
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 26 November 2019
Sec. Viral Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.02736
This article is part of the Research Topic Immune Responses to Chikungunya Virus View all 7 articles
 Natalie A. Prow1,2†
Natalie A. Prow1,2† Thiago D. C. Hirata3†
Thiago D. C. Hirata3† Bing Tang1
Bing Tang1 Thibaut Larcher4
Thibaut Larcher4 Pamela Mukhopadhyay1
Pamela Mukhopadhyay1 Tiago Lubiana Alves3
Tiago Lubiana Alves3 Thuy T. Le1
Thuy T. Le1 Joy Gardner1
Joy Gardner1 Yee Suan Poo1
Yee Suan Poo1 Eri Nakayama1,5
Eri Nakayama1,5 Viviana P. Lutzky1
Viviana P. Lutzky1 Helder I. Nakaya3
Helder I. Nakaya3 Andreas Suhrbier1,2*
Andreas Suhrbier1,2*Chikungunya virus (CHIKV) is a mosquito transmitted alphavirus associated with a robust systemic infection and an acute inflammatory rheumatic disease. A high fiber diet has been widely promoted for its ability to ameliorate inflammatory diseases. Fiber is fermented in the gut into short chain fatty acids such as acetate, propionate, and butyrate, which enter the circulation providing systemic anti-inflammatory activities. Herein we show that mice fed a high fiber diet show a clear exacerbation of CHIKV arthropathy, with increased edema and neutrophil infiltrates. RNA-Seq analyses illustrated that a high fiber diet, in this setting, promoted a range of pro-neutrophil responses including Th17/IL-17. Gene Set Enrichment Analyses demonstrated significant similarities with mouse models of inflammatory psoriasis and significant depression of macrophage resolution phase signatures in the CHIKV arthritic lesions from mice fed a high fiber diet. Supplementation of the drinking water with butyrate also increased edema after CHIKV infection. However, the mechanisms involved were different, with modulation of AP-1 and NF-κB responses identified, potentially implicating deoptimization of endothelial barrier repair. Thus, neither fiber nor short chain fatty acids provided benefits in this acute infectious disease setting, which is characterized by widespread viral cytopathic effects and a need for tissue repair.
A beneficial anti-inflammatory role for a high fiber diet is well-described for a large range of largely non-infectious disease settings in murine models (1–7). The use of high fiber diets to ameliorate human diseases is thus being actively pursued (8, 9), in particularly for autoimmune conditions (10, 11). Some evidence for benefit in humans has emerged, although results have often been inconclusive (12).
A high fiber diet changes the gut microbiome, with a number of studies in mice (3, 13, 14) and humans (15, 16) detailing the changes in bacterial species compositions. Bacterial fermentation of fiber (primarily undigested and/or indigestible carbohydrate) results in the production of short chain fatty acids (SCFA) such as acetate, propionate and butyrate, which enter the circulation and are believed to be the key players in dietary fiber-mediated systemic anti-inflammatory activities (8, 10, 17). SCFAs mediate effects on a number of cells including T cells (18, 19) particularly regulatory T cells (20), macrophages (21–23) and endothelial cells (24). Butyrate in particular has been shown to provide anti-inflammatory activities in a range of settings (5, 25–27), including non-infectious arthritic diseases (28–30). SCFAs are transported into cells via a series of receptors, with butyrate believed to act as an intracellular inhibitor of histone deacetylases (HDACs), with NF-κb (31), and AP-1 also targeted in some settings (32–34).
Chikungunya virus (CHIKV) belongs to a group of mosquito-borne arthritogenic alphaviruses that include the primarily Australian Ross River and Barmah Forest viruses, the African o'nyong-nyong virus, the Sindbis group of viruses, and the South American Mayaro virus (35). The largest documented outbreak of CHIKV disease ever recorded caused more than 10 million cases and began in 2004 in Africa and reached more than 100 countries in Africa, Asia, and the Americas, with small outbreaks also seen in Europe (36). Symptomatic infection of adults with CHIKV is nearly always associated with acute and often chronic polyarthralgia and/or polyarthritis, which can be debilitating and usually lasts weeks to months, occasionally longer. Other common symptoms include fever, rash, and myalgia (36). At present, no particularly effective drug or licensed vaccine is available for human use for any of these alphaviruses; although paracetamol/acetaminophen and non-steroidal anti-inflammatory drugs can provide relief from rheumatic symptoms and several CHIKV vaccines are in development (36).
CHIKV infection usually results in a 5–7 days long viremia, which is primarily controlled by a rapid type I IFN response (37, 38) and subsequently by anti-viral antibodies (36). A large range of cell types are infected in vivo including fibroblasts, muscle cells, endothelial cells, and macrophages (39). CHIKV infection usually results in cell death or cytopathic effects (CPE), mainly apoptosis and to a lesser extent necroptosis and pyroptosis, with connective tissue damage also evident during the viremic period in humans (36, 40). Infection drives a systemic pro-inflammatory response with the up-regulation of multiple mediators (36, 41, 42). CHIKV arthropathy is generally viewed as an immunopathology (43–45), with the pro-inflammatory arthritogenic response sharing similarities with rheumatoid arthritis (46). The inflammatory arthropathy is triggered by viral infection of joint tissues and is associated with a robust mononuclear cell infiltrate comprised primarily of monocytes, macrophages, NK cells, and T cells (47, 48). CD4 T cells are important for driving CHIKV arthritis (36), with Tregs associated with disease amelioration (49, 50). Macrophages/monocytes also play an important role in the arthritic immunopathology (36), with the pro-inflammatory response to CHIKV infection in peripheral blood shown to be monocyte centric (41, 51). However, macrophages are also required for resolution of inflammation, both generally (52–54) and specifically for CHIKV arthritic inflammation (45).
We have developed an adult C57BL/6J (wild-type) mouse model of acute and chronic CHIKV infection and hind foot arthritis that recapitulates many aspects of human disease (47, 55). RNA-Seq and bioinformatics studies in CHIKV patients (41) has also illustrated that this mouse model largely recapitulates (42) many of the inflammatory signatures seen in humans. CHIKV is able to replicate to high titers in humans with viremias up to 2.9 × 108 pfu/ml (56) and even higher in the elderly (1010 viruses per ml of blood) (57). Similar titers are reached in the feet in the mouse model (47), with up to 8% of the polyadenylated RNA in the infected feet being of viral origin (42). The mouse model has been widely exploited for testing new interventions (43, 58–65), and is used herein to determine the potential for modulating CHIKV arthropathy with high fiber diet and SCFAs. Only a few studies (66, 67) have addressed the question of whether high fiber diet and/or SCFAs can provide anti-inflammatory benefits in infectious disease settings.
C57BL/6J mice (6–8 weeks) were purchased from the Animal Resources Center (Canning Vale, WA, Australia). Female mice were inoculated with 104 CCID50 of the Reunion Island isolate (LR2006-OPY1) in 40 μl of medium (RPMI1640 supplemented with 2% fetal calf serum), s.c. into both hind feet as described previously (47, 55). The virus (GenBank KT449801) was prepared in C6/36 cells (55). Serum viremia was determined by CCID50 assay using C6/36 and Vero cells as described (37, 55). Foot swelling was measured using digital calipers and is presented as a group average of the percentage increase in foot height times width for each foot compared with the same foot on day 0 (55).
qRT PCR was undertaken as described (55) using CHIKV E1 primers. Each sample was analyzed in duplicate and normalized to RPL13A mRNA levels.
High and no fiber diets were supplied by Specialty Feeds (Glen Forrest, WA, Australia); the formulations are shown Supplementary Figures 1, 2 and were formulated to have similar digestible energy contents. Water for these later groups was acidified to pH 3–4 as per standard animal house practice. Drinking water was adjusted to pH 7 and supplemented with 200 mM of the sodium salt of the indicated SCFA (Sigma Aldrich, St Louis, MO, USA) (19, 68, 69). Drinking water was changed every 2–3 days. Mice were fed these diets and/or had SCFA supplementations in their drinking water for at least 3 weeks prior to CHIKV infection.
C57BL/6 mice were infected with CHIKV as described above, and whole feet (cut above the ankle) harvested on day 6.5 post infection. Tissues were placed in RNAlater (Life Technologies, Carlsbad, CA, USA) overnight at 4°C and then homogenized into TRIzol (Invitrogen) and RNA extracted as described (42). There were four groups; high fiber and no fiber diets (water pH 3–4), and butyrate and water (pH 7) no fiber diets. For each group three biological replicates were created by pooling equal amounts of RNA from 3 to 4 feet from 3 to 4 mice. A total of 12 pooled RNA samples were DNase treated using RNAse-Free DNAse Set (Qiagen, Hilden, Germany), purified using an RNeasy MinElute Kit following the manufacturers' instructions.
Library preparation and sequencing were conducted by the Australian Genome Research Facility (Melbourne, Australia). cDNA libraries were prepared using a TruSeq RNA Sample Prep Kit (v2) (Illumina Inc. San Diego, USA), which included isolation of poly-adenylated RNA using oligo-dt beads. cDNA libraries were sequenced from both ends (100 bp) using Illumina HiSeq 2000 Sequencer (Illumina Inc.). The CASAVA v1.8.2 pipeline was used to separate the bar-coded sequences and extract 100 base pair, paired end reads into FASTQ files.
The read counts were used to determine gene expression and identify differentially expressed genes (DEGs) using R packages (R version 3.2.0) “edgeR” (3.18.1) and “limma” (3.32.7). (https://bioconductor.org/packages/release/bioc/html/edgeR.html). The default TMM normalization method of edgeR was used to normalize the counts. The GLM model was used to perform differential expression comparison between the groups. Genes that had >1 CPM in at least three samples were retained for further analysis. Differentially gene expression was considered significant if the Benjamini–Hochberg corrected p-value (i.e., FDR or q value) was <0.05. DEGs (q < 0.05) were analyzed by Ingenuity Pathway Analysis and Integrated System for Motif Activity Response Analysis (ISMARA) (70) as described (42, 71). ISMARA was undertaken by uploading the RNA-Seq fastq files, identifying the replicates (allowing averaging, n = 3) and undertaking pair-wise comparisons; high vs. no fiber (normal water) and butyrate vs. water (no fiber diet).
Enrichment analyses were performed using GSEA from fgsea (v1.10.0) R package (72), using up and down-regulated DEGs from the high fiber vs. no fiber diet comparison. The program limma (v3.40.2) R package (73) was used to determine the fold change (in all the genes) in datasets from murine models of inflammatory psoriasis models (GSE27628). These gene sets were ranked by log2 fold-change and considered as gene ranks in the GSEA.
Macrophage resolution phase signature. The microarray data (Gene Express accession number E-MEXP-3189) posted by Stables et al. was analyzed as described (52) to determine DEGs up-regulated in resolution phase macrophages (rM) when compared with naïve macrophages and inflammatory macrophages (Stables et al. provided a list of rM DEGs, but not the direction of expression change). The 146 up-regulated DEGs in rM were used in a GSEA with the 27,537 gene set (ranked by log2 fold-change) obtained by RNA-Seq analysis comparing feet day 7 post CHIKV infection with feet from mock infected mice (42). The up-regulated DEGs in rM were also used in a GSEA with the 34,624 gene set (ranked by log2 fold-change) obtained by RNA-Seq comparing CHIKV arthritis in mice fed a high and no fiber diet (feet day 6.5 post CHIKV infection) (Supplementary Table 1a).
Histology, immunohistochemistry and quantitation were performed as described previously (45, 47, 71). Briefly, feet were fixed in paraformaldehyde, decalcified and embedded in paraffin, and sections stained with hematoxylin and eosin (H&E). Sections were scanned using Aperio AT Turbo (Aperio, Vista, CA) and analyzed using Aperio ImageScope software (v10) and the Positive Pixel Count v9 algorithm. Strong blue divided by total red pixels (default settings) represents a measure of cellular infiltration as leukocytes have a high nuclear to cytoplasmic area ratio.
For immunohistochemistry, sections were stained with rat anti-mouse Ly6G (catalog number NMP-R14; Abcam, Cambridge, MA, USA), with detection using Warp Red Chromogen Kit (Biocare Medical, Concord, CA, USA).
Statistics were performed using IBM SPSS Statistics (version 19). For mouse data the t-test was used if the difference in the variances was <4, skewness was >-2, and kurtosis was <2. Where the data was non-parametric and the distributions were similar the Kruskal–Wallis test was used, otherwise the Kolmogorov–Smirnov test was used. The Related Samples Wilcoxon Signed Rank test was used instead of a paired t-test as the paired data being compared was non-parametric.
To investigate the effects of a high fiber diet on CHIKV arthritis, adult C57BL/6J mice were fed a high fiber, standard and no fiber diet for 3 weeks and were then infected with CHIKV as described (42, 47). Foot swelling was significantly higher in mice on a high fiber diet than in mice on a low fiber diet, with mice on a standard diet showing an intermediate phenotype (Figure 1A). Viremia in these groups of animals was not significantly different (Figure 1B).
