- 1Division of Arthritis and Rheumatic Diseases, Department of Medicine, Oregon Health & Science University, Portland, OR, United States
- 2Departments of Ophthalmology, Medicine, and Cell Biology, Oregon Health & Science University, Portland, OR, United States
- 3Legacy Devers Eye Institute, Portland, OR, United States
Spondyloarthritis (SpA) is a group of immune mediated inflammatory diseases with a strong association to the major histocompatibility (MHC) class I molecule, HLA-B27. Although the association between HLA-B27 and AS has been known for almost 50 years, the mechanisms underlying disease pathogenesis are elusive. Over the years, three hypotheses have been proposed to explain HLA-B27 and disease association: 1) HLA B27 presents arthritogenic peptides and thus creates a pathological immune response; 2) HLA-B27 misfolding causes endoplasmic reticulum (ER) stress which activates the unfolded protein response (UPR); 3) HLA-B27 dimerizes on the cell surface and acts as a target for natural killer (NK) cells. None of these hypotheses explains SpA pathogenesis completely. Evidence supports the hypothesis that HLA-B27-related diseases have a microbial pathogenesis. In animal models of various SpAs, a germ-free environment abrogates disease development and colonizing these animals with gut commensal microbes can restore disease manifestations. The depth of microbial influence on SpA development has been realized due to our ability to characterize microbial communities in the gut using next-generation sequencing approaches. In this review, we will discuss various putative pathobionts in the pathogenesis of HLA-B27-associated diseases. We pursue whether a single pathobiont or a disruption of microbial community and function is associated with HLA-B27-related diseases. Furthermore, rather than a specific pathobiont, metabolic functions of various disease-associated microbes might be key. While the use of germ-free models of SpA have facilitated understanding the role of microbes in disease development, future studies with animal models that mimic diverse microbial communities instead of mono-colonization are indispensable. We discuss the causal mechanisms underlying disease pathogenesis including the role of these pathobionts on mucin degradation, mucosal adherence, and gut epithelial barrier disruption and inflammation. Finally, we review the various uses of microbes as therapeutic modalities including pre/probiotics, diet, microbial metabolites and fecal microbiota transplant. Unravelling these complex host-microbe interactions will lead to the development of new targets/therapies for alleviation of SpA and other HLA-B27 associated diseases.
Introduction
Spondyloarthritis (SpA) is an umbrella term used for various disorders including ankylosing spondylitis (AS), arthritis associated with inflammatory bowel disease (IBD), acute anterior uveitis, a subset of juvenile idiopathic arthritis (JIA), reactive arthritis (ReA), psoriatic arthritis (PsA), and undifferentiated spondyloarthritis (USpA) (Figure 1). These diseases share common clinical features (such as sacroiliitis, enthesitis and dactylitis) and overlapping extra-articular manifestations (i.e., uveitis, psoriasis, and bowel inflammation). Uveitis is the most common extra-articular manifestation of AS. In addition, many AS patients also have gut inflammation, such as Crohn’s disease (CD) and ulcerative colitis (UC). On the other hand, axial/peripheral arthritis is the most common extra-intestinal complications in IBD, especially in patients with CD (1, 2). These conditions may occur either simultaneously or sequentially, with almost 50% of AS patients having subclinical gut inflammation and around 15% of IBD patients have peripheral SpA (3, 4). In addition to similar and overlapping disease manifestation, there is a considerable overlap among the genetic risk factors for AS, CD, and PsA, such as IL23R, IL12B, STAT3, ORMDL3, and CARD9 (5), which are associated with IL-23 signaling. The immune and inflammatory response between AS and CD shows considerable overlap dominated by the Th17 helper cell pathways (6, 7). In addition, association with a non-major histocompatibility gene Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) has also been reported in patients with AS (8) and IBD (9) either alone or in combination with polymorphisms in HLA class-I alleles (10). Furthermore, other factors such as environment, host immune regulation, disruption of mucosal barrier, and gut microbial dysbiosis contribute toward pathogenesis of SpA [ (11, 12) Figure 2]. Host genetic susceptibility is associated with perturbed immune/inflammatory response, which may lead to microbial dysbiosis and pathobiont expansion and loss of barrier function, resulting in inflammation in the gut, joints, eye, and skin (13–16). While our focus for this review is on HLA-B27-associated microbes and their role in various SpAs, we will also discuss pathobionts and host-microbial relationships in IBD that are relevant to this topic.
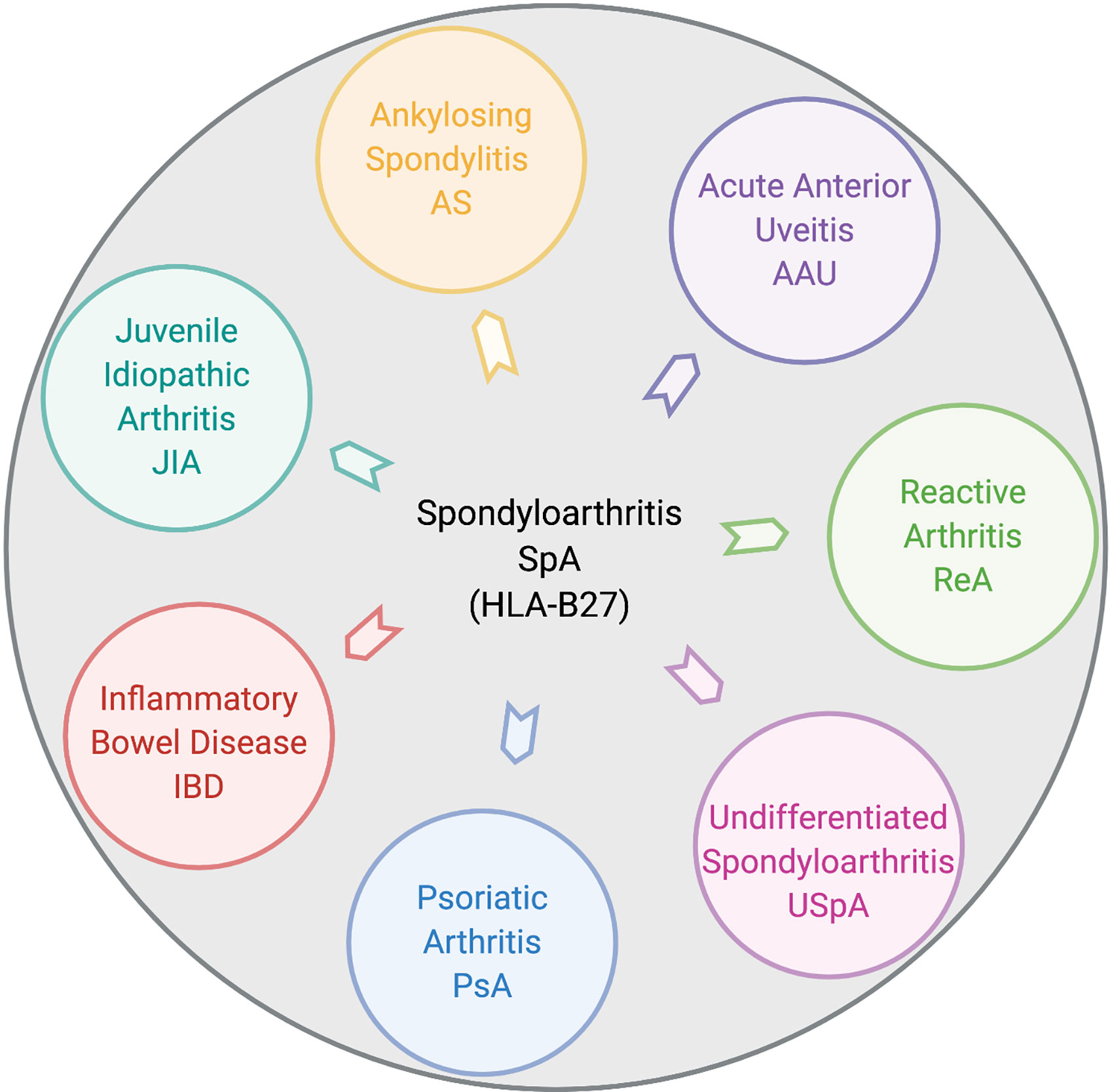
Figure 1 Disease overlap in HLA-B27-associated spondyloarthropathies. Pictorial representation of various disorders broadly included within HLA-B27-associated spondyloarthritis (SpA). These include ankylosing spondylitis (AS), acute anterior uveitis (AAU), reactive arthritis (ReA), juvenile idiopathic arthritis (JIA), inflammatory bowel disease (IBD), psoriatic arthritis (PsA), and undifferentiated spondyloarthritis (USpA). Figure created with Biorender.com.
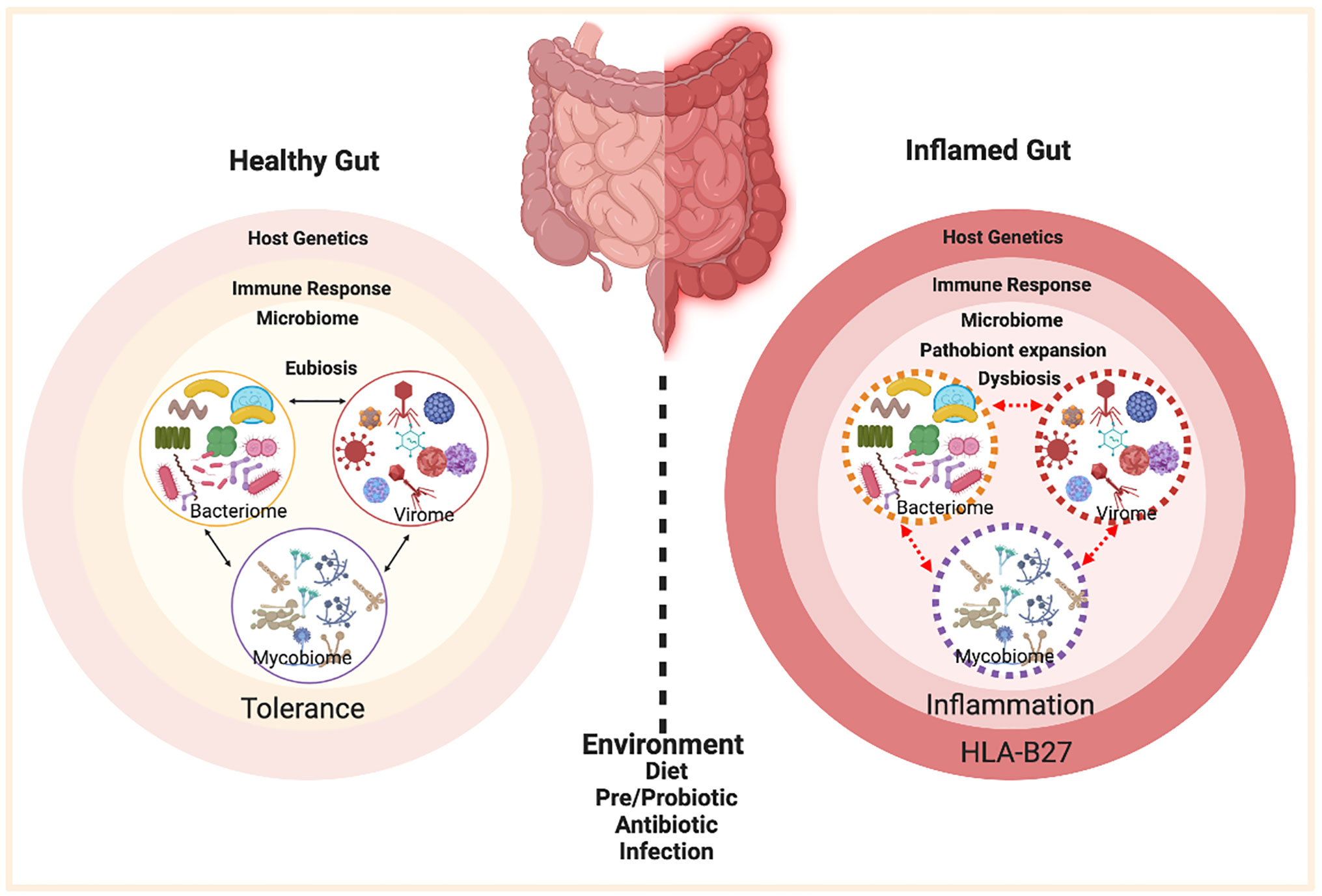
Figure 2 Factors contributing to HLA-B27 associated gut inflammation in SpA. In the healthy gut (left panel) there is microbial eubiosis/homeostasis within the members of bacteriome, mycobiome, and viruses in a stable interdependent microbial community. The gut immune response recognizes commensals and does not mount an inflammatory response. A non-susceptible host genetic background and a lack of environmental stress contribute toward favorable host-microbe interactions and microbial homeostasis. On the contrary, HLA-B27 associated gut (right panel) displays a microbial dysbiosis, which may be associated with pathobiont expansion, dysbiosis in various microbial components (bacteriome, mycobiome, virome) and loss of colonization resistance from the gut commensal microbes. HLA-B27 associated host immune dysregulation can cause loss of barrier protection and therefore bacterial components can activate an aberrant immune response, which in turn triggers an inflammatory response. Figure created with Biorender.com.
