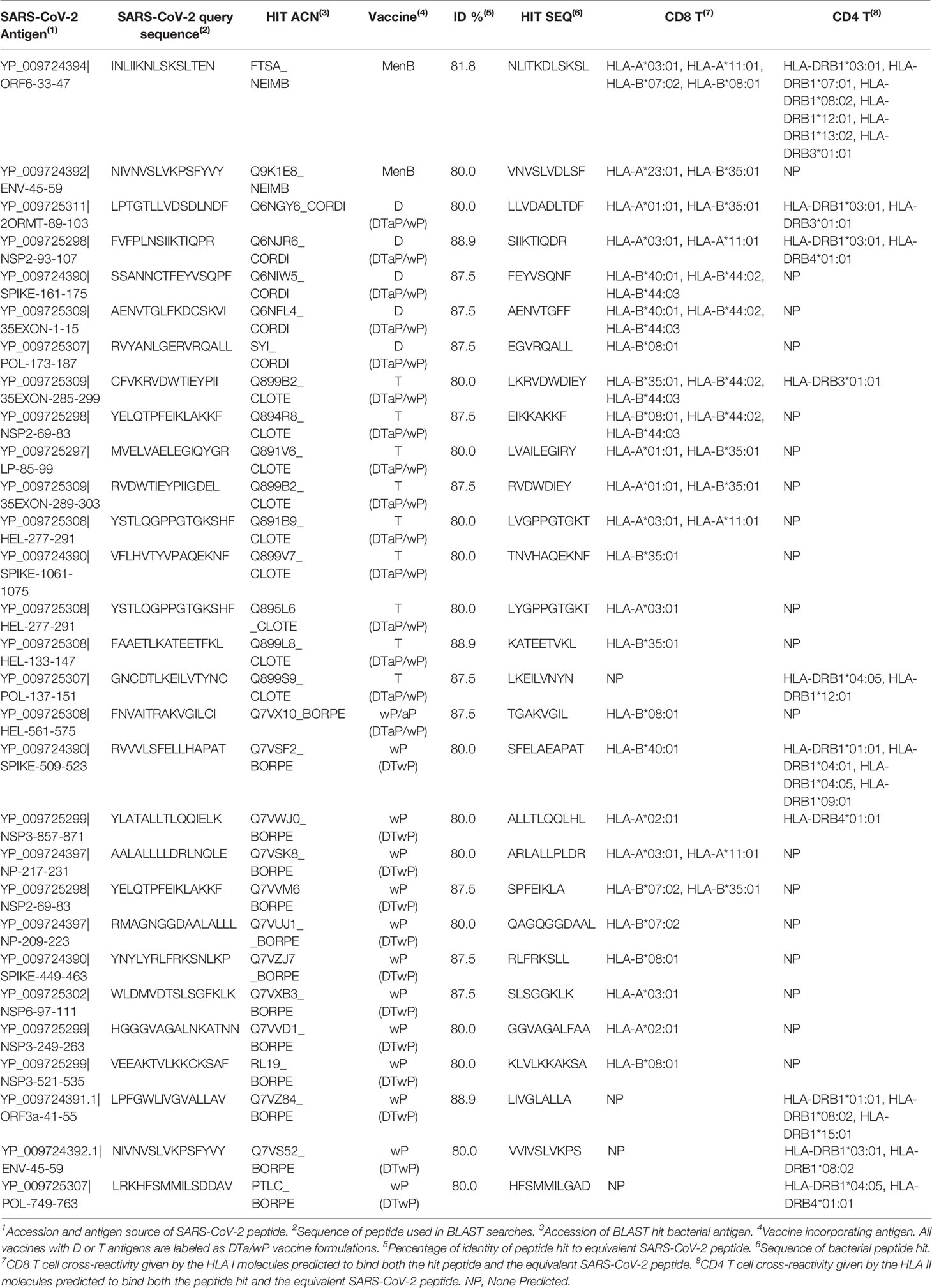
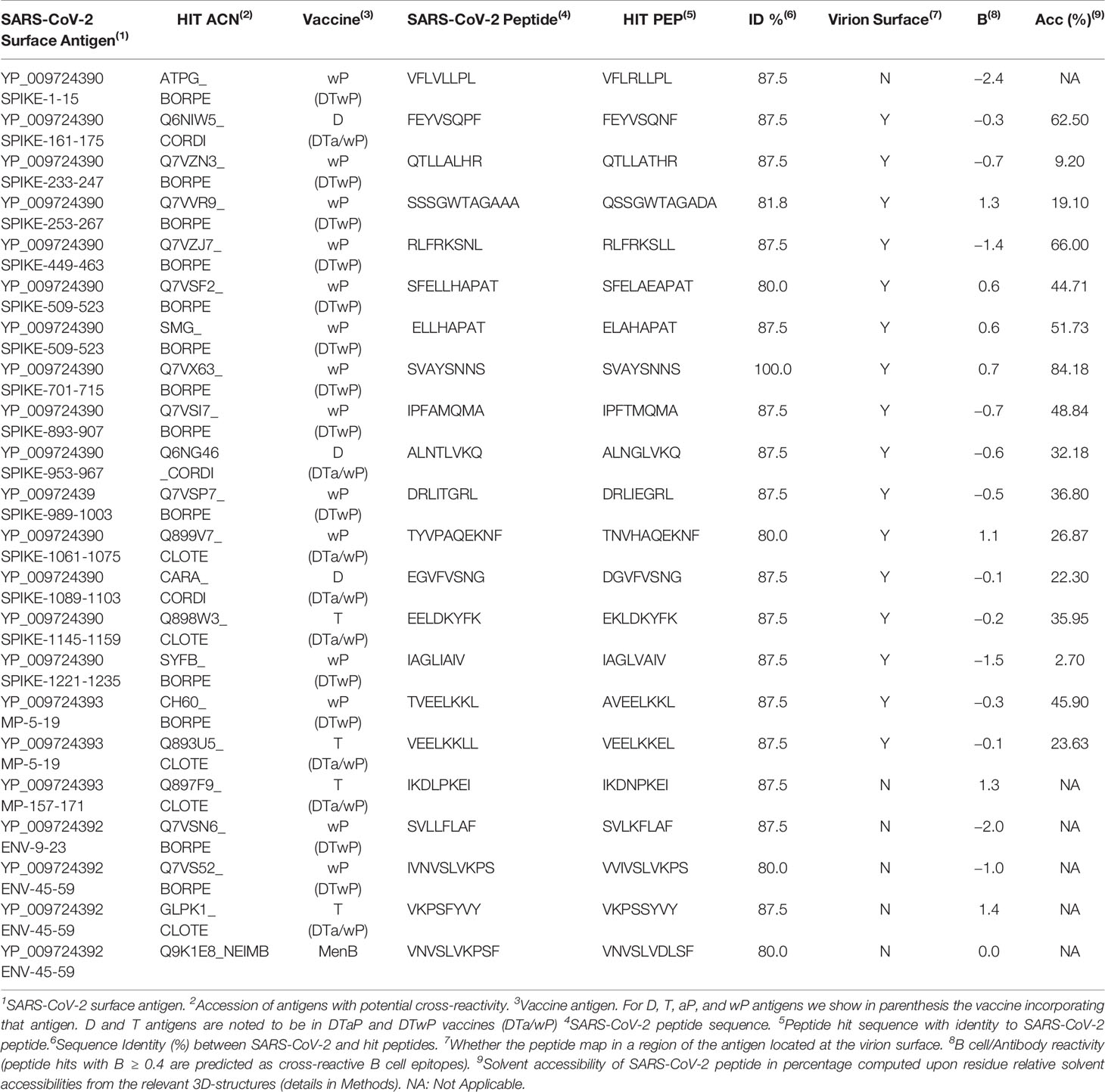
- Department of Immunology & O2, Faculty of Medicine, University Complutense of Madrid, Madrid, Spain
The recently emerged SARS-CoV-2 causing the ongoing COVID-19 pandemic is particularly virulent in the elderly while children are largely spared. Here, we explored the potential role of cross-reactive immunity acquired from pediatric vaccinations and exposure to common human pathogens in the protection and pathology of COVID-19. To that end, we sought for peptide matches to SARS-CoV-2 (identity ≥ 80%, in at least eight residues) in the proteomes of 25 human pathogens and in vaccine antigens, and subsequently predicted their T and B cell reactivity to identify potential cross-reactive epitopes. We found that viruses subject to pediatric vaccinations do not contain cross-reactive epitopes with SARS-CoV-2, precluding that they can provide any general protection against COVID-19. Likewise, common viruses including rhinovirus, respiratory syncytial virus, influenza virus, and several herpesviruses are also poor or null sources of cross-reactive immunity to SARS-CoV-2, discarding that immunological memory against these viruses can have any general protective or pathological role in COVID-19. In contrast, we found combination vaccines for treating diphtheria, tetanus, and pertussis infectious diseases (DTP vaccine) to be significant sources of potential cross-reactive immunity to SARS-CoV-2. DTP cross-reactive epitopes with SARS-CoV-2 include numerous CD8 and CD4 T cell epitopes with broad population protection coverage and potentially neutralizing B cell epitopes in SARS-CoV-2 Spike protein. Worldwide, children receive several DTP vaccinations, including three-four doses the first year of life and one at 4–6 years of age. Moreover, a low antigenic Tdap dose is also given at ages 9–14. Thereby, children may well be protected from SARS-CoV-2 through cross-reactive immunity elicited by DTP vaccinations, supporting testing in the general population to prevent COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new emergent virus identified in late 2019 as the etiological agent behind a mysterious outbreak of pneumonia in Wuhan, China (1, 2). The World Health Organization (WHO) later named the disease as coronavirus disease 19 (COVID-19) to account for the variety of clinical manifestations associated with SARS-CoV-2 infections (3, 4). The infectivity and pathogenicity SARS-CoV-2 have taken the world unguarded, leading to the ongoing COVID-19 pandemic.