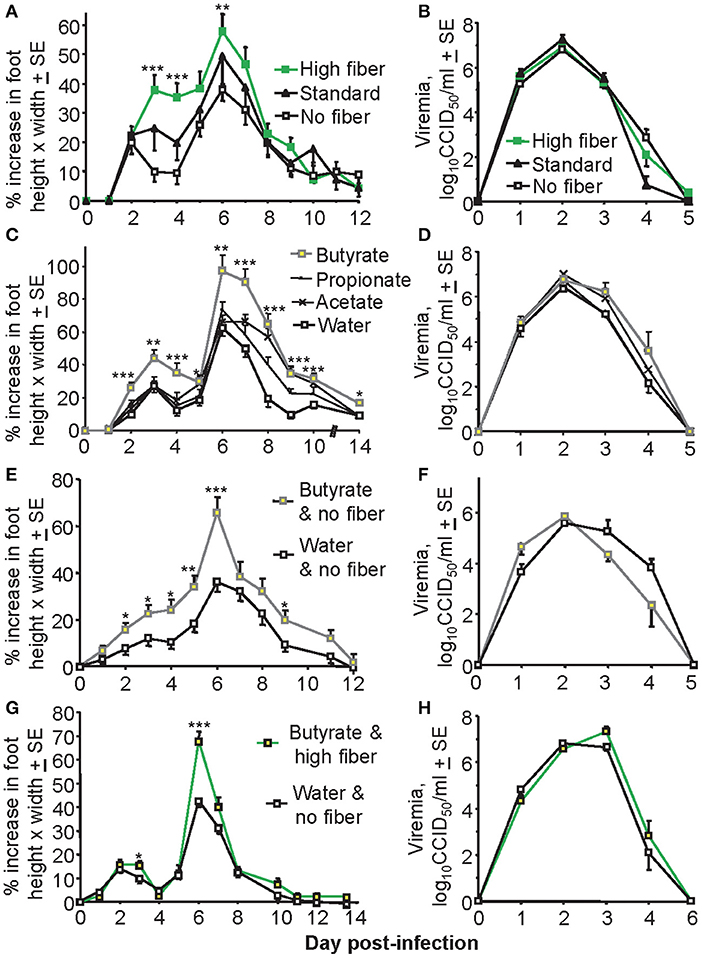
Figure 1. Diet affects foot swelling but not viremia. (A) Mice (C57BL/6J) were fed on either a high fiber diet, a standard diet or a no fiber diet for 3 weeks and were then infected with CHIKV and maintained on the same diet for the duration of the experiment. Foot swelling was measured over time and was higher in the high fiber group compared with the no fiber group; ***p < 0.001, **p = 0.008 (Day 7 approached significance p = 0.051), t-tests; n = 28–30 feet from 14 to 15 mice, with data from two independent experiments shown. (B) Viremia for the mice described in (A). (C) Mice were fed on a standard diet and water was supplemented with 200 mM of the indicated SCFA for the duration of the experiment. Foot swelling was measured over time. Mice drinking butyrate had significantly increased foot swelling than control mice (standard water); ***p < 0.001, **p ≤ 0.003, *p < 0.04, t-tests; n = 12 feet from six mice. (D) Viremia for the mice described in (C). (E) Mice were fed on a no fiber diet and the drinking water was supplemented with butyrate, or were fed a no fiber diet and supplied water for the duration of the experiment. After 3 weeks mice were infected with CHIKV and foot swelling measured. Butyrate significantly increased foot swelling; ***p < 0.001, **p = 0.01, *p < 0.04, t-tests; n = 22–24 feet from 11 to 12 mice, with data from two independent experiments shown. (F) Viremia for the mice treated as described in (E); n = 6 mice, one experiment. (G) Mice were fed on a high fiber diet and the drinking water was supplemented with butyrate, or mice were fed on a no fiber diet and supplied unsupplemented water. After 3 weeks mice were infected with CHIKV and foot swelling measured. Butyrate/high fiber showed significantly increased foot swelling; ***p < 0.001, *p < 0.04, t-tests; n = 24 feet from 12 mice, with data from two independent experiments shown. (H) Viremia for mice treated as described in (G); n = 6 mice, one experiment.
Dietary fiber is fermented into SCFA, with supply of SCFA in drinking water frequently used to try and recapitulate the anti-inflammatory effects of a high fiber diet (28–30). Butyrate supplied in the drinking water to animals on a standard diet significantly increased foot swelling, with propionate and acetate being less active (Figure 1C). Again viremia was unaffected (Figure 1D). A similar effect on foot swelling was observed when mice on a no fiber diet were given butyrate to drink (Figure 1E), with viremia again not significantly affected (Figure 1F). When butyrate and a high fiber diet were combined no overt additive effects were observed (Figure 1G); the increase in mean maximum foot swelling (on day 6) mediated by butyrate was about 30% in Figures 1C,E and was also about 30% in Figure 1G. Viremia was again unaffected (Figure 1H).
Overall the most consistent statistically significant increase in foot swelling mediated by fiber or butyrate was on day 6, with days 6–7 usually representing the period of peak foot swelling that corresponds with a pronounced inflammatory infiltrate (45, 47).
To determine whether the effects of fiber and butyrate on foot swelling were mediated by differences in viral loads in the foot tissues, CHIKV RNA levels were determined in feet by qRT PCR. CHIKV RNA levels were not significantly altered by a high fiber diet (Figure 2A) or butyrate in the drinking water (Figure 2B). This was true for both acute arthritis (day 6.5) and chronic arthritis, nominally measured on day 30 post infection (55) (Figures 2A,B). The differences in foot swelling seen in Figure 1 could thus not be accounted for by differences in viremia or viral loads in the feet.
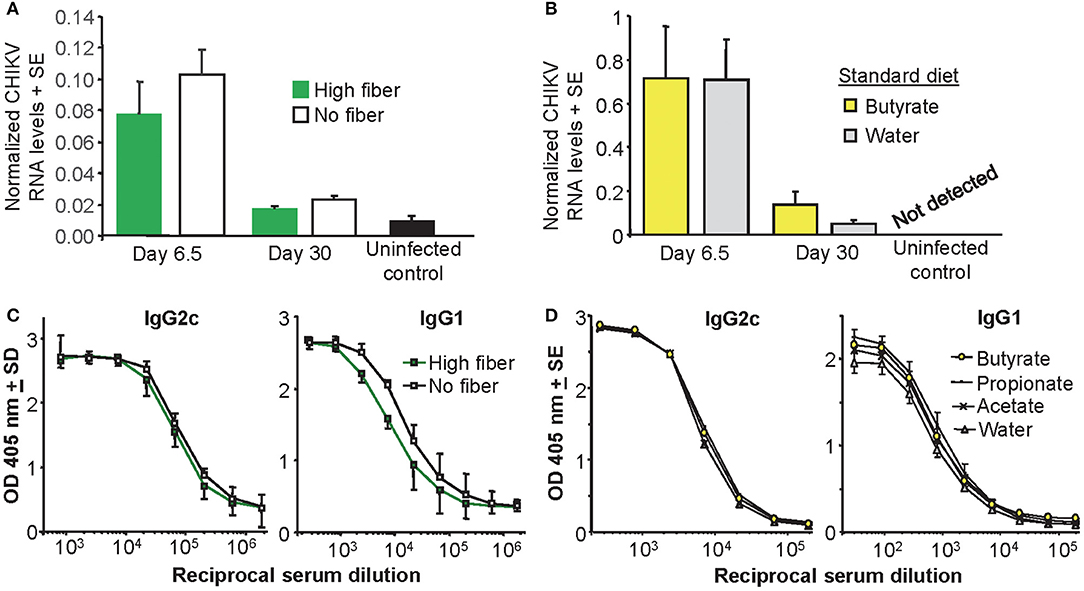
Figure 2. Viral loads and antibody responses. (A) Mice were fed on a high fiber or a no fiber diet for 3 weeks and were then infected with CHIKV (or left uninfected as controls). On day 6.5 and day 30 post infection feet were analyzed for CHIKV RNA by qRT PCR. Diet did not significantly affect CHIKV RNA levels (n = 4 mice and feet per group). (B) Mice were fed on a standard fiber and water was supplemented with butyrate or left unsupplemented (Water) and after 3 weeks mice were infected with CHIKV (or left uninfected as controls). Butyrate did not significant affect CHIKV RNA levels (n = 6 mice and feet per group). (C) Mice fed as for (A) with serum taken on day 30 post infection and assayed by ELISA for anti-CHIKV IgG2c or IgG1 antibody levels (n = 6 per group). (D) Mice with water supplementation as in (B), with serum taken on day 30 post infection and assayed by ELISA for anti-CHIKV IgG2c or IgG1 antibody levels (n = 6 per group).
Mice fed a high fiber diet showed no significant differences in their anti-CHIKV IgG2c or IgG1 responses post infection, when compared with mice fed a no fiber diet (Figure 2C). Although the IgG1 levels [associated with Th2 (74)] appeared a little lower (Figure 2C, IgG1), this did not reach significance when 50% end point titers in the two groups were compared. SCFA supplementation in the water supply also had no significant impact on anti-CHIKV IgG2c or IgG1 antibody titers (Figure 2D). These data failed to provide evidence of a significant effect of a high fiber diet or drinking butyrate on antibody responses and the Th1/Th2 balance in this CHIKV infection setting. These observations contrast with non-infectious disease settings where butyrate enhanced Th1 responses (75–77).
CHIKV arthritis is characterized by a pronounced infiltrate of mononuclear cells into joint tissues (35, 45, 47, 78). The extent of the infiltrate can be quantified using the Aperio Positive Pixel Count Algorithm on digital scans of H&E stained foot sections, with infiltrating leukocytes having a higher blue (nuclear) to red (cytoplasmic) ratio (42, 55). As expected, substantial and significant infiltrates were evident after CHIKV infection; however, the high fiber and no fiber groups were not significantly different (Figure 3A). A clear increase in edema was, however, evident by H&E staining in the high fiber group (Figure 3B; other examples are shown in Supplementary Figure 3). This increase in edema likely accounts for the increased foot swelling seen in this group (Figure 1A). CHIKV-induced edema around peripheral joints is well-described in humans (79–82) and is recapitulated in this adult wild-type mouse model (47).
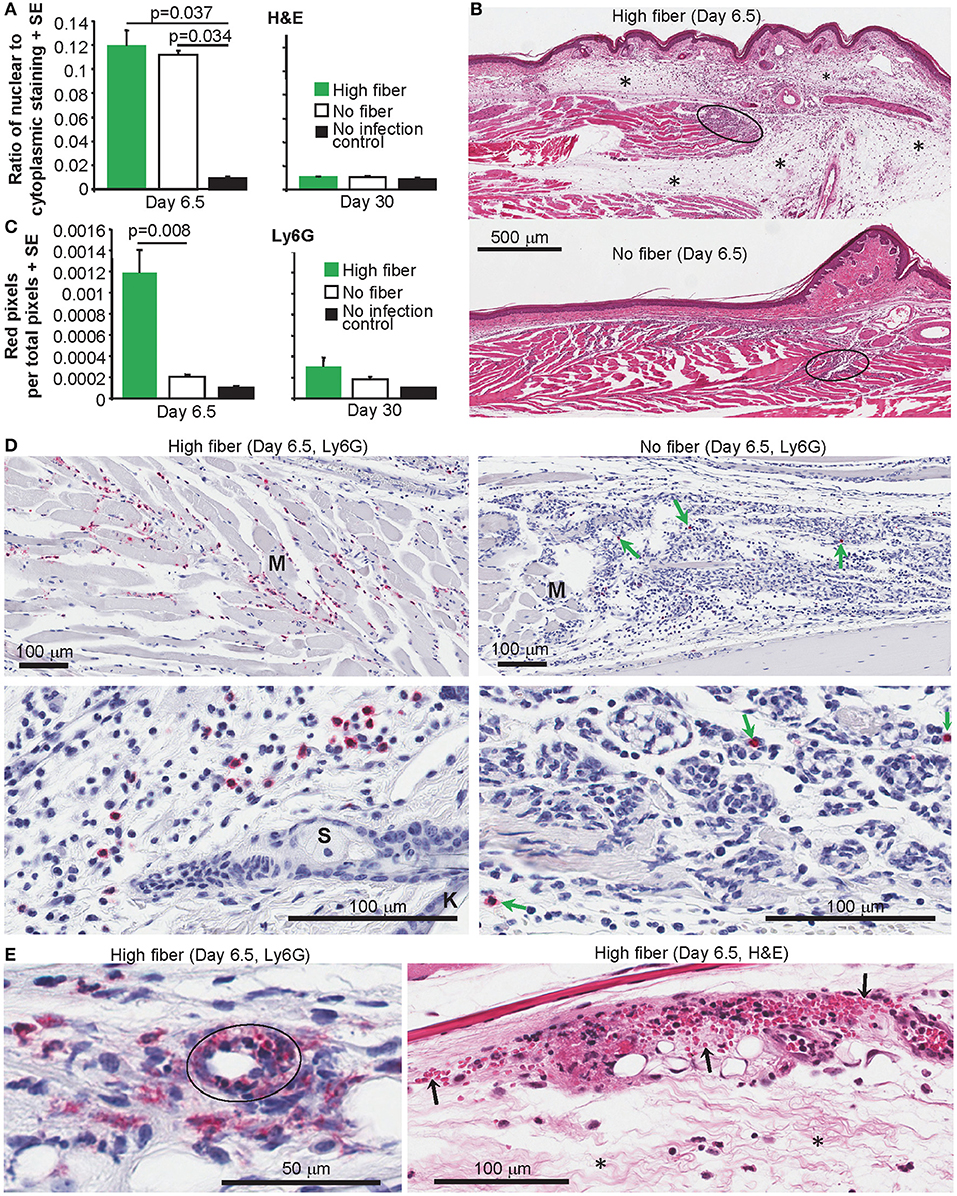
Figure 3. Histology and IHC of feet from CHIKV-infected mice fed high and no fiber diets. (A) Mice were fed with high fiber or no fiber diet and infected with CHIKV or left uninfected (No infection control) (as in Figure 1A). On day 6.5 and day 30 feet from separated groups of mice were examined by histology and H&E staining. Aperio pixel count was used to determine the ratio of strong blue (nuclear) to red (cytoplasmic) staining; a measure of the cellular infiltrate. Statistics by Kolmogorov Smirnov test (p = 0.037, n = 4 feet and mice per group) and Kruskal–Wallis test (p = 0.034, n = 3/4 feet and mice per group). There were no significant differences between any groups on day 30 post infection (n = 4 feet and mice per group). (B) Examples of H&E staining of mice examined in (A). Black ovals indicate areas of high density infiltrates in muscle tissues. Asterisks (*) indicate areas of edema. (C) Feet from groups of mice as in (A) were analyzed by IHC and the neutrophil-specific anti-Ly6G antibody. Anti-Ly6G staining was detected using Warp Red, with red staining quantified by Aperio pixel count. Statistics by Kolmogorov Smirnov test (Day 6.5, p = 0.008, n = 4–9 feet and mice per group). There were no significant differences between any groups on day 30 post infection (n = 5/6 feet and mice per group). (D) Examples of anti-Ly6G IHC (Red) in muscle (top panels) and subcutaneous tissues (bottom panels). M, a muscle bundle; K, keratinocyte epidermal skin layer; S, Sebaceous gland. (E) Anti-Ly6G IHC (Red) showing a blood vessel (black oval) with intramural neutrophils and indications of parietal necrosis. H&E staining showing hemorrhage (extravascular red blood cells indicated by arrows) and edema (*).
