- Consejo Nacional de Investigaciones Científicas y Técnicas (UNSAM-CONICET), Instituto de Investigaciones Biotecnológicas Dr. Rodolfo A. Ugalde, Universidad Nacional de San Martín, Buenos Aires, Argentina
Pathogenic microorganisms confront several proteolytic events in the molecular interplay with their host, highlighting that proteolysis and its regulation play an important role during infection. Microbial inhibitors, along with their target endogenous/exogenous enzymes, may directly affect the host's defense mechanisms and promote infection. Omp19 is a Brucella spp. conserved lipoprotein anchored by the lipid portion in the Brucella outer membrane. Previous work demonstrated that purified unlipidated Omp19 (U-Omp19) has protease inhibitor activity against gastrointestinal and lysosomal proteases. In this work, we found that a Brucella omp19 deletion mutant is highly attenuated in mice when infecting by the oral route. This attenuation can be explained by bacterial increased susceptibility to host proteases met by the bacteria during establishment of infection. Omp19 deletion mutant has a cell division defect when exposed to pancreatic proteases that is linked to cell-cycle arrest in G1-phase, Omp25 degradation on the cell envelope and CtrA accumulation. Moreover, Omp19 deletion mutant is more susceptible to killing by macrophage derived microsomes than wt strain. Preincubation with gastrointestinal proteases led to an increased susceptibility of Omp19 deletion mutant to macrophage intracellular killing. Thus, in this work, we describe for the first time a physiological function of B. abortus Omp19. This activity enables Brucella to better thrive in the harsh gastrointestinal tract, where protection from proteolytic degradation can be a matter of life or death, and afterwards invade the host and bypass intracellular proteases to establish the chronic infection.
Introduction
The intestinal mucosa is the largest interface between the external environment and the tissues of the human body. The first line of defense in the gastrointestinal tract is in the lumen, where microorganisms are degraded in a non-specific fashion by pH and gastric, pancreatic and biliary secretions. Pathogenic microorganisms confront several proteolytic events in the molecular interplay with their host, therefore proteolysis and its regulation play an important role during infection. Microbes synthetize protease inhibitors to control endogenous proteases. Some inhibitors can also interact with exogenous peptidases produced by other species and thus may directly affect host's defense mechanisms (1). Few works in the literature show the importance of bacterial protease inhibitors activity against host-proteases (2–4). Our hypothesis is that pathogenic bacteria synthesize protease inhibitors to evade the antimicrobial activity from host's proteases.
In our laboratory, we have been working on the use of a conserved Brucella spp. protein devoid of its lipid moiety called U-Omp19 as a vaccine against brucellosis (5–7). Omp19 has significant sequence identity with bacterial protease inhibitors from I38 family. Remarkably, recombinant U-Omp19 inhibits gastrointestinal and lysosomal proteases (8, 9). However, the physiological function of Omp19 in Brucella is still unknown.
Brucellosis is a worldwide re-emerging zoonotic disease that is transmitted from domestic and wild animals to humans. The human disease, mostly caused by Brucella abortus and B. melitensis, represents an important cause of morbidity worldwide whereas animal brucellosis is associated with serious economic losses caused mainly by elicited abortions and infertility (10, 11).
Oral infection is one of the principal ways of brucellosis transmission. Animals usually lick tissues from abortions or ingest contaminated pasture and humans acquire often the disease by consumption of infected, unpasteurized dairy products (10, 12–16). Few virulence factors required for food-borne infection by Brucella have been described: Urease and cholylglycine hydrolase that confer resistance to gastric acidity and bile salts, respectively (17, 18). Once inside the host, Brucella disseminate via infected phagocytic cells to different tissues and organs, developing foci of infection, surviving intracellularly and leading to a chronic disease (19).
Digestive enzymes, primarily proteases, contribute to the non-specific host defense system exerting a toxic action on microorganisms by destruction of their cell wall (20). Omp19 is a lipoprotein anchored in the Brucella outer membrane (7). This location together with its protease inhibitor activity suggest that it may play a protective role against host proteases.
In this work, we studied if Omp19 enables Brucella to better thrive in the harsh gastrointestinal tract, where protection from proteolytic degradation can be a matter of life or death, and thus promoting host invasion and intracellular infection.
Materials and Methods
Ethics Statement
Protocols of this study agreed with international ethical standards for animal experimentation (Helsinki Declaration and amendments, Amsterdam Protocol of welfare and animal protection and NIH guidelines for the Care and Use of Laboratory Animals). Protocols of this study were approved by the Institutional Committee for the Care and Use of Experimentation Animals from UNSAM (CICUAE-UNSAM_N°04/2014).
Bacterial Strains, Media, and Culture Conditions
Brucella strains were derived from the wild type (wt) 2308 biovar and were: (i) smooth virulent wt B. abortus; (ii) unmarked omp19 deletion mutant (Δomp19); and (iii) omp19 complemented Δomp19 mutant (Δomp19pBBR4omp19). All strains were grown as described in Czibener and Ugalde (21). When necessary, media were supplemented with the Ampicillin (100 μg/ml) or Nalidixic acid (5 μg/ml). CFU determination from intestine containing samples were performed in medium with following antibiotics to inhibit normal flora growth: Vancomycin (20 μg/ml), Cycloheximide (100 μg/ml), Bacitracin (10 U/ml), and Nalidixic acid. All work with live Brucella was performed in BSL3-laboratories and BSL3-animal facility at UNSAM. Escherichia coli strains were grown at 37°C in LB with Ampicillin.
Generation of Mutant Strains
(i) Δomp19 Strain
Omp19 (BAB1_1930) unmarked chromosomal mutant was generated as described in Herrmann et al. (22). Briefly, two DNA fragments of ~500 bp containing flanking regions of BAB1_1930 were amplified from B. abortus 2308 genomic DNA. Primers used to amplify omp19′s upstream regions were: omp19(EcoRI)_Up_Fw_5′-GAATTCTCGAAGGCTGTTTCGCTATCG-3′ and omp19_Up_Rv_5′- CAGGTTCTCCATTTGCGCATTT-3′; and omp19_Down_Fw_5′-CAAATGGAGAACCTGTCTGACCCGGAAACGATGAAC-3′ and omp19(BamHI)_Down_Rv_5′-GGATCCTTGTGCGCCTGACGATGC-3′ for downstream region. Fragments were ligated by overlapping PCR using omp19(EcoRI)_Up_Fw and omp19(BamHI)_Down_Rv. The resulting fragment was digested with EcoRI and BamHI, cloned into pK18mobSacB (23) and conjugated to B. abortus 2308 by biparental mating. Single recombinants selection, selection with sucrose, excision of plasmids, and generation of deletion mutants was performed as described previously described (21). Deletion of BAB1_1930 was confirmed by PCR and sequence analysis and western blot (Figure S1).
(ii) Complementation of Δomp19 Mutant
A 1000 bp DNA fragment containing the complete gene (BAB1_1930) and its promotor was amplified using primers Omp19(BamHI)_ATG_5′-ATGGATCCATGGGAATTTCAAAAGCAAGTCTGC-3′ and Omp19(SpeI)_TGA_5′-GAACTAGTTCAGCGCGACAGCGTCA-3′, digested with BamHI and SpeI and ligated into pBBR4 to generate the plasmid pBBR4omp19. This plasmid was electroporated into Δomp19 mutant. The resulting complemented strain was called Δomp19pBBR4omp19. Complementation was confirmed by PCR and western blot (not shown).
Recombinant Proteins, Enzymes, and Extracts
Mouse intestine- and stomach-extracts were obtained as previously described (8). Briefly, intestines and stomachs extracts were obtained from 6 to 12 weeks old female or male Balb/c mice (n = 10). Prior to fluid preparation, mice were fasted for 2.5 h (water ad lib.) and euthanized by CO2 inhalation. Stomachs and small intestines were resected, homogenized in PBS, and fluid separated by centrifugation (10 min, 13,200 × g at 4°C). Pooled Intestinal or stomach fluids were snap-frozen in liquid nitrogen and stored at −80°C. Protein concentration and proteolytic activity were determined as previously described by Ibañez et al. (8). Microsomes of J774 murine macrophages were obtained as described previously by Coria et al. (9). Pancreatin, Elastase, and Trypsin from pig and α-Chymotrypsin from bovine were from Sigma.
Recombinant U-Omp19 was produced as previously described by Pasquevich et al. (5). For Omp25 production, the complete sequence of B. abortus omp25 gene (GenBank_X79284.1) (24) was synthetized and subcloned into pET22(b)+ (Novagen) in frame with 6 × His-tag (genscript). Expression and purification was performed as described in Goel and Bhatnagar (25).
