
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 30 September 2020
Sec. Immunological Tolerance and Regulation
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.576255
This article is part of the Research Topic Novel Advances in Allergy Diagnosis and Treatment View all 21 articles
In the last decade single domain antibodies (nanobodies, VHH) qualified through their unique characteristics have emerged as accepted and even advantageous alternative to conventional antibodies and have shown great potential as diagnostic and therapeutic tools. Currently nanobodies find their main medical application area in the fields of oncology and neurodegenerative diseases. According to late-breaking information, nanobodies specific for coronavirus spikes have been generated these days to test their suitability as useful therapeutics for future outbreaks. Their superior properties such as chemical stability, high affinity to a broad spectrum of epitopes, low immunogenicity, ease of their generation, selection and production proved nanobodies also to be remarkable to investigate their efficacy for passive treatment of type I allergy, an exaggerated immune reaction to foreign antigens with increasing global prevalence.
Type I allergy, an IgE antibody mediated hypersensitivity disease, represents a common health problem affecting almost 30% of the population worldwide (1). The recognition of allergens by specific IgE antibodies that are generated after sensitization is a key event for the initiation of allergic inflammation (2). Allergic patients suffer from a variety of allergic symptoms including rhinoconjunctivitis and asthma (3) but also food allergy and skin inflammation (4). These clinical manifestations cause a major burden by reducing the quality of life of affected persons (5). While anti-inflammatory treatment based on pharmacotherapy reduces allergic symptoms and is the most commonly prescribed medication for treatment of allergic patients (6), only allergen-specific immunotherapy (AIT) represents a causative treatment of type I allergy. In fact, AIT induces a protective immunity in allergic patients based on the modification of cellular and humoral responses to the disease causing allergen (7). Besides the inhibition of IgE binding to their specific allergen, the immune deviation from a TH2 to TH1 response, and the decreases in numbers of effector cells in target organs, the generation and maintenance of allergen-specific regulatory T and B cells and the involvement of their suppressive cytokines are essential for the induction of allergen tolerance (8–10). Beyond doubt the improvement of allergic symptoms is further caused by AIT-induced IgG antibodies found in serum and nasal secretions (8, 11–14). For many years AIT was conducted with aqueous natural allergen extracts and patients experienced considerable side effects due to the unpredictable composition and poor quality of the injected extracts (1). Recent developments like next-generation forms of AIT based on molecular approaches may overcome the limitations of current forms of AIT (15, 16). The last generation of improved vaccines, i.e. peptide carrier vaccines, induces an IgG response that targets IgE binding sites on allergens. Induced IgG antibodies effectively block IgE binding and are termed blocking antibodies (1, 17).
However, the efficacy of such blocking antibodies was long questioned because it revealed to be cumbersome to isolate reproducible defined, i.e. monoclonal allergen-specific antibodies comprising the capacity to inhibit allergen-induced allergic reactions.
A recent proof of concept study re-stimulated the idea to generate monoclonal allergen-specific antibodies and to evaluate their feasibility for allergy treatment. The authors could show that a single subcutaneous injection of a mixture of two human monoclonal allergen-specific IgG4 antibodies significantly reduced allergic symptoms in allergic patients (18, 19). Moreover, validated in a PCA mouse model, the mixture of these two monoclonal antibodies proved to be more potent in inhibiting mast cell degranulation than IgG antibodies purified from patients’ sera who underwent successful AIT (18). Furthermore, these human monoclonal IgG4 antibodies recently completed the phase II clinical trial in treatment of cat allergic patients (https://clinicaltrials.gov/ct2/show/NCT03838731). These results proved for the first time that allergy treatment with monoclonal allergen-specific antibodies is a well-tolerated, rapid, and effective approach to reduce allergic inflammation and rekindled the blocking antibody concept (11, 20, 21).
Nevertheless, the generation and identification of blocking conventional human or humanized antibodies is connected with high costs for production, validation and application (22, 23). Therefore, cost-effective alternatives are currently sought.
The nanobody technology represents such an alternative implying a significant improvement to the laborious methods to obtain monoclonal blocking conventional antibodies. Due to their beneficial properties of small molecules and monoclonal antibodies, nanobodies in general are an attractive agent for development of novel therapeutic strategies (24, 25). The ease of their generation and production, the single domain organization, their beneficial biochemical properties and their feature to recognize small cavities on the surface of antigens and hence bind to epitopes inaccessible for conventional antibodies (26) have raised the particular interest of allergologists recently.
Can the nanobody technology provide enhanced opportunity to generate a panel of antigen-binding molecules with various epitope specificities for certain allergens different to conventional antibodies? Will these identified allergen-specific nanobodies be more efficient in blocking than conventional IgG antibodies due to their pronounced cleft recognition? Will it be possible with this technology to find single nanobodies that are able to abrogate IgE-mediated allergic inflammation? These questions and our wish to answer these questions attracted our attention. Within this review, we focus on the powerful nanobody technology to generate allergen-specific nanobodies and report on their evaluation for prospective application for passive allergy treatment.
If allergologists are asked why the search for effective protective allergen-specific monoclonal antibodies is complex and laborious, they will describe this issue by the typical quest for a needle in a haystack. Through intense and precise molecular and immunological exploration of available allergen-specific monoclonal antibodies in the past it was proven that epitope specificity and affinity are decisive for their inhibitory potential to block IgE binding and thus IgE-mediated reactions (21, 27–29). The commitment to find and isolate monoclonal antibodies with specificity and high affinity for certain allergens and even more for certain epitopes always started with several fundamental decisions. Amongst them the choice for the perfect source to gain DNA coding for antibodies and the applied technology to generate allergen-specific antibodies are two of the most critical ones. Regarding the DNA source both animals, mainly mice, and humans served as blood, spleen, tonsils and even bone marrow donors in the last decades to isolate B cells or plasma cells and thus DNA coding for antibodies (30–32). For the proof of principle, murine IgG antibodies overlapping with human IgE binding sites are valuable tools to investigate the effects to inhibit IgE epitope recognition on allergens and consequently to contribute to the design of hypoallergenic derivatives suitable for AIT (33). However, the direct therapeutic use of these murine monoclones in humans is limited by the high incidence of harmful immune responses against these administered foreign proteins (34). To mitigate this limitation numerous murine monoclonal antibodies have been re-engineered by chimerization and humanization technologies. These expensive procedures are justified for fatal diseases like different forms of cancer but were barely applied for allergen-specific murine antibodies so far with a few exceptions (35, 36). This was one of the main reasons why allergologists in the recent past endeavour to focus on human donors including allergic patients, AIT-treated patients and even healthy individuals depending on the research question (28, 37, 38).
Various methods were utilized to generate allergen-specific genuine, i.e. native antibodies with the preservation of the natural VH and VL pairing including hybridoma technology, Epstein-Barr-Virus (EBV) transformation, single B cell sorting and cloning and HumAb mice (transgenic mice that produce fully human antibodies) (18, 39–49). In parallel, versatile approaches were developed to generate non-genuine antibodies by random combination of VH and VL chains, i.e., combinatorial Fab/ScFv libraries or (semi-) synthetic libraries (37, 38, 50–60). Based on PCR amplification as strong tool to depict large antibody repertoires and phage display to screen these large repertoires, many recombinant allergen-specific antibody fragments (Fabs or ScFvs) were isolated (37, 38, 50–56, 58–64).
