (PIP3) together tune the growth of BCR micro-clusters by recruiting Dock2 to remodel F-actin cytoskeleton (65). Remarkably, this electrostatical interaction mechanism might not be suitable for mIgG-associated Igα and Igβ as they contain polyacidic regions (58, 60).
Mechanical Regulation and Its Coupling With Cholesterol and Negatively Charged Lipid
In general, the PM provides a platform for immune receptors to sense outside physical and biochemical cues. Unlike biomolecules in solution that can freely rotate and adopt many different possible orientations, immune receptors’ orientations are significantly restricted by the PM. They only can adopt limited protein topologies, which are essential for receptor-ligand binding and downstream signaling transduction. The membrane anchor pattern, extracellular length, and orientation of immune receptors all could influence the recognition of their ligands by affecting ligand accessibility and association kinetics. Meanwhile, the physical platform also tightly restricts immune receptors diffusion, which is distinct to that in solution, thereby drastically affecting immune receptor-ligand binding affinity (66). Moreover, the immune receptors also experience mechanical force induced by membrane tension and cytoskeleton contractions (23–25). The mechanical force has been reported to induce conformational changes of immune receptor and ligand to regulate their binding strength and immune functions. It is very likely that these mechanically regulated protein conformational changes could propagate across the PM and transduce toward the inside of the cell to allosterically regulate the conformation of the receptors’ cytoplasmic tails and potentially their associated kinases or other adaptor molecules. Such propagation could provide a rapid physical activation of receptor signal transduction, other than traditionally accepted biochemical ways. This mechanical regulation might be universal in immune receptors. During cross-membrane mechanical propagation, cholesterol could inevitably be integrated with force to collectively regulate the conformation of receptor’s TMD. For example, cholesterol can regulate PM tension, which in turn tunes cholesterol distribution, membrane stiffness and bending (67–69), thereby inducing immune receptors ectodomain conformational changes and TMD titling. Cholesterol could potentially prevent or facilitate the TMD tilting, which may be dependent on how cholesterol dynamically interacts with receptor’s TMD. Collectively, the mechano-biochemical coupling could contribute to the conformational changes, triggering and clustering of immune receptors.
The PM provides a physical platform for TCR/CD3 complex to sense antigens (66). On this platform, TCR inevitably experiences external mechanical forces when T cells contact the antigen-presenting cell (APC) or migrate on the APC and the extracellular matrix (ECM), and the internal mechanical force produced by dynamic cytoskeleton contraction during T-cell searching for foreign antigens on the APC or membrane bending tension upon T-cell/APC contact formation (23, 70–73). For TCR recognition of pMHCs, the mechanical force can prolong the bond lifetimes for agonistic antigens but not antagonists by selectively inducing conformational changes (Figure 1C) of the agonistic pMHC to initiate the formation of new hydrogen bonds (electrostatic attraction between the hydrogen atom and negatively charged nitrogen or oxygen atom) (72, 73). This force induced by pMHC/TCR binding inevitably increases local membrane tension and further induces membrane bending, which might disrupt the interactions between CD3 polybasic regions and negatively charged lipids of the inner leaflet to expose ITAM motifs for Lck to phosphorylate and further trigger downstream signaling.
PM stiffness can also regulate the antigen discrimination of BCRs. BCR has stringent affinity discrimination when contacting with rigid APC PM during the invagination of antigens (74). Also, PM shape can regulate BCR stimulation by affecting the formation of BCR microclusters (75). On the PM platform, the mechanical force provides multiple effects on different isotyped BCRs, which influences the activation sensitivity of BCR by pathological antigens (antigens that can induce a specific immune response to cause the infectious, allergic or autoimmune diseases) (76). Low mechanical force (<12 pN) is enough to trigger the activation (e.g. BCR, pSyk, pPLCγ2, and pTyr clusters) of IgG- or IgE-BCR on memory B cells, but not IgM-BCR on mature naive B cells (76).
