- Department of Infectious Diseases, Facultad de Medicina, Hospital Universitario Ramón y Cajal, Universidad de Alcalá (IRYCIS), Madrid, Spain
HIV infection exerts profound and perhaps irreversible damage to the gut mucosal-associated lymphoid tissues, resulting in long-lasting changes in the signals required for the coordination of commensal colonization and in perturbations at the compositional and functional level of the gut microbiota. These abnormalities in gut microbial communities appear to affect clinical outcomes, including T-cell recovery, vaccine responses, HIV transmission, cardiovascular disease, and cancer pathogenesis. For example, the microbial signature associated with HIV infection has been shown to induce tryptophan catabolism, affect the butyrate synthesis pathway, impair anti-tumoral immunity and affect oxidative stress, which have also been linked to the pathogenesis of cancer. Furthermore, some of the taxa that are depleted in subjects with HIV have proved to modulate the anti-tumor efficacy of various chemotherapies and immunotherapeutic agents. The aim of this work is to provide a broad overview of recent advances in our knowledge of how HIV might affect the microbiota, with a focus on the pathways shared with cancer pathogenesis.
Introduction
A hallmark of treated HIV infection is sustained, low-level viral inflammation. While the cause of this persistent activation of innate and adaptive immunity despite well-controlled HIV RNA replication is not completely understood, it is widely assumed that chronic defects of mucosal immunity are a major contributor (1). HIV targets the mucosa on structural and functional levels (2–4). Arguably, these disturbances will have consequences on the signals required for the coordination of commensal colonization, which may explain the shifts in microbial distributions and metabolic activity of gut microbial communities (5–7). In addition, these abnormalities caused by HIV infection have been shown to result in increased translocation of microbial products from the gut to the circulation in both animal models and HIV-infected individuals (8, 9). It has been repeatedly shown that biomarkers of bacterial translocation positively correlate with markers of T-cell activation, monocyte activation, and proinflammatory cytokines (10). It is widely accepted now that sustained low-level activation of the innate and adaptive immune systems is a major driver of AIDS and non-AIDS-related comorbidities (11–15). Collectively, these observations argue that microbial translocation, a phenomenon intrinsically linked to the gut microbiota, is a driver of inflammation, and adverse outcomes during treated HIV infection.
Influence of the Microbiota on HIV Immunopathogenesis During Treated Infection
The gut microbiota has been associated with HIV immunopathogenesis (5, 16–19). Defining the influence of HIV on the microbiota, however, is more difficult. Studies on the impact of SIV infection in the gut microbiota of non-human primates have found only modest differences in the fecal bacterial communities between SIV-infected macaques compared to uninfected macaques, suggesting that the development of immunosuppression, rather than SIV infection itself, may drive the differences (20, 21). In addition, induction of dysbiosis with vancomycin does not accelerate the progression of untreated SIV infection (22). The effects of HIV infection on microbial diversity appear to be confounded by a number of factors, including the nadir of CD4+ T-cells (23) and the risk factor for HIV acquisition (24, 25). While admittedly there are difficulties dissecting the specific effects of HIV disease on the microbial communities, there is wide consensus that the gut microbiomes of HIV-positive individuals exhibit specific compositional and functional shifts (5, 19, 26–29). Surprisingly, the microbiota associated with HIV infection shares traits with that associated with other proinflammatory conditions, such as the depletion of butyrate-producing bacteria observed in inflammatory bowel disease (30).
It is therefore tempting to assume that so-called “HIV-associated dysbiosis” may be implicated in the sustainment of systemic inflammation in treated HIV disease. Several taxa and their associated pathways (Figure 1) have been linked with persistent immune abnormalities (5, 7, 31). The real picture, however, may be far more complex. From an ecological point of view, the components of a rapidly evolving ecosystem will respond to environmental perturbations by adapting their composition and functions to achieve the optimal fitness within their changing habitat (32). For example, the fecal microbiota of people with HIV has been shown to harbor greater abundances of genes related to resistance to oxidative stress, such as the genetic machinery for glutathione metabolism or zeatin biosynthesis pathways (7, 31).
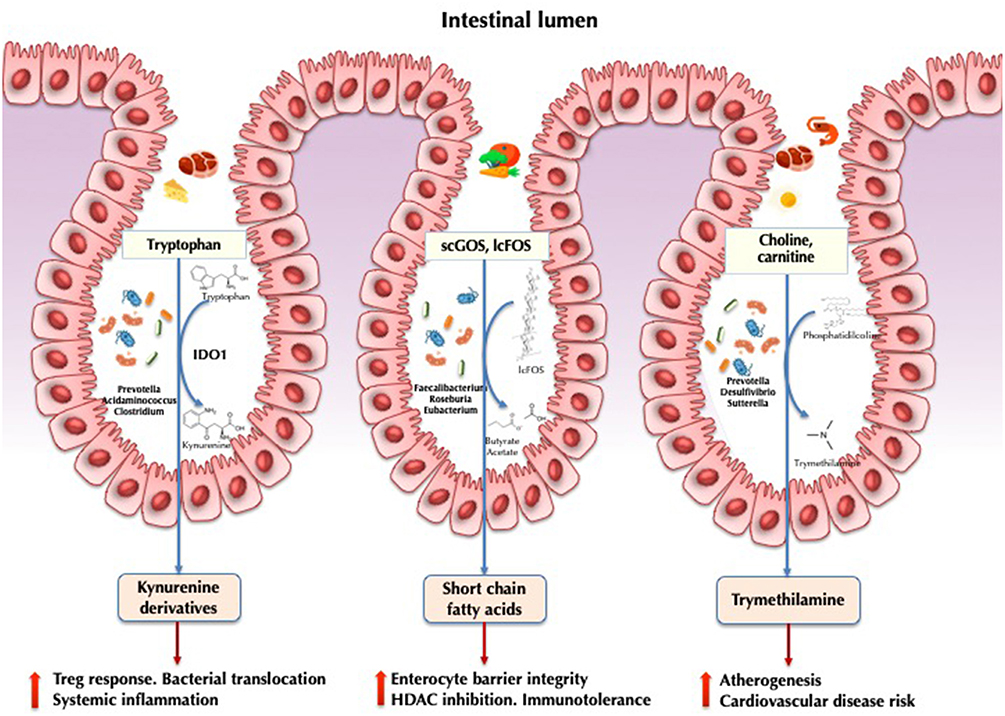
Figure 1. Implications of the gut microbiota in HIV pathogenesis. IDO1, indoleamine 2,3-dioxygenase 1; scGOS, short-chain galactooligosaccharides; lcFOS, long-chain fructooligosaccarides; HDAC, histone deacetylases.
