- 1Center for Infectious Medicine, Department of Medicine Huddinge, Karolinska Institutet, Stockholm, Sweden
- 2Department of Cancer Immunology, Institute for Cancer Research, Oslo University Hospital, Oslo, Norway
- 3The KG Jebsen Center for Cancer Immunotherapy, Institute of Clinical Medicine, University of Oslo, Oslo, Norway
Natural killer (NK) cells have a central role within the innate immune system, eliminating virally infected, foreign and transformed cells through their natural cytotoxic capacity. Release of their cytotoxic granules is tightly controlled through the balance of a large repertoire of inhibitory and activating receptors, and it is the unique combination of these receptors expressed by individual cells that confers immense diversity both in phenotype and functionality. The diverse, yet unique, NK cell repertoire within an individual is surprisingly stable over time considering the constant renewal of these cells at steady state. Here we give an overview of NK cell differentiation and discuss metabolic requirements, intra-lineage plasticity and transcriptional reprogramming during IL-15-driven homeostatic proliferation. New insights into the regulation of NK cell differentiation and homeostasis could pave the way for the successful implementation of NK cell-based immunotherapy against cancer.
NK Cell Development
Natural killer (NK) cells are granular lymphocytes able to unleash stored cytotoxic potential to kill foreign, transformed or infected cells. Compared to other cytotoxic cells, NK cells are not restricted by the need for prior sensitization and can further orchestrate the early phase of the adaptive immune response. NK cells are found in significant numbers in blood, bone marrow, liver, lymphoid organs, lung, and uterus (1) and develop from common lymphoid progenitors in the bone marrow (2). Identification of NK cell precursors outside the bone marrow, namely fetal thymocytes (CD34+CD3−CD4−CD8−) and fetal liver cells (CD34+CD38+) suggests that NK development is not restricted to the bone marrow (3–5). Commitment to the NK cell lineage requires the transcription factors ID2 and E4BP4 along with IL-15 signaling (6–11). The search for an NK-restricted precursor identified CD34+CD38+CD45RA+CD7+CD10+CD123−CD127− cells which can give rise to T-bet+ and Eomes+ NK cells, two transcription factors central for NK cell maturation in mice (12, 13). Expression of T-bet and Eomes induces CD122 (encoded by IL2RB) expression on NK cells, a component of both the IL-2 and IL-15 receptor allowing for survival and effector function signaling to occur (12, 14). Although NK cells belong to the innate immune system, many aspects of T cell biology share a striking similarity with NK cells (15).
NK Cell Differentiation and Functional Specialization
In humans, NK cells are characterized as CD56+CD3− cells. They can be broadly divided into CD56bright and CD56dim subpopulations based on clear functional and phenotypic differences (16–18). CD56bright NK cells are highly responsive to cytokine priming and fulfill an immunomodulatory role. Expression of CCR7, CD62L, CXCR3, CCR5, CCR2, and CXCR4 allows CD56bright cells to home to secondary lymphoid tissues, the liver, skin and bone marrow, where they represent the dominant NK cell subset (1, 19–22). Conversely, cytotoxic CD56dim NK cells, which prioritize activating and inhibitory receptor input, mainly express CX3CR1 and CXCR1 (22), and account for the majority of peripheral blood NK cells (23).
CD56bright NK cells have been suggested as precursors of CD56dim NK cells based on combinatorial approaches including transcriptional studies (24–27). CD56bright NK cells can acquire CD16 expression, effectively transitioning into CD56dim NK cells (18) and CD16+CD56bright NK cells exist as functional intermediates (28). Furthermore, CD56bright NK cells are the first lymphocyte population to reconstitute after stem cell transplantation, with CD16 acquisition, decreased surface expression of CD56 and cytotoxic effector functions following at a later time point (29–31). Conversely, in response to cytokine stimulation CD56dim NK cells can adopt a “bright-like” phenotype via upregulation of CD56 (32). CD56bright NK cells also have longer telomers compared to CD56dim NK cells, evidence for having undergone fewer cell divisions (18), and have an increased proliferative capacity compared to CD56dim NK cells (33).
Within the CD56dim NK cell population, further distinctions of individual subsets based on phenotypic and functional characteristics can be made (Figure 1) (34). Cells expressing the inhibitory receptor NKG2A are found on the immature end of the spectrum, whereas acquisition of killer cell immunoglobulin like receptors (KIR) gives rise to more differentiated educated and uneducated NK cells with varying functional potential (35). The inhibitory signal strength between self-MHC and NKG2A and KIR fine-tunes the functional potential in a process termed education (35, 36). Expression of CD57, a carbohydrate epitope of unknown binding, is associated with terminal maturation, reduced proliferative capacity, and increased functional potential (37). At the mature end of the spectrum is a unique group of NK cells termed adaptive or memory-like NK cells (38, 39) that can be found in approximately 40% of cytomegalovirus (CMV) seropositive individuals. Adaptive NK cells are characterized by single self-KIR expression, epigenetic downregulation of intracellular signaling molecules and expression of the activating receptor NKG2C and CD57 (40–45). Functionally, adaptive NK cells exhibit increased ADCC activity compared to their non-adaptive counterpart. Although the combination of NKG2A, KIR and CD57 expression is commonly used to define NK cell subsets in humans, this is a simplified model considering that up to 100,000 unique subsets exist within healthy individuals (46).
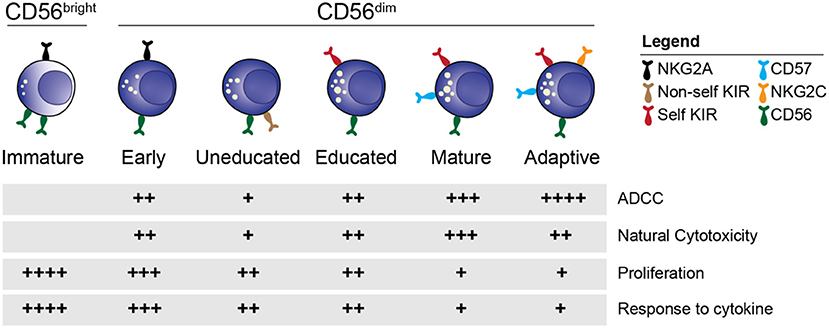
Figure 1. NK cell subsets. Overview of the distinct stages of NK cell differentiation based on phenotypic and functional properties.
Transcriptional Regulation of Human NK Cell Differentiation
Recently, several studies have shed light on the transcriptional regulation of NK cell differentiation. Mouse studies identified the importance of T-bet and Eomes in the differentiation step from immature CD27+CD11b− to mature CD27−CD11b+ NK cells22, as well as the role of ZBTB32, IRF2, and IKZF3 in NK cell differentiation (47–49). Bulk sequencing, combined with ChIP sequencing, of human CD56bright and CD56dim NK cells identified the TCF1-LEF-MYC axis within the CD56bright population and the PRDM1-MAF-ZEB2 axis within CD56dim NK cells (50). These transcription factor controlled regulatory schemes within effector cells (CD56dim NK cells) and proliferative precursor cells (CD56bright NK cells) dictated their functional programs as well as localization and trafficking. Expression of BACH2 in CD56bright NK cells repressed BLIMP1 expression while ZEB2 expression in CD56dim NK cells repressed TCF1 expression. The first single-cell RNA sequencing (scRNA seq) study in human NK cells was focused on characterizing the heterogeneity within peripheral blood and organs in both mice and humans (51), without detailing the gene regulatory circuit involved in NK cell differentiation. A recent study from our group (52) set out to delineate the temporal transcriptional regulation of human NK cell differentiation with the aid of scRNA seq. Confirming previous phenotypic and functional studies, we identified two main transcriptional islands, which corresponded to the CD56bright and CD56dim NK cell populations. Intriguingly, they were connected by a narrow bridge which, based on RNA velocity analysis (53), identified a transition from the CD56bright to CD56dim island. This gradual transition between the two main subsets was further corroborated by mapping a confined set of gene trends along pseudotime using Palantir (54).
