
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 30 November 2020
Sec. Microbial Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.615056
This article is part of the Research Topic The Role of Dietary Interventions in The Regulation of Host-Microbe Interactions View all 24 articles
The intestinal microbiota, composed of a large population of microorganisms, is often considered a “forgotten organ” in human health and diseases. Increasing evidence indicates that dysbiosis of the intestinal microbiota is closely related to colorectal cancer (CRC). The roles for intestinal microorganisms that initiated and facilitated the CRC process are becoming increasingly clear. Hypothesis models have been proposed to illustrate the complex relationship between the intestinal microbiota and CRC. Recent studies have identified Streptococcus bovis, enterotoxigenic Bacteroides fragilis, Fusobacterium nucleatum, Enterococcus faecalis, Escherichia coli, and Peptostreptococcus anaerobius as CRC candidate pathogens. In this review, we summarized the mechanisms involved in microbiota-related colorectal carcinogenesis, including inflammation, pathogenic bacteria, and their virulence factors, genotoxins, oxidative stress, bacterial metabolites, and biofilm. We also described the clinical values of intestinal microbiota and novel strategies for preventing and treating CRC.
Colorectal cancer (CRC) is one of the most common cancers and is a major global health burden. CRC ranks third in terms of incidence and second in mortality worldwide, accounting for 1.8 million new cases and 881,000 deaths in 2018 (1). With its continued progression in western countries, the incidence of CRC is predicted to increase to 2.2 million new cases and 1.1 million deaths worldwide by 2030 (2). In China, over 376,000 new cases and 191,000 deaths are estimated to occur annually (3). CRC incidence and mortality have decreased steadily in recent decades among adults aged ≥65 years because of increased colonoscopy screenings; however, the opposite has occurred in adults younger than 50 years (4). In the United States from the mid-1980s through 2013, the colon cancer incidence increased by 2.4% annually in adults aged 20–29 years and by 1.0% annually in adults aged 30–39 years. It has also increased by 1.3% annually in adults aged 40–49 years and by 0.5% annually in adults aged 50–54 years since the mid-1990s (5, 6). This trend in younger adults, along with the continued burden in the overall population, is alarming; therefore, new strategies for early detection and prevention of CRC are urgently needed. As with many diseases, the CRC etiology is highly complex and involves both genetic and environmental factors (7). Evidence from twin and family studies indicates that only a small fraction of CRCs, including familial adenomatous polyposis (FAP), hereditary nonpolyposis colorectal cancer (HNPCC or Lynch syndrome), Peutz-Jeghers syndrome, and other more rare disorders, are genetically predisposed (8–10). In addition, most CRCs are sporadic or non-inherited (11). Environmental factors, such as western dietary habits, smoking, weight gain and obesity, diabetes, and heavy alcohol consumption, play major roles in causing sporadic CRC. Among environmental risk factors, the intestinal microbiota is an important contributor (12–15). Increasing evidence indicates that the intestinal microbiota plays a vital role in CRC initiation, progression, and metastasis (16, 17).
The human intestinal microbiota is composed of 1013 to 1014 microbes, encompasses ∼10 times more bacterial cells than human cells and contains > 100 times as many genes as in the human genome (18, 19). A healthy human intestinal microbiota plays a crucial role in harvesting energy (20), shaping the intestinal epithelium (21), protecting against pathogens (22), and maintaining immunity (23). Contrariwise, intestinal microbiota dysbiosis alters host physiological functions, leading to various diseases (24). Emerging studies on the relationship between the intestinal microbiota and CRC have analyzed the role of the intestinal microbiota in colorectal carcinogenesis. These studies have shown the differences in the intestinal microbiota compositions between patients with CRC and healthy individuals as well as which microbes are enriched or depleted in patients with CRC. Interestingly, microbiome alterations also occur with colorectal adenoma, the early stage of CRC. Thus, microbiome alterations might be used as biomarkers for early CRC detection. On the other hand, previous research findings suggest that modulating the intestinal microbiome may be a new strategy for CRC prevention and treatment. In this review, we summarized recent advances in understanding the associations between the intestinal microbiota and CRC based on evidence from animal and human studies, especially in terms of mechanisms. We also partly describe the clinical value of the intestinal microbiota and novel strategies for preventing and treating CRC.
In as early as the 1970s, animal experiments showed that intestinal microbial populations were crucial in mediating the carcinogenic effects of different compounds in intestine (25–27). For example, Wynder group used germ-free and conventional rats to study the effects of the intestinal microbiota on colonic sensitivity to the carcinogenic effect of 1,2-dimethylhydrazine. They found that only 20% of germ-free rats developed colonic tumors, whereas 93% of conventional rats developed multiple colonic tumors (26), and subcutaneously injecting azoxymethane increased the incidence and multiplicity of the colonic tumors in germ-free rats compared with that in conventional rats (27). Intestinal microbial dysbiosis could be observed within the intestines of mice with both spontaneous and chemically induced colon tumorigenesis. For example, ApcMin/+ mice, a familial model of colonic tumor disease, spontaneously developed intestinal tumors due to a mutation in the adenomatous polyposis coli (APC) tumor-suppressor gene. The intestinal microbial diversity was reduced in C57BL/6J ApcMin/+ mice compared with that in wild-type C57BL/6J mice (28). Additionally, ApcMin/+ mice gavaged with feces from CRC patients exhibited enhanced intestinal adenoma progression (29).
High-throughput microbiome sequencing enables researchers to study microbial communities that colonize tumors as well as nontumor colonic sites and characterize individualized oncogenic microbiomes (13). Patients with CRC have shown reduced bacterial diversity and richness compared with those of healthy individuals (30). Additionally, although no unifying CRC-associated microbiota structure has yet been determined, accumulating human studies have shown that compared with matched microbiotas from healthy individuals, the intestinal microbiotas of CRC patients are structurally separate in both fecal (13, 31–35) and mucosal (12, 30, 36) samples. In addition, emerging studies have demonstrated the role of fungi in colorectal tumorigenesis. A previous study identified that fungal composition were different in tissue biopsies of 27 subjects with colorectal adenoma and adjacent tissues (37). Consistently, Yu et al. found that unlike the observation with bacteria, the fungal alpha diversities were not significantly different between CRC patients and healthy individuals, but the compositions were obviously altered. The Basidiomycota:Ascomycota ratio was increased in CRC patients compared with healthy subjects. And class Malasseziomycetes was enriched in CRC while classes Saccharomycetes and Pneumocystidomycetes were depleted (15). Overall, the CRC microbiota exhibits dysbiosis, reflecting a different ecological microenvironment in patients with CRC.
Despite the variations in intestinal microbiota, several individual bacterial species have been associated with CRC (Table 1). Streptococcus bovis (S. bovis), a gram-positive cocci, is a reported risk factor for CRC (38–40). Enterotoxigenic Bacteroides fragilis (ETBF), a bacterium producing B. fragilis toxin (BFT), causes diarrhea and inflammatory bowel disease (IBD) (41–44). Fusobacterium nucleatum (F. nucleatum) is enriched in human colorectal adenomas and carcinomas (45, 57) and may contribute to disease progression from adenoma to cancer (46). In our recent study, F. nucleatum was significantly increased in patients with early-stage CRC (49). The presence of F. nucleatum in CRC tissues indicated a worse prognosis (47, 48). Some studies reported that Enterococcus faecalis (E. faecalis) was significantly higher in patients with CRC compared with that in healthy controls (31, 51). E. faecalis infection induces superoxide production, which damages DNA in epithelial cells (50, 52). Although Escherichia coli (E. coli) is a gut commensal bacterium, studies have reported higher levels of colonic colonization by mucosa-associated E. coli in CRC patients compared with that in healthy people (53, 54, 58). Yu et al. found Peptostreptococcus anaerobius (P. anaerobius) was significantly enriched in fecal and mucosal microbiotas from patients with CRC (55, 56). Notably, it is not one specific microorganism that is responsible for CRC, but a group of bacteria whose detrimental actions surpass those of the beneficial commensals. On the other hand, some bacteria, mostly probiotics such as the butyrate-producer, Clostridium butyicum, and lactate-producer such as S. thermophilus, are depleted in CRC patients. These bacteria may exert a protective effect against CRC.
