- Experimental Immunology, Department of Biomedicine, University of Basel and University Hospital Basel, Basel, Switzerland
Non-polymorphic MHC class I-related molecule MR1 presents antigenic bacterial metabolites to mucosal-associated invariant T (MAIT) cells and self-antigens to MR1-restricted T (MR1T) cells. Both MR1-restricted T cell populations are readily identified in healthy individuals, with MAIT cells accounting for 1–10% of circulating T cells, while MR1T cells have frequencies comparable to peptide-specific T cells (<0.1%). Self-reactive MR1T cells display a heterogeneous phenotype, and are capable of releasing both TH1 and TH2 cytokines, supporting not only activation of inflammation but also contributing to its regulation. Importantly, MR1T cells recognize and kill a diverse range of MR1-expressing tumor cells. On the other hand, evidence suggests MAIT cells augment cancer growth and metastases. This review addresses the potential role of MR1-restricted T cells in controlling tumor cells, facilitating their elimination and regulating cancer immunity. We also discuss therapeutic opportunities surrounding MR1-restricted T cells in cancer.
Introduction
T cells restricted for antigen presenting molecules with limited or absent polymorphism, like cluster differentiation 1 (CD1) family of proteins or MHC class I-related molecule 1 (MR1), respectively, comprise adaptive-like T cells participating in immune homeostasis and in diseases (1). They express T cell receptors (TCRs) that sense non-peptide antigens and constitute discrete populations expressing either TCR αβ or γδ heterodimers. These TCRs can either be diverse or have strong expression bias being semi-invariant (1–3), such as in the case of mucosal-associated invariant T (MAIT) cells (4, 5). Since their discovery, major efforts have been made in exploring the role of MAIT cells in disease settings such as infection, autoimmunity and cancer. Due to the lack of human studies, understanding of MR1-associated antigens and mechanisms behind MR1 presentation, progress has been slow and sometimes inconclusive. More recently, a diverse, self-reactive, TCR αβ T cell population was identified which was MR1-restricted and exhibited anti-tumor responses (6). Seemingly present in all healthy donors tested, they represent an excellent candidate for further study in cancer. This review will discuss current knowledge regarding the roles of MR1-restricted T cells in cancer and their possible introduction in cell therapy.
MR1 is highly conserved among most mammals, in particular the α1 and α2 domains have 90 and 89% homology between mice and humans, respectively (7, 8). Furthermore, the MR1 sequence is the same between different individuals, and, as such, is termed non-polymorphic (9). The MR1 gene is encoded in chromosome 6 (8) and is ubiquitously transcribed in all human cells. The structural features of MR1 are very similar to that of MHC class I. A cleft between the α1 and α2 domains containing α-helices form a solvent exposed antigen-binding pocket atop a β-pleated sheet in a manner dissimilar to that of the narrow channels formed in the CD1 family. However, instead of a lining of polar residues in the case of MHC class I or indeed hydrophobic residues like CD1, the MR1 pocket is lined with largely aromatic residues to confer an environment that is both hydrophobic and charged (10). These features endow MR1 with the space and capacity to bind diverse small cyclic and bicyclic molecules (11). Moreover, the MR1 pocket seems not to accommodate peptide chains, nor does it exhibit open ends like MHC class II (10–12).
The first MR1 ligands to be characterized were vitamin B metabolites, in particular a metabolite of folic acid (vitamin B9) called 6-formylpterin (6-FP) and precursors of riboflavin (vitamin B2) as 6,7-dimethyl-8-D-ribityllumazine (RL-6,7-diMe) and 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) (10, 12). Later studies identified riboflavin itself and riboflavin adducts 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO), photolumazine I (PLI) and photolumazine III (PLIII) (13). In silico modeling allowed the discovery of further MR1-presented small molecules including: 3-formylsalicylic acid and diclofenac metabolites that were responsible for MAIT inhibition and weak activation of rare MAIT TCR, respectively (11). Furthermore, other studies implied bacterial antigens other than riboflavin metabolites (14) as well as tumor-associated antigens (1, 15). Therefore, the pocket of MR1 is highly plastic and might allow binding of other unknown antigens. Interestingly, all known antigens bind the A'-pocket leaving the F' unfilled. As the F' pocket is shared among MR1 molecules from different species, its evolutionary conservation suggests an important role. Although it could be possible that the F' pocket plays an important role in MR1 refolding and proper trafficking within the cell, like MHC class I molecules binding to tapasin and tapasin-related molecules, or MHC class II molecules binding to the invariant chain, there is the possibility that it can accommodate undiscovered ligands that are bigger than the small antigenic metabolites identified so far.
