- 1Institute of Mitochondrial Biology and Medicine, Key Laboratory of Biomedical Information Engineering of the Ministry of Education, School of Life Sciences and Technology, Xi’an Jiaotong University, Xi’an, China
- 2Institute of Human Virology and Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, Baltimore, MD, United States
Defensins are a major family of host defense peptides expressed predominantly in neutrophils and epithelial cells. Their broad antimicrobial activities and multifaceted immunomodulatory functions have been extensively studied, cementing their role in innate immunity as a core host-protective component against bacterial, viral and fungal infections. More recent studies, however, paint defensins in a bad light such that they are “alleged” to promote viral and bacterial infections in certain biological settings. This mini review summarizes the latest findings on the potential pathogenic properties of defensins against the backdrop of their protective roles in antiviral and antibacterial immunity. Further, a succinct description of both tumor-proliferative and -suppressive activities of defensins is also given to highlight their functional and mechanistic complexity in antitumor immunity. We posit that given an enabling environment defensins, widely heralded as the “Swiss army knife,” can function as a “double−edged sword” in host immunity.
Introduction to Human Defensins
Defensins are a family of small (2–5 kDa), cationic host defense peptides with a β-sheet core structure stabilized by three conserved intramolecular disulfide bonds. The first mammalian defensin, also termed microbicidal cationic protein, was isolated in 1980 by Lehrer and colleagues from rabbit lung macrophages (1, 2). It was not until 1985 when the same lab discovered homologous peptides in human neutrophils did Lehrer coin the term defensin (3, 4) to describe disulfide-stabilized cationic peptides of mammalian origins with broad antimicrobial activity against bacteria, viruses and fungi. Based on disulfide topology, mammalian defensins are classified into three subfamilies, α, β, and θ-defensins (5–8). In humans, there exist only α and β-defensins. θ-defensins, with a unique circular structure stabilized by three parallel disulfide bonds in a ladder pattern, are only found in leukocytes of rhesus macaques (9). Although RNA transcripts homologous to the rhesus θ-defensin gene are found in humans, they contain a premature stop codon in the upstream signal sequence that abolishes their subsequent translation (10).
To date, six human α-defensins have been identified, which are further divided into two major classes according to their expression patterns and gene structures: myeloid defensins or human neutrophil peptides (HNPs) 1 to 4 and human (enteric) defensins (HDs) 5 and 6 (11–13). HNPs are stored in the azurophilic granules of human neutrophils, of which HNPs 1–3 and their much less abundant fourth cousin HNP4 account, collectively, for 5–7% of the total neutrophil protein (4; 14; 15). HNPs-containing granules normally undergo restricted secretion and are commonly directed for fusion with phagolysosomes, where high concentrations of HNPs directly kill phagocytosed microbes (16, 17). Upon holocrine secretion and neutrophil infiltration during inflammation, HNPs are released into the extracellular milieu through degranulation of activated neutrophils (17–19). HD5 and HD6 are constitutively expressed in and secreted by Paneth cells at the bottom of the small intestinal crypt (12, 13, 20, 21). While the concentration of HD5 at the luminal surface of the small intestine is estimated to be as high as 50–250 μg/ml, it is significantly lower at the colonic mucosal surface due to the distance from secretion (21). HD5 ranging from 1 to 50 μg/ml is also found in vaginal fluid from healthy women (22) and induced in the male and female reproductive tract in response to sexually transmitted infections (STIs) (23–25). Although more than 30 β-defensin genes exist in the human genome, only a few have been extensively characterized at the genomic and functional levels (26). Unlike α-defensin expression, which is commonly regulated at the level of secretion, β-defensin expression is transcriptionally regulated and restricted to keratinocytes of the skin and epithelial cells. For instance, while human β-defensin 1 (HBD1) is constitutively expressed, HBD2 and HBD3 are induced by microbial insults and pro-inflammatory cytokines in various epithelial and mucosal tissues (27, 28).
Since their first discovery in the early 1980s, defensins have been intensively investigated for their broad antimicrobial activities and multifaceted immunomodulatory functions under both physiological and pathogenic conditions. Many excellent reviews have shed light on a multitude of sophisticated molecular and cellular mechanisms by which defensins act against bacteria, viruses and fungi and function as pleiotropic immune effectors in inflammation, development and cancer (5, 11, 26, 29–34). By and large, defensins are heralded as the “Swiss army knife” in innate immunity against microbial pathogens. Nevertheless, accumulating recent evidence has unveiled a potential pathogenic role defensins play in host-pathogen interactions and tumorigenesis, indicating that the mechanisms of action of defensins are far more complex than previously thought. The growing recognition that defensins can be both advantageous and detrimental, depending on their spatial-temporal settings, gives us the impetus to review the recent literature on their protective and pathogenic roles in health and disease.
Defensins in Viral Infection
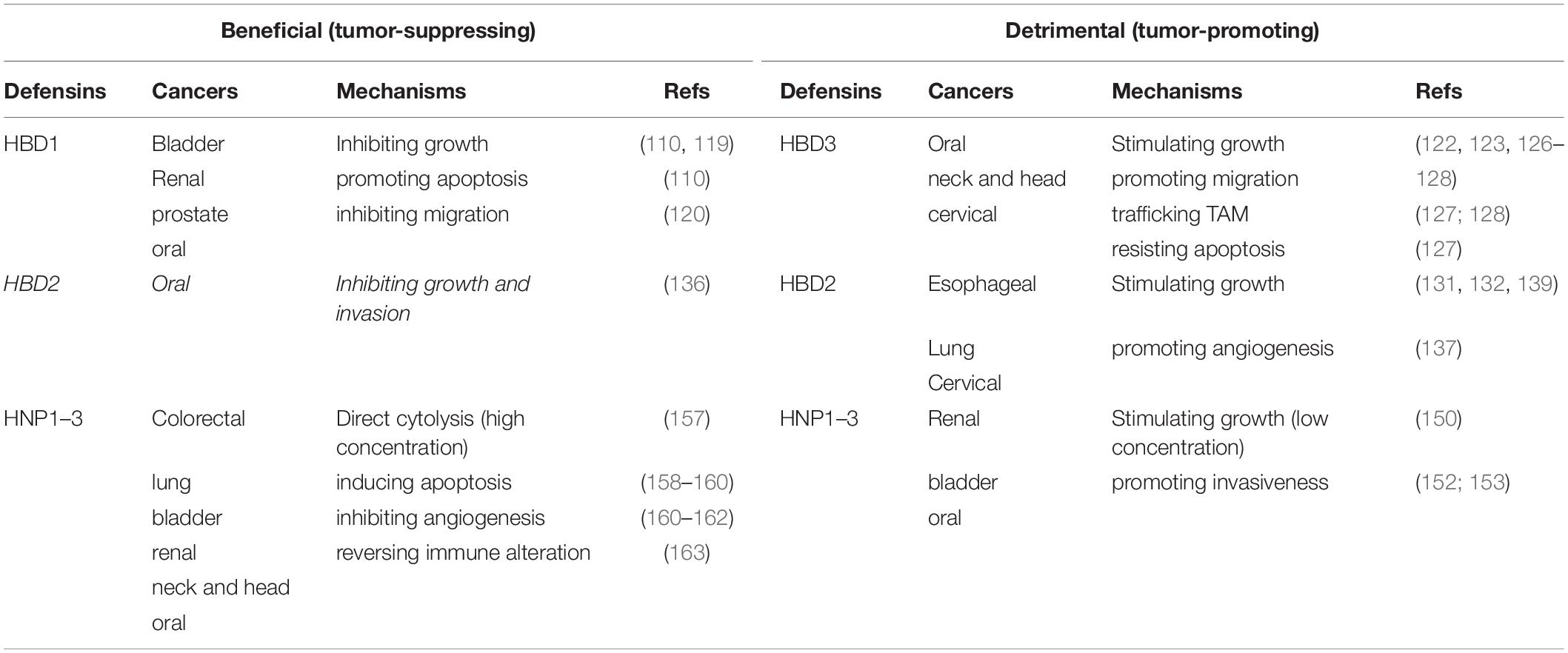
Defensins directly inactivate and inhibit the replication of a variety of viruses, and their multifaceted mechanisms of action have been elucidated (30, 31); the underlying mechanisms of the role of defensins in host-virus interactions are more complex as evidenced with HIV-1 (Figure 1). Early studies demonstrated that defensins are able to target multiple steps of host-virus interactions to reduce the infectivity of both enveloped and non-enveloped viruses. HNP1–3, HD5 and retrocyclins 1 and 3 deduced from human θ-defensin pseudogenes effectively block adhesion of enveloped herpes simplex virus 2 (HSV-2) to host cells by preventing HSV-2 gB interactions with its receptor HSPGs (35–37). Defensins also inhibit fusion of virions of several enveloped viruses with their host cells. Retrocyclin 2 and HBD3 interfere with viral fusion mediated by influenza virus hemagglutinin (HA) and other viral proteins such as baculovirus gp64 and Sindbis virus E1 protein (38). While HNP1 is well recognized for its direct anti-HIV activity (39, 40), it also restrains HIV-1 uptake by inhibiting Env-mediated viral fusion and downregulating host cell surface expression of CD4 and coreceptor CXCR4 (41), a controversial mechanism for HBD2 and HBD3 inhibition of HIV-1 infection (42–44).