Formation of the Functional Template for Education
NK cell education is the process whereby NK cells are functionally tuned via inhibitory interactions mediated between self-MHC and KIR or NKG2A. This is further fine-tuned by the signal strength determined by the number of inhibitory interactions (35, 36). As NK cells do not undergo positive or negative selection, it was initially assumed that they would express a minimum of one inhibitory receptor in order to maintain tolerance to self (55). However, the presence of NKG2A−KIR− cells and evidence of completely stochastic KIR repertoires in the developing immune system (56–59) suggested that alternative mechanisms are in play to ensure tolerance to self. Indeed, NK cells that lack self-specific inhibitory receptors circulate in a hypo-responsive state (56, 60, 61). Furthermore, NK cells have the ability to undergo re-education after transfer from one MHC class I environment to another, further validating the need for sustained inhibitory interactions in order to retain functionality (62, 63).
Despite education being a dynamic process that forms an important cornerstone in NK cell functionality, the intracellular mechanism underlying education remained elusive until recently. Multiple models have been proposed, including the arming, the disarming and the rheostat model without a general consensus being reached (35, 64, 65). Discriminating between educated and uneducated NK cells required a functional readout or sequencing of the HLA genes, as no phenotypic readout existed. Recent work from our lab identified granzyme B retention as a sensitive and specific phenotypic readout for education in resting NK cells, putting the core cytolytic machinery itself in the spotlight in the search for an underlying mechanism behind NK cell education (66). Transcriptionally, educated NK cells were identical to uneducated NK cells, but accumulated granzyme B in dense-core secretory lysosomes located close to the centrosome. After target cell interaction, these large granules containing granzyme B were released, in line with increased cytotoxicity compared to uneducated cells lacking these particular granules. Pharmacological inhibition of the protein kinase PIKfyve and genetic silencing of its downstream target, the lysosome-specific calcium channel TRPML1, suggested a model where unopposed activating receptor input leads to remodeling of the lysosomal compartment and loss of dense-core secretory lysosomes in cells that lack self-specific receptors. Downstream of such morphological changes, signaling from acidic calcium stores may fine-tune the cell's functional potential through inter-organelle communication with the endoplasmic reticulum.
Our recent scRNA-seq study (52) identified a gradual increase in expression of effector molecules and genes involved in lysosomal function within the CD56dim population. Furthermore, genes important for vesicle formation and trafficking, such as RAB27A, were higher expressed within the CD56dim NK cell subset. Mutations in RAB27A cause Griscelli syndrome type 2, resulting in a degranulation defect (67), as Rab27a is recruited to the lytic granules by LFA-1 stimulation, aiding the granule in docking to the plasma membrane (68, 69). Hence, CD56dim NK cells are poised for modulation of the lysosomal compartment mediated via inhibitory and activating receptor input received at the cell surface, resulting in fine tuning of their functionality.
NK Cell Homeostasis
IL-15 is the main cytokine required for NK cell development, but also for survival, proliferation, metabolism and functionality (70). The importance of IL-15 signaling in NK cell development is best observed through mutations in the receptor components and downstream signaling molecules which, together, present as immunodeficiencies characterized by a lack of NK cells (71–74). Immune cells, including DCs, monocytes and other non-hematopoietic cells trans-present IL-15 on the IL-15Rα chain, which binds to the heterodimer consisting of IL-2Rβ (CD122) and the common γ-chain (CD132) found on the NK cell surface. Downstream signaling is mediated via JAK1/3, allowing for recruitment and activation of the transcription factor STAT5, a survival signal for NK cells (73). A downstream target of STAT5 is the cytokine induced SH2-containing protein (CIS, encoded by CISH), which functions as a negative feedback loop by inhibiting the upstream JAK1 (75). Cish−/− knockout mice presented increased anti-tumor activity and proliferative capacity as a result of being hyper-responsive to IL-15 signaling (75). Mathematical modeling has been implemented in an attempt to better understand the impact of IL-15 receptor signaling on proliferation. The model predicted that increasing IL-15Rα expression on the cell surface will accelerate the formation of IL-15/IL-15R complexes, particularly at low IL-15 concentrations, until a saturation level is reached and no further proliferative response can be achieved (76). These results highlight the broad and central role for IL-15 in NK cell development, differentiation, homeostasis and priming of effector function.
Quorum sensing, which is a form of chemical communication in bacteria whereby sensing of an autoinducer is used to synchronize group behavior, has recently been proposed to also control immune cell homeostasis (77). For example, colony stimulating factor 1 (CSF1) produced by the surrounding stromal cells is proposed to function as the autoinducer in macrophages, whereby uptake of CSF1 controls the rate of proliferation and survival to maintain a steady population density at homeostasis (78). In T cells, IL-2 replaces CSF1 as the autoinducer, which together with IL-6 has been suggested to also modulate the differentiation from an effector T cell to a central memory T cell (79, 80). The logical autoinducer counterpart in NK cells is IL-15. The threshold for IL-15 induced proliferation is subset-dependent, as observed by the onset of proliferation across the maturation spectrum. This is in line with the concept of quorum sensing, whereby the level of IL-15 in the microenvironment dictates the degree of proliferation and overall size of the population.
The IL-15 mTOR Axis
The unique role of IL-15 in NK cell biology is largely attributed to the IL-15 mammalian target of rapamycin (mTOR) signaling axis and the metabolic regulation of NK cells. Mouse studies identified a dose-dependent downstream signaling pathway, where high dose IL-15 activated the mammalian target of rapamycin (mTOR) as well as STAT5. mTOR, a serine/threonine kinase consisting of the two complexes mTORC1 and mTORC2, is a master regulator in cells. mTORC1 senses for nutrients in the microenvironment to control metabolism while mTORC2 is involved in controlling the cytoskeletal organization of the cell (81–83). Metabolic reprogramming due to environmental cues has been identified as a key regulatory mechanism behind immune cell differentiation and function in NK cells and other immune cells (81–85). In mice, increased cytokine priming led to metabolic reprogramming, with increased metabolic activity, and a switch in energy source from oxidative phosphorylation (OXPHOS) to glycolysis. An increase in metabolism allowed for IFNγ and granzyme B production, conferring increased functionality which could be reversed through the use of rapamycin, an mTOR inhibitor (81). Viral infection can also activate mTOR leading to metabolic reprogramming, as observed in murine CMV infection122. It is possible that in a tumor setting, a lack of available glucose due to high glycolytic activity by the tumor cells could lead to functional inhibition due to lack of mTOR activation (81, 86).
In addition to mediating NK cell functionality via modulation of the cellular metabolism, mTOR may serve as a functional rheostat during NK cell education (82, 87). Educated NK cells exhibited higher basal mTOR activity, which was further increased upon activating receptor ligation and also correlated with the number of inhibitory receptors expressed (87). Expression of SHP-1, a phosphatase required to convert inhibitory receptor input into functional responsiveness, was required for increased mTOR activity in educated cells (88). Conversely, continuous activating receptor input in the absence of inhibitor input dampened mTOR activity. Although education is not transcriptionally regulated in human NK cells, mTOR activity is dependent on its localization to the lysosomal compartment which in turn can be negatively regulated by TRPML1 (89, 90). The connection between lysosomal remodeling during education and metabolic regulation through mTOR is an unexplored area in NK cell biology that warrants further investigation.
NK Cell Repertoire Dynamics and Intra-Lineage Plasticity
At the donor level, the NK cell repertoire is vastly diverse and unique (46). However, once the NK cell repertoire has been fully formed and in some cases further shaped by pathogens such as CMV, it is well-maintained over time considering the rather rapid turnover of the cells (44, 91). Proliferation therefore plays an important role in replenishing the NK cell pool at steady-state and in maintaining a stable repertoire. NK cell proliferation has mainly been examined in viral or disease settings, where it is associated with rapid cell turnover resulting in subset skewing toward immature NK cells with higher proliferative potential (92–94). In a recent study we asked the question of whether or not stable NK cell repertoires are maintained under homeostatic proliferation (95). We hypothesized that the observed stability was either the result of self-renewal from an immature pool of progenitor cells followed by differentiation, or the result of intra-lineage plasticity (BOX 1). This process has been observed in other immune cells (96, 97) but NK cell plasticity has largely remained unexplored (98).
