
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 17 December 2019
Sec. Microbial Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.02950
This article is part of the Research Topic Advances in The Immunology of Host Defense Peptide: Mechanisms and Applications of Antimicrobial Functions and Beyond View all 29 articles
The skin and intestine are active organs of the immune system that are constantly exposed to the outside environment. They support diverse microbiota, both commensal and pathogenic, which encompass bacteria, viruses, fungi, and parasites. The skin and intestine must maintain homeostasis with the diversity of commensal organisms present on epithelial surfaces. Here we review the current literature pertaining to epithelial barrier formation, microbial composition, and the complex regulatory mechanisms governing the interaction between the innate immune system and microbiota in the skin and intestine. We also compare and contrast the skin and intestine—two different organ systems responsible creating a protective barrier against the external environment, each of which has unique mechanisms for interaction with commensal populations and host repair.
The skin and intestine both rely on multifaceted mechanisms to maintain homeostasis and protect against invading microbes. Components essential for proper homeostasis between the external environments and the skin or intestine include the physical barrier formed by epithelial cells, the chemical barrier, the presence of beneficial commensal microbiota, and finally the tissue-resident and infiltrating immune cells. The barrier surfaces of the skin and intestine are not only habitats for commensal microbiota, but they also represent potential entry sites for pathogens, including bacteria, viruses, fungi, and parasites. The direct interface between the epithelial tissue barrier and microbiota poses a challenge for the barrier-lining epithelial cells and resident immune cells to distinguish dangerous pathogens from commensals and respond accordingly. Therefore, complex regulatory mechanisms have evolved to allow for delicate coordination between host tissues and their resident microbes. In this review, we provide an overview of the epithelial anatomy of the skin and intestine and interactions between host and microbiota at these surfaces. We focus on the role of microbiota and the innate immune system at homeostasis, in protection against infections, and in tissue repair of the skin and intestine.
The large surface areas of the skin and intestine—at least 30 m2 of skin in adults and about 400 m2 of intestinal epithelium—provides an expansive interface for interaction with the outside environment and increases the risk of invasion by pathogens (1–3). Given their extensive surface areas, the skin and intestine not only harbor millions of commensal microbiota, but they also must rely on multiple protective strategies to prevent entry of pathogens. As a result, the skin and intestine have developed site-specific physical, chemical, microbial, and immunologic barriers to maintain health and eradicate pathogenic bacteria.
The physical barrier of the skin and intestine provides the first line of defense against external perturbation at these sites. The physical barrier of the skin is formed by numerous layers of epidermal and dermal keratinocytes (Figure 1). The outermost layer of the epidermis is the stratum corneum, composed of as many as 100 layers of keratinized cell envelopes (corneocytes) that form a protective barrier (5). Barrier lipids, derived from lamellar bodies form an occlusive matrix between corneocytes (6). Deeper epidermal layers, including the stratum granulosum and stratum spinosum, are major producers of keratin filaments, which form a structural support for the epidermis (5). Finally, the basal layer of the epidermis contains stem cells that proliferate in homeostatic conditions and in response to injury in order to reconstitute the physical epidermal barrier. Epidermal keratinocytes maintain tight physical contact through tight junctions and adherens junctions, which form protective layer that is nearly impermeable to microbes. In addition to providing physical protection at the skin barrier, tight junction proteins, such as zona occludins proteins, play roles in proliferation and differentiation of keratinocytes in the skin, allowing re-establishment of the barrier against microbes after breach of the skin from wounding (7).
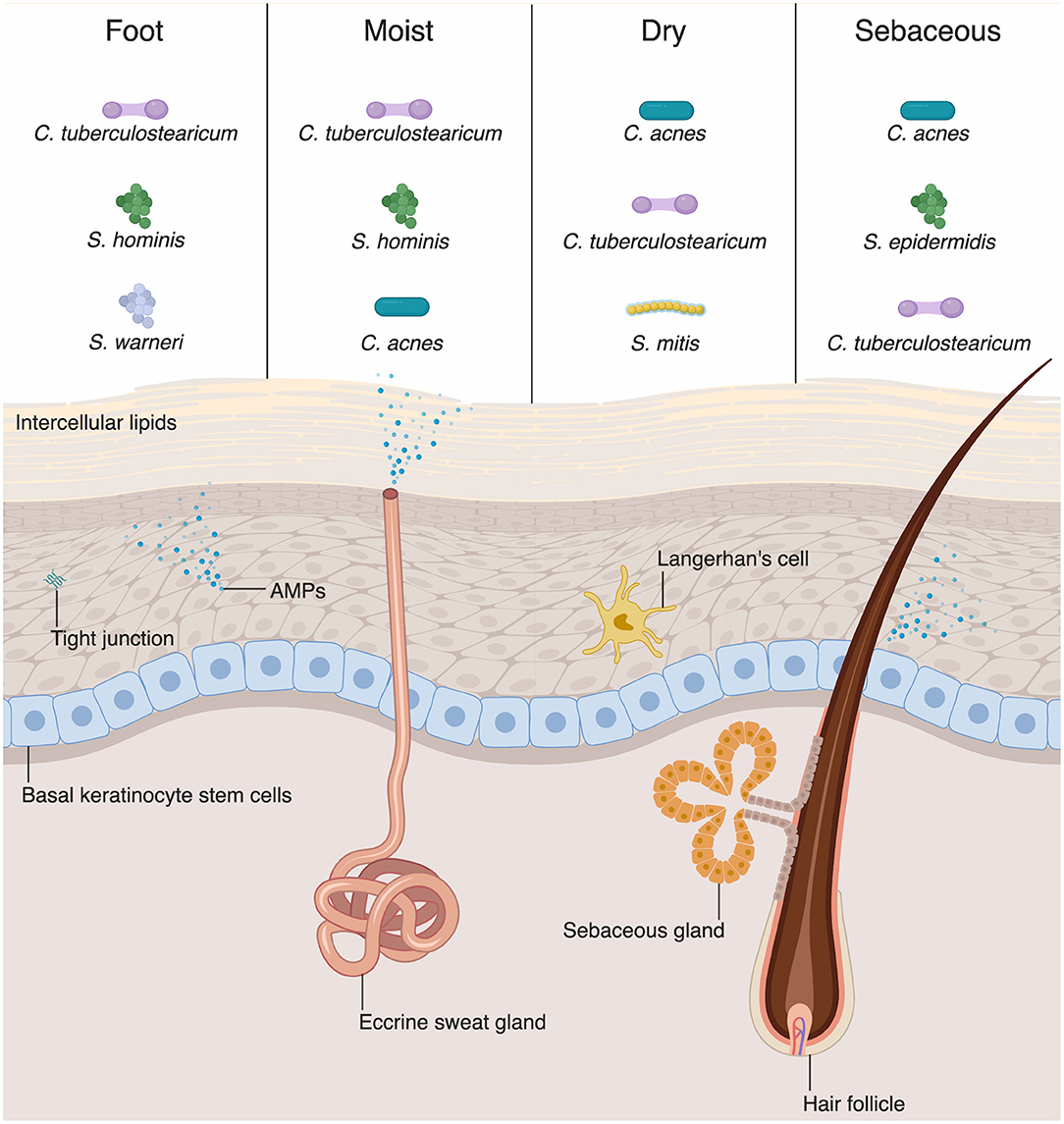
Figure 1. Skin-microbial interactions promote innate immune function. The skin is an active immune organ whose function is augmented by the presence of commensal microbiota. The epidermis is made up of numerous keratinocytes. The stratum corneum is sealed via intracellular lipids, and other epidermal keratinocytes are connected via tight junctions. Dermal appendages include sweat glands, hair follicles, and sebaceous glands, all of which contribute to immune function. Keratinocytes and dermal appendages release antimicrobial peptides and proteins (AMPs), which provide defense against pathogenic microbes. A number of bacteria species are commensal colonizers of the skin surface. The top three bacterial species for each skin site are shown (4). Dry and sebaceous sites are colonized predominantly by Cutibacterium acnes, whereas moist sites and the foot are colonized chiefly by Corynebacterium tuberculostearicum.