Besides, PM curvature causes the redistribution of FcϵRI (77, 78). FcϵRI bond with IgE always locates at the contact membrane regions which are less curved (79). Similarly, FcγRIIA also can be regulated by the mechanical force under the physiological flow conditions, which facilitates the capture of neutrophils directly by endothelial cells (80). Moreover, the recognition of FcγRII and FcγRIII to IgG is influenced by the anchor patterns (e.g. GPI and transmembrane domain) on the immune cell PM (81, 82).
It has been reported that high membrane tension or mechanical force helps CD28 (another costimulatory receptor of TCR complex) to facilitate TCR signaling on the PM platform (83, 84). Also, LFA-1/ICAM-1 interaction is affected by the mechanical force to regulate T cell migration (85–88). For natural killer cells, PM stiffness can regulate the NKG2D/MICA interaction to determine the cell cytotoxic activity (89).
Electrical Potential Regulation and Its Coupling With Mechanical and Biochemical Cues
The PM electrical potential, which is commonly overlooked in the immunology field, is another essential biophysical factor that also potentially regulates immune receptor functions. It is generally defined as the electric potential difference between the intracellular and extracellular solution. PM potential depolarization can facilitate the opening of Ca2+ channels and initiate the mitotic activity to regulate the activation and proliferation of lymphocyte cells (90–92). The regulation of PM potential on neural activity and the networks has been widely reported. Its molecular regulation mechanism mainly divides into three ways (93). First, TMD conformational change of the voltage-dependent ion channels is triggered by PM potential depolarization (94). Considering that the TCR complex TMD contains several charged residues buried in the lipid bilayer, we propose that PM potential depolarization, which is triggered by T cell activation (95), might tilt the conformation of TCR TMD to further allosterically regulate the dissociation of CD3 tails from the inner leaflet of PM and activate intracellular downstream signaling (Figure 1D). However, this depolarization-induced TCR allosteric activation needs to be further investigated with detailed biophysical investigation. Second, ion influx, an indirect effect of depolarization, regulates transmembrane proteins. The possible mechanism might be that ion influx regulates ligand binding and tyrosine phosphorylation (96, 97). It has been reported that Ca2+ influx disrupts the interactions between CD3 cytoplasmic polybasic regions and negatively charged lipids to favor CD3 ITAMs phosphorylation. However, whether PM potential might directly tune CD3 ITAMs phosphorylation is still unknown. Third, the electro-osmosis or electrophoresis induced by local electric fields, re-distributes transmembrane protein on the PM (98). Like neural synapses, the immune synapses might also exist this electromigration to regulate immune receptor distribution pattern, which favors the recognition of APCs by T cells.
Notably, PM surface usually exists a ~2 nm electrical double layer (EDL, a layer formed by freely diffusing electrolyte ions in the nanometer range of the charged surface), which is regulated by lipid distribution and intracellular/extracellular ions concentration (16). Key proximal regions of TMD, usually containing acidic or basic amino acids and locating in the EDL, might respond to the electrical potential change to regulate immune receptor conformations. For example, Ca2+ influx of T cell activation indeed changes the ion distribution and reduces the interaction between the cytoplasmic domain of CD3 or CD28 and EDL, activating downstream signal transduction.
PM potential can also affect other chemical and biophysical properties, such as lipid distribution, membrane fluidity, tension, and curvature of PM (99, 100), dynamically changing the mechanical and biochemical environment where immune receptors reside (23). For example, PM potential depolarization can induce changes in PM curvature and tension, which could further regulate the mechano-dependent behavior of immune receptors, such as the conformational changes and ligand binding kinetics (23, 100, 101). Meanwhile, the depolarization also reduces the lateral diffusion of membrane components (e.g. cholesterol, phospholipids, and protein) to affect the formation of lipid rafts, receptor microclusters, and microvilli (12, 99, 102, 103), all of which are crucial for immune receptor activation (12, 26, 28).
Inversely, the compositions of lipids and cholesterols can directly affect the charge distribution on the PM surface, which further determines the PM potential (12, 16, 30). PM tension and curvature might also tune immune cell PM potential through mechanosensitive Piezo1 channel (104), causing a series of related regulation on immune receptor activation. However, the detailed mechanism still needs further investigation.