Box 1. Cellular plasticity
Plasticity refers to phenotypic and functional changes occurring within populations of cells. Intra-lineage plasticity, also known as functional plasticity, refers to cells of a given lineage adapting to their surroundings in response to cytokine or receptor input which is translated into transcriptional changes resulting in an altered phenotype and modified functionality. An example of this is macrophages transition between an M1 and M2 phenotype, T cells transitioning from Th to Treg phenotype or ILC subsets transitioning between ILC1- 3 phenotypes.
We developed a simplified model that induced a linear onset of IL-15 induced proliferation with maximal retention of NK cell subsets (based on NKG2A, KIR and CD57) to mimic homeostatic conditions. We observed subset-specific proliferation kinetics, which correlated with mTOR activation. IL-15-induced mTORC1 upregulation prior to proliferation onset could predict downstream proliferation 3 days later at both the donor and subset level. Repeated sampling of the same blood donors over time confirmed stable NK cell repertoires, but also an intrinsic metabolic set point determining the level of mTOR activation in response to IL-15 stimulation.
Despite subset-specific proliferation kinetics, the actual subset frequencies at the population level remained largely stable, suggesting that the repertoires were maintained through intra-lineage plasticity. Indeed, sorting individual NK cell subsets prior to proliferation revealed a surprising degree of cellular plasticity in both immature and mature subsets. Acquisition of NKG2A in sorted KIR+ NK cells was associated with increased proliferative potential and decreased functionality, while the reverse was true for CD57 acquisition by the same subset. Surprisingly, a fraction of CD57+ NK cells lost CD57 expression, acquired NKG2A and started to proliferate, suggesting they may not be terminally differentiated. Rapidly cycling educated NK cells underwent transcriptional reprogramming, resulting in a more immature signature, while slowly cycling educated NK cells acquired a more mature signature when compared with baseline subsets.
Our simplified in vitro homeostatic NK cell proliferation model allowed us to examine the central role IL-15 plays in maintaining NK cell homeostasis (Figure 2). CD57 expression was associated with a negative influence on mTOR activation and proliferation but enhanced functional potential. Although it is used as a main marker for subset discrimination in NK cells, the function of CD57 remains unknown (37, 99). In neural cells, CD57 has mainly been associated with adhesion proteins, while binding to the IL-6 receptor has also been proposed (100). It would be interesting to further delineate whether CD57 plays a functional role, or if it is simply a surrogate marker for other ongoing cellular modifications.
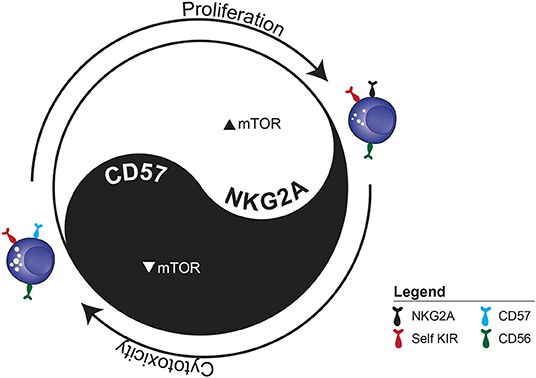
Figure 2. The functional dichotomy between proliferation and cytotoxicity observed during IL-15-induced homeostatic proliferation. The example illustrates the two distinct fates of sorted CD56dim KIR+ NKG2A−CD57− NK cell depending on whether they acquire NKG2A or CD57.
Due to their differential mTOR activation profile, it is tempting to speculate that NKG2A+ and CD57+ cells display distinct metabolic profiles. Metabolic reprogramming is responsible for the differentiation of naïve T cells into active effector and later into memory T cells (101–106). The transition of naïve into effector T cells depends on the upregulation of glycolysis and the TCA cycle to provide material for de-novo synthesis of proteins, nucleic acids and lipids, whereas formation of memory T cells rely on OXPHOS and fatty acid oxidation (FAO) (107). Such differential use of metabolic programs has also been observed in Th cell subsets (108). In addition, T cell memory formation is influenced through the reorganization of mitochondrial content (109). Interestingly, survival of memory-like NK cells in mice upon CMV infection is dependent on sufficient degradation of dysfunctional mitochondria via mitophagy upon virus clearance (110).
Differences in terms of proliferation speed, phenotype, and functionality between homeostatic and spontaneous proliferation have been investigated in murine T cells (111–113). Spontaneous proliferation, occurring in severely lymphopenic mice, was characterized by a rapid onset of cell division that was cytokine-independent. Homeostatic proliferation, on the other hand, occurred in mildly lymphopenic mice at a slower division rate and required both cytokine and T cell receptor (TCR) stimulation. The proliferation-induced phenotype was reverted after removal of the proliferation cues and cytotoxic capacity of CD8+ T cells was lost during the initial phase of intense proliferation (111–113).
Considering the asymmetric PI3K and mTOR activity post-cell division observed in T cells and its role in controlling differentiation fate and the functional dichotomy in proliferating vs. arrested NK cells (101–106), it would be of interest to do microscopy studies of cellular division or functional interactions with target cells. Based on the induced transcriptional signature in rapidly cycling NK cells, which included both RNA-modifying metabolic genes and actin filament organization genes (95), the loss of functionality in rapidly cycling cells may be due to underlying deficits at the immune synapse. Conjugate formation experiments combined with F-actin staining at the site of the immune synapse would further shed light on the loss of functionality observed during intense homeostatic proliferation.
NK Cell Homeostasis in vivo
Given the essential role of IL-15 on NK cells, stimulation of IL-15 signaling pathways has been explored in clinical settings (114–120). In this regard, three main strategies have been pursued; using recombinant-human IL-15 (116) generated by E. coli, an IL-15 superagonist, ALT-803 (114, 115) and transfection of an IL-15 containing CAR construct (121, 122). These have been thus far tested in Phase I and II clinical trials, with recombinant-human IL-15 and ALT-803 both showing moderate success in inducing NK cell proliferation and activation in vivo and in particular cases inducing disease remission. A limitation of this approach has been the induction of some minor side effects relating to an increased inflammatory environment. Subcutaneous delivery of the compounds has resulted in a partial reduction of these side effects (115). Recent pre-clinical studies have highlighted the potential of combination therapy using this IL-15 signaling stimulation and other immunotherapeutic agents such as monoclonal antibodies or check-point blockade (118, 119). Reflecting this, there are currently more than 100 registered clinical trials exploring IL-15 stimulation via either of these two methods in a series of different cancer settings (www.clinicaltrials.gov). In vitro, transfection with an IL-15 containing CAR construct sustained autonomous NK cell growth over 42 days and increased systemic IL-15 serum levels were observed in mouse studies (121). However, in 11 patients treated in a Phase I/II trial, the detection of infused CAR+ NK cells by flow cytometry was limited to the first 2 weeks post infusion (122).
In the setting of stem cell transplantation, NK cells are the first lymphocyte population that can be detected following engraftment (123). Their ability to mediate graft-vs.-leukemia (GVL) effects is vital for elimination of residual disease, as increased number of NK cells after transplantation result in better treatment outcome (124, 125). Insights into the specificity of NK cell alloreactivity, determined by specific combinations of KIR and HLA, paved the way for the ground-breaking discovery of a potential role of NK cells in mediating GVL in haploidentical HSCT against AML (126, 127). Studies aiming at harnessing NK cell alloreactivity in the context of HSCT have recently been reviewed (128, 129). The indication that NK cells may deliver a potent GVL effect in the setting of HSCT inspired the whole NK cell community to develop adoptive NK cell therapy based on transfer of “KIR ligand mismatched” NK cells across HLA barriers to promote missing self-recognition. Whereas many studies did not find a beneficial effect of genetic KIR ligand mismatch, calculation of the functional dose of KIR ligand mismatched NK cells was associated with less relapse after NK cell therapy against AML (130–132).
A recent series of Phase III clinical trials have brought mTOR inhibition to the forefront of transplantation (117, 120). In both of these studies a series of patients received Sirolimus, an alternate name for rapamycin, as a prophylactic against graft vs. host disease (GVHD). Sandmaier et al. reported that inclusion of Sirolimus to the standard calcineurin inhibitor treatment showed decreased incidence of grade 2–4 acute GVHD. Similarly, increased overall survival, and progression-free survival, as well as decreased non-relapse related mortality was observed in the sirolimus treated group. Due to the clear improved benefit of Sirolimus treatment, the trial carried out by Sandmaier et al. was terminated prior to complete patient recruitment.
