- Hunan Key Laboratory of Medical Epigenomics, Department of Dermatology, Second Xiangya Hospital, Central South University, Changsha, China
Autoimmune diseases can be chronic with relapse of inflammatory symptoms, but it can be also acute and life-threatening if immune cells destroy life-supporting organs, such as lupus nephritis. The etiopathogenesis of autoimmune diseases has been revealed as that genetics and environmental factors-mediated dysregulated immune responses contribute to the initiation and development of autoimmune disorders. However, the current understanding of pathogenesis is limited and the underlying mechanism has not been well defined, which lows the development of novel biomarkers and new therapeutic strategies for autoimmune diseases. To improve this, broadening and deepening our understanding of pathogenesis is an unmet need. As genetic susceptibility cannot explain the low accordance rate of incidence in homozygous twins, epigenetic regulations might be an additional explanation. Therefore, this review will summarize current progress of studies on epigenetic dysregulations contributing to autoimmune diseases, including SLE, rheumatoid arthritis (RA), psoriasis, type 1 diabetes (T1D), and systemic sclerosis (SSc), hopefully providing opinions on orientation of future research, as well as discussing the clinical utilization of potential biomarkers and therapeutic strategies for these diseases.
Introduction
Autoimmunity is a pathological condition that self-immune system cannot distinguish self-antigens and attacks self-tissues and organs, resulting in inflammation and organ damages. Autoimmune diseases, such as SLE, RA, and T1D are sometimes referred to as “invisible disabilities” or life-threatening diseases, with high incidence rate of 11%. Approximately 600 million people suffer from a breakdown of immune tolerance. Aberrant differentiation and function of immune cells are believed to be a key player in the pathogenesis of autoimmune diseases. However, the molecular mechanism remains unknown.
Genetic susceptibility can partially explain some of the abnormalities of immune imbalance. For instance, over 60 genes have been revealed in previous genetic studies as risk genes in lupus, and some of them have been found to be related to antibody production, complementary deficiency and renal involvements (1). However, genetic studies cannot completely explain the incidence rate of SLE in homozygous twins ranges from 24 to 58% (2), indicating that in addition to genetics, environment factors are also involved in the pathogenesis of SLE. As one of molecular mechanisms of environmental factors, epigenetics has been proposed as a critical player in the diseases by accumulating evidence, and it might provide additional explanation for the dysregulation of immune system. Therefore, this review focuses on the current understanding of epigenetic-mediated regulations on immune cell differentiation and functions, summarizes the contribution of dysregulated epigenetic modifications to autoimmune disorders, and discusses the possibility of utilization of unique and specific epigenetic modifications as potential biomarkers and novel therapeutic targets for these diseases.
Epigenetics in Pathogenesis of Autoimmune Diseases
All cells and tissues in our body share the same set of genomic DNA, however, cells display various morphology and phenotypes due to the gene transcription mediated by epigenetics. Epigenetics is a biological process that recruits or removes reversible and potentially heritable modifications in genomic DNA and/or chromatin but does not change DNA sequence. It is mainly comprised of DNA methylation, histone modifications, and non-coding RNAs-mediated regulations. Epigenetic regulations participate in numerous biological process, such as cell proliferation and differentiation. and increasing evidence has shown that dysregulated epigenetic modifications are involved in pathogenesis of several autoimmune diseases (3–6). The influence of environmental factors, such as UVB, and disease predominance in female emphasizing the importance of epigenetics in the pathogenesis of autoimmune disorders (7). In addition, 5-azacytidine and procainamide (8) are capable of inducing lupus via epigenetic alterations. Similar phenomena have been found in other autoimmune diseases: dysregulation of epigenetic modifications in RA synovial fibroblasts (RASF) leading to abnormal gene expression (9), Epstein-Barr virus (EBV) infection, sunlight (10, 11) and aberrantly expressed miRNAs (12, 13) contributing to the pathogenesis of multiple sclerosis (MS).
As the intensively studied epigenetic modification, DNA methylation refers to a well-known biological process which involves a recruitment of a methyl group to a cytosine or adenine residue at the 5th position on the pyrimidine ring, resulting in inhibiting the binding of transcription factors on the promoter region of gene, which will repress the gene transcription (14). This process is mainly regulated by methyltransferase, including DNA methyltransferase 1 (DNMT1), DNMT3a, and DNMT3b. Each of methyltransferase executes different functions. For instance, during cell replication DNMT1 maintains the methylation levels, whereas DNMT3a and DNMT3b promote methylation process (15). On the contrary, DNA hydroxymethylation, and demethylation are processes that re-activate transcription of silenced genes (16). DNA hydroxymethylation is an instable status and in the middle of demethylation process. DNA hydroxymethylation is mediated by hydroxymethylation transferases, such as ten-eleven translocation methylcytosine dioxygenase 1 (TET1), TET2, and TET3 (17).
Histone modification is a covalent post-translational regulation that modulates gene transcription by altering the structure of chromatin. Histone modifications include methylation, acetylation, ubiquitination, phosphorylation, sumoylation, etc. (18). Acetylation and deacetylation are intensively studied ones which can recruit or remove an acetyl group on histones, thereby activating, or inhibiting gene transcription. Mechanically, acetylation activates gene transcription by opening the chromatin structure and facilitating the binding of transcription factors, while methylation converts opened chromatins into a restrictive structure, inhibiting the binding of transcription factors via stereo hindrance, thereby resulting in the repression of gene expression. Acetylation is mediated by histone acetyltransferases (HATs), deacetylation is regulated by histone deacetylases (HDACs) (19). However, the effects of histone modifications vary depending on the modification positions and the number of modifications. For example, H3K4me3 promotes gene expression whereas H3K9me3 and H3K27me3 represses gene transcription (20, 21).
microRNAs (miRNAs) are small non-coding RNAs, which are usually 21-25 base pairs. It has been well established that miRNAs modulate gene expression at posttranscriptional and posttranslational level through binding to the 3′-UTRs of target mRNAs, resulting in blocking gene translation by mRNA cleavage and degradation (22–24). Besides, long ncRNAs are recently identified non-coding RNAs, with the length of >200 nt. Differing from miRNAs, lncRNAs can either promote or inhibit gene expression. LncRNAs usually act by complexes of lncRNA: RNA, lncRNA: protein or lncRNA: chromatin (25, 26). Accumulating evidence suggests that lncRNAs are involved in numerous human diseases, such as cancer, by changing the primary and secondary structure of DNA, thereby regulating gene expression (27, 28).
Dysregulated Epigenetic Regulations in Autoimmune Diseases
Abnormal DNA Methylation in Autoimmune Diseases
DNA Hypomethylation in SLE
SLE is a multi-organ involved autoimmune disease that is characterized by aberrant immune cells, such as dendritic cells, B and T lymphocytes. Although the pathogenesis of SLE has been studied for over a century, the exact cause of lupus remains unknown. Increasing evidence from varies groups, including our group, have reported that DNA methylation plays a critical role in immune cells hyper-active in lupus conditions.
DNA hypomethylation in lupus T cells
The first evidence of epigenetic regulation in lupus is from the observation that after a long-term administration of two DNA methylation inhibitors, procainamide and hydralazine, normal mice showed a lupus-like phenotype. In addition, cells from thymus and lymph nodules from MRL/lpr mice (spontaneous lupus mouse model) show lower DNA methylation level compared with cells from MRL/mpj control mice (29, 30). This evidence might provide an explanation for over-proliferated and over-activated immune cells in lupus mice.
