
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 11 February 2020
Sec. Vaccines and Molecular Therapeutics
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.00170
This article is part of the Research Topic Mycobacterial Glycolipids – Role in Immunomodulation and Targets for Vaccine Development View all 7 articles
Diseases due to mycobacteria, including tuberculosis, leprosy, and Buruli ulcer, rank among the top causes of death and disability worldwide. Animal studies have revealed the importance of T cells in controlling these infections. However, the specific antigens recognized by T cells that confer protective immunity and their associated functions remain to be definitively established. T cells that respond to mycobacterial peptide antigens exhibit classical features of adaptive immunity and have been well-studied in humans and animal models. Recently, innate-like T cells that recognize lipid and metabolite antigens have also been implicated. Specifically, T cells that recognize mycobacterial glycolipid antigens (mycolipids) have been shown to confer protection to tuberculosis in animal models and share some biological characteristics with adaptive and innate-like T cells. Here, we review the existing data suggesting that mycolipid-specific T cells exist on a spectrum of “innateness,” which will influence how they can be leveraged to develop new diagnostics and vaccines for mycobacterial diseases.
Mycobacterial diseases such as tuberculosis (TB), leprosy, and Buruli ulcer, are of high global health importance and disproportionately affect individuals in resource-limited settings. A major challenge to reducing the global burden of these diseases is the lack of effective diagnostics and vaccines. Several studies in murine and non-human primate models implicate T cells as necessary to control Mycobacterium tuberculosis (M.tb) (1, 2). Specifically, these studies led to a focus on T cells that recognize peptide antigens presented by major histocompatibility complex (MHC) molecules, in part because MHC class II (MHC-II) knockout mice infected with M.tb had a significantly shorter life-span than MHC-I knockout or wild-type mice (2). Consequently, the majority of new vaccine development efforts have been focused on eliciting robust CD4 T cell responses to mycobacterial protein antigens (3, 4).
However, T cells can also recognize non-peptide antigens, such as lipids and microbial metabolites, presented by cluster of differentiation 1 (CD1) and MHC-related protein 1 (MR1), respectively (5, 6) (Figure 1A). These CD1- and MR1-restricted T cells have been largely defined as “innate-like” T cells due to their unique biology, as we describe below. The two major populations of CD1- and MR1-restricted T cells are invariant natural killer T (iNKT) cells and mucosal associated invariant T (MAIT) cells, respectively. Notably, iNKT and MAIT cells represent only a subset of T cells that recognize non-peptide antigens. Here, we focus our comments on the potential role of T cells that respond to mycobacterial cell surface lipid (mycolipid) antigens. We describe their origins, phenotypes, and functions, with particular attention to data emerging from human studies. We also provide context for the existing data with respect to a large body of literature examining T cells with adaptive or innate-like phenotypes. Our analysis suggests that mycolipid-specific T cells lie somewhere in the middle of this spectrum, which will influence how we leverage their unique biology to improve the diagnosis and treatment of mycobacterial diseases.
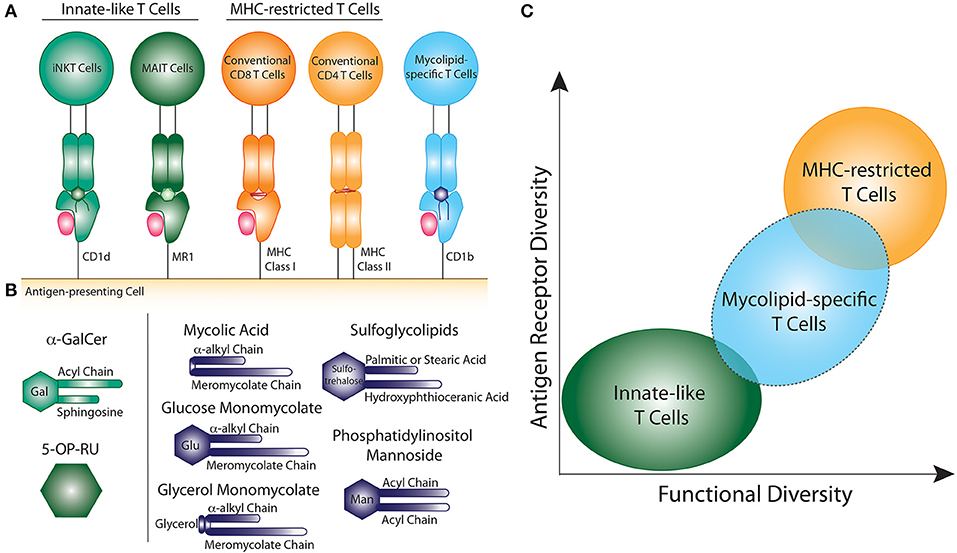
Figure 1. Comparison of innate-like, MHC-restricted, and mycolipid-specific T cells. (A) Graphical depiction of innate-like (green), MHC-restricted (orange), and mycolipid-specific T cells (blue). (B) Graphical depictions of canonical innate-like antigens or mycolipid antigens presented by CD1b. α-galactosyl ceramide (α-GalCer) is recognized by iNKT cells when presented by CD1d and is comprised of a galactose head group and a sphingosine and acyl chain. 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) is recognized by MAIT cells when presented by MR1. Mycolic acid is comprised of an α-alkyl and meromycolate chain. Glucose monomycolate and glycerol monomycolate are comprised of a mycolic acid base with a glucose and glycerol head group, respectively. Sulfoglycolipids are comprised of a sulfated trehalose headgroup and an hydroxyphthioceranic acid and palmitic acid or stearic acid chains. Phosphatidyl-myo-inositol mannoside is comprised of two acyl chains and two or more mannose residues as a headgroup. (C) Summary of the key determinants of T cell “innateness”. Innate-like T cells (green) have low antigen receptor diversity and fewer functional subsets. Peptide-specific T cells (orange) exhibit high antigen receptor diversity and higher functional diversity and plasticity. Mycolipid-specific T cells (blue) have qualities that overlap with both innate-like and MHC-restricted T cells, but are functionally more similar to MHC-restricted T cells.