In contrast to the stratified squamous epithelium of the skin, the intestinal barrier is composed of a single layer of columnar epithelial cells (Figure 2) (11). However, this single layer of intestinal epithelial cells (IECs) is made of diverse cell types with absorptive, secretory, and immune function (2). This includes not only the absorptive enterocytes, which encompass the majority of IECs, but also secretory goblet cells, Paneth cells, and enteroendocrine cells. All cells that make up the intestinal barrier are constantly renewed by intestinal epithelial stem cells located in the bases of mucosal crypts (Figure 2). As in the skin, IECs are connected via tight junctions, which form a strong physical barrier that impedes translocation of pathogenic microbes or toxins.
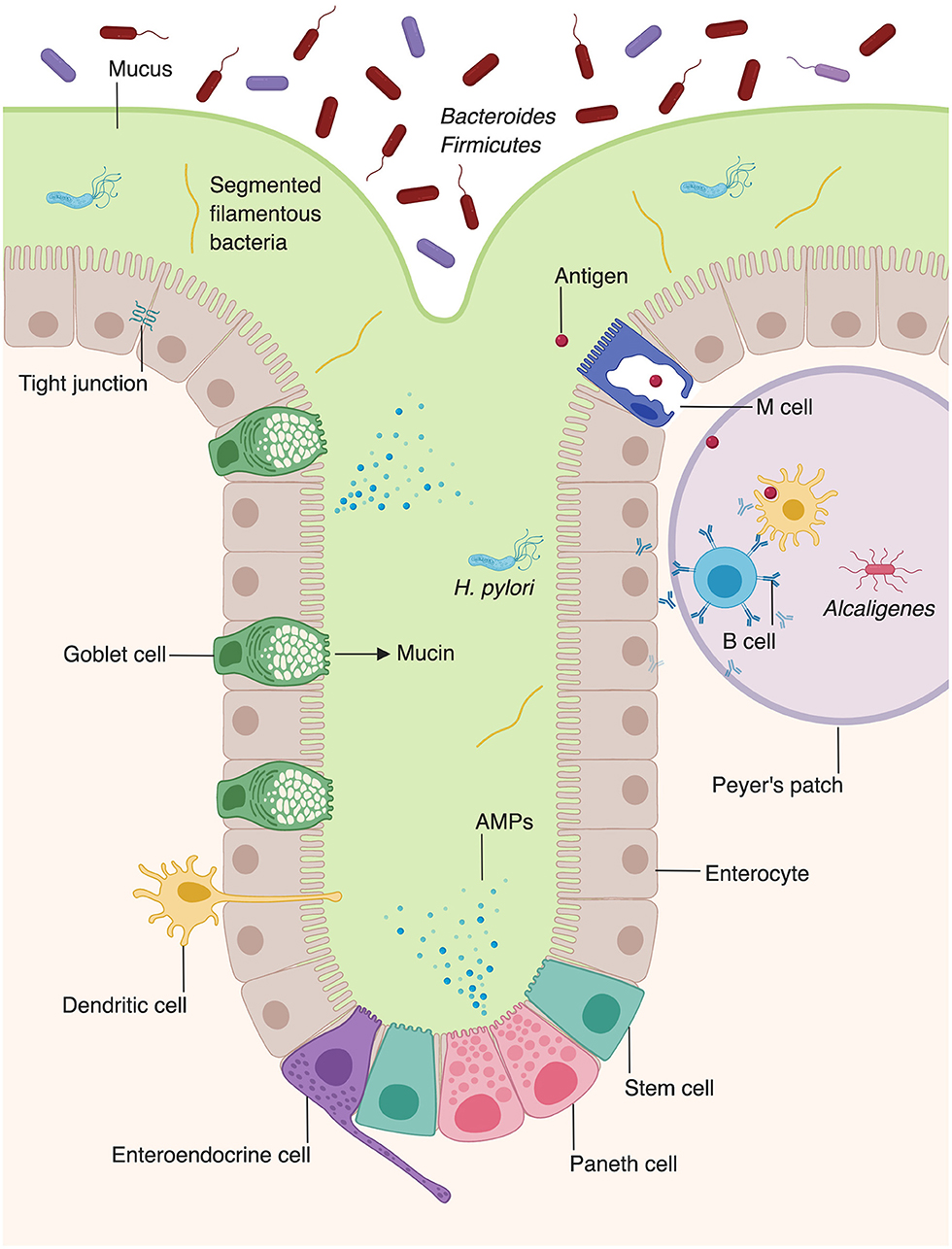
Figure 2. Microbiota augment intestinal innate immunity. Intestinal epithelial cells, which make up the physical barrier of the intestine, secrete antimicrobial peptides and proteins (AMPs). Goblet cells secrete mucus which forms an additional layer of protection against pathogens. Dendritic cells present antigen to B cells within Peyer's patches, stimulating them to secrete IgA. The intestine provides unique niches in which commensal bacteria thrive. Bacteroides and Firmicutes species comprise the majority of luminal bacteria, whereas segmented filamentous bacteria and Helicobacter pylori can penetrate into the mucus layer of the intestine (8, 9). Alcaligenes species are able to inhabit Peyer's patches (10).
