- Section of Cell and Developmental Biology, Division of Biological Sciences, University of California, San Diego, San Diego, CA, United States
Toll mediates a robust and effective innate immune response across vertebrates and invertebrates. In Drosophila melanogaster, activation of Toll by systemic infection drives the accumulation of a rich repertoire of immune effectors in hemolymph, including the recently characterized Bomanins, as well as the classical antimicrobial peptides (AMPs). Here we report the functional characterization of a Toll-induced hemolymph protein encoded by the bombardier (CG18067) gene. Using the CRISPR/Cas9 system to generate a precise deletion of the bombardier transcriptional unit, we found that Bombardier is required for Toll-mediated defense against fungi and Gram-positive bacteria. Assaying cell-free hemolymph, we found that the Bomanin-dependent candidacidal activity is also dependent on Bombardier, but is independent of the antifungal AMPs Drosomycin and Metchnikowin. Using mass spectrometry, we demonstrated that deletion of bombardier results in the specific absence of short-form Bomanins from hemolymph. In addition, flies lacking Bombardier exhibited a defect in pathogen tolerance that we trace to an aberrant condition triggered by Toll activation. These results lead us to a model in which the presence of Bombardier in wild-type flies enables the proper folding, secretion, or intermolecular associations of short-form Bomanins, and the absence of Bombardier disrupts one or more of these steps, resulting in defects in both immune resistance and tolerance.
Introduction
Innate immune pathways are found in plants, fungi, and animals and provide a rapid defense against a broad range of pathogens (1–3). In the fruit fly Drosophila melanogaster, the two major innate immune pathways are Toll and Imd (4–6). The Toll pathway is activated by Gram-positive bacteria with Lys-type peptidoglycan and by fungi, and is required for defense against these microbes (7–10). Conversely, the Imd pathway is activated by and plays a major role in survival against Gram-negative bacteria and Gram-positive bacteria with DAP-type peptidoglycan (11, 12). These pathways, which are both mediated by NF-κB transcription factors, are broadly conserved as initiators of innate immune responses. Activation of either pathway induces robust production of an array of immune molecules, including antimicrobial peptides (AMPs) (13–17).
AMPs are found in all kingdoms of life (18–22). These peptides have long been thought to play the principal effector role in innate immune defense due to their demonstrated in vitro antimicrobial activity and their marked upregulation after infection. However, recent research in D. melanogaster suggests that AMPs play a major role in Imd-mediated defense, but a relatively minor role in Toll-mediated immunity (23).
In contrast to the AMPs, the Drosophila-specific Bomanin peptides (Boms), which are highly induced after infection, are indispensable for resistance against pathogens controlled by the Toll pathway (24). BomΔ55C flies, which lack 10 of the 12 Bom genes, succumb to fungal and Gram-positive bacterial infections at rates indistinguishable from Toll-deficient flies (23, 24), suggesting that Boms rather than AMPs are the primary Toll effectors.
Bom peptides, like AMPs, are secreted from the fat body, the Drosophila immune organ, into the hemolymph, the Drosophila circulatory fluid. The family is comprised of three groups. The short-form peptides are 16–17 residues long and contain only the Bom motif. The tailed forms contain the Bom motif followed by a C-terminal tail. Finally, the bicipital forms consist of two Bom motifs connected by a linker region (24). BomΔ55C flies lack all six of the short-form Boms, two of the three tailed Boms, and two of the three bicipital Boms. High-level expression of short-form Boms is sufficient to rescue the sensitivity of BomΔ55C flies to C. glabrata infection (25). Furthermore, the absence of Toll-induced candidacidal activity in BomΔ55C hemolymph can be rescued by high-level expression of a short-form Bom (25). However, no in vitro antimicrobial activity has been observed with Bom peptides alone (25), suggesting that the Bomanins act in coordination with additional humoral effectors.
In this study, we demonstrate an essential role in Toll-mediated humoral defense for a previously uncharacterized hemolymph protein, Bombardier (one that deploys Boms).
Materials and Methods
CRISPR/Cas9 Deletion of bombardier Locus
The bombardier gene (CG18067) was deleted using CRISPR/Cas9 technology according to established protocols (26). Briefly, a pair of gRNAs designed to delete the region 2R: 20,534,248–20,536,154 were cloned into pU6-BbsI-chiRNA (Addgene plasmid #45946). Homology arms (1,017 bp left and 1,022 bp right) were cloned into pDsRed-attP (Addgene plasmid #51019). The plasmid pBS-Hsp70-Cas9 (Addgene plasmid #46294) was used as the Cas9 source. Constructs were injected into w1118 embryos. F1 progeny were screened for DsRed eyes and homozygous lines were established. See Supplemental Table 1 for gRNA and homology arm primer sequences.
Toll Activation, Drosophila Infection, and Survival Analysis
Flies were raised at 25°C on cornmeal molasses agar media1. The w1118 strain was used as the wild type. Microbial isolates, culture conditions, and conditions for infection for Enterococcus faecalis, Enterobacter cloacae, Fusarium oxysporum, and Candida glabrata were as described previously (24), except that C. glabrata was concentrated to OD600 = 100. Flies were incubated at 25°C after live bacterial infection and at 29°C after fungal infection. For heat-killed challenge, bacterial cultures were autoclaved and resuspended in 20% glycerol to OD600 = 10 for E. faecalis and OD600 = 300 for M. luteus. For both survival assays and hemolymph preparation, flies challenged with heat-killed bacteria were incubated at 29°C.
Hemolymph Antimicrobial Assays
Candidacidal activity of hemolymph was assayed as described previously (25), except that hemolymph was prepared from groups of 30 flies and all activity assays were carried out for 1 h at room temperature. The number of colonies representing zero percent killing was set as the value obtained by assaying uninduced w1118 hemolymph.
