
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 11 January 2021
Sec. Comparative Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.603089
 Selma Schmidt1
Selma Schmidt1 Elena L. Sassu2
Elena L. Sassu2 Eleni Vatzia1†
Eleni Vatzia1† Alix Pierron1†
Alix Pierron1† Julia Lagler1,3
Julia Lagler1,3 Kerstin H. Mair1
Kerstin H. Mair1 Maria Stadler1
Maria Stadler1 Christian Knecht2
Christian Knecht2 Joachim Spergser4
Joachim Spergser4 Marlies Dolezal5
Marlies Dolezal5 Sven Springer6
Sven Springer6 Tobias Theuß6
Tobias Theuß6 Vicky Fachinger6
Vicky Fachinger6 Andrea Ladinig2
Andrea Ladinig2 Armin Saalmüller1
Armin Saalmüller1 Wilhelm Gerner1*
Wilhelm Gerner1*The gram-negative facultative intracellular bacteria Salmonella Typhimurium (STM) often leads to subclinical infections in pigs, but can also cause severe enterocolitis in this species. Due to its high zoonotic potential, the pathogen is likewise dangerous for humans. Vaccination with a live attenuated STM strain (Salmoporc) is regarded as an effective method to control STM infections in affected pig herds. However, information on the cellular immune response of swine against STM is still scarce. In this study, we investigated the T-cell immune response in pigs that were vaccinated twice with Salmoporc followed by a challenge infection with a virulent STM strain. Blood- and organ-derived lymphocytes (spleen, tonsils, jejunal and ileocolic lymph nodes, jejunum, ileum) were stimulated in vitro with heat-inactivated STM. Subsequently, CD4+ T cells present in these cell preparations were analyzed for the production of IFN-γ, TNF-α, and IL-17A by flow cytometry and Boolean gating. Highest frequencies of STM-specific cytokine-producing CD4+ T cells were found in lamina propria lymphocytes of jejunum and ileum. Significant differences of the relative abundance of cytokine-producing phenotypes between control group and vaccinated + infected animals were detected in most organs, but dominated in gut and lymph node-residing CD4+ T cells. IL-17A producing CD4+ T cells dominated in gut and gut-draining lymph nodes, whereas IFN-γ/TNF-α co-producing CD4+ T cells were present in all locations. Additionally, the majority of cytokine-producing CD4+ T cells had a CD8α+CD27- phenotype, indicative of a late effector or effector memory stage of differentiation. In summary, we show that Salmonella-specific multifunctional CD4+ T cells exist in vaccinated and infected pigs, dominate in the gut and most likely contribute to protective immunity against STM in the pig.
Salmonella Typhimurium (STM) is a gram-negative facultative intracellular bacterium that belongs to the family of the Enterobacteriaceae and is able to infect a broad range of hosts. Non-typhoidal Salmonella serovars such as STM frequently cause food-borne gastroenteritis in humans due to their zoonotic properties and pose a permanent risk for food safety (1, 2). Apart from eggs and egg products, pigs and pork are among the top transmission vectors of STM to humans via the food chain (3, 4).
Salmonellosis in the pig can manifest as diarrhea and lethargy in weaned pigs; however, in many cases, pigs are infected subclinically and often become carrier animals with Salmonella persisting in tonsils, gut and gut-associated lymphoid tissues (5). In addition to hygiene measures and feed intervention strategies, vaccination is considered an effective tool in controlling Salmonella in affected pig farms (6). The live attenuated histidine-adenine auxotrophic vaccine Salmoporc (Ceva Santé Animale, Libourne, France, formerly IDT Biologika GmbH) is commercially available in Europe, and has already been proven to reduce clinical signs, shedding and tissue colonization in pigs in several studies (7–13).
So far, studies covering the immune response against STM in the pig have focused on detecting signs of antibody-mediated immunity. Vaccination of pigs with Salmoporc has been shown to induce Salmonella-specific IgM, IgG and IgA antibodies (11, 14). Studies in which pigs were infected with a virulent STM strain observed an increase of Salmonella-specific IgM antibodies with IgM-IgG class switch occurring 14 days post infection, as well as the induction of Salmonella-specific IgA antibodies (15, 16). In contrast to the humoral immune response, the cell-mediated immune response to STM in swine has not been investigated in detail so far. Existing studies have mainly focused on measuring mRNA expression of cytokines in STM-infected piglets (17–20).
STM infection has been more intensively studied in mice where CD4+ T cells were shown to have a dominant role in primary clearance of infection (21, 22). Especially the development of Th1 cells and their production of tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ), leading to the activation of macrophages, is required for successful bacterial killing (23–26). Additionally, interleukin-17A (IL-17A) produced by Th17 cells contributes to protection by recruiting neutrophils to the intestine and regulating the expression of epithelial tight junction proteins (27, 28). In fact, Salmonella-specific Th1 and Th17 cells have been shown to develop simultaneously in spleen and intestine of orally infected mice (29). More recent studies in mice have also demonstrated the importance of non-circulating tissue-resident memory Th1 cells which are able to react immediately in case of a re-infection (30). In contrast to mice in which STM leads to septicemia, STM colonization in the pig is mainly focused on the intestinal tract. The local cellular immune response mounted in gut and mesenteric lymph nodes as well as the generation of memory T cells within these tissues is therefore of great interest. Although the identification of tissue-resident T cells (Trm) in swine is not yet possible due to a lack of antibodies specific for Trm associated markers, differentiation of CD4+ T cells in the pig can be analyzed by the expression of CD8α and CD27 (31). In this way three major subsets within porcine CD4+ T helper cells can be identified: the subset of CD8α-CD27+ CD4+ T cells constitutes naïve cells, while the CD8α+CD27+ and CD8α+CD27- subsets represent central and effector memory CD4+ T cells, respectively (32).
Due to the lack of knowledge on T-cell mediated immunity in pigs in response to STM infections, we studied the antigen-specific CD4+ T-cell immune response of swine, both locally and systemically. CD4+ T cells and their production of IFN-γ, TNF-α and IL-17A were analyzed in various organs by intracellular cytokine staining (ICS) after in vitro stimulation with Salmonella antigen. Our results show the induction of STM-specific multifunctional CD4+ T cells after vaccination and infection with STM that predominantly possessed an effector memory phenotype and dominated in the porcine intestine.
Prior to the study, sows (Large White x Landrace) from a University-owned pig farm in Lower Austria were tested for Salmonella-specific antibodies by the IDEXX Swine Salmonella Ab Test (IDEXX Europe, Hoofddorp, The Netherlands). The five sows with the lowest sample to positive (S/P) ratios were selected and sixteen four-week-old male castrated pigs (Large White x Landrace x Pietrain) from those sows were included in the study. Piglets at this farm are routinely vaccinated against PCV-2 (Ingelvac CircoFLEX®, Boehringer-Ingelheim, Ingelheim am Rhein, Germany) at three weeks of age and Mycoplasma hyopneumoniae (M+PAC®, MSD Animal Health, Kenilworth, USA) in the first and third week of life. After arrival, the animals were weighed and the data was used to achieve a similar distribution of animals with different body weights in two groups: a control group of four animals (pig #1-4) and a group of twelve animals that was later vaccinated and challenge-infected (pig #5-16, V+I). Animals were housed in two separate rooms on straw bedding for the first six weeks. One week before the infection, all animals were moved into a biosafety level (BSL) 2 facility at the University of Veterinary Medicine Vienna with the control group and the V+I group accommodated in separate compartments of the facility. The Salmonella-free status of the piglets was validated by serological testing for Salmonella-specific antibodies by the IDEXX Swine Salmonella Ab Test (IDEXX Europe) (Figure 1). Moreover, fecal samples were collected 9, 7 and 6 days prior to the first vaccination and tested for Salmonella. The methodology for this microbiological testing is described below in chapter 2.4. All piglets tested negative for Salmonella in the feces at the indicated time points.
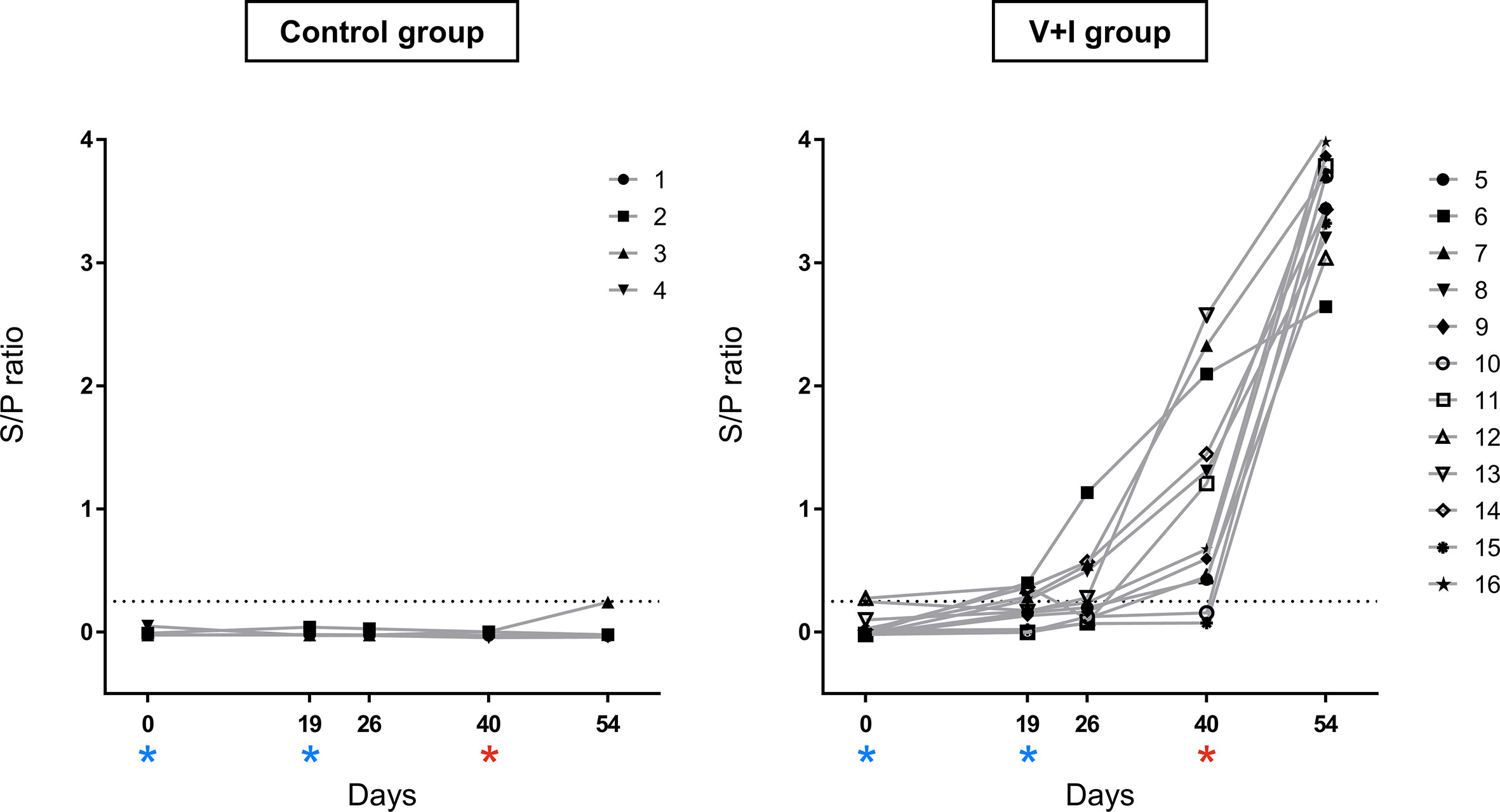
Figure 1 Salmonella-specific antibodies in serum over the course of the experiment. Salmonella-specific antibodies were measured by ELISA in serum of all animals. Sample to positive (S/P) ratios were calculated by dividing optical densities (OD) of samples by OD of the positive reference control. The dotted line represents the cut-off at a S/P ratio of 0.25. Results < 0.25 were defined as negative, samples ≥ 0.25 as positive. The left graph shows ratios for control animals, the right graph for vaccinated and infected animals. Dates of vaccinations (SD0, SD19) are indicated by blue asterisks, date of infection (SD40) by a red asterisk. The combination of different symbols and numbers represent values of individual animals.
Starting at five weeks of age, the V+I group was orally vaccinated twice in a 19-day interval [study days (SD) 0 and 19] with 1 ml of the live attenuated histidine-adenine auxotrophic STM vaccine (Salmoporc, Ceva Santé Animale, Dessau-Roßlau, Germany) containing a dose of 1.33 x 109 colony forming units (cfu). The control group received 1 ml of tap water orally. The vaccine was applied using a button cannula attached to a 5 ml syringe. Three weeks after the second immunization (SD40), animals of the V+I group were orally challenged with 5 ml per animal containing 1 x 109 cfu/ml of a virulent monophasic STM strain (DT193, no. RKI 06-1900, described by (33) and provided by Ceva Innovation Center GmbH). For the application via an oral drencher kit (Ceva Santé Animale) the challenge strain was mixed with sugar beet solution to enhance acceptance by the animals. The control group received 5 ml of sugar beet syrup diluted in water. Two weeks after infection, necropsy was performed over five consecutive days (SDs 52–56). All animals were anaesthetized by intramuscular injection of Ketaminhydrochlorid (Narketan®, 10 mg/kg body weight, Vétoquinol, Lure Cedex, France) and Azaperon (Stresnil®, 1.3 mg/kg body weight, Elanco, Greenfield, USA) followed by euthanasia via intracardial injection of T61® (tetracaine hydrochloride, mebezonium iodide and embutramide, 1 ml/10 kg body weight, MSD Animal Health). The animal experiment was approved by the institutional ethics committee, the Advisory Committee for Animal Experiments (§12 of Law for Animal experiments, Tierversuchsgesetz - TVG) and the Federal Ministry for Science, Research and Economy (BMWF-68.205/0241-WF/V/3b/2016).