Defining the clinical scope of the changes in gut microbial communities can be challenging because a big proportion of bacteria are dead, dormant, or inactive (33, 34). Expensive and time-consuming techniques are required to measure the proteins and metabolites synthesized by active bacteria. The extent of functional adaptation of microbial communities to the ecological perturbation induced by HIV might influence the different immunologic outcomes achieved during antiretroviral therapy (ART). In fact, HIV infection activates an important fraction of the gut microbiota. Although only 20% of the fecal microbiota is metabolically active in healthy controls, HIV infection is characterized by the activation of up to 50% of microbial communities (35). Among immunological ART responders, the metabolic activity of some taxa (Succinivibrionaceae family) is boosted, acting as anti-inflammatory buffers thanks to the accumulation of proinflammatory mediator. In addition, cannabinoid oleamide and biliverdin (a viral inhibitor) are also accumulated within bacteria and may contribute to health recovery by inhibiting viral replication, stimulating the immune system, and ultimately reducing inflammation. These findings are in sharp contrast to those observed in immunological non-responders whose gut bacteria metabolism is most similar to that of ART-naïve participants. The metabolic activity of their gut bacteria is characterized by the cleavage of the sialic and dolichol components necessary to maintain enterocyte integrity (19).
The Kynurenine Pathway
Indoleamine-2,3-dioxygenase-1 (IDO1) involved in tryptophan catabolism via the kynurenine pathway is correlated with epithelial barrier disruption and bacterial translocation in HIV infection (36). Induction results in the production of kynurenine derivatives with immunosuppressive effects, impairing mucosal immunity, and promoting bacterial translocation and higher mortality (37). In a seminal study, Vujkovic-Cvijin et al. (5) characterized 140 genera significantly correlated with tryptophan catabolism. Some of these taxa were found to encode the genetic machinery that reproduces the same tryptophan catabolism as human IDO1. This finding was further confirmed by metabolomic analysis in gut bacteria via the detection of the kynurenine subproduct 3-hydroxyanthranilate (34). In a subsequent study combining metagenomic and metatranscriptomic data, we showed that HIV-infected individuals exhibited increased anaerobic catabolism of tryptophan via tryptophanase anaerobic fermentation compared with healthy controls (23). This expression was upregulated in the Prevotella, Acidaminococcus, and Clostridium genera. It is likely that the HIV-associated microbiota exerts a strong influence on this critical pathway at the crossroads between metabolism and immunity.
Short-Chain Fatty Acids
Short-chain fatty acids (SCFAs) are the primary fermentation products of gut microbiota from dietary fibers. The most abundantly produced SCFAs include acetate, propionate, and butyrate (38, 39). Butyrate is a regulator of intestinal homeostasis and a modulator of immune cell response. It is involved in the maintenance of enterocyte barrier integrity and mucine production (40), induces transcription of human genes via histone deacetylase inhibition (41), and promotes immunotolerance to commensal bacteria (42). Several studies have demonstrated a decrease in butyrate-producing bacteria, including Roseburia, Coprococcus, Faecalibacterium, and Eubacterium, in both HIV-treated and ART-naïve individuals, in association with altered SCFAs profiles (17, 43). In patients with ulcerative colitis, depletion of both Faecalibacterium prausnitzii and Roseburia intestinalis has been proposed to be the hallmark of dysbiosis (44). It is increasingly accepted that the butyrate synthesis pathway supports intestinal inflammation and represents a potential therapeutic target for interventions aimed at mitigating chronic inflammation (45). Propionate and acetate have been less studied in HIV but have been linked to conferring protection against cardiovascular disease and playing other beneficial roles in other diseases (46).
Trimethylamine-N-Oxide
Trimethylamine-N-oxide (TMAO) is a gut microbiota-dependent choline and carnitine metabolite that is responsible for an increased risk of atherogenesis and cardiovascular disease risk (47), particularly in individuals who consume large quantities of meat and possess a specific microbiome signature with enriched proportions of the genus Prevotella (48). This metabolite has also been associated with atherosclerotic plaque burden in HIV in some (49, 50) but not all (51) studies. A recent cohort study comparing the fecal microbiota of HIV-infected individuals with and without ischemic heart disease showed that high TMAO plasma levels was a marker of cardiovascular heart disease and correlated with the fecal abundance of Phascolarctobacterium, Desulfovibrio, Sutterella, and Faecalibacterium (52).
Microbiota as a Tool for Precision Medicine for HIV
Hopefully, future studies will exploit these connections between microbiota and HIV immunopathogenesis to improve the clinical management of HIV infection. From a diagnostic point of view, one could utilize microbiota to identify individuals at higher risk of HIV acquisition (53–55), to anticipate the responsiveness to pre-exposure prophylaxis strategies with topical antiretroviral drugs (56), and to predict the risk of precancerous anal lesions (57). From a therapeutic point of view, we may gain the ability to manipulate the microbiota to enhance vaccine immunogenicity (58), boost immune recovery after ART initiation (59, 60), and attenuate chronic inflammation and bacterial translocation (61). A number of studies assessing HIV patients' dietary supplementation with prebiotics and probiotics have collectively suggested that dietary supplementation may exert some beneficial immunological effects, particularly in ART-naïve individuals (30, 59, 62–64). However, two recent controlled studies focused on ART-naive (60) and ART-suppressed (65) individuals have failed to detect significant parameters of inflammation, bacterial translocation or immune activation. These findings call into question the utility of these strategies. The first pilot study of fecal microbiota transplantation in HIV failed to demonstrate adequate engraftment of colonoscopy microbiota on the microbiota of the recipients (66). Ongoing studies (NCT02256592 and NCT03329560) are evaluating different modalities of fecal microbiota transplantation. Clinical trials assessing the use of postbiotics—metabolites or cell-wall components released by microbiota—and represent the future landscape of this fascinating field.
Influence of Microbiota in Cancer
Microbiota as a Trigger of Cancer Pathogenesis
Cancer is a multifaceted disease influenced by both genetic and environmental factors. Microorganisms are emerging as one of the contributors to carcinogenesis, and today we know that approximately 20% of the global cancer burden is directly attributable to infectious agents (67). Beyond the neoplasias directly linked to infectious agents, increasing evidence reveals that microbial communities as a whole play a key role in carcinogenesis by altering the balance of host cell proliferation and apoptosis; hindering anti-tumoral immunity; and influencing the metabolism of host-produced factors, ingested food components, and drugs (68, 69).
Barrier failure has been proposed to be the most relevant mechanism for bacterially driven carcinogenesis, resulting in increased host-microbiota interactions (70, 71). The failure of control mechanisms (e.g., barrier defects, immune defects, dysbiosis) is believed to represent the trigger of bacterial-driven carcinogenesis (72), leading to activation of different responses that converge in cell proliferation and cancer development. The microbiome itself represent a functional barrier by suppressing the growth of pathobionts via different mechanisms, including both resource competition and direct interference competition (73). Therefore, dysbiosis has also been associated with cancer (71). Alterations of gut bacteria have been linked to the development of colorrectal cancer (CRC) (74), but also to extraintestinal cancers, including liver (75), breast (76), and lung cancer (77, 78). While lung microbiome investigations are still in their infancy, the lung microbiotas of patients with lung cancer are distinct from those of other patients (e.g., individuals with emphysema) (79). The abundance of several types of bacteria in the lungs—including Granulicatella, Streptococcus, and Veillonella—has been proposed to be a hallmark of lung cancer (80). An association between the abundance of the Koriobacteriaceae family in the lungs and recurrence free survival has been reported (81). Furthermore, the fecal microbiota of individuals with lung cancer is depleted of Bifidobacteria (82), a commensal genus with known anti-tumoral effects. Bifidobacteria appears able to enhance the efficacy of anti-programmed cell death ligand 1 therapy (83).