SARS-CoV-2 infection can be asymptomatic, and the course of COVID-19 can range from mild to severe and death (5). COVID-19 cases and severity vary widely between different countries. For example, COVID-19 deaths per million reported in Spain are 606, while in Serbia, with a similar population density and geographical latitude are 30 (https://www.worldometers.info/coronavirus/). COVID-19 also varies between individuals, and medical conditions such as hypertension and obesity are risk factors (5). However, the strongest correlation factor in COVID-19 cases and severity is age. In fact, the majority of CODID-19 fatalities occur among the elderly (90% of the victims are over 70 years) while the pediatric population is largely spared (6). The large impact of COVID-19 in the geriatric population is somewhat expected, since their ability to mount immune responses to new pathogens has deteriorated (7). Yet, the virulence of the virus appears to be related to a disproportionate immune response, and indeed some patients appear to benefit from drugs with immunomodulatory properties (8). That newborns and infants, with undertrained adaptive immune systems, are safe from COVID-19 is unexpected, but telling.
Humans are not immunologically naïve, or not for long. When exposed to a new infection the host immune system preferentially engage existing cross-reactive memory B and T cells (9, 10). The promiscuity in antigen-recognition repertoires in memory B and T cells allows for quicker responses to multitude of antigens, saving energy and resources to the host. Activation of cross-reactive memory B and T cells can lead to protective immunity but also induce life-threatening immunopathology (9, 10). In this context, we reasoned that children are likely protected from SARS-CoV-2 by cross-reactive adaptive immunity elicited through vaccinations. Immunity elicited by vaccines wanes over time (11) which will leave adults more susceptible to COVID-19, and perhaps cross-reactive immunity from pathogens may in some cases protect or enhance the severity of the disease.
Adaptive immunity is driven by the recognition by T and B cells of small antigen fragments known as epitopes (12). B cell epitopes are also the targets of antibodies, and they need to be readily free and exposed to the solvent for recognition. In contrast, T cells recognize epitopes bound to major histocompatibility complex (MHC) molecules displayed in the cell surface of antigen presenting cells (12). Thereby, to test our hypothesis and identify potential sources of cross-reactive immunity to SARS-CoV-2, we conducted a systematic search for peptide matches to SARS-CoV-2 in 25 human pathogens including 18 viruses and 7 bacteria—the majority targeted by vaccinations—and in selected vaccine antigens, and predicted their T and B cell reactivity to identify cross-reactive epitopes. T cell reactivity of peptides (T cell epitopes) was predicted on the basis of their binding to MHC molecules frequently expressed in the human population using public bioinformatics tools available at IEDB (https://www.iedb.org/). B cell reactivity (B cell epitopes) was also predicted at IEDB (https://www.iedb.org/). Among the viruses without available vaccine, we considered herpes simplex virus 1 and 2, Epstein–Barr virus, human cytomegalovirus, human rhinovirus A, B, and C and respiratory syncytial virus A and B, which are all prevalent in the population. After these analyses, we found numerous cross-reactive epitopes between antigens in tetatus, diphteria, and pertussis (DTP) vaccines and SARS-CoV-2, including T cell epitopes with broad population protection coverage and potentially neutralizing B-cell epitopes. Overall, our results clearly support that cross-reactive immunity from DTP vaccines can be protecting children against SARS-CoV-2 and could protect the general population.
Methods
Pathogen and Vaccine Antigen Sequences
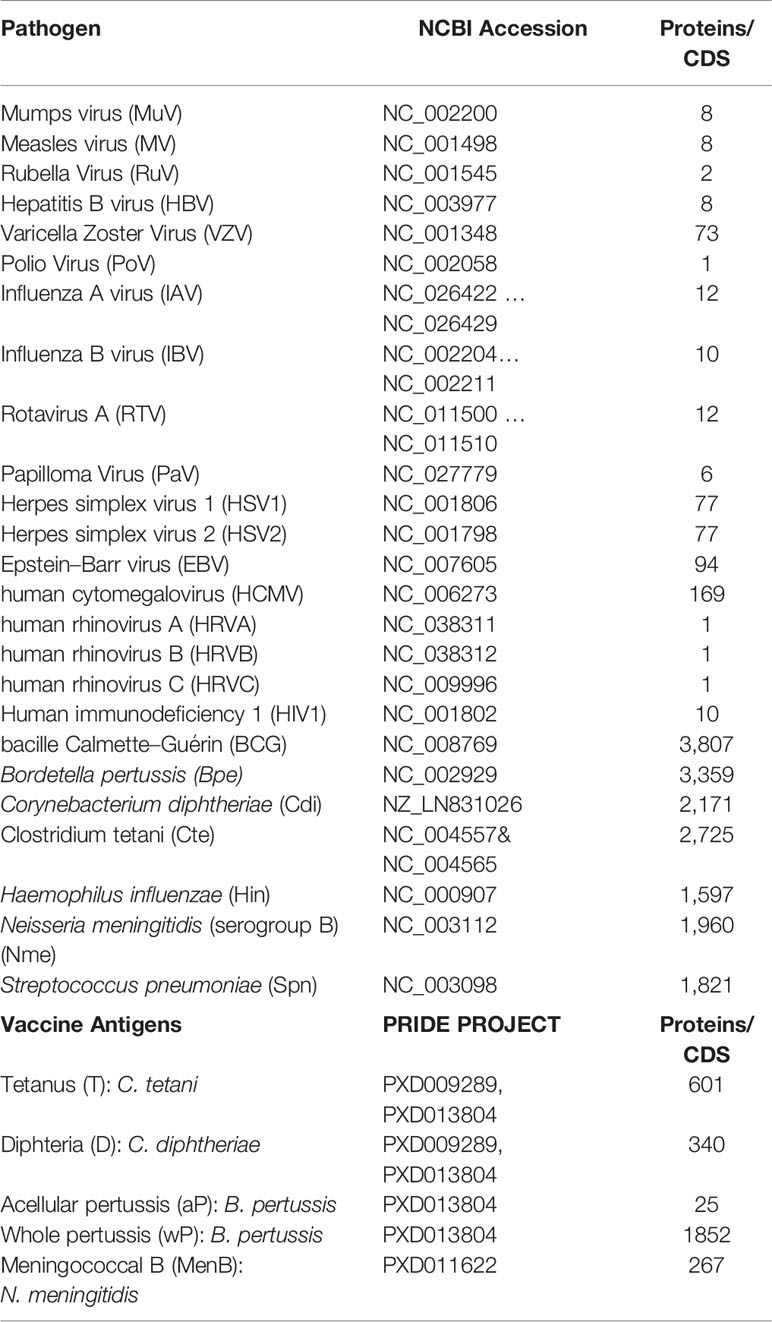
The amino acid sequences encoded by the genomes of 25 human pathogens in FASTA format were obtained from the GenBank entries indicated in Table 1. The amino acid sequences of antigens included in diphteria (D), tetanus (T), and pertussis cellular (wP) and acellular (aP) vaccines were obtained upon the proteomics projects PXD009289 and PXD013804 deposited in the Proteomics Identification Database (PRIDE) by the authors (13, 14) (Table 1). The amino acid sequence of the antigens included the meningococcal B vaccine Bexsero® (MenB) was obtained upon the PRIDE proteomic project PXD011622 (15).
Identification of SARS-CoV-2 Peptide Matches in Pathogens and Vaccine Antigens
Overlapping 15 mer peptides with 10 residue overlaps were generated in silico for each amino acid coding sequence (CDS) encoded by SARS-CoV-2 reference genome (NCBI accession NC_045512) covering their entire length. Peptides were used as queries in similarity searches using BLASTP (16) against custom made databases formatted for BLAST usage. BLAST formatted databases were generated upon FASTA files including the selected proteomes and vaccine antigens described elsewhere in Methods. BLAST searches were performed with default parameters, and the expected value set to 10,000. BLAST alignments were parsed using PERL scripts, and hit subject sequences consisting of eight or more residues with an identity ≥80% were selected as peptide matches with SARS-CoV-2.
Determining Peptide Identity With Human and Microbiome Proteins
Identity between SARS-CoV-2 peptides and hits with human and microbiome proteins was obtained through local BLAST searches using an expectation value of 10,000. Human microbiome proteins were obtained from the NIH Human Microbiome Project (17) and included 6,320,906 amino acid sequences from species colonizing any/all body parts. Human proteins included 113,275 amino acid sequences and consisted of reference proteins linked to the human GRCh38 genome ensemble (https://www.ncbi.nlm.nih.gov/genome/guide/human/). Amino acid sequences of human and microbiome proteins were downloaded as FASTA files and formatted for BLAST searches.