Clinical monitoring of the animals was performed daily and observations were recorded and evaluated by a scoring system, considering rectal temperature, diarrhea, vomiting and changes in behavior. In addition, body weight of all animals was recorded on a weekly basis. After euthanasia inner organs of all animals were evaluated for pathological changes by visual examination. Fecal samples were taken from all animals at the beginning of the study (SDs −9, −7, −6) as well as in two-week intervals after the first vaccination (SDs 12, 26) for bacteriological examination. Blood samples from the jugular vein (Vena jugularis) were taken from all animals prior to vaccination (SD0) and prior to second immunization and challenge infection (SDs 19, 26, 40). On necropsy days (SDs 52–56) blood was drawn by cardiac puncture from anaesthetized animals prior to euthanasia. On these days, samples were also collected from liver, spleen, tonsils, jejunal lymph nodes, ileocolic lymph nodes, jejunum, ileum and cecum. Tissue samples from liver, jejunum, ileum and cecum were always taken from the same area of the organ. The liver sample was taken from the dorsal portion of the left lobe. The jejunum was fully unrolled and the middle determined. From this point, 15 cm of tissue was dissected in the oral and aboral direction (i.e. total length of 30 cm). For the sample of the ileum, a 30 cm section orally of the Ostium ileale was collected. A ligation 5 cm from the cecal tip provided the sample of the cecum.
Serum was obtained after centrifugation of blood samples for 10 min and 1,900 × g at room temperature. For the detection of Salmonella-specific antibodies, serum samples were tested using a commercially available ELISA kit (IDEXX Swine Salmonella Ab Test, IDEXX Europe, Hoofddorp, The Netherlands) according to the manufacturer’s instructions. Results were recorded as S/P ratios determined by the ratio between mean optical density (OD) of each sample and mean OD of the positive control. According to the manufacturer’s recommendations, S/P ratios < 0.25 were defined as negative and samples ≥ 0.25 as positive.
Liver, spleen, tonsils, jejunal lymph nodes, ileocolic lymph nodes, jejunum, ileum and cecum from euthanized animals as well as fecal samples taken before and after both immunizations of the animals were investigated for the presence of Salmonella enterica. Samples were streaked onto Xylose-Lysine-Deoxycholate (XLD) agar plates (BBL™, Becton Dickinson (BD), Heidelberg, Germany) and incubated in ambient air at 37°C for 48 h. In addition, all samples were pre-enriched in buffered peptone water (BPW, Millipore™, Merck KGaA, Darmstadt, Germany) for 24 h at 37°C. After incubation 0.1 ml of each culture was transferred to Rappaport-Vassiliadis R10 and Selenite broth (both Difco™, BD), incubated for 24 h at 42°C, and subsequently sub-cultured onto XLD agar plates (BBL™, BD) and incubated aerobically at 37°C for 48 h. Presumptive Salmonella colonies were confirmed by MALDI TOF mass spectrometry.
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood sampled in monovettes (Kabe Labortechnik, Nümbrecht-Elsenroth, Germany) by density gradient centrifugation (Pancoll human, density: 1.077 g/ml, PAN Biotech, Aidenbach, Germany; 30 min at 920 x g). Lymphocytes from spleen, tonsil and mesenteric lymph nodes were isolated as described previously (31). To collect lamina propria lymphocytes (LPL) from jejunum and ileum, opened intestines were rinsed in sterile phosphate-buffered saline (PBS, PAN Biotech) and dissected into small pieces (approx. 2x2x2 mm). Subsequently, the tissue was incubated for 60 min at 37°C on a shaker in HBSS supplemented with 2 mM DTT (Carl Roth GmbH+Co.KG, Karlsruhe, Germany), 0.1 mM EDTA (Carl Roth) and 25 U/ml DNase Type 1 (ThermoFisher, Waltham, MA, USA) for the release of intra-epithelial lymphocytes. Supernatants from this incubation were discarded and the remaining tissue was placed in cell culture medium (RPMI 1640 supplemented with 100 IU/ml penicillin and 0.1 mg/ml streptomycin (all PAN Biotech)) to remove traces of DTT and EDTA for 15 min at 37°C. Thereafter, the tissue was transferred to cell culture medium supplemented with 25 U/ml DNase Type 1 and 300 U/ml Collagenase 1 (ThermoFisher) for enzymatic degradation. After incubation for 60 min on a shaker at 37°C, supernatants were collected, centrifuged at 4°C and 600 x g for 10 min and the cell pellet was re-suspended and filtered through a cell strainer. Cells were re-suspended in 40% Percoll® (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and under-layered with 70% Percoll before being centrifuged at room temperature for 30 min at 920 x g. Cells from the interphase were collected, washed twice with PBS and once with cell culture medium (as above but with 5% fetal calf serum (FCS, Merck, Darmstadt, Germany). Afterwards, cells were re-suspended in cell culture medium supplemented with gentamicin and 10% FCS. All cell preparations were counted in a Sysmex XP 300 hematology analyzer (Sysmex Europe GmbH, Norderstedt, Germany).
Antigens for in vitro stimulation of lymphocytes were prepared as follows. The vaccine strain (STM no. 421/125) and the challenge strain (STM no. RKI 06-1900) were cultured via two precultures in STM 6/83 medium (in-house) at 37°C and 150 rpm on a shaker. The culture was then centrifuged at 7000 x g for 10 min and the pellet re-suspended in PBS. After determination of colony forming units, the concentrate was heat-inactivated in a water bath at 60°C for 90 min. Subsequently, the antigen was aliquoted and stored at −80°C until use.
For intracellular staining of IFN-γ, TNF-α and IL-17A, 5 x 105 freshly isolated cells were cultivated in 200 µl/well of round-bottomed 96-well microtiter plates (Greiner Bio One, Frickenhausen, Germany) For stimulation, wells received either heat-inactivated 1.9 x 108 cfu/ml STM vaccine strain or heat-inactivated 2.7 x 108 cfu/ml STM challenge strain. In pilot experiments, we had established that frequencies of cytokine-producing CD4+ T cells plateaued at STM doses >108 cfu/ml (data not shown). The plates were then cultured for approximately 19 h at 37°C. Cells incubated in cell culture medium only served as negative controls. During the last 4 h of culture, Brefeldin A (BD GolgiPlug™, BD Biosciences, San Jose, CA, USA) was present in microcultures at a final concentration of 1 μg/ml. Cultivated cells were harvested and re-suspended in buffer containing PBS with 3% FCS. To analyze the phenotype of lymphocyte subsets by flow cytometry (FCM), cells were surface-stained with primary monoclonal antibodies directed to CD4 (mIgG2b, clone: 74-12-4), CD8α (mIgG2a, clone: 11/295/33, biotinylated) and CD27 (mIgG1, clone: b30c7), all produced and prepared in-house. Binding of primary antibodies was detected by the following secondary reagents: goat anti-mouse IgG2b-A488, rat anti-mouse IgG1-PE-Cy7 (both ThermoFisher) and Streptavidin-BV421 (BioLegend, San Diego, CA, USA). For exclusion of dead cells, Fixable Viability Dye eFlour780 (ThermoFisher) was used according to manufacturer’s protocol with 0.025 μl reactive dye per sample. Free binding sites of secondary antibodies were blocked with whole mouse IgG molecules (2 µg per sample; Jackson ImmunoResearch Laboratories, West Grove, PA, USA). Thereafter, samples were fixed and permeabilized with BD Cytofix/Cytoperm™ Fixation/Permeabilization Kit (BD Biosciences) according to manufacturer’s instructions. This was followed by intracellular staining with IFN-γ-PE (mIgG1, clone: P2G10, BD Biosciences), TNF-α-BV605 (mIgG1, clone: Mab11, BioLegend) and IL-17A-A647 (mIgG1, clone: SCPL1362, BD Biosciences). All incubation steps were performed in 96-well round-bottom plates for 20 min at 4°C with the exception of the intracellular staining step that was carried out for 30 min. FCM analyses were performed on a FACSCanto™ II (BD Biosciences). Data of at least 1 x 106 lymphocytes per sample for PBMC, spleen, tonsils and lymph nodes were recorded. For cells isolated from jejunum and ileum at least 3 x 105 lymphocytes were recorded. Data were analyzed with FlowJo™ Software for Windows (Version 10.4.1; FlowJo, Ashland, OR, USA).
Data for the graphs in Figures 1, 7, and S2 including calculation of mean, median and standard deviations were analyzed with GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). Statistical analysis was performed in R version 3.6.2 (R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/). We fitted univariate linear mixed models using function lmer in R package lme4 v1.1-21 (34) with log10 transformed frequencies of cytokine-producing CD4+ T cells as “response”. Main effects of animal treatment and in vitro stimulation and the interaction between them were modeled as fixed categorical effects with two levels (control versus infected) and three levels (medium, vaccine strain and challenge strain), respectively. A random intercept pig effect was included in the model to account for the covariance structure (multiple observations per pig) in our data. For the hypothesis testing, we used maximum likelihood estimation by setting option REML to false. All assumptions for linear mixed models were met. Residuals and random intercepts were normally distributed and residuals homoscedastic. We verified the absence of collinearity via generalized variance inflation factors (35) using function vif in package car v3.0-8 (36). We calculated contrasts between least square means of animal treatment and in vitro stimulation levels respectively with package emmeans v1.4.7 (37). Significance was declared at a multiple testing corrected 10% false discovery rate (38). We further performed Principal Component Analysis with package factoextra v1.0.7 (39). We produced biplots using function fviz_pca_biplot, which display PCA scores of samples (shown as dots) and loadings of each variable (shown as vectors) in the same graph. Dots that are close to each other represent samples with similar values. The longer a vector of a variable the bigger the influence of this variable on that principal component. Vectors pointing in a similar direction, forming small angles between them can be interpreted as positively correlated, vectors forming an angle of 90° as uncorrelated and vectors pointing in opposing directions as negatively correlated. We also produced heatmaps with function heatmap.2 in package gplots v3.0.3. (40). Hierarchical clustering using option method= “ward.D2”, based on Euclidian distances for rows and columns are shown as dendrograms. For these multivariate descriptive plots, centered and scaled frequencies of cytokine-producing CD4+ T cells corrected for individual pig effects were used. We calculated these residuals by subtracting restricted maximum likelihood BLUP animal effects, (option REML set to true), estimated from the same linear mixed model as used for hypothesis testing, from the log10 transformed raw frequencies of cytokine-producing CD4+ T cells. During statistical analysis, one animal from the control group (pig #4) was found to be an outlier in the majority of analyzed tissues and was therefore excluded from the ICS data set and subsequent multivariate plots as well as hypothesis testing of cytokine-producing phenotypes for all tissues.
Due to a lack of knowledge on the T-cell response of pigs against STM, the animal experiment for this study was designed to achieve a strong stimulation of the immune system of pigs in the V+I group by a two-time vaccination and subsequent challenge infection, based on previous observations in demonstrating the efficacy of the Salmoporc vaccine (11). With this study design, no adverse effects on the health of the pigs following infection were expected. Indeed, average daily gain of weight developed homogeneously for both groups for the duration of the study. After moving to the BSL2 animal facility, both groups showed an increase in rectal temperature for one day with temperatures ranging between 39.3°C and 40.4°C and an average increase of 0.35°C in the control group and 0.2°C in the V+I group. This rise in temperature can most likely be attributed to the stress of being moved and adapting to a new environment. After that, all animals showed rectal temperatures within a physiologic range until the end of the study. In addition, mild signs of diarrhea (e.g. pasty feces) were observed in a small number of V+I animals during a three-day interval after challenge infection (data not shown).
To confirm a successful STM infection of the animals, production of serum antibodies as well as STM presence in organs were investigated. The humoral response against STM was evaluated by use of the commercially available IDEXX Swine Salmonella Ab Test (Figure 1). Results of the Salmonella antibody ELISA confirmed the Salmonella-free status of the animals at the beginning of the study (SD0). All pigs from the control group remained serologically negative for Salmonella sp. throughout the whole course of the experiment. S/P ratios of animals from the V+I group rose moderately after the second immunization and a further substantial increase was observed after challenge infection on SD40.
Samples from spleen, liver, tonsils, jejunal lymph nodes, ileocolic lymph nodes, jejunum, ileum and cecum taken at necropsy for each animal were analyzed for the presence of Salmonella enterica. No Salmonella spp. could be detected in the control group in any of the sampled organs (Figure 2). Regarding the V+I group, Salmonella spp. could be isolated from all three selected parts of the gut, the jejunal lymph nodes and the ileocolic lymph nodes from at least two animals. Additionally, Salmonella spp. were also identified in the tonsils of all V+I animals. Spleen and liver were negative for both control group and V+I group. As the pigs were kept under isolated conditions and the vaccine strain has a limited persistence, it can be assumed that the detection of Salmonella spp. is the STM challenge strain. Fecal samples collected prior to (SDs -9, -7, -6) and after both vaccinations (SDs 12, 26) of all animals (control and V+I group) tested negative for Salmonella spp. as well (data not shown).