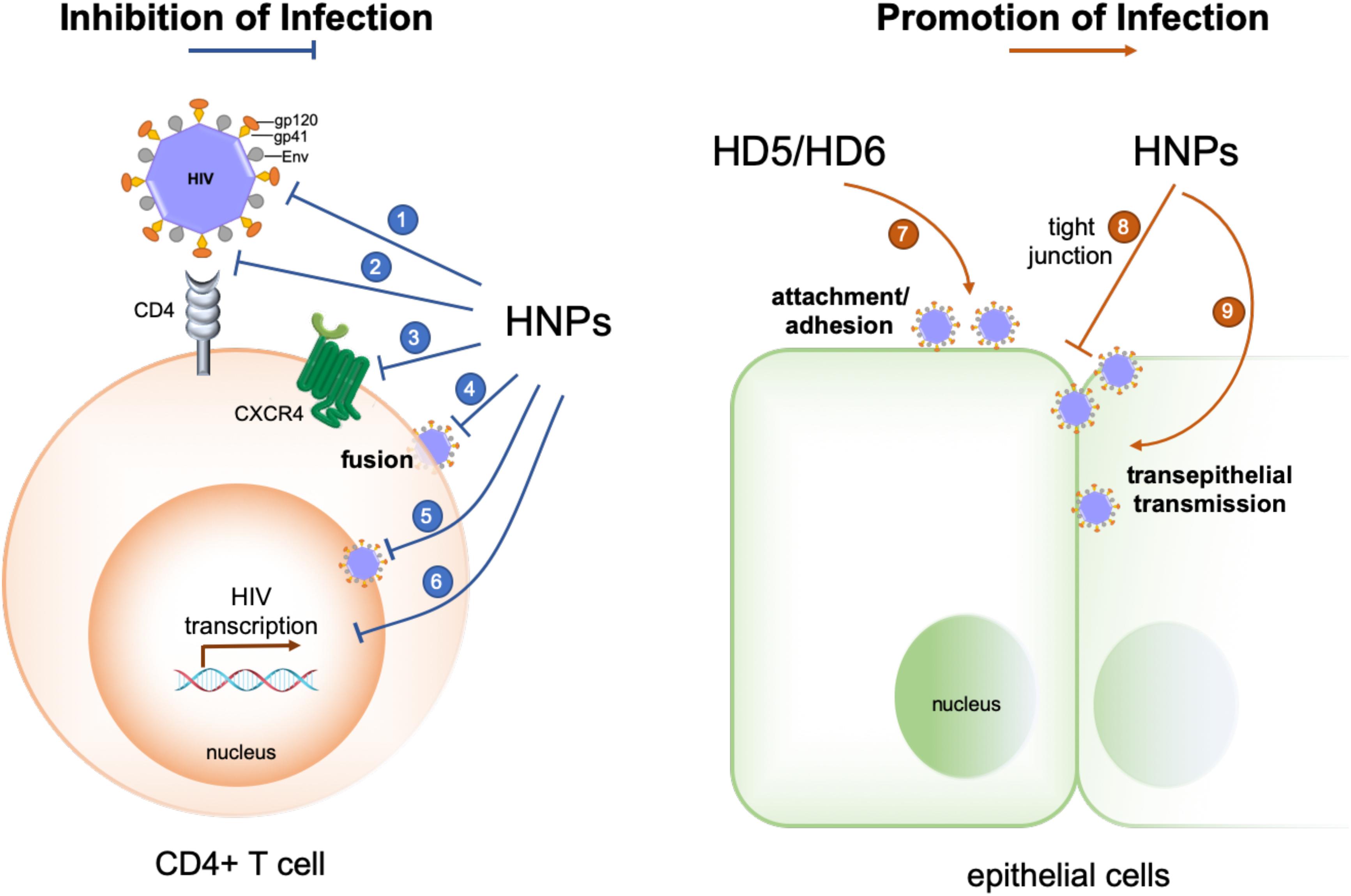
Figure 1. Opposing effects of human α-defensins on HIV-1 infection. Infection-inhibiting mechanisms include: (1) direct inactivation of the virus (39, 40), (2) blockade of gp120-CD4 interactions (164, 165), (3) coreceptor downregulation (41, 166), (4) inhibition of gp41- and Env-mediated viral fusion (41), (5) inhibition of nuclear import of viral RNA (30, 39), and (6) suppression of HIV transcription (30, 39, 167). Infection-promoting mechanisms include: (7) enhancing viral adhesion/attachment (25, 56, 57, 59), and (8, 9) disrupting tight junction to promote trans-epithelial transmission of HIV (60).
Post-entry inhibition of viral infection by defensins has been observed with several families of non-enveloped viruses, notably HPV (45). Without affecting the binding and entry steps, α-defensins effectively block intracellular uncoating of HPV and its escape from cytoplasmic vesicles by stabilizing its viral capsid structure to prevent interactions of viral proteins and genome with host factors essential for productive infection (45–49). This general inhibitory mechanism has been verified for other non-enveloped viruses such as human adenovirus (HAdV) and JC polyomavirus where α-defensins stabilize viral capsid proteins, thus diminishing subsequent intracellular infection (50–54). Of note, post-entry inhibition of enveloped viruses such as HIV-1 and influenza by HNP1 is mediated through interfering with cell signaling pathways such as PKC that are required for viral replication (39, 55).
More recent studies, mostly by the same research groups who demonstrated the beneficial role of defensins in controlling viral infection, unveil infection-promoting effects of defensins in HIV-1 and certain serotypes of HAdV infections (25, 51, 56–60). Chang and colleagues reported that HD5 and HD6, induced by Neisseria gonorrhoeae infection in a cervicovaginal tissue culture system, increase HIV infectivity in a CD4- and HIV coreceptor-independent manner (25). HD5 and HD6 promote HIV infection by acting on the virion to enhance viral attachment to its target cells (57). These defensins antagonize anti-HIV activity of polyanion microbicide candidates that block HIV entry (56). HNP1, the prototypic α-defensin extensively studied for its multifaceted anti-HIV activity, is also capable of disrupting epithelial integrity to promote HIV traversal across epithelial barriers, thus facilitating viral infection and dissemination (60). These findings by the Chang group are of particular interest since increased HNP1 and HD5 expression in the genitourinary tract upon STIs could potentially generate sufficiently high concentrations of defensins to enhance HIV-1 infection under physiological conditions. Other examples regarding the enhancing effect of defensins on enveloped virus infection have been reported. For example, cryptdin 3, one of several mouse α-defensins expressed in the small intestine (61) also enhances HIV infection in vitro presumably by facilitating viral entry (58). A recent study shows that an alphaherpesvirus, equine herpesvirus type 1, is resistant to equine β-defensins 2–3, which inhibit bacteria and viruses, and exploits these defensins to invade the host for viral spread (62).