The chemical barrier of the skin is formed by numerous secreted lipids and acids. As previously mentioned, the lipid layer secreted by lamellar bodies, is important for maintaining an occlusive matrix between cells and among layers of the stratum corneum (12, 13). Site-specific lipid content also influences the microbial composition of various cutaneous body sites (4, 14). In fact, microbial composition is relatively homogenous among multiple sebaceous sites but varies greatly between sebaceous and dry skin sites (4). Pathogenic microbes are also directly inhibited by some lipids or free fatty acids. For example, sapienic acid can efficiently inhibit pathogenic Staphylococcus aureus (S. aureus), but does not have sufficient activity against Staphylococcus epidermidis (S. epidermidis) (15). Overall, the chemical barrier formed by epidermal lipids and fatty acids is important for modulating microbial survival at the skin surface.
In addition, the stratum corneum of the epidermis maintains an acidic pH under homeostatic conditions. The term “acid mantle” has been used to describe the acidic condition of the stratum corneum (16). This acidic pH is important for skin barrier function and microbial defense by providing hostile environment for certain microorganisms (12). Furthermore, there are a number of pH-dependent enzymes that are critical for synthesis, production and maintenance of the lipid composition in the skin. Lipids, such as triglycerides and cholesterol, are hydrolyzed by skin-resident bacteria and yeasts into free fatty acids. Free fatty acids maintain a low pH that inhibits growth of pathogenic species such as Staphylococcus aureus (S. aureus), while allowing persistence of commensal bacteria such as coagulase negative Staphylococcus and Corynebacterium (1, 17).
The intestine relies on goblet cells to secrete a thick layer of jelly-like mucus made of glycoproteins to separate luminal bacteria from epithelial cells and create a distinct protected zone (Figure 2) (18). Mucins create both a chemical and a physical barrier between the intestinal lumen and EICs, and can even directly modulate expression of tolerogenic and inflammatory cytokines (19). In addition to providing physical protection, mucin layer is also rich in secretory IgA and antimicrobial proteins (AMPs) that provide a chemical immune defense against potential invading microorganisms (20, 21). Mucin synthesis is increased by short chain fatty acids (SCFAs), a fermentation product of bacterial metabolism (22). Furthermore, mucin production is decreased in germ-free mice, but production of mucin can be rescued by activation of microbe-sensing receptors, suggesting that commensal microbes enhance the intestinal barrier (2, 23). The composition of the mucin layer differs between the small and large intestine. The mucous layer of the small intestine is physically penetrable by bacteria, and epithelial cells are protected via secreted AMPs (24). In contrast, the large intestine contains both penetrable outer mucus layer and an impenetrable inner mucous layer (25).
With the rise of new techniques such as 16S and whole genome metagenomic shotgun sequencing, we have begun to understand in greater detail the diversity and functions of microbiota that colonize the skin and intestine (14, 26). The skin and intestine support a tremendous diversity and number of microbiota. In both the skin and intestine, commensal microbiota are important for maintaining epithelial homeostasis and overall health of the tissue (4, 27).
Although differing profoundly in taxonomic composition, the skin and intestine are similar in that the microbial composition varies among sites and niches. Recent sequencing studies have extensively mapped the species inhabiting various skin or body sites with different compositions, including wet, dry, and sebaceous sites (Figure 1) (14, 28). Distinct skin sites contain unique distribution of bacteria, in part governed by the lipid composition of a skin site (14). For example, sebaceous gland-rich areas, such as the glabella and back, are colonized most predominantly by Cutibacterium (formerly known as Propionibacterium) species, which are closely associated with the common condition acne vulgaris (14). Moist sites, such as the axilla and interdigital web spaces, are largely colonized by Corynebacteria and Staphylococci species (14).
In addition to bacteria, which are the most abundant kingdoms of organisms found on the skin, numerous fungi and viruses inhabit the skin (14). In contrast to bacteria, which are found in nearly all bodies sites and whose composition is governed by physiologic conditions, fungal distribution varies based on distinct body sites rather than physiologic conditions (29). The core body and arms have a relatively homogenous fungal composition and are predominantly colonized by Malassezia species, whereas the foot harbors a much greater fungal diversity (29). Viral composition, predominantly Polyomaviridae and Papillomaviridae, shows most diversity between individuals, rather than depending on body site or composition (28).
In contrast to the skin, which is inhabited by aerobic bacteria, aerotolerant anaerobes, or facultative anaerobes, the intestine is mostly colonized by anaerobes, such as bacteria of the phyla Bacteroidetes and Firmicutes (Figure 2) (8, 14). Whereas, the microbial composition of the skin is largely determined by environmental factors such as the presence or absence of sebum, the intestinal microbiota is dependent on location, niche, and external factors, such as diet (14, 30). The large intestine harbors a higher microbial diversity and density within individuals than the small intestine (31, 32). However, evidence suggests that the microbial composition of the small intestine is more dynamic than that of the large intestine, with large temporal fluctuations in ileal microbial constituents within a single day (33). Fewer studies have examined the microbial composition of the small intestine, compared to the large intestine. However, one study utilized 16s rRNA sequencing to examine the bacterial compositions of the jejunum, ileum, cecum, and recto-sigmoid colon (32). Facultative anaerobic bacteria were present in all four locations along the gastrointestinal tract. Lactobacilli, streptococci, and Enterococcus were detected at high frequencies in the jejunum and ileum. In addition to facultative anaerobes, which were the major operational taxonomic unit in both the small and large intestine, the large intestine was also found to contain obligate anaerobic bacteria (32).
Within the small or large intestine, the environmental niches can be luminal, mucus-associated, epithelial-associated, or lymphoid tissue-resident (30). Which phyla of bacteria inhabit a specific intestinal niche depends significantly on the characteristics of both the bacteria and the niche itself. Luminal bacteria are largely of the Bacteroidetes and Firmicutes phyla, and represent the largest percent of intestinal biomass (8). Recent studies have illuminated that the outer mucus layer of the large intestine forms a unique “mucus-associated” microbial niche with distinct bacterial communities (9). Specifically, bacteria of this niche are adapted to thrive in high levels of bioavailable iron and carbon, an ability conferred by their distinct genome-encoded metabolic and mucolytic activities. For example, Helicobacter pylori secretes urease to increase the pH of the mucin layer and disrupts the strong glycoprotein bonds, which allows it to burrow into the stomach mucosa (34).