Lupus T cell auto-reactivity is found to attribute to DNA hypomethylation (31). These findings were further confirmed by the evidence that induction of auto-reactive CD4+ T cells from healthy controls by the administration of 5-azacytidine (31, 32), which followed prior evidence of the induction of IL-2 and IFN-γ by the same drug (33). Accumulating evidence have revealed the regulatory effects of DNA methylation on individual genes during the T cell activation and differentiation. IFN-γ and IL-4 are signature cytokines for Th1 and Th2 program, respectively. During Th1 and Th2 differentiation processes, DNA hypomethylation level has been observed at Ifng and Il4 loci (34, 35). In addition, compared to naïve T cells, decreased DNA methylation level is found at the key transcription factor FOXP3 locus in regulatory T cells (Treg) (36). Furthermore, the key transcription factor Bcl6 in Tfh cell has been reported to be highly expressed but with a decreased level of 5hmC (37) during Tfh cell differentiation, suggesting that Tfh cell differentiation is also mediated by DNA methylation modification.
In addition, genomic DNA in lupus CD4+ T cells has been found to show DNA hypomethylation (38, 39). DNA hypomethylation has been observed on promoter region of lfa-1 in CD4+ T cells from active lupus patients and over-expressed LFA-1 has been found on an autoreactive subset of T cells, which produces perforin and granzyme B to lyse autologous cells (31, 40), thereby inducing inflammation and tissue damages. Epigenetic accessibility and transcriptional poising of interferon-regulated genes in Naïve CD4+ T cells from SLE patients have been shown in a genome-wide DNA methylation study (41). In this study, DNA hypomethylation is observed on interferon-regulated genes, such as IFI44L, which suggest that lupus T cell progenitors have abnormalities (41). More interesting is that our recent studies have proposed DNA hypomethylation level on IFI44L promoter as a biomarker for the diagnosis of lupus, which have both high sensitivity and specificity (42). In a consequent study, different DNA methylation patterns have been observed in organ-specific manner in lupus. For instance, different DNA methylation patterns have been on lupus patients with renal involvement vs. non-renal involvements, and malar rash vs. discoid rash (43). Interesting, some protein such as RFX1 (44), high mobility group box protein 1(HMGB1) (45) and DNA Damage-Inducible 45 alpha (Gadd45a) (46) have been revealed as regulators for this epigenetic regulation by our previous studies.
Besides, in lupus CD4+ T cells, 5-hmC binds in transcriptional regulatory regions of lineage-specific signature genes, such as IL-17 and IFN-gamma, which promote inflammation. Mechanically, TET2 protein, a hydroxymethylation transferase, is found to be recruited to 5-hmC-binding regions of Il17 and Ifnr, and then promotes the production of IL-17 and IFN-gamma (47). We have recently observed that lupus CD4+ T cells display an increased 5-hmC level on whole genomic DNA compared with normal controls, with the enhanced expression of TET2 and TET3. As a consequence of DNA demethylation, transcription activator CTCF binds to the promoter region of SOCS1 and therefore promotes SOCS1 over-expression in SLE CD4+ T cells (48).
DNA hypomethylation in lupus B cells
SLE is an autoantibody-mediated autoimmune disorder. As the main and unique origin of autoantibodies, numerous evidence has well document that B cell plays an essential role in the pathogenesis of SLE. Pre-clinical studies and clinical trials of B cell-targeting treatments have proven to be effective to some extent. Not to our surprise, DNA hypomethylation has been also shown in lupus B cells (49), which might regulate B cell development, differentiation, and auto-reactivity. For example, abnormally expressed HRES1/p28 by lupus B cells is reported to be regulated via DNA methylation (50). DNA hypomethylation on LINE1 gene has been shown in lupus B cells (51). The regulatory effect of DNA methylation in B cells is further supported by the evidence that enhanced levels of anti-nuclear antibodies can be induced by adoptive transferring of DNMT1 inhibitor-treated B cells (52). Although it is elucidated that antibody production is attributed to DNA hypomethylation in V(D)J region and Igh 3′-LCR (53), little has been revealed in this process in the lupus condition. Furthermore, in auto-reactive B cells, DNA hypomethylation might be a result of decreased level of DNMT1 and DNMT3b, or active DNA demethylation mediated by activation-induced cytidine deaminase (AID) (54).
Aberrant DNA Methylation in Psoriasis
Psoriasis is a chronic inflammatory autoimmune skin disease, which is characterized by hyper proliferation of keratinocytes and dysregulated T cells, especially Th17 cells (55). Similar with SLE, genetic susceptibility is not the only factor for the onset of this disease, due to that the concordance of psoriasis in monozygotic twins is 35–72% (56), suggesting that epigenetic regulations might be an additional factor. Increased evidence has shown the critical role of DNA methylation in the hyper-proliferated keratinocytes.
In our previous study, abnormal DNA methylation pattern has been observed in skin lesions and PBMCs of patients with psoriasis vulgaris (57, 58). On the gene specific level, the abnormal methylation pattern on the promoter of p16INK4a gene has been reported in psoriatic epidermis (59). Increased DNA methylation level on promotor of secreted frizzled-related protein (Sfrp4) has been observed in inflamed psoriatic skin and in the IL-23-induced psoriatic mice, thereby reducing the expression of Sfrp4, a negative regulator for keratinocyte proliferation (60). Hypomethylation of LINE-1 has been found in psoriatic keratinocytes. More importantly, manipulating LINE-1 methylation may change the gene expression, thereby resulting in a phenotypic alteration of psoriatic skin (61). In addition, aberrant DNA methylation pattern has also been revealed in CD4+ T cells from psoriatic patients (62), indicating that the epigenetic regulations on immune cells also attributing to psoriasis pathogenesis.
Aberrant DNA Methylation Status in RA
RA is an autoreactive immune cell-mediated inflammation which primarily affects joints. Autoreactive immune cells and synovial fibroblasts (SF) are well defined as the critical players in the pathogenesis of RA. Heterogeneity in RA patients is a hindrance for rheumatologists and dermatologists to diagnose and treat patients. The treatment of RA is always delayed due to the current criteria that in addition to meeting all diagnostic criteria, RA patients need to consistently display arthritic symptoms for at least 6 months (63). Early intervention is necessary because a clinical trial on BeSt have shown that BeSt can delay the onset of RA on several patients (64).
Increasing evidence has shown that DNA methylation contributes to the pathogenesis of RA. Increased DNA methylation variability has been observed in rheumatoid arthritis-discordant monozygotic twins (65), indicating the importance of DNA methylation in the pathogenesis of RA. Abnormal genome-wide DNA methylation patterns have been revealed in CD4+ T cells from Chinese Han patients with rheumatoid arthritis (66). In PBMCs from RA patients, decreased DNA methylation levels have been found at the promoter regions of Il6 and ERa, which may be associated with over-production of IL-6 and hyperactive ERa signaling (67–69). Global DNA hypomethylation is also found in T cells from RA patients (31, 70). On the gene specific level, CD40L gene has found to be demethylated on CD4+ T cells from RA patients (69). Moreover, DNA hypomethylation on promoter region of L1 retrotransposon gene has been observed in RA fibroblast-like synoviocytes (71, 72). Further, DNA hypomethylation on CXCL12 gene has been shown in synovial fibroblasts, that may result in cell infiltration in joints (73, 74). More interesting, DNA methylation status has been proposed as biomarkers to predict the drug responses (75).