Microbiota-Associated Pathways Linked to Carcinogenesis
Recent studies of CRC have identified different mechanisms of carcinogenesis. The bacterial driver-passenger model proposes that the colonic mucosa of patients at risk of CRC is colonized by pro-inflammatory bacteria that can produce genotoxins that lead to DNA mutations and increase cell proliferation (“drivers”). These changes facilitate the replacement of the commensal bacteria with opportunistic pathogens (“passengers”) with competitive advantage in this niche, which leads to tumor progression (72). From the 1990s onward, various studies have demonstrated an association between CRC and specific colonic bacterial species, which favor the development of cancer through different pathogenic pathways (Figure 2) (86). Very impressively, Fusobacterium nucleatum and certain co-occurring bacteria have been found not only in primary CRC but also in distant metastases. Antibiotic treatment of mice carrying xenografts of F. nucleatum-positive human CRC slowed tumor growth, demonstrating the causal role of this taxon in oncogenesis (87).
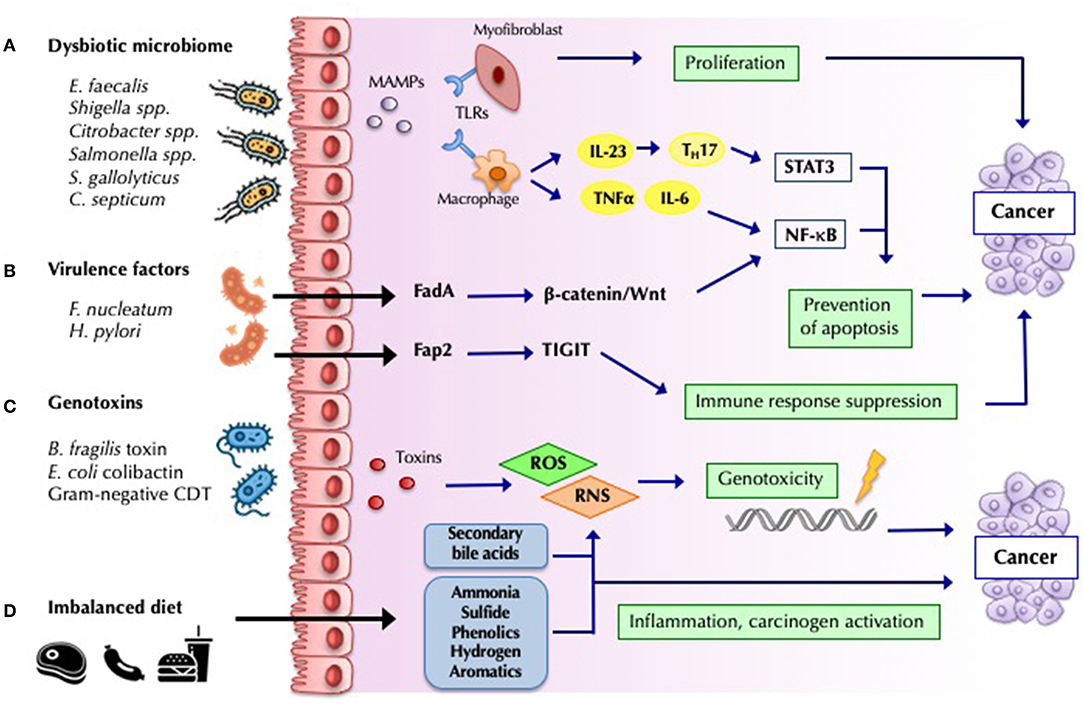
Figure 2. Mechanisms by which bacteria influence cancer development and progression. (A) Barrier loss and increased bacterial translocation engages pattern recognition by Toll-like receptors (TLRs) and activation of innate and adaptive responses. The interleukin-23 (IL-23)-IL-17 axis, IL-6, and tumor necrosis factor-α, lead to chronic inflammation mediated by nuclear factor-κB (NF-κB) and signal transducer and activator of transcription 3 (STAT3) activation, favoring tumor progression (68). (B) Bacterial virulence-factors promote carcinogenesis by engaging specific host pathways, which plays a decisive role in many malignancies. Fusobacterium nucleatum Fad-A binds host E-cadherin on colonic epithelial cells, and triggers Wnt/β-catenin pathway activation, resulting in increased NF-κB, and ultimately in increased tumor growth (84). Other virulence factor such as H.pylori CagA have been widely studied (68). (C) Some microorganisms modulate tumorigenesis through specific toxins, which induce host DNA damage. Cytolethal distending toxin (CDT) produced by Gram-negative bacteria, Bacteroides fragilis toxin and Escherichia coli colibactin constitute some of the most studied toxins identified as potential drivers of CRC (70). (D) Dietary residues determine the composition and metabolic activity of the microbiota. An imbalanced high-fat, high-meat, low-fiber diet, lead to a greater exposition to secondary bile acids, and protein fermentation metabolites (such as ammonia, phenols, sulfides, and nitrosamines), which have inflammatory and carcinogenic effects (85).
Among the carcinogenic mechanisms shown in Figure 2, microbial fermentation products of dietary fiber into SCFAs, including butyrate, propionate, and acetate, with known anti-inflammatory properties (85) likely play a major role. Butyrate is one of the primary sources of energy for enterocytes, and it has been associated with the downregulation of the WNT signaling pathway, inhibition of proliferation and migration of neoplastic cells, and apoptosis induction (88). Butyrate also reinforces mucosal health via Treg-cell activation and IL-10 expression (89). Butyrate producers (e.g., F. prausnitzii, Roseburia, and Bifidobacterium) are depleted in CRC patients (69).
Another mechanism related to the catabolism of dietary precursors strongly influenced by the microbiota is the production of the proatherogenic TMAO. While the implications of this derivative of choline metabolism appear clear for cardiovascular disease (47), this pathway has been rarely studied in the field of oncology. One investigation has suggested that alterations in choline metabolism may be associated with a higher risk of CRC (90).
