- 1Department of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy
- 2GlaxoSmithKline (GSK) Srl, Siena, Italy
- 3Pasteur Laboratory, Istituto Pasteur-Fondazione Cenci Bolognetti, Rome, Italy
Understanding Staphylococcus aureus (S. aureus)–host immune system interaction is crucial to meet the tremendous medical need associated with this life-threatening bacterial infection. Given the crucial role of dendritic cells (DC) in dictating immune responses upon microbial challenge, we investigated how the bacterial viability and the conservation of structural integrity influence the response of human DC to S. aureus. To this end, human primary DC were stimulated with the methicillin-resistant S. aureus USA300 live strain, USA300 inactivated by heat (HI), ultraviolet irradiation (UVI), or paraformaldehyde treatment (PFAI) and subsequently analyzed for cell phenotype and immune-modulatory properties. Although no differences in terms of DC viability and maturation were observed when DC were stimulated with live or inactivated bacteria, the production of IL-12, IL-23, and other cytokines differed significantly. The Th1 and Th17 expansion was also more pronounced in response to live vs. inactivated S. aureus. Interestingly, cytokine production in DC treated with live and inactivated USA300 required phagocytosis, whereas blocking endosomal Toll-like receptor signaling mainly reduced the cytokine release by live and HI USA300. A further analysis of IFN-β signaling revealed the induction of a cyclic GMP-AMP synthase stimulator of interferon genes (cGAS-STING)-independent and IRF3-dependent signaling pathway(s) in UVI-stimulated DC. This study underscores the capacity of human DC to discriminate between live and inactivated S. aureus and, further, indicates that DC may represent a valuable experimental setting to test different inactivation methods with regard to the retention of S. aureus immunoregulatory properties. These and further insights may be useful for the development of novel therapeutic and prophylactic anti-S. aureus vaccine strategies.
Introduction
Staphylococcus aureus (S. aureus) is an opportunistic pathogen that causes a wide spectrum of infections, ranging from superficial skin lesions to life-threatening conditions, such as sepsis, endocarditis, and pneumonia (1, 2). The large numbers of nosocomial and community-acquired infections make S. aureus a serious public health problem and, consequently, a major burden on society worldwide (3). In addition, the resistance or limited effectiveness of antibiotic treatment, as well as the lack of an effective vaccine against these bacteria reveals a global unmet medical need for effective therapies.
Analysis of immune response against S. aureus has revealed a complex and multifaceted process (4, 5). The wide repertoire of S. aureus virulence factors triggers multiple pattern recognition receptor (PRR) pathways, leading to the activation of different innate and, subsequently, adaptive immune responses. Activation of Toll-like receptors (TLRs) such as TLR2 (6), TLR9 (7), and TLR8 (8) after S. aureus infection has been demonstrated in human and mouse settings. In addition, S. aureus can escape from host cell endosomes to the cytoplasm (9), where it can activate cytosolic sensors, including the nucleotide oligomerization domain 2 (NOD-2) (10), the NOD-like receptor P3 (NLRP3) (11), Absent in melanoma 2 (AIM2) (12), and stimulator of interferon (IFN) genes (STING) (13). In different cell types, infection with S. aureus induces type I IFN signaling through the activation of diverse PRRs (7, 14). Recently, Scumpia et al. showed that both TLR and STING pathways are activated a few hours after macrophage infection with live S. aureus and compete for the regulation of ~95% of the induced genes; in particular, TLR signaling predominantly activated a proinflammatory program while STING signaling activated an antiviral/type I IFN response (13). Conversely, heat-killed bacterium activated mainly TLR signaling (13). This differential response may be related to the production of signaling nucleotides—cyclic di-AMP (c-di-AMP) synthesized in response to infection with live bacteria—that are able to activate the pro-inflammatory cGAS–STING–IRF3 response leading to type I IFN production (15, 16). However, the relative contribution of each pathway in orchestrating the immune response against S. aureus is not fully understood and is complicated by the existence of many bacterial strains and infection routes.
Dendritic cells (DC) are key components of the immune system for their extraordinary capacity to initiate primary immune responses and stimulate naïve T cells (17). DC play an important role in orchestrating and regulating immune responses against pathogens, including S. aureus (18, 19). In this respect, we recently demonstrated that the virulence factors Esx secreted by live S. aureus modulate human DC functions and their capacity to support a Th1/Th17 response (20). These results illustrated how human DC may sense small differences in S. aureus virulence to dynamically instruct both innate and adaptive immunity.
In the present study, we further interrogated the DC experimental setting to understand how the viability and the structural integrity of S. aureus may influence the interaction with human DC and, in turn, the outcome of the Th response. Indeed, because of the vita-pathogen associated molecular patterns (PAMPs)—structures associated only with live microorganisms (21)—innate immune responses can discriminate between live and dead microbes and rapidly evaluate the severity of the infectious threat and modulate the magnitude and the quality of the immune responses (22–24).
An in vitro model, based on human monocyte-derived DC, was used to analyze differences in the response to live or inactivated S. aureus preparations in terms of DC phenotype, immune-modulatory properties, and regulation of cytokine production. This study highlights important aspects of the complex and multifaceted interplay of different innate immune signaling pathways in human DC in response to the interaction with live and differently inactivated S. aureus, which could be exploited to design novel therapeutic and prophylactic anti-S. aureus vaccine strategies.
Materials and Methods
Antibodies and Other Reagents
Monoclonal antibodies (Abs), specific for cluster of differentiation (CD)1a, CD14, CD38, CD86, CD83, HLA-DR, CD40, IgG1, and IgG2a (BD Bioscience, San Diego, CA, USA), were directly conjugated to fluorescein isothiocyanate (FITC) or phycoerythrin (PE). To exclude dead cells from the analysis, Fixable Viability Dye eFluor®780 (FvDye) (eBioscience, San Diego, CA, USA) was used. For immunoblotting analysis, rabbit anti-STING (Cell Signaling, Danvers, MA, USA # 2775), anti-IRF3 (Santa Cruz, Santa Cruz, TX, USA # sc-9082), anti-IRF7 (Santa Cruz, # sc-9083), anti-STAT1 (BD Bioscience, # 610186), anti-phospho STAT1 (Cell Signaling Technology, Leiden, The Netherlands, # 7649), anti-STAT2 (BD Transduction Laboratories, # 610188), anti-phospho STAT2 (R&D Systems, Minneapolis, MN, USA, MAB2890), mouse anti-actin (Sigma-Aldrich, St. Louis, MO, USA #A0483), and horseradish peroxidase-conjugated secondary antibody anti mouse (Santa Cruz, # sc-2005) and anti rabbit (Santa Cruz, # sc-2004) were used. For phagocytosis and phagosomal acidification experiments, cytochalasin D 5 μM (Sigma-Aldrich, # C8723) and chloroquine 2 μM (Sigma-Aldrich, # C6628) were used.