CRC occurrence is related to changes in the overall intestinal flora structure as well as infection with one or several specific bacteria. To better understand the relationship between intestinal microbiota and CRC, researchers raised a few hypotheses as follows:
Sears and Pardoll proposed the “Alpha-bug” hypothesis based on previous work on ETBF carcinogenesis in ApcMin/+ mice (59). ETBF rapidly induced the exclusive activation of signal transducer and activator of transcription-3 (STAT3) with colitis characterized by T helper 17 (Th17) responses, which might promote cancer in cooperation with the modified colonic epithelium (42). The Alpha-bug hypothesis integrates the single intestinal bacterium and microbiome community views of microbial carcinogenesis. Alpha-bugs not only induce tumors directly, but also remodel the bacterial community to one that promotes Alpha-bug induction of intestinal mucosal immune responses and intestinal epithelial cell (IEC) alterations resulting in cancer. Additionally, Alpha-bugs can enhance carcinogenesis by selectively “crowding out” of cancer-protective intestinal bacteria. Potential Alpha-bug candidates include ETBF, S. bovis, superoxide-producing E. faecalis, and E. coli.
Next-generation sequencing technology has allowed us to explore the microbial composition of both healthy and diseased body sites. These studies have revealed that the intestinal microbiota associated with CRC contains bacterial species that differ in their temporal associations with developing tumors. Based on this, Tjalsma et al. first proposed a bacterial counterpart of the genetic “driver-passenger” model for CRC (60). This model classifies microbes into two categories. First, certain indigenous intestinal bacteria (termed driver bacteria) produce genotoxic substances to damage the epithelial cell DNA, thus initiating CRC. Second, tumor environmental alterations that favor proliferation of opportunistic bacteria (termed passenger bacteria), such as Fusobacterium spp., mediate colorectal tumorigenesis. Although the driver bacterial aspect of the driver-passenger model is related to the Alpha-bug hypothesis, it differs from the Alpha-bug model (61). The driver-passenger model highlights that although the driver bacteria initiate CRC, these bacteria will not always exist, and will be replaced by passenger bacteria as a loss of growth advantage, whereas the Alpha-bug hypothesis posits that driver bacteria persistently colonize developing tumors. Thus, these authors suggest that bacterial drivers and passengers have distinct temporal associations with CRC tissue. This model well explains the various results among different studies.
Colorectal carcinogenesis is highly complex and involves genetic and environmental factors. Emerging studies suggest that several mechanisms, including inflammation, pathogenic bacteria, genotoxins, oxidative stress, metabolites, and biofilm, are closely linked to the intestinal microbiota. Here, we review the known microbiota-associated mechanisms in CRC carcinogenesis (Figure 1).
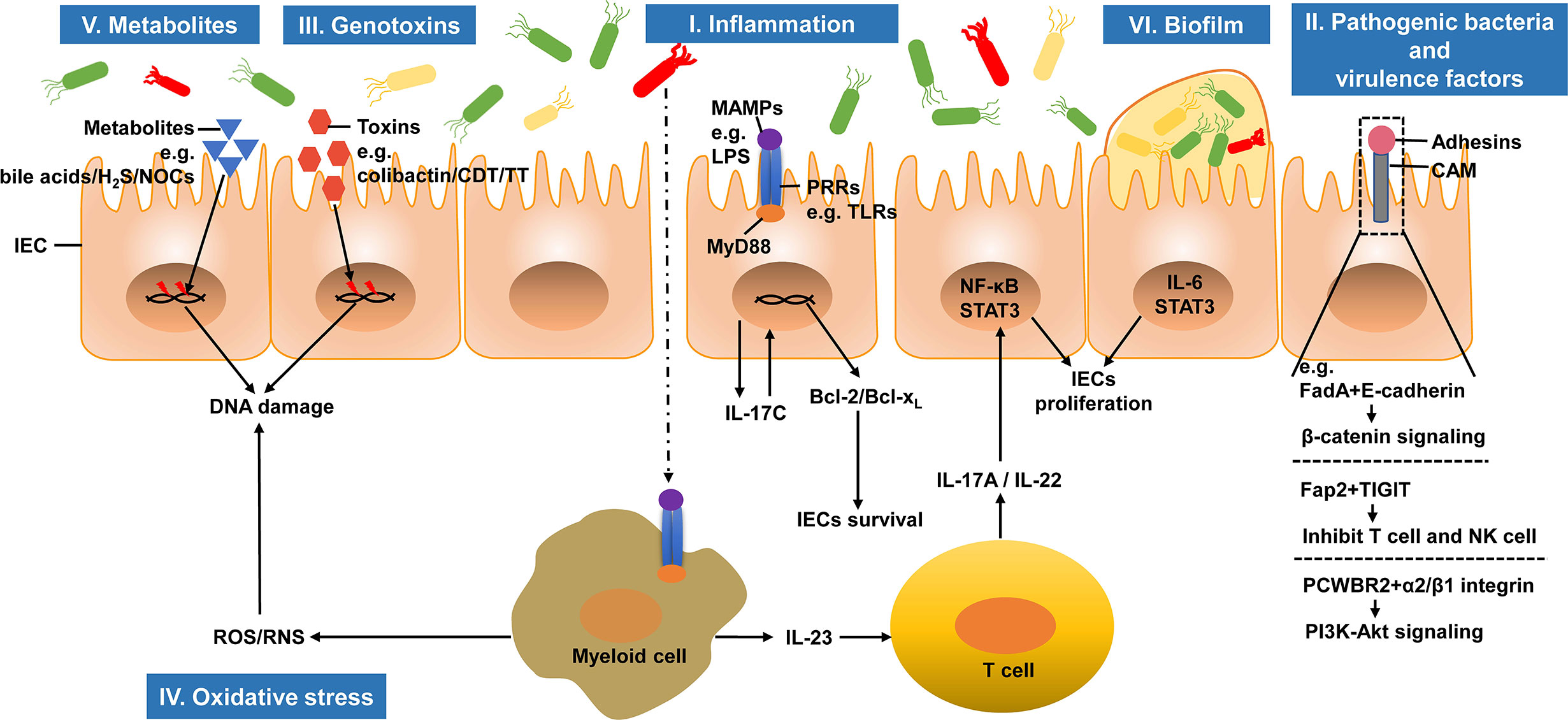
Figure 1 Microbiota-associated mechanisms in colorectal carcinogenesis. The intestinal microbiota can regulate the initiation and progression of CRC. I. The infiltration of commensal bacteria or their products activates tumor-associated myeloid cells and induces tumor promoting inflammation. II. Pathogenic bacteria and their virulence factors adhere to IECs and promote tumorigenesis. III. Genotoxins produced by bacteria induce DNA damages in IECs and initiate CRC development. IV. Under the stimulation of chronic inflammation, inflammatory cells can produce ROS and RNS, which in turn induce DNA damage. V. Several bacterial metabolites, including secondary bile acids, H2S and NOCs, can cause DNA damage, which promote CRC carcinogenesis. VI. Biofilm, microbial communities, promotes carcinogenesis through IL-6 and its downstream effector STAT3 activation. IEC, intestinal epithelial cell; H2S, hydrogen sulfide; NOCs, N-nitroso compounds; CDT, cytolethal distending toxin; TT, typhoid toxin; MAMP, microbe-associated molecular pattern; LPS, lipopolysaccharide; PRR, pattern recognition receptor; TLR, Toll-like receptor; MyD88, myeloid differentiation factor 88; NF-κB, nuclear factor-κB; STAT3, signal transducer and activator of transcription 3; CAM, cell adhesin molecule; FadA, Fusobacterium adhesin A; TIGIT, T-cell immunoglobulin and ITIM domain; PCWBR2, putative cell wall binding repeat2; ROS, reactive oxygen species; RNS, reactive nitrogen species.