MAIT cells classically express a Vα7.2-Jα33 (TRAV1-2-TRAJ33) TCR, paired to a limited number of β chains for example Vβ2 (TRBV20) or Vβ13 (TRBV6) (Figure 1) (4, 5, 16, 17). Alternative TRAJ genes are also used when maintaining a CDR3α loop conserved in length and with a Tyrosine in position 95, crucial for 5-OP-RU recognition (18). Furthermore, atypical TRAV1-2− MAIT cells have been described, that are stained with a 5-OP-RU-loaded MR1 tetramer and react to bacteria-infected cells (14, 19). In contrast to MAIT cells, MR1T cells are a novel population of self-reactive MR1-restricted T cells that are characterized by diverse TCR usage and are not stimulated by bacterial ligands (6, 20). MAIT cells have a very high frequency (1–10%) in the blood of healthy individuals (21, 22) compared to MR1T cells that are less abundant and found at a frequency of ~1:2500 of circulating T cells (6). Regarding localization, MAIT cells are enriched within barrier tissues and in particular in mucosa, gut lamina propria, liver (16, 17, 23, 24), lungs and skin (25, 26) and less frequently in lymph nodes (23). Less is known about MR1T cells except that they were found in the blood of each healthy individual studied and MR1T cell clones were activated by cancer cell lines in an MR1-dependant manner (6, 20).
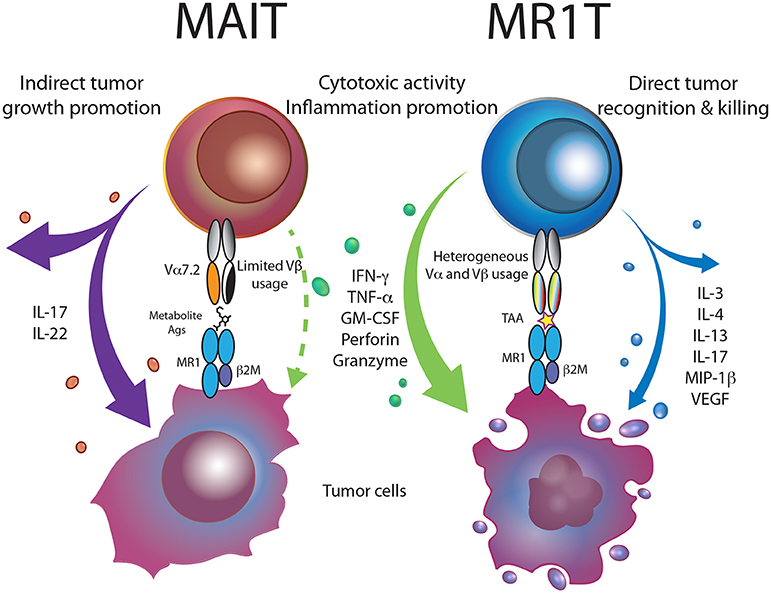
Figure 1. MR1-restricted T cells in cancer. Bacterial metabolite-reactive MAIT cells, within the tumor microenvironment, are skewed toward the production of Th17 cytokines, promoting tumor growth and metastasis. MR1T cells recognizing MR1-presented tumor-associated antigens (TAA), release a vast array of cytokines and kill tumor cells, thus supporting cancer immunity.
Development of MAIT cells is thought to occur after recognition of commensal bacteria-derived antigens presented by double-positive (DP) thymocytes (23, 26–28). A three-stage transcriptional program drives MAIT cells to acquire an innate-like phenotype, characterized by high expression of CD161 and transcription factors PLZF, T-bet and RORγT (21, 27, 29–31). Up to five different subsets of MAIT cells can be distinguished in humans based on the expression of TCR co-receptors. The most abundant subset in human blood consists of CD4−CD8αβ+ or CD8αα+ cells (approximately 80% of MAIT cells); double-negative (DN) CD4−CD8− represent about 15% of total MAIT cells, few CD4+CD8− and CD4+CD8+ are present (12, 30). So far, the analysis of a large number (>100) of MR1T cell clones showed that they were either CD8+ or DN (our unpublished studies) and only few of them expressed CD161 (6), suggesting that these cells are heterogeneous. MR1T cell functional heterogeneity is even more pronounced, with different clones displaying distinct TH1, TH2, or TH17 cytokine and transcriptional profiles upon stimulation (Figure 1) (6). MAIT cells do not express the lymph node-homing receptors CCR7 and CD62L, and only slight differences were observed in their expression of chemokine receptors and integrins, that dictate their likelihood for tissue residency (23, 30, 32). MR1T cells also show tissue-homing capacity, but lower expression of the chemokine receptors CCR4 and CCR6, compared to representative MAIT clones (6), suggesting comparatively different localization patterns.
MAIT cell activation occurs after TCR engagement with MR1-presented antigens on infected cells (33), as well as in a TCR-independent manner after stimulation by inflammatory cytokines such as IL-12 and IL-18 or type I interferons (34–37). Immediately after antigen recognition and activation, MAIT cells have the capacity to release granzyme B and perforin to promptly kill infected cells (17, 23, 24). In addition, TH1 and TH17 cytokines are secreted such as IFNγ, TNFα, IL-2, IL-17 that function against invading pathogens (26, 36, 38, 39). Sensitivity to cytokines underlies MAIT cells' ability to tailor their response to different scenarios despite bearing a semi-invariant TCR (40). Uncoupled TCR and cytokine stimulation induce distinct effector functions on MAIT cells, that become synergistic when occurring at the same time (36, 37). TCR engagement in the absence of cytokines induces homeostatic, tissue repair-oriented functions that instead, in presence of cytokine co-stimulation, become pro-inflammatory exerting an anti-microbial activity (36, 39). Cytokine modulation of MAIT cell function impacts the role of these cells in different pathological conditions, as discussed below.