Based on the above elaboration, the PM platform provides mechanical-electric-chemical coupling to synergistically regulate immune receptor-ligand recognition, conformational changes, and cross-membrane activation. This is also exciting to be investigated in the future.
Conclusion
In recent decades, the regulation of immune cell PM chemical properties on the receptor activation has been broadly investigated, and some of them have been revealed. However, many other PM biophysical effects (e.g. membrane tension and electrical potential) have not been clearly examined. Especially, whether and how biophysical-chemical cues couple together to tune receptors, and their molecular mechanism of these regulation patterns all need to be further investigated. Answering all these above questions will improve our understanding of immune receptor activation, especially TCR, thus contributing to immunotherapies development [e.g. chimeric antigen receptor (CAR) T-cell design].
Author Contributions
WC conceived the writing. TZ wrote the first version of the manuscript and prepared the figures. WH and WC revised the paper. All authors contributed to the article and approved the submitted version.
Funding
WC is funded by grants from the Ministry of Science and Technology of China (No. 2017ZX10203205), and the National Natural Science Foundation of China (No. 31971237). WH is funded by grants from The National Natural Science Foundation of China (No. 12002307) and China Postdoctoral Science Foundation (No. 2020M671697).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jizhong Lou for the thoughtful discussion.
References
1. Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol (2017) 18:361–74. doi: 10.1038/nrm.2017.16
2. Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev (2013) 93:1019–137. doi: 10.1152/physrev.00028.2012
3. Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys (2010) 39:407–27. doi: 10.1146/annurev.biophys.093008.131234
4. Mouritsen OG, Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys J (1984) 46:141–53. doi: 10.1016/S0006-3495(84)84007-2
5. Singer S. A fluid lipid-globular protein mosaic model of membrane structure. Ann N Y Acad Sci (1972) 195:16–23. doi: 10.1111/j.1749-6632.1972.tb54780.x
6. Nicolson GL. The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim Biophys Acta (2014) 1838:1451–66. doi: 10.1016/j.bbamem.2013.10.019
7. Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science (1972) 175:720–31. doi: 10.1126/science.175.4023.720
8. Nicolson GL. Cell membrane fluid-mosaic structure and cancer metastasis. Cancer Res (2015) 75:1169–76. doi: 10.1158/0008-5472.CAN-14-3216
9. Tank DW, Wu ES, Webb WW. Enhanced molecular diffusibility in muscle membrane blebs: release of lateral constraints. J Cell Biol (1982) 92:207–12. doi: 10.1083/jcb.92.1.207
10. Bernardino de la Serna J, Schütz GJ, Eggeling C. Cebecauer MJFic, and biology d. There is no simple model of the plasma membrane organization. Front Cell Dev Biol (2016) 4:106. doi: 10.3389/fcell.2016.00106
11. Pike LJ. Rafts defined: a report on the keystone symposium on lipid rafts and cell function. J Lipid Res (2006) 47:1597–8. doi: 10.1194/jlr.E600002-JLR200
12. Cebecauer M, Amaro M, Jurkiewicz P, Sarmento MJ, Sachl R, Cwiklik L, et al. Membrane Lipid Nanodomains. Chem Rev (2018) 118:11259–97. doi: 10.1021/acs.chemrev.8b00322
13. Rothman JE, Lenard J. Membrane asymmetry. Science (1977) 195:743–53. doi: 10.1126/science.402030
14. Baumgart T, Capraro BR, Zhu C, Das SL. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu Rev Phys Chem (2011) 62:483–506. doi: 10.1146/annurev.physchem.012809.103450