The epithelial-associated bacteria make up a smaller proportion of intestine bacteria since fewer bacteria are able to infiltrate through the mucous layer (30). Epithelial-associated bacteria express distinct genes that allow them to colonize epithelial cells. For example, expression of commensal colonization factor (Ccf ) genes allows Bacillus fragilis to metabolize carbohydrates present in the intestinal lumen and therefore promotes their colonization of intestinal epithelium, illustrating the importance of nutrient-specific factors in determining microbial composition (35). Furthermore, although B. fragilis is an anaerobic bacteria and thrives predominantly in the intestinal lumen, it also grows well in nanomolar oxygen concentrations, such as that found in intestinal crypts (36). Epithelial-associated bacteria are also important for proper function of the intestinal immune system. For example, segmented filamentous bacteria adhere tightly to EICs and induces a Th17 response, conferring protection against pathogenic mucosal bacteria (30). Intestine-associated lymphoid tissues, including Peyer's patches and lymphoid follicles, are colonized largely by Alcaligenes species (10). However, it should be noted that, under homeostatic conditions, these bacteria do not spread to the spleen or produce a systemic IgG response. Colonization of intestine-associated lymphoid tissues by these bacteria only results in the local production of Alcaligenes-specific IgA antibodies, highlighting the tolerogenic response to a lymphoid tissue-resident bacteria (10). Overall, the special distribution of intestinal bacteria is dependent on niche-specific factors, such as availability of nutrients or site specific microbial-host interactions.
Commensal species, which can vary according to topography and anatomic environments, also undergo temporal changes as humans develop over time. It was previously thought that in utero fetuses were in a germ-free environment. However, data have shown that bacteria can be cultured from the umbilical cord and meconium of healthy full term babies (37, 38). 16S rRNA gene sequencing recently confirmed the presence of microbiota in newborn meconium and amniotic fluid (39). Meconium samples contained bacterial DNA, the majority of which mapped to Pelomonas puraquae. Conversely, amniotic fluid bacterial DNA mapped to skin commensal species such as Cutibacterium acnes and Staphylococcus species (39). The neonatal skin is first colonized by microbes present in the birth canal. Subsequently, an infant's microbiome is shaped by contact with the outside environment. Studies have shown that the skin flora of a baby is largely shaped by the mother's microbiome at birth and that there are notable differences in both skin and intestinal microbiota between infants born naturally or by C-section (40). The infant can also be exposed to viruses, such as herpes simplex virus type 2, present in the mother's vaginal tract (41). Over the course of the first year of life, the infant's skin microbiome is established and begins to resemble that of adults (42).
The intestine similarly has a temporal shift in its microbial flora as the baby transitions from an exclusively milk diet to solid foods (42). An initial diet of breast milk results in high levels of facultative and obligate anaerobes, such as Escherichia coli, Streptococcus, and Bifidobacterium species (43). Breast milk provides a source of human milk oligosaccharides and milk glycoconjugates, which are consumed by Bifidobacterium species (44). Bacteroides and Clostridia species predominate as babies are weaned and ingest more complex carbohydrates (43). Clostridia species are particularly specialized in degrading plant polysaccharides and are therefore able to thrive in the intestine once complex carbohydrates are introduced into the infant diet (45).
Beyond the early years of life, both skin and intestinal microbiome become more stable, and within-individual variation in microbial communities over time is much less than between-individual variations (28, 46, 47). Despite the relative stability of the skin microbiome, it is less stable over time than the intestinal microbiome (48). Furthermore, the level of microbial stability over time is significantly different among individuals; some individuals have a very stable skin microbiome, whereas others do not. Skin sites that have extensive environmental contact, such as the palm, display the least stability in microbial composition. Interestingly, individuals with a more diverse intestinal microbiome (in terms of number of bacteria species) also have a more stable microbiome over time, whereas individuals with a more diverse skin microbiome have a less stable microbiome over time (48). Microbial diversity decreases in the elderly, coinciding with a decline in immunocompetence in older populations (49). The complex shifts in establishing a commensal population depending on skin and intestinal sites, physiologic conditions, and temporality highlight the importance of finely-tuned interactions between host and microbiota.
In both the skin and intestine, microbial diversity is influenced by a plethora of exogenous factors, including diet, antibiotic use, and obesity (50–52). In the skin, treatment with topical or systemic antibiotics has been linked to shifts in the cutaneous microbiome. For example, use of topical antibiotics, such a bacitracin, neomycin, and polymyxin B (found in the commonly-used triple antibiotic ointment) lead to decreased commensal Staphylococcus strain in mice (53). Oral isotretinoin or tetracycline treatment leads to decreased abundance of Cutibacterium on the skin and the microbiome of sebaceous areas shifts to mimic that of dry sites, containing a greater proportion of Staphylococcus and Streptococcus species (54).
Diet is a strong driver of microbial composition in the intestine. An animal-based diet increases the abundance of bacteria that are bile-tolerant, such as Alistipes, Bilophila, and Bacteroides (50). In contrast, a vegan or vegetarian diet is associated with an increased prevalence of lactic acid bacteria, including Ruminococcus, Eubacterium rectale, and Roseburia (55). Prevotella species predominate in humans whose diets are high in carbohydrates and simple sugars (56). High fiber diets lead to a higher abundance of bacteria that ferment fiber into SCFAs, which have a broad range of beneficial effects, including immunomodulatory properties (57). Diet can even influence the circadian dynamics of intestinal microbiota: diet-induced obesity causes a dampening of diurnal variations in microbial composition (58).
The skin and intestine have developed symbiotic relationships with commensal microbes and established a homeostasis that balances tolerating commensal microbes while defending against pathogens.
In the skin, the presence of commensal bacteria is crucial for maintenance of a healthy cutaneous environment. In development, skin immune tolerance begins developing in the post-natal period when T reg cells begin expressing the pathogen-specific FOXP3 transcription factor, coinciding with commensal colonization (59). Later in development, the continued presence of skin commensal bacteria modulates production of numerous cytokines and AMPs that help to protect the skin against pathogens. For example, commensal bacteria such as S. epidermidis can induce production of various cytokines by IL-17+CD8+ T cells (60). S. epidermidis can also produce ligands that suppress inappropriate immune activation by inhibiting production of tumor necrosis factor-α and IL-6 (61). Recent work demonstrated that germ-free mice have decreased expression of Toll-like receptors (TLRs), AMPs, complement cascades, and IL-1 cytokine signaling in the skin, when compared to specific pathogen free mice (62).
It is also well-established that bacterial colonization is essential for maturation of the intestinal innate immune system and, as in the skin, commensal microbiota work in tandem with the immune system to protect the host against pathogens (63, 64). For example, Bacteroides fragilis (B. fragilis) and commensal Clostridium cluster such as IV and XIVa can accumulate Foxp3+ Treg cells in mice and help build immune tolerance to the commensal microbiome (65, 66). In addition, the lamina propria harbors macrophages, whose function is phagocytosis of pathogens. However, lamina propria-associated macrophages do not express as strong proinflammatory phagocytic responses as macrophages at other sites (67). This suggests adaptation of the host in response to a large population of commensal microbiota, which minimizes unnecessary inflammation (34).