The Microbiota Modulates the Efficacy and Toxicity of Anticancer Therapies
The microbiota can modulate cancer initiation and progression, but it might also influence response to therapy and treatment-related toxicity (91). First, the bioavailability of many oral drugs depends on their biotransformation in the gut by local microbiota and may also indirectly affect the metabolism of systemically delivered drugs via the regulation of xenobiotic metabolism in distant organs such as the liver (92). Second, the immune response plays an essential role in anticancer activity, and the microbiome might affect chemotherapy response via this mechanism. There is evidence that oxaliplatin and cyclophosphamide activity is modulated by gut microbiota by priming myeloid cells for high-level reactive oxygen species (ROS) production (resulting in DNA damage) and enhancing T-helper cell-mediated anti-tumor responses, respectively (93, 94). Chemotherapy-related adverse events can also be managed via microbiome modulation. For example, diarrhea caused by irinotecan toxicity, which is mediated by microbial-produced β-glucuronidases, can be regulated by targeting microbial metabolism (95). The microbiota might also play a role in response and toxicity to radiotherapy. Radiation-related mucosal injury is associated with changes in the microbiome, and germ-free mice have been shown to be resistant to radiation enteritis (91). Lastly, recent pioneering studies have yielded paradigm shifts in our knowledge of the interactions between gut bacteria and cancer therapy. The gut microbiome has been shown to modulate the anti-tumor efficacy in pre-clinical models of various chemotherapies (93, 94) and immunotherapeutic agents (96–99), including antibodies against cytotoxic T lymphocyte-associated antigen 4 (CTLA4) and anti-programmed cell death protein 1 (PD-1) (92). Individuals with metastatic melanoma responding to anti-PD-1 were enriched with Faecalibacterium genus in intestinal microbiota; non-responding individuals had a higher abundance of Bacteroidales (97). Another study found an abundance of Bifidobacterium in responding individuals; Ruminococcus obecum and Roseburia intestinalis were associated with a lack of responsiveness (99). The role of the microbiota on treatment response is further supported by striking data showing poorer survival outcomes on patients with metastatic non-small cell lung cancer or renal cell carcinoma receiving antibiotics just before or just after initiation of treatment with immune checkpoint blockade (100). Converging data support a robust interaction between specific bacteria and the systemic immune response (97–99). In subjects with non-small cell lung cancer specific memory CD4+ and CD8+ T-cells against Akkermansia muciniphila predicted a longer progression-free survival (98). In subjects with melanoma the abundance of Faecalibacterium genes positively correlated with the with a higher frequency of cytotoxic CD8 T-cell infiltration in the tumor bed. Similarly, in mice intratumoral CD8+ T-cell infiltration after anti-PD-L1 treatment correlated the microbiota composition (100).
Is It Possible to Exploit the Microbiome to Improve Clinical Outcomes in Oncology?
Emerging evidence suggests that altering the microbiota might represent a therapeutic avenue for cancer management (101). Modulation of gut microbiota in preclinical models has been shown to enhance therapeutic response (102). Landmark studies have demonstrated that fecal microbiota transplantation from cancer patients who had responded to anti-PD-1 therapy improved the effects of PD-1 blockade in germ-free or antibiotic-treated mice (97–99). Several trials involving patients on immune checkpoint blockade undergoing fecal microbiota transplant are currently underway, but definitive data are lacking (91). Probiotics have been shown to boost anti-tumor immune responses in mice, but their off-trial use in humans is discouraged because there is still insufficient evidence to implement dietary guidelines or prebiotic administration in the setting of cancer therapy (91). Manipulation of the microbiome in cancer patients might result in novel indications for this intervention, as illustrated by the efficacy demonstrated in the first case series of patients with refractory immune checkpoint inhibitor-associated colitis successfully treated with fecal microbiota transplantation (103). Nearly 40 clinical trials assessing gut microbiota modulation in cancer are ongoing (91). The results of these investigations will inform best strategies and define indications of this therapeutic approach to improve clinical outcomes in oncology.
HIV, Cancer, and the Microbiota: Converging Pathways and Research Avenues
Can we learn anything from microbiome studies of HIV-positive patients that may be applicable to cancer? First, the vast majority of mechanistic studies regarding the influence of the microbiome in HIV are cross sectional in nature (104). The well-known limitations of these studies are magnified by underappreciated confounding factors related to microbiota studies. For example, it took several years for the field to recognize that the increased abundance of Prevotella spp. observed in the first studies of HIV-infected individuals (5, 7, 16, 18) was confounded by the lower proportion of men who had sex with men in the control groups (24). Given the particular clinical profile of patients undergoing anticancer treatment, these confounders may be even more pronounced in patients with cancer.
Several pathways strongly influenced by microbiota appear to affect pathogenic mechanisms present in different conditions. Gut microbial signatures associated with clinical outcomes in both HIV and cancer and the putative mechanisms are summarized in Table 1. For example, the major butyrate producers Faecalibacterium prausnitzii and Roseburia intestinalis are depleted in subjects with HIV (17, 43), intestinal bowel disease (44), and CRC (69). Because butyrate production has been shown to promote Treg-cell activation and IL-10 expression (89, 105), the butyrate synthesis pathway is a potential therapeutic target for conditions in which enterocyte barrier integrity and mucosal tolerogenic immune responses are implicated. The kynurenine pathway has been also implicated in both HIV (5) and cancer pathogenesis (129). IDO1 is frequently overexpressed in many malignancies, where it correlates with poor survival and prognosis. Besides its role in immunosuppression, IDO1 promotes cancer development by inducing inflammatory neovascularization, interacting with checkpoint inhibitors, and modulating gut microbiota (130). While it is still too soon to draw conclusions about the therapeutic potential of IDO1 inhibitors for HIV disease and cancer, an increasing number of IDO1 inhibitors are currently in preclinical development or under evaluation in clinical trials (131, 132).
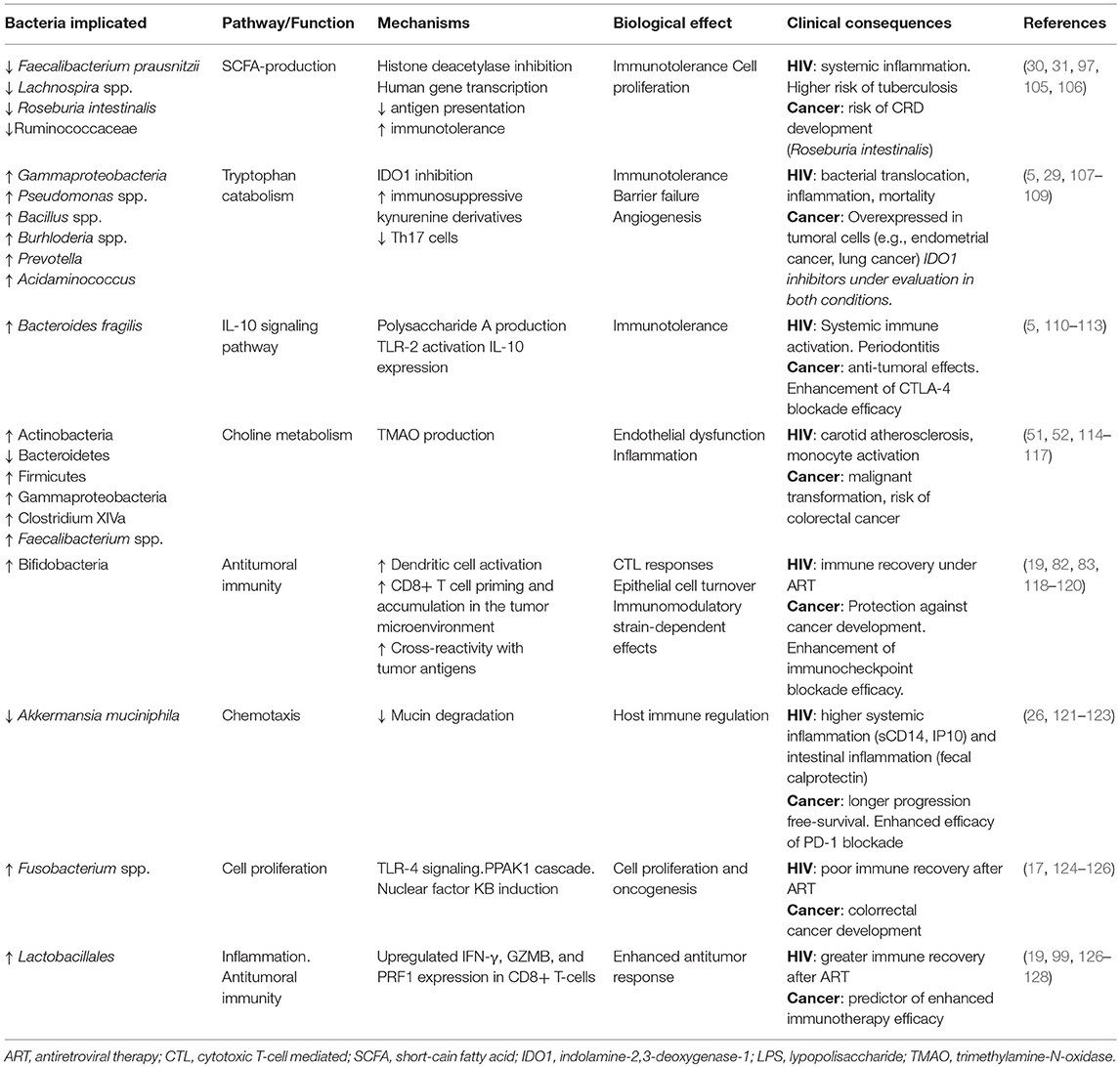
Table 1. Gut microbial signatures associated with clinical outcomes in both HIV and cancer and putative mechanisms.