Prediction of T Cell Reactivity
T cell reactivity (presence of T cell epitopes) of peptides was assessed by predicting their binding to human leukocyte antigen (HLA) molecules. Peptide binding was predicted to HLA class I (HLA I) molecules HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*23:01, HLA-A*24:02, HLA-B*07:02, HLA-B*08:01, HLA-B*35:01, HLA-B*40:01, HLA-B*44:02, HLA-B*44:03 using IEDB MHC I binding tool (http://tools.iedb.org/mhci/) with default recommended method through the RESTful interface. At least one of these HLA I molecules is expressed by ~95% of the world population regardless of the ethnic background as computed by the EPISOPT method (18). Models used to predict binding to HLA I molecules were the appropriated for peptides with eight and nine residues. For longer peptides, the predicted binding was obtained evaluating all encompassing 9mer peptides. A 2% percentile rank cutoff was chosen to consider that binding had occurred. Peptide binding was also predicted to HLA class II (HLA II) HLA-DR molecules encompassing the β chains HLA-DRB1*01:01, HLA-DRB1*03:01, HLA-DRB1*04:01, HLA-DRB1*04:05, HLA-DRB1*07:01, HLA-DRB1*08:02, HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*12:01, HLA-DRB1*13:02, HLA-DRB1*15:01 HLA-DRB3*01:01 HLA-DRB3*02:02 HLA-DRB4*01:01, HLA-DRB5*01:01 using the IEDB MHC II binding tool (http://tools.iedb.org/mhcii/) with default recommended method through the RESTful interface. A 10% percentile rank cutoff was chosen to consider that binding had occurred. HLA-DR molecules incorporate a non-polymorphic α chain, and the selected β chains are expressed by ~81% of the population as computed by the IEDB coverage tool (http://tools.iedb.org/population/) (19).
Prediction of B Cell Reactivity
B cell or antibody reactivity (presence of B cell epitopes) of peptides was predicted using BepiPred1.0 (19) at the IEDB Analysis Resource (http://http://tools.iedb.org/bcell/). BepiPred predicts linear B cell epitopes and reports antigenicity values per residue (ai), upon which we calculated a global value of peptide antigenicity or B cell/antibody reactivity (B) using Eq. 1.
Where l is the total number of residues in the peptide. Peptides with B values ≥0.4 were considered as antigenic or potential B cell epitopes.
Other Procedures
Virion surface ectodomains of SARS-CoV-2 Spike (S), membrane (M) and envelope (E) proteins were identified from UNIPROT annotation records P0DTC2, P0DTC5, and P0DTC4, respectively. PyMOL Molecular Graphics System, Version 1.8 Schrödinger, was used to map and visualize B cell epitopes on 3D structures. PDB files with three-dimensional coordinates of SARS-CoV-2 S, M, and E glyproteins were obtained from Zhong Lab (https://zhanglab.ccmb.med.umich.edu/COVID-19/) and residue relative solvent accessibility (RSA) of peptides mapping in these proteins was computed using NACCESS (20). We used the non-parametric Kruskal–Wallis test to compare COVID-19 cases and death rates in European countries, including Asian countries from former URSS, with regard to the inclusion of DTaP or DTwP vaccines. Vaccination data was obtained from (https://apps.who.int/immunization_monitoring/) and death rates from (https://www.worldometers.info/coronavirus/).
Results and Discussion
Identification Cross-Reactive Immunity to SARS-CoV-2
Adaptive immunity is mediated by B and T cells which recognize small fragments, epitopes in the target antigens. Thereby, to identify sources of potential cross-reactive immunity to SARS-CoV-2, we devised a method to search for peptide matches between SARS-CoV-2 and sources of interest (details in Methods). Briefly, we partitioned the entire SARS-CoV-2 into 15 mer peptides with 10 residue overlaps and used them as query in modified BLAST searches against subject proteomes. We then selected ungapped subject hits with eight or more residues and ≥80% identity to SARS-CoV-2 as potential sources of cross-reactive immunity. We next predicted B cell and T cell reactivity of peptide hits using relevant epitope prediction methods (details in Methods). T cells can be divided into CD4 and CD8 T cells which recognize peptides displayed in the cell surface of antigen presenting cells bound to class I and class II MHC molecules, respectively. Therefore, we anticipated CD4 and CD8 T cell epitopes by predicting peptide binding to class I and class II human MHC molecules (in human known as HLA molecules for Human Leukocyte Antigens). For realization of T cell cross-reactivity, both peptide hits and equivalent SARS-CoV-2 peptides have to bind to the same class I and/or class II HLA molecules. Peptide hits with B cell epitope scores ≥0.4 were considered to be cross-reactive for B cell/antibody mediated immunity.
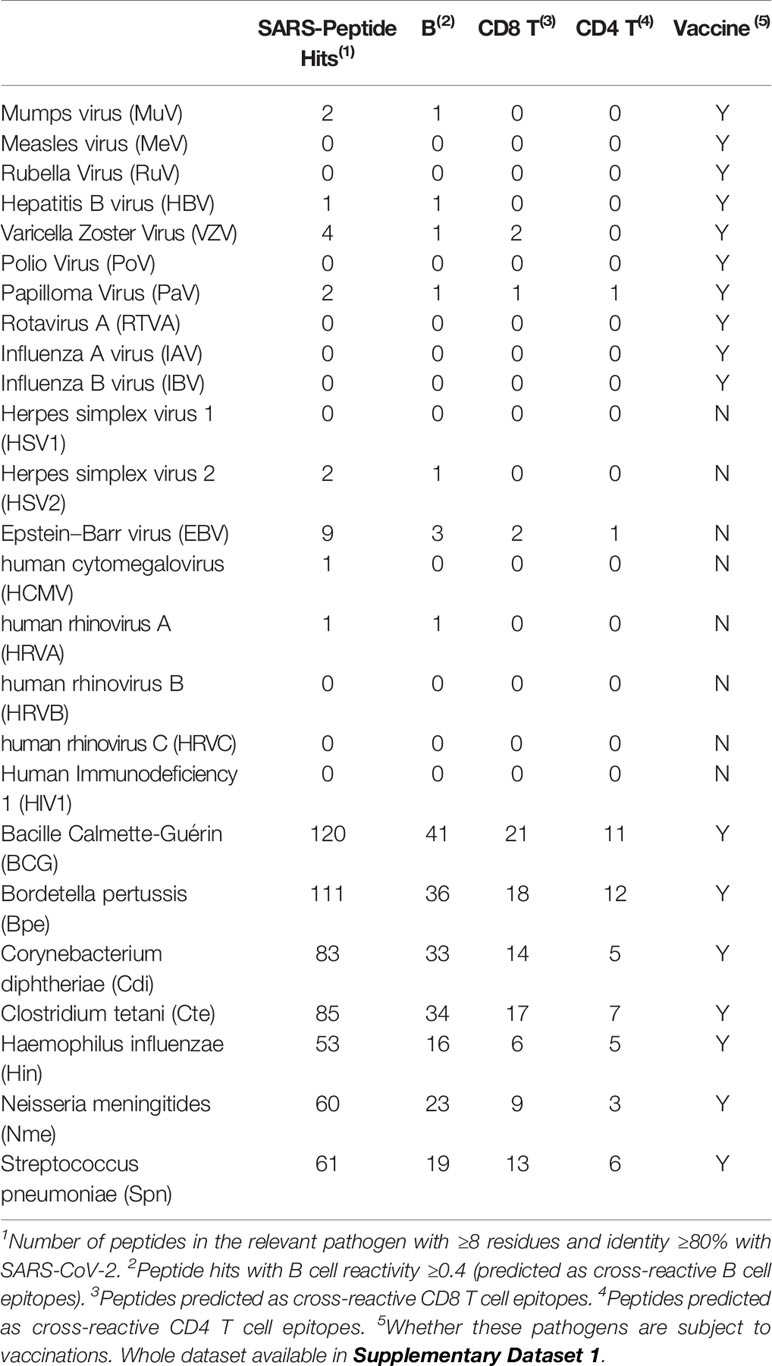
We first sought for cross-reactive immunity to SARS-CoV-2 in the proteomes of 25 human pathogens, including 18 viruses not related to SARS-CoV-2 and seven bacteria. Cross-reactive heterologous immunity is not limited to closely related pathogens, and in fact, it can occur between unrelated viruses (21) and between bacteria and viruses (22). As we age, there is a reduction in naive T cells and T cell repertoire diversity (23–25), likely prevailing memory T cells specific for pathogens causing common and/or persistent infections as shown for human cytomegalovirus (26). Therefore, in this study we considered viruses causing common, persistent and/or recurrent infections, including influenza (A and B), various herpesviruses (Epstein–Barr Virus, human cytomegalovirus and herpes simplex virus 1 and 2), human rhinovirus (HRV) A, B and C, human respiratory syncytial virus (RSV) A and B and human immunodeficiency virus 1 (HIV1). We also considered viruses targeted by vaccinations including poliovirus (PoV), mumps virus (MuV), rubella virus (RuV), papilloma virus (PaV), and rotavirus A (RTVA). Among the bacteria we considered bacille Calmette–Guérin (BCG), Bordetella pertussis (Bpe), Corynebacterium diphtheriae (Cdi), Clostridium tetani (Cte), Haemophilus influenzae (Hin), Neisseria meningitidis (serogroup B) and Streptococcus pneumoniae (Spn), which are all targeted by vaccinations. We found 595 unique peptides in the analyzed proteomes matching 551 peptides from SARS-CoV-2, indicating that the potential cross-reactive epitopes with SARS-CoV-2 differ for each pathogen. The complete data is provided in Supplementary Dataset 1, and in Table 2 we summarize the results. Overall, the number of SARS-CoV-2 peptide matches as well as that of potential cross-reactive B and T cell epitopes correlated with the size of the proteomes (Table 2). We cannot discard that some cross-reactive epitopes may be smaller and/or share less identity. In addition, there might be some cross-reactive epitopes rising from pathogen variants that we could not detect as we used single reference proteomes. We used state-of-the-art bioinformatics tools to predict cross-reactive T and B cell epitopes, but if we had used alternative prediction methods the cross-reactive T and B cell epitopes may be somewhat different. Overall, the cross-reactive epitopes detected through this approach will need experimental confirmation.