All mentioned technologies have definitely contributed to the isolation and evaluation of monoclonal allergen-specific IgG, IgE antibodies and fragments thereof and furthermore to assess their feasibility for allergy treatment. Nevertheless, all mentioned technologies are also reported to have some limitations. While the hybridoma technology and EBV transformation are generally unsuitable for a comprehensive screening of large antibody repertoires because of their inefficient fusion and transformation events, the single B cell sorting was long hampered by inadequate staining technologies to clearly identify allergen-specific antibody producing cells (32, 39). The main drawback of combinatorial libraries is that they usually rely on random combination and thus most likely unnatural VH and VL antibody pairings. Additionally, it turned out independent of the applied technology to be very difficult to isolate monoclonal IgG and IgE antibodies with a broad epitope spectrum for each allergen. It also revealed that besides several blocking antibodies also many non-blocking or even enhancing antibodies were isolated (44, 63–65). While all three types of monoclonal antibodies were unambiguously supportive to study the structural requirements for efficient effector cell activation and hence contribute to elucidate the underlying mechanisms of type I allergy, non-blocking and enhancing antibodies were fully useless for the prospective application as protective antibodies.
These insights forced allergologists to look beyond the conventional antibody horizon.
About 30 years ago, a group of Belgian scientists made an unexpected discovery, which was patented and later presented to the scientific community in the form of the well-known discovery publication in the journal Nature in 1993 (66). They found that a significant amount of non-canonical types of antibodies is naturally present in blood of Camelidae in addition to conventional antibodies. This exceptional type of antibody called Heavy Chain-only Antibody (HCAb) lacks light chains and consists of a homodimer of shortened (without CH1 domain) heavy chains. The antigen-recognition region in HCAbs is formed by only one variable domain (VHH) that is directly linked via a hinge region to the Fc-domain (66). Later on, similar non-canonical HCAbs were found in some cartilaginous fishes such as sharks and ratfish (67–69). The antigen-binding variable domain of these antibodies was named VNAR as opposed to VHH in camelids. A recombinant protein version of the VHH-or VNAR-domain is usually called “single domain antibody” or “nanobody”. The very popular term “nanobody” is the commercial name given by the Belgian biopharmaceutical company Ablynx, a pioneer in HCAb-based therapeutic applications that was acquired by Sanofi in 2018.
The nanobody generation technology was proven to be a very efficient machinery to generate nanobodies with required properties and offered crucial advantages compared to traditional techniques utilized to produce murine or human conventional antibodies. After the typical initial immunization (of camelids) step, the full repertoire of cDNA coding for functional nanobodies can be efficiently cloned from peripheral blood lymphocytes of immunized animals using PCR amplification and then a panel of nanobodies of required specificity can be easily selected using phage (or other type of) display-based methods (66, 70–72). In addition, there are different in vitro affinity maturation approaches to improve features of initially selected nanobodies (71, 73, 74). In some cases, especially if the antigen of interest is toxic, unstable, non-immunogenic or not available in sufficient quantity, other types of libraries (naive, semisynthetic or fully synthetic libraries) can be efficiently used instead of immune libraries for generation of nanobodies (75–79). Synthetic libraries can be made using special predesigned scaffolds such as humanized scaffolds optimized for intracellular stability (77) or optimized for bacterial expression (80). Non-immune libraries are typically much larger than immune libraries and a ribosome display was suggested for the initial selection round of such large libraries to work with higher concentrations of nanobody variants than in case of phage display (79, 80). Synthetic libraries combined with different selection procedures were successfully used to obtain conformationally selective nanobodies against G protein-coupled receptors (78), sybodies against very challenging targets such as the heterodimeric bacterial ABC exporterTM287/288 (81) or the intracellular KDEL receptor (82) to name a few examples from many others.
Nanobodies comprise unique features that distinguish them from classical antibodies. Nanobodies are the smallest known antibody fragments (4 × 2.5 x 3 nm, 12–15 kDa) of natural origin that are able to specifically bind their cognate antigens. Due to their often extended CDR3 loop they can form unusual paratopes, i.e. finger-like extensions and thus recognize special native antigenic epitopes (small cavities, concave surfaces, conformational epitopes, active sites of enzymes) that are hidden for conventional antibodies (Figure 1). Indeed, nanobodies have proven to be useful tools for modulating the activity of enzymes (26, 83, 84). It could be therefore speculated that allergen-specific nanobodies that modulate or inhibit the proteolytic activity of certain allergens (e.g., Phl p 1, Der p 1) might reduce their penetration capacity through mucosal surfaces. Furthermore, nanobodies are able to bind small peptides with high affinity (85–89). Their high affinity, solubility and stability over a wide range of temperatures and pH, ease of producing in bacteria or other expression systems make them convenient molecules for different applications, as well as for all possible engineering modifications e.g., development of complex constructs and conjugates. Nanobody-based tools are therefore increasingly used for research, molecular visualization, diagnostics and development of new treatment options for various pathologies, including cancer and other socially significant diseases (71, 72, 90–94).
