
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Immunol. , 23 June 2020
Sec. Vaccines and Molecular Therapeutics
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01531
This article is part of the Research Topic Coronavirus Disease (COVID-19): Pathophysiology, Epidemiology, Clinical Management and Public Health Response View all 400 articles
With more than 6.9 M confirmed cases and ~400 K deaths as on June 8, 2020 (1), COVID-19, ushered in by the SARS-CoV-2 has projected itself as a microscopic-holocaust, much more sinister than those portrayed in the SciFi movies. Asymptomatic transmission of the virus has been projected as the Achilles' heel in the context of the current control strategies of the pandemic (2, 3). Reports on undiagnosed deep vein thrombosis among patients, succumbing to the viral assault (4) and demonstration of direct infection of human blood vessel and kidney organoids (5) have triggered huge hue and cry. The extreme high transmissibility of the virus, bracketed together with current absence of population immunity and occurrence of stark clinical consequences projects the swift advancement in effective therapeutic stratagems as the need of the hour. Needless to say, researchers, across the globe, are beavering to devise appropriate diagnostic and therapeutic strategies. The various nucleic acid based detection-approaches like PCR, isothermal nucleic acid amplification-based methods, CRISPR/Cas platforms as well as immunoassay based point-of-care lateral flow tests are marked with respective pros and cons (6, 7). On the other hand, strategies of inhibiting the viral fusion/entry, disrupting the replication pathway, suppressing the inflammatory response, using convalescent plasma treatment and vaccine development have been at the forefront of recent research (8). The success lies in our comprehensive understanding of the “biochemically and genetically guileful” virus. At this juncture, it is relevant to mention that long-term development of appropriate antibody and other protein therapeutics to effectively bind and neutralize the viral infection is imperative. This would be significant in case the researchers need to buy excess time to ensure befitting vaccine discovery and development. Such therapeutics could possibly provide an alternative/additional way to assist those people who might show unresponsiveness to vaccines (as, exemplified by many in the elderly population) or do not obtain vaccine. Amidst the current hay-wired situation, the recent communiqué from Israeli Defense Minister Natfali Bennet about the successful isolation of a “monoclonal neutralizing antibody” with potency to “neutralize [disease] inside carriers” bodies' by the scientists in the Israel Institute for Biological Research has ushered in new waves of hope (9).
Prior to getting ahead, it would be prudent to recapitulate the general aspects of the lifecycle of the highly pathogenic human coronaviruses (CoVs) (10) (Figure 1A). Talking about the viral pathogenesis, the receptor binding domains (RBD) of the spike (S) glycoprotein interact with the human angiotensin-converting enzyme 2 (ACE2)- the receptor that invites SARS-CoV and SARS-CoV-2 into human cells (Figure 1Ba). The presence of a furin cleavage site at interfacial zone of the S1/S2 subunits of the SARS-CoV-2 S glycoprotein demarcates the virus from SARS-CoV and SARS-related CoVs (13). Precise understanding of the SARS-CoV-2 S ectodomain trimer is envisaged to be instrumental in developing vaccines, therapeutic antibodies and diagnostics. The prospective targets of neutralizing antibodies (nAbs) against human pathogenic CoVs are depicted in Figure 1Bb. Monoclonal antibodies (mAbs), functional antigen-binding fragment (Fab), single-chain variable region fragment (scFv), and single-domain antibodies (nanobodies or Nbs) have been assessed against various human CoVs (14–19). Jiang et al. (10) have recently reviewed the development of SARS-CoV- and MERS-CoV-specific nAbs, while literature reports on nAbs against SARS-CoV-2 are comparatively scanty. Previous studies on neutralization with anti-SARS-CoV-1 RBD and anti-MERS-CoV RBD antibodies had unveiled a premature switching from the pre-fusion to post-fusion conformation following a closure of the receptor binding site and trapping the RBD in “up” conformation (20–22). The structure of CR3022, an antibody derived from a convalescent SARS patient, in complex with the RBD of the S protein at a resolution of 3.1 Å was recently reported (23). Interestingly, a cross-reactive interaction between SARS-CoV-2 and SARS-CoV was evinced by the elucidation that a highly conserved but cryptic, epitope, distal from the receptor binding site is targeted by CR3022. However, at least two RBDs on the trimeric S protein in the “up” conformation and slight rotation are prerequisites to access the binding epitope by CR3022. The authors proposed that albeit, the CR3022 fails to neutralize SARS-CoV-2 in vitro, the epitope could plausibly confer in vivo protection. On a similar vein, researchers have resorted to the use of SARS-CoV-2 S murine polyclonal antibodies for the inhibition of SARS-CoV-2 S mediated entrance into cells (13). The study vouched that vaccination could elicit cross-neutralizing antibodies, targeting the conserved S epitopes.
