- 1Comparative Genetics and Refinement, Biomedical Primate Research Centre, Rijswijk, Netherlands
- 2Theoretical Biology and Bioinformatics, Utrecht University, Utrecht, Netherlands
The activity and function of natural killer (NK) cells are modulated through the interactions of multiple receptor families, of which some recognize MHC class I molecules. The high level of MHC class I polymorphism requires their ligands either to interact with conserved epitopes, as is utilized by the NKG2A receptor family, or to co-evolve with the MHC class I allelic variation, which task is taken up by the killer cell immunoglobulin-like receptor (KIR) family. Multiple molecular mechanisms are responsible for the diversification of the KIR gene system, and include abundant chromosomal recombination, high mutation rates, alternative splicing, and variegated expression. The combination of these genetic mechanisms generates a compound array of diversity as is reflected by the contraction and expansion of KIR haplotypes, frequent birth of fusion genes, allelic polymorphism, structurally distinct isoforms, and variegated expression, which is in contrast to the mainly allelic nature of MHC class I polymorphism in humans. A comparison of the thoroughly studied human and macaque KIR gene repertoires demonstrates a similar evolutionarily conserved toolbox, through which selective forces drove and maintained the diversified nature of the KIR gene cluster. This hypothesis is further supported by the comparative genetics of KIR haplotypes and genes in other primate species. The complex nature of the KIR gene system has an impact upon the education, activity, and function of NK cells in coherence with an individual’s MHC class I repertoire and pathogenic encounters. Although selection operates on an individual, the continuous diversification of the KIR gene system in primates might protect populations against evolving pathogens.
Introduction
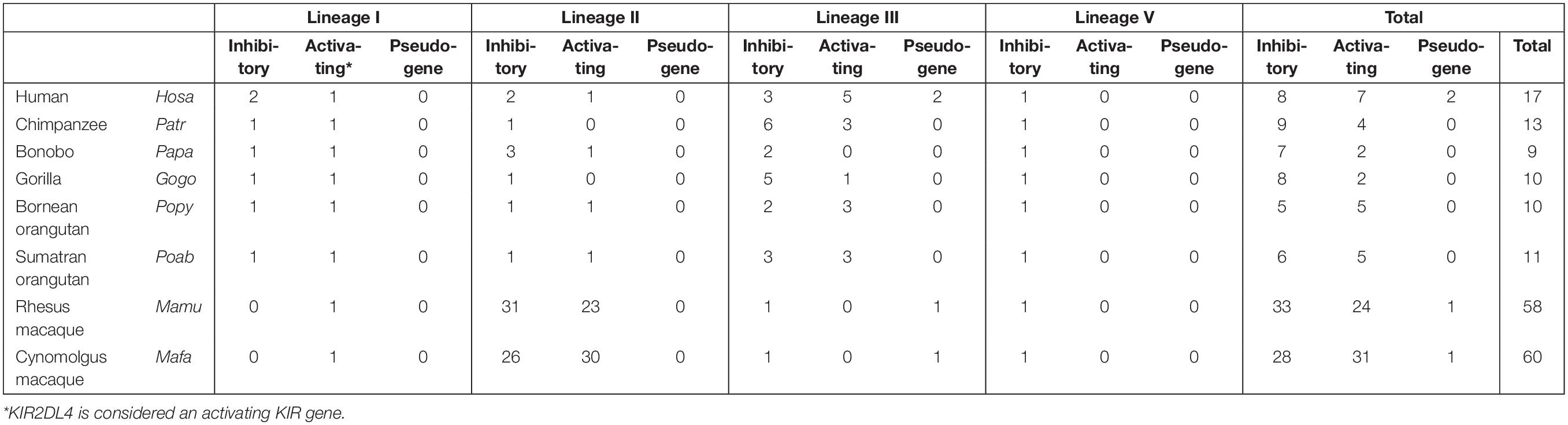
The innate and adaptive arms of the immune system are interconnected, and feature several effector functions that provide efficient and specific protection against infection and tumor formation. Major components of the adaptive arm comprise T and B lymphocytes characterized by rearranging antigen receptors, which exert cytotoxic and humoral immunity, respectively. The cytotoxicity mediated by T lymphocytes highly depends on the presentation of intracellular antigen segments derived from pathogens by MHC class I molecules and subsequent clonal expansion of cells with specific receptors. A third type of lymphocytes bridge the innate and adaptive immune response, and comprises natural killer (NK) cells, which participate, for instance, in the recognition and elimination of aberrant cells that down-regulate their MHC class I expression to evade detection by T lymphocytes (1). Without prior priming or clonal expansion, inhibitory and activating receptors on the NK cell surface interact with MHC class I molecules on nucleated cells to modulate NK cell effector functions, which include the killing of target cells by the release of cytolytic proteins and the regulation of other immune cells by the secretion of cytokines (2). The genes encoding the MHC class I molecules are considered the most polymorphic genes known in vertebrates, a phenomenon that resulted from selective pressure to adapt to the rapid diversification of pathogens. This extended repertoire of MHC class I genes and alleles requires the NK cell receptors to co-evolve to maintain a functional relation with their ligands. The recognition of MHC class I molecules by NK cells involves two receptor families: the conserved CD94:NKG2A receptors and the highly polymorphic and diverse killer cell immunoglobulin-like receptors (KIR). Both receptor families consist of inhibitory and activating members. Their engagement with MHC class I molecules calibrates the responsiveness of NK cells through a continuous educational process, which largely controls subsequent NK cell activity (3, 4). The KIR receptors are encoded within the Leukocyte Receptor Complex (LRC) on chromosome 19q13.4, and share this genomic region with other structurally similar immune-regulators, such as the leukocyte Ig-like receptors (LILRs) and the leukocyte-associated Ig-like receptors (LAIRs; Figure 1) (5). Based on different Alu elements that can be regarded as a molecular clock, the initial expansion of the primate KIR gene cluster is estimated to date back to approximately 31 to 44 million years ago. This process continued, and is currently reflected by extensive gene duplications and point mutations (6). Different diversifying mechanisms in combination with evolutionary selective factors propel the complex KIR gene content at the individual level but also at the population and species-specific level, which all together contribute to the heterogeneity of NK cell subsets and their activity. The KIR gene diversification is not limited to humans. Comparative analyses that include other primate species might help in gaining a thorough understanding of the evolutionary processes that resulted in the diversification of this gene system. In the following sections, we will discuss the different genetic mechanisms that drove the evolution of the highly plastic KIR gene system in hominoids (humans and great apes) and Old World monkeys, and how this might influence their NK cell response.
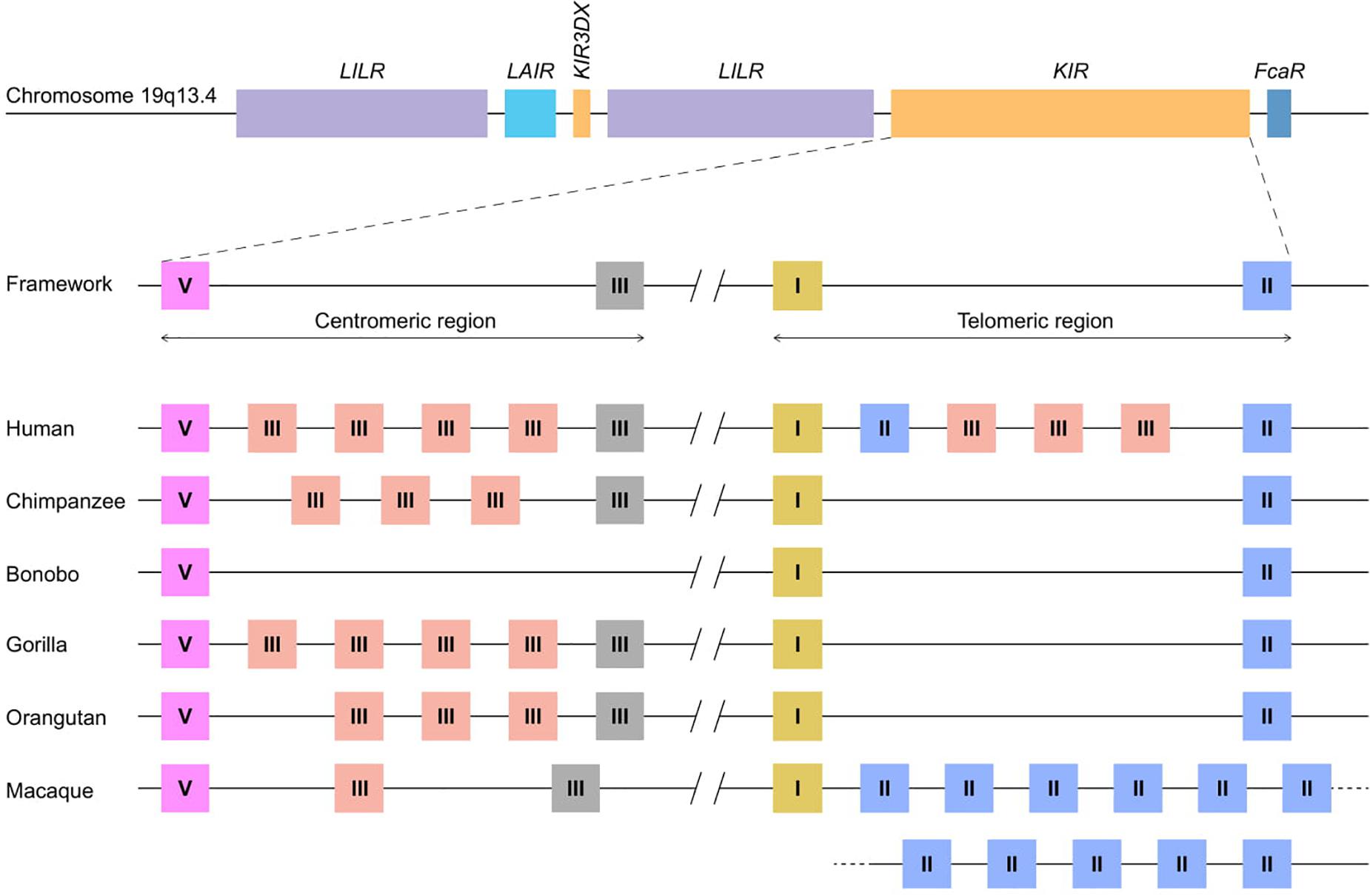
Figure 1. KIR haplotype organizations in different primate species. A schematic overview of the Leukocyte Receptor Complex (LRC) on chromosome 19q13.4 and KIR haplotype organizations in different primate species. A fixed copy of the KIR3DX gene is located within the primate LILR gene cluster, whereas the expanded KIR gene cluster is flanked by the LILR and FcaR genes. The expansion involved four different KIR gene lineages – I, II, III, and V – the members of which are indicated as yellow, blue, red/gray, and pink boxes, respectively. The gray lineage III boxes represent pseudogenes. In most hominoids, KIR haplotype organizations follow a standard framework, in which the centromeric and telomeric regions are bordered by genes from lineages V and III, and lineages I and II, respectively. A relatively large non-coding segment separates the centromeric and telomeric haplotype sections.
Co-Evolution of MHC and KIR Genes
The complex KIR gene system requires a comprehensive nomenclature guideline for the different genes and allotypes in order to distinguish the corresponding receptors by their structure and signaling potential (7–9). Receptors may contain one to three Ig-like domains, which are encoded by exon 3 (D0 domain), exon 4 (D1 domain), and exon 5 (D2 domain), and are referred to as KIR1D, KIR2D, and KIR3D in the official nomenclature. Further classification defines the inhibitory or activating signaling function of the KIR receptors, which is characterized by either a long or short cytoplasmic tail, respectively, and specified with an “L” or an “S” following the domain number denotation. The long cytoplasmic tail contains one or two immune tyrosine-based inhibitory motifs (ITIMs), whereas the signal transduction of activating KIR depends on the interaction with an adaptor molecule that includes an immune tyrosine-based activating motif (ITAM) such as DAP12. Pseudogenes are indicated with a “P” (e.g., KIR3DP). In addition, a four-character species designation is included in front of the KIR acronym (e.g., Mamu-KIR3DL20 in rhesus macaques; Macaca mulatta).