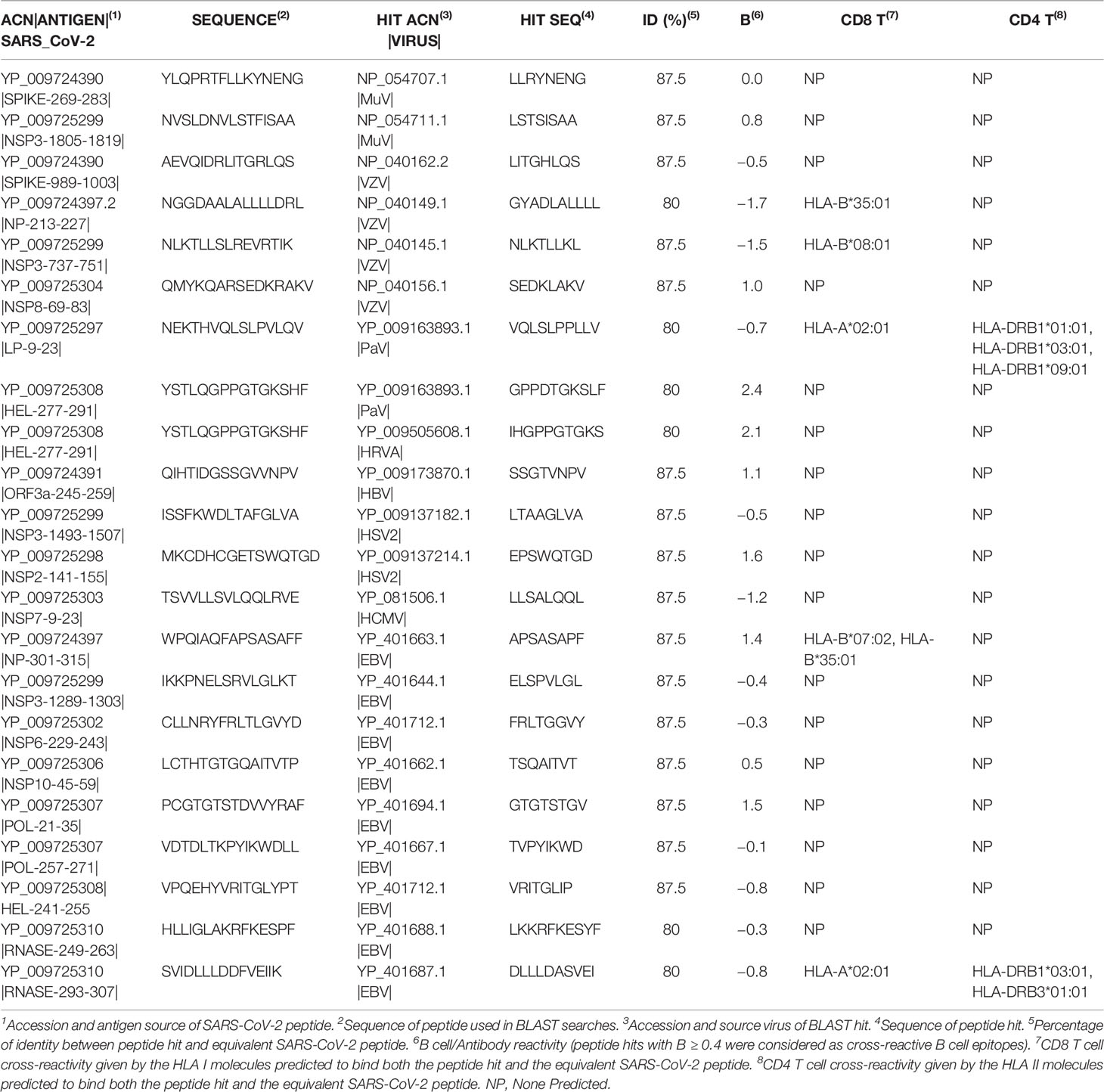
Cross-Reactive Immunity to SARS-CoV-2 From Prevalent Viruses and Viruses Targeted by Vaccinations
A recent study has detected SARS-CoV-2 reactive CD4 T cells in unexposed individuals, suggesting cross-reactive T cell recognition between circulating common cold coronaviruses and SARS-CoV-2 (27). T cell cross-reactivity between SARS-CoV-2 and other circulating coronaviruses is expected as they share high sequence similarity. However, whether infection by common cold coronaviruses leads to such cross-reactivity remains to be confirmed. In fact, common cold coronaviruses are responsible for less than 10% of all common colds of viral etiology (28). In the virus included in this study, there are only 22 SARS-CoV-2 peptide matches, including nine peptides with predicted B cell/antibody reactivity and five with predicted T cell cross-reactivity (Table 2). Two SARS-CoV-2 peptides in the Spike protein, the target of protective neutralizing antibodies (29), are present in MuV and VZV (Table 3). However, these two peptides have no predicted B cell reactivity, and more importantly do not map in accessible regions (data not shown). Of the 5 T cell cross-reactive epitopes, two are present in EBV; one of them (APSASAPF) is restricted by two HLA I molecules (B*07:02 and B*35:01) and the other (DLLLDASVEI) by one HLA I (A*02:01) and two HLAII (HLA-DRB1*03:01 and HLA-DRB3*01:01) molecules. No vaccine is available for EBV, and these cross-reactive epitopes may be drivers of protective or pathological immune responses in the individual expressing the noted HLA molecules. Such a role could be unraveled through functional recall assays (e.g. by ELISPOT assays) aimed to identify peptide-specific T cell responses in EBV+ and EBV− HLA type subjects with a history of SARS-CoV-2 infection, including asymptomatic and convalescents from severe disease. The other three potential cross-reactive T cell epitopes are found in two viruses targeted by vaccinations (Table 3). There are two CD8 T cell epitopes in VZV, one restricted by B*35:01 and another by B*08:01 and additional epitope in PaV that can be presented by class I (A*02:01) and class II (DRB1*01:01, HLA-DRB1*03:01, HLA-DRB1*09:01) HLA molecules (Table 3). PaV vaccines consist of recombinant L1 proteins (30) while the potentially cross-reactive PaV epitope maps in the E1 protein (Table 3). Thereby, PaV infection but not the vaccine can elicit T cell cross-reactivity to SARS-CoV-2. Overall, we can conclude that the selected viruses and particularly those targeted by vaccinations are poor sources of immune cross-reactivity to SARS-CoV-2 (Tables 2, 3). Thereby, we can discard that current pediatric vaccinations for viral diseases protect against SARS-CoV-2 infection, as recently suggested for RuV vaccine (31). Such suggestion was based on the distant homology observed between SARS-CoV-2 NS3P and Rubella virus p150 proteins, but not a single peptide is shared between SARS-CoV-2 and RuV.
Cross-Reactive Immunity to SARS-CoV-2 From Bacteria Targeted by Vaccinations
In contrast to viruses, the selected bacterial proteomes are considerable sources of cross-reactive immunity to SARS-CoV-2, correlating with their large proteomes (Table 2). However, the potential immune cross-reactivity rising from these bacteria shall be considered relevant. The match between SARS-CoV-2 and bacteria peptidomes is quite private; only 28 SARS-CoV-2 peptides out of 535 have a match with more than one bacterial proteome. Moreover, 88% of SARS-CoV-2 peptide matches with bacteria are absent in human or have an identity under that with bacteria (see Supplementary Dataset 1). The majority (68%) of cross-reactive SARS-CoV-2 peptides with bacteria do also match the human microbiome (44% with higher identity) (see Supplementary Dataset 1). This finding might look trivial, but it is actually very interesting. It has been noted that maintaining a diverse microbiota is essential for a competent immunity system (32, 33). Loss of microbiota diversity is associated with many diseases; it occurs in the elderly and immunocompromised subjects, and it can increase susceptibility to viral infections (34). How a diverse microbiota helps to fight viral infections is unclear, but it could be by increasing the chance for cross-reactive immunity. Interestingly, it has been shown that epitope similarity to the microbiota can enhance the immunogenicity of disease-associated antigenic epitopes (35).