HNP1- and HD5-promoted viral infection has also been observed with certain serotypes of HAdV as reported by the Smith group (51), who previously deciphered the capsid-stabilizing mechanism of defensins against HPV and HAdV and delineated their structural determinants of antiviral activity (46–54, 63). As is the case with HIV-1, HNP1- and HD5-dependent enhancement in infection by HAdV-D and -F correlates with increased viral attachment to target cells independently of receptor binding (51). To address the physiological relevance of defensin-enhanced adenovirus infection, Smith and colleagues utilized a murine enteric organoid (enteroid) to examine the impact of naturally secreted cryptdins on the infectivity of an enteric mouse pathogen, mouse adenovirus 2 (MAdV-2). MAdV-2 infection increases in the enteroids expressing mouse α-defensins but not in the ones devoid of them (64). This ex vivo study demonstrates that α-defensin-enhanced viral infection occurs not only in traditional cell cultures, but also under physiologic conditions.
Defensins in Bacterial Infection
Defensins are capable of killing bacteria or inhibiting bacterial growth through a multiplicity of antimicrobial mechanisms such as direct membrane disruption (11, 65, 66) and inhibition of bacterial cell wall synthesis (67–69). Defensins can also reduce bacterial infection by neutralizing secreted toxins (70–73). In general, human α-defensins are less cationic but more hydrophobic than β-defensins, and they can differ mechanistically in the killing of bacteria (11). While HBD1 and HBD2 are active preferably against Gram-negative bacteria (74), their significantly more cationic counterpart HBD3 is potently bactericidal against both Gram-positive and -negative strains (75). Due to its heavily cationic nature, HBD3 broadly kills bacteria in a structure-independent manner (76, 77). Notably, disulfide reduction of the weakly bactericidal HBD1 turns it into a potent antimicrobial peptide against opportunistic pathogenic fungi and Gram-positive commensal bacteria (78). Excellent reviews on the antifungal activity of defensins are also available (79, 80). Our review focuses on the role of human α-defensins in host-bacteria interactions to contrast their protective and pathogenic functions.
Bevins and colleagues demonstrated that HD5-transgenic mice are markedly resistant to oral challenge with virulent Salmonella typhimurium, consistent with the antibacterial activity of HD5 in vitro, whereas wild-type mice are susceptible to infection (81). An in vivo protective role against Salmonella infection is also illustrated for mouse intestinal α-defensins or cryptdins (82). Of note, enteric HD6, while exhibiting little bactericidal and membranolytic activity in vitro, protects mice from Salmonella infection by entrapping bacteria with a unique self-assembled “nanonets” structure to preclude the pathogen’s direct contact with the intestinal epithelium (83).
HNP1–3 secreted by infiltrating neutrophils in Staphylococcus aureus infection induce TNF-α and IFN-γ release from macrophages, which, in turn, increase phagocytosis of pathogens by macrophages – an essential step in bacterial clearance (84, 85). HNP1 also inhibits phagosomal escape and intracellular multiplication of Listeria monocytogenes and Mycobacterium tuberculosis in macrophages (86, 87), suggesting that the defensin, although not being expressed by macrophages, contributes to their antimicrobial function. Notably, HNP1 acts in the aftermath of Salmonella infection as a “molecular brake” on macrophage-driven inflammation by preventing protein translation to ensure both pathogen clearance and the resolution of inflammation with minimal bystander tissue damage (88).
While the protective roles of defensins in bacterial infection are widely reported in the field, we have made a surprising recent discovery that α-defensins can contribute to the pathogenicity of Shigella (89–92). Unlike other enteropathogenic bacteria, Shigella lacks general adhesion machinery such as fimbriae due presumably to pervasive genome reduction during the course of adaptation to the intracellular environment (93–95). As a result, Shigella is much less adhesive and invasive in vitro than other fimbriated enteropathogenic bacteria despite its extraordinary infectivity in humans. Further, although highly infectious in humans, Shigella hardly infects any other animals including mice with abundant enteric α-defensins (cryptdins) (96, 97). This seemingly paradoxical phenomenon or conundrum in Shigella pathogenesis has remained largely obscure mechanistically at the molecular and cellular levels (97–99). We found that the lack of fimbriae in Shigella affords the pathogen a unique bacterial surface, onto which HD5 forms multimeric structures to mediate Shigella adhesion to host epithelium; enhanced bacterial adhesion in turn strongly promotes Shigella invasion of host cells, ensuing dramatically augmented infection in vivo and ex vivo (Figure 2). These studies support the premise that Shigella exploits HD5 for virulence (89, 91), thereby explaining not only its extraordinary pathogenicity but also its restricted host selectivity.
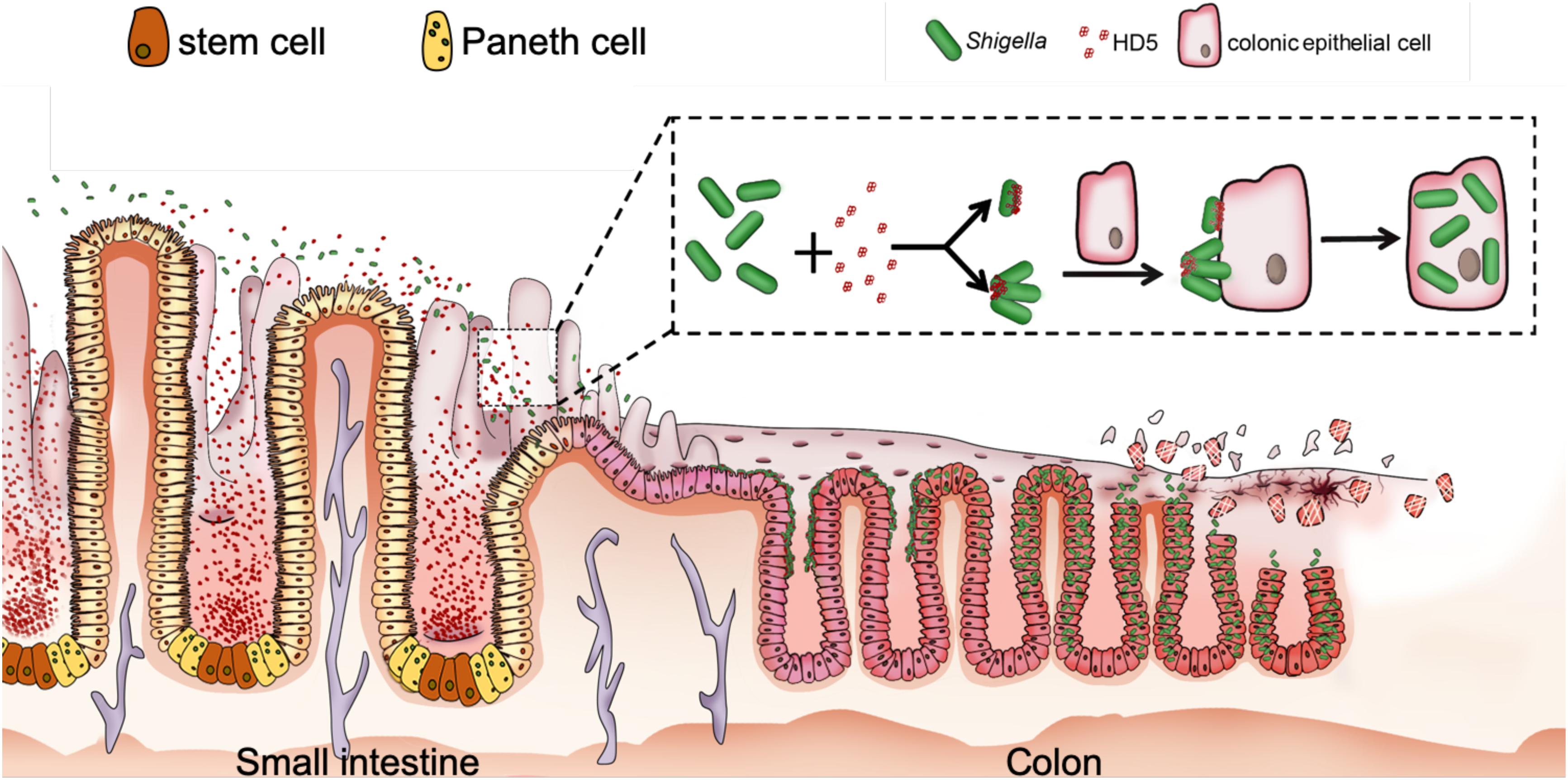
Figure 2. Proposed model for HD5-promoted Shigella infection of the colonic epithelium. HD5 in the lumen of the small intestine encounters poorly adhesive Shigella in transit, binds to Shigella surface, and promotes Shigella adhesion to the colonic epithelium by bridging single bacterium and host cell and/or clustering multiple bacteria for multivalent attachment to host cell, leading to increased bacterial infection from the apical surface (89, 91).