A characteristic feature of alphaviral arthritides is the general paucity of neutrophils in the arthritic lesions (45, 83, 84). In contrast, neutrophils are a prominent feature of autoimmune arthritides, such as rheumatoid arthritis (85). We have previously reported that in CCR2−/− mice CHIKV arthritis is exacerbated by an increase in infiltrating neutrophils (45). As neutrophils can promote edema in various settings (86–89), foot sections from arthritic feet of CHIKV-infected mice fed high and no fiber diets were analyzed by immunohistochemistry (IHC) using anti-Ly6G staining [a neutrophil specific marker (45)]. Sections were digitally scanned and analyzed by Aperio Positive Pixel Count Algorithm. Significantly more anti-Ly6G staining was evident in the feet of mice (on day 6.5 post infection) fed a high fiber diet when compared to feet of mice fed a no fiber diet (Figure 3C, Day 6.5). No significant difference between groups was seen on day 30 post infection (Figure 3C, Day 30). Examples of the anti-Ly6G staining (red) from the high fiber group are shown for muscle (Figure 3D, top left), subcutaneous tissues (Figure 3D, bottom left) and synovium (Supplementary Figure 4). Fewer anti-Ly6G staining cells were observed in the no fiber group; this was clearly evident even when viewing areas containing high numbers of infiltrating cells (Figure 3D, green arrows, right hand panels).
Anti-Ly6G staining of feet of mice fed a high fiber diet also revealed occasional blood vessels with intramural neutrophils and indications of parietal necrosis (Figure 3E, Ly6G); such features were not seen in feet of mice fed a no fiber diet. Consistent with this observation were areas of hemorrhage in feet of mice fed a high fiber diet (Figure 3E, H&E), which were much less apparent in feet of mice fed a no fiber diet.
RNA-Seq was undertaken using mRNA from feet taken day 6.5 post CHIKV infection for mice fed high and no fiber diets. Quality assurance data for the RNA-Seq data are shown in Supplementary Figure 5. The full RNA-Seq gene counts are shown in in Supplementary Table 1a and differentially express genes (DEGs) with false discovery rate (FDR) <0.05 (q < 0.05) are listed in Supplementary Table 1b. This DEG list (with a q < 0.05 filter) was used for all subsequent bioinformatics treatments, unless stated otherwise. Amongst these 571 DEGs were a series of up-regulated cytokines/chemokines associated with neutrophil recruitment, survival and/or activation (Table 1; Supplementary Table 1b). The DEG list was analyzed using the Ingenuity Pathway Analysis (IPA) Upstream Regulator (USR) feature. The top USR by positive activation z-score was CEBPA (CCAAT/enhancer binding protein alpha) (Supplementary Table 1c), a transcription factor that induces neutrophilic differentiation (granulopoiesis) (90) (Table 1). The top USR by negative activation z-score was TP73 (p73) (Supplementary Table 1c), a p53-related protein, with p73−/− mice showing massive neutrophil infiltrates and edema (112) (Table 1).
Rora (RORα) was present in the DEG list (Supplementary Table 1b), and represents a transcription factor this is associated inter alia with differentiation of Th17 cells (103) (Table 1).
The overall T cell infiltrate densities as measured by IHC for CD3 were not significantly different for high and no fiber groups (Supplementary Figure 6), arguing that Rora differences are not associated with differences in T cell recruitment. IL17A also identified as an USR when the IPA USR analysis was expanded to include both direct and indirect USR activities (Table 1, Supplementary Table 1e). IL-17 is a key cytokine that links T-cell activation to neutrophil mobilization and activation (117), with IL-17 playing an important role in promoting rheumatoid arthritis (115). The IPA USR analysis (direct and indirect) also returned (i) IL-1 and TNF, two cytokines known to increase endothelial permeability and edema (118–120) and (ii) PI3K and ERK, two kinases involved in survival and migration of neutrophils, including SCFA-induced chemotaxis (113) (Table 1, Supplementary Table 1e). These analyses support the IHC data (Figures 3C,D) and suggest that a range of inflammatory mediators and pathways that promote neutrophil-mediated inflammation were increased in mice fed a high fiber diet. Given the identification of IL17A as a USR (Table 1), the gene counts for the six IL-17 genes (IL-17a,b,c,d,f, and IL25) might be viewed as surprisingly low; although overall IL-17 counts were significantly higher for the high fiber group than the no fiber group (Supplemental Table 1a, see yellowed section, two way ANOVA, p = 0.028). The low counts may be due to an inherent feature of read alignment programs, which disregard reads that map to more than one gene. Although this process is important for accurate quantitation of transcripts, it results in loss of read counts for genes like IL-17 that exist as a family of homologous genes.
The RNA-Seq analysis showed down-regulation of 61 keratin and keratin-associated-protein genes (Figure 4A, Supplementary Table 1f, n = 61) amongst the 402 down-regulated DEGs identified in feet during peak CHIKV-arthritis in mice fed a high fiber diet (Supplementary Table 1b). Most of these genes are expressed in skin, hair follicles and hair (Supplementary Table 1f). About half of these genes were previously shown to be down-regulated in the same setting in mice fed normal chow (42), perhaps consistent with the intermediate phenotype seen in Figure 1A.
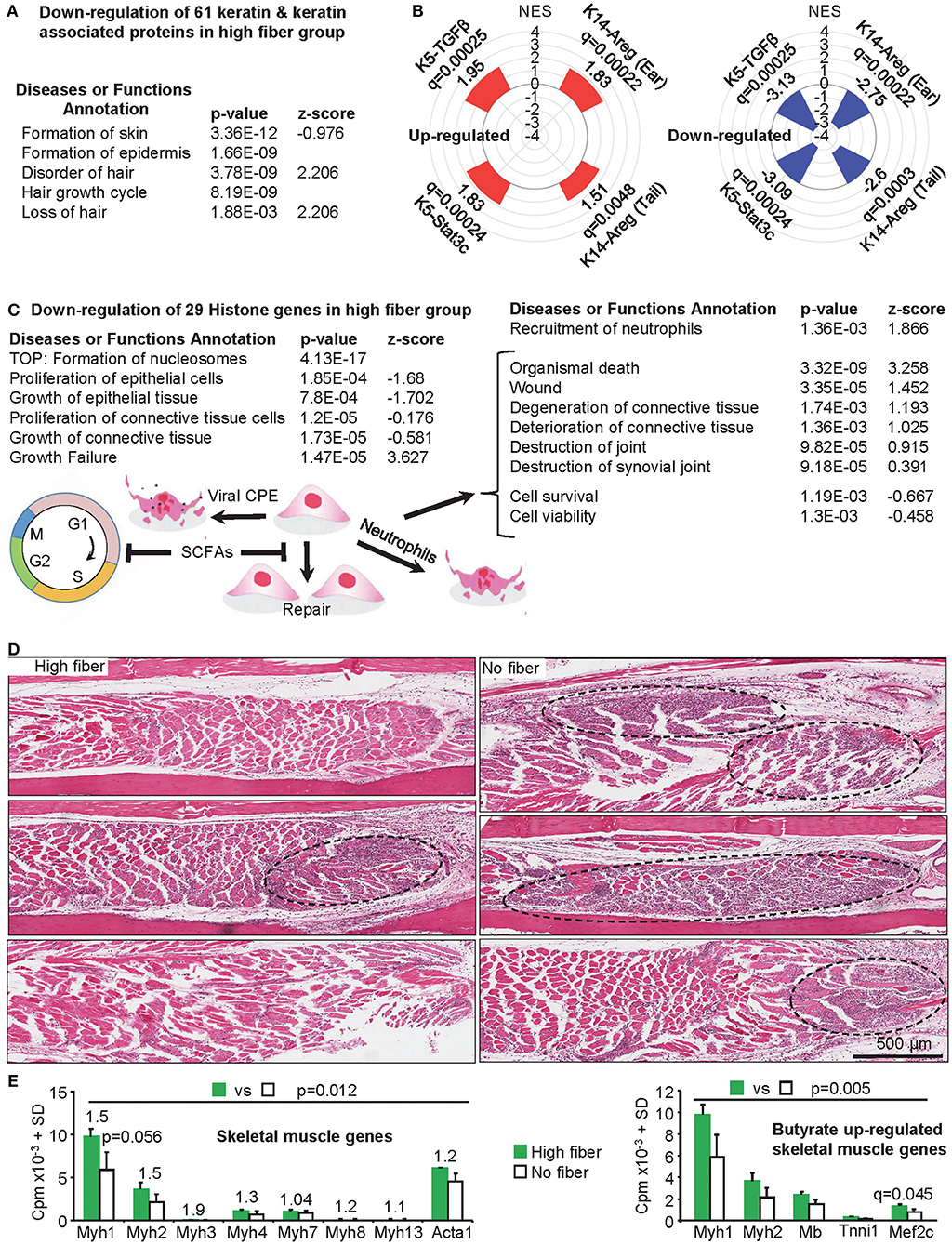
Figure 4. Additional changes mediated by a high fiber diet. (A) Skin and hair changes. The RNA-Seq data shows a significant down-regulation of 61 keratin and keratin associated proteins (Supplementary Table 1f). When the DEG list (Supplementary Table 1b, q < 0.05) was analyzed by IPA (direct only) using the Diseases or Functions Annotation feature, a range of annotations associated with skin and hair were identified (Supplementary Table 1d); the top four skin and hair annotations by p-value are shown. (B) Gene Set Enrichment Analyses was performed for up and down-regulated DEGs from high vs. no fiber diet (Supplementary Table 1b) and compared with gene sets pre-ranked by log2FC of inflammatory psoriasis mouse models (microarray study GSE27628): K14-Areg, over-expression of human amphiregulin in the basal epidermal layer (tail and skin); K5-Stat3c, basal keratinocyte-specific over-expression of a constitutively active mutant of signal transducer and activator of transcription 3; K5-TGFβ over-expression of the latent form of transforming growth factor β1 in basal keratinocyte. NES, Normalized Enrichment Score; q, FDR adjusted p-value. (C) Histone, proliferation and tissue destruction signatures. The RNA-Seq data shows a significant down-regulation of 29 histone genes in the high fiber group (Supplementary Table 1h). Formation of nucleosomes was the top Diseases or Functions Annotation by p-value (IPA analysis as in A), with a series of proliferation and growth annotations also returned with negative z-scores. The same analysis also returned a series of annotations associated with tissue destruction (right hand table) (Supplementary Table 1d). SCFAs (butyrate, propionate and valerate) can promote G1 cell cycle arrest and may thus inhibit proliferative replacement and repair of damaged tissues. CPE would contribute to tissue damage and neutrophils may also promote tissue destruction. (D) Skeletal muscle. H&E of skeletal muscle in feet of mice fed a high and no fiber diet day 6.5 post CHIKV infection are shown (three images from three mice per group). Muscle fibers (staining red/pink) appeared more extensively replaced by nuclear blue/purple staining of infiltrating inflammatory leukocytes in the no fiber group, with the latter also containing larger areas of dense blue/purple staining (dashed black ovals). (E) Left bar graph; mouse skeletal muscle myosin heavy chain and actin gene mean read counts (using read counts from Supplementary Table 1a). Numbers above the bars are fold change. No individual gene reached significance (q < 0.05); Myh1 approached significance by p-value. However, taken together significance was reached; statistics by Related Samples Wilcoxon Signed Rank Test. Right bar graph: skeletal muscle genes reported to be up-regulated by butyrate (121) were also up-regulated in the high fiber group, although only Mef2c reached significance. Taking the 5 genes together, differences between high fiber and no fiber reached significance; statistics by Related Samples Wilcoxon Signed Rank Test. For both graphs, SDs were derived from three biological replicates (Supplementary Table 1a).
Analyses of the down-regulated DEGs using Enrichr Disease Perturbations from GEO, suggested similarities with mouse models of psoriasis (Supplementary Table 1g). Gene Set Enrichment Analyses (GSEAs) were thus undertaken to compare both up and down-regulated DEGs from high vs. no fiber diet (Supplementary Table 1b) with publically available microarray data (GSE27628) from mouse models of inflammatory psoriasis (122). Genes up and down-regulated in CHIKV-infected mice fed a high fiber diet were also up and down-regulated in four mouse models of inflammatory psoriasis, respectively. All comparisons were highly significant by adjusted p-values (Figure 4B, Supplementary Table 1g). A high fiber diet thus increased skin inflammation after CHIKV infection with a signature similar to that seen in mouse models of inflammatory psoriasis.
Down-regulation of hair follicle genes (Supplementary Table 1f) is perhaps consistent with hair follicles being located next to areas of subcutaneous edema (Figure 3B, Supplementary Figure 7A) and adjacent to neutrophil infiltrates (Figure 3D, bottom left) (Supplementary Figure 7B). The presence of neutrophils in the epidermis or in the hair follicles, as has been described for psoriasis (123, 124), was not observed in our studies.
Another prominent feature of the RNA-Seq data was down-regulation of 29 histone genes (mean fold change −1.75) in the high fiber group (Figure 4C, Supplementary Table 1h). Butyrate has been shown in several settings to promote cell-cycle arrest in G0/G1 via p21 and/or p15 (125), the former often via a p53-independent mechanism (126, 127). Such an activity would reduce transcription of many histone genes during S-phase (128). A p53-independent mechanism may also operate in the current setting, as overall p53 activity was decreased (Supplementary Table 1c, TP53). Two other SCFAs, propionate and valerate, can also promote cell-cycle arrest in G0/G1 in certain settings (129–133). The IPA Diseases or Functions Annotations showed multiple epithelial and connective tissue proliferation and growth annotations with negative z-scores (Figure 4C, Supplementary Table 1d), consistent with G1 arrest. CHIKV infections generate high levels of type I IFNs (37, 42, 71), which can also promote cell cycle blockage at G0/G1 (134); however, these cytokines were not differentially regulated in the high fiber vs. no fiber groups.
The IPA Diseases or Functions Annotations suggests that the high fiber diet promoted tissue damage during peak CHIKV arthritis (Figure 4C, right) (Supplementary Table 1d). Neutrophil infiltration may account for the latter, as we have previously shown that recruitment of neutrophils into joints in CHIKV-infected CCR2−/− mice promotes cartilage damage (45), with neutrophils also associated with tissue damage in other settings (54). However, the overt histologically detectable cartilage damage seen in CCR2−/− mice was not observed herein, with CCR2−/− mice perhaps representing an extreme scenario given the complete absence of monocytes/macrophage infiltration during CHIKV arthritis (45). The tissue damage annotations (Figure 4C, right; Supplementary Table 1d) are unlikely to reflect differences in viral CPE, as viral loads were not different in the high fiber vs. no fiber groups (Figure 2B). However, given the high viral loads in feet (42, 47) extensive CPE would be expected in both groups, with reductions in cell proliferation in the high fiber group (Figure 4C, left) perhaps contributing to reduced proliferative tissue repair.