MALDI-TOF Analysis of Hemolymph
The Toll pathway was activated in flies using heat-killed M. luteus, then incubated at 29°C for 24 h. Hemolymph was extracted as in Lindsay et al. (25), with slight modifications. Hemolymph extracted with glass capillaries from five male flies was pooled and transferred into 0.1% trifluoroacetic acid (TFA)/50% acetonitrile (ACN). One μl of each mixture was spotted on a Bruker MSP 96 ground steel plate, mixed 1:1 with a saturated solution of Universal MALDI matrix (Sigma-Aldrich) in 0.1% TFA/78% ACN, and air-dried. MALDI-TOF spectra were acquired using a Bruker Autoflex mass spectrometer. Data were collected from 1,500 to 10,000 m/z in positive linear mode, and 1,000–5,000 m/z in positive reflectron mode. Peptide calibration standard II (Bruker) was mixed with Universal MALDI matrix and used as an external calibration standard. At least ten independent samples were collected for each genotype. For peptide identification, peaks were matched to those of corresponding peaks in prior studies (13, 25). Representative spectra were visualized using R 3.3.2 and ggplot2 2.2.1 (27, 28).
Gene Expression Quantitation
The Toll pathway was activated with heat-killed M. luteus. Using TRIzol (Ambion), total RNA was extracted 18 h after Toll activation from four to six adult flies (2–5 days old). Next, cDNA was synthesized from 500 ng total RNA using the SuperScript II Reverse Transcriptase kit (Invitrogen). Quantitative RT-PCR was performed on an iQ5 cycler (BioRad) using iQ SYBR Green Supermix (BioRad). Quantification of mRNA levels was calculated relative to levels of the ribosomal protein gene rp49 using the Pfaffl method (29). Three independent replicates were completed. See Supplemental Table 1 for qPCR primer sequences.
Hemolymph LC-MS
Flies were challenged with heat-killed M. luteus to activate the Toll pathway. Hemolymph was extracted from 100 to 110 each of w1118, Δbbd, and BomΔ55C flies using the same method as in the hemolymph antimicrobial assays, with 50–60 flies processed per Zymo-Spin IC column (Zymo Research) and yielding a total of ~10 μl hemolymph per genotype. Three independent biological replicates were processed for Δbbd and BomΔ55C, and two independent biological replicates were processed for w1118. Extracted hemolymph was mixed 1:1 (vol/vol) with denaturing buffer (8 M Urea, 50 mM Tris, pH 7.8, 150 mM NaCl, protease and phosphatase inhibitors) and protein concentration was determined using a BCA assay. For each sample, 40 μg of hemolymph was diluted to 1 M urea using 50 mM ammonium bicarbonate and digested overnight with trypsin (Promega, V511A) at a 1:100 (trypsin:protein) ratio. After digestion, peptides were reduced with 1 mM dithiothreitol at room temperature for 30 min and then alkylated with 5 mM iodoacetamide at room temperature in the dark for 30 min. Formic acid was added to a 0.1% final concentration and peptides were desalted using the C18-Stage-Tip method and then vacuum dried. The dried peptides were reconstituted in 5% formic acid/5% acetonitrile and 1 μg of total peptide for each sample was loaded for MS analysis. Samples were run in technical triplicates on a Q-Exactive mass spectrometer with instrument and chromatography settings as described previously (30), except for the following modifications: the RAW files were analyzed using Andromeda/MaxQuant (version 1.6.7.0) (31) with default settings (32) except the match between the run and LFQ quantitation settings was enabled for label free quantification. Data were searched against a concatenated target-decoy database comprised of forward and reversed sequences from the unreviewed UniprotKB/Swiss-Prot FASTA Drosophila database (2019). A mass accuracy of 20 ppm was assigned for the first search and 4.5 ppm for the main search. The statistical analysis was calculated using the DEP analysis R-package (33).
Bacterial Load Quantification
Bacterial load upon death (BLUD) was obtained as in Duneau et al. (34), with slight modifications. Briefly, flies were infected with E. faecalis and vials were monitored every 30 min for newly dead flies. These flies were then individually homogenized with a pestle in 400 μl LB media. Homogenates were also prepared from individual live w1118 flies 120 h post-infection (hpi). Homogenates were diluted serially in LB and spread on LB agar plates for incubation at 37°C overnight. Colonies were counted manually and the number of viable bacteria per fly was calculated. Data were obtained from three independent experiments.
Data Analysis
GraphPad Prism 5 was used for statistical tests. Survival data were plotted as Kaplan-Meier curves and were analyzed using the Gehan-Breslow-Wilcoxon test to determine statistical significance. Statistical differences in candidacidal activity were calculated using one-way ANOVA followed by Tukey's test. Multiple Mann-Whitney U tests were used to calculate differences between BLUD samples (p = 0.0085 after Šidák correction for multiple comparisons, α = 0.05, k = 6). Spearman rank correlation was used to assess the relationship between BLUD and time of death.
Results
The bombardier Gene Is Specifically Required for Toll-Mediated Defense
The bombardier (bbd) gene contains a consensus Toll-responsive NF-κB binding site within its promoter region and is strongly expressed upon Toll activation by Gram-positive bacterial infection or other inducers (14, 17, 35, 36). The encoded protein is predicted to be secreted and to generate a mature protein of 222 amino acids with a coiled coil near its C-terminus (37, 38). Orthologs of Bombardier are found across the Drosophila genus, but in no other genera (39).
We began our analysis of the bombardier gene by generating a null mutant, using CRISPR/Cas9 to delete 1,906 bp encompassing the annotated transcriptional unit. Flies homozygous for this deletion (hereafter Δbbd) were viable and morphologically wild-type. Given that bombardier is Toll-inducible, we assayed Δbbd flies for a potential loss-of-function phenotype in Toll-mediated immunity. Specifically, we infected adult Δbbd flies with various pathogens and then monitored survival. Two additional genotypes were used as controls: w1118 flies, which served as the wild type, and BomΔ55C flies, which lack Toll-mediated humoral defenses due to deletion of the 10 of the 12 Bom genes (24).
As shown in Figure 1, we observed a marked immunodeficiency when Δbbd flies were challenged with representative species for the three classes of microbes against which Toll provides defense. With the yeast Candida glabrata, more than 90% of w1118, but no Δbbd flies, survived 5 days after infection (Figure 1A). In the case of the filamentous fungus Fusarium oxysporum, 70% of w1118 adults, but fewer than 20% of Δbbd adults, were alive 5 days post-infection (Figure 1B). Finally, with Enterococcus faecalis, a Gram-positive bacterium, 50% of wild-type flies, but no Δbbd flies, were alive 5 days after infection (Figure 1C).