The mammalian KIR genes originate from two progenitor gene lineages: KIR3DX and KIR3DL. The KIR3DX lineage is represented by a single gene copy located in the center of the LILR gene cluster (Figure 1). The gene is fixed in most primate species, and its function is currently unknown (10). This lineage is, however, expanded in cattle, and encodes multiple inhibitory and a single activating functional KIR3DX receptor, which interact with an expanded repertoire of classical MHC molecules (11, 12). In contrast, the KIR3DL lineage expanded in primates and was diversified by duplications, deletions, and recombinations, which resulted in an elaborated KIR gene family. Based on their structure, ligand specificity, and/or phylogenetic analysis, the primate KIR receptors are divided into four lineages. Lineage I genes encode receptors with a D0-D2 domain configuration; lineage II (D0-D1-D2) is defined by the specificity for subtypes of HLA-A and -B in humans; lineage III includes receptors with D1-D2 and D0-D1-D2 domain configurations; and lineage V (D0-D1-D2) is represented by human KIR3DL3 and its orthologs. In the primate species studied, at least one KIR gene was discovered for each lineage, which indicates that gene duplication and diversification predates primate speciation. The subsequent lineage expansions are, however, species specific (Table 1).
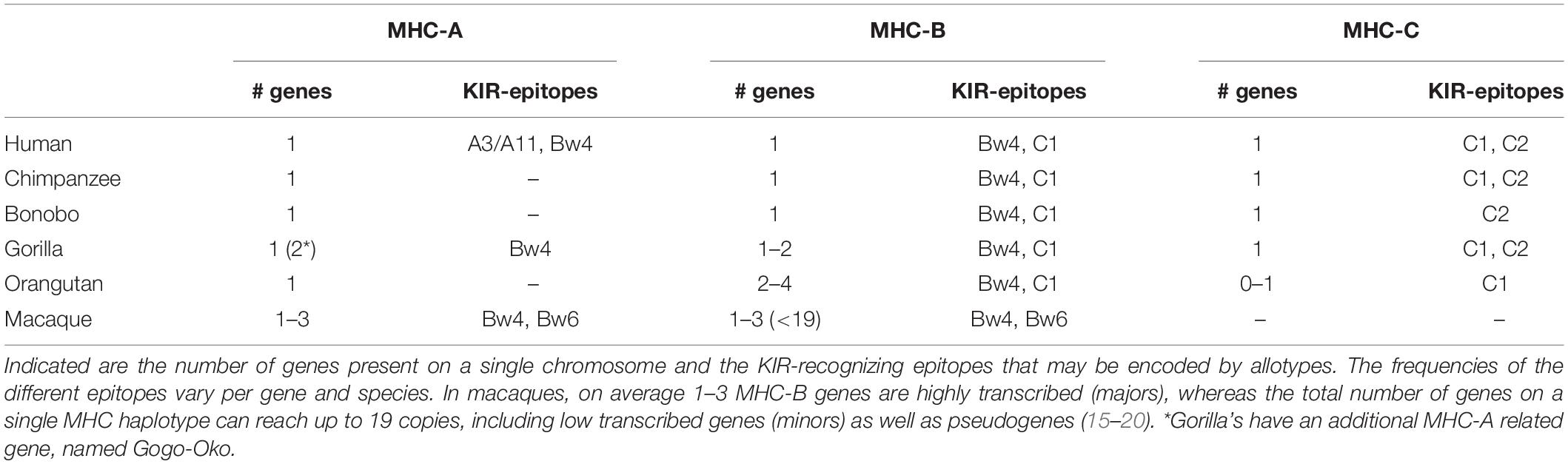
Lineage I and V KIR genes have a conserved nature in all primate species examined, and comprise, respectively, KIR2DL4 and KIR2DL5, and KIR3DL3, or a similar structure, such as Mamu-KIR3DL20 in rhesus macaques. More extensive and species-specific expansions are reported for KIR genes that cluster into lineages II and III (Table 1), and the data suggest that this coincides with the evolution of their MHC class I ligands. Therefore, diversification of the lineage II and III KIR genes might be indirectly propelled by the adaption of the MHC class I molecules to pathogenic encounters. For hominoids, this section of co-evolution of KIR and MHC has been comprehensively reviewed by Wroblewski and colleagues (13). In short, the MHC gene content in great apes displays to a limited extent a variable number of MHC-A, -B, and -C genes per haplotype (Table 2). MHC-C, which originated from a duplication of an MHC-B gene, is fixed in all hominoids except for orangutans, where it is present on about half of the haplotypes (14). In addition, the epitopes recognized by the relevant KIR are differentially distributed across the different MHC class I genes (Table 2). The C1 and C2 epitopes, for example, are absent in bonobos and orangutans, respectively, whereas the A3/A11 epitope is only defined on HLA-A molecules. The hominoid MHC class I evolution is accompanied by the reduction and refinement of KIR specific for MHC-A and -B, which is reflected in their limited number of lineage II KIR receptors, whereas the emergence and fixation of MHC-C in humans, chimpanzees, and gorillas drove the expansion and specialization of lineage III KIR (Table 1) (13).
Old World monkeys, including macaques, lack an MHC-C ortholog, but instead display extensive copy number variation regarding polymorphic MHC-A and -B genes, as opposed to the fixed number of MHC class I genes in hominoids (Table 2) (15–18). The expression level of the different MHC-A and -B molecules, however, varies considerably in macaques. It is generally accepted that per haplotype at least a single MHC-A and 1 to 3 MHC-B genes are characterized by high transcription, and are referred to as “majors,” whereas the other MHC class I genes have lower transcription levels (“minors”), or may be pseudogenes. The differential transcription suggests a more classical function for the major MHC molecules, such as antigen presentation, whereas the minors might exert more specialized functions (19, 20). Only a few interactions of macaque MHC and KIR are documented, and, so far, all interactions involved lineage II KIR that recognize Bw4 and Bw6 epitopes on MHC-A and -B allotypes (Table 2) (21–25). This putative lineage II specificity for the copious macaque MHC class I repertoire coincides with an extensive ligand expansion, and, thus far, 54 and 56 different lineage II KIR genes have been documented for rhesus and cynomolgus macaques, respectively (Table 1) (7). Like the majors and minors for the MHC system, the KIR genes, may display differential expression levels, which are modulated by sequence polymorphisms and by an individual’s MHC class I repertoire (26–28). Lineage III KIR genes, which encode ligands for MHC-C in hominoids and were subject to expansion, are represented in macaques by a single gene and encodes a receptor with only the D1 extracellular domain (KIR1D). Its presence on 22% and 82% of the rhesus and cynomolgus macaque KIR haplotypes, respectively, suggests a balancing selection for this structurally modified receptor, which might execute a function other than conventional MHC recognition (29).
The maximal expression of six distinct MHC class I genes in most hominoids and the specialization of MHC-C as ligand for lineage III KIR is in line with their modest KIR gene expansion (Tables 1, 2). Macaques may harbor over 20 distinct MHC class I genes in one individual, of which only a few are dominantly expressed and considered to be majors. The expanded MHC repertoire in macaques probably propelled the extensive expansion and differential expression of their lineage II KIR. The balanced expansion of the MHC and KIR gene systems in primates indicates co-evolution in order to maintain a functional relation.
Transposable Elements Facilitate Chromosomal Recombination
One of the mechanisms responsible for the extensive KIR gene diversification in macaques, and to a lesser extent in hominoids, involves chromosomal rearrangements that are accompanied by deletions and recombination, which may result in the generation of fusion genes (Figure 2A). This type of gene formation may shuffle the binding and signaling domains of different KIR receptors, thereby functionally altering the response potential of KIR family members. The dense head-to-tail arrangement of the KIR genes is likely to facilitate at least in part the chromosomal instability of this gene cluster. A KIR haplotype spans approximately 150 to 350 kb, depending on the number of genes present. Most KIR genes are separated by only 2.5 kb, as opposed to the wider haplotype configurations of more stable and less expanded gene families, such as the LILR gene cluster (6, 30–33). In addition, the presence of transposable elements, including Alu and LINE elements, in the intergenic and intragenic KIR sequences is another factor that further promotes genetic instability (6, 34–36). These repetitive elements are present in all primate KIR genes, although with species-specific variation, and drive recombination and genetic deletions (35, 37–39). For the few completely sequenced fusion KIR genes in humans, the chromosomal breakpoints indeed map in the intragenic transposable elements. This supports the idea that the abundant presence of transposons in the KIR cluster facilitates chromosome fragility, which is reflected by genetic expansion and contraction, and the formation of fusion genes (34, 40, 41). A considerable number of human fusion KIR genes were generated by reshuffling that involved segments of pseudogenes (34). The conservation of two pseudogenes in the human KIR repertoire, KIR2DP1 and KIR3DP1, might be explained by their role in promoting recombination events. The human KIR haplotypes that include an apparent fusion gene are represented by relatively low frequencies (42–45). Positive selection of fusion entities might, however, increase their frequencies in certain populations (45). Ancient recombination events and subsequent selection might have contributed substantially to the current human KIR repertoire, but the modest expansion of the human KIR genes nowadays indicates limited recent recombination events. In contrast, an excessive number of recombination events are recorded in rhesus and cynomolgus macaques, with the presence of at least one fusion KIR gene on 42% and 49% of their haplotypes, respectively (29, 43). The abundant presence of fusion genes indicates that in these species the reshuffling of KIR gene segments is an ongoing process that expands the macaque KIR repertoire. Although information on the non-coding regions in the macaque KIR cluster is limited at present, the chromosome instability and consequential recombinations in concert with selection are likely to have driven the extensive expansion of lineage II KIR genes. This fast mode of evolution is further reflected in the relatively low number of orthologs that are shared between the closely related rhesus and cynomolgus macaques and their populations (29).