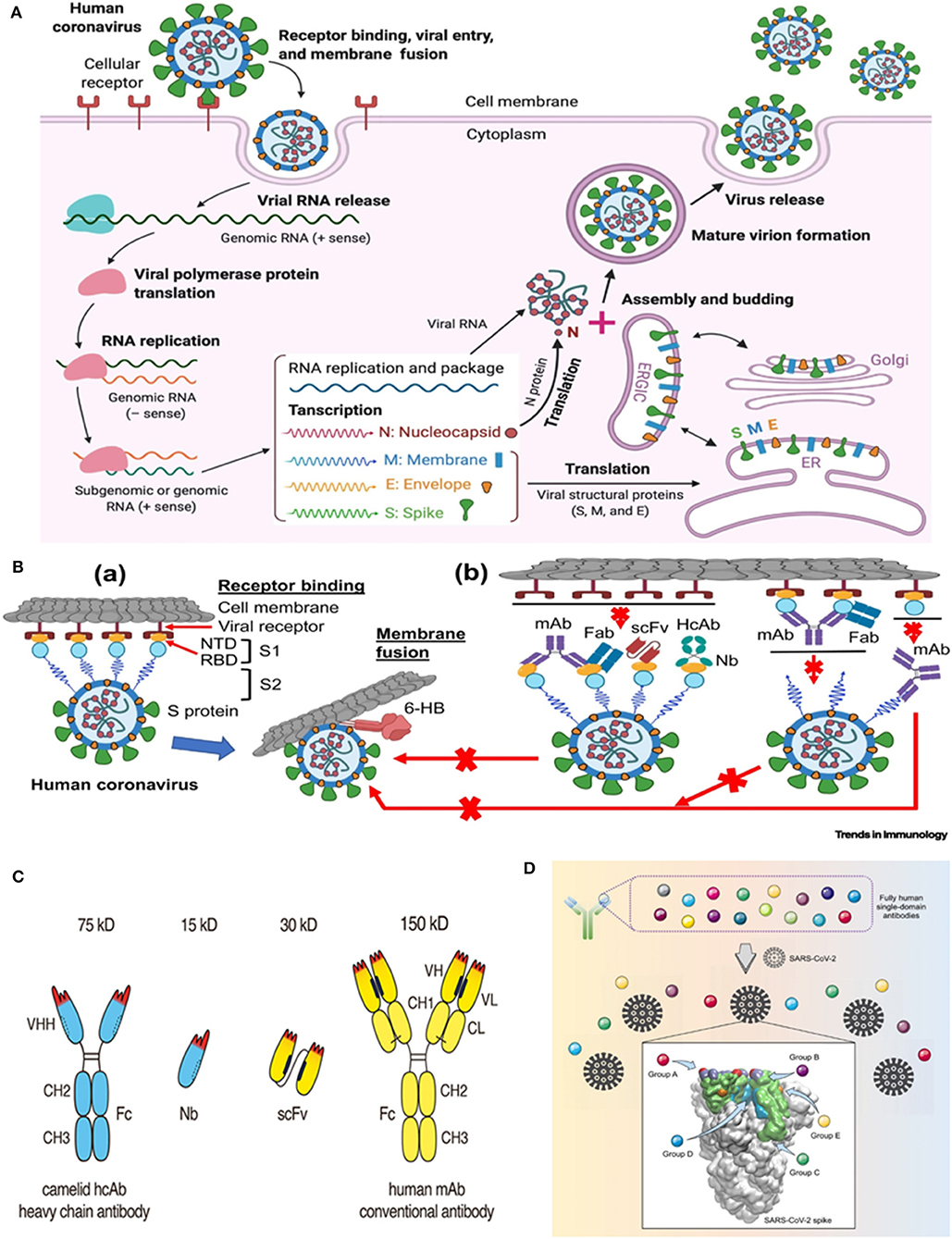
Figure 1. Life cycle of highly pathogenic human coronaviruses (CoVs) and specific neutralizing antibodies (nAbs) against these coronaviruses. (A) Life cycle of highly pathogenic human CoVs. These CoVs enter host cells by first binding to their respective cellular receptors [angiotensin-converting enzyme 2 (ACE2) for severe acute respiratory syndrome (SARS)-CoV-2 or SARS-CoV and dipeptidyl peptidase 4 (DPP4) for Middle East respiratory syndrome (MERS)-CoV] on the membranes of host cells expressing ACE2 (e.g., pneumocytes, enterocytes) or DPP4 (e.g., liver or lung cells including Huh-7, MRC-5, and Calu-3) via the surface spike (S) protein, which mediates virus–cell membrane fusion and viral entry. Viral genomic RNA is released and translated into viral polymerase proteins. The negative (–)-sense genomic RNA is synthesized and used as a template to form sub-genomic or genomic positive (+)-sense RNA. Viral RNA and nucleocapsid (N) structural protein are replicated, transcribed, or synthesized in the cytoplasm, whereas other viral structural proteins, including S, membrane (M), and envelope (E), are transcribed then translated in the endoplasmic reticulum (ER) and transported to the Golgi. The viral RNA-N complex and S, M, and E proteins are further assembled in the ER–Golgi intermediate compartment (ERGIC) to form a mature virion, then released from host cells. (B) Potential targets of nAbs against SARS-CoV-2 and other pathogenic human CoVs. (a) Human CoV receptor binding and membrane fusion process. The CoV first binds a viral receptor (ACE2 or DPP4) through the receptor-binding domain (RBD) in the S protein, followed by fusion of the virus with cell membranes via the formation of a six-helix bundle (6-HB) fusion core. NTD, N-terminal domain. (b) Potential targets of nAbs on the S protein of human CoVs. Monoclonal antibody (mAb), antigen-binding fragment (Fab), single-chain variable region fragment (scFv), or single-domain antibody [nanobody (Nb) or VHH derived from camelid heavy chain antibody (HcAb)] binds to the RBD, S1 subunit (non-RBD, including NTD), or S2 of the viral S protein, blocking binding between the RBD and the respective receptor (for RBD-targeting nAbs), interfering with the conformational change of S (for S1-targeting nAbs), or hindering S2-mediated membrane fusion (for S2-targeting nAbs), leading to the inhibition of infection with pathogenic human CoVs in the host cells. The figure was created using BioRender (https://biorender.com/). [Reproduced from (10), under the provisions of Creative Commons License, CC BY 4.0, Copyright © 2020 The Author(s). Published by Elsevier Ltd.]. (C) Advantageous features of camelid heavy chain antibodies. Heavy chain antibodies are composed of two heavy chains. The target-binding module is composed of a single VHH domain. A recombinant VHH domain, designated nanobody (Nb) is highly soluble and does not show any tendency to associate with other hydrophobic protein surfaces. Conventional antibodies are composed of two heavy and two light chains. The target-binding module is composed of two non-covalently associated variable domains VH and VL. In intact antibodies, the proper orientation of these domains is mediated by a hydrophobic interface and is further stabilized by the disulfide-linked CL and CH1 domains. A pair of VH and VL domains can be linked genetically into a single-chain variable fragment (scFv) in which the proper orientation of domains is mediated alone by the hydrophobic interface between the two V-domains. [Reproduced from (11), under the provisions of Creative Commons Attribution License (CC BY). Copyright © 2017 Bannas, Hambach and Koch-Nolte]. (D) Targeting of diverse epitopes within the SARS-CoV-2 spike protein receptor binding domain (RBD) by human single-domain antibodies, potential therapeutic candidates for COVID-19. [Reproduced from (12) Copyright ©2020 Elsevier Inc., based on the reuse-provisions of Elsevier's COVID-19 Resource Centre].