Mait Cells in Pathology
Several studies have addressed the relevance of MAIT cells in infectious disease, autoimmunity and cancer, both in humans and in mouse models. The frequency of MAIT cells in the blood is often reduced in patients suffering of bacterial or viral infections, autoimmune or metabolic diseases and several cancers, compared to that of healthy donors or before disease onset (41–47). A reduction in circulating MAIT cell frequency reflects their migration toward peripheral tissues, where MAIT cells are increased or redistributed in the proximity of inflamed areas. This is largely driven by the expression of several tissue-homing chemokine receptors and integrins (and absence of lymph node-homing receptors) whose expression is increased by inflammatory cytokines (34–36). Cytokine-dependent, TCR-independent activation of MAIT cells becomes prominent in the case of viral infections and autoimmune-diseases, where bacterial antigens are absent at the pathologic site. In addition, MAIT cells are necessary to maintain homeostatic conditions, mucosal barrier integrity and play beneficial roles in the context of acute bacterial infections (33, 48, 49). On the other hand, MAIT cells were found to be deleterious in chronic bacterial infections (50, 51) and a variety of autoimmune diseases where their effector functions are altered (43, 44, 49, 52–56). Therefore, finding unifying rules regarding the role of MAIT cells in disease is difficult, and is likely dependent on the local cytokine milieu, other immune cell populations and microbial species present (46, 47).
MAIT Cells in Cancer
The possibility of directing the host immune system against tumor cells is the rationale behind immunotherapy which has largely focused on local T cell activation by checkpoint blockade inhibitors (57, 58) or redirecting the specificity of immune cells against tumors (59). Many resources have been used in attempt to unleash the antitumor potential of T cells, whose infiltration inside the tumor positively correlates to a good prognosis (60, 61). Given the possibility to overcome the limitations of extensive HLA polymorphism by exploiting monomorphic MR1 molecules, priority should be given to understand the roles of MR1-restricted T cells in cancer.
While the presence of tumor-reactive MR1T cells in healthy or pathologic tissues has not been assessed yet, MAIT cell frequencies are altered in tumors (61). As observed for other diseases, MAIT cells seem to be subjected to modifications in numbers and phenotype in cancer. For example, in colorectal cancer, different studies show either similar, or decreased frequencies of peripheral blood MAIT cells in patients compared to healthy individuals (45, 62, 63). Nevertheless, MAIT cells were found to be increased in primary colorectal lesions and hepatic metastatic lesions, compared to unaffected tissues (45, 62, 64, 65). The phenotype of MAIT cells in the tumor niche is also altered, since they produce less IFN-γ (45, 62) (Figure 1), where MAIT cell infiltration correlated with a poor survival (64). As for liver-related diseases, MAIT cells accumulate locally (66–69), while their frequency decreased in the blood of hepatocellular carcinoma (HCC) patients, compared to healthy donors (68, 69). Indeed, within HCC tumor lesions, MAIT cell frequency was reduced compared to peritumoral hepatic area and the HCC microenvironment polarized tumor-infiltrating MAIT cells to an exhausted phenotype (PD-1high CTLA-4+ TIM-3+), leading to a reduced response to bacterial antigens and the production of tumor-promoting cytokines (69). Therefore, evidence suggests MAIT cells have a tumor-supporting function and their presence is associated with an unfavorable outcome (69).
A similar observation was made in multiple myeloma (MM), where MAIT cells in newly diagnosed MM patients have reduced effector functions and CD27 expression, indicative of a functionally impaired phenotype (70). MAIT cells were able to expand in response to gastrointestinal microbiota and promote gut integrity only after bone marrow transplantation (71, 72). Reconstitution of the MAIT cell population resulted in suppression of conventional T cell proliferation and prevention of graft-vs.-host disease, indicating a possible regulatory role for MAIT cells (71–73). This immune-suppressive regulatory role of MAIT cells was also recently described in mouse cancer models where MAIT cells promoted progression and metastases of melanomas and methylcholanthrene-induced fibrosarcoma (74). In these models, MAIT cells activated in vivo by MR1-expressing tumor cells suppressed NK cells effector functions, chiefly through IL-17A secretion (74). MAIT cell activation was blocked by treatment of tumor cells with 6-FP or administration of MR1 blocking antibodies, suggesting TCR-mediated activation of MAIT cells. Considering that in the described models tumor cells presented MAIT antigens (74), MAIT cells could also induce tissue-repair and perhaps tumor-promoting functions if TCR engagement was solely responsible for activation (36, 39). In sum, MAIT cells often accumulate within tumors and under the influence of the tumor microenvironment, acquire an exhausted phenotype and skewed effector functions to suppress immune anti-tumor functions (Figure 1).