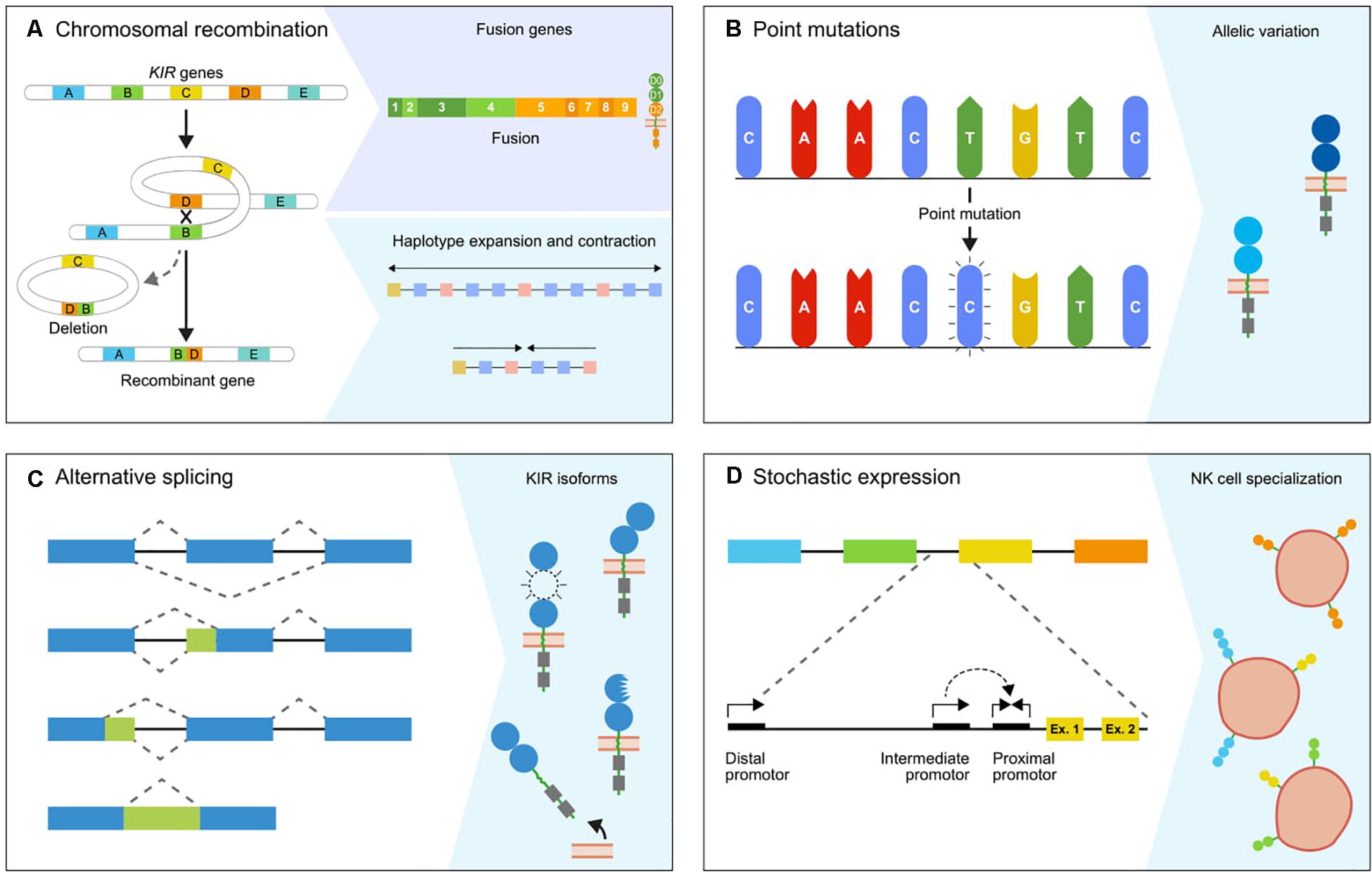
Figure 2. The genetic mechanisms propelling diversification. The primate KIR cluster diverged as a result of multiple molecular processes, which together modulate the KIR gene content and expression status. (A) The expansion and contraction of KIR haplotypes is mediated by chromosomal recombinations, which can introduce or remove one or multiple KIR genes. Occasionally, a recombination event is accompanied by the generation of a fusion gene, which functionally and structurally expands the gene repertoire. (B) The KIR genes are further diversified by point mutations in coding and non-coding regions, which generate alleles that encode receptors with different structures, localization, function, and expression. (C) Alternative splicing is another mechanism that has a similar impact on the function and structure of receptors. The blue and green boxes indicate exons and introns. The isoforms are generated by different splice events, which involve alternative splice sites and exon skipping. (D) The differential expression of subsets of KIR receptors on different NK cell clones forms another level of variation that is mediated and maintained by sequence variability in the complex promoter regions and epigenetic modifications. A conjunction of the proximal, intermediate, and distal promoters is required to induce KIR expression.
In all hominoids and Old World monkeys, the 5’ section of the KIR gene cluster is occupied by KIR3DL3 or similar structures, which are considered framework genes and might carry out essential functions. The structure and evolutionary pathway of these lineage V KIR genes is a complex outcome of multiple recombination events (46). Additional chromosomal rearrangements in rhesus macaques involved the exchange of the cytoplasmic tail of KIR3DL20 with the tails of KIR2DL04 (lineage I) and KIR1D (lineage III). These recombination events are not conserved in macaque populations, which implies the relatively recent formation of novel gene entities propelled by ancient recombination hotspots (29).
Chromosomal recombination events generate genetic variability in the KIR gene cluster by the formation of fusion genes. Subsequent selection of these novel genes might supply an adaptive and protective strategy in the arms race with rapidly evolving pathogens.
KIR Haplotype Diversity in Primate Species
Chromosomal rearrangements not only generate novel KIR gene entities by recombination but also diversify the haplotype gene content by insertions and deletions of genes (Figure 2A). In general, hominoid KIR haplotypes consist of two genomic regions that are bordered by four framework genes (Figure 1). The proximal half of the haplotype is termed the centromeric region and is defined by KIR3DL3 to KIR3DP1/KIRDP, whereas the distal part, or telomeric region, ranges from KIR2DL4 to KIR3DL2/KIR3DL1. Within these sections, KIR genes of different lineages expanded and contracted during hominoid speciation. In humans, the expansion involved lineage III KIR genes in their centromeric and telomeric regions, whereas expansion in chimpanzees, gorillas, and orangutans expansion took place in the same lineage in the centromeric region only (Figure 1). The human haplotype content ranges from 7 to 12 KIR genes, whereas the number in chimpanzee and orangutan haplotypes stretches from 5 to 11 and 5 to 10 functional KIR genes, respectively. In contrast to other hominoids, bonobos are characterized by a contraction of their KIR region, with only 3–7 KIR genes expressed on a haplotype. The shortest bonobo KIR haplotype consists of only the framework genes (47). The contracted bonobo KIR cluster coincides with a reduced nucleotide variation in their MHC class I repertoire, which might be caused by a bottleneck or pathogen-driven selective sweep after divergence from the chimpanzee’s lineage (48–51). In contrast, a highly variable KIR haplotype content is encountered in the macaque, with 4 to 17 functional KIR genes that mainly map to the telomeric region (Figure 1). The haplotype framework in macaques is less fixed than in hominoids, with only KIR3DL20 expressed on all haplotypes, whereas KIR2DL04 is present on 70% of the rhesus macaque haplotypes. A gene orthologous to hominoid KIR3DL2/KIR3DL1 that usually marks the telomeric region is absent.
The diversified KIR haplotypes in hominoids and Old World monkeys stem from a primordial configuration, for which a model has been proposed by Guethlein and colleagues (35). This model describes abundant duplications and recombination events that eventually formed a conserved haplotype framework in hominoids. The previously mentioned transposable elements are likely propagating these chromosomal rearrangements that continue to mediate the diversification of haplotype configurations. One major hotspot for recombination is mapping in between the centromeric and telomeric regions, which facilitates the reorganization of the different haplotype regions. In addition, KIR haplotypes also display the insertion and deletion of one or multiple KIR genes propelled by unequal crossing-over, which is occasionally accompanied by the formation of a fusion gene (29, 40, 42–44). In humans, these contractions and expansions, which are mediated by double-stranded breaks at intragenic and intergenic repetitive elements, resulted in haplotypes that expressed 3 to 15 KIR genes (40). The short haplotypes do not express all framework genes. For instance, the deletion of KIR2DL4 is commonly observed on genotypes defined across different populations (52, 53). Approximately 7% of the human KIR haplotypes are showing indications for contraction and expansion (42). Although the number of completely defined KIR haplotypes in other hominoids is low, several rare KIR configurations in chimpanzees and orangutans illustrate genetic footprints for insertion and deletion events, which is also occasionally accompanied by the formation of a fusion gene (54, 55). In macaques, only two completely sequenced haplotypes are available at present, whereas an abundant number of haplotypes are deduced at the transcription level by segregation studies (26, 29, 31, 43, 56, 57). The presence of multiple highly similar allotypes, encoded by highly similar KIR genes, on a single haplotype indicates an expansion by the insertion of one or more genes. Such events were recorded for 47% and 26% of the rhesus and cynomolgus macaque haplotypes, respectively (29). In contrast, a minimal KIR gene content and the presence of a fusion gene often are indicative of a haplotype contraction. An example of a prominent haplotype reduction in rhesus macaques involved the deletion of the complete centromeric region by an intragenic recombination of KIR3DL20 and KIR2DL04 (29). The variable haplotype content and the relatively high number of fusion genes indicate extensive recombination as a mechanism to diversify the macaque KIR gene system in a still ongoing process. This phenomenon is observed to a lesser extent for the KIR haplotypes in hominoids, where the process seems to have relaxed.
The KIR Gene Allelic Repertoire Is Expanded by Point Mutations
Another level of variation is displayed by allelic polymorphisms, which is explained to a large extent by the occurrence of single nucleotide polymorphisms (SNP; Figure 2B). These nucleotide variations have a wide-ranging impact, and may modulate the expression level at the cell surface, ligand specificity, interaction strength, and localization of the KIR receptor. Single nucleotide variations in the extracellular D0 and D1 domains of human KIR2DL2∗004 and KIR3DL1∗004, for example, retain the receptors within the cell, which might be caused by misfolding (58, 59). Polymorphisms in KIR2DL3 alleles affect the avidity of the receptor to bind their HLA-C ligands. The low-avidity KIR2DL3∗001 and the high-avidity KIR2DL3∗005 only differ at three nucleotides in their D1 domain, which alters the orientation of their extracellular domains and thereby their binding strength (60). Although most KIR disease association studies determine the gene content by the presence and absence of KIR gene sections, and thereby lack allele-level resolution, several studies demonstrated that the functional differences of KIR alleles might also impact health and disease. For example, two KIR2DL1 alleles in the African KhoeSan population evolved by single nucleotide mutations and are associated with a reduced risk for pregnancy disorders (61). Other associations demonstrated that the highly expressed KIR3DL1 alleles are more protective against disease progression in HIV-infected individuals than lower expressed allotypes, except for the intracellularly retained KIR3DL1∗004, which is low in expression but highly protective (62–64).
A total of 1110 human KIR alleles are cataloged in the Immuno Polymorphism Database (IPD-KIR, release 2.9.0), whereas the number of reported alleles for different non-human primate species ranges from 521 KIR alleles in rhesus macaques to 5 KIR alleles in Bornean orangutans (IPD-NHKIR, release 1.2.0.0). These allele numbers may give a distorted view of the actual levels of polymorphism within a species due to the differential number of individuals studied. The high level of allelic polymorphism appears to be at least comparable in humans and macaques. The thoroughly documented allelic polymorphism in humans and macaques reveals a varying number of alleles per KIR gene, with most nucleotide variation exhibited by the framework genes (7, 29, 44). In addition, a high number of alleles were reported for certain KIR genes located on the telomeric haplotype region in humans (KIR3DL1, KIR2DS4) and the highly frequent inhibitory KIR genes in macaques (KIR3DL01, KIR3DL07). An expansion of the allele numbers for the frequently expressed KIR genes might indicate a continuous role in co-evolution with particular pathogens. The less common KIR genes, which include mostly activating KIR, vary in gene content rather than allelic polymorphism and therefore seem to execute more specialized functions and/or might be involved in the recognition of conserved ligands and peptides (7, 29, 44).
For humans, KIR alleles are also distinguished by SNPs in their introns (IPD-KIR, release 2.9.0) (65), which might impact, for instance, the expression level and post-transcriptional splicing. A total of 353 human KIR alleles can only be distinguished from the reference gene based on intronic variations (IPD-KIR, release 2.9.0), and this number is likely to be underestimated (65). Sequence data on the non-coding KIR gene regions are lacking for non-human primate species, but a similar extent of intronic variations might be feasible and may impact their receptor functionality. However, there are no disease or health associations reported for intronic polymorphisms within the KIR genes, but abundant pathological conditions are described for intronic variations in many other genes mapping elsewhere in the genome (66). For example, a SNP in the human CYP2D6 gene is linked to a decreased expression of the functional transcript and correlates with a lower metabolic activity (67). For HLA-DP, a single nucleotide variation in the 3′ UTR modulates the expression level of different allotypes, which impacts the susceptibility to chronic hepatitis B virus infection (68).
Allele variation is mainly generated by synonymous and non-synonymous point mutations, and only the latter ones will impact the composition of the gene products. In sharp contrast to MHC class I polymorphisms, the allelic nucleotide variations of the KIR genes are evenly distributed over the coding regions. The high concentration of CpG islands located in the KIR gene cluster might contribute to an elevated mutation rate, as these islands are in general more prone to promote nucleotide transitions (69–71). In addition, chromosomal rearrangements are known as mutagenic events (69, 72–74). In particular, the regions that surround genomic insertions and deletions display an increased mutation rate, which might be induced by error-prone DNA replication (69, 75–77). The abundant recombination that is accompanied by insertions and deletions in the primate KIR cluster is likely to contribute to the extensive allelic KIR variation. Within two and three generations of human and macaque families studied, the birth of novel KIR alleles is described, which might further substantiate the rapid mutation rate in this gene cluster (29, 78). To our knowledge, such an event has not been recorded for the highly polymorphic MHC class I genes.