The physical and chemical barriers discussed above are crucial in preventing tissue penetration of the microbes by decreasing direct contact between them. However, pathogenic microbes may gain access to the tissue when there is a breach of these barriers. Disruption of the skin barrier may occur through physical cut or toxic chemical exposure. Barrier disruption is also associated with chronic intestinal diseases such as inflammatory bowel disease, obesity, and diabetes, all of which can increase the intestinal permeability (68). In the following section, we discuss innate immune mechanisms upon barrier breach and how they are modulated by commensal bacteria.
Prompt recognition and eradication of pathogens are necessary to prevent infection. The innate immune system provides a first line of defense against pathogens upon barrier breach. Recent evidence has illuminated the role of commensal microbes in strengthening innate immune defense against pathogens (69). Pattern recognition receptors (PRRs) interact with microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) such as lipopolysaccharide (LPS) and peptidoglycan (PG) on bacteria and nucleic acids from bacteria (70). Upon activation of PRRs, downstream signaling leads to release of inflammatory cytokines or activation of immune cells. In the skin, PRRs are present on immune cells and keratinocytes. TLR stimulation can mediate direct antimicrobial action through the stimulation of macrophages to undergo phagocytosis and can also induce cytokines that mediate the differentiation of monocytes into macrophages and dendritic cells (70, 71). Commensal microbes secrete molecules which may act directly as TLR ligands; S. epidermidis secretes multiple small molecules that act as TLR2 and EGFR agonists, stimulating production of AMPs that have activity against group A Streptococcus and S. aureus (72–74).
AMPs play a critical role in innate immunity by acting under homeostatic conditions and destroying pathogenic microbes through various mechanisms (75). Keratinocytes, the major cell type in the skin, produce various AMPs (Figure 1) (49, 76). Human β-defensin-1 (hBD-1) is constitutively expressed by keratinocytes whereas hBD-2 and −3 are upregulated in response to inflammation (77–79). Human cathelicidin (hCAP-18) is cleaved and processed to the active form of antimicrobial peptide, LL-37 which then disrupts microbial membranes (80). Some specialized keratinocytes that make up appendages such as hair follicles, sweat glands, and sebaceous glands have various AMPs pertinent to their microenvironments (Figure 1). For example, dermcidin is traditionally thought to be a sweat-gland specific AMP, yet there are emerging evidences that it is also produced by sebaceous gland in humans and mice (81, 82). Sebaceous glands also produce cathelicidin and hBD-2 (83, 84). Commensal bacteria have been shown to secrete AMPs. S. epidermidis secretes phenol-soluble modulin γ and δ that have antibiotic effects on S. aureus (85). Commensal bacteria can also act on lipids secreted from sebaceous glands and hydrolyze them to free fatty acids (FFAs) (86). FFAs have intrinsic antibacterial effects against various Gram-positive bacteria; sapienic acid has activity against methicillin-resistant S. aureus (MRSA) (87). Furthermore, FFAs can induce sebocytes to upregulate expression of hBD-2 (88).
Just as external environmental effects are known to modulate skin microbial composition, environmental factors also regulate microbial recognition and AMP production in the skin. Ligand-dependent activation of the vitamin D receptor (VDR) is required for recruitment of macrophages to the injury site after wounding (89, 90). Genes coding for TLR2 are induced by the presence of 1,25-dihydroxy vitamin D3, which is regulated in part by exposure to UV light (83). Furthermore, vitamin D3-induced expression of TLR2 leads to cathelicidin production upon exposure to microbial components. Conversely, TLR2 activation can lead to increased expression of the VDR, which can be activated by vitamin D3 to produce cathelicidin (91). Vitamin D3-eluting wound nanodressings have even been shown to increased cathelicidin expression in human skin wounded explants (92).
Similarly to epidermal keratinocytes, IECs express PRRs, such as TLRS, NOD-like receptors (NLRs), and RIG-I-like receptors (RLRs) (93). PRR signaling is important both in homeostasis and in response to pathogenic bacteria, highlighting the diverse functions of innate immunity at steady-state and under disease conditions (94–96). PRRs also respond differently under homeostatic vs. inflammatory conditions, in part because of the presence of damage-associated molecular patterns (DAMPs), which are released by injured epithelial cells (97). Interestingly, steady-state activation of TLRs by commensal intestinal microbiota is also important for proper intestinal homeostasis. For example, mice deficient in TLRs, downstream signaling components of the TLR pathway, or normal commensal microbiota all displayed greater morbidity and mortality following intestinal epithelial disruption (95). Furthermore, TLR activation by commensal bacteria can enhance the protective function of tight junctions against pathogens by strengthening the zonula occludens-1 protein (94). The activated macrophages also signal repair pathway that promotes rapid enterocyte proliferation to repair the tissue defect by producing growth factors (34, 43). This highlights the importance of synergistic activity of commensal microbiota and host innate immunity in maintenance of a healthy epithelium.
In addition to producing barrier-protective mucins, EICs are also a rich source of AMPs (Figure 2) (2). Enterocytes produce AMPs including regenerating islet-derived protein IIIγ (REGIIIγ) and numerous β-defensins, which play diverse antimicrobial roles, including spatial segregation of bacteria (98, 99). Beyond their roles in barrier formation and production of AMPs, enterocytes also facilitate the translocation of secretory immunoglobulins, particularly IgA, across the intestinal wall (100). Paneth cells, present in intestinal crypts, produce additional AMPs, including α-defensins, lysozyme, and phospholipase A2 (98, 101, 102).
Intestinal commensals are able to induce AMP production in the intestine. Lactobacillus and probiotic E. coli strains are able to induce secretion of hBD-2 from enterocytes (103, 104). Commensal microbiota in the intestine are also capable of producing molecules that protect the host from chronic inflammatory diseases. For example, polysaccharide A, produced by B. fragilis, prevents inflammatory bowel disease (IBD) via an IL-10-producing CD4+ T cell-dependent mechanism (105). SCFAs produced by commensals of the genuses Bifidobacterium and Bacteroides interact with the G-protein-coupled receptor 43 (GPCR43) (106). Mice deficient in GPCR43 have impaired resolution of inflammation in models of IBD, arthritis and asthma. Similarly to vitamin D3-dependent regulation of AMPs in the skin, butyrate regulates AMP production in the intestine. Butyrate is a SCFA that is produced by fermentation of carbohydrates in the lumen by intestinal bacteria (107). Butyrate strongly induces cathelicidin production in colonic epithelial cells, and moderately induces h-BD1 and h-BD2 (108). Factors produced by commensal bacteria in the intestine may also prevent injury to IECs or facilitate intestinal repair. Numerous commensal bacteria produce compounds that prevent damage by noxious stimuli. Competence and sporulation factor (CSF) produced by Bacillus subtilis activates the mitogen-activated protein kinase (MAPK) pathway to protect epithelial cells from oxidative stress (109). Similarly, Lactobacillus rhamnosus produces two compounds, p75 and p40, which prevent cytokine-induced apoptosis of IECs through epidermal growth factor receptor (EGFR) signaling, which activates anti-apoptotic Akt/protein kinase B (110–112). Tight junction assembly is promoted by Bifidobacterium and butyrate from various bacteria, underscoring the ability of commensal bacteria to promote intestinal barrier function (113–115).