Dysregulated DNA Methylation in Systemic Sclerosis (SSc)
SSc is a relatively rare disease which is characterized by damages of connective tissues mediated by autoreactive immune cells. Its etiopathogenesis remains unclear. Abnormal epigenetic modifications have been shown in SSc. In an integration study of Genome-Wide DNA Methylation and Transcription, several DNA methylation regulated-gene expression have been revealed in SSc PBMCs (76) and dermal fibroblasts (77). Decreased DNA methylation level has been observed in CD4+ T cells from SSc patients and reduced expression of DNMTs have been found in CD4+ T cells from these patients (78). DNA demethylation on promoter regions of CD11a, CD70, and CD40L genes have been found in CD4+ T cells from SSc patients (78–81). However, hypermethylated genes, such as PRF1, CDKN2A, Foxp3, CD11a, and CD70, have been observed in whole blood from black South African patients with SSc (82). Moreover, as the key transcription factor to Th17 cells, RORC1 and RORC2 have been found to show hypomethylation and be correlated with inflammatory status in SSc PBMCs (83). Furthermore, in dermal fibroblasts from SSc patients, hypermethylation has been found in FLl1 and TGF-beta-related genes, which are Wnt pathway antagonist genes (84–86), accompanied by increased levels of DNMT1 (87) and TET1 (88).
Abnormal DNA Methylation Levels in T1D
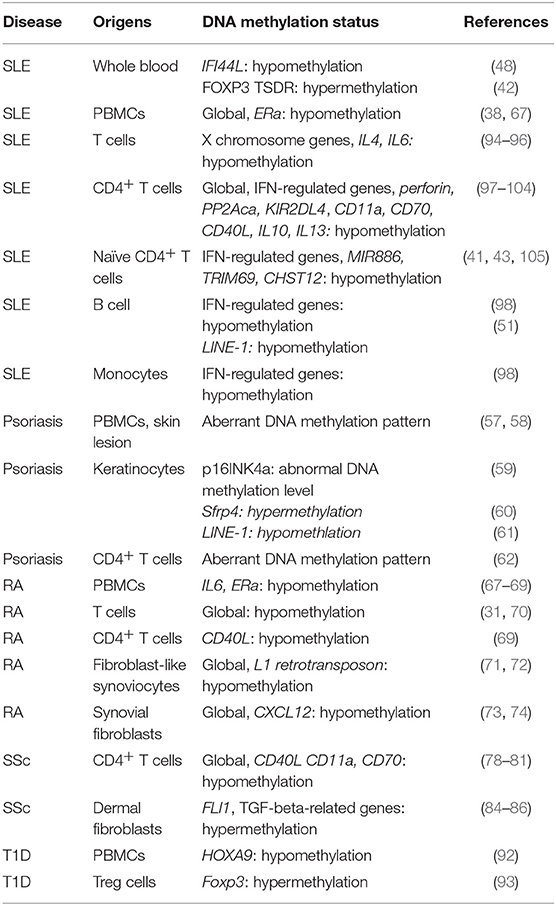
T1D is well-documented as an autoimmune disease, which is mainly mediated by T cells by attacking beta cells. In an epigenome-wide association study (GWAS) in 52 monozygotic twins, epigenetic modification patterns have been mapped in CD4+ T cells, CD19+ B cells, and CD14+ monocytes (89). This study has identified a substantial enrichment of differentially variable CpG positions (89), suggesting the involvement of DNA methylation in T1D. In addition, differential DNA methylation status on 88 CpG sites has been found in lymphoblast cell lines which are derived from 6 pairs of monozygotic twins concordant for T1D and 3 pairs of monozygotic twins discordant for T1D, separately. In these cells lines, the altered expression of genes, including Hla, Ins and Il2rb, are involved in immune responses (90). Furthermore, dysregulated DNA methylation have been found in Pdchb16, Magi2, and Fancc in T1D-discordant monozygotic twins (91). DNA demethylation on transcription factor HOXA9 has been observed in T1D patients (92). DNA hypermethylation has been found in the promoter region of Foxp3, which represses the binding of transcription factor IRF-7 to Foxp3, resulting in the reduced number of regulatory T cells in the peripheral blood from T1D patients (93). More interesting, the serum levels of unmethylated preproinsulin DNA might serve as a biomarker for T1D (91). Dysregulated DNA methylation are listed in Table 1.
Aberrant Histone Modifications in Autoimmune Diseases
Dysregulated Histone Modifications in Lupus
Lupus CD4+ T cells show global histone H3 and H4 hypoacetylation (106). Abnormal histone modifications have been found in the promoter region of TNFSF7 in T cells, resulting in overexpression of CD70, which might be the one cause of auto-reactivity of T cells (107). Administrating HDAC inhibitors on healthy T cells results in decreased CD3ς chain expression, thereby leading abnormalities in T cells (108). A transcription factor CREMα might be involved in the process of histone acetylation in active lupus T cells via inhibition of IL-2 production. This process might be mediated by recruiting HDAC to Cre binding sites in the promoter region of Il2 (109). Besides, abnormal H3K4me3 modification has been observed on lupus-related candidate genes in lupus PBMCs (110). Lupus monocytes show altered acetylation status of global H4. Among them, 63% of these H4 acetylated genes are potentially modulated by IFN regulatory factors (111), which are involved in the pathogenesis of SLE.
In addition to the whole genomic modifications, histone modification has been reported to modify specific gene expression. For example, increased H3 acetylation level has been found at the IL-17 locus and enhanced IL-10 production has been revealed to be mediated by chromatin remodeling. This process is further revealed to be mediated by Stat3 (112, 113). Moreover, histone hyperacetylation has been shown to be a cause for an increased serum level of TNF-α and an enhanced maturation status of monocytes from lupus patients (114). However, it is still unclear whether histone modifications are the initiator or results of immune disorders, even though the contribution of histone modifications in pathogenesis of lupus has been revealed in mouse studies.
Sirtuin-1 (Sirt-1) is a histone deacetylase, which has been observed to be overexpressed by T cells from MRL/lpr mice (115). Knocking down Sirt-1 in lupus mice leads to a temporary enhancement of H3 and H4 acetylation, accompanied by attenuated lupus symptoms such as reduced serum levels of anti-dsDNA, IgG deposition in glomerular and histological changes (38). Treating MRL/lpr mice with HDAC inhibitors can attenuate renal damage and decrease level of inflammatory cytokines (116). A recent progress has been made from a genetic and epigenetic mapping study which identifies candidate causal variants in 21 autoimmune diseases in different T cell subtypes, including Th1, Th2, Treg, and Th17 cells (117). In this study, unique H3K27 peaks are shown in the super-enhancer in Il2RA locus, particularly in Treg and Th17 cells.
In our previous study, we have demonstrated that RFX1 inhibits Th17 cell differentiation via increased histone H3 acetylation, decreased DNA methylation and H3K9 tri-methylation (118), thereby contributing to SLE pathogenesis. More recently, the downregulation of TNF-alpha-induced protein 3 (TNFAIP3), one of the major SLE susceptibility genes involving in the regulation of inflammatory responses through modulation of the nuclear factor-kappaB (NF-kappaB) pathway, has been observed in lupus patients. This downregulation may be mediated by reduced H3K4me3 in the gene promotor region (119), providing a promising target for the treatment of SLE in clinical practice.
In addition, some epigenetic targeting therapies also revealed the importance of histone modification in pathogenesis of autoimmune diseases. For example, selective HDAC6 inhibition has been shown to attenuate early stage of lupus nephritis via down-regulation both innate and adaptive immune responses (120). Selective HDAC6 inhibitor also showed therapeutic effects on lupus mice by improving renal function and survival (121). A novel histone deacetylase 3-selective inhibitor has been reported to inhibit IL-6 production by PBMCs from RA patients (122). As well as in T1D, histone deacetylase inhibitors have been shown to modify pancreatic cell fate determination and amplify endocrine progenitors (123).