The variation involving KIR genes at the allele level impacts the interactions with their highly polymorphic MHC class I ligands, and demonstrate that point mutations contribute to a diversified KIR gene system. The general lack of allele level characterization in the clinic might limit the number of associations reported for KIR allele heterogeneity and their functional and disease-related effects. Even intronic variations might impact the KIR receptor expression and function. These few associations highlight the need to further characterize the KIR gene content of humans and other primate species at an allele level resolution.
Alternative Splicing as a Mechanism for Structural Diversification
The complexity of the primate KIR gene cluster is further extended by alternative splicing (Figure 2C) (79–83). This post-transcriptional mechanism can generate multiple messenger RNA (mRNA) transcripts from a single gene, which are translated into different receptor isoforms. Constitutive splicing excludes the intronic sequences from the precursor mRNA (pre-mRNA) and ligates the coding exons. Alternative splicing deviates from this pattern by the use of alternative splice sites, the skipping of exons, and the retention of introns (Figure 2C) (84). The alternative splice events for human and macaque KIR transcripts are well documented, and demonstrated that both in- and out-of-frame transcripts are generated (79–83). The out-of-frame transcripts often have an early stop codon, and this results in early truncation of the transcript. Even though these out-of-frame transcripts appear as a redundant side effect of alternative splicing, it might reflect a regulatory pathway to rapidly down-regulate receptor expression. The functional impact of the in-frame generated KIR isoforms may be diverse. The skipping of exons generates transcripts that encode modified KIR isoforms, which lack one or two extracellular domains, the stem region, or the transmembrane region. These KIR isoforms probably exhibit differential binding properties or are secreted as soluble receptors (Figure 2C) (85). In-frame splice events that involve alternative splice sites might insert a partial intronic sequence into the transcript or delete a part of a coding exon. Although the functional and structural consequences of these KIR isoforms are harder to predict, they are likely to modify the receptor expression level, cellular localization, and ligand interactions.
Several splice events were frequently recorded or were defined for multiple KIR genes, and implicate the existence of conserved splice events that generate structurally and functionally distinct isoforms. For example, exon 4 (coding for the D1 domain) is frequently skipped from KIR3DL20 transcripts in macaques, thereby generating transcripts that encode both the complete receptor and receptors with a D0-D2 domain configuration (43, 57). This macaque isoform is termed KIR2DL05, as it displays an 89.5% sequence similarity with human KIR2DL5. Moreover, it demonstrates that alternative splicing expands the macaque KIR repertoire by generating a second two-domain structure (KIR2DL) additional to KIR2DL04. The most frequent KIR splice event in humans involved the skipping of exon 6, which encodes the stem region. Other frequent events included the skipping of exon 5 (D2 domain) and partial deletions in exons 4 and 5. These events result in isoforms that are likely to display altered binding properties, but their exact activity and localization remains elusive. Another common splice event in humans might function as a regulatory switch for expression of the 9A and 10A KIR2DL4 alleles by restoring or disrupting the open reading frame (ORF) (79). Less frequent alternative splicing events were often found to be gene specific, and were mainly out-of-frame events that encoded for truncated receptors. Except for most exon skipping events, only a single splice event was shared between humans and macaques. This event involved a partial deletion of exon 3 (D0 domain) mediated by an alternative 5’ splice site (79). Data on the alternative splicing in other hominoids are lacking, but a similar extent of alternative splicing is likely to diversify their KIR receptors and repertoire.
The splicing of pre-mRNA not only facilitates diversification of the KIR repertoire, but might also compensate for genomic alterations that result in out-of-frame transcripts. The expression of human and macaque lineage III KIR genes, for example, requires the constitutive skipping of exon 3 to maintain an ORF. This exon contains a deletion of 5 bp at the genomic DNA level, which would shift the reading frame that introduces an early stop codon (79, 86). The constitutive skipping of exon 3 suggests that the expanded repertoire of human KIR2D receptors evolved from a KIR3D gene. The absence of a conserved 33 bp sequence in intron 2 of all human and macaque lineage III KIR genes might relate to the constitutive exon skipping by, for example, disrupting the spliceosome recognition site (79).
The extensive levels of alternative splicing observed in humans and macaques defines another layer of complexity for the KIR gene cluster. This diversifying mechanism generates structurally and functionally distinct receptor isoforms, and might be involved in the regulation of receptor expression levels. Although not all isoforms might be functional, the frequency and consistency of several alternative splicing events suggest that alternative splicing is a rapid mechanism to diversify the KIR content in hominoids and Old World monkeys.
Differential NK Cell Populations Due to Variegated KIR Gene Expression
KIR gene plasticity is further reflected by the stochastic expression of a subset of KIR genes from the total gene repertoire in individual NK cells (Figure 2D). This selective transcriptional activation generates specialized NK cell populations, which express different numbers and combinations of KIR genes (87, 88). The stochastic KIR expression is activated during NK cell maturation, and the transcriptional pattern is maintained by the methylation of silenced KIR genes (28, 89). The different KIR receptor combinations are generated largely at random, but might be shaped by the individual KIR gene frequencies and the MHC class I repertoire. Therefore, KIR genes that are present on both chromosomes in heterozygous individuals, or genes that are present as two or more allotypes on a single haplotype (e.g., by duplication or gene insertion), could be expressed in a mono- and multi-allelic manner. This may generate NK cell subsets that transcribe two or more allelic copies of a certain KIR gene (28). Divergent expression patterns are documented for human KIR2DL4, which is expressed in all NK cells, and for KIR3DL3, which is expressed at low levels (90, 91).
The molecular regulation of KIR gene expression is well studied in humans, and involves multiple promoter regions in the intergenic sequences that control gene demethylation and transcription (27, 90–96). The proximal promoter is located directly in front of the first exon of a KIR gene and functions as a probabilistic switch (Figure 2D). Bi-directional transcription of this promoter generates forward and reverse transcripts that correlate with the activation and suppression of KIR gene transcription, respectively. Forward transcripts of a distal promoter are associated with activation of the proximal promoter region and appear to be required for eventual KIR gene expression. A third promoter upstream of the proximal promoter, also denoted as the intermediate promoter, modulates the bidirectional transcription of the proximal promoter directly or indirectly by mediating correct splicing of the forward proximal promoter transcripts (27, 94). In all human KIR genes, the promoter regions are highly conserved, with 91–99.6% sequence similarity. Exceptions are found for the promoters of KIR2DL4 and KIR3DL3, which substantiates their diverged expression profile (94). Three types of promoter regions are defined for human KIR2DL5, which display considerable differences in their nucleotide sequence and transcription factor binding sites. Types I and III control variegated expression, whereas transcripts of KIR2DL5 alleles that exhibit the type II promoter are undetectable (97, 98). These type II promoters are probably inactivated by a SNP in their Runt-related transcription factor (RUNX) transcription binding site, which is an important motif in the regulation of gene expression, and is generally conserved in all KIR genes (98). An identical SNP is identified in the proximal promoter of the pseudogene KIR3DP1, and might indicate that the inactive type II promoter is swapped to particular KIR2DL5 alleles by chromosomal recombination (98–100). Within the KIR promoter regions, multiple other transcription factor binding sites are identified, which can vary per KIR gene and thereby contribute to differential gene expression. Allelic variations of the different transcription factor binding sites modulate the expression levels of KIR alleles (27, 92). For example, a KIR2DL1 allele displayed low expression, which was associated with three SNPs in the distal promoter that generated a binding site for the Zinc finger E-box-binding homeobox 1 (ZEB1) protein (27). This transcription factor is associated with the down-regulation of IL2 expression, and might have a similar impact on the expression of this specific KIR2DL1 allele. Just like the variation in the KIR gene introns, the nucleotide polymorphisms in the promoter regions are grossly undervalued, despite the direct impact on the expression of KIR alleles.
The variegated expression pattern of the KIR genes defines NK cell subsets, of which several are tissue resident. These NK cell populations might execute specialized functions in particular tissues that could be mediated by specific sets of KIR receptors. For example, the KIR expression profile of NK cells that were derived from the lung, liver, and uterus deviates from the expression pattern observed in peripheral blood NK cells (101–103). Expression of KIR was also established for subsets of T cells, in particular terminally differentiated CD8 + T cells, of which 30% exhibited KIR expression (104–106). The majority of these T cells dominantly express a single inhibitory or activating KIR gene, which is generally distinct from the KIR gene expression pattern on NK cells within the same individual (104). The expression pattern of NK cells and CD8 + T cells can be erased by in vitro treatment with a methylation inhibitor (5-azacytidine), and thereby induce the expression of formerly silenced KIR genes (28, 96, 107). This demonstrates that the stochastic KIR gene expression is maintained by methylation in both types of lymphocytes.
The variability in the promoter regions that is mainly generated by point mutations and chromosomal recombination events contributes to the diversification of NK cell subsets by the stochastic methylation of KIR genes. The promoter regions and epigenetic regulation of the KIR gene cluster in non-human primate species are less well characterized, but their stochastic expression pattern indicates a similar genetic mechanism.
The Different Characters of Diversification in the KIR and MHC Clusters
The expansion of the primate KIR cluster was probably initiated by the integration of multiple retroviral elements near or in the founding KIR genes. Subsequent duplications were mediated by these transposable elements, and this process had an impact on the expansion of the KIR gene repertoire (35). These recombination events might have enhanced the mutation rate within this genomic region that generated a diverse set of KIR alleles, and subsequently some of these were positively selected during evolution. In the case of exons, the point mutations may affect the receptor structure, function, localization, and expression, whereas polymorphisms in the introns may enhance the level of alternative splicing by affecting existing or generating alternative splice sites. In addition, the high level of point mutations caused variation within the promoter regions, and thereby modulated the variegated expression pattern and expression level of KIR receptors. It appears that all the different molecular mechanisms are intertwined and enhanced by each other, which multiplies their diversifying impact on the primate KIR gene system.
The MHC class I gene family is considered one of the most polymorphic genomic regions in primates, but displays a different nature of diversity as compared to its KIR ligands. In hominoids, the fixed number of MHC-A, -B, and -C genes on a haplotype indicate low levels of recent duplications and chromosomal recombination, which is substantiated by an exceptionally low recombination rate for the MHC class I region (108, 109). Chromosomal rearrangements that are accompanied by the formation of an MHC class I fusion gene, as is determined for the KIR genes, is to our knowledge not known. In most hominoids, MHC class I polymorphism is mainly generated by point mutations in concert with a recombination of small segments. These genetic modifications are especially located in the exons encoding the peptide-binding site, and indicate a rigorous selection for a diverse array of allotypes. The functional impact is reflected in differential peptide presentation (18). Additional modification of the MHC repertoire is reflected at the transcription level by alternative splicing, which is reported for human and macaque MHC transcripts (110–114). Considering the high level of allelic polymorphism in the HLA genes, which may involve nucleotide substitutions that disrupt existing or generate novel alternative splice sites, one might expect abundant alternative splicing events in their transcripts. However, only a modest level of alternative splicing is demonstrated for several classical and non-classical HLA class I alleles, which mainly involved exon skipping that abrogated receptor surface expression (110). Specific isoforms of the non-classical HLA-G, however, are well known and are associated with cancer and inflammatory diseases (115–118). In contrast, alternative splicing in primate KIR was not limited to certain alleles, and also comprised conserved splice events that were common to multiple KIR genes and lineages (79). The classical MHC class I allotypes are constitutively expressed on all nucleated cells, and thereby lack a variegated expression pattern (119, 120). However, individual MHC allotypes may display a differential expression level, which is affected by sequence variation, tissue distribution, and pathogenic encounters (120, 121). In humans, the relative surface expression of HLA-A and -B is approximately ten times higher compared to HLA-C molecules (120, 122). This suggests that the HLA-C gene might slowly shift its main function from classical antigen presentation into the modulation of NK cell responses during infection and reproductive biology. In addition, the expression levels of different HLA-C alleles display variation, in which highly expressed allotypes correlated with a beneficial control of HIV infection (123). The differential expression pattern is also determined for the expanded MHC class I region in macaques, with only a few highly expressed MHC-A and -B allotypes (19, 124). The MHC expression levels are, however, not strictly maintained and can be modulated during infection by immune regulators such as interferon and tumor necrosis factor (TNF) (120).
