- 1College of Medicine, Graduate Institute of Biomedical Sciences, Chang Gung University, Taoyuan City, Taiwan
- 2Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom
- 3Nuffield Department of Primary Care Health Sciences, University of Oxford, Oxford, United Kingdom
We are living through an unprecedented accumulation of data on gene expression by macrophages, reflecting their origin, distribution, and localization within all organs of the body. While the extensive heterogeneity of the cells of the mononuclear phagocyte system is evident, the functional significance of their diversity remains incomplete, nor is the mechanism of diversification understood. In this essay we review some of the implications of what we know, and draw attention to issues to be clarified in further research, taking advantage of the powerful genetic, cellular, and molecular tools now available. Our thesis is that macrophage specialization and functions go far beyond immunobiology, while remaining an essential contributor to innate as well as adaptive immunity.
Introduction
Participation in several Ceppellini workshops by one of the authors (SG) provided an opportunity to examine and present to young investigators some aspects of the unique features of the macrophage, a cell type with an ancient origin in eukaryotic evolution. SG's attachment to the macrophage family has extended over 50 years, rejuvenated over every decade as methodological advances brought new insights and information. However, their biological role in the multicellular organism has remained incomplete, eclipsed as accessory to the specific recognition, and effector functions of lymphoid cells. Metchnikoff already appreciated their professional phagocytic capacity, their digestive proficiency, and potential role in antimicrobial defense (1), while Ehrlich and Wright (2) drew attention to the role of antibodies and opsonins, which enhance phagocytic uptake by monocytes, macrophages, and polymorphonuclear neutrophils (PMN). The discovery of complement and, decades later, the plasma membrane receptors for the Fc domain of IgG specific antibodies and for C3 activated by the classical and alternative pathways, initiated pioneering studies by many investigators [reviewed by Taylor et al. (3)]. Hortega recognized the special properties of microglia in the Central Nervous System (CNS) (4). The discovery of Dendritic cells(DC) by Steinman and Cohn (5, 6), demonstrated their superior role in antigen capture, processing, and presentation to naïve lymphocytes of peptides, in association with the highly polymorphic Major Histocompatibility (MHC) antigens, thus inducing specific T and B lymphocyte activation and expansion (7). DC-like cells can be readily produced in culture of mouse bone marrow or human monocytes in Granulocyte Macrophage Colony-Stimulating Factor (GM-CSF; CSF-2) and IL-4 (8). To some extent DC eclipsed the role of macrophages in adaptive immunity, although their role in innate immunity was secured by the discovery of Toll-like Receptors (TLR) (9). The discovery and characterization of cytokines produced by and acting on macrophages, such as Tumor necrosis factor(TNF) (10) and IL-1 (11), prepared the way for anti-TNF therapy (12), to ameliorate destructive immunopathologies such as rheumatoid arthritis. Activation of macrophages by cellular immunity, characterized by Mackaness (13), was shown to be antigen dependent, but non-specific, and lead to the characterization of Interferon (IFN) gamma (14) as the sole mediator of classical activation produced by antigen-specific T lymphocytes and Natural Killer (NK) cells. After setting the stage above, further relevant milestones of macrophage history will be introduced in subsequent sections. Selected historic figures important in the present understanding of tissue macrophage diversity are shown in Figure 1.

Figure 1. Historic figures associated with macrophages, related cells and their specialized functions.
The Mononuclear Phagocyte System, a Dispersed Organ
Metchnikoff recognized migratory and sessile, fixed tissue phagocytic cells in his early studies of invertebrate development, by microscopy, and intravital labeling. Direct observation of their recruitment to foreign particles injected in vivo lead to further studies in many vertebrate species on their role in host defense against bacteria. Tissue macrophages were subsequently shown to be widely distributed as a system of related cells during development, in the adult steady state, during inflammation, and infection. Aschoff introduced the term Reticulo-Endothelial System (RES), hallmarked by the efficient clearance of particles from the circulation, and extravascular space (15). The imprecise RES nomenclature was replaced by that of the Mononuclear Phagocyte System (MPS) (16), to distinguish mononuclear monocytes and macrophages from PMN, while sharing their highly active capacity as phagocytes. Although widely used till the present day, this terminology is not perfect, since other cell types phagocytose dying cells, and some macrophage-related cells are poorly or even non-phagocytic (17). The diverse cells of the MPS cannot all be characterized by single antigen markers or unique functions expressed at all stages of cell differentiation or activation. Nevertheless, their origin and diversification have common features which point to the valid concept of a specific, dispersed myeloid lineage.
During mammalian development, macrophages derive from haematopoietic precursors in para-aortic regions of the embryo, the yolk sac and fetal liver, seeding organs such as the brain and other tissues before birth (18, 19). A paradigm shift over recent decades has shown that after birth, in the absence of inflammation, resident macrophages in adult tissues derive from embryonic macrophages which can persist, and gradually turn over locally throughout adult life (20–22). This is especially the case for microglia in the (CNS) and Langerhans cells in the epidermis. The bone marrow, which develops as the main haematopoietic organ perinatally, and fumctions throughout adult life (23), serves to replenish resident tissue macrophages, for example in the gut (24), where macrophages turn over more actively, and provides blood monocytes (25) in response to increased demand, for example during inflammation and infection (26). The chemokines and receptors which mediate distribution of monocytes and macrophages in the fetus and adult are not completely defined, nor the adhesion molecules which determine organ-specific localization. Chemokines of resident macrophages include fractalkine and its receptor, CX3CR1 (27), and inflammatory, and immune monocyte recruitment mediated by CCL2 and its receptor, CCR2 [Figure 2; (29, 30)]. Apart from these and related chemokines, recent studies have uncovered macrophage axonal guidance by semaphorins, and plexinA (31, 32). While resident macrophage populations, for example in the peritoneal cavity, persist locally, they can be induced by inflammation, to enter lymphatic vessels for delivery to lymph nodes (33), or to enter neighboring organs such as liver, by sterile local injury (34). Blood monocytes of bone marrow origin may remain inside the circulation and interact with the luminal surface of vascular endothelium (35), become part of sinus-lining endothelium, as Kupffer cells, or diapedese into tissues. Such recruited monocytes are transient in blood (24–48 h) and shorter lived (4–7 days) after migration into tissues, compared with resident macrophages of yolk sac origin e.g., microglia, which can be extremely long-lived. Other reservoirs of precursors and mature macrophages are found in splenic red pulp (36) or in secondary haematopoietic organs, such as liver.