MR1T Cells in Cancer
Self-reactive MR1-restricted MR1T cells were isolated and identified by their ability to recognize tumor cells in a TCR-mediated manner. Given their heterogeneous functional profile, they could have a variety of functions and modulate the innate and adaptive responses to cancer. MR1T cells were originally described as capable of recognizing several tumor cell lines from different tissues expressing physiological levels of MR1 (6), thus revealing an unexpected broad crossreactivity toward tumor cells. A recent study confirmed this finding and by investigating one TCR showed the capacity of this T cell clone to kill several tumor cell lines (20). Such broad crossreactivity provides the bases for potential use of MR1T cells in immunotherapy of different tumors as recently discussed (75).
In contrast to MAIT cells, MR1T cells do not recognize bacteria-infected cells, and are not activated by healthy cells (6, 20). The self-antigens responsible for activation are as yet unknown, but they were shown to be displaced from MR1 by vitamin B derivatives (5-OP-RU and acetyl-6-formylpterin), indicating that they exploit the same binding pocket as of other MR1 ligands (6, 20). So far, evidence indicates that these hydrophilic compounds can be found in either freshly explanted mouse breast tumor or from lysed THP-1 cells (6). Evidence strongly suggests the presence of multiple self-antigens and antigen-specificity of MR1T cells, since two T cell clones bearing different TCRs distinguished between two different lysate fractions (6). Furthermore, a recently described MR1T clone is selectively activated by wild-type MR1 and not by the K43A mutant (20), possibly indicating that some of the antigens recognized by MR1T cells could covalently bind MR1 on lysine 43 as already described for 5-OP-RU (12). Alternatively, the lysine 43 mutant does not assume the conformation required for the stimulation of the single clone described by Crowther et al. In conclusion, unlike MAIT cells, MR1T cells are activated by MR1-bound self-antigens expressed by tumor cell lines grown in vitro or in vivo.
Given the recognition of multiple cancer cells lines by individual MR1T cell clones, it is feasible that cancers share common antigens presented by MR1. The frequency and the phenotype of MR1T cells in cancer patients are not yet known, however, given their ability to recognize in vivo expanded tumor cells, their role might be beneficial to patients. Considering the aforementioned features and the possibility to confer tumor reactivity to other T cells by TCR transfer (6, 20), MR1T cells represent potential candidates for broadly reactive T cell therapy in cancer (75). The functional dichotomy of MAIT and MR1T cells and their potential role in cancer immunity are outlined in Figure 1.
MR1-Restricted T-Cell Therapies: Current Opportunities and Future Perspectives
Three attributes are required for off-the-shelf anti-tumor T-cell therapies; (i) specificity toward antigens expressed by tumor and not normal cells; (ii) pan-recipient compatibility to avoid host-vs.-graft responses in infused patients, and (iii) availability of TCRs that recognize different tumor types.
Biochemical studies are required for the identification of antigens presented by MR1 and generated in tumors but not in normal cells. Understanding their nature will reveal the molecular basis of their apparent accumulation in tumor cells. In addition, these antigens will be ideal tools for the generation of MR1 tetramers to further study MR1T cells. Furthermore, TCR structural motifs that confer MR1-restriction will also be of relevance. Investigating large numbers of MR1-restricted TCRs will perhaps provide this type of information and will be key for engineering of pan-cancer MR1T cells for off-the-shelf therapy.
An intrinsic and optimal feature of TCR restriction to non-polymorphic antigen-presenting molecules is what can be defined as pan-recipient compatibility. Indeed, these TCRs are devoid of graft-vs.-host activity, which instead prevents the therapeutic use of HLA-restricted TCRs in the entire population. This feature opens the possibility of transferring the same MR1T TCRs to every patient, overcoming the need of isolating HLA-matched tumor-specific TCR for every recipient. Moreover, the non-polymorphic nature of MR1 allows the generation of engineered HLA-deficient MR1T cells for an off-the-shelf therapy, immediately available for on-demand infusion, also in patients with compromised T cell compartments. There is also added value of using such TCRs that are seemingly not influenced by the expression of CD4 or CD8 co-stimulatory molecules, thus redirecting responses in both CD4+, CD8+ and DN T cells required for efficient cytotoxic T cell priming and cancer immunity (76–78).
Off-the-shelf use may also require a combination of TCRs that recognize the antigens preferentially expressed in each individual tumor. Therefore, a novel classification of tumors according to their accumulation of MR1-presented antigens and their screening in each patient will represent a method to select the MR1T TCRs with relevant specificity in a personalized immunotherapy.