The skin and intestine are both active immune organs that are under constant environmental exposure. Therefore, complex regulatory mechanisms have evolved to maintain homeostasis. In addition to acting as physical barriers, epithelial cells of the skin and intestine produce AMPs, which act as endogenous antibiotics to protect against potential pathogens. Immune cells also constantly surveil both of these surfaces. More recently, it has been appreciated commensal microbiota may induce beneficial, tolerogenic immune responses under homeostasis or prime the immune system to fight against pathogens upon barrier breach. Some commensal bacteria may even produce AMPs on their own. An improved understanding of beneficial microbial-immune interactions has paved the way for new research involving exogenous supplementation of skin and intestinal microbial populations. For example, topical application of Gram-negative bacterial species obtained from healthy human volunteers improved atopic dermatitis in a mouse model (116). Manipulation of intestinal microbiota may be a promising therapeutic option for the treatment of numerous disease, including obesity, IBD, colorectal cancer, and liver disease (117). Although further studies will be needed to validate the safety and efficacy of a microbial-based therapeutic approach, it is clear that a healthy skin and intestinal microbiome is crucial for healthy epithelial homeostasis and immunity.
MC, ML, DN, and AM contributed to conception and design of the review. MC, ML, and DN performed initial literature search and wrote the first draft of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version. AM supervised all aspects of the review and manuscript writing and is the corresponding author.
AM was supported by R01AI139207 01, received funding from the Dermatology Foundation Research Grant and the Duke Physician-Scientist Strong Start Award. AM also received funding from Silab Inc. to support her laboratory, but the sponsor did not have any control over the content nor results of the study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Eyerich S, Eyerich K, Traidl-Hoffmann C, Biedermann T. Cutaneous barriers and skin immunity: differentiating a connected network. Trends Immunol. (2018) 39:315–27. doi: 10.1016/j.it.2018.02.004
2. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. (2014) 14:141–53. doi: 10.1038/nri3608
3. Gallo RL. Human skin is the largest epithelial surface for interaction with microbes. J Investig Dermatol. (2017) 137:1213–4. doi: 10.1016/j.jid.2016.11.045
4. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. (2011) 9:244–53. doi: 10.1038/nrmicro2537
6. Menon GK, Lee SE, Lee SH. An overview of epidermal lamellar bodies: novel roles in biological adaptations and secondary barriers. J Dermatol Sci. (2018) 92:10–7. doi: 10.1016/j.jdermsci.2018.03.005
7. Volksdorf T, Heilmann J, Eming SA, Schawjinski K, Zorn-Kruppa M, Ueck C, et al. Tight junction proteins claudin-1 and occludin are important for cutaneous wound healing. Am J Pathol. (2017) 187:1301–12. doi: 10.1016/j.ajpath.2017.02.006
8. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. (2005) 308:1635–8. doi: 10.1126/science.1110591
9. Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. (2015) 6:8292. doi: 10.1038/ncomms9292
10. Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci USA. (2010) 107:7419–24. doi: 10.1073/pnas.1001061107
11. Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. (2018) 50:103. doi: 10.1038/s12276-018-0126-x
12. Ali SM, Yosipovitch G. Skin pH: from basic science to basic skin care. Acta Derm Venereol. (2013) 93:261–7. doi: 10.2340/00015555-1531
13. Bouwstra JA, Gooris GS, Dubbelaar FE, Weerheim AM, Ponec M. pH, cholesterol sulfate, and fatty acids affect the stratum corneum lipid organization. J Investig Dermatol Symp Proc. (1998) 3:69–74. doi: 10.1038/jidsymp.1998.17
14. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. (2009) 324:1190–2. doi: 10.1126/science.1171700
15. Moran JC, Alorabi JA, Horsburgh MJ. Comparative transcriptomics reveals discrete survival responses of S. aureus and S. epidermidis to sapienic acid. Front Microbiol. (2017) 8:33. doi: 10.3389/fmicb.2017.00033
16. Korting HC. Das Säuremantelkonzept von Marchionini und die Beeinflussung der Resident-Flora der Haut durch Waschungen in Abhängigkeit vom pH-Wert. In: Hautreinigung mit Syndets. Berlin: Springer (1990). p. 93–103.
17. Fluhr JW, Kao J, Jain M, Ahn SK, Feingold KR, Elias PM. Generation of free fatty acids from phospholipids regulates stratum corneum acidification and integrity. J Investig Dermatol. (2001) 117:44–51. doi: 10.1046/j.0022-202x.2001.01399.x
18. McCauley HA, Guasch G. Three cheers for the goblet cell: maintaining homeostasis in mucosal epithelia. Trends Mol Med. (2015) 21:492–503. doi: 10.1016/j.molmed.2015.06.003
19. Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. (2013) 342:447–53. doi: 10.1126/science.1237910
20. Kurashima Y, Kiyono H. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu Rev Immunol. (2017) 35:119–47. doi: 10.1146/annurev-immunol-051116-052424
21. Knoop KA, Newberry RD. Goblet cells: multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. (2018) 11:1551–7. doi: 10.1038/s41385-018-0039-y
22. Finnie IA, Dwarakanath AD, Taylor BA, Rhodes JM. Colonic mucin synthesis is increased by sodium butyrate. Gut. (1995) 36:93–9. doi: 10.1136/gut.36.1.93
23. Fontaine N, Meslin JC, Lory S, Andrieux C. Intestinal mucin distribution in the germ-free rat and in the heteroxenic rat harbouring a human bacterial flora: effect of inulin in the diet. Br J Nutr. (1996) 75:881–92. doi: 10.1079/BJN19960194
24. Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastrointest Liver Physiol. (2013) 305:G341–7. doi: 10.1152/ajpgi.00046.2013
25. Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. (2016) 16:639–49. doi: 10.1038/nri.2016.88
26. Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. (2018) 16:143–55. doi: 10.1038/nrmicro.2017.157
27. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
28. Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. (2016) 165:854–66. doi: 10.1016/j.cell.2016.04.008
29. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. (2013) 498:367–70. doi: 10.1038/nature12171
30. Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev. (2014) 260:35–49. doi: 10.1111/imr.12186
31. Kastl AJ Jr, Terry NA, Albenberg LG, Wu GD. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol. (2019) 9:33–45. doi: 10.1016/j.jcmgh.2019.07.006
32. Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. (2005) 54:1093–101. doi: 10.1099/jmm.0.45935-0