HNP1 is also active in promoting Shigella infection of epithelial cells (90), consistent with an earlier finding that human neutrophil granular proteins (containing HNPs) enhance Shigella adhesion in vitro at sub-lethal concentrations (100). Although HNP1 is weaker than HD5 with respect to their ability to promote Shigella adhesion, its strong activity in disrupting the epithelial barrier contributes additionally to Shigella infection (90). It is worth noting that HD5 exacerbates the pathogenicity of Shigella in macrophages. Despite that HD5 boosts phagocytosis of Shigella by macrophages, an antimicrobial event generally unfavorable to invading pathogens, it fails to prevent subsequent phagosomal escape and intracellular multiplication of Shigella, resulting in necrosis of infected macrophages induced by multiplying Shigella and massive release of intracellular bacteria (92).
For human α-defensins, their hydrophobicity and selective cationicity segregated on a dimeric structure stabilized by intramolecular disulfides are critical for antimicrobial activity (11). Several mutational studies have identified the functional determinants of α-defensins in promoting viral and bacterial infections (25, 56, 57, 59, 60, 90–92). Briefly, disulfide bonding in defensins is absolutely required for their ability to enhance HIV-1 infection (25, 60) and to promote Shigella adhesion and invasion (91, 92); hydrophobic residues in α-defensins, i.e., Trp26 and Phe28 in HNP1, Leu16, Leu26, Tyr27 and Leu29 in HD5, and Phe2 and Phe29 in HD6, play a pivotal functional role (59, 90, 91); dimerization and/or oligomerization of α-defensins are functionally indispensable (59, 83, 91, 101, 102); selective cationicity, as exemplified by Arg28 in HD5, can be critical for promoting HIV and Shigella infection (59, 91, 92). Obviously, although α-defensins are highly variable in amino acid sequence, their functional determinants are rather conserved, irrespective of their pathogenic and protective roles in host immunity.
Defensins in Tumorigenesis
Most cancers develop from epithelial cells and tissues (carcinomas) where β-defensins are expressed for mucosal surface protection against microbial infection (26, 27, 103, 104). Since β-defensins are differentially expressed in normal tissues and tumors, their role in tumor development and progression has attracted considerable interest (32, 105–107). HBD1 is downregulated in most carcinomas (108–118), and the stimuli of this downregulation are yet to be identified. Growing evidence suggests that HBD1 functions as a tumor suppressor in most carcinomas (110, 119, 120). By contrast, HBD3 is frequently overexpressed in various carcinomas (121–124), and its upregulation has been ascribed to LPS-stimulated EGFR activation (121) or HPV co-infection-induced p53 degradation (125), among others. Importantly, HBD3 stimulates tumor growth and migration (122, 123, 126), confers resistance of tumor cells to apoptosis (127), and helps the recruitment of tumor-associated macrophages that promote tumor progression (127, 128). Consistent with the oncogenic role of upregulated H BD3, mouse β-defensin 14, the ortholog of HBD3, acts as a chemoattractant to enhance angiogenesis and tumor development in vivo (129). The regulation of HBD2 and its influence in tumorigenesis vary from cancer to cancer (106) and can be controversial at times (130, 131). HBD2 is upregulated in esophageal, lung and skin cancers (108, 109, 118, 132), but downregulated in oral and colon cancers (112, 114, 133). While the mechanisms of HBD2 regulation are only partially understood (132, 134, 135), HBD2 appears to play a suppressive role in tumor development and progression when it is downregulated (136), but a proliferative role when upregulated (131, 132, 137–139), in agreement with HBD1 and HBD3. The suppressive and proliferative properties of defensins in tumorigenesis are tabulated in Table 1.
The role of α-defensins in tumorigenesis has also been extensively examined (140, 141). Elevated levels of myeloid α-defensins, HNP1–3, are frequently detected in many different types of tumor tissues and in biological fluids from cancer patients (142–155). While tumor-infiltrating immune cells, and neutrophils in particular, are likely a major contributor to increased HNP1–3 in tumors (151), several studies also suggest that tumor cells themselves may produce HNP1–3 through a yet-to-be-identified mechanism (142, 150). HNP1–3 have been shown to promote tumor cell proliferation (150, 156), contributing to tumor progression and invasiveness (152, 153). Due to their membranolytic activity toward bacteria and limited sites of expression, much of the early studies of α-defensins have focused on their ability to lyse tumor cells at high concentrations (157). More recent work, however, has shed light on the mechanistic complexity of the antitumor activity of HNP1–3, including inducing apoptosis (158–160), inhibiting angiogenesis (160–162), and altering immune milieu in HPV-associated neoplasia by recruiting immature dendritic cells (163).
Concluding Remarks
Long recognized as a class of host defense peptides and immunomodulators important for innate immune responses to viral, bacterial and fungal infections, human defensins are widely thought to be host protective. Growing recent evidence suggests, however, that they can also be pathogenic under certain biological conditions by promoting viral and bacterial infections. The interchangeable roles between a “Swiss army knife” and a “double-edged sword” played by human α-defensins in host immunity are under-appreciated in the field, despite the well-recognized fact that defensins can be both suppressors and promotors in tumorigenesis, depending on which defensin and cancer type are studied. While the mechanisms of host protection by human defensins are well-understood, much remain obscure with respect to the molecular and cellular events dictating defensins’ pro-infective activity. A better understanding of how human defensins promote infection may ultimately lead to new therapeutic interventions of infectious diseases.
Author Contributions
DX and WL wrote the manuscript.
Funding
DX was supported by the National Natural Science Foundation of China (Grant No. 31770146).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Theresa L. Chang of the Rutgers University for incisive comments.
References
1. Patterson-Delafield J, Martinez RJ, Lehrer RI. Microbicidal cationic proteins in rabbit alveolar macrophages: a potential host defense mechanism. Infect Immun. (1980) 30:180–92.
2. Patterson-Delafield J, Szklarek D, Martinez RJ, Lehrer RI. Microbicidal cationic proteins of rabbit alveolar macrophages: amino acid composition and functional attributes. Infect Immun. (1981) 31:723–31.
3. Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, et al. Defensins natural peptide antibiotics of human neutrophils. J Clin Invest. (1985) 76:1427–35.
4. Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. Primary structures of three human neutrophil defensins. J Clin Invest. (1985) 76:1436–9.
8. Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. (2005) 6:551–7.
9. Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. (1999) 286:498–502.
10. Lehrer RI, Cole AM, Selsted ME. theta-Defensins: cyclic peptides with endless potential. J Biol Chem. (2012) 287:27014–9.
12. Bevins CL, Martin-Porter E, Ganz T. Defensins and innate host defence of the gastrointestinal tract. Gut. (1999) 45:911–5.
13. Bevins CL, Salzman NH. Paneth cells antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. (2011) 9:356–68.
14. Wilde CG, Griffith JE, Marra MN, Snable JL, Scott RW. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J Biol Chem. (1989) 264:11200–3.
15. Tongaonkar P, Golji AE, Tran P, Ouellette AJ, Selsted ME. High fidelity processing and activation of the human alpha-defensin HNP1 precursor by neutrophil elastase and proteinase 3. PLoS One. (2012) 7:e32469.