HLA-B27, Spondyloarthritis, and Gut Microbiome
Human leukocyte antigen (HLA)-B27, is a major histocompatibility complex (MHC)-class I molecule associated with various SpAs. Its association with the prototypic SpA- ankylosing spondylitis was discovered almost 50 years ago (17), and is the strongest association known between a genetic factor and a complex, genetic disease. Subsequently, the association of HLA-B27 was found with ReA, axial arthritis in association with IBD, AAU and the axial arthritis subset of PsA (18–20). The MHC complex is located on chromosome 6 (21), and plays a critical role in immunity and recognition of self in almost all cells of the body (22). MHC class I includes HLA-A, HLA-B, and HLA-C, and presents antigen to CD8 T cells. Despite HLA-B27 being a class I molecule, CD8 T-cells have not been associated with disease development; instead CD4+ T cells are thought to drive disease (23). While the actual role of HLA-B27 in triggering inflammation in various disorders remains unresolved, three different theories have been suggested. These include, i) presentation of arthritogenic peptides, which may activate a pathological immune response (24) ii) misfolding of heavy chain of HLA-B27 and its effects on ER-associated degradation (ERAD) and activation of unfolded protein response (UPR) (25–28); and iii) HLA-B27 dimerization during cell surface recycling and recognition by immune receptors on natural killer (NK) cells (29, 30). While these hypotheses may provide some explanation, such as the activation of the inflammatory IL-23/lL-17 axis through UPR or non-canonical activation of CD4+ T cells by HLA-B27 dimers, the exact mechanism by which HLA-B27 leads to development of SpA is unknown (31). In the last decade, HLA-B27-associated perturbation in the gut microbiota has emerged as an underlying mechanism in disease pathogenesis. Indeed, many of the risk genes common to IBD and SpA (e.g., IL23R, NOD2) are associated with innate immune response pathways (32, 33), consistent with the hypothesis that alteration in the host immune response to gut microbes may play a key role in these disorders.
More than twenty-five years ago, a new insight came from the experimental data revealing the role of gut microbiota in complex inflammatory disorders. In 1994, it was observed that HLA-B27 transgenic (TG) rats raised in a germ-free environment fail to develop either arthritis or colitis (34). Recolonizing the gut with either altered Schaedler’s flora (ASF) or just a few commensal microbes was sufficient for disease development (35, 36). Rosenbaum and Davey proposed a hypothesis that ‘HLA-B27 predisposes to ankylosing spondylitis by altering the microbiome’ (37). Since the last decade many studies in patients and rodent models have shown alteration of gut microbial communities in various HLA-B27 associated disorders (12, 38–40). Based on our extensive studies with the HLA-B27 TG rat model, we demonstrated that HLA-B27-associated microbial dysbiosis is subject to host genetics and environment. This lead to the proposal of an ecological model of microbial dysbiosis (12).
Putative Pathobionts in hla-b27 Associated Spondyloarthropathies
Through the advent of high throughput sequencing approaches, many putative pathobionts have been identified in various HLA-B27-associated SpAs and bowel inflammation. A pathobiont is defined as a microbe that can cause or promote disease only when specific genetic or environmental conditions are altered in the host such as dysregulated host immune response and microbial dysbiosis (41). The term “pathobiont” was first used for Helicobacter hepaticus, a bacterium associated with gut inflammation in immunocompromized mice, but not in wild-type animals (42, 43). While most of the sequencing studies have focused on the alterations in bacterial component of the gut microbiota, recent research is also focused to define the fungal and viral component of the gut microbiome (mycobiome and virome respectively) and interkingdom interactions in SpA. In this section, we will discuss important bacterial, fungal, and viral pathobionts in HLA-B27-associated SpAs including overlapping inflammatory diseases such as CD and colitis.
Bacterial Pathobionts
Pathobionts Associated With Reactive Arthritis
Reactive arthritis (ReA) is an immune mediated inflammation of the synovial tissue that usually develops after a urinary or gut infection (44). HLA-B27-associated ReA is a type of SpA triggered by bacteria such as Campylobacter, Chlamydia, Salmonella, Shigella, and Yersinia, resulting in oligoarthritis of the lower limbs and sometimes with urethritis and conjunctivitis (45). One of the earlier studies isolated Chlamydia trachomatis strains from the eye and urethra of a patient with ReA, previously known as Reiter’s Syndrome (46). Subsequently, in a study focused on endoplasmic reticulum (ER) stress, which is thought to be downstream to HLA-B27 protein misfolding, C. trachomatis was shown to induce IL-23 expression in infected myeloid cells (47). In addition, stimulation through TLR or ER stress can cause the activation of ER stress induced transcription factor CHOP, which in turn increases the expression of IL-23. However, TLR engagement in itself can trigger ER stress through activation of XBP1, essential for production of inflammatory cytokines in macrophages (48). HLA-B27 misfolding was associated with enhanced replication of Salmonella by the activation of the unfolded protein response (UPR), through the transcription factor XBP1 (49) Although this study employs HeLa cells that do not express TLR, it is possible that innate immune receptors may be involved in response to bacteria or bacterial products (50). In another study, peripheral blood and serum polymerase chain reaction (PCR) analysis in patients with Chlamydia induced ReA showed the presence of DNA from C. trachomatis in the peripheral blood cells, but not in the serum (51). This can be explained by the fact that C. trachomatis resides in the monocytic cells, prevents their apoptosis and stimulates the production of inflammatory mediators (51). In fact, it is a hallmark of Chlamydia induced ReA, in which bacteria causing synovitis persist in low quantities making it hard to detect using PCR or culture techniques. However, Freise and colleagues later standardized PCR detection for C. trachomatis from synovial fluid (52). This may explain why various attempts to cultivate other pathogenic bacteria such as Yersinia or Salmonella from affected joints have yielded negative results (53, 54). Instead, Salmonella and Yersinia antigens have been identified in synovial fluid and/or tissue by immunohistochemistry. Antibodies against LPS from these microbes has been shown to be present in the synovial fluid many years after the infection which explains the strong IgA responses in people who have had a bout of ReA (53, 54). In a mouse model of Salmonella enteritis induced joint inflammation, increased levels of inflammatory cytokines IL-17 and TNF-α were observed in the mesenteric lymph node and synovium respectively. Neutralizing IL-17 in mice infected with S. enteritis, prevented synovitis and curbed the increase in TNF-α, suggesting the role of IL-17 in gut and joint inflammation (55). In addition to these pathogenic microbes, diarrheagenic Escherichia coli (DEC) has also shown to increase the incidence of musculoskeletal symptoms in individuals who contracted DEC associated diarrhea during their international travels. Of these patients, a small number of patients also met the criteria for ReA (56). Thus, while ReA may be highly associated with various bacterial enteric pathogens such as Campylobacter and Salmonella (57), its incidence after DEC infections is low. Recent study in the Yersinia enterocolitica murine model of ReA (TNFRp55-/- mice) has shown an important role for mesenteric dendritic cells. Intestinal dendritic cells migrated to the regional lymph nodes and contributed toward the immunopathogenesis of ReA (58). It is important to note that in most cases, infections with enteric pathogens do not result in development of ReA. In a comprehensive review by Ajene and colleagues (59), the ReA incidence for Salmonella, Campylobacter, and Shigella ranged from 0.1%–29%, 0%–16%, and 0%–12% respectively. Nevertheless, in studies in which enteric pathogens do lead to the development of ReA, the ability of a bacterium or bacterial antigen to reach the joint or gain access to particular cells such as the macrophages and evade the host defense might play an important role. In view of these different microbes associated with ReA, disease pathogenesis is thought to involve host-microbe interactions as evident with the presence of bacteria or their products in the joint, followed by local immune response.
Altered Schaedler’s Flora (ASF)
In 1965, Russell Schaedler developed a model microbial community to colonize germ-free animals to prevent the colonization of opportunistic pathogens (60), which was later modified to be more representative of gut microbiota and renamed as Altered Schaedler’s Flora (ASF) (61). 16s rRNA sequencing of ASF was performed to define the phylogeny of the ASF microbes (namely two members of Clostridium sp., Lactobacillus intestinalis, Lactobacillus murinus, Mucispirillum schaedleri, Eubacterium plexicaudatum, Pseudoflavonifractor sp., and Parabacteroides goldsteinii) (62, 63). Even though the ASF is a reductionist model microbial community, functional analysis of the ASF metagenome compared with the wild mice metagenome showed the functional similarity between gut microbiome of ASF and wild type mice (64). Early experiments colonizing germ-free HLA-B27 TG rats with ASF played a pivotal role in establishing the role of gut microbiota (especially Bacteroides) in the development of gut inflammation (35). Another mouse model for SpA is the SKG model, which has a mutation in the ZAP-70 (T cell receptor signaling gene). Upon injecting with curdlan, a component of bacterial and fungal cell walls, these mice develop SpA with uveitis, arthritis, and CD like ileitis (65, 66). Germ-free SKG mice recolonized with ASF exhibit increased arthritis incidence, although the severity of arthritis was attenuated in comparison with the specific pathogen free (SPF) mice (66). SPF mice had the highest incidence of Ileitis followed by ASF recolonized mice, while the germ-free mice did not have Ileitis (66). This suggests that dysregulation of mucosal-microbe interface is necessary for the development of ileitis, which fails to occur in germ-free SKG mice. In comparison, ileitis is mild in SKG mice colonized with ASF, highlighting the importance of a diverse microbial community in disease development. ASF studies have paved a way to understand the host-microbe interaction in a measurable way and have emphasized the role of commensal microbes acting as pathobionts in disease development.
Dialister
Dialister is a saccharolytic bacteria, belongs to family Vellionelaceae (67), that can convert succinate to propionate (68). Tito and coworkers (69) studied the relationship between the intestinal microbial composition of ileal and colon biopsies from inflamed and non-inflamed tissues, and observed SpA-associated microbial dysbiosis. Of note, they found that Dialister was increased in the inflamed tissue and positively correlates with the disease score, whereas the non-inflamed tissue had low frequency of Dialister. Another study examining post-infectious SpA reported an increase in the relative abundance of Dialister. Subjects who developed enthesitis also had increased abundance of Campylobacter and subjects with uveitis and radiographic sacroiliitis had increased abundance of Erwinia and unclassified Ruminococcaceae, respectively (70). Interestingly, some species of Dialister such as D. pneumosintes and D. invisus are shown to be pathogenic in orthodontic infections (71). Oral pathobionts can colonize the gut during inflammation, as there is increased availability of oxygen and lack of colonization resistance during inflammatory conditions (72). This in turn drives the Th1 response primarily by interferon gamma (IFNγ), and exacerbates gut inflammation (72). Another study focused on microbial dysbiosis associated with HLA alleles in healthy subjects with AS, and rheumatoid arthritis (RA), a chronic autoimmune disease defined by inflammation of the synovium and joint destruction (73). However, the authors did not observe HLA-B27-associated changes in Dialister in healthy subjects with either AS or RA associated alleles. While one study focused on the microbiota from biopsies collected from post-infectious SpA patients (69), the latter study focused on the fecal samples from HLA-B27 positive healthy individuals, many of whom will not develop disease. Stated differently, while it is possible for patient cohorts from different geographical locations to have distinct microbes driving disease, differences due to sampling location and disease severity also contributes to the association with distinct pathobionts.
Blautia
Another pathobiont associated with AS is Blautia. It is a member of the family Lachnospiraceae, which has been associated with gut inflammation in an experimental model of SpA (48, 74). A recent study by Zhang and colleagues (75) on the fecal samples from AS patients in a Chinese cohort has shown Blautia, Megamonas and Dorea associated with AS patients with a concomitant decrease in Lachnospira, Ruminococcus, and Clostridium_XlVb. In a study on patients with ReA, the authors reported an increase in enteropathogens such as Erwinia and Pseudomonas as well as several other microbes including Blautia, Coprococcus, Roseburia, and Collinsella (70). Many of these microbes (e.g., Blautia) were thought to be gut commensals, but new studies have shown them to be increased specifically with disease and thus a pathobiont. In a rat model of SpA, Blautia has been associated with HLA-B27 and SpA on the Lewis background, but not on the Fischer background (12). Hablot and colleagues (76) compared the microbial dysbiosis in mice with dextran sodium sulfate (DSS) induced colitis from mice with arthritis and colitis (induced with collagen and DSS). They found that mice with arthritis and colitis had increased relative abundance of Blautia, Gemellaceae, and Ruminococcus gnavus as compared with the colitis only group. Both Blautia and Ruminococcus are closely associated members of the family Lachnospiraceae, which are among the main producers of short chain fatty acids (SCFAs), and many taxa of this family are associated with various inflammatory diseases (77). While the role of Blautia in HLA-B27 associated SpAs was discussed in this section, the role of Ruminococcus gnavus as a pathobiont is discussed below.
Ruminococcus gnavus
Ruminococcus gnavus is a known pathobiont associated with SpA and associated IBD (51, 78). In a cohort of SpA patients and related as well as unrelated healthy controls, there was an increase in the relative abundance of R. gnavus, which correlated with the disease activity and with patients having a history of IBD. This change was not observed in their subjects with RA (78). Increases in R. gnavus have been associated with other inflammatory diseases including inflammatory bowel disease (79, 80), CD (81), and pouchitis in UC patients (82). Another instance of increased abundance of R. gnavus comes from patients with systemic lupus erythematosus (SLE), an autoimmune disease characterized by a hyperactive immune system which causes inflammation in many tissues, as well as an aberrant antibody response. Most SLE patients will develop either arthritis or synovitis sometime during their disease. A study on SLE patients showed an increased amounts of R. gnavus that correlated with disease activity, which was highest in the patients with lupus nephritis (83–85). One plausible mechanism for the contribution of R. gnavus in an inflammatory disease, CD, has been shown by Henke and colleagues (86). They found that R. gnavus secretes a complex gluco-rhamnan polysaccharide, which can induce the production of inflammatory cytokines like TNFα by activation of TLR4 on the dendritic cells and may explain the mechanism underlying the association between gut inflammation in CD and R. gnavus. TNFα is a potent inflammatory mediator in both SpA and IBD. This may explain why an increase in the relative abundance of R. gnavus in patients correlates with disease severity in both arthritis and gut inflammation. R. gnavus is also shown to provide colonization resistance to the gut microbial community by the production of bacteriocin ruminococcin A, which is active against pathogenic members of class Clostridia, especially Clostridium perfringens, Clostridium difficile, and other members phylogenetically related with R. gnavus (87), thus providing a competitive edge in gut colonization and inflammation. These studies show the harmful role of R. gnavus through different mechanisms. Association of R. gnavus and other pathobionts with both SpA and IBD may partially explain the overlap between the mechanisms underlying these complex inflammatory diseases in many patients.
