- Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
Respiratory Syncytial Virus (RSV) causes serious respiratory tract illness and substantial morbidity and some mortality in populations at the extremes of age, i.e., infants, young children, and the elderly. To date, RSV vaccine development has been unsuccessful, a feature linked to the lack of biomarkers available to assess the safety and efficacy of RSV vaccine candidates. We examined microRNAs (miR) as potential biomarkers for different types of RSV vaccine candidates. In this study, mice were vaccinated with a live attenuated RSV candidate that lacks the small hydrophobic (SH) and attachment (G) proteins (CP52), an RSV G protein microparticle (GA2-MP) vaccine, a formalin-inactivated RSV (FI-RSV) vaccine or were mock-treated. Several immunological endpoints and miR expression profiles were determined in mouse serum and bronchoalveolar lavage (BAL) following vaccine priming, boost, and RSV challenge. We identified miRs that were linked with immunological parameters of disease and protection. We show that miRs are potential biomarkers providing valuable insights for vaccine development.
Introduction
Respiratory Syncytial Virus (RSV) is a cause of lower respiratory tract infection (LRTI) worldwide and is responsible for >30 million new LRTI episodes and up to 199,000 deaths in children under 5 years old resulting in more than 3.4 million hospital admission associated with severe RSV disease (1, 2). The elderly population is also markedly affected by RSV (3). Currently, the only approved RSV prophylactic is palivizumab which is used for high-risk patients, but such treatment has limited applicability due to cost and treatment logistics (4–9). Unfortunately, all efforts to develop a safe and effective RSV vaccine have been unsuccessful (10–16). Attempts in the 1960s to develop a formalin-inactivated RSV (FI-RSV) vaccine candidate were hampered by several factors, including lack of protection against RSV infection in infants and young children, and an association with vaccine enhanced disease that resulted in two deaths upon natural RSV infection of vaccinees (10, 17–19). Efforts to develop live attenuated RSV vaccine candidates using cold-passaging, chemical mutagenesis, or reverse genetics have also been unsuccessful largely due to over- or under-attenuation, which currently cannot be precisely predicted (20–30), and natural RSV infection does not provide long-term protective immunity. Several other RSV vaccine platforms have been developed including subunit (31–38), vectored (39–46), particle-based (47–57), or nucleic acid-based (58–63), but none are FDA-approved a feature linked to our incomplete understanding of the host immune response to RSV (10). There are several target populations for RSV vaccines: infants, young children, pregnant women, and the elderly (10). Due to the differences in these target populations, vaccine safety, efficacy, and platform strategies will need to be different (10, 11, 18, 33). By establishing measures of vaccine protection and disease, a wide range of promising vaccine candidates can be evaluated early in development.
The host immune response is important in the outcome of RSV infections (14, 60, 64, 65), and an imbalance between Th1- and Th2-type cytokines is understood to be responsible for a variety of inflammatory disorders (66, 67). Biomarkers can be surrogates for clinical endpoints and are needed to improve vaccine design and efficacy. Small regulatory microRNAs (miRs) have fundamental roles in regulating the expression and function of key immunological mediators such as cytokines (68–70). miR expression profiles have been identified and shown to be useful predictors for several allergic inflammatory diseases (71–74). In addition, specific miRs have been shown to function in regulating key pathogenic mechanisms in asthma and airway hyperresponsiveness, including polarization of adaptive immune responses, activation of T cells (75–78), regulation of eosinophil development (79–84) and modulation of cytokine-driven responses (68–70). miRs are stable, in a variety of tissues, bodily fluids, and sera allowing for sensitive and accurate measurements regarding the physiological state of the individual (72–74). miRs govern host gene expression by inducing mRNA degradation or translation inhibition and have a prominent role in determining the level of protein expression of host gene targets (85–90). Several miRs can also upregulate target gene expression via regulation of promoter function (91, 92). It has been shown that miR patterns of expression vary for numerous physiological processes that have been deemed useful for diagnosis of neurodegenerative disorders, autoimmune diseases, cardiovascular disease, and cancers; likewise miRs have also been implicated in infectious diseases (42, 93–98). Assessing circulating miRs in the sera of patients has supported miR profiling as a powerful non-invasive biomarker tool.
Previously, it was shown that RSV infection of normal tracheal epithelial cells (NTECs) with GFP-expressing RSV (rgRSV) downregulated the expression of multiple miRNAs (99). Of the 24 miRNAs, miR-221 was shown to regulate nerve growth factor (NGF), a key neurokinin that prevents apoptosis in respiratory cells (99). Later, RSV infection of type II respiratory epithelial cells was shown to induce expression of five and down-regulation of three microRNAs via an RSV G protein regulated mechanism (100). RSV deregulated miRNAs were demonstrated to regulate several key immunological pathways. In a follow-up study, RSV infection of normal human bronchoepithelial (NHBE) cells, miRNA deregulation was tied to mechanisms involving IFN beta and the transcription factor NF-κβ (101). We showed that RSV G and NS1/NS2 proteins can modulate miRNA expression (102, 103). Several studies have investigated differential expression of miRNAs in clinical RSV infections and shown deregulated patterns that can be used as potential biomarkers of infections (104–109). While these data show miRNA deregulation during infection, miRNA expression following vaccination with different RSV vaccine candidates under investigation is not well-understood and has the potential to identify safe vs. unsafe vaccine candidates.
As miRs regulate host gene responses, it is important to determine if miR profiles serve to predict safe, efficacious, or diseased vaccine outcomes, particularly since RSV lacks a licensed vaccine. To determine if patterns of miR expression may serve as a surrogate of RSV immunity or disease requires proof of biological relevance. Therefore, we identified miR biomarkers and immune correlates associated with RSV vaccination to establish baselines for biomarker expression across different vaccination types and strategies. Since serum miR profiles provide indications of how miRs may regulate the immune response induced by RSV vaccination or infection (50), serum miR profiles may also suggest vaccine disease outcomes. We hypothesized that RSV infection or vaccination would alter the pattern and tempo of miR expression and that this would be reflected in changes by the host immune or disease response. In the present study, we examined serum miRs in BALB/c mice at various time-points post-RSV vaccination or RSV challenge using several RSV vaccine types. A miR PCR array was used to identify miRs post-vaccination, post-boost, or post-RSV challenge, and correlated with immune parameters and markers of disease.
Materials and Methods
Mice
Specific-pathogen-free, 6-to-8 weeks old female BALB/c mice (The Jackson Laboratory) were used. Mice were maintained in microisolator cages with sterilized water and food ad libitum. All experiments were approved by and performed in accordance with the guidelines of the University of Georgia Institutional Animal Care and Use Committee (IACUC).
Viruses and Cell Culture
CP52 was a gift from Stephen Whitehead and Brian Murphy at LID, National Institute of Allergy and Infectious Diseases, Bethesda, MD. CP52 is a cold-passaged live attenuated vaccine strain that lacks the RSV G and SH genes and is derived from RSV B1. RSV A2 and CP52 were propagated in mycoplasma-free Vero E6 cells (ATCC CRL-1586) using DMEM (Gibco) containing 5% FBS (Hyclone) at 37°C/5% CO2 and 32°C/5% CO2, respectively (110). Viral titers were determined by plaque assay on Vero E6 cells, and plaques were enumerated by an anti-F protein (clone 131-2A) immunostaining assay (111, 112). Infections were performed in serum-free DMEM (SF-DMEM).
FI-RSV Preparation
The preparation of formalin-inactivated RSV (FI-RSV) vaccine was adapted from the FI-RSV Lot 100 method (113). Briefly, strain A2 was used to infect Vero E6 cells (MOI = 0.1), and at day 4 pi, the cells were lysed following scraping, sonicated, and clarified by centrifugation at 600 x g for 15 min at 4°C. The supernatant was transferred to a tube and filter sterilized using a 2 μm filter; the final protein concentration (determined by BCA) was adjusted to 1 mg/ml. Viral stocks were inactivated by the addition of 37% formalin (final dilution 1:4,000) and incubated at 37°C for 3 days in agitation. FI-RSV was pelleted by ultracentrifugation for 2 h at 25,000 rpm, re-suspended in SF-DMEM at 1/25th of the original volume and adsorbed overnight at room temperature in 4 mg/mL aluminum hydroxide. The compound material was pelleted by centrifugation and the pellet was suspended in SF-DMEM and total virus inactivation was confirmed via plaque assay on Vero cells. This procedure resulted in an FI-RSV vaccine that is concentrated 100-fold and contains 16 mg/ml alum. The vaccine was aliquoted in 1 ml volumes and stored at 4°C.
RSV GA2 Microparticle-Based Vaccine
A microparticle-based RSV G protein vaccine consisting of 3 μm CaCO3 cores was prepared using alternating poly-I-glutamic acid (PGA, negative charge) and poly-I-lysine (PLL, positive charge) layering to build up to seven layers with an RSV G peptide CX3C motif linked to a cationic sequence added as the outermost layer (4). The composition of the seven-layer film was determined using amino acid analysis, which showed that a comparable amount of the peptide component was present in each vaccine batch. Endotoxin levels by limulus amebocyte lysate (LAL) assay were <0.1 EU/μg. The dispersity of the particle vaccines was monitored by dynamic light scattering (DLS). DLS is used to determine the size distribution profile of small particles in suspension or polymers in solution. This layer-by-layer microparticle vaccine has an apparent diameter of ~150 nm for uncoated particles to about 400–500 nm for fully coated particles. Some particle aggregation was detected in each batch with a second population of particles in the 1,500–2,000 nm range.
Vaccine Delivery
We examined three vaccine types: (1) live-attenuated (CP52), (2) inactivated (FI-RSV), and (3) an RSV G peptide microparticle-based (GA2-MP). The GA2-MP vaccines were suspended in PBS and dispersed by water bath sonication immediately prior to immunization. Doses were adjusted to deliver 50 μg designed peptide (DP)/100 μl/mouse. Mice were subcutaneously (s.c.) immunized with GA2-MP without adjuvant between the shoulder blades. 106 PFU equivalents of FI-RSV was used to intramuscularly (i.m.) vaccinate mice. Mice received a 1:25 dilution of FI-RSV in PBS by i.m. injection in a final volume of 50 μL/mouse. FI-RSV was a positive control for vaccine enhanced disease. 106 PFU of live CP52 diluted in PBS was used to vaccinate mice by intranasal (i.n.) instillation in a final volume of 50 μL/mouse. CP52 was a positive control for vaccine protection. PBS vaccinated mice received 50 μL of PBS (vehicle control) by s.c. injection. Mice were anesthetized by i.p. administration of 2,2,2- tribromoethanol (Avertin; 200 μg/kg Sigma) and a portion of vaccinated mice were i.n. challenged with 106 PFU A2 diluted in PBS.
Lung Virus Titers and Disease Endpoints
Lung virus titers were determined in treatment and control mice by plaque assay on Vero E6 cells (111). Briefly, lungs were aseptically removed from mice at day 5 post-RSV (106 PFU/mouse) challenge, and individual lung specimens were homogenized at 4°C in 1 mL of SF-DMEM using a gentleMACS™ Dissociator (Miltenyi Biotec). Samples were clarified by centrifugation for 10 min at 200 × g and supernatants were transferred and stored at −80°C. For the plaque assay, 10-fold serial dilutions of the lung homogenates were adsorbed to 90% confluent Vero E6 cell monolayers for 2 h, at 37°C, overlaid with 1% methylcellulose medium and incubated at 37°C for 5 days. RSV plaques were enumerated by immunostaining with monoclonal antibodies against RSV F protein (clone 131-2A) as previously described (112). Lungs from vaccinated and challenged mice were examined for disease pathogenesis, and as anticipated (54, 114, 115), only the lungs from FI-RSV vaccinated mice challenged with RSV showed substantially enhanced disease (data not shown).