15. Yang M, Brackenbury WJ. Membrane potential and cancer progression. Front Physiol (2013) 4:185. doi: 10.3389/fphys.2013.00185
16. Savtchenko LP, Poo MM, Rusakov DA. Electrodiffusion phenomena in neuroscience: a neglected companion. Nat Rev Neurosci (2017) 18:598–612. doi: 10.1038/nrn.2017.101
17. Surls J, Nazarov-Stoica C, Kehl M, Olsen C, Casares S, Brumeanu TD. Increased membrane cholesterol in lymphocytes diverts T-cells toward an inflammatory response. PloS One (2012) 7:e38733. doi: 10.1371/journal.pone.0038733
18. Chyu KY, Lio WM, Dimayuga PC, Zhou J, Zhao X, Yano J, et al. Cholesterol lowering modulates T cell function in vivo and in vitro. PloS One (2014) 9:e92095. doi: 10.1371/journal.pone.0092095
19. Mailer RKW, Gistera A, Polyzos KA, Ketelhuth DFJ, Hansson GK. Hypercholesterolemia induces differentiation of regulatory T Cells in the liver. Circ Res (2017) 120:1740–53. doi: 10.1161/CIRCRESAHA.116.310054
20. Ma X, Bi E, Lu Y, Su P, Huang C, Liu L, et al. Cholesterol induces CD8(+) T cell exhaustion in the tumor microenvironment. Cell Metab (2019) 30:143–56. doi: 10.1016/j.cmet.2019.04.002
21. Elliott JI, Surprenant A, Marelli-Berg FM, Cooper JC, Cassady-Cain RL, Wooding C, et al. Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat Cell Biol (2005) 7:808–16. doi: 10.1038/ncb1279
22. Rysavy NM, Shimoda LM, Dixon AM, Speck M, Stokes AJ, Turner H, et al. Beyond apoptosis: the mechanism and function of phosphatidylserine asymmetry in the membrane of activating mast cells. Bioarchitecture (2014) 4:127–37. doi: 10.1080/19490992.2014.995516
23. Zhu C, Chen W, Lou J, Rittase W, Li K. Mechanosensing through immunoreceptors. Nuture Immunol (2019) 20:1269–78. doi: 10.1038/s41590-019-0491-1
24. Rossy J, Laufer JM, Legler DF. Role of mechanotransduction and tension in T cell function. Front Immunol (2018) 9:2638. doi: 10.3389/fimmu.2018.02638
25. Dupre L, Houmadi R, Tang C, Rey-Barroso J. T lymphocyte migration: an action movie starring the actin and associated actors. Front Immunol (2015) 6:586. doi: 10.3389/fimmu.2015.00586
26. Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science (2017) 356:6338. doi: 10.1126/science.aal3118
27. Jung YM, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci United States America (2016) 113:E5916–24. doi: 10.1073/pnas.1605399113
28. Greicius G, Westerberg L, Davey EJ, Buentke E, Scheynius A, Thyberg J, et al. Microvilli structures on B lymphocytes: inducible functional domains? Int Immunol (2004) 16:353–64. doi: 10.1093/intimm/dxh031
29. Lewis RS, Cahalan MD. Subset-specific expression of potassium channels in developing murine T lymphocytes. Science (1988) 239:771–5. doi: 10.1126/science.2448877
30. Ma Y, Poole K, Goyette J, Gaus K. Introducing membrane charge and membrane potential to T cell signaling. Front Immunol (2017) 8:1513. doi: 10.3389/fimmu.2017.01513
31. Baier CJ, Fantini J, Barrantes FJ. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci Rep (2011) 1:69. doi: 10.1038/srep00069
32. Hedger G, Koldso H, Chavent M, Siebold C, Rohatgi R, Sansom MSP. Cholesterol interaction sites on the transmembrane domain of the hedgehog signal transducer and class F G protein-coupled receptor smoothened. Structure (2019) 27:549–59. doi: 10.1016/j.str.2018.11.003
33. Swamy M, Beck-Garcia K, Beck-Garcia E, Hartl FA, Morath A, Yousefi OS, et al. A cholesterol-based allostery model of T cell receptor phosphorylation. Immunity (2016) 44:1091–101. doi: 10.1016/j.immuni.2016.04.011
34. Fantini J, Di Scala C, Evans LS, Williamson PTF, Barrantes FJ. A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes. Sci Rep (2016) 6:21906. doi: 10.1038/srep21907