Akkermansia muciniphila
Many studies on human and animal models of SpA have shown increased abundance of Akkermansia muciniphila, a mucin degrading bacteria found in human intestinal content (88). A study on pediatric SpA cohort, the microbiota from patients separated from healthy controls, and was divided subjects into two clusters each dominated by increased levels of either A. muciniphila or genus Bacteroides (89). Furthermore, to evaluate the pathogenicity of altered microbial composition in children with SpA, the group performed fecal microbial transplant to germ-free K/BxN mice. Transplanted mice displayed over-representation of Bacteroides and Akkermansia, and the latter positively correlated with disease activity. Addition of Akkermansia to ASF also increased the permissiveness to arthritis in these mice, when compared to mice that received ASF alone (90). Metagenomic analysis of fecal samples from patients with enthesitis related arthritis (ERA) showed decrease in the relative abundance of Faecalibacterium prausnitzii in both pediatric and adult SpA cohort, while the relative frequency of Bacteroides fragilis was increased in pediatric SpA cohort and decreased in adult SpA cohort (91). A. muciniphila has also been associated with disease in experimental SpA. In HLA-B27 TG Fischer rats, the relative abundance of A. muciniphila was increased along with elevated IgA coating of intestinal microbes (11). In a subsequent study, we compared the effect of host genetic background on microbial dysbiosis and found that the increased level of A. muciniphila was found in the Fischer HLA-B27 rats in comparison to Fischer wild-type controls; while in Lewis HLA-B27 TG rats there was an increase in the relative abundance of Prevotella when compared to Lewis wild type controls. Addition of A. muciniphila in germ-free and SPF IL10-/- mice is sufficient to exacerbate gut inflammation. IL10 is an anti-inflammatory cytokine and IL10-/- mice develop chronic colitis with marked increase in pathological type-I helper T cell response (92) in SPF but not in germ-free conditions. NLRP6 deficiency in these mice results in the enrichment of A. muciniphila, which then acts as a pathobiont in the development of colitis (93), and highlights the ability of NLRP6 in regulating colonization of colitogenic bacteria. Contrary to the role of A. muciniphila in inflammation, relative abundance of A. muciniphila has been shown to have an inverse correlation with obesity and metabolic diseases (94, 95). In fact, supplementation of A. muciniphila reversed high fat diet induced obesity in mice, which was mediated by altered adipocyte metabolism and improved gut barrier function (96). Akkermansia is a short chain fatty acid producer (97) and is thought to increase fatty acid oxidation in intestines and adipose tissues (98). Taken together, these distinct and opposite (pathobiont vs commensal) roles of A. muciniphila in various disorders highlight the tight regulation of the microbial abundance within the community and the effect of dysbiosis (increase or decrease in relative abundance) in health and disease.
Prevotella
Prevotella is another mucus degrading pathobiont that shows gut inflammation associated increased relative abundance in NLRP6-/- mice (99). Prevotella can reach to the crypt in the mucosal layer of the gastrointestinal tract, and are associated with SpA. Metagenomic analysis from the gut microbial DNA from a Chinese cohort of AS patients has revealed the abundance of Prevotella melaninogenica, Prevotella copri, and Prevotella spp. C561 and decreased abundance in Bacteroides spp (100). Interestingly, they also observed increased abundance of the Bifidobacterium genus, a commensal gut bacteria commonly found in probiotics. On the contrary, there was a decrease in the abundance of family Prevotellaceae in AS patients. This may be due to differences in the cohorts (Chinese vs caucasian), sampling location (fecal vs ileal biopsy), or it could also be due to the relationship between other members of the microbial community and/or the effect of host genetics (100). Scher and colleagues observed increased relative abundance of P. copri has also been reported in 16s microbiome sequencing of fecal samples from patients with new onset RA (101). They performed metagenomic sequencing of patient derived Prevotella strains and also compared the metagenomes between healthy controls and new-onset RA patients. Patients with new onset RA had decreased abundance of vitamin metabolism (i.e., biotin, pyroxidal, and folate) and pentose phosphate pathway which was consistent with Prevotella genomes lacking these functions. To determine whether Prevotella copri was sufficient to drive gut inflammation, they gavaged antibiotic treated mice with P- copri and after 2 weeks they found that P. copri had dominated the gut microbiota in these mice and exacerbated the susceptibility to DSS induced colitis (101). Consistent with these mouse studies, another study found enrichment of P. copri in patients during the pre-clinical phase of RA, before disease onset (102), which suggests a role of P. copri in intestinal dysbiosis and disease susceptibility. A recent cross-sectional study utilized data from a previous TwinsUK cohort, and used genotyping and microbiota data after excluding patients with RA and their twins. The authors found Prevotella spp in the gut microbiota of individuals who had RA associated genotype associated without the disease. again suggesting a role for host-microbe interactions prior to disease onset (103).
Mucispirillum schaedleri
Mucispirillum schaedleri, a Gram negative pathobiont, is a member of ASF, and is known to colonize the gut mucus layer in rodents (104). It was reported that in mice having a combined deficiency of two susceptibility genes for CD, namely nucleotide-binding oligomerization domain-containing protein 2 (NOD2) and NADPH oxidase, disease can be induced by M. schaedleri (105). NOD2 pays an important role in microbial regulation in the ileum (106), whereas NADPH oxidase is known to regulate the gut intestinal barrier through the production of reactive oxygen species (107). The authors demonstrated that in the absence of both NOD2 and phagocytic NADPH oxidase, there is accumulation of M. schaedleri in the gut lumen and mucosa associated with gut inflammation. Since, Mucispirillum is a bacterium found in rodents, it is an unlikely contributor to human diseases. However another bacterial taxa, Proteobacteria, which is closely related to Mucispirillum have been reported to have increased abundance in both patients with SpA (108) and CD (109). In contrast, another study reported that M. schaedleri can protect mice against Salmonella typhimurium virulence factors (110). This was supported by data from our study with the HLA-B27 TG rats, in which we found a decrease in the relative abundance of M. schaedleri as compared to the wild type rats (12), which may suggest a protective effect of the microbe. Since M. schaedleri is a mucolytic bacterium like A. muciniphila, we can hypothesize that it may be beneficial to the host at low relative abundance within the microbial community. However, increased abundance of M. schaedleri may compromise the spatial segregation by bringing luminal microbes close to the intestinal epithelial cells, thereby triggering an inflammatory response.
Adherent Invasive Escherichia coli
Invasive properties of various bacteria such as E. coli, S. typhimurium, and Citrobacter rodentium, may be critical for their ability to colonize the host. Of these, E coli has been associated with the induction of gut inflammation in CD (111). A study characterizing adherent invasive Escherichia coli (AIEC) found that these E. coli may harbor genes associated with bacterial adhesion and invasion and therefore regulate barrier permeability contributing to gut inflammation (112). Another study assessed the prevalence of AIEC associated with the intestinal mucosa of patients with CD, UC, and of healthy controls (113). They found that AIEC was found associated with the inflamed regions in the ileal mucosa in patients with CD, but was not observed in the ileal mucosa of patients with UC. In another study on the IgA coated bacterial fraction from patients with CD-associated SpA detected enrichment of E. coli in comparison to patients with CD alone. This IgA coated E. coli fraction displayed genotypic and phenotypic similarities to AIEC. Colonization of these AIEC in germ-free mice induced inflammatory Th17 mucosal immune response in comparison to colonization with non-AIEC strains of E. coli (114). This study identified immune reactive pathobionts that provide a link between mucosal immunity and systemic inflammation in CD-associated SpA and may guide future therapies.
Fungal Pathobionts
While microbial dysbiosis and pathobiont enrichment have been associated with SpA and associated CD for almost two decades, most of the work has been focused on the gut bacteria and some Archaea. However, fungal products such as β-glucan have been known to trigger SpA and ileal inflammation in SKG mice model (BALB/c ZAP-70W163C mutant) of SpA (115). Another study has shown the association of anti-Saccharomyces cerevisiae antibodies (ASCA) with intestinal inflammation in patients with Ax SpA and associated CD (116). In the last decade, with new and advanced approaches to characterize the fungal component of the gut microbiome (mycobiome), their role in disease pathogenesis is being studied more extensively. In a recent study in patients with SpA, treatment with IL-17 inhibitors was associated with a shift in bacterial and fungal taxa, specifically the bacteria from the family Clostridiales and the yeast Candida albicans (117). In these patients, the changes in the gut microbiome were associated with the perturbations in metabolic pathways and overexpression of IL-17/23 cytokines and the expansion of IL-25/17 producing tuft cells as well as type 2 innate lymphoid cells (ILC2), both of which are implicated in helminth immunity (117). A study investigated the mycobiome in IBD (CD and UC) and found increased intestinal fungal diversity in patients with CD in comparison with healthy controls (118). However, they did not observe any difference between the fungal species between the CD and UC groups. Another study focused on the relationship between Candida albicans and gut inflammation by using mice that lack Galectin 3 (Gal3-/-), an intestinal lectin that binds specifically to C. albicans, and showed that in Gal3-/- mice, DSS colitis was worse in comparison to wild type mice, with enhanced colonization by C. albicans (119). This revealed the role of C. albicans in augmenting DSS mediated colitis as well as the role of Gal-3 in preventing colonization by C. albicans. In another example of host fungal interaction, Iliev’s group (120) illustrated that the fungal community in the gut interacts with the immune system through innate immune receptor Dectin-1. In the DSS colitis model, mice deficient in Dec-1 had exacerbated colitis if challenged with Candida tropicalis whereas WT mice did not show an increase in colitis. In a study focused on both micro- and myco-biota in AS patients, the authors showed an increase in the levels of Ascomycota, where altered mycobiota was associated with the degree of radiographic damage (121). El Mouzan and colleagues (122, 123) investigated the gut fungi in treatment naive new onset CD in a pediatric Saudi Arabian cohort and found fungal dysbiosis associated with CD patients without the loss of fungal diversity between CD patients and healthy controls (HCs). They found that patients with CD had an increase in Psathyrellaceae, Cortinariaceae, Psathyrella, and Gymnopilus with a concomitant decrease in Monilinia. In a recent study, the authors found Malassezia restricta, a common skin commensal fungus, associated with the intestinal mucosa in CD patients (124). M. restricta was specifically associated with individuals carrying the IBD risk gene, CARD9-, a signaling adaptor protein with an antifungal role. The study showed that CARD9 variants present in these patients can induce the host immune cells to produce inflammatory cytokines against M. restricta. In a mouse model, M. restricta exacerbated colitis in germ-free as well as gnotobiotic mice. Taken together, these studies display the importance of fungi in HLA-B27-associated SpAs and highlight the importance of host-bacteria-fungal interactions in these diseases.
Fungi-Bacteria Functional Interaction
Recent studies have shown the role of inter-kingdom fungal-bacterial interactions contributing to various SpAs. One such study showed investigated the bacterial-fungal interkingdom networks in AS patients (121) and showed perturbed relations between gut bacteria and fungi as evident by decreased fungal to bacterial biodiversity ratios in these patients. Another study looking into the mycobiome of Japanese CD patients showed an increase in the abundance of C. albicans, Entyloma, and Trichosporon in the CD patients in comparison with the HCs. In contrast the HCs had increased abundance of Saccharomyces and Sarocladium in comparison to the CD patients. Microbial dysbiosis was also observed in CD patients as evident by decreased microbial diversity and increased abundance of Enterococcus in CD patients (125). Bacterial-fungi correlations showed positive correlation between Enterococcus and Malassezia. CD patients showed a positive correlation between Ruminococcus and Sarocladium and Ustilago (125). These associations are especially interesting as these bacteria and fungi have been separately associated with various spondyloarthropathies. In another study, it was shown that patients with CD are associated with increased levels of the fungus Candida tropicalis and two bacteria E. coli and Serratia marcescens. C. tropicalis positively correlated with E. coli and S. marcescens in these patients and was observed to associate closely in biofilms in comparison to other microbes (126). Studies are also investigating specific bacteria-fungi relationships in rodent models. Mice treated with DSS to induce colitis showed an increase in disease severity when supplemented with Candida albicans. On the other hand, colitis improved in these mice with the addition of Saccharomyces boulardii. Treatment with antibiotics affected the disease severity and the effects of fungi on colitis. While treatment with vancomycin that targets all Gram negative microbes protected mice from colitis, treatment with colistin to target Enterobacteraceae specifically retained the colitis phenotype (127). Disease was not affected by addition of either C. albicans or S. boulardii. Fungal-bacterial correlations were decreased severely in the colistin treated mice, suggesting that effect of fungi on colitis was due to its interaction with bacteria belonging to family Enterobacteraceae. Restoring the Enterobacteraceae in these mice restored the effect of both C. albicans and S. boulardii on the colitis (127). These studies highlight that microbial functions are considerably affected by various positive and negative trans-kingdom interactions between the members of the bacteria and fungal community.