Other important issues are the functions of MAIT cells and of MR1T cells with respect to their direct role in tumor immunity. In mice, the MAIT17 subset is more prominent than MAIT1 and although MAIT cells in humans have a mixed TH1-TH17 phenotype (79–81), in the case of cancer and other pathologies it can be skewed toward the production of the IL-17, a multi-functional cytokine that can support tumor growth (82–84). According to this evidence, potential therapies to block MAIT cell activity in cancer have been proposed. These proposals converge on the possible blockade of MR1 presentation rather than targeting MAIT cells, given their important role in the homeostasis of mucosal tissues against invading pathogens. Infusions of MR1-inhibitory ligands such as acetyl-6-FP or MR1-blocking antibodies could indeed block MAIT cell activation, but it is not known whether this could increase gut permeability as observed in the case of MR1-deficient NOD mice (49). There are two additional possible outcomes that detract from this strategy. The first, is that blocking MR1 would affect its antigen presentation to all MR1-restricted T cells, including tumor-reactive MR1T cells that likely have beneficial activity, stimulating rather than suppressing cancer immunity and directly killing tumor cells. Secondly, repeated infusions of MR1 ligands which do not stimulate MAIT cells, might lead to priming and expansion of other MR1-restricted T cells specific for the infused molecules, with potentially deleterious side effects.
A more appealing use of MR1 ligands might exploit drugs to increase the generation or the accumulation of MR1T antigens in tumor cells. This mechanism of action in immunotherapy has already been proposed with aminobisphosphonate drugs to increase tumor recognition by Vγ9Vδ2 T cells (85). Thus, characterization of the tumor-associated antigens recognized by MR1T cells will be important to decipher the regulation of metabolic pathways for pharmacological exploitation.
Discovery of these antigens could also stimulate the development of vaccines to expand and generate memory populations of tumor-reactive MR1T cells, a strategy discussed for other tumor-associated antigens (86). Broad tumor crossreactivity of MR1T TCRs (6, 20) will add value to this strategy and will represent an innovative type of tumor vaccination.
Concluding Remarks
Antigen-presentation by MR1 functions to report the metabolic programming of microbes (87) and undoubtedly the host, to T cells. Our knowledge of MAIT cells has developed over the last decade but less is known about the more recently discovered MR1T cells. Further work to identify MR1T-stimulatory molecules and a broader understanding of MR1T cell diversity is needed to fully appreciate the relationship of antigen recognition and their physiological functions. So far, evidence indicates a pro-tumoral activity of MAIT cells, whereas the published literature on MR1T cells indicates they have an anti-tumoral activity. Thanks to these features, MR1T cells represent an interesting population to exploit in cancer immunotherapy.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The University of Basel has filed patents on MR1T cells.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to present and past collaborators of the Experimental Immunology laboratory for their contribution to the topics of this review and to Paula Cullen for reading the manuscript. The authors thank the support of Swiss National Foundation (310030-173240), the Swiss Cancer League (KFS-4707-02-2019), Cancer League beider Basel (KLbB-4779-02-2019), University of Basel and D-BSSE ETH Zürich (PMB-02-17) to GD.
References
1. Lepore M, Mori L, De Libero G. The conventional nature of non-MHC-restricted T cells. Front Immunol. (2018) 9:1365. doi: 10.3389/fimmu.2018.01365
2. Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. (2014) 32:323–66. doi: 10.1146/annurev-immunol-032713-120243
3. Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. (2015) 16:1114–23. doi: 10.1038/ni.3298
4. Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8-alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. (1993) 178:1–16. doi: 10.1084/jem.178.1.1
5. Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. (1999) 189:1907–21. doi: 10.1084/jem.189.12.1907
6. Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife. (2017) 6:e24476. doi: 10.7554/eLife.24476
7. Yamaguchi H, Hirai M, Kurosawa Y, Hashimoto K. A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem Biophys Res Commun. (1997) 238:697–702. doi: 10.1006/bbrc.1997.7379
8. Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol. (1998) 161:4066–77.