The proteomes of BCG, B. pertussis, C. diphtheriae, C. tetani, H. influenzae, N. meningitidis and S. pneumoniae, contain numerous potentially cross-reactive epitopes, and make us wonder whether vaccination against these bacteria could elicit protective heterologous immunity against SARS-CoV-2 (Table 2). Such possibility has already been proposed for BCG (36–38) but through a different mechanism. BCG is a live attenuated strain of Mycobacterium bovis used to prevent tuberculosis (TB) (39), which is known to confer heterologous protective immunity to unrelated pathogens, including viruses, by inducing trained immunity (40, 41). Heterologous trained immunity is antigen non-specific and involves functional reprogramming of innate immune cells (42). It was the observation that there are less COVID-19 cases and fatalities in countries implementing BCG vaccination that led to postulate that trained immunity resulting from BCG vaccination could confer some protection against SARS-CoV-2 (36–38). Although we fully acknowledge the concept of trained immunity, our results also support that, if any, long-term protection against SARS-CoV-2 induced by BCG is most likely due to the presence of cross-reactive epitopes. We, however, find questionable that current BCG vaccinations are responsible for long-term protection against SARS-CoV-2.
BCG is only given once at birth, and long-term protection against any pathogen generally requires repeated antigen re-exposure to maintain memory T and B cells (43). Moreover, BCG vaccination is only included in some countries, while children and teenagers are largely unaffected by SARS-CoV-2 worldwide. In line with these arguments, a recent epidemiological study has discarded that BCG vaccination in childhood has a protective effect against COVID-19 in adulthood (44). Therefore, we next turned to analyze if immune cross-reactivity to SARS-CoV-2 from vaccines to C. diphtheriae, B. pertusis, C. tetani, H. influenzae, N. meningitidis and S. pneumoniae is what keeps children and young people safe from COVID-19.
Cross-Reactive Immunity to SARS-CoV-2 From Bacterial Vaccines: DTP and MenB
Pediatric vaccines for bacterial disease differ widely, and we first investigated their composition. Vaccines for H. influenzae (Hib), S. pneumoniae (PCV), and N. meningitides serotypes A, C, W, and Y (MenC and MenACWY) consist of bacteria cell wall polysaccharides, alone or conjugated with tetatus and/or diphteria toxoids (45–48). In contrast, vaccines for N. meningitides serotype B (MenB) consist of surface proteins from N. meningitides (15). Diphteria (D) and tetanus (T) vaccines contain inactivated toxins produced by the bacteria (toxoids) but in addition include a large array of proteins as shown by proteomics studies, which can be immunogenic (14, 49). D and T antigens are generally combined with pertussis (P) antigens as a combination DTP vaccine, which is available in two main formulations: DTaP and DTwP. DTaP contains selected P antigens without cells (acellular pertusis, aP), and DTwP includes inactivated whole pertussis bacteria (wP) (50).
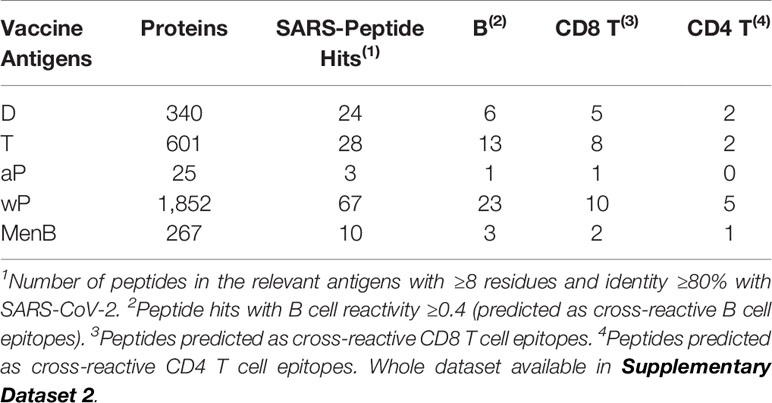
After considering the nature of bacterial vaccines, we discarded conjugate and polysaccharides vaccines and sought for potential sources of cross-reactive immunity in antigens identified by proteomics in DTa/wP and MenB vaccines. D and T antigens can vary between different vaccines, and here we considered the sum of all those reported elsewhere in (13, 14, 49). Our results, summarized in Table 4, indicate that combination DTP vaccines are significant sources of B and T cell cross-reactivity to SARS-CoV-2, while such cross-reactivity is small from MenB vaccine. In DTaP, P antigens do not contribute to increase the cross-reactive immunity provided by D and T antigens, but in DTwP, wP provide as much cross-reactive immunity as D and T antigens together (Table 4).
It is interesting to note that combination DTP vaccines, particularly DTwP, appear to be good inducers of cross-reactive CD8 T cells, which are key to clear viral infections by killing infected cells. The elicitation of virus cross-reactive CD8 T cells with bacterial antigens can occur through cross-presentation. This process enables antigen presenting cells to display endocytosed antigens in the context of HLA I molecules and prime CD8 T cells with anti-viral activity (51). We actually found more cross-reactive CD8 T cell epitopes than CD4 T cell epitopes (Table 5). However, this imbalance can be due to the fact that peptides with CD8 T cell epitopes can be shorter than those with CD4 T cell epitopes. In any case, the number of CD8 and CD4 T cell epitopes does not necessarily match with the magnitude of the CD8 and CD4 T cell response to SARS-CoV-2. In fact, SARS-CoV-2 specific CD4 T cell responses appear to be dominant when stimulating peripheral mononuclear blood cells with overlapping peptides (52). Interestingly, some cross-reactive T cell epitopes in DTP and MenB vaccines can be presented by HLA I and HLA II molecules, which will enable them to elicit both, CD8 and CD4 T cell responses. Moreover, many of the cross-reactive CD8 and CD4 T cell epitopes can be presented by various HLA I and HLA II molecules, respectively. T cell epitopes are only immunogenic in those individual expressing the HLA molecules that restrict the responses. With our data, we computed that the percentage of the world population that will have cross-reactive CD8 T cell responses to SARS-CoV-2 from MenB, DTaP, and DTwP vaccinations will be 46.54, 65.59, and 85.79%, respectively. Moreover, the percentage of the world population that will have cross-reactive CD8 and CD4 T cells to SARS-CoV-2 from MenB, DTaP, and DTwP vaccines will be 70.9, 77.47, and 94.67%, respectively. For populations expressing low frequency HLA molecules not included in this study, a more specific analysis considering such molecules will be required to uncover the relevant cross-reactive T cell epitopes and their coverage in that population. Overall, however, we can confirm that combination DTP vaccines, and particularly DTwP, would be able to elicit cross-reactive T cell responses against SARS-CoV-2 in most individuals.
Elicitation of effective cross-reactive T cell responses to SARS-CoV-2, particularly CD8 T cell responses, would allow the host to clear the virus quickly and exhibit milder courses of COVID-19. However, children appear to be largely unaffected by the virus, which is consistent with having protective antibody immunity blocking and limiting viral infection. Interestingly, such protective immunity could also result from cross-reactive antibodies elicited by combination DTP vaccines. Thus, we found 16 SARS-CoV-2 peptide matches with DTP antigens, mapping in ectodomains of virion surface antigens and of those, 14 are in SARS-CoV-2 Spike protein (Table 6). None of the SARS-CoV-2 peptide matches mapping in virion surface antigens are from aP antigens, while 11 are from wP, highlighting the superiority of DTwP vaccines in eliciting potentially protective cross-reactive immunity to SARS-CoV-2. It is worth noting that some SARS-CoV-2 peptide matches such as FEYVSQPF are not predicted as B cell epitopes (B < 0.4) but yet could be readily recognized by antibodies as shown by the computed solvent accessibilities (Table 6). B cell epitope predictions methods are in general quite unreliable, and actual solvent accessibilities values are better measures of potential B cell reactivity.
SARS-CoV-2 entry in the host cells is facilitated by angiotensin-converting enzyme 2 (ACE2), which is engaged by the receptor binding domain (RBD) of the Spike protein (53, 54). Antibodies that interfere with such interaction have been shown to be neutralizing and block viral entry (29). Interestingly, two of the potential cross-reactive B cell epitopes (RLFRKSNL and SFELLHAPAT) from wP antigens map in solvent exposed regions of the spike RBD domain (Figure 1), indicating that they can be neutralizing. There is also a cross-reactive B cell epitope (FEYVSQPF) from a D antigen, which is in close proximity (15–20 Å) to the RBD domain. Antibodies targeting this epitope will, by steric hindrance, block viral engagement with ACE2. Other accessible B cell epitopes from D, T, and wP antigens could also lead to the same effect (Table 6 and Figure 1). Antibody blocking of ACE2 interaction with Spike by steric hindrance is enhanced by the fact that SARS-CoV-2 Spike protein is a trimer.