Lipids found in the mycobacterial cell wall, collectively termed mycolipids here, have been shown to bind CD1 molecules and activate human T cells (5, 7, 8) (Figure 1B). Long-chain mycolipids preferentially bind to CD1b due to its large binding pocket, while comparatively shorter lipids, and lipopeptides are presented by CD1c and CD1a, respectively (9–11). Mycolic acid (MA) was the first mycobacterial lipid antigen shown to be recognized by T cells via the CD1b antigen presentation pathway (5). Since then, glucose monomycolate (GMM), and glycerol monomycolate (GroMM) were discovered as glycated forms of mycolic acid that are also recognized as T cell antigens (12, 13). In addition to the mycolates, lipoarabinomannan (LAM), and its constituent lipid phosphotidyl-myo-inositol mannoside (PIM), are abundant cell surface mycolipids recognized by human T cells (14, 15). Finally, diacylated sulfogylcolipids (SGLs), which are uniquely expressed in the cell wall of virulent strains of M.tb, are also T cell antigens (16). Thus, six structurally defined mycolipids are presented by CD1b to human T cells (Figure 1B). In addition, mannosyl-β-1-phosphoketide (MPM) is restricted by CD1c (17) and didehydroxymycobactin (DDM) is a lipopeptide antigen presented by CD1a (10).
T cell responses to many of these antigens have been detected among individuals infected with M.tb and Mycobacterium leprae, the causative agents of TB and leprosy, respectively. In vitro-derived T cell lines have been isolated from peripheral blood and lesion biopsies of individuals infected with M. leprae (15, 18). GMM- and SGL-specific T cells have been detected at a higher frequency in individuals infected with M.tb than healthy individuals (16, 19). Mycolipid-specific T cells have also been studied directly from the peripheral blood of individuals with active TB (20).
As a result of MHC polymorphism, MHC-restricted T cells exhibit diverse TCRs that are unlikely to be shared across individuals, even for a specific peptide antigen. By contrast, the CD1 and MR1 antigen presentation systems are highly conserved across human populations (21, 22). Thus, one defining feature of iNKT and MAIT cells is the expression of a semi-invariant TCR that is shared across genetically unrelated individuals. The canonical TCR that is expressed by iNKT cells includes an invariant TCR-α chain that is comprised of a TRAV10/TRAJ18 gene rearrangement (23, 24). The canonical TCR expressed by MAIT cells expresses a TCR-α chain comprised of a TRAV1-2/TRAJ33,12,22 gene rearrangements (24–26). While MHC-restricted T cells are typically activated by a single peptide antigen, iNKT and MAIT cells that express the canonical invariant TCR-α chain have the ability to recognize multiple ligands. iNKT cells are defined by their recognition of α-galactosylceramide (α-GalCer), which is a potent and high affinity antigen (27, 28). However, iNKT cells can also recognize a variety of other antigens presented by CD1d, such as lyso-phospholipids, glycosphingolipids, and sulfatides (29–32). The canonical ligand for MAIT cells is 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) presented by MR1, which is recognized with high affinity (6, 33). MAIT cells can also recognize chemically diverse small molecules such as drugs, drug metabolites, and drug-like molecules (34).
The development of CD1b tetramers for GMM, SGLs, and MA have enabled the isolation of mycolipid-specific T cells to study the T cell receptor (TCR) repertoire and functional diversity directly ex vivo (35–40). TCR diversity among T cells specific for mycolipids has been best studied using GMM as the model antigen. Like iNKT and MAIT cells, a subset of GMM-specific T cells present in genetically unrelated donors expresses a semi-invariant TCR. This TCR is termed the germline-encoded mycolyl-reactive (GEM) TCR and includes a conserved TCR-α that consists of a TRAV1-2/TRAJ9 gene rearrangement (38). However, recent data suggest that GMM-specific T cells expressing this conserved TCR may actually constitute the minority of GMM-specific T cells, which have been shown to express a more diverse TCR repertoire (35, 39). GMM-specific TCRs have also been shown to display varying levels of affinity and specificity for GMM. A GMM-specific T cell clone with a TCR that uses TRAV1-2 has a higher affinity for GMM-CD1b tetramer and expresses a similar but distinct cytokine profile than a T cell clone that does not use TRAV1-2 (39). Further, the GMM-specific T cell clone LDN5 displays such fine specificity that alterations to the sugar head group or lipid tail moieties distal to the head group can abrogate antigenicity (13). A different GMM-specific T cell clone expressing the semi-invariant TCR-α chain was shown to recognize both GMM and MA, which lacks a sugar head group (38, 41). The mechanism for this promiscuity is not fully understood but may relate to modulation of the CD1b surface by the embedded lipid tails (42). Future studies comparing TCR sequences may reveal whether a molecular or structural pattern is responsible for the TCR being permissive to multiple mycolate ligands.
Of note, NKT cells can also express diverse TCRs and are collectively referred to as Type II iNKT cells (43, 44). Similarly, T cells that are restricted by MR1 can also express TCRs that do not contain the TRAV1-2 gene segment (45, 46). This is similar to the pattern of GMM-specific TCR diversity that has been described above as some GMM-specific T cells express a semi-invariant TCR, but others exhibit diverse TCRs.
Thus, GMM-specific TCRs exhibit properties of both innate and adaptive T cells. On the one hand, they are characterized by diverse TCRs with varying affinity, but there is also a dominant and conserved semi-invariant TCR that is shared across individuals. While several mycolipid-restricted TCRs have been identified, further analysis of large groups of T cells for each lipid antigen is required to determine if common TCR genes or motifs are shared between mycolipid-specific T cells from unrelated donors. This can be accomplished by single cell RNA-sequencing or targeted TCR sequencing of mycolipid-specific T cells.