Infection of Mice
Six to eight-week-old female BALB/c mice were bred at IIB-UNSAM. Five mice/group were inoculated (i) wt, (ii) Δomp19, or (iii) Δomp19pBBR4omp19 Brucella strains either by gavage (i.g.) with 1 × 109 CFU in 0.2 ml PBS (18, 26) or with 1 × 1010 CFU directly into the oral cavity as previously described by von Bargen et al. (27). Infected mice were kept in cages within a BSL3 facility. At different times post-infection mice were euthanized by CO2 inhalation and organs were aseptically collected, homogenized, and plated for CFU determination. Intestinal samples were plated on TSA supplemented with Vancomycin, Cycloheximide, Bacitracin, and Nalidixic acid. In some experiments tissue samples from duodenum were obtained for immunofluorescence analysis.
Intestinal Tissue Immunofluorescence
Duodenum sections from mice infected i.g. either with wt or Δomp19 B. abortus were excised, fixed (4% paraformaldehyde), immersed in 30%-sucrose buffer, embedded in OCT-medium and frozen (-80°C). Cryosections (10 μm) were mounted on positively charged glass-slides (Biogenex), permeabilized with 0.2% Tween20 and blocked with 1% BSA and 5% horse serum in PBS. Brucella detection was performed as previously described (21). RNAse A (10 μg/ml) treated samples were counterstained with Alexa-Fluor555-WGA (ThermoFisher) and TO-PRO®-3 (Invitrogen). Sections were mounted using FluorSave reagent (Calbiochem) and images obtained on an IX-81 Olympus microscope with FV-1000 confocal module. A ROI was set for each treatment, background subtracted and images merged (RGB) (ImageJ software, NIH).
Bacterial Susceptibility to Proteases
(i) Agar Disk-Diffusion Method
Brucella strains (1 × 108 CFU) were spread on TSA plates supplemented with Vancomycin, Cycloheximide, Bacitracin, and Nalidixic acid. Five-mm filter disks impregnated with either PBS, intestine- or stomach-extract were placed on the agar surface. After 72 h of incubation (37°C) zones of inhibition were determined.
(ii) Protease Broth-Susceptibility Test
Brucella strains (1 × 105 CFU/ml) were incubated in 10% TSB plus buffer, intestine-extract (8.5 mg/ml), pancreatin (2 mg/ml), α-chymotrypsin (50 μM), trypsin (20 μM), pancreatic elastase (5 μM), or microsomes from J774 macrophages (2 mg/ml) for the different periods of time at 37°C. Negative control was buffer supplemented with 10% TSB. Buffer was PBS (intestine extract or microsomes), 0.5% ClNa (pancreatin), 10 mM Tris-HCl, pH7.8 (α-chymotrypsin and trypsin), or 10 mM Tris-HCl pH8.8 (pancreatic elastase). All protease solutions were sterilized by filtration before to incubation with the bacteria. Live bacteria (CFU/ml) were determined at different time points by serial dilutions plating.
Bacterial Growth Analysis
Brucella strains were labeled with TRSE (Invitrogen) as previously described by Brown et al. (28). Bacteria were spotted on 1% agarose pads with 10% TSB plus PBS or pancreatin (2 mg/ml). Images were obtained before and after 24 h of culture on an Olympus IX-81 microscope with FV-1000 confocal module. Images were subtracted the background and merged using RGB format (ImageJ software). Number of total bacteria (N) and initial number of bacteria (N0, number of labeled or partially labeled bacteria) were enumerated using Spot Detector plugin (ICY software, Institute Pasteur). Three to nine images/condition in duplicates were evaluated (50–150 colonies/condition). Then, assuming exponential growth, the average number of cell divisions (n) was calculated:
DNA Content on Individual Bacteria
Brucella in exponential phase (5 × 107 CFU/ml) were incubated with or without pancreatin (2 mg/ml). After 1.5–6 h, cells were washed, fixed, incubated with RNase A and labeled with SYTOX-Green (Invitrogen). Samples were analyzed in a FACS ARIA II (BD Biosciences) and analyzed with FlowJo7.6.2 software (Tree Star).
Western Blot
Brucella strains (5 × 108 CFU/ml) were cultured with 10% TSB buffer with or without pancreatic elastase (10 μM), washed and boiled in sample buffer (5 min). CFU/ml were determined in a sample taken prior to stop the reaction and 1 × 107 CFU/lane were subjected to SDS-PAGE and transferred onto nitrocellulose membranes. Immunoblotting was performed using mouse monoclonal antibodies against Omp1, Omp2b, Omp25, Omp10, Omp16, and Omp19 (29), rabbit polyclonal anti-CtrA (30) or mouse anti-GroEL serum, followed by incubation with anti-mouse-IgG-HRP (Sigma) or anti-mouse-IgG IRDye antibodies (Li-Cor Biosciences). Images were acquired with Odyssey image-scanner and band intensities (RFU) were quantified (Image-Studio-Lite Software). Omp16, Omp10, and GroEL were similar in all treatments and served as loading control. Percentage of digested Omp25 was calculated:
Omp25 Digestion
Purified Omp25 (1 μM) was incubated with pancreatic elastase (1 μM) or buffer (10 mM Tris-HCl, pH8.8) with or without U-Omp19 (45 μM). Reactions were stopped by sample buffer addition and boiling. Omp25 digestion was followed by western blot.
Cell Culture and Infection Assay
J774 macrophages were maintained in RPMI 1640 supplemented with 5% fetal bovine serum (FBS) and streptomycin (50 μg/ml)-penicillin (50 U/ml) (Gibco Life Technologies) in a humidify 5% CO2 atmosphere at 37°C. Cells (5 × 104 per well) were seeded on 24-well plates in antibiotic-free medium and were kept for 24 h. B. abortus infections were carried out at a multiplicity of infection (MOI) of 500:1 or 100:1. After a 1 h incubation with the bacteria, wells were washed three times with PBS and incubated with fresh medium containing 50 μg/ml of Gentamycin and 100 μg/ml streptomycin to kill non-internalized bacteria. At the indicated time points, infected cells were washed three times with PBS and lysed with 500 μl 0.1% Triton X-100 (Sigma-Aldrich). The intracellular CFU were determined by plating serial dilutions on TSA. In some experiments, prior to the infection of cell the bacteria were incubated at 5 × 107 CFU/ml with or without pancreatin (2 mg/ml) during 2 h. Afterwards bacteria were washed and suspended in medium to infect the cells.
Statistical Analysis
Statistical analysis and plotting were performed using Prism® 7.04 (GraphPad, Inc., USA). CFU data were logarithmically transformed. Unpaired two-tailed Student t-test was used for pairwise comparisons between means of two groups or one-way or two-way ANOVA followed by Bonferroni's posttest was used for comparing more than two means. Significance level was set at p < 0.05.
Results
Omp19 Expression Is Needed for Oral Acquired B. abortus Infection
To investigate the role of Omp19 in Brucella infection, a deletion mutant (Δomp19) and its complemented strain (Δomp19pBBR4omp19) were constructed in the B. abortus wild-type (wt) strain 2308.
Mutant and wt strains had similar growth curves, resistance to low pH and bile salts. Moreover, membrane permeability to hydrophobic substances, expression of main outer membrane proteins (Omps) (Omp1, Omp2b, Omp25, Omp10, and Omp16) and lipopolysaccharide O-antigen were similar between wt and Δomp19 strains (Figures S1A–F). The authenticity of the mutant was verified by PCR and immunoblot analysis on whole-cell extracts with an anti-Omp19 Mab (Figures S1E,F).
To evaluate the role of Omp19 in the establishment of B. abortus infection through the digestive tract in vivo, BALB/c mice were inoculated intragastrically (i.g.) with wt, Δomp19 or Δomp19pBBR4omp19 and 20 days post-infection bacterial loads at spleens and cervical lymph nodes (CLNs) were assessed. While wt and Δomp19pBBR4omp19 established infection, there were significant lower numbers of CFUs at spleens and CLNs from Δomp19 infected mice (p < 0.001 vs. wt) (Figures 1A,B).
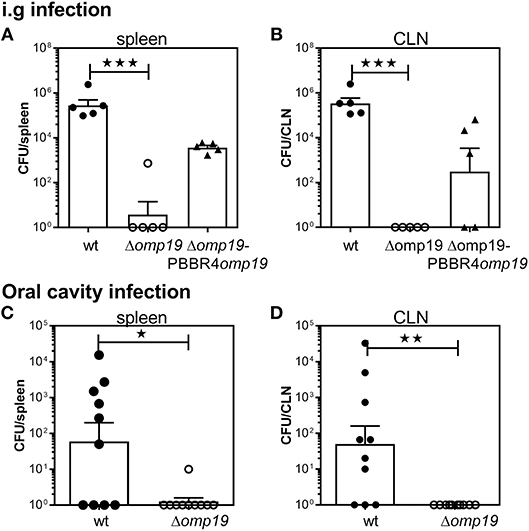
Figure 1. B. abortus Δomp19 is attenuated after oral infection. (A,B) BALB/c mice were i.g. inoculated with 1 × 109 CFU/mouse of wt, Δomp19 or Δomp19pBBR4omp19 B. abortus strains. Spleen and CLNs were homogenized and plated for CFU counting at 20 days post-infection (***p < 0.001 vs. wt B. abortus). Results are representative of three independent experiments with 5 mice per group. (C,D) BALB/c mice were inoculated at the oral cavity with wt or Δomp19 B. abortus (1 × 1010 CFU/mouse). Twenty days after infection the number of CFU at the spleens and CLNs were quantified (*p < 0.05 and **p < 0.01 vs. wt B. abortus). Pooled data from two independent experiments are shown. Each point represents data from an individual mouse. Horizontal lines and error bars represent the mean ± s.e.m. (For some mice, no live bacteria were recovered from spleens or CLNs. These were arbitrarily assigned with values of 1 CFU).