9. Parra-Cuadrado JF, Navarro P, Mirones I, Setien F, Oteo M, Martinez-Naves E. A study on the polymorphism of human MHC class I-related MR1 gene and identification of an MR1-like pseudogene. Tissue Antigens. (2000) 56:170–2. doi: 10.1034/j.1399-0039.2000.560211.x
10. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
11. Keller AN, Eckle SB, Xu W, Liu L, Hughes VA, Mak JY, et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat Immunol. (2017) 18:402–11. doi: 10.1038/ni.3679
12. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
13. Harriff MJ, McMurtrey C, Froyd CA, Jin H, Cansler M, Null M, et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci Immunol. (2018) 3:eaao2556. doi: 10.1126/sciimmunol.aao2556
14. Meermeier EW, Laugel BF, Sewell AK, Corbett AJ, Rossjohn J, McCluskey J, et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat Commun. (2016) 7:12506. doi: 10.1038/ncomms12506
15. McWilliam HE, Eckle SB, Theodossis A, Liu L, Chen Z, Wubben JM, et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat Immunol. (2016) 17:531–7. doi: 10.1038/ni.3416
16. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. (2003) 422:164–9. doi: 10.1038/nature01433
17. Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat Commun. (2014) 5:3866. doi: 10.1038/ncomms4866
18. Koay HF, Gherardin NA, Xu C, Seneviratna R, Zhao Z, Chen Z, et al. Diverse MR1-restricted T cells in mice and humans. Nat Commun. (2019) 10:2243. doi: 10.1038/s41467-019-10198-w
19. Gherardin NA, Keller AN, Woolley RE, Le Nours J, Ritchie DS, Neeson PJ, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. (2016) 44:32–45. doi: 10.1016/j.immuni.2015.12.005
20. Crowther MD, Dolton G, Legut M, Caillaud ME, Lloyd A, Attaf M, et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat Immunol. (2020) 21:178–85. doi: 10.1038/s41590-019-0578-8
21. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. (2009) 7:e54. doi: 10.1371/journal.pbio.1000054
22. Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. (2013) 210:2305–20. doi: 10.1084/jem.20130958
23. Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. doi: 10.1182/blood-2010-08-303339
24. Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, Premel V, et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. (2013) 9:e1003681. doi: 10.1371/journal.ppat.1003681
25. Li J, Reantragoon R, Kostenko L, Corbett AJ, Varigos G, Carbone FR. The frequency of mucosal-associated invariant T cells is selectively increased in dermatitis herpetiformis. Australas J Dermatol. (2017) 58:200–4. doi: 10.1111/ajd.12456
26. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366:6624. doi: 10.1126/science.aax6624
27. Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. (2014) 5:3143. doi: 10.1038/ncomms4143
28. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. (2019) 366:494–9. doi: 10.1126/science.aaw2719
29. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17:1300–11. doi: 10.1038/ni.3565
30. Gherardin NA, Souter MN, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. (2018) 96:507–25. doi: 10.1111/imcb.12021
31. Koay HF, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci Immunol. (2019) 4:eaay6039. doi: 10.1126/sciimmunol.aay6039
32. Sobkowiak MJ, Davanian H, Heymann R, Gibbs A, Emgard J, Dias J, et al. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur J Immunol. (2019) 49:133–43. doi: 10.1002/eji.201847759
33. Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11:701–8. doi: 10.1038/ni.1890
34. Jo J, Tan AT, Ussher JE, Sandalova E, Tang XZ, Tan-Garcia A, et al. Toll-like receptor 8 agonist and bacteria trigger potent activation of innate immune cells in human liver. PLoS Pathog. (2014) 10:e1004210. doi: 10.1371/journal.ppat.1004210
35. Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. (2014) 44:195–203. doi: 10.1002/eji.201343509
36. Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. TCR- or cytokine-activated CD8(+) mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. (2019) 28:3061–76. e3065. doi: 10.1016/j.celrep.2019.08.054
37. Lamichhane R, Galvin H, Hannaway RF, de la Harpe SM, Munro F, Tyndall JD, et al. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur J Immunol. (2020) 50:178–91. doi: 10.1002/eji.201948279
38. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. (2019) 28:3249–62. e3245. doi: 10.1016/j.celrep.2019.07.039
39. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. (2019) 28:3077–91. e3075. doi: 10.1016/j.celrep.2019.08.050
40. Slichter CK, McDavid A, Miller HW, Finak G, Seymour BJ, McNevin JP, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. (2016) 1:e86292. doi: 10.1172/jci.insight.86292
41. Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. (2013) 121:1124–35. doi: 10.1182/blood-2012-07-445429
42. Cho YN, Kee SJ, Kim TJ, Jin HM, Kim MJ, Jung HJ, et al. Mucosal-associated invariant T cell deficiency in systemic lupus erythematosus. J Immunol. (2014) 193:3891–901. doi: 10.4049/jimmunol.1302701