35. Bietz A, Zhu H, Xue M, Xu C. Cholesterol Metabolism in T Cells. Front Immunol (2017) 8:1664. doi: 10.3389/fimmu.2017.01664
36. Molnar E, Swamy M, Holzer M, Beck-Garcia K, Worch R, Thiele C, et al. Cholesterol and sphingomyelin drive ligand-independent T-cell antigen receptor nanoclustering. J Biol Chem (2012) 287:42664–74. doi: 10.1074/jbc.M112.386045
37. Roh K-H, Lillemeier BF, Wang F, Davis M. The coreceptor CD4 is expressed in distinct nanoclusters and does not colocalize with T-cell receptor and active protein tyrosine kinase p56lck. PNAS (2015) 112:E1604–13. doi: 10.1073/pnas.1503532112
38. Hu YS, Cang H, Lillemeier BF. Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes. Proc Natl Acad Sci United States America (2016) 113:7201–6. doi: 10.1073/pnas.1512331113
39. Krishnan S, Nambiar MP, Warke VG, Fisher CU, Mitchell J, Delaney N, et al. Alterations in lipid raft composition and dynamics contribute to abnormal T cell responses in systemic lupus erythematosus. J Immunol (2004) 172:7821–31. doi: 10.4049/jimmunol.172.12.7821
40. Deng GM, Tsokos GC. Cholera toxin B accelerates disease progression in lupus-prone mice by promoting lipid raft aggregation. J Immunol (2008) 181:4019–26. doi: 10.4049/jimmunol.181.6.4019
41. Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature (2016) 531:651–5. doi: 10.1038/nature17412
42. Wang F, Beck-Garcia K, Zorzin C, Schamel WW, Davis MM. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat Immunol (2016) 17:844–50. doi: 10.1038/ni.3462
43. Liu B, Chen W, Natarajan K, Li Z, Margulies DH, Zhu C. The cellular environment regulates in situ kinetics of T-cell receptor interaction with peptide major histocompatibility complex. Eur J Immunol (2015) 45:2099–110. doi: 10.1002/eji.201445358
44. Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat Immunol (2005) 6:1168–76. doi: 10.1038/ni1262
45. Tolar P, Hanna J, Krueger PD, Pierce SK. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity (2009) 30:44–55. doi: 10.1016/j.immuni.2008.11.007
46. Karnell FG, Brezski RJ, King LB, Silverman MA, Monroe JG. Membrane cholesterol content accounts for developmental differences in surface B cell receptor compartmentalization and signaling. J Biol Chem (2005) 280:25621–8. doi: 10.1074/jbc.M503162200
47. Brezski RJ, Monroe JG. B cell antigen receptor-induced Rac1 activation and Rac1-dependent spreading are impaired in transitional immature B cells due to levels of membrane cholesterol. J Immunol (2007) 179:4464–72. doi: 10.4049/jimmunol.179.7.4464
48. Maity PC, Blount A, Jumaa H, Ronneberger O, Lillemeier BF, Reth M. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci Signaling (2015) 8:ra93–3. doi: 10.1126/scisignal.2005887
49. Becker M, Hobeika E, Jumaa H, Reth M, Maity PC. CXCR4 signaling and function require the expression of the IgD-class B-cell antigen receptor. Proc Natl Acad Sci USA (2017) 114:5231–6. doi: 10.1073/pnas.1621512114
50. Gold MR, Reth MG. Antigen Receptor Function in the Context of the Nanoscale Organization of the B Cell Membrane. Annu Rev Immunol (2019) 37:97–123. doi: 10.1146/annurev-immunol-042718-041704
51. Blery M, Tze L, Miosge LA, Jun JE, Goodnow CC. Essential role of membrane cholesterol in accelerated BCR internalization and uncoupling from NF-kappa B in B cell clonal anergy. J Exp Med (2006) 203:1773–83. doi: 10.1084/jem.20060552
52. Kulma M, Kwiatkowska K, Sobota A. Raft coalescence and FcgammaRIIA activation upon sphingomyelin clustering induced by lysenin. Cell Signal (2012) 24:1641–7. doi: 10.1016/j.cellsig.2012.04.007