Aberrant Non-coding RNA Mediating Regulations in Autoimmune Diseases
Aberrant Non-coding RNA Mediating Regulations in Lupus
Dysregulated non-coding RNAs in lupus T cells
It has been well-documented that miRNAs can bind to various regions but modulate the same gene expression. A large number of miRNAs have been reported to be aberrantly expressed by T cells. Some of these miRNAs have been found to target lupus-related genes, such as Il10, Il17, and dnmt1. It has been reported that the expression level of miR-21, miR-126 and miR148a is observed to be reduced in lupus T cells and they are found to target DNMT1, although they bind to different regions of DNMT1(124, 125). Furthermore, the inhibition of miR-21, miR-29b, and miR-148a in SLE T cells has been found to be capable of attenuating lupus phenotypes, suggesting potential therapeutic roles in SLE (125, 126). In addition, miR-21 has been found to inhibit the expression of PDCD4 on lupus T cells, thereby promoting T cell proliferation and the expression of CD40L and IL-10 (127). Moreover, miR-142 (128) and miR-31 (129) have been demonstrate to modulate T cell activity by suppressing IL-4 and IL-10 production by T cells, inhibiting the expression of CD40L and ICOS and enhancing secretion of IL-2 by T cells. In addition to our previous studies on aberrantly expressed miR-146a and−241-3p/5p by lupus T cells, we have further found that mycophenolic acid, which has been commonly utilized in clinic for lupus treatment, attenuates the auto-reactivity of lupus T cells through miR-146a and−241-3p/5p, suggesting the pathogenic role of these two miRNAs in SLE (130).
More recently, in short time-series expression miner analysis, some lncRNAs from lupus T cells have been found to be correlated with SLE disease activity (131), suggesting that the aberrant expression profile of lncRNAs may play a role in SLE pathogenesis. In addition, large intergenic non-coding RNAs (lincRNAs), a specific type of lncRNAs, can also modulate gene expression and is involved in various biological processes and diseases. For example, lupus PBMCs show lower level of linc0597and Linc0949, compared to those from rheumatoid arthritis patients and normal subjects (132). More importantly, the decreased level of linc0949 is correlated with the level of C3, SLE disease activity index (SLEDAI), and the appearance of lupus-specific organ damages. More interesting is that the levels of linc0949 can increase significantly depending on efficiency of treatment in lupus patients, suggesting a role as a biomarker for SLEDAI and drug response (132, 133).
Aberrantly expressed microRNAs in lupus B cells
As critical regulators in B cell development and differentiation, miRNAs are also involved in the aberrant B cell expression and functions. Lupus B cells shows increased levels of miR-30a. The level of miR-30a in lupus B cells negatively correlates with Lyn, which negatively regulates B cell activation (134). It has been found that miR-155 and miR-181b negatively regulate AID expression, thereby modulating antibody diversity (135, 136). In lupus-prone mice, the levels of miR-15a in regulatory B cells are positively correlate with the serum level of anti-dsDNA antibodies (137). In our recent studies, increased expression of miR-1246 has been observed in lupus B cells, and it has been found to regulate EBF1 expression, thereby promoting the expression of CD40 and antibody production (138). Moreover, enhanced levels of miR-17-92 and miR-21 have been found in SLE B cells (139, 140). More interesting, miRNA profiling of B cell subsets has been proposed as a biomarker for lupus (141), indicating a critical role of miRNAs in lupus abnormal B cells. Moreover, miR-150 is found to be decreased in B cells from MRL-lpr mice, which might be a result of a decreased acetylation level and inhibition expression of the miR-150 host gene (142). miRNAs that are dysregulated in other autoimmune diseases are summarized in Table 2.
Aberrant Micro-RNA Mediating Regulations in Psoriasis
Similar, some abnormally miRNAs have been reported in psoriatic patients. In our previous studies, mir-210 is found to be overexpressed by T cells filtrating in the dermis of psoriatic lesions. Further, mir-210 is capable of inducing helper T (Th) 17 and Th1 cell differentiation but inhibiting Th2 differentiation by repressing expression of STAT6 and LYN (145). In addition, the upstream regulation has been revealed as that TGF-beta and IL-23 enhance miR-210 expression by inducing HIF-1alpha, which recruits P300 and promotes histone H3 acetylation in the miR-210 promoter region (145). As Th17 cells playing a critical role in pathogenesis of psoriasis, targeting mir-210 might provide potential therapeutic strategies for psoriasis patients. Besides, mir-17-92 cluster has been revealed to promotes the proliferation and the chemokine production of keratinocytes (146), mir-let-7b has been shown to inhibit keratinocyte differentiation by targeting IL-6 mediated ERK signaling in psoriasis (147), mir-194 has been demonstrated to regulate keratinocyte proliferation and differentiation via Grainyhead-like 2 in psoriasis (148).
Conclusion
As the epigenetic era approaches, more and more evidence has shown the importance of epigenetic regulations in the pathogenesis of autoimmune diseases. Newly discovered non-coding RNAs, such as LncRNA, extra RNAs and circle RNAs have begun to undergo significant research into their roles in disease pathogenesis. The specific epigenetic regulations in autoimmune diseases might provide potential biomarkers for diseases. For example, in our previous study, the DNA methylation level of the IFI44L promoter is both sensitive and specific in lupus patients and lower in nephritis patients than in patients without renal damage (164), indicating an organ-specific biomarker to predict LN. Another urgent need is to be able to translate research findings into clinical application. The most significant challenges include complex techniques, time consuming, and the high cost of DNA methylation arrays and bisulfite next-generation sequencing. To solve this problem, as in our study of IFI44L, rather than pyrosequencing of IFI44L DNA methylation levels, we have developed a high-resolution melting (HRM) analysis for detecting IFI44L DNA methylation levels, which can be easily completed with QPCR. This new technique may be more available for clinical use in the future. With regard to treatment, as our new finding on miR-210 in mouse psoriasis treatment (145), miRNAs might provide alternative options to currently used drugs. The application of Crisper-Cas9 may shed light by guiding epigenetic modifications on specific genes. Together, epigenetic modifications provide additional tools for broadening the understanding of autoimmune diseases, as well as development of potential biomarkers and therapies.