Viral Pathobionts
The human gut virome is another emerging component of the gut microbiome, which is thought to impact human health either directly or via the modulation of the bacteriome through bacteriophages. However, the virome has been more of a dark matter with studies on a limited number of known viruses. A recent study by Norman and others (128) showed that the virome was altered in IBD with a significant expansion of Caudovirales bacteriophages. They compared the bacterial and Caudovirales bacteriophage communities and found distinct relationships in CD and UC patients. It was associated with increased richness but decreased viral diversity and these changes were concomitant to the changes in the bacterial communities suggesting a role of viral perturbations in IBD. Studies on the human virome have shown that viruses display bacteria like inter-individual variability and also respond dynamically to various environmental influences like diet (129, 130). In the healthy gut the viral core is made of virulent phages. However, in patients with CD, the virome shifts toward a temperate viral core and the changes in the viral community affect the bacterial community (131). Determining the virome in HLA-B27 mediated diseases may shed light on pathogenesis and may be crucial for the development of phage biomarkers.
So far, we have focused on specific bacterial, fungal, and viral pathobionts associated with HLA-B27-assocated SpA and other immune/inflammatory disorders that overlap clinically with SpA. However, determination of disease associated microbes may also depend upon other environmental factors such as geographical location, diet, genetic factors (other than HLA-B27), as well as technical factors like sampling location, related vs unrelated controls, methods of sequencing and data analysis. Spatial heterogeneity of microbial community profile through the gastrointestinal tract has shown to vary immensely. For example, the fecal microbiota provides a view of the microbial diversity at a given time point, and is used in majority of the microbiome studies, it neglects the mucosa-associated microbes (132). In addition, effects of related and unrelated healthy controls along with related healthy controls that cohabit need to be considered, as they can introduce variability. While efforts are being made to standardize the microbiome studies (133), attention toward the host, environmental and technical difference issues will be highly valuable while inferring results from multiple studies.
Pathobionts vs Dysbiosis
The gut microbial community is diverse with enormous inter-individual variability due to host genetics and other environmental factors, which may explain why different studies on SpA with diverse patient cohorts have reported expansion of distinct disease-associated microbes or pathobionts. In healthy individuals, pathobionts are present in relatively low abundance and increase during dysbiosis in disease susceptible individuals, contributing to pathogenesis. This could be an active increase in their relative abundance due to changes in their microenvironment, or they can increase as the colonization pressure from gut commensals is lost due to inflammation. Therefore, it is vital to study these pathobionts in the context of their host genetics and microbial community structure. IL-2 knockout mice have dysregulated T cell functions and develop chronic immune mediated colitis in SPF mice (134), however these mice like the IL-10-/- mice discussed earlier fail to develop colitis under germ-free conditions (135). These studies emphasize the interaction between the host genetics and gut microbiome in disease development. This is exemplified in HLA-B27 rats, which also fail to develop colitis in germ-free conditions. Introduction of Bacteroides vulgatus in germ-free HLA-B27 TG rats is sufficient to induce colitis, but its introduction in SPF raised athymic HLA-B27-TG rats fails to induce colitis (136). In contrast, addition of B. vulgatus is not sufficient to induce colitis in IL-10-/- mice, which suggested that resident enteric bacteria are necessary for immune activation and this development of spontaneous colitis in this model. In fact, presence of B. vulgatus protects the IL-2-/- mice from developing E. coli induced colitis, which underscores that different microbes and their interactions may dictate their ability to trigger disease (36, 137). In TNFΔARE mice model, biosynthesis of TNF is dysregulated, leading to the development of chronic inflammatory arthritis and CD like ileitis. Germ-free TNFΔARE mice did not develop ileitis, even when colonized by a pathobiont E. coli LF82. Development of CD like gut inflammation occurs only when these mice were colonized with cecal content from inflamed mice raised in SPF condition (138). Collectively, these studies performed with different genetic susceptibility models pinpoint the joint role of diverse microbiota and genetic susceptibility as a requirement for development of SpA or associated gut inflammation.
Metchnikoff (1908) observed that intestinal microbes are dependent on our dietary intake, and therefore it may be possible to modify the gut microbiota. He defined “dysbiosis” as ecological imbalance in the gut microbial community. While the concept of microbial dysbiosis was forgotten in the following decades with the focus on antibiotics, and lack of ability to culture and classify gut microbes, which are mostly obligate anaerobes and refractory to culture. Recent advances in culture free determination of microbial community members using next generation sequencing abilities, the microbial dysbiosis and its role in human health and disease has been a focus. These studies have classified many distinct features of microbial dysbiosis, such as reduced microbial diversity, pathobiont expansion and loss/alteration of microbial community structure (139). Another important feature of microbial dysbiosis is the perturbation in the metabolic function, which has been associated with many immune and inflammatory disorders including IBD and SpA (139–142).
In our study with the HLA-B27 transgene (12) on three different rat genetic backgrounds namely Lewis, Fischer and Dark agouti (DA), we have demonstrated that the gut microbiome is dependent on the host genetics and environment. Only two of the backgrounds (Lewis and Fischer) were disease susceptible, whereas the DA rats were resistant to HLA-B27-associated SpA. In disease susceptible Lewis and Fischer backgrounds, HLA-B27-associated gut microbial dysbiosis was dependent on the host background, while the immune dysregulation was independent of the host genetics and environmental effects, and showed considerable overlap of inflammatory mediators between HLA-B27 TG Lewis and HLA-B27 TG Fischer rats. Analyzing the predictive metabolome and host-microbe inter-omic analysis suggested that in comparison with their respective wild type controls, HLA-B27-associated with different pathobionts in Lewis (Prevotella) and Fischer (Akkermansia, members of family Lachnospiraceae). However, the microbial functional/metabolic pathways perturbed in both backgrounds were similar. In both these backgrounds, these diverse pathobionts are associated with common host genes for immune/inflammatory pathways (74). This led us to propose an ecological model of dysbiosis where perturbation of the microbial community structure and function contributes to disease pathogenesis, instead of a single microbe driving disease (12). These studies suggest that gut microbial functions are highly dependent on their community structure and the gut microenvironment (143).
Pathobionts Are Context Dependent
Many microbes deemed as commensals can act as pathobionts under certain circumstances. In such cases, the pathogenicity of these pathobionts is dependent on the host genetics as well as the composition of the gut microbiome. In antibiotic treated mice, adding commensal Bacteroides spp. can induce the development of colitis in IBD-susceptible background, but not in IBD-non-susceptible background (144). This may explain why adding back commensal microbes such as ASF to germ-free HLA-B27 TG rats is sufficient to drive colitis as well as arthritis (35). Conversely, pathobionts have been shown to exert beneficial effects in certain disorders. As mentioned earlier, supplementation of A. Muciniphila in mice and humans has shown to improve various metabolic parameters in obesity (145, 146). These results suggest that a microbe can act as a pathobiont depending on the context of host genetics and their position in the microbial community structure (Figure 3). We reported that presence of segmented filamentous bacteria (SFB) in both Lewis and Fischer genetic backgrounds correlates with disease in the presence of HLA-B27 but not in wild type controls (12). SFB adheres to the epithelial cells in terminal ileum in rodents at the time of weaning and induces the development of Th17 cells (147). SFB is absent on the DA rat background and these animals are resistant to gut inflammation and arthritis in the presence of HLA-B27. In contrast to these studies, SFB was shown to protect against rotavirus infection and diarrheal disease (148). In another study, SFB was able to protect from Citrobacter rodentium induced colitis (147). This may suggest that the microbial community is complex, and the results from the mono-colonization studies, while important to determine mechanistic pathways, may not be sufficient to recapitulate fully functional microbial communities.
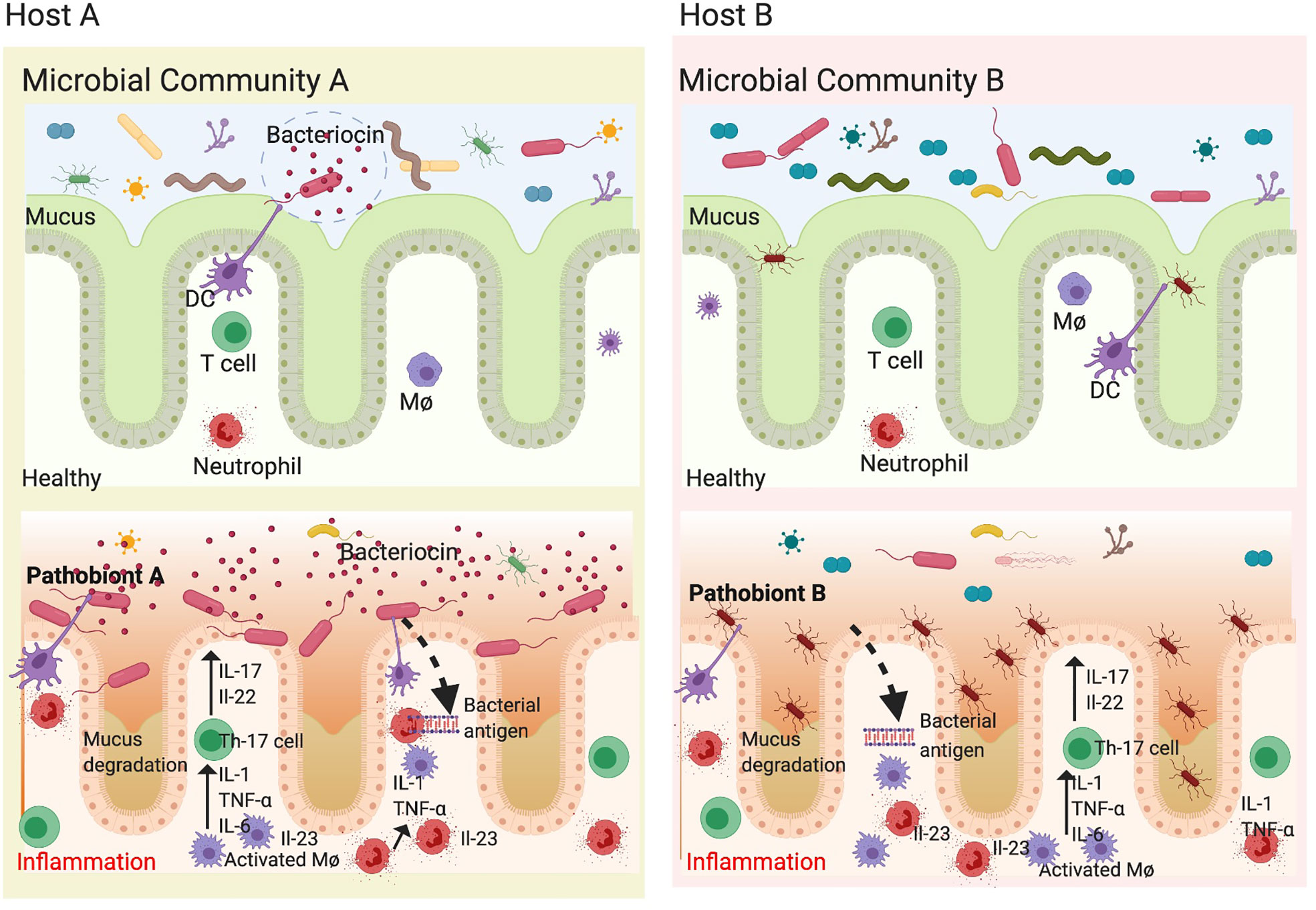
Figure 3 Pathobionts depend on host genetics and gut microbial community. The gut microbiota is highly diverse and varies between healthy individuals depending on host genetics, diet and environment. Each individual microbial community consists mostly of commensals which provide colonization resistance to opportunistic pathobionts. In genetically susceptible individuals (HLA-B27), there is microbial dysbiosis concomitant to loss of epithelial barrier resulting in an inflammatory microenvironment which further increases the loss in commensal microbes. This presents a unique opportunity for pathobionts to thrive and exacerbate inflammation. Since different individuals have different microbial community structure and different pathobionts depending on their genetics and environment, this may explain why we observe different pathobionts associated with various spondyloarthropathies. In microbial community A, only pathobiont A is present and when the conditions change (a trigger). In addition, certain pathobionts can make bactericidal compounds known as bacteriocins to avoids colonization resistance. When the pathobiont A is in bloom, it increases and exacerbates inflammation by degrading mucus and disrupting epithelial barrier. However, in another microbial community on a distinct host genetic background and in the presence of pathobiont B, which is mucous associated, an increased relative abundance can be associated with disease as most microbes are unable to cross the mucous barrier. In both cases, the dendritic cells (DCs) and Macrophages (Mφ) release inflammatory mediators such as IL-23, TNF-α, and IL-6, which activate T helper 17 cells to make IL-17, IL-22 causing inflammation and disrupting epithelial barrier thereby perpetuating the inflammatory cycle. Figure created with Biorender.com.
Association vs Causation
The association between the host species and their symbiotic microbes is a result of millions of years of coevolution, which has resulted in a homeostatic balance between the gut microbial community in host health and disease (149, 150). While imbalance in the symbiotic microbial communities inhabiting our body has been linked to various SpAs (151), it is not clear whether microbial dysbiosis is the cause or the effect of these disorders. Experimental models of various SpAs can help elucidate the causal microbes and microbial pathways, which can be confirmed in patients. One such example is Porphyromonas gingivalis, a periodontal bacterium, which has been shown to colonize synovial joints and exacerbate collagen induced arthritis (152). This suggests that P. gingivalis plays a mechanistic role in arthritis due to its translocation to the joints.