Bacterial USA300 Growth and Inactivation Conditions
Briefly, S. aureus USA300 was grown in tryptic soy broth (TSB, BD Bioscience # BA-25107.05) overnight at 37°C. The next day, bacterial broth was diluted 1:100 in fresh TSB, cultured until the exponential phase of growth (OD600 of 0.6), and then washed in RPMI 1640 and resuspended in RPMI 1640 supplemented with L-glutamine (2 mM) and 15% fetal bovine serum (FBS) for DC infection. To inactivate USA300, 10 ml of bacterial culture (0.6 OD600) was washed in PBS and either treated with UV-irradiation (254 nm) for 40 min on ice (UV-inactivated, UVI), heat-inactivated at 100°C for 15 min (HI) or fixed with 4% paraformaldehyde (PFA-inactivated, PFAI) (Panreac Quimica, Castellar del Valles, ES) for 30 min at room temperature (RT), and then washed three times with PBS. To confirm the lack of viable bacteria following UV irradiation, heat killing, or PFA fixation, 200 μl of the undiluted bacterial suspension was plated on tryptic soy agar (TSA, Oxoid, Basingstoke, Hants, UK) and incubated at 37°C overnight. The absence of growth indicated the efficacy of the inactivation procedures.
DC Preparation and Stimulation
Istituto Superiore di Sanità (ISS) Review Board approved the present research project (CE/13/387). Informed consent was obtained from all donors before collecting the blood samples. No specific analysis on S. aureus infectious status was performed on blood samples given for research studies since the agreement signed by ISS and the Blood Donation Center does not allow the monitoring of specific infectious diseases in addition to those routinely performed within the serology testing for suitability of blood donation. DC were prepared as previously described (25). DC were generated by culturing monocytes with 50 ng/ml GM-CSF (R&D Systems, Minneapolis, MN, USA) and 200 U/ml of IL-4 (Miltenyi, Bergisch Gladbach, DE) for 5 days at 0.5 × 106 cells/ml in RPMI 1640 (BioWhittaker Europe, Verviers, BE) supplemented with L-glutamine (2 mM) (Lonza, Basel, CH) and 15% FBS (Lonza). At day 5, cells were tested for their differentiation status by evaluating CD1a expression (>90% CD1a+- as apex; “CD14−”) and lack of CD14 (>95% CD14). Before stimulation, the medium was replaced with RPMI supplemented only with L-glutamine (2 mM) and 15% FBS. Cytokine deprivation did not affect DC survival rate, which was >90%. For live S. aureus infection, DC were infected as previously described (20), using a multiplicity of infection (MOI) of 0.1 bacterium/cell. For treatment with inactivated S. aureus preparations, a dose response with inactivated bacteria was used, monitoring CD86 expression on the surface of stimulated DC (data not shown). An MOI of 1 inactivated bacterium/cell was the chosen dose leading to a comparable stimulation to that observed in DC infected with live bacteria and was then used in all experiments. To assess the role of phagocytosis in the induction of cytokine response, prior to the stimulation with live and inactivated USA300 strain, DC were pre-treated with 5 μM Cytochalasin D for 30 min. Inhibition of endosomal acidification was performed by pre-treating DC with 2 μM chloroquine for 30 min before bacterial stimulation.
Flow Cytometry Analysis
Cells (105) were washed once in phosphate buffered saline (PBS) (Lonza) containing 2% FBS and incubated with indicated monoclonal Abs at 4°C for 30 min. DC were then washed and fixed with 2% formaldehyde (Panreac Quimica) before analysis on a Gallios cytometer (Beckman Coulter, Brea, CA, USA). A total of 30,000 events were analyzed per sample in FvDye negative live cells. In viable DC, the expression of cell surface molecules was evaluated using the median fluorescence intensity (MFI) after subtraction of the values of the isotype Ab controls.
Cell Viability Detection
Cell viability was analyzed by using FvDye according to manufacturing protocols. Twenty-four hours post infection, DC were stained with FvDye as previously described (26). Finally, cells were fixed overnight with 4% formaldehyde before analysis on a Gallios cytometer (Beckman Coulter). Data were analyzed by Kaluza software (Beckman Coulter).
T Cell Response
Total CD4+ T cells were isolated from autologous frozen peripheral blood mononuclear cells by indirect magnetic sorting with a CD4+ T-cell isolation kit (Miltenyi), as previously described (26). Purified cells were plated in 96-well U-bottomed tissue culture plates at the density of 0.4 × 106 cells/ml with DC previously stimulated for 24 h with the live and inactivated S. aureus at a density of 0.4 × 105/ml (ratio 1 DC: 10 CD4+ T cells). At day 5, supernatants were harvested for IFN-γ and IL-17 detection.
Cytokine Determination
Supernatants of DC cultures were harvested 24 h after stimulation with live and inactivated S. aureus, filtered (0.2 μm), and stored at −80°C. The production of IL-12, TNF-α, IL-10, IL-1β, IL-6, and IL-8 was measured by human Inflammatory Cytokine kit (Cytometric Bead Array, CBA, BD Bioscience). Release of IL-23, IFN-γ, and IL-17 was instead assayed by specific ELISA kits (R&D Systems) or by human Th1/Th2 Cytokine kit (Cytometric Bead Array, CBA, BD Bioscience).
RNA Isolation and Quantitative Real-Time PCR
Total RNA was extracted from DC (1 × 106) using TRIzol Reagent (Thermo Fisher Scientific, Palm beach, FL, USA) following the manufacturer's recommendations. Reverse transcriptions were performed as previously described (25). Quantitative PCR assays were performed at least in duplicate using the Platinum Taq DNA Polymerase (Invitrogen Life Technologies Frederick, MD) and the SYBR Green I (Lonza) on a LightCycler (Roche Diagnostics, Basel, CH). Primer pairs used to analyze GAPDH, IFN-β, and IFN-αs expression have been previously described (27). Transcript expression was normalized to the GAPDH level using the Equation 2−ΔCt.
Immunoblot Analysis
Western blotting was performed as previously described (25). Briefly, 25 μg of total protein extracts was separated on 10% SDS-PAGE gel and blotted onto nitrocellulose membranes (Merck–Millipore, Darmstadt, DE). Blots were incubated with rabbit polyclonal Abs against STING, IRF3, IRF7, STAT-1, and STAT-2. Detection was achieved using anti-rabbit horseradish peroxidase-conjugated secondary Ab (Santa Cruz), and visualized with Enhanced Chemiluminescence plus kit (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). A ChemiDoc XRS (Bio-Rad, Hercules, CA, USA) instrument and ImageLab software (Bio-Rad) were used to reveal and analyze the chemiluminescence signal. β-Actin levels were analyzed by mouse anti-β-actin Ab to verify the loaded protein amount.
Statistical Analysis
Statistical analysis was carried out using a two-tailed Student's t-test for paired data. A p-value < 0.05 was considered statistically significant.