Upon gavage administration, initial events of bacterial invasion and onset of infection in the oral cavity may be bypassed. Thus, BALB/c mice were administered directly into the oral cavity as described in von Bargen et al. (27) with wt or Δomp19. Twenty days post-infection B. abortus were isolated from spleens and CLNs from wt infected mice, whereas almost no CFUs were found in these organs of Δomp19 infected mice (p < 0.05 and p < 0.01 vs. wt, respectively) (Figures 1C,D).
Altogether, these results demonstrate that Omp19 plays a crucial role in the establishment of infection by Brucella through the oral route in mice.
Brucella abortus Reaches Intestinal Tissues Upon Oral Infection and Requires Omp19 to Evade the Bacteriostatic Action of Intestinal Content
To evaluate if Δomp19 attenuation after oral infection is due to higher susceptibility to gastrointestinal content, short-term gavage infection experiments were performed. BALB/c mice were i.g. inoculated with wt or Δomp19 strains and at different time points post-infection the stomach and intestinal sections were analyzed. After 15 min equal numbers of bacteria were isolated from the stomachs of both groups (Figure 2A). After 1 h both strains were present in the duodenum at the lumen as well as in the epithelium (Figure 2B).
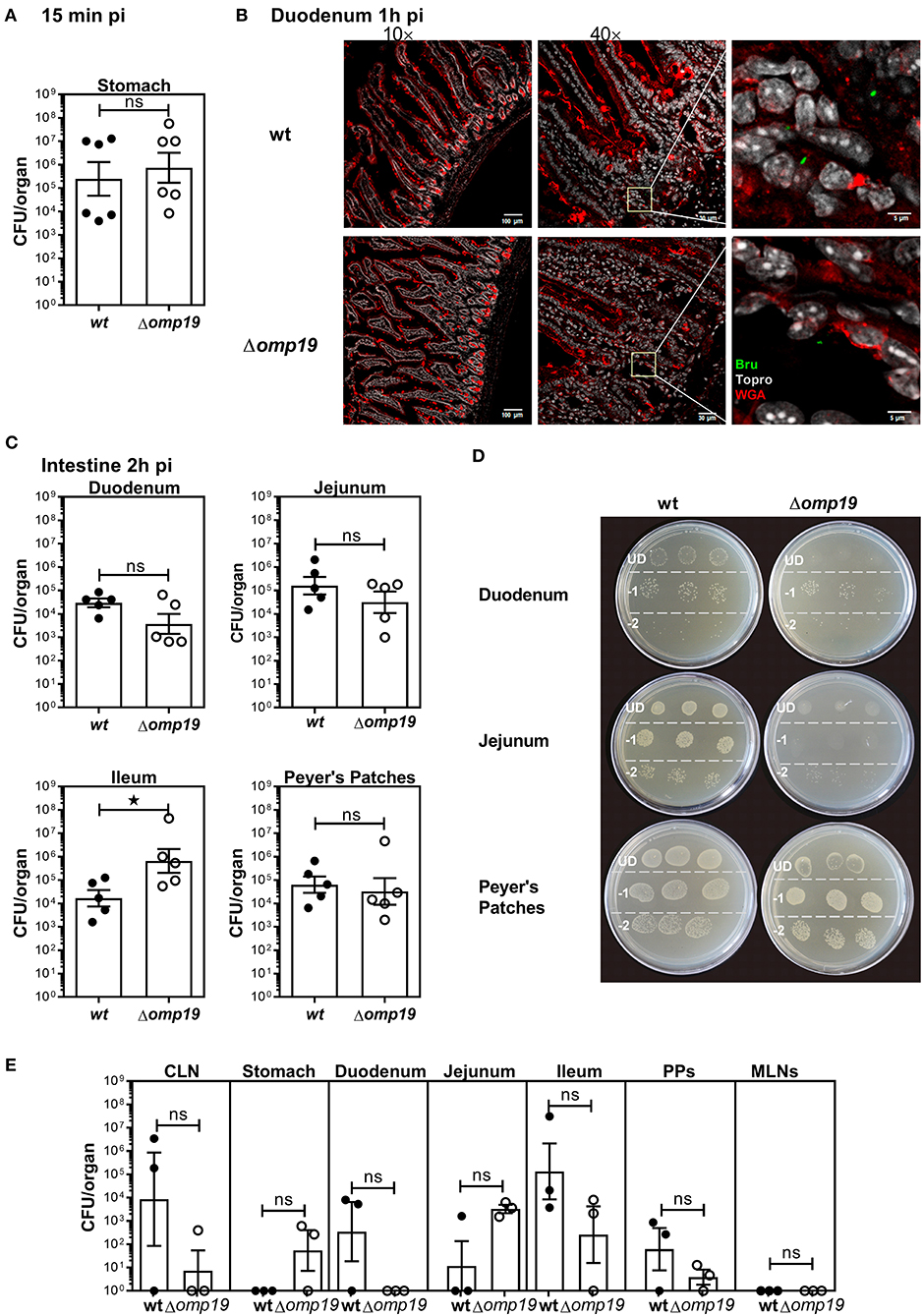
Figure 2. Brucella abortus requires Omp19 to evade the bacteriostatic action of intestinal content. BALB/c mice were i.g. inoculated with (1 × 109 CFU) of wt or Δomp19 B. abortus strains. (A) Total CFUs per stomach in animals sacrificed at 15 min post-infection. Each point represents an individual mouse, horizontal lines, and error bars represent the mean ± s.e.m. (B) Confocal microscopy images of duodenum of infected mice at 1 h post-infection. The images correspond to ROI merged signals for Brucella (green channel), mucin (WGA) (red channel) and nuclei (TO-PRO-3, NIR channel). The inset region of middle images (40×) was magnified and presented in the right, showing individual Brucellae in the epithelium. Scale bars are: 100 μm (left panels), 30 μm (middle panels) and 5 μm (right panels). (C) Total B. abortus CFUs recovered from duodenum, jejunum, Ileum or Peyer's Patches from infected mice sacrificed at 2 h post-infection. (D) Representative agar plates showing sequential 1:10 dilutions and drop plating from depicted tissues homogenates from wt or Δomp19 infected mice. UD (undiluted), −1: 1 to 10 dilution; −2: 1 to 100 dilution. Results are representative of two independent experiments. (E) BALB/c mice were inoculated at the oral cavity with wt or Δomp19 B. abortus (1 × 1010 CFU/mouse). Two hours after infection the number of CFU at CLNs, Stomach, Duodenum, Jejunum, Ileum, PPs, and MLNs were quantified. Each bar represents the mean CFU/organ (logarithmic sc±ale) and error bars represent the mean ± s.e.m. (For some mice, no live bacteria were recovered, these were arbitrarily assigned with values of 1 CFU). (Statistical analysis was performed by unpaired t-test to compare between the indicated groups: nsp > 0.05; *p < 0.05).
Next, B. abortus loads in different sections of the small intestine: duodenum, jejunum, ileum and Peyer's patches were evaluated. Almost no differences in wt and Δomp19 CFUs were detected at 2 h post-infection with a slight but significant increase in Δomp19 CFUs at Ileum (Figure 2C) that may not explain the attenuation of this strain when infecting by the oral route. However, when plated undiluted (direct plating from each tissue on TSA + Antibiotics) low-density bacterial growth and small colonies were found in the drops of Δomp19, indicating that the intestine content impaired Δomp19 strain's growth. This effect was temporarily and reversible, since upon dilution it disappeared and both, wt and Δomp19, showed similar numbers and phenotype of colonies (Figure 2D). These results indicate that the intestine content exerts a bacteriostatic action on Δomp19, suggesting that Omp19 protects Brucella from intestinal proteases.
Similar results were obtained when bacteria where inoculated directly at the oral cavity of mice. Both strains, wt and Δomp19, were recovered from intestinal tissues after 1 h of infection (Figure 2E), indicating that Brucella reaches the intestine after oral infection (by gavage or oral cavity delivery) and there it is exposed to the intestinal content that exerts a bacteriostatic effect.
Omp19 Protects B. abortus Against the Action of Pancreatic Proteases
To further assess the role of Omp19 against the action of gastric and gut content, in vitro bacterial susceptibility assays were performed.
An agar disk-diffusion test indicated that Δomp19 is more susceptible to the action of intestine content than the wt strain (p < 0.01 vs. wt + intestine extract) (Figure 3A), whereas stomach content did not affect bacterial growth.