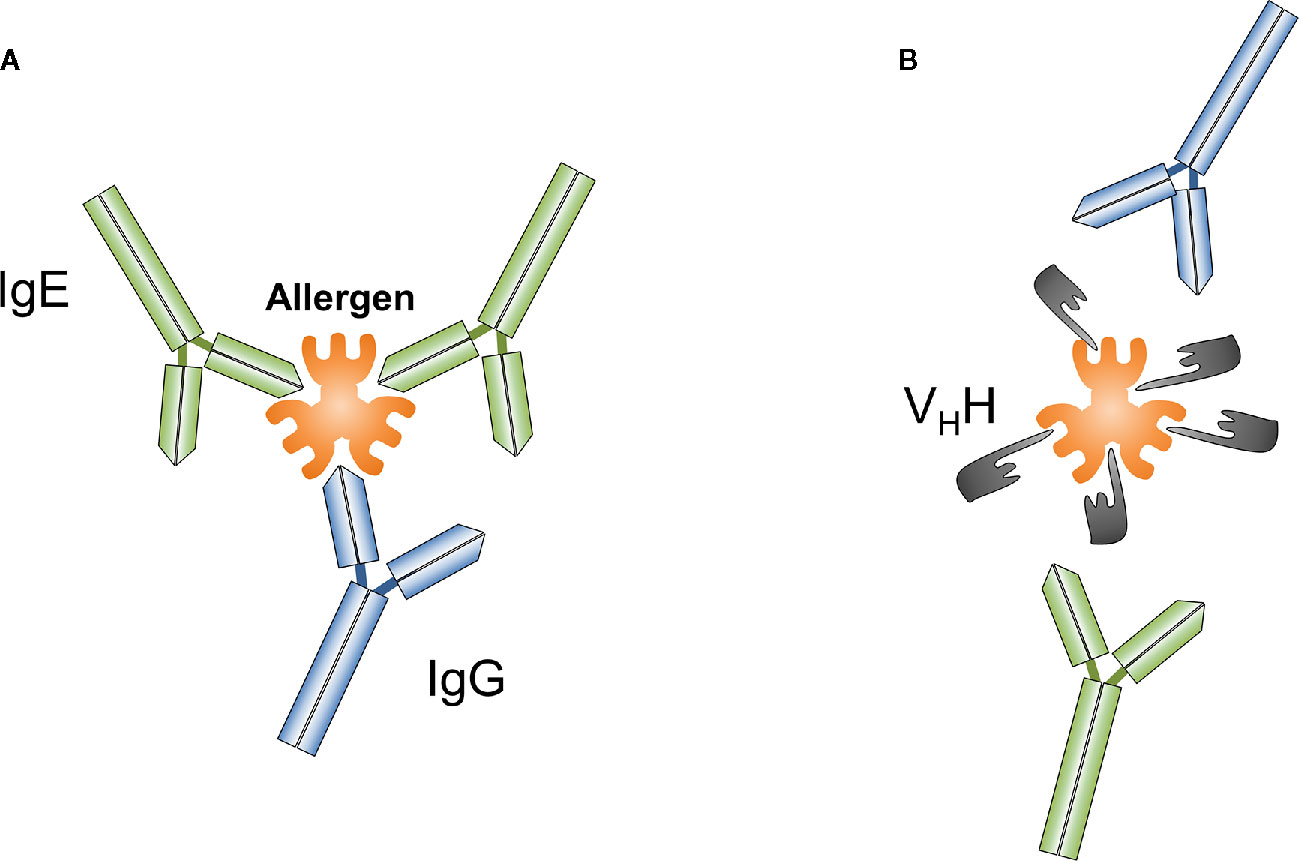
Figure 1 Conventional antibodies such as IgG or IgE (A) and nanobodies (VHH) (B) can be generated against different epitopes of targeted antigens, a particular allergen. Nanobodies overlapping with IgE binding sites on allergens prevent IgE-mediated allergic reactions.
So far, only one allergen-specific nanobody is described in the literature. This nanobody is reported to be specific for the major peanut allergen, Ara h 3 and was isolated from a synthetic library of humanized nanobodies via phage display (95). The interaction between Ara h 3 and the Ara h 3-specific nanobody resulted in a dissociation constant of 400 nM representing medium affinity binding and was further investigated by the structural determination of formed co-crystals (95). The authors acknowledged that additional work is needed to improve the affinity of the isolated nanobody to make it an attractive tool for the development of biosensors for peanut allergen detection. This finding clarifies that the selection procedure is only one part of the successful discovery of potent IgE-blocking nanobodies, thus the evaluation of selected nanobodies is critical as well.
Nevertheless, we are confident that soon more allergen-specific nanobodies will arise to be studied for their potential to abrogate IgE-mediated allergic inflammation.
Similar to the evaluation of conventional antibodies with the focus to identify effective protective monoclones, generated nanobodies have to be assessed first for their allergen specificity, epitope recognition, cross-reactivity to homologous allergens present in related species, for their affinity to their corresponding allergens and most importantly for their ability to inhibit patients´ IgE binding to these allergens (Figures 2A–C). After the allergen specificity of isolated nanobodies is confirmed, the proof for cross-reactivity (Figure 2A) is of great importance because IgE antibodies from allergic patients often displayed cross-reactivity to allergens from other allergen sources (28, 96). High affinity and slow dissociation of formed nanobody/allergen complexes will be critical prerequisites for allergen-specific nanobodies to be chosen as suitable candidate (Figure 2B). However, the pivotal characteristics for an allergen-specific nanobody to be attractive for further processing will be the determination of its potential to block patients’ IgE binding and hence IgE-mediated effector cell activation (Figure 2C). Additionally, specific nanobodies have to be tested as well for their cross-protectivity to homologous allergens. All these properties are crucial requirements for allergen-specific nanobodies to be selected for further essential investigations concerning half-life, clearance and safety.
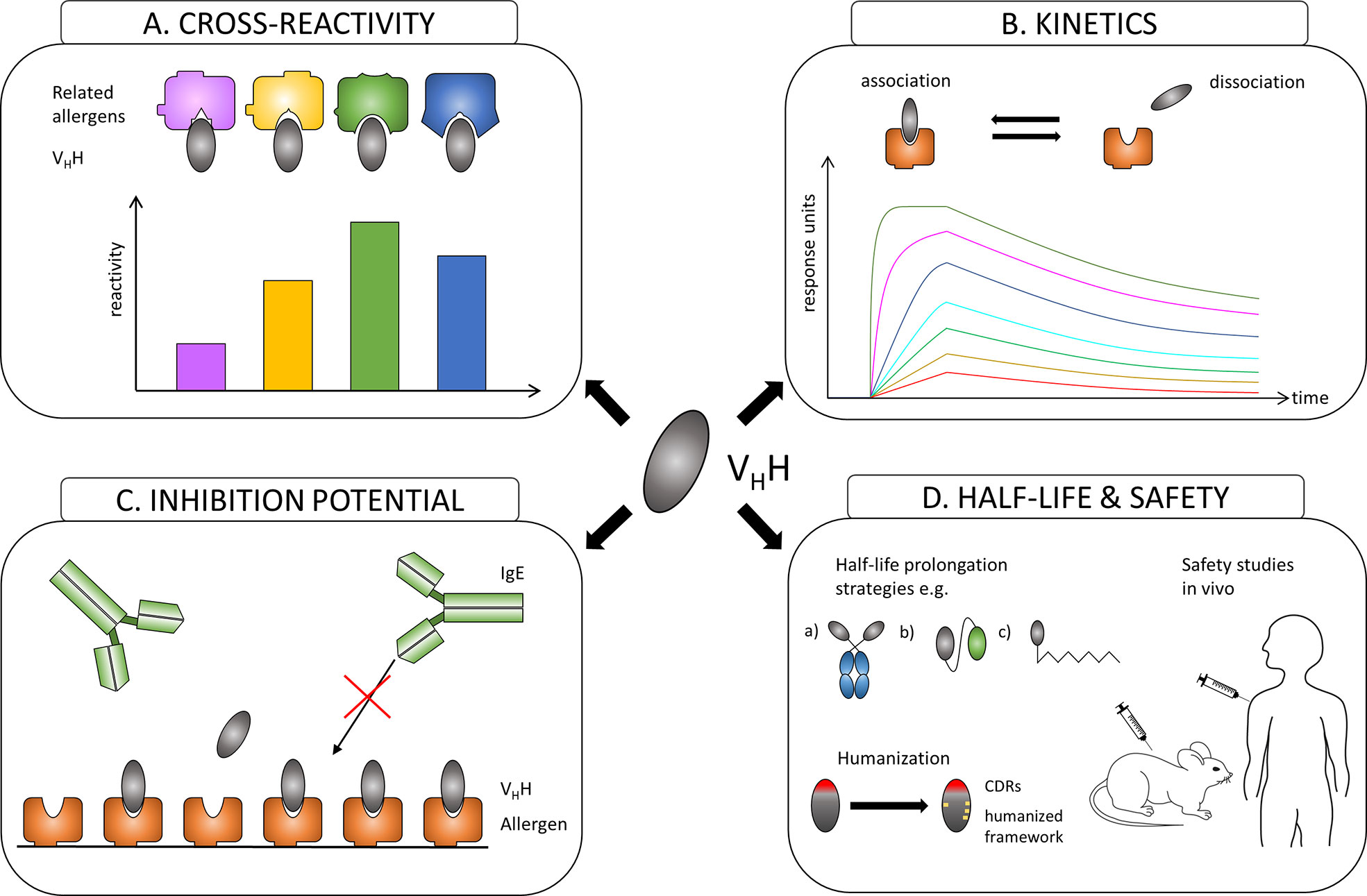
Figure 2 Overview of the evaluation process of the suitability of allergen-specific nanobodies for allergy treatment. (A) Evaluating cross-reactivity to related allergens. (B) Measuring affinities of selected candidates to the allergen. (C) Investigating the potential to block allergen-specific IgE from binding to the allergen. (D) Adjusting the half-life of a suitable nanobody by e.g.: a) linking to IgG Fc region; b) oligomerization to homomers or heteromers to facilitate linking to other proteins like human serum albumin (HSA); c) PEGylation. Increasing safety by humanization of the framework and performing safety studies in vivo.