At this juncture, the germaneness of antibody engineering may be comprehended in the context of continual search for high-affinity antibodies, effective against conserved targets as well as novel therapeutics with attributes like better tumor and tissue penetration and efficient launching of immune effector functions (24). Particularly, in the context of antitumor therapeutics, Bannas et al. (11) had raised concerns about the large-size (150 kDa) dictated practical snag of in vivo delivery of conventional antibodies to tumor cells. On the other hand, aggregation and/or mispairing of V-domains due to lower stability and solubility of engineered antibodies- a consequence of intrinsic hydrophobic interactions of VH and VL domains (that constitute the antigen binding fragment (Fab) of IgG antibodies) have been another pertinent issue. As plausible solutions, nanobodies (15 kDa) and nanobody based human heavy chain antibodies (75 kDa) (11) have instigated considerable research impetus. Besides conventional antibodies, camelids produce heavy-chain-only antibodies (HCAbs) with a single variable domain as the target recognition module (25, 26). This single variable domain without an effector domain functions as a single-domain antibody, VHH, or nanobody (Nb) (Figure 1C). Although the prospects of using nanobodies as research and diagnostic tools have been critically and comprehensively assessed (27, 28) and a plethora of nanobodies are currently being placed under pre-clinical or clinical assessments for various diseases like brain tumors, inflammation, lung diseases, as well as autoimmune diseases, paralleling the performance of classical antibodies with nanobodies for therapeutic applications could be bit fiddly (29). Nevertheless, studies have attested the advantages of nanobodies in contrast to conventional antibodies with respect to the former's smaller size, amenability for processing into multiple formats, desirable thermal and chemical stability, high solubility, commendable in vivo tissue penetration and targeting, lower susceptibility to steric hindrances (that may otherwise obstruct optimal binding) as well as ability to display antigenic affinity and specificity at par with conventional antibodies (11, 30–34). Prospects of genetically linking to Fc-domains, peptide tags, or other nanobodies as well as site-specific chemical fusion with nanoscale materials, radionuclides, photosensitizers, etc. widen the spectrum of their applications. Furthermore, the expedient attributes of nanobodies and human Fc domains may be combined in chimeric nanobody-heavy chain antibodies, half the size of the conventional antibodies, as mentioned before (11).
Post perusal of the afore-stated, harnessing VHHs as therapeutics against various viral infectious agents seems to be an interesting proposition (35). In this respect, use of VHH against dengue virus (36); hepatitis C virus (37); multiple VHH monovalent candidates against poliovirus (38) and norovirus (39); anti-CXCR4 monovalent and bivalent (40) as well as anti-p24 monovalent and bivalent (41) nanobodies against HIV; VHH bivalent/albumin-linked nanobody against rabies virus (42) and anti-VP6 VHH as an effective prophylactic treatment against rotavirus A-associated diarrhea (43) have been documented. Investigations on the application of nanobodies against respiratory pathogens has also gained pace in recent years. Use of H5N1-HA bivalent nanobody against influenza virus (44), as well as the application of multi-domain antibody MD3606 (generated using diverse camelid single-domain antibodies to influenza virus hemagglutinin) to protect mice against influenza A and B infection post intravenous administration or expression using recombinant adeno-associated vector (32), merit special mention. Similarly, two llama-derived single-domain antibodies with human respiratory syncytial virus (RSV)–neutralizing action have been reported to selectively bind to RSV fusion protein (F) in its pre-fusion state with picomolar affinity (45). Delivering a trimeric nanobody, ALX-0171 (that interacted with antigenic site II of RSV F protein at subnanomolar affinity), prophylactically or therapeutically directly to lungs of cotton rats was effective in down-scaling both nasal and lung RSV titers (46). Stalin Raj et al. (47) had resorted to direct cloning and expression of VHHs of HCAbs from the bone marrow of MERS-CoV–infected Arabian camels and identified several MERS-CoV–specific VHHs or nanobodies. With a prolonged half-life in serum, camel/human chimeric HCAbs were efficacious in endowing protection to mice against MERS-CoV challenge. In a similar vein, the efficacy to target MERS-CoV S RBD using novel neutralizing Nb (NbMS10) and its human-Fc-fused version (NbMS10-Fc) has been documented (48). Remarkably, the Nbs were able to cross-neutralize infections caused by diverse MERS-CoV strains isolated from humans and camels. The Fc-tagged Nb was able to confer complete protection of humanized mice from lethal MERS-CoV assault.