The primate KIR and MHC gene families are both reflected by great complexity, and seem to co-evolve to maintain a functional relationship. The MHC class I diversification mainly involved allelic polymorphism in the exons encoding the peptide binding site and recombination of small segments, which is driven by the arms race with rapidly evolving pathogens. The KIR genes, in contrast, are diverged by haplotype expansion and contraction, random point mutations, and the generation of novel fusion genes. The expression and structural variability of the KIR receptors are further modified at the epigenetic and post-transcriptional level, whereas a similar diversification of the MHC class I molecules is limited. The conjunction of different genetic mechanisms generates an extensive plasticity for the primate KIR gene cluster, which seems to exceed the diversity of the polymorphic MHC class I genes.
CD94:NKG2A- or KIR-Dependent Education in Different Primate Species
A comparison of the KIR gene system in primate species illustrates a variable degree of gene expansion, reflected in the differential expansion of gene lineages (Figure 1). This might be largely due to co-evolution with their diverse MHC class I repertoire. The variable extent of expansion, however, is emphasized by the number of functional genes per KIR haplotype and by the overall size of the KIR gene repertoire documented for a certain primate species. The extremes are represented by the heavily contracted KIR haplotypes in bonobos versus the widely expanded set of KIR genes in macaques (Figure 1). The flexibility to expand and contract KIR haplotypes and repertoires, apparently without compromising sufficient and protective immune responses, might be closely related to the nature of NK cell education in different primate species.
Natural killer cells require self-tolerance and a signal to activate, which are acquired through an educational process. NK cell education involves the recognition of self-MHC class I molecules or the presented peptides by at least one inhibitory NK cell receptor. Alternative educational pathways that are MHC-independent are reported, but their exact contribution to the acquiring of NK cell functions is elusive (125, 126). The MHC-dependent education is predominant and can be approached in two ways (Figure 3) (13, 127, 128). One strategy of NK cell education involves the interaction of inhibitory CD94:NKG2A NK cell receptors with the non-polymorphic MHC-E molecules, which are complexed with conserved signal peptides derived from the diversified classical MHC class I molecules (129–131). One could argue that this approach allows the immune system to scan in a crude way whether total MHC class I expression has been abrogated. In the complementary approach, however, NK cell education is established through interaction of the MHC class I molecules with polymorphic KIR receptors. This seems to reflect a more sophisticated strategy in which the immune system checks at the epitope level for a malfunctioning of MHC class I expression. KIR-dependent NK cell education is mainly conducted through the interactions of inhibitory KIR and MHC class I molecules. However, activating KIR contribute to the tuning of NK cell responsiveness by dampening NK cell activity upon MHC class I recognition (132). Currently, only for KIR2DS1 the effect on NK cell education is described. In the following sections, we mainly consider the educational impact of inhibitory KIR.
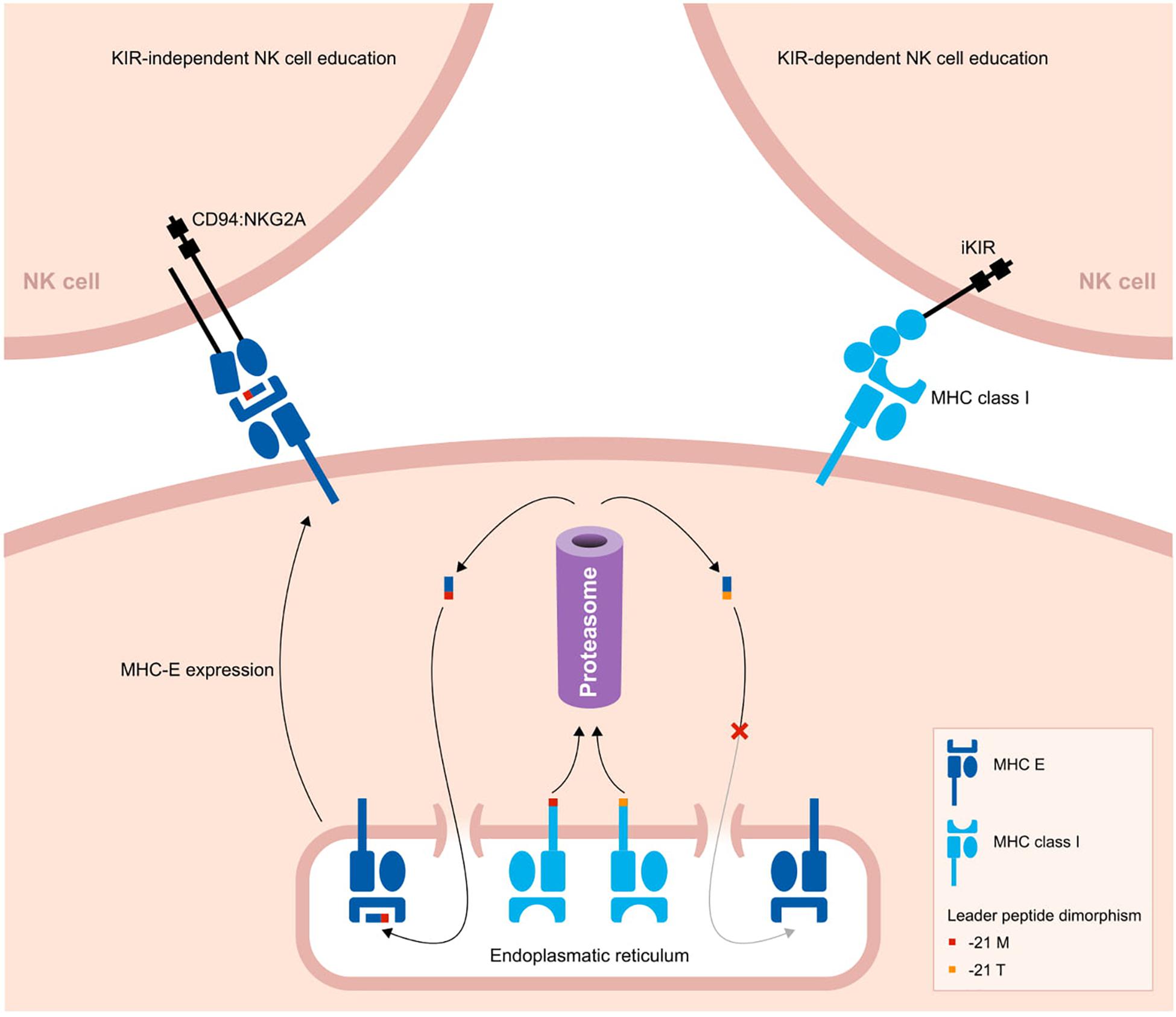
Figure 3. Two pathways to educate NK cells in primates. A schematic overview of two strategies to educate NK cells. The leader peptide of MHC class I molecules either contains a methionine (−21M) or threonine (−21T) residue. The -21M peptides strongly bind to MHC-E molecules, and these complexes display an increased cell surface level. Through the conserved CD94:NKG2A receptors, the MHC-E complexes educate and license NK cells. In contrast, -21T leader peptides, which are predominantly present in MHC-B allotypes of humans and chimpanzees, do not interact with MHC-E molecules. Therefore, in the presence of one or more -21T MHC class I allotypes, the cell surface level of MHC-E is decreased. In this case, NK cells are educated by MHC class I molecules that interact with their KIR ligands.
Whether the NK cells are educated by the CD94:NKG2A or KIR pathway might depend on a single nucleotide dimorphism at position 21 of the MHC class I leader sequences. Most MHC-A and -C molecules in hominoids have a methionine (−21M) residue present at this position, whereas in general this position is occupied by threonine (−21T) in MHC-B molecules. The -21M peptides strongly bind to MHC-E molecules and promote cell surface expression of MHC-E complexes (133). The presence of five or six classical MHC class I allotypes containing the -21M residue drives the NK cell education toward the more conserved MHC-E and CD94:NKG2A interactions. However, approximately 62% of human individuals display a -21T HLA-B homozygous genotype, with a variable distribution in different populations (127). In chimpanzees, -21T is near fixed in their MHC-B allotypes (13). The homozygous threonine genotype corresponds with a low MHC-E surface expression. As a consequence, human and chimpanzee NK cells are largely educated by their KIR repertoire (13, 127). In contrast, in macaque MHC-A and -B allotypes, methionine is the predominant residue at position 21 of the leader sequence, which results in an NK cell education that mostly relies on the conserved CD94:NKG2A pathway (127).
In primate species with a KIR-dependent NK cell education, one can envision that an expanded KIR repertoire may compromise NK cell activity. This might drive selection for a limited KIR expansion, as we will discuss in the next section. If this reasoning is true, the KIR-independent education of NK cells in macaques might result in an extensive expansion of their KIR gene system. We think that the primary function of macaque KIR is focused on the recognition and elimination of infected or malignant cells. This defense mechanism relies on the recognition of Bw4 and Bw6 epitopes, but KIR interactions are also sensitive to non-self peptides that can be presented by MHC class I molecules (134–138). A large genetic diversity of KIR genes provides a broader repertoire to scan all the variable MHC class I allotypes in combination with their peptides originating from pathogens. It has been proposed that up to seven distinct KIR receptors are required for successful peptide recognition (139). This optimal receptor count might even be higher when the Bw4 and Bw6 epitope specificity is considered for the different KIR allotypes. The high level of chromosomal recombination and the relatively frequent formation of fusion genes in macaques might indicate selection for a widely diversified KIR gene system. Considering their KIR-independent NK cell education, KIR expansion in macaques might be exempted from potential negative selection on large KIR gene repertoires.
However, not all macaque KIR haplotypes contain a large number of genes, and they even display indications for contraction by chromosomal recombination events. The formation of novel gene entities by the shuffling of head- and tail-encoding exons is achieved by recombination events, which are coherently accompanied both by contractions and expansions of KIR haplotypes. There might be a trade-off between the expansion of the overall KIR repertoire in a population by generating fusion genes and the contraction of KIR haplotypes in individuals. Rapid expansion and diversification generate a highly plastic macaque KIR gene system that appears to be maintained by selection to militate against rapidly evolving pathogens.
KIR Haplotype Expansion and Contraction: Finding the Equilibrium
As compared to macaques, hominoids appear to have a more limited haplotype content and overall KIR repertoire (Figure 1 and Table 1). These limitations might be maintained by selective pressure on an efficient KIR-dependent NK cell education, but should be balanced with protection against infections. This balance might be reflected in the slightly variable KIR gene content per haplotype.
A large KIR repertoire is likely to provide a broad array of MHC class I specificities that may result in the education of an increased fraction of NK cells (Figure 4A). Moreover, the expression of multiple self-specific inhibitory KIR receptors by NK cell clones enhances the magnitude of their effector response (140). Although only a small population of NK cells dominantly expresses more than one inhibitory KIR receptor, an expanded KIR repertoire might enlarge this NK cell population size and elevate the strength of the NK cell response (Figures 4A,C). A potential detrimental effect of an expanded KIR haplotype might emerge if the repertoire comprises only a few or abundant self-specific receptors. On the one hand, the variegated expression of a large KIR repertoire that consists of few self-specific receptors might thin out the educated NK cell population and provide an inefficient immune surveillance (Figure 4B). Indications for a biased expression of self-specific KIR suggest modulation of the KIR expression by an individual’s MHC class I repertoire (62, 140, 141), which would ensure a more robust immune response and might compensate for a large non-self-specific KIR expansion. On the other hand, a large repertoire of self-specific KIR might enlarge the fraction of educated NK cells that display increased activity, which might be protective in infections and cancer (Figure 4C). However, elevated NK cell activity, which might be further enhanced by the expression of multiple self-specific KIR on NK cell subsets, or excessive NK cell inhibition by abundant self-specific KIR interactions are also associated with implantation failure and recurrent miscarriages (142–145). Furthermore, overactivation might desensitize NK cells and result in hyporeactivity (146), which might weaken subsequent immune responses. Therefore, a large KIR repertoire that is used in NK cell education might act as a double-edged sword that can both enhance and compromise an individual’s immune response.