Analyzing gut microbiota from a functional perspective will be crucial to advancing knowledge about the role of the microbiome in the pathogenesis of cancer and understanding its interactions with immunotherapy. While bifidobacteria have not typically appeared to be compositionally relevant in most HIV studies reliant on 16S sequencing, its functional importance is clear when we assess the functional level of the microbiota. For example, while using 16S sequencing we only demonstrated modest changes in gut microbiota structure after a short prebiotic intervention, which did not include changes in the abundance of bifidobacteria (30). Using proteomics we demonstrated a 100-fold increase in the activity of the Bifidobacteriaceae family, which strongly correlated with the thymic output, a surrogate marker of the ability of the immune system to renew the T-cell pool (118). In a study aimed at identifying the bacterial biomarkers of precancerous anal lesions in HIV, Bifidobacterium spp. were also the most predictive taxa in stools of anal dysplasia (57). Because Bifidobacterium spp. enhance anti-tumor immunity and anti-PD-L1 efficacy (83), it is likely that the importance of this genus will remain underappreciated until researchers evaluate the functional level of the microbiota.
While the microbiome agenda is expanding, it is still unclear whether we can effectively manipulate the microbiome to treat HIV and cancer. Pilot studies analyzing the effects of fecal microbiota transplantation will provide powerful indications of our ability to modify clinical outcomes via microbiota manipulation. In the coming years, we look forward to learning to exploit the potential of the microbiota for precision medicine (e.g., predicting treatment responsiveness or toxicities). Gaining further insights into the mechanisms by which the microbiota influences HIV disease and cancer will help to leverage the microbiome to develop interventions for both conditions.
Author Contributions
SS-V conceived the paper. SS-V, SH, and JM-S draft the first version of the manuscript. All authors reviewed and approved the final version.
Funding
This work was supported by the Instituto de Salud Carlos III (Plan Estatal de I+D+i 2013–2016, projects PI15/00345 and PI18/00154, the Fundación Asociación Española contra el Cáncer within the ERA (European Research Area)-NET aligning national/regional translational cancer research programmes and activities (TRANSCAN)-2 program (Grant Number AC17/00019), and cofinanced by the European Development Regional Fund A way to achieve Europe (ERDF). Icons in figures were downloaded from www.flaticon.com.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. (2013) 39:3–45. doi: 10.1016/j.immuni.2013.10.001
2. Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. (2004) 200:749–59. doi: 10.1084/jem.20040874
3. Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. (2010) 6:e1001052. doi: 10.1371/journal.ppat.1001052
4. Somsouk M, Estes JD, Deleage C, Dunham RM, Albright R, Inadomi JM, et al. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS. (2015) 29:43–51. doi: 10.1097/QAD.0000000000000511
5. Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. (2013) 5:193ra91. doi: 10.1126/scitranslmed.3006438
6. Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. (2015) 211:19–27. doi: 10.1093/infdis/jiu409
7. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. (2015) 8:760–72. doi: 10.1038/mi.2014.107
8. Nazli A, Chan O, Dobson-Belaire WN, Ouellet M, Tremblay MJ, Gray-Owen SD, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. (2010) 6:e1000852. doi: 10.1371/journal.ppat.1000852
9. Hirao LA, Grishina I, Bourry O, Hu WK, Somrit M, Sankaran-Walters S, et al. Early mucosal sensing of SIV infection by paneth cells induces IL-1β production and initiates gut epithelial disruption. PLoS Pathog. (2014) 10:e1004311. doi: 10.1371/journal.ppat.1004311
10. Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS. (2016) 11:182–90. doi: 10.1097/COH.0000000000000234
11. Strategies for Management of Antiretroviral Therapy (SMART) Study Group, El-Sadr WM, Lundgren J, Neaton JD, Gordin F, Abrams D, et al. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med. (2006) 355:2283–96. doi: 10.1056/NEJMoa062360
12. INSIGHT START Study Group, Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, et al. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med. (2015) 373:795–807. doi: 10.1056/NEJMoa1506816
13. Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. (2008) 5:e203. doi: 10.1371/journal.pmed.0050203
14. Tenorio AR, Chan ES, Bosch RJ, Macatangay BJ, Read SW, Yesmin S, et al. Rifaximin has a marginal impact on microbial translocation, T-cell activation and inflammation in HIV-positive immune non-responders to antiretroviral therapy - ACTG A5286. J Infect Dis. (2015) 211:780–90. doi: 10.1093/infdis/jiu515
15. Angelidou K, Hunt PW, Landay AL, Wilson CC, Rodriguez B, Deeks SG, et al. Changes in inflammation but not in T-Cell activation precede non-AIDS-defining events in a case-control study of patients on long-term antiretroviral therapy. J Infect Dis. (2018) 218:239–48. doi: 10.1093/infdis/jix666
16. Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. (2013) 14:329–39. doi: 10.1016/j.chom.2013.08.006
17. McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. (2013) 1:26. doi: 10.1186/2049-2618-1-26
18. Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. (2014) 7:983–94. doi: 10.1038/mi.2013.116
19. Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Bargiela R, et al. Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine. (2016) 8:203–16. doi: 10.1016/j.ebiom.2016.04.033
20. Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, et al. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe. (2016) 19:323–35. doi: 10.1016/j.chom.2016.02.010
21. Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. (2012) 151:253–66. doi: 10.1016/j.cell.2012.09.024
22. Ortiz AM, Flynn JK, DiNapoli SR, Vujkovic-Cvijin I, Starke CE, Lai SH, et al. Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat Med. (2018) 24:1313–6. doi: 10.1038/s41591-018-0132-5
23. Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. (2015) 29:2409–18. doi: 10.1097/QAD.0000000000000869
24. Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. (2016) 5:135–46. doi: 10.1016/j.ebiom.2016.01.032
25. Tuddenham SA, Koay WLA, Zhao N, White JR, Ghanem KG, Sears CL, et al. The impact of HIV infection on gut microbiota alpha-diversity: an individual level meta-analysis. Clin Infect Dis. (2019). doi: 10.1093/cid/ciz258. [Epubh ahead of print].
26. Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. (2014) 10:e1003829. doi: 10.1371/journal.ppat.1003829
27. Dillon SM, Kibbie J, Lee EJ, Guo K, Santiago ML, Austin GL, et al. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS. (2017) 31:511–21. doi: 10.1097/QAD.0000000000001366
28. Neff CP, Krueger O, Xiong K, Arif S, Nusbacher N, Schneider JM, et al. Fecal microbiota composition drives immune activation in HIV-infected individuals. EBioMedicine. (2018) 30:192–202. doi: 10.1016/j.ebiom.2018.03.024
29. Vázquez-Castellanos JF, Serrano-Villar S, Jiménez-Hernández N, Soto Del Rio MD, Gayo S, Rojo D, et al. Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J. (2018) 12:1964–76. doi: 10.1038/s41396-018-0151-8
30. Serrano-Villar S, Vázquez-Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando-Martínez S, et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. (2017) 10:1279–93. doi: 10.1038/mi.2016.122
31. Guillén Y, Noguera-Julian M, Rivera J, Casadellà M, Zevin AS, Rocafort M, et al. Low nadir CD4+ T-cell counts predict gut dysbiosis in HIV-1 infection. Mucosal Immunol. (2018):119–29. doi: 10.1038/s41385-018-0083-7
32. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. (2012) 336:1255–62. doi: 10.1126/science.1224203
33. Rojo D, Méndez-García C, Raczkowska BA, Bargiela R, Moya A, Ferrer M, et al. Exploring the human microbiome from multiple perspectives: factors altering its composition and function. FEMS Microbiol Rev. (2017) 41:453–78. doi: 10.1093/femsre/fuw046
34. Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Sainz T, et al. HIV infection results in metabolic alterations in the gut microbiota different from those induced by other diseases. Sci Rep. (2016) 6:26192. doi: 10.1038/srep26192
35. Serrano-Villar S, Moreno S, Ferrer M. The functional consequences of the microbiome in HIV: insights from metabolomic studies. Curr Opin HIV AIDS. (2018) 13:88–94. doi: 10.1097/COH.0000000000000430
36. Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. (2010) 2:32ra36. doi: 10.1126/scitranslmed.3000632
37. Byakwaga H, Boum Y, Huang Y, Muzoora C, Kembabazi A, Weiser SD, et al. The kynurenine pathway of tryptophan catabolism, CD4+ T cell recovery and mortality in HIV-infected ugandans initiating ART. J Infect Dis. (2014) 210:383–91. doi: 10.1093/infdis/jiu115
38. Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. (2010) 12:304–14. doi: 10.1111/j.1462-2920.2009.02066.x
39. Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. (2014) 8:1323–35. doi: 10.1038/ismej.2014.14
40. Xiong H, Guo B, Gan Z, Song D, Lu Z, Yi H, et al. Butyrate upregulates endogenous host defense peptides to enhance disease resistance in piglets via histone deacetylase inhibition. Sci Rep. (2016) 6:27070. doi: 10.1038/srep27070
41. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. (2014) 111:2247–52. doi: 10.1073/pnas.1322269111
42. Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, et al. Recovery of the gut microbiome following fecal microbiota transplantation. MBio. (2014) 5:e00893–14. doi: 10.1128/mBio.00893-14
43. Serrano-Villar S, Zhou Y, Rodgers AJ, Moreno S. Different impact of raltegravir versus efavirenz on CD4/CD8 ratio recovery in HIV-infected patients. J Antimicrob Chemother. (2017) 72:235–9. doi: 10.1093/jac/dkw375
44. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. (2014) 63:1275–83. doi: 10.1136/gutjnl-2013-304833
45. Shen S, Wong CH. Bugging inflammation: role of the gut microbiota. Clin Transl Immunol. (2016) 5:e72. doi: 10.1038/cti.2016.12
46. Aldunate M, Srbinovski D, Hearps AC, Latham CF, Ramsland PA, Gugasyan R, et al. Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front Physiol. (2015) 6:164. doi: 10.3389/fphys.2015.00164
47. Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. (2013) 368:1575–84. doi: 10.1056/NEJMoa1109400
48. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. (2013) 19:576–85. doi: 10.1038/nm.3145
49. Miller PE, Haberlen SA, Brown TT, Margolick JB, DiDonato JA, Hazen SL, et al. Intestinal microbiota-produced trimethylamine-N-oxide and its association with coronary stenosis and HIV serostatus. J Acquir Immune Defic Syndr. (2016) 72:114–18. doi: 10.1097/QAI.0000000000000937
50. Shan Z, Clish CB, Hua S, Scott JM, Hanna DB, Burk RD, et al. Gut microbial-related choline metabolite trimethylamine-N-oxide is associated with progression of carotid artery atherosclerosis in HIV infection. J Infect Dis. (2018) 218:1474–9. doi: 10.1093/infdis/jiy356
51. Missailidis C, Neogi U, Stenvinkel P, Trøseid M, Nowak P, Bergman P. The microbial metabolite trimethylamine-N-oxide in association with inflammation and microbial dysregulation in three HIV cohorts at various disease stages. AIDS. (2018) 32:1589–98. doi: 10.1097/QAD.0000000000001813
52. Kehrmann J, Menzel J, Saeedghalati M, Obeid R, Schulze C, Holzendorf V, et al. Gut microbiota in human immunodeficiency virus-infected individuals linked to coronary heart disease. J Infect Dis. (2019) 219:497–508. doi: 10.1093/infdis/jiy524
53. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. (2015) 42:965–76. doi: 10.1016/j.immuni.2015.04.019
54. Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. (2017) 46:29–37. doi: 10.1016/j.immuni.2016.12.013
55. Zozaya M, Ferris MJ, Siren JD, Lillis R, Myers L, Nsuami MJ, et al. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome. (2016) 4:16. doi: 10.1186/s40168-016-0161-6
56. Klatt NR, Cheu R, Birse K, Zevin AS, Perner M, Noël-Romas L, et al. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science. (2017) 356:938–45. doi: 10.1126/science.aai9383
57. Serrano-Villar S, Vásquez-Domínguez E, Pérez-Molina JA, Sainz T, de Benito A, Latorre A, et al. HIV, HPV, and microbiota: partners in crime? AIDS. (2017) 31:591–4. doi: 10.1097/QAD.0000000000001352
58. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. (2014) 41:478–92. doi: 10.1016/j.immuni.2014.08.009
59. Gori A, Rizzardini G, Van't Land B, Amor KB, van Schaik J, Torti C, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol. (2011) 4:1–10. doi: 10.1038/mi.2011.15
60. Serrano-Villar S, de Lagarde M, Vázquez-Castellanos J, Vallejo A, Bernadino JI, Madrid N, et al. Effects of immunonutrition in advanced human immunodeficiency virus disease: a randomized placebo-controlled clinical trial (Promaltia Study). Clin Infect Dis. (2019) 68:120–30. doi: 10.1093/cid/ciy414