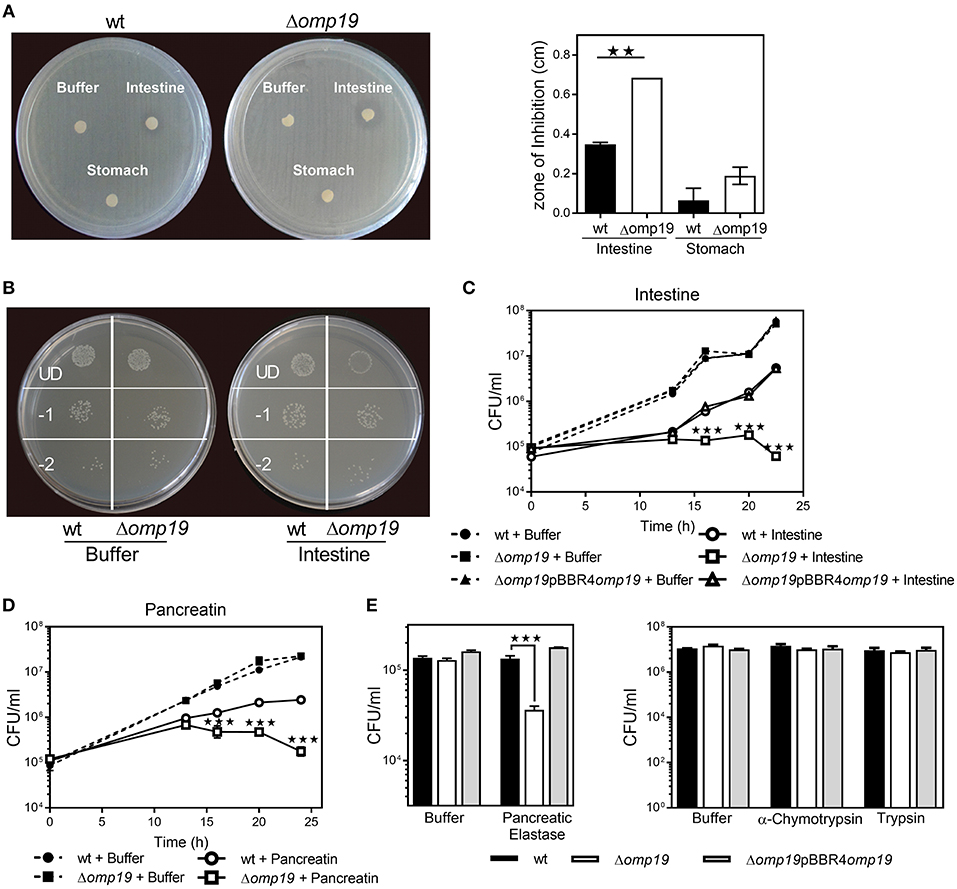
Figure 3. Omp19 protects B. abortus against the action of pancreatic proteases. (A) 1 × 108 CFU of wt and Δomp19 B. abortus were spread on TSA plates supplemented with antibiotics. Five-mm filter-disk were impregnated with either PBS, intestine- or stomach-extract and placed on the agar surface. The plates were incubated at 37°C for 72 h and afterwards the diameter of the zones of inhibition were determined (diameter of no growth zone minus diameter of the disk). (**p < 0.01 vs. wt B. abortus + intestine extract). (B) Representative picture of a plate with wt and Δomp19 B. abortus treated with buffer or intestine extract. Plated undiluted (UD) or after serial dilutions: 1/10 (−1) and 1/100 (−2). (C) wt, Δomp19 or Δomp19pBBR4omp19 B. abortus strains (1 × 105 CFU/ml) were incubated with PBS or intestine extract at 37°C. Live bacteria (CFU/ml) were determined after 12, 16, 20, and 24 h of incubation by serial dilutions plating (***p < 0.001 vs. wt B. abortus + intestine extract). (D) wt and Δomp19 B. abortus (1 × 105 CFU/ml) were incubated with buffer (0.5% ClNa) or pancreatin (2 mg/ml). Live bacteria (CFU/ml) were determined after 12, 16, 20, and 24 h of incubation by plating serial dilutions on TSA (***p < 0.001 vs. wt B. abortus + pancreatin at the same time point). (E) wt and Δomp19 B. abortus (1 × 105 CFU/ml) were incubated with buffer (10 mM Tris-HCl, pH8.8) or pancreatic elastase for 5 h or with buffer (10 mM Tris-HCl, pH7.8), α-chymotrypsin or trypsin for 24 h. Live bacteria (CFU/ml) were determined by plating serial dilutions on TSA (***p < 0.001 vs. wt B. abortus + pancreatic elastase). Results are representative of two or three independent experiments.
Incubation with intestine-extract inhibited Δomp19's growth and this action was bacteriostatic, since viable bacteria were recovered by dilution (Figure 3B). Viable bacteria determination over time indicated that in presence of intestine-extract Δomp19 was unable to grow (p < 0.001 vs. wt + intestine), whereas the wt and the complemented strains grew exponentially after 13 h of culture (Figure 3C). Similar results were obtained using pancreatin (a pig pancreatic extract) (Figure 3D), supporting that B. abortus requires Omp19 to grow when exposed to intestinal content.
As purified U-Omp19 inhibits main gastrointestinal proteases (8), the effect of individual proteases (pancreatic elastase, α-chymotrypsin, trypsin) on wt, Δomp19 and Δomp19pBBR4omp19 viability was assessed. Δomp19 was more susceptible in vitro to pancreatic elastase action than wt and Δomp19pBBR4omp19 (p < 0.001). In contrast, α-chymotrypsin and trypsin did not alter bacterial growth (Figure 3E).
These results together demonstrate that B. abortus requires the expression of Omp19 to resist the action of intestinal proteases.
Δomp19 B. abortus Stops Cell Division and Cell-Cycle Progression at G1-Phase After Incubation With Pancreatic Proteases
To evaluate if Δomp19's growth impairment when exposed to proteases is due to a cell division defect, Brucella's growth was studied by microscopy. Brucella Texas-red succinimidyl-ester (TRSE) labeling allows, after growth, the visualization of an unlabeled pole and subsequently unlabeled or partially labeled daughter cells. TRSE-labeled wt and Δomp19 were cultured on TSB-agarose pads containing buffer or pancreatin. After 24 h wt and Δomp19 in buffer-pads and wt in pancreatin-pads formed microcolonies with many unlabeled cells surrounding partially labeled cells. However, Δomp19 in pancreatin-pads formed no or small colonies (small chains) with few unlabeled sphere-shaped bacteria (Figure 4A), indicating a cell division defect. Quantitative analysis of labeled (or partially labeled) cells and unlabeled cells in each image revealed a significantly lower average number of cell divisions for Δomp19 in pancreatin-pads (p < 0.001 vs. wt in pancreatin) (Figure 4B).