A complication in determining causal microbial mechanisms is that host-microbiome interactions are complex and have a multitude of variables affecting them. This makes it challenging to determine the pathobiont or other microbial species responsible for the disease phenotype (153). While mono-colonization of germ-free animals is a simplistic tool for determining causation, it lacks complex inter-microbial interactions and may not recapitulate the complexities of a stable microbial community. Thus, instead of colonization with single commensal and/or pathobiont, development of complex model microbiomes or synthetic microbial communities (154) and better culture techniques to culture/characterize refractory microbes (155) may help determine disease mechanisms. Development of better animal models and/or methods to colonize selective microbes in a complex microbial community will enable us to directly approach the mechanisms and address causality (156).
Disease Mechanisms
Determination of the causal relationships and the underlying mechanisms with which these microbes interact with each other and their host is vital to develop therapeutic targets. Deciphering the mechanisms of microbial function in health and disease is crucial to ascertain cause and result relationships. Pathobionts are thought to exert their effects on host physiology through mucin degradation, disruption of epithelial barrier function as well as the loss of colonization resistance by commensals, therefore making them foremost targets to investigate disease mechanisms.
Mucin Degradation
The mucus layer provides the first layer of physical barrier by limiting the microbial contact with the host tissue. Mucin degradation by microbial metalloproteinases has been suggested to contribute to IBD pathogenesis (157). Pathobionts such as Akkermansia and Prevotella, which are implicated in HLA-B27-associated SpAs, are known to have mucin degrading capabilities albeit through different mechanisms. For example, Prevotella may contribute to inflammation by encoding enzymes (superoxide reductase and a phosphoadenosine phosphosulfate reductase), which enable Prevotella to resist host reactive oxygen species and outcompete microbes essential for mucosal homeostasis such as Bacteroides spp (68, 101); Akkermansia can exacerbate inflammation by degrading the mucus layer over epithelial cells, thereby weakening the epithelial barrier (158). Mucolytic bacteria could contribute to disruption of intestinal barrier and joint inflammation. One such bacteria, R. gnavus is known to express ß-glucuronidase, that can convert bile acids into inflammatory secondary bile acids (deoxycholic and lithocholic acids), associated with intestinal inflammation (78, 159).
Mucosal Adherence and Barrier Disruption
Almost 60 percent of patients with HLA-B27-associated SpAs have microscopic gut lesions, with one third having overt gut inflammation (160). First degree relatives of SpA and IBD patients also show signs of subclinical gut inflammation and impaired gut epithelial barrier (161, 162). While the underlying mechanism is not fully understood, animal models of SpAs and IBD have highlighted the importance of gut mucosa for host-microbe interactions. Many pathobionts associated with HLA-B27 such as Akkermansia, Prevotella and Mucispirillum have the ability to adhere and degrade the mucus layer (11, 12, 101). When epithelial barrier is disrupted, gut commensal microbes such as Bacteroides vulgatus are also sufficient to cause and perpetuate IBD in immunocompromized mice (163). During dysbiosis, increase in the pathobionts can degrade the mucus layer and may activate local inflammatory response, which could lead to disruption of the epithelial barrier. While we do not know if pathobionts cause barrier disruption, or if barrier disruption triggers pathobiont bloom, both events are related and critical to disease pathogenesis.
Loss of Colonization Resistance
Of the many symbiotic functions performed by gut commensals, formation of stable microbial communities is perhaps the most important function, since it provides colonization resistance to infections and pathobiont expansion. The gut microbial community is dynamic with constant struggle for niche and resources between the gut commensals and the opportunistic pathobionts for energy, resources and niche. In a healthy individual, the gut commensals can either kill the pathobionts by production of bacteriocins, outcompete them for resources or activate the immune response to produce antimicrobial peptides (164). One such mechanism is the fucosylation of epithelial cells that promotes colonization by commensals and resistance to pathogens (165). Under eubiosis, the host immune system has the ability to distinguish gut commensals from pathobionts, although the mechanisms are not clear. In a recent study, the authors demonstrate that pathobionts such as Citrobacter rodentium can trigger inflammatory Th17 cells, while gut commensals like SFB trigger tissue-resident homeostatic Th17 cells (166). These tissue resident Th17 cells do not make inflammatory cytokines or participate in inflammatory reactions and have a slow metabolism. On the contrary, Th17 cells in response to C. rodentium have an inflammatory effector potential (166). During dysbiosis, mucosa associated pathobiont/s can expand at the expense of gut commensal microbes, degrade mucus and activate host inflammatory response (11).
Therapeutic Implications
Gut microbe/s or microbial metabolites may provide novel treatment opportunities in HLA-B27-associated SpAs. These approaches aim to answer the basic question- what parameters affect the reorganization of a stable microbial community after dysbiosis and/or inflammatory insult? Here we focus on the role of pre and probiotics, short chain fatty acid, diet and fecal microbiota transplant in alleviation or amelioration of the disease.
Pre- and Pro-Biotics
Prebiotics and probiotics can contribute to maintaining healthy gut community and therefore play and important role in the overall health of the gastrointestinal tract (167). Prebiotics are plant-based fiber, which may enhance the activity of beneficial gut bacteria, thereby promoting host health (168). On the other hand, probiotics consists of live bacterial strains, which upon ingestion in adequate amounts may confer health benefit to the host (169). Studies on HLA-B27 TG rats treated with prebiotic compounds inulin and oligofructose, demonstrated reduced colitis severity, associated with increase in the relative abundance of Lactobacillus and Bifidobacterium species. Prebiotic treatment significantly decreased inflammatory cytokines such as IL-1β and increased the levels of TGF-β, an immunomodulatory cytokine in the cecal tissue of these rats (170). In another study by the same group, the prebiotic combination of inulin and oligofructose was also effective in partially preventing colitis. While the HLA-B27 TG rats showed an altered microbial community, concomitant with an increase in Bifidobacterium, they were unable to observe any changes to the luminal short chain fatty acid concentrations (171). A later study by another group tried to dissect the effects of prebiotic treatment by using either inulin or fructo-oligosaccharide in HLA-B27 TG. All of the HLA-B27 TG rats which were fed fructo-oligosaccharide showed significant reduction in colitis, whereas only half of the HLA-B27 TG rats fed inulin showed improvement in colitis. While both groups were associated with decrease in the Clostridium cluster XI, rats fed fructo-oligosaccharides showed increase in Bifidobacterium, and inulin fed HLA-B27 TG rats showed an increase in Bacteroides, Prevotella, Porphyromonas group (172). Another group also measured the impact of a probiotic and prebiotic combination (Lactobacilli, Bifidobacteria, and inulin) on the severity of colitis and microbial community in HLA-B27 TG rats. Colitis was attenuated in HLA-B27 rats which received the probiotic, and they showed increase in microbial diversity, specifically an increase in the Bifidobacterium animalis (173). These studies show the promise of prebiotic and probiotic supplements as therapy for gut inflammation associated with various SpAs by renewing and restoring host gut microbial communities.
Short Chain Fatty Acids
Another important contribution of gut microbiota is the production of microbial metabolites such as butyrate, propionate, and acetate; collectively known as short chain fatty acids (SCFA). These are the primary products of non-digestible carbohydrates/prebiotics, and their oxidation provides a major source of energy for the colonocytes (174). Microbial dysbiosis is accompanied by perturbations in the microbial metabolic function including changes in production/oxidation of short chain fatty acids (SCFA) and trimethylamine N-oxide (TMAO), which play an important role in modulation of host physiology [reviewed in (175)]. Asquith and colleagues showed that HLA-B27 expression alters the host and microbial metabolic profile with an increase in the levels of histidine, tyrosine, spermidine, N-acetylmuramate and glycerate in HLA-B27 TG rats (140). When supplemented with propionate (a SCFA), HLA-B27 TG demonstrated attenuation in the inflammatory disease. In another study, administration of propionate was shown to attenuate the severity of uveitis in an inducible model of experimental autoimmune uveitis (176). Propionate and other SCFAs such as butyrate can activate G protein-coupled receptors, GPR41 and GPR43 (177) (124). Mice deficient in GRP43 show exacerbation of inflammation in colitis, arthritis and asthma. The authors demonstrated that resolution of inflammatory response was dependent on the activation of GPR43 by SCFA (178). SCFAs such as butyrate have shown to effect the host innate immune function (179). They showed a reduction in inflammatory cytokines produced by macrophages in vitro when treated with butyrate, as well as in macrophages isolated from mice whose drinking water was supplemented with butyrate. Taken together, SCFAs play an important role in mucosal homeostasis by not only fueling the colonocytes, but also by suppressing the innate immune cells from mounting an inflammatory response. These diverse roles highlight the importance SCFAs as a therapeutic in various inflammatory (SpA, IBD) as well as metabolic (type I diabetes) diseases.
Diet
Diet plays a major role in microbial community maintenance since it is an important source of small molecules, which are converted to various metabolic products by gut bacteria. High fat westernized diet is thought to be associated with various inflammatory disorders, whereas high fiber diet is associated with amelioration of inflammation (180). In a study by Rodrigues-Cabezas and colleagues, amelioration of colitis was reported in HLA-B27 TG rats given fiber enriched diet (181). This was associated with increased production of SCFA (butyrate and propionate), which in turn acted synergistically to inhibit pro-inflammatory mediators (181). Supplementing the diet with butyrate in mice has been shown to mediate the homeostasis of regulatory T cells (182, 183). In a mouse model, butyrate supplementation increased serotonin derived activation of the aryl hydrocarbon receptor (AhR). AhR is a transcription factor involved in the sensing of environmental signals like the redox potential (184); and recognized as the mediator for various ligands from diet, commensal microbes and host metabolites [Reviewed in (185)]. While AhR has recently been discovered as a B cell transcription factor and induces the development of regulatory B cells (186), its effect on immune modulation has been shown in IBD. Indole derivatives from cruciferous vegetables activate the AhR, and plays an important role in the maintenance of innate lymphoid and intraepithelial lymphocytes (187, 188). AhR receptors are expressed by peripherally derived regulatory T cells in the gut, and their expression has been shown to play a key role in the gut homing and anti-inflammatory functions of gut regulatory T cells (189). They are known to be involved in the maintenance of epithelial barrier and dampen various inflammatory conditions (190, 191). A study by Maslowski et al. (178), showed that decreased intake of complex plant polysaccharide fiber perturbs the microbial community leading to decreased production of SCFAs. Thus, diet has a role in maintenance of gut microbial community, which promotes gut health by increasing host beneficial metabolites as well as by maintaining community resistance by gut commensals to pathogenic microbes.
Fecal Microbiota Transplant (FMT)
The role of microbiota in disease pathogenesis and evidence from FMT for Clostridium difficile infection makes FMT a promising treatment for HLA-B27 associated spondyloarthropathies. While FMTs are currently not available for spondyloarthropathies, FMT trials are underway in patients with RA (ClinicalTrials.gov identifier: NCT03944096), who are refractory to methotrexate treatment, as well as in a European cohort for psoriatic arthritis (ClinicalTrials.gov identifier: NCT03058900). Trials are also underway for the FMT treatment of AS patients (ASGUT- ClinicalTrials.gov Identifier: NCT03726645). In a recent report by (192), FMT treatment for C. difficile infection resulted in a decrease in disease activity for PsA. Recently, FMT has been proven to be effective for the induction of clinical remission associated with endoscopic improvement in active ulcerative colitis concomitant with persistent increase in microbial diversity (193). A major concern with FMT trials is development of a standardized fecal sample, and the concerns for long term colonization of a foreign microbial community in a different host microenvironment. While patients with C. difficile pseudomembranous colitis offer very little colonization resistance to FMT, in other inflammatory diseases the resident microbial community can prevent the colonization of the microbial community from FMT. To standardize the gut microbiota for FMT, researchers have developed methods to develop a synthetic microbial community which mimics the fecal microbiota (154). This synthetic microbial FMT is under trials for C. difficile infection (Clinical trial registration number: CinicalTrials.gov NCT01372943). Together, these studies show a remarkable role of FMT in repopulating the gut microbiota and its role in disease amelioration.
Conclusions and Future Directions
HLA-B27 has been recognized to associate with various SpAs, especially AS for almost 50 years. With the advent of culture free sequencing techniques, the role of microbiome and microbial dysbiosis in disease pathogenesis is now well accepted. However, we are beginning to appreciate the complexity of gut microbial communities which consist of bacteria, archaea, viruses, and fungi. If we include the inter-kingdom interactions as well the host-microbiota relationships as factors affecting the role of individual microbes, these interactions in a stable microbial community become very complex. However, spatial differences in microbial community structure along with difference due to host, environmental, and technological factors should be considered before we cement the role of certain microbes and their functions in HLA-B27 associated SpAs.
The emerging picture suggests an important role of pathobionts in contributing toward HLA-B27 associated SpAs. Mechanistic studies on these pathobionts in germ-free and gnotobiotic rodent models have provided fundamental insights into the role of microbes in disease pathogenesis. While this review has focused on bacteria and fungi in SpA and associated disorders, studies of viruses especially bacteriophages in the gut mucosal environment could help explain whether viruses play a role in development of SpA in genetically susceptible hosts. However, the gut microbiome is a complex ecosystem of polymicrobial communities and therefore understanding the functional/metabolic implications of these microbial perturbations in context of an established gut microbial community is of immense value in determining host-microbe causal relations in SpA. The mechanistic effect of pathobiont expansion and dysbiosis in the context of host genetics and environment, will provide opportunities to develop novel therapeutic targets for disease alleviation/amelioration in HLA-B27 associated SpAs.