The first connection between inflammation and cancer is attributed to Rudolf Virchow, who noted the presence of leukocytes in tumors in 1863 (62). Until the past decade that clear evidence has been obtained that inflammation acts as a critical factor in tumorigenesis. Chronic inflammation is widely accepted as a risk factor for CRC (63–65). Patients with IBD, including both ulcerative colitis (UC) and Crohn’s disease (CD), have higher risks of CRC. Previous meta-analyses summarized the 30-year cumulative risk of CRC, which is increased by up to 18.4% in UC patients (66) and 8.3% in CD patients (67).
The intestinal microbiota interacts closely with the host immune system. Bacterial stimulation of immune responses can cause continuous low-grade inflammation, resulting in tumorigenesis. Conversely, inflammation cannot induce CRC without the microbiota or bacteria-derived compounds (68). Therefore, increasing efforts have been made to understand tumor-elicited inflammation, which follows tumor development and can be detected in most solid malignancies (69).
Normally, the intestinal mucosal barrier segregates the intestinal microbiota from immune cells. The intestinal mucosal barrier is composed of a single layer of IECs joined by tight junctions (70). In both human and CRC mouse models, the intestinal mucosal barrier is highly permeable (71). Disrupting the intestinal mucosal barrier function via dextran sodium sulfate (DSS)-induced colitis leads to increased susceptibility to CRC (72). Specific ablation of matriptase, a membrane-anchored serine protease that strengthens the intestinal epithelial barrier by promoting tight junction formation, causes CRC development (73). All these suggest that transformed IECs fail to form an effective surface barrier, enabling commensal bacteria and their degradation products to invade the tumor stoma. Host recognizes the microbiota via various pattern recognition receptors [PRRs, such as Toll-like receptors (TLRs)], which control the inflammatory response to microorganism-associated molecular patterns, such as lipopolysaccharide (74). Invading commensal bacteria and their components engage TLRs on tumor-infiltrating myeloid cells following activation of myeloid differentiation factor 88 (MyD88)-mediated production of inflammatory cytokines, most notably interleukin (IL)-23. IL-23 then activates IL-17A, IL-6, and IL-22 production (71, 75, 76), eventually promoting tumor cell proliferation by activating nuclear factor-κB (NF-κB) and STAT3 signaling pathway (77, 78). Moreover, commensal bacteria and their components also upregulate IL-17C in transformed IECs through TLR/MyD88 dependent signaling. IL-17C induces B-cell lymphoma-2 (Bcl-2) and Bcl-xL expression in IECs in an autocrine manner to promote tumor cell survival and tumorigenesis (75).
F. nucleatum generates a pro-inflammatory environment that permits colorectal neoplasia progression by activating the NF-κB pathway and recruiting tumor-infiltrating immune cells in ApcMin/+ mice (45). IL-17A is highly expressed when F. nucleatum is enriched in human CRC (79).
ETBF secretes a 21 kDa BFT that cleaves E-cadherin on host IECs, thus disrupting the colonic barrier (80). Infecting ApcMin/+ mice with ETBF selectively induces STAT3 activation with CRC characterized by Th17 responses (42). ETBF also promotes colonic cancer by secreting particles that stimulate IECs to produce exosome-like nanoparticles containing elevated levels of chemokine C-C motif ligand 20 and prostaglandin E2, which are required for Th17 cell recruitment and proliferation of IL-17 signals to transformed IECs to support their growth and survival (81).
P. anaerobius can provoke a pro-inflammatory immune microenvironment to promote tumorigenesis. In ApcMin/+ mice, P. anaerobius broadly induces pro-inflammatory cytokine expression, which in turn recruits a series of tumor-infiltrating immune cells, especially immunosuppressive myeloid-derived suppressor cells, tumor-associated macrophages, and granulocytic tumor-associated neutrophils, to promote tumor progression (56).
Several candidate pathogenic bacteria play vital roles in colorectal carcinogenesis by attaching to the mucosal surface. Bacterial adherence is often a prerequisite step to tumor promotion. F. nucleatum, an oral commensal bacterium, acts at the early step of colorectal carcinogenesis. Researchers demonstrated that F. nucleatum adheres to and induces CRC through its unique Fusobacterium adhesin A (FadA), which selectively binds to E-cadherin and activates the β-catenin signaling pathway, thus inducing oncogenic and inflammatory responses (82). Additionally, F. nucleatum inhibits T cell activation and natural killer cell cytotoxicity through its surface adhesin, Fap2, which binds to the human immune inhibitory receptor T-cell immunoglobulin and ITIM domain (83). A recent study showed that Fap2-dependent invasion induced secretion of the proinflammatory cytokines, IL-8, and CXCL1, which promoted CRC cell migration (84). F. nucleatum also modulates autophagy in IECs by activating regulatory microRNAs (85, 86). P. anaerobius normally resides in the oral cavity and gut. Yu et al. found that P. anaerobius promoted CRC development in ApcMin/+ mice via its surface protein, putative cell wall binding repeat 2 (PCWBR2). PCWBR2 directly binds to intestinal epithelial cell receptor integrin α2/β1 to initiate an oncogenic PI3K-Akt signaling pathway, which promotes tumor cell proliferation (56). S. bovis, occasionally presents in the human gastrointestinal tract flora and is increased in patients with CRC (87, 88). Its role in CRC development is likely inflammation-driven carcinogenesis via, but not limited to, IL-1, cyclooxygenase-2 (COX-2), and IL-8 (89, 90). Salmonella infection in human can be chronic and increase the risk of cancer. Salmonella promotes colonic tumorigenesis dependent on its protein AvrA, which can activate both the Wnt/β-catenin and STAT3 signaling pathways in colonic tumor cells (91–93).
Bacterially produced genotoxins are related to colonic carcinogenesis because of their DNA-damaging effects. E. coli harbors the genomic island, polyketide synthase (pks), which codes for production of the polyketide-peptide genotoxin, colibactin (94, 95). Cultured mammalian epithelial cells exposed to pks+ E. coli exhibited transient DNA damage (94). In a xenograft model, researchers found that colibactin promoted cell senescence, followed by hepatocyte growth factor production and enhanced tumor cell proliferation (96). Campylobacter jejuni produces a genotoxin, cytolethal distending toxin, which causes double-stranded DNA breaks and promotes colorectal tumorigenesis (97). Salmonella also produces a genotoxin, typhoid toxin, which damages DNA via the PI3K pathway in colonic epithelial cells (98).
Oxidative stress is an imbalance between production of pro-oxidative molecules (e.g., reactive oxygen species (ROS) and reactive nitrogen species (RNS) and the effectiveness of anti-oxidative defenses. Oxidative stress is common in chronic inflammation caused by the intestinal microbiota. Under the stimulation of chronic inflammation, inflammatory cells produce many ROS and RNS, which can induce DNA damage and further activate oncogenes or inactivate tumor-suppressor genes, thus increasing CRC development. The gut microbiota can also directly produce ROS. E. faecalis infection in macrophages induces superoxide production, which damages DNA in epithelial cells via a bystander effect (50, 99). In vitro and in vivo studies demonstrated that E. faecalis can produce hydroxyl radicals (100, 101), which are powerful mutagens that cause DNA breaks, point mutations and protein-DNA crosslinking, thus contributing to chromosomal instability and CRC risk (102). P. anaerobius activates TLR2/TLR4 on IECs and boosts intracellular ROS levels, which promotes cholesterol synthesis and cell proliferation (55). E. coli and BFT of ETBF also promote ROS production by colonic epithelial cells (103, 104).
According to a recent study, 38.3% of CRC cases were related to poor diets with intake of food low in whole grains, low in dairy products and high in red and processed meats (105). Additionally, obesity, which increases CRC risk of by 19%, and being overweight have been recognized as significant risk factors for CRC (106). A report found that each 5-kg/m2 increase in body mass index is associated with a 5% increase in CRC risk (6). In a sense, these make CRC a somewhat preventable disease. Tilg et al. previously reviewed this topic, and they highlighted that the microbiota could indeed reflect a ‘‘missing link’’ in the close interaction between dietary factors and CRC (107). Diet affects CRC, partly by modulating the intestinal microbiome composition and diversity. For example, diets high in animal protein and fat yielded enterotypes dominated by Bacteroides, whereas diets high in carbohydrates yielded enterotypes dominated by Prevotella (108, 109). As a mediator between the diet and the host, the intestinal microbiota plays a considerable role in host metabolism. Undigested dietary components (e.g., fructo-oligosaccharides) reach the large intestine, and intestinal microbes ferment the host products (e.g., bile acids). Organic acids, particularly the three short-chain fatty acids (SCFAs) acetate, butyrate, and propionate, are the predominant fermentation products in healthy adults who consume balanced diets (7, 20). However, with unbalanced dietary patterns, microbial metabolism also generates pro-carcinogenic chemicals such as secondary bile acids, N-nitroso compounds (NOCs), and hydrogen sulfide (H2S) (110).