MHC-restricted T cells undergo positive selection in the thymus by thymic epithelial cells and egress as naive T cells (47, 48). By contrast, iNKT cells and MAIT cells undergo positive selection by other thymocytes that express CD1d or MR1 on their cell surface, respectively (49, 50). iNKT cells acquire an innate-like effector memory program during thymic development and egress from the thymus expressing the transcription factor promyelocytic leukemia zinc finger (PLZF), which regulates an innate-like phenotype in iNKT and MAIT cells, as well as innate lymphoid and natural killer cells (51–53). MAIT cells egress from the thymus with a naive surface phenotype and acquire an innate-like effector memory program, including PLZF expression, during expansion in the periphery following microbial colonization (52, 54). The development of mycolipid-specific T cells is largely unknown. In a human transgenic mouse model, MA-specific T cells were selected by thymocytes that express CD1b rather than thymic epithelial cells, similar to iNKT and MAIT cells (55). However, it has been shown that murine mycolic acid-specific T cells do not express PLZF (55). Whether human mycolipid-specific T cells express PLZF is unknown.
Naive MHC-restricted T cells are present at low frequency and require TCR ligation by their cognate antigen as well as additional stimulation by professional antigen presenting cells to become activated (56–58). These T cells will then undergo a rapid expansion, up to 25-fold in 72 h, and differentiate into effector T cells (59). Upon clearance of antigen, the effector T cell population will then contract and form long-lived memory T cells that have enhanced proliferative potential and more rapid activation and cytokine production upon subsequent encounter with antigen (60).
By contrast, iNKT and MAIT cells do not necessarily require cognate recognition of antigen to be activated in the periphery, and they acquire their effector memory phenotype during development, as described above. For example, bacterial infection and toll-like receptor (TLR) agonists can induce interferon gamma (IFN-γ) production in MAIT cells, through an interleukin (IL-)12- and IL-18-dependent mechanism (61). It was also shown that MAIT cells derived from human peripheral blood produce IFN-γ and tumor necrosis factor (TNF) when stimulated with IL-12, IL-15, and IL-18 (62). Further, TCR signaling is not sufficient for MAIT cell activation in the absense of these inflammatory signals (62). iNKT cells express high levels of IL-12 receptor, and IFN-γ production during in vitro stimulation is dependent on IL-12 signaling (63). In the absence of activation, iNKT and MAIT cells are present at high frequency, comprising up to 1 and 10% of circulating T cells, respectively (64–66). However, they have an impaired proliferative potential compared to MHC-restricted T cells after stimulation (67).
It is unknown what factors are required for activation of naive mycolipid-specific T cells. Mycolipid-specific T cells are found at a higher frequency in individuals with documented exposure to mycobacteria, suggesting that prior antigen exposure drives activation and proliferation (16, 19). There is evidence that immunization of cattle with GMM produces GMM-specific T cells with enhanced proliferative potential, also suggesting that exposure to antigen can lead to activation and enhanced recall responses (68). In addition, a study of GMM-specific T cells directly ex vivo showed that over 80% of this population expresses CD45RO, further suggesting that they are antigen experienced (69). The proportion of different memory subsets found in antigen-experienced individuals is currently unknown, and whether antigen experience is required for memory phenotype acquisition also remains to be determined (Table 1). This could be comprehensively examined by looking at the memory phenotype of mycolipid-specific T cells using CD1 tetramers in longitudinal cohort studies and in animal models pre- and post-antigen exposure. Other metrics of T cell memory should also be investigated, such as enhanced proliferative potential, clonal expansion, and enhanced secondary recall responses. Finally, it is unknown whether mycolipid-specific T cells can be activated in a cytokine-driven and TCR-independent manner (Table 1). This could be investigated using in vitro activation studies such as IFN-γ secretion or activation marker upregulation (e.g., CD69) as assessed by flow cytometry.
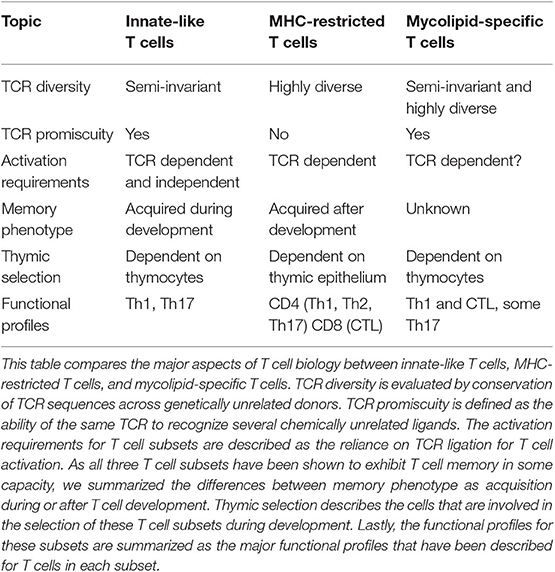
Table 1. Summary of properties exhibited by innate-like, MHC-restricted, and mycolipid-specific T cells.
The phenotypes of MHC-restricted T cells are broadly dictated by the inflammatory environment, TCR co-receptor expression, and in some cases, TCR affinity (70). Largely, these functional categories are pro-inflammatory (Th1, Th2, and Th17), cytotoxic, or regulatory (Treg). The pro-inflammatory subsets are regulated by the transcription factors T-bet, GATA3, and ROR-γt, respectively (71–73). Regulatory T cells are regulated by the transcription factor FoxP3 (74, 75). However, there is evidence that MHC-restricted T cells exhibit functional plasticity and can alter their functional program over the course of the T cell's life-span (76).
iNKT cells are largely characterized by constitutive IFN-γ expression and IL-4 production, a phenotype not commonly exhibited by MHC-restricted T cells (77, 78). MAIT cells are capable of secreting a variety of cytokines, such as IL-17, IFN-γ, IL-2, and TNF (64). The functional subsets of iNKT and MAIT cells are largely defined by the magnitude of PLZF expression and the expression of the master transcription factors that define MHC-restricted T cell functional subsets (77, 79, 80). However, no Th2 equivalent has been identified for MAIT cells (80, 81).