H&E staining of muscle tissues in the feet of CHIKV-infected mice fed a no fiber diet showed denser, more focal, inflammatory infiltrates in skeletal muscle tissues, when compared with CHIKV-infected mice fed a high fiber diet (Figure 4D, dashed black ovals). In these areas the red/pink staining of the muscle fibers was largely lost indicating more extensive muscle fiber degeneration in the no fiber group (Figure 4D). No compelling muscle signatures emerged from the bioinformatics analyses of the DEG list (Supplementary Table 1b). However, extracting mouse skeletal muscle myosin heavy chain (Myh1, 2, 3, 4, 7, 8, 13) and actin (Acta1) gene read counts from the full gene list (Supplementary Table 1a), illustrated that the mean read counts for these genes were always higher in the high fiber group; and taking all genes together, expression was significantly higher in the high vs. no fiber group (Figure 4E, left bar graph). Greater amounts of muscle specific mRNA species thus correlated with less muscle destruction in the high fiber group. Decreased skeletal muscle damage in the high fiber group, runs counter to the increased tissue damage or reduced tissue repair suggested by the analyses shown in Figure 4C. However, a recent report illustrated that butyrate supplementation promoted skeletal muscle formation in mice and up-regulated the expression of skeletal muscle genes, specifically, Myh1 and Myh2, myoglobin (Mb), and troponin-I, as well as the myocyte enhancer factor-2C (Mef2c), a key transcription factor for myogenesis (121). Extracting these genes from the full gene list (Supplementary Table 1a) illustrated that all these genes had higher mean read counts in the high fiber diet group, although only Mef2c reached significance (q = 0.045) (Figure 4E, right bar graph). When all five genes were taken together, their expression in the high fiber group was significantly higher (Figure 4E, right bar graph). Taken together, these results suggest that a high fiber diet is mildly myoprotective/myogenic in this CHIKV infection setting.
Tissue injury usually results in inflammation and neutrophil recruitment, with subsequent resolution of inflammation and initiation of tissue repair requiring an active process that involves adoption of a resolution phase phenotype by macrophages. The resolution phase is usually characterized by apoptosis of neutrophils, the removal of apoptotic neutrophils by macrophages (efferocytosis), the prevention of further neutrophil recruitment and the initiation of tissue repair (53, 54, 135). In contrast to autoimmune diseases (85), the cellular infiltrates in alphaviral arthritides usually have few neutrophils (45, 83, 84), perhaps arguing that a resolution phase phenotype is present during peak arthritis. In this model viremia peaks days 1–3 and is usually over by day 5 post infection, with peak arthritis occurring days 6–7 post infection (47).
To determine whether a resolution phase macrophage (rM) signature is present during peak CHIKV arthritis, a Gene Set Enrichment Analysis (GSEA) was undertaken to determine whether genes up-regulated in rM (52) were significantly represented during peak arthritis in feet of CHIKV-infected mice fed a normal diet (42). Using the microarray data posted by, and the methods described in, Stables et al. (52), we identified 146 genes that were up-regulated in rM (when compared with naïve macrophages and inflammatory macrophages) (Supplementary Table 1i). These DEGs were used in a GSEA using a pre-ranked (log2 fold change) list of the 18,517 genes obtained by RNA-Seq analysis of feet day 7 post CHIKV infection vs. mock infected feet (42) (Supplementary Table 1i). The GSEA provided a NES score of 3.57 and a high level of significance (q < 0.001), with 69/146 genes identified as core enriched genes (Figure 5A, Supplementary Table 1i). Genes up-regulated in rM were thus generally also up-regulated in arthritic feet day 7 post CHIKV infection, when compared with mock infected feet (Figure 5B). This analysis indicates that a significant rM signature is present in the feet of mice fed normal chow during peak CHIKV arthritis, perhaps explaining (at least in part) why there is a paucity of neutrophil infiltrates.
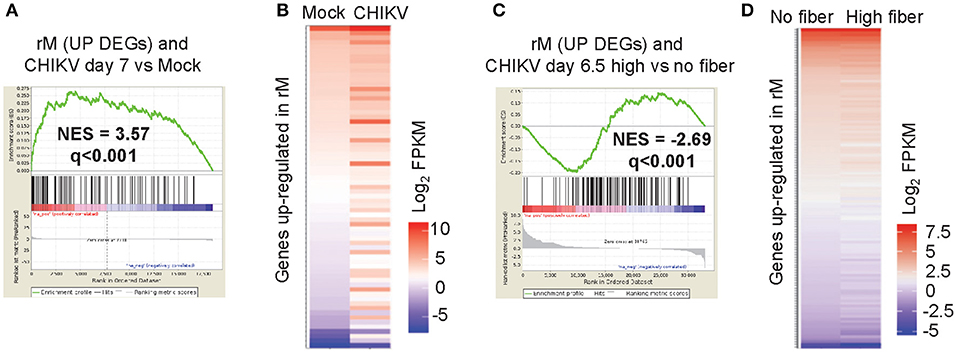
Figure 5. Macrophage resolution phase signature. The gene lists used in this figure are provided in Supplementary Table 1i. (A) A Gene Set Enrichment Analysis (GSEAs) comparing the 146 up-regulated DEGs associated with rM with the expression profile obtained from RNA-Seq analysis of feet day 7 post CHIKV infection vs. mock infected feet (18,517 genes pre-ranked by log2 expression). A normalized enrichment score (NES) and false discovery rate (q) is shown. (B) Heat map illustrating that the 69 core enriched genes identified by the GSEA (out of the 146 genes up-regulated in rM) are generally also up-regulated during peak CHIKV arthritis (day 7 post infection). (C) A GSEA comparing the up-regulated DEGs associated with rM with the expression profile obtained from RNA-Seq analysis of feet of mice fed a high fiber diet vs. feet of mice fed a no fiber diet day 6.5 post CHIKV infection (33,054 genes pre-ranked by log2 expression). (D) Heat map illustrating that the 126 core enriched genes identified by the GSEA (out of the 146 genes up-regulated in rM) are generally down-regulated during peak CHIKV arthritis in mice fed a high fiber diet.
To determine whether the rM signature is modulated by a high fiber diet, a GSEA was undertaken using the same 146 genes up-regulated in rM and the pre-ranked 33,054 genes from CHIKV-infected feet from the high fiber vs. no fiber diet group (Supplementary Table 1i). The GSEA again providing a high level of significance, with 128/146 genes identified as core enriched genes (Figure 5C, Supplementary Table 1i). However, the NES score was negative (−2.69), with ≈35% of the genes up-regulated in rM down-regulated in feet of mice fed a high fiber diet (Figure 5D). Thus, the high fiber diet was associated with a significant down-modulation of the rM gene signature during peak CHIKV arthritis, which might explain (at least in part) the delayed clearance of neutrophils in the high fiber group (Figure 3C).
The type 2 immunity genes associated with tissue repair (IL-4, IL-13, IL-25, IL-5, TSLP, IL-33, TGFB1, and IL-10) (136) were not significantly lower in the high fiber group (Supplementary Table 1b). The IPA USR actually suggested an up-regulation of TGFB1 activity (Supplementary Table 1e, activation z-score 1.1, p = 0.0014), although this analysis did show down-regulation of IL-10 (Table 1). The first five genes in the aforementioned list also showed very low expression levels (Supplementary Table 1a), consistent with the dominant Th1 bias associated with CHIKV infection in this model (45, 47). That fiber has no significant effect on Th1/Th2 is supported by Figure 2C.
The effects of high fiber diet are often associated with butyrate, with butyrate substitution in the drinking water (like high fiber diet) also able to exacerbate CHIKV-induced foot swelling significantly (Figures 1C,E,G). As observed after a high fiber diet (Figure 3A), drinking butyrate did not significantly increase the levels of cellular infiltrates in feet 6.5 days after CHIKV infection compared to drinking unsupplemented water (Figure 6A). This remained true when mice drank butyrate and were fed a standard diet (Figure 6B). Similar to the observations made for the high fiber diet group (Figure 3B), increased edema was again discernible by H&E in the feet of CHIKV-infected mice drinking butyrate (Figure 6C).
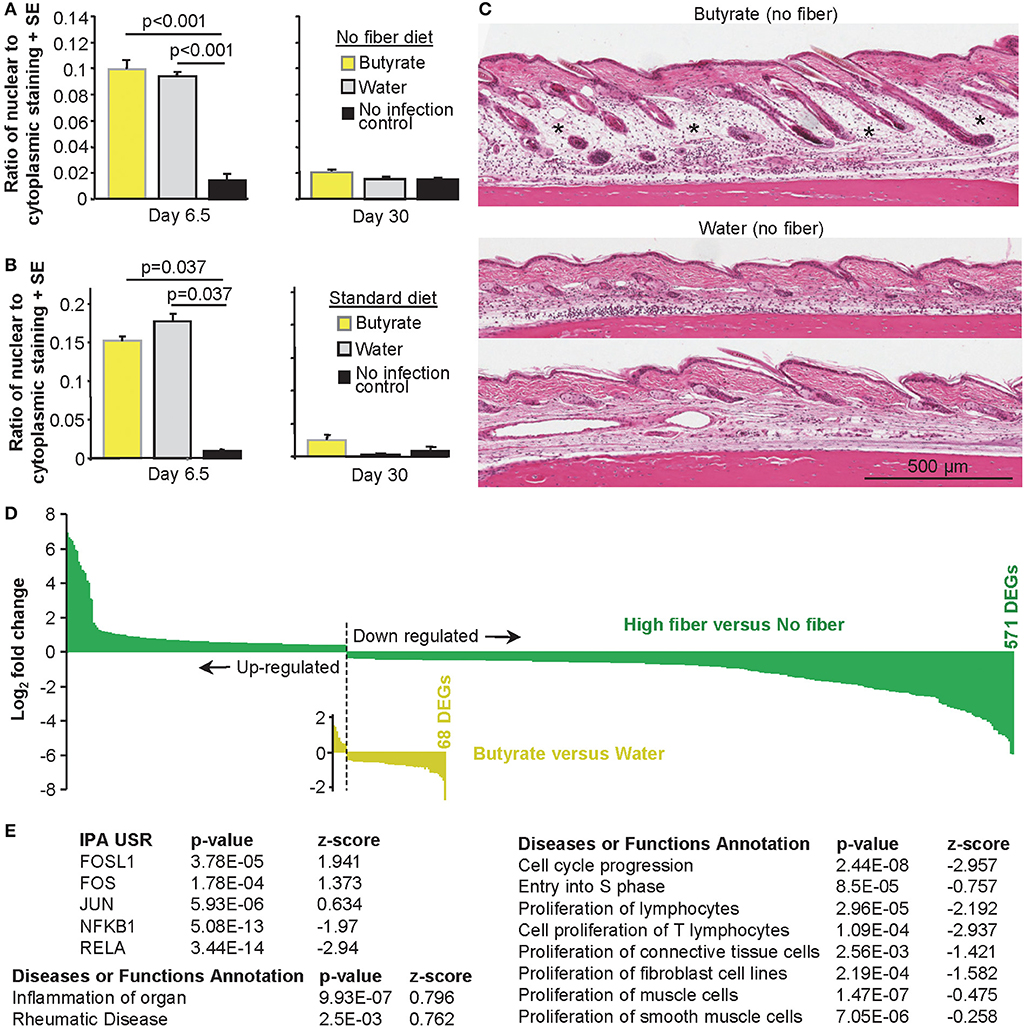
Figure 6. The effects of butyrate on CHIKV rheumatic disease. (A) Mice were fed a no fiber diet and given water supplemented with and without butyrate as in Figure 1E. On the indicated day, feet were examined by H&E and Aperio pixel count as in Figure 3A. Statistics by t-tests, n = 6 mice and feet per group. (B) As for (A), but mice were fed on a standard diet as in Figure 1C. Statistics by Kolmogorov–Smirnov tests n = 3–6 feet and mice per group. (C) H&E images of foot sections of mice treated as in (A). *Edema. (D) Bar graph of fold change of all 571 DEGs (q < 0.05) for high fiber vs. no fiber (green) and 68 DEGs for butyrate vs. water (yellow). Full data sets available in Supplementary Tables 1b,k. (E) IPA USR and Diseases or Functions Annotation analyses (direct only) of the 68 butyrate DEGs. Full data sets available in Supplementary Tables 1l,n.
IHC staining for neutrophils illustrated very low levels of neutrophils in both butyrate and water (no fiber) groups, with marginally more neutrophils in the butyrate group, but this was not significant (Supplementary Figure 8).
RNA-Seq was undertaken using mRNA from feet of mice harvested on day 6.5 post CHIKV infection after mice were given either butyrate or unsupplemented water to drink; both groups were on a no fiber diet (drink and diet conditions also used for Figures 1E, 6A). Quality assurance data for the RNA-Seq are shown in Supplementary Figure 5. The full RNA-Seq gene counts are provided in Supplementary Table 1j and differentially express genes (DEGs) with false discovery rate (FDR) q < 0.05 are listed in Supplementary Table 1k and were used for all subsequent bioinformatics treatments, unless stated otherwise.
The number of DEGs for butyrate vs. water was only 68 (compared with 571 for high vs. no fiber diet, q < 0.05 for both), illustrating that the perturbations in CHIKV rheumatic disease mediated by drinking butyrate were substantially less widespread than those engendered by a high fiber diet (Figure 6D). This observation is perhaps consistent with the more complex role played by fiber, which inter alia produces a range of different SCFAs (18, 137). In addition, drinking butyrate resulted in only two genes with a fold change (FC) >2.8 (log2 of 1.5) and a maximum fold change of 6.5 (log2 of 2.71). In contrast, a high fiber diet resulted in 144 genes with FC>2.8 and a maximum fold change 118 (log2 of 6.88). The overall magnitude of transcriptional perturbations were thus also substantially lower for butyrate (Figure 6D).