On the other hand, a parallel study by Gooptu et al. did not report significant differences compared to standard treatment regarding GVHD incidence, progression free survival nor overall survival (117). This discrepancy may be due to the differences in standard treatment and dosage of Sirolimus between the two studies. In the latter study, the immune reconstitution was evaluated at a series of timepoints up to 24 months. Sirolimus treatment led to a decreased lymphocyte cell count in the first 3 months of treatment, and an increased ratio of regulatory T cells to CD8+ T cells throughout the first 6 months of treatment. Lower NK cell counts were observed in the first month following Sirolimus treatment, although this recovered to similar levels compared to standard treatment by the 3rd month. Given the phenotypic and functional heterogeneity of NK cell subsets and the critical role of mTOR and IL-15 signaling in driving NK cell plasticity, it would be of great interest to further evaluate the precise composition of the NK cell compartment during such therapies.
Cytokine-Based Expansion Protocols for NK Cell Therapy
There are several up to date and comprehensive reviews describing the prospects of using various preparations of NK cells in cell-based immunotherapy (133, 134). These include strategies based on autologous and allogeneic NK cells that have been stimulated by various cytokines alone or in combination with irradiated feeder cells expressing membrane bound cytokines such as IL-21 or IL-15 (121, 135–138). Therefore, we will focus on a general discussion on how these protocols may drive dramatic phenotypic and functional changes to the NK cell repertoire (34, 95). To expand large numbers of cells for multi-dosing schemes, many strategies are based on supra-physiological levels of cytokines, including any combination of IL-2, IL-15, IL-12, and IL-18 (139, 140). However, this can result in severe and acute cytokine deprivation post-infusion as the cells become “addicted” to cytokines (BOX 2). Severe side-effects (141, 142) prevent patients from being treated with the same cytokines and persistence is further limited through clearance of infused NK cells by host immunity.
Box 2. Cellular addiction
Cytokine priming results in intracellular signaling changes occurring within cells. Continuous stimulation with non-physiological cytokine levels can result in an altered cellular state, which requires further cytokine stimulation to support survival. This can be referred to as cytokine-dependence or addiction, whereby cytokine withdrawal can lead to detrimental consequences to the cell.
The Balance Between Pro- and Anti-apoptotic Molecules During IL-15 Driven Proliferation
We recently set out to characterize the mechanism behind IL-15 addiction and withdrawal in expanded NK cells. NK cells exhibited a dose-dependent IL-15 addiction, where high-doses induced rapid proliferation, skewing toward a naïve phenotype, and subsequent crash upon cytokine withdrawal (143). Timing of IL-15 dosing is crucial for NK cell survival and effector function as chronic high-dose IL-15 stimulation leads to decreased viability of NK cells with reduced respiratory spare capacity and functional activity (144).
Numerous pro- and anti-apoptotic genes make up the apoptosis network balancing the outcome of the cell during various types of stimulations (70, 145–147). Within resting NK cells, BCL-2 has been identified as an important anti-apoptotic protein which can be further upregulated through IL-15 stimulation, leading to downstream STAT5, but not mTOR activation (82, 148). In actively proliferating NK cells, MCL-1 expression is vital to maintain viability (149). BIM is a pro-apoptotic molecule and its downstream target BAX is directly inhibited by BCL-2 (147). In murine effector CD8+ T cells, increased BIM levels are balanced by increased BCL-2 levels, expression of which dictates the amount of BIM that can be tolerated (150). Similarly, in murine NK cells, the BCL-2/BIM ratio was influenced by IL-15 stimulation and withdrawal, whereby changes in the ratio could render the cells sensitive to cell death (70, 150, 151). In line with these studies, we observed an IL-15 dose-dependent increase in BCL-2, MCL-1, and BIM expression. BCL-2 and MCL-1 were both crucial for survival in NK cells stimulated with high-dose IL-15 as shown through blocking experiments. Interestingly, rapidly cycling NK cells exhibited reduced BCL-2 levels compared to slowly or non-cycling NK cells during their expansion phase, in line with T cell proliferation studies (145).
After cytokine withdrawal, anti-apoptotic proteins decreased, and a potent apoptosis-inducing splice variant, BIM S, (152, 153) was preferentially upregulated in proliferating cells and remained highly expressed until 24 h after cytokine withdrawal. This severely altered the pro/anti-apoptotic ratio, exposing rapidly cycling cells to high levels of toxic BIM S within 24 h after cytokine withdrawal (Figure 3). The importance of these apoptotic proteins in IL-15 mediated survival and function has also been observed in murine studies (70). How homeostatic and induced proliferation affects NK cell cytotoxicity, and how apoptosis is induced in cycling cells upon cytokine withdrawal, has potentially important implications for current cell therapy expansion protocols.
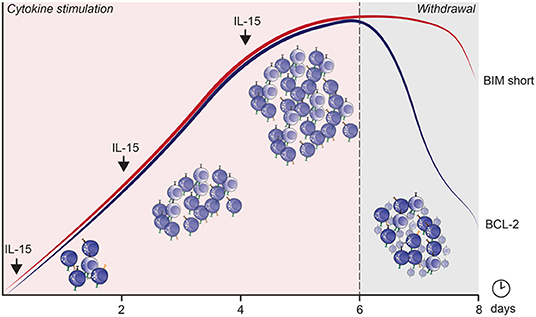
Figure 3. The mechanism by which apoptosis is induced in cycling NK cells after IL-15 induced cytokine dependence and subsequent withdrawal. The curves represent expression of BIM short (red) and BCL-2 (blue) over culture time.
Concluding Remarks
NK cells circulate in a pre-primed state full of effector molecules, such as granzyme B and perforin, and have a natural ability to kill cancer cells. Based on their cytotoxic capacity they hold great potential in the clinic as a cancer treatment, made evident by the number of ongoing clinical trials. However, to date most completed and ongoing clinical trials are based on the transfer of cytokine-activated polyclonal NK cell populations from donors with very variable NK cell repertoires. To fully harness the clinical potential of NK cells, future trials need to be founded on recent breakthroughs in our understanding of the vast repertoire diversity and the fundamental mechanisms that govern the intrinsic functional potential of distinct NK cell subsets at steady state and following cytokine stimulation.
Understanding how NK cells repertoires are formed and maintained over time, and what functional roles individual cell subsets perform in a homeostatic setting, are important to improve current therapies and develop future treatment strategies. Generating an “ideal” NK cell product for treatment could involve modifying existing cells to improve functionality, expanding highly cytotoxic subsets while ensuring retention of functionality or designing a “synthetic” genetically engineered killer cell from induced pluripotent stem cells.
Furthermore, we need to understand how NK cells are functionally shaped by their surroundings. The soluble factors, metabolic cues, fluctuations in oxygen levels, and pH encountered by an NK cell in the tumor microenvironment are very different from steady state and their impact on NK cell function and persistence cannot be underestimated. This is particularly difficult to study in the human setting, with mouse models only providing an approximation.
By understanding the basic biology, from development to differentiation to receptor and cytokine input, we will build up our tool kit that can then be applied to design and develop effective treatment strategies. After all, the “natural” killing capacity is there, we just need to understand how to harness it.
Author Contributions
AP made the figures. AP and K-JM wrote the paper. LK, AH-I, BJ and ES contributed to the writing of the paper.
Conflict of Interest
K-JM is a consultant at Fate Therapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This review including figures is partly based on the introduction of the PhD thesis by Dr Aline Pfefferle Dynamics of natural killer cell homeostasis: implications for cell-based cancer immunotherapy defended at the Karolinska Institute June 5, 2019.