Most of the data regarding mycolipid-specific T cells have been obtained through studies of in vitro-derived T cell clones, which have been shown to express IFN-γ, TNF, and IL-17, as well as lyse M.tb-infected cells and control M.tb replication in vitro (13, 16, 18, 38, 55, 82). T cells with different affinities for GMM-loaded CD1b tetramers have been shown to have different functional capabilities. The high affinity GMM-specific T cells expressed high levels of Th1 cytokines, whereas the lower affinity T cells expressed Th1 cytokines along with IL-17 (39). The most comprehensive study performed on cells stimulated directly ex vivo showed that CD4+ mycolipid-specific T cells secrete IFN-γ, TNF, IL-2, and IL-17 in polyfunctional combinations (83). Notably, different lipid antigens yielded unique functional profiles with GMM-specific T cells being the most polyfunctional, while glycerol monomycolate-specific T cells largely expressed IL-2 alone (83). Whether CD4+ mycolipid-specific T cells can be categorized into traditional T-helper lineages based on expression of canonical transcription factors is currently unknown, as are the factors that govern acquisition of specific functions. These questions can be investigated in human cohorts using tetramers and intranuclear flow cytometry or RNA sequencing as well as animal studies to determine how these functional profiles are acquired. In addition, the diversity of functions exhibited by mycolipid-specific T cells could be comprehensively examined using single-cell RNA sequencing.
iNKT cells and MAIT cells may contribute to protection from M.tb based on data derived from in vitro and in vivo models of TB. Patients with active TB have reduced levels of iNKT and MAIT cells in peripheral blood or bronchoalveolar lavage samples when compared to healthy donors and individuals with latent TB, suggesting that these cells home to the lung to mediate a protective immune response (84–87). In a murine model, pulmonary mycobacterial infection leads to recruitment of MAIT cells in the lung, and this was associated with reduced bacterial burden during infection (88). While MAIT cells from this study exhibited diverse functional profiles, no particular functional profile was associated with reduction in disease burden (88). iNKT cells can recognize infected macrophages and kill intracellular mycobacteria in a granulocyte-macrophage colony-stimulating factor (GM-CSF) dependent manner (89, 90). The role of mycolipid-specific T cells in mediating protection has been evaluated in two animal models. Guinea pigs immunized with purified or synthetic mycobacterial lipids showed reduced lung pathology upon subsequent challenge with M.tb (91, 92). In a humanized CD1 transgenic mouse model, MA-specific T cells were adoptively transferred, trafficked to the lung during M.tb challenge, and led to decreased bacterial burden in a CD1-dependent manner (55). Interestingly, MA-specific T cells were activated earlier in the course of M.tb infection than MHC class II-restricted T cells that recognized antigen 85B (Ag85B) (55). MA-specific T cells isolated from the lung secreted TNF, IFN-γ, IL-2, and expressed CD107a, a surrogate marker of cytotoxic activity (55). Taken together, these data suggest that mycolipid-specific T cells can provide protection from mycobacterial infection, given they efficiently traffic to the lung during the early stages of infection. However, future studies should investigate which among the various lipid antigens are recognized by protective T cells, what functional profiles these T cells exhibit, and how to elicit these T cell responses through vaccination in humans and animal models.
In summary, several factors suggest that mycolipid-specific T cells may be more like MHC-restricted T cells than iNKT or MAIT cells in their functional qualities (Figure 1C). These include a low precursor frequency, increased frequency in mycobacteria-infected individuals, lack of PLZF expression, and the preliminary evidence of antigen-dependent activation, proliferation, and memory phenotype acquisition. However, some studies have shown that a subset of mycolipid-specific T cells exhibit innate-like qualities as well, such as expression of a semi-invariant TCR and promiscuous ligand recognition. On the whole, mycolipid-specific T cells have qualities of both innate-like and MHC-restricted T cells and exist on a spectrum of “innateness” (Figure 1C). Future studies should be directed at understanding their development in humans and animal models, ex vivo functional studies in humans, and how memory phenotypes are modulated by antigen exposure (Table 1).
The diagnosis of mycobacterial disease could be improved by leveraging the biology of mycolipid-specific T cells. Latent TB is typically diagnosed by an interferon gamma release assay, which measures T cell responses to secreted protein antigens but is a poor predictor for progression to active disease (93). The diagnosis of leprosy and Buruli ulcer are established by identifying bacilli in affected skin lesions, but this often results in false negative results because of the paucibacillary nature of these diseases. Notably, diagnostics targeting cell surface mycolipids are already being used for both TB and leprosy. High-affinity IgG antibodies targeting LAM have been incorporated into a urine-based assay for TB and have shown clinical utility in the setting of HIV co-infection (94). IgM antibodies targeting phenolic glycolipids (PGLs) are being used for the diagnosis of leprosy (95). Even though the current standard for leprosy diagnosis incorporates IgM antibodies to PGL, this could improve as PGLs also likely contain T cell epitopes, which could be leveraged to develop higher affinity IgG antibodies to PGLs. This is also probably true of LAM, currently the target of antibody-based diagnostics for TB, which actually contains a T cell epitope in the form of PIM (14). Finally, diagnostics could be improved by targeting specific lipids, such as SGLs, which have been shown to be preferentially expressed by M.tb and are absent from BCG and environmental strains (96, 97).
One approach to further improve diagnostics would be to advance our understanding of how mycolipid-specific T cells influence the generation of these mycolipid-specific antibodies. Notably, CD4 GMM- and MA-specific T cells express CD40 ligand (CD40L), which suggests that these T cells may be involved in B cell maturation and immunoglobulin class-switching (83). iNKT cells have previously been shown to provide both cognate and non-cognate help to B cell class switching and affinity maturation (98–100). Similarly, supernatants from activated MAIT cells promote B cell differentiation into plasmablasts, and production of IgA, IgM, and IgG antibodies (101). It is possible that mycolipid-specific T cells provide both cognate and non-cognate help to B cells.
Though we have focused our comments on mycolipids as T cell antigens, it is important to note that these lipids may also have adjuvant properties. Indeed, many of these antigens are structurally similar to adjuvants in clinical use, such as GLA-SE and trehalose dibehenate (TDB) (102, 103). Some T cell antigens, such as phosphotidyl-myo-inositol mannosides, may fortuitously act as both antigen and adjuvant by activating Toll-like receptor pathways (14, 104).