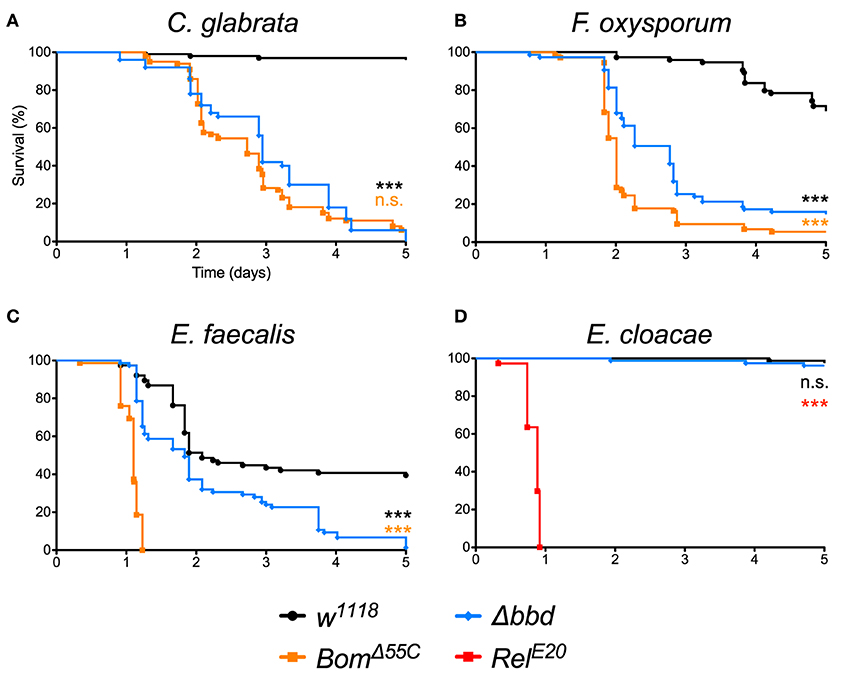
Figure 1. The bombardier gene is specifically required for Toll-mediated defense. (A–D) Survival curves of flies infected as indicated. The w1118 strain was the wild-type control; BomΔ55C and RelE20 were the susceptible controls (24, 40). Experiments were completed in triplicate with at least 25 flies per genotype in each replicate. Statistical significance was determined using the Gehan-Breslow-Wilcoxon test and Δbbd is shown relative to w1118 in black, relative to BomΔ55C in orange, and relative to RelE20 in red (***p < 0.0001; n.s., not significant, p > 0.05).
The impairment of Toll-mediated defenses by deletion of bombardier was significant for all three pathogens (p < 0.0001). In the case of C. glabrata, the immunodeficiency of Δbbd phenocopied that observed for BomΔ55C flies (n.s., p > 0.05). In contrast, with either F. oxysporum or E. faecalis, the rate of death was greater for BomΔ55C than for Δbbd (***p < 0.0001 for both infections). The Δbbd mutant thus displays a substantial, but not complete, loss of Toll-mediated defense.
The expression of bombardier is strongly induced by Toll, but not Imd activation (14). We therefore hypothesized that Imd-mediated defenses would not require bombardier function. To test this prediction, we infected Δbbd flies with Enterobacter cloacae, a Gram-negative bacterium. In this experiment, Δbbd flies were as immunocompetent as w1118 flies: more than 90% of both genotypes survived at least 5 days post-infection (Figure 1D). In contrast, 100% of RelE20 flies, which are deficient in Imd signaling (40), succumbed to infection within 1 day. Thus, bombardier functions in defense against a range of pathogens for which Toll mediates defense—yeast, filamentous fungi, and Lys-type Gram-positive bacteria—but not against Gram-negative bacteria, against which the Imd pathway is active.
The Candidacidal Activity of Hemolymph Requires Bombardier, but Neither Drosomycin Nor Metchnikowin
Next, we investigated the potential humoral role of Bombardier by preparing and assaying cell-free hemolymph. We have previously shown that hemolymph from wild-type flies exhibits a Toll-dependent and Bomanin-dependent candidacidal activity (25). However, we were also curious as to the identity of the active antifungal component. In particular, we considered the potential role of Metchnikowin (Mtk) and Drosomycin (Drs), two antimicrobial peptides (AMPs) that have documented antifungal activity in vitro and are strongly Toll-induced in vivo (14, 41, 42). We therefore took advantage of the recently described ΔAMPs strain, which is deficient for Mtk and Drs, as well as all other induced AMPs other than the Cecropins (23). Extracting and assaying Toll-induced hemolymph, we found that hemolymph from ΔAMPs flies had a killing activity against C. glabrata comparable to that of wild-type hemolymph (Figure 2). In contrast, we failed to detect any killing of C. glabrata by Δbbd hemolymph. We conclude that Boms and Bombardier, but neither Mtk nor Drs, are required for humoral defense against C. glabrata.
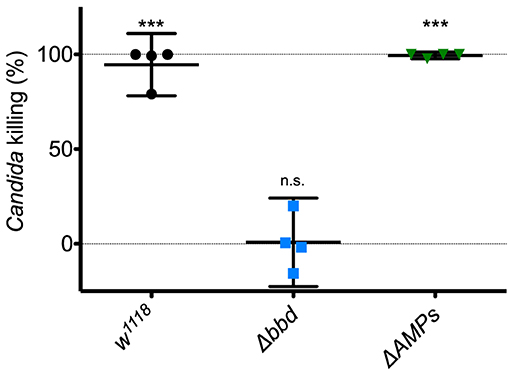
Figure 2. The Toll-induced candidacidal activity of hemolymph requires Bombardier, but neither Drosomycin nor Metchnikowin. Heat-killed M. luteus was used to activate the Toll pathway in flies. Hemolymph was extracted from flies 24 h after Toll induction, mixed with C. glabrata and incubated for 1 h to allow for killing. The surviving yeast cells were plated, and colonies were counted to determine the level of candidacidal activity in the extracted hemolymph. Colony counts from uninduced w1118 hemolymph were used as the control for no (0%) killing. Experiments were completed four times, with each point representing one replicate. One-way ANOVA was calculated followed by Tukey's test. Significance is shown relative to the null hypothesis of 0% killing (***p < 0.0001; n.s., not significant, p > 0.05). Error bars represent the 95% confidence interval.