There was no overlap in DEGs identified for “butyrate vs. water” and “high vs. no fiber” when a FDR q < 0.05 filter was applied to both. When the significance filter for “butyrate vs. water” was reduced to p < 0.05 (i.e., a p-value filter without FDR adjustment), only a small overlap of six genes for up-regulated genes (with 169 DEGs for high fiber q < 0.05 and 692 genes for butyrate p < 0.05) and 46 genes for down-regulated genes (402 DEGs for high fiber q < 0.05 and 1,220 genes for butyrate p < 0.05) became apparent. IPA USR analyses illustrated an overlap of (i) 2 USRs (FOS and JUN) for the 9 USRs (for butyrate vs. water) and the 34 USRs (for high vs. no fiber) with positive z-scores, and (ii) an overlap of 8 USRs (NOTCH1, CEBPB, STAT3, FOXO3, TP53, CREM, CTNNB1, KLF4, and PPARG) for the 45 USRs (for butyrate vs. water) and the 29 USRs (for high vs. no fiber) with negative z-score (Supplementary Tables 1c,l).
The “Integrated System for Motif Activity Response Analysis” (ISMARA) (70) provides an independent analysis of the RNA-Seq data and generates an activity score for usage of known transcription factor binding sites as determined from the promoter regions of genes identified by RNA-Seq. ISMARA provides a Z-value, with higher Z-values suggesting more significant differences between the two groups. ISMARA identified differential use (with Z > 1.5) of 11 transcription factor binding sites for “butyrate vs. water” and 60 for “high vs. no fiber.” Three of these sites were shared and showed the same direction of change (Supplementary Table 1m). Thus, although both butyrate and high fiber increased peak foot swelling and promoted edema after CHIKV infection, the mechanisms involved appear to be largely different.
Analysis of the 68 DEGs (Butyrate vs. Water; Supplementary Table 1k) using the USR feature of IPA (direct only), indicated that butyrate consumption up-regulated (positive z-score) pathways associated with FOSL1 (FRA-1), FOS (c-FOS), and JUN during peak CHIKV arthritis (Figure 6E, Supplementary Table 1l). c-JUN, JUNB and JUND, and c-FOS, FOSB, FRA-1 and FRA2 combine to form AP-1 transcription factor complexes, that can also include Maf and ATF family members (138). Modulation of AP-1 complexes was further supported by down-regulation of JUNB, FOSB, and FOS amongst the 68 DEGs (Supplementary Table 1k), with ATF4 down-regulation also identified by the IPA USR (Supplementary Table 1l). ISMARA also suggested modulation of AP-1 activities, although only Junb-Jund reached, and Fosl2 approached, significance (Supplementary Figure 9, Supplementary Table 1m). Butyrate has been shown to modulate AP-1 activities in several settings (32–34), with AP-1 modulation previously associated with edema (139, 140) and inflammation (141). Both FOS and JUN were also identified (with positive z-scores) in the IPA USR analysis of high vs. no fiber DEGs (Supplementary Table 1c).
The IPA USR analysis suggested that several NF-κB, STAT, and IRF pathways were down-regulated during peak CHIKV arthritis in mice drinking butyrate (Supplementary Table 1l). NF-κB inhibition was quite marked and highly significant (Figure 6E), consistent with this pathway being a major target of butyrate activity (31, 142). NFKBIA (NF-κB inhibitor alpha) was also identified (with positive z-score) in the IPA USR analysis of high vs. no fiber DEGs (Supplementary Table 1c).
Using the Diseases or Functions Annotation feature of IPA, significant annotations for inflammation and rheumatic disease were returned (Figure 6E), consistent with Figures 1E, 6C. However, the dominant signature (with multiple annotations; Supplementary Table 1n) was cell cycle arrest, reduced growth and/or reduced proliferation (Figure 6E, right hand table); an observation consistent with the ability of butyrate to promote cell cycle arrest (125–127). However, the lack of histones in the DEG list (Supplementary Table 1k) suggests this effect was less substantial than that seen after a high fiber diet, consistent with Figure 6D.
The IPA USR analysis (direct only) suggested the activities of several transcription factors associated with type I IFN responses (42, 71) are changed in the arthritic feet of mice fed high fiber diet, although activation z-scores were generally low (e.g., IRF8, STAT1, STAT3, NF-κB family, CTNNB1, ATF3; Supplementary Table 1c). No USRs central to the anti-CHIKV innate responses were identified [e.g., IRF3, IRF7, IFNAR (37)] and the DEG list (Supplementary Table 1b) had few, if any, effectors known to mediate anti-viral activity against CHIKV (42). These results are consistent with the lack of significant changes in viremia (Figure 1B) or viral loads in feet (Figure 2A). In addition, RNA-Seq reads can be mapped to the CHIKV genome (42), with the number of reads similar for high fiber verse no fiber groups (Supplementary Figure 10).
Butyrate substitution in the drinking water also did not affect viremia (Figure 1F) or viral loads in feet (Figure 2B) or RNA-Seq reads mapping to the CHIKV genome (Supplementary Figure 10). However, a number of innate genes/activities associated with antiviral activity against CHIKV (42) were down-regulated by butyrate (i) Mx1 (143), ≈1.5 fold down-regulated (Supplementary Table 1k), (ii) IRF3 and STAT1 activities by IPA USR analyses (Supplementary Table 1l, z-score −1.85 and −1.33, respectively), and (iii) Irf2_Irf1_Irf8_Irf9_Irf7 site usage (Supplementary Figure 9A, down in butyrate, Z-value 3.29). In contrast, the negative regulator of type I IFN responses, OASL1 (144) was down-regulated ≈1.5 fold (Supplementary Table 1k). The low magnitude of changes (Figure 6D), the counter-regulatory affects mediated by OASL1 and/or IRF2, and/or redundancy in the type I IFN system, may explain the lack of an effect on CHIKV loads.
To the best of our knowledge this paper represents the first study investigating the role of a high fiber diet and SCFAs on alphaviral rheumatic disease. Although a large body of work in non-infectious disease settings would argue that a high fiber diet should ameliorate inflammation, we illustrate herein that a high fiber diet modulated the CHIKV rheumatic immunopathology with increased edema, Th17/IL-17 activities, neutrophil infiltration, and psoriasis-like signatures, but reduced muscle damage. Although imbibing butyrate has also been shown to ameliorate inflammation outside the gut in multiple non-infectious disease settings (145–147), herein we show that butyrate consumption increased edema during CHIKV infection. Neither the high fiber diet nor drinking butyrate affected viral loads or anti-viral antibody responses; however, both clearly exacerbated CHIKV arthritic immunopathology.
The most dramatic change to CHIKV immunopathology mediated by a high fiber diet was the increase in edema and infiltrating neutrophils during peak arthritis. The high fiber diet mediated a range of changes to the CHIKV arthritic signature that supported neutrophil recruitment, survival, and activation (Table 1). Key drivers of neutrophil mediated inflammation are Th17 cell and IL-17, and although we have not formally demonstrated increased Th17 cells or IL-17 protein levels in the feet of CHIKV infected mice fed a high fiber diet, bioinformatic analyses suggest both are up-regulated in the high fiber group (Table 1, Supplementary Table 1a). Perhaps the most compelling bioinformatics support (for a fiber-mediated increase in pro-inflammatory and IL-17-mediated immunopathology after CHIKV infection) comes from the significant similarities between CHIKV arthritis in mice fed a high fiber diet and mouse models of inflammatory psoriasis (122) (Figure 4B). Although this was an unexpected and novel finding, there are salient parallels between CHIKV infection and psoriasis; (i) CHIKV after a high fiber diet and psoriasis both involve neutrophil recruitment (148), (ii) psoriasis can cause alopecia (149), and can have follicular involvement (124), with alopecia frequently reported by CHIKV patients (150–153) and also seen in some CHIKV mouse models (154, 155), (iii) although distinct from psoriatic lesions, skin manifestations (usually maculopapular rashes) are well-described for arthritogenic alphavirus infections in humans, but are rarely overt in mouse models, although they can be detected by IHC in certain settings (156), and (iv) perhaps most cogent, both psoriasis and CHIKV are associated with arthritis (157). The changes in skin immunopathology seen herein are consistent with the well-described ability of the gut microbiome to influence immunity in the skin via the so-called gut-skin-axis (158). A role for IL-17 in psoriasis has been established (159) and several studies in humans have also found an association between gut microbiota and Th17 responses (160, 161). Promotion of systemic Th17/IL-17 responses by acetate was also reported in another infectious disease setting where mice were infected orally with the gram-negative enteric bacteria Citrobacter rodentium (19). In addition, HDAC1 inhibition [an activity of butyrate (162)] has been shown to promote IL-17 transcription in human T cell lines in vitro (163). Whether a high fiber diet would predispose to more severe skin or rheumatic manifestations in humans after CHIKV infection remains to be determined, and any study seeing to establish a link would need to control for inter alia the multiple ways in which diet might affect arboviral infections (164).
The mechanisms whereby the high fiber diet would mediate the neutrophil-promoting changes during CHIKV arthritis are likely to be complex given the interaction of various SCFAs with multiple cell types (10, 18, 24, 66, 113), the infection of different cell types by CHIKV, and the complex interplay of anti-viral and inflammatory responses seen post infection (36, 41, 42, 51). Nevertheless, a likely key contributing mechanism highlighted herein is the down-modulation in the rM gene signature seen in feet during peak CHIKV arthritis in mice fed a high fiber diet (compared with peak CHIKV arthritis in mice fed a no fiber diet) (Figure 5). Adoption by infiltrating macrophages of a resolution phase phenotype is usually associated with loss of neutrophils and initiation of tissue repair (53, 54, 135). Both the presence of neutrophils and the GSEA argue that adoption of a rM phenotype is delayed in CHIKV arthropathy in mice fed a high fiber diet. A high fiber diet is believed generally to provide anti-inflammatory activities (10, 18) and anti-inflammatory drugs have been associated with delayed wound healing and/or tissue repair in a number of settings (165–167). Although a high fiber diet provided beneficial effects in influenza infections in mice, the mechanism also involved modulation of neutrophil-mediated tissue damage via the reshaping of macrophage functionality by SCFAs (66), perhaps arguing this is an important pathway whereby fiber mediates its effects on immunopathology.
Butyrate consumption increased edema during CHIKV arthritis, but the mechanisms involved appeared to be different from those seen for the high fiber diet. Bioinformatic analyses of RNA-Seq data suggested modulation of AP-1 and NF-κB activities and/or promotion of cell cycle arrest may be involved. The RNA-Seq and IHC analyses did not suggest any increases in pro-inflammatory cytokines (e.g., TNF, IL-1, and VEGF) or altered cellular infiltrates that might be associated with increased edema. Instead, we speculate that butyrate is promoting edema (at least in part) by acting on endothelial cells (24, 168). CHIKV infects endothelial cells in vivo (37, 169) and edema is a well-known symptom of CHIKV infection (47, 82). Endothelial barrier repair requires NF-κB activation (170, 171) and a NF-κB to AP-1 transition (172). Herein we provide evidence that butyrate promoted certain AP-1 activities and markedly inhibited canonical NF-κB pathways, consistent with previous studies on butyrate (31–34, 142). Such modulation of these key pathways may deoptimize endothelial barrier repair during CHIKV infection leading to increased edema. Cell cycle arrest may also play a role in promoting edema (both for butyrate Figure 6E and high fiber diet Figure 4C), as edema is a well-known side-effect of rapamycin (173, 174), a drug that also induces G1 arrest.
Perhaps curious is (i) the inability herein to recapitulate the effects of the high fiber diet with butyrate and (ii) the relatively minor effect butyrate had on transcription when compared with the high fiber diet (Figure 6D). For instance, butyrate has been widely reported to mediate G1 cell-cycle arrest via HDAC inhibition (125–127, 175). However, herein we see a more pronounced cell-cycle arrest signature in the high fiber group, as indicated by a widespread down-regulation of histone mRNAs (Supplementary Table 1h) that was not observed in the butyrate group (Supplementary Table 1k). The bioavailability of butyrate in vivo is actually quite low, so the circulating concentrations required for systemic HDAC inhibition may not be efficiently reached after drinking butyrate (175, 176). The more pronounced cell-cycle signature seen in the high fiber group may thus be due to additional or complimentary cell-cycle arrest activities mediated other SCFAs such as propionate and valerate (129–133).
There are a number of limitations for the studies described herein. Tissue levels of SCFAs were not assessed as part of this study; analyses which might provide some insights into the bioavailability issues discussed in the previous paragraph. RNA-Seq was also not performed on uninfected mice from the different dietary groups, limiting the interpretation of which changes in the transcriptome are caused by diet alone, and which are the result of the interplay of infection and diet. In addition, the different dietary regimens may mediate indirect effects on CHIKV arthritis, conceivably via changes in food/water uptake or altered physiology. We have also not addressed the question of whether a high fiber diet might change bone marrow neutrophil development or differentiation, or whether a high fiber diet affects circulating neutrophil numbers or differentiation states or the CHIKV infection-mediated transient leukopenia (169).
Arguably the most concerning aspect of CHIKV disease is the chronic arthralgia (joint pain), which can be protracted and is often difficult to manage with existing medications (36). We show herein that the significant differences in neutrophil infiltrates seen during peak arthritis (Figure 3C, Day 6.5), were not apparent during chronic arthritis [nominally deemed to be day 30 post infection in this model (55)] (Figure 3C, Day 30). Viral CPE and tissue damaging processes are largely over by the time the chronic phase of disease manifests (55), with the anti-inflammatory activity of fiber conceivably able to provide benefit during this later stage of CHIKV disease. Animal models of chronic arthralgia have not been established and so investigating the potential benefits of a high fiber diet for chronic alphaviral arthralgia would likely require human studies. Although chronic alphaviral arthralgia likely involves inflammatory pain, the arthralgia can have neuropathic characteristics (36), which are not usually considered to be effectively managed with anti-inflammatory interventions.
The observations made herein for acute CHIKV rheumatic immunopathology contrasts markedly with most reports in the field that describe disease amelioration by high fiber diets and SCFAs. However, the majority of such studies were conducted in non-infectious disease settings, with the role of fiber and SCFAs in infectious diseases largely unexplored, especially in scenarios where there is a robust systemic infection and widespread tissue damage, with a need for tissue repair. Some other studies have also shown deleterious effects from fiber and SCFAs (68, 177), reinforcing the notion that the health benefits of fiber and SCFAs may often be quite setting dependent.
The raw RNA-Seq data has been deposited to NCBI;BioProject ID, PRJNA555135; Submission ID, SUB5943019. SAMN12301865 to 7—High fiber diet, feet day 6.5 post CHIKV infection. SAMN12301868/69/70—No fiber diet, feet day 6.5 post CHIKV infection. SAMN12301871/2/3—Butyrate, no fiber diet, feet day 6.5 post CHIKV infection. SAMN12301874/5/6—Water, no fiber diet, feet day 6.5 post CHIKV infection. All gene sets are provided in Supplementary Table 1.