An emerging approach to the diagnosis of infectious diseases is using high-throughput sequencing of TCRs that are expanded in peripheral blood. Using machine learning, a recent study showed that a TCR “fingerprint” of cytomegalovirus (CMV) infection could be identified (105). This is a notable result because most CMV-specific TCRs would be private due to the highly polymorphic nature of MHC. On the other hand, CD1-restricted TCRs are expected to be public due to the non-polymorphic nature of CD1. Indeed, our group found that both shared and private TCR repertoires of GMM-specific T cells were associated with active TB disease in a South African cohort (35). Thus, TCRs specific for mycolipids could be developed into a blood-based diagnostic tool.
Mycolipid-specific T cells also hold significant promise for new vaccine approaches. There is no licensed vaccine for either leprosy or Buruli ulcer. The only licensed vaccine for TB is Mycobacterium bovis bacille Calmette-Guérin (BCG). This vaccine protects children from disseminated forms of TB but shows variable efficacy in preventing pulmonary TB in adults (106–108). As individuals with pulmonary TB are infectious, the epidemic cannot be controlled with BCG, and new TB vaccines are urgently needed. Immunizing with mycolipids has conferred modest protection to TB in rodent models (91, 92). The next step to increasing vaccine efficacy should be to ensure inclusion of a variety of antigenic types. One strategy of increasing antigenic breadth is by utilizing live-attenuated or killed whole cell vaccines (109). If T cell responses to mycolipid antigens are associated with positive outcomes in these studies, then using lipid-subunit vaccines to target a specific population can also be considered as a novel vaccine strategy. Natural candidates for lipid-subunit vaccine are SGLs, which have only been detected in virulent strains of M.tb (97). Further, SGL-specific T cells are expanded in individuals who have been exposed to M.tb (16).
Thus, mycolipid-specific T cells appear to bridge innate-like and adaptive T cell biology and should be considered for novel diagnostic and vaccination strategies targeting mycobacterial diseases.
CJ and CS wrote and edited the manuscript.
This work was supported by the U.S. National Institutes of Health (R01AI12518904 to CS) and the Doris Duke Charitable Foundation (Grant No. 2016103 to CS). CJ was supported by the Institute of Translational Health Sciences (5TL1TR00231803).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to apologize to colleagues whose work we have not cited due to space constraints or oversight. We also thank Krystle Yu, Annemike De Jong, and Martin Prlic for their critical reading and helpful discussion on the manuscript.
1. Lin PL, Rutledge T, Green AM, Bigbee M, Fuhrman C, Klein E, et al. CD4 T cell depletion exacerbates acute Mycobacterium tuberculosis while reactivation of latent infection is dependent on severity of tissue depletion in cynomolgus macaques. AIDS Res Hum Retroviruses. (2012) 28:1693–702. doi: 10.1089/aid.2012.0028
2. Mogues BT, Goodrich ME, Ryan L, Lacourse R, North RJ. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med. (2001) 193:271–80. doi: 10.1084/jem.193.3.271
3. Méndez-Samperio P. Development of tuberculosis vaccines in clinical trials: current status. Scand J Immunol. (2018) 88:e12710. doi: 10.1111/sji.12710
4. Rowland R, McShane H. Tuberculosis vaccines in clinical trials. Expert Rev Vaccines. (2011) 10:645–58. doi: 10.1586/erv.11.28
5. Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. (1994) 372:691–4. doi: 10.1038/372691a0
6. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
7. Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. (1989) 19:285–92. doi: 10.1002/eji.1830190211
8. Martin LH, Calabi F, Milstein C. Isolation of CD1 genes: a family of major histocompatibility complex-related differentiation antigens. Proc Natl Acad Sci USA. (1986) 83:9154–8. doi: 10.1073/pnas.83.23.9154
9. Beckman EM, Melián A, Behar SM, Sieling PA, Chatterjee D, Furlong ST, et al. CD1c restricts responses of mycobacteria-specific T cells. Evidence for antigen presentation by a second member of the human CD1 family. J Immunol. (1996) 157:2795–803.
10. Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O'Connor PB, et al. (2004). T cell activation by lipopeptide antigens. Science. 303:527–31. doi: 10.1126/science.1089353