53. Kwiatkowska K, Sobota A. The clustered Fcγ receptor II is recruited to Lyn-containing membrane domains and undergoes phosphorylation in a cholesterol-dependent manner. Eur J Immunol (2001) 31:989–98. doi: 10.1002/1521-4141(200104)31:4<989::aid-immu989>3.0.co;2-v
54. Kondadasula SV, Roda JM, Parihar R, Yu J, Lehman A, Caligiuri MA, et al. Colocalization of the IL-12 receptor and FcgammaRIIIa to natural killer cell lipid rafts leads to activation of ERK and enhanced production of interferon-gamma. Blood (2008) 111:4173–83. doi: 10.1182/blood-2007-01-068908
55. Fernandes MJ, Rollet-Labelle E, Pare G, Marois S, Tremblay ML, Teillaud JL, et al. CD16b associates with high-density, detergent-resistant membranes in human neutrophils. Biochem J (2006) 393:351–9. doi: 10.1042/BJ20050129
56. David A, Fridlich R, Aviram I. The presence of membrane Proteinase 3 in neutrophil lipid rafts and its colocalization with FcgammaRIIIb and cytochrome b558. Exp Cell Res (2005) 308:156–65. doi: 10.1016/j.yexcr.2005.03.034
57. Beekman JM, van der Linden JA, van de Winkel JG, Leusen JH. FcgammaRI (CD64) resides constitutively in lipid rafts. Immunol Lett (2008) 116:149–55. doi: 10.1016/j.imlet.2007.12.003
58. Chen X, Pan W, Sui Y, Li H, Shi X, Guo X, et al. Acidic phospholipids govern the enhanced activation of IgG-B cell receptor. Nat Commun (2015) 6:8552. doi: 10.1038/ncomms9552
59. Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell (2008) 135:702–13. doi: 10.1016/j.cell.2008.09.044
60. Li H, Yan C, Guo J, Xu C. Ionic protein-lipid interactions at the plasma membrane regulate the structure and function of immunoreceptors. Adv Immunol (2019) 144:65–85. doi: 10.1016/bs.ai.2019.08.007
61. Shi X, Bi Y, Yang W, Guo X, Jiang Y, Wan C, et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature (2013) 493:111–5. doi: 10.1038/nature11699
62. Kuhns MS, Davis MM. The safety on the TCR trigger. Cell (2008) 135:594–6. doi: 10.1016/j.cell.2008.10.033
63. Aivazian D, Stern LJ. Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat Struct Biol (2000) 7:1023–6. doi: 10.1038/80930
64. Xu C, Xie H, Guo X, Gong H, Liu L, Qi H, et al. A PIP2-derived amplification loop fuels the sustained initiation of B cell activation. Sci Immunol (2017) 2:eaan0787. doi: 10.1126/sciimmunol.aan0787
65. Wang J, Xu L, Shaheen S, Liu S, Zheng W, Sun X, et al. Growth of B cell receptor microclusters is regulated by PIP2 and PIP3 equilibrium and Dock2 recruitment and activation. Cell Rep (2017) 21:2541–57. doi: 10.1016/j.celrep.2017.10.117
66. Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature (2010) 464:932–6. doi: 10.1038/nature08944
67. Biswas A, Kashyap P, Datta S, Sengupta T, Sinha B. Cholesterol depletion by MβCD enhances cell membrane tension and Its variations-reducing integrity. Biophys J (2019) 116:1456–68. doi: 10.1016/j.bpj.2019.03.016
68. Petelska AD, Naumowicz M, Figaszewski ZA. The interfacial tension of the lipid membrane formed from lipid-cholesterol and lipid-lipid systems. Cell Biochem Biophys (2006) 44:205–11. doi: 10.1385/CBB:44:2:205
69. Nomoto T, Takahashi M, Fujii T, Chiari L, Toyota T, Fujinami M. Effects of cholesterol concentration and osmolarity on the fluidity and membrane tension of free-standing black lipid membranes. Anal Sci (2018) 34:1237–42. doi: 10.2116/analsci.18P200
70. Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature (2003) 423:190–3. doi: 10.1038/nature01605
71. Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell (2014) 157:357–68. doi: 10.1016/j.cell.2014.02.053