References
1. Björkström NK, Ljunggren HG, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol. (2016) 16:310–20. doi: 10.1038/nri.2016.34
2. Colucci F, Caligiuri MA, Di Santo JP. What does it take to make a natural killer? Nat Rev Immunol. (2003) 3:413–25. doi: 10.1038/nri1088
3. José Sanchez M, Muench MO, Grazia Roncarolo M, Lanier LL, Phillips JH. Identification of a Common T/Natural Killer Cell Progenitor in Human Fetal Thymus. J Exp Med. (1994) 180:569–76. doi: 10.1084/jem.180.2.569
4. Jaleco AC, Blom B, Res P, Weijer K, Lanier LL, Phillips JH, et al. Fetal liver contains committed NK progenitors, but is not a site for development of CD34+ cells into T cells. J Immunol. (1997) 159:694–702.
5. Renoux VM, Zriwil A, Peitzsch C, Michaëlsson J, Friberg D, Soneji S, et al. Identification of a human natural killer cell lineage-restricted progenitor in fetal and adult tissues. Immunity. (2015) 43:394–407. doi: 10.1016/j.immuni.2015.07.011
6. Mrozek E, Anderson P, Caligiuri MA. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. (1996) 87:2632–40. doi: 10.1182/blood.V87.7.2632.bloodjournal8772632
7. Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S-I, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix–loop–helix inhibitor Id2. Nature. (1999) 397:702–6. doi: 10.1038/17812
8. Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. (2007) 204:1119–30. doi: 10.1084/jem.20061959
9. Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. (2009) 10:1118–24. doi: 10.1038/ni.1787
10. Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. (2009) 206:2977–86. doi: 10.1084/jem.20092176
11. Male V, Nisoli I, Kostrzewski T, Allan DSJ, Carlyle JR, Lord GM, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. (2014) 211:635–42. doi: 10.1084/jem.20132398
12. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors t-bet and eomes control key checkpoints of natural killer cell maturation. Immunity. (2012) 36:55–67. doi: 10.1016/j.immuni.2011.11.016
13. Simonetta F, Pradier A, Roosnek E. T-bet and eomesodermin in NK cell development, maturation, and function. Front Immunol. (2016) 7:241. doi: 10.3389/fimmu.2016.00241
14. Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. (2005) 6:1236–44. doi: 10.1038/ni1268
15. Narni-Mancinelli E, Vivier E, Kerdiles YM. The “T-cell-ness” of NK cells: unexpected similarities between NK cells and T cells. Int Immunol. (2011) 23:427–31. doi: 10.1093/intimm/dxr035
16. Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. (2001) 22:633–40. doi: 10.1016/S1471-4906(01)02060-9
17. Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. (2006) 214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x
18. Romagnani C, Juelke K, Falco M, Morandi B, D'Agostino A, Costa R, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. (2007) 178:4947–55. doi: 10.4049/jimmunol.178.8.4947
19. Martín-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-γ for TH1 priming. Nat Immunol. (2004) 5:1260–5. doi: 10.1038/ni1138
20. Shiow LR, Rosen DB, Brdičková N, Xu Y, An J, Lanier LL, et al. CD69 acts downstream of interferon-α/β to inhibit S1P 1 and lymphocyte egress from lymphoid organs. Nature. (2006) 440:540–4. doi: 10.1038/nature04606
21. Shi FD, Ljunggren HG, La Cava A, Van Kaer L. Organ-specific features of natural killer cells. Nat Rev Immunol. (2011) 11:658–71. doi: 10.1038/nri3065
22. Bernardini G, Antonangeli F, Bonanni V, Santoni A. Dysregulation of chemokine/chemokine receptor axes and NK cell tissue localization during diseases. Front Immunol. (2016) 7:402. doi: 10.3389/fimmu.2016.00402
23. Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells : a unique innate immunoregulatory role for the CD56 bright subset. Blood. (2001) 97:3146–51. doi: 10.1182/blood.V97.10.3146
24. Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. (2006) 203:1033–43. doi: 10.1084/jem.20052507
25. Gineau L, Cognet C, Kara N, Lach F, Dunne J, Veturi U, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. (2012) 122:821–32. doi: 10.1172/JCI61014
26. Mace EM, Hsu AP, Monaco-Shawver L, Makedonas G, Rosen JB, Dropulic L, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56 bright subset. Blood. (2013) 121:2669–78. doi: 10.1182/blood-2012-09-453969
27. Maciejewski-Duval A, Meuris F, Bignon A, Aknin M-L, Balabanian K, Faivre L, et al. Altered chemotactic response to CXCL12 in patients carrying GATA2 mutations. J Leukoc Biol. (2016) 99:1065–76. doi: 10.1189/jlb.5MA0815-388R
28. Béziat V, Duffy D, Nguyen Quoc S, Le Garff-Tavernier M, Decocq J, Combadiere B, et al. CD56brightCD16+ NK cells: a functional intermediate stage of NK cell differentiation. J Immunol. (2011) 186:6753–61. doi: 10.4049/jimmunol.1100330
29. Jacobs R, Stoll M, Stratmann G, Leo R, Link H, Schmidt RE. CD16- CD56+ natural killer cells after bone marrow transplantation. Blood. (1992) 79:3239–44. doi: 10.1182/blood.V79.12.3239.bloodjournal79123239
30. Dulphy N, Haas P, Busson M, Belhadj SPeffault de Latour R, Robin M, et al. An Unusual CD56brightCD16low NK Cell Subset Dominates the Early Posttransplant Period following HLA-Matched Hematopoietic Stem Cell Transplantation. J Immunol. (2008) 181:2227–37. doi: 10.4049/jimmunol.181.3.2227
31. Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: Correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE. (2010) 5:1–12. doi: 10.1371/journal.pone.0011966
32. Freud AG, Yu J, Caligiuri MA. Human natural killer cell development in secondary lymphoid tissues. Semin Immunol. (2014) 26:132–7. doi: 10.1016/j.smim.2014.02.008
33. Baume DM, Robertson MJ, Levine H, Manley TJ, Schow PW, Ritz J. Differential responses to interleukin 2 define functionally distinct subsets of human natural killer cells. Eur J Immunol. (1992) 22:1–6. doi: 10.1002/eji.1830220102
34. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK cell differentiation uncoupled from NK-cell education. Blood. (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
35. Goodridge JP, Önfelt B, Malmberg K-J. Newtonian cell interactions shape natural killer cell education. Immunol Rev. (2015) 267:197–213. doi: 10.1111/imr.12325
36. Brodin P, Lakshmikanth T, Johansson S, Kärre K, Höglund P. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood. (2009) 113:2434–41. doi: 10.1182/blood-2008-05-156836
37. Lopez-Vergès S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, et al. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. (2010) 116:3865–74. doi: 10.1182/blood-2010-04-282301
38. O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. (2006) 7:507–16. doi: 10.1038/ni1332
39. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. (2009) 457:557–61. doi: 10.1038/nature07665
40. Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. (2004) 104:3664–71. doi: 10.1182/blood-2004-05-2058
41. Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. (2011) 108:14725–32. doi: 10.1073/pnas.1110900108
42. Della Chiesa M, Falco M, Podestà M, Locatelli F, Moretta L, Frassoni F, et al. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation : a role for human cytomegalovirus? Blood. (2012) 119:399–410. doi: 10.1182/blood-2011-08-372003
43. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C + natural killer cells with potent function. Blood. (2012) 119:2665–74. doi: 10.1182/blood-2011-10-386995