11. Rosat JP, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J Immunol. (1999) 162:366–71.
12. Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, et al. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol. (2009) 16:82–92. doi: 10.1016/j.chembiol.2008.11.008
13. Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, et al. Structural requirements for glycolipid antigen recognition by CD1b- restricted T cells. Science. (1997) 278:283–6. doi: 10.1126/science.278.5336.283
14. de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. (2005) 310:1321–4. doi: 10.1126/science.1115301
15. Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. (1995) 269:227–30. doi: 10.1126/science.7542404
16. Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Böhmer G, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. (2004) 199:649–59. doi: 10.1084/jem.20031097
17. Moody DB, Ulrichs T, Mühlecker W, Young DC, Gurcha SS, Grant E, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. (2000) 404:884–8. doi: 10.1038/35009119
18. Sieling PA, Ochoa M-T, Jullien D, Leslie DS, Sabet S, Rosat J-P, et al. Evidence for human CD4 + T cells in the CD1-restricted repertoire: derivation of Mycobacteria-reactive T cells from leprosy lesions. J Immunol. (2000) 164:4790–6. doi: 10.4049/jimmunol.164.9.4790
19. Layton ED, Yu KKQ, Smith MT, Scriba TJ, De Rosa SC, Seshadri C. Validation of a CD1b tetramer assay for studies of human mycobacterial infection or vaccination. J Immunol Methods. (2018) 458:44–52. doi: 10.1016/j.jim.2018.04.004
20. Ulrichs T, Moody DB, Grant E, Kaufmann SHE, Porcelli SA. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun. (2003) 71:3076–87. doi: 10.1128/IAI.71.6.3076-3087.2003
21. Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. (1999) 54:122–7. doi: 10.1034/j.1399-0039.1999.540202.x
22. Parra-Cuadrado JF, Navarro P, Mirones I, Setién F, Oteo M, Martínez-Naves E. A study on the polymorphism of human MHC class I-related MR1 gene and identification of an MR1-like pseudogene. Tissue Antigens. (2000) 56:170–2. doi: 10.1034/j.1399-0039.2000.560211.x
23. Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4-8- T cells. J Exp Med. (1994) 180:1171–6. doi: 10.1084/jem.180.3.1171
24. Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8– αβ T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med. (1993) 178:1–16. doi: 10.1084/jem.178.1.1
25. Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. (2014) 211:1601–10. doi: 10.1084/jem.20140507
26. Lepore M, Kalinichenko A, Kalinicenko A, Colone A, Paleja B, Singhal A, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun. (2014) 5:3866. doi: 10.1038/ncomms4866
27. Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. (1997) 278:1626–9. doi: 10.1126/science.278.5343.1626
28. Sidobre S, Naidenko OV, Sim B-C, Gascoigne NRJ, Garcia KC, Kronenberg M. The Vα14 NKT cell TCR exhibits high-affinity binding to a glycolipid/CD1d complex. J Immunol. (2002) 169:1340–8. doi: 10.4049/jimmunol.169.3.1340
29. Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. (2009) 7:e1000228. doi: 10.1371/journal.pbio.1000228
30. Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. (2004) 199:947–57. doi: 10.1084/jem.20031389
31. Kinjo Y, Wu D, Kim GS, Xing GW, Poles MA, Ho DD, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. (2005) 434:520–5. doi: 10.1038/nature03407
32. Zhou D, Mattner J, Cantu C, Schrantz N, Yin N, Gao Y, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. (2004) 306:1786–9. doi: 10.1126/science.1103440
33. Corbett AJ, Eckle SBG, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
34. Keller AN, Eckle SBG, Xu W, Liu L, Hughes VA, Mak JYW, et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat Immunol. (2017) 18:402–11. doi: 10.1038/ni.3679
35. DeWitt WS, Yu KKQ, Wilburn DB, Sherwood A, Vignali M, Day CL, et al. A Diverse lipid antigen–specific TCR repertoire is clonally expanded during active tuberculosis. J Immunol. (2018) 201:888–96. doi: 10.4049/jimmunol.1800186
36. James CA, Yu KKQ, Gilleron M, Prandi J, Yedulla VR, Moleda ZZ, et al. CD1b tetramers identify T cells that recognize natural and synthetic diacylated sulfoglycolipids from Mycobacterium tuberculosis. Cell Chem Biol. (2018) 25:392–402.e14. doi: 10.1016/j.chembiol.2018.01.006
37. Kasmar AG, van Rhijn I, Cheng T-Y, Turner M, Seshadri C, Schiefner A, et al. CD1b tetramers bind T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. (2011) 208:1741–7. doi: 10.1084/jem.20110665
38. Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. (2013) 14:706–13. doi: 10.1038/ni.2630
39. Van Rhijn I, Gherardin NA, Kasmar A, de Jager W, Pellicci DG, Kostenko L, et al. TCR bias and affinity define two compartments of the CD1b-glycolipid-specific T cell repertoire. J Immunol. (2014) 192:4054–60. doi: 10.4049/jimmunol.1400158
40. Van Rhijn I, Iwany SK, Fodran P, Cheng TY, Gapin L, Minnaard AJ, et al. CD1b-mycolic acid tetramers demonstrate T-cell fine specificity for mycobacterial lipid tails. Eur J Immunol. (2017) 47:1525–34. doi: 10.1002/eji.201747062
41. Gras S, Van Rhijn I, Shahine A, Cheng T-Y, Bhati M, Tan LL, et al. T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat Commun. (2016) 7:13257. doi: 10.1038/ncomms13257
42. Chancellor A, Tocheva AS, Cave-Ayland C, Tezera L, White A, Al Dulayymi JR, et al. CD1b-restricted GEM T cell responses are modulated by Mycobacterium tuberculosis mycolic acid meromycolate chains. Proc Natl Acad Sci USA. (2017) 114:E10956–64. doi: 10.1073/pnas.1708252114
43. Arrenberg P, Halder R, Dai Y, Maricic I, Kumar V. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a β-linked self-glycolipid. Proc Natl Acad Sci USA. (2010) 107:10984–9. doi: 10.1073/pnas.1000576107
44. Dhodapkar MV, Kumar V. Type II NKT cells and their emerging role in health and disease. J Immunol. (2017) 198:1015–21. doi: 10.4049/jimmunol.1601399
45. Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife. (2017) 6:e24476. doi: 10.7554/eLife.24476.020
46. Meermeier EW, Laugel BF, Sewell AK, Corbett AJ, Rossjohn J, McCluskey J, et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat Commun. (2016) 7:e12506. doi: 10.1038/ncomms12506
47. Kisielow P, Teh HS, Blüthmann H, Von Boehmer H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature. (1988) 335:730–3. doi: 10.1038/335730a0
48. Miller JFAP. The golden anniversary of the thymus. Nat Rev Immunol. (2011) 11:489–95. doi: 10.1038/nri2993
49. Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. (1995) 182:2091–6. doi: 10.1084/jem.182.6.2091
50. Seach N, Guerri L, Le Bourhis L, Mburu Y, Cui Y, Bessoles S, et al. Double positive thymocytes select mucosal-associated invariant T cells. J Immunol. (2013) 191:6002–9. doi: 10.4049/jimmunol.1301212
51. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. (2014) 508:397–401. doi: 10.1038/nature13047
52. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of mait cells in mouse and human. PLoS Biol. (2009) 7:0525–36. doi: 10.1371/journal.pbio.1000054
53. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. (2008) 29:391–403. doi: 10.1016/j.immuni.2008.07.011
54. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. (2019) 366:494–9. doi: 10.1126/science.aaw2719
55. Zhao J, Siddiqui S, Shang S, Bian Y, Bagchi S, He Y, et al. Mycolic acid-specific T cells protect against Mycobacterium tuberculosis infection in a humanized transgenic mouse model. Elife. (2015) 4:e08525. doi: 10.7554/eLife.08525.013
56. Gonzalo J-A, Delaney T, Corcoran J, Goodearl A, Gutierrez-Ramos JC, Coyle AJ. Cutting edge: the related molecules CD28 and inducible costimulator deliver both unique and complementary signals required for optimal T cell activation. J Immunol. (2001) 166:1–5. doi: 10.4049/jimmunol.166.1.1
57. London CA, Lodge MP, Abbas AK. Functional responses and costimulator dependence of memory CD4 + T cells. J Immunol. (2000) 164:265–72. doi: 10.4049/jimmunol.164.1.265
58. Schlienger K, Craighead N, Lee KP, Levine BL, June CH. Efficient priming of protein antigen-specific human CD4+T cells by monocyte-derived dendritic cells. Blood. (2000) 96:3490–8. doi: 10.1182/blood.V96.10.3490
59. Gudmundsdottir H, Wells AD, Turka LA. Dynamics and requirements of T cell clonal expansion in vivo at the single-cell level: effector function is linked to proliferative capacity. J Immunol. (1999) 162:5212–23.