SCFAs are a major class of metabolites produced via microbial metabolism of dietary components. Although acetate, butyrate and propionate have health-promoting effects, butyrate is the most potent with respect to cancer protection. Butyrate, produced predominantly by Firmicutes via fermentation of dietary fiber and resistant starches, is the chief energy source for colonocytes and regulates epithelial proliferation. Butyrate can inhibit histone deacetylase activity in colonocytes and immune cells, consequently downregulating proinflammatory cytokines and inducing apoptosis in CRC cells (111, 112). SCFAs (especially butyrate) can significantly lower fecal pH in the colon, thereby inhibiting pathogenic bacterial proliferation and DNA damage, and enhancing apoptosis and preventing cancer cell proliferation (113). In addition, butyrate and propionate shape the mucosal immune system by regulating colonic regulatory T-cell differentiation (42, 112, 114). Extracellular SCFAs also participate in this process by interacting with host cell surface receptors such as G protein-coupled receptors (GPCRs), GPR41, GPR43, and GPR109A (115–117). In turn, reduced SCFA levels are linked to a high CRC risk, such as in patients with advanced colorectal adenoma (118).
Bile acid metabolism is another main type of microbial metabolism. Primary bile acids are produced in the liver from cholesterol and metabolized to secondary forms by intestinal bacteria. Increasing evidence has shown that fat may affect CRC risk via its role in bile acid metabolism in the host and intestinal microbiota. High-fat diets lead to increased secondary fecal bile acid concentrations in populations with a CRC risk (119, 120). In a long-term diet study, mice fed a western-style diet high in fat developed significantly more colonic tumors than did mice on a control diet, correlating with higher cell proliferation in colonic crypts, impaired bile acid transport, and altered activity of the farnesoid X receptor (FXR), a nuclear bile acid receptor. These results suggest that western-style diets increase cancer risk via FXR inactivation, leading to bile acid deregulation and increased colonocyte proliferation (121). In a dietary intervention trial, healthy African Americans consuming a high-fat, low-fiber diet had more fecal bile acids than did healthy rural Africans who consumed a low-fat, high-fiber diet. However, when these dietary patterns were swapped, this phenomenon changed accordingly (122). Secondary bile acids have been shown to be genotoxic via oxidative stress from ROS generation causing oxidative DNA damage (123).
High protein intake increases detrimental metabolites in the colon, such as NOCs and H2S. NOCs are positively correlated with CRC in European populations and can promote cancer and exert carcinogenic effects via DNA alkylation (124, 125). Sulfate-reducing bacteria may use methionine and cysteine as substrates, leading to H2S generation. A study showed that sulfate-reducing bacterial abundance was increased in the stools of CRC patients compared with those of healthy individuals (126). H2S can stimulate CRC progression by inhibiting butyrate oxidation and inducing breakdown of the gut barrier. H2S can also induce DNA damage via ROS (7, 127).
Biofilm is an emerging concept in studying the relationship between the intestinal microbiota and CRC. Biofilms, aggregations of microbial communities encased in a polymeric matrix, invade the colonic mucosal layer and come into direct contact with mucosal epithelial cells. Dejea et al. reported that invasive polymicrobial bacterial biofilms were detected in most right-sided tumors (89%) but in only 12% of left-sided tumors and were accompanied by diminished IEC E-cadherin, increased epithelial permeability, and enhanced IL-6 and STAT3 activation. The IL-6 family and their downstream effector, STAT3, promote CRC through increased epithelial proliferation and diminished apoptosis. Consequently, Dejea et al. proposed a model that biofilm formation enhanced colonic epithelial permeability, which facilitates bacterial antigen translocation and promotes pro-carcinogenic tissue inflammation (128, 129).
Studies exploring CRC mechanisms share the ultimate goal of better CRC prevention and treatment. Studies on the metagenomic landscape of the CRC microbiota have enabled selecting useful biomarkers, and investigations of microbiota-related mechanisms can help develop effective strategies for CRC prevention and treatment. Since this part is not the focus of this review, we kindly refer to the previous reviews for details (130, 131).
Biomarkers represent a major translational application of the microbiota. Previous studies have shown that microbiota-related biomarkers may be used for screening and as prognostic tools for CRC treatment (Table 2).
Several studies found that alterations in the fecal microbiomes of patients with CRC also occurred in patients with colorectal adenoma, which is recognized as a precursor of most CRCs. Hence, these might be used to screen individuals at risk for CRC, who can be treated in time with excellent clinical outcomes. An effective screening biomarker leading to early detection would substantially reduce CRC-related mortality. The 5-year survival rate of patients with localized CRC is 90%, while that of patients with distant metastatic diseases is only 14% (138). In addition to classic invasive endoscopic approaches, several early noninvasive CRC screening tools, such as fecal immunohistochemical testing (FIT), have been widely used because of their effects on reducing both CRC incidence and mortality. However, these techniques have been criticized for their relatively low sensitivity. FIT has only 79% sensitivity for detecting CRC and 25%–27% sensitivity for detecting advanced colorectal adenomas (139, 140). Therefore, efficient, safe, affordable, and noninvasive screening tools with high sensitivity for CRC are needed, and accumulating metagenomic CRC datasets may enable this. Some studies have demonstrated the potential for combining fecal microbiome data with FIT to improve CRC detection (132, 133). For example, fecal F. nucleatum stands out as highly valuable among several candidate biomarkers (134–136). Adding fecal F. nucleatum quantitation to FIT increases the area under the receiver operating characteristic curve from 0.86 to 0.95 (136). Some groups also try to find oral biomarkers associated with CRC detection, such as Streptococcus and Prevotellas pp. (137). In this study, researchers developed an oral microbiota-based classifier that distinguished patients with CRC and adenomas from healthy individuals. Screening the fecal metabolome is another promising non-invasive procedure for obtaining a unique metabolic fingerprint to diagnose CRC, although few studies with different metabolomic methods have shown the diagnostic potential of metabolites such as SCFAs (141). Apart from the potential for CRC screening, bacterial biomarkers may also serve as prognostic biomarkers. For example, Mima et al. found that larger amounts of F. nucleatum in CRC tissue were associated with worse clinical outcomes, including shorter survival times and a worse prognosis (47).
Finally, many studies have explored associations between microbial markers and CRC, but to date, no universal microbial marker is defined for CRC detection. The complexity of the microbiome presents various challenges. First, the high variability of the intestinal microbiota compositions among different populations owing to sex, age, diet, drug use, genetic background, and geographic location make identifying a universal microbial marker impossible. Thus, validating CRC screening markers for different populations and identifying core biomarkers that are robust across populations may be a possible solution in future studies. Second, limitations in techniques, such as different sample collection and storage methods and various analysis processes should be considered. Hence, standardized methods of sample collection, standardized analysis processes and unified quantitative standards for the microbial markers are needed. Therefore, scientists’ unremitting efforts are needed to overcome these scientific and technical challenges to allow better clinical translation.
Another translational application is microbiota modulation for CRC prevention and treatment. As described previously, the intestinal microbiota plays a major role in CRC via several mechanisms. Therefore, intestinal microbiota modulation, which aims to reverse established microbial dysbiosis, is a novel strategy for CRC prevention and treatment. Different strategies, such as dietary intervention, probiotics, prebiotics, and fecal microbiota transplantation (FMT), have been employed.