72. Wu P, Zhang T, Liu B, Fei P, Cui L, Qin R, et al. Mechano-regulation of peptide-MHC class I conformations determines TCR antigen recognition. Mol Cell (2019) 73:1015–27. doi: 10.1016/j.molcel.2018.12.018
73. Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, et al. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA (2015) 112:1517–22. doi: 10.1073/pnas.1424829112
74. Spillane KM, Tolar P. B cell antigen extraction is regulated by physical properties of antigen-presenting cells. J Cell Biol (2017) 216:217–30. doi: 10.1083/jcb.201607064
75. Treanor B, Depoil D, Bruckbauer A, Batista FD. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J Exp Med (2011) 208:1055–68. doi: 10.1084/jem.20101125
76. Wan Z, Chen X, Chen H, Ji Q, Chen Y, Wang J, et al. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. Elife (2015) 4:e06925. doi: 10.7554/eLife.06925
77. Spendier K. N-terminal amphipathic helix of Amphiphysin can change the spatial distribution of immunoglobulin E receptors (FcepsilonRI) in the RBL-2H3 mast cell synapse. Results Immunol (2016) 6:1–4. doi: 10.1016/j.rinim.2015.11.001
78. Spendier K, Carroll-Portillo A, Lidke KA, Wilson BS, Timlin JA, Thomas JL. Distribution and dynamics of rat basophilic leukemia immunoglobulin E receptors (FcεRI) on planar ligand-presenting surfaces. Biophys J (2010) 99:388–97. doi: 10.1016/j.bpj.2010.04.029
79. Machado R, Bendesky J, Brown M, Spendier K, Hagen GM. Imaging membrane curvature inside a FcϵRI-centric synapse in RBL-2H3 cells using TIRF microscopy with polarized excitation. J Biol Chem (2019) 5:63. doi: 10.3390/jimaging5070063
80. Nishi H, Furuhashi K, Cullere X, Saggu G, Miller MJ, Chen Y, et al. Neutrophil FcgammaRIIA promotes IgG-mediated glomerular neutrophil capture via Abl/Src kinases. J Clin Invest (2017) 127:3810–26. doi: 10.1172/JCI94039
81. Williams TE, Nagarajan S, Selvaraj P, Zhu C. Concurrent and independent binding of Fcgamma receptors IIa and IIIb to surface-bound IgG. Biophys J (2000) 79:1867–75. doi: 10.1016/S0006-3495(00)76436-8
82. Hu W, Zhang Y, Sun X, Zhang T, Xu L, Xie H, et al. FcgammaRIIB-I232T polymorphic change allosterically suppresses ligand binding. Elife (2019) 8:e46689. doi: 10.7554/eLife.46689
83. Bashour KT, Gondarenko A, Chen H, Shen K, Liu X, Huse M, et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci USA (2014) 111:2241–6. doi: 10.1073/pnas.1315606111
84. Hong J, Ge C, Jothikumar P, Yuan Z, Liu B, Bai K, et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat Immunol (2018) 19:1379–90. doi: 10.1038/s41590-018-0259-z
85. Chen W, Zhu C. Mechanical regulation of T-cell functions. Immunol Rev (2013) 256:160–76. doi: 10.1111/imr.12122
86. Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem (2010) 285:35967–78. doi: 10.1074/jbc.M110.155770
87. Xiang X, Lee CY, Li T, Chen W, Lou J, Zhu C. Structural basis and kinetics of force-induced conformational changes of an alphaA domain-containing integrin. PloS One (2011) 6:e27946. doi: 10.1371/journal.pone.0027946
88. Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol (2012) 199:497–512. doi: 10.1083/jcb.201201091
89. Mordechay L, Edri A, Hadad U, Porgador A, Schvartzman M, Le Saux G. Mechanical regulation of the cytotoxic activity of natural killer cells. bioRxiv (2020) 7 (1):122–32. doi: 10.1021/acsbiomaterials.0c01121
90. Kiefer H, Blume AJ, Kaback HR. Membrane potential changes during mitogenic stimulation of mouse spleen lymphocytes. Proc Natl Acad Sci USA (1980) 77:2200–4. doi: 10.1073/pnas.77.4.2200
91. Hess SD, Oortgiesen M, Cahalan MD. Calcium oscillations in human-T and natural-killer-cells depend upon membrane-potential and calcium influx. J Immunol (1993) 150:2620–33.