All mouse work was conducted in accordance with the Australian code for the care and use of animals for scientific purposes as defined by the National Health and Medical Research Council of Australia. Mouse work was approved by the QIMR Berghofer Medical Research Institute animal ethics committee (P1060 A705603M) and was conducted in biosafety level-3 facility at the QIMR Berghofer. Mice were euthanized using carbon dioxide.
NP, BT, TTL, and JG: undertook the experiments and analyzed the data. TL: interpreted the histology. TH, TA, PM, YP, and HN: bioinformatics. EN and VL: methodology, interpretation, and manuscript review. NP and AS: funding acquisition. AS: conceptualized the study and wrote the manuscript.
This work was supported by a project grant from the National Health and Medical Research Council (NHMRC) of Australia (APP1078468) and intramural seed funding from the Australian Infectious Disease Research Center. NP was supported by an Advance Queensland Research Fellowship from the Queensland Government, Australia. AS holds a Principal Research Fellowship from the National Health and Medical Research Council of Australia. EN was supported in part by the Daiichi Sankyo Foundation of Life Science. TH was supported by FAPESP (São Paulo Research Foundation).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the QIMR B animal house staff for their assistance, Clay Winterford (QIMR B) and his team at for their help with histology and immunohistochemistry, and Dr. Gunter Hartel (Head Statistician, QIMR B) for help with statistics. We thank Dr. I. Anraku for his assistance in managing the BSL3 facility at QIMR B.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02736/full#supplementary-material
1. Vieira AT, Galvao I, Macia LM, Sernaglia EM, Vinolo MA, Garcia CC, et al. Dietary fiber and the short-chain fatty acid acetate promote resolution of neutrophilic inflammation in a model of gout in mice. J Leukoc Biol. (2017) 101:275–84. doi: 10.1189/jlb.3A1015-453RRR
2. Zhang Y, Dong A, Xie K, Yu Y. Dietary supplementation with high fiber alleviates oxidative stress and inflammatory responses caused by severe sepsis in mice without altering microbiome diversity. Front Physiol. (2018) 9:1929. doi: 10.3389/fphys.2018.01929
3. Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. (2017) 135:964–77. doi: 10.1161/CIRCULATIONAHA.116.024545
4. Sakanoi Y, E S, Yamamoto K, Ota T, Seki K, Imai M, et al. Simultaneous intake of euglena gracilis and vegetables synergistically exerts an anti-inflammatory effect and attenuates visceral fat accumulation by affecting gut microbiota in mice. Nutrients. (2018) 10:E1417. doi: 10.3390/nu10101417
5. Matt SM, Allen JM, Lawson MA, Mailing LJ, Woods JA, Johnson RW. Butyrate and dietary soluble fiber improve neuroinflammation associated with aging in mice. Front Immunol. (2018) 9:1832. doi: 10.3389/fimmu.2018.01832
6. Di Caro V, Cummings JL, Alcamo AM, Piganelli JD, Clark RSB, Morowitz MJ, et al. Dietary cellulose supplementation modulates the immune response in a murine endotoxemia model. Shock. (2019) 51:526–34. doi: 10.1097/SHK.0000000000001180
7. Hung TV, Suzuki T. Dietary fermentable fibers attenuate chronic kidney disease in mice by protecting the intestinal barrier. J Nutr. (2018) 148:552–61. doi: 10.1093/jn/nxy008
8. Saidane O, Courties A, Sellam J. Dietary fibers in osteoarthritis: what are the evidences? Joint Bone Spine. (2019) 86:411–4. doi: 10.1016/j.jbspin.2018.10.010
9. Telle-Hansen VH, Holven KB, Ulven SM. Impact of a healthy dietary pattern on gut microbiota and systemic inflammation in humans. Nutrients. (2018) 10:1783. doi: 10.3390/nu10111783
10. Meng X, Zhou HY, Shen HH, Lufumpa E, Li XM, Guo B, et al. Microbe-metabolite-host axis, two-way action in the pathogenesis and treatment of human autoimmunity. Autoimmun Rev. (2019) 18:455–75. doi: 10.1016/j.autrev.2019.03.006
11. Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. (2018) 360:j5145. doi: 10.1136/bmj.j5145
12. Veronese N, Solmi M, Caruso MG, Giannelli G, Osella AR, Evangelou E, et al. Dietary fiber and health outcomes: an umbrella review of systematic reviews and meta-analyses. Am J Clin Nutr. (2018) 107:436–44. doi: 10.1093/ajcn/nqx082
13. Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. (2015) 6:6734. doi: 10.1038/ncomms7734
14. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. (2015) 6:7320. doi: 10.1038/ncomms8320
15. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. (2010) 107:14691–6. doi: 10.1073/pnas.1005963107
16. Jefferson A, Adolphus K. The effects of intact cereal grain fibers, including wheat bran on the gut microbiota composition of healthy adults: a systematic review. Front Nutr. (2019) 6:33. doi: 10.3389/fnut.2019.00033
17. Gavin PG, Hamilton-Williams EE. The gut microbiota in type 1 diabetes: friend or foe? Curr Opin Endocrinol Diabetes Obes. (2019) 26:207–12. doi: 10.1097/MED.0000000000000483
18. Luu M, Visekruna A. Short-chain fatty acids: bacterial messengers modulating the immunometabolism of T cells. Eur J Immunol. (2019) 49:842–8. doi: 10.1002/eji.201848009
19. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. (2015) 8:80–93. doi: 10.1038/mi.2014.44
20. Haase S, Haghikia A, Wilck N, Muller DN, Linker RA. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. (2018) 154:230–8. doi: 10.1111/imm.12933
21. Nakajima A, Nakatani A, Hasegawa S, Irie J, Ozawa K, Tsujimoto G, et al. The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS ONE. (2017) 12:e0179696. doi: 10.1371/journal.pone.0179696
22. Park JW, Kim HY, Kim MG, Jeong S, Yun CH, Han SH. Short-chain fatty acids inhibit staphylococcal lipoprotein-induced nitric oxide production in murine macrophages. Immune Netw. (2019) 19:e9. doi: 10.4110/in.2019.19.e9
23. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. (2019) 50:432–45 e7. doi: 10.1016/j.immuni.2018.12.018
24. Li M, van Esch B, Wagenaar GTM, Garssen J, Folkerts G, Henricks PAJ. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur J Pharmacol. (2018) 831:52–9. doi: 10.1016/j.ejphar.2018.05.003
25. Theiler A, Barnthaler T, Platzer W, Richtig G, Peinhaupt M, Rittchen S, et al. Butyrate ameliorates allergic airway inflammation by limiting eosinophil trafficking and survival. J Allergy Clin Immunol. (2019) 144:764–76. doi: 10.1016/j.jaci.2019.05.002
26. Chen G, Ran X, Li B, Li Y, He D, Huang B, et al. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMed. (2018) 30:317–25. doi: 10.1016/j.ebiom.2018.03.030
27. Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.01486
28. Kim DS, Kwon JE, Lee SH, Kim EK, Ryu JG, Jung KA, et al. Attenuation of rheumatoid inflammation by sodium butyrate through reciprocal targeting of HDAC2 in osteoclasts and HDAC8 in T cells. Front Immunol. (2018) 9:1525. doi: 10.3389/fimmu.2018.01525
29. Lucas S, Omata Y, Hofmann J, Bottcher M, Iljazovic A, Sarter K, et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun. (2018) 9:55. doi: 10.1038/s41467-017-02490-4
30. Hui W, Yu D, Cao Z, Zhao X. Butyrate inhibit collagen-induced arthritis via Treg/IL-10/Th17 axis. Int Immunopharmacol. (2019) 68:226–33. doi: 10.1016/j.intimp.2019.01.018
31. Bach Knudsen KE, Laerke HN, Hedemann MS, Nielsen TS, Ingerslev AK, Gundelund Nielsen DS, et al. Impact of diet-modulated butyrate production on intestinal barrier function and inflammation. Nutrients. (2018) 10:E1499. doi: 10.3390/nu10101499
32. Nepelska M, Cultrone A, Beguet-Crespel F, Le Roux K, Dore J, Arulampalam V, et al. Butyrate produced by commensal bacteria potentiates phorbol esters induced AP-1 response in human intestinal epithelial cells. PLoS ONE. (2012) 7:e52869. doi: 10.1371/journal.pone.0052869
33. Kida Y, Shimizu T, Kuwano K. Sodium butyrate up-regulates cathelicidin gene expression via activator protein-1 and histone acetylation at the promoter region in a human lung epithelial cell line, EBC-1. Mol Immunol. (2006) 43:1972–81. doi: 10.1016/j.molimm.2005.11.014
34. Rivero JA, Adunyah SE. Sodium butyrate induces tyrosine phosphorylation and activation of MAP kinase (ERK-1) in human K562 cells. Biochem Biophys Res Commun. (1996) 224:796–801. doi: 10.1006/bbrc.1996.1102
35. Suhrbier A, Jaffar-Bandjee MC, Gasque P. Arthritogenic alphaviruses–an overview. Nat Rev Rheumatol. (2012) 8:420–9. doi: 10.1038/nrrheum.2012.64
36. Suhrbier A. Rheumatic manifestations of chikungunya: emerging concepts and interventions. Nature Rev Rheumatol. (2019) 15:597–611. doi: 10.1038/s41584-019-0276-9
37. Rudd PA, Wilson J, Gardner J, Larcher T, Babarit C, Le TT, et al. Interferon response factors 3 and 7 protect against chikungunya virus hemorrhagic fever and shock. J Virol. (2012) 86:9888–98. doi: 10.1128/JVI.00956-12
38. Carpentier KS, Morrison TE. Innate immune control of alphavirus infection. Curr Opin Virol. (2018) 28:53–60. doi: 10.1016/j.coviro.2017.11.006
39. Matusali G, Colavita F, Bordi L, Lalle E, Ippolito G, Capobianchi MR, et al. Tropism of the chikungunya Virus. Viruses. (2019) 11:175. doi: 10.3390/v11020175
40. Lokireddy S, Vemula S, Vadde R. Connective tissue metabolism in chikungunya patients. Virol J. (2008) 5:31. doi: 10.1186/1743-422X-5-31
41. Michlmayr D, Pak TR, Rahman AH, Amir ED, Kim EY, Kim-Schulze S, et al. Comprehensive innate immune profiling of chikungunya virus infection in pediatric cases. Mol Syst Biol. (2018) 14:e7862. doi: 10.15252/msb.20177862
42. Wilson JA, Prow NA, Schroder WA, Ellis JJ, Cumming HE, Gearing LJ, et al. RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathog. (2017) 13:e1006155. doi: 10.1371/journal.ppat.1006155
43. Long KM, Heise MT. Protective and pathogenic responses to chikungunya virus infection. Curr Trop Med Rep. (2015) 2:13–21. doi: 10.1007/s40475-015-0037-z
44. Petitdemange C, Wauquier N, Vieillard V. Control of immunopathology during chikungunya virus infection. J Allergy Clin Immunol. (2015) 135:846–55. doi: 10.1016/j.jaci.2015.01.039
45. Poo YS, Nakaya H, Gardner J, Larcher T, Schroder WA, Le TT, et al. CCR2 deficiency promotes exacerbated chronic erosive neutrophil-dominated chikungunya virus arthritis. J Virol. (2014) 88:6862–72. doi: 10.1128/JVI.03364-13
46. Nakaya HI, Gardner J, Poo YS, Major L, Pulendran B, Suhrbier A. Gene profiling of chikungunya virus arthritis in a mouse model reveals significant overlap with rheumatoid arthritis. Arthritis Rheum. (2012) 64:3553–63. doi: 10.1002/art.34631
47. Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, et al. Chikungunya virus arthritis in adult wild-type mice. J Virol. (2010) 84:8021–32. doi: 10.1128/JVI.02603-09
48. Gasque P, Couderc T, Lecuit M, Roques P, Ng LF. Chikungunya virus pathogenesis and immunity. Vector Borne Zoonotic Dis. (2015) 15:241–9. doi: 10.1089/vbz.2014.1710
49. Lee WW, Teo TH, Her Z, Lum FM, Kam YW, Haase D, et al. Expanding regulatory T cells alleviates chikungunya virus-induced pathology in mice. J Virol. (2015) 89:7893–904. doi: 10.1128/JVI.00998-15
50. Kulkarni SP, Ganu M, Jayawant P, Thanapati S, Ganu A, Tripathy AS. Regulatory T cells and IL-10 as modulators of chikungunya disease outcome: a preliminary study. Eur J Clin Microbiol Infect Dis. (2017) 36:2475–81. doi: 10.1007/s10096-017-3087-4
51. Soares-Schanoski A, Baptista Cruz N, de Castro-Jorge LA, de Carvalho RVH, Santos CAD, Ros ND, et al. Systems analysis of subjects acutely infected with the chikungunya virus. PLoS Pathog. (2019) 15:e1007880. doi: 10.1371/journal.ppat.1007880
52. Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood. (2011) 118:e192–208. doi: 10.1182/blood-2011-04-345330
53. Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat Rev Immunol. (2010) 10:427–39. doi: 10.1038/nri2779
54. Wang J. Neutrophils in tissue injury and repair. Cell Tissue Res. (2018) 371:531–9. doi: 10.1007/s00441-017-2785-7
55. Poo YS, Rudd PA, Gardner J, Wilson JA, Larcher T, Colle MA, et al. Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection. PLoS Negl Trop Dis. (2014) 8:e3354. doi: 10.1371/journal.pntd.0003354
56. Appassakij H, Khuntikij P, Kemapunmanus M, Wutthanarungsan R, Silpapojakul K. Viremic profiles in asymptomatic and symptomatic chikungunya fever: a blood transfusion threat? Transfusion. (2013) 53(10 Pt 2):2567–74. doi: 10.1111/j.1537-2995.2012.03960.x
57. Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, et al. Persistent chronic inflammation and infection by chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. (2010) 184:5914–27. doi: 10.4049/jimmunol.0900255
58. Garcia-Arriaza J, Cepeda V, Hallengard D, Sorzano CO, Kummerer BM, Liljestrom P, et al. A novel poxvirus-based vaccine, MVA-CHIKV, is highly immunogenic and protects mice against chikungunya infection. J Virol. (2014) 88:3527–47. doi: 10.1128/JVI.03418-13
59. Wang D, Suhrbier A, Penn-Nicholson A, Woraratanadharm J, Gardner J, Luo M, et al. A complex adenovirus vaccine against chikungunya virus provides complete protection against viraemia and arthritis. Vaccine. (2011) 29:2803–9. doi: 10.1016/j.vaccine.2011.01.108
60. Selvarajah S, Sexton NR, Kahle KM, Fong RH, Mattia KA, Gardner J, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis. (2013) 7:e2423. doi: 10.1371/journal.pntd.0002423
61. Goh LY, Hobson-Peters J, Prow NA, Gardner J, Bielefeldt-Ohmann H, Pyke AT, et al. Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin Immunol. (2013) 149:487–97. doi: 10.1016/j.clim.2013.10.004
62. Prow NA, Liu L, Nakayama E, Cooper TH, Yan K, Eldi P, et al. A vaccinia-based single vector construct multi-pathogen vaccine protects against both Zika and chikungunya viruses. Nat Commun. (2018) 9:1230. doi: 10.1038/s41467-018-03662-6
63. Hallengard D, Kakoulidou M, Lulla A, Kummerer BM, Johansson DX, Mutso M, et al. Novel attenuated chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol. (2014) 88:2858–66. doi: 10.1128/JVI.03453-13
64. Chen W, Foo SS, Zaid A, Teng TS, Herrero LJ, Wolf S, et al. Specific inhibition of NLRP3 in chikungunya disease reveals a role for inflammasomes in alphavirus-induced inflammation. Nat Microbiol. (2017) 2:1435–45. doi: 10.1038/s41564-017-0015-4
65. Kam YW, Lum FM, Teo TH, Lee WW, Simarmata D, Harjanto S, et al. Early neutralizing IgG response to chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med. (2012) 4:330–43. doi: 10.1002/emmm.201200213
66. Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, et al. Dietary fiber confers protection against flu by shaping Ly6c(-) patrolling monocyte hematopoiesis and CD8(+) T cell metabolism. Immunity. (2018) 48:992–1005 e8. doi: 10.1016/j.immuni.2018.04.022
67. Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, Briseno CG, et al. Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. (2018) 22:3440–53 e6. doi: 10.1016/j.celrep.2018.03.001
68. Park J, Goergen CJ, HogenEsch H, Kim CH. Chronically elevated levels of short-chain fatty acids induce T cell-mediated ureteritis and hydronephrosis. J Immunol. (2016) 196:2388–400. doi: 10.4049/jimmunol.1502046
69. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. (2009) 461:1282–6. doi: 10.1038/nature08530
70. Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. (2014) 24:869–84. doi: 10.1101/gr.169508.113
71. Prow NA, Tang B, Gardner J, Le TT, Taylor A, Poo YS, et al. Lower temperatures reduce type I interferon activity and promote alphaviral arthritis. PLoS Pathog. (2017) 13:e1006788. doi: 10.1371/journal.ppat.1006788
72. Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. (2016). doi: 10.1101/060012.
73. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
74. Schroder WA, Le TT, Major L, Street S, Gardner J, Lambley E, et al. A physiological function of inflammation-associated SerpinB2 is regulation of adaptive immunity. J Immunol. (2010) 184:2663–70. doi: 10.4049/jimmunol.0902187
75. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4(+) T cells. Front Immunol. (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
76. Chen L, Sun M, Wu W, Yang W, Huang X, Xiao Y, et al. Microbiota metabolite butyrate differentially regulates Th1 and Th17 cells' differentiation and function in induction of colitis. Inflamm Bowel Dis. (2019) 25:1450–61. doi: 10.1093/ibd/izz046
77. Wang J, Wen L, Wang Y, Chen F. Therapeutic effect of histone deacetylase inhibitor, sodium butyrate, on allergic rhinitis in vivo. DNA Cell Biol. (2016) 35:203–8. doi: 10.1089/dna.2015.3037
78. Morrison TE, Oko L, Montgomery SA, Whitmore AC, Lotstein AR, Gunn BM, et al. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: evidence of arthritis, tenosynovitis, myositis, and persistence. Am J Pathol. (2011) 178:32–40. doi: 10.1016/j.ajpath.2010.11.018
79. Huits R, De Kort J, Van Den Berg R, Chong L, Tsoumanis A, Eggermont K, et al. Chikungunya virus infection in Aruba: diagnosis, clinical features and predictors of post-chikungunya chronic polyarthralgia. PLoS ONE. (2018) 13:e0196630. doi: 10.1371/journal.pone.0196630
80. Valamparampil JJ, Chirakkarot S, Letha S, Jayakumar C, Gopinathan KM. Clinical profile of Chikungunya in infants. Indian J Pediatr. (2009) 76:151–5. doi: 10.1007/s12098-009-0045-x
81. Gerardin P, Barau G, Michault A, Bintner M, Randrianaivo H, Choker G, et al. Multidisciplinary prospective study of mother-to-child chikungunya virus infections on the island of La Reunion. PLoS Med. (2008) 5:e60. doi: 10.1371/journal.pmed.0050060
82. Jaffar-Bandjee MC, Ramful D, Gauzere BA, Hoarau JJ, Krejbich-Trotot P, Robin S, et al. Emergence and clinical insights into the pathology of chikungunya virus infection. Expert Rev Anti Infect Ther. (2010) 8:987–96. doi: 10.1586/eri.10.92
83. Fraser JR, Cunningham AL, Clarris BJ, Aaskov JG, Leach R. Cytology of synovial effusions in epidemic polyarthritis. Aust N Z J Med. (1981) 11:168–73. doi: 10.1111/j.1445-5994.1981.tb04226.x
84. Soden M, Vasudevan H, Roberts B, Coelen R, Hamlin G, Vasudevan S, et al. Detection of viral ribonucleic acid and histologic analysis of inflamed synovium in Ross River virus infection. Arthritis Rheum. (2000) 43:365–9. doi: 10.1002/1529-0131(200002)43:2<365::AID-ANR16>3.0.CO;2-E
85. Chen W, Wang Q, Ke Y, Lin J. Neutrophil function in an inflammatory milieu of rheumatoid arthritis. J Immunol Res. (2018) 2018:8549329. doi: 10.1155/2018/8549329
86. Suo J, Linke B, Meyer dos Santos S, Pierre S, Stegner D, Zhang DD, et al. Neutrophils mediate edema formation but not mechanical allodynia during zymosan-induced inflammation. J Leukoc Biol. (2014) 96:133–42. doi: 10.1189/jlb.3A1213-628R
87. Rosengren S, Arfors KE. Neutrophil-mediated vascular leakage is not suppressed by leukocyte elastase inhibitors. Am J Physiol. (1990) 259(4 Pt 2):H1288–94. doi: 10.1152/ajpheart.1990.259.4.H1288
88. Shiga Y, Onodera H, Kogure K, Yamasaki Y, Yashima Y, Syozuhara H, et al. Neutrophil as a mediator of ischemic edema formation in the brain. Neurosci Lett. (1991) 125:110–2. doi: 10.1016/0304-3940(91)90003-C
89. Liu YW, Li S, Dai SS. Neutrophils in traumatic brain injury (TBI): friend or foe? J Neuroinflammation. (2018) 15:146. doi: 10.1186/s12974-018-1173-x
90. Friedman AD, Keefer JR, Kummalue T, Liu H, Wang QF, Cleaves R. Regulation of granulocyte and monocyte differentiation by CCAAT/enhancer binding protein alpha. Blood Cells Mol Dis. (2003) 31:338–41. doi: 10.1016/S1079-9796(03)00135-9
91. De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. (2013) 121:4930–7. doi: 10.1182/blood-2013-02-486217
92. Greer A, Irie K, Hashim A, Leroux BG, Chang AM, Curtis MA, et al. Site-specific neutrophil migration and CXCL2 expression in periodontal tissue. J Dent Res. (2016) 95:946–52. doi: 10.1177/0022034516641036
93. Dienz O, Rud JG, Eaton SM, Lanthier PA, Burg E, Drew A, et al. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. (2012) 5:258–66. doi: 10.1038/mi.2012.2
94. Moots RJ, Sebba A, Rigby W, Ostor A, Porter-Brown B, Donaldson F, et al. Effect of tocilizumab on neutrophils in adult patients with rheumatoid arthritis: pooled analysis of data from phase 3 and 4 clinical trials. Rheumatology. (2017) 56:541–9. doi: 10.1093/rheumatology/kew370
95. Le TT, Skak K, Schroder K, Schroder WA, Boyle GM, Pierce CJ, et al. IL-1 contributes to the anti-cancer efficacy of ingenol mebutate. PLoS ONE. (2016) 11:e0153975. doi: 10.1371/journal.pone.0153975
96. Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, et al. IFN-lambda resolves inflammation via suppression of neutrophil infiltration and IL-1beta production. J Exp Med. (2015) 212:845–53. doi: 10.1084/jem.20140995
97. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. (2013) 13:159–75. doi: 10.1038/nri3399
98. Niki Y, Yamada H, Seki S, Kikuchi T, Takaishi H, Toyama Y, et al. Macrophage- and neutrophil-dominant arthritis in human IL-1 alpha transgenic mice. J Clin Invest. (2001) 107:1127–35. doi: 10.1172/JCI11530
99. Zhu X, Xiao L, Huo R, Zhang J, Lin J, Xie J, et al. Cyr61 is involved in neutrophil infiltration in joints by inducing IL-8 production by fibroblast-like synoviocytes in rheumatoid arthritis. Arthritis Res Ther. (2013) 15:R187. doi: 10.1186/ar4377
100. Reichel CA, Khandoga A, Anders HJ, Schlondorff D, Luckow B, Krombach F. Chemokine receptors Ccr1, Ccr2, and Ccr5 mediate neutrophil migration to postischemic tissue. J Leukoc Biol. (2006) 79:114–22. doi: 10.1189/jlb.0605337
101. Price PJ, Luckow B, Torres-Dominguez LE, Brandmuller C, Zorn J, Kirschning CJ, et al. Chemokine (C-C Motif) receptor 1 is required for efficient recruitment of neutrophils during respiratory infection with modified vaccinia virus Ankara. J Virol. (2014) 88:10840–50. doi: 10.1128/JVI.01524-14
102. Williams AE, Jose RJ, Mercer PF, Brealey D, Parekh D, Thickett DR, et al. Evidence for chemokine synergy during neutrophil migration in ARDS. Thorax. (2017) 72:66–73. doi: 10.1136/thoraxjnl-2016-208597
103. Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. (2008) 28:29–39. doi: 10.1016/j.immuni.2007.11.016
104. Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPalpha is required for induction of granulocytic differentiation. Blood. (2003) 102:1267–75. doi: 10.1182/blood-2003-02-0477
105. Huebener P, Pradere JP, Hernandez C, Gwak GY, Caviglia JM, Mu X, et al. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J Clin Invest. (2015) 125:539–50. doi: 10.1172/JCI76887
106. Qi Y, Ge J, Ma C, Wu N, Cui X, Liu Z. Activin A regulates activation of mouse neutrophils by Smad3 signalling. Open Biol. (2017) 7:160342. doi: 10.1098/rsob.160342
107. Coccia EM, Stellacci E, Valtieri M, Masella B, Feccia T, Marziali G, et al. Ectopic expression of interferon regulatory factor-1 potentiates granulocytic differentiation. Biochem J. (2001) 360(Pt 2):285–94. doi: 10.1042/bj3600285
108. Keeter WC, Moriarty A, Butcher MJ, Ma KW, Nadler JL, Galkina E. IL-12 induced STAT4 activation plays a role in pro-inflammatory neutrophil functions. J Immunol. (2018) 200 (Suppl. 1):166.59.