First, dietary factors are critical in CRC evolution. Dietary intervention is considered the most reasonable and economical approach to CRC treatment (142). Previous studies have shown the possibility of applying dietary strategies to modulate the intestinal microbiota. Populations consuming different diets have markedly different intestinal microbial compositions. Dietary intervention encourages the growth of certain bacterial strains that may convert indigestible dietary components into beneficial metabolites for the host. One systematic review of cohort studies showed that instead of western diets, adopting a healthy dietary pattern (high intake of fruits and vegetables, whole grain cereals, fish, white meats, and soy derivatives) decreased CRC risk (143). Consistently, in one study, 2-week food exchanges were performed between native Africans with low CRC rates and African Americans with high CRC rates. The African Americans consumed a high-fiber, low-fat diet, and the native Africans consumed a high-fat, low-fiber western diet. The dietary changes resulted in the African Americans exhibiting rapid and reciprocal changes in their intestinal microbiotas and mucosal biomarkers of CRC risk (122). Thus, higher fiber diets might be an effective method of treating CRC. Notably however, although short-term dietary intervention can rapidly reshape the intestinal microbiome, it cannot prevent CRC because once the original long-term diet is resumed, the intestinal microbiome returns to its previous composition (144). Moreover, obesity is positively correlated with CRC risk. Thus, reducing excessive dietary fat intake is extremely important for preventing CRC.
Second, the other ideal method for modulating the microbiota may be direct consumption of probiotics and/or prebiotics. The International Scientific Association for Probiotics and Prebiotics defines probiotics as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (145). Probiotics may function in CRC prevention and treatment by inactivating carcinogens or mutagens, modulating host immunity, inhibiting cell proliferation, and improving gut barrier function (131). Several chemical-induced animal model studies evidenced that administering probiotics exerted significant protective effects against CRC. Faecalibacterium prausnitzii, a potential probiotic, produces hydrophobic microbial anti-inflammatory molecules that can downregulate the NF-κB pathway in intestinal epithelial cells and prevent colitis in animal models (146). Treatment with a mixture of probiotics (Lactobacillus plantarum, L. acidophilus, and Bifidobacterium longum) in CRC patients increased the amount of cell junction proteins, thereby improving intestinal mucosal barrier integrity (147). Oral intake of L. casei reduced the atypia of colorectal tumors in patients who had undergone resection (148). One probiotic intervention study revealed that patients with CRC who received B. lactis Bl-04 and L. acidophilus NCFM had increased abundances of butyrate-producing bacteria, such as Faecalibacterium and Clostridiales spp., and decreased abundances of CRC-associated genera, including Fusobacterium and Peptostreptococcus (149). Prebiotics are nondigestible food ingredients that feed beneficial intestinal bacteria and improve host health. Synbiotics are the combination of prebiotics and probiotics. Numerous clinical trials have reported the effects of synbiotics on patients with CRC, including fewer postoperative infections and shorter hospital stays (150, 151). A synbiotic consisting of prebiotic inulin and the probiotics, L. rhamnosus GG (LGG) and B. lactis Bb12, reduced colorectal proliferation and improved epithelial barrier function in patients with histories of colonic polyps (152). Additionally, probiotic administration can ameliorate the adverse effects of chemotherapy and immunotherapy. Several studies have reported positive effects of probiotic use in CRC, including reduced diarrhea incidence, enhanced gut barrier integrity, and reduced inflammation (147, 153–155). For example, Osterlund et al. suggested that LGG supplementation might reduce the frequency of severe diarrhea and abdominal discomfort in CRC patients receiving 5-fluorouracil (5-FU) (156). Probiotics can also be useful in radiation therapy. A previous study found that probiotics could repair radiation-induced injuries (157). Hence, probiotics may be a potential complement in CRC prevention and treatment.
Third, FMT is an emerging biotherapeutic because of the increased understanding of how an altered intestinal microbiota affects diseases (158). Transferring stool transplants from healthy donors to patients believed to harbor a disease-inducing altered microbiota enables FMT to bring a healthy, disease-free microbiome into the patient’s gastrointestinal tract, which then restores eubiosis and may ameliorate various gastrointestinal disorders, including C. difficile infection (CDI), IBD and irritable bowel syndrome (159). Compared with other modulatory strategies, FMT has its own advantages and is the most direct method of shaping the microbiome with the most evidence of clinical efficacy. Currently, FMT is an established treatment for recurrent and refractory CDI, with cure rates of 80%–90% (160). Although its application in clinical CRC treatment is unexplored, a recent mouse study showed that introducing of fecal transplants from wild to laboratory mice promoted host fitness and improved resistance against DSS/azoxymethane (AOM)-induced colorectal tumorigenesis (161). Thus, FMT may be a novel CRC treatment strategy. Furthermore, modulating the intestinal microbiome via FMT may abrogate refractory colitis as an adverse effect of immunotherapy. Wang et al. reported the first human cases of immune checkpoint inhibitor-associated colitis successfully treated with FMT, with gut microbial reconstitution correlating with complete resolution of colitis for up to 53 days after one dose and 78 days after two doses (162).
In a word, modulating the intestinal microbiota in various ways may improve CRC prevention and treatment. Previous efforts to elucidate oncogenic mechanisms yielded an unprecedented opportunity to explore new strategies for diagnosing and treating CRC, with promising results. However, these strategies have controversies and challenges. For example, although dietary interventions (such as high-fiber intake) may potentially prevent CRC, more clinical and nutritional studies are needed to establish the most appropriate conditions for both dosage and duration (6). Further, not all probiotics are useful or work the same, and their benefit depends on the strain, dosage, intervention duration, and intestinal transit time. The safety of probiotics is also controversial, and some less characterized probiotics can alter the intestinal barrier. Therefore, further investigations are needed to identify safe and effective probiotics for CRC therapy and standardize the application and regulatory frameworks. Orally administered probiotics face an important technical problem that could minimize their efficacy. When they reach the colon, probiotics often lose their viability. Thus, new techniques, such as microencapsulation, must be developed to ensure their viability. Of course, some studies have already reported promising results for these techniques in animal models. More preclinical and clinical studies are needed to elucidate their availability in humans (163). Although FMT applications in treating recurrent and refractory CDI are highly successful, in CRC, they have only been used in animal models, and their clinical use in other diseases requires more supporting data from controlled trials. Currently, FMT is considered as a safe method with few adverse effects, but the long-term outcomes remain unclear. Furthermore, much information regarding the human intestines remains unclear, such as intestinal viral and fungal compositions and intestinal bacterial functions. Hence, disease transmission between the donor and recipient remains a risk. Accordingly, future research should focus on identifying the intestinal microbiota, defining its function, and developing defined microbial communities as alternatives to whole feces transplantation. A sound post-FMT follow-up system must be established to monitor the clinical efficacy and long-term adverse events. Additionally, FMT lacks a unified regulatory framework, and different countries have different regulations. Therefore, formalization of regulatory frameworks becomes other essential issue (164).
The intestinal microbiota, often referred to as a “forgotten organ”, is gradually unraveling its mysterious veil. Numerous studies suggest that the intestinal microbiota is crucial in the CRC pathogenesis. Studies on CRC mechanisms have provided many new ideas for CRC prevention and treatment. However, because of individual variations, tumor stages, and cross-species translation, many challenges remain to be overcome in clinical practice. Continuous efforts in preclinical and clinical research are needed to better understand the links between the intestinal microbiota and CRC. In the near future, the intestinal microbiota will likely become a powerful weapon in fighting CRC.