60. Rogers PR, Dubey C, Swain SL. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J Immunol. (2000) 164:2338–46. doi: 10.4049/jimmunol.164.5.2338
61. Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, et al. CD161++CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. (2014) 44:195–203. doi: 10.1002/eji.201343509
62. Slichter CK, McDavid A, Miller HW, Finak G, Seymour BJ, McNevin JP, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. (2016) 1:1–16. doi: 10.1172/jci.insight.86292
63. Brigl M, Tatituri RVV, Watts GFM, Bhowruth V, Leadbetter EA, Barton N, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med. (2011) 208:1163–77. doi: 10.1084/jem.20102555
64. Gherardin NA, Souter MNT, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. (2018) 96:507–25. doi: 10.1111/imcb.12021
65. Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A. Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest. (2002) 110:793–800. doi: 10.1172/JCI0215832
66. Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology. (2007) 122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x
67. Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, et al. Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat Commun. (2019) 10:687. doi: 10.1038/s41467-019-08604-4
68. Nguyen TKA, Koets AP, Santema WJ, van Eden W, Rutten VPMG, Van Rhijn I. The mycobacterial glycolipid glucose monomycolate induces a memory T cell response comparable to a model protein antigen and no B cell response upon experimental vaccination of cattle. Vaccine. (2009) 27:4818–25. doi: 10.1016/j.vaccine.2009.05.078
69. Kasprowicz VO, Cheng TY, Ndung'u T, Sunpath H, Moody DB, Kasmar AG. HIV disrupts human T cells that target mycobacterial glycolipids. J Infect Dis. (2016) 213:628–33. doi: 10.1093/infdis/jiv455
70. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. (2010) 28:445–89. doi: 10.1146/annurev-immunol-030409-101212
71. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. (2006) 126:1121–33. doi: 10.1016/j.cell.2006.07.035
72. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. (2000) 100:655–69. doi: 10.1016/S0092-8674(00)80702-3
73. Zhen W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene. Cell. (1997) 89:587–96. doi: 10.1016/S0092-8674(00)80240-8
74. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. (2003) 4:330–6. doi: 10.1038/ni904
75. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. (2003) 299:1057–61. doi: 10.1126/science.1079490
76. Sallusto F, Cassotta A, Hoces D, Foglierini M, Lanzavecchia A. Do memory CD4 T cells keep their cell-type programming: plasticity versus fate commitment?: T-cell heterogeneity, plasticity, and selection in humans. Cold Spring Harb Perspect Biol. (2018) 10:a029421. doi: 10.1101/cshperspect.a029421
77. Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci USA. (2008) 105:11287–92. doi: 10.1073/pnas.0801631105
78. Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. (2002) 195:625–36. doi: 10.1084/jem.20011786
79. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. (2013) 14:1146–54. doi: 10.1038/ni.2731
80. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SBG, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. (2015) 212:1095–108. doi: 10.1084/jem.20142110
81. Garner LC, Klenerman P, Provine NM. Insights into mucosal-associated invariant T cell biology from studies of invariant natural killer T cells. Front Immunol. (2018) 9:1478. doi: 10.3389/fimmu.2018.01478
82. Busch M, Herzmann C, Kallert S, Zimmermann A, Hofer C, Mayer D, et al. Lipoarabinomannan-responsive polycytotoxic T cells are associated with protection in human tuberculosis. Am J Respir Crit Care Med. (2016) 194:345–55. doi: 10.1164/rccm.201509-1746OC
83. Seshadri C, Lin L, Scriba TJ, Peterson G, Freidrich D, Frahm N, et al. T cell responses against mycobacterial lipids and proteins are poorly correlated in South African adolescents. J Immunol. (2015) 195:4595–603. doi: 10.4049/jimmunol.1501285
84. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. (2010) 8:1–14. doi: 10.1371/journal.pbio.1000407
85. Kee SJ, Kwon YS, Park YW, Cho YN, Lee SJ, Kim TJ, et al. Dysfunction of natural killer T cells in patients with active Mycobacterium tuberculosis infection. Infect Immun. (2012) 80:2100–8. doi: 10.1128/IAI.06018-11
86. Malka-Ruimy C, Youssef G, Ben Lambert M, Tourret M, Ghazarian L, Faye A, et al. Mucosal-associated invariant T cell levels are reduced in the peripheral blood and lungs of children with active pulmonary tuberculosis. Front Immunol. (2019) 10:206. doi: 10.3389/fimmu.2019.00206
87. Paquin-Proulx D, Costa PR, Silveira CGT, Marmorato MP, Cerqueira NB, Sutton MS, et al. Latent Mycobacterium tuberculosis infection is associated with a higher frequency of mucosal-associated invariant T and invariant natural killer T cells. Front Immunol. (2018) 9:1394. doi: 10.3389/fimmu.2018.01394