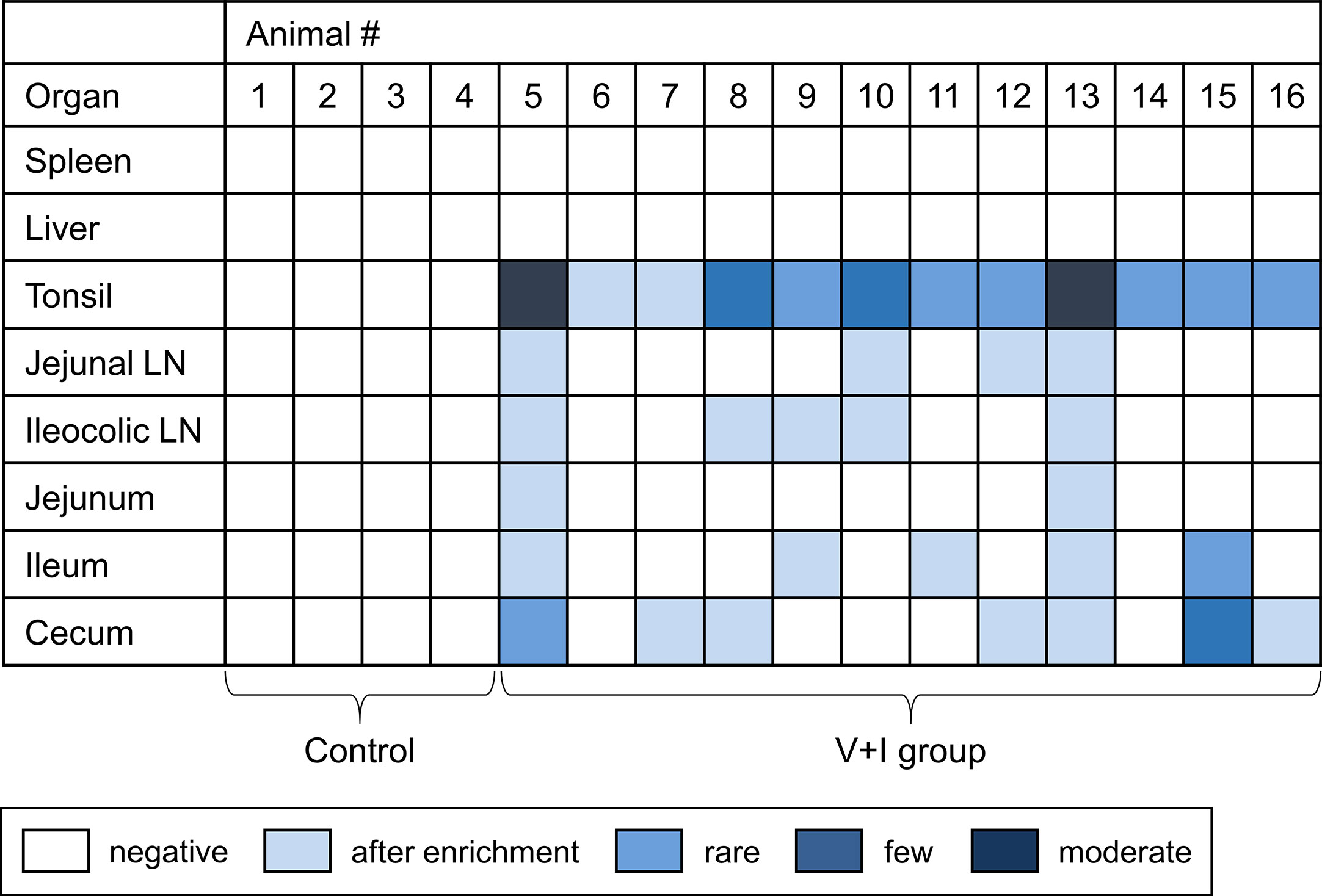
Figure 2 Detection of Salmonella Typhimurium on the day of necropsy. Microbiological analysis was performed for spleen, liver, tonsil, jejunal lymph node, ileocolic lymph node, jejunum, ileum and cecum. Results are displayed as a heat map: dark blue/blue boxes indicate detection of Salmonella Typhimurium by agar isolation. Light blue boxes indicate isolation after enrichment. White boxes display negative findings for Salmonella Typhimurium.
Main focus of the study was the analysis of a Salmonella-specific CD4+ T-cell response in regard to IFN-γ, TNF-α, and IL-17A production. To that end, PBMC as well as lymphocytes isolated from spleen, tonsils, jejunal lymph nodes, ileocolic lymph nodes, jejunum and ileum collected on final SDs were stimulated in vitro with either the vaccine strain or the challenge infection strain. Cells cultivated in medium-only served as negative controls. For analysis of cytokine-producing cells and their phenotype, multi-color FCM was performed. To identify Salmonella-specific CD4+ T cells, CD4+ cells were gated within live lymphocytes and further analyzed for IFN-γ, TNF-α and IL-17A production (Figure S1A). Subsequently, Boolean gating was applied, resulting in seven possible cytokine-producing phenotypes: IFN-γ single-producing, TNF-α single-producing, IL-17A single-producing, IFN-γ/TNF-α co-producing, IFN-γ/IL-17A co-producing, TNF-α/IL-17A co-producing and IFN-γ/TNF-α/IL-17A triple-producing cells. Raw frequencies of these cytokine-producing phenotypes for all organs and in vitro stimulations are listed in Table S1.
The frequencies of cytokine-producing CD4+ T cells varied between different animals and organs following stimulation with STM (Figures S1B–D). Comparing frequencies of cytokine-producing CD4+ T cells across organs, highest frequencies of Salmonella-specific CD4+ T cells for all cytokine-related phenotypes could be observed in LPL preparations of jejunum and ileum (Figure S2, N.B. the scaling of the y-axes in this figure varies between jejunum/ileum, lymphoid organs and blood/spleen).
To work out the significance of noted differences between control group and V+I animals, we chose to model the proportions of cytokine-producing CD4+ T cells for all phenotypes, animals and organs in a generalized linear mixed model. All samples from control animals as well as non-stimulated samples from V+I animals that were cultivated in medium-only served as reference values. The effect of each pig as an individual strongly influenced the data and therefore residuals were calculated for each sample to minimize the effect of the factor ‘animal’. Principal component analysis (PCA) and heat map analysis were carried out on residuals of cytokine-producing CD4+ T cells. With the exception of IFN-γ single-producing CD4+ T cells in blood, no significant differences in frequency of cytokine-producing CD4+ T cells were found between stimulation with the vaccine strain and the challenge infection strain (Table S2). Furthermore, no separate clustering could be observed between both stimulation variants in PCA and heat map analysis (data not shown). For these reasons, data from both stimulation variants were combined for all subsequent analyses.
STM primarily infects the gastrointestinal tract and does so by breaching the gut epithelial barrier. To examine the local T-cell immune response at this site of invasion, cluster analysis was first conducted for the two intestinal sections (jejunum, ileum, Figure 3). Dimension 1 of the PCA revealed a clear separation of the gut tissue-derived data points into two fractions: Samples from V+I animals stimulated with STM clustered together (orange dots) on one side. On the opposite side, all samples from control animals (blue dots if stimulated with STM antigen, green dots if cultivated in medium) as well as V+I samples cultivated in medium-only (remaining green dots) formed a cluster (Figures 3A, C). Hence, Dimension 1 resulted out of the animal treatment (control vs. vaccination and infection) as well as stimulation of the individual samples (medium vs. STM antigen). For both intestinal sections, Dimension 1 accounted for more than 80% of the variation. Dimension 2, which accounted for 8.1% and 10.1% of variation in jejunum and ileum, respectively, is associated with the type of cytokine production. Predominantly IFN-γ single- and IFN-γ/TNF-α co-producing cells clustered together in the upper quadrant while IL-17A containing phenotypes were grouped toward the other direction. Findings of the PCA were corroborated by results of the heat map analysis (Figures 3B, D). Heat maps show increases (red) and decreases (blue) of relative abundance of cytokine-producing CD4+ T cells. Relative abundances of phenotypes were markedly increased in samples from the V+I group that were stimulated with STM when comparing it to samples from control animals and medium-only samples. This resulted in two distinct clusters where STM-stimulated V+I samples are clearly separated from samples of control animals and samples from all animals that were cultivated in medium. Again similar to PCA results, hierarchical clustering of cytokine phenotypes in dendrograms showed that IL-17A+ phenotypes such as IFN-γ/TNF-α/IL-17A triple-producing, IFN-γ/IL-17A co-producing and TNF-α/IL-17A co-producing CD4+ T cells are closely related phenotypes while IFN-γ/TNF-α co-producing and IFN-γ single-producing CD4+ T cells clustered apart from them. To determine whether the observed differences between the control and the V+I group were significant, p-values were calculated for all phenotypes and organs and corrected for multiple testing where necessary (p-values below 0.1 were considered significant). Significant differences between control group and V+I group were found for 5 out of 7 phenotypes in both intestinal sections (Table 1). The phenotypes not reaching significant difference between groups were IL-17A single-producing CD4+ T cells in both intestinal tissues as well as IFN-γ single-producing CD4+ T cells in the jejunum and TNF-α/IL-17A co-producing CD4+ T cells in the ileum.
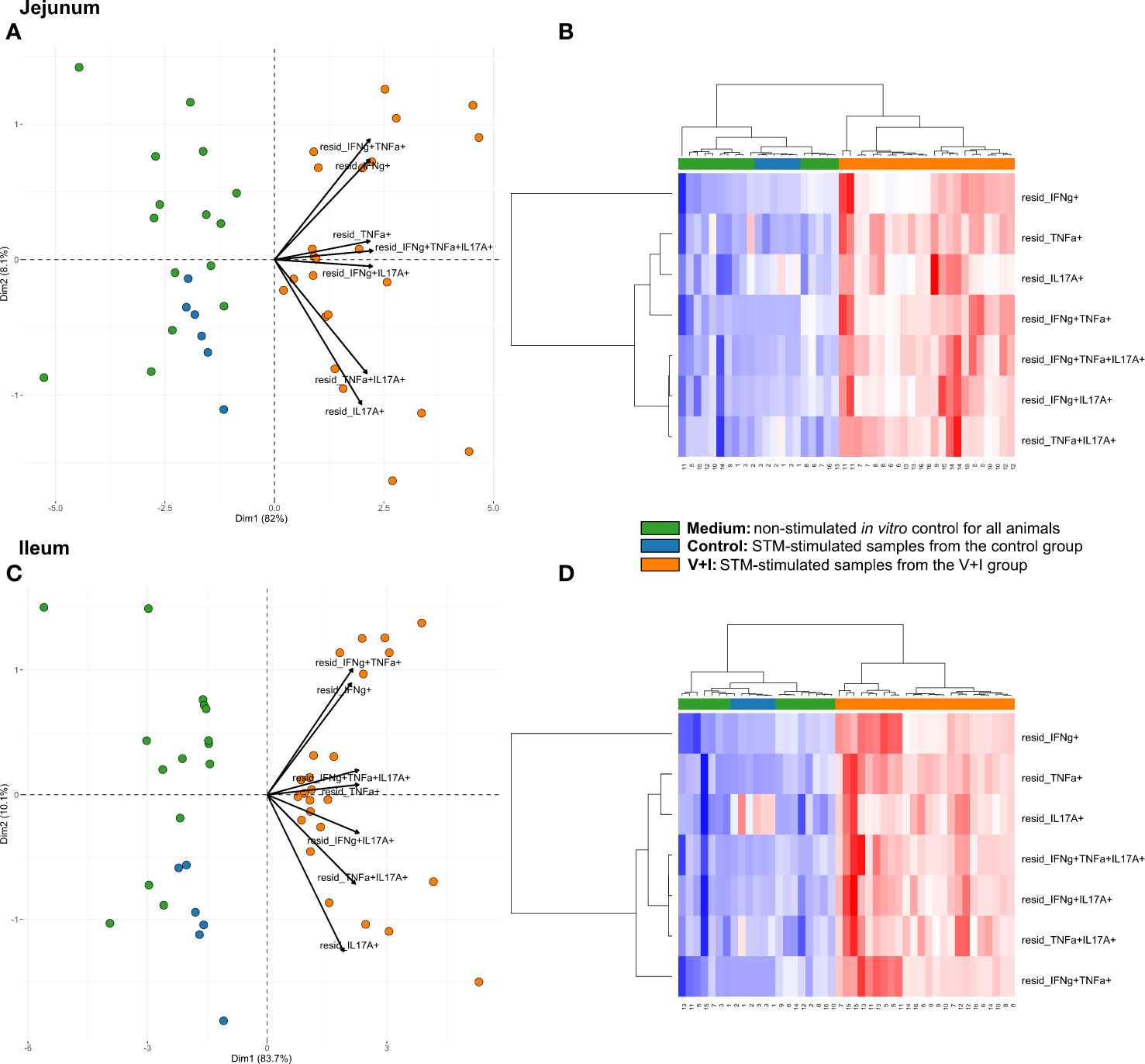
Figure 3 Unsupervised cluster analysis for Salmonella Typhimurium (STM)-stimulated interferon-γ (IFN-γ)/tumor necrosis factor-α (TNF-α)/interleukin-17A (IL-17A) producing CD4+ T cells isolated from jejunum and ileum. (A) Principal component analysis (PCA) based on residuals calculated from cytokine-producing CD4+ T cells derived from jejunum and stimulated in vitro with STM or medium-only. Each colored circle represents one of three samples derived from one animal. Samples stimulated with STM (vaccine or challenge strain) are depicted in orange for V+I animals and blue for control animals; samples from both groups cultivated in medium-only are shown in green. Arrows depict cytokine-producing phenotypes. (B) Heat map of residuals calculated from cytokine-producing CD4+ T cells derived from jejunum and stimulated in vitro with STM or medium-only. Heat maps, generated from data converted to Z-scores, show increase (red) or decrease (blue) of relative abundance of cytokine-producing CD4+ T cells. Each row represents a cytokine-producing phenotype and each column a sample. Numbers below columns indicate numbers of the individual animals. Samples stimulated with STM (vaccine or challenge strain) are indicated by orange boxes for V+I animals and blue boxes for control animals; samples from both groups cultivated in medium-only are indicated by green boxes. (C, D) Same as (A), (B) but for cytokine-producing CD4+ T cells derived from the ileum.