Results
Live or Inactivated S. aureus-Stimulated DC Drive a Differential Expansion of Th1 and Th17 Cells
We previously demonstrated that DC respond to S. aureus infection by acquiring a mature phenotype required for the conditioning of Th1/Th17 response (20). Here, we extend our analysis to the characterization of how different S. aureus inactivation methods impact the DC immune phenotype and regulatory properties. After the identification of the optimal S. aureus MOI and stimulation doses with inactivated bacteria (see Materials and Methods), we assayed the ability of DC stimulated with either live or killed bacteria to expand IFN-γ and IL-17 producing CD4+ T lymphocytes in a mixed lymphocyte reaction (MLR) setting. Interestingly, we observed a 2-fold and 4-fold higher production of IFN-γ and IL-17 in MLR cultures of DC infected with live S. aureus USA300, compared to non-replicating bacteria, respectively (Figures 1A,B). A Th1 promoting phenotype was conferred by killed S. aureus USA300, in particular by UVI and PFAI preparations (Figure 1A), whereas all inactivated bacteria poorly induced IL-17 production (Figure 1B). This differential response was not related to the maturation status of DC cultures, since the cytofluorimetric analysis of CD86, CD83, HLA-DR, CD40, and CD38 expression demonstrated that all markers were induced to a similar extent on DC surface after 24 h stimulation with live, HI-, UVI-, or PFAI-USA300 (Figure 1C). In addition no effect of different bacterial preparations on cell viability was observed (Figure 1D).
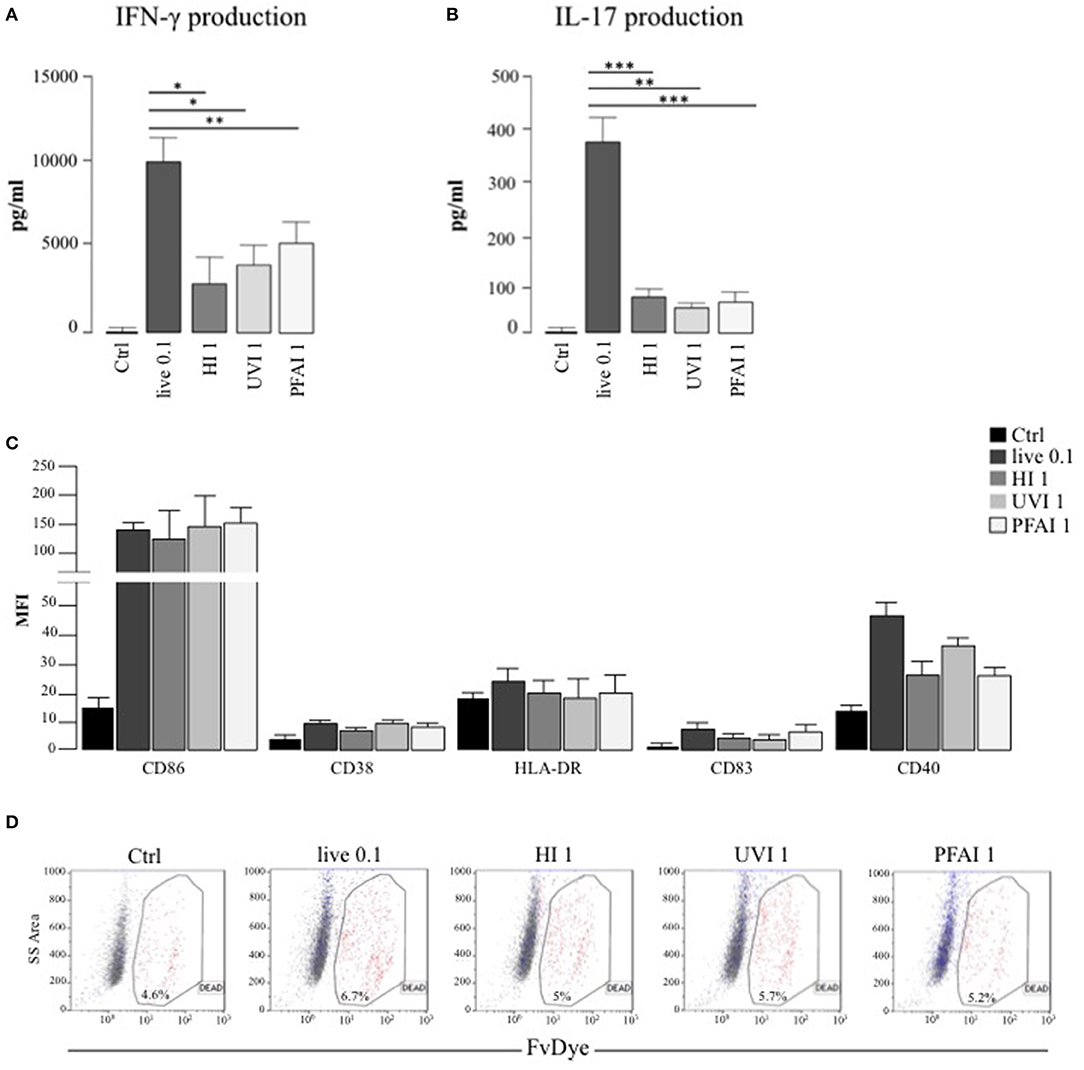
Figure 1. DC response to live and inactivated S. aureus. (A,B) Expansion of IFN-γ and IL-17 producing T cells driven by live and inactivated S. aureus-stimulated DC. Untreated DC (Ctrl) and DC stimulated for 24 h with live and differentially inactivated USA300 strain were co-cultured with autologous total CD4+ T cells for 5 days. (A) The level of IFN-γ was measured by human Th1/Th2 Cytokine array kit in harvested supernatants. (B) IL-17 secretion was instead evaluated by ELISA. The results represent means ± SEM of four independent experiments (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). (C,D) Analysis of DC maturation and viability in response to live, HI-, UVI-, or PFAI-USA300 stimulation. DC were left untreated (Ctrl) or stimulated for 24 h with live and differentially inactivated USA300 strain. (C) Surface expression of the indicated molecules was evaluated by cytofluorimetric analysis in three independent experiments and graphed by calculating the mean fluorescence intensity (MFI) after the subtraction of the isotype Ab controls. Mean MFI ± SEM are shown. (D) Cell viability was evaluated by DC staining with FvDye. Numbers in the dot plots correspond to the percentage of dead cells. A representative experiment, out of three independent experiments performed that yielded similar results, is shown.
Induction of a Different Cytokine Profile in DC Stimulated With Live or Inactivated S. aureus
To investigate whether a different profile of cytokine production could be responsible for the Th1/Th17 response, we analyzed cytokine release from DC stimulated for 24 h with live or HI, UVI-, and PFAI-USA300. A robust production of IL-12, TNF-α, and IL-6 was observed in response to live and UVI and PFAI S. aureus preparations. DC stimulated by HI USA300 produced 6- to 10-fold lower of those cytokines (Figure 2A). IL-10, IL-1β, and the chemokine IL-8 were mainly released from DC infected with live USA300 (Figure 2B). We also observed that IL-23 induction occurred with live and UVI bacteria, while no production was detected in DC cultured in the presence of HI- and PFAI S. aureus (Figure 2C).