Nanobodies are considered as proteins of weak immunogenicity due to the shared similarities with variable VH domains of human immunoglobulins (IgG3 subclass), and they can be further improved by a humanization approach (97) (Figure 2D). Consequently, no immune response against applied nanobodies was raised in mice or humans that were injected with nanobody-containing constructs (98–100). Safety of nanobody-based drugs is confirmed by several completed Phase 1 and Phase 2 clinical trials (101) and recent approval by the US Food and Drug Administration (FDA) and the European medicines agency (EMA) of the first therapeutic nanobody, Caplacizumab, a bivalent nanobody designed for the treatment of thrombotic thrombocytopenic purpura and thrombosis (102).
Though advantageous for in vivo imaging, the small size of nanobodies could be seen as a disadvantage for passive treatment of allergy due to a quick renal clearance of nanobodies from blood (approx. 30 min). Many different strategies to extend the in vivo half-life of nanobody-based construct have been developed (103). They include increasing the hydrodynamic radius of a protein by attaching highly flexible and hydrophilic molecules such as polyethylene glycol (PEG) and carbohydrates or by genetic fusion with polypeptide chains mimicking the biochemical properties of PEG, fusion of VHH to the Fc region of IgG, fusion or non-covalent binding to albumin (104) (Figure 2D). Nanobodies can also be used as modules to engineer larger molecules with several valencies and/or specificities, such as multivalent (105–108), bispecific (105, 109), and other (110, 111) constructs that may acquire considerably higher specificity, binding efficiency and biological activity (106, 107, 111). Nanobodies were also considered as possible ligands to design new highly specific immunosorbents (112–114).
Different types of nanobody-based tools/approaches can be envisaged to be potentially profitable for an allergy treatment: a) bispecific nanobodies for topical application to capture allergens before they penetrate epithelial mucosa in airways, b) very stable nanobodies to capture food allergen in gastrointestinal tract, c) anti-idiotypic nanobodies mimicking allergenic epitopes as a possible replacement for a complex natural allergen for a new kind of AIT vaccine development, d) multivalent nanobody-based constructs for systemical administration to efficiently block allergen interaction with IgE on mast or basophil cells, e) efficient immunosorbents to remove IgE from the blood by immune apheresis. Correspondingly, different administration approaches for nanobody-based constructs can be developed: aerosol or topical applications, oral route or subcutaneous administrations. Temporary blocking of allergen-IgE interaction (i.e. by topical or systemic administration of specific nanobodies) or a subtraction of IgE from the periphery blood (i.e. apheresis) may give a short-term treatment effect. For a long-term treatment effect we could hypothesize the use of anti-idiotypic nanobodies to IgE. Such nanobodies may represent “internal images” of an allergen and mimick hypoallergenic B cell epitopes. To efficiently induce IgG response that targets IgE binding sites on allergens, these nanobodies should be fused to a viral coat protein as it was described for next-generation forms of AIT (15).
The generation of allergen-specific nanobodies unambiguously represents a reasonable progress in the field of allergy. With their well-documented qualities including their ability to recognize unusual “hidden” epitopes, high affinity binding, solubility, extreme stability and low immunogenicity, nanobodies attracted the interest of allergologists to study their suitability for passive allergy treatment. The chance to find allergen-specific nanobodies with this powerful technology that ideally comprise high affinity and bind to epitopes partly or fully overlap with IgE binding sites on allergens is tempting. However, so far no allergen-specific nanobody fulfilling these criteria was reported indicating that it might be rather difficult to raise allergen-specific nanobodies of sufficient affinities. Whether the current lack of such nanobodies is owed to some inherent structural or functional properties of nanobodies and/or the camelid immune system or the simple reason that the current research focus in the allergy field is on AIT and its improvement has to be resolved. If allergen-specific nanobodies are identified that competitively block allergen binding to IgE and thus abrogate IgE-mediated allergic inflammation, we assume that they will represent appropriate tools for future allergy treatment. Their economic properties, i.e. low production costs encouraged researchers to elaborate antibody engineering of these single-domain antibodies for diverse applications in biotechnology and medicine. This gathered knowledge will facilitate the implementation of modified allergen-specific nanobodies tailored to the needs of allergy treatment. Nanobodies can be easily formatted for a particular application e.g., modified as recognition modules in large constructs or as bi- or oligo-specific, bi- or oligo-valent derivatives.
With the availability of allergen-specific nanobodies or their derivatives with inhibitory potential, it should be possible to examine engineered candidates in proof of concept testings for efficacy and safety in experimental animal models to identify promising nanobody-based drugs for clinically relevant allergens.
SF, IZ and ST reviewed the literature, IZ and ST generated figures. SF and ST wrote the manuscript draft. All authors contributed to the article and approved the submitted version.
SF was supported by Austria Science Fund (FWF) grants F4607 and I3946-B33 and ST was supported by the Russian Foundation for Basic Research (RFBR) grant 18-515-14003.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AIT, Allergen-specific ImmunoTherapy; EBV, Epstein-Barr Virus; Fab, antigen-binding Fragment of antibodies; HCAb, Heavy Chain-only Antibody; HumAb mice, transgenic mice that produce fully human antibodies; PCA, Passive Cutaneous Anaphylaxis; scFv, single chain Fragment variable (recombinant derivative of a classical antibody); VHH, Variable domain of Heavy chain of Heavy chain-only antibodies.
1. Valenta R, Karaulov A, Niederberger V, Gattinger P, van Hage M, Flicker S, et al. Molecular aspects of allergens and allergy. Adv Immunol (2018) 138:195–256. doi: 10.1016/bs.ai.2018.03.002
2. Murphy K, Weaver C. Allergy and allergic disease. In: Janeway’s Immunobiology, 9th edition. New York City, NY: Garland Science (2017). p. 601–41.