88. Sakala IG, Kjer-Nielsen L, Eickhoff CS, Wang X, Blazevic A, Liu L, et al. Functional heterogeneity and antimycobacterial effects of mouse mucosal-associated invariant T cells specific for riboflavin metabolites. J Immunol. (2015) 195:587–601. doi: 10.4049/jimmunol.1402545
89. Rothchild AC, Jayaraman P, Nunes-Alves C, Behar SM. iNKT cell production of GM-CSF controls Mycobacterium tuberculosis. PLoS Pathog. (2014) 10:e1003805. doi: 10.1371/journal.ppat.1003805
90. Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize Mycobacterium tuberculosis-infected macrophages, produce interferon-γ, and kill intracellular bacteria. PLoS Pathog. (2008) 4:e1000239. doi: 10.1371/journal.ppat.1000239
91. Dascher CC, Hiromatsu K, Xiong X, Morehouse C, Watts G, Liu G, et al. Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. Int Immunol. (2003) 15:915–25. doi: 10.1093/intimm/dxg091
92. Larrouy-Maumus G, Layre E, Clark S, Prandi J, Rayner E, Lepore M, et al. Protective efficacy of a lipid antigen vaccine in a guinea pig model of tuberculosis. Vaccine. (2017) 35:1395–402. doi: 10.1016/j.vaccine.2017.01.079
93. Abubakar I, Drobniewski F, Southern J, Sitch AJ, Jackson C, Lipman M, et al. Prognostic value of interferon-γ release assays and tuberculin skin test in predicting the development of active tuberculosis (UK PREDICT TB): a prospective cohort study. Lancet Infect Dis. (2018) 18:1077–87. doi: 10.1016/S1473-3099(18)30355-4
94. Broger T, Sossen B, du Toit E, Kerkhoff AD, Schutz C, Ivanova Reipold E, et al. Novel lipoarabinomannan point-of-care tuberculosis test for people with HIV: a diagnostic accuracy study. Lancet Infect Dis. (2019) 19:852–61. doi: 10.1016/S1473-3099(19)30001-5
95. Gurung P, Gomes CM, Vernal S, Leeflang MMG. Diagnostic accuracy of tests for leprosy: a systematic review and meta-analysis. Clin Microbiol Infect. (2019) 25:1315–27. doi: 10.1016/j.cmi.2019.05.020
96. Goren MB, Brokl O, Schaefer WB. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect Immun. (1974) 9:142–9. doi: 10.1128/IAI.9.1.142-149.1974
97. Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol. (2011) 18:1537–49. doi: 10.1016/j.chembiol.2011.10.013
98. Doherty DG, Melo AM, Moreno-Olivera A, Solomos AC. Activation and regulation of B cell responses by invariant natural killer T cells. Front Immunol. (2018) 9:1360. doi: 10.3389/fimmu.2018.01360
99. Leadbetter EA, Brigl M, Illarionov P, Cohen N, Luteran MC, Pillai S, et al. NK T cells provide lipid antigen-specific cognate help for B cells. Proc Natl Acad Sci USA. (2008) 105:8339–44. doi: 10.1073/pnas.0801375105
100. Tonti E, Galli G, Malzone C, Abrignani S, Casorati G, Dellabona P. NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood. (2009) 113:370–6. doi: 10.1182/blood-2008-06-166249
101. Bennett MS, Trivedi S, Iyer AS, Hale JS, Leung DT. Human mucosal-associated invariant T (MAIT) cells possess capacity for B cell help. J Leukoc Biol. (2017) 102:1261–9. doi: 10.1189/jlb.4a0317-116r
102. Coler RN, Bertholet S, Moutaftsi M, Guderian JA, Windish HP, Baldwin SL, et al. Development and characterization of synthetic glucopyranosyl lipid adjuvant system as a vaccine adjuvant. PLoS ONE. (2011) 6:e16333. doi: 10.1371/journal.pone.0016333
103. Werninghaus K, Babiak A, Groß O, Hölscher C, Dietrich H, Agger EM, et al. Adjuvanticity of a synthetic cord factor analogue for subunit Mycobacterium tuberculosis vaccination requires FclRγ-Syk- Card9-dependent innate immune activation. J Exp Med. (2009) 206:89–97. doi: 10.1084/jem.20081445
104. Jones BW, Means TK, Heldwein KA, Keen MA, Hill PJ, Belisle JT, et al. Different Toll-like receptor agonists induce distinct macrophage responses. J Leukoc Biol. (2001) 69:1036–44. doi: 10.1189/jlb.69.6.1036
105. Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. (2017) 49:659–65. doi: 10.1038/ng.3822
106. Fine PEM. Variation in protection by BCG: implications of and for heterologous immunity. Lancet. (1995) 346:1339–45. doi: 10.1016/S0140-6736(95)92348-9
107. Mangtani P, Abubakar I, Ariti C, Beynon R, Pimpin L, Fine PEM, et al. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin Infect Dis. (2014) 58:470–80. doi: 10.1093/cid/cit790
108. Roy A, Eisenhut M, Harris RJ, Rodrigues LC, Sridhar S, Habermann S, et al. Effect of BCG vaccination against Mycobacterium tuberculosis infection in children: systematic review and meta-analysis. BMJ. (2014) 349:g4643. doi: 10.1136/bmj.g4643
Keywords: mycobacteria, glycolipids, lipid antigen, T cell, CD1, tuberculosis, diagnostics, vaccines
Citation: James CA and Seshadri C (2020) T Cell Responses to Mycobacterial Glycolipids: On the Spectrum of “Innateness”. Front. Immunol. 11:170. doi: 10.3389/fimmu.2020.00170
Received: 01 October 2019; Accepted: 22 January 2020;
Published: 11 February 2020.
Edited by:
Margarida Correia-Neves, University of Minho, PortugalReviewed by:
Leandro J. Carreno, University of Chile, ChileCopyright © 2020 James and Seshadri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chetan Seshadri, c2VzaGFkcmlAdS53YXNoaW5ndG9uLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.