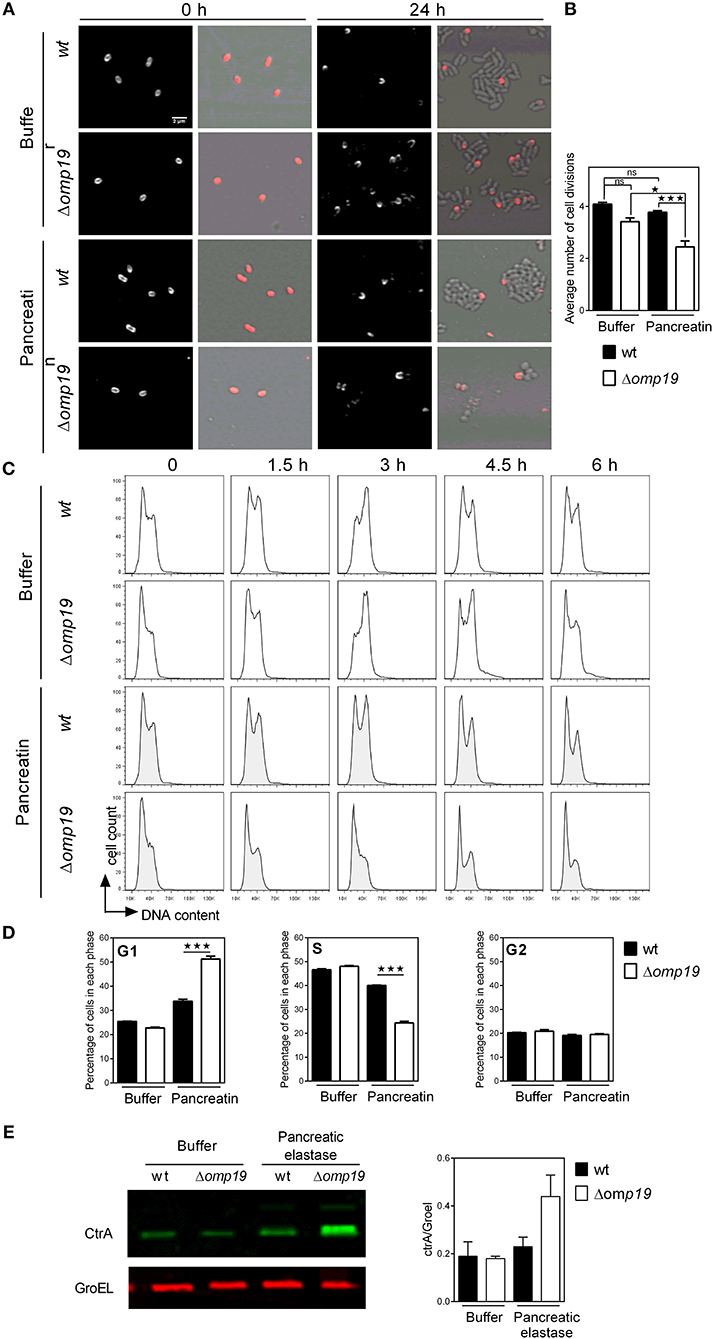
Figure 4. Δomp19 B. abortus has a cell division defect and cell cycle arrest upon incubation with pancreatic proteases. TRSE-labeled wt and Δomp19 B. abortus were dropped on TSB-agarose pads containing buffer or pancreatin and cultured for 24 h at 37°C. (A) Representative Texas Red fluorescence (left) and phase contrast microscopy images (right) from the beginning of incubation (0 h) and after 24 h of incubation are shown. (B) Average number of cell divisions after 24 h of culture obtained by quantification of the number of labeled (or partially labeled) cells and unlabeled cells in each individual colony (*p < 0.05 and ***p < 0.001 vs. wt B. abortus strain in pancreatin). Results are representative of two independent experiments. (C) Flow cytometry analysis of DNA content on individual bacteria. wt and Δomp19 B. abortus were incubated in buffer or pancreatin for the indicated time periods and the content of DNA was evaluated by Flow cytometry. Representative histograms are shown. Results are representative of two independent experiments. (D) Bar graphs indicate the percentage of cells in each phase of cell cycle after 3 h of culture. (***p < 0.001 vs. wt B. abortus strain in pancreatin). (E) B. abortus wt and Δomp19 strains were incubated with buffer or pancreatic elastase. Equal quantities of bacteria were subjected to SDS-page followed by western blot analysis using specific antibodies for CtrA and GroEL. Images are representative of two independent experiments. The ratio of CtrA and GroEL signals was evaluated by quantitative analysis of western blot images. Bar graph represent the mean ± s.e.m. of pooled results from two independent experiments.
Cell division requires critical regulation of the cell-cycle to coordinate genome replication and segmentation, therefore cell-cycle progression on individual bacteria was determined. While incubation of wt with pancreatin did not alter its progression along the cell-cycle, Δomp19 resulted in cell-cycle arrest at G1 (Figure 4C), that was evident after 3 h of incubation by the rate of cells accumulated in G1-phase (p < 0.001 vs. wt + pancreatin, Figure 4D). Besides, expression of cell-cycle master regulator CtrA and chaperonin GroEL were evaluated upon treatment with pancreatic elastase. Pancreatic elastase treatment increased CtrA signal in Δomp19, whereas GroEL expression was similar in both strains exposed or not to proteases (Figure 4E).
Together, these results reveal that Δomp19 exposed to pancreatic proteases has a cell division defect that is linked to impaired progression through G1-phase and CtrA accumulation.
Omp19 Protects Omp25 From Pancreatic Elastase Digestion
As cell envelope constitutes the first contact with host-proteases, cell envelope proteins were evaluated in wt and Δomp19 upon protease treatment. No changes between wt and Δomp19 were detected upon pancreatic elastase treatment in Omp1, Omp10, or Omp16. On the contrary, in both strains Omp25 presented a lower molecular weight band and reduced Omp2b intensity upon pancreatic elastase incubation that would correspond to digested Omp25 and Omp2b, respectively (Figure 5A). While no Omp19-dependent protection of Omp2b digestion was evidenced in wt strain compared to Δomp19 strain, the percentage of digested Omp25 was higher in Δomp19 (Figure 5B), highlighting Omp19's inhibitory role of pancreatic elastase. Omp19 inhibition of pancreatic elastase digestion of Omp25 was confirmed in vitro using recombinant purified proteins. Pancreatic elastase digestion of rOmp25 was evidenced by a reduced Omp25-specific signal in western blot compared with the signal of rOmp25 without protease. This reduction was lower when U-Omp19 was added, indicating that U-Omp19 inhibits Omp25 digestion by pancreatic elastase (Figure 5C). Differences in the digestion pattern between Omp25 expressed on the Brucella membrane and recombinant Omp25, may be due to differential accessibility of pancreatic elastase cleavage sites, since in membrane associated Omp25 most cleavage sites are in predicted transmembrane regions or in loops facing the periplasm, only one cleavage site would be accessible to the protease when Omp25 is in the context of the Brucella membrane (Figure 5D).
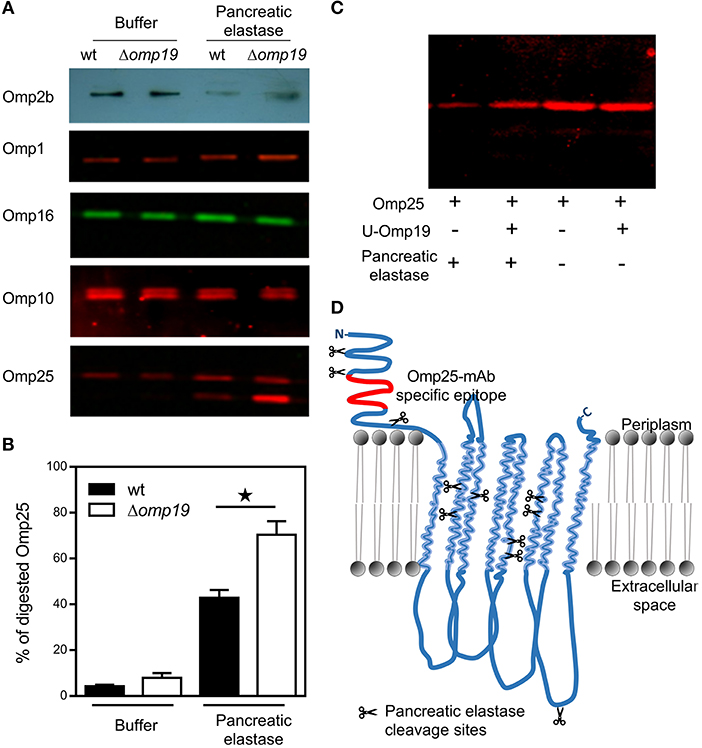
Figure 5. Omp19 on the B. abortus membrane protects Omp25 from pancreatic elastase digestion. Wt and Δomp19 B. abortus strains were incubated with buffer (10 mM Tris-HCl, pH8.8) or pancreatic elastase. (A) Equal quantities of bacteria were subjected to SDS-page followed by western blot analysis using specific antibodies for cell envelope proteins: Omp2b, Omp1, Omp16, Omp10, and Omp25. Images are representative from two or three independent experiments. (B) Percentage of digested Omp25 evaluated by quantitative analysis of western blot images. Data represent pooled results from two independent experiments (*p < 0.05 vs. wt B. abortus strain in pancreatic elastase). (C) Recombinant purified Omp25 was incubated with pancreatic elastase with or without U-Omp19. Following incubation, each mixture of reaction was separated on SDS-PAGE followed by western blot analysis with Omp25 specific antibodies. (D) Graphical representation of BOCTOPUS (31) or PRED-TMBB2 (32) transmembrane β-barrel predicted topology for Omp25 with respect to the lipid bilayer representation of the B. abortus outer membrane. Scissors indicate predicted pancreatic elastase cleavage sites (AA or AG) on Omp25 sequence. The position of the specific epitope for the anti-Omp25 mAb used is colored in red.
These results together indicate that when Omp19 is absent, pancreatic elastase gains access to the membrane following degradation of Omp25, on the contrary under physiologic condition where Omp19 is present, Brucella wt can withstand this protease activity.
Omp19 Impairs Macrophage Microsomal Proteolytic Killing of B. abortus
Reaching the intracellular replicative niche is the next step for establishment of infection. Therefore, the ability of Δomp19 mutants to enter cells and replicate intracellularly was studied in professional phagocytes (Figure 6). In agreement with previous studies significant lower amounts of Δomp19 were found after 6, 24, and 48 h of infection in comparison to wt strain (Figure 6A). Moreover, Δomp19 strain was significantly more susceptible to killing by microsomal content than wt or Δomp19pBBR4omp19 (p < 0.01 vs. wt + microsomes or Δomp19pBBR4omp19 + microsomes) (Figure 6B), suggesting that Omp19 may protect the bacteria from lysosomal proteolysis during intracellular traffic.