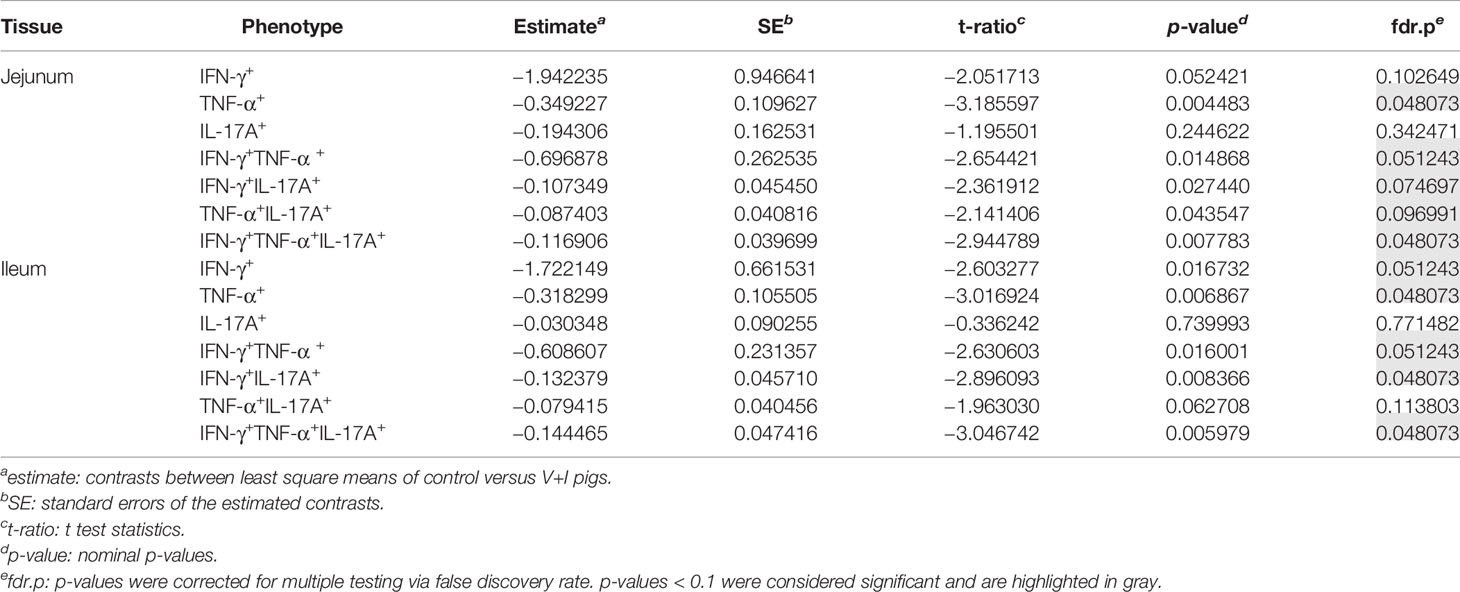
Table 1 Calculation of contrasts of Salmonella-specific CD4+ T-cell cytokine responses between control and V+I pigs in jejunum and ileum.
Looking at the set of lymphatic organs, clustering in both intestinal lymph nodes was similar to that observed in jejunum and ileum, for both PCA and heat maps (Figures 4A–D). In both mesenteric lymph nodes, STM-stimulated V+I samples clustered away from control and medium-only samples with Dimension 1 accounting for approximately 70% of the variation (Figures 4A, C). Of note, IFN-γ single-producing CD4+ T cells clustered separately from all other phenotypes in the PCA. This is most probably due to an increase in abundance of cytokine-producing cells in some control animals for this phenotype as displayed in the corresponding heat maps (Figures 4B, D). Accordingly, differences in relative abundance of IFN-γ single-producers between groups did not reach significance for either lymph node (Table 2). Comparable to observations in the intestine, phenotypes that included IL-17A also formed closely related clusters in both lymph nodes as illustrated by heat map dendrograms. P-values for both lymph nodes revealed significant differences between control conditions and STM-stimulated samples from V+I animals for TNF-α single-producing, IFN-γ/TNF-α co-producing and TNF-α/IL-17A co-producing CD4+ T cells. Additional significant differences were reached for IFN-γ/IL-17A co-producing and IFN-γ/TNF-α/IL-17A triple-producing CD4+ T cells in ileocolic lymph nodes. In the tonsil, however, separation of STM-stimulated V+I samples from control animals and medium-only samples was less distinct (Figures 4E, F). Correspondingly, no significant differences in abundance of cytokine-producing phenotypes could be detected between groups in the tonsil (Table 2).
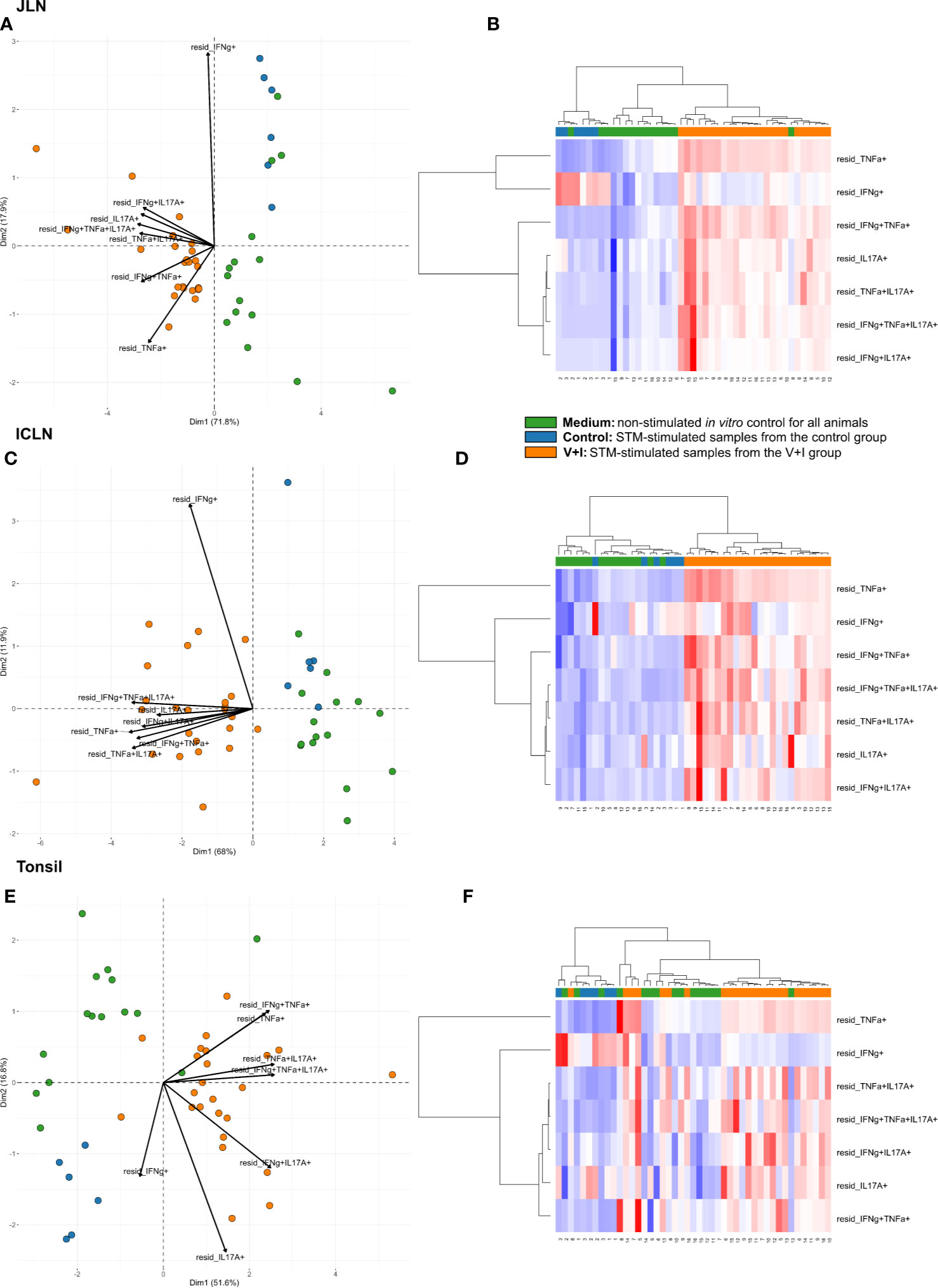
Figure 4 Unsupervised cluster analysis for Salmonella Typhimurium (STM)-stimulated interferon-γ (IFN-γ)/tumor necrosis factor-α (TNF-α)/interleukin-17A (IL-17A) producing CD4+ T cells isolated from jejunal lymph node (JLN), ileocolic lymph node (ICLN) and tonsil. (A) Principal component analysis (PCA) based on residuals calculated from cytokine-producing CD4+ T cells derived from jejunal lymph node and stimulated in vitro with STM or medium-only. Each colored circle represents one of three samples derived from one animal. Samples stimulated with STM (vaccine or challenge strain) are depicted in orange for V+I animals and blue for control animals; samples from both groups cultivated in medium-only are shown in green. Arrows depict cytokine-producing phenotypes. (B) Heat map of residuals calculated from cytokine-producing CD4+ T cells derived from jejunal lymph node and stimulated in vitro with STM or medium-only. Heat maps, generated from data converted to Z-scores, show increase (red) or decrease (blue) of relative abundance of cytokine-producing CD4+ T cells. Each row represents a cytokine-producing phenotype and each column a sample. Numbers below columns indicate numbers of the individual animals. Samples stimulated with STM (vaccine or challenge strain) are indicated by orange boxes for V+I animals and blue boxes for control animals; samples from both groups cultivated in medium-only are indicated by green boxes. (C–F) Same as (A), (B) but for CD4+ T cells derived from ileocolic lymph node (C, D) and tonsil (E, F).
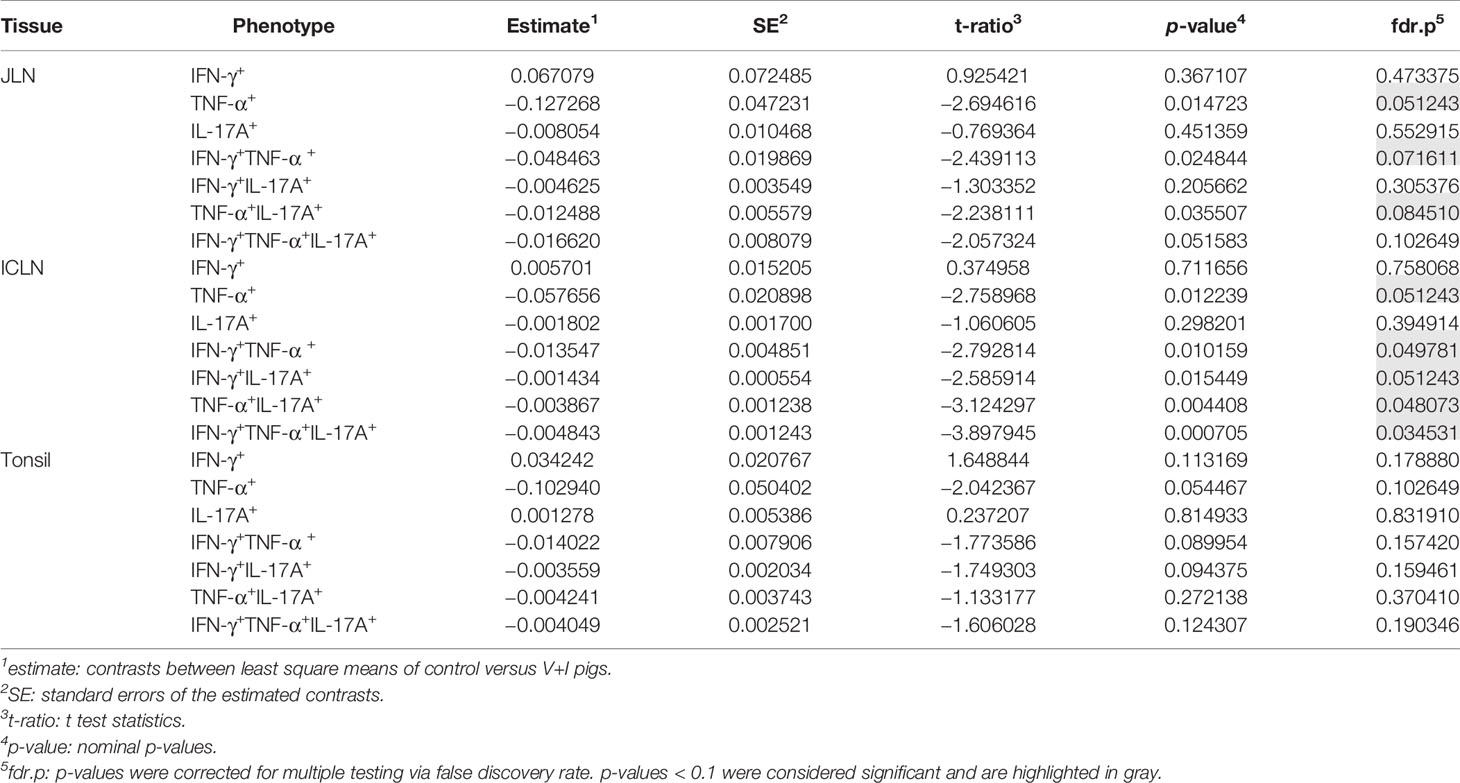
Table 2 Calculation of contrasts of Salmonella-specific CD4+ T-cell cytokine responses between control and V+I pigs in jejunal lymph node (JLN), ileocolic lymph node (ICLN) and tonsil.