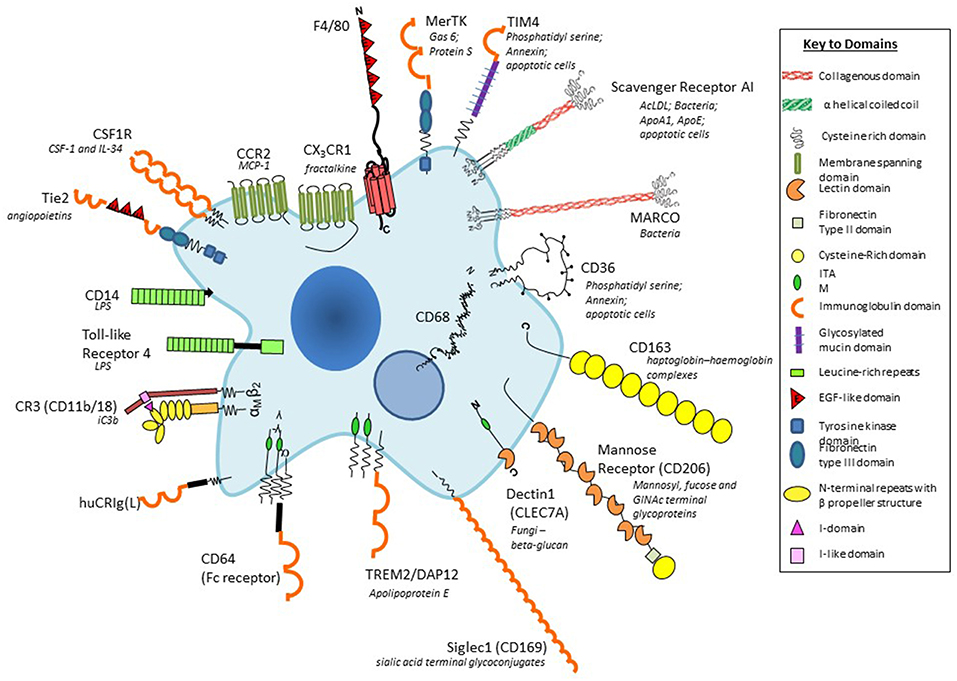
Figure 2. Plasma membrane antigens and receptors expressed by macrophages. Macrophages are able to express a large repertoire of membrane receptors implicated in the recognition and uptake of foreign and modified self-ligands, some of which are illustrated here. These receptors incorporate a range of structural domains, illustrated schematically; they serve as useful marker antigens for immunocytochemistry and FACS analysis (e.g., F4/80, CD68, CSF-1 receptor, Mer-TK, CD64). They function as opsonic (antibody and or complement coated particles to enhance uptake via Fc and complement receptors) or non-opsonic, carbohydrate-binding lectins, and scavenger receptors. The phagocytic receptors mediate clearance of microbes (e.g., MARCO), apoptotic cells (for example CD36, SR-A, TIM4), and circulating ligands; CCR2, and CX3CR1 are examples of GPCR receptors for the monocyte/macrophage chemokines MCP-1 and fractalkine, respectively; other receptors bind growth promoting and regulatory cytokines, for example, CSF-1, and angiopoietins (Tie-2), and CD163 for clearance of injurious haptoglobin–hemoglobin complexes. Toll-like receptor-4 and CD14 react with bacterial membrane components such as lipopolysaccharide (LPS) to induce pro-inflammatory signaling; Dectin-1 recognizes fungi through beta glucan in their wall, activating a range of innate immunological responses. Siglec-1 (CD169), a receptor for sialic acid terminal glycoconjugates, mediates adhesion of host cells and microbes, whereas CD206, a receptor for clearance of Mannosyl-, fucose-, GlcNAc-terminal glycoproteins, is a prototypical marker of M2-type activation. The scavenger receptor SR-A internalizes polyanionic ligands such as modified lipoproteins, as well as selected microbes, whereas CD36 mediates adhesion and M2-induced macrophage fusion and giant cell formation. TREM-2 mutations have been implicated in neurodegeneration and osteoclast function. For further details, see BMC, with permission (28).
While the dual origin of tissue resident macrophages is now widely accepted, there is still uncertainty about the relative contribution of the bone marrow in the adult steady state. In mouse liver, for example, early studies by van Furth and Cohn (37), before their embryonic origin was appreciated, argued for a major contribution of recently dividing bone marrow-derived blood monocytes to resident Kupffer cell populations. The pendulum has swung to yolk sac origin, perhaps too far, as acknowledged by more recent studies (38). The Geissmann group, investigating the origin of murine osteoclasts, showed that after initial perinatal formation of multinucleated cells in bone, monocytes of bone marrow origin are recruited and continue to fuse with osteoclasts throughout adult life (39).
Growth and Differentiation
Studies by Metcalf (Figure 1) on colony forming cells and lineage-specific growth factors contributed greatly to our understanding of haematopoietic stem cell growth and differentiation in vitro (40). Lineage tracing by several groups (41–43) built on studies by Stanley on CSF-1 [reviewed by Chitu and Stanley (44)] and on GM-CSF (45), the major growth/differentiation factors for monocytes, macrophages, and DC. After initial description by von Kolliker in 1873 (Figure 1) (46), Loutit (47) produced proof of the bone marrow origin of osteoclasts; CSF-1 –deficient osteopetrotic op/op mice (48) lacked many, but not all tissue macrophage populations (49). Residual tissue macrophage populations such as microglia, for example, may depend on IL-34, a second ligand for the CSF-1 Receptor, since patients with profound human CSF-1 R deficiency have grossly abnormal CNS development attributed to the absence of microglia (50). Collin et al. have identified mutations which affect monocyte and DC growth and differentiation in humans; bone marrow transplantation and adoptive transfer of haematopoietic stem cells provide clinical and experimental models of monocytopoietic differentiation in vivo (51). Recent studies by Olsson et al. (52) and Yanez et al. (53) have demonstrated a binary origin of monocytes in the mouse, exploiting single cell and population RNA seq analysis and adoptive cell transfer.
In spite of these basic discoveries, we need more quantitative information on the number of monocytes, macrophages, and DC in human tissues, and their life span in vivo. Yona et al. traced the relationship of human monocytes in the steady state and the kinetic response of monocyte subpopulations to endotoxin administration in vivo (54). The subset of monocytic precursors which gives rise to osteoclasts remains to be determined; osteoclasts can be readily produced in vitro by culture of monocytes in CSF-1, and Rank Ligand (46), which should facilitate such studies.