Microneutralization Assay
Two-fold serial dilutions (1:50-1:1,600) of mouse serum in SF-DMEM were incubated with 105 PFU of A2 for 1 h at 37°C, 5% CO2. Palivizumab (MedImmune) was used as positive control for neutralizing activity, and positive control wells of virus without sera and negative control wells without virus or sera were included in triplicate on each plate. The antibody-virus mixtures were transferred to 80–90% confluent monolayers of Vero E6 cells in 96-well-plates and incubated for 2 h at 37°C, 5% CO2. The virus overlay was aspirated, and 150 μl/well of DMEM-10% FBS was added and plates were incubated for 3–4 days at 37°C, 5% CO2, and the plates were fixed with cold 80% acetone in PBS for 10 min, rinsed twice with PBS followed by three washes with 150 μl/well of wash buffer (PBS + 0.1% Tween-20). A monoclonal antibody to the RSV F protein (clone 131-2A) was diluted in PBS with 0.5% gelatin + 0.15% Tween 20 and incubated for 1 h at 37°C, 5% CO2. RSV plaques were enumerated using horseradish peroxidase (HRP) conjugated goat anti-mouse IgG (Southern Biotech), developed using TMB substrate (Sigma), and absorbance measured at 450/650 nm dual-wavelength (BioTek Epoch™ microplate spectrophotometer) and Gen5 Data Analysis software. The percentage of neutralization was calculated, and all samples were normalized to the average value from the no serum control wells.
Indirect ELISA
RSV A2-specific and B1-specific IgG antibodies were detected by ELISA using 96-well-high binding plates (Corning) coated with 106 PFU/mL A2 or B1 in 0.05 M carbonate-bicarbonate buffer, pH 9.6. Sera were added to plates in serial dilutions. RSV-specific antibodies were detected with HRP-conjugated antibodies specific for mouse IgG (Southern Biotech) followed by the addition of SureBlue TMB-peroxidase substrate (KPL, Inc.) for 15 min. Antibody titers were determined as the last sample dilution that generated an OD450 reading of >0.2 (mean OD value of background plus 2 standard deviations of the mean).
ELISPOT Analysis
MultiScreen filter 96-well-plates (Millipore) were coated with the anti-mouse IL4 or anti-mouse IFNγ capture antibody (R&D Systems) and incubated overnight at 4°C. The plates were then blocked with 200 μL of RPMI-10 medium (RPMI 1640 supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 50 μM 2-mercaptoethanol, and 2 mM L-glutamine) and incubated for 2 h at 37°C. In parallel, spleens were harvested from mice at day 5 post-A2 challenge and prepared to a single cell suspension. The cell suspensions were collected by centrifugation for 10 min at 200 × g and suspended in RPMI-10 at 107 cells/mL. Spleen cell suspensions were added to the wells, and cells were stimulated with either 10 μg/mL RSV M2 (82−90) peptide, 10 μg/mL RSV F (51−66) peptide, 10 μg/mL RSV G (183−198) peptide, or 10 μg/mL eGFP (200−208) (irrelevant peptide control) for 24 h at 37°C and 5% CO2. Plates were washed 4 times with wash buffer (0.05% Tween-20 in PBS), anti-mouse IL4 or anti-mouse IFNγ detection antibody (R&D Systems) was added, and plates were incubated overnight at 4°C. Detection antibody was removed, plates were washed, and cytokine spots were developed using NBT/BCIP substrate (R&D Systems). Spots were enumerated using an ELISPOT reader (AID, San Diego).
Quantification of Cytokines
At day 3 post-A2 challenge, a subset of mice from each group was sacrificed and BAL and sera were collected. The mouse lungs were flushed three times with 1 ml of PBS and the retained BAL was centrifuged at 400 × g for 5 min at 4°C. The recovered supernatants were collected and stored at −80°C until assessed for cytokine concentration, and the cell pellet was suspended in 200 μL of FACS staining buffer (PBS containing 1% BSA). Total cell numbers were counted using a hemocytometer. The Luminex® xMAP system using a MILLIPLEX MAP mouse cytokine immunoassay (Millipore) was used to quantitate cytokines in cell-free BAL supernatants and sera according to the manufacturer protocol. Briefly, beads coupled with anti-IFNγ, anti-IL1α, anti-IL2, anti-IL4, anti-IL5, anti-IL6, anti-IL9, anti-IL10, anti-IL12p40, anti-IL13, anti-IL15, anti-IL17A, anti-MCP1, anti-RANTES, anti-TNFα, and anti-eotaxin monoclonal antibodies were sonicated, mixed, and diluted 1:50 in assay buffer. For the assay, 25 μL of beads were mixed with 25 μL of PBS (for BAL samples) or serum matrix (for serum samples), 25 μL of assay buffer and 25 μL of BAL supernatant or serum and incubated overnight at 4°C. After washing, beads were incubated with biotinylated detection antibodies for 1 h and the reaction mixture was then incubated with streptavidin-phycoerythrin (PE) conjugate for 30 min at room temperature, washed, and suspended in PBS. The assay was analyzed on a Luminex 200 instrument (Luminex Corporation, Austin, TX) using Luminex xPONENT 3.1 software.
RNA Isolation
Blood was collected from mice via axillary vessels in 1.5 ml microcentrifuge tubes (Fisher), allowed to clot for 30 min at room temperature, and centrifuged at 900 × g for 10 min and 4°C. Serum layer was transferred to a new microcentrifuge tube and centrifuged for 10 min at 16,000 × g and 4°C, and the cleared supernatant was transferred to a new microcentrifuge tube without disturbing the pellet. One hundred microliter of serum sample per mouse was processed for RNA isolation using miRNeasy Serum/Plasma Kit (Qiagen) as per manufacturer's recommended protocol or stored at −80 till processing. Serum/Plasma C. elegans miR-39 Spike-In Control (Qiagen) was spiked into each sample prior to RNA purification as an internal control for miR expression profiling in serum to allow for monitoring of RNA recovery and purity, and reverse transcription efficiency. The RNA concentration and purity were determined by Qubit RNA assay broad range (Qubit RNA assay BR, Invitrogen) fluorometry. This reagent specifically binds to RNA only and does not detect DNA, protein or free nucleotides. Additionally, spectrophotometric analysis of all samples using Epoch Gen 5 spectrophotometer (Biotek) showed that all RNA samples had A260/280 ratios ≥1.8.
miR PCR Arrays and Data Analysis
First-strand cDNA synthesis was performe4d with 200 ng/total RNA from each sample using the miScript II RT kit with miScript HiSpec Buffer (Qiagen) following manufacturer protocol. Briefly, cDNA synthesis was performed at 37°C for 60 min followed by inactivation at 95°C for 5 min. First-strand cDNA was diluted 1:10 in molecular grade water and expression of 84 miRs having a role in T or B cell function was assessed using a miScript miR PCR Array Mouse T cell and B cell Activation (Qiagen) array following the manufacturer's protocol on a Stratagene Mx3000P/3005P real-time instrument. Each array plate contains oligos specific to 84 mature miRs validated to regulate T cell or B cell development and function as well as oligos for spike in (C. elegans miR-39), six housekeeping genes [small nucleolar/nuclear RNA (snoRNAs) SNORD61, SNORD95, SNORD96A, SNORD68, SNORD72, and RNU6B] and positive and negative controls for reverse transcription and PCR. Data obtained were analyzed with miScript miR PCR analysis template (Qiagen) using the ΔΔCT method (116). miRs with fold change ≥2 were considered upregulated while miRs with fold change ≤ 0.5 were considered downregulated. RT-qPCR using miR-specific primers was then performed on differentially expressed miRs between treatment groups using a PCR array. The specificity of amplification was validated by dissociation curve analysis.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism (ver 5.0; GraphPad). Statistical significance was determined using a one-way ANOVA or two-way ANOVA followed by Bonferroni's post-hoc comparisons tests; a p ≤ 0.05 was considered significant.
Pathway Analysis
Analysis of pathways regulated by differentially expressed miRs was carried out using DIANA miRPath ver 3.0 (117–119) using the microT-CDS database. The significance of pathway association was determined using a p-value threshold ≤ 0.05 and microT threshold of 0.8 using Fisher's exact test. When multiple miRs were analyzed together, data were filtered to identify pathway intersections instead of unions to identify common core pathways regulated by the miRs. Pathway hits were corrected for false discovery rate.
Results
To assess the serum miR profiles in RSV-vaccinated or challenged mice, the mice were vaccinated (primed) with a live attenuated vaccine (CP52), an inactivated vaccine (FI-RSV), or a microparticle peptide-based vaccine carrying the G CX3C motif (GA2-MP) and boosted 3 weeks later. Three weeks post-boost vaccinated mice were i.n. challenged with 106 PFU A2, and the sera and lung tissues were collected from vaccinated and mock-treated mice at several time-points. Antibody responses to RSV were assessed to confirm that antigens induced a recall response upon vaccination and challenge. RSV A2 or B1-specific serum IgG was determined at 2 weeks post-boost and 5 days post-RSV challenge, and the levels of neutralizing antibody determined at day 5 pi. Lung viral titers were determined at 5 days post-challenge. Broadly, vaccination with CP52 or FI-RSV elicited a cross-reactive humoral response to either A2 or B1 relative to mock (Figures 1A,C, respectively). CP52 (but not FI-RSV) vaccination followed by A2 challenge elicited a strong antibody response (Figures 1B,D, respectively), which was neutralizing (Figure 1E) and correlated with a statistically significant (p ≤ 0.05) reduction in lung viral titers (Figure 1F). Despite the G protein CX3C motif having intra-strain conservation (120), it was less immunogenic as indicated by the lower anti-A2 and B1 IgG serum levels. These data show the prototypical responses associated with CP52, GA2-MP, and FI-RSV vaccination in mice (121, 122).
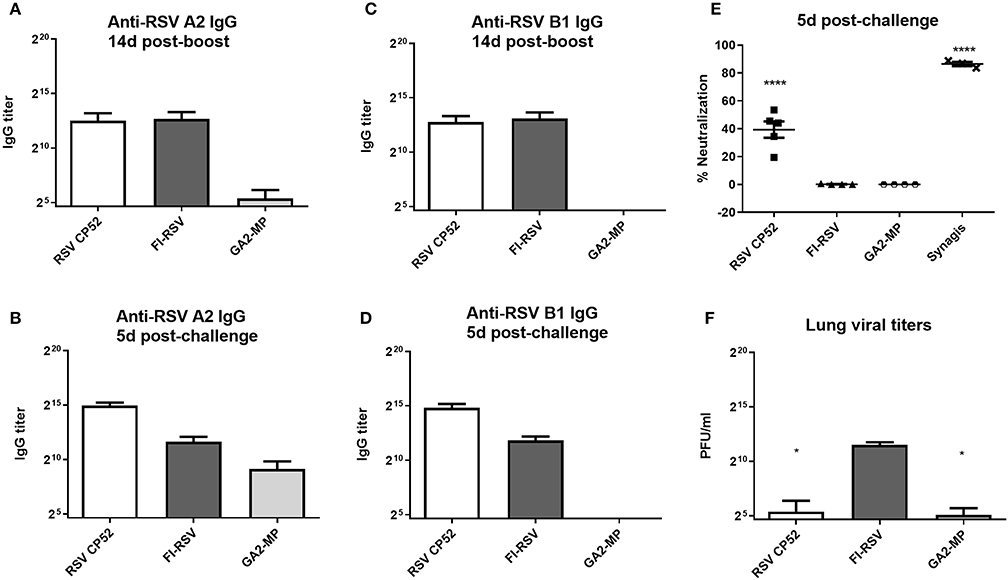
Figure 1. RSV vaccines types, serum IgG, and virus clearance. Sera at day 14 (A) and 5 (B) post-RSV A or B challenge of prime-boosted mice (C,D); IgG reactivity was determined against A2 (A,C) and B1 (B,D). Three weeks after the boost-vaccination mice were i.n. challenged with 106 PFU of A2. (E) RSV neutralizing antibody levels were measured by microneutralization assay at day 5 post-RSV challenge. (F) Lung virus titers were determined 5 days post-challenge by plaque assay. PBS only-treated groups treated had no detectable effect and are not included. All samples were assayed in duplicate and n = 4 mice/group. Error bars represent the SEM and results were considered significant with a *p ≤ 0.05 and ****p ≤ 0.0001 as determined by one-way ANOVA and Bonferroni's test.