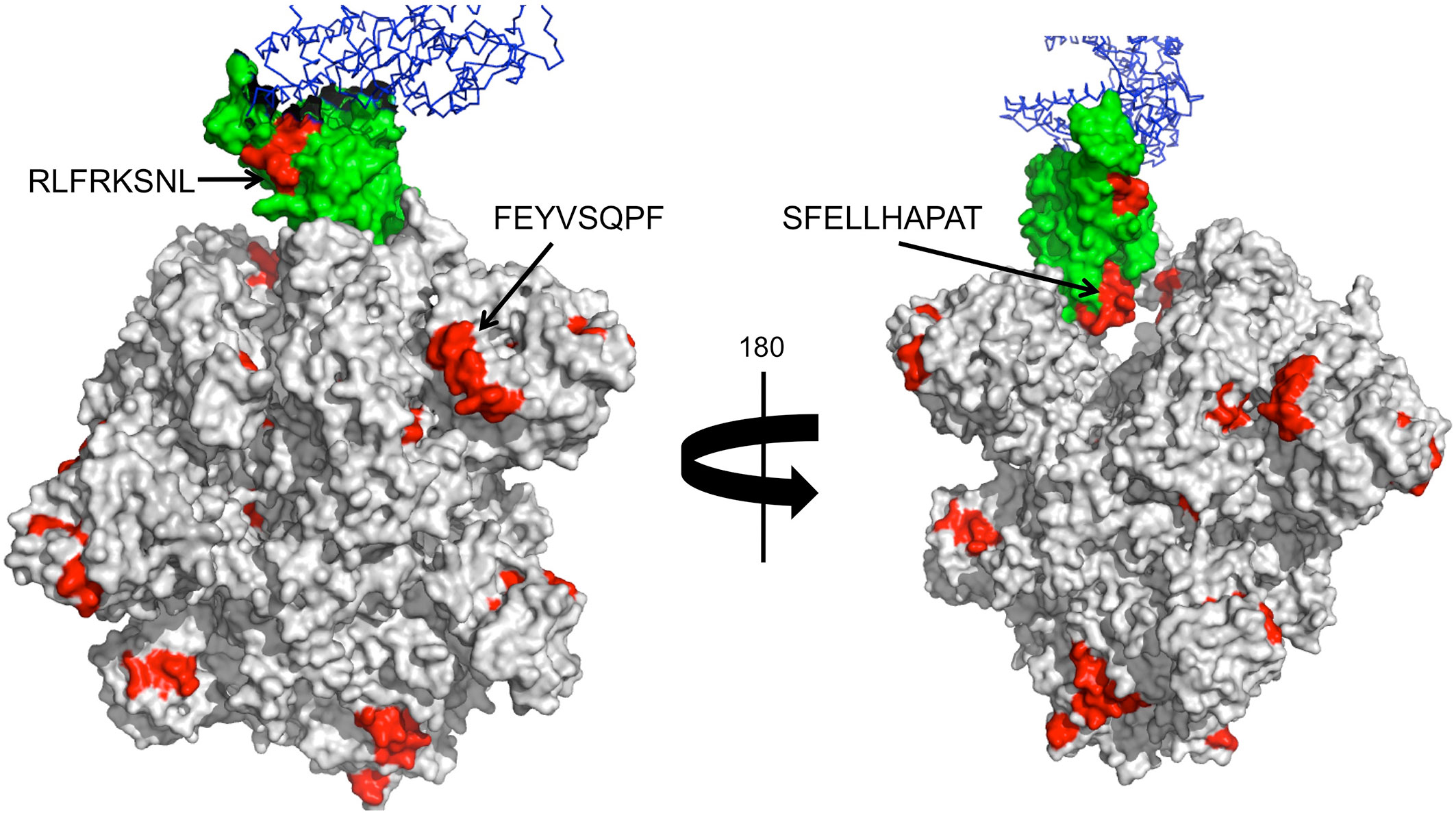
Figure 1 Cross-reactive B cell epitopes tagging SARS-CoV-2 Spike protein. Panel shows a surface rendering of the tertiary structure of the trimeric SARS-CoV-2 Spike protein in the open state with cross-reactive B cell epitopes in red. The Spike RBD is shown in green and in blue ACE2. We show the location and sequence of three B cell epitopes that are in RBD or nearby. These B cell epitopes can be the targets of neutralizing antibodies.
We realize that we did not analyze if the potential cross-reactive B cell epitopes in SARS-CoV-2 are also accessible in the source proteins. Although such analysis can be of interest, we found it unnecessary since our approach was aimed to identify continuous/linear B cell epitopes and not-conformational B cell epitopes. Actually, all identified cross-reactive B cell epitopes in SARS-CoV-2 Spike protein with solvent accessibility above 33% map in loop regions. Moreover, it is worth noting that extracellular proteases can degrade antigens as well as immune cells such as neutrophils, which phagocyte antigens and release them semi-degraded. Thus, during the course of immune response there are all sort of antigen fragments (intracellular, extracellular, accessible, buried etc) and antibodies against them are generated, as shown by B cell epitope mapping analysis (55–57). Of all the antibodies generated against a given pathogen, only those recognizing accessible B cell epitopes in accessible antigens may be protective against that particular pathogen. However, our results support that all the remaining antibodies still can also be useful for providing cross-reactive immunity.
DTP Vaccination and COVID-19 Epidemiology
Our data indicates that DTP vaccines can elicit ample T cell and B cell cross-reactive immunity against SARS-CoV-2. Likewise, MenB vaccines can also induce some substantial cross-reactive T cell immunity against SARS-CoV-2. However, MenB vaccines are unlikely responsible for the observed resilient of children to SARS-CoV-2 worldwide; MenB vaccines have been introduced very recently, in the last decade, and not everywhere (58, 59). In contrast, combination DTP vaccines are included in every single vaccination program since the 1940–50 (50). During infancy, children receive 3–4 doses (2, 3, 4, and 11 months of age) of either DTaP or DTwP, often in combination with inactivated poliovirus, conjugated Hib and recombinant antigens from HBV. Children also receive a pediatric DTaP booster vaccination at 4–6 years of age and a Tdap booster vaccination with lower antigenic load at 9–14 years of age. Moreover, pregnant women are also recommended to have a Tdap shot (https://www.who.int/topics/immunization/en/). Repeated vaccinations with DTP will therefore support that children and the youngest could be protected against SARS-CoV-2 through cross-reactive immunity elicited by these vaccines. Cross-reactive immunity elicited by DTP vaccines will eventually wane over time explaining why COVID-19 cases, and severity increases with age. We can also speculate that Tdap vaccination during pregnancy may also explain the lower incidence of COVID19 deaths in women. Moreover, Tdap vaccinated women could also pass passive cross-reactive immunity against SARS-CoV-2 to breast-feeding newborns.
The current vaccination trend has favored the use DTaP instead of DTwP vaccines (50). In Europe most countries use combination DTaP vaccines, but some still use combination DTwP vaccines. Taken in consideration that the P component in DTaP vaccines has little contribution to the cross-reactive immunity SARS-CoV-2, we can also conclude that cross-reactive immunity from the D and T components is likely what protects children against COVID-19 worldwide. This protection may be enhanced by conjugated vaccines using D and T toxoids, such as Hib and PCV (45, 46, 48, 60). However, our data also indicate that cross-reactive immunity to SARS-CoV-2 from the P component in combination DTwP vaccines rivals that of D and T antigens together, suggesting that DTwP vaccines may confer more protection against SARS-CoV-2. Interestingly, COVID-19 deaths and cases per million in Europe and Asian countries of the former URSS (Table S1) are significantly lower (p < 0.005) in those countries implementing DTwP vaccines (Figure 2). Median deaths and cases per million in countries with wP vaccines are 13 and 451, while these same figures in countries with aP vaccines are 52 and 1,387.