Author Contributions
TG and JR jointly created the review outline. TG wrote the review and JR provided editing. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by NIH Grant RO1 EY029266 and EY026572. JTR also receives support from the Grandmaison Fund for Autoimmunity Research, the Stan and Madelle Rosenfeld Family Trust, the William and Mary Bauman Family Foundation, Research to Prevent Blindness, and NIH Grant, P30 EY010572. TG is a Jane Bruckel early career investigator supported by Spondylitis Association of America.
Conflict of Interest
JTR is an unpaid advisor to Viome.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. D’Incà R, Podswiadek M, Ferronato A, Punzi L, Salvagnini M, Sturniolo GC. Articular manifestations in inflammatory bowel disease patients: a prospective study. Dig Liver Dis (2009) 41(8):565–9. doi: 10.1016/j.dld.2009.01.013
2. Kumar A, Lukin D, Battat R, Schwartzman M, Mandl LA, Scherl E, et al. Defining the phenotype, pathogenesis and treatment of Crohn’s disease associated spondyloarthritis. J Gastroenterol (2020) 55(7):667–78. doi: 10.1007/s00535-020-01692-w
3. Gracey E, Dumas E, Yerushalmi M, Qaiyum Z, Inman RD, Elewaut D. The ties that bind: skin, gut and spondyloarthritis. Curr Opin Rheumatol (2019) 31(1):62–9. doi: 10.1097/bor.0000000000000569
4. Karreman MC, Luime JJ, Hazes JMW, Weel A. The Prevalence and Incidence of Axial and Peripheral Spondyloarthritis in Inflammatory Bowel Disease: A Systematic Review and Meta-analysis. J Crohns Colitis (2017) 11(5):631–42. doi: 10.1093/ecco-jcc/jjw199
5. Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med (2012) 4(1):6–. doi: 10.1186/gm305
6. Smith JA, Colbert RA. Review: The interleukin-23/interleukin-17 axis in spondyloarthritis pathogenesis: Th17 and beyond. Arthritis Rheumatol (Hoboken NJ) (2014) 66(2):231–41. doi: 10.1002/art.38291
7. Gálvez J. Role of Th17 Cells in the Pathogenesis of Human IBD. ISRN Inflammation (2014) 2014:928461–. doi: 10.1155/2014/928461
8. Kadi A, Izac B, Said-Nahal R, Leboime A, Van Praet L, de Vlam K, et al. Investigating the genetic association between ERAP1 and spondyloarthritis. Ann Rheum Dis (2013) 72(4):608–13. doi: 10.1136/annrheumdis-2012-201783
9. Küçükşahin O, Ateş A, Türkçapar N, Törüner M, Turgay M, Duman T, et al. Association between single nucleotide polymorphisms in prospective genes and susceptibility to ankylosing spondylitis and inflammatory bowel disease in a single centre in Turkey. Turk J Gastroenterol (2016) 27(4):317–24. doi: 10.5152/tjg.2016.15466
10. Castro-Santos P, Moro-García MA, Marcos-Fernández R, Alonso-Arias R, Díaz-Peña R. ERAP1 and HLA-C interaction in inflammatory bowel disease in the Spanish population. Innate Immun (2017) 23(5):476–81. doi: 10.1177/1753425917716527
11. Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT. Perturbed Mucosal Immunity and Dysbiosis Accompany Clinical Disease in a Rat Model of Spondyloarthritis. Arthritis Rheumatol (2016) 68(9):2151–62. doi: 10.1002/art.39681
12. Gill T, Asquith M, Brooks SR, Rosenbaum JT, Colbert RA. Effects of HLA-B27 on Gut Microbiota in Experimental Spondyloarthritis Implicate an Ecological Model of Dysbiosis. Arthritis Rheumatol (2018) 70(4):555–65. doi: 10.1002/art.40405
13. Taurog JD, Chhabra A, Colbert RA. Ankylosing Spondylitis and Axial Spondyloarthritis. N Engl J Med (2016) 374(26):2563–74. doi: 10.1056/NEJMra1406182
14. Rosenbaum JT, Asquith M. The microbiome and HLA-B27-associated acute anterior uveitis. Nat Rev Rheumatol (2018) 14(12):704–13. doi: 10.1038/s41584-018-0097-2
15. Abdollahi-Roodsaz S, Abramson SB, Scher JU. The metabolic role of the gut microbiota in health and rheumatic disease: mechanisms and interventions. Nat Rev Rheumatol (2016) 12(8):446–55. doi: 10.1038/nrrheum.2016.68
16. Scher JU. The Microbiome in Psoriasis and Psoriatic Arthritis: Joints. J Rheumatol Suppl (2018) 94:32–5. doi: 10.3899/jrheum.180134
17. Brewerton DA, Hart FD, Nicholls A, Caffrey M, James DC, Sturrock RD. Ankylosing spondylitis and HL-A 27. Lancet (1973) 1(7809):904–7. doi: 10.1016/s0140-6736(73)91360-3
18. Brewerton DA, Nichols A, Caffrey M, Walters D, James DC. Acute anterior uveitis and HLA 27. Lancet (1973) 2:994s. doi: 10.1016/S0140-6736(73)91090-8
19. Brewerton DA, Nicholls A, Oates JK, Caffrey M, Walters D, James DCO. Reiter’s disease and HL-A 27. Lancet (1973) 302(7836):996–8. doi: 10.1016/S0140-6736(73)91091-X
20. Feng B-J, Sun L-D, Soltani-Arabshahi R, Bowcock AM, Nair RP, Stuart P, et al. Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PloS Genet (2009) 5(8):e1000606. doi: 10.1371/journal.pgen.1000606
21. The Mhc Sequencing C. Complete sequence and gene map of a human major histocompatibility complex. Nature (1999) 401:921. doi: 10.1038/44853
22. Benacerraf B. Role of MHC gene products in immune regulation. Science (1981) 212(4500):1229–38. doi: 10.1126/science.6165083
23. May E, Dorris ML, Satumtira N, Iqbal I, Rehman MI, Lightfoot E, et al. CD8 alpha beta T cells are not essential to the pathogenesis of arthritis or colitis in HLA-B27 transgenic rats. J Immunol (2003) 170(2):1099–105. doi: 10.4049/jimmunol.170.2.1099
24. Benjamin R, Parham P. Guilt by association: HLA-B27 and ankylosing spondylitis. Immunol Today (1990) 11(4):137–42. doi: 10.1016/0167-5699(90)90051-a
25. Mear JP, Schreiber KL, Münz C, Zhu X, Stevanović S, Rammensee HG, et al. Misfolding of HLA-B27 as a result of its B pocket suggests a novel mechanism for its role in susceptibility to spondyloarthropathies. J Immunol (1999) 163(12):6665–70.
26. Colbert RA. HLA-B27 misfolding: a solution to the spondyloarthropathy conundrum? Mol Med Today (2000) 6(6):224–30. doi: 10.1016/s1357-4310(00)01699-3
27. Dangoria NS, DeLay ML, Kingsbury DJ, Mear JP, Uchanska-Ziegler B, Ziegler A, et al. HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. J Biol Chem (2002) 277(26):23459–68. doi: 10.1074/jbc.M110336200
28. Colbert RA, Tran TM, Layh-Schmitt G. HLA-B27 misfolding and ankylosing spondylitis. Mol Immunol (2014) 57(1):44–51. doi: 10.1016/j.molimm.2013.07.013
29. Allen RL, O’Callaghan CA, McMichael AJ, Bowness P. Cutting edge: HLA-B27 can form a novel beta 2-microglobulin-free heavy chain homodimer structure. J Immunol (1999) 162(9):5045–8.
30. Kollnberger S, Bird L, Sun M-Y, Retiere C, Braud VM, McMichael A, et al. Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheum (2002) 46(11):2972–82. doi: 10.1002/art.10605
31. Colbert RA, Navid F, Gill T. The role of HLA-B*27 in spondyloarthritis | Elsevier Enhanced Reader. Best Pract Res Clin Rheumatol 31(6):797–815. doi: 10.1016/j.berh.2018.07.012
32. Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol (2019) 17(8):497–511. doi: 10.1038/s41579-019-0213-6
33. Laukens D, Peeters H, Marichal D, Vander Cruyssen B, Mielants H, Elewaut D, et al. CARD15 gene polymorphisms in patients with spondyloarthropathies identify a specific phenotype previously related to Crohn’s disease. Ann Rheum Dis (2005) 64(6):930–5. doi: 10.1136/ard.2004.028837
34. Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernández-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med (1994) 180(6):2359–64. doi: 10.1084/jem.180.6.2359
35. Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE Jr., Balish E, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest (1996) 98(4):945–53. doi: 10.1172/JCI118878
36. Rath HC, Wilson KH, Sartor RB. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect Immun (1999) 67(6):2969–74. doi: 10.1128/IAI.67.6.2969-2974.1999
37. Rosenbaum JT, Davey MP. Time for a gut check: evidence for the hypothesis that HLA-B27 predisposes to ankylosing spondylitis by altering the microbiome. Arthritis Rheumatol (2011) 63(11):3195–8. doi: 10.1002/art.30558
38. Costello M-E, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Brief Report: Intestinal Dysbiosis in Ankylosing Spondylitis. Arthritis Rheumatol (2015) 67(3):686–91. doi: 10.1002/art.38967
39. Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol (2015) 67(1):128–39. doi: 10.1002/art.38892
40. Lin P, Bach M, Asquith M, Lee AY, Akileswaran L, Stauffer P, et al. HLA-B27 and human β2-microglobulin affect the gut microbiota of transgenic rats. PloS One (2014) 9(8):e105684. doi: 10.1371/journal.pone.0105684
41. Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe (2010) 7(4):265–76. doi: 10.1016/j.chom.2010.03.004
42. Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun (1997) 65(8):3126–31. doi: 10.1128/IAI.65.8.3126-3131.1997
43. Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun (1998) 66(11):5157–66. doi: 10.1128/IAI.66.11.5157-5166.1998
44. Colmegna I, Cuchacovich R, Espinoza LR. HLA-B27-associated reactive arthritis: pathogenetic and clinical considerations. Clin Microbiol Rev (2004) 17(2):348–69. doi: 10.1128/cmr.17.2.348-369.2004
45. van der Linden S, van der Heijde D. Clinical aspects, outcome assessment, and management of ankylosing spondylitis and postenteric reactive arthritis. Curr Opin Rheumatol (2000) 12(4):263–8. doi: 10.1097/00002281-200007000-00005
46. Gordon FB, Quan AL, Steinman TI, Philip RN. Chlamydial isolates from Reiter’s syndrome. Br J Vener Dis (1973) 49(4):376–80. doi: 10.1136/sti.49.4.376
47. Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci USA (2010) 107(41):17698–703. doi: 10.1073/pnas.1011736107
48. Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol (2010) 11(5):411–8. doi: 10.1038/ni.1857
49. Antoniou AN, Lenart I, Kriston-Vizi J, Iwawaki T, Turmaine M, McHugh K, et al. Salmonella exploits HLA-B27 and host unfolded protein responses to promote intracellular replication. Ann Rheum Dis (2019) 78(1):74–82. doi: 10.1136/annrheumdis-2018-213532
50. Mendez JM, Kolora LD, Lemon JS, Dupree SL, Keestra-Gounder AM. Activation of the Endoplasmic Reticulum Stress Response Impacts the NOD1 Signaling Pathway. Infect Immun (2019) 87(8):1–15. doi: 10.1128/iai.00826-18
51. Kuipers JG, Jürgens-Saathoff B, Bialowons A, Wollenhaupt J, Köhler L, Zeidler H. Detection of Chlamydia trachomatis in peripheral blood leukocytes of reactive arthritis patients by polymerase chain reaction. Arthritis Rheumatol (1998) 41(10):1894–5. doi: 10.1002/1529-0131(199810)41:10<1894::AID-ART24>3.0.CO;2-E
52. Freise J, Bernau I, Meier S, Zeidler H, Kuipers JG. Detection of Chlamydia trachomatis-DNA in synovial fluid: evaluation of the sensitivity of different DNA extraction methods and amplification systems. Arthritis Res Ther (2009) 11(6):R175. doi: 10.1186/ar2864
53. Granfors K, Jalkanen S, Lindberg AA, Mäki-Ikola O, von Essen R, Lahesmaa-Rantala R, et al. Salmonella lipopolysaccharide in synovial cells from patients with reactive arthritis. Lancet (1990) 335(8691):685–8. doi: 10.1016/0140-6736(90)90804-e
54. Granfors K, Ogasawara M, Hill JL, Lahesmaa-Rantala R, Toivanen A, Yu DT. Analysis of IgA antibodies to lipopolysaccharide in Yersinia-triggered reactive arthritis. J Infect Dis (1989) 159(6):1142–7. doi: 10.1093/infdis/159.6.1142
55. Noto Llana M, Sarnacki SH, Vázquez MV, Gartner AS, Giacomodonato MN, Cerquetti MC. Salmonella enterica induces joint inflammation and expression of interleukin-17 in draining lymph nodes early after onset of enterocolitis in mice. Infect Immun (2012) 80(6):2231–9. doi: 10.1128/iai.00324-12
56. Tuompo R, Lääveri T, Hannu T, Pakkanen SH, Kirveskari J, Leirisalo-Repo M, et al. Reactive arthritis and other musculoskeletal symptoms associated with acquisition of diarrhoeagenic Escherichia coli (DEC). Ann Rheum Dis (2020) 79(5):605–11. doi: 10.1136/annrheumdis-2019-216736
57. Townes JM, Deodhar AA, Laine ES, Smith K, Krug HE, Barkhuizen A, et al. Reactive arthritis following culture-confirmed infections with bacterial enteric pathogens in Minnesota and Oregon: a population-based study. Ann Rheum Dis (2008) 67(12):1689–96. doi: 10.1136/ard.2007.083451
58. Silva JE, Mayordomo AC, Dave MN, Aguilera Merlo C, Eliçabe RJ, Di Genaro MS. Dendritic Cells of Mesenteric and Regional Lymph Nodes Contribute to Yersinia enterocolitica O:3-Induced Reactive Arthritis in TNFRp55-/- Mice. J Immunol (2020) 204(7):1859–68. doi: 10.4049/jimmunol.1901137
59. Ajene AN, Fischer Walker CL, Black RE. Enteric pathogens and reactive arthritis: a systematic review of Campylobacter, salmonella and Shigella-associated reactive arthritis. J Health Popul Nutr (2013) 31(3):299–307. doi: 10.3329/jhpn.v31i3.16515
60. Schaedler RW, Dubos R, Costello R. THE DEVELOPMENT OF THE BACTERIAL FLORA IN THE GASTROINTESTINAL TRACT OF MICE. J Exp Med (1965) 122:59–66. doi: 10.1084/jem.122.1.59
61. Trexler PC, Orcutt RP. Development of gnotobiotics and contamination control in laboratory animal science. In: Macpherson CW, Mattingly SF, editors. 50 Years of Laboratory Animal Science. Memphis: American Association for Laboratory Animal Science (1999). p. 121–8.
62. Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol (1999) 65(8):3287–92. doi: 10.1128/AEM.65.8.3287-3292.1999
63. Wannemuehler MJ, Overstreet A-M, Ward DV, Phillips GJ. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. (2014) 2(2):1–2. doi: 10.1128/genomeA.00287-14
64. Biggs MB, Medlock GL, Moutinho TJ, Lees HJ, Swann JR, Kolling GL, et al. Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J (2017) 11(2):426–38. doi: 10.1038/ismej.2016.130
65. Ruutu M, Thomas G, Steck R, Degli-Esposti MA, Zinkernagel MS, Alexander K, et al. $\beta$-glucan triggers spondylarthritis and Crohn’s disease–like ileitis in SKG mice. Arthritis Rheum (2012) 64(7):2211–22. doi: 10.1002/art.34423
66. Rehaume LM, Mondot S, Aguirre de Carcer D, Velasco J, Benham H, Hasnain SZ, et al. ZAP-70 genotype disrupts the relationship between microbiota and host, leading to spondyloarthritis and ileitis in SKG mice. Arthritis Rheumatol (2014) 66(10):2780–92. doi: 10.1002/art.38773
67. Rôças IN, Siqueira JF Jr. Characterization of Dialister species in infected root canals. J Endod (2006) 32(11):1057–61. doi: 10.1016/j.joen.2006.04.010
68. Morotomi M, Nagai F, Sakon H, Tanaka R. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol (2008) 58(Pt 12):2716–20. doi: 10.1099/ijs.0.2008/000810-0
69. Tito RY, Cypers H, Joossens M, Varkas G, Van Praet L, Glorieus E, et al. Brief Report: Dialister as a Microbial Marker of Disease Activity in Spondyloarthritis. Arthritis Rheumatol (2017) 69(1):114–21. doi: 10.1002/art.39802
70. Manasson J, Shen N, Garcia Ferrer HR, Ubeda C, Iraheta I, Heguy A, et al. Gut Microbiota Perturbations in Reactive Arthritis and Postinfectious Spondyloarthritis. Arthritis Rheumatol (2018) 70(2):242–54. doi: 10.1002/art.40359
71. Morio F, Jean-Pierre H, Dubreuil L, Jumas-Bilak E, Calvet L, Mercier G, et al. Antimicrobial susceptibilities and clinical sources of Dialister species. Antimicrob Agents Chemother (2007) 51(12):4498–501. doi: 10.1128/AAC.00538-07
72. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science (2017) 358(6361):359–65. doi: 10.1126/science.aan4526
73. Asquith M, Sternes PR, Costello M-E, Karstens L, Diamond S, Martin TM, et al. HLA Alleles Associated With Risk of Ankylosing Spondylitis and Rheumatoid Arthritis Influence the Gut Microbiome. Arthritis Rheumatol (2019) 71(10):1642–50. doi: 10.1002/art.40917
74. Gill T, Brooks SR, Rosenbaum JT, Asquith M, Colbert RA. Novel Inter-omic Analysis Reveals Relationships Between Diverse Gut Microbiota and Host Immune Dysregulation in HLA-B27-Induced Experimental Spondyloarthritis. Arthritis Rheumatol (2019) 71(11):1849–57. doi: 10.1002/art.41018
75. Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol (2019) 20(7):928–42. doi: 10.1038/s41590-019-0378-1
76. Hablot J, Peyrin-Biroulet L, Kokten T, El Omar R, Netter P, Bastien C, et al. Experimental colitis delays and reduces the severity of collagen-induced arthritis in mice. PloS One (2017) 12(9):e0184624. doi: 10.1371/journal.pone.0184624
77. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms (2020) 8(4):1–25. doi: 10.3390/microorganisms8040573
78. Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis (2017) 76(9):1614–22. doi: 10.1136/annrheumdis-2016-211064
79. Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med (2017) 9(1):103. doi: 10.1186/s13073-017-0490-5
80. Hoffmann TW, Pham H-P, Bridonneau C, Aubry C, Lamas B, Martin-Gallausiaux C, et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J (2016) 10(2):460–77. doi: 10.1038/ismej.2015.127
81. Levine JS, Burakoff R. Extraintestinal manifestations of inflammatory bowel disease. Gastroenterol Hepatol (2011) 7(4):235–41.
82. Machiels K, Sabino J, Vandermosten L, Joossens M, Arijs I, de Bruyn M, et al. Specific members of the predominant gut microbiota predict pouchitis following colectomy and IPAA in UC. Gut (2017) 66(1):79–88. doi: 10.1136/gutjnl-2015-309398
83. Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis (2019) 78(7):947–56. doi: 10.1136/annrheumdis-2018-214856
84. Silverman GJ. The microbiome in SLE pathogenesis. Nat Rev Rheumatol (2019) 15(2):72–4. doi: 10.1038/s41584-018-0152-z
85. Silverman GJ, Azzouz DF, Alekseyenko AV. Systemic Lupus Erythematosus and dysbiosis in the microbiome: cause or effect or both? Curr Opin Immunol (2019) 61:80–5. doi: 10.1016/j.coi.2019.08.007
86. Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci USA (2019) 116(26):12672–7. doi: 10.1073/pnas.1904099116
87. Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, Nardi M, et al. a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol (2001) 67(9):4111–8. doi: 10.1128/aem.67.9.4111-4118.2001
88. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol (2004) 54(Pt 5):1469–76. doi: 10.1099/ijs.0.02873-0
89. Stoll ML, Kumar R, Morrow CD, Lefkowitz EJ, Cui X, Genin A, et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther (2014) 16(6):486. doi: 10.1186/s13075-014-0486-0
90. Stoll ML, Pierce MK, Watkins JA, Zhang M, Weiss PF, Weiss JE, et al. Akkermansia muciniphila is permissive to arthritis in the K/BxN mouse model of arthritis. Genes Immun (2019) 20(2):158–66. doi: 10.1038/s41435-018-0024-1
91. Stoll ML, Weiss PF, Weiss JE, Nigrovic PA, Edelheit BS, Bridges SL Jr., et al. Age and fecal microbial strain-specific differences in patients with spondyloarthritis. Arthritis Res Ther (2018) 20(1):14. doi: 10.1186/s13075-018-1510-6
92. Davidson NJ, Leach MW, Fort MM, Thompson-Snipes L, Kühn R, Müller W, et al. T helper cell 1-type CD4+ T cells, but not B cells, mediate colitis in interleukin 10-deficient mice. J Exp Med (1996) 184(1):241–51. doi: 10.1084/jem.184.1.241
93. Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, et al. NLRP6 Protects Il10-/- Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep (2017) 19(4):733–45. doi: 10.1016/j.celrep.2017.03.080
94. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA (2013) 110(22):9066–71. doi: 10.1073/pnas.1219451110
95. de Groot PF, Belzer C, Aydin Ö, Levin E, Levels JH, Aalvink S, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PloS One (2017) 12(12):e0188475. doi: 10.1371/journal.pone.0188475
96. Cani PD, de Vos WM. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front Microbiol (2017) 8:1765. doi: 10.3389/fmicb.2017.01765
97. Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best Pract Res Clin Gastroenterol (2017) 31(6):637–42. doi: 10.1016/j.bpg.2017.10.001
98. Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio (2014) 5(4):1–10. doi: 10.1128/mBio.01438-14
99. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell (2011) 145(5):745–57. doi: 10.1016/j.cell.2011.04.022
100. Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol (2017) 18(1):142. doi: 10.1186/s13059-017-1271-6
101. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
102. Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis (2019) 78(5):590–3. doi: 10.1136/annrheumdis-2018-214514
103. Wells P.M. AAS, Bowyer RCE, Freidin MB, Finckh A, Strowig T, Lesker TR, et al. Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease: a cross-sectional study. Lancet Rheumatol (2020) 2:e418–e27. doi: 10.1016/S2665-9913(20)30064-3
104. Robertson BR, Rourke JL, Neilan BA, Vandamme P, On SLW, Fox JG, et al. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol (2005) 55(3):1199–204. doi: 10.1099/ijs.0.63472-0
105. Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, et al. A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol (2019) 4(34):1–14. doi: 10.1126/sciimmunol.aaw4341
106. Sidiq T, Yoshihama S, Downs I, Kobayashi KS. Nod2: A Critical Regulator of Ileal Microbiota and Crohn’s Disease. Front Immunol (2016) 7:367. doi: 10.3389/fimmu.2016.00367
107. Aviello G, Knaus UG. NADPH oxidases and ROS signaling in the gastrointestinal tract. Mucosal Immunol (2018) 11(4):1011–23. doi: 10.1038/s41385-018-0021-8
108. Bazin T, Hooks KB, Barnetche T, Truchetet M-E, Enaud R, Richez C, et al. Microbiota Composition May Predict Anti-Tnf Alpha Response in Spondyloarthritis Patients: an Exploratory Study. Sci Rep (2018) 8(1):5446. doi: 10.1038/s41598-018-23571-4
109. Gevers D, Kugathasan S, Denson Lee A, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe (2014) 15(3):382–92. doi: 10.1016/j.chom.2014.02.005
110. Herp S, Brugiroux S, Garzetti D, Ring D, Jochum LM, Beutler M, et al. Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe (2019) 25(5):681–94.e8. doi: 10.1016/j.chom.2019.03.004
111. Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect Immun (1999) 67(9):4499–509. doi: 10.1128/IAI.67.9.4499-4509.1999
112. Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest (2007) 87(10):1042–54. doi: 10.1038/labinvest.3700661
113. Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology (2004) 127(2):412–21. doi: 10.1053/j.gastro.2004.04.061
114. Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote T(H)17-dependent inflammation. Sci Transl Med (2017) 9(376):1–12. doi: 10.1126/scitranslmed.aaf9655
115. Ruutu M, Thomas G, Steck R, Degli-Esposti MA, Zinkernagel MS, Alexander K, et al. β-glucan triggers spondylarthritis and Crohn’s disease-like ileitis in SKG mice. Arthritis Rheum (2012) 64(7):2211–22. doi: 10.1002/art.34423
116. Rodrigues IK, Andrigueti M, de Oliveira Gil ID, de Lucca Schiavon L, de Andrade KR, Pereira IA, et al. An investigation into the relationship between anti-Helicobacter pylori and anti-Saccharomyces cerevisiae antibodies in patients with axial spondyloarthritis and Crohn disease. Rheumatol Int (2015) 35(2):359–66. doi: 10.1007/s00296-014-3088-x
117. Manasson J, Wallach DS, Guggino G, Stapylton M, Badri MH, Solomon G, et al. Interleukin-17 Inhibition in Spondyloarthritis Is Associated With Subclinical Gut Microbiome Perturbations and a Distinctive Interleukin-25-Driven Intestinal Inflammation. Arthritis Rheumatol (2020) 72(4):645–57. doi: 10.1002/art.41169
118. Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol (2007) 43(7):831–41. doi: 10.1080/00365520801935434
119. Jawhara S, Thuru X, Standaert-Vitse A, Jouault T, Mordon S, Sendid B, et al. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis (2008) 197(7):972–80. doi: 10.1086/528990
120. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science (2012) 336(6086):1314–7. doi: 10.1126/science.1221789
121. Li M, Dai B, Tang Y, Lei L, Li N, Liu C, et al. Altered Bacterial-Fungal Interkingdom Networks in the Guts of Ankylosing Spondylitis Patients. mSystems (2019) 4(2):1–14. doi: 10.1128/mSystems.00176-18
122. El Mouzan M, Wang F, Al Mofarreh M, Menon R, Al Barrag A, Korolev KS, et al. Fungal Microbiota Profile in Newly Diagnosed Treatment-naïve Children with Crohn’s Disease. J Crohns Colitis (2017) 11(5):586–92. doi: 10.1093/ecco-jcc/jjw197
123. El Mouzan MI, Korolev KS, Al Mofarreh MA, Menon R, Winter HS, Al Sarkhy AA, et al. Fungal dysbiosis predicts the diagnosis of pediatric Crohn’s disease. World J Gastroenterol (2018) 24(39):4510–6. doi: 10.3748/wjg.v24.i39.4510
124. Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe (2019) 25(3):377–88.e6. doi: 10.1016/j.chom.2019.01.007
125. Imai T, Inoue R, Kawada Y, Morita Y, Inatomi O, Nishida A, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol (2019) 54(2):149–59. doi: 10.1007/s00535-018-1530-7
126. Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. MBio (2016) 7(5):1–11. doi: 10.1128/mBio.01250-16
127. Sovran B, Planchais J, Jegou S, Straube M, Lamas B, Natividad JM, et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome (2018) 6(1):152. doi: 10.1186/s40168-018-0538-9
128. Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell (2015) 160(3):447–60. doi: 10.1016/j.cell.2015.01.002
129. Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res (2011) 21(10):1616–25. doi: 10.1101/gr.122705.111