RSV Vaccine Types and the Th1- and Th2-Type Response
Th1- or Th2-type responses were assessed by ELISPOT assays, and levels of IFNγ or IL4 expression (representing Th1- or Th2-type responses, respectively) were evaluated at day 14 post-boost vaccination by re-stimulated splenocytes with RSV M2, F or G peptides. As expected, CP52 vaccinated mice had the highest frequency of IFNγ expressing cells compared to IL4 expressing cells (Figure 2A), while splenocytes from FI-RSV vaccinated mice had the highest frequency of IL4 expressing cells compared to IFNγ expressing cells (Figure 2B). GA2-MP vaccinated mice had higher levels of G protein-specific IL4 secreting cells than IFNγ expressing cells, however this difference was not statically significant (p > 0.005). MCP1 and RANTES are chemokines involved in leukocyte recruitment to the airway, and to sites of inflammation in response to RSV infection (123–125). Given the role of these immune cell types in disease pathogenesis, levels of MCP1 (Figure 2C) and RANTES (Figure 2D) in sera and BAL were determined by multiplex cytokine/chemokine assays. MCP1 was localized to the lungs, with the highest level in the BAL from FI-RSV vaccinated mice although the level of expression was not substantially different between CP52 and GA2-MP vaccinated mice. In contrast, RANTES was expressed systemically, as evident by a higher level in the sera compared to BAL, with higher levels for FI-RSV and CP52 immune mice compared to GA2-MP vaccinated mice at day 3 post-RSV challenge. As anticipated, no detectable cytokine expression occurred following mock (PBS) treatment (data not shown). These data indicate that CP52 or FI-RSV vaccinated mice have an overall higher level of inflammation than mice vaccinated with GA2-MP upon RSV challenge. Other Th2-specific cytokines (IL4, IL5, IL6, IL10, and IL3) were higher in the sera (data not shown) and BAL of FI-RSV vaccinated mice compared to all other vaccinated groups which further confirm the biased Th2-type cytokine response associated with FI-RSV vaccination.
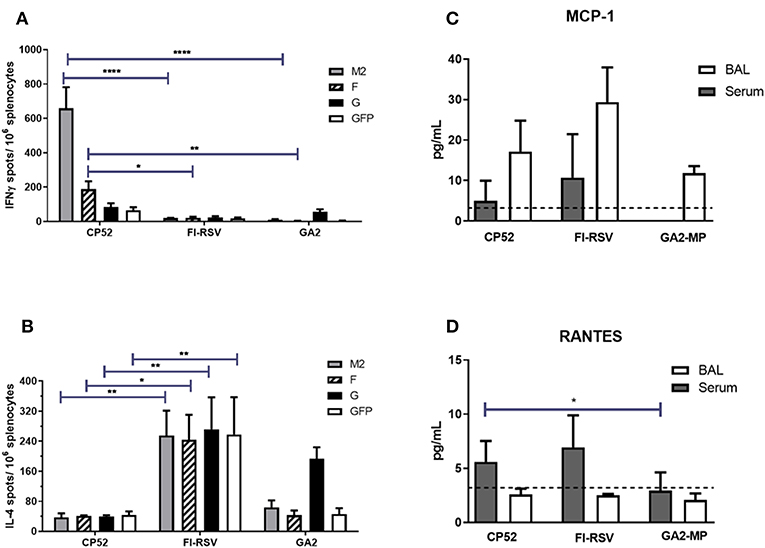
Figure 2. Vaccine types and Th1/ Th2 memory responses. The number of M282−90, F51−66, G183−198, and eGFP200−208-specific (irrelevant peptide control) IL4- and IFNγ- producing splenocytes were determined by ELISPOT harvested at 14 days post-boost vaccination. (A) IFNγ- producing splenocytes and (B) IL4-producing splenocytes. The data are presented as ELISPOTS/106 splenocytes. Three weeks after the boost mice were i.n. challenged with 106 PFU of A2. The level of (C) MCP1 and (D) RANTES were measured in sera and BAL supernatant by multiplex cytokine/chemokine assay and the data are presented as pg/mL of cytokine in BAL supernatant at day 3 post-challenge (n = 4–6 mice/group). The dashed line indicates the limit of detection (LOD) = 3.2 pg/ml. Error bars represent the SEM from n = 4 mice/group and results were considered significant with a *p ≤ 0.05, **p ≤ 0.01, and ****p ≤ 0.0001 as determined by two-way ANOVA and Bonferroni's test using GraphPad Prism ver. 8.0.
The Type of Vaccination Is Linked With Different miR Expression Patterns
Evaluating the Th1- or Th2-type cytokine response using accompanying assays is not efficient for testing of multiple vaccine candidates; however, the examination of miR biomarkers as correlates of the host immune response may aid vetting of safe or disease vaccine types, and considerably accelerate RSV vaccine research. The pattern of miRs can be readily evaluated using PCR (126) in a variety of fluids and tissues (127–133), there is sequence conservation across species (134), and miRs regulate key immunological processes (135). These features can be used to determine baseline data that may differentiate safe from disease risk vaccine types to aid the development of vaccine candidates. Since the memory T cell response is pivotal to development of immunity and disease, we analyzed 84 key miRs connected with T cell function in the sera from vaccinated mice, pre- and post-RSV A2 challenge, at several time-points, e.g., 1-week post-prime/boost or 3d post-challenge. Total RNA was isolated from sera, used for cDNA synthesis, and miR expression was assayed using optimized primer-probes. Fold-changes in miR expression was plotted after normalization.
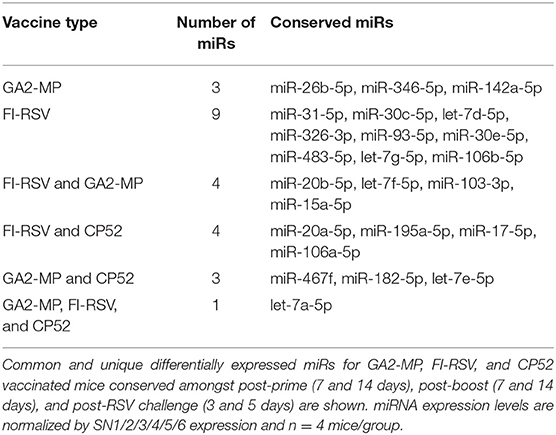
Analysis of the 84 miRs across all treatments identified miR expression signatures unique to prime-boost vaccination and RSV challenged mice, and those miR signatures conserved among all treatments (Figure 3). In general, each vaccine type, i.e., CP52, FI-RSV, or GA2-MP induced temporal and vaccine-specific miR expression patterns (Figure 3) where miR expression levels were heightened post-boost relative to prime and challenge. Given the differences in vaccine type and vaccination routes, differences were expected and emphasize miR patterns for their utility as potential vaccination biomarkers. Analogously, miR responses following A2 challenge of CP52 vaccinated mice compared to FI-RSV vaccinated mice correlated with safe vs. disease phenotypes of Th1- vs. Th2-type cytokines. Intergroup comparisons identified 58, 70 and 65 miRs differentially expressed in sera from CP52, FI-RSV, and GA2-MP vaccinated mice, respectively. Within groups, 8, 18, and 11 miRs were conserved between prime, boost and 3 days post-challenge for CP52, FI-RSV, and GA2-MP vaccinated mice, respectively. Of the miRs examined, let-7a-5p expression was upregulated >2SD in all vaccinated mice, thus this is likely not a biomarker for distinguishing vaccine-specific responses, but instead a general inflammatory biomarker (Table 1).
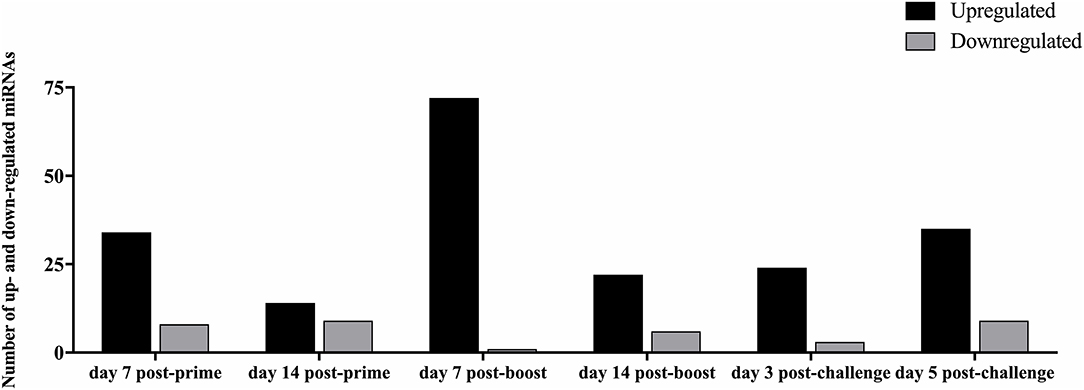
Figure 3. The number of differentially expressed miRNAs during vaccination and post-RSV challenge. Sera miRNA profiles of vaccinated mice (n = 4/group) were evaluated at day 7 post-prime, day 14 post-prime, day 7 post-boost, day 14 post-boost, day 3 post-challenge, and day 5 post-challenge using a miRNA PCR array. The y-axis indicates the number of differentially expressed miRNAs. Significance was determined using a fold-change threshold of >2, the result was reported as a fold-upregulation. If the fold change was <0.5, the result was reported as a fold-downregulation.
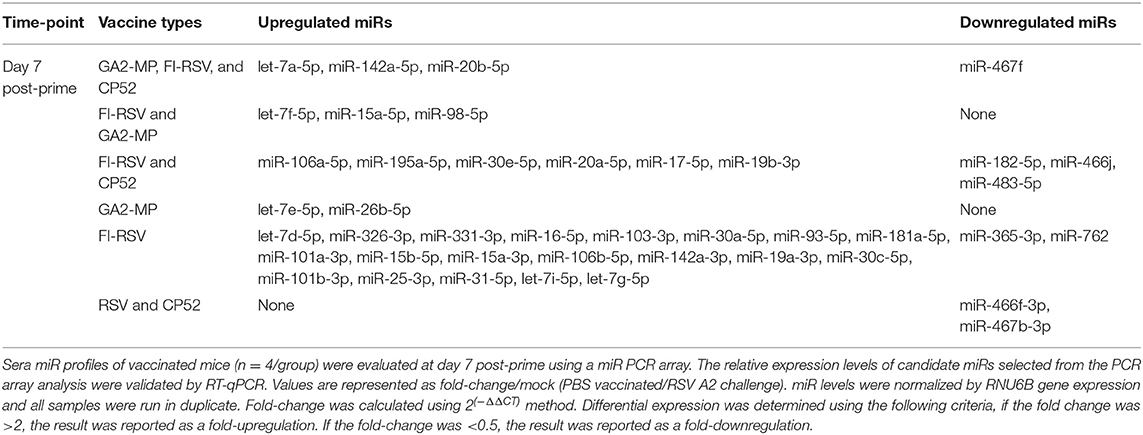
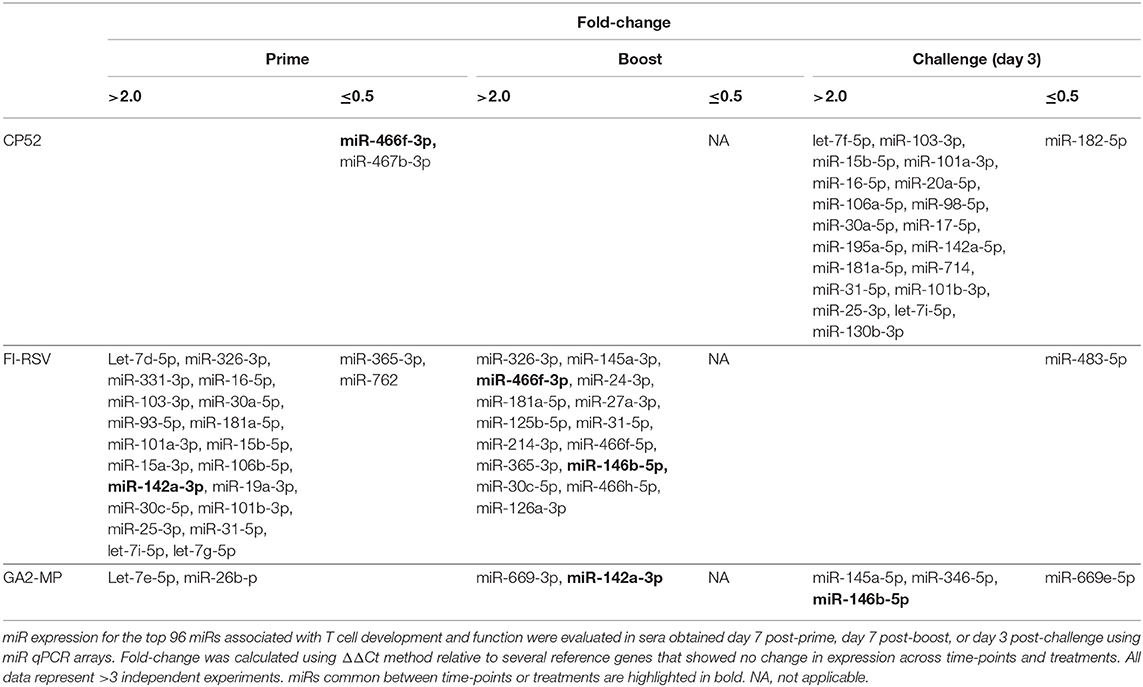
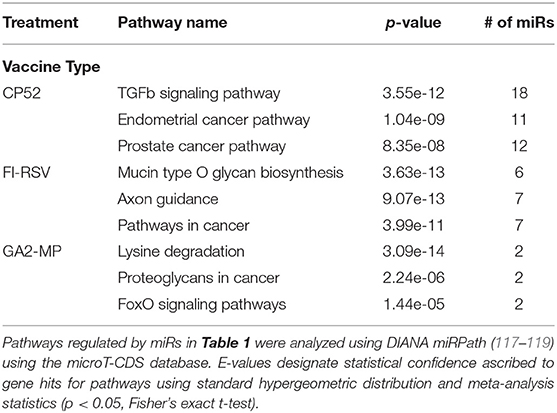
CP52 vaccination resulted in lower expression of miR-466f-3p and miR-467b-3p and did not induce any miRs. FI-RSV vaccination repressed miR-365-3p and miR-762 expression post-priming, but led to >2.0-fold induction of multiple miRs e.g., let-7d-5p, miR-326-3p, miR-331-3p, miR-16-5p, miR-103-3p, miR-30a-5p, miR-93-5p, miR-181a-5p, miR-101a-3p, miR-15b-5p, miR-15a-3p, miR-106b-5p, miR-142a-3p, miR-19a-3p, miR-30c-5p, miR-101b-3p, miR-25-3p, miR-31-5p, let-7i-5p, let-7g-5p post-prime (Table 2), and miR-326-3p, miR-145a-3p, miR-466f-3p, miR-24-3p, miR-181a-5p, miR-27a-3p, miR-125b-5p, miR-31-5p, miR-214-3p, miR-466f-5p, miR-365-3p, miR-146b-5p, miR-30c-5p, miR-466h-5p, miR-126a-3p post-boost (Table 3). In contrast, GA2-MP vaccination induced only miRs let-7e-5p and miR-26b-5p post-prime and miR-669f-3p, miR-142a-3p post-boost (Table 3). miR-466f-3p had divergent expression between CP52 and FI-RSV vaccinated mice, while miR-142a-3p showed early induction post-FI-RSV but was induced in GA2-MP vaccinated mice post-boost (Table 3).