33. Booijink CC, El-Aidy S, Rajilic-Stojanovic M, Heilig HG, Troost FJ, Smidt H, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. (2010) 12:3213–27. doi: 10.1111/j.1462-2920.2010.02294.x
34. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. (2010) 10:159–69. doi: 10.1038/nri2710
35. Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. (2013) 501:426–9. doi: 10.1038/nature12447
36. Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. (2004) 427:441–4. doi: 10.1038/nature02285
37. Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. (2005) 51:270–4. doi: 10.1007/s00284-005-0020-3
38. Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. (2008) 159:187–93. doi: 10.1016/j.resmic.2007.12.007
39. Stinson LF, Boyce MC, Payne MS, Keelan JA. The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front Microbiol. (2019) 10:1124. doi: 10.3389/fmicb.2019.01124
40. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
41. Malm G. Neonatal herpes simplex virus infection. Semin Fetal Neonatal Med. (2009) 14:204–8. doi: 10.1016/j.siny.2009.01.005
42. Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:852. doi: 10.1016/j.chom.2015.05.012
43. Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. (2004) 12:129–34. doi: 10.1016/j.tim.2004.01.001
44. Kirmiz N, Robinson RC, Shah IM, Barile D, Mills DA. Milk glycans and their interaction with the infant-gut microbiota. Annu Rev Food Sci Technol. (2018) 9:429–50. doi: 10.1146/annurev-food-030216-030207
45. Korpela K, Salonen A, Vepsalainen O, Suomalainen M, Kolmeder C, Varjosalo M, et al. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. (2018) 6:182. doi: 10.1186/s40168-018-0567-4
46. Jalanka-Tuovinen J, Salonen A, Nikkila J, Immonen O, Kekkonen R, Lahti L, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE. (2011) 6:e23035. doi: 10.1371/journal.pone.0023035
47. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
48. Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. (2014) 15:531. doi: 10.1186/s13059-014-0531-y
49. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. (2012) 488:178–84. doi: 10.1038/nature11319
50. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
51. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. (2013) 152:39–50. doi: 10.1016/j.cell.2012.10.052
52. Yallapragada SG, Nash CB, Robinson DT. Early-life exposure to antibiotics, alterations in the intestinal microbiome, and risk of metabolic disease in children and adults. Pediatr Ann. (2015) 44:e265–9. doi: 10.3928/00904481-20151112-09
53. SanMiguel AJ, Meisel JS, Horwinski J, Zheng Q, Grice EA. Topical antimicrobial treatments can elicit shifts to resident skin bacterial communities and reduce colonization by Staphylococcus aureus competitors. Antimicrob Agents Chemother. (2017) 61:e00774–17. doi: 10.1128/AAC.00774-17
54. Kelhala HL, Aho VTE, Fyhrquist N, Pereira PAB, Kubin ME, Paulin L, et al. Isotretinoin and lymecycline treatments modify the skin microbiota in acne. Exp Dermatol. (2018) 27:30–6. doi: 10.1111/exd.13397
55. Tomova A, Bukovsky I, Rembert E, Yonas W, Alwarith J, Barnard ND, et al. The effects of vegetarian and vegan diets on gut microbiota. Front Nutr. (2019) 6:47. doi: 10.3389/fnut.2019.00047
56. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. (2011) 334:105–8. doi: 10.1126/science.1208344
57. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
58. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. (2014) 20:1006–17. doi: 10.1016/j.cmet.2014.11.008
59. Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. (2015) 43:1011–21. doi: 10.1016/j.immuni.2015.10.016
60. Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. (2015) 520:104–8. doi: 10.1038/nature14052
61. Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. (2009) 15:1377–82. doi: 10.1038/nm.2062
62. Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, et al. Commensal microbiota modulate gene expression in the skin. Microbiome. (2018) 6:20. doi: 10.1186/s40168-018-0404-9
63. Fulde M, Hornef MW. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev. (2014) 260:21–34. doi: 10.1111/imr.12190
64. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. (2013) 14:685–90. doi: 10.1038/ni.2608
65. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. (2010) 107:12204–9. doi: 10.1073/pnas.0909122107
66. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
67. Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Investig. (2005) 115:66–75. doi: 10.1172/JCI200519229
68. Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke JD, Serino M, et al. Intestinal permeability–a new target for disease prevention and therapy. BMC Gastroenterol. (2014) 14:189. doi: 10.1186/s12876-014-0189-7
69. Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Investig Dermatol. (2011) 131:1974–80. doi: 10.1038/jid.2011.182
70. Krutzik SR, Tan B, Li H, Ochoa MT, Liu PT, Sharfstein SE, et al. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med. (2005) 11:653–60. doi: 10.1038/nm1246
71. Doyle SE, O'Connell RM, Miranda GA, Vaidya SA, Chow EK, Liu PT, et al. Toll-like receptors induce a phagocytic gene program through p38. J Exp Med. (2004) 199:81–90. doi: 10.1084/jem.20031237
72. Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Investig Dermatol. (2010) 130:2211–21. doi: 10.1038/jid.2010.123
73. Li D, Lei H, Li Z, Li H, Wang Y, Lai Y. A novel lipopeptide from skin commensal activates TLR2/CD36-p38 MAPK signaling to increase antibacterial defense against bacterial infection. PLoS ONE. (2013) 8:e58288. doi: 10.1371/journal.pone.0058288
74. Wanke I, Steffen H, Christ C, Krismer B, Gotz F, Peschel A, et al. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Investig Dermatol. (2011) 131:382–90. doi: 10.1038/jid.2010.328
75. Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. (2003) 55:27–55. doi: 10.1124/pr.55.1.2
76. Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun. (2005) 73:6771–81. doi: 10.1128/IAI.73.10.6771-6781.2005
77. Fulton C, Anderson GM, Zasloff M, Bull R, Quinn AG. Expression of natural peptide antibiotics in human skin. Lancet. (1997) 350:1750–1. doi: 10.1016/S0140-6736(05)63574-X
78. Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. (1997) 387:861. doi: 10.1038/43088
79. Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. (2001) 276:5707–13. doi: 10.1074/jbc.M008557200
80. Sorensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. (2001) 97:3951–9. doi: 10.1182/blood.V97.12.3951
81. Schittek B, Hipfel R, Sauer B, Bauer J, Kalbacher H, Stevanovic S, et al. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat Immunol. (2001) 2:1133–7. doi: 10.1038/ni732