43. Carolan E, Tobin LM, Mangan BA, Corrigan M, Gaoatswe G, Byrne G, et al. Altered distribution and increased IL-17 production by mucosal-associated invariant T cells in adult and childhood obesity. J Immunol. (2015) 194:5775–80. doi: 10.4049/jimmunol.1402945
44. Hiejima E, Kawai T, Nakase H, Tsuruyama T, Morimoto T, Yasumi T, et al. Reduced numbers and proapoptotic features of mucosal-associated invariant T cells as a characteristic finding in patients with inflammatory bowel disease. Inflamm Bowel Dis. (2015) 21:1529–40. doi: 10.1097/MIB.0000000000000397
45. Ling L, Lin Y, Zheng W, Hong S, Tang X, Zhao P, et al. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Sci Rep. (2016) 6:20358. doi: 10.1038/srep20358
46. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. (2019) 20:1110–28. doi: 10.1038/s41590-019-0444-8
47. Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. (2019) 19:643–57. doi: 10.1038/s41577-019-0191-y
48. Meierovics AI, Cowley SC. MAIT cells promote inflammatory monocyte differentiation into dendritic cells during pulmonary intracellular infection. J Exp Med. (2016) 213:2793–809. doi: 10.1084/jem.20160637
49. Rouxel O, Da Silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. (2017) 18:1321–31. doi: 10.1038/ni.3854
50. Booth JS, Salerno-Goncalves R, Blanchard TG, Patil SA, Kader HA, Safta AM, et al. Mucosal-associated invariant T cells in the human gastric mucosa and blood: role in helicobacter pylori infection. Front Immunol. (2015) 6:466. doi: 10.3389/fimmu.2015.00466
51. D'Souza C, Pediongco T, Wang H, Scheerlinck JY, Kostenko L, Esterbauer R, et al. Mucosal-associated invariant T cells augment immunopathology and gastritis in chronic helicobacter pylori infection. J Immunol. (2018) 200:1901–16. doi: 10.4049/jimmunol.1701512
52. Serriari NE, Eoche M, Lamotte L, Lion J, Fumery M, Marcelo P, et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin Exp Immunol. (2014) 176:266–74. doi: 10.1111/cei.12277
53. Teunissen MBM, Yeremenko NG, Baeten DLP, Chielie S, Spuls PI, de Rie MA, et al. The IL-17A-producing CD8+ T-cell population in psoriatic lesional skin comprises mucosa-associated invariant T cells and conventional T cells. J Invest Dermatol. (2014) 134:2898–907. doi: 10.1038/jid.2014.261
54. Magalhaes I, Pingris K, Poitou C, Bessoles S, Venteclef N, Kiaf B, et al. Mucosal-associated invariant T cell alterations in obese and type 2 diabetic patients. J Clin Invest. (2015) 125:1752–62. doi: 10.1172/JCI78941
55. Toussirot E, Laheurte C, Gaugler B, Gabriel D, Saas P. Increased IL-22- and IL-17A-producing mucosal-associated invariant T cells in the peripheral blood of patients with ankylosing spondylitis. Front Immunol. (2018) 9:1610. doi: 10.3389/fimmu.2018.01610
56. Cassius C, Branchtein M, Battistella M, Amode R, Lepelletier C, Jachiet M, et al. Persistent deficiency of mucosal-associated invariant T cells during dermatomyositis. Rheumatology. (2019) kez564. doi: 10.1093/rheumatology/kez564
57. Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol. (2007) 19:813–24. doi: 10.1093/intimm/dxm057
58. Demaria O, Cornen S, Daeron M, Morel Y, Medzhitov R, Vivier E. Harnessing innate immunity in cancer therapy. Nature. (2019) 574:45–56. doi: 10.1038/s41586-019-1593-5
59. June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. (2018) 359:1361–5. doi: 10.1126/science.aar6711
60. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. (2013) 39:1–10. doi: 10.1016/j.immuni.2013.07.012
61. Godfrey DI, Le Nours J, Andrews DM, Uldrich AP, Rossjohn J. Unconventional T cell targets for cancer immunotherapy. Immunity. (2018) 48:453–73. doi: 10.1016/j.immuni.2018.03.009
62. Sundstrom P, Ahlmanner F, Akeus P, Sundquist M, Alsen S, Yrlid U, et al. Human mucosa-associated invariant T cells accumulate in colon adenocarcinomas but produce reduced amounts of IFN-gamma. J Immunol. (2015) 195:3472–81. doi: 10.4049/jimmunol.1500258
63. Won EJ, Ju JK, Cho YN, Jin HM, Park KJ, Kim TJ, et al. Clinical relevance of circulating mucosal-associated invariant T cell levels and their anti-cancer activity in patients with mucosal-associated cancer. Oncotarget. (2016) 7:76274–90. doi: 10.18632/oncotarget.11187
64. Zabijak L, Attencourt C, Guignant C, Chatelain D, Marcelo P, Marolleau JP, et al. Increased tumor infiltration by mucosal-associated invariant T cells correlates with poor survival in colorectal cancer patients. Cancer Immunol Immunother. (2015) 64:1601–8. doi: 10.1007/s00262-015-1764-7
65. Shaler CR, Tun-Abraham ME, Skaro AI, Khazaie K, Corbett AJ, Mele T, et al. Mucosa-associated invariant T cells infiltrate hepatic metastases in patients with colorectal carcinoma but are rendered dysfunctional within and adjacent to tumor microenvironment. Cancer Immunol Immunother. (2017) 66:1563–75. doi: 10.1007/s00262-017-2050-7