Cluster analysis for STM-stimulated cytokine-producing CD4+ T cells was also performed for blood and spleen to gain more insight into the systemic CD4+ T-cell immune response against STM (Figure 5). With a few exceptions, control and medium-only samples largely clustered away from STM-stimulated V+I samples and Dimension 1 accounted for around 50% of the variation in blood and spleen (Figures 5A, C, respectively). In contrast to analyzed gut tissue and lymph nodes, relative abundances of cytokine-producing CD4+ T cells scattered across animal groups and the type of in vitro stimulation (medium versus STM antigen) (Figures 5B, D). Nonetheless, significant differences between control and V+I group could be detected for IFN-γ/TNF-α co-producing CD4+ T cells in blood and spleen, with additional significant differences for IFN-γ/IL-17A and IFN-γ/TNF-α/IL-17A co-producing CD4+ T cells in the blood (Table 3). Of note, IFN-γ/TNF-α co-producing CD4+ T cells constituted the only phenotype which was significantly different between control and V+I animals in all organs, with the exception of the tonsil.
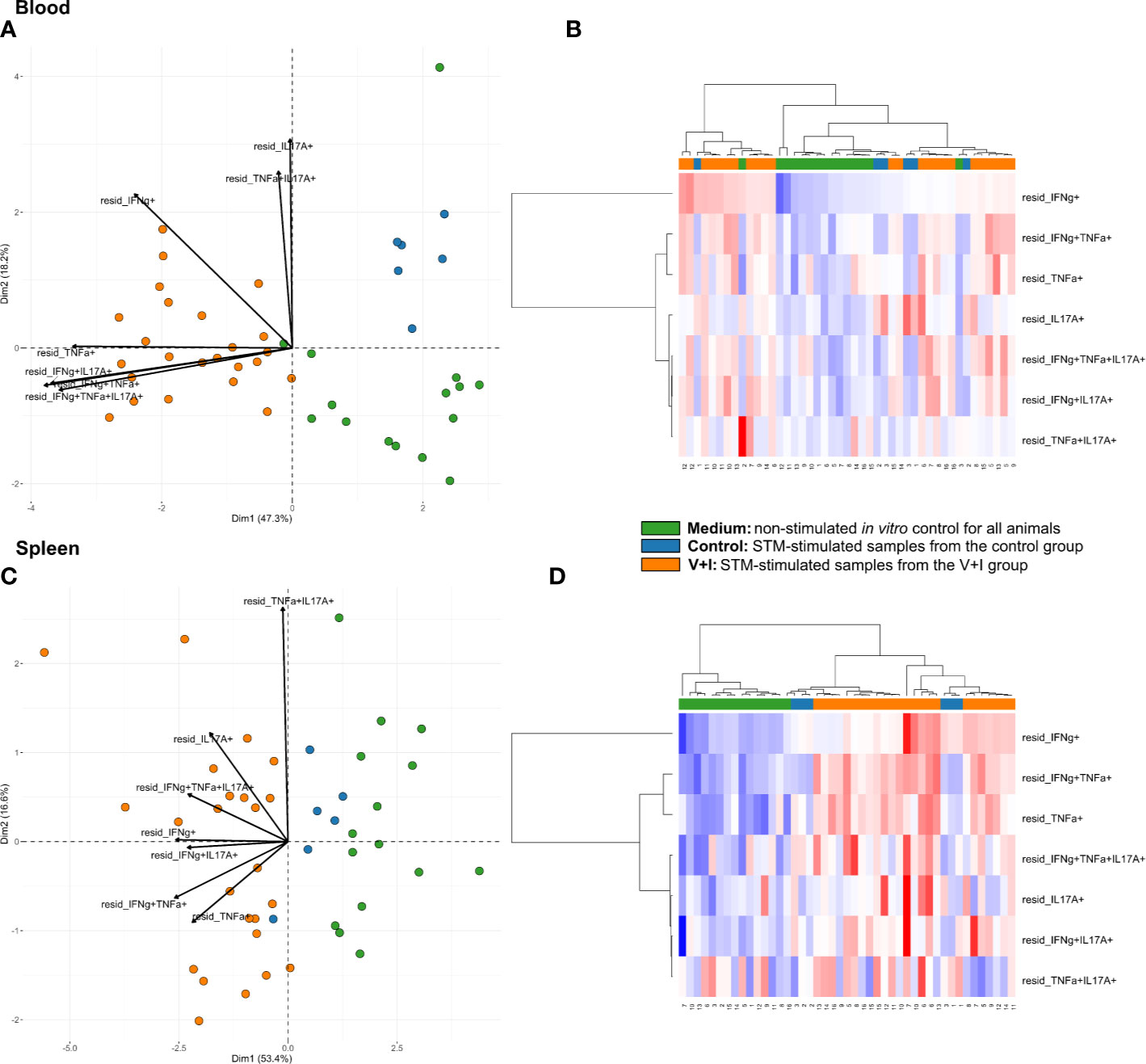
Figure 5 Unsupervised cluster analysis for Salmonella Typhimurium (STM)-stimulated interferon-γ (IFN-γ)/tumor necrosis factor-α (TNF-α)/interleukin-17A (IL-17A) producing CD4+ T cells isolated from blood and spleen. (A) Principal component analysis (PCA) based on residuals calculated from cytokine-producing CD4+ T cells derived from blood and stimulated in vitro with STM or medium-only. Each colored circle represents one of three samples derived from one animal. Samples stimulated with STM (vaccine or challenge strain) are depicted in orange for V+I animals and blue for control animals; samples from both groups cultivated in medium-only are shown in green. Arrows depict cytokine-producing phenotypes. (B) Heat map of residuals calculated from cytokine-producing CD4+ T cells derived from blood and stimulated in vitro with STM or medium-only. Heat maps, generated from data converted to Z-scores, show increase (red) or decrease (blue) of relative abundance of cytokine-producing CD4+ T cells. Each row represents a cytokine-producing phenotype and each column a sample. Numbers below columns indicate numbers of the individual animals. Samples stimulated with STM (vaccine or challenge strain) are indicated by orange boxes for V+I animals and blue boxes for control animals; samples from both groups cultivated in medium-only are indicated by green boxes. (C, D) Same as (A), (B) but for cytokine-producing CD4+ T cells derived from the spleen.
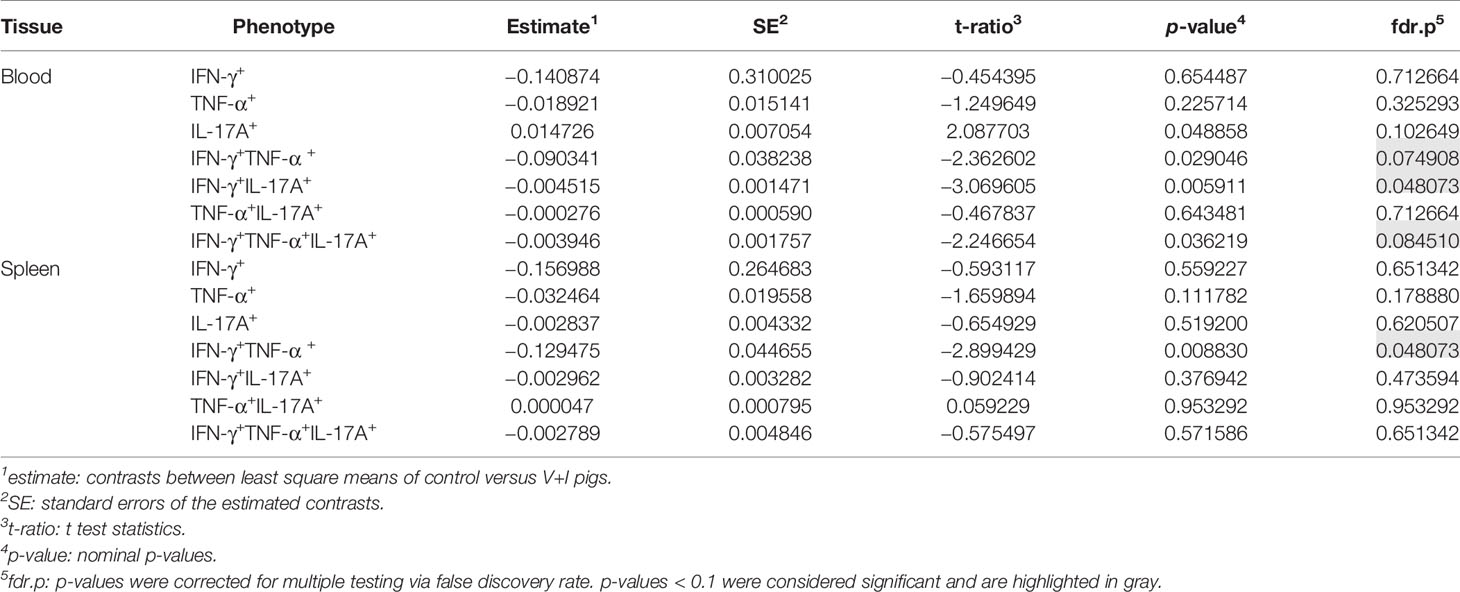
Table 3 Calculation of contrasts of Salmonella-specific CD4+ T-cell cytokine responses between control and V+I pigs in blood and spleen.
Due to marked differences between organs in terms of both frequencies of cytokine-producing CD4+ T cells and the type of cytokine production, we carried out a cluster analysis for STM-stimulated samples from V+I animals that encompassed all seven organs. Here, the formation of three clusters by PCA could be observed (Figure 6). Blood and spleen as the two systemic organs formed one cluster with tonsil and lymph nodes forming another tight cluster of lymphatic origin on the same side, while all samples from the gut tissue clustered separately. As can be seen from the raw data (Figure S2), the abundances of Salmonella-specific cytokine-producing CD4+ T cells in jejunum and ileum highly surmounted those detected in the other organs as indicated by the arrows for all cytokine-producing phenotypes pointing toward the cluster of gut tissue samples (Figure 6). Accordingly, Dimension 1 accounted for 88.7% variability in the data set. Of note, Dimension 2 accounted only for 5.2% of the variability, but still gave a clear separation of blood and spleen versus the lymphatic organs. Combining this with the cytokine production phenotypes, where IFN-γ-single producers and IFN-γ/TNF-α co-producing cells separated from the other phenotypes, this indicates that IL-17A-producing phenotypes are less prominent in the systemic immune response found in blood and spleen.
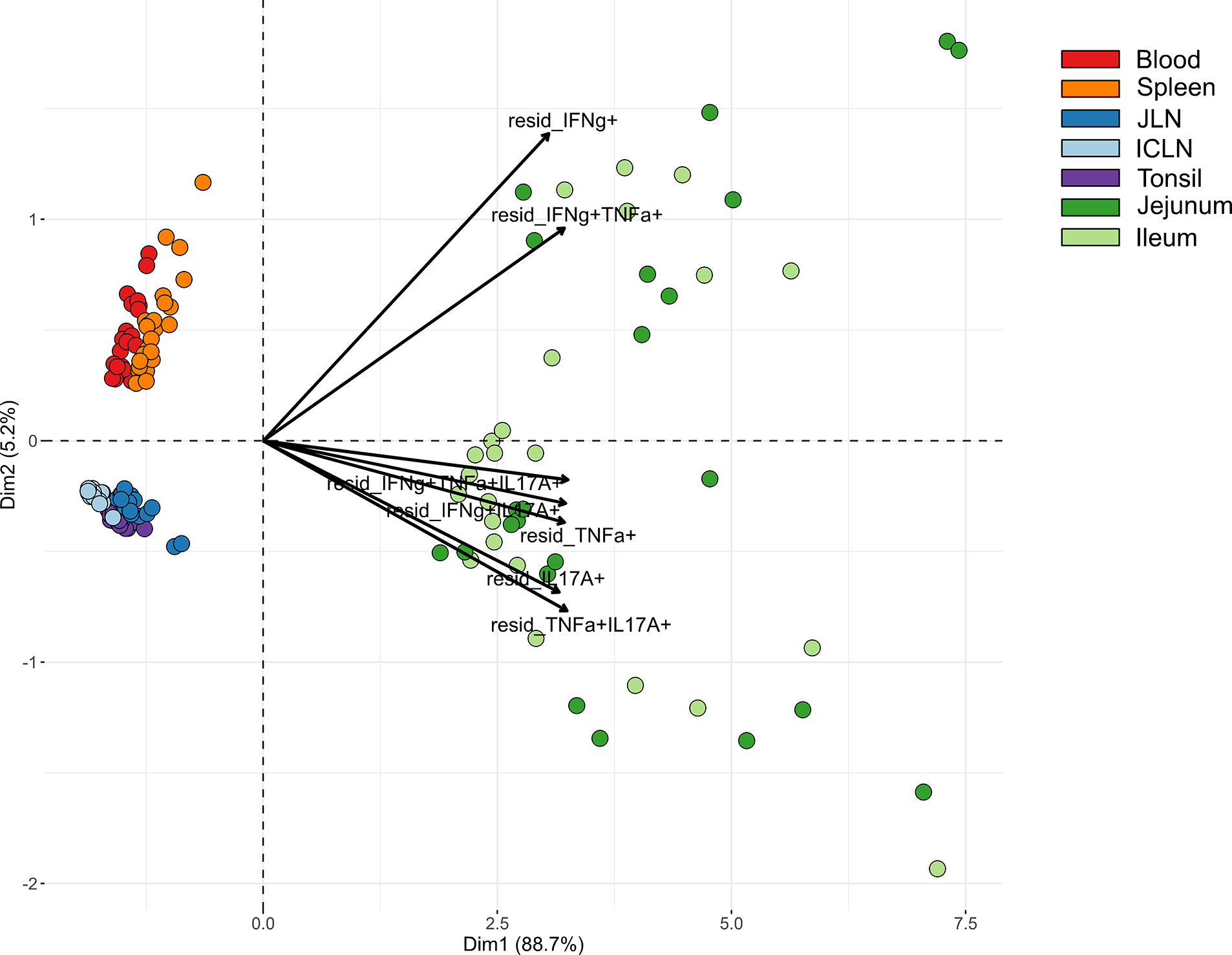
Figure 6 Principal component analysis (PCA) for STM-stimulated interferon-γ (IFN-γ)/tumor necrosis factor-α (TNF-α)/interleukin-17A (IL-17A) producing CD4+ T cells isolated from various organs. PCA based on residuals calculated from cytokine-producing CD4+ T cells derived from jejunum, ileum, jejunal lymph node (JLN), ileocolic lymph node (ICLN), tonsil, blood and spleen after in vitro stimulation with STM. Each colored circle represents one of two samples from one animal (stimulation with either vaccine or challenge strain). Samples are colored according to the organ of origin (blood: red, spleen: orange, JLN: blue, ICLN: light blue, tonsil: purple, jejunum: green, ileum: light green). Arrows depict cytokine-producing phenotypes.