61. Villar-García J, Hernández JJ, Güerri-Fernández R, González A, Lerma E, Guelar A, et al. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients. JAIDS J Acquir Immune Defic Syndr. (2015) 68:256–63. doi: 10.1097/QAI.0000000000000468
62. Asmuth DM, Ma ZM, Albanese A, Sandler NG, Devaraj S, Knight TH, et al. Oral serum-derived bovine immunoglobulin improves duodenal immune reconstitution and absorption function in patients with HIV enteropathy. AIDS. (2013) 27:2207–17. doi: 10.1097/QAD.0b013e328362e54c
63. Cahn P, Ruxrungtham K, Gazzard B, Diaz RS, Gori A, Kotler DP, et al. The immunomodulatory nutritional intervention NR100157 reduced CD4+ T-cell decline and immune activation: a 1-year multicenter randomized controlled double-blind trial in HIV-infected persons not receiving antiretroviral therapy (The BITE Study). Clin Infect Dis. (2013) 57:139–46. doi: 10.1093/cid/cit171
64. Arnbjerg CJ, Vestad B, Hov JR, Pedersen KK, Jespersen S, Johannesen HH, et al. Effect of Lactobacillus rhamnosus GG supplementation on intestinal inflammation assessed by PET/MRI scans and gut microbiota composition in HIV-infected individuals. JAIDS J Acquir Immune Defic Syndr. (2018) 78:450–7. doi: 10.1097/QAI.0000000000001693
65. Overton ET, Yeh E, Presti R, et al. Assessing the probiotic effect in treated HIV: results of ACTG A5350. Abstract #35. In: CROI 2019. Conference on Retroviruses and Opportunistic Infections. Seattle (2019).
66. Vujkovic-Cvijin I, Rutishauser RL, Pao M, Hunt PW, Lynch SV, McCune JM, et al. Limited engraftment of donor microbiome via one-time fecal microbial transplantation in treated HIV-infected individuals. Gut Microbes. (2017) 8:440–50. doi: 10.1080/19490976.2017.1334034
67. Goodman B, Gardner H. The microbiome and cancer. J Pathol. (2018) 244:667–76. doi: 10.1002/path.5047
69. Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. (2014) 20:908–22. doi: 10.3748/wjg.v20.i4.908
70. Brennan CA, Garrett WS. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol. (2016) 70:395–411. doi: 10.1146/annurev-micro-102215-095513
71. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. (2013) 13:800–12. doi: 10.1038/nrc3610
72. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. (2012) 10:575–82. doi: 10.1038/nrmicro2819
73. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. (2014) 124:4212–8. doi: 10.1172/JCI72333
74. Lin C, Cai X, Zhang J, Wang W, Sheng Q, Hua H, et al. Role of gut microbiota in the development and treatment of colorectal cancer. Digestion. (2018). doi: 10.1159/000494052. [Epub ahead of print].
75. Mima K, Nakagawa S, Sawayama H, Ishimoto T, Imai K, Iwatsuki M, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. (2017) 402:9–15. doi: 10.1016/j.canlet.2017.05.001
76. Kwa M, Plottel CS, Blaser MJ, Adams S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J Natl Cancer Inst. (2016) 108:djw029. doi: 10.1093/jnci/djw029
77. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. (2019) 176:998–1013.e16. doi: 10.1016/j.cell.2018.12.040
78. Zhang WQ, Zhao SK, Luo JW, Dong XP, Hao YT, Li H, et al. Alterations of fecal bacterial communities in patients with lung cancer. Am J Transl Res. (2018) 10:3171–85.
79. Liu Y, O'Brien JL, Ajami NJ, Scheurer ME, Amirian ES, Armstrong G, et al. Lung tissue microbial profile in lung cancer is distinct from emphysema. Am J Cancer Res. (2018) 8:1775–87. Available online at: http://www.ajcr.us/files/ajcr0073906.pdf
80. Mur LA, Huws SA, Cameron SJ, Lewis PD, Lewis KE. Lung cancer: a new frontier for microbiome research and clinical translation. Ecancermedicalscience. (2018) 12:866. doi: 10.3332/ecancer.2018.866
81. Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol Biomarkers Prev. (2019) 28:731–40. doi: 10.1158/1055-9965.EPI-18-0966
82. Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, et al. Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol. (2019) 9:112. doi: 10.3389/fcimb.2019.00112
83. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. (2015) 350:1084–9. doi: 10.1126/science.aac4255
84. Kelly D, Yang L, Pei Z. Gut microbiota, fusobacteria, and colorectal cancer. Diseases. (2018) 6:109. doi: 10.3390/diseases6040109
85. O'Keefe SJ. Diet, microorganisms, and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. (2016) 13:691–706. doi: 10.1038/nrgastro.2016.165
86. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. (2019) 25:679–86. doi: 10.1038/s41591-019-0406-6
87. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. (2017) 358:1443–8. doi: 10.1126/science.aal5240
88. Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. (2019) 11:11. doi: 10.1186/s13073-019-0621-2
89. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
90. Manor O, Zubair N, Conomos MP, Xu X, Rohwer JE, Krafft CE, et al. A multi-omic association study of trimethylamine N-oxide. Cell Rep. (2018) 24:935–46. doi: 10.1016/j.celrep.2018.06.096
91. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. (2019) 20:e77–91. doi: 10.1016/S1470-2045(18)30952-5
92. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. (2017) 17:271–85. doi: 10.1038/nrc.2017.13
93. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. (2013) 342:967–70. doi: 10.1126/science.1240527
94. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. (2013) 342:971–6. doi: 10.1126/science.1240537
95. Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. (2010) 330:831–5. doi: 10.1126/science.1191175
96. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. (2015) 350:1079–84. doi: 10.1126/science.aad1329
97. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
98. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. (2018) 359:91–7. doi: 10.1126/science.aan3706
99. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. (2018) 359:104–8. doi: 10.1126/science.aao3290
100. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. (2018) 29:1437–44. doi: 10.1093/annonc/mdy103
101. Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. (2018) 68:gutjnl-2018–317220. doi: 10.1136/gutjnl-2018-317220
102. Derosa L, Routy B, Kroemer G, Zitvogel L. The intestinal microbiota determines the clinical efficacy of immune checkpoint blockers targeting PD-1/PD-L1. Oncoimmunology. (2018) 7:e1434468. doi: 10.1080/2162402X.2018.1434468
103. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont H, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. (2018) 24:1804–8. doi: 10.1038/s41591-018-0238-9
104. Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS. (2016) 30:2737–51. doi: 10.1097/QAD.0000000000001289
105. Segal LN, Clemente JC, Li Y, Ruan C, Cao J, Danckers M, et al. Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe. (2017) 21:530–7.e4. doi: 10.1016/j.chom.2017.03.003
106. Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. (2017) 15:465–78. doi: 10.1038/nrmicro.2017.44
107. Ino K, Yoshida N, Kajiyama H, Shibata K, Yamamoto E, Kidokoro K, et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br J Cancer. (2006) 95:1555–61. doi: 10.1038/sj.bjc.6603477
108. Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, et al. Differential expression and significance of PD-L1, IDO-1, and B7-H4 in human lung cancer. Clin Cancer Res. (2017) 23:370–8. doi: 10.1158/1078-0432.CCR-16-0150
109. Lu J, Liu X, Liao YP, Wang X, Ahmed A, Jiang W, et al. Breast cancer chemo-immunotherapy through liposomal delivery of an immunogenic cell death stimulus plus interference in the IDO-1 pathway. ACS Nano. (2018) 12:11041–61. doi: 10.1021/acsnano.8b05189
110. Robles AI, Cooks T, Vega-Valle E, Vetizou M, Rose U, Miyanaga A, et al. Abstract PR07: role of the microbiota in inflammation and lung cancer. Clin Cancer Res. (2018) 24:PR07. doi: 10.1158/1557-3265.AACRIASLC18-PR07
111. Rams TE, Andriolo M, Feik D, Abel SN, McGivern TM, Slots J. Microbiological study of HIV-related periodontitis. J Periodontol. (1991) 62:74–81. doi: 10.1902/jop.1991.62.1.74
112. McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. (2014) 142:24–31. doi: 10.1111/imm.12231
113. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. (2008) 453:620–5. doi: 10.1038/nature07008
114. Bae S, Ulrich CM, Neuhouser ML, Malysheva O, Bailey LB, Xiao L, et al. Plasma choline metabolites and colorectal cancer risk in the Women's Health Initiative Observational Study. Cancer Res. (2014) 74:7442–52. doi: 10.1158/0008-5472.CAN-14-1835
115. Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. (2011) 11:835–48. doi: 10.1038/nrc3162
116. Guertin KA, Li XS, Graubard BI, Albanes D, Weinstein SJ, Goedert JJ, et al. Serum trimethylamine N-oxide, carnitine, choline, and betaine in relation to colorectal cancer risk in the alpha tocopherol, beta carotene cancer prevention study. cancer epidemiol. Biomarkers Prev. (2017) 26:945–52. doi: 10.1158/1055-9965.EPI-16-0948
117. Haissman JM, Haugaard AK, Ostrowski SR, Berge RK, Hov JR, Trøseid M, et al. Microbiota-dependent metabolite and cardiovascular disease marker trimethylamine-N-oxide (TMAO) is associated with monocyte activation but not platelet function in untreated HIV infection. BMC Infect Dis. (2017) 17:445. doi: 10.1186/s12879-017-2547-x
118. Deusch S, Serrano-Villar S, Rojo D, Martínez-Martínez M, Bargiela R, Vázquez-Castellanos JF, et al. Effects of HIV, ART, and prebiotics on the active fraction of the gut microbiota. AIDS. (2018) 32:1229–37. doi: 10.1097/QAD.0000000000001831
119. Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. (2017) 8:2345. doi: 10.3389/fmicb.2017.02345
120. Bessell CA. Abstract B060: commensal bacteria Bifidobacterium stimulates an antitumor response via cross-reactivity. Microb Metabol. (2019) 2019:B060. doi: 10.1158/2326-6074.CRICIMTEATIAACR18B060
121. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. (2013) 110:9066–71. doi: 10.1073/pnas.1219451110
122. Wu W, Lv L, Shi D, Ye J, Fang D, Guo F, et al. Protective effect of akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front Microbiol. (2017) 8:1804. doi: 10.3389/fmicb.2017.01804
123. Rocafort M, Noguera-Julian M, Rivera J, Pastor L, Guillén Y, Langhorst J, et al. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome. (2019) 7:73. doi: 10.1186/s40168-019-0687-5
124. Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, et al. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. (2017) 8:31802–14. doi: 10.18632/oncotarget.15992
125. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor–κB, and up-regulating expression of MicroRNA-21. Gastroenterology. (2017) 152:851–66.e24. doi: 10.1053/j.gastro.2016.11.018
126. Lee SC, Chua LL, Yap SH, Khang TF, Leng CY, Raja Azwa RI, et al. Enrichment of gut-derived Fusobacterium is associated with suboptimal immune recovery in HIV-infected individuals. Sci Rep. (2018) 8:14277. doi: 10.1038/s41598-018-32585-x
127. Pérez-Santiago J, Gianella S, Massanella M, Spina CA, Karris MY, Var SR, et al. Gut lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. (2013) 27:1921–31. doi: 10.1097/QAD.0b013e3283611816
128. Gui QF, Lu HF, Zhang CX, Xu ZR, Yang YH. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet Mol Res. (2015) 14:5642–51. doi: 10.4238/2015.May.25.16
129. Bishnupuri KS, Alvarado DM, Khouri AN, Shabsovich M, Chen B, Dieckgraefe BK, et al. IDO1 and kynurenine pathway metabolites activate PI3K-Akt signaling in the neoplastic colon epithelium to promote cancer cell proliferation and inhibit apoptosis. Cancer Res. (2019) 79:1138–50. doi: 10.1158/0008-5472.CAN-18-0668
130. Liu M, Wang X, Wang L, Ma X, Gong Z, Zhang S, et al. Targeting the IDO1 pathway in cancer: from bench to bedside. J Hematol Oncol. (2018) 11:100. doi: 10.1186/s13045-018-0644-y
131. Prendergast GC, Malachowski WP, DuHadaway JB, Muller AJ. Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. (2017) 77:6795–811. doi: 10.1158/0008-5472.CAN-17-2285
Keywords: HIV, cancer, microbiota, immunotherapy, dysbiosis
Citation: Herrera S, Martínez-Sanz J and Serrano-Villar S (2019) HIV, Cancer, and the Microbiota: Common Pathways Influencing Different Diseases. Front. Immunol. 10:1466. doi: 10.3389/fimmu.2019.01466
Received: 17 April 2019; Accepted: 11 June 2019;
Published: 27 June 2019.
Edited by:
Mirko Paiardini, Emory University School of Medicine, United StatesReviewed by:
Jason Brenchley, National Institute of Allergy and Infectious Diseases (NIAID), United StatesAndrew S. Neish, Emory University, United States
Copyright © 2019 Herrera, Martínez-Sanz and Serrano-Villar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergio Serrano-Villar, c2VyZ2lvLnNlcnJhbm9Ac2FsdWQubWFkcmlkLm9yZw==
†These authors have contributed equally to this work
 Sabina Herrera
Sabina Herrera Javier Martínez-Sanz
Javier Martínez-Sanz Sergio Serrano-Villar
Sergio Serrano-Villar