44. Beziat V, Liu LL, Malmberg J-A, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. (2013) 121:2678–88. doi: 10.1182/blood-2012-10-459545
45. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. (2015) 42:443–56. doi: 10.1016/j.immuni.2015.02.008
46. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. (2013) 5:1–12. doi: 10.1126/scitranslmed.3006702
47. Lohoff M, Duncan GS, Ferrick D, Mittrücker H-W, Bischof S, Prechtl S, et al. Deficiency in the transcription factor interferon regulatory factor (Irf)-2 leads to severely compromised development of natural killer and T helper type 1 Cells. J Exp Med. (2000) 192:325–36. doi: 10.1084/jem.192.3.325
48. Beaulieu AM, Zawislak CL, Nakayama T, Sun JC. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat Immunol. (2014) 15:546–53. doi: 10.1038/ni.2876
49. Holmes ML, Huntington ND, Thong RP, Brady J, Hayakawa Y, Andoniou CE, et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. (2014) 33:2721–34. doi: 10.15252/embj.201487900
50. Collins PL, Cella M, Porter SI, Li S, Gurewitz GL, Hong HS, et al. Gene regulatory programs conferring phenotypic identities to human NK cells. Cell. (2019) 176:348–60. doi: 10.1016/j.cell.2018.11.045
51. Crinier A, Milpied P, Escalière B, Piperoglou C, Galluso J, Balsamo A, et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity. (2018) 49:1–16. doi: 10.1016/j.immuni.2018.09.009
52. Pfefferle A, Netskar H, Ask EH, Lorenz S, Goodridge JP, Sohlberg E, et al. A temporal transcriptional map of human natural killer cell differentiation. BioRxiv. (2019). 1–38. doi: 10.1101/630657
53. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. (2018) 560:494–8. doi: 10.1038/s41586-018-0414-6
54. Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe'er D. Palantir characterizes cell fate continuities in human hematopoiesis. BioRxiv. (2018). doi: 10.1101/385328
55. Raulet D, Held W, Correa I, Dorfman J, Wu M, Corral L. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol Rev. (1997) 155:41–52. doi: 10.1111/j.1600-065X.1997.tb00938.x
56. Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. (2005) 105:4416–24. doi: 10.1182/blood-2004-08-3156
57. Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, et al. Identification of bidirectional promoters in the human KIR genes. Genes Immun. (2007) 8:245–53. doi: 10.1038/sj.gene.6364381
58. Andersson S, Fauriat C, Malmberg JA, Ljunggren HG, Malmberg KJ. KIR acquisition probabilities are independent of self-HLA class I ligands and increase with cellular KIR expression. Blood. (2009) 114:95–104. doi: 10.1182/blood-2008-10-184549
59. Schönberg K, Fischer JC, Kögler G, Uhrberg M. Neonatal NK-cell repertoires are functionally, but not structurally, biased toward recognition of self HLA class I. Blood. (2011) 117:5152–6. doi: 10.1182/blood-2011-02-334441
60. Höglund P, Öhlen C, Carbone E, Franksson L, Ljunggren HG, Latour A, et al. Recognition of β2-microglobulin-negative (β2m–) T-cell blasts by natural killer cells from normal but not from β2m– mice: nonresponsiveness controlled by β2m– bone marrow in chimeric mice. Proc Natl Acad Sci USA. (1991) 88:10332–6. doi: 10.1073/pnas.88.22.10332
61. Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. (2006) 25:331–42. doi: 10.1016/j.immuni.2006.06.013
62. Elliott JM, Wahle JA, Yokoyama WM. MHC class I–deficient natural killer cells acquire a licensed phenotype after transfer into an MHC class I–sufficient environment. J Exp Med. (2010) 207:2073–9. doi: 10.1084/jem.20100986
63. Joncker NT, Shifrin N, Delebecque F, Raulet DH. Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J Exp Med. (2010) 207:2065–72. doi: 10.1084/jem.20100570
64. Brodin P, Kärre K, Höglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. (2009) 30:143–9. doi: 10.1016/j.it.2009.01.006
65. Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. (2008) 224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x
66. Goodridge JP, Jacobs B, Saetersmoen ML, Clement D, Hammer Q, Clancy T, et al. Remodeling of secretory lysosomes during education tunes functional potential in NK cells. Nat Commun. (2019) 10:1–15. doi: 10.1038/s41467-019-08384-x
67. Ménasché G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet. (2000) 25:173–6. doi: 10.1038/76024
68. Stinchcombe JC, Bossi G, Booth S, Griffiths GM. The immunological synapse of CTL contains a secretory domain and membrane bridges. Immunity. (2001) 15:751–61. doi: 10.1016/S1074-7613(01)00234-5
69. Wood SM, Meeths M, Chiang SCC, Bechensteen AG, Boelens JJ, Heilmann C, et al. Different NK cell-activating receptors preferentially recruit Rab27a or Munc13-4 to perforin-containing granules for cytotoxicity. Blood. (2009) 114:4117–27. doi: 10.1182/blood-2009-06-225359
70. Huntington ND, Puthalakath H, Gunn P, Naik E, Michalak EM, Smyth MJ, et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat Immunol. (2007) 8:856–63. doi: 10.1038/ni1487
71. Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature. (1995) 377:65–8. doi: 10.1038/377065a0
72. Gilmour KC, Fujii H, Cranston T, Davies EG, Kinnon C, Gaspar HB. Defective expression of the interleukin-2/interleukin-15 receptor ß subunit leads to a natural killer cell-deficient form of severe combined immunodeficiency. Blood. (2001) 98:877–9. doi: 10.1182/blood.V98.3.877
73. Huntington ND, Legrand N, Alves NL, Jaron B, Weijer K, Plet A, et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J Exp Med. (2008) 206:25–34. doi: 10.1084/jem.20082013
74. Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, et al. Interleukin-2 Receptor γ Chain Mutation Results in X-Linked Severe Combined Immunodeficiency in Humans. J Immunol. (2008) 181:5817–27.
75. Delconte RB, Kolesnik TB, Dagley LF, Rautela J, Shi W, Putz EM, et al. CIS is a potent checkpoint in NK cell–mediated tumor immunity. Nat Immunol. (2016) 17:816–24. doi: 10.1038/ni.3470
76. Zhao YM, French AR. Mechanistic Model of Natural Killer Cell Proliferative Response to IL-15 Receptor Stimulation. PLoS Comput Biol. (2013) 9:e1003222. doi: 10.1371/journal.pcbi.1003222
77. Postat J, Bousso P. Quorum sensing by monocyte-derived populations. Front Immunol. (2019) 10:2140. doi: 10.3389/fimmu.2019.02140
78. Jenkins SJ, Hume DA. Homeostasis in the mononuclear phagocyte system. Trends Immunol. (2014) 35:358–67. doi: 10.1016/j.it.2014.06.006
79. Hart Y, Reich-Zeliger S, Antebi YE, Zaretsky I, Mayo AE, Alon U, et al. Paradoxical signaling by a secreted molecule leads to homeostasis of cell levels. Cell. (2014) 158:1022–32. doi: 10.1016/j.cell.2014.07.033
80. Polonsky M, Rimer J, Kern-Perets A, Zaretsky I, Miller S, Bornstein C, et al. Induction of CD4 T cell memory by local cellular collectivity. Science. (2018) 360:eaaj1853. doi: 10.1126/science.aaj1853
81. Donnelly RP, Loftus RM, Keating SE, Liou KT, Biron CA, Gardiner CM, et al. mTORC1-Dependent Metabolic Reprogramming Is a Prerequisite for NK Cell Effector Function. J Immunol. (2014) 193:4477–84. doi: 10.4049/jimmunol.1401558
82. Marçais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. (2014) 15:749–57. doi: 10.1038/ni.2936
83. Keating SE, Zaiatz-Bittencourt V, Loftus RM, Keane C, Brennan K, Finlay DK, et al. Metabolic Reprogramming Supports IFN-γ Production by CD56 bright NK Cells. J Immunol. (2016) 196:2552–60. doi: 10.4049/jimmunol.1501783