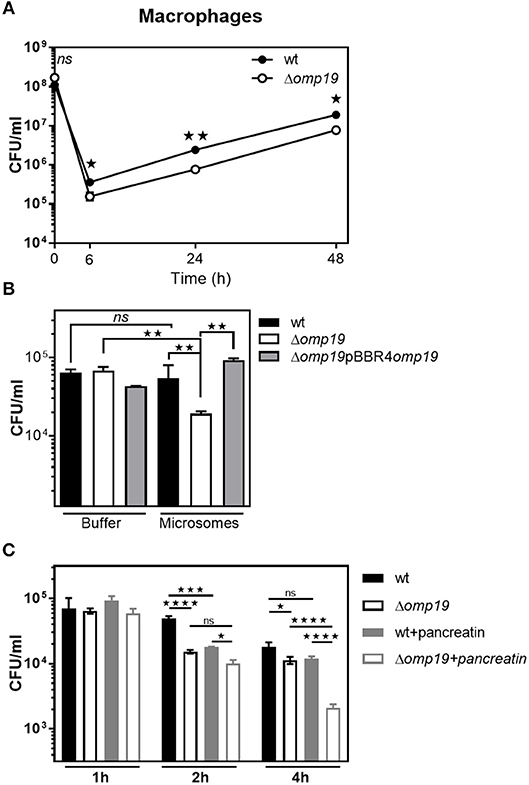
Figure 6. Omp19 impairs macrophage microsomal proteolytic killing of B. abortus. (A) J774 macrophages were infected (MOI 500:1) with the B. abortus wt or Δomp19 strains. At the indicated time points post-infection intracellular CFUs of B. abortus wt or Δomp19 strains were determined. (Statistical analysis was performed by two-way ANOVA followed by Bonferroni posttest to compare between the indicated groups: nsp > 0.05; *p < 0.05 or **p < 0.01). (B) wt, Δomp19 or Δomp19pBBR4omp19 B. abortus strains (1 × 105 CFU/ml) were incubated with PBS or J774 derived microsomes at 37°C. Live bacteria (CFU/ml) were determined after 8 h by serial dilutions plating (Statistical analysis was performed by one-way ANOVA followed by Bonferroni posttest to compare between the indicated groups: nsp > 0.05, **p < 0.01). (C) Δomp19 and wt strains were preincubated with buffer or pancreatin for 1 h prior to infection of J774 macrophages (MOI 100:1). Intracellular CFUs were determined at 1, 2, or 4 h post-infection. (Statistical analysis was performed by two-way ANOVA followed by Bonferroni posttest to compare between the indicated groups: nsp > 0.05; *p < 0.05, ***p < 0.001, ****p < 0.0001).
When infecting through the oral route Brucella will reach the intracellular compartment after facing with gastrointestinal proteases, thus Δomp19 and wt strains were preincubated with pancreatin or buffer for 2 h prior to infection of J774 macrophages and intracellular bacterial counts were determined after 1, 2, or 4 h of infection. Pre-incubation with pancreatin did not affect bacterial internalization, since similar amounts of intracellular bacteria of both strains were recovered after 1 h of infection. After 4 h of infection, preincubation with pancreatin led to an increased susceptibility of Δomp19 to intracellular killing by macrophages, compared to pancreatin pretreated wt (p < 0.0001) or buffer pretreated Δomp19 (p < 0.0001) (Figure 6C). These results indicate that the sequential action of intestinal proteases followed by intracellular microsome proteolytic killing has an important effect on hampering Δomp19 ability to establish an intracellular niche in macrophages. Altogether these results may explain the highly attenuated phenotype of this strain when infection occurs by the oral route.
Discussion
After consumption of infected milk or experimental oral infection, live Brucella are detected in fecal samples of natural host like cattle, bison, wolf and marine mammals, indicating that Brucella transits and pass the harsh environment of gastrointestinal tract (33).
Our results demonstrate that in mice, after oral infection (either by gavage or inoculation at the oral cavity) Brucella reaches the gut. After 1 h of infection brucellae were found at the lumen and epithelium of duodenum. This fast infection capacity of Brucella was shown in a calf ligated ileal-loop model, in which Brucella bacteremia was detected 30 min after intraluminal inoculation without histopathologic traces of lesions (34). Brucella may spread systemically from the digestive tract by transepithelial migration in mucosal epithelial barrier or through M cells (26, 34, 35).
As protease inhibitor activity against main gastrointestinal proteases was demonstrated for U-Omp19 and because of its strategic location on the outer membrane for interacting with host proteases (7–9), we speculated that Omp19 may allow Brucella to withstand the gastrointestinal proteolysis and infect orally. Omp19's protease inhibitor broad-specificity (8, 9) would also be advantageous regarding the different proteases that Brucella may encounter along infection. Like broad-spectrum serine-protease inhibitor from Tannerella forsythia, that may protect it from proteases from other bacteria and from the host (3).
In this work, Omp19's role in virulence in an oral infection murine model was examined. Our results showed that Omp19 expression is needed for establishment of oral acquired B. abortus infection. In contrast to wt, Δomp19 was cleared from the spleens and CLNs at 20 days post infection. Remarkably after intraperitoneal infection of mice, omp19 deletion resulted in significant loss of virulence but the bacteria were not cleared (36, 37), this difference highlights the importance of Omp19 for Brucella oral infection, probably due to the huge amounts of proteases encountered when infecting through this route. Attenuation upon systemic infection and intracellularly may be due to other host-proteases action, like lysosomal proteases, to which Omp19's inhibitory activity was demonstrated (9).
Intestinal content exerted a bacteriostatic action on Δomp19 in vivo and in vitro, revealing a protective role for Omp19 in Brucella against intestinal proteases. This is the first work demonstrating in vivo a role of a protease inhibitor in acquisition of a bacterial disease by the oral route, therefore these findings are highly relevant for foodborne infections. Interestingly, gut microbiota, that survive in this protease-rich medium, produce protease inhibitors to protect them self from exogenous proteases (38–41).
In vitro experiments with purified proteases shed light into the role of individual proteases in the bacteriostatic action of intestinal content. Δomp19's growth is hampered by the action of pancreatic elastase, indicating that inhibition of this protease by Omp19 on B. abortus membrane is important during the initial steps of infection. Trypsin and α-chymotrypsin have been shown to elicit antibacterial activities against E. coli, Proteus vulgaris, Pseudomonas aeruginosa, S. aureus, Streptococcus pyogenes, and Vibrio cholerae (42, 43), but have no effect on B. abortus. This resistance is Omp19-independent, indicating that it may be mediated by other mechanism.
Pancreatic proteases induce a cell division defect in Δomp19 that is linked to cell-cycle arrest in G1-phase. Interestingly, G1 arrest also occurs during intracellular trafficking of B. abortus and on starvation in Sinorhizobium meliloti (44, 45). Therefore, delaying initiation of DNA replication could be a common feature used by α-proteobacteria in response to harsh conditions such as infection or starvation.
In Caulobacter crescentus, degradation of the CtrA cell-cycle master regulator occurs at specific points in the cell-cycle. Clearance of active CtrA at the G1/S transition allows the initiation of DNA replication and cell-cycle progression (30, 46). Moreover, expression of a constitutively active stable CtrA derivative results in dominant G1 arrest (30). In B. abortus, the essential role of CtrA in cell division was recently confirmed (47). Thus, accumulation of CtrA in Δomp19 upon pancreatic protease treatment, agrees with the cell-cycle arrest in G1 induced in this strain upon treatment with proteases.
Antimicrobial functions of proteases can be due to the attack of Omps leading to loss of membrane integrity (42, 43, 48, 49). Since outer membrane proteins are exposed on the bacterial surface, they could be targets of pancreatic elastase. Among all Omps evaluated, our results indicate that Omp10, Omp16, and Omp1 of either wt or Δomp19 were resistant to the action of pancreatic elastase, whereas, Omp2b and Omp25 were digested by this protease. This result is consistent with protease digestion of Omps in E. coli or P. aeruginosa, in which the major Omps, OmpA, and OmpF, respectively, were degraded, while other Omps remained not affected (48, 49). Although Brucella Omp25 does not share identity with E. coli OmpA (50), topology predictions suggest that both contain similar secondary structural properties and may play a similar function (51). Notably, Omp19 expression in Brucella inhibited pancreatic elastase mediated Omp25 digestion. This role of Omp19 on inhibition of pancreatic elastase mediated Omp25 digestion was confirmed in vitro using recombinant purified proteins. Omp19 inhibition of pancreatic elastase digestion of Omp25 may explain the resistance of wt strain to the action of this protease. A similar role was described for the periplasmic protease inhibitor ecotin from E. coli, which reduces the bactericidal action of neutrophil elastase by protecting OmpA on the bacterial membrane from neutrophil elastase mediated digestion (2).
In this work, we found that a Brucella omp19 deletion mutant is highly attenuated in mice after oral infection. This attenuation can be explained by bacterial increased susceptibility to host proteases met by Brucella during establishment of infection. Δomp19 has a cell division defect when exposed to pancreatic proteases that is linked to cell-cycle arrest in G1-phase, Omp25 degradation on the cell envelope and CtrA accumulation. Interestingly, a link between these three molecules was found recently, in which CtrA can bind the promotor of omp25 and omp19. The same work demonstrates that CtrA controls the expression of Omp25 (47), therefore the increment in Omp25 intensity in Δomp19 upon pancreatic elastase treatment may be explained by the increment in CtrA expression.