Short-Form Bom Peptides Are Specifically Absent From Δbbd Hemolymph
MALDI-TOF provides a robust tool for characterizing small (<5,000 MW) peptides present in hemolymph after Toll activation. As shown in Figures 3A,B, such a readout includes the aforementioned AMPs (Mtk and Drs), several short-form Boms (BomS1, S2, S3, and S6; see Supplemental Table 2 for updated Bomanin nomenclature), and other induced peptides (e.g., IM4). We have previously shown that deleting the 55C Bom gene cluster removes the peaks attributable to the short-form Boms, while leaving the remaining signals unaffected (25). Remarkably, analysis of Δbbd hemolymph yielded a similar pattern. As shown in Figures 3C,D, the short-form Boms that were readily detectable in the wild type—S1, S2, S3, and S6—were absent in Δbbd hemolymph, whereas the remaining peptides, including Mtk, Drs, and IM4, displayed a wild-type profile.
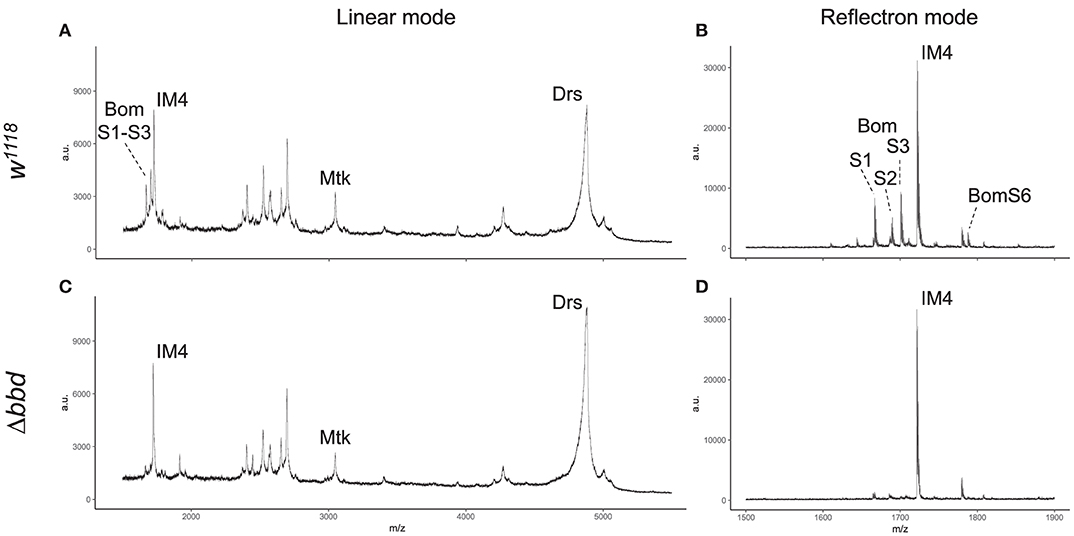
Figure 3. Short-form Bom peptides are specifically absent in hemolymph from Δbbd flies. MALDI-TOF mass spectra of w1118 (A,B) and Δbbd (C,D) hemolymph samples were collected in linear (A,C) and reflectron mode (B,D). For peptide identification, peaks were matched to those of corresponding peaks in prior studies (13, 25). Spectra were obtained from at least ten independent biological replicates and representative spectra are shown (a.u., arbitrary units; m/z, mass/charge).
Although Δbbd disrupts the accumulation of short-form Bom peptides in hemolymph, this effect does not reflect a disruption in transcription or stability of the corresponding Bom mRNAs: robust induction of Toll-regulated genes, including genes of short-form Boms, was readily detectable with qRT-PCR (Supplemental Figure 1).
Because proteins such as Bombardier and bicipital Boms are too large to be detected by our MALDI-TOF protocol, we used LC-MS to further characterize the relationship between Bombardier and the Boms in hemolymph. For these studies, we prepared Toll-induced hemolymph from three genotypes: w1118, Δbbd, and BomΔ55C. In wild-type hemolymph, we readily detected Bombardier protein (Figure 4), consistent with the presence of a canonical secretion signal sequence in the Bombardier coding sequence. Bombardier, like the Boms, is thus secreted into hemolymph upon Toll induction. We also detected all three bicipital Boms—BomBc1, BomBc2, and BomBc3. The LC-MS studies thus complemented the MALDI-TOF studies, with bicipital Boms detected by the former and short-form Boms by the latter (tailed Boms are not detected by either protocol). Next, we assayed Δbbd hemolymph. As expected, Bombardier was not detected. However, the three bicipital Boms were present at comparable levels in wild-type and Δbbd hemolymph (see Figure 4). Combined with the MALDI-TOF studies, these results demonstrate that Δbbd blocks accumulation in hemolymph of short-form, but not bicipital, Boms. Lastly, we analyzed hemolymph from BomΔ55C flies, which lack 10 of the 12 Bom genes. As expected, the products of the two deleted bicipital genes (BomBc1 and BomBc2) were absent, whereas the product of the remaining bicipital gene (BomBc3) was present at wild-type levels (see Figure 4). Turning our attention to Bombardier, we observed no effect of the 55C Bom deletion. Thus, Bombardier is required for the presence of short-form Boms in hemolymph, but the 55C Boms are not required for the presence of Bombardier.
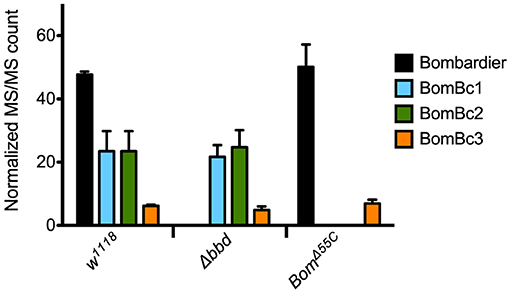
Figure 4. The presence of bicipital Bomanins in hemolymph is unaffected by loss of Bombardier. MS/MS counts for the indicated proteins as determined by Andromeda/MaxQuant were normalized to total MS/MS counts in each run. Error bars represent standard deviation for biological replicates (n = 3 for Δbbd and BomΔ55C, n = 2 for w1118). Supplemental Table 3 shows the full dataset.