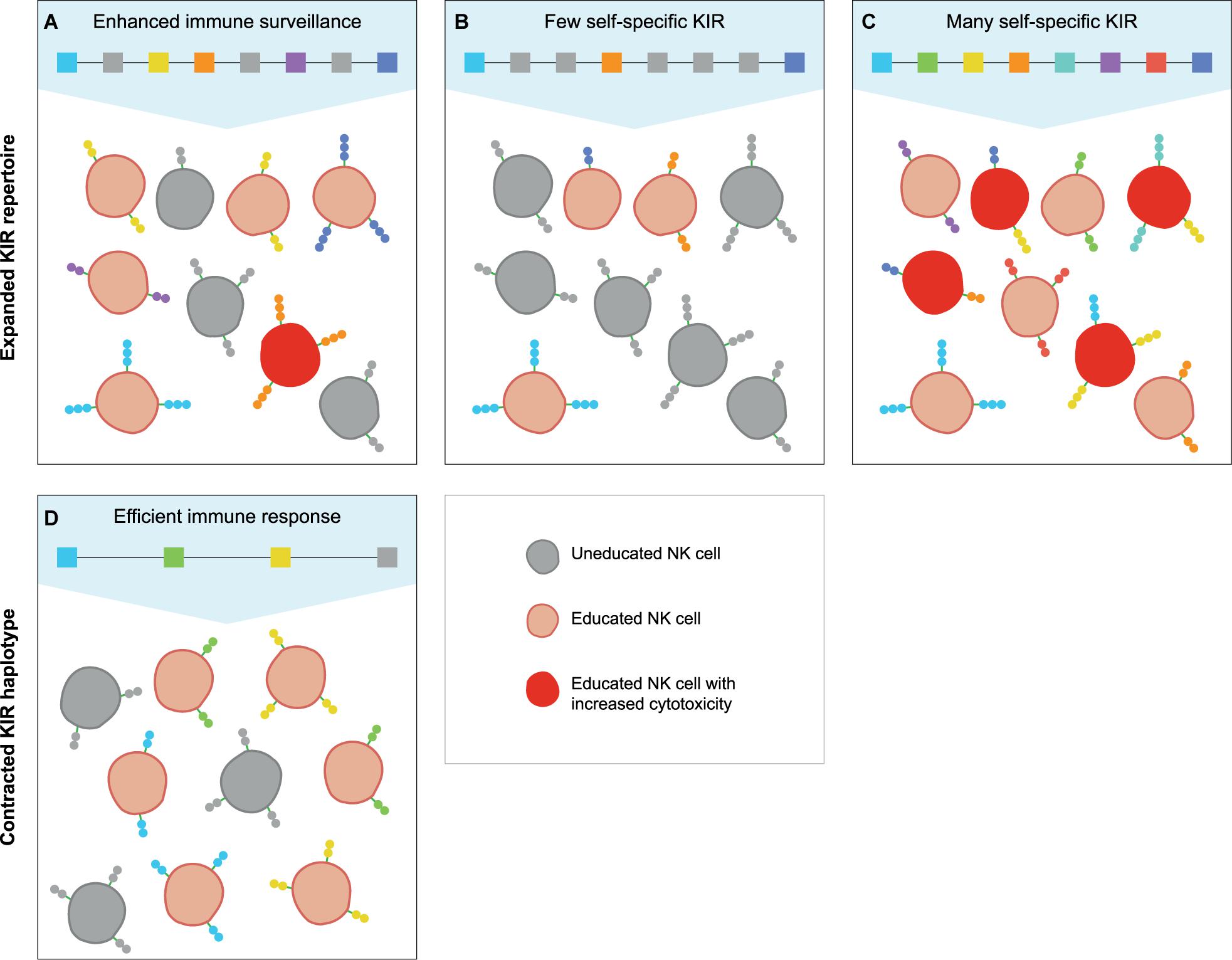
Figure 4. The education of NK cells by expanded and contracted KIR haplotypes. The proposed populations of NK cells that are educated (red cells) by self-specific KIR (colored boxes and receptors) on expanded and contracted haplotypes. The gray boxes represent non-self-specific KIR, and are displayed on the uneducated NK cell clones (gray cells). (A) An expanded KIR haplotype provides a broad MHC class I specificity, and might educate some NK cell clones through multiple self-specific receptors, which increases their cytotoxicity (dark red cells). (B) A large KIR repertoire with only a few self-specific KIR might lower the fraction of educated NK cells and thereby provide an insufficient immune response, (C) whereas abundant self-specific receptors increase the educated fraction and their cytotoxicity. (D) A small KIR repertoire might educate large fractions of NK cells with a limited MHC class I specificity, which might provide largely sufficient immune responses.
In contrast, individuals that have a limited KIR haplotype rely on only one or few self-specific KIR receptors to educate their NK cells (Figure 4D). Even though a sufficient percentage of NK cells might be educated by a limited KIR repertoire, the specificity is restricted, and specialised NK cell populations might be lacking. The complete absence of NK cell education occurs in MHC class I-deficient mice, which display a near normal NK cell count with an overall reduced responsiveness (147, 148). In humans and other hominoid species, individuals that completely lack self-specific KIR are not documented. This indicates that even minimal KIR haplotypes provide education, and suggests that framework KIR receptors could play a substantial role in the NK cell education of hominoids. In addition, the chance that an individual completely lacks self-specific KIR receptors is reduced by the heterozygous nature of the KIR gene cluster. As far as we know, only few human and no non-human primate individuals are documented that were homozygous for their KIR haplotypes at an allele level (149). In a rhesus macaque family studied, one individual was assumed to be KIR-homozygous according to segregation. However, more detailed analysis illustrated that one KIR gene copy appeared to have gained point mutations that resulted in the haplotypes diverging at an allele level (29). This individual macaque possessed a largely homozygous KIR content, but did not display an impaired immune system; it also produced healthy offspring, which suggests that KIR-heterozygosity is not vital. However, KIR haplotype diversity might compensate for limited KIR haplotypes and improve the immune surveillance, as is also described for MHC heterozygosity (150–152).
In contrast to non-self-specific T lymphocytes, which are depleted upon a failed positive or negative selection in the thymus, uneducated NK cells are present in the peripheral blood. The relatively high level of uneducated NK cells in individuals with small or large non-self-specific KIR repertoires could affect their immune surveillance, but does not preclude an efficient immune response during infection or tumor formation. In fact, unlicensed NK cells appear to be more efficient at eradicating infected or malignant cells that persistently express MHC class I molecules or viral mimic ligands through their reactivation by cytokines or NKG2D receptors (153–155). Therefore, a fraction of uneducated NK cells in combination with a largely educated NK cell population might be more protective than a completely educated NK cell pool with broad MHC class I specificity.
There could be another factor, however, that limits expansion of the KIR haplotypes and gene repertoire, in addition to their role in NK cell education. In orangutans, MHC-B allotypes contain a -21M leader peptide, which would suggest education via the conserved CD94:NKG2A pathway (127). In contrast to macaques, the orangutan KIR system is not extensively expanded, and is more in line with other hominoids that display a KIR-dependent NK cell education. The emergence of MHC-C as a specialized ligand for KIR might override the dimorphism and coherent increase in MHC-E expression, and drive NK cell education via the KIR receptors. In addition, the number of characterized MHC-B molecules in orangutans is relatively low (IPD-MHC, release 3.4.0.1) (156). A larger sample group of orangutans or additional functional studies would be required to test our hypothesis for the differential KIR expansion in primate species that exert a KIR-independent or -dependent NK cell education.
Nevertheless, the diverse KIR haplotype content and overall gene repertoire in hominoids and Old World monkeys are likely to affect the education, activity, and function of their NK cells, but the precise effect of the haplotype expansions and contractions remains ambiguous. The equal distribution of both small and large KIR repertoires in humans and macaques indicates a balancing selection, which might be an ongoing process to achieve a haplotype equilibrium that serves differential functions, such as fighting infections and promoting successful pregnancy.
Conclusion
The KIR gene system is well studied in humans, and reveals multiple mechanisms that contribute to the plasticity of this immunogenetic cluster (Figure 2). In other hominoid species, such as chimpanzees and orangutans, indications for a similar diversifying genetic toolset is evident, although robust data on some mechanisms are lacking, such as alternative splicing and variegated expression. The variability of the extensively diversified KIR gene cluster in macaques exceeds that observed in hominoids, with a prominent expansion of the lineage II KIR genes, which is largely mediated by recombination events. The rapid evolution of the KIR gene cluster may counteract the adaptive nature of pathogens. The species-specific diversification of the KIR gene cluster might be largely driven by co-evolution with their diversified MHC class I repertoire and thereby indirectly by the arms race with pathogens. In addition, a KIR-dependent or -independent NK cell education might impact the variable haplotype content and the extent of KIR gene expansion. Nevertheless, the different molecular mechanisms responsible for diversification of the KIR gene cluster are shared in Old World monkeys and hominoids, which suggests an evolutionary effort to diversify the KIR gene system.
Author Contributions
JB wrote the manuscript. NG and RB edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank D. Devine for editing the manuscript and F. van Hassel for preparing the figures.
References
1. Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. (1990) 11:237–44. doi: 10.1016/0167-5699(90)90097-s
2. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. (2008) 9:503–10.
3. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
4. Brodin P, Karre K, Hoglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. (2009) 30:143–9. doi: 10.1016/j.it.2009.01.006
5. Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. (2005) 1:129–39. doi: 10.1371/journal.pgen.0010027
6. Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. (2000) 51:268–80. doi: 10.1007/s002510050620
7. Bruijnesteijn J, de Groot NG, Otting N, Maccari G, Guethlein LA, Robinson J, et al. Nomenclature report for killer-cell immunoglobulin-like receptors (KIR) in macaque species: new genes/alleles, renaming recombinant entities and IPD-NHKIR updates. Immunogenetics. (2020) 72:37–47. doi: 10.1007/s00251-019-01135-8
8. Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. (2003) 55:220–6.