Upon entry into mammalian cells, the intracellular pathogen Brucella abortus resides within a membrane-bound compartment, the Brucella-containing vacuole (BCV), the maturation of which is controlled by the bacterium to generate a replicative organelle derived from the endoplasmic reticulum (ER). BCVs traffic along the endocytic pathway and fuse with lysosomes, and such fusion events are required for further maturation of BCVs into an ER-derived replicative organelle (52). Thus, the role of Omp19 for intracellular survival was studied. In agreement with previous work (36, 37), Δomp19 was attenuated inside macrophages. This attenuation may be due to increased susceptibility to intracellular proteases when lacking Omp19. This hypothesis is reinforced by the fact that Omp19 is able to inhibit lysosomal proteases (9) and here we demonstrated that Δomp19 is more susceptible to proteolytic killing by microsomes from macrophages. This increased susceptibility may explain the slight attenuation for systemic infections in mice, in which high persistence of Δomp19 was shown after 4 weeks of infection (36, 37). An additive effect in increasing susceptibility of Δomp19 was observed when the strains were preincubated with pancreatic proteases prior to infection of macrophages. This increased susceptibility may account for the high attenuation of Δomp19 after in vivo oral infection. Therefore, Omp19 would allow Brucella spp. to bypass lysosomal destruction thus enabling Brucella to survive inside macrophages and start a chronic infection.
Overall, this study demonstrates that the protease inhibitor Omp19 confers B. abortus the ability to resist the action of proteases. Together with urease that may protect Brucella from stomach low pH (17) and cholylglycine hydrolase that confers resistance to bile salts (18), Omp19 by inhibiting intestinal and intracellular proteases contributes to the establishment of chronic infection through the oral route.
Contribution to the Field Statement
Understanding how infectious pathogens spread is critical to prevent infectious diseases. One of the principal ways in which human and animal Brucellosis is acquired, is the oral route. This implies that Brucellae must survive the harsh conditions along the gastrointestinal tract before reaching the mononuclear phagocytes to form a replicative niche. In this work, we demonstrate that Brucella has a lipoprotein, called Omp19, which is a protease inhibitor, that enables it to survive the proteolytical action of gut digestive and microsomal derived proteases. The significance of our research is in identifying a new mechanism involved in virulence in oral acquired Brucellosis, that will enhance our understanding of Brucella pathogenesis and would serve as a model for other food-borne diseases.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
Protocols of this study agreed with international ethical standards for animal experimentation (Helsinki Declaration and amendments, Amsterdam Protocol of welfare and animal protection and NIH guidelines for the Care and Use of Laboratory Animals). Protocols of this study were approved by the Institutional Committee for the Care and Use of Experimentation Animals from UNSAM (CICUAE-UNSAM_N°04/2014).
Author Contributions
KP, MC, and JC designed the experiments. Funding acquisition was done by JC. MC performed most laboratory assays with assistance from KP, FG, LB, DR, and LC. MR performed susceptibility to bile salts assays and some of the J774 macrophage infection assays. KP, JC, and MC performed all statistical analysis. DC provided bacterial strains, materials and, together with JC and KP contributed with their expertise on the subject. KP, MC, and JC interpreted all results. KP and JC wrote the manuscript. All authors reviewed, commented, and approved the manuscript.
Funding
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT-Argentina): PICT 2013 No 1500, PICT 2016 No 1310 (to JC) and in part by grants from the Bill and Melinda Gates Foundation through the Grand Challenges Explorations Initiative (OPP1060394 and OPP1119024) (to JC). MC was supported by a fellowship of the National Research Council of Argentina (CONICET).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. L. Shapiro (Stanford University, California, USA) for generously providing rabbit polyclonal anti-CtrA serum. We gratefully acknowledge the technical assistance of Valeria Sanchez for performing intestinal cryosections as well as Ariel Billordo for operating the FACSCalibur and FACS ARIA II. We also thank Dr. Xavier De Bolle (University of Namur, Namur, Belgium) for constructive discussions and technical support regarding TRSE labeling of Brucella. KP, FG, MR, LC, DC, and JC are members of the National Research Council of Argentina (CONICET).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01436/full#supplementary-material
References
1. Kedzior M, Seredynski R, Gutowicz J. Microbial inhibitors of cysteine proteases. Med Microbiol Immunol. (2016) 205:275–96. doi: 10.1007/s00430-016-0454-1
2. Eggers T, Murray IA, Delmar VA, Day AG, Craik CS. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem J. (2004) 379 (Pt 1):107–18. doi: 10.1042/bj20031790
3. Ksiazek M, Mizgalska D, Enghild JJ, Scavenius C, Thogersen IB, Potempa J. Miropin, a novel bacterial serpin from the periodontopathogen Tannerella forsythia, inhibits a broad range of proteases by using different peptide bonds within the reactive center loop. J Biol Chem. (2015) 290:658–70. doi: 10.1074/jbc.M114.601716
4. Stapels DA, Ramyar KX, Bischoff M, von Kockritz-Blickwede M, Milder FJ, Ruyken M, et al. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc Natl Acad Sci USA. (2014) 111:13187–92. doi: 10.1073/pnas.1407616111
5. Pasquevich KA, Estein SM, Garcia Samartino C, Zwerdling A, Coria LM, Barrionuevo P, et al. Immunization with recombinant Brucella species outer membrane protein Omp16 or Omp19 in adjuvant induces specific CD4+ and CD8+ T cells as well as systemic and oral protection against Brucella abortus infection. Infect Immun. (2009) 77:436–45. doi: 10.1128/IAI.00123-09
6. Pasquevich KA, Ibanez AE, Coria LM, Garcia Samartino C, Estein SM, Zwerdling A, et al. An oral vaccine based on U-Omp19 induces protection against B. abortus mucosal challenge by inducing an adaptive IL-17 immune response in mice. PLoS ONE. (2011) 6:e16203. doi: 10.1371/journal.pone.0016203
7. Tibor A, Decelle B, Letesson JJ. Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect Immun. (1999) 67:4960–2.
8. Ibanez E, Coria LM, Carabajal MV, Delpino MV, Risso GS, Cobiello PG, et al. A bacterial protease inhibitor protects antigens delivered in oral vaccines from digestion while triggering specific mucosal immune responses. J Control Release. (2015) 220 (Pt A):18–28. doi: 10.1016/j.jconrel.2015.10.011
9. Coria LM, Ibanez AE, Tkach M, Sabbione F, Bruno L, Carabajal MV, et al. A Brucella spp. protease inhibitor limits antigen lysosomal proteolysis, increases cross-presentation, and enhances CD8+ T cell responses. J Immunol. (2016) 196:4014–29. doi: 10.4049/jimmunol.1501188
10. Moreno E. Retrospective and prospective perspectives on zoonotic brucellosis. Front Microbiol. (2014) 5:213. doi: 10.3389/fmicb.2014.00213
11. Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. The new global map of human brucellosis. Lancet Infect Dis. (2006) 6:91–9. doi: 10.1016/S1473-3099(06)70382-6
12. Chomel B, DeBess EE, Mangiamele DM, Reilly KF, Farver TB, Sun RK, et al. Changing trends in the epidemiology of human brucellosis in California from 1973 to 1992: a shift toward foodborne transmission. J Infect Dis. (1994) 170:1216–23. doi: 10.1093/infdis/170.5.1216
13. Garcell HG, Garcia EG, Pueyo PV, Martin IR, Arias AV, Alfonso Serrano RN. Outbreaks of brucellosis related to the consumption of unpasteurized camel milk. J Infect Public Health. (2016) 9:523–7. doi: 10.1016/j.jiph.2015.12.006
14. Rhodes HM, Williams DN, Hansen GT. Invasive human brucellosis infection in travelers to and immigrants from the Horn of Africa related to the consumption of raw camel milk. Travel Med Infect Dis. (2016) 14:255–60. doi: 10.1016/j.tmaid.2016.03.013
15. Leong KN, Chow TS, Wong PS, Hamzah SH, Ahmad N, Ch'ng CC. Outbreak of human brucellosis from consumption of raw goats' milk in Penang, Malaysia. Am J Trop Med Hyg. (2015) 93:539–41. doi: 10.4269/ajtmh.15-0246
16. Yoo JR, Heo ST, Lee KH, Kim YR, Yoo SJ. Foodborne outbreak of human brucellosis caused by ingested raw materials of fetal calf on Jeju Island. Am J Trop Med Hyg. (2015) 92:267–9. doi: 10.4269/ajtmh.14-0399
17. Sangari FJ, Seoane A, Rodriguez MC, Aguero J, Garcia Lobo JM. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect Immun. (2007) 75:774–80. doi: 10.1128/IAI.01244-06
18. Delpino MV, Marchesini MI, Estein SM, Comerci DJ, Cassataro J, Fossati CA, et al. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect Immun. (2007) 75:299–305. doi: 10.1128/IAI.00952-06
19. von Bargen K, Gorvel JP, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev. (2012) 36:533–62. doi: 10.1111/j.1574-6976.2012.00334.x
20. Salvo Romero E, Alonso Cotoner C, Pardo Camacho C, Casado Bedmar M, Vicario M. The intestinal barrier function and its involvement in digestive disease. Rev Esp Enferm Dig. (2015) 107:686–96. doi: 10.17235/reed.2015.3846/2015
21. Czibener C, Ugalde JE. Identification of a unique gene cluster of Brucella spp. that mediates adhesion to host cells. Microbes Infect. (2012) 14:79–85. doi: 10.1016/j.micinf.2011.08.012
22. Herrmann K, Bukata L, Melli L, Marchesini MI, Caramelo J J, Comerci DJ. Identification and characterization of a high-affinity choline uptake system of Brucella abortus. J Bacteriol. (2013) 195:493–501. doi: 10.1128/JB.01929-12
23. Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. (1994) 145:69–73. doi: 10.1016/0378-1119(94)90324-7
24. de Wergifosse P, Lintermans P, Limet JN, Cloeckaert A. Cloning and nucleotide sequence of the gene coding for the major 25-kilodalton outer membrane protein of Brucella abortus. J Bacteriol. (1995) 177:1911–4. doi: 10.1128/jb.177.7.1911-1914.1995
25. Goel D, Bhatnagar R. Intradermal immunization with outer membrane protein 25 protects Balb/c mice from virulent B. abortus 544. Mol Immunol. (2012) 51:159–68. doi: 10.1016/j.molimm.2012.02.126
26. Paixao TA, Roux CM, den Hartigh AB, Sankaran-Walters S, Dandekar S, Santos RL, et al. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect Immun. (2009) 77:4197–208. doi: 10.1128/IAI.00417-09
27. von Bargen K, Gagnaire A, Arce-Gorvel V, de Bovis B, Baudimont F, Chasson L, et al. Cervical lymph nodes as a selective niche for brucella during oral infections. PLoS ONE. (2014) 10:e0121790. doi: 10.1371/journal.pone.0121790
28. Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. (2012) 109:1697–701. doi: 10.1073/pnas.1114476109
29. Cloeckaert A, de Wergifosse P, Dubray G, Limet JN. Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect Immun. (1990) 58:3980–7.