16. Nordenfelt P, Tapper H. Phagosome dynamics during phagocytosis by neutrophils. J Leukoc Biol. (2011) 90:271–84.
17. Faurschou M, Sorensen OE, Johnsen AH, Askaa J, Borregaard N. Defensin-rich granules of human neutrophils: characterization of secretory properties. Biochim Biophys Acta. (2002) 1591:29–35.
18. Ganz T. Extracellular release of antimicrobial defensins by human polymorphonuclear leukocytes. Infect Immun. (1987) 55:568–71.
19. Gabay JE, Scott RW, Campanelli D, Griffith J, Wilde C, Marra MN, et al. Antibiotic proteins of human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. (1989) 86:5610–4.
20. Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. (2013) 75:289–311.
21. Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. (2002) 3:583–90.
22. Quayle AJ, Porter EM, Nussbaum AA, Wang YM, Brabec C, Yip KP, et al. Gene expression immunolocalization, secretion of human defensin-5 in human female reproductive tract. Am J Pathol. (1998) 152:1247–58.
23. Porter E, Yang H, Yavagal S, Preza GC, Murillo O, Lima H, et al. Distinct defensin profiles in Neisseria gonorrhoeae and Chlamydia trachomatis urethritis reveal novel epithelial cell-neutrophil interactions. Infect Immun. (2005) 73:4823–33.
24. Spencer JD, Hains DS, Porter E, Bevins CL, DiRosario J, Becknell B, et al. Human alpha defensin 5 expression in the human kidney and urinary tract. PLoS One. (2012) 7:e31712.
25. Klotman ME, Rapista A, Teleshova N, Micsenyi A, Jarvis GA, Lu W, et al. Neisseria gonorrhoeae-induced human defensins 5 and 6 increase HIV infectivity: role in enhanced transmission. J Immunol. (2008) 180:6176–85.
26. Semple F, Dorin JR. beta-Defensins: multifunctional modulators of infection inflammation and more? J Innate Immun. (2012) 4:337–48.
27. Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human beta-defensins. Cell Mol Life Sci. (2006) 63:1294–313.
28. O’Neil DA. Regulation of expression of beta-defensins: endogenous enteric peptide antibiotics. Mol Immunol. (2003) 40:445–50.
29. Bevins CL. Innate immune functions of alpha-defensins in the small intestine. Dig Dis. (2013) 31:299–304.
31. Holly MK, Diaz K, Smith JG. Defensins in viral infection and pathogenesis. In: L Enquist editor. Annual Review of Virology. (Vol. 4), Washington DC: Department of Microbiology University of Washington (2017) p. 369–91.
32. Jin G, Weinberg A. Human antimicrobial peptides and cancer. Semin Cell Dev Biol. (2019) 88:156–62.
33. Rehaume LM, Hancock RE. Neutrophil-derived defensins as modulators of innate immune function. Crit Rev Immunol. (2008) 28:185–200.
34. Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins cathelicidins, eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. (2004) 22:181–215.
35. Yasin B, Wang W, Pang M, Cheshenko N, Hong T, Waring AJ, et al. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J Virol. (2004) 78:5147–56.
36. Hazrati E, Galen B, Lu W, Wang W, Ouyang Y, Keller MJ, et al. Human α- and β-defensins block multiple steps in herpes simplex virus infection. J Immunol. (2006) 177:8658.
37. Wang A, Chen F, Wang Y, Shen M, Xu Y, Hu J, et al. Enhancement of antiviral activity of human alpha-defensin 5 against herpes simplex virus 2 by arginine mutagenesis at adaptive evolution sites. J Virol. (2013) 87:2835–45.
38. Leikina E, Delanoe-Ayari H, Melikov K, Cho M-S Chen, A Waring, AJ et al. Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nat Immunol. (2005) 6:995–1001.
39. Chang TL, Vargas J Jr., DelPortillo A, Klotman ME. Dual role of alpha-defensin-1 in anti-HIV-1 innate immunity. J Clin Invest. (2005) 115:765–73.
40. Mackewicz CE, Yuan J, Tran P, Diaz L, Mack E, Selsted ME, et al. alpha-Defensins can have anti-HIV activity but are not CD8 cell anti-HIV factors. AIDS. (2003) 17:F23–32.
41. Demirkhanyan LH, Marin M, Padilla-Parra S, Zhan C, Miyauchi K, Jean-Baptiste M, et al. Multifaceted mechanisms of HIV-1 entry inhibition by human alpha-defensin. J Biol Chem. (2012) 287:28821–38.
42. Quiñones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS. (2003) 17:F39–48.
43. Lafferty MK, Sun L, DeMasi L, Lu W, Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. (2010) 115:1564–71.
44. Sun L, Finnegan CM, Kish-Catalone T, Blumenthal R, Garzino-Demo P, La Terra Maggiore GM, et al. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol. (2005) 79:14318–29.
45. Buck CB, Day PM, Thompson CD, Lubkowski J, Lu W, Lowy DR, et al. Human α-defensins block papillomavirus infection. Proc Natl Acad Sci USA. (2006) 103:1516.
46. Tenge VR, Gounder AP, Wiens ME, Lu W, Smith JG. Delineation of interfaces on human alpha-defensins critical for human adenovirus and human papillomavirus inhibition. PLoS Pathog. (2014) 10:e1004360.
47. Wiens ME, Smith JG. Alpha-Defensin HD5 inhibits furin cleavage of human papillomavirus 16 L2 to block infection. J Virol. (2015) 89:2866–74.
48. Wiens ME, Smith JG. Alpha-Defensin HD5 inhibits human papillomavirus 16 infection via capsid stabilization and redirection to the lysosome. mBio. (2017) 8:e2304–16.
49. Gulati NM, Miyagi M, Wiens ME, Smith JG, Stewart PL. Alpha-defensin HD5 stabilizes human papillomavirus 16 capsid/core interactions. Pathog Immun. (2019) 4:196–234.
50. Smith JG, Nemerow GR. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe. (2008) 3:11–9.
51. Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. (2010) 6:e1000959.
52. Nguyen EK, Nemerow GR, Smith JG. Direct evidence from single-cell analysis that human alpha-defensins block adenovirus uncoating to neutralize infection. J Virol. (2010) 84:4041–9.
53. Gounder AP, Wiens ME, Wilson SS, Lu W, Smith JG. Critical determinants of human alpha-defensin 5 activity against non-enveloped viruses. J Biol Chem. (2012) 287:24554–62.
54. Zins SR, Nelson CDS, Maginnis MS, Banerjee R, O’Hara BA, Atwood WJ. The human alpha defensin HD5 neutralizes JC polyomavirus infection by reducing endoplasmic reticulum traffic and stabilizing the viral capsid. J Virol. (2014) 88:948–60.
55. Salvatore M, Garcia-Sastre A, Ruchala P, Lehrer RI, Chang T, Klotman ME. alpha-Defensin inhibits influenza virus replication by cell-mediated mechanism(s). J Infect Dis. (2007) 196:835–43.
56. Ding J, Rapista A, Teleshova N, Lu W, Klotman ME, Chang TL. Mucosal human defensins 5 and 6 antagonize the anti-HIV activity of candidate polyanion microbicides. J Innate Immun. (2011) 3:208–12.
57. Rapista A, Ding J, Benito B, Lo YT, Neiditch MB, Lu W, et al. Human defensins 5 and 6 enhance HIV-1 infectivity through promoting HIV attachment. Retrovirology. (2011) 8:45.
58. Tanabe H, Ouellette AJ, Cocco MJ, Robinson WEJr Differential effects on human immunodeficiency virus type 1 replication by alpha-defensins with comparable bactericidal activities. J Virol. (2004) 78:11622–31.
59. Valere K, Lu W, Chang TL. Key determinants of human alpha-defensin 5 and 6 for enhancement of HIV infectivity. Viruses. (2017) 9:244.
60. Valere K, Rapista A, Eugenin E, Lu W, Chang TL. Human alpha-defensin HNP1 increases HIV traversal of the epithelial barrier: a potential role in STI-mediated enhancement of HIV transmission. Viral Immunol. (2015) 28:609–15.