Tissue Distribution and Organ-Specific Properties
In the mouse, we have benefited from the availability of monoclonal antibody markers such as F4/80 to detect macrophages in the developing embryo, in the adult steady state and following a wide range of models of inflammation, infection, malignancy, and atherosclerosis. In addition, we used a panel of mab to identify tissue-specific heterogeneity of marker expression (3). Figure 2 provides a schematic cartoon of these and additional macrophage plasma membrane receptors (55). With the aid of these reagents we identified substantial morphologic and antigenic heterogeneity of resident murine macrophages in different tissue environments such as CNS, spleen and bone marrow (28). Further studies demonstrated heterogeneous antigen expression of monocyte-derived macrophages in BCG-induced granulomata (56), as well as in multinucleated macrophage giant cells (57) and osteoclasts (58). Knowledge of the in situ phenotypes of human tissue macrophages is still limited.
Heterogeneity of Tissue Macrophages: Antigen Markers
The F4/80 antigen(EMR1/ADGRE1), discovered by Austyn and Gordon (59), was used by Hume and others (60) to define monocytes, and macrophages in the mouse. F4/80 is mainly expressed on the plasma membrane, with minimal endocytosis, and is stable to aldehyde fixation; immunocytochemistry therefore can provide exquisite detail of plasma membrane processes in tissue macrophages, suggestive of potential interactions with neighboring cells. Regional variation in morphology and dendritic processes is particularly notable in the brain (61). F4/80, a member of a leukocyte 7-transmembrane, adhesion G protein-coupled receptor family, has been implicated in peripheral tolerance (62), but natural ligands have not been identified. It is also expressed by eosinophils in mouse and human; EMR1 has been identified in other species (63), but expression is transient in human monocyte-derived macrophages. A related molecule, EMR2 (CD312), discovered by Lin and Stacey (64), is expressed by human myeloid cells in blood and tissues, binds chondroitin sulfate B/dermatan sulfate and has been implicated in a human genetic syndrome, vibratory urticaria (65), associated with mast cell degranulation. EMR2 undergoes autoproteolytic cleavage of its extracellular domain to generate an N-terminal polypeptide agonist of GPCR activation.
The F4/80 antigen is expressed during mouse development from midgestation (19) and has been particularly useful in studies of microglia (61). It is also well-expressed in the adult mouse on resident tissue macrophages in the peritoneal cavity, red pulp of spleen, epidermal Langerhans cells, lamina propria of the gut, and Kupffer cells; expression is low on alveolar macrophages in lung, and absent or minimal in white pulp and T-cell rich areas. F4/80 is absent on osteoclasts, metallophilic macrophages in the splenic marginal zone and on subcapsular sinus macrophages in lymph nodes, which express the pan-macrophage endo/lysosomal marker, CD68. Bone marrow- derived monocytes and tissue macrophages recruited to sites of inflammation, infection and malignancy in the mouse express F4/80 strongly.
SIGLEC-1(CD169, sialoadhesin)is a macrophage-specific sialic acid-recognition lectin discovered by Crocker on bone marrow stromal macrophages, at the center of haematopoietic islands (66). It is strongly expressed by marginal metallophils in mouse spleen and by subcapsular sinusoidal macrophages in lymph nodes. It has been implicated in retention and release of monocytes from bone marrow into the circulation. Other lectins widely expressed by macrophages, especially after alternative activation by IL-4/-13, include the macrophage mannose receptor (CD206) (67), and Dectin-1 (CLEC7A), identified as a receptor for fungal beta –glucan by Brown and Gordon (68) and Taylor et al. (3). Scavenger receptors implicated in clearance of apoptotic cells (69), non-opsonic microbial phagocytosis and lipoprotein endocytosis (70), include SRA-I/II, constitutively present on many tissue macrophages (71) and the structurally related collagenous receptor, MARCO (72), which is constitutively expressed by macrophages in the outer marginal zone of rodent spleen (73), but is induced on many tissue macrophages by microbial Toll-like receptor stimulation.
In addition to the above antigens, macrophages express plasma membrane receptors (28) involved in opsonic recognition of IgG antibodies (FcR), complement components (e.g., CD 11b/18), and other opsonins such as milk fat globulin. Other adhesion molecules include various integrins and CD44; plasma membrane receptors that mediate apoptotic cell clearance include an adhesion GPCR BAI-1 (17) and immunoreceptor tyrosine-based activation motif (ITAM) receptors, Tyro, Axl, and MerTK (74). Immunoregulatory receptors include TREM 1 and 2, SIRP alpha, and PD-1. These and receptors for growth factors, cytokines and chemokines have served as useful reagents for FACS, lineage and functional analysis, contributing to our knowledge of macrophage heterogeneity in mouse and human. CD11b expression, for example, is well-expressed on microglia and peritoneal macrophages whereas it is downregulated on alveolar macrophages and Kupffer cells in situ.
Gene Expression
Advances in analysis of macrophage mRNA expression by bulk and single cell sequencing have begun to provide a great deal of new information which has not yet been fully validated by protein expression in situ (75–77). However, important conclusions can already be drawn. These studies confirm that macrophages from different tissues are biosynthetically highly active, expressing a large number of diverse, yet canonical macrophage genes. However, tissue macrophages from different organs also express distinctive antigen and mRNA signatures (77) (Figure 3). Recent publications have reported scRNA-seq analysis of blood mononuclear cells (80), embryonic and adult cell populations, including human placenta (81, 82), which contains both fetal, and maternal macrophages. Improved methods of in situ protein expression (83, 84) are required to validate heterogeneity of genomic and epigenomic expression by macrophages isolated from different tissues. Spatial reconstruction of immune niches has been proposed by combining photoactivatable reporters and sc RNA-seq(NICHE-seq) (85). Consortia of investigators are contributing to a human tissue atlas (86), which has already lead to discovery of novel cell types and functions. Open access to data will extend knowledge of variation in gene expression by macrophages from different sources. This will illustrate developmental, physiologic, and pathologic expression and functions of resident and monocyte-derived macrophages, as well as indicating the cells with which they interact locally. Striking results have already been reported on the overriding effect of phagocytosis of apoptotic cells on gene expression by macrophages in different sites. These have used in vivo models in gut (87), for example, and include parabiotic experiments (88). The microbiome of the gut does not only affect the macrophage phenotype in its local microenvironment, but also systemically (89, 90), through release of microbial products.