3. Siroux V, Boudier A, Nadif R, Lupinek C, Valenta R, Bousquet J. Association between asthma, rhinitis, and conjunctivitis multimorbidities with molecular IgE sensitization in adults. Allergy (2019) 74:824–7. doi: 10.1111/all.13676
4. Renz H, Allen KJ, Sicherer SH, Sampson HA, Lack G, Beyer K, et al. Food Allergy. Nat Rev Dis Primers (2018) 4:17098. doi: 10.1038/nrdp.2017.98
5. Kiotseridis H, Cilio CM, Bjermer L, Aurivillius M, Jacobsson H, Dahl Å, et al. Quality of life in children and adolescents with respiratory allergy, assessed with a generic and disease-specific instrument. Clin Respir J (2013) 7:168–75. doi: 10.1111/j.1752-699X.2012.00298.x
6. Criado PR, Criado RF, Maruta CW, Machado Filho Cd. Histamine, histamine receptors and antihistamines: new concepts. Bras Dermatol (2010) 85:195–210. doi: 10.1590/s0365-05962010000200010
7. Dorofeeva Y, Shilovsky I, Tulaeva I, Focke-Tejkl M, Flicker S, Kudlay D, et al. Past, presence, and future of allergen immunotherapy vaccines. Allergy (2020) 00:1–19. doi: 10.1111/all.14300
8. Shamji MH, Durham SR. Mechanisms of allergen immunotherapy for inhaled allergens and predictive biomarkers. J Allergy Clin Immunol (2017) 140:1485–98. doi: 10.1016/j.jaci.2017.10.010
9. Palomares O, Akdis M, Martín-Fontecha M, Akdis CA. Mechanisms of immune regulation in allergic diseases: the role of regulatory T and B cells. Immunol Rev (2017) 278:219–36. doi: 10.1111/imr.12555
10. Globinska A, Boonpiyathad T, Satitsuksanoa P, Kleuskens M, van de Veen W, Sokolowska M, et al. Mechanisms of allergen-specific immunotherapy: Diverse mechanisms of immune tolerance to allergens. Ann Allergy Asthma Immunol (2018) 121:306–12. doi: 10.1016/j.anai.2018.06.026
11. Flicker S, Valenta R. Renaissance of the blocking antibody concept in type I allergy. Int Arch Allergy Immunol (2003) 132:13–24. doi: 10.1159/000073260
12. Lupinek C, Wollmann E, Valenta R. Monitoring allergen immunotherapy effects by microarray. Curr Treat Options Allergy (2016) 3:189–203. doi: 10.1007/s40521-016-0084-2
13. Shamji MH, Kappen J, Abubakar-Waziri H, Zhang J, Steveling E, Watchman S, et al. Nasal allergen-neutralizing IgG4 antibodies block IgE-mediated responses: Novel biomarker of subcutaneous grass pollen immunotherapy. J Allergy Clin Immunol (2019) 143:1067–76. doi: 10.1016/j.jaci.2018.09.039
14. Reisinger J, Horak F, Pauli G, van Hage M, Cromwell O, König F, et al. Allergen-specific nasal IgG antibodies induced by vaccination with genetically modified allergens are associated with reduced nasal allergen sensitivity. J Allergy Clin Immunol (2005) 116:347–54. doi: 10.1016/j.jaci.2005.04.003
15. Curin M, Khaitov M, Karaulov A, Namazova-Baranova L, Campana R, Garib V, et al. Next-generation of allergen-specific immunotherapies: molecular approaches. Curr Allergy Asthma Rep (2018) 18:39. doi: 10.1007/s11882-018-0790-x2018
16. Zhernov Y, Curin M, Khaitov M, Karaulov A, Valenta R. Recombinant allergens for immunotherapy: state of the art. Curr Opin Allergy Clin Immunol (2019) 19:402–14. doi: 10.1097/ACI.0000000000000536
17. Marth K, Breyer I, Focke-Tejkl M, Blatt K, Shamji MH, Layhadi J, et al. A nonallergenic birch pollen allergy vaccine consisting of hepatitis PreS-fused Bet v 1 peptides focuses blocking IgG toward IgE epitopes and shifts immune responses to a tolerogenic and Th1 phenotype. J Immunol (2013) (2013) 190:3068–78. doi: 10.4049/jimmunol.1202441
18. Orengo JM, Radin AR, Kamat V, Badithe A, Ben LH, Bennett BL, et al. Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement. Nat Commun (2018) 9:1421. doi: 10.1038/s41467-018-03636-8
19. Durham S, Schwabe C, Robson R, Ahlström Emanuelsson C, Shamji M, Singh D, et al. A randomized clinical trial of passive immunotherapy with single-dose anti-Fel d 1 monoclonal antibodies REGN 1908-1909 in cat induced rhinoconjunctivitis: exploratory efficacy endpoints, safety, and pharmacokinetics. Allergy (2017) 72(S103):64–5.
20. Wachholz PA, Durham SR. Mechanisms of immunotherapy: IgG revisited. Curr. Opin. Allergy Clin Immunol (2004) 4:313–8. doi: 10.1097/01.all.0000136753.35948.c0
21. Flicker S, Gadermaier E, Madritsch C, Valenta R. Passive immunization with allergen-specific antibodies. Curr Top Microbiol Immunol (2011) 352:141–59. doi: 10.1007/82_2011_143
22. Hernandez I, Bott SW, Patel AS, Wolf CG, Hospodar AR, Sampathkumar S, et al. Pricing of monoclonal antibody therapies: higher if used for cancer? Am J Manag Care (2018) 24:109–12.