92. Monroe JG, Cambier JC. B cell activation. I. Anti-immunoglobulin-induced receptor cross-linking results in a decrease in the plasma membrane potential of murine B lymphocytes. J Exp Med (1983) 157:2073–86. doi: 10.1084/jem.157.6.2073
93. Zhang LI, Poo MM. Electrical activity and development of neural circuits. Nat Neurosci (2001) 4 Suppl:1207–14. doi: 10.1038/nn753
94. Zucker RS. Calcium- and activity-dependent synaptic plasticity. Curr Opin Neurobiol (1999) 9:305–13. doi: 10.1016/s0959-4388(99)80045-2
95. Felber SM, Brand MD. Early plasma-membrane-potential changes during stimulation of lymphocytes by concanavalin A. Biochem J (1983) 210:885–91. doi: 10.1042/bj2100885
96. Cooper DM, Schell MJ, Thorn P, Irvine RF. Regulation of adenylyl cyclase by membrane potential. J Biol Chem (1998) 273:27703–7. doi: 10.1074/jbc.273.42.27703
97. Reddy R, Smith D, Wayman G, Wu Z, Villacres EC, Storm DR. Voltage-sensitive adenylyl cyclase activity in cultured neurons. A calcium-independent phenomenon. J Biol Chem (1995) 270:14340–6. doi: 10.1074/jbc.270.24.14340
98. Poo M. In situ electrophoresis of membrane components. Annu Rev Biophys Bioeng (1981) 10:245–76. doi: 10.1146/annurev.bb.10.060181.001333
99. Oghalai JS, Zhao HB, Kutz JW, Brownell WE. Voltage- and tension-dependent lipid mobility in the outer hair cell plasma membrane. Science (2000) 287:658–61. doi: 10.1126/science.287.5453.658
100. Zhang PC, Keleshian AM, Sachs F. Voltage-induced membrane movement. Nature (2001) 413:428–32. doi: 10.1038/35096578
101. Todorov AT, Petrov AG, Fendler JH. Flexoelectricity of charged and dipolar bilayer-lipid membranes studied by stroboscopic interferometry. Langmuir (1994) 10:2344–50. doi: 10.1021/la00019a053
102. Poste G, Nicolson GL. Membrane reconstitution. North Holland: Elsevier Biomedical Press (1982).
103. Golan DE, Alecio MR, Veatch WR, Rando RR. Lateral mobility of phospholipid and cholesterol in the human erythrocyte membrane: effects of protein-lipid interactions. Biochemistry (1984) 23:332–9. doi: 10.1021/bi00297a024
Keywords: immune receptor, plasma membrane, biophysical-chemical coupling, electrical potential, mechanical force
Citation: Zhang T, Hu W and Chen W (2021) Plasma Membrane Integrates Biophysical and Biochemical Regulation to Trigger Immune Receptor Functions. Front. Immunol. 12:613185. doi: 10.3389/fimmu.2021.613185
Received: 01 October 2020; Accepted: 06 January 2021;
Published: 19 February 2021.
Edited by:
Harry W. Schroeder, University of Alabama at Birmingham, United StatesReviewed by:
Michael Reth, University of Freiburg, GermanyWenxia Song, University of Maryland, College Park, United States
Copyright © 2021 Zhang, Hu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Chen, amFja3dlaWNoZW5Aemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Tongtong Zhang1†
Tongtong Zhang1† Wei Chen
Wei Chen