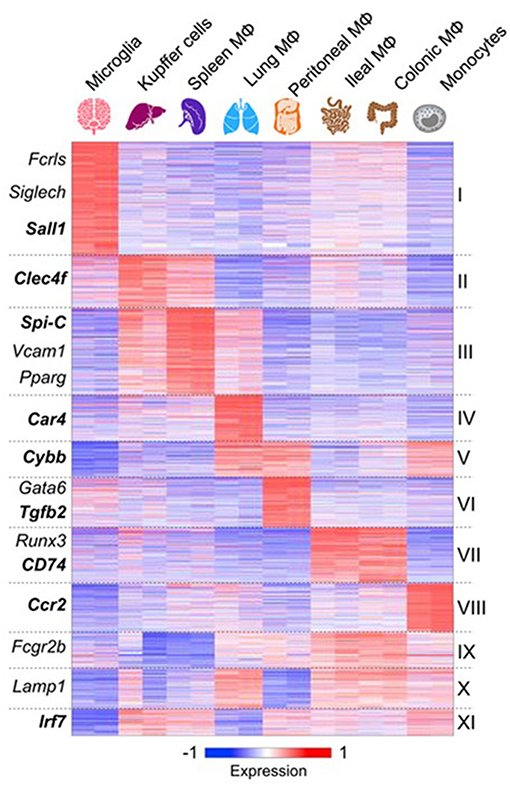
Figure 3. Macrophages express canonical and tissue-specific mRNA signatures. From (77) for further details, with permission. See also (78), ImmGen Consortium (79).
Polarization and Plasticity of Macrophages
We used selected membrane markers to examine the phenotype of mouse peritoneal and human monocyte-derived macrophages in culture, following exposure to Th1 and−2 associated cytokines. In the mouse, IL-4, and subsequently IL-13, was shown to enhance expression and function of mannose receptors (CD206), whereas Interferon gamma selectively downregulated this marker (91). Since MHC class II expression was upregulated by both types of cytokine, we termed this process, alternative, and classical activation, respectively. The terminology M2 and M1 was introduced to include other prototypic stimuli such as immune complexes and macrophage expressed-signatures of selected marker genes (92, 93). We found, using a range of in vitro and in vivo models, that transglutaminase 2 expression, which is not specific for macrophages, was a consistent marker of alternative macrophage activation in humans and mice (94). Subsequent studies by many investigators showed that macrophage polarization involved a spectrum of changes in gene expression (95); to be a useful concept, we proposed that the term alternative activation should be restricted to the prototypical Th-2 cytokines, IL-4, and IL-13 and their common and specific plasma membrane receptors (96). Microarray analysis of macrophage populations using a range of activation and regulatory stimuli, indicates that modules of genes can be identified as signatures to distinguish among different forms of activation. Further analyses of single cell RNA, and protein expression of gene signatures by yolk sac- and bone marrow-derived macrophages and their correlation with distinct functions such as cytotoxicity, and tissue repair, are required to refine polarization in individual organs.
Both classical and alternative macrophage activation can be divided into two distinct phases, an initial priming step by the appropriate cytokine, and completion by a phagocytic or microbial stimulus which induces further changes in gene expression and serves to localize macrophage effector activity. Microbial uptake enhances cytotoxic and pro-inflammatory activity of interferon-primed, classically activated macrophages, whereas uptake of apoptotic cells by IL-4 treated macrophages, enhances anti-inflammatory gene expression by alternatively primed macrophages (97). In experimental models, LPS can induce paradoxical enhancement of JNK activation following Scavenger receptor ligation of IL-4-primed macrophages, suggesting that the outcome will depend on the nature of the phagocytic receptor involved (98).
Priming of macrophages can also induce an adaptive enhancement of microbial phagocytosis and innate immune function. For example, LPS or microbial stimulation upregulates MARCO expression enabling subsequent enhanced uptake of Neisseria meningitidis via this receptor (99, 100). This observation harks back to the earlier studies of Mackaness on macrophage activation by BCG and Listeria monocytogenes, shown to be antigen dependent, but non-specific for the inducing organism (13). Netea et al. have extended this phenomenon, an example of “trained immunity” (101, 102), and have implicated epigenetic mechanisms in its imprinting.
These concepts are important in attempts to reverse polarization, for example of tumor associated macrophages, for potential immunotherapy. Evidence that the macrophage phenotype in vivo is plastic and reversible by adoptive transfer to different tissue microenvironments is sketchy. Van de Laar et al. have shown that yolk sac macrophages, fetal liver and adult monocytes efficiently colonize the empty alveolar niche of Csf2rb−/− mice, unlike mature liver peritoneal or colon macrophages (103). We have found that once macrophages have differentiated terminally, for example to a resident peritoneal phenotype, they cannot be induced to express adhesion receptors characteristic of other terminally differentiated macrophages such as those found in bone marrow haemopoietic clusters. Furthermore, experiments need to distinguish between changes in cell populations and individual cells. However, the phenomenon of induced pluripotency (104) indicates that transcription factors and chromatin conformation can enable true plasticity and the ability to give rise to embryonic stem cells, able to generate different somatic cell types, including macrophages (105) and microglia (106) de novo.
Generation of Diversity in Tissue Macrophages
The evidence that resident embryo or bone marrow-derived populations of tissue macrophages, distributed throughout organs in the steady state, acquire distinct phenotypes as well as expressing core macrophage properties, raises a fascinating problem of origin of their diversity. The extent of adaptation by monocytes recruited by infection to different tissue environments, for example in granuloma formation, requires further characterization. In order to establish a testable hypothesis to account for the generation of diversity, we have to keep in mind several properties which distinguish macrophages from T and B lymphocytes, in which antigen receptor gene rearrangement and clonal selection have provided unexpected solutions to account for repertoire diversity and antigen specificity. Macrophages express a broader range of receptors than lymphocytes to distinguish foreign, modified-self and self-ligands; these include proteins and peptides, carbohydrates, nucleic acids, and lipids. Macrophage receptors can be viewed as “hard wired,” unlike the more selective, antigen-specific receptors of adaptive lymphocytes. Tissue macrophages are terminally differentiated, capable of only a limited degree of proliferative capacity once they enter tissues. Clonal selection can therefore be ruled out. We do not know the size of the macrophage repertoire, but it must be substantial to accommodate interactions with other cell types within the body, including macrophages themselves, as well as so-called “pattern recognition receptors” for exogenous and endogenous ligands. Many investigators acknowledge that the local tissue as well as exogenous micro-environment must play a specifying role in inducing or selecting expression of a particular constellation of surface receptors and gene products [for example (75, 107)]. In addition, macrophages can recognize a host of intracellular ligands in their cytosolic, biosynthetic, secretory, or endocytic compartments. However, chromatin conformation, transcription factors and enhancers, in addition to epigenetic mechanisms, must also determine the programme of differential gene expression, and modulation of the macrophage phenotype (108–112). T'Jonck et al. have discussed the role of niche signals and transcription factors involved in tissue resident macrophage development in detail (113).