84. Nandagopal N, Ali AK, Komal AK, Lee SH. The critical role of IL-15-PI3K-mTOR pathway in natural killer cell effector functions. Front Immunol. (2014) 5:187. doi: 10.3389/fimmu.2014.00187
85. Man K, Kallies A. Synchronizing transcriptional control of T cell metabolism and function. Nat Rev Immunol. (2015) 15:574–84. doi: 10.1038/nri3874
86. Warburg O. The Metabolism of Carcinoma Cells. Cancer Res. (1925) 9:148–63. doi: 10.1158/jcr.1925.148
87. Marçais A, Marotel M, Degouve S, Koenig A, Fauteux-Daniel S, Drouillard A, et al. High mTOR activity is a hallmark of reactive natural killer cells and amplifies early signaling through activating receptors. Elife. (2017) 6:1–21. doi: 10.7554/eLife.26423
88. Viant C, Fenis A, Chicanne G, Payrastre B, Ugolini S, Vivier E. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Commun. (2014) 5:1–11. doi: 10.1038/ncomms6108
89. Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. (2011) 334:678–83. doi: 10.1126/science.1207056
90. Park S, Ahuja M, Kim MS, Brailoiu GC, Jha A, Zeng M, et al. Fusion of lysosomes with secretory organelles leads to uncontrolled exocytosis in the lysosomal storage disease mucolipidosis type IV. EMBO Rep. (2016) 17:266–78. doi: 10.15252/embr.201541542
91. Lutz CT, Karapetyan A, Al-Attar A, Shelton BJ, Holt KJ, Tucker JH, et al. Human NK Cells proliferate and die in vivo more rapidly than T Cells in healthy young and elderly adults. J Immunol. (2011) 186:4590–8. doi: 10.4049/jimmunol.1002732
92. Lünemann A, Vanoaica LD, Azzi T, Nadal D, Münz C. A Distinct Subpopulation of Human NK Cells Restricts B Cell Transformation by EBV. J Immunol. (2013) 191:4989–95. doi: 10.4049/jimmunol.1301046
93. Azzi T, Lünemann A, Murer A, Ueda S, Beziat V, Malmberg K-J, et al. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood. (2014) 124:2533–43. doi: 10.1182/blood-2014-01-553024
94. Liu LL, Pfefferle A, Yi Sheng VO, Björklund AT, Béziat V, Goodridge JP, et al. Harnessing adaptive natural killer cells in cancer immunotherapy. Mol Oncol. (2015) 9:1904–17. doi: 10.1016/j.molonc.2015.10.001
95. Pfefferle A, Jacobs B, Netskar H, Ask EH, Lorenz S, Clancy T, et al. Intra-lineage Plasticity and Functional Reprogramming Maintain Natural Killer cell repertoire Diversity. Cell Rep. (2019) 29:2284–94. doi: 10.1016/j.celrep.2019.10.058
96. Laurent P, Jolivel V, Manicki P, Chiu L, Contin-Bordes C, Truchetet ME, et al. Immune-mediated repair: a matter of plasticity. Front Immunol. (2017) 8:454. doi: 10.3389/fimmu.2017.00454
97. Colonna M. Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity. (2018) 48:1104–17. doi: 10.1016/j.immuni.2018.05.013
98. Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. (2017) 18:1004–15. doi: 10.1038/ni.3800
99. Nielsen CM, White MJ, Goodier MR, Riley EM. Functional significance of CD57 expression on human NK cells and relevance to disease. Front Immunol. (2013) 4:422. doi: 10.3389/fimmu.2013.00422
100. Cebo C, Durier V, Lagant P, Maes E, Florea D, Lefebvre T, et al. Function and molecular modeling of the interaction between human interleukin 6 and its HNK-1 oligosaccharide ligands. J Biol Chem. (2002) 277:12246–52. doi: 10.1074/jbc.M106816200
101. Liston A, Gray DHD. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol. (2014) 14:154–65. doi: 10.1038/nri3605
102. Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8+ T cell differentiation. Nat Immunol. (2016) 17:704–11. doi: 10.1038/ni.3438
103. Verbist KC, Guy CS, Milasta S, Liedmann S, Kaminski MM, Wang R, et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature. (2016) 532:389–93. doi: 10.1038/nature17442
104. Yassin M, Russell SM. Polarity and asymmetric cell division in the control of lymphocyte fate decisions and function. Curr Opin Immunol. (2016) 39:143–9. doi: 10.1016/j.coi.2016.02.004
105. Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, et al. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. (2017) 552:404–9. doi: 10.1038/nature25144
106. Chen YH, Kratchmarov R, Lin WHW, Rothman NJ, Yen B, Adams WC, et al. Asymmetric PI3K Activity in Lymphocytes Organized by a PI3K-Mediated Polarity Pathway. Cell Rep. (2018) 22:860–8. doi: 10.1016/j.celrep.2017.12.087
107. Patel CH, Powell JD. Targeting T cell metabolism to regulate T cell activation, differentiation and function in disease. Curr Opin Immunol. (2017) 46:82–8. doi: 10.1016/j.coi.2017.04.006
108. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4 + T Cell Subsets. J Immunol. (2011) 186:3299–303. doi: 10.4049/jimmunol.1003613
109. Buck MDD, O'Sullivan D, Klein Geltink RII, Curtis JDD, Chang CH, Sanin DEE, et al. Mitochondrial dynamics controls T Cell fate through metabolic programming. Cell. (2016) 166:63–76. doi: 10.1016/j.cell.2016.05.035
110. O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. (2015) 43:331–42. doi: 10.1016/j.immuni.2015.07.012
111. Goldrath AW, Bogatzki LY, Bevan MJ. Naive T cells transiently acquire a memory-like phenotype during homeostasis-driven proliferation. J Exp Med. (2000) 192:557–64. doi: 10.1084/jem.192.4.557
112. Boyman O, Létourneau S, Krieg C, Sprent J. Homeostatic proliferation and survival of naïve and memory T cells. Eur J Immunol. (2009) 39:2088–94. doi: 10.1002/eji.200939444
113. Min B. Spontaneous T cell proliferation: a physiologic process to create and maintain homeostatic balance and diversity of the immune system. Front Immunol. (2018) 9:547. doi: 10.3389/fimmu.2018.00547
114. Margolin K, Morishima C, Velcheti V, Miller JS, Lee SM, Silk AW, et al. Phase I trial of ALT-803, a novel recombinant IL15 complex, in patients with advanced solid tumors. Clin Cancer Res. (2018) 24:5552–61. doi: 10.1158/1078-0432.CCR-18-0945
115. Romee R, Cooley S, Berrien-Elliott MM, Westervelt P, Verneris MR, Wagner JE, et al. First-in-human phase 1 clinical study of the IL-15 superagonist complex ALT-803 to treat relapse after transplantation. Blood. (2018) 131:2515–27. doi: 10.1182/blood-2017-12-823757
116. Cooley S, He F, Bachanova V, Vercellotti GM, DeFor TE, Curtsinger J, et al. First-in-human trial of rhIL-15 and haploidentical natural killer cell therapy for advanced acute myeloid leukemia. Blood Adv. (2019) 3:1970–80. doi: 10.1182/bloodadvances.2018028332
117. Gooptu M, Kim HT, Howard A, Choi SW, Soiffer RJ, Antin JH, et al. Effect of sirolimus on immune reconstitution following myeloablative allogeneic stem cell transplantation: an ancillary analysis of a randomized controlled trial comparing tacrolimus/sirolimus and tacrolimus/methotrexate (Blood and Marrow Transplant Clinial Trials Network/BMT CTN 0402). Biol Blood Marrow Transplant. (2019) 25:2143–51. doi: 10.1016/j.bbmt.2019.06.029
118. Knudson KM, Hicks KC, Alter S, Schlom J, Gameiro SR. Mechanisms involved in IL-15 superagonist enhancement of anti-PD-L1 therapy. J Immunother Cancer. (2019) 7:1–16. doi: 10.1186/s40425-019-0551-y
119. Pinette A, McMichael E, Courtney NB, Duggan M, Benner BN, Choueiry F, et al. An IL-15-based superagonist ALT-803 enhances the NK cell response to cetuximab-treated squamous cell carcinoma of the head and neck. Cancer Immunol Immunother. (2019) 68:1379–89. doi: 10.1007/s00262-019-02372-2
120. Sandmaier BM, Kornblit B, Storer BE, Olesen G, Maris MB, Langston AA, et al. Addition of sirolimus to standard cyclosporine plus mycophenolate mofetil-based graft-versus-host disease prophylaxis for patients after unrelated non-myeloablative haemopoietic stem cell transplantation: a multicentre, randomised, phase 3 trial. Lancet Haematol. (2019) 6:e409–e418. doi: 10.1016/S2352-3026(19)30088-2