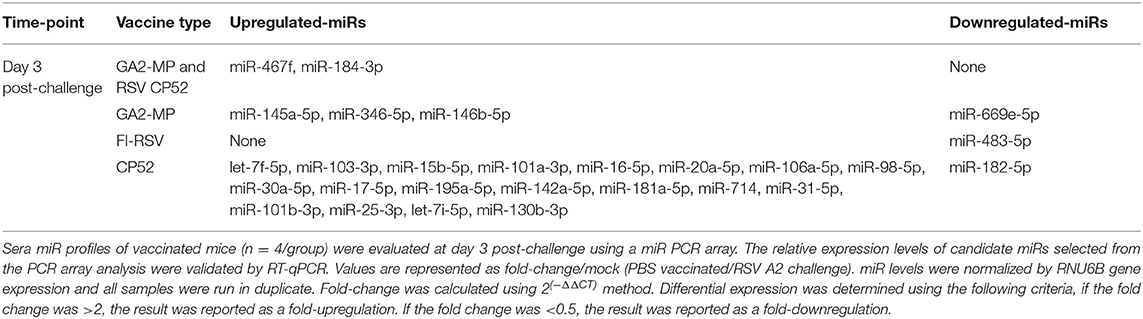
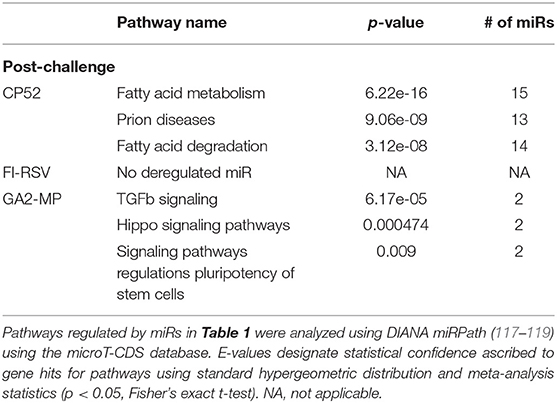
miR profiling for each vaccine type post-challenge showed unique patterns and tempos of expression. For example, CP52 vaccinated mice had higher expression of let-7f-5p, miR-103-3p, miR-15b-5p, miR-101a-3p, miR-16-5p, miR-20a-5p, miR-106a-5p, miR-98-5p, miR-30a-5p, miR-17-5p, miR-195a-5p, miR-142a-5p, miR-181a-5p, miR-714, miR-31-5p, miR-101b-3p, miR-25-3p, let-7i-5p, miR-130b-3p, and reduced miR-182-5p post-challenge (Table 4). In contrast, FI-RSV vaccinated mice showed repression of miR-483-5p post-challenge, while GA2-MP vaccinated mice had induction of miR-145a-5p, miR-346-5p, miR-146b-5p, and repression of miR-669e-5p post-challenge. Thus, comparing between vaccine groups at 7 days post-prime, 7 days post-boost, or 3 days post-RSV challenge showed miRs profiles as related to safe or disease responses (Table 5).
miR Patterns Specific to the Vaccine Type
To determine if serum miR profiles were specific to a vaccine type e.g., live (CP52) i.n. delivered, or killed (FI-RSV) i.m. delivered, or subunit (GA2-MP) s.c. delivered, the miRs were evaluated from vaccinated mice at day 7 post-vaccination. Interestingly, we observed nearly twice as many miRs expressed in the sera at day 7 post-vaccination compared to 14 days post-vaccination (Supplementary Table 1). The sera miR profiles showed that CP52 vaccinated mice had decreased miR-466f-3p and miR-467b-3p expression at 7 days post-vaccination, whereas sera miR expression from FI-RSV-vaccinated mice had two downregulated miRs (miR-365-3p and miR-62) while sera from GA2-MP vaccinated mice expressed higher miRs, i.e., let-7e-5p and miR-26b-5p at 7 days post-prime (Table 2). Interestingly, miR-467f expression was downregulated for all vaccine types at day 7 post-vaccination. Additionally, several miRs were identified in all vaccinated mice, e.g., let-7a-5p, miR-142a-5p, and miR-20b-5p which were upregulated (Table 1). The results showed that several miRs were expressed specifically to CP52 (miR-466f-3p, miR-467b-3p), to FI-RSV (let-7d-5p, miR-326-3p, miR-331-3p, miR-16-5p, miR-103-3p, miR-30a-5p, miR-93-5p, miR-181a-5p, miR-101a-3p, miR-15b-5p, miR-15a-3p, miR-106b-5p, miR-142a-3p, miR-19a-3p, miR-30c-5p, miR-101b-3p, miR-25-3p, miR-31-5p, let-7i-5p, let-7g-5p) and to GA-M2 vaccines types (let-7e-5p, miR-26b-5p).
miRs Patterns Induced by Post-Boost Vaccination and RSV Challenge
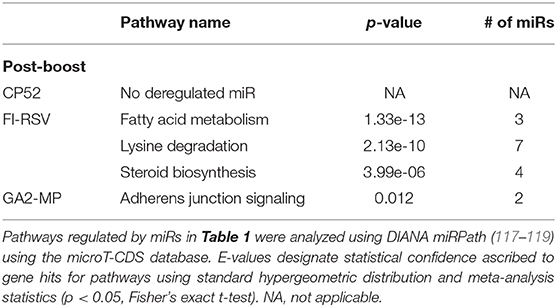
Serum miR profiles were examined to determine the miR profiles by the vaccine types post-boost (Table 3). Of 75 differentially expressed miRs evaluated (Table 3; Figure 3), the miRs commonly expressed were miR-195a-5p, miR-320-3p, let-7a-5p, miR-181b-5p, miR-672-5p, let-7e-5p, miR-17-5p, let-7c-5p, miR-714, let-7d-5p, let-7f-5p, miR-574-5p, miR-182-5p, miR-16-5p, miR-467f, miR-21a-5p, miR-130b-3p, miR-1187, miR-15b-5p, miR-26b-5p, miR-20a-5p, miR-184-3p, miR-762, miR-20b-5p, miR-25-3p, let-7i-5p, let-7g-5p which were induced by all vaccine types. As these miRs are commonly expressed it is likely their expression is linked to a general response, i.e., the pro-inflammatory response associated with vaccination. The serum miRs upregulated specific to CP52 vaccinated mice were miR-98-5p, miR-26a-5p, miR-155-5p, miR-223-3p, miR-669e-5p, for GA2-MP vaccinated mice miR-669f-3p and miR-142a-3p, and fifteen miRs were upregulated in the sera of FI-RSV vaccinated mice. Two miRs, miR-669f-3p and miR-142a-3p, were commonly expressed in CP52 and GA2-MP vaccinated mice, and 50 miRs were differentially expressed post-RSV challenge of vaccinated mice. All data are shown in Supplementary Tables 2, 3. At day 3 post-RSV challenge, 24 miRs were upregulated for all vaccinated mice types compared to mock-vaccinated (Table 4). For CP52-vaccinated mice, several serum miRs were upregulated (e.g., let-7f-5p, miR-103-3p, miR-15b-5p, miR-101a-3p, miR-16-5p, miR-20a-5p, miR-106a-5p, miR-98-5p, miR-30a-5p, miR-17-5p, miR-195a-5p, miR-142a-5p, miR-181a-5p, miR-714, miR-31-5p, miR-101b-3p, miR-25-3p, let-7i-5p, miR-130b-3p), for FI-RSV-vaccinated mice only miR-483-5p was upregulated, and for GA2-MP vaccinated mice miR-145a-5p, miR-346-5p, and miR-146b-5p were upregulated. Interestingly, miR-184-3p was expressed in the sera by all vaccine groups suggesting that this miR is not vaccine-specific. The data from these studies is summarized in Table 5.
miRs and the Host Pathway
miRs act as rheostats to subtly regulate aspects of the host immune response to virus infection and vaccination (64). They fine-tune responses, adjust functions, and bolster or dampen immune operations to maintain homeostasis. The pattern of miR expression highlights their function, i.e., constrain or enhance responses in a temporal fashion. As this study examines the pattern and tempo of miRs expressed in the response to vaccination and challenge, it is not surprising to identify unique and common miR profiles and those that are differentially expressed during and after vaccination or challenge. A goal of these studies is to determine if the miR expression patterns can be used to predict safe or unsafe responses to vaccination or challenge. Viral infection and vaccination induce inflammation and determining miR pathways that are induced or repressed in mice can help differentiate safe vs. disease risk vaccines. As we examined the miR expression pattern linked to the type of vaccine and the cytokine response to RSV vaccination and challenge, we analyzed the gene pathways that could be regulated by miRs that are induced or repressed following CP52, FI-RSV or GA2-MP priming, prime-boost, and RSV challenge using DIANA miRPath (117–119). The intersecting pathways were selected for examination having a p-value cutoff of p < 0.05 (Table 5). CP52 vaccination downregulated miR-466f-3p and miR467b-3p which is known to regulate genes of the TGFβ signaling which has been shown to have an important role in RSV replication and inflammation leading to lung injury, fibrosis, and remodeling (136–144). Additionally, these miRs are predicted to regulate cancer-related pathways (Table 6; Supplemental Table 1) which contain many of the top genes involved in cell cycle control, a feature live RSV infection is known to regulate (65, 102, 142, 145–147). FI-RSV priming induced miRs predicted to regulate mucin biosynthesis, axon guidance, and other pathways in cancer (Table 2) while GA2-MP primed miRs were predicted to regulate Lysine degradation, proteoglycan expression and function and FoxO signaling (Table 6). FoxO signaling pathway has been shown to regulate the innate immune pathways in respiratory epithelium following infection (148). Analysis of miR at 7d post-boost showed distinct miR pathway profiles among candidate vaccines. While both CP52 and GA2-MP boosting did not alter miR expression, FI-RSV boosting led to many deregulated miRs (Table 5). In particular, the fatty acid metabolism pathway is predicted to be regulated by these miRs. Fatty acid metabolism is essential for RSV replication (149). Additional pathways predicted to be regulated by miRs and linked to FI-RSV boosting include lysine degradation and steroid biosynthesis (Table 6). GA2-MP boosting affected miRs patterns predicted to regulate adherens junction signaling which are associated to disruption of the airway barrier during infection [Table 7; (150–152)]. miRs deregulated following RSV challenge in CP52 or GA2-MP vaccinated mice were predicted to regulate multiple pathways in fatty acid metabolism and pluripotency, likely related to cell cycle (Table 8). Additionally, TGFb and Hippo signaling pathways were also shown to likely regulated by the miR expression patterns (Table 8). The Hippo pathway is thought to be involved in modulating the potency of anti-viral response particularly in a nutritional deprivation state (153).