These considerations leave many questions as to how, when and where, and specifically by which intrinsic and environmental mechanisms, diversity is achieved. Surprisingly little consideration has been given to the nature of the diverse ligands in the extracellular matrix of different tissues (114); nor the role of various epithelia, endothelia, mesenchymal, and neuro-endocrine cells, all of which interact with macrophages as a result of their unique migration and organ distribution (83, 84, 115, 116).
Tissue-Specific Functions of Macrophages
Tissue macrophages express general, prototypic, functions throughout the body which contribute to homeostasis, recognition and responses to intrinsic and external perturbation, restoring physiologic stability, and contributing to repair after injury. In different organs they adapt to different micro-environments with variations on the themes of clearance of particles and soluble ligands, digestion or storage in lysosomes, constitutive, and induced biosynthesis, and secretion. They interact with living or dying cells and microbes, blood and lymph, undergoing metabolic adaptation, and altering adhesion to extracellular matrix as they migrate, through different locations over time. In the process, they may respond to injurious stimuli by autophagy, cell growth or death. Nevertheless, we can already discern remarkable variations in organ-specific functions to which they contribute; these include a central role in haematopoietic turnover, and haem degradation (117, 118), lymphoid trafficking of immune cells (33); mucosal physiology, for instance in the gut (119, 120); remodeling in the CNS (107, 121, 122); neural- adipose tissue metabolism (123), and adipose- sympathetic nervous interactions (124); and electrical activity in the heart (125). Current studies in single cell RNA and protein expression by tissue macrophages will provide more examples of trophic and defense functions, contributing to embryonic development, anatomic, physiologic, and pathologic processes. Returning to our earlier discussion of how such diversity might be generated, it seems likely that encounters with different ligands in their tissue microenvironment can exploit pre-existing or induce novel sensors to activate adaptive changes in transcription and epigenetic modification; this begs the question of the extent and mechanisms of initial tissue-specific receptor diversification. While differentiation can generate a core panel of recognition molecules on and within the macrophage, it may be necessary to postulate further induction, feedback amplification, or selection by as yet unknown somatic gene expression mechanisms. Investigating the details of osteoclast and DC development in vivo and in vitro may provide further clues to novel molecular mechanisms.
Conclusions
Recent progress in molecular and cellular biology have brought exciting insights into view, enabling us to characterize monocyte/macrophage heterogeneity in situ. Understanding the themes of their functions within multicellular organisms across a range of evolutionary stages will make it possible to discover a unifying pattern extending far beyond innate or adaptive, cellular and humoral immunity. The challenge will be to imagine the properties underlying the genes and molecules which can lead us to such knowledge. Finally, we need to consider the implications of monocyte/macrophage heterogeneity for therapy. Factors to be taken into account for macrophage-directed immunotherapy include the expression of target antigens on distinct subpopulations, the route of administration, risk of off-target effects and species differences. Similarly, for potential adoptive cell therapy, the origin, differentiation, proliferative capacity and activation status have to be defined, as well as genetic compatibility.
Author Contributions
SG conceived and wrote the manuscript. AP reviewed and edited the manuscript and designed the figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge stimulating discussions with numerous colleagues and fellow macrophage enthusiasts.
Abbreviations
ADGR, adhesion 7-transmembrane G-protein coupled receptor; BAI-1, brain-specific angiogenesis inhibitor-1; CSF1-R, macrophage colony stimulating factor receptor; DC, Dendritic cells; EMR, epidermal growth factor-like module-containing, mucin-like hormone receptor-like receptor; ITAM, immunoreceptor tyrosine-based activation motif; GM-CSF, granulocyte macrophage colony stimulating factor; GPCR, G-protein coupled receptor; IL, Interleukin; iPSC, induced pluripotent stem cell; MARCO, macrophage receptor with collagenous domain; MHC, major histocompatibility complex; MPS, Mononuclear phagocyte system; NK, natural killer cell; PD-1, Programmed cell death-1; PMN, polymorphonuclear leukocyte; RES, reticulendothelial system; scRNA seq, single cell ribonucleic acid sequencing; Sirp, signal regulatory protein alpha; SR-A, scavenger receptor A; Tie2, Tyrosine-protein kinase receptor for angiopoietins-2; TLR, toll-like receptor; TNF, tumor necrosis factor; TREM-2, Triggering receptor expressed on myeloid cells-2.
References
1. Teti G, Biondo C, Beninati C. The phagocyte, metchnikoff, and the foundation of immunology. Microbiol Spectr. (2016) 4:MCHD-0009-2015. doi: 10.1128/microbiolspec.MCHD-0009-2015
2. Kaufmann SHE. Immunology's coming of age. Front Immunol. (2019) 10:684. doi: 10.3389/fimmu.2019.00684
3. Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. (2005) 23:901–44. doi: 10.1146/annurev.immunol.23.021704.115816
4. Sierra A, De Castro F, Del Rio-Hortega J, Rafael Iglesias-Rozas J, Garrosa M, Kettenmann H. The “big-bang” for modern glial biology: translation and comments on pio del rio-hortega 1919 series of papers on microglia. Glia. (2016) 64:1801–40. doi: 10.1002/glia.23046
5. Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I morphology, quantitation, tissue distribution. J Exp Med. (1973) 137:1142–62. doi: 10.1084/jem.137.5.1142
6. Steinman RM, Moberg CL. Zanvil alexander cohn 1926-1993. J Exp Med. (1994) 179:1–30. doi: 10.1084/jem.179.1.1
7. Moberg CL. An appreciation of ralph marvin steinman (1943-2011). J Exp Med. (2011) 208:2337–42. doi: 10.1084/jem.20112294
8. Austyn JM. Dendritic cells in the immune system-history, lineages, tissues, tolerance, and immunity. Microbiol Spectr. (2016) 4:MCHD-0046-2016. doi: 10.1128/microbiolspec.MCHD-0046-2016
9. Medzhitov R. TLR-mediated innate immune recognition. Semin Immunol. (2007) 19:1–2. doi: 10.1016/j.smim.2007.02.001
10. Beutler B, Cerami A. The biology of cachectin/TNF–a primary mediator of the host response. Annu Rev Immunol. (1989) 7:625–55. doi: 10.1146/annurev.iy.07.040189.003205
11. Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. (2019) 50:778–95. doi: 10.1016/j.immuni.2019.03.012
12. Feldmann M, Maini RN. Lasker clinical medical research award. TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat Med. (2003) 9:1245–50. doi: 10.1038/nm939
13. Mackaness GB. The immunological basis of acquired cellular resistance. J Exp Med. (1964) 120:105–20. doi: 10.1084/jem.120.1.105
14. Lazear HM, Schoggins JW, Diamond MS. Shared and distinct functions of type I and type III interferons. Immunity. (2019) 50:907–23. doi: 10.1016/j.immuni.2019.03.025
15. Aschoff L. Das reticulo-endotheliale system. Ergeb Inn Med Kinderheilk. (1924) 26:1–118. doi: 10.1007/978-3-642-90639-8_1