9. Robinson J, Guethlein LA, Maccari G, Blokhuis J, Bimber BN, de Groot NG, et al. Nomenclature for the KIR of non-human species. Immunogenetics. (2018) 70:571–83. doi: 10.1007/s00251-018-1064-4
10. Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. (2006) 7:209. doi: 10.1186/1471-2164-7-209
11. Sanderson ND, Norman PJ, Guethlein LA, Ellis SA, Williams C, Breen M, et al. Definition of the cattle killer cell Ig-like receptor gene family: comparison with aurochs and human counterparts. J Immunol. (2014) 193:6016–30. doi: 10.4049/jimmunol.1401980
12. Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. (2007) 59:517–22. doi: 10.1007/s00251-007-0214-x
13. Wroblewski EE, Parham P, Guethlein LA. Two to tango: co-evolution of hominid natural killer cell receptors and MHC. Front Immunol. (2019) 10:177. doi: 10.3389/fimmu.2019.00177
14. de Groot NG, Heijmans CMC, van der Wiel MKH, Blokhuis JH, Mulder A, Guethlein LA, et al. Complex MHC class I gene transcription profiles and their functional impact in orangutans. J Immunol. (2016) 196:750. doi: 10.4049/jimmunol.1500820
15. Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, et al. Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3. (2013) 3:1195–201. doi: 10.1534/g3.113.006254
16. Doxiadis GGM, de Groot N, Otting N, Blokhuis JH, Bontrop RE. Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics. (2011) 63:73–83. doi: 10.1007/s00251-010-0486-4
17. Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. (2004) 14:1501–15. doi: 10.1101/gr.2134504
18. de Groot NG, Blokhuis JH, Otting N, Doxiadis GG, Bontrop RE. Co-evolution of the MHC class I and KIR gene families in rhesus macaques: ancestry and plasticity. Immunol Rev. (2015) 267:228–45. doi: 10.1111/imr.12313
19. Otting N, Heijmans CMC, Noort RC, de Groot NG, Doxiadis GGM, van Rood JJ, et al. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci USA. (2005) 102:1626–31. doi: 10.1073/pnas.0409084102
20. Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O’Connor DH. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J. (2013) 54:196–210. doi: 10.1093/ilar/ilt036
21. Schafer JL, Colantonio AD, Neidermyer WJ, Dudley DM, Connole M, O’Connor DH, et al. KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: maintenance of Bw4 specificity since the divergence of apes and Old World monkeys. J Immunol. (2014) 192:1907–17. doi: 10.4049/jimmunol.1302883
22. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. (2013) 13:133–44. doi: 10.1038/nri3370
23. Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus macaque inhibitory and activating KIR3D interact with Mamu-A-encoded ligands. J Immunol. (2011) 186:2156–63. doi: 10.4049/jimmunol.1002634
24. Bimber BN, Evans DT. The killer-cell immunoglobulin-like receptors of macaques. Immunol Rev. (2015) 267:246–58. doi: 10.1111/imr.12329
25. Hermes M, Weil S, Groth A, Dressel R, Koch J, Walter L. Characterisation of mouse monoclonal antibodies against rhesus macaque killer immunoglobulin-like receptors KIR3D. Immunogenetics. (2012) 64:845–8. doi: 10.1007/s00251-012-0640-2
26. Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P, et al. Characterization of killer immunoglobulin-like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genomics. (2011) 12:295. doi: 10.1186/1471-2164-12-295
27. Wright PW, Li H, Huehn A, O’Connor GM, Cooley S, Miller JS, et al. Characterization of a weakly expressed KIR2DL1 variant reveals a novel upstream promoter that controls KIR expression. Genes Immun. (2014) 15:440–8. doi: 10.1038/gene.2014.34
28. Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. (2003) 197:245–55. doi: 10.1084/jem.20021127
29. Bruijnesteijn J, de Groot N, van der Wiel MKH, Otting N, de Vos-Rouweler AJM, de Groot NG, et al. Unparalleled rapid evolution of KIR genes in rhesus and cynomolgus macaque populations. J Immunol. (2020) 204:1770–86. doi: 10.4049/jimmunol.1901140
30. Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. (2002) 23:81–8. doi: 10.1016/s1471-4906(01)02155-x
31. Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. (2005) 15:25–35. doi: 10.1101/gr.2381205
32. Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. (1999) 10:154–60. doi: 10.1007/s003359900961
33. Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. (2013) 14:301–23. doi: 10.1146/annurev-genom-091212-153455
34. Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. (2010) 19:737–51. doi: 10.1093/hmg/ddp538
35. Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. (2007) 179:491–504. doi: 10.4049/jimmunol.179.1.491
36. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. (2000) 97:4778. doi: 10.1073/pnas.080588597
37. Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. (2003) 73:823–34. doi: 10.1086/378594
38. Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. (2006) 79:41–53. doi: 10.1086/504600
39. Han K, Lee J, Meyer TJ, Wang J, Sen SK, Srikanta D, et al. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. (2007) 3:e184. doi: 10.1371/journal.pgen.0030184
40. Roe D, Vierra-Green C, Pyo CW, Eng K, Hall R, Kuang R, et al. Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes Immun. (2017) 18:127–34. doi: 10.1038/gene.2017.10
41. Rajalingam R, Gardiner CM, Canavez F, Vilches C, Parham P. Identification of seventeen novel KIR variants: fourteen of them from two non-Caucasian donors. Tissue Antigens. (2001) 57:22–31. doi: 10.1034/j.1399-0039.2001.057001022.x
42. Pyo C-W, Wang R, Vu Q, Cereb N, Yang SY, Duh F-M, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics. (2013) 14:89. doi: 10.1186/1471-2164-14-89
43. Bruijnesteijn J, van der Wiel MKH, Swelsen WTN, Otting N, de Vos-Rouweler AJM, Elferink D, et al. Human and rhesus macaque KIR haplotypes defined by their transcriptomes. J Immunol. (2018) 200:1692–701.
44. Vierra-Green C, Roe D, Hou L, Hurley CK, Rajalingam R, Reed E, et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR Loci in 506 European-American individuals. PLoS One. (2012) 7:e47491. doi: 10.1371/journal.pone.0047491
45. Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. (2009) 19:757–69. doi: 10.1101/gr.085738.108
46. Leaton LA, Shortt J, Kichula KM, Tao S, Nemat-Gorgani N, Mentzer AJ, et al. Conservation, extensive heterozygosity, and convergence of signaling potential all indicate a critical role for KIR3DL3 in higher primates. Front Immunol. (2019) 10:24. doi: 10.3389/fimmu.2019.00024
47. Rajalingam R, Hong M, Adams EJ, Shum BP, Guethlein LA, Parham P. Short KIR haplotypes in pygmy chimpanzee (Bonobo) resemble the conserved framework of diverse human KIR haplotypes. J Exp Med. (2001) 193:135–46. doi: 10.1084/jem.193.1.135
48. de Groot NG, Heijmans CMC, Helsen P, Otting N, Pereboom Z, Stevens JMG, et al. Limited MHC class I intron 2 repertoire variation in bonobos. Immunogenetics. (2017) 69:677–88. doi: 10.1007/s00251-017-1010-x
49. Maibach V, Vigilant L. Reduced bonobo MHC class I diversity predicts a reduced viral peptide binding ability compared to chimpanzees. BMC Evol Biol. (2019) 19:14. doi: 10.1186/s12862-019-1352-0
50. Wroblewski EE, Guethlein LA, Norman PJ, Li Y, Shaw CM, Han AS, et al. Bonobos maintain immune system diversity with three functional types of MHC-B. J Immunol. (2017) 198:3480. doi: 10.4049/jimmunol.1601955
51. de Groot NG, Stevens JMG, Bontrop RE. Does the MHC confer protection against malaria in bonobos? Trends Immunol. (2018) 39:768–71. doi: 10.1016/j.it.2018.07.004
52. Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. (2020) 48:D783–8.
53. Nakimuli A, Chazara O, Farrell L, Hiby SE, Tukwasibwe S, Knee O, et al. Killer cell immunoglobulin-like receptor (KIR) genes and their HLA-C ligands in a Ugandan population. Immunogenetics. (2013) 65:765–75. doi: 10.1007/s00251-013-0724-7
54. Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. (2010) 6:e1001192. doi: 10.1371/journal.pgen.1001192
55. Guethlein LA, Norman PJ, Heijmans CMC, de Groot NG, Hilton HG, Babrzadeh F, et al. Two orangutan species have evolved different KIR alleles and haplotypes. J Immunol. (2017) 198:3157–69. doi: 10.4049/jimmunol.1602163
56. NCBI Macaca Mulatta Isolate AG07107 Chromosome 19, Whole Genome Shotgun Sequence. (2018). Available online at: https://www.ncbi.nlm.nih.gov/nuccore/CM014354 (accessed July 01, 2020).
57. Blokhuis JH, van der Wiel MK, Doxiadis GGM, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics. (2010) 62:295–306. doi: 10.1007/s00251-010-0434-3
58. VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. (2006) 177:5347–57. doi: 10.4049/jimmunol.177.8.5347
59. Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1. (3DL1∗004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. (2003) 171:6640–9. doi: 10.4049/jimmunol.171.12.6640
60. Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. (2013) 190:6198–208. doi: 10.4049/jimmunol.1300464
61. Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, et al. Loss and gain of natural killer cell receptor function in an african hunter-gatherer population. PLoS Genet. (2015) 11:e1005439. doi: 10.1371/journal.pgen.1005439
62. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. (2006) 203:633–45. doi: 10.1084/jem.20051884
63. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. (2007) 39:733–40. doi: 10.1038/ng2035
64. Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. (2001) 166:2992–3001. doi: 10.4049/jimmunol.166.5.2992
65. Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SGE. IPD–the immuno polymorphism database. Nucleic Acids Res. (2013) 41:D1234–40.
66. Cooper DN. Functional intronic polymorphisms: buried treasure awaiting discovery within our genes. Hum Genomics. (2010) 4:284–8. doi: 10.1186/1479-7364-4-5-284
67. Toscano C, Klein K, Blievernicht J, Schaeffeler E, Saussele T, Raimundo S, et al. Impaired expression of CYP2D6 in intermediate metabolizers carrying the ∗41 allele caused by the intronic SNP 2988G>A: evidence for modulation of splicing events. Pharmacogenet Genomics. (2006) 16:755–66. doi: 10.1097/01.fpc.0000230112.96086.e0
68. Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. (2012) 86:6979–85. doi: 10.1128/jvi.00406-12
69. Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. (2011) 12:756–66. doi: 10.1038/nrg3098
70. Hwang DG, Green P. Bayesian markov chain monte carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. (2004) 101:13994–4001. doi: 10.1073/pnas.0404142101
71. Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. (2000) 156:297–304.
72. Nachman MW. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. (2001) 17:481–5. doi: 10.1016/s0168-9525(01)02409-x
73. Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. (2002) 18:337–40. doi: 10.1016/s0168-9525(02)02669-0
74. Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. (2008) 4:e1000071. doi: 10.1371/journal.pgen.1000071
75. Tian D, Wang Q, Zhang P, Araki H, Yang S, Kreitman M, et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. (2008) 455:105–8. doi: 10.1038/nature07175
76. Zhu L, Wang Q, Tang P, Araki H, Tian D. Genomewide association between insertions/deletions and the nucleotide diversity in bacteria. Mol Biol Evol. (2009) 26:2353–61. doi: 10.1093/molbev/msp144
77. McDonald MJ, Wang W-C, Huang H-D, Leu J-Y. Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. (2011) 9:e1000622. doi: 10.1371/journal.pbio.1000622
78. Maxwell LD, Williams F, Gilmore P, Meenagh A, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity: II. KIR2DS4. Hum Immunol. (2004) 65:613–21. doi: 10.1016/j.humimm.2004.02.028
79. Bruijnesteijn J, van der Wiel MKH, de Groot N, Otting N, de Vos-Rouweler AJM, Lardy NM, et al. Extensive alternative splicing of KIR transcripts. Front Immunol. (2018) 9:2846. doi: 10.3389/fimmu.2018.02846
80. Vilches C, Rajalingam R, Uhrberg M, Gardiner CM, Young NT, Parham P. KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig-like domains. J Immunol. (2000) 164:5797. doi: 10.4049/jimmunol.164.11.5797
81. Dohring C, Samaridis J, Colonna M. Alternatively spliced forms of human killer inhibitory receptors. Immunogenetics. (1996) 44:227–30. doi: 10.1007/s002510050116
82. Wilson MJ, Torkar M, Trowsdale J. Genomic organization of a human killer cell inhibitory receptor gene. Tissue Antigens. (1997) 49:574–9. doi: 10.1111/j.1399-0039.1997.tb02804.x
83. Blokhuis JH, Doxiadis GG, Bontrop RE. A splice site mutation converts an inhibitory killer cell Ig-like receptor into an activating one. Mol Immunol. (2009) 46:640–8. doi: 10.1016/j.molimm.2008.08.270