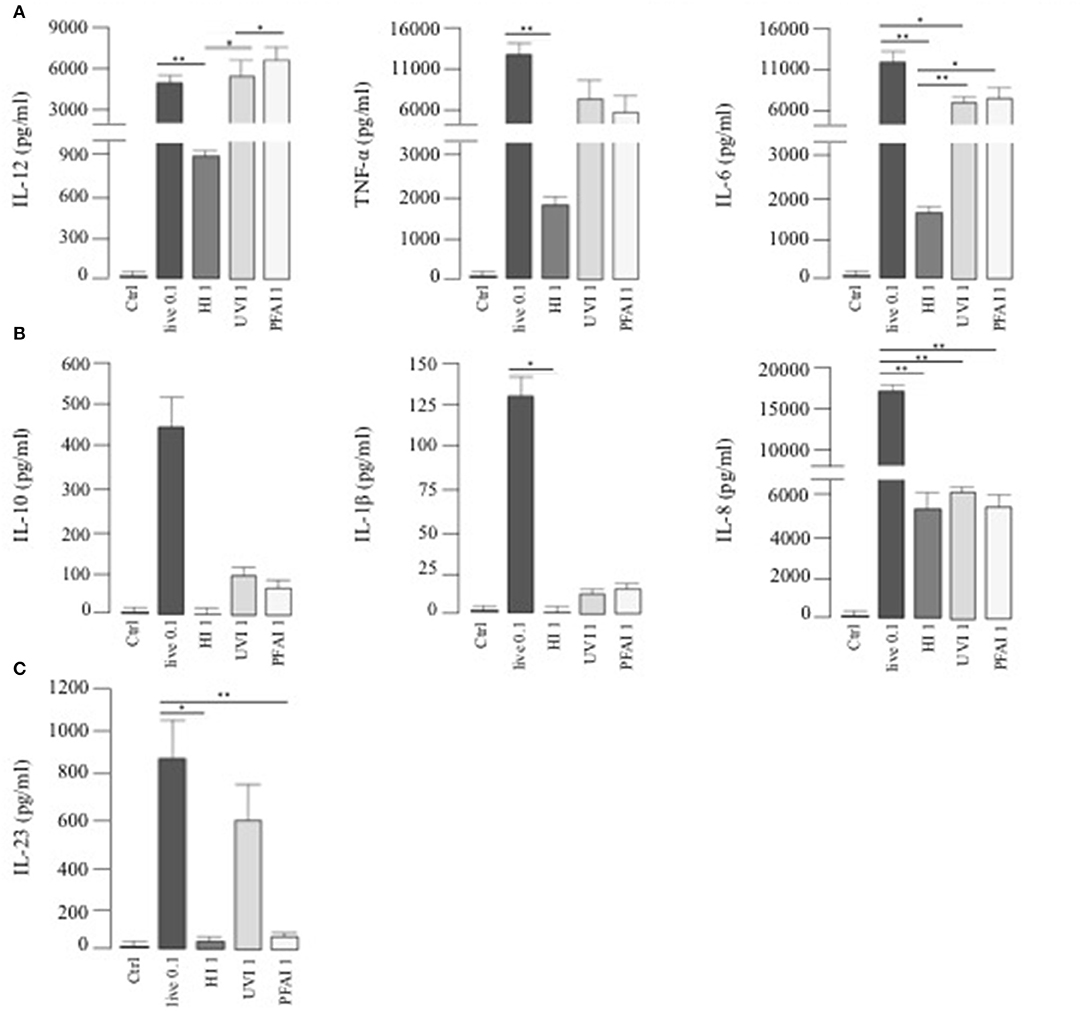
Figure 2. Cytokine release by DC stimulated with live, HI-, UVI-, and PFAI-USA300. DC were left untreated (Ctrl) or stimulated for 24 h with live and differentially inactivated USA300 strain. The secretion of IL-12, TNF-α, IL-6 (A), IL-10, IL-1β, IL-8 (B), and IL-23 (C) was measured in DC culture supernatants. The results represent means ± SEM of four independent experiments (*p ≤ 0.05; **p ≤ 0.01).
Characterization of Type I IFN Expression and Intracellular Pathway in DC Treated With Live or Inactivated S. aureus
We extended this analysis to type I IFNs (mainly IFN-αs and IFN-β), pivotal cytokines in the regulation of innate immune response against pathogens (28). Interestingly, stimulation with UVI S. aureus induced a significantly higher IFN-β mRNA level than observed in cells infected with live bacteria (Figure 3A). HI S. aureus induced 3-fold less IFN-β expression with respect to PFAI and live treatments (Figure 3A). With regard to IFN-α, only treatment with UVI S. aureus induced IFN-α expression after 5 h (Figure 3A), although at 24 h, live bacteria also drove IFN-α expression (Figure 3B).
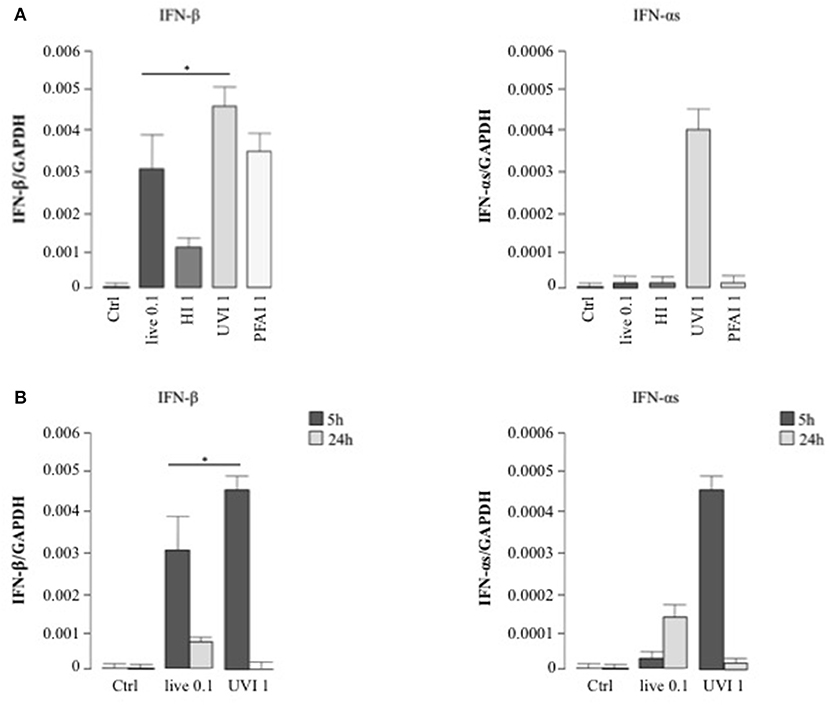
Figure 3. Type I IFN expression in DC stimulated with live and inactivated USA300 preparations. DC were left untreated (Ctrl) or stimulated for 5 h with live, HI, UVI, and PFAI S. aureus (A) and for 24 h with live and UVI S. aureus (B). The expression of IFN-β and IFN-αs was evaluated by real-time quantitative RT-PCR. mRNA levels were normalized by the 2−ΔCt formula using GAPDH as housekeeping gene. The results represent means ± SEM of four independent experiments (*p ≤ 0.05).
Next, the activation of STAT transcription factors involved in type I IFN signaling—STAT1 and STAT2—as well as expression of the IFN-inducible gene IRF7 were analyzed in DC stimulated for 5 and 24 h with live or UVI S. aureus to prove the functionality of the type I IFN expression analyzed by real-time PCR in Figure 3B. After 5 h stimulation, STAT1 and STAT2 displayed an increased level of phosphorylation in UVI-treated DC, compared to those present in DC cultures infected with live S. aureus. Similarly, IRF7 expression was stimulated by live S. aureus (Figure 4). Conversely, at 24 h, an inverse pattern of expression was observed, with higher activation in live USA300-challenged cells, which correlates with the long-lasting type I IFN induction in these cultures (as seen in Figure 3B).