The results indicate that the miR profiles and their tempos of expression are adjusted to the type of vaccine and challenge, an effect linked to both non-specific responses (e.g., inflammation) and specific immune responses (e.g., T cell activation or memory). It is important to note that some miRs are unaffected by vaccination while others undergo a global up- or down-regulation upon vaccination or challenge. For reasons of brevity, we have focused on understanding those miR expression patterns induced ≥2SD above the control. It is important to note that several immune regulatory molecules are miR targets. Specifically, cytokines/chemokines are immune effector molecules and are integrated in the net responses to vaccination and challenge. miRs control the activation and integration of the pathways to support T cell responses while maintaining homeostasis. Additional information regarding the miR host pathway analysis can be found in Data Sheet 1 of the Supplementary Material section.
Discussion
The development of safe and effective RSV vaccine candidates can be assisted by a better understanding of biomarker expression. Biomarkers may allow for the prediction of probable vaccine candidate outcomes. Additional analyses are needed to further aid decisions regarding vaccine candidates, but ways to improve RSV vaccine candidate selection has become paramount after more than 5 decades of unsuccessful research efforts. We hypothesized that assessment of miR profiles with general Th1/Th2 cytokine responses would enable correlations with safe, live vaccines (CP52), subunit vaccines (GA-M2), and disease-enhancing vaccines (FI-RSV) to help develop baselines for a better understanding of prospective RSV vaccine candidates. In this study, show vaccine-specific and temporal miRNA expression profiles relating to efficacy or vaccine-associated disease. We examined miR expression profiles of vaccinated mice pre- and post-RSV challenge were determined for 84 miRs associated with T cell responses and function. We showed that while both CP52 and FI-RSV vaccination induce a humoral response, only CP52 induced a neutralizing antibody response leading to reduction in RSV replication (Figure 1E). Further, splenocytes from CP52, FI-RSV, or GA2-MP vaccinated mice were stimulated with RSV-specific peptides then assayed for Th1-type or Th2-type cytokines. The cytokine and miR expression showed that M282−90 re-stimulation of splenocytes from CP52 but not FI-RSV vaccinated mice led to a strong induction of IFNg which is characteristic of a Th1 response. In contrast, peptide stimulation of splenocytes from FI-RSV vaccinated mice led to a strong induction of IL4, a cytokine characteristic of a Th2-type response. Peptide stimulation of splenocytes from GA2-MP vaccinated mice led to a higher number of G183−198 IL4- and IFN-specific secreting cells, characteristic of a balanced Th1-/Th2-type response to the G protein. Taken together, these results led to the assessment that CP52 vaccination primes for a safe response while FI-RSV primes for disease following vaccination and GA2-MP primes for a mostly balanced cytokine response.
The miR PCR array showed differential expression of a conserved set of miRs across prime-boost vaccination and RSV-challenge, more specifically 11 miRNAs in GA2-MP vaccinated mice, 18 miRs in FI-RSV vaccinated mice, and 8 miRs in RSV CP52 vaccinated mice. Several of these miRs have been shown to participate in the regulation of the immune response, and in some cases are associated with RSV infection. FI-RSV vaccinated mice had let-7d-5p, let-7f-5p, and let-7g-5p miR expression at post-prime, post-boost, and post-challenge. GA2-MP vaccinated mice had similar results with let-7e-5p and let-7f expression. Members of the let-7 family target IL-6 expression, and has an extensive list of other experimentally validated targets including SOCS4, caspase-3, p27, TLR4, IL-13, and IL10 (101). Let-7 could be a mechanism of IL-6 regulation during RSV infection (101). RSV infection induces secretion of numerous pro-inflammatory cytokines, including type I and types II IFNs, TNFα, IL-12, and IL-6 (101, 154–156). Mice vaccinated with CP52 or GA2-MP induced differential miR-467f expression during prime-boost vaccination and post-challenge. Previous miR screens for respiratory viruses have not previously identified miR-467f; however, a microarray-based approach to evaluate the miR profile of HIV-associated nephropathy in a mouse model showed that treatment with rapamycin (an mTOR inhibitor) to halt disease progression induced upregulation of miR-467f expression (157). Interestingly, rapamycin inhibits RSV-induced mTOR activation and increases the frequency of RSV-specific CD8 T cells and RSV-specific memory T cell precursors in mice (158). Therefore, miR-467f may have a role in cellular immunity during vaccination and RSV infection. miR-106a-5p and miR-106b-5p expression levels were upregulated in FI-RSV vaccinated mice during prime-boost vaccination and post-challenge with RSV. Interestingly, allergic airway inflammation in mice has been associated with increased miR-106a expression and decreased IL-10, suggesting that miR-106a may regulate IL-10 expression and Th2-type responses (159, 160). Expression levels of miR-30c-5p and miR-30e-5p (from the miR-30/384-5p family) were upregulated in FI-RSV vaccinated mice at prime and boost-vaccination, and post-challenge. Upregulation of miR-30c-5p expression in the airway wall has been shown in a BALB/c mouse model of chronic asthma (161). The miR profiles identified for vaccine-induced protection and vaccine-enhanced disease appear to correlate with protective immune functions and airway inflammation, respectively. Although this study produced a list of miRs that may regulate RSV vaccine efficacy, additional studies are warranted to clarify the mechanisms behind how these miRs mediate host-virus interactions.
Overall, the results from these studies show that vaccine candidates associated safe or disease responses exhibited differential miR profiles following boosting which were higher in magnitude compared to priming or RSV challenge sera specimens. The results demonstrate that a considerable number of miRs are different between vaccine types, and a common set of miRs is expressed for all vaccine treatments. Pathway analysis of miR targets identified pathways correlated with inflammation particularly those that may contribute to airway inflammation, leukocyte recruitment and alveolar infiltration (12, 162, 163). The miR profiles from vaccinated mice were linked to cytokine phenotypes of protection or disease and appear to correlate with miRs that regulate protective immune functions or airway inflammation. Additional studies are warranted to validate miR phenotypes to determine the mechanisms of action linked to host gene regulation, and the associated immune response to determine their value as predictive biomarkers. These studies show that serum miR profiles may offer a proxy to assist vaccine development and facilitate a better understanding of vaccine studies.
Ethics Statement
Specific-pathogen-free, 6-to-8 weeks old female BALB/c mice (The Jackson Laboratory) were used. Mice were maintained in microisolator cages with sterilized water and food ad libitum. All experiments were performed in accordance with the guidelines of the University of Georgia Institutional Animal Care and Use Committee (IACUC).
Author Contributions
LA, PJ, AB, and RT conceived, designed, performed, analyzed the experiments, and wrote the manuscript. RT contributed reagents, materials, and analysis tools.
Funding
This project was supported by an ARCS Foundation fellowship to LA, and the Georgia Research Alliance funding to RT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Jeff Powell (Artificial Cell Technologies, New Haven, CT) for providing the GA2-MP vaccine. We would also like to thank Colin Williams, Ali S. Maleki, and Jackelyn Murray for assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02323/full#supplementary-material
References
1. Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. (2017) 390:946–58. doi: 10.1016/S0140-6736(17)30938-8
2. Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med. (2009) 360:588–98. doi: 10.1056/NEJMoa0804877
3. Parikh RC, McLaurin KK, Margulis AV, Mauskopf J, Ambrose CS, Pavilack M, et al. Chronologic age at hospitalization for respiratory syncytial virus among preterm and term infants in the United States. Infect Dis Ther. (2017) 6:477–86. doi: 10.1007/s40121-017-0167-9
4. Anderson EJ, Carosone-Link P, Yogev R, Yi J, Simoes EAF. Effectiveness of palivizumab in high-risk infants and children: a propensity score weighted regression analysis. Pediatr Infect Dis J. (2017) 36:699–704. doi: 10.1097/INF.0000000000001533
5. Hoch HE, Collaco JM. Recurrent wheezing in childhood and palivizumab. Am J Respir Crit Care Med. (2017) 196:1–2. doi: 10.1164/rccm.201701-0256ED
6. Mochizuki H, Kusuda S, Okada K, Yoshihara S, Furuya H, Simoes EAF. Palivizumab prophylaxis in preterm infants and subsequent recurrent wheezing. Six-year follow-up study. Am J Respir Crit Care Med. (2017) 196:29–38. doi: 10.1164/rccm.201609-1812OC
7. Resch B. Product review on the monoclonal antibody palivizumab for prevention of respiratory syncytial virus infection. Hum Vaccin Immunother. (2017) 13:2138–49. doi: 10.1080/21645515.2017.1337614
8. Sanchez-Luna M, Burgos-Pol R, Oyaguez I, Figueras-Aloy J, Sanchez-Solis M, Martinon-Torres F, et al. Cost-utility analysis of Palivizumab for Respiratory Syncytial Virus infection prophylaxis in preterm infants: update based on the clinical evidence in Spain. BMC Infect Dis. (2017) 17:687. doi: 10.1186/s12879-017-2803-0
9. Wong SK, Li A, Lanctot KL, Paes B. Adherence and outcomes: a systematic review of palivizumab utilization. Expert Rev Respir Med. (2018) 12:27–42. doi: 10.1080/17476348.2018.1401926
10. Anderson LJ, Dormitzer PR, Nokes DJ, Rappuoli R, Roca A, Graham BS. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine. (2013) 31(Suppl. 2):B209–15. doi: 10.1016/j.vaccine.2012.11.106
11. Drysdale SB, Sande CJ, Green CA, Pollard AJ. RSV vaccine use–the missing data. Expert Rev Vaccines. (2016) 15:149–52. doi: 10.1586/14760584.2016.1114419
12. Graham BS. Vaccine development for respiratory syncytial virus. Curr Opin Virol. (2017) 23:107–12. doi: 10.1016/j.coviro.2017.03.012
13. Higgins D, Trujillo C, Keech C. Advances in RSV vaccine research and development - a global agenda. Vaccine. (2016) 34:2870–5. doi: 10.1016/j.vaccine.2016.03.109
14. Jorquera PA, Anderson L, Tripp RA. Understanding respiratory syncytial virus (RSV) vaccine development and aspects of disease pathogenesis. Expert Rev Vaccines. (2016) 15:173–87. doi: 10.1586/14760584.2016.1115353