Author Contributions
HW and YC wrote the manuscript. HZ edited the manuscript. MZ and QL revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81602767, No. 81430074, No. 81830097), National Basic Research Program of China (No. 2014CB541904), the Natural Science Foundation of Hunan Province (2017JJ3453, 2017SK2042, 2018JJ3756) the National Key research and Development Program of China (2016YFC0903900), and the Natural Key Clinical Specialty Construction Project of National Health and Family Planning Commission of the People's Republic of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Zeng J, Wu H, Zhao M, Lu Q. Novel biomarkers for systemic lupus erythematosus. Biomark Med. (2017) 11:677–86. doi: 10.2217/bmm-2016-0379
2. Hedrich CM, Mabert K, Rauen T, Tsokos GC. DNA methylation in systemic lupus erythematosus. Epigenomics. (2017) 9:505–25. doi: 10.2217/epi-2016-0096
3. Jeffries MA, Sawalha AH. Epigenetics in systemic lupus erythematosus: leading the way for specific therapeutic agents. Int J Clin Rheumtol. (2011) 6:423–39. doi: 10.2217/ijr.11.32
4. Ballestar E. Epigenetics lessons from twins: prospects for autoimmune disease. Clin Rev Allergy Immunol. (2010) 39:30–41. doi: 10.1007/s12016-009-8168-4
5. Brown CC, Wedderburn LR. Genetics: mapping autoimmune disease epigenetics: what's on the horizon? Nat Rev Rheumatol. (2015) 11:131–2. doi: 10.1038/nrrheum.2014.210
6. Jeffries MA, Sawalha AH. Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev Clin Immunol. (2015) 11:45–58. doi: 10.1586/1744666X.2015.994507
7. Rhodes B, Vyse TJ. The genetics of SLE: an update in the light of genome-wide association studies. Rheumatology. (2008) 47:1603–11. doi: 10.1093/rheumatology/ken247
8. Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. (1993) 92:38–53. doi: 10.1172/JCI116576
9. Sanchez-Pernaute O, Ospelt C, Neidhart M, Gay S. Epigenetic clues to rheumatoid arthritis. J Autoimmun. (2008) 30:12–20. doi: 10.1016/j.jaut.2007.11.006
10. Kragt J, van Amerongen B, Killestein J, Dijkstra C, Uitdehaag B, Polman C, et al. Higher levels of 25-hydroxyvitamin D are associated with a lower incidence of multiple sclerosis only in women. Mult Scler. (2009) 15:9–15. doi: 10.1177/1352458508095920
11. Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. (2008) 9:516–26. doi: 10.1038/nrg2395
12. Koch MW, Metz LM, Kovalchuk O. Epigenetics and miRNAs in the diagnosis and treatment of multiple sclerosis. Trends Mol Med. (2013) 19:23–30. doi: 10.1016/j.molmed.2012.10.008
13. Kucukali CI, Kurtuncu M, Coban A, Cebi M, Tuzun E. Epigenetics of multiple sclerosis: an updated review. Neuromolecular Med. (2014) 17:83–96. doi: 10.1007/s12017-014-8298-6
14. Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. (2007) 128:669–81. doi: 10.1016/j.cell.2007.01.033
15. Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. (2011) 12:647–56. doi: 10.1038/embor.2011.110
16. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. (2013) 502:472–9. doi: 10.1038/nature12750
17. Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. (2009) 114:144–7. doi: 10.1182/blood-2009-03-210039
18. Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. (2014) 1839:627–43. doi: 10.1016/j.bbagrm.2014.03.001
19. Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. (2011) 2011:371832. doi: 10.1155/2011/371832
20. Renaudineau Y, Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J Med. (2011) 60:10–6. doi: 10.2302/kjm.60.10
21. Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. (2012) 48:491–507. doi: 10.1016/j.molcel.2012.11.006
22. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. (2004) 303:83–6. doi: 10.1126/science.1091903
23. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. (2010) 79:351–79. doi: 10.1146/annurev-biochem-060308-103103
24. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. (2009) 11:228–34. doi: 10.1038/ncb0309-228
25. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. (2013) 493:231–5. doi: 10.1038/nature11661
26. Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. (2013) 20:440–6. doi: 10.1038/nsmb.2516
27. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. (2011) 21:354–61. doi: 10.1016/j.tcb.2011.04.001
28. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. (2014) 111:1002–7. doi: 10.1073/pnas.1313768111
29. Mizugaki M, Yamaguchi T, Ishiwata S, Shindo H, Hishinuma T, Nozaki S, et al. Alteration of DNA methylation levels in MRL lupus mice. Clin Exp Immunol. (1997) 110:265–9. doi: 10.1111/j.1365-2249.1997.tb08326.x
30. Zhou Y, Lu Q. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun Rev. (2008) 7:376–83. doi: 10.1016/j.autrev.2008.03.003
31. Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. (1990) 33:1665–73. doi: 10.1002/art.1780331109
32. Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. (1988) 140:2197–200.
33. Ballas ZK. The use of 5-azacytidine to establish constitutive interleukin 2-producing clones of the EL4 thymoma. J Immunol. (1984) 133:7–9.
34. Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. (1998) 9:765–75. doi: 10.1016/S1074-7613(00)80642-1
35. Bix M, Locksley RM. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. (1998) 281:1352–4. doi: 10.1126/science.281.5381.1352
36. Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. (2009) 182:259–73. doi: 10.4049/jimmunol.182.1.259
37. Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S, et al. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. (2016) 14:1735–47. doi: 10.1016/j.celrep.2016.01.038
38. Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. (2010) 20:170–9. doi: 10.1101/gr.100289.109
39. Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng W, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J Autoimmun. (2014) 54:127–36. doi: 10.1016/j.jaut.2014.07.002
40. Takeuchi T, Amano K, Sekine H, Koide J, Abe T. Upregulated expression and function of integrin adhesive receptors in systemic lupus erythematosus patients with vasculitis. J Clin Invest. (1993) 92:3008–16. doi: 10.1172/JCI116924
41. Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. (2013) 43:78–84. doi: 10.1016/j.jaut.2013.04.003
42. Zhao M, Zhou Y, Zhu B, Wan M, Jiang T, Tan Q, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis. (2016) 75:1998–2006. doi: 10.1136/annrheumdis-2015-208410
43. Renauer P, Coit P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, et al. DNA methylation patterns in naive CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus Sci Med. (2015) 2:e000101. doi: 10.1136/lupus-2015-000101
44. Zhao M, Sun Y, Gao F, Wu X, Tang J, Yin H, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J Autoimmun. (2010) 35:58–69. doi: 10.1016/j.jaut.2010.02.002
45. Li Y, Huang C, Zhao M, Liang G, Xiao R, Yung S, et al. A possible role of HMGB1 in DNA demethylation in CD4+ T cells from patients with systemic lupus erythematosus. Clin Dev Immunol. (2013) 2013:206298. doi: 10.1155/2013/206298
46. Li Y, Zhao M, Yin H, Gao F, Wu X, Luo Y, et al. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. (2010) 62:1438–47. doi: 10.1002/art.27363
47. Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. (2015) 42:613–26. doi: 10.1016/j.immuni.2015.03.005
48. Zhao M, Wang J, Liao W, Li D, Li M, Wu H, et al. Increased 5-hydroxymethylcytosine in CD4(+) T cells in systemic lupus erythematosus. J Autoimmun. (2016) 69:64–73. doi: 10.1016/j.jaut.2016.03.001
49. Garaud S, Le Dantec C, Jousse-Joulin S, Hanrotel-Saliou C, Saraux A, Mageed RA, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol. (2009) 182:5623–32. doi: 10.4049/jimmunol.0802412
50. Fali T, Le Dantec C, Thabet Y, Jousse S, Hanrotel C, Youinou P, et al. DNA methylation modulates HRES1/p28 expression in B cells from patients with Lupus. Autoimmunity. (2014) 47:265–71. doi: 10.3109/08916934.2013.826207
51. Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankarn N. Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin Chim Acta. (2011) 412:1457–61. doi: 10.1016/j.cca.2011.04.002
52. Mazari L, Ouarzane M, Zouali M. Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc Natl Acad Sci USA. (2007) 104:6317–22. doi: 10.1073/pnas.0610434104
53. Giambra V, Volpi S, Emelyanov AV, Pflugh D, Bothwell AL, Norio P, et al. Pax5 and linker histone H1 coordinate DNA methylation and histone modifications in the 3' regulatory region of the immunoglobulin heavy chain locus. Mol Cell Biol. (2008) 28:6123–33. doi: 10.1128/MCB.00233-08
54. Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. (2010) 11:607–20. doi: 10.1038/nrm2950
55. Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. (2014) 32:227–55. doi: 10.1146/annurev-immunol-032713-120225
56. Gervin K, Vigeland MD, Mattingsdal M, Hammero M, Nygard H, Olsen AO, et al. DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genes. PLoS Genet. (2012) 8:e1002454. doi: 10.1371/journal.pgen.1002454
57. Zhang P, Su Y, Chen H, Zhao M, Lu Q. Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J Dermatol Sci. (2010) 60:40–2. doi: 10.1016/j.jdermsci.2010.07.011
58. Zhang P, Zhao M, Liang G, Yin G, Huang D, Su F, et al. Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J Autoimmun. (2013) 41:17–24. doi: 10.1016/j.jaut.2013.01.001
59. Chen M, Chen ZQ, Cui PG, Yao X, Li YM, Li AS, et al. The methylation pattern of p16INK4a gene promoter in psoriatic epidermis and its clinical significance. Br J Dermatol. (2008) 158:987–93. doi: 10.1111/j.1365-2133.2008.08505.x
60. Bai J, Liu Z, Xu Z, Ke F, Zhang L, Zhu H, et al. Epigenetic downregulation of SFRP4 contributes to epidermal hyperplasia in psoriasis. J Immunol. (2015) 194:4185–98. doi: 10.4049/jimmunol.1403196
61. Yooyongsatit S, Ruchusatsawat K, Noppakun N, Hirankarn N, Mutirangura A, Wongpiyabovorn J. Patterns and functional roles of LINE-1 and Alu methylation in the keratinocyte from patients with psoriasis vulgaris. J Hum Genet. (2015) 60:349–55. doi: 10.1038/jhg.2015.33
62. Park GT, Han J, Park SG, Kim S, Kim TY. DNA methylation analysis of CD4+ T cells in patients with psoriasis. Arch Dermatol Res. (2014) 306:259–68. doi: 10.1007/s00403-013-1432-8
63. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. (1988) 31:315–24. doi: 10.1002/art.1780310302
64. Allaart CF, Goekoop-Ruiterman YP, de Vries-Bouwstra JK, Breedveld FC, Dijkmans BA, FARR study group. Aiming at low disease activity in rheumatoid arthritis with initial combination therapy or initial monotherapy strategies: the BeSt study. Clin Exp Rheumatol. (2006) 24(6 Suppl. 43):S−77–82.