84. Lee Y, Rio DC. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu Rev Biochem. (2015) 84:291–323.
85. Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. (2007) 37:199–211. doi: 10.1002/eji.200636316
86. Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. (2004) 172:356–69. doi: 10.4049/jimmunol.172.1.356
87. Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. (1997) 7:739–51. doi: 10.1016/s1074-7613(00)80393-3
88. Andersson S, Malmberg JA, Malmberg KJ. Tolerant and diverse natural killer cell repertoires in the absence of selection. Expe Cell Res. (2010) 316:1309–15. doi: 10.1016/j.yexcr.2010.02.030
89. Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. (2002) 169:4253–61. doi: 10.4049/jimmunol.169.8.4253
90. Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. (2003) 170:6073–81. doi: 10.4049/jimmunol.170.12.6073
91. Trompeter HI, Gomez-Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C, et al. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol. (2005) 174:4135–43. doi: 10.4049/jimmunol.174.7.4135
92. Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the Bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. (2008) 4:e1000254. doi: 10.1371/journal.pgen.1000254
93. Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, et al. Identification of bidirectional promoters in the human KIR genes. Genes Immun. (2007) 8:245–53. doi: 10.1038/sj.gene.6364381
94. Li H, Wright PW, McCullen M, Anderson SK. Characterization of KIR intermediate promoters reveals four promoter types associated with distinct expression patterns of KIR subtypes. Genes Immun. (2016) 17:66–74. doi: 10.1038/gene.2015.56
95. Parham P. Immunogenetics of killer-cell immunoglobulin-like receptors. Tissue Antigens. (2003) 62:194–200. doi: 10.1034/j.1399-0039.2003.00126.x
96. Gomez-Lozano N, Trompeter HI, de Pablo R, Estefania E, Uhrberg M, Vilches C. Epigenetic silencing of potentially functional KIR2DL5 alleles: implications for the acquisition of KIR repertoires by NK cells. Eur J Immunol. (2007) 37:1954–65. doi: 10.1002/eji.200737277
97. Cisneros E, Moraru M, Gómez-Lozano N, López-Botet M, Vilches C. KIR2DL5: an orphan inhibitory receptor displaying complex patterns of polymorphism and expression. Front Immunol. (2012) 3:289. doi: 10.3389/fimmu.2012.00289
98. Vilches C, Gardiner CM, Parham P. Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5. Gene. J Immunol. (2000) 165:6416. doi: 10.4049/jimmunol.165.11.6416
99. Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, et al. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol. (2005) 35:16–24. doi: 10.1002/eji.200425493
100. van Bergen J, Stewart CA, van den Elsen PJ, Trowsdale J. Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur J Immunol. (2005) 35:2191–9. doi: 10.1002/eji.200526201
101. Marquardt N, Scharenberg M, Mold JE, Hård J, Kekäläinen E, Buggert M, et al. High-dimensional analysis reveals a distinct population of adaptive-like tissue-resident NK cells in human lung. bioRxiv [Preprint]. (2019). doi: 10.1101/2019.12.20.883785
102. Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol. (2015) 194:2467–71. doi: 10.4049/jimmunol.1402756
103. Ivarsson MA, Stiglund N, Marquardt N, Westgren M, Gidlof S, Bjorkstrom NK. Composition and dynamics of the uterine NK cell KIR repertoire in menstrual blood. Mucosal Immunol. (2017) 10:322–31. doi: 10.1038/mi.2016.50
104. Björkström NK, Béziat V, Cichocki F, Liu LL, Levine J, Larsson S, et al. CD8 T cells express randomly selected KIRs with distinct specificities compared with NK cells. Blood. (2012) 120:3455–65. doi: 10.1182/blood-2012-03-416867
105. Mingari MC, Moretta A, Moretta L. Regulation of KIR expression in human T cells: a safety mechanism that may impair protective T-cell responses. Immunol Today. (1998) 19:153–7. doi: 10.1016/s0167-5699(97)01236-x
106. Huard B, Karlsson L. KIR expression on self-reactive CD8+ T cells is controlled by T-cell receptor engagement. Nature. (2000) 403:325–8. doi: 10.1038/35002105
107. Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. (2009) 130:213–24. doi: 10.1016/j.clim.2008.08.009
108. Lam TH, Shen M, Chia JM, Chan SH, Ren EC. Population-specific recombination sites within the human MHC region. Heredity. (2013) 111:131–8. doi: 10.1038/hdy.2013.27
109. Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. (2005) 76:634–46. doi: 10.1086/429393
110. Voorter CE, Gerritsen KE, Groeneweg M, Wieten L, Tilanus MG. The role of gene polymorphism in HLA class I splicing. Int J Immunogenet. (2016) 43:65–78. doi: 10.1111/iji.12256
111. Krangel MS. Secretion of HLA-A and -B antigens via an alternative RNA splicing pathway. J Exp Med. (1986) 163:1173–90. doi: 10.1084/jem.163.5.1173
112. Dunn PP, Hammond L, Coates E, Street J, Griner L, Darke C. A “silent” nucleotide substitution in exon 4 is responsible for the “alternative expression” of HLA-A∗01:01:38L through aberrant splicing. Hum Immunol. (2011) 72:717–22.
113. Dai Z-X, Zhang G-H, Zhang X-H, Xia H-J, Li S-Y, Zheng Y-T. The β2-microglobulin–free heterodimerization of rhesus monkey MHC Class I a with its normally spliced variant reduces the ubiquitin-dependent degradation of MHC class I A. J Immunol. (2012) 2012:1100665.
114. Dai ZX, Zhang GH, Zhang XH, Zheng YT. Identification and characterization of a novel splice variant of rhesus macaque MHC IA. Mol Immunol. (2013) 53:206–13. doi: 10.1016/j.molimm.2012.08.006
115. Ishitani A, Geraghty DE. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc Natl Acad Sci USA. (1992) 89:3947. doi: 10.1073/pnas.89.9.3947
116. Rouas-Freiss N, Bruel S, Menier C, Marcou C, Moreau P, Carosella ED. Switch of HLA-G alternative splicing in a melanoma cell line causes loss of HLA-G1 expression and sensitivity to NK lysis. Int J Cancer. (2005) 117:114–22. doi: 10.1002/ijc.21151
117. Kirszenbaum M, Moreau P, Gluckman E, Dausset J, Carosella E. An alternatively spliced form of HLA-G mRNA in human trophoblasts and evidence for the presence of HLA-G transcript in adult lymphocytes. Proc Natl Acad Sci USA. (1994) 91:4209. doi: 10.1073/pnas.91.10.4209
118. Rizzo R, Bortolotti D, Bolzani S, Fainardi E. HLA-G molecules in autoimmune diseases and infections. Front Immunol. (2014) 5:592. doi: 10.3389/fimmu.2014.00592
119. Fleming KA, McMichael A, Morton JA, Woods J, McGee JO. Distribution of HLA class 1 antigens in normal human tissue and in mammary cancer. J Clin Pathol. (1981) 34:779–84. doi: 10.1136/jcp.34.7.779
120. Wei X, Orr HT. HLA Class I. 2nd ed. In: PJ Delves editor. Encyclopedia of Immunology. Oxford: Elsevier (1998). p. 1108–11.
121. Dellgren C, Nehlin JO, Barington T. Cell surface expression level variation between two common human leukocyte antigen alleles, HLA-A2 and HLA-B8, is dependent on the structure of the C terminal part of the alpha 2 and the alpha 3 domains. PLoS One. (2015) 10:e0135385. doi: 10.1371/journal.pone.0135385
122. Apps R, Meng Z, Del Prete GQ, Lifson JD, Zhou M, Carrington M. Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J Immunol. (2015) 194:3594–600. doi: 10.4049/jimmunol.1403234
123. Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, et al. Influence of HLA-C expression level on HIV control. Science. (2013) 340:87.
124. Otting N, Heijmans CM, van der Wiel M, de Groot NG, Doxiadis GG, Bontrop RE. A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics. (2008) 60:507–14. doi: 10.1007/s00251-008-0311-5
125. He Y, Tian Z. NK cell education via nonclassical MHC and non-MHC ligands. Cell Mol Immunol. (2017) 14:321–30. doi: 10.1038/cmi.2016.26
126. He Y, Peng H, Sun R, Wei H, Ljunggren H-G, Yokoyama WM, et al. Contribution of inhibitory receptor TIGIT to NK cell education. J Autoimmun. (2017) 81:1–12. doi: 10.1016/j.jaut.2017.04.001
127. Horowitz A, Djaoud Z, Nemat-Gorgani N, Blokhuis J, Hilton HG, Béziat V, et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci Immunol. (2016) 1:eaag1672. doi: 10.1126/sciimmunol.aag1672
128. Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC Class I. Immunity. (2006) 25:331–42.
129. Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. (1998) 187:813–8. doi: 10.1084/jem.187.5.813
130. Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A. B and C. Nature. (1998) 391:795–9. doi: 10.1038/35869
131. Lee N, Llano M, Carretero M, Ishitani A, Navarro F, López-Botet M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. (1998) 95:5199–204. doi: 10.1073/pnas.95.9.5199
132. Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaëlsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. (2010) 115:1166–74. doi: 10.1182/blood-2009-09-245746
133. Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. (1998) 160:4951–60.
134. Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. (1995) 267:1016–8. doi: 10.1126/science.7863326
135. Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. (1997) 185:1523–8. doi: 10.1084/jem.185.8.1523
136. Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. (2004) 34:1673–9. doi: 10.1002/eji.200425089
137. Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci USA. (2010) 107:10160. doi: 10.1073/pnas.0913745107
138. Li Y, Mariuzza RA. Structural basis for recognition of cellular and viral ligands by NK cell receptors. Front Immunol. (2014) 5:123. doi: 10.3389/fimmu.2014.00123
139. Carrillo-Bustamante P, de Boer RJ, Keşmir C. Specificity of inhibitory KIRs enables NK cells to detect changes in an altered peptide environment. Immunogenetics. (2018) 70:87–97. doi: 10.1007/s00251-017-1019-1
140. Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol. (2007) 179:5977–89. doi: 10.4049/jimmunol.179.9.5977
141. Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, et al. Genetic control of human NK cell repertoire. J Immunol. (2002) 169:239–47. doi: 10.4049/jimmunol.169.1.239
142. Templer S, Sacks G. A blessing and a curse: is high NK cell activity good for health and bad for reproduction? Hum Fertil. (2016) 19:166–72. doi: 10.1080/14647273.2016.1219072
143. Shakhar K, Rosenne E, Loewenthal R, Shakhar G, Carp H, Ben-Eliyahu S. High NK cell activity in recurrent miscarriage: what are we really measuring? Hum Reprod. (2006) 21:2421–5. doi: 10.1093/humrep/del131
144. Colucci F. The role of KIR and HLA interactions in pregnancy complications. Immunogenetics. (2017) 69:557–65. doi: 10.1007/s00251-017-1003-9
145. Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. (2008) 23:972–6. doi: 10.1093/humrep/den011
146. Tripathy SK, Keyel PA, Yang L, Pingel JT, Cheng TP, Schneeberger A, et al. Continuous engagement of a self-specific activation receptor induces NK cell tolerance. J Exp Med. (2008) 205:1829–41. doi: 10.1084/jem.20072446
147. Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science. (1991) 253:199–202. doi: 10.1126/science.1853205
148. Hoglund P, Ohlen C, Carbone E, Franksson L, Ljunggren HG, Latour A, et al. Recognition of beta 2-microglobulin-negative (beta 2m-) T-cell blasts by natural killer cells from normal but not from beta 2m- mice: nonresponsiveness controlled by beta 2m- bone marrow in chimeric mice. Proc Natl Acad Sci USA. (1991) 88:10332–6. doi: 10.1073/pnas.88.22.10332
149. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, et al. Defining KIR and HLA Class I genotypes at highest resolution via high-throughput sequencing. Am J Hum Genet. (2016) 99:375–91. doi: 10.1016/j.ajhg.2016.06.023
150. Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. (1975) 256:50–2. doi: 10.1038/256050a0
151. McClelland EE, Penn DJ, Potts WK. Major histocompatibility complex heterozygote superiority during coinfection. Infect Immun. (2003) 71:2079–86. doi: 10.1128/iai.71.4.2079-2086.2003
152. Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci USA. (2002) 99:11260–4. doi: 10.1073/pnas.162006499
153. Tu MM, Mahmoud AB, Makrigiannis AP. Licensed and unlicensed NK cells: differential roles in cancer and viral control. Front Immunol. (2016) 7:166. doi: 10.3389/fimmu.2016.00166
154. Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Investig. (2012) 122:3260–70. doi: 10.1172/jci62749
155. Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nature Immunol. (2010) 11:321–7. doi: 10.1038/ni.1849
Keywords: Killer cell immunoglobin-like receptor, KIR, NK cell, NK cell education, human, macaque, non-human primates
Citation: Bruijnesteijn J, de Groot NG and Bontrop RE (2020) The Genetic Mechanisms Driving Diversification of the KIR Gene Cluster in Primates. Front. Immunol. 11:582804. doi: 10.3389/fimmu.2020.582804
Received: 13 July 2020; Accepted: 18 August 2020;
Published: 11 September 2020.
Edited by:
Jim Kaufman, University of Cambridge, United KingdomReviewed by:
Paul J. Norman, University of Colorado Denver, United StatesFrancesco Colucci, University of Cambridge, United Kingdom
Copyright © 2020 Bruijnesteijn, de Groot and Bontrop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jesse Bruijnesteijn, YnJ1aWpuZXN0ZWlqbkBicHJjLm5s
 Jesse Bruijnesteijn
Jesse Bruijnesteijn Natasja G. de Groot
Natasja G. de Groot Ronald E. Bontrop
Ronald E. Bontrop