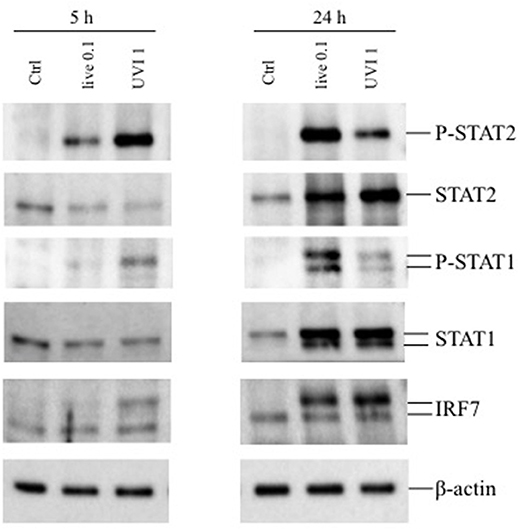
Figure 4. Expression of phosphorylated STAT1/2 and IRF7 in DC stimulated with live and UVI-USA300. DC were left untreated (Ctrl) or stimulated with live and UVI-USA300 for 5 and 24 h. Cell lysates were analyzed by Western blot to detect the expression and the phosphorylation of the indicated proteins. β-actin levels were analyzed as control for protein loading.
DC Response Is Triggered by Live and Inactivated S. aureus Through the Activation of Different Intracellular Signaling Pathways
Cytokine expression in response to pathogens is triggered by different signaling pathways, activated by either membrane or intracellular receptors as well as cytosolic sensors (29, 30). To determine if the cytokine profiles were driven by extracellular sensing or by phagocytosis, DC were pre-treated with cytochalasin D, an inhibitor of actin polymerization that blocks bacterial internalization, prior to stimulation with live and inactivated S. aureus (Figure 5 and Table 1). IFN-β expression was completely abrogated in DC stimulated with live or different inactivated USA300 preparations when cytochalasin D was added to DC cultures (Figure 5), indicating that internalization and DC interaction with surface molecules were necessary to trigger IFN-β expression.
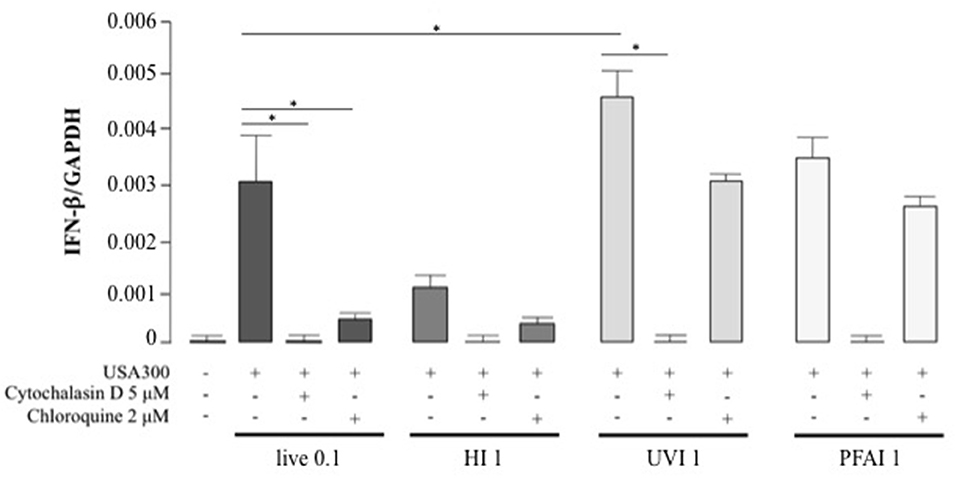
Figure 5. Effect of phagocytosis and phagosomal acidification inhibition on IFN-β expression in DC stimulated with live, HI, UVI, and PFAI USA300. DC were pre-treated with 5 μM Cytochalasin D or with 2 μM Chloroquine for 30 min and then stimulated for 5 h with live and inactivated USA300 strain. The expression of IFN-β was evaluated by real-time quantitative RT-PCR. mRNA levels were normalized by the 2−ΔCt formula using GAPDH as housekeeping gene. The results represent means ± SEM of four independent experiments (*p ≤ 0.05).
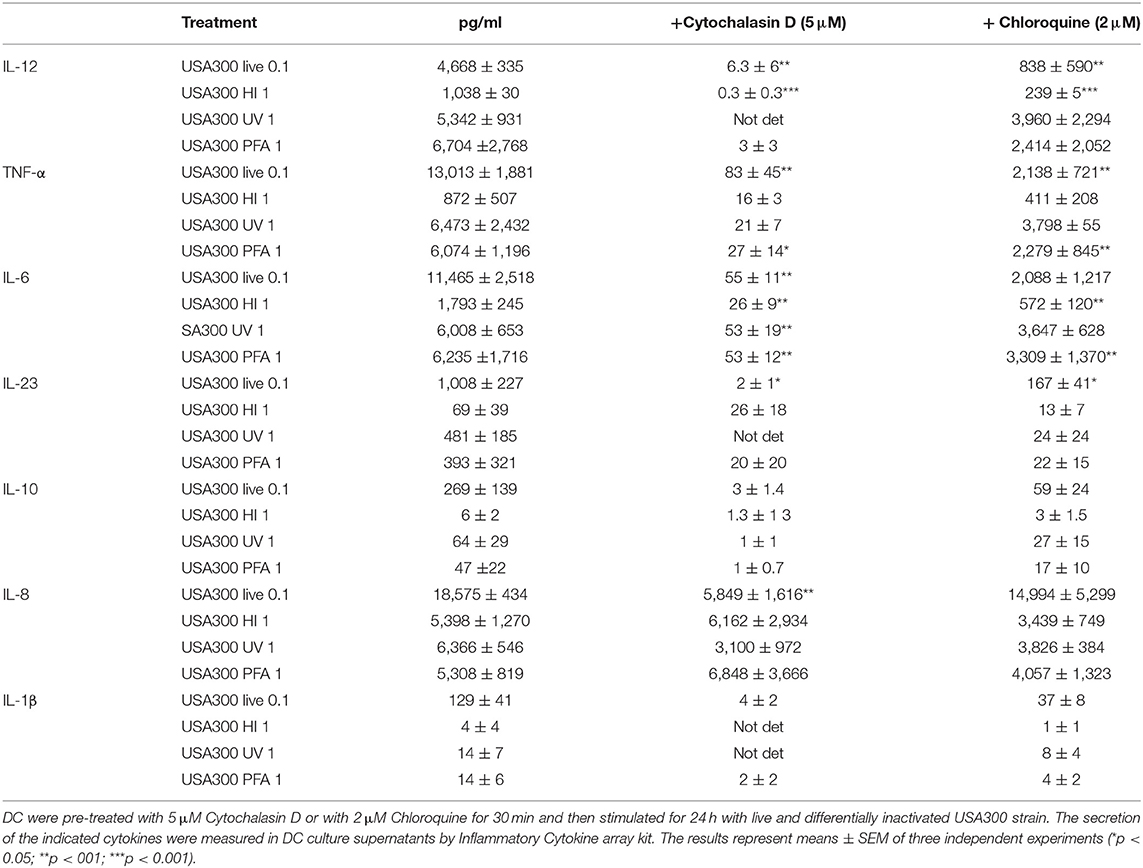
Table 1. Effect of phagocytosis and phagosomal acidification inhibition on DC cytokine expression induced by live and inactivated USA300 preparations.