65. Webster AP, Plant D, Ecker S, Zufferey F, Bell JT, Feber A, et al. Increased DNA methylation variability in rheumatoid arthritis-discordant monozygotic twins. Genome Med. (2018) 10:64. doi: 10.1186/s13073-018-0575-9
66. Guo S, Zhu Q, Jiang T, Wang R, Shen Y, Zhu X, et al. Genome-wide DNA methylation patterns in CD4+ T cells from Chinese Han patients with rheumatoid arthritis. Mod Rheumatol. (2017) 27:441–7. doi: 10.1080/14397595.2016.1218595
67. Liu HW, Lin HL, Yen JH, Tsai WC, Chiou SS, Chang JG, et al. Demethylation within the proximal promoter region of human estrogen receptor alpha gene correlates with its enhanced expression: implications for female bias in lupus. Mol Immunol. (2014) 61:28–37. doi: 10.1016/j.molimm.2014.05.002
68. Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. (2008) 58:2686–93. doi: 10.1002/art.23758
69. Liao J, Liang G, Xie S, Zhao H, Zuo X, Li F, et al. CD40L demethylation in CD4(+) T cells from women with rheumatoid arthritis. Clin Immunol. (2012) 145:13–8. doi: 10.1016/j.clim.2012.07.006
70. Corvetta A, Della Bitta R, Luchetti MM, Pomponio G. 5-Methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. J Chromatogr. (1991) 566:481–91. doi: 10.1016/0378-4347(91)80265-E
71. Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis. (2013) 72:110–7. doi: 10.1136/annrheumdis-2012-201526
72. Neidhart M, Rethage J, Kuchen S, Kunzler P, Crowl RM, Billingham ME, et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. (2000) 43:2634–47. doi: 10.1002/1529-0131(200012)43:12<2634::AID-ANR3>3.0.CO;2-1
73. Takami N, Osawa K, Miura Y, Komai K, Taniguchi M, Shiraishi M, et al. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. (2006) 54:779–87. doi: 10.1002/art.21637
74. Karouzakis E, Rengel Y, Jungel A, Kolling C, Gay RE, Michel BA, et al. DNA methylation regulates the expression of CXCL12 in rheumatoid arthritis synovial fibroblasts. Genes Immun. (2011) 12:643–52. doi: 10.1038/gene.2011.45
75. Nair N, Wilson AG, Barton A. DNA methylation as a marker of response in rheumatoid arthritis. Pharmacogenomics. (2017) 18:1323–32. doi: 10.2217/pgs-2016-0195
76. Zhu H, Zhu C, Mi W, Chen T, Zhao H, Zuo X, et al. Integration of genome-Wide DNA methylation and transcription uncovered aberrant methylation-regulated genes and pathways in the peripheral blood mononuclear cells of systemic sclerosis. Int J Rheumatol. (2018) 2018:7342472. doi: 10.1155/2018/7342472
77. Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis. (2015) 74:1612–20. doi: 10.1136/annrheumdis-2014-205303
78. Lei W, Luo Y, Lei W, Luo Y, Yan K, Zhao S, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol. (2009) 38:369–74. doi: 10.1080/03009740902758875
79. Wang Y, Shu Y, Xiao Y, Wang Q, Kanekura T, Li Y, et al. Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenetics. (2014) 6:25. doi: 10.1186/1868-7083-6-25
80. Jiang H, Xiao R, Lian X, Kanekura T, Luo Y, Yin Y, et al. Demethylation of TNFSF7 contributes to CD70 overexpression in CD4+ T cells from patients with systemic sclerosis. Clin Immunol. (2012) 143:39–44. doi: 10.1016/j.clim.2012.01.005
81. Lian X, Xiao R, Hu X, Kanekura T, Jiang H, Li Y, et al. DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum. (2012) 64:2338–45. doi: 10.1002/art.34376
82. Matatiele P, Tikly M, Tarr G, Gulumian M. DNA methylation similarities in genes of black South Africans with systemic lupus erythematosus and systemic sclerosis. J Biomed Sci. (2015) 22:34. doi: 10.1186/s12929-015-0142-2
83. Almanzar G, Klein M, Schmalzing M, Hilligardt D, El Hajj N, Kneitz H, et al. Disease manifestation and inflammatory activity as modulators of Th17/Treg balance and RORC/FoxP3 methylation in systemic sclerosis. Int Arch Allergy Immunol. (2016) 171:141–54. doi: 10.1159/000450949
84. Lleo A, Liao J, Invernizzi P, Zhao M, Bernuzzi F, Ma L, et al. Immunoglobulin M levels inversely correlate with CD40 ligand promoter methylation in patients with primary biliary cirrhosis. Hepatology. (2012) 55:153–60. doi: 10.1002/hep.24630
85. Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. (2006) 54:2271–9. doi: 10.1002/art.21948
86. Romero LI, Zhang DN, Cooke JP, Ho HK, Avalos E, Herrera R, et al. Differential expression of nitric oxide by dermal microvascular endothelial cells from patients with scleroderma. Vasc Med. (2000) 5:147–58. doi: 10.1191/135886300701568379
87. Qi Q, Guo Q, Tan G, Mao Y, Tang H, Zhou C, et al. Predictors of the scleroderma phenotype in fibroblasts from systemic sclerosis patients. J Eur Acad Dermatol Venereol. (2009) 23:160–8. doi: 10.1111/j.1468-3083.2008.03016.x
88. Hattori M, Yokoyama Y, Hattori T, Motegi S, Amano H, Hatada I, et al. Global DNA hypomethylation and hypoxia-induced expression of the ten eleven translocation (TET) family, TET1, in scleroderma fibroblasts. Exp Dermatol. (2015) 24:841–6. doi: 10.1111/exd.12767
89. Paul DS, Teschendorff AE, Dang MA, Lowe R, Hawa MI, Ecker S, et al. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat Commun. (2016) 7:13555. doi: 10.1038/ncomms13555
90. Stefan M, Zhang W, Concepcion E, Yi Z, Tomer Y. DNA methylation profiles in type 1 diabetes twins point to strong epigenetic effects on etiology. J Autoimmun. (2014) 50:33–7. doi: 10.1016/j.jaut.2013.10.001
91. Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. (2011) 7:e1002300. doi: 10.1371/journal.pgen.1002300
92. Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. (2008) 57:3189–98. doi: 10.2337/db08-0645
93. Wang Z, Zheng Y, Hou C, Yang L, Li X, Lin J, et al. DNA methylation impairs TLR9 induced Foxp3 expression by attenuating IRF-7 binding activity in fulminant type 1 diabetes. J Autoimmun. (2013) 41:50–9. doi: 10.1016/j.jaut.2013.01.009
94. Hewagama A, Gorelik G, Patel D, Liyanarachchi P, McCune WJ, Somers E, et al. Overexpression of X-linked genes in T cells from women with lupus. J Autoimmun. (2013) 41:60–71. doi: 10.1016/j.jaut.2012.12.006
95. Zhao M, Tang J, Gao F, Wu X, Liang Y, Yin H, et al. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J Biomed Biotechnol. (2010) 2010:931018. doi: 10.1155/2010/931018
96. Mi XB, Zeng FQ. Hypomethylation of interleukin-4 and−6 promoters in T cells from systemic lupus erythematosus patients. Acta Pharmacol Sin. (2008) 29:105–12. doi: 10.1111/j.1745-7254.2008.00739.x
97. Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. (2011) 6:593–601. doi: 10.4161/epi.6.5.15374
98. Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. (2013) 9:e1003678. doi: 10.1371/journal.pgen.1003678
99. Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. (2002) 46:1282–91. doi: 10.1002/art.10234
100. Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. (2004) 50:1850–60. doi: 10.1002/art.20255
101. Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. (2005) 174:6212–9. doi: 10.4049/jimmunol.174.10.6212
102. Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. (2004) 172:3652–61. doi: 10.4049/jimmunol.172.6.3652
103. Kozlowska A, Hrycaj P, Lacki JK, Jagodzinski PP. Perforin level in CD4+ T cells from patients with systemic lupus erythematosus. Rheumatol Int. (2010) 30:1627–33. doi: 10.1007/s00296-009-1329-1
104. Balada E, Castro-Marrero J, Felip L, Ordi-Ros J, Vilardell-Tarres M. Clinical and serological findings associated with the expression of ITGAL, PRF1, and CD70 in systemic lupus erythematosus. Clin Exp Rheumatol. (2014) 32:113–6.
105. Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, et al. Renal involvement in lupus is characterized by unique DNA methylation changes in naive CD4+ T cells. J Autoimmun. (2015) 61:29–35. doi: 10.1016/j.jaut.2015.05.003
106. Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, et al. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. (2008) 35:804–10.
107. Zhou Y, Qiu X, Luo Y, Yuan J, Li Y, Zhong Q, et al. Histone modifications and methyl-CpG-binding domain protein levels at the TNFSF7 (CD70) promoter in SLE CD4+ T cells. Lupus. (2011) 20:1365–71. doi: 10.1177/0961203311413412
108. Nambiar MP, Warke VG, Fisher CU, Tsokos GC. Effect of trichostatin A on human T cells resembles signaling abnormalities in T cells of patients with systemic lupus erythematosus: a new mechanism for TCR zeta chain deficiency and abnormal signaling. J Cell Biochem. (2002) 85:459–69. doi: 10.1002/jcb.10160
109. Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. (2011) 17:714–24. doi: 10.1016/j.molmed.2011.07.005
110. Dai Y, Zhang L, Hu C, Zhang Y. Genome-wide analysis of histone H3 lysine 4 trimethylation by ChIP-chip in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Clin Exp Rheumatol. (2010) 28:158–68.
111. Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. (2010) 11:124–33. doi: 10.1038/gene.2009.66
112. Apostolidis SA, Rauen T, Hedrich CM, Tsokos GC, Crispin JC. Protein phosphatase 2A enables expression of interleukin 17 (IL-17) through chromatin remodeling. J Biol Chem. (2013) 288:26775–84. doi: 10.1074/jbc.M113.483743
113. Hedrich CM, Rauen T, Apostolidis SA, Grammatikos AP, Rodriguez Rodriguez N, Ioannidis C, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci USA. (2014) 111:13457–62. doi: 10.1073/pnas.1408023111
114. Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, Petri MA. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin Immunol. (2007) 123:74–81. doi: 10.1016/j.clim.2006.12.008
115. Hu N, Long H, Zhao M, Yin H, Lu Q. Aberrant expression pattern of histone acetylation modifiers and mitigation of lupus by SIRT1-siRNA in MRL/lpr mice. Scand J Rheumatol. (2009) 38:464–71. doi: 10.3109/03009740902895750
116. Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. (2003) 111:539–52. doi: 10.1172/JCI16153
117. Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. (2015) 518:337–43. doi: 10.1038/nature13835
118. Zhao M, Tan Y, Peng Q, Huang C, Guo Y, Liang G, et al. IL-6/STAT3 pathway induced deficiency of RFX1 contributes to Th17-dependent autoimmune diseases via epigenetic regulation. Nat Commun. (2018) 9:583. doi: 10.1038/s41467-018-02890-0
119. Zhao H, Wang L, Luo H, Li QZ, Zuo X. TNFAIP3 downregulation mediated by histone modification contributes to T-cell dysfunction in systemic lupus erythematosus. Rheumatology. (2017) 56:835–43. doi: 10.1093/rheumatology/kew508
120. Ren J, Liao X, Vieson MD, Chen M, Scott R, Kazmierczak J, et al. Selective HDAC6 inhibition decreases early stage of lupus nephritis by down-regulating both innate and adaptive immune responses. Clin Exp Immunol. (2018) 191:19–31. doi: 10.1111/cei.13046
121. Choi EW, Song JW, Ha N, Choi YI, Kim S. CKD-506, a novel HDAC6-selective inhibitor, improves renal outcomes and survival in a mouse model of systemic lupus erythematosus. Sci Rep. (2018) 8:17297. doi: 10.1038/s41598-018-35602-1
122. Gillespie J, Savic S, Wong C, Hempshall A, Inman M, Emery P, et al. Histone deacetylases are dysregulated in rheumatoid arthritis and a novel histone deacetylase 3-selective inhibitor reduces interleukin-6 production by peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Rheum. (2012) 64:418–22. doi: 10.1002/art.33382
123. Haumaitre C, Lenoir O, Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. (2008) 28:6373–83. doi: 10.1128/MCB.00413-08
124. Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. (2011) 63:1376–86. doi: 10.1002/art.30196
125. Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. (2010) 184:6773–81. doi: 10.4049/jimmunol.0904060
126. Qin H, Zhu X, Liang J, Wu J, Yang Y, Wang S, et al. MicroRNA-29b contributes to DNA hypomethylation of CD4+ T cells in systemic lupus erythematosus by indirectly targeting DNA methyltransferase 1. J Dermatol Sci. (2013) 69:61–7. doi: 10.1016/j.jdermsci.2012.10.011
127. Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. (2011) 70:1496–506. doi: 10.1136/ard.2010.139857
128. Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S, et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. (2012) 64:2953–63. doi: 10.1002/art.34505
129. Fan W, Liang D, Tang Y, Qu B, Cui H, Luo X, et al. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. (2012) 64:3715–25. doi: 10.1002/art.34596
130. Tang Q, Yang Y, Zhao M, Liang G, Wu H, Liu Q, et al. Mycophenolic acid upregulates miR-142-3P/5P and miR-146a in lupus CD4+T cells. Lupus. (2015) 24:935–42. doi: 10.1177/0961203315570685
131. Li LJ, Zhao W, Tao SS, Li J, Xu SZ, Wang JB, et al. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell Immunol. (2017) 319:17–27. doi: 10.1016/j.cellimm.2017.06.004
132. Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, et al. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. (2015) 17:131. doi: 10.1186/s13075-015-0632-3
133. Duarte JH. Connective tissue diseases: Large intergenic noncoding RNA linked to disease activity and organ damage in SLE. Nat Rev Rheumatol. (2015) 11:384. doi: 10.1038/nrrheum.2015.82