61. Ouellette AJ. Paneth cell alpha-defensins in enteric innate immunity. Cell Mol Life Sci. (2011) 68:2215–29.
62. Van Cleemput J, Poelaert KCK, Laval K, Vanderheijden N, Dhaenens M, Daled S, et al. An alphaherpesvirus exploits antimicrobial beta-defensins to initiate respiratory tract infection. J Virol. (2020) 94:e1676–1619.
63. Wilson SS, Wiens ME, Smith JG. Antiviral mechanisms of human defensins. J Mol Biol. (2013) 425:4965–80.
64. Wilson SS, Bromme BA, Holly MK, Wiens ME, Gounder AP, Sul Y, et al. Alpha-defensin-dependent enhancement of enteric viral infection. Plos Pathogens. (2017) 13:e1006446.
65. Kagan BL, Selsted ME, Ganz T, Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc Natl Acad Sci USA. (1990) 87:210–4.
66. Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. (1989) 84:553–61.
67. de Leeuw E, Li C, Zeng P, Li C, Diepeveen-de Buin M, Lu WY, et al. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. (2010) 584:1543–8.
68. Munch D, Sahl HG. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria - Impact on binding and efficacy of antimicrobial peptides. Biochim Biophys Acta. (2015) 1848(11 Pt B):3062–71.
69. Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, et al. Plectasin a fungal defensin targets the bacterial cell wall precursor lipid II. Science. (2010) 328:1168.
70. Kim C, Gajendran N, Mittrücker H-W Weiwad, M Song, Y-H Hurwitz, R et al. Human alpha-defensins neutralize anthrax lethal toxin and protect against its fatal consequences. Proc Natl Acad Sci USA. (2005) 102:4830–5.
71. Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase a novel bacterial evasion mechanism. J Immunol. (2004) 172:1169–76.
72. Kudryashova E, Quintyn R, Seveau S, Lu W, Wysocki VH, Kudryashov DS. Human defensins facilitate local unfolding of thermodynamically unstable regions of bacterial protein toxins. Immunity. (2014) 41:709–21.
73. Lehrer RI, Jung G, Ruchala P, Wang W, Micewicz Waring, AJ et al. Human alpha-defensins inhibit hemolysis mediated by cholesterol-dependent cytolysins. Infect Immun. (2009) 77:4028–40.
74. Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. (1997) 387:861.
75. Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. (2001) 276:5707–13.
76. Wu Z, Hoover DM, Yang D, Boulegue C, Santamaria F, Oppenheim JJ, et al. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc Natl Acad Sci USA. (2003) 100:8880–5.
77. Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, et al. Through the looking glass mechanistic insights from enantiomeric human defensins. J Biol Chem. (2009) 284:29180–92.
78. Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature. (2011) 469:419–23.
79. Ordonez SR, Veldhuizen EJA, van Eijk M, Haagsman HP. Role of soluble innate effector molecules in pulmonary defense against fungal pathogens. Front Microbiol. (2017) 8:2098.
80. Parisi K, Shafee TMA, Quimbar P, van der Weerden NL, Bleackley MR, Anderson MA. The evolution function and mechanisms of action for plant defensins. Semin Cell Dev Biol. (2019) 88:107–18.
81. Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. (2003) 422:522–6.
82. Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, et al. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. (1999) 286:113.
83. Chu H, Pazgier M, Jung G, Nuccio S-P Castillo, PA de Jong MF, et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. (2012) 337:477–81.
84. Soehnlein O, Kai-Larsen Y, Frithiof R, Sorensen OE, Kenne E, Scharffetter-Kochanek K, et al. Neutrophil primary granule proteins HBP and HNP1–3 boost bacterial phagocytosis by human and murine macrophages. J Clin Invest. (2008) 118:3491–502.
85. Soehnlein O, Kenne E, Rotzius P, Eriksson EE, Lindbom L. Neutrophil secretion products regulate anti-bacterial activity in monocytes and macrophages. Clin Exp Immunol. (2008) 151:139–45.
86. Arnett E, Lehrer RI, Pratikhya P, Lu W, Seveau S. Defensins enable macrophages to inhibit the intracellular proliferation of Listeria monocytogenes. Cell Microbiol. (2011) 13:635–51.
87. Tan BH, Meinken C, Bastian M, Bruns H, Legaspi A, Ochoa MT, et al. Macrophages acquire neutrophil granules for antimicrobial activity against intracellular pathogens. J Immunol. (2006) 177:1864.
88. Brook M, Tomlinson GH, Miles K, Smith RWP, Rossi AG, Hiemstra PS, et al. Neutrophil-derived alpha defensins control inflammation by inhibiting macrophage mRNA translation. Proc Natl Acad Sci USA. (2016) 113:4350.
89. Murphy AG, Maloy KJ. Defens-IN! human α-defensin 5 Acts as an unwitting double agent to promote Shigella infection. Immunity. (2018) 48:1070–2.
90. Liao C, Fang K, Xiao J, Zhang W, Zhang B, Yuan W, et al. Critical determinants of human neutrophil peptide 1 for enhancing host epithelial adhesion of Shigella flexneri. Cell Microbiol. (2019) 21:e13069.
91. Xu D, Liao C, Zhang B, Tolbert WD, He W, Dai Z, et al. Human Enteric alpha-defensin 5 Promotes Shigella infection by enhancing bacterial adhesion and invasion. Immunity. (2018) 48:1233–44e6.
92. Xu D, Liao C, Xiao J, Fang K, Zhang W, Yuan W, et al. Human enteric defensin 5 promotes Shigella infection of macrophages. Infect Immun. (2019) 88:e769–719.
93. Bravo V, Puhar A, Sansonetti P, Parsot C, Toro CS. Distinct mutations led to inactivation of Type 1 fimbriae expression in Shigella spp. PLoS One. (2015) 10:e0121785.
94. Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling. Invasion and death by type III secretion. Clin Microbiol Rev. (2008) 21:134–56.
96. DuPont HL, Levine MM, Hornick RB, Formal SB. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. (1989) 159:1126–8.
97. Schnupf P, Sansonetti PJ. Shigella pathogenesis: new insights through advanced methodologies. Microbiol Spect. (2019) 7:BAI–0023–2019.
98. Brotcke Zumsteg A, Goosmann C, Brinkmann V, Morona R, Zychlinsky A. IcsA is a Shigella flexneri adhesin regulated by the type III secretion system and required for pathogenesis. Cell Host Microbe. (2014) 15:435–45.
99. Mahmoud RY, Stones DH, Li W, Emara M, El-domany RA, Wang D, et al. The multivalent adhesion molecule SSO1327 plays a key role in Shigella sonnei pathogenesis. Mol Microbiol. (2016) 99:658–73.
100. Eilers B, Mayer-Scholl A, Walker T, Tang C, Weinrauch Y, Zychlinsky A. Neutrophil antimicrobial proteins enhance Shigella flexneri adhesion and invasion. Cell Microbiol. (2010) 12:1134–43.
101. Rajabi M, Ericksen B, Wu X, de Leeuw E, Zhao L, Pazgier M, et al. Functional determinants of human enteric α-defensin HD5: crucial role for hydrophobicity at dimer interface. J Biol Chem. (2012) 287:21615–27.
102. Chairatana P, Nolan EM. Molecular basis for self-assembly of a human host-defense peptide that entraps bacterial pathogens. J Am Chem Soc. (2014) 136:13267–76.
103. Feng Z, Dubyak GR, Lederman MM, Weinberg A. Cutting Edge: human β defensin 3—A novel antagonist of the HIV-1 coreceptor CXCR4. J Immunol. (2006) 177:782.
104. Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharmaceut Design. (2009) 15:2377–92.
106. Ghosh SK, McCormick TS, Weinberg A. Human beta defensins and cancer: contradictions and common ground. Front Oncol. (2019) 9:341. doi: 10.3389/fonc.2019.00341
107. Weinberg A, Jin G, Sieg S, McCormick TS. The yin and yang of human Beta-defensins in health and disease. Front Immunol. (2012) 3:294. doi: 10.3389/fimmu.2012.00294
108. Gambichler T, Skrygan M, Huyn J, Bechara FG, Sand M, Altmeyer P, et al. Pattern of mRNA expression of β-defensins in basal cell carcinoma. BMC Cancer. (2006) 6:163.