YC, ZL, and LL discussed the contents, wrote, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
This present work was funded by the grants of the National S&T Major Project of China (2018YFC2000500), and the National Natural Science Foundation of China (81771724, 31700800, 81790631), the Foundation of China’s State Key Laboratory for Diagnosis and Treatment of Infectious Diseases.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut (2017) 66(4):683–91. doi: 10.1136/gutjnl-2015-310912
3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
4. Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin (2017) 67(3):177–93. doi: 10.3322/caac.21395
5. Siegel RL, Fedewa SA, Anderson WF, Miller KD, Ma J, Rosenberg PS, et al. Colorectal Cancer Incidence Patterns in the United States, 1974-2013. JNCI J Natl Cancer Inst (2017) 109(8):djw322. doi: 10.1093/jnci/djw322
6. Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology (2020) 158(2):322–40. doi: 10.1053/j.gastro.2019.06.048
7. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
8. Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer (2002) 99(2):260–6. doi: 10.1002/ijc.10332
9. Rustgi AK. The genetics of hereditary colon cancer. Genes Dev (2007) 21(20):2525–38. doi: 10.1101/gad.1593107
10. Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med (2008) 359(20):2143–53. doi: 10.1056/NEJMra0802968
11. Drewes JL, Housseau F, Sears CL. Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br J Cancer (2016) 115(3):273–80. doi: 10.1038/bjc.2016.189
12. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WKK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun (2015) 6:8727–7. doi: 10.1038/ncomms9727
13. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut (2017) 66(1):70–8. doi: 10.1136/gutjnl-2015-309800
14. Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology (2018) 155(2):529–541 e525. doi: 10.1053/j.gastro.2018.04.018
15. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut (2019) 68(4):654–62. doi: 10.1136/gutjnl-2018-317178
16. Jobin C. Colorectal cancer: looking for answers in the microbiota. Cancer Discovery (2013) 3(4):384–7. doi: 10.1158/2159-8290.CD-13-0042
17. Tlaskalova-Hogenova H, Vannucci L, Klimesova K, Stepankova R, Krizan J, Kverka M. Microbiome and colorectal carcinoma: insights from germ-free and conventional animal models. Cancer J (2014) 20(3):217–24. doi: 10.1097/ppo.0000000000000052
18. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Sci (N Y NY) (2006) 312(5778):1355–9. doi: 10.1126/science.1124234
19. Tsai Y-L, Lin T-L, Chang C-J, Wu T-R, Lai W-F, Lu C-C, et al. Probiotics, prebiotics and amelioration of diseases. J BioMed Sci (2019) 26(1):3–3. doi: 10.1186/s12929-018-0493-6
20. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res (2013) 54(9):2325–40. doi: 10.1194/jlr.R036012
21. Natividad JM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res (2013) 69(1):42–51. doi: 10.1016/j.phrs.2012.10.007
22. Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature (2016) 535(7610):85–93. doi: 10.1038/nature18849
23. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (2016) 352(6285):539–44. doi: 10.1126/science.aad9378
24. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med (2019) 25(3):377–88. doi: 10.1038/s41591-019-0377-7
25. Reddy BS, Weisburger JH, Narisawa T, Wynder EL. Colon carcinogenesis in germ-free rats with 1,2-dimethylhydrazine and N-methyl-n’-nitro-N-nitrosoguanidine. Cancer Res (1974) 34(9):2368–72.
26. Reddy BS, Narisawa T, Wright P, Vukusich D, Weisburger JH, Wynder EL. Colon carcinogenesis with azoxymethane and dimethylhydrazine in germ-free rats. Cancer Res (1975) 35(2):287–90.
27. Reddy BS, Narisawa T, Weisburger JH. Colon carcinogenesis in germ-free rats with intrarectal 1,2-dimethylhydrazine and subcutaneous azoxymethane. Cancer Res (1976) 36(8):2874–6.
28. Son JS, Khair S, Pettet D, Ouyang N, Tian X, Zhang Y, et al. Altered Interactions between the Gut Microbiome and Colonic Mucosa Precede Polyposis in APCMin/+ Mice. PloS One (2015) 10(6):e0127985. doi: 10.1371/journal.pone.0127985
29. Li L, Li X, Zhong W, Yang M, Xu M, Sun Y, et al. Gut microbiota from colorectal cancer patients enhances the progression of intestinal adenoma in Apc(min/+) mice. EBioMedicine (2019) 48:301–15. doi: 10.1016/j.ebiom.2019.09.021
30. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS One (2012) 7(6):e39743. doi: 10.1371/journal.pone.0039743
31. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J (2012) 6(2):320–9. doi: 10.1038/ismej.2011.109
32. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst (2013) 105(24):1907–11. doi: 10.1093/jnci/djt300
33. Zackular JP, Rogers MA, Ruffin MTT, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) (2014) 7(11):1112–21. doi: 10.1158/1940-6207.capr-14-0129
34. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528
35. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med (2019) 25(4):679–89. doi: 10.1038/s41591-019-0406-6
36. Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J (2014) 8(4):881–93. doi: 10.1038/ismej.2013.185
37. Luan C, Xie L, Yang X, Miao H, Lv N, Zhang R, et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep (2015) 5:7980. doi: 10.1038/srep07980
38. Gupta A, Madani R, Mukhtar H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis (2010) 12(3):164–71. doi: 10.1111/j.1463-1318.2009.01814.x
39. Srivastava A, Walter N, Atkinson P. Streptococcus bovis infection of total hip arthroplasty in association with carcinoma of colon. J Surg Orthop Adv (2010) 19(2):125–8.
40. Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis (2011) 53(9):870–8. doi: 10.1093/cid/cir609
41. Sears CL(. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev (2009) 22(2):349–69. doi: 10.1128/cmr.00053-08
42. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med (2009) 15(9):1016–22. doi: 10.1038/nm.2015
43. Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/jci72334
44. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe (2018) 23(2):203–214 e205. doi: 10.1016/j.chom.2018.01.007
45. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
46. Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev (2015) 24(5):373–85. doi: 10.1097/cej.0000000000000116
47. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
48. Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, et al. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget (2016) 7:46158–72. doi: 10.18632/oncotarget.10064
49. Liu X, Cheng Y, Shao L, Ling Z. Alterations of the Predominant Fecal Microbiota and Disruption of the Gut Mucosal Barrier in Patients with Early-Stage Colorectal Cancer. BioMed Res Int (2020) 2020:2948282. doi: 10.1155/2020/2948282
50. Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology (2007) 132(2):551–61. doi: 10.1053/j.gastro.2006.11.040
51. Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol (2008) 23(8 Pt 1):1298–303. doi: 10.1111/j.1440-1746.2008.05490.x
52. Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res (2008) 68(23):9909–17. doi: 10.1158/0008-5472.can-08-1551
53. Veziant J, Gagnière J, Jouberton E, Bonnin V, Sauvanet P, Pezet D, et al. Association of colorectal cancer with pathogenic Escherichia coli: Focus on mechanisms using optical imaging. World J Clin Oncol (2016) 7(3):293–301. doi: 10.5306/wjco.v7.i3.293
54. Chervy M, Barnich N, Denizot J. Adherent-Invasive E. coli: Update on the Lifestyle of a Troublemaker in Crohn’s Disease. Int J Mol Sci (2020) 21(10). doi: 10.3390/ijms21103734
55. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology (2017) 1521419-1433(6):e1415. doi: 10.1053/j.gastro.2017.01.009
56. Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
57. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
58. Denizot J, Desrichard A, Agus A, Uhrhammer N, Dreux N, Vouret-Craviari V, et al. Diet-induced hypoxia responsive element demethylation increases CEACAM6 expression, favouring Crohn’s disease-associated Escherichia coli colonisation. Gut (2015) 64(3):428–37. doi: 10.1136/gutjnl-2014-306944
59. Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis (2011) 203(3):306–11. doi: 10.1093/jinfdis/jiq061
60. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol (2012) 10(8):575–82. doi: 10.1038/nrmicro2819
61. Yu Y-N, Fang J-Y. Gut Microbiota and Colorectal Cancer. Gastrointest Tumors (2015) 2(1):26–32. doi: 10.1159/000380892
62. Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet (2001) 357(9255):539–45. doi: 10.1016/s0140-6736(00)04046-0
63. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
64. Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease. N Engl J Med (2015) 372(15):1441–52. doi: 10.1056/NEJMra1403718
65. Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol (2016) 70(1):395–411. doi: 10.1146/annurev-micro-102215-095513
66. Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut (2001) 48(4):526–35. doi: 10.1136/gut.48.4.526
67. Canavan C, Abrams KR, Mayberry J. Meta-analysis: colorectal and small bowel cancer risk in patients with Crohn’s disease. Aliment Pharmacol Ther (2006) 23(8):1097–104. doi: 10.1111/j.1365-2036.2006.02854.x
68. Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (2012) 338(6103):120–3. doi: 10.1126/science.1224820
69. Grivennikov SI, Greten FR, Karin M. Immunity, Inflammation, and Cancer. Cell (2010) 140(6):883–99. doi: 10.1016/j.cell.2010.01.025
70. Lee TC, Huang YC, Lu YZ, Yeh YC, Yu LC. Hypoxia-induced intestinal barrier changes in balloon-assisted enteroscopy. J Physiol (2018) 596(15):3411–24. doi: 10.1113/jp275277
71. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature (2012) 491(7423):254–8. doi: 10.1038/nature11465
72. Tanaka T, Kohno H, Suzuki R, Hata K, Sugie S, Niho N, et al. Dextran sodium sulfate strongly promotes colorectal carcinogenesis in Apc(Min/+) mice: inflammatory stimuli by dextran sodium sulfate results in development of multiple colonic neoplasms. Int J Cancer (2006) 118(1):25–34. doi: 10.1002/ijc.21282
73. Kosa P, Szabo R, Molinolo AA, Bugge TH. Suppression of Tumorigenicity-14, encoding matriptase, is a critical suppressor of colitis and colitis-associated colon carcinogenesis. Oncogene (2012) 31(32):3679–95. doi: 10.1038/onc.2011.545
74. Goodman B, Gardner H. The microbiome and cancer. J Pathol (2018) 244(5):667–76. doi: 10.1002/path.5047
75. Song X, Gao H, Lin Y, Yao Y, Zhu S, Wang J, et al. Alterations in the Microbiota Drive Interleukin-17C Production from Intestinal Epithelial Cells to Promote Tumorigenesis. Immunity (2014) 40(1):140–52. doi: 10.1016/j.immuni.2013.11.018
76. Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut (2018) 67(11):1984–94. doi: 10.1136/gutjnl-2016-313498
77. Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, et al. Regulation of the IL-23 and IL-12 Balance by Stat3 Signaling in the Tumor Microenvironment. Cancer Cell (2009) 15(2):114–23. doi: 10.1016/j.ccr.2008.12.018
78. Wang K, Min K, Di Caro G, Wong J, Shalapour S, Wan J, et al. Interleukin-17 Receptor A Signaling in Transformed Enterocytes Promotes Early Colorectal Tumorigenesis. Immunity (2014) 41(6):1052–63. doi: 10.1016/j.immuni.2014.11.009
79. Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila) (2017) 10(7):398–409. doi: 10.1158/1940-6207.capr-16-0178
80. Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci (2007) 120(Pt 11):1944–52. doi: 10.1242/jcs.03455
81. Deng Z, Mu J, Tseng M, Wattenberg B, Zhuang X, Egilmez NK, et al. Enterobacteria-secreted particles induce production of exosome-like S1P-containing particles by intestinal epithelium to drive Th17-mediated tumorigenesis. Nat Commun (2015) 6:6956–6. doi: 10.1038/ncomms7956
82. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
83. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
84. Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umaña A, Zhang Y, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal (2020) 13(641). doi: 10.1126/scisignal.aba9157
85. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology (2017) 152851-866(4):e824. doi: 10.1053/j.gastro.2016.11.018
86. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell (2017) 170(3):548–563.e516. doi: 10.1016/j.cell.2017.07.008
87. Ma HM, Shyu KG, Hwang JJ, Huang SH, Hsieh WC, Lien WP. Streptococcus bovis endocarditis associated with colonic adenocarcinoma: report of a case. J Formos Med Assoc (1992) 91(8):814–7.
88. Beebe JL, Koneman EW. Recovery of uncommon bacteria from blood: association with neoplastic disease. Clin Microbiol Rev (1995) 8(3):336–56. doi: 10.1128/CMR.8.3.336
89. Biarc J, Nguyen IS, Pini A, Gossé F, Richert S, Thiersé D, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis ). Carcinogenesis (2004) 25(8):1477–84. doi: 10.1093/carcin/bgh091
90. Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer (2010) 9:249–9. doi: 10.1186/1476-4598-9-249
91. Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis (2014) 3(6):e105. doi: 10.1038/oncsis.2014.20
92. Lu R, Wu S, Zhang Y-G, Xia Y, Zhou Z, Kato I, et al. Salmonella Protein AvrA Activates the STAT3 Signaling Pathway in Colon Cancer. Neoplasia (N Y NY) (2016) 18(5):307–16. doi: 10.1016/j.neo.2016.04.001
93. Wang J, Lu R, Fu X, Dan Z, Zhang Y-G, Chang X, et al. Novel Regulatory Roles of Wnt1 in Infection-Associated Colorectal Cancer. Neoplasia (N Y NY) (2018) 20(5):499–509. doi: 10.1016/j.neo.2018.03.001
94. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U.S.A. (2010) 107(25):11537–42. doi: 10.1073/pnas.1001261107
95. Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun (2014) 5:4724. doi: 10.1038/ncomms5724
96. Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut (2014) 63(12):1932–42. doi: 10.1136/gutjnl-2013-305257
97. Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol (2016) 17(3):230–40. doi: 10.1038/ni.3384
98. Martin OCB, Bergonzini A, D’Amico F, Chen P, Shay JW, Dupuy J, et al. Infection with genotoxin-producing Salmonella enterica synergises with loss of the tumour suppressor APC in promoting genomic instability via the PI3K pathway in colonic epithelial cells. Cell Microbiol (2019) 21(12):e13099–9. doi: 10.1111/cmi.13099
99. de Almeida CV, Taddei A, Amedei A. The controversial role of Enterococcus faecalis in colorectal cancer. Therap Adv Gastroenterol (2018) 11:1756284818783606. doi: 10.1177/1756284818783606
100. Huycke MM, Moore D, Joyce W, Wise P, Shepard L, Kotake Y, et al. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol (2001) 42(3):729–40. doi: 10.1046/j.1365-2958.2001.02638.x
101. Huycke MM, Moore DR. In vivo production of hydroxyl radical by Enterococcus faecalis colonizing the intestinal tract using aromatic hydroxylation. Free Radic Biol Med (2002) 33(6):818–26. doi: 10.1016/s0891-5849(02)00977-2
102. Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res (2004) 567(1):1–61. doi: 10.1016/j.mrrev.2003.11.001
103. Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U.S.A. (2011) 108(37):15354–9. doi: 10.1073/pnas.1010203108
104. Elatrech I, Marzaioli V, Boukemara H, Bournier O, Neut C, Darfeuille-Michaud A, et al. Escherichia coli LF82 differentially regulates ROS production and mucin expression in intestinal epithelial T84 cells: implication of NOX1. Inflammation Bowel Dis (2015) 21(5):1018–26. doi: 10.1097/mib.0000000000000365
105. Zhang FF, Cudhea F, Shan Z, Michaud DS, Imamura F, Eom H, et al. Preventable Cancer Burden Associated With Poor Diet in the United States. JNCI Cancer Spectr (2019) 3(2):pkz034. doi: 10.1093/jncics/pkz034
106. Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, et al. Body Fatness and Cancer–Viewpoint of the IARC Working Group. N Engl J Med (2016) 375(8):794–8. doi: 10.1056/NEJMsr1606602
107. Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell (2018) 33(6):954–64. doi: 10.1016/j.ccell.2018.03.004
108. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U.S.A. (2010) 107(33):14691–6. doi: 10.1073/pnas.1005963107
109. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Sci (N Y NY) (2011) 334(6052):105–8. doi: 10.1126/science.1208344
110. O’Keefe SJD. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol (2016) 13(12):691–706. doi: 10.1038/nrgastro.2016.165
111. Fung KYC, Cosgrove L, Lockett T, Head R, Topping DL. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br J Nutr (2012) 108(5):820–31. doi: 10.1017/S0007114512001948
112. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U.S.A. (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
113. Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, et al. The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response. Cancers (2020) 12(6):1406. doi: 10.3390/cancers12061406
114. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504(7480):446–50. doi: 10.1038/nature12721
115. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem (2003) 278(13):11312–9. doi: 10.1074/jbc.M211609200
116. Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol (2013) 13(6):869–74. doi: 10.1016/j.coph.2013.08.006
117. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007
118. Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr (2013) 97(5):1044–52. doi: 10.3945/ajcn.112.046607
119. Ou J, DeLany JP, Zhang M, Sharma S, O’Keefe SJ. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr Cancer (2012) 64(1):34–40. doi: 10.1080/01635581.2012.630164
120. Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr (2013) 98(1):111–20. doi: 10.3945/ajcn.112.056689
121. Dermadi D, Valo S, Ollila S, Soliymani R, Sipari N, Pussila M, et al. Western Diet Deregulates Bile Acid Homeostasis, Cell Proliferation, and Tumorigenesis in Colon. Cancer Res (2017) 77(12):3352–63. doi: 10.1158/0008-5472.can-16-2860
122. O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun (2015) 6:6342–2. doi: 10.1038/ncomms7342
123. Dvorak K, Payne CM, Chavarria M, Ramsey L, Dvorakova B, Bernstein H, et al. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett’s oesophagus. Gut (2007) 56(6):763–71. doi: 10.1136/gut.2006.103697
124. Gill CI, Rowland IR. Diet and cancer: assessing the risk. Br J Nutr (2002) 88 Suppl 1:S73–87. doi: 10.1079/bjn2002632
125. Loh YH, Jakszyn P, Luben RN, Mulligan AA, Mitrou PN, Khaw KT. N-Nitroso compounds and cancer incidence: the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am J Clin Nutr (2011) 93(5):1053–61. doi: 10.3945/ajcn.111.012377
126. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
127. Marquet P, Duncan SH, Chassard C, Bernalier-Donadille A, Flint HJ. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol Lett (2009) 299(2):128–34. doi: 10.1111/j.1574-6968.2009.01750.x
128. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U.S.A. (2014) 111(51):18321–6. doi: 10.1073/pnas.1406199111
129. Hold GL, Garrett WS. Gut microbiota. Microbiota organization–a key to understanding CRC development. Nat Rev Gastroenterol Hepatol (2015) 12(3):128–9. doi: 10.1038/nrgastro.2015.25
130. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol (2019) 16(11):690–704. doi: 10.1038/s41575-019-0209-8
131. Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene (2020) 39(26):4925–43. doi: 10.1038/s41388-020-1341-1
132. Baxter NT, Ruffin MTT, Rogers MAM, Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med (2016) 8(1):37–7. doi: 10.1186/s13073-016-0290-3
133. Suehiro Y, Zhang Y, Hashimoto S, Takami T, Higaki S, Shindo Y, et al. Highly sensitive faecal DNA testing of TWIST1 methylation in combination with faecal immunochemical test for haemoglobin is a promising marker for detection of colorectal neoplasia. Ann Clin Biochem (2018) 55(1):59–68. doi: 10.1177/0004563217691064
134. Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer (2017) 141(12):2528–36. doi: 10.1002/ijc.31011
135. Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, et al. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin Chem (2018) 64(9):1327–37. doi: 10.1373/clinchem.2018.289728
136. Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut (2017) 66(8):1441–8. doi: 10.1136/gutjnl-2016-312766
137. Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut (2018) 67(8):1454–63. doi: 10.1136/gutjnl-2017-314814
138. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
139. Hundt S, Haug U, Brenner H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med (2009) 150(3):162–9. doi: 10.7326/0003-4819-150-3-200902030-00005
140. Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med (2014) 160(3):171–1. doi: 10.7326/M13-1484
141. Villéger R, Lopès A, Veziant J, Gagnière J, Barnich N, Billard E, et al. Microbial markers in colorectal cancer detection and/or prognosis. World J Gastroenterol (2018) 24(22):2327–47. doi: 10.3748/wjg.v24.i22.2327
142. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature (2013) 500(7464):585–8. doi: 10.1038/nature12480
143. Yusof AS, Md Isa Z, Shah SA. Dietary patterns and risk of colorectal cancer: Asystematic review of cohort studies (2000-2011). Asian Pac J Cancer Prev (2012) 13(9):4713–7. doi: 10.7314/APJCP.2012.13.9.4713
144. Xu Z, Knight R. Dietary effects on human gut microbiome diversity. Br J Nutr (2014) 113(S1):S1–5. doi: 10.1017/s0007114514004127
145. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11(8):506–14. doi: 10.1038/nrgastro.2014.66
146. Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut (2016) 65(3):415–25. doi: 10.1136/gutjnl-2014-307649
147. Liu Z, Qin H, Yang Z, Xia Y, Liu W, Yang J, et al. Randomised clinical trial: the effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery - a double-blind study. Aliment Pharmacol Ther (2011) 33(1):50–63. doi: 10.1111/j.1365-2036.2010.04492.x
148. Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, Takeyama I, et al. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int J Cancer (2005) 116(5):762–7. doi: 10.1002/ijc.21115
149. Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol (2017) 4(1):e000145. doi: 10.1136/bmjgast-2017-000145
150. Sommacal HM, Bersch VP, Vitola SP, Osvaldt AB. Perioperative synbiotics decrease postoperative complications in periampullary neoplasms: a randomized, double-blind clinical trial. Nutr Cancer (2015) 67(3):457–62. doi: 10.1080/01635581.2015.1004734
151. Flesch AT, Tonial ST, Contu PC, Damin DC. Perioperative synbiotics administration decreases postoperative infections in patients with colorectal cancer: a randomized, double-blind clinical trial. Rev Col Bras Cir (2017) 44(6):567–73. doi: 10.1590/0100-69912017006004
152. Rafter J, Bennett M, Caderni G, Clune Y, Hughes R, Karlsson PC, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr (2007) 85(2):488–96. doi: 10.1093/ajcn/85.2.488
153. Demers M, Dagnault A, Desjardins J. A randomized double-blind controlled trial: impact of probiotics on diarrhea in patients treated with pelvic radiation. Clin Nutr (2014) 33(5):761–7. doi: 10.1016/j.clnu.2013.10.015
154. Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol (2019) 234(10):17127–43. doi: 10.1002/jcp.28473
155. Zaharuddin L, Mokhtar NM, Muhammad Nawawi KN, Raja Ali RA. A randomized double-blind placebo-controlled trial of probiotics in post-surgical colorectal cancer. BMC Gastroenterol (2019) 19(1):131. doi: 10.1186/s12876-019-1047-4
156. Osterlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, et al. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br J Cancer (2007) 97(8):1028–34. doi: 10.1038/sj.bjc.6603990
157. Packey CD, Ciorba MA. Microbial influences on the small intestinal response to radiation injury. Curr Opin Gastroenterol (2010) 26(2):88–94. doi: 10.1097/MOG.0b013e3283361927
158. Wang ZK, Yang YS, Chen Y, Yuan J, Sun G, Peng LH. Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World J Gastroenterol (2014) 20(40):14805–20. doi: 10.3748/wjg.v20.i40.14805
159. Smits LP, Bouter KEC, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic Potential of Fecal Microbiota Transplantation. Gastroenterology (2013) 145(5):946–53. doi: 10.1053/j.gastro.2013.08.058
160. Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, et al. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther (2017) 46(5):479–93. doi: 10.1111/apt.14201
161. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell (2017) 171(5):1015–1028 e1013. doi: 10.1016/j.cell.2017.09.016
162. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med (2018) 24(12):1804–8. doi: 10.1038/s41591-018-0238-9
163. Drago L. Probiotics and Colon Cancer. Microorganisms (2019) 7(3). doi: 10.3390/microorganisms7030066
Keywords: biomarker, colorectal cancer, dietary intervention, intestinal microbiota, inflammation, metabolites
Citation: Cheng Y, Ling Z and Li L (2020) The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 11:615056. doi: 10.3389/fimmu.2020.615056
Received: 08 October 2020; Accepted: 02 November 2020;
Published: 30 November 2020.
Edited by:
Qixiao Zhai, Jiangnan University, ChinaReviewed by:
Zhihong Liu, Guangdong Academy of Science, ChinaCopyright © 2020 Cheng, Ling and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanjuan Li, bGpsaUB6anUuZWR1LmNu; Zongxin Ling, bGluZ3pvbmd4aW5Aemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.