A concerted effort of biologist Michael Rout and chemist Brian Chait has been directed toward selecting high affinity and effective neutralizing nanobodies, interacting with the various non-overlapping target-epitopes of SARS-CoV-2 S (49). The researchers envisage to set-up the appropriate nanobodies as increased level multimers to augment affinity and eventually tune them at the molecular level to better their neutralizing potency. Similarly, researchers from Protein Production UK, a project hosted by the Rosalind Franklin Institute in association with Diamond Light Source, UK, have made nanobodies (exhibiting high affinity to the S protein of the SARS-CoV-2), available to scientist at the University of Oxford for deeper delving into the structure of the virus (50). On a stimulating note, scientists from the University of Texas (UT) at Austin, the National Institutes of Health and Ghent University in Belgium have documented the isolation of two potently neutralizing VHHs, targeting the SARS-CoV-1 and MERS-CoV RBDs, respectively (34). Wrapp et al. (34) had resorted to sequential immunization of a llama subcutaneously multiple times with SARS-CoV-1 S and MERS-CoV S protein. Two sequential rounds of panning were executed by phage display using either SARS-CoV-1 S or MERS-CoV S proteins to procure VHHs directed against the S proteins. The researchers successfully isolated seven unique MERS-CoV S and five SARS-CoV-1 S specific VHHs post-sequencing of the positive clones, multiple sequence alignment, and phylogenetic analysis. Following expression in Pichia pastoris and purification from yeast medium, the interaction of the purified VHHs with the perfusion-stabilized MERS-CoV S and SARS-CoV-1 S was attested by ELISA. Pertinently, the SARS-CoV-1 RBD-directed VHH could cross-react with the SARS-CoV-2 RBD. A fascinating dimension to the work was the neutralization of the SARS-CoV-2 S pseudotyped viruses by the cross reactive VHH, engineered as a bivalent human IgG Fc-fusion. The plausible scaled up production of the VHH-Fc fusion was attested in a commercial-standard CHO cell system. The MERS VHH-55, SARS VHH-72 and VHH-72-Fc, exhibiting desirable biophysical attributes and potent neutralization potency, could be prospective therapeutic candidates. However, appropriate in vivo experimentations as part of preclinical studies are prerequisite.
Retrieval of information from the preprint at BioRxiv evinces the successful endeavors of Swiss researchers Walter et al. (51) in identifying 63 unique anti-RBD synthetic nanobodies or sybodies, interacting in the context of the full-length SARS-CoV-2 spike ectodomain. Assisted by a prompt in vitro selection platform (encompassing ribosome and phage display), the task of selecting the sybodies was accomplished within 12 days. Six of the selected sybodies displayed double-digit nanomolar binding affinity with the viral spike while five of them could inhibit RBD interaction with ACE2. Furthermore, the researchers identified a pair of anti-RBD sybodies that could concomitantly interact with the RBD. It would be interesting to peruse the outcomes of the authors' previously reported NestLink technology (52) based delving of the selection pools to unearth unique sybodies with little off-rates and capacity to identify rare epitopes. The authors are upbeat about plausible therapeutic exploitation of the sybodies for the development of an inhalable drug as useful prophylaxis against COVID-19.
To speak about yet another development, Beroni Group (an international biopharmaceutical enterprise) in concert with Tianjin University in China has recently identified 24 types of nanobodies (post-screening a library with one billion-plus nanobody sequences) for prompt detection and treatment of SARS-CoV-2 (53). Eight of them are directed against the S protein while sixteen of them target the nucleocapsid (N) protein- the latter could find application as a marker in diagnostic assays. Based on approaches of structural biology, computational biology, and protein engineering, the researchers are gearing up to optimize the properties of the nanobodies besides endeavoring to reduce their immunogenicity and augment the therapeutic efficiency by humanizing them. By the same token, researchers from Fudan University and Biomissile Corporation, China have directed their endeavors toward the development of a phage-displayed single-domain antibody library based on embedding naive complementarity-determining regions (CDRs) into framework sites of a human germline immunoglobulin heavy chain variable region (IGHV) allele (12). Their study, encompassing the library-biopanning against SARS-CoV-2 RBD and S1 subunit led to the revelation of fully human single-domain antibodies, displaying low-nanomolar/subnanomolar range affinities toward five distinct epitopes on SARS-CoV-2 RBD (Figure 1D). Amongst the groups of A, B, C, D, and E neutralizing antibodies, the group D members, n3088 and n3130 could target a “cryptic” epitope, positioned in the spike trimeric interface, resulting in effective neutralization of SARS-CoV-2. The researchers are buoyant about the apt application of these, either alone or in synergy with other SARS-CoV-2 neutralizing antibodies, especially the ACE2-competing neutralizing antibodies. They may also be employed as integrant for creating bispecific or multispecific antibodies (12). Previously, He et al. (54) had demonstrated an augmented efficacy of oligomeric nanobodies, relative to monomeric nanobodies against MERS coronavirus RBD. Investigating the potential of such oligomeric nanobodies in the case of SARS-CoV-2 would be attention-grabbing.