23. Saluja R, Arciero VS, Cheng S, McDonald E, Wong WWL, Cheung MC, et al. Examining trends in cost and clinical benefit of novel anticancer drugs over time. J Oncol Pract (2018) 14:e280–94. doi: 10.1200/JOP.17.00058
24. Bannas P, Hambach J, Koch-Nolte F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol (2017) 8:1603. doi: 10.3389/fimmu.2017.01603
25. Jovceska I, Muyldermans S. The therapeutic potential of nanobodies. BioDrugs (2020) 34:11–26. doi: 10.1007/s40259-019-00392-z
26. De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA (2006) 103:4586–91. doi: 10.1073/pnas.0505379103
27. Dodev TS, Bowen H, Shamji MH, Bax HJ, Beavil AJ, McDonnel JM, et al. Inhibition of allergen-dependent IgE activity by antibodies of the same specificity but different class. Allergy (2015) 70:720–4. doi: 10.1111/all.12607
28. Gadermaier E, James LK, Shamji MH, Blatt K, Fauland K, Zieglmayer P, et al. Epitope specificity determines cross-protection of a SIT-induced IgG4 antibody. Allergy (2016) 71:36–46. doi: 10.1111/all.12710
29. James L. The cloning and expression of human monoclonal antibodies: implications for allergen immunotherapy. Curr Allergy Asthma Rep (2016) 16:15. doi: 10.1007/s11882-015-0588-z
30. Kurosawa N, Yoshioka M, Fujimoto R, Yamagishi F, Isobe M. Rapid production of antigen-specific monoclonal antibodies from a variety of animals. BMC Biol (2012) 10:80. doi: 10.1186/1741-7007-10-80
31. Lanzavecchia A, Sallusto F. Human B cell memory. Curr Opin Immunol (2009) 21:298–304. doi: 10.1016/j.coi.2009.05.019
32. Eckl-Dorna J, Villazala-Merino S, Campion NJ, Byazrova M, Filatov A, Kudlay D, et al. Tracing IgE-producing cells in allergic patients. Cells (2019) 8:994. doi: 10.3390/cells8090994
33. Glesner J, Kapingidza AB, Godzwon M, Offermann LR, Mueller GA, DeRose EF, et al. A human IgE antibody binding site on Der p 2 for the design of a recombinant allergen for immunotherapy. J Immunol (2019) 203:2545–56. doi: 10.4049/jimmunol.1900580
34. Hansel TT, Kropshofer H, Singer T, Mitchell JA, George AJT. The safety and side effects of monoclonal antibodies. Nat Rev Drug Discovery (2010) 9:325–38. doi: 10.1038/nrd3003
35. Babu KS, Arshad SH, Holgate ST. Omalizumab, a novel anti-IgE therapy in allergic disorders. Expert Opin Biol Ther (2001) 1:1049–58. doi: 10.1517/14712598.1.6.1049
36. Mitchell P, Leigh R. A drug safety review of treating eosinophilic asthma with monoclonal antibodies. Expert Opin Drug Saf (2019) 18:1161–70. doi: 10.1080/14740338.2019.1675634
37. Steinberger P, Kraft D, Valenta R. Construction of a combinatorial IgE library from an allergic patient. Isolation and characterization of human IgE Fabs with specificity for the major timothy grass pollen allergen, Phl p 5. J Biol Chem (1996) 271:10967–72. doi: 10.1074/jbc.271.18.10967
38. Gadermaier E, Marth K, Lupinek C, Campana R, Hofer G, Blatt K, et al. Isolation of a high-affinity Bet v 1-specific IgG-derived ScFv from a subject vaccinated with hypoallergenic Bet v 1 fragments. Allergy (2018) 73:1425–35. doi: 10.1111/all.13394
39. Tiller T. Single B cell antibody technologies. N Biotechnol (2011) 28:453–7. doi: 10.1016/j.nbt.2011.03.014
40. Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature (1975) 256:495. doi: 10.1038/256495a0
41. Olsson L, Kaplan HS. Human-human hybridomas producing monoclonal antibodies of predefined antigenic specificity. Proc Natl Acad Sci USA (1980) 77:5429–31. doi: 10.1073/pnas.77.9.5429
42. Wurth MA, Hadadianpour A, Horvath DJ, Daniel J, Bogdan O, Goleniewska K. Human IgE mAbs define variability in commercial Aspergillus extract allergen composition. JCI Insight (2018) 3:e123387. doi: 10.1172/jci.insight.123387
43. Pomés A, Glesner J, Richardson C, Chapman MD, Mueller GA, Smith SA. Human IgE monoclonal antibodies for epitope mapping of Der p 2. Allergy (2019) 74(Suppl. 106):222–3.
44. Visco V, Dolecek C, Denépoux S, Le Mao J, Guret C, Rousset F, et al. Human IgG monoclonal antibodies that modulate the binding of specific IgE to birch pollen Bet v 1. J Immunol (1996) 157:956–62.
45. Lebecque S, Dolecek C, Laffer S, Visco V, Denépoux S, Pin JJ, et al. Immunologic characterization of monoclonal antibodies that modulate human IgE binding to the major birch pollen allergen Bet v 1. J Allergy Clin Immunol (1997) 99:374–84. doi: 10.1016/s0091-6749(97)70056-3
46. James LK, Bowen H, Calvert RA, Dodev TS, Shamji MH, Beavil AJ, et al. Allergen specificity of IgG4-expressing B cells in patients with grass pollen allergy undergoing immunotherapy. J Allergy Clin Immunol (2012) 130:663–70. doi: 10.1016/j.jaci.2012.04.006
47. Heeringa JJ, Rijvers L, Arends NJ, Driessen GJ, Pasmans SG, van Dongen JJM, et al. IgE-expressing memory B cells and plasmablasts are increased in blood of children with asthma, food allergy and atopic dermatitis. Allergy (2018) 73:1331–6. doi: 10.1111/all.13421
48. Croote D, Darmanis S, Nadeau KC, Quake SR. High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science (2018) 362:1306. doi: 10.1126/science.aau2599
49. Jimenez-Saiz R, Ellenbogen Y, Bruton K, Spill P, Sommer DD, Lima H, et al. Human BCR analysis of single-sorted, putative IgE+ memory B cells in food allergy. J Allergy Clin Immunol (2019) 144:336–9. doi: 10.1016/j.jaci.2019.04.001
50. Flicker S, Steinberger P, Norderhaug L, Sperr WR, Majlesi Y, Valent P, et al. Conversion of grass pollen allergen-specific human IgE into a protective IgG1 antibody. Eur J Immunol (2002) 32:2156–62. doi: 10.1002/1521-4141(200208)32:8<2156::AID-IMMU2156>3.0.CO;2-A
51. Jakobsen CG, Bodtger U, Kristensen P, Poulsen LK, Roggen EL. Isolation of high-affinity human IgE and IgG antibodies recognising Bet v 1 and Humicola lanuginosa lipase from combinatorial phage libraries. Mol Immunol (2004) 41:941–53. doi: 10.1016/j.molimm.2004.05.009
52. Jylhä S, Mäkinen-Kiljunen S, Haahtela T, Söderlund H, Takkinen K, Laukkanen ML. Selection of recombinant IgE antibodies binding the beta-lactoglobulin allergen in a conformation-dependent manner. J Immunol Methods (2009) 350:63–70. doi: 10.1016/j.jim.2009.07.007
53. Laukkanen ML, Mäkinen-Kiljunen S, Isoherranen K, Haahtela T, Söderlund H, Takkinen K. Hevein-specific recombinant IgE antibodies from human single-chain antibody phage display libraries. J Immunol Methods (2003) 278:271–81. doi: 10.1016/s0022-1759(03)00070-x
54. Andréasson U, Flicker S, Lindstedt M, Valenta R, Greiff L, Korsgren M. The Human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J Mol Biol (2006) 362:212–27. doi: 10.1016/j.jmb.2006.06.062
55. Persson H, Flicker S, Sadegh MK, Greiff L, Valenta R, Ohlin M. A common idiotype in IgE and its relation to recognition of the grass pollen allergen Phl p 2. Mol Immunol (2008) 45:2715–20. doi: 10.1016/j.molimm.2008.01.004
56. Persson H, Sadegh MK, Greiff L, Ohlin M. Delineating the specificity of an IgE-encoding transcriptome. J Allergy Clin Immunol (2007) 120:1186–92. doi: 10.1016/j.jaci.2007.06.041
57. Persson J, Augustsson P, Laurell T, Ohlin M. Acoustic microfluidic chip technology to facilitate automation of phage display selection. FEBS J (2008) 275:5657–66. doi: 10.1111/j.1742-4658.2008.06691.x
58. Madritsch C, Gadermaier E, Roder UW, Lupinek C, Valenta R, Flicker S. High-density IgE recognition of the major grass pollen allergen Phl p 1 revealed with single-chain IgE antibody fragments obtained by combinatorial cloning. J Immunol (2015) 194:2069–78. doi: 10.4049/jimmunol.1402185
59. de Lalla C, Tamborini E, Longhi R, Tresoldi E, Manoni M, Siccardi AG, et al. Human recombinant antibody fragments specific for a rye-grass pollen allergen: characterization and potential applications. Mol Immunol (1996) 33:1049–58. doi: 10.1016/s0161-5890(96)00061-2
60. Braren I, Blank S, Seismann H, Deckers S, Ollert M, Grunwald T, et al. Generation of human monoclonal allergen-specific IgE and IgG antibodies from synthetic antibody libraries. Clin Chem (2007) 53:837–44. doi: 10.1373/clinchem.2006.078360
61. Flicker S, Vrtala S, Steinberger P, Vangelista L, Bufe A, Petersen A, et al. A human monoclonal IgE antibody defines a highly allergenic fragment of the major timothy grass pollen allergen, Phl p 5: molecular, immunological, and structural characterization of the epitope-containing domain. J Immunol (2000) 165:3849–59. doi: 10.4049/jimmunol.165.7.3849
62. Flicker S, Steinberger P, Ball T, Krauth MT, Verdino P, Valent P, et al. Spatial clustering of the IgE epitopes on the major timothy grass pollen allergen Phl p 1: importance for allergenic activity. J Allergy Clin Immunol (2006) 117:1336–43. doi: 10.1016/j.jaci.2006.02.012
63. Laffer S, Vangelista L, Steinberger P, Kraft D, Pastore A, Valenta R. Molecular characterization of Bip 1, a monoclonal antibody that modulates IgE binding to birch pollen allergen, Bet v 1. J Immunol (1996) 157:4953–62.