16. Van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ. (1972) 46:845–52.
17. Morioka S, Maueroder C, Ravichandran KS. Living on the edge: efferocytosis at the interface of homeostasis and pathology. Immunity. (2019) 50:1149–62. doi: 10.1016/j.immuni.2019.04.018
18. Perry VH, Andersson PB, Gordon S. Macrophages and inflammation in the central nervous system. Trends Neurosci. (1993) 16:268–73. doi: 10.1016/0166-2236(93)90180-T
19. Morris L, Graham CF, Gordon S. Macrophages in haemopoietic and other tissues of the developing mouse detected by the monoclonal antibody F4/80. Development. (1991) 112:517–26.
20. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. (2014) 14:571–8. doi: 10.1038/nri3712
21. Hoeffel G, Ginhoux F. Ontogeny of tissue-resident macrophages. Front Immunol. (2015) 6:486. doi: 10.3389/fimmu.2015.00486
22. Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. (2015) 15:731–44. doi: 10.1038/nri3920
23. Dzierzak E, De Pater E. Regulation of blood stem cell development. Curr Top Dev Biol. (2016) 118:1–20. doi: 10.1016/bs.ctdb.2016.01.001
24. Faria AMC, Reis BS, Mucida D. Tissue adaptation: implications for gut immunity and tolerance. J Exp Med. (2017) 214:1211–26. doi: 10.1084/jem.20162014
25. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. (2010) 116:e74–80. doi: 10.1182/blood-2010-02-258558
26. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. (2003) 19:71–82. doi: 10.1016/S1074-7613(03)00174-2
27. Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, et al. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. (2014) 211:1571–83. doi: 10.1084/jem.20140678
28. Gordon S, Pluddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. (2017) 15:53. doi: 10.1186/s12915-017-0392-4
29. Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. (2006) 354:610–21. doi: 10.1056/NEJMra052723
31. Kong Y, Janssen BJ, Malinauskas T, Vangoor VR, Coles CH, Kaufmann R, et al. Structural basis for plexin activation and regulation. Neuron. (2016) 91:548–60. doi: 10.1016/j.neuron.2016.06.018
32. Seiradake E, Jones EY, Klein R. Structural perspectives on axon guidance. Annu Rev Cell Dev Biol. (2016) 32:577–608. doi: 10.1146/annurev-cellbio-111315-125008
33. Zhang Y, Roth TL, Gray EE, Chen H, Rodda LB, Liang Y, et al. Migratory and adhesive cues controlling innate-like lymphocyte surveillance of the pathogen-exposed surface of the lymph node. Elife. (2016) 5:e18156. doi: 10.7554/eLife.18156
34. Wang J, Kubes P. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell. (2016) 165:668–78. doi: 10.1016/j.cell.2016.03.009
35. Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. (2007) 317:666–70. doi: 10.1126/science.1142883
36. Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. (2009) 325:612–6. doi: 10.1126/science.1175202
37. Van Furth R, Cohn ZA. The origin and kinetics of mononuclear phagocytes. J Exp Med. (1968) 128:415–35. doi: 10.1084/jem.128.3.415
38. Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity. (2018) 49:595–613. doi: 10.1016/j.immuni.2018.10.005
39. Jacome-Galarza CE, Percin GI, Muller JT, Mass E, Lazarov T, Eitler J, et al. Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature. (2019) 568:541–5. doi: 10.1038/s41586-019-1105-7
40. Metcalf D. Growth and differentiation factors. Microbiol Spectr. (2016) 4:MCHD-0004-2015. doi: 10.1128/microbiolspec.MCHD-0004-2015
41. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. (2016) 44:439–49. doi: 10.1016/j.immuni.2016.02.024
42. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. (2015) 518:547–51. doi: 10.1038/nature13989
43. Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. (2013) 38:79–91. doi: 10.1016/j.immuni.2012.12.001
44. Chitu V, Stanley ER. Regulation of embryonic and postnatal development by the CSF-1 receptor. Curr Top Dev Biol. (2017) 123:229–75. doi: 10.1016/bs.ctdb.2016.10.004
45. Dougan M, Dranoff G, Dougan SK. GM-CSF, IL-3, and IL-5 family of cytokines: regulators of inflammation. Immunity. (2019) 50:796–811. doi: 10.1016/j.immuni.2019.03.022
46. Martin TJ. Historically significant events in the discovery of RANK/RANKL/OPG. World J Orthop. (2013) 4:186–97. doi: 10.5312/wjo.v4.i4.186
47. Nisbet NW, Menage J, Loutit JF. Osteogenesis in osteopetrotic mice. Calcif Tissue Int. (1982) 34:37–42. doi: 10.1007/BF02411206
48. Wiktor-Jedrzejczak W, Bartocci A, Ferrante AWJr, Ahmed-Ansari A, Sell KW, Pollard JW, et al. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci USA. (1990) 87:4828–32. doi: 10.1073/pnas.87.12.4828
49. Witmer-Pack MD, Hughes DA, Schuler G, Lawson L, Mcwilliam A, Inaba K, et al. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. (1993) 104(Pt 4):1021–9.