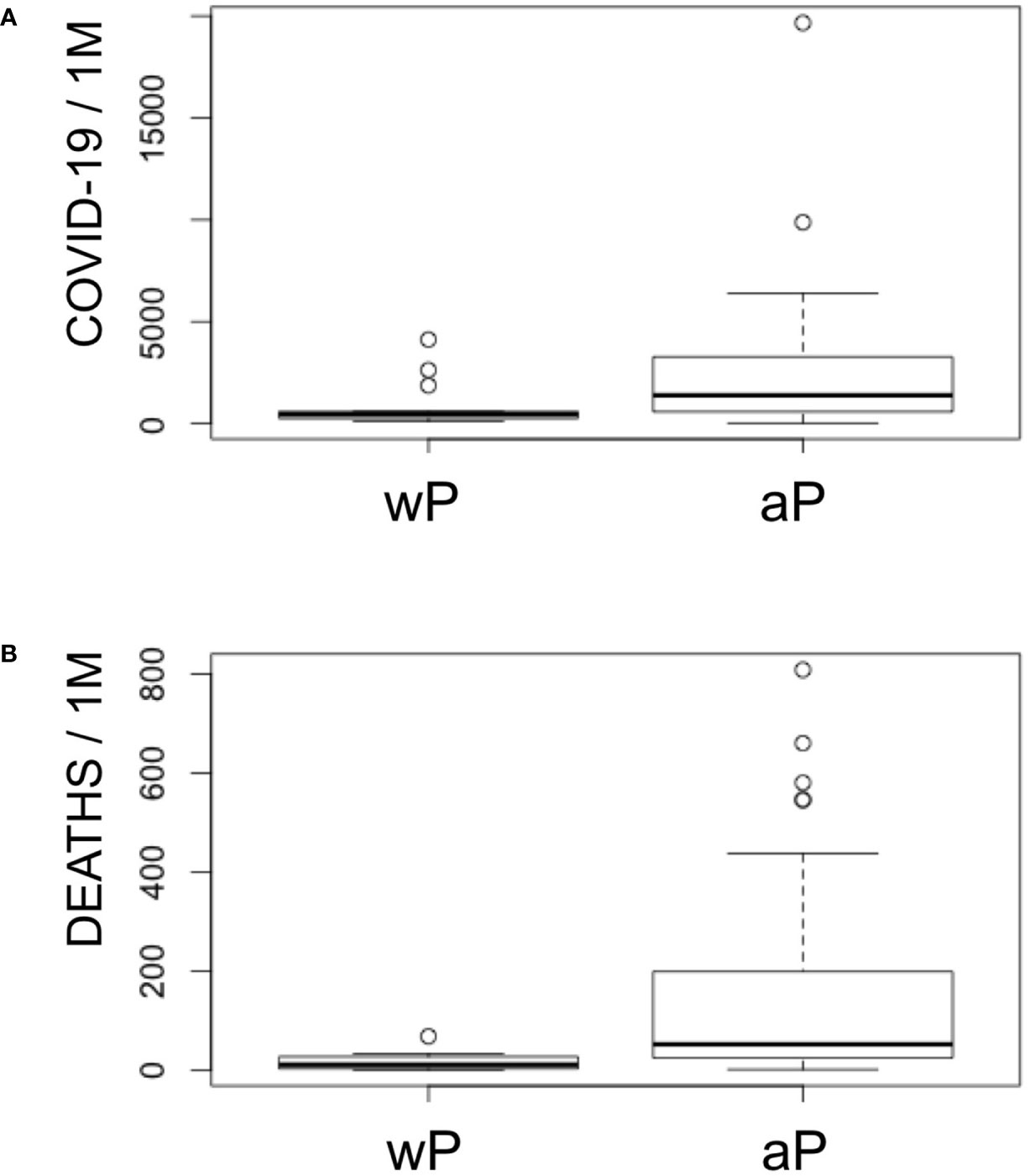
Figure 2 COVID-19 cases and deaths in Europe with regard to the P component in DTP vaccines. Panel shows a boxplot representation of COVID-19 cases (A) and deaths (B) per million in Europe including Asian countries from former URSS with regard to the inclusion of aP or wP in combination DTP vaccines. Non-parametric Kruskal–Wallis test indicates that both deaths and cases are significant lower in countries with DTwP vaccines (p < 0.005).
Epidemiological data show that adolescents primed with DTaP during infancy are also less protected against clinical pertussis than DTwP-primed children (61). Priming with DTwP or DTaP combination vaccines during infancy actually determine later immune memory responses at preadolescent age to all DTaP antigens, with wP-priming resulting in a beneficial Th1-dominated response (62). Most likely this Th1-dominated response is also beneficial against SARS-CoV-2.
Conclusion
Our findings indicate that cross-reactive immunity elicited by DT antigens in combination DTP vaccines is likely keeping children and the youngest safe from COVID-19 worldwide. Cross-reactive immunity elicited by these vaccines surely wanes over time explaining why COVID-19 cases and severity increases with age. The global health crisis caused by COVID-19 has prompted many efforts to develop a specific vaccine against SARS-CoV-2, which if successful will take a long time to achieve. However, we need a vaccine for SARS-CoV-2 today, and our results support that DTP vaccinations, and in particular DTwP vaccinations, could protect the general population against COVID-19 through cross-reactive immunity. Given the heterogenicity of DTP vaccines, our results also suggest that COVID-19 death rates and severity in different countries may be linked to differences in the composition of DTP vaccines, vaccination regimes, and initiation of DTP vaccination programs.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author wishes to thank the editor for handling the manuscript with care and professionalism. The author also wishes to thank the reviewers for their positive assessment and their thoughtful and insightful comments, which helped to improve the manuscript. The author also thanks Dr. Esther M. Lafuente for comments and motivation and the Complutense University for special grant action B AE21/20-23164 to PAR to cover potential publication costs. A preprint version of this manuscript is available at https://osf.io/sbgy3 (DOI: 10.31219/osf.io/sbgy3).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.586984/full#supplementary-material
Supplementary Data sheet 1 | Excel file including all SARS-CoV-2 peptide matches and predicted cross-reactive epitopes in 25 human pathogens, including 18 viruses and seven bacteria.
Supplementary Data sheet 2 | Excel file including all SARS-CoV-2 peptide matches and predicted cross-reactive epitopes from DTP and MenB vaccines.
Supplementary Table 1 | COVID-19 cases and fatalities per million in different countries with regard to the use of aP/wP vaccines.
References
1. She J, Jiang J, Ye L, Hu L, Bai C, Song Y. novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin Transl Med (2019) 2020(9):19. doi: 10.1186/s40169-40020-00271-z
2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (2020) 395:497–506. doi: 10.1016/S0140-6736(1020)30183-30185
3. Viruses, CSGotICoTo. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol (2020) 5:536–44. doi: 10.1038/s41564-41020-40695-z
4. Rodriguez-Morales AJ, Cardona-Ospina JA, Gutierrez-Ocampo E, Villamizar-Pena R, Holguin-Rivera Y, Escalera-Antezana JP, et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med Infect Dis (2020) 34:101623. doi: 10.1016/j.tmaid.2020.101623
5. Li R, Tian J, Yang F, Lv L, Yu J, Sun G, et al. Clinical characteristics of 225 patients with COVID-19 in a tertiary Hospital near Wuhan, China. J Clin Virol (2020) 127:104363. doi: 10.1016/j.jcv.2020.104363
6. Ge H, Wang X, Yuan X, Xiao G, Wang C, Deng T, et al. The epidemiology and clinical information about COVID-19. Eur J Clin Microbiol Infect Dis (2020) 39:1011–9. doi: 10.1007/s10096-10020-03874-z
7. Fulton RB, Varga SM. Effects of aging on the adaptive immune response to respiratory virus infections. Aging Health (2009) 5:775. doi: 10.2217/ahe.2209.2269
8. Zhao M. Cytokine storm and immunomodulatory therapy in COVID-19: Role of chloroquine and anti-IL-6 monoclonal antibodies. Int J Antimicrob Agents (2020) 55:105982. doi: 10.101016/j.ijantimicag.102020.105982
9. Agrawal B. Heterologous Immunity: Role in Natural and Vaccine-Induced Resistance to Infections. Front Immunol (2019) 10:2631. doi: 10.3389/fimmu.2019.02631. eCollection 02019.
10. Welsh RM, Fujinami RS. Pathogenic epitopes, heterologous immunity and vaccine design. Nat Rev Microbiol (2007) 5:555–63. doi: 10.1038/nrmicro1709
11. Gu XX, Plotkin SA, Edwards KM, Sette A, Mills KHG, Levy O, et al. Waning Immunity and Microbial Vaccines-Workshop of the National Institute of Allergy and Infectious Diseases. Clin Vaccine Immunol (2017) 24:e00034–00017. doi: 10.01128/CVI.00034-00017
12. Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J Immunol Res (2017) 2017:2680160. doi: 10.1155/2017/2680160
13. Moller J, Kraner ME, Burkovski A. Proteomics of Bordetella pertussis whole-cell and acellular vaccines. BMC Res Notes (2019) 12:329. doi: 10.1186/s13104-13019-14373-13102
14. Moller J, Kraner ME, Burkovski A. More than a Toxin: Protein Inventory of Clostridium tetani Toxoid Vaccines. Proteomes (2019) 7:15. doi: 10.3390/proteomes7020015
15. Rodrigues CMC, Chan H, Vipond C, Jolley K, Harrison OB, Wheeler J, et al. Typing complex meningococcal vaccines to understand diversity and population structure of key vaccine antigens. Wellcome Open Res (2019) 3:151. doi: 10.12688/wellcomeopenres.14859.12682. eCollection 12018.
16. Altschul SF, Madden TL, Sch√§ffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res (1997) 25:3389–402. doi: 10.1093/nar/3325.3317.3389
17. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449:804–10. doi: 10.1038/nature06244
18. Molero-Abraham M, Lafuente EM, Flower DR, Reche PA. Selection of conserved epitopes from hepatitis C virus for pan-populational stimulation of T-cell responses. Clin Dev Immunol (2013) 2013:601943. doi: 10.1155/2013/601943
19. Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf (2006) 7:153. doi: 10.1186/1471-2105-1187-1153
20. Hubbard SJ, Thornton JM. NACCESS, Computer Program. University College London: Department of Biochemistry and Molecular Biology (1993).
21. Che JW, Selin LK, Welsh RM. Evaluation of non-reciprocal heterologous immunity between unrelated viruses. Virology (2015) 482:89–97. doi: 10.1016/j.virol.2015.1003.1002
22. Mathurin KS, Martens GW, Kornfeld H, Welsh RM. CD4 T-cell-mediated heterologous immunity between mycobacteria and poxviruses. J Virol (2009) 83:3528–39. doi: 10.1128/JVI.02393-02308
23. Egorov ES, Kasatskaya SA, Zubov VN, Izraelson M, Nakonechnaya TO, Staroverov DB, et al. The Changing Landscape of Naive T Cell Receptor Repertoire With Human Aging. Front Immunol (2018) 9:1618. doi: 10.3389/fimmu.2018.01618. eCollection 02018.