30. Domian J, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. (1997) 90:415–24. doi: 10.1016/S0092-8674(00)80502-4
31. Hayat S, Peters C, Shu N, Tsirigos KD, Elofsson A. Inclusion of dyad-repeat pattern improves topology prediction of transmembrane beta-barrel proteins. Bioinformatics. (2016) 32:1571–3. doi: 10.1093/bioinformatics/btw025
32. Tsirigos D, Elofsson A, Bagos PG. PRED-TMBB2: improved topology prediction and detection of beta-barrel outer membrane proteins. Bioinformatics. (2016) 32:i665–71. doi: 10.1093/bioinformatics/btw444
33. Tessaro SV, Forbes LB. Experimental Brucella abortus infection in wolves. J Wildl Dis. (2004) 40:60–5. doi: 10.7589/0090-3558-40.1.60
34. Rossetti CA, Drake KL, Siddavatam P, Lawhon SD, Nunes JE, Gull T, et al. Systems biology analysis of Brucella infected Peyer's patch reveals rapid invasion with modest transient perturbations of the host transcriptome. PLoS ONE. (2013) 8:e81719. doi: 10.1371/journal.pone.0081719
35. Nakato G, Hase K, Suzuki M, Kimura M, Ato M, Hanazato M, et al. Cutting Edge: Brucella abortus exploits a cellular prion protein on intestinal M cells as an invasive receptor. J Immunol. (2012) 189:1540–4. doi: 10.4049/jimmunol.1103332
36. Tibor A, Wansard V, Bielartz V, Delrue R. M., Danese I, Michel P, et al. Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect Immun. (2002) 70:5540–6. doi: 10.1128/IAI.70.10.5540-5546.2002
37. de Souza Filho JA, de Paulo Martins V, Campos PC, Alves-Silva J, Santos NV, de Oliveira FS, et al. Mutant Brucella abortus membrane fusogenic protein induces protection against challenge infection in mice. Infect Immun. (2015) 83:1458–64. doi: 10.1128/IAI.02790-14
38. Turroni F, Foroni E, O'Connell Motherway M, Bottacini F, Giubellini V, Zomer A, et al. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol. (2010) 76:3206–19. doi: 10.1128/AEM.02938-09
39. Shiga Y, Yamagata H, Tsukagoshi N, Udaka S. BbrPI, an extracellular proteinase inhibitor of Bacillus brevis, protects cells from the attack of exogenous proteinase. Biosci Biotechnol Biochem. (1995) 59:2348–50. doi: 10.1271/bbb.59.2348
40. Mkaouar H, Akermi N, Mariaule V, Boudebbouze S, Gaci N, Szukala F, et al. Siropins, novel serine protease inhibitors from gut microbiota acting on human proteases involved in inflammatory bowel diseases. Microb Cell Fact. (2016) 15:201. doi: 10.1186/s12934-016-0596-2
41. Ivanov D, Emonet C, Foata F, Affolter M, Delley M, Fisseha M, et al. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem. (2006) 281:17246–52. doi: 10.1074/jbc.M601678200
42. Farouk A. Antibacterial activity of proteolytic enzymes. Int J Pharmaceut. (1982) 12:295–8. doi: 10.1016/0378-5173(82)90100-4
43. Felsenfeld O, Gyr K. Action of some pancreatic enzymes on Vibrio cholerae. Med Microbiol Immunol. (1977) 163:53–60. doi: 10.1007/BF02126709
44. Deghelt M, Mullier C, Sternon JF, Francis N, Laloux G, Dotreppe D, et al. G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat Commun. (2014) 5:4366. doi: 10.1038/ncomms5366
45. De Nisco NJ, Abo RP, Wu CM, Penterman J, Walker GC. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci USA. (2014) 111:3217–24. doi: 10.1073/pnas.1400421111
46. Jenal U. The role of proteolysis in the Caulobacter crescentus cell cycle and development. Res Microbiol. (2009) 160:687–95. doi: 10.1016/j.resmic.2009.09.006
47. Francis N, Poncin K, Fioravanti A, Vassen V, Willemart K, Ong TA, et al. CtrA controls cell division and outer membrane composition of the pathogen Brucella abortus. Mol Microbiol. (2017) 103:780–97. doi: 10.1111/mmi.13589
48. Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. (2000) 289:1185–8. doi: 10.1126/science.289.5482.1185
49. Hirche TO, Benabid R, Deslee G, Gangloff S, Achilefu S, Guenounou M, et al. Neutrophil elastase mediates innate host protection against Pseudomonas aeruginosa. J Immunol. (2008) 181:4945–54. doi: 10.4049/jimmunol.181.7.4945
50. Verstreate DR, Creasy MT, Caveney NT, Baldwin CL, Blab MW, Winter AJ. Outer membrane proteins of Brucella abortus: isolation and characterization. Infect Immun. (1982) 35:979–89.
51. Goolab S, Roth RL, van Heerden H, Crampton MC. Analyzing the molecular mechanism of lipoprotein localization in Brucella. Front Microbiol. (2015) 6:1189. doi: 10.3389/fmicb.2015.01189
Keywords: bacterial protease inhibitor, Omp19, gastrointestinal route of infection, brucellosis, intracellular proteases
Citation: Pasquevich KA, Carabajal MV, Guaimas FF, Bruno L, Roset MS, Coria LM, Rey Serrantes DA, Comerci DJ and Cassataro J (2019) Omp19 Enables Brucella abortus to Evade the Antimicrobial Activity From Host's Proteolytic Defense System. Front. Immunol. 10:1436. doi: 10.3389/fimmu.2019.01436
Received: 01 May 2019; Accepted: 07 June 2019;
Published: 26 June 2019.
Edited by:
Leopoldo Santos-Argumedo, Center for Research and Advanced Studies (CINVESTAV), MexicoReviewed by:
Araceli Contreras-Rodriguez, National Polytechnic Institute, MexicoEric Muraille, Free University of Brussels, Belgium
Copyright © 2019 Pasquevich, Carabajal, Guaimas, Bruno, Roset, Coria, Rey Serrantes, Comerci and Cassataro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karina A. Pasquevich, a3Bhc3F1ZXZpY2hAaWliLnVuc2FtLmVkdS5hcg==; Juliana Cassataro, anVjYXNzYXRhcm9AaWliLnVuc2FtLmVkdS5hcg==
†These authors have contributed equally to this work
 Karina A. Pasquevich
Karina A. Pasquevich Marianela V. Carabajal
Marianela V. Carabajal Francisco F. Guaimas
Francisco F. Guaimas Laura Bruno
Laura Bruno Mara S. Roset
Mara S. Roset Lorena M. Coria
Lorena M. Coria Diego J. Comerci
Diego J. Comerci Juliana Cassataro
Juliana Cassataro