50. Oosterhof N, Chang IJ, Karimiani EG, Kuil LE, Jensen DM, Daza R, et al. Homozygous mutations in CSF1R cause a pediatric-onset leukoencephalopathy and can result in congenital absence of microglia. Am J Hum Genet. (2019) 104:936–47. doi: 10.1016/j.ajhg.2019.03.010
51. Collin M, Bigley V. Monocyte, macrophage, and dendritic cell development: the human perspective. Microbiol Spectr. (2016) 4:MCHD-0015-2015. doi: 10.1128/microbiolspec.MCHD-0015-2015
52. Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. (2016) 537:698–702. doi: 10.1038/nature19348
53. Yanez A, Coetzee SG, Olsson A, Muench DE, Berman BP, Hazelett DJ, et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity. (2017) 47:890–902 e4. doi: 10.1016/j.immuni.2017.10.021
54. Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med. (2017) 214:1913–23. doi: 10.1084/jem.20170355
55. Gordon S. Phagocytosis: an immunobiologic process. Immunity. (2016) 44:463–75. doi: 10.1016/j.immuni.2016.02.026
56. Keshav S, Chung P, Milon G, Gordon S. Lysozyme is an inducible marker of macrophage activation in murine tissues as demonstrated by in situ hybridization. J Exp Med. (1991) 174:1049–58. doi: 10.1084/jem.174.5.1049
57. Milde R, Ritter J, Tennent GA, Loesch A, Martinez FO, Gordon S, et al. Multinucleated giant cells are specialized for complement-mediated phagocytosis and large target destruction. Cell Rep. (2015) 13:1937–48. doi: 10.1016/j.celrep.2015.10.065
58. Pereira M, Petretto E, Gordon S, Bassett JHD, Williams GR, Behmoaras J. Common signalling pathways in macrophage and osteoclast multinucleation. J Cell Sci. (2018) 131:jcs216267. doi: 10.1242/jcs.216267
59. Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. (1981) 11:805–15. doi: 10.1002/eji.1830111013
60. Hume DA, Gordon S. Mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. identification of resident macrophages in renal medullary and cortical interstitium and the juxtaglomerular complex. J Exp Med. (1983) 157:1704–9. doi: 10.1084/jem.157.5.1704
61. Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. (1990) 39:151–70. doi: 10.1016/0306-4522(90)90229-W
62. Lin HH, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J, et al. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med. (2005) 201:1615–25. doi: 10.1084/jem.20042307
63. Waddell LA, Lefevre L, Bush SJ, Raper A, Young R, Lisowski ZM, et al. ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front Immunol. (2018) 9:2246. doi: 10.3389/fimmu.2018.02246
64. Lin HH, Stacey M. G protein-coupled receptors in macrophages. Microbiol Spectr. (2016) 4:MCHD-0028-2016. doi: 10.1128/microbiolspec.MCHD-0028-2016
65. Boyden SE, Desai A, Cruse G, Young ML, Bolan HC, Scott LM, et al. Vibratory urticaria associated with a missense variant in ADGRE2. N Engl J Med. (2016) 374:656–63. doi: 10.1056/NEJMoa1500611
66. Klaas M, Crocker PR. Sialoadhesin in recognition of self and non-self. Semin Immunopathol. (2012) 34:353–64. doi: 10.1007/s00281-012-0310-3
67. Martinez-Pomares L. The mannose receptor. J Leukoc Biol. (2012) 92:1177–86. doi: 10.1189/jlb.0512231
68. Brown GD, Gordon S. Immune recognition. a new receptor for beta-glucans. Nature. (2001) 413:36–7. doi: 10.1038/35092620
69. Gordon S, Pluddemann A. Macrophage clearance of apoptotic cells: a critical assessment. Front Immunol. (2018) 9:127. doi: 10.3389/fimmu.2018.00127
70. Neyen C, Pluddemann A, Roversi P, Thomas B, Cai L, Van Der Westhuyzen DR, et al. Macrophage scavenger receptor A mediates adhesion to apolipoproteins A-I and E. Biochemistry. (2009) 48:11858–71. doi: 10.1021/bi9013769
71. Plüddemann A, Mukhopadhyay S, Gordon S. The interaction of macrophage receptors with bacterial ligands. Expert Rev Mol Med. (2006) 8:1–25. doi: 10.1017/S1462399406000159
72. Mukhopadhyay S, Chen Y, Sankala M, Peiser L, Pikkarainen T, Kraal G, et al. MARCO, an innate activation marker of macrophages, is a class A scavenger receptor for Neisseria meningitidis. Eur J Immunol. (2006) 36:940–9. doi: 10.1002/eji.200535389
73. Kraal G, Mebius R. New insights into the cell biology of the marginal zone of the spleen. Int Rev Cytol. (2006) 250:175–215. doi: 10.1016/S0074-7696(06)50005-1
74. Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol. (2015) 33:355–91. doi: 10.1146/annurev-immunol-032414-112103
75. Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. (2016) 17:18–25. doi: 10.1038/ni.3325
76. Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. (2014) 15:1181–9. doi: 10.1038/ni.3006
77. Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. (2014) 159:1312–26. doi: 10.1016/j.cell.2014.11.018
78. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. (2012) 13:1118–28. doi: 10.1038/ni.2419
79. Immgenconsortium. Open-source ImmGen: mononuclear phagocytes. Nat. Immunol. (2016) 17:741. doi: 10.1038/ni.3478
80. Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. (2017) 356:eaah4573. doi: 10.1126/science.aah4573
81. Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, et al. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv. (2018) 4:eaau4788. doi: 10.1126/sciadv.aau4788
82. Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. (2018) 563:347–53. doi: 10.1038/s41586-018-0698-6
83. Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. (2018) 174:968–81 e15. doi: 10.1016/j.cell.2018.07.010
84. Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell. (2018) 174:1373–87 e19. doi: 10.1016/j.cell.2018.08.039
85. Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science. (2017) 358:1622–6. doi: 10.1126/science.aao4277
86. Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. Elife. (2017) 6:e27041. doi: 10.7554/eLife.27041
87. Cummings RJ, Barbet G, Bongers G, Hartmann BM, Gettler K, Muniz L, et al. Different tissue phagocytes sample apoptotic cells to direct distinct homeostasis programs. Nature. (2016) 539:565–9. doi: 10.1038/nature20138
88. A-Gonzalez N, Quintana JA, Garcia-Silva S, Mazariegos M, Gonzalez De La Aleja A, Nicolas-Avila JA, et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J Exp Med. (2017) 214:1281–96. doi: 10.1084/jem.20161375