15. Rezaee F, Linfield DT, Harford TJ, Piedimonte G. Ongoing developments in RSV prophylaxis: a clinician's analysis. Curr Opin Virol. (2017) 24:70–8. doi: 10.1016/j.coviro.2017.03.015
16. Roberts JN, Graham BS, Karron RA, Munoz FM, Falsey AR, Anderson LJ, et al. Challenges and opportunities in RSV vaccine development: meeting report from FDA/NIH workshop. Vaccine. (2016) 34:4843–9. doi: 10.1016/j.vaccine.2016.07.057
17. Becker Y. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy–a review. Virus Genes. (2006) 33:235–52. doi: 10.1007/s11262-006-0064-x
18. Castilow EM, Olson MR, Varga SM. Understanding respiratory syncytial virus (RSV) vaccine-enhanced disease. Immunol Res. (2007) 39:225–39. doi: 10.1007/s12026-007-0071-6
19. Johnson TR, Graham BS. Contribution of respiratory syncytial virus G antigenicity to vaccine-enhanced illness and the implications for severe disease during primary respiratory syncytial virus infection. Pediatr Infect Dis J. (2004) 23:S46–57. doi: 10.1097/01.inf.0000108192.94692.d2
20. Boyoglu-Barnum S, Todd SO, Meng J, Barnum TR, Chirkova T, Haynes LM, et al. Mutating the CX3C motif in the G protein should make a live respiratory syncytial virus vaccine safer and more effective. J Virol. (2017) 91:e02059–16. doi: 10.1128/JVI.02059-16
21. Connors M, Crowe JE Jr, Firestone CY, Murphy BR, Collins PL. A cold-passaged, attenuated strain of human respiratory syncytial virus contains mutations in the F and L genes. Virology. (1995) 208:478–84. doi: 10.1006/viro.1995.1178
22. Gonzalez IM, Karron RA, Eichelberger M, Walsh EE, Delagarza VW, Bennett R, et al. Evaluation of the live attenuated cpts 248/404 RSV vaccine in combination with a subunit RSV vaccine (PFP-2) in healthy young and older adults. Vaccine. (2000) 18:1763–72. doi: 10.1016/S0264-410X(99)00527-7
23. Groppo R, DiNapoli J, Il Jeong K, Kishko M, Jackson N, Kleanthous H, et al. Effect of genetic background and delivery route on the preclinical properties of a live attenuated RSV vaccine. PLoS ONE. (2018) 13:e0199452. doi: 10.1371/journal.pone.0199452
24. Hsu KH, Crowe JE Jr, Lubeck MD, Davis AR, Hung PP, Chanock RM, et al. Isolation and characterization of a highly attenuated respiratory syncytial virus (RSV) vaccine candidate by mutagenesis of the incompletely attenuated RSV A2 ts-1 NG-1 mutant virus. Vaccine. (1995) 13:509–15. doi: 10.1016/0264-410X(94)00002-5
25. Jin H, Cheng X, Traina-Dorge VL, Park HJ, Zhou H, Soike K, et al. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine. (2003) 21:3647–52. doi: 10.1016/S0264-410X(03)00426-2
26. Karron RA, Luongo C, Thumar B, Loehr KM, Englund JA, Collins PL, et al. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci Transl Med. (2015) 7:312ra175. doi: 10.1126/scitranslmed.aac8463
27. Le Nouen C, Brock LG, Luongo C, McCarty T, Yang L, Mehedi M, et al. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc Natl Acad Sci USA. (2014) 111:13169–74. doi: 10.1073/pnas.1411290111
28. Malkin E, Yogev R, Abughali N, Sliman J, Wang CK, Zuo F, et al. Safety and immunogenicity of a live attenuated RSV vaccine in healthy RSV-seronegative children 5 to 24 months of age. PLoS ONE. (2013) 8:e77104. doi: 10.1371/journal.pone.0077104
29. Russell RF, McDonald JU, Ivanova M, Zhong Z, Bukreyev A, Tregoning JS. Partial attenuation of respiratory syncytial virus with a deletion of a small hydrophobic gene is associated with elevated interleukin-1beta responses. J Virol. (2015) 89:8974–81. doi: 10.1128/JVI.01070-15
30. Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE Jr, Boyce TG, et al. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis. (2000) 182:1331–42. doi: 10.1086/315859
31. Blanco JCG, Pletneva LM, McGinnes-Cullen L, Otoa RO, Patel MC, Fernando LR, et al. Efficacy of a respiratory syncytial virus vaccine candidate in a maternal immunization model. Nat Commun. (2018) 9:1904. doi: 10.1038/s41467-018-04216-6
32. Cayatte C, Snell Bennett A, Rajani GM, Hostetler L, Maynard SK, Lazzaro M, et al. Inferior immunogenicity and efficacy of respiratory syncytial virus fusion protein-based subunit vaccine candidates in aged versus young mice. PLoS ONE. (2017) 12:e0188708. doi: 10.1371/journal.pone.0188708
33. Esposito S, Pietro GD. Respiratory syncytial virus vaccines: an update on those in the immediate pipeline. Future Microbiol. (2016) 11:1479–90. doi: 10.2217/fmb-2016-0106
34. Garg R, Latimer L, Gomis S, Gerdts V, Potter A, van Drunen Littel-van den Hurk S. Maternal vaccination with a novel chimeric glycoprotein formulated with a polymer-based adjuvant provides protection from human parainfluenza virus type 3 in newborn lambs. Antiviral Res. (2019) 162:54–60. doi: 10.1016/j.antiviral.2018.12.010
35. Herve PL, Descamps D, Deloizy C, Dhelft V, Laubreton D, Bouguyon E, et al. Non-invasive epicutaneous vaccine against Respiratory Syncytial Virus: Preclinical proof of concept. J Control Release. (2016) 243:146–59. doi: 10.1016/j.jconrel.2016.10.003
36. Sarkar I, Zardini Buzatto A, Garg R, Li L, van Drunen Littel-van den Hurk S. Metabolomic and immunological profiling of respiratory syncytial virus infection after intranasal immunization with a subunit vaccine candidate. J Proteome Res. (2019) 18:1145–61. doi: 10.1021/acs.jproteome.8b00806
37. Schneider-Ohrum K, Cayatte C, Bennett AS, Rajani GM, McTamney P, Nacel K, et al. Immunization with low doses of recombinant postfusion or prefusion respiratory syncytial virus F primes for vaccine-enhanced disease in the cotton rat model independently of the presence of a Th1-biasing (GLA-SE) or Th2-biasing (Alum) adjuvant. J Virol. (2017) 91:e02180–16. doi: 10.1128/JVI.02180-16
38. Zhang L, Durr E, Galli JD, Cosmi S, Cejas PJ, Luo B, et al. Design and characterization of a fusion glycoprotein vaccine for Respiratory Syncytial Virus with improved stability. Vaccine. (2018) 36:8119–30. doi: 10.1016/j.vaccine.2018.10.032
39. Brock LG, Liu X, Liang B, Lingemann M, Liu X, Herbert R, et al. Murine pneumonia virus expressing the fusion glycoprotein of human respiratory syncytial virus from an added gene is highly attenuated and immunogenic in rhesus macaques. J Virol. (2018) 92:e00723–18. doi: 10.1128/JVI.00723-18
40. Green CA, Sande CJ, Scarselli E, Capone S, Vitelli A, Nicosia A, et al. Novel genetically-modified chimpanzee adenovirus and MVA-vectored respiratory syncytial virus vaccine safely boosts humoral and cellular immunity in healthy older adults. J Infect. (2019) 78:382–92. doi: 10.1016/j.jinf.2019.02.003
41. Liang B, Kabatova B, Kabat J, Dorward DW, Liu X, Surman S, et al. Effects of alterations to the CX3C motif and secreted form of human respiratory syncytial virus (RSV) G protein on immune responses to a parainfluenza virus vector expressing the RSV G protein. J Virol. (2019) 93:e02043–18. doi: 10.1128/JVI.02043-18
42. Lu X, Yang J, Wu H, Yang Z, Jin C, Wang J, et al. High-throughput sequencing identifies HIV-1-replication- and latency-related miRNAs in CD4(+) T cell lines. Arch Virol. (2017) 162:1933–42. doi: 10.1007/s00705-017-3305-5
43. Phan SI, Adam CM, Chen Z, Citron M, Liang X, Espeseth AS, et al. Genetic stability of parainfluenza virus 5-vectored human respiratory syncytial virus vaccine candidates after in vitro and in vivo passage. J Virol. (2017) 91:e00559–17. doi: 10.1128/JVI.00559-17
44. Phan SI, Zengel JR, Wei H, Li Z, Wang D, He B. Parainfluenza virus 5 expressing wild-type or prefusion respiratory syncytial virus (RSV) fusion protein protects mice and cotton rats from RSV challenge. J Virol. (2017) 91:e00560–17. doi: 10.1128/JVI.00560-17
45. Wang D, Phan S, DiStefano DJ, Citron MP, Callahan CL, Indrawati L, et al. A single-dose recombinant parainfluenza virus 5-vectored vaccine expressing respiratory syncytial virus (RSV) F or G protein protected cotton rats and African green monkeys from RSV challenge. J Virol. (2017) 91:e00066–17. doi: 10.1128/JVI.00066-17
46. Wiegand MA, Gori-Savellini G, Gandolfo C, Papa G, Kaufmann C, Felder E, et al. A Respiratory syncytial virus vaccine vectored by a stable chimeric and replication-deficient sendai virus protects mice without inducing enhanced disease. J Virol. (2017) 91:e02298–16. doi: 10.1128/JVI.02298-16
47. Harcourt JL, Anderson LJ, Sullender W, Tripp RA. Pulmonary delivery of respiratory syncytial virus DNA vaccines using macroaggregated albumin particles. Vaccine. (2004) 22:2248–60. doi: 10.1016/j.vaccine.2003.11.050
48. Murawski MR, McGinnes LW, Finberg RW, Kurt-Jones EA, Massare MJ, Smith G, et al. Newcastle disease virus-like particles containing respiratory syncytial virus G protein induced protection in BALB/c mice, with no evidence of immunopathology. J Virol. (2010) 84:1110–23. doi: 10.1128/JVI.01709-09
49. Schmidt MR, McGinnes LW, Kenward SA, Willems KN, Woodland RT, Morrison TG. Long-term and memory immune responses in mice against Newcastle disease virus-like particles containing respiratory syncytial virus glycoprotein ectodomains. J Virol. (2012) 86:11654–62. doi: 10.1128/JVI.01510-12
50. Rigter A, Widjaja I, Versantvoort H, Coenjaerts FE, van Roosmalen M, Leenhouts K, et al. A protective and safe intranasal RSV vaccine based on a recombinant prefusion-like form of the F protein bound to bacterium-like particles. PLoS ONE. (2013) 8:e71072. doi: 10.1371/journal.pone.0071072
51. Van Braeckel-Budimir N, Haijema BJ, Leenhouts K. Bacterium-like particles for efficient immune stimulation of existing vaccines and new subunit vaccines in mucosal applications. Front Immunol. (2013) 4:282. doi: 10.3389/fimmu.2013.00282
52. Lee S, Quan FS, Kwon Y, Sakamoto K, Kang SM, Compans RW, et al. Additive protection induced by mixed virus-like particles presenting respiratory syncytial virus fusion or attachment glycoproteins. Antiviral Res. (2014) 111:129–35. doi: 10.1016/j.antiviral.2014.09.005
53. McGinnes Cullen L, Schmidt MR, Kenward SA, Woodland RT, Morrison TG. Murine immune responses to virus-like particle-associated pre- and postfusion forms of the respiratory syncytial virus F protein. J Virol. (2015) 89:6835–47. doi: 10.1128/JVI.00384-15
54. Jorquera PA, Tripp RA. Synthetic biodegradable microparticle and nanoparticle vaccines against the respiratory syncytial virus. Vaccines. (2016) 4:E45. doi: 10.3390/vaccines4040045
55. Schwarz B, Morabito KM, Ruckwardt TJ, Patterson DP, Avera J, Miettinen HM, et al. Viruslike particles encapsidating respiratory syncytial virus M and M2 proteins induce robust T cell responses. ACS Biomater Sci Eng. (2016) 2:2324–32. doi: 10.1021/acsbiomaterials.6b00532
56. Hwang HS, Kim KH, Lee Y, Lee YT, Ko EJ, Park S, et al. Virus-like particle vaccines containing F or F and G proteins confer protection against respiratory syncytial virus without pulmonary inflammation in cotton rats. Hum Vaccin Immunother. (2017) 13:1031–9. doi: 10.1080/21645515.2016.1272743