Thus, to determine if endosomal TLR signaling was involved in IFN-β induction, before stimulation with live or inactivated USA300, DC were treated with chloroquine, a weak base that prevents the endosomal acidification and impairs the signaling of TLRs localized in the endosome, as TLR3, TLR7/8, and TLR9. Chloroquine pre-treatment abolished IFN-β expression driven by live or HI S. aureus USA300, with little effect on IFN-β induced by UVI and PFAI bacteria. Similarly, phagocytosis and endosomal TLR-signaling inhibition also decreased IL-12, TNF-α, IL-6, and IL-10 production (Table 1). Conversely, the expression of the chemokine IL-8 was not inhibited by endosomal TLR signaling blockade (Table 1).
Since the inhibition of phagosomal acidification by chloroquine impaired cytokine expression only in live or HI-stimulated DC, we postulated the involvement of other intracellular signaling pathways in UVI- and PFAI-stimulated DC. In particular, the activation of the cGAS–STING–IRF3 axis was previously shown to control IFN-β expression in murine and human macrophages infected with live S. aureus (13). Analysis of the STING/IRF3 pathway by Western blot revealed a faint band corresponding to phosphorylated STING only in live S. aureus-infected DC (Figure 6), whereas no STING phosphorylation was observed in UVI-treated cells despite IFN-β and IFN-α expression, which correlates with the appearance of a slower-migrating IRF3 phosphorylated form present in both live and UVI-stimulated DC (Figure 6). We also observed that inhibition of autophagy by E64D/PepA allowed the accumulation of phospho-STING in live S. aureus-infected DC (Figure 6), consistent with the observation that the autophagic molecule p62/SQSTM1 drives ubiquitinated STING to autophagosomes after its phosphorylation by TBK1 (31). Collectively, these data demonstrate that STING activation is likely required for IRF3 phosphorylation only in response to live S. aureus infection, thus suggesting the existence of a not fully characterized STING-independent and IRF3-dependent signaling pathway(s) in UVI-stimulated DC driving the IFN-β expression.
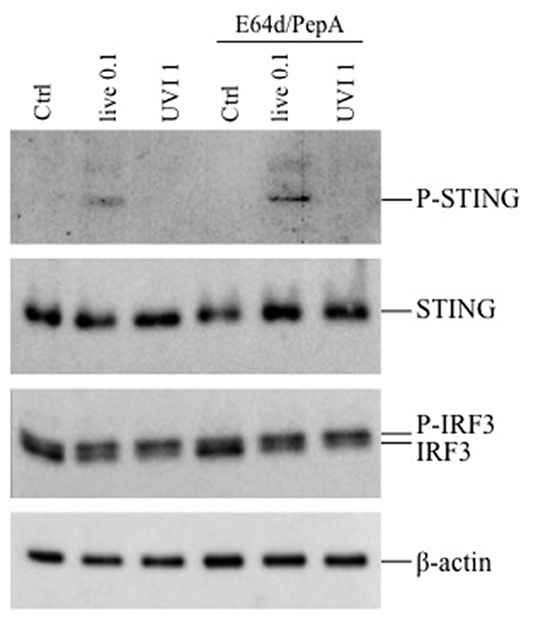
Figure 6. Activation of STING-IRF3 signaling pathway in response to live and UVI USA300. DC were left untreated (Ctrl) or stimulated with live and UVI USA300 strain for 5 h. Cell lysates were analyzed by Western blot to detect the expression and the phosphorylation of STING and IRF3. β-Actin levels were analyzed as control for protein loading.
Discussion
A full understanding of the complex interplay between S. aureus and the host immune responses represents a major goal for the development of an effective vaccine. Indeed, vaccine candidates tested to date have been ineffective in coping with the pathogenic complexity of S. aureus (32). Since vaccination relies on presentation of microbial antigens by antigen-presenting DC to naïve T cells, an understanding of the human DC response to S. aureus is essential for the development of preventive and therapeutic vaccine strategies.
In previous studies, we demonstrated that Esx virulence factors influence the DC response to S. aureus by modulating apoptosis, cytokine production, and, in turn, T cell differentiation (20). These data also demonstrated the utility of a DC-based in vitro model to study small differences in S. aureus virulence. Previously, it was shown that administration of heat-inactivated S. aureus failed to induce protection in mice, but rather skewed a beneficial IL-17 T cell response to a detrimental IL-10 producing T cell response (33). Also, immunization with S. aureus inactivated using UV irradiation, rather than HI, conferred protection to mice challenged with virulent methicillin-sensitive or methicillin-resistant strain by increasing survival and diminishing bacterial burden and kidney abscesses (34).
Based on these findings, we sought to investigate further the features of S. aureus–host interaction, focusing on the importance of both S. aureus viability and structural integrity in triggering DC immune responses. Our comparative analysis of the effects induced by live, HI, UVI, and PFAI USA300 sheds light on the capacity of DC to discriminate between live and inactivated S. aureus, and to fine-tune their responses according to the type and severity of the infectious threat (22). This feature mainly relies on the capacity of the immune cells to recognize the so-called vita-PAMPs, such as microbial RNA, bacterial metabolites, signaling molecules like second messengers, signal peptides (35), and quorum-sensing molecules (36), with the latter uniquely associated with live microorganisms (24).
Our analysis demonstrated that the different inactivation methods did not affect the capacity of USA300 to induce the expression of maturation markers, such as CD86, CD83, CD38, CD40, and HLA-DR; however, intriguing differences were found in the capacity of DC to drive the expansion of Th1 and Th17 producing CD4+ T cells in response to live or inactivated S. aureus. While both live and, at small extent, inactivated staphylococci induce IFN-γ production, only live USA300 promoted the expansion of IL-17 producing T cells, suggesting that Th1 and Th17 responses are induced by different mechanisms that might be independent (Th1) or dependent (Th17) from bacterial viability. However, the Th17 response seems to be related to the release of IL-1β, which occurs only from DC infected with live bacteria. IL-1β is a Th17-promoting cytokine that requires the proteolysis by caspase-1, for activation and secretion, which occurs only in response to live S. aureus infection (37).