Clustering among cytokine-producing phenotypes as seen in heat map dendrograms (Figures 3, 4, and 5B, C) also featured such a grouping. It could be observed across organs that IL-17A-containing phenotypes formed closely related clusters whereas IFN-γ single-, TNF-α single- and IFN-γ/TNF-α co-producing CD4+ T cells largely clustered separately. To study this from an alternate point of view, heat maps were calculated individually for all cytokine-producing phenotypes (Figure S3). For the majority of investigated phenotypes, distinct clusters emerged for STM-stimulated V+I samples on the one hand and control and medium-only samples on the other hand. Matching with observations from Figure 6, the dendrograms showed that organs predominantly form clusters according to their respective origins from systemic, lymphatic or gut tissue.
It has been shown previously that porcine CD4+ T cells can be differentiated into naïve CD4+ T cells with a CD8α-CD27+ phenotype, whereas CD8α+CD27+ and CD8α+CD27- CD4+ T cells constitute central memory (Tcm) and effector memory (Tem) subsets, respectively (32). We therefore analyzed CD8α alongside CD27 expression for all seven cytokine-producing phenotypes in all organs (Figure 7). The gating strategy for the identification of these phenotypes is shown in Figure S4. Representative raw data for the CD8α/CD27 expression of cytokine-producing CD4+ T cells following STM-stimulation is shown in Figure S5. As illustrated in Figure 7, hardly any Salmonella-specific cytokine-producing CD4+ T cells with the hitherto uncharacterized CD8α-CD27- phenotype were found. Also, CD8α-CD27+ naïve T cells showed only a low capacity for cytokine production with some exceptions such as for single TNF-α and IL-17A-producers in the blood. Instead, Salmonella-specific cytokine-producing CD4+ T cells predominantly had a CD8α+CD27- effector memory phenotype, especially in the gut tissue. While a CD8α+CD27+ central memory population was present primarily for IFN-γ/TNF-α double- and IFN-γ/TNF-α/IL-17A triple-producing CD4+ T cells within analyzed non-gut locations, cytokine phenotypes present in jejunum and ileum were almost exclusively effector memory T cells.
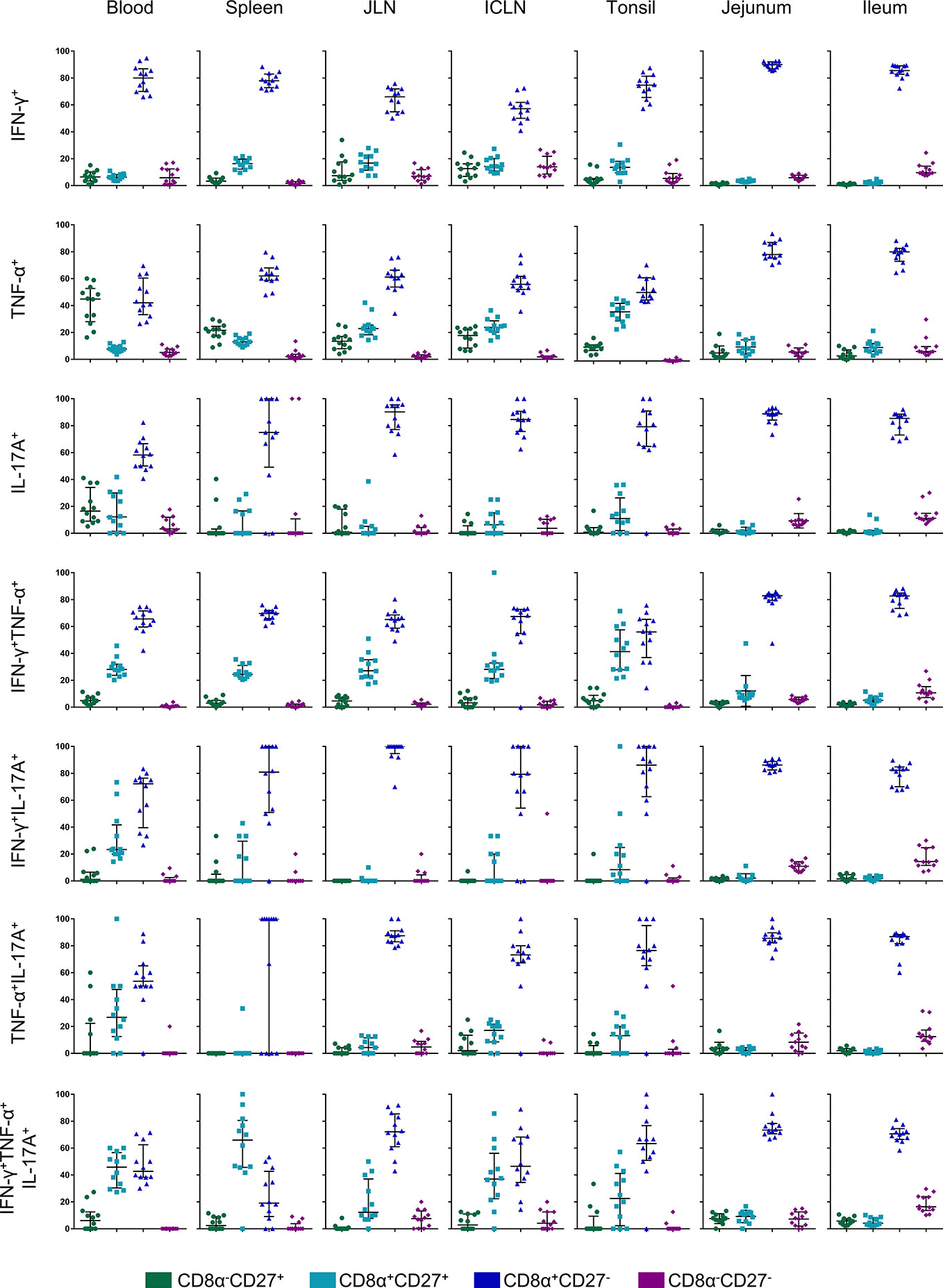
Figure 7 CD8α and CD27 expression of cytokine-producing CD4+ T cells in analyzed organs. Intracellular cytokine staining was performed on lymphocytes isolated from organ tissue following overnight in vitro stimulation with the challenge strain. CD4+ T cells were analyzed for expression of interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and interleukin-17A (IL-17A) and grouped into seven cytokine-producing phenotypes. Cytokine-producing CD4+ T cells were further subgated for CD8α and CD27 to identify four CD8a/CD27-defined cell populations: CD8α-CD27+ (green), CD8α+CD27+ (light blue), CD8α+CD27- (dark blue), CD8α-CD27- (violet). Individual graphs indicate percentages of CD8a/CD27-defined cell populations for each cytokine-producing phenotype from individual animals of the V+I group. Black bars indicate the median and whiskers show the interquartile range. Data was obtained from day of necropsy of the respective animals.
Information on the porcine T-cell response against STM is still scarce. For the most part, previous studies have focused on detecting local and systemic immune responses on mRNA level and discovered that STM infection in the pig causes upregulation of mRNA expression of cytokines like IFN-γ, TNF-α, and IL-1β (16–20). However, the phenotype of the cells producing these mRNA transcripts was not investigated. Only one study also measured the expression of transcription factors T-bet, Gata-3 and Foxp3 in CD4+ T cells following STM infection (41) but the results were not fully conclusive. Differently, in the murine model CD4+ T cells are widely recognized as the main immune cell subset responsible for protection against STM (21, 22). Hence, we decided to lay the focus of our study on CD4+ T cells.
Considering also the lack of knowledge on the magnitude of the T-cell response in swine to STM, we decided to stimulate the immune system by a double vaccination with Salmoporc and subsequent challenge infection with a virulent STM strain. We chose this regimen because it was also used in the past to demonstrate the efficacy of the vaccine. These studies showed that, in comparison to two vaccinations or single infection, STM-specific IgG titers in blood were highest after this combination of two vaccinations and subsequent challenge infection (11). Hence, we assumed that this might also indicate an activation of CD4+ T cells at the systemic level. We further reasoned that this would probably coincide with activation of T cells in the gut mucosa, since the vaccine and the challenge infection were applied orally.
Of the seven organs analyzed in this study, the highest colonization with STM was found in the tonsils. This is not surprising as the tonsil is known to be invaded by various bacteria, with STM frequently establishing a persistent infection in this location (42, 43). Of note, the tonsil was the tissue with the lowest frequencies of cytokine-producing CD4+ T cells and no significant differences for any cytokine phenotype were found between V+I animals and the control conditions. One possible explanation might be the location of the pathogen within the organ. Unlike jejunum and ileum where STM invades the epithelial cells, it mainly resides extracellularly in porcine tonsils (44), thus making it more difficult for the immune system to attack the pathogen. Studies in recent years also present evidence that bacterial gene expression related to survival and persistence of Salmonella in the tonsil is vastly different from gene expression related to these mechanisms in the gut and gut-associated lymph nodes (45–47). Another factor may be the anti-inflammatory nature of the tonsil. In humans, tonsil-derived dendritic cells only induce a weak T-cell response to mucosally encountered pathogens but rather maintain immunotolerance (48). This observation fits with studies in the pig where the presence of Actinobacillus pleuropneumoniae in the tonsil induced an increase in IL-10 expression (49). Although IL-10 production was not measured in our study, a similar scenario is conceivable which could explain the poor induction of Th1 and Th17 responses in this organ.
Surpassing all other organs, we found the highest frequencies of cytokine-producing Salmonella-specific CD4+ T cells within LPLs isolated from jejunum and ileum. Of note, STM infection in the pig is mostly limited to the intestine, whereas a potential systemic infection is not well studied (50). Mouse models have shown that dissemination to mesenteric lymph nodes, blood and systemic organs can take place (51). We did not find Salmonella in spleen and liver of any of the animals two weeks post infection. As the immune system of V+I animals was already primed by vaccination prior to infection, it is possible that the infection was either already cleared or did not even spread to these systemic locations. In accordance to this, only few phenotypes of cytokine-producing CD4+ T cells showed significant rises in blood and spleen of V+I animals (Table 3). More phenotypes significantly different between V+I and control pigs were found for the investigated gut-draining lymph nodes (Table 2), whereas in the gut sections nearly all of the seven possible cytokine-producing phenotypes were induced (Table 1). This complies with studies in mice and humans, where T cells residing in the lamina propria have already been recognized as an important element of the response against STM and Salmonella Typhi (52–54). The presence of a very high abundance of Salmonella-specific cytokine-producing CD4+ T cells in the intestinal lamina propria of V+I animals in our study suggests that they also play a vital role in the mucosal immune response against STM infection in the pig.
Looking more closely at the seven possible cytokine-producing phenotypes in the different organs, it became apparent that IL-17A-containing phenotypes formed closely related clusters, often separating from IFN-γ and IFN-γ/TNF-α co-producing CD4+ T cells. This separation is especially striking in jejunum and ileum. Th17 cells and their signature cytokine IL-17A are well known for contributing to mucosal immunity and protection against intracellular enteric pathogens (55–57). Infection with STM has previously been shown to induce the expression of Th17 cytokines in the intestinal mucosa of other species such as mice, calves and rhesus macaques (27, 58). A depletion of Th17 cells in the intestinal mucosa of rhesus macaques by infection with simian immunodeficiency virus was correlated with increased STM dissemination to mesenteric lymph nodes (27) suggesting that Th17 cells are important in disease containment. On the other hand, it has been discussed that IL-17A-induced recruitment of neutrophils and the resulting inflammation is exploited by pathogens like STM and ultimately promotes bacterial colonization (59). Since all V+I animals were vaccinated before infection and we did not detect any signs of inflammation in their intestines, a negative impact of the Salmonella-specific Th17 phenotypes observed in this study seems unlikely. Instead, we provide evidence that Th17 cells also seem to play a protective role in host defense against STM in swine.