66. Hengst J, Strunz B, Deterding K, Ljunggren HG, Leeansyah E, Manns MP, et al. Nonreversible MAIT cell-dysfunction in chronic hepatitis C virus infection despite successful interferon-free therapy. Eur J Immunol. (2016) 46:2204–10. doi: 10.1002/eji.201646447
67. Bolte FJ, O'Keefe AC, Webb LM, Serti E, Rivera E, Liang TJ, et al. Intra-hepatic depletion of mucosal-associated invariant T cells in hepatitis C virus-induced liver inflammation. Gastroenterology. (2017) 153:1392–1403.e1392. doi: 10.1053/j.gastro.2017.07.043
68. Di Blasi D, Boldanova T, Mori L, Terracciano L, Heim MH, De Libero G. Unique T-cell populations define immune-inflamed hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. (2019) 9:195–218. doi: 10.1016/j.jcmgh.2019.08.004
69. Duan M, Goswami S, Shi JY, Wu LJ, Wang XY, Ma JQ, et al. Activated and exhausted MAIT cells foster disease progression and indicate poor outcome in hepatocellular carcinoma. Clin Cancer Res. (2019) 25:3304–16. doi: 10.1158/1078-0432.CCR-18-3040
70. Gherardin NA, Loh L, Admojo L, Davenport AJ, Richardson K, Rogers A, et al. Enumeration, functional responses and cytotoxic capacity of MAIT cells in newly diagnosed and relapsed multiple myeloma. Sci Rep. (2018) 8:4159. doi: 10.1038/s41598-018-22130-1
71. Bhattacharyya A, Hanafi LA, Sheih A, Golob JL, Srinivasan S, Boeckh MJ, et al. Graft-derived reconstitution of mucosal-associated invariant T cells after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. (2018) 24:242–51. doi: 10.1016/j.bbmt.2017.10.003
72. Varelias A, Bunting MD, Ormerod KL, Koyama M, Olver SD, Straube J, et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J Clin Invest. (2018) 128:1919–36. doi: 10.1172/JCI91646
73. Kawaguchi K, Umeda K, Hiejima E, Iwai A, Mikami M, Nodomi S, et al. Influence of post-transplant mucosal-associated invariant T cell recovery on the development of acute graft-versus-host disease in allogeneic bone marrow transplantation. Int J Hematol. (2018) 108:66–75. doi: 10.1007/s12185-018-2442-2
74. Yan J, Allen S, McDonald E, Das I, Mak JYW, Liu L, et al. MAIT cells promote tumor initiation, growth, and metastases via tumor MR1. Cancer Discov. (2020) 10:124–41. doi: 10.1158/2159-8290.CD-19-0569
75. Mori L, De Libero G. Bohemian Rhapsody of MR1T cells. Nat Immunol. (2020) 21:108–10. doi: 10.1038/s41590-019-0588-6
76. Bennett SR, Carbone FR, Karamalis F, Flavell RA, Miller JF, Heath WR. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. (1998) 393:478–80. doi: 10.1038/30996
77. Schoenberger SP, Toes RE, van der Voort EI, Offringa R, Melief CJ. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. (1998) 393:480–3. doi: 10.1038/31002
78. Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. (2018) 18:635–47. doi: 10.1038/s41577-018-0044-0
79. Cui Y, Franciszkiewicz K, Mburu YK, Mondot S, Le Bourhis L, Premel V, et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J Clin Invest. (2015) 125:4171–85. doi: 10.1172/JCI82424
80. Dias J, Leeansyah E, Sandberg JK. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc Natl Acad Sci USA. (2017) 114:E5434–43. doi: 10.1073/pnas.1705759114
81. Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. (2019) 216:133–51. doi: 10.1084/jem.20181483
82. Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature. (2015) 522:345–8. doi: 10.1038/nature14282
83. McGeachy MJ, Cua DJ, Gaffen SL. The IL-17 family of cytokines in health and disease. Immunity. (2019) 50:892–906. doi: 10.1016/j.immuni.2019.03.021
84. Silva-Santos B, Mensurado S, Coffelt SB. gammadelta T cells: pleiotropic immune effectors with therapeutic potential in cancer. Nat Rev Cancer. (2019) 19:392–404. doi: 10.1038/s41568-019-0153-5
85. Gober HJ, Kistowska M, Angman L, Jeno P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. (2003) 197:163–8. doi: 10.1084/jem.20021500
86. Finn OJ. The dawn of vaccines for cancer prevention. Nat Rev Immunol. (2018) 18:183–94. doi: 10.1038/nri.2017.140
Keywords: MR1, MR1T, self-antigens, tumor recognition, T-cell therapy
Citation: Vacchini A, Chancellor A, Spagnuolo J, Mori L and De Libero G (2020) MR1-Restricted T Cells Are Unprecedented Cancer Fighters. Front. Immunol. 11:751. doi: 10.3389/fimmu.2020.00751
Received: 03 February 2020; Accepted: 02 April 2020;
Published: 28 April 2020.
Edited by:
Lucy Jane Walker, GlaxoSmithKline, United KingdomReviewed by:
Antonella Sistigu, Agostino Gemelli University Polyclinic, ItalyDr. Kawaljit Kaur, University of California, Los Angeles, United States
Copyright © 2020 Vacchini, Chancellor, Spagnuolo, Mori and De Libero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gennaro De Libero, R2VubmFyby5kZWxpYmVyb0B1bmliYXMuY2g=
 Alessandro Vacchini
Alessandro Vacchini Andrew Chancellor
Andrew Chancellor Julian Spagnuolo
Julian Spagnuolo Lucia Mori
Lucia Mori Gennaro De Libero
Gennaro De Libero