130. Mukhopadhya I, Segal JP, Carding SR, Hart AL, Hold GL. The gut virome: the ‘missing link’ between gut bacteria and host immunity? Therap Adv Gastroenterol (2019) 12:1756284819836620. doi: 10.1177/1756284819836620
131. Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe (2019) 26(6):764–78.e5. doi: 10.1016/j.chom.2019.10.009
132. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol (2016) 14(1):20–32. doi: 10.1038/nrmicro3552
133. Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol (2017) 35(11):1077–86. doi: 10.1038/nbt.3981
134. Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell (1993) 75(2):253–61. doi: 10.1016/0092-8674(93)80067-o
135. Schultz M, Tonkonogy SL, Sellon RK, Veltkamp C, Godfrey VL, Kwon J, et al. IL-2-deficient mice raised under germfree conditions develop delayed mild focal intestinal inflammation. Am J Physiol Gastrointest Liver Physiol (1999) 276(6):G1461–G72. doi: 10.1152/ajpgi.1999.276.6.G1461
136. Hoentjen F, Tonkonogy SL, Qian BF, Liu B, Dieleman LA, Sartor RB. CD4(+) T lymphocytes mediate colitis in HLA-B27 transgenic rats monoassociated with nonpathogenic Bacteroides vulgatus. Inflammation Bowel Dis (2007) 13(3):317–24. doi: 10.1002/ibd.20040
137. Waidmann M, Bechtold O, Frick J-S, Lehr H-A, Schubert S, Dobrindt U, et al. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology (2003) 125(1):162–77. doi: 10.1016/s0016-5085(03)00672-3
138. Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut (2016) 65(2):225–37. doi: 10.1136/gutjnl-2015-309333
139. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol (2014) 14(3):141–53. doi: 10.1038/nri3608
140. Asquith M, Davin S, Stauffer P, Michell C, Janowitz C, Lin P, et al. Intestinal Metabolites Are Profoundly Altered in the Context of HLA-B27 Expression and Functionally Modulate Disease in a Rat Model of Spondyloarthritis: HLA-B27 EXPRESSION PROFOUNDLY IMPACTS THE INTESTINAL METABOLOME. Arthritis Rheumatol (2017) 69(10):1984–95. doi: 10.1002/art.40183
141. Davenport M, Poles J, Leung JM, Wolff MJ, Abidi WM, Ullman T, et al. Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflammation Bowel Dis (2014) 20(4):723–31. doi: 10.1097/MIB.0000000000000011
142. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol (2012) 13(9):R79. doi: 10.1186/gb-2012-13-9-r79
143. Jochum L, Stecher B. Label or Concept - What Is a Pathobiont? Trends Microbiol (2020) 28(10):789–92. doi: 10.1016/j.tim.2020.04.011
144. Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe (2011) 9(5):390–403. doi: 10.1016/j.chom.2011.04.009
145. Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med (2019) 25(7):1096–103. doi: 10.1038/s41591-019-0495-2
146. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med (2017) 23(1):107–13. doi: 10.1038/nm.4236
147. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. doi: 10.1016/j.cell.2009.09.033
148. Shi Z, Zou J, Zhang Z, Zhao X, Noriega J, Zhang B, et al. Segmented Filamentous Bacteria Prevent and Cure Rotavirus Infection. Cell (2019) 179(3):644–58.e13. doi: 10.1016/j.cell.2019.09.028
149. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science (2008) 320(5883):1647–51. doi: 10.1126/science.1155725
150. Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol (2008) 6(10):776–88. doi: 10.1038/nrmicro1978
151. Gill T, Asquith M, Rosenbaum JT, Colbert RA. The intestinal microbiome in spondyloarthritis. Curr Opin Rheumatol (2015) 27(4):319–25. doi: 10.1097/BOR.0000000000000187
152. Chukkapalli S, Rivera-Kweh M, Gehlot P, Velsko I, Bhattacharyya I, Calise SJ, et al. Periodontal bacterial colonization in synovial tissues exacerbates collagen-induced arthritis in B10.RIII mice. Arthritis Res Ther (2016) 18(1):161. doi: 10.1186/s13075-016-1056-4
153. Fischbach MA. Microbiome: Focus on Causation and Mechanism. Cell (2018) 174(4):785–90. doi: 10.1016/j.cell.2018.07.038
154. Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome (2013) 1(1):3. doi: 10.1186/2049-2618-1-3
155. Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature (2016) 533(7604):543–6. doi: 10.1038/nature17645
156. Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature (2018) 557(7705):434–8. doi: 10.1038/s41586-018-0092-4
157. Pruteanu M, Hyland NP, Clarke DJ, Kiely B, Shanahan F. Degradation of the extracellular matrix components by bacterial-derived metalloproteases: implications for inflammatory bowel diseases. Inflammation Bowel Dis (2011) 17(5):1189–200. doi: 10.1002/ibd.21475
158. Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PloS One (2013) 8(9):e74963. doi: 10.1371/journal.pone.0074963
159. Beaud D, Tailliez P, Anba-Mondoloni J. Genetic characterization of the beta-glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology (2005) 151(Pt 7):2323–30. doi: 10.1099/mic.0.27712-0
160. Mielants H, De Keyser F, Baeten D, Van den Bosch F. Gut inflammation in the spondyloarthropathies. Curr Rheumatol Rep (2005) 7(3):188–94. doi: 10.1007/s11926-996-0038-y
161. Kevans D, Turpin W, Madsen K, Meddings J, Shestopaloff K, Xu W, et al. Determinants of intestinal permeability in healthy first-degree relatives of individuals with Crohn’s disease. Inflammation Bowel Dis (2015) 21(4):879–87. doi: 10.1097/MIB.0000000000000323
162. Secondulfo M, de Magistris L, Fiandra R, Caserta L, Belletta M, Tartaglione MT, et al. Intestinal permeability in Crohn’s disease patients and their first degree relatives. Dig Liver Dis (2001) 33(8):680–5. doi: 10.1016/s1590-8658(01)80045-1
163. Sydora BC, Macfarlane SM, Walker JW, Dmytrash AL, Churchill TA, Doyle J, et al. Epithelial barrier disruption allows nondisease-causing bacteria to initiate and sustain IBD in the IL-10 gene-deficient mouse. Inflammation Bowel Dis (2007) 13(8):947–54. doi: 10.1002/ibd.20155
164. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev (2017) 279(1):70–89. doi: 10.1111/imr.12567
165. Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature (2014) 514(7524):638–41. doi: 10.1038/nature13823
166. Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, et al. The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity (2019) 51(1):77–89.e6. doi: 10.1016/j.immuni.2019.05.004
167. Quigley EM. Prebiotics and probiotics: their role in the management of gastrointestinal disorders in adults. Nutr Clin Pract (2012) 27(2):195–200. doi: 10.1177/0884533611423926
168. Smith NM, Maloney NG, Shaw S, Horgan GW, Fyfe C, Martin JC, et al. Daily Fermented Whey Consumption Alters the Fecal Short-Chain Fatty Acid Profile in Healthy Adults.s. Front Nutr (2020) 7:165:165. doi: 10.3389/fnut.2020.00165
169. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11(8):506–14. doi: 10.1038/nrgastro.2014.66
170. Hoentjen F, Welling GW, Harmsen HJM, Zhang X, Snart J, Tannock GW, et al. Reduction of colitis by prebiotics in HLA-B27 transgenic rats is associated with microflora changes and immunomodulation. Inflammation Bowel Dis (2005) 11(11):977–85. doi: 10.1097/01.mib.0000183421.02316.d5
171. Hoentjen F, Tannock GW, Mulder CJ, Dieleman LA. The prebiotic combination inulin/oligofructose prevents colitis in HLA-B27 rats by immunomodulation and changes in intestinal microflora. Eur J Gastroenterol Hepatol (2006) 18(1):A27. doi: 10.1097/00042737-200601000-00102
172. Koleva PT, Valcheva RS, Sun X, Gänzle MG, Dieleman LA. Inulin and fructo-oligosaccharides have divergent effects on colitis and commensal microbiota in HLA-B27 transgenic rats. Br J Nutr (2012) 108(9):1633–43. doi: 10.1017/S0007114511007203
173. Schultz M, Munro K, Tannock GW, Melchner I, Göttl C, Schwietz H, et al. Effects of feeding a probiotic preparation (SIM) containing inulin on the severity of colitis and on the composition of the intestinal microflora in HLA-B27 transgenic rats. Clin Diagn Lab Immunol (2004) 11(3):581–7. doi: 10.1128/CDLI.11.3.581-587.2004
174. Ruppin H, Bar-Meir S, Soergel KH, Wood CM, Schmitt MG Jr. Absorption of short-chain fatty acids by the colon. Gastroenterology (1980) 78(6):1500–7. doi: 10.1016/S0016-5085(19)30508-6
175. Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. Specialized metabolites from the microbiome in health and disease. Cell Metab (2014) 20(5):719–30. doi: 10.1016/j.cmet.2014.10.016
176. Nakamura YK, Janowitz C, Metea C, Asquith M, Karstens L, Rosenbaum JT, et al. Short chain fatty acids ameliorate immune-mediated uveitis partially by altering migration of lymphocytes from the intestine. Sci Rep (2017) 7(1):11745. doi: 10.1038/s41598-017-12163-3
177. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem (2003) 278(13):11312–9. doi: 10.1074/jbc.M211609200
178. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature (2009) 461(7268):1282–6. doi: 10.1038/nature08530
179. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
180. Duan Y, Zeng L, Zheng C, Song B, Li F, Kong X, et al. Inflammatory Links Between High Fat Diets and Diseases. Front Immunol (2018) 9:2649. doi: 10.3389/fimmu.2018.02649
181. Rodrı́guez-Cabezas ME, Gálvez J, Camuesco D, Lorente MD, Concha A, Martinez-Augustin O, et al. Intestinal anti-inflammatory activity of dietary fiber (Plantago ovata seeds) in HLA-B27 transgenic rats. Clin Nutr (2003) 22(5):463–71. doi: 10.1016/S0261-5614(03)00045-1
182. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504(7480):446–50. doi: 10.1038/nature12721
183. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
184. Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol (2004) 36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5
185. Gutiérrez-Vázquez C, Quintana FJ. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity (2018) 48(1):19–33. doi: 10.1016/j.immuni.2017.12.012
186. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-Derived Metabolites Suppress Arthritis by Amplifying Aryl-Hydrocarbon Receptor Activation in Regulatory B Cells. Cell Metab (2020) 31(4):837–51.e10. doi: 10.1016/j.cmet.2020.03.003
187. Tilg H. Cruciferous vegetables: prototypic anti-inflammatory food components. Clin Phytoscience (2015) 1(1):10. doi: 10.1186/s40816-015-0011-2
188. Qiu J, Guo X, Chen Z-ME, He L, Sonnenberg GF, Artis D, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity (2013) 39(2):386–99. doi: 10.1016/j.immuni.2013.08.002
189. Ye J, Qiu J, Bostick JW, Ueda A, Schjerven H, Li S, et al. The Aryl Hydrocarbon Receptor Preferentially Marks and Promotes Gut Regulatory T Cells. Cell Rep (2017) 21(8):2277–90. doi: 10.1016/j.celrep.2017.10.114
190. Kimura A, Naka T, Nakahama T, Chinen I, Masuda K, Nohara K, et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med (2009) 206(9):2027–35. doi: 10.1084/jem.20090560
191. Di Meglio P, Duarte JH, Ahlfors H, Owens NDL, Li Y, Villanova F, et al. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity (2014) 40(6):989–1001. doi: 10.1016/j.immuni.2014.04.019
192. Selvanderan SP, Goldblatt F, Nguyen NQ, Costello SP. Faecal microbiota transplantation for Clostridium difficile infection resulting in a decrease in psoriatic arthritis disease activity. Clin Exp Rheumatol (2019) 37(3):514–5.
Keywords: pathobiont, HLA-B27, spondyloarthritis, gut inflammation, dysbiosis
Citation: Gill T and Rosenbaum JT (2021) Putative Pathobionts in HLA-B27-Associated Spondyloarthropathy. Front. Immunol. 11:586494. doi: 10.3389/fimmu.2020.586494
Received: 23 July 2020; Accepted: 02 December 2020;
Published: 18 January 2021.
Edited by:
Gregg Joshua Silverman, New York University, United StatesReviewed by:
Antony Nicodemus Antoniou, Northumbria University, United KingdomKristi Kuhn, University of Colorado Denver, United States
Copyright © 2021 Gill and Rosenbaum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James T. Rosenbaum, cm9zZW5iYWpAb2hzdS5lZHU=
 Tejpal Gill
Tejpal Gill James T. Rosenbaum
James T. Rosenbaum