109. Cullen EM, Brazil JC, O'Connor CM. Mature human neutrophils constitutively express the transcription factor EGR-1. Mol Immunol. (2010) 47:1701–9. doi: 10.1016/j.molimm.2010.03.003
110. Walmsley SR, Print C, Farahi N, Peyssonnaux C, Johnson RS, Cramer T, et al. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med. (2005) 201:105–15. doi: 10.1084/jem.20040624
111. Hwang J, Kita R, Kwon HS, Choi EH, Lee SH, Udey MC, et al. Epidermal ablation of Dlx3 is linked to IL-17-associated skin inflammation. Proc Natl Acad Sci USA. (2011) 108:11566–71. doi: 10.1073/pnas.1019658108
112. Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. (2000) 404:99–103. doi: 10.1038/35003607
113. Vinolo MA, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly YM, et al. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS ONE. (2011) 6:e21205. doi: 10.1371/journal.pone.0021205
114. Parsonage G, Filer A, Bik M, Hardie D, Lax S, Howlett K, et al. Prolonged, granulocyte-macrophage colony-stimulating factor-dependent, neutrophil survival following rheumatoid synovial fibroblast activation by IL-17 and TNFalpha. Arthritis Res Ther. (2008) 10:R47. doi: 10.1186/ar2406
115. Hashimoto M. Th17 in animal models of rheumatoid arthritis. J Clin Med. (2017) 6:E73. doi: 10.3390/jcm6070073
116. Bazzoni F, Tamassia N, Rossato M, Cassatella MA. Understanding the molecular mechanisms of the multifaceted IL-10-mediated anti-inflammatory response: lessons from neutrophils. Eur J Immunol. (2010) 40:2360–8. doi: 10.1002/eji.200940294
117. Zenobia C, Hajishengallis G. Basic biology and role of interleukin-17 in immunity and inflammation. Periodontol 2000. (2015) 69:142–59. doi: 10.1111/prd.12083
118. Finsterbusch M, Voisin MB, Beyrau M, Williams TJ, Nourshargh S. Neutrophils recruited by chemoattractants in vivo induce microvascular plasma protein leakage through secretion of TNF. J Exp Med. (2014) 211:1307–14. doi: 10.1084/jem.20132413
119. Royall JA, Berkow RL, Beckman JS, Cunningham MK, Matalon S, Freeman BA. Tumor necrosis factor and interleukin 1 alpha increase vascular endothelial permeability. Am J Physiol. (1989) 257(6 Pt 1):L399–410. doi: 10.1152/ajplung.1989.257.6.L399
120. Rodrigues SF, Granger DN. Blood cells and endothelial barrier function. Tissue Barriers. (2015) 3:e978720. doi: 10.4161/21688370.2014.978720
121. Chang S, Chen X, Huang Z, Chen D, Yu B, Chen H, et al. Dietary sodium butyrate supplementation promotes oxidative fiber formation in mice. Anim Biotechnol. (2018) 29:212–5. doi: 10.1080/10495398.2017.1358734
122. Swindell WR, Johnston A, Carbajal S, Han G, Wohn C, Lu J, et al. Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis. PLoS ONE. (2011) 6:e18266. doi: 10.1371/journal.pone.0018266
123. Zenz R, Eferl R, Kenner L, Florin L, Hummerich L, Mehic D, et al. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. (2005) 437:369–75. doi: 10.1038/nature03963
124. Souza BCE, Bandeira LG, Cunha T, Valente NYS. Follicular psoriasis: an underdiagnosed entity? An Bras Dermatol. (2019) 94:116–8. doi: 10.1590/abd1806-4841.20197987
125. Hitomi T, Matsuzaki Y, Yokota T, Takaoka Y, Sakai T. p15(INK4b) in HDAC inhibitor-induced growth arrest. FEBS Lett. (2003) 554:347–50. doi: 10.1016/S0014-5793(03)01186-4
126. Coradini D, Pellizzaro C, Marimpietri D, Abolafio G, Daidone MG. Sodium butyrate modulates cell cycle-related proteins in HT29 human colonic adenocarcinoma cells. Cell Prolif. (2000) 33:139–46. doi: 10.1046/j.1365-2184.2000.00173.x
127. Sowa Y, Orita T, Hiranabe-Minamikawa S, Nakano K, Mizuno T, Nomura H, et al. Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites. Ann N Y Acad Sci. (1999) 886:195–9. doi: 10.1111/j.1749-6632.1999.tb09415.x
128. Nelson DM, Ye X, Hall C, Santos H, Ma T, Kao GD, et al. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol Cell Biol. (2002) 22:7459–72. doi: 10.1128/MCB.22.21.7459-7472.2002
129. Bindels LB, Porporato P, Dewulf EM, Verrax J, Neyrinck AM, Martin JC, et al. Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br J Cancer. (2012) 107:1337–44. doi: 10.1038/bjc.2012.409
130. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. (2002) 132:1012–7. doi: 10.1093/jn/132.5.1012
131. Park JH, Noh SM, Woo JR, Kim JW, Lee GM. Valeric acid induces cell cycle arrest at G1 phase in CHO cell cultures and improves recombinant antibody productivity. Biotechnol J. (2016) 11:487–96. doi: 10.1002/biot.201500327
132. Kim K, Kwon O, Ryu TY, Jung CR, Kim J, Min JK, et al. Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer. Mol Med Rep. (2019) 20:1569–74. doi: 10.3892/mmr.2019.10431
133. Blottiere HM, Buecher B, Galmiche JP, Cherbut C. Molecular analysis of the effect of short-chain fatty acids on intestinal cell proliferation. Proc Nutr Soc. (2003) 62:101–6. doi: 10.1079/PNS2002215
134. Sangfelt O, Erickson S, Castro J, Heiden T, Gustafsson A, Einhorn S, et al. Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene. (1999) 18:2798–810. doi: 10.1038/sj.onc.1202609
135. Kim SY, Nair MG. Macrophages in wound healing: activation and plasticity. Immunol Cell Biol. (2019) 97:258–67. doi: 10.1111/imcb.12236
136. Gieseck RL 3rd, Wilson MS, Wynn TA. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol. (2018) 18:62–76. doi: 10.1038/nri.2017.90
137. Prasad KN, Bondy SC. Dietary fibers and their fermented short-chain fatty acids in prevention of human diseases. Mech Ageing Dev. (2018). doi: 10.1016/j.mad.2018.10.003. [Epub ahead of print].
138. Bejjani F, Evanno E, Zibara K, Piechaczyk M, Jariel-Encontre I. The AP-1 transcriptional complex: local switch or remote command? Biochim Biophys Acta Rev Cancer. (2019) 1872:11–23. doi: 10.1016/j.bbcan.2019.04.003
139. Nguyen C, Teo JL, Matsuda A, Eguchi M, Chi EY, Henderson WR Jr, et al. Chemogenomic identification of Ref-1/AP-1 as a therapeutic target for asthma. Proc Natl Acad Sci USA. (2003) 100:1169–73. doi: 10.1073/pnas.0437889100
140. Tegeder I, Niederberger E, Israr E, Guhring H, Brune K, Euchenhofer C, et al. Inhibition of NF-kappaB and AP-1 activation by R- and S-flurbiprofen. FASEB J. (2001) 15:595–7. doi: 10.1096/fasebj.15.3.595
141. Trop-Steinberg S, Azar Y. AP-1 expression and its clinical relevance in immune disorders and cancer. Am J Med Sci. (2017) 353:474–83. doi: 10.1016/j.amjms.2017.01.019
142. Silva JPB, Navegantes-Lima KC, Oliveira ALB, Rodrigues DVS, Gaspar SLF, Monteiro VVS, et al. Protective mechanisms of butyrate on inflammatory bowel disease. Curr Pharm Des. (2018) 24:4154–66. doi: 10.2174/1381612824666181001153605
143. Li YG, Siripanyaphinyo U, Tumkosit U, Noranate N, A-nuegoonpipat A, Pan Y, et al. Poly (I:C), an agonist of toll-like receptor-3, inhibits replication of the Chikungunya virus in BEAS-2B cells. Virol J. (2012) 9:114. doi: 10.1186/1743-422X-9-114
144. Lee MS, Kim B, Oh GT, Kim YJ. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. (2013) 14:346–55. doi: 10.1038/ni.2535
145. Xu Y, Gao C, Guo H, Zhang W, Huang W, Tang S, et al. Sodium butyrate supplementation ameliorates diabetic inflammation in db/db mice. J Endocrinol. (2018) 238:231–44. doi: 10.1530/JOE-18-0137
146. Folkerts J, Stadhouders R, Redegeld FA, Tam SY, Hendriks RW, Galli SJ, et al. Effect of dietary fiber and metabolites on mast cell activation and mast cell-associated diseases. Front Immunol. (2018) 9:1067. doi: 10.3389/fimmu.2018.01067
147. Sun B, Jia Y, Hong J, Sun Q, Gao S, Hu Y, et al. Sodium butyrate ameliorates high-fat-diet-induced non-alcoholic fatty liver disease through peroxisome proliferator-activated receptor alpha-mediated activation of beta oxidation and suppression of inflammation. J Agric Food Chem. (2018) 66:7633–42. doi: 10.1021/acs.jafc.8b01189
148. Blauvelt A, Chiricozzi A. The immunologic role of IL-17 in psoriasis and psoriatic arthritis pathogenesis. Clin Rev Allergy Immunol. (2018) 55:379–90. doi: 10.1007/s12016-018-8702-3
149. George SM, Taylor MR, Farrant PB. Psoriatic alopecia. Clin Exp Dermatol. (2015) 40:717–21. doi: 10.1111/ced.12715
150. Cunha RVD, Trinta KS. Chikungunya virus: clinical aspects and treatment - A review. Mem Inst Oswaldo Cruz. (2017) 112:523–31. doi: 10.1590/0074-02760170044
151. Sá-Caputo D, Paineiras-Domingos LL, Guedes-Aguiar EO, Carvalho-Lima RP, de Paoli S, Morel DS, et al. Could hair loss be a relevant symptom associated with Chikungunya? Trichol Cosmetol Open J. (2017) 1:25–30. doi: 10.17140/TCOJ-1-106
152. van Aalst M, Nelen CM, Goorhuis A, Stijnis C, Grobusch MP. Long-term sequelae of chikungunya virus disease: a systematic review. Travel Med Infect Dis. (2017) 15:8–22. doi: 10.1016/j.tmaid.2017.01.004
153. Soumahoro MK, Gerardin P, Boelle PY, Perrau J, Fianu A, Pouchot J, et al. Impact of Chikungunya virus infection on health status and quality of life: a retrospective cohort study. PLoS ONE. (2009) 4:e7800. doi: 10.1371/journal.pone.0007800
154. Patil DR, Hundekar SL, Arankalle VA. Expression profile of immune response genes during acute myopathy induced by chikungunya virus in a mouse model. Microbes Infect. (2012) 14:457–69. doi: 10.1016/j.micinf.2011.12.008
155. Ziegler SA, Lu L, da Rosa AP, Xiao SY, Tesh RB. An animal model for studying the pathogenesis of chikungunya virus infection. Am J Trop Med Hyg. (2008) 79:133–9. doi: 10.4269/ajtmh.2008.79.133
156. Goh LY, Hobson-Peters J, Prow NA, Gardner J, Bielefeldt-Ohmann H, Suhrbier A, et al. Monoclonal antibodies specific for the capsid protein of chikungunya virus suitable for multiple applications. J Gen Virol. (2015) 96(Pt 3):507–12. doi: 10.1099/jgv.0.000002
157. O'Rielly DD, Jani M, Rahman P, Elder JT. The genetics of psoriasis and psoriatic arthritis. J Rheumatol Suppl. (2019) 95:46–50. doi: 10.3899/jrheum.190119
158. Salem I, Ramser A, Isham N, Ghannoum MA. The gut microbiome as a major regulator of the gut-skin axis. Front Microbiol. (2018) 9:1459. doi: 10.3389/fmicb.2018.01459
159. Amin M, Darji K, No DJ, Bhutani T, Wu JJ. Review of IL-17 inhibitors for psoriasis. J Dermatolog Treat. (2018) 29:347–52. doi: 10.1080/09546634.2017.1395796
160. Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. (2017) 3:e1700492. doi: 10.1126/sciadv.1700492
161. Luo A, Leach ST, Barres R, Hesson LB, Grimm MC, Simar D. The microbiota and epigenetic regulation of T helper 17/regulatory T cells: in search of a balanced Immune system. Front Immunol. (2017) 8:417. doi: 10.3389/fimmu.2017.00417
162. Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, et al. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am J Physiol Gastrointest Liver Physiol. (2012) 302:G1405–15. doi: 10.1152/ajpgi.00543.2011
163. Wu Q, Nie J, Gao Y, Xu P, Sun Q, Yang J, et al. Reciprocal regulation of RORgammat acetylation and function by p300 and HDAC1. Sci Rep. (2015) 5:16355. doi: 10.1038/srep16355
164. Weger-Lucarelli J, Auerswald H, Vignuzzi M, Dussart P, Karlsson EA. Taking a bite out of nutrition and arbovirus infection. PLoS Negl Trop Dis. (2018) 12:e0006247. doi: 10.1371/journal.pntd.0006247
165. Zhao-Fleming H, Hand A, Zhang K, Polak R, Northcut A, Jacob D, et al. Effect of non-steroidal anti-inflammatory drugs on post-surgical complications against the backdrop of the opioid crisis. Burns Trauma. (2018) 6:25. doi: 10.1186/s41038-018-0128-x
166. Bootun R. Effects of immunosuppressive therapy on wound healing. Int Wound J. (2013) 10:98–104. doi: 10.1111/j.1742-481X.2012.00950.x
167. Wang AS, Armstrong EJ, Armstrong AW. Corticosteroids and wound healing: clinical considerations in the perioperative period. Am J Surg. (2013) 206:410–7. doi: 10.1016/j.amjsurg.2012.11.018
168. Morikawa A, Sugiyama T, Koide N, Mori I, Mu MM, Yoshida T, et al. Butyrate enhances the production of nitric oxide in mouse vascular endothelial cells in response to gamma interferon. J Endotoxin Res. (2004) 10:32–8. doi: 10.1179/096805104225003852
169. Labadie K, Larcher T, Joubert C, Mannioui A, Delache B, Brochard P, et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. (2010) 120:894–906. doi: 10.1172/JCI40104
170. Mao SZ, Ye X, Liu G, Song D, Liu SF. Resident endothelial cells and endothelial progenitor cells restore endothelial barrier function after inflammatory lung injury. Arterioscler Thromb Vasc Biol. (2015) 35:1635–44. doi: 10.1161/ATVBAHA.115.305519
171. Mao SZ, Ye X, Liu G, Song D, Liu SF. An obligatory role of NF-kappaB in mediating bone marrow derived endothelial progenitor cell recruitment and proliferation following endotoxemic multiple organ injury in mice. PLoS ONE. (2014) 9:e111087. doi: 10.1371/journal.pone.0111087
172. Liu G, Ye X, Miller EJ, Liu SF. NF-kappaB-to-AP-1 switch: a mechanism regulating transition from endothelial barrier injury to repair in endotoxemic mice. Sci Rep. (2014) 4:5543. doi: 10.1038/srep05543
173. Gharbi C, Gueutin V, Izzedine H. Oedema, solid organ transplantation and mammalian target of rapamycin inhibitor/proliferation signal inhibitors (mTOR-I/PSIs). Clin Kidney J. (2014) 7:115–20. doi: 10.1093/ckj/sfu001
174. Motse KG, Mashabane MJ. Sirolimus-induced lymphoedema. S Afr Med J. (2016) 106:886–7. doi: 10.7196/SAMJ.2016.v106i9.10636
175. Chen J, Zhao KN, Vitetta L. Effects of intestinal microbial(-)elaborated butyrate on oncogenic signaling pathways. Nutrients. (2019) 11:E1026. doi: 10.3390/nu11051026
176. Bultman SJ. Bacterial butyrate prevents atherosclerosis. Nat Microbiol. (2018) 3:1332–3. doi: 10.1038/s41564-018-0299-z
Keywords: chikungunya, immunopathology, arthritis, fiber, diet
Citation: Prow NA, Hirata TDC, Tang B, Larcher T, Mukhopadhyay P, Alves TL, Le TT, Gardner J, Poo YS, Nakayama E, Lutzky VP, Nakaya HI and Suhrbier A (2019) Exacerbation of Chikungunya Virus Rheumatic Immunopathology by a High Fiber Diet and Butyrate. Front. Immunol. 10:2736. doi: 10.3389/fimmu.2019.02736
Received: 26 July 2019; Accepted: 08 November 2019;
Published: 26 November 2019.
Edited by:
Lisa F. P. Ng, Singapore Immunology Network (A*STAR), SingaporeReviewed by:
Xenia Maria Ficht, San Raffaele Hospital (IRCCS), ItalyCopyright © 2019 Prow, Hirata, Tang, Larcher, Mukhopadhyay, Alves, Le, Gardner, Poo, Nakayama, Lutzky, Nakaya and Suhrbier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Suhrbier, YW5kcmVhcy5zdWhyYmllckBxaW1yYmVyZ2hvZmVyLmVkdS5hdQ==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.