82. Dahlhoff M, Zouboulis CC, Schneider MR. Expression of dermcidin in sebocytes supports a role for sebum in the constitutive innate defense of human skin. J Dermatol Sci. (2016) 81:124–6. doi: 10.1016/j.jdermsci.2015.11.013
83. Nagy I, Pivarcsi A, Kis K, Koreck A, Bodai L, McDowell A, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microb Infect. (2006) 8:2195–205. doi: 10.1016/j.micinf.2006.04.001
84. Lee DY, Yamasaki K, Rudsil J, Zouboulis CC, Park GT, Yang JM, et al. Sebocytes express functional cathelicidin antimicrobial peptides and can act to kill Propionibacterium acnes. J Investig Dermatol. (2008) 128:1863–6. doi: 10.1038/sj.jid.5701235
85. Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Investig Dermatol. (2010) 130:192–200. doi: 10.1038/jid.2009.243
86. Marples RR, Downing DT, Kligman AM. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J Investig Dermatol. (1971) 56:127–31. doi: 10.1111/1523-1747.ep12260695
87. Drake DR, Brogden KA, Dawson DV, Wertz PW. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res. (2008) 49:4–11. doi: 10.1194/jlr.R700016-JLR200
88. Nakatsuji T, Kao MC, Zhang L, Zouboulis CC, Gallo RL, Huang CM. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating beta-defensin-2 expression. J Investig Dermatol. (2010) 130:985–94. doi: 10.1038/jid.2009.384
89. Song L, Papaioannou G, Zhao H, Luderer HF, Miller C, Dall'Osso C, et al. The vitamin D receptor regulates tissue resident macrophage response to injury. Endocrinology. (2016) 157:4066–75. doi: 10.1210/en.2016-1474
90. Schauber J, Dorschner RA, Coda AB, Buchau AS, Liu PT, Kiken D, et al. Injury enhances TLR2 function and antimicrobial peptide expression through a vitamin D-dependent mechanism. J Clin Investig. (2007) 117:803–11. doi: 10.1172/JCI30142
91. Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. (2006) 311:1770–3. doi: 10.1126/science.1123933
92. Jiang J, Zhang Y, Indra AK, Ganguli-Indra G, Le MN, Wang H, et al. 1alpha,25-dihydroxyvitamin D3-eluting nanofibrous dressings induce endogenous antimicrobial peptide expression. Nanomedicine. (2018) 13:1417–32. doi: 10.2217/nnm-2018-0011
93. Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. (2010) 10:131–44. doi: 10.1038/nri2707
94. Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology. (2004) 127:224–38. doi: 10.1053/j.gastro.2004.04.015
95. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. (2004) 118:229–41. doi: 10.1016/j.cell.2004.07.002
96. Fukata M, Arditi M. The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol. (2013) 6:451–63. doi: 10.1038/mi.2013.13
97. Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and non-pathogenic microbes by the innate immune system. Cell Host Microbe. (2009) 6:10–21. doi: 10.1016/j.chom.2009.06.007
98. Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. (2012) 12:503–16. doi: 10.1038/nri3228
99. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. (2011) 334:255–8. doi: 10.1126/science.1209791
100. Kitamura T, Garofalo RP, Kamijo A, Hammond DK, Oka JA, Caflisch CR, et al. Human intestinal epithelial cells express a novel receptor for IgA. J Immunol. (2000) 164:5029–34. doi: 10.4049/jimmunol.164.10.5029
101. Ouellette AJ. Paneth cell alpha-defensins in enteric innate immunity. Cell Mol Life Sci. (2011) 68:2215–29. doi: 10.1007/s00018-011-0714-6
102. Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. (2011) 9:356–68. doi: 10.1038/nrmicro2546
103. Schlee M, Harder J, Koten B, Stange EF, Wehkamp J, Fellermann K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin Exp Immunol. (2008) 151:528–35. doi: 10.1111/j.1365-2249.2007.03587.x
104. Mondel M, Schroeder BO, Zimmermann K, Huber H, Nuding S, Beisner J, et al. Probiotic E. coli treatment mediates antimicrobial human beta-defensin synthesis and fecal excretion in humans. Mucosal Immunol. (2009) 2:166–72. doi: 10.1038/mi.2008.77
105. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. (2008) 453:620–5. doi: 10.1038/nature07008
106. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. (2009) 461:1282–6. doi: 10.1038/nature08530
107. Scheppach W. Effects of short chain fatty acids on gut morphology and function. Gut. (1994) 35(Suppl. 1):S35–8. doi: 10.1136/gut.35.1_Suppl.S35
108. Schauber J, Dorschner RA, Yamasaki K, Brouha B, Gallo RL. Control of the innate epithelial antimicrobial response is cell-type specific and dependent on relevant microenvironmental stimuli. Immunology. (2006) 118:509–19. doi: 10.1111/j.1365-2567.2006.02399.x
109. Okamoto K, Fujiya M, Nata T, Ueno N, Inaba Y, Ishikawa C, et al. Competence and sporulation factor derived from Bacillus subtilis improves epithelial cell injury in intestinal inflammation via immunomodulation and cytoprotection. Int J Colorectal Dis. (2012) 27:1039–46. doi: 10.1007/s00384-012-1416-8
110. Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. (2002) 277:50959–65. doi: 10.1074/jbc.M207050200
111. Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. (2007) 132:562–75. doi: 10.1053/j.gastro.2006.11.022
112. Yoda K, Miyazawa K, Hosoda M, Hiramatsu M, Yan F, He F. Lactobacillus GG-fermented milk prevents DSS-induced colitis and regulates intestinal epithelial homeostasis through activation of epidermal growth factor receptor. Eur J Nutr. (2014) 53:105–15. doi: 10.1007/s00394-013-0506-x
113. Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in CaCo-2 cell monolayers. J Nutr. (2009) 139:1619–25. doi: 10.3945/jn.109.104638
114. Hsieh CY, Osaka T, Moriyama E, Date Y, Kikuchi J, Tsuneda S. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol Rep. (2015) 3:e12327. doi: 10.14814/phy2.12327
115. Karczewski J, Troost FJ, Konings I, Dekker J, Kleerebezem M, Brummer RJ, et al. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol. (2010) 298:G851–9. doi: 10.1152/ajpgi.00327.2009
116. Myles IA, Williams KW, Reckhow JD, Jammeh ML, Pincus NB, Sastalla I, et al. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight. (2016) 1:86955. doi: 10.1172/jci.insight.86955
Keywords: skin, intestine, microbiome, innate immunity, AMPs
Citation: Coates M, Lee MJ, Norton D and MacLeod AS (2019) The Skin and Intestinal Microbiota and Their Specific Innate Immune Systems. Front. Immunol. 10:2950. doi: 10.3389/fimmu.2019.02950
Received: 07 September 2019; Accepted: 02 December 2019;
Published: 17 December 2019.
Edited by:
Emilio Luis Malchiodi, University of Buenos Aires, ArgentinaReviewed by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranCopyright © 2019 Coates, Lee, Norton and MacLeod. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amanda S. MacLeod, YW1hbmRhLm1hY2xlb2RAZHVrZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.