Bombardier Mediates Both Infection Resistance and Tolerance
The Δbbd survival phenotype could be due to an inability to control pathogen growth—a defect in resistance—or an inability to endure infection—a defect in tolerance. Because flies lacking Bombardier demonstrate an increased susceptibility to infection and decreased levels of known resistance factors, the short-form Boms, it seemed likely that Δbbd flies, like BomΔ55C flies, have a defect in infection resistance. In exploring this hypothesis, we found that the model recently developed by Duneau et al. provided a useful framework (34). Following infection of an individual fly, there are two stereotypic outcomes: either the pathogen replicates, reaches a lethal burden, and the fly dies; or the pathogen is controlled at a level below the lethal burden and the fly survives with a persistent infection. Variation in survival curves for different pathogens and fly genotypes reflects variation in both the time required to reach lethal burden and in the fraction of flies that are able to control the infection before it reaches such a threshold. In cases where a fraction of flies control infection, group survival typically drops after infection and then reaches a plateau (23).
The survival curve for Δbbd flies infected with E. faecalis does not plateau (see Figure 1C). Instead, it exhibits a profile that we hypothesize reflects two phases of death. In the first phase, extending roughly 2 days post-infection, some Δbbd flies reach a lethal burden of E. faecalis and die, as reflected in a sharp decline in survival; the remainder control the infection. In the second phase, from 2.5 days onward, those flies with a persistent infection die at a reduced but steady rate, due to a defect in tolerance. If this hypothesis is correct, flies dying in the first phase should have a bacterial load upon death (BLUD) comparable to that of wild-type flies dying from infection. Furthermore, those dying in the second phase should have a much lower pathogen burden, comparable to that of wild-type survivors with a persistent infection.
To test our predictions regarding pathogen burden, we measured the BLUD of individual flies after infection with live E. faecalis and divided the data into two time intervals (Figure 5). For the earlier interval (dead flies obtained between 17 and 51.5 hpi), both BomΔ55C and Δbbd bacterial loads upon death were not significantly different from w1118 (Figure 5, red, p > 0.05). For the later time interval (flies obtained between 68 and 120.5 hpi), Δbbd flies perished at significantly lower bacterial loads compared to that of Δbbd flies which died earlier (Figure 5, Δbbd early compared to Δbbd late, p < 0.0001), indicating that these two groups die from distinct causes. Importantly, late-death Δbbd flies perished at significantly lower bacterial loads than those of w1118 suffering early deaths (p < 0.0001), demonstrating that Δbbd flies have a defect in tolerance.
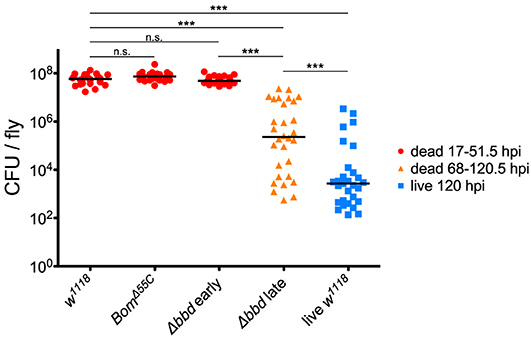
Figure 5. Bombardier mediates both infection resistance and tolerance. Bacterial load upon death (BLUD) of w1118, BomΔ55C, and Δbbd flies, plotted by early (17–51.5 hpi, red) or late (68–120.5 hpi, orange) time of death post-infection, as well as bacterial load of live w1118 flies 120 hpi (blue). Data was obtained and combined from three independent experiments totaling n = 26 for w1118, n = 30 for BomΔ55C, n = 33 for Δbbd red, n = 30 for Δbbd orange, and n = 29 for live w1118. Black bars indicate median values. Statistics were calculated using multiple Mann-Whitney U-tests. For significance, p = 0.0085 after Šidák correction for multiple comparisons (α = 0.05, k = 6). The pathogen loads of early deaths for BomΔ55C and Δbbd were not significantly different from w1118 (p > 0.05). The pathogen load of late Δbbd fly deaths is significantly different from that of the early-death Δbbd and w1118 groups (***p < 0.0001) and also significantly different from that of live w1118 flies 120 hpi (***p < 0.0001). Finally, the early-death w1118 pathogen load was significantly different from that of live w1118 flies 120 hpi (***p < 0.0001) (hpi, hours post-infection).
Together, the survival curve and BLUD data offer strong support for our two-phase-model: Δbbd flies died early in infection with high bacterial loads, due to a defect in resistance, and died later with lower bacterial loads, reflecting a deficiency in tolerance. However, we note that the bacterial loads of Δbbd flies dying in the later phase were still significantly greater than those of w1118 flies alive 120 hpi (Figure 5, Δbbd late compared to live w1118, p < 0.0001). This indicates that the later-death Δbbd group has not completely controlled infection compared to the live w1118 flies, and suggests that both resistance and tolerance contribute to the later Δbbd fly deaths. Although we cannot rule out a minor resurgence in bacterial proliferation preceding late death of bbd flies, we note that BLUD and time of death were not significantly correlated for these flies (Supplemental Figure 2, Spearman correlation test, r = −0.2654, p = 0.1564).
Immune Activation, Specifically Bom Expression, Is Deleterious in the Absence of Bombardier
What is the nature of the tolerance defect we observed in Δbbd flies? More specifically, is their health impaired by an excessive or toxic immune response, or is death due to another class of impaired tolerance (43)? To distinguish between these explanations, we assayed the effect of activating the immune response in Δbbd flies in the absence of infection.
When Δbbd flies were challenged with heat-killed E. faecalis, we observed a decrease in survival that first was apparent 3 days post-challenge followed by a steady decline in the number of live flies in the following days (Figure 6A), consistent with the timing of the late-phase deaths (see Figure 5). Overall, the death rate was slower than that of live infection, but the extent of killing was similar between heat-killed and live E. faecalis: fewer than 20% of flies survived (compare Figures 1C, 6A). In contrast, no effect on survival was observed upon challenge of either w1118 or BomΔ55C flies with heat-killed E. faecalis: >95% flies survived seven or more days post-challenge.