109. Scola N, Gambichler T, Saklaoui H, Bechara FG, Georgas D, Stücker M, et al. The expression of antimicrobial peptides is significantly altered in cutaneous squamous cell carcinoma and precursor lesions. Br J Dermatol. (2012) 167:591–7.
110. Sun CQ, Arnold R, Fernandez-Golarz C, Parrish AB, Almekinder T, He J, et al. Human β-defensin-1, a potential chromosome 8p tumor suppressor: control of transcription and induction of apoptosis in renal cell carcinoma. Cancer Res. (2006) 66:8542.
111. Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. (2001) 158:1639–51.
112. Joly S, Compton LM, Pujol C, Kurago ZB, Guthmiller JM. Loss of human β-defensin 1, 2, and 3 expression in oral squamous cell carcinoma. Oral Microbiol Immunol. (2009) 24:353–60.
113. Wenghoefer M, Pantelis A, Dommisch H, Reich R, Martini M, Allam JP, et al. Decreased gene expression of human β-defensin-1 in the development of squamous cell carcinoma of the oral cavity. Int J Oral Maxillof Surgery. (2008) 37:660–3.
114. Semlali A, Al Amri A, Azzi A, Al Shahrani O, Arafah M, Kohailan M, et al. Expression and new exon mutations of the human beta defensins and their association on colon cancer development. PLoS One. (2015) 10:e0126868.
115. Bonamy C, Sechet E, Amiot A, Alam A, Mourez M, Fraisse L, et al. Expression of the human antimicrobial peptide β-defensin-1 is repressed by the EGFR-ERK-MYC axis in colonic epithelial cells. Sci Rep. (2018) 8:18043.
116. Ling Y-M Chen, J-Y Guo, L Wang, C-Y Tan, W-T Wen, Q et al. β-defensin 1 expression in HCV infected liver/liver cancer: an important role in protecting HCV progression and liver cancer development. Sci Rep. (2017) 7:13404.
117. Donald CD, Sun CQ, Lim SD, Macoska J, Cohen C, Amin MB, et al. Cancer-specific loss of β-defensin 1 in renal and prostatic carcinomas. Labor Investigat. (2003) 83:501–5.
118. Arinura Y, Ashitani J, Yanagi S, Tokojima M, Abe K, Mukae H, et al. Elevated serum β-defensins concentrations in patients with lung cancer. Anticancer Res. (2004) 24:4051–8.
119. Bullard RS, Gibson W, Bose SK, Belgrave JK, Eaddy AC, Wright CJ, et al. Functional analysis of the host defense peptide human beta defensin-1: new insight into its potential role in cancer. Mol Immunol. (2008) 45:839–48.
120. Han Q, Wang R, Sun C, Jin X, Liu D, Zhao X, et al. Human beta-defensin-1 suppresses tumor migration and invasion and is an independent predictor for survival of oral squamous cell carcinoma patients. PLoS One. (2014) 9:e91867. doi: 10.1371/journal.pone.0091867
121. Shuyi Y, Feng W, Jing T, Hongzhang H, Haiyan W, Pingping M, et al. Human beta-defensin-3 (hBD-3) upregulated by LPS via epidermal growth factor receptor (EGFR) signaling pathways to enhance lymphatic invasion of oral squamous cell carcinoma. Oral Surgery Oral Med Oral Pathol Oral Radiol Endodontol. (2011) 112:616–25.
122. Kesting MR, Loeffelbein DJ, Hasler RJ, Wolff K-D Rittig, A Schulte, M et al. Expression profile of human beta-defensin 3 in oral squamous cell carcinoma. Cancer Investigat. (2009) 27:575–81.
123. Xu D, Zhang B, Liao C, Zhang W, Wang W, Chang Y, et al. Human beta-defensin 3 contributes to the carcinogenesis of cervical cancer via activation of NF-κB signaling. Oncotarget. (2016) 7:75902–13.
124. Fathy H, Amin MM, El-Gilany A-H. Upregulation of human β-defensin-3 and cathelicidin LL-37 in Kaposi’s sarcoma. F1000Res. (2012) 1:38.
125. DasGupta T, Nweze EI, Yue H, Wang L, Jin J, Ghosh SK, et al. Human papillomavirus oncogenic E6 protein regulates human β-defensin 3 (hBD3) expression via the tumor suppressor protein p53. Oncotarget. (2016) 7:27430–44.
126. Winter J, Pantelis A, Reich R, Martini M, Kraus D, Jepsen S, et al. Human beta-defensin-1, -2, and -3 exhibit opposite effects on oral squamous cell carcinoma cell proliferation. Cancer Investigat. (2011) 29:196–201.
127. Mburu YK, Abe K, Ferris LK, Sarkar SN, Ferris RL. Human β-defensin 3 promotes NF-κB-mediated CCR7 expression and anti-apoptotic signals in squamous cell carcinoma of the head and neck. Carcinogenesis. (2011) 32:168–74.
128. Jin G, Kawsar HI, Hirsch SA, Zeng C, Jia X, Feng Z, et al. An antimicrobial peptide regulates tumor-associated macrophage trafficking via the chemokine receptor CCR2, a model for tumorigenesis. PLoS One (2010) 5:e10993. doi: 10.1371/journal.pone.0010993
129. Röhrl J, Huber B, Koehl GE, Geissler EK, Hehlgans T. Mouse β-Defensin 14 Defb14 promotes tumor growth by inducing angiogenesis in a CCR6-dependent manner. J Immunol. (2012) 188:4931.
130. Lisovskiy ILSM, Lytvyn DI, Markeeva NV, Turchak OV, Nespryadko SV. Pattern of β-Defensin-2 (hBD-2) and EGFR mRNAs expression in cervical and vulval cancer cells. Exp Oncol. (2001) 23:248–52.
131. Markeeva NLI, Zhuravel E, Soldatkina M, Lyzogubov V, Usenko V. Involvement of human beta-defensin-2 in proliferation of transformed cells of human cervix. Exp Oncol. (2005) 27:308–13.
132. Shi N, Jin F, Zhang X, Clinton SK, Pan Z, Chen T. Overexpression of human β-defensin 2 promotes growth and invasion during esophageal carcinogenesis. Oncotarget. (2014) 5:11333–44.
133. Meyer JE, Harder J, Gorogh T, Weise JB, Schubert S, Janssen D, et al. Human beta-defensin-2 in oral cancer with opportunistic candida infection. Anticancer Res. (2004) 24:1025–30.
134. Boughan PK, Argent RH, Body-Malapel M, Park J-H Ewings, KE Bowie, AG et al. Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor: critical regulators of β-defensins during helicobacter pylori infection. J Biol Chem. (2006) 281:11637–48.
135. Kawsar HI, Ghosh SK, Hirsch SA, Koon HB, Weinberg A, Jin G. Expression of human β-defensin-2 in intratumoral vascular endothelium and in endothelial cells induced by transforming growth factor β. Peptides. (2010) 31:195–201.
136. Kamino Y, Kurashige Y, Uehara O, Sato J, Nishimura M, Yoshida K, et al. HBD-2 is downregulated in oral carcinoma cells by DNA hypermethylation and increased expression of hBD-2 by DNA demethylation and gene transfection inhibits cell proliferation and invasion. Oncol Rep. (2014) 32:462–8.
137. Baroni A, Donnarumma G, Paoletti I, Longanesi-Cattani I, Bifulco K, Tufano MA, et al. Antimicrobial human beta-defensin-2 stimulates migration proliferation and tube formation of human umbilical vein endothelial cells. Peptides. (2009) 30:267–72.
138. Conejo-Garcia JR, Benencia F, Courreges M-C Kang, E Mohamed-Hadley A, Buckanovich RJ, et al. Tumor-infiltrating dendritic cell precursors recruited by a β-defensin contribute to vasculogenesis under the influence of Vegf-A. Nat Med. (2004) 10:950–8.