64. Denépoux S, Eibensteiner PB, Steinberger P, Vrtala S, Visco V, Weyer A, et al. Molecular characterization of human IgG monoclonal antibodies specific for the major birch pollen allergen Bet v 1. Anti-allergen IgG can enhance the anaphylactic reaction. FEBS Lett (2000) 465:39–46. doi: 10.1016/s0014-5793(99)01703-2
65. Flicker S, Steinberger P, Eibensteiner PB, Lebecque S, Kraft D, Valenta R. Molecular characterization of a human immunoglobulin G4 antibody specific for the major birch pollen allergen, Bet v 1. Clin Exp Allergy (2008) 38:365–73. doi: 10.1111/j.1365-2222.2007.02883.x
66. Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Bajyana Songa E, et al. Naturally occurring antibodies devoid of light chains. Nature (1993) 363:446–8. doi: 10.1038/363446a0
67. Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature (1995) 374:168–73. doi: 10.1038/374168a0
68. Rast JP, Amemiya CT, Litman RT, Strong SJ, Litman GW. Distinct patterns of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics (1998) 47:234–45. doi: 10.1007/s002510050353
69. Nuttall SD, Krishnan UV, Hattarki M, De Gori R, Irving RA, Hudson PJ. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol Immunol (2001) 38:313–26. doi: 10.1016/s0161-5890(01)00057-8
70. Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkönig A, Ruf A, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc (2014) 9:674–93. doi: 10.1038/nprot.2014.039
71. Muyldermans S. Nanobodies: Natural single-domain antibodies. Annu Rev Biochem (2013) 82:775–97. doi: 10.1146/annurev-biochem-063011-092449
72. Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. Nanobodies and their potential applications. Nanomed (Lond) (2013) 8:1013–26. doi: 10.2217/nnm.13.86
73. Yau KYF, Dubuc G, Li S, Hirama T, MacKenzie CR, Jermutus L, et al. Affinity maturation of a V(H)H by mutational hotspot randomization. J Immunol Methods (2005) 297:213–24. doi: 10.1016/j.jim.2004.12.005
74. Koide A, Tereshko V, Uysal S, Margalef K, Kossiakoff AA, Koide S. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J Mol Biol (2007) 373:941–53. doi: 10.1016/j.jmb.2007.08.027
75. Goldman ER, Anderson GP, Liu JL, Delehanty JB, Sherwood LJ, Osborn LE, et al. Facile generation of heat-stable antiviral and antitoxin single domain antibodies from a semisynthetic llama library. Anal Chem (2006) 78:8245–55. doi: 10.1021/ac0610053
76. Monegal A, Ami D, Martinelli C, Huang H, Aliprandi M, Capasso P, et al. Immunological applications of single-domain llama recombinant antibodies isolated from a naïve library. Protein Eng Des Sel (2009) 22:273–80. doi: 10.1093/protein/gzp002
77. Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, et al. NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife (2016) 5:e16228. doi: 10.7554/eLife.16228
78. McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol (2018) 25(3):289–96. doi: 10.1038/s41594-018-0028-6
79. Zimmermann I, Egloff P, Hutter CAJ, Kuhn BT, Bräuer P, Newstead S, et al. Generation of synthetic nanobodies against delicate proteins. Nat Protoc (2020) 15:1707–41. doi: 10.1038/s41596-020-0304-x
80. Ferrari D, Garrapa V, Locatelli M, Bolchi A. A novel nanobody scaffold optimized for bacterial expression and suitable for the construction of ribosome display libraries. Mol Biotechnol (2020) 62:43–55. doi: 10.1007/s12033-019-00224-z
81. Hutter CAJ, Timachi MH, Hürlimann LM, Zimmermann I, Egloff P, Göddeke H, et al. The extracellular gate shapes the energy profile of an ABC exporter. Nat Commun (2019) 10:2260. doi: 10.1038/s41467-019-09892-6
82. Bräuer P, Parker JL, Gerondopoulos A, Zimmermann I, Seeger MA, Barr FA, et al. Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science (2019) 363:1103–7. doi: 10.1126/science.aaw2859
83. Lauwereys M, Ghahroudi MA, Desmyter A, Kinne J, Hölzer W, De Genst E, et al. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J (1998) 17:3512–20. doi: 10.1093/emboj/17.13.351
84. Unger M, Eichhoff AM, Schumacher L, Strysio M, Menzel M, Schwan C, et al. Selection of nanobodies that block the enzymatic and cytotoxic activities of the binary clostridium difficile toxin CDT. Sci Rep (2015) 5:7850. doi: 10.1038/srep07850
85. De Genst EJ, Guilliams T, Wellens J, O’Day EM, Waudby CA, Meehan S, et al. Structure and properties of a complex of alpha-synuclein and a single-domain camelid antibody. J Mol Biol (2010) 9402:326–43. doi: 10.1016/j.jmb.2010.07.001
86. Braun MB, Traenkle B, Koch PA, Emele F, Weiss F, Poetz O, et al. Peptides in headlock–a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci Rep (2016) 6:19211. doi: 10.1038/srep19211
87. Virant D, Traenkle B, Maier J, Kaiser PD, Bodenhöfer M, Schmees C, et al. A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat Commun (2018) 9:930. doi: 10.1038/s41467-018-03191-2
88. Götzke H, Kilisch M, Martínez-Carranza M, Sograte-Idrissi S, Rajavel A, Schlichthaerle T, et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat Commun (2019) 10:4403. doi: 10.1038/s41467-019-12301-7
89. Ling J, Cheloha RW, McCaul N, J Sun ZJ, Wagner G, Ploegh HL. A nanobody that recognizes a 14-residue peptide epitope in the E2 Ubiquitin-conjugating enzyme UBC6e modulates its activity. Mol Immunol (2019) 114:513–23. doi: 10.1016/j.molimm.2019.08.008
90. Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol Biotechnol (2007) 77:13–22. doi: 10.1007/s00253-007-1142-2
91. Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: Big opportunities for small antibodies. Drug Discovery Today (2016) 21:1076‒113. doi: 10.1016/j.drudis.2016.04.003