121. Liu E, Tong Y, Dotti G, Shaim H, Savoldo B, Mukherjee M, et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia. (2018) 32:520–31. doi: 10.1038/leu.2017.226
122. Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N Engl J Med. (2020) 382:545–53. doi: 10.1056/NEJMoa1910607
123. Ullah MA, Hill GR, Tey S-K. Functional Reconstitution of Natural Killer Cells in Allogeneic Hematopoietic Stem Cell Transplantation. Front Immunol. (2016) 7:144. doi: 10.3389/fimmu.2016.00144
124. Porrata LF, Inwards DJ, Ansell SM, Micallef IN, Johnston PB, Gastineau DA, et al. Early lymphocyte recovery predicts superior survival after autologous stem cell transplantation in non-hodgkin lymphoma: a prospective study. Biol Blood Marrow Transplant. (2008) 14:807–16. doi: 10.1016/j.bbmt.2008.04.013
125. Rueff J, Medinger M, Heim D, Passweg J, Stern M. Lymphocyte subset recovery and outcome after autologous hematopoietic stem cell transplantation for plasma cell myeloma. Biol Blood Marrow Transplant. (2014) 20:881–903. doi: 10.1016/j.bbmt.2014.03.007
126. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. (1997) 7:753–63. doi: 10.1016/S1074-7613(00)80394-5
127. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. (2002) 295:2097–100. doi: 10.1126/science.1068440
128. Cooley S, Parham P, Miller JS. Strategies to activate NK cells to prevent relapse and induce remission following hematopoietic stem cell transplantation. Blood. (2018) 131:1053–62. doi: 10.1182/blood-2017-08-752170
129. Mavers M, Bertaina A. High-Risk Leukemia : Past, Present, and Future Role of NK Cells. J Immunol Res. (2018) 2018:1–12. doi: 10.1155/2018/1586905
130. Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. (2005) 105:3051–7. doi: 10.1182/blood-2004-07-2974
131. Curti A, Ruggeri L, Parisi S, Bontadini A, Dan E, Motta MR, et al. Larger size of donor alloreactive NK cell repertoire correlates with better response to NK cell immunotherapy in elderly acute myeloid leukemia patients. Clin Cancer Res. (2016) 22:1–8. doi: 10.1158/1078-0432.CCR-15-1604
132. Björklund AT, Carlsten M, Sohlberg E, Liu LL, Clancy T, Karimi M, et al. Complete Remission with Reduction of High-Risk Clones following Haploidentical NK-Cell Therapy against MDS and AML. Clin Cancer Res. (2018) 24:1834–44. doi: 10.1158/1078-0432.CCR-17-3196
133. Hu W, Wang G, Huang D, Sui M, Xu Y. Cancer immunotherapy based on natural killer cells: current progress and new opportunities. Front Immunol. (2019) 10:1205. doi: 10.3389/fimmu.2019.01205
134. Shimasaki N., Jain A., Campana D. (2020). NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19, 200–218. doi: 10.1038/s41573-019-0052-1
135. Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. (2005) 106:376–83. doi: 10.1182/blood-2004-12-4797
136. Denman CJ, Senyukov VV, Somanchi SS, Phatarpekar PV, Kopp LM, Johnson JL, et al. Membrane-bound IL-21 promotes sustained Ex Vivo proliferation of human natural killer cells. PLoS ONE. (2012) 7:e30264. doi: 10.1371/journal.pone.0030264
137. Chang YH, Connolly J, Shimasaki N, Mimura K, Kono K, Campana D. A chimeric receptor with NKG2D specificity enhances natural killer cell activation and killing of tumor cells. Cancer Res. (2013) 73:1777–86. doi: 10.1158/0008-5472.CAN-12-3558
138. Li Y, Hermanson DL, Moriarity BS, Kaufman DS. Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell. (2018) 23:181–92. doi: 10.1016/j.stem.2018.06.002
139. Cheng M, Chen Y, Xiao W, Sun R, Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol. (2013) 10:230–52. doi: 10.1038/cmi.2013.10
140. Malmberg KJ, Sohlberg E, Goodridge JP, Ljunggren HG. Immune selection during tumor checkpoint inhibition therapy paves way for NK-cell “missing self” recognition. Immunogenetics. (2017) 69:547–56. doi: 10.1007/s00251-017-1011-9
141. Conlon KC, Lugli E, Welles HC, Rosenberg SA, Fojo AT, Morris JC, et al. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J Clin Oncol. (2015) 33:74–82. doi: 10.1200/JCO.2014.57.3329
142. Jiang T, Zhou C, Ren S. Role of IL-2 in cancer immunotherapy. Oncoimmunology. (2016) 5:e1163462. doi: 10.1080/2162402X.2016.1163462
143. Jacobs B, Pfefferle A, Clement D, Berg-Larsen A, Saetersmoen ML, Lorenz S, et al. Induction of the BIM short splice variant sensitizes proliferating NK Cells to IL-15 Withdrawal. J Immunol. (2019) 202:736–46. doi: 10.4049/jimmunol.1801146
144. Felices M, Lenvik AJ, Mcelmurry R, Chu S, Hinderlie P, Bendzick L, et al. Continuous treatment with IL-15 exhausts human NK cells via a metabolic defect. JCI Insight. (2018) 3:1–14. doi: 10.1172/jci.insight.96219
145. Hildeman DA, Mitchell T, Aronow B, Wojciechowski S, Kappler J, Marrack P. Control of Bcl-2 expression by reactive oxygen species. Proc Natl Acad Sci USA. (2003) 100:15035–40. doi: 10.1073/pnas.1936213100
146. Min-Oo G, Bezman NA, Madera S, Sun JC, Lanier LL. Proapoptotic Bim regulates antigen-specific NK cell contraction and the generation of the memory NK cell pool after cytomegalovirus infection. J Exp Med. (2014) 211:1289–96. doi: 10.1084/jem.20132459
147. Peña-Blanco A, García-Sáez AJ. Bax, Bak and beyond — mitochondrial performance in apoptosis. FEBS J. (2018) 285:416–31. doi: 10.1111/febs.14186
148. Mao Y, Van Hoef V, Zhang X, Wennerberg E, Lorent J, Witt K, et al. IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells. Blood. (2016) 128:1475–89. doi: 10.1182/blood-2016-02-698027
149. Viant C, Guia S, Hennessy RJ, Rautela J, Pham K, Bernat C, et al. Cell cycle progression dictates the requirement for BCL2 in natural killer cell survival. J Exp Med. (2017) 214:491–510. doi: 10.1084/jem.20160869
150. Kurtulus S, Tripathi P, Moreno-Fernandez ME, Sholl A, Katz JD, Grimes HL, et al. Bcl-2 allows effector and memory CD8+ T cells to tolerate higher expression of bim. J Immunol. (2011) 186:5729–37. doi: 10.4049/jimmunol.1100102
151. Wojciechowski S, Tripathi P, Bourdeau T, Acero L, Grimes HL, Katz JD, et al. Bim/Bcl-2 balance is critical for maintaining naive and memory T cell homeostasis. J Exp Med. (2007) 204:1665–75. doi: 10.1084/jem.20070618
152. O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: A novel member of the Bcl-2 family that promotes apoptosis. EMBO J. (1998) 17:384–95. doi: 10.1093/emboj/17.2.384
Keywords: natural killer cells (NK cells), IL-15, mTOR, homeostasis, NK cell differentiation
Citation: Pfefferle A, Jacobs B, Haroun-Izquierdo A, Kveberg L, Sohlberg E and Malmberg K-J (2020) Deciphering Natural Killer Cell Homeostasis. Front. Immunol. 11:812. doi: 10.3389/fimmu.2020.00812
Received: 28 January 2020; Accepted: 08 April 2020;
Published: 12 May 2020.
Edited by:
Aharon Freud, The Ohio State University, United StatesReviewed by:
Markus Uhrberg, Heinrich Heine University of Düsseldorf, GermanyStephen K. Anderson, National Cancer Institute at Frederick, United States
Copyright © 2020 Pfefferle, Jacobs, Haroun-Izquierdo, Kveberg, Sohlberg and Malmberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karl-Johan Malmberg, a2FsbGUubWFsbWJlcmdAa2kuc2U=
 Aline Pfefferle
Aline Pfefferle Benedikt Jacobs
Benedikt Jacobs Alvaro Haroun-Izquierdo
Alvaro Haroun-Izquierdo Lise Kveberg
Lise Kveberg Ebba Sohlberg
Ebba Sohlberg Karl-Johan Malmberg
Karl-Johan Malmberg