In contrast to other IL-17A+ populations, however, IL-17A single-producing CD4+ T cells were the only cytokine phenotype that did not reach significant difference between control and V+I pigs in any of the organs. Indeed, we observed across organs that CD4+ T-cell phenotypes consisting of more than one cytokine, such as IFN-γ/TNF-α, IFN-γ/IL-17A, TNF-α/IL-17A and IFN-γ/TNF-α/IL-17A, overall reached significant differences over the control group more frequently than single-cytokine producing CD4+ T cells. T cells producing several cytokines, also called multifunctional (MF) T cells, have been associated with protection in several bacterial and viral infections in humans and mice (60–63) and were found to be functionally superior to their single-producing counterparts (64). Studies in swine have also demonstrated the involvement of antigen-specific MF CD4+ T cells in response to various pathogens (65–68). While co-production of IFN-γ, TNF-α and IL-2 by T cells is frequently investigated, reports on these cytokines in combination with IL-17A in the context of infectious diseases are rather scarce. Concerning Salmonella, MF IL-17A T-cell responses have been reported in the blood and terminal ileum of humans. Vaccination or infection with Salmonella Typhi led to the induction of MF CD8+ and CD4+ T-cell responses, that, in case of MF CD8+ T cells in PBMC, were demonstrated to correlate with disease outcome (54, 69, 70). To our knowledge, this is the first description of simultaneous production of IL-17A with IFN-γ and TNF-α by antigen-specific CD4+ T cells in the pig and it can be speculated that MF CD4+ T cells may serve as a correlate of protection for STM infection in swine. Especially IFN-γ/TNF-α co-producing CD4+ T cells appear as a promising candidate as significant differences between groups were reached for six out of the seven analyzed organs for this phenotype, including the blood. Moreover, based on observations in the mouse model, it is likely that these cells contribute to STM clearance by a potent stimulation of macrophages that have engulfed the pathogen (23, 26).
Interestingly, it has recently been shown that non-cognate stimulation of Th1 cells contributes to resolution of Salmonella infection in mice (71, 72). This describes a mechanism where T cells are stimulated by T-cell receptor (TCR)-independent stimuli such as inflammatory cytokines without TCR recognition of cognate antigen presented by antigen presenting cells. Since we analyzed total CD4+ T cells in our study and used whole bacterial antigen for in vitro re-stimulation, it is possible that a fraction of the measured cytokine production by CD4+ T cells may be derived from non-cognate stimulation and not via direct stimulation of the TCR. So far, these bystander responses have mostly been described in effector or memory CD4+ T cells (73, 74). Considering the strong stimulation applied in our study with two vaccinations and a challenge infection, it is conceivable that some of the T cells in our analyses had already reached a differentiation status that may have enabled them to respond in a non-cognate manner. Although that could lead to slightly overestimated numbers of STM-specific CD4+ T cells in V+I animals, it might more accurately reflect the situation occurring in vivo.
For investigation of further functional differentiation, we looked at the expression of CD8α and CD27, which have been proposed for the distinction between central (Tcm) and effector memory (Tem) CD4+ T cells in the pig (32). Salmonella-specific cytokine-producing CD4+ T cells in all organs predominantly expressed CD8α while lacking CD27, corresponding to a Tem phenotype in the pig. Conversely, previous studies on viral infections in swine have shown that the CD4+ Tcm subset was capable of IFN-γ, TNF-α and/or IL-2 production (66, 75). However, we did detect two phenotypes, namely IFN-γ/TNF-α co-producing along with IFN-γ/TNF-α/IL-17A triple-producing CD4+ T cells, with a sizable cell population co-expressing CD8α and CD27 in non-gut tissues, indicating a Tcm subset. As IFN-γ/TNF-α co-producing CD4+ T cells were also present in significantly higher amounts in V+I compared to control animals in the blood, they might be exploited as a phenotype for STM T-cell immunity in the pig.
In jejunum and ileum, almost the entirety of all cells regardless of the cytokine phenotype displayed features of Tem cells. Indeed, it is mainly Tem cells that reach non-lymphoid tissues like the intestine (76, 77). The mucosal memory pool contains recirculating as well as resident memory T cells (Trm) that stay in the tissue long term to initiate an immediate immune response against enteric pathogens (77, 78). In humans, Salmonella Typhi-specific CD4+ Trm cells were induced in the ileal mucosa after Ty21a immunization (54). Moreover, resident memory Th1 cells have been shown to be indispensable for protection against STM infection in mice (30). Unfortunately, due to a lack of available antibodies for markers of tissue-residency, Trm cells cannot yet be identified in the pig. Nevertheless, we can speculate that a part of the population now identified as Tem cells in the gut may represent Trm cells. These cells may have been already induced by the vaccination, settled down permanently in the intestine and were re-activated upon challenge infection.
In conclusion, by vaccinating piglets with a live attenuated STM vaccine and challenging them with a virulent STM strain, we could show the induction of STM-specific multifunctional CD4+ T cells across organs with a strong enrichment in the intestinal mucosa. These cells predominantly possessed an effector memory phenotype. Their multifunctional cytokine profile suggests an involvement in protective immunity against STM infection and IFN-γ/TNF-α co-producing CD4+ T cells in the blood might be investigated further as a marker for long-term protective immunity against STM infections. Finally, we think that our study forms an important foundation for more in-depth studies on the T-cell response in pigs against STM following only vaccination or infection. In this way, the postulated correlates of protection will be further corroborated.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Advisory Committee for Animal Experiments, University of Veterinary Medicine, Vienna and the Federal Ministry for Science, Research and Economy (BMWF-68.205/0241-WF/V/3b/2016).
SSp, TT, VF, AS, and WG conceived the idea and designed the project. ES, CK, and AL organized the animal experiment and necropsy with sample collection. EV, AP, JL, KM, and MS performed lymphocyte isolation and in vitro stimulation. SSc performed flow cytometry experiments. JS conducted bacteriological analysis. MD performed statistical analysis. SSc and WG analyzed the experiments, interpreted the data, and wrote the manuscript. SSp, TT, VF, AL, and AS assisted with interpretation of the data. All authors contributed to the article and approved the submitted version.
This project was funded by Ceva Innovation Center GmbH (formerly belonging to IDT Biologika GmbH), Dessau-Roßlau, Germany.
SSp, TT, and VF are employed by Ceva Innovation Center GmbH.
The authors declare that this study received funding from Ceva Innovation Center GmbH (formerly belonging to IDT Biologika GmbH), Dessau-Roßlau, Germany. The funder had the following involvement in the study: The co-authors employed by the funder were involved in the study design and interpretation of the results as indicated in the “author contribution” statement. However, this did not influence the scientific integrity of the study and the presented findings.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Michaela Koch for performing the IDEXX Swine Salmonella Ab Test as well as Heiko Stein, Markus Cihar, and Sophie Dürlinger for their assistance with the animal work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.603089/full#supplementary-material
1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis (2010) 50:882–9. doi: 10.1086/650733
2. Cheng RA, Eade CR, Wiedmann M. Embracing Diversity: Differences in Virulence Mechanisms, Disease Severity, and Host Adaptations Contribute to the Success of Nontyphoidal Salmonella as a Foodborne Pathogen. Front Microbiol (2019) 10:1368. doi: 10.3389/fmicb.2019.01368
3. EFSA. European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC): The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFS2 (2018) 16:22–67. doi: 10.2903/j.efsa.2018.5500
4. Campos J, Mourão J, Peixe L, Antunes P. Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens (2019) 8. doi: 10.3390/pathogens8010019
5. Bonardi S. Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol Infect (2017) 145:1513–26. doi: 10.1017/S095026881700036X
6. Andres VM, Davies RH. Biosecurity Measures to Control Salmonella and Other Infectious Agents in Pig Farms: A Review. Compr Rev Food Sci Food Saf (2015) 14:317–35. doi: 10.1111/1541-4337.12137
7. Springer S, Lindner T, Steinbach G, Selbitz HJ. Investigation of the efficacy of a genetically-stabile live Salmonella typhimurium vaccine for use in swine. Berl Munch Tierarztl Wochenschr (2001) 114:342–5. doi: 10.31274/safepork-180809-1072
8. Eddicks M, Palzer A, Hörmansdorfer S, Ritzmann M, Heinritzi K. Uberprüfung der Verträglichkeit des Salmonella Typhimurium-Lebendimpfstoffes Salmoporc für drei Tage alte Saugferkel. DTW Dtsch Tierarztl Wochenschr (2009) 116:249–54.
9. Roesler U, Stief M, Leffler M, Truyen U, Lehmann J, Szabo I, et al. Persistence, excretion and efficacy of an attenuated Salmonella vaccine in suckling piglets. Prakt Tierarzt (2010) 91:59–65.
10. Davies R, Gosling RJ, Wales AD, Smith RP. Use of an attenuated live Salmonella Typhimurium vaccine on three breeding pig units: A longitudinal observational field study. Comp Immunol Microbiol Infect Dis (2016) 46:7–15. doi: 10.1016/j.cimid.2016.03.005
11. Theuß T, Ueberham E, Lehmann J, Lindner T, Springer S. Immunogenic potential of a Salmonella Typhimurium live vaccine for pigs against monophasic Salmonella Typhimurium DT 193. BMC Vet Res (2017) 13:343. doi: 10.1186/s12917-017-1271-5
12. Smith RP, Andres V, Martelli F, Gosling B, Marco-Jimenez F, Vaughan K, et al. Maternal vaccination as a Salmonella Typhimurium reduction strategy on pig farms. J Appl Microbiol (2018) 124:274–85. doi: 10.1111/jam.13609
13. Peeters L, Dewulf J, Boyen F, Brossé C, Vandersmissen T, Rasschaert G, et al. Bacteriological evaluation of vaccination against Salmonella Typhimurium with an attenuated vaccine in subclinically infected pig herds. Prev Vet Med (2020) 182:104687. doi: 10.1016/j.prevetmed.2019.04.016
14. Trepnau D, Ulrich E, Uhlig R, Lindner T, Selbitz H-J, Rösler U, et al. Antikörperantwort nach Impfung von Mastschweinen mit einem Salmonella-Typhimurium-Lebendimpfstoff in Abhängigkeit von der Applikationsform. Berl Munch Tierarztl Wochenschr (2008) 121:334–40.
15. Szabó I, Scherer K, Roesler U, Appel B, Nöckler K, Hensel A. Comparative examination and validation of ELISA test systems for Salmonella typhimurium diagnosis of slaughtering pigs. Int J Food Microbiol (2008) 124:65–9. doi: 10.1016/j.ijfoodmicro.2008.02.022
16. Matiasovic J, Stepanova H, Kudlackova H, Havlickova H, Sisak F, Rychlik I, et al. Immune response of pigs to Salmonella enterica serovar Derby and Typhimurium infections. Vet Microbiol (2014) 170:284–90. doi: 10.1016/j.vetmic.2014.02.003
17. Martins RP, Lorenzi V, Arce C, Lucena C, Carvajal A, Garrido JJ. Innate and adaptive immune mechanisms are effectively induced in ileal Peyer’s patches of Salmonella typhimurium infected pigs. Dev Comp Immunol (2013) 41:100–4. doi: 10.1016/j.dci.2013.04.020
18. Meurens F, Berri M, Auray G, Melo S, Levast B, Virlogeux-Payant I, et al. Early immune response following Salmonella enterica subspecies enterica serovar Typhimurium infection in porcine jejunal gut loops. Vet Res (2009) 40:5. doi: 10.1051/vetres:2008043
19. Knetter SM, Bearson SM, Huang T-H, Kurkiewicz D, Schroyen M, Nettleton D, et al. Salmonella enterica serovar Typhimurium-infected pigs with different shedding levels exhibit distinct clinical, peripheral cytokine and transcriptomic immune response phenotypes. Innate Immun (2015) 21:227–41. doi: 10.1177/1753425914525812
20. Collado-Romero M, Arce C, Ramírez-Boo M, Carvajal A, Garrido JJ. Quantitative analysis of the immune response upon Salmonella typhimurium infection along the porcine intestinal gut. Vet Res (2010) 41:23. doi: 10.1051/vetres/2009072
21. Kurtz JR, Goggins JA, McLachlan JB. Salmonella infection: Interplay between the bacteria and host immune system. Immunol Lett (2017) 190:42–50. doi: 10.1016/j.imlet.2017.07.006
22. Pham OH, McSorley SJ. Protective host immune responses to Salmonella infection. Future Microbiol (2015) 10:101–10. doi: 10.2217/fmb.14.98
23. Nauciel C, Espinasse-Maes F. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect Immun (1992) 60:450–4. doi: 10.1128/IAI.60.2.450-454.1992
24. Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol (1996) 156:3321–6.
25. Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol (2005) 175:4603–10. doi: 10.4049/jimmunol.175.7.4603
26. Tubo NJ, Jenkins MK. CD4+ T Cells: guardians of the phagosome. Clin Microbiol Rev (2014) 27:200–13. doi: 10.1128/CMR.00097-13
27. Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med (2008) 14:421–8. doi: 10.1038/nm1743
28. Abusleme L, Moutsopoulos NM. IL-17: overview and role in oral immunity and microbiome. Oral Dis (2017) 23:854–65. doi: 10.1111/odi.12598
29. Lee S-J, McLachlan JB, Kurtz JR, Fan D, Winter SE, Baumler AJ, et al. Temporal expression of bacterial proteins instructs host CD4 T cell expansion and Th17 development. PLoS Pathog (2012) 8:e1002499. doi: 10.1371/journal.ppat.1002499
30. Benoun JM, Peres NG, Wang N, Pham OH, Rudisill VL, Fogassy ZN, et al. Optimal protection against Salmonella infection requires noncirculating memory. Proc Natl Acad Sci USA (2018) 115:10416–21. doi: 10.1073/pnas.1808339115
31. Reutner K, Leitner J, Essler SE, Witter K, Patzl M, Steinberger P, et al. Porcine CD27: identification, expression and functional aspects in lymphocyte subsets in swine. Dev Comp Immunol (2012) 38:321–31. doi: 10.1016/j.dci.2012.06.011
32. Reutner K, Leitner J, Müllebner A, Ladinig A, Essler SE, Duvigneau JC, et al. CD27 expression discriminates porcine T helper cells with functionally distinct properties. Vet Res (2013) 44:18. doi: 10.1186/1297-9716-44-18
33. Trüpschuch S, Laverde Gomez JA, Ediberidze I, Flieger A, Rabsch W. Characterisation of multidrug-resistant Salmonella Typhimurium 4,5,12:i:- DT193 strains carrying a novel genomic island adjacent to the thrW tRNA locus. Int J Med Microbiol (2010) 300:279–88. doi: 10.1016/j.ijmm.2010.02.001
34. Bates D, Maechler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw (2015) 67. doi: 10.18637/jss.v067.i01
35. Fox J, Monette G. Generalized Collinearity Diagnostics. J Am Stat Assoc (1992) 87:178–83. doi: 10.1080/01621459.1992.10475190
37. Lenth R. Estimated Marginal Means, aka Least-Squares Means [R package emmeans version 1.4.8]. (2020). Available at: https://CRAN.R-project.org/package=emmeans.
38. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol (1995) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
39. Kassambara A, Mundt F. Extract and Visualize the Results of Multivariate Data Analyses [R package factoextra version 1.0.7]. (2020). Available at: https://CRAN.R-project.org/package=factoextra.
40. Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. Various R Programming Tools for Plotting Data [R package gplots version 3.0.4]. (2020). Available at: https://CRAN.R-project.org/package=gplots.
41. Kreuzer S, Rieger J, Strucken EM, Thaben N, Hünigen H, Nöckler K, et al. Characterization of CD4+ subpopulations and CD25+ cells in ileal lymphatic tissue of weaned piglets infected with Salmonella Typhimurium with or without Enterococus faecium feeding. Vet Immunol Immunopathol (2014) 158:143–55. doi: 10.1016/j.vetimm.2014.01.001
42. Wood RL, Pospischil A, Rose R. Distribution of persistent Salmonella typhimurium infection in internal organs of swine. Am J Vet Res (1989) 50:1015–21.
43. Fedorka-Cray PJ, Kelley LC, Stabel TJ, Gray JT, Laufer JA. Alternate routes of invasion may affect pathogenesis of Salmonella typhimurium in swine. Infect Immun (1995) 63:2658–64. doi: 10.1128/IAI.63.7.2658-2664.1995
44. van Parys A, Boyen F, Volf J, Verbrugghe E, Leyman B, Rychlik I, et al. Salmonella Typhimurium resides largely as an extracellular pathogen in porcine tonsils, independently of biofilm-associated genes csgA, csgD and adrA. Vet Microbiol (2010) 144:93–9. doi: 10.1016/j.vetmic.2009.12.021
45. Boyen F, Pasmans F, van Immerseel F, Morgan E, Adriaensen C, Hernalsteens J-P, et al. Salmonella Typhimurium SPI-1 genes promote intestinal but not tonsillar colonization in pigs. Microbes Infect (2006) 8:2899–907. doi: 10.1016/j.micinf.2006.09.008
46. van Parys A, Boyen F, Leyman B, Verbrugghe E, Haesebrouck F, Pasmans F. Tissue-specific Salmonella Typhimurium gene expression during persistence in pigs. PLoS One (2011) 6:e24120. doi: 10.1371/journal.pone.0024120
47. Verbrugghe E, van Parys A, Leyman B, Boyen F, Haesebrouck F, Pasmans F. HtpG contributes to Salmonella Typhimurium intestinal persistence in pigs. Vet Res (2015) 46:118. doi: 10.1186/s13567-015-0261-5
48. Hallissey CM, Heyderman RS, Williams NA. Human tonsil-derived dendritic cells are poor inducers of T cell immunity to mucosally encountered pathogens. J Infect Dis (2014) 209:1847–56. doi: 10.1093/infdis/jit819
49. Müllebner A, Sassu EL, Ladinig A, Frömbling J, Miller I, Ehling-Schulz M, et al. Actinobacillus pleuropneumoniae triggers IL-10 expression in tonsils to mediate colonisation and persistence of infection in pigs. Vet Immunol Immunopathol (2018) 205:17–23. doi: 10.1016/j.vetimm.2018.10.008
50. Boyen F, Haesebrouck F, Maes D, van Immerseel F, Ducatelle R, Pasmans F. Non-typhoidal Salmonella infections in pigs: a closer look at epidemiology, pathogenesis and control. Vet Microbiol (2008) 130:1–19. doi: 10.1016/j.vetmic.2007.12.017
51. Watson KG, Holden DW. Dynamics of growth and dissemination of Salmonella in vivo. Cell Microbiol (2010) 12:1389–97. doi: 10.1111/j.1462-5822.2010.01511.x
52. Gautreaux MD, Deitch EA, Berg RD. T lymphocytes in host defense against bacterial translocation from the gastrointestinal tract. Infect Immun (1994) 62:2874–84. doi: 10.1128/IAI.62.7.2874-2884.1994
53. Yu X, Zhang H, Yu L, Liu M, Zuo Z, Han Q, et al. Intestinal Lamina Propria CD4+ T Cells Promote Bactericidal Activity of Macrophages via Galectin-9 and Tim-3 Interaction during Salmonella enterica Serovar Typhimurium Infection. Infect Immun (2018) 86. doi: 10.1128/IAI.00769-17
54. Booth JS, Goldberg E, Barnes RS, Greenwald BD, Sztein MB. Oral typhoid vaccine Ty21a elicits antigen-specific resident memory CD4+ T cells in the human terminal ileum lamina propria and epithelial compartments. J Transl Med (2020) 18:102. doi: 10.1186/s12967-020-02263-6
55. Khader SA, Gopal R. IL-17 in protective immunity to intracellular pathogens. Virulence (2010) 1:423–7. doi: 10.4161/viru.1.5.12862
56. Dubin PJ, Kolls JK. Th17 cytokines and mucosal immunity. Immunol Rev (2008) 226:160–71. doi: 10.1111/j.1600-065X.2008.00703.x
57. Sellge G, Magalhaes JG, Konradt C, Fritz JH, Salgado-Pabon W, Eberl G, et al. Th17 cells are the dominant T cell subtype primed by Shigella flexneri mediating protective immunity. J Immunol (2010) 184:2076–85. doi: 10.4049/jimmunol.0900978
58. Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, et al. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect Immun (2007) 75:4342–50. doi: 10.1128/IAI.01571-06
59. Liu JZ, Pezeshki M, Raffatellu M. Th17 cytokines and host-pathogen interactions at the mucosa: dichotomies of help and harm. Cytokine (2009) 48:156–60. doi: 10.1016/j.cyto.2009.07.005
60. Casazza JP, Betts MR, Price DA, Precopio ML, Ruff LE, Brenchley JM, et al. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J Exp Med (2006) 203:2865–77. doi: 10.1084/jem.20052246
61. Darrah PA, Patel DT, de Luca PM, Lindsay RW, Davey DF, Flynn BJ, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med (2007) 13:843–50. doi: 10.1038/nm1592
62. Kosinska AD, Johrden L, Zhang E, Fiedler M, Mayer A, Wildner O, et al. DNA prime-adenovirus boost immunization induces a vigorous and multifunctional T-cell response against hepadnaviral proteins in the mouse and woodchuck model. J Virol (2012) 86:9297–310. doi: 10.1128/JVI.00506-12
63. Lindenstrøm T, Agger EM, Korsholm KS, Darrah PA, Aagaard C, Seder RA, et al. Tuberculosis subunit vaccination provides long-term protective immunity characterized by multifunctional CD4 memory T cells. J Immunol (2009) 182:8047–55. doi: 10.4049/jimmunol.0801592
64. Kannanganat S, Ibegbu C, Chennareddi L, Robinson HL, Amara RR. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single-cytokine-producing cells. J Virol (2007) 81:8468–76. doi: 10.1128/JVI.00228-07
65. Käser T, Pasternak JA, Delgado-Ortega M, Hamonic G, Lai K, Erickson J, et al. Chlamydia suis and Chlamydia trachomatis induce multifunctional CD4 T cells in pigs. Vaccine (2017) 35:91–100. doi: 10.1016/j.vaccine.2016.11.050
66. Koinig HC, Talker SC, Stadler M, Ladinig A, Graage R, Ritzmann M, et al. PCV2 vaccination induces IFN-γ/TNF-α co-producing T cells with a potential role in protection. Vet Res (2015) 46:20. doi: 10.1186/s13567-015-0157-4
67. Talker SC, Stadler M, Koinig HC, Mair KH, Rodríguez-Gómez IM, Graage R, et al. Influenza A Virus Infection in Pigs Attracts Multifunctional and Cross-Reactive T Cells to the Lung. J Virol (2016) 90:9364–82. doi: 10.1128/JVI.01211-16
68. Holzer B, Morgan SB, Matsuoka Y, Edmans M, Salguero FJ, Everett H, et al. Comparison of Heterosubtypic Protection in Ferrets and Pigs Induced by a Single-Cycle Influenza Vaccine. J Immunol (2018) 200:4068–77. doi: 10.4049/jimmunol.1800142
69. Fresnay S, McArthur MA, Magder L, Darton TC, Jones C, Waddington CS, et al. Salmonella Typhi-specific multifunctional CD8+ T cells play a dominant role in protection from typhoid fever in humans. J Transl Med (2016) 14:62. doi: 10.1186/s12967-016-0819-7
70. McArthur MA, Sztein MB. Heterogeneity of multifunctional IL-17A producing S. Typhi-specific CD8+ T cells in volunteers following Ty21a typhoid immunization. PLoS One (2012) 7:e38408. doi: 10.1371/journal.pone.0038408
71. Pham OH, O’Donnell H, Al-Shamkhani A, Kerrinnes T, Tsolis RM, McSorley SJ. T cell expression of IL-18R and DR3 is essential for non-cognate stimulation of Th1 cells and optimal clearance of intracellular bacteria. PLoS Pathog (2017) 13:e1006566. doi: 10.1371/journal.ppat.1006566
72. O’Donnell H, Pham OH, Li L-x, Atif SM, Lee S-J, Ravesloot MM, et al. Toll-like receptor and inflammasome signals converge to amplify the innate bactericidal capacity of T helper 1 cells. Immunity (2014) 40:213–24. doi: 10.1016/j.immuni.2013.12.013
73. Di Genova G, Savelyeva N, Suchacki A, Thirdborough SM, Stevenson FK. Bystander stimulation of activated CD4+ T cells of unrelated specificity following a booster vaccination with tetanus toxoid. Eur J Immunol (2010) 40:976–85. doi: 10.1002/eji.200940017
74. Bangs SC, Baban D, Cattan HJ, Li CK-F, McMichael AJ, Xu X-N. Human CD4+ memory T cells are preferential targets for bystander activation and apoptosis. J Immunol (2009) 182:1962–71. doi: 10.4049/jimmunol.0802596
75. Talker SC, Koinig HC, Stadler M, Graage R, Klingler E, Ladinig A, et al. Magnitude and kinetics of multifunctional CD4+ and CD8β+ T cells in pigs infected with swine influenza A virus. Vet Res (2015) 46:52. doi: 10.1186/s13567-015-0182-3
76. von Andrian UH, Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med (2000) 343:1020–34. doi: 10.1056/NEJM200010053431407
77. Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity (2014) 41:886–97. doi: 10.1016/j.immuni.2014.12.007
Keywords: interferon-γ, interleukin-17A, pig, Salmonella Typhimurium, multifunctional T cells, lamina propria lymphocytes, tumor necrosis factor-α
Citation: Schmidt S, Sassu EL, Vatzia E, Pierron A, Lagler J, Mair KH, Stadler M, Knecht C, Spergser J, Dolezal M, Springer S, Theuß T, Fachinger V, Ladinig A, Saalmüller A and Gerner W (2021) Vaccination and Infection of Swine With Salmonella Typhimurium Induces a Systemic and Local Multifunctional CD4+ T-Cell Response. Front. Immunol. 11:603089. doi: 10.3389/fimmu.2020.603089
Received: 05 September 2020; Accepted: 25 November 2020;
Published: 11 January 2021.
Edited by:
Javier Dominguez, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainCopyright © 2021 Schmidt, Sassu, Vatzia, Pierron, Lagler, Mair, Stadler, Knecht, Spergser, Dolezal, Springer, Theuß, Fachinger, Ladinig, Saalmüller and Gerner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wilhelm Gerner, d2lsaGVsbS5nZXJuZXJAdmV0bWVkdW5pLmFjLmF0
†Present address: Eleni Vatzia, The Pirbright Institute, Pirbright, United Kingdom Alix Pierron, Toxalim (Research Center in Food Toxicology), Université de Toulouse, INRAe, Toulouse, France
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.