92. Van Audenhove I, Gettemans J. Nanobodies as versatile tools to understand, diagnose, visualize and treat cancer. EBioMed (2016) 8:40‒48. doi: 10.1016/j.ebiom.2016.04.028
93. Iezzi ME, Policastro L, Werbajh S, Podhajcer O, Canziani GA. Single-domain antibodies and the promise of modular targeting in cancer imaging and treatment. Front Immunol (2018) 9:273. doi: 10.3389/fimmu.2018.00273
94. Tillib SV. Prospective applications of single-domain antibodies in biomedicine. Mol Biol (Mosk) (2020) 54:317–26. doi: 10.1134/S0026893320030164
95. Chen F, Ma H, Li Y, Wang H, Samad A, Zhou J, et al. Screening of nanobody specific for peanut major allergen Ara h 3 by phage display. J Agric Food Chem (2019) 67:11219–29. doi: 10.1021/acs.jafc.9b02388
96. Hecker J, Diethers A, Schulz D, Sabri A, Plum M, Michel Y, et al. An IgE epitope of Bet v 1 and fagales PR10 proteins as defined by a human monoclonal IgE. Allergy (2012) 67:1530–7. doi: 10.1111/all.12045
97. Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem (2009) 284:3273–84. doi: 10.1074/jbc.M806889200
98. Cortez-Retamozo V, Backmann N, Senter PD, Wernery U, De Baetselier P, Muyldermans S, et al. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res (2004) 64:2853–7. doi: 10.1158/0008-5472.can-03-3935
99. Baral TN, Magez S, Stijlemans B, Conrath K, Vanhollebeke B, Pays E, et al. Experimental therapy of African trypanosomiasis with a nanobody-conjugated human trypanolytic factor. Nat Med (2006) 12:580–4. doi: 10.1038/nm1395
100. Coppieters K, Dreier T, Silence K, de Haard H, Lauwereys M, Casteels P, et al. Formatted anti-tumor necrosis factor α VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheumatol (2006) 54:1856–66. doi: 10.1002/art.21827
101. Könning D, Zielonka S, Grzeschik J, Empting M, Valldorf B, Krah S, et al. Camelid and shark single domain antibodies: Structural features and therapeutic potential. Curr Opin Struct Biol (2017) 45:10‒16. doi: 10.1016/j.sbi.2016.10.019
102. Duggan S. Caplacizumab: first global approval. Drugs (2018) 78:1639–42. doi: 10.1007/s40265-018-0989-0
103. De Vlieger D, Ballegeer M, Rossey I, Schepens B, Saelens X. Single-domain antibodies and their formatting to combat viral infections. Antibodies (Basel) (2018) 8:1. doi: 10.3390/antib8010001
104. Kontermann RE. Strategies for extended serum half-life of protein therapeutics. Curr Opin Biotechnol (2011) 22:868–76. doi: 10.1016/j.copbio.2011.06.012
105. Stone E, Hirama T, Tanha J, Tong-Sevinc H, Li S, MacKenzie CR, et al. The assembly of single domain antibodies into bispecific decavalent molecules. J Immunol Methods (2007) 318:88‒94. doi: 10.1016/j.jim.2006.10.006
106. Hultberg A, Temperton NJ, Rosseels V, Koenders M, Gonzalez-Pajuelo M, Schepens B, et al. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PloS One (2011) 6:e17665. doi: 10.1371/journal.pone.0017665
107. Tillib SV, Ivanova TI, Vasilev LA, Rutovskaya MV, Saakyan SA, Gribova IY, et al. Formatted single-domain antibodies can protect mice against infection with influenza virus (H5N2). Antiviral Res (2013) 97:245‒54. doi: 10.1016/j.antiviral.2012.12.014
108. Huet HA, Growney JD, Johnson JA, Li J, Bilic S, Ostrom L, et al. Multivalent nanobodies targeting death receptor 5 elicit superior tumor cell killing through efficient caspase induction. MAbs (2014) 6:1560‒70. doi: 10.4161/19420862.2014.975099
109. Nosenko MA, Atretkhany KN, Mokhonov VV, Efimov GA, Kruglov AA, Tillib SV, et al. VHH-based bispecific antibodies targeting cytokine production. Front Immunol (2017) 8:1073. doi: 10.3389/fimmu.2017.01073
110. Garas MN, Tillib SV, Zubkova OV, Rogozhin VN, Ivanova TI, Vasilev LA, et al. Construction of a pIX-modified adenovirus vector able to effectively bind to nanoantibodies for targeting. Acta Naturae (2014) 6:95–105. doi: 10.32607/20758251-2014-6-2-95-105
111. Burmistrova DA, Tillib SV, Shcheblyakov DV, Dolzhikova IV, Shcherbinin DN, Zubkova OV, et al. Genetic passive immunization with adenoviral vector expressing chimeric nanobody-Fc molecules as therapy for genital infection caused by Mycoplasma hominis. PloS One (2016) 11:e0150958. doi: 10.1371/journal.pone.0150958
112. Hussack G, Luo Y, Veldhuis L, Hall JC, Tanha J, Mackenzie R. Multivalent anchoring and oriented display of single-domain antibodies on cellulose. Sensors (Basel) (2009) 9:5351–67. doi: 10.3390/s90705351
113. Tillib SV, Privezentseva ME, Ivanova TI, Vasilev LF, Efimov GA, Gursky YG, et al. Single-domain antibody-based ligands for immunoaffinity separation of recombinant human lactoferrin from the goat lactoferrin of transgenic goat milk. J Chromatogr B Analyt Technol BioMed Life Sci (2014) 949-950:48–57. doi: 10.1016/j.jchromb.2013.12.034
Keywords: allergy, allergen, nanobody, VHH, blocking antibody, allergy treatment
Citation: Flicker S, Zettl I and Tillib SV (2020) Nanobodies—Useful Tools for Allergy Treatment? Front. Immunol. 11:576255. doi: 10.3389/fimmu.2020.576255
Received: 25 June 2020; Accepted: 15 September 2020;
Published: 30 September 2020.
Edited by:
Christiane Hilger, Luxembourg Institute of Health, LuxembourgReviewed by:
Tomasz Uchanski, Luxembourg Institute of Health, LuxembourgCopyright © 2020 Flicker, Zettl and Tillib. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabine Flicker, c2FiaW5lLmZsaWNrZXJAbWVkdW5pd2llbi5hYy5hdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.