These data prompted us to identify and further dissect similarity and divergence in cell response as well as intracellular pathways activated in human DC by either live S. aureus or inactivated bacterial preparations. For instance, the comparison of cytokine production induced by live, HI, UVI, and PFAI staphylococci revealed a different profile depending on the type of inactivation. Indeed, only UVI and PFAI S. aureus preserved the ability to induce in DC a robust production of IL-12, IL-6, and TNF-α. IL-23 was instead released only in response to live and UVI bacteria stimulation. Moreover, IL-1β, IL-10, and IL-8 were produced mainly by live USA300-infected DC. These data suggest that DC can discriminate between the risk associated with live bacterial infection vs the stimulation with an inactivated S. aureus. In particular, DC respond to stimulus with live bacteria producing, in addition to IL-12, IL-6, TNF-α, and IL-23, also IL-1β and IL-8, which are involved in neutrophil recruitment and early anti-staphylococcal defense in the invaded tissue (5). Moreover, to dampen an excessive activation of T cells and inflammatory phagocytes, which lead to host tissue damage, by recognizing live S. aureus, DC secrete the anti-inflammatory cytokine IL-10 that is able to influence disease outcome during acute infection (38). Surprisingly, IFN-β expression in DC stimulated with UVI and PFAI bacteria is higher than that found in DC infected with live S. aureus; conversely, the IFN-α expression displayed a time-dependent regulation where UVI bacteria resulted in a stronger inducer at an early time point such as 5 h compared to live S. aureus while the opposite occurs at later time point.
To further dissect the key events of host–pathogen interaction, we analyzed the contribution of S. aureus phagocytosis and internalization on cytokine expression. Phagocytosis was found to be an essential step for the activation of an optimal cytokine response to both live and inactivated USA300 bacteria. These data are consistent with previous experiments in vivo showing that S. aureus internalization was required for peritoneal macrophage cytokine response (39); furthermore, our observations indicate that the majority of inflammatory and regulatory signaling pathways occurred after bacterial internalization, based on the loss of cytokine expression in the presence of cytochalasin D. Although internalization is crucial for both live and inactivated USA300, it is likely that different intracellular pathways are triggered in response to either live or inactivated S. aureus. In particular, chloroquine treatment, which blocks endosomal acidification and subsequent endosomal TLR signaling, impacted the expression of IFN-β, as well as IL-12, TNF-α, and IL-6 in response to live and HI bacteria while it only poorly interferes with the UVI and PFAI S. aureus-induced expression of these cytokines. Interestingly, in mouse macrophages, both TLR and STING signaling pathways contributed to the transcriptional response induced by live but not killed S. aureus (13).
Having determined that IFN-β expression in UVI-stimulated DC was not dependent on endosomal TLR signal, we investigated the role of the STING/IRF3 axis; interestingly, STING activation was required for IRF3 phosphorylation only in response to live S. aureus infection, as previously observed (13). Conversely, IRF3 phosphorylation was induced in a STING-independent manner by UVI stimulation, suggesting the involvement of another unknown IRF3-dependent cytoplasmic sensor activated by UVI S. aureus.
Given the differences observed in cytokine production and Th response induced by the different bacterial inactivation procedures, an important aspect emerging from this study is the confirmation that high-temperature inactivation profoundly affects the immune stimulatory properties of S. aureus. Such observation could be related to profound alterations of PAMPs and structures in HI bacterium, which are essential to promote an appropriate DC response. Conversely, PFA and UV treatment preserved the S. aureus capacity to stimulate a range of cytokines that well overlapped with those released from DC infected with live bacteria with the exception of IL-10 and IL-1β. In addition, it is likely that the different killing methods may profoundly impact the ratio between vita- and classical PAMPs. Despite S. aureus replication, the availability and/or release of bacterial molecules involved in DC immune recognition may differ, depending on the inactivation method; for instance, UV-treated bacteria may preserve, for a while, some metabolic activities that trigger specific intracellular pathway(s) distinct from those induced by HI bacteria. Taken together, our results demonstrate that live or inactivated S. aureus infection stimulates distinct pathways in DC. Such information may prove helpful in the design and testing of novel vaccination strategies to optimize a protective immune response against this pathogen.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by Istituto Superiore di Sanità - CE/13/387. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MC performed experiments, analyzed data, and prepared the manuscript. SS performed experiments and analyzed data. ME performed experiments and participated in experimental design and data analysis. EG, RC, MS, and FR performed experiments. FB revised the manuscript and contributed to data interpretation. JH discussed the data. EC participated in experimental design, data analysis, and manuscript writing. All authors approved the manuscript before it was submitted.
Funding
This work was supported by Istituto Superiore di Sanità.
Conflict of Interest
FB is an employee of GSK Vaccines and owns patents on S. aureus vaccine candidates as well as GSK stocks. FB has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Miller LS, Cho JS. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol. (2011) 11:505–18. doi: 10.1038/nri3010
2. Olaniyi R, Pozzi C, Grimaldi L, Bagnoli F. Staphylococcus aureus-associated skin and soft tissue infections: anatomical localization, epidemiology, therapy and potential prophylaxis. Curr Top Microbiol Immunol. (2017) 409:199–227. doi: 10.1007/82_2016_32
3. Monaco M, Pimentel De Araujo F, Cruciani M, Coccia EM, Pantosti A. Worldwide epidemiology and antibiotic resistance of Staphylococcus aureus. Curr Top Microbiol Immunol. (2016) 409:21–56. doi: 10.1007/82_2016_3
4. Karauzum H, Datta SK. Adaptive immunity against Staphylococcus aureus. Curr Top Microbiol Immunol. (2016) 409:419–39. doi: 10.1007/82_2016_1
5. Bekeredjian-Ding I, Stein C, Uebele J. The innate immune response against Staphylococcus aureus. Curr Top Microbiol Immunol. (2017) 409:385–418. doi: 10.1007/82_2015_5004
6. Hilmi D, Parcina M, Stollewerk D, Ostrop J, Josten M, Meilaender A, et al. Heterogeneity of host TLR2 stimulation by Staphylocoocus aureus isolates. PLoS ONE. (2014) 9:e96416. doi: 10.1371/journal.pone.0096416
7. Parker D, Prince A. Staphylococcus aureus induces type I IFN signaling in dendritic cells via TLR9. J Immunol. (2012) 189:4040–6. doi: 10.4049/jimmunol.1201055