24. Moro-Garcia MA, Alonso-Arias R, Lopez-Larrea C. Molecular mechanisms involved in the aging of the T-cell immune response. Curr Genomics (2012) 13:589–602. doi: 10.2174/138920212803759749
25. Nikolich-Zugich J. T cell aging: naive but not young. J Exp Med (2005) 201:837–40. doi: 10.1084/jem.20050341
26. Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med (2005) 202:673–85. doi: 10.1084/jem.20050882
27. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell (2020) 20:30610–3. doi: 10.1016/j.cell.2020.05.015
28. Makela MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol (1998) 36:539–42. doi: 10.1128/JCM.1136.1122.1539-1542.1998
29. Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature (2020) 26:020–2380. doi: 10.1101/2020.03.21.990770
30. Pandhi D, Sonthalia S. Human papilloma virus vaccines: Current scenario. Indian J Sex Transm Dis AIDS. (2011) 32:75–85. doi: 10.4103/0253-7184.85409
31. Franklin R, Young A, Neumann B, Fernandez R, Joannides A, Reyahi A, et al. Homologous protein domains in SARS-CoV-2 and measles, mumps and rubella viruses: preliminary evidence that MMR vaccine might provide protection against COVID-19. medRxiv (2020) 1:10. doi: 10.1101/2020.04.10.20053207
32. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell (2014) 157:121–41. doi: 10.1016/j.cell.2014.1003.1011
33. Roughan G, Nishida I. Concentrations of long-chain acyl-acyl carrier proteins during fatty acid synthesis by chloroplasts isolated from pea (Pisum sativum), safflower (Carthamus tinctoris), and amaranthus (Amaranthus lividus) leaves. Arch Biochem Biophys (1990) 276:38–46. doi: 10.1016/0003-9861(1090)90007-l
34. Mosca A, Leclerc M, Hugot JP. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front Microbiol (2016) 7:455. doi: 10.3389/fmicb.2016.00455. eCollection 02016.
35. Carrasco Pro S, Lindestam Arlehamn CS, Dhanda SK, Carpenter C, Lindvall M, Faruqi AA, et al. Microbiota epitope similarity either dampens or enhances the immunogenicity of disease-associated antigenic epitopes. PloS One (2018) 13:e0196551. doi: 10.0191371/journal.pone.0196551. eCollection 0192018.
36. Miller A, Reandelar MJ, Fasciglione K, Roumenova V, Li Y, GH O. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv (2020) 0:12. doi: 10.1101/2020.03.24.20042937
37. Curtis N, Sparrow A, Ghebreyesus TA, Netea MG. Considering BCG vaccination to reduce the impact of COVID-19. Lancet (2020) 395:1545–6. doi: 10.1016/S0140-6736(1520)31025-31024
38. O’Neill LAJ, Netea MG. BCG-induced trained immunity: can it offer protection against COVID-19? Nat Rev Immunol (2020) 20:335–7. doi: 10.1038/s41577-41020-40337-y
39. Mangtani P, Abubakar I, Ariti C, Beynon R, Pimpin L, Fine PE, et al. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin Infect Dis (2014) 58:470–80. doi: 10.1093/cid/cit1790
40. Arts RJW, Carvalho A, La Rocca C, Palma C, Rodrigues F, Silvestre R, et al. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep (2016) 17:2562–71. doi: 10.1016/j.celrep.2016.2511.2011
41. Moorlag S, Arts RJW, van Crevel R, Netea MG. Non-specific effects of BCG vaccine on viral infections. Clin Microbiol Infect (2019) 25:1473–8. doi: 10.1016/j.cmi.2019.1404.1020
42. Sanchez-Ramon S, Conejero L, Netea MG, Sancho D, Palomares Ì, Subiza JL. Trained Immunity-Based Vaccines: A New Paradigm for the Development of Broad-Spectrum Anti-infectious Formulations. Front Immunol (2018) 9:2936. doi: 10.3389/fimmu.2018.02936. eCollection 02018.
43. Zinkernagel RM. What if protective immunity is antigen-driven and not due to so-called “memory” B and T cells? Immunol Rev (2018) 283:238–46. doi: 10.1111/imr.12648
44. Hamiel U, Kozer E, Youngster I. SARS-CoV-2 Rates in BCG-Vaccinated and Unvaccinated Young Adults. Jama (2020) 323:2340–1. doi: 10.1001/jama.2020.8189
45. Ahmad H, Chapnick EK. Conjugated polysaccharide vaccines. Infect Dis Clin North Am (1999) 13:113–33, vii. doi: 10.1016/s0891-5520(1005)70046-70045
46. Choe YJ, Blatt DB, Lee HJ, Choi EH. Associations between geographic region and immune response variations to pneumococcal conjugate vaccines in clinical trials: A systematic review and meta-analysis. Int J Infect Dis (2020) 92:261–8. doi: 10.1016/j.ijid.2019.1012.1021
47. Cohn AC, MacNeil JR, Clark TA, Ortega-Sanchez IR, Briere EZ, Meissner HC, et al. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep (2013) 62:1–28.
48. Daniels CC, Rogers PD, Shelton CM. A Review of Pneumococcal Vaccines: Current Polysaccharide Vaccine Recommendations and Future Protein Antigens. J Pediatr Pharmacol Ther (2016) 21:27–35. doi: 10.5863/1551-6776-5821.5861.5827
49. Moller J, Kraner M, Sonnewald U, Sangal V, Tittlbach H, Winkler J, et al. Proteomics of diphtheria toxoid vaccines reveals multiple proteins that are immunogenic and may contribute to protection of humans against Corynebacterium diphtheriae. Vaccine (2019) 37:3061–70. doi: 10.1016/j.vaccine.2019.3004.3059
50. Guiso N, Meade BD. and Wirsing von K√∂nig, CH. Pertussis vaccines: The first hundred years. Vaccine (2020) 38:1271–6. doi: 10.1016/j.vaccine.2019.1211.1022
51. Embgenbroich M, Burgdorf S. Current Concepts of Antigen Cross-Presentation. Front Immunol (2018) 9:1643. doi: 10.3389/fimmu.2018.01643. eCollection 02018.
52. Habel JR, Nguyen THO, van de Sandt CE, Juno JA, Chaurasia P, Wragg K, et al. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A*02:01 phenotype. Proc Natl Acad Sci U S A (2020) 10:2015486117. doi: 10.1101/2020.08.17.20176370
53. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature (2020) 581:215–20. doi: 10.1038/s41586-41020-42180-41585
54. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature (2020) 581:221–4. doi: 10.1038/s41586-41020-42179-y
55. Azcarate IG, Marin-Garcia P, Abad P, Peez-Benavente S, Paz-Artal E, Reche PA, et al. Plasmodium falciparum immunodominant IgG epitopes in subclinical malaria. Sci Rep (2020) 10:9398. doi: 10.1038/s41598-41020-66384-41590
56. Quinzo MJ, Lafuente EM, Zuluaga P, Flower DR, Reche PA. Computational assembly of a human Cytomegalovirus vaccine upon experimental epitope legacy. BMC Bioinf (2019) 20:476. doi: 10.1186/s12859-12019-13052-12856
57. Alonso-Padilla J, Lafuente EM, Reche PA. Computer-Aided Design of an Epitope-Based Vaccine against Epstein-Barr Virus. J Immunol Res (2017) 2017:9363750. doi: 10.1155/2017/9363750
58. Kuhdari P, Stefanati A, Lupi S, Valente N, Gabutti G. Meningococcal B vaccination: real-world experience and future perspectives. Pathog Glob Health (2016) 110:148–56. doi: 10.1080/20477724.20472016.21195072
59. Soler-Garcia A, Fernandez de Sevilla M, Abad R, Esteva C, Alsina L, Vazquez J, et al. Meningococcal Serogroup B Disease in Vaccinated Children. J Pediatr Infect Dis Soc (2019) 21:454–9. doi: 10.1093/jpids/piz071
60. Goldblatt D. Conjugate vaccines. Clin Exp Immunol (2000) 119:1–3. doi: 10.1046/j.1365-2249.2000.01109.x
61. Sheridan SL, Ware RS, Grimwood K, Lambert SB. Reduced risk of pertussis in whole-cell compared to acellular vaccine recipients is not confounded by age or receipt of booster-doses. Vaccine (2015) 33:5027–30. doi: 10.1016/j.vaccine.2015.5008.5021
Keywords: coronavirus disease 19 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), epitope, cross-reactive immunity, DTP vaccine
Citation: Reche PA (2020) Potential Cross-Reactive Immunity to SARS-CoV-2 From Common Human Pathogens and Vaccines. Front. Immunol. 11:586984. doi: 10.3389/fimmu.2020.586984
Received: 24 July 2020; Accepted: 25 September 2020;
Published: 16 October 2020.
Edited by:
Moriya Tsuji, Columbia University Irving Medical Center, United StatesReviewed by:
Jordana Grazziela Coelho-dos-Reis, Oswaldo Cruz Foundation (Fiocruz), BrazilGabriel M. Gutierrez, Leidos, United States
Copyright © 2020 Reche. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pedro A. Reche, cGFyZWNoZWdAbWVkLnVjbS5lcw==
 Pedro A. Reche
Pedro A. Reche