These studies spark obvious anticipations and hopes for the potential application of nanobodies against COVID-19. The attributes of small size (almost one-fourth of the size of human antibodies) and simple structure, ease and comparatively lower cost, low immunogenicity and ability to display high affinity have endowed them with a special niche in the realm of therapeutics and rapid point-of-care diagnostics. Nanobodies seem to be quite efficient in trapping and stabilizing conformation-switchable targets in specific conformations, facilitating greater insight into biomolecular mechanisms and interactions. This could be of immense relevance to mine information on SARS-CoV-2 pathogenesis. Most importantly, highly stable VHHs could be nebulized and exploited for the development of inhalable prophylactic formulations, thereby ensuring straight delivery to the lungs- the combat zone. Another merit lies in the plausibility of stockpiling the VHHs without trade-off in their stability even after extended storages and using them as therapeutic choices in case of disasters like COVID-19. To conclude, I do hope that the incessant and concerted research endeavors would surely pave the way to a safer world, liberated from the grasp of SARS-CoV-2 and akin.
RK reviewed the literature, critically analyzed it and authored the article.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
A bouquet of gratitude is extended to all the researchers and frontline warriors, engaged in the battle against COVID-19.
1. World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. (2020). Available online at: https://covid19.who.int/?gclid=Cj0KCQjww_f2BRC-ARIsAP3zarEfohyPnXxXij4jnp412SSC_7kKj9J3pEly4tpE21VVLydZtb3KaZsaAlH-EALw_wcB (accessed June 9, 2020).
2. Day M. COVID-19: four fifths of cases are asymptomatic, China figures indicate. BMJ. (2020) 369:m1375. doi: 10.1136/bmj.m1375
3. Gandhi M, Yokoe DS, Havlir DV. Asymptomatic transmission, the Achilles' heel of current strategies to control COVID-19. N Engl J Med. (2020) 382:2158–60. doi: 10.1056/NEJMe2009758
4. Wichmann D, Sperhake JP, Lütgehetmann M, Steurer S, Edler C, Heinemann A, et al. Autopsy findings and venous thromboembolism in patients with COVID-19: A prospective cohort study. Ann Intern Med. (2020). doi: 10.7326/M20-2003. [Epub ahead of print].
5. Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. (2020) 181:905–13.e7. doi: 10.1016/j.cell.2020.04.004
6. Bai H, Cai X, Zhan X. Landscape coronavirus disease 2019 test (COVID-19 test) in vitro- A comparison of PCR vs Immunoassay vs CRISPR-based test. OSF [Preprints]. (2020). doi: 10.31219/osf.io/6eagn
7. Shen M, Zhou Y, Ye J, AL-maskri AAA, Kang Y, Zeng S, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. (2020) 10:97–101. doi: 10.1016/j.jpha.2020.02.010
8. Li H, Zhou Y, Zhang M, Wang H, Zhao Q, Liu J. Updated approaches against SARS-CoV-2. Antimicrob Agents Chemother. (2020) 64:e00483–20. doi: 10.1128/AAC.00483-20
9. Perper R. Israel's Government Research Agency Says it Successfully Isolated a Key Coronavirus Antibody, Paving the Way for a Possible Breakthrough Treatment. (2020). Available online at: https://www.pulselive.co.ke/bi/politics/israels-government-research-agency-says-it-successfully-isolated-a-key-coronavirus/fyfyz3q (accessed May 8, 2020).
10. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. (2020) 41:355–9. doi: 10.1016/j.it.2020.03.007
11. Bannas P, Hambach J, Koch-Nolte F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol. (2017) 8:1603. doi: 10.3389/fimmu.2017.01603
12. Wu Y, Li C, Xia S, Tian X, Kong Y, Wang Z, et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. (2020) 27:891–8.e5. doi: 10.1101/2020.03.30.015990
13. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. (2020) 181:281–92.e6. doi: 10.1016/j.cell.2020.02.058
14. Zhou Y, Yang Y, Huang J, Jiang S, Du L. Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. (2019) 11:60. doi: 10.3390/v11010060
15. Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. PNAS. (2007) 104:12123–8. doi: 10.1073/pnas.0701000104
16. Rockx B, Corti D, Donaldson E, Sheahan T, Stadler K, Lanzavecchia A, et al. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol. (2008) 82:3220–35. doi: 10.1128/JVI.02377-07
17. Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. PNAS. (2004) 101:2536–41. doi: 10.1073/pnas.0307140101
18. He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J Immunol. (2006) 176:6085–92. doi: 10.4049/jimmunol.176.10.6085
19. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. (2020) 9:382–5. doi: 10.1080/22221751.2020.1729069
20. Hwang WC, Lin Y, Santelli E, Sui J, Jaroszewski L, Stec B, et al. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem. (2006) 281:34610–6. doi: 10.1074/jbc.M603275200
21. Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. (2019) 176:1026–39. doi: 10.1016/j.cell.2018.12.028
22. Wang L, Shi W, Chappell JD, Joyce MG, Zhang Y, Kanekiyo M, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J Virol. (2018) 92:e02002–17. doi: 10.1128/JVI.02002-17
23. Yuan M, Wu NC, Zhu X, Lee CCD, So RT, Lv H, et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. (2020) 368:630–3. doi: 10.1101/2020.03.13.991570
24. Saeed AF, Wang R, Ling S, Wang S. Antibody engineering for pursuing a healthier future. Front. Microbiol. (2017) 8:495. doi: 10.3389/fmicb.2017.00495
25. Hamers-Casterman CTSG, Atarhouch T, Muyldermans S, Robinson G, Hammers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature. (1993) 363:446–8. doi: 10.1038/363446a0
26. Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. (2013) 82:775–97. doi: 10.1146/annurev-biochem-063011-092449
27. De Meyer T, Muyldermans S, Depicker A. Nanobody-based products as research and diagnostic tools. Trends Biotechnol. (2014) 32:263–70. doi: 10.1016/j.tibtech.2014.03.001
28. Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today. (2016) 21:1076–113. doi: 10.1016/j.drudis.2016.04.003
29. Jovčevska I, Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. (2019) 34:11–6. doi: 10.1007/s40259-019-00392-z
30. De Vlieger D, Ballegeer M, Rossey I, Schepens B, Saelens X. Single-domain antibodies and their formatting to combat viral infections. Antibodies. (2019) 8:1. doi: 10.3390/antib8010001
31. Govaert J, Pellis M, Deschacht N, Vincke C, Conrath K, Muyldermans S, et al. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J Biol Chem. (2012) 287:1970–9. doi: 10.1074/jbc.M111.242818
32. Laursen NS, Friesen RH, Zhu X, Jongeneelen M, Blokland S, Vermond J, et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. (2018) 362:598–602. doi: 10.1126/science.aaq0620
33. Fernandes JC. Therapeutic application of antibody fragments in autoimmune diseases: Current state and prospects. Drug Discov Today. (2018) 23:1996–2002. doi: 10.1016/j.drudis.2018.06.003
34. Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. (2020) 181:1–12 doi: 10.1101/2020.03.26.010165
35. Sroga P, Safronetz D, Stein DR. Nanobodies: a new approach for the diagnosis and treatment of viral infectious diseases. Future Virol. (2020) 15: 195–205. doi: 10.2217/fvl-2019-0167
36. Fatima A, Wang H, Kang K, Xia L, Wang Y, Ye W, et al. Development of VHH antibodies against dengue virus type 2 NS1 and comparison with monoclonal antibodies for use in immunological diagnosis. PLoS ONE. (2014) 9:95263. doi: 10.1371/journal.pone.0095263
37. Tarr AW, Lafaye P, Meredith L, Damier-Piolle L, Urbanowicz RA, Meola A, et al. An alpaca nanobody inhibits hepatitis C virus entry and cell-to-cell transmission. Hepatol. (2013) 58:932–9. doi: 10.1002/hep.26430