8. Bergstrom B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, et al. TLR8 senses Staphylococcus aureus RNA in human primary monocytes and macrophages and induces IFN-beta production via a TAK1-IKKbeta-IRF5 signaling pathway. J Immunol. (2015) 195:1100–11. doi: 10.4049/jimmunol.1403176
9. Grosz M, Kolter J, Paprotka K, Winkler AC, Schafer D, Chatterjee SS, et al. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin alpha. Cell Microbiol. (2014) 16:451–65. doi: 10.1111/cmi.12233
10. Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot JP, Karin M, et al. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci USA. (2009) 106:12873–8. doi: 10.1073/pnas.0904958106
11. Munoz-Planillo R, Franchi L, Miller LS, Nunez G. A critical role for hemolysins and bacterial lipoproteins in Staphylococcus aureus-induced activation of the Nlrp3 inflammasome. J Immunol. (2009) 183:3942–8. doi: 10.4049/jimmunol.0900729
12. Hanamsagar R, Aldrich A, Kielian T. Critical role for the AIM2 inflammasome during acute CNS bacterial infection. J Neurochem. (2014) 129:704–11. doi: 10.1111/jnc.12669
13. Scumpia PO, Botten GA, Norman JS, Kelly-Scumpia KM, Spreafico R, Ruccia AR, et al. Opposing roles of Toll-like receptor and cytosolic DNA-STING signaling pathways for Staphylococcus aureus cutaneous host defense. PLoS Pathog. (2017) 13:e1006496. doi: 10.1371/journal.ppat.1006496
14. Parker D, Planet PJ, Soong G, Narechania A, Prince A. Induction of type I interferon signaling determines the relative pathogenicity of Staphylococcus aureus strains. PLoS Pathog. (2014) 10:e1003951. doi: 10.1371/journal.ppat.1003951
15. Marinho FV, Benmerzoug S, Oliveira SC, Ryffel B, Quesniaux VFJ. The emerging roles of STING in bacterial infections. Trends Microbiol. (2017) 25:906–18. doi: 10.1016/j.tim.2017.05.008
16. Meade N, Furey C, Li H, Verma R, Chai Q, Rollins MG, et al. Poxviruses evade cytosolic sensing through disruption of an mTORC1-mTORC2 regulatory circuit. Cell. (2018) 174:1143–57. doi: 10.1016/j.cell.2018.06.053
17. Lutz MB. Induction of CD4(+) regulatory and polarized effector/helper T cells by dendritic cells. Immune Netw. (2016) 16:13–25. doi: 10.4110/in.2016.16.1.13
18. Wu X, Xu F. Dendritic cells during Staphylococcus aureus infection: subsets and roles. J Transl Med. (2014) 12:358. doi: 10.1186/s12967-014-0358-z
19. Darisipudi MN, Nordengrun M, Broker BM, Peton V. Messing with the sentinels-The interaction of Staphylococcus aureus with dendritic cells. Microorganisms. (2018) 6:E87. doi: 10.3390/microorganisms6030087
20. Cruciani M, Etna MP, Camilli R, Giacomini E, Percario ZA, Severa M, et al. Staphylococcus aureus Esx factors control human dendritic cell functions conditioning Th1/Th17 response. Front Cell Infect Microbiol. (2017) 7:330. doi: 10.3389/fcimb.2017.00330
21. Blander JM, Barbet G. Exploiting vita-PAMPs in vaccines. Curr Opin Pharmacol. (2018) 41:128–36. doi: 10.1016/j.coph.2018.05.012
22. Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. (2012) 12:215–25. doi: 10.1038/nri3167
23. Moretti J, Blander JM. Detection of a vita-PAMP STINGs cells into reticulophagy. Autophagy. (2018) 14:1102–4. doi: 10.1080/15548627.2018.1441471
24. Ugolini M, Sander LE. Dead or alive: how the immune system detects microbial viability. Curr Opin Immunol. (2018) 56:60–6. doi: 10.1016/j.coi.2018.09.018
25. Etna MP, Giacomini E, Pardini M, Severa M, Bottai D, Cruciani M, et al. Impact of Mycobacterium tuberculosis RD1-locus on human primary dendritic cell immune functions. Sci Rep. (2015) 5:17078. doi: 10.1038/srep17078
26. Etna MP, Giacomini E, Severa M, Pardini M, Aguilo N, Martin C, et al. A human dendritic cell-based in vitro model to assess Mycobacterium tuberculosis SO2 vaccine immunogenicity. ALTEX. (2014) 31:397–406. doi: 10.14573/altex.1311041
27. Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, et al. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. (2004) 34:796–805. doi: 10.1002/eji.200324610
28. Boxx GM, Cheng G. The roles of type I interferon in bacterial infection. Cell Host Microbe. (2016) 19:760–9. doi: 10.1016/j.chom.2016.05.016
29. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. (2011) 30:16–34. doi: 10.3109/08830185.2010.529976
30. Kimura H, Yoshizumi M, Ishii H, Oishi K, Ryo A. Cytokine production and signaling pathways in respiratory virus infection. Front Microbiol. (2013) 4:276. doi: 10.3389/fmicb.2013.00276
31. Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. (2018) 37:E97858. doi: 10.15252/embj.201797858
32. Pozzi C, Olaniyi R, Liljeroos L, Galgani I, Rappuoli R, Bagnoli F. Vaccines for Staphylococcus aureus and target populations. Curr Top Microbiol Immunol. (2017) 409:491–528. doi: 10.1007/82_2016_54
33. Schmaler M, Jann NJ, Ferracin F, Landmann R. T and B cells are not required for clearing Staphylococcus aureus in systemic infection despite a strong TLR2-MyD88-dependent T cell activation. J Immunol. (2011) 186:443–52. doi: 10.4049/jimmunol.1001407
34. Burnside K, Lembo A, Harrell MI, Klein JA, Lopez-Guisa J, Siegesmund AM, et al. Vaccination with a UV-irradiated genetically attenuated mutant of Staphylococcus aureus provides protection against subsequent systemic infection. J Infect Dis. (2012) 206:1734–44. doi: 10.1093/infdis/jis579
35. Bufe B, Schumann T, Kappl R, Bogeski I, Kummerow C, Podgorska M, et al. Recognition of bacterial signal peptides by mammalian formyl peptide receptors: a new mechanism for sensing pathogens. J Biol Chem. (2015) 290:7369–87. doi: 10.1074/jbc.M114.626747
36. Kendall MM, Sperandio V. What a dinner party! Mechanisms and functions of interkingdom signaling in host-pathogen associations. MBio. (2016) 7:e01748. doi: 10.1128/mBio.01748-15
37. Sokolovska A, Becker CE, Ip WK, Rathinam VA, Brudner M, Paquette N, et al. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nat Immunol. (2013) 14:543–53. doi: 10.1038/ni.2595
38. Leech JM, Lacey KA, Mulcahy ME, Medina E, Mcloughlin RM. IL-10 plays opposing roles during Staphylococcus aureus systemic and localized infections. J Immunol. (2017) 198:2352–65. doi: 10.4049/jimmunol.1601018
Keywords: DC, S. aureus, cytokines, signaling pathway, T cell response
Citation: Cruciani M, Sandini S, Etna MP, Giacomini E, Camilli R, Severa M, Rizzo F, Bagnoli F, Hiscott J and Coccia EM (2019) Differential Responses of Human Dendritic Cells to Live or Inactivated Staphylococcus aureus: Impact on Cytokine Production and T Helper Expansion. Front. Immunol. 10:2622. doi: 10.3389/fimmu.2019.02622
Received: 25 June 2019; Accepted: 22 October 2019;
Published: 12 November 2019.
Edited by:
Marina De Bernard, University of Padova, ItalyReviewed by:
Laura Maggi, University of Florence, ItalyO. Colin Stine, University of Maryland, Baltimore, United States
Copyright © 2019 Cruciani, Sandini, Etna, Giacomini, Camilli, Severa, Rizzo, Bagnoli, Hiscott and Coccia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eliana M. Coccia, ZWxpYW5hLmNvY2NpYUBpc3MuaXQ=
†These authors have contributed equally to this work
 Melania Cruciani
Melania Cruciani Silvia Sandini
Silvia Sandini Marilena P. Etna
Marilena P. Etna Elena Giacomini
Elena Giacomini Romina Camilli
Romina Camilli Martina Severa
Martina Severa Fabiana Rizzo
Fabiana Rizzo Fabio Bagnoli
Fabio Bagnoli John Hiscott
John Hiscott Eliana M. Coccia1*
Eliana M. Coccia1*