89. Abdel-Haq R, Schlachetzki JCM, Glass CK, Mazmanian SK. Microbiome-microglia connections via the gut-brain axis. J Exp Med. (2019) 216:41–59. doi: 10.1084/jem.20180794
90. Erny D, Hrabe De Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. (2015) 18:965–77. doi: 10.1038/nn.4030
91. Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. (1992) 176:287–92. doi: 10.1084/jem.176.1.287
92. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. (2002) 23:549–55. doi: 10.1016/S1471-4906(02)02302-5
93. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. (2008) 8:958–69. doi: 10.1038/nri2448
94. Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. (2013) 121:e57–69. doi: 10.1182/blood-2012-06-436212
95. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. (2014) 41:14–20. doi: 10.1016/j.immuni.2014.06.008
96. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. (2014) 6:13. doi: 10.12703/P6-13
97. Bosurgi L, Cao YG, Cabeza-Cabrerizo M, Tucci A, Hughes LD, Kong Y, et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science. (2017) 356:1072–6. doi: 10.1126/science.aai8132
98. Guo M, Hartlova A, Gierlinski M, Prescott A, Castellvi J, Losa JH, et al. Triggering MSR1 promotes JNK-mediated inflammation in IL-4-activated macrophages. EMBO J. (2019) 38:e100299. doi: 10.15252/embj.2018100299
99. Bowdish DM, Loffredo MS, Mukhopadhyay S, Mantovani A, Gordon S. Macrophage receptors implicated in the “adaptive” form of innate immunity. Microbes Infect. (2007) 9:1680–7. doi: 10.1016/j.micinf.2007.09.002
100. Locati M, Mantovani A, Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv Immunol. (2013) 120:163–84. doi: 10.1016/B978-0-12-417028-5.00006-5
101. Arts RJ, Netea MG. Adaptive characteristics of innate immune responses in macrophages. Microbiol Spectr. (2016) 4:MCHD-0023-2015. doi: 10.1128/microbiolspec.MCHD-0023-2015
102. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and adaptive immune memory: an evolutionary continuum in the host's response to pathogens. Cell Host Microbe. (2019) 25:13–26. doi: 10.1016/j.chom.2018.12.006
103. Van De Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. (2016) 44:755–68. doi: 10.1016/j.immuni.2016.02.017
104. Theunissen TW, Jaenisch R. Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development. (2017) 144:4496–509. doi: 10.1242/dev.157404
105. Mukherjee C, Hale C, Mukhopadhyay S. A simple multistep protocol for differentiating human induced pluripotent stem cells into functional macrophages. Methods Mol Biol. (2018) 1784:13–28. doi: 10.1007/978-1-4939-7837-3_2
106. Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports. (2017) 8:1727–42. doi: 10.1016/j.stemcr.2017.05.017
107. Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. (2017) 356:eaa3222. doi: 10.1126/science.aal3222
108. Glass CK. Genetic and genomic approaches to understanding macrophage identity and function. Arterioscler Thromb Vasc Biol. (2015) 35:755–62. doi: 10.1161/ATVBAHA.114.304051
109. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. (2010) 38:576–89. doi: 10.1016/j.molcel.2010.05.004
110. Hoeksema MA, Glass CK. Nature and nurture of tissue-specific macrophage phenotypes. Atherosclerosis. (2019) 281:159–67. doi: 10.1016/j.atherosclerosis.2018.10.005
111. Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. (2013) 34:216–23. doi: 10.1016/j.it.2012.11.001
112. Zhu YP, Thomas GD, Hedrick CC. 2014 Jeffrey M. hoeg award lecture: transcriptional control of monocyte development. Arterioscler Thromb Vasc Biol. (2016) 36:1722–33. doi: 10.1161/ATVBAHA.116.304054
113. T'jonck W, Guilliams M, Bonnardel J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell Immunol. (2018) 330:43–53. doi: 10.1016/j.cellimm.2018.02.005
114. Hynes RO. The extracellular matrix: not just pretty fibrils. Science. (2009) 326:1216–9. doi: 10.1126/science.1176009
115. Pearce OMT, Delaine-Smith RM, Maniati E, Nichols S, Wang J, Bohm S, et al. Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. (2018) 8:304–19. doi: 10.1158/2159-8290.CD-17-0284
116. Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. (2019) 20:928–942. doi: 10.1038/s41590-019-0378-1
117. Haldar M, Murphy KM. Origin, development, and homeostasis of tissue-resident macrophages. Immunol Rev. (2014) 262:25–35. doi: 10.1111/imr.12215
118. Haldar M, Kohyama M, So AY, Kc W, Wu X, Briseno CG, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. (2014) 156:1223–34. doi: 10.1016/j.cell.2014.01.069
119. Gabanyi I, Muller PA, Feighery L, Oliveira TY, Costa-Pinto FA, Mucida D. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell. (2016) 164:378–91. doi: 10.1016/j.cell.2015.12.023
120. Muller PA, Koscso B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. (2014) 158:300–13. doi: 10.1016/j.cell.2014.04.050
121. Hammond TR, Marsh SE, Stevens B. Immune signaling in neurodegeneration. Immunity. (2019) 50:955–74. doi: 10.1016/j.immuni.2019.03.016
122. Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. (2016) 352:712–6. doi: 10.1126/science.aad8373
123. Wolf Y, Boura-Halfon S, Cortese N, Haimon Z, Sar Shalom H, Kuperman Y, et al. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat Immunol. (2017) 18:665–74. doi: 10.1038/ni.3746
124. Pirzgalska RM, Seixas E, Seidman JS, Link VM, Sanchez NM, Mahu I, et al. Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nat Med. (2017) 23:1309–18. doi: 10.1038/nm.4422
Keywords: mononuclear phagocyte, macrophage, tissue-specific function, monocyte, plasticity, macrophage heterogeneity, macrophage receptors
Citation: Gordon S and Plüddemann A (2019) The Mononuclear Phagocytic System. Generation of Diversity. Front. Immunol. 10:1893. doi: 10.3389/fimmu.2019.01893
Received: 29 May 2019; Accepted: 26 July 2019;
Published: 09 August 2019.
Edited by:
Francesca Di Rosa, Consiglio Nazionale Delle Ricerche (CNR), ItalyReviewed by:
Antonio Sica, University of Eastern Piedmont, ItalyAndreas Wack, Francis Crick Institute, United Kingdom
Copyright © 2019 Gordon and Plüddemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siamon Gordon, c2lhbW9uLmdvcmRvbkBwYXRoLm94LmFjLnVr
 Siamon Gordon
Siamon Gordon Annette Plüddemann
Annette Plüddemann