57. Kim AR, Lee DH, Lee SH, Rubino I, Choi HJ, Quan FS. Protection induced by virus-like particle vaccine containing tandem repeat gene of respiratory syncytial virus G protein. PLoS ONE. (2018) 13:e0191277. doi: 10.1371/journal.pone.0191277
58. Smith TRF, Schultheis K, Morrow MP, Kraynyak KA, McCoy JR, Yim KC, et al. Development of an intradermal DNA vaccine delivery strategy to achieve single-dose immunity against respiratory syncytial virus. Vaccine. (2017) 35:2840–7. doi: 10.1016/j.vaccine.2017.04.008
59. Smith TRF, Schultheis K, Broderick KE. Nucleic acid-based vaccines targeting respiratory syncytial virus: delivering the goods. Hum Vaccin Immunother. (2017) 13:2626–9. doi: 10.1080/21645515.2017.1363134
60. Hua Y, Jiao YY, Ma Y, Peng XL, Fu YH, Zhang XJ, et al. Enhanced humoral and CD8+ T cell immunity in mice vaccinated by DNA vaccine against human respiratory syncytial virus through targeting the encoded F protein to dendritic cells. Int Immunopharmacol. (2017) 46:62–9. doi: 10.1016/j.intimp.2017.02.023
61. Hua Y, Jiao YY, Ma Y, Peng XL, Fu YH, Zheng YP, et al. DNA vaccine encoding central conserved region of G protein induces Th1 predominant immune response and protection from RSV infection in mice. Immunol Lett. (2016) 179:95–101. doi: 10.1016/j.imlet.2016.09.011
62. Eroglu E, Singh A, Bawage S, Tiwari PM, Vig K, Pillai SR, et al. Immunogenicity of RSV F DNA vaccine in BALB/c mice. Adv Virol. (2016) 2016:7971847. doi: 10.1155/2016/7971847
63. Wu H, Dennis VA, Pillai SR, Singh SR. RSV fusion (F) protein DNA vaccine provides partial protection against viral infection. Virus Res. (2009) 145:39–47. doi: 10.1016/j.virusres.2009.06.012
64. Barichievy S, Bakre A. Host-encoded miRNAs involved in host-pathogen interactions. In: Tripp RA, Karpilow J, editors. Frontiers in RNAi. Vol. 1. Oak Park, IL: Bentham Science Publishers (2014). p. 107–43. doi: 10.2174/9781608059409114010010
65. Mgbemena V, Segovia J, Chang T, Bose S. Kruppel-like factor 6 regulates transforming growth factor-beta gene expression during human respiratory syncytial virus infection. Virol J. (2011) 8:409. doi: 10.1186/1743-422X-8-409
66. Jadidi-Niaragh F, Mirshafiey A. The deviated balance between regulatory T cell and Th17 in autoimmunity. Immunopharmacol Immunotoxicol. (2012) 34:727–39. doi: 10.3109/08923973.2011.619987
67. Mukherjee M, Nair P. Autoimmune responses in severe asthma. Allergy Asthma Immunol Res. (2018) 10:428–47. doi: 10.4168/aair.2018.10.5.428
68. Arenas-Padilla M, Mata-Haro V. Regulation of TLR signaling pathways by microRNAs: implications in inflammatory diseases. Central Eur J Immunol. (2018) 43:482–9. doi: 10.5114/ceji.2018.81351
69. Salvi V, Gianello V, Tiberio L, Sozzani S, Bosisio D. Cytokine targeting by miRNAs in autoimmune diseases. Front Immunol. (2019) 10:15. doi: 10.3389/fimmu.2019.00015
70. Yan L, Liang M, Hou X, Zhang Y, Zhang H, Guo Z, et al. The role of microRNA-16 in the pathogenesis of autoimmune diseases: a comprehensive review. Biomed Pharmacother. (2019) 112:108583. doi: 10.1016/j.biopha.2019.01.044
71. Eyileten C, Wicik Z, De Rosa S, Mirowska-Guzel D, Soplinska A, Indolfi C, et al. MicroRNAs as diagnostic and prognostic biomarkers in ischemic stroke-a comprehensive review and bioinformatic analysis. Cells. (2018) 7:E249. doi: 10.3390/cells7120249
72. Huang X, Zhu Z, Guo X, Kong X. The roles of microRNAs in the pathogenesis of chronic obstructive pulmonary disease. Int Immunopharmacol. (2019) 67:335–47. doi: 10.1016/j.intimp.2018.12.013
73. Lorente-Cebrian S, Gonzalez-Muniesa P, Milagro FI, Martinez JA. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: emerging roles as biomarkers and therapeutic targets. Clin Sci. (2019) 133:23–40. doi: 10.1042/CS20180890
74. Pockar S, Globocnik Petrovic M, Peterlin B, Vidovic Valentincic N. MiRNA as biomarker for uveitis - a systematic review of the literature. Gene. (2019) 696:162–75. doi: 10.1016/j.gene.2019.02.004
75. Amaral AJ, Andrade J, Foxall RB, Matoso P, Matos AM, Soares RS, et al. miRNA profiling of human naive CD4 T cells links miR-34c-5p to cell activation and HIV replication. EMBO J. (2017) 36:346–60. doi: 10.15252/embj.201694335
76. Gutierrez-Vazquez C, Rodriguez-Galan A, Fernandez-Alfara M, Mittelbrunn M, Sanchez-Cabo F, Martinez-Herrera DJ, et al. miRNA profiling during antigen-dependent T cell activation: a role for miR-132-3p. Sci Rep. (2017) 7:3508. doi: 10.1038/s41598-017-03689-7
77. Torri A, Carpi D, Bulgheroni E, Crosti MC, Moro M, Gruarin P, et al. Extracellular MicroRNA signature of human helper T cell subsets in health and autoimmunity. J Biol Chem. (2017) 292:2903–15. doi: 10.1074/jbc.M116.769893
78. Zhou H, Wu L. The development and function of dendritic cell populations and their regulation by miRNAs. Protein Cell. (2017) 8:501–13. doi: 10.1007/s13238-017-0398-2
79. Johansson K, Malmhall C, Ramos-Ramirez P, Radinger M. MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation. J Allergy Clin Immunol. (2017) 139:1007–16.e9. doi: 10.1016/j.jaci.2016.06.035
80. Kim RY, Horvat JC, Pinkerton JW, Starkey MR, Essilfie AT, Mayall JR, et al. MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase-mediated suppression of histone deacetylase 2. J Allergy Clin Immunol. (2017) 139:519–32. doi: 10.1016/j.jaci.2016.04.038
81. Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, Schorl C, et al. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS ONE. (2012) 7:e40676. doi: 10.1371/journal.pone.0040676
82. Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. (2012) 129:1064–75.e9. doi: 10.1016/j.jaci.2012.01.060
83. Malmhall C, Johansson K, Winkler C, Alawieh S, Ekerljung L, Radinger M. Altered miR-155 expression in allergic asthmatic airways. Scand J Immunol. (2017) 85:300–7. doi: 10.1111/sji.12535
84. Yang M, Eyers F, Xiang Y, Guo M, Young IG, Rosenberg HF, et al. Expression profiling of differentiating eosinophils in bone marrow cultures predicts functional links between microRNAs and their target mRNAs. PLoS ONE. (2014) 9:e97537. doi: 10.1371/journal.pone.0097537
85. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
86. Cloonan N. Re-thinking miRNA-mRNA interactions: intertwining issues confound target discovery. Bioessays. (2015) 37:379–88. doi: 10.1002/bies.201400191
87. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. (2010) 79:351–79. doi: 10.1146/annurev-biochem-060308-103103
88. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
89. Jia S, Zhai H, Zhao M. MicroRNAs regulate immune system via multiple targets. Discov Med. (2014) 18:237–47.
90. Jost D, Nowojewski A, Levine E. Small RNA biology is systems biology. BMB Rep. (2011) 44:11–21. doi: 10.5483/BMBRep.2011.44.1.11
91. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. (2008) 105:1608–13. doi: 10.1073/pnas.0707594105
92. Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, et al. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. (2012) 40:1695–707. doi: 10.1093/nar/gkr934
93. Bellingham SA, Hill AF. Analysis of miRNA signatures in neurodegenerative prion disease. Methods Mol Biol. (2017) 1658:67–80. doi: 10.1007/978-1-4939-7244-9_6
94. Chandra S, Vimal D, Sharma D, Rai V, Gupta SC, Chowdhuri DK. Role of miRNAs in development and disease: lessons learnt from small organisms. Life Sci. (2017) 185:8–14. doi: 10.1016/j.lfs.2017.07.017
95. Dahiya N, Atreya CD. MicroRNAs and major blood-borne infectious viral diseases. Microrna. (2014) 2:212–8. doi: 10.2174/2211536602666131118224225
96. Diosa-Toro M, Echavarria-Consuegra L, Flipse J, Fernandez GJ, Kluiver J, van den Berg A, et al. MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression. PLoS Negl Trop Dis. (2017) 11:e0005981. doi: 10.1371/journal.pntd.0005981
97. Kumar H, Bot A. Role of MicroRNAs in shaping innate immunity and as therapeutic targets for autoimmune diseases. Int Rev Immunol. (2017) 36:123–4. doi: 10.1080/08830185.2017.1340043
98. Xu Z, Zhou A, Ni J, Zhang Q, Wang Y, Lu J, et al. Differential expression of miRNAs and their relation to active tuberculosis. Tuberculosis. (2015) 95:395–403. doi: 10.1016/j.tube.2015.02.043
99. Othumpangat S, Walton C, Piedimonte G. MicroRNA-221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS ONE. (2012) 7:e30030. doi: 10.1371/journal.pone.0030030
100. Bakre A, Mitchell P, Coleman JK, Jones LP, Saavedra G, Teng M, et al. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J Gen Virol. (2012) 93:2346–56. doi: 10.1099/vir.0.044255-0
101. Thornburg NJ, Hayward SL, Crowe JE Jr. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-kappaB. MBio. (2012) 3:e00220–12. doi: 10.1128/mBio.00220-12
102. Bakre A, Wu W, Hiscox J, Spann K, Teng MN, Tripp RA. Human respiratory syncytial virus non-structural protein NS1 modifies miR-24 expression via transforming growth factor-beta. J Gen Virol. (2015) 96:3179–91. doi: 10.1099/jgv.0.000261
103. Bakre AA, Harcourt JL, Haynes LM, Anderson LJ, Tripp RA. The central conserved region (CCR) of respiratory syncytial virus (RSV) G protein modulates host miRNA expression and alters the cellular response to infection. Vaccines. (2017) 5:E16. doi: 10.3390/vaccines5030016
104. Banos-Lara MDR, Zabaleta J, Garai J, Baddoo M, Guerrero-Plata A. Comparative analysis of miRNA profile in human dendritic cells infected with respiratory syncytial virus and human metapneumovirus. BMC Res Notes. (2018) 11:432. doi: 10.1186/s13104-018-3541-0
105. Eilam-Frenkel B, Naaman H, Brkic G, Veksler-Lublinsky I, Rall G, Shemer-Avni Y, et al. MicroRNA 146-5p, miR-let-7c-5p, miR-221 and miR-345-5p are differentially expressed in Respiratory Syncytial Virus (RSV) persistently infected HEp-2 cells. Virus Res. (2018) 251:34–9. doi: 10.1016/j.virusres.2018.05.006
106. Hasegawa K, Perez-Losada M, Hoptay CE, Epstein S, Mansbach JM, Teach SJ, et al. RSV vs. rhinovirus bronchiolitis: difference in nasal airway microRNA profiles and NFkappaB signaling. Pediatr Res. (2018) 83:606–14. doi: 10.1038/pr.2017.309
107. Inchley CS, Sonerud T, Fjaerli HO, Nakstad B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect Dis. (2015) 15:150. doi: 10.1186/s12879-015-0878-z
108. McCaskill JL, Ressel S, Alber A, Redford J, Power UF, Schwarze J, et al. Broad-spectrum inhibition of respiratory virus infection by MicroRNA mimics targeting p38 MAPK signaling. Mol Ther Nucleic Acids. (2017) 7:256–66. doi: 10.1016/j.omtn.2017.03.008
109. Wang S, Liu P, Yang P, Zheng J, Zhao D. Peripheral blood microRNAs expression is associated with infant respiratory syncytial virus infection. Oncotarget. (2017) 8:96627–35. doi: 10.18632/oncotarget.19364
110. Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. (2000) 74:6442–7. doi: 10.1128/JVI.74.14.6442-6447.2000
111. Tripp RA, Moore D, Jones L, Sullender W, Winter J, Anderson LJ. Respiratory syncytial virus G and/or SH protein alters Th1 cytokines, natural killer cells, and neutrophils responding to pulmonary infection in BALB/c mice. J Virol. (1999) 73:7099–107.