134. Liu Y, Dong J, Mu R, Gao Y, Tan X, Li Y, et al. MicroRNA-30a promotes B cell hyperactivity in patients with systemic lupus erythematosus by direct interaction with Lyn. Arthritis Rheum. (2013) 65:1603–11. doi: 10.1002/art.37912
135. Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. (2008) 28:630–8. doi: 10.1016/j.immuni.2008.04.002
136. de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. (2008) 205:2199–206. doi: 10.1084/jem.20080579
137. Yuan Y, Kasar S, Underbayev C, Vollenweider D, Salerno E, Kotenko SV, et al. Role of microRNA-15a in autoantibody production in interferon-augmented murine model of lupus. Mol Immunol. (2012) 52:61–70. doi: 10.1016/j.molimm.2012.04.007
138. Luo S, Liu Y, Liang G, Zhao M, Wu H, Liang Y, et al. The role of microRNA-1246 in the regulation of B cell activation and the pathogenesis of systemic lupus erythematosus. Clin Epigenetics. (2015) 7:24. doi: 10.1186/s13148-015-0063-7
139. Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, et al. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol Med. (2011) 3:605–15. doi: 10.1002/emmm.201100171
140. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. (2008) 9:405–14. doi: 10.1038/ni1575
141. Duroux-Richard I, Cuenca J, Ponsolles C, Pineiro AB, Gonzalez F, Roubert C, et al. MicroRNA profiling of B cell subsets from systemic lupus erythematosus patients reveals promising novel biomarkers. Int J Mol Sci. (2015) 16:16953–65. doi: 10.3390/ijms160816953
142. Forster N, Gallinat S, Jablonska J, Weiss S, Elsasser HP, Lutz W. p300 protein acetyltransferase activity suppresses systemic lupus erythematosus-like autoimmune disease in mice. J Immunol. (2007) 178:6941–8. doi: 10.4049/jimmunol.178.11.6941
143. Lashine YA, Salah S, Aboelenein HR, Abdelaziz AI. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus. (2015) 24:240–7. doi: 10.1177/0961203314552117
144. Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. (2009) 60:1065–75. doi: 10.1002/art.24436
145. Wu R, Zeng J, Yuan J, Deng X, Huang Y, Chen L, et al. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J Clin Invest. (2018) 128:2551–68. doi: 10.1172/JCI97426
146. Zhang W, Yi X, An Y, Guo S, Li S, Song P, et al. MicroRNA-17-92 cluster promotes the proliferation and the chemokine production of keratinocytes: implication for the pathogenesis of psoriasis. Cell Death Dis. (2018) 9:567. doi: 10.1038/s41419-018-0621-y
147. Wu Y, Liu L, Bian C, Diao Q, Nisar MF, Jiang X, et al. MicroRNA let-7b inhibits keratinocyte differentiation by targeting IL-6 mediated ERK signaling in psoriasis. Cell Commun Signal. (2018) 16:58. doi: 10.1186/s12964-018-0271-9
148. Yu X, An J, Hua Y, Li Z, Yan N, Fan W, et al. MicroRNA-194 regulates keratinocyte proliferation and differentiation by targeting Grainyhead-like 2 in psoriasis. Pathol Res Pract. (2017) 213:89–97. doi: 10.1016/j.prp.2016.11.020
149. Lu MC, Yu CL, Chen HC, Yu HC, Huang HB, Lai NS. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol. (2014) 177:641–51. doi: 10.1111/cei.12374
150. Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. (2010) 12:R81. doi: 10.1186/ar3006
151. Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. (2011) 63:373–81. doi: 10.1002/art.30115
152. Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. (2008) 58:1001–9. doi: 10.1002/art.23386
153. Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, et al. MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol. (2013) 33:1100–9. doi: 10.1007/s10875-013-9896-z
154. Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. (2010) 62:1733–43. doi: 10.1002/art.27443
155. Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, et al. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther. (2012) 20:1251–60. doi: 10.1038/mt.2012.36
156. Honda N, Jinnin M, Kajihara I, Makino T, Makino K, Masuguchi S, et al. TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol. (2012) 188:3323–31. doi: 10.4049/jimmunol.1100876
157. Cao YL, Jia YJ, Xing BH, Shi DD, Dong XJ. Plasma microRNA-16-5p,−17-5p and−20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J Obstet Gynaecol Res. (2017) 43:974–81. doi: 10.1111/jog.13317
158. Garcia-Contreras M, Shah SH, Tamayo A, Robbins PD, Golberg RB, Mendez AJ, et al. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci Rep. (2017) 7:5998. doi: 10.1038/s41598-017-05787-y
159. Sebastiani G, Ventriglia G, Stabilini A, Socci C, Morsiani C, Laurenzi A, et al. Regulatory T-cells from pancreatic lymphnodes of patients with type-1 diabetes express increased levels of microRNA miR-125a-5p that limits CCR2 expression. Sci Rep. (2017) 7:6897. doi: 10.1038/s41598-017-07172-1
160. Xu K, Bian D, Hao L, Huang F, Xu M, Qin J, et al. microRNA-503 contribute to pancreatic beta cell dysfunction by targeting the mTOR pathway in gestational diabetes mellitus. EXCLI J. (2017) 16:1177–87. doi: 10.17179/excli2017-738
161. Cui X, You L, Zhu L, Wang X, Zhou Y, Li Y, et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism. (2018) 78:95–105. doi: 10.1016/j.metabol.2017.09.006
162. Flowers E, Aouizerat BE, Abbasi F, Lamendola C, Grove KM, Fukuoka Y, et al. Circulating microRNA-320a and microRNA-486 predict thiazolidinedione response: Moving towards precision health for diabetes prevention. Metabolism. (2015) 64:1051–9. doi: 10.1016/j.metabol.2015.05.013
163. Argyropoulos C, Wang K, Bernardo J, Ellis D, Orchard T, Galas D, et al. Urinary MicroRNA profiling predicts the development of microalbuminuria in patients with type 1 diabetes. J Clin Med. (2015) 4:1498–517. doi: 10.3390/jcm4071498
Keywords: epigenetics, autoimmunity, SLE, DNA methylation, miRNAs
Citation: Wu H, Chen Y, Zhu H, Zhao M and Lu Q (2019) The Pathogenic Role of Dysregulated Epigenetic Modifications in Autoimmune Diseases. Front. Immunol. 10:2305. doi: 10.3389/fimmu.2019.02305
Received: 15 April 2019; Accepted: 11 September 2019;
Published: 27 September 2019.
Edited by:
Marzia Dolcino, University of Verona, ItalyReviewed by:
Ziaur S. M. Rahman, Penn State Milton S. Hershey Medical Center, United StatesOnkar Prakash Kulkarni, Birla Institute of Technology and Science, India
Copyright © 2019 Wu, Chen, Zhu, Zhao and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianjin Lu, cWlhbmx1NTg2MEBjc3UuZWR1LmNu
†These authors have contributed equally to this work
 Haijing Wu†
Haijing Wu† Qianjin Lu
Qianjin Lu