38. Thys B, Schotte L, Muyldermans S, Wernery U, Hassanzadeh-Ghassabeh G, Rombaut B. In vitro antiviral activity of single domain antibody fragments against poliovirus. Antivir Res. (2010) 87:257–64. doi: 10.1016/j.antiviral.2010.05.012
39. Koromyslova AD, Hansman GS. Nanobodies targeting norovirus capsid reveal functional epitopes and potential mechanisms of neutralization. PLoS Pathog. (2017) 13:e1006636 doi: 10.1371/journal.ppat.1006636
40. Jähnichen S, Blanchetot C, Maussang D, Gonzalez-Pajuelo M, Chow KY, Bosch L, et al. CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc Natl Acad Sci USA. (2010) 107:20565–70. doi: 10.1073/pnas.1012865107
41. Gray ER, Brookes JC, Caillat C, Turbé V, Webb BL, Granger LA, et al. Unravelling the molecular basis of high affinity nanobodies against HIV p24: in vitro functional, structural, in silico insights. ACS Infect Dis. (2017) 3:479–91. doi: 10.1021/acsinfecdis.6b00189
42. Terryn S, Francart A, Rommelaere H, Stortelers C, Van Gucht S. Post-exposure treatment with anti-rabies VHH and vaccine significantly improves protection of mice from lethal rabies infection. PLoS Neglect Trop D. (2016) 10:e0004902. doi: 10.1371/journal.pntd.0004902
43. Maffey L, Vega CG, Miño S, Garaicoechea L, Parreño V. Anti-VP6 VHH: an experimental treatment for rotavirus A-associated disease. PLoS ONE. (2016) 11:e0162351. doi: 10.1371/journal.pone.0162351
44. Ibanez LI, De Filette M, Hultberg A, Verrips T, Temperton N, Weiss RA, et al. Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. J Infect Dis. (2011) 203:1063–72. doi: 10.1093/infdis/jiq168
45. Rossey I, Gilman MS, Kabeche SC, Sedeyn K, Wrapp D, Kanekiyo M, et al. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat Commun. (2017) 8:14158. doi: 10.1038/ncomms14158
46. Detalle L, Stohr T, Palomo C, Piedra PA, Gilbert BE, Mas V, et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob Agents Chemother. (2016) 60:6–13. doi: 10.1128/AAC.01802-15
47. Stalin Raj V, Okba NM, Gutierrez-Alvarez J, Drabek D, van Dieren B, Widagdo W, et al. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Sci Adv. (2018) 4:eaas9667. doi: 10.1126/sciadv.aas9667
48. Zhao G, He L, Sun S, Qiu H, Tai W, Chen J, et al. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J Virol. (2018) 92:e00837–18. doi: 10.1128/JVI.00837-18
49. Rockefeller University. Research Program on COVID-19/SARS-CoV-2 (2020). Available online at: https://www.rockefeller.edu/coronavirus/research-program-covid-19-sars-cov-2/ (accessed May 8, 2020).
50. The Rosalind Franklin Institute. Science Minister Hails Nanobody Breakthrough at Franklin. (2020). Available online at: https://www.rfi.ac.uk/science-minister-hails-nanobody-breakthrough/ (accessed May 8, 2020).
51. Walter JD, Hutter CA, Zimmermann I, Earp J, Egloff P, Sorgenfrei M, et al. Synthetic nanobodies targeting the SARS-CoV-2 receptor-binding domain. BioRxiv [Preprint]. (2020). doi: 10.1101/2020.04.16.045419
52. Egloff P, Zimmermann I, Arnold FM, Hutter CA, Morger D, Opitz L, et al. Engineered peptide barcodes for in-depth analyses of binding protein libraries. Nat Methods. (2019) 16:421–8. doi: 10.1038/s41592-019-0389-8
53. Beroni Group. Beroni Group and Tianjin University Have Made a Significant Discovery in Developing a Medical Solution Using Nanobody Technology for the Novel Coronavirus. (2020). Available online at: https://www.beronigroup.com/2020/05/19/beroni-group-and-tianjin-university-have-made-a-significant-discovery-in-developing-a-medical-solution-using-nanobody-technology-for-the-novel-coronavirus/ (accessed June 3, 2020).
Keywords: COVID-19, SARS-CoV-2, neutralizing antibody, nanobodies, spike protein
Citation: Konwarh R (2020) Nanobodies: Prospects of Expanding the Gamut of Neutralizing Antibodies Against the Novel Coronavirus, SARS-CoV-2. Front. Immunol. 11:1531. doi: 10.3389/fimmu.2020.01531
Received: 12 May 2020; Accepted: 10 June 2020;
Published: 23 June 2020.
Edited by:
Abdul Qader Abbady, Atomic Energy Commission of Syria, SyriaReviewed by:
Serge Muyldermans, Vrije University Brussel, BelgiumCopyright © 2020 Konwarh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rocktotpal Konwarh, cm9ja3RvdHBhbC5rb253YXJoQGFhc3R1LmVkdS5ldA==; cm9jazEzMTFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.