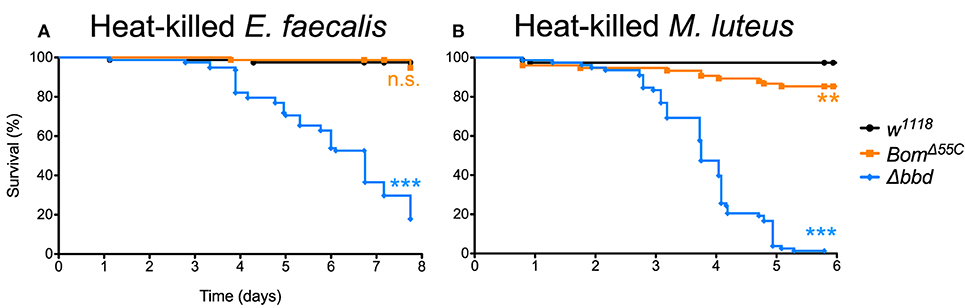
Figure 6. Immune activation is deleterious in absence of Bombardier. Fly survival after introduction of (A) heat-killed E. faecalis and (B) heat-killed M. luteus. Experiments were completed in triplicate with at least 25 flies per genotype in each replicate. Statistics were determined using the Gehan-Breslow-Wilcoxon test. Significance is shown relative to w1118 (***p < 0.0001; n.s., not significant, p > 0.05).
The effect of immune stimulation on Δbbd survival was not specific to E. faecalis. When we repeated the challenge experiments with heat-killed Micrococcus luteus, which activates the Toll response [see Supplemental Figure 1, as well as (25, 44)], the effect on Δbbd survival was again marked: 5 days after challenge, fewer than 5% of Δbbd flies were alive, compared to survival of >95% of w1118 and 85% of BomΔ55C flies over the same period of time (Figure 6B).
As both M. luteus and E. faecalis induce the Toll pathway, Toll activation could be the key factor in Δbbd mortality. To address this hypothesis, Δbbd flies were crossed with MyD88kra1 (Toll-deficient) flies to generate the MyD88kra1 Δbbd double mutant, and the resulting flies were challenged with heat-killed E. faecalis and M. luteus. Unlike Δbbd flies, MyD88kra1 Δbbd flies survived challenge with Toll activators (Figures 7A,B). Because blocking the Toll pathway with MyD88kra1 rescues the Δbbd phenotype triggered by heat-killed bacteria (p < 0.0001 compared to Δbbd, p > 0.05 compared to MyD88kra1 for both heat-killed bacteria), we conclude that Toll activation underlies the death of Δbbd flies in the absence of infection.
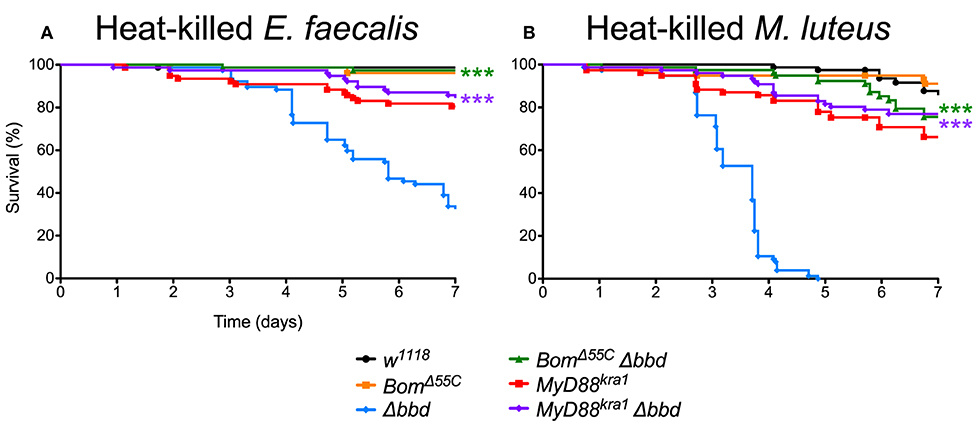
Figure 7. Toll-induced Bom expression is responsible for death in immune stimulated Δbbd flies. Survival of flies challenged with (A) heat-killed E. faecalis and (B) heat-killed M. luteus. Experiments were completed in triplicate with at least 25 flies per genotype in each replicate. Statistics were determined using the Gehan-Breslow-Wilcoxon test. Significance of double mutant survival curves is shown relative to the survival curve of Δbbd (***p < 0.0001).
As described above, Bom genes are transcribed in Δbbd flies (Supplemental Figure 1), but short-form Bom peptides do not appear in hemolymph (Figure 3). This suggests a mislocalization of these peptides, perhaps in an unprocessed or misfolded state. Given that short-form Bom genes are among the most abundantly transcribed genes after infection (17, 36), such mislocalized or misfolded Boms could rapidly accumulate to high levels in Δbbd flies. Could this explain the death of Δbbd flies upon immune stimulation? To address this question, we generated BomΔ55C Δbbd double mutants and assayed the effect of immune induction alongside both BomΔ55C and Δbbd flies (Figures 7A,B). The result was unequivocal: introducing BomΔ55C, which deletes all of the short-form Boms, eliminated the effect of Δbbd on survival following immune stimulation (p < 0.0001 compared to Δbbd, p > 0.05 compared to BomΔ55C for both heat-killed bacteria). The fact that BomΔ55C is epistatic to Δbbd demonstrates that Toll-driven expression of Bom genes is specifically responsible for the death of immune-stimulated Δbbd flies.
Discussion
The results presented in this study identify a key factor that regulates humoral and Bom-mediated defense in Drosophila. We demonstrate that Δbbd flies are defective in resistance to pathogens controlled by the Toll pathway. The results support the hypothesis that this defect results from the absence of short-form Boms in Δbbd hemolymph. Absence of Boms is sufficient to cause a defect in resistance (24) and Δbbd hemolymph appears to be lacking the short-form Boms but no other component, save Bombardier itself. Furthermore, Δbbd phenocopies BomΔ55C with regard to survival after C. glabrata infection, and resistance to C. glabrata can be restored in BomΔ55C flies by expression of short-form Boms (25). Finally, Δbbd hemolymph lacks candidacidal activity, which is dependent on short-form Bom peptides (25) and which we show here does not require Drs or Mtk.