112. Jorquera PA, Choi Y, Oakley KE, Powell TJ, Boyd JG, Palath N, et al. Nanoparticle vaccines encompassing the respiratory syncytial virus (RSV) G protein CX3C chemokine motif induce robust immunity protecting from challenge and disease. PLoS ONE. (2013) 8:e74905. doi: 10.1371/journal.pone.0074905
113. Prince GA, Curtis SJ, Yim KC, Porter DD. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J Gen Virol. (2001) 82:2881–8. doi: 10.1099/0022-1317-82-12-2881
114. Haynes LM, Jones LP, Barskey A, Anderson LJ, Tripp RA. Enhanced disease and pulmonary eosinophilia associated with formalin-inactivated respiratory syncytial virus vaccination are linked to G glycoprotein CX3C-CX3CR1 interaction and expression of substance P. J Virol. (2003) 77:9831–44. doi: 10.1128/JVI.77.18.9831-9844.2003
115. Anderson LJ. Respiratory syncytial virus vaccine development. Semin Immunol. (2013) 25:160–71. doi: 10.1016/j.smim.2013.04.011
116. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
117. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. (2015) 43:W460–6. doi: 10.1093/nar/gkv403
118. Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. (2012) 40:W498–504. doi: 10.1093/nar/gks494
119. Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. (2009) 25:1991–3. doi: 10.1093/bioinformatics/btp299
120. Jorquera PA, Oakley KE, Powell TJ, Palath N, Boyd JG, Tripp RA. Layer-by-layer nanoparticle vaccines carrying the G protein CX3C motif protect against RSV infection and disease. Vaccines. (2015) 3:829–49. doi: 10.3390/vaccines3040829
121. Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. (1997) 94:13961–6. doi: 10.1073/pnas.94.25.13961
122. Crowe JE Jr, Bui PT, Firestone CY, Connors M, Elkins WR, Chanock RM, et al. Live subgroup B respiratory syncytial virus vaccines that are attenuated, genetically stable, and immunogenic in rodents and nonhuman primates. J Infect Dis. (1996) 173:829–39. doi: 10.1093/infdis/173.4.829
123. Gotera J, Giuffrida M, Mavarez A, Pons H, Bermudez J, Maldonado M, et al. Respiratory syncytial virus infection increases regulated on activation normal T cell expressed and secreted and monocyte chemotactic protein 1 levels in serum of patients with asthma and in human monocyte cultures. Ann Allergy Asthma Immunol. (2012) 108:316–20. doi: 10.1016/j.anai.2012.03.006
124. Oshansky CM, Barber JP, Crabtree J, Tripp RA. Respiratory syncytial virus F and G proteins induce interleukin 1alpha, CC, and CXC chemokine responses by normal human bronchoepithelial cells. J Infect Dis. (2010) 201:1201–7. doi: 10.1086/651431
125. Conti P, DiGioacchino M. MCP-1 and RANTES are mediators of acute and chronic inflammation. Allergy Asthma Proc. (2001) 22:133–7. doi: 10.2500/108854101778148737
126. Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. (2016) 14:143. doi: 10.1186/s12967-016-0893-x
127. Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, et al. Circulating microRNAs as potential biomarkers of infectious disease. Front Immunol. (2017) 8:118. doi: 10.3389/fimmu.2017.00118
128. da Silva AM, de Araujo JN, de Freitas RC, Silbiger VN. Circulating MicroRNAs as potential biomarkers of atrial fibrillation. Biomed Res Int. (2017) 2017:7804763. doi: 10.1155/2017/7804763
129. Henshall DC, Hamer HM, Pasterkamp RJ, Goldstein DB, Kjems J, Prehn JHM, et al. MicroRNAs in epilepsy: pathophysiology and clinical utility. Lancet Neurol. (2016) 15:1368–76. doi: 10.1016/S1474-4422(16)30246-0
130. Iljas JD, Guanzon D, Elfeky O, Rice GE, Salomon C. Review: bio-compartmentalization of microRNAs in exosomes during gestational diabetes mellitus. Placenta. (2017) 54:76–82. doi: 10.1016/j.placenta.2016.12.002
131. Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci. (2017) 146:47–94. doi: 10.1016/bs.pmbts.2016.12.013
132. Liu X, Liu X, Wu Y, Wu Q, Wang Q, Yang Z, et al. MicroRNAs in biofluids are novel tools for bladder cancer screening. Oncotarget. (2017) 8:32370–9. doi: 10.18632/oncotarget.16026
133. Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging role of MicroRNAs as liquid biopsy biomarkers in gastrointestinal cancers. Clin Cancer Res. (2017) 23:2391–9. doi: 10.1158/1078-0432.CCR-16-1676
134. Mauri M, Kirchner M, Aharoni R, Ciolli Mattioli C, van den Bruck D, Gutkovitch N, et al. Conservation of miRNA-mediated silencing mechanisms across 600 million years of animal evolution. Nucleic Acids Res. (2017) 45:938–50. doi: 10.1093/nar/gkw792
135. Zhao Y, Song Y, Yao L, Song G, Teng C. Circulating microRNAs: promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. (2017) 36:77–94. doi: 10.1089/dna.2016.3426
136. Fujio K, Komai T, Inoue M, Morita K, Okamura T, Yamamoto K. Revisiting the regulatory roles of the TGF-beta family of cytokines. Autoimmun Rev. (2016) 15:917–22. doi: 10.1016/j.autrev.2016.07.007
137. Gagliardo R, Chanez P, Gjomarkaj M, La Grutta S, Bonanno A, Montalbano AM, et al. The role of transforming growth factor-beta1 in airway inflammation of childhood asthma. Int J Immunopathol Pharmacol. (2013) 26:725–38. doi: 10.1177/039463201302600316
138. Ganeshan K, Johnston LK, Bryce PJ. TGF-beta1 limits the onset of innate lung inflammation by promoting mast cell-derived IL-6. J Immunol. (2013) 190:5731–8. doi: 10.4049/jimmunol.1203362
139. Kang HR, Cho SJ, Lee CG, Homer RJ, Elias JA. Transforming growth factor (TGF)-beta1 stimulates pulmonary fibrosis and inflammation via a Bax-dependent, bid-activated pathway that involves matrix metalloproteinase-12. J Biol Chem. (2007) 282:7723–32. doi: 10.1074/jbc.M610764200
140. Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. (2008) 134:392–404. doi: 10.1016/j.cell.2008.07.025
141. Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. (2006) 25:455–71. doi: 10.1016/j.immuni.2006.07.011
142. McCann KL, Imani F. Transforming growth factor beta enhances respiratory syncytial virus replication and tumor necrosis factor alpha induction in human epithelial cells. J Virol. (2007) 81:2880–6. doi: 10.1128/JVI.02583-06
143. Oh SA, Li MO. TGF-beta: guardian of T cell function. J Immunol. (2013) 191:3973–9. doi: 10.4049/jimmunol.1301843
144. Tu E, Chia PZ, Chen W. TGFbeta in T cell biology and tumor immunity: angel or devil? Cytokine Growth Factor Rev. (2014) 25:423–35. doi: 10.1016/j.cytogfr.2014.07.014
145. Gibbs JD, Ornoff DM, Igo HA, Zeng JY, Imani F. Cell cycle arrest by transforming growth factor beta1 enhances replication of respiratory syncytial virus in lung epithelial cells. J Virol. (2009) 83:12424–31. doi: 10.1128/JVI.00806-09
146. Pokharel SM, Shil NK, Bose S. Autophagy, TGF-beta, and SMAD-2/3 signaling regulates interferon-beta response in respiratory syncytial virus infected macrophages. Front Cell Infect Microbiol. (2016) 6:174. doi: 10.3389/fcimb.2016.00174
147. Thornburg NJ, Shepherd B, Crowe JE Jr. Transforming growth factor beta is a major regulator of human neonatal immune responses following respiratory syncytial virus infection. J Virol. (2010) 84:12895–902. doi: 10.1128/JVI.01273-10
148. Seiler F, Hellberg J, Lepper PM, Kamyschnikow A, Herr C, Bischoff M, et al. FOXO transcription factors regulate innate immune mechanisms in respiratory epithelial cells. J Immunol. (2013) 190:1603–13. doi: 10.4049/jimmunol.1200596
149. Ohol YM, Wang Z, Kemble G, Duke G. Direct inhibition of cellular fatty acid synthase impairs replication of respiratory syncytial virus and other respiratory viruses. PLoS ONE. (2015) 10:e0144648. doi: 10.1371/journal.pone.0144648
150. Kast JI, McFarlane AJ, Globinska A, Sokolowska M, Wawrzyniak P, Sanak M, et al. Respiratory syncytial virus infection influences tight junction integrity. Clin Exp Immunol. (2017) 190:351–9. doi: 10.1111/cei.13042
151. Rezaee F, DeSando SA, Ivanov AI, Chapman TJ, Knowlden SA, Beck LA, et al. Sustained protein kinase D activation mediates respiratory syncytial virus-induced airway barrier disruption. J Virol. (2013) 87:11088–95. doi: 10.1128/JVI.01573-13
152. Rezaee F, Harford TJ, Linfield DT, Altawallbeh G, Midura RJ, Ivanov AI, et al. cAMP-dependent activation of protein kinase A attenuates respiratory syncytial virus-induced human airway epithelial barrier disruption. PLoS ONE. (2017) 12:e0181876. doi: 10.1371/journal.pone.0181876
153. Wong W. Hippo-sized antiviral defenses. Sci Signal. (2017) 10:eaan4043. doi: 10.1126/scisignal.aan4043
154. Garofalo R, Mei F, Espejo R, Ye G, Haeberle H, Baron S, et al. Respiratory syncytial virus infection of human respiratory epithelial cells up-regulates class I MHC expression through the induction of IFN-beta and IL-1 alpha. J Immunol. (1996) 157:2506–13.
155. Konig B, Streckert HJ, Krusat T, Konig W. Respiratory syncytial virus G-protein modulates cytokine release from human peripheral blood mononuclear cells. J Leukoc Biol. (1996) 59:403–6. doi: 10.1002/jlb.59.3.403
156. Krishnan S, Craven M, Welliver RC, Ahmad N, Halonen M. Differences in participation of innate and adaptive immunity to respiratory syncytial virus in adults and neonates. J Infect Dis. (2003) 188:433–9. doi: 10.1086/376530
157. Cheng K, Rai P, Plagov A, Lan X, Mathieson PW, Saleem MA, et al. Rapamycin-induced modulation of miRNA expression is associated with amelioration of HIV-associated nephropathy (HIVAN). Exp Cell Res. (2013) 319:2073–80. doi: 10.1016/j.yexcr.2013.04.011
158. de Souza AP, de Freitas DN, Antuntes Fernandes KE, D'Avila da Cunha M, Antunes Fernandes JL, Benetti Gassen R, et al. Respiratory syncytial virus induces phosphorylation of mTOR at ser2448 in CD8 T cells from nasal washes of infected infants. Clin Exp Immunol. (2016) 183:248–57. doi: 10.1111/cei.12720
159. Kumar M, Sharma A, Ahmad T, Mabalirajan U, Aich J, Agrawal A, et al. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. J Appl Physiol. (2012) 113:459–64. doi: 10.1152/japplphysiol.00001.2012
160. Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol. (2013) 132:3–13; quiz 14. doi: 10.1016/j.jaci.2013.04.039
161. Herbert C, Collison A, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm Med. (2011) 11:29. doi: 10.1186/1471-2466-11-29
162. Knudson CJ, Hartwig SM, Meyerholz DK, Varga SM. RSV vaccine-enhanced disease is orchestrated by the combined actions of distinct CD4 T cell subsets. PLoS Pathog. (2015) 11:e1004757. doi: 10.1371/journal.ppat.1004757
Keywords: RSV, miR, vaccines, immune, disease, microRNA
Citation: Atherton LJ, Jorquera PA, Bakre AA and Tripp RA (2019) Determining Immune and miRNA Biomarkers Related to Respiratory Syncytial Virus (RSV) Vaccine Types. Front. Immunol. 10:2323. doi: 10.3389/fimmu.2019.02323
Received: 16 December 2018; Accepted: 13 September 2019;
Published: 09 October 2019.
Edited by:
Alexis M. Kalergis, Pontifical Catholic University of Chile, ChileReviewed by:
Marina Boukhvalova, Sigmovir Biosystems, United StatesMarcelo Lopez-Lastra, Pontifical Catholic University of Chile, Chile
Copyright © 2019 Atherton, Jorquera, Bakre and Tripp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralph A. Tripp, cmF0cmlwcEB1Z2EuZWR1
 Lydia J. Atherton
Lydia J. Atherton Patricia A. Jorquera
Patricia A. Jorquera Abhijeet A. Bakre
Abhijeet A. Bakre Ralph A. Tripp
Ralph A. Tripp