139. Gao C, Yue W, Tian H, Li L, Li S, Si L. Human beta-defensin 2 promotes the proliferation of lung cancer cells through ATP-binding cassette transporter G2. Int J Clin Exp Pathol. (2016) 9:5944–9.
140. Hancock REW, Haney EF, Gill EE. The immunology of host defence peptides: beyond antimicrobial activity. Nat Rev Immunol. (2016) 16:321–34.
141. Droin N, Hendra J-B Ducoroy, P and Solary E. Human defensins as cancer biomarkers and antitumour molecules. J Proteom. (2009) 72:918–27.
142. Melle C, Ernst G, Schimmel B, Bleul A, Thieme H, Kaufmann R, et al. Discovery and identification of α-defensins as low abundant tumor-derived serum markers in colorectal cancer. Gastroenterology. (2005) 129:66–73.
143. Kemik O, Kemik AS, Sumer A, Begenik H, Purisa S, Tuzun S. Human neutrophil peptides 1, 2 and 3 (HNP 1–3): elevated serum levels in colorectal cancer and novel marker of lymphatic and hepatic metastasis. Hum Exp Toxicol. (2011) 32:167–71.
144. Albrethsen J, Møller CH, Olsen J, Raskov H, Gammeltoft S. Human neutrophil peptides 1, 2 and 3 are biochemical markers for metastatic colorectal cancer. Eur J Cancer. (2006) 42:3057–64.
145. Mothes H, Melle C, Ernst G, Kaufmann R, von Eggeling F, Settmacher U. Human Neutrophil Peptides 1-3–early markers in development of colorectal adenomas and carcinomas. Dis Markers. (2008) 25:123–9.
146. Zou H, Harrington JJ, Sugumar A, Klatt KK, Smyrk TC, Ahlquist DA. Detection of colorectal disease by stool defensin assay: an exploratory study. Clin Gastroenterol Hepatol. (2007) 5:865–8.
147. Albrethsen J, Bøgebo R, Gammeltoft S, Olsen J, Winther B, Raskov H. Upregulated expression of human neutrophil peptides 1, 2 and 3 (HNP 1-3) in colon cancer serum and tumours: a biomarker study. BMC Cancer. (2005) 5:8. doi: 10.1186/1471-2407-5-8
148. Bateman A, Singh A, Jothy S, Fraser R, Esch F, Solomon S. The levels and biologic action of the human neutrophil granule peptide HP-1 in lung tumors. Peptides. (1992) 13:133–9.
149. Li J, Zhao J, Yu X, Lange J, Kuerer H, Krishnamurthy S, et al. Identification of biomarkers for breast cancer in nipple aspiration and ductal lavage fluid. Clin Cancer Res. (2005) 11:8312.
150. Müller CA, Markovic-Lipkovski J, Klatt T, Gamper J, Schwarz G, Beck H, et al. Human α-defensins HNPs-1, -2, and -3 in renal cell carcinoma: influences on tumor cell proliferation. Am J Pathol. (2002) 160:1311–24.
151. Lundy FT, Orr DF, Gallagher JR, Maxwell P, Shaw C, Napier SS, et al. Identification and overexpression of human neutrophil α-defensins (human neutrophil peptides 1, 2 and 3) in squamous cell carcinomas of the human tongue. Oral Oncol. (2004) 40:139–44.
152. Holterman DA, Diaz JI, Blackmore PF, Davis JW, Schellhammer PF, Corica A, et al. Overexpression of α-defensin is associated with bladder cancer invasiveness. Urol Oncol Sem Orig Investigat. (2006) 24:97–108.
153. Gunes M, Gecit I, Pirincci N, Kemik AS, Purisa S, Ceylan K, et al. Plasma human neutrophil proteins-1, -2, and -3 levels in patients with bladder cancer. J Cancer Res Clin Oncol. (2013) 139:195–9.
154. Roesch-Ely M, Nees M, Karsai S, Ruess A, Bogumil R, Warnken U, et al. Proteomic analysis reveals successive aberrations in protein expression from healthy mucosa to invasive head and neck cancer. Oncogene. (2007) 26:54–64.
155. Escher N, Spies-Weißhart B, Kaatz M, Melle C, Bleul A, Driesch D, et al. Identification of HNP3 as a tumour marker in CD4+ and CD4- lymphocytes of patients with cutaneous T-cell lymphoma. Eur J Cancer. (2006) 42:249–55.
156. Nishimura M, Abiko Y, Kurashige Y, Takeshima M, Yamazaki M, Kusano K, et al. Effect of defensin peptides on eukaryotic cells: primary epithelial cells fibroblasts and squamous cell carcinoma cell lines. J Dermatol Sci. (2004) 36:87–95.
157. Lichtenstein AT, Ganz T, Selsted M, Lehrer R. In vitro tumor cell cytolysis mediated by peptide defensin of human and rabbit granulocytes. Blood. (1986) 68:1407–10.
158. Gaspar D, Freire JM, Pacheco TR, Barata JT, Castanho MARB. Apoptotic human neutrophil peptide-1 anti-tumor activity revealed by cellular biomechanics. Biochim Biophys Acta (BBA) Mol Cell Res. (2015) 1853:308–16.
159. Xu N, Wang Y-S Pan, W-B Xiao, B Wen, Y-J Chen, X-C et al. Human α-defensin-1 inhibits growth of human lung adenocarcinoma xenograft in nude mice. Mol Cancer Ther. (2008) 7:1588.
160. Wang Y-S Li, D Shi, H-S Wen, Y-J Yang, L Xu, N et al. Intratumoral expression of mature human neutrophil peptide-1 mediates antitumor immunity in mice. Clin Cancer Res. (2009) 15:6901.
161. Economopoulou M, Bdeir K, Cines DB, Fogt F, Bdeir Y, Lubkowski J, et al. Inhibition of pathologic retinal neovascularization by alpha-defensins. Blood. (2005) 106:3831–8.
162. Chavakis T, Cines DB, Rhee J-S Liang, OD Schubert, UWE Hammes, H-P et al. Regulation of neovascularization by human neutrophil peptides (α-defensins): a link between inflammation and angiogenesis. FASEB J. (2004) 18:1306–8.
163. Hubert P, Herman L, Maillard C, Caberg J-H Nikkels, A Pierard, G et al. Defensins induce the recruitment of dendritic cells in cervical human papillomavirus-associated (pre)neoplastic lesions formed in vitro and transplanted in vivo. FASEB J. (2007) 21:2765–75.
164. Wang W, Owen SM, Rudolph DL, Cole AM, Hong T, Waring AJ, et al. Activity of α- and θ-defensins against primary isolates of HIV-1. J Immunol. (2004) 173:515–20.
165. Furci L, Sironi F, Tolazzi M, Vassena L, Lusso P. α-defensins block the early steps of HIV-1 infection: interference with the binding of gp120 to CD4. Blood. (2006) 109:2928–36.
166. Seidel A, Ye Y, de Armas LR, Soto M, Yarosh W, Marcsisin RA, et al. Cyclic and acyclic defensins inhibit human immunodeficiency virus type-1 replication by different mechanisms. PLoS One. (2010) 5:e9737.
Keywords: antimicrobial peptide, host defense peptide, defensin, innate immunity, Shigella, host–pathogen interaction
Citation: Xu D and Lu W (2020) Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 11:764. doi: 10.3389/fimmu.2020.00764
Received: 27 February 2020; Accepted: 06 April 2020;
Published: 07 May 2020.
Edited by:
Mark Hulett, La Trobe University AustraliaReviewed by:
Francois Niyonsaba, Juntendo University JapanVignesh Ramachandran, University of Kuala Lumpur Malaysia
Copyright © 2020 Xu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use distribution or reproduction in other forums is permitted provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited in accordance with accepted academic practice. No use distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Xu, ZGFuLnh1QHhqdHUuZWR1LmNu; Wuyuan Lu, d2x1QGlodi51bWFyeWxhbmQuZWR1
 Dan Xu
Dan Xu Wuyuan Lu
Wuyuan Lu