For pathogens other than C. glabrata, the effect of deleting Bombardier is less severe than that of deleting the ten Bom genes clustered at 55C. Our mass spectrometry data suggest an explanation. Whereas, short-form Boms are absent from Δbbd hemolymph, bicipital Boms are present. (Tailed Boms were not detected with either mass spectrometry method.) Therefore, we postulate that the bicipital Boms, which are not required for resistance to C. glabrata (25), are functional against other pathogens. This would explain why Δbbd flies are more resistant than BomΔ55C flies upon infection with E. faecalis or F. oxysporum (Figure 1). In this regard, we note that Bombardier and all three forms of Bom proteins—short, tailed, and bicipital—are found across the Drosophila genus, supporting the notion that all three classes of Boms are immunoprotective and therefore maintained across the Drosophila genus.
It might appear that our discovery of Bombardier was serendipitous, given our role in defining the Bomanin gene family (24, 25). In hindsight, however, the link was forged in our approach. We selected CG18067 from the most strongly inducible Toll-regulated loci, a group that also includes eight of the Bomanin genes. Next, we engineered a CRISPR/Cas9 deletion of CG18067 and assayed this knockout with the identical set of pathogens that we had used for the BomΔ55C deletion, screening for loss of survival upon infection. Having examined a gene that is as strongly induced as the Bomanins, present in the same range of species as the Bomanins, and with a spectrum of loss-of-function phenotypes similar to that of the Bomanins, it is not particularly surprising that we would find ourselves studying a gene that affects the Bomanins.
Bombardier Function and Structure
What is the function of Bombardier? Deleting the gene results in the absence of short-form Boms from hemolymph, an effect we find is at the level of protein. Other mature immune peptides are present at normal levels in the hemolymph, and there is thus no general defect in translation, secretion, or processing. Based on these findings, we propose that Bombardier normally functions either to chaperone short Boms as they are secreted from the fat body into the hemolymph or, alternatively, to protect the Boms from misfolding or aggregation while in the hemolymph. We further hypothesize that it is the ectopic localization or aberrant form of short-form Boms in Δbbd flies that generates morbidity upon Toll pathway activation. In support of this idea, we showed that Bom expression underlies the lethality observed in Δbbd flies (Figure 7). Whether the short-form Boms physically interact with Bombardier, perhaps in the context of a larger antimicrobial complex, is currently unknown.
Activation of Toll-like receptor (TLR) signaling is important for innate immunity, but induction of the pathway can lead to autoimmune disorders and chronic inflammatory disease (45–48). Here we report an autoimmune activity driven by Toll-induced Bom expression in flies lacking a downstream pathway component, Bombardier. To what extent this parallel can be exploited in the context of understanding autoimmune disorders promises to be a significant focus for future investigation.
Data Availability Statement
The LC-MS datasets generated for this study can be accessed on the MassIVE data repository using the accession identifier: MSV000084509.
Author Contributions
SL and SW: conceptualization, writing—original draft, and visualization. SL, AF, and EB: methodology. SL and AF: formal analysis. SL, AF, and LC: investigation. SL, AF, LC, EB, and SW: writing—review and editing. EB and SW: supervision and funding acquisition.
Funding
This work was supported by National Institutes of Health (NIH) grant R01 GM050545 (to SW), NIH grant GM119132 (to EB), and NIH predoctoral training grant T32 GM008666 (to SL). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Scott Lindsay for a wealth of helpful discussions and technical assistance; Andres Mauricio Caraballo-Rodriguez and the Pieter Dorrestein Lab for guidance on and access to MALDI-TOF mass spectrometry equipment; Scott Rifkin for advice on statistical approaches; Emily Troemel, Bill McGinnis, Scott Lindsay, and Roland Liu for comments on the manuscript; and the laboratories of Bill McGinnis, Valentino Gantz, Ethan Bier, and James Posakony for access to equipment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.03040/full#supplementary-material
Footnote
References
1. Beutler B. Innate immunity: an overview. Mol Immunol. (2004) 40:845–59. doi: 10.1016/j.molimm.2003.10.005
2. Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. (2009) 324:742–3. doi: 10.1126/science.1171647
3. Kombrink A, Tayyrov A, Essig A, Stöckli M, Micheller S, Hintze J, et al. Induction of antibacterial proteins and peptides in the coprophilous mushroom coprinopsis cinerea in response to bacteria. ISME J. (2019) 13:588–602. doi: 10.1038/s41396-018-0293-8
4. Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. (2002) 3:121–6. doi: 10.1038/ni0202-121
5. Lemaitre B, Hoffmann J. The host defense of drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
6. Imler JL. Overview of drosophila immunity: a historical perspective. Dev Comp Immunol. (2014) 42:3–15. doi: 10.1016/j.dci.2013.08.018
7. Rutschmann S, Kilinc A, Ferrandon D. Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in drosophila. J Immunol. (2002) 168:1542–6. doi: 10.4049/jimmunol.168.4.1542
8. Gottar M, Gobert V, Matskevich AA, Reichhart J-M, Wang C, Butt TM, et al. Dual detection of fungal infections in drosophila via recognition of glucans and sensing of virulence factors. Cell. (2006) 127:1425–37. doi: 10.1016/j.cell.2006.10.046
9. Valanne S, Wang J-H, Rämet M. The drosophila toll signaling pathway. J Immunol. (2011) 186:649–56. doi: 10.4049/jimmunol.1002302
10. Lindsay SA, Wasserman SA. Conventional and non-conventional drosophila toll signaling. Dev Comp Immunol. (2014) 42:16–24. doi: 10.1016/j.dci.2013.04.011
11. Kleino A, Silverman N. The drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. (2014) 42:25–35. doi: 10.1016/j.dci.2013.05.014
12. Myllymäki H, Valanne S, Rämet M. The drosophila imd signaling pathway. J Immunol. (2014) 192:3455–62. doi: 10.4049/jimmunol.1303309
13. Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci USA. (1998) 95:11342–7. doi: 10.1073/pnas.95.19.11342
14. De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The toll and imd pathways are the major regulators of the immune response in drosophila. EMBO J. (2002) 21:2568–79. doi: 10.1093/emboj/21.11.2568
15. Levy F, Rabel D, Charlet M, Bulet P, Hoffmann JA, Ehret-Sabatier L. Peptidomic and proteomic analyses of the systemic immune response of drosophila. Biochimie. (2004) 86:607–16. doi: 10.1016/j.biochi.2004.07.007
16. Verleyen P, Baggerman G, D'Hertog W, Vierstraete E, Husson SJ, Schoofs L. Identification of new immune induced molecules in the haemolymph of drosophila melanogaster by 2D-nanoLC MS/MS. J Insect Physiol. (2006) 52:379–88. doi: 10.1016/j.jinsphys.2005.12.007
17. Troha K, Im JH, Revah J, Lazzaro BP, Buchon N. Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. PLoS Pathog. (2018) 14:e1006847. doi: 10.1371/journal.ppat.1006847
18. Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. (1980) 106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x
19. Imler JL, Bulet P. Antimicrobial peptides in drosophila: structures, activities and gene regulation. Chem Immunol Allergy. (2005) 86:1–21. doi: 10.1159/000086648
20. Radek K, Gallo R. Antimicrobial peptides: natural effectors of the innate immune system. Semin Immunopathol. (2007) 29:27–43. doi: 10.1007/s00281-007-0064-5
21. Maróti Gergely G, Kereszt A, Kondorosi É, Mergaert P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res Microbiol. (2011) 162:363–74. doi: 10.1016/j.resmic.2011.02.005
22. Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals. (2013) 6:1543–75. doi: 10.3390/ph6121543
23. Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife. (2019) 8:e44341. doi: 10.7554/eLife.44341
24. Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for drosophila toll-mediated immunity. PLoS Pathog. (2015) 11:e1004876. doi: 10.1371/journal.ppat.1004876
25. Lindsay SA, Lin SJH, Wasserman SA. Short-form bomanins mediate humoral immunity in drosophila. J Innate Immun. (2018) 10:306–14. doi: 10.1159/000489831
26. Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in drosophila. Genetics. (2014) 196:961–71. doi: 10.1534/genetics.113.160713
27. R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing (2016).
29. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. (2001) 29:e45. doi: 10.1093/nar/29.9.e45
30. Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. (2018) 172:590–604.e13. doi: 10.1016/j.cell.2017.12.032
31. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. (2008) 26:1367–72. doi: 10.1038/nbt.1511
32. Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. (2016) 11:2301–19. doi: 10.1038/nprot.2016.136
33. Zhang X, Smits AH, Van Tilburg GBA, Ovaa H, Huber W, Vermeulen M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat Protoc. (2018) 13:530–50. doi: 10.1038/nprot.2017.147
34. Duneau D, Ferdy J-B, Revah J, Kondolf H, Ortiz GA, Lazzaro BP, et al. Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. Elife. (2017) 6:e28298. doi: 10.7554/eLife.28298
35. Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. A kappaB sequence code for pathway-specific innate immune responses. EMBO J. (2007) 26:3826–35. doi: 10.1038/sj.emboj.7601798
36. Valanne S, Salminen TS, Järvelä-Stölting M, Vesala L, Rämet M. Immune-inducible non-coding RNA molecule lincRNA-IBIN connects immunity and metabolism in drosophila melanogaster. PLoS Pathog. (2019) 15:e1007504. doi: 10.1371/journal.ppat.1007504
37. Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. (2019) 37:420–3. doi: 10.1038/s41587-019-0036-z
38. Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. (1991) 252:1162–4. doi: 10.1126/science.252.5009.1162
39. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. (2008) 36:W5–9. doi: 10.1093/nar/gkn201
40. Hedengren M, Åsling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, et al. Relish, a central factor in the control of humoral but not cellular immunity in drosophila. Mol Cell. (1999) 4:827–37. doi: 10.1016/S1097-2765(00)80392-5
41. Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert WF, Hetru C, et al. Insect immunity: septic injury of drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. (1994) 269:33159–63.
42. Levashina EA, Ohresser S, Bulet P, Reichhart J, Hetru C, Hoffmann JA. Metchnikowin, a novel immune-inducible proline-rich peptide from drosophila with antibacterial and antifungal properties. Eur J Biochem. (1995) 233:694–700. doi: 10.1111/j.1432-1033.1995.694_2.x
43. Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. (2012) 30:271–94. doi: 10.1146/annurev-immunol-020711-075030
44. Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. (1997) 94:14614–9. doi: 10.1073/pnas.94.26.14614
45. Björkbacka H, Kunjathoor VV, Moore KJ, Koehn S, Ordija CM, Lee MA, et al. Reduced atherosclerosis in MyD88-null mice links elevated serum cholesterol levels to activation of innate immunity signaling pathways. Nat Med. (2004) 10:416–21. doi: 10.1038/nm1008
46. Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, et al. Regulation of lung injury and repair by toll-like receptors and hyaluronan. Nat Med. (2005) 11:1173–9. doi: 10.1038/nm1315
47. Kim S, Takahashi H, Lin W-W, Descargues P, Grivennikov S, Kim Y, et al. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. (2009) 457:102–6. doi: 10.1038/nature07623
Keywords: Drosophila melanogaster, immunity, Toll, Bomanins, humoral
Citation: Lin SJH, Fulzele A, Cohen LB, Bennett EJ and Wasserman SA (2020) Bombardier Enables Delivery of Short-Form Bomanins in the Drosophila Toll Response. Front. Immunol. 10:3040. doi: 10.3389/fimmu.2019.03040
Received: 18 October 2019; Accepted: 11 December 2019;
Published: 10 January 2020.
Edited by:
Susanna Valanne, Tampere University, FinlandReviewed by:
Anni Kleino, University of Helsinki, FinlandBrian P. Lazzaro, Pennsylvania State University (PSU), United States
Copyright © 2020 Lin, Fulzele, Cohen, Bennett and Wasserman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven A. Wasserman, c3RldmVud0B1Y3NkLmVkdQ==
 Samuel J. H. Lin
Samuel J. H. Lin Amit Fulzele
Amit Fulzele Steven A. Wasserman
Steven A. Wasserman